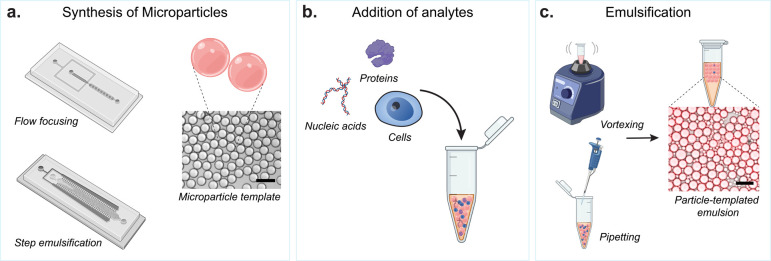
Introduction
From “Lab on a Chip” to “Lab on a Particle”
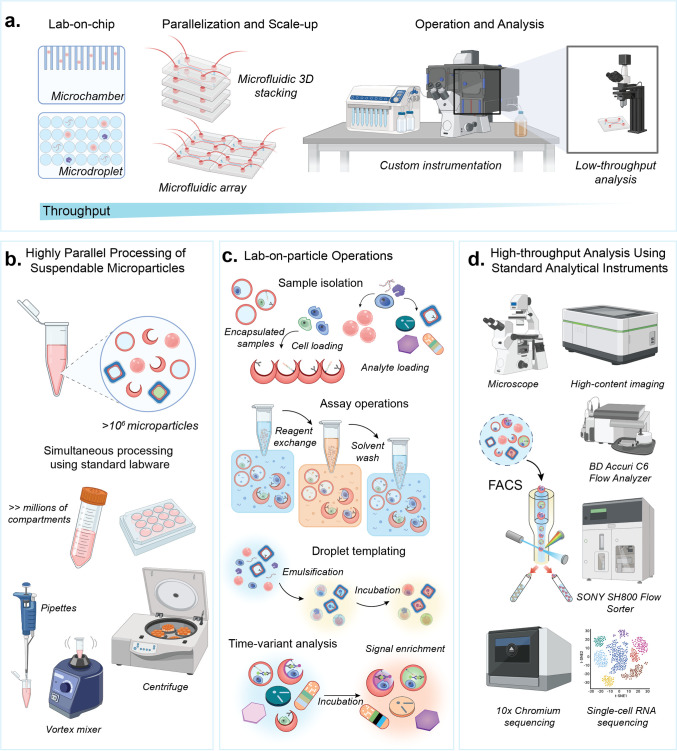
The miniaturization and automation of life science laboratory operations has leveraged fabrication advances in the integrated circuit industry, using planar substrates or “chips”. The term “lab on a chip” was coined to articulate this merger and has encompassed research on continuous flow, droplet, and digital microfluidic approaches (Figure 1a).1−7 Lab on a Particle (LoP) technologies are emerging as complementary approaches to perform microscale reactions to analyze molecules and cells.8−12 Unlike microfluidic chips, LoP platforms utilize microparticles to confine samples and facilitate reactions within compartments that are fully suspendable and can be highly parallelized (Figure 1b). These compartments can be analyzed using easily accessible instruments, like flow cytometers, fluorescence activated cell sorters (FACS), microscopes, and other imaging platforms (Figure 1d). The microparticles used for LoP assays are often manufactured with unique shapes and/or chemistries, enabling functions such as templating droplets; capturing molecules, cells, and secretions; or barcoding reactions (Figure 1c). Materials and structures essential for assay functions are imprinted in each particle, eliminating the need for custom chips or specialized instruments, thus making LoP platforms compatible with standard laboratory instrumentation and infrastructure. In this way, the microparticles in LoP technologies act in a similar manner as a software application, or App, running on ubiquitous life science instrument hardware, fueling more rapid adoption and expanded impact.
Figure 1.
From lab on a chip to lab on a particle. (a) Lab on a chip technology uses microchambers or droplets to confine reactions, enabling the analysis of target cells or molecules with high precision. Parallelization and scale-up rely on the 2D surfaces of chips and custom instrumentation, which often lead to reduced analysis throughput. (b) Lab on a particle technology enables millions of microparticle-based compartments to be scaled in 3D in standard tubes, where fluidic operations are performed using standard laboratory equipment. (c) Operations on particles include cell loading and encapsulation, analyte binding, reagent exchange and washing, and templating of water-in-oil emulsions. Signal enrichment can occur on particles through reactions that are either confined or locally bound. Microparticles are barcoded by shape, size, pattern, color, or other means to enable time-variant analysis as reactions or cell behavior progresses over time. (d) Microparticles are analyzed using standard analytical instruments compatible with cells, such as microscopes, flow cytometers, fluorescence activated cell sorters and single-cell sequencing instruments.
Advantages of Lab on a Particle Systems
One of the main advantages of LoP systems is facile integration into current laboratory instruments and experimental workflows, resulting in more rapid and easy adoption by scientists.13 These platforms simplify and parallelize complex workflows by replacing intricate microfluidic circuitry,14 valving,4 and other flow control operations with straightforward methods like pipetting, mixing, and centrifugation aligned with standard lab practices (Figure 1b). Following simple pipetting and centrifugation operations to prepare samples, the small size of microparticles ensures compatibility with commercial instruments that have been developed previously to analyze cells. Instead of developing new instruments, LoP workflows rely on a breadth of high-precision and widely available instruments, like flow cytometers,15 FACS,16 imaging flow cytometers,17 image-activated cell sorters,18 and single-cell sequencing instruments (Figure 1d).19
Additionally, microparticles in these systems can be suspended in oil-free aqueous environments, enabling reagents and unbound molecules to diffuse in and out and facilitate solution exchange that is often difficult to achieve using microdroplet20 or microwell technologies.21 Signals can be accumulated in or on the particles over time, which can aid in time-variant measurements or workflows where signal amplification is necessary.9,22 Emulsification of particles driven by simple mixing steps can further seal compartments if needed for an assay, rapidly generating millions of drops in parallel.11,23,24 By enabling millions of compartmentalized reactions within a few hundred microliters of assay volume, microparticles can improve efficiency in biological workflows by speeding up experiments, minimizing reagent consumption, and reducing costs while enabling high-throughput, massively parallelized analysis.11 The degree of flexibility and control in handling complex biological processes makes advanced assay systems more accessible via LoP technology.
Along with making assays easier to perform, LoP systems enable novel analytical measurements that were previously unattainable or challenging using Lab on a Chip-based methods. Microparticles in LoP platforms serve as multifunctional reaction substrates that can be adapted through different chemical modifications to integrate various cellular and biomolecular workflows. This includes the functionalization of the solid-phase compartments with small molecule affinity agents like biotin, peptides, extracellular matrix (ECM) proteins, antigens, antibodies, nucleic acids, or other biomolecules. Specifically, microparticle surfaces functionalized with peptides linked to major histocompatibility complex (MHC) proteins have been shown to mimic antigen-presenting cells, activating immune cells in an LoP-based workflow.25 Notably, compartments formed by microparticles can be tailored for single-cell profiling of both adherent and suspension cell types and their secretions.9,25−27 High-sensitivity detection of analytes is enabled by digitization of amplified signals generated by reactions in a confined volume.24,28 The range of detectable analytes encompasses proteins, enzymes, antibodies, and DNA/RNA molecules detected through traditional sandwich immune complexes,9,11 enzyme-linked immunosorbent assays (ELISA),22,29 or DNA/RNA hybridization30 and amplification methods.28 Additionally, microparticles can be barcoded for multiplexed detection, by shape12 or using unique oligonucleotide sequences.9,26,31 Oligonucleotide barcodes can also be incorporated into assays to link proteomic and transcriptomic analyses using standard single-cell sequencing instruments.9,26 Furthermore, with advancements in high-throughput and high-content instruments such as imaging flow cytometry32−34 and image-activated cell sorters,35−40 LoP platforms can unlock novel analytical functions, including observing morphological changes of cells adhered within particles or cell–cell interactions or binding. The ability to incorporate varied functionalities that are critical to numerous biological workflows enables a multifaceted approach to accelerate life science research using LoP technology.
Evolution of Hydrogels for Cell Encapsulation
Gel Microdrops as Early Lab on a Particle Systems
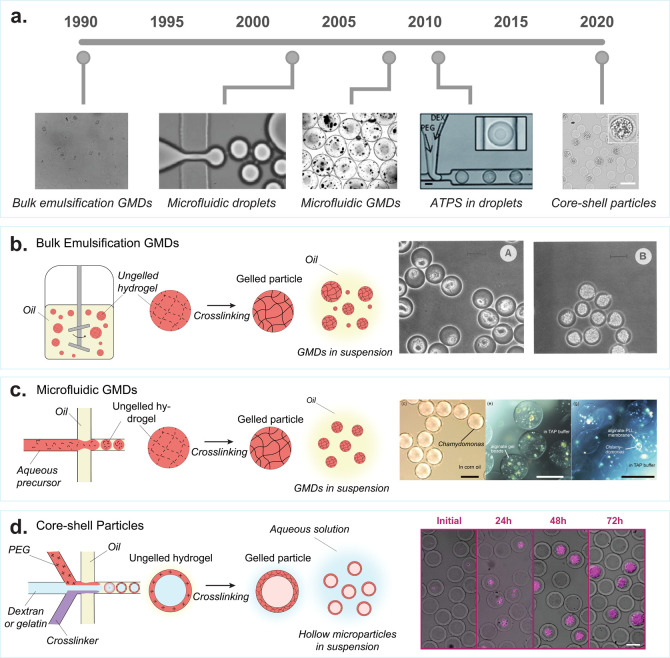
One of the earliest “lab on a particle” systems for analyzing biological materials was gel microdrops (GMDs, Figure 2a).41 During the 1990s, flow cytometers became widely accessible tools for high-throughput analysis and sorting of cells.49 However, they were primarily limited to characterizing cells based on surface labels and required fixation and permeabilization to interrogate intracellular markers. These approaches provided no intrinsic capabilities for growth and secretion-based characterization. Developing a method for long-term cell compartmentalization could expand the scope of flow cytometry, extending it to extracellular molecular and colony-level phenotypic analysis.
Figure 2.
Evolution of cell encapsulation in hydrogel drops for single-cell analysis. (a) Some of the first hydrogel microparticles for cell and protein screening were made using bulk emulsification methods, producing polydisperse agarose particles called gel microdrops (GMDs). [Reprinted with permission from Macmillan Publishers Ltd.: Nature, Weaver, J. C., et al. Nat Biotechnol1988, 6 (9), 1084–1089 (ref (41)) Copyright 1988.] Droplet microfluidics became popular in the early 2000s [Reprinted with permission from ref (42). Anna, S. L., et al. Applied Physics Letters2003, 82 (3), 364–366, 2003 licensed under a Creative Common Attribution (CC BY) license], which was later used to encapsulate cells in uniform GMDs [Reproduced from On-Chip Alginate Microencapsulation of Functional Cells Workman, V, et al. Macromol. Rapid Commun.2008, 29 (2), 165–170 (ref (43)) Copyright 2008 Wiley]. The first studies of aqueous two-phase systems in microfluidic droplets started in the early 2010s. Scale bar is 50 μm. [Reproduced from Vijayakumar, K., et al. Chemical Science2010, 1 (4), 447–452 (ref (44)) with permission from The Royal Society of Chemistry.] This led to the development of hollow hydrogel microparticles that form around encapsulated cells. Scale bar is 50 μm. [Reproduced from Leonaviciene, G., et al. Lab Chip2020, 20 (21), 4052–4062 (ref (45)) with permission from The Royal Society of Chemistry.] (b) Bulk emulsion GMDs: Formation of polydisperse GMDs by vigorously mixing an ungelled polymer solution in oil, to create water-in-oil emulsions. These emulsions, once stabilized, are gelled to form nonuniform GMDs. The oil is removed, and the GMDs are transferred to an aqueous solution. Scale bar is 20 μm. [Reprinted with permission from ref (46). Copyright, 1990 American Society for Microbiology.] (c) Microfluidic GMDs: A microfluidic droplet generator is used to create monodisperse water-in-oil droplet emulsions consisting of ungelled polymer precursors. Subsequently, the solution undergoes gelation to form uniform GMDs. After gelation, the oil is removed, and the GMDs are transferred into an aqueous solution. Scale bar is 100 μm. [Reproduced from Morimoto, Y., et al. Lab Chip2009, 9 (15), 2217–2223 (ref (47)) with permission from The Royal Society of Chemistry.] (d) Core–shell particles: An aqueous two-phase system is employed within a microfluidic droplet generator to produce core–shell microparticles featuring a hollow inner cavity. Polyethylene glycol (PEG), dextran, and a cross-linker are combined to form a water-in-oil emulsion using the microfluidic droplet generator. PEG and dextran undergo phase separation; dextran moves toward the center, while PEG aligns at the surface of the emulsion. Subsequently, the PEG-rich phase is cross-linked to create a solid outer shell. Afterward, the particle is transitioned from oil to water. During this transfer, the inert dextran escapes through the pores of the outer shell, resulting in a hollow interior. Scale bar is 50 μm. [Reproduced with permission from Proceedings of the National Academy of Sciences USA van Zee, M, et al. Proc. Natl. Acad. Sci. U.S.A.2022, 119 (4), e2109430119 (ref (48)).]
In the early 1990s, various research groups developed strategies to embed cells within agarose microparticles.41,46,50−52 By mixing molten agarose with mineral oil and vortexing, scientists were able to create 10–100 μm diameter emulsions that were subsequently gelled by cooling to 4 °C to form GMDs (Figure 2b). Single cells were entrapped in the agarose particles by including them in the molten agarose at a sufficiently low concentration prior to vortexing, following Poisson statistics. The GMDs were then separated from mineral oil for processing via FACS and analysis of the small fraction of GMDs containing cells.
Applications of Gel Microdrops
Early applications of GMDs were for growth and enzymatic activity-based assays. The Weaver group was one of the first to demonstrate that bacterial, fungal, and mammalian cell species can be grown in GMDs and used the system to characterize drug resistance and nutrient sensitivity of various cell types.53 Other groups used the ability to screen cells based on growth rates,54−57 speed up antibiotic susceptibility testing,58,59 and perform viability measurements.60 The Nir group used GMDs to grow monoclonal colonies of Escherichia coli (E. coli) expressing β-galactosidase (β-gal), exposed the enzymes to substrate, and sorted out colonies producing enzyme with high catalytic activity for sequencing and downstream analysis.50
Biotin affinity sites can also be attached to GMDs, enabling the embedding of probes that could be used for assay functions, such as capturing cell secretions. Several research groups have used GMDs to capture antibodies for antibody discovery and cell line development.61,62 GMDs have also been used to characterize cytokine secretions from T cells using similar workflows.63
One challenge with these initial systems was microparticle polydispersity, indicated by a high coefficient of variation (CV) in droplet diameter greater than 40%. The nonuniformity of particles made handling, processing in flow, and data analysis difficult. This difficulty was largely due to the challenge of controlling the size of the droplets formed by vortexing. Additionally, using filters to isolate particles below a certain size threshold would result in the loss of a significant fraction of particles and cells. Although attempts were made to use membrane emulsification to create more uniform GMDs, these methods were found to be inconsistent and challenging to implement.64
The emergence of droplet microfluidic technology allowed for the creation of monodisperse droplets enabling the formation of uniform GMDs (Figure 2c).43,47,65 Microfluidic droplet generators were used to create uniform water-in-oil emulsions. By introducing a hydrogel precursor into the aqueous phase and allowing it to gel after droplet formation, GMDs with consistent diameters can be fabricated. The uniformity improved the throughput and precision in analysis of GMDs via FACS-based readouts, opening up new avenues of research, including genetic applications,23,66,67 emulsion polymerase chain reaction (PCR),68,69 microbial community screening,66 biosensor screening,70 aptamer screening,71 growth selection,72 3D cell culture,73 secretion analysis,74 and coculture systems.75 GMDs also became adopted in the private sector by companies such as FGen (Ginkgo Bioworks) and Amyris to increase production of various bioproducts and coculture for biologic discovery.
Limitations of Gel Microdrops
Despite the successful exploration of a wide variety of applications, several limitations still restrict the potential implementation of traditional GMD workflows in cell-based analysis. A notable challenge is the premature release and escape of cells from particles during extended assays, particularly long-term growth studies.76 This issue is exacerbated in the case of microbes, which possess enzymatic machinery (proteases, amylases, etc.) capable of degrading biological hydrogels. Furthermore, cells initially encapsulated in GMDs often gravitate toward the edges due to buoyancy differences, rather than remaining centralized. Additionally, current strategies for controlled cell release predominantly involve elevating temperatures above 37 °C, detrimentally impacting cell viability for subsequent growth or molecular assessments.76 The encapsulation of cells in 3D hydrogels also disrupts the natural spreading morphology typical of adherent cell types, potentially altering cell behavior and hindering growth. This presents a significant barrier for studies aimed at elucidating genetic mechanisms or replicating real-world cell interactions. Moreover, engineering agarose hydrogels with capture probes, genetic barcodes, and other functional motifs essential for single-cell screening workflows is challenging. When achieved, there is a limited dynamic range and poor reproducibility, limiting the use of agarose-based particles for complex cell-based functional assays.
To address some of the limitations associated with GMDs, synthetic hydrogels such as poly ethylene glycol (PEG) can be utilized as alternatives to biobased hydrogels. Cells lack the machinery required to break down synthetic polymers, which circumvents the issue of premature degradation. While encapsulating cells in completely solid synthetic hydrogels may impede cellular growth and function compared to biological hydrogels, creating voids within the synthetic hydrogel particles allows for unrestricted cell growth and functionality, more closely mimicking a native environment. Furthermore, a variety of engineering tools can be leveraged to modify the porosity of synthetic hydrogels to enable optimal nutrient exchange as well as enable the linking of functional binding sites to aid in cell screening assays.
Fabrication of Core–Shell Particles
Aqueous two-phase systems (ATPS) have been utilized to create microstructured particles with unique morphologies, enabling the formation of functional compartments for discrete cellular and molecular analysis. This typically involves mixing a reactive polymeric phase with a nonreactive interpolymer, utilizing phase separation between these solutions to polymerize and form morphologically distinct particles. To produce core–shell particles featuring a hollow interior, a reactive PEG polymer is combined with dextran and a cross-linker in set concentrations within a microfluidic droplet generator (Figure 2d). Within the microfluidic channels, the individual solutions undergo phase separation and are subsequently cross-linked downstream to create hollow hydrogel particles that exhibit a core–shell architecture.44 When the PEG is gelled and the oil is removed, the dextran separates from the particle and leaves behind an empty void, which is filled by the aqueous culture media.77−80 The Weitz group has shown that the structure, core position, and pore size can all be modified by changing the microfluidic fabrication and hydrogel properties.81,82 This adjustment allows for the encapsulation and growth of cells within a synthetic matrix that avoids premature degradation (Figure 2d), offers easy functionalization, and enables solution transfer across the particles that enable multistep cell screening workflows. Moreover, the addition of cleavable functional groups, such as disulfide or peptide linkages, to the outer PEG shell enables the controlled breakdown of the particles, facilitating the release of encapsulated cells or their products.
Applications of Hollow Hydrogel Particles
Bacteria, yeast, microalgae, and mammalian cells have all been demonstrated to remain viable and proliferate within hollow microparticles. van Zee et al. have shown that the cell growth in hollow microparticles is significantly improved over aqueous-in-oil microfluidic droplets.48 The stability of these particles facilitates easy multistep processing, essential for exposing encapsulated cells or their products to various solutions. This is crucial, for instance, in determining the dynamic response range of encapsulated protein sensors to analytes. Additionally, the hollow interior offers a 2D surface that allows adherent cell types to attach, and the adherence can be further improved by incorporating various moieties, such as arginine-glycine-aspartate (RGD) and poly-l-lysine (PLL), into the interior shell surface.83 Due to their resilience under harsh physical conditions, such as vigorous shaking or high shear forces, these particles are suitable for use in novel environments like bioreactors. This allows scientists to select cells based on environmental conditions that previously could not be replicated in microfluidic microwell or droplet systems. Moreover, the ability of these particles to withstand high temperatures enables one to conduct PCR and loop-mediated isothermal amplification (LAMP) within intact particles, thereby opening up new possibilities in molecular screening.
The first applications of hollow microparticles have been the selection of cells based on growth,48 RNA cytometry,84 drug delivery,85 cell delivery,86,87 water treatment,88 and enzymatic analysis.89 Atrandi Biosciences has commercialized instruments and microfluidic kits for sequencing microbes using hollow shell particles.90 This underscores one of the limitations of hollow microparticles: the necessity for end-users to possess microfluidic equipment to encapsulate their cells during the droplet formation process. However, considering the unique advantages in characterizing growing cell colonies, it is expected that hollow microparticles will continue to find exciting applications in the coming years.
Nanovials: Cavity-Containing Hydrogel Particles
Microparticles designed with an open cavity exposed to the surrounding environment facilitate direct cell loading, making LoP platforms more accessible without necessitating microfluidic instruments and devices. Compared to hollow particle systems that depend on microfluidic-based encapsulation, nanovials offer an off-the-shelf solution for researchers aiming to conduct multistep cell screening workflows.
The fabrication of particles with a crescent-shaped cross section was first reported in 2012 by Ma et al. The open cavity was used to load smaller polymer beads matching the size of the cavity.91 Similar to the hollow microparticles, crescent-shaped particles were fabricated using an ATPS consisting of polymeric reactive and inert phases in a microfluidic cross-junction droplet generator followed by UV cross-linking of the photopolymerizable polymer. In 2019, Liu et al. also reported production of large crescent-shaped microparticles, referred to as “micro-buckets”, to use as living cell carriers owing to their propensity to self-orient with the cavity side facing upward.92 Since then, there have been innovations in the manufacturing, chemistry, and spatial patterning of such cavity-containing microparticles, leading to increasing functionality. de Rutte et al. reported functionalized crescent-shaped particles termed “nanovials” for single-cell screening and secretion analysis workflows using commercial FACS-based instruments for sorting of nanovial-containing cells and associated secretions.11 Functionalizing the nanovial surface with cell adhesion motifs, antigens, and/or antibodies through widely available biotin–streptavidin noncovalent interactions allows for the capture of individual cells and their corresponding secretions. The most recent iteration of nanovials uses a PEG-gelatin ATPS, which results in a localized gelatin layer in the nanovial cavity which can be functionalized to capture both cells and their secretions with reduced crosstalk.93
The general advantages of nanovials include a hydrogel surface and a confined volume to concentrate and capture molecules as well as compatibility with standard cell culture and analysis instruments.11 The small volume of the nanovial cavity and local capture surface allows for rapid concentration of secreted products, shortening the incubation time required to generate detectable levels of secreted molecules and reducing crosstalk to neighboring nanovials. Nanovials allow for a highly modular assay platform and simple workflow which relies on standard laboratory equipment, such as micropipettes, microscopes, and FACS. Nanovial assays are compatible with both adherent and suspension cells while providing protection against damage and high stresses during droplet-in-air sorting.9 Unlike droplet microfluidic assays leveraging water in oil emulsions, cells used in nanovial assays can be kept in their preferred media to maintain their viability over long periods of time (multiple days). Nanovials with adhered cells can be sorted based on their secretory function at rates of millions of events per hour.
Fabrication of Nanovials
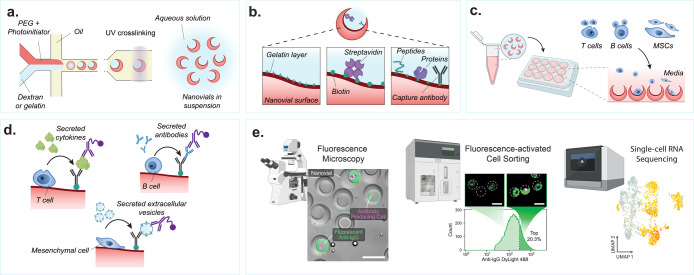
In order to fabricate millions of nanovial particles at scale, uniform droplets containing an ATPS surrounded by oil are generated using a microfluidic droplet generator (Figure 3a). The ATPS consists of two precursor polymer phases: (1) reactive PEG mixed with a photoinitiator and (2) inert dextran or gelatin. When mixed at predetermined ratios, the two solutions undergo phase separation into an outer PEG-rich and inner dextran- or gelatin-rich phase (Figure 3a). Hester et al. identified the role fluid flow and buoyancy play in achieving rapid phase separation of the two precursor polymers.94 Exposing these phase-separated droplets to a UV beam cross-links the PEG-rich layer, and subsequent washing steps remove the internal water-soluble dextran or gelatin. Millions of nanovials can be produced within hours and stored for later use over months to years, thus eliminating the need for microfluidic device fabrication and operation for each experiment.11 Nanovials have been shipped around the world at ambient temperature without any detectable loss of function, enabling laboratories without microfluidics experience to perform nanovial-based assays.
Figure 3.
Nanovial fabrication and experimental workflow. (a) An aqueous phase consisting of reactive PEG precursor and photoinitiator is coflowed with a second aqueous phase consisting of gelatin or dextran solution in a microfluidics droplet generator resulting in uniform monodispersed aqueous two-phase water-in-oil droplets. The phase-separated droplets are exposed to UV light downstream to polymerize the PEG phase. The dextran or gelatin sacrificial phase is removed during washing steps resulting in an open cavity and final crescent-shaped cross-sectional morphology. (b) If fabricated with gelatin, the nanovials will have a localized gelatin layer at the cavity surface. The gelatin or PEG surface can be functionalized with biotin and streptavidin moieties to attach peptides, proteins, or antibodies to localize cells and their secretions to individual nanovials. (c) Various cell types with a wide diversity of secreted products can be loaded onto and analyzed on nanovials. Cells are loaded onto nanovials in tubes or well plates in bulk, and unbound cells can be filtered out. (d) Fluorescent and/or oligo-barcode labeled detection antibodies are incubated with cells on nanovials to detect their secretions. (e) Single-cell secretion analysis is performed with microscopy, FACS, and/or single-cell sequencing techniques. Scale bar is 100 μm. [First two images reproduced from de Rutte, J., et al. Suspendable Hydrogel Nanovials for Massively Parallel Single-Cell Functional Analysis and Sorting. ACS Nano2022, 16 (5), 7242–7257 (ref (11)) Copyright 2022 American Chemical Society. Last image reprinted with permission from Macmillan Publishers Ltd.: Nature, Udani, S., et al. Nat. Nanotechnol. (ref (9)) Copyright 2023.]
Recent nanovial fabrication efforts have been focused on localizing cells and their secretions to the internal nanovial cavity.93 Initial PEG-dextran nanovials did not take full advantage of the particle geometry as cell binding motifs were decorated throughout the nanovial, allowing cells to bind and secrete outside of the cavity, leading to higher convective transport of secretions and increased binding to neighboring particles, resulting in nonspecific signal. The PEG-gelatin nanovials ameliorate this issue; a thin layer of cavity-localized gelatin is advantageously left behind after washing.93 This can then be leveraged to selectively functionalize the surface of the nanovial cavity using N-hydroxysuccinimide-ester (NHS-ester) bioconjugation to the lysine residues on the gelatin layer (Figure 3b). Therefore, the cavity of nanovials can be specifically decorated with NHS-conjugated biotin, allowing streptavidin binding, followed by immobilization of biotin-conjugated cell- and secretion-capture antibodies and/or molecules. In addition, the localized gelatin layer serves as a binding motif for some adherent cell lines, such as mesenchymal stromal cells, macrophages, CHO-K1, and 293T cells.9,93
General Workflow for Nanovial Secretion Assays
Cell Loading and Binding on Nanovials
Single cells are stochastically loaded into nanovials by simply mixing cells and nanovials or by seeding cells on top of nanovials resting on the bottom of a vessel.11 The number of cell-loaded nanovials follows a Poisson distribution. When the cavity size approaches that of a single cell, single-cell loading is favored, such that the fraction of nanovials that contain single cells exceeds the expectations from the Poisson distribution.93 Nanovials can be functionalized to bind specific cell types in a mixed population, e.g. using gelatin or RGD for integrin-based adhesion, antibodies against cell surface markers, or antigens/peptide-major histocompatibility complexes (p-MHC) molecules specific to chimeric antigen receptors (CARs) or T cell receptors (TCRs) (Figure 3c).
Secretion Capture and Labeling
Sandwich immunoassays are used to assess the secretion levels of cells loaded onto nanovials. Secretions are bound by capture antibodies immobilized on the cavity of the nanovial. Secreted analytes captured on the nanovial are then stained with fluorescently labeled and/or oligonucleotide-labeled detection antibodies (Figure 3d). Multiple capture antibodies against different secretions can also be immobilized on a single nanovial for multiplexed analysis.95
Readout, Sorting, and Downstream Analysis
Fluorescent signals from sandwich immunoassays on nanovials are analyzed and/or sorted by flow cytometry or FACS (Figure 3e). Nanovials have been successfully used with a wide range of commercially available cytometers such as the SONY SH800S, BD FACSAria, On-chip Biotechnologies On-chip Sort, Union Biometrica Biosorter, and Cytek Biosciences ImageStream MKII.13 Sorted cells in nanovials can then be recultured or used in further downstream analysis, such as single-cell and nucleic acid sequencing (Figure 3e).
Nanovials as a Platform for Antibody Discovery
Monoclonal antibody (mAb) discovery, whether utilizing immunized animals for hybridoma generation96 or antigen-baiting of memory B cells,97 or protein surface display platforms like phages or yeast,98 still necessitates multiple confirmation steps to ensure mAb specificity and function. Further screening of potential mAbs is a limited-throughput endeavor and bottleneck in the process. Typical hybridoma screening involves initially screening supernatant samples from pooled cells followed by serial dilution screening of the top performing pools. Pooling is necessary to screen the millions of hybridomas produced from a single animal. However, this approach can result in low-affinity, high-secreting cells masking the performance of higher-affinity antibodies secreted at lower rates.99 Additionally, antigen specificity screening is performed in microwell plate ELISAs, which are low-throughput compared to single-cell approaches like FACS.
Nanovials address these antibody discovery challenges by facilitating the screening of individual cells and their corresponding secretions, which are captured on discrete, solid-phase particles. These particles are designed to be compatible with analysis and sorting in a high-throughput manner using FACS.11,93 Sandwich immunoassays can be conducted on the nanovial surface to quantify antigen-specific immunoglobulin G (IgG) from antibody-secreting cells. Due to the small volume of the nanoliter-scale particle cavity, antibody accumulation occurs much more rapidly, allowing for a decrease in incubation time from days or weeks to 30–60 min (Figure 4a).
Figure 4.
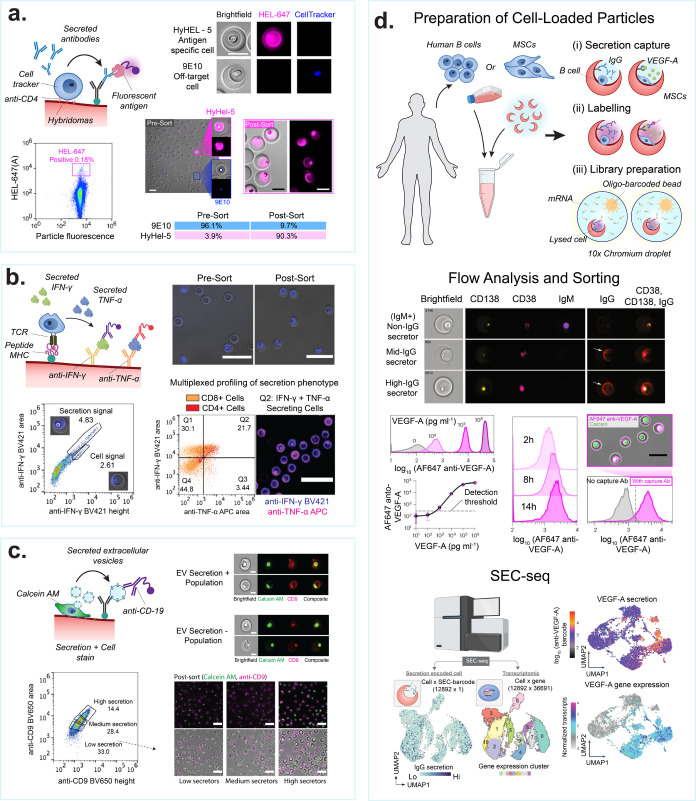
Screening secreted cellular products on nanovials. (a) Antibody secretions can be captured from hybridoma lines, producer cell lines, and primary antibody-secreting B cells. Schematic shows cells are captured, e.g., with antibodies specific to cell surface markers, and secreted antibodies are captured onto antigens or antibodies on the nanovial surface. Images show antigen-specific IgG bound on nanovials (magenta) secreted by HyHEL-5 cells, while 9E10 cells secreting an off-target IgG (blue) do not have corresponding signal on nanovials. Flow scatter plot highlighting the gate used to sort antigen-specific IgG secretors. Presort and postsort microscopy images are shown. The table shows sort enrichment of spiked HyHEL-5 cells. Scale bars are 100 μm. [Reproduced from de Rutte, J., et al. Suspendable Hydrogel Nanovials for Massively Parallel Single-Cell Functional Analysis and Sorting. ACS Nano2022, 16 (5), 7242–7257 (ref (11)). Copyright 2022 American Chemical Society.] (b) Schematic showing multiple cytokines can be captured in parallel from activated T cells that are engaged through TCR interactions with peptides loaded onto class I major histocompatibility complex (p-MHC). Images and FACS plots show T cells captured using p-MHC and screened for IFNγ and TNFα production. Fluorescence peak area vs height scatter plots showing gates used to differentiate nanovial staining vs cell staining of permeabilized cells. Scale bars are 100 μm. [Reprinted with permission from ref (95). Copyright, 2022 D. Koo.] (c) MSCs are captured based on binding to gelatin on nanovials and screened based on extracellular vesicle secretion. Scale bars are 20 μm (top) and 100 μm (bottom). [Reprinted with permission from ref (27). Copyright, 2023 D. Koo.] Imaging flow cytometry of MSCs and captured EVs, stained with an antibody against the tetraspanin, CD9 (red), showing EV secretion positive and negative populations. The viability dye, calcein AM, is used to stain live cells (green). Bottom panel images show calcein AM-stained MSCs (green) on nanovials stained with anti-CD9 (magenta) following FACS sorting based on EV-specific secretion signal gates (low, medium, high secretors). (d) Secretion is associated with single-cell RNA sequencing data (SEC-seq) by using oligo-barcoded detection antibodies and droplet single-cell barcoding of cDNA libraries. Imaging flow cytometry of nonsecreting and IgG-secreting cells. Flow cytometry histograms of VEGF-A signal on nanovials from a VEGF-A concentration sweep or signal from secreting cells over time. Signal is dependent on the presence of a VEGF-A capture antibody. Scale bar is 50 μm. [Reprinted with permission from Macmillan Publishers Ltd.: Nature, Udani, S., et al. Nat. Nanotechnol. (ref (9)). Copyright 2023.] SEC-seq data shows transcriptome-based clustering of single-cell expression profiles and corresponding IgG or VEGF-A secretion signal. [Reprinted with permission from Macmillan Publishers Ltd.: Nature, Cheng, R., et al. Nat. Commun. (ref (26)) Copyright 2023.]
Nanovials have been used for an antigen-specific hybridoma screening workflow yielding both high purity and high enrichment rates (Figure 4a). de Rutte et al. showcased the ability to isolate antigen-specific antibody-producing cells from a nonspecific background hybridoma.11 In their experiment, HyHEL5 hybridoma cells, which secrete antibodies specific to Hen Egg Lysozyme (HEL), were spiked into a 9E10 (antimyc tag secretors) at a ratio of 1:25. By employing an antimouse IgG capture antibody to collect secreted IgG antibodies and labeling with fluorescent recombinant HEL protein for detection, a distinct and specific signal was associated with the HyHEL5 subpopulation. Over 1 million nanovial events were analyzed within hours, yielding a postsort population enriched to over 90% HyHEL5 cells. Single cells sorted into 96 microwell plates could be clonally expanded and antibody sequence information from over 50% of the sorts could be amplified through single-cell reverse transcriptase PCR. The successful application of nanovials for characterizing hybridomas, as described in this study, lays the foundation for their potential utility in the discovery and enrichment of other antibody-secreting cells, such as plasma cells, which are a good target to discover affinity matured, high-affinity antibodies. Partillion Bioscience is commercializing antibody discovery kits for use with hybridoma and plasma cells, leveraging LoP-based nanovial technology.100
Nanovials as a Platform for Functional Characterization of Cell Therapies
Cell-based therapeutics are an emerging modality offering the potential to treat intractable diseases such as cancer. Recent advancements have expanded the scope of these therapies beyond oncology, into areas such as inflammation and autoimmune conditions. However, the efficacy of therapeutic cells is often driven by functions such as their secreted products, including cytokines, growth factors, and extracellular vesicles (EVs).8 Therefore, characterization and selection of functioning cell populations can lead to more potent cell therapies. Conventional potency assays for cell therapies typically rely on bulk cell assessments, which do not account for single-cell heterogeneity.
Nanovials have been demonstrated as a platform for high-throughput single-cell analysis to isolate and select cells based on secretory function while enabling the recovery of cells postanalysis. A study by Koo et al. demonstrated analysis of four markers simultaneously: two surface markers (CD4+ and CD8+) and two secreted proteins (interferon gamma (IFNγ) and interleukin-2 (IL-2) or tumor necrosis factor alpha (TNFα)).95 This approach demonstrated the ability to analyze and select viable polyfunctional primary T cells at a high throughput (up to 1 million nanovials were analyzed and sorted within 1 h). In addition, the cells with a polyfunctional phenotype were successfully recovered, regrown, and shown to proliferate for up to 7 days postsort. Therefore, the linkage between secretion phenotype and surface markers associated with specific T cell subsets can be explored with the nanovial platform.
The preceding approach can be applied and further expanded to sort antigen-specific T cells or CAR-T cells based on a combination of their affinity toward p-MHCs or antigens and the level of secreted proteins, to discover functional TCRs or CARs. Single cells can be loaded into nanovials prefunctionalized with p-MHC (TCR) or an antigen of interest (CAR-T) as cell binding and activation motifs (Figure 4b). As a result, nanovials can serve as artificial antigen-presenting cells to specifically bind and activate cells of interest. Activation through p-MHC or antigen induces bound cells to secrete cytokines which can then be accumulated and captured within the cavity of the nanovials. This approach enables high-throughput selection of cells based on both surface binding and function. Cell functionality can be quantified by classic T cell activation markers, such as IFNγ, IL-2, TNFα, and granzyme B (GrB). Koo et al. employed this approach to sort live cognate T cells based on CD3+ and CD8+ surface markers and IFNγ secretion utilizing p-MHC functionalized nanovials.25 Moreover, the authors were able to sequence the TCRs of sorted cells on nanovials and recover paired αβ-chains using existing 10X chromium microfluidics-based single-cell sequencing. Cell-containing nanovials with different p-MHCs were precoated with unique oligonucleotide-barcodes and were directly input into the 10X platform postsort. This approach enabled linking TCR sequences to targets with 100% accuracy and greatly improved the true positive rate for recovery of rare cancer-specific TCRs.
Nanovials can be employed to analyze and sort single cells based on their EV secretion levels showing the long-term maintenance of this therapeutic phenotype (Figure 4c). Koo et al. compared the gene expression differences between high EV secretors and low EV secretors from a population of mesenchymal stem cells (MSC), identifying pathways for EV biogenesis, cell proliferation, and regeneration through single-cell transcriptomic analysis.27 Moreover, the high-secreting MSCs exhibited enhanced EV secretion phenotypes and paracrine repair activity in vivo in a murine myocardial infarction model when compared to low-secreting cells, showing promise for enhancing cell therapies.
Nanovials as a Platform to Link Secretions to Transcriptomes
Secretion encoded single-cell sequencing (SEC-seq) is a novel method which leverages nanovials and standard single-cell RNA sequencing (10X Genomics) to retain and analyze both transcriptome and secretion information from single cells. In SEC-seq, mRNA can be captured and quantified from individual cells in the presence of oligo-barcode tagged secretions captured on nanovials (Figure 4d). Using SEC-seq, Udani et al. investigated the relationship between secretion of vascular endothelial growth factor splice variant A (VEGF-A) and the underlying transcriptome state in MSCs under normoxic and hypoxic conditions.9 VEGF-A secretion was found to be heterogeneous among the cell population and weakly correlated with the VEGF-A transcript level. Interestingly, it was also discovered that the VEGFA mRNA was not correlated with the level of secretion. Instead, another set of genes, including a few surface markers, were highly correlated with VEGF-A secretion and marked a small cell subpopulation with higher secretion levels largely independent of VEGFA expression. Cheng et al. also applied SEC-seq to analyze IgG secretion levels from human B cells.26 A new surface marker, CD59, associated with plasma cells secreting higher levels of IgG was uncovered, along with other pathways associated with higher IgG levels including metabolism, misfolded protein response, and endoplasmic reticulum generation. SEC-seq technology can link cell secretion to its surface markers and transcriptomes at the single-cell level which can lead to the discovery of previously unknown gene networks useful to modulate cellular secretion for therapeutic development.
While the protected cavity of nanovials is well-suited to capture cells and prevent convection of secretions from the captured cells, surrounding nanovials in an oil phase leads to further accumulation and segmentation of secretion signal.11 Due to the uniform outer diameter of nanovials, droplets formed that surround nanovials can be highly uniform following emulsification, with CVs of 3.2% and 4.0% for 35 and 85 μm nanovials, respectively.11 Similar templating of monodisperse emulsions can also be achieved with spherical hydrogel particles, allowing for accumulation of reactants and amplified molecular assays.
Spherical Particles as Substrates for Assays
LoP applications can extend beyond cell selection to molecular assays in diagnostics, life science research, and environmental monitoring. LoP-based molecular assays have several advantages over traditional assays, including ease-of-use, accessibility, additional multiplexing capabilities, and amplified readouts.
History of Particles as Molecular Sensors
Luminex pioneered the use of fluorescent dye-doped particles by developing one of the first particle-based molecular assays in 1997 known as Flowmetrix that could detect up to 64 distinct analytes.101,102 In FlowMetrix, a carboxylate-modified polystyrene microsphere is attached to a specific capture reagent, which when combined with its corresponding analyte and dye-labeled detection antibody creates a fluorescent sandwich. The microspheres are doped with various concentrations of orange- and red-emitting fluorochromes to create distinct microsphere classification profiles while a green fluorochrome is used for the analyte detection.101 When excited by a two-laser flow cytometry system, a quantitative signal of analyte binding associated with a specific particle type is created.
To extend multiplexing capabilities, Microplex microspheres were encoded with two or three fluorescent dyes at distinct ratios, expanding the gradient of unique colors that can be detected to 100 or 500 distinct color sets, respectively.103 Both Flowmetrix and Microplex were an improvement to precursor technology published in the late 1970s that used particles of different sizes and fluorescent dye compositions to detect analytes; however, they were limited to a small number of analytes and not able to differentiate between aggregates of different-sized microspheres. Although these Luminex technologies are widely referenced, there are limitations to the technology. Notably, the assays on these beads are mostly direct binding assays and incorporate no amplification chemistries, resulting in a higher limit of detection (LOD) for the analyte. Although dependent on the antibodies used, Luminex particle-based assays, as reported by the Vignali research group, can achieve a sensitivity of ∼3 pg/mL, whereas traditional ELISAs can detect cytokines down to ∼0.1 pg/mL.102 Another challenge for fluorescent particle-based assays is antibody cross-reactivity. Cross-reactivity for a multiplexed assay occurs when any of the affinity reagents bind to other target molecules or other capture molecules creating a false positive signal. This challenge can be seen when sample sera contains nonspecific antibodies such as heterophile antibodies.104 Lastly, because these assays employ fluorophores with some spectral overlap within and between particles, crosstalk can occur.105
Particle-Templated Amplification Assays
By encapsulating spherical particles in droplets surrounded by oil, reactions occurring on each particle can be partitioned, enabling signal amplification and improved limits of detection (picomolar scale).28 This expands the capabilities beyond measuring direct binding of an analyte to capture reagents on particles through labeled detection antibodies.106 The monodispersity of the particles used in the assay is crucial because each particle templates a droplet around it that ensures that there are uniform conditions for reactions on each particle enabling direct comparison or detection of amplified products.24 Compartmentalized reactions have included nucleic acid amplification tests and ELISAs.23,24,28 The compartmentalization reduces crosstalk between neighboring compartments as reaction products remain localized within the templated droplet. A significant characteristic of spherical particle-templated assays is the ability to rapidly generate numerous subnanoliter compartments in parallel, which is critical for digital assays. For example, Wang et al. showed the ability to create >500,000 compartments (drops) within 2 min using simple pipetting to create a particle-templated emulsion.24 This is an essential component of this technology as it allows for digital assays, where the presence of single molecules within a subset of the confined drops can be amplified and detected through counting compartments with signals above a threshold.
Principles of Particle Fabrication
Similar to hollow shell particles and nanovials, microfluidic droplet generators are used to create monodisperse spherical hydrogel particles suitable for particle-based emulsion assays (Figure 5a). By adjusting the flow rate of polymer precursor and oil phases, the viscosity of fluids, and the geometry of the channels, the size and stability of particle generation can be fine-tuned. Commonly used precursor solutions of the dispersed polymer precursor phase include acrylamide monomers,107 PEG-based derivatives,108 and agarose.23,68 Categorized by channel geometry, flow-focusing devices109 and step emulsifier110 devices are the most commonly used. In the step emulsifier, particle formation is driven by Laplace pressure differences arising from the geometric structure of the microchannel. The generation of particles can be further enhanced by parallelization of the dispersed phase channels, achieving high-throughput production of particles.111,112
Figure 5.
Schematic of spherical particle-templated assays. (a) Particle fabrication methods using microfluidic devices for creating spherical hydrogel microparticles. Commonly used designs include flow focusing and step emulsification devices. Brightfield image of example microparticles produced. (b) Analytes used in particle-templated assays including nucleic acids, proteins, and single cells. (c) Methods for creating particle-templated emulsions using readily available lab instruments, which include vortexing and pipetting, resulting in uniform emulsification. Brightfield image with fluorescent overlay of an example particle-templated emulsion. Scale bars are 100 μm.
Emulsification and Functionalization of Particles
Water-in-oil emulsions can be formed around spherical particles, making particle-templated emulsions (PTEs) that can be used for molecular assays. The commonly used method to form these templated emulsions is creating a mixture of particles, reagents, and immiscible oil phase, followed by vortexing or pipetting vigorously to achieve emulsification (Figure 5c).24,28 This method excludes the use of microfluidic systems, expanding access to particle-templated emulsification. With the large surface area of particles, more enzymes and substrates could be immobilized and concentrated locally to increase the efficiency of reaction compared to microwell systems. Furthermore, compared to agitation-based emulsification without particles, particle-templated emulsification creates more homogeneous droplets, enabling consistent reaction conditions.24,28
There are several strategies for functionalizing particles with enzymes, capture antibody, or aptamer.28,106 Since the functional groups in the polymer backbone can be easily manipulated, particles can be decorated with aldehyde, thiol, vinyl sulfone, or other groups to enable various chemical conjugation strategies such as Schiff base and thiol–ene reactions.113,114 Furthermore, chemically modified biotin and streptavidin can be bound to the particles, allowing for the particles to be coated with a wide range of biotinylated antigens and antibodies.
Applications of Particle-Templated Droplets
Particle-templated assays present advancements over direct sandwich assays on microparticles as they can detect as low as femtomolar concentrations of proteins and provide more accurate absolute quantification.115 Here, we summarize some of the applications for nucleic acid, protein, and single-cell analysis (Figure 6).
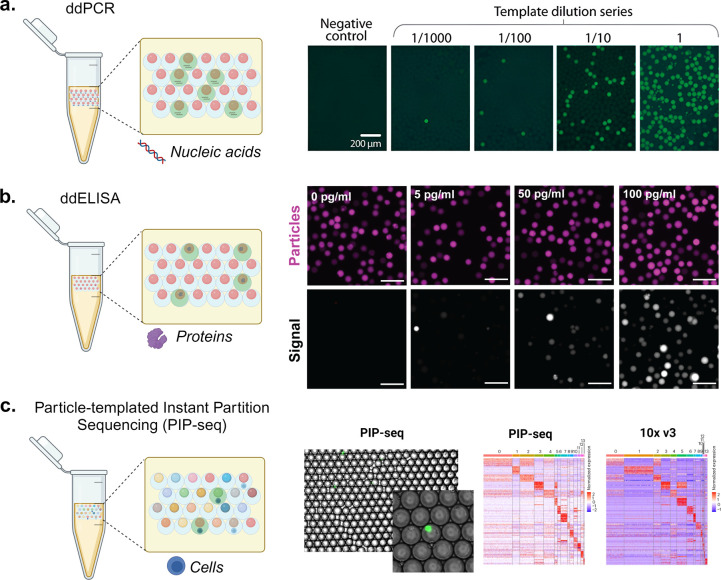
Figure 6.
Molecular and cellular assays using particle-templated emulsions. (a) Using PTEs to perform droplet digital PCR (ddPCR). The schematic shows droplets positive (green fluorescence) and negative (no fluorescence) for nucleic acid amplification using ddPCR. The fluorescence microscopy images show the amplification of yeast genomic DNA at varying dilutions in a digital regime where the fractions of positive droplets (with fluorescence) correspond with the DNA concentration. Scale bars are 200 μm. [Reproduced from Hatori, M., et al. Particle-Templated Emulsification for Microfluidics-Free Digital Biology. Anal. Chem.2018, 90 (16), 9813–9820 (ref (28)) Copyright 2018 American Chemical Society.] (b) Using PTE to perform ddELISA. The schematic shows droplets positive (green fluorescence) and negative (no fluorescence) for analyte binding and enzymatic amplification using ddELISA. The fluorescence microscopy images illustrate particles (magenta) and fluorescence signal from enzymatic turnover (grayscale) where an increasing fraction of positive droplets corresponds with an increasing concentration of a heart failure protein biomarker. Scale bars are 100 μm. [Reprinted with permission from ref (119). Copyright, 2023 V. Shah.] (c) Using PTE to perform single-cell RNA sequencing. The schematic illustrates coencapsulation of particles and cells in droplets, where color indicates different oligonucleotide barcodes on particles. Similar cell clustering and marker genes are observed for PIP-seq compared to 10X Chromium V3 workflows for cells from healthy breast tissue. [Reprinted with permission from Macmillan Publishers Ltd.: Nature, Clark, I. C., et al. Nat Biotechnol.2023, 41 (11), 1557–1566 (ref (31)). Copyright 2023.]
Particle-Templated Droplets for Nucleic Acid Quantification
Particle-templated droplets can be used for digital nucleic acid amplification to yield higher sensitivity and accuracy for targeted nucleic acid detection and quantification compared to conventional quantitative PCR (qPCR).116 Digital nucleic acid amplification (dNAA) uses compartments (e.g., chip chambers, droplets, and particles) to fractionate a sample resulting in zero or one nucleic acid sequence into each partition.117 This enables calculation of the sample concentration based on Poisson statistics. Chambers and droplets for dNAA are either limited in scale by the footprint of the chip or require a serial process of droplet generation. To resolve these issues, Hatori et al. employed particle-templated emulsification for microfluidics-free dNAA (Figure 6a).28 In contrast to qPCR, which can be used to detect low concentrations but relies on a standard curve for quantification and has challenges with measuring small concentration differences, particle-templated dNNA has the ability to quantify absolute concentrations via the digital readout of the positive and negative compartments, enabling the detection and quantification of low-concentration analytes in diverse biological samples.
Particle-Templated Droplets for Protein Quantification
Particle-templated droplets include both a solid-phase comprising high-specificity affinity reagents as well as a compartment that limits transport and promoting accumulation of amplification reactions, creating the opportunity for sensitive protein quantification assays (Figure 6b).24 This can be useful for clinical applications such as blood tests where the target protein may be of low abundance. Digital ELISA approaches apply similar Poisson loading statistics of individual analytes into compartments to directly quantify concentrations by counting amplified compartments. Early digital ELISA demonstrations by Rissin et al. yielded protein detection limits in serum at <10–15 M, which is much lower than the detectable concentrations of conventional ELISA.118 Microparticles were seeded into microchambers in these pioneering experiments, and custom optical analysis systems were used for readout, which has been a limitation for widespread adoption. Wang et al. were able to use particle-templated droplets to streamline the assay and increase the number of compartments while maintaining femtomolar detection limits for single enzymes. These assays can be multiplexed by changing the particle shape, size, or fluorescent label on each particle, allowing for the simultaneous measurement of multiple targets within a single sample, which is a step beyond the capabilities of plate-based, or even digital ELISAs.24,119
Particle-Templated Droplets for Analyzing Cells
Early work from Richard Mathies’ lab used particle-templated emulsions to perform nucleic acid amplification on individual cells encapsulated in solidified agarose droplets, although this was initially limited to a few target molecules.23 The advent of single-cell RNA sequencing (scRNA-seq) using microfluidic droplets or wells has enabled the study of the entire transcriptome of individual cells, significantly expanding the ability of templated droplets for nucleic acid analyis.23,31 Performing scRNA-seq has required microfluidic expertise or commercial instruments such as the 10X Chromium or BD Rhapsody systems. To improve ease of use and access, the Abate group developed particle-templated instant partition sequencing (PIP-seq, Figure 6c).31 This approach is scalable and is an instrument-free scRNA-seq method that can compartmentalize cells into oligonucleotide-barcoded hydrogel particle templated droplets without microfluidics. The accuracy of these systems is comparable to the current gold standard, the 10X Chromium workflows, as demonstrated by Clark et al., yielding similar results in classification, marker identification, and gene expression levels.31 Nevertheless, challenges persist with cells encapsulated in particle-templated emulsions (PTEs), notably the unclear impact on the yield of starting cell populations, potentially due to cell shearing from particle surfaces during emulsion formation. Moreover, high fluid shear stress may damage cells, leading to the risk of mRNA release and cross-contamination between compartments.120 Fluent BioSciences has developed a PIP-seq product that is currently commercially available to perform particle-templated sequencing.121
Strategies for Multiplexing of Assays on Spherical Particles
Labeling particles facilitates the simultaneous multiplexing of various assays on a singular particle platform. For instance, oligonucleotide barcoding, with its high fidelity and vast sequence possibilities, is particularly suited for extensive particle barcoding, such as encoding mRNA from thousands of single cells. Further, polyacrylamide-based hydrogel particles can be labeled using custom acrydite-modified primers, followed by the sequential assembly of the barcode sequences using split and pool techniques.121 Labeling particles with colors is beneficial for direct visualization and detection using optical approaches.106,122 Strategies to achieve this include embedding nanophosphors,123 quantum dots,124−126 fluorescent,127,128 or photonic nanoparticles129,130 into the particle templates. Notably, the Fordyce group has reported the most extensive spectral barcode library to date, comprising over a thousand unique codes, by using lanthanide nanophosphors (LNs) in varying ratiometric quantities.131 In constructing these barcoded particles, four different LNs are mixed in varying ratios, with one LN consistently integrated at a fixed concentration to serve as a baseline for comparing the relative intensities of the other colors. LNs offer distinct advantages over traditional spectral barcodes, such as excitation at a single wavelength, narrow emission spectra, and resistance to photobleaching and quenching unlike organic fluorophores. However, it is important to note that these advanced barcoding techniques often require specialized instruments for readout.
Multiplexing capabilities for spherical particles using standard instruments, like flow cytometers, are limited since, besides fluorophore spectral barcoding, the only additional tunable geometric factor is the spherical particle diameter. Thus, alternative particle barcoding methods are needed to further expand the multiplexing levels possible, such as particle-shape barcoding.
Flow-Lithography Particles and Shape-Based Barcoding
Shape-barcoded particles have emerged as a solution to extend multiplexing capacity, using physical shape as a barcode instead of—or in addition to—fluorophores. Strategies for shape-based barcoding have been developed to create uniquely identifiable structural or graphical variations, such as (1) particles with different outlines or through-hole shapes; (2) particles with distinct graphical features such as characters, pictograms, numbers, and “QR codes”; and (3) particles that incorporate a combination of shape, graphical, and spectral barcoding.
General Workflow for the Fabrication and Use of Shaped Particles
Fabrication of Shaped Particles
Flow lithography techniques are used to polymerize shaped particles within microfluidic channels. The process uses photopolymerizable polymer precursors that are formed into particles through UV cross-linking, initiated by photoinitiators. Depending on the channel structure and arrangements of coflowing streams, specific flow patterns can be generated. Coflowing streams can comprise single or multiple inert and/or reactive polymers to define particle shapes within the microfluidic channel under UV light projected through a photomask, forming predefined and potentially multimaterial structures (Figure 7a). Additionally, the use of multiple flow streams containing different cross-linkable polymer precursors allows for the creation of multimaterial substrates, important for spatially selective reactivity or hydrophilicity or chemical functionalization. The precursor solution can be modified to include probes and detection agents necessary for assay workflows;30,132 it can also incorporate binding sites for conjugating the required reagents and probes after particle fabrication.22,133,134 Similar to other LoP technologies, biotin–streptavidin chemistry provides modular binding sites for affinity assays or cell binding.22
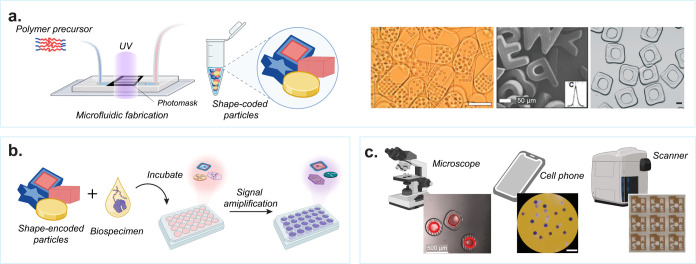
Figure 7.
Workflow for the fabrication and use of shaped particles. (a) Fabrication of shaped microparticles in microfluidic devices using photomasks and UV polymerization. Representative shape-encoded particles used for biomolecular detection. [From Pregibon, D. C., et al. Multifunctional Encoded Particles for High-Throughput Biomolecule Analysis. Science2007, 315 (5817), 1393–1396 (ref (30)) reprinted with permission from AAAS; Kim, L. N., et al. Chem. Commun.2015, 51 (60), 12130–12133 (ref (133)) with permission from The Royal Society of Chemistry; and Destgeer, G., et al. Lab Chip2020, 20 (19), 3503–3514 (ref (22)) with permission from The Royal Society of Chemistry.] (b) Schematic representation of assay workflow using shaped microparticles including incubation and signal amplification steps. (c) Readout of assay results on individual microparticles using standard instrumentation such as microscopes, cell phones, or scanners. [Reproduced from Destgeer, G., et al. Lab Chip2020, 20 (19), 3503–3514 (ref (22)) with permission from The Royal Society of Chemistry; Derveaux, S., et al. Anal. Bioanal. Chem.2008, 391 (7), 2453–2467 (ref (135)) with permission from The Royal Society of Chemistry; and Svedberg, G., et al. Lab Chip2017, 17 (3), 549–556 (ref (134)) with permission from The Royal Society of Chemistry.]
Incubating with Samples and Detection Reagents
Following fabrication, the probe-laden microparticles are combined with a biospecimen, enabling the selective binding of analytes to the corresponding probes on the particles (Figure 7b). The analyte is locally bound at the active binding sites on the particles, facilitating solution exchange and wash steps to introduce additional reagents or eliminate unwanted background, thereby enhancing specific signals. Multiple types of particles, each having a unique shape, can be processed in a single assay workflow enabling parallelization and multiplexing. Detection and quantification of nucleic acids30,133,135 and proteins29,134 have been demonstrated through the subsequent binding of fluorescent labels. Detection of analytes that are low in abundance is achieved through enzymatic amplification techniques or localization of signal to a small fraction in the particles to enhance the signal-to-noise. Moreover, shaped particles offer an advantage over other two-dimensional or well-based multiplexed assays due to their ability to be freely suspended in solution, resulting in improved binding kinetics.136 Multiplexed detection of a wide range of biomarkers of importance in health can help obtain comprehensive insights in a cost-effective manner.137
Optical Readout of Shaped Particles
The final step in using shaped microparticles for molecular diagnostics is optical measurement (Figure 7c), which is typically through fluorescence30,132 and/or brightfield134,135 imaging.29 This is accomplished using microscopes and cameras or low-cost readers, employing the economies of scale for consumer electronics devices like cell phones and flatbed scanners. Such readers can be deployed widely, making these technologies accessible in resource-limited regions. By capitalizing on the high multiplexing capacity of shape barcoding, multiplexed assays can be achieved using a single fluorescent channel without compromising the upper limit of the number of biomarkers of interest.30 The use of multiple particles to detect the same analyte also provides improved statistical certainty to measurements, what is also referred to as “swarm sensing”.29 This approach enhances the scale and accuracy of molecular diagnostics in a variety of biological applications.
Advances in Flow-Lithography Particle Fabrication Methods
The history of shape barcoding in LoP technologies is centered around the patterning of photopolymerizable materials. These materials transition from a liquid to a solid state through light-induced cross-linking.138 Early shaped particles were crafted using contact lithography, a technique involving the use of a physical mask onto which the photopolymerizable material was applied and cured.139,140 Replica molding, a process where a mold is used to pattern shaped particles, was also widely used.141 However, these methods are low-throughput as the speed of particle fabrication is limited by the two-dimensional area of the mask or mold.142 The Doyle group addressed this limitation with continuous-flow lithography. Continuous-flow lithography (Figure 8a) combines microscope projection photolithography with microfluidic flow of the precursor polymer stream and inhibition of polymerization at the channel walls, resulting in a high-throughput, precisely controlled, and adaptable approach to creating microparticles, offering flexibility in terms of size, shape, and composition.10 Due to continuous operation, there may be a compromise in the resolution of particles cured from the moving stream. The Kwon group developed optofluidic maskless lithography (OFML), in parallel, where a dynamic mask, composed of a digital micromirror device, could generate diverse patterns with single-pixel resolution (1.54 × 1.54 μm2) during a 0.1 s exposure time (Figure 8b).143 The dynamic mask also allowed for synchronizing exposure with a particle as it flowed downstream during curing, reducing flow-induced blurring of particle shapes.
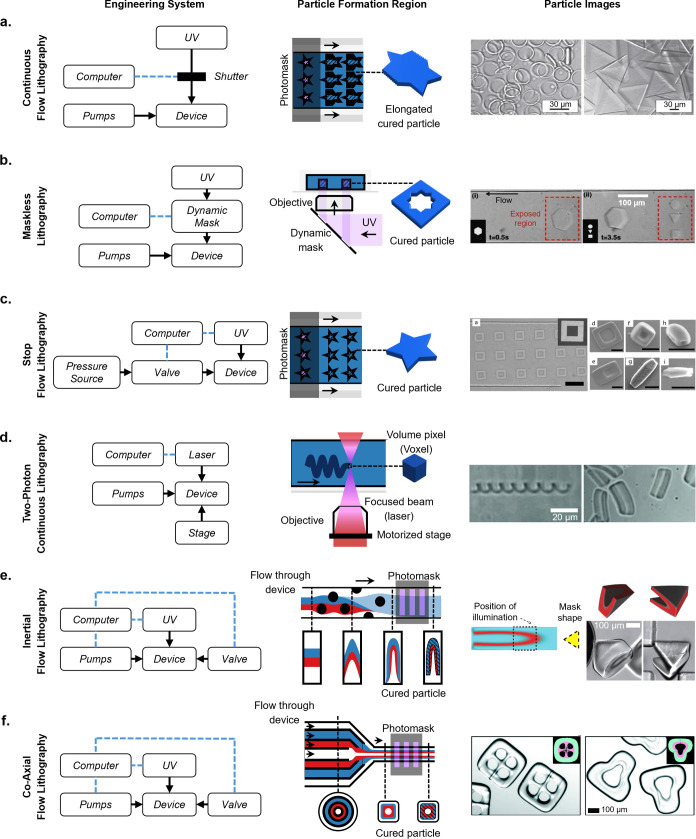
Figure 8.
Shaped particle fabrication methods by flow-lithography from oldest to newest. (a) Continuous flow lithography particles. The fabrication system requires a computer-controlled shutter and continuous flowing pumps. Scale bar is 30 μm. [Reprinted with permission from Macmillan Publishers Ltd.: Nature, Dendukuri, D., et al. Nature Mater.2006, 5 (5), 365–369 (ref (10)) Copyright 2006.] (b) Maskless lithography particles. The shutter is replaced with a dynamic mask. Scale bar is 100 μm. [Reprinted with permission from ref (143). Anna, S. L., et al. Applied Physics Letters2003, 82 (3), 364–366 licensed under a Creative Common Attribution (CC BY) license.] (c) Stop flow lithography particles. A computer-controlled valve is introduced to control flow, resulting in higher resolution particles. Scale bar is 30 μm. [Reproduced from Chung, S. E., et al. Applied Physics Letters2007, 91 (4), 041106 (ref (144)) with permission from The Royal Society of Chemistry.] (d) Two-photon continuous lithography particles. This fabrication system requires both a focused laser and a motorized stage. Scale bar is 20 μm. [Reproduced from Stop-Flow Lithography in a Microfluidic Device. Dendukuri, D., et al. Lab Chip20127 (7), 818–828 (ref (145)) Copyright 2012 Wiley.] (e) Inertial flow lithography particles. Before fabrication, the device used to shape flow and the necessary photomask is predesigned using uFlow. Pillars in the device create complex flow geometries, and a shaped photomask produces particles of the desired shape. Scale bar is 100 μm. [Reproduced from Two-Photon Continuous Flow Lithography, Advanced Materials201524 (10), 1304–1308 (ref (146)) Copyright 2015 Wiley.] (f) Coaxial flow lithography particles. A 3D-printed device shapes the coaxial flow. Scale bar is 100 μm. [Reproduced from Destgeer, G., et al. Lab Chip2020, 20 (19), 3503–3514 (ref (22)) with permission from The Royal Society of Chemistry.] Engineering systems and close-up views of particle fabrication region adapted from Lewis, C. L., et al. Anal. Chem.2010, 82 (13), 5851–5858 (ref (142)) with permission from The Royal Society of Chemistry.
The Doyle group advanced their previous technology by introducing stop-flow lithography (SFL). This configuration involves the use of a valve either upstream or downstream of the device to halt the flow of the prepolymer solution before polymerization (Figure 8c).144 This meticulous control leads to an improvement in the resolution of the fabricated particles. However, this resolution enhancement comes at the expense of fabrication throughput. The need for the fluid to alternate between stationary and flowing states introduces an inherent limitation in the overall time and material efficiency of the process.
By merging an older technology with microfluidics, the Pisignano group introduced continuous-flow two-photon flow lithography in 2012. This method combines two-photon lithography with continuous flow, where an ultrafast laser focused through an objective excites the polymer precursor fluid and polymerizes it as the fluid flows (Figure 8d).145 Unlike other fabrication methods, this technique allows for the creation of high-resolution particles with highly complex 3D shapes without the need for a sheath or inert layer. It can generate particle geometries not previously demonstrated by other methods, such as springs, and can achieve submicrometer features in three dimensions (with surface roughness as low as 10 nm). However, this sequential polymerization method is inherently lower throughput and comes with more intensive technology requirements: an ultrafast laser and a programmable stage capable of movement in at least two dimensions (y and z).
Complex Multimaterial Particles
In 2015, the Di Carlo group developed a technology capable of fabricating multimaterial three-dimensional structures in a flow-lithography format. This technology, known as optical transient liquid molding (OTLM), combines flow shaping using inertial microfluidics and stop-flow lithography (Figure 8e).146 The group introduced a flow design software named “uFlow”, where users can adjust the optical mask and microchannel structure, and the software predicts a final flow shape and particle geometry. Multimaterial particles with inner cavities, 3D structures, and spatially selective patterning of chemistry and hydrophilicity were fabricated using these flow configurations. Early applications of amphiphilic particles, featuring a hydrophilic inner material and a hydrophobic outer material, included spontaneous formation of uniform emulsions through stabilizing aqueous droplets.12 However, the topology of particles achievable using this technique is limited by the initial side-by-side configuration of coflowing prepolymer streams.
Coaxial flow lithography enhances previously discussed fabrication methods by employing a 3D printed microfluidic device that shapes the flow, allowing multiple fluid streams to flow coaxially and create streams within streams (Figure 8f).22 This enables the production of multimaterial particles with inner cavities. Changing the 3D printed microfluidic device allows the creation of multiple shapes with different inner and outer geometries. Additionally, the particle surface can be patterned using a photomask during the curing step. The overall workflow is similar to a stop-flow lithography setup, making it accessible to laboratories already capable of stop-flow lithography, with the introduction of 3D printed devices for particle production.
Shaped Particles for Nucleic Acid Quantification
The diversity of the microparticle types available increases dramatically with shape.22,134,147 Utilizing the geometric properties of microparticles to barcode diverse analytes unlocks an almost unlimited number of possibilities for molecular30,126,148 and cellular multiplexing applications.149 For instance, the Kwon group used magnetic color and shape-barcoded microparticles to raise the multiplexing limit to the level of billions.147 The scale is suitable for the tremendous diversity of DNA and RNA sequences that are present in living cells.
Shaped Particles for DNA Detection
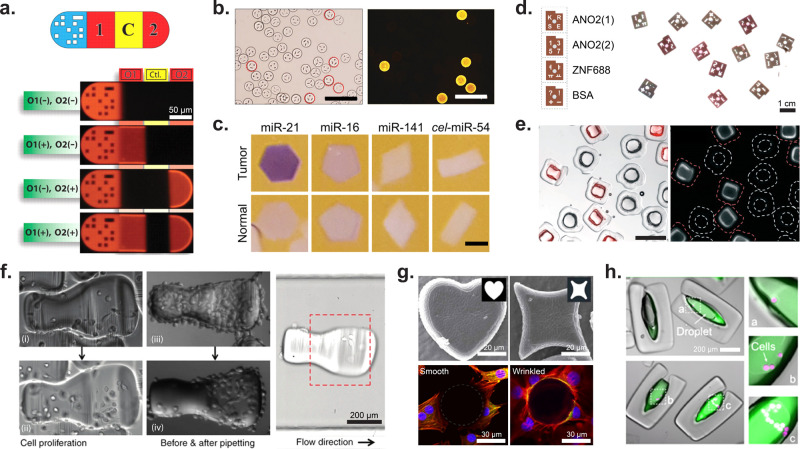
The Doyle group reported a graphical barcoding method to generate dot patterns on a microcarrier particle encoding different DNA targets for hybridization assays. As shown in Figure 9a, the particle consisted of a dot pattern for barcoding, two DNA oligomer probes (O1 and O2), and a negative control area.30 The results showed that O1 and O2 targets can be detected simultaneously with no crosstalk. This method allows for multiplexed, single-channel fluorescence detection of DNA with high simplicity, providing high-density information using a single sample.
Figure 9.
Application of shaped microparticles for multiplexed and cellular assays. (a) DNA detection on barcoded particles. The particle is subdivided as follows: graphical barcode portion, detection section for DNA oligomer 1, control (should always be dark), detection for DNA oligomer 2. Particles are approximately 90 μm in width and 180–270 μm in length. [From Pregibon, D. C., et al. Multifunctional Encoded Particles for High-Throughput Biomolecule Analysis. Science2007, 315 (5817), 1393–1396 (ref (30)) reprinted with permission from AAAS.] (b) HPV DNA mutant detection where each differently shaped particle is conjugated to probes for a unique HPV DNA mutant. Images of positive particles corresponding to the barcode for HPV mutant 33. Scale bars are 200 μm. [Reproduced from Kim, L. N., et al. Chem. Commun.2015, 51 (60), 12130–12133 (ref (133)) with permission from The Royal Society of Chemistry.] (c) Images showing miRNA detection (purple) from normal and tumor cells. The oncogenic miRNA, miR-21, is seen in higher concentrations in the particles incubated with tumor lysate. Slightly elevated levels of miR-16, an endogenous standard in miRNA analysis of colon cancer, was seen in the particles incubated with tumor lysate. miR-141 is a marker of poor prognosis associated with advanced colon cancer. The probe cel-miR-54 was used as a negative control. Scale bars are 200 μm. [Reproduced from Derveaux, S., et al. Anal. Bioanal Chem.2008, 391 (7), 2453–2467 (ref (135)) with permission from The Royal Society of Chemistry.] (d) ELISA read out on optical scanner for the detection of autoantibodies from multiple sclerosis patients. ANO2 (1) serves as a positive control for MS autoantibodies, ANO2 (2) serves as negative control, and ZFN688 is a secondary negative control. Particles have a diameter of 900 μm and a thickness of 150 μm. [Reproduced from Svedberg, G., et al. Lab Chip2017, 17 (3), 549–556 (ref (134)) with permission from The Royal Society of Chemistry.] (e) Enzyme-linked assay in particle-templated drops where the fluorescent product within a droplet supported by square particles accumulates while no fluorescent product accumulated in negative control circular particles. Particles range in size from 340–400 μm with cavity dimensions of 100–200 μm. Scale bar is 500 μm. [Reproduced from Destgeer, G., et al. Lab Chip2020, 20 (19), 3503–3514 (ref (22)) with permission from The Royal Society of Chemistry.] (f) MDA-MB-231GFP cells on collagen patterned microcarriers. The first column shows cell proliferation on a microcarrier over time; the second column shows how cells on the nonshelter region were removed via pipetting while cells in the shelter area were protected from pipetting shear forces. The third image shows how the microcarrier orients in flow. Scale bar is 200 μm. [Reprinted with permission from Macmillan Publishers Ltd.: Nature, Wu, C., et al. Microsyst. Nanoeng.2018, 4 (1), 21 (ref (157)) Copyright 2018.] (g) Wrinkled, nonspherical particles. The top row shows two different wrinkled particle shapes, while the bottom row shows cell adhesion differences between a spherical nonwrinkled particle (left) and a wrinkled particle (right). Scale bar is 20 μm (top) and 30 μm (bottom). [Reprinted with permission from Macmillan Publishers Ltd.: Nature, Li, M., et al. Sci. Rep.2016, 6 (1) 30463 (ref (159)) Copyright 2016.] (h) LNCaP cells (magenta) encapsulated in amphiphilic particles. The fluorescence intensity of MMP-cleavable fluorogenic substrate (green) increases with increasing number of encapsulated cells. Scale bar is 200 μm. [From Wu, C., et al. Monodisperse Drops Templated by 3D-Structured Microparticles. Science Advances2020, 6 (45), eabb9023 (ref (12)) reprinted with permission from AAAS.]
The Kwon group developed a method to fabricate shaped-encoded silica microparticles (SSMs) with OFML, coating polymeric microparticles with a silica layer, and conjugating hybridization probes.133 By harnessing the huge encoding capacity of SSMs, they performed 10-plexed in vitro human papillomavirus (HPV) genotyping (Figure 9b). Only the SSM with probes complementary to the target sequence show strong fluorescence signals by R-phycoerythrin conjugated streptavidin that bound to biotinylated detection probes. The SSMs were able to detect target HPV sequences with high specificity and showed minimal noise from cross-hybridization or adsorption.
Shaped Particles for RNA Quantification
The Doyle group demonstrated the detection of multiple human mRNA targets using a single hydrogel particle. To enhance the sensitivity of shape-barcoded particles to the femtomolar level, they increased mRNA transport into the particles by enhancing particle porosity through the incorporation of porogens.148 LoP-based systems offer the advantage of tunable particle properties beyond just shape and size, as factors like particle porosity, hydrophilicity or hydrophobicity, and availability of active binding sites can be modified during the fabrication process.
Shape-barcoded microparticles have shown significant utility in the multiplexed detection of miRNAs, which are key in understanding and diagnosing various diseases (Figure 9c).135 The Doyle group demonstrated a platform that uses shape-encoded hydrogel particles, synthesized using stop-flow lithography, for multiplexed miRNA detection, achieving detection limits as low as 0.22 femtomoles for colon cancer clinically relevant miRNAs.150 Expanding on this concept, the limit of detection is further improved through a microfluidic platform that isolates functional hydrogel microparticles in monodisperse droplets, integrating an enzymatic amplification scheme that enhances sensitivity by an order of magnitude compared to direct labeling.150
Another advancement in RNA detection was the introduction of a shape-barcoded particle-based RNA quality assay. The Yi group pioneered this assay to assess bacterial rRNA titer and RNA sample integrity by targeting rRNA sequences common to all E. coli ribosomes. Their research demonstrated that the fluorescence intensity of the shape-barcoded particles declined as rRNA degradation increased.151
Shaped Particles for Protein Quantification
Shaped microparticles are a promising solution to expand multiplexing of protein immunoassays. Doyle’s group reported hydrogel microparticles with graphical codes for multiplexed detection of protein panels (IL-2, interleukin-4, and TNF-α) with very low detection limits (1–8 pg/mL).132 This protein panel has clinical value since cytokine signaling initiates immune responses and maintains immune system memory. The technology laid the foundation for the Fireplex multiplexed particle-based assays, currently commercialized by Abcam. FirePlex immunoassays offer high multiplexing capacity, sensitivity, and specificity (a detection range of 0.5 pg/mL over a 5-log dynamic range).152,153 This technique allows up to 70 analytes with a single 12.5 μL sample. The FirePlex particles consist of three functional regions separated by two inert spacers: both ends serve for encoding by varying the combination of green and yellow, fluorescent dyes, and the antigen-specific capture antibodies are bonded in the center of the particles, as well as the spacers for reducing fluorescent code interference to the fluorescent analysis of central parts using a flow cytometer. The human cytokine panels can detect 17 cytokines with high sensitivity.152,154
To reach even higher multiplexing, the Kwon group developed lithographic encoded particles (LEPs), which achieve up to two million codes through the patterning of four unique numbers, characters, and symbols on the particles.134 Their proof-of-concept panel utilized four different barcoded particles for multiplexed detection of autoantibodies (ANO2(1) and ANO2(2)) toward the ultimate goal of diagnosing multiple sclerosis (MS) (Figure 9d). Using MS plasma and healthy controls, the Kwon group demonstrated the ability to detect the target autoantibodies with high specificity. As the assay was not enzymatically amplified, the limit of detection was limited by the direct detection of antigens.
Amphiphilic particles, as a class of shaped microparticles, are composed of both a hydrophobic and hydrophilic region and can spontaneously stabilize aqueous droplets when submerged in oil.22,29,155 These particles can be fabricated using either OTLM or coaxial flow lithography and are well-suited for amplified immunoassays due to their amphiphilic nature.22,146,155 During initial incubation steps, the particles can be freely exposed to aqueous solutions sequentially, in the same manner as a traditional ELISA. However, during signal amplification, each particle can act as an individual droplet compartment when submerged in oil by harnessing the inner layer hydrophilicity to form uniform picoliter- to nanoliter-volume droplets within the inner hydrophilic region. Amphiphilic particles were fabricated with more than 12 shapes by changing the 3D-printed nozzle designs, demonstrating capacity for three-dimensional shape-based multiplexing.22 Initial proof-of-concept assays demonstrated that particles with horseradish peroxidase (HRP) bound to their inner cavity would form a fluorescent substrate localized to the particle inner cavity droplet with minimal crosstalk to their non-HRP-containing neighbors (Figure 9e).22 Further studies demonstrated that these particles could be used to measure the heart failure biomarker, N-terminal pro-B-type natriuretic peptide (NT-proBNP), with lower limits of detection than traditional binding assays.29
Shaped Particles for Cellular Analysis
Shaped particles, especially cavity-containing shaped particles, can be used for holding and analyzing cells and their products. Adherent cells that spread over spherical particles were recently analyzed using imaging flow cytometry, demonstrating unique morphological features not observable for cells in a suspended state.156 However, the orientation of a spherical particle is not controllable and affects the imaging plane for the adhered cells. Furthermore, cells remain on the exposed particle surface and are subject to fluid shear stresses. Shaped particles have been used as cell carriers for adherent cells. Cell carriers designed using inertial flow lithography were designed not only to shield cells from shear forces with a recessed cavity but also to align in flow, ensuring that cells are aligned properly in the channel for consistent imaging of cell morphology (Figure 9f).157 Beyond modifying particle shape, other groups have developed ways to increase the adhesive properties of the particle surface. The Irimia group used SFL to create anisotropic particles that contain sections functionalized with ECM favorable to cell adhesion. They demonstrated their ability to culture both endothelial and breast cancer cells on the ECM-functionalized particles.149 Another method of generating adhesive particles is to use the acrylate groups unpolymerized after SFL and conduct an aza-Michael reaction between these acrylate groups and amines in cell adhesion promoters (such as PLL or RGD).158 Beyond adding proteins to the surface of particles, the surface morphology of the particles can be modified to increase roughness and aid cell adhesion (Figure 9g).159
Amphiphilic particles that template the formation of droplets within oil were also used to quantify the secretions from cells encapsulated within. A fluorogenic assay for matrix metalloproteases (MMPs) enabled the quantification of MMP activity from single cells over time using a fluorescence microscopy readout (Figure 9h).12 The fluorogenic substrate was cleaved to a fluorescent product by MMPs, and this product accumulated within the particle-templated drops to detectable levels over hours.
Conclusions and Future Directions
Lab on a particle technologies represent a next step in scaling of biological and chemical assays. Microparticles are sized to be well-suited to analyze single cells and single molecules, the ultimate “quanta” of biology. The small size of particles also means millions of barcoded assays or separate compartments can be analyzed in parallel even from a small sample volume. For example, millions of nanovials can fit into a 500 μL volume. Compared to the most highly parallelized multiwell plates, the volume supported by each particle is more than a thousand times smaller. These particle systems also forego the robotic automation of microwell plates or engineered instruments that interface with lab on a chip devices through fluidic ports or electrodes and instead use standard liquid handling processes in life science laboratories. Automation of analysis leverages equipment suited to analyze single cells at high throughput and with high precision (Figure 1d).
Building on knowledge from the biomaterials field, there are numerous opportunities to modify particles to interface with cells and molecules. The porosity of particles can be engineered to selectively entrap and enrich particular biological entities.148 Particle stiffness and viscoelastic properties can play a role in modulating the behavior and function of bound cells.160 Local biochemical functionalization can be used to isolate specific cells or molecules, where large surface areas of fractal surfaces and porous hydrogels give unique opportunities for local concentration enhancement and avidity effects.161 These effects are expected to drive improved molecular assays and capture of molecules released from cells.
Particles can also be modified to act as artificial cells. Adding specific cell surface proteins can allow researchers to deconstruct the interactions between cells and characterize secretions or morphological changes that occur upon interacting with an artificial cell. For some membrane proteins that are not easily produced in a recombinant form, we anticipate extracellular vesicles or virus-like particles can be linked to particle surfaces instead to present the correct epitopes in a phospholipid bilayer membrane. These approaches could be particularly interesting for studying the immune synapse between T cells and antigen-presenting cells and critical elements leading to functional responses, such as secretion of effector molecules. Similarly, the immune synapse by which B cells interact with antigen through clustering of B cell receptors could be further explored. These platforms are ideal for functional screening and discovery of the next generation of cell therapies based on engineered receptors (TCRs, CARs, T cell engagers, etc.). Because particles are compatible with flow cytometry and single-cell sequencing, multiple different analysis modes can be pursued, or linked modes, e.g., secretion and sequencing as demonstrated with SEC-seq.9,26
Functionalization and design of particles will continue to be an active area of research, opening up new applications. Ongoing studies on the nanovial platform include screening of other secreted products, such as MMPs, growth factors, and non-IgG antibodies, to better understand how to engineer cells to secrete specific therapeutic molecules. Particles can also be functionalized with other ECM proteins to study cell–ECM interactions in disease models, such as fibrosis. Including ECM matrix components in the nanovials can further mimic the cellular microenvironment. Methods to link and barcode a huge array of biofunctional particle types in a simple workflow are still needed.
Most studies to date have focused on analyzing individual cells on particles; however, characterizing how cells interact is also feasible. By loading multiple cells into a particle cavity in a sequential manner, multicell assays are possible. Applications include characterizing (1) how cells interact and secrete molecules during development; (2) how immune cells (T cells, NK cells, neutrophils, macrophages) recognize target cells, secrete molecules, and ultimately kill target cells; and (3) how cells respond when exposed to pathogenic or commensal microbes. Cell growth phenotypes in pure or mixed populations are also able to be explored massively in parallel using these techniques. A specific application of widespread interest is in the antibody discovery field. Screening of antibody-secreting cells to identify antibodies that bind to target cells expressing antigen can leverage a multicell workflow. As a caveat, particles with larger diameters may be needed for some of these multicell assays, which may reduce the compatibility with some flow cytometers that have narrow nozzles or certain single-cell sequencing chips.
Beyond cellular assays, numerous molecular assays, especially digital assays (nucleic acid and proteins) should benefit from LoP approaches. In recent years, digital LAMP has become a promising approach for precise and sensitive quantitation of nucleic acids.162 These assays have been successful within droplet formats and can potentially be adopted for the particle-templated formats that will allow for further flexibility in multiplexing and smaller partitions for higher specificity. One challenge with these digital approaches is the current need to encapsulate particles in an emulsion. This reduces the compatibility with most flow cytometers. Techniques like double emulsions applied to LoP systems may address this challenge, or amplification chemistries or particle designs such that the product of amplification is trapped on or within the particle to preserve the reaction without formation of an emulsion or after an emulsion is broken.
In total, the application space for LoP technologies should span a similar breadth as lab on a chip technologies: single-cell analysis, multicellular organoid models, to molecular diagnostics and drug discovery—and the research is just getting started. Finally, although a key advantage of current LoP technologies is the ability to make use of standard instruments, the tuning or design of instruments that are better suited to analyze and sort large particles, and perform additional high-content analysis of those particles (e.g., imaging, fluorescence lifetime, proteomic assays, multiomic assays), will also accelerate the field and expand the richness of data collected—optimized to feed the data-hungry AI models that will shepherd future biological discovery.
Acknowledgments
We thank Alice Matsuda, Andrew Tran, and Boyi Wang for procuring permissions for use of figures. Portions of the figures were generated using biorender.com.
Biographies
Rajesh Ghosh received his Bachelor of Technology in Chemical Engineering from SRM Institute of Science and Technology, India, in 2018. Following his graduation, he worked as a research associate at the Indian Institute of Technology Madras, where he focused on developing democratized point-of-care diagnostic tools using low-cost paper-based devices. In 2020, he began his doctoral research in Bioengineering at the University of California, Los Angeles (UCLA), under the supervision of Prof. Dino Di Carlo. His doctoral research centers on developing novel cell and protein screening platforms using lab-on-a-particle technology and directed evolution.
Alyssa Arnheim earned her Bachelor of Science in Biomedical Engineering from Boston University in 2018. Following her graduation, she worked as a research associate at Sanaria, a malaria vaccine pharmaceutical company, researching malarial parasite infectivity and coordinating vaccine extraction operations. In 2021, she started her doctoral studies in Bioengineering at the University of California, Los Angeles (UCLA), under the supervision of Prof. Dino Di Carlo. Her doctoral research focuses on developing novel lab-on-a-particle point-of-care diagnostics for heart failure and neurotoxin detection.
Mark van Zee received his Bachelor of Science and Art in Biochemistry and Physics from the University of California, Los Angeles in 2018. During his undergraduate studies, he worked as a research assistant in the Bioengineering Department at the University of California, Los Angeles (UCLA) under the mentorship of Prof. Dino Di Carlo developing gel microdrop (GMD) technologies for the isolation of fast growing microalgae colonies. He continued his research as a Junior Engineer, Ph.D. student (graduated in 2022), and postdoctoral scholar within the same lab and mentor where he developed the innovative PicoShell technology for the selection of rare clonal populations.
Lily Shang earned her Bachelor of Science in Bioengineering from University of California, Berkeley in 2021. In 2021, she started her doctoral studies in Bioengineering at the University of California, Los Angeles (UCLA), under the supervision of Prof. Dino Di Carlo. Her doctoral research includes the development and implementation of particle-based hydrogel scaffolds for wound healing and vaccine applications.
Citradewi Soemardy earned her Bachelor of Science in Biotechnology from University of California, San Diego in 2016. Following her graduation, she worked as a research associate at the Center for Gene Therapy at City of Hope exploring CAR T cells for HIV treatment. In 2021, she started her doctoral studies in Bioengineering at University of California, Los Angeles, under the mentorship of Prof. Dino Di Carlo. Her doctoral research focuses on development of particle-based single-cell screening for cell therapy applications.
Rui Chian Tang received his Bachelor of Science in Biochemical Science and Technology from National Taiwan University in 2021. Following his graduation, he worked for a year as a research assistant at the Institute of Nanomedicine and Medical Engineering, National Health Research Institutes in Taiwan. In 2022, he embarked on his doctoral studies in Bioengineering at the University of California, Los Angeles (UCLA), under the mentorship of Prof. Dino Di Carlo. His doctoral research focuses on developing and implementing innovative particle-based granular hydrogel systems for applications in tissue engineering and cell delivery.
Michael Mellody earned his Bachelor of Science in Bioengineering from the University of California, Los Angeles in 2019. He started his doctoral studies in 2019 as a National Science Foundation Graduate Research Fellow at the University of California, Irvine. He continued his doctoral work at the University of California, Los Angeles in 2021. His doctoral research includes developing automated cell culture systems and creating novel single cell assays to study fibrotic conditions.
Sevana Baghdasarian studied chemical engineering and received her Bachelor of Science degree from California State Polytechnic University, Pomona (Cal Poly Pomona) in 2015 and her Master of Science degree from University of California, Los Angeles (UCLA) in 2018. Her recent work leverages biomaterials and microfluidics to fabricate research tools to interface with cells and tissues at the cellular scale. Her doctoral thesis under the mentorship of Prof. Dino Di Carlo is focused on employing the lab-on-a-particle platform to enable immune cell profiling and secretion-based multiplex screening of T cells for TCR discovery.
Edwin Sanchez Ochoa earned his Bachelor of Science in Biomedical Engineering with a Minor in Spanish from San Jose State University in Spring 2023. In Fall 2023, Edwin started a Master in Bioengineering program at the University of California, Los Angeles. His research includes advancements in the fabrication of 3D-printed microfluidic devices and the encapsulation of organoids in large-sized particles.
Shun Ye received his B.Sc. in Physical Medicine and Rehabilitation and M.Sc. in Biomedical Engineering from Beijing Sport University and Pennsylvania State University in 2020 and 2023, respectively. He finished a research fellow training at Chinese Academy of Sciences in 2021. In 2023, he started his doctoral research in Bioengineering at the University of California, Los Angeles, under the cosupervision of Prof. Dino Di Carlo, Aydogan Ozcan, and Sam Emaminejad. His doctoral research focuses on developing novel point-of-care diagnostic platforms, including ferrobotics platforms for cancer diagnostics and vertical flow devices for cardiovascular diagnostics.
Siyu Chen earned her Bachelor of Engineering in Chemical and Biological Engineering from the University of Queensland in 2022. She started her doctoral studies and research in Bioengineering at the University of California, Los Angeles (UCLA) in 2023. Her research primarily explores the effects of matrix viscoelasticity and stiffness on muscle stem cell fate and epigenetic modifications during the aging process.
Cayden Williamson graduated from the University of California, Berkeley in 2017 with a B.S. in Bioengineering. In 2019, he began his bioengineering doctoral work at UCLA under the supervision of Professor Dino Di Carlo. His work focuses on developing novel platforms for the analysis and engineering of microbial populations.
Amrith Karunaratne received his Bachelor of Science in Biomedical Engineering from the University of California, Irvine in 2019. Since his graduation, Amrith has worked in the medical device industry as a Senior Research and Development Engineer. In 2021, he began his graduate studies at the University of California, Los Angeles under the guidance of Prof. Dino Di Carlo. His research focuses on hollow microparticle-based scaffold systems and high-resolution 3D-printed microfluidics for tissue engineering and scalable manufacturing applications, respectively.
Dino Di Carlo is the Armond and Elena Hairapetian Professor of Bioengineering at UCLA, serial entrepreneur, and inventor. He serves in academic leadership roles as the Vice Chair of the Bioengineering Department and Deputy Director of a National Science Foundation Engineering Research Center. He is an author on >190 peer-reviewed articles and an inventor on >70 issued patents in the U.S. and across the world. He has received numerous awards, including the Presidential Early Career Award for Scientists and Engineers (PECASE), the highest honor bestowed upon young scientists and engineers by the United States government. He also has served in business leadership roles. He cofounded several companies in life science research tools, diagnostics, medical device, and pharmaceutical industries over the last 10 years and continues to serve on the board of directors of many of these companies. He is a cofounder and director at Cytovale, which developed the first early sepsis diagnostic test for rapid triage in the emergency department. He is a Scientific Advisor to several life sciences companies, including Cue Health (NASDAQ:HLTH), which developed the first at-home molecular diagnostic test to receive regulatory clearance by the United States Food and Drug Administration.
Author Contributions
Dino Di Carlo (D.D.) outlined the manuscript, contributed writing to the introduction and conclusion sections, and edited the final manuscript and figures. Alyssa Arnheim (A.A.) created the figures, contributed to the writing for the flow-lithography particles and shape-based barcoding section and edited the final manuscript and figures. Sevana Baghdasarian (S.B.) contributed to the figures and writing for the nanovial section. Siyu Chen (S.C.) created the figures and contributed to the context writing for the flow-lithography particles and shape-based barcoding section. Rajesh Ghosh (R.G.) created the figures, contributed writing to the introduction and evolution of hydrogels for the cell encapsulation section, and edited the final manuscript and figures. Amrith Karunaratne (A.K.) contributed to the figures and writing for the evolution of hydrogels for the cell encapsulation section. Michael Mellody (M.M.) created the figures and contributed to the writing for the nanovial section. Edwin Sanchez Ochoa (E.S.O.) contributed to the writing for Spherical Particles as Substrates for Assays section. Lily Shang (L.S.) created the figures and contributed to the writing for the Spherical Particles as Substrates for Assays section. Citradewi Soemardy (C.S.) contributed to the figures and writing for the nanovial section. Rui Chian Tang (R.C.T.) created the figures and contributed to the writing for the spherical particle-templated emulsification section. Mark van Zee (M.v.Z.) contributed to the figures and writing for the evolution of hydrogels for the cell encapsulation section. Cayden Williamson (C.W.) contributed to the figures and writing for the evolution of hydrogels for the cell encapsulation section. Shun Ye (S.Y.) created the figures and contributed to the writing for the flow-lithography particles and shape-based barcoding section.
Author Contributions
† R.G. and A.A. contributed equally to this work.
The authors declare the following competing financial interest(s): D.D. and the Regents of the University of California have financial interests in Partillion Bioscience which is commercializing Lab on a Particle technology.
References
- Reyes D. R.; Iossifidis D.; Auroux P.-A.; Manz A. Micro Total Analysis Systems. 1. Introduction, Theory, and Technology. Anal. Chem. 2002, 74 (12), 2623–2636. 10.1021/ac0202435. [DOI] [PubMed] [Google Scholar]
- Folch A.; Ayon A.; Hurtado O.; Schmidt M. A.; Toner M. Molding of Deep Polydimethylsiloxane Microstructures for Microfluidics and Biological Applications. Journal of Biomechanical Engineering 1999, 121 (1), 28–34. 10.1115/1.2798038. [DOI] [PubMed] [Google Scholar]
- Link D. R.; Grasland-Mongrain E.; Duri A.; Sarrazin F.; Cheng Z.; Cristobal G.; Marquez M.; Weitz D. A. Electric Control of Droplets in Microfluidic Devices. Angew. Chem., Int. Ed. 2006, 45 (16), 2556–2560. 10.1002/anie.200503540. [DOI] [PubMed] [Google Scholar]
- Unger M. A.; Chou H.-P.; Thorsen T.; Scherer A.; Quake S. R. Monolithic Microfabricated Valves and Pumps by Multilayer Soft Lithography. Science 2000, 288 (5463), 113–116. 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- Duffy D. C.; McDonald J. C.; Schueller O. J. A.; Whitesides G. M. Rapid Prototyping of Microfluidic Systems in Poly(Dimethylsiloxane). Anal. Chem. 1998, 70 (23), 4974–4984. 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- Service R. F. The Incredible Shrinking Laboratory. Science 1995, 268 (5207), 26–27. 10.1126/science.7701335. [DOI] [PubMed] [Google Scholar]
- Srinivasan V.; Pamula V. K.; Fair R. B. An Integrated Digital Microfluidic Lab-on-a-Chip for Clinical Diagnostics on Human Physiological Fluids. Lab Chip 2004, 4 (4), 310–315. 10.1039/b403341h. [DOI] [PubMed] [Google Scholar]
- Miwa H.; Dimatteo R.; de Rutte J.; Ghosh R.; Di Carlo D. Single-Cell Sorting Based on Secreted Products for Functionally Defined Cell Therapies. Microsyst. Nanoeng. 2022, 8, 84. 10.1038/s41378-022-00422-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udani S.; Langerman J.; Koo D.; Baghdasarian S.; Cheng B.; Kang S.; Soemardy C.; de Rutte J.; Plath K.; Di Carlo D. Associating Growth Factor Secretions and Transcriptomes of Single Cells in Nanovials Using SEC-Seq. Nat. Nanotechnol. 2024, 19, 354. 10.1038/s41565-023-01560-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendukuri D.; Pregibon D. C.; Collins J.; Hatton T. A.; Doyle P. S. Continuous-Flow Lithography for High-Throughput Microparticle Synthesis. Nat. Mater. 2006, 5 (5), 365–369. 10.1038/nmat1617. [DOI] [PubMed] [Google Scholar]
- de Rutte J.; Dimatteo R.; Archang M.; van Zee M.; Koo D.; Lee S.; Sharrow A.; Krohl P.; Mellody M.; Zhu S.; Eichenbaum J.; Kizerwetter M.; Udani S.; Ha K.; Willson R.; Bertozzi A.; Spangler J.; Damoiseaux R.; Di Carlo D. Suspendable Hydrogel Nanovials for Massively Parallel Single-Cell Functional Analysis and Sorting. ACS Nano 2022, 16 (5), 7242–7257. 10.1021/acsnano.1c11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-Y.; Ouyang M.; Wang B.; de Rutte J.; Joo A.; Jacobs M.; Ha K.; Bertozzi A. L.; Di Carlo D. Monodisperse Drops Templated by 3D-Structured Microparticles. Science Advances 2020, 6 (45), eabb9023 10.1126/sciadv.abb9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rutte J.; Dimatteo R.; Zhu S.; Archang M. M.; Di Carlo D. Sorting Single-Cell Microcarriers Using Commercial Flow Cytometers. SLAS Technology 2022, 27 (2), 150–159. 10.1016/j.slast.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin J.; Quake S. R. Microfluidic Large-Scale Integration: The Evolution of Design Rules for Biological Automation. Annu. Rev. Biophys. Biomol. Struct. 2007, 36 (1), 213–231. 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- BD AccuriTM C6 Plus | Flow Cytometer. BD Biosciences. https://www.bdbiosciences.com/en-us/products/instruments/flow-cytometers/research-cell-analyzers/bd-accuri-c6-plus (accessed 2024-03-12).
- Sony SH800S Cell Sorter. Sony Biotechnology. https://www.sonybiotechnology.com/us/instruments/sh800s-cell-sorter/system/ (accessed 2024-03-12).
- ImageStream. Cytek Biosciences. https://cytekbio.com/pages/imagestream (accessed 2024-03-12).
- BD FACSDiscoverTM S8 Cell Sorter. BD Biosciences. https://www.bdbiosciences.com/en-us/products/instruments/flow-cytometers/research-cell-sorters/bd-facsdiscover-s8 (accessed 2024-03-12).
- Chromium X Series. 10x Genomics. https://www.10xgenomics.com/instruments/chromium-x-series (accessed 2024-03-12).
- Mazutis L.; Gilbert J.; Ung W. L.; Weitz D. A.; Griffiths A. D.; Heyman J. A. Single-Cell Analysis and Sorting Using Droplet-Based Microfluidics. Nat. Protoc 2013, 8 (5), 870–891. 10.1038/nprot.2013.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.; Chen J. J.; Mu L.; Xue Q.; Wu Y.; Wu P.-H.; Li J.; Vortmeyer A. O.; Miller-Jensen K.; Wirtz D.; Fan R. High-Throughput Secretomic Analysis of Single Cells to Assess Functional Cellular Heterogeneity. Anal. Chem. 2013, 85 (4), 2548–2556. 10.1021/ac400082e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Destgeer G.; Ouyang M.; Wu C.-Y.; Di Carlo D. Fabrication of 3D Concentric Amphiphilic Microparticles to Form Uniform Nanoliter Reaction Volumes for Amplified Affinity Assays. Lab Chip 2020, 20 (19), 3503–3514. 10.1039/D0LC00698J. [DOI] [PubMed] [Google Scholar]
- Novak R.; Zeng Y.; Shuga J.; Venugopalan G.; Fletcher D. A.; Smith M. T.; Mathies R. A. Single-Cell Multiplex Gene Detection and Sequencing with Microfluidically Generated Agarose Emulsions. Angew. Chem. Int. Ed 2011, 50 (2), 390–395. 10.1002/anie.201006089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Shah V.; Lu A.; Pachler E.; Cheng B.; Di Carlo D. Counting of Enzymatically Amplified Affinity Reactions in Hydrogel Particle-Templated Drops. Lab Chip 2021, 21 (18), 3438–3448. 10.1039/D1LC00344E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo D.; Mao Z.; Dimatteo R.; Noguchi M.; Tsubamoto N.; McLaughlin J.; Tran W.; Lee S.; Cheng D.; de Rutte J.; Burton Sojo G.; Witte O. N.; Di Carlo D. Defining T Cell Receptor Repertoires Using Nanovial-Based Binding and Functional Screening. Proc. Natl. Acad. Sci. U. S. A. 2024, 121 (14), e2320442121 10.1073/pnas.2320442121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng R. Y.-H.; de Rutte J.; Ito C. E.; Ott A.; Bosler L.; Kuo W.-Y.; Liang J.; Hall B.; Rawlings D.; Di Carlo D.; James R. SEC-Seq: Association of Molecular Signatures with Antibody Secretion in Thousands of Single Human Plasma Cells. Nat. Commun. 2023, 14, 3567. 10.1038/s41467-023-39367-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo D.; Cheng X.; Udani S.; Zhu D.; Li J.; Hall B.; Tsubamoto N.; Hu S.; Ko J.; Cheng K.; Di Carlo D. Optimizing Cell Therapy by Sorting Cells with High Extracellular Vesicle Secretion. bioRxiv 2023, 10.1101/2023.05.29.542772. [DOI] [Google Scholar]
- Hatori M. N.; Kim S. C.; Abate A. R. Particle-Templated Emulsification for Microfluidics-Free Digital Biology. Anal. Chem. 2018, 90 (16), 9813–9820. 10.1021/acs.analchem.8b01759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V.; Yang X.; Arnheim A.; Udani S.; Tseng D.; Luo Y.; Ouyang M.; Destgeer G.; Garner O. B.; Koydemir H. C.; Ozcan A.; Di Carlo D. Amphiphilic Particle-Stabilized Nanoliter Droplet Reactors with a Multimodal Portable Reader for Distributive Biomarker Quantification. ACS Nano 2023, 17 (20), 19952–19960. 10.1021/acsnano.3c04994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pregibon D. C.; Toner M.; Doyle P. S. Multifunctional Encoded Particles for High-Throughput Biomolecule Analysis. Science 2007, 315 (5817), 1393–1396. 10.1126/science.1134929. [DOI] [PubMed] [Google Scholar]
- Clark I. C.; Fontanez K. M.; Meltzer R. H.; Xue Y.; Hayford C.; May-Zhang A.; D’Amato C.; Osman A.; Zhang J. Q.; Hettige P.; Ishibashi J. S. A.; Delley C. L.; Weisgerber D. W.; Replogle J. M.; Jost M.; Phong K. T.; Kennedy V. E.; Peretz C. A. C.; Kim E. A.; Song S.; Karlon W.; Weissman J. S.; Smith C. C.; Gartner Z. J.; Abate A. R. Microfluidics-Free Single-Cell Genomics with Templated Emulsification. Nat. Biotechnol. 2023, 41 (11), 1557–1566. 10.1038/s41587-023-01685-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda K.; Ayazi A.; Gossett D. R.; Sadasivam J.; Lonappan C. K.; Sollier E.; Fard A. M.; Hur S. C.; Adam J.; Murray C.; Wang C.; Brackbill N.; Di Carlo D.; Jalali B. High-Throughput Single-Microparticle Imaging Flow Analyzer. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (29), 11630–11635. 10.1073/pnas.1204718109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei C.; Kobayashi H.; Wu Y.; Li M.; Isozaki A.; Yasumoto A.; Mikami H.; Ito T.; Nitta N.; Sugimura T.; Yamada M.; Yatomi Y.; Di Carlo D.; Ozeki Y.; Goda K. High-Throughput Imaging Flow Cytometry by Optofluidic Time-Stretch Microscopy. Nat. Protoc 2018, 13 (7), 1603–1631. 10.1038/s41596-018-0008-7. [DOI] [PubMed] [Google Scholar]
- Muñoz H. E.; Li M.; Riche C. T.; Nitta N.; Diebold E.; Lin J.; Owsley K.; Bahr M.; Goda K.; Di Carlo D. Single-Cell Analysis of Morphological and Metabolic Heterogeneity in Euglena Gracilis by Fluorescence-Imaging Flow Cytometry. Anal. Chem. 2018, 90 (19), 11280–11289. 10.1021/acs.analchem.8b01794. [DOI] [PubMed] [Google Scholar]
- Ota S.; Horisaki R.; Kawamura Y.; Ugawa M.; Sato I.; Hashimoto K.; Kamesawa R.; Setoyama K.; Yamaguchi S.; Fujiu K.; Waki K.; Noji H. Ghost Cytometry. Science 2018, 360 (6394), 1246–1251. 10.1126/science.aan0096. [DOI] [PubMed] [Google Scholar]
- Nitta N.; Sugimura T.; Isozaki A.; Mikami H.; Hiraki K.; Sakuma S.; Iino T.; Arai F.; Endo T.; Fujiwaki Y.; Fukuzawa H.; Hase M.; Hayakawa T.; Hiramatsu K.; Hoshino Y.; Inaba M.; Ito T.; Karakawa H.; Kasai Y.; Koizumi K.; Lee S.; Lei C.; Li M.; Maeno T.; Matsusaka S.; Murakami D.; Nakagawa A.; Oguchi Y.; Oikawa M.; Ota T.; Shiba K.; Shintaku H.; Shirasaki Y.; Suga K.; Suzuki Y.; Suzuki N.; Tanaka Y.; Tezuka H.; Toyokawa C.; Yalikun Y.; Yamada M.; Yamagishi M.; Yamano T.; Yasumoto A.; Yatomi Y.; Yazawa M.; Di Carlo D.; Hosokawa Y.; Uemura S.; Ozeki Y.; Goda K. Intelligent Image-Activated Cell Sorting. Cell 2018, 175 (1), 266–276. 10.1016/j.cell.2018.08.028. [DOI] [PubMed] [Google Scholar]
- Isozaki A.; Mikami H.; Tezuka H.; Matsumura H.; Huang K.; Akamine M.; Hiramatsu K.; Iino T.; Ito T.; Karakawa H.; Kasai Y.; Li Y.; Nakagawa Y.; Ohnuki S.; Ota T.; Qian Y.; Sakuma S.; Sekiya T.; Shirasaki Y.; Suzuki N.; Tayyabi E.; Wakamiya T.; Xu M.; Yamagishi M.; Yan H.; Yu Q.; Yan S.; Yuan D.; Zhang W.; Zhao Y.; Arai F.; Campbell R. E.; Danelon C.; Carlo D. D.; Hiraki K.; Hoshino Y.; Hosokawa Y.; Inaba M.; Nakagawa A.; Ohya Y.; Oikawa M.; Uemura S.; Ozeki Y.; Sugimura T.; Nitta N.; Goda K. Intelligent Image-Activated Cell Sorting 2.0. Lab Chip 2020, 20 (13), 2263–2273. 10.1039/D0LC00080A. [DOI] [PubMed] [Google Scholar]
- Nitta N.; Iino T.; Isozaki A.; Yamagishi M.; Kitahama Y.; Sakuma S.; Suzuki Y.; Tezuka H.; Oikawa M.; Arai F.; Asai T.; Deng D.; Fukuzawa H.; Hase M.; Hasunuma T.; Hayakawa T.; Hiraki K.; Hiramatsu K.; Hoshino Y.; Inaba M.; Inoue Y.; Ito T.; Kajikawa M.; Karakawa H.; Kasai Y.; Kato Y.; Kobayashi H.; Lei C.; Matsusaka S.; Mikami H.; Nakagawa A.; Numata K.; Ota T.; Sekiya T.; Shiba K.; Shirasaki Y.; Suzuki N.; Tanaka S.; Ueno S.; Watarai H.; Yamano T.; Yazawa M.; Yonamine Y.; Di Carlo D.; Hosokawa Y.; Uemura S.; Sugimura T.; Ozeki Y.; Goda K. Raman Image-Activated Cell Sorting. Nat. Commun. 2020, 11 (1), 3452. 10.1038/s41467-020-17285-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schraivogel D.; Kuhn T. M.; Rauscher B.; Rodríguez-Martínez M.; Paulsen M.; Owsley K.; Middlebrook A.; Tischer C.; Ramasz B.; Ordoñez-Rueda D.; Dees M.; Cuylen-Haering S.; Diebold E.; Steinmetz L. M. High-Speed Fluorescence Image–Enabled Cell Sorting. Science 2022, 375 (6578), 315–320. 10.1126/science.abj3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang R.; Xia L.; Gutierrez B.; Gagne I.; Munoz A.; Eribez K.; Jagnandan N.; Chen X.; Zhang Z.; Waller L.; Alaynick W.; Cho S. H.; An C.; Lo Y.-H. Low-Latency Label-Free Image-Activated Cell Sorting Using Fast Deep Learning and AI Inferencing. Biosens. Bioelectron. 2023, 220, 114865 10.1016/j.bios.2022.114865. [DOI] [PubMed] [Google Scholar]
- Weaver J. C.; Williams G. B.; Klibanov A.; Demain A. L. Gel Microdroplets: Rapid Detection and Enumeration of Individual Microorganisms by Their Metabolic Activity. Nat. Biotechnol. 1988, 6 (9), 1084–1089. 10.1038/nbt0988-1084. [DOI] [Google Scholar]
- Anna S. L.; Bontoux N.; Stone H. A. Formation of Dispersions Using “Flow Focusing” in Microchannels. Appl. Phys. Lett. 2003, 82 (3), 364–366. 10.1063/1.1537519. [DOI] [Google Scholar]
- Workman V. L.; Dunnett S. B.; Kille P.; Palmer D. D. On-Chip Alginate Microencapsulation of Functional Cells. Macromol. Rapid Commun. 2008, 29 (2), 165–170. 10.1002/marc.200700641. [DOI] [Google Scholar]
- Vijayakumar K.; Gulati S.; deMello A. J.; Edel J. B. Rapid Cell Extraction in Aqueous Two-Phase Microdroplet Systems. Chemical Science 2010, 1 (4), 447–452. 10.1039/c0sc00229a. [DOI] [Google Scholar]
- Leonaviciene G.; Leonavicius K.; Meskys R.; Mazutis L. Multi-Step Processing of Single Cells Using Semi-Permeable Capsules. Lab Chip 2020, 20 (21), 4052–4062. 10.1039/D0LC00660B. [DOI] [PubMed] [Google Scholar]
- Nir R.; Lamed R.; Gueta L.; Sahar E. Single-Cell Entrapment and Microcolony Development within Uniform Microspheres Amenable to Flow Cytometry. Appl. Environ. Microbiol. 1990, 56 (9), 2870–2875. 10.1128/aem.56.9.2870-2875.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto Y.; Tan W.; Tsuda Y.; Takeuchi S. Monodisperse Semi-Permeable Microcapsules for Continuous Observation of Cells. Lab Chip 2009, 9 (15), 2217–2223. 10.1039/b900035f. [DOI] [PubMed] [Google Scholar]
- van Zee M.; de Rutte J.; Rumyan R.; Williamson C.; Burnes T.; Radakovits R.; Sonico Eugenio A.; Badih S.; Lee S.; Lee D.-H.; Archang M.; Di Carlo D. High-Throughput Selection of Cells Based on Accumulated Growth and Division Using PicoShell Particles. Proc. Natl. Acad. Sci. U. S. A. 2022, 119 (4), e2109430119 10.1073/pnas.2109430119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayat C.; Ratinaud M.-H. Cell Cycle Analysis by Flow Cytometry: Principles and Applications. Biology of the Cell 1993, 78 (1–2), 15–25. 10.1016/0248-4900(93)90110-Z. [DOI] [PubMed] [Google Scholar]
- Nir R.; Yisraeli Y.; Lamed R.; Sahar E. Flow Cytometry Sorting of Viable Bacteria and Yeasts According to Beta-Galactosidase Activity. Appl. Environ. Microbiol. 1990, 56 (12), 3861–3866. 10.1128/aem.56.12.3861-3866.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. B.; Weaver J. C.; Demain A. L. Rapid Microbial Detection and Enumeration Using Gel Microdroplets and Colorimetric or Fluorescence Indicator Systems. J. Clin Microbiol 1990, 28 (5), 1002–1008. 10.1128/jcm.28.5.1002-1008.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J. C.; Bliss J. G.; Harrison G. I.; Powell K. T.; Williams G. B. Microdrop Technology: A General Method for Separating Cells by Function and Composition. Methods 1991, 2 (3), 234–247. 10.1016/S1046-2023(05)80066-0. [DOI] [Google Scholar]
- Weaver J. C.; Bliss J. G.; Powell K. T.; Harrison G. I.; Williams G. B. Rapid Clonal Growth Measurements at the Single-Cell Level: Gel Microdroplets and Flow Cytometry. Nat. Biotechnol. 1991, 9 (9), 873–877. 10.1038/nbt0991-873. [DOI] [PubMed] [Google Scholar]
- Gift E. A.; Park H. J.; Paradis G. A.; Demain A. L.; Weaver J. C. FACS-Based Isolation of Slowly Growing Cells: Double Encapsulation of Yeast in Gel Microdrops. Nat. Biotechnol. 1996, 14 (7), 884–887. 10.1038/nbt0796-884. [DOI] [PubMed] [Google Scholar]
- Katsuragi T.; Tanaka S.; Nagahiro S.; Tani Y. Gel Microdroplet Technique Leaving Microorganisms Alive for Sorting by Flow Cytometry. J. Microbiol. Methods 2000, 42 (1), 81–86. 10.1016/S0167-7012(00)00179-2. [DOI] [PubMed] [Google Scholar]
- Zengler K.; Toledo G.; Rappé M.; Elkins J.; Mathur E. J.; Short J. M.; Keller M. Cultivating the Uncultured. Proc. Natl. Acad. Sci. U.S.A. 2002, 99 (24), 15681–15686. 10.1073/pnas.252630999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akselband Y.; Cabral C.; Castor T. P.; Chikarmane H. M.; McGrath P. Enrichment of Slow-Growing Marine Microorganisms from Mixed Cultures Using Gel Microdrop (GMD) Growth Assay and Fluorescence-Activated Cell Sorting. Journal of Experimental Marine Biology and Ecology 2006, 329 (2), 196–205. 10.1016/j.jembe.2005.08.018. [DOI] [Google Scholar]
- Ryan C.; Nguyen B. T.; Sullivan S. J. Rapid Assay for Mycobacterial Growth and Antibiotic Susceptibility Using Gel Microdrop Encapsulation. J. Clin Microbiol 1995, 33 (7), 1720–1726. 10.1128/jcm.33.7.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akselband Y.; Cabral C.; Shapiro D. S.; McGrath P. Rapid Mycobacteria Drug Susceptibility Testing Using Gel Microdrop (GMD) Growth Assay and Flow Cytometry. J. Microbiol. Methods 2005, 62 (2), 181–197. 10.1016/j.mimet.2005.02.012. [DOI] [PubMed] [Google Scholar]
- Pereira T.; Millar T. J.; Chuck J.-A. Viability Analysis of Alginate Encapsulated Micro-Organisms Using Fluorescent Stains. J. Microencapsulation 2005, 22 (7), 787–792. 10.1080/02652040500273829. [DOI] [PubMed] [Google Scholar]
- Weaver J. C.; McGrath P.; Adams S. Gel Microdrop Technology for Rapid Isolation of Rare and High Producer Cells. Nat. Med. 1997, 3 (5), 583–585. 10.1038/nm0597-583. [DOI] [PubMed] [Google Scholar]
- Hammill L.; Welles J.; Carson G. R. The Gel Microdrop Secretion Assay: Identification of a Low Productivity Subpopulation Arising during the Production of Human Antibody in CHO Cells. Cytotechnology 2000, 34 (1), 27–37. 10.1023/A:1008186113245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcanu V.; Williams N. A. Cell Identification and Isolation on the Basis of Cytokine Secretion: A Novel Tool for Investigating Immune Responses. Nat. Med. 2001, 7 (3), 373–376. 10.1038/85533. [DOI] [PubMed] [Google Scholar]
- Zhou Q.-Z.; Liu X.-Y.; Liu S.-J.; Ma G.-H.; Su Z.-G. Preparation of Uniformly Sized Agarose Microcapsules by Membrane Emulsification for Application in Sorting Bacteria. Ind. Eng. Chem. Res. 2008, 47 (17), 6386–6390. 10.1021/ie800011r. [DOI] [Google Scholar]
- Luo D. Cell Encapsules with Tunable Transport and Mechanical Properties. Biomicrofluidics 2007, 034102. 10.1063/1.2757156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamminen M. V.; Virta M. P. J. Single Gene-Based Distinction of Individual Microbial Genomes from a Mixed Population of Microbial Cells. Frontiers in Microbiology 2015, 6, 195. 10.3389/fmicb.2015.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X.; Chen B.; He M.; Hu B. Agarose-Droplet-Based Digital LAMP Assay for Counting Virus DNA in Single-Particle ICP-MS. Anal. Chem. 2022, 94 (17), 6582–6590. 10.1021/acs.analchem.2c00427. [DOI] [PubMed] [Google Scholar]
- Leng X.; Zhang W.; Wang C.; Cui L.; Yang C. J. Agarose Droplet Microfluidics for Highly Parallel and Efficient Single Molecule Emulsion PCR. Lab Chip 2010, 10 (21), 2841–2843. 10.1039/c0lc00145g. [DOI] [PubMed] [Google Scholar]
- Zhu Z.; Zhang W.; Leng X.; Zhang M.; Guan Z.; Lu J.; Yang C. J. Highly Sensitive and Quantitative Detection of Rare Pathogens through Agarose Droplet Microfluidic Emulsion PCR at the Single-Cell Level. Lab Chip 2012, 12 (20), 3907–3913. 10.1039/c2lc40461c. [DOI] [PubMed] [Google Scholar]
- Buffi N.; Merulla D.; Beutier J.; Barbaud F.; Beggah S.; van Lintel H.; Renaud P.; Meer J. R. van der Development of a Microfluidics Biosensor for Agarose-Bead Immobilized Escherichia Coli Bioreporter Cells for Arsenite Detection in Aqueous Samples. Lab Chip 2011, 11 (14), 2369–2377. 10.1039/c1lc20274j. [DOI] [PubMed] [Google Scholar]
- Zhang W. Y.; Zhang W.; Liu Z.; Li C.; Zhu Z.; Yang C. J. Highly Parallel Single-Molecule Amplification Approach Based on Agarose Droplet Polymerase Chain Reaction for Efficient and Cost-Effective Aptamer Selection. Anal. Chem. 2012, 84 (1), 350–355. 10.1021/ac2026942. [DOI] [PubMed] [Google Scholar]
- Eun Y.-J.; Utada A. S.; Copeland M. F.; Takeuchi S.; Weibel D. B. Encapsulating Bacteria in Agarose Microparticles Using Microfluidics for High-Throughput Cell Analysis and Isolation. ACS Chem. Biol. 2011, 6 (3), 260–266. 10.1021/cb100336p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utech S.; Prodanovic R.; Mao A. S.; Ostafe R.; Mooney D. J.; Weitz D. A. Microfluidic Generation of Monodisperse, Structurally Homogeneous Alginate Microgels for Cell Encapsulation and 3D Cell Culture. Adv. Healthcare Materials 2015, 4 (11), 1628–1633. 10.1002/adhm.201500021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokkalingam V.; Tel J.; Wimmers F.; Liu X.; Semenov S.; Thiele J.; Figdor C. G.; Huck W. T. S. Probing Cellular Heterogeneity in Cytokine-Secreting Immune Cells Using Droplet-Based Microfluidics. Lab Chip 2013, 13 (24), 4740–4744. 10.1039/c3lc50945a. [DOI] [PubMed] [Google Scholar]
- Ohan J.; Pelle B.; Nath P.; Huang J.-H.; Hovde B.; Vuyisich M.; Dichosa A. E.; Starkenburg S. R. High-Throughput Phenotyping of Cell-to-Cell Interactions in Gel Microdroplet Pico-Cultures. BioTechniques 2019, 66 (5), 218–224. 10.2144/btn-2018-0124. [DOI] [PubMed] [Google Scholar]
- Li M.; Van Zee M.; Riche C. T.; Tofig B.; Gallaher S. D.; Merchant S. S.; Damoiseaux R.; Goda K.; Di Carlo D. A Gelatin Microdroplet Platform for High-Throughput Sorting of Hyperproducing Single-Cell-Derived Microalgal Clones. Small 2018, 14 (44), 1803315 10.1002/smll.201803315. [DOI] [PubMed] [Google Scholar]
- Ziemecka I.; van Steijn V.; Koper G. J. M.; Rosso M.; Brizard A. M.; van Esch J. H.; Kreutzer M. T. Monodisperse Hydrogel Microspheres by Forced Droplet Formation in Aqueous Two-Phase Systems. Lab Chip 2011, 11 (4), 620–624. 10.1039/C0LC00375A. [DOI] [PubMed] [Google Scholar]
- Watanabe T.; Motohiro I.; Ono T. Microfluidic Formation of Hydrogel Microcapsules with a Single Aqueous Core by Spontaneous Cross-Linking in Aqueous Two-Phase System Droplets. Langmuir 2019, 35 (6), 2358–2367. 10.1021/acs.langmuir.8b04169. [DOI] [PubMed] [Google Scholar]
- Wang H.; Liu H.; Liu H.; Su W.; Chen W.; Qin J. One-Step Generation of Core–Shell Gelatin Methacrylate (GelMA) Microgels Using a Droplet Microfluidic System. Adv. Materials Technologies 2019, 4 (6), 1800632 10.1002/admt.201800632. [DOI] [Google Scholar]
- Seiffert S.; Thiele J.; Abate A. R.; Weitz D. A. Smart Microgel Capsules from Macromolecular Precursors. J. Am. Chem. Soc. 2010, 132 (18), 6606–6609. 10.1021/ja102156h. [DOI] [PubMed] [Google Scholar]
- Wu Q.; Huang X.; Liu R.; Yang X.; Xiao G.; Jiang N.; Weitz D. A.; Song Y. Multichannel Multijunction Droplet Microfluidic Device to Synthesize Hydrogel Microcapsules with Different Core–Shell Structures and Adjustable Core Positions. Langmuir 2024, 40 (3), 1950–1960. 10.1021/acs.langmuir.3c02579. [DOI] [PubMed] [Google Scholar]
- Werner J. G.; Lee H.; Wiesner U.; Weitz D. A. Ordered Mesoporous Microcapsules from Double Emulsion Confined Block Copolymer Self-Assembly. ACS Nano 2021, 15 (2), 3490–3499. 10.1021/acsnano.1c00068. [DOI] [PubMed] [Google Scholar]
- Han J.-H.; Kim J.; Acter S.; Kim Y.; Lee H.-N.; Chang H.-K.; Suh K.-D.; Kim J. W. Uniform Hollow-Structured Poly(Vinyl Amine) Hydrogel Microparticles with Controlled Mesh Property and Enhanced Cell Adhesion. Polymer 2014, 55 (5), 1143–1149. 10.1016/j.polymer.2014.01.017. [DOI] [Google Scholar]
- Leonaviciene G.; Mazutis L. RNA Cytometry of Single-Cells Using Semi-Permeable Microcapsules. Nucleic acids research 2023, 51 (1), e2–e2. 10.1093/nar/gkac918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windbergs M.; Zhao Y.; Heyman J.; Weitz D. A. Biodegradable Core–Shell Carriers for Simultaneous Encapsulation of Synergistic Actives. J. Am. Chem. Soc. 2013, 135 (21), 7933–7937. 10.1021/ja401422r. [DOI] [PubMed] [Google Scholar]
- Siltanen C.; Diakatou M.; Lowen J.; Haque A.; Rahimian A.; Stybayeva G.; Revzin A. One Step Fabrication of Hydrogel Microcapsules with Hollow Core for Assembly and Cultivation of Hepatocyte Spheroids. Acta Biomaterialia 2017, 50, 428–436. 10.1016/j.actbio.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J. Microfluidic-Based Synthesis of Hydrogel Particles for Cell Microencapsulation and Cell-Based Drug Delivery. Polymers 2012, 4 (2), 1084–1108. 10.3390/polym4021084. [DOI] [Google Scholar]
- Abbaspourrad A.; Carroll N. J.; Kim S.-H.; Weitz D. A. Surface Functionalized Hydrophobic Porous Particles Toward Water Treatment Application. Adv. Mater. 2013, 25 (23), 3215–3221. 10.1002/adma.201300656. [DOI] [PubMed] [Google Scholar]
- Di Girolamo S.; Puorger C.; Lipps G. Stable and Selective Permeable Hydrogel Microcapsules for High-Throughput Cell Cultivation and Enzymatic Analysis. Microb Cell Fact 2020, 19 (1), 170. 10.1186/s12934-020-01427-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atrandi Biosciences. https://atrandi.com/application/single-cell-metagenomics/technology (accessed 2024-03-10).
- Ma S.; Thiele J.; Liu X.; Bai Y.; Abell C.; Huck W. Fabrication of Microgel Particles with Complex Shape via Selective Polymerization of Aqueous Two-Phase Systems. small 2012, 8 (15), 2356–2360. 10.1002/smll.201102715. [DOI] [PubMed] [Google Scholar]
- Liu Q.; Zhao M.; Mytnyk S.; Klemm B.; Zhang K.; Wang Y.; Yan D.; Mendes E.; van Esch J. H. Self-Orienting Hydrogel Micro-Buckets as Novel Cell Carriers. Angew. Chem. Int. Ed 2019, 58 (2), 547–551. 10.1002/anie.201811374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.; de Rutte J.; Dimatteo R.; Koo D.; Di Carlo D. Scalable Fabrication and Use of 3D Structured Microparticles Spatially Functionalized with Biomolecules. ACS Nano 2022, 16 (1), 38–49. 10.1021/acsnano.1c05857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester E.; Carney S.; Shah V.; Arnheim A.; Patel B.; Di Carlo D.; Bertozzi A. Fluid Dynamics Alters Liquid–Liquid Phase Separation in Confined Aqueous Two-Phase Systems. Proc. Natl. Acad. Sci. U.S.A. 2023, 120 (49), e2306467120. 10.1073/pnas.2306467120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo D.; Dimatteo R.; Lee S.; de Rutte J.; Di Carlo D.. Sorting Single T Cells Based on Secreted Cytokines and Surface Markers Using Hydrogel Nanovials. bioRxiv 2022. 10.1101/2022.04.28.489940. [DOI] [Google Scholar]
- Mitra S.; Tomar P. C. Hybridoma Technology; Advancements, Clinical Significance, and Future Aspects. Journal of Genetic Engineering and Biotechnology 2021, 19 (1), 159 10.1186/s43141-021-00264-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah H. B.; Smith K.; Wren J. D.; Webb C. F.; Ballard J. D.; Bourn R. L.; James J. A.; Lang M. L. Insights From Analysis of Human Antigen-Specific Memory B Cell Repertoires. Front. Immunol. 2019, 9, 3064. 10.3389/fimmu.2018.03064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledsgaard L.; Kilstrup M.; Karatt-Vellatt A.; McCafferty J.; Laustsen A. H. Basics of Antibody Phage Display Technology. Toxins (Basel) 2018, 10 (6), 236. 10.3390/toxins10060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraes J. Z.; Hamaguchi B.; Braggion C.; Speciale E. R.; Cesar F. B. V.; Soares G. de F. da S.; Osaki J. H.; Pereira T. M.; Aguiar R. B. Hybridoma Technology: Is It Still Useful?. Current Research in Immunology 2021, 2, 32–40. 10.1016/j.crimmu.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partillion Bioscience - Unlocking Functional Single Cell Biology
- Fulton R. J.; McDade R. L.; Smith P. L.; Kienker L. J.; Kettman J. R. Jr. Advanced Multiplexed Analysis with the FlowMetrixTM System. Clinical Chemistry 1997, 43 (9), 1749–1756. 10.1093/clinchem/43.9.1749. [DOI] [PubMed] [Google Scholar]
- Vignali D. A. A. Multiplexed Particle-Based Flow Cytometric Assays. Journal of Immunological Methods 2000, 243 (1), 243–255. 10.1016/S0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- Graham H.; Chandler D. J.; Dunbar S. A. The Genesis and Evolution of Bead-Based Multiplexing. Methods 2019, 158, 2–11. 10.1016/j.ymeth.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Waterboer T.; Sehr P.; Pawlita M. Suppression of Non-Specific Binding in Serological Luminex Assays. Journal of Immunological Methods 2006, 309 (1), 200–204. 10.1016/j.jim.2005.11.008. [DOI] [PubMed] [Google Scholar]
- Sukhanova A.; Nabiev I. Fluorescent Nanocrystal-Encoded Microbeads for Multiplexed Cancer Imaging and Diagnosis. Critical Reviews in Oncology/Hematology 2008, 68 (1), 39–59. 10.1016/j.critrevonc.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Roh Y. H.; Lee H. J.; Bong K. W. Microfluidic Fabrication of Encoded Hydrogel Microparticles for Application in Multiplex Immunoassay. BioChip J. 2019, 13 (1), 64–81. 10.1007/s13206-019-3104-z. [DOI] [Google Scholar]
- Di Lorenzo F.; Seiffert S. Effect of Droplet Size in Acrylamide-Based Microgel Formation by Microfluidics. Macromol. React. Eng. 2016, 10 (3), 201–205. 10.1002/mren.201500061. [DOI] [Google Scholar]
- Choi C.-H.; Jung J.-H.; Hwang T.-S.; Lee C.-S. In Situ Microfluidic Synthesis of Monodisperse PEG Microspheres. Macromol. Res. 2009, 17 (3), 163–167. 10.1007/BF03218673. [DOI] [Google Scholar]
- Stan C. A.; Tang S. K. Y.; Whitesides G. M. Independent Control of Drop Size and Velocity in Microfluidic Flow-Focusing Generators Using Variable Temperature and Flow Rate. Anal. Chem. 2009, 81 (6), 2399–2402. 10.1021/ac8026542. [DOI] [PubMed] [Google Scholar]
- Li Z.; Leshansky A. M.; Pismen L. M.; Tabeling P. Step-Emulsification in a Microfluidic Device. Lab Chip 2015, 15 (4), 1023–1031. 10.1039/C4LC01289E. [DOI] [PubMed] [Google Scholar]
- Wu J.; Yadavali S.; Lee D.; Issadore D. A. Scaling up the Throughput of Microfluidic Droplet-Based Materials Synthesis: A Review of Recent Progress and Outlook. Applied Physics Reviews 2021, 8 (3), 031304 10.1063/5.0049897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rutte J. M.; Koh J.; Di Carlo D. Scalable High-Throughput Production of Modular Microgels for In Situ Assembly of Microporous Tissue Scaffolds. Adv. Funct. Mater. 2019, 29 (25), 1900071 10.1002/adfm.201900071. [DOI] [Google Scholar]
- Thiele J.; Ma Y.; Foschepoth D.; Hansen M. M. K.; Steffen C.; Heus H. A.; Huck W. T. S. DNA-Functionalized Hydrogels for Confined Membrane-Free in Vitro Transcription/Translation. Lab Chip 2014, 14 (15), 2651. 10.1039/c3lc51427g. [DOI] [PubMed] [Google Scholar]
- Le Goff G. C.; Srinivas R. L.; Hill W. A.; Doyle P. S. Hydrogel Microparticles for Biosensing. Eur. Polym. J. 2015, 72, 386–412. 10.1016/j.eurpolymj.2015.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C.; Liu L.; Vasudevan H. N.; Chang K.-C.; Abate A. R. Accurate Bulk Quantitation of Droplet Digital Polymerase Chain Reaction. Anal. Chem. 2021, 93 (29), 9974–9979. 10.1021/acs.analchem.1c00877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M. Digital PCR Hits Its Stride. Nat. Methods 2012, 9 (6), 541–544. 10.1038/nmeth.2027. [DOI] [Google Scholar]
- Hou Y.; Chen S.; Zheng Y.; Zheng X.; Lin J.-M. Droplet-Based Digital PCR (ddPCR) and Its Applications. TrAC Trends in Analytical Chemistry 2023, 158, 116897 10.1016/j.trac.2022.116897. [DOI] [Google Scholar]
- Rissin D. M.; Kan C. W.; Campbell T. G.; Howes S. C.; Fournier D. R.; Song L.; Piech T.; Patel P. P.; Chang L.; Rivnak A. J.; Ferrell E. P.; Randall J. D.; Provuncher G. K.; Walt D. R.; Duffy D. C. Single-Molecule Enzyme-Linked Immunosorbent Assay Detects Serum Proteins at Subfemtomolar Concentrations. Nat. Biotechnol. 2010, 28 (6), 595–599. 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah V.Democratizing Droplet Based Assays for Protein Measurement. Ph.D. Dissertation, University of California, Los Angeles, 2023. [Google Scholar]
- Papoutsakis E. T. Fluid-Mechanical Damage of Animal Cells in Bioreactors. Trends Biotechnol. 1991, 9 (1), 427–437. 10.1016/0167-7799(91)90145-8. [DOI] [PubMed] [Google Scholar]
- Particle-Templated Instant Partitions. Fluent BioSciences. https://www.fluentbio.com/technology/ (accessed 2024-03-11).
- Cai L.; Wang H.; Yu Y.; Bian F.; Wang Y.; Shi K.; Ye F.; Zhao Y. Stomatocyte Structural Color-Barcode Micromotors for Multiplex Assays. National Science Review 2020, 7 (3), 644–651. 10.1093/nsr/nwz185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerver R. E.; Gomez-Sjoberg R.; Baxter B. C.; Thorn K. S.; Fordyce P. M.; Diaz-Botia C. A.; Helms B. A.; DeRisi J. L. Programmable Microfluidic Synthesis of Spectrally Encoded Microspheres. Lab Chip 2012, 12 (22), 4716–4723. 10.1039/C2LC40699C. [DOI] [PubMed] [Google Scholar]
- Liu H.; Qian X.; Wu Z.; Yang R.; Sun S.; Ma H. Microfluidic Synthesis of QD-Encoded PEGDA Microspheres for Suspension Assay. J. Mater. Chem. B 2016, 4 (3), 482–488. 10.1039/C5TB02209F. [DOI] [PubMed] [Google Scholar]
- Ji X.-H.; Cheng W.; Guo F.; Liu W.; Guo S.-S.; He Z.-K.; Zhao X.-Z. On-Demand Preparation of Quantum Dot-Encoded Microparticles Using a Droplet Microfluidic System. Lab Chip 2011, 11 (15), 2561–2568. 10.1039/c1lc20150f. [DOI] [PubMed] [Google Scholar]
- Roh Y. H.; Sim S. J.; Cho I.-J.; Choi N.; Bong K. W. Vertically Encoded Tetragonal Hydrogel Microparticles for Multiplexed Detection of miRNAs Associated with Alzheimer’s Disease. Analyst 2016, 141 (15), 4578–4586. 10.1039/C6AN00188B. [DOI] [PubMed] [Google Scholar]
- Shao C.; Chi J.; Shang L.; Fan Q.; Ye F. Droplet Microfluidics-Based Biomedical Microcarriers. Acta Biomaterialia 2022, 138, 21–33. 10.1016/j.actbio.2021.10.037. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Vagin S. I.; Rieger B.; Destgeer G.. Fabrication of Crescent Shaped Microparticles for Particle Templated Droplet Formation. bioRxiv 2023; 10.1101/2023.10.06.561257. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Xie Z.; Gu H.; Jin L.; Zhao X.; Wang B.; Gu Z. Multifunctional Photonic Crystal Barcodes from Microfluidics. NPG Asia Mater. 2012, 4 (9), e25–e25. 10.1038/am.2012.46. [DOI] [Google Scholar]
- Kim J. H.; Kim J. B.; Choi Y. H.; Park S.; Kim S.-H. Photonic Microbeads Templated by Oil-in-Oil Emulsion Droplets for High Saturation of Structural Colors. Small 2022, 18 (8), 2105225 10.1002/smll.202105225. [DOI] [PubMed] [Google Scholar]
- Nguyen H. Q.; Baxter B. C.; Brower K.; Diaz-Botia C. A.; DeRisi J. L.; Fordyce P. M.; Thorn K. S. Programmable Microfluidic Synthesis of Over One Thousand Uniquely Identifiable Spectral Codes. Advanced Optical Materials 2017, 5 (3), 1600548 10.1002/adom.201600548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appleyard D. C.; Chapin S. C.; Doyle P. S. Multiplexed Protein Quantification with Barcoded Hydrogel Microparticles. Anal. Chem. 2011, 83 (1), 193–199. 10.1021/ac1022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim L. N.; Kim M.; Jung K.; Bae H. J.; Jang J.; Jung Y.; Kim J.; Kwon S. Shape-Encoded Silica Microparticles for Multiplexed Bioassays. Chem. Commun. 2015, 51 (60), 12130–12133. 10.1039/C5CC02048D. [DOI] [PubMed] [Google Scholar]
- Svedberg G.; Jeong Y.; Na H.; Jang J.; Nilsson P.; Kwon S.; Gantelius J.; Svahn H. A. Towards Encoded Particles for Highly Multiplexed Colorimetric Point of Care Autoantibody Detection. Lab Chip 2017, 17 (3), 549–556. 10.1039/C6LC01358A. [DOI] [PubMed] [Google Scholar]
- Juthani N.; Doyle P. S. A Platform for Multiplexed Colorimetric microRNA Detection Using Shape-Encoded Hydrogel Particles. Analyst 2020, 145 (15), 5134–5140. 10.1039/D0AN00938E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh Y. H.; Lee H. J.; Bong K. W. Microfluidic Fabrication of Encoded Hydrogel Microparticles for Application in Multiplex Immunoassay. BioChip J. 2019, 13 (1), 64–81. 10.1007/s13206-019-3104-z. [DOI] [Google Scholar]
- Derveaux S.; Stubbe B. G.; Braeckmans K.; Roelant C.; Sato K.; Demeester J.; De Smedt S. C. Synergism between Particle-Based Multiplexing and Microfluidics Technologies May Bring Diagnostics Closer to the Patient. Anal Bioanal Chem. 2008, 391 (7), 2453–2467. 10.1007/s00216-008-2062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Su C.; Chen Y.; Tian S.; Lu C.; Huang W.; Lv Q. Current Understanding of the Applications of Photocrosslinked Hydrogels in Biomedical Engineering. Gels 2022, 8 (4), 216. 10.3390/gels8040216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez C. J.; Mason T. G. Colloidal Alphabet Soup: Monodisperse Dispersions of Shape-Designed LithoParticles. J. Phys. Chem. C 2007, 111 (12), 4477–4480. 10.1021/jp0672095. [DOI] [Google Scholar]
- Rolland J. P.; Maynor B. W.; Euliss L. E.; Exner A. E.; Denison G. M.; DeSimone J. M. Direct Fabrication and Harvesting of Monodisperse, Shape-Specific Nanobiomaterials. J. Am. Chem. Soc. 2005, 127 (28), 10096–10100. 10.1021/ja051977c. [DOI] [PubMed] [Google Scholar]
- Lewis C. L.; Choi C.-H.; Lin Y.; Lee C.-S.; Yi H. Fabrication of Uniform DNA-Conjugated Hydrogel Microparticles via Replica Molding for Facile Nucleic Acid Hybridization Assays. Anal. Chem. 2010, 82 (13), 5851–5858. 10.1021/ac101032r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin M. A.; Werner H.; Udani S.; Di Carlo D.; Destgeer G. Flow Lithography for Structured Microparticles: Fundamentals, Methods and Applications. Lab Chip 2022, 22 (21), 4007–4042. 10.1039/D2LC00421F. [DOI] [PubMed] [Google Scholar]
- Chung S. E.; Park W.; Park H.; Yu K.; Park N.; Kwon S. Optofluidic Maskless Lithography System for Real-Time Synthesis of Photopolymerized Microstructures in Microfluidic Channels. Appl. Phys. Lett. 2007, 91 (4), 041106 10.1063/1.2759988. [DOI] [Google Scholar]
- Dendukuri D.; Gu S. S.; Pregibon D. C.; Hatton T. A.; Doyle P. S. Stop-Flow Lithography in a Microfluidic Device. Lab Chip 2007, 7 (7), 818–828. 10.1039/b703457a. [DOI] [PubMed] [Google Scholar]
- Laza S. C.; Polo M.; Neves A. A. R.; Cingolani R.; Camposeo A.; Pisignano D. Two-Photon Continuous Flow Lithography. Adv. Mater. 2012, 24 (10), 1304–1308. 10.1002/adma.201103357. [DOI] [PubMed] [Google Scholar]
- Wu C.-Y.; Owsley K.; Di Carlo D. Rapid Software-Based Design and Optical Transient Liquid Molding of Microparticles. Adv. Mater. 2015, 27 (48), 7970–7978. 10.1002/adma.201503308. [DOI] [PubMed] [Google Scholar]
- Lee H.; Kim J.; Kim H.; Kim J.; Kwon S. Colour-Barcoded Magnetic Microparticles for Multiplexed Bioassays. Nat. Mater. 2010, 9 (9), 745–749. 10.1038/nmat2815. [DOI] [PubMed] [Google Scholar]
- Choi N. W.; Kim J.; Chapin S. C.; Duong T.; Donohue E.; Pandey P.; Broom W.; Hill W. A.; Doyle P. S. Multiplexed Detection of mRNA Using Porosity-Tuned Hydrogel Microparticles. Anal. Chem. 2012, 84 (21), 9370–9378. 10.1021/ac302128u. [DOI] [PubMed] [Google Scholar]
- Bong K. W.; Kim J. J.; Cho H.; Lim E.; Doyle P. S.; Irimia D. Synthesis of Cell-Adhesive Anisotropic Multifunctional Particles by Stop Flow Lithography and Streptavidin–Biotin Interactions. Langmuir 2015, 31 (48), 13165–13171. 10.1021/acs.langmuir.5b03501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. J.; Chen L.; Doyle P. S. Microparticle Parking and Isolation for Highly Sensitive MicroRNA Detection. Lab Chip 2017, 17 (18), 3120–3128. 10.1039/C7LC00653E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duenas Y.; Lee J.; Jung S.; Yi H. Multiplexed Hydrogel Microparticle Suspension Arrays for Facile Ribosomal RNA Integrity Assays. Biotechnol Bioproc E 2015, 20 (5), 956–964. 10.1007/s12257-015-0265-z. [DOI] [Google Scholar]
- Introducing FirePlex®-HT High-throughput Multiplex··· ABCAM. https://www.abcam.com/index.html?pageconfig=news&rid=16918 (accessed 2024-03-14).
- Platchek M.; Lu Q.; Tran H.; Xie W. Comparative Analysis of Multiple Immunoassays for Cytokine Profiling in Drug Discovery. SLAS Discovery 2020, 25 (10), 1197–1213. 10.1177/2472555220954389. [DOI] [PubMed] [Google Scholar]
- Perea A.; Heinrich B.; Austin C.; Rafferty M.; Camilleri L.; To L.; Atabakhsh E.; Pregibon D.. Accelerating High-Throughput Screening with FirePlex®-HT: An Automatable, Multiplex Immunoassay Using FirePlex® Particle Technology, 2019. https://docs.abcam.com/pdf/fireplex/fireplex-ht-poster.pdf.
- Destgeer G.; Ouyang M.; Di Carlo D. Engineering Design of Concentric Amphiphilic Microparticles for Spontaneous Formation of Picoliter to Nanoliter Droplet Volumes. Anal. Chem. 2021, 93 (4), 2317–2326. 10.1021/acs.analchem.0c04184. [DOI] [PubMed] [Google Scholar]
- Yamashita M.; Tamamitsu M.; Kirisako H.; Goda Y.; Chen X.; Hattori K.; Ota S. High-Throughput 3D Imaging Flow Cytometry of Suspended Adherent 3D Cell Cultures. Small Methods 2023, 2301318. 10.1002/smtd.202301318. [DOI] [PubMed] [Google Scholar]
- Wu C.-Y.; Stoecklein D.; Kommajosula A.; Lin J.; Owsley K.; Ganapathysubramanian B.; Di Carlo D. Shaped 3D Microcarriers for Adherent Cell Culture and Analysis. Microsyst Nanoeng 2018, 4 (1), 21. 10.1038/s41378-018-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W.; Kim D. Y.; Mun S. J.; Choi J. H.; Roh Y. H.; Bong K. W. Direct Functionalization of Cell-Adhesion Promoters to Hydrogel Microparticles Synthesized by Stop-Flow Lithography. J. Polym. Sci. 2022, 60 (11), 1767–1777. 10.1002/pol.20210934. [DOI] [Google Scholar]
- Li M.; Joung D.; Hughes B.; Waldman S. D.; Kozinski J. A.; Hwang D. K. Wrinkling Non-Spherical Particles and Its Application in Cell Attachment Promotion. Sci. Rep 2016, 6 (1), 30463 10.1038/srep30463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A. J.; Sen S.; Sweeney H. L.; Discher D. E. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell 2006, 126 (4), 677–689. 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- D’Elia A. M.; Jones O. L.; Canziani G.; Sarkar B.; Chaiken I.; Rodell C. B. Injectable Granular Hydrogels Enable Avidity-Controlled Biotherapeutic Delivery. ACS Biomater Sci. Eng. 2024, 10 (3), 1577–1588. 10.1021/acsbiomaterials.3c01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S.; Li C.; Zheng X.; Huang W.; Tao Y.; Yu Y.; Yang L.; Lan Y.; Ma L.; Bian S.; Du W. OsciDrop: A Versatile Deterministic Droplet Generator. Anal. Chem. 2022, 94 (6), 2918–2925. 10.1021/acs.analchem.1c04852. [DOI] [PubMed] [Google Scholar]