Abstract
The generation of specialized neural cells in the developing and postnatal central nervous system is a highly regulated process, whereby neural stem cells divide to generate committed neuronal progenitors, which then withdraw from the cell cycle and start to differentiate. Cell cycle checkpoints play a major role in regulating the balance between neural stem cell expansion and differentiation. Loss of tumor suppressors involved in checkpoint control can lead to dramatic alterations of neurogenesis, thus contributing to neoplastic transformation. Here we summarize and critically discuss the existing literature on the role of tumor suppressive pathways and their regulatory networks in the control of neurogenesis and transformation.
Keywords: Stem cells, Tumor suppressor, Neurogenesis, Transformation
Introduction
Neurogenesis is the process by which functional neurons are generated from neural stem and progenitor cells (NPCs). Neurogenesis occurs in both developing and adult brains [1–6].
Control of cell division and cell death during neurogenesis is crucial for the generation of new neuronal cells. Neural stem cells (NSCs) have the ability to self-renew (expansion) or generate specialized cells, either directly or via generation of transit-amplifying (or intermediate) progenitors (differentiation) [4, 7]. Significant progress has been made over the last decade in the study of almost all the aspects of mammalian neurogenesis both at embryonic and perinatal stages [5].
Early in nervous system development, NSCs form a polarized epithelium, named the ventricular zone (VZ), whose apical domain delimits the lumen of the neural tube. As development proceeds, an increased proportion of stem cells start to switch from symmetrical division, which generates additional stem cells, to asymmetrical division [4, 7]. In particular, the switch to asymmetrical division in the cerebral cortex leads to the generation of neurons in the cortical plate (CP) and of intermediate progenitors that leave the ventricular zone and form a second germinal region, called the sub-ventricular zone (SVZ) [4, 7–11].
Despite the fact that most precursors are exhausted by the end of the development to generate neurons and glia, a minor proportion of NPCs have also been found within two well-described neurogenic niches: the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) in the dentate gyrus [12–14]. In addition, recent studies suggest that NSCs may also reside in non-canonical neurogenic niche of the adult brain, including the cerebellum [15], substantia nigra [16, 17], and retina [18, 19].
Within the nervous system, cellular and environmental determinants tightly control the expansion and the differentiation of NSCs [7]. In this respect, self-renewal programs rely on complex regulatory networks that balance the function of proto-oncogenes (promoting self-renewal), gate-keeping tumors suppressor (limiting self-renewal), and care-taking tumors suppressors (maintaining genomic integrity) [11, 20, 21]. As a result, proliferation, self-renewal, and genomic stability are tightly controlled in both embryonic and adult stem cells [22, 23]. A defect in self-renewal, division, or differentiation of stem cells can lead to developmental defects, premature aging phenotype, failure to repair injured tissue, as well neoplastic transformation [24, 25].
The focus of this review is on the role of tumor suppressive pathways in regulation of neurogenesis in the mammalian brain. Alterations of tumor suppressive control in NSCs can lead to transformation and tumor development in both the embryonic and adult nervous system. It is therefore fundamental to dissect mechanisms underlying tumor suppressive function in the CNS as they may provide insights into its pathophysiology.
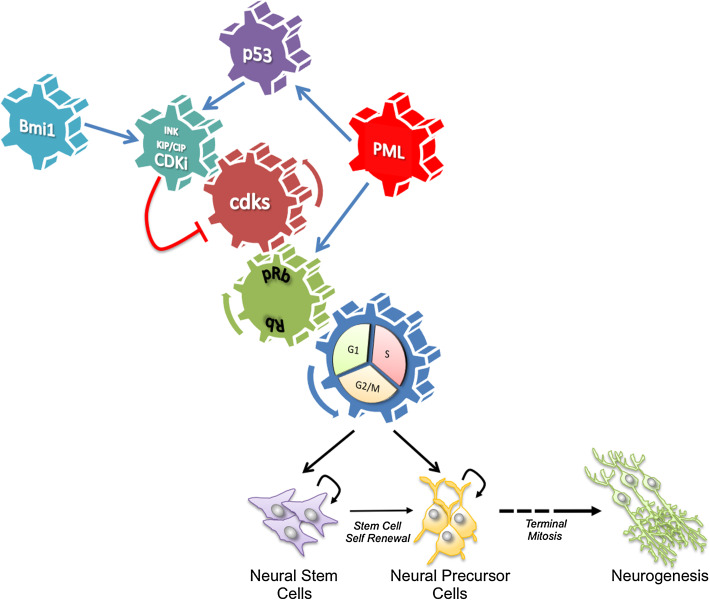
We will critically review the existing literature on two main tumor suppressors, retinoblastoma (Rb) and p53, and the stem cell factor Bmi1 (Fig. 1). As shown in Fig. 1, the main point of intersection between the two tumor suppressive pathways is represented by cyclin/cyclin-dependent kinases (CDKs) and their inhibitors (CKIs). p53 positively regulates CKIs, whereas CDK-complexes negatively regulate Rb. The Polycomb protein Bmi1 controls stem cell self-renewal by suppressing expression of CKIs, thus leading to Rb inactivation. Integration of these regulatory modules regulates the balance between expansion and differentiation in the developing CNS and adult neurogenic niches.
Fig. 1.
Tumor suppressive factors and cell cycle control in the developing CNS and adult neurogenic niches. In neural/progenitor stem cells, a complex interplay of different tumor suppressors, downstream effectors, and regulatory networks controls self-renewal and differentiation
G1/S checkpoint: cyclins, cyclin-dependent kinases, and their inhibitors
In mammalian cells, proper progression through the cell cycle is monitored by checkpoints that sense possible defects during DNA duplication and chromosome segregation. Physiological activation of these checkpoints induces cell cycle arrest through modulation of cyclins and cyclin-dependent kinases (CDKs; Fig. 1) [34, 35]. The control of G1/S transition is primarily achieved through the coordinated action of two main cyclin/CDK complexes containing D-type and E-type cyclins. Cyclin D bind CDK4/6 early during the G1/S transition, whereas Cyclin E/Cdk2 complexes are active slightly later. Cyclin/cdk complexes are negatively regulated by CKIs, the main executers of the G1/S checkpoint. These are divided into two families: the Ink4/Arf type (p16Ink4a, p15Ink4b, p18Ink4c and p19Ink4d) and the Cip/Kip type (p21Waf1/Cip1, p27Kip1 and p57Kip2) [36]. For more in-depth analysis of the role of CDK and CKIs in regulation of the cell cycle, the reader can refer to a number of comprehensive review articles by leading laboratories in the field [8, 37, 38].
With respect to the role of cyclin/CDK complexes and CKIs in cell-fate regulation during neurogenesis, extensive literature suggests that the latter promote differentiation, whereas the former play an inhibitory role [11, 39, 40]. For example, the anti-proliferative gene p27 has long been proposed to act as part of clock control for oligodendrocyte differentiation by limiting the number of divisions before the onset of differentiation [41]. Other studies have shown that CDK2 is required for S-phase entry and cell progression in NPCs isolated from both embryonic and early post-natal brain [42, 43]. In this contest, CDK2 has also been shown to be dispensable for NPC proliferation, differentiation, and survival of adult-born DG granule neurons in vivo [44]. More recently, work by Calegari and coworkers showed that CDK4/cyclin D1 overexpression inhibited neurogenesis, but at the same time it promoted the generation and expansion of basal progenitors, resulting in thicker SVZ and overall larger cortical surface area [9]. A subsequent study from the same group reported an important role of CDK4/Cyclin D1 also in the control of adult neurogenesis in the hippocampus [45]. Finally, Beukelaers and colleagues implicated CDK6 as a major regulator of adult neurogenesis [46]. Specifically, CDK6 deficiency prevents the expansion of neuronally committed precursors by lengthening duration of the G1 phase, thus reducing concomitantly the number of newborn neurons [46]. Further studies are needed to fully dissect the role and interplay of G1/S checkpoint members in regulation of neurogenesis.
p53 and its family
p53 (also known as tumor protein 53, tp53) was first identified over 30 years ago as a cellular protein associated with the SV40 T-antigen oncoprotein [47–49]. Although it was originally described as an oncogene, p53 is a tumor suppressor inactivated in more than 50 % of human cancers [50–55]. As a “guardian of the genome”, p53 induces cell cycle arrest and cell death after DNA damage, thus contributing to the maintenance of genomic stability [56]. p53 is found in both the nucleus and the cytoplasm [57]. Nuclear p53 acts as a sequence-specific transcription factor, which regulates the expression of a vast number of genes involved in many different cellular processes from cell cycle checkpoint control to programmed cell death and metabolism (for extensive review of the literature see [57–61]. In particular, the CKI p21 is a major p53 target and plays an important role in p53-mediated regulation of G1/S transition (Fig. 1). In the cytoplasm, p53 promotes the activation of programmed cell death by directly regulating proapoptotic players such as Puma [57]. In addition, cytoplasmic p53 can repress autophagy, an intracellular degradation pathway involved in regulation of metabolism and survival [62]. In contrast, nuclear p53 has been proposed to induce autophagy via upregulation of the autophagy-associated factor DRAM1 [63]. Thus, p53-dependent regulation of autophagy varies dependent on its subcellular distribution.
In the CNS, p53 plays a key role in regulating programmed cell death in postmitotic neurons [64]. Evidence is emerging that p53 controls neurogenesis in the developing as well as in adult brain [65, 66]. In this respect, the tumor suppressor p53 has been shown to be abundant, both at RNA and protein levels, in regions of active proliferation during development and in adult neurogenic niches such as the SVZ and the SGZ [66–68] (Box 1). While p53 is found in post-mitotic neurons of the cerebral cortex and hippocampus of the developing brain, its expression is enriched in cycling progenitor/stem cells (NPCs) [65, 67, 68]. These data suggested that p53 might play an important role in the control of NPC proliferation and potentially regulation of CNS development. Indeed, p53-deficent mice exhibit high-frequency developmental abnormalities of the nervous system, in particular exencephaly and/or mild to moderate hyperplasia in approximately a quarter of p53-KO embryos [69, 70]. The partial penetrance of these phenotypes may be related to the existence of compensatory mechanisms probably mediated by the other members of the p53 family, p63 and p73 [71–75]. In the postnatal SVZ, p53 loss leads to over-production of NPCs, along with an expansion of neuronal and glial lineages [65, 76]. In accordance with the in vivo phenotypes, NPCs derived from the p53-null mice display enhanced self-renewal capacity, altered differentiation and reduction in apoptosis [65, 76].
Box 1.
Neurogenic niche
| Stem cell niche | References |
| It is defined as the microenvironment in which stem cells are found. There are two neurogenic niches in the adult brain, where under physiological conditions NPCs give rise to new neurons and macroglia: (1) the subventricular zone (SVZ), a germinal area situated within the walls of the lateral ventricles where NPCs generate cells that migrate into the olfactory bulb (OB); (2) the subgranular zone (SGZ) of the dentate gyrus (DG), where newly generated granule cells become integrated into the local network. These two areas have similar structural organization and tight association with the local vasculature | [29] and [30–32] |
| Adult progenitors | |
| A population of multipotent neural cells mainly present in the two specialized niches of the adult mammalian brain (SVZ and SGZ). They maintain neurogenesis throughout adult life. SVZ progenitors derive directly from radial glia (RG) and are called type-B NSCs (astrocyte-like GFAP-positive NSCs). Type-B NSCs are in intimate contact with all the other SVZ cell types, including the rapidly dividing transit-amplifying type-C cells and the lineage-committed migratory neuronal type-A cells. The SGZ contains type-B NSCs (astrocyte-like GFAP-positive NSCs) and GFAP-negative type-D cells. Thus, in both germinal regions of the mammalian postnatal brain, astrocyte-like cells represent the main pool of multipotent progenitors | [29, 33] |
Analysis of the transcriptome of p53 KO NPCs provided cues on the underlying mechanisms. In this respect, the known p53 targets p21Cip1/Waf1 and p27Kip1 are down-regulated in p53-null neurospheres [65]. These CKIs are regulated by p53 in a direct [77] as well as indirect manner [27, 78]. Their loss was shown to significantly affect NPC proliferation/differentiation [25]. In the absence of either p21Cip1/Waf1 or p27Kip1, proliferating NPCs fail to exit the cell cycle, thus eventually leading to their exhaustion [79, 80]. Subsequent studies have suggested that p53-dependent induction of p21Cip1/Waf1 results in lengthening of the cell cycle and decreased expansion of NPCs within the developing [81] as well as the adult brain [82, 83]. With respect to p27Kip1, Doetsch and coworkers [79] showed that its loss does not affect the number of NPCs in the adult SVZ. Instead, the pool of transient amplifying cells increased, accompanied by a reduction in neuroblast number [79]. Thus, in the adult brain, p27Kip1 appears essential for the transition from transit-amplifying cells to neuroblasts. Other studies have confirmed a role for p27Kip1 in regulation of neurogenesis in the RMS, olfactory bulb (OB), and cerebellum of the postnatal mouse [84, 85]. In the developing cerebral cortex, p27Kip1 is expressed strongly by the post-mitotic neurons of the CP, and in mature granule cells (GCs) of the cerebellum but only weakly in proliferating neuroblasts of the SVZ [86] and in granule cell precursors (GCPs) of the cerebellum [85]. The different phenotypes observed in p27 Kip1 and p21 Cip1/Waf1 KO animals suggest that they may function at different stages of neurogenesis. Alternatively, they may control cell fate through different mechanisms. In this respect, it is important to note that p27 regulates neuronal migration in the developing cortex in a cyclin-independent manner [87, 88]. Finally, a recent study by Gil-Perotin and coworkers [89] demonstrated a non-redundant role of p27Kip1 and p53 in regulation of proliferation of SVZ NPCs and even an antagonistic effect on the generation of adult neuroblasts. Overall, these data suggest that the role of p27Kip1 in regulation of neurogenesis may be more complex than previously thought.
Evidence in the literature suggests that p53 cooperates with the phosphatase and tension homolog (PTEN) in regulation of NPC expansion. In particular, proliferation of NPCs lacking p53 is further enhanced by concomitant PTEN inactivation [90]. Phosphatase and tension homolog is a major tumor suppressor antagonizing the oncogenic function of class I phosphoinositide 3-kinase (PI3K). Loss of PTEN results in increased PI3K signaling and augmented proliferation of NPCs [91]. Interestingly, although p53-deficient NPCs are still able to respond to differentiation cues, this property is completely abrogated when both p53 and PTEN are deleted in GFAP+ astrocytic progenitors, thus leading to neoplastic transformation and tumor development [92, 93]. Mechanistically, increased c-Myc expression and activity in double-KO NPCs contributes to the tumorigenic phenotype [92, 93]. Inactivation of the neurofibromatosis gene 1 (NF1) also cooperates with p53 loss in promoting overproliferation, inhibition of differentiation, and ultimately glioma development (Fig. 2) [94]. Notably, a recent study by Zhu and colleagues [95] revealed that loss of p53 transcriptional function via deletion of part of its DNA binding domain induces pleiotropic accumulation of cooperative oncogenic alterations that in turn drive gliomagenesis (Fig. 2). It remains to be determined why a loss of p53-dependent transcription is a stronger tumorigenic signal than its complete genetic loss. One potential explanation would be that cytosolic, transcription-independent function of p53 could contribute to cancer development. Alternatively, the above-mentioned p53 mutant could bear gain-of-function activity and regulate transcription via DNA binding-independent mechanisms.
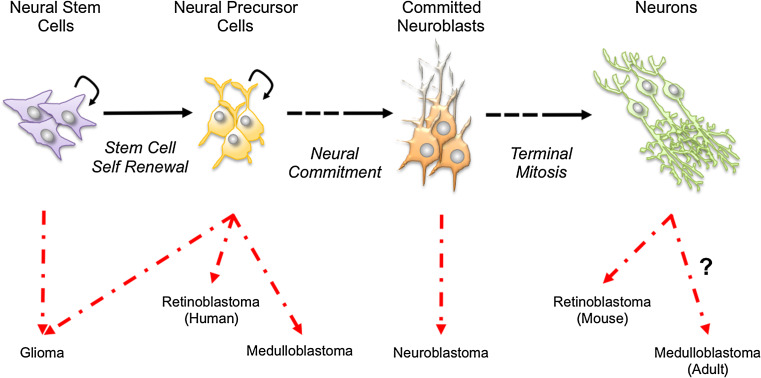
Fig. 2.
Neurogenesis and transformation. Nervous system neoplasms are believed to originate from different stages of the neurogenesis process. The figure summarizes the current knowledge. Please refer to the text for more details information and references
It is worth noting that accumulation of mutant p53 within the SVZ gives rise to uncontrolled proliferation of Olig2+ transient-amplifying progenitor-like cells, thus suggesting that this subpopulation, rather than more truly stem cells, may drive brain cancer development (Fig. 2) [95]. A recent study using a mouse genetic mosaic system showed that indeed Olig2+ Nf1/p53 DKO progenitors act as cell of origin for the proneural subtype of glioma (Fig. 2) [96]. It is therefore possible that acquisition of an oligodendrocytic precursors fate is required for development of this glioma subtype. Overall, these data implicate the tumor suppressor p53 in the control of NPC expansion, differentiation, and transformation. One of the remaining outstanding questions is whether p53 could control neurogenesis also via mechanisms not related to its role in cell cycle regulation. In particular, it would be important to study the impact of p53 metabolic function on neural stem cell fate and transformation. This could extend beyond regulation of survival. In this respect, it is conceivable that p53-mediated regulation of metabolism and cellular redox could contribute to the control of self-renewal, which has been shown to be sensitive to changes in ROS levels.
However, p53 is not the whole story. In the late 1990s, two novel p53 family members were identified, termed respectively p63 [97, 98] and p73 [99]. The full-length isoforms of these proteins, called TAp63 and TAp73, respectively, function as pro-apoptotic proteins [98, 100–104], whereas naturally occurring N-terminal truncated variants of p63 (ΔNp63) and p73 (ΔNp73) act as pro-survival proteins, at least partially by antagonizing the full-length family members [105–109]. Could the TA isoforms bear tumor-suppressive properties like p53? Research in this area has been in part disappointing, as only a small percentage of human cancers display mutations in p63 and p73 [110]. However, Tyler Jack’s laboratory proposed that p63 and p73, rather than being redundant, contribute to p53 activation and tumor suppressive function [111]. A subsequent study showed that dual inactivation of p63 and p73 results in increased tumor susceptibility, which is further enhanced upon inactivation of p53 [112]. Although p53 family members appear to act synergistically in controlling tumor suppression, their inactivation leads to distinct developmental phenotypes [106, 113, 114]. In this respect, mice lacking all p73 isoforms display severe defects in CNS development [114], including congenital hydrocephalus and hippocampal dysgenesis, but fail to show exencephalic outgrowth. Loss of p73 results in decreased NPC self-renewal during both embryonic and adult neurogenesis, thus suggesting that p73 is a novel regulator of NPC function [102, 115–117]. One of these studies [102] also demonstrates a correlation between the reduced proliferation capacity of p73 −/− NPCs and expression levels of the transcription factor Sox2, an essential player in the maintenance of NSC self-renewal property. It is currently unclear what is the effect of p73 loss on p53 activation in NPCs. Finally, isoform-specific inactivation of either TA- or ΔN-p73 fails to fully recapitulate the CNS phenotypes observed in p73−/−, thus suggesting a degree of redundancy between the two isoforms [118].
The third member of the family, p63, is a known regulator of self-renewal during development/homeostasis of epithelial tissues. With respect to its role in neurogenesis, p63 shorter isoform, ΔN-p63 has been implicated in regeneration of the olfactory bulb (OB) epithelium [119]. In particular, it regulates quiescence in horizontal basal (stem) cells (HBC), thus inhibiting their differentiation to fully mature olfactory neurons and/or other cell types [120]. These observations suggest that ΔN-p63 acts as a molecular switch controlling self-renewal and differentiation of OB stem cells. ΔN-p63 may also regulate survival in embryonic cortical NPCs by antagonizing p53 pro-apoptotic function [121]. Thus, tuning the balance between ΔN-p63 and p53 may dictate the survival of embryonic NPCs. However, a separate study failed to report abnormal development of either brain or spinal cord in p63 −/− mouse embryos [117]. Further work is needed to address these discrepancies between the abovementioned studies and fully dissect the role and interplay of p53 family members in regulation of neurogenesis.
Rb and the pocket protein family
The retinoblastoma gene (Rb or Rb1) was the first tumor suppressor to be identified on the basis of its involvement in tumorigenesis [122–125]. In this respect, germline mutations in the Rb gene (chromosomal location 13q14) predispose to the ocular tumor retinoblastoma (Fig. 2), a rare childhood cancer of the retina that initiates during development [126–128]. Subsequent studies led to the demonstration that affected individuals develop retinal tumors as a consequence of the loss of the second Rb allele [125]. Additional evidence for a tumor suppressive role of the Rb gene came from the discovery of the absence of functional Rb in virtually all retinoblastoma tumors examined and its frequent inactivation in other human cancers, such as brain tumors, small-cell lung carcinoma, and osteosarcoma [129–131].
Studies examining the transformation mechanisms of DNA tumor viruses started to shed new light on the role of Rb as a major regulator of cell proliferation. Rb is a target of several viral oncoproteins, such as adenovirus E1A [129, 132], simian virus 40 (SV40) large T antigen (Tag) [133, 134], and the E7 proteins of human papillomavirus (HPV-16) [135]. Importantly, mutation of the Rb binding region of any of these viral oncoproteins abrogated viral transformation [136]. Subsequent studies revealed a key role played by Rb in regulation of the G1/S transition (Fig. 1). In particular, Rb encodes a 110-kDa nuclear phosphoprotein (Rb) that is able to bind and inhibit members of the E2F transcription factor family, thus repressing transcription of genes important for S-phase entry and progression [128, 137, 138]. Retinoblastoma phosphorylation by CDKs relieves repression of E2F transcriptional activity, thus promoting cell cycle progression [139, 140]. Retinoblastoma can actively repress transcription [141] of E2F target genes through recruitment of histone deacetylase (HDAC), thus directly affecting their epigenetic status [141].
More recently, in addition to its role in regulating the G1-to-S-phase transition, Rb has been shown to mediate both accurateness and timing of DNA replication and to control proper segregation of chromosomes in mitosis [142–144]. Using gene targeting, Coschi and coworkers demonstrated that Rb facilitates chromosome condensation, independently to its role in DNA replication [142]. Further studies reported that Rb is also required for regulation of centromeres. In this respect, Rb mediates the proper condensation and orientation of centromeres on duplicated chromosomes and thus their attachment to the spindle [143]. When Rb function is compromised, the level of chromosome mis-segregation increases dramatically and becomes comparable to tumor cells [143]. Finally, mouse embryo fibroblasts (MEFs) lacking functional Rb and its related proteins p107 and p130 (see also below) have been shown to be more prone to chromosome breakage [144]. Together, these data suggest that the chromosome instability (CIN) resulting from the inability of Rb to maintain sister chromatid cohesion represents another important function of Rb as tumor suppressor [143, 145].
Insights into the importance of Rb as a regulator of cell cycle and differentiation in vivo and in particular in the central nervous system came from the analysis of Rb knockout animals. Rb is necessary for embryo development, as Rb −/− embryos die by embryonic day 15 (E15), exhibiting dramatic neural tube defects and skeletal muscle defects [146–149]. In particular, these abnormalities were accompanied by a partial failure of differentiation, increased apoptosis, as well as ectopic cell cycle [146–148, 150]. Further studies showed that part of the described neural phenotypes, including increased apoptosis, were due to a non-cell autonomous effect of Rb inactivation [151]. Conditional inactivation of Rb in the CNS confirmed the increased ectopic mitosis in the developing cortex. However, Rb −/− NPCs were able to survive and differentiate, thus leading to increased brain cellularity [152]. Conditional inactivation of Rb in the cerebellum and the retina revealed its key role in regulation of development also in these CNS structures [153, 154]. Finally, Rb was shown to regulate migration of newborn interneurons in the embryonic cortex in a cell-autonomous fashion, thus suggesting a role of Rb beyond cell cycle control [155]. Interestingly, this phenotype is observed also in p27−/− embryos, although it is still unclear whether [87] p27Kip1 function in this context appears independent of cell cycle regulation [87]. It is presently unclear whether p27Kip1 and Rb regulate neuronal migration through similar mechanisms.
The Rb protein is a member of a family of three closely related mammalian proteins that includes p107 (also known as retinoblastoma-like 1) and p130 (retinoblastoma-like 2). Together they are known as “pocket proteins” due to shared sequence homology in the pocket A/B domain, which mediates interactions with transcription factors and oncoproteins [138, 156–160]. Although the three “pocket protein” family members have been shown to bear distinct binding properties for the various E2Fs [161], overexpression experiments have indicated functional similarities in the regulation of the cell cycle as well as development [127, 162]. For example when overexpressed, all three family members cause G1 phase arrest via repression of E2F-mediated gene transcription. Moreover, all Rb proteins are phosphorylated by CDKs [127, 163]. p130 −/− and p107 −/− mice survive to adulthood without overt phenotypes, unlike those observed in Rb-deficient animals [164, 165]. The lack of severe phenotype in the p107 −/− and p130 −/− mice may be due to the compensatory activity of Rb. Vice versa, Rb loss was accompanied by compensatory upregulation of p107 [166, 167]. Additional evidence for redundancy within the pocket family came from the observation that inactivation of Rb only is not sufficient to cause retinoblastoma in mice. In contrast, loss of all three members is sufficient to transform retinal cells and lead to metastatic retinoblastoma [168]. As human retinoblastoma is predominantly associated with loss of Rb only, it is possible that mouse retinoblastoma pathogenesis differs from human retinoblastoma (Fig. 2). In this respect, human retinoblastoma is believed to originate from cone cell precursors (Fig. 2), whereas postmitotic neurons act as cell of origin of retinoblastoma in pocket family knockout mice [128, 169]. Interestingly, in human retinoblastoma, Rb deficiency is associated with Mdm2 amplification, which in turn impairs p53 activation downstream Rb loss, thus providing an additional oncogenic signal for full transformation [169]. Notably, Rb loss cooperates with p53 loss in promoting glioma as well as medulloblastoma development in NPCs [170, 171], two tumors displaying genetic and/or functional inactivation of both tumor suppressive pathways in humans (Fig. 2). Finally, glioma can be induced by inactivation of pocket family members via overexpression of SV40 [172]. Overall, these studies suggest that redundancy among pocket family members act as tumor suppressive barrier, unless the p53 pathway is inactivated. Mechanisms underlying this phenomenon remain unexplored.
Despite the observed redundancy, pocket family members may have specialized function. In this respect, expression of p107, unlikely Rb and p130, appears restricted to cycling NPCs in the SVZ, and is downregulated as NPCs differentiate into neurons [173]. p107 may regulate the decision of NPCs to exit the cell cycle and commit to neuronal fate [174]. Accordingly, p107-deficent cortices display increased proliferation associated with augmented Hes1 expression and impaired neurogenesis [173, 174]. p107 may also be acting through the FGF growth factor signaling pathway. Like Hes1, the expression of Fgf2 is increased in neural progenitor of p107-deficent mice to promote proliferation of NPCs [175].
Regulators of the Rb and p53 in the CNS
PML
Tumor suppressors appear to work in a complex network of functional interactions, which finely tune their activities in regulation of cell fate and transformation. Among them, the promyelocytic leukemia protein (PML) and the Polycomb group protein Bmi1 have been shown to play important roles in regulation of the p53 and Rb pathways.
The PML gene was originally identified at the t(15;17) chromosomal translocation breakpoint of acute promyelocytic leukemia (APL), a subtype of acute myeloid leukemia [176–181]. This translocation generates the oncogenic fusion gene PML/retinoic acid receptor-α (PML/RARα), which drives APL in humans and mice [177, 178, 182, 183]. Promyelocytic leukemia protein/retinoic acid receptor-α exerts its oncogenic effects in part by blocking normal PML function [184, 185]. Promyelocytic leukemia protein is localized within discrete nuclear structures referred to as PML-nuclear bodies (PML-NBs), of which it is the essential component [184, 186–188].
To date, PML has been shown to influence and/or regulate several cellular processes, including transcription, cell cycle and cellular senescence. In this respect, we will largely refer to the many comprehensive review articles by our laboratory and others. Several studies have implicated PML in the regulation of Rb and p53 through multiple mechanisms (Fig. 1) [184, 189–194].
Our more recent work has described a functional interaction between Rb and PML in NPCs during neocortex development. We proposed that PML-mediated regulation of Rb occurs at least in part via its direct targeting, along with the protein phosphatase 1 alpha (PP1α), to the PML-NBs, thus favoring Rb dephosphorylation [195]. As a result, loss of PML leads to Rb hyperphosphorylation and inactivation of G1–S checkpoint. In vivo, PML loss promotes increased cycling in cortical NPCs, thus leading to the expansion of the VZ. This effect appears restricted to a specific NPC subtype, radial glial cells (RGCs). Expansion of RGCs is accompanied by a reduced number of more committed intermediate progenitor cells (IPCs), thus suggesting that PML via Rb regulates transition from RGCs to IPCs. This is particularly interesting as little is known abut the molecular mechanisms regulating the generation of IPCs from RGCs. Promyelocytic leukemia protein-deficient embryos show smaller brains with a prominent reduction in neocortical wall thickness, although the overall layered structure of the cortex remains unaffected. Thus, PML plays an important role in regulation of cell fate and corticogenesis.
It remains to be investigated whether PML works solely through Rb or if other Rb family members are involved. In this respect, the phenotype of PML-deficient brains resembles the one observed in p107 −/− mice [173, 174], thus suggesting that PML could affect the function of other pocket proteins. In addition, it is currently unclear whether PML regulates p53 in NPCs. Finally, our recent work has shown that the PML-interacting protein, DAXX, acts as chaperone for the histone variant H3.3 in neurons, thus regulating chromatin remodeling and transcription upon neuronal activation [196]. As both H3.3 and DAXX are found in PML-NBs in cycling cells [197], these new findings raise the exciting possibility that PML may control cell fate in neural stem cells via regulation of DAXX-mediated H3.3 loading [196]. Interestingly, PML also colocalizes with ATRX, which is part of the DAXX chaperone complex [198]. ATRX has been shown to regulate transition through mitosis in the developing brain. Mutations of ATRX cause the ATR-X syndrome, which is characterized by brain retardation and thalassemia. Furthermore, a number of disease-associated mutants of ATRX are impaired in their ability to localize to PML-NBs [199].
A number of recent studies have also highlighted a more general role of PML in the control of stem cell function. For instance, recent work from Pandolfi’s group has implicated PML in the regulation of stem cell quiescence in the hematopoietic system [200]. As a result, PML loss affects self-renewal of hematopoietic stem cells (HSCs) potentially through its action on the mTOR pathway. Interestingly, the tumor suppressor p53 and its target gene p21 have recently been shown to control the cell fate in HSCs [201]. Another recent study has shown that PML loss skews the balance between different progenitor subtypes in the mammary gland and affects its development, although the underlying mechanisms have not been investigated [202].
The role of PML in the regulation of NPC expansion and differentiation could have obvious implications for cancer research, as alterations of these processes can lead to the development of brain tumors [203]. In this respect, PML expression is substantially decreased in human cancers of multiple histologic origins, including a small subset of tumor medulloblastoma [204]. However, a comprehensive study of PML expression in brain cancer is still missing. PML inactivation is not sufficient to cause cancer [11, 95, 193], but it can promote tumor progression in the context of other oncogenic lesions, such as loss of PTEN. It is therefore conceivable that also in brain cancer PML downregulation could act in concert with loss of other tumor suppressor and/or oncogenic activation. One could hypothesize that decreased PML expression could affect tumors where the p53 or Rb pathways are not genetically inactivated. In this respect, a number of medulloblastomas and gliomas do not present genetic loss of either tumor suppressive pathway [205]. In these tumors, PML could contribute to p53 and/or Rb functional inactivation potentially in conjunction with other regulators, such as Mdm2 or G1/S cyclins.
Finally, modulation of PML expression could be used to modulate expansion and differentiation of neural stem cells for regenerative medicine-related applications. In this respect, regulation of PML stability can be achieved also via pharmacological means using arsenic trioxide, which targets PML for proteasomal degradation either directly or indirectly via increased PML oxidation [206, 207]. However, it is important to note that arsenic trioxide has rather pleiotropic effects in the cell, thus suggesting that PML could be only one of its targets.
The Polycomb group protein Bmi1
The embryonic and postnatal development of the CNS requires a fine balance between cell proliferation and differentiation. This is in part achieved by maintenance of an active or repressed state at discrete loci through epigenetic regulation. The family of Polycomb group (PcG) proteins regulate gene expression by forming large complexes at specific chromosomal sites called Polycomb response elements (PREs) and inducing chromatin remodeling. There are two main PcG complexes, Polycomb regulatory complex 1 and 2 (PRC1 and PRC2). The B cell-specific Moloney murine leukemia virus integration 1 (Bmi1) Polycomb ring finger oncogene is the first functional PcG member identified [208]. The B cell-specific Moloney murine leukemia virus integration 1 is part of PRC1, which mediates histone ubiquitylation. The activity of PRC1 is regulated by interaction with H3K27me3, which is catalyzed by PRC2. However, recent findings suggest that the PRC1 can also work in an H3K27me3-independent manner [209]. In mammals, the main targets of PcG proteins during embryogenesis are homeobox (Hox) genes, which regulate key developmental processes including self-renewal and maintenance of developmental potential of neural stem/progenitor cells [210]. Polycomb-group proteins generally act as repressors, but based on a recent study, they can also promote transcriptional activation of genes involved in metabolism through association with a specific RNA polymerase II variant [211].
Bmi1 is ubiquitously expressed in mammalian tissues, but higher levels are detected in thymus, heart, testis, embryonic stem cells (ES), and embryonic and postnatal neural stem cells [212, 213]. The first evidence of a possible role of Bmi1 in the CNS was revealed by the study of Bmi1 knockout mice, which showed, in addition to hematopoietic and skeletal abnormalities, neurological symptoms manifested by an ataxic gait and sporadic seizures within a month after birth [213]. In this respect, histopathological analysis of the Bmi1 −/− brain revealed an overall reduced size, in particular a reduced cellularity of the granular and molecular layer of the cerebellum [73]. Despite these changes in brain size, the overall brain architecture was not affected [73]. To gain insight into the molecular mechanism underlying this phenotype, Jacobs and colleagues [73] used primary mouse embryonic fibroblasts (MEFs) derived from Bmi1 −/− embryos and found that expression of the tumor suppressors p16Ink4a and p19Ink4d (p14Arf in human), which are encoded by the ink4a locus [214], was markedly raised in Bmi1-deficient cells (Fig. 1). As discussed above, p16Arf4a directly inhibits the cyclin D1 and cyclin-dependent kinase 4/6 (Cdk4/6) complex, thus leading to Rb activation and inhibition of G1/S progression [215]. In contrast, p19Ink4d expression leads to p53-mediated cell cycle arrest and apoptosis [216, 217]. p19Ink4d attenuates mouse double minute 2 (Mdm2)-mediated degradation of p53, which then activates the transcription of several growth suppressive and apoptotic genes such as Bax, Puma, and another CDKi, p21Waf1/Cip1 [216, 218]. Conversely, increased expression of Bmi1 leads to fibroblast immortalization and down-regulation of p16Ink4a and p19Ink4d [219]. ink4a loss dramatically reduced the neurological defects observed in Bmi1-deficient mice, indicating that ink4a was a critical in vivo target of Bmi1 [73]. The B cell-specific Moloney murine leukemia virus integration 1 also represses the expression of p21Waf1/Cip1 through direct binding to its promoter [220, 221] or via cooperation with Forkhead box protein G1 (FoxG1; [222]—see also below). In agreement with its role downstream Shh [223, 224], Bmi1 is essential for cerebellar development and contributes to medulloblastoma genesis [223, 224] (see below). In conclusion, these in vitro and in vivo studies identify the CKIs p16Ink4a, p19Ink4d, and p21Waf1/Cip1 as critical downstream targets of the PcG protein Bmi1 for regulation of development and tumorigenesis.
Stem cells persist throughout life in numerous tissues including the central nervous system (CNS) [6, 225–227] and peripheral nervous system (PNS) [228]. Interestingly, reduction of self-renewal in Bmi1-deficient neural stem cells leads to their postnatal depletion via up-regulation of the p16Ink4a and p19Ink4d CKIs [212]. Arf deficiency, but not Ink4a deficiency, partially rescued cerebellum development, thus demonstrating regional differences in the sensitivity to loss of p16Ink4a or p19Ink4d [229].
Loss of Bmi1 in hematopoietic stem cells (HSCs) and thymocytes results in a marked increase of the intracellular levels of reactive oxygen species (ROS) and subsequent engagement of the DNA damage response pathway [230]. The increased levels of ROS occur through de-repression of targets genes (other than the Ink4a/Arf locus or p21) involved in ROS generation and via impaired mitochondrial function. Similarly, it has been shown that in Bmi1-deficient neurons increased p19Ink4d/p53 levels and co-repressors at promoter regions of antioxidant genes results in a repressed chromatin state and antioxidant gene down-regulation, which in turn leads to increased ROS levels and cell death [231]. Interestingly, NPCs and HSCs seem to differ with respect of dependence on ROS levels. In this respect, HSCs appear to require low ROS to self-renew [232]. In contrast, NPCs seem to thrive in higher ROS levels [233]. It would be important to determine whether the Bmi1-mediated regulation of genes involved in redox control is as pronounced in NPCs as in HSCs. Interestingly, a context-specific role of Bmi1 has been reported in the nervous system. In particular, the dramatic increase in NPC self-renewal seen in CNS NPCs upon Bmi1 overexpression is not observed in spinal cord progenitors, which do not express FoxG1, thus suggesting that the Bmi1/FoxG1/p21 axis is critical for Bmi1 function [222]. In addition, one could hypothesize that Bmi1-mediated suppression of Ink4a/Arf locus is more important for postnatal NPCs as opposed to inhibition of p21Waf1/Cip1 and/or cooperation with FoxG1 during development.
As mentioned above, Bmi1 can contribute to CNS cancer development. In addition to medulloblastoma [224], Bmi1 is highly expressed in human glioma [234, 235], where its copy numbers are increased [236]. In particular, Bmi1 accumulates in glioma stem cells and promotes their self-renewal [237]. While no studies have reported that Bmi1 overexpression is sufficient to induce tumor initiation in the CNS, Bmi1 genetic loss impairs Shh-driven medulloblastoma initiation [238]. Furthermore, Bmi1 downregulation in primary medulloblastoma [239] and glioma cells [240, 241] limits their tumorigenic capacity in vivo. Interestingly, Bmi1 appears to regulate glioma stem cell-induced tumorigenesis also in an Ink4a/Arf-independent manner, thus suggesting that other Bmi1 targets are important for glioma progression [240]. In this respect, the role of Bmi1 in regulation of ROS levels in glioma has not been investigated. Overall, these studies suggest that Bmi1 may contribute to tumor development, but it remains to be established why Bmi1 overexpression is not sufficient to promote transformation. One possibility is that higher levels of Bmi1 may activate tumor suppressive checkpoints, which can in turn lead to differentiation and/or death instead of self-renewal. Furthermore, there is an urgent need to produce genome-wide studies analyzing Bmi1 target genes in normal and neoplastic stem cells and their association with selected epigenetic traits. This fundamental work will shed light on the role of Bmi1 in normal development/tissue homeostasis and cancer pathogenesis in the CNS.
Conclusions
Several lines of evidence indicate that the p53 and Rb tumor suppressive pathways are key regulators of neural stem cell function. Importantly, their regulatory networks (Bmi1, PML, and others) have several points of intersection, thus further strengthening the concept that a complex integration of intracellular (and extracellular) cues is what dictates the final outcome, i.e., expansion, death, or differentiation. Among the many outstanding questions, we believe it will be crucial to determine the role of the p53 and Rb pathways and their regulatory networks at the different stages of neurogenesis. This could have implications for our understanding of the transformation process in the central nervous system. For instance, inactivation of a growth suppressive checkpoint could have completely different effects on self-renewal and differentiation in a stem cell versus a committed progenitor. Another important question is to define the effect of defective tumor suppressive control on genomic stability and its consequences on cell fate decisions, particularly in view of the differentiation-inducing effect of DNA damage in non-neural stem cells. Future research aimed at addressing these questions will have to rely more and more on an integrated approach to tackle the complexity of cell fate regulation in neural stem cells as well as other stem cell types.
Acknowledgments
We would like to acknowledge members of the PS lab for critical discussion. PS is supported by the Samantha Dickson Brain Tumour Trust (SDBTT). Special thanks are due to the Brian Cross family for their generous support of the Samantha Dickson Brain Cancer Unit. SB is supported by the SDBTT. We also thank David Hunter and Wendy Tansey for a generous donation in memory of Peter Clark.
Glossary
- Neural stem cells (NSCs)
Neural stem cells are the self-renewing, multi-potent stem cells of the nervous system. Neural stem cells have the potential to give rise to offspring cells that grow and differentiate in neurons and glia cells.
- Transit amplifying cells
Transit amplifying cells are mitotic cells of the neural lineage with a fast dividing cell cycle that retain the ability to proliferate and to give rise to terminally differentiated cells but are not capable of indefinite self-renewal.
- Neural precursor cells (NPCs)
Neural precursor cells (NPCs) general term to identify any neural stem or progenitor cells.
- Note
Controversy about the exact definition remains and the concept is still evolving [1, 26–28].
- Self-renewal
The process by which a stem cell divides asymmetrically or symmetrically over an extended period of time (for example, during the life-span of an animal) to generate one or two daughter stem cells that have a developmental potential similar to the mother cell. Self-renewal is used by neural stem cells to expand during development, within niche of the adult brain, and upon brain injury.
- Quiescent cell
A cell whose cell cycle has been temporary arrested, although, strictly speaking, it might still be cycling but with a particular long cell cycle. Note! Adult neural stem cells can be quiescent.
- Post-mitotic cells
A cell that is incapable of proliferation, such as a neuron, in which cell cycle is irreversibly blocked.
References
- 1.Gage FH. Mammalian neural stem cells. Science. 2000;287(5457):1433–1438. doi: 10.1126/science.287.5457.1433. [DOI] [PubMed] [Google Scholar]
- 2.Imayoshi I, Sakamoto M, Ohtsuka T, Kageyama R. Continuous neurogenesis in the adult brain. Dev Growth Differ. 2009;51(3):379–386. doi: 10.1111/j.1440-169X.2009.01094.x. [DOI] [PubMed] [Google Scholar]
- 3.Imayoshi I, Sakamoto M, Ohtsuka T, Takao K, Miyakawa T, Yamaguchi M, Mori K, Ikeda T, Itohara S, Kageyama R. Roles of continuous neurogenesis in the structural and functional integrity of the adult forebrain. Nat Neurosci. 2008;11(10):1153–1161. doi: 10.1038/nn.2185. [DOI] [PubMed] [Google Scholar]
- 4.Kriegstein A, Alvarez-Buylla A. The glial nature of embryonic and adult neural stem cells. Annu Rev Neurosci. 2009;32:149–184. doi: 10.1146/annurev.neuro.051508.135600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ming GL, Song H. Adult neurogenesis in the mammalian central nervous system. Annu Rev Neurosci. 2005;28:223–250. doi: 10.1146/annurev.neuro.28.051804.101459. [DOI] [PubMed] [Google Scholar]
- 6.Temple S. The development of neural stem cells. Nature. 2001;414(6859):112–117. doi: 10.1038/35102174. [DOI] [PubMed] [Google Scholar]
- 7.Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6(10):777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- 8.Lange C, Calegari F. Cdks and cyclins link G1 length and differentiation of embryonic, neural and hematopoietic stem cells. Cell Cycle. 2010;9(10):1893–1900. doi: 10.4161/cc.9.10.11598. [DOI] [PubMed] [Google Scholar]
- 9.Lange C, Huttner WB, Calegari F. Cdk4/cyclinD1 overexpression in neural stem cells shortens G1, delays neurogenesis, and promotes the generation and expansion of basal progenitors. Cell Stem Cell. 2009;5(3):320–331. doi: 10.1016/j.stem.2009.05.026. [DOI] [PubMed] [Google Scholar]
- 10.Miller FD, Gauthier-Fisher A. Home at last: neural stem cell niches defined. Cell Stem Cell. 2009;4(6):507–510. doi: 10.1016/j.stem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 11.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20(5):233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Kempermann G, Kuhn HG, Gage FH. Genetic influence on neurogenesis in the dentate gyrus of adult mice. Proc Natl Acad Sci USA. 1997;94(19):10409–10414. doi: 10.1073/pnas.94.19.10409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 14.Richards LJ, Kilpatrick TJ, Bartlett PF. De novo generation of neuronal cells from the adult mouse brain. Proc Natl Acad Sci USA. 1992;89(18):8591–8595. doi: 10.1073/pnas.89.18.8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee A, Kessler JD, Read TA, Kaiser C, Corbeil D, Huttner WB, Johnson JE, Wechsler-Reya RJ. Isolation of neural stem cells from the postnatal cerebellum. Nat Neurosci. 2005;8(6):723–729. doi: 10.1038/nn1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lie DC, Dziewczapolski G, Willhoite AR, Kaspar BK, Shults CW, Gage FH. The adult substantia nigra contains progenitor cells with neurogenic potential. J Neurosci. 2002;22(15):6639–6649. doi: 10.1523/JNEUROSCI.22-15-06639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao M, Momma S, Delfani K, Carlen M, Cassidy RM, Johansson CB, Brismar H, Shupliakov O, Frisen J, Janson AM. Evidence for neurogenesis in the adult mammalian substantia nigra. Proc Natl Acad Sci USA. 2003;100(13):7925–7930. doi: 10.1073/pnas.1131955100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amato MA, Arnault E, Perron M. Retinal stem cells in vertebrates: parallels and divergences. Int J Dev Biol. 2004;48(8–9):993–1001. doi: 10.1387/ijdb.041879ma. [DOI] [PubMed] [Google Scholar]
- 19.Raymond PA, Barthel LK, Bernardos RL, Perkowski JJ. Molecular characterization of retinal stem cells and their niches in adult zebrafish. BMC Dev Biol. 2006;6:36. doi: 10.1186/1471-213X-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 21.Zhao T, Xu Y. p53 and stem cells: new developments and new concerns. Trends Cell Biol. 2010;20(3):170–175. doi: 10.1016/j.tcb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 22.Hong Y, Cervantes RB, Stambrook PJ. DNA damage response and mutagenesis in mouse embryonic stem cells. Methods Mol Biol. 2006;329:313–326. doi: 10.1385/1-59745-037-5:313. [DOI] [PubMed] [Google Scholar]
- 23.Hong Y, Cervantes RB, Tichy E, Tischfield JA, Stambrook PJ. Protecting genomic integrity in somatic cells and embryonic stem cells. Mutat Res. 2007;614(1–2):48–55. doi: 10.1016/j.mrfmmm.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 24.Krtolica A. Stem cell: balancing aging and cancer. Int J Biochem Cell Biol. 2005;37(5):935–941. doi: 10.1016/j.biocel.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 25.Cunningham JJ, Roussel MF. Cyclin-dependent kinase inhibitors in the development of the central nervous system. Cell Growth Differ. 2001;12(8):387–396. [PubMed] [Google Scholar]
- 26.Doetsch F, Caille I, Lim DA, Garcia-Verdugo JM, Alvarez-Buylla A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell. 1999;97(6):703–716. doi: 10.1016/S0092-8674(00)80783-7. [DOI] [PubMed] [Google Scholar]
- 27.Doetsch F, Petreanu L, Caille I, Garcia-Verdugo JM, Alvarez-Buylla A. EGF converts transit-amplifying neurogenic precursors in the adult brain into multipotent stem cells. Neuron. 2002;36(6):1021–1034. doi: 10.1016/S0896-6273(02)01133-9. [DOI] [PubMed] [Google Scholar]
- 28.Ming GL, Song H. Adult neurogenesis in the mammalian brain: significant answers and significant questions. Neuron. 2011;70(4):687–702. doi: 10.1016/j.neuron.2011.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alvarez-Buylla A, Lim DA. For the long run: maintaining germinal niches in the adult brain. Neuron. 2004;41(5):683–686. doi: 10.1016/S0896-6273(04)00111-4. [DOI] [PubMed] [Google Scholar]
- 30.Mirzadeh Z, Merkle FT, Soriano-Navarro M, Garcia-Verdugo JM, Alvarez-Buylla A. Neural stem cells confer unique pinwheel architecture to the ventricular surface in neurogenic regions of the adult brain. Cell Stem Cell. 2008;3(3):265–278. doi: 10.1016/j.stem.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shen Q, Wang Y, Kokovay E, Lin G, Chuang SM, Goderie SK, Roysam B, Temple S. Adult SVZ stem cells lie in a vascular niche: a quantitative analysis of niche cell–cell interactions. Cell Stem Cell. 2008;3(3):289–300. doi: 10.1016/j.stem.2008.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tavazoie M, Van der Veken L, Silva-Vargas V, Louissaint M, Colonna L, Zaidi B, Garcia-Verdugo JM, Doetsch F. A specialized vascular niche for adult neural stem cells. Cell Stem Cell. 2008;3(3):279–288. doi: 10.1016/j.stem.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doetsch F. The glial identity of neural stem cells. Nat Neurosci. 2003;6(11):1127–1134. doi: 10.1038/nn1144. [DOI] [PubMed] [Google Scholar]
- 34.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30(11):630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Wuarin J, Nurse P. Regulating S phase: CDKs, licensing and proteolysis. Cell. 1996;85(6):785–787. doi: 10.1016/S0092-8674(00)81261-1. [DOI] [PubMed] [Google Scholar]
- 36.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 37.Besson A, Dowdy SF, Roberts JM. CDK inhibitors: cell cycle regulators and beyond. Dev Cell. 2008;14(2):159–169. doi: 10.1016/j.devcel.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 38.Santamaria D, Ortega S. Cyclins and CDKS in development and cancer: lessons from genetically modified mice. Front Biosci. 2006;11:1164–1188. doi: 10.2741/1871. [DOI] [PubMed] [Google Scholar]
- 39.Bally-Cuif L, Hammerschmidt M. Induction and patterning of neuronal development, and its connection to cell cycle control. Curr Opin Neurobiol. 2003;13(1):16–25. doi: 10.1016/S0959-4388(03)00015-1. [DOI] [PubMed] [Google Scholar]
- 40.Dehay C, Kennedy H. Cell-cycle control and cortical development. Nat Rev Neurosci. 2007;8(6):438–450. doi: 10.1038/nrn2097. [DOI] [PubMed] [Google Scholar]
- 41.Durand B, Raff M. A cell-intrinsic timer that operates during oligodendrocyte development. BioEssays. 2000;22(1):64–71. doi: 10.1002/(SICI)1521-1878(200001)22:1<64::AID-BIES11>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 42.Belachew S, Aguirre AA, Wang H, Vautier F, Yuan X, Anderson S, Kirby M, Gallo V. Cyclin-dependent kinase-2 controls oligodendrocyte progenitor cell cycle progression and is downregulated in adult oligodendrocyte progenitors. J Neurosci. 2002;22(19):8553–8562. doi: 10.1523/JNEUROSCI.22-19-08553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferguson KL, Callaghan SM, O’Hare MJ, Park DS, Slack RS. The Rb-CDK4/6 signaling pathway is critical in neural precursor cell cycle regulation. J Biol Chem. 2000;275(43):33593–33600. doi: 10.1074/jbc.M004879200. [DOI] [PubMed] [Google Scholar]
- 44.Vandenbosch R, Borgs L, Beukelaers P, Foidart A, Nguyen L, Moonen G, Berthet C, Kaldis P, Gallo V, Belachew S, Malgrange B. CDK2 is dispensable for adult hippocampal neurogenesis. Cell Cycle. 2007;6(24):3065–3069. doi: 10.4161/cc.6.24.5048. [DOI] [PubMed] [Google Scholar]
- 45.Artegiani B, Lindemann D, Calegari F. Overexpression of cdk4 and cyclinD1 triggers greater expansion of neural stem cells in the adult mouse brain. J Exp Med. 2011;208(5):937–948. doi: 10.1084/jem.20102167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beukelaers P, Vandenbosch R, Caron N, Nguyen L, Belachew S, Moonen G, Kiyokawa H, Barbacid M, Santamaria D, Malgrange B. Cdk6-dependent regulation of G(1) length controls adult neurogenesis. Stem Cells. 2011;29(4):713–724. doi: 10.1002/stem.616. [DOI] [PubMed] [Google Scholar]
- 47.Lane DP, Crawford LV. T antigen is bound to a host protein in SV40-transformed cells. Nature. 1979;278(5701):261–263. doi: 10.1038/278261a0. [DOI] [PubMed] [Google Scholar]
- 48.Linzer DI, Maltzman W, Levine AJ. The SV40 A gene product is required for the production of a 54,000 MW cellular tumor antigen. Virology. 1979;98(2):308–318. doi: 10.1016/0042-6822(79)90554-3. [DOI] [PubMed] [Google Scholar]
- 49.Prives C. Signaling to p53: breaking the MDM2-p53 circuit. Cell. 1998;95(1):5–8. doi: 10.1016/S0092-8674(00)81774-2. [DOI] [PubMed] [Google Scholar]
- 50.Attardi LD, Jacks T. The role of p53 in tumour suppression: lessons from mouse models. Cell Mol Life Sci. 1999;55(1):48–63. doi: 10.1007/s000180050269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Isobe M, Emanuel BS, Givol D, Oren M, Croce CM. Localization of gene for human p53 tumour antigen to band 17p13. Nature. 1986;320(6057):84–85. doi: 10.1038/320084a0. [DOI] [PubMed] [Google Scholar]
- 52.Jacks T. Lessons from the p53 mutant mouse. J Cancer Res Clin Oncol. 1996;122(6):319–327. doi: 10.1007/BF01220798. [DOI] [PubMed] [Google Scholar]
- 53.Kern SE, Kinzler KW, Bruskin A, Jarosz D, Friedman P, Prives C, Vogelstein B. Identification of p53 as a sequence-specific DNA-binding protein. Science. 1991;252(5013):1708–1711. doi: 10.1126/science.2047879. [DOI] [PubMed] [Google Scholar]
- 54.Matlashewski G, Lamb P, Pim D, Peacock J, Crawford L, Benchimol S. Isolation and characterization of a human p53 cDNA clone: expression of the human p53 gene. EMBO J. 1984;3(13):3257–3262. doi: 10.1002/j.1460-2075.1984.tb02287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McBride OW, Merry D, Givol D. The gene for human p53 cellular tumor antigen is located on chromosome 17 short arm (17p13) Proc Natl Acad Sci USA. 1986;83(1):130–134. doi: 10.1073/pnas.83.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9(5):402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 57.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458(7242):1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Belyi VA, Ak P, Markert E, Wang H, Hu W, Puzio-Kuter A, Levine AJ. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol. 2010;2(6):a001198. doi: 10.1101/cshperspect.a001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lane D, Levine A. p53 Research: the past thirty years and the next thirty years. Cold Spring Harb Perspect Biol. 2010;2(12):a000893. doi: 10.1101/cshperspect.a000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mills AA. p53: link to the past, bridge to the future. Genes Dev. 2005;19(18):2091–2099. doi: 10.1101/gad.1362905. [DOI] [PubMed] [Google Scholar]
- 61.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9(10):691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 62.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22(2):181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Crighton D, Wilkinson S, O’Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126(1):121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 64.Miller FD, Pozniak CD, Walsh GS. Neuronal life and death: an essential role for the p53 family. Cell Death Differ. 2000;7(10):880–888. doi: 10.1038/sj.cdd.4400736. [DOI] [PubMed] [Google Scholar]
- 65.Meletis K, Wirta V, Hede SM, Nister M, Lundeberg J, Frisen J. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133(2):363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 66.van Lookeren Campagne M, Gill R. Tumor-suppressor p53 is expressed in proliferating and newly formed neurons of the embryonic and postnatal rat brain: comparison with expression of the cell cycle regulators p21Waf1/Cip1, p27Kip1, p57Kip2, p16Ink4a, cyclin G1, and the proto-oncogene Bax. J Comp Neurol. 1998;397(2):181–198. doi: 10.1002/(SICI)1096-9861(19980727)397:2<181::AID-CNE3>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 67.Poulaki V, Benekou A, Bozas E, Bolaris S, Stylianopoulou F. p53 expression and regulation by NMDA receptors in the developing rat brain. J Neurosci Res. 1999;56(4):427–440. doi: 10.1002/(SICI)1097-4547(19990515)56:4<427::AID-JNR10>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 68.Rinon A, Molchadsky A, Nathan E, Yovel G, Rotter V, Sarig R, Tzahor E. p53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/delamination processes. Development. 2011;138(9):1827–1838. doi: 10.1242/dev.053645. [DOI] [PubMed] [Google Scholar]
- 69.Armstrong JF, Kaufman MH, Harrison DJ, Clarke AR. High-frequency developmental abnormalities in p53-deficient mice. Curr Biol. 1995;5(8):931–936. doi: 10.1016/S0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 70.Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet. 1995;10(2):175–180. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 71.De Laurenzi V, Melino G. Evolution of functions within the p53/p63/p73 family. Ann NY Acad Sci. 2000;926:90–100. doi: 10.1111/j.1749-6632.2000.tb05602.x. [DOI] [PubMed] [Google Scholar]
- 72.Dotsch V, Bernassola F, Coutandin D, Candi E, Melino G. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2(9):a004887. doi: 10.1101/cshperspect.a004887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397(6715):164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 74.Levrero M, De Laurenzi V, Costanzo A, Gong J, Melino G, Wang JY. Structure, function and regulation of p63 and p73. Cell Death Differ. 1999;6(12):1146–1153. doi: 10.1038/sj.cdd.4400624. [DOI] [PubMed] [Google Scholar]
- 75.Levrero M, De Laurenzi V, Costanzo A, Gong J, Wang JY, Melino G. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J Cell Sci. 2000;113(Pt 10):1661–1670. doi: 10.1242/jcs.113.10.1661. [DOI] [PubMed] [Google Scholar]
- 76.Gil-Perotin S, Marin-Husstege M, Li J, Soriano-Navarro M, Zindy F, Roussel MF, Garcia-Verdugo JM, Casaccia-Bonnefil P. Loss of p53 induces changes in the behaviour of subventricular zone cells: implication for the genesis of glial tumors. J Neurosci. 2006;26(4):1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-P. [DOI] [PubMed] [Google Scholar]
- 78.Cheng T, Rodrigues N, Dombkowski D, Stier S, Scadden DT. Stem cell repopulation efficiency but not pool size is governed by p27(kip1) Nat Med. 2000;6(11):1235–1240. doi: 10.1038/81335. [DOI] [PubMed] [Google Scholar]
- 79.Doetsch F, Verdugo JM, Caille I, Alvarez-Buylla A, Chao MV, Casaccia-Bonnefil P. Lack of the cell-cycle inhibitor p27Kip1 results in selective increase of transit-amplifying cells for adult neurogenesis. J Neurosci. 2002;22(6):2255–2264. doi: 10.1523/JNEUROSCI.22-06-02255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kippin TE, Martens DJ, van der Kooy D. p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 2005;19(6):756–767. doi: 10.1101/gad.1272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci USA. 2006;103(9):3232–3237. doi: 10.1073/pnas.0508476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Medrano S, Burns-Cusato M, Atienza MB, Rahimi D, Scrable H. Regenerative capacity of neural precursors in the adult mammalian brain is under the control of p53. Neurobiol Aging. 2009;30(3):483–497. doi: 10.1016/j.neurobiolaging.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pechnick RN, Zonis S, Wawrowsky K, Pourmorady J, Chesnokova V. p21Cip1 restricts neuronal proliferation in the subgranular zone of the dentate gyrus of the hippocampus. Proc Natl Acad Sci USA. 2008;105(4):1358–1363. doi: 10.1073/pnas.0711030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li X, Tang X, Jablonska B, Aguirre A, Gallo V, Luskin MB. p27(KIP1) regulates neurogenesis in the rostral migratory stream and olfactory bulb of the postnatal mouse. J Neurosci. 2009;29(9):2902–2914. doi: 10.1523/JNEUROSCI.4051-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyazawa K, Himi T, Garcia V, Yamagishi H, Sato S, Ishizaki Y. A role for p27/Kip1 in the control of cerebellar granule cell precursor proliferation. J Neurosci. 2000;20(15):5756–5763. doi: 10.1523/JNEUROSCI.20-15-05756.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee MH, Nikolic M, Baptista CA, Lai E, Tsai LH, Massague J. The brain-specific activator p35 allows Cdk5 to escape inhibition by p27Kip1 in neurons. Proc Natl Acad Sci USA. 1996;93(8):3259–3263. doi: 10.1073/pnas.93.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nguyen L, Besson A, Heng JI, Schuurmans C, Teboul L, Parras C, Philpott A, Roberts JM, Guillemot F. p27kip1 independently promotes neuronal differentiation and migration in the cerebral cortex. Genes Dev. 2006;20(11):1511–1524. doi: 10.1101/gad.377106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen L, Besson A, Roberts JM, Guillemot F. Coupling cell cycle exit, neuronal differentiation and migration in cortical neurogenesis. Cell Cycle. 2006;5(20):2314–2318. doi: 10.4161/cc.5.20.3381. [DOI] [PubMed] [Google Scholar]
- 89.Gil-Perotin S, Haines JD, Kaur J, Marin-Husstege M, Spinetta MJ, Kim KH, Duran-Moreno M, Schallert T, Zindy F, Roussel MF, Garcia-Verdugo JM, Casaccia P. Roles of p53 and p27(Kip1) in the regulation of neurogenesis in the murine adult subventricular zone. Eur J Neurosci. 2011;34(7):1040–1052. doi: 10.1111/j.1460-9568.2011.07836.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW. Regulation of PTEN transcription by p53. Mol Cell. 2001;8(2):317–325. doi: 10.1016/S1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 91.Chalhoub N, Zhu G, Zhu X, Baker SJ. Cell type specificity of PI3K signaling in Pdk1- and Pten-deficient brains. Genes Dev. 2009;23(14):1619–1624. doi: 10.1101/gad.1799609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, DePinho RA. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455(7216):1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, Stommel JM, Dunn KL, Wiedemeyer R, You MJ, Brennan C, Wang YA, Ligon KL, Wong WH, Chin L, dePinho RA. Pten and p53 converge on c-Myc to control differentiation, self-renewal, and transformation of normal and neoplastic stem cells in glioblastoma. Cold Spring Harb Symp Quant Biol. 2008;73:427–437. doi: 10.1101/sqb.2008.73.047. [DOI] [PubMed] [Google Scholar]
- 94.Alcantara Llaguno S, Chen J, Kwon CH, Jackson EL, Li Y, Burns DK, Alvarez-Buylla A, Parada LF. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. doi: 10.1016/j.ccr.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y, Yang J, Zheng H, Tomasek GJ, Zhang P, McKeever PE, Lee EY, Zhu Y. Expression of mutant p53 proteins implicates a lineage relationship between neural stem cells and malignant astrocytic glioma in a murine model. Cancer Cell. 2009;15(6):514–526. doi: 10.1016/j.ccr.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146(2):209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schmale H, Bamberger C. A novel protein with strong homology to the tumor suppressor p53. Oncogene. 1997;15(11):1363–1367. doi: 10.1038/sj.onc.1201500. [DOI] [PubMed] [Google Scholar]
- 98.Yang A, Kaghad M, Wang Y, Gillett E, Fleming MD, Dotsch V, Andrews NC, Caput D, McKeon F. p63, a p53 homolog at 3q27-29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell. 1998;2(3):305–316. doi: 10.1016/S1097-2765(00)80275-0. [DOI] [PubMed] [Google Scholar]
- 99.Kaghad M, Bonnet H, Yang A, Creancier L, Biscan JC, Valent A, Minty A, Chalon P, Lelias JM, Dumont X, Ferrara P, McKeon F, Caput D. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell. 1997;90(4):809–819. doi: 10.1016/S0092-8674(00)80540-1. [DOI] [PubMed] [Google Scholar]
- 100.Jost CA, Marin MC, Kaelin WG., Jr p73 is a simian (correction of human) p53-related protein that can induce apoptosis. Nature. 1997;389(6647):191–194. doi: 10.1038/38298. [DOI] [PubMed] [Google Scholar]
- 101.Murray-Zmijewski F, Lane DP, Bourdon JC. p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress. Cell Death Differ. 2006;13(6):962–972. doi: 10.1038/sj.cdd.4401914. [DOI] [PubMed] [Google Scholar]
- 102.Talos F, Nemajerova A, Flores ER, Petrenko O, Moll UM. p73 suppresses polyploidy and aneuploidy in the absence of functional p53. Mol Cell. 2007;27(4):647–659. doi: 10.1016/j.molcel.2007.06.036. [DOI] [PubMed] [Google Scholar]
- 103.Tomasini R, Tsuchihara K, Tsuda C, Lau SK, Wilhelm M, Ruffini A, Tsao MS, Iovanna JL, Jurisicova A, Melino G, Mak TW. TAp73 regulates the spindle assembly checkpoint by modulating BubR1 activity. Proc Natl Acad Sci USA. 2009;106(3):797–802. doi: 10.1073/pnas.0812096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung CC, Khan F, Itie-Youten A, Wakeham A, Tsao MS, Iovanna JL, Squire J, Jurisica I, Kaplan D, Melino G, Jurisicova A, Mak TW. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22(19):2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fillippovich I, Sorokina N, Gatei M, Haupt Y, Hobson K, Moallem E, Spring K, Mould M, McGuckin MA, Lavin MF, Khanna KK. Transactivation-deficient p73alpha (p73Deltaexon2) inhibits apoptosis and competes with p53. Oncogene. 2001;20(4):514–522. doi: 10.1038/sj.onc.1204118. [DOI] [PubMed] [Google Scholar]
- 106.Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289(5477):304–306. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 107.Ravni A, Tissir F, Goffinet AM. DeltaNp73 transcription factors modulate cell survival and tumor development. Cell Cycle. 2010;9(8):1523–1527. doi: 10.4161/cc.9.8.11291. [DOI] [PubMed] [Google Scholar]
- 108.Tissir F, Ravni A, Achouri Y, Riethmacher D, Meyer G, Goffinet AM. DeltaNp73 regulates neuronal survival in vivo. Proc Natl Acad Sci USA. 2009;106(39):16871–16876. doi: 10.1073/pnas.0903191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yang A, McKeon F. P63 and P73: P53 mimics, menaces and more. Nat Rev Mol Cell Biol. 2000;1(3):199–207. doi: 10.1038/35043127. [DOI] [PubMed] [Google Scholar]
- 110.Irwin MS, Kaelin WG., Jr Role of the newer p53 family proteins in malignancy. Apoptosis. 2001;6(1–2):17–29. doi: 10.1023/A:1009663809458. [DOI] [PubMed] [Google Scholar]
- 111.Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416(6880):560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 112.Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7(4):363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 113.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398(6729):708–713. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 114.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, McKeon F, Caput D. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404(6773):99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 115.Agostini M, Tucci P, Killick R, Candi E, Sayan BS, di Val Rivetti, Cervo P, Nicotera P, McKeon F, Knight RA, Mak TW, Melino G. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci USA. 2011;108(52):21093–21098. doi: 10.1073/pnas.1112061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fujitani M, Cancino GI, Dugani CB, Weaver IC, Gauthier-Fisher A, Paquin A, Mak TW, Wojtowicz MJ, Miller FD, Kaplan DR. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol. 2010;20(22):2058–2065. doi: 10.1016/j.cub.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 117.Holembowski L, Schulz R, Talos F, Scheel A, Wolff S, Dobbelstein M, Moll U. While p73 is essential, p63 is completely dispensable for the development of the central nervous system. Cell Cycle. 2011;10(4):680–689. doi: 10.4161/cc.10.4.14859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wilhelm MT, Rufini A, Wetzel MK, Tsuchihara K, Inoue S, Tomasini R, Itie-Youten A, Wakeham A, Arsenian-Henriksson M, Melino G, Kaplan DR, Miller FD, Mak TW. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24(6):549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Packard A, Schnittke N, Romano RA, Sinha S, Schwob JE. DeltaNp63 regulates stem cell dynamics in the mammalian olfactory epithelium. J Neurosci. 2011;31(24):8748–8759. doi: 10.1523/JNEUROSCI.0681-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fletcher RB, Prasol MS, Estrada J, Baudhuin A, Vranizan K, Choi YG, Ngai J. p63 regulates olfactory stem cell self-renewal and differentiation. Neuron. 2011;72(5):748–759. doi: 10.1016/j.neuron.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Dugani CB, Paquin A, Fujitani M, Kaplan DR, Miller FD. p63 antagonizes p53 to promote the survival of embryonic neural precursor cells. J Neurosci. 2009;29(20):6710–6721. doi: 10.1523/JNEUROSCI.5878-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Friend SH, Bernards R, Rogelj S, Weinberg RA, Rapaport JM, Albert DM, Dryja TP. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323(6089):643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 123.Fung YK, Murphree AL, T’Ang A, Qian J, Hinrichs SH, Benedict WF. Structural evidence for the authenticity of the human retinoblastoma gene. Science. 1987;236(4809):1657–1661. doi: 10.1126/science.2885916. [DOI] [PubMed] [Google Scholar]
- 124.Lee WH, Bookstein R, Hong F, Young LJ, Shew JY, Lee EY. Human retinoblastoma susceptibility gene: cloning, identification, and sequence. Science. 1987;235(4794):1394–1399. doi: 10.1126/science.3823889. [DOI] [PubMed] [Google Scholar]
- 125.Weinberg RA. Tumor suppressor genes. Science. 1991;254(5035):1138–1146. doi: 10.1126/science.1659741. [DOI] [PubMed] [Google Scholar]
- 126.Blanquet V, Turleau C, Gross MS, Goossens M, Besmond C. Identification of germline mutations in the RB1 gene by denaturant gradient gel electrophoresis and polymerase chain reaction direct sequencing. Hum Mol Genet. 1993;2(7):975–979. doi: 10.1093/hmg/2.7.975. [DOI] [PubMed] [Google Scholar]
- 127.Classon M, Harlow E. The retinoblastoma tumour suppressor in development and cancer. Nat Rev Cancer. 2002;2(12):910–917. doi: 10.1038/nrc950. [DOI] [PubMed] [Google Scholar]
- 128.Dyer MA, Bremner R. The search for the retinoblastoma cell of origin. Nat Rev Cancer. 2005;5(2):91–101. doi: 10.1038/nrc1545. [DOI] [PubMed] [Google Scholar]
- 129.Egan C, Bayley ST, Branton PE. Binding of the Rb1 protein to E1A products is required for adenovirus transformation. Oncogene. 1989;4(3):383–388. [PubMed] [Google Scholar]
- 130.Harbour JW, Lai SL, Whang-Peng J, Gazdar AF, Minna JD, Kaye FJ. Abnormalities in structure and expression of the human retinoblastoma gene in SCLC. Science. 1988;241(4863):353–357. doi: 10.1126/science.2838909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Horowitz JM, Park SH, Bogenmann E, Cheng JC, Yandell DW, Kaye FJ, Minna JD, Dryja TP, Weinberg RA. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc Natl Acad Sci USA. 1990;87(7):2775–2779. doi: 10.1073/pnas.87.7.2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Whyte P, Buchkovich KJ, Horowitz JM, Friend SH, Raybuck M, Weinberg RA, Harlow E. Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature. 1988;334(6178):124–129. doi: 10.1038/334124a0. [DOI] [PubMed] [Google Scholar]
- 133.DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54(2):275–283. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- 134.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243(4893):934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 135.Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. Embo J. 1989;8(13):4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hu QJ, Dyson N, Harlow E. The regions of the retinoblastoma protein needed for binding to adenovirus E1A or SV40 large T antigen are common sites for mutations. Embo J. 1990;9(4):1147–1155. doi: 10.1002/j.1460-2075.1990.tb08221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2(4):E65–E67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- 138.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81(3):323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 139.Buchkovich K, Duffy LA, Harlow E. The retinoblastoma protein is phosphorylated during specific phases of the cell cycle. Cell. 1989;58(6):1097–1105. doi: 10.1016/0092-8674(89)90508-4. [DOI] [PubMed] [Google Scholar]
- 140.Cooper S, Shayman JA. Revisiting retinoblastoma protein phosphorylation during the mammalian cell cycle. Cell Mol Life Sci. 2001;58(4):580–595. doi: 10.1007/PL00000883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Luo RX, Postigo AA, Dean DC. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92(4):463–473. doi: 10.1016/S0092-8674(00)80940-X. [DOI] [PubMed] [Google Scholar]
- 142.Coschi CH, Martens AL, Ritchie K, Francis SM, Chakrabarti S, Berube NG, Dick FA. Mitotic chromosome condensation mediated by the retinoblastoma protein is tumor-suppressive. Genes Dev. 2010;24(13):1351–1363. doi: 10.1101/gad.1917610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Manning AL, Dyson NJ. RB: mitotic implications of a tumour suppressor. Nat Rev Cancer. 2012;12(3):220–226. doi: 10.1038/nrc3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Harn T, Foijer F, van Vugt M, Banerjee R, Yang F, Oostra A, Joenje H, te Riele H. Loss of Rb proteins causes genomic instability in the absence of mitogenic signaling. Genes Dev. 2010;24(13):1377–1388. doi: 10.1101/gad.580710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sage J, Straight AF. RB’s original CIN? Genes Dev. 2010;24(13):1329–1333. doi: 10.1101/gad.1948010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Clarke AR, Maandag ER, van Roon M, van der Lugt NM, van der Valk M, Hooper ML, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359(6393):328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 147.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359(6393):295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 148.Lee EY, Chang CY, Hu N, Wang YC, Lai CC, Herrup K, Lee WH, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359(6393):288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 149.Zacksenhaus E, Jiang Z, Chung D, Marth JD, Phillips RA, Gallie BL. pRb controls proliferation, differentiation, and death of skeletal muscle cells and other lineages during embryogenesis. Genes Dev. 1996;10(23):3051–3064. doi: 10.1101/gad.10.23.3051. [DOI] [PubMed] [Google Scholar]
- 150.Lee EY, Hu N, Yuan SS, Cox LA, Bradley A, Lee WH, Herrup K. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 1994;8(17):2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 151.Lipinski MM, Macleod KF, Williams BO, Mullaney TL, Crowley D, Jacks T. Cell-autonomous and non-cell-autonomous functions of the Rb tumor suppressor in developing central nervous system. Embo J. 2001;20(13):3402–3413. doi: 10.1093/emboj/20.13.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ferguson KL, Vanderluit JL, Hebert JM, McIntosh WC, Tibbo E, MacLaurin JG, Park DS, Wallace VA, Vooijs M, McConnell SK, Slack RS. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. Embo J. 2002;21(13):3337–3346. doi: 10.1093/emboj/cdf338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.MacPherson D, Sage J, Kim T, Ho D, McLaughlin ME, Jacks T. Cell type-specific effects of Rb deletion in the murine retina. Genes Dev. 2004;18(14):1681–1694. doi: 10.1101/gad.1203304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Marino S, Hoogervoorst D, Brandner S, Berns A. Rb and p107 are required for normal cerebellar development and granule cell survival but not for Purkinje cell persistence. Development. 2003;130(15):3359–3368. doi: 10.1242/dev.00553. [DOI] [PubMed] [Google Scholar]
- 155.Ferguson KL, McClellan KA, Vanderluit JL, McIntosh WC, Schuurmans C, Polleux F, Slack RS. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. Embo J. 2005;24(24):4381–4391. doi: 10.1038/sj.emboj.7600887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ewen ME, Faha B, Harlow E, Livingston DM. Interaction of p107 with cyclin A independent of complex formation with viral oncoproteins. Science. 1992;255(5040):85–87. doi: 10.1126/science.1532457. [DOI] [PubMed] [Google Scholar]
- 157.Ewen ME, Xing YG, Lawrence JB, Livingston DM. Molecular cloning, chromosomal mapping, and expression of the cDNA for p107, a retinoblastoma gene product-related protein. Cell. 1991;66(6):1155–1164. doi: 10.1016/0092-8674(91)90038-Z. [DOI] [PubMed] [Google Scholar]
- 158.Hannon GJ, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 1993;7(12A):2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 159.Li Y, Graham C, Lacy S, Duncan AM, Whyte P. The adenovirus E1A-associated 130-kD protein is encoded by a member of the retinoblastoma gene family and physically interacts with cyclins A and E. Genes Dev. 1993;7(12A):2366–2377. doi: 10.1101/gad.7.12a.2366. [DOI] [PubMed] [Google Scholar]
- 160.Mayol X, Grana X, Baldi A, Sang N, Hu Q, Giordano A. Cloning of a new member of the retinoblastoma gene family (pRb2) which binds to the E1A transforming domain. Oncogene. 1993;8(9):2561–2566. [PubMed] [Google Scholar]
- 161.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24(17):2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 162.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3(1):11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- 163.Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14(6):223–229. doi: 10.1016/S0168-9525(98)01470-X. [DOI] [PubMed] [Google Scholar]
- 164.Cobrinik D, Lee MH, Hannon G, Mulligan G, Bronson RT, Dyson N, Harlow E, Beach D, Weinberg RA, Jacks T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10(13):1633–1644. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 165.Lee MH, Williams BO, Mulligan G, Mukai S, Bronson RT, Dyson N, Harlow E, Jacks T. Targeted disruption of p107: functional overlap between p107 and Rb. Genes Dev. 1996;10(13):1621–1632. doi: 10.1101/gad.10.13.1621. [DOI] [PubMed] [Google Scholar]
- 166.Callaghan DA, Dong L, Callaghan SM, Hou YX, Dagnino L, Slack RS. Neural precursor cells differentiating in the absence of Rb exhibit delayed terminal mitosis and deregulated E2F 1 and 3 activity. Dev Biol. 1999;207(2):257–270. doi: 10.1006/dbio.1998.9162. [DOI] [PubMed] [Google Scholar]
- 167.Donovan SL, Schweers B, Martins R, Johnson D, Dyer MA. Compensation by tumor suppressor genes during retinal development in mice and humans. BMC Biol. 2006;4:14. doi: 10.1186/1741-7007-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ajioka I, Martins RA, Bayazitov IT, Donovan S, Johnson DA, Frase S, Cicero SA, Boyd K, Zakharenko SS, Dyer MA. Differentiated horizontal interneurons clonally expand to form metastatic retinoblastoma in mice. Cell. 2007;131(2):378–390. doi: 10.1016/j.cell.2007.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Xu XL, Fang Y, Lee TC, Forrest D, Gregory-Evans C, Almeida D, Liu A, Jhanwar SC, Abramson DH, Cobrinik D. Retinoblastoma has properties of a cone precursor tumor and depends upon cone-specific MDM2 signaling. Cell. 2009;137(6):1018–1031. doi: 10.1016/j.cell.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jacques TS, Swales A, Brzozowski MJ, Henriquez NV, Linehan JM, Mirzadeh Z, O’Malley C, Naumann H, Alvarez-Buylla A, Brandner S. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. Embo J. 2010;29(1):222–235. doi: 10.1038/emboj.2009.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14(8):994–1004. [PMC free article] [PubMed] [Google Scholar]
- 172.Xiao A, Wu H, Pandolfi PP, Louis DN, Van Dyke T. Astrocyte inactivation of the pRb pathway predisposes mice to malignant astrocytoma development that is accelerated by PTEN mutation. Cancer Cell. 2002;1(2):157–168. doi: 10.1016/S1535-6108(02)00029-6. [DOI] [PubMed] [Google Scholar]
- 173.Vanderluit JL, Ferguson KL, Nikoletopoulou V, Parker M, Ruzhynsky V, Alexson T, McNamara SM, Park DS, Rudnicki M, Slack RS. p107 regulates neural precursor cells in the mammalian brain. J Cell Biol. 2004;166(6):853–863. doi: 10.1083/jcb.200403156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vanderluit JL, Wylie CA, McClellan KA, Ghanem N, Fortin A, Callaghan S, MacLaurin JG, Park DS, Slack RS. The Retinoblastoma family member p107 regulates the rate of progenitor commitment to a neuronal fate. J Cell Biol. 2007;178(1):129–139. doi: 10.1083/jcb.200703176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.McClellan KA, Vanderluit JL, Julian LM, Andrusiak MG, Dugal-Tessier D, Park DS, Slack RS. The p107/E2F pathway regulates fibroblast growth factor 2 responsiveness in neural precursor cells. Mol Cell Biol. 2009;29(17):4701–4713. doi: 10.1128/MCB.01767-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Borrow J, Goddard AD, Sheer D, Solomon E. Molecular analysis of acute promyelocytic leukemia breakpoint cluster region on chromosome 17. Science. 1990;249(4976):1577–1580. doi: 10.1126/science.2218500. [DOI] [PubMed] [Google Scholar]
- 177.de The H, Chomienne C, Lanotte M, Degos L, Dejean A. The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor alpha gene to a novel transcribed locus. Nature. 1990;347(6293):558–561. doi: 10.1038/347558a0. [DOI] [PubMed] [Google Scholar]
- 178.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66(4):675–684. doi: 10.1016/0092-8674(91)90113-D. [DOI] [PubMed] [Google Scholar]
- 179.Longo L, Pandolfi PP, Biondi A, Rambaldi A, Mencarelli A, Lo Coco F, Diverio D, Pegoraro L, Avanzi G, Tabilio A, et al. Rearrangements and aberrant expression of the retinoic acid receptor alpha gene in acute promyelocytic leukemias. J Exp Med. 1990;172(6):1571–1575. doi: 10.1084/jem.172.6.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Melnick A, Licht JD. Deconstructing a disease: RARalpha, its fusion partners, and their roles in the pathogenesis of acute promyelocytic leukemia. Blood. 1999;93(10):3167–3215. [PubMed] [Google Scholar]
- 181.Pandolfi PP, Grignani F, Alcalay M, Mencarelli A, Biondi A, LoCoco F, Pelicci PG. Structure and origin of the acute promyelocytic leukemia myl/RAR alpha cDNA and characterization of its retinoid-binding and transactivation properties. Oncogene. 1991;6(7):1285–1292. [PubMed] [Google Scholar]
- 182.Kakizuka A, Miller WH, Jr, Umesono K, Warrell RP, Jr, Frankel SR, Murty VV, Dmitrovsky E, Evans RM. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66(4):663–674. doi: 10.1016/0092-8674(91)90112-C. [DOI] [PubMed] [Google Scholar]
- 183.Pandolfi PP, Alcalay M, Fagioli M, Zangrilli D, Mencarelli A, Diverio D, Biondi A, Lo Coco F, Rambaldi A, Grignani F, et al. Genomic variability and alternative splicing generate multiple PML/RAR alpha transcripts that encode aberrant PML proteins and PML/RAR alpha isoforms in acute promyelocytic leukaemia. Embo J. 1992;11(4):1397–1407. doi: 10.1002/j.1460-2075.1992.tb05185.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Salomoni P, Pandolfi PP. The role of PML in tumor suppression. Cell. 2002;108(2):165–170. doi: 10.1016/S0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 185.Wang ZG, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi PP. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20(3):266–272. doi: 10.1038/3030. [DOI] [PubMed] [Google Scholar]
- 186.Ishov AM, Sotnikov AG, Negorev D, Vladimirova OV, Neff N, Kamitani T, Yeh ET, Strauss JF, 3rd, Maul GG. PML is critical for ND10 formation and recruits the PML-interacting protein DAXX to this nuclear structure when modified by SUMO-1. J Cell Biol. 1999;147(2):221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2(5):a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Shen TH, Lin HK, Scaglioni PP, Yung TM, Pandolfi PP. The mechanisms of PML-nuclear body formation. Mol Cell. 2006;24(3):331–339. doi: 10.1016/j.molcel.2006.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Borden KL, Culjkovic B. Perspectives in PML: a unifying framework for PML function. Front Biosci. 2009;14:497–509. doi: 10.2741/3258. [DOI] [PubMed] [Google Scholar]
- 190.Bourdeau V, Baudry D, Ferbeyre G. PML links aberrant cytokine signaling and oncogenic stress to cellular senescence. Front Biosci. 2009;14:475–485. doi: 10.2741/3256. [DOI] [PubMed] [Google Scholar]
- 191.Hofmann TG, Will H. Body language: the function of PML nuclear bodies in apoptosis regulation. Cell Death Differ. 2003;10(12):1290–1299. doi: 10.1038/sj.cdd.4401313. [DOI] [PubMed] [Google Scholar]
- 192.Pearson M, Pelicci PG. PML interaction with p53 and its role in apoptosis and replicative senescence. Oncogene. 2001;20(49):7250–7256. doi: 10.1038/sj.onc.1204856. [DOI] [PubMed] [Google Scholar]
- 193.Salomoni P, Ferguson BJ, Wyllie AH, Rich T. New insights into the role of PML in tumour suppression. Cell Res. 2008;18(6):622–640. doi: 10.1038/cr.2008.58. [DOI] [PubMed] [Google Scholar]
- 194.Takahashi Y, Lallemand-Breitenbach V, Zhu J, de The H. PML nuclear bodies and apoptosis. Oncogene. 2004;23(16):2819–2824. doi: 10.1038/sj.onc.1207533. [DOI] [PubMed] [Google Scholar]
- 195.Regad T, Bellodi C, Nicotera P, Salomoni P. The tumor suppressor PML regulates cell fate in the developing neocortex. Nat Neurosci. 2009;12(2):132–140. doi: 10.1038/nn.2251. [DOI] [PubMed] [Google Scholar]
- 196.Michod D, Bartesaghi S, Khelifi A, Bellodi C, Berliocchi L, Nicotera P, Salomoni P. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron. 2012;74(1):122–135. doi: 10.1016/j.neuron.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24(12):1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. DAXX is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci USA. 2010;107(32):14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Berube NG, Mangelsdorf M, Jagla M, Vanderluit J, Garrick D, Gibbons RJ, Higgs DR, Slack RS, Picketts DJ. The chromatin-remodeling protein ATRX is critical for neuronal survival during corticogenesis. J Clin Invest. 2005;115(2):258–267. doi: 10.1172/JCI22329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453(7198):1072–1078. doi: 10.1038/nature07016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Milyavsky M, Gan OI, Trottier M, Komosa M, Tabach O, Notta F, Lechman E, Hermans KG, Eppert K, Konovalova Z, Ornatsky O, Domany E, Meyn MS, Dick JE. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7(2):186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 202.Li W, Ferguson BJ, Khaled WT, Tevendale M, Stingl J, Poli V, Rich T, Salomoni P, Watson CJ. PML depletion disrupts normal mammary gland development and skews the composition of the mammary luminal cell progenitor pool. Proc Natl Acad Sci USA. 2009;106(12):4725–4730. doi: 10.1073/pnas.0807640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Salomoni P, Betts-Henderson J. The role of PML in the nervous system. Mol Neurobiol. 2011;43(2):114–123. doi: 10.1007/s12035-010-8156-y. [DOI] [PubMed] [Google Scholar]
- 204.Gurrieri C, Capodieci P, Bernardi R, Scaglioni PP, Nafa K, Rush LJ, Verbel DA, Cordon-Cardo C, Pandolfi PP. Loss of the tumor suppressor PML in human cancers of multiple histologic origins. J Natl Cancer Inst. 2004;96(4):269–279. doi: 10.1093/jnci/djh043. [DOI] [PubMed] [Google Scholar]
- 205.Huse JT, Holland EC. Targeting brain cancer: advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 206.Jeanne M, Lallemand-Breitenbach V, Ferhi O, Koken M, Le Bras M, Duffort S, Peres L, Berthier C, Soilihi H, Raught B, de The H. PML/RARA oxidation and arsenic binding initiate the antileukemia response of As2O3. Cancer Cell. 2010;18(1):88–98. doi: 10.1016/j.ccr.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 207.Zhang XW, Yan XJ, Zhou ZR, Yang FF, Wu ZY, Sun HB, Liang WX, Song AX, Lallemand-Breitenbach V, Jeanne M, Zhang QY, Yang HY, Huang QH, Zhou GB, Tong JH, Zhang Y, Wu JH, Hu HY, de The H, Chen SJ, Chen Z. Arsenic trioxide controls the fate of the PML-RARalpha oncoprotein by directly binding PML. Science. 2010;328(5975):240–243. doi: 10.1126/science.1183424. [DOI] [PubMed] [Google Scholar]
- 208.Alkema MJ, Wiegant J, Raap AK, Berns A, van Lohuizen M. Characterization and chromosomal localization of the human proto-oncogene BMI-1. Hum Mol Genet. 1993;2(10):1597–1603. doi: 10.1093/hmg/2.10.1597. [DOI] [PubMed] [Google Scholar]
- 209.Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. RYBP-PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3 K27me3. Cell. 2012;148(4):664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat Rev Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 211.Brookes E, de Santiago I, Hebenstreit D, Morris KJ, Carroll T, Xie SQ, Stock JK, Heidemann M, Eick D, Nozaki N, Kimura H, Ragoussis J, Teichmann SA, Pombo A. Polycomb associates genome-wide with a specific RNA polymerase II variant, and regulates metabolic genes in ESCs. Cell Stem Cell. 2012;10(2):157–170. doi: 10.1016/j.stem.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Bmi-1 dependence distinguishes neural stem cell self-renewal from progenitor proliferation. Nature. 2003;425(6961):962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev. 1994;8(7):757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 214.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 215.Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9(1):22–30. doi: 10.1016/S0959-437X(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 216.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6(9):663–673. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 217.Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113(2):175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Marine JC. p53 stabilization: the importance of nuclear import. Cell Death Differ. 2010;17(2):191–192. doi: 10.1038/cdd.2009.183. [DOI] [PubMed] [Google Scholar]
- 219.Vonlanthen S, Heighway J, Altermatt HJ, Gugger M, Kappeler A, Borner MM, van Lohuizen M, Betticher DC. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84(10):1372–1376. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1(1):87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 221.Subkhankulova T, Zhang X, Leung C, Marino S. Bmi1 directly represses p21Waf1/Cip1 in Shh-induced proliferation of cerebellar granule cell progenitors. Mol Cell Neurosci. 2010;45(2):151–162. doi: 10.1016/j.mcn.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 222.Fasano CA, Phoenix TN, Kokovay E, Lowry N, Elkabetz Y, Dimos JT, Lemischka IR, Studer L, Temple S. Bmi-1 cooperates with Foxg1 to maintain neural stem cell self-renewal in the forebrain. Genes Dev. 2009;23(5):561–574. doi: 10.1101/gad.1743709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, Van Lohuizen M, Marino S. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature. 2004;428(6980):337–341. doi: 10.1038/nature02385. [DOI] [PubMed] [Google Scholar]
- 224.Marino S. Medulloblastoma: developmental mechanisms out of control. Trends Mol Med. 2005;11(1):17–22. doi: 10.1016/j.molmed.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 225.Alvarez-Buylla A, Kohwi M, Nguyen TM, Merkle FT. The heterogeneity of adult neural stem cells and the emerging complexity of their niche. Cold Spring Harb Symp Quant Biol. 2008;73:357–365. doi: 10.1101/sqb.2008.73.019. [DOI] [PubMed] [Google Scholar]
- 226.Alvarez-Buylla A, Temple S. Stem cells in the developing and adult nervous system. J Neurobiol. 1998;36(2):105–110. doi: 10.1002/(SICI)1097-4695(199808)36:2<105::AID-NEU1>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 227.Morshead CM, Craig CG, van der Kooy D. In vivo clonal analyses reveal the properties of endogenous neural stem cell proliferation in the adult mammalian forebrain. Development. 1998;125(12):2251–2261. doi: 10.1242/dev.125.12.2251. [DOI] [PubMed] [Google Scholar]
- 228.Kruger GM, Mosher JT, Bixby S, Joseph N, Iwashita T, Morrison SJ. Neural crest stem cells persist in the adult gut but undergo changes in self-renewal, neuronal subtype potential, and factor responsiveness. Neuron. 2002;35(4):657–669. doi: 10.1016/S0896-6273(02)00827-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19(12):1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, Ahn BH, Kumar NG, Rovira II, Xu XL, van Lohuizen M, Motoyama N, Deng CX, Finkel T. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459(7245):387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chatoo W, Abdouh M, David J, Champagne MP, Ferreira J, Rodier F, Bernier G. The Polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci. 2009;29(2):529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, Ikeda Y, Mak TW, Suda T. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431(7011):997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- 233.Le Belle JE, Orozco NM, Paucar AA, Saxe JP, Mottahedeh J, Pyle AD, Wu H, Kornblum HI. Proliferative neural stem cells have high endogenous ROS levels that regulate self-renewal and neurogenesis in a PI3K/Akt-dependant manner. Cell Stem Cell. 2011;8(1):59–71. doi: 10.1016/j.stem.2010.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hayry V, Tynninen O, Haapasalo HK, Wolfer J, Paulus W, Hasselblatt M, Sariola H, Paetau A, Sarna S, Niemela M, Wartiovaara K, Nupponen NN. Stem cell protein BMI-1 is an independent marker for poor prognosis in oligodendroglial tumours. Neuropathol Appl Neurobiol. 2008;34(5):555–563. doi: 10.1111/j.1365-2990.2008.00949.x. [DOI] [PubMed] [Google Scholar]
- 235.Tirabosco R, De Maglio G, Skrap M, Falconieri G, Pizzolitto S. Expression of the Polycomb-group protein BMI1 and correlation with p16 in astrocytomas an immunohistochemical study on 80 cases. Pathol Res Pract. 2008;204(9):625–631. doi: 10.1016/j.prp.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 236.Hayry V, Tanner M, Blom T, Tynninen O, Roselli A, Ollikainen M, Sariola H, Wartiovaara K, Nupponen NN. Copy number alterations of the Polycomb gene BMI1 in gliomas. Acta Neuropathol. 2008;116(1):97–102. doi: 10.1007/s00401-008-0376-0. [DOI] [PubMed] [Google Scholar]
- 237.Abdouh M, Facchino S, Chatoo W, Balasingam V, Ferreira J, Bernier G. BMI1 sustains human glioblastoma multiforme stem cell renewal. J Neurosci. 2009;29(28):8884–8896. doi: 10.1523/JNEUROSCI.0968-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Michael LE, Westerman BA, Ermilov AN, Wang A, Ferris J, Liu J, Blom M, Ellison DW, van Lohuizen M, Dlugosz AA. Bmi1 is required for Hedgehog pathway-driven medulloblastoma expansion. Neoplasia. 2008;10(12):1343–1349. doi: 10.1593/neo.81078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Wiederschain D, Chen L, Johnson B, Bettano K, Jackson D, Taraszka J, Wang YK, Jones MD, Morrissey M, Deeds J, Mosher R, Fordjour P, Lengauer C, Benson JD. Contribution of Polycomb homologues Bmi-1 and Mel-18 to medulloblastoma pathogenesis. Mol Cell Biol. 2007;27(13):4968–4979. doi: 10.1128/MCB.02244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bruggeman SW, Hulsman D, Tanger E, Buckle T, Blom M, Zevenhoven J, van Tellingen O, van Lohuizen M. Bmi1 controls tumor development in an Ink4a/Arf-independent manner in a mouse model for glioma. Cancer Cell. 2007;12(4):328–341. doi: 10.1016/j.ccr.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 241.Dirks P. Bmi1 and cell of origin determinants of brain tumor phenotype. Cancer Cell. 2007;12(4):295–297. doi: 10.1016/j.ccr.2007.10.003. [DOI] [PubMed] [Google Scholar]