Abstract
The identification of neurological symptoms caused by vitamin A deficiency pointed to a critical, early developmental role of vitamin A and its metabolite, retinoic acid (RA). The ability of RA to induce post-mitotic, neural phenotypes in various stem cells, in vitro, served as early evidence that RA is involved in the switch between proliferation and differentiation. In vivo studies have expanded this “opposing signal” model, and the number of primary neurons an embryo develops is now known to depend critically on the levels and spatial distribution of RA. The proneural and neurogenic transcription factors that control the exit of neural progenitors from the cell cycle and allow primary neurons to develop are partly elucidated, but the downstream effectors of RA receptor (RAR) signaling (many of which are putative cell cycle regulators) remain largely unidentified. The molecular mechanisms underlying RA-induced primary neurogenesis in anamniote embryos are starting to be revealed; however, these data have been not been extended to amniote embryos. There is growing evidence that bona fide RARs are found in some mollusks and other invertebrates, but little is known about their necessity or functions in neurogenesis. One normal function of RA is to regulate the cell cycle to halt proliferation, and loss of RA signaling is associated with dedifferentiation and the development of cancer. Identifying the genes and pathways that mediate cell cycle exit downstream of RA will be critical for our understanding of how to target tumor differentiation. Overall, elucidating the molecular details of RAR-regulated neurogenesis will be decisive for developing and understanding neural proliferation–differentiation switches throughout development.
Keywords: Neurogenesis, Retinoic acid receptor, Proliferation-differentiation switch
Introduction
The role of retinoic acid (RA) in neurogenesis has been known indirectly for as long as haliver (halibut) and cod liver oils were used to remedy neurological and ophthalmic disorders. Vitamin A, from which RA is derived, was discovered to be fat-soluble in 1891, labeled as vitamin A in 1920, chemically described in 1931, and synthesized in 1946 (reviewed in [1]). Prior to receiving its name, vitamin A had been known for years to be essential to life—the ancient Egyptians used extracts of (vitamin A rich) beef liver to treat night blindness more than 3,500 years ago and the Greeks prescribed the eating of liver to cure night blindness since at least 300 BC [2]. The link between vitamin A and vision was firmly established in the 1930s when George Wald identified vitamin A in the retinas and melanin-containing choroid layers of eye [3]. “Pigs born without eyeballs” was the title of one researcher’s report about a pregnant gilt (young female pig) that received a vitamin A-deficient (VAD) diet and gave birth to 11 piglets without eyes [4]. The etymology of retin-ol, -al, -oic, -oid was derived from the retina, where these molecules were first discovered [5].
The first VAD animals possessed many other neuropathies in addition to blindness. Early studies found spinal cord abnormalities in swine fed a diet consisting nearly entirely of wheat [6]. Although the authors noted that the abnormalities disappeared when the diet was supplemented with “Fat soluble A” (later identified as vitamin A), they attributed neural degeneration to wheat toxicity rather than a dietary deficiency [6]. A more definitive study was later conducted in pigs fed an otherwise nutritious diet that was solely deficient in vitamin A [7]. Nerve degeneration was found in the spinal cord, optic, femoral, and sciatic nerves, as well as the lateral geniculate body [7]. A severe neuromuscular phenotype, complete with hind limb paralysis, was observed in rats deprived of vitamin A prenatally, whereas only partial paralysis was seen in rats deprived of vitamin A postnatally [8]. Indeed, replicating paralysis in other animal systems proved difficult since vitamin A is readily stored in the fetus [8]. Therefore, historical examples of neurological symptoms from VAD pointed to a critical, early developmental role of vitamin A, and by association, RA.
In the early 1980s, it was discovered that RA induced multipotent P19 embryonal carcinoma cells to differentiate into neuronal and glial tissue, in vitro [9]. F9 embryonal carcinoma cells differentiated into neurons with the addition of RA and dibutyryl cAMP [10]. The ability of RA to induce neural phenotypes on various stem cells in vitro is summarized in [11]. Shortly after the cloning of the first retinoic acid receptors (RARs) [12, 13], it was observed that the P19-derived cell line, RAC65, was incapable of neuronal differentiation due to a 70-amino acid truncation in the RARα ligand-binding domain [14, 15]. Since DNA-binding was still intact, but activation by ligand was inhibited, the receptor acted as a dominant transcriptional repressor, aka, a dominant negative (DN) receptor. Further investigation showed that the RAC65 line was also unable to up-regulate p27Kip1, a negative cell cycle regulator, and key effector of RA-mediated inhibition of cell cancer growth [16, 17]. These studies demonstrate that RARα is an essential factor in neuronal differentiation, in vitro, and links RA signaling to the cell cycle and a proliferation–differentiation switch.
Studies from several groups showed that both the number of primary neurons an embryo develops and the time that those neurons appear depends critically on the level of RA signaling [18–21]. Whole mount in situ hybridization studies revealed that both RARα and RARγ are localized in the neural plate and neural tube in neurula-stage embryos; hence, the receptors are expressed at the correct time and place to regulate primary neurogenesis [22]. DN-RARα injected embryos lacked primary neurons and were paralyzed and unresponsive to touch; microinjection of the constitutively active VP16-RARα, or xRXRβ together with xRARα2 created ectopic neurons [19, 21]. Reduced RA signaling in VAD quail led to a paucity of neurons in the spinal cord, concurrent with loss of proneural genes, such as Neurogenin1/2 [23, 24]. Increasing RA levels in chick spinal cord explants resulted in increased expression of NeuroD, a basic helix–loop–helix protein that promotes neural differentiation [23]. Treatment of Xenopus embryos with RA or the RAR-selective agonist TTNPB led to ectopic primary neuron formation in the neural plate; antagonist treatment, or loss of either RARα or RARγ led to the loss of primary neurons and subsequent paralysis of embryos [22].
In this review, we discuss neurogenesis primarily in terms of how RA facilitates differentiation of neurons at the expense of proliferation. We explore the evolution and mechanism of RARs in neurogenesis and identify key molecular cell cycle regulators of neuronal development as well as potential downstream effectors of RAR signaling. Finally, we propose how cancer differentiation therapy can benefit from knowledge of RA and proliferation–differentiation switches.
Neurogenesis as a model to study proliferation–differentiation switches
Developing systems exhibit a dynamic balance between cell proliferation and differentiation. The molecular mechanisms regulating this equilibrium remain an important, yet poorly understood question in development. The opposing signal model is a significant conceptual advance in developmental patterning [23]. In its most general terms, the model holds that mutually inhibitory interactions between factors promoting proliferation, and those promoting differentiation, regulate developmental patterning processes. For example, fibroblast growth factors (FGFs) promote proliferation while largely inhibiting differentiation; RA is a differentiation-inducing molecule that inhibits cell proliferation. Examples of RA mediating proliferation–differentiation switches occur frequently in developmental biology. RA-regulated processes include somitogenesis and axial elongation, cardiogenesis, neurogenesis (e.g., primary neurogenesis, hindbrain patterning, eye morphogenesis), limb development, and visceral organ formation [23, 25–28]. Regulating the switch between proliferation and differentiation is fundamental to vertebrate neurogenesis.
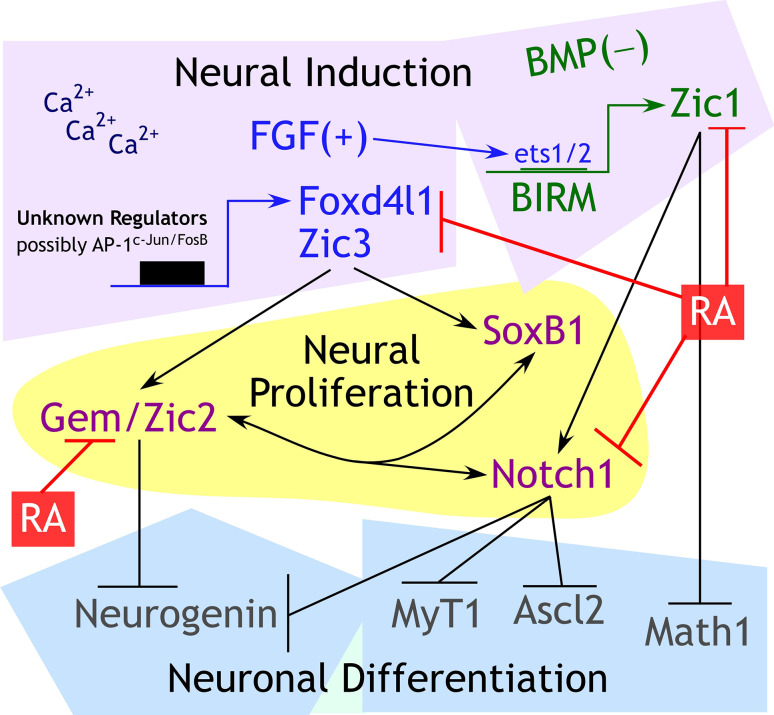
Vertebrate neural induction requires inhibition of bone morphogenetic protein (BMP) signaling [29]. FGFs and other growth factors play important, but incompletely understood roles in facilitating neural induction or A-P patterning (reviewed in [30]). Neural induction is associated with the expression of a suite of pro-proliferation transcription factors, downstream of FGF signaling (Foxd4l1 and Zic3) or BMP inhibition (Zic1) [29, 31–36] (Fig. 1). Zic1 and Zic3 stabilize the neural fate immediately after neural induction, promoting proliferation of neural progenitors, up-regulating Notch signaling, and inhibiting differentiation [37–41]. Zic3 is a direct target of the pluripotency factors Oct4, Nanog, and Sox2 [42–44], and its expression is diminished after differentiation with RA [43]. Calcium signaling through L-type calcium channels is also a major player in neural induction and is required for the expression of Zic3 and Geminin [45–49]. Geminin, a gene that postpones lineage commitment of cells [50], is associated with proliferating neural progenitors [51] and interacts with Brahma-related gene 1 (Brg1) to inhibit neuronal differentiation [52]. Zic2 is downstream of Foxd4l1 and is expressed in an alternating pattern with Neurogenin; Zic2 inhibits Neurogenin, and therefore, neurons do not differentiate where Zic2 is expressed [53]. FoxD4L1, Geminin, Zic1 and Zic3 promote Notch, Sox2 and Sox3 expression [39, 50, 54–56]. The concerted action of this group of genes promotes proliferation and maintenance of immature neural precursors (Fig. 1).
Fig. 1.
Important proliferation factors that mediate the early transcriptional response of BMP inhibition and FGF signaling downstream of neural induction. After neural tissue is induced through active FGF and Ca2+ signaling and BMP inhibition, Zic1, Zic3 and Foxd4l1 are up-regulated [29, 34–36, 48, 49]. Foxd4l1 and Zic3 are downstream targets of FGF signaling, possibly mediated by AP-1 [32–34, 263, 264], whereas Zic1 is an immediate early gene of BMP inhibition and is driven by a BMP inhibitor-responsive promoter module (BIRM) [34, 35]. Geminin (Gem) and Zic2 are regulated by Foxd4l1 and promote Notch signaling and inhibition of proneural gene Neurogenin [31, 52–55]. Zic1 also promotes Notch signaling and directly represses proneural gene Math1 [40, 41]. Cross-regulation between Geminin, Zic, SoxB1 (Sox2 and Sox3), Sox11, and Notch maintains proliferation in the neuroectoderm [39, 50, 54–56]. Potential inhibitory interactions between Sox11 and Zic genes are explored in Moody 2013, but not displayed here. Collectively, proneural genes Neurogenin and Math1 are repressed by these proliferative signals, and primary neurogenesis is inhibited as a result. RA inhibits neural proliferation quite early in this process by downregulating the expression of Geminin, Zic1/2/3, and Notch [22]
Through an as yet unclear mechanism, neural progenitors exit from the cell cycle and differentiate into primary neurons under the control of the proneural and neurogenic transcription factors such as Neurogenin, Math1, Ascl2, MyT1 (Fig. 1) (reviewed in [57, 55]). Primary neurons are defined as four sets of neurons (sensory, interneurons, motor, and trigeminal) visible in the open neural plate stage. Primary neurons differentiate from the deep neuroectoderm layer of the embryo, whereas the superficial neuroectodermal layer maintains an immature, proliferative state [58]. We and others showed that RA is required for primary neurogenesis [19, 21, 59]. RA inhibits the expression of Zic, Geminin, Notch, and Foxd4l1 while promoting expression of proneural and neurogenic genes [18, 22] (Fig. 1).
Primary neurogenesis in anamniotes versus amniotes
Primary neurogenesis is an important model for understanding proliferation–differentiation switches in all species throughout development; it also describes how the adult brain can regenerate new neurons, and how cancer cells arise. However, primary neurogenesis, per se, only occurs in anamniote embryos, which develop neurons as early as the neural plate stage, later enabling the larvae to swim and feed precociously [60, 61]. Since amniote (reptilian, avian and mammalian) embryos are protected from the external environment and develop completely before hatching, primary neurons are not required for survival and were probably lost during evolution [62]. The earliest born neurons of the cortex are often referred to as secondary neurons in anamniotes, although this process corresponds to primary neurogenesis in amniotes. Terminology aside, it is quite likely that the first cortical neurons in amniotes will be initiated using a mechanism similar to primary neurogenesis in anamniotes.
Before any developmental mechanism can be elucidated, these incipient neurons for amniotes, secondary neurons for anamniotes, must be defined, yet this has remained elusive. Early-born axon tracts, nerve fibers that establish a scaffold for which other axons can follow, are predicted to serve as “pioneer” neurons in the developing brain (reviewed in [63]). Unlike primary neurons, these pioneer neurons exist not for basic survival, but rather, because the embryo is sufficiently small that guidance cues (e.g., chemoattractants) are close enough for axon trajectories to be established [64]. Recent labeling studies in aborted human embryos identified so-called “predecessor neurons” in the preplate, which are the earliest identified neurons to date [65]. Like primary neurons, these “predecessor neurons” are created prior to neural tube closure and are likely to be transient in nature [65]. Whether RA is involved in the differentiation of predecessor neurons from proliferating neuroepithelial cells is unknown. However, considering that evolution often conserves developmental mechanisms, the possibility is very likely and remains an interesting open question.
Although there is no clear picture on how RA might foster differentiation of the earliest neurons in amniotes, and secondary neurons in anamniotes, RA does play other important, evolutionarily conserved roles in CNS development across most chordates, including three major patterning processes: (1) posteriorization of neuroectoderm; (2) D-V patterning of the spinal cord; (3) A-P patterning of the hindbrain. Excellent reviews have been written about these well-known patterning events [11, 25–27, 66, 67]. Briefly, RA together with Wnts and FGF signaling posteriorizes neuroectoderm, which would otherwise be anterior in character by default [19, 68]. Similarly, RA signaling is an important component of the neural posteriorizing pathway in the hindbrain—RA determines the identity and delineates borders of posterior rhombomeric segments (reviewed in [69]). During neurulation, RA secreted from paraxial mesoderm functions in the specification of nerve cell types in the spinal cord, in particular, motor neurons ([70], reviewed in [67]).
Most examples in amniotic systems concern the function of RA in patterning neural tissue—there is a paucity of information about how RA promotes neuronal differentiation. We consider three areas where RA plays a role in differentiation: in photoreceptors, hippocampus, and cortical neurons. RA can cause dissociated, neonatal rat retinas in culture to differentiate into photoreceptor cells expressing rhodopsin and/or recoverin [71]. Human, mouse, and monkey embryonic stem cells can also be differentiated into rod photoreceptors, albeit more laboriously, due to the requirement of an intermediate step of Notch inactivation, followed by a cocktail containing RA, Shh, FGFs, and taurine [72]. This suggests that RA is more important in the final steps of rod photoreceptor differentiation than in the early process of differentiating ES cells into retinal progenitors. In contrast, both mature (NeuN+) and immature (dcx+) neurons of the hippocampal dentate gyrus are reduced in retinoid-deficient mice, indicating that RA affects very early steps of the neuronal differentiation pathway in the hippocampus [73]. Neural stem cells in the proliferative ventricular zone of the cortex also require RA to differentiate into intermediate progenitor cells of the sub-ventricular zone and post-mitotic neurons of the cortical plate [74]. Meninges are thought to be the source of RA due to high levels of RA-synthesis enzymes, ALDH1A2 and RDH10 [74, 75]. Foxc1 mutant mice that fail to form meninges normally exhibit increased proliferation, and are deficient in mature, Ctip2+ neurons [74]. However, when forebrain explants from Foxc1 hypomorphs were co-cultured with wild-type meninges (from which RA diffuses), cell cycle exit was restored [74]. How RA regulates differentiation in other aspects of cortical development, and how these processes can be related to the gene networks observed in vertebrate primary neurogenesis remain to be explored.
What was the first RAR-regulated nervous system?
A neural plate or neural tube need not be present for neurogenesis to occur. For example, the primitive acoelomorph flatworm, Symsagittifera roscoffensis (aka the mint sauce worm), which is considered to represent the earliest extant bilaterian organism, lacks a nerve cord, but possesses neurite bundles that span the A-P axis of the body [76]. Prior to the emergence of more complex bilaterians, nerve “cords”, “nets”, and “rings” already existed in cnidarians [77, 78] and ctenophores [79, 80]. Secretory apparatus resembling synaptic vesicles required for neurotransmitter communication can be found in single-celled choanoflagellates [81]. Therefore, it is clear that neural tissue can adopt a variety of forms, yet retain the function of communication between one part of the organism and others. Where then do retinoids fit into this process?
Data from vertebrate embryology support an essential role for retinoid signaling in the development of primary neurons [19, 21, 22, 24]. However, it is equally clear that nervous systems of considerable complexity can be found in organisms for which RARs have not been identified (e.g., Drosophila melanogaster). The larvacean urochordate, Oikopleura dioica can form a functional nervous system that expresses homologs of marker genes for vertebrate forebrain, hindbrain and spinal cord (but not midbrain) in their CNS [82], yet it lacks important components of RAR signaling (RARs, CYP26), while retaining RXR and Adh3 [83]. Intriguingly, other urochordates, hemichordates, and cephaolochordates express RARs and RXRs, as do echinoderms [84, 85]. It is clear that the RA signaling machinery has been lost in Oikopleura [85], although, it is not known to what extent their nervous system function is altered compared with other urochordates.
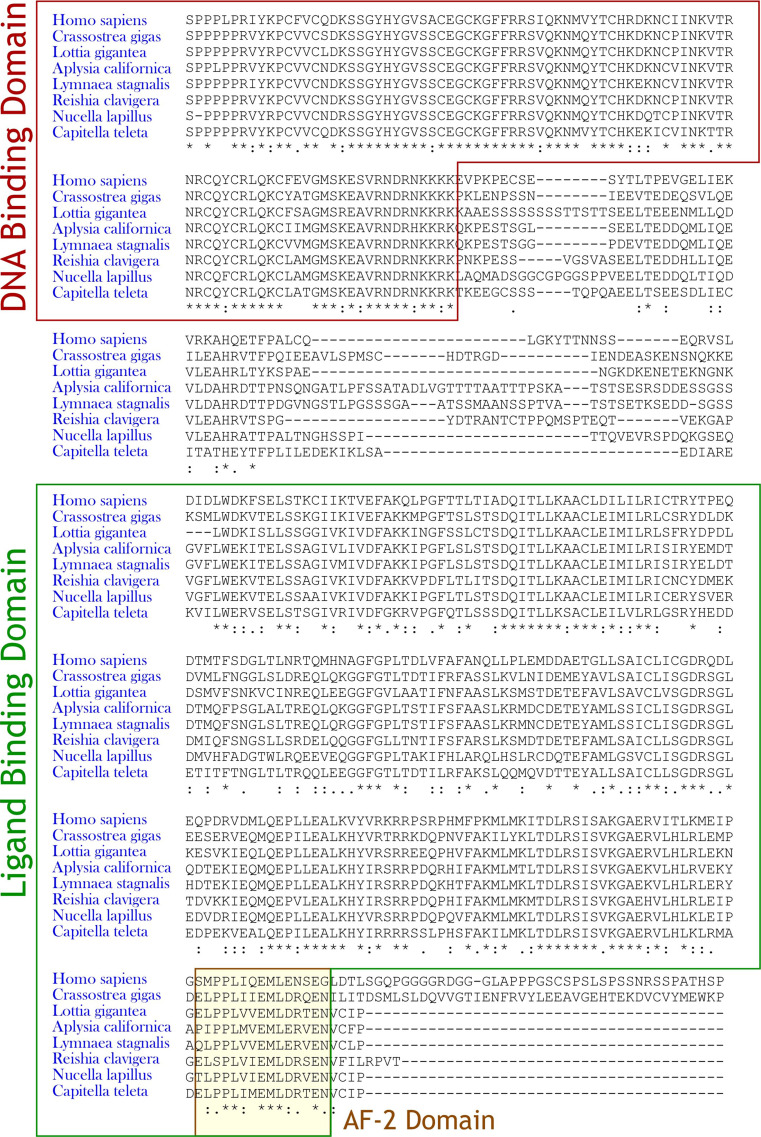
The recent explosion in genome sequences from taxonomically diverse organisms reveals the presence of RARs and RXRs in a variety of invertebrates beyond the deuterostome superphylum. Components of the RAR signaling machinery have been reported from cnidarians, mollusks and nematodes [86] and 9cRA and other RXR activators perturb development in mollusks [87–90]. Uncovering RAR homologs in lower organisms is relatively straightforward in silico, and numerous examples have been identified [86]. A quick BLAST search for this review using the RARα ligand-binding domain, identified putative RARs in Pacific oyster, Crassostrea gigas and the California sea hare, Aplysia californica, among others (Fig. 2). Bioactive retinoids and RARs exist in some Lophotrochozoan species, e.g., the owl limpet, Lottia gigantean, the bristleworm, Capitella telata, and the giant pond snail, Lymnaea stagnalis [86, 91]. Lymnaea were shown to contain atRA and 9cRA in the hundreds of nanomolar range, and treatment with either chemical induced neurite outgrowth and growth cone turning in cell culture [92]. Putative RAR [93, 94] and RXR [87, 95] orthologs were recently cloned in the rock shell, Reishia clavigera and dog whelk, Nucella lapillus. Both species of gastropods are susceptible to imposex induced by RXR activators (rexinoids) [87, 89].
Fig. 2.
MAFFT alignment of RARα2 in Homo sapiens versus Lophotrochozoan species. Alignment begins with the conserved DNA-binding domain of RARα2 (no conservation is observed in the N-terminal region)
It is important to note that the presence of an apparent RAR or RXR in a particular species does not conclusively show that the receptor will bind to RA and regulate gene expression in an RA-dependent manner. For example, Reishia clavigera and Nucella lapillus RARs and RXRs heterodimerize as do their vertebrate counterparts, but tcRAR and nlRAR appear not to be responsive to RA [87, 93]. Some investigators have speculated that many invertebrate nuclear receptors could have functions different from than regulating hormonal responses [96], although, others take the opposite position, that hormone-responsiveness was the ancestral state for nuclear receptors [97, 98]. Taken together, the evidence suggests that RA signaling is an ancient process that has been repeatedly lost in a variety of lineages [99]. While important for neural development and differentiation in vertebrates and most chordates, there is a considerable knowledge gap regarding requirements for RA signaling during invertebrate neurogenesis. It is tempting to speculate that advanced cephalopod mollusks with complex nervous systems, such as the octopus, have retained RA signaling in neurogenesis.
Neuronal differentiation and cell cycle genes
Proliferation within neural tissue is a direct consequence of self-renewal via symmetric (giving rise to two daughter stem/progenitor cells) or asymmetric (giving rise to one stem/progenitor cell and one lineage-restricted cell) cell division (reviewed in [100]). There are two schools of thought with respect to how cells make the proliferation–differentiation decision during the cell cycle. In the first, differentiation of neural stem cells is characterized by asymmetrical cell divisions leading initially to fate-restricted progenitors, and finally to terminally differentiated daughter cells that are incapable of dividing (reviewed in [101]). Critically, the proliferation/differentiation decision does not rely on cell cycle arrest, but rather a terminally differentiated cell is simply born in G0 and never makes a decision to proliferate or differentiate (reviewed in [101]). An alternative view is that cell cycle arrest turns a proliferative cell into a post-mitotic cell that differentiates into its final form. A checkpoint towards the end of G1 phase serves as a major restriction point where cells can continue to divide or enter the quiescent G0 phase [102, 103]. Signals favoring neuronal differentiation (e.g., RA) increase the expression of CDK inhibitors which promote cell cycle exit of G1 phase cells [104, 105]. While these two views appear to be mechanistically distinct, they are really quite similar because the genes promoting cell cycle exit and those responsible for stabilizing the G0 phase are identical. Thus, irrespective of whether the terminally differentiated cell was naturally born in G0 or came to G0 from G1, the molecular factors that got them to G0 are the same.
Chromatin remodeling processes control chromosome assembly and segregation, regulating DNA accessibility throughout the cell cycle by condensing or decondensing DNA, thereby manipulating the inactivation or activation of the replication machinery. Chromatin remodelers are therefore, key determinants in proliferation–differentiation decisions. Brahma-related gene 1 (Brg1) is the catalytic subunit that provides ATPase activity to the chromatin remodeling complex SWI/SNF for nucleosome disruption [106]. Brg1 is required for neuronal differentiation as demonstrated by loss-of-function studies facilitated by morpholino (MO) injection in Xenopus embryos [107]. Loss of Brg1 function leads to an expansion of the neural progenitor population and decreased expression of the neurogenic basic helix–loop–helix (bHLH) genes Neurogenin and NeuroD as well as the neural differentiation marker N-Tubulin [107]. Brg1 activity is antagonized by Geminin which suppresses neurogenesis and inhibits Neurogenin and NeuroD transcriptional activity [52]. Eyes absent homolog 1 (Eya1) and Six homeobox1 (Six1) also interact with Brg1, recruiting SWI/SNF to mediate the transcription of Neurog1 and Neurod1 in the otocyst and cochlea [108]. The role of Brg1 in mammalian systems is more complex. Conditional deletion of murine Brg1 led to reduced mitotic index in cultured neural progenitors [109]. However, another study found that deletion of Brg1 did not alter cell cycle length or cause an increase in proliferation [110]. Rather, Brg1 controls neuronal fate decisions: neural stem cells derived from Brg1-knockout mice differentiate towards the ependymal (glial) lineage, at the expense of the neuronal lineage [110].
Cyclin-dependent kinase inhibitors (CIKs), such as proteins belonging to the Ink4 and Cip/Kip families, inhibit different cyclin-CDK complexes at different time points, and govern cell cycle progression/withdrawal [111–115]. Cip/Kip proteins mediate the assembly of CyclinD-CDK4/6 in early G1 phase [111]. However, in the presence of Ink4, the CyclinD-CDK4/6 complex is disassembled and Cyclin D is targeted for degradation, thus freeing Cip/Kip proteins to bind and inhibit CyclinE-CDK2 [111]. The inhibition of CDK2 by Cip/Kip and CDK4/6 by Ink4 prevents the cell from progressing through the G1-S transition, causing G1 arrest [116, 117]. CKIs play important roles in neurogenesis, influencing cell fate decisions by controlling the timing of onset of neural determination gene expression [118]. In the developing mouse neocortex, ectopic expression of p27 Kip1 was shown to prolong G1 phase [119], a hallmark of cells proceeding to neuronal differentiation [101]. Loss of p27 Kip1 causes increased proliferation in the adult dentate gyrus due to a delay in cell cycle exit of immature neurons. p27 xic1, a Xenopus Cip/Kip protein, is required for primary neurogenesis [120–122] and functions by CDK inhibition which prevents phosphorylation and promotes stabilization of the Neurogenin protein [120, 123]. Recently, p27 xic1, was shown to be phosphorylated by atypical protein kinase C (aPKC), which prevents CDK inhibition, shortens the G1 phase of neural progenitors, and encourages their proliferation [124]. The p21-activated serine/threonine protein kinase Pak3 also promotes cell cycle withdrawal and causes premature primary neurogenesis in Xenopus [125].
Another aspect of cell cycle control in neurogenesis concerns calcium (Ca2+) influx. Barth and colleagues first identified a role for Ca2+ in neural induction by demonstrating that dorsal ectodermal explants could be differentiated into neural cells in the presence of Ca2+ and LiCl [126]. Later, it was demonstrated in Xenopus laevis and Pleurodeles waltl that explants exposed to dissociation medium (Mg2+ and Ca2+-free) also differentiated into neural cells [127, 128]. The mere act of dissociation was enough to release Ca2+ into the cells [129]. Neuralization of explants could also be induced by Concanavalin A [46, 130, 131], which promotes Ca2+ influx/uptake [132, 133], and this was inhibited by Ca2+ channel antagonists [46]. However, during normal embryonic development cells are neither dissociated into individual cells nor is Concanavalin A (a legume glycoprotein) an endogenous factor. Hemmati-Brivanlou and Melton proposed the default neural model wherein inhibition of BMP signaling, per se, is necessary and sufficient for neural induction [134, 135]. Subsequent experiments showed that neural induction also requires FGF signaling [136] and that FGF4 activates Ca2+ channels required for neural gene expression [137]. BMP-antagonists such as Noggin trigger intracellular Ca2+ release [46], and high Ca2+ levels inhibit BMP signaling and simultaneously activate the FGF/Erk signaling pathway [138]. Critically, the presence of Ca2+ signaling is required for cells to become neural tissue; in the absence of Ca2+, cells adopt an ectodermal fate [139].
Although Ca2+ plays a role in neuralizing tissue, it is unclear whether Ca2+ promotes proliferation of neuroectoderm, fosters differentiation, or both [140]. The role of Ca2+ in cell cycle is clear. Ca2+ is required in all growth and division stages of the cell cycle, encouraging cyclin D1/CDK4 accumulation in early G1, CDK2 activation in G1/S, and CDC2 activity in G2/M (reviewed in [141]). Proliferating fibroblast cells exposed to low Ca2+ media arrest in G1 and do not synthesize DNA [142]. Proliferation can be restored in these cells by the addition of Ca2+ [142]. The proliferative ventricular zone of the neocortex is reliant on ATP-dependent intracellular Ca2+ waves [143]. When Ca2+ waves were inhibited with an ATP receptor antagonist, cells failed to enter S phase, as indicated by reduced BrdU staining ([143], reviewed in [144]). Loss of L-type Ca2+ channels, which are differentially expressed in ectodermal tissues during early development [145], causes the down-regulation of Zic3 and Geminin [129], two genes that are associated with the immature, proliferative phase of neurogenesis [57]. Much needs to be learned about other regulators of Ca2+ signaling, and which signaling pathway components that control proliferation/differentiation switches are sensitive to Ca2+ influx. For example, it has been two decades since Ca2+ release during neural induction was shown to increase protein kinase C and cyclic AMP activity, two pathways that are relatively disconnected from BMP and FGF signaling [146–149]. Curiously, this result has not been followed up despite its interest to the field [150].
Members of the Id (Inhibitor of DNA-binding/differentiation) family are helix–loop–helix proteins that inhibit the ability of bHLH proteins to homo- or heterodimerize and bind to DNA [151, 152]; they foster proliferation, in most (but not all) biological systems. Id1−/−Id3−/− mouse embryos exhibit premature neurogenesis characterized by ectopic and early onset of expression of neurogenic bHLH proteins, such as MATH1/2/3 and NeuroD1 [153]. Id3 promotes proliferation of neural crest precursors in Xenopus [154]. Loss of Id4 compromises the proliferative capacity of ventricular zone stem cells in the mammalian cortex [155]. Id proteins are intrinsically linked to the cell cycle in neurogenesis. Id2 inhibits bHLH factor E47 [156], which prevents heterodimerization of E47 with NeuroD1 [157]. An important downstream target of E47 is the CIK p57Kip2; hence, Id2 effectively down-regulates p57 Kip2 via E47 inhibition, thus promoting proliferation in neuroblastoma cells [156]. Activation of the anaphase promoting complex with Cdh1 coactivator (APC Cdh1) by differentiation signals (such as RA [158]) causes Id2 to be targeted for degradation, thus promoting neuronal differentiation [159]. When Cdh1 is ablated in mice, increased proliferation in neurospheres and stabilization of substrates such as Geminin is observed [160]. Id1 also antagonizes a different CIK, p16Ink4a [161], which is found specifically in adult nervous tissue [162]. Taken together, these data indicate that neurogenic transcription factors interact specifically with the cell cycle machinery to modulate proliferation during neurogenesis.
ETS proteins are nuclear targets for extracellular signaling pathways [163] and are modified by mitogen-activated protein kinases (MAPK), integrins, or Ca+2/calmodulin-dependent protein kinases, often downstream of growth factor pathways [163, 164]. Members of the Ets family of transcriptional activators, such as Ets1, Ets2, Elk1 and repressors, such as Ets2 Repressor Factor (ERF) and Etv3, are important cell cycle regulators; their function is tightly regulated by MAPK-mediated phosphorylation. The phosphorylation state of ERF varies at different points in the cell cycle and this affects ERF subcellular localization. ERF shuttles between the nucleus, where it functions as a repressor, and the cytoplasm where it is inactive [165]. Loss of ERF results in the complete depletion of primary neurons in Xenopus embryos [22]. Nuclear localization of ERF occurs in cells arrested in G0 or G1 [165]. Growth factor or serum stimulation of proliferation causes ERK to phosphorylate ERF, which leads to its export from the nucleus [165]. Phosphate mutant ERF proteins cannot be exported and cause cell cycle arrest in the G0/G1 phase [165] and increases the number of Xenopus primary neurons [22]. Thus, ERF is critical for negatively regulating transcription factors required for cell cycle re-entry from G0 or quite possibly, elongation of G1 (unpublished data from [166]), which is associated with neuronal differentiation [101]. The direct targets of ERF repression in neurogenesis are currently unknown. However, since Myc has been identified as an ERF target in fibroblasts [167], it is likely that ERF would also repress n-Myc, a gene that is functionally interchangeable with c-Myc [168]. n-Myc promotes expansion of neural progenitor populations, and would be a good candidate to be repressed by ERF to foster differentiation [169].
Many more cell cycle control genes that regulate the differentiation of neural progenitors have yet to be identified. Furthermore, which cell cycle genes are downstream mediators of known differentiation cues remain to be deciphered. Regulators of cell cycle progression during G0 and G1 phase and the G0/G1 transition are likely to be key determinants of the proliferation-differentiation decision. However, it is still controversial whether cells make that decision from G1, or if cells are simply born in G0 due to asymmetrical factors they inherited, or the niche in which they are residing. Various events during the cell cycle such as chromatin remodeling, the opposing effect of cyclin-CDK and CIKs, and the phosphorylation and nuclear shuttling of Ets and Ets repressors form a network that controls when and where neural progenitors commit to differentiate.
Downstream effectors of retinoic acid signaling related to neurogenesis
Throughout neurogenesis, RA is readily available due to the presence of retinaldehyde dehydrogenase 2 (ALDH1A2), which synthesizes RA in the paraxial mesoderm of the developing embryo. RA then diffuses to the neural plate and spinal cord to promote the differentiation of neural progenitors [23]. Due to the availability of RA, and known action of the RARs, most of the direct gene targets of RAR during neurogenesis are expected to be transcriptionally up-regulated; although, the possible recruitment of ligand-dependent transcriptional co-repressors cannot be completely excluded [170]. Very few direct targets of RA that are definitively involved in the neural proliferation–differentiation switch have been identified. The known direct RAR target, HoxA1, [171, 172], is required for the differentiation of embryonic stem (ES) cells into neurons [173]. HoxA1-null ES cells are refractory to treatment with RA as measured by reduced expression of post-mitotic neuron markers (e.g., β-tubulin III, Nestin); RA sensitivity can be restored by rescue with HoxA1 cDNA [173]. Btg2 is also a putative direct target of RAR [174] and is induced by RA in neurula-stage embryos [175, 176] and in various cell lines [174, 177]. Btg2 decreases arginine methylation and lysine acetylation of histone H4 at RAR target genes, thus increasing the transcriptional activity of RAR [177]. Btg2 is expressed in differentiating neuroblasts [178] and promotes neuronal differentiation in PC12 cells [179]. Loss of Btg2 increases proliferation in the neural plate of Xenopus embryos [180], likely via repression of Cyclin D1 transcription [181].
Evidence from a variety of cell culture systems shows that RA directly and indirectly regulates the expression of many other genes, in addition to Btg2, that facilitate cell cycle exit and differentiation [182–185]. Considering that RAR activation promotes differentiation at the expense of proliferation, the most likely downstream effectors of RAR are inhibitors of the cell cycle. We previously showed that RA-induced ETS repressors are key components of the proliferation–differentiation switch during primary neurogenesis, in vivo [22]. ERF and ETV3 inhibit proliferative signals by displacing activating ETS proteins from promoters of cell cycle control genes while recruiting co-repressor complexes to facilitate cell cycle arrest [186, 187]. ETV3 was identified as an anti-proliferative factor induced during macrophage differentiation [186, 188] and neuronal differentiation [22]. ERF mediates the switch between proliferation and differentiation in macrophages, fibroblasts, extraembryonic ectoderm, and neuroectoderm [22, 167, 187, 189]. Importantly, both Erf and Etv3 are upregulated by RAR agonists, down-regulated by RAR antagonists, and knockdown of ERF or ETV3 results in paralysis, loss of primary neurons and increased proliferation of undifferentiated neural progenitors. Thus, these Ets-repressors are key effectors that inhibit neural progenitor identity and promote differentiation.
Multiple genomics-based studies have elucidated some genes that respond to RA under a variety of differentiation conditions [175, 176, 190–196]. A subset of the genes identified in these analyses will be candidate neurogenic genes regulated by RAR. The pro-neural gene Ascl1 (Mash1), a bHLH transcription factor and activator of Notch signaling, is an interesting downstream effector because Ascl1 regulates both proliferation and differentiation in a temporally and spatially restricted manner [197]. Ascl1 promotes positive cell cycle regulators in the proliferating ventricular zone of the cortex, but later fosters differentiation in the post-mitotic cortical plate [197]. It is unknown whether there are inherent temporal and spatial differences in RAR signaling in the developing cortex, but RAR is known to create graded, spatially restricted expression of Ascl1 in other systems. Low Ascl1 expression is observed in the presence of RA in ventral spinal cord progenitors; high Ascl1 is observed in the absence of RA in hindbrain serotonergic progenitors [198]. Numb, another gene in the Notch pathway, is also a potentially intriguing downstream effector of RA. Numb homologs were recently characterized in Xenopus—knockdown of Numb-like causes the complete loss of primary neurons, expansion of neural progenitor markers, and increased proliferation [199]. Numb can promote proliferation or differentiation, depending on which isoform is expressed [200]. A sharp change in Numb isoform expression was observed in P19 embryonal carcinoma cells when neuronal differentiation was stimulated by RA [201]. The molecular mechanism of Numb alternative splicing remains an open question, although RA-induced differentiation alters splicing machinery in P19 cells [202] and in SH-SY5Y neuroblastoma cells [203].
Other potential downstream effectors of RA include ATP7A, Elongin A, Reelin, and Prdm12. RARβ2 induces the expression of ATP7A, a Golgi-associated protein that removes copper from cells [204]. Knock down of RARβ2 inhibits expression of ATP7A, reducing copper efflux. Copper levels are apparently critical for the response of neuroblastoma cells to RA because copper supplementation induced proliferation, and copper chelation promoted differentiation [204]. Elongin A is an elongation factor that is essential for neuronal differentiation, in vivo [205]. Elongin A −/− embryos have widespread CNS defects, and ES cells derived from these embryos fail to differentiate into neurons in response to RA treatment [205]. The hypothesis is that Elongin A improves the processivity of RNA Polymerase II on genes that are upregulated by RA [205]. Increased RNAPII occupancy was observed on Neurogenin1/2 and HoxA7 genes in response to RA in Elongin A +/+ embryonic stem cells; however, this was not observed in Elongin A −/− cells [205]. Reelin is an extracellular matrix glycoprotein that regulates the number of newborn neurons during development [206]. De novo neurogenesis in regions of the adult brain was decreased in Reelin-mutant mice [206]. RA increases occupancy of Sp1 and Pax3 promoters, and concomitant demethylation at the Reelin promoter in NT2 cells [207, 208]. In summary, some of the important players downstream of RA have been identified, but many of the detailed molecular interactions required for the proliferation of neural progenitors and their differentiation into neurons remain obscure.
Differentiation therapy for cancer
RA has been known to inhibit growth of many tumor-cell lines derived from cancers of different origin (e.g., neuroblastomas, adenocarcinomas, lymphomas, sarcomas, melanomas) for at least 40 years [209]. Neuroblastoma cell lines were commonly used to demonstrate the differentiation and anti-tumorigenic potential of RA. LA–N-1/2/5, CHP-134, KA [210, 211], SH-SY5Y [212], Neuro-2a [213], SK-N-BE2 [214], IMR-32 [215], SMS-KCNR [216], and D283 [217] can all be differentiated by retinoids into cells expressing neuronal markers and exhibiting neurite morphology. Although many cell lines can be differentiated in response to RA, the clinical response to RA treatment is variable. Neuroblastomas represent 11 % of all pediatric cancers [218]. The potential for neuroblastomas to be differentiated is so important that pathological classifications have been created to assess the degree of tumor differentiation—the higher the differentiation state, the better the prognosis [219].
The molecular mechanisms underlying RA-stimulated differentiation in neuroblastomas have not been resolved, nor is the ability of these tumors to become RA resistant completely understood. Some evidence indicates that RARs regulate the expression of microRNAs (miRNAs) that support neurite outgrowth and decrease cellular motility (e.g., invasion and metastasis) and proliferation in neuroblastoma cells (reviewed in [220]). miRNA profiles can predict survival of patients with neuroblastoma, therefore, making targeting miRNAs with antagomirs (oligonucleotides that block miRNA activity) a promising therapy [221]. In addition to altering miRNA expression, RA can induce genome-wide changes in DNA methylation by increasing expression of DNA methyltransferases and concomitant hypomethylation of promoters during neuroblastoma differentiation [222]. These changes in miRNA and DNA methylation alter the epigenetic landscape and ultimately affect the expression of oncogenes and tumor-suppressor genes. For example, microarray data demonstrate that expression of tumor-suppressors such as Erf and Etv3 (see section above) are down-regulated in human medulloblastoma (a type of neuroblastoma) [223–225]. Whether this results from increased methylation of these genes, or if treatment with RA would up-regulate Erf and Etv3 to accelerate differentiation in human neuroblastoma tissue is an intriguing, open question.
Another possible mechanism to explain the success (or lack of success) of tumor differentiation involves RAR coregulators. PRAME is a human tumor antigen that is overexpressed in a variety of cancers and is a prognostic indicator for poor survival in neuroblastomas [226]. PRAME functions as a dominant repressor of RAR signaling that renders cancer cells refractory to RA treatment [227]. Furthermore, a synthetic lethal screen recently found that stimulating differentiation of neuroblastoma cells is dependent on the transcription factor, ZNF423, a ligand-independent, coactivator of RAR signaling [228, 229]. HDAC inhibitors increase RA sensitivity by promoting dissociation of repressive complexes from RAR, thus, accelerating the differentiation process [230, 231]. Taken together, these studies indicate that the presence or absence of RAR-modulators, such as ZNF423 and PRAME, in tumor cells is critically important for sensitivity of cells to RA differentiation therapy and disease outcome [227, 228].
The identification of neural stem cells (NSCs) [232–234] and CNS stem cells [235, 236] has expanded the possible applications of RA in differentiating tumors of neural origin, particularly in aggressive brain tumors such as glioma, meningioma, and neuroma. It is currently unknown whether most brain tumors originate from mutated NSCs within the perivascular niche [237, 238] or if normal cells acquire mutations that cause their dedifferentiation into immature, carcinogenic neural progenitors [239]. Tumorigenesis could take a hierarchical or linear pathway from cancer stem cells to malignant tumor cells or could result from normal tissue losing differentiation markers (e.g., β-Tubulin) and gaining proliferation markers (e.g., Sox2, Nestin) (reviewed in [240]). Within the last few years, dedifferentiation as a mode of action in tumorigenesis has been re-evaluated and re-popularized in a variety of cancers including intestinal [241, 242], respiratory [243], breast [244], and brain [239, 245]. Molecular evidence from Drosophila revealed that dedifferentiation is associated with the loss of a neural-specific zinc-finger protein Lola-N that normally functions to repress cell cycle genes like cdc25 in post-mitotic neurons [245]. Loss of lola results in brain tumors, and lola mutant neurons express neuroblast genes and proliferate in regions of the brain where proliferation usually does not occur [245]. Similar mechanisms might be at work in human tumors.
Considering this renewed interest in dedifferentiation, one might expect RA to play an important role in the treatment of brain tumors. RA was once a prospective treatment for malignant glioma [246–249], but has not proved to be an effective treatment, mostly due to side effects and resistance. There is no doubt that RA is successful in cell culture models of gliomas. In glioblastoma progenitors [250], RA quickly induces cell cycle arrest, inhibiting growth and decreasing clonogenic capacity [251]. RA down-regulates CD133, Msi-1, Nestin and Sox-2 while increasing differentiation markers in these cells [251]. However, retinoid signaling is more complex in human gliomas, in vivo, and resistance and side effects are common. One possible hypothesis for resistance is that RA can be channeled to a pro-proliferative, oncogenic pathway depending on the relative abundance of the RA-transporting proteins CRABP2 and FABP5 (reviewed in [252]). In high-grade, undifferentiated, metastasized glioma, CRABP2 is down-regulated and FABP5 is up-regulated [253, 254], which may divert RA towards the PPARβ/δ pathway (promoting cell survival) and away from RARs (which promote differentiation) [255, 256]. This hypothesis is intriguing, but much more research is needed to test it.
A new differentiation therapy for gliomas using IDH1 (isocitrate dehydrogenase 1) inhibitors [257, 258] has provided another link between retinoid signaling and differentiation. IDH1 mutant tumors produce 2-hydroxyglutarate which is associated with genome-wide hypermethylation of a select group of cancer genes which are reproducible and recognizable as a “glioma methylome” [259, 260]. Intriguingly, IDH1 inhibitor therapy absolutely requires retinoid signaling. All IDH1-mutant tumors have retinol binding protein 1 (RBP1) promoter hypermethylation, and decreased levels of RBP1 [261]. Decreased RBP1 ultimately implies that RA bioavailability is reduced, and thus tumors cannot differentiate. RBP1 hypermethylation serves as a unique biomarker of glioma, and might correlate with improved sensitivity to RA differentiation therapy [261]. This result has immediately produced a pre-clinical trial whereby RA has been repurposed in the treatment of IDH-mutant tumors [262].
Summary and future directions
As described above, it is now well-established that the timing of primary neuron appearance and the number of primary neurons produced is regulated by the levels of RA signaling during embryonic development [18–21]. RA acts in opposition to growth factor signaling to halt the proliferation of neural progenitors and stimulate neuronal differentiation. The effects of RA on primary neurogenesis are at least partly mediated by induction of Ets repressors that act at mid- to late gastrula stages to induce cell cycle exit of neuronal progenitors and their differentiation into primary neurons [22]. Although much is known about neuronal differentiation, we still know relatively little about the molecular mechanisms through which RA and its receptors regulate neuronal differentiation and many questions remain to be answered. Which receptors are the primary players in neurogenesis? Knockdown or antagonism of either RARα or RARγ blocks primary neurogenesis, suggesting that both may be required for primary neurogenesis [22]. Do these receptors act independently, or is one required for expression or maintenance of the other? While it appears that cell cycle genes are the most likely targets for RAR, it is unclear which of these are critical and whether RARs act indirectly as has been suggested [22], or if RARs recruit ligand-dependent transcriptional repressors to directly repress expression of cell-cycle genes required for proliferation. Some candidates for downstream effectors of RA signaling can be identified from the literature (see above), but only whole genome approaches are likely to fully elucidate the RA-regulated gene network functional in neurogenesis. The rapid increase in the quality of the available databases from Xenopus (laevis and tropicalis) will facilitate the study of these important questions, in vivo.
While a role for RA signaling in primary neurogenesis in anamniote embryos has been demonstrated convincingly, much less is known about its potential roles in neuronal differentiation in amniote embryos. Is RA required for the development of “predecessor neurons” that may correspond to primary neurons, or is this process independent of RA? Apparently bona fide RARs have been identified from mollusks and other invertebrates—what is their function in neurogenesis? Can a role for RA in neuronal differentiation be demonstrated in the invertebrates that have apparent RARs, or is the situation more like that in Drosophila where neurogenesis does not require RA? Considering that Oikopleura has some components of RA signaling (RXRs, Adh3 but not RARs or CYP26), is it possible that RXR signaling plays an important role in neurogenesis when RARs are absent? Or does RXR play a more fundamental role in neurogenesis of both vertebrates and invertebrates that has, so far, remained unknown?
Although RA was first identified as a differentiation agent that inhibited the growth of numerous tumor-derived cell lines in the 1970s and 1980s, the early promise of RA differentiation therapy was not realized. Recent studies have rekindled interest in RA differentiation therapy for cancers, particularly in aggressive brain tumors such as gliomas. A more complete understanding of how RA regulates cell-cycle exit may provide therapeutic targets for future generations of tumor-selective retinoids. The identification of novel components in RA-regulated signaling pathways in neuronal differentiation may also provide important cancer diagnostic and prognostic markers.
Acknowledgments
This study was supported by grants from the National Science Foundation (IOS-0719576, IOS-1147236) to B.B.
References
- 1.Semba RD. On the ‘discovery’ of vitamin A. Ann Nutr Metab. 2012;61(3):192–198. doi: 10.1159/000343124. [DOI] [PubMed] [Google Scholar]
- 2.Wolf G. A historical note on the mode of administration of vitamin A for the cure of night blindness. Am J Clin Nutr. 1978;31(2):290–292. doi: 10.1093/ajcn/31.2.290. [DOI] [PubMed] [Google Scholar]
- 3.Wald G. Vitamin A in the retina. Nature. 1933;132:316–317. [Google Scholar]
- 4.Hale F. Pigs born without eyeballs. J Hered. 1933;24(3):105–106. [Google Scholar]
- 5.Wald G. The molecular basis of visual excitation. Nature. 1968;219(5156):800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- 6.Hart EB, Miller WS, McCollum EV. Further studies on the nutritive deficiencies of wheat and grain mixtures and the pathological conditions produced in swine by their use. J Biol Chem. 1916;25:239–259. [Google Scholar]
- 7.Hughes JS, Lienhardt HF, Aubel CE. Nerve degeneration resulting from avitaminosis A. J Nutr. 1929;2(2):183–186. [Google Scholar]
- 8.Aberle SBD. Neurological disturbances in rats reared on diets deficient in vitamin A. J Nutr. 1933;7(4):445–461. [Google Scholar]
- 9.Jones-Villeneuve EM, McBurney MW, Rogers KA, Kalnins VI. Retinoic acid induces embryonal carcinoma cells to differentiate into neurons and glial cells. J Cell Biol. 1982;94(2):253–262. doi: 10.1083/jcb.94.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuff EL, Fewell JW. Induction of neural-like cells and acetylcholinesterase activity in cultures of F9 teratocarcinoma treated with retinoic acid and dibutyryl cyclic adenosine monophosphate. Dev Biol. 1980;77(1):103–115. doi: 10.1016/0012-1606(80)90459-5. [DOI] [PubMed] [Google Scholar]
- 11.Maden M. Retinoic acid in the development, regeneration and maintenance of the nervous system. Nat Rev Neurosci. 2007;8(10):755–765. doi: 10.1038/nrn2212. [DOI] [PubMed] [Google Scholar]
- 12.Petkovich M, Brand NJ, Krust A, Chambon P. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature. 1987;330(6147):444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 13.Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330(6149):624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 14.Kruyt FA, van der Veer LJ, Mader S, van den Brink CE, Feijen A, Jonk LJ, Kruijer W, van der Saag PT. Retinoic acid resistance of the variant embryonal carcinoma cell line RAC65 is caused by expression of a truncated RAR alpha. Differentiation. 1992;49(1):27–37. doi: 10.1111/j.1432-0436.1992.tb00766.x. [DOI] [PubMed] [Google Scholar]
- 15.Pratt MA, Kralova J, McBurney MW. A dominant negative mutation of the alpha retinoic acid receptor gene in a retinoic acid-nonresponsive embryonal carcinoma cell. Mol Cell Biol. 1990;10(12):6445–6453. doi: 10.1128/mcb.10.12.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuo T, Thiele CJ. p27Kip1: a key mediator of retinoic acid induced growth arrest in the SMS-KCNR human neuroblastoma cell line. Oncogene. 1998;16(25):3337–3343. doi: 10.1038/sj.onc.1201830. [DOI] [PubMed] [Google Scholar]
- 17.Sasaki K, Tamura S, Tachibana H, Sugita M, Gao Y, Furuyama J, Kakishita E, Sakai T, Tamaoki T, Hashimoto-Tamaoki T. Expression and role of p27(kip1) in neuronal differentiation of embryonal carcinoma cells. Brain Res Mol Brain Res. 2000;77(2):209–221. doi: 10.1016/s0169-328x(00)00053-x. [DOI] [PubMed] [Google Scholar]
- 18.Franco PG, Paganelli AR, Lopez SL, Carrasco AE. Functional association of retinoic acid and hedgehog signaling in Xenopus primary neurogenesis. Development. 1999;126(19):4257–4265. doi: 10.1242/dev.126.19.4257. [DOI] [PubMed] [Google Scholar]
- 19.Blumberg B, Bolado J, Jr, Moreno TA, Kintner C, Evans RM, Papalopulu N. An essential role for retinoid signaling in anteroposterior neural patterning. Development. 1997;124(2):373–379. doi: 10.1242/dev.124.2.373. [DOI] [PubMed] [Google Scholar]
- 20.Papalopulu N, Kintner C. A posteriorising factor, retinoic acid, reveals that anteroposterior patterning controls the timing of neuronal differentiation in Xenopus neuroectoderm. Development. 1996;122(11):3409–3418. doi: 10.1242/dev.122.11.3409. [DOI] [PubMed] [Google Scholar]
- 21.Sharpe CR, Goldstone K. Retinoid receptors promote primary neurogenesis in Xenopus. Development. 1997;124(2):515–523. doi: 10.1242/dev.124.2.515. [DOI] [PubMed] [Google Scholar]
- 22.Janesick A, Abbey R, Chung C, Liu S, Taketani M, Blumberg B. ERF and ETV3L are retinoic acid-inducible repressors required for primary neurogenesis. Development. 2013;140(15):3095–3106. doi: 10.1242/dev.093716. [DOI] [PubMed] [Google Scholar]
- 23.Diez del Corral R, Olivera-Martinez I, Goriely A, Gale E, Maden M, Storey K. Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron. 2003;40(1):65–79. doi: 10.1016/s0896-6273(03)00565-8. [DOI] [PubMed] [Google Scholar]
- 24.Maden M, Gale E, Kostetskii I, Zile M. Vitamin A-deficient quail embryos have half a hindbrain and other neural defects. Curr Biol. 1996;6(4):417–426. doi: 10.1016/s0960-9822(02)00509-2. [DOI] [PubMed] [Google Scholar]
- 25.Maden M. Retinoid signalling in the development of the central nervous system. Nat Rev Neurosci. 2002;3(11):843–853. doi: 10.1038/nrn963. [DOI] [PubMed] [Google Scholar]
- 26.Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139(5):843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 27.Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9(7):541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 28.Cunningham TJ, Zhao X, Sandell LL, Evans SM, Trainor PA, Duester G. Antagonism between retinoic acid and fibroblast growth factor signaling during limb development. Cell Rep. 2013;3(5):1503–1511. doi: 10.1016/j.celrep.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wills AE, Choi VM, Bennett MJ, Khokha MK, Harland RM. BMP antagonists and FGF signaling contribute to different domains of the neural plate in Xenopus. Dev Biol. 2010;337(2):335–350. doi: 10.1016/j.ydbio.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dorey K, Amaya E. FGF signalling: diverse roles during early vertebrate embryogenesis. Development. 2010;137(22):3731–3742. doi: 10.1242/dev.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan B, Neilson KM, Moody SA. Microarray identification of novel downstream targets of FoxD4L1/D5, a critical component of the neural ectodermal transcriptional network. Dev Dyn. 2010;239(12):3467–3480. doi: 10.1002/dvdy.22485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HC, Tseng WA, Lo FY, Liu TM, Tsai HJ. FoxD5 mediates anterior-posterior polarity through upstream modulator Fgf signaling during zebrafish somitogenesis. Dev Biol. 2009;336(2):232–245. doi: 10.1016/j.ydbio.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Branney PA, Faas L, Steane SE, Pownall ME, Isaacs HV. Characterisation of the fibroblast growth factor dependent transcriptome in early development. PLoS One. 2009;4(3):e4951. doi: 10.1371/journal.pone.0004951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchal L, Luxardi G, Thome V, Kodjabachian L. BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc Natl Acad Sci USA. 2009;106(41):17437–17442. doi: 10.1073/pnas.0906352106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tropepe V, Li S, Dickinson A, Gamse JT, Sive HL. Identification of a BMP inhibitor-responsive promoter module required for expression of the early neural gene zic1. Dev Biol. 2006;289(2):517–529. doi: 10.1016/j.ydbio.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 36.Rogers CD, Ferzli GS, Casey ES. The response of early neural genes to FGF signaling or inhibition of BMP indicate the absence of a conserved neural induction module. BMC Dev Biol. 2011;11:74. doi: 10.1186/1471-213X-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aruga J, Mikoshiba K. Role of BMP, FGF, calcium signaling, and Zic proteins in vertebrate neuroectodermal differentiation. Neurochem Res. 2011;36(7):1286–1292. doi: 10.1007/s11064-011-0422-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merzdorf CS. Emerging roles for zic genes in early development. Dev Dyn. 2007;236(4):922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- 39.Rogers CD, Harafuji N, Archer T, Cunningham DD, Casey ES. Xenopus Sox3 activates sox2 and geminin and indirectly represses Xvent2 expression to induce neural progenitor formation at the expense of non-neural ectodermal derivatives. Mech Dev. 2009;126(1–2):42–55. doi: 10.1016/j.mod.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aruga J, Tohmonda T, Homma S, Mikoshiba K. Zic1 promotes the expansion of dorsal neural progenitors in spinal cord by inhibiting neuronal differentiation. Dev Biol. 2002;244(2):329–341. doi: 10.1006/dbio.2002.0598. [DOI] [PubMed] [Google Scholar]
- 41.Ebert PJ, Timmer JR, Nakada Y, Helms AW, Parab PB, Liu Y, Hunsaker TL, Johnson JE. Zic1 represses Math1 expression via interactions with the Math1 enhancer and modulation of Math1 autoregulation. Development. 2003;130(9):1949–1959. doi: 10.1242/dev.00419. [DOI] [PubMed] [Google Scholar]
- 42.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, Gifford DK, Melton DA, Jaenisch R, Young RA. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lim LS, Loh YH, Zhang W, Li Y, Chen X, Wang Y, Bakre M, Ng HH, Stanton LW. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol Biol Cell. 2007;18(4):1348–1358. doi: 10.1091/mbc.E06-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, Wong KY, Sung KW, Lee CW, Zhao XD, Chiu KP, Lipovich L, Kuznetsov VA, Robson P, Stanton LW, Wei CL, Ruan Y, Lim B, Ng HH. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 45.Papanayotou C, De Almeida I, Liao P, Oliveira NM, Lu SQ, Kougioumtzidou E, Zhu L, Shaw A, Sheng G, Streit A, Yu D, Wah Soong T, Stern CD. Calfacilitin is a calcium channel modulator essential for initiation of neural plate development. Nat Commun. 2013;4:1837. doi: 10.1038/ncomms2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moreau M, Leclerc C, Gualandris-Parisot L, Duprat AM. Increased internal Ca2+ mediates neural induction in the amphibian embryo. Proc Natl Acad Sci USA. 1994;91(26):12639–12643. doi: 10.1073/pnas.91.26.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leclerc C, Daguzan C, Nicolas MT, Chabret C, Duprat AM, Moreau M. L-type calcium channel activation controls the in vivo transduction of the neuralizing signal in the amphibian embryos. Mech Dev. 1997;64(1–2):105–110. doi: 10.1016/s0925-4773(97)00054-3. [DOI] [PubMed] [Google Scholar]
- 48.Leclerc C, Lee M, Webb SE, Moreau M, Miller AL. Calcium transients triggered by planar signals induce the expression of ZIC3 gene during neural induction in Xenopus. Dev Biol. 2003;261(2):381–390. doi: 10.1016/s0012-1606(03)00298-7. [DOI] [PubMed] [Google Scholar]
- 49.Batut J, Vandel L, Leclerc C, Daguzan C, Moreau M, Neant I. The Ca2+-induced methyltransferase xPRMT1b controls neural fate in amphibian embryo. Proc Natl Acad Sci USA. 2005;102(42):15128–15133. doi: 10.1073/pnas.0502483102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim JW, Hummert P, Mills JC, Kroll KL. Geminin cooperates with Polycomb to restrain multi-lineage commitment in the early embryo. Development. 2011;138(1):33–44. doi: 10.1242/dev.059824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spella M, Britz O, Kotantaki P, Lygerou Z, Nishitani H, Ramsay RG, Flordellis C, Guillemot F, Mantamadiotis T, Taraviras S. Licensing regulators Geminin and Cdt1 identify progenitor cells of the mouse CNS in a specific phase of the cell cycle. Neuroscience. 2007;147(2):373–387. doi: 10.1016/j.neuroscience.2007.03.050. [DOI] [PubMed] [Google Scholar]
- 52.Seo S, Herr A, Lim JW, Richardson GA, Richardson H, Kroll KL. Geminin regulates neuronal differentiation by antagonizing Brg1 activity. Genes Dev. 2005;19(14):1723–1734. doi: 10.1101/gad.1319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brewster R, Lee J, i Altaba AR. Gli/Zic factors pattern the neural plate by defining domains of cell differentiation. Nature. 1998;393(6685):579–583. doi: 10.1038/31242. [DOI] [PubMed] [Google Scholar]
- 54.Yan B, Neilson KM, Moody SA. Notch signaling downstream of foxD5 promotes neural ectodermal transcription factors that inhibit neural differentiation. Dev Dyn. 2009;238(6):1358–1365. doi: 10.1002/dvdy.21885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moody SA, Klein SL, Karpinski BA, Maynard TM, Lamantia AS. On becoming neural: what the embryo can tell us about differentiating neural stem cells. Am J Stem Cells. 2013;2(2):74–94. [PMC free article] [PubMed] [Google Scholar]
- 56.Papanayotou C, Mey A, Birot AM, Saka Y, Boast S, Smith JC, Samarut J, Stern CD. A mechanism regulating the onset of Sox2 expression in the embryonic neural plate. PLoS Biol. 2008;6(1):e2. doi: 10.1371/journal.pbio.0060002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogers CD, Moody SA, Casey ES. Neural induction and factors that stabilize a neural fate. Birth Defects Res C Embryo Today. 2009;87(3):249–262. doi: 10.1002/bdrc.20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chalmers AD, Welchman D, Papalopulu N. Intrinsic differences between the superficial and deep layers of the Xenopus ectoderm control primary neuronal differentiation. Dev Cell. 2002;2(2):171–182. doi: 10.1016/s1534-5807(02)00113-2. [DOI] [PubMed] [Google Scholar]
- 59.Sharpe C, Goldstone K. The control of Xenopus embryonic primary neurogenesis is mediated by retinoid signalling in the neurectoderm. Mech Dev. 2000;91(1–2):69–80. doi: 10.1016/s0925-4773(99)00273-7. [DOI] [PubMed] [Google Scholar]
- 60.Hartenstein V. Early neurogenesis in Xenopus: the spatio-temporal pattern of proliferation and cell lineages in the embryonic spinal cord. Neuron. 1989;3(4):399–411. doi: 10.1016/0896-6273(89)90200-6. [DOI] [PubMed] [Google Scholar]
- 61.Carrasco AE, Blumberg B (2004) A Critical Role for Retinoid Receptors in Axial Patterning and Neuronal Differentiation. In: Grunz H (ed) The Vertebrate Organizer. Springer Science & Business Media, New York, pp 279–298
- 62.Wullimann MF, Rink E, Vernier P, Schlosser G. Secondary neurogenesis in the brain of the African clawed frog, Xenopus laevis, as revealed by PCNA, Delta-1, Neurogenin-related-1, and NeuroD expression. J Comp Neurol. 2005;489(3):387–402. doi: 10.1002/cne.20634. [DOI] [PubMed] [Google Scholar]
- 63.Hevner RF, Zecevic N (2006) Pioneer Neurons and Interneurons in the Developing Subplate: Molecular Markers, Cell Birthdays, and Neurotransmitters. In: Erzurumlu R, Guido W, Molnár Z (eds) Development and Plasticity in Sensory Thalamus and Cortex. Springer US, New York, pp 1–18
- 64.Raper J, Mason C. Cellular strategies of axonal pathfinding. Cold Spring Harb Perspect Biol. 2010;2(9):a001933. doi: 10.1101/cshperspect.a001933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bystron I, Rakic P, Molnar Z, Blakemore C. The first neurons of the human cerebral cortex. Nat Neurosci. 2006;9(7):880–886. doi: 10.1038/nn1726. [DOI] [PubMed] [Google Scholar]
- 66.Hyatt GA, Schmitt EA, Marsh-Armstrong N, McCaffery P, Drager UC, Dowling JE. Retinoic acid establishes ventral retinal characteristics. Development. 1996;122(1):195–204. doi: 10.1242/dev.122.1.195. [DOI] [PubMed] [Google Scholar]
- 67.Diez del Corral R, Morales A. Retinoic Acid Signaling during Early Spinal Cord Development. J Dev Biol. 2014;2(3):174–197. [Google Scholar]
- 68.Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129(18):4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- 69.Glover JC, Renaud JS, Rijli FM. Retinoic acid and hindbrain patterning. J Neurobiol. 2006;66(7):705–725. doi: 10.1002/neu.20272. [DOI] [PubMed] [Google Scholar]
- 70.Wilson L, Gale E, Chambers D, Maden M. Retinoic acid and the control of dorsoventral patterning in the avian spinal cord. Dev Biol. 2004;269(2):433–446. doi: 10.1016/j.ydbio.2004.01.034. [DOI] [PubMed] [Google Scholar]
- 71.Kelley MW, Turner JK, Reh TA. Retinoic acid promotes differentiation of photoreceptors in vitro. Development. 1994;120(8):2091–2102. doi: 10.1242/dev.120.8.2091. [DOI] [PubMed] [Google Scholar]
- 72.Osakada F, Ikeda H, Mandai M, Wataya T, Watanabe K, Yoshimura N, Akaike A, Sasai Y, Takahashi M. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26(2):215–224. doi: 10.1038/nbt1384. [DOI] [PubMed] [Google Scholar]
- 73.Jacobs S, Lie DC, DeCicco KL, Shi Y, DeLuca LM, Gage FH, Evans RM. Retinoic acid is required early during adult neurogenesis in the dentate gyrus. Proc Natl Acad Sci USA. 2006;103(10):3902–3907. doi: 10.1073/pnas.0511294103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Siegenthaler JA, Ashique AM, Zarbalis K, Patterson KP, Hecht JH, Kane MA, Folias AE, Choe Y, May SR, Kume T, Napoli JL, Peterson AS, Pleasure SJ. Retinoic acid from the meninges regulates cortical neuron generation. Cell. 2009;139(3):597–609. doi: 10.1016/j.cell.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McCaffery PJ, Adams J, Maden M, Rosa-Molinar E. Too much of a good thing: retinoic acid as an endogenous regulator of neural differentiation and exogenous teratogen. Eur J Neurosci. 2003;18(3):457–472. doi: 10.1046/j.1460-9568.2003.02765.x. [DOI] [PubMed] [Google Scholar]
- 76.Semmler H, Chiodin M, Bailly X, Martinez P, Wanninger A. Steps towards a centralized nervous system in basal bilaterians: insights from neurogenesis of the acoel Symsagittifera roscoffensis . Dev Growth Differ. 2010;52(8):701–713. doi: 10.1111/j.1440-169X.2010.01207.x. [DOI] [PubMed] [Google Scholar]
- 77.Burnett AL, Diehl NA. The nervous system of Hydra. I. Types, distribution and origin of nerve elements. J Exp Zool. 1964;157:217–226. doi: 10.1002/jez.1401570205. [DOI] [PubMed] [Google Scholar]
- 78.Galliot B, Quiquand M, Ghila L, de Rosa R, Miljkovic-Licina M, Chera S. Origins of neurogenesis, a cnidarian view. Dev Biol. 2009;332(1):2–24. doi: 10.1016/j.ydbio.2009.05.563. [DOI] [PubMed] [Google Scholar]
- 79.Simmons DK, Pang K, Martindale MQ. Lim homeobox genes in the Ctenophore Mnemiopsis leidyi: the evolution of neural cell type specification. Evodevo. 2012;3(1):2. doi: 10.1186/2041-9139-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jager M, Chiori R, Alie A, Dayraud C, Queinnec E, Manuel M. New insights on ctenophore neural anatomy: immunofluorescence study in Pleurobrachia pileus (Muller, 1776) J Exp Zool B Mol Dev Evol. 2011;316B(3):171–187. doi: 10.1002/jez.b.21386. [DOI] [PubMed] [Google Scholar]
- 81.Burkhardt P, Stegmann CM, Cooper B, Kloepper TH, Imig C, Varoqueaux F, Wahl MC, Fasshauer D. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc Natl Acad Sci USA. 2011;108(37):15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Canestro C, Bassham S, Postlethwait J. Development of the central nervous system in the larvacean Oikopleura dioica and the evolution of the chordate brain. Dev Biol. 2005;285(2):298–315. doi: 10.1016/j.ydbio.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 83.Canestro C, Albalat R, Postlethwait JH. Oikopleura dioica alcohol dehydrogenase class 3 provides new insights into the evolution of retinoic acid synthesis in chordates. Zoolog Sci. 2010;27(2):128–133. doi: 10.2108/zsj.27.128. [DOI] [PubMed] [Google Scholar]
- 84.Marletaz F, Holland LZ, Laudet V, Schubert M. Retinoic acid signaling and the evolution of chordates. Int J Biol Sci. 2006;2(2):38–47. doi: 10.7150/ijbs.2.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Canestro C, Postlethwait JH, Gonzalez-Duarte R, Albalat R. Is retinoic acid genetic machinery a chordate innovation? Evol Dev. 2006;8(5):394–406. doi: 10.1111/j.1525-142X.2006.00113.x. [DOI] [PubMed] [Google Scholar]
- 86.Albalat R, Canestro C. Identification of Aldh1a, Cyp26 and RAR orthologs in protostomes pushes back the retinoic acid genetic machinery in evolutionary time to the bilaterian ancestor. Chem Biol Interact. 2009;178(1–3):188–196. doi: 10.1016/j.cbi.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 87.Castro LF, Lima D, Machado A, Melo C, Hiromori Y, Nishikawa J, Nakanishi T, Reis-Henriques MA, Santos MM. Imposex induction is mediated through the Retinoid × Receptor signalling pathway in the neogastropod Nucella lapillus. Aquat Toxicol. 2007;85(1):57–66. doi: 10.1016/j.aquatox.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 88.Horiguchi T. Masculinization of female gastropod mollusks induced by organotin compounds, focusing on mechanism of actions of tributyltin and triphenyltin for development of imposex. Environ Sci. 2006;13(2):77–87. [PubMed] [Google Scholar]
- 89.Horiguchi T, Ohta Y, Nishikawa T, Shiraishi F, Shiraishi H, Morita M. Exposure to 9-cis retinoic acid induces penis and vas deferens development in the female rock shell, Thais clavigera. Cell Biol Toxicol. 2008;24(6):553–562. doi: 10.1007/s10565-007-9051-9. [DOI] [PubMed] [Google Scholar]
- 90.Nishikawa J, Mamiya S, Kanayama T, Nishikawa T, Shiraishi F, Horiguchi T. Involvement of the retinoid × receptor in the development of imposex caused by organotins in gastropods. Environ Sci Technol. 2004;38(23):6271–6276. doi: 10.1021/es049593u. [DOI] [PubMed] [Google Scholar]
- 91.Campo-Paysaa F, Marletaz F, Laudet V, Schubert M. Retinoic acid signaling in development: tissue-specific functions and evolutionary origins. Genesis. 2008;46(11):640–656. doi: 10.1002/dvg.20444. [DOI] [PubMed] [Google Scholar]
- 92.Dmetrichuk JM, Carlone RL, Jones TR, Vesprini ND, Spencer GE. Detection of endogenous retinoids in the molluscan CNS and characterization of the trophic and tropic actions of 9-cis retinoic acid on isolated neurons. J Neurosci. 2008;28(48):13014–13024. doi: 10.1523/JNEUROSCI.3192-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Urushitani H, Katsu Y, Ohta Y, Shiraishi H, Iguchi T, Horiguchi T. Cloning and characterization of the retinoic acid receptor-like protein in the rock shell, Thais clavigera. Aquat Toxicol. 2013;142–143:403–413. doi: 10.1016/j.aquatox.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 94.Gutierrez-Mazariegos J, Nadendla EK, Lima D, Pierzchalski K, Jones JW, Kane M, Nishikawa JI, Hiromori Y, Nakanishi T, Santos MM, Castro LF, Bourguet W, Schubert M, Laudet V. A mollusk retinoic acid receptor (RAR) ortholog sheds light on the evolution of ligand binding. Endocrinology. 2014;155(11):4275–4286. doi: 10.1210/en.2014-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Urushitani H, Katsu Y, Ohta Y, Shiraishi H, Iguchi T, Horiguchi T. Cloning and characterization of retinoid X receptor (RXR) isoforms in the rock shell, Thais clavigera. Aquat Toxicol. 2011;103(1–2):101–111. doi: 10.1016/j.aquatox.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 96.Markov GV, Laudet V. Origin and evolution of the ligand-binding ability of nuclear receptors. Mol Cell Endocrinol. 2011;334(1–2):21–30. doi: 10.1016/j.mce.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 97.Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312(5770):97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- 98.Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301(5640):1714–1717. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- 99.Albalat R. The retinoic acid machinery in invertebrates: ancestral elements and vertebrate innovations. Mol Cell Endocrinol. 2009;313(1–2):23–35. doi: 10.1016/j.mce.2009.08.029. [DOI] [PubMed] [Google Scholar]
- 100.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441(7097):1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 101.Salomoni P, Calegari F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 2010;20(5):233–243. doi: 10.1016/j.tcb.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 102.Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246(4930):603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 103.Bertoli C, Skotheim JM, de Bruin RA. Control of cell cycle transcription during G1 and S phases. Nat Rev Mol Cell Biol. 2013;14(8):518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Galderisi U, Jori FP, Giordano A. Cell cycle regulation and neural differentiation. Oncogene. 2003;22(33):5208–5219. doi: 10.1038/sj.onc.1206558. [DOI] [PubMed] [Google Scholar]
- 105.Herrup K, Yang Y. Cell cycle regulation in the postmitotic neuron: oxymoron or new biology? Nat Rev Neurosci. 2007;8(5):368–378. doi: 10.1038/nrn2124. [DOI] [PubMed] [Google Scholar]
- 106.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Seo S, Richardson GA, Kroll KL. The SWI/SNF chromatin remodeling protein Brg1 is required for vertebrate neurogenesis and mediates transactivation of Ngn and NeuroD. Development. 2005;132(1):105–115. doi: 10.1242/dev.01548. [DOI] [PubMed] [Google Scholar]
- 108.Ahmed M, Xu J, Xu PX. EYA1 and SIX1 drive the neuronal developmental program in cooperation with the SWI/SNF chromatin-remodeling complex and SOX2 in the mammalian inner ear. Development. 2012;139(11):1965–1977. doi: 10.1242/dev.071670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55(2):201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ninkovic J, Steiner-Mezzadri A, Jawerka M, Akinci U, Masserdotti G, Petricca S, Fischer J, von Holst A, Beckers J, Lie CD, Petrik D, Miller E, Tang J, Wu J, Lefebvre V, Demmers J, Eisch A, Metzger D, Crabtree G, Irmler M, Poot R, Gotz M. The BAF complex interacts with Pax6 in adult neural progenitors to establish a neurogenic cross-regulatory transcriptional network. Cell Stem Cell. 2013;13(4):403–418. doi: 10.1016/j.stem.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13(12):1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 112.Harper JW, Adami GR, Wei N, Keyomarsi K, Elledge SJ. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 113.Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, Massague J. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78(1):59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- 114.Dulic V, Kaufmann WK, Wilson SJ, Tlsty TD, Lees E, Harper JW, Elledge SJ, Reed SI. p53-dependent inhibition of cyclin-dependent kinase activities in human fibroblasts during radiation-induced G1 arrest. Cell. 1994;76(6):1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 115.Kato JY, Matsuoka M, Polyak K, Massague J, Sherr CJ. Cyclic AMP-induced G1 phase arrest mediated by an inhibitor (p27Kip1) of cyclin-dependent kinase 4 activation. Cell. 1994;79(3):487–496. doi: 10.1016/0092-8674(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 116.Hengst L, Gopfert U, Lashuel HA, Reed SI. Complete inhibition of Cdk/cyclin by one molecule of p21(Cip1) Genes Dev. 1998;12(24):3882–3888. doi: 10.1101/gad.12.24.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Reynisdottir I, Polyak K, Iavarone A, Massague J. Kip/Cip and Ink4 Cdk inhibitors cooperate to induce cell cycle arrest in response to TGF-beta. Genes Dev. 1995;9(15):1831–1845. doi: 10.1101/gad.9.15.1831. [DOI] [PubMed] [Google Scholar]
- 118.Cunningham JJ, Roussel MF. Cyclin-dependent kinase inhibitors in the development of the central nervous system. Cell Growth Differ. 2001;12(8):387–396. [PubMed] [Google Scholar]
- 119.Mitsuhashi T, Aoki Y, Eksioglu YZ, Takahashi T, Bhide PG, Reeves SA, Caviness VS., Jr Overexpression of p27Kip1 lengthens the G1 phase in a mouse model that targets inducible gene expression to central nervous system progenitor cells. Proc Natl Acad Sci USA. 2001;98(11):6435–6440. doi: 10.1073/pnas.111051398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vernon AE, Devine C, Philpott A. The cdk inhibitor p27Xic1 is required for differentiation of primary neurones in Xenopus. Development. 2003;130(1):85–92. doi: 10.1242/dev.00193. [DOI] [PubMed] [Google Scholar]
- 121.Carruthers S, Mason J, Papalopulu N. Depletion of the cell-cycle inhibitor p27(Xic1) impairs neuronal differentiation and increases the number of ElrC(+) progenitor cells in Xenopus tropicalis . Mech Dev. 2003;120(5):607–616. doi: 10.1016/s0925-4773(03)00010-8. [DOI] [PubMed] [Google Scholar]
- 122.Su JY, Rempel RE, Erikson E, Maller JL. Cloning and characterization of the Xenopus cyclin-dependent kinase inhibitor p27XIC1. Proc Natl Acad Sci USA. 1995;92(22):10187–10191. doi: 10.1073/pnas.92.22.10187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ali F, Hindley C, McDowell G, Deibler R, Jones A, Kirschner M, Guillemot F, Philpott A. Cell cycle-regulated multi-site phosphorylation of Neurogenin 2 coordinates cell cycling with differentiation during neurogenesis. Development. 2011;138(19):4267–4277. doi: 10.1242/dev.067900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sabherwal N, Thuret R, Lea R, Stanley P, Papalopulu N. aPKC phosphorylates p27Xic1, providing a mechanistic link between apicobasal polarity and cell-cycle control. Dev Cell. 2014;31(5):559–571. doi: 10.1016/j.devcel.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Souopgui J, Solter M, Pieler T. XPak3 promotes cell cycle withdrawal during primary neurogenesis in Xenopus laevis . EMBO J. 2002;21(23):6429–6439. doi: 10.1093/emboj/cdf644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barth LG, Barth LJ. Sequential Induction of the Presumptive Epidermis of the Rana pipiens gastrula. Biol Bull. 1964;127(3):413–427. doi: 10.2307/1539912. [DOI] [PubMed] [Google Scholar]
- 127.Grunz H, Tacke L. Neural differentiation of Xenopus laevis ectoderm takes place after disaggregation and delayed reaggregation without inducer. Cell Differ Dev. 1989;28(3):211–217. doi: 10.1016/0922-3371(89)90006-3. [DOI] [PubMed] [Google Scholar]
- 128.Saint-Jeannet JP, Huang S, Duprat AM. Modulation of neural commitment by changes in target cell contacts in Pleurodeles waltl. Dev Biol. 1990;141(1):93–103. doi: 10.1016/0012-1606(90)90104-q. [DOI] [PubMed] [Google Scholar]
- 129.Leclerc C, Rizzo C, Daguzan C, Neant I, Batut J, Auge B, Moreau M. Neural determination in Xenopus laevis embryos: control of early neural gene expression by calcium. J Soc Biol. 2001;195(3):327–337. [PubMed] [Google Scholar]
- 130.Takata K, Yamamoto KY, Ishii I, Takahashi N. Glycoproteins responsive to the neural-inducing effect of concanavalin A in Cynops presumptive ectoderm. Cell Differ. 1984;14(1):25–31. doi: 10.1016/0045-6039(84)90005-8. [DOI] [PubMed] [Google Scholar]
- 131.Gualandris L, Rouge P, Duprat AM. Target cell surface glycoconjugates and neural induction in an amphibian. J Embryol Exp Morphol. 1985;86:39–51. [PubMed] [Google Scholar]
- 132.Ozato K, Huang L, Ebert JD. Accelerated calcium ion uptake in murine thymocytes induced by concanavalin A. J Cell Physiol. 1977;93(1):153–160. doi: 10.1002/jcp.1040930119. [DOI] [PubMed] [Google Scholar]
- 133.Greenberg DA, Carpenter CL, Messing RO. Lectin-induced enhancement of voltage-dependent calcium flux and calcium channel antagonist binding. J Neurochem. 1987;48(3):888–894. doi: 10.1111/j.1471-4159.1987.tb05600.x. [DOI] [PubMed] [Google Scholar]
- 134.Hemmati-Brivanlou A, Kelly OG, Melton DA. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell. 1994;77(2):283–295. doi: 10.1016/0092-8674(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 135.Hemmati-Brivanlou A, Melton DA. Inhibition of activin receptor signaling promotes neuralization in Xenopus. Cell. 1994;77(2):273–281. doi: 10.1016/0092-8674(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 136.Delaune E, Lemaire P, Kodjabachian L. Neural induction in Xenopus requires early FGF signalling in addition to BMP inhibition. Development. 2005;132(2):299–310. doi: 10.1242/dev.01582. [DOI] [PubMed] [Google Scholar]
- 137.Lee KW, Moreau M, Neant I, Bibonne A, Leclerc C. FGF-activated calcium channels control neural gene expression in Xenopus. Biochim Biophys Acta. 2009;1793(6):1033–1040. doi: 10.1016/j.bbamcr.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 138.Lin HH, Bell E, Uwanogho D, Perfect LW, Noristani H, Bates TJ, Snetkov V, Price J, Sun YM. Neuronatin promotes neural lineage in ESCs via Ca(2+) signaling. Stem Cells. 2010;28(11):1950–1960. doi: 10.1002/stem.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Moreau M, Neant I, Webb SE, Miller AL, Leclerc C. Calcium signalling during neural induction in Xenopus laevis embryos. Philos Trans R Soc Lond B Biol Sci. 2008;363(1495):1371–1375. doi: 10.1098/rstb.2007.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rebellato P. Calcium signaling in neurogenesis: regulation of proliferation, differentiation and migration of neural stem cells. Stockholm: Karolinska Institutet; 2013. [Google Scholar]
- 141.Kahl CR, Means AR. Regulation of cell cycle progression by calcium/calmodulin-dependent pathways. Endocr Rev. 2003;24(6):719–736. doi: 10.1210/er.2003-0008. [DOI] [PubMed] [Google Scholar]
- 142.Boynton AL, Whitfield JF, Isaacs RJ. The different roles of serum and calcium in the control of proliferation of BALB/c 3T3 mouse cells. In Vitro. 1976;12(2):120–123. doi: 10.1007/BF02796358. [DOI] [PubMed] [Google Scholar]
- 143.Weissman TA, Riquelme PA, Ivic L, Flint AC, Kriegstein AR. Calcium waves propagate through radial glial cells and modulate proliferation in the developing neocortex. Neuron. 2004;43(5):647–661. doi: 10.1016/j.neuron.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 144.Leclerc C, Neant I, Moreau M. The calcium: an early signal that initiates the formation of the nervous system during embryogenesis. Front Mol Neurosci. 2012;5:3. doi: 10.3389/fnmol.2012.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Drean G, Leclerc C, Duprat AM, Moreau M. Expression of L-type Ca2+ channel during early embryogenesis in Xenopus laevis . Int J Dev Biol. 1995;39(6):1027–1032. [PubMed] [Google Scholar]
- 146.Otte AP, Kramer IM, Mannesse M, Lambrechts C, Durston AJ. Characterization of protein kinase C in early Xenopus embryogenesis. Development. 1990;110(2):461–470. doi: 10.1242/dev.110.2.461. [DOI] [PubMed] [Google Scholar]
- 147.Otte AP, van Run P, Heideveld M, van Driel R, Durston AJ. Neural induction is mediated by cross-talk between the protein kinase C and cyclic AMP pathways. Cell. 1989;58(4):641–648. doi: 10.1016/0092-8674(89)90099-8. [DOI] [PubMed] [Google Scholar]
- 148.Otte AP, Koster CH, Snoek GT, Durston AJ. Protein kinase C mediates neural induction in Xenopus laevis . Nature. 1988;334(6183):618–620. doi: 10.1038/334618a0. [DOI] [PubMed] [Google Scholar]
- 149.Otte AP, Moon RT. Protein kinase C isozymes have distinct roles in neural induction and competence in Xenopus. Cell. 1992;68(6):1021–1029. doi: 10.1016/0092-8674(92)90074-m. [DOI] [PubMed] [Google Scholar]
- 150.Stern CD. Neural induction: old problem, new findings, yet more questions. Development. 2005;132(9):2007–2021. doi: 10.1242/dev.01794. [DOI] [PubMed] [Google Scholar]
- 151.Ling F, Kang B, Sun XH. Id proteins: small molecules, mighty regulators. Curr Top Dev Biol. 2014;110:189–216. doi: 10.1016/B978-0-12-405943-6.00005-1. [DOI] [PubMed] [Google Scholar]
- 152.Perk J, Iavarone A, Benezra R. Id family of helix-loop-helix proteins in cancer. Nat Rev Cancer. 2005;5(8):603–614. doi: 10.1038/nrc1673. [DOI] [PubMed] [Google Scholar]
- 153.Lyden D, Young AZ, Zagzag D, Yan W, Gerald W, O’Reilly R, Bader BL, Hynes RO, Zhuang Y, Manova K, Benezra R. Id1 and Id3 are required for neurogenesis, angiogenesis and vascularization of tumour xenografts. Nature. 1999;401(6754):670–677. doi: 10.1038/44334. [DOI] [PubMed] [Google Scholar]
- 154.Kee Y, Bronner-Fraser M. To proliferate or to die: role of Id3 in cell cycle progression and survival of neural crest progenitors. Genes Dev. 2005;19(6):744–755. doi: 10.1101/gad.1257405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131(21):5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- 156.Rothschild G, Zhao X, Iavarone A, Lasorella A. E Proteins and Id2 converge on p57Kip2 to regulate cell cycle in neural cells. Mol Cell Biol. 2006;26(11):4351–4361. doi: 10.1128/MCB.01743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Longo A, Guanga GP, Rose RB. Crystal structure of E47-NeuroD1/beta2 bHLH domain-DNA complex: heterodimer selectivity and DNA recognition. Biochemistry. 2008;47(1):218–229. doi: 10.1021/bi701527r. [DOI] [PubMed] [Google Scholar]
- 158.Cuende J, Moreno S, Bolanos JP, Almeida A. Retinoic acid downregulates Rae1 leading to APC(Cdh1) activation and neuroblastoma SH-SY5Y differentiation. Oncogene. 2008;27(23):3339–3344. doi: 10.1038/sj.onc.1210987. [DOI] [PubMed] [Google Scholar]
- 159.Lasorella A, Stegmuller J, Guardavaccaro D, Liu G, Carro MS, Rothschild G, de la Torre-Ubieta L, Pagano M, Bonni A, Iavarone A. Degradation of Id2 by the anaphase-promoting complex couples cell cycle exit and axonal growth. Nature. 2006;442(7101):471–474. doi: 10.1038/nature04895. [DOI] [PubMed] [Google Scholar]
- 160.Eguren M, Porlan E, Manchado E, Garcia-Higuera I, Canamero M, Farinas I, Malumbres M. The APC/C cofactor Cdh1 prevents replicative stress and p53-dependent cell death in neural progenitors. Nat Commun. 2013;4:2880. doi: 10.1038/ncomms3880. [DOI] [PubMed] [Google Scholar]
- 161.Ohtani N, Zebedee Z, Huot TJ, Stinson JA, Sugimoto M, Ohashi Y, Sharrocks AD, Peters G, Hara E. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409(6823):1067–1070. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 162.Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443(7110):448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2(11):827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 164.Oikawa T, Yamada T. Molecular biology of the Ets family of transcription factors. Gene. 2003;303:11–34. doi: 10.1016/s0378-1119(02)01156-3. [DOI] [PubMed] [Google Scholar]
- 165.Le Gallic L, Virgilio L, Cohen P, Biteau B, Mavrothalassitis G. ERF nuclear shuttling, a continuous monitor of Erk activity that links it to cell cycle progression. Mol Cell Biol. 2004;24(3):1206–1218. doi: 10.1128/MCB.24.3.1206-1218.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sgouras DN, Athanasiou MA, Beal GJ, Jr, Fisher RJ, Blair DG, Mavrothalassitis GJ. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 1995;14(19):4781–4793. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Verykokakis M, Papadaki C, Vorgia E, Le Gallic L, Mavrothalassitis G. The RAS-dependent ERF control of cell proliferation and differentiation is mediated by c-Myc repression. J Biol Chem. 2007;282(41):30285–30294. doi: 10.1074/jbc.M704428200. [DOI] [PubMed] [Google Scholar]
- 168.Malynn BA, de Alboran IM, O’Hagan RC, Bronson R, Davidson L, DePinho RA, Alt FW. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14(11):1390–1399. [PMC free article] [PubMed] [Google Scholar]
- 169.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16(20):2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.White JH, Fernandes I, Mader S, Yang XJ. Corepressor recruitment by agonist-bound nuclear receptors. Vitam Horm. 2004;68:123–143. doi: 10.1016/S0083-6729(04)68004-6. [DOI] [PubMed] [Google Scholar]
- 171.Kolm PJ, Sive HL. Regulation of the Xenopus labial homeodomain genes, HoxA1 and HoxD1: activation by retinoids and peptide growth factors. Dev Biol. 1995;167(1):34–49. doi: 10.1006/dbio.1995.1005. [DOI] [PubMed] [Google Scholar]
- 172.Langston AW, Thompson JR, Gudas LJ. Retinoic acid-responsive enhancers located 3′ of the Hox A and Hox B homeobox gene clusters. Functional analysis. J Biol Chem. 1997;272(4):2167–2175. doi: 10.1074/jbc.272.4.2167. [DOI] [PubMed] [Google Scholar]
- 173.Martinez-Ceballos E, Gudas LJ. Hoxa1 is required for the retinoic acid-induced differentiation of embryonic stem cells into neurons. J Neurosci Res. 2008;86(13):2809–2819. doi: 10.1002/jnr.21729. [DOI] [PubMed] [Google Scholar]
- 174.Donato LJ, Suh JH, Noy N. Suppression of mammary carcinoma cell growth by retinoic acid: the cell cycle control gene Btg2 is a direct target for retinoic acid receptor signaling. Cancer Res. 2007;67(2):609–615. doi: 10.1158/0008-5472.CAN-06-0989. [DOI] [PubMed] [Google Scholar]
- 175.Arima K, Shiotsugu J, Niu R, Khandpur R, Martinez M, Shin Y, Koide T, Cho KW, Kitayama A, Ueno N, Chandraratna RA, Blumberg B. Global analysis of RAR-responsive genes in the Xenopus neurula using cDNA microarrays. Dev Dyn. 2005;232(2):414–431. doi: 10.1002/dvdy.20231. [DOI] [PubMed] [Google Scholar]
- 176.Janesick A, Nguyen TT, Aisaki K, Igarashi K, Kitajima S, Chandraratna RA, Kanno J, Blumberg B. Active repression by RARgamma signaling is required for vertebrate axial elongation. Development. 2014;141(11):2260–2270. doi: 10.1242/dev.103705. [DOI] [PubMed] [Google Scholar]
- 177.Passeri D, Marcucci A, Rizzo G, Billi M, Panigada M, Leonardi L, Tirone F, Grignani F. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol Cell Biol. 2006;26(13):5023–5032. doi: 10.1128/MCB.01360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Iacopetti P, Barsacchi G, Tirone F, Maffei L, Cremisi F. Developmental expression of PC3 gene is correlated with neuronal cell birthday. Mech Dev. 1994;47(2):127–137. doi: 10.1016/0925-4773(94)90085-x. [DOI] [PubMed] [Google Scholar]
- 179.el-Ghissassi F, Valsesia-Wittmann S, Falette N, Duriez C, Walden PD, Puisieux A. BTG2(TIS21/PC3) induces neuronal differentiation and prevents apoptosis of terminally differentiated PC12 cells. Oncogene. 2002;21(44):6772–6778. doi: 10.1038/sj.onc.1205888. [DOI] [PubMed] [Google Scholar]
- 180.Sugimoto K, Okabayashi K, Sedohara A, Hayata T, Asashima M. The role of XBtg2 in Xenopus neural development. Dev Neurosci. 2007;29(6):468–479. doi: 10.1159/000097320. [DOI] [PubMed] [Google Scholar]
- 181.Canzoniere D, Farioli-Vecchioli S, Conti F, Ciotti MT, Tata AM, Augusti-Tocco G, Mattei E, Lakshmana MK, Krizhanovsky V, Reeves SA, Giovannoni R, Castano F, Servadio A, Ben-Arie N, Tirone F. Dual control of neurogenesis by PC3 through cell cycle inhibition and induction of Math1. J Neurosci. 2004;24(13):3355–3369. doi: 10.1523/JNEUROSCI.3860-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Georgopoulou N, Hurel C, Politis PK, Gaitanou M, Matsas R, Thomaidou D. BM88 is a dual function molecule inducing cell cycle exit and neuronal differentiation of neuroblastoma cells via cyclin D1 down-regulation and retinoblastoma protein hypophosphorylation. J Biol Chem. 2006;281(44):33606–33620. doi: 10.1074/jbc.M602689200. [DOI] [PubMed] [Google Scholar]
- 183.Kosaka C, Sasaguri T, Komiyama Y, Takahashi H. All-trans retinoic acid inhibits vascular smooth muscle cell proliferation targeting multiple genes for cyclins and cyclin-dependent kinases. Hypertens Res. 2001;24(5):579–588. doi: 10.1291/hypres.24.579. [DOI] [PubMed] [Google Scholar]
- 184.Luo P, Wang A, Payne KJ, Peng H, Wang JG, Parrish YK, Rogerio JW, Triche TJ, He Q, Wu L. Intrinsic retinoic acid receptor alpha-cyclin-dependent kinase-activating kinase signaling involves coordination of the restricted proliferation and granulocytic differentiation of human hematopoietic stem cells. Stem Cells. 2007;25(10):2628–2637. doi: 10.1634/stemcells.2007-0264. [DOI] [PubMed] [Google Scholar]
- 185.Sueoka N, Lee HY, Walsh GL, Hong WK, Kurie JM. Posttranslational mechanisms contribute to the suppression of specific cyclin:CDK complexes by all-trans retinoic acid in human bronchial epithelial cells. Cancer Res. 1999;59(15):3838–3844. [PubMed] [Google Scholar]
- 186.Klappacher GW, Lunyak VV, Sykes DB, Sawka-Verhelle D, Sage J, Brard G, Ngo SD, Gangadharan D, Jacks T, Kamps MP, Rose DW, Rosenfeld MG, Glass CK. An induced Ets repressor complex regulates growth arrest during terminal macrophage differentiation. Cell. 2002;109(2):169–180. doi: 10.1016/s0092-8674(02)00714-6. [DOI] [PubMed] [Google Scholar]
- 187.Hester KD, Verhelle D, Escoubet-Lozach L, Luna R, Rose DW, Glass CK. Differential repression of c-myc and cdc2 gene expression by ERF and PE-1/METS. Cell Cycle. 2007;6(13):1594–1604. doi: 10.4161/cc.6.13.4336. [DOI] [PubMed] [Google Scholar]
- 188.Sawka-Verhelle D, Escoubet-Lozach L, Fong AL, Hester KD, Herzig S, Lebrun P, Glass CK. PE-1/METS, an antiproliferative Ets repressor factor, is induced by CREB-1/CREM-1 during macrophage differentiation. J Biol Chem. 2004;279(17):17772–17784. doi: 10.1074/jbc.M311991200. [DOI] [PubMed] [Google Scholar]
- 189.Papadaki C, Alexiou M, Cecena G, Verykokakis M, Bilitou A, Cross JC, Oshima RG, Mavrothalassitis G. Transcriptional repressor erf determines extraembryonic ectoderm differentiation. Mol Cell Biol. 2007;27(14):5201–5213. doi: 10.1128/MCB.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Shi Z, Lou M, Zhao Y, Zhang Q, Cui D, Wang K. Effect of all-trans retinoic acid on the differentiation of U87 glioma stem/progenitor cells. Cell Mol Neurobiol. 2013;33(7):943–951. doi: 10.1007/s10571-013-9960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Arisi MF, Starker RA, Addya S, Huang Y, Fernandez SV. All trans-retinoic acid (ATRA) induces re-differentiation of early transformed breast epithelial cells. Int J Oncol. 2014;44(6):1831–1842. doi: 10.3892/ijo.2014.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Su D, Gudas LJ. Gene expression profiling elucidates a specific role for RARgamma in the retinoic acid-induced differentiation of F9 teratocarcinoma stem cells. Biochem Pharmacol. 2008;75(5):1129–1160. doi: 10.1016/j.bcp.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Oliveira E, Casado M, Raldua D, Soares A, Barata C, Pina B. Retinoic acid receptors’ expression and function during zebrafish early development. J Steroid Biochem Mol Biol. 2013;138:143–151. doi: 10.1016/j.jsbmb.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 194.Akanuma H, Qin XY, Nagano R, Win-Shwe TT, Imanishi S, Zaha H, Yoshinaga J, Fukuda T, Ohsako S, Sone H. Identification of Stage-Specific Gene Expression Signatures in Response to Retinoic Acid during the Neural Differentiation of Mouse Embryonic Stem Cells. Front Genet. 2012;3:141. doi: 10.3389/fgene.2012.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ishibashi T, Usami T, Fujie M, Azumi K, Satoh N, Fujiwara S. Oligonucleotide-based microarray analysis of retinoic acid target genes in the protochordate, Ciona intestinalis . Dev Dyn. 2005;233(4):1571–1578. doi: 10.1002/dvdy.20486. [DOI] [PubMed] [Google Scholar]
- 196.Coyle DE, Li J, Baccei M. Regional differentiation of retinoic acid-induced human pluripotent embryonic carcinoma stem cell neurons. PLoS One. 2011;6(1):e16174. doi: 10.1371/journal.pone.0016174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Castro DS, Martynoga B, Parras C, Ramesh V, Pacary E, Johnston C, Drechsel D, Lebel-Potter M, Garcia LG, Hunt C, Dolle D, Bithell A, Ettwiller L, Buckley N, Guillemot F. A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets. Genes Dev. 2011;25(9):930–945. doi: 10.1101/gad.627811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Jacob J, Kong J, Moore S, Milton C, Sasai N, Gonzalez-Quevedo R, Terriente J, Imayoshi I, Kageyama R, Wilkinson DG, Novitch BG, Briscoe J. Retinoid acid specifies neuronal identity through graded expression of Ascl1. Curr Biol. 2013;23(5):412–418. doi: 10.1016/j.cub.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nieber F, Hedderich M, Jahn O, Pieler T, Henningfeld KA. NumbL is essential for Xenopus primary neurogenesis. BMC Dev Biol. 2013;13:36. doi: 10.1186/1471-213X-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Verdi JM, Bashirullah A, Goldhawk DE, Kubu CJ, Jamali M, Meakin SO, Lipshitz HD. Distinct human NUMB isoforms regulate differentiation vs. proliferation in the neuronal lineage. Proc Natl Acad Sci USA. 1999;96(18):10472–10476. doi: 10.1073/pnas.96.18.10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bani-Yaghoub M, Kubu CJ, Cowling R, Rochira J, Nikopoulos GN, Bellum S, Verdi JM. A switch in numb isoforms is a critical step in cortical development. Dev Dyn. 2007;236(3):696–705. doi: 10.1002/dvdy.21072. [DOI] [PubMed] [Google Scholar]
- 202.Alam AH, Suzuki H, Tsukahara T. Retinoic acid treatment and cell aggregation independently regulate alternative splicing in P19 cells during neural differentiation. Cell Biol Int. 2010;34(6):631–643. doi: 10.1042/CBI20090332. [DOI] [PubMed] [Google Scholar]
- 203.Meseguer S, Mudduluru G, Escamilla JM, Allgayer H, Barettino D. MicroRNAs-10a and -10b contribute to retinoic acid-induced differentiation of neuroblastoma cells and target the alternative splicing regulatory factor SFRS1 (SF2/ASF) J Biol Chem. 2011;286(6):4150–4164. doi: 10.1074/jbc.M110.167817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Bohlken A, Cheung BB, Bell JL, Koach J, Smith S, Sekyere E, Thomas W, Norris M, Haber M, Lovejoy DB, Richardson DR, Marshall GM. ATP7A is a novel target of retinoic acid receptor beta2 in neuroblastoma cells. Br J Cancer. 2009;100(1):96–105. doi: 10.1038/sj.bjc.6604833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Yasukawa T, Bhatt S, Takeuchi T, Kawauchi J, Takahashi H, Tsutsui A, Muraoka T, Inoue M, Tsuda M, Kitajima S, Conaway RC, Conaway JW, Trainor PA, Aso T. Transcriptional elongation factor elongin A regulates retinoic acid-induced gene expression during neuronal differentiation. Cell Rep. 2012;2(5):1129–1136. doi: 10.1016/j.celrep.2012.09.031. [DOI] [PubMed] [Google Scholar]
- 206.Won SJ, Kim SH, Xie L, Wang Y, Mao XO, Jin K, Greenberg DA. Reelin-deficient mice show impaired neurogenesis and increased stroke size. Exp Neurol. 2006;198(1):250–259. doi: 10.1016/j.expneurol.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 207.Chen Y, Kundakovic M, Agis-Balboa RC, Pinna G, Grayson DR. Induction of the reelin promoter by retinoic acid is mediated by Sp1. J Neurochem. 2007;103(2):650–665. doi: 10.1111/j.1471-4159.2007.04797.x. [DOI] [PubMed] [Google Scholar]
- 208.Chen Y, Sharma RP, Costa RH, Costa E, Grayson DR. On the epigenetic regulation of the human reelin promoter. Nucleic Acids Res. 2002;30(13):2930–2939. doi: 10.1093/nar/gkf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lotan R, Nicolson GL. Inhibitory effects of retinoic acid or retinyl acetate on the growth of untransformed, transformed, and tumor cells in vitro. J Natl Cancer Inst. 1977;59(6):1717–1722. doi: 10.1093/jnci/59.6.1717. [DOI] [PubMed] [Google Scholar]
- 210.Sidell N. Retinoic acid-induced growth inhibition and morphologic differentiation of human neuroblastoma cells in vitro. J Natl Cancer Inst. 1982;68(4):589–596. [PubMed] [Google Scholar]
- 211.Sidell N, Altman A, Haussler MR, Seeger RC. Effects of retinoic acid (RA) on the growth and phenotypic expression of several human neuroblastoma cell lines. Exp Cell Res. 1983;148(1):21–30. doi: 10.1016/0014-4827(83)90184-2. [DOI] [PubMed] [Google Scholar]
- 212.Encinas M, Iglesias M, Liu Y, Wang H, Muhaisen A, Cena V, Gallego C, Comella JX. Sequential treatment of SH-SY5Y cells with retinoic acid and brain-derived neurotrophic factor gives rise to fully differentiated, neurotrophic factor-dependent, human neuron-like cells. J Neurochem. 2000;75(3):991–1003. doi: 10.1046/j.1471-4159.2000.0750991.x. [DOI] [PubMed] [Google Scholar]
- 213.Wu PY, Lin YC, Chang CL, Lu HT, Chin CH, Hsu TT, Chu D, Sun SH. Functional decreases in P2X7 receptors are associated with retinoic acid-induced neuronal differentiation of Neuro-2a neuroblastoma cells. Cell Signal. 2009;21(6):881–891. doi: 10.1016/j.cellsig.2009.01.036. [DOI] [PubMed] [Google Scholar]
- 214.Hammerle B, Yanez Y, Palanca S, Canete A, Burks DJ, Castel V, Font de Mora J. Targeting neuroblastoma stem cells with retinoic acid and proteasome inhibitor. PLoS One. 2013;8(10):e76761. doi: 10.1371/journal.pone.0076761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sumantran VN, Brederlau A, Funa K. BMP-6 and retinoic acid synergistically differentiate the IMR-32 human neuroblastoma cells. Anticancer Res. 2003;23(2B):1297–1303. [PubMed] [Google Scholar]
- 216.Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature. 1985;313(6001):404–406. doi: 10.1038/313404a0. [DOI] [PubMed] [Google Scholar]
- 217.Hallahan AR, Pritchard JI, Chandraratna RA, Ellenbogen RG, Geyer JR, Overland RP, Strand AD, Tapscott SJ, Olson JM. BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat Med. 2003;9(8):1033–1038. doi: 10.1038/nm904. [DOI] [PubMed] [Google Scholar]
- 218.Patterson DM, Shohet JM, Kim ES (2011) Preclinical models of pediatric solid tumors (neuroblastoma) and their use in drug discovery. Curr Protoc Pharmacol. Chapter 14:Unit 14.17 [DOI] [PubMed]
- 219.Shimada H, Umehara S, Monobe Y, Hachitanda Y, Nakagawa A, Goto S, Gerbing RB, Stram DO, Lukens JN, Matthay KK. International neuroblastoma pathology classification for prognostic evaluation of patients with peripheral neuroblastic tumors: a report from the Children’s Cancer Group. Cancer. 2001;92(9):2451–2461. doi: 10.1002/1097-0142(20011101)92:9<2451::aid-cncr1595>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 220.Stallings RL, Foley NH, Bray IM, Das S, Buckley PG. MicroRNA and DNA methylation alterations mediating retinoic acid induced neuroblastoma cell differentiation. Semin Cancer Biol. 2011;21(4):283–290. doi: 10.1016/j.semcancer.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Stallings RL. MicroRNA involvement in the pathogenesis of neuroblastoma: potential for microRNA mediated therapeutics. Curr Pharm Des. 2009;15(4):456–462. doi: 10.2174/138161209787315837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Das S, Foley N, Bryan K, Watters KM, Bray I, Murphy DM, Buckley PG, Stallings RL. MicroRNA mediates DNA demethylation events triggered by retinoic acid during neuroblastoma cell differentiation. Cancer Res. 2010;70(20):7874–7881. doi: 10.1158/0008-5472.CAN-10-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ramaswamy S, Tamayo P, Rifkin R, Mukherjee S, Yeang CH, Angelo M, Ladd C, Reich M, Latulippe E, Mesirov JP, Poggio T, Gerald W, Loda M, Lander ES, Golub TR. Multiclass cancer diagnosis using tumor gene expression signatures. Proc Natl Acad Sci USA. 2001;98(26):15149–15154. doi: 10.1073/pnas.211566398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, Kim JY, Goumnerova LC, Black PM, Lau C, Allen JC, Zagzag D, Olson JM, Curran T, Wetmore C, Biegel JA, Poggio T, Mukherjee S, Rifkin R, Califano A, Stolovitzky G, Louis DN, Mesirov JP, Lander ES, Golub TR. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415(6870):436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- 225.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, Varambally S, Ghosh D, Chinnaiyan AM. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Oberthuer A, Hero B, Spitz R, Berthold F, Fischer M. The tumor-associated antigen PRAME is universally expressed in high-stage neuroblastoma and associated with poor outcome. Clin Cancer Res. 2004;10(13):4307–4313. doi: 10.1158/1078-0432.CCR-03-0813. [DOI] [PubMed] [Google Scholar]
- 227.Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122(6):835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 228.Huang S, Laoukili J, Epping MT, Koster J, Holzel M, Westerman BA, Nijkamp W, Hata A, Asgharzadeh S, Seeger RC, Versteeg R, Beijersbergen RL, Bernards R. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15(4):328–340. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Holzel M, Huang S, Koster J, Ora I, Lakeman A, Caron H, Nijkamp W, Xie J, Callens T, Asgharzadeh S, Seeger RC, Messiaen L, Versteeg R, Bernards R. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142(2):218–229. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Altucci L, Gronemeyer H. The promise of retinoids to fight against cancer. Nat Rev Cancer. 2001;1(3):181–193. doi: 10.1038/35106036. [DOI] [PubMed] [Google Scholar]
- 231.Ulrich T. Curing neuroblastoma by making it grow up. Boston: Boston Children’s Hospital; 2013. [Google Scholar]
- 232.Temple S. Division and differentiation of isolated CNS blast cells in microculture. Nature. 1989;340(6233):471–473. doi: 10.1038/340471a0. [DOI] [PubMed] [Google Scholar]
- 233.Reynolds BA, Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255(5052):1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- 234.Snyder EY, Deitcher DL, Walsh C, Arnold-Aldea S, Hartwieg EA, Cepko CL. Multipotent neural cell lines can engraft and participate in development of mouse cerebellum. Cell. 1992;68(1):33–51. doi: 10.1016/0092-8674(92)90204-p. [DOI] [PubMed] [Google Scholar]
- 235.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 236.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Cancerous stem cells can arise from pediatric brain tumors. Proc Natl Acad Sci USA. 2003;100(25):15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Calabrese C, Poppleton H, Kocak M, Hogg TL, Fuller C, Hamner B, Oh EY, Gaber MW, Finklestein D, Allen M, Frank A, Bayazitov IT, Zakharenko SS, Gajjar A, Davidoff A, Gilbertson RJ. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 238.Liu C, Sage JC, Miller MR, Verhaak RG, Hippenmeyer S, Vogel H, Foreman O, Bronson RT, Nishiyama A, Luo L, Zong H. Mosaic analysis with double markers reveals tumor cell of origin in glioma. Cell. 2011;146(2):209–221. doi: 10.1016/j.cell.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, Singer O, Ellisman MH, Verma IM. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338(6110):1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Friedmann-Morvinski D, Verma IM. Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 2014;15(3):244–253. doi: 10.1002/embr.201338254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.van Es JH, Sato T, van de Wetering M, Lyubimova A, Nee AN, Gregorieff A, Sasaki N, Zeinstra L, van den Born M, Korving J, Martens AC, Barker N, van Oudenaarden A, Clevers H. Dll1 + secretory progenitor cells revert to stem cells upon crypt damage. Nat Cell Biol. 2012;14(10):1099–1104. doi: 10.1038/ncb2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, Rupec RA, Gerhard M, Schmid R, Barker N, Clevers H, Lang R, Neumann J, Kirchner T, Taketo MM, van den Brink GR, Sansom OJ, Arkan MC, Greten FR. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1–2):25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 243.Tata PR, Mou H, Pardo-Saganta A, Zhao R, Prabhu M, Law BM, Vinarsky V, Cho JL, Breton S, Sahay A, Medoff BD, Rajagopal J. Dedifferentiation of committed epithelial cells into stem cells in vivo. Nature. 2013;503(7475):218–223. doi: 10.1038/nature12777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, Arendt LM, Kuperwasser C, Bierie B, Weinberg RA. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 2011;108(19):7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Southall TD, Davidson CM, Miller C, Carr A, Brand AH. Dedifferentiation of neurons precedes tumor formation in Lola mutants. Dev Cell. 2014;28(6):685–696. doi: 10.1016/j.devcel.2014.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Yung WK, Kyritsis AP, Gleason MJ, Levin VA. Treatment of recurrent malignant gliomas with high-dose 13-cis-retinoic acid. Clin Cancer Res. 1996;2(12):1931–1935. [PubMed] [Google Scholar]
- 247.See SJ, Levin VA, Yung WK, Hess KR, Groves MD. 13-cis-retinoic acid in the treatment of recurrent glioblastoma multiforme. Neuro Oncol. 2004;6(3):253–258. doi: 10.1215/S1152851703000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kaba SE, Kyritsis AP, Conrad C, Gleason MJ, Newman R, Levin VA, Yung WK. The treatment of recurrent cerebral gliomas with all-trans-retinoic acid (tretinoin) J Neurooncol. 1997;34(2):145–151. doi: 10.1023/a:1005743707803. [DOI] [PubMed] [Google Scholar]
- 249.Wismeth C, Hau P, Fabel K, Baumgart U, Hirschmann B, Koch H, Jauch T, Grauer O, Drechsel L, Brawanski A, Bogdahn U, Steinbrecher A. Maintenance therapy with 13-cis retinoid acid in high-grade glioma at complete response after first-line multimodal therapy–a phase-II study. J Neurooncol. 2004;68(1):79–86. doi: 10.1023/b:neon.0000024748.26608.2f. [DOI] [PubMed] [Google Scholar]
- 250.Galli R, Binda E, Orfanelli U, Cipelletti B, Gritti A, De Vitis S, Fiocco R, Foroni C, Dimeco F, Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64(19):7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- 251.Ying M, Wang S, Sang Y, Sun P, Lal B, Goodwin CR, Guerrero-Cazares H, Quinones-Hinojosa A, Laterra J, Xia S. Regulation of glioblastoma stem cells by retinoic acid: role for Notch pathway inhibition. Oncogene. 2011;30(31):3454–3467. doi: 10.1038/onc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Wolf G. Retinoic acid as cause of cell proliferation or cell growth inhibition depending on activation of one of two different nuclear receptors. Nutr Rev. 2008;66(1):55–59. doi: 10.1111/j.1753-4887.2007.00006.x. [DOI] [PubMed] [Google Scholar]
- 253.Campos B, Centner FS, Bermejo JL, Ali R, Dorsch K, Wan F, Felsberg J, Ahmadi R, Grabe N, Reifenberger G, Unterberg A, Burhenne J, Herold-Mende C. Aberrant expression of retinoic acid signaling molecules influences patient survival in astrocytic gliomas. Am J Pathol. 2011;178(5):1953–1964. doi: 10.1016/j.ajpath.2011.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Barbus S, Tews B, Karra D, Hahn M, Radlwimmer B, Delhomme N, Hartmann C, Felsberg J, Krex D, Schackert G, Martinez R, Reifenberger G, Lichter P. Differential retinoic acid signaling in tumors of long- and short-term glioblastoma survivors. J Natl Cancer Inst. 2011;103(7):598–606. doi: 10.1093/jnci/djr036. [DOI] [PubMed] [Google Scholar]
- 255.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129(4):723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Schug TT, Berry DC, Toshkov IA, Cheng L, Nikitin AY, Noy N. Overcoming retinoic acid-resistance of mammary carcinomas by diverting retinoic acid from PPARbeta/delta to RAR. Proc Natl Acad Sci USA. 2008;105(21):7546–7551. doi: 10.1073/pnas.0709981105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Rohle D, Popovici-Muller J, Palaskas N, Turcan S, Grommes C, Campos C, Tsoi J, Clark O, Oldrini B, Komisopoulou E, Kunii K, Pedraza A, Schalm S, Silverman L, Miller A, Wang F, Yang H, Chen Y, Kernytsky A, Rosenblum MK, Liu W, Biller SA, Su SM, Brennan CW, Chan TA, Graeber TG, Yen KE, Mellinghoff IK. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340(6132):626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Garrett-Bakelman FE, Melnick AM. Differentiation therapy for IDH1/2 mutant malignancies. Cell Res. 2013;23(8):975–977. doi: 10.1038/cr.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, Verhaak RG, Hoadley KA, Hayes DN, Perou CM, Schmidt HK, Ding L, Wilson RK, Van Den Berg D, Shen H, Bengtsson H, Neuvial P, Cope LM, Buckley J, Herman JG, Baylin SB, Laird PW, Aldape K. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, Thompson CB, Kaufman A, Guryanova O, Levine R, Heguy A, Viale A, Morris LG, Huse JT, Mellinghoff IK, Chan TA. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Chou AP, Chowdhury R, Li S, Chen W, Kim AJ, Piccioni DE, Selfridge JM, Mody RR, Chang S, Lalezari S, Lin J, Sanchez DE, Wilson RW, Garrett MC, Harry B, Mottahedeh J, Nghiemphu PL, Kornblum HI, Mischel PS, Prins RM, Yong WH, Cloughesy T, Nelson SF, Liau LM, Lai A. Identification of retinol binding protein 1 promoter hypermethylation in isocitrate dehydrogenase 1 and 2 mutant gliomas. J Natl Cancer Inst. 2012;104(19):1458–1469. doi: 10.1093/jnci/djs357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Liau L, Cloughesy T, Lai A (2013) Pre-Clinical Studies Investigating the Use of Isotretinoin for the Treatment of IDH1 Mutant Glioma Patients. Accelerate Brain Cancer Cure, Inc., Neuro-Oncology & Neurosurgery, UCLA
- 263.Lee SY, Lee HS, Moon JS, Kim JI, Park JB, Lee JY, Park MJ, Kim J. Transcriptional regulation of Zic3 by heterodimeric AP-1(c-Jun/c-Fos) during Xenopus development. Exp Mol Med. 2004;36(5):468–475. doi: 10.1038/emm.2004.59. [DOI] [PubMed] [Google Scholar]
- 264.Yoon J, Kim JH, Lee OJ, Lee SY, Lee SH, Park JB, Lee JY, Kim SC, Kim J. AP-1(c-Jun/FosB) mediates xFoxD5b expression in Xenopus early developmental neurogenesis. Int J Dev Biol. 2013;57(11–12):865–872. doi: 10.1387/ijdb.130163jk. [DOI] [PubMed] [Google Scholar]