Abstract
Dps proteins are members of an extensive family of proteins that oxidise and deposit iron in the form of ferric oxide, and are also able to bind DNA. Ferroxidation centres are formed at the interface of anti-parallel dimers, which further assemble into dodecameric nanocages with a hollow core where ferric oxide is deposited. Streptomyces coelicolor encodes three Dps-like proteins (DpsA, B and C). Despite sharing the conserved four-helix bundle organisation observed in members of the Dps family, they display significant differences in the length of terminal extensions, or tails. DpsA possess both N- and C-terminal tails of different lengths, and their removal affects quaternary structure assembly to varying degrees. DpsC quaternary structure, on the other hand, is heavily dependent on its N-terminal tail as its removal abolishes correct protein folding. Analysis of the crystal structure of dodecamers from both proteins revealed remarkable differences in the position of tails and interface surface area; and provides insight to explain the differences in biochemical behaviour observed while comparing DpsA and DpsC.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-014-1658-4) contains supplementary material, which is available to authorized users.
Keywords: Protein interface, PISA, Dodecamer, Hetero-oligomeric, Ferroxidation
Introduction
Bacteria have evolved sophisticated and often elaborate mechanisms to efficiently detect and combat potentially damaging environmental changes and stresses such as nutrient limitations, temperature instabilities, pH changes and osmotic pressures [1]. The generation of reactive oxygen species (ROS) as by-products of metabolic processes also leads to oxidative damage [2]. The respiratory chain enzymes can produce up to 87 % of hydrogen peroxide created within the E. coli cell [3], and although the potency of hydrogen peroxide as a damaging compound itself is limited, it can give rise to reactive hydroxyl radicals (·OH) through Fenton chemistry. This reaction, involving the oxidation of transition metals such as ferrous iron by H2O2 [4], leads to products capable of inducing DNA strand breaks, peroxidation of lipids and damage to other intracellular macromolecules [5].
Iron is indispensable to all forms of life, but properties like the redox potential of the Fe(II)/Fe(III) couple can potentially lead to fatal side effects. Bacteria have accordingly developed several strategies to maintain iron homeostasis [6]. Ferritins are members of an extensive superfamily of iron-related proteins that are fundamental to the metabolism and control of cellular iron. This protein family can be broken into three sub-families: classical ferritins (Ftn), the haem-binding bacterioferritins (Bfr), and DNA protection proteins from starved cells (Dps). Whilst there is divergence in biological roles, architectures and phylogenetic distribution, they all share a common role in iron homeostasis. Not only are these proteins capable of acting as a bioavailable iron reservoir, but by sequestering and detoxifying the intracellular free ferrous iron they provide protection against oxidative damage [7].
Dps are a major group within the ferritin superfamily sharing structural and functional homologies to Ftn. These proteins’ monomeric assembly generates the archetypal ferritin four-helix bundle (A, B, C and D helices) with an additional short helix (BC). The four-helix bundle adopts an “up-down-down-up” arrangement allowing the fully folded helix pairs (A + B and C + D) to remain anti-parallel. This also allows the N and C termini to be placed at opposing ends of the bundle [8, 9]. The BC helix is located at the loop connecting the B and C helices and is positioned centrally and orthogonally to the four-helix bundle, this being one of the differentiating features between Dps and Ftn. Rather than the 24-mer oligomer observed in Ftn [10], Dps-like proteins assemble into smaller dodecameric nanocages [9]. The conserved ferroxidase centre of Dps is located at the interface between two antiparallel subunits that form the twofold symmetry dimers [11]. The multimeric assembly of Dps proteins results in a twofold symmetry axis and two sets of threefold symmetry axis (23-point symmetry) allowing each subunit to interface with five other subunits. Along with the six dimer interfaces responsible for the establishment of the inter-subunit ferroxidase centre, there is also the formation of four “ferritin-like” trimer interfaces and four “Dps-like” trimer interfaces. The “ferritin-like” dimer resembles the arrangement of the threefold symmetry-related subunits of Ftn and forms a pore associated with iron translocation [12]. The “Dps-like” trimer defines an interface common only to Dps within the ferritin superfamily [13].
Most Dps proteins possess terminal extensions or tails at their N and C termini, extending from the four-helix bundle and protruding from the surface of the dodecameric cage. These tails display poor sequence conservation, and have been implicated mainly in DNA binding. The heterogeneity of roles played by the tails is illustrated by numerous studies involving tail-deleted mutants. In E. coli Dps, removal of the N-terminal tail abolishes DNA binding, although the role of the tail in dodecameric assembly is unclear [14]. Similarly, a Dps from L. lactis (PDB: 1ZUJ) possesses a long N-tail mediating DNA binding but apparently not required for oligomerisation, as a tail-deleted mutant behaves like the parental protein dodecamer upon gel filtration and therefore the tail was considered not essential for dodecameric assembly [15]. On the other hand, D. radiodurans encodes for two Dps proteins, DrDps1 (PDB: 2C2U) and DrDps2 (PDB: 2C2J), both displaying interesting tail features. DrDps1 has a long N-tail containing a short helix, and its removal abolished dodecameric assembly [16], while DrDps2 possess a C-terminal tail that contains a small helix and a novel iron binding site. Deletion of this tail renders the protein unable to oligomerise or even to assemble into a dimer, suggesting a role for the tail on the assembly of this very stable interface [17].
The terminal tails in M. smegmatis Dps proteins have been extensively explored. This organism encodes two Dps (MsDps1 and MsDps1). MsDps1 is characterised by a C-terminal tail that is involved in both DNA binding and oligomeric assembly [18, 19]. The 26-residue C-terminal tail can be split into a bi-functional entity. Removal of the last 16 residues abolishes DNA binding but maintains dodecameric assembly, while complete removal of the tail leads to an unexpected open decameric structure [20]. The N-terminal tail was also implicated in oligomeric assembly, as the first four residues of the N-tail interact with 5 residues of the C-terminal tail at the ferritin-like interface. Loss of the N-terminal tail disrupts assembly and also interrupts DNA interaction, although this is thought to be an indirect effect affecting the C-terminal tail configuration. The MsDps2 possesses a longer N-terminal tail that is also involved in bracing the dodecamer, as the C-terminal tail of MsDps1 does. This N-tail not only braces an adjacent subunit, but extends on to interact with the adjacent subunit’s dimer partner [21].
We have previously described the three Dps proteins encoded by Streptomyces coelicolor (DpsA, B and C), focusing on their in vivo roles and the regulation of their expression [22, 23]. Furthermore, our extensive phylogenetic analyses revealed that Dps proteins in Actinobacteria can be segregated into three distinct groups, illustrative of a unique evolutionary history for each of the ScDps. The diversity of Dps proteins encoded by Actinobacteria can be explained by gene duplication or lateral acquisition events. Orthologs for DpsA and DpsC are rare in Streptomyces, and the latter in particular is part of a narrow clade of proteins populated by orthologs expressed by organisms found in extreme environments. Intriguingly, the tail composition of Dps proteins mirrors their phylogenetic distribution, namely DpsA-like proteins possess both short N- and C-terminal tails, while DpsC-like proteins have long N-terminal tails and short C-tails [24]. The above facts triggered our interest into elucidating the crystal structure of DpsA and DpsC, and to assign functional roles to their respective tails in terms of oligomeric assembly and biochemical properties.
Experimental procedures
Cloning of S. coelicolor dps genes
The genes encoding Streptomyces coelicolor DpsA (SCO0596), DpsB (SCO5756) and DpsC (SCO1050) were extracted from pDpsA4, pDpsB4 and pDpsC1, respectively [22], by digesting with NdeI/BglII, and subsequently cloned into NdeI/BamHI-digested pET26b+. The resulting plasmids pDpsA14, pDpsB14 and pDpsC14 encode, respectively, C-terminally His-tagged DpsA, DpsB and DpsC. DpsA-DCT was created by digesting pDpsA14 with XhoI followed by religation, which deleted the coding sequence for 25 residues of the C-terminal tail. DpsA-DNT and DpsA-DTM coding sequences were supplied as synthetic constructs in pCR2.1 from Eurofins (sequences provided in Supplementary Fig. 1B). The genes were excised with NdeI/BglII and cloned into NdeI/BamHI-digested pET26b+ to create pDpsA-DNT and pDpsA-DTM (both containing a C-terminal His tag). All recombinant plasmids were verified by sequencing.
Expression and purification of Dps recombinant proteins
BL21 (DE3) cells containing the expression plasmids were grown in 1-l cultures of 2× YT containing kanamycin (25 µg/ml) at 37 °C to an optical density of 0.9 at 600 nm. Recombinant protein expression was induced with the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and cultures were incubated at 30 °C for a further 2 h prior to harvesting cells by centrifugation. Cells were resuspended in Buffer 1 [20 mM Tris/HCl, 500 mM NaCl, 50 mM imidazole, Complete protease inhibitor cocktail (Roche Diagnostics), pH 7.5] and disrupted by sonication. Cellular debris was removed by centrifugation and the supernatant containing soluble proteins applied to a Ni Sepharose High Performance column (HisTrap HP 5 ml, GE Healthcare) pre-equilibrated with the same buffer and purified according to manufacturer’s guidelines. Protein solutions were subjected to ion exchange chromatography (RESOURCE Q 1 ml, GE Healthcare). Fractions containing pure Dps proteins were pooled and buffer exchanged using gel filtration (HiTrap 5 ml, GE Healthcare) into 20, 200 mM NaCl and 5 % glycerol.
Native PAGE
7.5 % native polyacrylamide gel electrophoresis was carried out according to the Laemmli method, but with the omission of SDS. Samples were loaded with a native sample buffer (Invitrogen) and electrophoresis was carried out at 90 V at 4 °C. Gels were then fixed and stained with Coomassie R250.
DNA oxidative protection
In vitro DNA protection against oxidative damage was assessed using 40 ng pUC18 (2,686 bp) in 20, 200 mM NaCl, 5 % glycerol. Dps protein was added to a final concentration of 0.5 µM and the DNA/protein mixture incubated at room temperature for 10 min prior to the addition of [NH4]2[Fe][SO4]2·6H2O to reach 50 μM, subsequently, incubation for 5 min was followed by addition of H2O2 in molar excess (5 mM) and a further incubation for 10 min before reactions were resolved with a 1 % agarose gel and stained with ethidium bromide.
DNAse protection
Protection of DNA against DNase-mediated enzymatic cleavage was analysed by first incubating Dps protein (0.5 µM) with DNA (50 ng) in a 20 µl final volume at room temperature for 30 min. To the Dps/DNA reaction mixture 1 U of DNase was added which is an amount in excess of that required for complete degradation of the DNA. After additional incubation for 5 min at room temperature, reactions were resolved by agarose electrophoresis and stained with ethidium bromide.
Ferroxidase activity
Dps-mediated ferric iron oxidation was followed at 25 °C by monitoring the absorbance at a wavelength of 310 nm. Air oxygen or H2O2 were used as oxidants, in the case of the latter combined with fully degassed solutions. Protein solutions were prepared in 10 mM Tris/HCl, pH 7.5 at 0.2 mg/ml (~0.74 μM), and reactions started by addition of freshly prepared anaerobic ammonium iron (II) sulphate at final concentrations of 138, 207 and 276 μM. The Fe2+:cage ratios were 186, 279 and 372, respectively. The reactions were monitored at 10-s intervals for 20 min. Fe(II) auto oxidation was monitored in parallel with reactions lacking protein. Specific activities were calculated as change in absorbance per second/mg of protein used in the reaction, using the linear portion of the curves. The data used were collected from three independent replicates using 276 μM substrate concentration.
Staining for Dps protein-bound iron
Iron-loaded Dps (20 μg), using the ferroxidation reactions described above, were resolved by Native PAGE and subsequently stained for ferric iron [25]. Following electrophoresis, gels were incubated in potassium ferricyanide solution (100 mM potassium ferricyanide, 50 mM Tris–HCl, 100 mM NaCl, pH 7.5) in the dark for 10 min prior to destaining with a 10 % methanol, 10 % tri-chloroacetic acid prepared immediately before use. After required colour development, gels were imaged and then stained with Coomassie R250.
Protein crystallisation
Sitting drop vapour diffusion 96-well plate screens were made using an Innovadyne Screenmaker 96 + 8, and the JCSG-plus HT96 screen (Molecular Dimensions). Sitting drops included 100 μl of precipitant with 100 nl protein, and 100 nl of precipitant with 200 nl protein. Best crystals formed at room temperature within 2 weeks in condition E2 for DpsA (0.2 M NaCl, 0.1 m sodium cocodylate pH 6.5, 0.2 M ammonium sulphate), C10 for DpsA-DCT (0.1 M bicine pH 9.0, 10 % w/v PEG 20 000 and 2 % v/v/dioxane), and G12 for DpsC (0.1 M Bis–Tris pH 5.5, and 3 M NaCl).
X-ray data collection and structural refinement
Crystals were cryoprotected with 25 % v/v glycerol and frozen in liquid nitrogen. Diffraction data were collected on beamline I02 (DpsC) and I04 (DpsA and DpsA-DCT) at the Diamond Light Source (Harwell Science and Innovation Campus, Didcot, Oxford, UK). All datasets were integrated and scaled with XDS and XSCALE [26], through xia2 [27].
Structure determination and validation
A search model was built using Clustal Omega [28] to align and Chainsaw (within ccp4 package) [29] to chain-trace S. coelicor DpsA onto the M. smegmatis Dps structure (1UVH). The structure of DpsA was solved by molecular replacement with Phaser [30] using the aforementioned search model. The structures of DpsA-DCT and DpsC were solved using the refined structure of DpsA as a search model. The structures were refined iteratively with Refmac5 [31] and model building in Coot [32]. For structure validation PDB files were submitted to the ERRAT server [Protein Structure and Validation Server (http://services.mbi.ucla.edu/SAVES/); 33] for the identification of mistraced protein regions due to errors in model building. Ramachandran plot analyses were carried out using RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php; [34]). Structural figures were generated using Pymol. Superpositions were generated using the align function in Pymol, with the alignment restricted to backbone carbons only. UCSF Chimera [35] was used to visualise the molecular structures of the Dps proteins from their PDB files.
Proteins, interface, structure and assemblies analysis (PISA)
Full atomic co-ordinates of DpsA and DpsC in PDB format were uploaded, independently, to the PDBePISA interactive tool (http://www.ebi.ac.uk/msdsrv/prot_int/pistart.html; [36]). The cell parameters and space symmetry group boxes were cross-referenced, process ligands boxes unchecked, processing mode was set to automatic and interfaces analysis selected.
Computational alanine scanning of protein–protein interfaces
A computational approach was used for the prediction of energetically important amino acid residues involved in protein interfaces (http://www.robetta.org/alascansubmit.jsp, [37]). For the prediction, the polypeptide chains that form the interface under study are required to be defined. Thus in the case of DpsA and DpsC, individual submissions were utilised for the dimer interfaces, “ferritin-like” interfaces and “Dps-like” interfaces.
Results
Terminal tails contribute differently to the oligomeric assembly and biochemical properties of S. coelicolor DpsA
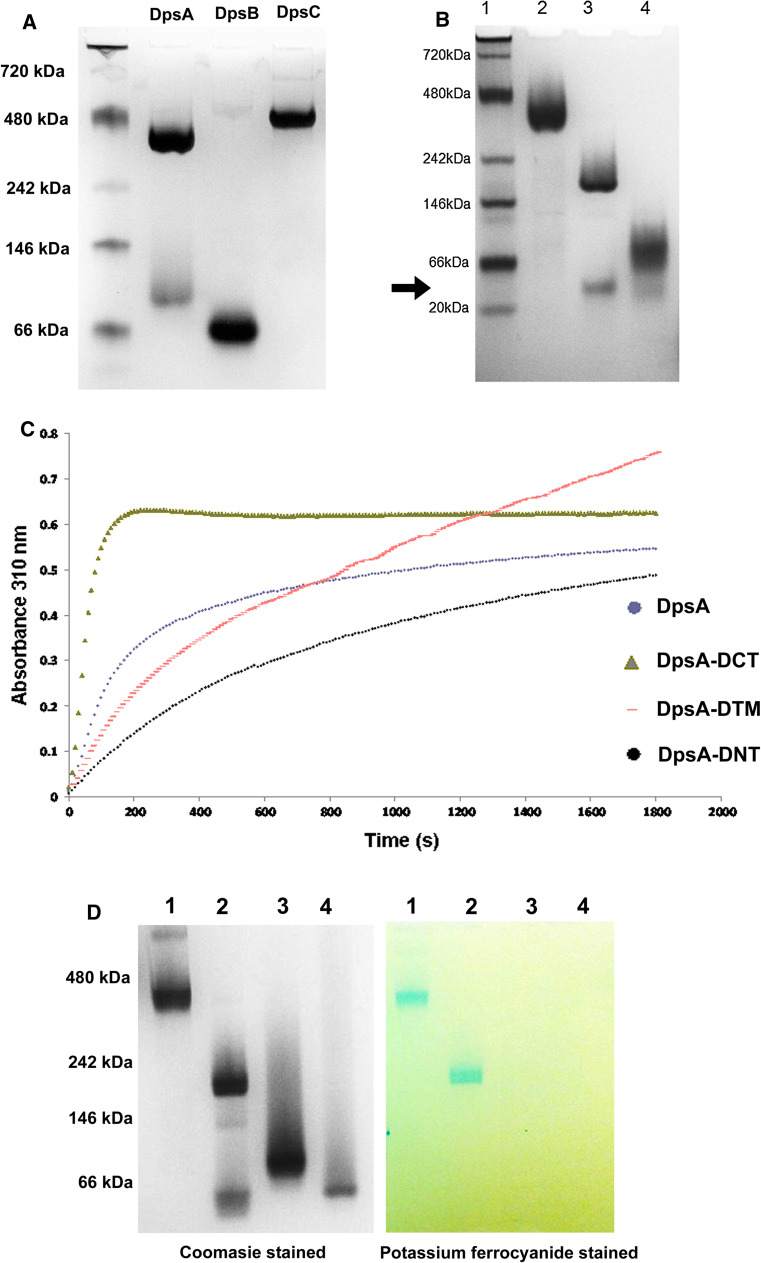
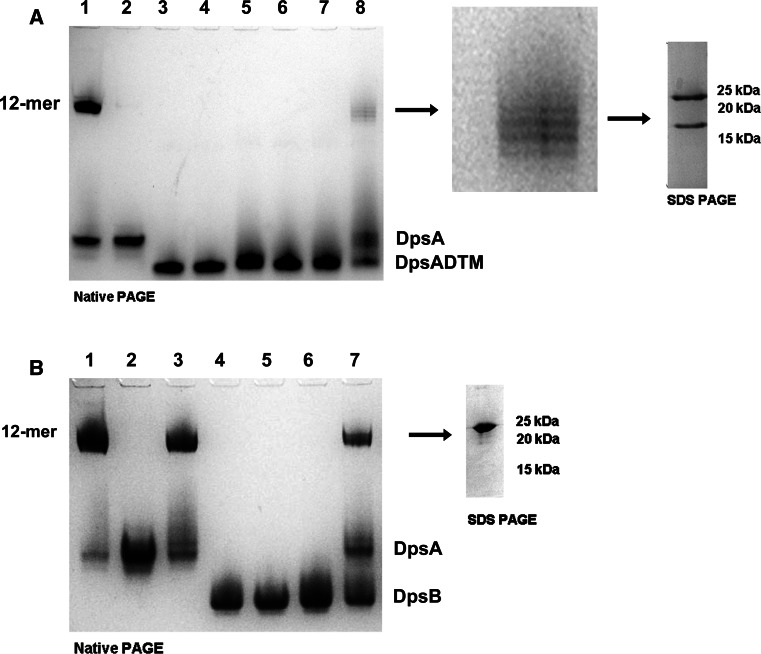
DpsA monomers have a molecular weight of around 20 kDa (187 residues) and possess two terminal extensions or tails of different lengths, extending from the conserved four-helix bundle typical of Dps proteins. The N-terminal tail is 15 amino acids long while the C-terminal tail is 25 amino acids long (Supplementary Fig. 1A). The ability of recombinant DpsA, alongside the other Dps proteins encoded by S. coelicolor dpsB and dpsC, to form dodecamers in solution was first explored by Blue Native PAGE. Both DpsA and DpsC can form dodecamers, while DpsB only assembles into a dimer as estimated by its gel migration (Fig. 1a).
Fig. 1.
Terminal tails contribute to DpsA dodecameric assembly and influence ferroxidation. a Recombinant DpsA and DpsC assemble into dodecamers, but DpsB remains in dimeric form. b Removal of terminal tails affects DpsA oligomerisation. Samples in lanes are size markers (1), DpsA (2), DpsA-DCT (3) and DpsA-DNT (4). DpsA-DCT forms a low-order oligomer thought to be a dimer (arrow). Native molecular weight reference proteins used were Apoferritin band 2 (480 kDa), β-phycoerythrin (242 kDa), lactate dehydrogenase (146 kDa), bovine serum albumin (66 kDa) and soybean trypsin inhibitor (20 kDa). c Fe(II) oxidation by DpsA and mutant variants was monitored by changes in absorbance at 310 nm every 10 s. Air oxygen was used as oxidant with addition of 276 μM Fe(II). Data were averaged from three independent experiments. d DpsA and DpsA-DCT can mineralise iron. After ferroxidation using 200 µM H2O2 as oxidant, gels were stained for protein with coomassie (left gel) and with potassium ferrocyanide for ferric oxide (right gel). Samples in lanes are DpsA (1), DpsA-DCT (2), DpsA-DNT (3) and DpsA-DTM (4). The DpsA-DCT low-order oligomer observed in lane 2 (arrow) is unable to deposit iron oxide. Position of native size markers is indicated. All protein samples were electrophoresed in Blue Native PAGE (7 %) gels
Various DpsA mutant variants lacking tails were generated and assessed for their ability to oligomerise into dodecamers. Interestingly, removal of the C-tail affects oligomerisation partially, although the mutant protein DpsA-DCT (monomer mass 17.8 kDa) can still form a high-order oligomer judged to be a hexamer as estimated by Native PAGE migration. This oligomeric organisation seems partially unstable in the conditions used, as some minor dissociation into a probable dimeric form is observed (Fig. 1b, arrow). Removal of the N-tail, on the other hand abolishes high-order oligomerisation as DpsA-DNT (monomer mass 18.47 kDa, lacking the N-tail) was unable to oligomerise beyond what appears to be a dimeric complex. It is relevant to point out that during Native PAGE all Dps variants migrated slower than would be predicted from their molecular weight.
The ability of DpsA and mutant variants to perform ferroxidation was assessed by monitoring changes in absorbance at 310 nm, in the presence of O2 and H2O2 as oxidants. A further mutant variant, DpsA-DTM (monomer mass 16.3 kDa) lacking both N- and C-tails, was also incorporated into the study. Ferroxidase reactions using 200 µM H2O2 and various Fe(II) concentrations resulted in high absorbance values indicative of Fe(III) formation, that did not increase even after extended incubation. This suggests that with these conditions the reactions went to completion too early for their progress to be recorded. All protein species tested were able to oxidise Fe(II) using air O2 (Fig. 1c). This confirmed that dimeric interfaces were formed and therefore ferroxidase centres were created between anti-parallel monomers even in those species unable to form a dodecamer (DpsA-DNT and DpsA-DTM). DpsA and DpsA-DCT displayed the typical profile observed for other Dps proteins, namely a steep increase in absorbance leading towards a plateau indicative of saturation of the internal cavity of the protein nanocage. Interestingly, DpsA-DCT reached saturation earlier than full-length DpsA (Fig. 1c). Comparison of the protein’s specific activities (Supplementary Table 1) showed a reduction in both DpsA-DNT and DpsA-DTM when compared to DpsA and DpsA-DCT, although DpsA-DTM was able to sustain ferroxidase activity without reaching saturation, indicative of continuous oxidation of Fe(II).
Protein samples used in the above experiment were subsequently assessed for their ability to deposit ferric oxide. In-gel staining of protein-bound ferric iron revealed that only full-length DpsA and DpsA-DCT are able to deposit ferric oxide, indicating the formation of a protein cage cavity (Fig. 1d). The presence of a cavity where ferric iron is deposited formed by DpsA-DCT suggests that its oligomeric state may be underestimated by Native PAGE. As expected, the low-order oligomer described for DpsA-DNT cannot deposit an iron core. Non-oligomerising species like DpsA-DNT and DpsA-DTM are unable to form a hollow core and therefore cannot deposit ferric oxide, supporting our assumption that they assemble as dimers.
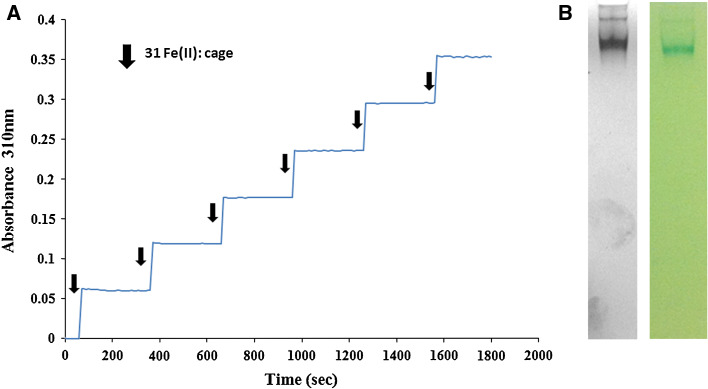
Full-length DpsC was also assessed for its ability to oxidise Fe(II). Attempts using O2 as oxidant failed to show any ferroxidase activity in solution, which indicates an inability of DpsC to use air oxygen as an oxidant. When using H2O2 as oxidant, addition of substrate (ammonium ferrous sulphate) in an excess resulted in instantaneous protein precipitation. This could not be prevented by modifying ionic strength or the concentration of buffer components. To avoid this, we set reactions using 200 µM H2O2 as oxidant, and initiated them by adding a lower substrate concentration (23 µM, 31 Fe(II): protein cage) in a step-wise manner. The absorbance measured at 310 nm showed an increase after each addition of substrate (Fig. 2a). One minute prior to the next addition of Fe(II), further H2O2 was added. A lack of increase in absorbance at this point indicated that all the iron from the previous addition was successfully oxidised, and that the oxidant was not a limiting factor. Iron was oxidised upon addition and the pattern of addition and growth in absorbance continued for the entirety of the reaction.
Fig. 2.
DpsC can mineralise iron using H2O2 as oxidant in a step-wise manner. a Ferroxidation reaction where Fe(II) was added in a step-wise manner, at a 31 Fe(II) atoms: cage ratio, and 200 µM H2O2. b DpsC forms a dodecamer able to deposit ferric oxide in the presence of H2O2 as oxidant. Blue Native PAGE (7 %) followed by coomassie (left) and iron staining (right)
This result is reminiscent of published observations on Dps proteins from S. solfataricus [38] and P. furiosus (39), where only H2O2 can efficiently mediate ferroxidation and ferrous iron was fed in a step-wise manner. The protein samples subjected to the above procedure were separated by Blue Native gel and stained with both Coomassie and potassium ferrocyanide. DpsC assembles as a dodecamer and deposits ferric oxide (Fig. 2b). We constructed a mutated DpsC protein lacking its long N-terminal tail, but further studies were unsuccessful as it was expressed in the form of an insoluble aggregate (not shown).
Full-length DpsA and C, and mutant DpsA variants were able to confer protection to DNA against free radical damage. Plasmid DNA incubated in the presence of Fe(II) and H2O2 suffered degradation, while the addition of protein preserved the integrity of DNA. This protection is mediated solely by the ability of the Dps proteins tested to inhibit Fenton chemistry, as no DNA gel retardation was observed (Supplementary Fig. 2). Although this suggests that no direct physical interaction takes place between the proteins and DNA in the conditions tested, further studies are needed to conclusively determine whether DpsA and C are unable to bind DNA, the association is of a transient nature or specific DNA topological conformations are required for the association to occur.
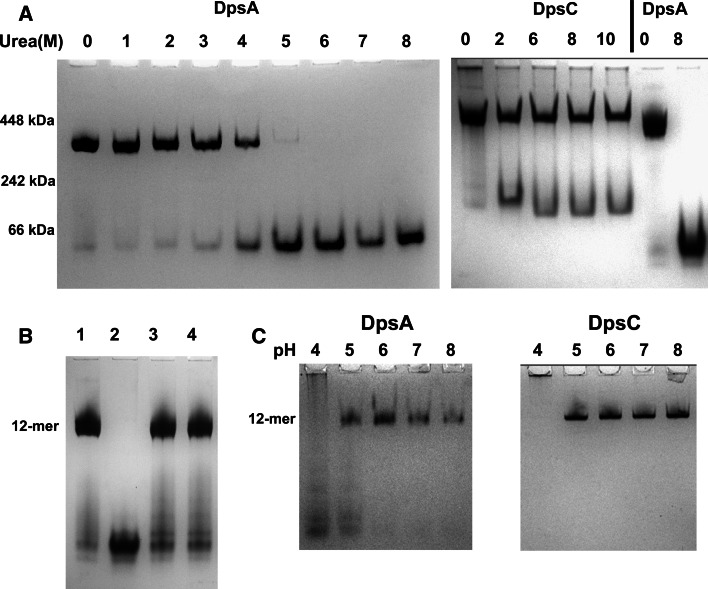
The stability of DpsA and DpsC dodecamers differs significantly. We proceeded to explore the conditions required to dissociate the dodecameric complex of both DpsA and DpsC. Remarkably, while DpsA can be fully dissociated by treatment with 5 M urea or higher, the DpsC dodecamer is less susceptible to this treatment and only a minor proportion of protein dissociates from the dodecameric complex throughout the range of concentrations tested, even when using high urea concentration (Fig. 3a). This partial dissociation can be explained by the presence of oligomers within the mixture where the interfaces responsible for holding the dodecameric structure are not well established, and therefore urea can induce conformational changes leading to dissociation into lower order oligomers like dimers.
Fig. 3.
DpsC is more stable than DpsA. DpsC and DpsA were incubated with various concentration of urea overnight and subjected to Blue Native PAGE (a). Urea molar concentrations are indicated, as well as position of molecular weight markers. Urea-denatured DpsA can be induced to refold and assemble as a dodecamer after buffer exchange (b). Lane 1 DpsA, lane 2 DpsA/8 M Urea, lanes 3, 4 DpsA after removal of urea by buffer exchange. DpsA dodecameric assembly is affected by low pH, while DpsC seems unaffected (c). pH is indicated over each lane. Notice that DpsC remains in the well at pH 4, probably due to aggregation at the low pH
The dissociation of DpsA following treatment with urea can be reversed, and dodecameric assembly can be restored by buffer exchange (Fig. 3b). Acidic pH also affects DpsA oligomeric status, as some dissociation occurs at pH 5 and lower, while DpsC is stable at low pH values (Fig. 3c).
Crystal structure provides clues to explain the differences in stability observed between DpsA and DpsC
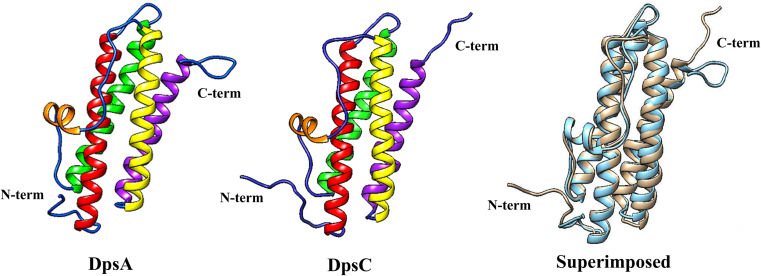
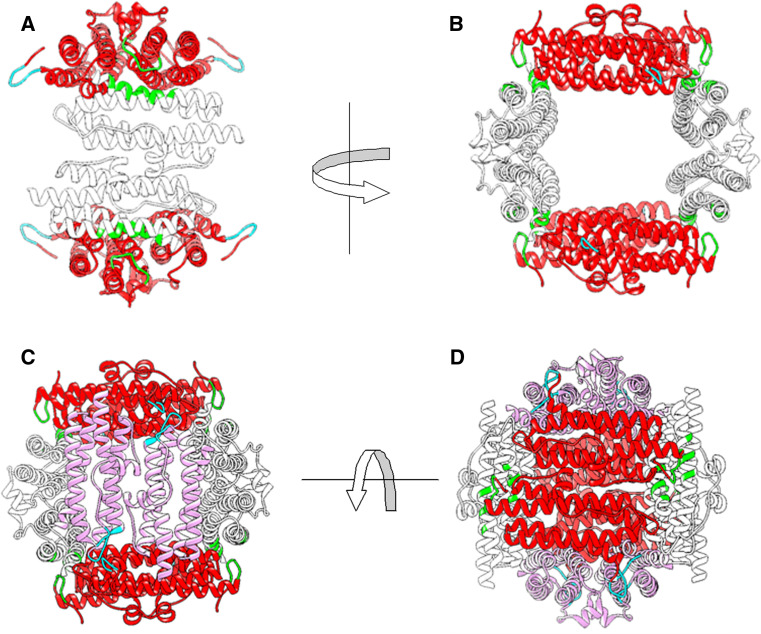
DpsA crystals diffracted to 1.78 Å and yielded one monomer per asymmetric unit with amino acids corresponding to 3–166 of the 187 amino acids present in the native DpsA protein sequence. Similarly to other Dps-like structures, the DpsA monomer shown in Fig. 4 is a square classed four-helix bundle with helices denoted as A, B, C and D which correspond to residues 16–46 (A), 52–80 (B), 107–134 (C) and 137–161 (D). There is an additional small BC helix, which is located perpendicular to the cylindrical four-helix bundle in the long loop between the B and C helices. This BC helix is composed of residues 88–94. There are non-helical regions located at the N-terminal (12 residues long) and C-terminal (9 residues long) regions which make up the features known as “tails”, in addition to loops located between helices. There are 18 residues from the C-terminal tail missing from the crystallographic model, with 3 amino acid residues missing from the N-terminal tail (Fig. 4). This inability to locate the terminal residues has been previously documented with other Dps protein crystal structures such as the Deinococcus radiodurans Dps, in which 163 out of 207 residues were modelled [40].
Fig. 4.
DpsA and DpsC monomer structures. Helices are shown in colour (a red, b green, bc orange, c yellow and d magenta). Superimposition of DpsA (blue) and DpsC (brown) is shown (RMSD 1.047, 136 atom pairs)
In the case of DpsC (diffracted to 1.78 Å Bragg spacing), helix A is composed of 31 amino acids (Met-45 to Leu-75) and helix B is 28 residues in length (Phe-81 to Gln-107). The BC helix runs from residues 117–123, the C helix is composed of 28 residues (Val-136 to Glu-163), while the D helix is 26 residues long (Asp-167 to Leu-192). Based on secondary structure prediction, DpsC possesses a very long N-terminal tail of approximately 44 residues. Analysis of DpsC crystals revealed that only 17 amino acids from this tail were visible in the electron density map, with the other 27 amino acids disordered in the crystal structure. DpsC is predicted to have an 8-residue C-terminal tail and seven of the eight residues were resolved (Fig. 4).
All Dps protein monomer structures available from the PDB were superimposed on top of DpsA and DpsC monomeric structures and the root mean square deviation (RMSD) of Cαs reported (data shown on Supplementary Table 2, superimposition of DpsA and DpsC in Fig. 4). The RMSD of DpsC superimposed on DpsA is 1.047 Å between 136 atom pairs. DpsA was found to be most similar to the M. smegmatis Dps (1VEI; [12]) with an extremely low RMSD of 0.68 Å between 160 atom pairs. Conversely, the Dps from B. anthracis (1JI5; [41]), when compared to DpsA, results in a higher RMSD (1.23 Å between 128 atoms). DpsC has a structure very similar to a Dps from the thermophilic cyanobacterium T. elongatus (2C41; [42]), with a very low RMSD of 0.761 Å between 130 atom pairs. The Dps from D. radiodurans (2C2F, [39]) was the most distinct from DpsC with a RMSD between 120 atoms of 1.16 Å.
DpsA and DpsC PDB files were submitted to the PDBePISA interactive tool for a detailed analysis of macromolecular interactions and interfaces [36]. Atomic co-ordinates were submitted with the intention of identifying interfaces present in the multimeric assemblies and residues involved in these interfaces. These interfacing regions may govern the strength of the oligomeric assembly as a whole. Three major types of interfaces highly conserved within the Dps family were predicted, namely the dimer interfaces at the twofold symmetry axes, the ferritin-like trimer interface and the Dps-like interface (Supplementary Table 3). It is noticeable that the area of the dimer interface in DpsC (1870.5 Å2, 51 residues per monomer) is significantly larger than that for DpsA (1310.3 Å2, 36 residues per monomer), correlating with the increased dodecamer stability of the former. Additionally, residues 28–33 from DpsC N-terminal tail participate in the dimer interface, extending over the opposite subunit of the dimer (discussed later).
The DpsA ferritin-like interface predominantly comprises residues from the CD loop and residues along the length of the D helix. Both N and C-tails have sections that contribute to the interface, with residues 8–14 of the N-terminal tail and 165–168 of the C-terminal tail (discussed in next section). The ferritin-like trimer interface of DpsC is the smallest of its interfaces in terms of area (735 Å2) and accounts for just 7.9 % of the solvent-accessible area of the protein monomer, but similarly to DpsA contains 42 residues. This interface is also predominantly hydrophilic in nature as evidenced by the high number of salt bridges and hydrogen bonds present at the interface: 9 and 8 in quantity, respectively. The N and C-terminal tails of DpsC also play a role in this interface, as observed for DpsA.
The C-terminal trimer complex, commonly referred to as the Dps-like interface, is the smallest interface found within the DpsA protein assembly in terms of surface area, at 512 Å2 per monomer. The interface is created by 26 residues from each monomer. The Dps-like trimer interface of DpsC is predicted to play a more significant role in maintaining oligomeric assembly than its equivalent in DpsA. In DpsC, all four major helices lend residues towards this interface. Furthermore, the entire C-terminal tail is involved in the interface, bridging over the adjacent subunit. The interface has an area of 820.4 Å2 per monomer and is created with 33 residues per subunit. Of significant interest is the hydrogen bond between His-199 (C-terminal tail) and Pro137 (start of the C helix), of adjacent subunits. This anchors the tail to the scaffold of the dodecamer, allowing it to support and stabilise its assembly. Since this is a threefold symmetry-related structure, the tails triangulate the entire interfacing region adding weight to their function as ‘braces’ for the dodecameric assembly. Additionally, the CSS score for this interface is also the highest of all the CSS scores, highlighting its significance.
Terminal tails participate in interfaces likely to contribute to dodecameric stability in both DpsA and DpsC
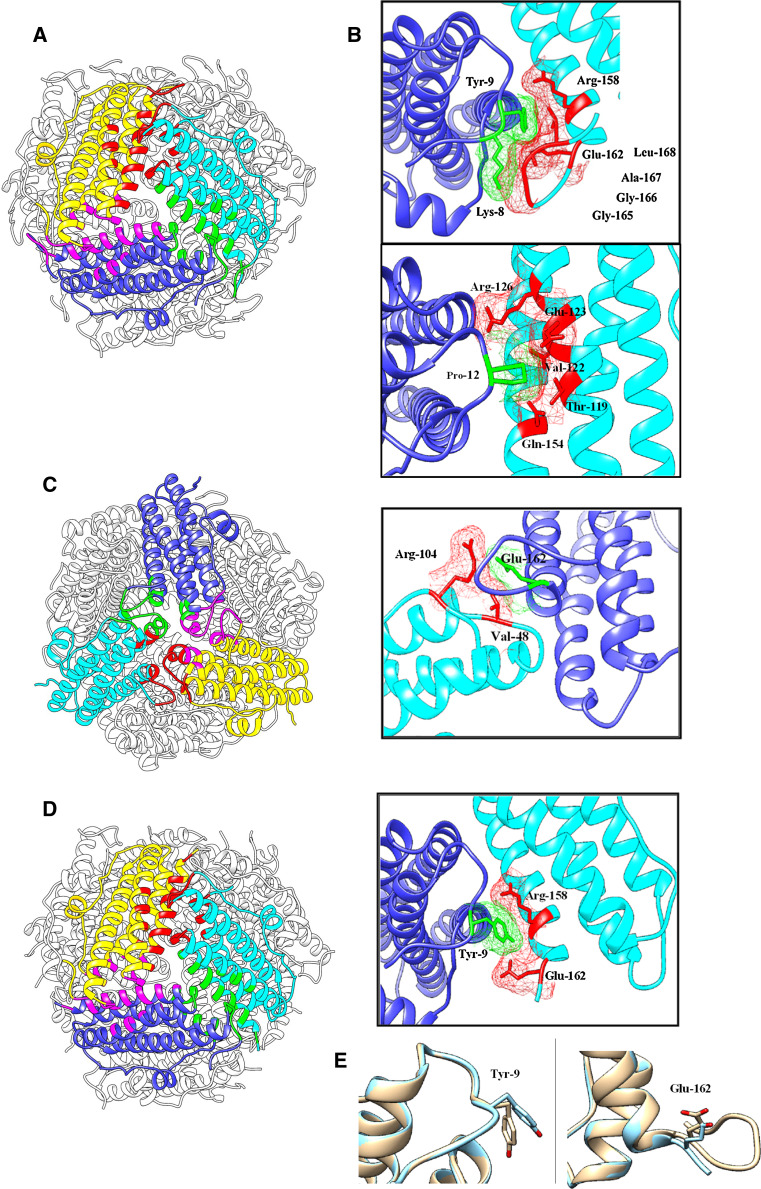
The structural analysis of both DpsA and DpsC revealed key roles for their respective terminal tails in terms of quaternary structure assembly, as briefly mentioned earlier. DpsA N-terminal tail is involved in two of the three structural organisations defined by the PISA analysis; the dimer interface and the ferritin-like trimer interface. Supplementary Table 4 highlights the interface in which each DpsA N-terminal residue contributes to and the amino acids with which they interface from a single symmetry-related subunit, as well as amino acids with which they form predicted hydrogen bonding interactions (Hb). Only one N-tail residue (Pro-7) is modelled to interact in the dimer interface, with Arg-104 within the symmetry-related partner. In the case of the ferritin-like trimer interface, although three subunits are involved, the N-tail only interacts with a single symmetry-related subunit, in a clockwise manner around the N-terminal pore. To paraphrase this, the N-terminal tail of one subunit interfaces with the C helix, the D helix and the C-tail from an adjacent subunit in a DpsA dodecamer.
Thr-10 interacts with Arg-158, linking the N-tail with the D helix of its symmetry-related partner at the N-terminal trimer interface, while Pro-12 and Arg-126 provide a link between the N-terminal tail and the C helix. Both Lys-8 and Tyr-9 are also heavily involved in interacting with the C-terminal tail of an adjacent subunit within the ferritin-like trimer. Lys-8 interfaces with three C-terminal tail residues (Gly-165, Gly-166 and Ala-167) and hydrogen bonds with Gly-165. Tyr-9 interfaces with residues Arg-158, Glu-162, Gly-166 and Leu-168, all but Arg-158 being part of the C-tail. Additionally, Pro-12 interfaces with 5 different residues (Thr-119, Val-122, Glu-123, Arg-126 and Gln-154) also accepting hydrogen bonding from Arg-126. Pro-12 is a hydrophobic residue and thus its location on the surface of the protein would be stabilised by the shielding achieved by the five residues with which it interfaces (Fig. 5a).
Fig. 5.
Interfaces contributed by DpsA N-terminal tails. Subunits involved in interfaces are shown in yellow, blue and cyan. Interfacing surfaces between corresponding subunits are shown in red, green and pink. Electron density in the vicinity of molecules is shown. a View of DpsA ferritin-like trimer interface. b Detailed view of N-tail residues participating in the interface. c Dps-like trimer interface and detailed view of interfaces involving C-terminal residues in adjacent panel. d DpsA-DCT ferritin-like interface. Adjacent to it a detailed view of the interface between Tyr-9, from one subunit, and Arg-158 and Glu-162 from a symmetry-related subunit. A hydrogen bond also occurs between Tyr-9 and Glu-162 that does not happen in DpsA. e Conformation of Tyr-9 differs in DpsA-DCT (blue) when superimposed with DpsA (brown). Glu-162 also shows conformation alterations in DpsA-DCT (blue) to DpsA (brown)
A similar analysis of DpsA C-terminal tail reveals interfaces involved in two oligomeric assemblies; the ferritin-like trimer and the Dps-like trimer interfaces (Supplementary Table 5). The former has already been mentioned above, while within the Dps-like trimer interface five C-terminal tail residues interface with Arg-104, which is situated in the loop between the BC helix and the C helix of a partner subunit. Glu-162, the first residue in the C-terminal tail, deserves special attention. It is involved in the formation of one hydrogen bond and two salt bridges with Arg-104, in addition to interfacing with Tyr-9 on the N-terminal tail (Fig. 5b). This suggests that interfaces involving Glu-162 may be important in maintaining the positioning of the C-terminal tail.
Removal of DpsA C-terminal tails seemed to affect oligomeric assembly in solution as described earlier for DpsA-DCT, although a cavity was formed by the mutant protein as revealed by ferric oxide deposition (Fig. 1d). The crystal structure of DpsA-DCT was obtained (PDB 4CYA) and its analysis showed, surprisingly, a dodecameric assembly highly similar to DpsA (Fig. 5d). A PISA analysis showed that the Dps-like trimer interface formed by DpsA-DCT is slightly smaller than its equivalent in DpsA, which is expected considering that C-terminal residues involved in the interface are missing in the former (Supplementary Table 3). In this mutated protein Glu-162 was the only C-terminal tail residue maintained. The interfaces between Lys-8 in the N-terminal tail and various C-terminal tail residues are missing, in particular a hydrogen bond with Gly-165. This suggests that these interfaces may not be as relevant in stabilising a dodecameric assembly. Interestingly, in DpsA-DCT the interfaces formed by Glu-162 are all preserved, and an additional hydrogen bond is predicted to form between Glu-162 and Tyr-9. This is likely a result of a slight change in conformations of Tyr-9 and Glu-162 in DpsA-DCT, and highlights the relevance of preserving a strong N-tail:C-tail interaction for the stabilisation of the dodecameric assembly (Fig. 5c; Supplementary Table 6).
As for DpsA, the DpsC N-terminal tail contributes to the formation and stabilisation of both the dimer interface and the ferritin-like trimer interface. In contrast to the DpsA N-tail, five N-tail residues are found to interface with the opposing subunit in the DpsC dimer, with a total of five hydrogen bonding interactions and a salt bridge predicted to form across the interface. Notably, Arg-28 forms two hydrogen bonds with Glu-135 from its dimer partner. Furthermore, Arg-28 also forms a salt bridge with the aforementioned Glu-135. Since the N-terminal tail extends across its partner in the dimer, this suggests that the tail is anchored in place across the surface of the dodecamer, acting as a ‘brace’ to stabilise the structure (Fig. 6a). Val-133 would also appear to be a key residue in this interface as it is involved with three of the five N-tail residues interfacing (Supplementary Table 7).
Fig. 6.
DpsC terminal tails contribution to dimer and Dps-like interfaces. Comparison of DpsA and DpsC dimer interfaces confirms different configurations for their respective N-terminal tails (a). The interaction between Arg-28 and Glu-135 contributes to the positioning of the N-tail in DpsC (b), strengthening the dimer interface. Dps-like trimer interface of DpsC (c) and detailed view (d) of interfacing residues from the C-terminal tail and contributing to the stability of this interface. Subunits involved in interfaces are shown in yellow, blue and cyan. Interfacing surfaces between corresponding subunits are shown in red, green and pink. Electron density in the vicinity of molecules is shown
Seven residues from DpsC N-tail play a role in the ferritin-like trimer interface, although they are different from the residues involved at the dimer interface. Ile-30 interfaces with a total of five amino acids and, due to its hydrophobic nature it would require shielding. This could in fact provide a method for correctly positioning the tail in place on the surface of the dodecamer allowing other vital interactions to occur. The DpsC C-terminal tail is shorter than the N-tail, and seven of the eight residues resolved within the structure interface with 18 interfacing residues. The interfaces within the ferritin-like trimer are on the boundary of distances qualifying as an interface and therefore were not considered significant to the stability of the interface. The C-terminal tail seems critical to the stability of the Dps-like trimer though, as each C-tail interfaces with independent subunits in an anti-clockwise circular manner around the C-terminal pore. One of such interactions is the predicted hydrogen bond between His-199 and Ser-141, pinning the tail to the surface of the threefold symmetry-related subunit (Fig. 6b; Supplementary Table 8). Additionally, Gln-83 takes part in a three-partite interface engaging the same residue from the three subunits. This interface is of high relevance for the stability of the Dps-like trimer, as it results in hydrogen bonding between these residues and other residues within the region.
Non-assembling DpsA mutant variants can be incorporated into a dodecameric structure when combined with full-length DpsA
Our earlier observations exploring the contribution of tails to DpsA dodecameric assembly revealed that the N-tail must be present to ensure stable oligomerisation. We then explored whether tail-less mutant variants, unable to assemble as dodecamers in solution under the conditions tested, could be incorporated within dodecamers containing full-length DpsA monomers. This hypothesis was tested by devising a ‘two dimension’ electrophoresis approach. Full-length DpsA and the truncated variant DpsA-DTM were denatured with 8 M urea. Equal concentrations of the denatured proteins were then mixed and subjected to refolding by buffer exchange, leading to the assembly of dodecamers. The refolded protein samples were loaded in adjacent lanes and separated by Native PAGE (first dimension). One gel lane was cut and stained with Coomassie, and aligned to the unstained lane to identify the position of high-order oligomers. The band corresponding to the high-order oligomer in the unstained lane was cut from the gel and placed on top of an SDS-PAGE gel (second dimension), followed by electrophoresis. The denaturing conditions provided by the SDS causes dissociation of the dodecamers, and as a result the monomers are separated by size.
When DpsA and DpsA-DTM were subjected to the above procedure, two distinct monomers can be recovered from the dodecamer resulting from refolding. One monomer corresponds to full-length DpsA, while the other is DpsA-DTM. Since both species were recovered from a band migrating at the size corresponding to a DpsA dodecamer, we can conclude that both DpsA and DpsA-DTM were able to associate into a hetero-oligomeric complex (Fig. 7a). As a negative control, we performed a similar experiment mixing DpsA and DpsB. The latter is unable to assemble into dodecamers in the conditions used [24]. Only DpsA was recovered from the high-order oligomer following SDS-PAGE, confirming that our earlier observations are not the result of non-specific aggregation of DpsA dodecamers to non-assembling variants, but that hetero-oligomeric assembly took place. A further evidence of this is the ‘ladder like’ appearance of the high-order oligomers observed after Native Page (Fig. 7, panel a lane 6), We interpret this heterogeneity in size a consequence of varying ratios of full-length and mutant DpsA variants within the hetero-oligomeric dodecamers formed. We observed no ‘ladder like’ migration when DpsA and DpsB were mixed (Fig. 7b lane 7).
Fig. 7.
Non-assembling DpsA mutant monomers can be incorporated into a dodecameric structure when combined with full-length DpsA. Proteins were denatured in 8 M Urea, and refolded by buffer exchange. Refolded protein mixtures were separated by Native PAGE, and high-order oligomers were cut out of the gel and subjected to SDS-PAGE (indicated by arrow). a Lanes 1 Native DpsA, 2 denatured DpsA, 3 Native DpsA-DTM, 4 denatured DpsA-DTM, 5–7 refolded DpsA-DCT and 8 refolded DpsA and DpsA-DTM mixture. A zoomed view of this region is shown, as well as the bands resolved from this complex after SDS-PAGE. b Lanes 1 Native DpsA, 2 denatured DpsA, 3 refolded DpsA, 4 native DpsB, 5 denatured DpsB, 6 refolded DpsB and 7 refolded DpsA and DpsB mixture. Position of DpsA, DpsA-DCT, DpsA-DTM and DpsB non-oligomerised monomers is indicated
Discussion
Dps-like proteins share a quaternary protein architecture which involves the self-assembly of 12 subunits to form a spherical protein cage [9]. The cage structure affords an environment suitable for the oxidation of ferrous iron with the internal cavity acting as a vessel for the formation and storage of a bioavailable iron oxide core. This detoxification process nullifies Fenton chemistry and consequently serves as the main cellular protective feature of Dps [43]. The study of Dps proteins has been largely confined to unicellular rod bacteria such as E. coli; however, the Dps from Streptomyces coelicolor represent an opportunity to explore their physiological contribution to a complex filamentous bacteria. Differential functional roles have been proposed for each ScDps [22], therefore the structural and functional characterisation of these proteins may shed light on their in vivo roles.
This work focused on the ScDps proteins (DpsA and DpsC) that assemble into high-order oligomers. DpsB can only assemble into likely dimers both in vivo and in vitro [24, this work], and therefore was not explored in detail. In particular, we aimed at defining the contribution of terminal tails to oligomeric assembly. Native PAGE and crystal structure studies confirmed the dodecameric organisation of DpsA and DpsC, and both proteins displayed the ability to oxidise ferrous iron and deposit ferric iron within the protein cage cavity, although interesting differences were observed. DpsA showed a preference for H2O2 as oxidant as described for other Dps proteins [44], although it can use air oxygen efficiently. The very fast rate of oxidation observed with H2O2 as oxidant is reminiscent of observations described for B. anthracis Dps (Dlp-1 and Dlp-2) [45]. Unexpectedly, DpsC precipitated upon addition of ferrous iron to the reaction mixture in similar conditions used for DpsA. Oxidation was eventually achieved by sequential addition of reduced quantities of iron to the reactions, together with H2O2. The precipitation could be attributed to a “salting out” effect as seen when using ammonium sulphate precipitation. The competition for water between ammonium sulphate and hydrophilic amino acid side chains on the surface of the protein results in a decrease of protein hydration. DpsC is enriched with an increased numbers of salt bridges, particularly at the dimer interface. However, the screening effect of ammonium ferrous sulphate may be enough to disrupt the bridges leading to secondary or higher structure disruption and thus formation of insoluble protein salt. Both DpsA and DpsC were capable of preserving the integrity of DNA by inhibiting Fenton chemistry reactions, although neither was able to bind DNA in the conditions tested. Furthermore, the oxidative protection assays did not reveal any band shifts suggesting that Dps–DNA interaction was not triggered by the presence of Fe2+ and H2O2 as required for the Campylobacter jejuni Dps–DNA interaction [46]. The ability to bind DNA by Dps proteins is usually mediated by charged residues at the terminal tails. In the case of DpsA and DpsC, the involvement of the tails in interfaces securing oligomeric assembly could explain a lack of flexibility in tail conformation, needed for DNA binding and reminiscent of the lack of DNA binding properties of A. tumefaciens Dps [47].
Removal of terminal tails affected oligomeric assembly in both DpsA and DpsC, although this was assessed in more depth in the former. Removal of DpsA C-terminal tail induces the formation of an oligomer of smaller size than full-length DpsA, although the mutated protein DpsA-DCT displayed ferroxidation and iron core deposition capabilities, indicative of the formation of a protein cage cavity. It is worth highlighting that Native PAGE does not provide an accurate assessment of molecular weight and therefore we limit our interpretations using this technique to whether the proteins under study were able to form high-order oligomers deemed to be dodecamers or not. The fivefold increase in specific activity displayed by DpsA-DCT, when compared to DpsA, suggests that an open cage structure is being formed in solution, such as the decamer observed in a M. smegmatis deleted tail mutant Dps [20]. Indeed our Native PAGE shows some degree of dissociation in DpsA-DCT, and indicates a less stable oligomeric assembly in this mutant protein while in solution. This may facilitate substrate access to ferroxidation sites in DpsA-DCT, accounting for its increase activity.
The DpsA N-terminal tail in particular plays a key role in maintaining dodecameric assembly, as its removal in DpsA-DNT prevented the formation of a high-order oligomer. Ferroxidase activity was preserved in this mutant variant, indicating that dimer formation took place. As expected, a mutant protein like DpsA-DTM lacking both tails is unable to oligomerise, although dimers are formed as evidenced by its ferroxidase activity. All DpsA mutant variants were able to protect DNA form oxidative damage, further confirming their ferroxidation potential.
These tail mutants, unable to oligomerise beyond a dimer, also displayed abnormal migration patterns in Native PAGE. Since size estimations based on Native PAGE are questionable, alternative techniques are usually required. The dimer interface formation is mediated by strong interactions, and therefore possesses considerable stability. Resistance of ferritin dimer dissociation has been observed in a range of treatments from low pH to incubation with denaturing agents such as urea [48]. Our data indicate that the dimer interface is stable even after urea treatment (Fig. 7a, DpsA-DTM lanes), as native and urea-denatured DpsA-DTM both migrate at the same position in the gel, and therefore we are witnessing dimeric organisation of the DpsA mutant variants unable to form high-order oligomers.
We consider that dimers are the ‘building blocks’ upon which dodecamers are assembled, by the incorporation of further dimers. Previous work has used Native PAGE to propose the occurrence of trimers as intermediate elements in the dodecameric assembly of a M. smegmatis Dps [18]. This interpretation arises from the observation of changes in oligomeric status, from a ‘trimer’ to a dodecamer after heat treatment. We believe this to be a misinterpretation of Native PAGE experiments, and the so-called trimers are in fact dimers migrating abnormally and displaying a larger molecular weight (curiously on par with BSA, as our experiments show for dimeric forms). Additionally, further studies of the same protein failed to show the so-called ‘trimer’ species [19]. A second argument could be made, inquiring about the very existence of a ‘free monomers’ available to associate into ‘trimers’. Considering the stability of the dimer interface, it seems unlikely that in the same solution dimers and monomers can coexist, as dimeric status will always be favoured.
The critical contribution of the terminal tails to dodecameric assembly is justified by their involvement in important interfaces contributing to oligomer assembly. In DpsA, the N-terminal tail seems to stabilise both the dimer and ferritin-like trimer interfaces. Each N-tail at the “ferritin-like” interface of DpsA overlaps the C-terminal region of a symmetry-related subunit four-helix bundle. Indeed the N-tail critical role is further supported by the inability of N-terminal deleted mutants to form dodecamers, and our inability to express a soluble N-terminal His-tagged DpsA (not shown). The presence of 6 histidines on this tail is likely to interfere with correct tail positioning and the correct assembly of the protein complex. This positioning is mediated, among others, by the interfacing between Tyr-9 at the N-tail and Glu-162 at the C-tail. The single residue left on the C-tail (Glu-162) of the DpsA-DCT mutant was found to be responsible for maintaining the interaction with the N-terminal tail. The preservation of the N-tail: C-tail interaction in the deleted C-tail mutant indicates how critical it is that the N-tail conformation is maintained. However, this interaction may not be sufficient to maintain a strong dodecameric assembly in solution, as our data show a small proportion of putative dimer dissociation in DpsA-DCT. Mutation of this apparently critical residue at the C-tail will conclusively confirm the role of this particular interface for overall assembly. In contrast with its interaction with the N-tail, the role played by the DpsA C-tail at the Dps-like interface seems less critical, as this interface is reliant on hydrophobic interactions that occur mainly between the four helix bundles. Thus the loss of the C-tail may only weaken this interaction without preventing its assembly, leading to successful assembly of dodecamers in DpsA-DCT.
In DpsC, the N-terminal tail is heavily involved in the dimer and ferritin-like interfaces, probably contributing to their respective stabilities. The crystal structure confirmed that tail residues were involved with numerous interactions across the surface of the dodecamer, acting as a brace to the dimer interface. The tail also assumes a role at the “ferritin-like” interface similarly interacting with C-tail residues. Combined together, these interface interactions support the central role for this tail in overall protein folding and dodecamer assembly, as reflected by the lack of success in preparing a soluble N-terminal DpsC mutant. Although we did not explore the role of the DpsC C-terminal tail in vitro, valuable observations may be inferred from the structure. Although only eight residues long, the PISA analysis of this tail showed that the seven residues resolved in the structure contribute to important interfaces. In particular, each tail braces its adjacent subunit in the Dps-like trimer interface, reinforcing it.
All the above structural features provide an explanation for the increased in vitro stability of DpsC, which we infer is a consequence of the increased interface areas between subunits and unique interactions mediated by tail residues. All the above supports our previous hypothesis, based on phylogenetic analyses, that DpsC has an extremophile origin and was laterally acquired by Streptomyces species [24]. Indeed, PISA analyses on a range of Dps proteins revealed a divide in terms of structural architecture, correlated with the mesophilic and thermophilic nature of the host organisms (Supplementary Table 9). The latter are characterised by an extended dimer interface, enriched with inter-subunit hydrogen bonds and salt bridges, thought to contribute to the protein stability differential between thermophilic and mesophilic species [49].
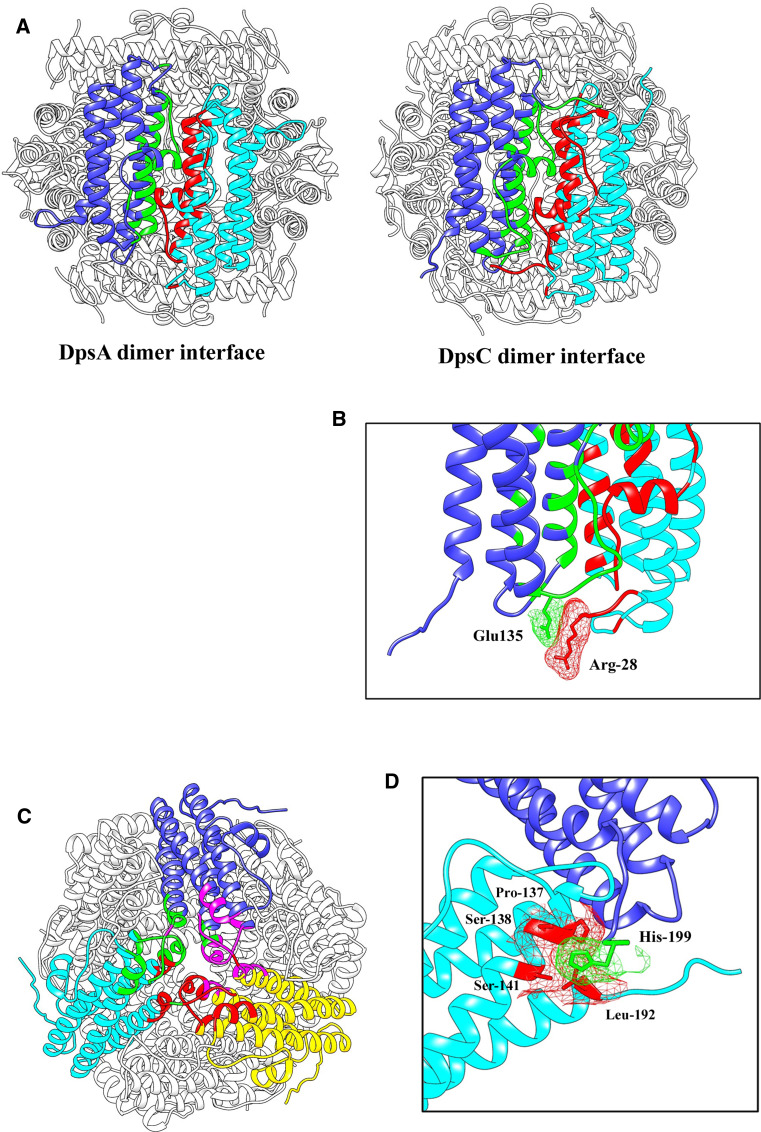
Probably the most interesting observation described here is the ability of non-assembling, tail-less DpsA variants to be incorporated into dodecamers when combined with full-length DpsA. While the structure analysis revealed the important role of the N-terminal tail for assembly, the formation of hetero-oligomers indicates that only a limited number of N-terminal tails is sufficient to stabilise a dodecameric assembly in DpsA. Although quantifying accurately the ratio of full-length:tail-less species in the hetero-oligomers formed lies outside the aims of this paper, we can speculate on how this is achieved. Based on our interpretation that urea denaturation preserves dimeric organisation, hetero-oligomeric assembly can only be supported by the formation of ferritin-like and ‘Dps-like’ trimer interfaces between full-length and tail-less dimers, with the tails stabilising such hybrid interfaces. A minimum of two full-length dimers would be sufficient to provide the required interfaces to stably hold a chimeric dodecamer. These dimers provide a total of eight terminal tails: 4 N-tails and 4 C-tails. Each N-tail can interact with a single DpsA tail-less monomer, while the C-tail can interact with a separate DpsA tail-less monomer, providing stability for a total of 4 DpsA tail-less dimers (Fig. 8).
Fig. 8.
Model representing the putative interfaces responsible for the stabilisation of DpsA: DpsA-DTM hetero-oligomers. Full-length DpsA dimers (red) are required to provide tails which stabilise a total of 4 DpsA-DTM dimers (white and pink). a Highlighted by the arrows are the N-tails of DpsA interfacing with the helix bundles of a DpsA-DTM dimer (green region). b Rotated view of image in a showing how the interfaces with the N-tails occur at both sides of the structure due to symmetry of the structure. c A further DpsA-DTM dimer (pink) interfaces with C-tails of DpsA (cyan region). d The dodecamer is completed with a fourth DpsA-DTM dimer (pink) which also interfaces with the C-tails of DpsA. All the interfaces are strengthened by inter-subunit hydrophilic interactions between the DpsA tails and the DpsA-DTM helical bundles. Images were created using UCFS Chimera 1.7rc
Interfaces that form at the ferritin-like interface between DpsA-DTM subunits and the N-tail of the DpsA subunits (green sections) are strengthened by inter-subunit hydrogen bonding interactions; namely Thr-10 at the N-tail to Arg-158 at the DpsA-DTM four-helix bundle, and Pro-12 at the N-tail to Arg-126 at the DpsA-DTM bundle (Fig. 8a, b). Because of the anti-parallel nature of the dimers and the symmetry that exists within the dodecamer, these interfaces also occur on the opposite side of the assembly between the same DpsA dimer and a different DpsA-DTM dimer. The incorporation of a further two DpsA-DTM dimers (pink) complete the dodecameric assembly and are held in place by hydrophilic interactions between Glu-162 of the C-tail from DpsA and Arg-104 located on the BC to C loop of the corresponding DpsA-DTM subunit. These represent the interactions that occur at the “Dps-like” interface (cyan sections) (Fig. 8c, d).
The above model does not attempt to infer the stages of the assembly and can only be confirmed by determining the crystal structure of individual chimeric species, which is not a trivial task due to the difficulties in isolation of different hetero-oligomeric combinations. Nevertheless, it provides a valid model to guide further exploration to confirm that tail mediated interactions are ultimately responsible for dodecameric assembly in S. coelicolor DpsA, as our data suggest. Additionally, we provide an explanation for the resilience of DpsC against denaturing agents, mediated by substantial subunit interactions further supported by both terminal tails. Considering the putative extremophile origin of DpsC and its orthologs, our observations suggest that such organisms are a promising source of Dps proteins, suitable for the engineering of nanocage-based scaffolds for biotechnological application requiring increased stability under harsh chemical environments.
Electronic supplementary material
Below is the link to the electronic supplementary material.
References
- 1.Groves M, Lucana DO (2010) Adaptation to oxidative stress by gram-positive bacteria: the redox sensing system HbpS-SenS-SenR from Streptomyces reticuli: Microbiology Series No. 2, vol. 1, Formatex, Spain, pp 1620
- 2.Mailloux RJ, Harper ME. Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic Biol Med. 2011;51:1106–1115. doi: 10.1016/j.freeradbiomed.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 3.González-Flecha B, Demple B. Metabolic sources of hydrogen peroxide in aerobically growing Escherichia coli . J Biol Chem. 1995;270:13681–13687. doi: 10.1074/jbc.270.23.13681. [DOI] [PubMed] [Google Scholar]
- 4.Luo Y, Han Z, Chin SM, Linn S. Three chemically distinct types of oxidants formed by iron-mediated Fenton reactions in the presence of DNA. Proc Natl Acad Sci USA. 1994;91:12438–12442. doi: 10.1073/pnas.91.26.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. doi: 10.1016/S0168-6445(03)00055-X. [DOI] [PubMed] [Google Scholar]
- 7.Abdul-Tehrani H, Hudson AJ, Chang YS, Timms AR, Hawkins C, Williams JM, Harrison PM, Guest JR, Andrews SC. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J Bacteriol. 1999;181:1415–1428. doi: 10.1128/jb.181.5.1415-1428.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gauss GH, Benas P, Wiedenheft B, Young M, Douglas T, Lawrence CM. Structure of the DPS-like protein from Sulfolobus solfataricus reveals a bacterioferritin-like dimetal binding site within a DPS-like dodecameric assembly. Biochemistry. 2006;45:10815–10827. doi: 10.1021/bi060782u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant RA, Filman DJ, Finkel SE, Kolter R, Hogle JM. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 10.Carrondo MA. Ferritins, iron uptake and storage from the bacterioferritin viewpoint. EMBO J. 2003;22:1959–1968. doi: 10.1093/emboj/cdg215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellapadrona G, Ardini M, Ceci P, Stefanini S, Chiancone E. Dps proteins prevent Fenton-mediated oxidative damage by trapping hydroxyl radicals within the protein shell. Free Radic Biol Med. 2010;48:292–297. doi: 10.1016/j.freeradbiomed.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 12.Roy S, Gupta S, Das S, Sekar K, Chatterji D, Vijayan M. X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules. J Mol Biol. 2004;339:1103–1113. doi: 10.1016/j.jmb.2004.04.042. [DOI] [PubMed] [Google Scholar]
- 13.Chiancone E, Ceci P. The multifaceted capacity of Dps proteins to combat bacterial stress conditions: detoxification of iron and hydrogen peroxide and DNA binding. Biochim Biophys Acta. 2010;1800:798–805. doi: 10.1016/j.bbagen.2010.01.013. [DOI] [PubMed] [Google Scholar]
- 14.Ceci P, Cellai S, Falvo E, Rivetti C, Rossi GL, Chiancone E. DNA condensation and self-aggregation of Escherichia coli Dps are coupled phenomena related to the properties of the N-terminus. Nucleic Acids Res. 2004;32:5935–5944. doi: 10.1093/nar/gkh915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stillman TJ, Upadhyay M, Norte VA, Sedelnikova SE, Carradus M, Tzokov S, Bullough PA, Shearman CA, Gasson MJ, Williams CH, Artymiuk PJ, Green J. The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding. Mol Microbiol. 2005;57:1101–1112. doi: 10.1111/j.1365-2958.2005.04757.x. [DOI] [PubMed] [Google Scholar]
- 16.Grove A, Wilkinson SP. Differential DNA binding and protection by dimeric and dodecameric forms of the ferritin homolog Dps from Deinococcus radiodurans . J Mol Biol. 2005;347:495–508. doi: 10.1016/j.jmb.2005.01.055. [DOI] [PubMed] [Google Scholar]
- 17.Reon BJ, Nguyen KH, Bhattacharyya G, Grove A. Functional comparison of Deinococcus radiodurans Dps proteins suggests distinct in vivo roles. Biochem J. 2012;447:381–391. doi: 10.1042/BJ20120902. [DOI] [PubMed] [Google Scholar]
- 18.Gupta S, Chatterji D. Bimodal protection of DNA by Mycobacterium smegmatis DNA-binding protein from stationary phase cells. J Biol Chem. 2003;278:5235–5241. doi: 10.1074/jbc.M208825200. [DOI] [PubMed] [Google Scholar]
- 19.Ceci P, Ilari A, Falvo E, Giangiacomo L, Chiancone E. Reassessment of protein stability, DNA binding, and protection of Mycobacterium smegmatis Dps. J Biol Chem. 2005;280:34776–34785. doi: 10.1074/jbc.M502343200. [DOI] [PubMed] [Google Scholar]
- 20.Roy S, Saraswathi R, Gupta S, Sekar K, Chatterji D, Vijayan M. Role of N and C-terminal tails in DNA binding and assembly in Dps: structural studies of Mycobacterium smegmatis Dps deletion mutants. J Mol Biol. 2007;370:752–767. doi: 10.1016/j.jmb.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Roy S, Saraswathi R, Chatterji D, Vijayan M. Structural studies on the second Mycobacterium smegmatis Dps: invariant and variable features of structure, assembly and function. J Mol Biol. 2008;375:948–959. doi: 10.1016/j.jmb.2007.10.023. [DOI] [PubMed] [Google Scholar]
- 22.Facey PD, Hitchings MD, Saavedra-Garcia P, Fernandez-Martinez L, Dyson PJ, Del Sol R. Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Mol Microbiol. 2009;73:1186–1202. doi: 10.1111/j.1365-2958.2009.06848.x. [DOI] [PubMed] [Google Scholar]
- 23.Facey PD, Sevcikova B, Novakova R, Hitchings MD, Crack JC, Kormanec J, Dyson PJ, Del Sol R. The dpsA gene of Streptomyces coelicolor: induction of expression from a single promoter in response to environmental stress or during development. PLoS One. 2011;6:e25593. doi: 10.1371/journal.pone.0025593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Facey PD, Hitchings MD, Williams JS, Skibinski DO, Dyson PJ, Del Sol R. The evolution of an osmotically inducible dps in the genus Streptomyces. PLoS One. 2013;8:e60772. doi: 10.1371/journal.pone.0060772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leong LM, Tan BH, Ho KK. A specific stain for the detection of nonheme iron proteins in polyacrylamide gels. Anal Biochem. 1992;207:317–320. doi: 10.1016/0003-2697(92)90018-3. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winter G, Lobley CM, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2010;69:1260–1273. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein N. CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J Appl Crystallogr. 2008;41:641–643. doi: 10.1107/S0021889808006985. [DOI] [Google Scholar]
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Colovos C, Yeates TO. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein–protein interfaces. Sci STKE. 2004;2004:l2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 38.Wiedenheft B, Mosolf J, Willits D, Yeager M, Dryden KA, Young M, Douglas T. An archaeal antioxidant: characterization of a Dps-like protein from Sulfolobus solfataricus. Proc Natl Acad Sci USA. 2005;102:10551–10556. doi: 10.1073/pnas.0501497102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramsay B, Wiedenheft B, Allen M, Gauss GH, Lawrence CM, Young M, Douglas T. Dps-like protein from the hyperthermophilic archaeon Pyrococcus furiosus . J Inorg Biochem. 2006;100:1061–1068. doi: 10.1016/j.jinorgbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Romao CV, Mitchell EP, McSweeney S. The crystal structure of Deinococcus radiodurans Dps protein (DR2263) reveals the presence of a novel metal centre in the N terminus. J Biol Inorg Chem. 2006;11:891–902. doi: 10.1007/s00775-006-0142-5. [DOI] [PubMed] [Google Scholar]
- 41.Papinutto E, Dundon WG, Pitulis N, Battistutta R, Montecucco C, Zanotti G. Structure of two iron-binding proteins from Bacillus anthracis. J Biol Chem. 2002;277:15093–15098. doi: 10.1074/jbc.M112378200. [DOI] [PubMed] [Google Scholar]
- 42.Franceschini S, Ceci P, Alaleona F, Chiancone E, Ilari A. Antioxidant Dps protein from the thermophilic cyanobacterium Thermosynechococcus elongatus. FEBS J. 2006;273:4913–4928. doi: 10.1111/j.1742-4658.2006.05490.x. [DOI] [PubMed] [Google Scholar]
- 43.Zeth K. Dps biomineralizing proteins: multifunctional architects of nature. Biochem J. 2012;445:297–311. doi: 10.1042/BJ20120514. [DOI] [PubMed] [Google Scholar]
- 44.Bellapadrona G, Stefanini S, Zamparelli C, Theil EC, Chiancone E. Iron translocation into and out of Listeria innocua Dps and size distribution of the protein-enclosed nanomineral are modulated by the electrostatic gradient at the 3-fold “ferritin-like” pores. J Biol Chem. 2009;284:19101–19109. doi: 10.1074/jbc.M109.014670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Kim K, Leighton T, Theil EC. Paired Bacillus anthracis Dps (miniferritin) have different reactivities with peroxide. J Biol Chem. 2006;281:27827–27835. doi: 10.1074/jbc.M601398200. [DOI] [PubMed] [Google Scholar]
- 46.Huergo LF, Rahman H, Ibrahimovic A, Day CJ, Korolik V. Campylobacter jejuni Dps protein binds DNA in the presence of iron or hydrogen peroxide. J Bacteriol. 2013;195:1970–1978. doi: 10.1128/JB.00059-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ceci P, Ilari A, Falvo E, Chiancone E. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage: x-ray crystal structure, iron binding, and hydroxyl-radical scavenging properties. J Biol Chem. 2003;278:20319–20326. doi: 10.1074/jbc.M302114200. [DOI] [PubMed] [Google Scholar]
- 48.Linder MC, Kakavandi HR, Miller P, Wirth PL, Nagel GM. Dissociation of ferritins. Arch Biochem Biophys. 1989;269:485–496. doi: 10.1016/0003-9861(89)90132-X. [DOI] [PubMed] [Google Scholar]
- 49.Kumar S, Nussinov R. Close-range electrostatic interactions in proteins. Chembiochem. 2002;3:604–617. doi: 10.1002/1439-7633(20020703)3:7<604::AID-CBIC604>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.