Abstract
Exosomes are secreted membrane vesicles of endosomal origin present in biological fluids. Exosomes may serve as shuttles for amyloidogenic proteins, notably infectious prions, and may participate in their spreading in vivo. To explore the significance of the exosome pathway on prion infectivity and release, we investigated the role of the endosomal sorting complex required for transport (ESCRT) machinery and the need for ceramide, both involved in exosome biogenesis. Silencing of HRS-ESCRT-0 subunit drastically impairs the formation of cellular infectious prion due to an altered trafficking of cholesterol. Depletion of Tsg101-ESCRT-I subunit or impairment of the production of ceramide significantly strongly decreases infectious prion release. Together, our data reveal that ESCRT-dependent and -independent pathways can concomitantly regulate the exosomal secretion of infectious prion, showing that both pathways operate for the exosomal trafficking of a particular cargo. These data open up a new avenue to regulate prion release and propagation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1945-8) contains supplementary material, which is available to authorized users.
Keywords: Prions, PrPres, PrPSc, Exosomes, ESCRT, Ceramide, Cholesterol
Introduction
Misfolding and aggregation of proteins are responsible for numerous neurodegenerative diseases, including Alzheimer’s and Parkinson’s diseases, amyotrophic lateral sclerosis (ALS) and infectious prion diseases [1]. In each case, one single or very few specific host protein(s) or peptides like Tau, α-synuclein, superoxide dismutase 1 (SOD1), Tar DNA-binding protein 43 (TDP-43), prion protein (PrP) or β-amyloid peptide (Aβ) misfolds from a soluble native conformation into a harmful aggregated state, rich in beta sheets [2–12]. Amplification of misfolding through seeded/nucleated polymerization leads to not fully defined toxic species and neuronal destruction. In transmissible spongiform encephalopathies (TSEs), a group of fatal neurodegenerative disorders that affect humans and animals, the conversion of the normal cellular prion protein (PrPC) into its pathological misfolded isoform PrPSc corresponds to the central event responsible for the pathology [13, 14]. PrPSc differs from its normal isoform by its high content in β-sheet structure, its insolubility and its partial resistance to proteinase K (PK) digestion. Although PrPres (the PK-resistant form of PrPSc) is tightly associated with prion infectivity, PK-sensitive PrPSc might be infectious as well [15]. The mechanisms that initiate the conversion of PrPC into its pathological isoform are still not fully understood but metabolism and trafficking of cholesterol were found both to play pivotal roles in prion conversion and/or degradation [16–20]. Both PrPC and PrPSc isoforms are associated with lipid rafts/membrane microdomains (DRMs) enriched in cholesterol and sphingolipids [20–22] and preventing PrPC raft association or destabilization of rafts by cholesterol depletion blocks PrPC to PrPSc conversion [19, 23].
The cellular pathways by which infectious prions are disseminated throughout the organism are also a matter of debate [24]. We and others showed that prion-infected cultured cells actively secrete exosomes bearing misfolded PrP protein and prion infectivity [25–31]. Similarly, several pathological misfolded proteins like Tau, α-synuclein, SOD1, TDP-43 or the Aβ peptide involved in the neurodegenerative diseases cited above were found to be released in association with exosomes, thus highlighting that a range of prion-like proteins with seeding activity may gain access to the extracellular space through their association with exosomes that can participate to their spreading [9, 24, 32–38]. In this context, infectious prions appear a promising paradigm to study exosome-mediated secretion of misfolded proteins.
Exosomes correspond to intra-luminal vesicles (ILVs) of endosomal/multivesicular bodies (MVBs) that are secreted upon MVB fusion with the plasma membrane [39]. The endosomal sorting complex required for transport (ESCRT) machinery is involved in MVB and ILV biogenesis [40, 41]. ESCRT machinery comprises approximately 20 proteins that assemble into four complexes (ESCRT-O, I, II and III) and additional regulatory proteins (Vps4A,B; Alix and VTA1) (reviewed in [42, 43]). Recently, selected components of the ESCRT machinery have been shown to modulate exosome secretion and composition [44, 45]. However, in mouse oligodendroglial cell line (oli-neu-cells), the release of exosome containing the proteolipid protein PLP was found to be independent of ESCRT function but rather requires ceramide synthesis through the hydrolysis of sphingomyelin by neutral sphingomyelinases (n-SMases) [46]. Additional players in ILV/exosome generation have also been recently identified [47, 48], indicating that host cell subpopulations of MVBs containing ILVs are generated by distinct mechanisms leading to the secretion of heterogeneous vesicles in terms of size and cargo composition.
Using RNA interference and pharmacological strategies, we investigated herein the participation of ESCRTs and ceramide on prion multiplication and release in Mov and Rov cellular models infected with the ovine prion strain 127S. Our data show that selected ESCRTs control prion multiplication and reveal for the first time that ESCRT-dependent and-independent pathways concomitantly modulate the exosomal secretion of infectious prions, indicating that both pathways operate for the exosomal trafficking of a particular cargo and opening novel insights to regulate prion release and propagation.
Results
Mov neuroglial cells as a model to study exosome-mediated prion release
The Mov neuroglial cell line was isolated from a transgenic mouse (see “Materials and methods”). These cells express the ovine, but not the murine PrPC protein and we previously showed that Mov cells efficiently replicate the ovine prion strain 127S [49] and release infectious prions in association with exosomes in the extracellular medium [28]. Extracellular vesicles/exosomes secreted from non-infected (Mov NI) and chronically infected Mov (Mov 127S) cells were harvested by differential ultracentrifugation as 100,000×g exosomal pellets (100 K pellet) and analyzed by Western blotting and electron microscopy (see Fig. 1). Western blotting analyses of the 100 K pellets indicated they are positive for exosomal proteins (including PrPC, Alix and Tsg101) but negative for the ER calnexin, a protein absent from exosomes (see lanes 2 and 4 in Fig. 1a and lanes 5 and 6 in Fig. 1b). To confirm that PrPres was associated with exosomes released by infected cells, the 100 K pellets from Mov NI and Mov 127S cells (lanes 5 an 6 in Fig. 1b) were PK-digested (see lanes 7 and 8) and analyzed by Western blotting. The data presented in Fig. 1b show the presence of PrPres only in cell lysates (lane 4) and 100 K pellets (lane 8) from Mov 127S cells. Morphological analysis of the corresponding 100 K pellets using transmission electron microscopy (TEM) reveals typical exosomal structures with a size ranging from 20 to 120 nm (Fig. 1c) [50]. Infectivity associated with 127S exosomes was then analyzed through a scrapie cell assay (SCA). In this procedure, the ovine RK13 (ovRK13) cell line permissive to the 127S prion strain [51, 52] was challenged by exosomal preparations from Mov 127S cells. Inoculated cells were maintained during four weeks and cell lysates were PK-digested and analyzed by Western blotting (Fig. 1d). The dose-dependent accumulation of aggregated PrPres in the challenged cultures (see lanes 1–4) indicates that infectivity in the 100 K pellets can readily be assessed with this cell-based assay. Importantly, no PrPres was detected when the cell-based assay was performed in the absence of doxycycline (-dox see lane 5) (i.e., when ovine PrPC was not expressed), demonstrating that PrPres was produced de novo by the inoculated cells and did not originate from the infectious inoculum.
Fig. 1.
Scrapie infected Mov cells release PrPres and infectivity in association with exosomes. a Cell lysates (20 μg) and the 100 K pellets (equivalent to 12 ml of conditioned medium) of uninfected Mov (NI) or infected Mov (127S) cells were analyzed by Western blotting using antibodies against PrP or Alix and Tsg101 as exosomal proteins or against Calnexin, an endoplasmic reticulum protein absent from exosomes. b PrPres is detected in the lysate (300 μg) and in the 100 K pellets (equivalent to 20 ml of conditioned medium) from Mov 127S cells. Cell lysates or 100 K pellets from Mov NI (lanes 1, 3, 5 and 7) or Mov 127S (lanes 2, 4, 6 and 8) cells were treated (+) or not (–) with PK before immunoblotting with antibodies directed against PrP or Alix as exosomal marker. c Electron microscopy analysis of the 100 K pellets from Mov NI (100 K NI) and Mov 127S (100 K 127S) cells. Scale bars represent 100 nm. d Cell-based assay of prion infectivity in the 100 K pellet from Mov 127S cells: increasing volumes of the 100 K pellet (corresponding to 17–450 μl of conditioned medium) of infected Mov 127S cultures were used to inoculate recipient permissive uninfected ovRK13 cells. Four weeks later, the challenged cultures were collected, solubilized and lysates (1000 μg) were digested with PK and processed for PrPres detection by immunoblotting. The dose-dependent presence of PrPres indicates that the 100 K pellet is infectious. No PrPres was detected when recipient ovRK13 cells that did not express the PrPC protein (-dox) were inoculated
HRS/Vps27 knockdown (KD) strongly inhibits PrPres and prion formation
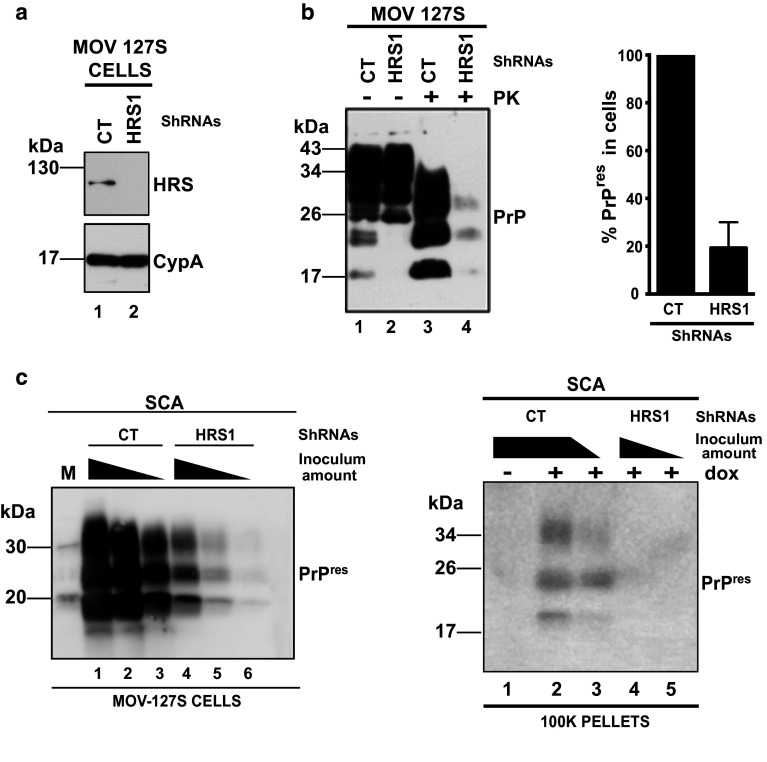
HRS/Vps27, a subunit of ESCRT-0 was previously shown to modulate exosome secretion [44, 45] in two different cellular models. To investigate the role of HRS on the release of infectious prions by Mov 127S cells, lentiviral vectors encoding HRS-ShRNAs (Sh-HRS) and a non-specific ShRNA target (Sh-CT), were used to transduce Mov 127S cells. Efficient HRS silencing with each of three different ShRNAs (see Fig. 2a and Fig. S1a left panel) leads to a strong reduction (80 %) of PrPres in the transduced cells (see Fig. 2b in left panel and graph in right panel; Fig. S1a right panel and see also the strong decrease of abnormal PrP staining after guanidine treatment in immunofluorescence analysis in Fig. S1b). This dramatic effect was not due to inhibition of PrP expression as total PrP levels were similar in Sh-CT and Sh-HRS cells (compare lanes 1 and 2 in Fig. 2b). Similar observations were made in neuroblastoma N2a cells infected with the murine prion strain 22L (scN2a#22L, see Fig. S1c) where HRS depletion did not change PrP levels (compare lanes 5 and 6) but strongly reduces PrPres in scN2a#22L (compare lanes 7 and 8).
Fig. 2.
HRS knock down (KD) inhibits accumulation of PrPres and infectivity. a Mov 127S cells were transduced with lentivectors encoding a non-specific shRNA (CT, lane 1) or a ShRNA directed against HRS (HRS1, lane 2). After puromycin selection cells were harvested and HRS depletion was detected by immunoblotting. Loading control was assessed with the cyclophilin A antibody. b HRS KD decreases PrPres. Cell lysates from infected Mov cells transduced with Sh-CT (lanes 1 and 3) and Sh-HRS1 (lanes 2 and 4) were analyzed for PrP before (lanes 1 and 2) or after (lanes 3 and 4) PK treatment. Note that HRS KD inhibits PrPres accumulation but has no effect on PrP expression. The graph on the right panel illustrates the PrPres decrease upon HRS silencing. The data shown are from six independent experiments. Values are given as means ± standard deviation (SD). c Inhibition of prion multiplication and release upon HRS KD, as assessed by Scrapie Cell Assay. Serial 1/3 dilutions of cell homogenates (left panel, corresponding to 30, 10 and 3.3 μg of proteins) from SH-CT (lanes 1–3) and Sh-HRS1 (lanes 4–6) or of the corresponding 100 K pellets (right panel, the equivalent of 150 and 50 μl of conditioned medium) were inoculated to target ovRK13 cells. PrPres in the infected cultures was analyzed 4 weeks later by immunoblotting. No PrPres was detected when recipient ovRK13 cells that did not express the PrPC protein (-dox) were inoculated. Note the dramatic decrease of infectivity in cells and 100 K pellets upon KD of HRS. M corresponds to molecular weight markers
To determine if inhibition of PrPres was associated with lower levels of infectivity, cell homogenates from Sh-CT and Sh-HRS transduced cells were tested using SCA. For this purpose, ovRK13 permissive cultures were challenged with cell homogenates from Sh-CT or HRS-depleted cells. Inoculated cells were maintained during four weeks and cell lysates were PK-digested and analyzed by Western blotting. The data revealed that ovRK13 cells inoculated with HRS-depleted cell homogenate accumulated much less PrPres, indicating that HRS-depleted cells were much less infectious (see Fig. 2c left panel, compare lanes 1–3 with lanes 4–6). Infectivity of the corresponding 100 K pellets was also measured by SCA. Four weeks after inoculation, PrPres analysis by Western blotting in the challenged ovRK13 cultures revealed that infectivity associated with the 100 K pellets was no longer detected upon silencing of HRS (see Fig. 2c right panel, compare lanes 2, 3 with lanes 4, 5). To determine if HRS depletion negatively affects exosomes release from Mov 127S cells, exosomes from Mov 127S-Sh-CT and Mov 127S-Sh-HRS1 cells (Fig. S2a) were isolated and analyzed by Western blotting using antibodies directed against Tsg101 and Flotillin-1 exosomal markers or Calnexin as negative exosomal control. The data presented in Fig S2b (representative of two independent experiments carried out with independently transduced cells) revealed no modification of Tsg101 and Flotillin-1 signals in the 100 K pellets from HRS-depleted cells (compare lane 3 with lane 4 in panel b) suggesting that HRS depletion does not affect exosomal release in the Mov 127S cellular model. This first set of data demonstrates that in various cell models, HRS is required for prion multiplication.
HRS silencing causes endosomal accumulation of cholesterol in infected cells
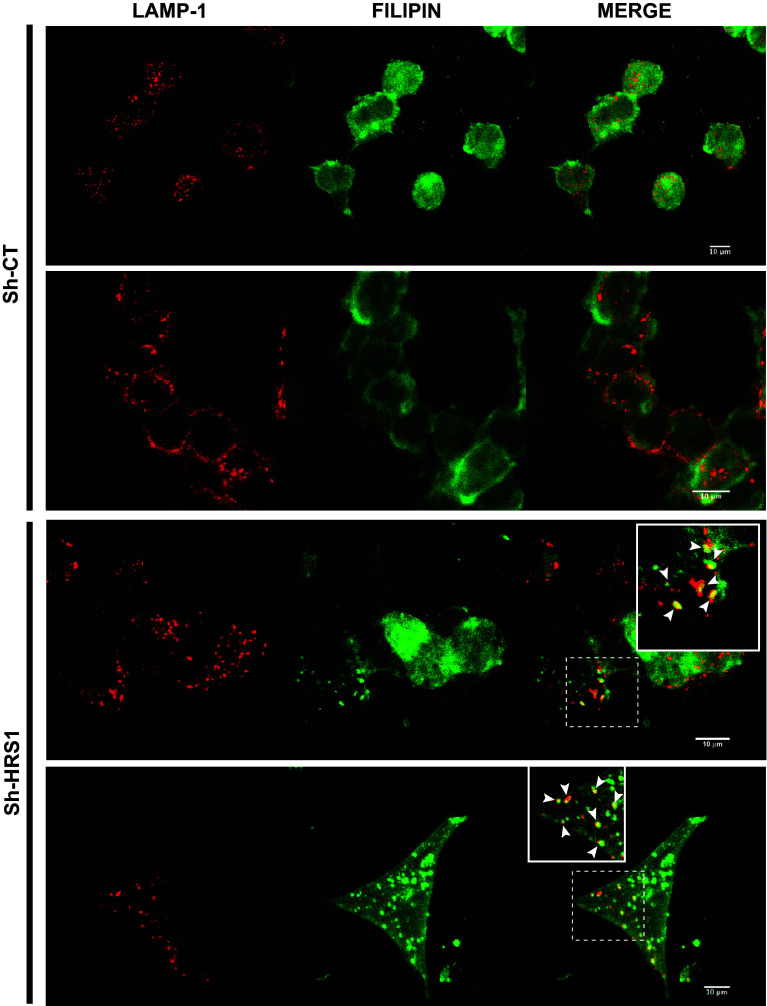
Recent data revealed that HRS is required for the transport of low-density lipoprotein-derived cholesterol from endosomes to the endoplasmic reticulum, providing evidence that HRS plays an essential role in cholesterol trafficking [53]. In HeLa cells, depletion of HRS and also of the 2 key cholesterol-sorting proteins NPC1 and NPC2, causes late endosome/lysosome (LE/LY) accumulation of LDL-derived cholesterol. In persistently infected neuroblastoma N2a or hypothalamic GT1-7 cells (scN2a#22L or scGT1-7#RML, respectively), depletion of NPC1 or treatment with the cholesterol synthesis/trafficking inhibitor U18666A leads to cholesterol accumulation in LE/LY and also to a strong decrease of PrPres [17, 54]. These observations prompted us to investigate free cholesterol trafficking in HRS-depleted Mov 127S cells. As shown in Fig. S3, Filipin (that labels free cholesterol) accumulated in intracellular compartments of HRS-depleted cells (see white arrows), whereas a diffuse signal was observed in Sh-CT cells. To determine if Filipin positive compartments colocalize with LE/LY compartments, co-labeling experiments using Filipin and the anti-Lamp1 antibody that labels LE/LY compartments, were carried out. The data presented in Fig. 3 revealed that Filipin positive compartments colocalize with those which are Lamp1 positive (see white arrowheads) confirming the previously published data in the HeLa cellular system [53]. Previous studies revealed that strong inhibition of PrPres observed in U18666A-treated cells or NPC1 knockdown cells can be attributed to increase PrPres degradation through the autophagy and/or lysosomal degradation processes [17, 54, 55]. Ammonium chloride (NH4Cl) is commonly used to impair lysosomal degradation, acting by alkalinizing lysosomes and preventing autophagosome fusion with lysosomes resulting in the accumulation of LC3-II (an autophagic marker) and p62 (an LC3 interactor known to be associated with cytoplasmic ubiquitin-positive structures). In this context, Sh-CT and Sh-HRS transduced cells were treated with NH4Cl (20 mM) for 16 h. The data presented in Fig. S4 confirm the efficient inhibition of lysosomal, autophagy and degradation processes as assessed by the accumulation of LC3-II and p62 in both Sh-CT and Sh-HRS transduced cells (compare lanes 1 and 2 with lanes 3 and 4, Fig. S4a in medium panels and lanes 1 and 3 with lanes 2 and 4 Fig. S4b). Analysis of PrP expression in treated cells also revealed an increase of total PrP (compare lanes 1 and 2 with 3 and 4 in Fig. S4a upper panel and lanes 1 and 3 with lanes 2 and 4 in Fig. S4b) indicating that inhibition of the lysosomal/autophagy process affects total PrP life cycle. However, analyses of PrPres when homogenates were PK treated, showed that NH4Cl treatment had no effect on the levels of PrPres that remained unchanged in Sh-CT and Sh-HRS treated cells, respectively (see Fig. S4b, compare lane 5 with lane 6 for Sh-CT and lane 7 with lane 8 for Sh-HRS1). Similar results were also obtained using other autophagy and lysosomal inhibitors like Chloroquine and Bafilomycin A1, an inhibitor of serine proteases like Leupeptine or by treating the cells with the proteasome inhibitor MG132 (data not shown). PrPres was also not recovered when cells were treated with NH4Cl, leupeptine or chloroquine during one week indicating that longer treatment is not sufficient to restore PrPres in HRS depleted cells (data not shown). Together, using this battery of drugs blocking lysosomal, autophagic or proteasomal degradation processes, and oppositely to what it was described in cholesterol synthesis and trafficking deficient infected N2a cells [17], we were unable to obtain in our system direct evidence that HRS silencing might stimulate PrPres degradation. We hypothesized that HRS depletion could negatively impact PrPres through an alternative mechanism.
Fig. 3.
HRS depletion causes accumulation of cholesterol in late endosomes/lysosomes in Mov 127S cells. Mov 127S cells transduced with lentivectors encoding Sh-CT (upper panels) or Sh-HRS1 (bottom panels) were fixed and labeled for Lamp1 (red) and for free cholesterol (Filipin, green). Note the co-localization between cholesterol and Lamp1 positive compartments (white arrowheads in the insets) in HRS-depleted but not in CT cells. Representative confocal images are shown. Scale bar represents 10 μm
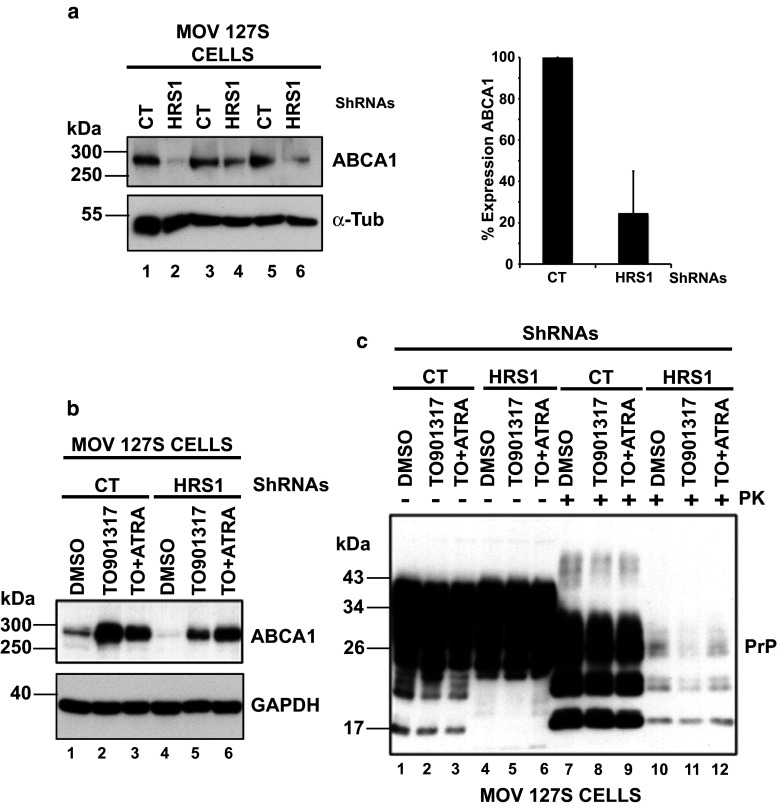
The expression of the ATP-binding cassette transporter A1 (ABCA1) is strongly decreased in HRS-depleted cells
The ABCA1 transporter, a key factor involved in cholesterol homeostasis, is involved in cholesterol efflux via its trafficking between the plasma membrane and intracellular compartments such as LE/LY. Mutations in this transporter result in Tangier Disease characterized by the accumulation of cholesterol in LE/LY [56]. Interestingly, ABCA1 expression is enhanced upon prion infection and its inhibition impairs prion multiplication in infected N2a cells [57]. Similarly, curing prion-infected cells with pentosan sulfate decreases ABCA1 expression [58], suggesting that ABCA1 expression and prion replication are tightly linked. The relationship between ABCA1 expression and prion multiplication prompted us to investigate ABCA1 expression in our HRS-depleted cells. Western blotting analyses indicated that ABCA1 expression was decreased by 70 % upon HRS depletion (compare lanes 1, 3 and 5 with lanes 2, 4 and 6 in Fig. 4a left panel and see the right panel graph from 5 independent experiments carried out with independent transduced cells) suggesting that ABCA1 could be a potential factor involved in PrPres decrease in our cellular system. To investigate if ABCA1 decrease could be causally linked to the strong reduction of PrPres in HRS-depleted cells, we used TO-901317 and all-trans retinoic acid (ATRA), two molecules that were previously found to stimulate the expression of ABCA1 [59]. Sh-CT and Sh-HRS depleted cells were treated with TO-901317 or a combination of TO-901317 + ATRA. The data presented in Fig. 4b (representative of three independent experiments carried out with independent transduced cells) showed that both treatments indeed enhanced ABCA1 expression in Sh-CT and HRS-depleted cells (compare lane 1 with lanes 2 and 3 for CT and lane 4 with lanes 5 and 6 for HRS-depleted cells). To determine if enhanced ABCA1 expression in HRS-depleted cells can restore PrPres, lysates of treated and untreated cells were PK digested and analyzed by Western blotting. The data presented in Fig. 4c failed to reveal any increase of PrPres in Sh-CT or Sh-HRS cells thus indicating that decrease of ABCA1 is not instrumental in PrPres inhibition (compare lane 7 with lanes 8, 9 and lane 10 with lanes 11, 12).
Fig. 4.
HRS depletion strongly reduces ABCA1 transporter expression. a HRS depletion decreases ABCA1 expression in Mov 127S cells. Sh-CT (lanes 1, 3 and 5) and Sh-HRS (lanes 2, 4 and 6) transduced Mov 127S cells were immunoblotted (left panel) for ABCA1 (upper panel) or for α-Tubulin (bottom panel) as a loading control. Note the strong decrease of ABCA1 expression in HRS-depleted cells in 3 independent experiments. The percentage of ABCA1 expression (right panel) in five independent experiments is shown. Values are given as means ± standart deviation (SD). b Mov 127S Sh-CT and Sh-HRS were treated with TO-901317 (TO, lanes 2, 5), TO + ATRA (lanes 3, 6) or with the DMSO (diluent) as negative control (lanes 1, 4) during 4 days. Cells were collected and lysates analyzed by Western blotting using anti-ABCA1 or GAPDH (lower panel, loading control) antibodies. c For PrP and PrPres detection, cell lysates from (b) were treated (+) or not (−) with PK before immunoblotting with anti-PrP antibodies
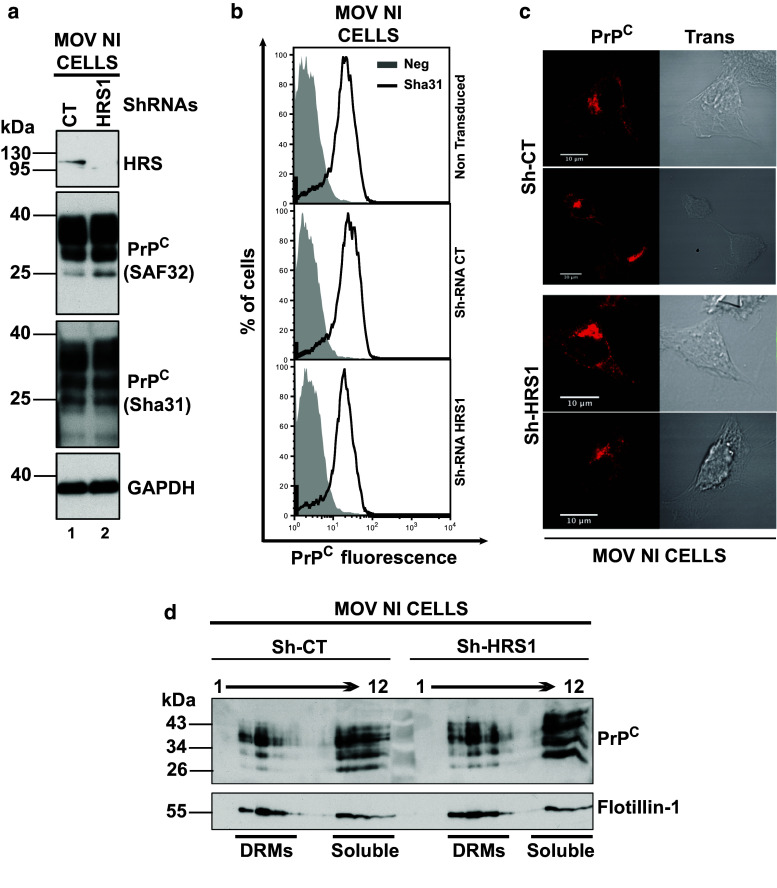
Cholesterol is incorporated into detergent-resistant microdomains (DRMs, i.e., rafts) that are essential for prion propagation [60]. To determine if HRS-mediated impairment of cholesterol trafficking would result in an altered association of total PrP to raft microdomains, DRMs were isolated from Sh-CT versus HRS-depleted Mov 127S cells. Data presented in Fig. S5a did not reveal any major effect of HRS depletion on the association of PrP to DRMs. Similar data were also observed when scN2a#22L infected cells were treated with cholesterol synthesis inhibitor U18666A [17]. The cellular distribution of PrP upon HRS silencing was also assessed by immunofluorescence microscopy and the data presented in Fig. S5b show that silencing of HRS did not induce any detectable alteration in the distribution of total PrP (compare total PrP signal in Sh-CT versus Sh-HRS1). A similar conclusion was also obtained upon silencing of HRS in Mov NI cells (Fig. 5). Immunoblotting analysis (Fig. 5a), cytometry (Fig. 5b) and confocal imaging (Fig. 5c) failed to reveal any detectable effect of HRS silencing on the levels (compare lane 1 with lane 2 in Fig. 5a and compare surface PrPC expression in Sh-RNA-CT and ShRNAs-HRS1 panels in FACS analysis in Fig. 5b) and on the subcellular distribution of PrPC (compare Sh-CT and Sh-HRS1 panels in immunofluorescence experiments in Fig. 5c). Similarly, association of PrPC to rafts remained similar in HRS-depleted cells (compare PrPC distribution in Sh-CT at the left with Sh-HRS1 at the right in Fig. 5d). Therefore, HRS depletion does not clear abnormal PrP by negatively affecting PrPC expression or trafficking. Since HRS silencing has no obvious impact on the expression and on the subcellular distribution of PrPC, nor on the PrPres degradation, we hypothesized that HRS depletion could negatively affect the conversion process itself (see “Discussion”).
Fig. 5.
Expression and distribution of PrPC in Mov NI cells is not affected by HRS depletion. a Mov NI cells cells were transduced with lentivectors encoding ShRNAs Sh-CT (lane 1) or Sh-HRS1 (lane 2). After puromycin selection, cells were harvested and cell lysates were immunoblotted with anti-HRS (upper panel), two different anti-PrP antibodies (SAF32, in second panel and Sha31 in third panel) or with anti-GAPDH for loading control (see bottom panel) antibodies. Note that PrPC expression is not altered in HRS-depleted cells. Immunoblot is a representative of 4 independent experiments carried out with independent transduced cells. b Expression of PrPC at the cell surface. Non-transduced Mov NI and Mov NI-ShRNA-CT/Mov NI-ShRNA-HRS1 transduced cells were collected in PBS-EDTA buffer and cell surface expression of PrPC was measured by FACS using the Sha31 anti-PrP antibody. Gray histograms represent not labeled cells. Solid black lines represent cell surface PrPC labeled cells. Note that HRS depletion does not affect PrPC cell surface expression. The data are representative of 3 independent experiments with independent transduced cells. c PrPC expression and distribution in Mov NI-ShRNA-CT and Mov NI-ShRNA-HRS1 transduced cells. Transduced cells were analyzed by confocal imaging using anti-PrP (SAF32) antibody. Left panels correspond to PrPC labelings (red) and right panels correspond to the transmission analyses of ShRNA-CT (upper panels) and ShRNA-HRS1 (bottom panels). Images are single sections through the middle of the cells. Representative images are shown. Scale bar 10 μm. d DRMs isolation from Mov NI-ShRNA-CT and -ShRNA-HRS1 transduced cells. Sh-CT and Sh-HRS1 cells were lysed in buffer containing 1 % triton ×100 at 4 °C. Equal amount of lysates were fractionated by flotation on a 5–30–40 % sucrose step gradient. Twelve fractions were collected from the top of the gradient and were analyzed by Western blotting using anti-PrP and anti-Flotillin-1 (as a DRM-associated protein) antibodies. DRMs are in fractions 3–5 while fractions 9–12 correspond to soluble proteins
Silencing of Tsg101 impairs the release of prion infectivity
The strong inhibition of prion multiplication upon HRS silencing led us to investigate additional components of the ESCRT machinery. It was recently reported that depletion of the ESCRT-I Tumor Susceptibility gene 101 (Tsg101) in HeLa CIITA cells, impairs the release of MHC class II-positive exosomes [44]. In addition, our previous studies showed that Tsg101 was associated with PrP-positive exosomes released by Mov cells [28]. This prompted us to investigate the role of Tsg101 on the release of PrPres and infectivity. For this purpose, Mov 127S cells were transduced with lentiviral vectors encoding negative control Sh-RNA or two different Tsg101 Sh-RNAs to silence Tsg101 expression. Western blotting indicated that the two-selected Tsg101 ShRNAs efficiently reduce the expression of Tsg101, while levels of total PrP did not change in the cells (compare lane 1 with lanes 2 and 3 in Fig. 6a and lane 1 with lanes 2 and 3 in Fig. 6b). Similar data were observed for PrPres when lysates were PK digested (compare lane 4 with lanes 5 and 6 in Fig. 6b). Similarly, SCA analysis of Sh-CT and Sh-Tsg101 cell homogenates also indicated that prion infectivity was similar in Tsg101-depleted cells (compare lanes 1–4 with lanes 5–8 in Fig. 6d), highlighting that HRS and Tsg101 depletions have strikingly different outcomes on prion multiplication. Biochemical analysis of the 100 K exosomal pellets shows, in absence of PK treatment, that total PrP as well as the exosomal Alix were strongly reduced in the 100 K pellets from Tsg101-depleted cells (compare lane 7 with lanes 8 and 9 in Fig. 6b and see the graphs in Fig. 6c left panel from 3 independent experiments). When exosome lysates from Sh-CT and Sh-Tsg101 were PK-digested, we also observed that Tsg101 depletion reduces PrPres by 70 % (compare lane 10 with lanes 11 and 12 in Fig. 6b and see associated graph in Fig. 6c right panel). SCA analyses carried out with the 100 K exosomal pellets revealed a 83 % decrease of infectivity release by Tsg101-depleted cells (see Fig. 6e, compare lane 2 with lane 3 in the left panel and see the associated graph in the right panel from 5 independent experiments). Altogether, these data indicate that Tsg101-mediated inhibition of exosomes release strongly inhibits infectious prion secretion.
Fig. 6.
Tsg101 depletion decreases infectivity release. a Mov 127S cells were transduced with lentivectors encoding Sh-CT (negative control, lane 1) or two different ShRNAs against Tsg101 (Sh-Tsg101-1, lane 2 and Sh-Tsg101-2, lane 3) and were puromycin-selected. Western blotting analysis of cell lysates (20 μg of proteins) shows that efficient inhibition of Tsg101 expression (top panel) does not affect PrP levels (middle panel). GAPDH (lower panel) was used as a loading control. b 100 K pellets were harvested from Mov 127S cells transduced with Sh-CT, Sh-Tsg101-1 or Sh-Tsg101-(1 + 2). Transduced cells and the corresponding 100 K pellets were analyzed before (−) and after (+) PK digestion for Tsg101 (upper panel), Alix (middle panel) and PrP (lower panel). As shown in (a), depletion of Tsg101 did not affect PrP expression (compare lane 1 with lanes 2–3) nor PrPres accumulation (compare lane 4 with lanes 5–6) in transduced cells. In sharp contrast, PrP and PrPres were strongly reduced in 100 K pellets from Tsg101-depleted cells (compare lane 7 with lanes 8–9 and lane 10 with lanes 11–12, respectively). c Left panel percentage of Alix release from Sh-CT versus Sh-Tsg101 cells. Right panel percentage of PrPres release from Sh-CT versus Sh-Tsg101 cells. The data are from three independent experiments. Values are given as mean ± SD. d Analysis of cell-associated infectivity by the scrapie cell assay. Serial 1/3 dilutions of cell homogenates (corresponding to 30, 10, 3.3 and 1.1 μg of proteins) from SH-CT (lanes 1–4) and Sh-Tsg101-1 (lanes 5–8) transduced Mov 127S cells were inoculated to target ovRK13 cells. PrPres in the challenged cultures was analyzed 4 weeks later by immunoblotting. Note that PrPres accumulation was similar in ovRK13 challenged with SH-CT or Sh-Tsg101 cell homogenates (compare lanes 1–4 with lanes 5–8) indicating that depletion of Tsg101 did not affect prion multiplication in the cells. M corresponds to molecular weight markers. e Analysis of 100 K pellet-associated infectivity by SCA. 100 K pellets from Sh-CT or Sh-Tsg101-1 transduced Mov 127S cells were inoculated to target ovRK13 cells and PrPres in the challenged target cells was analyzed 4 weeks later by immunoblotting. Note the strong decrease of PrPres signal indicating that the 100 K pellets from Tsg101-depleted cells are much less infectious (lane 3). No PrPres was detected when recipient ovRK13 cells that did not express the PrPC protein (-dox) were inoculated (lane 1). The graphic representation (right panel) shows the percentage of released infectivity in five independent experiments. Values are given as mean ± SD
Inhibition of the ceramide biosynthesis pathway decreases the release of infectious prions
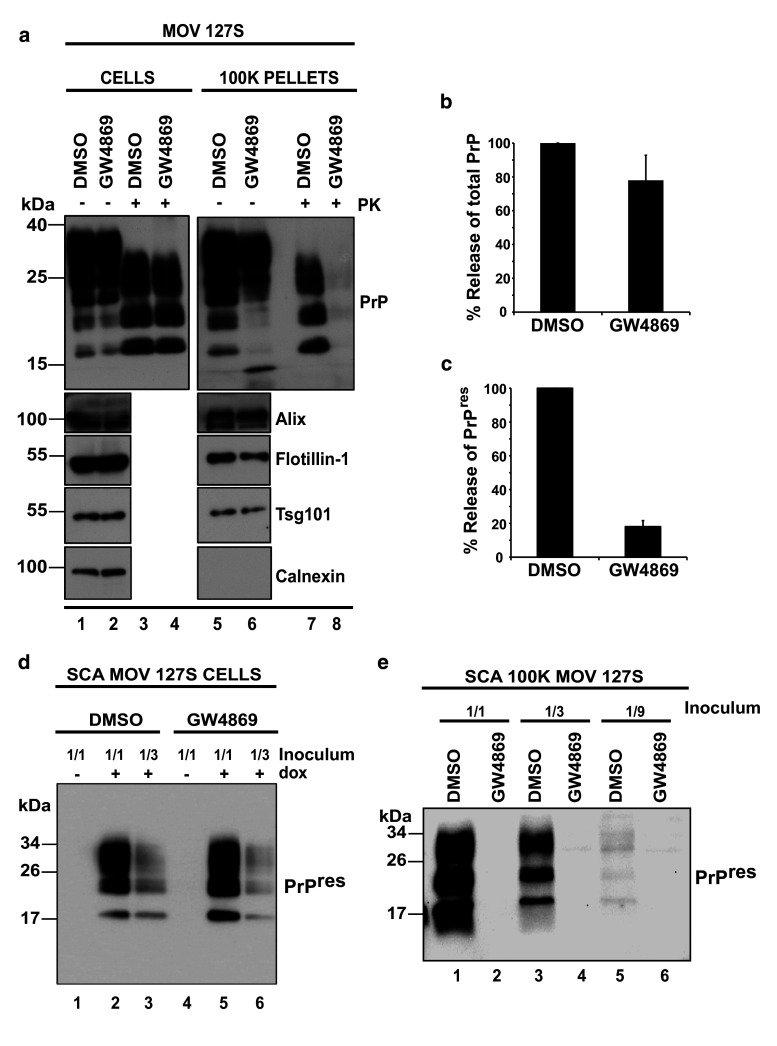
Ceramides are enriched in exosomes and their synthesis inhibition leads to reduction of exosome release [46]. To investigate whether infectious prions could also be targeted to exosomes through the ceramide pathway, Mov 127S cells were treated with the neutral sphingomyelinase (nSMase) inhibitor GW4869 (10 μM for 26 h). Cells and culture media were collected, the 100 K exosomal pellets were isolated and the samples were processed for total PrP, PrPres and infectivity analyses as described before. While cellular PrPres (compare lanes 3 and 4 in Fig. 7a) and infectivity (compare lanes 2 and 3 with lanes 5 and 6 in Fig. 7d) were essentially unaffected, GW4869 treatment dramatically affects the release of both PrPres (compare lanes 7 and 8 in Fig. 7a and see the graph in Fig. 7c from 3 independent experiments) and infectivity (compare lanes 1, 3 and 5 with lanes 2, 4 and 6, respectively, in Fig. 7e). In sharp contrast, release of total PrP or exosomal proteins such as Alix, Flotillin-1 or Tsg101 were marginally (20–30 % of decrease) or not affected (compare lane 5 with lane 6 in Fig. 7a and see the graph in Fig. 7b from 3 independent experiments for total PrP in exosomes or graphs in Fig. S6 for exosomal markers from 4 independent experiments) by GW4869 treatment. These data suggest that PrPres and associated infectivity were selectively excluded from exosomes when cells are treated with the GW4869 (see “Discussion”).
Fig. 7.
The neutral Sphingomyelinase inhibitor GW4869 strongly reduces prion infectivity release. a Mov 127S cells were incubated with the diluent (DMSO, negative control; lanes 1 and 3) or with the neutral Sphingomyelinase inhibitor GW4869 (10 μM; lanes 2 and 4) for 26 h. Cells were recovered and the 100 K pellets were harvested from the corresponding culture supernatants. Cell lysates and the 100 K pellets were immunoblotted before (−) and after (+) PK digestion for PrP, Alix, Flotillin-1 or Tsg101 (exosomal proteins) and Calnexin (non-exosomal protein). PrPres was no longer detected in the 100 K pellets from GW4869-treated cells (compare lanes 7 and 8), suggesting inhibition of release. b Percentage of total PrP in the 100 K pellets from DMSO/GW4869-treated cells. The data are representative of three independent experiments. Values are given as mean ± SD. Note that total PrP release is marginally affected in GW4869 treated cells. (c) Percentage of PrPres in the 100 K pellets from DMSO/GW4869 treated cells. The data are from three independent experiments. Values are given as mean ± SD. Note the strong decrease of PrPres release in GW4869-treated cells. d Prion infectivity associated with cells, as revealed by the SCA. SCA was performed with 1/1 and 1/3 homogenate dilutions (corresponding to 30 and 10 μg of cellular proteins) of DMSO- and GW4869-treated Mov 127S cells. DMSO- (lanes 2 and 3) or GW4869-treated cells (lanes 5 and 6) were used to infect ovRK13 target cells. Four weeks later, inoculated cells were recovered and lysates were PK-digested and analyzed for PrPres. No PrPres was detected when recipient ovRK13 cells that did not express the PrPC protein (-dox) were inoculated (lanes 1 and 3). e Experiments similar to (d) were carried out with the 100 K pellets. Decreasing amounts (1/1, 1/3 and 1/9) of the 100 K pellets (corresponding to 450, 150 and 50 μl of conditioned medium) from DMSO (lanes 1, 3 and 5) or GW4869 (lanes 2, 4 and 6) treated Mov 127S cultures were used to inoculate recipient permissive uninfected ovRK13 cells. Four weeks later, inoculated cells were recovered and lysates were PK-digested and analyzed for PrPres. Note that GW4869 strongly impairs the release of infectivity
To extend these findings, we used ovRK13 cells as another cell model permissive to the ovine 127S prion strain. Chronically infected ovRK13 epithelial cells (Rov 127S) were treated with GW4869, 100 K exosomal pellets and the corresponding secreting cells were isolated for biochemical analysis and infectivity assessment by the SCA. As observed with Mov 127S cells, GW4869 treatment did not affect infectivity levels in ovRK13 target cells (compare lanes 2, 4, 6 with lanes 3, 5, 7 in left panel of Fig. S7a) but dramatically inhibited infectivity released in the extracellular medium (compare lane 8 with lane 9 in right panel of Fig. S7a). In sharp contrast, and in line with data obtained with Mov 127S cells (Fig. 7), GW4869 treatment did not or marginally inhibit the release of other exosomal proteins (Alix, Flotilin-1 or Tsg101) (Fig. S7b). These data indicate that inhibition of the neutral sphingomyelinase pathway strongly impairs the release of infectious prion in two different cellular models.
Discussion
The cellular mechanisms by which infectious prions are disseminated throughout the organism are not fully understood. Infectious prions can be transferred by direct cell–cell contacts [61, 62] and via nanotubes [63] but also in a contact independent manner via exosomes [25–30, 51, 64, 65] or microvesicles [66]. Interestingly, exosomes also transport several other prionoids (SOD1, TDP-43, α-synuclein, Tau and the β-amyloid peptide) involved in neurodegenerative diseases such as ALS, PD and AD, respectively, and have been proposed to play a role in their in vivo spreading [9, 33, 35, 36, 38] for review [24, 37].
One may envision that inhibition of the function of cellular machineries involved in exosome biogenesis and/or release could impair the release of these pathological proteins thus limiting their dissemination throughout the organism. In this study, we chose infectious prions as models to establish the role of the exosomal pathway in the release of pathological misfolded proteins.
Here, we present data highlighting that both ESCRT-dependent and -independent pathways participate in the multiplication of infectious prions and/or their secretion. We found that silencing of HRS ESCRT-0 strongly decreases the formation of the cellular PrPres and infectivity and consequently impacts on the release of infectious prions. It was recently reported that HRS depletion affects cholesterol trafficking and leads to its accumulation in LE/LY [53]. Interestingly, cholesterol was previously well characterized as a key factor involved in the conversion of PrPC into PrPSc (for review [19]). Perturbation of cholesterol synthesis and trafficking prevents prion conversion or its degradation. Inhibition of intracellular biosynthesis and/or transport of LDL-derived cholesterol by drugs (U18666A and hydroxy tamoxifen, i.e., OHT) or inhibition of NPC1 (Niemann Pick C1), a protein involved in the export of cholesterol from LE/LY to the endoplasmic reticulum and plasma membrane, induces a drastic decrease of PrPres in infected culture cells [17, 54, 55]. The strong decrease of PrPres in U18666A-treated cells was linked to a stimulated PrPres degradation in a lysosomal/autophagosomal-dependent manner [17]. On the opposite, treatment with 4-hydroxy tamoxifen (4-OHT) leads to prion degradation independently of an autophagic process [55]. We observed a strong decrease of PrPres upon HRS depletion. Using a battery of drugs known to block lysosomal, autophagic, or proteasome degradation processes, we failed to restore PrPres in our HRS-depleted cells despite that total PrP signals, much alike autophagic markers like p62 and LC3-II were significantly accumulated in treated cells. These data suggest that these degradation pathways have only minor impacts on PrPres half-life in Mov cells. Similar data were also observed for Rov 127S cells [67]. However, these data also suggest that HRS depletion could negatively impact PrPres through a mechanism distinct from its degradation.
Filimonenko et al. (2007) [68] revealed that functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. In their study, they found that depletion of ESCRT components inhibits the autophagic degradation. Therefore, in HRS-depleted cells, the decrease of abnormal PrP is most likely unrelated to the degradation process and rather points to a defect in the conversion of PrPC to abnormal PrP.
During the course of this study, we also found that HRS-depleted cells show a decreased expression of the ABCA1 transporter. ABCA1 is essential for cholesterol homeostasis and when mutated, is responsible of the Tangier Disease characterized by the accumulation of cholesterol [56]. Other diseases characterized by cholesterol accumulation in LE/LY with a link to decreased ABCA1 expression and activity are Niemann Pick disease (NPD) type C and B [69, 70]. Noteworthy, two independent studies revealed that ABCA1 expression is tightly linked with level of PrPres [57, 58]. Depletion of ABCA1 by RNA interference decreases PrPres [57] and on the opposite, curing prion infection with pentosan sulfate decreases ABCA1 expression [58], thus suggesting that ABCA1 could be a potential candidate involved in the strong decrease of PrPres we observed in HRS-depleted cells. The mechanism by which ABCA1 could be regulated by HRS is not yet resolved. To investigate if the decrease of ABCA1 expression could explain the strong reduction of PrPres, we stimulated ABCA1 expression with two molecules (TO-901317 and ATRA) previously shown to enhance ABCA1 expression [59]. Treatment of HRS-depleted cells with these molecules failed to restore PrPres, thus indicating that ABCA1 is not the only factor responsible for the observed decrease of PrPres in HRS-depleted cells.
Because rafts/DRMs are enriched in cholesterol and play a crucial role in the conversion of PrPC to abnormal PrP, we analyzed the expression level, the cellular distribution and the association of total PrP and PrPC with DRMs in infected and non-infected Mov cells, respectively. Our data failed to reveal evident differences for these three parameters suggesting that neither the expression level nor the distribution of PrPC are key factors involved in the observed PrPres decrease in HRS-depleted cells. However, at the present, we cannot exclude that some raft-associated co-factors essential for the conversion process might be affected by HRS depletion.
In contrast to the observations made after HRS silencing, depletion of the Tsg101-ESCRT-I subunit impairs the release of PrPC, PrPres and infectivity without affecting expression and formation of PrPC and PrPSc in transduced cells. Although HRS and Tsg101 are essential players in MVB biogenesis, they play distinct roles in cargo selection and ILVs formation [71]. We previously showed that Tsg101 depletion decreases exosome secretion [44]. We confirm these findings in Mov cells and further show that targeting Tsg101 strongly impaired prion infectivity release. Thus, our data suggest that release of infectious prions associated with exosomes requires Tsg101/ESCRT-I. Interestingly, we observed that PrPC release was also negatively affected in Sh-Tsg101-Mov NI cells, indicating that the effect of Tsg101 is not specific to PrPSc (data not shown). Although we obtained evidence for an ESCRT-dependent secretion of prions, it is of interest that inhibition of the ceramide biosynthesis pathway using the nSMase GW4869 inhibitor did not affect prion multiplication in the cells but dramatically inhibited their release. In Mov and Rov cells, but in contrast to other cellular models [46, 64], the release of several exosome-associated proteins (Alix, flotillin-1 and Tsg101) was only marginally (20–30 %) decreased by GW4869 treatment. Our data suggest that ceramide may participate into the differential exosomal recruitment of total PrP and PrPres/infectivity. During the preparation of this manuscript, Guo et al. (2014) also reported that the neutral sphingomyelinase pathway was involved in the release of PrPres by GT1-7 cells infected by the M1000 prion strain [64]. Together, all these observations indicate that different prion strains can hijack the ceramide pathway in several cell types to promote their exosomal release. Interestingly, we observed that recruitment of total PrP in exosomes was marginally affected in GW4869 treated cells in contrast to the findings in Tsg101-depleted cells (see above). These data indicate that ESCRT-dependent and -independent pathways can concomitantly regulate the packaging and the release of infectious prions in a different manner suggesting that the simultaneous targeting of these different pathways could efficiently impair prion multiplication and spreading.
It is increasingly recognized that exosomes are in fact a heterogeneous population of vesicles, possibly released from different types of MVBs [44]. More specifically, we don’t know yet whether all secreted vesicles harbor prion or if infectivity is specifically associated to a subset of these vesicles. Interestingly, in a preliminary set of experiments, we found that Mov 127S cells release smaller vesicles compared to those from uninfected cells, suggesting that the set of secreted vesicles is affected by prion infection. Future investigations (Nanoparticle Tracking Analysis NTA-Nanosight technology and Electron Microscopy associated with immunogold labeling for abnormal PrP detection) will be necessary to confirm this observation and to determine whether GW4869 treatment inhibits the release of a subset of vesicles carrying infectious prions.
Recent studies indicated that prion infection of cells is extremely rapid and that the plasma membrane is the primary site of prion conversion of PrPC into PrPSc [72] when cells are exposed to exogenous prions. Other studies indicated that conversion might be initiated in recycling endosomes in the neuroblastoma and hypothalamic cellular models [54, 73]. Our data on HRS silencing supports an additional site of conversion as cellular components involved in the late endosomal pathway play key roles in the formation of PrPSc and infectivity. After submission of our study, Yim et al. (2015) proposed multivesicular bodies as the major internal site of prion conversion [74]. In their study, in perfect agreement with inhibition of PrPres by HRS silencing in Mov 127S cells, they observed that HRS depletion strongly decreases PrPres in infected N2a and SMB cellular models. They further found that affecting proper maturation of the MVB is detrimental for abnormal PrP propagation. The exact mechanisms by which HRS controls MVB maturation and hence the PrPC to abnormal PrP conversion process are not known. Proper targeting to MVB of cofactor(s) essential for conversion might be involved as well as (in addition or in conjunction) with defects in cholesterol distribution observed in HRS-depleted cells. However, depending on the cell type, conversion may preferentially occur in one or several subcellular sites. The conversion process can also differ among prion strains and thus be differentially dependent on the recycling or the late endosomal pathways. On this line, we recently found that prion strains can be differentially released through the exosomal pathway [75].
An emerging view is that a growing number of pathological misfolded proteins can amplify inside the cells through seeded/nucleated polymerization. However, how misfolded proteins gain access to the extracellular space and to new cells for further amplification is a critical issue under intense investigations. Cell culture studies recently identified exosomes/EV as potential shuttles for cell-to-cell spreading of several protein aggregates. However, in the absence of experimental tools to specifically inhibit exosome release, the biological relevance of exosome-mediated release of misfolded proteins in disease progression remains uncertain. In this study, we provide experimental evidence that extracellular release of actively multiplying prions can be substantially inhibited. Even if prions can potentially be transferred by cell contacts [61, 62] and/or nanotubes [63], impairing prion release through the exosomal pathway opens up a new avenue for interventional strategies.
Materials and methods
Cells
Mov cells are immortalized neuroglial cells isolated from a transgenic mouse expressing ovine VRQ allele of PrP [49] and were kindly provided by Hubert Laude and Vincent Béringue (INRA Jouy-en-Josas). Rov cells are derived from the rabbit kidney epithelial RK13 cell line and express the ovine VRQ allele of PrP in a doxycycline-dependent manner [52]. N2a cells (clone N2a#58) and scN2a#22L lines have been described previously. N2a#58 cells overexpress murine PrPC and scN2a#22L are N2a#58 cells infected with brain homogenates from mouse adapted scrapie strain 22L [25, 76]. Mov and Rov cells are permissive to the 127S ovine prion strain.
Cells were cultured in Opti-MEM-1 (Invitrogen) in the presence of 10 % fetal calf serum, 2 mM l-glutamine and 50 units/ml penicillin/50 μg/ml streptomycin (fullOptiMEM medium). For exosome production, cells were cultured in bovine exosome depleted medium (exofree medium). The exofree medium was obtained after ultracentrifugation of the fullOptiMEM medium at 100,000×g during 16 h and filtrated through a 0.20 μm unit.
Antibodies
Primary antibodies: the monoclonal SAF32 and Sha31 antibodies directed against prion proteins were provided by J. Grassi and the SpiBio company (Bertin Technologies). The anti-HRS was kindly provided by Sylvie Urbé and was from Millipore (cat#ABC35). The anti-TSG101 was from Aviva Systems Biology (ARP37310_T100) or from Santa Cruz (C2 sc7964). The anti-Cyclophilin A (CypA) was kindly provided by P. Gallay (Scripps, California). The anti-Alix (pab0204) was from Covalab, the anti-ABCA1 (AB.H10) (ab18180) from abcam, the anti-Calnexin from Stressgen (SPA-865), the anti-Flotillin-1 from Sigma Aldrich (F1180), the anti-SQSTM1/p62 (GP62-C) from Progen Biotechnik, the anti-LC3I,II (LC3B) from Cell Signaling and the anti-Lamp1-488 (sc19992-488) from Santa Cruz Biotechnology. Flotillin-1 and Alix (H-270) antibodies used in Fig. S7b were from BD transduction laboratories and Santa Cruz Biotechnology, respectively.
Secondary antibodies: the anti-Mouse-HRP (NA931) and the anti-Rabbit-HRP (NA934) were from Amersham and were used in Western blotting. The APC-linked goat anti-mouse used in FACS analyses was from Southern Biot (ref 1012–11). For immunofluorescence experiments, the donkey anti-mouse alexa fluor 555 and the donkey anti-rabbit alexa fluor 488 were from Molecular Probes.
Plasmids
The psPAX2 plasmid is a 2nd generation lentiviral packaging construct encoding the HIV-1 Gag, GagPol, Tat and Rev accessory proteins, the pMDG2 plasmid encoding the vesicular stomatitis envelope glycoprotein (VSVg) were provided by Didier Trono. The pLKO1 constructs encoding Small hairpin RNAs directed against HRS (TRCN0000088688; TRCN313945; TRCN314016), TSG101 (TRCN0000054603; TRCN0000054607), were from Sigma-Aldrich (Mission ShRNA plasmid DNA). The negative ShRNA control was a gift from Clotilde Thery and was described in [77]. The mix of psPAX2, pVSVg, pLKO1 constructs were used to generate lentiviral vectors encoding ShRNAs.
Lentivector vector production and RNA interference
The lentiviral and retroviral vector particles were produced by transient transfection of the packaging construct (HIV-1 psPAX2), a minimal genome (HIV-1 pLKO1) bearing the expression cassette encoding the ShRNAs control (CT) or against ESCRT components and the plasmid encoding the VSV-G-envelope expressing plasmid pMDG2 (DNA ratio 8:8:4 μg) into 293T cells (3.5 × 106 cells plated 1 day before transfection in 100-mm dishes) by the calcium phosphate method [78]. Viral particles were normalized by an exogenous reverse transcriptase assay and titrated on non-infected Mov cells. Transductions of Mov NI and 127S cells were carried out with different MOI in 6 well plates (200,000 cells plated 1 day before) in presence of 6 μg/ml of polybrene overnight. One day after transduction, cells were passaged and cultured in presence of puromycin (3 μg/ml) during 3 days until the death of non-transduced control cells was complete. To avoid clonal selection, we selected Mov NI or 127S cells transduced with MOI (>1). In these conditions, less than 10 % of transduced cells died after puromycin selection compared to 100 % of non-transduced cells. Efficiency of the knockdown was monitored by Western blotting using the anti-HRS and anti-TSG101 antibodies.
Drugs and treatments
Infected Mov 127S cells were treated with 10 μM of GW4869 (D1692, Sigma-Aldrich) diluted in DMSO or with DMSO alone in exofree medium for 26 h. For lysosomal/autophagy and proteasomal degradation inhibition, Mov-ShCT and Mov-ShHRS cells were treated with NH4Cl (20 mM), BafilomycinA1 (10 or 40 nM) or Chloroquine (10 or 100 μM), leupeptine (15 or 100 μM) or MG132 (8 μM) for 16 h to 7 days. Cells were recovered and the presence of total PrP and PrPres was determined by Western blotting. Lysosomal and proteasomal degradation inhibition was validated by measuring the accumulation of p62 and LC3-I,II ratio by Western blotting.
TO-901317 (T2320) and the all-trans retinoic acid (ATRA; T1850000) were purchased from Sigma Aldrich and were resuspended in DMSO. Mov Sh-CT and Mov Sh-HRS were treated for 4 days with TO-901317 (4 μM) and ATRA (5 μM) or with DMSO alone as negative control.
Exosome preparation: differential centrifugation
The Mov cells (2 millions at day 0) were cultured in 20 ml of exofree medium for 4 days in 2xT162 cm2 flasks. Media (40 ml) were subjected to differential centrifugations (3,000×g 5 min, 4,500×g 10 min, 10,000×g 30 min and 100,000×g 70 min). The resulting exosomal pellet was washed in 1× PBS and ultracentrifuged at 100,000×g during 70 min. The final pellets (100 K pellets) were resuspended in 100 μl of 1× PBS. 50 μl (equivalent to 17.5–20 ml of conditioned medium) were used to PK treatment and 20–30 μl for exosomal markers by Western blotting. 1–2 μl of exosomal resuspension (equivalent to 0.4–0.8 ml of conditioned medium) was used for the Scrapie Cell Assay (SCA) and 5–10 μl (equivalent to 2–4 ml of condionned medium) for EM analyses.
Proteinase K (PK) treatment
Cells and exosomes (100 K pellets) were resuspended in PK lysis buffer (0.5 % sodium deoxycholate, 0.5 % Triton X-100, 150 mM NaCl and 50 mM Tris–HCl, pH7.5.) and submitted to PK digestion (16 μg of PK/mg of protein) during 30 min at 37 °C. For cellular lysates, 300 μg of proteins were used. For 100 K pellets, 50 μl were mixed with an uninfected cellular lysate containing 300 μg of proteins. PK digestion was stopped at 4 °C by addition of 0.1 M PMSF. PrPres was pelleted at 20,000×g during 1 h at 4 °C. The pellet was resuspended in loading buffer and analyzed by WB.
Detergent-resistant microdomains (DRMs) isolation
DRMs from Mov127S-Sh-CT versus Mov127S-Sh-HRS1 cells or Mov NI-Sh-CT versus Mov NI-Sh-HRS1 were isolated as previously described [79]. Briefly, cells were washed and scraped with ice-cold PBS. Pelleted cells were lysed on ice for 20 min in TNE (25 mM Tris HCl pH7.5; 150 mM NaCl; 5 mM EDTA) containing 1 % Triton X-100 and protease inhibitor cocktail. The post-nuclear supernatant was recovered and protein concentration was determined. Equal amount of protein lysate was diluted in 60 % sucrose (wt/wt) to obtain a 45 % sucrose concentration in 3 ml. The lysate was overlaid with 6 ml of 30 % TNE sucrose and 3 ml of 5 % TNE sucrose in an SW41 Ti ultracentrifuge tube. The step gradient was centrifuged for 22 h at 180,000×g (avg rcf) at 4 °C. Twelve 1 ml fractions (excluding the pellet) were collected from the top of the gradient. 30 μl of each fraction was analyzed by Western blotting using the anti-PrP mix (Saf32 and Sha31) and the anti-Flotillin-1.
Immunofluorescence and confocal microscopy imaging
Mov Sh-CT and Sh-HRS cells were grown on 12-mm-diameter coverslips in six-well plates. Immunofluorescence (IF) staining was performed at room temperature. The cells were washed with PBS, fixed with 4 % paraformaldehyde in 1× PBS for 15 min, quenched with 50 mM NH4Cl, permeabilized by 0.1 % Triton X-100 for 5 min. For total PrP or PrPC detection in Mov 127S and Mov NI, respectively, cells were labeled for 1 h in 1× PBS containing 1 % BSA with the primary antibody (SAF32 or Sha31). Cells were washed and stained for 45 min with the anti-mouse alexa fluor 555 secondary antibody. For PrPSc detection, cells were treated with guanidine thiocyanate 3 M during 5 min. After extensive washing in 1× PBS, cells were blocked 1 h in 1× PBS containing 1 % BSA and were labeled for 1 h with the primary antibody mix (Sha31 anti-PrP and anti-Caveolin-1) in PBS-BSA. Cells were washed and stained for 45 min with the corresponding fluorescent secondary antibody in PBS-BSA (anti-mouse alexa fluor 555 for Sha31 and anti-rabbit alexa fluor 488 for the anti-Caveolin-1).
For Filipin staining, cells were rinsed with cold 1× PBS and fixed 20 min with 3 % PFA at room temperature. Cells were washed in PBS and treated 10 min with PBS-glycine (1.5 mg/ml) at room temperature. Cells were washed with PBS and stained with a Filipin-III solution (0.05 mg/ml; FluoProbes) in 1× PBS containing 10 % of FCS during 75 min at room temperature.
Cells were then washed with PBS and stained for 1 h with the anti-Lamp1-Alexa488. After washing with PBS, coverslips were mounted with immu-mount medium (Thermo Scientific). Images were acquired using the Spectral Confocal Microscope (TCS SP5 AOBS) or with the confocal microscope Zeiss LSM-510 (PLATIM platform from ENS-Lyon).
Western blotting
Cells were collected after treatment with 1× PBS-EDTA 1 mM on ice. Cell lysis was realized in PK buffer for 20 min at 4 °C. Post-nuclear supernatant was obtained after centrifugation of lysates at 800×g during 10 min at 4 °C. Protein concentration was determined with the Bradford reagent (Biorad). Western blotting procedures have been previously described [79].
In the absence of PK treatment, 20–30 μg of cell lysates and 30 μl of 100 K pellets (equivalent to 12 ml of conditioned medium) were loaded for analysis by SDS-PAGE of exosomal and non-exosomal markers.
Electron microscopy
The 100 K pellets (5–10 μl of exosomal resuspension) were gently resuspended and fixed with a mixture of 2 % paraformaldehyde/0.065 % glutaraldehyde in 0.2 M phosphate buffer at 4 °C. The suspension (5–10 μl) was loaded on to formvar–carbon-coated EM grids and fixed a second time. Samples were contrasted and embedded in a mixture of uranyl acetate and methylcellulose, and viewed under a CM120 electron microscope (Philips, Eindhoven, The Netherlands).
Scrapie cell assay (SCA)
Cell pellets to be assayed were resuspended in sterile PBS and homogenized with a high-speed homogenizer (TeSeE Precess 48 system). The cell-based assay procedure has been described in details previously [51]. Briefly, the assayed samples, i.e. identical quantities of cell homogenates and 100 K pellets harvested from identical quantities of cell culture media were diluted to 3 ml with cell culture medium containing 1 μg/ml dox. All these samples were incubated with ovRK13 cells grown in individual wells of 6-well plates. One week later, the media were renewed. Infection was allowed to proceed for 3 more weeks with one medium change per week. At the end, cell cultures of each well were rinsed with cold PBS and solubilized for 10 min at 4 °C in 1 ml of Triton-DOC lysis buffer (50 mM Tris/HCl (pH 7.4), 0.5 % Triton-X100, 0.5 % sodium deoxycholate). The lysates were clarified by low speed centrifugation (425×g, 1 min) and cellular proteins in the post-nuclear supernatants were quantified by bicinchoninic acid (BCA, Pierce). Digestion of 1000 μg of proteins with PK (recombinant grade, Roche) was performed for 2 h at 37 °C with a mass ratio of 4 μg of PK per mg of cellular proteins and the reaction was stopped by addition of Pefabloc (Sigma–Aldrich) to 4 mM. PK-digested samples were centrifuged for 30 min at 20,000×g and pellets were analyzed for PrPres by Western blotting.
FACS analysis
Mov NI cells transduced by ShRNA-CT and ShRNA-HRS1 lentivectors were collected using 1× PBS containing 1 mM EDTA, counted and fixed 15 min at room temperature in 4 % paraformaldehyde and then extensively washed in 1× PBS. The surface staining against PrPC was performed by the mouse anti-PrP Sha31 antibody (1 h at room temperature; dilution 1/100 in 1× PBS, 1 % BSA). After extensive washing steps in 1× PBS, the primary antibody was revealed by APC-linked anti-mouse (1/500 in PBS-1 %BSA) during 45 min at room temperature. For negative cells, transduced cells were labeled with the secondary antibody. After three washing steps, cells were analyzed on a FACScalibur (Becton–Dickinson Biosciences) collecting 10,000 events. The data were analyzed using the FlowJo software.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material Figure S1: HRS depletion inhibits PrP res formation. (a) Mov 127S cells were transduced with lentivectors encoding ShRNAs Sh-CT (lane 1) or three different Sh-HRS (1, 2 and 3; lanes 2-4). Cell lysates (20 μg) were immunoblotted using anti-HRS (upper, left panel) or anti-PrP antibodies (bottom, left panel). Cell lysates (300 μg) were PK digested and immunoblotted using anti-PrP antibodies (right panel). Note the strong decrease of PrPres in Sh-HRS1, 2 and 3 (lanes 2-4) compared to Sh-CT (lane 1). Non-PK digested Sh-CT lysate was used as a negative control of PK digestion (lane 5). (b) Mov 127S cells transduced with lentivectors encoding Sh-CT or Sh-HRS1 were fixed and analyzed by confocal microscopy using anti-PrP (red) and anti-Caveolin1 (green) antibodies. Images are single sections through the middle of the cells. Abnormal PrP was detected after incubating the cells 5 min with 3 M guanidine thiocyanate. Note the strong decrease of abnormal PrP signal in Sh-HRS1 cells. Nuclei were stained with DAPI. Scale bars represent 10 μm. (c) Infected scN2a#22L cells were transduced with ShRNAs Sh-CT (non-specific target negative control; lanes 5 and 7) or Sh-HRS1 (lanes 6 and 8). After puromycin selection, cells were harvested and HRS depletion was assessed by immunoblotting (upper panel). Note the strong decrease of HRS signal (lane 6) compared to CT ShRNA (lane 5). Loading control was assessed using coomassie staining (bottom panel). Cell lysates from Sh-CT (lane 7) and Sh-HRS KD (lane 8) cells were PK digested and PrPres was detected by Western blotting. Note the strong decrease of PrPres signal in HRS-depleted cells (lane 8) compared to control cells (lane 7). The normal N2a#58 (lanes 1 and 3) and infected N2a#22L cells (lanes 2 and 4) were used as negative and positive controls for PK digestion and PrPres detection. (EPS 13072 kb)
Supplementary material Figure S2: HRS depletion does affect exosomal release. (a) Mov 127S cells were transduced with lentivectors encoding ShRNAs Sh-CT (lane 1) or Sh-HRS1 (lane 2). Cell lysates were immunoblotted using anti-HRS (upper panel) or anti-GAPDH antibodies (bottom panel) for loading control. (b) Cell lysates (lanes 1&2) and the 100 K pellets (lanes 3&4) from supernatants of Mov 127S cells transduced with ShRNA-CT (lanes 1 and 3) and ShRNA-HRS1 (lanes 2 and 4) lentivectors were analyzed by Western blotting using antibodies against Tsg101 or Flotillin-1 as exosomal proteins and against Calnexin as negative exosomal control or anti-GAPDH for cell lysate loading control. Note that Tsg101 and Flotillin-1 signals were similar in 100 K pellets from ShRNA-HRS1 and shRNA-CT. The data are representative of two experiments carried out with independent transduced cells. (EPS 2230 kb)
Supplementary material Figure S3: HRS depletion causes accumulation of cholesterol in infected Mov cells. Mov 127S cells transduced with lentivectors encoding Sh-CT or Sh-HRS1 were stained with Filipin (green) to visualize the distribution of free cholesterol. Note the punctuate signals of free cholesterol in Sh-HRS1 cells (see white arrows) compared to the diffuse signal observed in Sh-CT cells. Representative images are shown. Scale bar represents 10 μm. (EPS 9726 kb)
Supplementary material Figure S4: Decrease of PrP res in HRS-depleted cells is not inhibited by NH4Cl treatment. (a) Mov 127S cells transduced with Sh-CT (lanes 1 and 3) or Sh-HRS1 (lanes 2 and 4) were treated for 16 h with ammonium chloride (NH4Cl) (20 mM) (lanes 3 and 4) or with PBS (lanes 1 and 2). Cell lysates (20 μg) were analyzed by immunoblotting with anti-PrP, anti-p62 and anti-LC3 antibodies, as indicated. Cyclophilin A (Cyp A) was used as a loading control. Note the accumulation of PrP, p62 and LC3-II as evidence of successful inhibition of the degradation processes in NH4Cl-treated cells. (b) Mov 127S cells transduced with Sh-CT (lanes 1, 2, 5, 6) or Sh-HRS1 (lanes 3, 4, 7, 8) were treated for 16 h with ammonium chloride (NH4Cl) (20 mM) (lanes 5 to 8) or not (lanes 1 to 4) as in a). Cell lysates were analyzed by immunoblotting for PrP before (-) or after (+) PK digestion. Note that PrPres did not raise in NH4Cl-treated Mov 127S cells. Undigested lysates were also analyzed for p62 and for GAPDH (as a loading control). (EPS 3144 kb)
Supplementary material Figure S5: Total PrP distribution in Mov 127S cells is not affected by HRS depletion. (a) DRMs isolation from Mov 127S cells transduced with lentivectors encoding Sh-CT and Sh-HRS1. Sh-CT and Sh-HRS1 cells were lysed in buffer containing 1 % triton X100 at 4 °C. Equivalent amount of cell lysates were fractionated by flotation on a 5-30-40 % sucrose step gradient. Twelve fractions were collected from the top of the gradient and were analyzed by Western blotting using anti-PrP and anti-Flotillin-1 (as a DRM-associated protein) antibodies. DRMs are in fractions 3-4 while fractions 9-12 correspond to soluble proteins. (b) Mov 127S cells transduced with lentivectors encoding Sh-CT or Sh-HRS1 were fixed and analyzed by confocal microscopy using anti-PrP (red) and anti-Caveolin1 (green) antibodies. Images are single sections through the middle of the cells. Nuclei were stained with DAPI. Scale bars represent 10 μm. (EPS 12567 kb)
Supplementary material Figure S6: Release of the exosomal markers Alix and Flotillin-1 were marginally decreased in GW4869 treated cells. Quantifications of band signals for Alix (left panel) and Flotillin-1 (right panel) exosomal markers from 4 independent experiments. Values are given as mean ± SD. *P value < 0.05. Statistics and calculation of the P value were done by the GraphPad PRISM software with the Mann–Whitney test. (EPS 373 kb)
Supplementary material Figure S7: The neutral Sphingomyelinase inhibitor GW4869 strongly reduces prion infectivity release in ovRK13 127S (Rov 127S) cellular model. Rov 127S cells were incubated with the diluent (DMSO, negative control) or with the neutral Sphingomyelinase inhibitor GW4869 (10 μM) for 26 h. Cells were collected, homogenized in PBS and 100 K pellets were harvested from the corresponding culture supernatants. a) Analysis of prion infectivity in DMSO- and GW4869-treated Rov 127S cells and in their corresponding 100 K pellets. For SCA experiments, recipient ovRK13 target cells were inoculated with 1/1, 1/3 and 1/9 inoculum dilutions (corresponding to 30, 10 and 3.3 μg of cellular proteins, respectively), of DMSO- and GW4869-treated Rov cells (lanes 1-7) and with the corresponding 100 K pellets (equivalent to 450 μl of conditioned medium) (lanes 8 to 10). No PrPres was detected when recipient ovRK13 cells that did not express the PrPC protein (-dox) were inoculated with cell homogenate and 100 K pellet (lanes 1 and 9). Note that GW4869 treatment did not affect cell infectivity but strongly inhibited the infectivity in the 100 K pellet. (b) Biochemical analysis of 100 K pellets from DMSO- (lane 1) and GW4869-treated (lane 2) Rov127S cells. 100 K pellets corresponding to 20 ml of culture medium were analyzed by immunoblotting for Alix, TSG101 and Flotillin-1 exosomal proteins. Note that Alix, Tsg101 and Flotillin-1 signals are not or marginally affected by the GW4869 treatment as observed in the Mov 127S cellular model. (EPS 1937 kb)
Acknowledgments
This work was supported by CNRS, INSERM, INRA and the ANR program (ExoPrion AO2008). KL obtained a one-year FINOVI fellowship. We thank Jennifer T. Miller (NCI-Frederick) for carefully reading the manuscript. We acknowledge the PLATIM microscope platform at ENS-Lyon (SFR BioSciences Gerland–Lyon Sud UMS3444/US8, France).
Contributor Information
Didier Vilette, Email: d.vilette@envt.fr.
Pascal Leblanc, Email: Pascal.Leblanc@ens-lyon.fr.
References
- 1.Jucker M, Walker LC. Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature. 2013;501(7465):45–51. doi: 10.1038/nature12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64(6):783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- 3.Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11(4):301–307. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grad LI, Cashman NR. Prion-like activity of Cu/Zn superoxide dismutase: implications for amyotrophic lateral sclerosis. Prion. 2014;8(1):33–41. doi: 10.4161/pri.27602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo JL, Lee VM. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J Biol Chem. 2011;286(17):15317–15331. doi: 10.1074/jbc.M110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herva ME, Zibaee S, Fraser G, Barker RA, Goedert M, Spillantini MG. Anti-amyloid compounds inhibit alpha-synuclein aggregation induced by protein misfolding cyclic amplification (PMCA) J Biol Chem. 2014;289(17):11897–11905. doi: 10.1074/jbc.M113.542340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer V, Dinkel PD, Rickman Hager E, Margittai M. Amplification of Tau fibrils from minute quantities of seeds. Biochemistry. 2014;53(36):5804–5809. doi: 10.1021/bi501050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munch C, O’Brien J, Bertolotti A. Prion-like propagation of mutant superoxide dismutase-1 misfolding in neuronal cells. Proc Natl Acad Sci USA. 2011;108(9):3548–3553. doi: 10.1073/pnas.1017275108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nonaka T, Masuda-Suzukake M, Arai T, Hasegawa Y, Akatsu H, Obi T, Yoshida M, Murayama S, Mann DM, Akiyama H, Hasegawa M. Prion-like properties of pathological TDP-43 aggregates from diseased brains. Cell Rep. 2013;4(1):124–134. doi: 10.1016/j.celrep.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Polymenidou M, Cleveland DW. The seeds of neurodegeneration: prion-like spreading in ALS. Cell. 2011;147(3):498–508. doi: 10.1016/j.cell.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watts JC, Giles K, Oehler A, Middleton L, Dexter DT, Gentleman SM, DeArmond SJ, Prusiner SB. Transmission of multiple system atrophy prions to transgenic mice. Proc Natl Acad Sci USA. 2013;110(48):19555–19560. doi: 10.1073/pnas.1318268110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yonetani M, Nonaka T, Masuda M, Inukai Y, Oikawa T, Hisanaga S, Hasegawa M. Conversion of wild-type alpha-synuclein into mutant-type fibrils and its propagation in the presence of A30P mutant. J Biol Chem. 2009;284(12):7940–7950. doi: 10.1074/jbc.M807482200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95(23):13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prusiner SB. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet. 2013;47:601–623. doi: 10.1146/annurev-genet-110711-155524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sajnani G, Silva CJ, Ramos A, Pastrana MA, Onisko BC, Erickson ML, Antaki EM, Dynin I, Vazquez-Fernandez E, Sigurdson CJ, Carter JM, Requena JR. PK-sensitive PrP is infectious and shares basic structural features with PK-resistant PrP. PLoS Pathog. 2012;8(3):e1002547. doi: 10.1371/journal.ppat.1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bate C, Salmona M, Diomede L, Williams A. Squalestatin cures prion-infected neurons and protects against prion neurotoxicity. J Biol Chem. 2004;279(15):14983–14990. doi: 10.1074/jbc.M313061200. [DOI] [PubMed] [Google Scholar]
- 17.Gilch S, Bach C, Lutzny G, Vorberg I, Schatzl HM. Inhibition of cholesterol recycling impairs cellular PrP(Sc) propagation. Cell Mol Life Sci. 2009;66(24):3979–3991. doi: 10.1007/s00018-009-0158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilch S, Kehler C, Schatzl HM. The prion protein requires cholesterol for cell surface localization. Mol Cell Neurosci. 2006;31(2):346–353. doi: 10.1016/j.mcn.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Hannaoui S, Shim SY, Cheng YC, Corda E, Gilch S. Cholesterol balance in prion diseases and Alzheimer’s disease. Viruses. 2014;6(11):4505–4535. doi: 10.3390/v6114505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol. 1995;129(1):121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naslavsky N, Stein R, Yanai A, Friedlander G, Taraboulos A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J Biol Chem. 1997;272(10):6324–6331. doi: 10.1074/jbc.272.10.6324. [DOI] [PubMed] [Google Scholar]
- 22.Vey M, Pilkuhn S, Wille H, Nixon R, DeArmond SJ, Smart EJ, Anderson RG, Taraboulos A, Prusiner SB. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci USA. 1996;93(25):14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campana V, Sarnataro D, Zurzolo C. The highways and byways of prion protein trafficking. Trends Cell Biol. 2005;15(2):102–111. doi: 10.1016/j.tcb.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Bellingham SA, Guo BB, Coleman BM, Hill AF. Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases? Front Physiol. 2012;3:124. doi: 10.3389/fphys.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alais S, Simoes S, Baas D, Lehmann S, Raposo G, Darlix JL, Leblanc P. Mouse neuroblastoma cells release prion infectivity associated with exosomal vesicles. Biol Cell. 2008;100(10):603–615. doi: 10.1042/BC20080025. [DOI] [PubMed] [Google Scholar]
- 26.Castro-Seoane R, Hummerich H, Sweeting T, Tattum MH, Linehan JM, Fernandez de Marco M, Brandner S, Collinge J, Klohn PC. Plasmacytoid dendritic cells sequester high prion titres at early stages of prion infection. PLoS Pathog. 2012;8(2):e1002538. doi: 10.1371/journal.ppat.1002538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coleman BM, Hanssen E, Lawson VA, Hill AF. Prion-infected cells regulate the release of exosomes with distinct ultrastructural features. FASEB J. 2012;26(10):4160–4173. doi: 10.1096/fj.11-202077. [DOI] [PubMed] [Google Scholar]
- 28.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci USA. 2004;101(26):9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leblanc P, Alais S, Porto-Carreiro I, Lehmann S, Grassi J, Raposo G, Darlix JL. Retrovirus infection strongly enhances scrapie infectivity release in cell culture. EMBO J. 2006;25(12):2674–2685. doi: 10.1038/sj.emboj.7601162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211(5):582–590. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
- 31.Properzi F, Logozzi M, Abdel-Haq H, Federici C, Lugini L, Azzarito T, Cristofaro I, di Sevo D, Ferroni E, Cardone F, Venditti M, Colone M, Comoy E, Durand V, Fais S, Pocchiari M. Detection of exosomal prions in blood by immunochemistry techniques. J Gen Virol. 2015 doi: 10.1099/vir.0.000117. [DOI] [PubMed] [Google Scholar]
- 32.Basso M, Pozzi S, Tortarolo M, Fiordaliso F, Bisighini C, Pasetto L, Spaltro G, Lidonnici D, Gensano F, Battaglia E, Bendotti C, Bonetto V. Mutant copper-zinc superoxide dismutase (SOD1) induces protein secretion pathway alterations and exosome release in astrocytes: implications for disease spreading and motor neuron pathology in amyotrophic lateral sclerosis. J Biol Chem. 2013;288(22):15699–15711. doi: 10.1074/jbc.M112.425066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danzer KM, Kranich LR, Ruf WP, Cagsal-Getkin O, Winslow AR, Zhu L, Vanderburg CR, McLean PJ. Exosomal cell-to-cell transmission of alpha synuclein oligomers. Mol Neurodegener. 2012;7:42. doi: 10.1186/1750-1326-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emmanouilidou E, Melachroinou K, Roumeliotis T, Garbis SD, Ntzouni M, Margaritis LH, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J Neurosci. 2010;30(20):6838–6851. doi: 10.1523/JNEUROSCI.5699-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gomes C, Keller S, Altevogt P, Costa J. Evidence for secretion of Cu, Zn superoxide dismutase via exosomes from a cell model of amyotrophic lateral sclerosis. Neurosci Lett. 2007;428(1):43–46. doi: 10.1016/j.neulet.2007.09.024. [DOI] [PubMed] [Google Scholar]
- 36.Grad LI, Yerbury JJ, Turner BJ, Guest WC, Pokrishevsky E, O’Neill MA, Yanai A, Silverman JM, Zeineddine R, Corcoran L, Kumita JR, Luheshi LM, Yousefi M, Coleman BM, Hill AF, Plotkin SS, Mackenzie IR, Cashman NR. Intercellular propagated misfolding of wild-type Cu/Zn superoxide dismutase occurs via exosome-dependent and -independent mechanisms. Proc Natl Acad Sci USA. 2014;111(9):3620–3625. doi: 10.1073/pnas.1312245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Properzi F, Logozzi M, Fais S. Exosomes: the future of biomarkers in medicine. Biomark Med. 2013;7(5):769–778. doi: 10.2217/bmm.13.63. [DOI] [PubMed] [Google Scholar]
- 38.Rajendran L, Honsho M, Zahn TR, Keller P, Geiger KD, Verkade P, Simons K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc Natl Acad Sci USA. 2006;103(30):11172–11177. doi: 10.1073/pnas.0603838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200(4):373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21(1):77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458(7237):445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 42.Babst M. A protein’s final ESCRT. Traffic. 2005;6(1):2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt O, Teis D. The ESCRT machinery. Curr Biol. 2012;22(4):R116–R120. doi: 10.1016/j.cub.2012.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colombo M, Moita C, van Niel G, Kowal J, Vigneron J, Benaroch P, Manel N, Moita LF, Thery C, Raposo G. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J Cell Sci. 2013 doi: 10.1242/jcs.128868. [DOI] [PubMed] [Google Scholar]
- 45.Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K, Ueno Y, Shimosegawa T, Sugamura K. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem Biophys Res Commun. 2010;399(3):384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- 46.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319(5867):1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Hernandez D, Gutierrez-Vazquez C, Jorge I, Lopez-Martin S, Ursa A, Sanchez-Madrid F, Vazquez J, Yanez-Mo M. The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J Biol Chem. 2013;288(17):11649–11661. doi: 10.1074/jbc.M112.445304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell. 2011;21(4):708–721. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Archer F, Bachelin C, Andreoletti O, Besnard N, Perrot G, Langevin C, Le Dur A, Vilette D, Baron-Van Evercooren A, Vilotte JL, Laude H. Cultured peripheral neuroglial cells are highly permissive to sheep prion infection. J Virol. 2004;78(1):482–490. doi: 10.1128/JVI.78.1.482-490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeringer E, Barta T, Li M, Vlassov AV. Strategies for isolation of exosomes. Cold Spring Harb Protoc. 2015;4:319–323. doi: 10.1101/pdb.top074476. [DOI] [PubMed] [Google Scholar]
- 51.Arellano-Anaya ZE, Savistchenko J, Mathey J, Huor A, Lacroux C, Andreoletti O, Vilette D. A simple, versatile and sensitive cell-based assay for prions from various species. PLoS One. 2011;6(5):e20563. doi: 10.1371/journal.pone.0020563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vilette D, Andreoletti O, Archer F, Madelaine MF, Vilotte JL, Lehmann S, Laude H. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc Natl Acad Sci USA. 2001;98(7):4055–4059. doi: 10.1073/pnas.061337998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Du X, Kazim AS, Brown AJ, Yang H. An essential role of Hrs/Vps27 in endosomal cholesterol trafficking. Cell Rep. 2012;1(1):29–35. doi: 10.1016/j.celrep.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 54.Marijanovic Z, Caputo A, Campana V, Zurzolo C. Identification of an intracellular site of prion conversion. PLoS Pathog. 2009;5(5):e1000426. doi: 10.1371/journal.ppat.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marzo L, Marijanovic Z, Browman D, Chamoun Z, Caputo A, Zurzolo C. 4-hydroxytamoxifen leads to PrPSc clearance by conveying both PrPC and PrPSc to lysosomes independently of autophagy. J Cell Sci. 2013;126(Pt 6):1345–1354. doi: 10.1242/jcs.114801. [DOI] [PubMed] [Google Scholar]
- 56.Puntoni M, Sbrana F, Bigazzi F, Sampietro T. Tangier disease: epidemiology, pathophysiology, and management. Am J Cardiovasc Drugs. 2012;12(5):303–311. doi: 10.1007/BF03261839. [DOI] [PubMed] [Google Scholar]
- 57.Kumar R, McClain D, Young R, Carlson GA. Cholesterol transporter ATP-binding cassette A1 (ABCA1) is elevated in prion disease and affects PrPC and PrPSc concentrations in cultured cells. J Gen Virol. 2008;89(Pt 6):1525–1532. doi: 10.1099/vir.0.83358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cui HL, Guo B, Scicluna B, Coleman BM, Lawson VA, Ellett L, Meikle PJ, Bukrinsky M, Mukhamedova N, Sviridov D, Hill AF. Prion infection impairs cholesterol metabolism in neuronal cells. J Biol Chem. 2014;289(2):789–802. doi: 10.1074/jbc.M113.535807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang H, Badralmaa Y, Yang J, Lempicki R, Hazen A, Natarajan V. Retinoic acid and liver X receptor agonist synergistically inhibit HIV infection in CD4 + T cells by up-regulating ABCA1-mediated cholesterol efflux. Lipids Health Dis. 2012;11:69. doi: 10.1186/1476-511X-11-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilch S, Nunziante M, Ertmer A, Schatzl HM. Strategies for eliminating PrP(c) as substrate for prion conversion and for enhancing PrP(Sc) degradation. Vet Microbiol. 2007;123(4):377–386. doi: 10.1016/j.vetmic.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 61.Kanu N, Imokawa Y, Drechsel DN, Williamson RA, Birkett CR, Bostock CJ, Brockes JP. Transfer of scrapie prion infectivity by cell contact in culture. Curr Biol. 2002;12(7):523–530. doi: 10.1016/S0960-9822(02)00722-4. [DOI] [PubMed] [Google Scholar]
- 62.Paquet S, Langevin C, Chapuis J, Jackson GS, Laude H, Vilette D. Efficient dissemination of prions through preferential transmission to nearby cells. J Gen Virol. 2007;88(Pt 2):706–713. doi: 10.1099/vir.0.82336-0. [DOI] [PubMed] [Google Scholar]
- 63.Gousset K, Schiff E, Langevin C, Marijanovic Z, Caputo A, Browman DT, Chenouard N, de Chaumont F, Martino A, Enninga J, Olivo-Marin JC, Mannel D, Zurzolo C. Prions hijack tunnelling nanotubes for intercellular spread. Nat Cell Biol. 2009;11(3):328–336. doi: 10.1038/ncb1841. [DOI] [PubMed] [Google Scholar]
- 64.Guo BB, Bellingham SA, Hill AF. The neutral sphingomyelinase pathway regulates packaging of the prion protein into exosomes. J Biol Chem. 2014 doi: 10.1074/jbc.M114.605253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vella LJ, Sharples RA, Nisbet RM, Cappai R, Hill AF. The role of exosomes in the processing of proteins associated with neurodegenerative diseases. Eur Biophys J. 2008;37(3):323–332. doi: 10.1007/s00249-007-0246-z. [DOI] [PubMed] [Google Scholar]
- 66.Mattei V, Barenco MG, Tasciotti V, Garofalo T, Longo A, Boller K, Lower J, Misasi R, Montrasio F, Sorice M. Paracrine diffusion of PrP(C) and propagation of prion infectivity by plasma membrane-derived microvesicles. PLoS One. 2009;4(4):e5057. doi: 10.1371/journal.pone.0005057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dron M, Moudjou M, Chapuis J, Salamat MK, Bernard J, Cronier S, Langevin C, Laude H. Endogenous proteolytic cleavage of disease-associated prion protein to produce C2 fragments is strongly cell- and tissue-dependent. J Biol Chem. 2010;285(14):10252–10264. doi: 10.1074/jbc.M109.083857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179(3):485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi HY, Karten B, Chan T, Vance JE, Greer WL, Heidenreich RA, Garver WS, Francis GA. Impaired ABCA1-dependent lipid efflux and hypoalphalipoproteinemia in human Niemann-Pick type C disease. J Biol Chem. 2003;278(35):32569–32577. doi: 10.1074/jbc.M304553200. [DOI] [PubMed] [Google Scholar]
- 70.Lee CY, Ruel I, Denis M, Genest J, Kiss RS. Cholesterol trapping in Niemann-Pick disease type B fibroblasts can be relieved by expressing the phosphotyrosine binding domain of GULP. J Clin Lipidol. 2013;7(2):153–164. doi: 10.1016/j.jacl.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 71.Razi M, Futter CE. Distinct roles for Tsg101 and Hrs in multivesicular body formation and inward vesiculation. Mol Biol Cell. 2006;17(8):3469–3483. doi: 10.1091/mbc.E05-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goold R, Rabbanian S, Sutton L, Andre R, Arora P, Moonga J, Clarke AR, Schiavo G, Jat P, Collinge J, Tabrizi SJ. Rapid cell-surface prion protein conversion revealed using a novel cell system. Nat Commun. 2011;2:281. doi: 10.1038/ncomms1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Godsave SF, Wille H, Kujala P, Latawiec D, DeArmond SJ, Serban A, Prusiner SB, Peters PJ. Cryo-immunogold electron microscopy for prions: toward identification of a conversion site. J Neurosci. 2008;28(47):12489–12499. doi: 10.1523/JNEUROSCI.4474-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yim YI, Park BC, Yadavalli R, Zhao X, Eisenberg E, Greene LE. The multivesicular body is the major internal site of prion conversion. J Cell Sci. 2015;128(7):1434–1443. doi: 10.1242/jcs.165472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arellano-Anaya ZE, Huor A, Leblanc P, Lehmann S, Provansal M, Raposo G, Andreoletti O, Vilette D. Prion strains are differentially released through the exosomal pathway. Cell Mol Life Sci. 2014 doi: 10.1007/s00018-014-1735-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nishida N, Harris DA, Vilette D, Laude H, Frobert Y, Grassi J, Casanova D, Milhavet O, Lehmann S. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J Virol. 2000;74(1):320–325. doi: 10.1128/JVI.74.1.320-325.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12(1)(19–30):11–13. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 78.Alais S, Soto-Rifo R, Balter V, Gruffat H, Manet E, Schaeffer L, Darlix JL, Cimarelli A, Raposo G, Ohlmann T, Leblanc P. Functional mechanisms of the cellular prion protein (PrP(C)) associated anti-HIV-1 properties. Cell Mol Life Sci. 2012;69(8):1331–1352. doi: 10.1007/s00018-011-0879-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leblanc P, Baas D, Darlix JL. Analysis of the interactions between HIV-1 and the cellular prion protein in a human cell line. J Mol Biol. 2004;337(4):1035–1051. doi: 10.1016/j.jmb.2004.02.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Figure S1: HRS depletion inhibits PrP res formation. (a) Mov 127S cells were transduced with lentivectors encoding ShRNAs Sh-CT (lane 1) or three different Sh-HRS (1, 2 and 3; lanes 2-4). Cell lysates (20 μg) were immunoblotted using anti-HRS (upper, left panel) or anti-PrP antibodies (bottom, left panel). Cell lysates (300 μg) were PK digested and immunoblotted using anti-PrP antibodies (right panel). Note the strong decrease of PrPres in Sh-HRS1, 2 and 3 (lanes 2-4) compared to Sh-CT (lane 1). Non-PK digested Sh-CT lysate was used as a negative control of PK digestion (lane 5). (b) Mov 127S cells transduced with lentivectors encoding Sh-CT or Sh-HRS1 were fixed and analyzed by confocal microscopy using anti-PrP (red) and anti-Caveolin1 (green) antibodies. Images are single sections through the middle of the cells. Abnormal PrP was detected after incubating the cells 5 min with 3 M guanidine thiocyanate. Note the strong decrease of abnormal PrP signal in Sh-HRS1 cells. Nuclei were stained with DAPI. Scale bars represent 10 μm. (c) Infected scN2a#22L cells were transduced with ShRNAs Sh-CT (non-specific target negative control; lanes 5 and 7) or Sh-HRS1 (lanes 6 and 8). After puromycin selection, cells were harvested and HRS depletion was assessed by immunoblotting (upper panel). Note the strong decrease of HRS signal (lane 6) compared to CT ShRNA (lane 5). Loading control was assessed using coomassie staining (bottom panel). Cell lysates from Sh-CT (lane 7) and Sh-HRS KD (lane 8) cells were PK digested and PrPres was detected by Western blotting. Note the strong decrease of PrPres signal in HRS-depleted cells (lane 8) compared to control cells (lane 7). The normal N2a#58 (lanes 1 and 3) and infected N2a#22L cells (lanes 2 and 4) were used as negative and positive controls for PK digestion and PrPres detection. (EPS 13072 kb)
Supplementary material Figure S2: HRS depletion does affect exosomal release. (a) Mov 127S cells were transduced with lentivectors encoding ShRNAs Sh-CT (lane 1) or Sh-HRS1 (lane 2). Cell lysates were immunoblotted using anti-HRS (upper panel) or anti-GAPDH antibodies (bottom panel) for loading control. (b) Cell lysates (lanes 1&2) and the 100 K pellets (lanes 3&4) from supernatants of Mov 127S cells transduced with ShRNA-CT (lanes 1 and 3) and ShRNA-HRS1 (lanes 2 and 4) lentivectors were analyzed by Western blotting using antibodies against Tsg101 or Flotillin-1 as exosomal proteins and against Calnexin as negative exosomal control or anti-GAPDH for cell lysate loading control. Note that Tsg101 and Flotillin-1 signals were similar in 100 K pellets from ShRNA-HRS1 and shRNA-CT. The data are representative of two experiments carried out with independent transduced cells. (EPS 2230 kb)
Supplementary material Figure S3: HRS depletion causes accumulation of cholesterol in infected Mov cells. Mov 127S cells transduced with lentivectors encoding Sh-CT or Sh-HRS1 were stained with Filipin (green) to visualize the distribution of free cholesterol. Note the punctuate signals of free cholesterol in Sh-HRS1 cells (see white arrows) compared to the diffuse signal observed in Sh-CT cells. Representative images are shown. Scale bar represents 10 μm. (EPS 9726 kb)
Supplementary material Figure S4: Decrease of PrP res in HRS-depleted cells is not inhibited by NH4Cl treatment. (a) Mov 127S cells transduced with Sh-CT (lanes 1 and 3) or Sh-HRS1 (lanes 2 and 4) were treated for 16 h with ammonium chloride (NH4Cl) (20 mM) (lanes 3 and 4) or with PBS (lanes 1 and 2). Cell lysates (20 μg) were analyzed by immunoblotting with anti-PrP, anti-p62 and anti-LC3 antibodies, as indicated. Cyclophilin A (Cyp A) was used as a loading control. Note the accumulation of PrP, p62 and LC3-II as evidence of successful inhibition of the degradation processes in NH4Cl-treated cells. (b) Mov 127S cells transduced with Sh-CT (lanes 1, 2, 5, 6) or Sh-HRS1 (lanes 3, 4, 7, 8) were treated for 16 h with ammonium chloride (NH4Cl) (20 mM) (lanes 5 to 8) or not (lanes 1 to 4) as in a). Cell lysates were analyzed by immunoblotting for PrP before (-) or after (+) PK digestion. Note that PrPres did not raise in NH4Cl-treated Mov 127S cells. Undigested lysates were also analyzed for p62 and for GAPDH (as a loading control). (EPS 3144 kb)
Supplementary material Figure S5: Total PrP distribution in Mov 127S cells is not affected by HRS depletion. (a) DRMs isolation from Mov 127S cells transduced with lentivectors encoding Sh-CT and Sh-HRS1. Sh-CT and Sh-HRS1 cells were lysed in buffer containing 1 % triton X100 at 4 °C. Equivalent amount of cell lysates were fractionated by flotation on a 5-30-40 % sucrose step gradient. Twelve fractions were collected from the top of the gradient and were analyzed by Western blotting using anti-PrP and anti-Flotillin-1 (as a DRM-associated protein) antibodies. DRMs are in fractions 3-4 while fractions 9-12 correspond to soluble proteins. (b) Mov 127S cells transduced with lentivectors encoding Sh-CT or Sh-HRS1 were fixed and analyzed by confocal microscopy using anti-PrP (red) and anti-Caveolin1 (green) antibodies. Images are single sections through the middle of the cells. Nuclei were stained with DAPI. Scale bars represent 10 μm. (EPS 12567 kb)
Supplementary material Figure S6: Release of the exosomal markers Alix and Flotillin-1 were marginally decreased in GW4869 treated cells. Quantifications of band signals for Alix (left panel) and Flotillin-1 (right panel) exosomal markers from 4 independent experiments. Values are given as mean ± SD. *P value < 0.05. Statistics and calculation of the P value were done by the GraphPad PRISM software with the Mann–Whitney test. (EPS 373 kb)
Supplementary material Figure S7: The neutral Sphingomyelinase inhibitor GW4869 strongly reduces prion infectivity release in ovRK13 127S (Rov 127S) cellular model. Rov 127S cells were incubated with the diluent (DMSO, negative control) or with the neutral Sphingomyelinase inhibitor GW4869 (10 μM) for 26 h. Cells were collected, homogenized in PBS and 100 K pellets were harvested from the corresponding culture supernatants. a) Analysis of prion infectivity in DMSO- and GW4869-treated Rov 127S cells and in their corresponding 100 K pellets. For SCA experiments, recipient ovRK13 target cells were inoculated with 1/1, 1/3 and 1/9 inoculum dilutions (corresponding to 30, 10 and 3.3 μg of cellular proteins, respectively), of DMSO- and GW4869-treated Rov cells (lanes 1-7) and with the corresponding 100 K pellets (equivalent to 450 μl of conditioned medium) (lanes 8 to 10). No PrPres was detected when recipient ovRK13 cells that did not express the PrPC protein (-dox) were inoculated with cell homogenate and 100 K pellet (lanes 1 and 9). Note that GW4869 treatment did not affect cell infectivity but strongly inhibited the infectivity in the 100 K pellet. (b) Biochemical analysis of 100 K pellets from DMSO- (lane 1) and GW4869-treated (lane 2) Rov127S cells. 100 K pellets corresponding to 20 ml of culture medium were analyzed by immunoblotting for Alix, TSG101 and Flotillin-1 exosomal proteins. Note that Alix, Tsg101 and Flotillin-1 signals are not or marginally affected by the GW4869 treatment as observed in the Mov 127S cellular model. (EPS 1937 kb)