Abstract
The yeast SUMO (small ubiquitin-like modifier) orthologue SMT3 was initially discovered in a genetic suppressors screen for the centromeric protein Mif2 (Meluh and Koshland in Mol Bio Cell 6:793–807, 1). Later, it turned out that the homologous mammalian proteins SUMO1 to SUMO4 are reversible protein modifiers that can form isopeptide bonds with lysine residues of respective target proteins (Mahajan et al. in Cell 88:97–107, 2). This was the discovery of a post-translational modification called sumoylation, which enzymatically resembles ubiquitination. However, very soon it became clear that SUMO attachments served a far more diverse role than ubiquitination. Meanwhile, numerous cellular processes are known to be subject to the impact of SUMO modification, including transcription, protein targeting, protein solubility, apoptosis or activity of various enzymes. In many instances, SUMO proteins create new protein interaction surfaces or block existing interaction domains (Geiss-Friedlander and Melchior in Nat Rev in Mol Cell Biol 8:947–956, 3). For the past few years, sumoylation attracted increasing attention as a versatile regulator of toxic protein properties in neurodegenerative diseases. In this review, we summarize the growing knowledge about the involvement of sumoylation in neurodegeneration, and discuss the underlying molecular principles affected by this multifaceted and intriguing post-translational modification.
Keywords: Sumo, Neurodegeneration, Protein aggregation, Protein solubility
Introduction
The yeast SUMO (small ubiquitin-like modifier) orthologue SMT3 was initially discovered in a genetic suppressors screen for the centromeric protein Mif2 [1]. Later, it turned out that the homologous mammalian proteins SUMO1 to SUMO4 are reversible protein modifiers that can form isopeptide bonds with lysine residues of respective target proteins [2]. This was the discovery of a post-translational modification called sumoylation, which enzymatically resembles ubiquitination. However, very soon it became clear that SUMO attachments served a far more diverse role than ubiquitination. Meanwhile, numerous cellular processes are known to be subject to the impact of SUMO modification, including transcription, protein targeting, protein solubility, apoptosis or activity of various enzymes. In many instances, SUMO proteins create new protein interaction surfaces or block existing interaction domains (for review see [3]).
For the past few years, sumoylation attracted increasing attention as a versatile regulator of toxic protein properties in neurodegenerative diseases. In this review, we summarize the growing knowledge about the involvement of sumoylation in neurodegeneration, and discuss the underlying molecular principles affected by this multifaceted and intriguing post-translational modification.
The SUMO protein family
SUMO proteins are members of the ubiquitin-like protein family, which get covalently conjugated to substrate proteins, altering their properties and thus hugely increasing the proteome complexity at a post-translational level. Since the discovery of ubiquitin more than 25 years ago, a dozen ubiquitin-like proteins were discovered, among those NEDD8 (neural precursor cell expressed, developmentally down-regulated eight) [4], ISG 15 (interferon inducible gene 15) [5], ATG 8 (autophagic ubiquitin-like protein) [6] and SUMO (small ubiquitin-like modifier). SUMO was first identified in mammals where it was found to be covalently conjugated to the GTPase activating protein RanGAP1 [2, 7]. The SUMO family members are ubiquitously expressed in all types of eukaryotic cells. The human genome encodes four distinct SUMO isoforms, named SUMO1 to SUMO4 [8–10].
SUMO modification is an essential process for most eukaryotic organisms [11, 12]. A few exceptions have been reported so far, including the fission yeast S. pombe and the filamentous fungus A. nidulans. Deletion of the gene coding for the single SUMO paralog leads to severe growth deficiencies as well as self-impaired conidation and self-sterility in S. pombe and A. nidulans, respectively, but is not lethal [13, 14]. Two SUMO1 knockout studies in mice have shown controversial results and it is still under debate whether individual SUMO isoforms are essential or SUMO1 functions can be compensated by SUMO2/3 and vice versa [15, 16]. Nevertheless, the complete abrogation of the SUMO conjugation by Ubc9 knockout causes embryonic lethality in mice, which dispels any doubts that sumoylation is an essential cellular process [11].
Similar to all ubiquitin-related modifiers, SUMO proteins are synthesized as immature pro-forms. Removal of the C-terminal extension and revelation of the conserved Gly–Gly motif is done by a family of SUMO-specific proteases and is a prerequisite for SUMO conjugation to substrates.
SUMO1 is about 11 kDa, 101-amino-acid protein that shares only ≈18 % sequence homology with ubiquitin. SUMO2 and SUMO3 are almost identical, differing among each other in only three amino-terminal residues, but having only ≈47 % homology with SUMO1. SUMO1–3 are ubiquitously expressed, while SUMO4 mRNA expression is confined primarily to the lymph node, kidney and spleen [10]. Despite limited sequence homology, all SUMO isoforms resemble the three-dimensional structure of ubiquitin [17].
The term sumoylation usually implies the covalent binding of SUMO proteins to acceptor lysine residues of target proteins. However, SUMO proteins can also non-covalently bind to SUMO interaction motifs (SIM) of other proteins or even to SIMs present in SUMO molecules itself. SIMs are characterized by a hydrophobic core flanked by acidic and/or serine residues [18, 19]. SIM-mediated non-covalent SUMO binding can have tremendous regulatory impact on the respective target protein. For instance, the transcriptional repressor DAXX requires SUMO binding for its repressive function [20]. Components of the enzymatic sumoylation machinery also contain SUMO binding sites that fulfill the requirement for a SIM, and require non-covalent binding of the SUMO protein for their function.
Sumoylation machinery
Sumoylation is a post-translational modification which requires formation of an isopeptide bond between the C-terminal Gly residue of SUMO and the ε-amino group of a Lys acceptor in the target protein [2]. Biochemically, sumoylation resembles ubiquitination, and a similar enzymatic pathway is used for sumoylation. However, specific enzymes of the ubiquitination and sumoylation machineries do not overlap. Sumoylation employs an enzymatic pathway with three classes of enzymes: E1-activating enzyme (Aos1/Uba2), E2-conjugase (Ubc9) and E3-ligase (Fig. 1; for review see [3]). Following activation of the mature SUMO C-terminus by the heterodimer Aos1/Uba2, SUMO is passed to a Cys residue in the catalytic pocket of the single E2–Ubc9. SUMO is conjugated predominantly to acceptor Lys residues, which are part of a Ψ-K-X-[D/E] consensus motif, where Ψ can be any large hydrophobic residue (I, V or L), K is the target lysine, X is any residue and D/E is an aspartate or glutamate [21, 22]. The presence of a consensus site within the amino acid sequence of a protein is often not sufficient for its efficient modification. For example, consensus Lys residues, buried in stable helices are not recognized by Ubc9 [23]. For an acceptor site to be SUMO modified it needs to be in an extended conformation or part of an unstructured area [24]. Amino acid stretches surrounding the target lysine can have an impact on the conjugation, e.g., downstream clusters of acidic residues or pre-existing phosphorylation [25, 26].
Fig. 1.
SUMO conjugation cycle. SUMO is expressed as a precursor protein and processed by a SUMO-specific protease (SENP) to expose the C-terminal di-glycine motif (maturation). Mature SUMO is activated in an ATP-dependent manner by the SUMO activating enzyme (E1) SAE1/SAE2 and is transferred through a trans-esterification process to the SUMO conjugating enzyme (E2) Ubc9. SUMO can then be conjugated to the target lysine of a substrate, defined in many cases by the consensus motif ΨKXE. For most substrates conjugation is facilitated by a SUMO E3 ligase. Sumoylation is a very dynamic reversible modification due to specific class proteases that can remove SUMO moieties from modified targets
Ubc9 can recognize and bind directly to consensus SUMO motifs [22]. Although many SUMO targets are conjugated within a consensus site, there are an increasing number of proteins that are modified at Lys residues that do not fall into a consensus motif, e.g., human E2-25K [23], PCNA [27] and axin [28]. It is currently not known how Ubc9 recognizes non-consensus sumoylation sites. Nevertheless, for efficient modification of many targets the action of an E3-ligase, which transfers SUMO from Ubc9 to the acceptor Lys, is required. In addition, the activity of E3 ligases confers limited target specificity. However, only a relatively small number of E3 ligases, seemingly responsible for a large quantity of targets, have been identified to date. There are three groups of E3 ligases described in the SUMO pathway. The largest one, the SP-RING-finger-like E3 ligases, function as adaptor proteins that directly bind Ubc9 and the SUMO target. SP-RING E3 ligases bind SUMO non-covalently via the SIMs. Such SUMO E3-ligases are the enzymes from the PIAS family (protein inhibitor of activated signal transducer and activator of transcription) [29–31]. In mammals five members of this family have been identified so far: PIAS1, PIAS3, PIASxα, PIASxβ and PIASy. MMS21 (known also as NSE2), which is part of a multimeric complex involved in DNA repair also belongs to the SP-RING ligases [32–34].
The second group of E3 ligases is represented by Ran binding protein 2 (RanBP2) [35]. RanBP2 is part of the nuclear pore complex; it binds stably to Ubc9, but not to targets. This is an E3 ligase unique for the sumoylation machinery, since it shares no sequence homology with any ubiquitin E3 and is neither a RING-finger nor a HECT-type E3 ligase. RanBP2 is facilitating sumoylation by placing the Ubc9-SUMO complex in an orientation favorable for an attack by acceptor Lys residue. Finally, E3 ligase activity has been ascribed to the human Polycomb protein 2 (Pc2) (reviewed in [36]). The exact mechanism how Pc2 exerts its SUMO E3-ligase activity remains unknown (for a review of SUMO ligases and isopeptidases see [37]).
Although SUMO1 and SUMO2/3 use the same enzymatic conjugation pathway [38], they most probably serve different functions as the different isoforms can be conjugated to different target proteins [39, 40]. A subset of substrates can be modified by both SUMO1 and SUMO2/3, but there is an increasing amount of evidence that the regulatory mechanisms for SUMO1 and SUMO2/3 conjugation and deconjugation are different [41]. Finally, the pool of non-conjugated SUMO2/3 is much bigger than that of SUMO1.
SUMO1 gets attached to substrates as a single molecule, while SUMO2/3 can form polymeric chains due to the presence of a consensus SUMO motif (VKTE) at the N-terminus of SUMO2/3. Indication for a distinct function of the poly-SUMO2/3 chains comes from genetic studies in yeast, showing deficits in chromosome segregation, DNA-repair and mitosis [42]. PolySUMO chains can be recognized by proteins containing several SIMs, such as the RING-finger 4 (RNF4) ubiquitin E3 ligase. RNF4 contains four SIMs and a RING domain that together facilitate polySUMO-specific ubiquitination [43].
The de-sumoylation machinery
Sumoylation is a reversible and highly dynamic post-translational modification. Following covalent binding to acceptor lysine residues, SUMO modifications can be removed from their targets in rapid cycles of sumoylation and de-sumoylation. Therefore, the biochemical detection of sumoylation is more challenging than the demonstration of ubiquitination, which is probably one of the reasons why sumoylation escaped its discovery for a much longer time than ubiquitination. A class of enzymes called SENPs (sentrin-specific proteases) is responsible for reversing the SUMO conjugation ([41, 44]; reviewed in [33]). SENPs are SUMO-specific Cys-proteases which cleave the isopeptide bond formed between SUMO and the target. There are six members of the SENP family in humans SENP1-3 and SENP5-7, which deconjugate SUMO paralogs. SENP enzymes differ in their preference for cleavage of different SUMO isoforms. SENP5 and SENP3 specifically cleave SUMO 2/3 conjugates and are thus possibly involved in SUMO-chain editing [41, 45]. Most SENPs can also cleave SUMO C-terminally to reveal the Gly–Gly motif and are thus involved in SUMO maturation. For SENP6 it has been shown in vitro that it can primarily act as a C-terminal SUMO hydrolase [46]. The majority of the SUMO proteases localize to different nuclear regions—nuclear periphery [47], nucleolus [48], nuclear bodies [47, 49], and nuclear pore complexes [50]. Others like SENP6 are also found in the cytoplasm [46]. The subcellular localization differs between members of the SENP family and therefore they target different subsets of substrates for deconjugation. Interestingly, the overall expression in whole rat brain extracts as well as the time-dependent subcellular localization of SENP1 and SENP6 in cultured rat hippocampal neurons are differentially regulated during development and neuronal maturation. This further supports that different SENPs have non-redundant roles, and alterations in their subcellular distribution contributes to regulating the balance between sumoylation and de-sumoylation [51].
Very recently, a second class of SUMO proteases has been discovered with different substrate specificity compared to the SENP family: De-sumoylating isopeptidase 1 and 2 (DeSI-1 and -2) belong to the PPPDE (permuted papain fold peptidases of Ds-RNA viruses and eukaryotes) superfamily that comprises cysteine proteases with a papain-like fold. While SENPs are predominantly localized in the nucleus, allocated to different nuclear structures, DeSI-1 and -2 are localized in both the nucleus and cytoplasm or predominantly localized in the cytoplasm, respectively. Furthermore, De-SI-1 and -2 recognize different substrates when compared to SENPs [52]. Therefore, DeS1 and -2 add further complexity to the dynamic regulation of sumoylation. Moreover, considering the growing number, diversity and differentially regulated SUMO target proteins, it seems plausible that further SUMO isopeptidases will be discovered in the future.
SUMO in neuronal function
Post-translational modifications are essential parts of all signaling pathways and regulatory mechanisms which enable cells to adapt to the rapidly changing environmental conditions. Their tight control in the central nervous system is of critical importance for maintaining neuronal cell viability, function and connectivity. While the contribution of other post-translational modifications, especially phosphorylation and ubiquitination, to neuronal function and dysfunction have been under investigation for a long time, sumoylation of neuron-specific targets remained unknown until recently, since most assays for identification of SUMO conjugated proteins were carried out in non-neuronal cell lines. In addition, covalent SUMO modification in general escaped discovery until 1996, partly due to the unstable and hard-to-detect nature of this protein modification. However, the increasing evidence for the involvement of sumoylation in various cellular processes provoked a number of studies aiming to disclose the role of SUMO in the nervous system. This led to the identification of multiple intriguing aspects of SUMO conjugation in the CNS. The first evidence that sumoylation occurs at the synapse was provided by Martin et al. Modification of the kainate receptor subunit GluR6a induces receptor channel internalization by endocytosis and indicates a role of sumoylation in modulating synaptic transmission and plasticity [53].
Activity of voltage gated K+ channels Kv1.5 has been shown to be modulated by sumoylation in myocytes [54]. The expression of Kv channels in neurons and the presence of SUMO consensus motifs in Kv1.1 and Kv1.2 potassium channels led to the speculation that SUMO might be involved in the regulation of their function in the CNS [55]. The later was then confirmed by Plant et al. [56, 57] showing that a maximum of two out of four subunits of the potassium channel Kv2.1 are concurrently sumoylated, which provides a reversible and graded regulation mechanism of hippocampal neuron excitability.
Sumoylation has been implicated in proper repair of DNA single-strand breaks in neurons. DNA single-strand breaks, which occur continuously and are an inherent feature of chromosome metabolism to overcome torsional barriers, are resealed by tyrosyl DNA phosphodiesterase 1 (TDP1) [58]. In 2002, it was shown that mutations in TDP1 and subsequent loss of TDP1 DNA repair activity are the cause of a familial form of ataxia (spinocerebellar ataxia with axonal neuropathy; SCAN1). Deficiency of this DNA repair pathway affects primarily the post-mitotic, terminally differentiated, large neuronal cells (e.g., cerebellar pyramidal cells) [58]. Very recently, it was shown that SUMO modification of TDP1 is necessary for proper sub-nuclear TDP1 targeting in neurons in order to achieve a sufficiently high local TDP1 protein concentration at sites of DNA damage [59]. Of note, also topoisomerase 1, which is responsible for a large proportion of protein-linked single-strand DNA breaks, is itself a SUMO target and rapidly modified upon cellular stress [60].
Intriguingly, expression of sumoylation machinery components as well as sumoylated substrates is temporally and spatially regulated in the developing rat brain. Overall decrease in sumoylated proteins and sumoylation enzymes redistribution to dendritic sites has been observed during maturation of neurons [51]. The developmental regulation of sumoylation at synaptic sites is one of the most recent pieces of evidence suggesting that SUMO proteins play an important role in CNS development and neuron-specific functions. It further underscores that sumoylation is not just a nuclear affair as considered in the early days of SUMO research when most known SUMO targets were nuclear proteins.
Sumoylation was also implicated in axonal mRNA trafficking. The mRNA binding protein La assists local axonal protein synthesis by transporting mRNAs. La has been found to be modified by SUMO and wild-type La acts in anterograde and retrograde RNA transport, while sumoylation-deficient La participates only in anterograde transport [61]. By modulating the interactions between the cytoskeleton and the extracellular matrix, focal adhesion kinase (FAK) regulates axonal growth and path-finding [62, 63]. SUMO1 modification of FAK leads to its autophosphorylation and subsequent binding to Src kinases which enables FAK full activation. In the context of FAK, sumoylation influences neuronal cell mobility and axonal guidance [64]. These and studies on other SUMO substrates not mentioned here have suggested diverse roles of sumoylation in the CNS, ranging from neuronal development and synapse formation to synaptic transmission and plasticity.
Considering the diversity of functions that SUMO proteins play in CNS development and function, it is expected that there will be an increase in the number of reports implicating SUMO in CNS disorders. In this context, the role of SUMO in synaptic function is especially intriguing, as dysfunction and degeneration is thought to start at axonal terminals in many neurodegenerative diseases.
SUMO proteins may affect common pathogenic aspects in different neurodegenerative diseases
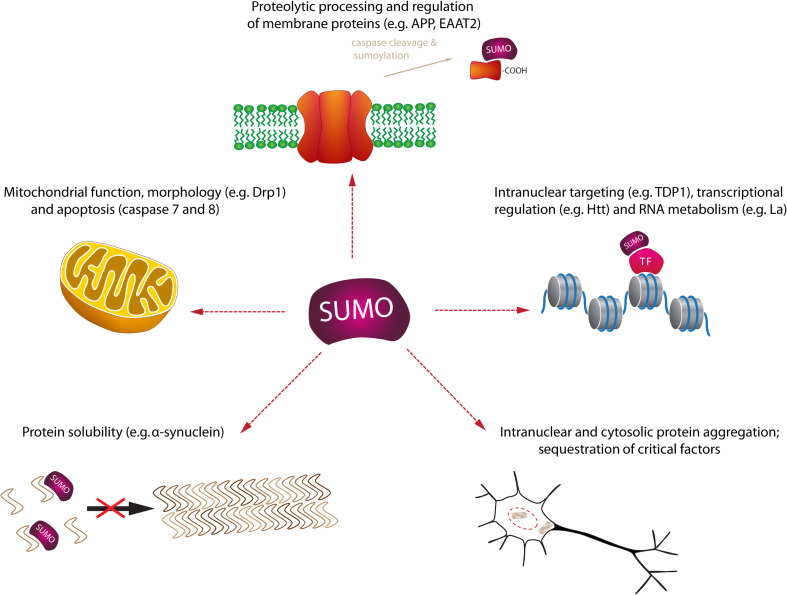
The field of neurodegenerative disease comprises a group of neurological disorders that is characterized by the progressive loss or dysfunction of specific neuronal systems. The type of neurons affected determines the spectrum of clinical symptoms observed. For example, degeneration of dopaminergic neurons in the substantia nigra (besides other neuronal nuclei) of Parkinson disease patients results in the typical cardinal motor symptoms of rigor (increased muscle tone), hypokinesia (reduced frequency and ability to instigate and perform voluntary movements) and tremor [65]. Another common neurodegenerative disease is amyotrophic lateral sclerosis (ALS), characterized by a continuous loss of motor neurons that results in progressive paresis of voluntarily innervated muscles. Death usually ensues due to respiratory failure within a few years [66]. Thus the diverse clinical appearance of different neurodegenerative diseases, e.g., Parkinson’s disease, Alzheimer’s disease, ALS, Huntington disease reflects the differential involvement of different neuronal systems. Nevertheless, molecular mechanisms and pathogenic principles of the underlying structural or functional impairment at the cellular levels are partially overlapping between these clinically quite diverse diseases. For example, disturbances in the transcription, transport, function and degradation of mRNAs and micro RNAs emerged as central research field in neurodegeneration. Several proteins involved in RNA function and metabolism, e.g., TDP-43 and FUS [67, 68], were identified as the main components of pathological protein deposits in both ALS and fronto-temporal lobar degeneration (FTLD). The causal relevance of these proteins for disease was further supported by the discovery of mutations, in genes for TDP-43 [69] and FUS [70] co-segregating with familial neurodegenerative diseases. Other cell biological aspects, ranging from mitochondrial protein targeting and mitochondrial (dys)function to oxidative stress and RNA transcription and metabolism, commonly involved in neurodegeneration are all typically affected by sumoylation (Fig. 2).
Fig. 2.
Roles of SUMO in neurodegeneration. Depicted are different functional aspects of sumoylation in neurodegeneration along with example substrates. APP amyloid precursor protein, DRP1 dynamin-related protein 1, EAAT2 excitatory amino-acid transporter 2, Htt huntingtin, TDP1 tyrosyl DNA phosphodiesterase 1
Maybe the most prominent common feature of both familial and sporadic forms of neurodegenerative disease is a decreased solubility of specific disease-associated proteins [71, 72].
This alteration in solubility is connected to a pathological propensity to form intracellular protein aggregates. In addition, several aggregation-prone proteins implicated in neurodegeneration were found to be sumoylated [73–78]. Based on the presumed function of SUMO attachments as a “protein solubility enhancer”, is seems plausible that regulation of protein aggregation is another way how sumoylation may influence neurodegenerative diseases, as outlined in detail below.
Neurodegeneration as aggregopathy—SUMO as a regulator of disease protein solubility?
Pathological protein aggregation is a biochemical and histopathological hallmark of most neurodegenerative diseases. Misfolding, reduced solubility and a pathological propensity to aggregate is thus a common property of such neurodegeneration-associated proteins [71, 79]. It is not fully understood if large aggregates or oligomeric species that emerge in the process of protein aggregation are indeed the toxic agent. Most likely, specific types of oligomeric precursors that precede the formation of large protein aggregates are the culprit [80]. Generally, interfering with these aggregation cascades at a certain stage is likely to alter the toxic properties of an aggregation-prone protein, and studying the regulators and mechanisms of protein solubility and aggregation is expected to provide crucial insights into the apical pathogenic mechanisms of neurodegenerative diseases.
Improper folding and reduced solubility of proteins can be the result of point mutations in the disease-associated gene. Alternatively, it may be related to oxidative protein damage or also excessive production of specific proteins, as in patients with a rare familial form of PD due to α-synuclein gene duplications or triplications [81]. The latter instance provides evidence that altered properties of α-synuclein lead to a gain of (toxic) function, not a loss of physiological function. It is conceivable that post-translational modifications play an important role in regulating the aggregation propensity of disease proteins. For example, phosphorylation has been shown to influence the aggregation and toxicity of α-synuclein, although there is an ongoing controversy whether phosphorylation of α-synuclein reduces or enhances the formation of pathological protein deposits [82–84]. In contrast to phosphorylation, the role of sumoylation for protein aggregation has been assessed only in a few instances.
SUMO proteins belong to the most soluble proteins known [85]. The extreme solubility of the SUMO proteins has even established the small ubiquitin-like molecule as a valuable tag for recombinant expression and purification of proteins in bacterial and eukaryotic systems. Thus SUMO may play a role in circumventing challenges like proper protein folding and poor solubility of heterologous proteins in expression hosts [85, 86]. This makes sumoylation in principle a superior candidate regulator in disease conditions characterized by reduced protein solubility and pathological aggregation, such as neurodegenerative diseases. However, only in a few instances the direct impact of SUMO on protein solubility has been documented, among which α-synuclein, DJ-1, huntingtin and STAT1.
A study of Shinbo et al. [74] shows that Parkinson’s disease-related L166P mutation in the multifunctional protein DJ-1 leads to improper SUMO conjugation and implicates decreased solubility of the L166P DJ1 mutant.
The C. elegans cytoplasmic intermediate filament IFB-1 is another striking example of SUMO’s role as a solubility enhancer. Kaminsky et al. [87] have shown that SUMO is required for the cytoplasmic mobile pool of IFb-1 and absence of SUMO caused accumulation of IFb-1 into filaments and cytoplasmic aggregates.
Recently, SUMO was shown to dissolve paracrystals of STAT transcription factors. The implication of this finding is twofold: it firstly provides an interesting example how protein aggregates are not only implicated in disease pathogenesis but also in physiological aspects of cell biology, as STAT assembled in “paracrystals” could not be de-phosphorylated anymore and therefore remained active. Moreover, in this case SUMO’s impact on protein solubility was mediated by its interplay with another post-translational modification, because sumoylated STAT protein could not be fully phosphorylated anymore and “semi-phosphorylated” STAT dimers blocked STAT assembly into crystalline structures [88, 89]. The effect of sumoylation on α-synuclein and huntingtin aggregation and toxicity is discussed in detail in the next sections.
Sumoylation in Parkinson’s disease and other synucleinopathies
Synucleinopathies are a group of disorders characterized by formation of inclusions called Lewy bodies, major component of which is α-synuclein [90]. They include Parkinson’s disease (PD), dementia with Lewy bodies (DLB), and multiple system atrophy (MSA).
Covalent modification of α-synuclein by SUMO has first been reported in non-neuronal cells [91]. More detailed analysis has shown that α-synuclein is SUMO conjugated in brain tissue of His-SUMO transgenic mice [75]. Synuclein is predominantly mono-sumoylated, however, conjugation can occur at multiple sites the major of which are K96 and K102. To investigate the effect of sumoylation on α-synuclein aggregation, the authors have used two different approaches: in vitro fibril formation and inclusion formation in a cell line-based assay [75]. Under physiological salt concentrations, non-modified wild-type α-synuclein forms mature amyloid fibrils similar to those found in synucleinopathy patients [92, 93]. Under those conditions, SUMO conjugation prevented α-synuclein fibril formation. The presence of free, non-conjugated SUMO in the α-synuclein-SUMO protein mixture did not inhibit fibrillization, but resulted in formation of fibrils of smaller size [75]. This points out that only SUMO which is covalently conjugated to α-synuclein is able to completely inhibit fibril formation. Intriguingly, a minor proportion of sumoylated α-synuclein was already sufficient to substantially delay α-synuclein fibril formation in vitro [75] The finding prompts considerations how the SUMO modification might prevent “polymerization” or aggregation of proteins: (1) Sumoylation could simply be a general “solubility enhancer” and thereby counteract aggregation; (2) SUMO attachments might sterically hinder the formation or elongation of longitudinal fibrils normally formed by many aggregation-prone proteins involved in neurodegeneration. It could thus act as a “chain terminator”; (3) the SUMO attachments could act in a chaperone-like manner and enhance proper protein folding of target proteins. For example, the α-synuclein, a protein with a central pathogenic role in Parkinson’s disease, contains two closely neighbored lysine residues that account for the majority of its sumoylation, but mass spectrometry revealed another nine lysines out of the 15 lysine residues in this protein subject to SUMO modification as well [75]. Thus, the highly dynamic and soluble SUMO modification might undergo rapid cycles of sumoylation and de-sumoylation at various target sites, and thereby influence “local solubility” and protein folding (Table 1).
Table 1.
Sumoylation in neurodegenerative disorders
| Disease | Substrate | Modified residue | Functional impact | Reference |
|---|---|---|---|---|
| Polyglutamine diseases | ||||
| HD | Huntingtin | K6, K10, K15 | Negative regulation of Htt97Q aggregation when fused to SUMO; sumoylation increases nuclear targeting and transcriptional repression | Steffan et al. [76] |
| SBMA | Androgen receptor | K385, K518 | Attenuates polyglutamine-mediated aggregation | Poukka et al. [130], Mukherjee et al. [131] |
| SCA type 1 | Ataxin-1 | Multiple sites | Mutant ataxin-1-82Q is sumoylated to a lesser extent than WT | Riley et al. [73], Ryu et al. [99] |
| SCA type 7 | Ataxin-7 | K257 | Negative regulation of mutant ataxin-7 aggregation | Janer et al. [100] |
| SCAN1 | TDP1 | K111 | Proper sub-nuclear targeting | Hudson et al. [59] |
| DRPLA | Atrophin 1 | Co-expression of SUMO1 and atrophin with expanded poly-glutamine stretches increases its nuclear aggregation | Terashima et al. [132] | |
| AD | APP | K587, K595 | Negative regulation of Abeta levels | Li et al. [77], Zhang and Sarge [104] |
| Tau | K340 | Phosphatase inhibition and MT depolarization increase tau sumoylation | Dorval and Fraser [91] | |
| ALS | SOD1 | K75 | Increases protein stability and aggregation | Fei et al. [78] |
| EAAT2 | NA | Accumulation of sumoylated proteolytic fragment of EAAT2 in the nuclei of spinal cord neurons from SOD-G93A mice | Gibb et al. [105], Foran et al. [107] | |
| Synucleinopathies | ||||
| PD | α-Synuclein | K96, K102 | Impaired sumoylation increases aggregation and toxicity | Krumova et al. [75], Dorval and Fraser [91] |
| DJ-1 | K130 | Dysregulated sumoylation decreases DJ1 solubility | Shinbo et al. [74] | |
| MSA | α-Synuclein | SUMO-positive brain inclusions | Pountney et al. [94] | |
| DLB | α-Synuclein | SUMO-positive brain inclusions | Pountney et al. [94] | |
| NIID | Various | SUMO-positive neuronal intranuclear inclusions of sporadic and familial NIID | Pountney et al. [112], Fujigasaki et al. [113] | |
| Hypoxia | Various | Cimarosti et al. [108] | ||
AD Alzheimer’s disease, ALS amyotrophic lateral sclerosis, APP amyloid precursor protein, APP amyloid precursor protein, DLB dementia with Lewy bodies, DRPLA dentatorubral-pallidoluysian atrophy, EAAT2 excitatory amino-acid transporter 2, HD Huntington’s disease, MSA multiple system atrophy, MT microtubule, NIID neuronal intranuclear inclusion disorder, PD Parkinson’s disease, SBMA spinal and bulbar muscular atrophy, SCA spinocerebellar ataxia, SCAN1 spinocerebellar ataxia with axonal neuropathy, SOD1 superoxide dismutase 1, TDP1 tyrosyl DNA phosphodiesterase 1
The role of SUMO in modulating α-synuclein aggregation was confirmed additionally in a cell-based assay. Moreover, impaired α-synuclein sumoylation by mutation of the two consensus SUMO sites exacerbated α-synuclein toxicity and reduced the number of surviving dopaminergic cells in a rat PD model [75]. This implicates a beneficial effect of α-synuclein sumoylation on neuronal cell survival.
In post-mortem studies, the majority of aggregated α-synuclein did not overlap with SUMO immunoreactive brain tissue from patients with α-synuclein-related neurodegenerative diseases, namely multiple system atrophy (MSA) and dementia with Lewy bodies (DLB) or Parkinson’s disease [94]. Lack of overlapping SUMO/α-synuclein immunoreactivity suggests that aggregated α-synuclein was mostly not SUMO-modified in these patients. Such a mutual exclusive staining pattern of aggregated proteins and SUMO immunoreactivity is in agreement with the idea of sumoylation as a “solubility-enhancer” and soluble, sumoylated α-synuclein predominantly present outside aggregated protein deposits.
Another potential role for sumoylation in PD involves the transcriptional regulator DJ1, which is mutated in few cases of recessively inherited PD. DJ-1 is SUMO modified at K130 and the conjugation is stimulated by the E3 ligases PIASxα and PIASy. Mutation of the SUMO conjugation site disrupted functions like ras-dependent transformation and cell growth [74]. Interestingly, the L166P PD-linked DJ-1 mutation caused multi or poly-SUMOylation of DJ-1. Improper SUMO-conjugation was also implicated in decreased DJ-1 solubility and higher susceptibility to UV-induced cell apoptosis [74].
An example of SUMO regulating proteins involved in PD not via conjugation, but rather protein–protein interaction is parkin. Mutations in the gene coding for the E3-ubiquitin ligase parkin are associated with autosomal recessive juvenile Parkinsonism. Parkin binds selectively to SUMO1 in a SIM (SUMO interaction motif)—independent manner. This interaction promotes Parkin’s nuclear transport and auto-ubiquitination [95].
Polyglutamine disorders
Polyglutamine diseases are neurodegenerative disorders caused by a trinucleotide expansion. They include Huntington’s disease (HD), spinobulbar muscular atrophy (SBMA) and spinocerebellar ataxias (SCA) and are characterized by progressive loss of certain neuronal subtypes. The encoded CAG expansions lead to the synthesis of toxic protein species bearing extended glutamine repeats [96].
In 2004 Steffan et al. [76] showed for the first time that a protein involved in neurodegeneration is SUMO conjugated. A pathogenic fragment of Huntingtin (Httex1p) was found to be modified by SUMO1 at the N-terminal lysines 6, 10, and 15. Sumoylation led to the increase of transcriptional repression mediated by Htt. Moreover, by generating a SUMO-Htt97QP fusion protein, where SUMO was N-terminally fused to Htt, it was demonstrated that SUMO reduces Htt inclusion formation. Huntingtin’s cytotoxicity is reduced in flies heterozygous for SUMO and in transgenic Drosophila, expressing SUMO-deficient Htt97QP-K6,10,15R. This indicates that SUMOylation of Htt contributes to the disease pathology possibly by stabilizing toxic Htt species [76]. Moreover, it is an example that SUMO-dependent reduction in aggregation is not always associated with a reduction in toxicity, and that SUMO’s net effect on toxicity can be positive or negative, in line with the generally high target dependency of its effect.
Spinocerebellar ataxia (SCA)
Spinocerebellar ataxia encompasses multiple disease forms, several of which fall under the category of polyglutamine diseases [97]. SCAs are progressive disorders characterized by degeneration of cerebellar Purkinje cells and often additional neuronal subsets of the CNS or PNS. By showing an increase in sumoylated proteins in the cortex of human patients and SCA1 transgenic mice (expressing mutant ataxin-1 with 82 glutamine repeats) Ueda and colleagues for the first time speculated about a potential link between SUMO modification and polyglutamine disorders [98]. Later, ataxin-1 was found to be SUMO conjugated at multiple lysine residues upstream of its polyglutamine tract. Mutant ataxin-1-82Q is to a lesser extent SUMO conjugated than wild-type ataxin-1-30Q. Additionally, an intact nuclear localization signal (NLS) is needed for efficient SUMO modification of ataxin-1 [73]. Overexpression of SUMO or sumoylation enzymes (Ubc9) seems to increase aggregation of polyQ ataxin-1 suggesting that SUMO modification of the mutant ataxin enhances its toxicity [99]. Oxidative stress induces sumoylation of both WT and mutant ataxin-1. Interestingly, phospho-deficient ataxin S776A or JNK inhibition prevented the oxidative-stress-induced sumoylation of ataxin and reduced aggregate formation, suggesting interplay between phosphorylation and sumoylation in the regulation of polyQ ataxin-1 aggregation [99].
Recent work has linked sumoylation to SCA7. Ataxin-7 has been shown to be modified by both SUMO1 and 2 in SCA7 cell culture models as well as patient tissues [100]. Furthermore, sumoylation-deficient polyQ expanded ataxin-7 shows increased inclusion formation, implying a role for SUMO in regulating ataxin-7 aggregation propensity [100].
Very recently, a protein implicated in a form of spinocerebellar ataxia that is not associated with polyglutamine expansions, spinocerebellar ataxia with axonal neuropathy (SCAN1), was shown to be subject to sumoylation: tyrosyl DNA phosphodiesterase 1 (TDP1), which is mutated in SCAN1 [58], is necessary for the repair of single-strand DNA breaks in neurons. Sufficiently high sub-nuclear concentrations of TDP1 at sites of DNA damage are only achieved when TDP1 can be sumoylated [59]. Thus in this case the role of sumoylation in neurodegeneration seems to be related to SUMO’s classical protein targeting function.
Spinal and bulbar muscular atrophy (SBMA)
Additional indication that sumoylation could be involved in polyglutamine toxicity came from the work of Chan et al. [101] in a Drosophila model of spinobulbar muscular atrophy SBMA, where polyglutamine repeats expand within the androgen receptor (AR) protein. Flies expressing the catalytically inactive SUMO E1-enzyme (Uba2 C175S) in addition to the disease mutant protein ARtrQ112 showed enhanced degeneration, manifested by loss of eye pigment cells [101]. This suggests that downregulation of sumoylation has deleterious effect on cells expressing toxic polyglutamine species.
Alzheimer’s disease (AD)
Alzheimer’s disease is the most common form of dementia, histopathologically characterized by massive neuronal loss in the hippocampus and cortical as well as subcortical brain regions. AD is yet another protein misfolding disease in which abnormal accumulations of Aβ and tau protein are found in the forms of amyloid plaques and neurofibrillary tangles, respectively [102]. Aβ protein is a product of the proteolysis of the transmembrane amyloid precursor protein (APP) by β-secretase (BACE), which cleaves the extracellular domain, and γ-secretase, cleaving in the membrane spanning region. The first two studies implicating a role of sumoylation in APP processing and secretion present contradicting results based on SUMO2/3 overexpression [77, 103]. A recent study by Zhang and Sarge [104] shows that is APP is conjugated by SUMO at residues K587 and K595 adjacent to the β-secretase cleavage site. Sumoylation at these sites is associated with reduced levels of Aβ production. Further studies focusing on the role of sumoylation for individual targets involved in APP cleavage, like BACE and other secretase factors are required for the full understanding of the regulation of APP processing by sumoylation.
Another player in AD is the microtubule-associated protein tau. Hyperphosphorylation is associated with tau self-assembly and formation of intracellular tangles. Tau was shown to be SUMO1 conjugated at Lys340 and blocking of the proteasome increased tau sumoylation levels. Although no direct proof that sumoylated tau is also phosphorylated was provided, the authors observed an increase of SUMO1-tau conjugates upon treatment with phosphatase inhibitors and speculated that tau sumoylation is induced by phosphorylation [91]. However, the relevance of sumoylation for tau function and tau-related diseases remains an open question so far.
In addition to sumoylation of AD-related proteins, an intriguing genetic link between AD and sumoylation has been suggested. Ahn and others have identified a single-nucleotide polymorphism (SNP) in intron seven of the Ubc9 gene to be associated with late-onset AD in a Korean population compared to controls [79].
Amyotrophic lateral sclerosis
Amyotrophic lateral sclerosis (ALS), also referred to as Lou Gehrig’s disease, is a form of motor neuron disease caused by the degeneration of upper and lower neurons. Mutations in the superoxide dismutase (SOD1) gene are associated with about 20 % of the familial ALS cases that make up approximately 10 % of all ALS patients. SOD1 is a SUMO substrate and sumoylation of both wild-type and mutant SOD1 increases its steady-state levels. Co-transfection of SUMO has been suggested to increase SOD1 aggregate formation [78].
Dysregulation of the astroglial glutamate transporter EAAT2 has been implicated as a contributor to excitotoxicity in ALS: caspase-3 cleaves EAAT2 at its cytosolic carboxy-terminal domain, generating a proteolytic CTE fragment and resulting in impaired glutamate uptake [105, 106]. Foran et al. [107] have recently shown that CTE is sumoylated and this promotes its accumulation in the nuclei of spinal cord astrocytes from SOD-G93A transgenic mice. Expression of CTE-SUMO1 fusion protein recapitulates the nuclear localization. Moreover, astrocytes expressing CTE-SUMO1 exhibit toxic effect for co-cultured motor neurons which is subsequently detrimental for the glial cells as well [107].
Mass spectrometry studies of cellular systems have suggested interplay between sumoylation and ubiquitination to play a role in the regulation of another ALS- and FTLD-associated protein, TDP43 (transactive response (TAR) DNA-binding protein 43) [108]. TDP-43 is an RNA-binding protein that translocates from the nucleus to the cytoplasm, and forms cytoplasmic inclusions in ALS and FTLD patients. It was recently found to be the main component of the ubiquitin-positive inclusion bodies in most motor neuron disease cases as well as other neurodegenerative diseases [68]. Moreover, mutations in TDP-43 are a rare cause for familial forms of ALS and in few instances also FTLD [68, 109, 110]. Neither SUMO acceptor lysines in TDP-43 nor the consequence of sumoylation for TDP-43 function have been identified so far. Therefore, whether sumoylation influences TDP-43′s RNA binding properties, nuclear localization, aggregation or all three probably disease-relevant features of the protein can only be speculated at this point.
Neuronal intranuclear inclusion disease (NIID)
Neuronal intranuclear inclusion disease (NIID) is a rare neurodegenerative condition characterized by inclusion formation in neuronal nuclei [111]. The clinical disease characteristics include progressive development of ataxia.
The first report to connect SUMO and NIID showed SUMO1 co-localization with markers for nuclear inclusions in brain autopsy sections from familial NIID cases [112]. A study by Takahashi-Fujigasaki et al. [113] confirmed that neuronal intranuclear inclusions of familial and sporadic NIID were immunoreactive with a SUMO1 antibody. Components of the intranuclear inclusion bodies like HDAC4, PML and ubiquitin colocalized with SUMO. A major SUMO substrate RanGAP1 was also found associated to the nuclear inclusions in the familial case of NIID [113]. The role of SUMO in the inclusion body formation and the disease pathology of NIID remains unknown. So far, it has not been shown whether free SUMO is sequestered into the inclusion bodies or some of the components forming these inclusions are sumoylated prior or after their aggregation.
Neurodegeneration and “classical” mitochondrial apoptosis pathways
The above-described sumoylated disease proteins are linked to specific neurological conditions and are situated at apical positions of the degenerative cascade. However, many protein components of the more downstream events that are part of the classical apoptosis pathways [114] are affected by SUMO modification. For most neurodegenerative diseases, the execution of neuronal apoptosis pathways during the course of disease has been demonstrated in animal models and in in vitro studies. Results from post-mortem human brain tissue were contradictory [115]. However, difficulties demonstrating apoptosis in human post-mortem samples may be due to the fact that cell death in chronic neurodegenerative diseases takes place over years while apoptosis in single neurons is completed within few hours. Aspects of the so-called intrinsic, mitochondrial death pathway are mostly encountered in neuronal apoptosis. Apoptotic release of mitochondrial pro-apoptotic factors, e.g., cytochrome c, results in downstream activation of effector caspases with death executing function [114]. Several years ago, it was shown that the execution of mitochondrial apoptosis requires mitochondrial fission [116]. The morphology of the mitochondrial network results from continuously balanced fission and fusion events of mitochondria, and blocking mitochondrial fission prevented cytochrome c release and completion of neuronal apoptosis [117]. Mitochondrial fission at steady state and under apoptotic conditions requires the GTPase dynamin-related protein (DRP1). The later is recruited from the cytosol to the mitochondrial outer membrane at the foci of fission via a mechanism that was largely unclear until recently. Harder et al. [118] provided the first evidence for the involvement of the SUMO conjugation machinery in mitochondrial dynamics: the authors show that DRP1 interacts with Ubc9 and is a substrate for SUMO conjugation [118]. Furthermore, SUMO is found at the sites of mitochondrial fission and co-localizes with DRP1 when associated with mitochondria. This raised the first speculations of an involvement of SUMO in targeting DRP1 to the mitochondrial membrane. The observed increase in mitochondrial fission upon SUMO1 overexpression in COS-7 cells further supports this idea [119]. Similarly, the SUMO protease SENP5 cleaves SUMO1 from proteins of the mitochondrial compartment and rescues the SUMO1-driven increase in fission events, which results in more elongated mitochondrial structures [119]. This regulation is at least partially achieved via DRP1 de-sumoylation.
Moreover, the mitochondrially anchored protein ligase (MAPL) was described as the first potential SUMO E3 ligase associated to mitochondria [120]. MAPL is a mitochondrial outer membrane protein with a RING-finger domain that can facilitate both ubiquitination as well as sumoylation in vitro with some evidence for increased specificity for SUMO in vivo. MAPL conjugates SUMO1 to HeLa-derived DRP1 ex vivo [120].
Sumoylation has also been shown to regulate DRP1 function and mitochondrial dynamics under apoptotic conditions. Wasiak et al. [121] have shown that the proapoptotic Bcl-2 family member Bax/Bak stimulates sumoylation of DRP1, which correlates with its stable mitochondrial membrane association in the later stages of apoptotic cell death.
Altogether, these data support the role of sumoylation in the maintenance of mitochondrial homeostasis in physiological and apoptotic conditions.
In addition, the role of SUMO protein modification might not be restricted to the mitochondrial aspects of apoptosis, but might be involved in other pathways of apoptosis induction. For example, caspase-7 and -8 turned out to be SUMO targets as well [122, 123]. For both, caspase-7 and -8, sumoylation is associated with their nuclear localization. Caspase-8 activation is not impaired by sumoylation [122], and no data addressing the effect of sumoylation on the activity of caspase-7 have been reported to date [124].
Sumoylation links oxidative stress and neurodegeneration
As outlined above, many pivotal players in neurodegenerative conditions are targets of SUMO modification. The SUMO attachment often modifies properties related to the respective protein’s toxicity, e.g., solubility and oligomerization/fibrillization, subcellular translocation or modulatory activity on transcription in cell free systems, cell culture or animal models. Nevertheless, dysregulation of the sumoylation machinery has not been convincingly documented in the respective human disease conditions. This lack of direct evidence for a dysregulated sumoylation machinery in human post-mortem tissue is not surprising considering that the detection of SUMO modification is often severely hampered by its unstable nature, under “ideal” experimental conditions and even more by the post-mortem delay in studies on human brain tissue.
However, in some instances, mutant disease-causing protein related to familial neurodegeneration is differentially sumoylated compared to its wild-type counterpart, as in the case of DJ-1 or ataxin-1 [73, 74], providing a direct link between sumoylation and disease. Other disease proteins, for example PD causing α-synuclein or ALS-related mutant SOD1, were sumoylated to the same extents as the respective wild-type proteins. Nevertheless, it is highly likely that also in sporadic disease cases, which make up the vast majority of neurodegenerative diseases (e.g., for AD, PD or ALS [125]), dysregulation of sumoylation occurs. One of most direct general links between neurodegenerative disease and impaired sumoylation is oxidative stress. In general, the activity of key enzymes of the SUMO pathway responds to a lot of different external stimuli. Most interestingly, the enzymatic centre of the E1 and E2 enzymes of the sumoylation cascade is reversibly oxidized and thereby efficiently inhibited by low levels of oxidative stress [126]. In a similar manner, oxidative stress enhances sumoylation by activation of the de-conjugating SENP3 isopeptidase [127]. In contrary, higher levels of oxidative stress lead to inactivation of de-sumoylating SENP isopeptidases [126]. The net result is reflected in a bi-phasic shift of the cellular sumoylation balance, depending on the degree of oxidative stress. It seems plausible that the deleterious consequences of oxidative stress frequently encountered in disease conditions, either as a primary insult or a secondary contributor to a vicious cycle of toxicity, are mediated by dysregulated sumoylation and subsequently increased toxicity of pathogenic proteins. In more acute cases of brain insults sumoylation may serve a protective mechanism. In a stroke in vivo model insult-dependent upregulation of sumoylation has been observed [128], while elevated global sumoylation levels in Ubc9 transgenic animals turned out to be protective against ischemic damage [124], as well as sumoylation of hypoxia-inducible factor-1α (HIF-1α) in an experimental brain death model [129].
Conclusions
In contrast to ubiquitination, which was already discovered at the end of the 1970s, sumoylation has been related to neurodegeneration in a very limited number of publications for the last few years. Ubiquitin-modification has been recognized as one of the first steps in the degradation of misfolded disease proteins, leading to proteasomal overload at least in some instances. The consequences of dysregulated sumoylation are much more diverse, and often not related to protein stability. One of the most intriguing aspects of SUMO proteins may be their role as a “solubility regulator” in aggregopathies. Dynamic cycles of sumoylation and de-sumoylation at several lysine residues may even serve as a non-conventional rapidly regulatable chaperone-like function, and help to re-fold or prevent aggregation of proteins [75]. In addition, due to the high target-dependency of SUMO-mediated effects, many other cellular functions can be affected, related to nuclear protein targeting [59] and transcriptional regulation (e.g., in the case of Huntington’s disease [76]), mitochondrial protein targeting and mitochondrial dysfunction as in the case of the PD-related protein DJ-1 [74] or modulation of proteolytic processing as observed with sumoylated A-beta protein [77].
Consequently, while it was discovered long after the description of its biochemically close relative ubiquitination, sumoylation is attracting more and more attention as an important player in neurodegenerative diseases, and disease mechanisms in general. Due to its very dynamic nature, studies on sumoylation are technically more challenging than investigating the consequences of ubiquitination. Nevertheless, with the increasing number of pivotal functions SUMO turns out to have in the regulation of central cellular functions and specific disease mechanisms, eventually it may well catch up with ubiquitination as a central post-translational regulator of the proteome both in physiological and disease conditions.
Finally, the question remains whether SUMO proteins or components of the (de-)sumoylation machinery could serve as targets for therapeutic approaches. At first glance, considering the variety of essential cellular functions involving sumoylation, it does not seem to be an ideal target candidate for disease-specific interventions. Small molecule inhibitors blocking the E1 or E2 enzymes of the sumoylation pathway would probably interfere with vital aspects of cell metabolism. However, sufficient target specificity might be achieved if inhibitors of single E3 ligases acting on a limited subset of target proteins were available. Furthermore, because of the very dynamic nature of the SUMO modification, sumoylation could be enhanced by inhibition of de-sumoylating enzymes. So far, less then ten different SUMO-specific isopeptidases are known, however, with a differential subcellular distribution and target specificity and new classes of SUMO isopeptidases still being discovered [52]. Isoform-specific inhibitors of SUMO isopeptidases may therefore serve as pharmacological means to enhance sumoylation of specific disease-relevant proteins in the future.
Acknowledgments
This work was supported by the Charcot Foundation for ALS Research (JHW).
References
- 1.Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mahajan R, Delphin C, Guan T, Gerace L, Melchior F. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 1997;88:97–107. doi: 10.1016/S0092-8674(00)81862-0. [DOI] [PubMed] [Google Scholar]
- 3.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 4.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 5.Kessler DS, Levy DE, Darnell JE., Jr Two interferon-induced nuclear factors bind a single promoter element in interferon-stimulated genes. Proc Natl Acad Sci USA. 1988;85:8521–8525. doi: 10.1073/pnas.85.22.8521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 7.Matunis MJ, Coutavas E, Blobel G. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J Cell Biol. 1996;135:1457–1470. doi: 10.1083/jcb.135.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melchior F. SUMO–nonclassical ubiquitin. Annu Rev Cell Dev Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 9.Bohren KM, Nadkarni V, Song JH, Gabbay KH, Owerbach D. A M55V polymorphism in a novel SUMO gene (SUMO-4) differentially activates heat shock transcription factors and is associated with susceptibility to type I diabetes mellitus. J Biol Chem. 2004;279:27233–27238. doi: 10.1074/jbc.M402273200. [DOI] [PubMed] [Google Scholar]
- 10.Guo D, Li M, Zhang Y, Yang P, Eckenrode S, Hopkins D, Zheng W, Purohit S, Podolsky RH, Muir A, Wang J, Dong Z, Brusko T, Atkinson M, Pozzilli P, Zeidler A, Raffel LJ, Jacob CO, Park Y, Serrano-Rios M, Larrad MT, Zhang Z, Garchon HJ, Bach JF, Rotter JI, She JX, Wang CY. A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat Genet. 2004;36:837–841. doi: 10.1038/ng1391. [DOI] [PubMed] [Google Scholar]
- 11.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y. Characterization of a fission yeast SUMO-1 homologue, pmt3p, required for multiple nuclear events, including the control of telomere length and chromosome segregation. Mol Cell Biol. 1999;19:8660–8672. doi: 10.1128/mcb.19.12.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wong KH, Todd RB, Oakley BR, Oakley CE, Hynes MJ, Davis MA. Sumoylation in Aspergillus nidulans: SUMO inactivation, overexpression and live-cell imaging. Fungal Genet Biol. 2008;45:728–737. doi: 10.1016/j.fgb.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alkuraya FS, Saadi I, Lund JJ, Turbe-Doan A, Morton CC, Maas RL. SUMO1 haploinsufficiency leads to cleft lip and palate. Science. 2006;313:1751. doi: 10.1126/science.1128406. [DOI] [PubMed] [Google Scholar]
- 16.Zhang FP, Mikkonen L, Toppari J, Palvimo JJ, Thesleff I, Janne OA. Sumo-1 function is dispensable in normal mouse development. Mol Cell Biol. 2008;28:5381–5390. doi: 10.1128/MCB.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 18.Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117–16127. doi: 10.1074/jbc.M512757200. [DOI] [PubMed] [Google Scholar]
- 20.Lin DY, Huang YS, Jeng JC, Kuo HY, Chang CC, Chao TT, Ho CC, Chen YC, Lin TP, Fang HI, Hung CC, Suen CS, Hwang MJ, Chang KS, Maul GG, Shih HM. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 22.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–21669. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 23.Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Korner R, Olsen JV, Jentsch S, Melchior F, Sixma TK. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol. 2005;12:264–269. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- 24.Macauley MS, Errington WJ, Scharpf M, Mackereth CD, Blaszczak AG, Graves BJ, McIntosh LP. Beads-on-a-string, characterization of ETS-1 sumoylated within its flexible N-terminal sequence. J Biol Chem. 2006;281:4164–4172. doi: 10.1074/jbc.M510488200. [DOI] [PubMed] [Google Scholar]
- 25.Yang SH, Galanis A, Witty J, Sharrocks AD. An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 2006;25:5083–5093. doi: 10.1038/sj.emboj.7601383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 28.Rui HL, Fan E, Zhou HM, Xu Z, Zhang Y, Lin SC. SUMO-1 modification of the C-terminal KVEKVD of Axin is required for JNK activation but has no effect on Wnt signaling. J Biol Chem. 2002;277:42981–42986. doi: 10.1074/jbc.M208099200. [DOI] [PubMed] [Google Scholar]
- 29.Johnson ES, Gupta AA. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/S0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi Y, Kahyo T, Toh E, Yasuda H, Kikuchi Y. Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J Biol Chem. 2001;276:48973–48977. doi: 10.1074/jbc.M109295200. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc Natl Acad Sci USA. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Nse2, a component of the Smc5-6 complex, is a SUMO ligase required for the response to DNA damage. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potts PR, Yu H. Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol Cell Biol. 2005;25:7021–7032. doi: 10.1128/MCB.25.16.7021-7032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao X, Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc Natl Acad Sci USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/S0092-8674(01)00633-X. [DOI] [PubMed] [Google Scholar]
- 36.Wotton D, Merrill JC. Pc2 and SUMOylation. Biochem Soc Trans. 2007;35:1401–1404. doi: 10.1042/BST0351401. [DOI] [PubMed] [Google Scholar]
- 37.Melchior F, Schergaut M, Pichler A. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem Sci. 2003;28:612–618. doi: 10.1016/j.tibs.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]
- 39.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 40.Vertegaal AC, Andersen JS, Ogg SC, Hay RT, Mann M, Lamond AI. Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics. 2006;5:2298–2310. doi: 10.1074/mcp.M600212-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- 42.Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, Yeh CH, Huang HY, Wang TF. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20:2067–2081. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatham MH, Geoffroy MC, Shen L, Plechanovova A, Hattersley N, Jaffray EG, Palvimo JJ, Hay RT. RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol. 2008;10:538–546. doi: 10.1038/ncb1716. [DOI] [PubMed] [Google Scholar]
- 44.Di BA, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Di BA, Gill G. SUMO-specific proteases and the cell cycle. An essential role for SENP5 in cell proliferation. Cell Cycle. 2006;5:2310–2313. doi: 10.4161/cc.5.20.3367. [DOI] [PubMed] [Google Scholar]
- 46.Kim KI, Baek SH, Jeon YJ, Nishimori S, Suzuki T, Uchida S, Shimbara N, Saitoh H, Tanaka K, Chung CH. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J Biol Chem. 2000;275:14102–14106. doi: 10.1074/jbc.275.19.14102. [DOI] [PubMed] [Google Scholar]
- 47.Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- 48.Nishida T, Tanaka H, Yasuda H. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur J Biochem. 2000;267:6423–6427. doi: 10.1046/j.1432-1327.2000.01729.x. [DOI] [PubMed] [Google Scholar]
- 49.Best JL, Ganiatsas S, Agarwal S, Changou A, Salomoni P, Shirihai O, Meluh PB, Pandolfi PP, Zon LI. SUMO-1 protease-1 regulates gene transcription through PML. Mol Cell. 2002;10:843–855. doi: 10.1016/S1097-2765(02)00699-8. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H, Saitoh H, Matunis MJ. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol. 2002;22:6498–6508. doi: 10.1128/MCB.22.18.6498-6508.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loriol C, Parisot J, Poupon G, Gwizdek C, Martin S. Developmental regulation and spatiotemporal redistribution of the sumoylation machinery in the rat central nervous system. PLoS ONE. 2012;7:e33757. doi: 10.1371/journal.pone.0033757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shin EJ, Shin HM, Nam E, Kim WS, Kim JH, Oh BH, Yun Y. DeSUMOylating isopeptidase: a second class of SUMO protease. EMBO Rep. 2012;13:339–346. doi: 10.1038/embor.2012.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martin S, Nishimune A, Mellor JR, Henley JM. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–325. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benson MD, Li QJ, Kieckhafer K, Dudek D, Whorton MR, Sunahara RK, Iniguez-Lluhi JA, Martens JR. SUMO modification regulates inactivation of the voltage-gated potassium channel Kv1.5. Proc Natl Acad Sci USA. 2007;104:1805–1810. doi: 10.1073/pnas.0606702104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin S, Wilkinson KA, Nishimune A, Henley JM. Emerging extranuclear roles of protein SUMOylation in neuronal function and dysfunction. Nat Rev Neurosci. 2007;8:948–959. doi: 10.1038/nrn2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Plant LD, Dementieva IS, Kollewe A, Olikara S, Marks JD, Goldstein SA. One SUMO is sufficient to silence the dimeric potassium channel K2P1. Proc Natl Acad Sci USA. 2010;107:10743–10748. doi: 10.1073/pnas.1004712107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plant LD, Dowdell EJ, Dementieva IS, Marks JD, Goldstein SA. SUMO modification of cell surface Kv2.1 potassium channels regulates the activity of rat hippocampal neurons. J Gen Physiol. 2011;137:441–454. doi: 10.1085/jgp.201110604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takashima H, Boerkoel CF, John J, Saifi GM, Salih MA, Armstrong D, Mao Y, Quiocho FA, Roa BB, Nakagawa M, Stockton DW, Lupski JR. Mutation of TDP1, encoding a topoisomerase I-dependent DNA damage repair enzyme, in spinocerebellar ataxia with axonal neuropathy. Nat Genet. 2002;32:267–272. doi: 10.1038/ng987. [DOI] [PubMed] [Google Scholar]
- 59.Hudson JJ, Chiang SC, Wells OS, Rookyard C, El-Khamisy SF. SUMO modification of the neuroprotective protein TDP1 facilitates chromosomal single-strand break repair. Nat Commun. 2012;3:733. doi: 10.1038/ncomms1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mao Y, Sun M, Desai SD, Liu LF. SUMO-1 conjugation to topoisomerase I: a possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Niekerk EA, Willis DE, Chang JH, Reumann K, Heise T, Twiss JL. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc Natl Acad Sci USA. 2007;104:12913–12918. doi: 10.1073/pnas.0611562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ivankovic-Dikic I, Gronroos E, Blaukat A, Barth BU, Dikic I. Pyk2 and FAK regulate neurite outgrowth induced by growth factors and integrins. Nat Cell Biol. 2000;2:574–581. doi: 10.1038/35023515. [DOI] [PubMed] [Google Scholar]
- 63.Ren XR, Ming GL, Xie Y, Hong Y, Sun DM, Zhao ZQ, Feng Z, Wang Q, Shim S, Chen ZF, Song HJ, Mei L, Xiong WC. Focal adhesion kinase in netrin-1 signaling. Nat Neurosci. 2004;7:1204–1212. doi: 10.1038/nn1330. [DOI] [PubMed] [Google Scholar]
- 64.Kadare G, Toutant M, Formstecher E, Corvol JC, Carnaud M, Boutterin MC, Girault JA. PIAS1-mediated sumoylation of focal adhesion kinase activates its autophosphorylation. J Biol Chem. 2003;278:47434–47440. doi: 10.1074/jbc.M308562200. [DOI] [PubMed] [Google Scholar]
- 65.Vekrellis K, Xilouri M, Emmanouilidou E, Rideout HJ, Stefanis L. Pathological roles of alpha-synuclein in neurological disorders. Lancet Neurol. 2011;10:1015–1025. doi: 10.1016/S1474-4422(11)70213-7. [DOI] [PubMed] [Google Scholar]
- 66.Kiernan MC, Vucic S, Cheah BC, Turner MR, Eisen A, Hardiman O, Burrell JR, Zoing MC. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. [DOI] [PubMed] [Google Scholar]
- 67.Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IR. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 69.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, Baralle F, de BJ, Mitchell JD, Leigh PN, Al-Chalabi A, Miller CC, Nicholson G, Shaw CE. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, Ganesalingam J, Williams KL, Tripathi V, Al-Saraj S, Al-Chalabi A, Leigh PN, Blair IP, Nicholson G, de BJ, Gallo JM, Miller CC, Shaw CE. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schulz JB, Dichgans J. Molecular pathogenesis of movement disorders: are protein aggregates a common link in neuronal degeneration? Curr Opin Neurol. 1999;12:433–439. doi: 10.1097/00019052-199908000-00010. [DOI] [PubMed] [Google Scholar]
- 72.Kabashi E, Durham HD. Failure of protein quality control in amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:1038–1050. doi: 10.1016/j.bbadis.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 73.Riley BE, Zoghbi HY, Orr HT. SUMOylation of the polyglutamine repeat protein, ataxin-1, is dependent on a functional nuclear localization signal. J Biol Chem. 2005;280:21942–21948. doi: 10.1074/jbc.M501677200. [DOI] [PubMed] [Google Scholar]
- 74.Shinbo Y, Niki T, Taira T, Ooe H, Takahashi-Niki K, Maita C, Seino C, Iguchi-Ariga SM, Ariga H. Proper SUMO-1 conjugation is essential to DJ-1 to exert its full activities. Cell Death Differ. 2006;13:96–108. doi: 10.1038/sj.cdd.4401704. [DOI] [PubMed] [Google Scholar]
- 75.Krumova P, Meulmeester E, Garrido M, Tirard M, Hsiao HH, Bossis G, Urlaub H, Zweckstetter M, Kugler S, Melchior F, Bahr M, Weishaupt JH. Sumoylation inhibits alpha-synuclein aggregation and toxicity. J Cell Biol. 2011;194:49–60. doi: 10.1083/jcb.201010117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steffan JS, Agrawal N, Pallos J, Rockabrand E, Trotman LC, Slepko N, Illes K, Lukacsovich T, Zhu YZ, Cattaneo E, Pandolfi PP, Thompson LM, Marsh JL. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 77.Li Y, Wang H, Wang S, Quon D, Liu YW, Cordell B. Positive and negative regulation of APP amyloidogenesis by sumoylation. Proc Natl Acad Sci USA. 2003;100:259–264. doi: 10.1073/pnas.0235361100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fei E, Jia N, Yan M, Ying Z, Sun Q, Wang H, Zhang T, Ma X, Ding H, Yao X, Shi Y, Wang G. SUMO-1 modification increases human SOD1 stability and aggregation. Biochem Biophys Res Commun. 2006;347:406–412. doi: 10.1016/j.bbrc.2006.06.092. [DOI] [PubMed] [Google Scholar]
- 79.Ahn K, Song JH, Kim DK, Park MH, Jo SA, Koh YH. Ubc9 gene polymorphisms and late-onset Alzheimer’s disease in the Korean population: a genetic association study. Neurosci Lett. 2009;465:272–275. doi: 10.1016/j.neulet.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 80.Karpinar DP, Balija MB, Kugler S, Opazo F, Rezaei-Ghaleh N, Wender N, Kim HY, Taschenberger G, Falkenburger BH, Heise H, Kumar A, Riedel D, Fichtner L, Voigt A, Braus GH, Giller K, Becker S, Herzig A, Baldus M, Jackle H, Eimer S, Schulz JB, Griesinger C, Zweckstetter M. Pre-fibrillar alpha-synuclein variants with impaired beta-structure increase neurotoxicity in Parkinson’s disease models. EMBO J. 2009;28:3256–3268. doi: 10.1038/emboj.2009.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, Hulihan M, Peuralinna T, Dutra A, Nussbaum R, Lincoln S, Crawley A, Hanson M, Maraganore D, Adler C, Cookson MR, Muenter M, Baptista M, Miller D, Blancato J, Hardy J, Gwinn-Hardy K. Alpha-synuclein locus triplication causes Parkinson’s disease. Science. 2003;302:841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 82.Paleologou KE, Oueslati A, Shakked G, Rospigliosi CC, Kim HY, Lamberto GR, Fernandez CO, Schmid A, Chegini F, Gai WP, Chiappe D, Moniatte M, Schneider BL, Aebischer P, Eliezer D, Zweckstetter M, Masliah E, Lashuel HA. Phosphorylation at S87 is enhanced in synucleinopathies, inhibits alpha-synuclein oligomerization, and influences synuclein-membrane interactions. J Neurosci. 2010;30:3184–3198. doi: 10.1523/JNEUROSCI.5922-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gorbatyuk OS, Li S, Sullivan LF, Chen W, Kondrikova G, Manfredsson FP, Mandel RJ, Muzyczka N. The phosphorylation state of Ser-129 in human alpha-synuclein determines neurodegeneration in a rat model of Parkinson disease. Proc Natl Acad Sci USA. 2008;105:763–768. doi: 10.1073/pnas.0711053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sato H, Arawaka S, Hara S, Fukushima S, Koga K, Koyama S, Kato T. Authentically phosphorylated alpha-synuclein at Ser129 accelerates neurodegeneration in a rat model of familial Parkinson’s disease. J Neurosci. 2011;31:16884–16894. doi: 10.1523/JNEUROSCI.3967-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marblestone JG, Edavettal SC, Lim Y, Lim P, Zuo X, Butt TR. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15:182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kaminsky R, Denison C, Bening-Abu-Shach U, Chisholm AD, Gygi SP, Broday L. SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans . Dev Cell. 2009;17:724–735. doi: 10.1016/j.devcel.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Droescher M, Begitt A, Vinkemeier U. Paracrystals of STAT proteins and their dissolution by SUMO: how reduced transcription factor solubility increases cytokine signaling. Oncotarget. 2011;2:527–528. doi: 10.18632/oncotarget.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Droescher M, Begitt A, Marg A, Zacharias M, Vinkemeier U. Cytokine-induced paracrystals prolong the activity of signal transducers and activators of transcription (STAT) and provide a model for the regulation of protein solubility by small ubiquitin-like modifier (SUMO) J Biol Chem. 2011;286:18731–18746. doi: 10.1074/jbc.M111.235978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 91.Dorval V, Fraser PE. Small ubiquitin-like modifier (SUMO) modification of natively unfolded proteins tau and alpha-synuclein. J Biol Chem. 2006;281:9919–9924. doi: 10.1074/jbc.M510127200. [DOI] [PubMed] [Google Scholar]
- 92.Crowther RA, Daniel SE, Goedert M. Characterisation of isolated alpha-synuclein filaments from substantia nigra of Parkinson’s disease brain. Neurosci Lett. 2000;292:128–130. doi: 10.1016/S0304-3940(00)01440-3. [DOI] [PubMed] [Google Scholar]
- 93.Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Fiber diffraction of synthetic alpha-synuclein filaments shows amyloid-like cross-beta conformation. Proc Natl Acad Sci USA. 2000;97:4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pountney DL, Chegini F, Shen X, Blumbergs PC, Gai WP. SUMO-1 marks subdomains within glial cytoplasmic inclusions of multiple system atrophy. Neurosci Lett. 2005;381:74–79. doi: 10.1016/j.neulet.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 95.Um JW, Chung KC. Functional modulation of parkin through physical interaction with SUMO-1. J Neurosci Res. 2006;84:1543–1554. doi: 10.1002/jnr.21041. [DOI] [PubMed] [Google Scholar]
- 96.La Spada AR, Paulson HL, Fischbeck KH. Trinucleotide repeat expansion in neurological disease. Ann Neurol. 1994;36:814–822. doi: 10.1002/ana.410360604. [DOI] [PubMed] [Google Scholar]
- 97.Orr HT. Cell biology of spinocerebellar ataxia. J Cell Biol. 2012;197:167–177. doi: 10.1083/jcb.201105092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ueda H, Goto J, Hashida H, Lin X, Oyanagi K, Kawano H, Zoghbi HY, Kanazawa I, Okazawa H. Enhanced SUMOylation in polyglutamine diseases. Biochem Biophys Res Commun. 2002;293:307–313. doi: 10.1016/S0006-291X(02)00211-5. [DOI] [PubMed] [Google Scholar]
- 99.Ryu J, Cho S, Park BC, Lee dH. Oxidative stress-enhanced SUMOylation and aggregation of ataxin-1: implication of JNK pathway. Biochem Biophys Res Commun. 2010;393:280–285. doi: 10.1016/j.bbrc.2010.01.122. [DOI] [PubMed] [Google Scholar]
- 100.Janer A, Werner A, Takahashi-Fujigasaki J, Daret A, Fujigasaki H, Takada K, Duyckaerts C, Brice A, Dejean A, Sittler A. SUMOylation attenuates the aggregation propensity and cellular toxicity of the polyglutamine expanded ataxin-7. Hum Mol Genet. 2010;19:181–195. doi: 10.1093/hmg/ddp478. [DOI] [PubMed] [Google Scholar]
- 101.Chan HY, Warrick JM, Andriola I, Merry D, Bonini NM. Genetic modulation of polyglutamine toxicity by protein conjugation pathways in Drosophila . Hum Mol Genet. 2002;11:2895–2904. doi: 10.1093/hmg/11.23.2895. [DOI] [PubMed] [Google Scholar]
- 102.Selkoe DJ. The cell biology of beta-amyloid precursor protein and presenilin in Alzheimer’s disease. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/S0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- 103.Dorval V, Mazzella MJ, Mathews PM, Hay RT, Fraser PE. Modulation of Abeta generation by small ubiquitin-like modifiers does not require conjugation to target proteins. Biochem. J. 2007;404:309–316. doi: 10.1042/BJ20061451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang YQ, Sarge KD. Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels. Biochem Biophys Res Commun. 2008;374:673–678. doi: 10.1016/j.bbrc.2008.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gibb SL, Boston-Howes W, Lavina ZS, Gustincich S, Brown RH, Jr, Pasinelli P, Trotti D. A caspase-3-cleaved fragment of the glial glutamate transporter EAAT2 is sumoylated and targeted to promyelocytic leukemia nuclear bodies in mutant SOD1-linked amyotrophic lateral sclerosis. J Biol Chem. 2007;282:32480–32490. doi: 10.1074/jbc.M704314200. [DOI] [PubMed] [Google Scholar]
- 106.Boston-Howes W, Gibb SL, Williams EO, Pasinelli P, Brown RH, Jr, Trotti D. Caspase-3 cleaves and inactivates the glutamate transporter EAAT2. J Biol Chem. 2006;281:14076–14084. doi: 10.1074/jbc.M600653200. [DOI] [PubMed] [Google Scholar]
- 107.Foran E, Bogush A, Goffredo M, Roncaglia P, Gustincich S, Pasinelli P, Trotti D. Motor neuron impairment mediated by a sumoylated fragment of the glial glutamate transporter EAAT2. Glia. 2011;59:1719–1731. doi: 10.1002/glia.21218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Seyfried NT, Gozal YM, Dammer EB, Xia Q, Duong DM, Cheng D, Lah JJ, Levey AI, Peng J. Multiplex SILAC analysis of a cellular TDP-43 proteinopathy model reveals protein inclusions associated with SUMOylation and diverse polyubiquitin chains. Mol Cell Proteomics. 2010;9:705–718. doi: 10.1074/mcp.M800390-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 110.Geser F, Lee VM, Trojanowski JQ. Amyotrophic lateral sclerosis and frontotemporal lobar degeneration: a spectrum of TDP-43 proteinopathies. Neuropathology. 2010;30:103–112. doi: 10.1111/j.1440-1789.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lieberman AP, Robitaille Y, Trojanowski JQ, Dickson DW, Fischbeck KH. Polyglutamine-containing aggregates in neuronal intranuclear inclusion disease. Lancet. 1998;351:884. doi: 10.1016/S0140-6736(05)70296-8. [DOI] [PubMed] [Google Scholar]
- 112.Pountney DL, Huang Y, Burns RJ, Haan E, Thompson PD, Blumbergs PC, Gai WP. SUMO-1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp Neurol. 2003;184:436–446. doi: 10.1016/j.expneurol.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 113.Takahashi-Fujigasaki J, Arai K, Funata N, Fujigasaki H. SUMOylation substrates in neuronal intranuclear inclusion disease. Neuropathol Appl Neurobiol. 2006;32:92–100. doi: 10.1111/j.1365-2990.2005.00705.x. [DOI] [PubMed] [Google Scholar]
- 114.Weishaupt JH, Bahr M. Degeneration of axotomized retinal ganglion cells as a model for neuronal apoptosis in the central nervous system—molecular death and survival pathways. Restor Neurol Neurosci. 2001;19:19–27. [PubMed] [Google Scholar]
- 115.Kermer P, Liman J, Weishaupt JH, Bahr M. Neuronal apoptosis in neurodegenerative diseases: from basic research to clinical application. Neurodegener Dis. 2004;1:9–19. doi: 10.1159/000076665. [DOI] [PubMed] [Google Scholar]
- 116.Frank S, Gaume B, Bergmann-Leitner ES, Leitner WW, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/S1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 117.Meuer K, Suppanz IE, Lingor P, Planchamp V, Goricke B, Fichtner L, Braus GH, Dietz GP, Jakobs S, Bahr M, Weishaupt JH. Cyclin-dependent kinase 5 is an upstream regulator of mitochondrial fission during neuronal apoptosis. Cell Death Differ. 2007;14:651–661. doi: 10.1038/sj.cdd.4402087. [DOI] [PubMed] [Google Scholar]
- 118.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 119.Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
- 120.Braschi E, Zunino R, McBride HM. MAPL is a new mitochondrial SUMO E3 ligase that regulates mitochondrial fission. EMBO Rep. 2009;10:748–754. doi: 10.1038/embor.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Besnault-Mascard L, Leprince C, Auffredou MT, Meunier B, Bourgeade MF, Camonis J, Lorenzo HK, Vazquez A. Caspase-8 sumoylation is associated with nuclear localization. Oncogene. 2005;24:3268–3273. doi: 10.1038/sj.onc.1208448. [DOI] [PubMed] [Google Scholar]
- 123.Hayashi N, Shirakura H, Uehara T, Nomura Y. Relationship between SUMO-1 modification of caspase-7 and its nuclear localization in human neuronal cells. Neurosci Lett. 2006;397:5–9. doi: 10.1016/j.neulet.2005.11.057. [DOI] [PubMed] [Google Scholar]
- 124.Lee Y-J, Mou Y, Maric D, Klimanis D, Auh S, Hallenbeck JM. Elevated global SUMOylation in Ubc9 transgenic mice protects their brains against focal cerebral ischemic damage. PLoS ONE. 2011;6(10):e25852. doi: 10.1371/journal.pone.0025852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–615. doi: 10.1038/nrneurol.2011.150. [DOI] [PubMed] [Google Scholar]
- 126.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 127.Han Y, Huang C, Sun X, Xiang B, Wang M, Yeh ET, Chen Y, Li H, Shi G, Cang H, Sun Y, Wang J, Wang W, Gao F, Yi J. SENP3-mediated de-conjugation of SUMO2/3 from promyelocytic leukemia is correlated with accelerated cell proliferation under mild oxidative stress. J Biol Chem. 2010;285:12906–12915. doi: 10.1074/jbc.M109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cimarosti H, Lindberg C, Bomholt SF, Ronn LC, Henley JM. Increased protein SUMOylation following focal cerebral ischemia. Neuropharmacology. 2008;54:280–289. doi: 10.1016/j.neuropharm.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 129.Chan JY, Tsai CY, Wu CH, Li FC, Dai KY, Sun EY, Chan SH, Chang AY. Sumoylation of hypoxia-inducible factor-1alpha ameliorates failure of brain stem cardiovascular regulation in experimental brain death. PLoS ONE. 2011;6:e17375. doi: 10.1371/journal.pone.0017375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc Natl Acad Sci USA. 2000;97(26):14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Mukherjee S, Thomas M, Dadgar N, Lieberman AP, Iñiguez-Lluhí JA. Small ubiquitin-like modifier (SUMO) modification of the androgen receptor attenuates polyglutamine-mediated aggregation. J Biol Chem. 2009;284(32):21296–21306. doi: 10.1074/jbc.M109.011494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Terashima T, Kawai H, Fujitani M, Maeda K, Yasuda H. SUMO-1 co-localized with mutant atrophin-1 with expanded polyglutamines accelerates intranuclear aggregation and cell death. Neuroreport. 2002;13(17):2359–2364. doi: 10.1097/00001756-200212030-00038. [DOI] [PubMed] [Google Scholar]