Abstract
Apoptosis is a common mode of cell death that contributes to neuronal loss associated with neurodegeneration. Single-nucleotide polymorphisms (SNPs) in chromosomal DNA are contributing factors dictating natural susceptibility of humans to disease. Here, the most common SNPs affecting neuronal vulnerability to apoptosis are reviewed in the context of neurological disorders. Polymorphic variants in genes encoding apoptotic proteins, either from the extrinsic (FAS, TNF-α, CASP8) or the intrinsic (BAX, BCL2, CASP3, CASP9) pathways could be highly valuable in the diagnosis of neurodegenerative diseases and stroke. Interestingly, the Arg72Pro SNP in TP53, the gene encoding tumor suppressor p53, was recently revealed a biomarker of poor prognosis in stroke due to its ability to modulate neuronal apoptotic death. Search for new SNPs responsible for genetic variability to apoptosis will ensure the implementation of novel diagnostic and prognostic tools, as well as therapeutic strategies against neurological diseases.
Keywords: Genetic, Polymorphism, p53, Neuron, Apoptosis, Neurodegeneration
Introduction
Programmed cell death (PCD) is an essential and genetically regulated process of cell elimination that maintains the physiological function of all organs and tissues, including the brain [1–3]. Type I or apoptotic cell death is one of the modes of PCD that can contribute to the pathogenesis of many diseases, including cancer, ischemic vascular disease, and neurodegenerative disorders [4, 5]. In neurological diseases, the relative contribution of apoptosis to the degenerative process is yet controversial; however, it appears to be a contributing factor [5, 6]. This review specifically focuses on the genetic determinants to apoptosis as an attempt to better understand the human variability to neurological disorders.
Human natural variability and vulnerability to disease can be determined by single-nucleotide polymorphisms (SNPs) in chromosomal DNA. In this context, SNPs located within pro-apoptotic and antiapoptotic genes have been identified, and their function in tumor susceptibility widely investigated [7, 8]. Notably, cancer is associated with enhanced resistance to cell death, whereas neurodegeneration is caused by premature cell death [2, 9]. It is therefore conceivable that SNPs of genes involved in the negative regulation of apoptotic (i.e., with oncogenic potential) and pro-apoptotic (i.e., with tumor suppression potential) pathways might dictate the risk of developing certain neurological diseases. Here, the most common SNPs affecting neuronal vulnerability to apoptosis are reviewed. Interestingly, variants in genes encoding apoptotic proteins are highly associated with neurodegeneration and stroke. Unraveling novel associations of SNPs responsible for genetic variability to apoptosis with neurological disorders will help in identifying new targets and therapeutic strategies against these devastating diseases.
Neuronal apoptotic pathways
Apoptotic cell death is executed by cysteinyl aspartyl proteases (called caspases) that trigger a coordinated cascade of events leading to the cleavage of crucial substrates, which rapidly dismantle the cell. Caspases are present as inactive pro-enzymes in healthy cells, and are converted into their active forms by proteolytic cleavage at internal aspartic acid residues. This cleavage splits the caspase protein into a small and a large subunit. The caspase cascade involves: the initiator caspases and the executioner caspases. Initiator caspases contain a long pro-domain that provides a protein–protein interaction platform that allows the recruitment of pro-caspases into an activating protein complex. Long pro-domain caspases include caspases-1, -2, -4, -5, -9, -11, and -12 (with an N-terminal caspase-activating recruitment domain), and caspases-8 and -10 (with an N-terminal death effector domain). Executioner caspases-3, -6, and -7, which lack the large N-terminal non-enzymatic domain but posses a short pro-domain, are responsible for most of the cell destruction during apoptosis. Initiator caspases are activated in response to particular stimuli, whereas executioner caspases are particularly important for the ordered dismantling of vital cellular structures [10, 11].
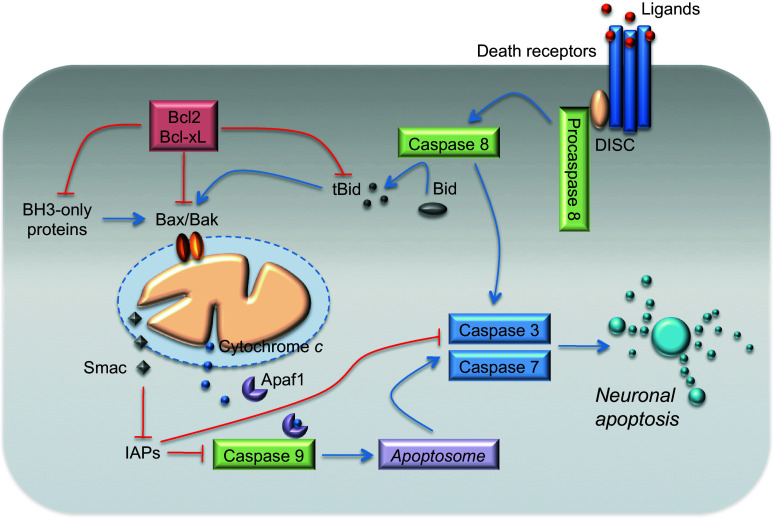
Apoptosis typically proceeds through one of the two signaling cascades, known as extrinsic and intrinsic pathways, both of which converge to activate the executioner caspases-3 and -7 (Fig. 1). The extrinsic pathway is initiated by binding of death receptors, such as Fas or tumor necrosis factor (TNF), with their respective ligands [Fas ligand (FasL) and TNF-α]. This ligand-receptor binding causes receptor trimerization and recruitment of several adaptor proteins at the cytosolic death-domain receptor. FAS recruits FAS-associated death domain protein (FADD), whereas TNF receptor recruits TNFR-associated death domain protein (TRADD), which then recruits FADD. These proteins further bind initiator pro-caspase-8 (or -10) and form the death-inducing signaling complex (DISC) thus enabling their autoactivation [5, 12, 13]. Depending on the efficiency of DISC formation, activated caspase-8 can either directly activate the downstream executioner caspases-3 and -7 [14], or initiate the cleavage of the pro-apoptotic BH3-interacting domain death agonist (Bid), which in turn engages the mitochondrial apoptotic cascade [15, 16] (Fig. 1).
Fig. 1.
The extrinsic and mitochondrial (intrinsic) apoptotic pathways. Apoptosis proceeds through one of two signaling cascades termed the extrinsic and mitochondrial (intrinsic) pathways. The extrinsic pathway is initiated by the ligation of transmembrane death receptors with their ligands, leading to the recruitment of adaptor molecules and to the formation of the pro-caspase-8 DISC. This results in the dimerization and activation of caspase-8, which can either directly cleave and activate caspase-3 and caspase-7, or cleave BH3-interacting domain death agonist (Bid), which in turn engages mitochondrial apoptotic signaling. Truncated Bid promotes the activation of Bax (Bcl-2-associated X protein) and Bak (Bcl-2 antagonist/killer-1), causing outer mitochondrial membrane (OMM) permeabilization. Antiapoptotic proteins, Bcl-2 (B cell lymphoma-2) and Bcl-xL (Bcl-2-like1), prevent OMM permeabilization by sequestering BH3-only proteins, or active Bax and Bak. BH3-only proteins promote Bax and Bak activation. The mitochondrial (intrinsic) pathway is activated by stimuli that trigger the OMM permeabilization and the release of proapoptotic proteins, such as Smac and cytochrome c, from the mitochondrial intermembrane space into the cytosol. Cytochrome c binds to apoptotic protease-activating factor 1 (Apaf-1), inducing its heptamerization and forming the apoptosome that recruits and activates caspase-9. Caspase-9 cleaves and activates the executioner caspase-3 and caspase-7. Smac inhibits the inhibitors of apoptosis proteins (IAPs). See text for a detailed description
The intrinsic (mitochondrial) pathway is activated by stimuli that trigger the permeabilization of the outer mitochondrial membrane (OMM) followed by the release of proapoptotic proteins from the mitochondrial intermembrane space, leading to executioner caspase activation [17, 18]. This pathway is regulated by members of the Bcl-2 family of proteins that contain one or more Bcl-2 homology (BH) domains [3, 19, 20]. The anti-apoptotic subfamily comprises proteins that contain BH domains 1–4, such as Bcl-2, Bcl-2-like 1 (Bcl-xL), and myeloid cell leukemia-1 (Mcl-1), whereas the pro-apoptotic Bax and Bak contain BH domains 1–3 [21, 22]. A larger group of pro-apoptotic proteins, including Bcl-2-associated death promoter (Bad), Bcl-2-interacting mediator of cell death (Bim), and Bid, among others, contain only the BH3 domain.
Activator proteins, including truncated Bid (tBid), Bim, and other factors, such as p53, directly interact with and activates Bax and Bak, resulting in OMM permeabilization [23]. However, antiapoptotic proteins bind and sequester BH3-only proteins, and also bind any monomeric, activated Bax and Bak proteins that might be present [24]. BH3-only proteins provoke death by either binding to antiapoptotic proteins [22] or displacing activators and Bax and Bak from anti-apoptotic proteins, permitting progression of the death signal [25].
Following activation, Bax and Bak form homo-oligomers and participate in the formation of pores in the OMM The OMM permeabilization promotes the release of pro-apoptotic proteins from the mitochondria, including cytochrome c [26]; second mitochondria-derived activator of caspase (Smac) and HtrA2/Omi; and apoptosis-inducing factor (AIF) and endonuclease G, which can induce caspase-independent chromosomal DNA cleavage [27]. Once in the cytosol, cytochrome c joins the adaptor protein apoptotic peptidase-activating factor (Apaf-1) and deoxyATP, and form the apoptosome that recruits pro-caspase-9, resulting in the allosteric activation of caspase-9 and the subsequent activation of effector caspases-3 and -7 [28]. In parallel, cytosolic Smac neutralizes the inhibitors of apoptosis proteins, the key inhibitor of caspases-9, -3, and -7 [29] (Fig. 1).
Although neurological disorders are clinically and neuropathologically very different, studies in transgenic mouse models and in vitro have revealed that neuronal loss by apoptosis is one of the pathological hallmarks of neurodegenerative disorders, including Alzheimer’s disease (AD), Parkinson’s disease (PD), HD, amyotrophic lateral sclerosis (ALS), stroke, brain trauma, spinal cord injury, and diabetic neuropathy [5, 6]. However, its relative importance and role in human disease is still controversial, since other modes of cell death, including type II or autophagy and type III or necrotic cell death, also participate to neuronal loss, in some cases even with higher quantitative relevance than apoptosis [4, 5, 30]. In addition, it is difficult to demonstrate apoptosis convincingly in the brains of patients because of methodological limitations and the imposition of strict criteria for the recognition of apoptosis. Despite these limitations, it is widely accepted that apoptosis participates in neuronal cell death and in the neurodegenerative process [5, 6].
In this context, different proteins involved in the mitochondrial apoptotic pathway have been detected in the brain of patients suffering from several neurological disorders [5]. Activated caspases-9 and -3, or their specific proteolytic products, have been reported in degenerating nerve cells of brain tissue affected by PD [31, 32], AD [33, 34], and ischemic stroke [35, 36]. Moreover, the downregulation of antiapoptotic members of the Bcl-2 family (Bcl-2 and Bcl-xL), and/or the upregulation of their pro-apoptotic counterparts (Bax and Bak), have been documented in patients affected by distinct neurodegenerative disorders, including AD and PD [11, 32, 37–39].
The Fas–FasL system is expressed in the CNS, including glial cells and neurons, and is associated with the maintenance of the immune suppressed status in normal brain [40]. In developing brains, the Fas system participates in neurite branching through its death domain without caspase activation [41]. On the other hand, expression of Fas and FasL is significantly elevated in a variety of neurologic disorders, suggesting that this system may play roles in neurodegenerative and neuroinflammatory responses [40, 42, 43]. Disruption of Fas signaling pathways attenuates neurological damage in experimental cellular and animal models of neuropathological diseases by limiting cell death, including traumatic brain injury [44, 45], multiple sclerosis (MS) [46, 47], stroke [48, 49], ALS [50], AD [51, 52], and PD [53–55].
In the CNS, TNF-α is produced by astrocytes, microglia, and neurons in response to several extrinsic and intrinsic stimuli. Exogenous signals triggered by experimental exposure to bacterial and viral proteins induce inflammatory responses within the CNS and stimulates the expression of TNF-α, among other cytokines [56, 57]. TNF-α expression is also induced by cell-intrinsic stimuli relating to acute brain injury, including cerebral ischemia and trauma, and abnormal release and/or uptake of neurotransmitters, such as glutamate [57], which occurs in several neurodegenerative diseases and stroke [58]. Under these pathological conditions, elevated TNF-α production and TNF-α receptor expression were shown to contribute to neuronal cell death [57, 59, 60].
Polymorphisms in apoptotic genes
SNPs are genetic variations in chromosomal DNA sequences in which a single nucleotide is substituted by one of the other three nucleotides. To be considered a SNP, the least common allele should have a ≥1 % frequency in the human population. The human genome contains about 10–20 million SNPs. The protein coding sequences contain approximately 100,000–300,000 common SNPs, and additional SNPs lie within putative regulatory regions of genes that might be relevant for human health and disease. Although only 1 % of SNPs have the potential to affect directly gene function, this can account for the wide natural genetic diversity of humans [61].
The ability to execute apoptosis is subjected to interindividual variations in humans, which is attributed to genetic factors [8, 62]. Studies in twin pairs support the role of genetic portrait in determining the activity of apoptotic pathways [63, 64]. There are only a limited number of known coding SNPs within PCD pathways; however, numerous non-coding gene polymorphisms should also be considered due to their possible regulatory role. Several of these SNPs have potential functional significance in apoptotic cell death [8]. It is important to note that genetic determinants of apoptotic susceptibility may not be limited to protein-coding genes. The recently described involvement of microRNAs in major apoptosis-mediated pathological processes, including cancer and neurodegeneration [65–67], makes SNPs located within microRNAs and their binding sites to be considered as genetic determinants of disease risk. In this sense, the expression of miRNA molecules is altered in the brain of patients with neurodegenerative diseases, such as AD, PD, HD, and ALS, suggesting that microRNAs might have a crucial regulatory role in these disorders. Thus, polymorphisms in microRNA target sites may constitute an important determinant of neurodegenerative disease risk. The function of microRNA and miRNA target sites SNPs in neurodegeneration has been recently reviewed [65, 68, 69] hence will not be further discussed in this review. Here we review the most common SNPs in genes encoding proteins that modulate neuronal vulnerability to apoptosis. Interestingly, theses SNPs are associated with neurodegenerative diseases and stroke. SNPs with functional significance and clinical association with neurological disorders are summarized in Table 1.
Table 1.
Clinical association with neurological disorders and functional significance of single-nucleotide polymorphisms in apoptotic genes
| Gene, SNP | Molecular description | Functional significance | Clinical association |
|---|---|---|---|
| FAS, −670A > G | A to G substitution within signal transducers and activators of transcription binding sites in the FAS promoter [73] | The −670 A allele is associated with higher level of gene transcription [73] |
Associated with AD risk [74, 75]; other studies failed to show this association [78, 79, 83] Associated with MS risk [72, 80, 81]; Niino et al. [82] failed to show this association |
| FAS, −1377G > A | G to A substitution within Sp1 binding site in the FAS promoter [73] | The −1377 G allele is associated with higher level of gene transcription [73] | Associated with AD risk [83] |
| TNF-α, −308G > A | G to A substitution in the TNF-α promoter | The −308 A allele has higher transcriptional activity and gene expression than the G allele [113] |
Associated with AD risk; however, data have led to disparate results [105, 109, 110] Increases the risk to develop early onset of sporadic PD [119]. Other studies failed to find this association [118] Associated with ischemic stroke risk, but the results have not been consistent across populations [121–125] |
| TNF-α, −850C > T | C to T substitution in the TNF-α promoter | The −850 T allele is associated with a higher level of gene transcription [113] | The TNF −850 T allele synergistically with carriage of the APOE ε4 alleles increase the risk of AD [105, 106]. No positive associations were found in an Italian population [113] |
| TNF-α, −1031C > T | C to T substitution in the TNF-α promoter | The −1031 C allele increases TNF expression [105] | Increases the risk of developing an early onset of sporadic PD [120] |
| CASP8, −652 6N ins/del | Six-nucleotide insertion/deletion in the CASP8 promoter region | The −652 6N del variant destroys the Sp1-binding site and decreases RNA expression and caspase-8 apoptotic activity [130] | Not studied in neurological disorders |
| CASP9, −1263A > G | A to G substitution in the promoter of CASP9 | The −1263 GG genotype enhances the transcriptional activity [141] | Not studied in neurological disorders |
| BCL2, −938C > A | C to A substitution in the inhibitory P2 BCL2 promoter | The −938 C allele increases promoter activity and binding of nuclear proteins [147] | Not studied in neurological disorders |
| BCL2, rs956572A > G | A to G substitution in the intronic region of BCL2 | The AA genotype is associated with lower Bcl-2 mRNA, protein concentrations and greater cellular sensitivity to stress-induced apoptosis [148] |
Modulates grey matter volume in the ventral striatum of healthy subjects [148] Increases the risk to develop bipolar disorder [149] |
| BAX, −248G > A | G to A substitution in the 5′-UTR of BAX | The −248 G variant decreases constitutive Bax expression and increases the Bcl-2/Bax ratio [151, 152] | Not studied in neurological diseases |
| TP53, Arg72Pro | Codon 72 of human TP53 has either the sequence CCC, which encodes proline, or CGC, which encodes arginine within a segment that encodes the proline-rich domain, which is important for p53-induced apoptosis [177, 206] | The Arg72 variant has enhanced capacity to trigger apoptosis in neurons [185] and proliferating cells [208–210] and increases neuronal vulnerability to ischemia-induced apoptosis [185] |
The Arg/Arg genotype is associated with poor functional outcome after stroke [185] and traumatic brain injury [212] The Arg/Arg genotype is associated with higher risk for HD [214]; however, a replication study contradicts this association [215] |
| MDM2, 309T > G | T to G substitution in the first intron of MDM2, which acts as a transcriptional enhancer region [216] | The 309 T allele increase the affinity of the transcriptional activator Sp1, resulting in higher levels of MDM2 RNA and protein and the subsequent attenuation of the p53 pathway [216, 218] | Not studied in neurological disorders |
SNP single-nucleotide polymorphism
Fas polymorphisms
Fas and FasL are expressed in the normal CNS, and their expression is increased in inflamed and degenerated brains. Therefore, the FasL–Fas system has a dual function in the CNS: maintaining the immune suppressed status in normal brain, and inducing neuronal cell death and inflammation in a variety of neurological disorders [40, 42, 43].
The FAS gene is placed in the 10q24 locus, which is involved in the risk of AD [70, 71] and MS [72], as judged by several linkage studies. Interestingly, activation of the FasL–Fas signaling pathway participates in both neuronal and immune cell apoptosis of AD and, as a negative regulator, in the inflammatory component of MS. Moreover, FasL-mediated apoptosis may balance immune cell access to the brain of patients suffering from AD and MS [42]. The FAS promoter is polymorphic, including a G to A substitution at −1,377 bp (−1377G > A, rs2234767) and an A to G substitution at −670 bp (−670A > G, rs1800682), which occur within Sp1 and signal transducers and activators of transcription 1 transcription factor binding sites, respectively, thus modulating gene expression and apoptotic signaling [73]. The most widely studied FAS polymorphism is the −670A > G SNP. Several studies have shown a significant association between this polymorphism and AD risk; but others have not. An association between this polymorphism and non-familial early onset AD was reported in a Scottish population [74] and in subjects from Scotland and Sweden [75], the homozygous GG genotype being enriched in the AD patients. In both studies, the strongest association occurs in carriers of the apolipoprotein E ε4 (APOE ε4) allele, which is strongly associated with age at onset, risk, and unfavorable outcome in AD [76, 77]. These results imply that −670A > G SNP, in interaction with APOE ε4, is a genetic risk factor for sporadic early onset AD. However, other studies failed to validate these findings [78, 79].
Several population studies have also associated the −670A > G SNP with the risk of developing MS. In particular, the FAS −670 G allele decreases the risk of MS, but does not affect the course of the disease [72, 80]. Whereas Katarci et al. [81] described that the association of FAS −670A > G SNP with MS risk is restricted to women, other studies failed to show any association [82].
Sibley et al. [73] demonstrated that the −1377 A allele has reduced ability to bind transcription factor Sp1, while the −1377 G allele is associated with a higher level of gene transcription. One study in subjects from Italy reported an association between −1377G > A SNP and risk of developing AD and a differential rate of cognitive decline during a 2-year follow-up. However, this study did not show an association with either AD risk or rate of cognitive decline and the FAS −670A > G SNP [83].
Recently, Erten-Lyons et al. [84] described that a 3′UTR SNP in FAS gene (rs1468063) is associated with progression of AD and brain volume. In particular, the CT genotype was significantly associated with faster disease progression, smaller total brain, and larger ventricular volumes. While authors speculated that this SNP could cause functional changes in the mRNA structure or its stability, they do not show experimental data demonstrating a functional effect of this SNP. In the study, the authors concluded that rs1468063 SNP might modulate the response to Alzheimer’s pathology by controlling Fas-mediated neuronal apoptosis.
The function of FAS SNPs on PD risk and outcome has been poorly explored. Fas-associated protein 1 (FAF1) was originally identified as a member of the Fas DISC that enhances apoptosis initiated through FasL [85]. FAF1 participates in diverse biological processes, including brain development and neural cell death [86]. FAF1 gene is placed at PARK10 locus on human chromosome 1p32, which is associated with late-onset PD [87]. Recently, Betarbet et al. [88] described that FAF1 levels are increased in the frontal cortex of PD patients, and increased FAF1 levels induces cell death and significantly potentiates the toxic effects of PD-related stressors, including oxidative stress and inhibitors of the mitochondrial complex I and the proteasome. Functional loss of FAF1 may provide a pro-survival signal to cells in a disease state, such as cancer and PD [89]. In this sense, SNP testing has revealed that FAF1 is associated with a genetic locus implicated in susceptibility to Crohn’s disease, increased risk for colorectal cancer [90], and papillary thyroid cancer [91]. However, the effect on PD risk remains unknown.
Dominant missense mutations in leucine-rich repeat kinase 2 (LRRK2) are the most common genetic cause of PD [92]. LRRK2 interacts with FADD and triggers neuronal death through the extrinsic apoptotic pathway. This pathway is activated by disease-triggering mutations, which strengthens the LRRK2–FADD association and the consequent recruitment and activation of caspase-8. The kinase domain of LRRK2 is essential for LRRK2–FADD interaction, since inhibition of the kinase function eliminates the increased FADD binding caused by PD mutations [93], and prevents neurodegeneration [94, 95]. In this context, PD patients have a higher frequency of a common recurrent mutation (G2019S, a glycine-to-serine substitution at codon 2019) located in the kinase domain of LRRK2 [96–98]. However, the effect of this mutation on LRKK2–FADD interaction is currently unknown.
TNF-α polymorphisms
The TNF-α gene, which maps within the class III region of human leukocyte antigen (HLA), is an important mediator of apoptotic and inflammatory responses. In humans, constitutive TNF-α and stimulated TNFR expression depend on two genetic biallelic polymorphisms in the TNF gene locus, localized in the promoter region and in the first intron. In one study, no relationship was found between TNF-α polymorphisms and elevated circulating TNF-α [99]; however, others demonstrated that SNPs of the promoter region of TNF-α gene affect binding to nuclear factors and influence the rate of transcription [100]. Nonetheless, it is unclear whether these variations are relevant for the in vivo regulation of gene activity [29, 100–102].
Circulating TNF-α levels are increased in the brain of AD patients [57, 103], and in vitro studies demonstrated that TNF-α induces the generation of the neurotoxic β-amyloid [104]. It has also been hypothesized that SNPs affecting levels of TNF-α affect the risk of developing AD [105]. Although several TNF-α SNPs have been described, only two promoter gene SNPs, −308G > A (rs1800629) and −850C > T (rs1799724), which modulates binding to transcription factors and affects gene expression, have been associated with increased risk of AD [105–110]. The TNF-α −850 T allele, synergistically with carriage of the APOE ε4 alleles, increase the risk of AD in Caucasian Australians and Northern Europeans [105, 106]. Laws et al. [111] corroborated the interaction of TNF-α −850 T allele with APOE ε4 alleles and its association with AD risk, and further reported an association with lower cerebrospinal fluid β-amyloid levels. However, no positive associations between TNF-α promoter haplotypes and AD disease were found in Italian population [112]. Regarding the TNF-α −308G > A SNP, the A allele has a higher transcriptional activity than the −308 G allele [113] and increases susceptibility for developing AD [108]. Although some case–control studies have associated −308G > A SNP with AD risk, data have led to disparate results [105, 109, 110]. It was suggested that discordant findings might indicate linkage with another locus nearby, different in the diverse population studied. In fact, the TNF-α −308G > A SNP is linked to different transcriptional activities depending on the different HLA haplotypes carrying this SNP [114]. In centenarians, the TNF −308G > A SNP has also been associated with the risk in age-related dementia and longevity. The few centenarians with AA genotype had increase prevalence of dementia and higher mortality risk and tended to show higher plasma levels of TNF-α than GA or GG genotypes. However, these findings need to be validated due to the low number of subjects with AA genotype included in the study [115]. Other SNPs of TNF-α, such as −863C > A, −238G > A, and −1031T > C, were also found to modulate transcriptional activity of TNF-α gene and to be associated with the risk of developing AD in different populations. However, the results have shown a high degree of heterogeneity, probably due to racial differences; besides that they were drawn from a limited number of studies. Thus, further investigations will be required to validate the impact of these SNPs on the risk of AD [105, 116].
Inflammatory processes play an important role in the pathogenesis of PD. Mogi et al. [117] demonstrated that TNF-α levels were enhanced in CSF and in the striatum of PD patients. Thus, genetically determined differences in cytokine production might influence the risk of developing sporadic PD [57, 118]. A significant increase in the frequency of TNF −308 A allele is found in PD patients, and the TNF −308 AA genotype increases the risk to develop early onset sporadic PD [119]. However, other studies failed to find this association [118]. Other genetic analysis of PD patients revealed that the occurrence of the −1031 C allele within the TNF-α promoter was markedly predictive of the early onset form of sporadic PD [120]. These findings suggest that modulation of TNF-α expression enhances the risk of developing PD, and further support the implication of dysfunctional TNF-α signaling in neurodegeneration [57].
The TNF-α gene has also been studied in stroke genomics. Several studies have reported a TNF −308G > A SNP association with ischemic stroke risk, but the results have not been consistent across populations [121–125]. While some studies found that the −308 A allele confers an increased risk of ischemic stroke in young patients [122] and in patients with a preceding febrile episode [124], other studies reported a protective role in adults with ischemic stroke [121] and lacunar infarcts [123]. Moreover, two case–control studies have documented that TNF −308 A allele is associated with resistance against ischemic stroke in males [126, 127]. However, Karahan et al. [128] found no association between the TNF −308G > A SNP and stroke. Whether population-specific differences in TNF −308 allelic frequencies, allelic heterogeneity, and variability in stroke classification criteria explain these contradictory results remains to be elucidated. Nevertheless, together, theses results suggest that TNF-α promoter region polymorphisms are responsible for the susceptibility against stroke and neurodegenerative diseases, but further studies are required to validate these findings.
Polymorphisms in the CASP8 gene
Caspase-8 is an effector protein of the death receptor family pathway, thus mediating neuroinflammatory and neurodegenerative processes. Immunohistochemistry studies revealed that activated caspase-8-positive sites were localized in the brains of AD [129, 130] and PD [131] patients. The CASP8 gene resides on chromosome 2q33. The most frequent references to CASP8 SNPs in literature are (1) the CASP8 promoter region six-nucleotide deletion/insertion (−652 6N ins/del; rs3834129) variant, and (2) a D302H SNP (rs1045485) coding region that results in aspartic acid-to-histidine substitution at codon 302. The −652 6N del variant in the CASP8 promoter destroys the Sp1-binding site that results in decreased RNA expression and lower caspase-8 apoptotic activity [130]. While CASP8 SNPs are strongly associated with the risk of developing several types of cancer [132–136], their association with neurological disorder development is yet unknown.
Gene expression profiling studies revealed that the CASP8 gene is found in close proximity to a susceptibility region for MS located in chromosome 2 [137, 138]. Furthermore, the caspase-8 gene is differentially expressed in studies performed in MS, as well as in the experimental autoimmune encephalomyelitis animal model of MS [138]. Recently, Camiña-Tato et al. [139] described that GG homozygosity for the intronic SNP rs2037815 and CT heterozygosity for the intronic SNP rs12990906 are associated with primary progressive MS when compared with relapse-onset MS and control groups. Moreover, the G allele for SNP rs2037815 was associated with a trend towards faster disease progression in primary progressive MS patients. The authors [139] concluded that CASP8 SNPs are not only associated with increased risk but also with disease susceptibility in primary progressive MS patients. However, Sand [140] described contradictory findings in this study, thus further studies remain to fully support the role of rs2037815 SNP on MS risk and progression.
Polymorphism in genes involved in the mitochondrial apoptotic pathway
Only a few mitochondrial apoptotic genes (BAX, BCL2, CASP3, CASP9) with potential functional significance have been genotyped in cancer patients versus healthy controls. Unfortunately, the majority of theses apoptosis-related SNPs have not yet been tested in appropriate case–control investigations [8].
Polymorphisms in the promoter region of the CASP9 gene may influence the promoter activity of this gene, thereby modulating susceptibility to apoptotic cell death. In particular, the A to G substitution at −1,263A bp (rs4645978) in the promoter region of CASP9 enhances the transcriptional activity of this gene [141]. The functionality of this SNP has been established in cancer risk and intervertebral disc degeneration. In this context, the CASP9 −1263 GG genotype, with lower transcriptional activity, was associated with a decreased risk of lung cancer when compared to that of −1263 AA [141]. On the other hand, subjects with the −1263 GG genotype have a higher risk of developing lumbar disc disease than those with the −1263 AA genotype [142, 143].
Polymorphisms in the CASP3 gene also influence caspase-3 production and/or activity, thereby modulating the susceptibility to lung cancer [144, 145]. Accordingly, Jang et al. recently reported, in a case–control study, an association of G-containing genotype for −928A > G, A-containing genotype for 77G > A, and C-containing genotype for 17532A > C SNPs with reduced risk for lung cancer.
The BCL2 gene, located on chromosome 18q21.3, consists of one large intron, three exons and two promoters with different functional properties. The second promoter (P2) is located 1,400 bp upstream of the translation initiation site and decreases the activity of the P1 promoter, thus functioning as a negative regulatory element [146]. Nückel et al. [147] described that the regulatory −938C > A SNP in the inhibitory P2 BCL2 promoter affects promoter activity, binding of nuclear proteins, and Bcl-2 protein level in B cells from chronic lymphocytic leukemia patients. The C allele displayed significantly increased BCL2 promoter activity and binding of nuclear proteins compared with the A allele. Concomitantly, the AA genotype is associated with increased Bcl-2 protein expression and unfavorable prognosis and overall survival in patients with chronic lymphocytic leukemia. Thus, the −938C > A SNP could be a good candidate to modulate neuronal survival.
Salvadore et al. [148] recently described an intronic SNP (rs956572) in the BCL-2 gene that exerts functional effects on Bcl-2 expression, since the AA homozygous genotype is associated with lower Bcl-2 mRNA, protein concentrations and greater cellular sensitivity to stress-induced apoptosis in human lymphoblasts. The A allele directly affects the brain by significantly reducing grey matter volume in the ventral striatum of healthy subjects [148]. In addition, the rs956572 SNP has been associated with the risk of developing bipolar disorder. Machado-Vieira et al. [149] reported in a recent study that bipolar disorder patients carrying the AA genotype exhibited elevated basal cytosolic Ca2+, in comparison to GG homozygotes, and suggested that it might be a good candidate for a risk allele in mood disorders.
Finally, experiments in mice have revealed a link between a SNP in the BAX promoter (−515 SNP) with the quantitative difference in Bax expression that affects neuronal cell death. Neurons harboring the −515 T polymorphic variant have twice the level of endogenous Bax protein and higher susceptibility to apoptosis than −515 C neurons [150]. Interestingly, a similar phenotype has been described in humans. Saxena et al. [151] demonstrated that high Bcl-2/Bax protein ratio contributes to death resistance in chronic lymphocytic leukemia. They described a novel −248G > A SNP in the 5′-UTR of the human BAX gene that decreases constitutive Bax expression and increases the Bcl-2/Bax protein ratio. In consequence, chronic lymphocytic leukemia patients that possess the −248G > A SNP (GA/GG genotype) show higher resistance to treatment, and a shorter overall survival, than those patients with a wild-type genotype [151, 152]. Given the relatively high prevalence in the normal population [152] and the central role of Bax in neuronal apoptosis [153–155], the BAX −248G > A SNP might also contribute to susceptibility of neurons in genetically complex neurodegenerative diseases.
Bcl-2 protein levels are modulated by the nuclear transcription factor p53. In particular, p53 trans-activate Bax [156], among other pro-apoptotic proteins, leading to Bcl-2 downregulation and apoptotic cell death. Furthermore, p53 mediates neuronal apoptosis in acute and chronic neurological disorders, including experimental cerebral ischemia [157–160], ischemic stroke [161, 162], AD [163–165], and PD [166, 167]. Due to the high impact of p53 in the control of cell survival, the role of TP53 polymorphisms on neuronal susceptibility to apoptosis is discussed in a separate section below.
Polymorphisms in the TP53 gene
The TP53 is a tumor suppressor gene encoding the nuclear transcription factor p53 that regulate several major cellular functions including gene transcription, DNA synthesis, DNA repair, cell cycle regulation, senescence, and cell death. In neurons, p53 mediates apoptosis induced by a range of insults including DNA damage, hypoxia/ischemia, trophic withdrawal, hypoglycemia, oxidative stress, and viral infections [168].
The p53 is short-lived and constitutively expressed, at low levels, in most cell types including neurons [169]. The levels of p53 protein are key for its activity, and are tightly controlled in the cell by covalent modifications [170]. Under unstressed conditions, the interaction of p53 with murine double minute-2 (MDM2) or the human homolog (HDM2) regulates continuous and rapid p53 degradation [171]. In the nucleus, HDM2/MDM2 binds to the transactivation domain of p53, ubiquitylates the protein and mediates its transport into the cytosol, where it is ubiquitylated and then degraded by the proteasome [172, 173]. In turn, p53 transcriptionally regulates MDM2 expression and hence the levels of both p53 and MDM2 are balanced through a negative feedback loop [174]. The related MDM4 protein, also known as MDMX, also modulates p53 activity [175]. The relationship between p53, MDM2 and MDM4 at the molecular level is complex; MDM2 binds to TP53 mRNA, controlling the rate of translation [176], and MDM2 regulates its own levels as well as those of MDM4 and p53 [177].
Under stress conditions, including DNA damage, p53 is stabilized and activated by phosphorylation or acetylation of p53, uncoupling p53 from its negative regulators, mainly the HDM2/MDM2 system, and by proteasome inhibition [178]. The activation of p53 triggers apoptosis by transcriptional activation of pro-apoptotic genes, and by transcriptional-independent mechanisms. On the one hand, p53 can mediate apoptosis by inducing the expression of Bax [156] and other pro-apoptotic proteins, including Bid [179], NADPH oxidase activator 1 (Noxa) [180], and p53-upregulated modulator of apoptosis (PUMA) [181], among other pro-apoptotic proteins that directly act on mitochondria and induce apoptosis. Furthermore, p53 can promote apoptosis through transcription-independent pathways that may involve translocation to mitochondria and direct protein–protein interactions [182, 183]. Recent studies have shown that p53 directly induces OMM permeabilization either by forming an inhibitory complex with protective Bcl-2 or Bcl-xL [160, 184, 185] or by transcriptionally independent activating of Bax, resulting in cytochrome c release and apoptosis [186].
Accumulation of p53 is essential for neuronal apoptosis in response to DNA damage, oxidative damage, cellular calcium overload, and excitotoxicity. In vivo and in vitro studies reveals that increased expression of p53 is associated with neuronal apoptosis following stroke [157–159, 187, 188] and traumatic brain injury [189–191]. Inhibition of p53 activity by systemic administration of pifithrin-α increases brain resistance to ischemic and excitotoxic injury [160, 192, 193]. Accordingly, mice lacking the TP53 gene show decreased neuronal damage after ischemic injury [194].
In addition to acute neurological disorders, progressive neuronal death associated with enhanced p53 levels has been detected in chronic neurodegenerative diseases including AD [165] and PD [167]. Accumulation of p53 in brain tissue has been detected in toxin-induced animal models of PD [195, 196] and in AD patients [197, 198]. Moreover, inhibition of p53 either with pharmacological inhibition or dominant negative constructs is neuroprotective against PD and AD [192, 198, 199].
Most of SNPs in the TP53 gene occur in non-coding sequences. The best characterized intronic TP53 polymorphism is a 16 base pair insertion in intron 3 (Ins16bp, rs17878362), which has been associated with an increase in the risk of several types of cancer; however this association might be a consequence of the close proximity of this polymorphism to the codon 72 polymorphism in exon 4 (Arg72Pro; rs1042522) [177, 200]. So far, the Arg72Pro SNP of TP53 gene is the only apoptosis-associated SNP that has been validated and subjected to a systematic analysis.
The TP53 codon 72 polymorphism (Arg72Pro)
Codon 72 is located in exon 4 in the segment of TP53 that encodes the proline-rich domain, which is important for p53 function, particularly for its ability to induce apoptosis [201, 202]. This domain contains the common polymorphic site, specific to humans, and has either the CCC sequence, which encodes proline (Pro72), or CGC, which encodes arginine (Arg72) [177, 203].
The SNP in codon 72 (Arg72Pr) was first described as a non-tumor-derived amino-acid change that altered the mobility and affected the structure of the proline-rich domain [204, 205]. In 1999, Thomas et al. [206] described for the first time that the Arg72 variant of p53 was more efficient than the Pro72 one at both suppressing transformation by oncogenes and initiating apoptosis. Noteworthy, the frequency of the Pro allele changes with latitude, the areas with intense exposure to the sunlight being the highest, and northern European territories the lowest [207]. All these observations demonstrate that the two alleles might produce functionally distinct proteins. It has been repeatedly demonstrated by different groups that the Pro allele is associated with reduced apoptotic potential of the protein in proliferating normal and tumor cells. Thus, the Arg72 variant of p53 is more effective at inducing apoptosis and protecting stressed cells from neoplastic development than the Pro72. This determines that Arg72Pro SNP has been associated to cancer progression, the age of its onset, and the overall survival of individuals [177, 203, 208–210].
Although one study presented data supporting a transcriptional-dependent mechanism to explain the different apoptotic potentials of the p53 polymorphic variants [211], most studies have reported the absence of differences in specific DNA binding or transcriptional activity between them. This led to demonstrate that the enhanced apoptotic capacity of the Arg72 variant was a consequence of its transport to the mitochondria, resulting to cytochrome c release to the cytosol and caspase-dependent apoptotic cell death [185, 209, 210].
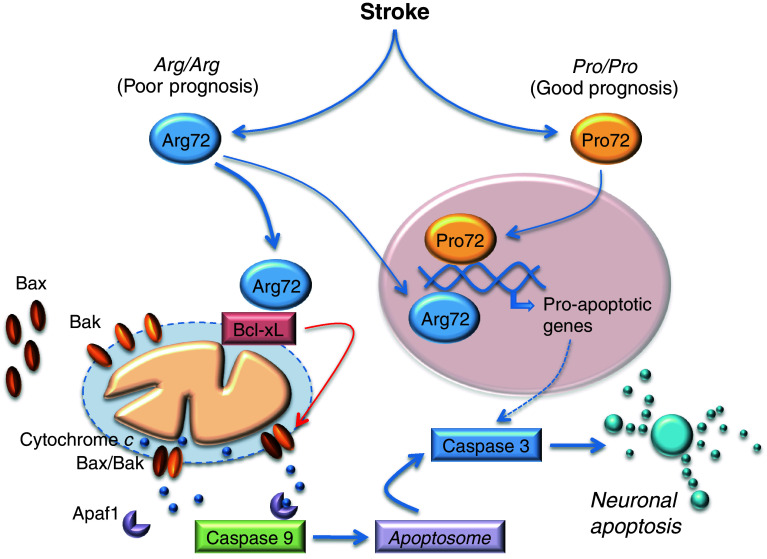
Recently, we [185] revealed that the Arg72 variant of p53 has a higher capacity to trigger neuronal apoptosis than the Pro72 one. Moreover, Arg72 increased the vulnerability of neurons to ischemia-induced apoptotic cell death through the activation of the mitochondrial pathway. We found that Arg72 translocates to the mitochondria and directly binds to, and inactivates, Bcl-xL, thus inducing cytochrome c release to promote caspase-9 activation and neuronal apoptosis (Fig. 2). Moreover, in two independent hospital-based prospective cohorts of patients we also described that the TP53 Arg72Pro SNP strongly determines functional outcome after stroke, regardless whether the origin is ischemic or hemorrhagic [185]. The Arg/Arg genotype was strongly associated to poor functional outcome after stroke, good prognosis being mainly restricted to stroke patients harboring the Pro allele (Fig. 2). Thus, Arg/Arg genotype governs neuronal vulnerability to apoptosis and can be considered as a genetic marker predicting poor functional outcome after either ischemic or hemorrhagic stroke [185].
Fig. 2.
The human TP53 Arg72Pro SNP modulates neuronal susceptibility to ischemia-induced neuronal apoptosis and dictates functional prognosis after stroke. Functional prognosis after stroke is associated with TP53 Arg72Pro SNP. The Arg/Arg genotype is associated with poor functional prognosis after either ischemic or hemorrhagic stroke; the good prognosis is mainly restricted to stroke patients harboring the Pro allele. While both Arg72 and Pro72 polymorphic variants can identically transactivate p53-downstream pro-apoptotic genes, Arg72, but not Pro72, translocates to the mitochondria, where it directly binds to, and inactivates, Bcl-xL, thus inducing mitochondrial outer membrane permeabilization. Subsequent cytochrome c release from the mitochondria to the cytosol promotes the activation of caspase-9 and -3, leading to neuronal apoptosis
Similarly, Martinez-Lucas et al. [212] found that TP53 Arg72Pro SNP is linked to functional outcome after traumatic brain injury in humans. They found that the TP53 Arg/Arg genotype is associated with increased risk of a bad outcome at discharge from the surgical intensive care unit.
The association of Arg72Pro SNP with the risk and progression of chronic neurodegenerative diseases is less clear. A case–control association study between sporadic AD and this SNP found no association between this locus and the risk, age of onset, and progression for AD [213]. Chattopadhyay et al. [214] conducted a case–control study for Arg72Pro SNP and found the Arg/Arg genotype in the TP53 gene to be a significant risk factor for HD. However, in a replication study, Arning et al. [215] contradicted this finding and found no association between Arg72Pro SNP and the age of onset in HD.
The MDM2 309T > G polymorphism
The E3 ubiquitin ligase MDM2 directly binds to and inhibits p53 by regulating its location, stability, and activity as a transcriptional activator [173, 176]. The essential role of MDM2 in the regulation of p53 activity argues that SNPs at this locus should be considered as a potential modulation of p53 function. In fact, polymorphisms in the MDM2 gene have been reported to influence cancer risk, or synergize with, p53 SNPs—or mutations—to modify cancer risk [177]. Although several polymorphisms have been identified in the MDM2 gene, the most intensively characterized SNP is a T to G substitution found at position 309 (309T > G, rs2279744) in the first intron of the MDM2 oncogene, which serves as a transcriptional enhancer region [216]. This SNP increases the affinity of the transcriptional activator Sp1, resulting in higher levels of MDM2 RNA and protein, with the subsequent attenuation of the p53 pathway [216–218]. Furthermore, the elevated levels of MDM2 in cells carrying the homozygous 309 GG genotype result in the inability to properly stabilize p53 and the attenuation of the p53 response to cellular stresses like DNA damage. Although the 309T > G SNP has been extensively associated with accelerated tumor formation in both hereditary and sporadic cancers in humans [216–218], its possible role in stroke and neurodegenerative diseases is largely unknown.
Concluding remarks
Apoptosis regulates selective cell elimination, and SNPs in genes encoding apoptosis regulatory proteins contributes to individual vulnerability to both cancer and neurological disease. Polymorphic variants in genes encoding apoptotic proteins that are associated with high risk of developing neurodegenerative diseases and stroke can reduce the risk of cancer. An interesting example is the Arg72Pro SNP of TP53, the gene encoding tumor suppressor p53, a key regulatory protein in apoptosis. Humans harboring the Arg/Arg allele of TP53 are more vulnerable to poor prognosis in stroke, and less susceptible to cancer, than those bearing the Pro/Pro allele. Thus, unraveling new associations between the SNPs dictating genetic variability to apoptosis and neurological diseases may help to identify novel diagnostic and prognostic tools, targets and therapeutic strategies against these disorders.
Acknowledgments
This work was supported by FEDER (European regional development fund), Instituto de Salud Carlos III (PS09/0366 and RD06/0026/1008) and Junta de Castilla y León (GREX206).
Abbreviations
- AD
Alzheimer’s disease
- AIF
Apoptosis-inducing factor
- ALS
Amyotrophic lateral sclerosis
- Apaf-1
Adaptor protein apoptotic peptidase-activating factor
- APOE ε4
Apolipoprotein E ε4
- Bad
Bcl-2-associated death promoter
- Bak
Bcl-2 antagonist/killer-1
- Bax
Bcl-2-associated X protein
- Bcl-xL
Bcl-2-like 1
- Bcl-2
B-cell lymphoma-2
- BH
Bcl-2 homology
- Bid
BH3-interacting domain death agonist
- Bim
Bcl-2-interacting mediator of cell death
- CASP3
Caspase-3
- CASP8
Caspase-8
- CASP9
Caspase-9
- CNS
Central nervous system
- DISC
Death-inducing signaling complex
- FAF1
Fas-associated protein 1
- FasL
Fas ligand
- HDM2
Human double minute-2
- HLA
Human leukocyte antigen
- LRRK2
Leucine-rich repeat kinase 2
- Mcl-1
Myeloid cell leukemia-1
- MDM2
Murine double minute-2
- MS
Multiple sclerosis
- Noxa
NADPH oxidase activator 1
- OMM
Outer mitochondrial membrane
- PCD
Programmed cell death
- PD
Parkinson disease
- PUMA
p53-upregulated modulator of apoptosis
- SNP
single-nucleotide polymorphisms
- tBid
Truncated Bid
- Sp1
Stimulatory protein 1
- TNF
Tumor necrosis factor
- TRADD
TNFR-associated death domain protein
References
- 1.Levi-Montalcini R. The nerve growth factor: its mode of action on sensory and sympathetic nerve cells. Harvey Lect. 1966;60:217–259. [PubMed] [Google Scholar]
- 2.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yuan J, Yankner BA. Apoptosis in the nervous system. Nature. 2000;407:802–809. doi: 10.1038/35037739. [DOI] [PubMed] [Google Scholar]
- 4.Bredesen DE, Rao RV, Mehlen P. Cell death in the nervous system. Nature. 2006;443:796–802. doi: 10.1038/nature05293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okouchi M, Ekshyyan O, Maracine M, Aw TY. Neuronal apoptosis in neurodegeneration. Antioxid Redox Signal. 2007;9:1059–1096. doi: 10.1089/ars.2007.1511. [DOI] [PubMed] [Google Scholar]
- 6.Cavallucci V, D’Amelio M. Matter of life and death: the pharmacological approaches targeting apoptosis in brain diseases. Curr Pharm Des. 2011;17:215–229. doi: 10.2174/138161211795049705. [DOI] [PubMed] [Google Scholar]
- 7.Zhivotovsky B, Orrenius S. Carcinogenesis and apoptosis: paradigms and paradoxes. Carcinogenesis. 2006;27:1939–1945. doi: 10.1093/carcin/bgl035. [DOI] [PubMed] [Google Scholar]
- 8.Imyanitov EN. Gene polymorphisms, apoptotic capacity and cancer risk. Hum Genet. 2009;125:239–246. doi: 10.1007/s00439-009-0636-7. [DOI] [PubMed] [Google Scholar]
- 9.Plun-Favreau H, Lewis PA, Hardy J, Martins LM, Wood NW. Cancer and neurodegeneration: between the devil and the deep blue sea. PLoS Genet. 2010;6:1–8. doi: 10.1371/journal.pgen.1001257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmer JC, Salvesen GS. Caspase substrates. Cell Death Differ. 2007;14:66–72. doi: 10.1038/sj.cdd.4402059. [DOI] [PubMed] [Google Scholar]
- 11.Ola MS, Nawaz M, Ahsan H. Role of Bcl-2 family proteins and caspases in the regulation of apoptosis. Mol Cell Biochem. 2011;351:41–58. doi: 10.1007/s11010-010-0709-x. [DOI] [PubMed] [Google Scholar]
- 12.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 13.Wajant H. The Fas signaling pathway: more than a paradigm. Science. 2002;296:1635–1636. doi: 10.1126/science.1071553. [DOI] [PubMed] [Google Scholar]
- 14.Peter ME, Krammer PH. The CD95(APO-1/Fas) DISC and beyond. Cell Death Differ. 2003;10:26–35. doi: 10.1038/sj.cdd.4401186. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 16.Billen LP, Shamas-Din A, Andrews DW. Bid: a Bax-like BH3 protein. Oncogene. 2008;27(Suppl 1):S93–104. doi: 10.1038/onc.2009.47. [DOI] [PubMed] [Google Scholar]
- 17.Bolanos JP, Moro MA, Lizasoain I, Almeida A. Mitochondria and reactive oxygen and nitrogen species in neurological disorders and stroke: therapeutic implications. Adv Drug Deliv Rev. 2009;61:1299–1315. doi: 10.1016/j.addr.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 19.Chittenden T, Flemington C, Houghton AB, Ebb RG, Gallo GJ, Elangovan B, Chinnadurai G, Lutz RJ. A conserved domain in Bak, distinct from BH1 and BH2, mediates cell death and protein binding functions. EMBO J. 1995;14:5589–5596. doi: 10.1002/j.1460-2075.1995.tb00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinou JC, Youle RJ. Mitochondria in apoptosis: bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 22.Willis SN, Fletcher JI, Kaufmann T, van Delft MF, Chen L, Czabotar PE, Ierino H, Lee EF, Fairlie WD, Bouillet P, Strasser A, Kluck RM, Adams JM, Huang DC. Apoptosis initiated when BH3 ligands engage multiple Bcl-2 homologs, not Bax or Bak. Science. 2007;315:856–859. doi: 10.1126/science.1133289. [DOI] [PubMed] [Google Scholar]
- 23.Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. Interaction with a membrane surface triggers a reversible conformational change in Bax normally associated with induction of apoptosis. J Biol Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- 24.Lovell JF, Billen LP, Bindner S, Shamas-Din A, Fradin C, Leber B, Andrews DW. Membrane binding by tBid initiates an ordered series of events culminating in membrane permeabilization by Bax. Cell. 2008;135:1074–1084. doi: 10.1016/j.cell.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Shamas-Din A, Brahmbhatt H, Leber B, Andrews DW. BH3-only proteins: orchestrators of apoptosis. Biochim Biophys Acta. 2011;1813:508–520. doi: 10.1016/j.bbamcr.2010.11.024. [DOI] [PubMed] [Google Scholar]
- 26.Ow YP, Green DR, Hao Z, Mak TW. Cytochrome c: functions beyond respiration. Nat Rev Mol Cell Biol. 2008;9:532–542. doi: 10.1038/nrm2434. [DOI] [PubMed] [Google Scholar]
- 27.Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- 28.Riedl SJ, Salvesen GS. The apoptosome: signalling platform of cell death. Nat Rev Mol Cell Biol. 2007;8:405–413. doi: 10.1038/nrm2153. [DOI] [PubMed] [Google Scholar]
- 29.Eckelman BP, Salvesen GS, Scott FL. Human inhibitor of apoptosis proteins: why XIAP is the black sheep of the family. EMBO Rep. 2006;7:988–994. doi: 10.1038/sj.embor.7400795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green DR, Galluzzi L, Kroemer G. Mitochondria and the autophagy-inflammation-cell death axis in organismal aging. Science. 2011;333:1109–1112. doi: 10.1126/science.1201940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartmann A, Hunot S, Michel PP, Muriel MP, Vyas S, Faucheux BA, Mouatt-Prigent A, Turmel H, Srinivasan A, Ruberg M, Evan GI, Agid Y, Hirsch EC. Caspase-3: a vulnerability factor and final effector in apoptotic death of dopaminergic neurons in Parkinson’s disease. Proc Natl Acad Sci USA. 2000;97:2875–2880. doi: 10.1073/pnas.040556597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tatton NA. Increased caspase 3 and Bax immunoreactivity accompany nuclear GAPDH translocation and neuronal apoptosis in Parkinson’s disease. Exp Neurol. 2000;166:29–43. doi: 10.1006/exnr.2000.7489. [DOI] [PubMed] [Google Scholar]
- 33.Stadelmann C, Deckwerth TL, Srinivasan A, Bancher C, Bruck W, Jellinger K, Lassmann H. Activation of caspase-3 in single neurons and autophagic granules of granulovacuolar degeneration in Alzheimer’s disease. Evidence for apoptotic cell death. Am J Pathol. 1999;155:1459–1466. doi: 10.1016/S0002-9440(10)65460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohn TT, Rissman RA, Davis MC, Kim YE, Cotman CW, Head E. Caspase-9 activation and caspase cleavage of tau in the Alzheimer’s disease brain. Neurobiol Dis. 2002;11:341–354. doi: 10.1006/nbdi.2002.0549. [DOI] [PubMed] [Google Scholar]
- 35.Duan SR, Wang JX, Wang J, Xu R, Zhao JK, Wang DS. Ischemia induces endoplasmic reticulum stress and cell apoptosis in human brain. Neurosci Lett. 2010;475:132–135. doi: 10.1016/j.neulet.2010.03.058. [DOI] [PubMed] [Google Scholar]
- 36.Sairanen T, Szepesi R, Karjalainen-Lindsberg ML, Saksi J, Paetau A, Lindsberg PJ. Neuronal caspase-3 and PARP-1 correlate differentially with apoptosis and necrosis in ischemic human stroke. Acta Neuropathol. 2009;118:541–552. doi: 10.1007/s00401-009-0559-3. [DOI] [PubMed] [Google Scholar]
- 37.Satou T, Cummings BJ, Cotman CW. Immunoreactivity for Bcl-2 protein within neurons in the Alzheimer’s disease brain increases with disease severity. Brain Res. 1995;697:35–43. doi: 10.1016/0006-8993(95)00748-f. [DOI] [PubMed] [Google Scholar]
- 38.Jarskog LF, Gilmore JH. Developmental expression of Bcl-2 protein in human cortex. Brain Res Dev Brain Res. 2000;119:225–230. doi: 10.1016/s0165-3806(99)00176-5. [DOI] [PubMed] [Google Scholar]
- 39.Levy OA, Malagelada C, Greene LA. Cell death pathways in Parkinson’s disease: proximal triggers, distal effectors, and final steps. Apoptosis. 2009;14:478–500. doi: 10.1007/s10495-008-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi C, Benveniste EN. Fas ligand/Fas system in the brain: regulator of immune and apoptotic responses. Brain Res Brain Res Rev. 2004;44:65–81. doi: 10.1016/j.brainresrev.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 41.Zuliani C, Kleber S, Klussmann S, Wenger T, Kenzelmann M, Schreglmann N, Martinez A, del Rio JA, Soriano E, Vodrazka P, Kuner R, Groene HJ, Herr I, Krammer PH, Martin-Villalba A. Control of neuronal branching by the death receptor CD95 (Fas/Apo-1) Cell Death Differ. 2006;13:31–40. doi: 10.1038/sj.cdd.4401720. [DOI] [PubMed] [Google Scholar]
- 42.Ethell DW, Buhler LA. Fas ligand-mediated apoptosis in degenerative disorders of the brain. J Clin Immunol. 2003;23:439–446. doi: 10.1023/b:joci.0000010420.96419.a8. [DOI] [PubMed] [Google Scholar]
- 43.Reich A, Spering C, Schulz JB. Death receptor Fas (CD95) signaling in the central nervous system: tuning neuroplasticity? Trends Neurosci. 2008;31:478–486. doi: 10.1016/j.tins.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Demjen D, Klussmann S, Kleber S, Zuliani C, Stieltjes B, Metzger C, Hirt UA, Walczak H, Falk W, Essig M, Edler L, Krammer PH, Martin-Villalba A. Neutralization of CD95 ligand promotes regeneration and functional recovery after spinal cord injury. Nat Med. 2004;10:389–395. doi: 10.1038/nm1007. [DOI] [PubMed] [Google Scholar]
- 45.Beier CP, Kolbl M, Beier D, Woertgen C, Bogdahn U, Brawanski A. CD95/Fas mediates cognitive improvement after traumatic brain injury. Cell Res. 2007;17:732–734. doi: 10.1038/cr.2007.60. [DOI] [PubMed] [Google Scholar]
- 46.Sabelko-Downes KA, Russell JH, Cross AH. Role of Fas–FasL interactions in the pathogenesis and regulation of autoimmune demyelinating disease. J Neuroimmunol. 1999;100:42–52. doi: 10.1016/s0165-5728(99)00191-5. [DOI] [PubMed] [Google Scholar]
- 47.Hovelmeyer N, Hao Z, Kranidioti K, Kassiotis G, Buch T, Frommer F, von Hoch L, Kramer D, Minichiello L, Kollias G, Lassmann H, Waisman A. Apoptosis of oligodendrocytes via Fas and TNF-R1 is a key event in the induction of experimental autoimmune encephalomyelitis. J Immunol. 2005;175:5875–5884. doi: 10.4049/jimmunol.175.9.5875. [DOI] [PubMed] [Google Scholar]
- 48.Martin-Villalba A, Herr I, Jeremias I, Hahne M, Brandt R, Vogel J, Schenkel J, Herdegen T, Debatin KM. CD95 ligand (Fas-L/APO-1L) and tumor necrosis factor-related apoptosis-inducing ligand mediate ischemia-induced apoptosis in neurons. J Neurosci. 1999;19:3809–3817. doi: 10.1523/JNEUROSCI.19-10-03809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rosenbaum DM, Gupta G, D’Amore J, Singh M, Weidenheim K, Zhang H, Kessler JA. Fas (CD95/APO-1) plays a role in the pathophysiology of focal cerebral ischemia. J Neurosci Res. 2000;61:686–692. doi: 10.1002/1097-4547(20000915)61:6<686::AID-JNR12>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 50.Raoul C, Buhler E, Sadeghi C, Jacquier A, Aebischer P, Pettmann B, Henderson CE, Haase G. Chronic activation in presymptomatic amyotrophic lateral sclerosis (ALS) mice of a feedback loop involving Fas, Daxx, and FasL. Proc Natl Acad Sci USA. 2006;103:6007–6012. doi: 10.1073/pnas.0508774103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Su JH, Anderson AJ, Cribbs DH, Tu C, Tong L, Kesslack P, Cotman CW. Fas and Fas ligand are associated with neuritic degeneration in the AD brain and participate in beta-amyloid-induced neuronal death. Neurobiol Dis. 2003;12:182–193. doi: 10.1016/s0969-9961(02)00019-0. [DOI] [PubMed] [Google Scholar]
- 52.Chen Z, Duan RS, Lepecheur M, Paly E, London J, Zhu J. SOD-1 inhibits FAS expression in cortex of APP transgenic mice. Apoptosis. 2005;10:499–502. doi: 10.1007/s10495-005-1879-y. [DOI] [PubMed] [Google Scholar]
- 53.Hartmann A, Mouatt-Prigent A, Faucheux BA, Agid Y, Hirsch EC. FADD: a link between TNF family receptors and caspases in Parkinson’s disease. Neurology. 2002;58:308–310. doi: 10.1212/wnl.58.2.308. [DOI] [PubMed] [Google Scholar]
- 54.Landau AM, Luk KC, Jones ML, Siegrist-Johnstone R, Young YK, Kouassi E, Rymar VV, Dagher A, Sadikot AF, Desbarats J. Defective Fas expression exacerbates neurotoxicity in a model of Parkinson’s disease. J Exp Med. 2005;202:575–581. doi: 10.1084/jem.20050163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gomez-Sintes R, Lucas JJ. NFAT/Fas signaling mediates the neuronal apoptosis and motor side effects of GSK-3 inhibition in a mouse model of lithium therapy. J Clin Invest. 2010;120:2432–2445. doi: 10.1172/JCI37873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uesugi M, Nakajima K, Tohyama Y, Kohsaka S, Kurihara T. Nonparticipation of nuclear factor kappa B (NFkappaB) in the signaling cascade of c-Jun N-terminal kinase (JNK)- and p38 mitogen-activated protein kinase (p38MAPK)-dependent tumor necrosis factor alpha (TNFalpha) induction in lipopolysaccharide (LPS)-stimulated microglia. Brain Res. 2006;1073–1074:48–59. doi: 10.1016/j.brainres.2005.12.043. [DOI] [PubMed] [Google Scholar]
- 57.Park KM, Bowers WJ. Tumor necrosis factor-alpha mediated signaling in neuronal homeostasis and dysfunction. Cell Signal. 2010;22:977–983. doi: 10.1016/j.cellsig.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y, Qin ZH. Molecular and cellular mechanisms of excitotoxic neuronal death. Apoptosis. 2010;15:1382–1402. doi: 10.1007/s10495-010-0481-0. [DOI] [PubMed] [Google Scholar]
- 59.Veglianese P, Lo Coco D, Bao Cutrona M, Magnoni R, Pennacchini D, Pozzi B, Gowing G, Julien JP, Tortarolo M, Bendotti C. Activation of the p38MAPK cascade is associated with upregulation of TNF alpha receptors in the spinal motor neurons of mouse models of familial ALS. Mol Cell Neurosci. 2006;31:218–231. doi: 10.1016/j.mcn.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 60.Tuttolomondo A, Di Raimondo D, di Sciacca R, Pinto A, Licata G. Inflammatory cytokines in acute ischemic stroke. Curr Pharm Des. 2008;14:3574–3589. doi: 10.2174/138161208786848739. [DOI] [PubMed] [Google Scholar]
- 61.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, Ziaugra L, Friedland L, Rolfe A, Warrington J, Lipshutz R, Daley GQ, Lander ES. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 62.Schmitz A, Bayer J, Dechamps N, Goldin L, Thomas G. Heritability of susceptibility to ionizing radiation-induced apoptosis of human lymphocyte subpopulations. Int J Radiat Oncol Biol Phys. 2007;68:1169–1177. doi: 10.1016/j.ijrobp.2007.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Camplejohn RS, Hodgson S, Carter N, Kato BS, Spector TD. Heritability of DNA-damage-induced apoptosis and its relationship with age in lymphocytes from female twins. Br J Cancer. 2006;95:520–524. doi: 10.1038/sj.bjc.6603257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finnon P, Robertson N, Dziwura S, Raffy C, Zhang W, Ainsbury L, Kaprio J, Badie C, Bouffler S. Evidence for significant heritability of apoptotic and cell cycle responses to ionising radiation. Hum Genet. 2008;123:485–493. doi: 10.1007/s00439-008-0500-1. [DOI] [PubMed] [Google Scholar]
- 65.Junn E, Mouradian MM. MicroRNAs in neurodegenerative disorders. Cell Cycle. 2010;9:1717–1721. doi: 10.4161/cc.9.9.11296. [DOI] [PubMed] [Google Scholar]
- 66.Lau P, de Strooper B. Dysregulated microRNAs in neurodegenerative disorders. Semin Cell Dev Biol. 2010;21:768–773. doi: 10.1016/j.semcdb.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 67.Lima RT, Busacca S, Almeida GM, Gaudino G, Fennell DA, Vasconcelos MH. MicroRNA regulation of core apoptosis pathways in cancer. Eur J Cancer. 2011;47:163–174. doi: 10.1016/j.ejca.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 68.Harraz MM, Dawson TM, Dawson VL. MicroRNAs in Parkinson’s disease. J Chem Neuroanat. 2011;42:127–130. doi: 10.1016/j.jchemneu.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Junn E, Mouradian MM. MicroRNAs in neurodegenerative disorders. Cell Cycle. 2011;9:1717–1721. doi: 10.4161/cc.9.9.11296. [DOI] [PubMed] [Google Scholar]
- 70.Ertekin-Taner N, Graff-Radford N, Younkin LH, Eckman C, Baker M, Adamson J, Ronald J, Blangero J, Hutton M, Younkin SG. Linkage of plasma Abeta42 to a quantitative locus on chromosome 10 in late-onset Alzheimer’s disease pedigrees. Science. 2000;290:2303–2304. doi: 10.1126/science.290.5500.2303. [DOI] [PubMed] [Google Scholar]
- 71.Myers A, Holmans P, Marshall H, Kwon J, Meyer D, Ramic D, Shears S, Booth J, DeVrieze FW, Crook R, Hamshere M, Abraham R, Tunstall N, Rice F, Carty S, Lillystone S, Kehoe P, Rudrasingham V, Jones L, Lovestone S, Perez-Tur J, Williams J, Owen MJ, Hardy J, Goate AM. Susceptibility locus for Alzheimer’s disease on chromosome 10. Science. 2000;290:2304–2305. doi: 10.1126/science.290.5500.2304. [DOI] [PubMed] [Google Scholar]
- 72.Huang WX, Huang MP, Gomes MA, Hillert J. Apoptosis mediators fasL and TRAIL are upregulated in peripheral blood mononuclear cells in MS. Neurology. 2000;55:928–934. doi: 10.1212/wnl.55.7.928. [DOI] [PubMed] [Google Scholar]
- 73.Sibley K, Rollinson S, Allan JM, Smith AG, Law GR, Roddam PL, Skibola CF, Smith MT, Morgan GJ. Functional FAS promoter polymorphisms are associated with increased risk of acute myeloid leukemia. Cancer Res. 2003;63:4327–4330. [PubMed] [Google Scholar]
- 74.Feuk L, Prince JA, Breen G, Emahazion T, Carothers A, St Clair D, Brookes AJ. Apolipoprotein-E dependent role for the FAS receptor in early onset Alzheimer’s disease: finding of a positive association for a polymorphism in the TNFRSF6 gene. Hum Genet. 2000;107:391–396. doi: 10.1007/s004390000383. [DOI] [PubMed] [Google Scholar]
- 75.Feuk L, Prince JA, Blennow K, Brookes AJ. Further evidence for role of a promoter variant in the TNFRSF6 gene in Alzheimer disease. Hum Mutat. 2003;21:53–60. doi: 10.1002/humu.10148. [DOI] [PubMed] [Google Scholar]
- 76.Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 77.Saunders AM, Strittmatter WJ, Schmechel D, George-Hyslop PH, Pericak-Vance MA, Joo SH, Rosi BL, Gusella JF, Crapper-MacLachlan DR, Alberts MJ, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 78.Theuns J, Feuk L, Dermaut B, Del-Favero J, Roks G, Van den Bossche D, Corsmit E, Van den Broeck M, van Duijn CM, Cruts M, Brookes AJ, Van Broeckhoven C. The TNFRSF6 gene is not implicated in familial early-onset Alzheimer’s disease. Hum Genet. 2001;108:552–553. doi: 10.1007/s004390100508. [DOI] [PubMed] [Google Scholar]
- 79.He XM, Zhang ZX, Zhang JW, Zhou YT, Tang MN, Wu CB, Hong Z. The Fas gene A-670G polymorphism is not associated with sporadic Alzheimer disease in a Chinese Han population. Brain Res. 2006;1082:192–195. doi: 10.1016/j.brainres.2006.01.086. [DOI] [PubMed] [Google Scholar]
- 80.van Veen T, Kalkers NF, Crusius JB, van Winsen L, Barkhof F, Jongen PJ, Pena AS, Polman CH, Uitdehaag BM. The FAS-670 polymorphism influences susceptibility to multiple sclerosis. J Neuroimmunol. 2002;128:95–100. doi: 10.1016/s0165-5728(02)00163-7. [DOI] [PubMed] [Google Scholar]
- 81.Kantarci OH, Hebrink DD, Achenbach SJ, Atkinson EJ, de Andrade M, McMurray CT, Weinshenker BG. CD95 polymorphisms are associated with susceptibility to MS in women. A population-based study of CD95 and CD95L in MS. J Neuroimmunol. 2004;146:162–170. doi: 10.1016/j.jneuroim.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 82.Niino M, Kikuchi S, Fukazawa T, Miyagishi R, Yabe I, Tashiro K. An examination of the Apo-1/Fas promoter Mva I polymorphism in Japanese patients with multiple sclerosis. BMC Neurol. 2002;2:8. doi: 10.1186/1471-2377-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiappelli M, Nasi M, Cossarizza A, Porcellini E, Tumini E, Pinti M, Troiano L, Franceschi M, Licastro F. Polymorphisms of fas gene: relationship with Alzheimer’s disease and cognitive decline. Dement Geriatr Cogn Disord. 2006;22:296–300. doi: 10.1159/000095160. [DOI] [PubMed] [Google Scholar]
- 84.Erten-Lyons D, Jacobson A, Kramer P, Grupe A, Kaye J. The FAS gene, brain volume, and disease progression in Alzheimer’s disease. Alzheimers Dement. 2010;6:118–124. doi: 10.1016/j.jalz.2009.05.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chu K, Niu X, Williams LT. A Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc Natl Acad Sci USA. 1995;92:11894–11898. doi: 10.1073/pnas.92.25.11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.De Zio D, Ferraro E, D’Amelio M, Simoni V, Bordi M, Soroldoni D, Berghella L, Meyer BI, Cecconi F. Faf1 is expressed during neurodevelopment and is involved in Apaf1-dependent caspase-3 activation in proneural cells. Cell Mol Life Sci. 2008;65:1780–1790. doi: 10.1007/s00018-008-8075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hicks AA, Petursson H, Jonsson T, Stefansson H, Johannsdottir HS, Sainz J, Frigge ML, Kong A, Gulcher JR, Stefansson K, Sveinbjornsdottir S. A susceptibility gene for late-onset idiopathic Parkinson’s disease. Ann Neurol. 2002;52:549–555. doi: 10.1002/ana.10324. [DOI] [PubMed] [Google Scholar]
- 88.Betarbet R, Anderson LR, Gearing M, Hodges TR, Fritz JJ, Lah JJ, Levey AI. Fas-associated factor 1 and Parkinson’s disease. Neurobiol Dis. 2008;31:309–315. doi: 10.1016/j.nbd.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Menges CW, Altomare DA, Testa JR. FAS-associated factor 1 (FAF1): diverse functions and implications for oncogenesis. Cell Cycle. 2009;8:2528–2534. doi: 10.4161/cc.8.16.9280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Weersma RK, Stokkers PC, Cleynen I, Wolfkamp SC, Henckaerts L, Schreiber S, Dijkstra G, Franke A, Nolte IM, Rutgeerts P, Wijmenga C, Vermeire S. Confirmation of multiple Crohn’s disease susceptibility loci in a large Dutch–Belgian cohort. Am J Gastroenterol. 2009;104:630–638. doi: 10.1038/ajg.2008.112. [DOI] [PubMed] [Google Scholar]
- 91.Neta G, Brenner AV, Sturgis EM, Pfeiffer RM, Hutchinson AA, Aschebrook-Kilfoy B, Yeager M, Xu L, Wheeler W, Abend M, Ron E, Tucker MA, Chanock SJ, Sigurdson AJ. Common genetic variants related to genomic integrity and risk of papillary thyroid cancer. Carcinogenesis. 2011;32:1231–1237. doi: 10.1093/carcin/bgr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Berwick DC, Harvey K. LRRK2 signaling pathways: the key to unlocking neurodegeneration? Trends Cell Biol. 2011;21:257–265. doi: 10.1016/j.tcb.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 93.Ho CC, Rideout HJ, Ribe E, Troy CM, Dauer WT. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J Neurosci. 2009;29:1011–1016. doi: 10.1523/JNEUROSCI.5175-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 95.Liu Z, Hamamichi S, Lee BD, Yang D, Ray A, Caldwell GA, Caldwell KA, Dawson TM, Smith WW, Dawson VL. Inhibitors of LRRK2 kinase attenuate neurodegeneration and Parkinson-like phenotypes in Caenorhabditis elegans and Drosophila Parkinson’s disease models. Hum Mol Genet. 2011;20:3933–3942. doi: 10.1093/hmg/ddr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lesage S, Durr A, Tazir M, Lohmann E, Leutenegger AL, Janin S, Pollak P, Brice A. LRRK2 G2019S as a cause of Parkinson’s disease in North African Arabs. N Engl J Med. 2006;354:422–423. doi: 10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 97.Ozelius LJ, Senthil G, Saunders-Pullman R, Ohmann E, Deligtisch A, Tagliati M, Hunt AL, Klein C, Henick B, Hailpern SM, Lipton RB, Soto-Valencia J, Risch N, Bressman SB. LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2006;354:424–425. doi: 10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- 98.Sierra M, Gonzalez-Aramburu I, Sanchez-Juan P, Sanchez-Quintana C, Polo JM, Berciano J, Combarros O, Infante J. High frequency and reduced penetrance of lRRK2 g2019S mutation among Parkinson’s disease patients in Cantabria (Spain) Mov Disord. 2011;26:2343–2346. doi: 10.1002/mds.23965. [DOI] [PubMed] [Google Scholar]
- 99.Kubota T, McNamara DM, Wang JJ, Trost M, McTiernan CF, Mann DL, Feldman AM. Effects of tumor necrosis factor gene polymorphisms on patients with congestive heart failure. VEST Investigators for TNF Genotype Analysis. Vesnarinone Survival Trial. Circulation. 1998;97:2499–2501. doi: 10.1161/01.cir.97.25.2499. [DOI] [PubMed] [Google Scholar]
- 100.Bayley JP, Ottenhoff TH, Verweij CL. Is there a future for TNF promoter polymorphisms? Genes Immun. 2004;5:315–329. doi: 10.1038/sj.gene.6364055. [DOI] [PubMed] [Google Scholar]
- 101.Higuchi T, Seki N, Kamizono S, Yamada A, Kimura A, Kato H, Itoh K. Polymorphism of the 5′-flanking region of the human tumor necrosis factor (TNF)-alpha gene in Japanese. Tissue Antigens. 1998;51:605–612. doi: 10.1111/j.1399-0039.1998.tb03002.x. [DOI] [PubMed] [Google Scholar]
- 102.Skoog T, van’t Hooft FM, Kallin B, Jovinge S, Boquist S, Nilsson J, Eriksson P, Hamsten A. A common functional polymorphism (C–A substitution at position −863) in the promoter region of the tumour necrosis factor-alpha (TNF-alpha) gene associated with reduced circulating levels of TNF-alpha. Hum Mol Genet. 1999;8:1443–1449. doi: 10.1093/hmg/8.8.1443. [DOI] [PubMed] [Google Scholar]
- 103.Tarkowski E, Liljeroth AM, Nilsson A, Ricksten A, Davidsson P, Minthon L, Blennow K. TNF gene polymorphism and its relation to intracerebral production of TNFalpha and TNFbeta in AD. Neurology. 2000;54:2077–2081. doi: 10.1212/wnl.54.11.2077. [DOI] [PubMed] [Google Scholar]
- 104.Blasko I, Marx F, Steiner E, Hartmann T, Grubeck-Loebenstein B. TNFalpha plus IFNgamma induce the production of Alzheimer beta-amyloid peptides and decrease the secretion of APPs. Faseb J. 1999;13:63–68. doi: 10.1096/fasebj.13.1.63. [DOI] [PubMed] [Google Scholar]
- 105.Di Bona D, Candore G, Franceschi C, Licastro F, Colonna-Romano G, Camma C, Lio D, Caruso C. Systematic review by meta-analyses on the possible role of TNF-alpha polymorphisms in association with Alzheimer’s disease. Brain Res Rev. 2009;61:60–68. doi: 10.1016/j.brainresrev.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 106.McCusker SM, Curran MD, Dynan KB, McCullagh CD, Urquhart DD, Middleton D, Patterson CC, McIlroy SP, Passmore AP. Association between polymorphism in regulatory region of gene encoding tumour necrosis factor alpha and risk of Alzheimer’s disease and vascular dementia: a case–control study. Lancet. 2001;357:436–439. doi: 10.1016/s0140-6736(00)04008-3. [DOI] [PubMed] [Google Scholar]
- 107.Alvarez V, Mata IF, Gonzalez P, Lahoz CH, Martinez C, Pena J, Guisasola LM, Coto E. Association between the TNFalpha-308 A/G polymorphism and the onset-age of Alzheimer disease. Am J Med Genet. 2002;114:574–577. doi: 10.1002/ajmg.10515. [DOI] [PubMed] [Google Scholar]
- 108.Candore G, Balistreri CR, Colonna-Romano G, Lio D, Caruso C. Major histocompatibility complex and sporadic Alzheimer’s disease: a critical reappraisal. Exp Gerontol. 2004;39:645–652. doi: 10.1016/j.exger.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 109.Lio D, Annoni G, Licastro F, Crivello A, Forte GI, Scola L, Colonna-Romano G, Candore G, Arosio B, Galimberti L, Vergani C, Caruso C. Tumor necrosis factor-alpha −308A/G polymorphism is associated with age at onset of Alzheimer’s disease. Mech Ageing Dev. 2006;127:567–571. doi: 10.1016/j.mad.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 110.Yang L, Lu R, Jiang L, Liu Z, Peng Y. Expression and genetic analysis of tumor necrosis factor-alpha (TNF-alpha) G −308A polymorphism in sporadic Alzheimer’s disease in a Southern China population. Brain Res. 2009;1247:178–181. doi: 10.1016/j.brainres.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 111.Laws SM, Perneczky R, Wagenpfeil S, Muller U, Forstl H, Martins RN, Kurz A, Riemenschneider M. TNF polymorphisms in Alzheimer disease and functional implications on CSF beta-amyloid levels. Hum Mutat. 2005;26:29–35. doi: 10.1002/humu.20180. [DOI] [PubMed] [Google Scholar]
- 112.Tedde A, Putignano AL, Nacmias B, Bagnoli S, Cellini E, Sorbi S. Lack of association between TNF-alpha polymorphisms and Alzheimer’s disease in an Italian cohort. Neurosci Lett. 2008;446:139–142. doi: 10.1016/j.neulet.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 113.Wilson AG, de Vries N, Pociot F, di Giovine FS, van der Putte LB, Duff GW. An allelic polymorphism within the human tumor necrosis factor alpha promoter region is strongly associated with HLA A1, B8, and DR3 alleles. J Exp Med. 1993;177:557–560. doi: 10.1084/jem.177.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Candore G, Lio D, Romano GC, Caruso C. Pathogenesis of autoimmune diseases associated with 8.1 ancestral haplotype: effect of multiple gene interactions. Autoimmun Rev. 2002;1:29–35. doi: 10.1016/s1568-9972(01)00004-0. [DOI] [PubMed] [Google Scholar]
- 115.Bruunsgaard H, Benfield TL, Andersen-Ranberg K, Hjelmborg JB, Pedersen AN, Schroll M, Pedersen BK, Jeune B. The tumor necrosis factor alpha −308G > A polymorphism is associated with dementia in the oldest old. J Am Geriatr Soc. 2004;52:1361–1366. doi: 10.1111/j.1532-5415.2004.52369.x. [DOI] [PubMed] [Google Scholar]
- 116.Qidwai T, Khan F. Tumour necrosis factor gene polymorphism and disease prevalence. Scand J Immunol. 2011;74:522–547. doi: 10.1111/j.1365-3083.2011.02602.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mogi M, Harada M, Riederer P, Narabayashi H, Fujita K, Nagatsu T. Tumor necrosis factor-alpha (TNF-alpha) increases both in the brain and in the cerebrospinal fluid from parkinsonian patients. Neurosci Lett. 1994;165:208–210. doi: 10.1016/0304-3940(94)90746-3. [DOI] [PubMed] [Google Scholar]
- 118.Infante J, Garcia-Gorostiaga I, Sanchez-Juan P, Sanchez-Quintana C, Gurpegui JL, Rodriguez-Rodriguez E, Mateo I, Berciano J, Combarros O. Inflammation-related genes and the risk of Parkinson’s disease: a multilocus approach. Eur J Neurol. 2008;15:431–433. doi: 10.1111/j.1468-1331.2008.02092.x. [DOI] [PubMed] [Google Scholar]
- 119.Bialecka M, Klodowska-Duda G, Kurzawski M, Slawek J, Gorzkowska A, Opala G, Bialecki P, Sagan L, Drozdzik M. Interleukin-10 (IL10) and tumor necrosis factor alpha (TNF) gene polymorphisms in Parkinson’s disease patients. Parkinsonism Relat Disord. 2008;14:636–640. doi: 10.1016/j.parkreldis.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 120.Nishimura M, Mizuta I, Mizuta E, Yamasaki S, Ohta M, Kaji R, Kuno S. Tumor necrosis factor gene polymorphisms in patients with sporadic Parkinson’s disease. Neurosci Lett. 2001;311:1–4. doi: 10.1016/s0304-3940(01)02111-5. [DOI] [PubMed] [Google Scholar]
- 121.Um JY, Kim HM. Tumor necrosis factor alpha gene polymorphism is associated with cerebral infarction. Brain Res Mol Brain Res. 2004;122:99–102. doi: 10.1016/j.molbrainres.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 122.Rubattu S, Speranza R, Ferrari M, Evangelista A, Beccia M, Stanzione R, Assenza GE, Volpe M, Rasura M. A role of TNF-alpha gene variant on juvenile ischemic stroke: a case–control study. Eur J Neurol. 2005;12:989–993. doi: 10.1111/j.1468-1331.2005.01136.x. [DOI] [PubMed] [Google Scholar]
- 123.Harcos P, Laki J, Kiszel P, Szeplaki Z, Szolnoki Z, Kovacs M, Melegh B, Szeplaki G, Fust G, Blasko B. Decreased frequency of the TNF2 allele of TNF-alpha −308 promoter polymorphism is associated with lacunar infarction. Cytokine. 2006;33:100–105. doi: 10.1016/j.cyto.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 124.Lalouschek W, Schillinger M, Hsieh K, Endler G, Greisenegger S, Marculescu R, Lang W, Wagner O, Cheng S, Mannhalter C. Polymorphisms of the inflammatory system and risk of ischemic cerebrovascular events. Clin Chem Lab Med. 2006;44:918–923. doi: 10.1515/CCLM.2006.165. [DOI] [PubMed] [Google Scholar]
- 125.Hoppe C, Klitz W, D’Harlingue K, Cheng S, Grow M, Steiner L, Noble J, Adams R, Styles L. Confirmation of an association between the TNF(−308) promoter polymorphism and stroke risk in children with sickle cell anemia. Stroke. 2007;38:2241–2246. doi: 10.1161/STROKEAHA.107.483115. [DOI] [PubMed] [Google Scholar]
- 126.Lee BC, Ahn SY, Doo HK, Yim SV, Lee HJ, Jin SY, Hong SJ, Lee SH, Kim SD, Seo JC, Leem KH, Chung JH. Susceptibility for ischemic stroke in Korean population is associated with polymorphisms of the interleukin-1 receptor antagonist and tumor necrosis factor-alpha genes, but not the interleukin-1beta gene. Neurosci Lett. 2004;357:33–36. doi: 10.1016/j.neulet.2003.12.041. [DOI] [PubMed] [Google Scholar]
- 127.Kim OJ, Lee JH, Choi JK, Oh SH, Hong SH, Oh D, Kim NK. Association between tumor necrosis factor-alpha (−308G–A and −238G–A) polymorphisms and homocysteine levels in patients with ischemic strokes and silent brain infarctions. Cerebrovasc Dis. 2010;30:483–490. doi: 10.1159/000319023. [DOI] [PubMed] [Google Scholar]
- 128.Karahan ZC, Deda G, Sipahi T, Elhan AH, Akar N. TNF-alpha −308G/A and IL-6 −174 G/C polymorphisms in the Turkish pediatric stroke patients. Thromb Res. 2005;115:393–398. doi: 10.1016/j.thromres.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 129.Rohn TT, Head E, Nesse WH, Cotman CW, Cribbs DH. Activation of caspase-8 in the Alzheimer’s disease brain. Neurobiol Dis. 2001;8:1006–1016. doi: 10.1006/nbdi.2001.0449. [DOI] [PubMed] [Google Scholar]
- 130.Sun T, Gao Y, Tan W, Ma S, Shi Y, Yao J, Guo Y, Yang M, Zhang X, Zhang Q, Zeng C, Lin D. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter is associated with susceptibility to multiple cancers. Nat Genet. 2007;39:605–613. doi: 10.1038/ng2030. [DOI] [PubMed] [Google Scholar]
- 131.Hartmann A, Troadec JD, Hunot S, Kikly K, Faucheux BA, Mouatt-Prigent A, Ruberg M, Agid Y, Hirsch EC. Caspase-8 is an effector in apoptotic death of dopaminergic neurons in Parkinson’s disease, but pathway inhibition results in neuronal necrosis. J Neurosci. 2001;21:2247–2255. doi: 10.1523/JNEUROSCI.21-07-02247.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.MacPherson G, Healey CS, Teare MD, Balasubramanian SP, Reed MW, Pharoah PD, Ponder BA, Meuth M, Bhattacharyya NP, Cox A. Association of a common variant of the CASP8 gene with reduced risk of breast cancer. J Natl Cancer Inst. 2004;96:1866–1869. doi: 10.1093/jnci/dji001. [DOI] [PubMed] [Google Scholar]
- 133.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, Scollen S, Baynes C, Ponder BA, Chanock S, Lissowska J, Brinton L, Peplonska B, Southey MC, Hopper JL, McCredie MR, Giles GG, Fletcher O, Johnson N, dos Santos Silva I, Gibson L, Bojesen SE, Nordestgaard BG, Axelsson CK, Torres D, Hamann U, Justenhoven C, Brauch H, Chang-Claude J, Kropp S, Risch A, Wang-Gohrke S, Schurmann P, Bogdanova N, Dork T, Fagerholm R, Aaltonen K, Blomqvist C, Nevanlinna H, Seal S, Renwick A, Stratton MR, Rahman N, Sangrajrang S, Hughes D, Odefrey F, Brennan P, Spurdle AB, Chenevix-Trench G, Beesley J, Mannermaa A, Hartikainen J, Kataja V, Kosma VM, Couch FJ, Olson JE, Goode EL, Broeks A, Schmidt MK, Hogervorst FB, Van’t Veer LJ, Kang D, Yoo KY, Noh DY, Ahn SH, Wedren S, Hall P, Low YL, Liu J, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Sigurdson AJ, Stredrick DL, Alexander BH, Struewing JP, Pharoah PD, Easton DF. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 134.Wang M, Zhang Z, Tian Y, Shao J, Zhang Z. A six-nucleotide insertion-deletion polymorphism in the CASP8 promoter associated with risk and progression of bladder cancer. Clin Cancer Res. 2009;15:2567–2572. doi: 10.1158/1078-0432.CCR-08-2829. [DOI] [PubMed] [Google Scholar]
- 135.Ma X, Zhang J, Liu S, Huang Y, Chen B, Wang D. Polymorphisms in the CASP8 gene and the risk of epithelial ovarian cancer. Gynecol Oncol. 2011;122:554–559. doi: 10.1016/j.ygyno.2011.05.031. [DOI] [PubMed] [Google Scholar]
- 136.Olsson M, Zhivotovsky B. Caspases and cancer. Cell Death Differ. 2011;18:1441–1449. doi: 10.1038/cdd.2011.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Goertsches R, Villoslada P, Comabella M, Montalban X, Navarro A, de la Concha EG, Arroyo R, Lopez de Munain A, Otaegui D, Palacios R, Perez-Tur J, Jonasdottir A, Benediktsson K, Fossdal R, Sawcer S, Setakis E, Compston A. A genomic screen of Spanish multiple sclerosis patients reveals multiple loci associated with the disease. J Neuroimmunol. 2003;143:124–128. doi: 10.1016/j.jneuroim.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 138.Comabella M, Martin R. Genomics in multiple sclerosis—current state and future directions. J Neuroimmunol. 2007;187:1–8. doi: 10.1016/j.jneuroim.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 139.Camina-Tato M, Fernandez M, Morcillo-Suarez C, Navarro A, Julia E, Edo MC, Montalban X, Comabella M. Genetic association of CASP8 polymorphisms with primary progressive multiple sclerosis. J Neuroimmunol. 2010;222:70–75. doi: 10.1016/j.jneuroim.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 140.Sand PG. CASP8 in MS. J Neuroimmunol. 2011;230:192. doi: 10.1016/j.jneuroim.2010.09.018. [DOI] [PubMed] [Google Scholar]
- 141.Park JY, Park JM, Jang JS, Choi JE, Kim KM, Cha SI, Kim CH, Kang YM, Lee WK, Kam S, Park RW, Kim IS, Lee JT, Jung TH. Caspase 9 promoter polymorphisms and risk of primary lung cancer. Hum Mol Genet. 2006;15:1963–1971. doi: 10.1093/hmg/ddl119. [DOI] [PubMed] [Google Scholar]
- 142.Wang H, Liu H, Zheng ZM, Zhang KB, Wang TP, Sribastav SS, Liu WS, Liu T. Role of death receptor, mitochondrial and endoplasmic reticulum pathways in different stages of degenerative human lumbar disc. Apoptosis. 2011;16:990–1003. doi: 10.1007/s10495-011-0644-7. [DOI] [PubMed] [Google Scholar]
- 143.Guo TM, Liu M, Zhang YG, Guo WT, Wu SX. Association between caspase-9 promoter region polymorphisms and discogenic low back pain. Connect Tissue Res. 2011;52:133–138. doi: 10.3109/03008207.2010.487621. [DOI] [PubMed] [Google Scholar]
- 144.Jang JS, Kim KM, Choi JE, Cha SI, Kim CH, Lee WK, Kam S, Jung TH, Park JY. Identification of polymorphisms in the caspase-3 gene and their association with lung cancer risk. Mol Carcinog. 2008;47:383–390. doi: 10.1002/mc.20397. [DOI] [PubMed] [Google Scholar]
- 145.Lee SY, Choi YY, Choi JE, Kim MJ, Kim JS, Jung DK, Kang HG, Jeon HS, Lee WK, Jin G, Cha SI, Kim CH, Jung TH, Park JY. Polymorphisms in the caspase genes and the risk of lung cancer. J Thorac Oncol. 2010;5:1152–1158. doi: 10.1097/JTO.0b013e3181e04543. [DOI] [PubMed] [Google Scholar]
- 146.Young RL, Korsmeyer SJ. A negative regulatory element in the bcl-2 5′-untranslated region inhibits expression from an upstream promoter. Mol Cell Biol. 1993;13:3686–3697. doi: 10.1128/mcb.13.6.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nuckel H, Frey UH, Bau M, Sellmann L, Stanelle J, Durig J, Jockel KH, Duhrsen U, Siffert W. Association of a novel regulatory polymorphism (−938C > A) in the BCL2 gene promoter with disease progression and survival in chronic lymphocytic leukemia. Blood. 2007;109:290–297. doi: 10.1182/blood-2006-03-007567. [DOI] [PubMed] [Google Scholar]
- 148.Salvadore G, Nugent AC, Chen G, Akula N, Yuan P, Cannon DM, Zarate CA, Jr, McMahon FJ, Manji HK, Drevets WC. Bcl-2 polymorphism influences gray matter volume in the ventral striatum in healthy humans. Biol Psychiatry. 2009;66:804–807. doi: 10.1016/j.biopsych.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Machado-Vieira R, Pivovarova NB, Stanika RI, Yuan P, Wang Y, Zhou R, Zarate CA, Jr, Drevets WC, Brantner CA, Baum A, Laje G, McMahon FJ, Chen G, Du J, Manji HK, Andrews SB. The Bcl-2 gene polymorphism rs956572AA increases inositol 1,4,5-trisphosphate receptor-mediated endoplasmic reticulum calcium release in subjects with bipolar disorder. Biol Psychiatry. 2011;69:344–352. doi: 10.1016/j.biopsych.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Semaan SJ, Li Y, Nickells RW. A single nucleotide polymorphism in the Bax gene promoter affects transcription and influences retinal ganglion cell death. ASN Neuro. 2010;2:87–101. doi: 10.1042/AN20100003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saxena A, Moshynska O, Sankaran K, Viswanathan S, Sheridan DP. Association of a novel single nucleotide polymorphism, G(−248)A, in the 5′-UTR of BAX gene in chronic lymphocytic leukemia with disease progression and treatment resistance. Cancer Lett. 2002;187:199–205. doi: 10.1016/s0304-3835(02)00378-6. [DOI] [PubMed] [Google Scholar]
- 152.Starczynski J, Pepper C, Pratt G, Hooper L, Thomas A, Milligan D, Bentley P, Fegan C. Common polymorphism G(−248)A in the promoter region of the bax gene results in significantly shorter survival in patients with chronic lymphocytic leukemia once treatment is initiated. J Clin Oncol. 2005;23:1514–1521. doi: 10.1200/JCO.2005.02.192. [DOI] [PubMed] [Google Scholar]
- 153.Deckwerth TL, Elliott JL, Knudson CM, Johnson EM, Jr, Snider WD, Korsmeyer SJ. BAX is required for neuronal death after trophic factor deprivation and during development. Neuron. 1996;17:401–411. doi: 10.1016/s0896-6273(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 154.White FA, Keller-Peck CR, Knudson CM, Korsmeyer SJ, Snider WD. Widespread elimination of naturally occurring neuronal death in Bax-deficient mice. J Neurosci. 1998;18:1428–1439. doi: 10.1523/JNEUROSCI.18-04-01428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Uo T, Kinoshita Y, Morrison RS. Neurons exclusively express N-Bak, a BH3 domain-only Bak isoform that promotes neuronal apoptosis. J Biol Chem. 2005;280:9065–9073. doi: 10.1074/jbc.M413030200. [DOI] [PubMed] [Google Scholar]
- 156.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 157.Crumrine RC, Thomas AL, Morgan PF. Attenuation of p53 expression protects against focal ischemic damage in transgenic mice. J Cereb Blood Flow Metab. 1994;14:887–891. doi: 10.1038/jcbfm.1994.119. [DOI] [PubMed] [Google Scholar]
- 158.Li Y, Chopp M, Zhang ZG, Zaloga C, Niewenhuis L, Gautam S. p53-immunoreactive protein and p53 mRNA expression after transient middle cerebral artery occlusion in rats. Stroke. 1994;25:849–855. doi: 10.1161/01.str.25.4.849. [DOI] [PubMed] [Google Scholar]
- 159.Saito A, Hayashi T, Okuno S, Nishi T, Chan PH. Modulation of p53 degradation via MDM2-mediated ubiquitylation and the ubiquitin–proteasome system during reperfusion after stroke: role of oxidative stress. J Cereb Blood Flow Metab. 2005;25:267–280. doi: 10.1038/sj.jcbfm.9600028. [DOI] [PubMed] [Google Scholar]
- 160.Endo H, Kamada H, Nito C, Nishi T, Chan PH. Mitochondrial translocation of p53 mediates release of cytochrome c and hippocampal CA1 neuronal death after transient global cerebral ischemia in rats. J Neurosci. 2006;26:7974–7983. doi: 10.1523/JNEUROSCI.0897-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cheng T, Liu D, Griffin JH, Fernandez JA, Castellino F, Rosen ED, Fukudome K, Zlokovic BV. Activated protein C blocks p53-mediated apoptosis in ischemic human brain endothelium and is neuroprotective. Nat Med. 2003;9:338–342. doi: 10.1038/nm826. [DOI] [PubMed] [Google Scholar]
- 162.Mitsios N, Gaffney J, Krupinski J, Mathias R, Wang Q, Hayward S, Rubio F, Kumar P, Kumar S, Slevin M. Expression of signaling molecules associated with apoptosis in human ischemic stroke tissue. Cell Biochem Biophys. 2007;47:73–86. doi: 10.1385/cbb:47:1:73. [DOI] [PubMed] [Google Scholar]
- 163.LaFerla FM, Hall CK, Ngo L, Jay G. Extracellular deposition of beta-amyloid upon p53-dependent neuronal cell death in transgenic mice. J Clin Invest. 1996;98:1626–1632. doi: 10.1172/JCI118957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kitamura Y, Shimohama S, Kamoshima W, Matsuoka Y, Nomura Y, Taniguchi T. Changes of p53 in the brains of patients with Alzheimer’s disease. Biochem Biophys Res Commun. 1997;232:418–421. doi: 10.1006/bbrc.1997.6301. [DOI] [PubMed] [Google Scholar]
- 165.Castro RE, Santos MM, Gloria PM, Ribeiro CJ, Ferreira DM, Xavier JM, Moreira R, Rodrigues CM. Cell death targets and potential modulators in Alzheimer’s disease. Curr Pharm Des. 2010;16:2851–2864. doi: 10.2174/138161210793176563. [DOI] [PubMed] [Google Scholar]
- 166.da Costa CA, Sunyach C, Giaime E, West A, Corti O, Brice A, Safe S, Abou-Sleiman PM, Wood NW, Takahashi H, Goldberg MS, Shen J, Checler F. Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson’s disease. Nat Cell Biol. 2009;11:1370–1375. doi: 10.1038/ncb1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Alves da Costa C, Checler F. Apoptosis in Parkinson’s disease: is p53 the missing link between genetic and sporadic Parkinsonism? Cell Signal. 2011;23:963–968. doi: 10.1016/j.cellsig.2010.10.020. [DOI] [PubMed] [Google Scholar]
- 168.Culmsee C, Mattson MP. p53 in neuronal apoptosis. Biochem Biophys Res Commun. 2005;331:761–777. doi: 10.1016/j.bbrc.2005.03.149. [DOI] [PubMed] [Google Scholar]
- 169.Soussi T. The p53 tumor suppressor gene: from molecular biology to clinical investigation. Ann N Y Acad Sci. 2000;910:121–137. doi: 10.1111/j.1749-6632.2000.tb06705.x. [DOI] [PubMed] [Google Scholar]
- 170.Kruse JP, Gu W. SnapShot: p53 posttranslational modifications. Cell. 2008;133(930–930):e931. doi: 10.1016/j.cell.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Michael D, Oren M. The p53–Mdm2 module and the ubiquitin system. Semin Cancer Biol. 2003;13:49–58. doi: 10.1016/s1044-579x(02)00099-8. [DOI] [PubMed] [Google Scholar]
- 172.Prives C, Hall PA. The p53 pathway. J Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 173.Vucic D, Dixit VM, Wertz IE. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 2011;12:439–452. doi: 10.1038/nrm3143. [DOI] [PubMed] [Google Scholar]
- 174.Hu Z, Jin G, Wang L, Chen F, Wang X, Shen H. MDM2 promoter polymorphism SNP309 contributes to tumor susceptibility: evidence from 21 case–control studies. Cancer Epidemiol Biomarkers Prev. 2007;16:2717–2723. doi: 10.1158/1055-9965.EPI-07-0634. [DOI] [PubMed] [Google Scholar]
- 175.Marine JC, Jochemsen AG. Mdmx and Mdm2: brothers in arms? Cell Cycle. 2004;3:900–904. [PubMed] [Google Scholar]
- 176.Candeias MM, Malbert-Colas L, Powell DJ, Daskalogianni C, Maslon MM, Naski N, Bourougaa K, Calvo F, Fahraeus R. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098–1105. doi: 10.1038/ncb1770. [DOI] [PubMed] [Google Scholar]
- 177.Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer. 2009;9:95–107. doi: 10.1038/nrc2584. [DOI] [PubMed] [Google Scholar]
- 178.Meek DW. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 179.Sax JK, Fei P, Murphy ME, Bernhard E, Korsmeyer SJ, El-Deiry WS. BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol. 2002;4:842–849. doi: 10.1038/ncb866. [DOI] [PubMed] [Google Scholar]
- 180.Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science. 2000;288:1053–1058. doi: 10.1126/science.288.5468.1053. [DOI] [PubMed] [Google Scholar]
- 181.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–694. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 182.Moll UM, Zaika A. Nuclear and mitochondrial apoptotic pathways of p53. FEBS Lett. 2001;493:65–69. doi: 10.1016/s0014-5793(01)02284-0. [DOI] [PubMed] [Google Scholar]
- 183.Chipuk JE, Green DR. p53’s believe it or not: lessons on transcription-independent death. J Clin Immunol. 2003;23:355–361. doi: 10.1023/a:1025365432325. [DOI] [PubMed] [Google Scholar]
- 184.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 185.Gomez-Sanchez JC, Delgado-Esteban M, Rodriguez-Hernandez I, Sobrino T, Perez de la Ossa N, Reverte S, Bolanos JP, Gonzalez-Sarmiento R, Castillo J, Almeida A. The human Tp53 Arg72Pro polymorphism explains different functional prognosis in stroke. J Exp Med. 2011;208:429–437. doi: 10.1084/jem.20101523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 187.Hong LZ, Zhao XY, Zhang HL. p53-mediated neuronal cell death in ischemic brain injury. Neurosci Bull. 2010;26:232–240. doi: 10.1007/s12264-010-1111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Galluzzi L, Blomgren K, Kroemer G. Mitochondrial membrane permeabilization in neuronal injury. Nat Rev Neurosci. 2009;10:481–494. doi: 10.1038/nrn2665. [DOI] [PubMed] [Google Scholar]
- 189.Napieralski JA, Raghupathi R, McIntosh TK. The tumor-suppressor gene, p53, is induced in injured brain regions following experimental traumatic brain injury. Brain Res Mol Brain Res. 1999;71:78–86. doi: 10.1016/s0169-328x(99)00155-2. [DOI] [PubMed] [Google Scholar]
- 190.Ng I, Yeo TT, Tang WY, Soong R, Ng PY, Smith DR. Apoptosis occurs after cerebral contusions in humans. Neurosurgery. 2000;46:949–956. doi: 10.1097/00006123-200004000-00034. [DOI] [PubMed] [Google Scholar]
- 191.Raghupathi R. Cell death mechanisms following traumatic brain injury. Brain Pathol. 2004;14:215–222. doi: 10.1111/j.1750-3639.2004.tb00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Culmsee C, Zhu X, Yu QS, Chan SL, Camandola S, Guo Z, Greig NH, Mattson MP. A synthetic inhibitor of p53 protects neurons against death induced by ischemic and excitotoxic insults, and amyloid beta-peptide. J Neurochem. 2001;77:220–228. doi: 10.1046/j.1471-4159.2001.t01-1-00220.x. [DOI] [PubMed] [Google Scholar]
- 193.Luo Y, Kuo CC, Shen H, Chou J, Greig NH, Hoffer BJ, Wang Y. Delayed treatment with a p53 inhibitor enhances recovery in stroke brain. Ann Neurol. 2009;65:520–530. doi: 10.1002/ana.21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Yonekura I, Takai K, Asai A, Kawahara N, Kirino T. p53 potentiates hippocampal neuronal death caused by global ischemia. J Cereb Blood Flow Metab. 2006;26:1332–1340. doi: 10.1038/sj.jcbfm.9600293. [DOI] [PubMed] [Google Scholar]
- 195.Mandir AS, Simbulan-Rosenthal CM, Poitras MF, Lumpkin JR, Dawson VL, Smulson ME, Dawson TM. A novel in vivo post-translational modification of p53 by PARP-1 in MPTP-induced Parkinsonism. J Neurochem. 2002;83:186–192. doi: 10.1046/j.1471-4159.2002.01144.x. [DOI] [PubMed] [Google Scholar]
- 196.Nair VD. Activation of p53 signaling initiates apoptotic death in a cellular model of Parkinson’s disease. Apoptosis. 2006;11:955–966. doi: 10.1007/s10495-006-6316-3. [DOI] [PubMed] [Google Scholar]
- 197.de la Monte SM, Sohn YK, Wands JR. Correlates of p53- and Fas (CD95)-mediated apoptosis in Alzheimer’s disease. J Neurol Sci. 1997;152:73–83. doi: 10.1016/s0022-510x(97)00131-7. [DOI] [PubMed] [Google Scholar]
- 198.Mattson MP. Pathways towards and away from Alzheimer’s disease. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Trimmer PA, Smith TS, Jung AB, Bennett JP., Jr Dopamine neurons from transgenic mice with a knockout of the p53 gene resist MPTP neurotoxicity. Neurodegeneration. 1996;5:233–239. doi: 10.1006/neur.1996.0031. [DOI] [PubMed] [Google Scholar]
- 200.Lazar V, Hazard F, Bertin F, Janin N, Bellet D, Bressac B. Simple sequence repeat polymorphism within the p53 gene. Oncogene. 1993;8:1703–1705. [PubMed] [Google Scholar]
- 201.Walker KK, Levine AJ. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc Natl Acad Sci USA. 1996;93:15335–15340. doi: 10.1073/pnas.93.26.15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sakamuro D, Sabbatini P, White E, Prendergast GC. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997;15:887–898. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- 203.Pietsch EC, Humbey O, Murphy ME. Polymorphisms in the p53 pathway. Oncogene. 2006;25:1602–1611. doi: 10.1038/sj.onc.1209367. [DOI] [PubMed] [Google Scholar]
- 204.Harris N, Brill E, Shohat O, Prokocimer M, Wolf D, Arai N, Rotter V. Molecular basis for heterogeneity of the human p53 protein. Mol Cell Biol. 1986;6:4650–4656. doi: 10.1128/mcb.6.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Buchman VL, Chumakov PM, Ninkina NN, Samarina OP, Georgiev GP. A variation in the structure of the protein-coding region of the human p53 gene. Gene. 1988;70:245–252. doi: 10.1016/0378-1119(88)90196-5. [DOI] [PubMed] [Google Scholar]
- 206.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemically and biologically. Mol Cell Biol. 1999;19:1092–1100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Beckman G, Birgander R, Sjalander A, Saha N, Holmberg PA, Kivela A, Beckman L. Is p53 polymorphism maintained by natural selection? Hum Hered. 1994;44:266–270. doi: 10.1159/000154228. [DOI] [PubMed] [Google Scholar]
- 208.Biros E, Kohut A, Biros I, Kalina I, Bogyiova E, Stubna J. A link between the p53 germ line polymorphisms and white blood cells apoptosis in lung cancer patients. Lung Cancer. 2002;35:231–235. doi: 10.1016/s0169-5002(01)00446-9. [DOI] [PubMed] [Google Scholar]
- 209.Dumont P, Leu JI, Della Pietra AC, III, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 210.Bonafe M, Salvioli S, Barbi C, Trapassi C, Tocco F, Storci G, Invidia L, Vannini I, Rossi M, Marzi E, Mishto M, Capri M, Olivieri F, Antonicelli R, Memo M, Uberti D, Nacmias B, Sorbi S, Monti D, Franceschi C. The different apoptotic potential of the p53 codon 72 alleles increases with age and modulates in vivo ischaemia-induced cell death. Cell Death Differ. 2004;11:962–973. doi: 10.1038/sj.cdd.4401415. [DOI] [PubMed] [Google Scholar]
- 211.Sullivan A, Syed N, Gasco M, Bergamaschi D, Trigiante G, Attard M, Hiller L, Farrell PJ, Smith P, Lu X, Crook T. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004;23:3328–3337. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- 212.Martinez-Lucas P, Moreno-Cuesta J, Garcia-Olmo DC, Sanchez-Sanchez F, Escribano-Martinez J, del Pozo AC, Lizan-Garcia M, Garcia-Olmo D. Relationship between the Arg72Pro polymorphism of p53 and outcome for patients with traumatic brain injury. Intensive Care Med. 2005;31:1168–1173. doi: 10.1007/s00134-005-2715-0. [DOI] [PubMed] [Google Scholar]
- 213.Rosenmann H, Meiner Z, Kahana E, Aladjem Z, Friedman G, Ben-Yehuda A, Grenader T, Wertman E, Abramsky O. An association study of the codon 72 polymorphism in the pro-apoptotic gene p53 and Alzheimer’s disease. Neurosci Lett. 2003;340:29–32. doi: 10.1016/s0304-3940(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 214.Chattopadhyay B, Baksi K, Mukhopadhyay S, Bhattacharyya NP. Modulation of age at onset of Huntington disease patients by variations in TP53 and human caspase activated DNase (hCAD) genes. Neurosci Lett. 2005;374:81–86. doi: 10.1016/j.neulet.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 215.Arning L, Kraus PH, Saft C, Andrich J, Epplen JT. Age at onset of Huntington disease is not modulated by the R72P variation in TP53 and the R196K variation in the gene coding for the human caspase activated DNase (hCAD) BMC Med Genet. 2005;6:35. doi: 10.1186/1471-2350-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, Onel K, Yip L, Hwang SJ, Strong LC, Lozano G, Levine AJ. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 217.Dharel N, Kato N, Muroyama R, Moriyama M, Shao RX, Kawabe T, Omata M. MDM2 promoter SNP309 is associated with the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Clin Cancer Res. 2006;12:4867–4871. doi: 10.1158/1078-0432.CCR-06-0111. [DOI] [PubMed] [Google Scholar]
- 218.Knappskog S, Bjornslett M, Myklebust LM, Huijts PE, Vreeswijk MP, Edvardsen H, Guo Y, Zhang X, Yang M, Ylisaukko-Oja SK, Alhopuro P, Arola J, Tollenaar RA, van Asperen CJ, Seynaeve C, Staalesen V, Chrisanthar R, Lokkevik E, Salvesen HB, Evans DG, Newman WG, Lin D, Aaltonen LA, Borresen-Dale AL, Tell GS, Stoltenberg C, Romundstad P, Hveem K, Lillehaug JR, Vatten L, Devilee P, Dorum A, Lonning PE. The MDM2 promoter SNP285C/309G haplotype diminishes Sp1 transcription factor binding and reduces risk for breast and ovarian cancer in Caucasians. Cancer Cell. 2011;19:273–282. doi: 10.1016/j.ccr.2010.12.019. [DOI] [PubMed] [Google Scholar]