Abstract
The inhibitor of differentiation Id2, a protein lacking the basic DNA-binding domain, is involved in the modulation of a number of biological processes. The molecular mechanisms explaining Id2 pleiotropic functions are poorly understood. Id2 and E2F4 are known to bind simultaneously to c-myc promoter. To study whether Id2 plays a global role on transcriptional regulation, we performed in vivo genome-wide ChIP/chip experiments for Id2 and E2F4 in adult mouse liver. An Id2-containing complex was bound to a common sequence downstream from the TSS on a subset of 442 E2F4 target genes mainly related to cell development and chromatin structure. We found a positive correlation between Id2 protein levels and the expression of E2F4/Id2 targets in fetal and adult liver. Id2 protein stability increased in fetal liver by interaction with USP1 de-ubiquitinating enzyme, which was induced during development. In adult liver, USP1 and Id2 levels dramatically decreased. In differentiated liver tissue, when Id2 concentration was low, E2F4/Id2 was bound to the same region as paused Pol II and target genes remained transcriptionally inactive. Conversely, in fetal liver when Id2 levels were increased, Id2 and Pol II were released from gene promoters and target genes up-regulated. During liver regeneration after partial hepatectomy, we obtained the same results as in fetal liver. Our results suggest that Id2 might be part of a reversible development-related program involved in the paused-ON/OFF state of Pol II on selected genes that would remain responsive to specific stimuli.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-014-1588-1) contains supplementary material, which is available to authorized users.
Keywords: Development, RNA polymerase II, Gene promoter, Fetal liver, Gene expression, Liver regeneration, Chip assay
Introduction
The inhibitor of differentiation Id2 is a HLH protein that lacks the basic region essential for specific binding to E boxes on DNA [1]. It can dimerize with either class-I or -II proteins, inhibiting their DNA binding activity in a dominant-negative fashion [2]. Id2 is a pleiotropic protein involved in the modulation of a variety of biological processes such as cell differentiation, cell proliferation, cell cycle control, senescence, apoptosis or angiogenesis, and metastasis [3–5].
The role of Id2 as a proliferative factor is now recognized as one of the most significant. Indeed, ectopic over-expression of Id2 in different cell types enhances cell proliferation [4, 6]. However, although deregulated expression of Id2 maintains a highly proliferative state, this is not sufficient for cell transformation [3]. Id2 was thought to control cell proliferation by repressing the expression of cell cycle inhibitors such as p21Cip1 or p57Kip2 that are modulated by bHLH factors [4, 7, 8]. Nevertheless, Id2 activity and function can also be dependent on its concentration, dimerization with non-bHLH proteins, or post-translational modifications and sub-cellular distribution [4, 7, 9–12]. Independently of the cell type or mitogenic stimulus, increased Id2 levels and nuclear translocation are key events for Id2 function.
In general, Id2 expression is high in undifferentiated proliferating cells, and low or almost absent in non-proliferating cells such as terminally differentiated cells [13]. On the other hand, Id2 is subjected to polyubiquitination and subsequent protein degradation by the proteasome [12]. It has recently been shown that USP1 de-ubiquitinating enzyme directly binds to and stabilizes Id2 in osteosarcoma cells inhibiting cell differentiation and promoting proliferation [10].
Once into the nucleus, the mechanisms by which Id2 modulates cell proliferation are poorly understood. We have previously shown in experimental models of liver regeneration the putative role played by Id2 as part of a repressor complex on c-myc promoter. On quiescent hepatocytes. a complex containing Id2/mSin3A (HDAC)/E2F4/p130 remains bound to the c-myc promoter while the gene is repressed [9, 14]. It has been postulated that E2F4 and retinoblastoma-binding protein 2 (RBP2) recruit Sin3 to cell-cycle genes during cell differentiation [15]. Id2 could be part of that differentiation program to establish permanent silencing of E2F4 target genes.
On the other hand, a model has been suggested in which E2F4/pocket proteins periodically and reversibly recruit Sin3 to gene promoters during cell-cycle progression [16, 17]. In agreement with this, upon mitogenic stimulation by partial hepatectomy (PHx), Id2 and mSin3A are released from the c-myc promoter, inducing a short c-myc up-regulation [9]. Interestingly, c-myc transcriptional initiation increases soon after PHx in mice liver, but it is compensated for by a concomitant block of transcriptional elongation [18]. The puzzling observation that both pausing and transcriptional initiation of c-myc are enhanced in the regenerating liver leads to the hypothesis that increased transcriptional initiation of c-myc in this growth process might be driven by a component of a more general response. Several genes that share target sequences could be simultaneously activated by a common mechanism, but only additional and specific factors (i.e. transcription or elongation factors) will render the final pattern of gene expression [18].
These data made us wonder whether Id2 could play such a role as part of a common mechanism for transcription initiation/pausing of E2F-driven genes, as has been shown for c-myc. Most data about the role of Id2 refer to either tumors or cultured and/or transformed cells that might reflect conditions far away from a physiological situation. This is especially relevant since Id2 activity depends on the cell microenvironment. Therefore, to explore whether Id2 plays a global role on transcriptional regulation of E2F4 target genes, we performed in vivo genomewide ChIP/chip experiments for Id2 and E2F4 in quiescent mouse liver. Our experiments revealed 442 E2F4/Id2 target genes mostly involved in tissue development and chromatin remodeling. A common E2F4/Id2-binding sequence downstream from TSS was identified in 95 % of genes. Upon differentiation in adult liver, Id2 was indirectly bound to gene promoters and the expression of target genes were repressed, while, in fetal liver, the opposite occurred. In addition, we found a positive correlation between Id2 protein levels and the expression of E2F4/Id2 targets. Id2 bound to the same region and showed the same dynamics as paused RNA-polymerase II (Pol II) on gene promoters upon differentiation. These results were also observed during liver regeneration after partial hepatectomy compared to sham operated mice. From the data presented here, one could infer that E2F4 and Id2 take part in a reversible development-related program on a subset of E2F4 target genes.
Materials and methods
Reagents
General reagents were purchased from Sigma Chemical (St Louis, MO, USA) Secondary antibodies HRP-conjugated were purchased from DAKO and those against Id2, E2F4 and Pol II were from SantaCruz Biotechnology (Santa Cruz, CA, USA). Other antibodies used were USP1 (Cell Signaling), H3-COOH terminal domain (Millipore), and GAPDH (Abcam). Reagents for qPCR were from Applied Biosystems (Foster City, CA, USA).
Animals
Mice (C57BL6) were from Taconic. Animals were kept in individual cages in a controlled environment (12 h dark/12 h light cycle) and received water and food ad libitum. Embryos were collected from sacrificed pregnant mice at day 14 gestation (E14 with the day of appearance of vaginal plug being taken as day 0). Fetal livers, dissected from embryos coming from the same pregnant mouse, were pooled in one sample. Two-thirds of the PHx or sham operations were performed in males as previously described [9] and liver tissue was collected 3 h later. Liver tissue samples were formaldehyde-cross-linked, cell-fractionated, or snap frozen in liquid nitrogen, and stored at −80 °C for future analysis.
Ethics statement
The studies satisfied the guidelines for experimental animals and were approved by the Research Committee of the School of Medicine at the University of Valencia. The mice were handled in agreement with the National Institutes of Health guidelines and the Guiding Principles for Research Involving Animals and Humans approved by the Council of the American Physiological Society.
RNA isolation and expression analysis by qPCR
Total RNA was extracted using TRIzol® reagent (Invitrogen Life Technologies) followed by additional column purification (RNeasy; Qiagen). The concentration and purity of RNA were assessed as described [14].
RNA (0.5 μg) was reverse-transcribed using the High Capacity RNA-to-cDNA kit (Applied Biosystems) and the cDNA products amplified by qPCR using the TaqMan Universal PCR Master Mix (Applied Biosystems). qPCR reactions were carried out with 1 μL cDNA in a final volume of 10 μL, using a 7900HT Fast Real-Time PCR system (Applied Biosystems). Specific primers (pre-developed Taqman primers) for Id2 (Mm00711781_m1), c-myc (Mm00487803_m1), ep400 (Mm00551485_m1), lsm1l (Mm00659493_m1), Jmjd6 (Mm00466679_m1), Suv39h2 (Mm00469689_m1), and 18S were purchased from Applied Biosystems. Results were normalized according to 18S quantification in the same sample reaction.
ChIP assay
Formaldehyde-fixed tissue samples were immunoprecipitated according to Torres et al. [14]. Briefly, liver tissue was homogenized and chromatin extracted as described. cross-linked chromatin was subjected to sonication with a Vibra-Cell VCX-500 sonicator. The diluted chromatin was pre-cleared and supernatants incubated with 2 μg of the corresponding antibody at 4 °C overnight. Immunocomplexes were incubated with blocked protein A/G-Sepharose (GE Healthcare) for an additional period of 4 h and recovered as previously described. An aliquot of cross-linked chromatin was subjected to the same immunoprecipitation reaction in the presence of normal serum IgG (DAKO) and used as negative control. An aliquot of whole chromatin was collected and labeled as input.
Analysis of ChIP assays by qPCR
DNA from the input, immunoprecipitated, and IgG fractions was purified with a PCR purification kit (Qiagen, Hilden, Germany) and analyzed by qPCR with specific primers labeled with Sybergreen. Primers were designed flanking that region matched by probes in the Affymetrix GeneChip Mouse Promoter Array. Amplification products were also size-fractionated by 2 % agarose gel electrophoresis and stained with ethidium bromide. Primers sequences are included in Online resource 1. Fold enrichment was calculated as percentage versus input.
ChIP/chip
Amplification and labeling
The products of three independent ChIP assays were amplified, labeled, and hybridized to mouse promoter 1.0 Array (Affymetrix UK, High Wycombe, UK). Immunoprecipitated and input DNA were purified and amplified by ligation-mediated PCR as described [19]. PCR-amplified ChIP targets were purified with Affymetrix cDNA cleanup columns, provided in the Genechip Sample Cleanup Module. Amplified DNA (1 μg) was used for the fragmentation reaction and labeling using the Affymetrix GeneChip WT Double-Stranded DNA Terminal Labeling Kit protocol.
Microarray hybridization
We used Affymetrix promoter arrays that contained approximately 28,000 known mouse genes centered on the region from −6 to +2.5 kb relative to the TSS at an average resolution of 35 bp. Array hybridization was performed according to the Affymetrix-recommended protocol and GeneChip Operating Software supplied by Affymetrix was used to generate CEL. files.
Quality control of microarray data (3 Id2 arrays, 3 E2F4 arrays, and 3 input DNA arrays) was performed using Affymetrix Tiling Analysis Software and R/Bioconductor. Plotted histograms of the raw data intensity suggested that enrichment by both Id2 and E2F4 was optimal in all the experiments performed. A detailed explanation is included in Online resource 2.
Data analysis
The automated Genomatix ChipInspector package was used to determine the E2F4 and Id2 binding sites on mouse promoters. CEL files were imported into ChipInspector, for linear total intensity normalization and Log2 transformation. The procedure combines one-class statistical analysis (input vs. bound) with exhaustive matching and six different experiments (comparisons). A single t test related to the significance analysis of microarrays algorithm was performed at the single probe level as described [20]. A minimum of three consecutive probes in a window size of 300 bp was used as the standard setting based on our experience with Affymetrix GeneChip Mouse Promoter Array. In order to diminish the possibility of including falsely detected probes we selected a high stringent Delta value so that we obtained a FDR of 0 % for both, E2F4 and Id2. Although this approach decreased the number of detected probes, the danger to get false discovery probes was dramatically reduced compared to higher FDR values typically used in most papers. This analysis resulted in 9,307 significant positive probes for E2F4 and 871 for Id2 (Online resource 3, 4).
The E2F4, Id2 or E2F4/Id2-enriched regions from ChipInspector analysis were directly subjected to downstream sequence analysis using the RegionMiner tool (Genomatix). GePS (Genomatix), which combines literature analysis with genome annotation, was used to create subgroups of ChIP/chip enriched genes included into different categories. Sequences spanning TSS were extracted using the Gene2promoter module of the Genomatix Software Suite.
Subcellular fractionation
Fresh liver tissue samples from either fetal or adult mice were used for cell fractionation. Nuclear and cytosolic fractions were extracted with the nuclear extract kit (Active Motif) following the manufacturer’s recommended protocol.
Immunoblot analysis
Tissue samples were homogenized and protein extraction was performed as previously described [14]. Equal amount of proteins were loaded into a SDS-PAGE gel and subjected to electrophoresis. Proteins were transferred to nitrocellulose membranes. The specific proteins were detected using the corresponding antibodies and secondary horseradish-conjugated antibodies. Additionally, to confirm the specificity of Id2 antibody, this was neutralized with blocking peptide (sc-489P; SantaCruz, Biotech). The neutralized antibody was then used side-by-side with the antibody alone, and the results were compared. Blots were developed by enhanced chemoluminiscence (GE Healthcare). Protein levels were quantified and normalized as described in figure legends.
Immunoprecipitation
Aliquots of 200 μl of proteins from whole liver tissue were immunoprecipitated overnight with α-Id2, α-USP-1 antibodies, or normal serum IgG at 4 °C. The protein–antibody complexes were pulled down adding 50 μl of 50 % (v/w) protein A/G-Sepharose. Sample pellets were then subjected to several washes, and immunocomplexes recovered by boiling the samples in electrophoresis loading buffer. The immunocomplexes were analyzed by western blot as previously described.
Nucleosome positioning by micrococcal nuclease protection assay
Nuclei were obtained from cross-linked livers, as described [21]. To estimate the position of nucleosomes, isolated nuclei from cross-linked tissues were incubated with micrococcal nuclease (MNase) to obtain mononucleosomes. After purification of the mononucleosomal DNA, qPCR analysis was performed using primers that amplified short tiled fragments covering the region included between 0 and +600 bp relative to the transcription start site (TSS) of genes. Amplification products represent protected regions from nuclease digestion by the presence of a nucleosome. The Input sample (an aliquot of the samples randomly fragmented by sonication to mononucleosomal size) was included to normalize each amplicon in the qPCR reaction.
Nuc-ChIP assay
To detect H3 or Pol II at MNase protected areas, the sonication step of the standard ChIP protocol was replaced by extensive micrococcal nuclease digestion of isolated cross-linked nuclei. The resulting chromatin fragments are thus enriched in mononucleosomes, which were immunoprecipitated with the desired antibody as described elsewhere. The detection of the immunoprecipitated DNA was carried out by qPCR (primers sequences are shown in Online resource 1).
Statistical analysis
Statistical analysis was performed with the Graph Pad Prism v.5.00 software (Graph Pad Software, San Diego, CA, USA). Data are the mean ± SEM of at least three independent experiments. Representative blots and ChIPs are shown. Statistical significance was estimated with one-sample Student’s t test. A p value of <0.05 was considered significant. The Mann–Whitney test was used to analyze the expression of datasets from fetal and adult liver.
Results
Identification of E2F4/Id2 binding sites in liver tissue by ChIP/chip analysis
To elucidate whether Id2 plays a global role in transcriptional regulation or if it is restricted to E2F4 target genes in mice liver, we combined chromatin immunoprecipitation and mouse whole genome promoter array profiling.
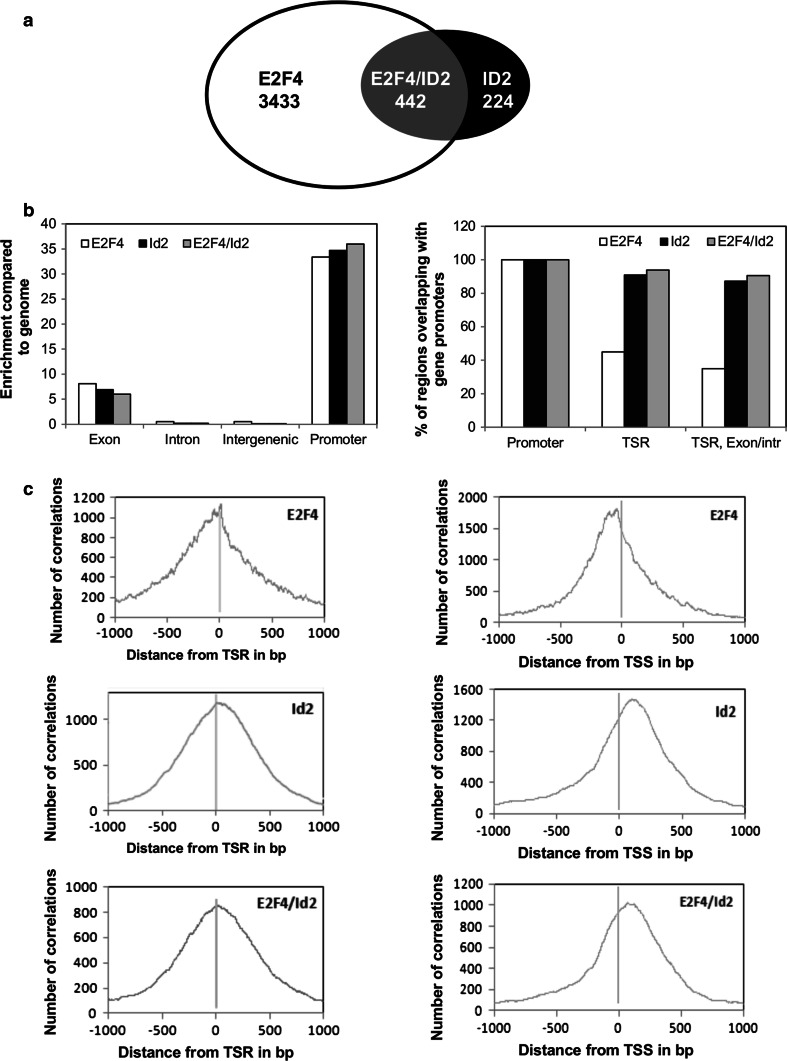
The analysis of at least three independent experiments in liver tissue samples identified 3,875 target genes for E2F4 and 666 for Id2 (Online resource 3, 4). As shown in Fig. 1, the merge analysis of both groups of target genes revealed a limited overlap of 442 genes bound by both proteins, E2F4 and Id2 (Online resource 5). These data confirm that, although the vast majority of E2F4 target genes (89 %) were not bound by Id2, there is an important subset of genes in which Id2 could have a role as a transcriptional regulator. On the other hand, we also found that there is a set of genes bound by Id2 alone (33 % of total Id2-bound genes).
Fig. 1.
Identification of E2F4/Id2 target genes and binding site localization on gene promoters in mice liver. a Venn diagrams showing E2F4, Id2 and overlapping E2F4/Id2 target genes identified by ChIP/chip experiments in adult mice liver. Target genes were the result of at least three independent experiments analyzed by ChipInspector program. b E2F4 and Id2 region classification analysis. General annotation and statistics for total number of regions covered by E2F4 (9307), Id2 (871), or E2F4/Id2 (550). The type of genomic element and region enrichment compared to the genome is shown in the left graph for each protein. Details for regions overlapping with gene promoters are shown in the right graph as the percentage of input regions overlapping with either, at least one transcriptional start region (TSR), or at least one TSR and the first exon or intron of alternative transcripts (TSR, exon/intron). Gene promoter regions were extracted with RegionMiner program and subjected to an exhaustive analysis. Information of overlapping regions and gene IDs is shown in Online resource 3–5. c Distribution and location of E2F4, Id2, and E2F4/Id2 binding regions relative to the middle of a TSR (left) or a TSS (right). GenomeInspector analysis of tiling array data were plotted as the number of correlations found between target regions and the distance from the indicated genomic elements (labeled as 0) flanked by ±1,000 bp
A genomewide factor location analysis with general annotation and statistics for genomic regions (Fig. 1b) revealed that positive regions for E2F4, Id2, or E2F4/Id2 binding had a homogeneous distribution with no apparent difference among the three groups. Indeed, in all groups, binding-enrichment was mainly observed in promoter regions (Fig. 1b, left panel). However, a more exhaustive analysis of regions distribution overlapping with gene promoters, revealed important differences among the groups. While 87 % of Id2 and 90.6 % of E2F4/Id2 regions included within promoters contained at least one transcriptional start region (TSR, defined as a region with more than one TSS at a distance of less than 40 bp) overlapping with an exon or an intron, just 35 % of E2F4-bound regions fell into this classification (Fig. 1b, right panel).
To further establish the binding position of E2F4 and Id2, we analyzed the frequency of distance correlations between tiling array positive regions and the middle of TSR or the TSS of annotated target genes. The three groups of target regions were centered at their respective TSRs (Fig. 1c). However, a dramatic difference was observed between E2F4 and Id2 binding positions related to the TSS of target genes: E2F4 was mostly restricted to positions immediately upstream of the TSS (most frequent distance at −38 bp from TSS), while Id2 bound to a downstream region (most frequent distance at +125 bp from TSS). We also observed that Id2 exhibited a more scattered binding on gene promoters than E2F4, the former showing a wider peak of distance correlations from the TSS. In addition, our genomewide factor location analysis showed that, while the average region length covered by E2F4 was 24 bp, total Id2 or E2F4/Id2 were bound to a larger region with an average length of 277 bp. Since Id2 lacks a DNA binding domain, this result could reflect the binding position of a larger protein complex to which Id2 could be bound.
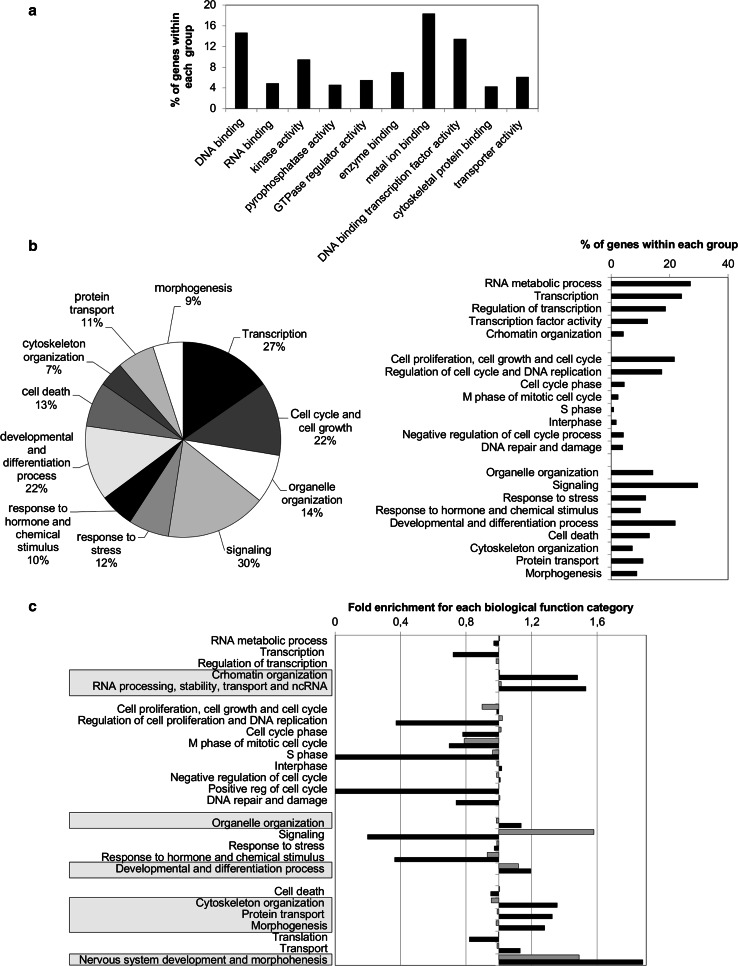
Molecular and biological functions of E2F4/Id2-bound genes
To study whether E2F4/Id2 has a role in a specific set of genes upon binding to their promoters, we functionally analyzed E2F4/Id2 target genes. As can be observed in Fig. 2a, the most representative molecular functions assigned to E2F4/Id2-bound genes was related to DNA binding, metal ion binding, and transcription factor activity. On the other hand, the higher percentages of E2F4/Id2-bound genes were clustered into four main biological functions (Fig. 2b): signaling, gene transcription, cell cycle and growth, and developmental and differentiation processes. We represented the molecular and biological functions of E2F4/Id2 and E2F4 alone in terms of fold enrichment versus total E2F4-bound genes. This analysis showed a clear difference between the two groups of genes (Fig 2c, Online resource 6). Re-clustering genes, so that they were annotated as unique genes within a particular category, showed that the overall profile of E2F4/Id2 targets are mostly related to cell development (Online resource 7). However, our ChIP/chip data are the result of experiments performed in adult mice liver. To study the impact of E2F4/Id2 versus E2F4 alone in the modulation of gene expression in adult mice liver, we analyzed [22] the in silico pattern of gene expression in both sets of genes (Online resource 8). As expected, most of genes bound by E2F4 were repressed or showed a very low expression level in adult liver (97 % of total genes) independently on the presence or absence of Id2. However, there is a subset of E2F4-bound genes exhibiting moderate to high gene expression. Interestingly, none of these highly-expressed genes was also bound by Id2. The biological functions of E2F4 targets showing a high expression are mainly related to metabolic functions and liver homeostasis. These data strongly suggest that E2F4/Id2 might be part of a repressor complex in quiescent adult liver.
Fig. 2.
Biological and functional characterization of E2F4/Id2 targets in adult liver. a Distribution of GO annotations assigned to a molecular function for E2F4/Id2 targets. The percentage is referred to the number of genes found within a particular category in relation to the total number of genes bound by E2F4/Id2 with an annotated GO molecular function. In some cases, genes may be included in more than one category. b Distribution of GO annotations assigned to E2F4/Id2-bound genes biological functions. Percentage of the most significant biological processes in which target genes have been described to be involved (left). Histogram depicts the percentage of genes found within most significant groups in relation to total E2F4/Id2-bound genes after an extensive and manual curated analysis (right). c Distribution of GO categories among genes bound by E2F4/Id2 vs. those bound by E2F4 alone. Black and gray bars represent E2F4/Id2 and E2F4 alone enrichment, respectively. E2F4/Id2, E2F4 alone, and total E2F4-bound regions showing a higher number of distance correlations from TSS were extracted and analyzed. Fold enrichment for each biological function was calculated compared to total E2F4-bound genes. Those categories found to be enriched in E2F4/Id2 vs. E2F4 targets are highlighted
Dynamics of Id2 binding to gene promoters during liver development and correlation with gene expression
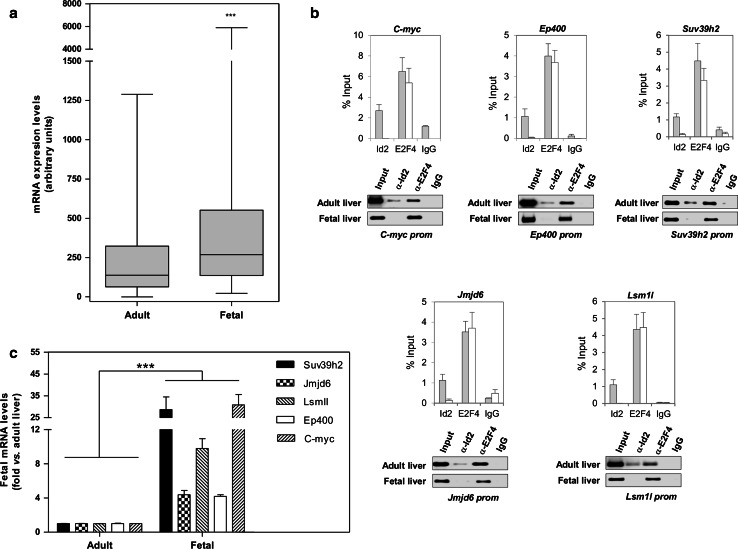
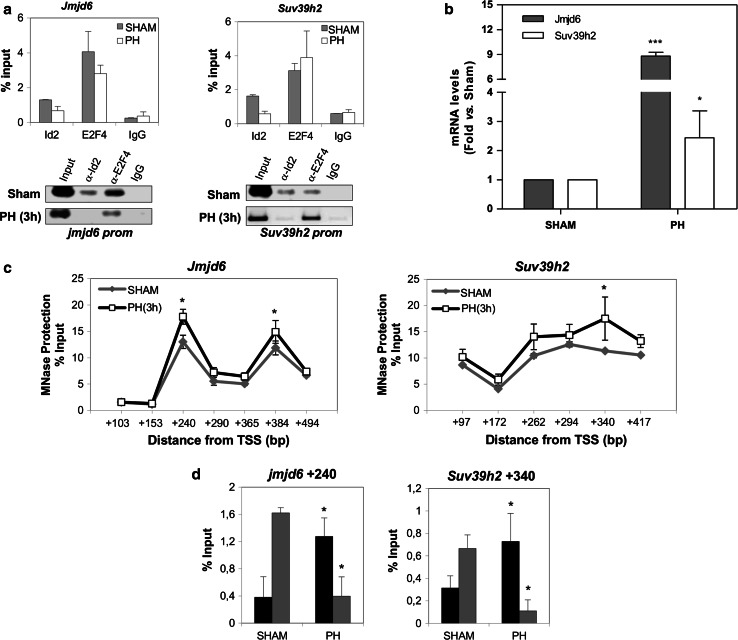
Our data suggest that in adult liver E2F4/Id2 might be part of a repressor complex on a subset of genes playing a major role during liver development. In agreement with this hypothesis, an in silico analysis of gene expression [23] showed that E2F4/Id2 target genes were dramatically up-regulated in fetal liver (Fig 3a). Therefore, we tested whether the presence or absence of Id2 bound to the complex correlates with the expression of target genes during liver development. We analyzed E2F4/Id2 binding to the region downstream from the TSS in a set of genes related to chromatin organization and/or RNA processing in liver samples from adult and fetal mice. As seen in Fig. 3b, and validating our ChIP/chip data, Id2 was bound to all the genes tested in quiescent adult liver. Interestingly, in fetal liver, Id2 was released from the TSS-downstream region of all genes. Moreover, the absence of Id2 from this region correlated with a high expression of target genes (Fig. 3c). The strong up-regulation of gene expression in fetal versus adult liver would further support the hypothesis of a repressive role for Id2 in a subset of E2F4 target genes in differentiated cells.
Fig. 3.
Dynamics of Id2 binding and expression of EF4/Id2 targets in differentiated and undifferentiated liver tissue. a In-silico expression analysis of E2F4/Id2 target genes in adult and fetal liver. The expression of those E2F4/Id2 target genes showing a biological function enrichment vs. E2F4 alone targets was analyzed (www.bidmcgenomics.org/LiverDevReg/index.html). A box and whiskers graph representing mRNA levels in both liver tissues is shown. ***P < 0.0001 vs. adult liver. b Validation of ChIP/chip data and analysis of Id2 and E2F4 binding to selected genes in adult (gray bars) and E14 fetal liver (white bars) by ChIP assay. qChIPs (upper) and semiquantitative ChIPs (lower) carried out with specific primers for the indicated genes are shown. As a positive control in quiescent liver, E2F/Id2 binding to the c-myc promoter was analyzed. Data are the result of at least three independent experiments where the P value for Id2 binding was never higher than P < 0.05 vs. adult liver. c mRNA levels of selected genes in fetal vs. adult liver. RT-qPCR data, shown as fold (n = 5) are mean ± SEM. ***P < 0.0001 vs. adult mice
Modulation of Id2 levels during liver development
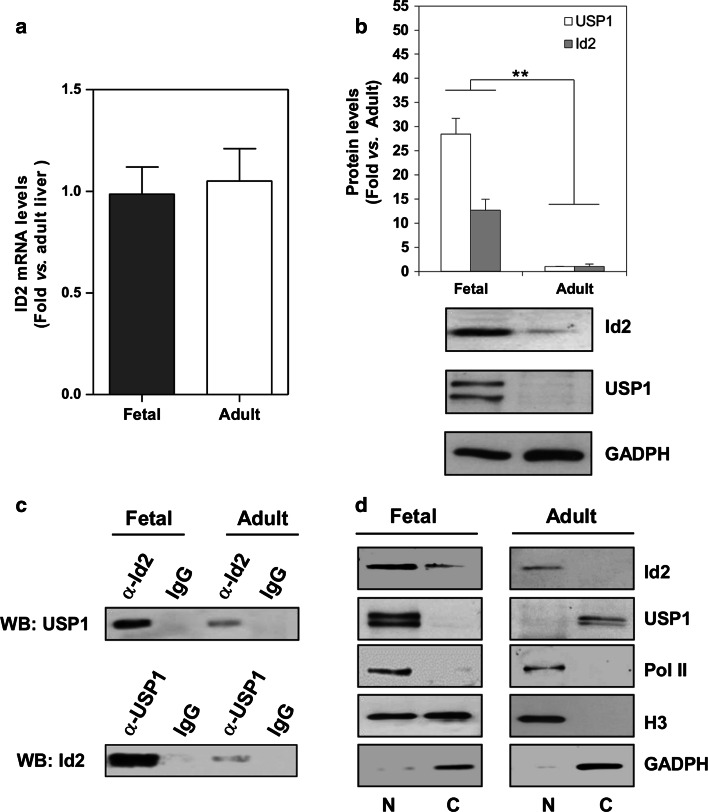
As mentioned above, data in the literature point out to the transcriptional control of Id2 as a key event in the modulation of its function. Aberrant Id2 expression has been reported in a long list of proliferating tumor cells [3–5]. Following this rationale, we might expect a dramatic up-regulation of Id2 mRNA levels during liver development. Surprisingly, Id2 fetal expression was not significantly different from adult liver (Fig. 4a).
Fig. 4.
Id2 expression and subcellular distribution in fetal and adult mice liver. a Id2 mRNA levels were determined by RT-qPCR in liver tissue samples from adult and E14 fetal mice and plotted as fold vs. adult liver. Data (n = 5) are mean ± SEM. No significant difference was found. b Id2 and USP1 protein levels were analyzed by western blot. Protein levels were quantified, normalized by GAPDH levels, and plotted. Data (n ≥ 5) are mean ± SEM. **P < 0.001 vs. adult liver. c Id2-USP1 coimmunoprecipitation analysis. Samples from fetal and adult liver tissue were immunoprecipitated with either α-Id2 or α-USP1 and normal IgG antibodies. The immunoprecipitated Id2 (upper panel) was analyzed by western blot with α-USP1, Immunoprecipitated USP1 (lower panel) was analyzed by western blot with α-Id2. Inputs of either Id2 or USP1 are shown in (b). d Nuclear (N) and cytosolic (C) distribution of Id2 and USP1. Western blot analysis of Id2 and USP1 in nuclear and cytosolic fractions from adult and fetal liver samples. Fraction purity was assessed by western blot with specific antibodies for both Pol II and histone H3 (used as nuclear marker of non-proliferating tissues) and GAPDH (used as a cytosolic marker). Representative blots (n = 5) for C and D are shown
USP1 has been shown to stabilize Id2 protein levels increasing its abundance in osteosarcoma tumors and in several other cell types [10, 24]. As shown in Fig. 4b, both Id2 and USP1 protein levels were strongly up-regulated during liver development. The specificity of the band recognized by the Id2 antibody was confirmed by the use of a blocking peptide (data not shown). Id2 stabilization by USP1 depends on their mutual recognition and interaction [10]. We confirmed by coimmunoprecipitation experiments that indeed, USP1 associates to Id2 during liver development (Fig. 4c).
Data in the literature indicate that Id2 and USP1 functions are primarily related to nuclear processes. Id2 does not contain a nuclear localization signal (NLS), thus translocation from the cytosolic compartment into the nucleus should be mediated by its interaction with a NLS-containing protein [7, 25, 26]. Conversely, USP1 contains two nuclear localization signals (NLS1 and NLS2) that mediate both its own transport into the nucleus and that for its interacting proteins [27]. Consistently, Id2 and USP1 subcellular distribution was analyzed in both fetal and adult liver tissues. The purity of subcellular fractions was confirmed by the analysis of nuclear (Pol II) and cytosolic (GAPDH) specific markers. H3 is usually the nuclear marker of choice when analyzing the purity of subcellular fractions. Nevertheless, it has been described that H3 is also present in the cytosolic compartment of highly proliferating cells [28]. As expected in fetal liver, a tissue with a high rate of proliferation, we can find H3 in both cell compartments. Conversely, H3 is restricted to the nuclear fraction of quiescent adult liver. Interestingly, we observed that Id2 and USP1 were mainly located within the nucleus of fetal liver (Fig 4d). A small amount of Id2 was observed in the cytosol, most probably reflecting the result of an active protein synthesis. Interestingly, in adult liver, we not only found a considerable reduction of total USP1 but also a different subcellular distribution that was restricted to the cytosol. Id2 was exclusively found in the nucleus of quiescent liver cells, when its levels were almost undetectable (Fig 4d, right panel). These data strongly suggest that Id2 nuclear functions and binding to gene promoters might be concentration-dependent. Indeed, Id2 was bound to gene promoters only in differentiated cells when its nuclear concentration was low.
Characterization of E2F4/Id2 binding sequence
To further explore the role of Id2 in quiescent liver, we characterized the E2F4/Id2 binding region on gene promoters. Highly conserved DNA sequences across species are thought to have regulatory value. The statistics of the 550 input regions for E2F4/Id2 show that 83.3 % of regions have an ortholog in Homo sapiens and in Rattus norvegicus. We found five putative motifs with a matrix similarity threshold of 0.80 in at least 75 % of orthologous regions, and a putative core element with a high GC content present in at least 95 % of input sequences (Online resource 9). In addition, we found that the E2FF binding site is present in 89 % of sequences, confirming the specificity of our experiments. Nevertheless, we could not find a conclusive module (combination of binding sites for E2FF and other transcription factor) present in a significant number of regions (Online resource 10). However, the regions always contained binding sites for factors related to transcription initiation.
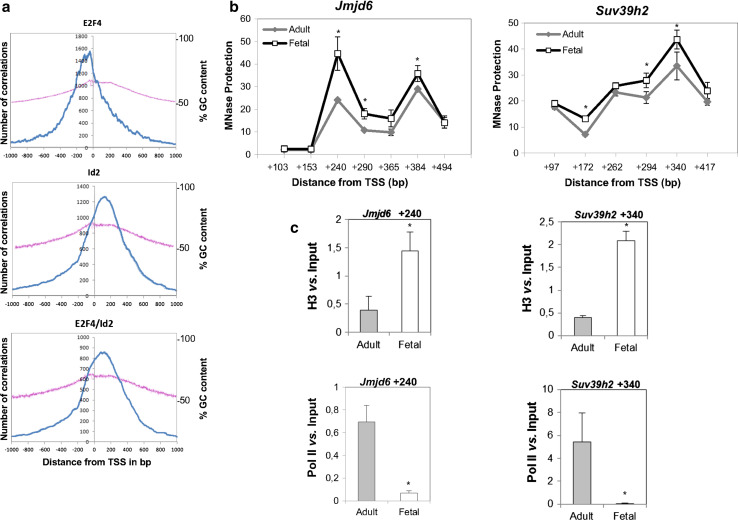
It has been described that transcription initiation is favored in CpG-island promoters [29]. Therefore, we studied whether the E2F4/Id2 complex preferentially bound to that region with a higher GC content on gene promoters. As can be observed in Fig. 5a, the percentage of GC content in gene promoters was not substantially different among groups. However, a careful analysis revealed that the highest number of distance correlations from TSS overlapped with the highest % GC only on gene promoters bound by either total Id2 or E2F4/Id2. In the total E2F4 group, the peak of distance correlations from TSS was centered out from the crest of maximal GC content. These data suggest that the core element found in E2F4/Id2 targets most probably is not a regular transcription factor-binding site, but a CpG region present in many gene promoters.
Fig. 5.
Impact of Id2 complex on nucleosome positioning at a high GC-content region downstream from TSS. a GC-content in total E2F4, Id2, and E2F4/Id2 binding regions relative to TSS (0). GenomeInspector analysis of tiling array data were plotted as the number of correlations found between target regions and the distance from TSS (labeled in blue) flanked by +/-1000 bp at the left axis. The percentage of GC content (pink line) in these regions is shown at the right axis. b qPCR analysis of nucleosome positioning at E2F4/Id2 binding region in fetal and adult liver. MNase protected regions at Jmjd6 (left) and Suv39h2 (right) were plotted as fold enrichment vs. distance from TSS (at the center of amplicons). Data (n = 3) are mean ± SEM. *P < 0.05 vs. adult liver. c Identification of complexes bound to MNase protected regions on Jmjd6 (left) and Suv39h2 (right) in liver samples from adult and fetal mice by Nuc-ChIP assay. Mononucleosome-enriched chromatin fragments were immunoprecipitated with either histone H3 (upper) or Pol II (lower) antibodies. Amplicons are centered at +240 and +340 bp from TSS for Jmjd6 and Suv39h2, respectively. Data (n = 3) are mean ± SEM. *P < 0.05 vs. adult liver
Nucleosome occupancy and Pol II pausing at the TSS-downstream region during liver development
Since Id2 lacks a DNA binding domain, it should be bound to a protein complex on DNA. In addition, the protein complex seems to be a big complex, since it covers a larger region than the one covered by E2F4 alone. The functional relevance of GC regions is poorly understood and controversial. It has been shown that most of these promoters have pre-assembled Pol II, although others report that a high GC content thermodynamically favors nucleosome assembly [29]. Importantly, pausing is enriched among genes that require a synchronous and precise control of expression. These genes tend to encode highly-regulated components of developmental and stimulus-responsive pathways [27, 30]. Taking these data together, we hypothesized that Id2 could favor nucleosomal occupancy or Pol II pausing, both known to preferentially bind regions with a high GC content.
The histone arginine demethylase, Jmjd6, and the histone methyltransferase, Suv39h2, are a good representation of the E2F4/Id2 target genes found in enriched functional categories since they are known to be essential for the differentiation of multiple tissues and cells during embryogenesis [31, 32]. Therefore, we selected these genes to study whether Id2 could favor nucleosomal occupancy or Pol II pausing in E2F4/Id2 targets at the TSS-downstream region.
In order to analyze in vivo the dynamics of this complex under the presence or absence of Id2 on gene promoters, MNase protection assays were performed in liver samples from adult and fetal mice. As shown in Fig. 5c, we found a protected region centered at +240 and +340 bp for Jmjd6 and Suv39h, respectively. Interestingly, the protected region is significantly increased in fetal liver when both genes are transcriptionally active. These results suggest that either a nucleosome or the Pol II complex may differentially change their occupancy on gene promoters in a development-related manner.
The MNase protection assay does not discriminate between nucleosomal occupancy and paused Pol II binding, therefore we studied the identity of proteins bound to the protected region by Nuc-ChIP assay with antibodies against Pol II and histone H3. Fig. 5d show that Pol II was bound to the protected region in quiescent adult liver in both genes. However, when both genes are transcriptionally active in fetal liver, the protected region was mainly occupied by a nucleosome, as can be deduced by the presence of histone H3.
Reversibility of Pol II pausing and Id2 binding to gene promoters at the onset of liver regeneration
It is generally believed that paused Pol II, maintaining an open chromatin structure, facilitates the expression of genes [33]. The oncogene c-myc is largely known to have a paused Pol II bound to its promoter [34]. We have already described the transcriptional up-regulation of Id2 and its release from c-myc promoter during liver regeneration after PHx [9]. Our results suggest that Id2 takes part in a development-related program. We wondered whether such a program could be reversible upon stimulation. Although liver regeneration is not a recapitulation of the embryonic program, both processes share key factors to promote cell proliferation [23]. Therefore, we analyzed E2F4/Id2 binding to Jmjd6 and Suv39h2 in liver samples at the early onset of liver regeneration after PHx.
As can be observed in Fig. 6a, b, the pattern of Id2 binding to these E2F4 target promoters correlated with the expression of both genes. Both were up-regulated when Id2 was released from their promoters. Moreover, we observed the same MNase protection profile in liver samples after PHx as in fetal liver (Fig. 6c). In both cases, as can be seen in Fig. 6d, Pol II was replaced by a nucleosome when gene expression was induced. These results suggest that the release of pausing triggered by liver resection might favor nucleosomal reassembly in the region previously occupied by Pol II.
Fig. 6.
Id2 and Pol II coordinated release from gene promoters at the onset of liver regeneration. a Analysis of Id2 and E2F4 binding to Jmjd6 (left) and Suv39h2 (right) by qChIP in liver samples from sham operated mice and 3 h after PHx. qChIPs (upper) and semiquantitative ChIPs (lower) carried out with specific primers for the indicated genes are shown. Data are the result of at least three independent experiments where the P value for Id2 binding was never higher than P < 0.05 vs. sham operated mice. b mRNA levels of Jmjd6 (black) and Suv39h2 (gray) in liver tissue from 3 h PHx vs. sham operated animals. RT-qPCR data, shown as fold (n = 5) are mean ± SEM. ***P < 0.0001 vs. sham operated mice. c qPCR analysis of nucleosome positioning at E2F4/Id2 binding region in sham (SH) and 3 h PHx liver samples. Data were plotted as fold enrichment of normalized-MNase protected regions vs. distance from TSS (at the center of amplicons). Data (n = 3) are mean ± SEM. *P < 0.05 vs. sham. d Identification of complexes bound to MNase protected regions on Jmjd6 (left) and Suv39h2 (right) in liver samples from sham and 3 h PHx. Mononucleosome-enriched chromatin fragments were immunoprecipitated with either histone H3 (black) or Pol II (gray) antibodies. Amplicons are centered at +240 and +340 bp from TSS for Jmjd6 and Suv39h2, respectively. Data (n = 3) are mean ± SEM. *P < 0.05 vs. sham operated liver samples
Discussion
Id2 belongs to a family of HLH proteins identified and named over two decades ago for its dual role as both inhibitors of the differentiation process and inhibitors of DNA binding. Id2 proteins lack the basic DNA-binding domain and, consequently, they cannot bind to DNA [1]. The molecular mechanism for the role of Id2 proteins in the transcriptional control was initially thought to be only dependent on its ability to dimerize with bHLH factors acting as dominant-negative [2]. Nevertheless, it has recently been demonstrated that Id2 could bind to proteins different from bHLH factors, such as pRB, and pocket proteins, such as p130, on myc promoter [9, 14], connecting in this way Id2 and E2F target genes.
We show here for the first time that Id2 is part of a more general mechanism of transcriptional regulation than was at first thought. Our ChIP/chip data show that Id2 binds to a subset of 442 E2F4 target genes (Fig. 1a; Online resource 5). We also found a low percentage of genes bound by Id2 alone (Fig. 1a; Online resource 3-5). These data might suggest that Id2, although preferentially binding to a E2F4-containing complex, does not necessarily need E2F4 to bind to a protein complex on gene promoters. Nevertheless, since we performed a very restrictive analysis to prevent false positive probes, we cannot rule out the possibility that this short percentage of Id2-bound genes could also bind E2F4 but were not detected.
When we compared E2F4’s most frequent localization (−38 bp from TSS) with that of Id2/E2F4 target genes (+125 bp from TSS), our high-density tiling array data suggested that E2F4 binding could be influenced by Id2 and thus behave like a complex on chromatin (Fig. 1c). In addition, since Id2 lacks a DNA binding domain, the more scattered binding profile of Id2 could reflect the binding position of a larger protein complex to which Id2 could be bound. This idea is in agreement with our genomewide factor location analysis which showed a larger average region length covered by total Id2 or E2F4/Id2 versus E2F4 alone. Indeed, Id2 alone and E2F4/Id2 show the same binding position. Moreover, these data further support the idea that our stringent analysis criteria, although enhancing our confidence in the target genes, most likely underestimates the actual number of E2F4/Id2 targets. Most probably, all Id2 targets represent E2F4/Id2 target genes.
Although we cannot deny the unquestionable role of Id2 as a proliferative factor, it has been proposed that there is a threshold of Id2 concentration which is crucial for its pleiotropic functions [3–6]. In agreement with this, here it was found that, in differentiated liver tissue, Id2 is bound to a selection of E2F4 target genes associated with cell development and chromatin structure (Fig. 2c). We observed that these genes, while transcriptionally inactive in adult liver, are up-regulated during liver development (Fig. 3a, c). Finally, Id2 concentration dramatically decreases in liver tissue upon differentiation.
Regarding the molecular mechanism involved in the modulation of Id2 protein levels, USP1 has been described to have an important role during embryonic development [35]. While we did not demonstrate directly that Id2 is stabilized by USP1, our co-immunoprecipitation experiments might suggest it (Fig. 4). Nevertheless, the de-ubiquitinating activity of USP1 is not the only function recognized for this protein. USP1 is also involved in the nuclear import of its interacting proteins [27]. Therefore, we cannot exclude a putative role for USP1 on Id2 transport into the nuclear compartment. The precise role of USP1 on Id2 stability or transport during liver development should be further explored. In addition, USP1 might also have a role interacting with and stabilizing other proteins during liver development.
We show that, in differentiated tissue, Id2 levels decrease and its localization is restricted to the nuclear compartment (Fig. 4b, d). In adult liver, nuclear Id2 binds to a protein complex together with Pol II on a selection of gene promoters (Fig 3b, c,5c, 6). We hypothesize that Id2 binding might keep target genes in a paused/ON state, or in other words be responsive to specific stimuli. In support of this idea, we observed that Id2 and Pol II bound to a region with a high GC content in differentiated liver tissue (Fig. 5a; Online resource 9b). CG content is higher in gene promoters whose activation is remodeling-independent and therefore are primed for gene transcription. In unstimulated bone marrow-derived cells most of these promoters have pre-assembled Pol II [29]. These data might suggest that E2F4/Id2 are bound to a set of genes characterized by a constitutively active chromatin structure.
Although it has been predicted that CG promoters are not compatible with nucleosomal assembly [36] others reported that a high GC content thermodynamically favors nucleosome assembly [29]. We observed that, when genes were transcriptionally active in both fetal or regenerating liver, nucleosome assembly was favored at the region downstream from TSS (Fig. 3c, 5c, e, 6b–d). This behavior has already been observed in genes with paused Pol II. Upon stimulation of target genes, paused Pol II is released from gene promoters and nucleosomes are reassembled in this region [33]. This observation is in agreement with the hypothesis in which a high GC content favors nucleosome assembly at the site previously occupied by Pol II [29]. In addition, it has been suggested that the competition between Pol II and nucleosomes for binding to gene promoters could be a mechanism to modulate inducibility of gene transcription [33]. Therefore, our results suggest that Id2 could be involved in the modulation of Pol II pausing at specific genes. It is worth highlighting that increased nucleosomal reassembly at the E2F4/Id2 binding region does not mean a repressive condition for gene transcription. The presence of a nucleosome will represent a physical barrier for gene transcription depending on the nucleosome modifications and, as a result, on how tightly this nucleosome is bound to DNA.
On the whole, Id2 binding to gene promoters seem to be part of a reversible development-related program on a subset of E2F4 target genes, since Id2 and paused Pol II are released from gene promoters during liver regeneration after PHx. Liver is a special organ in terms of its tremendous capacity to trigger an adaptation program when receiving an injury in order to recover its homeostasis. To maintain this ability is essential to assure the organ’s survival. Based on our data, it is tempting to speculate with the idea of Id2 as a factor involved in the maintenance of liver plasticity. Loss of pausing can drive genes to an unresponsiveness condition and permanent silencing. Permanent loss of pausing does not induce gene expression, but on the contrary, genes are down-regulated [37]. We cannot rule out the possibility that Id2 could act as an elongating factor competing with pausing factor/s for binding to Pol II. Therefore, the stoichiometric concentration of Id2 and the pausing-inducing factor/s would determine the final transcriptional result. In the future, it will be important to examine Id2 and Pol II distribution throughout the gene body upon stimulatory signals to determine whether Id2 remains bound to the elongating form of Pol II as has been described for elongating factors.
Following this model, although the effectiveness of transcription will surely rely on the specific combination of additional factors and events, several genes with E2F4/Id2 target sequences could be simultaneously activated by a common sensitization mechanism to induce transcription initiation. Accordingly, it has been reported that genes encoding developmental regulators fall into the group of genes with paused Pol II, experiencing transcriptional initiation and no elongation in differentiated cells [38]. As suggested in other reports [33, 39], in the long term, the DNA sequence by itself could favor nucleosome occupancy of that region, promoting the permanent silencing of selected E2F4-driven genes. This could determine, in a condition-specific manner, which promoters are able to enter the signal-responsive paused ON/OFF state.
This concept, which has been poorly sketched to explain the role of Id2 on c-myc transcription [9, 14], as a more general mechanism for the modulation of gene expression would open an important issue toward understanding the molecular mechanisms of transcriptional regulation under physiological conditions. Moreover, much of the transcriptional reprogramming induced in hepatocellular carcinoma mimics that of the developing liver [40, 41], and therefore the role of Id2 in tumor progression could also be related to the deregulation of this program. However, testing this model in the near future will require a more detailed understanding of the interplay between the paused Pol II complex and Id2.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was supported by grants from Spanish government: PN I+D+I 2008-2011 [BFU2010-18253 to J.R.V] and ISCIII including FEDER funding [PI12/02394 to E.R.G-T], Consellería de Educación [GVPROMETEO 2010-075] and Fundación INCLIVA to RZ. T.A is the recipient of a pre-doctoral fellowship from Ministerio de Educación and I.F-V is funded by Consellería de Educación [GVPROMETEO 2010-075].
References
- 1.Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- 2.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-Y. [DOI] [PubMed] [Google Scholar]
- 3.Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/S0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- 4.Lasorella A, Uo T, Iavarone A. Id proteins at the cross-road of development and cancer. Oncogene. 2001;20:8326–8333. doi: 10.1038/sj.onc.1205093. [DOI] [PubMed] [Google Scholar]
- 5.Coma S, Amin DN, Shimizu A, Lasorella A, Iavarone A, et al. Id2 promotes tumor cell migration and invasion through transcriptional repression of semaphorin 3F. Cancer Res. 2010;70:3823–3832. doi: 10.1158/0008-5472.CAN-09-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Norton JD, Atherton GT. Coupling of cell growth control and apoptosis functions of Id proteins. Mol Cell Biol. 1998;18:2371–2381. doi: 10.1128/mcb.18.4.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsumura ME, Lobe DR, McNamara CA. Contribution of the helix-loop-helix factor Id2 to regulation of vascular smooth muscle cell proliferation. J Biol Chem. 2002;277:7293–7297. doi: 10.1074/jbc.M108986200. [DOI] [PubMed] [Google Scholar]
- 8.Rothschild G, Zhao X, Iavarone A, Lasorella A. E Proteins and Id2 converge on p57Kip2 to regulate cell cycle in neural cells. Mol Cell Biol. 2006;26:4351–4361. doi: 10.1128/MCB.01743-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodríguez JL, Sandoval J, Serviddio G, Sastre J, Morante M, et al. Id2 leaves the chromatin of the E2F4-p130-controlled c-myc promoter during hepatocyte priming for liver regeneration. Biochem J. 2006;398:431–437. doi: 10.1042/BJ20060380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams SA, Maecker HL, French DM, Liu J, Gregg A, et al. USP1 de-ubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell. 2011;146:918–930. doi: 10.1016/j.cell.2011.07.040. [DOI] [PubMed] [Google Scholar]
- 11.Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/S0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 12.Bounpheng MA, Dimas JJ, Dodds SG, Christy BA. Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J. 1999;13:2257–2264. [PubMed] [Google Scholar]
- 13.Chen XS, Zhang YH, Cai QY, Yao ZX. ID2: a negative transcription factor regulating oligodendroglia differentiation. J Neurosci Res. 2012;90:925–932. doi: 10.1002/jnr.22826. [DOI] [PubMed] [Google Scholar]
- 14.Torres L, Sandoval J, Penella E, Zaragozá R, García C, et al. In vivo GSH depletion induces c-myc expression by modulation of chromatin protein complexes. Free Radic Biol Med. 2009;46:1534–1542. doi: 10.1016/j.freeradbiomed.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 15.van Oevelen C, Wang J, Asp P, Yan Q, Kaelin WG, Jr, et al. A role for mammalian Sin3 in permanent gene silencing. Mol Cell. 2008;32:359–370. doi: 10.1016/j.molcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayman JB, Takahashi Y, Indjeian VB, Dannenberg JH, Catchpole S, et al. E2F mediates cell cycle-dependent transcriptional repression in vivo by recruitment of an HDAC1/mSin3B corepressor complex. Genes Dev. 2002;16:933–947. doi: 10.1101/gad.969202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balciunaite E, Spektor A, Lents NH, Cam H, Te Riele H, et al. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol Cell Biol. 2005;25:8166–8178. doi: 10.1128/MCB.25.18.8166-8178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morello D, Fitzgerald MJ, Babinet C, Fausto N. c-myc, c-fos, and c-jun regulation in the regenerating livers of normal and H-2 K/c-myc transgenic mice. Mol Cell Biol. 1990;10:3185–3193. doi: 10.1128/mcb.10.6.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres L, Serna E, Bosch A, Zaragozá R, García C, et al. NF-ĸB as node for signal amplification during weaning. Cell Physiol Biochem. 2011;28:833–846. doi: 10.1159/000335797. [DOI] [PubMed] [Google Scholar]
- 20.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sacilotto N, Espert A, Castillo J, Franco L, López-Rodas G. Epigenetic transcriptional regulation of the growth arrest-specific gene 1 (Gas1) in hepatic cell proliferation at mononucleosomal resolution. PLoS ONE. 2011;6:e23318. doi: 10.1371/journal.pone.0023318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 23.Otu HH, Naxerova K, Ho K, Can H, Nesbitt N, et al. Restoration of liver mass after injury requires proliferative and not embryonic transcriptional patterns. J Biol Chem. 2007;282:11197–11204. doi: 10.1074/jbc.M608441200. [DOI] [PubMed] [Google Scholar]
- 24.Mistry H, Hsieh G, Buhrlage SJ, Huang M, Park E, et al. Small-molecule inhibitors of USP1 target ID1 degradation in leukemic cells. Mol Cancer Ther. 2013;12:2651–2662. doi: 10.1158/1535-7163.MCT-13-0103-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- 26.Lasorella A, Iavarone A. The protein ENH is a cytoplasmic sequestration factor for Id2 in normal and tumor cells from the nervous system. Proc Natl Acad Sci USA. 2006;103:4976–4981. doi: 10.1073/pnas.0600168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Santisteban I, Zorroza K, Rodriguez JA. Two nuclear localization signals in USP1 mediate nuclear import of the USP1/UAF1 complex. PLoS ONE. 2012;7:e38570. doi: 10.1371/journal.pone.0038570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson LJ, Gu Y, Yakovleva T, Tong K, Barrows C, et al. Modifications of H3 and H4 during chromatin replication, nucleosome assembly, and histone exchange. J Biol Chem. 2006;281:9287–9296. doi: 10.1074/jbc.M512956200. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, et al. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell. 2009;138:114–128. doi: 10.1016/j.cell.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee YF, Miller LD, Chan XB, Black MA, Pang B, et al. JMJD6 is a driver of cellular proliferation and motility and a marker of poor prognosis in breast cancer. Breast Cancer Res. 2012;14:R85. doi: 10.1186/bcr3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters AH, O’Carroll D, Scherthan H, Mechtler K, Sauer S, et al. Loss of the Suv39 h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 33.Gilchrist DA, Adelman K. Coupling polymerase pausing and chromatin landscapes for precise regulation of transcription. Biochim Biophys Acta. 2012;1819:700–706. doi: 10.1016/j.bbagrm.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krumm A, Meulia T, Brunvand M, Groudine M. The block to transcriptional elongation within the human c-myc gene is determined in the promoter-proximal region. Genes Dev. 1992;6:2201–2213. doi: 10.1101/gad.6.11.2201. [DOI] [PubMed] [Google Scholar]
- 35.Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, et al. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell. 2009;16(2):314–320. doi: 10.1016/j.devcel.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Segal E, Fondufe-Mittendorf Y, Chen L, Thåström A, Field Y, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilchrist DA, Nechaev S, Lee C, Ghosh SK, Collins JB, et al. NELF-mediated stalling of Pol II can enhance gene expression by blocking promoter-proximal nucleosome assembly. Genes Dev. 2008;22:1921–1933. doi: 10.1101/gad.1643208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, et al. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coulouarn C, Derambure C, Lefebvre G, Daveau R, Hiron M, et al. Global gene repression in hepatocellular carcinoma and fetal liver, and suppression of dudulin-2 mRNA as a possible marker for the cirrhosis-to-tumor transition. J Hepatol. 2005;42:860–869. doi: 10.1016/j.jhep.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 41.Li T, Wan B, Huang J, Zhang X. Comparison of gene expression in hepatocellular carcinoma, liver development, and liver regeneration. Mol Genet Genomics. 2010;283:485–492. doi: 10.1007/s00438-010-0530-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.