Abstract
Autophagy is an evolutionarily conserved process that degrades cytoplasmic components, thus contributing to cell survival and tissue homeostasis. Recent studies have demonstrated that autophagy maintains stem cells in relatively undifferentiated states (stemness) and also contributes to differentiation processes. Autophagy likewise plays a crucial role in somatic cell reprogramming, a finely regulated process that resets differentiated cells to a pluripotent state and that requires comprehensive alterations in transcriptional activities and epigenetic signatures. Autophagy assists in manifesting the functional consequences that arise from these alterations by modifying cellular protein expression profiles. The role of autophagy appears to be particularly relevant for early phases of cell reprogramming during the generation of induced pluripotent stems cells (iPSCs). In this review, we provide an overview of the core molecular machinery that constitutes the autophagic degradation system, describe the roles of autophagy in maintenance, self-renewal, and differentiation of stem cells, and discuss the autophagic process and its regulation during cell reprogramming.
Keywords: Autophagy, Stem cells, Self-renewal, Differentiation, Cell reprogramming
Introduction
In eukaryotic cells, the ubiquitin–proteasome and the lysosome systems constitute two major pathways that coordinate the degradation of proteins, other biological macromolecules, as well as organelles [1]. While proteasome-dependent degradation is predominantly limited to short-lived proteins, lysosomal degradation possesses a much broader substrate repertoire. Both extracellular materials as well as cytosolic components, including cellular organelles, can be targeted for lysosomal degradation [2]. Extracellular materials that are taken up by endocytosis are delivered by the fusion of late endosomes with lysosomes. Intracellular materials, such as long-lived proteins or old or damaged organelles, are engulfed by double-membrane vesicles termed autophagosomes [3, 4], and these autophagosomes subsequently fuse with lysosomes into autophagolysosomes [3, 4]. Degradation by autophagy (“self-eating”) not only allows for the recycling of cellular materials and the reshaping of the cellular proteome, as for example relevant during early embryonic development, but also provides a nutrient source during periods of starvation, hypoxic stress and other physiological stress situations [5]. Autophagy is therefore of fundamental importance for cell survival and for the maintenance of tissue homeostasis. The manifestation of pathophysiologies and disorders upon loss-of-function in autophagy genes and proteins further demonstrates the relevance of autophagic degradation processes. These for example include embryonic lethality [6, 7], excessive neurodegeneration [8], increased carcinogenesis [9], and a higher susceptibility to microbial infections [10]. In addition, evidence has also accumulated for an essential role of autophagy in somatic cell reprogramming [11, 12].
Somatic cell reprogramming is a finely regulated process that resets differentiated cells to a pluripotent state [13]. Cell reprogramming is of physiological importance during the zygote-to-embryo transition and for tissue regeneration, but also contributes to tumorigenesis and tumor progression through the establishment and maintenance of tumor stem cells [14–16]. The process of cell reprogramming is thus relevant during the entire life span of an organism. The reprogramming of somatic cells into a pluripotent state can be induced extrinsically by the transduction of combinations of transcriptions factors, such as Oct4, Sox2, Klf4, and c-Myc (OSKM). OSKMs and other transcription factors now serve as the foundation for cell-reprogramming approaches that begin to have significant translational potential for clinical applications [17, 18]. The transcriptional and epigenetic modifications induced by OSKM transcription factors cause complex changes in gene expression profiles and ultimately evoke the transition into a pluripotent cell state [19–21]. It would be expected that this transition is associated with changes in cellular protein profiles, and that such changes contribute to successful reprogramming. Indeed, initial studies have confirmed that the cellular proteome is significantly altered during cell reprogramming and that these alterations are fundamental for establishing or maintaining pluripotency [11, 12, 22, 23]. Both the ubiquitin–proteasome systems as well as autophagic degradation have been implicated in these processes, with autophagy initiation displaying prominently in the early stages of cell reprogramming [11, 12, 22, 23]. Following on from the fundamental research studies in this field, it will now become important to understand the role, the regulation, and the functional consequences of autophagy within the context of cell reprogramming. Next, we will therefore provide a brief overview of the core autophagic degradation machinery, will outline the known roles of autophagy in stem cells, as well as describe and discuss the relevance of autophagic signaling for successful cell reprogramming.
The autophagic degradation system
About 50 years ago, de Duve et al. [24] identified an intracellular degradation mechanism by which double-membrane structures enclose unwanted organelles and intracellular aggregates and subsequently degrade their content upon fusion with lysosomes. This process of self-eating was termed “autophagy”, with later studies specifying three major routes of autophagic degradation, including macroautophagy (in the following referred to as autophagy), microautophagy, and chaperone-mediated autophagy [25]. Over the past decades, the identification of autophagy-related proteins (Atgs) and of autophagy-related macromolecular complexes provided insight into the composition, complexity, and regulation of the core autophagic degradation system [3, 26]. In parallel to this, the community of autophagy researchers began to establish guidelines for the analysis and interpretation of autophagic signaling and autophagic flux, including the definition of a common nomenclature [27].
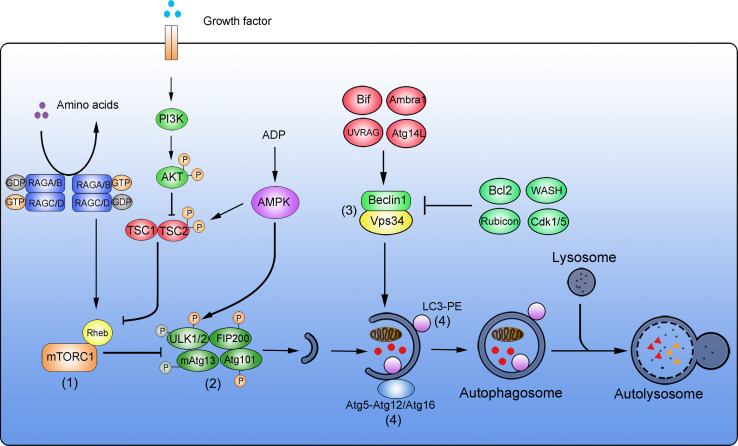
The processes from autophagosome formation to autolysosomal degradation can be divided into four major steps: initiation of the autophagosomal membrane, autophagosomal membrane elongation, target engulfment and autophagosome formation, and finally lysosomal fusion and degradation [28]. For each of these steps, specific multi-protein complexes and signaling platforms assemble to coordinate autophagy signal transduction and flux (Fig. 1).
Fig. 1.
Simplified overview of the autophagic degradation system. Four major complexes are key components of the autophagic molecular machinery. (1) mTORC1 complex. In nutrient-rich conditions, amino acids are sensed and result in the conversion of RAG GTPase heterodimers from RAGA/B-GDP-RAGC/D-GTP to RAGA/B-GTP-RAGC/G-GDP. This facilitates the binding of the small GTPase Rheb to mTOR, resulting in mTOR activation. Upon stimulation with growth factors, the PI3K-AKT pathway is activated to phosphorylate TSC2. TSC2-P inhibits Rheb and promotes the activation of mTORC1. High ADP: ATP ratios activate AMPK, promote TSC1-TSC2 activation by phosphorylation of TSC2, followed by inhibition of Rheb and inhibition of mTORC1. (2) ULK1 complex. During nutrient starvation or other metabolic stresses, the activation of mTORC1 is inhibited to block mTOR-dependent phosphorylation of ULK1, leading to ULK1 autophosphorylation. ULK1 is activated by AMPK-mediated phosphorylation. Phosphorylated ULK1 subsequently phosphorylates and activates FIP200 and mAtg13, resulting in autophagy induction. (3) Vps34 complex. Beclin 1 and Vps34 form the core of this complex. Bif, Ambra1, UVRAG, and Atg14L positively regulate the activity of the Beclin 1-Vps34 complex. Bcl2, WASH, Rubicon, and Cdk1/5 negatively regulate this complex to suppress autophagy. (4) Ubiquitin-like (Ubl) conjugation complex. Two Ubl systems are relevant during autophagy. Atg5 is covalently conjugated to Atg12, and the Atg5-Atg12 conjugate forms an active multimeric complex with Atg16. This first complex then localizes to autophagosomes. The second complex is the LC3-PE system. During autophagy induction, LC3 is conjugated to phosphatidylethanolamine (PE) to form LC3-II. LC3-II then locates to autophagosomes. Autophagosomes then enclose target materials that are degraded upon fusion with lysosomes and the formation of autolysosomes
Regulation of autophagy initiation through the mTOR complex
Autophagy can be initiated in response to nutrient deprivation and various other stresses, and in most cases serves to promote cell survival by stress alleviation and in particular in starvation conditions by providing an intracellular source of nutrients [29, 30]. As a central player in cellular nutrient sensing, the mammalian target of rapamycin (mTOR) acts as a crucial hub for autophagy induction or repression [31, 32]. Two distinct complexes contain mTOR as a catalytic subunit, namely mTORC1 and mTORC2 [33, 34]. Active mTORC1 suppresses autophagy and supports cell-mass accumulation by inhibitory phosphorylation of unc-51-like kinase 1 (ULK1), the mammalian homologue of yeast protein Atg1 [35, 36]. A lack of nutrients or growth factors inhibits mTORC1 and consequently results in reduced inhibitory phosphorylation of ULK1, thereby promoting autophagy induction [35–37]. Three major signaling pathways sense amino acid supply, growth factor concentrations, and the amounts of ATP converge at the level of mTORC1 [32]. In nutrient-rich conditions, the sensing of amino acids converts RAG GTPase heterodimers from the RAGA/B-GDP-RAGC/D-GTP state to the RAGA/B-GTP-RAGC/G-GDP state. Upon this GDP-GTP exchange, the small GTPase Ras homologue enriched in brain (Rheb) binds to and activates mTORC1 [38, 39]. In the presence of growth factors, the PI3K-AKT pathway is activated and phosphorylates TSC2, which in turn inhibits GTPase-activating protein (GAP) and promotes mTORC1 activation [40]. High ratios of ATP:ADP maintain AMP-activated protein kinase (AMPK) in its inactive state and prevent an AMPK-mediated inhibition of mTORC1 [41, 42]. Consequently, withdrawal of amino acids, growth factors, or declines in ATP initiate autophagy through mTORC1 inhibition [37, 43, 44]. Interestingly, inhibition of mTORC1 not only promotes autophagy but also contributes to preventing cellular senescence and overcoming cell-reprogramming barriers [45]. In this context, it was recently shown that the transient downregulation of mTOR expression is essential for successful cell reprogramming during iPSC formation and zygote-to-embryo transitions [11].
The ULK complex regulates phagophore formation
Upon mTORC1 inhibition, the unc-51-like kinase (ULK) complex is activated and plays a vital role in autophagy induction [35, 46]. The ULK complex is assembled of mammalian ULK1/ULK2 (redundant mammalian homologues of yeast Atg1), mammalian (m)Atg13, FIP200, and Atg101 [36]. Unlike the rapid changes in Atg1 complex abundance during autophagy induction in yeast, the ULK complex is expressed at rather stable amounts in mammalian cells and is activated by posttranslational modifications [4]. As mentioned earlier, inhibition of mTORC1 reduces the mTOR-dependent phosphorylation of ULK1, leading to an activating ULK1 autophosphorylation [35]. In parallel, AMPK can also phosphorylate and thereby activate ULK1 [35, 37]. Active ULK1 then phosphorylates FIP200 and mAtg13, giving rise to the final ULK1/2-mAtg13-FIP200-Atg101 complex, which continues to form the initial phagophore during the early stages of autophagosome formation [35, 36, 47]. Disruption of the ULK complex due to FIP200 gene deficiency is embryonically lethal at E13.5-E16.5 as a consequence of heart and liver developmental failures, suggesting a role of the complex in cell renovation during embryo development. A recent study showed that FIP200 deficiency causes loss of neural stem cells (NSCs) and impairs neuronal differentiation, indicating that the ULK complex is required for NSC maintenance and differentiation [48]. Whether the ULK complex contributes to other aspects and scenarios of stem cell maintenance or cell reprogramming is currently poorly understood and will need to be investigated further.
The Vps34 complex initiates the formation of pre-autophagosomal structures
Subsequent to phagophore assembly, the Vps34 complex facilitates the further formation of autophagosomes by promoting nucleation and growth of the pre-autophagosomal isolation membrane [26, 49]. The core components of the Vps34 complex comprise Vps34, a class III PI3K, as well as Vps15 and Beclin 1 [50]. Vps34 phosphorylates position 3 of phosphatidylinositol to produce phosphatidylinositol-(3)-phosphate (PI(3)P), a glycerophospholipid required for autophagosomal membrane elongation [51, 52]. Beclin 1 is the mammalian homologue of yeast Atg6 and recruits additional regulators to the Vps34 complex, including Atg14L, Rubicon, Ambra1, UVRAG, and Bif [4]. The activation of Vps34 within the complex is subject to multi-factorial control by these binding partners as well as by additional external modulators (Fig. 1). Beclin 1, Bif, Ambra1, UVRAG, and Atg14L promote the activation of the Vps34 complex, resulting in autophagy induction [53–58]. In contrast, Rubicon, as well as additional external regulators such as Bcl-2, Cdk1/Cdk5, and WASH, negatively regulate autophagy through the Vps34 complex [7, 56, 57, 59, 60]. Deficiencies in activating and inhibiting components of the Vps34 complex suppress or over-activate autophagy and lead to lethal developmental defects during embryonic stages. For example, Beclin 1 −/− ESCs fail to undergo normal differentiation, resulting in embryonic lethality at E6.0 [61, 62]. Ambra1 −/− mice die around E13.5 due to defects in neural tube development [54]. WASH-deficient embryos display extensive autophagy and die at E7.5 [7]. Therefore, Vps34 complex activity is essential during embryonic development and needs to be carefully controlled by both positive and negative regulators. Whether the Vps34 complex and its components are involved in the regulation of cell reprogramming is unknown at present, but the use of somatic cells, which lack these genes, is expected to provide answers to this question in the near future.
Ubiquitin-like (Ubl) conjugation complex
Two ubiquitin-like conjugation systems, the Atg5-Atg12-Atg16 and the LC3-PE conjugation system, are required for the further formation of autophagosomal membranes and for autophagosome maturation. To assemble the Atg5-Atg12-Atg16 complex, Atg5 is covalently conjugated to Atg12 [63]. The Atg5-Atg12 conjugate then forms an active multimer with Atg16, which localizes to autophagosomes [64]. The LC3-PE conjugation system contributes to autophagosomal signaling downstream of Atg5-Atg12-Atg16 complex formation. LC3, the mammalian homologue of yeast Atg8, is conjugated to phosphatidylethanolamine (PE) to form LC3-II, which then locates to autophagosomal membranes [65]. After fusion of autophagosomes with lysosomes, the intra-autolysosomal LC3-II is degraded and extra-vesicular LC3-II recycles back into the cytoplasm. LC3-I to -II conversion, translocation of LC3 from the cytoplasm into autophagosomes, as well as turnover of fusion proteins comprising GFP variants and LC3 belong to the most frequently used approaches to determine changes in autophagy and autophagic flux [27]. Both conjugation systems not only play key roles during development and adulthood but also appear to be implicated in the processes of cell reprogramming and differentiation. For example, deletion of Atg5 in MEFs or murine zygotes impairs the efficiency of cell reprogramming [11, 12], and Atg5 −/− ESCs display defects in differentiation [61]. Conditional knockout of Atg12 in hematopoietic stem cells (HSCs) sensitizes to apoptosis under stress conditions, indicating a role for autophagy proteins in promoting stem cell survival and fitness [66]. Indications for altered autophagic signaling in stem cells were also obtained at the level of the LC3-PE conjugation system. Increased numbers of LC3-positive autophagosomes and a pronounced accumulation of LC3-II were observed in human ESCs, neural stem cells (NSC), and cancer stem cells (CSCs) [48, 67–69]. Likewise, LC3-II accumulation is found during stem cell differentiation, HSC starvation, as well as during MEF and zygote reprogramming [11, 12, 66, 70, 71]. Since systemic deletions of many autophagy-related genes are embryonically or neonatally lethal, the use of conditional knockout models may assist in further understanding the role of autophagy and the ubiquitin-like conjugation complex in stem cell survival, maintenance, and in cell reprogramming [66, 72].
The role of autophagy in stem cell maintenance, self-renewal, and differentiation
As described above, multiple facets of the core autophagic machinery are involved in stem cell signaling. Stem cells possess two key abilities that distinguish them from somatic cells, namely the capacity for self-renewal and the capacity for differentiation. Factors promoting cellular senescence are known to inhibit the pluripotency of stem cells, with oxidative stress being one of the main contributors [73], and autophagy appears to play a role in avoidance of senescence. In order to maintain their stemness, especially long-lived stem cells actively reduce senescence signals by establishing low reactive oxygen species (ROS) environments [74]. A primary source for ROS are old or damaged mitochondria, which in stem cells are efficiently removed by autophagy (a process also referred to as mitophagy) [75]. Besides senescence, ROS and oxidative stress can also sensitize to apoptotic cell death (mediated through apoptotic signals which trigger the formation of Bax/Bak pores in the outer mitochondrial membrane) or to regulated necrotic cell death (mediated through opening of the mitochondrial permeability transition pore) [76, 77]. Autophagy therefore can also protect from ROS-induced cell death modalities. Besides maintaining stemness, autophagy also assists in establishing stemness after oocyte fertilization and also contributes to paving the way for the differentiation of stem cells into specific lineage cells by degrading unwanted organelles and proteins, simultaneously providing the building blocks for the neosynthesis of biomolecules [6, 78].
Taken together, autophagy is required to establish and maintain stemness, to avoid unwanted cell death, and to facilitate cell differentiation. We will describe and discuss the role of autophagy in stem cells (embryonic stem cells, adult stem cells, and cancer stem cells) in the following section in more detail.
The role of autophagy in embryonic stem cells (ESCs)
Pluripotent ESCs emerge from the inner cell mass of blastocysts and can differentiate into the three primary germ layers. Early studies demonstrated that mouse ESCs are autophagy competent since Atg5 −/− ESCs displayed impaired LC3-II conversion and bulk protein degradation upon amino acid starvation [79]. Autophagy was shown to be required for ESC differentiation in vitro, since ESCs lacking Atg5 or Beclin 1 failed to form cavities of embryonic bodies upon leukemia inhibitory factor (LIF) removal [61]. In human ESCs (hESCS), basal autophagic flux was observed in undifferentiated hESC cell lines [67, 80]. During hESC differentiation, autophagic activity markedly increases after treatment with type I TGF-β receptor inhibitor or by removal of MEF-secreted maintenance factors [80], as was determined by monitoring intracellular GFP-LC3 redistribution into punctate autophagosomal staining patterns. Cytosolic and nuclear pluripotency proteins such as Oct4, Sox2, and Nanog are degraded autophagically in hESCs, as was shown by their accumulation in the presence of autophagy inhibitors bafilomycin A1 (BafA1) or 3-MA, and it was proposed that autophagy thereby controls the homeostasis of pluripotency-associated proteins [67]. In mESCs, the failure to induce autophagy during differentiation prevents the formation of embryonic bodies [61]. In this context, autophagy is required to fulfill the energy requirements for the engulfment and clearance of apoptotic cells, indicating a role for autophagy that goes beyond the single-cell level. While autophagy seems essential for the differentiation of both mESCs and hESCs in vitro, in vivo studies have suggested that autophagy is essential only for early stage ESC formation and less so for the later stages of differentiation. For example, Atg5 −/− zygotes fail to develop beyond the four-to eight-cell stage when oocytes are obtained from mice with oocyte-specific Atg5 deficiency [12]. In contrast, Atg5 −/− and other Atg gene knockout zygotes (Atg3 −/−, Atg7 −/− , Atg16 l −/−) inherit sufficient maternal Atg proteins to develop normally pre-natally [6, 81–84]. Atg5 is essential for the formation of autophagosomes, as shown by the absence of GFP-LC3 punctae after fertilization in Atg5 −/− zygotes and at the four-to eight-cell stage prior to implantation [11, 12]. Data providing direct evidence for the regulation of autophagy post-implantation is currently not available. However, Beclin 1 deficiency results in embryonic lethality at E6.5, and this is associated with a prominent developmental failure of the amniotic fold [62]. This might indicate that appropriate autophagy regulation is crucial during post-implantation differentiation. Given the intricate interplay of autophagy regulators and the associated complexity of autophagy control described earlier in this manuscript, the above findings warrant further investigations into the role and relevance of autophagy control factors in both ESC formation and differentiation.
The role of autophagy in adult stem cells
Stem cell populations present in adult tissues can differentiate into specific and limited numbers of cell types. Autophagy appears to play an important role in adult stem cell maintenance as well as during their differentiation [85]. Autophagy inhibition was shown to manifest in functional defects in hematopoietic stem cells (HSCs) and causes severe myeloproliferation [86]. Targeted deletion of Atg7 results in impaired mitophagy, as evidenced by abnormally increased numbers of mitochondria in Atg7 −/− HSCs [86]. Under these conditions, HSCs produce high amounts of ROS and accumulate DNA damage [86]. The inducible deletion of Atg12 in adult mouse HSCs likewise causes prominent autophagy defects and furthermore sensitizes HSCs to undergo apoptosis upon cytokine withdrawal or upon caloric restriction [66]. Here, autophagic flux, monitored by GFP-LC3, increased upon withdrawal of cytokines, and this increase could be blocked by BafA1 and deficiency in FoxO3A. Wild-type HSCs instead are protected by the induction of autophagy through FoxO3A-mediated transcriptional responses [66]. Besides these roles for autophagy in HSC maintenance, autophagy additionally eliminates mitochondria during the differentiation of HSCs into erythrocytes [87].
In human mesenchymal stem cells (hMSCs) and in epidermal stem cells (EpiSCs), autophagic flux is constitutively elevated, as evidenced by conversion of LC3-I to -II and by GFP-LC3 accumulation [85, 88]. Similar to the findings in HSCs, elevated autophagy protects hMSCs from apoptosis, as was shown for conditions of hypoxic stress and for serum deprivation [89]. During the osteogenic differentiation of MSCs, an additional increase in autophagic activity can be observed, which is a consequence of reduced mTOR activity [71]. Osteogenic differentiation of MSCs is accompanied by early activation of AMPK and Raptor, which results in mTOR inhibition. Here, autophagy was detected by enhanced conversion of LC3-I to LC3-II, upregulation of Beclin 1, and a concomitant decrease of p62 [71]. A role for autophagy during differentiation was also identified for neural stem cells (NSCs), since the inhibition of autophagy with 3-MA or Wortmannin impairs neurogenesis [90]. The expression of autophagy genes prominently increases with neuronal differentiation at E15.5 and serves to satisfy high NSC energy demands during the differentiation process [90]. For this scenario, prominent LC3 conversion was observed in the subventricular zone (SVZ) where NSCs locate. Ablation of ULK complex protein FIP200 results in a progressive loss of NSCs and in defects in neuronal differentiation in postnatal mice [48], which provides evidence for the role of autophagy in both maintenance and differentiation of NSCs. Autophagy is also upregulated during the differentiation of cardiac stem cells. Here, fibroblast growth factor (FGF) signaling initially suppresses autophagy to prevent a premature differentiation of progenitor cells [70, 91]. FGF receptor inhibition increases the conversion of LC3-I to LC3-II and stimulates the transcription of genes contributing to autophagy regulation (Beclin1 and p27) [91]. Ablation of Frs2a (an adaptor protein of FGFR) promotes autophagic flux, as monitored by GFP-LC3 punctae, and the differentiation into cardiomyocytes [91].
Taken together, autophagy plays a critical role both in the maintenance and in the differentiation of adult stem cells. Strikingly, very little is known about the differences in the involvement of autophagy in the commitment of adult stem cells to different lineages. In the light of the above findings, further studies in this direction will be of significant interest, since they may elucidate specific and distinct substrate repertoires that need to be targeted for autophagic degradation.
The role of autophagy in cancer stem cells (CSCs)
Cancer stem cells (CSCs) constitute cancer cell sub-populations that can drive tumor propagation, growth, and recurrence. Like other types of stem cells, CSCs possess the capacity for self-renewal and differentiation. Within solid tumors, CSCs typically experience hypoxic and often nutrient-limited microenvironments. In such conditions, the induction of autophagy and the resulting capacity to recycle intracellular materials and to generate nutrients contributes to the survival of CSCs. This was for example shown to manifest in higher autophagic flux, measured by enhanced LC3 conversion and autophagosome formation in breast cancer stem cells found within mammospheres [68]. The inhibition of autophagy can sensitize CSCs to apoptosis and has been shown to decrease CSC tumorigenic capacity in various cancer entities, including breast and liver cancers [92–94]. The notorious treatment resistance of CSCs appears to be linked to autophagic signaling as well, since the inhibition of autophagy in colorectal CSCs significantly enhances the efficacy of photodynamic therapies and cytotoxic drugs [95, 96]. Similarly, silencing of Atg5 or Beclin 1 sensitizes CD133+ glioma stem cells to γ-radiation and abrogates their ability to form neurospheres [97]. Besides the direct effect on pre-existing CSCs, autophagy positively regulates the epithelial-to-mesenchymal transition (EMT) of cancer cells, and thereby facilitates the generation and migration of mesenchymal CSCs [98, 99].
Interestingly, autophagy induction can also be employed to eliminate CSCs. Rottlerin, a potassium channel opener, promotes the transcription of Atg5, Atg7, Atg12, and Beclin 1 and drives autophagy induction through the mTOR/AMPK axis, causing enhanced LC3 conversion [100, 101]. This subsequently can lead to apoptosis induction in breast and prostate cancer stem cells [100, 101]. Autophagy may therefore also suppress CSC generation, a conclusion that is supported by Beclin 1 +/− mice spontaneously developing tumors [62], and the inactivation of Beclin 1 by AKT promoting oncogenesis [9]. Autophagy may therefore have roles in the regulation of CSCs that may differ between tumorigenesis and progressed tumor stages. Autophagy seems to inhibit the generation of early tumors but supports the maintenance and migration of tumor (stem) cells in later disease stages, as was recently convincingly described for melanoma [102]. However, due to the scarcity of quantitative studies on the regulation and role of autophagy in CSCs in different tumor stages, further studies will be required to understand whether interference with autophagy can be exploited to devise novel strategies for CSC-targeted cancer therapies.
The role of autophagy in cell reprogramming
The reprogramming of differentiated cells to stem cells can occur physiologically but can also be induced extrinsically by complex cell manipulations [13]. The above-described changes of and differences in autophagic activities between stem cell and differentiated cell states indicate that the cell reprogramming process may be tightly linked to autophagy signaling. We will therefore next discuss the role of autophagy in different reprogramming processes, including iPSC reprogramming, zygote-to-embryo reprogramming, and adult stem cell reprogramming.
The role of autophagy in iPSC reprogramming
Differentiated somatic cells can be de-differentiated into induced pluripotent stem cells (iPSCs) by three major methodologies: nuclear transfer, cell fusion, and pluripotency-inducing transcription factor transduction. Transcription factor transductions are particularly convenient approaches for obtaining pluripotent cells and for studying the molecular processes involved in cell reprogramming. A striking feature of iPSCs is that the mass and number of their mitochondria are significantly reduced when compared to somatic cells, a feature that likewise has been observed for ES cells [74, 103, 104]. In ES cells, the reduced amount of mitochondria is associated with declines in cellular ROS, and low-ROS environments are a key requirement for maintaining stemness over prolonged periods of time [74]. Vessoni et al. therefore recently promoted the idea that mitophagy might play an important role in cell reprogramming [78]. The requirement for autophagy in cell reprogramming was indeed verified using an autophagy-deficient iPSC induction system [11]. Deficiency in major autophagy proteins such as Atg5, Atg3, and Atg7 abrogates OSKM-induced reprogramming of fibroblasts into iPSCs. Interestingly, OSKM transduction triggers a transient pulse of increased autophagy, which is initiated on day one, peaks on the following day, and then declines back to basal levels on day three. As expected, mitophagy ensues, reduces ROS levels, and contributes to creating suitable conditions for the establishment of pluripotency [11]. Whether autophagy has essential roles during iPSC generation that go beyond mitophagy, such as the degradation of specific groups of proteins, now poses one of the most interesting questions in this field of research.
Given the complexity of autophagy signaling and the possibility to induce or suppress autophagy through the mTOR axis by both extrinsic and intrinsic signals [105, 106], it is notable that during iPSC induction mTOR itself is transiently downregulated at both mRNA and protein levels [11]. This guarantees a robust induction of autophagy, since autophagy-suppressing signals upstream of mTOR are efficiently circumvented or neutralized by mTOR removal. It needs to be noted that even though the activities of both mTORC1 and mTORC2 are reduced during cell reprogramming, only the downregulation of mTORC1 is indispensable [11]. The relevance of autophagy induction and the requirement for this to be a transient process has also been shown in other studies. For example, the presence of mTOR inhibitor rapamycin during the first 3 days after OSKM transduction increases the efficiency of cell reprogramming, whereas treatment at later stages fails to increase the rates of iPSC generation [107]. The requirement to revert autophagy induction during cell reprogramming is further supported by evidence from TSC2-deficient cells and from experiments in the presence of very high concentrations of rapamycin, both of which completely abolish the generation of iPSCs [108]. An initial burst of autophagy may also be required to assist in erasing cellular content that contributes to maintaining the maternal imprint which blocks the reprogramming process, and a subsequent recovery of mTOR levels could be required to establish the pluripotency signaling network for cell reprogramming. Of note, these transient bursts of autophagy differ from those observed during osteogenic differentiation. During the latter, autophagy is triggered by AMPK/Akt-mediated mTOR inhibition [71], a conventional posttranslational mechanism for regulating mTOR activity (see Fig. 1). This potentially highlights that the targeted mTOR depletion may constitute a reprogramming-specific mechanism of autophagy induction.
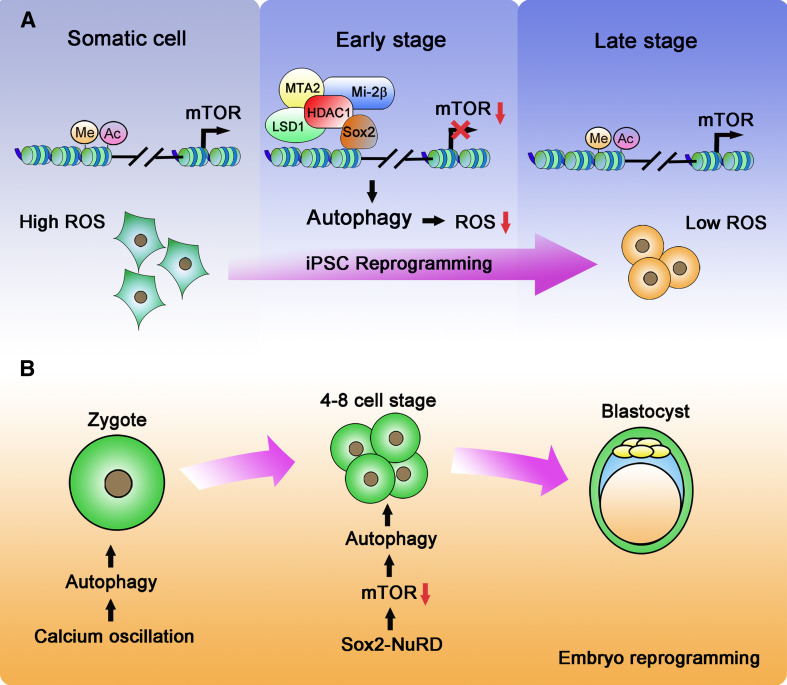
What is the molecular mechanism that downregulates mTOR expression and induces autophagy during cell reprogramming? The cellular transcriptome is comprehensively altered by the transduction of OSKM factors [109, 110], but mTOR suppression appears to rely solely on Sox2, as was found when introducing Oct4, Sox2, Klf4, and Myc individually [11]. While Sox2 is a well-known activating transcription factor [111], a suppression of transcription by Sox2 has rarely been reported. Sox2-dependent repression of mTOR transcription is facilitated by the NuRD (nucleosome remodeling and deacetylase) complex [11]. The NuRD complex belongs to the CHD (chromodomain, helicase, DNA binding) family of chromatin-remodeling complexes and can repress gene transcription by removal of histone acetylation and methylation [112]. The NuRD complex crucially co-regulates transcription during embryonic development [113], cancer progression [114], and is also involved in the control of senescence [115]. During the early stages of cell reprogramming, the NuRD complex, which comprises Mi-2β, LSD1, MTA2, and HDAC1, interacts with Sox2 at the mTOR promoter and demethylates and deacetylates H3K4me1 and H3K9K14, respectively (Fig. 2a). Corresponding to a transient suppression of mTOR expression, the NuRD complex dissociates from the mTOR promoter 2 days after the induction of reprogramming. In agreement with a role for the NuRD complex in cell reprogramming, it was described that expression of the NuRD subunit MBD3 is also essential to facilitate the early step of reprogramming into neural stem cells [116]. In contrast, Hanna and colleagues recently reported that a lack of MBD3 significantly promotes the efficiency of iPSC generation from primed murine pluripotent epiblast stem cells, Oct4+ primordial germ cells, as well as from murine and human fibroblasts [117]. It therefore seems that NuRD complexes may participate in the regulation of cell reprogramming in a context-dependent manner. Notably, little is known about the feedback mechanisms that must exist to shut down NuRD complex activity so as to resume transcription and translation of mTOR at later stages of iPSC induction.
Fig. 2.
Autophagy induction during cell reprogramming. a Autophagy in iPSC reprogramming. In somatic cells, the histones in the promoter region of mTOR are modified by acetylation and methylation (H3K9K14ac, H3K4me1), facilitating mTOR transcription. During early stages of the reprogramming process, Sox2 recruits the NuRD complex to the mTOR promoter, resulting in removal of the epigenetic modifications and causing the suppression of mTOR transcription. Downregulation of mTOR initiates autophagy, which is an indispensable event for cell iPSC reprogramming. Autophagy minimizes ROS levels in somatic cells and provides a suitable microenvironment for reprogramming. b Autophagy in embryonic reprogramming. After fertilization, calcium oscillations trigger an initial induction of autophagy. During the four-to eight-cell stage, Sox2 recruits the NuRD complex to downregulate mTOR expression, thereby leading to the continued induction of autophagy as needed for further development to the blastocyst
The role of autophagy in zygote-to-embryo reprogramming
After fertilization, the zygote is reprogrammed to form pluripotent cells that are located in the inner cell mass of blastocysts. During this process, maternal and paternal genomes are epigenetically modified, pluripotency genes are expressed, and inherited maternal proteins are erased [118, 119]. The ubiquitin–proteasome system is essential for the degradation of short-lived proteins during zygote reprogramming, whereas long-lived proteins and organelles are removed by autophagy [12, 119]. Autophagy is triggered nearly immediately after fertilization and is required for development beyond the four-to-eight cell stage [11, 12]. Autophagic flux measurements at the four-to eight-cell stage may have potential as early markers for subsequent embryonic viability, with significant application potential in the fields of in vitro fertilization and assisted reproductive technologies [120].
The physiological role of autophagy in zygote reprogramming has been investigated intensely in recent years and has also provided insight into the underlying molecular signaling events. The general relevance of autophagy was demonstrated first by studies in Atg5-deficient oocytes, which fail to develop beyond the four-to eight-cell stage following fertilization with Atg5 −/− sperms [12]. In reverse, over-activation of autophagy by mTOR inhibitor rapamycin accelerates embryonic reprogramming, and blastocysts appear as early as embryo day E3.0 [11]. Absence of autophagy impairs the embryonal capacity for protein neo-synthesis, a consequence that likely arises from the lack of maternal protein removal and the associated incapacity to recycle amino acids [12]. It is widely assumed that autophagy also plays a role in the removal of paternal mitochondria and mitochondrial DNA (mtDNA), since mitochondria and mtDNA are inherited maternally [121, 122], and this likewise may contribute to providing nutrients. While this is indeed the case in the nematode C. elegans [123, 124], this role of mitophagy is not evolutionarily conserved. In mice, paternal mitochondria in the zygote are not removed but instead fail to replicate due to the degradation of their mtDNA prior to fertilization [125].
The molecular mechanisms underlying autophagy induction during embryonic reprogramming appear to change throughout early embryonic development. Fertilization induces the initial pulse of autophagy independently of mTORC1 activity, since suppression of mTORC1 is neither essential nor sufficient for autophagy induction [12, 126]. Instead, calcium oscillations triggered by fertilization may initiate autophagic responses [12]. However, once the four-to-eight cell stage is reached, the Sox2- and NuRD-mediated downregulation of mTOR expression becomes indispensable for autophagy induction, and this mechanism is of equivalent relevance for iPSC reprogramming (Fig. 2b) [11]. It seems obvious that a transition between mTOR-independent post-fertilization autophagy to the subsequent mTOR-dependent autophagic signaling at the four-to-eight cell stage must be accurately coordinated, given the tight temporal coupling of these developmental steps. However, how this is achieved and whether this requires additional molecular control processes is presently unknown.
The role of autophagy in adult stem cell reprogramming
Differentiated somatic cells can be reprogrammed into adult stem cells with limited pluripotency. We now begin to understand if and how autophagy may play a role in adult stem cell reprogramming, and in the following discuss indications for such roles in the context of stem cell reprogramming approaches.
The hematopoietic system has been intensely investigated to identify strategies by which blood progenitor cells and HSCs can be generated from somatic cells. The efficacies of cell reprogramming in the hematopoietic system have improved significantly within the past years [127–129]. Multi-lineage blood progenitors can be generated from human fibroblasts without the need to pass through a stem cell pluripotency state. This can be achieved by the ectopic expression of Oct4 and cultivation in the presence of a specific cocktail of cytokines, which supports the development of hematopoietic progenitor cells [130]. HSC-like cells can also be generated from mouse fibroblasts or from human lineage-restricted myeloid precursors by transduction of specific sets of transcription factors (Gata2, Gfi1b, cFos, Etv6 and HOXA9, ERG, RORA, respectively) [127, 129]. Similarly, pre-B cells can be induced to transform into iHSCs that display functional and molecular features found in native HSCs using a set of six transcription factors (Run1t1, Hlf, Lmo2, Prdm5, Pbx1, and Zfp37) [128]. HSCs reside in a hypoxic niche in the bone marrow [131] and maintain reduced amounts of mitochondria to limit ROS generation in quiescent conditions [132, 133]. The transition of HSCs from quiescence to proliferation/differentiation is accompanied by increased mTOR activity and elevated ROS amounts [132], a possible consequence of autophagy repression. Since HSCs in aged mice can be rejuvenated by mTOR inhibition, which displays in restored self-renewal and hematopoiesis, autophagy may be required to regain pluripotency [134]. Correspondingly, it is possible that establishing conditions of reduced ROS and mitochondrial counts, as were previously discussed for ES cells, are required for efficient reprogramming of differentiated cells into HSCs. However, formal proof that the induction of autophagy is pivotal for this is currently outstanding. Considering the comprehensive experimental approaches and tools that are available to study autophagic signaling [27], the fact that still very little is known about if and how autophagy is involved in HSC cell reprogramming seems surprising. If autophagy is linked to establishing the quiescence state of HSCs, transcription factors Pbx1 and Meis 1 may be candidates that induce this function. Pbx1 was shown to promote iHSC generation, Meis1 enhances the efficiency of HSC reprogramming, and together they are involved in regulating HSC self-renewal through maintaining HSC quiescence [128, 135, 136]. Besides a potential role during reprogramming, autophagy also contributes to the differentiation of reprogrammed cells. During human granulosa cell differentiation into muscular cells upon 5-azacytidine treatment, the formation of numerous autophagosomes was observed by electron microscopy [137]. It will therefore be of interest to investigate whether changes in autophagic signaling or flux are generally detected and required during the re-differentiation of de-differentiated adult stem cells.
Cancer stem cells (CSCs) can develop from fully differentiated cells by various triggers and signals. For example, inflammatory signaling and activation of the NF-κB pathway can trigger the de-differentiation of intestinal epithelial cells with constitutive Wnt activation into tumor-initiating cells, as was shown in a model of intestinal tumorigenesis [138]. Tumor suppressor p53, which is known to limit the reprogramming of iPSCs, also restricts the self-renewal capacity of tumor stem cells [139, 140]. Myeloid progenitor cells that express oncogenic Kras and that lack p53 can transform into acute myeloid leukemia-initiating cells [141]. In glioblastoma multiforme (GBM), double silencing of neurofibromatosis type I (NF1) and p53 in astrocytes and in mature neurons generates stem-cell like progenitor cells, which promote GBM progression [142]. Similar to transcription factor transduction strategies for the generation of iPSCs, groups of transcription factors that promote the reprogramming into cancer stem cells were described. For example, the core transcription factors for GBM cell reprogramming have recently been identified as Pou3f2, Sox2, Sall2, and Olig2 [143]. The role of autophagy in CSC reprogramming overall remains largely elusive in this context. This may be partially due to the potentially distinct roles of autophagy in early stage and progressed, metastatic cancers. While autophagy reduces tumorigenesis and is associated with good prognosis in early stage cancers [62, 102, 144, 145], in advanced tumors elevated autophagy confers higher chemoresistance and tolerance to metabolic stresses [92, 96]. The generation of CSCs is typically accompanied by a switch from oxidative phosphorylation to glycolysis [146, 147]. The Kras oncogene can facilitate glycolysis during oncogenic transformation [148, 149] and intriguingly also promotes autophagic flux, as indicated by enhanced LC3 conversion [150, 151]. This promotes mitophagy and reduces respiratory activity, with a recovery of oxygen consumption rates observed upon autophagy inhibition by BafA1 and 3-MA [151]. As Kras is involved in the reprogramming of myeloid leukemia-initiating cells [141], it is conceivable that Kras-induced autophagy contributes to cancer cell reprogramming and CSC generation also during progressed tumor stages. While CSCs may inherit their metabolic adaptation from more differentiated progenitor cells, metabolic reprogramming from oxidative phosphorylation to glycolysis has also been described during the dedifferentiation process itself [146, 147]. It may therefore be postulated that autophagy could occur during the reprogramming of differentiated cells to CSCs in parallel to or as a consequence of metabolic adaptation or reprogramming. Sox2, which we showed to be involved in iPSC reprogramming and which is critical for transient autophagy induction [11], also promotes the reprogramming of GBM stem-like cells [143]. A Sox2-focused approach towards clarifying the role of autophagy during CSC reprogramming therefore poses an attractive avenue of investigation.
Concluding remarks
Autophagy is essential for cell reprogramming, as has been comprehensively documented during the zygote-to-embryo transition and also during iPSC generation. Autophagy not only contributes to establishing a suitable environment for successful reprogramming but also provides a source of intracellular building blocks for cell renovation. Notably, detailed insight into the role and relevance of autophagy during adult cell reprogramming and especially CSC generation are still outstanding. Benefiting from the deep molecular understanding of autophagy signaling that was obtained over the past decades, targeted manipulations of autophagic flux can already be exploited to improve the efficiency of cell reprogramming and will undoubtedly support the further development of translational stem cell-based applications in regenerative medicine. Likewise, targeting the contribution of autophagy to reprogramming cells into CSCs may allow preventing the further malignant transformation of tumor cells. We hope that our article will help the readership to obtain a representative overview of the current state of knowledge in this research field, and that the identification of knowledge gaps may attract further investigations into currently underexplored areas.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31300645, 81330047), 973 Program of the MOST of China (2010CB911902), and the Strategic Priority Research Programs of the Chinese Academy of Sciences (XDA01010407). MR and ZF are supported by the Science Foundation Ireland International Strategic Collaboration Programme: China (ISCP China).
Abbreviations
- Ambra1
Activating molecule in Beclin1-regulated autophagy 1
- AMPK
AMP-activated protein kinase
- Atg
Autophagy-related proteins
- Beclin 1
Coiled-coil, myosin-like BCL2-interacting protein
- BafA1
Bafilomycin A1
- CSC
Cancer stem cell
- CHD
Chromodomain, helicase, DNA binding
- EMT
Epithelial–mesenchymal transition
- ESC
Embryonic stem cell
- FGF
Fibroblast growth factor
- GBM
Glioblastoma
- GAP
GTPase-activating protein
- HSC
Hematopoietic stem cell
- iPSC
Induced pluripotent stem cell
- LC3
Microtubule-associated protein light chain 3
- LIF
Leukemia inhibitory factor
- MBD3
Methyl-CpG binding domain protein 3
- mTOR
Mammalian target of rapamycin
- NuRD
Nucleosome remodeling and deacetylase
- NSC
Neural stem cell
- PE
Phosphatidylethanolamine
- Rheb
Ras homologue enriched in brain
- ROS
Reactive oxygen species
- Sox2
SRY (sex determining region Y)-box 2
- SVZ
Subventricular zone
- TSC1/2
Tuberous sclerosis1/2
- ULK
Unc-51-like kinase
- WASH
Wiskott–Aldrich syndrome protein and SCAR homologue
References
- 1.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 2.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 3.Kundu M, Thompson CB. Autophagy: basic principles and relevance to disease. Annu Rev Pathol. 2008;3:427–455. doi: 10.1146/annurev.pathmechdis.2.010506.091842. [DOI] [PubMed] [Google Scholar]
- 4.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 6.Mizushima N, Levine B. Autophagy in mammalian development and differentiation. Nat Cell Biol. 2010;12:823–830. doi: 10.1038/ncb0910-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xia P, Wang S, Du Y, Zhao Z, Shi L, et al. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 2013;32:2685–2696. doi: 10.1038/emboj.2013.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong E, Cuervo AM. Autophagy gone awry in neurodegenerative diseases. Nat Neurosci. 2010;13:805–811. doi: 10.1038/nn.2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang RC, Wei Y, An Z, Zou Z, Xiao G, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13:722–737. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Xia P, Ye B, Huang G, Liu J, et al. Transient activation of autophagy via Sox2-mediated suppression of mTOR is an important early step in reprogramming to pluripotency. Cell Stem Cell. 2013;13:617–625. doi: 10.1016/j.stem.2013.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, et al. Autophagy is essential for preimplantation development of mouse embryos. Science. 2008;321:117–120. doi: 10.1126/science.1154822. [DOI] [PubMed] [Google Scholar]
- 13.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143:508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jopling C, Boue S, Izpisua Belmonte JC. Dedifferentiation, transdifferentiation and reprogramming: three routes to regeneration. Nat Rev Mol Cell Biol. 2011;12:79–89. doi: 10.1038/nrm3043. [DOI] [PubMed] [Google Scholar]
- 16.Abollo-Jimenez F, Jimenez R, Cobaleda C. Physiological cellular reprogramming and cancer. Semin Cancer Biol. 2010;20:98–106. doi: 10.1016/j.semcancer.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 18.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 19.Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koche RP, Smith ZD, Adli M, Gu H, Ku M, et al. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golipour A, David L, Liu Y, Jayakumaran G, Hirsch CL, et al. A late transition in somatic cell reprogramming requires regulators distinct from the pluripotency network. Cell Stem Cell. 2012;11:769–782. doi: 10.1016/j.stem.2012.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11:783–798. doi: 10.1016/j.stem.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, et al. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. J Cell Biol. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.C11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizushima N, Yoshimori T, Ohsumi Y. The role of atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 26.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol. 2009;10:458–467. doi: 10.1038/nrm2708. [DOI] [PubMed] [Google Scholar]
- 27.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, et al. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/S0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- 34.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, et al. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- 35.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hosokawa N, Hara T, Kaizuka T, Kishi C, Takamura A, et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol Biol Cell. 2009;20:1981–1991. doi: 10.1091/mbc.E08-12-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–657. doi: 10.1038/ncb839. [DOI] [PubMed] [Google Scholar]
- 41.Hardie DG. AMP-activated/SNF1 protein Kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 42.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell. 2006;126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 43.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, et al. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature. 2010;465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van der Vos KE, Coffer PJ. Glutamine metabolism links growth factor signaling to the regulation of autophagy. Autophagy. 2012;8:1862–1864. doi: 10.4161/auto.22152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menendez JA, Vellon L, Oliveras-Ferraros C, Cufi S, Vazquez-Martin A. mTOR-regulated senescence and autophagy during reprogramming of somatic cells to pluripotency: a roadmap from energy metabolism to stem cell renewal and aging. Cell Cycle. 2011;10:3658–3677. doi: 10.4161/cc.10.21.18128. [DOI] [PubMed] [Google Scholar]
- 46.Egan D, Kim J, Shaw RJ, Guan KL. The autophagy initiating kinase ULK1 is regulated via opposing phosphorylation by AMPK and mTOR. Autophagy. 2011;7:643–644. doi: 10.4161/auto.7.6.15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung CH, Jun CB, Ro SH, Kim YM, Otto NM, et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol Biol Cell. 2009;20:1992–2003. doi: 10.1091/mbc.E08-12-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C, Liang CC, Bian ZC, Zhu Y, Guan JL. FIP200 is required for maintenance and differentiation of postnatal neural stem cells. Nat Neurosci. 2013;16:532–542. doi: 10.1038/nn.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- 50.Funderburk S F, Wang Q J, Yue Z The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, et al. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett. 2010;584:1313–1318. doi: 10.1016/j.febslet.2010.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, et al. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol. 2007;9:1142–1151. doi: 10.1038/ncb1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. doi: 10.1038/nature05925. [DOI] [PubMed] [Google Scholar]
- 55.He S, Ni D, Ma B, Lee JH, Zhang T, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol. 2013;15:1206–1219. doi: 10.1038/ncb2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 57.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Furuya T, Kim M, Lipinski M, Li J, Kim D, et al. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol Cell. 2010;38:500–511. doi: 10.1016/j.molcel.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia P, Wang S, Huang G, Du Y, Zhu P, et al. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 2014;24:943–958. doi: 10.1038/cr.2014.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qu X, Zou Z, Sun Q, Luby-Phelps K, Cheng P, et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell. 2007;128:931–946. doi: 10.1016/j.cell.2006.12.044. [DOI] [PubMed] [Google Scholar]
- 62.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakoh-Nakatogawa M, Matoba K, Asai E, Kirisako H, Ishii J, et al. Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat Struct Mol Biol. 2013;20:433–439. doi: 10.1038/nsmb.2527. [DOI] [PubMed] [Google Scholar]
- 64.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Warr MR, Binnewies M, Flach J, Reynaud D, Garg T, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cho YH, Han KM, Kim D, Lee J, Lee SH, et al. Autophagy regulates homeostasis of pluripotency-associated proteins in hESCs. Stem Cells. 2014;32:424–435. doi: 10.1002/stem.1589. [DOI] [PubMed] [Google Scholar]
- 68.Gong C, Bauvy C, Tonelli G, Yue W, Delomenie C, et al. Beclin 1 and autophagy are required for the tumorigenicity of breast cancer stem-like/progenitor cells. Oncogene. 2013;32:2261–2272. doi: 10.1038/onc.2012.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bellodi C, Lidonnici MR, Hamilton A, Helgason GV, Soliera AR, et al. Targeting autophagy potentiates tyrosine kinase inhibitor-induced cell death in Philadelphia chromosome-positive cells, including primary CML stem cells. J Clin Invest. 2009;119:1109–1123. doi: 10.1172/JCI35660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang J, Liu J, Liu L, McKeehan WL, Wang F. The fibroblast growth factor signaling axis controls cardiac stem cell differentiation through regulating autophagy. Autophagy. 2012;8:690–691. doi: 10.4161/auto.19290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pantovic A, Krstic A, Janjetovic K, Kocic J, Harhaji-Trajkovic L, et al. Coordinated time-dependent modulation of AMPK/Akt/mTOR signaling and autophagy controls osteogenic differentiation of human mesenchymal stem cells. Bone. 2013;52:524–531. doi: 10.1016/j.bone.2012.10.024. [DOI] [PubMed] [Google Scholar]
- 72.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 73.Haines DD, Juhasz B, Tosaki A. Management of multicellular senescence and oxidative stress. J Cell Mol Med. 2013;17:936–957. doi: 10.1111/jcmm.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pan H, Cai N, Li M, Liu GH, Izpisua Belmonte JC. Autophagic control of cell ‘stemness’. EMBO Mol Med. 2013;5:327–331. doi: 10.1002/emmm.201201999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 77.Green DR, Galluzzi L, Kroemer G. Cell biology. Metabolic control of cell death. Science. 2014;345:1250256. doi: 10.1126/science.1250256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vessoni AT, Muotri AR, Okamoto OK. Autophagy in stem cell maintenance and differentiation. Stem Cells Dev. 2012;21:513–520. doi: 10.1089/scd.2011.0526. [DOI] [PubMed] [Google Scholar]
- 79.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, et al. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tra T, Gong L, Kao LP, Li XL, Grandela C, et al. Autophagy in human embryonic stem cells. PLoS One. 2011;6:e27485. doi: 10.1371/journal.pone.0027485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, et al. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 82.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sou YS, Waguri S, Iwata J, Ueno T, Fujimura T, et al. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol Biol Cell. 2008;19:4762–4775. doi: 10.1091/mbc.E08-03-0309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 85.Salemi S, Yousefi S, Constantinescu MA, Fey MF, Simon HU. Autophagy is required for self-renewal and differentiation of adult human stem cells. Cell Res. 2012;22:432–435. doi: 10.1038/cr.2011.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, et al. The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J Exp Med. 2011;208:455–467. doi: 10.1084/jem.20101145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mortensen M, Ferguson DJ, Edelmann M, Kessler B, Morten KJ, et al. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA. 2010;107:832–837. doi: 10.1073/pnas.0913170107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oliver L, Hue E, Priault M, Vallette FM. Basal autophagy decreased during the differentiation of human adult mesenchymal stem cells. Stem Cells Dev. 2012;21:2779–2788. doi: 10.1089/scd.2012.0124. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Q, Yang YJ, Wang H, Dong QT, Wang TJ, et al. Autophagy activation: a novel mechanism of atorvastatin to protect mesenchymal stem cells from hypoxia and serum deprivation via AMP-activated protein kinase/mammalian target of rapamycin pathway. Stem Cells Dev. 2012;21:1321–1332. doi: 10.1089/scd.2011.0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vazquez P, Arroba AI, Cecconi F, de la Rosa EJ, Boya P, et al. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy. 2012;8:187–199. doi: 10.4161/auto.8.2.18535. [DOI] [PubMed] [Google Scholar]
- 91.Zhang J, Liu J, Huang Y, Chang JY, Liu L, et al. FRS2alpha-mediated FGF signals suppress premature differentiation of cardiac stem cells through regulating autophagy activity. Circ Res. 2012;110:e29–e39. doi: 10.1161/CIRCRESAHA.111.255950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song YJ, Zhang SS, Guo XL, Sun K, Han ZP, et al. Autophagy contributes to the survival of CD133+ liver cancer stem cells in the hypoxic and nutrient-deprived tumor microenvironment. Cancer Lett. 2013;339:70–81. doi: 10.1016/j.canlet.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 93.Yue W, Hamai A, Tonelli G, Bauvy C, Nicolas V, et al. Inhibition of the autophagic flux by salinomycin in breast cancer stem-like/progenitor cells interferes with their maintenance. Autophagy. 2013;9:714–729. doi: 10.4161/auto.23997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Espina V, Mariani BD, Gallagher RI, Tran K, Banks S, et al. Malignant precursor cells pre-exist in human breast DCIS and require autophagy for survival. PLoS One. 2010;5:e10240. doi: 10.1371/journal.pone.0010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wei MF, Chen MW, Chen KC, Lou PJ, Lin SY, et al. Autophagy promotes resistance to photodynamic therapy-induced apoptosis selectively in colorectal cancer stem-like cells. Autophagy. 2014;10:1179–1192. doi: 10.4161/auto.28679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu S, Wang X, Chen J, Chen Y. Autophagy of cancer stem cells is involved with chemoresistance of colon cancer cells. Biochem Biophys Res Commun. 2013;434:898–903. doi: 10.1016/j.bbrc.2013.04.053. [DOI] [PubMed] [Google Scholar]
- 97.Lomonaco SL, Finniss S, Xiang C, Decarvalho A, Umansky F, et al. The induction of autophagy by gamma-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125:717–722. doi: 10.1002/ijc.24402. [DOI] [PubMed] [Google Scholar]
- 98.Kiyono K, Suzuki HI, Matsuyama H, Morishita Y, Komuro A, et al. Autophagy is activated by TGF-beta and potentiates TGF-beta-mediated growth inhibition in human hepatocellular carcinoma cells. Cancer Res. 2009;69:8844–8852. doi: 10.1158/0008-5472.CAN-08-4401. [DOI] [PubMed] [Google Scholar]
- 99.Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178:437–451. doi: 10.1083/jcb.200611146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kumar D, Shankar S, Srivastava RK. Rottlerin-induced autophagy leads to the apoptosis in breast cancer stem cells: molecular mechanisms. Mol Cancer. 2013;12:171. doi: 10.1186/1476-4598-12-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kumar D, Shankar S, Srivastava RK. Rottlerin induces autophagy and apoptosis in prostate cancer stem cells via PI3 K/Akt/mTOR signaling pathway. Cancer Lett. 2014;343:179–189. doi: 10.1016/j.canlet.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 102.Liu H, He Z, von Rutte T, Yousefi S, Hunger RE, et al. Down-regulation of autophagy-related protein 5 (ATG5) contributes to the pathogenesis of early-stage cutaneous melanoma. Sci Transl Med. 2013;5:202ra123. doi: 10.1126/scitranslmed.3005864. [DOI] [PubMed] [Google Scholar]
- 103.Armstrong L, Tilgner K, Saretzki G, Atkinson SP, Stojkovic M, et al. Human induced pluripotent stem cell lines show stress defense mechanisms and mitochondrial regulation similar to those of human embryonic stem cells. Stem Cells. 2010;28:661–673. doi: 10.1002/stem.307. [DOI] [PubMed] [Google Scholar]
- 104.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 105.Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nat Rev Mol Cell Biol. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ravikumar B, Vacher C, Berger Z, Davies JE, Luo S, et al. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet. 2004;36:585–595. doi: 10.1038/ng1362. [DOI] [PubMed] [Google Scholar]
- 107.Chen T, Shen L, Yu J, Wan H, Guo A, et al. Rapamycin and other longevity-promoting compounds enhance the generation of mouse induced pluripotent stem cells. Aging Cell. 2011;10:908–911. doi: 10.1111/j.1474-9726.2011.00722.x. [DOI] [PubMed] [Google Scholar]
- 108.He J, Kang L, Wu T, Zhang J, Wang H, et al. An elaborate regulation of Mammalian target of rapamycin activity is required for somatic cell reprogramming induced by defined transcription factors. Stem Cells Dev. 2012;21:2630–2641. doi: 10.1089/scd.2012.0015. [DOI] [PubMed] [Google Scholar]
- 109.Cantone I, Fisher AG. Epigenetic programming and reprogramming during development. Nat Struct Mol Biol. 2013;20:282–289. doi: 10.1038/nsmb.2489. [DOI] [PubMed] [Google Scholar]
- 110.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fong YW, Cattoglio C, Yamaguchi T, Tjian R. Transcriptional regulation by coactivators in embryonic stem cells. Trends Cell Biol. 2012;22:292–298. doi: 10.1016/j.tcb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hu G, Wade PA. NuRD and pluripotency: a complex balancing act. Cell Stem Cell. 2012;10:497–503. doi: 10.1016/j.stem.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reynolds N, Latos P, Hynes-Allen A, Loos R, Leaford D, et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem Cell. 2012;10:583–594. doi: 10.1016/j.stem.2012.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, et al. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009;11:1261–1267. doi: 10.1038/ncb1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dos Santos RL, Tosti L, Radzisheuskaya A, Caballero IM, Kaji K, et al. MBD3/NuRD facilitates induction of pluripotency in a context-dependent manner. Cell Stem Cell. 2014;15:102–110. doi: 10.1016/j.stem.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502:65–70. doi: 10.1038/nature12587. [DOI] [PubMed] [Google Scholar]
- 118.Stitzel ML, Seydoux G. Regulation of the oocyte-to-zygote transition. Science. 2007;316:407–408. doi: 10.1126/science.1138236. [DOI] [PubMed] [Google Scholar]
- 119.DeRenzo C, Seydoux G. A clean start: degradation of maternal proteins at the oocyte-to-embryo transition. Trends Cell Biol. 2004;14:420–426. doi: 10.1016/j.tcb.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 120.Tsukamoto S, Hara T, Yamamoto A, Kito S, Minami N, et al. Fluorescence-based visualization of autophagic activity predicts mouse embryo viability. Sci Rep. 2014;4:4533. doi: 10.1038/srep04533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hutchison CA, 3rd, Newbold JE, Potter SS, Edgell MH. Maternal inheritance of mammalian mitochondrial DNA. Nature. 1974;251:536–538. doi: 10.1038/251536a0. [DOI] [PubMed] [Google Scholar]
- 122.Ankel-Simons F, Cummins JM. Misconceptions about mitochondria and mammalian fertilization: implications for theories on human evolution. Proc Natl Acad Sci USA. 1996;93:13859–13863. doi: 10.1073/pnas.93.24.13859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Al Rawi S, Louvet-Vallee S, Djeddi A, Sachse M, Culetto E, et al. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. doi: 10.1126/science.1211878. [DOI] [PubMed] [Google Scholar]
- 124.Sato M, Sato K. Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science. 2011;334:1141–1144. doi: 10.1126/science.1210333. [DOI] [PubMed] [Google Scholar]
- 125.Luo SM, Ge ZJ, Wang ZW, Jiang ZZ, Wang ZB, et al. Unique insights into maternal mitochondrial inheritance in mice. Proc Natl Acad Sci USA. 2013;110:13038–13043. doi: 10.1073/pnas.1303231110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yamamoto A, Mizushima N, Tsukamoto S. Fertilization-induced autophagy in mouse embryos is independent of mTORC1. Biol Reprod. 2014;91:7. doi: 10.1095/biolreprod.113.115816. [DOI] [PubMed] [Google Scholar]
- 127.Doulatov S, Vo LT, Chou SS, Kim PG, Arora N, et al. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 2013;13:459–470. doi: 10.1016/j.stem.2013.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Riddell J, Gazit R, Garrison BS, Guo G, Saadatpour A, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157:549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pereira CF, Chang B, Qiu J, Niu X, Papatsenko D, et al. Induction of a hemogenic program in mouse fibroblasts. Cell Stem Cell. 2013;13:205–218. doi: 10.1016/j.stem.2013.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 131.Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chen C, Liu Y, Liu R, Ikenoue T, Guan KL, et al. TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J Exp Med. 2008;205:2397–2408. doi: 10.1084/jem.20081297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, et al. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12:446–451. doi: 10.1038/nm1388. [DOI] [PubMed] [Google Scholar]
- 134.Chen C, Liu Y, Liu Y, Zheng P. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal. 2009;2:ra75. doi: 10.1126/scisignal.2000559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2:484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Unnisa Z, Clark JP, Roychoudhury J, Thomas E, Tessarollo L, et al. Meis1 preserves hematopoietic stem cells in mice by limiting oxidative stress. Blood. 2012;120:4973–4981. doi: 10.1182/blood-2012-06-435800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brevini TA, Pennarossa G, Rahman MM, Paffoni A, Antonini S, et al. Morphological and molecular changes of human granulosa cells exposed to 5-azacytidine and addressed toward muscular differentiation. Stem Cell Rev. 2014;10:633–642. doi: 10.1007/s12015-014-9521-4. [DOI] [PubMed] [Google Scholar]
- 138.Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Goktuna SI, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152:25–38. doi: 10.1016/j.cell.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 139.Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Meletis K, Wirta V, Hede SM, Nister M, Lundeberg J, et al. p53 suppresses the self-renewal of adult neural stem cells. Development. 2006;133:363–369. doi: 10.1242/dev.02208. [DOI] [PubMed] [Google Scholar]
- 141.Zhao Z, Zuber J, Diaz-Flores E, Lintault L, Kogan SC, et al. p53 loss promotes acute myeloid leukemia by enabling aberrant self-renewal. Genes Dev. 2010;24:1389–1402. doi: 10.1101/gad.1940710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Friedmann-Morvinski D, Bushong EA, Ke E, Soda Y, Marumoto T, et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science. 2012;338:1080–1084. doi: 10.1126/science.1226929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Suva ML, Rheinbay E, Gillespie SM, Patel AP, Wakimoto H, et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Marino G, Salvador-Montoliu N, Fueyo A, Knecht E, Mizushima N, et al. Tissue-specific autophagy alterations and increased tumorigenesis in mice deficient in Atg4C/autophagin-3. J Biol Chem. 2007;282:18573–18583. doi: 10.1074/jbc.M701194200. [DOI] [PubMed] [Google Scholar]
- 145.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab . 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 148.Blum R, Jacob-Hirsch J, Amariglio N, Rechavi G, Kloog Y. Ras inhibition in glioblastoma down-regulates hypoxia-inducible factor-1alpha, causing glycolysis shutdown and cell death. Cancer Res. 2005;65:999–1006. [PubMed] [Google Scholar]
- 149.Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lock R, Roy S, Kenific CM, Su JS, Salas E, et al. Autophagy facilitates glycolysis during ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim JH, Kim HY, Lee YK, Yoon YS, Xu WG, et al. Involvement of mitophagy in oncogenic K-Ras-induced transformation: overcoming a cellular energy deficit from glucose deficiency. Autophagy. 2011;7:1187–1198. doi: 10.4161/auto.7.10.16643. [DOI] [PMC free article] [PubMed] [Google Scholar]