Abstract
The DNA sequence largely defines gene expression and phenotype. However, it is becoming increasingly clear that an additional chromatin-based regulatory network imparts both stability and plasticity to genome output, modifying phenotype independently of the genetic blueprint. Indeed, alterations in this “epigenetic” control layer underlie, at least in part, the reason for monozygotic twins being discordant for disease. Functionally, this regulatory layer comprises post-translational modifications of DNA and histones, as well as small and large noncoding RNAs. Together these regulate gene expression by changing chromatin organization and DNA accessibility. Successive technological advances over the past decade have enabled researchers to map the chromatin state with increasing accuracy and comprehensiveness, catapulting genetic research into a genome-wide era. Here, aiming particularly at the genomics/epigenomics newcomer, we review the epigenetic basis that has helped drive the technological shift and how this progress is shaping our understanding of complex disease.
Keywords: Complex diseases, Chromatin, Epigenomics
Epigenetic potential in disease
One of the first movements to follow completion of the Human Genome Project, just over a decade ago, was an immense investment in large-scale association studies. Aimed at unravelling the genetic bases of common phenotypic variation and disease, these genome-wide association studies (GWAS) successfully identified a vast number of DNA variants associated with disease risk. To date, though, few if any of the identified variants have been shown to play a causative role. This finding highlights the fact that phenotypic setpoints are not fully genetically predetermined, and that environment, particularly during development, plays a significant role. Indeed this variability of phenotype has attracted attention of late, including an increased discussion of phenotypic plasticity, phenotypic noise, variable penetrance, cellular memory, and developmental reprogramming. Collectively, these notions embody the net product of an entire regulatory level that, while still poorly understood, is proving vital to understanding complex disease aetiology.
There is ample evidence that a complex network of chromatin-based systems exists that cause stable changes to genetically predetermined phenotypes. We know that multiple mechanisms exist to impose and maintain long-term changes in gene expression. Included, are protein networks that covalently modify DNA and histones, and a wide variety of regulatory RNAs and proteins that associate with chromatin [1]. Some of the chromatin state patterns established by these systems are transmissible through cell division, and even through the germline. To date though, we know very little about how these processes define disease risk and outcome.
Simply put, the potential for impacting aetiology is immense. Chromatin-based paradigms have been reported to underpin endocrine regulation, learning, memory, neurological and physiological abnormalities, autism, type 2 diabetes, autoimmune diseases, and cancer [2–6]. Virtually every physiological process is amenable to epigenetic regulation. Add to the cellular complexity a layer of physiological complexity, for instance the potential to alter the number or connectivity of feeding neurons, and the number of ways to stably modify phenotype is vast. A case in point, “developmental reprogramming”, a process in which external cues during development cause phenotypic changes later in life, is thought to be chromatin-based; numerous molecular pathways and organ systems have been implicated in what is still a crude understanding of the topic.
Here, we review the recent explosion of genomic technologies and their potential in unravelling the epigenetic basis of disease aetiology. We summarize the essence of epigenomic analysis, the basis of its methodology, and the doors and challenges it is opening for the next generation of molecular geneticists.
Chromatin: from form to function
The first step in understanding how epigenomics will impact contemporary molecular medicine is understanding the epigenome itself. Eukaryotic DNA is present in the cell in either chromosomal or extrachromosomal forms. Whereas the latter, primarily mitochondrial DNA, exists in a relatively disorganized circular form, chromosomal DNA is packaged in multiple hierarchical levels of a repetitive structure collectively known as chromatin. In it’s most basic form, chromatin comprises a 147-bp nucleic acid stretch wrapped one and a half times around a nucleosome. Like pearls on a string, chains of nucleosomes organize into 30-nm fibres and higher order structures eventually forming chromatids at the peak of the cell cycle. Besides packaging metres of DNA into a picolitre volume, chromatin sub-compartmentalizes DNA for efficient regulatory processing of about 23,000 genes in a cell-type and stimulus-specific manner. Active areas of the genome are found in regions of euchromatin, loosely packed, and more or less accessible to regulatory factors, while inactive areas are found as more densely packed heterochromatin. Heterochromatin can be further divided either constitutive heterochromatin, silenced in all cell types (e.g. pericentric or telomeric regions), and facultative heterochromatin, regions that need to be on in some cell types and off in others. Importantly, because of its physical form, chromatin state information can be copied and passed on from mother cell to daughter cell, and in some circumstances from one generation to the next.
Broadly speaking, the chromatin state results from the convergence of dynamic interactions between DNA, RNA, and an estimate of about 1,200 proteins. Core chromatin-defining components, however, are typically described as falling into one of three categories: DNA modification, post-translational histone modification, and noncoding RNA.
DNA modification
Our genomes comprise relatively equal proportions of the four DNA bases A, G, C, T. In addition to their native states, some of these bases can be found in modified forms. The most common modification by far is cytosine methylation. It is found on approximately 1 % of all cytosine residues. Of this, about 30 % is distributed among CA, CT, and CC dinucleotides, with no known functional role to date and the remaining majority of about 70 % occurs at CG or CpG (C-phosphate-G) dinucleotides (not to be confused with C-G base pairs) [7–9]. Though historically thought to be stable, DNA methylation has been recently shown to undergo active demethylation, yielding multiple intermediate forms of CpG modification, including hydroxymethylation, formylation and carboxylation [10–12]. As early as 1975, DNA methylation was linked to X-inactivation, and later identified as a hallmark of genomic imprinting [13] and tissue specificity of gene expression [14]. Seminal works include the demonstrations that tissue-specific genes are undermethylated in their tissue of expression [15] and that housekeeping genes are controlled by a unique family of nonmethylated CpG island promoters [16].
A cursory examination of CpG methylation across the genome will immediately reveal four striking features: first, CpGs are not found randomly through the genome, but rather clustered in what are known as CpG islands; second, these CpG islands are highly enriched at or near promoter regions, implicating them in transcriptional regulation; third, one female X-chromosome, the inactive X, is broadly and heavily DNA-methylated; and fourth, about 100 loci across the genome display methylation patterns as direct copies of either the maternal or paternal methylation status, a phenomenon known as imprinting.
CpG islands are short (almost 1 kb) genomic regions present in more than half of the genes in the human genome. They are mainly localized at gene promoters and remain mostly unmethylated in somatic cells. The smaller fraction of CpGs not found clustered in islands, but rather located on gene bodies or distal regulatory regions, are less well understood. A subset of these elements, CpG-poor, low-methylated regions (LMRs; about 30 % methylation) can be found mostly methylated in somatic cells and historically have been associated with actively transcribed genes [17]. Recently, convincing evidence has emerged that these regions mark enhancers and insulators, inducible, cell-type specific elements formed by transcription factor binding to the underlying DNA.
Noncoding RNA
In recent years pioneering genomic studies have shown that as much as 90 % of the mammalian genome is transcribed [18]. While it is still debated as to how many of these transcripts are simply unannotated or real non-protein-coding RNAs (ncRNA), the findings indicate that a substantially larger part of the genome is transcribed than can be accounted for by annotated genes [19, 20]. Indeed, a growing number of ncRNAs are being assigned important regulatory functions, suggesting the existence of a substantial regulatory layer. Considering the number and diversity of ncRNAs, it is beyond the scope of this review to describe all of them and their associated functions. Below, however, we provide a cursory description of each of the major classes and touch briefly upon their potential in rerouting phenotypes.
miRNAs
miRNAs, also known as micro-RNAs, are the best-characterized family of small ncRNAs. Approximately 19–24 nucleotides in length in their mature processed form, they have been shown to regulate hundreds of genes by sequence-specific post-translational gene silencing. miRNAs in metazoans do not need to form a perfect base-pair match to their target site, and thus one miRNA can regulate many genes. It has been estimated that they could regulate 74–92 % of all protein-coding mRNAs in one cell type or another [21]. miRNAs function by recruiting a protein complex, called RISC (RNA-induced silencing complex), to target gene transcripts leading to silencing via degradation of the messenger RNA or by preventing translation. Alterations in specific miR levels and/or in miR machinery have been shown to be a hallmark of processes such as cell growth and proliferation, development, differentiation, organogenesis, metabolism, immunity and multiple diseases including obesity, cancer, cardiovascular disease, and diabetes [22] (for review see Esteller 2011).
piRNAs
piRNAs, or piwi-interacting RNAs (25–30 nt), are the largest class of small ncRNAs. Less conserved, more diverse, and more distinct in their biogenesis than miRNAs, piRNAs maintain genome integrity by silencing transposable elements in gametes of the germline [23, 24]. Importantly, this role, which involves guiding deposition of silencing marks such as DNA methylation, has also been suggested to regulate function in somatic stem cells [25, 26]. A recent study in Aplysia linked piRNAs to the establishment of a stable long-term change in neurons for the persistence of memory [27].
snoRNAs
Moving ever larger, a family of intermediate sized small nucleolar RNAs (snoRNA; 60–300 nt) can be found preferentially located in the nucleolus [28], the ribosome assembly compartment of the cell. snoRNAs comprise part of the snoRNP complex responsible for post-transcriptional modification (methylation and pseudouridylation) of ribosomal RNA [29, 30] and snRNAs [31]. The role of the snoRNA is to guide by sequence homology. These modifications facilitate folding and stability of the RNA molecules [32]. It is worth mentioning though that snoRNAs showing no sequence homology with the above-mentioned targets have been discovered [33, 34]. These so-called orphan snoRNAs are predicted to direct mRNA modification, but involvement in alternative splicing has also been suggested [35].
lncRNAs
Most variable in length, long ncRNAs (lncRNAs; 300 nt to several kb) are a heterogeneous group of RNAs some of which have been implicated in epigenetic regulation of gene expression. They are diverse and the majority are without functional annotation. The most well-known examples of the involvement of lncRNA in epigenetic regulation of gene expression are X-chromosome inactivation (Xist RNA), genomic imprinting (Air, H19 RNA), dosage compensation (Rox1/2 RNA), nuclear organization and compartmentalization, and nuclear–cytoplasmic trafficking [36, 37]. Intriguingly, a subset of lncRNA has also been implicated in the maintenance of pluripotency by serving as a guide or scaffold for chromatin-modifying enzymes at pluripotency genes [38].
Post-translational modifications of histones and nonhistone proteins
The fundamental repeating structural unit of chromatin, the nucleosome, comprises about 147 bp of DNA wrapped around a histone octamer, called a nucleosome. In most cells, the octamer is made up of dimers of the tetrameric H2A, H2B, H3 and H4. Histone H1 binds to the linker DNA region between the nucleosomes and helps stabilize higher order chromatin structure. Protruding away from the globular domain octamer of each nucleosome are relatively unstructured N-terminal histone tails. These tails can be post-translationally modified, thus altering their structural and functional properties. Two obvious examples are the creation of novel motifs for recognition by proteins or enzyme complexes and altered accessibility of DNA for transcription factor binding. Histone modifications come in a wide range of flavours, from the well-studied acetylation, methylation, ubiquitination and phosphorylation, to the more recently uncovered crotonylation, butyrylation, palmitoylation etc. In 2011, Tan et al. [39] identified 67 previously undescribed histone modifications in a single study. It is likely that at least a portion of these novel modifications will have specific enzyme complements for their deposition and removal, implying that we understand only the tip of the iceberg.
Post-translational histone modifications play a crucial role in many biological processes, including organismal development. A number of them are substantially dysregulated in cancer [40]. The best-characterized histone modifications are lysine methylations and lysine acetylations. Albeit oversimplified, acetylation of lysines can be thought of as relaxing the structure of chromatin and is therefore mainly associated with transcriptional activation. Histone methylation can have both silencing and activating effects, depending on the number of methyl groups attached (up to three) and the type of histone that carries the modification. Heterochromatin, for instance, is enriched in H3 lysine 9 di- and trimethylation and poor in acetylation. Euchromatin, typically more gene-rich, shows enrichment in H3 and H4 acetylation and H3K4 methylation. Importantly, a number of histone-modifying enzymes have been shown to play a role in the control of metabolism and metabolic adaptation [41–45]. Studies have similarly linked intracellular energy status to the activity of histone deacetylases (HDACs). In particular, calorie restriction has been shown to activate the sirtuin class of HDACs [46], a NAD+-sensitive enzyme class which exerts broad functions in chromatin organization, development, metabolism and cellular proliferation.
Equally important, metabolism itself, via metabolite–substrate links, directly impinges not only upon acetylation and methylation of histones but also on all protein substrates. Lysine acetylation is a prevalent modification in proteins involved in disparate cellular functions. Several disease-associated proteins have been shown to be regulated by post-translational modifications like the products of the oncosuppressor genes pRb [47] and p53 [48], the master regulator of mitochondrial biogenesis and function PGC-1 [49], and enzymes that catalyse intermediate metabolism [50]. Three independent proteomic analyses have shown that virtually every enzyme in glycolysis, gluconeogenesis, the tricarboxylic acid (TCA) cycle, the urea cycle, fatty acid metabolism, and glycogen metabolism is acetylated in human liver tissue [50–52]. Interestingly, comparison of these three acetylome datasets indicates that there is extremely high similarity in the spectrum of acetylated proteins between mouse and human liver, but there is a big variation between liver and, for example, leukaemia cells [50]. Intriguingly, Zhao et al. also showed in this study [50] that the concentration of metabolic fuels, such as glucose, amino acids and fatty acids, influences the acetylation status of metabolic enzymes.
Epigenomics: from techniques to technologies
The three biological paradigms above, DNA modification, ncRNA, and histone modification, combine functionally to impose a vast regulatory complexity in the form of chromatin dynamics. They create site-specific docking for enzyme complexes and modify accessibility to DNA regulatory elements and open reading frames. The last decade has seen the rapid emergence of key technologies that are redefining molecular genetics as a field, in many cases posing as many new challenges as the advantages they bring. Unfortunately, because of explosive rapidity in the emergence of these technologies and the mind-boggling amounts of data and insights spewed out of the technology platforms, the assumptions and technical limitations upon which the final conclusions are based are too often discounted or, simply put, too complicated for the newcomer. Below, we summarize the most common experimental approaches based on next- or second-generation sequencing (NGS) from their biochemical basis to the technologies that are boosting their potential.
Three techniques: harnessing mother nature
Epigenomics comprises the interrogation of protein–DNA–RNA interactions on a genome-wide scale to gain insight into the chromatin state. Our ability to read and analyse chromatin patterns gigabase-genomes at a time relies wholly on a few key biochemical techniques developed over about the last 100 years. Together they provide the stability, sensitivity, and specificity required for modern genomics.
PCR, reverse transcription, and restriction digests
One foundation of contemporary genetics is polymerase chain reaction (PCR). Developed in 1983 by Mullis it comprises the successive duplication of the desired DNA fragments a million-fold. Together with restriction digestion (the use of unique bacterial enzymes to sequence specifically cut DNA) and reverse transcription (the ability to reverse copy RNA back into a DNA template), PCR has revolutionized our ability to detect, read, modify and manipulate even the smallest amounts of genetic material. Together they set the stage for modern molecular genetics, including the vast potential of the current NGS boom.
Immunoprecipitation and pull-downs
The techniques mentioned above focus on the nucleic acids. Chromatin though is composed of DNA and a wide array of protein constituents either directly or indirectly bound to or forming chromatin itself. In order to achieve the necessary specificity to investigate single protein species we have to go back to the 1890s when Buchner and Ehrlich, two German scientists, discovered a substance in cell-free blood serum which was able to kill bacteria. Ehrlich was the first one to use the word “Antikoerper” in one of his publications in 1891. It took almost another 40 years until it was shown by Heidelberger that antibodies are able to precipitate antigens, a study that would pave the way for protein biochemistry. This discovery is the foundation of all immunoprecipitation methods. In 1984 Gilmour and Lis [53] expanded upon this idea and used UV-light to crosslink proteins to DNA. This pioneering work led to the development of chromatin immunoprecipitation (ChIP) [54] and the use of immunoprecipitation to gather protein-bound DNA target sequences, and thus laid the foundation for the current chromatin mapping era.
Simple chemistry
The last set of techniques are little more than chemical entities. The first is sodium bisulphite, which was recognized by Frommer et al. in 1992 [55] for its utility in detecting DNA methylation. Treatment of DNA with bisulphite converts cytosines into uracil, but it does not affect methylated cytosines. Thus the changes in the DNA sequence introduced by bisulphite treatment reflect the methylation status of the DNA. Two decades later, this stable reproducible technique serves as the basis of gold-standard measures of DNA methylation. The second is simple fixation chemistry. The final piece of the puzzle needed to produce reproducible reliable results in large-scale epigenomics was stability. Borrowing from the knowledge of histochemistry and biochemistry, multiple fixatives including the commonly used formalin and paraformaldehyde added the key element of consistent stabilization necessary for genome-wide upscaling.
Together this small handful of everyday biochemistry and molecular biology techniques represent the real foundation of the epigenomic era.
Three technologies: next generation sequencing
Sequencing
With the above techniques at hand, one simply needs the capacity to read hundreds of millions of nucleotides of DNA sequence per day, and we have “epigenomics”. Sequencing technology as we know it was first developed by Sanger et al. in 1975 [56, 57]. This first process, called the chain-termination method, forced labelled, terminating nucleotides into a PCR reaction. The result, a series of DNA fragments of increasing length, terminated at every possible position, and were readily detectable according to the incorporated terminating nucleotide. Four reactions were run, each with a different terminating nucleotide. When run and detected in parallel, the four reactions combined to reveal the aligned underlying sequence, and history was made. Since the inception of the technique using radioactive terminating nucleotides and long polyacrylamide gels, it has undergone several rounds of improvement, increasing both efficiency and reducing cost. As a testament to the invention, however, the current setup now based on fluorescent nucleotides and polymer-filled capillaries, still remains essentially the same. Indeed, Sanger sequencing formed the technical basis of the 1990 initiation of Human Genome Project, a projected 15 years, 3 billion dollar expedition. With unpredicted genomics and computing advances the project eventually outperformed expectations, computing the first draft of the human genome 5 years ahead of schedule.
Next generation sequencing
Perhaps surprisingly, the biggest challenge to increasing the practical productivity of Sanger-type sequencing was spatial, a challenge that led to the pursuit of micro-sizing sequencing reactions and ultimately implementation of a solid support towards this end. Coupled with improvements in sample preparation, sequencing and imaging this led to the development of NGS, a technology platform capable of concomitant optical analyses of millions of clonal sequencing reactions occurring as “spots” on a glass slide. The advent of this technology, almost a decade old now, led to much higher throughput and dramatically reduced per-base sequencing costs. The first commercial NGS device was released in 2005. To give a quantitative value to the sum of the technological leap, NGS sequencing of the human genome on new platforms could have been accomplished in days to weeks with a cost in the tens of thousands.
The third/fourth generation
In part due to the immense commercial potential of such platforms (Illumina’s estimated value about 6 billion USD in 2012), the “next” NGS was sure to be a short time coming. Given the number of approaches being pursued, as well as the pace and proprietary nature of development, there is actually no consensus on what exact technologies represent third-generation sequencing. Some consider the sequencing of individual DNA molecules (vs. groups of PCR-amplified clones) sufficient grounds [58] and indeed, numerous efforts are underway in this regard. Others, however, feel this is simply optimization of the current mode and falls short of a major advance in capability [59, 60]. A clear step forward will certainly be the use of nanopore technology to directly sequence individual raw biological DNA molecules themselves. In theory, this would “bypass” the need for costly and highly variable amplification procedures, thus alone eliminating one of the greatest bioinformatics hurdles of data interpretation from NGS platforms, that is the filtering and normalization of data for amplification artefacts. For a concise review on the latest technologies in DNA sequencing see Schadt et al. [60].
Epigenomics: back to biology
Like many rapid biotechnological advances, a major driving force for the epigenomics movement has been the posing of pertinent biological questions. In general these can be placed into five categories, some simply building upon already well-developed areas of “omic” investigation (e.g. transcriptomics), and others tapping into the unknown (e.g. the omics of chromatin conformation and chromatin state). In short, the driving force has been pursuit of insight into (1) 3-D cis/trans interactions such as understanding distal regulatory element function, (2) the number, nature and distribution of chromatin states, (3) the distribution and targeting principles of chromatin-associated proteins, including transcription factors and chromatin readers, writers and erasers, (4) the frequency and identity of RNA–protein interactions, and (5) RNA abundance and transcriptional control, both coding and noncoding. In addition to covering the majority of genomics as a field, these questions impact all of medical and cellular biology and thus underscore their potential for medicine. Below, in an effort to highlight their potential, we group and summarize the dozens of NGS techniques that have popped up over the last 5–10 years and highlight some of the contributions they have already made to the field.
3-D organization, looping, and macroregulatory domains
It has been known for decades that many cell types can be identified under the microscope solely on the basis of the gross ultrastructure of their nuclei. This is a testament to the highly defined chromatin organization that occurs during cell-type specification and differentiation. Indeed, active and inactive chromatins localize differently within the nucleus and establish much of this reproducible variation [61]. How this complex structure–function dynamic is established and maintained has become a recent focus of attention.
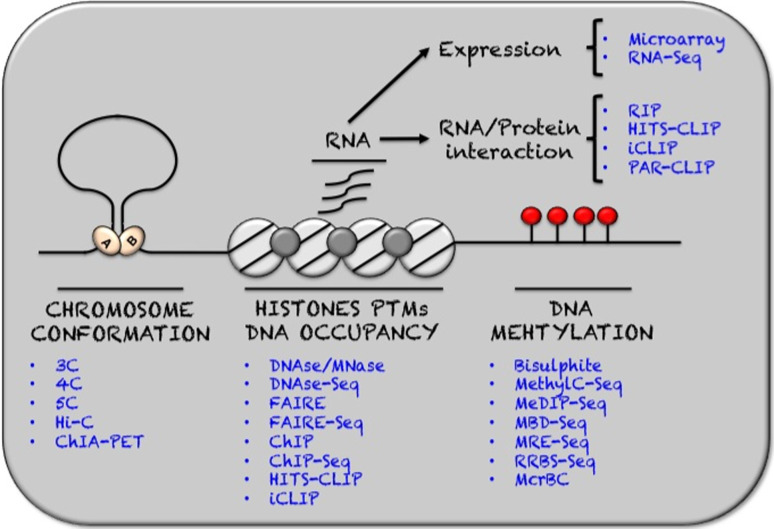
Both histone post-translational modifications and ncRNAs affect genome conformation and its spatial organization in the nucleus. A myriad of enhancers, promoters and insulators together with transcriptional and splicing machinery assemble large, defined, 3-D mega-complexes critical to proper gene expression. To evaluate the spatial organization of chromatin, chromosome conformation capture (3C) [62] arose, a chemical fixation-dependent interrogation of inter- and intrachromosomal proximity. DNA fragments covalently bound through fixation are captured, amplified, sequenced and aligned to the genome, yielding intra- and interchromosomal chromatin interaction maps. Since its inception, the single-gene/single-target focused 3C technique has undergone an evolutionary process in its own right. First, to 4C or circular chromosome conformation capture, broadening the output to all interacting regions for a specific DNA locus [63], then to 5C (carbon-copy chromosome conformation capture) providing a matrix of interaction frequencies for many pairs of sites [64], and finally to Hi-C, which allows concomitant examination of all DNA–DNA interactions in the genome at any one time [65]. Along similar lines, alterations in DNA purification principles and increases in signal to noise ratio achieved by coupling 3C and chromatin immunoprecipitation (ChIA-PET) have allowed genome-wide interaction analysis for any given protein of interest [66].
Together these developments have taught us the basic principles of the genomic “interactome”, how regulatory modules are physically bounded and defined, and provided a template for understanding coregulation of gene expression, cell-type-specific enhancer function and locus evolution.
Chromatin state, binding profiles/distribution
Perhaps the biggest impact of epigenomics has been the large-scale mapping of epigenetic marks and DNA-binding proteins. Because of their multitude in number and function, the effort has led to monumental additions to our understanding of gene regulation. Biologically, these techniques can be grouped very loosely into those examining DNA methylation, those examining DNA–protein interactions, and those interrogating enzyme sensitivity. Technically they all rely on either immunoprecipitation of protein-associated DNA, methylation-specific restriction/conversion/PCR, or specific enzyme digestion of chromatin. Since sequencing technologies have essentially replaced hybridization to tiled microarrays we only mention sequencing below. Importantly, though, citations have been given to those who made first use of the technique whether the readout was based on hybridization or sequencing.
DNA methylation
The most definitive tools to assess DNA methylation levels have certainly been based upon sodium bisulphite treatment of DNA, a process that converts unmethylated cytosines into uracil. By using different primers recognizing either cytosine or uracil one can distinguish between unmethylated and methylated DNA sequences in a targeted fashion. Coupling to random amplification and sequencing (bisulphite sequencing) subsequently gave insight into the DNA methylome as a whole [67]. Until very recently, the price of such studies was prohibitive and an avalanche of analytical tools emerged as an interim solution. These include but are not limited to the following techniques. Methylated DNA immunoprecipitation sequencing (MeDIP-seq) that, as the name implies, involves immunoprecipitation of fragmented DNA with an antibody selective for methyl cytosine [68, 69]. Similarly methyl-CpG binding domain protein sequencing (MBD-seq) pulls down methylated DNA using biotinylated methyl-CpG binding domain protein rather than an antibody [70]. Methylation-sensitive restriction enzyme sequencing (MRE-seq) [71] uses restriction enzymes, which cut only unmethylated DNA, and the resulting short fragments are analysed by sequencing. A method based on the original bisulphite sequencing is reduced representation bisulphite sequencing (RRBS-seq) [72], which is a combination of restriction enzyme digestion with bisulphite treatment of the short fragments, which increases the efficiency of the bisulphite reaction, leading to less false-negatives in the analysis. An alternative enzyme-based method to map the methylome uses McrBC, a restriction enzyme [73] which cleaves DNA containing methylcytosine but not unmethylated DNA. Each of these tools has its own independent set of pros and cons that have been extensively reviewed.
DNA–protein interactions
The field-defining ENCODE project relied heavily on mapping binding profiles of dozens of transcription factors, histone marks and other chromatin-associated proteins by ChIP sequencing (ChIP-seq). Originally used in a targeted approach coupled to quantitative real-time PCR detection [54], ChIP uses protein-specific antibodies to pull down DNA associated with the protein of interest. Whereas qPCR is still used as a gold standard for verification in large-scale studies, ChIP has been used extensively with microarrays (ChIP-chip) [74] and more recently sequencing [75] as the readout. The result provides a genome-wide picture of binding sites. Collectively, this tool has revealed a marking system for identification of enhancer and promoters genome-wide, has identified “all” the cis-regulatory sequences of the mouse genome [76], and has shown the genome to be organized into at least about 15 distinct chromatin states [77, 78]. The key caveats of ChIP-seq as an approach are twofold. First, as an antibody-based system, the results are only as good as the antibody used. It is essentially impossible to rule out the possibility that an antibody crossreacts with unknown factors. Second, the approach provides a snapshot, and therefore highly dynamic processes can easily be missed, and the user receives no information about the kinetics of change over time. While more cumbersome to establish, and based on chimeric proteins rather than endogenous ones, some techniques such as DamID have evolved to circumvent some of these issues. Because of their own substantial drawbacks such techniques have failed to gain general acceptance however. Perhaps most importantly, due to its reliance on expression of chimeric protein, the DamID tool cannot be used to examine raw patient material.
DNA accessibility
Considered as readouts or the “sum consequence” of the many DNA–protein interactions targeted by ChIP-seq, several technologies have been developed to assess genome accessibility. The parameter is used to define regions that are active or open, and thus presumably important for the cell type in question. Most promoters, transcriptional start sites, and regulatory enhancers, for instance, are highly “accessible”, and DNA accessibility can therefore be used in an unbiased manner to predict or support evidence for regulatory region function. Three techniques are primarily used, two based on restriction enzymes and one on simple fixation chemistry.
DNaseI and MNase (micrococcal nuclease) hypersensitivity analyses rely on exposure of chromatin to endonucleases that cut naked DNA more or less at random. The methods take advantage of the fact that in condensed chromatin DNA is less accessible to the enzymes. The right enzyme titration reveals DNA cleavage only where no nucleosomes are bound. Sequencing of the short DNA fragments produced, DNase-seq [79] and MNase-seq [80], provides a detailed map of accessibility. Similarly, FAIRE-seq (formaldehyde-assisted isolation of regulatory elements) [81] exploits chemical fixation and sedimentation to very simply purify open DNA that cannot be crosslinked to a nucleosome, i.e. the most accessible part of chromatin, and has been shown to correlate highly with DNaseI hypersensitive sites.
Unravelling the world of RNA
Perhaps the most obvious and widespread use of NGS technology has been a switch from microarray-based quantification of the transcriptome, to RNA-seq. RT-PCR-coupled sequencing allows unprecedented quantitation of coding and noncoding transcriptomes within days. In addition to a much greater quantitative dynamic range, the greatest advantage of RNA-seq is its unbiased nature. Whereas cDNA microarrays only interrogate specific probes placed on the array, RNA-seq more or less reads whatever is there. Of course, preparation biases do exist for RNA-seq. One of the most recent findings that could not have been made using hybridization technologies has been the identification of chimeric proteins and their transcripts [82].
Transcriptomics aside, NGS technology has been integral to the emergence of a series of new regulatory RNA paradigms. To help understand these processes, ChIP techniques have been developed to investigate RNA–protein interactions. RIP (RNA immunoprecipitation) was developed by Niranjanakumari et al. in 2002 [83], and involves performing a pull down of an RNA binding protein and its associated RNAs. Using the technique, the user can identify many of the RNAs interacting with a given protein of interest. Suffering from low resolution and the ability to only examine very stable complexes, RIP has now been largely replaced by techniques such as CLIP (crosslinking and immunoprecipitation) [84]. CLIP uses in vivo UV crosslinking to covalently stabilize RNA–protein interactions, allowing more stringent purification procedures and resolutions up to 30 nucleotides. Coupled with high-throughput sequencing, this method is called (HITS-CLIP or CLIP-seq) [85, 86]. Two recent modifications of the CLIP protocol led to an advance towards single nucleotide resolution. One of these methods is called PAR-CLIP (photoactivatable ribonucleoside-enhanced CLIP) and uses a photoactivatable nucleotide, which leads to a base transition during reverse transcription at the crosslinked nucleotide [87]. Similarly, iCLIP (Individual nucleotide resolution CLIP) [88] exploits the apparent limitation of the CLIP methods that the vast majority of cDNAs prematurely truncate immediately before the crosslinked nucleotide (Fig. 1).
Fig. 1.
Representative collection of the most widely used epigenomic techniques
Chromatin biology and pathogenesis
Chromatin to disease to chromatin
Research indicates shared roles for the genome and epigenome in the pathogenesis of complex diseases. Phenotypic variability associated with diseases such as cancer, obesity and diabetes, cannot be explained by genetic variation alone. More than a decade of GWAS has uncovered more than 800 single nucleotide polymorphisms (SNPs) associated with disease predisposition, development and progression. Few of these have, however, been shown to play a causative role. Given the pivotal role of the chromatin state in phenotypic plasticity and presumably disease susceptibility, there is now increasing interest in exploring nongenetic variation on a genome-wide scale, representing a shift from genomic to epigenomic research [89–91].
The epigenome represents a platform for stably translating gene–environment interactions into phenotype. Environmental signals are transferred to the epigenome directly or via signalling cascades and metabolic circuitry, and stabilized through modifications of both histones and DNA and potentially in stable alterations in ncRNAs. One of the first examples of a definitive disease-relevant gene–environment interaction came from studies on the Agouti mouse model [92]. This mouse model is characterized by a continuum in the coat colour phenotype, which ranges from completely yellow to degrees of yellow/agouti mottling to completely agouti. The extent of yellow coat colour correlates with adiposity, with yellow mice being obese and completely agouti mice lean. The difference in coat colour, and in body weight, reflect the methylation state in a retrotransposon (the IAP element) inserted in the promoter of the Agouti locus—with the unmethylated IAP giving rise to yellow/obese animals as a result of ectopic Agouti expression. This model clearly shows that a simple external intervention, for example a diet poor in methyl donors, can drive phenotype in isogenic animals through a defined epigenetic mechanism.
Following some of these proof-of-principle studies, maternal care and multiple aspects of maternal environment have been shown to alter DNA methylation patterns yielding long-term phenotypic consequence [93]. Both mouse [94] and human studies [95] have shown that maternal nutrition during gestation can leave epigenomic footprints in the form of DNA methylation causing obesity in offspring. Some of these effects have more recently been shown to cross the generational divide [96–98]. Evidence has also been accumulating for ageing-associated changes in CpG island methylation. One study [99] showed that although monozygotic twins exhibit far less variation in CpG methylation and histone acetylation in early life and the epigenome appears to significantly diverge with time.
DNA methylation aside, chromatin-modifying enzymes (such as histone methyltransferases/histone demethylases or histone acetyltransferases/HDACs) are sensitive to environmental perturbations. Their activity relies on the availability of small metabolites (such as AcCoA, SAM, NAD, FAD, and KG), generated by primary metabolism (glycolysis, Krebs cycle and mitochondrial oxidative phosphorylation). Evidence has emerged that alteration in the activity of chromatin-modifying enzymes is associated with substantially impaired metabolic phenotypes, as it is for the SET domain-containing methyltransferase SET7 [100, 101], the FAD-dependent demethylase LSD-1 [100] and the KG-dependent Jumonji domain-containing demethylase Jhdm2a/KDM3a [102]. Besides histone methyltransferases and histone demethylases, the role of histone acetyltransferases and HDACs is also well established and already extensively reviewed [41, 42, 46]. These studies, and others, highlight a direct link between metabolism and epigenetics. They underscore how epigenomics will move forward our understanding of metabolic disease aetiology.
The recent technological explosion has already had an impact on how we view pathogenesis. The use of ChIP-Seq to monitor chromatin dynamics during human adipogenesis [103], for instance, has provided a list of novel transcriptional regulators relevant for obesity. The genome-wide profiling of transcription factor 7 like 2 (TCF7L2) binding [104] in hepatocytes provided novel insight into the regulation of hepatic glucose production. TCF7L2 is the most significantly scoring type-2 diabetes SNP to emerge from GWAS [105, 106]. Also, liver-specific genome-wide circadian distribution of HDAC3 [107], whose deletion (as well as the deletion of the transcription factor Rev-erb) in mouse liver causes hepatic steatosis, has stimulated substantial interest in the circadian variation in epigenome and transcriptome regulation and function.
Epigenomics and epigenome-wide association studies (EWAS)
One future direction for epigenomic research is population-based studies, and the determination of phenotypic variation attributable to interindividual epigenomic variation. Most attempts to do so have so far failed because of either inadequate genome coverage or inadequate sample size. One approach to solving this problem is by large-scale, systematic epigenomic equivalents of GWAS—epigenome-wide association studies (EWAS). Technology has just become available that is comparable in resolution and throughput to the highly successful GWAS chips that allow genotyping of around 500,000 (500K) SNPs. One major challenge in EWAS, however, relative to GWAS (cost–benefit ratio aside) is that the signals obtained, beyond being simply correlative, may actually be consequences of the condition. Although any human disease epiallele association represents advanced knowledge with potentially diagnostic applicability, identifying causal potentially targetable variants must be the major goal.
To date, early EWASs of DNA methylation variation in complex disease have mainly been focused on cancer, where both gain and loss of DNA methylation at CpG islands and satellite DNA have been associated with tumour development [108]. For nonmalignant, common complex diseases such as diabetes and autoimmunity, the epigenetic component is only just beginning to be investigated. Some observations, such as MZ twin discordance and rising incidence of some complex diseases in migrant populations, e.g. type-1 diabetes [109], support the involvement of epigenetics. Given the predominant confounding role of the environment in generating epigenetic variation and phenotypic plasticity among populations and individuals of the same population, the most critical step in approaching EWAS is study design. A myriad of background genetic and life-history parameters will need to be critically catalogued, and tissue specificity purity and consistency will be of paramount importance as profound cell-type specificity exists for virtually all epigenetic marks examined to date. These problems have been systematically addressed in a recent review [110].
One strategy pursued by the same authors is to perform retrospective follow-up using sample banks [110]. Guthrie Cards, a biobanking resource created from blood samples from newborns and used since the late 1960s for life-threatening infantile metabolic disease screening, represents an immediate, well-preserved source of DNA for mapping with decades of follow-up data already at hand. Since blood cells are found in highly defined ratios, some of the critical factors regarding cell type etc. are already addressed. Designing follow-up studies, starting from selected populations going back to their newborn states will help researchers distinguish epigenomic variables introduced during life (and potentially important for age-related diseases) from those present at birth, defining both causal and disease-unrelated epigenomic fingerprints.
International human epigenome consortium
In pursuit of a more comprehensive understanding of our epigenomes, global efforts are underway. The International Human Epigenome Consortium (IHEC) has been established with the key goals of producing and analysing at least 1,000 reference maps of human epigenomes across multiple cellular states. These aim to catalyse basic biology and focus primarily on disease-relevant tissues and models and include multiple metabolic and inflammatory systems, human and murine, and critically, have been designed with public resource creation in mind (http://www.ihec-epigenomes.org/ and http://www.roadmapepigenomics.org). An immediate product of the effort will be generation of large sets of highly comparable datasets to define internal epigenomic variability of “healthy” people.
Back to the bedside
Disease epigenetics is moving faster than ever expected a decade ago, with achievements as important as the approval by the FDA of the first epigenetic-based drug in 2004. Three more drugs were approved between 2004 and 2009 and many others are in different developmental stages [111]. Most have been identified as potential therapeutic agents for cancers and belong to methyltransferase and deacetylase inhibitor classes. Further, there has been an explosive growth in commercial diagnostic solutions based, for example, on DNA methylation patterns. Until now, both diagnostics and therapeutics derived from the epigenomic research have been focused on malignancies and some inflammatory diseases (such as arthritis). The expectation though, is that, as far as research goes, this field of application will expand and involve other nonmendelian diseases, including metabolic diseases in which the stability of epigenetic signatures will be their power.
The next frontier will certainly be merging epigenomics with deep sequencing of patient DNA and inevitably much more personalized medicine. The dramatic reduction in both the cost per experiment and the time-to-results for NGS-based sequencing holds the promise of translating epigenomic research into clinical practice in the next few years and bringing personalized medicine one step closer to reality.
References
- 1.Mattick JS. A new paradigm for developmental biology. J Exp Biol. 2007;210(Pt 9):1526–1547. doi: 10.1242/jeb.005017. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447(7143):433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 3.Hewagama A, Richardson B. The genetics and epigenetics of autoimmune diseases. J Autoimmun. 2009;33(1):3–11. doi: 10.1016/j.jaut.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462(7275):868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling C, Groop L. Epigenetics: a molecular link between environmental factors and type 2 diabetes. Diabetes. 2009;58(12):2718–2725. doi: 10.2337/db09-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006;15(Spec No 2):R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 7.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462(7271):315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA. 2000;97(10):5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziller MJ, Müller F, Liao J, Zhang Y, Gu H, Bock C, et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet. 2011;7(12):e1002389. doi: 10.1371/journal.pgen.1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires aid-dependent DNA demethylation. Nature. 2010;463(7284):1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470(7334):419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 12.Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466(7310):1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366(6453):362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 14.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 15.Razin A, Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984;782(4):331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- 16.Bird AP. DNA methylation versus gene expression. J Embryol Exp Morphol. 1984;83 Suppl:31–40. [PubMed] [Google Scholar]
- 17.Wolf SF, Jolly DJ, Lunnen KD, Friedmann T, Migeon BR. Methylation of the hypoxanthine phosphoribosyltransferase locus on the human X chromosome: implications for X-chromosome inactivation. Proc Natl Acad Sci USA. 1984;81(9):2806–2810. doi: 10.1073/pnas.81.9.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol. 2010;8(5):e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, et al. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 22.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. Cell Cycle. 2005;4(9):1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 23.Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318(5851):761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- 24.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322(5906):1387–1392. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.King FJ, Szakmary A, Cox DN, Lin H. Yb modulates the divisions of both germline and somatic stem cells through piwi- and hh-mediated mechanisms in the Drosophila ovary. Mol Cell. 2001;7(3):497–508. doi: 10.1016/S1097-2765(01)00197-6. [DOI] [PubMed] [Google Scholar]
- 26.Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood. 2001;97(2):426–434. doi: 10.1182/blood.V97.2.426. [DOI] [PubMed] [Google Scholar]
- 27.Rajasethupathy P, Antonov I, Sheridan R, Frey S, Sander C, Tuschl T, Kandel ER. A role for neuronal piRNAs in the epigenetic control of memory-related synaptic plasticity. Cell. 2012;149(3):693–707. doi: 10.1016/j.cell.2012.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nielsen H, Orum H, Engberg J. A novel class of nucleolar RNAs from tetrahymena. FEBS Lett. 1992;307(3):337–342. doi: 10.1016/0014-5793(92)80708-O. [DOI] [PubMed] [Google Scholar]
- 29.Kiss-László Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell. 1996;85(7):1077–1088. doi: 10.1016/S0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 30.Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell. 1997;89(4):565–573. doi: 10.1016/S0092-8674(00)80238-X. [DOI] [PubMed] [Google Scholar]
- 31.Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell. 1998;2(5):629–638. doi: 10.1016/S1097-2765(00)80161-6. [DOI] [PubMed] [Google Scholar]
- 32.King TH, Liu B, McCully RR, Fournier MJ. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell. 2003;11(2):425–435. doi: 10.1016/S1097-2765(03)00040-6. [DOI] [PubMed] [Google Scholar]
- 33.Li SG, Zhou H, Luo YP, Zhang P, Qu LH. Identification and functional analysis of 20 box H/ACA small nucleolar RNAs (snoRNAs) from Schizosaccharomyces pombe. J Biol Chem. 2005;280(16):16446–16455. doi: 10.1074/jbc.M500326200. [DOI] [PubMed] [Google Scholar]
- 34.Yang JH, Zhang XC, Huang ZP, Zhou H, Huang MB, Zhang S, et al. Snoseeker: an advanced computational package for screening of guide and orphan snoRNA genes in the human genome. Nucleic Acids Res. 2006;34(18):5112–5123. doi: 10.1093/nar/gkl672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311(5758):230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 36.Nagano T, Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145(2):178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 37.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22(1):50–55. doi: 10.1016/j.gde.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17(6):292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 44.Smith CL, O’Malley BW. Coregulator function: a key to understanding tissue specificity of selective receptor modulators. Endocr Rev. 2004;25(1):45–71. doi: 10.1210/er.2003-0023. [DOI] [PubMed] [Google Scholar]
- 45.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119(2):157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 46.Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes Dev. 2009;23(16):1849–1869. doi: 10.1101/gad.1807009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan HM, Krstic-Demonacos M, Smith L, Demonacos C, La Thangue NB. Acetylation control of the retinoblastoma tumour-suppressor protein. Nat Cell Biol. 2001;3(7):667–674. doi: 10.1038/35083062. [DOI] [PubMed] [Google Scholar]
- 48.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90(4):595–606. doi: 10.1016/S0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 49.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 50.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327(5968):1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 52.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23(4):607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 53.Gilmour DS, Lis JT. Detecting protein-DNA interactions in vivo: distribution of RNA polymerase on specific bacterial genes. Proc Natl Acad Sci USA. 1984;81(14):4275–4279. doi: 10.1073/pnas.81.14.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solomon MJ, Larsen PL, Varshavsky A. Mapping protein-DNA interactions in vivo with formaldehyde: evidence that histone H4 is retained on a highly transcribed gene. Cell. 1988;53(6):937–947. doi: 10.1016/S0092-8674(88)90469-2. [DOI] [PubMed] [Google Scholar]
- 55.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanger F, Coulson AR. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J Mol Biol. 1975;94(3):441–448. doi: 10.1016/0022-2836(75)90213-2. [DOI] [PubMed] [Google Scholar]
- 57.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Munroe DJ, Harris TJ. Third-generation sequencing fireworks at Marco Island. Nat Biotech. 2010;28(5):426–428. doi: 10.1038/nbt0510-426. [DOI] [PubMed] [Google Scholar]
- 59.Metzker ML. Sequencing technologies – the next generation. Nat Rev Genet. 2010;11(1):31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 60.Schadt EE, Turner S, Kasarskis A. A window into third-generation sequencing. Hum Mol Genet. 2010;19(R2):R227–R240. doi: 10.1093/hmg/ddq416. [DOI] [PubMed] [Google Scholar]
- 61.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struct Mol Biol. 2007;14(11):1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 62.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 63.Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38(11):1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 64.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fullwood MJ, Ruan Y. Chip-based methods for the identification of long-range chromatin interactions. J Cell Biochem. 2009;107(1):30–39. doi: 10.1002/jcb.22116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Herman JG, Graff JR, Myöhänen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, et al. A bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26(7):779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 70.Serre D, Lee BH, Ting AH. MBD-isolated Genome Sequencing provides a high-throughput and comprehensive survey of DNA methylation in the human genome. Nucleic Acids Res. 2010;38(2):391–399. doi: 10.1093/nar/gkp992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 2010;28(10):1097–1105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33(18):5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sutherland E, Coe L, Raleigh EA. McrBC: a multisubunit GTP-dependent restriction endonuclease. J Mol Biol. 1992;225(2):327–348. doi: 10.1016/0022-2836(92)90925-A. [DOI] [PubMed] [Google Scholar]
- 74.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290(5500):2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 75.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4(8):651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 76.Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488(7409):116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28(8):817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Crawford GE, Holt IE, Whittle J, Webb BD, Tai D, Davis S, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16(1):123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20(1):90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17(6):877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Djebali S, Lagarde J, Kapranov P, Lacroix V, Borel C, Mudge JM, et al. Evidence for transcript networks composed of chimeric RNAs in human cells. PLoS One. 2012;7(1):e28213. doi: 10.1371/journal.pone.0028213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26(2):182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- 84.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies nova-regulated RNA networks in the brain. Science. 2003;302(5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 85.Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1(2):266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16(2):130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, et al. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Feinberg AP, Irizarry RA. Evolution in health and medicine Sackler colloquium: stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proc Natl Acad Sci USA. 2010;107(Suppl 1):1757–1764. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kulis M, Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 91.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465(7299):721–727. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 92.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23(3):314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 93.Weaver IC, Cervoni N, Champagne FA, D’Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 94.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8(4):253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18(21):4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467(7318):963–966. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 97.Chong S, Vickaryous N, Ashe A, Zamudio N, Youngson N, Hemley S, et al. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet. 2007;39(5):614–622. doi: 10.1038/ng2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, et al. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2010;143(7):1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102(30):10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brasacchio D, Okabe J, Tikellis C, Balcerczyk A, George P, Baker EK, et al. Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes. 2009;58(5):1229–1236. doi: 10.2337/db08-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.El-Osta A, Brasacchio D, Yao D, Pocai A, Jones PL, Roeder RG, et al. Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med. 2008;205(10):2409–2417. doi: 10.1084/jem.20081188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458(7239):757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mikkelsen TS, Xu Z, Zhang X, Wang L, Gimble JM, Lander ES, Rosen ED. Comparative epigenomic analysis of murine and human adipogenesis. Cell. 2010;143(1):156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Norton L, Fourcaudot M, Abdul-Ghani MA, Winnier D, Mehta FF, Jenkinson CP, Defronzo RA. Chromatin occupancy of transcription factor 7-like 2 (TCF7L2) and its role in hepatic glucose metabolism. Diabetologia. 2011;54(12):3132–3142. doi: 10.1007/s00125-011-2289-z. [DOI] [PubMed] [Google Scholar]
- 105.Duggirala R, Blangero J, Almasy L, Dyer TD, Williams KL, Leach RJ, et al. Linkage of type 2 diabetes mellitus and of age at onset to a genetic location on chromosome 10q in Mexican Americans. Am J Hum Genet. 1999;64(4):1127–1140. doi: 10.1086/302316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 107.Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, et al. A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science. 2011;331(6022):1315–1319. doi: 10.1126/science.1198125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lechner M, Boshoff C, Beck S. Cancer epigenome. Adv Genet. 2010;70:247–276. doi: 10.1016/B978-0-12-380866-0.60009-5. [DOI] [PubMed] [Google Scholar]
- 109.Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347(12):911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 110.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12(8):529–541. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mack GS. To selectivity and beyond. Nat Biotechnol. 2010;28(12):1259–1266. doi: 10.1038/nbt.1724. [DOI] [PubMed] [Google Scholar]