Abstract
Life-threatening invasive fungal infections are becoming increasingly common, at least in part due to the prevalence of medical interventions resulting in immunosuppression. Opportunistic fungal pathogens of humans exploit hosts that are immunocompromised, whether by immunosuppression or genetic predisposition, with infections originating from either commensal or environmental sources. Fungal pathogens are armed with an arsenal of traits that promote pathogenesis, including the ability to survive host physiological conditions and to switch between different morphological states. Despite the profound impact of fungal pathogens on human health worldwide, diagnostic strategies remain crude and treatment options are limited, with resistance to antifungal drugs on the rise. This review will focus on the global burden of fungal infections, the reservoirs of these pathogens, the traits of opportunistic yeast that lead to pathogenesis, host genetic susceptibilities, and the challenges that must be overcome to combat antifungal drug resistance and improve clinical outcome.
Keywords: Opportunistic, Fungi, Yeast, Pathogen, Candida, Cryptococcus, Histoplasma, Pneumocystis
Introduction
The emergence of fungi occurred approximately 1.6 billion years ago [1, 2]. With an estimated 1.5 million species occupying a range of environments [3], fungal species are extraordinarily diverse. Fungi are a major cause of disease in insects, amphibians, plants and even other fungi, with incidences of infection reaching unprecedented levels in recent years. Strikingly, millions of acres of the world’s forests have become victim to fungal infections. The blue stain fungus Grosmannia clavigera has devastated North American pine forests [4], and elm trees have been in significant decline since the twentieth century due to Dutch elm blight. Fungi also pose a major threat to food supplies, being responsible for the destruction of wheat and barley crops by wheat leaf rust and rice by rice blast fungus. Additionally, fungi pose a significant threat to animals, crippling bat and amphibian species worldwide [5].
Of the 1.5 million fungal species, around 300 are reported to be pathogenic in humans, and only a minority of these are common human pathogens [6]. Many fungi are commensal, forming part of our natural microbiota. Indeed, a recent study by Findley et al. [7] illustrated the diversity of fungal species on three different foot sites, and there is a growing appreciation that fungi have an important role in defining commensal microbial communities [8]. This commensal role allows fungi to infect humans in multiple ways. Ranging from cutaneous infections affecting 29 million North Americans each year [9], to superficial skin infections, to more than 2 million invasive fungal infections per year worldwide [10], commensal fungi have important roles in disease, being capable of switching to opportunistic pathogens. It is the opportunistic nature of fungal pathogens that triggered a stark rise in fungal infections in the late twentieth century, primarily in hosts with impaired immunity due to medical interventions such as chemotherapy for cancer, organ transplantation, or infection with HIV [11]. Overall, fungi are the cause of billions of infections worldwide, killing over 1.5 million people annually [10, 12]. Species of Aspergillus, Candida, Cryptococcus and Pneumocystis are at the forefront of fungal infections, accounting for approximately 90 % of human mortality cases [10]. Poor diagnostic tools and antifungal drug resistance accounts for a 50 % or higher mortality rate in patients with systemic fungal infections [11].
It has been postulated that global warming is in part responsible for the drastic increase in fungal infections, leading to the devastating loss of forests and crops worldwide. Remarkably, the common denominator of all human pathogenic fungi is the ability to grow at host temperatures [13, 14]. The steady rise in temperature upon climate change will selectively enable adaptation of fungi, broadening the array of species able to survive at host temperature [15]. Indeed, clinical isolates of the generally benign yeast Saccharomyces cerevisiae that exhibit enhanced capacity to grow at higher temperature (41 °C) compared to laboratory strains, were able to survive in mice [16]. The common fungal pathogens Cryptococcus neoformans, Histoplasma capsulatum and Aspergillus fumigatus are found in environments as diverse as pigeon excreta and soil, yet each retains the capacity to grow at 37 °C. Many of the genes required for growth at high temperatures in these pathogens are necessary for virulence and some are required for survival [17–19]. Consequently, high temperature growth is essential for pathogenesis. This review will focus on the reservoirs and mechanisms of pathogenic yeast infections, the challenges faced upon infection and the hurdles that must be overcome to combat antifungal drug resistance.
Global burden of fungal infections
Superficial and mucosal fungal infections are extremely common, but life-threatening systemic fungal infections are generally limited to immunocompromised individuals [20, 21]. The population of vulnerable individuals experiencing some form of altered immune function due to the HIV/AIDS epidemic, hospitalization, chronic illnesses, antibiotic-mediated microbiota alteration, or chemotherapies continues to increase [10, 22, 23], and has led to billions of cases of invasive fungal infections worldwide.
Cryptococcus
Cryptococcus neoformans is the leading cause of deaths due to fungal infections, with a global burden of nearly 1 million cases annually, and more than 620,000 deaths worldwide [23]. Cryptococcus is ubiquitous and globally distributed, with more than 70 % of children older than 5 demonstrating serum reactivity against cryptococcal proteins [24]. A major risk factor for cryptococcal infections is immunosuppression due to HIV/AIDS. In resource-limited areas with high rates of HIV/AIDS, the mortality rate approaches 70 % [23], and cryptococcal meningitis is considered an AIDS-defining illness [25]. Left untreated, cryptococcal meningitis is uniformly fatal.
In Western Europe and North America, the overall mortality rate of cryptococcal infections is 10 %, with approximately 700 deaths per year [23]. However, nearly 20 % of cryptococcosis cases in the US are in non-HIV patients [26], and the mortality rate of these infections can be greater than 30 % [27]. Many of these infections are in patients with conditions associated with immunosuppression or with solid organ transplants, although there are some incidences of underlying genetic susceptibility to fungal infections [26–28].
Another source of cryptococcosis in immunocompetent individuals is infections due to Cryptococcus gattii, which has caused an outbreak in the Pacific Northwest over the last 15 years [29–31]. The majority of these cases have been in immunocompetent individuals, and many cases were fatal despite antifungal therapies [31]. Although the origin of the outbreak was Vancouver Island, cases of C. gattii infections have been documented along the pacific coast and in Florida [32, 33].
An additional complication in cryptococcal treatment is the occurrence of immune reconstitution inflammatory syndrome (IRIS), where restoration of immune function after starting highly active antiretroviral therapies (HAART) results in enhanced and destructive immune responses to subclinical or cleared microbial infections [34]. Recent studies have reported that between 8 and 50 % of AIDS patients develop cryptococcal IRIS after treatment with HAART, and the mortality rate ranges between 30 and 60 % [35, 36].
Candida
Candida species are normal members of the human mucosal microbiota; studies have estimated between 25 and 40 % of people are colonized with Candida albicans at any given point [37, 38]. However, C. albicans also causes more than 400,000 deaths per year due to invasive candidiasis [39]. Disseminated Candida infections have an attributable mortality rate approaching 40 %, even with antifungal therapies [39].
Patients with HIV/AIDS or other forms of impaired immunity, especially neutropenia, are vulnerable to disseminated candidiasis [25, 39, 40]. Additionally, Candida species are the fourth most common cause of hospital-acquired bloodstream infections, and the incidence of Candida infections is increasing [41]. Candida albicans is the most common causal agent of nosocomial infection among the Candida species, followed by C. glabrata, C. parapsilosis, and C. tropicalis [41]. Many of these infections may be due to breakdown of mucosal surfaces during hospitalization or treatment, which then allows the superficial Candida colonization access to the bloodstream [42, 43]. Finally, C. albicans can form biofilms on medically implanted devices and catheters, creating a reservoir for bloodstream infections [44, 45].
Recently, mortality rates due to C. albicans infections in high-risk patients, such as stem cell transplant recipients or those undergoing therapy for leukemia, have decreased due to prophylactic fluconazole therapies [39, 42]. However, resistance can occur [46, 47], and the Centers for Disease Control and Prevention (CDC) has ranked fluconazole-resistant Candida as a serious threat [48].
Pneumocystis
Pneumocystis is an almost ubiquitous colonizer of human lungs, with approximately 80 % of children demonstrating serum antibodies to the organism [49–51]. However, it can also cause pneumocystis pneumonia (PCP) in immunocompromised hosts. In the early years of the AIDS epidemic, PCP was the most common clinical manifestation of decreased immune function [52]. Before HAART, approximately 65 % of AIDS patients in the United States exhibited PCP [52, 53]. Recent studies in developing nations have suggested that up to 40 % of HIV-infected patients exhibit PCP [52, 54]. An extrapolation of the current epidemiological reports suggests that there are more than 400,000 cases per year [10, 54].
Pneumocystis pneumonia remains one of the leading causes of morbidity and mortality worldwide [50]. In developed nations with readily available HAART, the mortality rate has dropped to approximately 10 % [55, 56]. However, in a recent study of pulmonary disease in AIDS patients in Uganda, the number of cases of PCP was lower than for tuberculosis or cryptococcal pneumonia, but the mortality was higher, with 75 % of the patients dying within 2 months of admission [57]. Additionally, Pneumocystis infections may also contribute to impaired lung function and increased mortality of patients with chronic obstructive pulmonary disease (COPD) [50, 58].
Aspergillus
Aspergillus is a ubiquitous filamentous fungus, and people are faced with continuous exposure to Aspergillus spores. As an opportunistic pathogen, Aspergillus mainly causes disease in patients with impaired immune function. Most infections are acquired exogenously instead of through reactivation of latent infections [59]. The most vulnerable population includes patients with neutropenia, organ or bone marrow transplant, or those undergoing immunosuppressive therapies [42, 59, 60]. Invasive aspergillosis is uniformly fatal if untreated, and even with current treatment options, mortality remains at 50 % [42, 61]. Current reports suggest that there are more than 200,000 cases of invasive aspergillosis per year [10].
Aspergillus can also cause allergic bronchopulmonary aspergillosis (ABPA), and recent estimates suggest that the global burden of ABPA is 4,837,000 patients [62, 63]. Patients with lung disease, such as COPD, cystic fibrosis, or emphysema are especially susceptible to ABPA [59, 60, 63]. Infection with Aspergillus can increase the mortality of these underlying diseases, contributing to the 100,000 deaths per year [10].
Histoplasma
Histoplasma capsulatum is endemic to the Midwest United States and Central America, though it has a global distribution [64]. Histoplasma is the most prevalent cause of systemic mycosis in Central America, and it infects several hundred thousand individuals annually [64, 65]. Recent studies estimate nearly 80 % of the adult population in the endemic areas show previous infections by Histoplasma [64]. Most of these infections occur in immunocompetent hosts, but the majority does not display symptoms. However, individuals who are exposed to large numbers of spores can suffer from acute pulmonary histoplasmosis. Additionally, the disease is more severe in patients with underlying lung disease [64, 65]. Disseminated histoplasmosis usually only occurs in individuals with compromised immune systems [64, 66]. Mortality due to disseminated histoplasmosis remains at 30 %, but it can be reduced with HAART [65].
Paracoccidioides
Paracoccidioides is an endemic dimorphic fungal pathogen in South America, and it is the most common cause of invasive mycosis in this region [65]. Nearly 10 million people are infected with this organism, but in most cases, the host immune system can prevent the conidia from causing disease [65]. However, individuals with compromised immune systems or those with decreased lung function, such as smokers, are vulnerable to expansion of the yeast from the lungs into disseminated disease [22, 65]. Interestingly, there is not a strong association of paracoccidiomycosis with HIV [67]. In some cases (between 5 and 13 %), patients experience relapse of paracoccidiomycosis due to reactivation of quiescent yeast cells [68].
Penicillium
Penicillium marneffei is an endemic fungal pathogen in Southeast Asia, causing disease primarily in immunocompromised patients, although infections in immunocompetent hosts also occur [69, 70]. In a study of AIDS patients in northern Thailand, it was the third most common cause of infection, after Mycobacterium tuberculosis and C. neoformans [71]. As an opportunistic pathogen, the prevalence of P. marneffei infections is increasing with the expansion of the HIV epidemic in Southeast Asia [72].
Penicillium marneffei infections are more common in the rainy season and are positively correlated with increased humidity [73, 74]. This suggests that disease occurs from primary infections shortly after exposure to the fungus [73]. These disseminated infections are uniformly fatal without antifungal treatments, but even with rapid administration of therapeutics, the mortality rate remains approximately 20 % [71, 74]. In children with immune deficiencies, the mortality rate is higher, at 55 % [70].
Coccidioides
Coccidiomycosis, also known as valley fever, is endemic to the Southwestern United States and Central America, and it causes thousands of hospitalizations and approximately ~400 deaths per year in the United States [75]. Coccidiomycosis is caused by Coccidioides immitis and Coccidioides posadasii, which occupy different geographic regions [76]. Although most infections are self-limiting lung infections, Coccidioides can also cause disseminated disease. Risk factors include compromised immune systems, but agricultural or construction work, outdoor activity, African or Asian ethnicity, increased age, or pregnancy also increase vulnerability to Coccidioides. Additionally, uncontrolled diabetes increases the risk of disseminated infections [77]. In patients with impaired T cell function and disseminated infection, mortality rates were 50 % [76].
Blastomyces
Blastomycosis is caused by infection with Blastomyces dermatitidis, a dimorphic fungal pathogen that is endemic in the Midwestern United States. Most infections are due to spore inhalation during outdoor activities, and approximately 30–50 % of infections are asymptomatic [78]. Of the symptomatic infections, most can be treated with antifungal drugs. However, mortality ranges between 4 and 22 % [78]. In a recent outbreak in Wisconsin, 70 % of the cases were hospitalized and 5 % died [79]. Only 25 % of the infected patients had any form of compromised immune system. However, there appears to be genetic risk factors for blastomycosis as the incidence rate is 12 times higher for Asians [79].
Rhizopus and Mucor
Mucormycosis is due to infections by basal fungi, most often by species of Rhizopus and Mucor [80]. Vulnerable populations include patients with hematopoietic stem cell transplants or other hematological malignancies, those with diabetes, and those with compromised immune systems [80, 81]. The association between mucormycosis and diabetes is particularly strong; a recent study showed that 70 % of mucormycosis patients had diabetic ketoacidosis [81]. The mortality rate can be as high as 90 % for disseminated infections [82, 83]. Cutaneous mucormycosis can also occur after natural disasters, when environmental spores are disturbed and patients have open wounds [84]. The case-fatality rate for cutaneous mucormycosis can be as high as 80 % [84, 85].
Sporothrix
Sporotrichosis is usually caused by traumatic inoculation by material contaminated with Sporothrix schenckii, although there are some cases of inhalational sporotrichosis [86]. Although the infections are restricted to the site of infection, there are cases of disseminated sporotrichosis, mostly in immunocompromised individuals [67]. A recent study of cases in HIV-infected patients in Brazil found that the incidence of sporotrichosis was increasing to the point where there the incidence was comparable to histoplasmosis and cryptococcosis [67]. Of the patients with both HIV and sporotrichosis, almost 40 % were hospitalized for disseminated infection [67].
Trichosporon
Like Candida, Trichosporon can exist as a member of the normal human microbiota [87]. However, patients with malignant hematological disorders are vulnerable to infection with Trichosporon [87]. Trichosporon infections are the second most common cause of disseminated fungal infections in these patients, although the overall rate is low [88]. The mortality rate of patients with hematological disorders and Trichosporon remains at 11 %, even with antifungal therapies [88].
Reservoirs and nature of infection
Here we discuss the reservoirs and nature of infection, focusing on the predominant opportunistic yeast and yeast-like pathogens, Cryptococcus, Candida, Histoplasma, and Pneumocystis.
Cryptococcus
Cryptococcus species are found ubiquitously in nature, but only C. neoformans and C. gattii are considered serious human pathogens [89, 90]. The source of human infections are thought to be exclusively environmental, as there is no evidence of human to human transmission other than through contaminated medical devices [91]. Furthermore, concordance in phenotypic and genotypic characterization between environmental and clinical isolates within the same geographical region suggests they belong to the same fungal population [89, 92, 93].
Environmental isolates of these yeasts have been reported all around the globe, recovered from soil, dust, avian excreta, trees and other plants, domestic and wild animals, as well as marine mammals [94]. Cryptococcus neoformans is most often found in excreta from pigeons and other birds. In southern Africa, isolates are commonly found in the decayed hollows of the mopane tree [95]. Cryptococcus gattii is mostly isolated from tropical and subtropical regions in association with eucalyptus trees, as well as from soil, trees, and animals in the Pacific Northwest [94, 96]. Plant materials have also been shown to promote fertility and virulence of C. gattii [97].
Cryptococcal infection occurs upon inhalation of airborne fungal cells, generally spores or desiccated yeast [98]. Once inside the host lung, the yeast particles will encounter either alveolar macrophages or dendritic cells and trigger an immune response [99]. This can lead to clearance of the infection, or result in a localized asymptomatic latent infection that is contained within a granuloma, where the yeast is enveloped by immune cells [99, 100]. This latency period could last for years before reactivation of dormant infection occurs and disease symptoms develop [89, 101]. In the case of C. neoformans, reactivation occurs when host immunity is compromised, such as in patients with HIV/AIDS [89]. Cryptococcus gattii infections appear to be new events as they occur in both immunocompromised and immunocompetent individuals [102].
Active infection in the lung leads to pneumonia-like illness, with common symptoms including cough, fever, and dyspnea, among others [103, 104]. Fungal cells can subsequently disseminate from the lung through vascular or lymphatic systems to cause systemic infections, with the central nervous system (CNS) being the preferred destination [91, 100]. Cryptococcus can cross the blood–brain barrier either directly by transcytosis through the endothelial cells or with the aid of host monocytes [105–107]. Infection of the CNS and brain parenchyma is life threatening and results in cryptococcal meningitis and meningoencephalitis [104, 108]. Neurological symptoms include headaches, visual and hearing impairment, seizures, and mental status change [108, 109].
Candida
Although a large number of Candida species have been documented, only a few are known to cause disease in humans [11, 110]. Over 90 % of all Candida infections are caused by five species: C. albicans, Candida glabrata, Candida parapsilosis, Candida tropicalis, and Candida krusei. Among these, C. albicans is by far the most prevalent, responsible for 90–100 % of superficial mucosal infections and 40–70 % of disseminated infections [11, 110].
Unlike most other pathogenic fungi, Candida species are a natural commensal of the human microbiome [111], found rarely in the soil and external environments, suggesting adaptation to a parasitic lifestyle [112]. The majority of Candida infections are from endogenous sources, derived from commensal populations acquired prior to disease development [11, 113]. Exogenous sources of infection are also common, especially in healthcare settings where transmission can occur from healthcare workers, other patients, and contaminated medical devices [11, 114].
Normally, Candida species exist harmlessly on our skin and mucosal surfaces as part of the commensal microbiota, colonizing the skin, oral cavity, gastrointestinal tract, and reproductive tract [111, 115]. However, disruption of the normal microbial flora or compromising the immune system may enable this fungus to overgrow, resulting in symptomatic infections. Due to the long evolutionary history with the human host, C. albicans is well adapted to survive and proliferate in the host environment. The switch from commensalism to pathogenesis involves significant changes in the fungus, including regulation of key virulence genes that allows it to quickly sense and adjust to different sites of infection in the body. Candida species must overcome different stresses, such as changes in temperature, pH, osmolarity, oxygen and nutrient availability [14, 112, 115–118].
Inflammations of the genitourinary tract are common clinical manifestations of Candida infections, which include vulvovaginal candidiasis (VVC) in women, balanitis and balanoposthitis in men, and candiduria in both sexes [119]. VVC, commonly referred to as thrush or yeast infection, typically presents as isolated occurrences of mild to moderate infection in otherwise healthy women, and are generally easily controlled with a single-dose therapy [110]. Episodes can occur either sporadically or due to predisposition resulting from a number of risk factors, including antibiotic use, pregnancy, diabetes, and immunosuppression [21, 120, 121]. A subgroup of patients will experience severe or recurrent VVC, in which case more rigorous and long-term antifungal therapies are required [110]. It is estimated that the majority of women develop at least one episode of VVC in their lifetime [122]. Candida balanitis occurs at a lower frequency than VVC, though the exact incidence is unclear due to the lack of population studies [119]. The disease is generally sexually transmitted and has been associated with diabetes and antibiotic use [123]. Candiduria, the presence of Candida species in the urinary tract, is commonly diagnosed in hospitalized patients, especially those with a urinary catheter [124–126]. Most cases of candiduria are asymptomatic or self-limiting, though it can cause increased vulnerability to bloodstream infections in high-risk patients, as well as increased mortality rates and hospital costs [119, 127].
Candida infections can also develop in the mouth or throat, resulting in oropharyngeal candidiasis. Clinical manifestations are typically characterized by white or red lesions on the surfaces of the tongue and oropharynx, which result in pain and burning sensations, alteration in taste, and tissue damage [128, 129]. Infection is generally associated with immunosuppression, antibiotic use, endocrine alterations, and denture use [128, 130]. It is estimated that over 90 % of HIV-infected patients will develop oropharyngeal candidiasis at some point during the progression of their illness [131].
Invasive candidiasis occurs when Candida species break the mucosal barrier, penetrating into deeper tissue and gaining access to the bloodstream [132]. Dissemination via the bloodstream allows the fungus to invade almost all body sites and organs, resulting in lethal systemic disease [110]. Major risk factors include immunosuppression, invasive medical procedures, and extended stay in the intensive care unit [11, 133, 134]. Early clinical symptoms of invasive candidiasis are non-specific and resemble other nosocomial infections, which impede accurate diagnosis and delays treatment, contributing to increased mortality [132, 135, 136].
Pneumocystis
Pneumocystis species are ubiquitous in nature, infecting a wide variety of mammalian hosts [11, 137]. While rodents were originally thought to be a natural reservoir, it is now known that Pneumocystis is not zoonotic. Current phenotypic and genetic evidence indicates that each mammalian species that contracts the disease has its own species of Pneumocystis [137–139]. Pneumocystis jirovecii is responsible for human infections. However, study of the organism has been hindered by the inability to culture Pneumocystis in vitro. Much of what we now know about P. jirovecii is based on direct clinical evidence or extrapolated from related animal models [50, 140].
PCR-based strategies have been used to type and track the spread of P. jirovecii [141, 142]. Children and immunocompromised individuals are identified as the most important sources of P. jirovecii infection [142–145]. While some environmental sources have been identified, transmission is thought to mostly occur through inhalation of airborne pathogens that spread from human to human [146, 147].
Pneumocystis has strong tropism for the lung, where it exists as an alveolar pathogen without invading the host [140]. Infections are typically asymptomatic or subclinical in the healthy host, but can develop into pneumocystic pneumonia in immunocompromised individuals, especially those with HIV infections [50]. Common clinical symptoms include progressive dyspnea, nonproductive cough, low-grade fever, and eventual respiratory failure with disease progression [140].
Histoplasma
Histoplasma capsulatum exists as a mold in soil environments and is often associated with bird and bat droppings, which enhance proliferation of the organism by accelerating sporulation [148]. Once contaminated, soil can yield the organism for years after the birds are gone [64]. Infections occur through inhalation of the aerosolized H. capsulatum from disturbed soils. Air currents can also carry the fungal spore for miles, resulting in infections far away from contaminated sites [148].
Most infections by H. capsulatum are asymptomatic in healthy individuals [148]. In children who were exposed for the first time and in individuals who experienced heavy exposure to the organism, pulmonary histoplasmosis may develop [64]. Common symptoms include fever, malaise, cough, and chest pain. Pulmonary infections can be complicated with infection of the mediastinal lymph nodes, resulting in Granulomatous mediastinitis.
In immunocompromised patients, such as those infected with HIV, disseminated histoplasmosis may occur [64, 149, 150]. Initial clinical presentation often includes fever, anorexia, and weight loss. Subsequent severe disease manifestation includes sepsis, as well as failure of respiratory, renal, and multi-organ systems.
Mechanisms of pathogenesis
Pathogenicity is the ability of a microbe to cause damage to the host [151]. The ability of an opportunistic pathogen to cause disease requires the expression of virulence factors. Here, we outline some of the traits that enable pathogenesis of the major opportunistic yeast pathogens of humans, C. albicans, C. neoformans, and H. capsulatum.
High temperature growth
Fungi must be capable of surviving in human host conditions. One trait absolutely required for pathogenicity in humans is the ability to grow at human body temperature, 37 °C [14]. The most prevalent fungal pathogen, C. albicans, is a natural commensal of mammals and is thus intrinsically competent for growth at 37 °C. Despite this thermal adaptation, the heat shock response has been retained in C. albicans and influences virulence [152], presumably reflecting the importance of febrile temperatures during systemic infection [153]. In C. albicans, the heat shock response is governed by the transcription factor Hsf1, and the molecular chaperones Hsp70 and Hsp90 [14]. Mutant C. albicans strains containing inactive Hsf1 are thermosensitive and display reduced virulence in a murine model of systemic candidiasis [154]. Furthermore, the membrane dynamics of C. albicans are crucial for sensing changes in temperature, with depletion of OLE1, encoding a fatty acid desaturase, impairing activation of Hsf1 [155]. Temperature is a central cue that enables the morphogenetic transition of C. albicans from yeast to filamentous growth through complex cellular circuitry [156].
Although the saprobe C. neoformans is usually found in environmental locales, the ability to grow at human physiological temperatures allows it to cause disease. Several signaling pathways have been identified as essential for C. neoformans growth at elevated temperature, including Ras1 and Cdc42 signaling [157], the unfolded protein response [158], the histone acetyltransferase Gcn5 [159], the cell wall integrity pathway [160], and calcineurin signaling [19]. In addition, increased expression of C. neoformans Hsp90 and other heat-shock proteins is observed upon infection of a murine lung [161], suggesting a role for Hsp90 in adapting to host conditions. Mutants that are unable to grow at 37 °C are unable to cause disease in murine models of cryptococcosis.
Like C. neoformans, the dimorphic fungus H. capsulatum can be found in the soil. However, H. capsulatum forms mycelia in the environment and changes to the yeast form upon inhalation by a mammalian host. The increase in temperature to 37 °C is key for this transition, and membrane dynamics may be involved in sensing this cue [162]. For example, addition of saturated fatty acids with a concurrent increase in temperature amplifies the H. capsulatum heat shock response [163].
Adaptation to pH
Opportunistic fungal pathogens must not only adapt to variations in temperature, but also pH. In the host, the pH can range from acidic (in the vaginal and gastrointestinal tracts and the immune cell phagolysosome) to basic (such as in the blood or saliva) [164]. Therefore, pH is a powerful signal for entry into the host bloodstream or other microenvironments. Additionally, environmental pH can lead to alterations in nutrient bioavailability.
In C. albicans, alkaline conditions result in cleavage and activation of the transcription factor Rim101, which leads to the upregulation of genes required for growth in alkaline conditions, including genes involved in iron acquisition and morphogenesis [165]. Additionally, Rim101 regulates expression of genes with roles in adhesion and modulating the fungal cell wall [166]. PHR1 and PHR2 are pH-responsive genes regulated by Rim101, with PHR2 being required for virulence at acidic infection sites (such as the vaginal tract), whereas PHR1 is required at alkaline sites of infection (such as the bloodstream) [167]. Mutants lacking Rim101 have reduced virulence in a mouse model of systemic candidiasis [168].
Cryptococcus neoformans also uses the Rim101 transcription factor to adapt to varying environmental cues. Similar to C. albicans, CnRim101 is required for growth in neutral/alkaline conditions, and also regulates expression of genes required for iron and metal homeostasis [169]. However, in C. neoformans, Rim101 is also activated by the cyclic AMP (cAMP)/protein kinase A (PKA) pathway [169]. Rim101 is required to modulate many important C. neoformans virulence traits, such as attachment of the polysaccharide capsule [169], the morphological change to titan cells [170], and modulation of the host immune system [171, 172]. Upon phagocytosis by macrophages, C. neoformans enters into an acidic phagolysosome; as a facultative intracellular pathogen, C. neoformans is well adapted for growth in acidic conditions [173].
Nutrient limitation
Key to survival is growth. Organisms are often nutrient starved within the host and must adapt to changes in available host nutrients. One mechanism by which C. albicans can respond to limited glucose is by secreting ammonia and autoalkalizing its surroundings to induce filamentation [174]. For C. neoformans, phagocytosis by macrophages induces a starvation stress response that includes upregulation of amino acid and sugar transporters, gluconeogenesis and fatty acid metabolism [175].
As iron levels are tightly controlled within the host, microbes must have mechanisms to acquire this essential cofactor [176]. Although C. albicans and C. neoformans do not appear to produce their own siderophores, which are high-affinity iron-binding proteins, they are capable of transporting xenosiderophores produced by other organisms [177, 178]. Additionally, both C. neoformans and C. albicans are able to use iron from hemoglobin and ferritin as iron sources [179, 180]. Cryptococcus neoformans utilizes a ferric reductase system involving the iron permease Cft1 and ferroxidase Cfo1 to acquire iron from host transferrin [181, 182]. Mutants lacking these enzymes show a reduced virulence in a mouse model of cryptococcosis. Iron levels are sensed by a master transcription factor, Cir1, which directs the expression of genes for iron acquisition. Cir1 is also involved in regulating growth at physiological temperature, capsule formation, and melanin production [183], highlighting the central role of iron regulation in C. neoformans virulence.
Candida albicans is able to acquire iron from hemoglobin by first binding hemoglobin to a receptor on the cell wall and subsequently endocytosing this complex [184]. The heme oxygenase Hmx1 then releases ferrous iron [185], carbon monoxide, and biliverdin [186]. As carbon monoxide has immunosupressive properties [187], its production by Hmx1 may decrease the ability of the host immune system to clear a C. albicans infection. The hmx1 mutant is unable to cause disease as this mutant is unable to grow in iron-limited conditions and has a defect in carbon monoxide production [186, 188]. Additionally, the transcriptional activator Sef1 induces iron-uptake genes and enables virulence, whereas the transcriptional repressor Sfu1 diminishes expression of iron-uptake genes and enables gastrointestinal commensalism [116]. These genetic programs may be fundamental to enabling C. albicans to survive iron depletion in the bloodstream while minimizing iron toxicity in the gut.
Histoplasma capsulatum is able to both produce siderophores and utilize reductive iron assimilation to obtain the iron necessary for growth [189]. Histoplasma capsulatum produces hydroxyamate-type siderophores, and these siderophores are required for proliferation within macrophages and infection in mice [189, 190]. Production of siderophores is negatively regulated by the GATA-type transcription factor SRE1 [191]. Histoplasma capsulatum can also take up ferrioxamine B, a xenosiderophore, via ferric reductases [192].
Changes in cellular shape and size
Host physiological temperature induces a filament-to-yeast transition in dimorphic fungi, such as H. capsulatum (Fig. 1). This switch from mycelia to yeast growth at host temperatures is a requirement for H. capsulatum virulence, and the histidine kinase Drk1 is a key regulator of this transition [193]. DRK1-silenced strains show a drastic reduction in virulence in a mouse model of histoplasmosis [193]. In addition, Drk1 regulates expression of two important H. capsulatum virulence genes, CBP1 and AGS1 [193]. The DNA-binding protein Ryp1 is also essential for yeast growth, and is related to the C. albicans master transcriptional regulator of phenotypic switching, Wor1. Ryp1 is a master regulator of morphogenesis, as it is required for the expression of yeast-phase-specific virulence genes and repression of mycelial-specific genes [194]. Recently, a temperature-responsive regulatory circuit composed of four Ryp proteins was identified that controls both the transition from filamentous to yeast forms and the expression of virulence genes such as YPS3 and CBP1 [195].
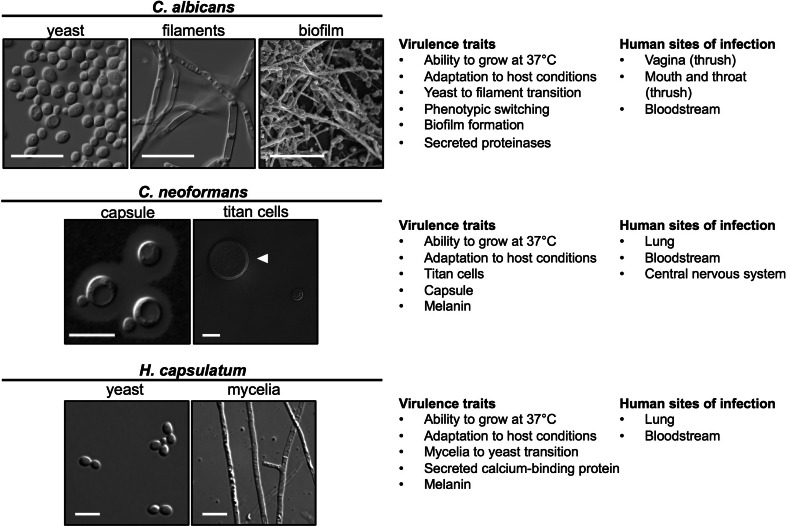
Fig. 1.
Yeast virulence traits. Top, C. albicans can exist as yeast, filaments, or as a biofilm. Wild-type yeast cells were grown in rich medium at 30 °C for 24 h. Filaments were grown in RPMI at 37 °C for 24 h. The biofilm is fluconazole treated and was formed in a rat central venous catheter. Scale bar is 20 μm. Middle, C. neoformans cells stained with India ink to highlight the capsule. Arrow indicates the size of a titan cell as compared to a normal cell. Scale bar 10 μm. Bottom, H. capsulatum can exist as either yeast or as mycelia. Scale bar is 5 μm
Strikingly, host physiological temperature induces the opposite morphological transition in the commensal fungus, C. albicans, from yeast to filament compared to the filament-to-yeast transition initiated in dimorphic fungi (Fig. 1). Candida albicans is able to undergo various morphological transitions that facilitate its ability to colonize and infect different niches within the human host. The most striking of these morphological transitions is that from yeast to filamentous forms. Typically, chains or branches of elongated connected cells are referred to as pseudohyphae [196], whereas cells containing defined septa between cells and no constrictions in their cell walls are known as hyphae [197]. Various different environmental cues induce the morphological transition from yeast to filaments. Many of these cues require a concurrent increase in temperature to 37 °C, and many of the cues mimic host physiological conditions. For example, serum is a potent inducer of filamentation at 37 °C, with key effectors being bacterial peptidoglycans and glucose [198–200]. At host physiological temperature, deprivation of nutrients such as carbon or nitrogen can induce a filamentous program, as can alkaline conditions (pH >6.5) or elevated CO2 [201]. The requirement of 37 °C for filamentation to occur in many cues is due at least in part to the Hsp90-mediated repression of filamentation, which is relieved at elevated temperature due to global problems in protein folding that can overwhelm the functional capacity of Hsp90 [202]. Many cues induce filamentation in C. albicans via the cAMP–PKA signaling pathway, which activates transcription factors such as Efg1 [203] and Flo8 [204]. Filamentation is also governed by repressors such as Nrg1, whose downregulation and degradation is required for both hyphal initiation and maintenance [205].
The ability of C. albicans to transition between yeast and filamentous states is key for its dissemination and virulence in different niches within the host. Invasion of epithelial cells by C. albicans can occur by either endocytosis or by active penetration by hyphal cells [206]. Cells locked in the yeast form are reduced in virulence [207, 208], and defective in adhering to and invading oral epithelial cells [209]. Similarly, cells unable to revert to the yeast form also have attenuated virulence and show a decrease in kidney fungal burden in a murine model of systemic candidiasis [210]. It is thought that yeast cells are required for dissemination throughout the body, while filamentous cells are essential for penetrating tissues, establishing infection and causing mortality [211]. A recent study has shown that a clinical isolate locked in the yeast form is more fit in a murine model of commensal infection [212]. Furthermore, another recent study demonstrated that cells locked in the yeast form are better able to colonize the gastrointestinal tract in a mouse model, while constitutively filamentous cells showed a decrease in colonization [213]. Taken together, this demonstrates that the polymorphic nature of C. albicans is a key virulence trait that allows it to adapt to the diverse conditions encountered within the host.
While both yeast and filamentous morphologies contribute to the pathogenesis of C. albicans, filamentous cells do possess specific virulence characteristics that increase their pathogenicity. For example, the sustained polarized growth exhibited by a filamentous cell provides a physical force that allows the fungal cell to penetrate host tissues, allowing for further invasion of tissues and organs [214]. Hyphal cells also express Als3, a hypha-specific protein found on the fungal cell surface [215]. Als3 is an adhesin that mediates binding to host cells, and induces endocytosis by binding to the host cell receptors E-cadherin and N-cadherin [215]. Hwp1 is another hyphal-specific fungal cell surface protein required to mediate attachment to epithelial cells [216]. In addition, the expression of genes encoding secreted aspartyl proteinases (SAPs) is coordinated with morphological state with SAP4 and SAP6 being expressed by hyphal cells [217, 218]. Additionally, connections exist between C. albicans filamentation and drug resistance. Certain proteins such as Hsp90 [219], O-mannosyltransferases [220], and the transcription factor Ndt80 [221] have roles in both morphogenesis and drug resistance.
In addition to the striking morphological differences between yeast and filamentous forms, C. albicans can also undergo distinct phenotypic switches. One prominent example is the switch between white and opaque cells. The more common white cells have a round or oval shape, while opaque cells are oblong and dimpled. This transition is governed by the regulator Wor1, with expression of WOR1 leading to opaque cell formation [222]. Opaque cells are mating competent, with progeny showing recombination between chromosomes and alterations in ploidy [223, 224]. White and opaque cells show distinct preferences for different host environments and niches. Since opaque cells are not stable at 37 °C, they preferentially colonize surface environments such as the skin [225]. Furthermore, while white cells require the elevated temperature of 37 °C for filamentation, opaque cells filament preferentially at 25 °C [226]. Finally, white-opaque switching may aid in the evasion of host defenses, where white phase cells are preferentially recognized by neutrophils in certain environmental conditions [227, 228]. A novel phenotypic switch was recently discovered upon passage of wild-type white cells through the mouse gastrointestinal tract [117]. Induction of WOR1 in this environment resulted in morphologically and functionally distinct GUT (gastrointestinally induced transition) cells, which are optimized for the digestive tract. A third phenotypic variant, the ‘gray’ phenotype has recently been identified [229]. This variant shows elevated levels of SAP expression and the propensity to colonize cutaneous tissue, similar to opaque cells. The master regulators Wor1 and Efg1 may act coordinately to regulate switching between white, gray and opaque cellular states [229].
Distinct morphological transitions are also important for virulence in C. neoformans (Fig. 1). While a typical C. neoformans cell is approximately 5–10 μM, specific conditions can induce the formation of titan cells, which can be as large as 100 μM [230]. Distinct features of titan cells include a thicker cell wall and denser capsule [231], as well as a tetraploid or octaploid DNA content [230]. Although the exact mechanism for titan cell formation is unclear, in part due to the difficulty in culturing titan cells in vitro [232], several signaling pathways have been implicated. The G protein-coupled receptors Ste3a (a pheromone receptor) and Gpr5 are required for titan cell production in vivo [170, 230] as is signaling through the cAMP/PKA pathway [170, 231]. The Rim101 transcription factor downstream of PKA signaling is also required for titan cell production in vivo [170]. Titan cells play a central role in C. neoformans pathogenicity, in that they interact with the host immune system. The large size of titan cells may prevent their phagocytosis [231, 233]; titan cells may also protect typical-sized cells from being phagocytosed [233], and promote dissemination from the lungs [234].
Biofilms
Biofilms are complex three-dimensional surface-associated communities of yeast and hyphal cells within an extracellular matrix (ECM), and can be found on medical devices such as catheters and artificial joints [235], or on mucosal surfaces [236], contributing to virulence (Fig. 1). The first step in biofilm formation is adherence to either an abiotic or biotic surface. In C. albicans, cell wall adhesins such as Eap1 and Als1, and other cell wall proteins have roles in attachment of the biofilm to the surface, which then stimulates changes in gene expression [237]. Adherence is followed by proliferation of yeast cells. This basal layer of yeast cells may contribute to anchoring the biofilm to the surface [238]. Next, yeast cells undergo the morphogenetic transition to filamentous growth. The formation of hyphae is important, as strains defective in filamentation have a vastly reduced ability to form biofilms, as with mutants lacking a major transcriptional regulator of morphogenesis, Efg1 [238]. Accumulation of the ECM is part of the biofilm maturation process. The ECM is composed of mainly protein and carbohydrates, including mannans and glucans; however, lipids and nucleic acids are also found in the ECM [239]. Finally, yeast cells are dispersed from the biofilm. These cells have elevated pathogenicity, and an increased ability to adhere [237].
Biofilms are notoriously difficult to treat with antifungals, and have especially high levels of resistance to azoles and polyenes [237]. Resistance to azoles is dependent on the molecular chaperone Hsp90 [45], glucan modification enzymes that are required for delivery and organization of the ECM [240], and β-1,3 glucan in the ECM that may sequester azoles [241]. Increased expression of efflux pumps early in biofilm development, and the presence of persister cells also contribute to antifungal resistance [242].
Secreted factors
Secretion of proteinases such as SAPs is crucial for both pathogenesis and nutrient acquisition. As mentioned above, C. albicans SAP4 and SAP6 expression is coordinated with hyphal formation [217, 218], while expression of SAP1 is regulated by the white-to-opaque phenotypic switch [243]. During infection, the SAP proteins have roles in adherence to host cells [244] and degradation of host proteins such as mucin [245], perhaps allowing penetration and colonization of tissues. SAP proteins may also degrade host antimicrobial factors such as lactoferrin, complement and the proteinase inhibitor cystatin A [244]. Secreted phospholipases cleave ester linkages in glycophospholipids, and can have roles in host cell penetration and invasion [246]. Mutants lacking the phospholipase Plb1 have reduced virulence in a murine model of Candida infection [247].
Histoplasma also secretes several factors important for pathogenesis. The calcium-binding protein Cbp1 is secreted from yeast-phase H. capsulatum, and is required for growth in calcium-depleted conditions and for virulence in a murine model of pulmonary histoplasmosis [248]. Resolution of the structure by NRM revealed that Cbp1 may be a lipid-binding protein, and possibly interacts with glycolipids in the host [249]. Yps3 is both a secreted factor as well as a cell wall protein in H. capsulatum, and while its exact function is yet to be elucidated, silencing of YPS3 results in reduced organ colonization in a murine model of infection [250].
Evasion of the host immune system
Capsule and cell wall
The capsule of C. neoformans is an important virulence determinant, playing key roles in surviving environmental conditions and modulating the host immune response (Fig. 1). The capsule is primarily composed of the polysaccharides glucuronoxylomannan (GXM) and glucuronoxylomannogalactan (GXMGal) [251]. Monomers of simple sugars are modified and polymerize within the cell before being secreted and attached to the cell wall surface. An important modification of capsule monomers is xylosylation, such that deletion of the xylosyltransferase gene CXT1 results in an altered capsule structure and reduced virulence [252]. O-acetylation contributes to the antigenicity of the capsule by affecting binding of antibodies and activation of the complement cascade [253]. Hyaluronic acid, synthesized by Cps1, is also added to the capsule and may play a role in facilitating passage of C. neoformans across the blood–brain barrier [254]. Finally, antigenic variation of capsule monomers can lead to differential binding of antibodies [255].
Just as conditions that mimic physiologically relevant host conditions induce filamentation in C. albicans, similar cues also induce capsule formation in C. neoformans. Upon infection of a host, the capsule increases in size with the thickness being determined by the location of infection. Infection of the lungs results in a thicker capsule than does infection of the brain [256], demonstrating that regulation of capsule synthesis is dependent on the environmental niche. Nutrient-poor conditions such as low glucose and low nitrogen can contribute to capsule induction by signaling through the cAMP–PKA pathway [257–259]. Similarly, as iron levels are tightly controlled within the host, conditions of low iron are a potent inducer of capsule formation [260]. The iron-sensing transcription factor Cir1 is one of the main regulators of capsule induction in response to iron poor conditions [183]. Induction of capsule formation in low iron conditions is also signaled through the cAMP–PKA pathway [251]. Additionally, the cAMP pathway is used to signal capsule formation in the presence of high CO2 levels [251, 261]. Physiological pH is another cue that is sensed by C. neoformans to regulate capsule formation, and the conserved Rim101 pH-sensing pathway integrates signals from the cAMP–PKA pathway to regulate attachment of the capsule to the fungal cell wall [169]. However, it appears that alkaline pH alone is insufficient to induce an enlarged capsule, and instead this condition must be combined with others, such as nutrient deprivation or the presence of serum [262]. Finally, stress response pathways can have a repressive effect on capsule induction, such as the Hog1 and protein kinase C (PKC) pathways that respond to osmotic stress [251].
The capsule is essential for C. neoformans interactions with the host immune system. The polysaccharides GXM and GXMGal have immunosuppressive properties such as modulation of macrophage, neutrophil and dendritic cell activities, and also inhibition of pro-inflammatory cytokine production [263]. GXMGal reduces B cell activity and inhibits activation of T cells. The capsule also has antiphagocytic properties, inhibiting phagocytosis in vitro due to the large size of the capsule, and possibly by hiding pathogen-associated molecular patterns (PAMPs) on the cell surface [264].
To avoid interaction with host phagocytes that would initiate the production of reactive oxygen species and the secretion of pro-inflammatory cytokines, H. capsulatum antigenic cell surface β glucans are hidden under a layer of α-(1,3)-glucan. Silencing of AGS1, encoding the α-glucan synthase, severely attenuates virulence of H. capsulatum in mouse lungs [265]. Loss of α-(1,3)-glucan allows for recognition of the β glucans by the host receptor dectin-1 [266]. Additionally, heat shock protein 60 (HSP60) on the cell surface binds to the CR3 receptor on macrophages, followed by phagocytosis [267]. However, interactions with CR3 do not typically result in a strong immune response unless other costimulatory signals are present [268, 269], allowing H. capsulatum to grow and survive within macrophages.
Escape from macrophages
Fungi have evolved multiple mechanisms to avoid being killed upon phagocytosis by immune cells such as macrophages. One mechanism is to escape the phagocyte, either by inducing macrophage lysis or by non-lytic exocytosis. Non-lytic expulsion of fungal cells was first described in C. neoformans, where live yeasts were able to escape macrophages without killing the host cell [270–272]. This prevents release of pro-inflammatory cytokines and potentially aids in transversal of cryptococcal cells across the blood–brain barrier [105]. This phenomenon of non-lytic expulsion has also been observed in C. albicans, although at lower frequency than for C. neoformans [273]. Candida albicans can also escape from macrophages via filamentation. These filaments can induce pyroptosis, a programmed pro-inflammatory macrophage death [274, 275]. This process is hypothesized to be dependent on the transition to hyphae, which stimulate host pyroptotic caspases and result in macrophage death [274, 275].
Nitric oxide detoxification
Nitric oxide (NO) is a nitrogen radical and antimicrobial effector produced by the host immune system in response to fungal infections. Candida albicans responds to this stress by increasing the expression of an NO dioxygenase flavohemoglobin, YHB1 [276, 277], which metabolizes NO. Candida albicans cells lacking YHB1 have increased sensitivity to NO and reduced virulence in a murine model of systemic candidiasis [276, 277].
Cryptococcus neoformans also employs an enzymatic defense against NO produced by the host. The flavohemoglobin denitrosylase Fhb1 consumes NO, and cells in which FHB1 is disrupted have reduced survival in macrophages and decreased virulence in a murine inhalation model [278]. A global analysis of the C. neoformans response to nitrosative stress revealed that the glutathione reductase Glr1 is upregulated in response to NO [279]. Deletion of GLR1 renders C. neoformans sensitive to nitrosative stress, and avirulent in a mouse inhalation model [279].
Although the presence of a flavohemoglobin in H. capsulatum is not apparent, a shotgun genomic microarray did identify NOR1, a P450 nitric oxide reductase homologue, whose expression is increased by nitrosative stress [280]. Overexpression of this gene decreased the sensitivity of H. capsulatum to reactive nitrogen species [280].
Melanin
In C. neoformans, melanins are pigmented molecules with antioxidant properties, synthesized by the laccase enzymes Lac1 and Lac2 [264]. Melanization occurs during infection [281], and overexpression of melanin results in decreased recognition by the host and modulation of host cell immune responses [282]. Melanized cells are also less susceptible to microbicidal peptides and recognition by macrophages, as well as being less susceptible to antifugal drugs in vitro [283]. Together, this demonstrates an important role for melanin in the virulence of C. neoformans.
Urea metabolism
In addition to being used as a nitrogen source by many fungi, C. albicans and C. neoformans also use urea metabolism to modulate the host immune system and alter dissemination [284]. Urease is considered a cryptococcal virulence factor as urease expression is linked with increased invasion across the blood–brain barrier [285], increased fungal burden, and altered host immune responses [286–288]. Urease-positive C. neoformans induce a strong non-protective Th2 response as demonstrated by an increased accumulation of eosinophils in the lung, increased serum IgE and higher levels of Th2 cytokines [288], demonstrating a role for urease in influencing the host immune response.
While C. albicans does not contain a urease enzyme, it does contain a urea amidolyase, encoded by DUR1,2 [289]. Deletion of DUR1,2 results in cells that are less virulent than wild-type cells in a murine model of disseminated candidiasis and results in decreased kidney fungal burden [290]. Additionally, deletion of DUR1,2 causes a decreased inflammatory response in the kidneys, reduced neutrophil infiltration and altered host cytokine and chemokine production, suggesting a role for urea metabolism in modulating host immune responses [290]. A dur1,2 mutant also shows impaired escape from host macrophages as it is unable to form filaments in the presence of urea or arginine, a urea-precursor [291]. This may be coupled to the defect of the dur1,2 strain in auto-alkalinization of the environment [174].
Host immune responses
The interactions between opportunistic fungal infections and the host immune system are complex and only beginning to be understood. Outcomes of infections are dependent on interactions between the host immune response and the intrinsic virulence of the pathogen. Opportunistic fungi such as C. albicans constantly interact with the host immune system, and perturbations to this balance can result in a switch from commensalism to pathogenesis, as discussed above. Alterations to the innate or adaptive host immune responses, either due to immunosuppression or genetic predisposition, can result in severe and often deadly fungal infections. The interplay between fungi and the host immune system has been the subject of several recent reviews [8, 292–294]. Here we summarize this interaction, and describe host factors that can increase susceptibility to fungal infections.
Host response to fungal pathogens
Innate
Physical barriers such as the skin and mucosal epithelial cells at sites continuously in contact with fungal pathogens are the first line of defense against fungal pathogens. The next barrier is the innate immune response, which is crucial for preventing invasive and systemic infections. The innate immune response is required for recognition of fungal PAMPs. The fungal cell wall is composed of many PAMPs, including both α- and β-glucans, chitin, chitosan, and mannans [295]. These PAMPs can be recognized by host pattern recognition receptors (PRRs) on phagocytic cells such as macrophages, dendritic cells and neutrophils [292, 293]. Specific PAMPs on different fungal species can be recognized by specific PRRs, including complement components, toll-like receptors (TLRs), C-type lectin receptors (CLRs), and mannose receptors (MR), among others [296–299]. These interactions will shape the resulting immune response, including phagocytosis of the pathogen, production of inflammatory cytokines, and activation of T cells.
Upon phagocytosis, fungal cells can be killed by effector molecules such as proteases, defensins, and cationic peptides as well as by the production of reactive oxygen and nitrogen species [294]. Neutrophils are highly effective at containing fungal infections, including inhibition of the C. albicans morphological transition [300]. Neutropenic patients, including those with acute leukemia, are at high risk of developing invasive Candida and Aspergillus infections, and are often given prophylactic antifungal therapies as a preventative measure [301]. However, increased neutrophils in mouse kidneys are associated with pathogenesis of C. albicans at late time points [302].
Adaptive
The adaptive immune system involves stimulation of T cells by antigen-presenting cells [292, 303]. These antigen-presenting cells can generate specific cytokine profiles that can bias the immune system towards Th1-, Th2-, Treg-, or Th17-responses [303]. The balance between these responses is crucial for the ability to clear the pathogen without causing autoimmune damage.
The adaptive immune response plays a pivotal role in preventing fungal infections, as highlighted by the incidence of fungal disease in AIDS patients [10, 304]. HIV infection depletes the pool of CD4+ T cells, consequently causing a loss of antifungal immunity. This results in impaired production of interferon-γ (IFN-γ) and tumor necrosis factor-α (TNF-α) [304]. Additionally, the population of memory B cells is depleted [305], and macrophage and dendritic cell function is impaired upon HIV infection [306], contributing to increased susceptibility to fungal infections.
Th1 responses include the production of IFN-γ, which is key for controlling many disseminated fungal infections [303]. Cryptococcus neoformans strains that induce Th1-biased responses are generally cleared from the lung and do not cause disease [307, 308]. The decrease in Th1-type cell-mediated immune responses in AIDS patients may explain their increased susceptibility to cryptococcal infections compared to patients with other types of immune deficiencies [309].
Th17 responses appear to be critical for host defense against fungal infections, especially Candida [310]. In HIV patients, susceptibility to mucosal candidiasis can be attributed to a loss of mucosal Th17 cells, which is accompanied by a decrease in the integrity of epithelial cells and alterations to the intestinal environment [311]. Decreases in the Th17 response also leads to increased severity of Pneumocystis infections in mice [312].
Regulatory T (Treg) cells continually survey the host for signs of mucosal infections and aid in preventing reinfection. However, Treg cells are able to suppress inflammation during systemic candidiasis by producing interleukin 4 (IL-4), IL-10, and transforming growth factor-β (TGF-β), which inhibit inflammatory Th1 and Th17 responses making them detrimental for clearing the infection [313]. Depletion of Treg cells results in decreased C. albicans growth in a murine model of candidiasis [313], and a decrease in Treg cells is also associated with an increase in the Th17 responses that are beneficial in clearing Histoplasma infections [314].
In contrast, Th2 responses to fungal infections are often deleterious [292]. During C. neoformans infections, the fungi are able to induce a strong Th2-biased immune response, with increased levels of IL-4 and IL-5, thus favoring persistence of the infection [309]. Mice with increased IL-4 levels also demonstrated increased Histoplasma infections [315]. In addition to favoring persistence of the infection, Th2 responses are also linked with allergic bronchopulmonary aspergillosis and increased susceptibility to invasive aspergillosis [316]. In contrast, Th2 responses may be protective against Pneumocystis pneumonia, with affected HIV individuals showing low levels of Th2 cytokines [317].
Immune reconstitution inflammatory syndrome
The recovered immune system in AIDS patients treated with antiretrovirals can overreact to PAMPs exposed during fungal infections, even if the infections have been cleared by antifungal therapies. This overactive inflammation is known as immune reconstitution inflammatory syndrome (IRIS), and it can cause significant mortality due to damage by the host immune system. IRIS is a major consideration in the treatment of AIDS patients with cryptococcosis. Recent studies have noted that between 8 and 50 % of AIDS patients develop cryptococcal IRIS, even when no live cryptococcal cells are recovered from the patient [35, 318]. IRIS can also occur in transplant patients after reduction in the immunosuppresive therapies used to prevent organ rejection [319]. The inflammatory immune response in these IRIS patients consists of increased IL-6 and C-reactive protein (CRP) levels. Additionally, many patients who go on to develop IRIS have decreased TNF-α and other Th1 cytokines prior to antiretroviral therapy [320]. The current standard of care for IRIS patients includes administration of steroids to decrease systemic inflammatory responses and prevent further damage to host tissues [36, 90].
Genetic susceptibility to fungal infections
Chronic mucocutaneous candidiasis
Recurrent Candida infections of cutaneous or mucosal surfaces in the absence of immunosuppressive conditions (such as HIV), is described as chronic mucocutaneous candidiasis (CMC). IL-17 signaling and activation of T cells are essential for defending against mucocutaneous C. albicans infections and genetic mutations that perturb this signaling pathway can lead to CMC [321].
Numerous single nucleotide polymorphisms have been associated with increased susceptibility to CMC (see [8, 292, 321]). For example, loss of function of the CLR dectin-1 can lead to increased onychomycosis and mucocutaneous candidiasis [322, 323]. This is the result of defective recognition of β glucan in the C. albicans cell wall and impaired cytokine production (including IL-1β [323] and IL-17 [322]). Neutrophil function remains unaffected, and thus systemic candidiasis is not associated with dectin-1 mutations [323]. The caspase recruitment domain-containing protein 9 (CARD9) is a signaling protein downstream of many CLRs, including dectin-1 [321] and activates the nuclear factor-κB (NF-κB) pathway [324]. Autosomal recessive mutations in CARD9 can result in decreased development of Th17 cells and is associated with CMC [324]. Autosomal dominant gain-of-function mutations such as those in STAT1 (signal transducer and activator of transcription 1) can also lead to inhibition of Th17 development, and increased susceptibility to CMC [325, 326]. Additionally, autosomal recessive mutations in the IL-17 receptor IL-17RA, and deficiencies in the cytokine IL-17F can result in CMC [327]. Mice with deficient IL-17A production show an impaired ability to clear a cutaneous C. albicans infection [328], suggesting that both IL-17A and IL-17F are required for the defense against chronic mucocutaneous candidiasis.
Additional syndromes can lead to CMC. One of these is hyper-IgE syndrome (HIES), in which patients experience recurrent pulmonary infections, eczema, staphylococcal infections, and CMC, among other symptoms. Mutations in STAT3 have been identified as a cause of HIES [329], in which an inability of CD4+ T lymphocytes to differentiate into Th17 cells results in impaired IL-17 production [330]. Mutations in DOCK8 (dedicator of cytokinesis 8) have been identified in the autosomal recessive form of this syndrome, in which there is defective T cell activation and Th17 cell differentiation [331]. Finally, the rare autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy (APECED) syndrome is characterized by development of CMC in almost all patients, and is caused by mutations in the autoimmune regulator AIRE [321, 332]. The strong prevalence of CMC in these patients may be due to the production of neutralizing autoantibodies against Th17-produced cytokines, specifically IL-17A, IL-17F and IL-22 [333].
Invasive fungal infections
While mucocutaneous Candida infections are primarily caused by defects in the adaptive immune system, deficiencies in the innate immune response, such as those caused by primary immunodefiencies can lead to invasive yeast infections. A few such cases are outlined below. Chronic granulomatous disease (CGD) is the result of mutations in genes encoding components of the NADPH oxidase complex, impairing the ability of phagocytes to produce reactive oxygen species [334]. While the primary fungal infection associated with CGD is invasive aspergillosis, [335] infections by Candida and Trichosporon species have also been noted [321]. Myeloperoxidase (MPO) deficiency is a disorder in which the production of antimicrobial hypochlorous acid by phagocytic cells is impaired, and mice deficient for either NADPH or MPO show increased mortality in response to a high dose of C. albicans [336]. While the majority of MPO patients do not suffer from fungal infections, those who do acquire infections usually also have diabetes [337]. Patients presenting with monocytopenia are more susceptible to histoplasmosis and cryptococcal infections [338], and cryptococcosis has also been observed in patients with hyper-IgE or hyper-IgM syndromes and idiopathic CD4 lymphocytopenia [321]. Finally, patients with severe combined immunodeficiency commonly acquire Pneumocystis pneumonia, as do those with hyper-IgM syndrome [321].
Management of fungal infections
Diagnosis
The first step in achieving the proper management of infectious disease is the effective diagnosis and species identification of the pathogen. Unfortunately, shortcomings in current diagnostic techniques remain one of the greatest challenges in the field. In most cases, diagnosis still relies on traditional culture methods and histopathology [10, 87, 339, 340]. Culturing the organism and subsequent identification is a slow process that takes several days, leading to considerable delays in treatment. Such delays have direct and significant impact on disease outcome and patient mortality, especially for severe invasive diseases [136, 340]. A number of novel molecular diagnostic approaches have been developed to improve the current system, including PCR-based assays and antigen detection systems [339, 341]. However, these are not regular practice in the clinic, and still need to be standardized and tested in large patient cohorts before they can be incorporated into clinical guidelines [87, 90, 110, 339].
Antifungal drugs
Due to the close evolutionary relatedness between humans and fungi, the number of targets that can be selectively exploited for drug development is limited [10, 342]. There are also very few drugs currently being developed, primarily because antifungals are not predicted to generate a large enough financial return for pharmaceutical companies [10]. Polyenes, azoles, and echinocandins represent the three most common classes of antifungals currently used in the clinic, each with their own advantages and limitations. Overall, host toxicity, cross reactivity with other drugs, and development of drug resistance pose great challenges to current antifungals.
Polyenes
Polyenes are broad-spectrum natural product antifungals discovered in the early 1950s from the bacterial genus Streptomyces [342, 343]. Polyenes bind to ergosterol in the fungal membrane, generating aqueous pores that result in leakage of fungal cell content and eventually cell death [344, 345]. Amphotericin B, one of the most successful polyene derivatives, was a gold standard in treating serious fungal infections. However, amphotericin B is known to cause severe systemic toxicity and nephrotoxicity [342, 346]. Various drug delivery systems have been developed to improve its safety profile, including the popular lipid formulation [347, 348]. While its clinical use decreased with the development of azoles in the late 1980s, it is still widely deployed to treat life-threatening disseminated and invasive mycoses [90, 110]. Fortunately, despite its long history of clinical use, resistance to polyenes remains a rare occurrence [349, 350].
Azoles
Azoles are synthetic compounds that were first introduced as antifungals in the late 1980s and early 1990s [344]. Their low toxicity led to extensive use in the clinic. Azoles disrupt the biosynthesis of ergosterol by inhibiting the cytochrome P-450-dependent enzyme lanosterol demethylase (also referred to as 14α-sterol demethylase) [351]. Azoles are chemically classified as either imidazoles if they have two nitrogen atoms in the azole ring, or triazoles if they have three [343, 351]. Imidazoles are typically limited to topical treatment of superficial infections, while triazoles are used more broadly in both superficial and systemic infections due to their superior pharmacokinetic and safety profile. The most commonly used azoles in the clinic include fluconazole, itraconazole, voriconazole, and posaconazole.
Azoles remain the drug of choice as initial therapy for most fungal infections and are often recommended as prophylaxis for high-risk patients [90, 110, 352]. Widespread use of azoles has led to increasing reports of azole resistance in the clinic, which is associated with greater treatment difficulties and patient mortality [353]. There has also been an increased incidence of infections caused by intrinsically azole-resistant fungal species, including C. glabrata and C. krusei, creating major challenges for future treatments [87].
Echinocandins
Echinocandins first entered the market in 2001, representing the newest class of antifungals to reach the clinic [354]. Echinocandins are large semi-synthetic lipopeptides that inhibit the cell wall enzyme complex β-1,3-d-glucan synthase [355], thereby decreasing the concentration of β-(1,3)-glucan in the fungal cell wall and causing the subsequent loss of cell wall integrity. Echinocandins are generally well tolerated with little to no side effects [356].
Echinocandins are active against Candida and Aspergillus species, including azole-resistant Candida isolates, but show no in vivo activity against Cryptococcus [90, 110, 342]. Currently available echinocandins include caspofungin, anidulafungin, and micafungin [354]. All three are not orally bioavailable and need to be administered intravenously. Echinocandin resistance has been reported in the clinic, and the incidence appears to be on the rise [357].
Vaccines
An ideal solution to fungal management is the prevention of disease through vaccination. However, there is currently no clinically available vaccine for any fungal pathogen. Over the years, a number of vaccines have been in development against different fungi, with promising results in animal models [10, 358]. However, only a few have been translated to human clinical trials, with the major barrier being the lack of funding [359]. Efficacy trials are also difficult to conduct as high-risk patients often routinely receive antifungal prophylaxis [10]. Nonetheless, two subunit vaccines against Candida have recently demonstrated success in Phase I clinical trials, one of which is currently being tested in a Phase II trial in the United States [360]. This vaccine represents a promising advance and may stimulate further development of much needed immunotherapeutics and vaccines [358, 361, 362].
Concluding remarks
A significant advance in our understanding of invasive fungal infections has been made in recent years, motivated at least in part by the increasing incidence of these infections. Given the growing number of transplant patients, those in receipt of immunosuppressive therapy and the global HIV pandemic, this comes as no surprise. Host niches are complex, dynamic and distinct to each individual, as are the pathogens’ response strategies for coexisting with or evading the host immune response. Taking steps towards understanding the underlying immunopathogenesis of fungal infections permits early and accurate diagnosis and treatment. We must continue to seek out novel diagnostics that will allow for rapid treatment of fungal infections with the appropriate antifungal. With the rise in antifungal drug resistance now a real threat, researchers must strive to discover novel antifungals and antifungal drug combinations, as well as effective immunotherapeutic strategies that combat resistance, improve patient outcome, shorten hospital stays and ease the economic burden.
Acknowledgments
We thank the J. Andrew Alspaugh and Chad Rappleye labs for images and Cowen lab members for helpful discussions. EJP is supported by a Canadian Institutes of Health Research (CIHR) Frederick Banting and Charles Best CGS Doctoral Award, XL by a University of Toronto Fellowship, MDL by a Sir Henry Wellcome Postdoctoral Fellowship (Wellcome Trust 096072), and LEC by a Ministry of Research and Innovation Early Researcher Award, Canada Research Chair in Microbial Genomics and Infectious Disease, Natural Sciences and Engineering Research Council Discovery Grant #355965, and by Canadian Institutes of Health Research Grants MOP-86452 and MOP-119520.
Abbreviations
- ABPA
Allergic bronchopulmonary aspergillosis
- APECED
Autoimmune polyendocrinopathy–candidiasis–ectodermal dystrophy
- cAMP
Cyclic AMP
- CDC
Centers for Disease Control and Prevention
- CGD
Chronic granulomatous disease
- CLR
C-type lectin receptors
- CMC
Chronic mucocutaneous candidiasis
- CNS
Central nervous system
- COPD
Chronic obstructive pulmonary disease
- CRP
C-reactive protein
- ECM
Extracellular matrix
- GUT
Gastrointestinally induced transition
- GXM
Glucuronoxylomannan
- GXMGal
Glucuronoxylomannogalactan
- HAART
Highly active antiretroviral therapies
- HIES
Hyper-IgE syndrome
- HSP
Heat shock protein
- IFN-γ
Interferon-γ
- IL
Interleukin
- IRIS
Immune reconstitution inflammatory syndrome
- MPO
Myeloperoxidase
- MR
Mannose receptors
- NF-κB
Nuclear factor-κB
- NO
Nitric oxide
- PAMP
Pathogen-associated molecular pattern
- PCP
Pneumocystis pneumonia
- PKA
Protein kinase A
- PKC
Protein kinase C
- PRR
Pattern recognition receptors
- SAP
Secreted aspartyl proteinase
- TGF-β
Transforming growth factor-β
- TLR
Toll-like receptors
- TNF-α
Tumor necrosis factor-α
- Treg cell
Regulatory T cell
- VVC
Vulvovaginal candidiasis
Footnotes
E. J. Polvi and X. Li contributed equally to this work.
References
- 1.Wang DY, Kumar S, Hedges SB. Divergence time estimates for the early history of animal phyla and the origin of plants, animals and fungi. Proc Biol Sci. 1999;266:163–171. doi: 10.1098/rspb.1999.0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butterfield NJ. Probable proterozoic fungi. Paleobiology. 2005;31:165–182. [Google Scholar]
- 3.Hawksworth DL. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol Res. 2001;105(12):1422–1432. [Google Scholar]
- 4.DiGuistini S, Wang Y, Liao NY, Taylor G, Tanguay P, Feau N, Henrissat B, Chan SK, Hesse-Orce U, Alamouti SM, Tsui CK, Docking RT, Levasseur A, Haridas S, Robertson G, Birol I, Holt RA, Marra MA, Hamelin RC, Hirst M, Jones SJ, Bohlmann J, Breuil C. Genome and transcriptome analyses of the mountain pine beetle-fungal symbiont Grosmannia clavigera, a lodgepole pine pathogen. Proc Natl Acad Sci USA. 2011;108:2504–2509. doi: 10.1073/pnas.1011289108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186–194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:983–989. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Program NIHISCCS, Kong HH, Segre JA. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14:405–416. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bickers DR, Lim HW, Margolis D, Weinstock MA, Goodman C, Faulkner E, Gould C, Gemmen E, Dall T, American Academy of Dermatology A, Society for Investigative D The burden of skin diseases: 2004 a joint project of the American Academy of Dermatology Association and the Society for Investigative Dermatology. J Am Acad Dermatol. 2006;55:490–500. doi: 10.1016/j.jaad.2006.05.048. [DOI] [PubMed] [Google Scholar]
- 10.Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012;4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 11.Pfaller MA, Diekema DJ. Epidemiology of invasive mycoses in North America. Crit Rev Microbiol. 2010;36:1–53. doi: 10.3109/10408410903241444. [DOI] [PubMed] [Google Scholar]
- 12.Brown GD, Denning DW, Levitz SM. Tackling human fungal infections. Science. 2012;336:647. doi: 10.1126/science.1222236. [DOI] [PubMed] [Google Scholar]
- 13.Bergman A, Casadevall A. Mammalian endothermy optimally restricts fungi and metabolic costs. MBio. 2010;1(5):e00212-10. doi: 10.1128/mBio.00212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leach MD, Cowen LE. Surviving the heat of the moment: a fungal pathogens perspective. PLoS Pathog. 2013;9:e1003163. doi: 10.1371/journal.ppat.1003163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Solache MA, Casadevall A. Global warming will bring new fungal diseases for mammals. MBio. 2010;1(1):e00061-10. doi: 10.1128/mBio.00061-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCusker JH, Clemons KV, Stevens DA, Davis RW. Saccharomyces cerevisiae virulence phenotype as determined with CD-1 mice is associated with the ability to grow at 42 °C and form pseudohyphae. Infect Immun. 1994;62:5447–5455. doi: 10.1128/iai.62.12.5447-5455.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhabhra R, Miley MD, Mylonakis E, Boettner D, Fortwendel J, Panepinto JC, Postow M, Rhodes JC, Askew DS. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect Immun. 2004;72:4731–4740. doi: 10.1128/IAI.72.8.4731-4740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamoth F, Juvvadi PR, Fortwendel JR, Steinbach WJ. Heat shock protein 90 is required for conidiation and cell wall integrity in Aspergillus fumigatus . Eukaryot Cell. 2012;11:1324–1332. doi: 10.1128/EC.00032-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans . EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havlickova B, Czaika VA, Friedrich M. Epidemiological trends in skin mycoses worldwide. Mycoses. 2008;51:2–15. doi: 10.1111/j.1439-0507.2008.01606.x. [DOI] [PubMed] [Google Scholar]
- 21.Sobel JD. Vulvovaginal candidosis. Lancet. 2007;369:1961–1971. doi: 10.1016/S0140-6736(07)60917-9. [DOI] [PubMed] [Google Scholar]
- 22.Marques SA, Robles AM, Tortorano AM, Tuculet MA, Negroni R, Mendes RP. Mycoses associated with AIDS in the Third World. Med Mycol. 2000;38:269–279. [PubMed] [Google Scholar]
- 23.Park BJ, Wannemuehler KA, Marston BJ, Govender N, Pappas PG, Chiller TM. Estimation of the current global burden of cryptococcal meningitis among persons living with HIV/AIDS. AIDS. 2009;23(4):525–530. doi: 10.1097/QAD.0b013e328322ffac. [DOI] [PubMed] [Google Scholar]
- 24.Goldman DL, Khine H, Abadi J, Lindenberg DJ, Pirofski LA, Niang R, Casadevall A. Serologic evidence for Cryptococcus neoformans infection in early childhood. Pediatrics. 2001;107(5):E66. doi: 10.1542/peds.107.5.e66. [DOI] [PubMed] [Google Scholar]
- 25.Schneider E, Whitmore S, Glynn KM, Dominguez K, Mitsch A, Centers for Disease Control and Prevention (CDC) McKenna MT (2008) Revised surveillance case definitions for HIV infection among adults, adolescents, and children aged <18 months and for HIV infection and aids among children aged 18 months to <13 years—United States. MMWR Recomm Rep. 2008;57(10):1–12. [PubMed] [Google Scholar]
- 26.Pyrgos V, Seitz AE, Steiner CA, Prevots DR, Williamson PR. Epidemiology of Cryptococcal meningitis in the US: 1997–2009. PLoS ONE. 2013;8(2):e56269. doi: 10.1371/journal.pone.0056269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bratton EW, El Husseini N, Chastain CA, Lee MS, Poole C, Stürmer T, Juliano JJ, Weber DJ, Perfect JR. Comparison and temporal trends of three groups with cryptococcosis: HIV-infected, solid organ transplant, and HIV-negative/non-transplant. PLoS ONE. 2012;7(8):e43582. doi: 10.1371/journal.pone.0043582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh N, Dromer F, Perfect JR, Lortholary O. Immunocompromised hosts: cryptococcosis in solid organ transplant recipients: current state of the science. Clin Infect Dis. 2008;47(10):1321–1327. doi: 10.1086/592690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartlett KH, Cheng P-Y, Duncan C, Galanis E, Hoang L, Kidd S, Lee M-K, Lester S, MacDougall L, Mak S, Morshed M, Taylor M, Kronstad JW. A decade of experience: Cryptococcus gattii in British Columbia. Mycopathologia. 2012;173(5–6):311–319. doi: 10.1007/s11046-011-9475-x. [DOI] [PubMed] [Google Scholar]
- 30.Byrnes EJ, Li W, Lewit Y, Ma H, Voelz K, Ren P, Carter DA, Chaturvedi V, Bildfell RJ, May RC, Heitman J. Emergence and pathogenicity of highly virulent Cryptococcus gattii genotypes in the northwest United States. PLoS Pathog. 2010;6(4):e1000850. doi: 10.1371/journal.ppat.1000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galanis E, Macdougall L, Kidd S, Morshed M, British Colombia Cryptococcus gattii Working Group Epidemiology of Cryptococcus gattii, British Columbia, Canada, 1999–2007. Emerging Infect Dis. 2010;16(2):251–257. doi: 10.3201/eid1602.090900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Byrnes I, Edmond J, Bildfell RJ, Frank SA, Mitchell TG, Marr KA, Heitman J. Molecular evidence that the range of the Vancouver Island outbreak of Cryptococcus gattii infection has expanded into the pacific northwest in the United States. J Infect Dis. 2009;199(7):1081–1086. doi: 10.1086/597306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kunadharaju R, Choe U, Harris JR, Lockhart SR, Greene JN. Cryptococcus gattii, Florida, USA, 2011. Emerg Infect. 2013;19(3):519–521. doi: 10.3201/eid1903.121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perfect JR, Bicanic T. Cryptococcosis diagnosis and treatment: what do we know now. Fungal Genet Biol. 2014 doi: 10.1016/j.fgb.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boulware DR, Bonham SC, Meya DB, Wiesner DL, Park GS, Kambugu A, Janoff EN, Bohjanen PR. Paucity of initial cerebrospinal fluid inflammation in cryptococcal meningitis is associated with subsequent immune reconstitution inflammatory syndrome. J Infect Dis. 2010;202(6):962–970. doi: 10.1086/655785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shelburne SA, 3rd, Darcourt J, White AC, Jr, Greenberg SB, Hamill RJ, Atmar RL, Visnegarwala F. The role of immune reconstitution inflammatory syndrome in AIDS-related Cryptococcus neoformans disease in the era of highly active antiretroviral therapy. Clin Infect Dis. 2005;40:1049–1052. doi: 10.1086/428618. [DOI] [PubMed] [Google Scholar]
- 37.Fetter A, Partisani M, Koenig H, Kremer M, Lang J-M. Asymptomatic oral Candida albicans carriage in HIV-infection: frequency and predisposing factors. J Oral Pathol Med. 1993;22(2):57–59. doi: 10.1111/j.1600-0714.1993.tb00043.x. [DOI] [PubMed] [Google Scholar]
- 38.Martins MD, Lozano-Chiu M, Rex JH. Declining rates of oropharyngeal candidiasis and carriage of Candida albicans associated with trends toward reduced rates of carriage of fluconazole-resistant C. albicans in human immunodeficiency virus-infected patients. Clin Infect Dis. 1998;27(5):1291–1294. doi: 10.1086/515006. [DOI] [PubMed] [Google Scholar]
- 39.Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang C-H, Webster KM. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis. 2009;48(12):1695–1703. doi: 10.1086/599039. [DOI] [PubMed] [Google Scholar]
- 40.Koh AY, Kohler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008;4(2):e35. doi: 10.1371/journal.ppat.0040035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39(3):309–317. doi: 10.1086/421946. [DOI] [PubMed] [Google Scholar]
- 42.Freifeld AG, Bow EJ, Sepkowitz KA, Boeckh MJ, Ito JI, Mullen CA, Raad II, Rolston KV, Young JAH, Wingard JR. Clinical practice guideline for the use of antimicrobial agents in neutropenic patients with cancer: 2010 update by the Infectious Diseases Society of America. Clin Infect Dis. 2011;52(4):e56–e93. doi: 10.1093/cid/cir073. [DOI] [PubMed] [Google Scholar]
- 43.Nucci M, Anaissie E. Revisiting the source of candidemia: skin or gut? Clin Infect Dis. 2001;33(12):1959–1967. doi: 10.1086/323759. [DOI] [PubMed] [Google Scholar]
- 44.Nett J, Andes DR. Candida albicans biofilm development, modeling a host–pathogen interaction. Curr Opin Microbiol. 2006;9(4):340–345. doi: 10.1016/j.mib.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 45.Robbins N, Uppuluri P, Nett J, Rajendran R, Ramage G, Lopez-Ribot JL, Andes D, Cowen LE. Hsp90 governs dispersion and drug resistance of fungal biofilms. PLoS Pathog. 2011;7:e1002257. doi: 10.1371/journal.ppat.1002257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hill JA, Ammar R, Torti D, Nislow C, Cowen LE. Genetic and genomic architecture of the evolution of resistance to antifungal drug combinations. PLoS Genet. 2013;9(4):e1003390. doi: 10.1371/journal.pgen.1003390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mansfield BE, Oltean HN, Oliver BG, Hoot SJ, Leyde SE, Hedstrom L, White TC. Azole drugs are imported by facilitated diffusion in Candida albicans and other pathogenic fungi. PLoS Pathog. 2010;6(9):e1001126. doi: 10.1371/journal.ppat.1001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Centres for Disease Control and Prevention . Antibiotic resistance threats in the United States, 2013. U.S. Deparment of Health and Human Services. Atlanta: Centres for Disease Control and Prevention; 2013. [Google Scholar]
- 49.Djawe K, Huang L, Daly KR, Levin L, Koch J, Schwartzman A, Fong S, Roth B, Subramanian A, Grieco K, Jarlsberg L, Walzer PD. Serum antibody levels to the Pneumocystis jirovecii major surface glycoprotein in the diagnosis of P. jirovecii pneumonia in HIV+ patients. PLoS ONE. 2010;5(12):e14259. doi: 10.1371/journal.pone.0014259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morris A, Norris KA. Colonization by Pneumocystis jirovecii and its role in disease. Clin Microbiol Rev. 2012;25:297–317. doi: 10.1128/CMR.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pifer LL, Hughes WT, Stagno S, Woods D. Pneumocystis carinii infection: evidence for high prevalence in normal and immunosuppressed children. Pediatrics. 1978;61(1):35–41. [PubMed] [Google Scholar]
- 52.Morris A, Lundgren JD, Masur H, Walzer PD, Hanson DL, Frederick T, Huang L, Beard CB, Kaplan JE. Current epidemiology of Pneumocystis pneumonia . Emerging Infect Dis. 2004;10(10):1713–1720. doi: 10.3201/eid1010.030985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Masur H, Michelis MA, Greene JB, Onorato I, Stouwe RA, Holzman RS, Wormser G, Brettman L, Lange M, Murray HW, Cunningham-Rundles S. An outbreak of community-acquired Pneumocystis carinii pneumonia: initial manifestation of cellular immune dysfunction. N Engl J Med. 1981;305(24):1431–1438. doi: 10.1056/NEJM198112103052402. [DOI] [PubMed] [Google Scholar]
- 54.Fisk DT, Meshnick S, Kazanjian PH. Pneumocystis carinii pneumonia in patients in the developing world who have acquired immunodeficiency syndrome. Clin Infect Dis. 2003;36(1):70–78. doi: 10.1086/344951. [DOI] [PubMed] [Google Scholar]
- 55.Huang L, Cattamanchi A, Davis JL, Boon Sd, Kovacs J, Meshnick S, Miller RF, Walzer PD, Worodria W, Masur H, International HIV-associated Opportunistic Pneumonias (IHOP) Study; Lung HIV Study HIV-associated Pneumocystis pneumonia . Prom Am Thorac Soc. 2011;8(3):294–300. doi: 10.1513/pats.201009-062WR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kelley CF, Checkley W, Mannino DM, Franco-Paredes C, Del Rio C, Holguin F. Trends in hospitalizations for AIDS-associated Pneumocystis jirovecii Pneumonia in the United States (1986 to 2005) Chest. 2009;136(1):190–197. doi: 10.1378/chest.08-2859. [DOI] [PubMed] [Google Scholar]
- 57.Kyeyune R, den Boon S, Cattamanchi A, Davis JL, Worodria W, Yoo SD, Huang L. Causes of early mortality in HIV-infected TB suspects in an East African referral hospital. J Acquir Immune Defic Syndr. 2010;55(4):446–450. doi: 10.1097/qai.0b013e3181eb611a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sivam S, Sciurba FC, Lucht LA, Zhang Y, Duncan SR, Norris KA, Morris A. Distribution of Pneumocystis jirovecii in lungs from colonized COPD patients. Diagn Microbiol Infect Dis. 2011;71(1):24–28. doi: 10.1016/j.diagmicrobio.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wald A, Leisenring W, van Burik JA, Bowden RA. Epidemiology of Aspergillus infections in a large cohort of patients undergoing bone marrow transplantation. J Infect Dis. 1997;175:1459–1466. doi: 10.1086/516480. [DOI] [PubMed] [Google Scholar]
- 60.Marr KA, Carter RA, Crippa F, Wald A, Corey L. Epidemiology and outcome of mould infections in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2002;34:909–917. doi: 10.1086/339202. [DOI] [PubMed] [Google Scholar]
- 61.Baddley JW, Andes DR, Marr KA, Kauffman CA, Kontoyiannis DP, Ito JI, Schuster MG, Brizendine KD, Patterson TF, Lyon GM, Boeckh M, Oster RA, Chiller T, Pappas PG. Antifungal therapy and length of hospitalization in transplant patients with invasive aspergillosis. Med Mycol. 2013;51(2):128–135. doi: 10.3109/13693786.2012.690108. [DOI] [PubMed] [Google Scholar]
- 62.Agarwal R, Chakrabarti A. Allergic bronchopulmonary aspergillosis in asthma: epidemiological, clinical and therapeutic issues. Future Microbiol. 2013;8(11):1463–1474. doi: 10.2217/fmb.13.116. [DOI] [PubMed] [Google Scholar]
- 63.Denning DW, Pleuvry A, Cole DC. Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Med Mycol. 2013;51(4):361–370. doi: 10.3109/13693786.2012.738312. [DOI] [PubMed] [Google Scholar]
- 64.Kauffman CA. Histoplasmosis: a clinical and laboratory update. Clin Microbiol Rev. 2007;20(1):115–132. doi: 10.1128/CMR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colombo AL, Tobón A, Restrepo A, Queiroz-Telles F, Nucci M. Epidemiology of endemic systemic fungal infections in Latin America. Med Mycol. 2011;49(8):785–798. doi: 10.3109/13693786.2011.577821. [DOI] [PubMed] [Google Scholar]
- 66.Goodwin RAJ, Shapiro JL, Thurman GH, Thurman SS, des Prez RM. Disseminated Histoplasmosis: clinical and pathologic correlations. Medicine (Baltimore) 1980;59(1):1–33. [PubMed] [Google Scholar]
- 67.Freitas DFS, Valle ACFD, da Silva MBT, Campos DP, Lyra MR, de Souza RV, Veloso VG, Zancopé-Oliveira RM, Bastos FI, Galhardo MCG. Sporotrichosis: an emerging neglected opportunistic infection in HIV-infected patients in Rio de Janeiro, Brazil. PLoS Negl Trop Dis. 2014;8(8):e3110. doi: 10.1371/journal.pntd.0003110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sylvestre TF, Silva LRF, Cavalcante RDS, Moris DV, Venturini J, Vicentini AP, Carvalho LRD, Mendes RP. Prevalence and serological diagnosis of relapse in Paracoccidioidomycosis patients. PLoS Negl Trop Dis. 2014;8(5):e2834. doi: 10.1371/journal.pntd.0002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duong TA. Infection due to Penicillium marneffei, an emerging pathogen: review of 155 reported cases. Clin Infect Dis. 1996;23(1):125–130. doi: 10.1093/clinids/23.1.125. [DOI] [PubMed] [Google Scholar]
- 70.Lee PPW, Chan K-W, Lee T-L, Ho MH-K, Chen X-Y, Li C-H, Chu K-M, Zeng H-S, Lau Y-L. Penicilliosis in children without HIV infection–are they immunodeficient? Clin Infect Dis. 2012;54(2):e8–e19. doi: 10.1093/cid/cir754. [DOI] [PubMed] [Google Scholar]
- 71.Supparatpinyo K, Chiewchanvit S, Hirunsri P, Uthammachai C, Nelson KE, Sirisanthana T. Penicillium marneffei infection in patients infected with human immunodeficiency virus. Clin Infect Dis. 1992;14(4):871–874. doi: 10.1093/clinids/14.4.871. [DOI] [PubMed] [Google Scholar]
- 72.Vanittanakom N, Cooper CR, Fisher MC, Sirisanthana T. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin Microbiol Rev. 2006;19(1):95–110. doi: 10.1128/CMR.19.1.95-110.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bulterys PL, Le T, Quang VM, Nelson KE, Lloyd-Smith JO. Environmental predictors and incubation period of AIDS-associated Penicillium marneffei infection in Ho Chi Minh City, Vietnam. Clin Infect Dis. 2013;56(9):1273–1279. doi: 10.1093/cid/cit058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Le T, Wolbers M, Chi NH, Quang VM, Chinh NT, Lan NPH, Lam PS, Kozal MJ, Shikuma CM, Day JN, Farrar J. Epidemiology, seasonality, and predictors of outcome of AIDS-associated Penicillium marneffei infection in Ho Chi Minh City, Vietnam. Clin Infect Dis. 2011;52(7):945–952. doi: 10.1093/cid/cir028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Flaherman VJ, Hector R, Rutherford GW. Estimating severe coccidioidomycosis in California. Emerg Infect Dis. 2007;13(7):1087–1090. doi: 10.3201/eid1307.061480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown J, Benedict K, Park B, Thompson G., 3rd Coccidioidomycosis: epidemiology. Clin Epidemiol. 2013;5:185–197. doi: 10.2147/CLEP.S34434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santelli AC, Blair JE, Roust LR. Coccidioidomycosis in patients with diabetes mellitus. Am J Med. 2006;119(11):964–969. doi: 10.1016/j.amjmed.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 78.Khuu D, Shafir S, Bristow B, Sorvillo F. Blastomycosis mortality rates, United States, 1990–2010. Emerg Infect Dis. 2014;20(11):1789–1794. doi: 10.3201/eid2011.131175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Roy M, Benedict K, Deak E, Kirby MA, McNiel JT, Sickler CJ, Eckardt E, Marx RK, Heffernan RT, Meece JK, Klein BS, Archer JR, Theurer J, Davis JP, Park BJ. A large community outbreak of blastomycosis in Wisconsin with geographic and ethnic clustering. Clin Infect Dis. 2013;57(5):655–662. doi: 10.1093/cid/cit366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mendoza L, Vilela R, Voelz K, Ibrahim AS, Voigt K, Lee SC. Human fungal pathogens of Mucorales and Entomophthorales. Cold Spring Harb Perspect Med. 2014 doi: 10.1101/cshperspect.a019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chakrabarti A, Das A, Mandal J, Shivaprakash MR, George VK, Tarai B, Rao P, Panda N, Verma SC, Sakhuja V. The rising trend of invasive zygomycosis in patients with uncontrolled diabetes mellitus. Med Mycol. 2006;44(4):335–342. doi: 10.1080/13693780500464930. [DOI] [PubMed] [Google Scholar]
- 82.Lanternier F, Sun H-Y, Ribaud P, Singh N, Kontoyiannis DP, Lortholary O. Mucormycosis in organ and stem cell transplant recipients. Clin Infect Dis. 2012;54(11):1629–1636. [Google Scholar]
- 83.Roden MM, Zaoutis TE, Buchanan WL, Knudsen TA, Sarkisova TA, Schaufele RL, Sein M, Sein T, Chiou CC, Chu JH, Kontoyiannis DP, Walsh TJ. Epidemiology and outcome of zygomycosis: a review of 929 reported cases. Clin Infect Dis. 2005;41(5):634–653. doi: 10.1086/432579. [DOI] [PubMed] [Google Scholar]
- 84.Green JP, Karras DJ. Update on emerging infections: news from the Centers for Disease Control and Prevention. Notes from the field: fatal fungal soft-tissue infections after a tornado–Joplin, Missouri, 2011. Ann Emerg Med. 2012;59(1):53–55. doi: 10.1016/j.annemergmed.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 85.Bonifaz A, Vázquez-González D, Tirado-Sánchez A, Ponce-Olivera RM. Cutaneous zygomycosis. Clin Dermatol. 2013;30(4):413–419. doi: 10.1016/j.clindermatol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 86.Barros MBDL, de Almeida Paes R, Schubach AO. Sporothrix schenckii and Sporotrichosis. Clin Microbiol Rev. 2011;24(4):633–654. doi: 10.1128/CMR.00007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Miceli MH, Diaz JA, Lee SA. Emerging opportunistic yeast infections. Lancet Infect Dis. 2011;11:142–151. doi: 10.1016/S1473-3099(10)70218-8. [DOI] [PubMed] [Google Scholar]
- 88.Girmenia C, Pagano L, Martino B, D’Antonio D, Fanci R, Specchia G, Melillo L, Buelli M, Pizzarelli G, Venditti M, Martino P, GIMEMA Infection Program Invasive infections caused by Trichosporon species and Geotrichum capitatum in patients with hematological malignancies: a retrospective multicenter study from Italy and review of the literature. J Clin Microbiol. 2005;43(4):1818–1828. doi: 10.1128/JCM.43.4.1818-1828.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin X, Heitman J. The biology of the Cryptococcus neoformans species complex. Annu Rev Microbiol. 2006;60:69–105. doi: 10.1146/annurev.micro.60.080805.142102. [DOI] [PubMed] [Google Scholar]
- 90.Perfect JR, Dismukes WE, Dromer F, Goldman DL, Graybill JR, Hamill RJ, Harrison TS, Larsen RA, Lortholary O, Nguyen MH, Pappas PG, Powderly WG, Singh N, Sobel JD, Sorrell TC. Clinical practice guidelines for the management of cryptococcal disease: 2010 update by the infectious diseases society of America. Clin Infect Dis. 2010;50:291–322. doi: 10.1086/649858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Idnurm A, Bahn YS, Nielsen K, Lin X, Fraser JA, Heitman J. Deciphering the model pathogenic fungus Cryptococcus neoformans . Nat Rev Microbiol. 2005;3:753–764. doi: 10.1038/nrmicro1245. [DOI] [PubMed] [Google Scholar]
- 92.Sorrell TC, Chen SC, Ruma P, Meyer W, Pfeiffer TJ, Ellis DH, Brownlee AG. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J Clin Microbiol. 1996;34:1253–1260. doi: 10.1128/jcm.34.5.1253-1260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamamoto Y, Kohno S, Koga H, Kakeya H, Tomono K, Kaku M, Yamazaki T, Arisawa M, Hara K. Random amplified polymorphic DNA analysis of clinically and environmentally isolated Cryptococcus neoformans in Nagasaki. J Clin Microbiol. 1995;33:3328–3332. doi: 10.1128/jcm.33.12.3328-3332.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cogliati M. Global molecular epidemiology of Cryptococcus neoformans and Cryptococcus gattii: an atlas of the molecular types. Scientifica (Cairo) 2013;2013:675213. doi: 10.1155/2013/675213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Litvintseva AP, Carbone I, Rossouw J, Thakur R, Govender NP, Mitchell TG. Evidence that the human pathogenic fungus Cryptococcus neoformans var. grubii may have evolved in Africa. PLoS ONE. 2011;6:e19688. doi: 10.1371/journal.pone.0019688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kidd SE, Chow Y, Mak S, Bach PJ, Chen H, Hingston AO, Kronstad JW, Bartlett KH. Characterization of environmental sources of the human and animal pathogen Cryptococcus gattii in British Columbia, Canada, and the Pacific Northwest of the United States. Appl Environ Microbiol. 2007;73:1433–1443. doi: 10.1128/AEM.01330-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen SC, Meyer W, Sorrell TC. Cryptococcus gattii infections. Clin Microbiol Rev. 2014;27:980–1024. doi: 10.1128/CMR.00126-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Velagapudi R, Hsueh YP, Geunes-Boyer S, Wright JR, Heitman J. Spores as infectious propagules of Cryptococcus neoformans . Infect Immun. 2009;77:4345–4355. doi: 10.1128/IAI.00542-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Coelho C, Bocca AL, Casadevall A. The intracellular life of Cryptococcus neoformans. Annu Rev Pathol. 2014;9:219–238. doi: 10.1146/annurev-pathol-012513-104653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sabiiti W, May RC. Mechanisms of infection by the human fungal pathogen Cryptococcus neoformans . Future Microbiol. 2012;7:1297–1313. doi: 10.2217/fmb.12.102. [DOI] [PubMed] [Google Scholar]
- 101.Garcia-Hermoso D, Janbon G, Dromer F. Epidemiological evidence for dormant Cryptococcus neoformans infection. J Clin Microbiol. 1999;37:3204–3209. doi: 10.1128/jcm.37.10.3204-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kronstad JW, Attarian R, Cadieux B, Choi J, D’Souza CA, Griffiths EJ, Geddes JM, Hu G, Jung WH, Kretschmer M, Saikia S, Wang J. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol. 2011;9:193–203. doi: 10.1038/nrmicro2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nadrous HF, Antonios VS, Terrell CL, Ryu JH. Pulmonary cryptococcosis in nonimmunocompromised patients. Chest. 2003;124:2143–2147. doi: 10.1016/s0012-3692(15)31671-8. [DOI] [PubMed] [Google Scholar]
- 104.Voelz K, May RC. Cryptococcal interactions with the host immune system. Eukaryot Cell. 2010;9:835–846. doi: 10.1128/EC.00039-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Charlier C, Nielsen K, Daou S, Brigitte M, Chretien F, Dromer F. Evidence of a role for monocytes in dissemination and brain invasion by Cryptococcus neoformans . Infect Immun. 2009;77:120–127. doi: 10.1128/IAI.01065-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen SH, Stins MF, Huang SH, Chen YH, Kwon-Chung KJ, Chang Y, Kim KS, Suzuki K, Jong AY. Cryptococcus neoformans induces alterations in the cytoskeleton of human brain microvascular endothelial cells. J Med Microbiol. 2003;52:961–970. doi: 10.1099/jmm.0.05230-0. [DOI] [PubMed] [Google Scholar]
- 107.Chang YC, Stins MF, McCaffery MJ, Miller GF, Pare DR, Dam T, Paul-Satyaseela M, Kim KS, Kwon-Chung KJ. Cryptococcal yeast cells invade the central nervous system via transcellular penetration of the blood-brain barrier. Infect Immun. 2004;72:4985–4995. doi: 10.1128/IAI.72.9.4985-4995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bicanic T, Harrison TS. Cryptococcal meningitis. Br Med Bull. 2004;72:99–118. doi: 10.1093/bmb/ldh043. [DOI] [PubMed] [Google Scholar]
- 109.Chen SC, Slavin MA, Heath CH, Playford EG, Byth K, Marriott D, Kidd SE, Bak N, Currie B, Hajkowicz K, Korman TM, McBride WJ, Meyer W, Murray R, Sorrell TC, Australia, New Zealand Mycoses Interest Group-Cryptococcus S Clinical manifestations of Cryptococcus gattii infection: determinants of neurological sequelae and death. Clin Infect Dis. 2012;55:789–798. doi: 10.1093/cid/cis529. [DOI] [PubMed] [Google Scholar]
- 110.Pappas PG, Kauffman CA, Andes D, Benjamin DK, Jr, Calandra TF, Edwards JE, Jr, Filler SG, Fisher JF, Kullberg BJ, Ostrosky-Zeichner L, Reboli AC, Rex JH, Walsh TJ, Sobel JD, Infectious Diseases Society of A Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis. 2009;48:503–535. doi: 10.1086/596757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Odds FC. Candida and candidosis. 2. London: Baillière Tindall; 1988. [Google Scholar]
- 112.Hube B. From commensal to pathogen: stage- and tissue-specific gene expression of Candida albicans . Curr Opin Microbiol. 2004;7:336–341. doi: 10.1016/j.mib.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 113.Taylor BN, Harrer T, Pscheidl E, Schweizer A, Rollinghoff M, Schroppel K. Surveillance of nosocomial transmission of Candida albicans in an intensive care unit by DNA fingerprinting. J Hosp Infect. 2003;55:283–289. doi: 10.1016/s0195-6701(03)00295-0. [DOI] [PubMed] [Google Scholar]
- 114.Asmundsdottir LR, Erlendsdottir H, Haraldsson G, Guo H, Xu J, Gottfredsson M. Molecular epidemiology of candidemia: evidence of clusters of smoldering nosocomial infections. Clin Infect Dis. 2008;47:e17–e24. doi: 10.1086/589298. [DOI] [PubMed] [Google Scholar]
- 115.Gow NA, Hube B. Importance of the Candida albicans cell wall during commensalism and infection. Curr Opin Microbiol. 2012;15:406–412. doi: 10.1016/j.mib.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 116.Chen C, Pande K, French SD, Tuch BB, Noble SM. An iron homeostasis regulatory circuit with reciprocal roles in Candida albicans commensalism and pathogenesis. Cell Host Microbe. 2011;10:118–135. doi: 10.1016/j.chom.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013;45:1088–1091. doi: 10.1038/ng.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Perez JC, Kumamoto CA, Johnson AD. Candida albicans commensalism and pathogenicity are intertwined traits directed by a tightly knit transcriptional regulatory circuit. PLoS Biol. 2013;11:e1001510. doi: 10.1371/journal.pbio.1001510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Achkar JM, Fries BC. Candida infections of the genitourinary tract. Clin Microbiol Rev. 2010;23:253–273. doi: 10.1128/CMR.00076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.de Leon EM, Jacober SJ, Sobel JD, Foxman B. Prevalence and risk factors for vaginal Candida colonization in women with type 1 and type 2 diabetes. BMC Infect Dis. 2002;2:1. doi: 10.1186/1471-2334-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Spinillo A, Capuzzo E, Acciano S, De Santolo A, Zara F. Effect of antibiotic use on the prevalence of symptomatic vulvovaginal candidiasis. Am J Obstet Gynecol. 1999;180:14–17. doi: 10.1016/s0002-9378(99)70141-9. [DOI] [PubMed] [Google Scholar]
- 122.Sobel JD, Faro S, Force RW, Foxman B, Ledger WJ, Nyirjesy PR, Reed BD, Summers PR. Vulvovaginal candidiasis: epidemiologic, diagnostic, and therapeutic considerations. Am J Obstet Gynecol. 1998;178:203–211. doi: 10.1016/s0002-9378(98)80001-x. [DOI] [PubMed] [Google Scholar]
- 123.Edwards S. Balanitis and balanoposthitis: a review. Genitourin Med. 1996;72:155–159. doi: 10.1136/sti.72.3.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kauffman CA, Vazquez JA, Sobel JD, Gallis HA, McKinsey DS, Karchmer AW, Sugar AM, Sharkey PK, Wise GJ, Mangi R, Mosher A, Lee JY, Dismukes WE. Prospective multicenter surveillance study of funguria in hospitalized patients. The National Institute for Allergy and Infectious Diseases (NIAID) Mycoses Study Group. Clin Infect Dis. 2000;30:14–18. doi: 10.1086/313583. [DOI] [PubMed] [Google Scholar]
- 125.Bougnoux ME, Kac G, Aegerter P, D’Enfert C, Fagon JY, CandiRea Study G Candidemia and candiduria in critically ill patients admitted to intensive care units in France: incidence, molecular diversity, management and outcome. Intensive Care Med. 2008;34:292–299. doi: 10.1007/s00134-007-0865-y. [DOI] [PubMed] [Google Scholar]
- 126.Lundstrom T, Sobel J. Nosocomial candiduria: a review. Clin Infect Dis. 2001;32:1602–1607. doi: 10.1086/320531. [DOI] [PubMed] [Google Scholar]
- 127.Alvarez-Lerma F, Nolla-Salas J, Leon C, Palomar M, Jorda R, Carrasco N, Bobillo F, Group ES Candiduria in critically ill patients admitted to intensive care medical units. Intensive Care Med. 2003;29:1069–1076. doi: 10.1007/s00134-003-1807-y. [DOI] [PubMed] [Google Scholar]
- 128.Coronado-Castellote L, Jimenez-Soriano Y. Clinical and microbiological diagnosis of oral candidiasis. J Clin Exp Dent. 2013;5:e279–e286. doi: 10.4317/jced.51242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Samaranayake LP, Keung Leung W, Jin L. Oral mucosal fungal infections. Periodontol. 2009;49:39–59. doi: 10.1111/j.1600-0757.2008.00291.x. [DOI] [PubMed] [Google Scholar]
- 130.Pankhurst CL. Candidiasis (oropharyngeal) Clin Evid (Online) 2013;2013:1304. [PMC free article] [PubMed] [Google Scholar]
- 131.de Repentigny L, Lewandowski D, Jolicoeur P. Immunopathogenesis of oropharyngeal candidiasis in human immunodeficiency virus infection. Clin Microbiol Rev. 2004;17:729–759. doi: 10.1128/CMR.17.4.729-759.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Eggimann P, Garbino J, Pittet D. Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect Dis. 2003;3:685–702. doi: 10.1016/s1473-3099(03)00801-6. [DOI] [PubMed] [Google Scholar]
- 133.Pappas PG, Alexander BD, Andes DR, Hadley S, Kauffman CA, Freifeld A, Anaissie EJ, Brumble LM, Herwaldt L, Ito J, Kontoyiannis DP, Lyon GM, Marr KA, Morrison VA, Park BJ, Patterson TF, Perl TM, Oster RA, Schuster MG, Walker R, Walsh TJ, Wannemuehler KA, Chiller TM. Invasive fungal infections among organ transplant recipients: results of the transplant-associated infection surveillance network (TRANSNET) Clin Infect Dis. 2010;50:1101–1111. doi: 10.1086/651262. [DOI] [PubMed] [Google Scholar]
- 134.Eggimann P, Bille J, Marchetti O. Diagnosis of invasive candidiasis in the ICU. Ann Intensive Care. 2011;1:37. doi: 10.1186/2110-5820-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ellepola AN, Morrison CJ. Laboratory diagnosis of invasive candidiasis. J Microbiol. 2005;43:65–84. [PubMed] [Google Scholar]
- 136.Morrell M, Fraser VJ, Kollef MH. Delaying the empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: a potential risk factor for hospital mortality. Antimicrob Agents Chemother. 2005;49:3640–3645. doi: 10.1128/AAC.49.9.3640-3645.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gigliotti F, Wright TW. Pneumocystis: where does it live? PLoS Pathog. 2012;8:e1003025. doi: 10.1371/journal.ppat.1003025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Durand-Joly I, El Aliouat M, Recourt C, Guyot K, Francois N, Wauquier M, Camus D, Dei-Cas E. Pneumocystis carinii f. sp. hominis is not infectious for SCID mice. J Clin Microbiol. 2002;40:1862–1865. doi: 10.1128/JCM.40.5.1862-1865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sinclair K, Wakefield AE, Banerji S, Hopkin JM. Pneumocystis carinii organisms derived from rat and human hosts are genetically distinct. Mol Biochem Parasitol. 1991;45:183–184. doi: 10.1016/0166-6851(91)90042-5. [DOI] [PubMed] [Google Scholar]
- 140.Thomas CF, Jr, Limper AH. Pneumocystis pneumonia. N Engl J Med. 2004;350:2487–2498. doi: 10.1056/NEJMra032588. [DOI] [PubMed] [Google Scholar]
- 141.Beard CB, Roux P, Nevez G, Hauser PM, Kovacs JA, Unnasch TR, Lundgren B. Strain typing methods and molecular epidemiology of Pneumocystis pneumonia. Emerg Infect Dis. 2004;10:1729–1735. doi: 10.3201/eid1010.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Peterson JC, Cushion MT. Pneumocystis: not just pneumonia. Curr Opin Microbiol. 2005;8:393–398. doi: 10.1016/j.mib.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 143.Wakefield AE, Lindley AR, Ambrose HE, Denis CM, Miller RF. Limited asymptomatic carriage of Pneumocystis jiroveci in human immunodeficiency virus-infected patients. J Infect Dis. 2003;187:901–908. doi: 10.1086/368165. [DOI] [PubMed] [Google Scholar]
- 144.Larsen HH, von Linstow ML, Lundgren B, Hogh B, Westh H, Lundgren JD. Primary Pneumocystis infection in infants hospitalized with acute respiratory tract infection. Emerg Infect Dis. 2007;13:66–72. doi: 10.3201/eid1301.060315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morris A, Wei K, Afshar K, Huang L. Epidemiology and clinical significance of pneumocystis colonization. J Infect Dis. 2008;197:10–17. doi: 10.1086/523814. [DOI] [PubMed] [Google Scholar]
- 146.Choukri F, Menotti J, Sarfati C, Lucet JC, Nevez G, Garin YJ, Derouin F, Totet A. Quantification and spread of Pneumocystis jirovecii in the surrounding air of patients with Pneumocystis pneumonia. Clin Infect Dis. 2010;51:259–265. doi: 10.1086/653933. [DOI] [PubMed] [Google Scholar]
- 147.Wakefield AE. DNA sequences identical to Pneumocystis carinii f. sp. carinii and Pneumocystis carinii f. sp. hominis in samples of air spora. J Clin Microbiol. 1996;34:1754–1759. doi: 10.1128/jcm.34.7.1754-1759.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wheat LJ, Freifeld AG, Kleiman MB, Baddley JW, McKinsey DS, Loyd JE, Kauffman CA, Infectious Diseases Society of America Clinical practice guidelines for the management of patients with histoplasmosis: 2007 update by the Infectious Diseases Society of America. Clin Infect Dis. 2007;45:807–825. doi: 10.1086/521259. [DOI] [PubMed] [Google Scholar]
- 149.Adenis A, Nacher M, Hanf M, Vantilcke V, Boukhari R, Blachet D, Demar M, Aznar C, Carme B, Couppie P. HIV-associated histoplasmosis early mortality and incidence trends: from neglect to priority. PLoS Negl Trop Dis. 2014;8:e3100. doi: 10.1371/journal.pntd.0003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Adenis AA, Aznar C, Couppie P. Histoplasmosis in HIV-infected patients: a review of new developments and remaining gaps. Curr Trop Med Rep. 2014;1:119–128. doi: 10.1007/s40475-014-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Casadevall A, Pirofski LA. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun. 1999;67:3703–3713. doi: 10.1128/iai.67.8.3703-3713.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Nicholls S, Leach MD, Priest CL, Brown AJ. Role of the heat shock transcription factor, Hsf1, in a major fungal pathogen that is obligately associated with warm-blooded animals. Mol Microbiol. 2009;74:844–861. doi: 10.1111/j.1365-2958.2009.06883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Leach MD, Tyc KM, Brown AJ, Klipp E. Modelling the regulation of thermal adaptation in Candida albicans, a major fungal pathogen of humans. PLoS ONE. 2012;7:e32467. doi: 10.1371/journal.pone.0032467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nicholls S, MacCallum DM, Kaffarnik FA, Selway L, Peck SC, Brown AJ. Activation of the heat shock transcription factor Hsf1 is essential for the full virulence of the fungal pathogen Candida albicans . Fungal Genet Biol. 2011;48:297–305. doi: 10.1016/j.fgb.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Leach MD, Cowen LE. Membrane fluidity and temperature sensing are coupled via circuitry comprised of Ole1, Rsp5, and Hsf1 in Candida albicans . Eukaryot Cell. 2014;13:1077–1084. doi: 10.1128/EC.00138-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.O’Meara T, Cowen LE. Hsp90-dependent regulatory circuitry controlling temperature-dependent fungal development and virulence. Cell Microbiol. 2014;16(4):473–481. doi: 10.1111/cmi.12266. [DOI] [PubMed] [Google Scholar]
- 157.Nichols CB, Perfect ZH, Alspaugh JA. A Ras1-Cdc24 signal transduction pathway mediates thermotolerance in the fungal pathogen Cryptococcus neoformans . Mol Microbiol. 2007;63:1118–1130. doi: 10.1111/j.1365-2958.2006.05566.x. [DOI] [PubMed] [Google Scholar]
- 158.Cheon SA, Jung KW, Chen YL, Heitman J, Bahn YS, Kang HA. Unique evolution of the UPR pathway with a novel bZIP transcription factor, Hxl1, for controlling pathogenicity of Cryptococcus neoformans . PLoS Pathog. 2011;7:e1002177. doi: 10.1371/journal.ppat.1002177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.O’Meara TR, Hay C, Price MS, Giles S, Alspaugh JA. Cryptococcus neoformans histone acetyltransferase Gcn5 regulates fungal adaptation to the host. Eukaryot Cell. 2010;9:1193–1202. doi: 10.1128/EC.00098-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gerik KJ, Bhimireddy SR, Ryerse JS, Specht CA, Lodge JK. PKC1 is essential for protection against both oxidative and nitrosative stresses, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans . Eukaryot Cell. 2008;7:1685–1698. doi: 10.1128/EC.00146-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Hu G, Cheng PY, Sham A, Perfect JR, Kronstad JW. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol. 2008;69:1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Leach M, Cowen L. To sense or die: mechanisms of temperature sensing in fungal pathogens. Curr Fungal Infect Rep. 2014;8:185–191. [Google Scholar]
- 163.Carratu L, Franceschelli S, Pardini CL, Kobayashi GS, Horvath I, Vigh L, Maresca B. Membrane lipid perturbation modifies the set point of the temperature of heat shock response in yeast. Proc Natl Acad Sci USA. 1996;93:3870–3875. doi: 10.1073/pnas.93.9.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Selvig K, Alspaugh JA. pH response pathways in fungi: adapting to host-derived and environmental signals. Mycobiology. 2011;39:249–256. doi: 10.5941/MYCO.2011.39.4.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol. 2004;54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- 166.Nobile CJ, Solis N, Myers CL, Fay AJ, Deneault JS, Nantel A, Mitchell AP, Filler SG. Candida albicans transcription factor Rim101 mediates pathogenic interactions through cell wall functions. Cell Microbiol. 2008;10:2180–2196. doi: 10.1111/j.1462-5822.2008.01198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.De Bernardis F, Muhlschlegel FA, Cassone A, Fonzi WA. The pH of the host niche controls gene expression in and virulence of Candida albicans . Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Davis D, Edwards JE, Jr, Mitchell AP, Ibrahim AS. Candida albicans RIM101 pH response pathway is required for host-pathogen interactions. Infect Immun. 2000;68:5953–5959. doi: 10.1128/iai.68.10.5953-5959.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.O’Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 2010;6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Okagaki LH, Wang Y, Ballou ER, O’Meara TR, Bahn YS, Alspaugh JA, Xue C, Nielsen K. Cryptococcal titan cell formation is regulated by G-protein signaling in response to multiple stimuli. Eukaryot Cell. 2011;10:1306–1316. doi: 10.1128/EC.05179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.O’Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio. 2013;4(1):e00522-12. doi: 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.O’Meara TR, Xu W, Selvig KM, O’Meara MJ, Mitchell AP, Alspaugh JA. The Cryptococcus neoformans Rim101 transcription factor directly regulates genes required for adaptation to the host. Mol Cell Biol. 2014;34:673–684. doi: 10.1128/MCB.01359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. MBio. 2011;2:e00055-11. doi: 10.1128/mBio.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Derengowski Lda S, Paes HC, Albuquerque P, Tavares AH, Fernandes L, Silva-Pereira I, Casadevall A. The transcriptional response of Cryptococcus neoformans to ingestion by Acanthamoeba castellanii and macrophages provides insights into the evolutionary adaptation to the mammalian host. Eukaryot Cell. 2013;12:761–774. doi: 10.1128/EC.00073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Weinberg ED. Iron availability and infection. Biochim Biophys Acta. 2009;1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 177.Tangen KL, Jung WH, Sham AP, Lian T, Kronstad JW. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans . Microbiology. 2007;153:29–41. doi: 10.1099/mic.0.2006/000927-0. [DOI] [PubMed] [Google Scholar]
- 178.Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, Ernst JF. The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun. 2002;70:5246–5255. doi: 10.1128/IAI.70.9.5246-5255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Almeida RS, Wilson D, Hube B. Candida albicans iron acquisition within the host. FEMS Yeast Res. 2009;9:1000–1012. doi: 10.1111/j.1567-1364.2009.00570.x. [DOI] [PubMed] [Google Scholar]
- 180.Jung WH, Saikia S, Hu G, Wang J, Fung CK, D’Souza C, White R, Kronstad JW. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans . PLoS Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Jung WH, Sham A, Lian T, Singh A, Kosman DJ, Kronstad JW. Iron source preference and regulation of iron uptake in Cryptococcus neoformans . PLoS Pathog. 2008;4:e45. doi: 10.1371/journal.ppat.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Jung WH, Hu G, Kuo W, Kronstad JW. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans . Eukaryot Cell. 2009;8:1511–1520. doi: 10.1128/EC.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans . PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Weissman Z, Shemer R, Conibear E, Kornitzer D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans . Mol Microbiol. 2008;69:201–217. doi: 10.1111/j.1365-2958.2008.06277.x. [DOI] [PubMed] [Google Scholar]
- 185.Santos R, Buisson N, Knight S, Dancis A, Camadro JM, Lesuisse E. Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase. Microbiology. 2003;149:579–588. doi: 10.1099/mic.0.26108-0. [DOI] [PubMed] [Google Scholar]
- 186.Navarathna DH, Roberts DD. Candida albicans heme oxygenase and its product CO contribute to pathogenesis of candidemia and alter systemic chemokine and cytokine expression. Free Radic Biol Med. 2010;49:1561–1573. doi: 10.1016/j.freeradbiomed.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Otterbein LE, Bach FH, Alam J, Soares M, Tao LuH, Wysk M, Davis RJ, Flavell RA, Choi AM. Carbon monoxide has anti-inflammatory effects involving the mitogen-activated protein kinase pathway. Nat Med. 2000;6:422–428. doi: 10.1038/74680. [DOI] [PubMed] [Google Scholar]
- 188.Pendrak ML, Chao MP, Yan SS, Roberts DD. Heme oxygenase in Candida albicans is regulated by hemoglobin and is necessary for metabolism of exogenous heme and hemoglobin to alpha-biliverdin. J Biol Chem. 2004;279:3426–3433. doi: 10.1074/jbc.M311550200. [DOI] [PubMed] [Google Scholar]
- 189.Hwang LH, Mayfield JA, Rine J, Sil A. Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog. 2008;4:e1000044. doi: 10.1371/journal.ppat.1000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hilty J, George Smulian A, Newman SL. Histoplasma capsulatum utilizes siderophores for intracellular iron acquisition in macrophages. Med Mycol. 2011;49:633–642. doi: 10.3109/13693786.2011.558930. [DOI] [PubMed] [Google Scholar]
- 191.Chao LY, Marletta MA, Rine J. Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum . Biochemistry. 2008;47:7274–7283. doi: 10.1021/bi800066s. [DOI] [PubMed] [Google Scholar]
- 192.Timmerman MM, Woods JP. Potential role for extracellular glutathione-dependent ferric reductase in utilization of environmental and host ferric compounds by Histoplasma capsulatum . Infect Immun. 2001;69:7671–7678. doi: 10.1128/IAI.69.12.7671-7678.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nemecek JC, Wuthrich M, Klein BS. Global control of dimorphism and virulence in fungi. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 194.Nguyen VQ, Sil A. Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc Natl Acad Sci USA. 2008;105:4880–4885. doi: 10.1073/pnas.0710448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Beyhan S, Gutierrez M, Voorhies M, Sil A. A temperature-responsive network links cell shape and virulence traits in a primary fungal pathogen. PLoS Biol. 2013;11:e1001614. doi: 10.1371/journal.pbio.1001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Warenda AJ, Konopka JB. Septin function in Candida albicans morphogenesis. Mol Biol Cell. 2002;13:2732–2746. doi: 10.1091/mbc.E02-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans . Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 198.Xu XL, Lee RT, Fang HM, Wang YM, Li R, Zou H, Zhu Y, Wang Y. Bacterial peptidoglycan triggers Candida albicans hyphal growth by directly activating the adenylyl cyclase Cyr1p. Cell Host Microbe. 2008;4:28–39. doi: 10.1016/j.chom.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 199.Brown V, Sexton JA, Johnston M. A glucose sensor in Candida albicans . Eukaryot Cell. 2006;5:1726–1737. doi: 10.1128/EC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hudson DA, Sciascia QL, Sanders RJ, Norris GE, Edwards PJ, Sullivan PA, Farley PC. Identification of the dialysable serum inducer of germ-tube formation in Candida albicans . Microbiology. 2004;150:3041–3049. doi: 10.1099/mic.0.27121-0. [DOI] [PubMed] [Google Scholar]
- 201.Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol Mol Biol Rev. 2011;75:213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, Heitman J, Cowen LE. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Curr Biol. 2009;19:621–629. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Bockmuhl DP, Ernst JF. A potential phosphorylation site for an A-type kinase in the Efg1 regulator protein contributes to hyphal morphogenesis of Candida albicans . Genetics. 2001;157:1523–1530. doi: 10.1093/genetics/157.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Cao F, Lane S, Raniga PP, Lu Y, Zhou Z, Ramon K, Chen J, Liu H. The Flo8 transcription factor is essential for hyphal development and virulence in Candida albicans . Mol Biol Cell. 2006;17:295–307. doi: 10.1091/mbc.E05-06-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lu Y, Su C, Liu H. Candida albicans hyphal initiation and elongation. Trends Microbiol. 2014;22:707–714. doi: 10.1016/j.tim.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Dalle F, Wachtler B, L’Ollivier C, Holland G, Bannert N, Wilson D, Labruere C, Bonnin A, Hube B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol. 2010;12:248–271. doi: 10.1111/j.1462-5822.2009.01394.x. [DOI] [PubMed] [Google Scholar]
- 207.Rocha CR, Schroppel K, Harcus D, Marcil A, Dignard D, Taylor BN, Thomas DY, Whiteway M, Leberer E. Signaling through adenylyl cyclase is essential for hyphal growth and virulence in the pathogenic fungus Candida albicans . Mol Biol Cell. 2001;12:3631–3643. doi: 10.1091/mbc.12.11.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Leberer E, Harcus D, Dignard D, Johnson L, Ushinsky S, Thomas DY, Schroppel K. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans . Mol Microbiol. 2001;42:673–687. doi: 10.1046/j.1365-2958.2001.02672.x. [DOI] [PubMed] [Google Scholar]
- 209.Park H, Myers CL, Sheppard DC, Phan QT, Sanchez AA, Edwards JE, Filler SG. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell Microbiol. 2005;7:499–510. doi: 10.1111/j.1462-5822.2004.00476.x. [DOI] [PubMed] [Google Scholar]
- 210.Murad AM, Leng P, Straffon M, Wishart J, Macaskill S, MacCallum D, Schnell N, Talibi D, Marechal D, Tekaia F, d’Enfert C, Gaillardin C, Odds FC, Brown AJ. NRG1 represses yeast-hypha morphogenesis and hypha-specific gene expression in Candida albicans . EMBO J. 2001;20:4742–4752. doi: 10.1093/emboj/20.17.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell. 2003;2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hirakawa MP, Martinez DA, Sakthikumar S, Anderson MZ, Berlin A, Gujja S, Zeng Q, Zisson E, Wang JM, Greenberg JM, Berman J, Bennett RJ, Cuomo CA. Genetic and phenotypic intra-species variation in Candida albicans . Genome Res. 2014 doi: 10.1101/gr.174623.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Vautier S, Drummond RA, Chen K, Murray GI, Kadosh D, Brown AJ, Gow NA, MacCallum DM, Kolls JK, Brown GD. Candida albicans colonization and dissemination from the murine gastrointestinal tract: the influence of morphology and Th17 immunity. Cell Microbiol. 2014 doi: 10.1111/cmi.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Brand A. Hyphal growth in human fungal pathogens and its role in virulence. Int J Microbiol. 2012 doi: 10.1155/2012/517529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell. 2011;10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Staab JF, Bradway SD, Fidel PL, Sundstrom P. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science. 1999;283:1535–1538. doi: 10.1126/science.283.5407.1535. [DOI] [PubMed] [Google Scholar]
- 217.Hube B, Monod M, Schofield DA, Brown AJ, Gow NA. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans . Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 218.White TC, Agabian N. Candida albicans secreted aspartyl proteinases: isoenzyme pattern is determined by cell type, and levels are determined by environmental factors. J Bacteriol. 1995;177:5215–5221. doi: 10.1128/jb.177.18.5215-5221.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Cowen LE. The fungal Achilles’ heel: targeting Hsp90 to cripple fungal pathogens. Curr Opin Microbiol. 2013;16(4):377–384. doi: 10.1016/j.mib.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 220.Prill SK, Klinkert B, Timpel C, Gale CA, Schroppel K, Ernst JF. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol Microbiol. 2005;55:546–560. doi: 10.1111/j.1365-2958.2004.04401.x. [DOI] [PubMed] [Google Scholar]
- 221.Sellam A, Askew C, Epp E, Tebbji F, Mullick A, Whiteway M, Nantel A. Role of transcription factor CaNdt80p in cell separation, hyphal growth, and virulence in Candida albicans . Eukaryot Cell. 2010;9:634–644. doi: 10.1128/EC.00325-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white-opaque switching in Candida albicans . Proc Natl Acad Sci USA. 2006;103:12813–12818. doi: 10.1073/pnas.0605270103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302. doi: 10.1016/s0092-8674(02)00837-1. [DOI] [PubMed] [Google Scholar]
- 224.Forche A, Alby K, Schaefer D, Johnson AD, Berman J, Bennett RJ. The parasexual cycle in Candida albicans provides an alternative pathway to meiosis for the formation of recombinant strains. PLoS Biol. 2008;6:e110. doi: 10.1371/journal.pbio.0060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kvaal C, Lachke SA, Srikantha T, Daniels K, McCoy J, Soll DR. Misexpression of the opaque-phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect Immun. 1999;67:6652–6662. doi: 10.1128/iai.67.12.6652-6662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Si H, Hernday AD, Hirakawa MP, Johnson AD, Bennett RJ. Candida albicans white and opaque cells undergo distinct programs of filamentous growth. PLoS Pathog. 2013;9:e1003210. doi: 10.1371/journal.ppat.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sasse C, Hasenberg M, Weyler M, Gunzer M, Morschhauser J. White-opaque switching of Candida albicans allows immune evasion in an environment-dependent fashion. Eukaryot Cell. 2013;12:50–58. doi: 10.1128/EC.00266-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Kolotila MP, Diamond RD. Effects of neutrophils and in vitro oxidants on survival and phenotypic switching of Candida albicans WO-1. Infect Immun. 1990;58:1174–1179. doi: 10.1128/iai.58.5.1174-1179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Tao L, Du H, Guan G, Dai Y, Nobile CJ, Liang W, Cao C, Zhang Q, Zhong J, Huang G. Discovery of a “white-gray-opaque” tristable phenotypic switching system in Candida albicans: roles of non-genetic diversity in host adaptation. PLoS Biol. 2014;12:e1001830. doi: 10.1371/journal.pbio.1001830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Okagaki LH, Strain AK, Nielsen JN, Charlier C, Baltes NJ, Chretien F, Heitman J, Dromer F, Nielsen K. Cryptococcal cell morphology affects host cell interactions and pathogenicity. PLoS Pathog. 2010;6:e1000953. doi: 10.1371/journal.ppat.1000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Zaragoza O, Garcia-Rodas R, Nosanchuk JD, Cuenca-Estrella M, Rodriguez-Tudela JL, Casadevall A. Fungal cell gigantism during mammalian infection. PLoS Pathog. 2010;6:e1000945. doi: 10.1371/journal.ppat.1000945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zaragoza O, Nielsen K. Titan cells in Cryptococcus neoformans: cells with a giant impact. Curr Opin Microbiol. 2013;16:409–413. doi: 10.1016/j.mib.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Okagaki LH, Nielsen K. Titan cells confer protection from phagocytosis in Cryptococcus neoformans infections. Eukaryot Cell. 2012;11:820–826. doi: 10.1128/EC.00121-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Crabtree JN, Okagaki LH, Wiesner DL, Strain AK, Nielsen JN, Nielsen K. Titan cell production enhances the virulence of Cryptococcus neoformans . Infect Immun. 2012;80:3776–3785. doi: 10.1128/IAI.00507-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kojic EM, Darouiche RO. Candida infections of medical devices. Clin Microbiol Rev. 2004;17:255–267. doi: 10.1128/CMR.17.2.255-267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Dongari-Bagtzoglou A, Kashleva H, Dwivedi P, Diaz P, Vasilakos J. Characterization of mucosal Candida albicans biofilms. PLoS ONE. 2009;4:e7967. doi: 10.1371/journal.pone.0007967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Finkel JS, Mitchell AP. Genetic control of Candida albicans biofilm development. Nat Rev Microbiol. 2011;9:109–118. doi: 10.1038/nrmicro2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Douglas LJ. Candida biofilms and their role in infection. Trends Microbiol. 2003;11:30–36. doi: 10.1016/s0966-842x(02)00002-1. [DOI] [PubMed] [Google Scholar]
- 239.Zarnowski R, Westler WM, Lacmbouh GA, Marita JM, Bothe JR, Bernhardt J, Lounes-Hadj Sahraoui A, Fontaine J, Sanchez H, Hatfield RD, Ntambi JM, Nett JE, Mitchell AP, Andes DR. Novel entries in a fungal biofilm matrix encyclopedia. MBio. 2014;5:e01333-14. doi: 10.1128/mBio.01333-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Taff HT, Nett JE, Zarnowski R, Ross KM, Sanchez H, Cain MT, Hamaker J, Mitchell AP, Andes DR. A Candida biofilm-induced pathway for matrix glucan delivery: implications for drug resistance. PLoS Pathog. 2012;8(8):e1002848. doi: 10.1371/journal.ppat.1002848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Nett J, Lincoln L, Marchillo K, Massey R, Holoyda K, Hoff B, VanHandel M, Andes D. Putative role of β-1,3 glucans in Candida albicans biofilm resistance. Antimicrob Agents Chemother. 2007;51:510–520. doi: 10.1128/AAC.01056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Taff HT, Mitchell KF, Edward JA, Andes DR. Mechanisms of Candida biofilm drug resistance. Future Microbiol. 2013;8:1325–1337. doi: 10.2217/fmb.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Morrow B, Srikantha T, Soll DR. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans . Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67:400–428. doi: 10.1128/MMBR.67.3.400-428.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.de Repentigny L, Aumont F, Bernard K, Belhumeur P. Characterization of binding of Candida albicans to small intestinal mucin and its role in adherence to mucosal epithelial cells. Infect Immun. 2000;68:3172–3179. doi: 10.1128/iai.68.6.3172-3179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Schaller M, Borelli C, Korting HC, Hube B. Hydrolytic enzymes as virulence factors of Candida albicans . Mycoses. 2005;48:365–377. doi: 10.1111/j.1439-0507.2005.01165.x. [DOI] [PubMed] [Google Scholar]
- 247.Leidich SD, Ibrahim AS, Fu Y, Koul A, Jessup C, Vitullo J, Fonzi W, Mirbod F, Nakashima S, Nozawa Y, Ghannoum MA. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans . J Biol Chem. 1998;273:26078–26086. doi: 10.1074/jbc.273.40.26078. [DOI] [PubMed] [Google Scholar]
- 248.Sebghati TS, Engle JT, Goldman WE. Intracellular parasitism by Histoplasma capsulatum: fungal virulence and calcium dependence. Science. 2000;290:1368–1372. doi: 10.1126/science.290.5495.1368. [DOI] [PubMed] [Google Scholar]
- 249.Beck MR, Dekoster GT, Cistola DP, Goldman WE. NMR structure of a fungal virulence factor reveals structural homology with mammalian saposin B. Mol Microbiol. 2009;72:344–353. doi: 10.1111/j.1365-2958.2009.06647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Bohse ML, Woods JP. RNA interference-mediated silencing of the YPS3 gene of Histoplasma capsulatum reveals virulence defects. Infect Immun. 2007;75:2811–2817. doi: 10.1128/IAI.00304-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.O’Meara TR, Alspaugh JA. The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev. 2012;25:387–408. doi: 10.1128/CMR.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Klutts JS, Doering TL. Cryptococcal xylosyltransferase 1 (Cxt1p) from Cryptococcus neoformans plays a direct role in the synthesis of capsule polysaccharides. J Biol Chem. 2008;283:14327–14334. doi: 10.1074/jbc.M708927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kozel TR, Levitz SM, Dromer F, Gates MA, Thorkildson P, Janbon G. Antigenic and biological characteristics of mutant strains of Cryptococcus neoformans lacking capsular O acetylation or xylosyl side chains. Infect Immun. 2003;71:2868–2875. doi: 10.1128/IAI.71.5.2868-2875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Jong AY, Wu C-H, Chen H-M, Luo F, Kwon-Chung KJ, Chang YC, LaMunyon CW, Plaas A, Huang S-H. Identification and characterization of CPS1 as a hyaluronic acid synthase contributing to the pathogenesis of Cryptococcus neoformans infection. Eukaryot Cell. 2007;6(8):1486–1496. doi: 10.1128/EC.00120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.McFadden DC, Fries BC, Wang F, Casadevall A. Capsule structural heterogeneity and antigenic variation in Cryptococcus neoformans . Eukaryot Cell. 2007;6:1464–1473. doi: 10.1128/EC.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Rivera J, Feldmesser M, Cammer M, Casadevall A. Organ-dependent variation of capsule thickness in Cryptococcus neoformans during experimental murine infection. Infect Immun. 1998;66:5027–5030. doi: 10.1128/iai.66.10.5027-5030.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Palmer DA, Thompson JK, Li L, Prat A, Wang P. Gib2, a novel Gβ-like/RACK1 homolog, functions as a Gβ subunit in cAMP signaling and is essential in Cryptococcus neoformans . J Biol Chem. 2006;281:32596–32605. doi: 10.1074/jbc.M602768200. [DOI] [PubMed] [Google Scholar]
- 258.Xue C, Hsueh YP, Chen L, Heitman J. The RGS protein Crg2 regulates both pheromone and cAMP signalling in Cryptococcus neoformans . Mol Microbiol. 2008;70:379–395. doi: 10.1111/j.1365-2958.2008.06417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Xue C, Bahn YS, Cox GM, Heitman J. G protein-coupled receptor Gpr4 senses amino acids and activates the cAMP-PKA pathway in Cryptococcus neoformans . Mol Biol Cell. 2006;17:667–679. doi: 10.1091/mbc.E05-07-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Vartivarian SE, Anaissie EJ, Cowart RE, Sprigg HA, Tingler MJ, Jacobson ES. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis. 1993;167:186–190. doi: 10.1093/infdis/167.1.186. [DOI] [PubMed] [Google Scholar]
- 261.Klengel T, Liang W-J, Chaloupka J, Ruoff C, Schröppel K, Naglik JR, Eckert SE, Mogensen EG, Haynes K, Tuite MF, Levin LR, Buck J, Muhlschlegel FA. Fungal adenylyl cyclase integrates CO2 sensing with cAMP signaling and virulence. Curr Biol. 2005;15:2021–2026. doi: 10.1016/j.cub.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zaragoza O, Casadevall A. Experimental modulation of capsule size in Cryptococcus neoformans . Biol Proced Online. 2004;6:10–15. doi: 10.1251/bpo68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Vecchiarelli A, Pericolini E, Gabrielli E, Kenno S, Perito S, Cenci E, Monari C. Elucidating the immunological function of the Cryptococcus neoformans capsule. Future Microbiol. 2013;8:1107–1116. doi: 10.2217/fmb.13.84. [DOI] [PubMed] [Google Scholar]
- 264.Coelho C, Bocca AL, Casadevall A. The tools for virulence of Cryptococcus neoformans . Adv Appl Microbiol. 2014;87:1–41. doi: 10.1016/B978-0-12-800261-2.00001-3. [DOI] [PubMed] [Google Scholar]
- 265.Rappleye CA, Engle JT, Goldman WE. RNA interference in Histoplasma capsulatum demonstrates a role for α-(1,3)-glucan in virulence. Mol Microbiol. 2004;53:153–165. doi: 10.1111/j.1365-2958.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- 266.Rappleye CA, Eissenberg LG, Goldman WE. Histoplasma capsulatum α-(1,3)-glucan blocks innate immune recognition by the β-glucan receptor. Proc Natl Acad Sci USA. 2007;104:1366–1370. doi: 10.1073/pnas.0609848104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Long KH, Gomez FJ, Morris RE, Newman SL. Identification of heat shock protein 60 as the ligand on Histoplasma capsulatum that mediates binding to CD18 receptors on human macrophages. J Immunol. 2003;170:487–494. doi: 10.4049/jimmunol.170.1.487. [DOI] [PubMed] [Google Scholar]
- 268.Ehlers MR. CR3: a general purpose adhesion-recognition receptor essential for innate immunity. Microbes Infect. 2000;2:289–294. doi: 10.1016/s1286-4579(00)00299-9. [DOI] [PubMed] [Google Scholar]
- 269.Mihu MR, Nosanchuk JD. Histoplasma virulence and host responses. Int J Microbiol. 2012 doi: 10.1155/2012/268123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ma H, Croudace JE, Lammas DA, May RC. Expulsion of live pathogenic yeast by macrophages. Curr Biol. 2006;16:2156–2160. doi: 10.1016/j.cub.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 271.Nicola AM, Robertson EJ, Albuquerque P, Derengowski Lda S, Casadevall A. Nonlytic exocytosis of Cryptococcus neoformans from macrophages occurs in vivo and is influenced by phagosomal pH. MBio. 2011;2:e00167-11. doi: 10.1128/mBio.00167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Alvarez M, Casadevall A. Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol. 2006;16:2161–2165. doi: 10.1016/j.cub.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 273.Bain JM, Lewis LE, Okai B, Quinn J, Gow NA, Erwig LP. Non-lytic expulsion/exocytosis of Candida albicans from macrophages. Fungal Genet Biol. 2012;49:677–678. doi: 10.1016/j.fgb.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Uwamahoro N, Verma-Gaur J, Shen HH, Qu Y, Lewis R, Lu J, Bambery K, Masters SL, Vince JE, Naderer T, Traven A. The pathogen Candida albicans hijacks pyroptosis for escape from macrophages. MBio. 2014;5:e00003–e00014. doi: 10.1128/mBio.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Wellington M, Koselny K, Sutterwala FS, Krysan DJ. Candida albicans triggers NLRP3-mediated pyroptosis in macrophages. Eukaryot Cell. 2014;13:329–340. doi: 10.1128/EC.00336-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hromatka BS, Noble SM, Johnson AD. Transcriptional response of Candida albicans to nitric oxide and the role of the YHB1 gene in nitrosative stress and virulence. Mol Biol Cell. 2005;16:4814–4826. doi: 10.1091/mbc.E05-05-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Ullmann BD, Myers H, Chiranand W, Lazzell AL, Zhao Q, Vega LA, Lopez-Ribot JL, Gardner PR, Gustin MC. Inducible defense mechanism against nitric oxide in Candida albicans . Eukaryot Cell. 2004;3:715–723. doi: 10.1128/EC.3.3.715-723.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.de Jesus-Berrios M, Liu L, Nussbaum JC, Cox GM, Stamler JS, Heitman J. Enzymes that counteract nitrosative stress promote fungal virulence. Curr Biol. 2003;13:1963–1968. doi: 10.1016/j.cub.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 279.Missall TA, Pusateri ME, Donlin MJ, Chambers KT, Corbett JA, Lodge JK. Posttranslational, translational, and transcriptional responses to nitric oxide stress in Cryptococcus neoformans: implications for virulence. Eukaryot Cell. 2006;5:518–529. doi: 10.1128/EC.5.3.518-529.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Nittler MP, Hocking-Murray D, Foo CK, Sil A. Identification of Histoplasma capsulatum transcripts induced in response to reactive nitrogen species. Mol Biol Cell. 2005;16:4792–4813. doi: 10.1091/mbc.E05-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Nosanchuk JD, Valadon P, Feldmesser M, Casadevall A. Melanization of Cryptococcus neoformans in murine infection. Mol Cell Biol. 1999;19:745–750. doi: 10.1128/mcb.19.1.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Huffnagle GB, Chen GH, Curtis JL, McDonald RA, Strieter RM, Toews GB. Down-regulation of the afferent phase of T cell-mediated pulmonary inflammation and immunity by a high melanin-producing strain of Cryptococcus neoformans . J Immunol. 1995;155:3507–3516. [PubMed] [Google Scholar]
- 283.Casadevall A, Rosas AL, Nosanchuk JD. Melanin and virulence in Cryptococcus neoformans . Curr Opin Microbiol. 2000;3:354–358. doi: 10.1016/s1369-5274(00)00103-x. [DOI] [PubMed] [Google Scholar]
- 284.Rutherford JC. The emerging role of urease as a general microbial virulence factor. PLoS Pathog. 2014;10:e1004062. doi: 10.1371/journal.ppat.1004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Olszewski MA, Noverr MC, Chen GH, Toews GB, Cox GM, Perfect JR, Huffnagle GB. Urease expression by Cryptococcus neoformans promotes microvascular sequestration, thereby enhancing central nervous system invasion. Am J Pathol. 2004;164:1761–1771. doi: 10.1016/S0002-9440(10)63734-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infect Immun. 2000;68:443–448. doi: 10.1128/iai.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Singh A, Panting RJ, Varma A, Saijo T, Waldron KJ, Jong A, Ngamskulrungroj P, Chang YC, Rutherford JC, Kwon-Chung KJ. Factors required for activation of urease as a virulence determinant in Cryptococcus neoformans . MBio. 2013;4:e00220-13. doi: 10.1128/mBio.00220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Osterholzer JJ, Surana R, Milam JE, Montano GT, Chen GH, Sonstein J, Curtis JL, Huffnagle GB, Toews GB, Olszewski MA. Cryptococcal urease promotes the accumulation of immature dendritic cells and a non-protective T2 immune response within the lung. Am J Pathol. 2009;174:932–943. doi: 10.2353/ajpath.2009.080673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Navarathna DH, Das A, Morschhauser J, Nickerson KW, Roberts DD. Dur3 is the major urea transporter in Candida albicans and is co-regulated with the urea amidolyase Dur1,2. Microbiology. 2011;157:270–279. doi: 10.1099/mic.0.045005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Navarathna DH, Lionakis MS, Lizak MJ, Munasinghe J, Nickerson KW, Roberts DD. Urea amidolyase (DUR1,2) contributes to virulence and kidney pathogenesis of Candida albicans . PLoS ONE. 2012;7:e48475. doi: 10.1371/journal.pone.0048475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Ghosh S, Navarathna DH, Roberts DD, Cooper JT, Atkin AL, Petro TM, Nickerson KW. Arginine-induced germ tube formation in Candida albicans is essential for escape from murine macrophage line RAW 264.7. Infect Immun. 2009;77:1596–1605. doi: 10.1128/IAI.01452-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2011;11:275–288. doi: 10.1038/nri2939. [DOI] [PubMed] [Google Scholar]
- 293.Netea MG, Brown GD, Kullberg BJ, Gow NA. An integrated model of the recognition of Candida albicans by the innate immune system. Nat Rev Microbiol. 2008;6:67–78. doi: 10.1038/nrmicro1815. [DOI] [PubMed] [Google Scholar]
- 294.Romani L. Immunity to fungal infections. Nat Rev Immunol. 2004;4:1–23. doi: 10.1038/nri1255. [DOI] [PubMed] [Google Scholar]
- 295.Latge JP. Tasting the fungal cell wall. Cell Microbiol. 2010;12:863–872. doi: 10.1111/j.1462-5822.2010.01474.x. [DOI] [PubMed] [Google Scholar]
- 296.Lionakis MS, Netea MG. Candida and host determinants of susceptibility to invasive candidiasis. PLoS Pathog. 2013;9:e1003079. doi: 10.1371/journal.ppat.1003079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Gow N, Netea M, Munro CA, Ferwerda G, Bates S, Mora-Montes H, Walker LA, Jansen T, Jacobs L, Tsoni V, Brown G, Odds FC, Van der Meer JW, Brown A, Kullberg BJ. Immune recognition of Candida albicans β-glucan by dectin 1. J Infect Dis. 2007;196(10):1565–1571. doi: 10.1086/523110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Netea MG, Gow NA, Munro CA, Bates S, Collins C, Ferwerda G, Hobson RP, Bertram G, Hughes HB, Jansen T, Jacobs L, Buurman ET, Gijzen K, Williams DL, Torensma R, McKinnon A, MacCallum DM, Odds FC, Van der Meer JW, Brown AJ, Kullberg BJ. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and toll-like receptors. J Clin Invest. 2006;116:1642–1650. doi: 10.1172/JCI27114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Dan JM, Wang JP, Lee CK, Levitz SM. Cooperative stimulation of dendritic cells by Cryptococcus neoformans mannoproteins and CpG oligodeoxynucleotides. PLoS ONE. 2008;3(4):e2046. doi: 10.1371/journal.pone.0002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Brown GD. Innate antifungal immunity: the key role of phagocytes. Annu Rev Immunol. 2011;29:1–21. doi: 10.1146/annurev-immunol-030409-101229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Bhatt VR, Viola GM, Ferrajoli A. Invasive fungal infections in acute leukemia. Ther Adv Hematol. 2011;2:231–247. doi: 10.1177/2040620711410098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Lionakis MS, Fischer BG, Lim JK, Swamydas M, Wan W, Richard Lee CC, Cohen JI, Scheinberg P, Gao JL, Murphy PM. Chemokine receptor Ccr1 drives neutrophil-mediated kidney immunopathology and mortality in invasive candidiasis. PLoS Pathog. 2012;8:e1002865. doi: 10.1371/journal.ppat.1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.van de Veerdonk FL, Netea MG. T-cell subsets and antifungal host defenses. Curr Fungal Infect Rep. 2010;4:238–243. doi: 10.1007/s12281-010-0034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Armstrong-James D, Meintjes G, Brown GD. A neglected epidemic: fungal infections in HIV/AIDS. Trends Microbiol. 2014;22:120–127. doi: 10.1016/j.tim.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 305.Pensieroso S, Galli L, Nozza S, Ruffin N, Castagna A, Tambussi G, Hejdeman B, Misciagna D, Riva A, Malnati M, Chiodi F, Scarlatti G. B-cell subset alterations and correlated factors in HIV-1 infection. AIDS. 2013;27:1209–1217. doi: 10.1097/QAD.0b013e32835edc47. [DOI] [PubMed] [Google Scholar]
- 306.Collini P, Noursadeghi M, Sabroe I, Miller RF, Dockrell DH. Monocyte and macrophage dysfunction as a cause of HIV-1 induced dysfunction of innate immunity. Curr Mol Med. 2010;10:727–740. doi: 10.2174/156652410793384141. [DOI] [PubMed] [Google Scholar]
- 307.Olszewski MA, Huffnagle GB, McDonald RA, Lindell DM, Moore BB, Cook DN, Toews GB. The role of macrophage inflammatory protein-1 α/CCL3 in regulation of T cell-mediated immunity to Cryptococcus neoformans infection. J Immunol. 2000;165:6429–6436. doi: 10.4049/jimmunol.165.11.6429. [DOI] [PubMed] [Google Scholar]
- 308.Wormley FL, Jr, Perfect JR, Steele C, Cox GM. Protection against cryptococcosis by using a murine gamma interferon-producing Cryptococcus neoformans strain. Infect Immun. 2007;75:1453–1462. doi: 10.1128/IAI.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Hole CR, Wormley FL., Jr Vaccine and immunotherapeutic approaches for the prevention of cryptococcosis: lessons learned from animal models. Front Microbiol. 2012;3:291. doi: 10.3389/fmicb.2012.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Conti HR, Shen F, Nayyar N, Stocum E, Sun JN, Lindemann MJ, Ho AW, Hai JH, Yu JJ, Jung JW, Filler SG, Masso-Welch P, Edgerton M, Gaffen SL. Th17 cells and IL-17 receptor signaling are essential for mucosal host defense against oral candidiasis. J Exp Med. 2009;206:299–311. doi: 10.1084/jem.20081463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Bixler SL, Mattapallil JJ. Loss and dysregulation of Th17 cells during HIV infection. Clin Dev Immunol. 2013;2013:852418. doi: 10.1155/2013/852418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Rudner XL, Happel KI, Young EA, Shellito JE. Interleukin-23 (IL-23)-IL-17 cytokine axis in murine Pneumocystis carinii infection. Infect Immun. 2007;75:3055–3061. doi: 10.1128/IAI.01329-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Netea MG, Sutmuller R, Hermann C, Van der Graaf CA, Van der Meer JW, van Krieken JH, Hartung T, Adema G, Kullberg BJ. Toll-like receptor 2 suppresses immunity against Candida albicans through induction of IL-10 and regulatory T cells. J Immunol. 2004;172:3712–3718. doi: 10.4049/jimmunol.172.6.3712. [DOI] [PubMed] [Google Scholar]
- 314.Kroetz DN, Deepe GS., Jr CCR5 dictates the equilibrium of proinflammatory IL-17+ and regulatory Foxp3+ T cells in fungal infection. J Immunol. 2010;184:5224–5231. doi: 10.4049/jimmunol.1000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Gildea LA, Gibbons R, Finkelman FD, Deepe GS., Jr Overexpression of interleukin-4 in lungs of mice impairs elimination of Histoplasma capsulatum . Infect Immun. 2003;71:3787–3793. doi: 10.1128/IAI.71.7.3787-3793.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Cenci E, Mencacci A, Del Sero G, Bacci A, Montagnoli C, d’Ostiani CF, Mosci P, Bachmann M, Bistoni F, Kopf M, Romani L. Interleukin-4 causes susceptibility to invasive pulmonary aspergillosis through suppression of protective type I responses. J Infect Dis. 1999;180:1957–1968. doi: 10.1086/315142. [DOI] [PubMed] [Google Scholar]
- 317.Myers RC, Dunaway CW, Nelson MP, Trevor JL, Morris A, Steele C. STAT4-dependent and -independent Th2 responses correlate with protective immunity against lung infection with Pneumocystis murina . J Immunol. 2013;190:6287–6294. doi: 10.4049/jimmunol.1300431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Bicanic T, Meintjes G, Rebe K, Williams A, Loyse A, Wood R, Hayes M, Jaffar S, Harrison TS. Immune reconstitution inflammatory syndrome in HIV-associated cryptococcal meningitis: a prospective study. J Acquir Immune Defic Syndr. 2009;51(2):130–134. doi: 10.1097/QAI.0b013e3181a56f2e. [DOI] [PubMed] [Google Scholar]
- 319.Perfect JR. The impact of the host on fungal infections. Am J Med. 2012;125:S39–S51. doi: 10.1016/j.amjmed.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 320.Boulware DR, Meya DB, Bergemann TL, Wiesner DL, Rhein J, Musubire A, Lee SJ, Kambugu A, Janoff EN, Bohjanen PR. Clinical features and serum biomarkers in HIV immune reconstitution inflammatory syndrome after cryptococcal meningitis: a prospective cohort study. PLoS Med. 2010;7:e1000384. doi: 10.1371/journal.pmed.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Lionakis MS. Genetic susceptibility to fungal infections in humans. Curr Fungal Infect Rep. 2012;6:11–22. doi: 10.1007/s12281-011-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, Venselaar H, Elbers CC, Johnson MD, Cambi A, Huysamen C, Jacobs L, Jansen T, Verheijen K, Masthoff L, Morre SA, Vriend G, Williams DL, Perfect JR, Joosten LA, Wijmenga C, van der Meer JW, Adema GJ, Kullberg BJ, Brown GD, Netea MG. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med. 2009;361:1760–1767. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Plantinga TS, van der Velden WJ, Ferwerda B, van Spriel AB, Adema G, Feuth T, Donnelly JP, Brown GD, Kullberg BJ, Blijlevens NM, Netea MG. Early stop polymorphism in human DECTIN-1 is associated with increased Candida colonization in hematopoietic stem cell transplant recipients. Clin Infect Dis. 2009;49:724–732. doi: 10.1086/604714. [DOI] [PubMed] [Google Scholar]
- 324.Glocker EO, Hennigs A, Nabavi M, Schaffer AA, Woellner C, Salzer U, Pfeifer D, Veelken H, Warnatz K, Tahami F, Jamal S, Manguiat A, Rezaei N, Amirzargar AA, Plebani A, Hannesschlager N, Gross O, Ruland J, Grimbacher B. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009;361:1727–1735. doi: 10.1056/NEJMoa0810719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Liu L, Okada S, Kong XF, Kreins AY, Cypowyj S, Abhyankar A, Toubiana J, Itan Y, Audry M, Nitschke P, Masson C, Toth B, Flatot J, Migaud M, Chrabieh M, Kochetkov T, Bolze A, Borghesi A, Toulon A, Hiller J, Eyerich S, Eyerich K, Gulacsy V, Chernyshova L, Chernyshov V, Bondarenko A, Grimaldo RM, Blancas-Galicia L, Beas IM, Roesler J, Magdorf K, Engelhard D, Thumerelle C, Burgel PR, Hoernes M, Drexel B, Seger R, Kusuma T, Jansson AF, Sawalle-Belohradsky J, Belohradsky B, Jouanguy E, Bustamante J, Bue M, Karin N, Wildbaum G, Bodemer C, Lortholary O, Fischer A, Blanche S, Al-Muhsen S, Reichenbach J, Kobayashi M, Rosales FE, Lozano CT, Kilic SS, Oleastro M, Etzioni A, Traidl-Hoffmann C, Renner ED, Abel L, Picard C, Marodi L, Boisson-Dupuis S, Puel A, Casanova JL. Gain-of-function human STAT1 mutations impair IL-17 immunity and underlie chronic mucocutaneous candidiasis. J Exp Med. 2011;208:1635–1648. doi: 10.1084/jem.20110958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.van de Veerdonk FL, Plantinga TS, Hoischen A, Smeekens SP, Joosten LA, Gilissen C, Arts P, Rosentul DC, Carmichael AJ, Smits-van der Graaf CA, Kullberg BJ, van der Meer JW, Lilic D, Veltman JA, Netea MG. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011;365:54–61. doi: 10.1056/NEJMoa1100102. [DOI] [PubMed] [Google Scholar]
- 327.Puel A, Cypowyj S, Bustamante J, Wright JF, Liu L, Lim HK, Migaud M, Israel L, Chrabieh M, Audry M, Gumbleton M, Toulon A, Bodemer C, El-Baghdadi J, Whitters M, Paradis T, Brooks J, Collins M, Wolfman NM, Al-Muhsen S, Galicchio M, Abel L, Picard C, Casanova JL. Chronic mucocutaneous candidiasis in humans with inborn errors of interleukin-17 immunity. Science. 2011;332:65–68. doi: 10.1126/science.1200439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Kagami S, Rizzo HL, Kurtz SE, Miller LS, Blauvelt A. IL-23 and IL-17A, but not IL-12 and IL-22, are required for optimal skin host defense against Candida albicans . J Immunol. 2010;185:5453–5462. doi: 10.4049/jimmunol.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Minegishi Y, Saito M, Tsuchiya S, Tsuge I, Takada H, Hara T, Kawamura N, Ariga T, Pasic S, Stojkovic O, Metin A, Karasuyama H. Dominant-negative mutations in the DNA-binding domain of STAT3 cause hyper-IgE syndrome. Nature. 2007;448:1058–1062. doi: 10.1038/nature06096. [DOI] [PubMed] [Google Scholar]
- 330.Milner JD, Brenchley JM, Laurence A, Freeman AF, Hill BJ, Elias KM, Kanno Y, Spalding C, Elloumi HZ, Paulson ML, Davis J, Hsu A, Asher AI, O’Shea J, Holland SM, Paul WE, Douek DC. Impaired T(H)17 cell differentiation in subjects with autosomal dominant hyper-IgE syndrome. Nature. 2008;452:773–776. doi: 10.1038/nature06764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Engelhardt KR, McGhee S, Winkler S, Sassi A, Woellner C, Lopez-Herrera G, Chen A, Kim HS, Lloret MG, Schulze I, Ehl S, Thiel J, Pfeifer D, Veelken H, Niehues T, Siepermann K, Weinspach S, Reisli I, Keles S, Genel F, Kutukculer N, Camcioglu Y, Somer A, Karakoc-Aydiner E, Barlan I, Gennery A, Metin A, Degerliyurt A, Pietrogrande MC, Yeganeh M, Baz Z, Al-Tamemi S, Klein C, Puck JM, Holland SM, McCabe ER, Grimbacher B, Chatila TA. Large deletions and point mutations involving the dedicator of cytokinesis 8 (DOCK8) in the autosomal-recessive form of hyper-IgE syndrome. J Allergy Clin Immunol. 2009;124:1289–1302. doi: 10.1016/j.jaci.2009.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Mathis D, Benoist C. Aire. Annu Rev Immunol. 2009;27:287–312. doi: 10.1146/annurev.immunol.25.022106.141532. [DOI] [PubMed] [Google Scholar]
- 333.Kisand K, Boe Wolff AS, Podkrajsek KT, Tserel L, Link M, Kisand KV, Ersvaer E, Perheentupa J, Erichsen MM, Bratanic N, Meloni A, Cetani F, Perniola R, Ergun-Longmire B, Maclaren N, Krohn KJ, Pura M, Schalke B, Strobel P, Leite MI, Battelino T, Husebye ES, Peterson P, Willcox N, Meager A. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010;207:299–308. doi: 10.1084/jem.20091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Segal BH, Leto TL, Gallin JI, Malech HL, Holland SM. Genetic, biochemical, and clinical features of chronic granulomatous disease. Medicine (Baltimore) 2000;79:170–200. doi: 10.1097/00005792-200005000-00004. [DOI] [PubMed] [Google Scholar]
- 335.Winkelstein JA, Marino MC, Johnston RB, Jr, Boyle J, Curnutte J, Gallin JI, Malech HL, Holland SM, Ochs H, Quie P, Buckley RH, Foster CB, Chanock SJ, Dickler H. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 2000;79:155–169. doi: 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- 336.Aratani Y, Kura F, Watanabe H, Akagawa H, Takano Y, Suzuki K, Dinauer MC, Maeda N, Koyama H. Critical role of myeloperoxidase and nicotinamide adenine dinucleotide phosphate-oxidase in high-burden systemic infection of mice with Candida albicans . J Infect Dis. 2002;185:1833–1837. doi: 10.1086/340635. [DOI] [PubMed] [Google Scholar]
- 337.Cech P, Papathanassiou A, Boreux G, Roth P, Miescher PA. Hereditary myeloperoxidase deficiency. Blood. 1979;53:403–411. [PubMed] [Google Scholar]
- 338.Vinh DC, Patel SY, Uzel G, Anderson VL, Freeman AF, Olivier KN, Spalding C, Hughes S, Pittaluga S, Raffeld M, Sorbara LR, Elloumi HZ, Kuhns DB, Turner ML, Cowen EW, Fink D, Long-Priel D, Hsu AP, Ding L, Paulson ML, Whitney AR, Sampaio EP, Frucht DM, DeLeo FR, Holland SM. Autosomal dominant and sporadic monocytopenia with susceptibility to mycobacteria, fungi, papillomaviruses, and myelodysplasia. Blood. 2010;115:1519–1529. doi: 10.1182/blood-2009-03-208629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Arvanitis M, Anagnostou T, Fuchs BB, Caliendo AM, Mylonakis E. Molecular and nonmolecular diagnostic methods for invasive fungal infections. Clin Microbiol Rev. 2014;27:490–526. doi: 10.1128/CMR.00091-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Richardson MD, Warnock DW. Fungal infection diagnosis and management. 4. Chichester, West Sussex, UK: Wiley-Blackwell; 2012. [Google Scholar]
- 341.Nguyen MH, Wissel MC, Shields RK, Salomoni MA, Hao B, Press EG, Shields RM, Cheng S, Mitsani D, Vadnerkar A, Silveira FP, Kleiboeker SB, Clancy CJ. Performance of Candida real-time polymerase chain reaction, β-d-glucan assay, and blood cultures in the diagnosis of invasive candidiasis. Clin Infect Dis. 2012;54:1240–1248. doi: 10.1093/cid/cis200. [DOI] [PubMed] [Google Scholar]
- 342.Denning DW, Hope WW. Therapy for fungal diseases: opportunities and priorities. Trends Microbiol. 2010;18:195–204. doi: 10.1016/j.tim.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 343.Gomez-Lopez A, Zaragoza O, Rodriguez-Tudela JL, Cuenca-Estrella M. Pharmacotherapy of yeast infections. Expert Opin Pharmacother. 2008;9:2801–2816. doi: 10.1517/14656566.9.16.2801. [DOI] [PubMed] [Google Scholar]
- 344.Ghannoum MA, Rice LB. Antifungal agents: mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin Microbiol Rev. 1999;12:501–517. doi: 10.1128/cmr.12.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Masia Canuto M, Gutierrez Rodero F. Antifungal drug resistance to azoles and polyenes. Lancet Infect Dis. 2002;2:550–563. doi: 10.1016/s1473-3099(02)00371-7. [DOI] [PubMed] [Google Scholar]
- 346.Deray G. Amphotericin B nephrotoxicity. J Antimicrob Chemother. 2002;49(Suppl 1):37–41. doi: 10.1093/jac/49.suppl_1.37. [DOI] [PubMed] [Google Scholar]
- 347.Dupont B. Overview of the lipid formulations of amphotericin B. J Antimicrob Chemother. 2002;49(Suppl 1):31–36. doi: 10.1093/jac/49.suppl_1.31. [DOI] [PubMed] [Google Scholar]
- 348.Wong-Beringer A, Jacobs RA, Guglielmo BJ. Lipid formulations of amphotericin B: clinical efficacy and toxicities. Clin Infect Dis. 1998;27:603–618. doi: 10.1086/514704. [DOI] [PubMed] [Google Scholar]
- 349.Vincent BM, Lancaster AK, Scherz-Shouval R, Whitesell L, Lindquist S. Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 2013;11:e1001692. doi: 10.1371/journal.pbio.1001692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Ellis D. Amphotericin B: spectrum and resistance. J Antimicrob Chemother. 2002;49(Suppl 1):7–10. doi: 10.1093/jac/49.suppl_1.7. [DOI] [PubMed] [Google Scholar]
- 351.Sheehan DJ, Hitchcock CA, Sibley CM. Current and emerging azole antifungal agents. Clin Microbiol Rev. 1999;12:40–79. doi: 10.1128/cmr.12.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Walsh TJ, Anaissie EJ, Denning DW, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Segal BH, Steinbach WJ, Stevens DA, van Burik JA, Wingard JR, Patterson TF, Infectious Diseases Society of America Treatment of aspergillosis: clinical practice guidelines of the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:327–360. doi: 10.1086/525258. [DOI] [PubMed] [Google Scholar]
- 353.Pfaller MA. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am J Med. 2012;125:S3–S13. doi: 10.1016/j.amjmed.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 354.Pound MW, Townsend ML, Drew RH. Echinocandin pharmacodynamics: review and clinical implications. J Antimicrob Chemother. 2010;65:1108–1118. doi: 10.1093/jac/dkq081. [DOI] [PubMed] [Google Scholar]
- 355.Denning DW. Echinocandin antifungal drugs. Lancet. 2003;362:1142–1151. doi: 10.1016/S0140-6736(03)14472-8. [DOI] [PubMed] [Google Scholar]
- 356.Holt SL, Drew RH. Echinocandins: addressing outstanding questions surrounding treatment of invasive fungal infections. Am J Health Syst Pharm. 2011;68:1207–1220. doi: 10.2146/ajhp100456. [DOI] [PubMed] [Google Scholar]
- 357.Arendrup MC, Perlin DS. Echinocandin resistance: an emerging clinical problem? Curr Opin Infect Dis. 2014;27:484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Nanjappa SG, Klein BS. Vaccine immunity against fungal infections. Curr Opin Immunol. 2014;28:27–33. doi: 10.1016/j.coi.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Spellberg B. Vaccines for invasive fungal infections. F1000 Med Rep. 2011;3:13. doi: 10.3410/M3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.NovaDigm Therapeutics, Inc (2013) Safety, tolerability, immunogenicity and efficacy of NDV-3A vaccine in preventing recurrent vulvovaginal candidiasis. Identifier: NCT01926028
- 361.Schmidt CS, White CJ, Ibrahim AS, Filler SG, Fu Y, Yeaman MR, Edwards JE, Jr, Hennessey JP., Jr NDV-3, a recombinant alum-adjuvanted vaccine for Candida and Staphylococcus aureus, is safe and immunogenic in healthy adults. Vaccine. 2012;30:7594–7600. doi: 10.1016/j.vaccine.2012.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.De Bernardis F, Amacker M, Arancia S, Sandini S, Gremion C, Zurbriggen R, Moser C, Cassone A. A virosomal vaccine against candidal vaginitis: immunogenicity, efficacy and safety profile in animal models. Vaccine. 2012;30:4490–4498. doi: 10.1016/j.vaccine.2012.04.069. [DOI] [PubMed] [Google Scholar]