Abstract
Genome organization into linear chromosomes likely represents an important evolutionary innovation that has permitted the development of the sexual life cycle; this process has consequently advanced nuclear expansion and increased complexity of eukaryotic genomes. Chromosome linearity, however, poses a major challenge to the internal cellular machinery. The need to efficiently recognize and repair DNA double-strand breaks that occur as a consequence of DNA damage presents a constant threat to native chromosome ends known as telomeres. In this review, we present a comparative survey of various solutions to the end protection problem, maintaining an emphasis on DNA structure. This begins with telomeric structures derived from a subset of prokaryotes, mitochondria, and viruses, and will progress into the typical telomere structure exhibited by higher organisms containing TTAGG-like tandem sequences. We next examine non-canonical telomeres from Drosophila melanogaster, which comprise arrays of retrotransposons. Finally, we discuss telomeric structures in evolution and possible switches between canonical and non-canonical solutions to chromosome end protection.
Keywords: Telomeres, Telomerase, Chromosomes, Genome evolution, DNA repair, Retrotransposons
Introduction
With no exception, eukaryotic nuclear genomes are organized into a variable number of chromosomes each consisting of a single linear DNA molecule. Linear genome organization may relate back to primordial protogenes that were likely formed by short RNA/DNA chains [1, 2]. In contrast, the genomes of all archaebacteria and the majority of bacteria are retained in circular DNA replicons. This difference in genome configuration may reflect distinct evolutionary strategies of prokaryotic and eukaryotic lineages. While the r-selection strategy in prokaryotes favors miniaturization and genomic streamlining, K-selection in ancestral eukaryotic lineages may have promoted a sexual life cycle featuring alteration of diploid and haploid cell generations. This represents an important evolutionary innovation as it permitted genomic expansion and an increase in organismal complexity. Circular chromosome topology is incompatible with the mode of chromosome recombination and segregation as we know it in eukaryotic meiosis. An odd number of crossovers between two circular chromosomes would yield unstable dicentric concatenates (dimers); this is not a problem with linear chromosomes where the resolution of crossovers always gives rise to two separate entities. Hence, linear genome organization is a prerequisite of meiosis and a defining feature of eukaryotic life. It also eliminates formation of dicentric chromosomes by DNA repair reactions that lead to sister chromatid exchanges in mitotic cells [3].
Linear chromosomes are not exclusive to eukaryotic nuclear genomes and have been found to be present in a number of viruses [4]; linear DNA forms were also detected in a subset of bacteria, plant plastids, and mitochondria, where they likely arose secondarily from a circular ancestral state [5, 6]. The evolutionary benefit of genome linearity in these settings is still not clear; one line of thought suggests that linear chromosomes may improve evolvability of an organism. Genetic loci located in the vicinity of natural chromosome ends are prone to frequent structural rearrangements and represent the most rapidly evolving parts of a genome. It has been proposed that this inherent instability of linear chromosome ends may be of an adaptive value as it may promote rapid evolution of new gene variants and increase survival in rapidly changing environments [7, 8].
While genome organization in the form of linear DNA molecules conveys, at least in the case of nuclear genomes of eukaryotes, undisputable evolutionary benefits, it also poses two major challenges for cellular metabolism that require unique solutions. First, the conventional DNA replication machinery lacks a mechanism to fill in the gap left by removal of the final RNA primer in the most terminal Okazaki fragment; it is therefore unable to fully replicate 3′ ends of linear chromosomes [9, 10]. This process has been dubbed “the end replication problem” and ultimately leads to the gradual attrition of telomeric DNA over multiple cell divisions. Telomere shortening beyond a threshold length causes telomere dysfunction and eventually leads to chromosome fusions and subsequent cell death.
The second challenge originates in the formal similarity between the natural chromosome ends (telomeres) and DNA double-strand breaks (DSBs) that arise as a consequence of DNA damage. Breakage of chromosomal DNA creates a serious threat to genome integrity and eukaryotic cells possess elaborate mechanisms to efficiently detect and repair DSBs [11]. Broken DNA triggers very strong cellular responses that on one hand activate DNA repair mechanisms and on the other hand signal the presence of DNA damage to the cell cycle machinery. This may lead to a temporal cell cycle arrest or even to activation of specialized programs such as cellular senescence or apoptosis. Such a response to exposed DNA ends is not inherent only to eukaryotes. DNA damage or replication stress prevents cell division in bacteria [12] and broken ends of chloroplast DNA are subject to rapid degradation and end-to-end fusions [13]. Thus, masking natural chromosome ends from eliciting DNA damage response and aberrant DNA repair is absolutely imperative for stability of nuclear as well as organellar genomes and for long-term cell survival.
Formation of telomeres from linearized chromosomes: prokaryotes, mitochondria, and viruses
The term “telomere” is often used exclusively to describe the specialized nucleoprotein structures that are found at the end of linear, nuclear chromosomes. However, telomeres are also present at linear, double-stranded molecules of several viruses, bacteria, and mitochondrial (mt) genomes. It was previously accepted that mitochondrial, as well as bacterial genomes, comprise solely of circular dsDNA. This central dogma was challenged over 40 years ago when it was shown that mitochondria genomes of the ciliate Tetrahymena pyriformis harbor exclusively linear DNA molecules [14]. Around 20 years after this discovery, the first prokaryotic linear chromosomes were also identified in the bacterium Borrelia burgdorferi [15]. Viral DNA molecules with telomeres could also be identified among poxviruses, adenoviruses, herpes viruses, and numerous bacteriophages (reviewed in [4, 16]). The presence of such genomes could represent an evolutionary snapshot of alternative solutions to the end protection and replication problems besides classical telomeres exhibited by the majority of eukaryotes.
While chromosome linearization is observed within bacteria, it is clearly not the most prevalent system of genome organization. Linear bacterial chromosomes have only been described in some eubacteria including Agrobacterium tumefaciens and a few members of the genus Borrelia as well as the order Actinomycetales (reviewed in [16–18]). Therefore, linear genomes are clearly the exception in eubacteria. In contrast, linearization of mitochondrial DNA is more common and widely spread across the phylogenetic tree of eukaryotes [19]. Linear mitochondrial genomes are found in many species from various taxonomical groups including algae, ciliates, apicomplexan protozoa, jakobids, slime molds, oomycetes, fungi, cnidarians, isopods, and Scombridae [20–22]. It is noteworthy that in some species, the mt genome is segmented into multiple linear molecules [5, 23, 24]; linear mt DNA can even co-exist with circular mt DNA within a single cell [21, 25].
Telomeres found in viruses, bacteria, and mitochondria are structurally distinct from their counterparts in higher organisms (see Fig. 1 for overview). However, they must still fulfill similar functions to ensure genome stability. Despite their conserved function, several distinct telomeric structures have been found, highlighting the development of several successful replication and end-protection strategies during evolution. It is, however, striking that similar telomere structures appear to arise in phylogenetically unrelated organisms. In the following section, we will summarize telomere structures that have been described so far in viruses, bacteria, and mitochondria, while maintaining focus on their protective functions. For deeper insights into the proposed replication strategies, see other excellent reviews on this topic [20, 26].
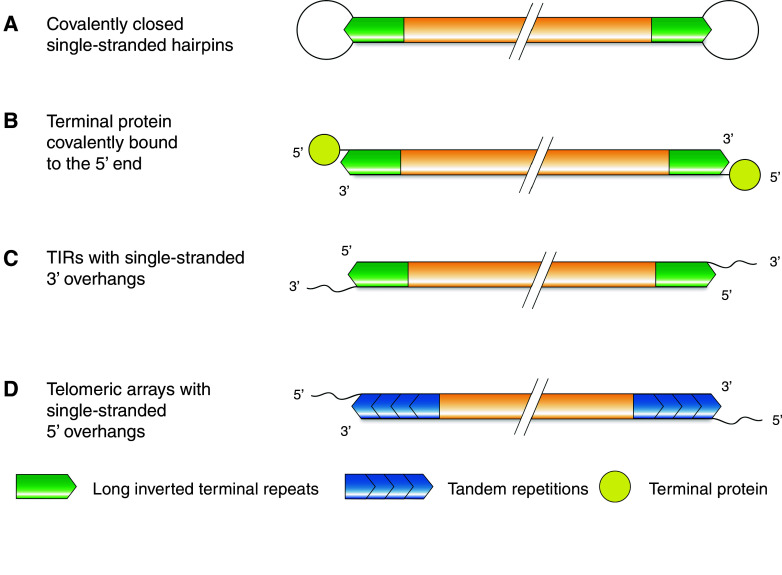
Fig. 1.
Telomeric structures from prokaryotes, mitochondria, and viruses. The presence of covalently closed hairpin structures at terminal regions has been observed within B. burgdorferi, A. tumefaciens, and the Vaccinia virus (VACV) (a). 5′ overhangs of the human Ad2/5 adenovirus are bound by a terminal protein (TP) (b). Mitochondrial telomeres from the green algae C. reinhardtii comprise long inverted terminal repeat regions with a 3′ overhang (c) whereas C. parapsilosis exhibits tandem repeats with 5′ overhangs (d)
Covalently closed single-stranded hairpins
The Vaccinia virus (VACV), a member of the poxvirus family, consists of a large dsDNA molecule (~200 kb), which terminates in a covalently closed, single-stranded hairpin on either side (Fig. 1a). This terminal hairpin is composed of a single-stranded loop and an A-T-rich stem flanked by an inverted repeat [27, 28]. Due to the presence of covalently closed hairpins, the ends of the viral genome are effectively hidden from DNA repair activities and exonucleolytic attacks. The terminal hairpins also allow for the complete replication of the viral genome within the host [29]. A similar telomere structure has also been found in several bacteriophages like the Escherichia coli phage N15 [30] and the Klebsiella oxytoca phage ΦKO2 [31] as well as in all members of the bacterial Borrelia spirochete group [32] and in Agrobacterium tumefaciens C58 [33].
Borrelia burgdorferi, the bacterial agent of Lyme disease in humans, has a segmented genome that consists of one linear chromosome (~900 kb) along with 12 linear and ten circular plasmids [34]. All linear dsDNA molecules of B. burgdorferi terminate with covalently closed hairpins flanked by inverted repeats. The replication of the linear chromosome is driven by an internal origin toward the terminus, whereas the terminal hairpin enables the complete replication of the genome since the replication fork can pass through the loop sequence. After fusion of the lagging and leading strand, the duplicated telomeres are converted on each side into terminal hairpins. This step is mediated by the telomere resolvase ResT, which cuts and re-ligates the telomere to form new covalently closed hairpins at chromosome termini [34, 35]. A similar enzyme can also be found in many organisms harboring linear double-stranded molecules with terminal hairpins such as the plant pathogen A. tumefaciens C58.
The genome of A. tumefaciens C58 comprises both a circular and linear chromosome; the enzyme responsible for the formation of new terminal hairpins is known as protelomerase TelA [36]. Besides its essential function in telomere replication, the Agrobacterium protelomerase TelA may also exhibit an additional role in telomere protection. Huang et al. [36] showed that TelA can efficiently bind to Agrobacterium terminal hairpins in vitro. This contrasts to TelK, the protelomerase of K. oxytoca phage ΦKO2, which is quickly released from the newly formed telomeric hairpins after replication. It has been suggested that TelA protects the telomeres, but its modification during chromosome replication promotes hairpin cleavage and religation. This leads to a switch from the potentially protective function towards a role in telomere replication. Telomeres protected by covalently closed hairpins have also been found in yeast mitochondria of Pichia pijperii, Williopsis mrakii [37], and in several Candida species like C. viswanathii and C. frijolesensis [5].
Terminal protein covalently bound to the 5′ end
A second type of telomere structure is found in several adenoviruses such as the human Ad2/5. The genome of Ad2/5 comprises a small dsDNA molecule (36 kb) with termini consisting of long ~100 nt terminal inverted repeats (TIRs) harboring the replication origins [38]. Each 5′ end of the molecule is covalently bound by a terminal protein (TP) (Fig. 1b) [39]. The 55-kDa TP of Ad2/5 protects the viral genome from exonuclease activity and also serves as a primer during replication. To initiate DNA replication, the viral DNA polymerase forms a heterodimer with TP and couples the first deoxynucleotide (dNTP) to the terminal protein. This “protein priming” replication strategy allows the complete duplication of the viral genome [38]. Linear double-stranded chromosomes with terminal proteins covalently bound to the 5′ end have also been found in several bacteriophages like the Bacillus subtilis phage Φ29 [40] and the E. coli phage PRD1 [41], as well as in bacteria of the genus Streptomyces [42]. It is noteworthy that two of these Streptomyces strains contain a functional promoter in their telomeric region. The role of this promoter region has not been resolved so far. However, it has been proposed that the promoter may be involved in priming of DNA replication during conjugation through priming of the last Okazaki fragment or in transcription of downstream genes [43].
The protection of linear replicons against nucleolytic resection in several Streptomyces species is mediated by the terminal protein Tpg [44, 45]. However, other factors seem to be additionally involved in protecting the ends. During replication, a 3′ overhang is created at the termini of the linear double-stranded molecules. This single-stranded protrusion contains several palindromic sequences and can fold into a complex secondary structure made out of several hairpins that are closed by a YGNAR sequence (N being mostly C). The G and A residues of the hairpins have the potential to form “sheared purine–purine pairing” giving rise to a single nucleotide loop that may protect the overhang from nucleolytic resection [46]. The linear mitochondrial genome of the yeast Candida subhashtii also ends with a long terminal inverted repeat and a terminal protein covalently bound to the 5′ terminus [47].
Terminal inverted repeats with single-stranded 3′ overhangs
The mitochondrial telomeres of the green algae Chlamydomonas reinhardtii also contain terminal inverted repeats. However, in contrast to the previously described structure, the TIRs in C. reinhardtii terminate in 3′ single-stranded overhangs (Fig. 1c) with an identical sequence [48]. Since the 3′ overhangs are non-complementary, the possibility that the linear genome can circularize should be excluded [48]. The mechanism of how these ends are protected against nucleases and the DNA repair machinery is unknown.
Telomeric arrays with single-stranded 5′ overhangs
Candida parapsilosis harbors arrays of tandemly repeated sequences (738 bp) at the ends of their linear mt chromosomes. The number of tandem repeats can vary ranging from one incomplete repeat to at least eight repeat units. All molecules end with an incomplete repeat forming a 5′ single-stranded protrusion (Fig. 1d) [49]. The 5′ overhang is approximately 110 nt in size and can be bound by the mitochondrial telomere-binding protein mtTBP. It has been shown that mtTBP protects an oligonucleotide identical to the last 51 nucleotides of the 5′ overhang from the action of several DNA modifying enzymes in vitro [50]. Therefore, mtTBP is proposed to be involved in the protection of mitochondrial telomeres from nucleases and other DNA repair activities. mtTBP shares homology with mitochondrial and bacterial single-strand binding proteins (SSB). While mtTBP has a binding preference for the telomeric single-stranded (ss) 5′ overhang, it can also associate with other ssDNA substrates [51]. Hence, in addition to its telomeric function, mtTBP may also play a role outside of telomeres. Another end protection mechanism ascribed to the presence of ss 5′ DNA protrusions is the formation of T-loops. These lariat structures formed by invasion of ssDNA into the duplex telomeric region were for the first time observed at canonical telomeres in mammals [52]. Nevertheless, they were detected by electron microscopy at the ends of mt telomeres in C. parapsilosis [53]. Nuclear T-loops are proposed to mediate protection of linear chromosomes [52, 54]. This may also apply to the mitochondrial telomeres of C. parapsilosis where they may, together with mtTBP, protect the termini of linear mt DNA from nucleases and the DNA repair machinery [55].
Other telomeric structures
Further telomeric structures that consist of direct repeats, complex repetitions, or a combination of the former discussed structures, have been found at the ends of linear mt DNA molecules and some viruses of the herpes family [56–59]. For example, mitochondria of the fungus Fusarium oxysporum exhibit linear plasmids that terminate with an array of small tandem repeats at one telomere, and a hairpin at the other terminus [59]. Furthermore, the linear mt plasmid (mF) of the slime mold Physarum polycephalum ends with TIRs comprised of complex repetitive motifs and potentially a protein component, which attaches to its 5′ termini [58].
The Epstein–Barr virus (EBV) carries a dsDNA genome that terminates with direct tandem repeats [60]. It seems that, in this case, the left terminus is blunt ended, while the right terminus has a 3′ extension consisting of a single base [61]. However, since the viral genome circularizes after entry into the host cell, the direct terminal repeats are deemed important for the circularization itself. This is thought to allow for packing of the virion DNA and occasional integration into the host DNA rather than for protecting the ends of the linear DNA molecule [61–63]. This also appears to be true for the human herpes virus 6 (HHV-6) as both sides of the linear HHV-6 genome terminate in identical direct repeats containing a TTAGGG sequence motif corresponding to the human telomeric repeat unit [64, 65]. Since the TTAGGG repeat array is several hundred nucleotides away from the actual terminus, it is unlikely that the virus uses the host telomerase to fully replicate its extrachromosomal genome [66]. In addition, it has been shown that the human telomere repeat array is not needed for the stable nuclear retention of the non-integrated HHV-6 genome [67]. The TTAGGG repeat array is rather important for the integration of the viral DNA into the host telomeric region during latency [68]. Furthermore, Arbuckle and Medveczky [69] hypothesize that the shelterin components TRF1/TRF2 may bind to the TTAGGG repeat array and help to mediate the integration of HHV-6 into the host telomere. Through the integration of HHV-6 into the host genome, the viral DNA can be efficiently maintained and mobilized after dormancy [70]. The integration into the host telomere also protects the viral genome from nucleases and the DNA repair machinery.
Canonical TTAGG telomeres
Since the discovery of TTGGGG repeats at chromosome ends in the ciliated protozoan Tetrahymena thermophila [71], the presence of G-rich sequences at telomeres has indicated a common sequence theme at chromosome ends over most eukaryotic organisms. Ten years after this discovery, the human telomeric sequence was cloned and found to comprise a very similar sequence of TTAGGG repeats [72]. Variations on the TTAGG telomeric sequence motive are found in many model species across the entire eukaryotic phylogenetic tree including mouse [73], zebrafish [74], Caenorhabditis elegans [75], fission and budding yeast [76–78], Arabidopsis thaliana [79], and C. reinhardtii [80] (for extensive list see http://telomerase.asu.edu/sequences.html). Due to their sheer abundance in the natural world, TTAGG-type sequences are therefore considered to represent canonical telomeres. While this telomere sequence is highly conserved, the length of the telomeric array is known to vary dramatically throughout nature; telomeres in budding yeast tend to average ~350 bp [81], whereas telomeres from higher eukaryotes can range from 2–15 kb in humans [82] to 150 kb in tobacco [83].
Telomere attrition by the end replication problem is, in the majority of eukaryotes, solved by telomerase, a reverse transcriptase that mediates de novo synthesis of telomeric DNA. First discovered in Tetrahymena [84], telomerase utilizes an internal RNA subunit (TER) as a template for elongation of telomeres by the catalytic reverse transcriptase (TERT) (reviewed in [85]). Recruitment of telomerase to telomere ends, however, needs to be maintained in a controlled manner to coordinate telomere synthesis with genome replication. For detailed discussion on telomere replication by telomerase, we refer to numerous reviews that extensively cover this topic [85–87]. In the following section, we will focus mainly on describing mechanisms employed for protecting canonical telomeres. We maintain a particular emphasis on the structural features of telomeric DNA and how they predetermine chromosome end protection mechanisms.
T-Loops
Eukaryotic chromosomes usually terminate with 3′ single-stranded DNA protrusions known as G-overhangs, which are formed by removal of the final RNA primer during DNA synthesis of the lagging strand. G-overhangs are believed to play a fundamental function in telomere protection through sequestering the exposed DNA end in the form of a T-loop. This blocks accessibility of DNA repair proteins, exonucleases, and the actions of telomerase at chromosome termini (Fig. 2a). T-loops have been observed by electron microscopy in a number of organisms including human, chicken, mouse, and pea [52, 88, 89]. The T-loop structure, however, has to exist in a dynamic state and must be constantly disbanded and re-established through numerous cell divisions and rounds of replication. RTEL1 was recently found to be essential for the disassembly of T-loops during DNA replication [90], therefore formation of a new T-loop must begin after replication in the late S-G2 phase of the cell cycle. It is known that DNA repair proteins play an essential role in recognition of newly replicated telomeres and the establishment of a new telomere structure [91–93].
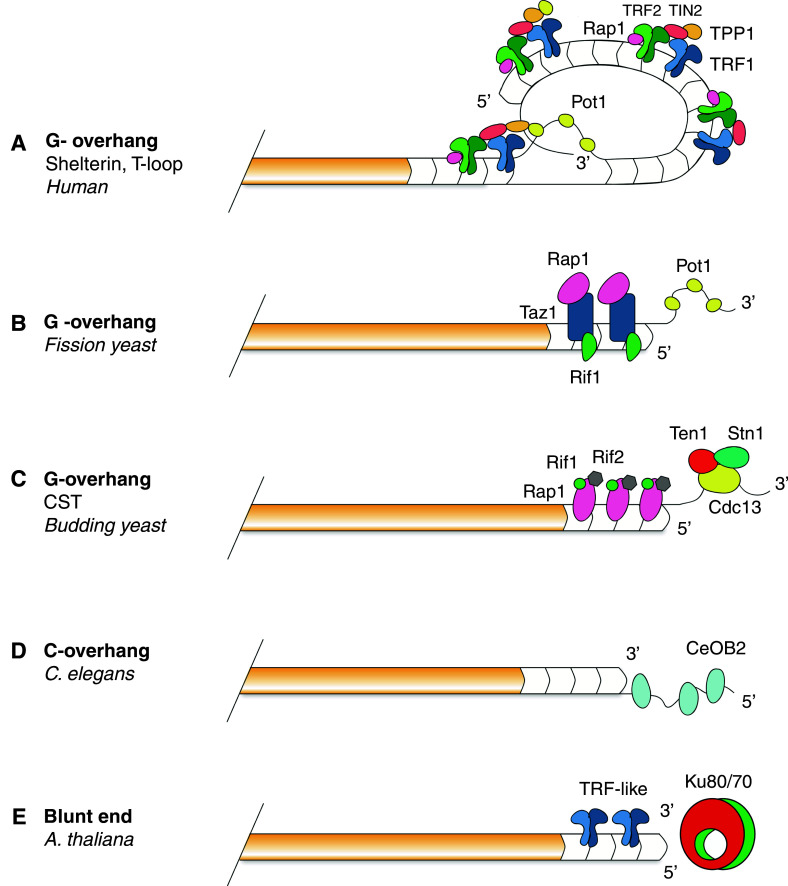
Fig. 2.
Canonical telomeres exhibiting TTAGG-like sequences. The shelterin complex is responsible for sequestration of the G-overhang into duplex double-stranded telomeric DNA (a). Once hidden, the G-overhang is no longer susceptible to DNA repair processes or the actions of telomerase. A similar complex is also found in fission yeast with the conserved functions of Pot1 (b). Budding yeast telomeres are protected by the CST (Cdc13/Stn1/Ten1) complex which associates with the G-overhang (c). C. elegans telomeres contain 5′ overhangs that are bound by CeOB2 (d). Blunt-ended telomeres present at a subset of A. thaliana chromosome ends are protected by the Ku heterodimer (e)
The nature of utilizing DNA repair proteins to establish protective caps at chromosome ends seems paradoxical given their main function in end joining and recombination, processes that need to be inhibited at natural chromosome ends. Key players in these responses include the MRN (Mre11/Rad50/Nbs1) complex, which is essential for the recognition of DNA breaks and is required for activation of DNA repair processes via key signaling intermediates ATM and ATR (reviewed in [94]). The MRN complex was initially found to co-localize with telomeres during S phase [93]. Analysis of the dynamics of Mre11 and Nbs1 at telomeres during the cell cycle also showed a distinct peak at early G2 phase [92]. The MRN complex was also shown to play an essential role in the processing and formation of G-overhangs in yeast and human cells [91, 95]. This suggests telomeres are recognized as unrepaired DNA ends directly after replication and are processed by the MRN complex to form an appropriate platform for T-loop formation.
T-loop formation and stabilization is to a large extent facilitated by the Shelterin complex. Shelterin is the major telomere-associated complex in mammals that consists of six proteins: TRF1 and TRF2, POT1, TIN2, TPP1, and the human homolog of yeast telomeric protein RAP1 (Fig. 2a). TRF1 and TRF2 both interact directly with double-stranded telomeric repeats via a C-terminal Myb/SANT-type domain [96–98]. Pot1 interacts with the single-stranded overhang via two oligosaccharide/oligonucleotide binding (OB) folds [99]. Connection between the DNA-interacting proteins is facilitated by TIN2, which stabilizes TRF1 and TRF2 on duplex telomeric DNA [100]; TPP1 is then responsible for an interaction between TIN2 and Pot1 [100–102]. Rap1 is known to interact directly with TRF2 although its function is not telomere-specific [103].
The DNA remodeling properties of TRF2 are able to mediate the formation of T-loop structures at chromosome ends. Incubation of recombinant TRF2 with a short telomeric fragment in vitro (also processed with a 5′ exonuclease to generate a 150–200 nt 3′ overhang) showed TRF2 was able to form lasso-like structures at terminal ends [52]. TRF2 was also found to localize to T-loop junctions, indicating a role in stabilizing the T-loop structure [104, 105]. Characterization of the basic domain of TRF2 has shown specific binding to four-way DNA junctions in vitro, although this binding was not telomere sequence-specific [106]. TRF1 has also shown to display DNA bending properties [107], although it was not shown to influence telomere loop formation in the same manner as TRF2 [52]. Proteins responsible for repair of DNA via homologous recombination (HR) were found to play a role in mediating the invasion of the single-stranded overhang into duplex telomeric DNA [108]. An in vitro assay to monitor incorporation of a labeled telomeric fragment exhibiting a 3′ overhang into a plasmid containing telomeric repeats showed RAD51 and RAD52 were essential for strand invasion.
The current model for T-loop formation therefore suggests that telomeres are recognized by DNA repair machinery immediately after DNA replication and are also accessible to DNA modification [92]. Exonucleases such as Mre11 are then able to play a part in processing the C-rich strand to generate sufficient 3′ overhangs at telomeres; shelterin then associates to initiate DNA looping, and finally HR proteins RAD51 and RAD52 mediate invasion of the single-stand overhang completing T-loop formation.
G-overhang: protection by OB fold proteins
Telomeres in some organisms such as Saccharomyces cerevisiae and Schizosaccharomyces pombe apparently do not form T-loops. This is either due to their short length (minichromosomes in ciliates), or to irregular telomeric sequences that may impede efficient homology-driven invasion of the G-overhang into the duplex telomere (S. cerevisiae and S. pombe). Chromosome end protection in these organisms is largely mediated by proteins that tightly bind to G-overhangs using the OB fold [109]. The archetype for OB-fold protein-mediated telomere protection is the heterodimeric telomere end-binding complex (TEBP) from the ciliate Oxytricha nova. TEBP consists of α and β subunits and is known to bind to the two repeats of a single-stranded TTTTGGGG sequence that forms G-overhangs on macronuclear minichromosomes [110]. The crystal structure of OnTEBP in association with ssDNA revealed that 3′ end of G-overhang is buried within the complex establishing a physical basis for chromosome end protection [111].
A protein related to TEBPα has been identified by homology searches in S. pombe. This protein binds ss telomeric DNA and was named protection of telomeres 1 (Pot1), as its inactivation in fission yeast leads to rapid telomere degradation and chromosome end-to-end fusions (Fig. 2b) [112]. Pot1 is an evolutionary conserved subunit of the shelterin complex and its deletion in vertebrates elicits acute DNA damage responses at telomeres [113–115]. It has been suggested that Pot1 outcompetes RPA, the major eukaryotic ss DNA binding complex that acts as the mediator of DNA damage activation for binding to telomeric ssDNA during telomere processing and replication [116].
Telomere integrity in budding yeast relies on yet another set of OB-fold proteins that form the heterotrimeric Cdc13/Stn1/Ten1 (CST) complex (Fig. 2c). Although Cdc13 specifically binds to yeast ss telomeric DNA through its OB fold, it is not related to TEBP/Pot1. It has been proposed that CST rather represents a telomere-specific RPA-like particle [109, 117]. The CST complex is essential in yeast and its function impacts several aspects of telomere maintenance. On one hand, Cdc13 is central for telomere replication as it interacts with the telomerase subunit Est1 and the catalytic subunit of polymerase α, and hence promotes elongation of 3′ chromosome end by telomerase and synthesis of the complementary DNA by lagging strand mechanism [118]. On the other hand, CST mediates telomere protection from nucleolytic degradation and DNA damage checkpoint activation [119, 120]. A complex analogous to the yeast CST has recently been discovered in plants and mammals, although its function in telomere protection appears to be less critical than in yeast [121–123]. Although telomere function is impaired in Arabidopsis stn1/ctc1 null mutants, plants are still viable and fertile [122, 123]. A series of recent studies performed in human and mouse cell lines indicate that mammalian CST may be primarily involved in facilitating replication of telomeres and not directly in chromosome end protection [124–126].
C-overhangs
Although chromosomes in most eukaryotes terminate with 3′ G-overhangs, telomeres containing ss 5′ C-rich overhangs were described to co-exist with G-overhang containing telomeres in C. elegans [127]. Two OB fold proteins, CeOB1 and CeOB2, were described to bind specifically to each overhang (Fig. 2d). CeOB1 binds to the G-overhang and its deletion causes elongation of telomeres and extension of G-overhangs. CeOB2 binds C-overhangs and its deficiency leads to telomere length heterogeneity and increased telomeric recombination [127, 128]. This data indicates that these proteins are involved in telomere protection. It has been suggested that 5′ C-overhangs are capable of forming T-loops. CeOB proteins may therefore protect chromosome termini either via direct sequestering of ss telomeric DNA, or through promoting T-loop formation and stability. A low abundance of C-overhangs has also been detected in human and mouse differentially terminated or G1/S arrested cells. Furthermore, they were found to be more prevalent in tumor cells exhibiting alternative telomere lengthening [129]. In such cells, C-overhangs may be products of T-loop resolution events that lead to rapid telomere deletions [130].
Blunt ends: role of the Ku complex
Sequestering G-overhangs into the T-loop structure provides an ideal way to hide exposed chromosome ends. G-overhangs, however, arise after processing of the last Okazaki fragment only on telomeres replicated by the lagging strand mechanism; telomeres synthesized by the leading strand mechanism are expected to be fully replicated producing a blunt DNA end. Despite this, G-overhangs have been detected at both leading and lagging strands in yeast and mammals [131, 132]. In mammalian cells, telomeres replicated by the leading strand mechanism are known to be processed by exonucleases in order to generate a new G-overhang capable of forming a T-loop. Apollo was the first 5′ exonuclease suggested to play such a role at telomeres due to interaction and co-localization with TRF2 [133]. The role of Apollo specifically at leading end telomeres was confirmed through examination of G-overhang abundance in mouse embryo fibroblasts with a conditional knockout of Apollo, which leads to a 30–40 % decrease in frequency [134]. However, Apollo is not the only player in this process and more recently it was found that POT1b, CST, and the 5′ exonuclease Exo1 also contribute to resection of the C-rich strand [135]. The model proposed by Wu et al. [135] suggests that TRF2 is responsible for recruitment of Apollo to leading end telomeres to perform 5′ resection shortly after replication. After resection, Pot1b associates with the single-stranded overhang, inhibiting extensive resection by Apollo. Exo1 can also transiently resect 5′ ends after Pot1b is recruited on both leading and lagging telomeres, the CST complex is then thought to control the correction of over-resected telomeres through an interaction with Pot1b.
Plant telomeres are also known to contain G-overhangs and are capable of forming T-loops [88, 136]. Interestingly, processing of leading strand telomeres does not seem to occur in angiosperm plants. G-overhangs were detected only on ~50 % of telomeres in Silene latifolia and A. thaliana, suggesting that the remaining telomeres are blunt-ended [136]. Recently, the presence of telomeric blunt ends was confirmed in A. thaliana, S. latifolia, and maize [137]. This provides evidence that telomere end protection may also be carried out in the absence of G-overhangs, an idea far removed from traditional telomeric structures. Interestingly, the integrity of the blunt ends requires presence of the Ku heterodimer, a complex that is normally involved in the activation of non-homologous end joining DNA repair (Fig. 2e). This ambiguity leads us to question why a protein complex usually involved in the initial steps of NHEJ-mediated DNA repair acts in protection of chromosome ends. One theory is that Ku associates with exposed blunt ends by default, but is restricted in activating DNA repair specifically at telomeres, potentially via telomere binding proteins. Mechanisms, by which Ku can isolate blunt ends from resection, and how blunt ends are protected from DNA damage responses, are currently unknown.
Noncanonical nuclear telomeres in eukaryotes
Retrotransposons/terminin
The discovery of telomeres in Drosophila by Muller [138] first highlighted the presence of protective caps at natural chromosome ends to distinguish them from X-ray-induced DNA breaks. However, in contrast to canonical TTAGG-like telomeres, Drosophila telomeres differ considerably in both their structure and maintenance processes. One striking difference is that Drosophila telomeric DNA does not contain canonical TTAGG-like repeats, but instead consists of tandem arrangements of three non-LTR (long terminal repeat) retrotransposons dubbed HeT-A, TART, and TAHRE [139–142]. HeT-A is the most abundant of this group and contains a single open reading frame (ORF) that encodes for a GAG-like protein [143–145]. TART and TAHRE also contain a GAG-like ORF along with a region encoding a reverse transcriptase (RT) (Fig. 3a).
Fig. 3.
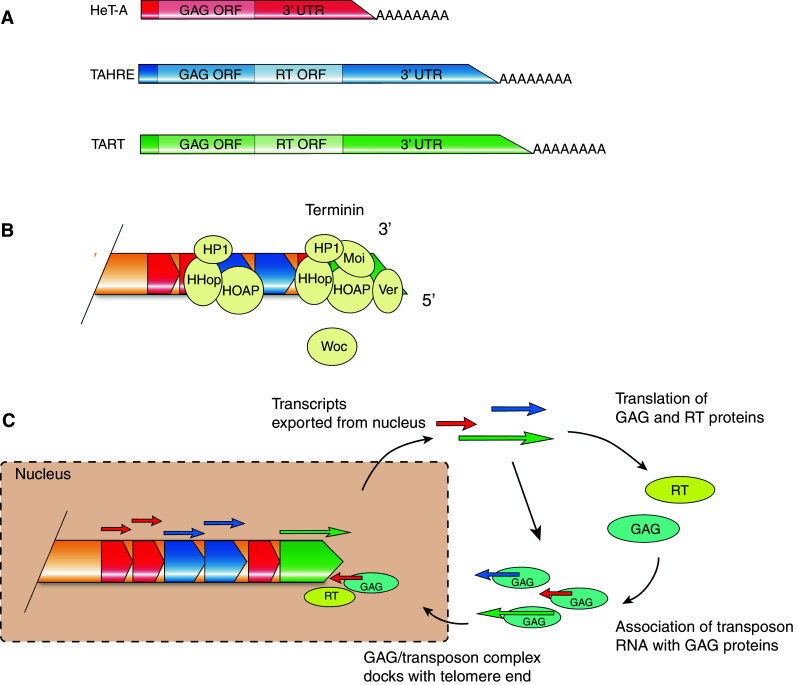
Telomeric retrotransposons in Drosophila melanogaster. Telomeric DNA from D. melanogaster comprises tandem repeats of retrotransposons HeT-A, TAHRE, and TART (a). Telomeric retrotransposons are capped by the terminin complex (b, figure adapted from Raffa et al. [159]). Telomeric retrotransposition results in telomere elongation (c). Transcripts are exported from the nucleus where GAG and RT proteins are translated. These proteins are then responsible for localization of transcripts to chromosome termini where they contribute to telomere elongation
In addition to lacking TTAGG-type telomeric sequences, telomerase activity is also absent from Drosophila [146]. Telomeric DNA is maintained at a long length in comparison to human telomeres, bringing about the question of how Drosophila counteracts the end replication problem. Drosophila instead relies on transposition of telomeric retrotransposons to chromosome ends in order to target attrition of telomeric DNA [147, 148]. This mechanism is thought to have arisen from a recent evolutionary loss of telomerase providing a new means of telomere maintenance and protection [149].
Telomere maintenance in Drosophila
At canonical telomeres, telomerase functions in the de novo elongation of telomeric DNA. The model for chromosome elongation in Drosophila is, however, dependent on the transcription and transposition of telomeric retrotransposons within the telomere to the chromosome terminus. Initial transcription of HeT-A and TAHRE is not initiated within the 5′ region, but is instead initiated from the 3′ UTR of the upstream element [150]. There are also several promoters for TART that function on both strands [151]. In the current model, as illustrated in Fig. 3c, transcripts are exported to the cytoplasm where GAG-like and RT proteins are translated. GAG proteins then bind to these transcripts to facilitate re-entry into the nucleus where they are directed to chromosome ends [143–145]. Transcripts then localize with the terminal chromosome end; this is possibly mediated by an interaction between GAG and capping proteins [152]. The poly(A) tail of the Het-A element is then thought to associate with the chromosome terminus where it is used as a template for reverse transcription. This creates a DNA overhang at the telomere in a similar way as described for telomerase [153]. Synthesis of the second strand completes the addition of a new retrotransposon to the telomere end. Addition of a single retrotransposon would presumably lead to substantial elongation of the telomere as telomeric retrotransposons range in size from 6 to 13 kb [152]. This suggests telomeric retrotransposition is a relatively rare event, otherwise massive overextension would occur.
Telomere capping in Drosophila
One surprising factor of chromosome end protection in Drosophila is that telomeric retrotransposons are not required for recruitment of capping factors. In TTAGG-like telomeres, sequence-specific binding of telomere binding proteins facilitates the protective capping of chromosome ends. This does not appear to be the case in Drosophila where studies have shown that loss of HTT repeats does not affect viability for a few generations [154, 155]. Stocks containing terminal telomere deletions still continued to show evidence of protected ends which were also transmitted to future generations [156, 157]. In fact, population studies have similarly shown a loss of telomeric DNA in natural populations [158]. Eventually, it was discovered that a capping complex known as terminin protects chromosome ends in Drosophila in a sequence-independent manner (for extensive review see [159]).
Terminin consists of the core telomere binding and associated proteins HP1, HOAP, HipHop, Modigliani, and Verrocchio [160–164] (Fig. 3b). Heterochromatin Protein 1 (HP1) was initially identified due to its localization to heterochromatin, although enrichment of HP1 was found to occur at telomeres [161]. HOAP was then found to be the first DNA binding component of the terminin complex essential for chromosome end protection. HOAP associates with HP1 to form the core capping complex [160, 165]. HipHop was later identified through affinity co-immunopurification with HOAP protein and was found to interact with both HP1 and HOAP [162]. Moi and Ver are recruited to telomeres through an interaction with HOAP and form a stable component of the terminin complex [163, 164, 166]. HOAP, HipHop, and HP1 have the ability to bind to the ends of terminally deleted chromosomes demonstrating sequence-unspecific protection [160–162, 167]. Together, these proteins form a complex reminiscent of that of the shelterin complex found in mammalian telomeres containing TTAGG repeats.
Additional proteins are known to associate with Drosophila telomeres, although they seem to play a more general role in genome maintenance. The first identified protein thought to be involved in telomere protection in Drosophila was UbcD1, an E2 ubiquitin ligase [168]. Mutants of UbcD1 were shown to display telomeric fusions. UbcD1 was therefore suggested to be involved in the ubiquitination of telomeric proteins; telomeric targets of UbcD1 ubiquitination still remain to be identified. HP1 is also not considered as solid member of terminin as it displays functions outside of telomere maintenance. Mutations in the without children (woc) gene, thought to encode for a putative transcription factor, also result in telomeric fusions [169]. Woc was found to localize to many euchromatic sites within the genome as well as telomeric DNA. However, Woc functions independently of HP1-HOAP and is not important for HP1 and HOAP localization to telomeres. Therefore, due to colocalization and association with Pol II, it was proposed that Woc acts as a transcription factor closely involved with Pol II-dependent transcription. The role of Woc at telomeres, however, is not understood. It was suggested that Woc could control transcription of telomere binding proteins or play a direct role in contributing to the structure of telomeric DNA.
Functions of DNA repair proteins in establishing the terminin complex
As shown from other studies, the actions of DNA repair proteins in late G2 phase of the cell cycle is important to establish functional telomere cap [91–93]. This also holds true in Drosophila where components of the MRN complex are important for recruitment of telomere capping factors. Mutants defective in components of the MRN complex have exhibited extensive chromosomal fusions, mitotic bridges, and reduced association of HP1 and HOAP at chromosome ends [170–174]. Single ATM knockouts (but not ATR single mutants) have also shown extensive end-to-end fusions, but recruitment of HOAP/HP1 and overall abundance of HeT-A repeats was unaffected [171]. In a similar study, ATM mutants were found to display reduced HP1 localization to telomeres [174]. Analysis of single and double ATM/ATR mutants, however, revealed functional redundancy between ATM and ATR [170]. While single ATM and ATR mutations have almost no effect on HOAP localization to telomeres, HOAP is completely absent at telomeres in double mutants. Because of the lack of a telomere-related phenotype in ATR single mutants, and a strong fusion phenotype in ATM single knockouts, it is thought ATM plays a primary role in recruitment of telomere capping factors and ATR only complements ATM activity when necessary.
Silkworm TTAGG-type telomeres are interspersed with retrotransposons TRAS1 and SART1
Silkworm telomeres were originally found to exhibit canonical TTAGG-type telomeres. Telomere restriction fragment analysis (TRF) confirmed that extreme ends contain approximately 6–8 kb of TTAGGn sequences; in situ hybridization also showed localization of TTAGG25 probes to chromosome ends [175]. During this study, analysis of the telomeric sequence indicated presence of a poly(A) tract; this was eventually shown to comprise the non-LTR retrotransposon TRAS1. TRAS1 was later found to be interspersed within silkworm TTAGG telomeric sequences [176]. Another family of non-LTR retrotransposons, SART1, was also found to integrate into TTAGG although in the opposite orientation to TRAS1 [177].
In Drosophila, retrotransposons contribute to telomere length elongation through transposition to chromosome ends. Although these elements are also found within silkworm telomeres, they are not similarly transported to chromosome ends, but instead insert between specific nucleotides of the TTAGG repeats. Retrotransposons that insert into specific sequences contain self-encoded endonuclease domains to create a nick in target sequences; isolation and characterization of the TRAS-EN domain showed an ability to specifically nick TTAGG sites between A and T, and also CCTAA sites between T and C [178]. This raises the question of how TTAGG repeats contribute to telomere maintenance in a retrotransposon-based system when the telomere structure is so different to that of Drosophila. As was already described, telomeric DNA in canonical TTAGG telomeres is elongated by the actions of telomerase. Telomerase is also present within silkworm, although it has very low activity [179]. It could be that retrotransposition is compensating for low telomerase activity at the telomere [180]. As for components of a capping complex, no proteins have yet been found to control capping activities in silkworm. Silkworm telomeres possess TTAGG sequences at extreme ends; it is therefore unlikely that terminin-like components, which bind in a sequence-unspecific manner, would be involved in chromosome capping. It is perhaps more likely that sequence-specific capping complexes similar to the mammalian shelterin or yeast CST complex would instead protect exposed chromosome ends.
Satellite sequences
Recombination-dependent telomere maintenance in the absence of telomerase was suggested to occur in some lower Diptera species (such as Chironomus and Anopheles) and plants (Allium and a subset of Solanaceae species). Chironomus contain telomere-associated DNA tandem repeats that are 170–350 bp long and are likely maintained by gene conversion [181]. In Anopheles gambiae, gene conversion between complex terminal satellite repeats present at natural telomeres seem to play a major role in the telomere elongation mechanism [182–184]. Alliaceae and some related liliaceous species have at the very ends of chromosomes, a repetitive satellite and/or rDNA sequences [185]. The Cestrum subgroup of Solanaceae species has also been shown to exhibit satellite sequences at telomeres; this does not include the model plants Nicotiana and Solanum, which have been shown to exhibit Arabidopsis TTTAGGG-type telomeres [186, 187]. It has been suggested that satellite sequences were adopted after the loss of TTTAGGG-type telomeres within the Cestrum subgroup. Ancestral telomere binding proteins were found in Cestrum species that displayed the ability to bind Arabidopsis-type telomeres. The presence of these proteins could represent a remnant of previous canonical telomeres in these species [188].
Evolution of telomeres
Evolution of linear genomes in mitochondria and bacteria
The origin of the linear genome in mitochondria and bacteria has not yet been fully uncovered. However, it seems unlikely in both cases that linear and circular genomes have developed independently. In the case of mitochondria, there are several indications that circular and linear DNA originated from a common ancestor harboring a circular genome. Fukuhara et al. [189] demonstrated that related species of the yeast genera Pichia and Williopsis can have either linear or circular mt DNA and that both forms harbor a similar set of genes. Moreover, Kosa et al. [190] discovered that two isogenic strains of Candida metapsilosis differ in the nature of their mt genome: PL448 (circular-mapping genome) and MCO448 (linear genome). Further analysis revealed that PL448 is missing telomeric regions and may be derived from MCO448 through intramolecular end-to-end fusions, which lead to the circularization of the former linear genome [190]. In addition to this, a comparative analysis of yeast mitochondrial genomes revealed that species with linear mt genomes are randomly distributed on the phylogenetic tree, making an independent evolution of circular and linear mt DNA unlikely [5]. The same seems to be true for linear replicon in bacteria [35, 46].
Linear genomes in mitochondria as well as in bacteria may have been created through accidental linearization of the ancestral circular DNA followed by the establishment of telomeric structures. Another possibility is that invasion of mobile genetic elements caused the linearization of bacterial and mitochondrial circular genomes. It seems that the latter hypothesis is more widely accepted in either case. It has been proposed that linear chromosomes in Actinomycetales and Agrobacterium originated from recombination events between linear plasmids and the ancestral circular genome [6, 33, 42, 191]. Such recombination events could give rise to linear chromosomes that terminate with the plasmid telomeric sequences and that use the plasmid-encoded proteins for their replication [42].
In the case of Borrelia linear chromosomes, a bacteriophage origin is discussed. The borrelian enzyme telomere resolvase/protelomerase, which is essential for telomere resolution, may be derived from an altered phage recombinase that was transferred to the bacterium. The presence of such an enzyme could have induced the linearization of the bacterial genome and the stabilization of terminal hairpins [34, 35]. Nosek and Tomaska [20, 192] propose that mitochondrial linear genomes are derived from resolution events taking place after transposons or plasmids have invaded the circular mt DNA. Support for this theory came from studies showing that mitochondrial plasmids can coexist with the major mt genome in several species [193–195]. Schardl et al. [193] demonstrated in cytoplasmatic male sterile mutants of maize that the linear mt plasmids S1 and S2 can invade the circular mt genome. This leads to the generation of several linear molecules ending with the plasmid termini and capped by the same terminal proteins (TP) [193]. In addition to this, it has been shown that mitochondria of the ciliate Oxytricha trifallax contain two linear molecules: a ~70-kb chromosome and a ~5-kb linear plasmid (mO plasmid). The 3′ end of the mO plasmid contains the same telomeric sequences as the mitochondrial chromosome. It is likely that the mO plasmid invaded the mt chromosome at least once since a 251-bp region on the plasmid shows 82 % identity to the major mt genome [194]. Swart et al. [194] suggested that plasmids like the mO plasmid might be used as vehicles to transfer telomeric regions between mitochondria of different species. The horizontal transfer of telomeric sequences via plasmids could explain the presence of similar telomere structures in mitochondria of unrelated species. In fact, horizontal transfer of linear mt plasmids has been shown between unrelated fungi [196]. Moreover, it was even suggested that linear plasmids found in the mitochondria of plants might be derived from phytopathogenic fungi [197]. However, direct evidence for the transfer of linear mt plasmids between plants and fungi are still missing.
A horizontal transfer of telomeric sequences between organisms could also explain the presence of similar telomeric structures in viruses and bacteria. Such a scenario has been proposed for hairpin telomeres found at linear DNA molecules of phages and bacteria [16]. Interestingly, protelomerase-like proteins have also been found in viruses capable of infecting eukaryotic brown algae and microalgae [198–200]. Alternatively, similar telomeric structures could have evolved independently in viruses, bacteria, and mitochondria, as a response to the same evolutionary constraint. It seems that convergent evolution is more likely in case of TP-capped telomeres found in Actinomycetales and Agrobacterium [16, 201, 202].
Retrotransposons vs. canonical telomeres in evolution
The formation of T-loop structures at chromosome ends is thought to have arisen early in the evolution of linear genomes. One model proposed by de Lange [203] suggests that freshly linearized circular genomes containing a limited number of repeat sequences formed T-loops through the prokaryotic recombination-dependent-replication (RDR) pathway. Formation of this structure would have permitted protection from resection and DNA repair activities. Nosek et al. [204] suggests that short repeat sequences generated at newly acquired linear chromosomes would not be sufficient to generate the T-loop structure and that longer sequences would be required. Therefore, telomere evolution was proposed to result from insertion of a selfish element into likely circular genomes, which drove the linearization of chromosomes, this selfish repeat sequence was subsequently amplified by rolling circle amplification that produced elongated telomeres capable of forming T-loops. The evolution of telomerase was then suggested to occur later, likely from reverse transcriptases that deal with non-LTR retrotransposons [203]. Still, the presence of a telomerase-based system of telomere elongation has been proposed to represent an ancient means of counterbalancing the end protection problem [149]. The presence of telomerase has also been traced back to Giardia lamblia, an early eukaryote that was found to contain a homologous telomerase sequence [205]. Despite this, it could be possible that other methods to counteract the end replication problem existed before the advent of telomerase, such as the retrotransposon-dependent telomere elongation system. While most insects have maintained a telomerase/TTAGG-based system, some Diptera species seem to have lost it as exemplified by Drosophila [149]; the same also appears to have occurred in some plant species that have adopted minisatellite repeat sequences at chromosome ends [206]. It could be therefore that losing telomerase could allow an organism to revert back to its previous state before the existence of telomerase, or that retrotransposons simply took over when telomerase was lost in evolution.
Evidence that telomerase predates telomeric retrotransposons is strong; for example, loss of canonical telomeres due to the inactivation of telomerase in S. pombe can be rescued by the amplification of blocks of rDNA or subtelomeric heterochromatin (HAATI survivors) [207]. Progressive loss of DNA from chromosomal termini is counterbalanced by continual addition and rearrangement of heterochromatic sequences. DNA end protection is carried out by the canonical end-protection protein Pot1, which is recruited to non-telomeric heterochromatin by Ccq1 and possibly by the presence of a 3′ overhang. This approach is thought to mirror that of Drosophila, which uses similar mechanisms to mediate de novo addition of retrotransposons to chromosome ends. In addition, studies from silkworm show that retrotransposons TRAS1 and SART1 insert into TTAGG-like sequences, suggesting the presence of a dual mechanism for telomere length elongation. Telomerase is present in Silkworm, although its expression is reduced [180]. It is possible that retrotransposition has arisen to complement the low telomerase activity exhibited in Silkworm allowing both pathways coexist in this organism. This may also represent a turning point in the choice between a telomerase versus retrotransposition-based system. The difference is, however, that retrotransposons in Silkworm are inserted into internal telomere sequences and are not transposed to terminal ends. Together, both studies suggest telomerase was lost in recent evolution in Drosophila, which in turn adopted retrotransposition to instead maintain DNA to cap chromosome ends.
It can be argued that studies in budding yeast also support this theory; it was previously shown that deletion of the Est1 component of telomerase in S. cerevisiae results in the survival of a subset of cells with altered telomere structure. These survivors can be split into two categories with distinct telomeric structures; type 1 survivors show amplification of the subtelomeric Y′ element whereas others represent telomere–telomere recombination products [208–210]. In the case of type 1 survivors, transcription of Y′ elements was initially found to occur in the absence of telomerase, the possibility of a retrotransposition-based system, however, seemed unlikely as Y′ elements lack ORFs encoding for GAG and RT proteins [211]. Mobility of Y′ elements was later found to occur in the absence of a functional telomerase and cDNA synthesis of transcribed Y′ elements was found to be dependent on Ty1 retrotransposon activity [212]. Ty1 retrotransposition was previously found to be induced at shortened telomeres [213]. The model proposed by Maxwell et al. [212] therefore suggests that Y′ elements are transcribed and converted to cDNA by the Ty1 retrotransposon machinery, which is incorporated into the genome via recombination. The presence of the Y′ element within the subtelomeric regions of budding yeast could represent an ancient element with the ability to mobilize, which has been made redundant through development of telomerase.
It is also interesting to note that the most abundant retrotransposon present at Drosophila telomeres, Het-A, lacks a reverse transcriptase ORF. In the model proposed for telomere elongation in Drosophila, telomeric retrotransposons are mobilized to chromosome ends via the actions of internal RT and GAG proteins. It is, however, puzzling why the most abundant retrotransposon does this without an internal RT ORF. It is possible that HeT-A uses RT activity from the two other known retrotransposons that collect at telomeres, their abundance is, however, much lower than HeT-A. Another possibility is that HeT-A utilizes a different RT in order to amplify itself at terminal ends. This mechanism is, in essence, a similar technique to that employed by telomerase using an RNA template to add telomeric sequences to chromosome ends [153]. Telomerase uses the TER template to extend the G-overhang, conventional DNA synthesis machinery then fills in the C-strand to complete elongation of the telomere. In the case of Drosophila, the retrotransposon would presumably also operate as a template for addition to the G-overhang in S-G2 phase directly after completion of DNA synthesis [153]. The use of an independent RT for this purpose is so far not described in Drosophila.
Conclusions
Comparison of telomeric structures over many organisms ranging from bacteria to higher eukaryotes provides an interesting overview on the variation of chromosome capping mechanisms over a number of species. This can range from simple capping components displayed in viruses including hairpins and single telomere binding proteins to higher nucleoprotein structures such as the T-loop. It appears that chromosome linearity has evolved independently from circular precursors in bacteria and mitochondrial genomes resulting in several distinct mechanisms of chromosome end protection and replication. Canonical (G-rich) telomeres represent the most common form in eukaryotic nuclear genomes, suggesting that this solution to the chromosome end-protection problem co-evolved together with telomerase from the last common eukaryotic ancestor. This is reflected in the high evolutionary conservation of the telomere protein components as most eukaryotes use a very similar set of telomere-binding proteins, which feature OB-folds and SANT/Myb motifs. There is, however, remarkable flexibility in the exact utilization of these proteins to achieve chromosome end protection across species. This is most obvious in budding yeast where the function of the classical shelterin-like complex was substituted by Rap1/CST proteins. Another example is the absence of G-overhangs on a subset of telomeres in angiosperm plants indicating existence of a capping mechanism that is not based on T-loop formation or ssDNA-binding proteins. Yet other alternative pathways have developed when telomerase is lost through evolution. Transposition or mobility of elements to chromosome ends appears to take over when this occurs as shown from studies in Drosophila, yeast, plants, and lower Diptera species. It is unknown whether this results from “rolling back the clock” to a previous state before the presence of telomerase or simply as a consequence of an emergency recovery system when telomerase is somehow rendered non-functional. Regardless of different structural features, telomere biogenesis and maintenance in all organisms has been intimately tied to mechanisms involved in DSB repair and recognition. In conclusion, comparative analysis of telomere capping structures in different organisms provides insights into the evolution of end-protection mechanisms that exclude DNA repair processes from natural chromosome ends.
Acknowledgments
Our research on telomeres is supported by the Austrian Science Fund (FWF, Y418-B03) and Austrian Academy of Sciences.
References
- 1.Egel R. Primal eukaryogenesis: on the communal nature of precellular states, ancestral to modern life. Life. 2012;2:170–212. doi: 10.3390/life2010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lode T. For quite a few chromosomes more: the origin of eukaryotes. J Mol Biol. 2012;423(2):135–142. doi: 10.1016/jjmb201207005S0022-2836(12)00557-8. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa F, Naito T. Why do we have linear chromosomes? A matter of Adam and Eve. Mutat Res. 1999;434(2):99–107. doi: 10.1016/S0921-8777(99)00017-8. [DOI] [PubMed] [Google Scholar]
- 4.Deng Z, Wang Z, Lieberman PM. Telomeres and viruses: common themes of genome maintenance. Front Oncol. 2012;2:201. doi: 10.3389/fonc.2012.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Valach M, Farkas Z, Fricova D, Kovac J, Brejova B, Vinar T, Pfeiffer I, Kucsera J, Tomaska L, Lang BF, Nosek J. Evolution of linear chromosomes and multipartite genomes in yeast mitochondria. Nucleic Acids Res. 2011;39(10):4202–4219. doi: 10.1093/nar/gkq1345gkq1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volff JN, Altenbuchner J. A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol Lett. 2000;186(2):143–150. doi: 10.1111/j.1574-6968.2000.tb09095.x. [DOI] [PubMed] [Google Scholar]
- 7.McEachern MJ. Telomeres: guardians of genomic integrity or double agents of evolution? In: Nosek J, Tomaska L, editors. Origin and evolution of telomeres. Austin: Land Bioscience; 2008. pp. 100–113. [Google Scholar]
- 8.Tomaska L, Nosek J. Telomere heterogeneity: taking advantage of stochastic events. FEBS Lett. 2009;583(7):1067–1071. doi: 10.1016/j.febslet.2009.02.032S0014-5793(09)00147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olovnikov AM. Principle of marginotomy in template synthesis of polynucleotides. Dokl Akad Nauk SSSR. 1971;201(6):1496–1499. [PubMed] [Google Scholar]
- 10.Watson JD. Origin of concatemeric T7 DNA. Nat New Biol. 1972;239(94):197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 11.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408(6811):433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 12.Lenhart JS, Schroeder JW, Walsh BW, Simmons LA. DNA repair and genome maintenance in Bacillus subtilis. Microbiol Mol Biol Rev. 2012;76(3):530–564. doi: 10.1128/MMBR.05020-1176/3/530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon T, Huq E, Herrin DL. Microhomology-mediated and nonhomologous repair of a double-strand break in the chloroplast genome of Arabidopsis. Proc Natl Acad Sci USA. 2010;107(31):13954–13959. doi: 10.1073/pnas.10043261071004326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suyama Y, Miura K. Size and structural variations of mitochondrial DNA. Proc Natl Acad Sci USA. 1968;60(1):235–242. doi: 10.1073/pnas.60.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbour AG, Garon CF. Linear plasmids of the bacterium Borrelia burgdorferi have covalently closed ends. Science. 1987;237(4813):409–411. doi: 10.1126/science.3603026. [DOI] [PubMed] [Google Scholar]
- 16.Casjens S, Huang WM. Prokaryotic telomeres: replication mechanisms and evolution. In: Nosek J, Tomaska L, editors. Origin and evolution of telomeres. Austin: Landes Bioscience; 2008. pp. 154–162. [Google Scholar]
- 17.McLeod MP, Warren RL, Hsiao WW, Araki N, Myhre M, Fernandes C, Miyazawa D, Wong W, Lillquist AL, Wang D, Dosanjh M, Hara H, Petrescu A, Morin RD, Yang G, Stott JM, Schein JE, Shin H, Smailus D, Siddiqui AS, Marra MA, Jones SJ, Holt R, Brinkman FS, Miyauchi K, Fukuda M, Davies JE, Mohn WW, Eltis LD. The complete genome of Rhodococcus sp. RHA1 provides insights into a catabolic powerhouse. Proc Natl Acad Sci USA. 2006;103(42):15582–15587. doi: 10.1073/pnas.0607048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Redenbach M, Scheel J, Schmidt U. Chromosome topology and genome size of selected actinomycetes species. Antonie Van Leeuwenhoek. 2000;78(3–4):227–235. doi: 10.1023/A:1010289326752. [DOI] [PubMed] [Google Scholar]
- 19.Bendich AJ. Reaching for the ring: the study of mitochondrial genome structure. Curr Genet. 1993;24(4):279–290. doi: 10.1007/BF00336777. [DOI] [PubMed] [Google Scholar]
- 20.Nosek J, Tomaska L (2002) Mitochondrial telomeres: alternative solutions to the end-replication problem. In: Krupp G, Parwaresch R (eds) Telomeres, telomerases and cancer. Kluwer Academic/Plenum Publishers, New York, pp 396–417
- 21.Raimond R, Marcade I, Bouchon D, Rigaud T, Bossy JP, Souty-Grosset C. Organization of the large mitochondrial genome in the isopod Armadillidium vulgare. Genetics. 1999;151(1):203–210. doi: 10.1093/genetics/151.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morgan JA, Macbeth M, Broderick D, Whatmore P, Street R, Welch DJ, Ovenden JR. Hybridisation, paternal leakage and mitochondrial DNA linearization in three anomalous fish (Scombridae) Mitochondrion. 2013 doi: 10.1016/j.mito.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 23.Smith DR, Kayal E, Yanagihara AA, Collins AG, Pirro S, Keeling PJ. First complete mitochondrial genome sequence from a box jellyfish reveals a highly fragmented linear architecture and insights into telomere evolution. Genome Biol Evol. 2012;4(1):52–58. doi: 10.1093/gbe/evr127evr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voigt O, Erpenbeck D, Worheide G. A fragmented metazoan organellar genome: the two mitochondrial chromosomes of Hydra magnipapillata. BMC Genomics. 2008;9:350. doi: 10.1186/1471-2164-9-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin FN. Linear mitochondrial genome organization in vivo in the genus Pythium. Curr Genet. 1995;28(3):225–234. doi: 10.1007/BF00309781. [DOI] [PubMed] [Google Scholar]
- 26.Nosek J, Tomaska L, Fukuhara H, Suyama Y, Kovac L. Linear mitochondrial genomes: 30 years down the line. Trends Genet. 1998;14(5):184–188. doi: 10.1016/S0168-9525(98)01443-7. [DOI] [PubMed] [Google Scholar]
- 27.Baroudy BM, Venkatesan S, Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982;28(2):315–324. doi: 10.1016/0092-8674(82)90349-x. [DOI] [PubMed] [Google Scholar]
- 28.Baroudy BM, Venkatesan S, Moss B. Structure and replication of vaccinia virus telomeres. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):723–729. doi: 10.1101/SQB.1983.047.01.083. [DOI] [PubMed] [Google Scholar]
- 29.Beaud G. Vaccinia virus DNA replication: a short review. Biochimie. 1995;77(10):774–779. doi: 10.1016/0300-9084(96)88195-8. [DOI] [PubMed] [Google Scholar]
- 30.Rybchin VN, Svarchevsky AN. The plasmid prophage N15: a linear DNA with covalently closed ends. Mol Microbiol. 1999;33(5):895–903. doi: 10.1046/j.1365-2958.1999.01533.x. [DOI] [PubMed] [Google Scholar]
- 31.Casjens SR, Gilcrease EB, Huang WM, Bunny KL, Pedulla ML, Ford ME, Houtz JM, Hatfull GF, Hendrix RW. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J Bacteriol. 2004;186(6):1818–1832. doi: 10.1128/JB.186.6.1818-1832.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Casjens S. Evolution of the linear DNA replicons of the Borrelia spirochetes. Curr Opin Microbiol. 1999;2(5):529–534. doi: 10.1016/s1369-5274(99)00012-0. [DOI] [PubMed] [Google Scholar]
- 33.Goodner B, Hinkle G, Gattung S, Miller N, Blanchard M, Qurollo B, Goldman BS, Cao Y, Askenazi M, Halling C, Mullin L, Houmiel K, Gordon J, Vaudin M, Iartchouk O, Epp A, Liu F, Wollam C, Allinger M, Doughty D, Scott C, Lappas C, Markelz B, Flanagan C, Crowell C, Gurson J, Lomo C, Sear C, Strub G, Cielo C, Slater S. Genome sequence of the plant pathogen and biotechnology agent Agrobacterium tumefaciens C58. Science. 2001;294(5550):2323–2328. doi: 10.1126/science.1066803. [DOI] [PubMed] [Google Scholar]
- 34.Chaconas G, Kobryn K. Structure, function, and evolution of linear replicons in Borrelia. Annu Rev Microbiol. 2010;64:185–202. doi: 10.1146/annurev.micro.112408.134037. [DOI] [PubMed] [Google Scholar]
- 35.Kobryn K, Briffotaux J, Karpov V. Holliday junction formation by the Borrelia burgdorferi telomere resolvase, ResT: implications for the origin of genome linearity. Mol Microbiol. 2009;71(5):1117–1130. doi: 10.1111/j.1365-2958.2008.06584. [DOI] [PubMed] [Google Scholar]
- 36.Huang WM, DaGloria J, Fox H, Ruan Q, Tillou J, Shi K, Aihara H, Aron J, Casjens S. Linear chromosome-generating system of Agrobacterium tumefaciens C58: protelomerase generates and protects hairpin ends. J Biol Chem. 2012;287(30):25551–25563. doi: 10.1074/jbc.M112.369488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dinouel N, Drissi R, Miyakawa I, Sor F, Rousset S, Fukuhara H. Linear mitochondrial DNAs of yeasts: closed-loop structure of the termini and possible linear-circular conversion mechanisms. Mol Cell Biol. 1993;13(4):2315–2323. doi: 10.1128/mcb.13.4.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Jong RN, van der Vliet PC. Mechanism of DNA replication in eukaryotic cells: cellular host factors stimulating adenovirus DNA replication. Gene. 1999;236(1):1–12. doi: 10.1016/s0378-1119(99)00249-8. [DOI] [PubMed] [Google Scholar]
- 39.Rekosh DM, Russell WC, Bellet AJ, Robinson AJ. Identification of a protein linked to the ends of adenovirus DNA. Cell. 1977;11(2):283–295. doi: 10.1016/0092-8674(77)90045-9. [DOI] [PubMed] [Google Scholar]
- 40.Meijer WJ, Serna-Rico A, Salas M. Characterization of the bacteriophage phi29-encoded protein p16.7: a membrane protein involved in phage DNA replication. Mol Microbiol. 2001;39(3):731–746. doi: 10.1046/j.1365-2958.2001.02260.x. [DOI] [PubMed] [Google Scholar]
- 41.Grahn AM, Bamford JK, O’Neill MC, Bamford DH. Functional organization of the bacteriophage PRD1 genome. J Bacteriol. 1994;176(10):3062–3068. doi: 10.1128/jb.176.10.3062-3068.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen CW, Huang CH, Lee HH, Tsai HH, Kirby R. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends Genet. 2002;18(10):522–529. doi: 10.1016/s0168-9525(02)02752-x. [DOI] [PubMed] [Google Scholar]
- 43.Lin YR, Hahn MY, Roe JH, Huang TW, Tsai HH, Lin YF, Su TS, Chan YJ, Chen CW. Streptomyces telomeres contain a promoter. J Bacteriol. 2009;191(3):773–781. doi: 10.1128/JB.01299-08JB.01299-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CW, Yu TW, Lin YS, Kieser HM, Hopwood DA. The conjugative plasmid SLP2 of Streptomyces lividans is a 50 kb linear molecule. Mol Microbiol. 1993;7(6):925–932. doi: 10.1111/j.1365-2958.1993.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 45.Hirochika H, Sakaguchi K. Analysis of linear plasmids isolated from Streptomyces: association of protein with the ends of the plasmid DNA. Plasmid. 1982;7(1):59–65. doi: 10.1016/0147-619x(82)90027-0. [DOI] [PubMed] [Google Scholar]
- 46.Kirby R, Chen CW. Genome architecture. In: Dyson P, editor. Streptomyces: molecular biology and biotechnology. Norfolk: Caister Academic Press; 2011. pp. 5–26. [Google Scholar]
- 47.Fricova D, Valach M, Farkas Z, Pfeiffer I, Kucsera J, Tomaska L, Nosek J. The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5′ termini. Microbiology. 2010;156(Pt 7):2153–2163. doi: 10.1099/mic.0.038646-0mic.0.038646-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vahrenholz C, Riemen G, Pratje E, Dujon B, Michaelis G. Mitochondrial DNA of Chlamydomonas reinhardtii: the structure of the ends of the linear 15.8-kb genome suggests mechanisms for DNA replication. Curr Genet. 1993;24(3):241–247. doi: 10.1007/BF00351798. [DOI] [PubMed] [Google Scholar]
- 49.Nosek J, Dinouel N, Kovac L, Fukuhara H. Linear mitochondrial DNAs from yeasts: telomeres with large tandem repetitions. Mol Gen Genet. 1995;247(1):61–72. doi: 10.1007/BF00425822. [DOI] [PubMed] [Google Scholar]
- 50.Tomaska L, Nosek J, Fukuhara H. Identification of a putative mitochondrial telomere-binding protein of the yeast Candida parapsilosis. J Biol Chem. 1997;272(5):3049–3056. doi: 10.1074/jbc.272.5.3049. [DOI] [PubMed] [Google Scholar]
- 51.Nosek J, Tomaska L, Pagacova B, Fukuhara H. Mitochondrial telomere-binding protein from Candida parapsilosis suggests an evolutionary adaptation of a nonspecific single-stranded DNA-binding protein. J Biol Chem. 1999;274(13):8850–8857. doi: 10.1074/jbc.274.13.8850. [DOI] [PubMed] [Google Scholar]
- 52.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97(4):503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 53.Tomaska L, Makhov AM, Griffith JD, Nosek J. T-loops in yeast mitochondria. Mitochondrion. 2002;1(5):455–459. doi: 10.1016/s1567-7249(02)00009-0. [DOI] [PubMed] [Google Scholar]
- 54.de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21(4):532–540. doi: 10.1038/sj.onc.1205080. [DOI] [PubMed] [Google Scholar]
- 55.Nosek J, Tomaska L. Mitochondrial telomeres: an evolutionary paradigm for the emergence of telomeric structures and their replication strategies. In: Nosek J, Tomaska L, editors. Origin and evolution of telomeres. Austin: Landes Bioscience; 2008. pp. 163–171. [Google Scholar]
- 56.Burger G, Forget L, Zhu Y, Gray MW, Lang BF. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc Natl Acad Sci USA. 2003;100(3):892–897. doi: 10.1073/pnas.03361151000336115100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shukla GC, Nene V. Telomeric features of Theileria parva mitochondrial DNA derived from cycle sequence data of total genomic DNA. Mol Biochem Parasitol. 1998;95(1):159–163. doi: 10.1016/s0166-6851(98)00098-x. [DOI] [PubMed] [Google Scholar]
- 58.Takano H, Kawano S, Kuroiwa T. Genetic organization of a linear mitochondrial plasmid (mF) that promotes mitochondrial fusion in Physarum polycephalum. Curr Genet. 1994;26(5–6):506–511. doi: 10.1007/BF00309941. [DOI] [PubMed] [Google Scholar]
- 59.Walther TC, Kennell JC. Linear mitochondrial plasmids of F. oxysporum are novel, telomere-like retroelements. Mol Cell. 1999;4(2):229–238. doi: 10.1016/s1097-2765(00)80370-6. [DOI] [PubMed] [Google Scholar]
- 60.Given D, Yee D, Griem K, Kieff E. DNA of Epstein–Barr virus. V. Direct repeats of the ends of Epstein–Barr virus DNA. J Virol. 1979;30(3):852–862. doi: 10.1128/jvi.30.3.852-862.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zimmermann J, Hammerschmidt W. Structure and role of the terminal repeats of Epstein–Barr virus in processing and packaging of virion DNA. J Virol. 1995;69(5):3147–3155. doi: 10.1128/jvi.69.5.3147-3155.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kintner CR, Sugden B. The structure of the termini of the DNA of Epstein–Barr virus. Cell. 1979;17(3):661–671. doi: 10.1016/0092-8674(79)90273-3. [DOI] [PubMed] [Google Scholar]
- 63.Matsuo T, Heller M, Petti L, O’Shiro E, Kieff E. Persistence of the entire Epstein–Barr virus genome integrated into human lymphocyte DNA. Science. 1984;226(4680):1322–1325. doi: 10.1126/science.6095452. [DOI] [PubMed] [Google Scholar]
- 64.Gompels UA, Macaulay HA. Characterization of human telomeric repeat sequences from human herpesvirus 6 and relationship to replication. J Gen Virol. 1995;76(Pt 2):451–458. doi: 10.1099/0022-1317-76-2-451. [DOI] [PubMed] [Google Scholar]
- 65.Martin ME, Thomson BJ, Honess RW, Craxton MA, Gompels UA, Liu MY, Littler E, Arrand JR, Teo I, Jones MD. The genome of human herpesvirus 6: maps of unit-length and concatemeric genomes for nine restriction endonucleases. J Gen Virol. 1991;72(Pt 1):157–168. doi: 10.1099/0022-1317-72-1-157. [DOI] [PubMed] [Google Scholar]
- 66.Thomson BJ, Dewhurst S, Gray D. Structure and heterogeneity of the a sequences of human herpesvirus 6 strain variants U1102 and Z29 and identification of human telomeric repeat sequences at the genomic termini. J Virol. 1994;68(5):3007–3014. doi: 10.1128/jvi.68.5.3007-3014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bulboaca GH, Deng H, Dewhurst S, Calos MP. Telomeric sequences from human herpesvirus 6 do not mediate nuclear retention of episomal DNA in human cells. Arch Virol. 1998;143(3):563–570. doi: 10.1007/s007050050312. [DOI] [PubMed] [Google Scholar]
- 68.Arbuckle JH, Medveczky MM, Luka J, Hadley SH, Luegmayr A, Ablashi D, Lund TC, Tolar J, De Meirleir K, Montoya JG, Komaroff AL, Ambros PF, Medveczky PG. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc Natl Acad Sci USA. 2010;107(12):5563–5568. doi: 10.1073/pnas.09135861070913586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arbuckle JH, Medveczky PG. The molecular biology of human herpesvirus-6 latency and telomere integration. Microbes Infect. 2011;13(8–9):731–741. doi: 10.1016/j.micinf.2011.03.006S1286-4579(11)00089-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kaufer BB, Jarosinski KW, Osterrieder N. Herpesvirus telomeric repeats facilitate genomic integration into host telomeres and mobilization of viral DNA during reactivation. J Exp Med. 2011;208(3):605–615. doi: 10.1084/jem.20101402jem.20101402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120(1):33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- 72.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85(18):6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meyne J, Ratliff RL, Moyzis RK. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA. 1989;86(18):7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gornung E, Gabrielli I, Sola L. Localization of the (TTAGGG)n telomeric sequence in zebrafish chromosomes. Genome. 1998;41(1):136–138. doi: 10.1139/g97-098. [DOI] [Google Scholar]
- 75.Cangiano G, La Volpe A. Repetitive DNA sequences located in the terminal portion of the Caenorhabditis elegans chromosomes. Nucleic Acids Res. 1993;21(5):1133–1139. doi: 10.1093/nar/21.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McEachern MJ, Blackburn EH. A conserved sequence motif within the exceptionally diverse telomeric sequences of budding yeasts. Proc Natl Acad Sci USA. 1994;91(8):3453–3457. doi: 10.1073/pnas.91.8.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murray AW, Schultes NP, Szostak JW. Chromosome length controls mitotic chromosome segregation in yeast. Cell. 1986;45(4):529–536. doi: 10.1016/0092-8674(86)90284-9. [DOI] [PubMed] [Google Scholar]
- 78.Shampay J, Szostak JW, Blackburn EH. DNA sequences of telomeres maintained in yeast. Nature. 1984;310(5973):154–157. doi: 10.1038/310154a0. [DOI] [PubMed] [Google Scholar]
- 79.Richards EJ, Ausubel FM. Isolation of a higher eukaryotic telomere from Arabidopsis thaliana. Cell. 1988;53(1):127–136. doi: 10.1016/0092-8674(88)90494-1. [DOI] [PubMed] [Google Scholar]
- 80.Petracek ME, Lefebvre PA, Silflow CD, Berman J. Chlamydomonas telomere sequences are A+T-rich but contain three consecutive G–C base pairs. Proc Natl Acad Sci USA. 1990;87(21):8222–8226. doi: 10.1073/pnas.87.21.8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gatbonton T, Imbesi M, Nelson M, Akey JM, Ruderfer DM, Kruglyak L, Simon JA, Bedalov A. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2006;2(3):e35. doi: 10.1371/journal.pgen.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Lange T. How shelterin solves the telomere end-protection problem. Cold Spring Harb Symp Quant Biol. 2010;75:167–177. doi: 10.1101/sqb.2010.75.017. [DOI] [PubMed] [Google Scholar]
- 83.Fajkus J, Kovarik A, Kralovics R, Bezdek M. Organization of telomeric and subtelomeric chromatin in the higher plant Nicotiana tabacum. Mol Gen Genet. 1995;247(5):633–638. doi: 10.1007/BF00290355. [DOI] [PubMed] [Google Scholar]
- 84.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43(2 Pt 1):405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 85.Osterhage JL, Friedman KL. Chromosome end maintenance by telomerase. J Biol Chem. 2009;284(24):16061–16065. doi: 10.1074/jbc.R900011200R900011200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nandakumar J, Cech TR. Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol. 2013;14(2):69–82. doi: 10.1038/nrm3505nrm3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Watson JM, Riha K. Comparative biology of telomeres: where plants stand. FEBS Lett. 2010;584(17):3752–3759. doi: 10.1016/j.febslet.2010.06.017S0014-5793(10)00507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cesare AJ, Quinney N, Willcox S, Subramanian D, Griffith JD. Telomere looping in P. sativum (common garden pea) Plant J. 2003;36(2):271–279. doi: 10.1046/j.1365-313X.2003.01882.x. [DOI] [PubMed] [Google Scholar]
- 89.Nikitina T, Woodcock CL. Closed chromatin loops at the ends of chromosomes. J Cell Biol. 2004;166(2):161–165. doi: 10.1083/jcb.200403118jcb.200403118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149(4):795–806. doi: 10.1016/j.cell.2012.03.030S0092-8674(12)00418-7. [DOI] [PubMed] [Google Scholar]
- 91.Chai W, Sfeir AJ, Hoshiyama H, Shay JW, Wright WE. The involvement of the Mre11/Rad50/Nbs1 complex in the generation of G-overhangs at human telomeres. EMBO Rep. 2006;7(2):225–230. doi: 10.1038/sj.embor.7400600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol Cell. 2005;20(4):551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 93.Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat Genet. 2000;25(3):347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 94.Czornak K, Chughtai S, Chrzanowska KH. Mystery of DNA repair: the role of the MRN complex and ATM kinase in DNA damage repair. J Appl Genet. 2008;49(4):383–396. doi: 10.1007/BF03195638473. [DOI] [PubMed] [Google Scholar]
- 95.Larrivee M, LeBel C, Wellinger RJ. The generation of proper constitutive G-tails on yeast telomeres is dependent on the MRX complex. Genes Dev. 2004;18(12):1391–1396. doi: 10.1101/gad.119940418/12/1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Broccoli D, Smogorzewska A, Chong L, de Lange T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat Genet. 1997;17(2):231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 97.Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17(2):236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 98.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270(5242):1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 99.Loayza D, De Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423(6943):1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 100.Ye JZ, Donigian JR, van Overbeek M, Loayza D, Luo Y, Krutchinsky AN, Chait BT, de Lange T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J Biol Chem. 2004;279(45):47264–47271. doi: 10.1074/jbc.M409047200. [DOI] [PubMed] [Google Scholar]
- 101.Houghtaling BR, Cuttonaro L, Chang W, Smith S. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2. Curr Biol. 2004;14(18):1621–1631. doi: 10.1016/j.cub.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 102.Liu D, Safari A, O’Connor MS, Chan DW, Laegeler A, Qin J, Songyang Z. PTOP interacts with POT1 and regulates its localization to telomeres. Nat Cell Biol. 2004;6(7):673–680. doi: 10.1038/ncb1142ncb1142. [DOI] [PubMed] [Google Scholar]
- 103.Li B, Oestreich S, de Lange T. Identification of human Rap1: implications for telomere evolution. Cell. 2000;101(5):471–483. doi: 10.1016/s0092-8674(00)80858-2. [DOI] [PubMed] [Google Scholar]
- 104.Stansel RM, de Lange T, Griffith JD. T-loop assembly in vitro involves binding of TRF2 near the 3′ telomeric overhang. EMBO J. 2001;20(19):5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wang RC, Smogorzewska A, de Lange T. Homologous recombination generates T-loop-sized deletions at human telomeres. Cell. 2004;119(3):355–368. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 106.Fouche N, Cesare AJ, Willcox S, Ozgur S, Compton SA, Griffith JD. The basic domain of TRF2 directs binding to DNA junctions irrespective of the presence of TTAGGG repeats. J Biol Chem. 2006;281(49):37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- 107.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16(7):1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127(4):709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 109.Lewis KA, Wuttke DS. Telomerase and telomere-associated proteins: structural insights into mechanism and evolution. Structure. 2012;20(1):28–39. doi: 10.1016/j.str.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gottschling DE, Zakian VA. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47(2):195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- 111.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single-strand DNA. Cell. 1998;95(7):963–974. doi: 10.1016/S0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 112.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292(5519):1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 113.Churikov D, Price CM. Pot1 and cell cycle progression cooperate in telomere length regulation. Nat Struct Mol Biol. 2008;15(1):79–84. doi: 10.1038/nsmb1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hockemeyer D, Sfeir AJ, Shay JW, Wright WE, de Lange T. POT1 protects telomeres from a transient DNA damage response and determines how human chromosomes end. EMBO J. 2005;24(14):2667–2678. doi: 10.1038/sj.emboj.7600733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wu L, Multani AS, He H, Cosme-Blanco W, Deng Y, Deng JM, Bachilo O, Pathak S, Tahara H, Bailey SM, Behringer RR, Chang S. Pot1 deficiency initiates DNA damage checkpoint activation and aberrant homologous recombination at telomeres. Cell. 2006;126(1):49–62. doi: 10.1016/j.cell.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 116.Baumann P, Price C. Pot1 and telomere maintenance. FEBS Lett. 2010;584(17):3779–3784. doi: 10.1016/j.febslet.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14(3):208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 118.Giraud-Panis MJ, Teixeira MT, Geli V, Gilson E. CST meets shelterin to keep telomeres in check. Mol Cell. 2010;39(5):665–676. doi: 10.1016/j.molcel.2010.08.024. [DOI] [PubMed] [Google Scholar]
- 119.Maringele L, Lydall D. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70Delta mutants. Genes Dev. 2002;16(15):1919–1933. doi: 10.1101/gad.225102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274(5285):249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 121.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, Ishikawa F. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36(2):193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 122.Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105(50):19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, Shippen DE. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36(2):207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gu P, Min JN, Wang Y, Huang C, Peng T, Chai W, Chang S. CTC1 deletion results in defective telomere replication, leading to catastrophic telomere loss and stem cell exhaustion. EMBO J. 2012;31(10):2309–2321. doi: 10.1038/emboj.2012.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang C, Dai X, Chai W. Human Stn1 protects telomere integrity by promoting efficient lagging-strand synthesis at telomeres and mediating C-strand fill-in. Cell Res. 2012;22(12):1681–1695. doi: 10.1038/cr.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang F, Stewart JA, Kasbek C, Zhao Y, Wright WE, Price CM. Human CST has independent functions during telomere duplex replication and C-strand fill-in. Cell Rep. 2012;2(5):1096–1103. doi: 10.1016/j.celrep.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Raices M, Verdun RE, Compton SA, Haggblom CI, Griffith JD, Dillin A, Karlseder J. C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell. 2008;132(5):745–757. doi: 10.1016/j.cell.2007.12.039. [DOI] [PubMed] [Google Scholar]
- 128.Lackner DH, Raices M, Maruyama H, Haggblom C, Karlseder J. Organismal propagation in the absence of a functional telomerase pathway in Caenorhabditis elegans. EMBO J. 2012;31(8):2024–2033. doi: 10.1038/emboj.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Oganesian L, Karlseder J. Mammalian 5′ C-rich telomeric overhangs are a mark of recombination-dependent telomere maintenance. Mol Cell. 2011;42(2):224–236. doi: 10.1016/j.molcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oganesian L, Karlseder J. 5′ C-rich telomeric overhangs are an outcome of rapid telomere truncation events. DNA Repair (Amst) 2013;12(3):238–245. doi: 10.1016/j.dnarep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McElligott R, Wellinger RJ. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16(12):3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88(5):657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 133.Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr Biol. 2006;16(13):1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 134.Wu P, van Overbeek M, Rooney S, de Lange T. Apollo contributes to G overhang maintenance and protects leading-end telomeres. Mol Cell. 2010;39(4):606–617. doi: 10.1016/j.molcel.2010.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wu P, Takai H, de Lange T. Telomeric 3′ overhangs derive from resection by Exo1 and Apollo and fill-in by POT1b-associated CST. Cell. 2012;150(1):39–52. doi: 10.1016/j.cell.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Riha K, McKnight TD, Fajkus J, Vyskot B, Shippen DE. Analysis of the G-overhang structures on plant telomeres: evidence for two distinct telomere architectures. Plant J. 2000;23(5):633–641. doi: 10.1046/j.1365-313x.2000.00831.x. [DOI] [PubMed] [Google Scholar]
- 137.Kazda A, Zellinger B, Rossler M, Derboven E, Kusenda B, Riha K. Chromosome end protection by blunt-ended telomeres. Genes Dev. 2012;26(15):1703–1713. doi: 10.1101/gad.194944.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Muller HJ. The remaking of chromosomes. Collecting Net. 1938;8:182–198. [Google Scholar]
- 139.Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. TAHRE, a novel telomeric retrotransposon from Drosophila melanogaster, reveals the origin of Drosophila telomeres. Mol Biol Evol. 2004;21(9):1620–1624. doi: 10.1093/molbev/msh180. [DOI] [PubMed] [Google Scholar]
- 140.Levis RW, Ganesan R, Houtchens K, Tolar LA, Sheen FM. Transposons in place of telomeric repeats at a Drosophila telomere. Cell. 1993;75(6):1083–1093. doi: 10.1016/0092-8674(93)90318-k. [DOI] [PubMed] [Google Scholar]
- 141.Rubin GM. Isolation of a telomeric DNA sequence from Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):1041–1046. doi: 10.1101/SQB.1978.042.01.104. [DOI] [PubMed] [Google Scholar]
- 142.Young BS, Pession A, Traverse KL, French C, Pardue ML. Telomere regions in Drosophila share complex DNA sequences with pericentric heterochromatin. Cell. 1983;34(1):85–94. doi: 10.1016/0092-8674(83)90138-1. [DOI] [PubMed] [Google Scholar]
- 143.Rashkova S, Athanasiadis A, Pardue ML. Intracellular targeting of gag proteins of the Drosophila telomeric retrotransposons. J Virol. 2003;77(11):6376–6384. doi: 10.1128/JVI.77.11.6376-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rashkova S, Karam SE, Kellum R, Pardue ML. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J Cell Biol. 2002;159(3):397–402. doi: 10.1083/jcb.200205039jcb.200205039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Rashkova S, Karam SE, Pardue ML. Element-specific localization of Drosophila retrotransposon gag proteins occurs in both nucleus and cytoplasm. Proc Natl Acad Sci USA. 2002;99(6):3621–3626. doi: 10.1073/pnas.032071999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sasaki T, Fujiwara H. Detection and distribution patterns of telomerase activity in insects. Eur J Biochem. 2000;267(10):3025–3031. doi: 10.1046/j.1432-1033.2000.01323.x. [DOI] [PubMed] [Google Scholar]
- 147.Biessmann H, Mason JM, Ferry K, d’Hulst M, Valgeirsdottir K, Traverse KL, Pardue ML. Addition of telomere-associated HeT DNA sequences “heals” broken chromosome ends in Drosophila. Cell. 1990;61(4):663–673. doi: 10.1016/0092-8674(90)90478-w. [DOI] [PubMed] [Google Scholar]
- 148.Sheen FM, Levis RW. Transposition of the LINE-like retrotransposon TART to Drosophila chromosome termini. Proc Natl Acad Sci USA. 1994;91(26):12510–12514. doi: 10.1073/pnas.91.26.12510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nakamura TM, Cech TR. Reversing time: origin of telomerase. Cell. 1998;92(5):587–590. doi: 10.1016/s0092-8674(00)81123-x. [DOI] [PubMed] [Google Scholar]
- 150.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88(5):647–655. doi: 10.1016/s0092-8674(00)81907-8. [DOI] [PubMed] [Google Scholar]
- 151.Danilevskaya ON, Traverse KL, Hogan NC, DeBaryshe PG, Pardue ML. The two Drosophila telomeric transposable elements have very different patterns of transcription. Mol Cell Biol. 1999;19(1):873–881. doi: 10.1128/MCB.19.1.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Capkova Frydrychova R, Biessmann H, Mason JM. Regulation of telomere length in Drosophila. Cytogenet Genome Res. 2008;122(3–4):356–364. doi: 10.1159/000167823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pardue ML, Danilevskaya ON, Lowenhaupt K, Slot F, Traverse KL. Drosophila telomeres: new views on chromosome evolution. Trends Genet. 1996;12(2):48–52. doi: 10.1016/0168-9525(96)81399-0. [DOI] [PubMed] [Google Scholar]
- 154.Levis RW. Viable deletions of a telomere from a Drosophila chromosome. Cell. 1989;58(4):791–801. doi: 10.1016/0092-8674(89)90112-8. [DOI] [PubMed] [Google Scholar]
- 155.Mason JM, Strobel E, Green MM. μ-2: mutator gene in Drosophila that potentiates the induction of terminal deficiencies. Proc Natl Acad Sci USA. 1984;81(19):6090–6094. doi: 10.1073/pnas.81.19.6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Biessmann H, Carter SB, Mason JM. Chromosome ends in Drosophila without telomeric DNA sequences. Proc Natl Acad Sci USA. 1990;87(5):1758–1761. doi: 10.1073/pnas.87.5.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Biessmann H, Mason JM. Progressive loss of DNA sequences from terminal chromosome deficiencies in Drosophila melanogaster. EMBO J. 1988;7(4):1081–1086. doi: 10.1002/j.1460-2075.1988.tb02916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kern AD, Begun DJ. Recurrent deletion and gene presence/absence polymorphism: telomere dynamics dominate evolution at the tip of 3L in Drosophila melanogaster and D. simulans. Genetics. 2008;179(2):1021–1027. doi: 10.1534/genetics.107.078345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Raffa GD, Ciapponi L, Cenci G, Gatti M. Terminin: a protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus. 2011;2(5):383–391. doi: 10.4161/nucl.2.5.17873. [DOI] [PubMed] [Google Scholar]
- 160.Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol. 2003;5(1):82–84. doi: 10.1038/ncb902ncb902. [DOI] [PubMed] [Google Scholar]
- 161.Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2(5):527–538. doi: 10.1016/s1097-2765(00)80152-5. [DOI] [PubMed] [Google Scholar]
- 162.Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, Rong YS. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. EMBO J. 2010;29(4):819–829. doi: 10.1038/emboj.2009.394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Raffa GD, Raimondo D, Sorino C, Cugusi S, Cenci G, Cacchione S, Gatti M, Ciapponi L. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 2010;24(15):1596–1601. doi: 10.1101/gad.57481024/15/1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, Gatti M. The Drosophila Modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci USA. 2009;106(7):2271–2276. doi: 10.1073/pnas.08127021060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell. 2001;12(6):1671–1685. doi: 10.1091/mbc.12.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Komonyi O, Schauer T, Papai G, Deak P, Boros IM. A product of the bicistronic Drosophila melanogaster gene CG31241, which also encodes a trimethylguanosine synthase, plays a role in telomere protection. J Cell Sci. 2009;122(Pt 6):769–774. doi: 10.1242/jcs.035097jcs.035097. [DOI] [PubMed] [Google Scholar]
- 167.Titen SW, Golic KG. Healing of euchromatic chromosome breaks by efficient de novo telomere addition in Drosophila melanogaster. Genetics. 2010;184(1):309–312. doi: 10.1534/genetics.109.109934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cenci G, Rawson RB, Belloni G, Castrillon DH, Tudor M, Petrucci R, Goldberg ML, Wasserman SA, Gatti M. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev. 1997;11(7):863–875. doi: 10.1101/gad.11.7.863. [DOI] [PubMed] [Google Scholar]
- 169.Raffa GD, Cenci G, Siriaco G, Goldberg ML, Gatti M. The putative Drosophila transcription factor woc is required to prevent telomeric fusions. Mol Cell. 2005;20(6):821–831. doi: 10.1016/j.molcel.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 170.Bi X, Srikanta D, Fanti L, Pimpinelli S, Badugu R, Kellum R, Rong YS. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci USA. 2005;102(42):15167–15172. doi: 10.1073/pnas.0504981102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bi X, Wei SC, Rong YS. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr Biol. 2004;14(15):1348–1353. doi: 10.1016/j.cub.2004.06.063. [DOI] [PubMed] [Google Scholar]
- 172.Ciapponi L, Cenci G, Ducau J, Flores C, Johnson-Schlitz D, Gorski MM, Engels WR, Gatti M. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr Biol. 2004;14(15):1360–1366. doi: 10.1016/j.cub.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 173.Ciapponi L, Cenci G, Gatti M. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics. 2006;173(3):1447–1454. doi: 10.1534/genetics.106.058081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Oikemus SR, McGinnis N, Queiroz-Machado J, Tukachinsky H, Takada S, Sunkel CE, Brodsky MH. Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes Dev. 2004;18(15):1850–1861. doi: 10.1101/gad.12025041202504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Okazaki S, Tsuchida K, Maekawa H, Ishikawa H, Fujiwara H. Identification of a pentanucleotide telomeric sequence, (TTAGG)n, in the silkworm Bombyx mori and in other insects. Mol Cell Biol. 1993;13(3):1424–1432. doi: 10.1128/mcb.13.3.1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Okazaki S, Ishikawa H, Fujiwara H. Structural analysis of TRAS1, a novel family of telomeric repeat-associated retrotransposons in the silkworm Bombyx mori. Mol Cell Biol. 1995;15(8):4545–4552. doi: 10.1128/MCB.15.8.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Takahashi H, Okazaki S, Fujiwara H. A new family of site-specific retrotransposons, SART1, is inserted into telomeric repeats of the silkworm Bombyx mori. Nucleic Acids Res. 1997;25(8):1578–1584. doi: 10.1093/nar/25.8.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Anzai T, Takahashi H, Fujiwara H. Sequence-specific recognition and cleavage of telomeric repeat (TTAGG)(n) by endonuclease of non-long terminal repeat retrotransposon TRAS1. Mol Cell Biol. 2001;21(1):100–108. doi: 10.1128/MCB.21.1.100-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Osanai M, Kojima KK, Futahashi R, Yaguchi S, Fujiwara H. Identification and characterization of the telomerase reverse transcriptase of Bombyx mori (silkworm) and Tribolium castaneum (flour beetle) Gene. 2006;376(2):281–289. doi: 10.1016/j.gene.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 180.Fujiwara H, Osanai M, Matsumoto T, Kojima KK. Telomere-specific non-LTR retrotransposons and telomere maintenance in the silkworm Bombyx mori. Chromosome Res. 2005;13(5):455–467. doi: 10.1007/s10577-005-0990-9. [DOI] [PubMed] [Google Scholar]
- 181.Cohn M, Edstrom JE. Telomere-associated repeats in Chironomus form discrete subfamilies generated by gene conversion. J Mol Evol. 1992;35(2):114–122. doi: 10.1007/BF00183222. [DOI] [PubMed] [Google Scholar]
- 182.Biessmann H, Donath J, Walter MF. Molecular characterization of the Anopheles gambiae 2L telomeric region via an integrated transgene. Insect Mol Biol. 1996;5(1):11–20. doi: 10.1111/j.1365-2583.1996.tb00035.x. [DOI] [PubMed] [Google Scholar]
- 183.Roth CW, Kobeski F, Walter MF, Biessmann H. Chromosome end elongation by recombination in the mosquito Anopheles gambiae. Mol Cell Biol. 1997;17(9):5176–5183. doi: 10.1128/MCB.17.9.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Walter MF, Bozorgnia L, Maheshwari A, Biessmann H. The rate of terminal nucleotide loss from a telomere of the mosquito Anopheles gambiae. Insect Mol Biol. 2001;10(1):105–110. doi: 10.1046/j.1365-2583.2001.00245.x. [DOI] [PubMed] [Google Scholar]
- 185.Pich U, Fuchs J, Schubert I. How do Alliaceae stabilize their chromosome ends in the absence of TTTAGGG sequences? Chromosome Res. 1996;4(3):207–213. doi: 10.1007/BF02254961. [DOI] [PubMed] [Google Scholar]
- 186.Fulneckova J, Hasikova T, Fajkus J, Lukesova A, Elias M, Sykorova E. Dynamic evolution of telomeric sequences in the green algal order Chlamydomonadales. Genome Biol Evol. 2012;4(3):248–264. doi: 10.1093/gbe/evs007evs007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sykorova E, Lim KY, Chase MW, Knapp S, Leitch IJ, Leitch AR, Fajkus J. The absence of Arabidopsis-type telomeres in cestrum and closely related genera Vestia and Sessea (Solanaceae): first evidence from eudicots. Plant J. 2003;34(3):283–291. doi: 10.1046/j.1365-313X.2003.01731.x. [DOI] [PubMed] [Google Scholar]
- 188.Peska V, Sykorova E, Fajkus J. Two faces of Solanaceae telomeres: a comparison between Nicotiana and Cestrum telomeres and telomere-binding proteins. Cytogenet Genome Res. 2008;122(3–4):380–387. doi: 10.1159/000167826000167826. [DOI] [PubMed] [Google Scholar]
- 189.Fukuhara H, Sor F, Drissi R, Dinouel N, Miyakawa I, Rousset S, Viola AM. Linear mitochondrial DNAs of yeasts: frequency of occurrence and general features. Mol Cell Biol. 1993;13(4):2309–2314. doi: 10.1128/mcb.13.4.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Kosa P, Valach M, Tomaska L, Wolfe KH, Nosek J. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 2006;34(8):2472–2481. doi: 10.1093/nar/gkl327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wood DW, Setubal JC, Kaul R, Monks DE, Kitajima JP, Okura VK, Zhou Y, Chen L, Wood GE, Almeida NF, Jr, Woo L, Chen Y, Paulsen IT, Eisen JA, Karp PD, Bovee D, Sr, Chapman P, Clendenning J, Deatherage G, Gillet W, Grant C, Kutyavin T, Levy R, Li MJ, McClelland E, Palmieri A, Raymond C, Rouse G, Saenphimmachak C, Wu Z, Romero P, Gordon D, Zhang S, Yoo H, Tao Y, Biddle P, Jung M, Krespan W, Perry M, Gordon-Kamm B, Liao L, Kim S, Hendrick C, Zhao ZY, Dolan M, Chumley F, Tingey SV, Tomb JF, Gordon MP, Olson MV, Nester EW. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294(5550):2317–2323. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- 192.Nosek J, Tomaska L. Mitochondrial genome diversity: evolution of the molecular architecture and replication strategy. Curr Genet. 2003;44(2):73–84. doi: 10.1007/s00294-003-0426-z. [DOI] [PubMed] [Google Scholar]
- 193.Schardl CL, Lonsdale DM, Pring DR, Rose KR. Linearization of maize mitochondrial chromosomes by recombination with linear episomes. Nature. 1984;310:292–296. doi: 10.1038/310292a0. [DOI] [Google Scholar]
- 194.Swart EC, Nowacki M, Shum J, Stiles H, Higgins BP, Doak TG, Schotanus K, Magrini VJ, Minx P, Mardis ER, Landweber LF. The Oxytricha trifallax mitochondrial genome. Genome Biol Evol. 2012;4(2):136–154. doi: 10.1093/gbe/evr136evr136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Takano H, Kawano S, Kuroiwa T. Constitutive homologous recombination between mitochondrial DNA and a linear mitochondrial plasmid in Physarum polycephalum. Curr Genet. 1992;22(3):221–227. doi: 10.1007/BF00351729. [DOI] [PubMed] [Google Scholar]
- 196.Kempken F. Horizontal transfer of a mitochondrial plasmid. Mol Gen Genet. 1995;248(1):89–94. doi: 10.1007/BF02456617. [DOI] [PubMed] [Google Scholar]
- 197.Handa H. Linear plasmids in plant mitochondria: peaceful coexistences or malicious invasions? Mitochondrion. 2008;8(1):15–25. doi: 10.1016/j.mito.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 198.Delaroque N, Boland W, Muller DG, Knippers R. Comparisons of two large phaeoviral genomes and evolutionary implications. J Mol Evol. 2003;57(6):613–622. doi: 10.1007/s00239-003-2501-y. [DOI] [PubMed] [Google Scholar]
- 199.Delaroque N, Muller DG, Bothe G, Pohl T, Knippers R, Boland W. The complete DNA sequence of the Ectocarpus siliculosus Virus EsV-1 genome. Virology. 2001;287(1):112–132. doi: 10.1006/viro.2001.1028. [DOI] [PubMed] [Google Scholar]
- 200.Wilson WH, Schroeder DC, Allen MJ, Holden MT, Parkhill J, Barrell BG, Churcher C, Hamlin N, Mungall K, Norbertczak H, Quail MA, Price C, Rabbinowitsch E, Walker D, Craigon M, Roy D, Ghazal P. Complete genome sequence and lytic phase transcription profile of a Coccolithovirus. Science. 2005;309(5737):1090–1092. doi: 10.1126/science.1113109. [DOI] [PubMed] [Google Scholar]
- 201.Kirby R. Chromosome diversity and similarity within the Actinomycetales. FEMS Microbiol Lett. 2011;319(1):1–10. doi: 10.1111/j.1574-6968.2011.02242.x. [DOI] [PubMed] [Google Scholar]
- 202.Chen CW. Streptomyces linear plasmids: replication and telomeres. In: Meinhardt F, Klassen R, editors. Microbial linear plasmids. Berlin Heidelberg New York: Springer; 2007. pp. 33–61. [Google Scholar]
- 203.de Lange T. T-loops and the origin of telomeres. Nat Rev Mol Cell Biol. 2004;5(4):323–329. doi: 10.1038/nrm1359nrm1359. [DOI] [PubMed] [Google Scholar]
- 204.Nosek J, Kosa P, Tomaska L. On the origin of telomeres: a glimpse at the pre-telomerase world. BioEssays. 2006;28(2):182–190. doi: 10.1002/bies.20355. [DOI] [PubMed] [Google Scholar]
- 205.Malik HS, Burke WD, Eickbush TH. Putative telomerase catalytic subunits from Giardia lamblia and Caenorhabditis elegans. Gene. 2000;251(2):101–108. doi: 10.1016/s0378-1119(00)00207-9. [DOI] [PubMed] [Google Scholar]
- 206.Fajkus J, Sykorova E, Leitch AR. Telomeres in evolution and evolution of telomeres. Chromosome Res. 2005;13(5):469–479. doi: 10.1007/s10577-005-0997-2. [DOI] [PubMed] [Google Scholar]
- 207.Jain D, Hebden AK, Nakamura TM, Miller KM, Cooper JP. HAATI survivors replace canonical telomeres with blocks of generic heterochromatin. Nature. 2010;467(7312):223–227. doi: 10.1038/nature09374. [DOI] [PubMed] [Google Scholar]
- 208.Lundblad V, Blackburn EH. An alternative pathway for yeast telomere maintenance rescues est1- senescence. Cell. 1993;73(2):347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 209.Teng SC, Chang J, McCowan B, Zakian VA. Telomerase-independent lengthening of yeast telomeres occurs by an abrupt Rad50p-dependent Rif-inhibited recombinational process. Mol Cell. 2000;6(4):947–952. doi: 10.1016/s1097-2765(05)00094-8. [DOI] [PubMed] [Google Scholar]
- 210.Teng SC, Zakian VA. Telomere-telomere recombination is an efficient bypass pathway for telomere maintenance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19(12):8083–8093. doi: 10.1128/MCB.19.12.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yamada M, Hayatsu N, Matsuura A, Ishikawa F. Y’-Help1, a DNA helicase encoded by the yeast subtelomeric Y’ element, is induced in survivors defective for telomerase. J Biol Chem. 1998;273(50):33360–33366. doi: 10.1074/jbc.273.50.33360. [DOI] [PubMed] [Google Scholar]
- 212.Maxwell PH, Coombes C, Kenny AE, Lawler JF, Boeke JD, Curcio MJ. Ty1 mobilizes subtelomeric Y’ elements in telomerase-negative Saccharomyces cerevisiae survivors. Mol Cell Biol. 2004;24(22):9887–9898. doi: 10.1128/MCB.24.22.9887-9898.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Scholes DT, Kenny AE, Gamache ER, Mou Z, Curcio MJ. Activation of a LTR-retrotransposon by telomere erosion. Proc Natl Acad Sci USA. 2003;100(26):15736–15741. doi: 10.1073/pnas.2136609100. [DOI] [PMC free article] [PubMed] [Google Scholar]