Abstract
The regulation of splice site (SS) usage is important for alternative pre-mRNA splicing and thus proper expression of protein isoforms in cells; its disruption causes diseases. In recent years, an increasing number of novel regulatory elements have been found within or nearby the 3′SS in mammalian genes. The diverse elements recruit a repertoire of trans-acting factors or form secondary structures to regulate 3′SS usage, mostly at the early steps of spliceosome assembly. Their mechanisms of action mainly include: (1) competition between the factors for RNA elements, (2) steric hindrance between the factors, (3) direct interaction between the factors, (4) competition between two splice sites, or (5) local RNA secondary structures or longer range loops, according to the mode of protein/RNA interactions. Beyond the 3′SS, chromatin remodeling/transcription, posttranslational modifications of trans-acting factors and upstream signaling provide further layers of regulation. Evolutionarily, some of the 3′SS elements seem to have emerged in mammalian ancestors. Moreover, other possibilities of regulation such as that by non-coding RNA remain to be explored. It is thus likely that there are more diverse elements/factors and mechanisms that influence the choice of an intron end. The diverse regulation likely contributes to a more complex but refined transcriptome and proteome in mammals.
Keywords: Alternative splicing, RNA element, Splicing factor, Secondary structure, Evolution
Introduction
Alternative pre-mRNA splicing allows the generation of up to thousands of variants from a gene, greatly contributing to the proteomic diversity of metazoans [1, 2]. In humans, about 92–95 % of multi-exon genes undergo alternative splicing [3, 4], with functional consequences ranging from altered mRNA stability and fine-tuning of protein functions to antagonistic effects. Aberrant splicing, resembling alternative splicing but in abnormal conditions, causes diseases [5, 6]. Therefore, it is important to understand how the splice sites (SS) are alternatively or aberrantly used in cell physiology or diseases.
In mammals, the two splice sites at the start and end (5′ and 3′) of introns have distinctly different arrangements of particular motifs. The 5′SS is mostly comprised of a GU dinucleotide within a short consensus sequence. In contrast, the 3′SS consists of tripartite consensus motifs: the branch point (BP), polypyrimidine tract (Py) and 3′AG dinucleotide (Fig. 1, upper diagram). The three motifs are differently conserved, required or spaced among different metazoan species S. cerevisiae [7], S. pombe [8], C. elegans [9–11], and mammals [12], as summarized by Hollins et al. [9]. Particularly, the mammalian tripartite motifs have more permissive in-between spaces and different requirements for the 3′AG [12, 13], likely providing more targets for splicing regulation than the 5′SS, as well as more than the 3′SS in other species.
Fig. 1.
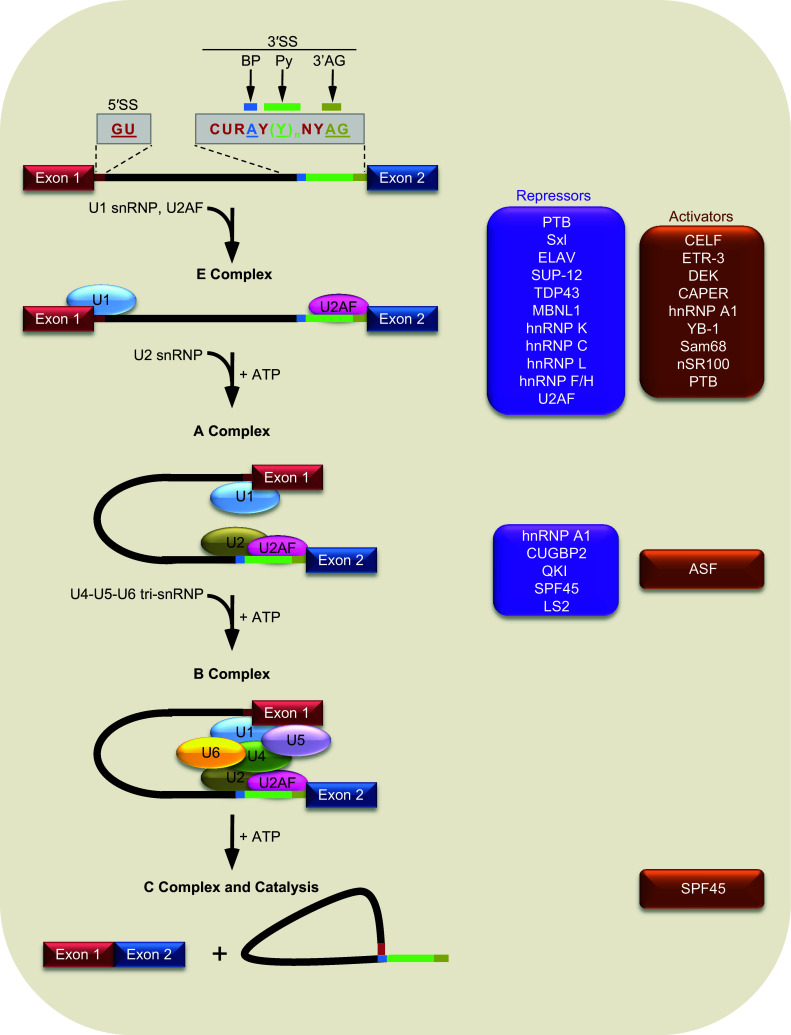
Diagram of pre-mRNA splicing steps and trans-acting 3′SS regulators. Shown at the top is splice sites motifs shown in different colors with consensus sequence and represented in the intron with respective colors. 5′SS (maroon line), BP (blue line), Py (green line) and 3′AG (olive line). Below is the stepwise assembly of constitutive splicing factors (ovals) for excision of intron between exon 1 (red) and exon 2 (blue). Boxes on the right list trans-acting alternative splicing factors that regulate the corresponding steps of spliceosome assembly for exon 2 splicing
The splice sites are recognized sequentially by a set of small nuclear ribonucleoproteins (snRNPs) during spliceosome assembly on pre-mRNA introns. This process has been reviewed recently by Will and Luhrmann [14], to which and references therein readers are referred for more details. Briefly, after the 5′SS is bound by the U1 snRNP, the 3′SS Py and 3′AG are bound by the U2 auxiliary factor (U2AF) heterodimer to form an E (early) complex (Fig. 1). Then the BP is base-paired with the U2 small nuclear ribonucleic acid (snRNA) of the U2 snRNP to form the A complex in an ATP-dependent manner. Subsequent recruitment of U4-U5-U6 tri-snRNP and complex rearrangements form the catalytic C complex where the intron is removed and exons are joined through two transesterification steps. In total, a spliceosome has over 170 proteins that assemble on pre-mRNA introns in in vitro systems using HeLa nuclear extract. It should be noted that, in the above systems relatively short introns have been used, mostly for intron definition and spliceosome formation; for longer introns and certain regulated exons, exon definition has been proposed as well [15–17].
The spliceosome assembly or splicing can be affected by a wide variety of cis-acting regulatory RNA elements called splicing enhancers or silencers [18, 19]. Many elements overlap with or are in-between the consensus motifs of splice sites though others are away from the sites. The elements often recruit trans-acting splicing factors and/or form RNA secondary structures to regulate splicing [18–20]. In addition to the commonly known regulatory factors, more and more so-called “constitutive splicing factors”, which were identified mostly from the HeLa nuclear extract system and model pre-mRNA transcripts, have also been demonstrated to regulate alternative splicing, for example the U2AF65 [21, 22], SmB/B′ [23] or U1C [24]. The list is probably going to extend to more factors in the future. Thus, before a complete survey of these factors is available, here we will still follow the tradition to call them regulatory or constitutive factors respectively.
Recent studies on the regulation of 3′SS usage have uncovered a diverse group of elements and corresponding factors previously unidentified in this region and suggested novel mechanisms beyond the earlier competition model. Here we review the recently characterized 3′SS RNA elements, their binding factors and try to categorize their mechanisms of action into several models according to their common features. Other layers of splicing regulation including chromatin modification and rate of transcription elongation, as well as posttranslational modification of splicing factors and upstream signaling are also discussed regarding their effects on 3′SS usage. Focus is mainly on progresses in the last decade in mammalian systems.
Regulation of 3′ splice site usage by RNA elements/splicing factors
Table 1 lists representative 3′SS regulatory RNA elements (silencers or enhancers) and trans-acting factors in the control of alternative splicing. Their regulatory targets during spliceosome assembly are shown in Fig. 1. Here we describe the elements/factors according their mechanisms of actions.
Table 1.
List of 3′SS RNA elements and protein factors that regulate 3′SS usage, grouped according to their target constitutive factors
| Target constitutive factor | Location | Regulatory element | Trans-acting factor | Regulated exon | Gene | Mechanism of regulation | References |
|---|---|---|---|---|---|---|---|
| Silencers/repressors | |||||||
| U2AF | Py | UCUU and UCUCU | PTB | 6B | β-tropomyosin | i | [25] |
| UCUU | PTB | 3 | α-tropomyosin | i | [26–28] | ||
| CUCUCU | PTB | N1 | c-Src | i | [16, 29, 30] | ||
| UCUU | PTB | SM | α-actinin | i | [31, 32] | ||
| CUCU like | PTB | 24 nt exon, EN and 5 | GABA receptor, Clathrin light chain B and NMDAR1 | i | [33] | ||
| C/U-rich motifs | PTB | Groupb | Groupb | ia | [34] | ||
| UUUUUGUUGUUUUUUUU | Sxl | 2 | Tra | i | [35, 36] | ||
| 8/10 U | ELAV | Terminal | Neuroglian | ia | [37] | ||
| UG repeats | SUP-12 | 2A | ADF/Cofilin | ia | [38] | ||
| UG | TDP43 | 9 | CFTR | ia | [39, 40] | ||
| YGCU(U/G)Y | MBNL1 | 5 | cTNT | i | [41, 42] | ||
| UCCCU | hnRNP K | 5a, 6 | Snap25, Runx1 | i | [43] | ||
| UUUs | hnRNP C | 10 | CD55 | i | [44] | ||
| Between Py and 3′ AG | CA repeats | hnRNP L | V10 | CD44 | ii | [45] | |
| CaRRE | hnRNP L | STREX | Slo gene | ii | [46–48] | ||
| CA repeats | hnRNPL | 20 | TJPI | ii | [49] | ||
| GGGUGGGGG | hnRNP F/H | 3 | PRMT5 (group)b | ii | [50, 51] | ||
| U2 | Near BP | UAGGG(A/U) | hnRNP A1 | 3 | Tat (HIV) | ii | [52] |
| UGUGU and GU | CUGBP2 | N1 | NMDA R1 | ii | [53] | ||
| UGU motifs | CUGBP2 | CUGBP2 | ii | [53] | |||
| AUUAAC | QKI | 12 | NUMB | i | [54] | ||
| ULM of SF1 | SPF45 | 6 | Fas | iia | [55] | ||
| G rich | LS2 | Minigene | Ftc and PEP | iia | [56] | ||
| U2AF of weaker 3′SSa | Upstream of 3′SS | UUUCUU, UUUUUC, UUUUCU | U2AF | Groupb | Groupb | iii | [22] |
| ISS in IVS9 | SF2/ASF, SRp40 | 9 | CFTR | iii | [57] | ||
| Constitutive factors e.g. U1, U2AF | Py | Stem loop | MBNL1 | 5 | cTNT | iv | [41, 42] |
| U runs | Sxl | 3 | Sxl | iv | [58–60] | ||
| CUCUCU | PTB | N1 | c-Src | iv | [16, 29, 30] | ||
| UUUs | hnRNP C | 10 | CD55 (groupb) | iva | [61] | ||
| Enhancers/activators | |||||||
| U2AF, U2 snRNA | Py | CUG and UG motifs | CELF | SM, NM | Actinin | I | [62] |
| U/G motifs | ETR-3 | 5 | cTNT | I | [63] | ||
| U2AF interaction (Py) | DEK | Minigene | IgM-AdML | II | [64] | ||
| Py | CAPER | 6 | VEGF | IIa | [65, 66] | ||
| Py with 3′ CG or AG | hnRNP A1 | Groupb | Groupb | II | [67] | ||
| CAUC | YB-1 | V5 | CD44 | II | [68] | ||
| 3′AG | 3′AG | Urp | Minigene | AdML | II | [69] | |
| BP | AUUAAA | Sam68 | V5 | CD44 | II | [70] | |
| Purine-rich ESE and BP | ASF/RS domain | Minigene | Actin, IgM | II | [71, 72] | ||
| Between Py and 3′AG | UGC | nSR100 | Groupb | Groupb | II | [34] | |
| U2AF of competing 3′SSa | Py | CU rich | PTB | Groupb | Groupb | III | [73] |
| Constitutive factors e.g. U1, U2AF | Near 3′SS | Docking/selector | hrp36a | Minigene | Dscam | IV | [74–76] |
| SR binding | U1, U2AF35 | Minigene | dsx | IV | [77–80] | ||
| Artificial binding sites | hnRNP A/B or hnRNP F/H | Minigene | hnRNPA1 | IV | [81–84] | ||
For mechanisms of silencing/repression, i: competition, ii: steric hindrance, iii: competing 3′SS, or iv: local RNA secondary structure/looping out. For enhancement/activation, I: competition with a repressor for the same or overlapping RNA element, II: interaction with U2AF or U2 snRNA, III: competing 3′SS, or IV: local RNA secondary structure or longer range RNA loops
aPredicted
bGroup of regulated exons as detailed in the text
Repression of 3′ splice site usage
There is a diverse group of silencers and repressors, but they mostly interfere with the early spliceosome factors (Fig. 2).
Fig. 2.
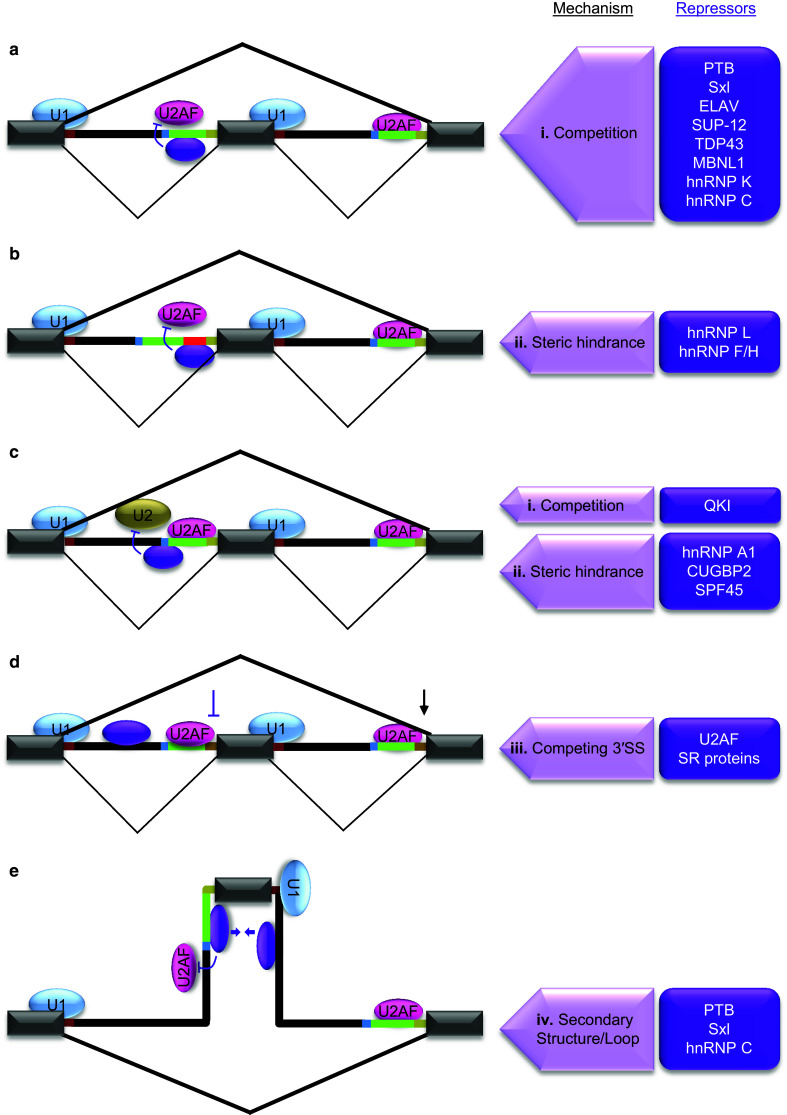
Mechanisms of splicing repression at 3′SS. The regulated exon shown in the middle is flanked by introns and exons. Splicing patterns in bold lines indicate the outcome of regulation. Purple oval represents one of the repressors listed in purple boxes on the right side that are involved in the regulation through respective mechanisms indicated on the left side. a Splicing repression through cis-acting elements within Py. The location of a major class of silencers within 3′SS overlaps with Py. These elements recruit trans-acting factors to inhibit U2AF binding to Py through competition (i). b Splicing repression through cis-acting element insertions between Py and 3′AG. A group of 3′SS silencers is uniquely positioned between Py and 3′AG (red line in intron). Such elements also inhibit U2AF mainly through steric hindrance (ii) caused by specific binding of repressors. c Splicing repression through cis-acting elements near BP. Some 3′SS silencers are located near or in overlap with BP. The binding of trans-acting factors to these elements results in inhibition of U2 snRNP interaction with BP either through competition (i) or steric hindrance (ii). d Inhibition of weakened 3′SS by competition of downstream stronger 3′SS. The weakening of upstream 3′SS by intronic binding of U2AF (purple oval) results in inhibition by competing 3′SS (iii). e Splicing repression through ‘RNA secondary structures or loops’. In some cases Py binding trans-acting alternative splicing factors interact cooperatively with another RNA-binding protein bound to downstream intron (purple ovals and arrows). This interaction results in looping out (iv) of the regulated exon and interferes with U1 and U2AF binding to its splice sites leading to inhibition of exon definition. RNA secondary structures also trap 3′SS motifs in a similar way but without proteins to repress splicing
Inhibition of U2AF binding to the Py
Inhibition of U2AF65 binding was observed upon repressor binding to RNA silencers overlapping with the Py (Fig. 2a). Well-studied early examples include the inhibition of exon 6B of beta-tropomyosin [25, 85], exon 3 of alpha-tropomyosin [27, 86], N1 exon of Rouse sarcoma oncogene (c-Src) [16, 29, 30], SM exon of alpha-actinin [32], and gamma amino-butyric acid (GABA) receptor, Clathrin light chain B and N-methyl-d-aspartate receptor subunit NR1 (NMDAR1) exons [33], by polypyrimidine tract-binding protein (PTB) binding to elements such as CUCU to compete with U2AF65 binding to the Py in mammalian systems. A more recent example of PTB-mediated repression of U2AF65 binding is its inhibition of neuron-enriched exons in non-neuronal cells [34]. A similar mechanism represses the male-specific alternative 3′SS of drosophila transformer (Tra) exon 2 by sex lethal (sxl) [35, 36].
Other elements include the U-rich and UG repeat elements that are bound by embryonic lethal abnormal visual system protein (ELAV) and SUP-12, respectively, to repress splicing of the neuroglian 3′ terminal exon and ADF/Cofilin exon 2A [37, 38], or UG motifs by TAR DNA-binding protein 43 (TDP43) to repress cystic fibrosis transmembrane conductance regulator (CFTR) exon 9 [39, 40]. These probably target U2AF65 as well. A similar YGCU(U/G)Y motif also inhibits U2AF65 when bound by muscleblind-like 1 (MBNL1), to repress the splicing of cardiac troponin T (cTNT) exon 5 [41, 42]. In this case, it also involves a mutually exclusive secondary structure encompassing the Py, to which U2AF65 binding is competed by MBNL1 [42].
A recently reported regulatory element within the Py is the UCCCU motif that is bound by heterogeneous ribonucleoprotein particle K (hnRNP K) to repress the exon 5a of neural SNARE complex component synaptosomal-associated protein 25 (Snap25) and exon 6 of runt-related transcription factor 1 (Runx1) [43]. Mutation of the hnRNP K consensus motifs abolished its binding and addition of hnRNP K reduces U2AF65 binding to the 3′SS in a dosage-dependent way in UV-crosslinking assays. The hnRNP K target motif appears to be enriched in the 3′SS of a group of neuronal alternative exons, implying a wider impact of this regulation [43].
Another recent study has found that hnRNP C binds a U-rich element within Py, to compete with U2AF65 causing complement decay-accelerating factor 55 (CD55) exon 10 skipping [44, 61]. The binding of U2AF65 to Py and exon inclusion increased upon hnRNP C knockdown as shown in individual-nucleotide resolution UV-crosslinking and immunoprecipitation (iCLIP) assays and RNA sequencing analysis. Direct competition of U2AF65 binding by hnRNP C was observed in UV cross-linking assays using U10 RNA oligonucleotides. This hnRNP C competition with U2AF65 similarly inhibits cryptic 3′SS to avoid the exonization of Alu elements [44].
Besides the Py, a second location of 3′SS elements and their binding factors that interfere with U2AF65 binding is between the Py and 3′AG (Fig. 2b). We have called these elements REPA (regulatory elements between the Py and 3′AG) [45–49, 51]. These include CA repeats and purine-rich elements particularly G-tracts.
CA repeat elements at this location are bound by hnRNP L to inhibit U2AF65 binding to upstream Py and splicing of a group of exons [45–49, 87]. The binding of hnRNP L and inhibition of U2AF65 were demonstrated in UV cross-linking/immunoprecipitation assays [45, 46, 49, 87]. Moreover, hnRNP L loss of function/rescue assays confirmed its role in the regulation of stress hormonal axis-regulated exon (STREX) [46].
One may argue that the boundary of the CA repeats actually could not be clearly distinguished from the Py, but the recently identified G-tracts between the Py and 3′AG in a group of genes should have defined the REPA elements without any doubt [50, 51]. G-tracts are recognized by the quasi RNA recognition motifs (RRMs) of hnRNP F/H [88–90]. A ‘GGGTGGGGG’ REPA element is identified in the upstream 3′SS of exon 3 of the protein arginine methyltransferase 5 (PRMT5) gene. It is bound by hnRNP F/H to inhibit U2AF65 binding to the Py in UV cross-linking and immunoprecipitation assays. Depleting hnRNP F/H enhanced U2AF65 binding to the Py and adding-back hnRNP H decreased it in a dosage-dependent manner. Moreover, hnRNP F/H knockdown increased the endogenous exon inclusion [50]. Therefore, unlike the enhancer effect of G-tracts at other intronic locations [91–93], the uniquely positioned REPA G-tracts between the Py and 3′AG are splicing silencers. The presence of similarly positioned G-tract elements in over a thousand of human 3′SS suggests a wider impact of these elements in diversifying the protein products and functions [51].
The 3′SS inhibition by the REPA elements could be better explained by steric hindrance between the trans-acting repressor and U2AF65/35 heterodimer, which could be co-purified stably from nuclear extracts [94]. The inhibition could be due to the space constraint imposed by the element ‘insertion’ between the Py and 3′AG, which are often bound by the heterodimer for splicing of AG-dependent 3′SS [95–98]. If so, the ‘insertion’ represents an opposite way to cause steric hindrance from that by the shortened distances between splice site motifs such as BP and 3′AG or 5′SS in repressing the 3′SS of the (mutually exclusive) SM exon of alpha-actinin [28, 32, 99].
It should be noted that a REPA might not necessarily always inhibit splicing. In the case of a UGC motif at the REPA location [34], it is bound by nSR100 (neural-specific Ser/Arg repeat-related protein of 100 kDa) in neurons to directly interact with U2AF65 and enhance its binding, which will be detailed in the enhancement section. Therefore, whether a REPA causes steric hindrance between the splicing regulators might depend on the particular factor. Moreover, since such trans-acting factors can also receive upstream signaling [46], the effect by even the same factor could be changed depending on the cell environment.
Taken together, these recent cases demonstrate that: (1) the py-rich elements and bound factors could be diverse, and (2) location of the elements can also be out of the Py: “inserted” between the Py and 3′AG (REPA) to recruit more trans-acting factors, to interfere with U2AF65 binding.
Inhibition of U2 snRNP interaction with the BP
The interaction of U2 snRNP with the BP is a critical step in spliceosome assembly. It is regulated by a number of trans-acting repressors binding to RNA motifs near or directly to the BP (Fig. 2c). An earlier example showed that the U2 snRNP could be interfered sterically by the arginine/serine-rich SR protein ASF/SF2 (SRSF1) binding several nucleotides upstream of the BP [100]. In the splicing repression of HIV Tat exon 3, hnRNP A1 binding to the −26nt does not interfere with U2AF binding but inhibits the subsequent U2 snRNP interaction with the BP [52]. The N1 exon of NMDA receptor subunit NMDAR1 and exon 6 of CELF RNA-binding protein family member CUG triplet repeat RNA-binding protein 2 (CUGBP2) are also regulated in this manner [53, 101–103]. NMDAR1 pre-mRNA contains UGUGU and GU motifs around the predicted BP of N1 exon upstream 3′SS and UGU motifs are similarly located for CUGBP2 exon 6 [53]. Such UG-rich motifs are one of the two groups of sequences recognized by CUGBP2 to regulate alternative splicing, the other group being CUG triplet repeats [102–111]. CUGBP2 bound to NMDAR1 and its own 3′SS motifs in chemical modification footprinting assays [53], and its inhibition of lariat formation was confirmed by using recombinant CUGBP2 or mutagenesis of its binding sites under in vitro splicing conditions [53]. Nuclear extract complementation and mutagenesis analyses revealed that CUGBP2 binding interferes with U2 snRNP association [53]. These studies support the model of branch site-perimeter-binding and interference with U2 snRNP for exon skipping.
A more recent example is QKI (quaking homolog, KH domain) competition with the BP binding constitutive factor SF1 (splicing factor 1) to repress the exon 12 of NUMB [54], a Notch pathway regulator [112–115]. QKI is a member of STAR (signal transduction and activation of RNA) family of proteins and binds ACUAAY motif [116, 117], which is similar to the consensus ACUNAC of SF1 [118]. Both QKI and SF1 bind the AUUAAC motif immediately upstream of Py in NUMB and QKI competes with SF1 in UV-crosslinking and mutagenesis assays [54].
In case of FAS (TNF receptor superfamily member 6) exon 6, the splicing repression requires the interactions between U2AF-homology domain (UHM) of SPF45 (splicing factor 45) with UHM-ligand motifs (ULMs) of constitutive splicing factors including SF1 and U2AF65 [55]. SPF45 crosslinked to RNA in a U2AF dependent manner [119]. The interactions of SPF45 with SF1 and U2AF likely compete with UHM-ULM interactions required for FAS exon 6 splicing, possibly by inhibiting the interaction of U2AF65 with SF1 and subsequent U2 snRNP binding to BP [55].
There are also G rich motifs that are enriched in the upstream 60nt of 3′SS to repress the exon usage of some testis-specific genes in Drosophila [56]. These motifs are bound by the LS2 protein; a homologue of the U2AF larger subunit emerged through gene duplication but has diverged in binding specificity for G-rich motifs, as determined by systematic evolution of ligands by exponential enrichment (SELEX) and electrophoretic mobility shift assays (EMSA) [56]. This change in binding specificity switched the protein function to a repressor. The motifs appear to be around the usual location of BP but whether it represses U2 snRNP remains unknown.
Together, the current evidence for the splicing repression of a number of these exons indicates that regulatory motifs overlapping or near BP can specifically inhibit the interaction of SF1 or U2 snRNP with the essential 3′SS motif. The observations in the above examples strongly support this model, particularly NUMB exon 12 which is repressed by a direct competition between QKI and SF1 to inhibit BP recognition.
Competition between 3′ splice sites
Competition between 3′SSs of two consecutive exons in splicing repression has been described recently [22]. In this scenario, the competitiveness of a downstream 3′SS is enhanced upon ‘weakening’ of the upstream 3′SS due to U2AF65 binding to an upstream intronic sequence. This mechanism has been proposed for a group of exons for which U2AF binding events in the intronic regions upstream of 3′SS were identified by CLIP-seq experiments. U2AF binding to these sites inhibits the usage of the 3′SS of these introns and promotes the usage of competing 3′SSs (Fig. 2d).
3′SS scanning and competition have been proposed for more than two decades but on the basis of a common BP shared by two competing 3′AGs [99]. The above-mentioned cases do not share the same BP between the two sites; therefore they are likely regulated differently. In this regard, a very interesting study is the biochemical characterization of the regulation of c-Src N1 exon by PTB [16, 17]. Here intron-bound PTB blocks the upstream 3′SS of N1 exon [91], prevents definition of the downstream intron but allows usage of the downstream 3′SS with the constitutive 5′SS further upstream of the N1 exon [17]. According to these effects, splicing of a weak middle exon like N1 is repressed from both sides by splicing repressors; therefore, it could not splice efficiently to its downstream 3′SS. However, the latter could still be spliced efficiently with the upstream constitutive 5′SS, thereby promoting skipping of the middle exon. This provides molecular details for the dynamic transition of spliceosome recognition from alternative to constitutive splice sites.
Another way that could be considered as competition between 3′SSs involves the SR proteins activating decoy 3′SS. The role of SR proteins in the regulation of alternative splicing has been largely based on their binding to exonic splicing enhancers (ESEs) but their binding sites have also been found within introns as well as intron–exon and exon–exon junctions [120–123]. Interestingly, the intronic binding of SR proteins is more frequently associated with splicing repression [57, 121, 124, 125]. Particularly, in regulation of CFTR exon 9, the SR proteins SF2/ASF and SRp40 interact with an intronic splicing silencer (ISS) to inhibit splicing by favoring a ‘decoy’ sequence through an alternative 3′SS usage [57]. A similar effect of SR proteins has been observed in the preferential usage of proximal or distal 3′SS in β-globin, hnRNP A1 and Adenovirus pre-mRNA substrates [100, 125, 126]. Besides the splicing repression through an ISS, the SR proteins in some cases antagonize the effect of each other to modulate the choice of splice sites [127, 128].
Together, these provide examples for the inhibition of 3′SS by enhancing the relative strength of competing 3′SSs.
Inhibition of 3′ splice site usage by local RNA secondary structure or longer range loops
Another possible mechanism of splicing repression is trapping a splicing motif inside the double stranded region of a stem loop or ‘looping out’ an entire exon through interaction between complementary regions of pre-mRNA, RNA-binding proteins or different domains of a protein bound to the flanking introns (Fig. 2e).
The effect of pre-mRNA secondary structure on splicing was first observed three decades ago followed by more examples later on [129–132]. Large-scale analyses indicate that stable secondary structures based on free energy prediction are enriched at alternative 3′ or 5′ splice sites and are conserved across species [133, 134]. Complementary mutagenesis analyses of particular sites indicate that RNA stem loops can trap motifs including the Py or 3′AG, or loop out the target exon, thereby inhibiting 3′SS usage [42, 130, 135, 136]. Regulation of cTNT exon 5 is a clear example of the formation of a splicing inhibitory stem loop in 3′SS [42].
For those secondary structures/loops bridged by RNA-binding proteins, two well-studied examples involving 3′SS elements are in the repression of Drosophila Sxl exon 3 and c-Src exon N1 [16, 58–60]. In the case of Sxl exon 3, an autoregulation through binding of SXL in the upstream 3′SS and downstream intron mediates the skipping. SXL exists as a monomer in solution but it can form higher order complexes through RNA-binding-dependent multimerization [58]. In regulation of Sxl exon 3, it binds to the two sites and interacts cooperatively through N terminus to ensure the splicing repression [58–60]. The amino terminus of SXL is essential for cooperative protein–protein interaction and plays a vital role in splicing regulation. Therefore, the multimerization of SXL suggests the formation of protein–protein bridges in the looping out model. For c-Src exon N1, PTB binds to both CU elements within the upstream 3′SS and in the downstream intron to repress exon inclusion, likely by looping out the N1 exon [30]. Whether PTB binds as a monomer or multimer remains unknown though PTB is a monomer in experimental solutions [137]. A model with different RRM domains of PTB contacting different target sites within the pre-mRNA has also been proposed [138]. However, they do self-interact in yeast two hybrid assays [139]. Therefore, it remains undetermined how the loop is formed if it does.
A more recent study proposed the looping out mechanism for the regulation of CD55 exon 10 by hnRNP C binding to upstream Py as described above [44], and downstream intron [61]. The binding of hnRNP C at these positions with 165 nucleotides in-between was determined by iCLIP. The spacer sequence between the two binding sites of hnRNP C flanking CD55 alternative exon is proposed to allow the cooperative particle formation and splicing repression through a looping out mechanism.
Although observations in the above cases suggest the protein-RNA or protein–protein interactions between the splicing factors bound to the flanking introns but direct physical evidence for the “looping out” of an exon in the pre-mRNA remains to be seen. Moreover, for the secondary structures formed by complementary RNA regions, how the structures are regulated in cells to determine the usage or skipping of a 3′SS is unclear. It could be imagined that trans-acting factors (protein or RNA) might be involved to receive the upstream signals.
From the above examples, it is clear that the elements and factors now appear to be quite diverse in sequence and location in the repression of 3′SS usage. Their mechanisms of action mainly involve RNA–RNA, RNA–protein or protein–protein interactions. The binding of trans-acting factors inhibits the recruitment of constitutive splicing factors (Fig. 2; Table 1). Such inhibition could possibly be caused by protein competition (i), steric hindrance (ii), 3′SS competition (iii) or RNA secondary structure/looping out (iv), for which the first one is the simplest and best characterized.
Enhancement of 3′ splice site usage
The enhancement of 3′SS usage mainly involves the inhibition of repressor binding, binding of trans-acting activator to enhancers or direct interaction with U2AF, to promote the early steps of spliceosome assembly (Figs. 1, 3; Table 1).
Fig. 3.
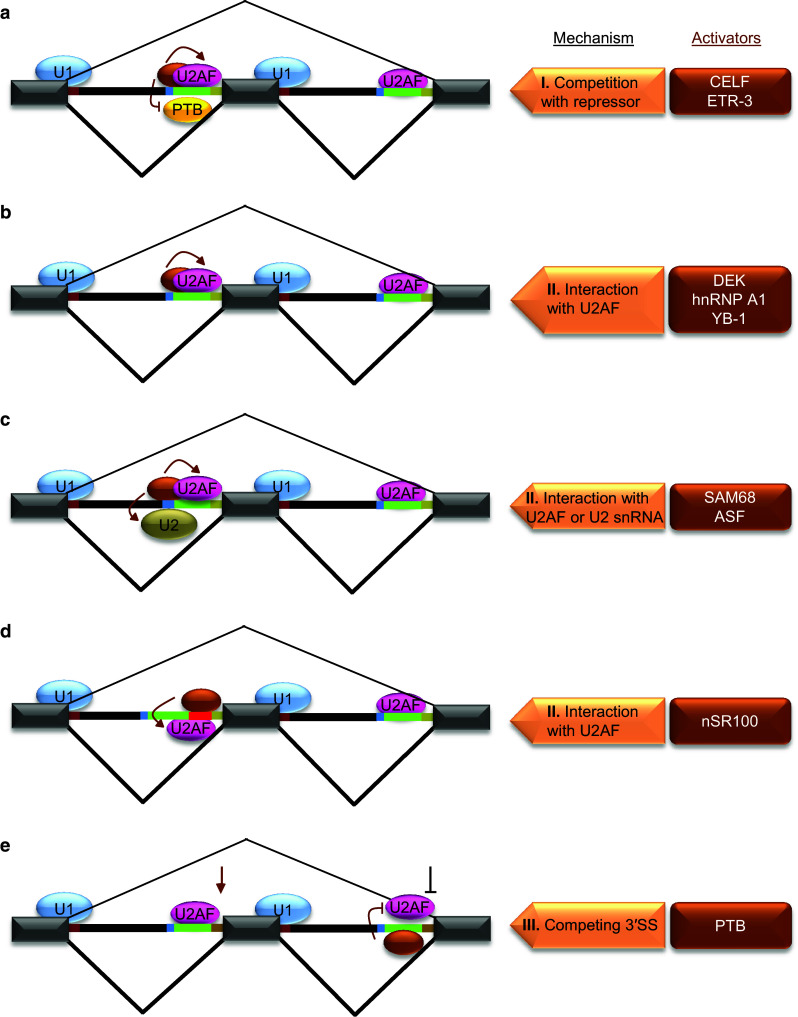
Mechanisms of splicing activation at 3′SS. Individual activators are shown as brown ovals and they are listed on the right side in brown boxes along with respective mechanisms for the regulation shown on the left side. a Splicing activation through inhibition of a repressor by enhancer motifs in Py. The binding of a trans-acting factor to an enhancer motif within Py results in competition with a repressor (yellow oval/PTB) binding (I) and facilitates U2AF binding to Py to promote 3′SS usage. b–d Splicing activation through cis-acting elements located at different positions within 3′SS. The locations of these elements are within Py (b), near BP (c) or between Py and 3′AG (red line) (d). A common feature of 3′SS regulation by these elements is their recognition by respective trans-acting factors for interaction with U2AF or U2 snRNA (II) to facilitate their RNA-binding leading to activation of splicing. e Activation of 3′SS by repression of downstream 3′SS. The repression of a downstream 3′SS through binding of PTB to Py inhibiting U2AF results in activation of a competing 3′SS (III)
Inhibition of repressor binding
The binding of a trans-acting factor to enhancer element within Py to interfere with the binding of a trans-acting repressor is a well-known mechanism of splicing activation at 3′SS (Fig. 3a). The CUG and UG enhancer motifs in the upstream 3′SS of actinin exons SM, NM and cTNT exon 5 are such examples [62, 63]. In this regulation, these motifs are bound by CELF proteins leading to competitive displacement of splicing repressor PTB that binds to adjacent sites, thereby enhancing the usage of the downstream exon. A recent example involves the displacement of hnRNP L by hnRNP LL due to a genetic mutation in the CHRNA1 (cholinergic receptor, nicotinic, alpha polypeptide 1) gene [140]. The latter, unlike hnRNP L, does not interact with PTB, thereby allowing U2AF65 interaction with a cryptic 3′SS and inclusion of an intronic sequence.
Interaction with U2AF or U2 snRNA by activators
Other regulatory elements at different positions of 3′SS are recognized by RNA-binding proteins that interact with constitutive splicing factors for splicing activation (Fig. 3b–d). A well-known example of such regulation is the splicing activation of minigene constructs of IgM and AdML pre-mRNAs through interaction of U2AF subunits with DEK (DEK proto-oncogene) [64]. The minigene activation is achieved by the interaction of a chromatin- and RNA-associating protein DEK with U2AF35 (Fig. 3b). However, the binding of DEK to the pre-mRNA is not demonstrated. Moreover, a similar mechanism is likely involved in the splicing activation of vascular endothelial growth factor (VEGF) exons 6 and 7 by CAPER proteins that share sequence homology with U2AF65 [65, 66, 141]. Although the molecular details for this mechanism are not known, it was postulated that interaction of CAPER with EWS/FLI-1, which interacts with some 3′SS constitutive factors, is involved [66].
A more recent example of similar regulation is the interaction between RNA-bound hnRNP A1 with U2AF to proofread the splice site and promote its recognition [67]. HnRNP A1 is often observed as a splicing repressor but this study explained a novel role in proofreading of 3′SS. The pre-mRNAs with pyrimidine rich sequences followed by AG or CG were utilized to study the ability of constitutive splicing factor U2AF to discriminate between the two dinucleotides. HnRNP A1 formed a complex with U2AF heterodimer facilitating it to discriminate between these 3′SS (Fig. 3b). Moreover, upon knockdown of hnRNP A1 in cells, the binding of U2AF to U-rich sequences of an intronless gene c-Jun as well as to 5′UTRs (5′ untranslated region) of three other transcripts increased, which suggests a role of hnRNP A1 for preventing the binding of U2AF to U-rich sequences beyond 3′SS in cells. However, whether hnRNPA1 has an enhancing effect on U2AF65 binding to the 3′SS of an endogenous exon in cells is not known.
A similar mechanism may also contribute to the activation of CD44 (CD44 molecule) exon V5 splicing [68], in addition to a previously known model [70]. A recent study reported CAUC splicing enhancer motifs within the weak Py of V5 upstream 3′SS that are bound by the Y box-binding protein-1 (YB-1) to recruit U2AF65 and U2AF35 through protein–protein interaction [68] (Fig. 3b). The YB-1 interacts through its C-terminal domain with the arginine-serine-rich (RS) domain of the U2AFs and enhances the recruitment of U2AF65 to Py, to which the U2AF65 alone does not bind [68]. Another RNA motif (AUAAA) immediately upstream of Py is bound by Sam68 (Src-associated in mitosis, 68 kDa protein) and enhances the 3′SS usage by facilitating U2AF65 binding to Py [70] (Fig. 3c).
The recently identified UGC motifs between Py and 3′AG also promote the inclusion of a group of exons specifically in neuronal cells [34]. nSR100 binds to these motifs and interacts with early spliceosome components including U2AF65 to promote exon inclusion (Fig. 3d). The nSR100 binding motifs were identified in the upstream introns of a group of regulated neural exons. The in vivo binding of nSR100 to these motifs was determined by photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP). Mutagenesis of minigenes in splicing reporter assays confirmed their critical role in splicing activation. Furthermore, immunoaffinity purification coupled to mass spectrometry analysis showed that nSR100 interacts with a number of constitutive splicing factors including U2AF subunits that bind in a close proximity to nSR100 binding motifs. Moreover, higher nSR100 expression was able to restore exon inclusion after U2AF65 knockdown, which suggests a compensatory effect when U2AF65 is rate limiting. This provides an interesting example of tissue specific splicing activation through the binding of a trans-acting factor to the motif uniquely positioned between Py and 3′AG, where locates the REPA elements.
Several examples involve the interaction of activators with the 3′AG, which is important for introns with weak Py tracts. In activating the c-Src N1 exon, ASF/SF2 binds to an ESE in the 5′ half of the exon but has two nucleotides overlapping with the 3′AG [142]. In regulating the Sxl exon 3, SPF45 also binds 3′AG to enhance its usage by enhancing the second catalytic step of splicing [119]. A U2AF35-related protein Urp contacts the 3′AG (or AC of the minor U12 type introns) and interacts with U2AF65 to promote splicing [69, 143], particularly the second step for U2 introns [69]. Another U2AF35-related protein U2AF26 also interacts with U2AF65 and promotes splicing [144]. The detailed molecular mechanisms of the splicing activation by ASF/SF2, SPF45, Urp or U2AF26 in these cases remain unclear. Interestingly, both ASF and Urp contain RS domains. SPF45 also contains RS dipeptides that can be phosphorylated by a SR protein kinase Clk1 in its regulation of splicing [145]. It was shown that ASF binding to or phosphorylated RS domain tethered to an ESE contacts the doubled stranded U2 snRNA-BP region to promote early spliceosome assembly and splicing [71, 72].
Taken together, these examples indicate that interaction with U2AF or U2 snRNA is another way for activators to enhance 3′SS usage.
Competition between 3′ splice sites
In this case, weakening of a downstream site activates an upstream site (Fig. 3e). This was observed in the control of alternative splicing by the commonly known repressor PTB [73]. This mechanism was inferred from the genome-wide PTB binding map based on CLIP-seq, which allowed the identification of a group of regulated exons in the context of PTB binding. Similar competitive activation of upstream 3′SSs has also been described for U2AF65, which binds an intronic site to weaken the downstream 3′SS [22]. A group of exons are regulated through this mechanism but the molecular details of this competition remain unknown.
Enhancement of weaker 3′ splice site by local RNA secondary structure or longer range looping
Besides the direct involvement of 3′SS elements and trans-acting factors in its regulation, another mechanism is the enhancement of an otherwise weaker 3′SS by RNA secondary structures or looping, to bring motifs or splice sites closer, particularly in splicing of long introns [84].
The usage of a distal 3′SS could be facilitated by a hairpin structure that brings it closer to the BP in an E1A intron [131], and more examples have been found recently in yeast introns [146]. A similar hairpin structure also blocks cryptic 3′AG to facilitate correct 3′AG usage [147]. Besides the 3′SS motifs, the 5′ and 3′ splice sites can also be brought closer by intra-molecular base-pairing, for instance between two complementary regions near the 5′SS and BP to aid the early spliceosome assembly in the yeast rp5l B pre-mRNA [148], as supported by both in vitro and in vivo evidence [148].
Studies on drosophila Dscam pre-mRNAs suggest a similar model of interaction between two intronic regions to bring splice sites closer in selecting a downstream 3′SS of mutually exclusive exons [75, 76] (Fig. 4a). The selective enhancement of 3′SS usage in the exon 6 cluster of Dscam requires base-pairing interaction between one of the selector sequences near 3′SS and a docking site [75, 76] (Fig. 4a). However, how a specific 3′SS among that of the other mutually exclusive exons is chosen in a particular cell or cell state remains unknown. It likely involves trans-acting factors such as hrp36, an hnRNP A1 homologue [74].
Fig. 4.
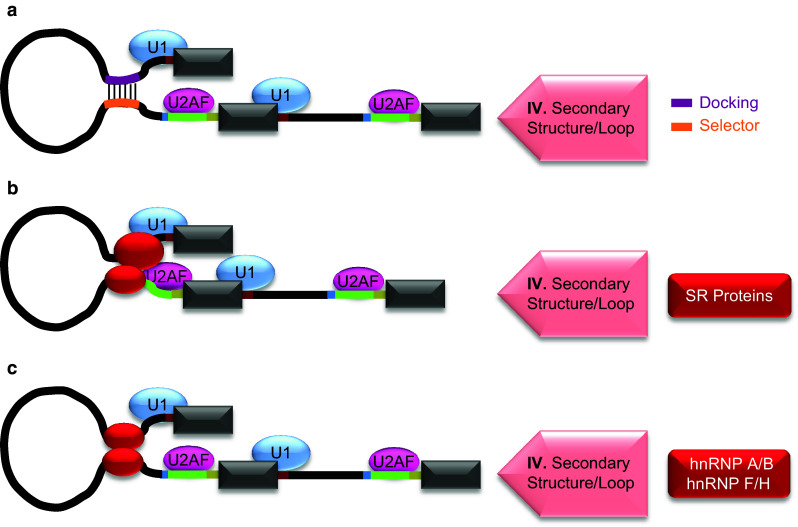
Mechanisms of weaker 3′SS activation through RNA secondary structures/loops. a Base-pairing interaction between intronic RNA motifs. The activation of a 3′SS through looping of pre-mRNA can also result from base-pairing of an intronic motif (purple line) with a second motif located near regulated splice site (orange line). These motifs are referred to as docking or selector sites as indicated on the right side. The formation of such secondary structures or loops (IV) promotes 3′SS usage. b Interaction between a complex of splicing factors and 3′SS spliceosome components. In this mechanism a protein complex (red oval) binds to an intron upstream of regulated exon and interacts with spliceosome proteins of 3′SS to promote its usage. SR proteins shown in red box on the right side form such known complexes. c Cooperative interaction of a splicing factor. This mechanism of pre-mRNA looping involves the binding of a trans-acting splicing factor (red oval) near intron ends. Examples of such factors are shown in red box on the right side. The cooperative interaction between the factors bound near intron ends creates a loop of spanning intronic sequence that brings its splice sites along with their spliceosomal components in close proximity to activate 3′SS
Besides RNA intramolecular base-pairing, protein–protein interaction may also bring the 5′ and 3′ splice sites closer (Fig. 4b). Proteins such as SRSF1 (ASF/SF2) and SRSF2 (SC35) specifically interact with both the U170K, a component of the 5′SS-binding U1 snRNP, and the 3′AG-binding U2AF35 [77, 78]. Moreover, SC35 interacts with Tra and Tra2 which also shows interaction with U2AF35 and suggested to facilitate interaction between splice sites in regulation of dsx sex specific splicing [77–80]. Studies with hnRNP A/B and F/H binding sites near introns ends indicate that their protein-RNA interactions could bring the splice sites closer as well [81–84] (Fig. 4c). Insertion of the binding sites near intron ends significantly increased the splicing of long introns suggesting looping out of a portion of pre-mRNA to bring two splice sites in close proximity. This model was further explained in vivo by hybridizing the ends of introns with oligonucleotides containing hnRNP A/B binding sites within their non-hybridizing tails. Computational analysis of hnRNP A/B and F/H binding sites at the end of introns supports this model as well [84].
Taken together, the current evidence on 3′SS activation by different elements/factors suggests that the mechanisms mainly involve the inhibition of a repressor (Fig. 3a), protein–protein interactions between an RNA-binding protein and U2AF (Fig. 3b–d), activation of a 3′SS by weakening of competing 3′SS (Fig. 3e), or local RNA secondary structures, longer range RNA loops through protein–protein bridge or RNA base-pairing to bring closer two motifs or splice sites to promote early spliceosome assembly (Fig. 4). However, many questions still remain, for instance, how does the activator protein help 3′SS recognition by interacting with the U2AF? Does it need U2AF65/35 binding to the RNA in such a constrained space or simply serve to bring the U2AFs close, like that by YB-1 [68], to help SF1 binding to the BP? Moreover, how do two splice sites compete and is it limited only to nearby 3′SSs?
Overall, according to the mode of protein/RNA interaction in the above examples, the mechanisms of 3′SS regulation (repression or activation) can be divided into: (1) protein competition for a common RNA element/sequence (repression i; enhancement I), (2) steric hindrance between protein factors (repression ii), (3) direct interaction between activators and U2AF or U2 snRNA (enhancement II), (4) competition between splice sites (repression iii, enhancement III), and (5) local RNA secondary structures or RNA loops (repression iv; enhancement IV).
Regulation of 3′ splice site usage by chromatin configuration/transcription rate
Chromatin-remodeling and the rate of transcription elongation also modulate exon usage with enhancing or repressing effect depending on the target exon [149–154]. However, the positional role of chromatin/nucleosome modifications and remodeling with respect to the precise splice sites remains largely unknown. Studies based on computational models revealed that the nucleosome occupancy at approximately 25 bp upstream of 3′AG is lowest as compared to other intronic or exonic regions [155], a position generally corresponding to BP [155, 156].
SWI/SNF is a chromatin-remodeling complex that uses ATP to facilitate conformational changes for transcriptional regulation [157–159]. The ATPase activity lies in the Brahma (Brm) or Brahma-related protein (Brg1). Overexpression and knockdown assays of Brm strongly suggest its role in splicing regulation [160]. Its most profound specific effect was observed for CD44 exon V5. However, the chromatin-remodeling activity of Brm does not seem to be required for this regulation as mutations or deletions of the ATPase domain did not substantially change splicing whereas deletion of its chromatin binding C-terminal part resulted in decreased exon inclusion. RNA immunoprecipitation assays of Brm revealed that it interacts with U1 and under conditions that favor the inclusion of CD44 exon V5 it also interacts with U5 snRNA [160].
The splicing of CD44 exon V5 is enhanced by Sam68 binding in the exon [161], as well as near BP as described above [70]. Co-immunoprecipitation assays revealed that Brm also associates with Sam68 and they cooperatively enhance splicing of this exon [160]. Moreover, ChIP (chromatin immunoprecipitation) walking using antibodies against phospho-CTD (C-terminal domain) of PolII (RNA polymerase II) revealed higher occupancy in the CD44 variant region including V5, which suggests a slower transcription rate for enhanced exon usage of CD44 V5 [160]. These data support a role for a factor of the chromatin-remodeling complex in the regulation of splicing, likely involving its interaction with those of the 3′SS.
Role of posttranslational modifications and upstream signaling in 3′ splice site regulation
Besides the cis-acting RNA elements, trans-acting factors and chromatin/transcriptional machinery, posttranslational modifications of trans-acting splicing factors by cellular signaling pathways add another layer of regulation. Table 2 lists the posttranslational modifications of 3′SS trans-acting factors that influence splicing regulation. The effects on splicing regulation through these factors are described below.
Table 2.
Posttranslational modifications of 3′SS factors
| Splicing factor | Modification | Residue | Effect on splicing | References |
|---|---|---|---|---|
| U2AF65 | Hydroxylation | Lys-15, Lys-276 | Activating | [162] |
| SF1 | Phosphorylation | Ser-80, Ser-82, Ser-20 | Activating/repressing | [163–166] |
| hnRNP K | Phosphorylation | Ser/Thr (predicted) | Repressing | [43] |
| hnRNP L | Phosphorylation | Ser-513 | Repressing | [46] |
| DEK | Phosphorylation | Ser-19, Ser-32 | Activating | [64] |
| Sm proteins | Methylation | Arg rich motifs | Activating | [167] |
| SRSF2 | Acetylation | Lys-52 | Activating | [168, 169] |
| U2AF35 | Ubiquitination | Within residues 43–146 | Repressing | [170] |
| Prp3 | Ubiquitination | Two Lys residues | Repressing | [171] |
| SF3ba | Spliceostatin binding | NA | Repressing | [172] |
NA not applicable
aNon-covalent binding
Hydroxylation
U2AF65 undergoes posttranslational hydroxylation by the Fe(II) and 2-oxoglutarate–dependent dioxygenase jumonji domain-6 protein (JMJD6) in the presence of oxygen [162]. The hydroxylation of Lys15 and Lys276 of the endogenous U2AF65 was detected by liquid chromatography-tandem mass spectrometry (LC–MS/MS) analysis of HeLa cells. U2AF65 co-immunoprecipitates with JMJD6 in an RNAse-sensitive manner. Knockdown of JMJD6 resulted in increased exon 19 skipping of endogenous MGEA6 pre-mRNA and a similar effect was observed for a α-tropomyosin minigene construct comprised of exons 1, 3 and 4. This indirectly shows the role of U2AF65 hydroxylation in splicing activation of these exons as JMJD6 catalyzes it. The effect of hydroxylation on MGEA6 splicing is further supported by treatment of cells with oxygenase inhibitor desferrioxamine (DFO), which showed a similar effect as JMJD6 knockdown. These data demonstrate a unique posttranslational modification of a splicing factor in the presence of oxygen, likely related to splicing regulation under hypoxia conditions. However, a direct effect of U2AF65 hydroxylation on splicing, the underlying molecular mechanism and the hydroxylation status of other splicing factors under similar conditions remain unknown.
Phosphorylation
Phosphorylation of 3′SS trans-acting factors such as SF1 also contributes to alternative splicing regulation [163–165]. The protein–protein interaction of SF1 and U2AF65 and their cooperative binding to 3′SS motifs is a critical step for further spliceosome assembly. In vitro kinase assays followed by mass spectrometry analysis revealed the phosphorylation of SF1 at serines 80 and 82 [165]. In vivo metabolic labeling of overexpressed SF1 or its mutants confirmed this phosphorylation. The phosphorylation of SF1 facilitates its ternary complex formation with U2AF65 and slightly enhances the RNA-binding in EMSA [163, 165], hence likely to promote the spliceosome assembly and splicing. Contrary to the activating effect of Ser80 and Ser82 phosphorylation, previous studies of SF1 Ser20 phosphorylation by protein kinase G (PKG) suggest a negative effect on protein–protein interaction as well as spliceosome assembly [164, 166]. Thus, phosphorylation of different residues of SF1 may have different effects on spliceosome assembly and interaction of SF1 with U2AF.
Two other examples of splicing factor phosphorylation are of hnRNPs K and L [43, 46]. The hnRNP K is phosphorylated in the PKA-regulated splicing of SNAP25 exon 5a. Its binding to the 3′SS Py is abrogated by pre-treatment with lambda protein phosphatase, suggesting an essential role of its phosphorylation in its regulation of the 3′SS. The serines 284 and 353 of hnRNP K are phosphorylated by the MAP/ERK pathway [173], but their role in the splicing regulation is unknown. In case of hnRNP L, depolarization activated CaMKIV (Calcium/calmodulin-dependent kinase IV) phosphorylates serine 513 to regulate splicing of STREX [46–48, 174]. The phosphorylation enhances its binding to the CaRRE1 (Calcium/calmodulin-dependent kinase IV-responsive RNA element) pre-mRNA and inhibition of U2AF65 [46].
The phosphorylation of DEK is required for intron removal [64]. This protein interacts with U2AF35 to form a complex that is disrupted upon phosphatase treatment. Mutagenesis analysis revealed that the complex formation is dependent on the phosphorylation of serines 19 and 32.
Other regulatory factors acting on the 3′SS are also phosphorylated but the phosphorylation effect on 3′SS usage has not been reported. For instance, hnRNP A1 is phosphorylated upon activation of the p38 MAP kinase pathway by osmotic or UV stress, which caused its redistribution from nucleus to the cytoplasm and accompanied enhanced proximal 5′SS usage of E1A reporter minigenes [175]. The redistribution requires phosphorylation of a number of serines within a C-terminal peptide that interacts with transportin 1 [176]. PTBP1 is phosphorylated by protein kinase A upon forskolin stimulation [177]. The target Ser16 is essential for its redistribution from the nucleus to the cytoplasm and neurites [177, 178].
The phosphorylation of SR proteins, which also influence 3′SS selection [57, 100, 126], regulates their subcellular redistribution, RNA-binding and effect on splicing [179–183]. Their specific kinase yeast Sky1 is implicated in 3′AG recognition [184]. Importantly, both the hypo- and hyperphosphorylation of SR proteins can inhibit splicing suggesting the role of their phosphorylation levels in alternative splicing [185–187].
Methylation
Many constitutive and alternative splicing factors are methylated including the spliceosome core Sm proteins [167, 188]. The reduction in methylation of Sm proteins leads to aberrant splicing of specific pre-mRNAs. For instance, the splicing of Mdm4 (MDM4, p53 regulator) exon 7 is repressed in the absence of PRMT5 (protein arginine methyltransferase 5) that methylates Sm proteins [167].
Acetylation
Acetylation inhibitors also affect various steps of spliceosome assembly, suggesting that acetylation of multiple components/steps of the spliceosome assembly are involved in this process [169]. SRSF2 acetylation on lysine 52 in its RRM destabilizes the protein through proteosomal degradation in the regulation of the cell growth/apoptosis variants of caspase-8 gene [168]. A large number of splicing regulator proteins including most of hnRNPs mentioned above are also acetylated in cells [150]. Particularly lysines 87, 98 and 224 of hnRNP F that are potential targets of acetylation and ubiquitination are critical for increased protein stability by a deacetylase inhibitor trichostatin A (TSA) [189]. Their acetylation effect on 3′SS usage remains to be examined.
Ubiquitination
Ubiquitination of U2AF35 contributes to splicing regulation. One of the Shigella factors IpaH9.8 specifically interacts with U2AF35 to interfere with splicing of IgM pre-mRNA [190]. IpaH9.8 catalyzes U2AF35 ubiquitination in vitro [170].
U4 snRNP component Prp3 is modified by Prp19 complex with nonproteolytic ubiquitin chains [171]. This ubiquitination facilitates the interaction of Prp3 with U5 snRNP and therefore promotes the stability of U4-U5-U6 tri-snRNP. Usp4 deubiquitinates Prp3 leading to disruption of the U4-U5-U6 tri-snRNP in HeLa splicing extracts, which inhibits in vitro splicing of a substrate ftz pre-mRNA. However, knockdown of Usp4 decreased the levels of correctly spliced products of a group of pre-mRNAs [171]. This suggests the role of Prp3 ubiquitination in maintenance of the U4-U5-U6 tri-snRNP and its reversal by Usp4 in spliceosomal rearrangements, particularly release of Prp3 and other U4 proteins, required for U6 to participate in active spliceosome [171, 191]. Moreover, a component of U5 snRNP called Prp8p is also ubiquitinated and this modification has been suggested to be involved in the maintenance of U4-U5-U6 tri-snRNP assembly [192, 193]. Therefore, more than one spliceosomal factors are ubiquitinated during splicing.
Spliceostatin binding to SF3b
Spliceostatin is a methylated derivative of a natural product FR901464 [172, 194]. Originally this product was identified for its anti-cancer effect through enhanced transcription activity of SV40, which promotes cell cycle arrest [195]. Later on it was discovered that spliceostatin binds to SF3b [172], an important component of U2 snRNP critical for its interaction with U2AF [196–199]. The binding of spliceostatin to SF3b is not a posttranslational modification. However, this binding results in inhibition of U2 snRNP recognition of the BP, in complex formation and in vitro splicing assays [172, 200, 201]. Spliceostatin treatment of HeLa cells leads to nonsense-mediated mRNA decay (NMD) of splice variants and downregulation of genes important for cell division, such as cyclin A2 and Aurora A kinase [200].
Taken together, current evidence has demonstrated that various modifications of trans-acting factors regulate protein level, protein–protein or protein-RNA interactions to control the 3′SS usage. However, the modifications of many more 3′SS splicing factors remain unknown.
Evolutionary emergence of regulatory elements and factors
Global insights into alternative exon creation revealed that over the course of mammalian evolution the abundance of alternative splicing significantly increased as compared to lower vertebrates [202]. This is further consolidated by independent alternative splicing data sets from different species [203, 204]. However, the evolutionary changes that contributed to the splicing regulatory mechanisms involved in the emergence of alternative exons remain largely unknown.
Evolutionary mechanisms for the de novo creation of alternative exons include tandem exon duplication, exonization of transposable elements and transition of a constitutive to alternative exon [205–209]. The transitions are achieved through accumulation of mutations at splice sites or splicing regulatory elements [209–212]. The splice sites of lower vertebrates such as fish shows remarkably low prevalence of regulatory elements such as GGG [213]. This implies that the 3′SS regulatory elements perhaps are largely characteristic of mammals.
Sequence alignment of the upstream 3′SS of Snap25 exon 5a from different species showed that the hnRNP K binding motifs are mostly conserved in vertebrates and expanded into multiple copies in mammals whereas they are absent in invertebrates [43]. Similar sequence alignment of STREX upstream 3′SS of different species indicated that the CA contents of CaRRE1 increased from lower to higher vertebrates [46, 214]. For the PRMT5 G-tract element [51], it clearly emerged in mammals over the course of evolution [215]. Human and several other mammals contain two copies of G-tract that are reduced to one copy in lower mammals and marsupials; in lower vertebrates including fish and amphibians this element is absent. The skipping of exon 3 is significantly increased in human PRMT5 transcripts. Moreover, the critical role of the G-tracts in splicing repression is confirmed by mutagenesis and splicing assays [51]. The evolutionary emergence of this splicing regulatory element provides a clear example of the transition of a mainly constitutive to an alternative exon. Together, these evolved regulatory elements may have contributed to the proteomic diversity in mammals [214].
Further considerations
Of the regulation mechanisms, the competition model is perhaps by far the simplest and most well supported for the 3′SS regulation. However, many more examples have been accumulating that cannot be simply explained by this model. Therefore, other mechanisms such as steric hindrance, splice site competition and local RNA secondary structure or looping have also been proposed. Yet, more direct evidence and molecular details are still needed for some of the latter models. Moreover, how the inhibition or activation happens after recruitment of the trans-acting factors remains unknown in most cases.
Despite the above progresses, further study is necessary for insights into the regulatory mechanisms. The regulation of splicing takes elaborate interplay of multiple factors to determine the fate of an exon. The examples discussed in this review mainly focus on the role of a particular element/factor within 3′SS. How they collaborate with other elements/factors or splice sites to determine the choice of a particular splice site among the many other sites remains largely unknown. For example in the regulation of STREX splicing, hnRNP L is a repressor through binding to CaRRE1 [46–48], but its interaction with other factors such as PTBP1, hnRNP K and LL in the regulation remains unknown [46, 216]. Moreover, the regulation of trans-acting factors by upstream signals has just started to be unveiled; more effort is needed to fully understand the impact of this regulation on 3′SS choice and splicing in general. A number of other modifications of splicing factors such as acetylation and methylation in the regulation of alternative splicing have just emerged recently [149, 150, 167–169, 189, 217, 218]. Furthermore, chromatin modification and the transcriptional machinery also have important impact on splice site choice; however, their detailed mechanisms of action remain to be answered, though models based on transcription rate and splice site strength have been proposed.
The functions of novel so-called non-coding RNAs (ncRNA) have just started to be revealed as well but already shown to impact many steps of gene expression [219]. Their role in splicing regulation particularly through 3′SS remains unexplored. One study showed that snoRNA HBII-52 regulates the splicing of serotonin receptor 5HT2C by base-pairing with a silencer element in its exon to influence the splicing of Vb through alternative usage of a 5′SS [220]. In another study, a natural anti-sense transcript (NAT) that overlaps with 5′SS is associated with intron retention in the 5′ UTR of Zeb2 pre-mRNA [221]. This intron retention preserves the IRES hence allowing the translation of Zeb2 protein. Several other studies also suggest a link between anti-sense RNAs and posttranscriptional processing particularly splicing [222–224]. Such mechanisms for 3′SS regulation remain to be explored.
Beyond the expected regulatory elements/factors, how do the constitutive (or even “core”) spliceosome factors regulate alternative splicing? Besides differences in the splice site sequences and strengths of target transcripts, the role of the homologous factors such as PUF60 and CAPER to U2AF65 [141, 225], U2AF26 to U2AF35 [144] in this process remains to be intensively explored. Moreover, many splicing factors themselves are alternatively spliced. Though some of their variant transcripts including those of the core spliceosome factors are subject to NMD [226], in other cases variant protein products of splicing regulators do have differential effects on splicing [227]. Furthermore, pseudogenes have also come into play with gene expression [228]. As analysis of the genome/transcriptome accelerates, more such homologous factors, splice variants, pseudogene or even non-homologous factors but with overlapping functions might be identified beyond the previously known constitutive or “core” spliceosome factors.
In addition to studying the details of molecular mechanisms regulating individual exons, genome/transcriptome-wide analyses among different species have identified conserved splicing regulatory elements within/nearby splice sites and reveal their evolutionary emergence [213, 229]. A human genome search identified several potential splicing regulatory elements including evolutionarily emerged splicing silencer G-tract element between Py and 3′AG [51]. Application of these approaches to more elements/factors will help to reveal the diverse regulation of 3′SS choice and evolutionary changes that contributed to the increased proteomic diversity and organismal complexity in mammals. The knowledge of large-scale analysis of splicing regulation will also help us understand the pathological conditions that arise from aberrant splicing due to genetic mutations. For instance, the mutations of several 3′SS spliceosome factors are associated with different clinical outcomes [230–232], suggesting their cooperation with other genetic lesions [233], or different target exon profiles.
In conclusion, the diversity of characterized 3′SS regulation has been increasing rapidly in the past decade. With more possibilities of regulation remaining to be explored, this diversity is likely going to continue to increase. The diverse regulation likely contributes to the higher transcriptome and proteome diversity in mammals than lower vertebrates. Further studies will help us understand not only this important step of gene regulation in normal cell physiology but also the pathophysiology and development of therapeutic approaches for aberrant splicing-caused diseases.
Acknowledgments
This work is supported by the Natural Sciences and Engineering Council of Canada (NSERC, #RGPIN/385807-2010), the Canadian Institutes of Health Research (CIHR, FRN_106608) and Research Manitoba.
References
- 1.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418(6894):236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 4.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends Mol Med. 2012;18(8):472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng D, Xie J. Aberrant splicing in neurological diseases. Wiley Interdisc Rev RNA. 2013;4(6):631–649. doi: 10.1002/wrna.1184. [DOI] [PubMed] [Google Scholar]
- 7.Umen JG, Guthrie C. The second catalytic step of pre-mRNA splicing. RNA. 1995;1(9):869–885. [PMC free article] [PubMed] [Google Scholar]
- 8.Huang T, Vilardell J, Query CC. Pre-spliceosome formation in S. pombe requires a stable complex of SF1-U2AF(59)-U2AF(23) EMBO J. 2002;21(20):5516–5526. doi: 10.1093/emboj/cdf555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollins C, Zorio DA, MacMorris M, Blumenthal T. U2AF binding selects for the high conservation of the C. elegans 3′ splice site. RNA. 2005;11(3):248–253. doi: 10.1261/rna.7221605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blumenthal T, Steward K, et al. RNA processing and gene structure. In: Riddle DL, Blumenthal T, Meyer BJ, et al., editors. C. elegans II. 2. New York: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 11.Kent WJ, Zahler AM. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res. 2000;10(8):1115–1125. doi: 10.1101/gr.10.8.1115. [DOI] [PubMed] [Google Scholar]
- 12.Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989;3(12B):2113–2123. doi: 10.1101/gad.3.12b.2113. [DOI] [PubMed] [Google Scholar]
- 13.Smith CW, Porro EB, Patton JG, Nadal-Ginard B. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature. 1989;342(6247):243–247. doi: 10.1038/342243a0. [DOI] [PubMed] [Google Scholar]
- 14.Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harbor Perspect Biol. 2011;3:a003707. doi: 10.1101/cshperspect.a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270(6):2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- 16.Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell. 2005;19(4):485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol. 2008;15(2):183–191. doi: 10.1038/nsmb.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 19.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24(24):10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T, Fu XD. Genomic functions of U2AF in constitutive and regulated splicing. RNA Biol. 2015;12(5):479–485. doi: 10.1080/15476286.2015.1020272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, Li H, et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol. 2014;21(11):997–1005. doi: 10.1038/nsmb.2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 2011;25(4):373–384. doi: 10.1101/gad.2004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosel TD, Hung LH, Medenbach J, Donde K, Starke S, Benes V, Ratsch G, Bindereif A. RNA-Seq analysis in mutant zebrafish reveals role of U1C protein in alternative splicing regulation. EMBO J. 2011;30(10):1965–1976. doi: 10.1038/emboj.2011.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauliere J, Sureau A, Expert-Bezancon A, Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol Cell Biol. 2006;26(23):8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gooding C, Roberts GC, Moreau G, Nadal-Ginard B, Smith CW. Smooth muscle-specific switching of alpha-tropomyosin mutually exclusive exon selection by specific inhibition of the strong default exon. EMBO J. 1994;13(16):3861–3872. doi: 10.1002/j.1460-2075.1994.tb06697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perez I, Lin CH, McAfee JG, Patton JG. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3(7):764–778. [PMC free article] [PubMed] [Google Scholar]
- 28.Smith CW, Nadal-Ginard B. Mutually exclusive splicing of alpha-tropomyosin exons enforced by an unusual lariat branch point location: implications for constitutive splicing. Cell. 1989;56(5):749–758. doi: 10.1016/0092-8674(89)90678-8. [DOI] [PubMed] [Google Scholar]
- 29.Chan RC, Black DL. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol Cell Biol. 1997;17(8):4667–4676. doi: 10.1128/MCB.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou MY, Underwood JG, Nikolic J, Luu MH, Black DL. Multisite RNA binding and release of polypyrimidine tract binding protein during the regulation of c-src neural-specific splicing. Mol Cell. 2000;5(6):949–957. doi: 10.1016/S1097-2765(00)80260-9. [DOI] [PubMed] [Google Scholar]
- 31.Matlin AJ, Southby J, Gooding C, Smith CW. Repression of alpha-actinin SM exon splicing by assisted binding of PTB to the polypyrimidine tract. RNA. 2007;13(8):1214–1223. doi: 10.1261/rna.219607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Southby J, Gooding C, Smith CW. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of alpha-actinin mutually exclusive exons. Mol Cell Biol. 1999;19(4):2699–2711. doi: 10.1128/MCB.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L, Liu W, Grabowski PJ. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA (New York) 1999;5(1):117–130. doi: 10.1017/S1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raj B, Irimia M, Braunschweig U, Sterne-Weiler T, O’Hanlon D, Lin ZY, Chen GI, Easton LE, Ule J, Gingras AC, et al. A global regulatory mechanism for activating an exon network required for neurogenesis. Mol Cell. 2014;56(1):90–103. doi: 10.1016/j.molcel.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh R, Banerjee H, Green MR. Differential recognition of the polypyrimidine-tract by the general splicing factor U2AF65 and the splicing repressor sex-lethal. RNA. 2000;6(6):901–911. doi: 10.1017/S1355838200000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valcarcel J, Singh R, Zamore PD, Green MR. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature. 1993;362(6416):171–175. doi: 10.1038/362171a0. [DOI] [PubMed] [Google Scholar]
- 37.Lisbin MJ, Qiu J, White K. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 2001;15(19):2546–2561. doi: 10.1101/gad.903101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anyanful A, Ono K, Johnsen RC, Ly H, Jensen V, Baillie DL, Ono S. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans . J Cell Biol. 2004;167(4):639–647. doi: 10.1083/jcb.200407085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayala YM, Pagani F, Baralle FE. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 2006;580(5):1339–1344. doi: 10.1016/j.febslet.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 40.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle FE. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20(7):1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23(15):3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci USA. 2009;106(23):9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao W, Razanau A, Feng D, Lobo VG, Xie J. Control of alternative splicing by forskolin through hnRNP K during neuronal differentiation. Nucleic Acids Res. 2012;40(16):8059–8071. doi: 10.1093/nar/gks504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell. 2013;152(3):453–466. doi: 10.1016/j.cell.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loh TJ, Cho S, Moon H, Jang HN, Williams DR, Jung DW, Kim IC, Ghigna C, Biamonti G, Zheng X, et al. hnRNP L inhibits CD44 V10 exon splicing through interacting with its upstream intron. Biochim Biophys Acta. 2015;1849(6):743–750. doi: 10.1016/j.bbagrm.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 46.Liu G, Razanau A, Hai Y, Yu J, Sohail M, Lobo VG, Chu J, Kung SK, Xie J. A conserved serine of heterogeneous nuclear ribonucleoprotein L (hnRNP L) mediates depolarization-regulated alternative splicing of potassium channels. J Biol Chem. 2012;287(27):22709–22716. doi: 10.1074/jbc.M112.357343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xie J, Jan C, Stoilov P, Park J, Black DL. A consensus CaMK IV-responsive RNA sequence mediates regulation of alternative exons in neurons. RNA. 2005;11(12):1825–1834. doi: 10.1261/rna.2171205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J, Hai Y, Liu G, Fang T, Kung SK, Xie J. The heterogeneous nuclear ribonucleoprotein L is an essential component in the Ca2+/calmodulin-dependent protein kinase IV-regulated alternative splicing through cytidine-adenosine repeats. J Biol Chem. 2009;284(3):1505–1513. doi: 10.1074/jbc.M805113200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heiner M, Hui J, Schreiner S, Hung LH, Bindereif A. HnRNP L-mediated regulation of mammalian alternative splicing by interference with splice site recognition. RNA Biol. 2010;7(1):56–64. doi: 10.4161/rna.7.1.10402. [DOI] [PubMed] [Google Scholar]
- 50.Sohail M, Xie J. Evolutionary emergence of a novel splice variant with an opposite effect on the cell cycle. Mol Cell Biol. 2015;35(12):2203–2214. doi: 10.1128/MCB.00190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sohail M, Cao W, Mahmood N, Myschyshyn M, Hong SP, Xie J. Evolutionarily emerged G tracts between the polypyrimidine tract and 3′AG are splicing silencers enriched in genes involved in cancer. BMC Genom. 2014;15:1143. doi: 10.1186/1471-2164-15-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tange TO, Damgaard CK, Guth S, Valcarcel J, Kjems J. The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J. 2001;20(20):5748–5758. doi: 10.1093/emboj/20.20.5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dembowski JA, Grabowski PJ. The CUGBP2 splicing factor regulates an ensemble of branchpoints from perimeter binding sites with implications for autoregulation. PLoS Genet. 2009;5(8):e1000595. doi: 10.1371/journal.pgen.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H, et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10(4):e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Corsini L, Bonnal S, Basquin J, Hothorn M, Scheffzek K, Valcarcel J, Sattler M. U2AF-homology motif interactions are required for alternative splicing regulation by SPF45. Nat Struct Mol Biol. 2007;14(7):620–629. doi: 10.1038/nsmb1260. [DOI] [PubMed] [Google Scholar]
- 56.Taliaferro JM, Alvarez N, Green RE, Blanchette M, Rio DC. Evolution of a tissue-specific splicing network. Genes Dev. 2011;25(6):608–620. doi: 10.1101/gad.2009011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Buratti E, Stuani C, De Prato G, Baralle FE. SR protein-mediated inhibition of CFTR exon 9 inclusion: molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 2007;35(13):4359–4368. doi: 10.1093/nar/gkm444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Bell LR. The Sex-lethal amino terminus mediates cooperative interactions in RNA binding and is essential for splicing regulation. Genes Dev. 1994;8(17):2072–2085. doi: 10.1101/gad.8.17.2072. [DOI] [PubMed] [Google Scholar]
- 59.Horabin JI, Schedl P. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol Cell Biol. 1993;13(12):7734–7746. doi: 10.1128/MCB.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakamoto H, Inoue K, Higuchi I, Ono Y, Shimura Y. Control of Drosophila Sex-lethal pre-mRNA splicing by its own female-specific product. Nucleic Acids Res. 1992;20(21):5533–5540. doi: 10.1093/nar/20.21.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol. 2010;17(7):909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gromak N, Matlin AJ, Cooper TA, Smith CW. Antagonistic regulation of alpha-actinin alternative splicing by CELF proteins and polypyrimidine tract binding protein. RNA. 2003;9(4):443–456. doi: 10.1261/rna.2191903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Charlet BN, Logan P, Singh G, Cooper TA. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol Cell. 2002;9(3):649–658. doi: 10.1016/S1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- 64.Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3′ splice site recognition by DEK. Science. 2006;312(5782):1961–1965. doi: 10.1126/science.1128659. [DOI] [PubMed] [Google Scholar]
- 65.Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell. 2005;17(3):429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 66.Huang G, Zhou Z, Wang H, Kleinerman ES. CAPER-alpha alternative splicing regulates the expression of vascular endothelial growth factor(1)(6)(5) in Ewing sarcoma cells. Cancer. 2012;118(8):2106–2116. doi: 10.1002/cncr.26488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcarcel J. hnRNP A1 proofreads 3′ splice site recognition by U2AF. Mol Cell. 2012;45(3):314–329. doi: 10.1016/j.molcel.2011.11.033. [DOI] [PubMed] [Google Scholar]
- 68.Wei WJ, Mu SR, Heiner M, Fu X, Cao LJ, Gong XF, Bindereif A, Hui J. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res. 2012;40(17):8622–8636. doi: 10.1093/nar/gks579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen H, Zheng X, Luecke S, Green MR. The U2AF35-related protein Urp contacts the 3′ splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev. 2010;24(21):2389–2394. doi: 10.1101/gad.1974810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tisserant A, Konig H. Signal-regulated Pre-mRNA occupancy by the general splicing factor U2AF. PLoS ONE. 2008;3(1):e1418. doi: 10.1371/journal.pone.0001418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen H, Green MR. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev. 2006;20(13):1755–1765. doi: 10.1101/gad.1422106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen H, Kan JL, Green MR. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell. 2004;13(3):367–376. doi: 10.1016/S1097-2765(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 73.Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36(6):996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Olson S, Blanchette M, Park J, Savva Y, Yeo GW, Yeakley JM, Rio DC, Graveley BR. A regulator of Dscam mutually exclusive splicing fidelity. Nat Struct Mol Biol. 2007;14(12):1134–1140. doi: 10.1038/nsmb1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.May GE, Olson S, McManus CJ, Graveley BR. Competing RNA secondary structures are required for mutually exclusive splicing of the Dscam exon 6 cluster. RNA. 2011;17(2):222–229. doi: 10.1261/rna.2521311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Graveley BR. Mutually exclusive splicing of the insect Dscam pre-mRNA directed by competing intronic RNA secondary structures. Cell. 2005;123(1):65–73. doi: 10.1016/j.cell.2005.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75(6):1061–1070. doi: 10.1016/0092-8674(93)90316-I. [DOI] [PubMed] [Google Scholar]
- 78.Fu XD, Maniatis T. The 35-kDa mammalian splicing factor SC35 mediates specific interactions between U1 and U2 small nuclear ribonucleoprotein particles at the 3′ splice site. Proc Natl Acad Sci USA. 1992;89(5):1725–1729. doi: 10.1073/pnas.89.5.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hertel KJ, Maniatis T. The function of multisite splicing enhancers. Mol Cell. 1998;1(3):449–455. doi: 10.1016/S1097-2765(00)80045-3. [DOI] [PubMed] [Google Scholar]
- 80.Graveley BR, Hertel KJ, Maniatis T. A systematic analysis of the factors that determine the strength of pre-mRNA splicing enhancers. EMBO J. 1998;17(22):6747–6756. doi: 10.1093/emboj/17.22.6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nasim FU, Hutchison S, Cordeau M, Chabot B. High-affinity hnRNP A1 binding sites and duplex-forming inverted repeats have similar effects on 5′ splice site selection in support of a common looping out and repression mechanism. RNA. 2002;8(8):1078–1089. doi: 10.1017/S1355838202024056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chabot B, Blanchette M, Lapierre I, La Branche H. An intron element modulating 5′ splice site selection in the hnRNP A1 pre-mRNA interacts with hnRNP A1. Mol Cell Biol. 1997;17(4):1776–1786. doi: 10.1128/MCB.17.4.1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Blanchette M, Chabot B. Modulation of exon skipping by high-affinity hnRNP A1-binding sites and by intron elements that repress splice site utilization. EMBO J. 1999;18(7):1939–1952. doi: 10.1093/emboj/18.7.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4(2):e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mulligan GJ, Guo W, Wormsley S, Helfman DM. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of beta-tropomyosin pre-mRNA. J Biol Chem. 1992;267(35):25480–25487. [PubMed] [Google Scholar]
- 86.Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science. 1995;268(5214):1173–1176. doi: 10.1126/science.7761834. [DOI] [PubMed] [Google Scholar]
- 87.Li H, Liu G, Yu J, Cao W, Lobo VG, Xie J. In vivo selection of kinase-responsive RNA elements controlling alternative splicing. J Biol Chem. 2009;284(24):16191–16201. doi: 10.1074/jbc.M900393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caputi M, Zahler AM. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J Biol Chem. 2001;276(47):43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- 89.Dominguez C, Allain FH. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res. 2006;34(13):3634–3645. doi: 10.1093/nar/gkl488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Garneau D, Revil T, Fisette JF, Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J Biol Chem. 2005;280(24):22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- 91.Chou MY, Rooke N, Turck CW, Black DL. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol Cell Biol. 1999;19(1):69–77. doi: 10.1128/MCB.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schaub MC, Lopez SR, Caputi M. Members of the heterogeneous nuclear ribonucleoprotein H family activate splicing of an HIV-1 splicing substrate by promoting formation of ATP-dependent spliceosomal complexes. J Biol Chem. 2007;282(18):13617–13626. doi: 10.1074/jbc.M700774200. [DOI] [PubMed] [Google Scholar]
- 93.Wang E, Dimova N, Cambi F. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res. 2007;35(12):4164–4178. doi: 10.1093/nar/gkm387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zamore PD, Green MR. Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci USA. 1989;86(23):9243–9247. doi: 10.1073/pnas.86.23.9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Moore MJ. Intron recognition comes of AGe. Nature structural biology. 2000;7(1):14–16. doi: 10.1038/71207. [DOI] [PubMed] [Google Scholar]
- 96.Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999;402(6763):838–841. doi: 10.1038/45602. [DOI] [PubMed] [Google Scholar]
- 97.Zorio DA, Blumenthal T. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999;402(6763):835–838. doi: 10.1038/45597. [DOI] [PubMed] [Google Scholar]
- 98.Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999;402(6763):832–835. doi: 10.1038/45590. [DOI] [PubMed] [Google Scholar]
- 99.Smith CW, Chu TT, Nadal-Ginard B. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol Cell Biol. 1993;13(8):4939–4952. doi: 10.1128/MCB.13.8.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kanopka A, Muhlemann O, Akusjarvi G. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature. 1996;381(6582):535–538. doi: 10.1038/381535a0. [DOI] [PubMed] [Google Scholar]
- 101.Moriyoshi K, Masu M, Ishii T, Shigemoto R, Mizuno N, Nakanishi S. Molecular cloning and characterization of the rat NMDA receptor. Nature. 1991;354(6348):31–37. doi: 10.1038/354031a0. [DOI] [PubMed] [Google Scholar]
- 102.Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21(4):1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol Cell Biol. 2005;25(3):879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suzuki H, Takeuchi M, Sugiyama A, Alam AK, Vu LT, Sekiyama Y, Dam HC, Ohki SY, Tsukahara T. Alternative splicing produces structural and functional changes in CUGBP2. BMC Biochem. 2012;13:6. doi: 10.1186/1471-2091-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280(5364):737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 106.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24(22):4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81(3):403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 108.Suzuki H, Jin Y, Otani H, Yasuda K, Inoue K. Regulation of alternative splicing of alpha-actinin transcript by Bruno-like proteins. Genes Cells. 2002;7(2):133–141. doi: 10.1046/j.1356-9597.2001.00506.x. [DOI] [PubMed] [Google Scholar]
- 109.Paillard L, Omilli F, Legagneux V, Bassez T, Maniey D, Osborne HB. EDEN and EDEN-BP, a cis element and an associated factor that mediate sequence-specific mRNA deadenylation in Xenopus embryos. EMBO J. 1998;17(1):278–287. doi: 10.1093/emboj/17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Timchenko NA, Welm AL, Lu X, Timchenko LT. CUG repeat binding protein (CUGBP1) interacts with the 5′ region of C/EBPbeta mRNA and regulates translation of C/EBPbeta isoforms. Nucleic Acids Res. 1999;27(22):4517–4525. doi: 10.1093/nar/27.22.4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res. 2009;37(15):5151–5166. doi: 10.1093/nar/gkp546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo M, Jan LY, Jan YN. Control of daughter cell fates during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17(1):27–41. doi: 10.1016/S0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 113.Pece S, Serresi M, Santolini E, Capra M, Hulleman E, Galimberti V, Zurrida S, Maisonneuve P, Viale G, Di Fiore PP. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol. 2004;167(2):215–221. doi: 10.1083/jcb.200406140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17(1):21–26. doi: 10.1016/S0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 115.Feng Y, Bankston A. The star family member QKI and cell signaling. Adv Exp Med Biol. 2010;693:25–36. doi: 10.1007/978-1-4419-7005-3_2. [DOI] [PubMed] [Google Scholar]
- 116.Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12(8):691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 117.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Corioni M, Antih N, Tanackovic G, Zavolan M, Kramer A. Analysis of in situ pre-mRNA targets of human splicing factor SF1 reveals a function in alternative splicing. Nucleic Acids Res. 2011;39(5):1868–1879. doi: 10.1093/nar/gkq1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcarcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002;109(3):285–296. doi: 10.1016/S0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 120.Graveley BR, Maniatis T. Arginine/serine-rich domains of SR proteins can function as activators of pre-mRNA splicing. Mol Cell. 1998;1(5):765–771. doi: 10.1016/S1097-2765(00)80076-3. [DOI] [PubMed] [Google Scholar]
- 121.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M, Jr, Fu XD. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50(2):223–235. doi: 10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang J, Smith PJ, Krainer AR, Zhang MQ. Distribution of SR protein exonic splicing enhancer motifs in human protein-coding genes. Nucleic Acids Res. 2005;33(16):5053–5062. doi: 10.1093/nar/gki810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Anko ML, Muller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13(3):R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Erkelenz S, Mueller WF, Evans MS, Busch A, Schoneweis K, Hertel KJ, Schaal H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19(1):96–102. doi: 10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Simard MJ, Chabot B. SRp30c is a repressor of 3′ splice site utilization. Mol Cell Biol. 2002;22(12):4001–4010. doi: 10.1128/MCB.22.12.4001-4010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fu XD, Mayeda A, Maniatis T, Krainer AR. General splicing factors SF2 and SC35 have equivalent activities in vitro, and both affect alternative 5′ and 3′ splice site selection. Proc Natl Acad Sci USA. 1992;89(23):11224–11228. doi: 10.1073/pnas.89.23.11224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gallego ME, Gattoni R, Stevenin J, Marie J, Expert-Bezancon A. The SR splicing factors ASF/SF2 and SC35 have antagonistic effects on intronic enhancer-dependent splicing of the beta-tropomyosin alternative exon 6A. EMBO J. 1997;16(7):1772–1784. doi: 10.1093/emboj/16.7.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Jumaa H, Nielsen PJ. The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 1997;16(16):5077–5085. doi: 10.1093/emboj/16.16.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Solnick D. Alternative splicing caused by RNA secondary structure. Cell. 1985;43(3 Pt 2):667–676. doi: 10.1016/0092-8674(85)90239-9. [DOI] [PubMed] [Google Scholar]
- 130.Clouet d’Orval B, d’Aubenton Carafa Y, Sirand-Pugnet P, Gallego M, Brody E, Marie J. RNA secondary structure repression of a muscle-specific exon in HeLa cell nuclear extracts. Science. 1991;252(5014):1823–1828. doi: 10.1126/science.2063195. [DOI] [PubMed] [Google Scholar]
- 131.Chebli K, Gattoni R, Schmitt P, Hildwein G, Stevenin J. The 216-nucleotide intron of the E1A pre-mRNA contains a hairpin structure that permits utilization of unusually distant branch acceptors. Mol Cell Biol. 1989;9(11):4852–4861. doi: 10.1128/MCB.9.11.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Eperon LP, Graham IR, Griffiths AD, Eperon IC. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54(3):393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 133.Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14(8):1463–1469. doi: 10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang J, Kuo CC, Chen L. GC content around splice sites affects splicing through pre-mRNA secondary structures. BMC Genom. 2011;12:90. doi: 10.1186/1471-2164-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Meyer M, Plass M, Perez-Valle J, Eyras E, Vilardell J. Deciphering 3′SS selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell. 2011;43(6):1033–1039. doi: 10.1016/j.molcel.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 136.Hiller M, Zhang Z, Backofen R, Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3(11):e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Monie TP, Hernandez H, Robinson CV, Simpson P, Matthews S, Curry S. The polypyrimidine tract binding protein is a monomer. RNA. 2005;11(12):1803–1808. doi: 10.1261/rna.2214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309(5743):2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 139.Kim JH, Hahm B, Kim YK, Choi M, Jang SK. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J Mol Biol. 2000;298(3):395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- 140.Rahman MA, Masuda A, Ohe K, Ito M, Hutchinson DO, Mayeda A, Engel AG, Ohno K. HnRNP L and hnRNP LL antagonistically modulate PTB-mediated splicing suppression of CHRNA1 pre-mRNA. Sci Rep. 2013;3:2931. doi: 10.1038/srep02931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Mollet I, Barbosa-Morais NL, Andrade J, Carmo-Fonseca M. Diversity of human U2AF splicing factors. FEBS J. 2006;273(21):4807–4816. doi: 10.1111/j.1742-4658.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 142.Rooke N, Markovtsov V, Cagavi E, Black DL. Roles for SR proteins and hnRNP A1 in the regulation of c-src exon N1. Mol Cell Biol. 2003;23(6):1874–1884. doi: 10.1128/MCB.23.6.1874-1884.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tronchere H, Wang J, Fu XD. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature. 1997;388(6640):397–400. doi: 10.1038/41137. [DOI] [PubMed] [Google Scholar]
- 144.Shepard J, Reick M, Olson S, Graveley BR. Characterization of U2AF(6), a splicing factor related to U2AF(35) Mol Cell Biol. 2002;22(1):221–230. doi: 10.1128/MCB.22.1.221-230.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu Y, Conaway L, Rutherford Bethard J, Al-Ayoubi AM, Thompson Bradley A, Zheng H, Weed SA, Eblen ST. Phosphorylation of the alternative mRNA splicing factor 45 (SPF45) by Clk1 regulates its splice site utilization, cell migration and invasion. Nucleic Acids Res. 2013;41(9):4949–4962. doi: 10.1093/nar/gkt170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Gahura O, Hammann C, Valentova A, Puta F, Folk P. Secondary structure is required for 3′ splice site recognition in yeast. Nucleic Acids Res. 2011;39(22):9759–9767. doi: 10.1093/nar/gkr662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Deshler JO, Rossi JJ. Unexpected point mutations activate cryptic 3′ splice sites by perturbing a natural secondary structure within a yeast intron. Genes Dev. 1991;5(7):1252–1263. doi: 10.1101/gad.5.7.1252. [DOI] [PubMed] [Google Scholar]
- 148.Charpentier B, Rosbash M. Intramolecular structure in yeast introns aids the early steps of in vitro spliceosome assembly. RNA. 1996;2(6):509–522. [PMC free article] [PubMed] [Google Scholar]
- 149.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327(5968):996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 151.Ip JY, Schmidt D, Pan Q, Ramani AK, Fraser AG, Odom DT, Blencowe BJ. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21(3):390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12(2):525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 153.Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res. 1998;26(24):5568–5572. doi: 10.1093/nar/26.24.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu XD, Bentley DL. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev. 2014;28(23):2663–2676. doi: 10.1101/gad.252106.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chen W, Luo L, Zhang L. The organization of nucleosomes around splice sites. Nucleic Acids Res. 2010;38(9):2788–2798. doi: 10.1093/nar/gkq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Huang H, Yu S, Liu H, Sun X. Nucleosome organization in sequences of alternative events in human genome. Biosystems. 2012;109(2):214–219. doi: 10.1016/j.biosystems.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 157.Reisman DN, Strobeck MW, Betz BL, Sciariotta J, Funkhouser W, Jr, Murchardt C, Yaniv M, Sherman LS, Knudsen ES, Weissman BE. Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene. 2002;21(8):1196–1207. doi: 10.1038/sj.onc.1205188. [DOI] [PubMed] [Google Scholar]
- 158.Kadam S, Emerson BM. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol Cell. 2003;11(2):377–389. doi: 10.1016/S1097-2765(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 159.Corey LL, Weirich CS, Benjamin IJ, Kingston RE. Localized recruitment of a chromatin-remodeling activity by an activator in vivo drives transcriptional elongation. Genes Dev. 2003;17(11):1392–1401. doi: 10.1101/gad.1071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat Struct Mol Biol. 2006;13(1):22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 161.Matter N, Herrlich P, Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420(6916):691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 162.Webby CJ, Wolf A, Gromak N, Dreger M, Kramer H, Kessler B, Nielsen ML, Schmitz C, Butler DS, Yates JR, 3rd, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325(5936):90–93. doi: 10.1126/science.1175865. [DOI] [PubMed] [Google Scholar]
- 163.Zhang Y, Madl T, Bagdiul I, Kern T, Kang HS, Zou P, Mausbacher N, Sieber SA, Kramer A, Sattler M. Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3′-splice site recognition. Nucleic Acids Res. 2013;41(2):1343–1354. doi: 10.1093/nar/gks1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang X, Bruderer S, Rafi Z, Xue J, Milburn PJ, Kramer A, Robinson PJ. Phosphorylation of splicing factor SF1 on Ser20 by cGMP-dependent protein kinase regulates spliceosome assembly. EMBO J. 1999;18(16):4549–4559. doi: 10.1093/emboj/18.16.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Manceau V, Swenson M, Le Caer JP, Sobel A, Kielkopf CL, Maucuer A. Major phosphorylation of SF1 on adjacent Ser-Pro motifs enhances interaction with U2AF65. FEBS J. 2006;273(3):577–587. doi: 10.1111/j.1742-4658.2005.05091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Rain JC, Rafi Z, Rhani Z, Legrain P, Kramer A. Conservation of functional domains involved in RNA binding and protein-protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA. 1998;4(5):551–565. doi: 10.1017/S1355838298980335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bezzi M, Teo SX, Muller J, Mok WC, Sahu SK, Vardy LA, Bonday ZQ, Guccione E. Regulation of constitutive and alternative splicing by PRMT5 reveals a role for Mdm4 pre-mRNA in sensing defects in the spliceosomal machinery. Genes Dev. 2013;27(17):1903–1916. doi: 10.1101/gad.219899.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Edmond V, Moysan E, Khochbin S, Matthias P, Brambilla C, Brambilla E, Gazzeri S, Eymin B. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J. 2011;30(3):510–523. doi: 10.1038/emboj.2010.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Luhrmann R. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA. 2009;15(1):153–175. doi: 10.1261/rna.1332609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Seyedarabi A, Sullivan JA, Sasakawa C, Pickersgill RW. A disulfide driven domain swap switches off the activity of Shigella IpaH9.8 E3 ligase. FEBS Lett. 2010;584(19):4163–4168. doi: 10.1016/j.febslet.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 171.Song EJ, Werner SL, Neubauer J, Stegmeier F, Aspden J, Rio D, Harper JW, Elledge SJ, Kirschner MW, Rape M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010;24(13):1434–1447. doi: 10.1101/gad.1925010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9):576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 173.Habelhah H, Shah K, Huang L, Ostareck-Lederer A, Burlingame AL, Shokat KM, Hentze MW, Ronai Z. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nat Cell Biol. 2001;3(3):325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 174.Razanau A, Xie J. Emerging mechanisms and consequences of calcium regulation of alternative splicing in neurons and endocrine cells. Cell Mol Life Sci. 2013;70(23):4527–4536. doi: 10.1007/s00018-013-1390-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.van der Houven van Oordt W, Diaz-Meco MT, Lozano J, Krainer AR, Moscat J, Caceres JF. The MKK(3/6)-p38-signaling cascade alters the subcellular distribution of hnRNP A1 and modulates alternative splicing regulation. J Cell Biol. 2000;149(2):307–316. doi: 10.1083/jcb.149.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Allemand E, Guil S, Myers M, Moscat J, Caceres JF, Krainer AR. Regulation of heterogenous nuclear ribonucleoprotein A1 transport by phosphorylation in cells stressed by osmotic shock. Proc Natl Acad Sci USA. 2005;102(10):3605–3610. doi: 10.1073/pnas.0409889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci USA. 2003;100(15):8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ma S, Liu G, Sun Y, Xie J. Relocalization of the polypyrimidine tract-binding protein during PKA-induced neurite growth. Biochim Biophys Acta. 2007;1773(6):912–923. doi: 10.1016/j.bbamcr.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 179.Caceres JF, Screaton GR, Krainer AR. A specific subset of SR proteins shuttles continuously between the nucleus and the cytoplasm. Genes Dev. 1998;12(1):55–66. doi: 10.1101/gad.12.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci USA. 1997;94(4):1148–1153. doi: 10.1073/pnas.94.4.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Cao W, Jamison SF, Garcia-Blanco MA. Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA. 1997;3(12):1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 182.Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 1998;17(21):6359–6367. doi: 10.1093/emboj/17.21.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ma CT, Ghosh G, Fu XD, Adams JA. Mechanism of dephosphorylation of the SR protein ASF/SF2 by protein phosphatase 1. J Mol Biol. 2010;403(3):386–404. doi: 10.1016/j.jmb.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Dagher SF, Fu XD. Evidence for a role of Sky1p-mediated phosphorylation in 3′ splice site recognition involving both Prp8 and Prp17/Slu4. RNA. 2001;7(9):1284–1297. doi: 10.1017/S1355838201016077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Prasad J, Colwill K, Pawson T, Manley JL. The protein kinase Clk/Sty directly modulates SR protein activity: both hyper- and hypophosphorylation inhibit splicing. Mol Cell Biol. 1999;19(10):6991–7000. doi: 10.1128/MCB.19.10.6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shin C, Manley JL. The SR protein SRp38 represses splicing in M phase cells. Cell. 2002;111(3):407–417. doi: 10.1016/S0092-8674(02)01038-3. [DOI] [PubMed] [Google Scholar]
- 187.Shin C, Feng Y, Manley JL. Dephosphorylated SRp38 acts as a splicing repressor in response to heat shock. Nature. 2004;427(6974):553–558. doi: 10.1038/nature02288. [DOI] [PubMed] [Google Scholar]
- 188.Boisvert FM, Cote J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2(12):1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 189.Koumbadinga G, Mahmood N, Lei L, Kan YC, Cao WG, Lobo VG, Yao XJ, Zhang SZ. Xie J (2015) Increased stability of heterogeneous ribonucleoproteins by a deacetylase inhibitor. BBA-Gene Regul Mech. 1849;8:1095–1103. doi: 10.1016/j.bbagrm.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 190.Okuda J, Toyotome T, Kataoka N, Ohno M, Abe H, Shimura Y, Seyedarabi A, Pickersgill R, Sasakawa C. Shigella effector IpaH9.8 binds to a splicing factor U2AF(35) to modulate host immune responses. Biochem Biophys Res Commun. 2005;333(2):531–539. doi: 10.1016/j.bbrc.2005.05.145. [DOI] [PubMed] [Google Scholar]
- 191.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 192.Bellare P, Kutach AK, Rines AK, Guthrie C, Sontheimer EJ. Ubiquitin binding by a variant Jab1/MPN domain in the essential pre-mRNA splicing factor Prp8p. RNA. 2006;12(2):292–302. doi: 10.1261/rna.2152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bellare P, Small EC, Huang X, Wohlschlegel JA, Staley JP, Sontheimer EJ. A role for ubiquitin in the spliceosome assembly pathway. Nat Struct Mol Biol. 2008;15(5):444–451. doi: 10.1038/nsmb.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3(9):570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- 195.Nakajima H, Sato B, Fujita T, Takase S, Terano H, Okuhara M. New antitumor substances, FR901463, FR901464 and FR901465. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J Antibiot (Tokyo) 1996;49(12):1196–1203. doi: 10.7164/antibiotics.49.1196. [DOI] [PubMed] [Google Scholar]
- 196.Gozani O, Potashkin J, Reed R. A potential role for U2AF-SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol. 1998;18(8):4752–4760. doi: 10.1128/MCB.18.8.4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Konarska MM, Sharp PA. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49(6):763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 198.Pikielny CW, Rymond BC, Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature. 1986;324(6095):341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- 199.Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA. 1999;5(6):819–831. doi: 10.1017/S1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Corrionero A, Minana B, Valcarcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev. 2011;25(5):445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Roybal GA, Jurica MS. Spliceostatin A inhibits spliceosome assembly subsequent to prespliceosome formation. Nucleic Acids Res. 2010;38(19):6664–6672. doi: 10.1093/nar/gkq494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Alekseyenko AV, Kim N, Lee CJ. Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA. 2007;13(5):661–670. doi: 10.1261/rna.325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2012;338(6114):1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 204.Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338(6114):1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Kondrashov FA, Koonin EV. Origin of alternative splicing by tandem exon duplication. Hum Mol Genet. 2001;10(23):2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- 206.Krull M, Brosius J, Schmitz J. Alu-SINE exonization: en route to protein-coding function. Mol Biol Evol. 2005;22(8):1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 207.Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. 2007;13(10):1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12(7):1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet. 2010;11(5):345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 210.Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21(2):73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 211.Lev-Maor G, Goren A, Sela N, Kim E, Keren H, Doron-Faigenboim A, Leibman-Barak S, Pupko T, Ast G. The “alternative” choice of constitutive exons throughout evolution. PLoS Genet. 2007;3(11):e203. doi: 10.1371/journal.pgen.0030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Koren E, Lev-Maor G, Ast G. The emergence of alternative 3′ and 5′ splice site exons from constitutive exons. PLoS Comput Biol. 2007;3(5):e95. doi: 10.1371/journal.pcbi.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci USA. 2004;101(44):15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Xie J. Differential evolution of signal-responsive RNA elements and upstream factors that control alternative splicing. Cell Mol Life Sci. 2014;71(22):4347–4360. doi: 10.1007/s00018-014-1688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sohail M, Xie J. Evolutionary emergence of a novel splice variant of opposite effect on cell cycle. Mol Cell Biol. 2015;35(12):2203–2214. doi: 10.1128/MCB.00190-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Liu G, Lei L, Yu J, Kung S, Xie J. Refinement of the spectra of exon usage by combined effects of extracellular stimulus and intracellular factors. Biochim Biophys Acta. 2014;1839(7):537–545. doi: 10.1016/j.bbagrm.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 217.Schor IE, Rascovan N, Pelisch F, Allo M, Kornblihtt AR. Neuronal cell depolarization induces intragenic chromatin modifications affecting NCAM alternative splicing. Proc Natl Acad Sci USA. 2009;106(11):4325–4330. doi: 10.1073/pnas.0810666106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sanchez SE, Petrillo E, Beckwith EJ, Zhang X, Rugnone ML, Hernando CE, Cuevas JC, Godoy Herz MA, Depetris-Chauvin A, Simpson CG, et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature. 2010;468(7320):112–116. doi: 10.1038/nature09470. [DOI] [PubMed] [Google Scholar]
- 219.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157(1):77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 220.Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311(5758):230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 221.Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008;22(6):756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hastings ML, Milcarek C, Martincic K, Peterson ML, Munroe SH. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997;25(21):4296–4300. doi: 10.1093/nar/25.21.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Morrissy AS, Griffith M, Marra MA. Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res. 2011;21(8):1203–1212. doi: 10.1101/gr.113431.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet. 2005;14(11):1465–1474. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- 225.Kralovicova J, Knut M, Cross NC, Vorechovsky I. Identification of U2AF(35)-dependent exons by RNA-Seq reveals a link between 3′ splice-site organization and activity of U2AF-related proteins. Nucleic Acids Res. 2015;43(7):3747–3763. doi: 10.1093/nar/gkv194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28(13):4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wollerton MC, Gooding C, Robinson F, Brown EC, Jackson RJ, Smith CW. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7(6):819–832. doi: 10.1017/S1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Pink RC, Wicks K, Caley DP, Punch EK, Jacobs L, Carter DR. Pseudogenes: pseudo-functional or key regulators in health and disease? RNA. 2011;17(5):792–798. doi: 10.1261/rna.2658311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Yeo GW, Van Nostrand EL, Liang TY. Discovery and analysis of evolutionarily conserved intronic splicing regulatory elements. PLoS Genet. 2007;3(5):e85. doi: 10.1371/journal.pgen.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 231.Patnaik MM, Lasho TL, Finke CM, Hanson CA, Hodnefield JM, Knudson RA, Ketterling RP, Pardanani A, Tefferi A. Spliceosome mutations involving SRSF2, SF3B1, and U2AF35 in chronic myelomonocytic leukemia: prevalence, clinical correlates, and prognostic relevance. Am J Hematol. 2013;88(3):201–206. doi: 10.1002/ajh.23373. [DOI] [PubMed] [Google Scholar]
- 232.Cazzola M, Della Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122(25):4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, Yoon CJ, Ellis P, Wedge DC, Pellagatti A, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood. 2013;122(22):3616–3627. doi: 10.1182/blood-2013-08-518886. [DOI] [PMC free article] [PubMed] [Google Scholar]


