Abstract
Syntrophins are a family of cytoplasmic membrane-associated adaptor proteins, characterized by the presence of a unique domain organization comprised of a C-terminal syntrophin unique (SU) domain and an N-terminal pleckstrin homology (PH) domain that is split by insertion of a PDZ domain. Syntrophins have been recognized as an important component of many signaling events, and they seem to function more like the cell’s own personal ‘Santa Claus’ that serves to ‘gift’ various signaling complexes with precise proteins that they ‘wish for’, and at the same time care enough for the spatial, temporal control of these signaling events, maintaining overall smooth functioning and general happiness of the cell. Syntrophins not only associate various ion channels and signaling proteins to the dystrophin-associated protein complex (DAPC), via a direct interaction with dystrophin protein but also serve as a link between the extracellular matrix and the intracellular downstream targets and cell cytoskeleton by interacting with F-actin. They play an important role in regulating the postsynaptic signal transduction, sarcolemmal localization of nNOS, EphA4 signaling at the neuromuscular junction, and G-protein mediated signaling. In our previous work, we reported a differential expression pattern of alpha-1-syntrophin (SNTA1) protein in esophageal and breast carcinomas. Implicated in several other pathologies, like cardiac dys-functioning, muscular dystrophies, diabetes, etc., these proteins provide a lot of scope for further studies. The present review focuses on the role of syntrophins in membrane targeting and regulation of cellular proteins, while highlighting their relevance in possible development and/or progression of pathologies including cancer which we have recently demonstrated.
Keywords: Adaptor proteins, Syntrophins, Santa Claus, Signal transduction, Protein targeting
Introduction
Cell signaling is a very complex system of cell communication that governs and coordinates the basic activities of a cell and coordinates its varied cellular actions. The ability of a cell to perceive and correctly respond to its microenvironment is the basis of its development and normal tissue homeostasis. Despite the multiple studies carried out all over the world, it is still unclear how exactly cells regulate the mechanisms that control the cross-talk between signaling cascades and how specificity in these signaling events is achieved. However, it is quite evident that an efficient cellular response would ask for a precise localization of its various participating components, such as cell receptors, signal transduction proteins, and the secondary effectors, and a coordinated coupling of these components would result into a functional signaling event. Any subversion in such a delicate and highly controlled signaling system may result in errors in cellular processing, responsible for serious pathologies. Adaptor proteins, e.g., Grb2, Sos1, Erk, etc., that play a crucial role in co-ordination of signaling events may act as the rescuers in such situations. Responsible for the proper clustering and anchoring of proteins at specific sub-cellular sites, adaptor proteins are thus emerging as the major contributors and regulators of cell signaling processes [1].
Adaptor proteins are an important class of proteins that lack any intrinsic enzymatic activity but may possess several binding domains, such as collagen homology 2 domain (CH2), pleckstrin homology domain (PH), Src homology 2 domain (SH2), Src homology 3 domain (SH3), etc., which confer on them the property of binding to and interacting with their specific binding partners, which are usually other cellular proteins, lipids, lipo-proteins, or glycoproteins, etc. Adaptor proteins thus serve to act as a scaffold for the formation of various signaling complexes, facilitating the correct sub-cellular localization and proximity of their partners, and thereby regulating and coordinating the respective signaling events spatially as well as temporally. Syntrophin, as a family of cytoplasmic adaptor proteins, provides an excellent example of such rescue proteins that in recent times have been shown to emerge as key players in the membrane targeting of several proteins and the regulation of many important intracellular signaling events [2]. In general, these proteins, via their multiple protein–protein interaction motifs, serve to dock cellular proteins to their specific working sites, i.e. regulate their intracellular targeting and thereby also their functioning. This postman’s nature of syntrophins, particularly alpha-1-syntrophin, co-incidentally also coined as ‘SNTA1’, relates more to a ‘Santa Claus’-like functioning of protein delivery within the cell.
What are syntrophins?
Syntrophins are a multigene family of membrane-associated adaptor proteins. They represent a biochemically heterogeneous group of 58- to 60-kDa intracellular membrane-associated proteins that are characterized by the presence of a PDZ (postsynaptic density protein-95/disc large/zona occludens-1) split PH (pleckstrin homology) domain and a SU (syntrophin unique) domain [3, 4] within their structure. The term ‘syntrophin’ has been derived from the greek word ‘syntrophos’ meaning companion or associate [5]. Syntrophins were first identified in the postsynaptic membranes of the Torpedo electric organ [6] and later shown to be present in many mammalian tissues [7, 8]. Interest in the protein first came from its location at the neuromuscular junction and later from the demonstration that it is directly associated with dystrophin, the protein product of Duchenne muscular dystrophy gene locus [9–11], within the dystrophin signaling complex, implicating its involvement in some important cellular mechanisms such as cell synapses and signal transduction.
The syntrophin family of scaffold proteins serves to provide a platform for the formation of signal transduction complexes to direct the various participating signaling components to their specialized membrane domains, and to link various cell surface receptors, ion channels, and downstream effectors to these signaling complexes. Primarily, they serve to link several cell components to the dystrophin protein complex via a direct interaction with the dystrophin protein [10, 11] and dystrophin-related proteins like utrophin and dystrobrevin [9, 12–14]. The presence of multiple binding domains like PH, SU, and PDZ in syntrophins allows them to interact simultaneously with varied cellular components, clustering cell proteins, neurotransmitters, receptors, glycoproteins, and lipids, etc. into appropriate functional networks at specific subcellular compartments and orchestrating these signal transduction complexes. By controlling their specific spatial and temporal distribution, these proteins thus help in coordinating and regulating various crucial cellular processes and intracellular signaling events [2].
Syntrophin family
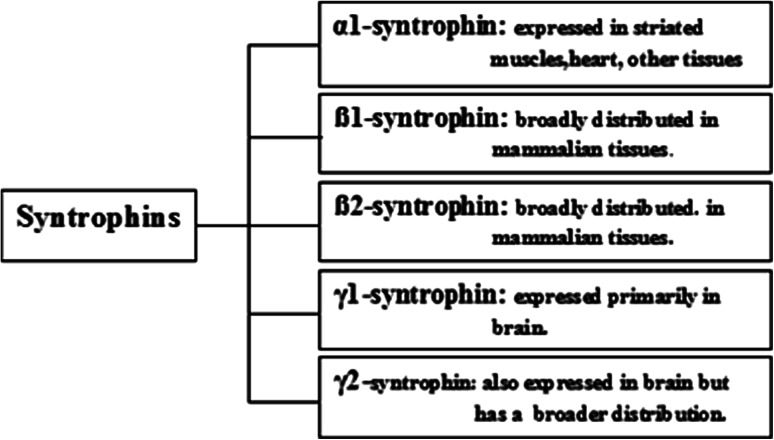
The syntrophin family of proteins consists of five known homologous isoforms: alpha-1- (α-1-) syntrophin, beta-1- (ß-1-) syntrophin, beta-2- (ß-2-) syntrophin, gamma-1- (γ-1-) syntrophin, and gamma-2- (γ-2-) syntrophin [7, 12, 15]. The nomenclature of these isoforms is based on the fact that, while α-1-syntrophin is acidic, ß-1-syntrophin and ß-2-syntrophin are the basic forms of this protein [5]. Of all these known isoforms, α-1-syntrophin was the first to be discovered in postsynaptic membranes of torpedo, followed by ß-1- and ß-2-syntrophin [3, 6, 7], while the two syntrophin isoforms, γ-1-syntrophin, and γ-2-syntrophin, have been recently identified [16]. All isoforms of syntrophin family may exist as monomers or dimers within the cell and almost all of these isoforms have been shown to bind to each other.
Alpha-1-syntrophin encoded in humans by the SNTA1 gene, represents a 58-kDa, acidic isoform with PI 6.7 and 505 amino acids length (unprocessed protein). This membrane protein is found mostly as a peripheral cytoplasmic membrane protein associated with dystrophin protein and dystrophin-related proteins, e.g., utrophin, dystrobrevin, glycoproteins, etc. in a complex called dystrophin-associated protein complex (DAPC) or dystrophin glycoprotein complex (DGC) [9, 10, 14] in muscle cells or concentrated at the neuromuscular junction in brain. The beta isoforms, i.e. ß-1- and ß-2-syntrophins, are encoded by the SNTB1 and SNTB2 genes, respectively, in humans. While ß-1-syntrophin is a 538 amino acids long protein, ß-2-syntrophin is 540 amino acids in length. The complete proteins represent a nearly 58- to 59-kDa dystrophin-associated protein A1 basic components 1 and 2, respectively, with PI in the range of 8.3–8.6. Both these ß-1- and ß-2-syntrophin isoforms are ubiquitously expressed in mammalian tissues; however, ß-1 is the predominant isoform [14]. Weak levels of isoform ß-2 are present in all mammalian tissues, except in liver and heart, where it is highly expressed. Both these isoforms have been shown to bind to dystrophin and the related proteins of DAPC [14, 17, 18]. The gamma isoforms, i.e. γ-1- and γ-2-syntrophin proteins, are encoded by SNTG1 and SNTG2 genes in humans. These γ-1- and γ-2-syntrophin genes encode for 517 and 539 amino acids long proteins of nearly 58 and 60 kDa molecular weight, respectively. While γ-1-syntrophin has been shown to interact with the dystrophin protein [16] and a very few of its related proteins, the γ-2 isoform has not been shown to bind to dystrophin or its related proteins. Both these isoforms have only been recently identified and, though they have been found to be present in abundance in brain tissues, only a few of their interacting partners have yet been identified and there is huge scope for further investigation into their possible interacting partners or the functioning of these syntrophin isoforms.
Each of the five syntrophin isoforms has been found to have a unique tissue expression (Fig. 1) and developmental pattern [4, 7, 12], and has also been shown to selectively interact with different dystrophin family proteins [9, 13, 19]. Alpha-1-syntrophin is primarily expressed in skeletal muscles [7] in addition to being expressed in other mammalian tissues, like heart, brain, stomach, breasts, colorectal tissues, etc. [20], ß-1-syntrophin and ß-2-syntrophin are ubiqutiously expressed and are present in almost all mammalian tissues [7], while γ-1-syntrophin has been shown to be expressed uniquely in the brain, specifically localized in hippocampal pyramidal neurons, purkinje neurons in cerebellum, and cortical neurons [21], and γ-2-syntrophin is also present in the brain but is considered to have a broader distribution in mammalian tissues including skeletal muscles, liver, testis, etc. [16, 21]. In addition to their difference in expression in mammalian tissues, these syntrophin isoforms have also been shown to reveal a marked difference in their subcellular location and distribution within the cells [18]. Thus, although more than one syntrophin isoform protein has been shown to be expressed in a single cell type, their subcellular localizations are effectively different and very tightly regulated [22]. For instance, in neurons/muscles, both the γ-isoforms are found to be localized to the endoplasmic reticulum, while in skeletal muscle, α-1-syntrophin has been shown to be distributed over the entire sarcolemma, present throughout the folds at neuromuscular junctions, whereas ß-1 has found to be present only at the neuromuscular junction folds and ß-2-syntrophin is found to be almost exclusively restricted to the neuromuscular junctions and confined to the lower portion of these folds [18, 21–23], while the γ-2-syntrophin localizes in the subsynaptic space beneath the neuromuscular junction in skeletal muscles [21]. The tightly regulated cellular and subcellular localization of the different isoforms of syntrophin proteins is indicative of distinct functional roles for these different isoforms within the cell [4, 12–14, 23]. For example, α-1-syntrophin, via its association with DAPC in skeletal muscles, has been shown to serve as a link between the extracellular matrix and internal cell signaling apparatus and cell cytoskeleton [24], and is also important for the proper targeting of proteins like utrophin, nNOS, etc. to the postsynaptic membrane of skeletal muscle [22–24]. Similarly, α-1-syntrophin, ß-1- and ß-2-syntrophins have been shown to bind to neuronal nitric oxide synthase (nNOS) and microtubule-associated serine/threonine kinases (MAST) for the membrane association of these proteins in brain cells [24–28], and the γ-1-syntrophin has been shown to bind to and regulate the subcellular localization of diacylglycerol kinase ζ (DGK-ζ) and is involved in cellular synaptic function [29].
Fig. 1.
Schematic representation of the various members of syntrophin family proteins and their general distribution in mammalian tissues
Chromosomal location and homology
All the isoforms of syntrophin family have been identified as the products of different genes [12, 13]. The full-length cDNA of α-1-syntrophin is 2,136 bp long and encodes a single large open reading frame that maps to the chromosome 20q11.2 [7]. The chromosomal location of β-2-syntrophin maps to chromosome 16q22-23 while β-1-syntrophin is located at 8q23-24. The γ-1 isoform of syntrophin maps to 8q11-12 and γ-2 isoform is located at 2p25.3. While α-1-syntrophin is encoded by a single mRNA, multiple species of mRNA have been shown to encode for the β-syntrophins [7, 12]. The deduced amino acid sequences of human α-1- and β-2-syntrophins are nearly identical to their homologues in mouse, suggesting a strong functional conservation among the individual isoforms. At the amino acid level, the human α-1-syntrophin isoform is 94 % identical to its mouse sequence and 93 % identical to the corresponding rabbit sequence [4, 7]. Also, for a particular isoform of syntrophin proteins, a high degree of interspecies conservation has been observed, with 96 % identity for β-2-syntrophin and at least 93 % for α-1-syntrophin [7, 12]. However it has been observed that the three human syntrophins, i.e. α-1, β-1 and β-2, are less strongly conserved with respect to each other, e.g., the human α-1-syntrophin peptide sequence is nearly 54 and 50 % identical to human β-1- and β-2-syntrophins, respectively, while the human β-2-syntrophin peptide has been shown to be 57 % identical to human β-1-syntrophin [3, 4]. The amino acid sequence of the two γ-syntrophin isoforms is found to be more similar to each other than to the α- or β-syntrophin isoforms. In fact, the γ-1-syntrophin and γ-2-syntrophin isoforms have been shown to possess a C-terminal tail of nearly 23 and 14 amino acids, respectively, extending their SU domain, and their amino acid sequence shows only 15–22 % substantial homology to the amino acid sequence of α- and β-syntrophin isoforms [21]. The difference between the γ-syntrophin isoforms and the α-/β-syntrophins in their amino acid sequence and in their interaction/association with other proteins, in fact qualifies them to be considered as a subfamily of syntrophin isoforms.
Structure
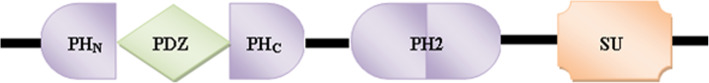
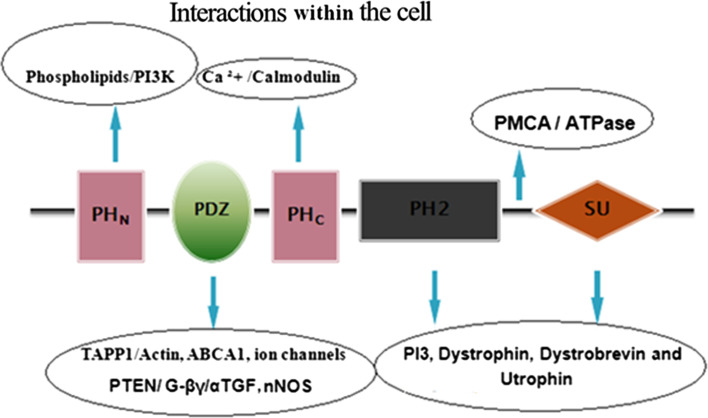
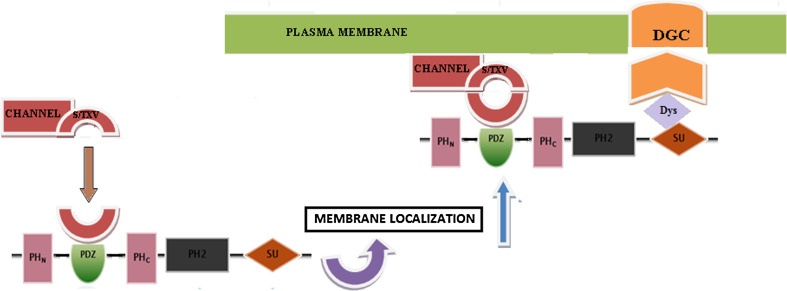
Structurally, all the members of syntrophin family share a common domain organization (Fig. 2). All the syntrophin isoforms contain two tandem PH domains, i.e. one N-terminal split PH domain (PH1) and one central PH domain (PH2), as well as a C-terminal SU domain. However, the γ-1- and γ-2-syntrophin isoforms differ slightly in their structure from the other syntrophin isoforms in the sense that, in addition to these conserved domains, they possess an extended 23 and 14 amino acids tail at their C-terminals, respectively. This C-terminal tail has been shown to form a putative P-loop that could be important in their dystrophin binding [21, 30]. All these domains play an important role in cytoskeletal dynamics as well as in diverse signal transduction mechanisms. The first PH domain is interrupted by a PDZ domain [3], and this split PH1 domain, and the C-terminal syntrophin-specific domain called the syntrophin unique domain, are responsible for the interactions between syntrophin and the DGC [4, 12–17]. Since none of these domains contains any intrinsic enzyme activity, syntrophins are therefore thought to function as adaptors that serve to target their binding partners to specific locations (Fig. 3) on the cell membrane [31].
Fig. 2.
Common structural organization of the syntrophin proteins
Fig. 3.
Diagrammatic representation of the various interactions shown within the cell
PH1 domain
Pleckstrin homology domains are abundant protein modules shown to play critical roles in cellular signaling and cytoskeletal organization [32]. They are called pleckstrin homology domains as they show homology to a region repeated in the pleckstrin protein. PH domains are regarded as lipid-binding modules that are capable of targeting several PH-containing proteins to the cell membrane and are involved in recognition of lipid signal transduction. There are two PH domains present within the syntrophin proteins. Although it has been shown that the first PH domain of the two γ-isoforms of syntrophin proteins does not fit the consensus sequence for PH domains and is different from the PH1 domain sequence of their α- or the β-counterparts, the basic key elements of a PH domain, such as the presence of certain charged/hydrophobic amino acid residues at specific positions, are found to be conserved in the PH1 domain of both these γ-syntrophin isoforms [33]. Several signaling proteins are known to possess these PH domains; however, the structural organization of the first PH1 domain in syntrophins with this PDZ-split-PH domain organization is quite unique. Protein domain searching using SMART program identifies phospholipace Cγ1 (PLCγ1), myosin X, as some of the proteins having a split PH domain organization with one or more intact modules in the middle of their PH domains [34, 35], e.g., the C-terminal domain of PLCγ1 contains an insert of a tendem array of the SH2-SH2-SH3 domains [32, 36–38]. The PH-1 domain of syntrophin protein is split into two halves with one half, i.e. the N-terminal half of the PH1 domain extending from amino acids 1–77, and the other half, i.e. the C-terminal half of the PH1 domain extending from amino acids 162–271. Between these two PH1 halves lies a highly conserved globular structure, of nearly 80 amino acids in length, called PDZ domain [2, 3, 32]. The insertion of the PDZ domain in the middle of the N-terminal PH domain is a highly conserved molecule characteristic of all isoforms of syntrophin proteins and signifies the functional importance of this peculiar structural arrangement [37–39]. The NH2 terminus of this pleckstrin homology 1 domain and the NH2 terminus of the PDZ domain have been reported to bind calmodulin [39] in a Ca2+-independent manner [40–42]. This domain has also been shown to be involved in the oligomerization of syntrophin proteins in vitro in a Ca2+-dependent manner. Calmodulin inhibits oligomerization in a Ca2+-independent manner [41–43]. The PH1 domain of syntrophins is also involved in binding to the membrane phospho-lipids and is implicated in lipid signaling mechanisms [43], e.g., the PH1 domain of alpha-1-syntrophin has been shown to interact with phosphotidylinositol 4,5-bisphosphate for its own membrane localization [43].
Structurally, PHN–PDZ–PHC is composed of two separate domains, i.e. PHN and PHC of the PH domain, and the PDZ domain. The NMR studies reveal that the PHN half is composed of three β-strands while the PHC half contains four β-strands and a C-terminal α-helix. The PDZ domain is inserted in the β3/β4-loop of the PH domain Thus, the PHN–PDZ–PHC tandem forms a functionally unique supra-module structure in which the split PH domain and the PDZ domain function synergistically in binding to membrane lipids, e.g., inositol phospholipid [39, 43]. The split PH domain of syntrophin proteins adopts a canonical PH domain fold that has been implicated in several interactions with the PH and PDZ domains of several other proteins, and may also interact with the complementary partial PH/PH-like domains present hidden in the primary sequence of many proteins [7, 39, 44]. In α-1-syntrophin, this intra-molecular association between the two PH domain halves has been observed [36, 39]. The insertion of the PDZ domain does not affect the conformation or interactions of the PH1 domain in syntrophin. The two complementary partial PH domains within one or between two different proteins or the two halves of a split PH domain have been shown to associate with each other to form an intact complete PH domain [39, 42–44]. However, the PHN–PDZ–PHC tandem has a higher avidity in binding to lipids as compared to the PHN-PHC or PDZ domains [37–39], and the lipid binding affinity of this PHN–PDZ–PHC tandem has been shown to depend on the position of the PDZ domain insertion [36, 39].
PDZ Domain
The name PDZ comes from the first three proteins found to contain repeats of this domain, i.e. postsynaptic density protein-95, Drosophila discs large protein, and the zona occludens protein 1. PDZ domains represent the most abundant interaction domains in the human genome, being present in bacteria to plants as well as in vertebrates [45]. The PDZ domain-containing proteins are essentially involved in the dynamic organization of the molecular architectures and anchoring of the cellular proteins, kinases, enzymes, etc. at specific membrane sites, thereby regulating the signal transduction between the extracellular and intracellular spaces [38–41]. To date, approximately 250 PDZ domains present in more than 150 different human proteins have been recognized [45]. These evolutionarily conserved domains have been shown to recognize certain amino acid regions for their interaction with other proteins, known as PDZ binding motifs (PBM). Interactions between these PDZ domains and their binding motifs play a very significant role in signal transduction mechanisms. The PDZ binding motifs are more often present at or near the C-terminal end of proteins, as these C-terminals are thought to aid in the binding between PDZ and PBM. However, in some cases, these PBM’s are also present in the internal regions of their binding partners [2, 45–48].
The PDZ domain stretches nearly inbetween amino acids 75–170 of the PH1 domain of syntrophin proteins. Consisting of seven β-strands and two α-helices, the well-folded PDZ domain is inserted at the β3–β4 loop within two long linker regions of the PH1 domain of syntrophins [37–39]. The PDZ domain of syntrophins acts as an adaptor for recruiting several membrane proteins, ion channels, receptors, and kinases, etc. to the membrane [31], and these interactions play a central role in organizing diverse signaling pathways. Studies reveal that the majority of PDZ interactions occur through the recognition of the C-terminal PBM and that the terminal carboxylate group is important for binding to take place [45–48]. The structural studies of the PDZ domain have revealed that this canonical mode of PDZ recognition is achieved through an extended binding groove that terminates in a conserved carboxylate binding loop [49–51]; however, there are still many queries regarding the selectivity or promiscuity of their binding that remain to be investigated. Generally, on the basis of the two amino acids present at positions 0 and -2, these C-terminal PBM’s are categorized into three major groups, i.e. Type I PBM, Type II, and Type III PBM [2, 48–52]. Type-I PBM is of S/T-X-I/L/V type, where the -2 position is occupied by either serine (S) or threonine (T), the -1 position can be occupied by any amino acid (X) while the 0 position is occupied by any of the three amino acids, i.e. isoluecine (I), leucine (L), or valine (V). Type-II PBM is of φ-X-φ type, where the -2 and 0 positions are occupied by any hydrophobic amino acid (φ) and the -1 position is occupied by any amino acid X. Type- III PBM is of D/E-X-φ type, where the -2 position is occupied by either by aspartate (D) or glutamate (E) residue, the -1 position is occupied by any amino acid (X) and the 0 position is occupied by any hydrophobic amino acid (φ) [47–52]. A classical example of PDZ domain binding with the C-terminal PBMs is exhibited by the PDZ domain of the α-1-syntrophin protein. Its PDZ domain interacts with the carboxyl termini type-I consensus sequence -E(S/T)XV of stress-activated protein kinase-3 (SAPK-3) [52–54]. A large number of other signaling proteins possessing C-terminal PBM that bind to the PDZ domain of alpha-1-syntrophin proteins have been identified [55]. These proteins may include aquaporin-4 [56], voltage-gated sodium channels [57, 58], potassium channels [59, 60], calcium channel TRPC1 [61], serine/threonine protein kinases [28, 62], the ATP-binding cassette transporter A1 [63], etc. Also, the PDZ domain of β2-syntrophin has been recently shown to bind to the microtubule-associated serine/threonine kinase [28], whereas the PDZ domain of the γ-1 and γ-2-syntrophin proteins has been shown to bind to diacylglycerol kinase ξ and sodium channel SCN5A [29, 58]. The PDZ domain of β-2-syntrophin has been reported to show some unusual features, e.g., the thermal unfolding of the PDZ domain of β2-syntrophin at different pH and denaturing conditions reveals that, unlike the PDZ domains of other proteins, it is marginally stable. The higher stability at pH >9, conformational plasticity, and comparatively more stable behavior of the PDZ domain of β-2-syntrophin has been related to its charge distribution. These properties of the β-2-syntrophin PDZ domain play an important role in the insulin granule mobilization in pancreatic beta cells via its interaction with the protein tyrosine phosphatase known as ICA512 (discussed later) [64, 65].
In addition to the C-terminal PBM, PDZ domains have also been reported to bind to certain internal motifs of their binding partners; however, such interactions are governed by very strict pre-requisites, e.g., proteins with internal sequences, structurally resembling a C-terminal end, such as a beta-hairpin finger-like structure or an aspartate residue, have been shown to bind to the PDZ domains [45–48]. In other words, internal PBMs are recognized by PDZ domains if they are presented with a secondary structure that is similar to a carboxylate group or sterically compatible with the PDZ binding groove [49]. Intermolecular binding of such an internal PBM with a PDZ domain has been characterized for the interaction between neuronal nitric oxide synthase, i.e. nNOS and the PDZ domain of alpha-1-syntrophin [66]. Although the PDZ domain of α-1-syntrophin interacts with the PDZ domain of nNOS and causes its membrane localization, crystallographic studies have revealed that an internal peptide sequence-ETTF-flanking the PDZ domain of nNOS, that forms a hairpin β-finger structure, is actually responsible for binding to the PDZ domain of α-1-syntrophin, although the presence of the PDZ domain near this internal motif of nNOS is also considered to be very important for the stabilization of the interaction between the two proteins [49, 67]. This internal motif having a β-finger structure is thus thought to replace the carboxylate group in binding to the pocket of the PDZ domain used for -COOH terminal binding [49]. Another example of proteins exhibiting such PDZ domain binding internal sequences is that of G-protein-coupled receptors (GPCR). Several GPCRs have been shown to possess type-I as well as type-II PBM in their sequence and, in addition to these typical C-terminal PBMs, the presence of internal PBM in nearly 27 different GPCRs has also been confirmed [66–69]. In addition to these PDZ–PBM interactions, PDZ–PDZ interactions between PDZ domain containing proteins has also been observed. An example of such an interaction is that of the β-2-syntrophin with microtubule-associated serine/threonine kinase-205 (MAST205) and syntrophin-associated serine/threonine kinase (SAST) [28]. The binding between the β-2-syntrophin isoform and MAST/SAST has been shown to involve a PDZ–PDZ domain interaction. Both these kinases are thought to link the dystrophin/utrophin proteins in DGC with microtubule filaments via syntrophins [28]. Several proteins containing PDZ domains have been shown to play a fundamental organizational role at both pre- and postsynaptic plasma membrane [39], thus further advocating the role of syntrophin proteins in cell synapse. All these findings indicate that, via the PDZ domain, syntrophins recruit several signal transduction molecules to the membrane, regulate the efficiency of intracellular signaling of G-proteins, phopholipids or the channel proteins, and have an involvement in the cell synaptic function which we will discuss further in the later sections of this review.
PH-2 domain
The second Pleckstrin homology, i.e. the PH-2 domain, lies at the C-terminus of syntrophin proteins. Unlike PH-1, it is not split by any embedded PDZ domain and exists as an intact PH domain. The PH-2 domain stretches between amino acids 281 to 406 of syntrophin proteins. The PH-2 domain consists of a conserved core structure composed of a partially open, two-sheeted β-barrel with one end of the barrel capped with a C-terminal α-helix [70–73]. The first β-sheet is composed of four anti-parallel β-strands (β1–β4), and the second β-sheet contains three strands (β5–β7). The PH2 domain of syntrophins has been shown to bind several lipid/proteins bound to the plasma membrane, and the best characterized function of PH2 domains is binding to inositol phospholipids [32, 36–39]. The PH-2 domain in syntrophins is mostly involved in the targeting of the PH domain containing proteins via PH–PH interaction, in detecting the lipid signals generated in cellular processes, and in the recruitment of signaling proteins to the sarcolemma [73, 74].
SU domain
The C-terminal 57 amino acids, within all isoforms of this family of proteins, exhibit a region of strong homology, while lacking homology to other characterized proteins. This highly conserved 57-amino acid sequence is hence known as the syntrophin unique or SU domain [4, 12, 13] and has been predicted to consist of from three to five strands of β-sheet separated by as many turns [7]. The PH2 and SU domains of syntrophins are responsible for binding to dystrophin, utrophin, and dystrobrevin, and it is this SU domain that links syntrophin proteins to the membrane via its interaction with the DGC [9, 17, 75, 76]. In addition to this, the two γ-syntrophins possess an additional amino acid tail at their C-terminus that encodes for a P-loop that is considered to be a potential nucleotide (ATP/GTP) binding site [21, 30]. Also, this P-loop is considered to aid in the binding of γ-syntrophins to the dystrophin protein in DAPC, and it has been shown that the splice variant of γ-1-syntrophins lacking this P-loop fails to bind dystrophin in vitro [21, 30]. Many details regarding the functional importance of the SU domains are currently poorly understood and thus leave scope for further investigations.
Syntrophins in cytoskeletal organization:
In skeletal muscles, the DAPC has been shown to form a link between the extracellular matrix and actin cytoskeleton, and is thought to have a role in providing structural stability to the cell membrane. In muscle cells, the DAPC provides stability against the forces of contraction or relaxation [77] and has been implicated in modulating the actin cytoskeleton by effecting the recruitment of proteins involved in actin cytoskeleton organization at the membrane [77–80]. Syntrophins being an important component of DAPC have been shown to play a critical role in executing this function of DAPC, while these proteins, in themselves, have been characterized as actin binding proteins whose subcellular localization is in turn regulated through cytoskeletal reorganization within the muscle cells [81]. α-, β-1- and β-2-syntrophins have been shown to bind to F-actin [81]. Although this binding involves several sites on syntrophin proteins, the second PH domain and the SU carboxyl-terminal are especially considered to be important. These proteins have also been implicated in the actin stabilization at the membrane and the inhibition of the actin-activated myosin ATPase activity [81].
Syntrophins serves to regulate the intracellular localization and activity of the downstream signaling targets of DAPC, e.g., the lipid proteins such as phosphoinositol-3-phosphate (PI3K) and diacylglycerol-ζ (DGK-ζ) have been shown to bind to syntrophins on the inner surface of the cell membrane [43, 78]. Both these lipid proteins are involved in the regulation of cytoskeletal organization. The γ-1-syntrophin isoform has been shown to bind to and regulate the subcellular localization of diacylglycerol kinase-ζ (DGK-ζ). DGK-ζ is an enzyme that phosphorylates diacylglycerol (DAG) to yield phosphatidic acid [29] and has also been implicated in the functioning of the actin remodeling and cytoskeletal dynamism. The C-terminal consensus PDZ-binding sequence of DGK-ζ has been shown to bind to the PDZ domain of γ-1-syntrophin. The interaction between DGK-ζ and γ-1-syntrophin thus provides a mechanism for the localization of DGK-ζ at the membrane. This membrane localization of DGK-ζ is in turn important for regulating the DAG concentration and the production of phospholipids at the membrane. DAG, on the other hand, is considered to be an important intermediate in the synthesis of many phospholipids and triglycerides [78, 82]. DGK-ζ is also seen to be highly enriched in PDGF-induced circular ruffles in fibroblasts. Also, γ-1-syntrophin and DGK-ζ have been shown to co-localize along with F-actin in the peripheral ruffles and lamellipodia of skeletal muscle cells [29, 79].
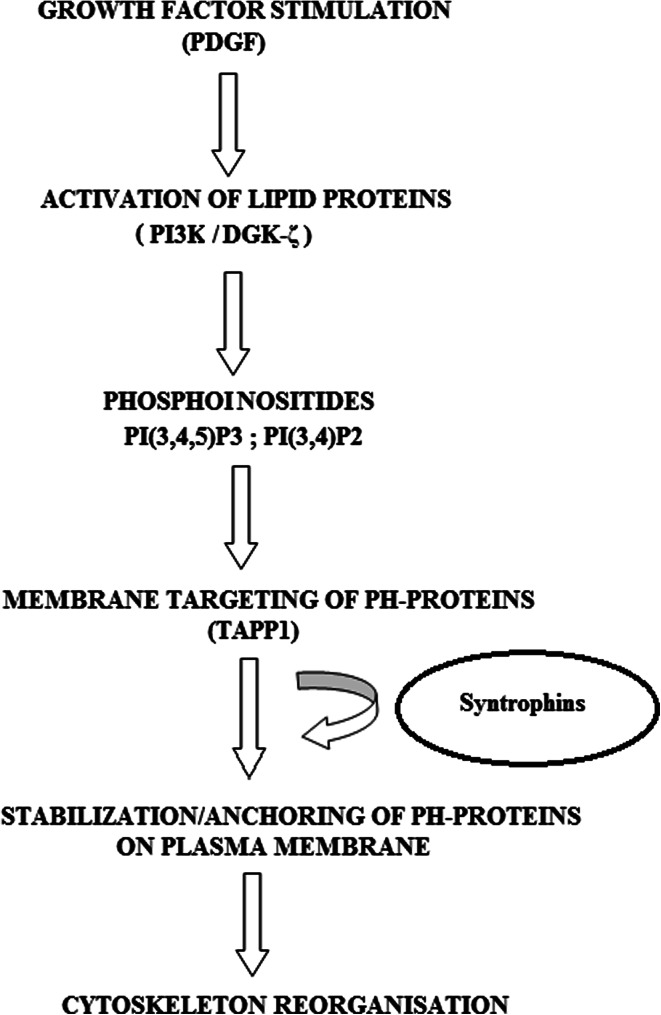
Changes in the actin organization are tightly linked to the production of signaling phospho-inositides such as phosphoinositol 3,4-bisphosphate [PI(3,4)P2] and phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3], which recruit PH domain-containing proteins to the sites of receptor activation at the plasma membrane [79]. These lipid signaling proteins control the actin cytoskeleton organization of the cells in response to growth factor stimulation. PI3K is a key component of multiple signaling pathways, including those that regulate cell survival, growth, and motility [80]. PI3K catalyzes the transient production of phosphatidylinositols like phosphatidylinositol 3,4,5-trisphosphate [PI(3,4,5)P3], which triggers the activation of downstream signaling pathways by recruiting PH domain-containing signaling proteins to the plasma membrane. TAPP1 (i.e. tandom PH domain-containing proteins1) is one of the PH domain-containing adapter proteins recruited to the plasma membrane of cells in response to phosphoinositol 3-kinase (PI3K) activation by growth factors like PDGF. The C-terminal PH domain of TAPP1 has been shown to interact with PI(3,4)P2 and translocate it along with itself to the plasma membrane [83]. At the membrane, these signaling proteins are then activated and thereby initiate various local responses including reorganization of the actin cytoskeleton. This phosphoinositol 3,4-bisphosphate-binding protein, i.e. TAPP1, has also been shown to interact with syntrophins and to regulate the actin cytoskeletal organization of the cell [83–87]. The C-terminal PDZ binding sequence of TAPP1 has been shown to preferentially bind to the PDZ domain of α-1-, β-2- and γ-1-syntrophins suggesting that these syntrophins contribute to the regulation of its intracellular localization [83]. The PI3K-driven signaling pathways downstream of PDGF stimulation have been shown to induce a rapid reorganization of the actin cytoskeleton, which may manifest as plasma membrane specializations such as lamellipodia, filopodia, and membrane ruffles, and it has been shown that DGC components are involved in the regulation of actin organization in response to PDGF signaling [88–90].
Thus, syntrophins and DGC together help in organizing signaling components that remodel the actin cytoskeleton. It has been shown that four different syntrophin proteins may associate with the DGC at a time and help in recruiting signaling proteins having PDZ binding motifs to the PDGF-induced circular ruffles on the membrane [19, 88]. Together, DGC and syntrophins work to act as a scaffold to promote the formation of a signaling complex, which includes TAPP1 and the other downstream effectors of pathway, and stabilize the association of TAPP1 with the membrane [79, 89]. Syntrophins, thereby regulating the localization and activity of the lipid proteins like PI3K, PI(3,4)P2 and DGK-ζ, TAPP1, play a central role in modulating the cytoskeleton organization within the cell (Fig. 4).
Fig. 4.
Regulation of cytoskeleton organization by syntrophins
Regulation of channel proteins
Syntrophin proteins have been implicated in regulation and localization of various ionic channels and membrane binding proteins (Fig. 5). The presence of a PDZ domain bestows syntrophins the ability to interact with several receptor molecules, muscle, and nerve voltage-gated Na+ channels and link them to the actin cytoskeleton and extracellular matrix via the DAPC [57]. The PDZ domain interacts with the C-terminal consensus sequence motif E(S/T)XV, where X represents any amino acid present in various skeletal muscle Na+ channels [56–58]. Two voltage-gated sodium channels of skeletal and cardiac muscle, i.e. SkM1 and SkM2 possessing this consensus sequence in their C-terminal, have been shown to bind to the PDZ domain of α-1-, ß-1-, and ß-2-syntrophin [57]. Moreover, the sodium channels in brain (NaChs) which lack this E(S/T)XV consensus sequence have also been shown to interact with syntrophins, although this interaction is not mediated via the PDZ domain of syntrophins.
Fig. 5.
Role of syntrophins in the membrane localization of ionic channel
Nav1.5 is the main cardiac voltage-gated sodium channel that generates the fast depolarization of the cardiac action potential, and plays a key role in cardiac conduction. Nav1.5 is part of a multi-protein complex in which dystrophin and syntrophin proteins play an important role in determining its expression levels [90]. The Nav1.5 C-terminus consists of the PDZ domain-binding motif formed by its last three residues (Ser-Ile-Val). The C-terminus of Nav1.5 thus binds with the dystrophin of DGC and this interaction is mediated by α- and β-syntrophin proteins [90]. Syntrophin proteins are thus required for the proper localization, expression, and functioning of Nav1.5 (Fig. 5).
Aquaporin-4 (AQP4) is a type of integral membrane channel protein that regulates the homeostasis and flow of water in brain. It belongs to the family of major intrinsic proteins (MIP) that form pores in the cell membranes and is concentrated in the astrocyte membranes facing the blood–brain barrier in brain and the brain–cerebrospinal fluid interfaces in the mammalian central nervous system (CNS) [91–93]. AQP4 has also been shown to express in the sarcolemma of fast-twitch fibers in skeletal muscle [91] and in the basolateral membranes of collecting duct epithelium [94] sites. The cellular distribution of this channel protein is thus in close agreement with the distribution of the dystrophin protein complex, which is also found in abundance at these sites [95–97]. Also, AQP4 has been shown to possess a type-I C-terminal PDZ domain binding sequence, i.e. Ser-Ser-Val (-SSV) that associates with the PDZ domain of the α-1-syntrophin protein [97, 98]. It has been observed that the subcellular localization of AQP4 is altered in α-1-syntrophin knocked out skeletal muscles (α-Syn−/−) but the total level of AQP4 protein in α-Syn−/− skeletal muscles does not change [56, 99], implicating that the subcellular localization and thus the functioning of these AQP4 channel proteins depends on the C-terminal-SSV/PDZ interaction between AQP4 and α-1-syntrophin protein. However α-1-syntrophin has no effect on the expression of AQP4 protein and the interaction between these two proteins represents a potential mechanism to sequester AQP4 at the plasma membrane by affecting its association with the dystrophin complex. α-1-syntrophin, via its PDZ domain, thus serves to regulate the functioning of AQP4 by retaining it on the membrane [55]. A further understanding of this interaction could help us in further elucidation of important cellular processes like the polarization mechanism, as well as the general functioning of astrocytes in CNS.
Inward rectifier potassium channels (Kir) represent a family of channel proteins, involved in the transport of potassium ions that play important roles in the maintenance and control of cell excitability. Many members of this family, e.g., Kir2.1, Kir2.2, Kir2.3, and Kir4.1, have been shown to bind to the components of DAPC [60]. One of the major physiological roles of these Kir potassium channels in glial cells is to promote potassium spatial buffering in the central nervous system, a process necessary to maintain an optimal potassium concentration in the extracellular environment [100–103]. Among these, Kir4.1, an important potassium channel expressed in the CNS tissues, primarily expressed in glial cells, astrocytes within the brain [100], and muller cells of the retina. Kir4.1 has been shown to be localized in glial cells by its association with the dystrophin DGC through a PDZ domain-mediated interaction with its α-1-syntrophin component. The presence of consensus PDZ domain binding motif (SNV) at the last three amino acids of the Kir4.1 potassium channel allows it to interact with the DGC complex, via the PDZ domain of α-1-syntrophin [59, 101]. Similarly, α-1-, β-1-, and β-2-syntrophin have been shown to interact with Kir2 channels via PDZ–PBM interactions [60]. The interaction between Kir4.1 and syntrophins is considered to be very important for their precise distribution in discrete subdomains of cell membranes as well as for the stability and proper functioning of these channels at the cell membranes. Syntrophins are thus involved in both the intracellular trafficking as well as the modulation of Kir channel activity. These interactions are thus suggestive of an important role for the DGC and syntrophins in nervous system physiology.
Transient receptor potential canonical channels (TRPC) are a family of membrane channels that belong to the TRP channel superfamily [104]. There are seven TRPC channels (TRPC1–TRPC7), each consisting of six transmembrane domains surrounding a central membrane pore. These TRPC channels play an important role in maintaining calcium homeostasis in striated muscles and in cardiac muscle cells [105]. TRPC channels (TRPC1, TRPC4, and TRPC5) are involved in the formation of certain store-operated channels and stretch-activated Ca2+ channels in the muscle cell membrane [104, 105] that are activated by the depletion of calcium stores and membrane stretching, respectively. These TRPC-involved channels help in the regulation of capacitative calcium entries (CCE) in muscles and long-term calcium homeostasis [61]. In skeletal muscles TRPC channels, particularly TRPC1 and TRPC4 have been shown to form a costameric macromolecular complex along with α-1-syntrophin and dystrophin proteins. This complex has been implicated in regulating the normal calcium entry into the skeletal muscle cells [106]. The PDZ domain of α-1-syntrophin is thought to mediate the association between α-1-syntrophin and TRPC channels as these TRPC channels possess a C-terminal consensus PDZ binding domains. Within this macromolecular complex, α-1-syntrophin serves as an adaptor that anchors theTRPC1/TRPC4 channels to the dystrophin-associated protein complex (DAPC) which in turn may connect them to the cell cytoskeleton or extra-cellular matrix, assuring the normal activation and modulation of these cation channels. α-1-syntrophin, via its multiple binding domains, may also aid in recruiting other signaling proteins to this complex, thus building a whole signalosomes responsible for regulating the activity of these TRPC channels and maintaining normal calcium homeostasis in muscle cells.
Role within the cell
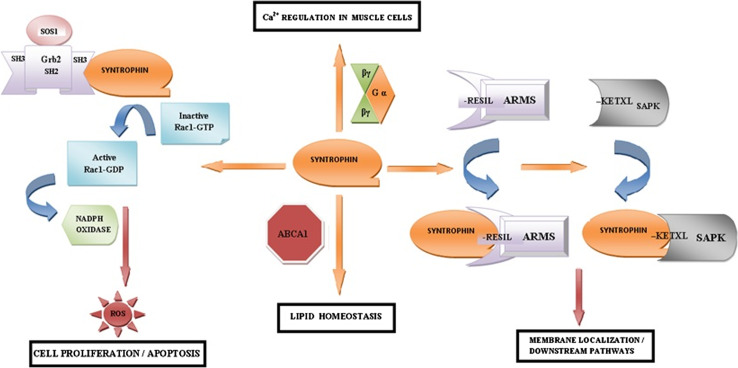
The dystrophin glycoprotein complex is thought to be a signal transduction complex that serves as a link between the extra-cellular matrix and inner signaling pathways and cell cytoskeleton [107]. The presence of as many as four syntrophin binding sites in close proximity within a single DGC indicates that syntrophins play an important role in tethering of multiple signaling molecules together to form signalosomes at specific membrane sites [3]. Syntrophin proteins have been shown to play a central role in trafficking and organization of molecular architectures containing several signal transduction proteins. They have been seen to be involved in the precise localization and/or regulation of many transduction proteins such as stress-activated protein kinase 3, ankyrin repeat-rich membrane spanning protein, Grb2, calmodulin, G-proteins, n-NOS, ABCA1, and many other signaling proteins within the cell (Fig. 6). All these associations have led to the conclusion that the syntrophins function to recruit signaling proteins to the dystrophin-based cytoskeletal complex and thereby regulate its downstream signaling.
Fig. 6.
Schematic representation of the many roles of syntrophin proteins in cell signaling
Stress-activated protein kinase 3 (SAPK-3) belongs to the family of mitogen-activated protein kinases (MAPK). These MAP Kinases are essentially protein kinases that are activated by a dual threonine/tyrosine phosphorylation elicited by certain conditions such as cellular stresses, bacterial endotoxins, or inflammatory cytokines [108]. SAPK-3 (also called ERK6/P38γ) is a more recently identified stress-activated protein kinase that is abundantly expressed in the skeletal muscles. The downstream targets of this MAP kinase are yet to be identified. However, α-1-syntrophin has been recently identified as a substrate for SAPK-3 [62]. In skeletal muscles, both these proteins are found in abundance at the neuromuscular junctions. SAPK-3 has been shown to bind to the PDZ domain of α-1-syntrophin through its C-terminal-KETXL sequence. SAPK-3 has been shown to phosphorylate α-1-syntrophin at two different serine residues, S193 and S201, in vitro [62]. The SAP kinases regulate the cell proliferation, gene expression, differentiation, mitosis, cell survival, and apoptosis within the cell, and the finding that SAPK-3 binds to the PDZ domain of α-1-syntrophin and that the phosphorylation of α-1-syntrophin depends on this interaction thus supports the involvement of α-1-syntrophin in these important cellular processes [62, 108]. Also, SAPK-3 is unique in the sense that it is the only member of its family having a carboxyl-terminal PDZ binding motif. The presence of a C-terminal PDZ binding sequence (-KETPL) in SAPK-3 thus provides a mechanism for its selective targeting to the specific subcellular sites, like cell synaptic junctions in skeletal muscles, as well as its specificity for α-1-syntrophin protein as a substrate pointing towards a more distinct physiological role for this isoform of stress-activated protein kinases within the skeletal muscles [62].
Ankyrin repeat-rich membrane spanning (ARMS) is a novel downstream substrate for protein kinase D and receptor tyrosine kinases, like ephrin (Eph) receptors and tropomyosin-related kinase (Trk) receptors that play a central role in neural development and cell synaptic funtion [109–111]. ARMS is a 220-kDa multidomain protein and, just like certain syntrophins isoforms, it is also expressed in muscles and found concentrated at the neuromuscular junction (NMJ) [112]. The C-terminus of ARMS possesses a typical class-I PDZ-domain binding conserved motif (-RESIL) that has been shown to bind to the PDZ domain of α-1-syntrophin and β-2-syntrophin isoforms. These syntrophin isoforms thus help to anchor ARMS proteins to the synaptic DGC and stabilize ARMS protein clusters at the NMJ and are also implicated in regulating the localization of ARMS during NMJ differentiation. Since ARMS functions as an important RTK downstream target and has been shown to enhance the Eph receptor signaling by increasing EphA4-induced Jak and Stat phosphorylation, it is likely that these syntrophins are also involved in enhancement of Eph signaling by effecting the specific subcellular localization of these protein complexes [110–112]. Over-expression of α-1-syntrophin induces ARMS clustering in a PDZ domain-dependent manner, while co-expression of ARMS enhances EphA4 signaling [112]. EphA4 signaling has recently been implicated in the regulation of synapse formation and plasticity, and, recently, ARMS has been shown to mediate sustained MAPK signaling, elicited by neurotrophins, implicating ARMS as an important target for receptor tyrosine kinase (RTK) signaling [113]. Thus, syntrophins play an important role in RTK-signaling events by aiding the localization of ARMS [112] which is essential for the NMJ development.
Growth factor receptor bound 2 (Grb2) is an important adaptor protein identified as a potentially important mediator of several signaling pathways that may elicit crucial cellular responses during cellular growth and development. Grb2 consists of two SH3 (src homology 3) domains flanking a SH2 (Src homology 2) domain within its structure. The presence of these binding domains, SH2 and SH3, has important functional implications for Grb2. Grb2, via its SH3 domains, can interact with proteins possessing proline-rich motif (PXXP), e.g., p66shc, while its SH2 domain allows it to interact with phosphor–tyrosine proteins. The Grb2 SH2 domain binds to different growth factors and receptors directly through tyrosine phosphorylated motifs as well as binding to other adapter proteins which mediate the indirect docking of Grb2 to cell receptors. α-1-syntrophin, via its proline-rich regions, has been shown to bind to this important adaptor protein [114]. In syntrophins, there are two different PXXP sequence-containing regions between amino acids 44–75 and 181–229, and both of these have been shown to bind each of the two Grb2 SH3 domains in vitro with high affinity [114]. Interaction of syntrophins with Grb2 has served as the basis of its involvement in the activation of an important Rho family GTPase Ras-related C3 botulinum toxin substrate 1, i.e. the Rac1 protein activation pathway. It has been shown that α-1-syntrophin phosphorylation on a tyrosine residue may result in a conformation change in its structure leading to an altered Grb2 binding. The C-terminal SH3 domain of Grb2 (C-SH3) binds to non-phosphorylated α-1-syntrophin, and this complex is envisioned as being unable to bind Son of Sevenless homolog 1 protein (Sos1), while phosphorylation causes it to bind to the SH2 domain of Grb2 and prevents its association with SH3 domain and activates Sos1, which in turn may activate Rac1 and the remainder of the pathway [107].
Recently, a signaling pathway has been described [112] that links the matrix laminin binding on the outside of the sarcolemma to the binding of Grb2 to α-1-syntrophin on the inside surface of the sarcolemma. The downstream signaling pathway has been elucidated as Grb2-Sos1-Rac1-PAK1-JNK. This leads to the activation of c-jun via the phosphorylation of its Serine residue, S65, and the downstream pathways thereby. The DGC connects to the extracellular matrix laminin via α-dystroglycan (αDG) and spans the sarcolemma by way of proteins such as β-DG, α-, β-, γ-, and δ-sarcoglycan (δ-SG), and sarcospan (SSPN) [115–118]. In cell signaling mediated by Grb2, normally Grb2 binds to a phosphorylated tyrosine-containing sequence, via its SH2 domain, while the N-terminal SH3 (N-SH3) domain of Grb2 binds Sos1 and activates it [112]. Syntrophin binding to Grb2 has been shown to activate the Sos1 protein and Rac1 protein [114–120]. The Grb2 protein has also been shown to bind to both α-1-syntrophin protein [113] and βDG via its SH3 domain [121]. Laminin binding on the outer sarcolemma has been shown to induce a conformational change in α-1-syntrophin on the inner surface of the membrane leading to the phosphorylation of α-1-syntrophin on a tyrosine residue. When syntrophin is tyrosine-phosphorylated, it no longer binds to the C-SH3 domain but this phophorylated syntrophin now binds to the SH2 domain of the Grb2 adaptor protein, [115] leaving the Sos1 protein free from Grb2 to interact with the E3B1/EPS8 protein complex which results in Rac1 activation. It has also been confirmed that the presence of syntrophin proteins is important for the recruitment of Rac1 to this complex as well as the functioning of this signaling complex [114–121]. Earlier, we have shown that p66shc, an adaptor protein that promotes oxidative stress, increases Rac1 activity through the mediation of the Sos1 protein [122, 123]. P66shc decreases Sos1 bound to the Grb2 protein and increases the formation of the Sos1-eps8-e3b1 tri-complex which eventually leads to Rac1 activation. This P66shc-induced dissociation of Sos1 from Grb2, formation of the Sos1-eps8-e3b1 complex and Rac1 activation have been shown to be mediated by the proline-rich (PPLP) motif in the CH2 domain of the P66shc protein. The relationship between p66shc, Grb2, and Sos1 provides us chances of proposing a novel mechanism for the activation of Rac1 through this review. Thus, though the exact mechanism of Rac1 activation by syntrophin has not been deduced completely, it is possible that the syntrophin-mediated Rac1 activity might involve the role of p66shc which we are trying to investigate.
Guanine nucleotide binding proteins (G-proteins) are an important class of signal transducing proteins that are bound to the inner surface of the cell membrane and are associated with a transmembrane receptor. These proteins along with their coupled receptors, i.e. G-protein-coupled receptors (GPCR), function by transmitting the outer cell signals, e.g., hormones, neurotransmitters, etc., to the inner cell compartment and by communicating these signals to downstream signaling targets, leading to the desired changes within the cell [124]. Heterotrimeric G-proteins, also belonging to G-protein family, are composed of two subunits, i.e. one Gα subunit and a Gβγ heterodimeric subunit. Studies have revealed that syntrophin proteins, particularly α-1-syntrophin, plays a major role in the regulation of these heterotrimeric G-protein signaling mechanisms. Interaction of the α-1-syntrophin protein with heterotrimeric G-proteins has been confirmed before [115, 125]. Also, laminin α1, an extracellular matrix component, has been shown to induce G-protein binding to the PDZ domain of the α-1-syntrophin protein which has the consequences of altering the intracellular Ca2+ concentration in the muscle cells. In the absence of laminin, when the heterotrimeric G-proteins hydrolysis into its constituting subunits, Gα and Gβγ, Gα remains free to be activated by guanine nucleotide exchange factors (GEF). In its active form, i.e. GTP-bound form, the Gα subunit can bind to and cause activation of the L-type Ca2+ channels called dihydropyridine receptors (DHPR), thereby stimulating the Ca2+ influx into the muscles cells [126]. However, laminin binding to the α-dystroglycan protein of the dystrophin complex on the outer membrane induces an association of these trimeric G-proteins with the PDZ domain of α-1-syntrophin. The Gβγ heterodimeric subunit in G-proteins has been shown to bind to the PDZ domain of the α-1-syntrophin protein. The β component of the Gβγ subunit has a binding motif -LWL- or -IWN- at its C-terminus, while several Gγ components of this subunit, e.g., Gγ4, Gγ12, etc., have TIL similar to the S/TXL type class-I binding motif as their C-terminus. The binding between the PDZ domain of α-1-syntrophin and Gβγ may thus be involving the C-terminus of the Gγ component or Gβ component or may even involve some internal PDZ binding motif present within any of the two components of the Gβγ subunit. Laminin binding increases the Gβγ binding to α-1-syntrophin, sequestering the G protein in its inactive form on the cell membrane. Thus, the Gα subunit is no longer free to bind to or activate DHPR Ca2+ channels, due to which the Ca2+ concentration within the muscle cell decreases drastically. This regulation of G-proteins by α-1-syntrophin has important implications in muscular dystrophies, such as ducchennes muscular dystrophy, which predominantly involve irregularities in cell Ca2+ concentrations. Further, the Gβ subunit is in itself involved in modulating the activities of several protein kinases, cyclases, or ion channels, e.g., it regulates the activation of some isoforms of the PI3K (PI3Kγ, PI3Kβ). Thus, G-protein binding to α-1-syntrophin could be regulating several other kinds of cell signaling pathways such as the Akt/PI3K pathway.
Interaction of α-1-syntrophin protein with the Gα subunit of heterotrimeric G-proteins has also been confirmed [125]. The Gαi, Gαs, Gαo and Gαq subtypes of the Gα component of the G-protein have been shown to associate with the α-1-syntrophin protein, which suggests that α-1-syntrophin may be involved in regulating G-protein signaling via a wide variety of GPCRs. The N-terminal half of the first PH domain in α-1-syntrophin as well as the C-terminal SU domain have been shown to contribute to the binding with the Gα subunit of heterotrimeric G-proteins [125]. An ordinary PH domain has a typical sandwich structure with seven β strands and an α-helix. However, the PH1 domain of syntrophins is split into an N-terminal half and a C-terminal half and each half forms a stable structure with PHN, possessing the first three β strands and PHC having the other three β strands and a helix. Thus, the N-terminal three β-strands of PHN are thought to be involved in binding to the Gα subunit of G-proteins. Along with PHN, the SU domain of α-1-syntrophin has also been shown to bind to the Gαs subunit, pointing towards a possible cooperative binding mechanism. Also, the binding has been found to be independent of the activation state of the Gα subunit, i.e. both the GTP- as well as the GDP-bound forms of Gα can form a complex with the α-1-syntrophin protein in the presence or absence of the GPCR ligands. Alpha-1-syntrophin may thus form a complex with G-proteins, by interacting with either its Gα or Gβγ heterodimeric subunit, and regulate the efficiency of signal transduction mediated by the GPCRs [125, 126]. The fact that the PDZ domain binds to both the C-terminal as well as the internal PBM sequences in GPCRs points towards the possibility that syntrophin proteins could possibly have diverse functions in the regulation of GPCR trafficking. Taken together, these findings suggest that syntrophins have multiple functions in G-protein-mediated intracellular signaling.
Further syntrophins have also been shown to bind calmodulin (CaM), which is a calcium binding messenger protein that mediates the function of Ca2+ in the cell by regulating its concentration within the cell. Calmodulin transduces calcium signals by binding to Ca2+ ions and then interacting with its downstream target proteins [127, 128]. These interactions thus suggest a potential role for syntrophins in cell signaling.
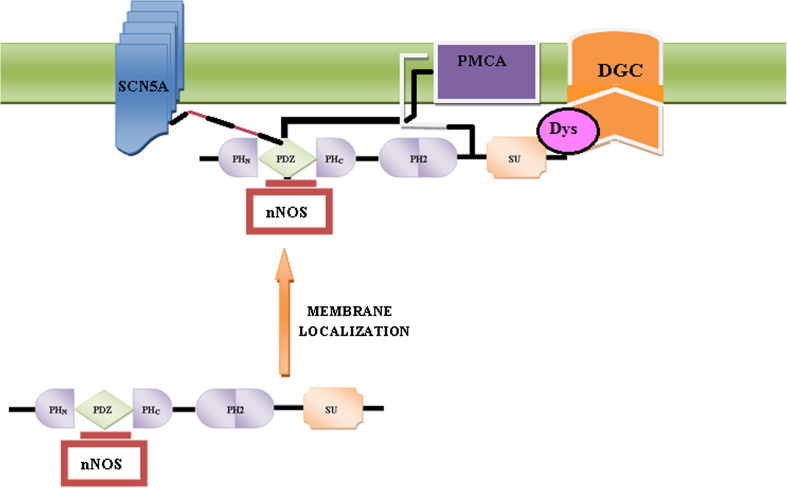
Nitric oxide (NO) is an endogenous signaling molecule synthesized from l-arginine by the enzymes called nitric oxide synthases (NOS). These NO synthases have been characterised as neural NOS, i.e. nNOS or NOS-1, inducible NOS, i.e. iNOS or NOS-2, and endothelial NOS, i.e. eNOS or NOS3 [129]. While nNOS and eNOS are expressed constitutively throughout the heart, the inducible one, iNOS, is expressed under patho-physiological conditions. In the human body in general, and within the heart in particular, NO has several functions, e.g., it affects contractility and normal muscle functioning [130], and thus the importance of NOS-1 signaling in the heart is well established. NOS-1 ablation enhances basal contractility [130, 131] and NOS-1-derived NO has been shown to increase as a consequence of experimental and pathological human heart failure [132]. In skeletal muscle cells, the muscle-specific isoform of neuronal nitric oxide synthase (nNOS), i.e. NOS-1μ, has been shown to bind to α-1-syntrophin protein which ensure its localization to the sarcolemma via the dystrophin complex (DGC) [132]. In fact, these proteins are thought to be a part of a macromolecular complex in which the sarcolemmal calcium pump, syntrophin proteins, and the neuronal nitric oxide synthase form a ternary association with each other. A physical interaction between the plasma membrane Ca2+/calmodulin-dependent ATPase 4b (PMCA4b), α-1-syntrophin, and NOS-1 in cardiac cells has been confirmed [133, 134]. The interaction between nNOS and α-1-syntrophin is, however, not dependant on the recognition of a COOH-terminal motif, rather it involves a PDZ–PDZ interaction [49]. The PDZ domain of nNOS specifically heterodimerizes with the PDZ domain of α-1-syntrophin protein [26, 133], and this interaction is a typical representative of an alternate class of PDZ domain binding, i.e. via the recognition of internal PBMs [133–135]. The interaction between the PDZ domains of these two proteins takes place in a linear head-to-tail fashion having a two-faced unusual polarized structure with a receptor face and a ligand face. The ligand face has a β-hairpin structure that acts as the PDZ ligand, binding the receptor face, i.e. the peptide binding site of the syntrophin PDZ domain [49], while the peptide binding groove of the nNOS PDZ domain remains available for binding with additional proteins. Such PDZ–PDZ interactions are proof that a terminal carboxylate group is not an absolute requirement for binding to the PDZ domains. Also, α-1-syntrophin interacts with the distal region of the large intracellular loop within PMCA4b, and the linker region between the PH2 domain and SU domain, corresponding to amino acids 399–447 of α-1-syntrophin, have been shown to be very important for this interaction to take place [135]. Thus, a ternary complex interaction between PMCA, α-1-syntrophin, and NOS-1 in cardiac cells has been proposed [134]. In this complex, PMCA acts as a negative regulator of NOS-1, and an over-expression of α-1-syntrophin and PMCA4b has been shown to strongly inhibit the NO production [132–134]. PMCA4b is shown to be linked to NOS-1 through interactions between the COOH-terminal tail of PMCA and the PDZ domain of NOS-1 [136], and α-1-syntrophin is shown to be bound to this complex via an interactions between its linker region (i.e. the region inbetween the PH2 and SU domains) and the large intracellular loop (between regions four and five) of PMCA4 (Fig. 7) [134–136]. PMCA and α-1-syntrophin negatively regulate the NOS-1 activity in a synergistic manner (Fig. 7), which further supports some significant functional implications for this complex formation on nitric oxide-regulated signaling pathways.
Fig. 7.
Diagrammatic representation of the multi-complex formation between PMCA, α-1-syntrophin, NOS, and the SCNA5 sodium channel. PMCA via its intracellular loop, located between trans-membrane domains 4 and 5, binds to the linker region of syntrophin, and via its COOH terminus binds to the PDZ domain of NOS-1. Syntrophin also connects the SCN5A sodium channel to the nNOS–PMCA complex and regulates NO concentration at the membrane
The ATP-binding cassette transporter A1 (ABCA1) mediates the release of cellular cholesterol and phospholipids to form high density lipoprotein [137, 138]. ApoA-I-mediated cholesterol efflux is a major event in reverse cholesterol transport which serves to generate HDL and transports the excess cholesterol from the peripheral tissues to the liver for biliary secretion [139–141]. The PDZ domain of the α-1-syntrophin protein has been shown to interact strongly with ABCA1 via the C-terminal three amino acids SYV of ABCA1. Co-expression of α-1-syntrophin has been shown to increase the half-life of ABCA1, down-regulating the degradation process of ABCA1, and also to increase the apoA-I-mediated release of cholesterol in human cell lines [142–145]. Binding of α-1-syntrophin to the C terminus of ABCA1 has been proposed to cause a conformational change in ABCA1 making it resistant to proteolysis by calpain [142–144]. Syntrophins have thus been implicated to be involved in lipid homeostasis in the brain [143]. Also, a novel interaction of ABCA1 with the PDZ protein β1-syntrophin has been identified. SiRNA-mediated down-regulation of β1-syntrophin has been shown to decrease ABCA1 protein levels, whereas over-expression of β1-syntrophin increases ABCA1 cell-surface expression and stimulated efflux to apoA-I [140–143]. These findings indicate that syntrophins are involved in the intracellular signaling pathways that regulate the cellular distribution, stability, and activity of this important transporter protein ABCA1 within cells.
Syntrophin-associated disorders/pathologies
Although the function of syntrophins is not yet fully understood, they have been implicated to have a regulatory role in diverse biological processes such as muscular development, neuronal and immunological cell synapses, etc., and are involved in several important signaling pathways like SAPK/MAPK pathway, Ca2+ concentration regulation in muscle cells, Rac1 activation, generation of reactive oxygen species (ROS), apoptosis, etc. The absence or aberrant/altered expression of syntrophins has been associated with several disorders particularly those related to muscles or neuromuscular development [146, 147], e.g., α-1-syntrophin-deficient skeletal muscles have been shown to exhibit hypertrophy and aberrant formation of neuromuscular junctions. Several structural abnormalities in the formation of neuromuscular junctions or synaptic function have been described in mice models [148], and the importance of α-, β-2-syntrophin proteins in the formation of these neuromuscular junctions (NMJ) is evident in α-1-/β-2 null mice which have been shown to develop aberrant NMJ with few or irregular junction folds. It has been seen that α-1-syntrophin and α-dystrobrevin are either absent or markedly reduced at the sarcolemmal membranes in Duchenne muscular dystrophy (DMD) or in denervation disorders [148–150]. α-1-syntrophin has been shown to play an important role in regulating the myogenesis, by modulating the myogenin expression, and the absence of this isoform has been shown to cause reduced and delayed myogenin expression in differentiating myoblasts [150]. In several muscle dystrophies, the expression profile of syntrophin proteins show a difference from the normal pattern, e.g., in Duchenne muscular dystrophy (DMD) and fukuyama muscular dystrophies (FMD), and altered/decreased expression of α1-syntrophin has been observed in myofibers [150–153]. β-1-syntrophin has been found to be the predominant isoform in developing muscle membranes, and its expression up-regulated in response to the loss of α-1-syntrophin in DMD and in other denervation disorders [149].
A sustained increase in the Ca2+ concentration has been observed in several muscular dystrophies, e.g., DMD. Due to this sustained increase in the cellular Ca2+ concentration, various Ca2+-sensitive proteolytic and phospholipolytic enzymes are activated that may lead to the degradation of muscle fibers in themselves [61, 153]. Calcium mishandling in DMD is thought to be due to the uncontrolled stretch-activated and store-operated Ca2+ influx supported by TRPC cationic channels in muscle cells. The molecular association between the TRPC channels and the α-1-syntophin/dystrophin complex has been implicated in the development of the calcium mishandling observed in several dystrophies, including DMD. Also, normal regulation of capacitative calcium entries (CCE), which is considered to be very important for maintaining Ca2+ homeostasis and long-term activity of muscle cells, is thought to depend on the association between TRPC channels (TRPC1, TRPC4) and the α-1-syntrophin protein. As has been discussed earlier, α-1-syntrophin anchors these cationic channels to the DAPC by interacting with dystrophin, and via dystrophin they can associate with the cell cytoskeleton. Any loss or disruption in this molecular association may result in calcium alterations, characteristic of many muscular dystrophies [61]. Similarly, it has been shown that in multisystemic disorder myotonic dystrophy type 1 (DM1), the dysregulation in alternative splicing of the α-dystrobrevin protein results in the localization of an aberrantly spliced α-dystrobrevin isoform to the sarcolemma, which shows increased binding to α-1-syntrophin and thus results in changes in the α-1-syntrophin concentration at the sarcolemma. This accounts for the increased levels of α-1-syntrophin associated with DGC, as has been observed in DM1 muscles [152].
In addition to muscle dystrophies, syntrophins have also been linked with some cardiac pathologies, e.g., long QT syndrome (LQTS) [154] and sudden infant death syndrome (SIDS). SIDS is a leading cause of deaths of infants below 1 year of age, and about 5–10 % of SIDS stem from LQTS [155]. LQTS is an inherited cardiac disorder characterized by QT phase prolongation and syncope or death from arrhythmia [156]. Alpha-1-syntrophin acts as molecular scaffold for complex formation between nNOS and plasma membrane Ca/calmodulin-dependant ATPase (PMCA) isoforms 1 and 4b (discussed earlier), and this association is very important for the proper membrane localization of NOS-1 and the regulation of NO synthesis [157]. α-1-syntrophin and PMCA have been shown to work synergistically to negatively regulate NOS-1 activity. The importance of NOS-1 signaling is well established in heart and muscle cells [41, 42, 134]. Association of nNOS with α-1-syntrophin in muscles localizes nNOS to the DGC, coupling NO production to the muscle contraction [133], which exerts a protective effect by increasing the blood flow to cope with the high metabolic requirements of contracting muscles. It has recently been shown that NOS-1 is important in protecting the heart against myocardial infarction, and its expression is up-regulated in the final stages of late-phase ischemic preconditioning [158]. In dystrophic muscles, that show a very low or an irregular α-1-syntrophin expression pattern, or in α-1-syntrophin knock-out mice, an irregular nNOS distribution at the sarcolemma has been observed, although the cytoplasmic reserve of nNOS is not affected. Since, in the sarcolemma, nNOS regulates the homeostasis of reactive free radical species (NO), any alteration in α-1-syntrophin protein expression would thus result in the alteration of syntrophin-mediated membrane localization of nNOS, having serious functional implications contributing to muscle degeneration, and may be responsible for the oxidative damage to muscle protein in diseases such as Duchenne muscular dystrophy [90, 158]. SNTA1 has also been shown to interact with the pore-forming α subunit SCN5A, also known as NaV1.5, of the cardiac sodium channel macromolecular complex [90]. It has been shown that SNTA1 connects SCN5A to the nNOS–PMCA complex in the heart, and that a perturbation in SNTA1 [159] stemming from a rare missense mutation (A390V-SNTA1) disrupts this association with PMCA4b causing increased late sodium current (INa), via S-nitrosylation of the cardiac sodium channel. These findings thus suggest that SNTA1 be considered as a rare LQTS-susceptibility gene [90, 159].
In recent times, β-2-syntrophin has been implicated in type-2 diabetes mellitus. Type-2 diabetes mellitus, or non-insulin-dependent diabetes mellitus (NIDDM), is a metabolic disorder that is often referred to as adult onset diabetes. Certain genetic factors, and in particular the sedentary lifestyle or excessive food consumption, are primarily responsible for the increasing rate of this metabolic disorder all over the world. Characterized by high blood glucose levels, i.e. hyperglycemia, NIDDM occurs due to an insufficient insulin production from the β cells in the pancreas to meet the metabolic demand, i.e. a relative insulin deficiency. Very often, NIDDM leads to insulin resistance in the body, i.e. the body cells are unable to respond to the normal actions of the insulin hormone or to maintain the normal glucose levels in blood [160, 161]. Insulin production from the pancreatic β cells occurs via insulin granules exocytosis and is stimulated by the raised levels of glucose in blood. This mechanism of insulin granule secretion by pancreatic β cells is not yet completely understood. However, recently, an islet cell auto-antigen 512 (ICA512), which is an intrinsic granule membrane protein in association with the β-2-syntrophin isoform, is thought to regulate the insulin granule turnover in pancreatic β cells. The β 2 isoform of syntrophins plays an important role in regulation of insulin granule mobilization in pancreatic β cells, as it interacts with the ICA512 protein connecting it to the β cell cytoskeletal (F-actin) via its interaction with the utrophin protein in the dystrophin complex, and thereby restrains the mobility of these insulin granules [162].
Islet cell auto-antigen 512 is a receptor protein tyrosine phosphatase, lacking any phosphatase activity that serves to tether the insulin granules to actin microfilaments via its association with the β-2-syntrophin protein [163]. During granule maturation, cleavage of ICA512 is caused by furin-like convertases, which generates an ICA512-trans-membrane fragment (ICA512-TF) [162, 163]. The ICA512-cytoplasmic fragment (ICA512-CF) binds to the PDZ domain of β-2-syntrophin, which is in turn associated with F-actin through utrophin [31]. The phosphorylation pattern of β-2-syntrophin has been shown to change in relation to stimulation of insulin secreation [65]. Hyperglycemia conditions stimulate phosphorylation of the β-2-syntrophin protein which causes the dissociation of ICA512-TF from β-2-syntrophin/F-actin and leads to an increase in the mobility of insulin granules, promoting β cell insulin production [164]. The phosphorylation of serine residues S75 and the dephosphorylation of S90 within β-2-syntrophin has been shown to weaken its interaction with ICA512, causing an increased granule mobility and insulin secretion [65]. The dissociation of ICA512-TF from β-2-syntrophin is further enhanced by the intracellular cleavage of ICA512 at the plasma membrane by Ca2+-activated protease calpian-1 and the generation of an ICA512-cytosolic fragment (ICA512-CF). This ICA512-CF can bind to both the components of the ICA512-TF/β-2-syntrophin complex and thereby disrupt their association. This leads to the increased displacement and increased mobility of insulin granules from cytoskeletal filaments and also increases their chances of reaching the apt membrane sites for exocytosis. However, with the increase in the insulin granule exocytosis, a feedback loop is thought to exist, whereby the newly generated ICA512-CF are targeted to the nuclease for enhancing the transcription of insulin gene, insulin granule genes, granule biogenesis, and β cell proliferation [161–163]. By this feed-back mechanism, the β cells are thought to balance the insulin granule storage and consumption as per the blood glucose levels in the body. Also, β-2-syntrophin has been confirmed as a Cdk5 substrate which has been shown to cause the phosphorylation of S75 in β-2-syntrophin [65]. Thus, the Cdk5-mediated phosphorylation state of β-2-syntrophin, especially S75, allosterically modulates the interaction between its PDZ domain and ICA512 and regulates the intracellular compartmentalization and functioning of β-2-syntrophin, while the disruption of this interaction stimulates the mobilization of insulin granules and regulation of insulin secreation from β cells [65, 162].
Also, syntrophins are thought to be involved in regulating blood pressure by mediating the formation of a α1D-adrenergic receptors/dystrophin signalosome. α1-Adrenergic receptors (α1-ARs)2 are G protein-coupled receptors that mediate various important physiological functions of nor-epinephrine (NE) and epinephrine, particularly in the cardiovascular system where they are responsible for regulating vascular tone and peripheral resistance [165, 166]. Alpha-1-syntrophin specifically interacts with the C-terminal domain (containing the amino acids sequence -RETDI) of α1D-ARs through a PDZ domain-mediated interaction [167] and serves to anchor α1D-AR to the dystrophin–utrophin cytoskeleton via its SU domain [75]. This dystrophin–utrophin complex can bind up to four syntrophins, allowing other syntrophins in the complex to scaffold additional regulators of signal transduction in close proximity to α1D-AR (i.e. plasma membrane calcium ATPase, nNOS, and/or TRPC) [134, 148] resulting in the formation of a complete signalosome to enable efficient functioning. The α1-adrenergic receptors are important clinical targets for the treatment of cardiovascular disease and benign prostatic hypertrophy, and also play a critical role in the regulation of blood pressure and cardiac function [168–170].
Autism is a neuronal developmental disorder that is characterized by an impaired reciprocal social interaction and communication, a restricted stereotyped pattern of interests, and often a repetitive behavior [171]. Although the real causes for this disorder are not yet clear, certain genetic factors are believed to play a role and certain rare types of mutations have been implicated with autism, e.g., in recent times, neuroglins have been implicated as autism-related genes. Neuroligins (NL) are certain neuronal cell membrane-adhesion molecules that have been recognized as the ligands for neurixins, e.g., NL-1 has been shown to be a splice site-specific ligand for β-neuroxins [172]. Neurexins in association with neuriligins are thought to play a crucial role in the assembly, differentiation, and maturation of pre- and postsynaptic terminals, and mutations in these neuroxins have been implicated in autism and other neuronal disorders [173–175]. Certain autism-related isoforms of these neuroglins, like neuroligin 3 (NL3), neuroligin 4x (NL4X), and neuroligin 4y (NL4Y), have been shown to interact with the γ-2-isoform of syntrophin. This γ-2-isoform of syntrophin is thus considered to be a putative inhibitory synaptic scaffolding protein for neuroligins. The PDZ domain of γ-2-syntrophin has been shown to bind to the PDZ domain binding sequence present within neuroligins. Further, the interaction between these neuroligins and γ-2-syntrophin have been shown to be influenced by some autism-related mutations, i.e. the binding between NL3 and γ-2-syntrophin is impaired in autism-related mutations [171, 172]. That is why the interaction between NLs and γ-2-syntrophin is considered to be very important in terms of etiology of autism, as the impaired interaction between these two seems to be contributing to the etiology of autism [171]. However, much work needs to be done before we can completely understand the association between γ-2-syntrophin and neuroligins or the role played by these proteins in autism.
The expression of several PDZ-containing proteins has been shown to alter in diverse human diseases, as these proteins are often involved in the regulation of such crucial intracellular pathways whose abnormal regulation can result in the development of pathologies, including various types of cancers [45, 176]. Interestingly, the presence of a PDZ domain binding sequence at the C-terminal of viral oncoproteins, such as E6 oncoproteins [177] found in the cancer causing human papillomavirus (HPV), further provides us a proof for the involvement of PDZ domain-containing proteins in cancer development/progression. Differential expression of the syntrophin protein has been implicated in human cancers like breast carcinomas, esophageal squamous cell carcinoma, or colon cancers, and it is suspected that the protein might be involved in regulating cancer cell proliferation or involved in carcinogenesis. In our study, we reported, for the first time, the expression profile of α-1-syntrophin protein in histologically confirmed human esophageal, stomach, lung, colon, rectal, and breast cancerous tissue samples [20]. Our results suggest a significant decrease in the expression level of the SNTA1 protein in both esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC) compared with their respective normal controls, while a significant increase in expression of the SNTA1 protein compared with normal tissue was observed in the case of breast carcinoma samples. In one of the studies of the American Association for Cancer Research, it has been shown that syntrophin expression predicts colon cancer outcome [178]. The study was done by construction of tissue microarrays, using tumor samples and metastatic lymph nodes, to evaluate the prognostic significance of syntrophin expression in colon carcinoma. In another such study, several genes differentially expressed in tumor progression, which can potentially help in better classifying pre-malignant lesions, and tongue squamous cell carcinomas (SCCs) were identified [179]. Among these potential markers of metastases, α-1-syntrophin was found to be one of those identified proteins. All these studies point towards a possible role of syntrophin proteins in cancer development and/or its progression. There is a strong possibility that the proteins could be used as a novel diagnostic or prognostic marker. However, further studies are required to be done regarding this issue that may help us to depict the possible signaling mechanism of this protein in cancer development and its possible use as a therapeutic target.
Concluding remarks
Since the discovery of syntrophin proteins, a number of the details of their function have been elucidated. It is now well established that these adaptor proteins help to recruit various signaling molecules into a complex. Unraveling the relationships between syntrophin proteins and their binding partners will help us to better understand these cellular signaling pathways. It can be anticipated that further progress in the field will allow us to understand the sequential molecular events that may include the regulation of cellular growth, muscle development, synapses, survival, proliferation, etc., and the functions that are presumably elicited by the coordination of signaling events by these adaptor proteins. In the next few years, we are likely to see further progress in defining the molecular mechanisms of the cellular processes involving syntrophin proteins. A complete understanding of the functioning of these proteins would be of great value for the design of novel therapeutics, so that these proteins could possibly be used as a biological marker for scientific purposes or possibly be used as a therapeutic target aimed at the prevention of common pathologies such as cancer.
Acknowledgment
This work was supported by the Department of Biotechnology, Ministry of Science and Technology, Government of India, under the scheme of Development of RNAi Technology No. PR12894/AGR/36/629/2009.
References
- 1.Lee SH, Sheng M. Development of neuron–neuron synapses. Curr Opin Neurobiol. 2000;19(1):125–131. doi: 10.1016/S0959-4388(99)00046-X. [DOI] [PubMed] [Google Scholar]
- 2.Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Adams ME, Dwyer TM, Dowler LL, White RA, Froehner SC. Mouse alpha 1- and beta 2-syntrophin gene structure, chromosome localization, and homology with a discs large domain. J Biol Chem. 1995;270(43):25859–25865. doi: 10.1074/jbc.270.43.25859. [DOI] [PubMed] [Google Scholar]
- 4.Ahn AH, Yoshida M, Anderson MS, Freener CA, Selig S, Hagiwara Y, Ozawa E, Kunkel LM. Cloning of human basic A1, a distinct 59-kDa dystrophin-associated protein encoded on chromosome 8q23–24. Proc Natl Acad Sci USA. 1994;91:4446–4450. doi: 10.1073/pnas.91.10.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Froehner SC, Adams ME, Peters MF, Gee SH. Syntrophins: modular adapter proteins at the neuromuscular junction and the sarcolemma. Soc Gen Physiol Ser. 1997;52:197–207. [PubMed] [Google Scholar]
- 6.Froehner SC. Peripheral proteins of postsynaptic membranes from torpedo electric organ identified with monoclonal antibodies. J Cell Biol. 1984;99:88–96. doi: 10.1083/jcb.99.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahn AH, Freener CA, Gussoni E, Yoshida M, Ozawa E, Kunkel LM. The three human syntrophin genes are expressed in diverse tissues, have distinct chromosomal location, and each bind to dystrophin and its relatives. J Biol Chem. 1996;271:2724–2730. doi: 10.1074/jbc.271.30.18100. [DOI] [PubMed] [Google Scholar]
- 8.Froehner SC, Murnane AA, Tobler M, Peng HB, Sealock R. A postsynaptic Mr 58,000 (58 K) protein concentrated at acetylcholine receptor-rich sites in Torpedo electroplaques and skeletal muscle. J Cell Biol. 1987;104:1633–1646. doi: 10.1083/jcb.104.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kramarcy NR, Vidal A, Froehner SC, Sealock R. Association of utrophin and multiple dystrophin short forms with the mammalian M(r) 58,000 dystrophin-associated protein (syntrophin) J Biol Chem. 1994;269:2870–2876. [PubMed] [Google Scholar]
- 10.Hoffman EP, Brown RH, Kunkel LM. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 11.Campbell KP, Kahl SD. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- 12.Adams ME, Butler MH, Dwyer TM, Peters MF, Murnane AA, Froehner SC. Two forms of mouse syntrophin, a 58 kd dystrophin-associated protein, differ in primary structure and tissue distribution. Neuron. 1993;11:531–540. doi: 10.1016/0896-6273(93)90157-M. [DOI] [PubMed] [Google Scholar]
- 13.Yang B, Ibraghimov-Beskrovnaya O, Moomaw CR, Slaughter CA, Campbell KP. Heterogeneity of the 59-kDa dystrophin associated protein revealed by cDNA cloning and expression. J Biol Chem. 1994;269:6040–6044. [PubMed] [Google Scholar]
- 14.Butler MH, Douville K, Murnane AA, Kramarcy NR, Cohen JB, Sealock R, Froehner SC. Association of the Mr 58,000 postsynaptic protein of electric tissue with Torpedo dystrophin and the Mr 87,000 postsynaptic protein. J Biol Chem. 1992;267:6213–6218. [PubMed] [Google Scholar]
- 15.Ahn AH, Kunkel LM. The structural and functional diversity of dystrophin. Nat Genet. 1993;3:283–291. doi: 10.1038/ng0493-283. [DOI] [PubMed] [Google Scholar]
- 16.Piluso G, Mirabella M, Ricci E, Belsito A, Abbondanza C, Servidei S, Puca AA, Tonali P, Puca GA, Nigro V. Gamma-1 and gamma-2-syntrophins, two novel dystrophin-binding proteins localized in neuronal cells. J Biol Chem. 2000;275:15851–15860. doi: 10.1074/jbc.M000439200. [DOI] [PubMed] [Google Scholar]
- 17.Ahn AH, Kunkel LM. Syntrophin binds to an alternatively spliced exon of dystrophin. J Cell Biol. 1995;128:363–371. doi: 10.1083/jcb.128.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramarcy NR, Sealock R. Syntrophin isoforms at the neuromuscular junction: developmental time course and differential localization. Mol Cell Neurosci. 2000;15:262–274. doi: 10.1006/mcne.1999.0823. [DOI] [PubMed] [Google Scholar]
- 19.Peters MF, Adams ME, Froehner SC. Differential association of syntrophin pairs with the dystrophin complex. J Cell Biol. 1997;138(1):81–93. doi: 10.1083/jcb.138.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhat HF, Baba RA, Bashir M, Saeed S, Kirmani D, Wani MM, Wani NA, Wani KA, Khanday FA. Alpha-1-syntrophin protein is differentialy expressed in human cancers. Biomarkers. 2010;16:31–36. doi: 10.3109/1354750X.2010.522731. [DOI] [PubMed] [Google Scholar]
- 21.Alessi A, Bragg AD, Percival JM, Yoo J, Albrecht DE, Froehner SC, Adams ME. γ-Syntrophin scaffolding is spatially and functionally distinct from that of the α/β syntrophins. Exp Cell Res. 2006;312:3084–3095. doi: 10.1016/j.yexcr.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Peters MF, Kramarcy NR, Sealock R, Froehner SC. β-2 syntrophin: localization at the neuromuscular junction in skeletal muscle. Neuro Report. 1994;5:1577–1580. [PubMed] [Google Scholar]
- 23.Sealock R, Butler MH, Kramarcy NR, Gao K, Murnane AA, Douville K, Froehner SC. Localization of dystrophin relative to acetylcholine receptor domains in electric tissue and adult and cultured skeletal muscle. J Cell Biol. 1991;113:1133–1144. doi: 10.1083/jcb.113.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams ME, Kramarcy N, Krall SP, Rossi SG, Rotundo RL, Sealock R, Froehner SC. Absence of alpha-syntrophin leads to structurally aberrant neuromuscular synapses deficient in utrophin. J Cell Biol. 2000;150:1385–1398. doi: 10.1083/jcb.150.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Way M, Pope B, Cross RA, Kendrick-Jones J, Weeds AG. Expression of the N-terminal domain of dystrophin in E.coli and demonstration of binding to F-actin. FEBS Lett. 1992;301:243–245. doi: 10.1016/0014-5793(92)80249-G. [DOI] [PubMed] [Google Scholar]
- 26.Brenman JE, Chao DS, Gee SH, McGee AW, Craven SE, Santillano DR, Wu Z, Huang F, Xia H, Peters MF, Froehner SC, Bredt DS. Interaction of nitric oxide synthase with the postsynaptic density protein PSD-95 and alpha1-syntrophin mediated by PDZ domains. Cell. 1996;84:757–767. doi: 10.1016/S0092-8674(00)81053-3. [DOI] [PubMed] [Google Scholar]
- 27.Okumura H, Okumura N, Iwamatsu A, Buijs RM, Romijn HJ, et al. Interaction of neuronal nitric oxide synthase with alpha-1-syntrophin in rat brain. J Biol Chem. 1999;247:11736–11741. doi: 10.1074/jbc.274.17.11736. [DOI] [PubMed] [Google Scholar]
- 28.Lumeng C, Phelps S, Crawford GE, Walden PD, Barald K, Chamberlain JS. Interactions between beta 2-syntrophin and a family of microtubule-associated serine/threonine kinases. Nat Neurosci. 1999;2:611–617. doi: 10.1038/10165. [DOI] [PubMed] [Google Scholar]
- 29.Hogan A, Shepherd L, Chabot J, Quenneville S, Prescott SM, Topham MK, Gee SH. Interaction of gamma 1-syntrophin with diacylglycerol kinase-zeta. Regulation of nuclear localization by PDZ interactions. J Biol Chem. 2001;276(28):26526–26533. doi: 10.1074/jbc.M104156200. [DOI] [PubMed] [Google Scholar]
- 30.Saraste M, Sibbald PR, Wittinghofer A. The P-loop-a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-F. [DOI] [PubMed] [Google Scholar]
- 31.Albrecht DE, Froehner SC. Syntrophins and dystrobrevins: defining the dystrophin scaffold at synapses. Neurosignals. 2002;11:123–129. doi: 10.1159/000065053. [DOI] [PubMed] [Google Scholar]
- 32.Lemmon MA, Ferguson KM. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem J. 2000;350(Part 1):1–18. [PMC free article] [PubMed] [Google Scholar]
- 33.Musacchio A, Gibson T, Rice P, Thompson J, Saraste M. The PH-domain: a common piece in the structural patchwork of signaling proteins. Trends Biochem Sci. 1993;18:343–348. doi: 10.1016/0968-0004(93)90071-T. [DOI] [PubMed] [Google Scholar]
- 34.Berg JS, Derfler BH, Pennisi CM, Corey DP, Cheney RE. Myosin-X, a novel myosin with pleckstrin homology domains, associated with regions of dynamic actin. J Cell Sci. 2000;113:3439–3451. doi: 10.1242/jcs.113.19.3439. [DOI] [PubMed] [Google Scholar]
- 35.Schultz J, Milpetz F, Bork P, Ponting CP. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci USA. 1998;95:5857–5864. doi: 10.1073/pnas.95.11.5857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gibson TJ, Hyvonen M, Musacchio A, Saraste M, Birney E. PH domain: the first anniversary. Trends Biochem Sci. 1994;19:349–353. doi: 10.1016/0968-0004(94)90108-2. [DOI] [PubMed] [Google Scholar]
- 37.Chang JS, Kim SK, Kwon TK, Bae SS, Min DS, Lee YH, Kim SO, Seo JK, Choi JH, Suh PG. Pleckstrin homology domains of phospholipase C-{gamma}1 directly interact with {beta}-tubulin for activation of phospholipase C-{gamma}1 and reciprocal modulation of {beta}-tubulin function in microtubule assembly. J Biol Chem. 2005;280:6897–6905. doi: 10.1074/jbc.M406350200. [DOI] [PubMed] [Google Scholar]
- 38.Chang JS, Seok H, Kwon TK, Min DS, Ahn BH, Lee YH, Suh JW, Kim JW, Iwashita S, Omori A, Ichinose S, Numata O, Seo JK, Oh YS, Suh PG. Interaction of elongation factor-1alpha and pleckstrin homology domain of phospholipase C-gamma 1 with activating its activity. J Biol Chem. 2002;277:19697–19702. doi: 10.1074/jbc.M111206200. [DOI] [PubMed] [Google Scholar]
- 39.Jing Y, Wenyu W, Weiguang X, Jia-fu L, Marvin EA, Froehner SC, Mingjie Z. Structure of the split PH domain and distinct lipid binding properties of the PH–PDZ supramodule of α-syntrophin. EMBO J. 2005;24:3985–3995. doi: 10.1038/sj.emboj.7600858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwata Y, Pan Y, Yoshida T, Hanada H, Shigekawa M. Bidirectional signaling between sarcoglycans and the integrin adhesion system in cultured L6 Myocytes. FEBS Lett. 1998;423:173–177. doi: 10.1016/S0014-5793(98)00085-4. [DOI] [PubMed] [Google Scholar]
- 41.Newbell BJ, Anderson JT, Jarrett HW. Ca2+-calmodulin binding to mouse α-1 syntrophin: syntrophin is also a Ca2+-binding protein. Biochemistry. 1997;36:1295–1305. doi: 10.1021/bi962452n. [DOI] [PubMed] [Google Scholar]
- 42.Oak SA, Jarrett HW. The oligomerization of mouse α-1-syntrophin and self-association of its pleckstrin homology domain 1. Biochemistry. 2000;39:8870–8877. doi: 10.1021/bi0000824. [DOI] [PubMed] [Google Scholar]
- 43.Chockalingam PS, Gee SH, Jarrett HW. Pleckstrin homology domain 1 of mouse α-1-syntrophin binds phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1999;38:5596–5602. doi: 10.1021/bi982564+. [DOI] [PubMed] [Google Scholar]
- 44.Van-Rossum DB, Patterson RL, Sharma S, Barrow RK, Kornberg M, Gill DL, Snyder SH. Phospholipase C gamma1 controls surface expression of TRPC3 through an intermolecular PH domain. Nature. 2005;434:99–104. doi: 10.1038/nature03340. [DOI] [PubMed] [Google Scholar]
- 45.Gardiol D. PDZ-containing proteins as targets in human pathologies. FEBS J. 2012;279:3529. doi: 10.1111/j.1742-4658.2012.08685.x. [DOI] [PubMed] [Google Scholar]
- 46.Feng W, Zhang M. Organization and dynamicsof PDZ-domain-related supramodules in the postsynaptic density. Nat Rev Neurosci. 2009;10:87–99. doi: 10.1038/nrn2540. [DOI] [PubMed] [Google Scholar]
- 47.Nourry C, Grant SG, Borg JP (2003) PDZ domain proteins: plug and play. Sci STKE2003 RE7 [DOI] [PubMed]
- 48.Chimura T, Launey T, Ito M. Evolutionarily conserved bias of amino acid usage refines the definition of PDZ-binding motif. BMC Genomics. 2011;12:300. doi: 10.1186/1471-2164-12-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hillier BJ, Christopherson KS, Prehoda KE, Bredt DS, Lim WA. Unexpected modes of PDZ domain scaffolding revealed by structure of nNOS-syntrophin complex. Science. 1999;284:812–815. doi: 10.1126/science.284.5415.812. [DOI] [PubMed] [Google Scholar]
- 50.Doyle DA, Lee A, Lewis J, Kim E, Sheng M, MacKinnon R. Crystal structures of a complexed and peptide-free membrane proteinbinding domain: molecular basis of peptide recognition by PDZ. Cell. 1996;85:1067–1076. doi: 10.1016/S0092-8674(00)81307-0. [DOI] [PubMed] [Google Scholar]
- 51.Schultz J, Hoffmuuller U, Krause G, Ashurst J, Macias MJ, Schmieder P, Schneider-Mergener J, Oschkinat H. Specific interactions between the syntrophin PDZ domain and voltage-gated sodium channels. Nature Struct Biol. 1998;5:19–24. doi: 10.1038/nsb0198-19. [DOI] [PubMed] [Google Scholar]
- 52.Wiedemann U, Boisguerin P, Leben R, Leitner D, Kraus G, Moelling K, Volkmer-Engert R, Oschkinat H. Quantification of PDZ domain specificity, prediction of ligand affinity and rational design of super binding peptides. J Mol Biol. 2004;343:703–718. doi: 10.1016/j.jmb.2004.08.064. [DOI] [PubMed] [Google Scholar]
- 53.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Clustering of Shaker-type K1 channels by direct interaction with the PSD-95/SAP90 family of membrane associated guanylate kinases. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 54.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 55.Adams ME, Mueller HA, Froehner SC. In vivo requirement of the {alpha}-syntrophin PDZ domain for the sarcolemmal localization of nNOS and aquaporin-4. J Cell Biol. 2001;155:113–122. doi: 10.1083/jcb.200106158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neely JD, Amiry-Moghaddam M, Ottersen OP, Froehner SC, Agre P, Adams ME. Syntrophin-dependent expression and localization of Aquaporin-4 water channel protein. Proc Natl Acad Sci USA. 2001;98:14108–14113. doi: 10.1073/pnas.241508198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gee SH, Madhavan R, Levinson SR, Caldwell JH, Sealock R, Froehner SC. Interaction of muscle and brain sodium channels with multiple members of the syntrophin family of dystrophin-associated proteins. J Neurosci. 1998;18:128–137. doi: 10.1523/JNEUROSCI.18-01-00128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ou Y, Strege P, Miller SM, Makielski J, Ackerman M, Gibbons SJ, Farrugia G. Syntrophin gamma 2 regulates SCN5A gating by a PDZ domain-mediated interaction. J Biol Chem. 2003;278:1915–1923. doi: 10.1074/jbc.M209938200. [DOI] [PubMed] [Google Scholar]
- 59.Connors NC, Adams ME, Froehner SC, Kofuji P. The potassium channel Kir4.1 associates with the dystrophin–glycoprotein complex via alpha-syntrophin in glia. J Biol Chem. 2004;279:28387–28392. doi: 10.1074/jbc.M402604200. [DOI] [PubMed] [Google Scholar]
- 60.Leonoudakis D, Conti LR, Anderson S, Radeke CM, McGuire LM, Adams ME, Froehner SC, Yates JR, Vandenberg CA. Protein trafficking and anchoring complexes revealed by proteomic analysis of inward rectifier potassium channel (Kir2.x)- associated proteins. J Biol Chem. 2004;279:22331–22346. doi: 10.1074/jbc.M400285200. [DOI] [PubMed] [Google Scholar]
- 61.Vandebrouck A, Sabourin J, Rivet J, Balghi H, Sebille S, Kitzis A, Raymond G, Cognard C, Bourmeyster N, Constantin B. Regulation of capacitative calcium entries by α-1-syntrophin: association of TRPC1 with dystrophin complex and the PDZ domain of α-1-syntrophin. FASEB J. 2007;21:608–617. doi: 10.1096/fj.06-6683com. [DOI] [PubMed] [Google Scholar]
- 62.Hasegawa M, Cuenda A, Spillantini MG, Thomas GM, Buee-Scherrer V, Cohen P, Goedert M. Stress-activated protein kinase-3 interacts with the PDZ domain of alpha1-syntrophin. A mechanism for specific substrate recognition. J Biol Chem. 1999;274:12626–12631. doi: 10.1074/jbc.274.18.12626. [DOI] [PubMed] [Google Scholar]
- 63.Buechler C, Boettcher A, Bared SM, Probst MC, Schmitz G. The carboxy terminus of the ATP-binding cassette transporter A1 interacts with a beta-2-syntrophin/utrophin complex. Biochem Biophys Res Commun. 2002;293:759–765. doi: 10.1016/S0006-291X(02)00303-0. [DOI] [PubMed] [Google Scholar]
- 64.Torchio GM, Ermacora MR, Sica MP. Equilibrium unfolding of the PDZ domain of β-2-syntrophin. Biophys J. 2012;102(12):2835–2844. doi: 10.1016/j.bpj.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schubert S, Knoch KP, Ouwendijk J, Mohammed S, Bodrov Y, Jager M, Altkruger A, Wegbrod C, Adams ME, Kim Y, Froehner SC, Jensen ON, Kalaidzidis Y, Solimena M. β-2-Syntrophin is a Cdk5 substrate that restrains the motility of insulin secretory granules. PLoS ONE. 2010;5(9):e12929. doi: 10.1371/journal.pone.0012929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Trejo JA. Internal PDZ ligands: novel endocytic recycling motifs for G protein-coupled receptors. Mol Pharmacol. 2005;67:1388–1390. doi: 10.1124/mol.105.011288. [DOI] [PubMed] [Google Scholar]
- 67.Paasche JD, Attramadal T, Kristiansen K, Oksvold MP, Johansen HK, Huitfeldt HS, Dahl SG, Attramadal H. Subtype-specific sorting of the ETA endothelin receptor by a novel endocytic recycling signal for G protein-coupled receptors. Mol Pharmacol. 2005;67:1581–1590. doi: 10.1124/mol.104.007013. [DOI] [PubMed] [Google Scholar]
- 68.Gage RM, Matveeva EA, Whiteheart SW, Von-Zastrow M. Type I PDZ ligands are sufficient to promote rapid recycling of G-protein coupled receptors independent of binding to NSF. J Biol Chem. 2005;280:3305–3313. doi: 10.1074/jbc.M406934200. [DOI] [PubMed] [Google Scholar]
- 69.Heydorn A, Sondergaard BP, Ersboll B, Holst B, Nielson FC, Haft CR, Whistler J, Schwartz TW. A library of 7TM receptor C-terminal tails. J Biol Chem. 2004;279:54291–54303. doi: 10.1074/jbc.M406169200. [DOI] [PubMed] [Google Scholar]
- 70.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 71.Fushman D, Cahill S, Lemmon MA, Schlessinger J, Cowburn D. Solution structure of pleckstrin homology domain of dynamin by heteronuclear NMR spectroscopy. Proc Natl Acad Sci USA. 1995;92:816–820. doi: 10.1073/pnas.92.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Macias MJ, Musacchio A, Ponstingl H, Nilges M, Saraste M, Oschkinat H. Structure of the pleckstrin homology domain from beta-spectrin. Nature. 1994;369:675–677. doi: 10.1038/369675a0. [DOI] [PubMed] [Google Scholar]
- 73.Yoon HS, Hajduk PJ, Petros AM, Olejniczak ET, Meadows RP, Fesik SW. Solution structure of a pleckstrin-homology domain. Nature. 1994;369:672–675. doi: 10.1038/369672a0. [DOI] [PubMed] [Google Scholar]
- 74.Zhao C, Yu DH, Shen R, Feng GS. Gab2, a new pleckstrin homology domain-containing adapter protein, acts to uncouple signaling from ERK kinase to Elk-1. J Biol Chem. 1999;274:19649–19654. doi: 10.1074/jbc.274.28.19649. [DOI] [PubMed] [Google Scholar]
- 75.Kachinsky AM, Froehner SC, Milgram SL. A PDZ-containing scaffold related to the dystrophin complex at the basolateral membrane of epithelial cells. J Cell Biol. 1999;145:391–402. doi: 10.1083/jcb.145.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suzuki A, Yoshida M, Ozawa E. Mammalian alpha 1- and beta 1-syntrophin bind to the alternative splice-prone region of the dystrophin COOH terminus. J Cell Biol. 1995;128:373–381. doi: 10.1083/jcb.128.3.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petrof BJ, Shrager JB, Stedman HH, Kelly AM, Sweeney HL. Dystrophin protects the sarcolemma from stresses developed during muscle contractions. Proc Natl Acad Sci. 1993;90:3710–3714. doi: 10.1073/pnas.90.8.3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ambramovici H, Hogan AB, Obagi C, Topham MK, Gee SH. Diacylglycerol kinase-ζ localization in skeletal muscles is regulated by phosphorylation and interaction with syntrophins. Mol Biol Cell. 2003;14:4499–4511. doi: 10.1091/mbc.E03-03-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hogan A, Yakubchyk Y, Josee-Chabot J, Obagi C, Daher E, Maekawa K, Gee SH. The phosphoinositol 3,4-bisphosphate-binding protein TAPP1 interacts with syntrophins and regulates actin cytoskeletal organization. J Biol Chem. 2004;51:53717–53724. doi: 10.1074/jbc.M410654200. [DOI] [PubMed] [Google Scholar]
- 80.Cantley LC. The phosphoinositide 3-kinase Pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- 81.Iwata Y, Sampaolesi M, Shigekawa M, Wakabayashi S. Syntrophin is an actin-binding protein the cellular localization of which is regulated through cytoskeletal reorganization in skeletal muscle cells. Eur J Cell Biol. 2004;83:555–565. doi: 10.1078/0171-9335-00415. [DOI] [PubMed] [Google Scholar]
- 82.Topham MK, Bunting M, Zimmerman GA, McIntyre TM, Blackshear PJ, Prescott SM. Protein kinase C regulates the nuclear localization of diacylglycerol kinase-ζ. Nature. 1998;394:697–700. doi: 10.1038/29337. [DOI] [PubMed] [Google Scholar]
- 83.Kimber WA, Trinkle-Mulcahy L, Cheung PC, Deak M, Marsden LJ, Kieloch A, Watt S, Javier RT, Gray A, Downes CP, Lucocq JM, Alessi DR. Evidence that the tandem-pleckstrin-homology-domain-containing protein TAPP1 interacts with Ptd(3,4)P2 and the multi-PDZ-domain-containing protein MUPP1 in vivo. Biochem J. 2002;361:525–536. doi: 10.1042/0264-6021:3610525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dowler S, Currie RA, Downes CP, Alessi DR. DAPP1: a dual adaptor for phosphotyrosine and 3-phosphoinositides. Biochem J. 1999;342:7–12. doi: 10.1042/0264-6021:3420007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marshall AJ, Krahn AK, Ma K, Duronio V, Hou S. TAPP1 and TAPP2 Are Targets of Phosphatidylinositol 3-Kinase Signaling in B Cells: sustained Plasma Membrane Recruitment Triggered by the B-Cell Antigen Receptor. Mol Cell Biol. 2002;22:5479–5491. doi: 10.1128/MCB.22.15.5479-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Marshall AJ, Niiro H, Lerner CG, Yun TJ, Thomas S, Disteche CM, Clark EA. Dual adapter for phosphotyrosine and 3-phosphotyrosine and 3-phosphoinositide (hDAPP1) (B cell adapter molecule of 32 kDa) (Blymphocyte adapter protein Bam32) J Exp Med. 2000;191:1319–1332. doi: 10.1084/jem.191.8.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Newey SE, Benson MA, Ponting CP, Davies KE, Blake DJ. Alternative splicing of dystrobrevin regulates the stoichiometry of syntrophin binding to the dystrophin protein complex. Curr Biol. 2000;10:1295–1298. doi: 10.1016/S0960-9822(00)00760-0. [DOI] [PubMed] [Google Scholar]
- 89.Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;5:647–657. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- 90.Bruno G, Jean-Sebastien R, Andrea AD, Romina B, Christophe B, Patrick R, Hans-Anton L, Thierry P, Hugues A. Cardiac sodium channel Nav1.5 is regulated by a multiprotein complex composed of syntrophins and dystrophin. Circ Res. 2006;99:407–414. doi: 10.1161/01.RES.0000237466.13252.5e. [DOI] [PubMed] [Google Scholar]
- 91.Frigeri A, Gropper MA, Umenishi F, Kawashima M, Brown D, Verkman AS. Localization of MIWC and GLIP water channel homologs in neuro-muscular, epithelial and glandular tissues. J Cell Sci. 1995;108:2993–3002. doi: 10.1242/jcs.108.9.2993. [DOI] [PubMed] [Google Scholar]
- 92.Nielsen S, Nagelhus EA, Amiry-Moghaddam M, Bourque C, Agre P, Ottersen OP. Specialized membrane domains for water transport in glial cells: high-resolution immunogold cytochemistry of aquaporin-4 in rat brain. J Neurosci. 1997;17:171–180. doi: 10.1523/JNEUROSCI.17-01-00171.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Agre P. The aquaporin water channels. Proc Am Thorac Soc. 2006;3(1):5–13. doi: 10.1513/pats.200510-109JH. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Terris J, Ecelbarger CA, Marples D, Knepper MA, Nielsen S. Distribution of aquaporin-4 water channel expression within rat kidney. Am J Physiol. 1995;269:F775–F785. doi: 10.1152/ajprenal.1995.269.6.F775. [DOI] [PubMed] [Google Scholar]
- 95.Blake DJ, Hawkes R, Benson MA, Beesley PW. Different dystrophin-like complexes are expressed in neurons and glia. J Cell Biol. 1999;147:645–658. doi: 10.1083/jcb.147.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Durbeej M, Campbell KP. Biochemical characterization of the epithelial dystroglycan complex. J Biol Chem. 1999;274:26609–26616. doi: 10.1074/jbc.274.37.26609. [DOI] [PubMed] [Google Scholar]
- 97.Tian M, Jacobson C, Gee SH, Campbell KP, Carbonetto S, Jucker M. Dystroglycan in the cerebellum is a laminin alpha 2-chain binding protein at the glial-vascular interface and is expressed in Purkinje cells. Eur J Neurosci. 1996;8:2739–2747. doi: 10.1111/j.1460-9568.1996.tb01568.x. [DOI] [PubMed] [Google Scholar]
- 98.Fanning AS, Anderson JM. PDZ domains: fundamental building blocks in the organization of protein complexes at the plasma membrane. J Clin Invest. 1999;103:767–772. doi: 10.1172/JCI6509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neely JD, Christensen BM, Nielsen S, Agre P. Heterotetrameric composition of aquaporin-4 water channels. Biochemistry. 1999;38(34):11156–11163. doi: 10.1021/bi990941s. [DOI] [PubMed] [Google Scholar]
- 100.Poopalasundaram S, Knott C, Shamotienko OG, Foran PG, Dolly O, Ghiani CA, Gallo V, Wilkin GP. Glial heterogeneity in expression of the inwardly rectifying K(+) channel, Kir4.1, in adult rat CNS. Glia. 2000;30:362–372. doi: 10.1002/(SICI)1098-1136(200006)30:4<362::AID-GLIA50>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 101.Ishii M, Horio Y, Tada Y, Hibino H, Inanobe A, Ito M, Yamada M, Gotow T, Uchiyama Y, Kurachi Y. Expression and clustered distribution of an inwardly rectifying potassium channel, KAB2/Kir4.1, on mammalian retinal Muller cell membrane: their regulation by insulin and laminin signals. J Neurosci. 1997;17:7725–7735. doi: 10.1523/JNEUROSCI.17-20-07725.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Amedee T, Robert A, Coles JA. Potassium homeostasis and glial energy metabolism. Glia. 1997;21:46–55. doi: 10.1002/(SICI)1098-1136(199709)21:1<46::AID-GLIA5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 103.Kofuji P, Connors NC. Molecular substrates of potassium spatial buffering in glial cells. Mol Neurobiol. 2003;28:195–208. doi: 10.1385/MN:28:2:195. [DOI] [PubMed] [Google Scholar]
- 104.Sabourin J, Cognard C, Constantin B. Regulation by scaffolding proteins of canonical transient receptor potential channels in striated muscle. J Muscle Res Cell Motil. 2009;30:289–297. doi: 10.1007/s10974-010-9206-9. [DOI] [PubMed] [Google Scholar]
- 105.Vandebrouck C, Martin D, Colson-Van SM, Debaix H, Gailly P. Involvement of TRPC in the abnormal calcium influx observed in dystrophic (mdx) mouse skeletal muscle fibres. J Biol Chem. 2002;158:1089–1096. doi: 10.1083/jcb.200203091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stiber JA, Zhang ZS, Burch J, Eu JP, Zhang S, Truskey GA, Seth M, Yamaguchi N, Meissner G, Shah R, Worley PF, Williams RS, Rosenberg PB. Mice lacking Homer-1 exhibit a skeletal myopathy characterized by abnormal transient receptor potential channel activity. Mol Cell Biol. 2008;28:2637–2647. doi: 10.1128/MCB.01601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Madhavan R, Massom LR, Jarrett HW. Calmodulin specifically binds three proteins of the dystrophin-glycoprotein complex. Biochem Biophys Res Commun. 1992;185:753–759. doi: 10.1016/0006-291X(92)91690-R. [DOI] [PubMed] [Google Scholar]
- 108.Cohen P. The search for physiological substrates of MAP and SAP kinases in mammalian cells. Trends Cell Biol. 1997;7:353–361. doi: 10.1016/S0962-8924(97)01105-7. [DOI] [PubMed] [Google Scholar]
- 109.Huang EJ, Reichardt LF. Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem. 2003;72:609–642. doi: 10.1146/annurev.biochem.72.121801.161629. [DOI] [PubMed] [Google Scholar]
- 110.Iglesias T, Cabrera-Poch N, Mitchell MP, Naven TJ, Rozengurt E, Schiavo G. Identification and cloning of Kidins220, a novel neuronal substrate of protein kinase D. J Biol Chem. 2000;275:40048–40056. doi: 10.1074/jbc.M005261200. [DOI] [PubMed] [Google Scholar]
- 111.Kong HY, Boulter J, Weber JL, Lai C, Chao MV. An evolutionarily conserved transmembrane protein that is a novel downstream target of neurotrophin and ephrin receptors. J Neurosci. 2001;21:176–185. doi: 10.1523/JNEUROSCI.21-01-00176.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shuo L, Yu C, Kwok OL, Juan CA, Froehner SC, Marvin E, Adams ME, Moses V, Chao NY. α-1-syntrophin regulates ARMS localization at the neuromuscular junction and enhances EphA 4 signaling in an ARMS-dependent manner. J Cell Biol. 2005;169(5):813–824. doi: 10.1083/jcb.200412008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Arevalo JC, Yano H, Teng KK, Chao MV. A unique pathway for sustained neurotrophin signaling through an ankyrin-rich membrane-spanning protein. EMBO J. 2004;23:2358–2368. doi: 10.1038/sj.emboj.7600253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oak SA, Russo K, Petrucci TC, Jarrett HW. Mouse alpha1-syntrophin binding to Grb2: further evidence of a role for syntrophin in cell signaling. Biochemistry. 2001;40:11270–11278. doi: 10.1021/bi010490n. [DOI] [PubMed] [Google Scholar]
- 115.Zhou YW, Oak SA, Senogles SE, Jarrett HW. Laminin-R1 globular domains three and four induce heterotrimeric G-protein binding to R-syntrophin’s PDZ domain. Am J Physiol Cell Physiol. 2005;288:C377–C388. doi: 10.1152/ajpcell.00279.2004. [DOI] [PubMed] [Google Scholar]
- 116.Madhavan R, Jarrett HW. Interactions between dystrophin glycoprotein complex proteins. Biochemistry. 1995;34:12204–12209. doi: 10.1021/bi00038a014. [DOI] [PubMed] [Google Scholar]
- 117.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Jarrett HW, Foster JL. Alternate binding of actin and calmodulin to multiple sites on dystrophin. J Biol Chem. 1995;270:5578–5586. doi: 10.1074/jbc.270.10.5578. [DOI] [PubMed] [Google Scholar]
- 119.Vidal M, Goudreau N, Cornille F, Cussac D, Gincel E, Garbay C. Molecular and cellular analysis of Grb2 SH3 domain mutants: interaction with Sos and dynamin. J Mol Biol. 1999;290:717–730. doi: 10.1006/jmbi.1999.2899. [DOI] [PubMed] [Google Scholar]
- 120.Oak SA, Zhou YW, Jarrett HW. Skeletal muscle signaling pathway through the dystrophin glycoprotein complex and Rac1. J Biol Chem. 2003;278:39287–39295. doi: 10.1074/jbc.M305551200. [DOI] [PubMed] [Google Scholar]
- 121.Yang B, Jung D, Rafael JA, Chamberlain JS, Campbell KP. Identification of alpha-syntrophin binding to syntrophin triplet, dystrophin, and utrophin. J Biol Chem. 1995;270(10):4975–4978. doi: 10.1074/jbc.270.10.4975. [DOI] [PubMed] [Google Scholar]
- 122.Khanday FA, Yamamori T, Singh IM, Zhang Z, Bugayenko A, Naqvi A, Santhanum L, Nabi N, Kasuno K, Day BW, Irani K. Rac1 Leads to Phosphorylation-dependent increase in stability of the p66shc adaptor protein: role in Rac1-induced oxidative stress. Mol Biol Cell. 2006;17:122–129. doi: 10.1091/mbc.E05-06-0570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khanday FA, Santhanam L, Kasuno K, Yamamori T, Naqvi A, Dericio J, Bugazenko A, Mattagajasingh I, Disanza A, Scita G, Irani K. SOS-mediated activation of Rac1 by p66shc. J Cell Biol. 2006;172:817–822. doi: 10.1083/jcb.200506001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Reece J, Campbell N. Biology. San Francisco: Benjamin Cummings; 2002. [Google Scholar]
- 125.Akiko O, Katsuya N, Nobuaki O. Interaction of α-1-syntrophin with multiple isoforms of heterotrimeric G protein a subunits. FEBS J. 2008;275:22–33. doi: 10.1111/j.1742-4658.2007.06174.x. [DOI] [PubMed] [Google Scholar]
- 126.Yatani A, Codina J, Imoto Y, Reeves JP, Birnbaumer L, Brown AM. A G-protein directly regulates mammalian cardiac calcium channels. Science. 1987;238:1288–1292. doi: 10.1126/science.2446390. [DOI] [PubMed] [Google Scholar]
- 127.Stevens FC. Calmodulin: an introduction. Can J Biochem Cell Biol. 1983;61(8):906–910. doi: 10.1139/o83-115. [DOI] [PubMed] [Google Scholar]
- 128.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10(8):322–328. doi: 10.1016/S0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 129.Nathan C, Xie QW. Nitric oxide synthases: roles, tolls, and controls. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- 130.Barouch LA, Harrison RW, Skaf MW, Rosas GO, Cappola TP, Kobeissi ZA, Hobai IA, Lemmon CA, Burnett AL, O’Rourke B, Rodriguez ER, Huang PL, Lima JA, Berkowitz DE, Hare JM. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature. 2002;416(6878):337–339. doi: 10.1038/416337a. [DOI] [PubMed] [Google Scholar]
- 131.Sears CE, Bryant SM, Ashley EA, Lygate CA, Rakovic S, Wallis HL, Neubauer S, Terrar DA, Casadei B. Cardiac neuronal nitric oxide synthase isoform regulates myocardial contraction and calcium handling. Circ Res. 2003;92:e52–e59. doi: 10.1161/01.RES.0000064585.95749.6D. [DOI] [PubMed] [Google Scholar]
- 132.Damy T, Ratajczak P, Shah AM, Camors E, Marty I, Hasenfuss G, Marotte F, Samuel JL, Heymes C. Increased neuronal nitric oxide synthase-derived NO production in the failing human heart. Lancet. 2004;363:1365–1367. doi: 10.1016/S0140-6736(04)16048-0. [DOI] [PubMed] [Google Scholar]
- 133.Brenman JE, Chao DS, Xia H, Aldape K, Bredt DS. Nitric oxide synthase complexed with dystrophin and absent from skeletal muscle sarcolemma in Duchenne muscular dystrophy. Cell. 1995;82:743–752. doi: 10.1016/0092-8674(95)90471-9. [DOI] [PubMed] [Google Scholar]
- 134.Judith CW, Angel LA, Tamer MA, Cassandra LH, Fiona HS, Aly OZ, Delvac O, Elizabeth JC, Mamta HB, Michael E, Ludwig N. The sarcolemmal calcium pump, α-1-syntrophin, and neuronal nitric-oxide synthase are parts of a macromolecular protein complex. J Biol Chem. 2006;281:23341–23348. doi: 10.1074/jbc.M513341200. [DOI] [PubMed] [Google Scholar]
- 135.Xu XZ, Choudhury A, Li X, Montell C. Coordination of an array of signaling proteins through homo- and heteromeric interactions between PDZ domains and target proteins. J Cell Biol. 1998;142:545–555. doi: 10.1083/jcb.142.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schuh K, Uldrijan S, Telkamp M, Rothlein N, Neyses L. The plasma membrane calmodulin-dependent calcium pump: a major regulator of nitric oxide synthase. J Cell Biol. 2001;155:201–205. doi: 10.1083/jcb.200104131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang Y, Oram JF. Unsaturated fatty acids inhibit cholesterol efflux from macrophages by increasing degradation of atp-binding cassette transporter a1. J Biol Chem. 2002;277:5692–5697. doi: 10.1074/jbc.M109977200. [DOI] [PubMed] [Google Scholar]
- 138.Attie AD, Kastelein JP, Hayden MR. Pivotal role of ABCA1 in reverse cholesterol transport influencing HDL levels and susceptibility to atherosclerosis. J Lipid Res. 2001;42:1717–1726. [PubMed] [Google Scholar]
- 139.Francis GA, Knopp RH, Oram JF. Defective removal of cellular cholesterol and phospholipids by apolipoprotein A-I in Tangier Disease. J Clin Investig. 1995;96:78–87. doi: 10.1172/JCI118082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Remaley AT, Schumacher UK, Stonik JA, Farsi BD, Nazih H, Brewer HB. Decreased reverse cholesterol transport from Tangier disease fibroblasts: acceptor specificity and effect of Brefeldin on lipid efflux. Arterioscler Thromb Vasc Biol. 1997;17:1813–1821. doi: 10.1161/01.ATV.17.9.1813. [DOI] [PubMed] [Google Scholar]
- 141.Takahashi Y, Smith JD. Cholesterol efflux to apolipoprotein AI involves endocytosis and resecretion in a calcium-dependent pathway. Proc Natl Acad Sci USA. 1999;96:11358–11363. doi: 10.1073/pnas.96.20.11358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Youichi M, Tomohiro O, Shinobu K, Akiko F, Kenya S, Michihiro I, Toshifumi Y, Shin’ichi T, Teruo A, Michinori M, Noriyuki K, Kazumitsu U. α-1-Syntrophin modulates turnover of ABCA1. J Biol Chem. 2004;279:15091–15095. doi: 10.1074/jbc.M313436200. [DOI] [PubMed] [Google Scholar]
- 143.Rigot V, Hamon Y, Chambenoit O, Aliber M, Duverger N, Chimini G. Distinct sites on ABCA1 control distinct steps required for cellular release of phospholipids. J Lipid Res. 2002;43:2077–2086. doi: 10.1194/jlr.M200279-JLR200. [DOI] [PubMed] [Google Scholar]
- 144.Wang N, Chen W, Linsel-Nitschke P, Martinez LO, Agerholm-Larsen B, Silver DL, Tall AR. A PEST sequence in ABCA1 regulates degradation by calpain protease and stabilization of ABCA1 by apoA-I. J Clin Investig. 2003;111:99–107. doi: 10.1172/JCI16808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Okuhira K, Fitzgerald ML, Sarracino DA, Manning JJ, Bell SA, Goss JL, Freeman MW. Purification of ATP-binding cassette transporter A1 and associated binding proteins reveals the importance of beta1-syntrophin in cholesterol efflux. J Biol Chem. 2005;280(47):39653–39664. doi: 10.1074/jbc.M510187200. [DOI] [PubMed] [Google Scholar]
- 146.Straub V, Campbell KP. Muscular dystrophies and the dystrophin glycoprotein complex. Curr Opin Neurol. 1997;10:168–175. doi: 10.1097/00019052-199704000-00016. [DOI] [PubMed] [Google Scholar]
- 147.Matsumura K, Tome FM, Collin H, Azibi K, Chaouch M, Kaplan JC, Fardeau M, Campbell KP. Deficiency of the 50K dystrophin associated glycoprotein in severe childhood autosomal recessive muscular dystrophy. Nature. 1992;359:320–322. doi: 10.1038/359320a0. [DOI] [PubMed] [Google Scholar]
- 148.Adams ME, Kramarcy N, Fukuda T, Engel AG, Sealock R, Froehner SC. Structural abnormalities at neuromuscular synapses lacking multiple syntrophin isoforms. J Neurosci. 2004;24:10302–10309. doi: 10.1523/JNEUROSCI.3408-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Compton AG, Cooper ST, Hill PM, Yang N, Froehner SC, North KN. The syntrophin-dystrobrevin sub-complex in human neuromuscular disorders. J Neuropathol Exp Neurol. 2005;64(4):350–361. doi: 10.1093/jnen/64.4.350. [DOI] [PubMed] [Google Scholar]
- 150.Kim MJ, Hwang SH, Lim JA, Froehner SC, Adams ME, Kim HS. α-Syntrophin modulates myogenin expression in differentiating myoblasts. PLoS ONE. 2010;5(12):e15355. doi: 10.1371/journal.pone.0015355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wakayama Y, Inoue, Kojima M, Jimi T, Yamashita S, Kumagai T, Shibuya S, Hara H, Oniki H. Altered alpha1-syntrophin expression in myofibers with Duchenne and Fukuyama muscular dystrophies. Histol Histopathol. 2006;21(1):23–34. doi: 10.14670/HH-21.23. [DOI] [PubMed] [Google Scholar]
- 152.Nakamori M, Kimura T, Kubota T, Matsumura T, Sumi H, Fujimura H, Takahashi MP, Sakoda S. Aberrantly spliced alpha-dystrobrevin alters alpha-syntrophin binding in myotonic dystrophy type 1. Neurology. 2008;70(9):677–685. doi: 10.1212/01.wnl.0000302174.08951.cf. [DOI] [PubMed] [Google Scholar]
- 153.Alderton JM, Steinhardt RA. Calcium influx through leak channels is responsible for the elevated levels of calcium-dependent proteolysis in dystrophic myotubes. J Biol Chem. 2000;275:9452–9460. doi: 10.1074/jbc.275.13.9452. [DOI] [PubMed] [Google Scholar]
- 154.Wu G, Ai T, Kim JJ, Mohapatra B, Xi Y, Li Z, Abbasi S, Purevjav E, Samani K, Ackerman MJ, Qi M, Moss AJ, Shimizu W, Towbin JA, Cheng J, Vatta M. Alpha-1-syntrophin mutation and the long-QT syndrome: a disease of sodium channel disruption. Circ Arrhythm Electrophysiol. 2008;1(3):193–201. doi: 10.1161/CIRCEP.108.769224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cheng J, Norstrand DW, Medeiros-Domingo A, Valdivia C, Tan B, Ye B, Kroboth S, Vatta M, Tester DJ, January CT, Makielski JC, Ackerman MJ. α1-Syntrophin mutations identified in sudden infant death syndrome cause an increase in late cardiac sodium current. Circ Arrhythm Electrophysiol. 2009;2:667–676. doi: 10.1161/CIRCEP.109.891440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lehnart SE, Ackerman MJ, Benson DW, Brugada R, Clancy CE, Donahue JK, George AL, Grant AO, Groft SC, January CT, Lathrop DA, Lederer WJ, Makielski JC, Mohler PJ, Moss A, Jeanne M, Olson TM, Przywara DA, Towbin JA, Wang LH, Marks AR. Inherited arrhythmias: A National Heart, Lung, and Blood Institute and Office of Rare Diseases workshop consensus report about the diagnosis, phenotyping, molecular mechanisms, and therapeutic approaches for primary cardiomyopathies of gene mutations affecting ion channel function. Circulation. 2007;116:2325–2345. doi: 10.1161/CIRCULATIONAHA.107.711689. [DOI] [PubMed] [Google Scholar]
- 157.Oceandy D, Cartwright EJ, Emerson M, Prehar S, Baudoin FM, Zi M, Alatwi N, Venetucci L, Schuh K, Judith CW, Armesilla AL, Neyses L. Neuronal nitric oxide synthase signaling in the heart is regulated by the sarcolemmal calcium pump 4b. Circulation. 2007;115:483–492. doi: 10.1161/CIRCULATIONAHA.106.643791. [DOI] [PubMed] [Google Scholar]
- 158.Wang Y, Kodani E, Wang J, Zhang SX, Takano H, Tang XL, Bolli R. Cardioprotection during the final stage of the late phase of ischemic preconditioning is mediated by neuronal NO synthase in concert with cyclooxygenase-2. Circ Res. 2004;95:84–91. doi: 10.1161/01.RES.0000133679.38825.a6. [DOI] [PubMed] [Google Scholar]
- 159.Ueda K, Valdivia C, Medeiros-Domingo A, David J, Tester DJ. Syntrophin mutation associated with long QT syndrome through activation of the nNOS–CN5A macromolecular complex. Proc Natl Acad Sci USA. 2008;105(27):9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Zimmet P, Alberti KG, Shaw J. Global and societal implications of diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 161.Diamond J. The double puzzle of diabetes. Nature. 2003;423:599–602. doi: 10.1038/423599a. [DOI] [PubMed] [Google Scholar]
- 162.Trajkovski M, Mziaut H, Schubert S, Kalaidzidis Y, Altkruger A, Solimena M. Regulation of insulin granule turnover in pancreatic β-cells by cleaved ICA512. J Biol Chem. 2008;283:33719–33729. doi: 10.1074/jbc.M804928200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ort T, Maksimova E, Dirkx R, Kachinsky AM, Berghs S, Froehner SC, Solimena M. The receptor tyrosine phosphatase-like protein ICA512 binds the PDZ domains of β2-syntrophin and nNOS in pancreatic β-cells. Eur J Cell Biol. 2000;79:621–630. doi: 10.1078/0171-9335-00095. [DOI] [PubMed] [Google Scholar]
- 164.Ort T, Voronov S, Guo J, Zawalich K, Freohner SC, Zawalich W, Solimena M. Dephosphorylation of β-2-syntrophin and Ca2+/μ-calpain-mediated cleavage of ICA512 upon stimulation of insulin secretion. EMBO J. 2001;20:4013–4023. doi: 10.1093/emboj/20.15.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Zhong H, Minneman KP. Alpha1-adrenoceptor subtypes. Eur J Pharmacol. 1999;375:261–276. doi: 10.1016/S0014-2999(99)00222-8. [DOI] [PubMed] [Google Scholar]
- 166.Hague C, Chen Z, Uberti M, Minneman KP. Alpha1-adrenergic receptor subtypes: non-identical triplets with different dancing partners? Life Sci. 2003;74(4):411–418. doi: 10.1016/j.lfs.2003.07.008. [DOI] [PubMed] [Google Scholar]
- 167.Zhongjian C, Chris H, Randy AH, Kenneth PM. Syntrophins regulate α_1D-adrenergic receptors through a PDZ domain-mediated interaction. J Biol Chem. 2006;281:12414–12420. doi: 10.1074/jbc.M508651200. [DOI] [PubMed] [Google Scholar]
- 168.McCloskey DT, Rokosh DG, O’Connell TD, Keung EC, Simpson PC, Baker AJ. α1-adrenoceptor subtypes mediate negative inotropy in myocardium from α1A/C-knockout and wild type mice. J Mol Cell Cardiol. 2002;34:1007–1017. doi: 10.1006/jmcc.2002.2049. [DOI] [PubMed] [Google Scholar]
- 169.Tanoue A, Nasa Y, Koshimizu T, Shinoura H, Oshikawa S, Kawai T, Sunada S, Takeo S, Tsujimoto G. The α1D-adrenergic receptor directly regulates arterial blood pressure via vasoconstriction. J Chromatogr A. 2002;109:765–775. doi: 10.1172/JCI14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tanoue A, Koba M, Miyawaki S, Koshimizu TA, Hosoda C, Oshikawa S, Tsujimoto G. Role of the α1D-adrenergic receptor in the development of salt-induced hypertension. Hypertension. 2002;40:101–106. doi: 10.1161/01.HYP.0000022062.70639.1C. [DOI] [PubMed] [Google Scholar]
- 171.Yamakawa H, Oyama S, Mitsuhashi H, Sasagawa N, Uchino S, Kohsaka S, Ishiura S. Neuroligins 3 and 4X interact with syntrophin-γ2 and the interactions are affected by autism-related mutations. Biochem Biophy Res Commun. 2007;355:41–46. doi: 10.1016/j.bbrc.2007.01.127. [DOI] [PubMed] [Google Scholar]
- 172.Ichtchenko K, Halta Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Sudhof TC. Neuroligin 1: a splice site-specific ligand for beta-neuroxins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- 173.Graf ER, Zhang X, Jin SX, Linhoff MW, Criag AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via Neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Dean C, Scholl FG, Choih J, DeMaria S, Berger J, Isacoff E, Scheiffele P. Neurexin mediates the assembly of presynaptic terminals. Nat Neurosci. 2003;6:708–716. doi: 10.1038/nn1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gilberg IC, Soderstrom H, Giros B, Leboyer M, Gilberg C, et al. Mutations of the X-linked genes encoding Neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Facciuto F, Cavatorta AL, Valdano MB, Marziali F, Gardiol D. Differential expression of PDZ domain containing proteins in human diseases-challenging topics and novel issues. FEBS J. 2012;279:3538–3548. doi: 10.1111/j.1742-4658.2012.08699.x. [DOI] [PubMed] [Google Scholar]
- 177.Pim D, Bergant M, Boon SS, Ganti K, Kranjec C, Massimi P, Subbaiah VK, Thomas M, Tomaic V, Banks L. Human papillomaviruses and the specificity of PDZ domain targeting. FEBS J. 2012;279:3530–3537. doi: 10.1111/j.1742-4658.2012.08709.x. [DOI] [PubMed] [Google Scholar]
- 178.Wiseman SM, Griffith O, Leung S, Masoudi H, Phang PT, Jones SJ, Hakama M (2008) Syntrophin expression predicts colon cancer outcome. Am Assoc Cancer Res
- 179.Carinci F, Lo Muzio L, Piattelli A, Rubini C, Chiesa F, Ionna F, Palmieri A, Maiorano E, Pastore A, Laino G, Dolci M, Pezzetti F. Potential markers of tongue tumor progression selected by cDNA microarray. Int J Immunopathol Pharmacol. 2005;18(3):513–524. doi: 10.1177/039463200501800311. [DOI] [PubMed] [Google Scholar]