Abstract
Gene fusion is a common mechanism of protein evolution that has mainly been discussed in the context of multidomain or symmetric proteins. Less is known about fusion of ancestral genes to produce small single-domain proteins. Here, we show with a domain-swapped mutant Plasmodium profilin that this small, globular, apparently single-domain protein consists of two foldons. The separation of binding sites for different protein ligands in the two halves suggests evolution via an ancient gene fusion event, analogous to the formation of multidomain proteins. Finally, the two fragments can be assembled together after expression as two separate gene products. The possibility to engineer both domain-swapped dimers and half-profilins that can be assembled back to a full profilin provides perspectives for engineering of novel protein folds, e.g., with different scaffolding functions.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-015-1932-0) contains supplementary material, which is available to authorized users.
Keywords: Actin-binding protein, Crystal structure, β-Hairpin, Modular, Protein interaction, Sequence analysis, Small-angle X-ray scattering
Introduction
Proteins have evolved during ~3.8 billion years to become complex and versatile molecular machines responsible for most life-supporting functions. Despite glimpses into the formation and evolution of certain protein fold families, the complicated processes of acquiring new protein folds are poorly understood. Most certainly, present proteins have formed from small and simple ancestors [1], and modular evolution of proteins from small fragments has been suggested already in connection with the discovery of introns in eukaryotic genes [2, 3]. Multimerization provides several structural and functional advantages, such as increased stability, possibilities for fine-tuning active site architectures and allosteric regulation, formation of larger binding surfaces, and simplified construction of large interaction networks [4–7]. Many of the reasons favoring oligomerization in the course of evolution are similarly applicable to both homo- and hetero-oligomers. However, assembly of complicated multimeric complexes is costly and error-prone. Thus, in many cases, oligomerization of functionally closely related entities has further led to gene duplication or fusion.
The formation of new proteins via gene duplication has been studied especially in the context of proteins of the triosephosphate isomerase (TIM) barrel superfamily. TIM barrel proteins consist of eight repeating α/β modules organized into a symmetric barrel and have likely formed via sequential gene duplication events [8–10]. Proteins catalyzing consecutive reactions in metabolic pathways can be products of gene duplication or fusion [11, 12]. In fact, fusion of non-identical genes is the most common evolutionary pathway for the formation of multidomain proteins [13–15]. It has been favored especially in the evolution of protein–protein interaction modules, providing the advantage of simplified assembly and topology of protein complexes [16]. As yet, there is little evidence for ancient gene fusion events in the case of small, asymmetric single-domain proteins.
Actin was long held to be a hallmark of eukaryotes, but upon the identification of bacterial actin homologs, it has emerged as a genuinely ancient protein [17, 18]. Actin has more interaction partners than any other eukaryotic protein, and its large filaments are a remarkable example of self-assembly involving a plethora of regulatory proteins [19]. These actin-binding proteins must have evolved hand in hand with the ability of actin to self-assemble [20], which has imposed the need for additional regulation. Accordingly, Apicomplexa, which have poorly polymerizing actins, harbor only few actin regulators [21].
Among the core set of actin-regulating proteins are profilins. They are small single-domain proteins that bind monomeric actin and proline-rich sequence motifs on opposite faces [22, 23]. Furthermore, profilin binding to protein ligands is regulated by membrane binding via polyphosphoinositides [24]. Most profilins are strictly monomeric, although tetramerization has been reported for at least human and plant profilins, and some functional relevance of these tetramers has been suggested [25, 26]. Profilins are evolutionarily widespread and present in not only all eukaryotes, but also in viruses and cyanobacteria, which probably acquired them via horizontal gene transfer from vertebrates [27, 28]. Despite the high degree of conservation at the level of 3D structure, the conservation of profilin primary sequences is remarkably low. This is expected for an ancient protein, whose structure is preserved for functional reasons and has had enough evolutionary time to explore a vast range of compatible sequences.
Apicomplexan parasites have a single profilin that has one of the most divergent structures within the protein family [29]. These organisms have presumably branched out from a common eukaryotic ancestor as early as the three kingdoms of animals, plants, and fungi [30, 31]. Here, we show that Plasmodium falciparum profilin consists of two independently folding units and suggest that evolution via fusion of two ancestral genes has led to the present profilin fold.
Materials and methods
Protein production and characterization
A synthetic, codon-optimized P. falciparum profilin gene (Mr. Gene) was used for the construction of three mutants, where residues 64–69 (Δ6), 62–71 (Δ10), and 59–75 (Δ17) were deleted. The Stratagene QuikChange Lightning kit was used for site-directed mutagenesis. The constructs were cloned into the pET-M11 vector, which contains an N-terminal 6xHis tag, followed by a tobacco etch virus (TEV) protease [53] cleavage site. For co-expression of the profilin halves, cDNA fragments encoding the N- and C-terminal halves of P. falciparum profilin [amino acids 1-67 (M1A…NG67) and 68-171 (T68K…SQ171), respectively] were cloned into the pETDuet vector (Novagen). The C-terminal fragment included an additional methionine as the first amino acid and a non-cleavable hexa-histidine tag after the last residue Gln171.
Wild-type profilin was expressed and purified as described [54]. All mutants were expressed in BL21(DE3) RIPL cells, cell pellets resuspended in lysis buffer [10 mM 2-amino-2-hydroxymethyl-propane-1,3-diol–HCl (Tris–HCl; pH 7.5), 300 mM NaCl, 5–10 % (v/v) glycerol], and lysed by sonication. The clarified supernatant was passed through a HisTrap Ni-affinity column (GE Healthcare), which was washed with the lysis buffer with 500 mM NaCl alone and supplemented with 10 and 25 mM imidazole. The cell pellets containing the half-profilin fragments were lysed and washed in the lysis buffer containing 20 mM imidazole. Finally, the His-tagged proteins were eluted with 300 mM imidazole in the lysis buffer. The affinity-purified wild-type, Δ6, Δ10, and Δ17 proteins were subjected to TEV digestion during an overnight dialysis and then passed through a HisTrap column to remove the His-tagged TEV protease and uncleaved protein. The proteins were then concentrated, and final purification was performed using size-exclusion chromatography (SEC) on a Superdex 75 16/60 column (GE Healthcare) in 10 mM Tris–HCl (pH 7.5) and 50 mM NaCl. SEC for the profilin halves was performed in 10 mM Tris–HCl (pH 7.5) and 100 mM NaCl. Peak fractions were pooled together, concentrated, and stored on ice.
To assess the role of oxidation on the oligomeric state, the wild-type profilin, Δ6, and Δ10 were incubated in the presence or absence of 10 mM dithiothreitol (DTT) on ice for 2 h and subsequently analyzed by analytical SEC using a Superdex 75 10/300 GL column (GE Healthcare) in 10 mM Tris–HCl (pH 7.5), 50 mM NaCl with or without 10 mM DTT.
Circular dichroism spectroscopy
The wild-type profilin, Δ6, and Δ10 were dialyzed in 10 mM Tris–HCl (pH 7.5), 50 mM NaF and diluted to 1.04, 0.86, and 1.40 mg ml−1, respectively. Synchrotron radiation (SR) circular dichroism (CD) spectra were recorded on the CD1 beamline at the ASTRID storage ring, ISA, Århus (Denmark) in the wavelength range of 180–280 nm in a 100-µm path length quartz cuvette. The DichroWeb server [55] was used for secondary structure determination using the CDSSTR [56] and SELCON3 [57] algorithms and the SP175 reference set optimized for 190–240 nm [58]. The Δ17 mutant and the half-profilin fragments were dialyzed in 10 mM phosphate buffer (pH 7.5) and 10 mM phosphate buffer (pH 7.5) with 150 mM NaF, respectively, and diluted to 0.05 and 0.04 mg ml−1, respectively. CD spectra were measured on an Applied Photophysics Chirascan spectropolarimeter for the final measurements in a 1-mm path length quartz cuvette.
Thermal denaturation curves were measured using an Applied Photophysics Chirascan Plus spectropolarimeter equipped with a thermal control unit (Quantum Northwest, TC125) and a direct temperature probe. Proteins were dialyzed into 10 mM Tris (pH 7.5), 10 mM NaF and diluted to 0.16 mg ml−1 (wild type) and 0.20 mg ml−1 (Δ6). CD spectra were recorded between 190 and 260 nm, using a quartz cuvette with a 0.5-mm path length, a temperature range of 20–80 °C, and a heating rate of 1 °C/min.
Small-angle X-ray scattering and light scattering
SAXS experiments were carried out on beamlines X33 at EMBL/DESY, Hamburg (Germany), and I911-4 at MAX-Lab, Lund (Sweden). The wild-type, Δ6, and Δ10 were concentrated to 1–5 mg ml−1, in either 25 mM sodium phosphate (pH 7.5) or 10 mM Tris–HCl (pH 7.5), 50 mM NaCl with or without 2 mM DTT. Analysis of the data was carried out using the ATSAS package [59]. Ab initio models were built using GASBOR [60]. Coupled rigid body and ab initio modeling of the dimers was done using the available crystal structures and BUNCH [61].
Static light scattering to determine the exact molecular mass of the SEC peak containing both profilin fragments was measured using a mini-DAWN TREOS multi-angle static light scattering detector (Wyatt Technology, Europe), coupled to a refractive index detector (Shodex), after separating the proteins over a Superdex S200 increase 10/300GL column (GE Healthcare), equilibrated with 10 mM Tris–HCl (pH 7.5) and 100 mM NaCl. The molecular mass was determined based on the measured light scattering and refractive index using the Astra v. 5.3.4 software (Wyatt Technology).
Crystallographic methods
The Δ6 mutant was crystallized at a concentration of 13 mg ml−1 in 1.8 M ammonium sulfate, 0.1 M 2-(N-morpholino)ethanesulfonic acid (pH 6) at room temperature. A 3.3-Å data set was collected from a single crystal on a PILATUS 6 M detector at the EMBL-Hamburg beamline P13 at PETRA III/DESY. The data were processed and scaled using the XDS package [62] and XDSi [63]. The wild-type profilin monomer (PDB code 2JKF; [29]) was used as a model for molecular replacement in PHASER [64]. The structure was refined using phenix.refine [65] to final R/Rfree factors of 0.240 and 0.263, respectively. The structure was validated using Molprobity [66]. Data collection and refinement statistics are shown in Table 1. All 3D structure diagrams were prepared using Pymol.
Table 1.
Data collection and refinement statistics
| Data collectiona | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 84.1, 246.6, 256.8 |
| Resolution (Å) | 20.0–3.30 (3.39–3.30) |
| No. of unique reflections | 154,448 (11,332) |
| Redundancy | 3.5 (3.4) |
| ‹I/σ(I)› | 8.1 (0.7) |
| R bmeas | 0.142 (2.066) |
| Completeness (%) | 99.6 (98.3) |
| CCc1/2 | 0.997 (0.365) |
| Refinementa | |
| Resolution (Å) | 20.0–3.30 (3.34–3.30) |
| R work/R free | 0.240/0.263 (0.389/0.453) |
| No. atoms | |
| Protein | 20,236 |
| Ligand/ion | 40 |
| Average B factors (Å2) | |
| Protein | 125 |
| Ligand/ion | 157 |
| Rms deviations | |
| Bond lengths (Å) | 0.004 |
| Bond angles (°) | 0.925 |
| Ramachandran plot (%) | |
| Most favored regions | 93.7 |
| Outliers | 0.6 |
Sequence analyses
Sequence searches were made against the non-redundant database using PSI-BLAST with three iterations, accepting hits with an e-value ≤10−9, then one round with e-values ≤10−6, and a last round with e-values ≤10−9 [67]. The full-length sequence for each database hit was retrieved, and these were re-aligned using MAFFT in its most accurate mode with up to 200 iterations [68]. Structure-based sequence alignments to 2JKF were used as hard restraints for the sequences from PDB entries 1A0K [69], 1CQA [70], 1F2K, 1PRQ [71], 1YPR [72], 3D9Y [73], 3LEQ, 3NEC [74]. These seeds were removed before calculations of conservation. Sequence entropy S at each site in the alignment was based on only those homologs with an e value less than 10−9 and calculated from where the summation runs over the 20 amino acid types and p i is the frequency of amino acid type i at the position. SALAMI [75] was used for structure searches and structural alignments.
Results
Dimerization of mutant Plasmodium profilins
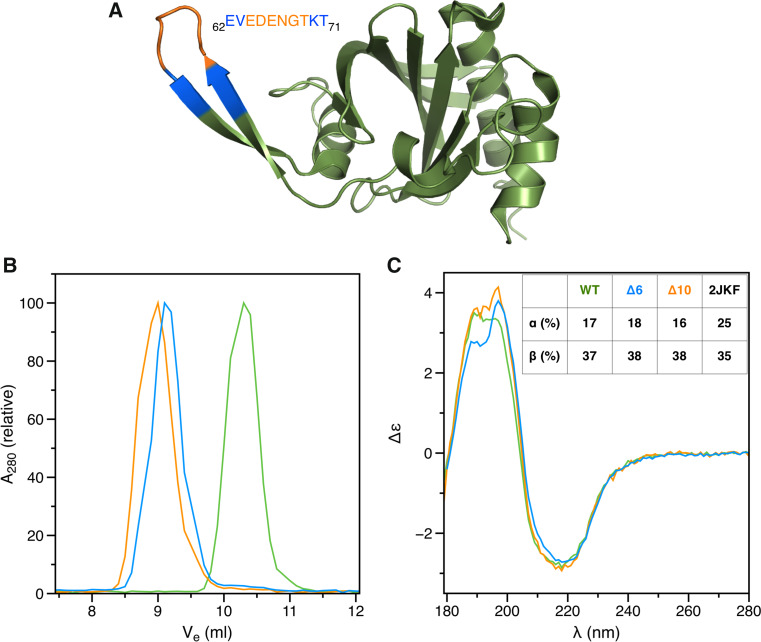
Despite low sequence similarity, P. falciparum profilin shares the overall fold with canonical profilins, containing a 7-stranded β sheet sandwiched between two α helices on each side (Fig. 1a). The most striking difference compared to higher eukaryotic profilins is a large β-hairpin extension, which we proposed participates in actin binding [29]. During the course of our work in characterizing the function of this motif, we created mutants that lack 6 (Δ6), 10 (Δ10), or 17 (Δ17) residues of the hairpin loop (Figs. 1a, S1A). Surprisingly, Δ6 and Δ10 form dimers, whereas Δ17 elutes in SEC as three peaks; one corresponding to a monomer, one to a dimer, and a third one in between the two (Figs. 1b, S1B). SRCD spectra indicated nearly identical secondary structure contents for the wild-type and the dimeric mutant proteins (Fig. 1c), while spectral features between 190 and 200 nm indicate minor differences between the structures. The middle peak of the Δ17 mutant is also folded with somewhat less β strands than the wild-type and the dimeric mutant proteins (Fig. S1C).
Fig. 1.
Dimerization of profilin mutants. a Structure of the wild-type P. falciparum profilin (2JKF; [29]). Residues deleted in the different mutants are indicated with colors: orange Δ6; blue Δ10. b Size-exclusion chromatogram showing dimerization of the deletion mutants (green wild-type profilin; orange Δ6; blue Δ10). c SRCD spectra of the wild-type and mutant profilins (colors as above). Secondary structure contents calculated from the spectra using DichroWeb and as in the wild-type crystal structure (2JKF) are shown in the inset
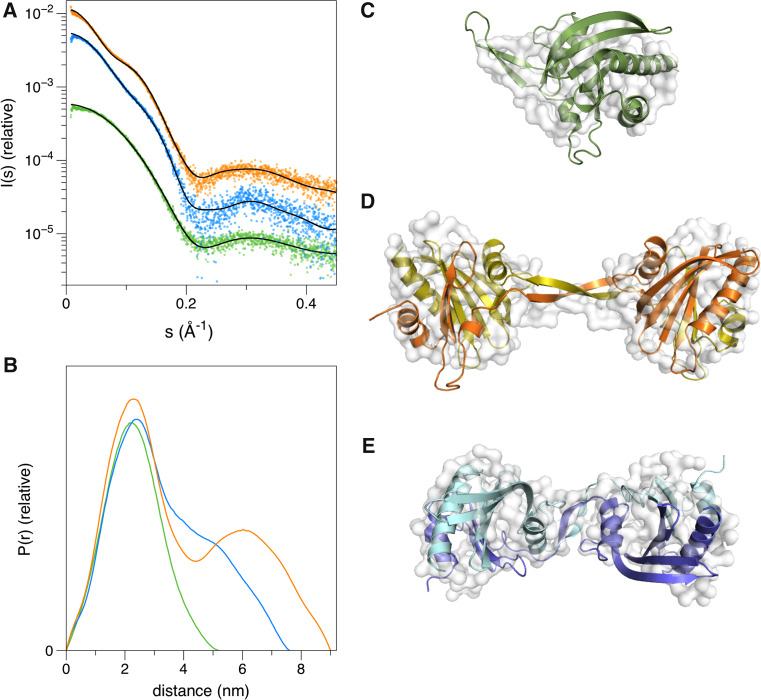
Small-angle X-ray scattering (SAXS) was further used to analyze the shape of the two dimers (Δ6 and Δ10) in solution (Fig. 2). Whereas the monomeric wild-type profilin has a globular shape similar to the crystal structure, Δ6 and Δ10 form elongated, dumbbell- or peanut-shaped structures that seem to have only a small contact interface between the monomers (Fig. 2). The Δ6 variant is more elongated than the Δ10 dimer, with a larger separation between the globular domains. Up to 10 mM DTT had no effect on the dimerization (Fig. S1D), suggesting that oligomerization does not occur via disulfide bridge formation due to exposure of the single buried cysteine residue (Fig. S1A).
Fig. 2.
Solution structures of the wild-type and mutant profilins. a SAXS curves and fits of the crystal structures (wild-type, Δ6)/model (Δ10) to the data. b Distance distribution curves. c–e SAXS models (white surface) calculated using GASBOR and fits of the crystal structures/model (colored cartoon representation) in them. Coloring in all panels follows that of Fig. 1
Dimerization occurs via domain swapping
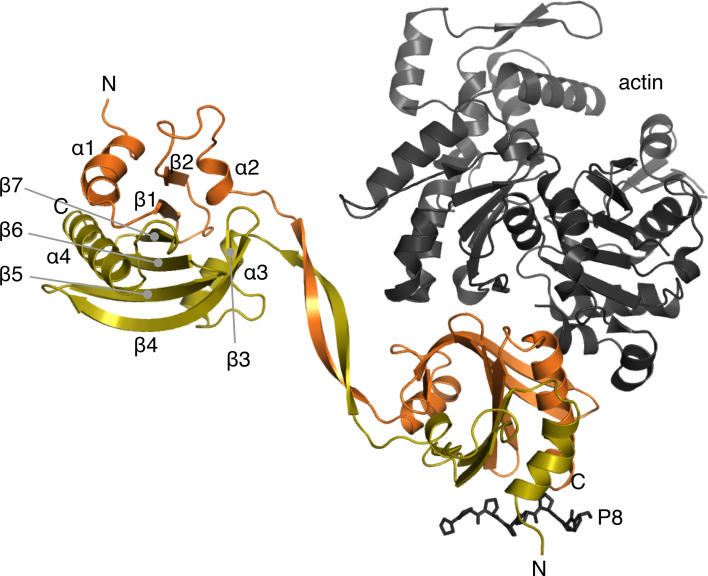
Intrigued by the mutagenesis-induced homodimerization of this normally strictly monomeric protein and the peculiar shape of the dimers in solution, we determined the crystal structure of the Δ6 mutant. The protein crystallized with 8 dimers in the asymmetric unit. Dimerization occurs via domain swapping at the β-hairpin extension, such that the N-terminal half of one monomer combines with the C-terminal half of a second one (Fig. 3). Hence, the dimer is formed of two globular domains composed of two polypeptide chains, connected by a 2-stranded β sheet bridge. The crystal structure of the Δ6 mutant fits well to the SAXS data, explaining the tight, reducing agent insensitive dimers with seemingly small inter-subunit contact areas (Fig. 2). Modeling of the Δ10 mutant based on the SAXS data indicates a similar arrangement, with a shorter linker between the globular domains. The structure of the globular domain formed through this domain swapping is, with an rmsd 0.8–1.0 Å, essentially indistinguishable from that of the wild-type monomeric profilin (Fig. 3).
Fig. 3.
Dimerization mode of the domain-swapped Δ6 mutant. The two chains are colored orange and yellow. The secondary structure elements of the core profilin fold are labeled in the upper left subunit. The N and C termini are labeled in both subunits. Actin and an octa-proline peptide are depicted on the lower right subunit as they are bound to a canonical profilin (PDB code: 2PAV [76]) and the wild-type P. falciparum profilin (PDB code: 2JKG [29]), respectively
The dimeric assembly highlights a feature that has passed unnoticed in previous comparisons of different profilin structures: Plasmodium profilin consists of two subdomains, of which the N-terminal one contains the proline-rich peptide-binding site and the C-terminal one the expected actin-binding site (Fig. 3). The central 7-stranded β sheet is assembled such that β7 contacts β1, and the N- and C-terminal subdomains both have a linear topology (Fig. 3). Such assembly suggests that this small, single-domain protein initially folds as two independent units that are only subsequently put together to form the full 3D fold. Thus, the intermediate form of the Δ17 mutant that elutes in SEC in between the monomer and dimer may represent an ‘open monomer’, which due to its elongated shape and longer hydrodynamic radius elutes earlier than the globular monomer of the same molecular weight (Fig. S1).
Plasmodium profilin unfolds in multiple steps
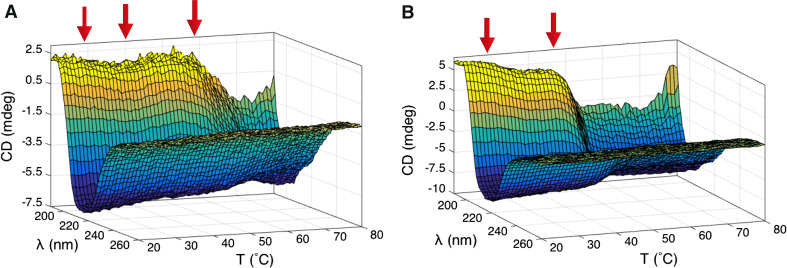
We used CD spectroscopy to study thermal unfolding of the wild-type P. falciparum profilin and the Δ6 mutant (Fig. 4). Upon heating, the wild-type protein unfolds via a 3-step pathway with two small transitions at 29 and 40 °C, involving only minor changes in the secondary structure contents, and a final unfolding step at 58 °C. The dimeric mutant profilin has a lower overall thermal stability and shares the first transition at 29 °C with the wild-type protein. However, instead of the two latter steps, there is only one major unfolding step at 45 °C.
Fig. 4.
Wild-type and mutant Plasmodium profilins unfold in multiple steps. a A thermal unfolding plot of the wild-type Plasmodium profilin shows three distinct transitions (indicated by red arrows) at 29, 40, and 58 °C. b The Δ6 mutant unfolds in two steps with transitions at 29 and 45 °C
Sequence conservation of the half-profilins
Profilin is an abundant protein, and many eukaryotes have several profilin isoforms. Even with a conservative database search, thousands of related sequences, spanning the whole spectrum of eukaryotic organisms, are found. For conservation calculations, the 828 nearest homologs were used. Because of the large insertions in the apicomplexan sequences, we compared only residues at positions occupied in ≥95 % of the sequences (Fig. 5a). Overall, there is no large difference in the conservation of each of the subdomains. However, whereas a naïve BLAST search with the full-length P. falciparum profilin sequence immediately finds 76 related proteins, nearly all of them from apicomplexan parasites, and a search with the C-terminal region (residues 75–173) gives almost the same results, the same search with the N-terminal 39 residues only identifies sequences from Plasmodium spp. and one sequence from Sarcocystis neurona, also an apicomplexan parasite. A search with the N-terminal part of a canonical profilin, e.g., human profilin 1, results in a wide range of other different profilins, and searching with the N terminus of Toxoplasma profilin also identifies a number of other apicomplexan profilins, excluding Plasmodium spp. Thus, based on the N terminus, there is a striking lack of sequence connection from Plasmodium profilin to other known proteins, even within the same phylum.
Fig. 5.
Conservation and gene structure. a Conservation within 828 profilin sequences. P. falciparum profilin residue numbers are marked on the X axis. Residue entropy (S) on the Y axis (black dots) was only calculated at sites present in >95 % of the sequences. S = 0 means 100 % conservation; higher S means more variation. The dots are connected with a thin black line to guide the eye. The orange horizontal line shows the mean residue entropy (0.40 for the N-terminal and 0.43 for the C-terminal fragment). The secondary structure elements of P. falciparum profilin are depicted as orange and black bars below the residue numbers. b Gene structure of Plasmodium profilin (PfPfn) and human profilin 4 (HsPfn4) in relation to the protein structure. Exons 1–4 of the Plasmodium profilin gene are shown as bars with different colors, and their lengths are in relation to the protein sequence in panel a. The protein segments encoded by the exons are colored accordingly in the 3D structure presentation of the Plasmodium protein. In the human profilin 4 gene, exon 1 corresponds to exons 1 and 2 in the Plasmodium profilin gene and encodes the entire N-terminal segment (black), while the C-terminal fragment (light gray) is encoded by 3, instead of 2, exons
Exon/intron boundaries coincide with the profilin subdomains
The gene structure of several profilins has been analyzed before [27], but not in relation to the corresponding protein structures. Vertebrate profilins 1–3 have likely diverged from a common profilin 1-like ancestor from sea urchin [27]. Profilin 4 is an unconventional family member that is thought to be the most ancient of the present animal profilins and, therefore, closest to a common ancestor. Both human profilin 4 and P. falciparum profilin are encoded by four exons (Fig. 5b). Interestingly, the segment encoded by the second exon in P. falciparum profilin ends at the tip of the β-hairpin extension, separating the two subdomains. In human profilin 4, the first exon codes for the entire N-terminal fragment, ending exactly where the β-hairpin insertion in apicomplexan profilins would start. Most profilin genes contain fewer exons/introns, and have likely lost introns during evolution, as has been suggested for the gene encoding profilin 3 in mammals and birds [27]. Taken further, the region encoded by exons 1 and 2 in P. falciparum profilin (α1β1/β2α2) could be a result of duplication and permutation of an αβ fragment.
Half-profilins can be co-expressed and assembled in vivo
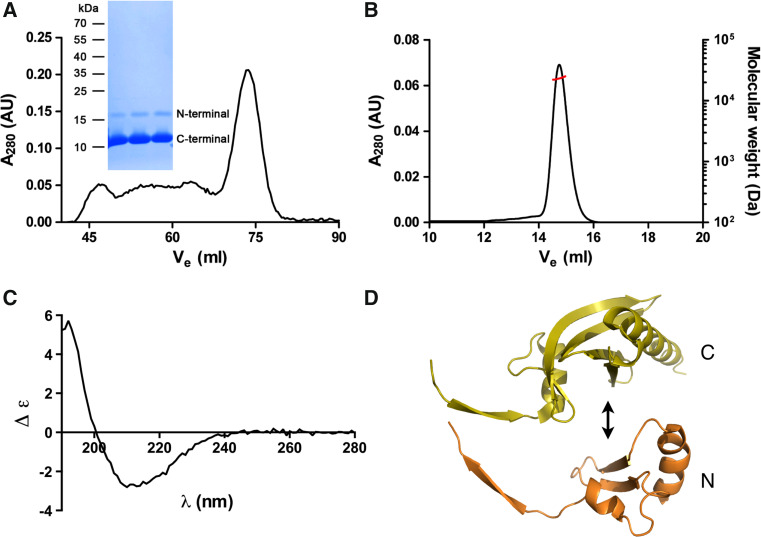
To test whether the two subdomains could be assembled together when expressed as separate gene products, we cloned both fragments and co-expressed them in bacteria. The fragments were expressed in soluble form and eluted in SEC together as a symmetric peak corresponding to the elution volume of the wild-type profilin monomer (Fig. 6a). Static light scattering gave a mass of 23.3 ± 0.2 kDa for the peak, which is close to the calculated molecular mass (20.0 kDa) of a complex of the two fragments (Fig. 6b). Both fragments were also identified from the sample by mass spectrometry after SDS-PAGE. CD spectroscopy confirms that the two profilin fragments are folded with a slightly lower β-strand content than the wild-type protein (Fig. 6c). The reduced fraction of β strands probably reflects absent β-hairpin interactions due to the cutting of the protein at the tip of the Apicomplexa-specific insertion and this motif turning into mostly unstructured tails. Thus, the two separate half-profilin fragments, indeed, seem to assemble together into a dimer that resembles the normal profilin fold (Fig. 6d).
Fig. 6.
Purification of half-profilins. a The co-expressed profilin fragments co-elute from SEC in a single symmetric peak, which corresponds to the full-length wild-type Plasmodium profilin. Both N- and C-terminal profilin fragments were identified by mass spectrometry from a Coomassie-stained SDS polyacrylamide gel (inset) of the SEC peak fraction. b Static light scattering gives a molecular weight of 23 kDa for the complex of the profilin fragments. The left Y axis and the black line show the UV absorbance at 280 nm and the right Y axis the molecular weight (in Da) from the light scattering signal (red line). c CD spectrum of the complex of the two profilin fragments. d Putative assembly of the two profilin fragments. It seems likely that the long β extensions of each half (on the left) adopt a more disordered conformation
Discussion
Domain swapping, even when an artifact caused by extreme conditions or mutations, can tell one about the behavior of proteins in their natural state and environment, e.g., by revealing folding intermediates or independently folding units, also termed foldons [32–36]. Here, domain-swapped dimers indicate that the conserved, globular, seemingly single-domain profilin fold is actually composed of two foldons that are assembled into a complete profilin also when co-expressed as separate gene products. Simple topologies that enable fast and efficient folding and minimize the possibility of misfolding have been suggested to be a result of evolutionary optimization [37]. In profilins, the two β strands of the N-terminal and the five strands of the C-terminal subdomain have such simple topologies (Fig. 3) and seem to, indeed, fold separately before being assembled together. Unfolding happens in 2–3 steps. The acid-induced unfolding pathway of human platelet profilin also includes a stable intermediate that has been proposed to represent a physiological state relevant for the release of both actin and proline-rich ligands [38].
Proteins capable of domain swapping may be predisposed to evolving toward oligomers [39]. Conversely, we can ask: Are proteins that have evolved by fusion of two proteins likely to display domain swapping under certain conditions or due to small changes in the polypeptide chain? In this case, most likely the loss of a glycine at a tight turn leads to the observed domain swapping. The hairpin has to be flexible enough to allow for the chain to fold back during the assembly of the monomer. When this does not happen, the halves seek the closest partners from neighboring monomers. For example, the evolutionary loss of a conserved glycine residue in the PDZ domains of the giant scaffolding proteins periaxin and AHNAK2 is linked to the formation of highly intertwined, domain-swapped dimers [40]. In general, domain-swapped proteins tend to oligomerize, when hinge loops are shortened [7, 41]. On the other hand, making such connecting loops longer may generate monomers instead of domain-swapped dimers [7, 42].
Relating protein folding happening on a ms–s timescale to evolution taking place during billions of years prompted us to question the early origin of profilins. Do the folding intermediates represent smaller ancestral proteins that have been fused to form the currently widespread and structurally conserved eukaryotic profilin family? Proteins must have evolved from smaller, simple ancestral units. Have the two foldons seen in Plasmodium profilin once been present as two separate primitive proteins: one harboring an actin-binding site and a second one that could bind to other (proline-rich) regulatory partners? Assembly of large complexes with multiple pieces contains several error-prone steps. Therefore, permanent fusion of partners that work as common ‘hubs’ in multiprotein complexes has been favored during evolution [16]. Profilin can be seen as one such hub, connecting actin monomers to multiple regulatory proteins and networks, as well as to membranes. The actin-binding site in profilins involves residues from the C-terminal subdomain only. The N-terminal subdomain harbors most of the proline-rich peptide-binding residues. However, the C-terminal helix also participates in peptide binding in canonical profilins [22, 43]. This is not the case for Plasmodium profilin, which binds to octa-proline solely via its N-terminal subdomain [29]. The involvement of the C terminus in peptide binding may, thus, be a later adaptation to different regulatory proteins requiring higher affinity interfaces. Curiously, the supposedly ancestral mammalian profilin 4 binds neither actin nor proline-rich sequences. It is not clear whether it has never had these properties or whether they were lost at some point in time.
Profilins are remarkably widespread proteins, spanning all branches of the eukaryotic tree of life. In addition, considering close structural homologs, the profilin fold family would reach bacteria, with, e.g., the roadblock proteins from Streptomyces, and cover a vast variety of tasks from scaffolding to enzymatic functions [44]. Therefore, if a fusion event is responsible for the modern profilin protein fold, it must have been a truly ancient event—or similar recombinations must have happened independently in different branches. Despite the high level of structural similarity, it seems from the sequence data that the N-terminal region of Plasmodium profilin may have an unusual evolutionary history that does not fit into a simple model of mutations with infrequent insertions. Rather, it has suffered large insertions and deletions.
While it has become clear that extensive building of new protein folds has occurred in Nature via duplication and recombination of simple, stable fragments, our ability to engineer functional protein folds de novo from small building blocks is still limited [45–48]. The feasibility of engineering novel proteins and activities by recombination of homologous fragments within the same fold family has been demonstrated [49, 50]. However, only the first steps have been taken in combining fragments from different, but still related, folds to create new functional proteins [51, 52]. Here, we demonstrate that also small, single-domain, asymmetric proteins can be split and engineered to adopt different topologies. This provides the possibility to create new scaffolding proteins and combine these with, e.g., enzymatic activities.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Moon Chatterjee and Esa-Pekka Kumpula for help with SRCD and SAXS data collection and the beamline staff at the CD1 beamline on ASTRID/ISA, Århus, the I911-4 beamline at MAX-Lab, Lund, and the EMBL-Hamburg beamlines X33 at DORIS and P13 at PETRA III, DESY, Hamburg, for excellent user support. The atomic coordinates and structure factor amplitudes have been submitted to the Protein Data Bank under the accession code 4D60. This work was supported by the Academy of Finland, the Sigrid Jusélius Foundation, the Emil Aaltonen Foundation, and the German Ministry of Education and Research.
Abbreviations
- Tris
2-amino-2-hydroxymethyl-propane-1,3-diol
- CD
Circular dichroism
- DTT
Dithiothreitol
- SEC
Size-exclusion chromatography
- SAXS
Small-angle X-ray scattering
- SR
Synchrotron radiation
- TEV
Tobacco etch virus
- TIM
Triosephosphate isomerase
Footnotes
S. P. Bhargav and J. Vahokoski contributed equally.
References
- 1.Soding J, Lupas AN. More than the sum of their parts: on the evolution of proteins from peptides. BioEssays. 2003;25:837–846. doi: 10.1002/bies.10321. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 3.Blake CCF. Do genes-in-pieces imply proteins-in-pieces? Nature. 1978;273:267. doi: 10.1038/273267a0. [DOI] [Google Scholar]
- 4.Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem Sci. 2004;29:618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 5.Ispolatov I, Yuryev A, Mazo I, Maslov S. Binding properties and evolution of homodimers in protein–protein interaction networks. Nucleic Acids Res. 2005;33:3629–3635. doi: 10.1093/nar/gki678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jee J, Byeon IL, Louis JM, Gronenborn AM. The point mutation A34F causes dimerization of GB1. Proteins Struct Funct Bioinf. 2008;71:1420–1431. doi: 10.1002/prot.21831. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Eisenberg D. 3D domain swapping: as domains continue to swap. Protein Sci. 2002;11:1285–1299. doi: 10.1110/ps.0201402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lang D, Thoma R, Henn-Sax M, Sterner R, Wilmanns M. Structural evidence for evolution of the beta/alpha barrel scaffold by gene duplication and fusion. Science. 2000;289:1546–1550. doi: 10.1126/science.289.5484.1546. [DOI] [PubMed] [Google Scholar]
- 9.Hocker B, Beismann-Driemeyer S, Hettwer S, Lustig A, Sterner R. Dissection of a (betaalpha)8-barrel enzyme into two folded halves. Nat Struct Biol. 2001;8:32–36. doi: 10.1038/83021. [DOI] [PubMed] [Google Scholar]
- 10.Richter M, Bosnali M, Carstensen L, Seitz T, Durchschlag H, Blanquart S, Merkl R, Sterner R. Computational and experimental evidence for the evolution of a (beta alpha)8-barrel protein from an ancestral quarter-barrel stabilised by disulfide bonds. J Mol Biol. 2010;398:763–773. doi: 10.1016/j.jmb.2010.03.057. [DOI] [PubMed] [Google Scholar]
- 11.Horowitz NH. On the evolution of biochemical syntheses. Proc Natl Acad Sci USA. 1945;31:153–157. doi: 10.1073/pnas.31.6.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.List F, Sterner R, Wilmanns M. Related (betaalpha)8-barrel proteins in histidine and tryptophan biosynthesis: a paradigm to study enzyme evolution. ChemBioChem. 2011;12:1487–1494. doi: 10.1002/cbic.201100082. [DOI] [PubMed] [Google Scholar]
- 13.Buljan M, Frankish A, Bateman A. Quantifying the mechanisms of domain gain in animal proteins. Genome Biol. 2010;11:R74. doi: 10.1186/gb-2010-11-7-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marsh JA, Teichmann SA. How do proteins gain new domains? Genome Biol. 2010;11:126. doi: 10.1186/gb-2010-11-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasek S, Risler JL, Brezellec P. Gene fusion/fission is a major contributor to evolution of multi-domain bacterial proteins. Bioinformatics. 2006;22:1418–1423. doi: 10.1093/bioinformatics/btl135. [DOI] [PubMed] [Google Scholar]
- 16.Marsh JA, Hernandez H, Hall Z, Ahnert SE, Perica T, Robinson CV, Teichmann SA. Protein complexes are under evolutionary selection to assemble via ordered pathways. Cell. 2013;153:461–470. doi: 10.1016/j.cell.2013.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ettema TJ, Lindas AC, Bernander R. An actin-based cytoskeleton in archaea. Mol Microbiol. 2011;80:1052–1061. doi: 10.1111/j.1365-2958.2011.07635.x. [DOI] [PubMed] [Google Scholar]
- 18.van den Ent F, Amos LA, Lowe J. Prokaryotic origin of the actin cytoskeleton. Nature. 2001;413:39–44. doi: 10.1038/35092500. [DOI] [PubMed] [Google Scholar]
- 19.Dominguez R. Actin-binding proteins—a unifying hypothesis. Trends Biochem Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Pollard TD. Actin-binding protein evolution. Nature. 1984;312:403. doi: 10.1038/312403a0. [DOI] [PubMed] [Google Scholar]
- 21.Sattler JM, Ganter M, Hliscs M, Matuschewski K, Schuler H. Actin regulation in the malaria parasite. Eur J Cell Biol. 2011;90:966–971. doi: 10.1016/j.ejcb.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Metzler WJ, Bell AJ, Ernst E, Lavoie TB, Mueller L. Identification of the poly-l-proline-binding site on human profilin. J Biol Chem. 1994;269:4620–4625. [PubMed] [Google Scholar]
- 23.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin–beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 24.Sohn RH, Chen J, Koblan KS, Bray PF, Goldschmidt-Clermont PJ. Localization of a binding site for phosphatidylinositol 4,5-bisphosphate on human profilin. J Biol Chem. 1995;270:21114–21120. doi: 10.1074/jbc.270.36.21114. [DOI] [PubMed] [Google Scholar]
- 25.Babich M, Foti LRP, Sykaluk LL, Clark CR. Profilin forms tetramers that bind to G-actin. Biochem Biophys Res Commun. 1996;218:125–131. doi: 10.1006/bbrc.1996.0022. [DOI] [PubMed] [Google Scholar]
- 26.Korupolu RV, Achary MS, Aneesa F, Sathish K, Wasia R, Sairam M, Nagarajaram HA, Singh SS. Profilin oligomerization and its effect on poly (l-proline) binding and phosphorylation. Int J Biol Macromol. 2009;45:265–273. doi: 10.1016/j.ijbiomac.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Polet D, Lambrechts A, Vandepoele K, Vandekerckhove J, Ampe C. On the origin and evolution of vertebrate and viral profilins. FEBS Lett. 2007;581:211–217. doi: 10.1016/j.febslet.2006.12.013. [DOI] [PubMed] [Google Scholar]
- 28.Guljamow A, Jenke-Kodama H, Saumweber H, Quillardet P, Frangeul L, Castets AM, Bouchier C, de Tandeau Marsac N, Dittmann E. Horizontal gene transfer of two cytoskeletal elements from a eukaryote to a cyanobacterium. Curr Biol. 2007;17:R757–R759. doi: 10.1016/j.cub.2007.06.063. [DOI] [PubMed] [Google Scholar]
- 29.Kursula I, Kursula P, Ganter M, Panjikar S, Matuschewski K, Schuler H. Structural basis for parasite-specific functions of the divergent profilin of Plasmodium falciparum. Structure. 2008;16:1638–1648. doi: 10.1016/j.str.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Douzery EJ, Snell EA, Bapteste E, Delsuc F, Philippe H. The timing of eukaryotic evolution: does a relaxed molecular clock reconcile proteins and fossils? Proc Natl Acad Sci USA. 2004;101:15386–15391. doi: 10.1073/pnas.0403984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Escalante AA, Ayala FJ. Evolutionary origin of Plasmodium and other Apicomplexa based on rRNA genes. Proc Natl Acad Sci USA. 1995;92:5793–5797. doi: 10.1073/pnas.92.13.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fersht AR. Nucleation mechanisms in protein folding. Curr Opin Struct Biol. 1997;7:3–9. doi: 10.1016/S0959-440X(97)80002-4. [DOI] [PubMed] [Google Scholar]
- 33.Hayes MV, Sessions RB, Brady RL, Clarke AR. Engineered assembly of intertwined oligomers of an immunoglobulin chain. J Mol Biol. 1999;285:1857–1867. doi: 10.1006/jmbi.1998.2415. [DOI] [PubMed] [Google Scholar]
- 34.Murray AJ, Head JG, Barker JJ, Brady RL. Engineering an intertwined form of CD2 for stability and assembly. Nat Struct Biol. 1998;5:778–782. doi: 10.1038/1816. [DOI] [PubMed] [Google Scholar]
- 35.Newcomer ME. Protein folding and three-dimensional domain swapping: a strained relationship? Curr Opin Struct Biol. 2002;12:48–53. doi: 10.1016/S0959-440X(02)00288-9. [DOI] [PubMed] [Google Scholar]
- 36.Zegers I, Deswarte J, Wyns L. Trimeric domain-swapped barnase. Proc Natl Acad Sci USA. 1999;96:818–822. doi: 10.1073/pnas.96.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Debes C, Wang M, Caetano-Anolles G, Grater F. Evolutionary optimization of protein folding. PLoS Comput Biol. 2013;9:e1002861. doi: 10.1371/journal.pcbi.1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLachlan GD, Cahill SM, Girvin ME, Almo SC. Acid-induced equilibrium folding intermediate of human platelet profilin. Biochemistry. 2007;46:6931–6943. doi: 10.1021/bi0602359. [DOI] [PubMed] [Google Scholar]
- 39.Canals A, Pous J, Guasch A, Benito A, Ribo M, Vilanova M, Coll M. The structure of an engineered domain-swapped ribonuclease dimer and its implications for the evolution of proteins toward oligomerization. Structure. 2001;9:967–976. doi: 10.1016/S0969-2126(01)00659-1. [DOI] [PubMed] [Google Scholar]
- 40.Han H, Kursula P. Periaxin and AHNAK nucleoprotein 2 form intertwined homodimers through domain swapping. J Biol Chem. 2014;289:14121–14131. doi: 10.1074/jbc.M114.554816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Green SM, Gittis AG, Meeker AK, Lattman EE. One-step evolution of a dimer from a monomeric protein. Nat Struct Biol. 1995;2:746–751. doi: 10.1038/nsb0995-746. [DOI] [PubMed] [Google Scholar]
- 42.Albright RA, Mossing MC, Matthews BW. High-resolution structure of an engineered Cro monomer shows changes in conformation relative to the native dimer. Biochemistry. 1996;35:735–742. doi: 10.1021/bi951958n. [DOI] [PubMed] [Google Scholar]
- 43.Kursula P, Kursula I, Massimi M, Song YH, Downer J, Stanley WA, Witke W, Wilmanns M. High-resolution structural analysis of mammalian profilin 2a complex formation with two physiological ligands: the formin homology 1 domain of mDia1 and the proline-rich domain of VASP. J Mol Biol. 2008;375:270–290. doi: 10.1016/j.jmb.2007.10.050. [DOI] [PubMed] [Google Scholar]
- 44.Aravind L, Mazumder R, Vasudevan S, Koonin EV. Trends in protein evolution inferred from sequence and structure analysis. Curr Opin Struct Biol. 2002;12:392–399. doi: 10.1016/S0959-440X(02)00334-2. [DOI] [PubMed] [Google Scholar]
- 45.Hocker B. Design of proteins from smaller fragments—learning from evolution. Curr Opin Struct Biol. 2014;27C:56–62. doi: 10.1016/j.sbi.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 46.Riechmann L, Winter G. Novel folded protein domains generated by combinatorial shuffling of polypeptide segments. Proc Natl Acad Sci USA. 2000;97:10068–10073. doi: 10.1073/pnas.170145497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riechmann L, Winter G. Early protein evolution: building domains from ligand-binding polypeptide segments. J Mol Biol. 2006;363:460–468. doi: 10.1016/j.jmb.2006.08.031. [DOI] [PubMed] [Google Scholar]
- 48.Zhang D, Iyer LM, Burroughs AM, Aravind L. Resilience of biochemical activity in protein domains in the face of structural divergence. Curr Opin Struct Biol. 2014;26:92–103. doi: 10.1016/j.sbi.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Claren J, Malisi C, Hocker B, Sterner R. Establishing wild-type levels of catalytic activity on natural and artificial (beta alpha)8-barrel protein scaffolds. Proc Natl Acad Sci USA. 2009;106:3704–3709. doi: 10.1073/pnas.0810342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hocker B, Claren J, Sterner R. Mimicking enzyme evolution by generating new (betaalpha)8-barrels from (betaalpha)4-half-barrels. Proc Natl Acad Sci USA. 2004;101:16448–16453. doi: 10.1073/pnas.0405832101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eisenbeis S, Proffitt W, Coles M, Truffault V, Shanmugaratnam S, Meiler J, Hocker B. Potential of fragment recombination for rational design of proteins. J Am Chem Soc. 2012;134:4019–4022. doi: 10.1021/ja211657k. [DOI] [PubMed] [Google Scholar]
- 52.Shanmugaratnam S, Eisenbeis S, Hocker B. A highly stable protein chimera built from fragments of different folds. Protein Eng Des Sel. 2012;25:699–703. doi: 10.1093/protein/gzs074. [DOI] [PubMed] [Google Scholar]
- 53.van den Berg S, Lofdahl PA, Hard T, Berglund H. Improved solubility of TEV protease by directed evolution. J Biotechnol. 2006;121:291–298. doi: 10.1016/j.jbiotec.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 54.Ignatev A, Bhargav SP, Vahokoski J, Kursula P, Kursula I. The lasso segment is required for functional dimerization of the Plasmodium formin 1 FH2 domain. PLoS One. 2012;7:e33586. doi: 10.1371/journal.pone.0033586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lobley A, Whitmore L, Wallace BA. DICHROWEB: an interactive website for the analysis of protein secondary structure from circular dichroism spectra. Bioinformatics. 2002;18:211–212. doi: 10.1093/bioinformatics/18.1.211. [DOI] [PubMed] [Google Scholar]
- 56.Compton LA, Johnson WCJ. Analysis of protein circular dichroism spectra for secondary structure using a simple matrix multiplication. Anal Biochem. 1986;155:155–167. doi: 10.1016/0003-2697(86)90241-1. [DOI] [PubMed] [Google Scholar]
- 57.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 58.Lees JG, Miles AJ, Wien F, Wallace BA. A reference database for circular dichroism spectroscopy covering fold and secondary structure space. Bioinformatics. 2006;22:1955–1962. doi: 10.1093/bioinformatics/btl327. [DOI] [PubMed] [Google Scholar]
- 59.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889806004699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kursula P. XDSi: a graphical interface for the data processing program XDS. J Appl Crystallogr. 2004;37:347–348. doi: 10.1107/S0021889804000858. [DOI] [Google Scholar]
- 64.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thorn KS, Christensen HE, Shigeta R, Huddler D, Shalaby L, Lindberg U, Chua NH, Schutt CE. The crystal structure of a major allergen from plants. Structure. 1997;5:19–32. doi: 10.1016/S0969-2126(97)00163-9. [DOI] [PubMed] [Google Scholar]
- 70.Fedorov AA, Ball T, Mahoney NM, Valenta R, Almo SC. The molecular basis for allergen cross-reactivity: crystal structure and IgE-epitope mapping of birch pollen profilin. Structure. 1997;5:33–45. doi: 10.1016/S0969-2126(97)00164-0. [DOI] [PubMed] [Google Scholar]
- 71.Liu S, Fedorov AA, Pollard TD, Lattman EE, Almo SC, Magnus KA. Crystal packing induces a conformational change in profilin-I from Acanthamoeba castellanii. J Struct Biol. 1998;123:22–29. doi: 10.1006/jsbi.1998.4009. [DOI] [PubMed] [Google Scholar]
- 72.Eads JC, Mahoney NM, Vorobiev S, Bresnick AR, Wen KK, Rubenstein PA, Haarer BK, Almo SC. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37:11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- 73.Ezezika OC, Younger NS, Lu J, Kaiser DA, Corbin ZA, Nolen BJ, Kovar DR, Pollard TD. Incompatibility with formin Cdc12p prevents human profilin from substituting for fission yeast profilin: insights from crystal structures of fission yeast profilin. J Biol Chem. 2009;284:2088–2097. doi: 10.1074/jbc.M807073200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kucera K, Koblansky AA, Saunders LP, Frederick KB, De La Cruz EM, Ghosh S, Modis Y. Structure-based analysis of Toxoplasma gondii profilin: a parasite-specific motif is required for recognition by Toll-like receptor 11. J Mol Biol. 2010;403:616–629. doi: 10.1016/j.jmb.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Margraf T, Schenk G, Torda AE. The SALAMI protein structure search server. Nucleic Acids Res. 2009;37:W480–W484. doi: 10.1093/nar/gkp431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin–actin complexes during filament elongation by Ena/VASP. EMBO J. 2007;26:4597–4606. doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nat Struct Biol. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- 78.Weiss MS, Hilgenfeld R. On the use of the merging R factor as a quality indicator for X-ray data. J Appl Crystallogr. 1997;30:203–205. doi: 10.1107/S0021889897003907. [DOI] [Google Scholar]
- 79.Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.