Abstract
Peroxisomes constitute a dynamic compartment of almost all eukaryotic cells. Depending on environmental changes and cellular demands peroxisomes can acquire diverse metabolic roles. The compartmentalization of peroxisomal matrix enzymes is a prerequisite to carry out their physiologic function. The matrix proteins are synthesized on free ribosomes in the cytosol and are ferried to the peroxisomal membrane by specific soluble receptors. Subsequent to cargo release into the peroxisomal matrix, the receptors are exported back to the cytosol to facilitate further rounds of matrix protein import. This dislocation step is accomplished by a remarkable machinery, which comprises enzymes required for the ubiquitination as well as the ATP-dependent extraction of the receptor from the membrane. Interestingly, receptor ubiquitination and dislocation are the only known energy-dependent steps in the peroxisomal matrix protein import process. The current view is that the export machinery of the receptors might function as molecular motor not only in the dislocation of the receptors but also in the import step of peroxisomal matrix protein by coupling ATP-dependent removal of the peroxisomal import receptor with cargo translocation into the organelle. In this review we will focus on the architecture and function of the peroxisomal receptor export machinery, the peroxisomal exportomer.
Keywords: Peroxisome, Exportomer, Peroxin, Protein import, Ubiquitination, AAA-type ATPases
Versatile functions of peroxisomes
Peroxisomes are single-membrane bound organelles present in all eukaryotic cells with the exception of mature erythrocytes and spermatozoa [1]. They are often structurally characterized by crystalline inclusions indicating the sometimes very high concentration of certain matrix proteins. Peroxisomes are characterized by a unique variability in metabolic tasks and enzyme content that is adjusted according to cellular needs. The β-oxidation of fatty acids and the detoxification of the hydrogen peroxide produced are the central and conserved functions of peroxisomes [2, 3]. In mammals, the breakdown of different types of fatty acids is distributed between peroxisomes and mitochondria whereas in fungi and plants the fatty acid degradation pathway is exclusively localized in the peroxisomal compartment [4]. Additionally, the core enzymes of the β-oxidation pathway are involved in the synthesis of signaling molecules, like phytohormones in plants [5, 6] and pheromones in Caenorabditis elegans and insects [7, 8] as well as the generation of H2O2 for signaling in hypothalamic neurons in mice [9]. Mammalian peroxisomes have an important function in the biosynthesis of bile acid and ether lipids such as plasmalogens [10]. Peroxisomes house important steps of penicillin biosynthesis in some filamentous fungi [11, 12] or certain reactions required for the composition of vitamin K1 in plants [13]. Peroxisomes also perform metabolic functions in concert with other organelles. In the context of β-oxidation and scavenging of ROS (reactive oxygen species), they exchange metabolites with mitochondria in mammals [14], while plant peroxisomes stand in close contact with mitochondria and chloroplasts during photorespiration [15].
Because of the central role of peroxisomes in lipid metabolism, several inherited diseases caused by mutation of single metabolic enzymes [16] or the defects peroxisome biogenesis [17] have been identified. The peroxisomal biogenesis disorders (PBDs) represent a spectrum of autosomal recessive metabolic disorders that are collectively characterized by abnormal peroxisome assembly and impaired peroxisomal function. The importance of this ubiquitous organelle for human health is highlighted by the fact that PBDs are multisystemic disorders often leading to death in early infancy. Recent data demonstrate that peroxisomes also provide a significant contribution to the antiviral innate immunity and that they can promote a rapid response to viral infection via peroxisomal antiviral signaling proteins [18]. In recent years, it has become clear that peroxisomal function counteracts the progressive brain damage and cognitive decline caused by Alzheimer`s disease, opening the possibility for new therapeutic targets [19, 20]. Furthermore, the peroxisomal reactive oxygen metabolism and redox homeostasis ties this organelle to age-associated diseases and the molecular process of aging [21].
The formation of functional peroxisomes depends on specific peroxisomal biogenesis factors, so called peroxins [22]. To date, 33 different peroxins have been described (Table 1) [23, 24]. Because of their distinct physical and functional interactions they can be divided into sub-networks that facilitate the four key stages of peroxisomal biogenesis, which comprise the formation of the peroxisomal membrane [25], fission [26], inheritance [27] as well as the sorting of peroxisomal matrix proteins to this organelle [28].
Table 1.
Peroxisomal biogenesis factors
| Role | S. cerevisiae peroxin | Human peroxin | Gene deletion phenotype in S. cerevisiae (or mammals)— mislocalization of matrix proteins | |
|---|---|---|---|---|
| PTS1 | PTS2 | |||
| Import peroxins | ||||
| PTS1 receptor | Pex5p | PEX5 | Severe | None |
| PTS2 receptor | Pex7p | PEX7 | None | Severe |
| PTS2 co-receptor | Pex18p | PEX5L | None | Severe |
| Pex21p | None | Severe | ||
| Docking complex | Pex13p | PEX13 | Severe | Severe |
| Pex14p | PEX14 | Severe | Severe | |
| Pex17p | Severe | Severe | ||
| Importomer assembly | Pex8p | Severe | Severe | |
| RING finger ligase complex | Pex2p | PEX2 | Severe | Severe |
| Pex10p | PEX10 | Severe | Severe | |
| Pex12p | PEX12 | Severe | Severe | |
| Receptor ubiquitin conjugation | Pex4p | UbcH5a/b/c (E2D1/2/3) | Severe | Severe |
| Pex22p | Severe | Severe | ||
| Ubc1p, 4p, 5p | Partial | Unknown | ||
| Receptor deubiquitination | Ubp15p | Partial | Unknown | |
| USP9X | None | None | ||
| AAA export complex | Pex1p | PEX1 | Severe | Severe |
| Pex6p | PEX6 | Severe | Severe | |
| Pex15p | PEX26 | Severe | Severe | |
| AWP1 | None | None | ||
| Membrane biogenesis and regulatory peroxins | Gene deletion phenotype in S. cerevisiae | |||
| Membrane biogenesis | Pex3p | PEX3 | Absence of peroxisomes | |
| Pex19p | PEX19 | Absence of peroxisomes | ||
| (Pex16p)a | PEX16 | None | ||
| Peroxisome proliferation | Pex11p, 25p, 27p | PEX11α/β/γ | Reduced number of peroxisomes | |
| Regulation of size, number and distribution | Pex28p–32p, Pex34p | Altered peroxisome number and/or morphology | ||
The table lists the known S. cerevisiae and human peroxisomal proteins required for peroxisomal biogenesis. In this table, the S. cerevisiae and human proteins required for peroxisome biogenesis are arranged according to their function. The superscript “a” points out that the protein is absent in S. cerevisiae and other yeast species with the exception of Yarrowia lipolytica
In this review, we will present a general overview on the dynamics of the peroxisomal matrix protein import receptors. Moreover, we will focus especially on the function of the peroxisomal biogenesis factors that act late in the receptor cycle and highlight their concerted function as receptor export machinery, the peroxisomal exportomer.
The import cycle of peroxisomal matrix protein receptors
The function of peroxisomes relies on the import of enzymes into their matrix. These matrix proteins need to be compartmentalized in order to carry out their biochemical reactions and thus connect the peroxisome to its different physiologic tasks. Because peroxisomes do not contain DNA, all peroxisomal gene products are encoded in the nucleus. Recent work demonstrates that mRNAs coding for peroxisomal proteins target to the proximity of peroxisomes [29], followed by the posttranslational import of the newly synthesized proteins into the organelle. A remarkable feature of peroxisomes is their capability to accommodate the import of fully folded proteins, which sometimes even maintain an oligomeric or co-factor bound state during import [30]. This distinguishes them from other organelles like mitochondria, chloroplasts and endoplasmic reticulum, which all import unfolded proteins, but is reminiscent of the secretion pathways of bacteria, archaea and thylakoid membranes [31]. However, peroxisomal matrix protein import is accomplished by dynamic receptors, which cycle between a soluble state in the cytosol and a membrane associated state at the peroxisomal membrane. This import process can conceptually be divided into five steps, comprising cargo recognition in the cytosol, receptor-cargo docking at the peroxisome, cargo translocation across the membrane, cargo release into the matrix and, receptor ubiquitination and export back to the cytosol (Fig. 1).
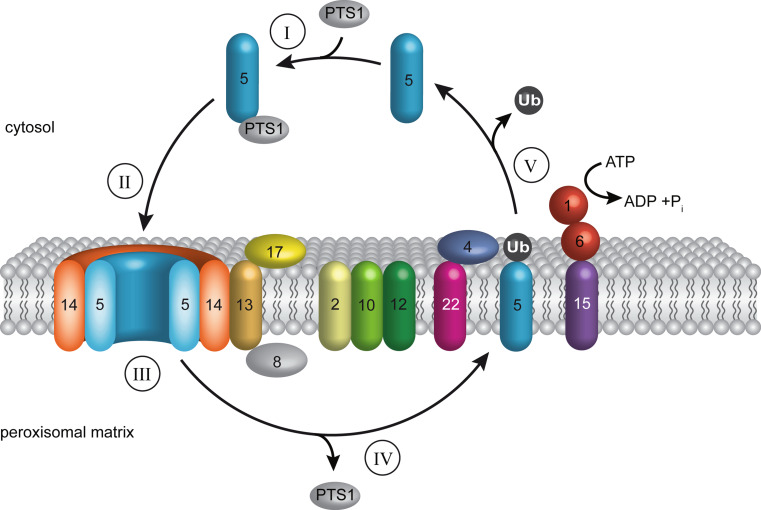
Fig. 1.
Peroxisomal matrix protein import in S. cerevisiae. Peroxisomal matrix proteins are imported into the peroxisomes via soluble receptors, which shuttle between the cytosol and the peroxisomal membrane. I The receptor Pex5p recognizes matrix proteins via their peroxisomal targeting signal 1 (PTS1) in the cytosol. II This receptor-cargo-complex binds to the docking-complex (Pex13p, Pex14p and Pex17p) at the peroxisomal membrane. It is assumed that the binding between the docking complex and cargo-loaded Pex5p leads to the formation of a transient pore, whose exact molecular constitution is still under discussion but at least contains Pex5p and Pex14p. III The cargo is translocated into the peroxisomal lumen in an unknown manner. IV Receptor-cargo dissociation, which might involve Pex8p. V After formation of the import pore and cargo translocation, the receptor is recycled, thus released from the peroxisomal membrane back to the cytosol for another round of import. For this, Pex5p is monoubiquitinated by the Pex22p-anchored ubiquitin-conjugating Pex4p (E2) and the ubiquitin ligase Pex12p (E3), which forms the RING-finger complex together with the other ubiquitin ligases Pex2p and Pex10p. The ubiquitin signal leads to an ATP-dependent dislocation of Pex5p from the peroxisomal membrane. This process is performed by the AAA-peroxins Pex1p and Pex6p, which are anchored to the peroxisomal membrane via Pex15p. Before starting a new round of import, the ubiquitin is removed
Targeting signal-dependent cargo recognition in the cytosol
The majority of the peroxisomal matrix proteins contains a peroxisomal targeting signal type 1 (PTS1) at the C-terminus, consisting mainly of the tripeptide sequence SKL or variations thereof as well as additional adjacent residues. The soluble PTS1-receptor Pex5p interacts with this dodecameric PTS1-signal via six tetratricopeptide repeats (TPRs) within its C-terminal half (Fig. 2) [32–36]. Crystal structures of the cargo-loaded and unloaded Pex5p revealed that cargo binding induces major conformational changes within the receptor, which might generate a docking-competent state of the receptor [37, 38].
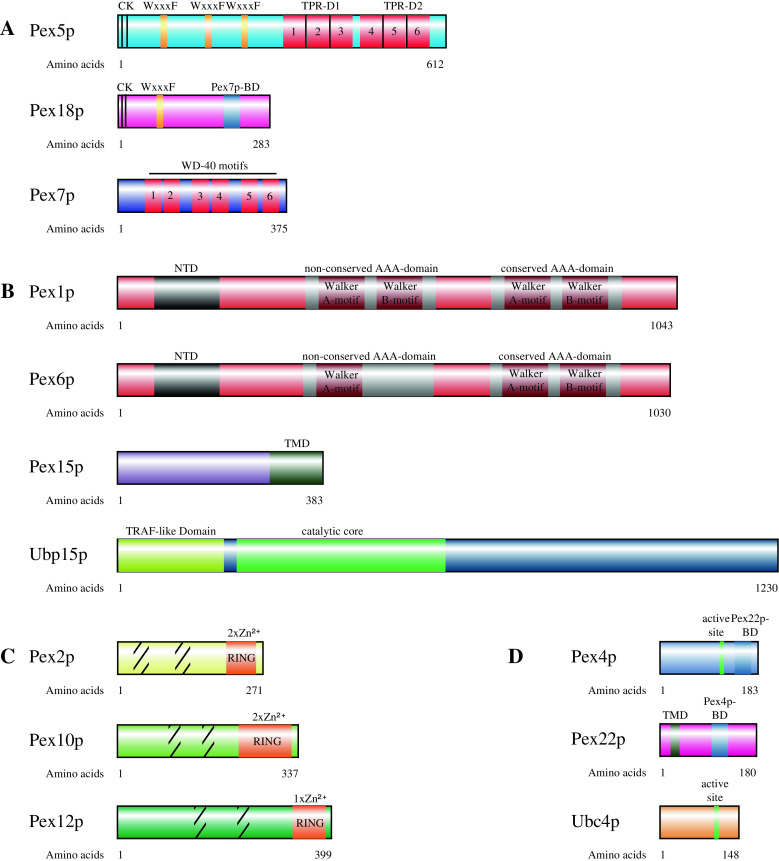
Fig. 2.
S. cerevisiae proteins involved in peroxisomal receptor export and their important domains. a Structural features of the receptors and co-receptors of the peroxisomal protein import machinery. These are required for cargo recognition and guidance of the cargo proteins along the peroxisomal import cascade. b Structural features of peroxisomal proteins necessary for the ATP-dependent export of ubiquitinated Pex5p. These are the AAA-peroxins which are responsible for the ATP-dependent dislocation of the ubiquitinated receptors from the peroxisomal membrane to the cytosol. The tail-anchored peroxisomal membrane protein Pex15p (Pex26p in mammals) anchors the AAA-peroxins Pex1p and Pex6p to the peroxisomal membrane. Ubp15p is a ubiquitin hydrolase which can remove ubiquitin chains from the receptor Pex5p. c Structural features of the membrane-bound RING-finger E3-ligases. The hatched areas are predicted transmembrane domains. d Structural features of the peroxisomal proteins with E2-ligase function; Pex22p is a membrane anchor for Pex4p. C Cysteine, K Lysine, WxxxF WxxxF-motif, TPR-D Tetratricopeptid repeats domain, BD Binding domain, NTD N-terminal domain, TMD transmembrane domain
A subset of matrix proteins contains the peroxisomal targeting signal type 2 (PTS2). The PTS2 represents a non-apeptide with the consensus sequence (R/K)-(L/V/I)-X5-(H/Q)-(L/A) found near the N-terminus of matrix proteins. PTS2-harbouring proteins are recognized by the WD40-protein Pex7p [39–41]. In contrast to the PTS1-receptor, Pex7p requires co-receptors. In most fungi and yeasts, Pex7p associates with the co-receptor Pex20p or the redundant Pex18p and Pex21p, whereas in mammalian and plant cells a splice variant of Pex5p, the Pex7p-binding domain containing Pex5L, was shown to cooperate with Pex7p [42]. It is interesting to note that also some specialized variants of these import pathways exist. For example, Arabidopsis thaliana and Trypanosoma brucei Pex7p are also required for PTS1-dependent import [43, 44]. At least in the case of methylotrophic yeasts, the PTS2-co-receptor Pex20p has been reported to recognize special cargo proteins on its own [45, 46]. In Caenorhabditis elegans [47] and in the protist Phaeodactylum tricornutum [48] the PTS2-import pathway is completely absent and all matrix proteins are imported by the PTS1-receptor Pex5p.
Even though the PTS1- and PTS2-import factors differ in some aspects, they share a comparable modular architecture. Accordingly, Pex7p is functionally related to the C-terminal part of Pex5p, as both comprise the corresponding cargo binding region while in terms of structure and function the PTS2 co-receptors are related to the N-terminal part of Pex5p, as this region is responsible for the peroxisomal targeting, association with the translocation machinery and, as discussed below, for ubiquitination. The resemblance of the corresponding factors is underscored by a chimera consisting of Pex18p (without its Pex7p-binding site) fused to the TPR-domains of Pex5p which can rescue PTS1-import in a PEX5-deficient yeast strain [49].
Alternatively, the targeting of a small number of peroxisomal proteins does not essentially rely on one of the two targeting signals and they can either be co-imported via an association with canonical PTS-cargo proteins (“piggy-back import”) or via the binding to the N-terminal region of Pex5p (“non-PTS import”) [50].
Docking of the receptor-cargo complex at the membrane
Subsequent to the formation of the receptor-cargo complex in the cytosol, the receptor guides the cargo protein to the docking-complex at the peroxisomal membrane. The membrane-bound proteins Pex13p and Pex14p are required for the initial docking step. Their genomic deletion drastically affects the membrane association of the receptor-cargo complex and, therefore, also the import of the cargo protein. Pex13p is an integral membrane protein which interacts via its N-terminus with the PTS2-receptor Pex7p, while its Src homology (SH3) domain can bind to the PTS1-receptor Pex5p as well as to the proline-rich SH3-ligand motif (PXXP) of Pex14p [51].
Pex14p is an integral membrane protein, but in some species it is also described to be peripherally associated. Pex14p interacts with the SH3-domain of Pex13p and with the WXXXF/Y motifs in the N-terminal part of Pex5p [52]. The NMR and crystal structures of the Pex14p-Pex5p assembly have revealed the structural basis for these transient interactions [53–55]. It is interesting to note that Pex14p is a multifunctional protein, which is not only involved in matrix protein import, but also autophagic degradation of peroxisomes [56] as well as movement of the organelle [57].
The docking complex in several species comprises a third protein in addition to Pex13p and Pex14p. Yeasts contain Pex17p, which is a peripheral membrane protein that associates to peroxisomes via Pex14p [58, 59]. In some filamentous fungi, a chimeric protein consisting of Pex14p-like N-terminal domain and a Pex17p-like C-terminal domain has been described, called Pex14/17p or Pex33p [60, 61]. A homolog of these Pex17p-like proteins has not yet been identified in higher eukaryotes. The function of the Pex17p-like proteins is not clear because they are not essential for receptor docking but still important for cargo import into the matrix.
Translocation across the membrane
Several hypotheses have been proposed how the cargo proteins traverse the membrane (as discussed in [28]). One concept (“transient pore hypothesis”) suggests that components of the docking complex as well as the import receptors themselves might become at least temporally an integral part of a dynamic import pore [62]. This hypothesis takes into account that the PTS1-receptor Pex5p can spontaneously insert into phospholipid membranes in vitro and changes its membrane topology during the protein import cascade with a portion of the peroxisomal Pex5p pool behaving like an integral membrane protein in vivo [63–66]. During the import cycle, Pex5p enters the luminal side of the peroxisomal membrane, although it is still a matter of debate, whether the whole receptor-cargo-complex (“extended shuttle hypothesis”) or just a part of Pex5p (“simple shuttle hypothesis”) reaches the peroxisomal matrix [67, 68]. Equally, Pex7p has been demonstrated to behave like a cycling receptor [69] and it is possible also that its co-receptors, P. pastoris Pex20p [70] and S. cerevisiae Pex18p [71], traverse the peroxisomal membrane. According to the “transient pore hypothesis”, the receptors might contribute to a dynamic import pore which opens the membrane dynamically for cargo-loaded Pex5p and Pex7p. In line with the assumption that the PTS1-receptor temporally becomes an important structural component of the translocon, Pex5p together with Pex14p represent the minimal unit for the import of the intraperoxisomal protein Pex8p in P. pastoris [72]. Moreover, experiments using planar lipid bilayer techniques provided first evidence that an isolated protein complex from S. cerevisiae, which mainly consisted of Pex5p and Pex14p, contained pore-forming activity with features expected for a protein-conducting channel [73]. The physical properties and the dynamic behavior of this pore, which displays a diameter of up to 9 nm, appear to match the criteria required for the passage of folded proteins. However, the exact protein composition of this translocon, the detailed mechanism of cargo translocation as well as the driving force for this event are yet to be elucidated.
Cargo release into the matrix
The mechanism of cargo release into the peroxisomal lumen subsequent to the translocation step is still not well characterized. Recent data from a mammalian in vitro system suggest that an alteration of the Pex5p–Pex14p binding mode may accompany the release of the cargo [74]. There is some evidence for a potential role of an additional protein in the disassociation of the PTS1-receptor-cargo complex, namely Pex8p of H. polymorpha [75]. Pex8p is an intra-peroxisomal peripheral membrane protein whose PTS1 and PTS2 signals could hypothetically compete with the cargo for the binding to Pex5p and, therefore, could trigger the dissociation of the receptor-cargo complex. Using proteins expressed in E. coli, a fluorescence correlation spectroscopy (FCS) study found that Pex5p-bound PTS1-cargo dissociates from Pex5p when Pex8p is interacting with this complex [75]. However, it has to be mentioned that in vivo PTS1-proteins can still be imported in yeast strains that express a Pex8p without its PTS1-sequence [76]. Another function assigned to S. cerevisiae Pex8p is that it is required for the physical connection of the docking complex with the RING-complex [77]. However, because this structural function has been suggested to be taken over by Pex3p in P. pastoris [78] and because Pex8p is not conserved in humans [24], the general mechanism of cargo liberation remains elusive.
Subsequent to the release into the peroxisomal matrix of mammals and plants the signal sequence of a subset of the imported proteins is proteolytically removed [79–81]. At least in the case of mammalian β-oxidation enzymes, the intraperoxisomal protease Tysnd1, which itself is a PTS1-protein, is responsible both for the removal of the leader peptide from PTS2 proteins as well as for the processing of PTS1 proteins and controls thereby the proper activity of these enzymes in β-oxidation [80].
Receptor export back to the cytosol
The PTS-receptors are exported back to the cytosol for further rounds of matrix protein import. This release is facilitated by the peroxisomal receptor export machinery. A central event in this process is the monoubiquitination of the receptor which depends on the ubiquitin-conjugating enzyme Pex4p and the presence of the peroxisomal RING-complex. This modification is thought to prime the receptor molecule for the recogntion by the AAA-type ATPase complex, which functions as dislocase by pulling the receptor from the membrane back to the cytosol. Moreover, the recent export-driven-import model suggests that the ATP-dependent export of the receptor is mechanistically linked to the translocation of the receptor-bound cargo protein into the matrix. If the monoubiquitination-dependent recycling pathway is impaired, the PTS-receptors enter a polyubiquitination-dependent alternative pathway (Fig. 3), which promotes their extraction from the membrane and degradation by the proteasome.
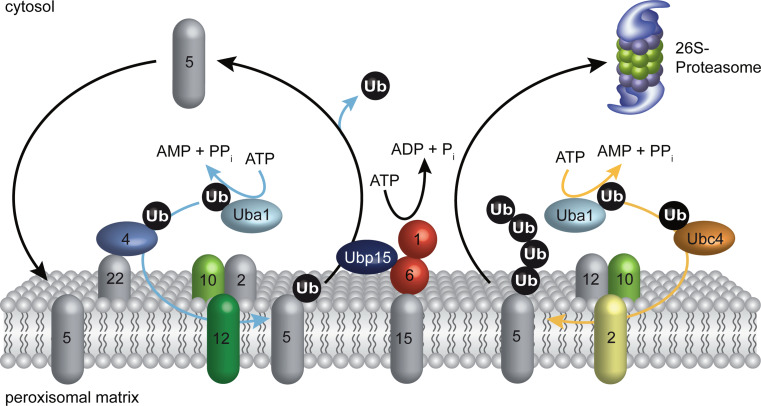
Fig. 3.
Ubiquitination cascades at the peroxisomal membrane in S. cerevisiae. The PTS1-receptor Pex5p is either mono- or polyubiquitinated at the peroxisomal membrane by two different ubiquitination pathways. For the mono- as well as for the polyubiquitination of Pex5p the cascades are initiated by the ATP-dependent ubiquitin-activating enzyme Uba1p (E1). For monoubiquitination of Pex5p (blue arrows) the activated ubiquitin is transferred to the ubiquitin-conjugating enzyme Pex4p (E2) and then delivered to Pex5p by assistance of the RING-ligase Pex12p (E3). The bound ubiquitin leads to a Pex1p/Pex6p-dependent export of Pex5p. Before starting a new import cycle, the ubiquitin is removed by the deubiquitinating enzyme Ubp15p. For polyubiquitination of the receptor (orange arrows), the activated ubiquitin is transferred to the ubiquitin-conjugating enzyme Ubc4p (E2) and then attached to Pex5p by the help of the RING-ligase Pex2p (E3). Also, polyubiquitination designates the receptor to its Pex1p/Pex6p-dependent export. As the polyubiquitated Pex5p is degraded by the 26S proteasome, this cascade is considered to be a part of a quality control system
The architecture of the the peroxisomal receptor export machinery: the exportomer
The cycle of the PTS-receptors and, therefore, the proper import of matrix proteins rely on the function of each single constituent of the membrane-bound peroxins as described above (2.1–2.5). Previous work defined the composition and characteristics of certain subcomplexes, like the docking- and the RING-complex [77, 78]. Based on the fact that both are physically connected by Pex8p in S. cerevisiae and both are needed for matrix protein import, the term “importomer” has been defined [77]. However, later studies disclosed that constituents of the AAA-complex can be co-purified with the importomer, strongly suggesting that all membrane-associated peroxin complexes required for matrix protein import are dynamically interconnected [82, 83]. Taking this into account, another attempt to define functionally related subcomplexes could be based on the membrane-associated steps of the PTS-receptor cycle in general and on the energy-dependence in particular. The current view is that the docking of the PTS-receptors at the site of the classically defined importomer is ATP-independent [84–87]. The RING-complex (Pex2p, Pex10p, Pex12p) as well as the Ubc-complexes (Pex22p, Pex4p, Ubc4p-family, UbcH5-family) are integral parts of the ATP-dependent ubiquitination cascade [88–93] and constitute together with the AAA-type ATPase complex (Pex1p, Pex6p, Pex15p, AWP1) [70, 84, 87, 94] the receptor export machinery, or alternatively, peroxisomal exportomer (Table 1; Fig. 2).
The ubiquitin-conjugating enzymes required for receptor monoubiquitination
Ubiquitination is a post-translational protein modification that is mediated by a three-step enzyme-cascade and results in the covalent attachment of ubiquitin to a substrate protein. The ubiquitin activating enzyme (E1) activates ubiquitin in an ATP-dependent manner and transfers it then to an ubiquitin-conjugating enzyme (E2). In the final third step, a ubiquitin-protein ligase (E3) binds both the ubiquitin-loaded E2 and the substrate, thereby facilitating the conjugation of the ubiquitin moiety to the target residue of the substrate protein [95]. In most cases, the ε-amino group of a lysine residue within the target protein is covalently linked to ubiquitin via a isopeptide bond. However, there are a few examples where ubiquitin is linked via a peptide bond to the α-amino group to the N-terminal amino acid, or via a oxyester bond to a serine or threonine, or even via a thioester bond to a cysteine [96].
Peroxisomal matrix protein import relies on the seldom found cysteine-dependent monoubiquitination of the PTS-receptors Pex5p [93, 97, 98] and Pex18p [71]. The E2-enzyme that has been demonstrated to catalyze this modification of Pex5p in S. cerevisiae is Pex4p (Ubc10p) [91, 93].
Pex4p is a soluble ubiquitin conjugating enzyme that is essential for the import of both PTS1 and PTS2 proteins and, therefore, has been the first E2 enzyme shown to be required for the biogenesis of an organelle [99–102]. It is anchored to the peroxisomal membrane by Pex22p [102–104]. A recent crystal structure of the S. cerevisiae Pex4p–Pex22p complex reveals that the Pex22p-binding site in Pex4p does not resemble a common substrate-binding motif and, therefore, it has been suggested that Pex22p may act as a co-activator of this E2 enzyme [104]. This finding is very similar to the situation of Ubc7p, the only other known E2-enzyme that has a membrane-anchor. This soluble E2-enzyme is involved in ERAD and binds to the membrane-protein Cue1p at the ER [105]. This interaction is thought to enhance the activity of Ubc7p [106, 107]. This regulatory feature may insure that Pex4p and Ubc7p can only exert full activity when they have reached the location of their physiological substrates.
The current view is that Pex4p monoubiquitinates membrane-bound Pex5p in order to prime the receptor for the export reaction [91], thereby enabling further rounds of matrix protein import. Interestingly, Pex4p and Pex22p are well conserved in yeast and plants but seemingly are absent in mammals [24]. Instead, members of the mammalian E2D family of conjugating enzymes took over the role of Pex4p [89]. UbcH5a, UbcH5b and UbcH5c catalyze the monoubiquitination of mammalian Pex5p at the conserved cysteine and are required for the export of the PTS1-receptor in an in vitro system [89]. Even though these three UbcH5-proteins carry out an essential function in peroxisome biogenesis, their task area is not restricted to this organelle as they are known to have several other cellular target proteins apparently not related to peroxisomes s, like IκBα or p53 [108–110]. It is not yet clear why the monoubiquitination of the PTS1-receptor in evolution has been attributed to the abundant and promiscuous UbcH5-proteins. However, it might be possible that the physiologic function of peroxisomes is interconnected to one of the cellular factors that is controlled by the same E2-enzymes, which would then allow them to regulate both processes in a concerted manner.
The peroxisomal RING-ligase complex
Peroxisomal matrix protein import depends on the presence of the three conserved RING-type ubiquitin-protein ligases Pex2p, Pex10p and Pex12p, which are involved in the ubiquitination of the PTS1-receptor Pex5p [88, 90, 92]. A fourth peroxisomal RING-ligase, TRIM37, has been reported for mammals [111]. Mutations in TRIM37, whose substrates are not known, can cause Mulibrey Nanism and Wilm’s tumor but do not interfere with peroxisomal matrix protein import [112]. In contrast, functional loss of Pex2p, Pex10p and Pex12p are the second most common cause of PBD [17, 113]. Importantly, mammalian Pex2p (formerly PAF-1) was the first gene that could be linked to PBDs [114, 115].
In general, E3 enzymes represent an important determinant of substrate specificity of ubiquitination reactions because they can be regarded as binding platform for the ubiquitin-charged E2 enzyme and the substrate, thereby insuring a specific transfer of ubiquitin to the target amino acid. E3 enzymes can be divided into several classes based on their structural features and mode of catalysis. Most of them fall into the two main classes: the HECT-type ligases, which form a thioester-intermediate with ubiquitin prior to the transfer to the target protein [116], as well as the RING-type ligases, that catalyze the direct transfer of ubiquitin from the E2 enzyme to the target [117]. RING-finger E3 ligases belong to the super-family of Treble-Clef fold containing proteins. This scaffold structure, whose fold is stabilized by a Zn2+-ion, is involved in the binding of interaction partners and can also be found in a wide variety of proteins outside the ubiquitin system [118]. The typical RING-finger, which was first described in 1991 [119], binds two Zn2+-ions through its conserved Cys and His residues in a unique “cross-brace” arrangement of its Zn2+-coordination sites [117].
The RING-domains of the peroxisomal integral membrane proteins Pex2p, Pex10p, and Pex12p are located in their C-termini and are exposed to the cytosol (Fig. 2). While the RING-domains of Pex2p and Pex10p coordinate two Zn2+-ions, the RING-motif of Pex12p is degenerated and binds only a single Zn2+-ion [120]. Interestingly, not just Pex12p but also several members of the RBR-(RING-between-RING) family of ubiquitin ligases, e.g., the Parkin-like Ariadne, contain a RING-domain at their C-terminus that has a single Zn2+-ion [121].
The RING-peroxins Pex2p, Pex10p and Pex12p form a distinct complex and stabilize each other in vivo [77, 78]. Based on earlier binary interaction studies [122–125], and on recent interaction data on all three RING-domains [88], the RING-peroxins form a trimeric complex. At least S. cerevisiae Pex10p (RING) functions as central component and directly binds to Pex2p (RING) and Pex12p (RING) while bridging the indirect interaction between these two RING-domains [88]. Possibly due to the reciprocal stabilization, the presence of each RING-peroxin is required for matrix protein import and ubiquitination of PTS1-receptor [126–128]. Nevertheless, the heteromeric architecture of the RING-complex seems to have a direct influence on the ligase activity of these E3 enzymes as the ubiquitination activity of the Pex10p/Pex12p RING-domains is enhanced in presence of Pex4p in vitro [88]. This finding reflects in vivo data on the role of the E2/E3 pair Pex4p/Pex12p in monoubiquitination of Pex5p [90]. Likewise, Pex10p (RING) also stimulates the ligase activity of Pex2p (RING) and the functional implication thereof will be discussed below (5).
While most RING-ligases act as a monomer, some utilize non-RING adaptor proteins or function as homo-dimer or hetero-dimer [117]. Interestingly, there is only one other example of a trimeric complex of RING-ligases, which is the polycomb complex [129]. Here, the oncogenic RING-protein Bmi1 stimulates the ligase activity of Ring1b and partially Ring1a for ubiquitination of histone H2A, while Bmi1 also stimulates the Ring1a-dependent ubiquitination of the topoisomerase Top2a [129–132]. The molecular mechanism underlying the regulation of heteromeric RING-complexes still is not fully understood. It has been reported that the autoubiquitination status of the substrate-targeting RING-ligase is regulated by the interacting RING-protein [131, 133–135]. Whether the in vitro observed autoubiquitination of the peroxisomal RING-domains also occurs in vivo is not clear [88, 90, 92].
Because Pex10p represents the central component of the RING-complex, it is interesting to note that it may also have additional functions that are distinct from Pex2p and Pex12p. A systematic functional screen of all peroxins in A. thaliana revealed that only Pex10p has a pleiotropic growth phenotype [136]. Furthermore, overexpression experiments of proteins with mutated RING-domain in wild-type background suggested that A. thaliana Pex10p but not Pex2p or Pex12p are required for the contact of peroxisomes to chloroplasts during photorespiration [137, 138]. In the filamentous fungus Podospora anserina, the entire RING-complex, in contrast to Pex5p, Pex7p and Pex14p, is implicated in the meiotic development [139–141]. However, whether the involvement of the RING-complex in meiocyte formation in P. anserina or the Pex10p-dependent association of peroxisomes with chloroplasts in A. thaliana is due to physical interactions or due to a putative functional interaction via ubiquitination-events is not known.
In the case of peroxisomal matrix protein import, different studies link the function of the RING-peroxins to the export of the PTS-receptors as they find Pex5p [123, 142] and the PTS2-co-receptor Pex20p [70] to accumulate inside the peroxisomal lumen in cells with disrupted RING-complex. Because the Pex4p/UbcH5-dependent monoubiquitination of Pex5p is reported to be a prerequisite for the export of Pex5p [89, 91] and as Pex12p cooperates with Pex10p in vitro [88] and catalyzes this Pex4p-dependent modification in vivo [90], the Pex10p/Pex12p heteromer may represent the active ligase-complex specificially dedicated to the monoubiquitination-mediated dislocation of the PTS1-receptor.
The peroxisomal AAA-type ATPases
The monoubiquitinated PTS1-receptor is substrate for the AAA-complex, which functions as dislocase that exports Pex5p from the membrane back to the cytosol [84, 87, 91, 94, 98, 143–145]. The peroxisomal AAA-type ATPases Pex1p and Pex6p are both equally required for peroxisomal biogenesis and, therefore, have a non-redundant function [146–150] which depends on the presence of their membrane anchor, Pex15p in yeast and its orthologues Pex26p in mammalian cells as well as APEM9 in plants [151–155]. A proper assembly of the AAA-complex is important as disruption of the interaction between human Pex1p and Pex6p is the most common cause of Zellweger syndrome [17, 156]. Pex1p (formerly PAS1) was the first peroxin to be identified and one of the founding members of the AAA-family [157–159].
AAA-proteins are characterized by a modular architecture. They belong to the class of P-loop NTPases characterized by conserved motifs for NTP-binding (Walker A motif) and hydrolysis (Walker B motif) [160]. They are defined by the evolutionary conserved AAA-domain, which contains the Walker A and B motifs, as well as other conserved regions like the Second Region of Homology (SRH) [157, 161, 162]. Pex1p and Pex6p display two AAA-domains, termed D1 and D2, post-positioned to the N-terminal domain (NTD) (Fig. 2). They share this double AAA-domain structure with other AAA-protein, including Cdc48p (p97/VCP) and Sec18p (NSF) [163, 164]. However, in the case of the AAA-peroxins the second AAA-domain is better conserved than the first [146, 150, 151, 165], whereas for NSF the first is better conserved that the second AAA-domain, and for Cdc48 both AAA-domains are equally well conserved. The binding of ATP and its hydrolysis is thought to result in conformational changes, as shown for p97 [166].
Most AAA-proteins form active oligomers with predominantly hexameric constitution [167]. The best analyzed example is p97, for which structural data, based on X-ray crystal analysis and cryoelectron microscopy [168, 169] provide some insight into the reaction cycle of this ATPase. However, the knowledge on the structural arrangement and molecular mechanism of the AAA-peroxins Pex1p and Pex6p is still scarce. They are expected to form a hetero-hexamer, but this has not yet been demonstrated. Furthermore, there are indications that cytosolic mammalian Pex1p might form homo-trimers prior to its interaction with Pex6p at the peroxisome [150]. Based on the analysis of point-mutations in the Walker A and B motifs of the AAA-peroxins (Fig. 2), it is clear that distinct ATP-binding and hydrolysis sites contribute to the assembly [146, 150, 151, 165]. In yeast, ATP-binding and hydrolysis in Pex6p govern the assembly/disassembly rates of the Pex6p/Pex15p-complex [151]. The Pex1p–Pex6p interaction is modulated by ATP-binding in D2 of Pex1p, while hydrolysis in D2 seems not to be involved [146]. However, ATP-hydrolysis in D2 is essential for the export of Pex5p [87], suggesting that ATP-hydrolysis in D2 of Pex1p is specifically required to generate the pulling force for extraction of Pex5p from the peroxisomal membrane. In this context, it is interesting to note that the release of the AAA-peroxins from the membrane might be regulated by the above described E2-enzyme Pex4p. Pex1p and Pex6p accumulate at the importomer in Pex4p-deficient yeast cells, which raises the possibility that the ubiquitin-dependent PTS1-receptor cycle and the dynamic recruitment and release of the AAA-peroxins are interconnected [83].
As indicated by the acronym AAA (ATPases associated with diverse cellular activities), several AAA-proteins carry out different tasks. In general, the common theme for the mode of action of the AAA-proteins might be that they act as mechanoenzymes that disassemble protein complexes linked to different cellular functions. An important example is again Cdc48p (p97/VCP) which is involved in ERAD, MAD, transcription factor processing, Golgi cisternae rearrangement, chromatin condensation, endosomal sorting and autophagy [170]. The AAA-peroxins have been implicated in the fusion of pre-peroxisomal vesicular structures in yeasts [171, 172] and especially Pex6p seems to be involved in the suppression of different cell death mechanisms [173–175]. However, the best understood function of Pex1p and Pex6p so far is their role in peroxisomal matrix protein import. The binding and hydrolysis of ATP by the AAA-peroxins is believed to induce conformational changes that finally generate the driving force to release the ubiquitinated PTS1-receptor from the peroxisomal membrane [84, 87].
However, while there is accumulating evidence that the purpose of monoubiquitination is to prime Pex5p for AAA-peroxin mediated export, the direct mechanistic influence of this modification remains to be investigated. The exact mechanism of substrate recognition and extraction from the membrane is not solved. To date, it is not clear if the ubiquitination alters the conformation of Pex5p and makes it accessible for a still unproven-direct interaction with the AAA-peroxins, or alternatively, if the ubiquitin-moiety itself may bind the AAA-peroxins. In the case of Cdc48p (p97/VCP), evidence for several mode for recognition of the ubiquitinated target have been published. This includes a direct interaction to ubiquitin [176], as well as a simultaneous binding to ubiquitin and a non-ubiquitinated part of the substrate [177]. Furthermore, an indirect interaction of Cdc48p (p97/VCP) with the ubiquitin-moieties of the target has been described, which is bridged by ubiquitin-binding adaptors [178]. It is interesting to note that the X-ray structure of the N-domain of murine Pex1p displays a double-ψ-β-barrel fold [179], which also is present in the N-domain of p97, where it functions as binding module for mono- and polyubiquitin [180]. But whether the domain found in Pex1p carries out such a function still has to be investigated.
The recent discovery of AWP1 (Associated with PRK1), a novel adaptor protein of human Pex6p [94] could add to our understanding of the mechanism of ubiquitin recognition by the AAA-peroxins. AWP1 has been described earlier as ubiquitin-binding NF-κB modulator [181]. However, AWP1 is also required for peroxisomal biogenesis and is able to interact with both Pex6p as well as with monoubiquitinated Pex5p [94]. Thus, AWP1 might function as specific linker, which enables the AAA-peroxins to transfer their suggested pulling force to the monoubiquitinated receptor molecule thereby driving its export. AWP1 interacts with monoubiquitinated Pex5p via an A20 zinc-finger domain, which is also found in the GTPase effector Rabex5 and the deubiquitinating enzymes A20 as well as Cezanne [182–184].
Deubiquitination of the receptor
The ubiquitin-moiety is removed from the receptor prior to a new round of matrix protein import. In general, the cleavage of ubiquitin from a substrate is carried out by ubiquitin hydrolases, also called deubiquitinating enzymes [185]. Recently, the hydrolase Ubp15p was identified as a novel interaction partner of Pex6p in S. cerevisiae [186]. Ubp15p functions as deubiquitinating enzyme acting on Pex5p, which represents the first characterized target of this enzyme [186]. Data obtained from a mammalian in vitro system suggest that the monoubiquitin moiety of Pex5p can be cleaved in two ways: a non-enzymatic cleavage of the thioester bond between Pex5p and ubiquitin by a nucleophilic attack of glutathione or, as the major pathway, enzyme-catalyzed by ubiquitin hydrolases [187]. USP9X has been described as the main ubiquitin hydrolase responsible for the enzymatic pathway [188]. USP9X is a cytosolic protein and is not specific for peroxisomal protein import as it has been linked to other ubiquitin-regulated events as well, like regulation of the transforming growth factor beta (TGFβ)-pathway [189], the anti apoptotic protein Mcl1 [190] or the ubiquitin-ligase MARCH7 [191]. USP9X is not an ortholog of Ubp15p. While in the mammalian in vitro system the ubiquitin moiety of Pex5p is removed by the cytosolic USP9X after completed export, the findings that membrane-associated Ub-Pex5p can be cleaved by the Pex6p-bound Ubp15p suggests that in yeast deubiquitination might occur during or shortly after membrane extraction by the AAA-peroxins [186, 188]. Furthermore, both studies do not exclude the possibility that additional or redundant ubiquitin hydrolases might exist.
Functional link between receptor export and cargo release: the export-driven import model
Early work defined peroxisomal matrix protein import as an energy-dependent process that requires the hydrolysis of ATP [192]. Later studies revealed that not the cargo recognition nor the docking of the receptor to the membrane but the export of the receptor back to the cytosol represents the energy-dependent step [86]. In recent years, the combined work of several groups has uncovered the ubiquitination machinery [89, 91, 97, 98, 187] as well as the AAA-complex [70, 84, 87, 94] as the ATP-consuming factors. Thus, energy-consumption, matrix-protein import and PTS-receptor export merge at the exportomer.
In principle, the protein composition of the peroxisomal receptor export machinery is evolutionary related to the proteins of the endoplasmic reticulum associated degradation (ERAD) machinery [193, 194]. ERAD represents a mechanism by which misfolded proteins are polyubiquitinated and extracted from the ER for their subsequent degradation by the proteasome in the cytosol [195]. A similar quality control system has been described for proteins of the outer mitochondrial membrane, called mitochondria associated degradation (MAD) [196]. A mechanistic parallel can be drawn between the ERAD- and MAD-substrates and the membrane-bound PTS-receptors, because all are extracted by mechanoenzymes of the AAA-type ATPase family in an ubiquitination-dependent reaction [197].
A recent model proposes a tight interconnection of ERAD-like receptor export to the cytosol and matrix protein import across the peroxisomal membrane [197]. This export-driven import model is supported by the fact that the presence of a functional receptor export complex is a prerequisite for the import of matrix proteins into peroxisomes. This mode of protein translocation requires ATP for the ubiquitin- and AAA-driven extraction of the receptor from the import pore and might be mechanically coupled to the movement and translocation of the cargo across the membrane. Two conclusions can be drawn from this model which could explain the import defects seen in mutants of the exportomer. First, the binding capacity for functional PTS-receptors at the peroxisomal membrane is limited. Indeed, attenuation of receptor export by functional impairment of the export machinery results in an accumulation of PTS-receptors at the membrane [70, 128] and would prevent the docking of newly-formed receptor-cargo complexes arriving from the cytosol. Data from experiments designed to address this question originate from work in A. thaliana [198]. The physiologic defects of mutated and only insufficiently active Pex6p were partially restored when combined with a weakly expressed allele of the docking protein Pex13p [198]. This fosters the assumption that PTS-receptor import and export rates have to be balanced to a certain level.
Second, this model concerns the question whether the export of the receptor and the release of the cargo might be mechanistically linked. Recent work on the ubiquitination of the PTS2-co-receptor Pex18p in S. cerevisiae provided first direct evidence for this model [71]. Cysteine-dependent monoubiquitination of Pex18p was found to be a prerequisite for the import of Pex7p into peroxisomes. Protease-protection assays demonstrated that Pex7p is partially protease-protected in wild-type cells, while Pex18p is accessable. This topology is reversed when the conserved cysteine of Pex18p is mutated or the AAA-peroxins are deleted [71], strongly indicating that monoubiquitination of Pex18p as well as AAA-complex activity trigger the import of cargo-loaded Pex7p. This mechanism would imply that cargo translocation into the matrix is also ATP-dependent. A CHO-cell based in vitro system supports the idea that cargo translocation requires ATP-hydrolysis [85], however, another study with a rat liver based in vitro system suggests that cargo release may occur independently of ATP and therefore presumably before the ubiquitination step [199]. In conclusion, the ATP-consuming receptor export machinery is thought to function as import motor for matrix proteins, either indirectly via balanced receptor import/export rates or directly via a linkage of cargo translocation with receptor export.
Receptor polyubiquitination: quality control and RADAR pathway
Alternatively to monoubiquitination on the conserved cysteine residue, the import receptors can be modified by the attachment of polyubiquitin chains on lysine residues, resulting in proteasomal degradation. In S. cerevisiae, polyubiquitinated Pex5p acculumates at the peroxisomal membrane when constituents of the Pex4p- or AAA-complexes are deleted [126–128]. This modification is mainly catalyzed by Ubc4p in conjunction with the partial redundant Ubc5p and Ubc1p [126–128]. These E2 enzymes also take part in diverse other cellular processes and share a high sequence similarity [200, 201]. Both Pex2p [90] and Pex10p [92] have been implicated as E3 enzymes for the generation of K48-linked polyubiquitin chains [126, 128] on Pex5p. Because receptor polyubiquitination predominantly takes place when the export machinery is affected in its function, the purpose of this modification is to remove the receptor from the peroxisomal membrane when the regular dislocation reaction is blocked. Site-directed mutagenesis of the conserved target lysine residues for polyubiquitination does not result in a growth defect of S. cerevisiae Pex5p on oleate medium [91, 93], and, therefore, the polyubiquitination of Pex5p is likely to represent a quality control pathway. However, mechanistically, polyubiquitination of Pex5p can be regarded as an alternative export signal. In vitro export assays demonstrated that a fraction of Pex5p can still be exported in a Pex4p-deficient system and that this residual export was strictly dependent on the presence of the two conserved lysine residues [91]. Moreover, in vivo evidence for the function of the polyubiquitination as alternative export signal comes from studies in P. pastoris Pex20p [202]. Mutation of the conserved cysteine induces polyubiquitination of Pex20p but still retains a partial functional receptor molecule which displays partial complementation in growth tests. However, only in the situation when the non-essential lysine residues are mutated in addition to the cysteine, the receptor completely looses its functionality. This result indicates that enhanced degradation of Pex20p can restore the protein import to a certain extent because the receptors are removed fast enough to allow the docking of further cargo-bound receptors. This mechanism has been described as RADAR (receptor accumulation and degradation the absence of recycling) [30, 70] in order to distinguish it from non-essential quality control. However, it should be noted that the mutation of the conserved cysteine of S. cerevisiae Pex5p and Pex18p alone is already sufficient to inhibit the function of these receptors completely [71, 93] in vivo and the export of mammalian Pex5p in vitro [97, 98, 203]. The latter might be explained by the assumption that the factors required for polyubiquitination could be too diluted in the in mammalian in vitro systems. It is interesting to note that the degradation of Pex5p in vivo apparently occurs much slower in S. cerevisiae than in P. pastoris and most of the tested species [100, 102, 142, 204, 205]. Thus, the instability of Pex5p observed in most species is most likely due to rapid degradtion via K48-linked polyubiquitination as described for the PTS1-receptor of H. polymorpha [206] and Pex20p in P. pastoris [70].
A possible additional function of the peroxisomal ubiquitination machinery is suggested by data from A. thaliana, indicating a potential role of the Ubc- and AAA-complex as regulator of matrix protein composition [102, 207]. Leaf peroxisomes undergo a partial remodeling of their enzyme content when photosynthesis is initiated. Isocitrate lyase is an enzyme that is active early in development and is usually is not anymore present in green leaf peroxisomes. This protein remains stable in peroxisomes in the absence of members of the Ubc- and AAA-complex indicating that these complexes may be important during the remodeling of peroxisome matrix content. However, these studies were solely based on protein stability and did not monitor a possible ubiquitination of the potential substrates.
Thus, while there might be potentially additional species-specific functions for the peroxisomal polyubiquitination machinery, the best studied function is the removal of the PTS-receptors when the monoubiquitination-dependent recycling pathway is blocked.
Conclusions and open questions concerning the export mechanism and the role of the cysteine-ubiquitination
The monoubiquitination of the PTS-receptor is a central event during peroxisomal matrix protein import. From this perspective, the components of the exportomer can be divided into the upstream factors of the E2- and E3-enzyme complexes on the one hand and the downstream factors of the AAA-complex and deubiquitinating enzymes (DUBs) on the other hand. These conclusions are mainly based on research on the PTS1-receptor Pex5p from different species, for which the ubiquitination cascade and AAA-mediated dislocation has been characterized. These studies define the AAA-complex as ubiquitin-dependent dislocase for the modified PTS-receptors [145].
The comparison to the structural, functional and evolutionary related Cdc48p (p97/VCP) has been of great importance to clarify of the functional role of the AAA-peroxins. However, it is important to investigate the fact that Pex1p and Pex6p, unlike p97, form a heteromer. This may disclose the functional differences of the non-redundant Pex1p and Pex6p, which might be related to ATP-dependent structural changes or the ubiquitin-dependent recognition of the PTS-receptors. A special focus will be on the mode of ubiquitin binding, especially whether orthologs of the adaptor AWP1 also exist in other species or whether alternative mechanisms are realized, like e.g., direct binding by either Pex1p, Pex6p or both.
The role of the exportomer in the cycling of receptors in the PTS2-pathway has not yet been studied in great detail. However, The membrane topology of the co-receptor Pex18p [71] as well as the distribution between peroxisome and cytosol of the orthologous co-receptor Pex20p [70, 202] depend on the AAA-peroxins. The finding that Pex18p [71, 208] is constitutively instable and polyubiquitinated, while its corresponding receptor and binding partner Pex7p is stable and does not appear to be ubiquitinated at all deserves further attention. Furthermore, it was not until recently that the cysteine-dependent monoubiquitinated form of Pex18p has been identified [71]. Because the conserved cysteine of Pex20p is important for its recycling [202], it can be anticipated that Pex20p might be monoubiquitinated as well. However, it will be important to elucidate if the same combination of E2 and E3 enzymes that is acting on Pex5p is also responsible for the poly- and monoubiquitination of the PTS2-co-receptors. Apart from the export mechanism, it will be a significant advance to clarify if the PTS2-receptor complex forms a PTS2-selective import pore.
Finally, one of the most intriguing questions concerns the fact that Pex5p and Pex18p are modified on a cysteine via a thioester bond and not by a more common isopeptide bond to a lysine. First evidence that ubiquitin can be attached to cysteine, serine or threonine residues came from studies of viral MARCH (Membrane-associated RING-CH) E3 ligases [209, 210]. These ligases polyubiquitinate cellular MHC I (major histocompatibility complex I) molecules, which results in their degradation with the consequence that the infected cell is not recognized by the immune system [211]. Recent data demonstrate that non-conventional ubiquitination plays also a role in cellular processes like ERAD, where unassembled TCRα and non-secreted NS-1 light chain are polyubiquitinated and degraded [212, 213]. The specific determinants of these non-lysine ubiquitination reactions are not known to date [96]. The E2 and E3 enzymes involved in the described examples, like e.g., the viral mK3 or the ERAD-ligase HRD1 [209, 213], are not restricted to this kind of modification because they can also ubiquitinate lysine residues. The modification depends on the position of the corresponding amino acid within the target protein [214]. This is also true for the PTS-receptors, because the cysteine of mammalian Pex5p [187, 188] and P. pastoris Pex20p [202] can be replaced by a lysine, resulting in a still largely functional protein. Thus, although highly conserved in evolution, the cysteine-dependent thioester-bond to ubiquitin is not essential for the basic export mechanisms and therefore may represent a central regulatory device. However, until now it is not clear if the same ubiquitination machinery targets the cysteine-to-lysine mutant and if this protein is efficiently recycled after it has been exported or if it is a substrate for a RADAR-related pathway.
There are several different hypotheses that could function as working models to solve the function of the cysteine. (1) The first idea is related to the fact that certain E3 enzymes, like HECT-type ligases [116] or the RBR-type ligases [215], form a ubiquitin-thioester intermediate on a cysteine before this ubiquitin molecule is finally transferred to the substrate. Pex5p is supposed to form oligomers at the membrane in the context of the transient import pore [62], which have to be rapidly disassembled after the cargo import process because the cytosolic form of Pex5p is a monomer. One hypothesis could be that not only Pex12p/Pex10p may modifiy the receptor but that also Pex5p itself could catalyze an intra-oligomeric ubiquitin-transfer in a relay-like system in order to accelerate the decomposition of the pore. Eventhough this idea seems highly speculative at this moment, there is an example of an intra-molecular ubiquitin-transfer, which has been described for the E2 enzyme E2-230K [216]. Here, the ubiquitination activity requires the intra-molecular transfer from the first cysteine to a second cysteine of E2-230K before the ubiquitin is delivered to the target protein, a process which the authors call a thiol-relay. (2) Another idea is based on the fact that thioester bonds are more instable than isopeptide bonds. This opens the posibility that the duration of the ubiquitin-moiety staying attached to the PTS-receptor might be shortened in order to prevent formation of a polyubiquitin chain or recognition by proteasomal adaptors. However, while the cysteine-ubiquitination of the mentioned examples of target proteins, like e.g., MHC I, elicits a faster degradation of the target, the thioester-based modification of Pex5p and Pex18p seems to prevent proteolysis. The rapid non-enzymatic disruption of the thioester bond of Ub-Pex5p in a mammalian in vitro system supports the assumption that the labile monoubiquitination protects the receptors for polyubiquitination [187, 217]. 3) Another possibility is based on the recent work on the control of the redoxbalance of peroxisomes [218], which adds to the previously described contribution of peroxisomes to the cellular reactive oxygen species levels [219]. In this context, it is considered that the cysteine residue of Pex5p required for monoubiquitination might be accessable for redox-changes, which might directly influence the availability of the residue for ubiquitination [220]. This redox-modification might regulate the fine-tuning of matrix protein import by controlling the import/export rates of the PTS1-receptor.
Certainly, many open questions remain to be answered concerning the matrix protein import into peroxisomes. In recent years, enough evidence regarding the ATP-dependent late stages of the receptor cycle has been collected from several groups and organisms to define the concept the exportomer. The understanding of the late acting peroxins as concerted acting components of the exportomer will be instrumental to define experimental concepts dedicated to tackle the molecular mechanism underlying peroxisomal protein import and therefore approach the central function of peroxisomes in health and disease.
Acknowledgments
We apologize to all the scientists whose work could not be cited due to space limitations. This work was supported by grants of the Deutsche Forschungsgemeinschaft (SFB 642) to RE and HWP.
Contributor Information
Harald W. Platta, Phone: +49-234-32224968, FAX: +49-234-3214266, Email: harald.platta@rub.de
Ralf Erdmann, Phone: +49-234-3224943, FAX: +49-234-3214266, Email: ralf.erdmann@rub.de.
References
- 1.Novikoff AB, Novikoff PM, Davis C, Quintana N. Studies on microperoxisomes. V. Are microperoxisomes ubiquitous in mammalian cells? J Histochem Cytochem. 1973;21(8):737–755. doi: 10.1177/21.8.737. [DOI] [PubMed] [Google Scholar]
- 2.Cooper TG, Beevers H. Beta oxidation in glyoxysomes from castor bean endosperm. J Biol Chem. 1969;244(13):3514–3520. [PubMed] [Google Scholar]
- 3.Lazarow PB, DeDuve C. A fatty acyl-CoA oxidazing system in rat liver peroxisomes; enhancement by clofibrate, a hypolipidemic drug. Proc Natl Acad Sci USA. 1976;73:2043–2046. doi: 10.1073/pnas.73.6.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunau W-H, Bühne S, Moreno de la Garza M, Kionka C, Mateblowski M, Schultz-Borchard U, Thieringer R. Comparative enzymology of β-oxidation. Biochem Soc Trans. 1988;16:418–420. doi: 10.1042/bst0160418. [DOI] [PubMed] [Google Scholar]
- 5.Baker A, Graham IA, Holdsworth M, Smith SM, Theodoulou FL. Chewing the fat: beta-oxidation in signalling and development. Trends Plant Sci. 2006;11(3):124–132. doi: 10.1016/j.tplants.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Kienow L, Schneider K, Bartsch M, Stuible HP, Weng H, Miersch O, Wasternack C, Kombrink E. Jasmonates meet fatty acids: functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana . J Exp Bot. 2008;59(2):403–419. doi: 10.1093/jxb/erm325. [DOI] [PubMed] [Google Scholar]
- 7.Joo HJ, Kim KY, Yim YH, Jin YX, Kim H, Kim MY, Paik YK. Contribution of the peroxisomal acox gene to the dynamic balance of daumone production in Caenorhabditis elegans . J Biol Chem. 2010;285(38):29319–29325. doi: 10.1074/jbc.M110.122663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spiegel CN, Batista-Pereira LG, Bretas JA, Eiras AE, Hooper AM, Peixoto AA, Soares MJ. Pheromone gland development and pheromone production in lutzomyia longipalpis (Diptera: psychodidae: phlebotominae) J Med Entomol. 2011;48(3):489–495. doi: 10.1603/ME10133. [DOI] [PubMed] [Google Scholar]
- 9.Diano S, Liu ZW, Jeong JK, Dietrich MO, Ruan HB, Kim E, Suyama S, Kelly K, Gyengesi E, Arbiser JL, Belsham DD, Sarruf DA, Schwartz MW, Bennett AM, Shanabrough M, Mobbs CV, Yang X, Gao XB, Horvath TL. Peroxisome proliferation-associated control of reactive oxygen species sets melanocortin tone and feeding in diet-induced obesity. Nat Med. 2011;17(9):1121–1127. doi: 10.1038/nm.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wanders RJ, Waterham HR. Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 11.Meijer WH, Gidijala L, Fekken S, Kiel JA, van den Berg MA, Lascaris R, Bovenberg RA, van der Klei IJ. Peroxisomes are required for efficient penicillin biosynthesis in Penicillium chrysogenum . Appl Environ Micobiol. 2010;76(17):5702–5709. doi: 10.1128/AEM.02327-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Müller WH, van der Krift TP, Krouwer AJ, Wösten HA, van der Voort LH, Smaal EB, Verkleij AJ. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum . EMBO J. 1991;10(2):489–495. doi: 10.1002/j.1460-2075.1991.tb07971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Widhalm JR, Ducluzeau AL, Buller NE, Elowsky CG, Olsen LJ, Basset GJ. Phylloquinone (Vitamin K(1)) Biosynthesis in plants: two peroxisomal thioesterases of lactobacillales origin hydrolyze 1,4-dihydroxy-2-naphthoyl-coa. Plant J. 2012;71(2):205–215. doi: 10.1111/j.1365-313X.2012.04972.x. [DOI] [PubMed] [Google Scholar]
- 14.Camões F, Bonekamp NA, Delille HK, Schrader M. Organelle dynamics and dysfunction: a closer link between peroxisomes and mitochondria. J Inherit Metab Dis. 2009;32(2):163–180. doi: 10.1007/s10545-008-1018-3. [DOI] [PubMed] [Google Scholar]
- 15.Reumann S, Weber AP. Plant peroxisomes respire in the light: some gaps of the photorespiratory C2 cycle have become filled–others remain. Biochim Biophys Acta. 2006;1763(12):1496–1510. doi: 10.1016/j.bbamcr.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 16.Wanders RJ, Waterham HR. Peroxisomal disorders: the single peroxisomal enzyme deficiencies. Biochim Biophys Acta. 2006;1763(12):1707–1720. doi: 10.1016/j.bbamcr.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Steinberg SJ, Dodt G, Raymond GV, Braverman NE, Moser AB, Moser HW. Peroxisome biogenesis disorders. Biochim Biophys Acta. 2006;1763(12):1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, Nibert ML, Superti-Furga G, Kagan JC. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141(4):668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kou J, Kovacs GG, Höftberger R, Kulik W, Brodde A, Forss-Petter S, Hönigschnabl S, Gleiss A, Brügger B, Wanders R, Just W, Budka H, Jungwirth S, Fischer P, Berger J. Peroxisomal alterations in Alzheimer’s disease. Acta Neuropathol. 2011;122(3):271–283. doi: 10.1007/s00401-011-0836-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lizard G, Rouaud O, Demarquoy J, Cherkaoui-Malki M, Iuliano L. Potential roles of peroxisomes in Alzheimer’s disease and in dementia of the Alzheimer’s type. J Alzheimers Dis. 2012;29(2):241–254. doi: 10.3233/JAD-2011-111163. [DOI] [PubMed] [Google Scholar]
- 21.Titorenko VI, Terlecky SR. Peroxisome metabolism and cellular aging. Traffic. 2011;12(3):252–259. doi: 10.1111/j.1600-0854.2010.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Distel B, Erdmann R, Gould SJ, Blobel G, Crane DI, Cregg JM, Dodt G, Fujiki Y, Goodman JM, Just WW, Kiel JA, Kunau WH, Lazarow PB, Mannaerts GP, Moser HW, Osumi T, Rachubinski RA, Roscher A, Subramani S, Tabak HF, Tsukamoto T, Valle D, van der Klei I, van Veldhoven PP, Veenhuis M. A unified nomenclature for peroxisome biogenesis factors. J Cell Biol. 1996;135(1):1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Islinger M, Grille S, Fahimi HD, Schrader M. The peroxisome: an update on mysteries. Histochem Cell Biol. 2012;137(5):547–574. doi: 10.1007/s00418-012-0941-4. [DOI] [PubMed] [Google Scholar]
- 24.Kiel JA, Veenhuis M, van der Klei IJ. PEX genes in fungal genomes: common, rare or redundant. Traffic. 2006;7(10):1291–1303. doi: 10.1111/j.1600-0854.2006.00479.x. [DOI] [PubMed] [Google Scholar]
- 25.Opaliński L, Veenhuis M, van der Klei IJ. Peroxisomes: membrane events accompanying peroxisome proliferation. Int J Biochem Cell Biol. 2011;43(6):847–851. doi: 10.1016/j.biocel.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Nagotu S, Veenhuis M, Van der Klei IJ. Divide et imera: the dictum of peroxisomes. Traffic. 2010;11(2):175–184. doi: 10.1111/j.1600-0854.2009.01019.x. [DOI] [PubMed] [Google Scholar]
- 27.Fagarasanu A, Mast FD, Knoblach B, Rachubinski RA. Molecular mechanism of organelle inheritance: lessons from peroxisomes in yeast. Nat Rev Mol Cell Biol. 2010;11(9):644–654. doi: 10.1038/nrm2960. [DOI] [PubMed] [Google Scholar]
- 28.Girzalsky W, Platta HW, Erdmann R. Protein transport across the peroxisomal membrane. Biol Chem. 2009;390(8):745–751. doi: 10.1515/BC.2009.104. [DOI] [PubMed] [Google Scholar]
- 29.Zipor G, Haim-Vilmovsky L, Gelin-Licht R, Gadir N, Brocard C, Gerst JE. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2009;106(47):19848–19853. doi: 10.1073/pnas.0910754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leon S, Goodman JM, Subramani S. Uniqueness of the mechanism of protein import into the peroxisome matrix: transport of folded, co-factor-bound and oligomeric proteins by shuttling receptors. Biochim Biophys Acta. 2006;1763(12):1552–1564. doi: 10.1016/j.bbamcr.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 31.Schnell DJ, Hebert DN. Protein translocons: multifunctional mediators of protein translocation across membranes. Cell. 2003;112(4):491–505. doi: 10.1016/S0092-8674(03)00110-7. [DOI] [PubMed] [Google Scholar]
- 32.Brocard C, Hartig A. Peroxisome targeting signal 1: is it really a simple tripeptide? Biochim Biophys Acta. 2006;1763(12):1565–1573. doi: 10.1016/j.bbamcr.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 33.Gosh D, Berg JM. A proteome-wide perspective on peroxisome targeting signal 1(PTS1)-Pex5p affinities. J Am Chem Soc. 2010;132(11):3973–3979. doi: 10.1021/ja9109049. [DOI] [PubMed] [Google Scholar]
- 34.Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108(5):1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lingner T, Kataya AR, Antonicelli GE, Benichou A, Nilssen K, Chen XY, Siemsen T, Morgenstern B, Meinicke P, Reumann S. Identification of novel plant peroxisomal targeting signals by a combination of machine learning methods and in vivo subcellular targeting analyses. Plant Cell. 2011;23(4):1556–1572. doi: 10.1105/tpc.111.084095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Leij I, van den Berg M, Boot R, Franse MM, Distel B, Tabak HF. Isolation of peroxisome assembly mutants from Saccharomyces cerevisiae with different morphologies using a novel positive selection procedure. J Cell Biol. 1992;119:153–162. doi: 10.1083/jcb.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fodor K, Wolf J, Erdmann R, Schliebs W, Wilmanns M. Molecular requirements for peroxisomal targeting of alanine-glyoxylate aminotransferase as an essential determinant in primary hyperoxaluria type 1. PLoS Biol. 2012;10(4):e1001309. doi: 10.1371/journal.pbio.1001309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanley WA, Filipp FV, Kursula P, Schüller N, Erdmann R, Schliebs W, Sattler M, Wilmanns M. Recognition of a functional peroxisome type 1 target by the dynamic import receptor pex5p. Mol Cell. 2006;24(5):653–663. doi: 10.1016/j.molcel.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazarow PB. The import receptor Pex7p and the PTS2 targeting sequence. Biochim Biophys Acta. 2006;1763(12):1599–1604. doi: 10.1016/j.bbamcr.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 40.Marzioch M, Erdmann R, Veenhuis M, Kunau W-H. PAS7 encodes a novel yeast member of the WD-40 protein family essential for import of 3-oxoacyl-CoA thiolase, a PTS2-containing protein, into peroxisomes. EMBO J. 1994;13(30):4908–4918. doi: 10.1002/j.1460-2075.1994.tb06818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO J. 1991;10(11):3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schliebs W, Kunau W-H. PTS2 co-receptors: diverse proteins with common features. Biochim Biophys Acta. 2006;1763(12):1605–1612. doi: 10.1016/j.bbamcr.2006.08.051. [DOI] [PubMed] [Google Scholar]
- 43.Galland N, Demeure F, Hannaert V, Verplaetse E, Vertommen D, Van der Smissen P, Courtoy PJ, Michels PA. Characterization of the role of the receptors PEX5 and PEX7 in the import of proteins into glycosomes of Trypanosoma brucei . Biochim Biophys Acta. 2007;1773(4):521–535. doi: 10.1016/j.bbamcr.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 44.Ramon NM, Bartel B. Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell. 2010;21(7):1263–1271. doi: 10.1091/mbc.E09-08-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otzen M, Wang D, Lunenborg MG, van der Klei IJ. Hansenula polymorpha Pex20p is an oligomer that binds the peroxisomal targeting signal 2 (PTS2) J Cell Sci. 2005;118(Pt 15):3409–3418. doi: 10.1242/jcs.02463. [DOI] [PubMed] [Google Scholar]
- 46.Zhang L, Léon S, Subramani S. Two independent pathways traffic the intraperoxisomal peroxin PpPex8p into peroxisomes: mechanism and evolutionary implications. Mol Biol Cell. 2006;17(2):690–699. doi: 10.1091/mbc.E05-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Motley AM, Hettema EH, Ketting R, Plasterk R, Tabak HF. Caenorhabditis elegans has a single pathway to target matrix proteins to peroxisomes. EMBO Rep. 2000;1(1):40–46. doi: 10.1093/embo-reports/kvd010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez NH, Felsner G, Schramm FD, Klingl A, Maier UG, Bolte K. A single peroxisomal targeting signal mediates matrix protein import in diatoms. PLoS ONE. 2011;6(9):e25316. doi: 10.1371/journal.pone.0025316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schäfer A, Kerssen D, Veenhuis M, Kunau WH, Schliebs W. Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol Cell Biol. 2004;24(20):8895–8906. doi: 10.1128/MCB.24.20.8895-8906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Klei IJ, Veenhuis M. PTS1-independent sorting of peroxisomal matrix proteins by Pex5p. Biochim Biophys Acta. 2006;1763(12):1794–1800. doi: 10.1016/j.bbamcr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 51.Williams C, Distel B. Pex13p: docking or cargo handling protein? Biochim Biophys Acta. 2006;1763(12):1585–1591. doi: 10.1016/j.bbamcr.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 52.Azevedo JE, Schliebs W. Pex14p, more than just a docking protein. Biochim Biophys Acta. 2006;1763(12):1574–1584. doi: 10.1016/j.bbamcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Neufeld C, Filipp FV, Simon B, Neuhaus A, Schüller N, David C, Kooshapur H, Madl T, Erdmann R, Schliebs W, Wilmanns M, Sattler M. Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19. EMBO J. 2009;28(6):745–754. doi: 10.1038/emboj.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiozawa K, Konarev PV, Neufeld C, Wilmanns M, Svergun DI. Solution structure of human Pex5.Pex14.PTS1 protein complexes obtained by small angle X-ray scattering. J Biol Chem. 2009;284(37):25334–25342. doi: 10.1074/jbc.M109.002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Su JR, Takeda K, Tamura S, Fujiki Y, Miki K. Crystal structure of the conserved N-terminal domain of the peroxisomal matrix protein import receptor, Pex14p. Proc Natl Acad Sci USA. 2009;106(2):417–421. doi: 10.1073/pnas.0808681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zutphen T, Veenhuis M, van der Klei IJ. Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy. 2008;4(1):63–66. doi: 10.4161/auto.5076. [DOI] [PubMed] [Google Scholar]
- 57.Bharti P, Schliebs W, Schievelbusch T, Neuhaus A, David C, Kock K, Herrmann C, Meyer HE, Wiese S, Warscheid B, Theiss C, Erdmann R. PEX14 is required for microtubule-based peroxisome motility in human cells. J Cell Sci. 2010;124(10):1759–1768. doi: 10.1242/jcs.079368. [DOI] [PubMed] [Google Scholar]
- 58.Huhse B, Rehling P, Albertini M, Blank L, Meller K, Kunau W-H. Pex17p of Saccharomyces cerevisiae is a novel peroxin and component of the peroxisomal protein translocation machinery. J Cell Biol. 1998;140(1):49–60. doi: 10.1083/jcb.140.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smith JJ, Szilard RK, Marelli M, Rachubinski RA. The peroxin Pex17p of the yeast Yarrowia lipolytica is associated peripherally with the peroxisomal membrane and is required for the import of a subset of matrix proteins. Mol Cell Biol. 1997;17(5):2511–2520. doi: 10.1128/mcb.17.5.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Managadze D, Würtz C, Wiese S, Schneider M, Girzalsky W, Meyer HE, Erdmann R, Warscheid B, Rottensteiner H. Identification of PEX33, a novel component of the peroxisomal docking complex in the filamentous fungus Neurospora crassa. Eur J Cell Biol. 2010;89(12):955–964. doi: 10.1016/j.ejcb.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 61.Opaliński L, Kiel JA, Homan TG, Veenhuis M, van der Klei IJ. Penicillium chrysogenum Pex14/17p–a novel component of the peroxisomal membrane that is important for penicillin production. FEBS J. 2010;277(15):3203–3218. doi: 10.1111/j.1742-4658.2010.07726.x. [DOI] [PubMed] [Google Scholar]
- 62.Erdmann R, Schliebs W. Peroxisomal matrix protein import: the transient pore model. Nat Rev Mol Cell Biol. 2005;6(9):738–742. doi: 10.1038/nrm1710. [DOI] [PubMed] [Google Scholar]
- 63.Gouveia AM, Guimaraes CP, Oliveira ME, Reguenga C, Sa-Miranda C, Azevedo JE. Characterization of the peroxisomal cycling receptor, Pex5p, using a cell-free in vitro import system. J Biol Chem. 2002;278(1):226–232. doi: 10.1074/jbc.M209498200. [DOI] [PubMed] [Google Scholar]
- 64.Gouveia AM, Guimaraes CP, Oliveira ME, Sa-Miranda C, Azevedo JE. Insertion of Pex5p into the peroxisomal membrane is cargo protein-dependent. J Biol Chem. 2002;278(7):4389–4392. doi: 10.1074/jbc.C200650200. [DOI] [PubMed] [Google Scholar]
- 65.Gouveia AM, Reguenga C, Oliveira ME, Sa-Miranda C, Azevedo JE. Characterization of peroxisomal Pex5p from rat liver: Pex5p in the Pex5p-Pex14p membrane complex is a transmembrane protein. J Biol Chem. 2000;275(42):32444–32451. doi: 10.1074/jbc.M004366200. [DOI] [PubMed] [Google Scholar]
- 66.Kerssen D, Hambruch E, Klaas W, Platta HW, de Kruijff B, Erdmann R, Kunau WH, Schliebs W. Membrane association of the cycling peroxisome import receptor Pex5p. J Biol Chem. 2006;281(37):27003–27015. doi: 10.1074/jbc.M509257200. [DOI] [PubMed] [Google Scholar]
- 67.Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105:187–196. doi: 10.1016/S0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- 68.Kunau W. Peroxisomes: the extended shuttle to the peroxisome matrix. Curr Biol. 2001;11(16):R659–R662. doi: 10.1016/S0960-9822(01)00386-4. [DOI] [PubMed] [Google Scholar]
- 69.Nair DM, Purdue PE, Lazarow PB. Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol. 2004;167(4):599–604. doi: 10.1083/jcb.200407119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Leon S, Zhang L, McDonald WH, Yates J, 3rd, Cregg JM, Subramani S. Dynamics of the peroxisomal import cycle of PpPex20p: ubiquitin-dependent localization and regulation. J Cell Biol. 2006;172(1):67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hensel A, Beck S, El Magraoui F, Platta HW, Girzalsky W, Erdmann R. Cysteine-dependent ubiquitination of Pex18p is linked to cargo translocation across the peroxisomal membrane. J Biol Chem. 2011;286:43495–43505. doi: 10.1074/jbc.M111.286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ma C, Schumann U, Rayapuram N, Subramani S. The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol Biol Cell. 2009;20(16):3680–3689. doi: 10.1091/mbc.E09-01-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meinecke M, Cizmowski C, Schliebs W, Kruger V, Beck S, Wagner R, Erdmann R. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010;12(3):273–277. doi: 10.1038/ncb2027. [DOI] [PubMed] [Google Scholar]
- 74.Freitas MO, Francisco T, Rodrigues TA, Alencastre IS, Pinto MP, Grou CP, Carvalho AF, Fransen M, Sá-Miranda C, Azevedo JE. PEX5 protein binds monomeric catalase blocking its tetramerization and releases it upon binding the N-terminal domain of PEX14. J Biol Chem. 2011;286(47):40509–40519. doi: 10.1074/jbc.M111.287201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang D, Visser NV, Veenhuis M, Van Der Klei IJ. Physical interactions of the peroxisomal targeting signal 1-receptor, Pex5p, studied by fluorescence correlation spectroscopy. J Biol Chem. 2003;278:43340–43345. doi: 10.1074/jbc.M307789200. [DOI] [PubMed] [Google Scholar]
- 76.Rehling P, Skaletz-Rorowski A, Girzalsky W, Voorn-Brouwer T, Franse MM, Distel B, Veenhuis M, Kunau W-H, Erdmann R. Pex8p, an intraperoxisomal peroxin of Saccharomyces cerevisiae required for protein transport into peroxisomes binds the PTS1 receptor pex5p. J Biol Chem. 2000;275(5):3593–3602. doi: 10.1074/jbc.275.5.3593. [DOI] [PubMed] [Google Scholar]
- 77.Agne B, Meindl NM, Niederhoff K, Einwächter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH. Pex8p. An intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell. 2003;11(3):635–646. doi: 10.1016/S1097-2765(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 78.Hazra PP, Suriapranata I, Snyder WB, Subramani S. Peroxisome remnants in pex3delta cells and the requirement of Pex3p for interactions between the peroxisomal docking and translocation subcomplexes. Traffic. 2002;3(8):560–574. doi: 10.1034/j.1600-0854.2002.30806.x. [DOI] [PubMed] [Google Scholar]
- 79.Kurochkin IV, Mizuno Y, Konagaya A, Sakaki Y, Schönbach C, Okazaki Y. Novel peroxisomal protease Tysnd1 processes PTS1- and PTS2-containing enzymes involved in beta-oxidation of fatty acids. EMBO J. 2007;26(3):835–845. doi: 10.1038/sj.emboj.7601525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Okumoto K, Kametani Y, Fujiki Y. Two proteases, Tysnd1 and PsLon, cooperatively regulate fatty-acid {beta}-oxidation in the peroxisomal matrix. J Biol Chem. 2011;286(52):44367–44379. doi: 10.1074/jbc.M111.285197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schuhmann H, Huesgen PF, Gietl C, Adamska I. The DEG15 serine protease cleaves peroxisomal targeting signal 2-containing proteins in Arabidopsis. Plant Physiol. 2008;148(4):1847–1856. doi: 10.1104/pp.108.125377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oeljeklaus S, Reinartz BS, Wolf J, Wiese S, Tonillo J, Podwojski K, Kuhlmann K, Stephan C, Meyer HE, Schliebs W, Brocard C, Erdmann R, Warscheid B. Identification of core components and transient interactors of the peroxisomal importomer by dual-track stable isotope labeling with amino acids in cell culture analysis. J Proteome Res. 2012;11(4):2567–2580. doi: 10.1021/pr3000333. [DOI] [PubMed] [Google Scholar]
- 83.Rosenkranz K, Birschmann I, Grunau S, Girzalsky W, Kunau WH, Erdmann R. Functional association of the AAA complex and the peroxisomal importomer. FEBS J. 2006;273(16):3804–3815. doi: 10.1111/j.1742-4658.2006.05388.x. [DOI] [PubMed] [Google Scholar]
- 84.Miyata N, Fujiki Y. Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol Cell Biol. 2005;25(24):10822–10832. doi: 10.1128/MCB.25.24.10822-10832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miyata N, Hosoi K, Mukai S, Fujiki Y. In vitro import of peroxisome-targeting signal type 2 (PTS2) receptor Pex7p into peroxisomes. Biochim Biophys Acta. 2009;1793(5):860–870. doi: 10.1016/j.bbamcr.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 86.Oliveira ME, Gouveia AM, Pinto RA, Sa-Miranda C, Azevedo JE. The energetics of Pex5p-mediated peroxisomal protein import. J Biol Chem. 2003;278(41):39483–39488. doi: 10.1074/jbc.M305089200. [DOI] [PubMed] [Google Scholar]
- 87.Platta HW, Grunau S, Rosenkranz K, Girzalsky W, Erdmann R. Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat Cell Biol. 2005;7(8):817–822. doi: 10.1038/ncb1281. [DOI] [PubMed] [Google Scholar]
- 88.El Magraoui F, Bäumer BE, Platta HW, Baumann JS, Girzalsky W, Erdmann R. The RING-type ubiquitin ligases Pex2p, Pex10p and Pex12p form a heteromeric complex that displays enhanced activity in an ubiquitin conjugating enzyme-selective manner. FEBS J. 2012;279(11):2060–2070. doi: 10.1111/j.1742-4658.2012.08591.x. [DOI] [PubMed] [Google Scholar]
- 89.Grou CP, Carvalho AF, Pinto MP, Wiese S, Piechura H, Meyer HE, Warscheid B, Sa-Miranda C, Azevedo JE. Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J Biol Chem. 2008;283(21):14190–14197. doi: 10.1074/jbc.M800402200. [DOI] [PubMed] [Google Scholar]
- 90.Platta HW, El Magraoui F, Baumer BE, Schlee D, Girzalsky W, Erdmann R. Pex2 and Pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol. 2009;29(20):5505–5516. doi: 10.1128/MCB.00388-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol. 2007;177(2):197–204. doi: 10.1083/jcb.200611012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Williams C, van den Berg M, Geers E, Distel B. Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun. 2008;374(4):620–624. doi: 10.1016/j.bbrc.2008.07.054. [DOI] [PubMed] [Google Scholar]
- 93.Williams C, van den Berg M, Sprenger RR, Distel B. A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem. 2007;282(31):22534–22543. doi: 10.1074/jbc.M702038200. [DOI] [PubMed] [Google Scholar]
- 94.Miyata N, Okumoto K, Mukai S, Noguchi M, Fujiki Y. AWP1/ZFAND6 functions in Pex5 export by interacting with Cys-mono ubiquitinated Pex5 and Pex6 AAA ATPase. Traffic. 2011;13(1):168–183. doi: 10.1111/j.1600-0854.2011.01298.x. [DOI] [PubMed] [Google Scholar]
- 95.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 96.Wang X, Herr RA, Hansen TH. Ubiquitination of substrates by esterification. Traffic. 2012;13(1):19–24. doi: 10.1111/j.1600-0854.2011.01269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Carvalho AF, Pinto MP, Grou CP, Alencastre IS, Fransen M, Sa-Miranda C, Azevedo JE. Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J Biol Chem. 2007;282(43):31267–31272. doi: 10.1074/jbc.M706325200. [DOI] [PubMed] [Google Scholar]
- 98.Okumoto K, Misono S, Miyata N, Matsumoto Y, Mukai S, Fujiki Y. Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic. 2011;12(8):1067–1083. doi: 10.1111/j.1600-0854.2011.01217.x. [DOI] [PubMed] [Google Scholar]
- 99.Crane DI, Kalish JE, Gould SJ. The Pichia pastoris PAS4 gene encodes a ubiquitin-conjugation enzyme required for peroxisome assembly. J Biol Chem. 1994;269(34):21835–21844. [PubMed] [Google Scholar]
- 100.van der Klei IJ, Hilbrands RE, Kiel JAKW, Rasmussen SW, Cregg JM, Veenhuis M. The ubiquitin-conjugating enzyme Pex4p of Hansenula polymorpha is required for efficient functioning of the PTS1 import machinery. EMBO J. 1998;17(13):3608–3618. doi: 10.1093/emboj/17.13.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wiebel FF, Kunau W-H. The PAS2 protein essential for peroxisome biogenesis is related to ubiquitin-conjugating enzymes. Nature. 1992;359(6390):73–76. doi: 10.1038/359073a0. [DOI] [PubMed] [Google Scholar]
- 102.Zolman BK, Monroe-Augustus M, Silva ID, Bartel B. Identification and functional characterization of Arabidopsis PEROXIN4 and the interacting protein PEROXIN22. Plant Cell. 2005;17(12):3422–3435. doi: 10.1105/tpc.105.035691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koller A, Snyder WB, Faber KN, Wenzel TJ, Rangell L, Keller GA, Subramani S. Pex22p of Pichia pastoris, essential for peroxisomal matrix protein import, anchors the ubiquitin-conjugating enzyme, Pex4p, on the peroxisomal membrane. J Cell Biol. 1999;146(1):99–112. doi: 10.1083/jcb.146.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Williams C, van den Berg M, Panjikar S, Stanley WA, Distel B, Wilmanns M. Insights into ubiquitin-conjugating enzyme/co-activator interactions from the structure of the Pex4p:pex22p complex. EMBO J. 2011;31(2):391–402. doi: 10.1038/emboj.2011.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 106.Bazirgan OA, Hampton RY. Cue1p is an activator of Ubc7p E2 activity in vitro and in vivo. J Biol Chem. 2008;283(19):12797–12810. doi: 10.1074/jbc.M801122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kostova Z, Mariano J, Scholz S, Koenig C, Weissman AM. A Ubc7p-binding domain in Cue1p activates ER-associated protein degradation. J Cell Sci. 2009;122(9):1374–1381. doi: 10.1242/jcs.044255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brzovic PS, Klevit RE. Ubiquitin transfer from the E2 perspective: why is UbcH5 so promiscuous? Cell Cycle. 2006;5:2867–2873. doi: 10.4161/cc.5.24.3592. [DOI] [PubMed] [Google Scholar]
- 109.Gonen H, Bercovich B, Orian A, Carrano A, Takizawa C, Yamanaka K, Pagano M, Iwai K, Ciechanover A. Identification of the ubiquitin carrier proteins, E2 s, involved in signal-induced conjugation and subsequent degradation of IkappaBalpha. J Biol Chem. 1999;274(21):14823–14830. doi: 10.1074/jbc.274.21.14823. [DOI] [PubMed] [Google Scholar]
- 110.Saville MK, Sparks A, Xirodimas DP, Wardrop J, Stevenson LF, Bourdon JC, Woods YL, Lane DP. Regulation of p53 by the ubiquitin-conjugating enzymes UbcH5B/C in vivo. J Biol Chem. 2004;279(40):42169–42181. doi: 10.1074/jbc.M403362200. [DOI] [PubMed] [Google Scholar]
- 111.Kallijärvi J, Lahtinen U, Hämäläinen R, Lipsanen-Nyman M, Palvimo JJ, Lehesjoki AE. TRIM37 defective in mulibrey nanism is a novel RING finger ubiquitin E3 ligase. Exp Cell Res. 2005;308(1):146–155. doi: 10.1016/j.yexcr.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 112.Kallijarvi J, Avela K, Lipsanen-Nyman M, Ulmanen I, Lehesjoki AE. The TRIM37 gene encodes a peroxisomal ring-b-box-coiled-coil protein: classification of Mulibrey Nanism as a New Peroxisomal Disorder. Am J Hum Genet. 2002;70(5):1215–1228. doi: 10.1086/340256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ebberink MS, Mooijer PA, Gootjes J, Koster J, Wanders RJ, Waterham HR. Genetic classification and mutational spectrum of more than 600 patients with a Zellweger syndrome spectrum disorder. Hum Mutat. 2011;32(1):59–69. doi: 10.1002/humu.21388. [DOI] [PubMed] [Google Scholar]
- 114.Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Shirayoshi Y, Mori T, Fujiki Y. A human gene responsible for Zellweger syndrome that affects peroxisome assembly. Science. 1992;255(5048):1132–1134. doi: 10.1126/science.1546315. [DOI] [PubMed] [Google Scholar]
- 115.Tsukamoto T, Miura S, Fujiki Y. Restoration by a 35 K membrane protein of peroxisome assembly in a peroxisome-deficient mammalian cell mutant. Nature. 1991;350(6313):77–81. doi: 10.1038/350077a0. [DOI] [PubMed] [Google Scholar]
- 116.Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun. 2007;354(2):329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 118.Burroughs AM, Iyer LM, Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol BioSyst. 2011;7(7):2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Freemont PS, Hanson IM, Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991;64(3):483–484. doi: 10.1016/0092-8674(91)90229-r. [DOI] [PubMed] [Google Scholar]
- 120.Koellensperger G, Daubert S, Erdmann R, Hann S, Rottensteiner H. Characterisation of zinc-binding domains of peroxisomal RING finger proteins using size exclusion chromatography/inductively coupled plasma-mass spectrometry. Biol Chem. 2007;388(11):1209–1214. doi: 10.1515/BC.2007.125. [DOI] [PubMed] [Google Scholar]
- 121.Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8(3):209. doi: 10.1186/gb-2007-8-3-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Albertini M, Girzalsky W, Veenhuis M, Kunau W-H. Pex12p of Saccharomyces cerevisiae is a component of a multi-protein complex essential for peroxisomal matrix protein import. Eur J Cell Biol. 2001;80(4):257–270. doi: 10.1078/0171-9335-00164. [DOI] [PubMed] [Google Scholar]
- 123.Chang CC, Warren DS, Sacksteder KA, Gould SJ. PEX12 interacts with PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J Cell Biol. 1999;147(4):761–774. doi: 10.1083/jcb.147.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eckert JH, Johnsson N. Pex10p links the ubiquitin conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J Cell Sci. 2003;116(17):3623–3634. doi: 10.1242/jcs.00678. [DOI] [PubMed] [Google Scholar]
- 125.Okumoto K, Abe I, Fujiki Y. Molecular Anatomy of the Peroxin Pex12p: rING Finger Domain Is Essential for the Pex12p Function and Interacts with the Peroxisome Targeting Signal Type 1-Receptor Pex5p and a RING Peroxin, Pex10p. J Biol Chem. 2000;275(33):25700–25710. doi: 10.1074/jbc.M003303200. [DOI] [PubMed] [Google Scholar]
- 126.Kiel JA, Emmrich K, Meyer HE, Kunau WH. Ubiquitination of the peroxisomal targeting signal type 1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J Biol Chem. 2005;280(3):1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 127.Kragt A, Voorn-Brouwer T, van den Berg M, Distel B. The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is mono ubiquitinated in wild type cells. J Biol Chem. 2005;280(9):7867–7874. doi: 10.1074/jbc.M413553200. [DOI] [PubMed] [Google Scholar]
- 128.Platta HW, Girzalsky W, Erdmann R. Ubiquitination of the peroxisomal import receptor Pex5p. Biochem J. 2004;384(Pt 1):37–45. doi: 10.1042/BJ20040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wei J, Zhai L, Xu J, Wang H. Role of Bmi1 in H2A ubiquitylation and Hox gene silencing. J Biol Chem. 2006;281(32):22537–22544. doi: 10.1074/jbc.M600826200. [DOI] [PubMed] [Google Scholar]
- 130.Alchanati I, Teicher C, Cohen G, Shemesh V, Barr HM, Nakache P, Ben-Avraham D, Idelevich A, Angel I, Livnah N, Tuvia S, Reiss Y, Taglicht D, Erez O. The E3 ubiquitin-ligase Bmi1/Ring1A controls the proteasomal degradation of Top2alpha cleavage complex—a potentially new drug target. PLoS ONE. 2009;4:e8104. doi: 10.1371/journal.pone.0008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24(5):701–711. doi: 10.1016/j.molcel.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 132.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25(11):2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Linares LK, Hengstermann A, Ciechanover A, Müller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100(21):12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mallery DL, Vandenberg CJ, Hiom K. Activation of the E3 ligase function of the BRCA1/BARD1 complex by poly ubiquitin chains. EMBO J. 2002;21(24):6755–6762. doi: 10.1093/emboj/cdf691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Okamoto K, Taya Y, Nakagama H. Mdmx enhances p53 ubiquitination by altering the substrate preference of the Mdm2 ubiquitin ligase. FEBS Lett. 2009;583(17):2710–2714. doi: 10.1016/j.febslet.2009.07.021. [DOI] [PubMed] [Google Scholar]
- 136.Nito K, Kamigaki A, Kondo M, Hayashi M, Nishimura M. Functional classification of Arabidopsis peroxisome biogenesis factors proposed from analyses of knockdown mutants. Plant Cell Physiol. 2007;48(6):763–774. doi: 10.1093/pcp/pcm053. [DOI] [PubMed] [Google Scholar]
- 137.Prestele J, Hierl G, Scherling C, Hetkamp S, Schwechheimer C, Isono E, Weckwerth W, Wanner G, Gietl C. Different functions of the C3HC4 zinc RING finger peroxins PEX10, PEX2, and PEX12 in peroxisome formation and matrix protein import. Proc Natl Acad Sci USA. 2010;107(33):14915–14920. doi: 10.1073/pnas.1009174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schumann U, Prestele J, O’Geen H, Brueggeman R, Wanner G, Gietl C. Requirement of the C3HC4 zinc RING finger of the Arabidopsis PEX10 for photorespiration and leaf peroxisome contact with chloroplasts. Proc Natl Acad Sci USA. 2007;104(3):1069–1074. doi: 10.1073/pnas.0610402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Berteaux-Lecellier V, Picard M, Thompson-Coffe C, Zickler D, Panvier-Adoutte A, Simonet JM. A nonmammalian homolog of the PAF1 gene (Zellweger syndrome) discovered as a gene involved in caryogamy in the fungus Podospora anserina . Cell. 1995;81(7):1043–1051. doi: 10.1016/S0092-8674(05)80009-1. [DOI] [PubMed] [Google Scholar]
- 140.Peraza-Reyes L, Arnaise S, Zickler D, Coppin E, Debuchy R, Berteaux-Lecellier V. The importomer peroxins are differentially required for peroxisome assembly and meiotic development in Podospora anserina: insights into a new peroxisome import pathway. Mol Microbiol. 2011;82(2):365–377. doi: 10.1111/j.1365-2958.2011.07816.x. [DOI] [PubMed] [Google Scholar]
- 141.Peraza-Reyes L, Zickler D, Berteaux-Lecellier V. The peroxisome RING-finger complex is required for meiocyte formation in the fungus Podospora anserina . Traffic. 2008;9(11):1998–2009. doi: 10.1111/j.1600-0854.2008.00812.x. [DOI] [PubMed] [Google Scholar]
- 142.Dodt G, Gould SJ. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol. 1996;135:1763–1774. doi: 10.1083/jcb.135.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fujiki Y, Nashiro C, Miyata N, Tamura S. Okumoto K (2012) New insights into dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p in shuttling of PTS1-receptor Pex5p during peroxisome biogenesis. Biochim Biophys Acta. 1823;1:145–149. doi: 10.1016/j.bbamcr.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 144.Grimm I, Saffian D, Platta HW. Erdmann R (2012) The AAA-type ATPases Pex1p and Pex6p and their role in peroxisomal matrix protein import in Saccharomyces cerevisiae . Biochim Biophys Acta. 1823;1:150–158. doi: 10.1016/j.bbamcr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 145.Platta HW, Debelyy MO, El Magraoui F, Erdmann R. The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p. Biochem Soc Trans. 2008;36:99–104. doi: 10.1042/BST0360099. [DOI] [PubMed] [Google Scholar]
- 146.Birschmann I, Rosenkranz K, Erdmann R, Kunau WH. Structural and functional analysis of the interaction of the AAA-peroxins Pex1p and Pex6p. FEBS J. 2005;272(1):47–58. doi: 10.1111/j.1432-1033.2004.04393.x. [DOI] [PubMed] [Google Scholar]
- 147.Kiel JA, Hilbrands RE, Bovenberg RA, Veenhuis M. Isolation of Penicillium chrysogenum PEX1 and PEX6 encoding AAA proteins involved in peroxisome biogenesis. Appl Microbiol Biotechnol. 2000;54(2):238–242. doi: 10.1007/s002530000378. [DOI] [PubMed] [Google Scholar]
- 148.Kiel JA, Hilbrands RE, van der Klei IJ, Rasmussen SW, Salomons FA, van der Heide M, Faber KN, Cregg JM, Veenhuis M. Hansenula polymorpha Pex1p and Pex6p are peroxisome-associated AAA proteins that functionally and physically interact. Yeast. 1999;15(11):1059–1078. doi: 10.1002/(SICI)1097-0061(199908)15:11<1059::AID-YEA434>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 149.Tamura S, Shimozawa N, Suzuki Y, Tsukamoto T, Osumi T, Fujiki Y. A cytoplasmic AAA family peroxin, Pex1p, interacts with Pex6p. Biochem Biophys Res Commun. 1998;245(3):883–886. doi: 10.1006/bbrc.1998.8522. [DOI] [PubMed] [Google Scholar]
- 150.Tamura S, Yasutake S, Matsumoto N, Fujiki Y. Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J Biol Chem. 2006;281(38):27693–27704. doi: 10.1074/jbc.M605159200. [DOI] [PubMed] [Google Scholar]
- 151.Birschmann I, Stroobants AK, Van Den Berg M, Schäfer A, Rosenkranz K, Kunau WH, Tabak HF. Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol Biol Cell. 2003;14(6):2226–2236. doi: 10.1091/mbc.E02-11-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Furuki S, Tamura S, Matsumoto N, Miyata N, Moser A, Moser HW, Fujiki Y. Mutations in the peroxin Pex26p responsible for peroxisome biogenesis disorders of complementation group 8 impair its stability, peroxisomal localization, and interaction with the Pex1p x Pex6p complex. J Biol Chem. 2006;281(3):1317–1323. doi: 10.1074/jbc.M510044200. [DOI] [PubMed] [Google Scholar]
- 153.Goto S, Mano S, Nakamori C, Nishimura M. Arabidopsis ABERRANT PEROXISOME MORPHOLOGY9 is a peroxin that recruits the PEX1-PEX6 complex to peroxisomes. Plant Cell. 2011;23(4):1573–1587. doi: 10.1105/tpc.110.080770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Matsumoto N, Tamura S, Fujiki Y. The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. NatCellBiol. 2003;5(5):454–460. doi: 10.1038/ncb982. [DOI] [PubMed] [Google Scholar]
- 155.Matsumoto N, Tamura S, Furuki S, Miyata N, Moser A, Shimozawa N, Moser HW, Suzuki Y, Kondo N, Fujiki Y. mutations in novel peroxin gene PEX26 that cause peroxisome-biogenesis disorders of complementation group 8 provide a genotype-phenotype correlation. Am J Hum Genet. 2003;73(2):233–246. doi: 10.1086/377004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Geisbrecht BV, Collins CS, Reuber BE, Gould SJ. Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci U S A. 1998;95(15):8630–8635. doi: 10.1073/pnas.95.15.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Beyer A. Sequence analysis of the AAA protein family. Protein Sci. 1997;6(10):2043–2058. doi: 10.1002/pro.5560061001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Erdmann R, Wiebel FF, Flessau A, Rytka J, Beyer A, Fröhlich KU, Kunau W-H. PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell. 1991;64(3):499–510. doi: 10.1016/0092-8674(91)90234-P. [DOI] [PubMed] [Google Scholar]
- 159.Kunau W-H, Beyer A, Franken T, Götte K, Marzioch M, Saidowsky J, Skaletz-Rorowski A, Wiebel FF. Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: forward and reversed genetics. Biochimie. 1993;75:209–224. doi: 10.1016/0300-9084(93)90079-8. [DOI] [PubMed] [Google Scholar]
- 160.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA + : a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9(1):27–43. [PubMed] [Google Scholar]
- 162.Wendler P, Ciniawsky S, Kock M. Kube S (2012) Structure and function of the AAA + nucleotide binding pocket. Biochim Biophys Acta. 1823;1:2–14. doi: 10.1016/j.bbamcr.2011.06.014. [DOI] [PubMed] [Google Scholar]
- 163.Dargemont C, Ossareh-Nazari B. Cdc48/p97, a key actor in the interplay between autophagy and ubiquitin/proteasome catabolic pathways. Biochim Biophys Acta. 2012;1:138–414. doi: 10.1016/j.bbamcr.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 164.Zhao C, Smith EC. Whiteheart SW (2012) Requirements for the catalytic cycle of the N-ethylmaleimide-sensitive factor (NSF) Biochim Biophys Acta. 1823;1:159–171. doi: 10.1016/j.bbamcr.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nashiro C, Kashiwagi A, Matsuzaki T, Tamura S, Fujiki Y. recruiting mechanism of the AAA peroxins, Pex1p and Pex6p, to Pex26p on peroxisome membrane. Traffic. 2011;12(6):774–788. doi: 10.1111/j.1600-0854.2011.01182.x. [DOI] [PubMed] [Google Scholar]
- 166.Beuron F, Flynn TC, Ma J, Kondo H, Zhang X, Freemont PS. Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantised elastic deformational model. J Mol Biol. 2003;327(3):619–629. doi: 10.1016/S0022-2836(03)00178-5. [DOI] [PubMed] [Google Scholar]
- 167.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA + ATPases. J Struct Biol. 2004;146(1–2):11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 168.Bebeacua C, Förster A, McKeown C, Meyer HH, Zhang X, Freemont PS. Distinct conformations of the protein complex p97-Ufd1-Npl4 revealed by electron cryomicroscopy. Proc Natl Acad Sci USA. 2012;109(4):1098–1103. doi: 10.1073/pnas.1114341109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16(5):715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 170.Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14(2):117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- 171.Titorenko VI, Rachubinski RA. peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J Cell Biol. 2000;150(4):881–886. doi: 10.1083/jcb.150.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.van der Zand A, Gent J, Braakman I, Tabak HF. Biochemically distinct vesicles from the endoplasmic reticulum fuse to form peroxisomes. Cell. 2012;149(2):397–409. doi: 10.1016/j.cell.2012.01.054. [DOI] [PubMed] [Google Scholar]
- 173.Jungwirth H, Ring J, Mayer T, Schauer A, Buttner S, Eisenberg T, Carmona-Gutierrez D, Kuchler K, Madeo F. Loss of peroxisome function triggers necrosis. FEBS Lett. 2008;582(19):2882–2886. doi: 10.1016/j.febslet.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 174.Seo JG, Lai CY, Miceli MV, Jazwinski SM. A novel role of peroxin PEX6: suppression of aging defects in mitochondria. Aging Cell. 2007;6(3):405–413. doi: 10.1111/j.1474-9726.2007.00291.x. [DOI] [PubMed] [Google Scholar]
- 175.Warner DR, Roberts EA, Greene RM, Pisano MM. Identification of novel Smad binding proteins. Biochem Biophys Res Commun. 2003;312(4):1185–1190. doi: 10.1016/j.bbrc.2003.11.049. [DOI] [PubMed] [Google Scholar]
- 176.Dai RM, Li CC. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol. 2001;3(8):740–744. doi: 10.1038/35087056. [DOI] [PubMed] [Google Scholar]
- 177.Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and poly ubiquitin chains. J Cell Biol. 2003;162(1):71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65(15):2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shiozawa K, Maita N, Tomii K, Seto A, Goda N, Akiyama Y, Shimizu T, Shirakawa M, Hiroaki H. Structure of the N-terminal domain of PEX1 AAA-ATPase. Characterization of a putative adaptor-binding domain. J Biol Chem. 2004;279(48):50060–50068. doi: 10.1074/jbc.M407837200. [DOI] [PubMed] [Google Scholar]
- 180.Park S, Isaacson R, Kim HT, Silver PA, Wagner G. Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure. 2005;13(7):995–1005. doi: 10.1016/j.str.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 181.Fenner BJ, Scannell M, Prehn JH. Identification of poly ubiquitin binding proteins involved in NF-kappaB signaling using protein arrays. Biochim Biophys Acta. 2009;1794(7):1010–1016. doi: 10.1016/j.bbapap.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 182.Evans PC, Smith TS, Lai MJ, Williams MG, Burke DF, Heyninck K, Kreike MM, Beyaert R, Blundell TL, Kilshaw PJ. A novel type of deubiquitinating enzyme. J Biol Chem. 2003;278(25):23180–23186. doi: 10.1074/jbc.M301863200. [DOI] [PubMed] [Google Scholar]
- 183.Raiborg C, Slagsvold T, Stenmark H. A new side to ubiquitin. Trends Biochem Sci. 2006;31(10):541–544. doi: 10.1016/j.tibs.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 184.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 185.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 186.Debelyy MO, Platta HW, Saffian D, Hensel A, Thoms S, Meyer HE, Warscheid B, Girzalsky W, Erdmann R. Ubp15p, a ubiquitin hydrolase associated with the peroxisomal export machinery. J Biol Chem. 2011;286(32):28223–28234. doi: 10.1074/jbc.M111.238600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Grou CP, Carvalho AF, Pinto MP, Huybrechts SJ, Sa-Miranda C, Fransen M, Azevedo JE. Properties of the ubiquitin-Pex5p thiol ester conjugate. J Biol Chem. 2009;284(16):10504–10513. doi: 10.1074/jbc.M808978200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Grou CP, Francisco T, Rodrigues TA, Freitas MO, Pinto MP, Carvalho AF, Domingues P, Wood SA, Rodríguez-Borges JE, Sá-Miranda C, Fransen M, Azevedo JE. Identification of ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting on the ubiquitin-peroxin 5 (PEX5) thioester conjugate. J Biol Chem. 2012;287(16):12815–12827. doi: 10.1074/jbc.M112.340158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, Martello G, Stinchfield MJ, Soligo S, Morsut L, Inui M, Moro S, Modena N, Argenton F, Newfeld SJ, Piccolo S. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 mono ubiquitination. Cell. 2009;136(1):123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 190.Schwickart M, Huang X, Lill JR, Liu J, Ferrando R, French DM, Maecker H, O’Rourke K, Bazan F, Eastham-Anderson J, Yue P, Dornan D, Huang DC, Dixit VM. Deubiquitinase USP9X stabilizes MCL1 and promotes tumour cell survival. Nature. 2010;463:103–107. doi: 10.1038/nature08646. [DOI] [PubMed] [Google Scholar]
- 191.Nathan JA, Sengupta S, Wood SA, Admon A, Markson G, Sanderson C, Lehner PJ. The ubiquitin E3 ligase MARCH7 is differentially regulated by the deubiquitylating enzymes USP7 and USP9X. Traffic. 2008;9(7):1130–1145. doi: 10.1111/j.1600-0854.2008.00747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Imanaka T, Small GM, Lazarow PB. Translocation of acyl-CoA oxidase into peroxisomes requires ATP hydrolysis but not a membrane potential. J Cell Biol. 1987;105:2915–2922. doi: 10.1083/jcb.105.6.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gabaldon T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA. Origin and evolution of the peroxisomal proteome. Biol Direct. 2006;1:8. doi: 10.1186/1745-6150-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Schluter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A. The evolutionary origin of peroxisomes: an ER-peroxisome connection. Mol Biol Evol. 2006;23(4):838–845. doi: 10.1093/molbev/msj103. [DOI] [PubMed] [Google Scholar]
- 195.Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- 196.Taylor EB, Rutter J. Mitochondrial quality control by the ubiquitin-proteasome system. Biochem Soc Trans. 2011;39(5):1509–1513. doi: 10.1042/BST0391509. [DOI] [PubMed] [Google Scholar]
- 197.Schliebs W, Girzalsky W, Erdmann R. Peroxisomal protein import and ERAD: variations on a common theme. Nat Rev Mol Cell Biol. 2010;11(12):885–890. doi: 10.1038/nrm3008. [DOI] [PubMed] [Google Scholar]
- 198.Ratzel SE, Lingard MJ, Woodward AW, Bartel B. Reducing PEX13 expression ameliorates physiological defects of late-acting peroxin mutants. Traffic. 2011;12(1):121–134. doi: 10.1111/j.1600-0854.2010.01136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Alencastre IS, Rodrigues TA, Grou CP, Fransen M, Sá-Miranda C, Azevedo JE. Mapping the cargo protein membrane translocation step into the PEX5 cycling pathway. J Biol Chem. 2009;284(40):27243–27251. doi: 10.1074/jbc.M109.032565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Seufert W, Jentsch S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 1990;9(2):543–550. doi: 10.1002/j.1460-2075.1990.tb08141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Seufert W, McGrath JP, Jentsch S. UBC1 encodes a novel member of an essential subfamily of yeast ubiquitin-conjugating enzymes involved in protein degradation. EMBO J. 1990;9(13):4535–4541. doi: 10.1002/j.1460-2075.1990.tb07905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Leon S, Subramani S. A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol. J Biol Chem. 2007;282(10):7424–7430. doi: 10.1074/jbc.M611627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Carvalho AF, Grou CP, Pinto MP, Alencastre IS, Costa-Rodrigues J, Fransen M, Sa-Miranda C, Azevedo JE. Functional characterization of two missense mutations in Pex5p - C11S and N526 K. Biochim Biophys Acta. 2007;1773(7):1141–1148. doi: 10.1016/j.bbamcr.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 204.Collins CS, Kalish JE, Morrell JC, McCaffery JM, Gould SJ. The peroxisome biogenesis factors Pex4p, Pex22p, Pex1p, and Pex6p Act in the terminal steps of peroxisomal matrix protein import. Mol Cell Biol. 2000;20(20):7516–7526. doi: 10.1128/MCB.20.20.7516-7526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zolman BK, Bartel B. An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci USA. 2004;101(6):1786–1791. doi: 10.1073/pnas.0304368101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kiel JA, Otzen M, Veenhuis M, van der Klei IJ. Obstruction of poly ubiquitination affects PTS1 peroxisomal matrix protein import. Biochim Biophys Acta. 2005;1745(2):176–186. doi: 10.1016/j.bbamcr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 207.Lingard MJ, Monroe-Augustus M, Bartel B. Peroxisome-associated matrix protein degradation in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(11):4561–4566. doi: 10.1073/pnas.0811329106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Purdue PE, Lazarow PB. Pex18p is constitutively degraded during peroxisome biogenesis. J Biol Chem. 2001;276(50):47684–47689. doi: 10.1074/jbc.M106823200. [DOI] [PubMed] [Google Scholar]
- 209.Cadwell K, Coscoy L. Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase. Science. 2005;309:127–130. doi: 10.1126/science.1110340. [DOI] [PubMed] [Google Scholar]
- 210.Wang X, Herr RA, Chua WJ, Lybarger L, Wiertz EJ, Hansen TH. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. J Cell Biol. 2007;177(4):613–624. doi: 10.1083/jcb.200611063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Randow F, Lehner PJ. Viral avoidance and exploitation of the ubiquitin system. Nat Cell Biol. 2009;11(5):527–534. doi: 10.1038/ncb0509-527. [DOI] [PubMed] [Google Scholar]
- 212.Ishikura S, Weissman AM, Bonifacino JS. Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem. 2010;285(31):23916–23924. doi: 10.1074/jbc.M110.127936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Shimizu Y, Okuda-Shimizu Y, Hendershot LM. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell. 2010;40(6):917–926. doi: 10.1016/j.molcel.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Cadwell K, Coscoy L. The specificities of Kaposi’s sarcoma-associated herpesvirus-encoded E3 ubiquitin ligases are determined by the positions of lysine or cysteine residues within the intracytoplasmic domains of their targets. J Virol. 2008;82(8):4184–4189. doi: 10.1128/JVI.02264-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–108. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Berleth ES, Pickart CM. Mechanism of ubiquitin conjugating enzyme E2–230 K: catalysis involving a thiol relay? Biochemistry. 1996;35(5):1664–1671. doi: 10.1021/bi952105y. [DOI] [PubMed] [Google Scholar]
- 217.Grou CP, Carvalho AF, Pinto MP, Alencastre IS, Rodrigues TA, Freitas MO, Francisco T, Sa-Miranda C, Azevedo JE. The peroxisomal protein import machinery–a case report of transient ubiquitination with a new flavor. Cell Mol Life Sci. 2009;66(2):254–262. doi: 10.1007/s00018-008-8415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ivashchenko O, Van Veldhoven PP, Brees C, Ho YS, Terlecky SR, Fransen M. Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Mol Biol Cell. 2011;22(9):1440–1451. doi: 10.1091/mbc.E10-11-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bonekamp NA, Völkl A, Fahimi HD, Schrader M. Reactive oxygen species and peroxisomes: struggling for balance. BioFactors. 2009;35(4):346–355. doi: 10.1002/biof.48. [DOI] [PubMed] [Google Scholar]
- 220.Fransen M, Nordgren M, Wang B. Apanasets O (2012) Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim Biophys Acta. 1822;9:1363–1373. doi: 10.1016/j.bbadis.2011.12.001. [DOI] [PubMed] [Google Scholar]