Abstract
Melanocytes that lack the GTPase Rab27a (ashen) are disabled in myosin Va-dependent melanosome capture because the association of the myosin with the melanosome surface depends on the presence of this resident melanosomal membrane protein. One interpretation of these observations is that Rab27a functions wholly or in part as the melanosome receptor for myosin Va (Myo5a). Herein, we show that the ability of the myosin Va tail domain to localize to the melanosome and generate a myosin Va null (dilute) phenotype in wild-type melanocytes is absolutely dependent on the presence of exon F, one of two alternatively spliced exons present in the tail of the melanocyte-spliced isoform of myosin Va but not the brain-spliced isoform. Exon D, the other melanocyte-specific tail exon, is not required. Similarly, the ability of full-length myosin Va to colocalize with melanosomes and to rescue their distribution in dilute melanocytes requires exon F but not exon D. These results predict that an interaction between myosin Va and Rab27a should be exon F dependent. Consistent with this, Rab27a present in detergent lysates of melanocytes binds to beads coated with purified, full-length melanocyte myosin Va and melanocyte myosin Va lacking exon D, but not to beads coated with melanocyte myosin Va lacking exon F or brain myosin Va. Moreover, the preparation of melanocyte lysates in the presence of GDP rather than guanosine-5′-O-(3-thio)triphosphate reduces the amount of Rab27a bound to melanocyte myosin Va-coated beads by approximately fourfold. Finally, pure Rab27a does not bind to myosin Va-coated beads, suggesting that these two proteins interact indirectly. Together, these results argue that Rab27a is an essential component of a protein complex that serves as the melanosome receptor for myosin Va, suggest that this complex contains at least one additional protein capable of bridging the indirect interaction between Rab27a and myosin Va, and imply that the recruitment of myosin Va to the melanosome surface in vivo should be regulated by factors controlling the nucleotide state of Rab27a.
INTRODUCTION
We have been studying the role of myosin Va, whose heavy chain is the product of the mouse coat color gene dilute (Mercer et al., 1991), in the transport and distribution of melanosomes within mouse melanocytes. Visible pigmentation in mammals requires that melanocytes donate these specialized pigment-producing organelles to keratinocytes, which make up the bulk of hair and skin. For this intercellular transfer to be effective, melanosomes must first be accumulated at the distal ends of the melanocytes' dendritic extensions, the site of transfer (Hearing and King, 1993). Melanocytes generate this peripheral accumulation of melanosomes via a cooperative transport mechanism in which fast, long-range, bidirectional, microtubule-dependent melanosome movements along the length of dendrites are coupled to myosin Va-dependent capture and local movement of the organelles within distal, actin-rich regions of the dendrite (Wu et al., 1998). When the capture mechanism is missing, as in melanocytes from mice homozygous for a functional null allele at dilute (dl20J) (Strobel et al., 1990), melanosomes redistribute according to microtubule density, leading to their accumulation in the central cytoplasm where the bulk of microtubules reside (Koyama and Takeuchi, 1981; Provance et al., 1996; Wei et al., 1997; Wu et al., 1998). Long-range, microtubule-based melanosome movements within dendrites continue in the absence of myosin Va, but the bidirectional nature of these movements prevents them from generating a peripheral accumulation of the organelles on their own (Wu et al., 1998). In wild-type melanocytes, myosin Va-dependent capture at the periphery prevents a fraction of melanosomes delivered there by centrifugal microtubule-dependent movements from being returned to the cell center by centripetal microtubule movements, thereby causing their peripheral accumulation.
Valuable insight into the mechanism by which myosin Va may attach to the melanosome surface has come from recent studies of mouse (ashen) and human (Griscelli) melanocytes that lack the Rab family GTPase Rab27a (Menasche et al., 2000; Wilson et al., 2000; Bahadoran et al., 2001; Hume et al., 2001; Wu et al., 2001). These melanocytes exhibit the exact same defect in melanosome distribution as dilute melanocytes, suggesting that Rab27a-deficient melanocytes are also defective in peripheral melanosome capture, that the capture mechanism requires Rab27a as well as myosin Va, and that Rab27a serves in some way to enable myosin Va-dependent melanosome capture. Importantly, both endogenous Rab27a and epitope-tagged Rab27a in fixed cells (Bahadoran et al., 2001; Hume et al., 2001; Wu et al., 2001), as well as green fluorescent protein (GFP)-tagged Rab27a in living cells (Hume et al., 2001), colocalize extensively with black, end-stage melanosomes. Furthermore, a large percentage of Rab27a-positive melanosomes is also myosin Va positive (Bahadoran et al., 2001; Hume et al., 2001; Wu et al., 2001), consistent with previous studies showing the colocalization of this myosin with end-stage melanosomes (Nascimento et al., 1997; Wu et al., 1997; Lambert et al., 1998). Finally, neither endogenous myosin Va, nor a GFP-tagged myosin Va tail domain fusion protein that also targets to melanosomes in wild-type melanocytes (Wu et al., 1998), associate with melanosomes in Rab27a-deficient melanocytes (Hume et al., 2001; Wu et al., 2001). Together, these results indicate that Rab27a is a resident melanosomal membrane protein and that it enables myosin Va-dependent melanosome capture by recruiting the myosin to the melanosome surface. Rab27a might accomplish this latter task by activating and/or unmasking another protein on the melanosome surface that functions as the receptor for myosin Va. Alternatively, Rab27a itself might function wholly or in part as the melanosome receptor for the myosin. Consistent with this latter idea, myosin Va is found in immunoprecipitates made from detergent lysates of melanocytes by using a Rab27a polyclonal antibody (Hume et al., 2001).
Herein, we sought to obtain additional support for the idea that myosin Va and Rab27a function as a motor–receptor pair by seeking definitive evidence of a physical interaction between them. In an effort to design a rigorous control for this study, we initially sought to identify the regions in myosin Va that are required for it to colocalize with melanosomes and to influence their distribution in vivo. These experiments focused in particular on exons D and F, two alternatively spliced exons of 27 and 25 amino acids, respectively, that are present in the tail domain of the melanocyte-spliced isoform of myosin Va but not the brain-spliced isoform (Seperack et al., 1995). These two exons, together with exon B, a three-residue exon specific to the tail domain of the brain isoform, comprise the total differences in sequence between the brain- and melanocyte-spliced isoforms of myosin Va and, as such, have been postulated to play important roles in those myosin Va functions that are specific to melanocytes and neurons (Seperack et al., 1995). Consistent with this idea, we found that exon F is absolutely required for myosin Va to colocalize with and to influence the position of melanosomes in the context of both dominant negative and rescue experiments. Armed with this information, we then tested the ability of beads coated with purified, full-length myosin Va with and without exon F to bind Rab27a present in detergent lysates of melanocytes. In precise agreement with the in vivo data, only beads coated with versions of myosin Va that contain exon F were found to bind Rab27a. Moreover, this interaction, like Rab–effector interactions in general, was found to be GTP dependent. Finally, experiments in which purified, GTP-loaded Rab27a was used in place of Rab27a present in melanocyte lysates were consistent with the idea that the interaction between myosin Va and Rab27a requires at least one more lysate-derived factor. We conclude, therefore, that Rab27a is an essential component of a protein complex that serves as the melanosome receptor for myosin Va, and that probably contains at least one additional protein capable of linking Rab27a to myosin Va.
MATERIALS AND METHODS
Melanocyte Cultures
The nontransformed, wild-type (D/D) melanocyte cell line melan-a was a generous gift of Dr. Dorothy Bennett (St. George's Hospital Medical School, London, United Kingdom) and was maintained as described previously (Wu et al., 1997). Wild-type primary melanocytes were obtained from C57BL/6J (D/D) mice, whereas dilute primary melanocytes were obtained from C57BL/6J mice that were homozygous for a true null allele at dilute, dl20J (Strobel et al., 1990; Wu et al. 1997). These latter mice were obtained by crossing heterozygous parents of the genotype dl20J/dvse (a generous gift of Drs. Neal Copeland and Nancy Jenkins, National Cancer Institute, National Institutes of Health, Bethesda, MD), and were distinguished from their dl20J/dvse and dvse/dvse littermates by Western blot analysis as described previously (Wu et al., 1998). These primary cultures were prepared from the skin of newborn mice as described previously (Wu et al., 2001), and they were grown on gelatin-coated Labtek chamber slides with coverglass bottoms (#155380; Nalge Nunc International, Naperville, IL) or #1.5 round coverslips in Ham's F-10 medium supplemented with 10% horse serum, 2% fetal calf serum, 1% penicillin streptomycin, 0.1 μM dibutryl cAMP, and 85 nM phorbol 12-myristate 13-acetate (#P-8139; Sigma-Aldrich, St. Louis, MO).
Transfection, Microinjection, and Image Acquisition
Melan-a melanocytes were transiently transfected with purified plasmid DNAs (#12662; QIAGEN, Valencia, CA) by using either FuGENE 6 transfection reagent (#1-814-443; Roche Applied Science, Indianapolis, IN) as described previously (Wu et al., 1998), or LipofectAMINE 2000 (#11668-019; Invitrogen, Carlsbad, CA) as described by the manufacturer. Primary wild-type and dilute melanocytes, as well as melan-a cells, were microinjected with purified plasmid DNAs exactly as described previously (Wu et al., 2001). Cells were fixed for 30 min at 21°C in freshly prepared 4% paraformaldehyde (#00380; Polysciences, Warrington, PA) in phosphate-buffered saline (PBS), washed in PBS, and mounted using Slow Fade (#S-2828; Molecular Probes, Eugene, OR). For scoring dominant negative constructs introduced by transfection, cells were fixed 36 h after transfection. For scoring dominant negative and rescue constructs introduced by microinjection, cells were fixed 10 h after microinjection. Cells were scored as having a dominant negative phenotype if there was a pronounced central accumulation of melanosomes within cells exhibiting a polarized shape (the small percentage of cells that round-up was excluded). All of the values for dominant negative constructs reported in the text were corrected for the 12% of melan-a cells, and 10% of wild-type primary cells, that exhibit an abnormal concentration of melanosomes in the cell center before transfection/microinjection. Expression of unfused GFP had no significant effect on melanosome distribution (our unpublished data). Dilute melanocytes were scored as being rescued if there was a pronounced shift in melanosome distribution from the cell center to the periphery. For imaging the localization of GFP-tagged proteins, images were acquired as 1.0-μm optical sections by using a 63× (1.4 numerical aperture) objective on a Zeiss LSM 510 confocal microscope. For determination of the relative total fluorescence in individual transfected cells, images were collected using a Zeiss TV135 inverted microscope equipped with a cooled charge-coupled device camera (#1300 Y/ES; Princeton Instruments, Princeton, NJ) and a 40× phase objective (0.75 numerical aperture). The GFP signal was collected through the total thickness of the cell under exposure conditions where the intensity of the fluorescent signal was within the linear response range of the camera. An outline of a cell was drawn based on the phase image, and the relative fluorescence per unit area of the cell was then quantitated using MetaMorph software (Universal Imaging, West Chester, PA).
Antibody Generation
To generate a polyclonal antibody against rat Rab27a, the full-length sequence was amplified using Pfu polymerase and the following 5′ and 3′ primers: 5′CTAGGATCCATGTCGGATGGAGATTATGAC3′ and 5′TGCGGATCCTCAACAGCCGCATAACCCCTTCTC3′. The polymerase chain reaction (PCR) product was then digested with BamHI, cloned into pGEX 2TK (#27-4587-01; Pharmacia, Peapack, NJ), and antibodies against the purified glutathione S-transferase (GST) fusion protein were generated in rabbits as described in detail previously (Jung et al., 2001)
Dominant Negative Constructs
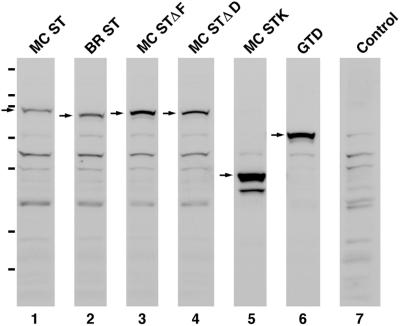
For these and all other myosin Va constructs described below, the nucleotide numbering corresponds to a modification of the numbering in accession no. X57377 (the myosin Va sequence in X57377 contains exon B [12 base pairs] and exon F [75 base pairs], but lacks exon D [81 base pairs]; our numbering takes into account the fact that the major spliced isoform present in brain contains exon B but lacks exons D and F, whereas the one in melanocytes contains exons D and F, but lacks exon B). The GFP-tagged fusion protein melanocyte short tail (MC ST), which contains the C-terminal 620 residues of the melanocyte-spliced isoform of myosin Va fused to the C terminus of enhanced green fluorescent protein (EGFP), was constructed by restriction enzyme digestion of the corresponding full-length cDNA as described previously (Wu et al., 1998). (Note that description failed to include the fact that the presence of a second SacI site contributed by exon D requires two sequential ligations, where a 2118-base pair fragment beginning at the SacI site within exon D and ending at a SalI site just 3′ of the polyadenylation sequence was inserted into plasmid pEGFP C3 [#6082-1; CLONTECH, Palo Alto, CA] first, followed by a 220-base pair SacI fragment beginning at nucleotide 3812 and ending at the SacI site within exon D). The GFP-tagged fusion protein brain short tail (BR ST), which contains the C-terminal 571 residues of the brain-spliced isoform of myosin Va fused to the C terminus of EGFP C3, was constructed by digestion of the corresponding full-length cDNA with SacI and SalI and insertion of the resulting 2190-base pair SacI/SalI fragment (nucleotides 3812–6002; 6077 in X57377) into pEGFP C3. The version of MC ST lacking exon D (MC STΔD) was constructed by overlap extension PCR by using Pfu polymerase (#600154; Stratagene, La Jolla, CA) and plasmid MC ST pEGFP C3 as a template. The 5′ fragment, which begins in the polylinker of pEGFP C3 just 5′ of the BglII site and ends with a sequence that skips exon D and contains the 21 base pairs that follow exon D, was amplified using the following 5′ and 3′ primers: 5′CTCTCGGCATGGACGAGCTGTACAAG3′ and 5′CTGTAGCTGG GATTCTAAGAGCCTGTTTGTTTCTTTCAAACCAATATA3′. The 3′ fragment, which begins with a sequence containing the 27 base pairs preceding exon D and then skips exon D, and ends just 3′ of the natural BglII site at nucleotide 4927 (4857 in X57377), was amplified using the following 5′ and 3′ primers: 5′TATATTGGTTTGAAAGAAACAAACAGGCTCTTAGAATCCCAGCTACAG3′ and 5′GTTCTCT-AACACCCTCACAAGCTGCTG3′. These two fragments were then mixed and amplified using the outside primers. The resulting product was digested with BglII and used to replace the corresponding BglII fragment in plasmid MC ST pEGFP C3, generating MC STΔD pEGFP C3. The version of MC ST lacking exon F (MC STΔF) was created in exactly the same manner except that the 3′ primer for the 5′ fragment was 5′GCTTTTCAAGTTGTTCCATCAGATCCAGGTTTTCATTGGTCAGCCGGG3′, whereas the 5′ primer for the 3′ fragment was 5′CGGCTGACCAATGAAAACCTGGATCTGATG-GAACAACT TGAAAAGCAG3′. The GFP-tagged fusion protein melanocyte stalk (MC STK), which contains the stalk portion of MC ST (residues 1238–1470), was generated by PCR by using Pfu polymerase, the corresponding full-length cDNA as a template, and the following 5′ and 3′ primers: 5′CTAGGATCCACTGCGCCAGGTGCGCCTGCTTAC3′ and 5′TGAGAATTCTTACTGCCCCACTTCTAGTTCACCA AT3′. The resulting fragment was digested with BamHI and EcoRI and cloned into plasmid pEGFP C1 (#6084-1; CLONTECH) that had been cut with BglII and EcoRI. The GFP-tagged fusion protein globular tail domain (GTD), which contains the C-terminal 410 residues of the myosin Va heavy chain that are common to both isoforms, was generated by PCR by using Pfu polymerase, the full-length cDNA for the brain-spliced isoform as a template, and the following 5′ and 3′ primers: 5′TCAGGATCCATTGGTGAACTAGAAGTGGGGCAG3′ and 5′CATGAATTCTCAGA-CCCGTGCGATGAAGCCCAGGCC3′. The resulting fragment was digested with BamHI and EcoRI and cloned into plasmid pEGFP C1 that had been cut with BglII and EcoRI. All dominant negative constructs were confirmed by sequencing and by identification of stable proteins of the expected molecular weight in Western blots of transiently transfected COS cells (our unpublished data) and melan-a melanocytes (Figure 3) probed with an antibody against GFP (#8367-1; CLONTECH).
Figure 3.
All six myosin Va tail domain GFP fusions are stable proteins in vivo. Shown are Western blots of whole cell extracts prepared from melan-a melanocytes transfected with each of the six myosin Va tail domain GFP fusion protein constructs described in the text and probed with an antibody to GFP (lanes 1–6). The arrows indicate the positions of the fusion proteins. The remaining bands can be largely if not entirely accounted for by bands appearing in the blot of untransfected melan-a cells (lane 7). The hash marks to the left indicate the migration of the following markers (top to bottom): 200, 116, 97, 66, 55, 36, and 31 kDa. The molecular masses of all six fusion proteins calculated from these blots are within a few percentage points of their estimated molecular masses based on sequence (MC ST, 95 kDa; BR ST, 89 kDa; MC STΔF, 92 kDa; MC STΔD, 92 kDa; MC STK, 52 kDa; and GTD, 72 kDa). Moreover, all six proteins are largely if not entirely intact (only MC STK shows one stable breakdown product of ∼44 kDa). The differences in the intensities of the background bands in lanes 1–6 are due to differences in the amounts of whole cell extract loaded. The differences in the amount of fusion protein per volume of extract were due largely to differences in transfection efficiencies for the different plasmids.
Rescue Constructs
Construction of all vectors containing full-length versions of myosin Va began with either of two cDNAs, one encoding the complete heavy chain of the brain-spliced isoform (BR MV; +B, −D, −F), and the other the complete heavy chain of the melanocyte-spliced isoform (MC MV; −B, +D, +F) (Seperack et al. 1995; Wu et al., 1998). Both cDNAs begin 60 base pairs 5′ of the ATG (or 19 base pairs 5′ of the start of the myosin Va sequence in X57377) and end at the first polyadenylation signal in the 3′-untranslated sequence (nucleotides 6072–6077 in X57377). To facilitate subsequent manipulations, both cDNAs were blunt-ended, Not-linkered, and cloned into Bluescript SK. To generate versions of MC MV and BR MV that are fused at their N terminus to GFP, we took advantage of a unique, 983-base pair StuI fragment that is identical in both cDNAs and that begins 56 base pairs 5′ of the ATG (and just inside the Not linker) and ends in the coding sequence at nucleotide 969 (in X57377). This natural fragment was excised from both full-length cDNAs and replaced with a StuI-digested PCR product that had been amplified using Pfu polymerase, full-length MC MV cDNA as template, and the following primers: 5′AGTAGGCCTATGGCCGCGTCCGAGCTCTACAC-C3′ and 5′TGCAGGCCTGC CTGGTGTGCGCCATCTCCTTCGC3′. MC MV and BR MV clones with the correct orientation were confirmed by sequencing the amplified region, digested with NotI, and the corresponding ∼6-kb NotI inserts cloned into an pEGFP C1 plasmid that had previously been digested with EcoRI, filled in with T4 DNA polymerase, ligated with a NotI linker (#1127; New England Biolabs, Beverly, MA), digested with NotI, and treated with phosphatase. The resulting fusion introduced five extra amino acids (AAARP) between the C terminus of GFP and the ATG of the myosin Va heavy chain. The ∼6-kb inserts described above were also cloned into plasmid pFLAG CMV-2 (IBI; Eastman Kodak, Rochester, NY) that had been digested with NotI and treated with phosphatase to create full-length versions of MC MV and BR MV with the nine-residue FLAG epitope tag at their N termini. To create unfused versions of MC MV and BR MV, the NotI inserts from the Bluescript clones described above were cloned directly into plasmid pcDNA 3.1 (+) (#V790–20; Invitrogen). All full-length clones were confirmed by sequencing the region of the in-frame fusion, and by the identification of proteins of the expected molecular weight in Western blots of transiently transfected COS cells probed with antibodies against myosin Va (DIL 2; Wu et al., 1997), the FLAG tag (#IB13091; Eastman Kodak), and/or GFP.
To create full-length, GFP-tagged versions of MC MV lacking either exon D (MC MVΔD) or exon F (MC MVΔF), we made use of several clones described above. First, the ∼6-kb NotI insert from a correct, full-length, GFP-tagged MC MV clone was transferred into a Bluescript plasmid in which the poly linker from the XbaI site to the KpnI site had been deleted, and in which the SacI site had been removed by digestion with SacI, followed by treatment with T4 DNA polymerase. The resulting plasmid (plasmid A) was then digested with BglII, releasing an internal 2647-base pair BglII fragment (nucleotides 2280–4927; 4857 in X57377) that spans the region of alternative splicing, and generating plasmid AΔ2.6 BglII. The 2647-base pair BglII fragment was then cloned into the BglII site of an pEGFP C1 plasmid in which the SacI and XbaI sites in the polylinker had been destroyed by digestion and T4 DNA polymerase treatment, generating plasmid B. Plasmid B was then digested with SacI and XbaI, releasing the 220-base pair SacI and the 551-base pair SacI/XbaI fragments that span exons D and F, and generating plasmid BΔSacI/XbaI. Plasmid BΔSacI/XbaI was then ligated with either 1) a 690-base pair SacI/XbaI fragment obtained from a correct, GFP-tagged MC STΔD clone, generating plasmid BΔexon D (which contains exon F but lacks exon D); or 2) a 476-base pair SacI/XbaI fragment, followed by a 220-base pair SacI fragment, both of which were obtained from a correct, GFP-tagged MC STΔF clone, generating plasmid BΔexon F (which contains exon D but lacks exon F). The ∼2.6-kb BglII inserts in plasmids BΔexon D and BΔexon F were then released by digestion with BglII and cloned into plasmid AΔ2.6 BglII, generating plasmids AΔexon D and AΔexon F. The 6-kb NotI inserts in these latter two plasmids were then released by digestion with NotI and cloned into the EGFP CI plasmid that had been modified to accept NotI inserts, generating plasmids MC MVΔD and MC MVΔF. Clones with the correct orientation were confirmed by sequencing and by transfection in COS cells as described above.
Baculovirus Constructs
We used the strategy described above for introducing full-length myosin Va cDNAs into GFP vectors to introduce full-length, FLAG-tagged versions of MC MV and BR MV into the baculovirus transfer vector pVL1392 (#V1392-20; Invitrogen). Specifically, the natural 983-base pair StuI fragment in both full-length cDNAs was replaced with a StuI-digested PCR product that had been amplified using Pfu polymerase, full-length MC MV cDNA as template, and the following 5′ and 3′ primers: 5′AGTAGGCCTCGCCACCATGGACTACAAAGACGATGACGACAAGGCCGCGTCCGAGCTCTACACC3′ and 5′TGCAGGCCTGCCTGGTGTGCGCCATCTCCTTCGC3′. This fragment introduces the nine-residue FLAG epitope tag at the N terminus of the heavy chains as well as a Kozak sequence 5′ of the ATG. MC MV and BR MV clones with the correct orientation were confirmed by sequencing the amplified region, digested with NotI, and the corresponding ∼6-kb NotI inserts cloned in the proper orientation into pVL1392 that had been digested with NotI and treated with phosphatase. To create similar transfer vectors for the MC MVΔD and MC MVΔF isoforms, we cloned the 6-kb NotI insert from a correct MC MV pVL1392 clone into Bluescript, cut this plasmid with BglII, discarded the 2.6-kb BglII insert, and replaced it with 2.6-kb BglII fragments released from either a correct GFP-tagged MC MVΔD clone or a correct GFP-tagged MC MVΔF clone. The 6-kb NotI inserts from Bluescript clones with the proper orientation were then cloned back into pVL1392. We also constructed the pVL1392 vector mDLC8A, which contains a full-length, unfused version of the A isoform (Wilson et al., 2001) of the mouse 8-kDa dynein light chain (Lu and Hammer, unpublished data).
Expression and Purification of Full-Length, FLAG-tagged Myosin Va Isoforms
Recombinant baculoviruses containing mDLC8A and each of the four myosin Va heavy chain isoforms (MC MV, BR MV, MC MVΔD, and MC MVΔF) were produced in Sf9 insect cells, plaque purified, and amplified as described previously (Wang et al., 2000). For protein expression, Sf9 cells were coinfected as described previously (Wang et al., 2000) with one of the four myosin Va heavy chain viruses, a recombinant virus driving the expression of calmodulin (Wang et al., 2000), and the mDLC8A virus (Note: We did not include a virus containing the 17-kDa essential myosin light chain, a third light chain identified in purified samples of myosin Va from chicken brain [Espindola et al., 2000], because myosin Va purified from mouse brain [Wang et al., 2000], as well as myosin Va immunoprecipitated from mouse melanocytes [Hammer and Sellers, unpublished data], do not contain this light chain). All four myosin Va isoforms were purified to homogeneity from high-speed supernatants of lysed Sf9 cells by affinity chromatography on Anti-FLAG M2 Antibody Affinity resin (#A2220; Sigma-Aldrich), followed by elution with excess free FLAG peptide (#F3290; Sigma-Aldrich), exactly as described previously for FLAG-tagged HMM and S1 fragments of myosin Va (Wang et al., 2000). The myosins were then concentrated and separated from FLAG peptide by ion exchange chromatography on Mono Q resin, and dialyzed into storage buffer [500 mM KCl, 10 mM 3-(N-morpholino)propanesulfonic acid pH 7.0, 0.1 mM EGTA, 1 mM DTT (dithiothreitol), and 0.1 mM phenylmethylsulfonyl fluoride]. Protein concentrations, as determined by the Bio-Rad protein assay (#500-0001; Bio-Rad, Hercules, CA) by using chicken gizzard smooth muscle myosin HMM as a standard, were adjusted to 1 mg/ml.
Binding of Rab27a to Beads Coated with Myosin Va
These experiments involved charging the Anti-FLAG M2 Antibody Affinity resin with purified myosin Va; incubating the charged resin with a detergent lysate of melanocytes; washing the resin; and eluting the myosin Va, together with any interacting proteins, by using excess FLAG peptide and Western blot analysis. To charge the resin, 300 μl (settled volume) of Anti-FLAG M2 Antibody Affinity resin was washed three times with 20 volumes of TBS (150 mM NaCl and 10 mM Tris pH 7.5) by centrifugation at 1000 × g for 2 min, mixed with 600 μl of 0.5 mg/ml purifed myosin Va in storage buffer plus 400 μl of TBS, incubated for 2 h at 4°C on a rotating mixer, and washed five times with 20 volumes per wash of TBS at 4°C. Beads prepared in this way contained ∼40 μg of bound myosin Va per 300 μl of settled volume. The melanocyte detergent lysate was prepared using mouse B16 F10 mouse melanocytes, which were grown to 90% confluence in DMEM supplemented with 10 nM α-melanophore–stimulating hormone. Ten 150-mm dishes were harvested by incubating each plate in 10 ml of Hanks' balanced salt solution supplemented with 10 mM EDTA, pH 8.0, for 10 min at room temperature followed by trituration. Cells were pooled, collected by centrifugation at 1500 × g for 8 min at 4°C, and lysed in 6 ml of lysis buffer containing 80 mM NaCl, 20 mM Tris pH 7.5, 0.75% NP-40 (#28324; Pierce Chemical, Rockford, IL), 100 μM guanosine-5′-O-(3-thio)triphosphate (GTPγS) (#G8634; Sigma-Aldrich), 0.3 mM MgCl2, 0.5 mM DTT, and a protease inhibitor mix (#1836170; Roche Applied Science) by brief vortexing and incubation at 4°C for 20 min on a rotating mixer. After centrifugation at 15,000 × g for 20 min at 4°C, the supernatant was diluted with two volumes of lysis buffer lacking NP-40, giving a resin-ready lysate containing 0.25% detergent. This extraction procedure released >98% of total cellular Rab27a into the 15,000 × g supernatant. Five milliliters of this lysate was then mixed with the 300 μl of charged resin for 3 h at 4°C on a rotating mixer in a 5-ml screw cap cryotube (#5000.0050; Nalgene). For washing, the resin from each sample was divided into two 2-ml Eppendorf tubes and washed six times at 4°C with 15 volumes of wash buffer containing 80 mM NaCl, 20 mM Tris pH 7.5, 0.1% NP-40, 30 μM GTPγS, 0.3 mM MgCl2, and 0.5 mM DTT by centrifugation at 2000 × g for 30 s. Myosin Va, together with any interacting proteins, were eluted from the washed resin by incubation for 1 h at 4°C with 400 μl per 300 μl of settled resin of elution buffer containing 200 mM NaCl, 10 mM Tris pH 7.5, 0.1 mM MgCl2, and 0.4 mg/ml purified FLAG peptide. After four consecutive centrifugations to remove the resin completely, the supernatant was mixed with 200 μl of SDS sample buffer containing 10% SDS, 0.15 M Tris pH 7.5, 20 mM EDTA pH 8.0, 20 mM B-mercaptoethanol, and 10% sucrose, and boiled for 5 min. Before Western blotting, samples were adjusted for slight differences in the amount of myosin Va heavy chain in the final eluate of those samples to be directly compared (e.g., resins charged with brain vs. melanocyte myosin Va) by resolution on 8% SDS-PAGE gels, staining with Coomassie blue, and quantitative laser densitometry. We also note that the amount of myosin Va bound to the resin did not drop significantly over the course of the incubation with cell lysate and the subsequent washing steps. To look for Rab27a in eluates, samples were resolved on 10% SDS-PAGE gels, transferred to nitrocellulose (BA 85; Schleicher & Schuell, Keene, NH) by using a semidry blotter (model Trans-Blot SD; Bio-Rad), and probed with either a monoclonal antibody (mAb) to human Rab27a (#R52320; Transduction Laboratories, Lexington, KY) (1:2000 dilution), which specifically recognizes Rab27a in mouse melanocytes (Wu et al., 2001), or a rabbit polyclonal antibody to rat Rab27a (see above) (1:10,000 dilution). To detect the mAb, a peroxidase-labeled goat anti-mouse secondary antibody that is specific for the heavy chain of IgG (#A9309; Sigma-Aldrich) was used (1:5000 dilution), because secondary antibodies that also see the IgG light chain detect a small amount of light chain that leaches off of the M2 Anti-FLAG antibody, producing a signal that runs just below Rab27a. The polyclonal antibody was detected using a peroxidase-labeled goat anti-rabbit secondary antibody (#RPN 2108; Amersham Biosciences, Piscataway, NJ). Blots were developed using enhanced chemiluminescence reagents from Amersham Biosciences (#RPN 2134) and exposed using Hyperfilm (#RPN 1674K; Amersham Biosciences). Signals in samples to be directly compared were quantified by laser densitometry within the linear response range of the instrument. In some experiments, GDP (#G7127; Sigma-Aldrich) was substituted for GTPγS in both lysis and wash buffers. Western blots showed that this change did not reduce the amount of cellular Rab27a solubilized by lysis buffer.
To look for evidence of a direct interaction between myosin Va and Rab27a, we used a modification of the procedure developed by Christofordis and Zerial (2000) to identify Rab5 effectors, where glutathione-Sepharose resin charged with GST-Rab5 was incubated sequentially in buffers designed to exchange GTP for GDP on Rab5 (high EDTA, low Mg2+, GTPγS) and then to stabilize the GTP-bound form of Rab5 (low EDTA, high Mg2+, GTPγS). To test the feasibility of this approach, we assessed the ability of a purified, GST fusion of Rab27a to load with GTPγS by using a procedure described previously (Terui et al., 1994). As has been seen for a subset of other Rabs, the addition of GST to the N terminus of Rab27a abrogated its ability to bind GTP. In contrast, a purified, His-tagged version of Rab27a loaded to at least ∼0.25 mol of GTPγS per mole of Rab27a. Given this, we subjected His-tagged Rab27a to the exact same set of buffer incubations designed to remove bound GDP and load GTPγS (Christoforidis and Zerial, 2000), except that these buffer changes were accomplished by dialysis of the purified fusion protein, because nickel-Sepharose resin is not compatible with EDTA. After this, the GTPγS-loaded, His-tagged Rab27a was clarified by centrifugation at 50,000 × g for 15 min at 4°C, and incubated at a final concentration of 4 μM with 300 μl of myosin Va-coated beads in a buffer containing 20 mM Tris pH 7.5, 80 mM NaCl, 4 mM MgCl2, 1 mM DTT, 0.25% NP-40, 100 μM GTPγS, and 0.5% bovine serum albumin. After a 90-min incubation at 4°C, the beads were washed six times (5 ml/wash) with a buffer containing 20 mM Tris pH 7.5, 80 mM NaCl, 4 mM MgCl2, 1 mM DTT, 25 μM GTPγS, and 0.1% NP-40. Myosin Va and any bound Rab27a were eluted with excess FLAG peptide, and the eluates probed for Rab27a by Western blot analysis.
Yeast Two-Hybrid Analysis
Rab27a and the tail domain of myosin Va were tested for physical interaction using an enhanced GAL4-based system (#K1612-1; CLONTECH). To clone the dominant active, Q78L version of Rab27a into pGBKT7 (#K1612B; CLONTECH), the complete coding sequence for Rab27a was amplified from the corresponding point mutant (Wu et al., 2001) by using the primers 5′TCACAGCCATGGCCTCGGATGGAGATTATGACTACCTCATC′3 and 5′ACTGGATCCTCAACAGCCGCATAACCCCTTCTCCTTCTCCTCACT′3 and digested with NcoI and BamHI. To clone the tail domain of myosin Va into pGADT7, the C-terminal 620 residues of the melanocyte-spliced isoform of myosin Va were amplified off of the corresponding full-length construct by using the primers 5′TCAGAATTCGAAGACATTGCACCAAGAACAGAGGAGCCA′3 and 5′GTGATACTCGAGTCAGACCCGTGCGATGAAGCCCAGG-CC′3 and then digested with EcoRI and XhoI. Both PCR products were confirmed by sequencing. The pGBKT7 plasmid containing Rab27a was transformed into yeast strain AH109 and selected on SD/Glu/Trp− plates, whereas the pGADT7 plasmid containing the myosin Va tail was transformed into yeast strain Y187 and selected on SD/Glu/Leu− plates. Single colonies of AH109 and Y187 transformants were mixed in 1 ml of YPAD media, grown for 48 h at 30°C, diluted 10- and 100-fold, and plated on SD/Glu/Trp− Leu− plates to determine mating efficiency, and SD/Glu/Trp− Leu−His−Ade− plates containing α-Xgal to test for physical interaction.
RESULTS
Ability of Myosin Va Tail Domain to Localize to Melanosomes and to Generate a Dominant Negative Phenotype Depends on Presence of exon F
We showed previously that the GFP fusion protein MC ST, which contains the distal stalk and globular tail portions of the melanocyte-spliced heavy chain isoform of myosin Va (Figure 1), targets to melanosomes and, in wild-type melanocytes such as the cell line melan-a, causes the organelles to redistribute to the cell center (Wu et al., 1998). Generation of this dilute-like phenotype was attributed to the ability of MC ST to displace endogenous myosin Va from the melanosome surface, thereby uncoupling the organelles from the peripheral actin cytoskeleton and allowing them to redistribute according to microtubule density as in dilute melanocytes. In contrast to MC ST, which has consistently produced this dominant negative phenotype (Figure 2, A and B), introduction of the GFP fusion protein BR ST, which contains the corresponding portion of the brain-spliced isoform of myosin Va (Figure 1), consistently failed to generate the dominant negative phenotype (Figure 2, C and D). This was corroborated by scoring transfected cells, where the dominant negative phenotype was present in 73% of cells expressing MC ST (n = 290), but only 3% of cells expressing BR ST (n = 310). Similar results were obtained when the plasmids were introduced by microinjection rather than transfection (74% for MC ST, n = 46; 4% for BR ST, n = 45), and when primary wild-type melanocytes were microinjected rather than melan-a melanocytes (72% for MC ST, n = 39; 4% for BR ST, n = 45). Furthermore, quantification of the total fluorescence per cell (see MATERIALS AND METHODS) indicated that the inability of BR ST to generate a dilute-like phenotype was not due to lower levels of expression relative to MC ST (our unpublished data). Finally, Western blots showed that the inability of BR ST to generate a dominant negative phenotype was not due to the intracellular degradation of the fusion protein (Figure 3, lane 2).
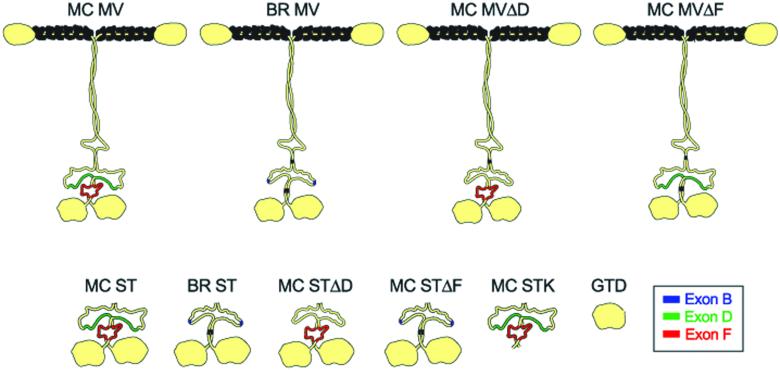
Figure 1.
Schematic of the structures of the full-length myosin Va isoforms and the myosin Va tail domains used in rescue and dominant negative experiments, respectively. Exon B, which is specific to the brain-spliced isoform, is shown in blue, whereas exons D and F, which are specific to the melanocyte-spliced isoform, are shown in green and red, respectively. At the top are the structures of the melanocyte-spliced isoform (MC MV), the brain-spliced isoform (BR MV), the melanocyte-spliced isoform engineered to lack exon D (MC MVΔD), and the melanocyte-spliced isoform engineered to lack exon F (MC MVΔF). At the bottom are the structures of MC ST, BR ST, MC STΔD, MC STΔF, MC STK, and the GTD. These schematics are based on a schematic published previously (Cheney et al., 1993), which depicted the shape of myosin Va as determined by electron microscopy.
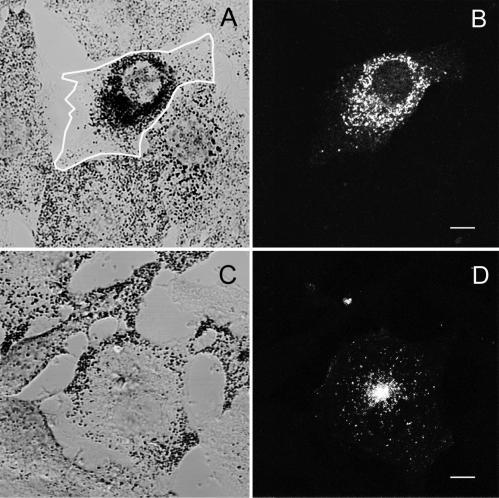
Figure 2.
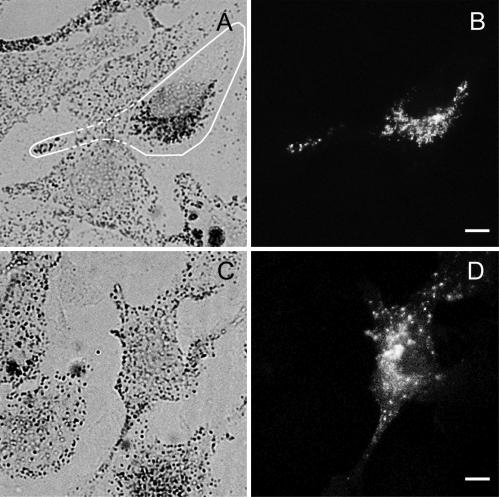
MC ST, but not BR ST, colocalizes with melanosomes and generates a dilute-like phenotype in wild-type melanocytes. Shown is the distribution of melanosomes (A and C) and GFP fluorescence (B and D) in melan-a melanocytes transfected with either GFP-MC ST (A and B) or GFP-BR ST (C and D). The edges of the transfected cell in A are marked with a white line. Bars, 6.5 μm.
This functional difference between MC ST and BR ST was mirrored by an equally striking difference in their abilities to associate with melanosomes in vivo. Specifically, although MC ST showed extensive colocalization with end-stage melanosomes in the majority of transfected cells, including virtually 100% of cells exhibiting the dominant negative phenotype (Figure 2, A and B; Wu et al., 1998, Figures 6–8), BR ST never showed anything but occasional coincidental colocalization with black melanosomes (Figure 2, C and D). Indeed, BR ST consistently labeled vesicles that were not pigmented, did not stain with melanosome markers such as tyrosinase-related protein-1, and were often accumulated at the microtubule-organizing center (Figure 2, C and D; our unpublished data). These results are consistent with the proposed mechanism for the generation of the dominant negative phenotype, wherein the ability to target to the melanosome would be a prerequisite for the tail domain to influence melanosome position.
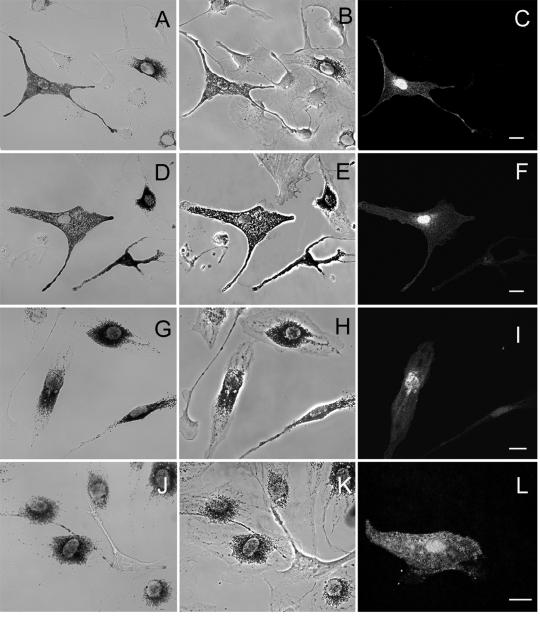
Figure 6.
MC MV and MC MVΔD, but not MC MVΔF and BR MV, rescue melanosome distribution in dilute melanocytes. Shown is the distribution of melanosomes in brightfield (A, D, G, and J), the cell shape in phase contrast (B, E, H, and K), and the distribution of GFP fluorescence due primarily to unfused GFP expressed from plasmid pEGFP C1 (C, F, I, and L), which was coinjected to ensure accurate identification of all injected cells (see text), in dilute melanocytes microinjected with MC MV (A–C), MC MVΔD (D–F), MC MVΔF (G–I), or BR MV (J–L). Bars, 11.5 μm.
Figure 8.
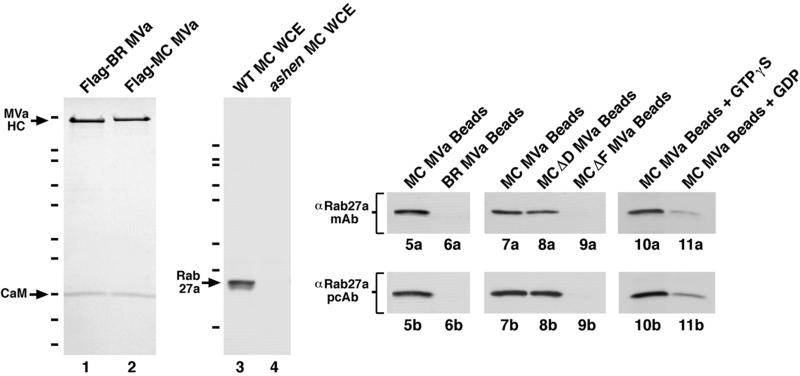
Beads coated with myosin Va interact with Rab27a in an exon F- and GTP-dependent manner. Lanes 1 and 2 show Coomassie blue-stained SDS-PAGE gels of BR MV (lane 1) and MC MV (lane 2) purified from baculovirus-infected insect cells via the FLAG epitope tag introduced at their N termini. The positions of their heavy chains (HC) and calmodulin (CaM) light chains are indicated. The HC of the melanocyte-spliced isoform is slightly higher than that of the brain-spliced isoform because it contains 49 more residues. Although not visible in this loading, the brain-spliced isoform, as well as the melanocyte-spliced isoform lacking exon D, possess the DLC8A light chain (Sellers and Hammer, unpublished observations). Similar purities were obtained for the MC MVΔD and MC MVΔF isoforms. The hash marks to the left indicate the migration of the following markers (top to bottom): 200, 116, 97, 66, 55, 36, 31, 22, 14 and 6 kDa. Lanes 3 and 4 show Western blots of whole cell extracts (WCE) made from wild-type (WT) melanocytes (lane 3) and from ashen melanocytes (lane 4), which are devoid of Rab27a (Wu et al., 2001), and probed with the polyclonal antibody against Rab27a generated in this study (the hash marks correspond to the migration of the markers listed above, except that the 14- and 6-kDa markers are not present). Lanes 5a–11a show Western blots of the material eluted from beads coated with the indicated myosin Va isoforms and probed with the mAb against Rab27a. Lanes 5b–11b show Western blots of the same FLAG-peptide eluates shown in 5a–11a but probed with the polyclonal antibody to Rab27a.
The sequences of BR ST and MC ST are identical except for three relatively small differences within their distal stalk domains, where the melanocyte isoform contains two insertions (exons D and F) that are not present in the brain isoform, whereas the brain isoform contains one insertion (exon B) that is not present in the melanocyte isoform (Figure 1) (Seperack et al., 1995). The 27-amino acid insertion corresponding to exon D, which is colored green in Figure 1, inserts into the second of the two major loops that disrupt the central coiled coil stalk domain (Loop 2 in Cheney et al., 1993), making this loop correspondingly bigger. The 25-amino acid insertion corresponding to exon F, which is colored red in Figure 1, inserts C-terminal of exon D into the third and final section of coiled coil in the stalk. Programs that calculate the likelihood of sequences forming coiled coils indicate that the insertion of exon F, in combination with a small proline-containing discontinuity located just N-terminal of the exon F insertion site, would introduce a full-fledged loop into the middle of the third section of coiled coil (Figure 1; our unpublished data). Finally, the three-amino acid insertion corresponding to exon B, which is colored blue in Figure 1, inserts into Loop 2 thirty-four residues N-terminal of where exon D inserts.
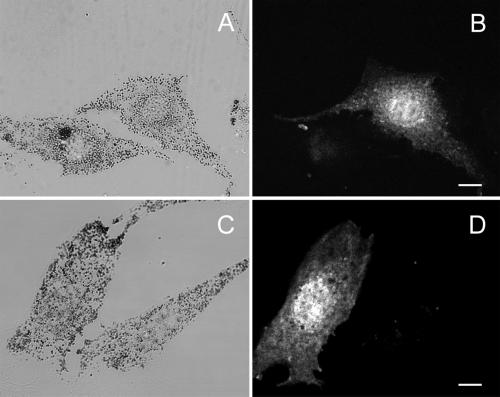
Although exon B could be a negative regulator of myosin Va–melanosome interaction, the most likely explanation for the difference between BR ST and MC ST is that exon D, exon F, or both are required for MC ST to target to melanosomes and cause their redistribution. To test this, we constructed the GFP fusion proteins MC STΔD, which lacks exon D but retains exon F (Figure 1), and MC STΔF, which lacks exon F but retains exon D (Figure 1). MC STΔD was essentially indistinguishable from MC ST, showing striking colocalization with melanosomes and generating a dominant negative phenotype in 71% of transfected melan-a melanocytes (n = 220) (Figure 4, A and B). In contrast, MC STΔF was essentially indistinguishable from BR ST, showing no propensity to colocalize with melanosomes and generating a dominant negative phenotype in only 3% of transfected cells (n = 245) (Figure 4, C and D). Furthermore, the inability of MC STΔF to generate a dilute-like phenotype was not due to lower levels of expression relative to MC ST (our unpublished data), or to the intracellular degradation of the fusion protein (Figure 3, lane 3). Therefore, exon F is required for MC ST to target to melanosomes and to disrupt myosin Va function in a wild-type background, but exon D is not.
Figure 4.
MC STΔD, but not MC STΔF, colocalizes with melanosomes and generates a dominant negative phenotype in wild-type melanocytes. Shown is the distribution of melanosomes (A and C) and GFP fluorescence (B and D) in melan-a melanocytes transfected with either GFP-MC STΔD (A and B) or GFP-MC STΔF (C and D). The edges of the transfected cell in A are marked with a white line. Bars, 6 μm.
exon F, Although Required, Is Not Sufficient
To determine whether exon F is sufficient to target to melanosomes and influence their position, we constructed the GFP fusion protein MC STK, which contains the distal stalk portion of MC ST, including exon F (Figure 1). This fusion protein did not exhibit any tendency to colocalize with melanosomes and generated a dominant negative phenotype in only 2% of transfected melan-a melanocytes (n = 210) (Figure 5, A and B). These results indicate that exon F, although required, is not sufficient, and imply, along with the data on BR ST, that the globular tail domain, which encompasses the remainder of MC ST not present in MC STK, and which has been implicated in cargo binding in other studies (reviewed in Reck-Peterson et al., 2000), must also be required but not sufficient. Consistent with these deductions, the GFP fusion protein GTD, which contains the globular tail domain portion of MC ST (Figure 1), also exhibited no tendency to colocalize with melanosomes and generated a dominant negative phenotype in only 2% of transfected cells (n = 190) (Figure 5, C and D). Furthermore, the inability of MC STK and GTD to generate a dilute-like phenotype was not due to lower levels of expression relative to MC ST (our unpublished data), or to the intracellular degradation of these fusion proteins (Figure 3, lanes 5 and 6). We conclude, therefore, that both exon F and the globular tail domain are required and that neither is sufficient.
Figure 5.
Neither MC STK, nor GTD, colocalizes with melanosomes or generate a dilute-like phenotype in wild-type melanocytes. Shown is the distribution of melanosomes (A and C) and the GFP fluorescence (B and D) in melan-a melanocytes transfected with either GFP-MC STK (A and B) or GFP-GTD (C and D). Bars, 9.5 μm (A and B) and 10.5 (C and D) μm.
Full-Length Myosin Va Also Requires exon F to Colocalize with Melanosomes and to Rescue Melanosome Distribution in Dilute Melanocytes
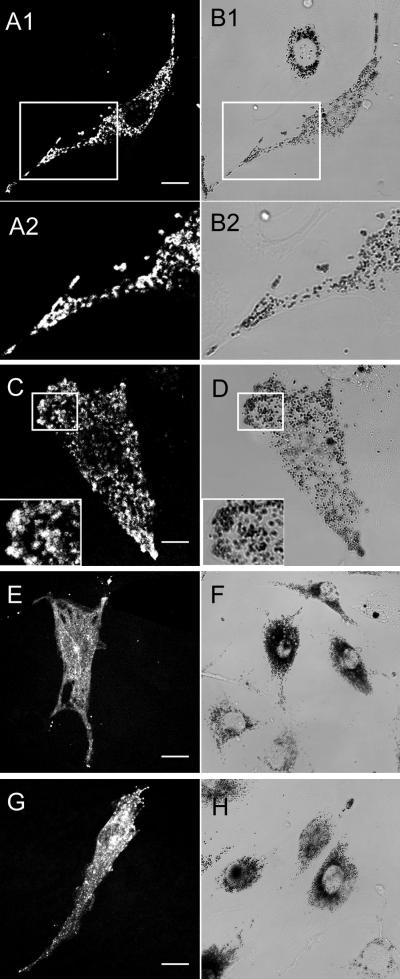
To confirm and extend the results obtained with tail domains, we assessed the ability of full-length myosin Va with and without exons D and F to rescue the abnormal perinuclear distribution of melanosomes characteristic of dilute melanocytes. To accomplish this, we microinjected primary melanocytes cultured from the skin of newborn mice homozygous for a true null allele at dilute (dl20J) (Strobel et al., 1990; Wu et al., 1997) with plasmids encoding various full-length myosin Va heavy chain isoforms fused at their N terminus to GFP, and scored microinjected cells for the restoration of melanosome distribution to the periphery. Four specific full-length myosin Va heavy chain constructs were tested: melanocyte myosin Va (MC MV); brain myosin Va (BR MV); melanocyte myosin Va without exon D (MC MVΔD), which lacks exon D but retains exon F; and melanocyte myosin Va without exon F (MC MVΔF), which lacks exon F but retains exon D (Figure 1). In exact agreement with the functional data obtained for tails, MC MV (Figure 6, A–C) and MC MVΔD (Figure 6, D–F) rescued melanosome distribution in 81% (n = 87) and 83% (n = 91) of dilute melanocytes, respectively, whereas MC MVΔF (Figure 6, G–I) and BR MV (Figure 6, J–L), which lack both exons D and F, did not rescue a single mutant cell (n = 74 and 72 for MC MVΔF and BR MV, respectively) (given that the fluorescence signal from these GFP-tagged myosins, in contrast to the signal from the GFP-tagged dominant negative constructs, was often very weak, the melanocytes in these experiments were coinjected with plasmid EGFP C1, which allowed for unequivocal identification of all microinjected cells, and, therefore, accurate scoring of rescue; the fluorescence images in C, F, I, and L reveal primarily the distribution of the unfused GFP expressed from this plasmid). Furthermore, in dilute melanocytes that were injected with just the plasmids encoding GFP-tagged myosin Va isoforms, and that happened to exhibit a strong fluorescence signal, myosin MC MV (Figure 7, A1–B2) and MC MVΔD (Figure 7, C and D, plus inset) showed extensive colocalization with black, end-stage organelles in rescued cells, whereas MC MVΔF (Figure 7, E and F) and BR MV (Figure 7, G and H) showed no tendency to associate with the melanosomes clustered in the perinuclear region. Together, these results show that the ability of full-length myosin Va to colocalize with melanosomes and rescue their distribution in myosin Va-deficient melanocytes is absolutely dependent on the presence of exon F, and that exon D is not required. These results also show that the addition of the GFP moiety to the N terminus of myosin Va does not interfere in any obvious way with its function in vivo. Indeed, the fact that the timing and extent of rescue with both FLAG-tagged and unfused versions of MC MV was no different than with the GFP-tagged molecule (our unpublished data) argues that the latter protein is fully functional.
Figure 7.
MC MV and MC MVΔD, but not MC MVΔF and BR MV, colocalize with melanosomes. Shown are the distributions of various GFP-tagged myosin Va isoforms (A1, A2, C, E, and G), together with the distributions of black, end-stage melanosomes in brightfield (B1, B2, D, F, and H), in dilute melanocytes microinjected with MC MV (A1–B2; B1 and B2 are an enlargement of a portion of the image in A1 and A2), MC MVΔD (C and D plus insets), MC MVΔF (E and F), and BR MV (G and H). These cells represent examples where the fluorescence signals for the four GFP-tagged myosin Va isoforms were much stronger than usual. Bars, 11.5 μm (A1 and B1), 5.5 μm (C and D), 16.5 μm (E and F), and 13 μm (G and H).
Beads Coated with Purified Myosin Va Bind Rab27a in an exon F-dependent Manner
The functional data presented above predict that a physical association between myosin Va and Rab27a should be absolutely dependent on the presence of exon F. To test this prediction, we asked whether beads coated with purified myosin Va bind Rab27a present in detergent lysates of melanocytes, and whether this binding is exon F dependent. To accomplish this, we expressed all four full-length myosin Va heavy chain constructs as N-terminal, FLAG-tagged fusions in baculovirus-infected insect cells (together with viruses driving the expression of calmodulin and the 8-kDa light chain of dynein; see MATERIALS AND METHODS), and purified them to homogeneity by affinity chromatography on agarose beads coated with the M2 mAb against the FLAG epitope tag (see MATERIALS AND METHODS). Figure 8, lanes 1 and 2, show typical examples of purified BR MV and MC MV, respectively. These two proteins, together with purified MC MVΔD and MC MVΔF, were then rebound to M2 antibody-coated beads; incubated with detergent lysates of B16 mouse melanocytes in the presence of GTPγS; washed repeatedly; and the myosin Va, together with any interacting proteins, eluted from the beads by incubation with excess FLAG peptide (see MATERIALS AND METHODS). To look for Rab27a as an interacting protein, the FLAG peptide eluates were resolved on 10% SDS-PAGE gels, blotted, and probed with a mAb against human Rab27a, which we have shown is specific for Rab27a in the context of mouse melanocytes (Wu et al., 2001). Figure 8, lanes 5a and 6a, show the Western blots obtained using beads coated with equal amounts of MC MV and BR MV, respectively. The beads coated with MC MV clearly bind Rab27a, whereas those coated with BR MV, which lacks both exons D and F, do not. To provide additional evidence that the ∼27-kDa band in lane 5A is Rab27a, we generated a rabbit polyclonal antibody against a GST fusion protein containing full-length rat Rab27a. This antibody, like the monoclonal α-Rab27a antibody, is absolutely specific for Rab27a in the context of the melanocyte, because Western blots of whole cell extracts from wild-type (Figure 8, lane 3) and ashen melanocytes (Figure 8, lane 4) show a ∼27-kDa band in the former, and nothing in the latter. Blots of the material eluted from the MC MV- and BR MV-coated beads and probed with this α-Rab27a polyclonal antibody confirmed that the former bind Rab27a (Figure 8, lane 5b), whereas the latter do not (Figure 8, lane 6b).
To determine whether the association of MC MV with Rab27a, like the association of MC MV with the melanosome, requires exon F, but not exon D, we compared the yields of Rab27a bound to beads coated with equal amounts of MC MV, MC MVΔD, and MC MVΔF (Figure 8, lanes 7–9) by using both the monoclonal (7a–9a) and polyclonal (7b–9b) α-Rab27a antibodies. Both antibodies showed that MC MVΔD-coated beads (8a and 8a) bound approximately the same amount of Rab27a as MC MV-coated beads (7a and 7b). Densitometry of blots made using the polyclonal antibody revealed that MC MVΔD bound 91 ± 6% (n = 3) as much Rab27a as MC MV. In contrast to this, both antibodies showed that MC MVΔF-coated beads (9a and 9b) bound negligible amounts of Rab27a. Densitometry of blots made using the polyclonal antibody revealed that MC MVΔF bound <0.05% as much Rab27a as MC MV (n = 3). Based in large part on the strength of these controls, which are in precise agreement with the in vivo data regarding the significance of exons D and F, we conclude that myosin Va and Rab27a interact in either a direct or indirect manner.
Interaction of Rab27a with Myosin Va-coated Beads Is GTP Dependent
All of the experiments described above were done with melanocytes lysed in buffer containing the nonhydrolyzable GTP analog GTPγS, so as to drive Rab27a toward its active, GTP-bound state. Given that Rab GTPases typically exhibit a significant reduction in affinity for their effectors when in the GDP-bound state (Zerial and McBride, 2001), and that the T23N mutant of Rab27a, which should bias the Rab toward its GDP-bound state, does not rescue ashen melanocytes and disables myosin Va-dependent melanosome capture when over expressed in wild-type melanocytes (Hume et al., 2001; Wu et al., 2001), we compared the yields of Rab27a bound to MC MV that were incubated with melanocyte detergent lysates prepared in either 100 μM GTPγS or 100 μM GDP. Figure 8 shows that the amount of Rab27a bound by MC MV-coated beads was significantly reduced by the substitution of GDP (lanes 11a and 11b) for GTPγS (lanes 10a and 10b) in the lysis and wash buffers. Densitometry of blots made using the polyclonal antibody revealed that MC MV-coated beads bound 23 ± 8% (n = 3) as much Rab27a when incubated with GDP-containing lysates as when incubated with GTPγS-containing lysates. Given the likelihood that the ratio of GTP-bound to GDP-bound Rab27a is lower in lysates prepared in the presence of excess GDP compared with excess GTPγS, we conclude that the interaction between myosin Va and Rab27a is GTP dependent.
Myosin Va and Rab27a Seem to Interact Indirectly
Although the results in Figure 8 reveal the existence of a reasonably stable association between myosin Va and Rab27a, they do not indicate whether these two proteins interact directly or indirectly. In an effort to resolve this issue, we expressed Rab27a as a histidine-tagged fusion protein in Escherichia coli, purified it to homogeneity, loaded it with GTPγS, incubated it at a concentration of 4 μM with beads coated with purified MC MV or with purified BR MV as a control, washed the beads, and subjected the materials eluted using excess FLAG peptide to Western blot analysis with the polyclonal antibody to Rab27a. Although Rab27a could be detected upon long exposure in the eluate from MC MV-coated beads, the amount was no greater in two separate experiments than that found in the eluate from BR MV-coated beads, which we consider to represent background (our unpublished data). Moreover, efforts to demonstrate a direct interaction between Rab27a and myosin Va by yeast two-hybrid analysis (see MATERIALS AND METHODS) were negative (our unpublished data), despite the fact that both constructs used in the pairwise assay (the dominant active Q78L mutant of Rab27a and the short tail portion of the melanocyte-spliced isoform of myosin Va) have been used successfully in other two hybrid screens (Wu and Hammer, unpublished data). We conclude, therefore, that the interaction between myosin Va and Rab27a is probably indirect.
DISCUSSION
Toward a Complete Molecular Description of Melanosome Receptor for Myosin Va
In this article, we show that exon F is required for myosin Va to associate with and influence the position of melanosomes in the context of both dominant negative and rescue experiments. Furthermore, exon D, the other alternatively spliced exon present in the distal stalk domain of the melanocyte-spliced isoform of myosin Va, is not required. Although these results have inherent value in defining the portions of the myosin Va heavy chain that are required for its association with the melanosome (see below), they served an even greater purpose as stringent controls for experiments designed to verify a physical association between myosin Va and Rab27a: only beads coated with myosin Va isoforms that contain exon F were found to bind Rab27a present in melanocyte lysates. Importantly, experiments using purified Rab27a were consistent with the idea that this interaction is indirect. Although we cannot exclude the possibility that the lack of interaction between purified Rab27a and myosin Va was due to problems with bacterially expressed Rab27a (e.g., a direct, stable interaction between Rab27a and myosin Va might require the presence of geranylgeranyl groups on the Rab), the fact that Rab27a and myosin Va also did not interact in the yeast two-hybrid assay, together with the growing evidence that another protein (melanophilin) is required for myosin Va-dependent melanosome capture (see below), leads us to conclude that Rab27a does not function alone as the melanosome receptor for myosin Va, but rather as an essential component of a multiprotein complex that serves as the receptor. Given that Rab27a, like other Rab GTPases, is tightly associated with the lipid bilayer by virtue of two C20 geranylgeranyl groups present near its C terminus (Seabra et al., 1995), and that the two cysteine residues to which these hydrophobic groups are attached posttranslationally are required for Rab27a to rescue ashen melanocytes (Wu et al., 2001), we also conclude that the minimal role played by Rab27a within this receptor complex is to tether myosin Va, and all other proteins that are required for their stable association, to the limiting membrane of the melanosome.
The minimum additional complexity required to generate a functional receptor would be the addition of a single protein that serves to bridge the indirect interaction between myosin Va and Rab27a. A strong candidate for such a bridging protein is melanophilin, the protein encoded by the leaden locus (Matesic et al., 2001), because leaden mice exhibit a coat color defect that is indistinguishable from that of ashen and dilute mice, mice harboring mutations in all three genes (on a nonagouti background) are no more diluted than the individual mutants (Matesic et al., 2001), and all three mutations are suppressed by the extragenic, semidominant suppressor dsu (Moore et al., 1988). Moreover, melanophilin is predicted to be a Rab binding protein based on its sequence similarity to the Rab3a-binding domain of Rabphilin 3a, Rab3a effector protein. Based on these observations, and on the fact that leaden melanocytes seem to be defective in myosin Va-dependent melanosome capture (Provance et al., 1996), we speculate that melanophilin binds to both Rab27a and myosin Va to bridge their interaction.
Identification of Myosin Va Heavy Chain Sequences Required for Targeting to Melanosome
In a previous study, we showed that MC ST contains all of the sequence information required for targeting myosin Va to the melanosome (Wu et al., 1998). In this study, dominant negative experiments performed using the two independently folded portions of MC ST (the stalk and globular tail domains), as well as versions of MC ST containing all of the normal splice variations within the stalk (+D, +F; −D, −F; +D, −F; −D, +F), indicated that MC ST requires both exon F and the GTD to localize to melanosomes and influence their position. This result is equally consistent with the presence of separate binding sites on the myosin Va receptor for exon F and the GTD, both of which must be occupied for stable association, or the presence of a single, conformation-dependent binding interface whose formation requires both exon F and the GTD.
Our observation that the GTD is required for melanosome targeting agrees with a number of recent studies implicating this domain in cargo binding, most strikingly those involving the yeast type V myosin Myo2p and its interactions with secretory vesicles (Schott et al., 1999) and the vacuole membrane (Catlett et al., 2000). Our demonstration that the GTD is not sufficient for melanosome targeting contrasts, however, with the work on Myo2p, where the ability of its GTD to associate with cargo and to generate a dominant negative phenotype indicates that its GTD is sufficient for targeting (Schott et al., 1999; Catlett et al., 2000). Similarly, the GTD of myosin Va is sufficient for targeting to the centrosome (Espreafico et al., 1998) and for binding to melanosomes in frog melanophores (Karchar et al., 2001), a result that seems to contradict our findings. We are, nevertheless, confident in our conclusions regarding the GTD, in large part because our extensive data on the requirement for exon F, which is supported by the identification of two dilute alleles where the sole defect in myosin Va is the exclusion of exon F from the melanocyte-spliced isoform (Huang et al., 1998), indicate that the GTD cannot be sufficient. The discrepancy between our results and those reported by Karchar et al. (2001) may reflect the difference in the way the GTD targeting was scored [in this study by colocalization and the ability to create a dominant negative phenotype in vivo, and in Karchar et al. (2001) by Western blot analysis of melanosome fractions isolated from melanophores transfected with the GTD]. Although we cannot eliminate the possibility that some GTD associates with melanosomes in mouse melanocytes, we can conclude that the amount is not sufficient to detect functionally or by GFP fluorescence. Along these lines, we note that previous experiments in melanophores showed that MC ST colocalizes with melanosomes and generates a dominant negative phenotype (Rodgers et al., 1999). Finally, the discrepancy regarding the GTD could be due to species-specific differences in the mechanism by which myosin V binds to melanosomes.
Regulation of Myosin Va–Melanosome Interaction
The interactions of Rab GTPases with their effectors are regulated by the Rabs' intrinsic and catalyzed rates of GTP hydrolysis and nucleotide exchange, because Rabs exhibit reasonable affinities for their effectors only when in the GTP-bound state (Zerial and McBride, 2001). Consistent with this, the T23N mutant of Rab27a, which should bias the Rab toward its GDP-bound or “off” state, does not rescue ashen melanocytes, and disables myosin Va-dependent melanosome capture when overexpressed in wild-type melanocytes (Hume et al., 2001; Wu et al., 2001). Herein, we have shown that the preparation of melanocyte lysates in the presence of excess GDP rather than excess GTPγS reduces the amount of Rab27a bound to beads coated with purified melanocyte myosin Va by approximately fourfold. Although we did not determine the ratio of GTP-bound to GDP-bound Rab27a in the lysates, the likelihood that the ratio is significantly lower in the GDP lysate leads us to conclude that the indirect interaction between myosin Va and Rab27a is GTP dependent. Although an in-depth understanding of the nature of this GTP dependence must await the identification of the additional protein or proteins that, together with Rab27a, comprise the receptor, our results suggest that the Rab27a-dependent recruitment of myosin Va to the melanosome surface, and, by extension, pigmentation in vivo, should be regulated by factors controlling the nucleotide state of the Rab27a, such as Rab27a-specific guanine nucleotide exchange proteins and GTPase-activating proteins. Direct tests of this hypothesis in living cells should be possible once reagents that perturb or enhance the activity of these factors become available.
Myosin Va–melanosome association in mammalian melanocytes may also be regulated by phosphorylation of the myosin Va heavy chain, especially given the recent work with frog melanophores, where the cell cycle-dependent phosphorylation of a serine residue within the GTD by calcium/calmodulin-dependent protein kinase II causes the dissociation of myosin Va from melanosomes during mitosis (Rodgers et al., 1999; Karcher et al., 2001). Whether phosphorylation at this site affects myosin Va–melanosome interaction in mammalian melanocytes, which are postmitotic, remains to be determined.
Generality of Our Results
Evidence is growing that Rab GTPases regulate motor proteins responsible for vesicle motility as well as the machinery governing vesicle docking and fusion (reviewed in Zerial and McBride, 2001; Hammer and Wu, 2002). For example, the GTP-bound form of Rab6 interacts physically with the kinesin-like protein Rabkinesin-6 (RB6K/Rab6-KIFL) (Echard et al., 1998; Hill et al., 2000; Fontijn et al., 2001); and Rab5, which regulates the formation and homotypic fusion of early endosomes, regulates the movement of these endosomes toward the minus end of microtubules (Nielsen et al., 1999). Furthermore, myosin Vb, the product of a second vertebrate myosin V heavy chain gene (Zhao et al., 1996), has recently been linked both physically and functionally to Rab11a (Lapierre et al., 2001), a Rab GTPase that regulates the recycling of membrane proteins after their internalization by endocytosis. Interestingly, although the tail of myosin Vb binds directly to Rab11a in a two-hybrid screen, a Rab11a-binding protein has been identified (Rab11-FIP2) that also interacts with the myosin tail (Hales et al., 2001). Perhaps the closest parallel with the work presented herein can be found in recent studies of the yeast type V myosin Myo2p (Schott et al., 1999), and the Rab GTPase Sec4p (Walch-Solimena et al., 1997), which indicate that secretory vesicle transport requires not only Myo2p but also Sec2p, a nucleotide exchange factor for vesicle-bound Sec4p. Moreover, mutations in Sec2p block vesicle transport at the restrictive temperature without blocking the translocation of Myo2p on actin, indicating that Sec2p is required for the coupling of secretory vesicles to Myo2p (Schott et al., 1999). Together, these results suggest that Sec2p, GTP-bound Sec4p, or a complex of the two, recruit Myo2p onto the secretory vesicle surface by serving as essential components of the vesicle receptor for Myo2p (reviewed in Pruyne and Bretscher, 2000).
In conclusion, we have provided definitive biochemical evidence of an association between myosin Va and Rab27a and presented additional evidence that their interaction is indirect. We conclude, therefore, that the dependence of myosin Va–melanosome association on Rab27a that was demonstrated by the analysis of ashen melanocytes (Bahadoran et al., 2001; Hume et al., 2001; Wu et al., 2001) is due to the fact that the Rab serves as an essential component of the melanosome receptor for myosin Va, and not as an activator of another component that serves as the receptor. Our data also point to the existence of an additional protein that would serve to bridge the indirect interaction between Rab27a and myosin Va. Given that Rab GTPases in general rely on the GTP-dependent recruitment of effector proteins to drive their downstream functions, we speculate that melanophilin, the product of the leaden locus, and a postulated Rab27a effector (Matesic et al., 2001), serves as the bridging protein. Although it remains to be seen whether our findings represent a general mechanism for motor–organelle association, the properties of Rab GTPases (reviewed in Zerial and McBride, 2001) do fit nicely with the requirements for motor protein receptors, including the fact that they are firmly attached to organelle membranes, partition as resident proteins within specific membrane compartments, and are present in sufficient numbers to allow specific pairing with most if not all members of the actin- and microtubule-based motor protein superfamilies. Moreover, Rabs are already committed to controlling, along with cognate soluble N-ethylmaleimide-sensitive factor attachment protein receptors, the specificity of fusion between the vesicle and its target membrane, so it makes sense that they might also control the movement of the vesicle to its site of fusion by specifying the motor responsible for this movement. Finally, the regulation of the GTP/GDP ratio on the Rab would provide an excellent way to regulate motor–Rab interaction during this phase of the vesicle's life cycle (reviewed in Hammer and Wu, 2002).
ACKNOWLEDGMENTS
We thank Dr. Paul Randazzo for assaying the Rab27a fusion proteins for GTP binding, and Dr. Edward D. Korn for comments on the manuscript.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–12–0595. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–12–0595.
REFERENCES
- Bahadoran P, Aberdam E, Mantoux F, Busca R, Bille K, Yalman N, de Saint Basile G, Casaroli-Marano R, Ortonne J-P, Ballotti R. Rab27a: a key to melanosome transport in human melanocytes. J Cell Biol. 2001;152:843–849. doi: 10.1083/jcb.152.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlett NL, Duex JE, Tang F, Weisman LS. Two distinct regions in a yeast myosin-V tail domain are required for the movement of different cargoes. J Cell Biol. 2000;150:513–525. doi: 10.1083/jcb.150.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheney RE, O'Shea MK, Heuser JE, Coelho MV, Wolenski JS, Espreafico EM, Forscher P, Larson RE, Mooseker MS. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993;75:13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- Christofordis S, Zerial M. Purification and identification of novel Rab effectors using affinity chromatography. Methods. 2000;20:403–410. doi: 10.1006/meth.2000.0953. [DOI] [PubMed] [Google Scholar]
- Echard A, Jollivet F, Martinez O, Lacapere J-J, Rousselet A, Janoueix-Lerosey I, Goud B. Interaction of a Golgi-associated kinesin-like protein with Rab6. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- Espindola FS, Suter DM, Partata LB, Cao T, Wolenski JS, Cheney RE, King SM, Mooseker MS. The light chain composition of chicken brain myosin-Va:calmodulin, myosin-II essential light chains, and 8-kDa dynein light chain/PIN. Cell Motil Cytoskeleton. 2000;47:269–281. doi: 10.1002/1097-0169(200012)47:4<269::AID-CM2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Espreafico EM, Coling DE, Tsakraklides V, Krough K, Wolenski JS, Kalinec G, Kachar B. Localization of myosin-V in the centrosome. Proc Natl Acad Sci USA. 1998;95:8636–8541. doi: 10.1073/pnas.95.15.8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontijn RD, Goud B, Echard A, Jollivet F, Van Marle J, Pannekoek H, Horrevoets AJG. The human kinesin-like protein RB6K is under tight cell cycle control and is essential for cytokinesis. Mol Cell Biol. 2001;21:2944–2955. doi: 10.1128/MCB.21.8.2944-2955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EKL, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Hammer III JA, Wu XS. Rabs grab motors: defining the connections between Rab GTPases and motor proteins. Curr Opin Cell Biol. 2002;14:69–75. doi: 10.1016/s0955-0674(01)00296-4. [DOI] [PubMed] [Google Scholar]
- Hearing VJ, King RA. Determinants of skin color: melanocytes and melanization. In: Levine N, editor. Pigmentation and Pigmentary Disorders. London, UK: CRC Press; 1993. pp. 4–18. [Google Scholar]
- Hill E, Clarke M, Barr FA. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 2000;21:5711–5719. doi: 10.1093/emboj/19.21.5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JD, Mermall V, Strobel MC, Russel LB, Mooseker MS, Copeland NG, Jenkins NA. Molecular genetic dissection of mouse unconventional myosin-Va: tail region mutations. Genetics. 1998;148:1963–1972. doi: 10.1093/genetics/148.4.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume AN, Collinson LM, Rapak A, Gomes AQ, Hopkins CR, Seabra MC. Rab27a regulates the peripheral distribution of melanosomes in melanocytes. J Cell Biol. 2001;152:795–808. doi: 10.1083/jcb.152.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer III JA. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153:1479–1497. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcher RL, Roland JT, Zappacosta F, Huddleston MJ, Annan RS, Carr SA, Gelfand VI. Cell cycle regulation of myosin-V by calcium/calmodulin-dependent protein kinase II. Science. 2001;293:1317–1320. doi: 10.1126/science.1061086. [DOI] [PubMed] [Google Scholar]
- Koyama Y, Takeuchi T. Ultrastructural observations on melanosome aggregation in genetically defective melanocytes of the mouse. Anat Rec. 1981;201:599–611. doi: 10.1002/ar.1092010404. [DOI] [PubMed] [Google Scholar]
- Lambert J, Onderwater J, Haeghen YV, Vancoille G, Koerten HK, Mommas AM, Naeyaert JM. Myosin V colocalizes with melanosomes and subcortical actin bundles not associated with stress fibers in human epidermal melanocytes. J Invest Dermatol. 1998;111:835–840. doi: 10.1046/j.1523-1747.1998.00395.x. [DOI] [PubMed] [Google Scholar]
- Lapierre LA, Kumar R, Hales C, Navarre J, Bhartur SG, Burnette JO, Provance DW, Jr, Mercer JA, Bahler M, Goldering JR. Myosin Vb is associated with the plasma membrane recycling system. Mol Biol Cell. 2001;12:1843–1857. doi: 10.1091/mbc.12.6.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matesic LE, Yip R, Reuss AE, Swing DA, O'Sullivan TN, Fletcher CF, Copeland NG, Jenkins NA. Mutations in MLPH, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice. Proc Natl Acad Sci USA. 2001;98:10238–10243. doi: 10.1073/pnas.181336698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menasche G, Pastural E, Feldmann J, Certain S, Ersoy F, Dupuis S, Wulffrat N, Bianchi D, Fischer A, Le Deist F, de Saint Basile G. Mutations in R.A.B27A cause Griscelli syndrome associated with hemophagocytic syndrome. Nat Genet. 2000;25:173–176. doi: 10.1038/76024. [DOI] [PubMed] [Google Scholar]
- Mercer JA, Seperack PK, Strobel MC, Copeland NG, Jenkins NA. Novel myosin heavy chain encoded by murine dilute coat color locus. Nature. 1991;349:709–713. doi: 10.1038/349709a0. [DOI] [PubMed] [Google Scholar]
- Moore KJ, Swing DA, Rinchik EM, Mucenski ML, Buchberg AM, Copeland NG, Jenkins NA. The murine dilute suppressor gene dsu suppresses the coat color phenotype of three pigment mutations that alter melanocyte morphology, d, ash, and ln. Genetics. 1988;119:933–941. doi: 10.1093/genetics/119.4.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento AAC, Amaral RG, Bizario JCS, Larson RE, Espreafico EM. Subcellular localization of myosin V in the B16 melanoma cells, a wild type cell line for the dilute gene. Mol Biol Cell. 1997;8:1971–1988. doi: 10.1091/mbc.8.10.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Severin F, Backer JM, Hyman AA, Zerial M. Rab5 regulates motility of early endosomes on microtubules. Nat Cell Biol. 1999;1:376–382. doi: 10.1038/14075. [DOI] [PubMed] [Google Scholar]
- Provance DWJ, Wei W, Ipe V, Mercer JA. Cultured melanocytes from dilute mice exhibit dendritic morphology and altered melanosome distribution. Proc Natl Acad Sci USA. 1996;93:14554–14558. doi: 10.1073/pnas.93.25.14554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. II. The role of the cortical actin cytoskeleton. J Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Provance DW, Jr, Mooseker MS, Mercer JA. Class V myosins. Biochim Biophys Acta. 2000;11496:36–51. doi: 10.1016/s0167-4889(00)00007-0. [DOI] [PubMed] [Google Scholar]
- Rodgers SL, Karchar RL, Roland JT, Minin AA, Steffen W, Gelfand VI. Regulation of melanosome movement in the cell cycle by reversible association with myosin V. J Cell Biol. 1999;146:1265–1275. doi: 10.1083/jcb.146.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D, Ho J, Pruyne D, Bretscher A. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J Cell Biol. 1999;147:791–807. doi: 10.1083/jcb.147.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabra MC, Ho YK, Anant JS. Deficient geranylgeranylation of Ram/Rab27 in choroideremia. J Biol Chem. 1995;270:24420–24427. doi: 10.1074/jbc.270.41.24420. [DOI] [PubMed] [Google Scholar]
- Seperack PK, Mercer JA, Strobel MC, Copeland NG, Jenkins NA. Retroviral sequences located within introns of the dilute gene alter expression in a tissue-specific manner. EMBO J. 1995;14:2326–2332. doi: 10.1002/j.1460-2075.1995.tb07227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel MC, Seperack PK, Copeland NG, Jenkins NA. Molecular analysis of two mouse dilute locus deletion mutations: spontaneous dilute lethal20J and radiation-induced dilute prenatal Aa2 alleles. Mol Cell Biol. 1990;10:501–509. doi: 10.1128/mcb.10.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terui T, Kahn RA, Randazzo PA. Effects of acid phospholipids on nucleotide exchange properties of ADP-ribosylation factor 1. J Biol Chem. 1994;269:28130–28135. [PubMed] [Google Scholar]
- Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Chen L, Arcucci O, Harvey EV, Bowers B, Xu Y, Hammer III JA, Sellers JR. Effect of ADP and ionic strength on the kinetic and motile properties of recombinant mouse myosin V. J Biol Chem. 2000;275:4329–4335. doi: 10.1074/jbc.275.6.4329. [DOI] [PubMed] [Google Scholar]
- Wei Q, Wu X, Hammer III JA. The predominant defect in dilute melanocytes is in melanosome distribution and not cell shape, supporting a role for myosin V in melanosome transport. J Muscle Res Cell Motil. 1997;18:517–527. doi: 10.1023/a:1018659117569. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Salata MW, Susalka SJ, Pfister KK. Light chains of mammalian cytoplasmic dynein: identification and characterization of a family of LC8 light chains. Cell Motil Cytoskeleton. 2001;49:229–240. doi: 10.1002/cm.1036. [DOI] [PubMed] [Google Scholar]
- Wilson S, Yip R, Swing DA, O'Sullivan N, Zhang Y, Novak ED, Swank RT, Russell LB, Copeland NG, Jenkins NA. A mutation in Rab27a causes the vesicle transport defects observed in ashen mice. Proc Natl Acad Sci USA. 2000;97:7933–7938. doi: 10.1073/pnas.140212797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bowers B, Rao K, Wei Q, Hammer III JA. Visualization of melanosome dynamics within wild-type and dilute melanocytes suggests a paradigm for myosin V function in vivo. J Cell Biol. 1998;143:1899–1918. doi: 10.1083/jcb.143.7.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bowers B, Wei Q, Kocher B, Hammer III JA. Myosin V associates with melanosomes in mouse melanocytes: evidence that myosin V is an organelle motor. J Cell Sci. 1997;110:847–859. doi: 10.1242/jcs.110.7.847. [DOI] [PubMed] [Google Scholar]
- Wu X, Rao K, Bowers MB, Copeland NG, Jenkins NA, Hammer III JA. Rab27a enables myosin Va-dependent melanosome capture by recruiting the myosin to the organelle. J Cell Sci. 2001;114:1091–1100. doi: 10.1242/jcs.114.6.1091. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhao LP, Koslovsky JS, Reihard J, Bahler M, Witt AE, Provance DW, Jr, Mercer JA. Cloning and characterization of myr6, an unconventional myosin of the dilute/myosin-V family. Proc Natl Acad Sci USA. 1996;93:10826–10831. doi: 10.1073/pnas.93.20.10826. [DOI] [PMC free article] [PubMed] [Google Scholar]