Abstract
Transgenic farm animals are attractive alternative mammalian models to rodents for the study of developmental, genetic, reproductive and disease-related biological questions, as well for the production of recombinant proteins, or the assessment of xenotransplants for human patients. Until recently, the ability to generate transgenic farm animals relied on methods of passive transgenesis. In recent years, significant improvements have been made to introduce and apply active techniques of transgenesis and genetic engineering in these species. These new approaches dramatically enhance the ease and speed with which livestock species can be genetically modified, and allow to performing precise genetic modifications. This paper provides a synopsis of enzyme-mediated genetic engineering in livestock species covering the early attempts employing naturally occurring DNA-modifying proteins to recent approaches working with tailored enzymatic systems.
Keywords: Active transgenesis, Livestock, Binary transposon system, Designer nuclease, Recombinase, Integrase, Synthetic biology
Introduction
Transgenic farm animals can serve as excellent models of human diseases, of biopharming, and of basic research [1–5]. During the past few years, transgenic farm animals have gained renewed popularity, because of the availability of annotated genome depositories (http://www.ensembl.org; http://www.ncbi.nlm.nih.gov/genome) and because of the introduction of active methods of transgenesis. Active transgenesis refers to the introduction of exogenously provided enzymes or nucleic acids encoding them [2, 6, 7], which catalyze specific gain-of-function or loss-of-function genetics in an unprecedented pace. The exogenous enzymes are only transiently present; however, by carefully selecting highly active or hyperactive variants [8–11], the desired genetic modification can be performed in individual cells, such as the mammalian zygote. Prominent examples are hyperactive transposon systems, such as Sleeping Beauty [9, 10], piggyBac [11], as well as designer nucleases, including zinc finger nucleases (ZFNs), transcription activator-like element nucleases (TALENs) and RNA-guided nucleases [1–3, 12]. In addition, Cre recombinase and ΦC31 integrase found some interest for farm animal transgenesis [13, 14]. Viral integrases (retrovirus, lentivirus, adeno-associated virus) apply similar mechanisms; however, viral transgenesis is already covered by a number of excellent reviews [15–18], and will not be discussed in this paper. The repertoire of molecular tools now allows the precise modification of large mammalian genomes at rapid pace and has led to a recent boost in this field [2, 19–22].
Brief time course of livestock transgenesis
Since the isolation of class II restriction enzymes (RE), the hypothesis that simultaneous delivery of a RE in combination with a transgene would increase the efficiency of foreign DNA incorporation was postulated. First evidence for this hypothesis came from a study in which illegitimate integration events of non-homologous DNA fragments into yeast genome was several fold enhanced when a standard RE was included in the transformation mixture [23]. The restriction enzyme-mediated integration (REMI) was subsequently used in unicellular organisms, fungi [24, 25] and xenopus [26]. The catalytic activity of RE in cultured mammalian cells [27, 28] prompted researchers to apply REMI in combination with pronuclear microinjection of mouse zygotes [29]. In the mouse model, the rate of transgenic embryos and live pups by PCR analysis was doubled (18 vs 9 %) in REMI versus standard pronuclear injection [29]. However, no information regarding number of copies, genomic sites of transgene incorporation, expression and transmission to progeny were given [29]. Further independent replications of this approach are warranted to unequivocally establish the usefulness of REMI for animal transgenesis.
It has been postulated that the co-delivery of transgene and a site-specific RE could increase efficiency of integration into the host genome by three non-excluding ways: (1) protecting the ends of the transgene constructs, (2) inducing DNA breaks, and (3) stimulating endogenous DNA repair mechanisms [30].
It has been well established that frequently cutting RE may pose a potential risk of causing genotoxic damage [31, 32]. In fact, introduction of a RE by electroporation into mammalian cells has been shown to induce genomic rearrangements such as deletions, duplications, and translocations [28, 33, 34]. To avoid these detrimental effects, REMI can be performed with rare-cutting meganucleases. Meganucleases or homing endonucleases (HE) are naturally occurring enzymes that recognize long consensus sequences spanning 12–40 bp [35]. One of the best characterized is I-SceI from Saccharomyces cerevisiae [36]. Albeit the consensus sequence spans 18 bp, I-SceI seems to allow some ambiguity in the recognition site. Co-injection of a transgene flanked by two I-SceI restriction sites with purified I-SceI into fertilized eggs of Oryzias latipes (medaka fish) and Xenopus tropicalis resulted in improved transgenic efficiencies [37, 38]. Preliminary studies assessed I-SceI for transgenesis in livestock [39]. I-SceI-injected bovine zygotes resulted in an increased proportion of embryos expressing the reporter gene and a reduced percentage of mosaic embryos. In a preliminary report [40], the I-SceI approach significantly increased ratio of transgenic bovine fibroblasts, suggesting that I-SceI can enhance transgene integration into the cattle genome. Recently, the first reporter transgenic pigs were generated by an I-SceI approach; however, no details about copy numbers, integration sites and transgene silencing have been reported [41]. Further studies are warranted to reveal the mechanistic role of I-SceI during mammalian transgenesis.
The first evidence for sperm-mediated transport of native simian virus 40 (SV40) DNA into rabbit oocytes came from a study by Brackett et al. [42]. The SV40 DNA encoded the complete genome of the virus, and infective SV40 virions could be recovered by coculture of fertilized embryos with a permissive kidney cell line from African green monkey. Eighteen years later Lavitrano et al. [43] used sperm mixed with plasmid DNA to produce transgenic mice. This report of sperm-mediated gene transfer (SMGT) was soon challenged by an independent study, which failed to replicate the experiment [44]. Since then, SMGT has been assessed for transgenesis in several invertebrates as well as vertebrates [45–47], including domestic species [48–50]. However, most of the studies in mammalian species provided poor evidences for transgene integration or recombinant protein expression.
In 1999, an alternate technique called intracytoplasmic sperm injection-mediated transgenesis (ICSI-Tr) was published [51]. Developed originally to produce transgenic mice, the method was later translated to other mammalian species [52–56] and birds [57]. In this methodology, double-stranded DNA molecules are complexed with membrane-damaged (dead) spermatozoa, which were subsequently microinjected into the cytoplasm of metaphase II oocytes. In this modification of SMGT, the physical or chemical disruption of sperm cell membranes is a prerequisite for successful gene transfer, which then requires the troublesome ICSI procedure. With ICSI-Tr, high percentages of transgenic offspring with low incidence of mosaicism have been reported [51]. Despite the success of this technique in terms of transgenic ratios, it does not escape from the numerous drawbacks of methodologies that rely on passive integration of transgenes, such as concatemeric transgene integration, silencing, and variegated transgene expression [58, 59].
To address some of these concerns, approaches to combine ICSI-Tr with the delivery of ectopic enzymes were assessed [60]. Initial experiments addressed the effect of a bacterial recombinase (RecA) [60] and Tn5 transposase [61] on mouse and livestock transgenesis [62, 63]. Both enzymes were able to increase the proportion of live transgenic animals compared to classic pronuclear microinjection and ICSI-Tr methods [64], but seemed to suffer from sub-optimal activities of the employed enzymes [29, 30].
The need for advanced transgenic methodologies that permit precise genetic and highly efficient modifications in preselected DNA sequences has driven research efforts to develop hyperactive and codon-optimized transposases (SB, PB, Tol2) [10, 11, 65–68], recombinases (Cre, flippase) [13, 69] and customized programmable nucleases, like zinc finger nucleases (ZFN), transcription activator-like element nucleases (TALEN), and RNA-guided nucleases [2, 19, 22, 70], which already initiated a revolution in the field of animal transgenesis.
Application of transposon systems for genetic engineering
Transposons or jumping genes belong to a diverse family of genetic elements that are able to move horizontally in genomes. Transposons were originally described in maize [71], but later identified as widespread components in the genomes of prokaryotes and eukaryotes [72]. Interestingly, transposable elements comprise high proportions of eukaryotic genomes (i.e., about 45 % of the human genome [73]) and the vast majority of them are inactive due to accumulated deleterious mutations [74].
Transposons are grouped in two distinct categories according to the mechanism used for mobilization (transposition). Class I transposons also called retrotransposons rely on a RNA intermediate, which is reverse-transcribed in a new genomic locus. As consequence the number of genomic units increases by a mechanism that can be characterized as “copy and paste” [75]. Retrotransposon mobilization is capable of inducing random mutations at high frequency, disrupting endogenous genes, and therefore it has been held responsible for causing several genetic disorders [75, 76].
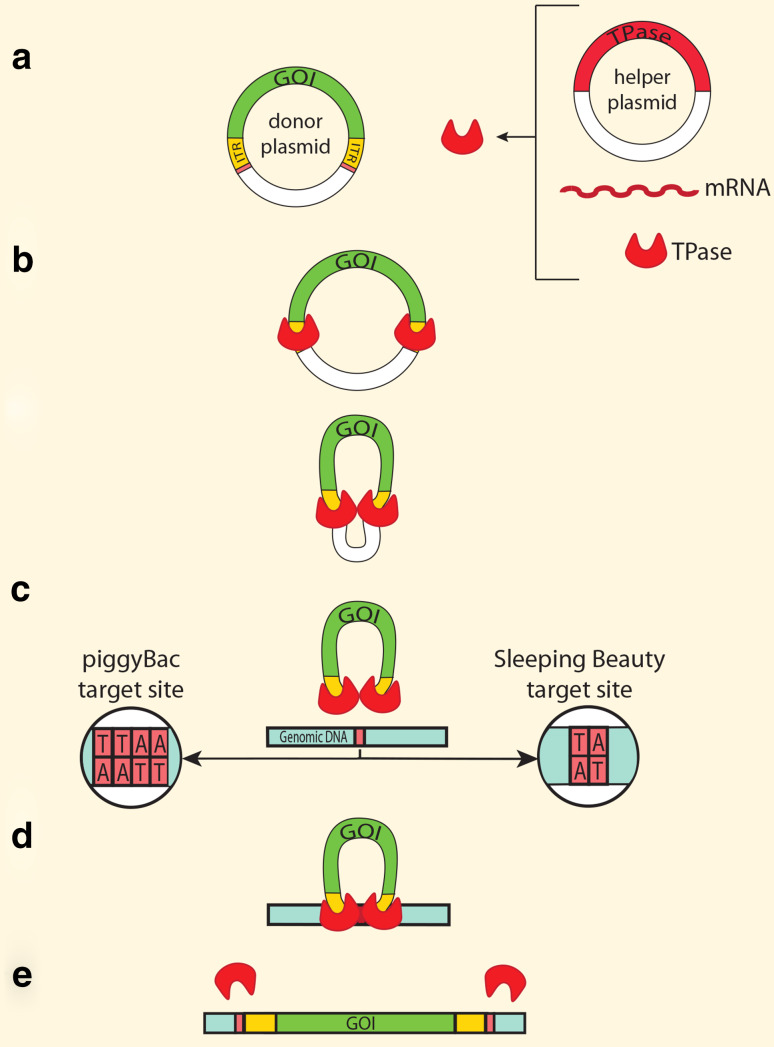
Class II transposons or DNA transposons are mobilized by a process that operates by a “cut and paste” mechanism [10, 77]. A transposase specifically recognizes inverted terminal repeats (ITRs), and precisely removes and relocates the ITR-flanked DNA segment to a different genome position [10, 77] (Fig. 1).
Fig. 1.
General mechanism for transposase-mediated transgene integration into the host genome. Transposase (TPase) is commonly synthesized from an expression vector (helper plasmid), mRNA or it is (rarely) provided as protein. The gene of interest (GOI) flanked by ITRs is delivered on an independent vector (donor plasmid) (a). TPase recognizes/binds ITRs flanking the GOI and catalyzes staggered double-strand breaks at the ends of ITRs (b). The resulting transposon–transposase complex binds at specific target sites in the host genomic DNA (c), and integrates the ITR-flanked transposon (d–e)
DNA transposons have been manipulated as useful gene transfer vectors for germline transgenesis, insertional mutagenesis and somatic cell transgenesis (gene therapy) (reviewed in [78]). Bicomponent transgenic systems have been developed [79] in which the gene of interest (GOI) is flanked by ITRs, and the transposase is provided in trans as mRNA or as an expression plasmid (helper plasmid) (Fig. 1). The excised transposon plus transposase proteins binds to a target DNA, where the insertion takes place [80]. Most transposases catalyze integration at short consensus sequences, for example TC1/mariner transposases recognize TA dinucleotides, and PB transposase recognizes TTAA tetranucleotides (Fig. 1). Through this mechanism, one monomeric copy of a transposon is integrated in the genome, leaving the empty backbone of the donor plasmid, which is eventually degraded (Fig. 1) [77].
Due to the development of hyperactive transposase variants, two-component transposon system has been adopted as an improved tool for germline transgenesis in a broad range of invertebrate and vertebrate species (reviewed in [81]). Integration efficiencies reported for various transposon systems (Table 1) rival the high integration rates of viral-based methods. In contrast to viral methods, transposon systems are characterized by being safe and capable of delivering large cargos [68, 82–85]. An advantage of transposase-mediated transgenesis is the monomeric transgene integration, making these constructs less prone to silencing in transposon transgenic animals [65].
Table 1.
Transposon transgenesis approaches in livestock
| Transposon | Species | Construct | Antibiotic selection | Overall efficiency* | Expression pattern | Unspecific integrations | Germline transmission generations | Method | References |
|---|---|---|---|---|---|---|---|---|---|
| Sleeping Beauty | Rabbit | CAGGS-Venus | AB-free | 1.4 % | Ubiquitous | No | F0, F1, F2 | PNI | [258] |
| Pig | CAGGS-Venus | AB-free | 6.8 % | Ubiquitous | ~5 % | F0, F1, F2 | CPI | [65] | |
| Pig | floxedUbi-GIN | G418 | NA | Ubiquitous | ~25 % | F0 | SCNT (HMC) | [13] | |
| Pig | DIV PuroΔtk APOBEC3G | Puromycin | ~3 % | Ubiquitous | No | F0 | CT | [87] | |
| Pig | INV-hITGA2/NV-hITGB1 | G418 | NA | Keratinocyte | ND | F0. | SCNT (HMC) | [259] | |
| Pig | HCR-hAAT-D374Y-PCSk9 | Puromycin | NA | Liver | ND | F0 | SCNT (HMC) | [260] | |
| PiggyBac | Chicken | CAGGS-EGFP-IRES-Puro | Puromycin | NA | Ubiquitous | ND | Prefounder, F1 | PGC transfection in vitro | [261] |
| Chicken | CMV-EGFP SV40-Neo | G418 | 49.6 % | Ubiquitous | ND | Prefounder, F1, F2 | PGC transfection in vitro | [262] | |
| Chicken | IRES-LacZ-CAGGS-EGFP-PGK-Neo | G418 | NA | Ubiquitous | ND | Prefounder | Embryo microinjection and electroporation | [263] | |
| Pig | CMV-Neo-EGFP | G418 | 1.3 % | Ubiquitous | ND | F0 | SCNT | [111] | |
| Pig | CAA–tdTomato | AB-free | 7.0 % | Eye lens | 5 % | F0 | CPI | Unpub. (KW) | |
| Pig | CAGGS-EGFP, SV40-Hygro | none | 5.7 % | Ubiquitous | ND | F0 | CPI | [106] | |
| Tol2 | Chicken | CAGGS-EGFP-IRES-Puro | Puromycin | NA | Ubiquitous | ND | Prefounder | PGC transfection in vitro | [261] |
| Chicken | CAGGS-EGFP | AB-free | 1.5 % | Ubiquitous | ND | Prefounder, F1 | PGC transfection in vivo | [264] |
AB-free antibiotic selection marker-free, NA Efficiencies as transgenic offspring per treated embryos are not applicable, ND not determined, CAGGS cytomegalovirus early enhancer/chicken beta-actin promoter, CMV cytomegalo virus (immediate early) promoter, Ubi ubiquitin C promoter, SV40 simian virus 40 promoter, DIV diverse promoters were tested, PGK phosphoglycerate kinase promoter, CAA crystallin Aα promoter, GIN EGFP-IRES-neomycin, APOBEC3G apolipoprotein B mRNA-editing enzyme, INV involucrin promoter, hITGB1 human beta1 integrin, hD374-PCSK9 D374Y gain-of-function mutation in the proprotein convertase subtilisin/kexin type 9, HCR-hAAT hepatocyte control region and human α1-antitrypsin promoter, hITGA2 human Integrin α2 (CD49b), HMC hand made cloning, PGC primordial germ cell
* Transgenic offspring per treated oocytes or embryos
The reported instability of transgene expression from sequences inserted by non-facilitating mechanisms has been linked to methylation of CpG-rich vector sequences [86] that flank the transgene and are co-inserted with the transgene. An added advantage of transposition transgenesis is that each event can be later segregated in the descendants [65, 79, 87]. Segregation of independently inserted sequences by transposition would maximize the overall efficiency of the methodology. The same segregation process can serve to recycle marker/antibiotic selection cassettes to comply with current regulatory guidelines regarding transgenic animals.
Under certain scenarios, intentional removal of the stably inserted sequences is required to turn on or off transgene expression, being the conditional transgenesis an illustrative example of such applications [88]. Since transposition does not change the ITRs [80], the transposon is susceptible to be remobilized, and eventually removed, if the transposase is reintroduced in the system. This can be exploited to excise unwanted genomic DNA sequences flanked by transposon ITRs. Proof of principle for this potentially useful strategy has come from experiments with induced pluripotent stem (iPS) cells [89, 90]. The recent development of transposase variants, which are excision competent, but integration deficient will facilitate the seamless removal of transposons [91]. Thus, transposon systems combine high delivery rates of transgenes and the possibility of seamless transposon removal.
The use of DNA transposons to engineer vertebrate genomes began in 1997, when an active transposase, SB, was reconstructed from non-functional transposon sequences isolated from several salmonid species [79]. It was demonstrated that the original SB variant can transpose DNA sequences in a broad range of vertebrate species [65, 92–95] with moderate activity [9, 96–98]. Using an in vitro evolutionary approach, Mátés et al. [10] finally came up with a hyperactive version: SB100X. Since then, this hyperactive transposase has become the gold standard for transposition approaches in animals.
Successful implementation of SB-mediated integration for germline transgenesis in small animal models was followed by translational research aimed to produce transgenic livestock animals (Table 2). There are two established methodologies to generate transgenic large animals, DNA microinjection of zygotes or somatic cell nuclear transfer (SCNT) (Fig. 2).
Table 2.
Recombinase superfamily divisions (adapted from [126])
| Superfamily | Family | Subdivisions | Recognition sites | Activity | Representative members |
|---|---|---|---|---|---|
| Site-specific recombinases | Tyrosine recombinases | Bidirectional | Identical | Reversible | Cre |
| Inversion, excision and integration | FLP | ||||
| R | |||||
| Unidirectional | Non-identical | Irreversible | Lambda | ||
| Inversion, excision and integration | HK101 | ||||
| pSAM2 | |||||
| Serine recombinases | Small | Identical | Irreversible | Beta-six | |
| Excision | CinH | ||||
| ParA | |||||
| Large | Non-identical | Irreversible | Bxb1 | ||
| Inversion, excision and integration | ΦC31 | ||||
| TP901 |
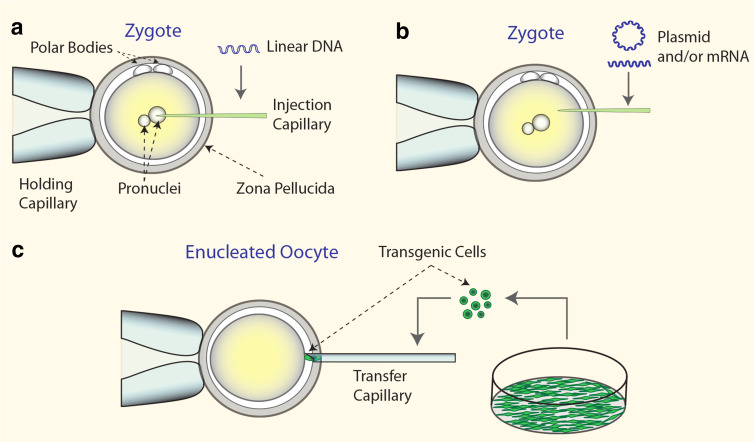
Fig. 2.
Genome engineering via in vivo and in vitro approaches. Injection of nucleic acids and/or protein into the pronucleus of a zygote (a) or into the cytoplasm (b). Genetic modification in primary cells, which are subsequently used in SCNT (c)
Microinjection of pronuclear stage embryos, developed by Gordon et al. [99], became a routine technique to produce transgenic mice. Later, pronuclear microinjection (PNI) was adapted to livestock zygotes [100, 101]. Unlike mouse oocytes, the porcine and bovine counterparts are darkened by lipid droplets precluding the visualization of pronuclei. Therefore, high-speed centrifugation of zygotes is mandatory to visualize the pronuclei [100, 101]. PNI is a technically demanding methodology, which has been characterized by low efficiency in terms of number of transgenic offerings per injected embryo, and variable and instable expression of the recombinant protein in the transgenic animals. The cytoplasmic injection (CPI) of plasmids into the cytoplasm of one-cell embryos [102] represents a simplified alternative, making it suitable for species with opaque zygotes (Fig. 2). Both PNI and the CPI methods were successfully employed with SB, PB and Tol2 transposon components for germline transgenesis in fish [103], frogs [92], mice [10, 104], rats [105] and domestic pigs [65, 87]. A significant increase in the ratio of transgenic animals per microinjected zygotes has been consistently reported. The feasibility and efficiency of transposon-mediated transgene integration into the pig genome are supported by the high proportion of born animals carrying at least one copy of the transgene (>40 %) [65, 66, 106]. Reported overall efficiency was also very impressive reaching 5.7 and 6.8 % of transgenic live pigs per microinjected zygote for PB and SB transposon systems, respectively [65, 106] (Table 1). Moreover, most transposon integrations corresponded to monomeric integrations and very low incidence of passive incorporation of vector backbone or SB transposase vector sequences (Table 1). Interestingly, all transgenic pigs stably expressed the transgene in a promoter-dependent manner in SB transgenic animals and only one case of variegated reporter expression was observed in the PB transgenic group [106]. This can be interpreted as transposase prefers safe harbor loci for integration.
Transposition transgenesis is also compatible with SCNT [87, 107]. SCNT involves the introduction of a somatic cell into an enucleated metaphase II-arrested oocyte, followed by activation by chemical or electric stimulation, and subsequent transfer to synchronized surrogate females for development to term [108]. Since the first report of the successful cloning of sheep from cultured cells [109], SCNT has become a major method to produce transgenic livestock. Advantages of SCNT are: (1) the high rate of transgenic animals per born animals, which often reaches 100 % (but low overall efficiency), and (2) the possibility to characterize the genotype of the somatic cells before use as nuclear donor [110]. High likelihood of obtaining a transgenic animal by SCNT with known genetic makeup would reduce costs associated with producing a transgenic animal. This is particularly relevant for monotocous species with long-generation interval like cattle, in which husbandry expenditures of surrogate females negatively impact the sustainability of transgenic endeavors.
Donor cells can be transfected with linear or circular DNA transgenes for passive integration or with the components of an active system, such as SB or PB (Fig. 2) [107, 111]. Although donor cells are not considered a limiting resource for SCNT, the use of transposons is associated with significantly enhanced proportion of stably transfected cells. Delivery of pmGENIE-3, a helper independent PB transposon, to bovine primary fibroblast cells in culture caused an impressive 42-fold increase in the number of resistant cell colonies over controls [112]; similar results were reported for an established immortalized porcine cell line, as well as in primary porcine cells transfected with the SB, PB, Tol2 or Passport transposon systems [67, 68, 87, 95]. A disadvantage of the SCNT approach is that an antibiotic selection cassette is usually needed for isolation of transgenic cell clones and therefore, it is carried over into SCNT transgenic animals. This drawback, which is strongly discouraged by current regulatory guidelines, can be overcome if the antibiotic gene is supplied on a separate vector and consequently is genomically integrated independently from the GOI. Under this circumstance, the antibiotic selection cassette may be removed by segregation of the GOI from unwanted sequences by an additional round of breeding [87]. Alternatively, unwanted sequences can be removed by Cre or Flp recombinase systems as addressed in depth in the next section.
Clean and stable genomic insertion events mediated by transposases are the most striking features of these systems, making transposons first choice when gene addition for gain of function or loss of function by expression of a dominant negative allele or RNA interference is sought. The availability of different transposon systems with distinct characteristics regarding their recognition sites and/or biased genome distribution confer versatility to the system by offering the possibility of choosing a specific transposon according to the application or particular goal. For instance, PB transposase has a slight tendency to land in or close to transcriptionally active regions of the chromatin [113], so it may be more appropriate for insertional mutagenesis studies. On the other hand, SB transposase shows no predilection for transcription units, it rather prefers intergenic chromosomal regions [114, 115], which makes it the system of choice for safe delivery of transgenes. As new transposable elements are discovered and recombinant transposases with optimum enzymatic activity in the mammalian environment and improved targeting activity are developed, it is expected that transposon-based systems will gain ground in the field of large animal transgenesis.
Application of site-specific recombinases
Site-specific recombinase systems occur naturally in prokaryotes and fungi, where they perform several biological functions such as bacterial phase variation, plasmid copy number regulation, bacteriophage integration/excision from bacterial genome and amplification of yeast plasmids [116–118]. Site-specific recombinases have in common the capacity to bring together two DNA partners, catalyze double-strand cleavage at specific sites, and rejoin reciprocal strands (Fig. 3).
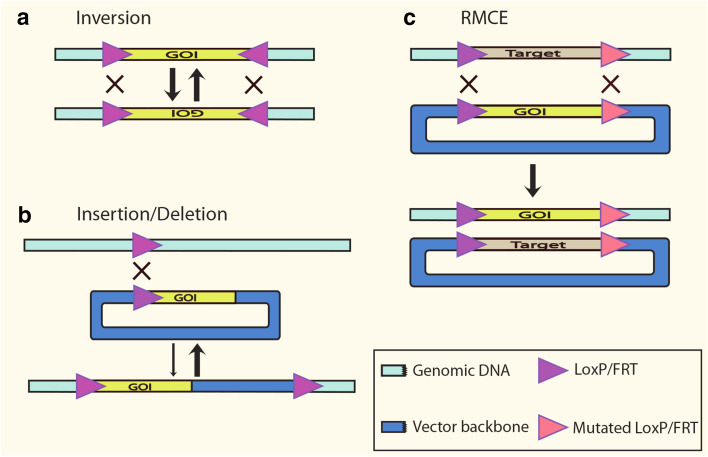
Fig. 3.
Recombination reactions catalyzed by Cre and Flp. The outcome of the recombination reaction is determined by the relative orientation of target sites. Inverted target sites in the same linear molecule dictate inversion of intervening DNA (a). Recombination between a target site located in a genomic address and an identical target site present in a circular DNA molecule results in insertion of circular DNA (b; thin arrow). The intramolecular recombination (thick arrow) is favored over the intermolecular reaction (thin arrow) (b). RMCE involves recombinase-mediated insertion and excision reactions, which lead to mutual exchange of the DNA between target sites (c). The inclusion of mutated target sites makes the recombination reaction irreversible
Importantly, site-specific recombination can proceed in heterologous environments, opening new avenues to engineer genomes in a predictable manner [119, 120], overcoming many problems associated with traditional methods, namely silencing or unpredictable expression of transgenes [121, 122] and unwanted remnant sequences left behind after genome manipulations.
Two basic elements comprise a site-specific recombinase system: two short consensus sequences and an enzyme that specifically recognizes those motifs and mediates strand exchange between the two DNA partner molecules [123, 124]. This process may lead to insertion, inversion, deletion or translocation of a DNA fragment in a reversible or irreversible manner [123, 125].
Members of the recombinase superfamily can be grouped according to the active amino acid present within the catalytic site in tyrosine or serine recombinases [126]. The mechanism of strand breakage, exchange and reunion markedly differs for each family (for details see [123]). The former group is further classified according to the mechanism of action in bidirectional or unidirectional [126] (Table 2).
From the tyrosine recombinase family, the Cre-loxP and Flp-FRT systems are by far the most extensively characterized members [127]. The minimum requirements for the recombination process to take place are two specific 34 base pair recognition sites and the recombinase [123]. In addition, based on the length of the recognition sites (34 bp) the probability that an identical sequence occurs by chance is extremely low (p ~ 10−21), and it is conveniently short enough as to be normally neutral toward gene expression when positioned in the genome.
Each loxP or FRT site comprises two inverted 13-bp symmetry elements, which serve as recombinase binding motif, flanking an 8-bp non-palindromic core element where strand recombination is catalyzed. The core nucleotide sequence asymmetry gives directionality to the reaction and therefore determines the type of modification [128]. Recombination of two identical target sites located on a circular and a linear DNA molecule each one will cause integration into the linear molecule. Inverted target sites in the same linear molecule dictates inversion of the DNA between recognition target sites (Fig. 3). If each identical target site is located on different linear DNA molecules, Cre or Flp recombination causes mutual exchange of sequences distal to the two recognition sites. Interestingly, it is known that recombination reaction occurs with the same efficiency regardless of the DNA topology (supercoiled or relaxed), and circular or linear molecules [128]. This versatility has positioned these members of tyrosine recombinase family at the vanguard of tools for genetic studies as well as biotechnological developments [125].
Based on the general properties of site-specific recombinase systems, different strategies for genome engineering have been developed to tailor diverse objectives, initially applied to classic model mammals and later to large domestic species. Early reports on the successful use of Flp and Cre recombinase systems for mammalian genome modifications employed recombinase-mediated DNA insertion (RMDI) [119, 120]. RMDI relies on recombination between two identical target sites, one inserted in a genomic address and the other one provided in a donor plasmid carrying the sequence to be inserted [129, 130] (Fig. 3). Upon recombination, the newly integrated DNA sequence is flanked by tandem-oriented target sites; therefore, it is prone to be excised by another round of recombination, explaining the low efficiency of this transgenic approach [119, 120]. One alternative is the use of heteromeric target sites, which recombine into inactive double mutant site and wild-type site, thus precluding another round of recombination [131–134].
Subsequently, an alternative methodology that addressed some pitfalls of RMDI, known as recombinase-mediated cassette exchange (RMCE), was introduced [135]. RMCE requires the host genome being previously tagged with compatible docking sites [132–134]. Once the appropriate tagged cell line is obtained and characterized, a site-specific recombinase mediates the exchange of the genomic-tagged cassette with the sequence of interest (Fig. 3). Recombinase may be provided as protein [136, 137], mRNA [138] or most commonly as an expression plasmid [129]. The GOI is delivered in a donor vector flanked by target sites that are homologous to the ones that were previously inserted in the genome. In summary, RMCE offers the precise engineering of animal genomes.
Inclusion of a positive selectable marker is normally required to enrich cell populations in which the desire transgenic event has occurred. However, retention of strong promoter and enhancer sequences associated with the selectable marker may have unpredictable effects on expression of linked genes [139–142], even those located at long distances from the inserted cassette [143]. From the perspective of the future introduction of transgenic animal products or derivatives into the food chain, production of selectable marker-free animals will be a mandatory condition to comply with regulatory agency guidelines raising concern regarding the possibility that antibiotic resistance genes being transferred to intestinal or environmental bacteria [144]. Therefore, deletion of selectable genes after in vitro selection of clonal cell lines is of upmost importance for both research and commercial application of transgenesis.
Removal of selectable marker genes introduced as part of the transgenic strategy can be accomplished by homologous recombination (HR), using the so-called “hit-and-run” or “tag-and-exchange” approaches [145–147]. Although successful, these methods subject the cell line to a second cycle of selection, which is not only time consuming but also may compromise the proliferative capacity of primary cell cultures. Therefore, the use of floxed or flirted selection cassettes provides the opportunity of marker deletion or replacement by site-specific recombination. Several reports have provided proof of concept for the potential of site-specific recombination technology for livestock genome engineering [65, 148–150]. Generation of marker-free cattle [14, 151] and goats [152] originated from cells that were selected and subsequently subjected to recombinase-mediated marker removal has been documented (Table 3).
Table 3.
Recombinase and integrase approaches in livestock
| Enzyme | Species | Recombination site | Targeted modification | Selection | Recombinase activity | Comments | Method | Germline transmission, generation | References |
|---|---|---|---|---|---|---|---|---|---|
| Cre | Goat | LoxP | Deletion of Neo and tk | G418 | Excision | SCNT | Yes | [152] | |
| Pig | LoxP/loxP257 | Venus >mCherry | FACS | RMCE | SCNT | Yes, F1 | [65] | ||
| Pig | LoxP/LoxP? | GFP >PSEN1M146I | G418/Puromycin | RMCE | SCNT | Yes | [13] | ||
| Cattle | LoxP | Deletion of Neo | FACS | Excision | SCNT | Preimplantation embryos | [151] | ||
| Flp | Goat | FRT | Puromycin >GFP | Puromycin/hygromycin | Gene replacement | SCNT | 35-day fetus | [150] | |
| ΦC31/Cre | Cattle | AttB-pseudo-attP/loxP | Artificial locus integration/deletion of Neo and DsRed | G418/FACS | Excision | Ambiguity | SCNT | ND | [14] |
| ΦC31/Cre/Dre | Cattle | AttB-pseudo-attP/loxP/rox | Deletion of Neo, tk, EGFP and plasmid backbone | G418/GCV/FACS | Excision | SCNT | Preimplantation embryos | [149] |
RMCE recombinase-mediated cassette exchange, ND not determined, FACS fluorescence-activated cell sorting, GCV ganciclovir, PSEN1M146I mutated presenilin-1 gene (Alzheimer’s disease-causing gene), Neo neomycin gene, tk thymidine kinase
The potential of site-specific recombinase technology is not limited to deletion of resistance marker sequences from the manipulated genome. In mice, strategies that combine marker gene removal with complex targeted sequence modifications have been developed [125]. These include large deletions [153], non-selectable subtle mutations [154], large-scale chromosomal rearrangements (translocation, duplication, inversion, deletion, or chromosomal gain or loss) [155] and swapping gene endogenous sequences for heterologous sequences [156]. Another application of site-specific recombinases that has revolutionized mouse genetic studies is the so-called conditional gene targeting. With the conditional gene targeting methodology the specific genetic modification is triggered in a specific cell type (tissue-specific) [157] or at a particular stage of development (temporal-specific) [158, 159].
Applying the basic principles of site-specific recombination and conditional gene targeting system, a myriad of novel strategies for mouse genomic manipulation have been developed, which are revolutionizing genetic research in the post-genomic era (reviewed in [159]). Equivalent conditional methods for genetic engineering of livestock are not yet available. Albeit, some steps toward establishing conditional gene targeting methods in large animals, like generation of pigs with Cre-induced expression [160, 161] have been undertaken, the complete conditional system has not been validated in domestic species.
Further flexibility to site-specific recombination applications has come with the introduction of the large family of serine recombinases. For example, the integrase ΦC31 [162, 163] induces recombination between two different target sites known as attP (39 bp minimal size) and attB (43 bp minimum size) [162]. Upon recombination, it originates two sequence hybrid sites, attL and attR, making the reaction unidirectional [164]. Depending on the configuration, ΦC31 can induce inversion, excision or integration of DNA sequences in heterologous genomes [165, 166]. For ΦC31-based strategies, an attP-tagged genome has to be generated by random integration or HR. Once the genomic single-copy tagging is achieved, unidirectional recombination between the genomic attP site and a vector attB site is catalyzed by ΦC31 [163].
An alternative approach to accomplish chromosomal targeting with ΦC31 integrase involves recombination at cryptic endogenous genomic recognition sites, also known as pseudo-attP sites. Recombination occurs at these pseudo-sites, because of their similarity in nucleotide sequence with the wild-type attP [167]. Pseudo-attP sites have been reported to be present not only in invertebrates [168, 169], lower vertebrates [170], but also in mouse [163], human [167, 171], cattle [172, 173], sheep [174], goat [175] and pig [176] genomes. Accumulating experimental evidence indicates that these pseudo-target sites reside in genomic locations that conform to the definition of “safe harbors” [177, 178]. Further improvement to this transgenic method was achieved by the introduction of evolved and mutated ΦC31 integrases, showing enhanced sequence specificity and integration frequency at preintegrated and pseudo-attP sites [179, 180].
Although ΦC31 has been the integrase that has received most attention, new members of the large serine subfamily [181] are constantly discovered. Such novel recombinase systems include: R4 [182], TP901-1 [183, 184], and Bxb1 [181, 185]. The wide spectrum of site-specific recombinases identified so far offers a set of tools to tailor controlled and sophisticated genome modifications for basic and applied research endeavors.
Application designer nucleases
Designer nucleases, also known as programmable nucleases, are regarded as a new generation of transgenic tools characterized by being efficient, customizable and capable of precise targeted genome modifications for a broad spectrum of applications [186–191]. Programmable nucleases include ZFNs, TALENs and a RNA-guided genome modification system termed CRISPR/Cas9 [192]. The CRISPR/Cas9 has recently emerged as a powerful and facile alternative to ZFNs and TALENs for inducing targeted genetic alterations in cells and embryos [2, 20, 22]. Generically, these chimeric proteins harbor a domain (protein or RNA) that recognizes and interacts with a specific genomic sequence and an associated catalytic module that induces site-specific DNA single- or double-strand breaks (DSBs). Enzyme-catalyzed DNA cleavage in turn activates host repair mechanisms through error-prone non-homologous end joining (NHEJ) and/or homology-directed repair (HDR) [188, 189, 193], which are ultimately responsible for the targeted genome modification. Depending on the system configuration, programmable nucleases can predictably alter nucleotide sequences to achieve gene knockout, gene insertion, gene correction or point mutations at predefined endogenous loci [189–191]. Moreover, long-range chromosomal rearrangements, including deletions, inversions and translocations can be accomplished by nuclease-induced DSBs.
The introduction of SCNT opened the possibility of conventional HR-based gene targeting in somatic cells of livestock [194, 195], an approach that has been inapplicable before because of the lack of germline competent stem cells in these species [196–199]. However, the extremely low rate of HR in somatic cells (10- to 100-fold lower than that in murine ES cells) [200–202], along with the inherent inefficiency of the SCNT technique [203] made this approach cumbersome and tedious. In consequence, only few loci were knocked out by conventional HR in livestock species since the establishment of SCNT in 1997.
Designer nucleases rapidly changed the scene in livestock transgenesis, evidenced by a burst in the number of published reports since their recent introduction (summarized in Table 4). The most appealing features of programmable nucleases are that their DNA-binding domain can be engineered to target almost any predefined DNA sequence in a particular genome [12]. During NHEJ, the break ends are ligated and small base pair deletions or insertions (indels) are commonly introduced at the site of breakage. Indels in coding exons frequently result in reading frameshift mutations and inactivation of the allele.
Table 4.
Selected designer nuclease approaches in livestock and monkeys
| Designer nuclease | Species | Target gene/gene KO | Deletion | Generation | Overall efficiency* | Method | Off-target effects/comments | Selection of somatic cells | References |
|---|---|---|---|---|---|---|---|---|---|
| ZFN | Rabbit | IgM | NHEJ | F0, F1 | 3.1 % | CPI | Mosaic F0; 14–22 % without mod. | NA | [265] |
| Pig | EGFP | NHEJ, m | F0 | Not given | SCNT | 80 % without mod. | FACS | [209] | |
| Pig | PPAR-γ | NHEJ, m | F0 | 0.15 % | SCNT | 10 % cotransfection | G418 | [210] | |
| Pig | GGTA1 | NHEJ, b | F0 | 1.4 % | SCNT | Resorbed fetuses and neonatal mortality | α-gal− cells counter selection | [186] | |
| Pig | GGTA1 | NHEJ, b | F0 | 1.6 % | SCNT | Resorbed fetuses | α-gal− cells counter selection | [266] | |
| Pig | GGTA/CMAH | NHEJ, b | F0 | 0.8 % | SCNT | Resorbed fetuses | Clonal exp. | [267] | |
| Pig | CMAH | NHEJ, b | F0 | 2.6 % | SCNT | Clonal exp. | [268] | ||
| Pig | CMAH | HDR, b, m | F0 | 0.4 % | SCNT | No integration of ZFN constructs | G418 | [268] | |
| Pig | RELA | NHEJ, b | F0 | 11 % | CPI | NA | [269] | ||
| Pig | IL2rg | NHEJ, b, m | Fetuses | 2 % | SCNT | High neonatal mortality | [270] | ||
| Cattle | BLG | NHEJ, b | F0 | 0.8 % | SCNT | Weak off-target effect | Clonal exp. | [211] | |
| Cattle | Beta-casein | Nick + HR | F0 | <0.5 % | SCNT | High levels of off-target mutations | Clonal exp. | [223] | |
| Rabbit | APOE | NHEJ, b | F1 | 25 % | PNI | No selection | [271] | ||
| TALEN | Cattle | Several | NHEJ, m, b | Preimplant | 50 % | CPI | High deletion/insertion | NA | [187] |
| Cattle | MSTN | NHEJ | F0 | 15 % | CPI | Mosaicism | NA | [233] | |
| Sheep | MSTN | NHEJ | F0 | 3.8 % | CPI | NA | [233] | ||
| Pig | RELA | NHEJ, m, b | Preimplant | 29 % | CPI | High deletion/insertion | NA | [187] | |
| Pig | LDLR | NHEJ, m, b | F0 | Not given | CT | Puromycin | [187] | ||
| Pig | RELA | NHEJ, m, b | F0 | 21 % | CPI | NA | [269] | ||
| Mini-pig | GGTA | NHEJ, b | F0 | 0.16 % | SCNT | High deletion | G418 | [272] | |
| Pig, | DAZL, APC | HDR, NHEJ | F0 | Not given | CT | No selection | [19] | ||
| Monkey | MECP2 | NHEJ | F0 | 1.2 % | CPI | High male fetal mortality, chimeric F0 | NA | [273] | |
| Chicken | OV | NHEJ | Founder, F1 | 8 % | Microinjection of PGC | No off-target effects | FACS | [274] | |
| CRISPR/Cas9 | Pig | vWF | NHEJ, b, m | F0 | 13.2 % | CPI | Small effect on embryo development | NA | [248] |
| Pig | RELA | HDR | F0 | Not given | CT | Off-target cleavage | No selection | [19] | |
| Pig | CD163, CD1D | NHEJ | F0 | 1.2–2.1 % | SCNT | Very low HDR | No selection or G418 | [246] | |
| Pig | CD163, CD1D | NHEJ | F0 | 4.2–3.6 % | CPI | NA | [246] | ||
| Pig | GGTA1, CMAH, iGb3S | NHEJ | F0 | 3.9 % | SCNT | α-gal− cells counter selection | [245] | ||
| Mini-pig | TYR | NHEJ, b | F0 | 0.9 % | SCNT | No off-target effects | G418? | [244] | |
| Mini-pig | PARK2, PINK1 | NHEJ, b | F0 | 1.2 % | SCNT | No off-target effects | G418? | [244] | |
| Goat | Several | NHEJ, b | F0 | 1.1 % | SCNT | No off-target effects | No selection | [247] | |
| Sheep | MSTN | NHEJ | F0 | 5.7 % | CPI | No off-target effects | NA | [249] | |
| Monkey | PPAR-γ, RAG1 | NHEJ | F0 | Not given | CPI | No off-target effects | NA | [275] | |
| Rabbit | TYR | NHEJ | F0, F1 | 3 % | PNI | No off-target effects | No selection | [276] | |
| Rabbit | IL2rg; Tiki1; IL2rg + RAG1 a | NHEJ b | F0 | 7.5–16.6 % | CPI | Low off-target effects | No selection | [238] |
PPAR-γ peroxisome proliferator-activated receptor gamma, GGTA1 α1,3-galactosyltransferase gene, CMAH cytidine monophosphate-N-acetylneuraminic acid hydroxylase, PRNP prion protein, RELA v-rel reticuloendotheliosis viral oncogene homolog A (avian), LDLR LDL receptor, IL2rg interleukin 2 receptor gamma gene, BLG beta-lactoglobulin, MECP2 methyl-CpG-binding protein, vWF Von Willebrand disease gene, MSTN myostatin gene, RAG1 recombination-activating gene 1, DAZL deleted in azoospermia-like, APC adenomatous polyposis coli, CD163 porcine reproductive and respiratory syndrome virus (PRRSV) receptor, CD1D non-classical major histocompatibility complex protein, iGb3S iGb3 synthase candidate glycosyltransferase, TYR tyrosinase gene, PARK2 parkin, RBR E3 ubiquitin protein ligase, PINK1 PTEN-induced putative kinase, OV ovalbumin gene, m mono-allelic, b bi-allelic, NA not applicable
* Transgenic offspring per treated oocytes or embryos
aMultiplex gene targeting
The other cell pathway triggered by specific nuclease cleavage of the genome is HDR. The likelihood of a HR increases several orders of magnitude in the vicinity of a DSB [204, 205]. Therefore designer nuclease-mediated DNA scission will favor precise modification of the target sequence by HR between the endogenous sequence and the provided donor template [206, 207]. Genome editing through HR is highly versatile allowing for targeted introduction of large genetic segments to precise single-base mutations.
Such improved efficiencies associated with programmed nucleases make it feasible to target both alleles of a gene simultaneously, to perform one-step multiplex gene targeting and to omit selectable markers [19, 186]. With this emerging transgenic technology it is now possible to achieve biallelic targeting in livestock [187, 208]. Perhaps one of the major advantages of engineered nuclease-mediated gene editing for commercial purposes is that beneficial traits or mutations can be introduced in livestock genomes without inserting surplus genetic material, which is one of the concerns associated with genetically modified organisms [144].
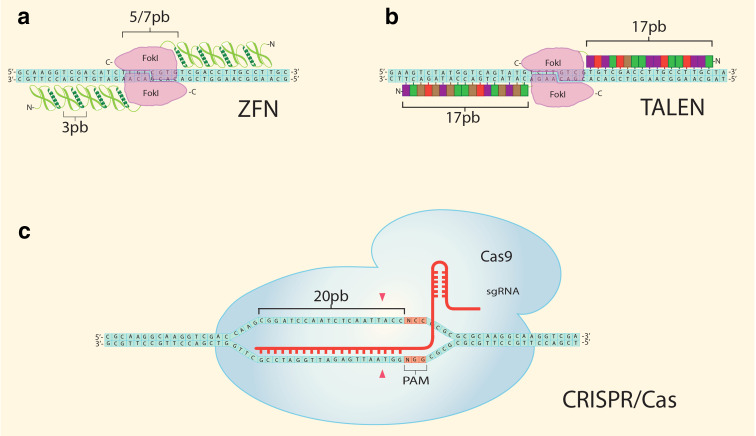
The first designer nuclease platform to be used in livestock was the ZFN [186, 209–211], followed by TALEN [187, 190, 212] and more recently by CRISPR/Cas9 [19, 213]. ZFNs utilize zinc finger motifs tethered to a non-specific nuclease (Fok1) to bind chromosomal DNA and perform a double cut [214]. This system is comprised of an array of zinc fingers (which recognizes 12 nucleotides in the target sequence; Fig. 4). As FokI nuclease requires dimerization to cleave DNA, two ZFN monomers are necessary to interact with specific sequences in opposite DNA strands to form an active nuclease dimer. This configuration doubles the length of the recognition site, which substantially increases the specificity of ZFNs. Despite the length of the recognition site, off-target cleavages may still occur [215, 216].
Fig. 4.
Schematic representation of components of ZFNs, TALENs and CRISPR/Cas9 systems. a Structure of ZFNs. A ZFN enzyme comprise a DNA-binding domain formed by zinc finger modules (ZF), each recognizing a unique 3-base pair sequence on the target DNA, and a DNA-cleaving domain composed of FokI. Two ZFN are designed to recognize DNA sequences that flank the desired cleavage site. In the example, each ZFN comprises four ZF which recognize 12 bp on opposite strands. Upon interaction of ZF with the target site, a FokI dimer catalyzes a targeted double-strand break. b Model of TALEN system. Targeted DNA cleavage is achieved by a pair of TALEN molecules. Each TALEN is comprised by a TALE and a FokI catalytic domain. TALE-targeting domain comprise a variable number of tandem arrays of repeats of typically 34 amino acids each (shown as colored boxes in b). The RVD is responsible for the repeat specificity to associate with a particular base pair on the target DNA. c Model of CRISPR/Cas9. Cas9 nuclease is directed to a specific sequence in the genomic DNA by the first 20 nucleotides of the sgRNA, which hybridizes with the target genomic DNA, which has to be followed by a mandatory protospacer adjacent motif (PAM; 5′-NGG for Cas9 system derived from Streptococcus pyogenes). Cas9 catalyzes a DSB upstream of the PAM (red arrowheads)
The first report on ZFN-mediated gene knockout in porcine somatic cells came from the Nagashima laboratory in 2010 [217], followed by the ZFN-mediated knockout of a single-copy GFP transgene in cloned pigs [218]. These initial studies were rapidly extended by the ZFN-mediated mono-allelic disruption of an endogenous gene (PPARγ) in cloned piglets [210] and the biallelic disruption of the α1,3-galactosyltransferase (GGTA1) gene in pig fibroblasts, subsequently employed in SCNT to produce knockout pigs (Table 4; [186, 219]). In cattle, ZFN-mediated disruption of the beta-lactoglobulin (BLG) gene in bovine fibroblasts and production of cloned cows have been recently reported (Table 4, [211]). Variants of ZFNs are the zinc finger nickases (ZFNickases), which induce site-specific single-strand breaks in genomic DNA. This modified version of ZFNs can be engineered by mutating the FokI catalytic domain in one of the ZFN monomers [220–222]. ZFNickases, and other programmable nucleases with single-strain cleavage capability, are better suited for situations in which HDR-based genome edition is sought. Single-strand break stimulates resolution by HDR; therefore the faulty prone NHEJ pathway is avoided [220, 221]. A ZFNickase was used to stimulate gene addition (lysostaphin) by HDR into the endogenous beta-casein (CSN2) locus of bovine fibroblasts. Treated cells were subsequently used to generate cloned cows that produced the antimicrobial transgene product in milk [223].
Although ZFNs have been exceptionally effective for knocking out genes in farm animal genomes [186], the lack of proprietary algorithms to predict active ZFN molecules has restricted their use [190, 224].
TALENs are fusion proteins that comprise an assembled DNA-targeting domain coupled to a DNA cleavage motif. The DNA-binding domain is derived from proteins secreted by a plant pathogen belonging to the genus Xanthomonas. The DNA-binding domain is tethered to a catalytic domain of the non-specific restriction endonuclease FokI. DNA recognition and binding is mediated by tandem repeats of typically 34 amino acids, except for the last module, called half-repeat, which comprises 20 amino acids [212]. Thirty-two of the amino acids that comprise the repeat are highly conserved, whereas variable residues at positions 12 and 13, repeat variable di-residues (RVDs), dictate the binding specificity to a single nucleotide [225, 226]. Based on this code, arrays of tandem repeats can be assembled to target almost any DNA sequence of choice. Similar to ZFNs, TALEN-mediated cleavage depends on the dimerization of a pair of TALEN monomers binding to opposite DNA strands, which activates the FokI nuclease domains. Typical a TALEN monomer contains up to twenty tandem repeats, such that upon dimerization a 40-bp target sequence is recognized. Despite this theoretical high specificity, there exists evidence that TALEN can bind to degenerate sequences, and induce DNA cleavage at off-target sites [227, 228].
Several particular features of TALENs make them easier to develop and use than ZFNs [229]. Tan et al. [19] assessed the potential of TALENs (and CRISPR/Cas9 nucleases, see below) to edit the genome of many commercially important species. The authors demonstrated that TALENs can efficiently target a variety of alleles involved in food production, reproductive efficiency and external traits (hornlessness) [230] in the genomes from different livestock species. In the same study, TALENs were used to induce NHEJ- or HDR-directed edits at specific loci to mimic mutations that are known to be associated with genetic diseases in humans. From the edited cells, live pigs carrying the induced mutations were generated by chromatin transfer (CT) and these animals promise to become valuable large mammalian models in translational medicine. The targeting efficiency ranged from approximately 10 % for single-nucleotide polymorphisms to >50 % for some larger alterations. According to recent data, the TALEN system is also functional and efficient in the preimplantation embryo context, since microinjection of TALEN mRNA directed to the GDF-8 gene [231, 232] into bovine and ovine zygotes resulted in correctly edited cattle and sheep [233]. In light of these encouraging findings, it is conceivable that genome edition by designer nucleases will become a practical strategy to introduce or suppress genetic characteristics in livestock populations to accelerate the genetic progress in harmony with classic breeding strategies.
The RNA-guided CRISPR/Cas9 system [234, 235] was recently discovered in bacteria and archaea, in which the RNA-guided foreign-DNA cleavage process provides adaptive immunity against invading phages or plasmids [236, 237]. The CRISPR/Cas9 sequence specificity is determined by Watson–Crick base complementarity with a single-guide RNA (sgRNA). The induced DNA damage is repaired by either HR or error-prone NHEJ that normally causes indels at the cleavage site. Design and generation of the synthetic sgRNA is markedly easier compared with the cumbersome protein engineering required to produce ZFNs and TALENs. By changing the nucleotide sequence of the sgRNA it is possible to target almost any site in the genome. The activity of the CRISPR/Cas9 system allows the implementation of high-throughput methodologies and multiplex editing of genomic loci in preimplantation mammalian embryos [238].
From 2013, the CRISPR/Cas9 system bursts into the genome engineering scenario through several independent reports providing encouraging evidence for simplicity and effectiveness of this system to engineer large animal genomes [2, 239–243]. Tan et al. [19] were the first researchers to apply CRISPR/Cas9 technology to target endogenous genes (P65 and APC) in primary pig cells in culture. Despite the demonstration that CRSPR/Cas9 works in livestock cells, recovery of CRISPR mutant cell clones was much lower than that with TALENs, suggesting that CRISPR/Cas9 system needs further optimization to achieve targeting efficiencies comparable to TALENs. Follow-up studies have demonstrated that CRISPR/Cas9 genome-edited cells can support development to term when used as nuclear donor in SCNT in pigs [244–246] and goats [247]. An appealing and straightforward alternative to SCNT to generate genome-edited animals is the injection of the CRISPR/Cas9 components in livestock one-cell embryos produced in vivo or in vitro. Using the CPI approach, Hai et al. [248] managed to obtain live pigs with mono- and biallelic mutations in the vWF gene to generate a relevant large animal model for hemophilia. The reported efficiency is quite impressive for a zygote microinjection-based method; with 10 out of 16 born piglets (~63 %) carrying one or both vWF alleles mutated. Another study with sheep zygotes microinjected with sgRNA/Cas9 mRNA [249] produced modest results in terms of the number of mutated offspring to born animals (2/32); however, no off-target mutations were detected. A recurrent problem of zygote microinjections is the high incidence of mosaic animals [227, 246, 249, 250], which is believed to originate when the nuclease remains active beyond the first embryo cleavage. Another potential problem associated with the use CRISPR/Cas9 editing system is the introduction of undesired mutations at off-target genomic sites [251, 252]. Based on the relatively short CRISPR/Cas9 recognition site (20 nt) and the known mismatch tolerance, especially at the 5′ region of the target sequence, occurrence of off-target mutations should not be disregarded [253]. A strategy to minimize off-target activity is to use a mutated version of Cas9 (D10A mutation [192, 254]) with nickase activity. DNA single-strand break stimulates HDR with negligible NHEJ-mediated mutations. Moreover, when required, a DSB can be simultaneously induced at the target site using a pair of appropriately spaced and oriented sgRNAs along with Cas9 nickase, that enhances genome editing specificity [255]. More research is warranted to ascertain if off-target DNA cleavage rates induced by CRISPR/Cas9 are a concern in the context of genome edition in large animals.
Conclusions
Transgenic methodologies are constantly evolving, providing researchers and biotechnologists with advanced tools for efficient and controlled genome modifications. Initial transgenic interventions in livestock were confined to simple gene insertion at random places in the genome. Thanks to constant advances in the area of genetic engineering, today it is possible to achieve precise genome modifications by inserting, replacing or removing predefined DNA sequences. In this regard, introduction of methodologies that enable enzymatic manipulation of animal genomes have opened new possibilities to create genetically modified animals for agriculture or biomedicine. Although the transgenic toolbox for large animals is currently equipped with powerful methodologies there are many aspects to improve in the associated reproductive technologies required to generate a transgenic animal. Low success rates of SCNT and zygote microinjection, two of the most commonly used methods to generate transgenic large animals, still represent a bottleneck. Efficiency of SCNT has remained low in spite of considerable efforts invested in developing more successful protocols, while embryo microinjection has been invariably associated with undesired chimerism. Thus, further improvements in surpassing the limitations of these techniques may impact favorably on the overall efficiency of transgenic methods.
Transposon-based systems are a straightforward alternative to achieve transgene integrations with persistent transgene expression and germline transmission. These characteristics along with increased transgenic efficiencies will certainly reduce costs and contribute to animal welfare by reducing the number of animal required to produce the desired genotype and by avoiding unwanted phenotypes. Transposon-based methods alone or combined with site-directed recombinases will simplify the production of marker-free animals to comply with regulatory guidelines for animal transgenesis.
The ground-breaking feature of designer nucleases is that they brought the possibility of purposely directing the genomic modification to a specific and unique chromosomal locus. Among the members of the engineered nucleases, RNA-guided nucleases are the ones that promise to change the paradigm of genome editing in large animals. The CRISPR/Cas9 system combines facile design and construction with high specificity, effectivity and real possibility of multiplex gene edition. However, there is still room for improvement in particular areas like minimizing off-target effects of designer nucleases, enhancement of nuclease activity, and development of methods to enrich cell population with targeted genome edits. Another avenue to improve engineered nuclease-based methods is through the genetic or pharmacologic manipulation of the DSB repair pathway. For instance, for many applications, enhancement of low-frequency HDR over the NHEJ would be convenient.
The launching of high-throughput genome sequencing at accessible prices will make it possible to improve the quality of current genome data in farm animals and it will become a valuable tool to verify transgenic lines at genome scale. It is anticipated that new generation transgenic tools in concert with updated genomic data will facilitate the production of large animal models for translational medicine. These large animal models will be instrumental for understanding disease pathogenesis and development of better therapeutic approaches of severe human pathologic conditions.
It is foreseen that similar opportunities will arise in agricultural applications of transgenic livestock. Genome sequencing and phenotyping will provide unprecedented opportunities for the identification of molecular markers that affect livestock performance, which can be readily addressed and manipulated at will by site-directed nucleases to improve productive traits. Experimental evidence has provided proof of principle that non-meiotic introgression of natural or novel genetic variants in livestock genomes is attainable using designer nucleases. Numerous reports cited in this review strongly indicate that designer nucleases have earned enough merit as genome engineering tools as to be considered in the near future in selection programs to advance genetic improvement when selective breeding is impracticable or inefficient. Importantly, the toolbox for genome engineering is still expanding, as new enzymatic systems are constantly discovered. One recent example are the bacterial casposons, which seem to combine the features of CRISPR/Cas and transposons [256, 257], suggesting that more sophisticated options for genome engineering will become feasible in the near feature.
Acknowledgments
Authors acknowledge the financial support from CONICET, FONCyT, UNRC (Republica Argentina), ICAR (India), as well as from DAAD and DFG (Germany).
Abbreviations
- Cas9
CRISPR-associated protein 9
- CPI
Cytoplasmic injection
- Ct
Chromatin transfer
- CRISPR
Clustered regularly interspaced short palindromic repeats
- DSB
Double-strand break
- GOI
Gene of interest
- HR
Homologous recombination
- HDR
Homology-directed repair
- ICSI
Intracytoplasmic sperm injection
- ICSI-Tr
Intracytoplasmic sperm injection-mediated transgenesis
- iPS
Induced pluripotent stem (cell)
- I-SceI
Homing endonuclease
- ITR
Inverted terminal repeat
- KO
Knockout
- NHEJ
Non-homologous end joining
- PB
piggyBac transposon system
- PNI
Pronuclear injection
- RE
Restriction enzyme
- RecA
Recombinase A
- REMI
Restriction enzyme-mediated integration
- RMCE
Recombinase-mediated cassette exchange
- RMDI
Recombinase-mediated DNA insertion
- SB
Sleeping Beauty transposon system
- SCNT
Somatic cell nuclear transfer
- sgRNA
Single-guide RNA
- SMGT
Sperm-mediated gene transfer
- SV40
Simian virus 40
- TALEN
Transcription activator-like element nuclease
- Tol2
Tol2 transposon system
- ZFN
Zinc finger nuclease
References
- 1.Miao X. Recent advances in the development of new transgenic animal technology. Cell Mol Life Sci. 2013;70(5):815–828. doi: 10.1007/s00018-012-1081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laible G, Wei J, Wagner S. Improving livestock for agriculture: technological progress from random transgenesis to precision genome editing heralds a new era. Biotechnol J. 2014 doi: 10.1002/biot.201400193. [DOI] [PubMed] [Google Scholar]
- 3.Prather RS. Pig genomics for biomedicine. Nat Biotechnol. 2013;31(2):122–124. doi: 10.1038/nbt.2490. [DOI] [PubMed] [Google Scholar]
- 4.Segal DJ, Meckler JF. Genome engineering at the dawn of the golden age. Annu Rev Genomics Hum Genet. 2013;14:135–158. doi: 10.1146/annurev-genom-091212-153435. [DOI] [PubMed] [Google Scholar]
- 5.Cibelli J, Emborg ME, Prockop DJ, Roberts M, Schatten G, Rao M, Harding J, Mirochnitchenko O. Strategies for improving animal models for regenerative medicine. Cell Stem Cell. 2013;12(3):271–274. doi: 10.1016/j.stem.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shinohara ET, Kaminski JM, Segal DJ, Pelczar P, Kolhe R, Ryan T, Coates CJ, Fraser MJ, Handler AM, Yanagimachi R, Moisyadi S. Active integration: new strategies for transgenesis. Transgenic Res. 2007;16(3):333–339. doi: 10.1007/s11248-007-9077-z. [DOI] [PubMed] [Google Scholar]
- 7.Garrels W, Ivics Z, Kues WA. Precision genetic engineering in large mammals. Trends Biotechnol. 2012;30(7):386–393. doi: 10.1016/j.tibtech.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Keravala A, Liu D, Lechman ER, Wolfe D, Nash JA, Lampe DJ, Robbins PD. Hyperactive Himar1 transposase mediates transposition in cell culture and enhances gene expression in vivo. Hum Gene Ther. 2006;17(10):1006–1018. doi: 10.1089/hum.2006.17.1006. [DOI] [PubMed] [Google Scholar]
- 9.Zayed H, Izsvák Z, Walisko O, Ivics Z. Development of hyperactive sleeping beauty transposon vectors by mutational analysis. Mol Ther. 2004;9(2):292–304. doi: 10.1016/j.ymthe.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 10.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, Grzela DP, Schmitt A, Becker K, Matrai J, Ma L, Samara-Kuko E, Gysemans C, Pryputniewicz D, Miskey C, Fletcher B, VandenDriessche T, Ivics Z, Izsvak Z. Molecular evolution of a novel hyperactive sleeping beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41(6):753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 11.Yusa K, Zhou L, Li MA, Bradley A, Craig NL. A hyperactive piggyBac transposase for mammalian applications. Proc Natl Acad Sci USA. 2011;108(4):1531–1536. doi: 10.1073/pnas.1008322108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15(5):321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 13.Jakobsen JE, Johansen MG, Schmidt M, Dagnaes-Hansen F, Dam K, Gunnarsson A, Liu Y, Kragh PM, Li R, Holm IE, Callesen H, Mikkelsen JG, Nielsen AL, Jorgensen AL. Generation of minipigs with targeted transgene insertion by recombinase-mediated cassette exchange (RMCE) and somatic cell nuclear transfer (SCNT) Transgenic Res. 2013;22(4):709–723. doi: 10.1007/s11248-012-9671-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu Y, Wang Y, Tong Q, Liu X, Su F, Quan F, Guo Z, Zhang Y. A site-specific recombinase-based method to produce antibiotic selectable marker free transgenic cattle. PLoS ONE. 2013;8(5):e62457. doi: 10.1371/journal.pone.0062457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitelaw CB, Lillico SG, King T. Production of transgenic farm animals by viral vector-mediated gene transfer. Reprod Domest Anim. 2008;43(Suppl 2):355–358. doi: 10.1111/j.1439-0531.2008.01184.x. [DOI] [PubMed] [Google Scholar]
- 16.Park F. Lentiviral vectors: are they the future of animal transgenesis? Physiol Genomics. 2007;31(2):159–173. doi: 10.1152/physiolgenomics.00069.2007. [DOI] [PubMed] [Google Scholar]
- 17.Pfeifer A, Hofmann A. Lentiviral transgenesis. Methods Mol Biol. 2009;530:391–405. doi: 10.1007/978-1-59745-471-1_21. [DOI] [PubMed] [Google Scholar]
- 18.Lillico S, Vasey D, King T, Whitelaw B. Lentiviral transgenesis in livestock. Transgenic Res. 2011;20(3):441–442. doi: 10.1007/s11248-010-9448-8. [DOI] [PubMed] [Google Scholar]
- 19.Carlson DF, Tan W, Hackett PB, Fahrenkrug SC. Editing livestock genomes with site-specific nucleases. Reprod Fertil Dev. 2013;26(1):74–82. doi: 10.1071/RD13260. [DOI] [PubMed] [Google Scholar]
- 20.Tan W, Carlson DF, Lancto CA, Garbe JR, Webster DA, Hackett PB, Fahrenkrug SC. Efficient nonmeiotic allele introgression in livestock using custom endonucleases. Proc Natl Acad Sci USA. 2013;110(41):16526–16531. doi: 10.1073/pnas.1310478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gun G, Kues WA. Current progress of genetically engineered pig models for biomedical research. Biores Open Access. 2014;3(6):255–264. doi: 10.1089/biores.2014.0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carroll D. Genome engineering with targetable nucleases. Annu Rev Biochem. 2014;83:409–439. doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 23.Schiestl RH, Petes TD. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88(17):7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black M, Seeber F, Soldati D, Kim K, Boothroyd JC. Restriction enzyme-mediated integration elevates transformation frequency and enables co-transfection of Toxoplasma gondii. Mol Biochem Parasitol. 1995;74(1):55–63. doi: 10.1016/0166-6851(95)02483-2. [DOI] [PubMed] [Google Scholar]
- 25.Maier FJ, Schafer W. Mutagenesis via insertional- or restriction enzyme-mediated-integration (REMI) as a tool to tag pathogenicity related genes in plant pathogenic fungi. Biol Chem. 1999;380(7–8):855–864. doi: 10.1515/BC.1999.105. [DOI] [PubMed] [Google Scholar]
- 26.Marsh-Armstrong N, Huang H, Berry DL, Brown DD. Germ-line transmission of transgenes in Xenopus laevis . Proc Natl Acad Sci USA. 1999;96(25):14389–14393. doi: 10.1073/pnas.96.25.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cavolina P, Agnese C, Maddalena A, Sciandrello G, Di Leonardo A. Induction of CAD gene amplification by restriction endonucleases in V79, B7 Chinese hamster cells. Mutat Res. 1989;225(1–2):61–64. doi: 10.1016/0165-7992(89)90034-1. [DOI] [PubMed] [Google Scholar]
- 28.Costa ND, Masson WK, Thacker J. The effectiveness of restriction endonucleases in cell killing and mutation. Somat Cell Mol Genet. 1993;19(5):479–490. doi: 10.1007/BF01233253. [DOI] [PubMed] [Google Scholar]
- 29.Seo BB, Kim CH, Yamanouchi K, Takahashi M, Sawasaki T, Tachi C, Tojo H. Co-injection of restriction enzyme with foreign DNA into the pronucleus for elevating production efficiencies of transgenic animals. Anim Reprod Sci. 2000;63(1–2):113–122. doi: 10.1016/s0378-4320(00)00148-2. [DOI] [PubMed] [Google Scholar]
- 30.Wall RJ. Transgenic livestock: progress and prospects for the future. Theriogenology. 1996;45:57–68. [Google Scholar]
- 31.Abella Columna E, Giaccia AJ, Evans JW, Yates BL, Morgan WF. Analysis of restriction enzyme-induced chromosomal aberrations by fluorescence in situ hybridization. Environ Mol Mutagen. 1993;22(1):26–33. doi: 10.1002/em.2850220106. [DOI] [PubMed] [Google Scholar]
- 32.Dewey WC, Miller HH, Leeper DB. Chromosomal aberrations and mortality of x-irradiated mammalian cells: emphasis on repair. Proc Natl Acad Sci USA. 1971;68(3):667–671. doi: 10.1073/pnas.68.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Obe G, Von der Hude W, Scheutwinkel-Reich M, Basler A. The restriction endonuclease Alu I induces chromosomal aberrations and mutations in the hypoxanthine phosphoribosyltransferase locus, but not in the Na+/K+ ATPase locus in V79 hamster cells. Mutat Res. 1986;174(1):71–74. doi: 10.1016/0165-7992(86)90079-5. [DOI] [PubMed] [Google Scholar]
- 34.Singh B, Bryant PE. Induction of mutations at the thymidine kinase locus in CHO cells by restriction endonucleases. Mutagenesis. 1991;6(3):219–223. doi: 10.1093/mutage/6.3.219. [DOI] [PubMed] [Google Scholar]
- 35.Hafez M, Hausner G. Homing endonucleases: DNA scissors on a mission. Genome. 2012;55(8):553–569. doi: 10.1139/g2012-049. [DOI] [PubMed] [Google Scholar]
- 36.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41(2):383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 37.Ogino H, McConnell WB, Grainger RM. Highly efficient transgenesis in Xenopus tropicalis using I-SceI meganuclease. Mech Dev. 2006;123(2):103–113. doi: 10.1016/j.mod.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Thermes V, Grabher C, Ristoratore F, Bourrat F, Choulika A, Wittbrodt J, Joly JS. I-SceI meganuclease mediates highly efficient transgenesis in fish. Mech Dev. 2002;118(1–2):91–98. doi: 10.1016/s0925-4773(02)00218-6. [DOI] [PubMed] [Google Scholar]
- 39.Bevacqua RJ, Canel NG, Hiriart MI, Sipowicz P, Rozenblum GT, Vitullo A, Radrizzani M, Fernandez Martin R, Salamone DF. Simple gene transfer technique based on I-SceI meganuclease and cytoplasmic injection in IVF bovine embryos. Theriogenology. 2013;80(2):104–113. doi: 10.1016/j.theriogenology.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 40.Ortega MN, Benítez SB, Barrionuevo BE, Olmos Nicotra MF, Alessio AP, Fili AE, Forcato DO, Stice SL, Bosch P. Meganuclease I-SceI enhances stable transgene integration in cultured bovine fetal fibroblasts. Reprod Fertil Dev. 2012;25(1):170–171. [Google Scholar]
- 41.Wang Y, Zhou XY, Xiang PY, Wang LL, Tang H, Xie F, Li L, Wei H. The meganuclease I-SceI containing nuclear localization signal (NLS-I-SceI) efficiently mediated mammalian germline transgenesis via embryo cytoplasmic microinjection. PLoS ONE. 2014;9(9):e108347. doi: 10.1371/journal.pone.0108347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brackett BG, Baranska W, Sawicki W, Koprowski H. Uptake of heterologous genome by mammalian spermatozoa and its transfer to ova through fertilization. Proc Natl Acad Sci USA. 1971;68(2):353–357. doi: 10.1073/pnas.68.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lavitrano M, Camaioni A, Fazio VM, Dolci S, Farace MG, Spadafora C. Sperm cells as vectors for introducing foreign DNA into eggs: genetic transformation of mice. Cell. 1989;57(5):717–723. doi: 10.1016/0092-8674(89)90787-3. [DOI] [PubMed] [Google Scholar]
- 44.Brinster RL, Sandgren EP, Behringer RR, Palmiter RD. No simple solution for making transgenic mice. Cell. 1989;59(2):239–241. doi: 10.1016/0092-8674(89)90282-1. [DOI] [PubMed] [Google Scholar]
- 45.Tsai HJ. Electroporated sperm mediation of a gene transfer system for finfish and shellfish. Mol Reprod Dev. 2000;56(2 Suppl):281–284. doi: 10.1002/(SICI)1098-2795(200006)56:2+<281::AID-MRD15>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 46.Shamila Y, Mathavan S. Sperm/DNA interaction: DNA binding proteins in sperm cell of silkworm Bombyx mori . Mol Reprod Dev. 2000;56(2 Suppl):289–291. doi: 10.1002/(SICI)1098-2795(200006)56:2+<289::AID-MRD17>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 47.Fernandez MA, Mani SA, Rangarajan PN, Seshagiri PB. Sperm-mediated gene transfer into oocytes of the golden hamster: assessment of sperm function. Indian J Exp Biol. 1999;37(11):1085–1092. [PubMed] [Google Scholar]
- 48.Cappello F, Stassi G, Lazzereschi D, Renzi L, Di Stefano C, Marfe G, Giancotti P, Wang HJ, Stoppacciaro A, Forni M, Bacci ML, Turchi V, Sinibaldi P, Rossi M, Bruzzone P, Pretagostini R, Della Casa G, Cortesini R, Frati L, Lavitrano M. hDAF expression in hearts of transgenic pigs obtained by sperm-mediated gene transfer. Transplant Proc. 2000;32(5):895–896. doi: 10.1016/s0041-1345(00)01176-3. [DOI] [PubMed] [Google Scholar]
- 49.Shemesh M, Gurevich M, Harel-Markowitz E, Benvenisti L, Shore LS, Stram Y. Gene integration into bovine sperm genome and its expression in transgenic offspring. Mol Reprod Dev. 2000;56(S2):306–308. doi: 10.1002/(SICI)1098-2795(200006)56:2+<306::AID-MRD21>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 50.Sciamanna I, Piccoli S, Barberi L, Zaccagnini G, Magnano AR, Giordano R, Campedelli P, Hodgson C, Lorenzini R, Spadafora C. DNA dose and sequence dependence in sperm-mediated gene transfer. Mol Reprod Dev. 2000;56(2 Suppl):301–305. doi: 10.1002/(SICI)1098-2795(200006)56:2+<301::AID-MRD20>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 51.Perry AC, Wakayama T, Kishikawa H, Kasai T, Okabe M, Toyoda Y, Yanagimachi R. Mammalian transgenesis by intracytoplasmic sperm injection. Science. 1999;284(5417):1180–1183. doi: 10.1126/science.284.5417.1180. [DOI] [PubMed] [Google Scholar]
- 52.Umeyama K, Saito H, Kurome M, Matsunari H, Watanabe M, Nakauchi H, Nagashima H. Characterization of the ICSI-mediated gene transfer method in the production of transgenic pigs. Mol Reprod Dev. 2012;79(3):218–228. doi: 10.1002/mrd.22015. [DOI] [PubMed] [Google Scholar]
- 53.Hirabayashi M, Kato M, Ito J, Hochi S. Viable rat offspring derived from oocytes intracytoplasmically injected with freeze-dried sperm heads. Zygote. 2005;13(1):79–85. doi: 10.1017/s096719940500300x. [DOI] [PubMed] [Google Scholar]
- 54.Pereyra-Bonnet F, Fernandez-Martin R, Olivera R, Jarazo J, Vichera G, Gibbons A, Salamone D. A unique method to produce transgenic embryos in ovine, porcine, feline, bovine and equine species. Reprod Fertil Dev. 2008;20(7):741–749. doi: 10.1071/rd07172. [DOI] [PubMed] [Google Scholar]
- 55.Bevacqua RJ, Pereyra-Bonnet F, Fernandez-Martin R, Salamone DF. High rates of bovine blastocyst development after ICSI-mediated gene transfer assisted by chemical activation. Theriogenology. 2010;74(6):922–931. doi: 10.1016/j.theriogenology.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 56.Chan AW, Luetjens CM, Dominko T, Ramalho-Santos J, Simerly CR, Hewitson L, Schatten G. Foreign DNA transmission by ICSI: injection of spermatozoa bound with exogenous DNA results in embryonic GFP expression and live rhesus monkey births. Mol Hum Reprod. 2000;6(1):26–33. doi: 10.1093/molehr/6.1.26. [DOI] [PubMed] [Google Scholar]
- 57.Mizushima S, Takagi S, Ono T, Atsumi Y, Tsukada A, Saito N, Sasanami T, Okabe M, Shimada K. Novel method of gene transfer in birds: intracytoplasmic sperm injection for green fluorescent protein expression in quail blastoderms. Biol Reprod. 2010;83(6):965–969. doi: 10.1095/biolreprod.110.085860. [DOI] [PubMed] [Google Scholar]
- 58.Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20(7):532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 59.Kues WA, Schwinzer R, Wirth D, Verhoeyen E, Lemme E, Herrmann D, Barg-Kues B, Hauser H, Wonigeit K, Niemann H. Epigenetic silencing and tissue independent expression of a novel tetracycline inducible system in double-transgenic pigs. FASEB J. 2006;20(8):1200–1202. doi: 10.1096/fj.05-5415fje. [DOI] [PubMed] [Google Scholar]
- 60.Kaneko T, Moisyadi S, Suganuma R, Hohn B, Yanagimachi R, Pelczar P. Recombinase-mediated mouse transgenesis by intracytoplasmic sperm injection. Theriogenology. 2005;64(8):1704–1715. doi: 10.1016/j.theriogenology.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Suganuma R, Pelczar P, Spetz JF, Hohn B, Yanagimachi R, Moisyadi S. Tn5 transposase-mediated mouse transgenesis. Biol Reprod. 2005;73(6):1157–1163. doi: 10.1095/biolreprod.105.044669. [DOI] [PubMed] [Google Scholar]
- 62.Maga EA, Sargent RG, Zeng H, Pati S, Zarling DA, Oppenheim SM, Collette NM, Moyer AL, Conrad-Brink JS, Rowe JD, BonDurant RH, Anderson GB, Murray JD. Increased efficiency of transgenic livestock production. Transgenic Res. 2003;12(4):485–496. doi: 10.1023/a:1024257906647. [DOI] [PubMed] [Google Scholar]
- 63.Mason JB, Najarian JG, Anderson GB, Murray JD, Maga EA. The effect of coating single- and double-stranded DNA with the recombinase A protein of Escherichia coli on transgene integration in mice. Transgenic Res. 2006;15(6):703–710. doi: 10.1007/s11248-006-9005-7. [DOI] [PubMed] [Google Scholar]
- 64.Moisyadi S, Kaminski JM, Yanagimachi R. Use of intracytoplasmic sperm injection (ICSI) to generate transgenic animals. Comp Immunol Microbiol Infect Dis. 2009;32(2):47–60. doi: 10.1016/j.cimid.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Garrels W, Mates L, Holler S, Dalda A, Taylor U, Petersen B, Niemann H, Izsvak Z, Ivics Z, Kues WA. Germline transgenic pigs by sleeping beauty transposition in porcine zygotes and targeted integration in the pig genome. PLoS ONE. 2011;6(8):e23573. doi: 10.1371/journal.pone.0023573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ivics Z, Garrels W, Mátés L, Yau TY, Bashir S, Zidek V, Landa V, Geurts A, Pravenec M, Rülicke T, Kues WA, Izsvák Z. Germline transgenesis in pigs by cytoplasmic microinjection of sleeping beauty transposons. Nat Protoc. 2014;9(4):810–827. doi: 10.1038/nprot.2014.010. [DOI] [PubMed] [Google Scholar]
- 67.Kawakami K. Tol2: a versatile gene transfer vector in vertebrates. Genome Biol. 2007;8(Suppl 1):S7. doi: 10.1186/gb-2007-8-s1-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suster ML, Abe G, Schouw A, Kawakami K. Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc. 2011;6(12):1998–2021. doi: 10.1038/nprot.2011.416. [DOI] [PubMed] [Google Scholar]
- 69.Schnutgen F, Stewart AF, von Melchner H, Anastassiadis K. Engineering embryonic stem cells with recombinase systems. Methods Enzymol. 2006;420:100–136. doi: 10.1016/S0076-6879(06)20007-7. [DOI] [PubMed] [Google Scholar]
- 70.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Methods. 2013;10(10):957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci USA. 1950;36(6):344–355. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Makałowski W, Pande A, Gotea V, Makałowska I. Transposable elements and their identification. Methods Mol Biol. 2012;855:337–359. doi: 10.1007/978-1-61779-582-4_12. [DOI] [PubMed] [Google Scholar]
- 73.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, Consortium IHGS Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 74.Muñoz-López M, García-Pérez JL. DNA transposons: nature and applications in genomics. Curr Genomics. 2010;11(2):115–128. doi: 10.2174/138920210790886871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ostertag EM, Kazazian HH. Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 76.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110(3):315–325. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 77.Plasterk RH, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15(8):326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 78.Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposon-mediated genome manipulation in vertebrates. Nat Methods. 2009;6(6):415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91(4):501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 80.van Luenen HG, Colloms SD, Plasterk RH. The mechanism of transposition of Tc3 in C. elegans . Cell. 1994;79(2):293–301. doi: 10.1016/0092-8674(94)90198-8. [DOI] [PubMed] [Google Scholar]
- 81.Mátés L, Izsvák Z, Ivics Z. Technology transfer from worms and flies to vertebrates: transposition-based genome manipulations and their future perspectives. Genome Biol. 2007;8(Suppl 1):S1. doi: 10.1186/gb-2007-8-s1-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122(3):473–483. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 83.Horie K, Yusa K, Yae K, Odajima J, Fischer SE, Keng VW, Hayakawa T, Mizuno S, Kondoh G, Ijiri T, Matsuda Y, Plasterk RH, Takeda J. Characterization of Sleeping Beauty transposition and its application to genetic screening in mice. Mol Cell Biol. 2003;23(24):9189–9207. doi: 10.1128/MCB.23.24.9189-9207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Balciunas D, Wangensteen KJ, Wilber A, Bell J, Geurts A, Sivasubbu S, Wang X, Hackett PB, Largaespada DA, McIvor RS, Ekker SC. Harnessing a high cargo-capacity transposon for genetic applications in vertebrates. PLoS Genet. 2006;2(11):e169. doi: 10.1371/journal.pgen.0020169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rostovskaya M, Naumann R, Fu J, Obst M, Mueller D, Stewart AF, Anastassiadis K. Transposon mediated BAC transgenesis via pronuclear injection of mouse zygotes. Genesis. 2013;51(2):135–141. doi: 10.1002/dvg.22362. [DOI] [PubMed] [Google Scholar]
- 86.Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. 2002;22(9):3157–3173. doi: 10.1128/MCB.22.9.3157-3173.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carlson DF, Garbe JR, Tan W, Martin MJ, Dobrinsky JR, Hackett PB, Clark KJ, Fahrenkrug SC. Strategies for selection marker-free swine transgenesis using the sleeping beauty transposon system. Transgenic Res. 2011;20(5):1125–1137. doi: 10.1007/s11248-010-9481-7. [DOI] [PubMed] [Google Scholar]
- 88.Ryding AD, Sharp MG, Mullins JJ. Conditional transgenic technologies. J Endocrinol. 2001;171(1):1–14. doi: 10.1677/joe.0.1710001. [DOI] [PubMed] [Google Scholar]
- 89.Woltjen K, Hämäläinen R, Kibschull M, Mileikovsky M, Nagy A. Transgene-free production of pluripotent stem cells using piggyBac transposons. Methods Mol Biol. 2011;767:87–103. doi: 10.1007/978-1-61779-201-4_7. [DOI] [PubMed] [Google Scholar]
- 90.Yusa K, Rad R, Takeda J, Bradley A. Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat Methods. 2009;6(5):363–369. doi: 10.1038/nmeth.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li X, Burnight ER, Cooney AL, Malani N, Brady T, Sander JD, Staber J, Wheelan SJ, Joung JK, McCray PB, Bushman FD, Sinn PL, Craig NL. piggyBac transposase tools for genome engineering. Proc Natl Acad Sci USA. 2013;110(25):E2279–E2287. doi: 10.1073/pnas.1305987110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sinzelle L, Vallin J, Coen L, Chesneau A, Du Pasquier D, Pollet N, Demeneix B, Mazabraud A. Generation of trangenic Xenopus laevis using the Sleeping Beauty transposon system. Transgenic Res. 2006;15(6):751–760. doi: 10.1007/s11248-006-9014-6. [DOI] [PubMed] [Google Scholar]
- 93.Yergeau DA, Johnson Hamlet MR, Kuliyev E, Zhu H, Doherty JR, Archer TD, Subhawong AP, Valentine MB, Kelley CM, Mead PE. Transgenesis in Xenopus using the sleeping beauty transposon system. Dev Dyn. 2009;238(7):1727–1743. doi: 10.1002/dvdy.21994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kitada K, Ishishita S, Tosaka K, Takahashi R, Ueda M, Keng VW, Horie K, Takeda J. Transposon-tagged mutagenesis in the rat. Nat Methods. 2007;4(2):131–133. doi: 10.1038/nmeth1002. [DOI] [PubMed] [Google Scholar]
- 95.Clark KJ, Carlson DF, Foster LK, Kong BW, Foster DN, Fahrenkrug SC. Enzymatic engineering of the porcine genome with transposons and recombinases. BMC Biotechnol. 2007;7:42. doi: 10.1186/1472-6750-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Baus J, Liu L, Heggestad AD, Sanz S, Fletcher BS. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol Ther. 2005;12(6):1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- 97.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24(20):9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Geurts AM, Yang Y, Clark KJ, Liu G, Cui Z, Dupuy AJ, Bell JB, Largaespada DA, Hackett PB. Gene transfer into genomes of human cells by the sleeping beauty transposon system. Mol Ther. 2003;8(1):108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- 99.Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proc Natl Acad Sci USA. 1980;77(12):7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bondioli KR, Biery KA, Hill KG, Jones KB, De Mayo FJ. Production of transgenic cattle by pronuclear injection. Biotechnology. 1991;16:265–273. [PubMed] [Google Scholar]
- 101.Hammer RE, Pursel VG, Rexroad CE, Jr, Wall RJ, Bolt DJ, Ebert KM, Palmiter RD, Brinster RL. Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985;315(6021):680–683. doi: 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- 102.Iqbal K, Barg-Kues B, Broll S, Bode J, Niemann H, Kues W. Cytoplasmic injection of circular plasmids allows targeted expression in mammalian embryos. Biotechniques. 2009;47(5):959–968. doi: 10.2144/000113270. [DOI] [PubMed] [Google Scholar]
- 103.Clark KJ, Urban MD, Skuster KJ, Ekker SC. Transgenic zebrafish using transposable elements. Methods Cell Biol. 2011;104:137–149. doi: 10.1016/B978-0-12-374814-0.00008-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marh J, Stoytcheva Z, Urschitz J, Sugawara A, Yamashiro H, Owens JB, Stoytchev I, Pelczar P, Yanagimachi R, Moisyadi S. Hyperactive self-inactivating piggyBac for transposase-enhanced pronuclear microinjection transgenesis. Proc Natl Acad Sci USA. 2012;109(47):19184–19189. doi: 10.1073/pnas.1216473109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jang CW, Behringer RR. Transposon-mediated transgenesis in rats. CSH Protoc. 2007;2007:pdb.prot4866. doi: 10.1101/pdb.prot4866. [DOI] [PubMed] [Google Scholar]
- 106.Li Z, Zeng F, Meng F, Xu Z, Zhang X, Huang X, Tang F, Gao W, Shi J, He X, Liu D, Wang C, Urschitz J, Moisyadi S, Wu Z. Generation of transgenic pigs by cytoplasmic injection of piggyBac transposase based pmGENIE-3 plasmids. Biol Reprod. 2014;90(5):93–102. doi: 10.1095/biolreprod.113.116905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jakobsen JE, Li J, Kragh PM, Moldt B, Lin L, Liu Y, Schmidt M, Winther KD, Schyth BD, Holm IE, Vajta G, Bolund L, Callesen H, Jorgensen AL, Nielsen AL, Mikkelsen JG. Pig transgenesis by sleeping beauty DNA transposition. Transgenic Res. 2011;20(3):533–545. doi: 10.1007/s11248-010-9438-x. [DOI] [PubMed] [Google Scholar]
- 108.Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- 109.Campbell KH, McWhir J, Ritchie WA, Wilmut I. Sheep cloned by nuclear transfer from a cultured cell line. Nature. 1996;380(6569):64–66. doi: 10.1038/380064a0. [DOI] [PubMed] [Google Scholar]
- 110.Bosch P, Hodges CA, Stice SL. Generation of transgenic livestock by somatic cell nuclear transfer. Biotecnologia Aplicada. 2004;21(3):128–136. [Google Scholar]
- 111.Wu Z, Xu Z, Zou X, Zeng F, Shi J, Liu D, Urschitz J, Moisyadi S, Li Z. Pig transgenesis by piggyBac transposition in combination with somatic cell nuclear transfer. Transgenic Res. 2013;22(6):1107–1118. doi: 10.1007/s11248-013-9729-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alessio A, Fili A, Forcato D, Olmos-Nicotra F, Alustiza F, Rodriguez N, Owens J, Moisyad S, Kues WA, Bosch P. Efficient piggyBac transposon-mediated transgene integration into bovine fetal fibroblast genome. Reprod Dom Anim. 2014;49(S1):8. [Google Scholar]
- 113.Wilson MH, Coates CJ, George AL., Jr PiggyBac transposon-mediated gene transfer in human cells. Mol Ther. 2007;15(1):139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- 114.Ikeda R, Kokubu C, Yusa K, Keng VW, Horie K, Takeda J. Sleeping beauty transposase has an affinity for heterochromatin conformation. Mol Cell Biol. 2007;27(5):1665–1676. doi: 10.1128/MCB.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25(6):2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Blakely G, Colloms S, May G, Burke M, Sherratt D. Escherichia coli XerC recombinase is required for chromosomal segregation at cell division. New Biol. 1991;3(8):789–798. [PubMed] [Google Scholar]
- 117.Gellert M, Nash H. Communication between segments of DNA during site-specific recombination. Nature. 1987;325(6103):401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- 118.Sadowski P. Site-specific recombinases: changing partners and doing the twist. J Bacteriol. 1986;165(2):341–347. doi: 10.1128/jb.165.2.341-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fukushige S, Sauer B. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci USA. 1992;89(17):7905–7909. doi: 10.1073/pnas.89.17.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.O’Gorman S, Fox DT, Wahl GM. Recombinase-mediated gene activation and site-specific integration in mammalian cells. Science. 1991;251(4999):1351–1355. doi: 10.1126/science.1900642. [DOI] [PubMed] [Google Scholar]
- 121.Bestor TH. Gene silencing as a threat to the success of gene therapy. J Clin Invest. 2000;105(4):409–411. doi: 10.1172/JCI9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Whitelaw E, Sutherland H, Kearns M, Morgan H, Weaving L, Garrick D. Epigenetic effects on transgene expression. Methods Mol Biol. 2001;158:351–368. doi: 10.1385/1-59259-220-1:351. [DOI] [PubMed] [Google Scholar]
- 123.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Annu Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 124.Stark WM, Boocock MR, Sherratt DJ. Catalysis by site-specific recombinases. Trends Genet. 1992;8(12):432–439. [PubMed] [Google Scholar]
- 125.Dymecki SM. Site-specific recombination in cells and mice. In: Joyner AL, editor. Gene targeting: a practical approach. The practical approach. 2. New York: Oxford University Press; 2000. pp. 36–99. [Google Scholar]
- 126.Wang Y, Yau YY, Perkins-Balding D, Thomson JG. Recombinase technology: applications and possibilities. Plant Cell Rep. 2011;30(3):267–285. doi: 10.1007/s00299-010-0938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grainge I, Jayaram M. The integrase family of recombinase: organization and function of the active site. Mol Microbiol. 1999;33(3):449–456. doi: 10.1046/j.1365-2958.1999.01493.x. [DOI] [PubMed] [Google Scholar]
- 128.Branda CS, Dymecki SM. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6(1):7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- 129.Turan S, Zehe C, Kuehle J, Qiao J, Bode J. Recombinase-mediated cassette exchange (RMCE): a rapidly-expanding toolbox for targeted genomic modifications. Gene. 2013;515(1):1–27. doi: 10.1016/j.gene.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 130.Jones JR, Shelton KD, Magnuson MA. Strategies for the use of site-specific recombinases in genome engineering. Methods Mol Med. 2005;103:245–257. doi: 10.1385/1-59259-780-7:245. [DOI] [PubMed] [Google Scholar]
- 131.Turan S, Galla M, Ernst E, Qiao J, Voelkel C, Schiedlmeier B, Zehe C, Bode J. Recombinase-mediated cassette exchange (RMCE): traditional concepts and current challenges. J Mol Biol. 2011;407(2):193–221. doi: 10.1016/j.jmb.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 132.Araki K, Araki M, Yamamura K. Targeted integration of DNA using mutant lox sites in embryonic stem cells. Nucleic Acids Res. 1997;25(4):868–872. doi: 10.1093/nar/25.4.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Araki K, Imaizumi T, Sekimoto T, Yoshinobu K, Yoshimuta J, Akizuki M, Miura K, Araki M, Yamamura K. Exchangeable gene trap using the Cre/mutated lox system. Cell Mol Biol (Noisy-le-grand) 1999;45(5):737–750. [PubMed] [Google Scholar]
- 134.Senecoff JF, Rossmeissl PJ, Cox MM. DNA recognition by the FLP recombinase of the yeast 2 mu plasmid. A mutational analysis of the FLP binding site. J Mol Biol. 1988;201(2):405–421. doi: 10.1016/0022-2836(88)90147-7. [DOI] [PubMed] [Google Scholar]
- 135.Schlake T, Bode J. Use of mutated FLP recognition target (FRT) sites for the exchange of expression cassettes at defined chromosomal loci. Biochemistry. 1994;33(43):12746–12751. doi: 10.1021/bi00209a003. [DOI] [PubMed] [Google Scholar]
- 136.Kolb AF, Siddell SG. Genomic targeting with an MBP-Cre fusion protein. Gene. 1996;183(1–2):53–60. doi: 10.1016/s0378-1119(96)00470-2. [DOI] [PubMed] [Google Scholar]
- 137.Baubonis W, Sauer B. Genomic targeting with purified Cre recombinase. Nucleic Acids Res. 1993;21(9):2025–2029. doi: 10.1093/nar/21.9.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.de Wit T, Drabek D, Grosveld F. Microinjection of cre recombinase RNA induces site-specific recombination of a transgene in mouse oocytes. Nucleic Acids Res. 1998;26(2):676–678. doi: 10.1093/nar/26.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Braun T, Bober E, Rudnicki MA, Jaenisch R, Arnold HH. MyoD expression marks the onset of skeletal myogenesis in Myf-5 mutant mice. Development. 1994;120(11):3083–3092. doi: 10.1242/dev.120.11.3083. [DOI] [PubMed] [Google Scholar]
- 140.Fiering S, Kim CG, Epner EM, Groudine M. An “in-out” strategy using gene targeting and FLP recombinase for the functional dissection of complex DNA regulatory elements: analysis of the beta-globin locus control region. Proc Natl Acad Sci USA. 1993;90(18):8469–8473. doi: 10.1073/pnas.90.18.8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim CG, Epner EM, Forrester WC, Groudine M. Inactivation of the human beta-globin gene by targeted insertion into the beta-globin locus control region. Genes Dev. 1992;6(6):928–938. doi: 10.1101/gad.6.6.928. [DOI] [PubMed] [Google Scholar]
- 142.Olson EN, Arnold HH, Rigby PW, Wold BJ. Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell. 1996;85(1):1–4. doi: 10.1016/s0092-8674(00)81073-9. [DOI] [PubMed] [Google Scholar]
- 143.Pham CT, MacIvor DM, Hug BA, Heusel JW, Ley TJ. Long-range disruption of gene expression by a selectable marker cassette. Proc Natl Acad Sci USA. 1996;93(23):13090–13095. doi: 10.1073/pnas.93.23.13090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Howard TH, Homan EJ, Bremel RD. Transgenic livestock: regulation and science in a changing environment. J Anim Sci. 2001;79:E1–E11. [Google Scholar]
- 145.Askew GR, Doetschman T, Lingrel JB. Site-directed point mutations in embryonic stem cells: a gene-targeting tag-and-exchange strategy. Mol Cell Biol. 1993;13(7):4115–4124. doi: 10.1128/mcb.13.7.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Stacey A, Schnieke A, McWhir J, Cooper J, Colman A, Melton DW. Use of double-replacement gene targeting to replace the murine alpha-lactalbumin gene with its human counterpart in embryonic stem cells and mice. Mol Cell Biol. 1994;14(2):1009–1016. doi: 10.1128/mcb.14.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wu H, Liu X, Jaenisch R. Double replacement: strategy for efficient introduction of subtle mutations into the murine Col1a-1 gene by homologous recombination in embryonic stem cells. Proc Natl Acad Sci USA. 1994;91(7):2819–2823. doi: 10.1073/pnas.91.7.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Graham C, Cole S, Laible G. Site-specific modification of the bovine genome using Cre recombinase-mediated gene targeting. Biotechnol J. 2009;4(1):108–118. doi: 10.1002/biot.200800200. [DOI] [PubMed] [Google Scholar]
- 149.Yu Y, Tong Q, Li Z, Tian J, Wang Y, Su F, Liu J, Zhang Y. Improved site-specific recombinase-based method to produce selectable marker- and vector-backbone-free transgenic cells. Sci Rep. 2014;4:4240. doi: 10.1038/srep04240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Yu H, Wang X, Zhu L, He Z, Liu G, Xu X, Chen J, Cheng G. Establishment of a rapid and scalable gene expression system in livestock by site-specific integration. Gene. 2013;515(2):367–371. doi: 10.1016/j.gene.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 151.Wang S, Sun X, Ding F, Zhang K, Zhao R, Li S, Li R, Tang B, Zhang L, Liu Y, Li J, Gao F, Wang H, Wang L, Dai Y, Li N. Removal of selectable marker gene from fibroblast cells in transgenic cloned cattle by transient expression of Cre recombinase and subsequent effects on recloned embryo development. Theriogenology. 2009;72(4):535–541. doi: 10.1016/j.theriogenology.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 152.Xu Y, Liu S, Yu G, Chen J, Xu X, Wu Y, Zhang A, Dowdy SF, Cheng G. Excision of selectable genes from transgenic goat cells by a protein transducible TAT-Cre recombinase. Gene. 2008;419(1–2):70–74. doi: 10.1016/j.gene.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 153.Zhang H, Hasty P, Bradley A. Targeting frequency for deletion vectors in embryonic stem cells. Mol Cell Biol. 1994;14(4):2404–2410. doi: 10.1128/mcb.14.4.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nagy A, Moens C, Ivanyi E, Pawling J, Gertsenstein M, Hadjantonakis AK, Pirity M, Rossant J. Dissecting the role of N-myc in development using a single targeting vector to generate a series of alleles. Curr Biol. 1998;8(11):661–664. doi: 10.1016/s0960-9822(98)70254-4. [DOI] [PubMed] [Google Scholar]
- 155.Ramírez-Solis R, Liu P, Bradley A. Chromosome engineering in mice. Nature. 1995;378(6558):720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 156.Zou YR, Müller W, Gu H, Rajewsky K. Cre-loxP-mediated gene replacement: a mouse strain producing humanized antibodies. Curr Biol. 1994;4(12):1099–1103. doi: 10.1016/s0960-9822(00)00248-7. [DOI] [PubMed] [Google Scholar]
- 157.Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265(5168):103–106. doi: 10.1126/science.8016642. [DOI] [PubMed] [Google Scholar]
- 158.Kühn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 159.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26(2):99–109. [PubMed] [Google Scholar]
- 160.Chen L, Li L, Pang D, Li Z, Wang T, Zhang M, Song N, Yan S, Lai LX, Ouyang H. Construction of transgenic swine with induced expression of Cre recombinase. Animal. 2010;4(5):767–771. doi: 10.1017/S1751731109991571. [DOI] [PubMed] [Google Scholar]
- 161.Li S, Flisikowska T, Kurome M, Zakhartchenko V, Kessler B, Saur D, Kind A, Wolf E, Flisikowski K, Schnieke A. Dual fluorescent reporter pig for Cre recombination: transgene placement at the ROSA26 locus. PLoS ONE. 2014;9(7):e102455. doi: 10.1371/journal.pone.0102455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Groth AC, Olivares EC, Thyagarajan B, Calos MP. A phage integrase directs efficient site-specific integration in human cells. Proc Natl Acad Sci USA. 2000;97(11):5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Belteki G, Gertsenstein M, Ow DW, Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing phiC31 integrase. Nat Biotechnol. 2003;21(3):321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- 164.Smith MC, Brown WR, McEwan AR, Rowley PA. Site-specific recombination by phiC31 integrase and other large serine recombinases. Biochem Soc Trans. 2010;38(2):388–394. doi: 10.1042/BST0380388. [DOI] [PubMed] [Google Scholar]
- 165.Thorpe HM, Wilson SE, Smith MC. Control of directionality in the site-specific recombination system of the Streptomyces phage phiC31. Mol Microbiol. 2000;38(2):232–241. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- 166.Farruggio AP, Chavez CL, Mikell CL, Calos MP. Efficient reversal of phiC31 integrase recombination in mammalian cells. Biotechnol J. 2012;7(11):1332–1336. doi: 10.1002/biot.201200283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol Cell Biol. 2001;21(12):3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166(4):1775–1782. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yin Y, Cao G, Xue R, Gong C. Construction of transformed, cultured silkworm cells and transgenic silkworm using the site-specific integrase system from phage φC31. Mol Biol Rep. 2014;41(10):6449–6456. doi: 10.1007/s11033-014-3527-5. [DOI] [PubMed] [Google Scholar]
- 170.Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Methods. 2005;2(12):975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hu ZP, Chen LS, Jia CY, Zhu HZ, Wang W, Zhong J. Screening of potential pseudo att sites of Streptomyces phage ΦC31 integrase in the human genome. Acta Pharmacol Sin. 2013;34(4):561–569. doi: 10.1038/aps.2012.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ma QW, Sheng HQ, Yan JB, Cheng S, Huang Y, Chen-Tsai Y, Ren ZR, Huang SZ, Zeng YT. Identification of pseudo attP sites for phage phiC31 integrase in bovine genome. Biochem Biophys Res Commun. 2006;345(3):984–988. doi: 10.1016/j.bbrc.2006.04.145. [DOI] [PubMed] [Google Scholar]
- 173.Qu L, Ma Q, Zhou Z, Ma H, Huang Y, Huang S, Zeng F, Zeng Y. A profile of native integration sites used by φC31 integrase in the bovine genome. J Genet Genomics. 2012;39(5):217–224. doi: 10.1016/j.jgg.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 174.Ni W, Hu S, Qiao J, Wang Y, Shi H, He Z, Li G, Chen C. ΦC31 integrase mediates efficient site-specific integration in sheep fibroblasts. Biosci Biotechnol Biochem. 2012;76(11):2093–2095. doi: 10.1271/bbb.120439. [DOI] [PubMed] [Google Scholar]
- 175.Ma H, Ma Q, Lu Y, Wang J, Hu W, Gong Z, Cai L, Huang Y, Huang SZ, Zeng F. PhiC31 integrase induces efficient site-specific recombination in the Capra hircus genome. DNA Cell Biol. 2014;33(8):484–491. doi: 10.1089/dna.2013.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Bi Y, Liu X, Zhang L, Shao C, Ma Z, Hua Z, Li L, Hua W, Xiao H, Wei Q, Zheng X. Pseudo attP sites in favor of transgene integration and expression in cultured porcine cells identified by Streptomyces phage phiC31 integrase. BMC Mol Biol. 2013;14:20. doi: 10.1186/1471-2199-14-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Sadelain M, Papapetrou EP, Bushman FD. Safe harbours for the integration of new DNA in the human genome. Nat Rev Cancer. 2012;12(1):51–58. doi: 10.1038/nrc3179. [DOI] [PubMed] [Google Scholar]
- 178.Papapetrou EP, Lee G, Malani N, Setty M, Riviere I, Tirunagari LM, Kadota K, Roth SL, Giardina P, Viale A, Leslie C, Bushman FD, Studer L, Sadelain M. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat Biotechnol. 2011;29(1):73–78. doi: 10.1038/nbt.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Keravala A, Lee S, Thyagarajan B, Olivares EC, Gabrovsky VE, Woodard LE, Calos MP. Mutational derivatives of PhiC31 integrase with increased efficiency and specificity. Mol Ther. 2009;17(1):112–120. doi: 10.1038/mt.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sclimenti CR, Thyagarajan B, Calos MP. Directed evolution of a recombinase for improved genomic integration at a native human sequence. Nucleic Acids Res. 2001;29(24):5044–5051. doi: 10.1093/nar/29.24.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu Z, Thomas L, Davies B, Chalmers R, Smith M, Brown W. Accuracy and efficiency define Bxb1 integrase as the best of fifteen candidate serine recombinases for the integration of DNA into the human genome. BMC Biotechnol. 2013;13:87. doi: 10.1186/1472-6750-13-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Olivares EC, Hollis RP, Calos MP. Phage R4 integrase mediates site-specific integration in human cells. Gene. 2001;278(1–2):167–176. doi: 10.1016/s0378-1119(01)00711-9. [DOI] [PubMed] [Google Scholar]
- 183.Stoll SM, Ginsburg DS, Calos MP. Phage TP901-1 site-specific integrase functions in human cells. J Bacteriol. 2002;184(13):3657–3663. doi: 10.1128/JB.184.13.3657-3663.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Keravala A, Groth AC, Jarrahian S, Thyagarajan B, Hoyt JJ, Kirby PJ, Calos MP. A diversity of serine phage integrases mediate site-specific recombination in mammalian cells. Mol Genet Genomics. 2006;276(2):135–146. doi: 10.1007/s00438-006-0129-5. [DOI] [PubMed] [Google Scholar]
- 185.Russell JP, Chang DW, Tretiakova A, Padidam M. Phage Bxb1 integrase mediates highly efficient site-specific recombination in mammalian cells. Biotechniques. 2006;40(4):462. doi: 10.2144/000112150. [DOI] [PubMed] [Google Scholar]
- 186.Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R, Cost GJ, Niemann H. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci USA. 2011;108(29):12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci USA. 2012;109(43):17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188(4):773–782. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA with fingers and TALENs. Mol Ther Nucleic Acids. 2012;1:e3. doi: 10.1038/mtna.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife (Cambridge) 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sun N, Abil Z, Zhao H. Recent advances in targeted genome engineering in mammalian systems. Biotechnol J. 2012;7(9):1074–1087. doi: 10.1002/biot.201200038. [DOI] [PubMed] [Google Scholar]
- 194.Dai Y, Vaught TD, Boone J, Chen SH, Phelps CJ, Ball S, Monahan JA, Jobst PM, McCreath KJ, Lamborn AE, Cowell-Lucero JL, Wells KD, Colman A, Polejaeva IA, Ayares DL. Targeted disruption of the alpha1,3-galactosyltransferase gene in cloned pigs. Nat Biotechnol. 2002;20(3):251–255. doi: 10.1038/nbt0302-251. [DOI] [PubMed] [Google Scholar]
- 195.McCreath KJ, Howcroft J, Campbell KH, Colman A, Schnieke AE, Kind AJ. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405(6790):1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- 196.Yu G, Chen J, Yu H, Liu S, Xu X, Sha H, Zhang X, Wu G, Xu S, Cheng G. Functional disruption of the prion protein gene in cloned goats. J Gen Virol. 2006;87(Pt 4):1019–1027. doi: 10.1099/vir.0.81384-0. [DOI] [PubMed] [Google Scholar]
- 197.Zhu C, Li B, Yu G, Chen J, Yu H, Xu X, Wu Y, Zhang A, Cheng G. Production of Prnp−/− goats by gene targeting in adult fibroblasts. Transgenic Res. 2009;18(2):163–171. doi: 10.1007/s11248-008-9220-5. [DOI] [PubMed] [Google Scholar]
- 198.Denning C, Burl S, Ainslie A, Bracken J, Dinnyes A, Fletcher J, King T, Ritchie M, Ritchie WA, Rollo M, de Sousa P, Travers A, Wilmut I, Clark AJ. Deletion of the alpha(1,3)galactosyl transferase (GGTA1) gene and the prion protein (PrP) gene in sheep. Nat Biotechnol. 2001;19(6):559–562. doi: 10.1038/89313. [DOI] [PubMed] [Google Scholar]
- 199.Zhou ZR, Zhong BS, Jia RX, Wan YJ, Zhang YL, Fan YX, Wang LZ, You JH, Wang ZY, Wang F. Production of myostatin-targeted goat by nuclear transfer from cultured adult somatic cells. Theriogenology. 2013;79(2):225–233. doi: 10.1016/j.theriogenology.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 200.Thomas KR, Capecchi MR. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51(3):503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 201.Porter AC, Itzhaki JE. Gene targeting in human somatic cells. Complete inactivation of an interferon-inducible gene. Eur J Biochem. 1993;218(2):273–281. doi: 10.1111/j.1432-1033.1993.tb18375.x. [DOI] [PubMed] [Google Scholar]
- 202.Brown JP, Wei W, Sedivy JM. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts. Science. 1997;277(5327):831–834. doi: 10.1126/science.277.5327.831. [DOI] [PubMed] [Google Scholar]
- 203.Smith LC, Suzuki J, Goff AK, Filion F, Therrien J, Murphy BD, Kohan-Ghadr HR, Lefebvre R, Brisville AC, Buczinski S, Fecteau G, Perecin F, Meirelles FV. Developmental and epigenetic anomalies in cloned cattle. Reprod Domest Anim. 2012;47(Suppl 4):107–114. doi: 10.1111/j.1439-0531.2012.02063.x. [DOI] [PubMed] [Google Scholar]
- 204.Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23(24):5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Rouet P, Smih F, Jasin M. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol Cell Biol. 1994;14(12):8096–8106. doi: 10.1128/mcb.14.12.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, Zeitler B, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory PD, Urnov FD, Jaenisch R. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29(8):731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lombardo A, Cesana D, Genovese P, Di Stefano B, Provasi E, Colombo DF, Neri M, Magnani Z, Cantore A, Lo Riso P, Damo M, Pello OM, Holmes MC, Gregory PD, Gritti A, Broccoli V, Bonini C, Naldini L. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8(10):861–869. doi: 10.1038/nmeth.1674. [DOI] [PubMed] [Google Scholar]
- 208.Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, Rebar EJ, Gregory PD, Klug A, Collingwood TN. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105(15):5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Whyte JJ, Zhao J, Wells KD, Samuel MS, Whitworth KM, Walters EM, Laughlin MH, Prather RS. Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Mol Reprod Dev. 2011;78(1):2. doi: 10.1002/mrd.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Yang D, Yang H, Li W, Zhao B, Ouyang Z, Liu Z, Zhao Y, Fan N, Song J, Tian J, Li F, Zhang J, Chang L, Pei D, Chen YE, Lai L. Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Res. 2011;21(6):979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Yu S, Luo J, Song Z, Ding F, Dai Y, Li N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res. 2011;21(11):1638–1640. doi: 10.1038/cr.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting. Science. 2011;333(6051):1843–1846. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- 213.Seruggia D, Montoliu L. The new CRISPR-Cas system: RNA-guided genome engineering to efficiently produce any desired genetic alteration in animals. Transgenic Res. 2014;23(5):707–716. doi: 10.1007/s11248-014-9823-y. [DOI] [PubMed] [Google Scholar]
- 214.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci USA. 1996;93(3):1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, Gregory PD, Pabo CO, Rebar EJ. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25(7):778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 216.Szczepek M, Brondani V, Büchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25(7):786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 217.Watanabe M, Umeyama K, Matsunari H, Takayanagi S, Haruyama E, Nakano K, Fujiwara T, Ikezawa Y, Nakauchi H, Nagashima H. Knockout of exogenous EGFP gene in porcine somatic cells using zinc-finger nucleases. Biochem Biophys Res Commun. 2010;402(1):14–18. doi: 10.1016/j.bbrc.2010.09.092. [DOI] [PubMed] [Google Scholar]
- 218.Whyte JJ, Prather RS. Genetic modifications of pigs for medicine and agriculture. Mol Reprod Dev. 2011;78(10–11):879–891. doi: 10.1002/mrd.21333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Bao L, Chen H, Jong U, Rim C, Li W, Lin X, Zhang D, Luo Q, Cui C, Huang H, Zhang Y, Xiao L, Fu Z. Generation of GGTA1 biallelic knockout pigs via zinc-finger nucleases and somatic cell nuclear transfer. Sci China Life Sci. 2014;57(2):263–268. doi: 10.1007/s11427-013-4601-2. [DOI] [PubMed] [Google Scholar]
- 220.Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22(7):1327–1333. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ramirez CL, Certo MT, Mussolino C, Goodwin MJ, Cradick TJ, McCaffrey AP, Cathomen T, Scharenberg AM, Joung JK. Engineered zinc finger nickases induce homology-directed repair with reduced mutagenic effects. Nucleic Acids Res. 2012;40(12):5560–5568. doi: 10.1093/nar/gks179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wang J, Friedman G, Doyon Y, Wang NS, Li CJ, Miller JC, Hua KL, Yan JJ, Babiarz JE, Gregory PD, Holmes MC. Targeted gene addition to a predetermined site in the human genome using a ZFN-based nicking enzyme. Genome Res. 2012;22(7):1316–1326. doi: 10.1101/gr.122879.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Liu X, Wang Y, Guo W, Chang B, Liu J, Guo Z, Quan F, Zhang Y. Zinc-finger nickase-mediated insertion of the lysostaphin gene into the beta-casein locus in cloned cows. Nat Commun. 2013;4:2565. doi: 10.1038/ncomms3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Camenisch TD, Brilliant MH, Segal DJ. Critical parameters for genome editing using zinc finger nucleases. Mini Rev Med Chem. 2008;8(7):669–676. doi: 10.2174/138955708784567458. [DOI] [PubMed] [Google Scholar]
- 225.Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335(6069):720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335(6069):716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, Santiago Y, Vincent AI, Meng X, Zhang L, Gregory PD, Anegon I, Cost GJ. Knockout rats generated by embryo microinjection of TALENs. Nat Biotechnol. 2011;29(8):695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- 228.Mussolino C, Cathomen T. On target? Tracing zinc-finger-nuclease specificity. Nat Methods. 2011;8(9):725–726. doi: 10.1038/nmeth.1680. [DOI] [PubMed] [Google Scholar]
- 229.Cade L, Reyon D, Hwang WY, Tsai SQ, Patel S, Khayter C, Joung JK, Sander JD, Peterson RT, Yeh JR. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 2012;40(16):8001–8010. doi: 10.1093/nar/gks518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Tan WS, Carlson DF, Walton MW, Fahrenkrug SC, Hackett PB. Precision editing of large animal genomes. Adv Genet. 2012;80:37–97. doi: 10.1016/B978-0-12-404742-6.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kambadur R, Sharma M, Smith TP, Bass JJ. Mutations in myostatin (GDF8) in double-muscled belgian blue and piedmontese cattle. Genome Res. 1997;7(9):910–916. doi: 10.1101/gr.7.9.910. [DOI] [PubMed] [Google Scholar]
- 232.Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Ménissier F, Massabanda J, Fries R, Hanset R, Georges M. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet. 1997;17(1):71–74. doi: 10.1038/ng0997-71. [DOI] [PubMed] [Google Scholar]
- 233.Proudfoot C, Carlson DF, Huddart R, Long CR, Pryor JH, King TJ, Lillico SG, Mileham AJ, McLaren DG, Whitelaw CB, Fahrenkrug SC. Genome edited sheep and cattle. Transgenic Res. 2014 doi: 10.1007/s11248-014-9832-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 238.Yan Q, Zhang Q, Yang H, Zou Q, Tang C, Fan N, Lai L. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen (Lond) 2014;3(1):12. doi: 10.1186/2045-9769-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Huang J, Guo X, Fan N, Song J, Zhao B, Ouyang Z, Liu Z, Zhao Y, Yan Q, Yi X, Schambach A, Frampton J, Esteban MA, Yang D, Yang H, Lai L. RAG1/2 knockout pigs with severe combined immunodeficiency. J Immunol. 2014;193(3):1496–1503. doi: 10.4049/jimmunol.1400915. [DOI] [PubMed] [Google Scholar]
- 240.Reyes LM, Estrada JL, Wang ZY, Blosser RJ, Smith RF, Sidner RA, Paris LL, Blankenship RL, Ray CN, Miner AC, Tector M, Tector AJ. Creating class I MHC-null pigs using guide RNA and the Cas9 endonuclease. J Immunol. 2014;193(11):5751–5757. doi: 10.4049/jimmunol.1402059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Boulanger L, Pannetier M, Gall L, Allais-Bonnet A, Elzaiat M, Le Bourhis D, Daniel N, Richard C, Cotinot C, Ghyselinck NB, Pailhoux E. FOXL2 is a female sex-determining gene in the goat. Curr Biol. 2014;24(4):404–408. doi: 10.1016/j.cub.2013.12.039. [DOI] [PubMed] [Google Scholar]
- 242.Chen Y, Cui Y, Shen B, Niu Y, Zhao X, Wang L, Wang J, Li W, Zhou Q, Ji W, Sha J, Huang X. Germline acquisition of Cas9/RNA-mediated gene modifications in monkeys. Cell Res. 2014 doi: 10.1038/cr.2014.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Wan H, Feng C, Teng F, Yang S, Hu B, Niu Y, Xiang AP, Fang W, Ji W, Li W, Zhao X, Zhou Q. One-step generation of p53 gene biallelic mutant Cynomolgus monkey via the CRISPR/Cas system. Cell Res. 2014 doi: 10.1038/cr.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, Yi X, Guo L, Esteban MA, Zeng Y, Yang H, Lai L. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2014 doi: 10.1007/s00018-014-1744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Li P, Estrada JL, Burlak C, Montgomery J, Butler JR, Santos RM, Wang ZY, Paris LL, Blankenship RL, Downey SM, Tector M, Tector AJ. Efficient generation of genetically distinct pigs in a single pregnancy using multiplexed single-guide RNA and carbohydrate selection. Xenotransplantation. 2014 doi: 10.1111/xen.12131. [DOI] [PubMed] [Google Scholar]
- 246.Whitworth KM, Lee K, Benne JA, Beaton BP, Spate LD, Murphy SL, Samuel MS, Mao J, O’Gorman C, Walters EM, Murphy CN, Driver J, Mileham A, McLaren D, Wells KD, Prather RS. Use of the CRISPR/Cas9 system to produce genetically engineered pigs from in vitro-derived oocytes and embryos. Biol Reprod. 2014;91(3):78. doi: 10.1095/biolreprod.114.121723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ni W, Qiao J, Hu S, Zhao X, Regouski M, Yang M, Polejaeva IA, Chen C. Efficient gene knockout in goats using CRISPR/Cas9 system. PLoS ONE. 2014;9(9):e106718. doi: 10.1371/journal.pone.0106718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24(3):372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ma Y, Ma J, Zhang X, Chen W, Yu L, Lu Y, Bai L, Shen B, Huang X, Zhang L. Generation of eGFP and Cre knockin rats by CRISPR/Cas9. FEBS J. 2014;281(17):3779–3790. doi: 10.1111/febs.12935. [DOI] [PubMed] [Google Scholar]
- 250.Sung YH, Baek IJ, Kim DH, Jeon J, Lee J, Lee K, Jeong D, Kim JS, Lee HW. Knockout mice created by TALEN-mediated gene targeting. Nat Biotechnol. 2013;31(1):23–24. doi: 10.1038/nbt.2477. [DOI] [PubMed] [Google Scholar]
- 251.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, Zhang F. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Hickman AB, Dyda F. CRISPR-Cas immunity and mobile DNA: a new superfamily of DNA transposons encoding a Cas1 endonuclease. Mob DNA. 2014;5:23. doi: 10.1186/1759-8753-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Krupovic M, Makarova KS, Forterre P, Prangishvili D, Koonin EV. Casposons: a new superfamily of self-synthesizing DNA transposons at the origin of prokaryotic CRISPR-Cas immunity. BMC Biol. 2014;12:36. doi: 10.1186/1741-7007-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Katter K, Geurts AM, Hoffmann O, Mates L, Landa V, Hiripi L, Moreno C, Lazar J, Bashir S, Zidek V, Popova E, Jerchow B, Becker K, Devaraj A, Walter I, Grzybowksi M, Corbett M, Filho AR, Hodges MR, Bader M, Ivics Z, Jacob HJ, Pravenec M, Bosze Z, Rulicke T, Izsvak Z. Transposon-mediated transgenesis, transgenic rescue, and tissue-specific gene expression in rodents and rabbits. Faseb J. 2013;27(3):930–941. doi: 10.1096/fj.12-205526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Staunstrup NH, Madsen J, Primo MN, Li J, Liu Y, Kragh PM, Li R, Schmidt M, Purup S, Dagnaes-Hansen F, Svensson L, Petersen TK, Callesen H, Bolund L, Mikkelsen JG. Development of transgenic cloned pig models of skin inflammation by DNA transposon-directed ectopic expression of human beta1 and alpha2 integrin. PLoS ONE. 2012;7(5):e36658. doi: 10.1371/journal.pone.0036658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Al-Mashhadi RH, Sorensen CB, Kragh PM, Christoffersen C, Mortensen MB, Tolbod LP, Thim T, Du Y, Li J, Liu Y, Moldt B, Schmidt M, Vajta G, Larsen T, Purup S, Bolund L, Nielsen LB, Callesen H, Falk E, Mikkelsen JG, Bentzon JF. Familial hypercholesterolemia and atherosclerosis in cloned minipigs created by DNA transposition of a human PCSK9 gain-of-function mutant. Sci Transl Med. 2013;5(166):166. doi: 10.1126/scitranslmed.3004853. [DOI] [PubMed] [Google Scholar]
- 261.Macdonald J, Taylor L, Sherman A, Kawakami K, Takahashi Y, Sang HM, McGrew MJ. Efficient genetic modification and germ-line transmission of primordial germ cells using piggyBac and Tol2 transposons. Proc Natl Acad Sci USA. 2012;109(23):E1466–E1472. doi: 10.1073/pnas.1118715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Park TS, Han JY. piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc Natl Acad Sci USA. 2012;109(24):9337–9341. doi: 10.1073/pnas.1203823109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Liu X, Li N, Hu X, Zhang R, Li Q, Cao D, Liu T, Zhang Y. Efficient production of transgenic chickens based on piggyBac. Transgenic Res. 2013;22(2):417–423. doi: 10.1007/s11248-012-9642-y. [DOI] [PubMed] [Google Scholar]
- 264.Tyack SG, Jenkins KA, O’Neil TE, Wise TG, Morris KR, Bruce MP, McLeod S, Wade AJ, McKay J, Moore RJ, Schat KA, Lowenthal JW, Doran TJ. A new method for producing transgenic birds via direct in vivo transfection of primordial germ cells. Transgenic Res. 2013;22(6):1257–1264. doi: 10.1007/s11248-013-9727-2. [DOI] [PubMed] [Google Scholar]
- 265.Flisikowska T, Thorey IS, Offner S, Ros F, Lifke V, Zeitler B, Rottmann O, Vincent A, Zhang L, Jenkins S, Niersbach H, Kind AJ, Gregory PD, Schnieke AE, Platzer J. Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS ONE. 2011;6(6):e21045. doi: 10.1371/journal.pone.0021045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Li P, Estrada JL, Burlak C, Tector AJ. Biallelic knockout of the alpha-1,3 galactosyltransferase gene in porcine liver-derived cells using zinc finger nucleases. J Surg Res. 2013;181(1):e39–e45. doi: 10.1016/j.jss.2012.06.035. [DOI] [PubMed] [Google Scholar]
- 267.Lutz AJ, Li P, Estrada JL, Sidner RA, Chihara RK, Downey SM, Burlak C, Wang ZY, Reyes LM, Ivary B, Yin F, Blankenship RL, Paris LL, Tector AJ. Double knockout pigs deficient in N-glycolylneuraminic acid and galactose alpha-1,3-galactose reduce the humoral barrier to xenotransplantation. Xenotransplantation. 2013;20(1):27–35. doi: 10.1111/xen.12019. [DOI] [PubMed] [Google Scholar]
- 268.Kwon DN, Lee K, Kang MJ, Choi YJ, Park C, Whyte JJ, Brown AN, Kim JH, Samuel M, Mao J, Park KW, Murphy CN, Prather RS. Production of biallelic CMP-Neu5Ac hydroxylase knock-out pigs. Sci Rep. 2013;3:1981. doi: 10.1038/srep01981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lillico SG, Proudfoot C, Carlson DF, Stverakova D, Neil C, Blain C, King TJ, Ritchie WA, Tan W, Mileham AJ, McLaren DG, Fahrenkrug SC, Whitelaw CB. Live pigs produced from genome edited zygotes. Sci Rep. 2013;3:2847. doi: 10.1038/srep02847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Watanabe M, Nakano K, Matsunari H, Matsuda T, Maehara M, Kanai T, Kobayashi M, Matsumura Y, Sakai R, Kuramoto M, Hayashida G, Asano Y, Takayanagi S, Arai Y, Umeyama K, Nagaya M, Hanazono Y, Nagashima H. Generation of interleukin-2 receptor gamma gene knockout pigs from somatic cells genetically modified by zinc finger nuclease-encoding mRNA. PLoS ONE. 2013;8(10):e76478. doi: 10.1371/journal.pone.0076478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ji D, Zhao G, Songstad A, Cui X, Weinstein EJ. Efficient creation of an APOE knockout rabbit. Transgenic Res. 2014 doi: 10.1007/s11248-014-9834-8. [DOI] [PubMed] [Google Scholar]
- 272.Xin J, Yang H, Fan N, Zhao B, Ouyang Z, Liu Z, Zhao Y, Li X, Song J, Yang Y, Zou Q, Yan Q, Zeng Y, Lai L. Highly efficient generation of GGTA1 biallelic knockout inbred mini-pigs with TALENs. PLoS ONE. 2013;8(12):e84250. doi: 10.1371/journal.pone.0084250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Liu H, Chen Y, Niu Y, Zhang K, Kang Y, Ge W, Liu X, Zhao E, Wang C, Lin S, Jing B, Si C, Lin Q, Chen X, Lin H, Pu X, Wang Y, Qin B, Wang F, Wang H, Si W, Zhou J, Tan T, Li T, Ji S, Xue Z, Luo Y, Cheng L, Zhou Q, Li S, Sun YE, Ji W. TALEN-mediated gene mutagenesis in rhesus and cynomolgus monkeys. Cell Stem Cell. 2014;14(3):323–328. doi: 10.1016/j.stem.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Park TS, Lee HJ, Kim KH, Kim JS, Han JY. Targeted gene knockout in chickens mediated by TALENs. Proc Natl Acad Sci USA. 2014;111(35):12716–12721. doi: 10.1073/pnas.1410555111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 276.Honda A, Hirose M, Sankai T, Yasmin L, Yuzawa K, Honsho K, Izu H, Iguchi A, Ikawa M, Ogura A. Single-step generation of rabbits carrying a targeted allele of the tyrosinase gene using CRISPR/Cas9. Exp Anim. 2014 doi: 10.1538/expanim.14-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]