Abstract
The molecular definition of tumor antigens recognized by cytolytic T lymphocytes (CTL) started in the late 1980s, at a time when the MHC class I antigen processing field was in its infancy. Born together, these two fields of science evolved together and provided each other with critical insights. Over the years, stimulated by the potential interest of tumor antigens for cancer immunotherapy, scientists have identified and characterized numerous antigens recognized by CTL on human tumors. These studies have provided a wealth of information relevant to the mode of production of antigenic peptides presented by MHC class I molecules. A number of tumor antigenic peptides were found to result from unusual mechanisms occurring at the level of transcription, translation or processing. Although many of these mechanisms occur in the cell at very low level, they are relevant to the immune system as they determine the killing of tumor cells by CTL, which are sensitive to low levels of peptide/MHC complexes. Moreover, these unusual mechanisms were found to occur not only in tumor cells but also in normal cells. Thereby, the study of tumor antigens has illuminated many aspects of MHC class I processing. We review here those insights into the MHC I antigen processing pathway that result from the characterization of human tumor antigens recognized by CTL.
Keywords: CTL, Tumor antigens, Proteasome, Aminopeptidase, Processing
Introduction
The immune system ensures constant surveillance against viral infections and cancers. In this process, cytolytic T lymphocytes (CTL) play a key role by detecting and clearing cells bearing abnormal, mutant or infectious proteins. The molecular nature of the viral antigens detected by CTL was uncovered in 1985 by Townsend et al., who demonstrated that influenza-specific CTL recognized short peptides corresponding to fragments of the viral nucleoprotein that are presented at the surface of infected cells by MHC class I molecules [1]. Four years later, Lurquin et al. [2] extended those findings to non-viral proteins, by showing that mouse tumor-specific CTL recognized a peptide derived from a self-protein that was mutated in cancer cells. This finding, which represented the first contribution of tumor immunology to the antigen processing field, led to the notion that MHC class I molecules continuously display antigenic peptides of 8–10 amino acids derived from intracellular self-proteins [3]. This allows the immune system to continuously check the integrity of the genome and detect any cell bearing abnormal genes or proteins.
The classical MHC class I processing pathway
Antigenic peptides presented to CTL are mostly produced in the cytosol from the degradation of cellular proteins by the multicatalytic complex called proteasome (Fig. 1). Peptides produced from proteasomal degradation are then transported by the transporter associated with antigen processing (TAP) into the lumen of the endoplasmic reticulum (ER), where they are further trimmed by aminopeptidases to a size that is suitable for loading onto MHC class I molecules. Stable peptide binding to MHC class I-β2 m dimers is facilitated by the peptide loading complex (PLC), which is composed of TAP, tapasin, ERp57 and calreticulin (CRT). Once a peptide binds to the MHC class I molecule, the fully assembled MHC class I/peptide complex is transported to the cell surface where it is available for CTL recognition. In order to achieve optimal immune surveillance, cells display a large array of peptides, sampling a variety of proteins expressed. In healthy cells, those peptides originate from normal autologous proteins and are ignored by the immune system as a result of self-tolerance. However, after viral infection or cell transformation, this repertoire also includes peptides derived from viral or tumor-associated proteins, which can be the target of an immune response. Because of their potential interest for cancer immunotherapy, the antigens recognized by anti-tumor CTL have been extensively studied, and to date, a large array of tumor antigenic peptides have been identified. A number of cancer patients have been included in clinical trials involving some of these antigens, and tumor regressions have been observed in some cases [4–7]. The identification of potential immunotherapeutic targets on human cancers has brought a lot of information on the various steps that are involved in the processing and the presentation of MHC-class I restricted antigenic peptides. This review will focus on those insights into the MHC I antigen processing that result from the characterization of human tumor antigens recognized by CTL.
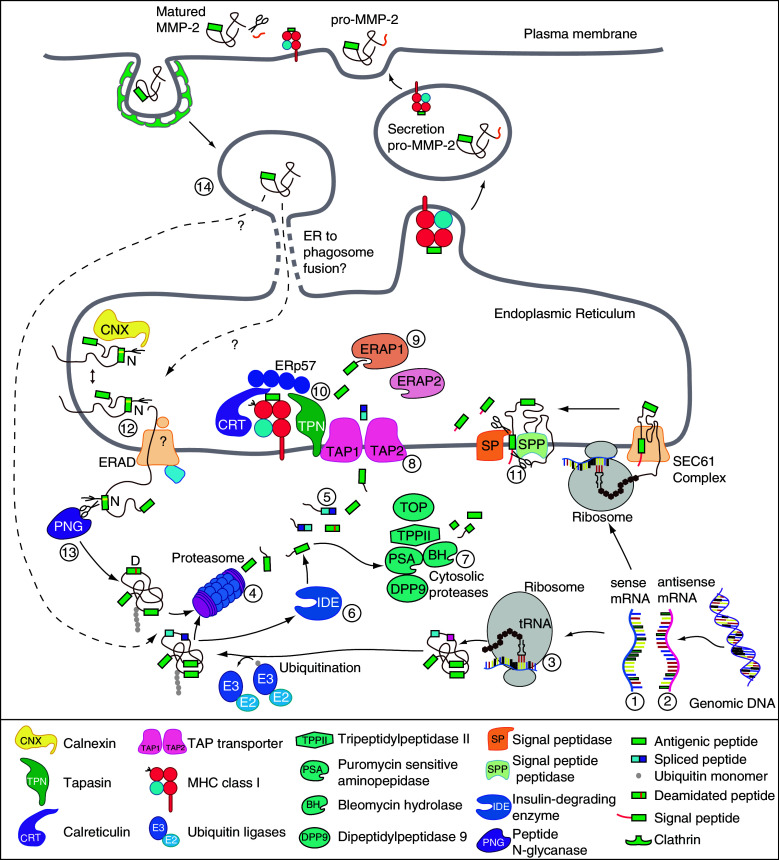
Fig. 1.
Pathways for processing and presentation of class I-restricted tumor antigens. CTL recognize on tumor cells antigenic peptides that are produced through a variety of unconventional processes, including aberrant transcription or mRNA splicing (1, 2), translation of alternative or cryptic open reading frames (3) or post-translational modifications (12). Proteins are targeted for degradation by the proteasome after the addition of ubiquitin chains by ubiquitin ligases (4). Surprisingly, proteasomes also produce antigenic peptides by splicing fragments that are originally non-contiguous in the parental protein (5). In the cytosol, additional proteases were found to produce or destroy antigenic peptides (6, 7). Peptides produced in the cytosol are then transported in the ER lumen by the transporter TAP (8), where they can be further trimmed by ER aminopeptidases such as ERAP1 or 2 (9). Peptides with the appropriate length and HLA binding motif are loaded onto MHC I molecules with the help of the peptide loading complex (10). The signal sequences of ER-targeted proteins also contain antigenic peptides: following cleavage of the signal sequence by the signal peptidase and the signal peptide peptidase (11), those peptides are released in the ER lumen and loaded onto MHC I independently of TAP. Membrane or secreted proteins also produce MHC class I binding peptides. Tyrosinase is a type I transmembrane protein, which supplies an antigenic peptide containing an aspartate instead of the genetically encoded asparagine. This peptide is produced after glycosylation of the tyrosinase protein in the ER, its retrotranslocation into the cytosol where it is deglycosylated and degraded by proteasome. Removal of the glycan moiety by peptide N-glycanase leads to deamidation of the asparagine into an aspartate residue, which is contained in the antigenic peptide presented by melanoma cells (12, 13). A peptide originating from secreted matrix metalloprotease-2 was found to be processed by the proteasome after endocytosis. After secretion, activation of pro-MMP-2 into mature MMP2 appears to be required for adequate processing of the antigenic peptide. Following endocytosis, the MMP-2 protein would be released in the cytosol and degraded by the proteasome (14)
Methods for the identification of tumor antigens
CTL clones displaying anti-tumor activity have often been isolated by cultivating blood or tumor-infiltrating lymphocytes of cancer-bearing patients with irradiated tumor cells [7–10]. Very often, the antigen recognized by such CTL was identified through an expression cloning approach, which aimed at isolating the encoding gene by transfecting a cDNA library and screening the transfected cells for their ability to activate the CTL clone [11, 12]. Subgenic fragments were then transfected similarly to define the region encoding the antigenic peptide, and candidate peptides bearing adequate HLA binding motifs were then tested for their ability to sensitize target cells to lysis by the CTL. This approach initially led to the identification of numerous tumor antigens [7, 11–17]. Later on, the use of mini-libraries containing sets of the previously identified antigenic cDNA facilitated the identification of additional peptides recognized by CTL derived from cancer patients [18, 19].
Additionally, the development of efficient techniques of mass spectrometry made it possible to identify peptides eluted from immunopurified MHC class I molecules [20–22]. Fractions of eluted peptides obtained by high performance liquid chromatography (HPLC) can be directly analyzed by tandem mass spectrometry or tested individually for recognition by anti-tumor CTL (see also the review by Mester et al. in this issue). Screening of protein databases generally helps defining the protein from which these peptides might originate. The complexity of the peptide repertoire eluted from MHC class I made it very demanding to identify new peptides by this approach, which, in turn, proved uniquely useful for the characterization of post-translational modifications such as serine/threonine phosphorylation [23] or glycosylation-dependent asparagine deamidation [24].
Finally, the “reverse immunology” approach relies on the prediction of peptides based on the screening of tumor protein sequences for peptides bearing HLA class I binding motifs [25–27]. A number of these cancer-associated sequences were identified by the screening of cDNA expression libraries using the serum of cancer-bearing patients [28–32], by cDNA representational difference analysis [33–35], by mRNA differential display [36] or by molecular probing of DNA libraries [14, 37]. Candidate peptides that bind the HLA of interest are then loaded on antigen presenting cells, which are used in vitro to stimulate lymphocytes isolated from cancer patients or healthy donors. The major drawback of this approach is that peptide-specific CTL may prove unable to recognize tumor cells that express the relevant gene and HLA allele. This is generally because the peptide is poorly processed or not at all. As an alternative, dendritic cells transduced with a viral vector encoding the antigen of interest were also used to stimulate blood lymphocytes [38–40].
Tumor antigens recognized by cytolytic T lymphocytes
So far, a large number of antigenic peptides presented on tumors have been identified using these various approaches, and classified according to the expression profile of the encoding gene [7, 41].
Antigens encoded by cancer-germline genes
A first category of antigenic peptides are those derived from cancer-germline genes (also called cancer-testis genes), which are expressed in cancer cells of different histological types and not in normal tissues except male germline cells [11, 37]. Since male germline cells do not display MHC class I molecules on their surface [42], the antigenic peptides derived from these cancer-germline genes are strictly tumor-specific and are therefore targets of choice for cancer immunotherapy. The prototypic cancer-germline gene family is the melanoma antigen (MAGE) gene family. It is composed of 24 functional genes grouped in three clusters found on different regions of the X chromosome: MAGE-A, -B, and -C [33, 43, 44]. Expression of the MAGE genes appears to take place following the demethylation of their promoter that occurs as a result of a genome-wide demethylation in male germ cells and in some advanced cancers [45–49]. Additional families of more distant MAGE genes expressed in normal tissues were found, but these genes do not encode any of the antigenic peptides so far identified [41, 50]. Other cancer-germline gene families include BAGE [15], GAGE [14, 19], LAGE/NY-ESO1 [30, 34] and SSX [29, 51, 52].
Antigens encoded by melanoma differentiation genes
A second group of antigens recognized by CTL isolated from melanoma patients correspond to peptides derived from differentiation proteins that are specific of the melanocyte lineage. The prototype for these melanoma differentiation proteins is tyrosinase, an enzyme involved in the biosynthesis of melanin [12, 53]. Even though these antigens are not tumor-specific, they are attractive targets for immunotherapy of melanoma, because they are expressed on most melanomas and are recognized by CTL in a large majority of patients. Differentiation antigens also include peptides derived from gp100/pmel17 [8, 9, 54], Melan-A/MART-1 [13, 55], gp75/TRP1 [56] and TRP2 [57]. Despite their expression in normal cells, melanocyte differentiation antigens are quite immunogenic and CTL can be obtained from the blood of melanoma patients as well as healthy donors [58]. Moreover, the occurrence of vitiligo, a partial skin depigmentation, has often been observed in melanoma patients, and was correlated with tumor regression and longer survival [59–61].
Antigens encoded by mutated genes
Several tumor antigens recognized by autologous CTL arise from point mutations in ubiquitously expressed genes. Some of these mutations affect genes of undefined function, while others appear to be related to tumor development. For example, a mutation in CDK4 was shown to affect binding of CDK4 to its inhibitor p16/INK4a, and could thereby favor uncontrolled cell division [62]. A mutation identified in the β-catenin gene [63] induces stabilization of the protein and formation of constitutive complexes between β-catenin and transcription factor Lef-Tcf, which may result in the persistent transactivation of target genes potentially involved in melanoma progression [64]. A mutation identified in gene CASP-8 appears to decrease cell sensitivity to apoptosis [65]. Because in many cases such mutations are not shared by distinct tumors, the corresponding antigenic peptides are unique to an individual tumor and therefore not useful for cancer immunotherapy. Surprisingly, only very few examples of peptides derived from genes often mutated in cancer have been shown to be antigenic. CD8 T cells against a mutated p53, K-ras or N-ras peptide could be isolated after in vitro stimulation of T cells originating from healthy donor or patients injected with peptide pulsed APC or autologous irradiated tumor cells [66–68]. Peptides derived from frameshift mutations [69, 70] or chromosomal translocations [71, 72] were also identified.
Antigens encoded by genes overexpressed in tumors
One of the first examples of antigenic peptides identified from a gene overexpressed in cancer is a peptide derived from the protooncogene HER-2/Neu [73], which is often overexpressed in breast and ovarian cancers as a result of gene amplification and/or upregulation of transcription [74, 75]. The characterization of an antigen recognized by CTL on renal carcinoma led to the identification of gene RAGE-1, which is expressed in several tumor samples of different histological types, but is silent in normal tissues except retina, where low expression is observed [76]. Because the eye is an immunologically privileged site [77, 78], and because retina appears not to express MHC class I molecules [79], immunization against RAGE-1 antigen can be considered, more specifically in the case of renal cell carcinoma, which usually do not express cancer-germline genes. A third example of gene overexpressed in cancer is PRAME, which encodes at least five peptides recognized by CTL in melanoma patients [17, 80]. PRAME is upregulated in a number of tumor types, but expressed at low levels in various normal tissues [17]. Interestingly, the CTL recognizing peptide PRAME301–309 was found to express a NK inhibitory receptor that blocks lysis upon interaction with HLA-Cw7 [17]. The presence of this inhibitory receptor restricts the action of the CTL to tumor cells that show partial HLA loss. Since then, a large number of other genes overexpressed in cancers were found to encode tumor-associated antigens [41]. Examples of such peptides are those derived from the inhibitor of apoptosis protein survivin [81, 82], the wild-type p53 protein [83, 84] or the Wilms tumor 1 transcription factor [85, 86]. Although such peptides are attractive because they are shared by numerous tumors, their use as vaccine targets is not devoid of the risk of developing autoimmune reactions due to the low but still detectable expression of these genes in healthy tissues.
Peptides derived from non-classical transcription and translation processes
Although antigenic peptides recognized by CTL generally derive from primary open reading frames, a number of reports have described cryptic antigenic peptides that were generated from non-classical events associated with either gene transcription or protein translation (see also the review by Starck and Shastri in this issue).
Transcriptional events leading to antigen presentation
The early observation that antigenic peptides corresponding to murine tumor antigens could be produced from transfected promoterless sequences [87–89] raised the possibility that transcription of short subgenic regions might lead to the expression of antigenic peptides. The ‘pepton hypothesis’ [90], which was put forward by Boon and Van Pel in 1989, suggested that antigenic peptides were encoded by short dedicated transcripts, produced by a different RNA polymerase not requiring conventional promoter sequences. Although this hypothesis was not substantiated at the transcriptional level, it was, somehow, at the translational level, and there are several examples of antigenic peptides translated from short open reading frames encoding just the antigenic peptide (see next section). Nevertheless, several aberrant transcriptional events were found to result in the expression of antigenic peptides on tumor cells. Studying human melanoma-specific CTL, Guilloux and co-workers identified a peptide generated from the aberrant transcription of an intron of the gene encoding N-acetylglucosaminyltransferase V (GnT-V) [91]. Interestingly, characterization of the antigen-coding messenger RNA suggested the presence of a cryptic promoter positioned in the intronic sequence. The RNA variant produced by activation of this putative promoter could be detected in about 50% of melanomas. Even more surprising was the peptide identified as the target of an HLA-B7-restricted CTL clone in a renal cell carcinoma patient [92]. Here, the antigen-coding sequence corresponded to the reverse strand of the housekeeping gene RU2 and was transcribed from a cryptic promoter located on the reverse strand of the first intron of this gene (Fig. 1). Although the sense transcript of RU2 was ubiquitously expressed, the reverse strand mRNA encoding the peptide was found in tumors of various histological types, but was absent in most normal tissues except testis, kidney, bladder and liver.
In addition to the presence of cryptic promoters, alternative splicing of messenger RNA also permits the translation of sequences that are usually non-coding and thereby provides new peptides to the MHC class I loading machinery. For example, intron retention is responsible for the generation of tumor antigenic peptides derived from genes MUM-1, SILV/gp100 or TRP2 [16, 93, 94].
Peptides derived from alternative open reading frames
The peptide derived from the translation of the third open reading frame (ORF) of melanoma differentiation gene TRP1/gp75 was one of the first examples of peptide encoded by an alternative ORF in human cancer [56]. The in vivo immunological relevance of this peptide was supported by the objective tumor regression observed in a melanoma patient infused with the corresponding autologous TIL and IL2 [95]. Peptides generated from the translation of alternative open reading frames were also described for cancer-germline genes NY-ESO1 [96] and LAGE-1 [97], from intestinal carboxyl esterase [98] and from macrophage colony-stimulating factor (M-CSF) [99]. The latter peptide, which is presented by HLA-B*3501, is 14 amino acids long and thus, to our knowledge, the longest class I-binding peptide known to date. The crystal structure of the peptide/HLA-B*3501 complex showed that the N- and C-terminus of the antigenic peptide were embedded inside the A and F pockets of the HLA molecule, while the central part of the peptide bulged flexibly out of the groove [100]. This showed that sufficient HLA binding is provided by anchoring the N-and C-terminal residues [100]. Using antibodies recognizing either the primary or the alternative ORF of M-CSF, it was shown that translation of both ORF was regulated independently [99].
More recently, peptides were identified from two distinct alternative open reading frames of MELOE, a gene overexpressed in melanoma [101, 102]. Surprisingly, in this case, the mRNA holds multiple short ORF of unknown function. Interestingly, both MELOE-derived antigenic peptides were broadly shared amongst melanomas, and in a clinical trial of adoptive cell immunotherapy, the infusion of patients with anti-MELOE-1 containing TIL was found to prevent relapses [101]. Another peptide produced from a very short ORF is derived from BING4, a gene overexpressed in some melanomas [103]. In this case, the antigen-coding ORF produces a peptide fragment that is only 10 amino acids long. Similarly, another peptide presented by human melanoma is encoded by gene HERV-K-MEL and appears to be translated from an ORF that comprises exclusively the peptide-coding sequence [104]. Interestingly, this gene belongs to the HERV-K family of human endogenous retroviruses and likely corresponds to a retroviral pseudogene sequence originating from an ancient retroviral infection. It is expressed in most melanoma samples tested, but not in normal tissues except testis and some skin samples. It is also transcribed in a majority of naevi and in some carcinomas and sarcomas.
Despite the growing number of reports describing CTL epitopes encoded from non-primary ORF, the mechanisms involved in the translation of these ORF have not yet been fully clarified. Bullock and Eisenlohr [105] have proposed the ‘initiation codon scanthrough model’ as a likely explanation for the use of some alternative open reading frames. Using frameshift mutants of the influenza nucleoprotein (NP) gene, they showed that production of an NP peptide can take place after the ribosome bypasses the conventional initiation codon, initiating translation at a downstream initiation site. Additional mechanisms that could account for the generation of non-primary ORF-derived peptides include reinitiation of translation at a downstream AUG following translation of an upstream ORF, ribosomal frameshifting in the course of elongation or internal initiation of translation using an internal ribosome entry site (IRES) (reviewed in [106]). Surprisingly, cryptic antigenic peptides were generated in tumors from alternative ORF despite the absence of conventional AUG codons [98, 107]. In both cases, mutating these non-AUG codons (CUG and ACG) completely abolished the presentation of the antigenic peptides to the respective T cells, demonstrating that such non-AUG codons can initiate the translation of the antigen-coding ORF. In vitro translation models have suggested for a long time that initiation at a non-AUG start codon involves the mismatched binding of anti-AUG tRNA loaded with methionine [108]. This mispairing results in the incorporation of a methionine residue at the non-AUG codon site. However, the initiation codon CUG was later shown to be decoded as leucine, suggesting that the translation initiation mechanism generating these peptides might use a leucine-loaded initiator tRNA rather than a methionine at the non-AUG initiation site [109, 110]. This is not peculiar to tumor cells, as Schwab et al. [110] demonstrated, using a transgenic mouse model, that translation of non-coding regions also occurs constitutively in normal cells. Initiation at a CUG codon differs from initiation at a conventional AUG codon. Indeed, the leucine start at a CUG initiation site depends on a specific set of ribosomes that scan sequences for CUG initiation sites, does not depend on initiation factor eIF2α [111], and diverges in its sensitivity to inhibitors acting on the P-site of the ribosomes [112]. Such cryptic initiation events could therefore allow the presentation of class I-binding peptides when conventional eIF2α-dependent AUG initiation is impaired, for example upon viral infection [112].
Another interesting example of non-conventional translational event is the melanoma peptide generated from the NA88-A pseudogene. This pseudogene originates from genomic insertion of a retrotranscript corresponding to the mRNA coding for the homeoprotein HPX42B [113]. The NA88-A pseudogene exhibits premature stop codons, deletions and insertions relative to the original HPX42B gene. The peptide recognized by the CTL is derived from a very short ORF that partially corresponds to the 3′ UTR of the HPX42B mRNA. Interestingly, the antigenic peptide can only be produced from the pseudogene sequence but not from HPX42B mRNA, because the latter lacks a stop codon located directly downstream of the C-terminus of the antigenic peptide.
Processing of tumor antigens by the proteasome
Proteolysis is essential to preserve cell homeostasis in a number of ways. It is involved in processes such as elimination of misfolded or damaged proteins [114], cell cycle progression [115], activation of transcription factors and signal transduction [116]. In eukaryotic cells, proteolysis is mainly dependent on the 26S proteasome, a large barrel-shape particle that breaks down poly-ubiquitinated protein substrates. The central core particle of the proteasome, namely the 20S proteasome, consists of four stacked heptameric rings that delimit a central catalytic chamber where the proteins are degraded (see also the review by Sijts and Kloetzel in this issue). The 20S proteasome associates with subunits of the 19S regulatory particle, which bind poly-ubiquitin chains, catalyze their removal from the protein substrate, unfold proteins, and help their translocation into the catalytic chamber in an ATP-dependent manner (reviewed in [117, 118]). The two outer rings of the 20S proteasome each contain seven structural α-subunits that control access to the catalytic chamber (α1–7). The two inner rings are composed of β-subunits (β1–7) and delineate the catalytic chamber [119–121]. In mammalian and yeast proteasome, the active sites of the protease complex are confined to the two inner rings and are dependent on three different β-subunits: β1, β2 and β5. Mutational studies and analysis of the crystal structure of proteasomes complexed with proteasome inhibitors identified the hydroxyl group of the N-terminal threonine of the active subunits as the nucleophile responsible for the hydrolysis of the peptide bond [119, 120, 122]. Three major catalytic activities were described based on the degradation of small fluorogenic substrates by the proteasome: PGPH (peptidyl-glutamyl peptide bond hydrolysing) or caspase-like activity, trypsin-like activity and chymotrypsin-like activity, which respectively cleave after acidic, basic and hydrophobic residues [123]. Based on the study of yeast proteasome mutants, each of these catalytic activities was associated with a given proteasome catalytic subunit [124–129]. However, the catalytic specificity of individual subunits is more complex than originally expected: some subunits have overlapping specificities [128] and proteasome digestion experiments using a protein substrate suggest that amino acids located up to five residues upstream or downstream of the cleavage site also influence cleavage strength [130, 131].
Protein degradation by the proteasome leads to the production of smaller size peptides, which constitute the major source of ligands for presentation onto MHC class I molecules (Fig. 1) [117]. One of the earliest hint that the proteasome was involved in the processing of peptides presented by MHC class I was the fact that ubiquitination, and therefore potentially the 26S proteasome, was necessary for presentation of the model antigen ovalbumin [132]. Direct evidence for the role of the proteasome in peptide processing was obtained using cell permeable proteasome inhibitors [133]. Such inhibitors, which reversibly or irreversibly bind the hydroxyl group of catalytic threonines, efficiently block presentation of a number of antigenic peptides [134–136] and limit the overall peptide supply to MHC class I molecules [133].
The standard proteasome and the immunoproteasome
In the presence of IFNγ, three additional proteasome catalytic subunits, β1i (LMP-2), β2i (MECL-1), and β5i (LMP-7), are induced in vertebrates. These subunits preferentially incorporate into the proteasome in place of their homologous counterparts, β1, β2, and β5, to create homogeneous particles of a different type, called the immunoproteasome. The immunoproteasome can be expressed in most cell types upon IFNγ induction, but is also constitutively present in some lymphoid tissues and dendritic cells [134, 137, 138]. Because of the difference in the nature of their active subunits, the standard proteasome and the immunoproteasome have different cleavage specificities. The analysis of digests obtained with peptide precursors, entire protein substrates or fluorogenic substrates showed that the immunoproteasome has a higher propensity to cleave after hydrophobic residues (chymotrypsin-like activity) and basic residues (trypsin-like activity), but a reduced propensity to cleave after acidic residues, as compared to the standard proteasome [131, 139, 140]. Peptides that bind efficiently to HLA class I molecules usually have a hydrophobic or a basic residue at their C-terminus, while they virtually never display an acidic C-terminus. Therefore, immunoproteasomes were predicted to be generally more efficient than standard proteasomes to produce good MHC-binding peptides [139, 140]. This idea was corroborated by the study of LMP2- and LMP7-knock-out mice, in which presentation of certain epitopes was decreased [141, 142]. It was challenged, however, by the work of Morel et al. [134] who studied a human CTL clone that recognized a renal cell carcinoma but did not recognize autologous EBV-transformed B cells. The gene encoding this antigen was identified, named RU1, and found to be ubiquitously expressed. Surprisingly, it was also expressed in the autologous EBV-B cells that were not recognized by the CTL. The authors explained this paradox by showing that the antigenic peptide recognized by their CTL was processed efficiently by the standard proteasome, but not by the immunoproteasome, which is present in EBV-transformed B cells. Given the abundant presence of immunoproteasome in the thymus, this may also explain the lack of negative selection of such CTL directed against a peptide derived from a ubiquitous protein [143]. The immunoproteasome is also abundant in dendritic cells, which therefore do not efficiently present such peptides, as observed by Morel et al. [134] for the RU1 peptide. This may explain the absence of autoimmunity related to the presence of CTL against such peptides in the periphery. It follows that the activation of the RU1-specific CTL in this cancer patient should result from direct priming by tumor cells.
A number of tumor antigenic peptides, often derived from melanoma differentiation antigens, have also been found to be poorly processed by the immunoproteasome. This is the case for the HLA-A2-restricted peptides Melan-A/MART-126–35 [134], gp100209–217 [144] and tyrosinase369–377 [144], which have been used in melanoma vaccination trials. In contrast, tumor peptides produced exclusively by the immunoproteasome have also been described; among these, the HLA-B40-restricted peptide derived from MAGE-A3 [145] and the HLA-A2-restricted peptide MAGE-C2336–344 [144]. In vitro digestions of precursor peptides with purified proteasomes have indicated that these differences in processing efficiency usually result from a prominent internal cleavage of the peptide by one of the proteasome types, resulting in the destruction of the antigenic peptide [144]. It is striking to note that, so far, all the peptides that were found to be poorly processed by the immunoproteasome are derived from self-ubiquitous (RU1) or differentiation (Melan-A, gp100, tyrosinase) proteins. As discussed above for RU1, this may contribute to the lack of negative selection of CTL against such peptides and the absence of autoimmunity. In contrast, all tumor antigens found to depend on the immunoproteasome for their processing correspond to tumor-specific antigens encoded by cancer-germline genes, whose very limited expression in tissues may suffice to explain the lack of negative selection and absence of autoimmunity.
Such differences in processing efficiency by the two proteasome types have major implications for the induction of immune responses against the corresponding tumor epitopes [143]. Immunoproteasomes are abundant in dendritic cells, which, therefore, cannot efficiently present those antigens that are destroyed by immunoproteasomes [134]. To immunize against such antigens, vaccine strategies based on short peptides or the corresponding minigenes, which are not dependent on the processing machinery, should be more efficient than full-length constructs. This prediction was confirmed in a study comparing minigene and full-length Melan-A constructs in wild-type and LMP2-knock-out mice [146]. Stronger CTL responses were induced against the HLA-A2-restricted Melan-A26–35 peptide in HLA-A2 transgenic mice using minigene retroviral constructs as compared to full-length constructs. This difference was abolished in LMP2-knock-out HLA-A2 transgenic mice, which developed stronger CTL responses to the full-length construct than wild-type mice, confirming that the immunoproteasome was responsible for the poor responses in the wild-type mice. The differential processing of some antigens also provides the tumor cells with the possibility of modulating their expression of some antigenic peptides by modifying their proteasome content, e.g. upon exposure to IFNγ.
Recent work from our laboratory has confirmed the existence of additional forms of proteasomes, which are intermediate between the standard and the immunoproteasome [147]. Two types of intermediate proteasomes were identified: intermediate proteasomes that contain β1, β2 and β5i and intermediate proteasomes with β1i, β2 and β5i. Interestingly, a high proportion of these intermediate proteasomes was observed in several normal tissues and in dendritic cells. Tumor cell lines mainly express standard proteasomes, but can contain 10–20% intermediate proteasomes. Some tumor antigens are processed exclusively by intermediate proteasomes containing β5i or intermediate proteasomes containing β1i and β5i.
Other peptidases involved in the processing of tumor antigens
The proteasome is the main protease involved in the production of MHC class I ligands. It is particularly required for the production of the C-terminus of most antigenic peptides, which can be produced with a N-terminal extension that is subsequently trimmed, either in the cytosol or in the endoplasmic reticulum (ER), which contains a dedicated aminopeptidase named ERAAP or ERAP1 (for a detailed review of trimming peptidases, see the article by van Endert in this issue). ERAAP is involved in the processing of a considerable fraction of MHC class I ligands (Fig. 1) [148–152]. ERAAP-deficient cells have reduced levels of surface MHC class I, due to a lower stability of peptide/MHC class I complexes, suggesting that ERAAP is involved in shaping the peptide repertoire associated with class I [152, 153]. In addition, ERAAP-deficient cells present unstable but unique MHC/peptide complexes that are absent from ERAAP-expressing cells and can elicit robust CD8 T cell responses [153]. In human cells, another ER aminopeptidase called ERAP2 can complement the action of ERAP1 in trimming N-terminally extended antigenic peptides [151].
N-terminal trimming can also occur in the cytosol, through the action of tripeptidyl peptidase II (TPPII), a large cytosolic serine protease that sequentially trims tripeptidic fragments from the N-terminus of polypeptides (Fig. 1). In addition to this aminopeptidase activity, authors have suggested that TPPII also displays some endopeptidase activity [154]. This led to the suggestion that TPPII might play a more general role in the production of the MHC class I presented peptides, particularly when proteasome activity was impaired [154, 155]. This concept, however, was not supported by more recent studies using more potent proteasome inhibitors or TPPII-knock-out mice [156–162]. Nevertheless, TPPII appears to be involved in the processing of at least four viral antigens [163–165]. But TPPII may also limit the presentation of some peptides, by degrading them in the cytosol: studying the processing of a peptide derived from the anti-apoptotic protein survivin, Preta et al. observed that serine peptidase inhibitor AAF-CMK increased the sensitivity of a colon carcinoma cell line to lysis by the corresponding CTL. Moreover, overexpression of TPPII in these colon-carcinoma cells seemed to correlate with resistance to the CTL, suggesting that TPPII might limit the presentation of this antigen and contribute to tumor immune escape [166].
Other cytosolic proteases such as puromycin aminopeptidase, bleomycin hydrolase, or leucine aminopeptidase are potentially involved in the N-terminal trimming of MHC class I ligands (Fig. 1) [167, 168]. Indeed, although studies using puromycin aminopeptidase, bleomycin hydrolase, or leucine aminopeptidase knock-out mice suggested that these proteases were not involved in the generation of the overall pool of MHC class I ligands [169–171], a recent study performed in human cells using RNAi silencing showed that some of these cytosolic aminopepidases influence surface expression of MHC class I in an allele-dependent manner [172]. Moreover, some of these proteases are apparently involved in the processing of specific antigenic peptides [168, 173], but also limit presentation of other peptides by contributing to their degradation, as previously suggested for the thimet oligopeptidase [170, 172, 174, 175].
In human tumors, the involvement of cytosolic aminopeptidases in the processing of class I tumor epitopes was brought forward by the analysis of the processing of antigenic peptide RU134–42, which was mentioned in the previous section [173]. In vitro studies using precursor peptides encompassing the antigenic peptide have shown that the exact C-terminus but not the final N-terminus can be produced by standard proteasomes, suggesting that, after proteasome digestion, the peptide is released as an N-terminally extended precursor. Fractionation of the cytosolic extracts identified PSA and TPPII as two enzymes able to produce the final N-terminus of the RU1 peptide. Their involvement was confirmed by treating cells with AAF-CMK, an inhibitor of both these proteases. This study suggests that the N-terminally extended RU1 peptide is trimmed in the cytosol by TPPII or PSA and then transported into the ER by TAP, despite the presence of a subaminoterminal proline, which is not optimal for TAP transport [176].
Geiss-Friedlander et al. [177] recently described the role of cytosolic prolyl-peptidase DPP9 in destroying the RU134–42 peptide (Fig. 1). DPP9 is one of the few peptidases able to cleave after proline residues, which are poor substrates for most peptidases because of their pyrrolidine ring. DPP9 was found to destroy the RU134–42 peptide by cleaving after the subaminoterminal proline. Downregulation of DPP9 increased recognition of target cells by RU1-specific CTL. DPP9 therefore appears to limit the presentation of proline-containing peptides. Potentially, however, it could help the processing of peptides with a proline at the C-terminus.
Parmentier et al. recently studied the processing of a proteasome-independent peptide derived from human tumor protein MAGE-A3. This peptide has been widely used in peptide vaccination trials in melanoma patients [178]. They found that this peptide was processed by a cytosolic Zn-metalloendopeptidase named insulysin or insulin-degrading enzyme (IDE) (Fig. 1) [136]. Known substrates of IDE include insulin, glucagon, atrial natriuretic peptide, transforming growth factor-alpha, amyloid-β peptide and oxydized proteins [179–182]. Remarkably, in vitro digestions of precursor peptides indicated that IDE produced directly both the C-terminus and the N-terminus of the MAGE-A3 antigenic peptide, which therefore did not require N-terminal trimming. Metalloprotease inhibitors or IDE silencing reduced recognition of melanoma cells by the specific CTL. Other antigenic peptides derived from MAGE-A3 are produced by the proteasome [145]. The analysis of MAGE-A3 protein degradation in melanoma cells treated with proteasome or metallopeptidase inhibitors suggests that a fraction of the MAGE-A3 proteins is degraded by the proteasome and another fraction is degraded by IDE. This supports the hypothesis that IDE might be part of a second cytosolic protein breakdown pathway with preference for substrates that are not efficiently targeted to or degraded by the proteasome [183]. In the course of MAGE-A3 protein breakdown, IDE- and the proteasome would each produce a distinct set of antigenic peptides presented to CTL by HLA class I molecules.
Post-translational modifications
Peptide splicing in the proteasome
Studying a CTL clone recognizing an autologous human renal cell carcinoma, Hanada et al. [184] found that the antigen recognized by this CTL was derived from fibroblast growth factor 5 (FGF-5), which is overexpressed in some cancers. Surprisingly, the shortest FGF-5 cDNA construct that conferred COS-7 cells the ability to express the antigenic peptide encoded a 49 amino acid-long peptide. Moreover, this ability was maintained after deletion of an internal segment encoding 24 amino acids in the middle of the 49-mer fragment. This led the authors to test peptides made of two short fragments corresponding to both ends of the 49-mer segment, spliced together to form a new peptide. Indeed, the peptide recognized by the CTL, was made of two fragments of 4 and 5 amino acids initially non-contiguous in the parental protein, but separated by a 40 amino acid intervening segment. Production of the antigenic peptide required excision of this intervening segment and splicing of the two fragments.
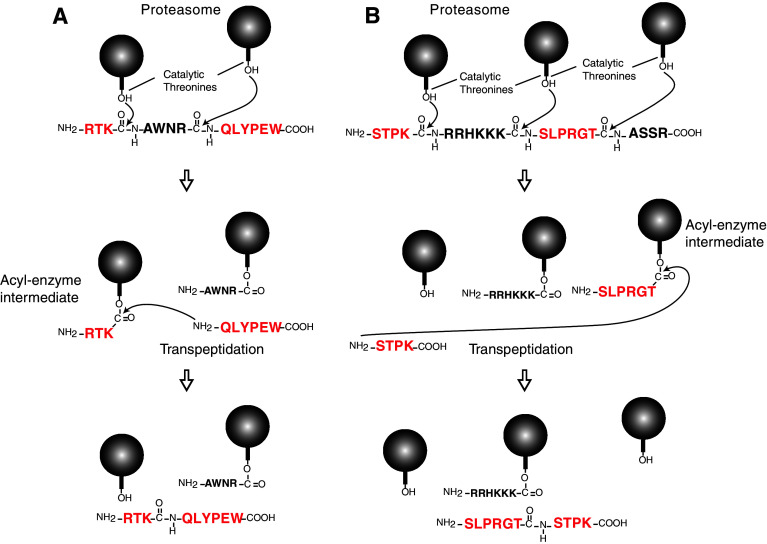
In parallel, studying the antigen recognized by a human anti-melanoma CTL clone, we described a second peptide produced by peptide splicing. In this case, the peptide was derived from the melanocyte differentiation protein gp100 and was produced after the splicing of two non-contiguous fragments of the gp100 protein and excision of a four-amino acid intervening segment (Fig. 1) [185]. The antigenic peptide, RTKQLYPEW, could be produced in EBV-B cells after electroporation of the unspliced 13-amino acid precursor RTKAWNRQLYPEW. The peptide splicing reaction could be reproduced in vitro, by incubating 20S proteasomes with the unspliced precursor peptide RTKAWNRQLYPEW, as measured by CTL activation and mass spectrometry. In order to dissect the mechanism of the peptide splicing reaction, we incubated pairs of peptides containing portions of the precursor peptide RTKAWNRQLYPEW with purified proteasomes. Although the antigenic peptide could be produced after incubation of proteasomes with peptides RTKAWNR and AWNRQLYPEW, no peptide was detected when using peptides RTK and QLYPEW. This demonstrates that the production of the spliced peptide was dependent on the cleavage of a peptide bond. Because the spliced peptide could be produced when incubating proteasomes with peptides RTKAWNR and QLYPEW, but not with peptides RTK and AWNRQLYPEW, we concluded that formation of the new peptide bond of the spliced peptide used the energy liberated by cleavage of the bond between K42 and A43. Thus, the splicing reaction appears to occur by transpeptidation involving the formation of an acyl-enzyme intermediate between RTK and the proteasome (Fig. 2a). Normally, this acyl-enzyme intermediate is rapidly hydrolyzed. However, inside the confined catalytic chamber of the proteasome, the intermediate is surrounded by peptide fragments, such as QLYPEW, which results from cleavage after residue 46. We proposed that the N-terminus of fragment QLYPEW could compete with water molecules and occasionally produce a nucleophilic attack on the ester bond of the intermediate, thereby creating a new peptide bond forming the spliced antigenic peptide RTKQLYPEW (Fig. 2a). This model was supported by the fact that N-α-acetylation of QLYPEW incubated with RTKAWNR and proteasomes completely prevented production of the spliced peptide.
Fig. 2.
Model of the peptide splicing reaction inside the proteasome. Mechanism of production of the spliced the peptide RTKQLYPEW derived from gp100 (a) and the spliced peptide SLPRGTSTPK derived from SP110 (b). The balls represent the catalytic β-subunits of proteasomes with the hydroxyl group of the side chain of the N-terminal threonine
A few years later, Warren et al. [186] studied the minor histocompatibility antigen recognized by a CTL clone isolated from a myeloma patient after MHC-matched allogeneic hematopoietic cell transplantation. They found that this CTL recognized an antigenic peptide encompassing a polymorphism in the SP110 protein. Again, the antigenic peptide contained two initially non-contiguous peptide fragments that were spliced together. However, in this case, the fragments were spliced together in the reverse order to that in which they occur in the predicted SP110 protein (Fig. 1). The antigenic peptide SLPRGTSTPK could be produced in EBV-B cells after electroporation of the 20 amino acid precursor STPKRRHKKKSLPRGTASSR. It was also produced in vitro after incubation of this precursor with purified 20S proteasomes. Again, the splicing reaction appears to occur by transpeptidation and involves the formation of an acyl-enzyme intermediate between peptide SLPRGT and the proteasome (Fig. 2b).
Recently, Dalet et al. [187] showed that the spliced FGF-5 peptide initially described by Hanada et al. was also produced by the proteasome through a transpeptidation reaction, despite the length of the intervening segment (40 amino acids). Shortening the length of the intervening peptide fragment improved the efficiency of the peptide splicing reaction [187]. By co-transfecting pairs of FGF-5 constructs mutated in either fragment of the final peptide, they showed that splicing of fragments originating from two distinct proteins can also occur, but at very low level, which is probably below physiological significance [187].
Both standard proteasomes and immunoproteasomes are able to splice peptides [186]. Some spliced peptides are better produced by standard proteasomes, while others are better produced by immunoproteasomes [188]. This is related to differences in cleavage specificity resulting in differential production of the fragments to splice. Because splicing likely occurs also in the thymus, spliced peptides derived from self proteins do belong to the normal peptide repertoire and should not be more immunogenic than regular peptides.
The peptide splicing reaction is a low efficiency process: about 104 molecules of precursor peptide were required to produce in vitro one molecule of spliced gp100 peptide [185]. The extreme sensitivity of CTL, which can recognize target cells presenting fewer than ten MHC/peptide complexes [189, 190], explains how this low efficiency process is immunologically relevant and can elicit a CD8 T cell response.
Deamidation and processing of membrane and secreted proteins
One thing that was clear from the beginning of the study of class I antigen processing was that it took place in the cytosol [191]. How peptides were produced from membrane or secreted proteins was completely unclear and was clarified by the elegant story of a peptide derived from tyrosinase, the first differentiation antigen identified on human melanoma (Fig. 1) [12, 53]. The sequence of the HLA-A2 binding peptide, as predicted from that of the tyrosinase gene, is YMNGTMSQV. However, when the group of Hunt and Engelhard eluted the peptides associated with HLA-A2 on melanoma, they identified a peptide with sequence YMDGTMSQV, which is identical to the predicted tyrosinase peptide except for the N in position 191, which is replaced by a D [24]. Transfection experiments further showed that the YMDGTMSQV peptide was indeed encoded by tyrosinase. Moreover, the YMNGTMSQV was not detected at the cell surface. The only possible explanation was that the peptide underwent a post-translational modification resulting in the conversion of N to D. The authors noticed that this asparagine residue corresponded to a N-glycosylation site. They proposed that the deamidation resulting in N to D conversion resulted from the removal of the glycan chain, which would occur in the cytosol after retrotranslocation of glycosylated unfolded tyrosinase from the ER to the cytosol. The peptide would then be produced in the cytosol by the proteasome and transported back into the ER via TAP to bind class I [192]. Subsequent studies fully confirmed this model, and a cytosolic peptide N-glycanase able to remove glycan chains while deamidating asparagines was found to be required for the production of the antigenic peptide [192–195]. Thus, the study of the deamidation of the tyrosinase peptide, which depends on glycosylation in the ER and subsequent deglycosylation in the cytosol, allowed to uncover retrotranslocation as a pathway for producing peptides from proteins that do not reside in the cytosol. Even though the detailed mechanism of retrotransport is not yet clear, retrotranslocation, as part of the ER-associated degradation (ERAD) pathway, is today considered as the main route whereby membrane and secreted proteins make their way to the cytosol to be processed into class I-binding peptides.
Another pathway for processing peptides from secreted proteins was described recently by Godefroy et al. [196], who studied a peptide derived from matrix metallopeptidase-2 (MMP-2) and recognized by CTL on melanoma cells. MMP-2 is secreted by tumor cells as an inactive proenzyme (pro-MMP-2), which is activated by removal of its prodomain through the coordinated action of MMP-14 (MT1-MMP) and the tissue inhibitor of metalloproteases 2 (TIMP-2) [197]. This activation usually occurs after secretion, and allows MMP-2 to degrade components of the extracellular matrix and basal membranes (Fig. 1) [198]. Surprisingly, the mere expression of pro-MMP-2 is not sufficient to produce the antigenic peptide. Rather, antigen presentation appears to require both the secretion of pro-MMP-2, which allows the removal of the pro-domain, and the endocytosis of secreted MMP2 through the αvβ3 integrin. Production of the MMP-2 epitope depends on both clathrin-coated pits and the proteasome. This implies that, after endocytosis of the secreted MMP-2, the protein is transferred into the cytosol where the peptide is processed by proteasome. This pathway could be similar to the cross-presentation pathway described in dendritic cells. The reason why non-secreted pro-MMP-2 cannot give rise to the antigenic peptide through the classical endogenous pathway was recently clarified [199]. Indeed, elimination of disulphide bonds located in the prodomain or in the fibronectin domain of MMP-2 was sufficient to restore production of the antigenic peptide through the endogenous pathway, suggesting that the presence of these disulphide bonds in the pro-MMP-2 protein might prevent its retrotranslocation and/or degradation by the proteasome.
TAP-independent presentation
The presentation of most MHC class I-binding peptides depends on the presence of TAP, which allows their transport from the cytosol to the ER where they can associate with MHC class I molecules [200–203]. Tumors could therefore reduce their TAP expression to escape immune recognition. Some peptides, however, are still presented by tumors in the absence of TAP. These include peptides derived from the signal peptide of secreted or membrane proteins, which are delivered to the ER during translation [204–206]. A classical example is the peptide derived from the signal sequence of tyrosinase, which is presented by HLA-A2 on melanomas [53, 207]. Another example is the antigenic peptide derived from the signal sequence of preprocalcitonin [208], a gene overexpressed in cancers. Presentation of these peptides is TAP-independent and proteasome-independent [207, 208]. The processing of the peptide derived from the signal sequence of preprocalcitonin was studied in details and found to involve both signal peptidase (SP) and signal peptide peptidase (SPP), an aspartic protease located in the ER membrane (Fig. 1) [208]. Cleavage of the signal peptide by SP and SPP releases the antigenic peptide into the ER lumen, where it can be directly loaded on HLA-A*0201. This is similar to the processing of peptides loaded on HLA-E molecules, which are derived from MHC class I signal sequences and are also released by SP and SPP. In the case of HLA-E, however, TAP transport of the peptides is required to enable surface expression of the HLA-E molecules, suggesting that, after SPP cleavage, the peptide is released in the cytosol and transported by TAP into the ER lumen [209–211].
Tapasin
As part of the peptide loading complex, tapasin plays multiple roles, which are essential for optimal loading of MHC class I molecules [212]. Tapasin stabilizes TAP and recruits TAP and ERp57 to the loading complex, thereby favoring peptide transport into the ER and exposure of MHC to the translocated peptides [213, 214]. Additionally, tapasin stabilizes empty MHC molecules and promotes their loading with high affinity peptides, a process known as “peptide editing” [215–217]. In the absence of tapasin, HLA alleles such as HLA-B*4402 fail to load stabilizing peptides and reach the cell surface [218]. Other alleles, such as HLA-B27, reach the cell surface but contain a suboptimal peptide repertoire [219]. In a recent study, one of us observed that presentation of two HLA-B*4402-restricted peptides derived from tumor proteins MAGE-A1 and MUM-1 was abolished in the presence of tapasin variants lacking the ability to recruit TAP and ERp57 to the peptide loading complex [220]. These results agreed with previous in vitro studies using recombinant proteins [217, 221], and further confirmed that, in the ER environment, the interaction of tapasin with both TAP and ERp57 is required for optimal loading of MHC class I molecules. In addition, exogenous loading of one of these tapasin-dependent tumor peptides on wild-type cells was completely inefficient, presumably because exogenous loading takes place at the cell surface in the absence of tapasin (unpublished results).
Acknowledgments
We thank S. Depelchin and J. Klein for editorial assistance. N.V. is a post-doctoral researcher with the Fonds National de la Recherche Scientifique.
References
- 1.Townsend AR, Gotch FM, Davey J. Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell. 1985;42(2):457–467. doi: 10.1016/0092-8674(85)90103-5. [DOI] [PubMed] [Google Scholar]
- 2.Lurquin C, Van Pel A, Mariame B, et al. Structure of the gene of tum—transplantation antigen P91A: the mutated exon encodes a peptide recognized with Ld by cytolytic T cells. Cell. 1989;58(2):293–303. doi: 10.1016/0092-8674(89)90844-1. [DOI] [PubMed] [Google Scholar]
- 3.Falk K, Rotzschke O, Stevanovic S, et al. Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature. 1991;351(6324):290–296. doi: 10.1038/351290a0. [DOI] [PubMed] [Google Scholar]
- 4.Marchand M, van Baren N, Weynants P, et al. Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE-3 and presented by HLA-A1. Intl J Cancer. 1999;80(2):219–230. doi: 10.1002/(sici)1097-0215(19990118)80:2<219::aid-ijc10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 5.Rosenberg SA, Yang JC, Schwartzentruber DJ, et al. Recombinant fowlpox viruses encoding the anchor-modified gp100 melanoma antigen can generate antitumor immune responses in patients with metastatic melanoma. Clin Cancer Res. 2003;9(8):2973–2980. [PMC free article] [PubMed] [Google Scholar]
- 6.van Baren N, Bonnet MC, Dreno B, et al. Tumoral and immunologic response after vaccination of melanoma patients with an ALVAC virus encoding MAGE antigens recognized by T cells. J Clin Oncol. 2005;23(35):9008–9021. doi: 10.1200/JCO.2005.08.375. [DOI] [PubMed] [Google Scholar]
- 7.Boon T, Coulie PG, Van den Eynde BJ, van der Bruggen P. Human T cell responses against melanoma. Annu Rev Immunol. 2006;24:175–208. doi: 10.1146/annurev.immunol.24.021605.090733. [DOI] [PubMed] [Google Scholar]
- 8.Kawakami Y, Eliyahu S, Jennings C, et al. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J Immunol. 1995;154(8):3961–3968. [PubMed] [Google Scholar]
- 9.Zarour H, De Smet C, Lehmann F, et al. The majority of autologous cytolytic T-lymphocyte clones derived from peripheral blood lymphocytes of a melanoma patient recognize an antigenic peptide derived from gene Pmel17/gp100. J Invest Dermatol. 1996;107(1):63–67. doi: 10.1111/1523-1747.ep12298177. [DOI] [PubMed] [Google Scholar]
- 10.Germeau C, Ma W, Schiavetti F, et al. High frequency of antitumor T cells in the blood of melanoma patients before and after vaccination with tumor antigens. J Exp Med. 2005;201(2):241–248. doi: 10.1084/jem.20041379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Bruggen P, Traversari C, Chomez P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254(5038):1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 12.Brichard V, Van Pel A, Wolfel T, et al. The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1993;178(2):489–495. doi: 10.1084/jem.178.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulie PG, Brichard V, Van Pel A, et al. A new gene coding for a differentiation antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med. 1994;180(1):35–42. doi: 10.1084/jem.180.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van den Eynde B, Peeters O, De Backer O, et al. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med. 1995;182(3):689–698. doi: 10.1084/jem.182.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boel P, Wildmann C, Sensi ML, et al. BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity. 1995;2(2):167–175. doi: 10.1016/s1074-7613(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 16.Coulie PG, Lehmann F, Lethe B, et al. A mutated intron sequence codes for an antigenic peptide recognized by cytolytic T lymphocytes on a human melanoma. Proc Natl Acad Sci USA. 1995;92(17):7976–7980. doi: 10.1073/pnas.92.17.7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda H, Lethe B, Lehmann F, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997;6(2):199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 18.Gaugler B, Van den Eynde B, van der Bruggen P, et al. Human gene MAGE-3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med. 1994;179(3):921–930. doi: 10.1084/jem.179.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Backer O, Arden KC, Boretti M, et al. Characterization of the GAGE genes that are expressed in various human cancers and in normal testis. Cancer Res. 1999;59(13):3157–3165. [PubMed] [Google Scholar]
- 20.Henderson RA, Michel H, Sakaguchi K, et al. HLA-A2.1-associated peptides from a mutant cell line: a second pathway of antigen presentation. Science. 1992;255(5049):1264–1266. doi: 10.1126/science.1546329. [DOI] [PubMed] [Google Scholar]
- 21.Cox AL, Skipper J, Chen Y, et al. Identification of a peptide recognized by five melanoma-specific human cytotoxic T cell lines. Science. 1994;264(5159):716–719. doi: 10.1126/science.7513441. [DOI] [PubMed] [Google Scholar]
- 22.Schirle M, Keilholz W, Weber B, et al. Identification of tumor-associated MHC class I ligands by a novel T cell-independent approach. Eur J Immunol. 2000;30(8):2216–2225. doi: 10.1002/1521-4141(2000)30:8<2216::AID-IMMU2216>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 23.Zarling AL, Polefrone JM, Evans AM, et al. Identification of class I MHC-associated phosphopeptides as targets for cancer immunotherapy. Proc Natl Acad Sci USA. 2006;103(40):14889–14894. doi: 10.1073/pnas.0604045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skipper JC, Hendrickson RC, Gulden PH, et al. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J Exp Med. 1996;183(2):527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celis E, Tsai V, Crimi C, et al. Induction of anti-tumor cytotoxic T lymphocytes in normal humans using primary cultures and synthetic peptide epitopes. Proc Natl Acad Sci USA. 1994;91(6):2105–2109. doi: 10.1073/pnas.91.6.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Bruggen P, Bastin J, Gajewski T, et al. A peptide encoded by human gene MAGE-3 and presented by HLA-A2 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE-3. Eur J Immunol. 1994;24(12):3038–3043. doi: 10.1002/eji.1830241218. [DOI] [PubMed] [Google Scholar]
- 27.Herman J, van der Bruggen P, Luescher IF, et al. A peptide encoded by the human MAGE3 gene and presented by HLA-B44 induces cytolytic T lymphocytes that recognize tumor cells expressing MAGE3. Immunogenetics. 1996;43(6):377–383. doi: 10.1007/BF02199806. [DOI] [PubMed] [Google Scholar]
- 28.Sahin U, Tureci O, Schmitt H, et al. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci USA. 1995;92(25):11810–11813. doi: 10.1073/pnas.92.25.11810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tureci O, Sahin U, Schobert I, et al. The SSX-2 gene, which is involved in the t(X;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM-MEL-40. Cancer Res. 1996;56(20):4766–4772. [PubMed] [Google Scholar]
- 30.Chen YT, Scanlan MJ, Sahin U, et al. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci USA. 1997;94(5):1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tureci O, Sahin U, Zwick C, et al. Identification of a meiosis-specific protein as a member of the class of cancer/testis antigens. Proc Natl Acad Sci USA. 1998;95(9):5211–5216. doi: 10.1073/pnas.95.9.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen YT, Gure AO, Tsang S, et al. Identification of multiple cancer/testis antigens by allogeneic antibody screening of a melanoma cell line library. Proc Natl Acad Sci USA. 1998;95(12):6919–6923. doi: 10.1073/pnas.95.12.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas S, De Smet C, Arden KC, et al. Identification of a new MAGE gene with tumor-specific expression by representational difference analysis. Cancer Res. 1998;58(4):743–752. [PubMed] [Google Scholar]
- 34.Lethe B, Lucas S, Michaux L, et al. LAGE-1, a new gene with tumor specificity. Intl J Cancer. 1998;76(6):903–908. doi: 10.1002/(sici)1097-0215(19980610)76:6<903::aid-ijc22>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 35.Martelange V, De Smet C, De Plaen E, et al. Identification on a human sarcoma of two new genes with tumor-specific expression. Cancer Res. 2000;60(14):3848–3855. [PubMed] [Google Scholar]
- 36.Lin C, Mak S, Meitner PA, et al. Cancer/testis antigen CSAGE is concurrently expressed with MAGE in chondrosarcoma. Gene. 2002;285(1–2):269–278. doi: 10.1016/s0378-1119(02)00395-5. [DOI] [PubMed] [Google Scholar]
- 37.De Plaen E, Arden K, Traversari C, et al. Structure, chromosomal localization, and expression of 12 genes of the MAGE family. Immunogenetics. 1994;40(5):360–369. doi: 10.1007/BF01246677. [DOI] [PubMed] [Google Scholar]
- 38.Chaux P, Luiten R, Demotte N, et al. Identification of five MAGE-A1 epitopes recognized by cytolytic T lymphocytes obtained by in vitro stimulation with dendritic cells transduced with MAGE-A1. J Immunol. 1999;163(5):2928–2936. [PubMed] [Google Scholar]
- 39.Luiten R, van der Bruggen P. A MAGE-A1 peptide is recognized on HLA-B7 human tumors by cytolytic T lymphocytes. Tissue Antigens. 2000;55(2):149–152. doi: 10.1034/j.1399-0039.2000.550206.x. [DOI] [PubMed] [Google Scholar]
- 40.Luiten RM, Demotte N, Tine J, van der Bruggen P. A MAGE-A1 peptide presented to cytolytic T lymphocytes by both HLA-B35 and HLA-A1 molecules. Tissue Antigens. 2000;56(1):77–81. doi: 10.1034/j.1399-0039.2000.560110.x. [DOI] [PubMed] [Google Scholar]
- 41.van der Bruggen P, Stroobant V, Van Pel A et al. (2010) Peptide database of T-cell defined tumor antigens. Cancer Immunity. http://www.cancerimmunity.org/peptidedatabase/Tcellepitopes.htm [PMC free article] [PubMed]
- 42.Haas GG, Jr, D’Cruz OJ, De Bault LE. Distribution of human leukocyte antigen-ABC and -D/DR antigens in the unfixed human testis. Am J Reprod Immunol Microbiol. 1988;18(2):47–51. doi: 10.1111/j.1600-0897.1988.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 43.Lurquin C, De Smet C, Brasseur F, et al. Two members of the human MAGEB gene family located in Xp21.3 are expressed in tumors of various histological origins. Genomics. 1997;46(3):397–408. doi: 10.1006/geno.1997.5052. [DOI] [PubMed] [Google Scholar]
- 44.Chomez P, De Backer O, Bertrand M, et al. An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res. 2001;61(14):5544–5551. [PubMed] [Google Scholar]
- 45.Ehrlich M, Gama-Sosa MA, Huang LH, et al. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues of cells. Nucleic Acids Res. 1982;10(8):2709–2721. doi: 10.1093/nar/10.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gama-Sosa MA, Slagel VA, Trewyn RW, et al. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983;11(19):6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diala ES, Cheah MS, Rowitch D, Hoffman RM. Extent of DNA methylation in human tumor cells. J Natl Cancer Inst. 1983;71(4):755–764. [PubMed] [Google Scholar]
- 48.Weber J, Salgaller M, Samid D, et al. Expression of the MAGE-1 tumor antigen is up-regulated by the demethylating agent 5-aza-2′-deoxycytidine. Cancer Res. 1994;54(7):1766–1771. [PubMed] [Google Scholar]
- 49.De Smet C, Lurquin C, Lethe B, et al. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19(11):7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lucas S, Brasseur F, Boon T. A new MAGE gene with ubiquitous expression does not code for known MAGE antigens recognized by T cells. Cancer Res. 1999;59(16):4100–4103. [PubMed] [Google Scholar]
- 51.Gure AO, Tureci O, Sahin U, et al. SSX: a multigene family with several members transcribed in normal testis and human cancer. Intl J Cancer. 1997;72(6):965–971. doi: 10.1002/(sici)1097-0215(19970917)72:6<965::aid-ijc8>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 52.Gure AO, Wei IJ, Old LJ, Chen YT. The SSX gene family: characterization of 9 complete genes. Intl J Cancer. 2002;101(5):448–453. doi: 10.1002/ijc.10634. [DOI] [PubMed] [Google Scholar]
- 53.Wolfel T, Van Pel A, Brichard V, et al. Two tyrosinase nonapeptides recognized on HLA-A2 melanomas by autologous cytolytic T lymphocytes. Eur J Immunol. 1994;24(3):759–764. doi: 10.1002/eji.1830240340. [DOI] [PubMed] [Google Scholar]
- 54.Vigneron N, Ooms A, Morel S, et al. A peptide derived from melanocytic protein gp100 and presented by HLA-B35 is recognized by autologous cytolytic T lymphocytes on melanoma cells. Tissue Antigens. 2005;65(2):156–162. doi: 10.1111/j.1399-0039.2005.00365.x. [DOI] [PubMed] [Google Scholar]
- 55.Kawakami Y, Eliyahu S, Sakaguchi K, et al. Identification of the immunodominant peptides of the MART-1 human melanoma antigen recognized by the majority of HLA-A2-restricted tumor infiltrating lymphocytes. J Exp Med. 1994;180(1):347–352. doi: 10.1084/jem.180.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang RF, Parkhurst MR, Kawakami Y, et al. Utilization of an alternative open reading frame of a normal gene in generating a novel human cancer antigen. J Exp Med. 1996;183(3):1131–1140. doi: 10.1084/jem.183.3.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang RF, Appella E, Kawakami Y, et al. Identification of TRP-2 as a human tumor antigen recognized by cytotoxic T lymphocytes. J Exp Med. 1996;184(6):2207–2216. doi: 10.1084/jem.184.6.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Visseren MJ, van Elsas A, van der Voort EI, et al. CTL specific for the tyrosinase autoantigen can be induced from healthy donor blood to lyse melanoma cells. J Immunol. 1995;154(8):3991–3998. [PubMed] [Google Scholar]
- 59.Nordlund JJ, Kirkwood JM, Forget BM, et al. Vitiligo in patients with metastatic melanoma: a good prognostic sign. J Am Acad Dermatol. 1983;9(5):689–696. doi: 10.1016/s0190-9622(83)70182-9. [DOI] [PubMed] [Google Scholar]
- 60.Rosenberg SA, White DE. Vitiligo in patients with melanoma: normal tissue antigens can be targets for cancer immunotherapy. J Immunother Emphasis Tumor Immunol. 1996;19(1):81–84. [PubMed] [Google Scholar]
- 61.Yee C, Thompson JA, Roche P, et al. Melanocyte destruction after antigen-specific immunotherapy of melanoma: direct evidence of T cell-mediated vitiligo. J Exp Med. 2000;192(11):1637–1644. doi: 10.1084/jem.192.11.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolfel T, Hauer M, Schneider J, et al. A p16INK4a-insensitive CDK4 mutant targeted by cytolytic T lymphocytes in a human melanoma. Science. 1995;269(5228):1281–1284. doi: 10.1126/science.7652577. [DOI] [PubMed] [Google Scholar]
- 63.Robbins PF, El-Gamil M, Li YF, et al. A mutated beta-catenin gene encodes a melanoma-specific antigen recognized by tumor infiltrating lymphocytes. J Exp Med. 1996;183(3):1185–1192. doi: 10.1084/jem.183.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rubinfeld B, Robbins P, El-Gamil M, et al. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275(5307):1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 65.Mandruzzato S, Brasseur F, Andry G, et al. A CASP-8 mutation recognized by cytolytic T lymphocytes on a human head and neck carcinoma. J Exp Med. 1997;186(5):785–793. doi: 10.1084/jem.186.5.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gjertsen MK, Bjorheim J, Saeterdal I, et al. Cytotoxic CD4+ and CD8+ T lymphocytes, generated by mutant p21-ras (12Val) peptide vaccination of a patient, recognize 12Val-dependent nested epitopes present within the vaccine peptide and kill autologous tumour cells carrying this mutation. Intl J Cancer. 1997;72(5):784–790. doi: 10.1002/(sici)1097-0215(19970904)72:5<784::aid-ijc14>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 67.Linard B, Bezieau S, Benlalam H, et al. A ras-mutated peptide targeted by CTL infiltrating a human melanoma lesion. J Immunol. 2002;168(9):4802–4808. doi: 10.4049/jimmunol.168.9.4802. [DOI] [PubMed] [Google Scholar]
- 68.Ito D, Visus C, Hoffmann TK, et al. Immunological characterization of missense mutations occurring within cytotoxic T cell-defined p53 epitopes in HLA-A*0201 + squamous cell carcinomas of the head and neck. Intl J Cancer. 2007;120(12):2618–2624. doi: 10.1002/ijc.22584. [DOI] [PubMed] [Google Scholar]
- 69.Linnebacher M, Gebert J, Rudy W, et al. Frameshift peptide-derived T-cell epitopes: a source of novel tumor-specific antigens. Intl J Cancer. 2001;93(1):6–11. doi: 10.1002/ijc.1298. [DOI] [PubMed] [Google Scholar]
- 70.Huang J, El-Gamil M, Dudley ME, et al. T cells associated with tumor regression recognize frameshifted products of the CDKN2A tumor suppressor gene locus and a mutated HLA class I gene product. J Immunol. 2004;172(10):6057–6064. doi: 10.4049/jimmunol.172.10.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yotnda P, Garcia F, Peuchmaur M, et al. Cytotoxic T cell response against the chimeric ETV6-AML1 protein in childhood acute lymphoblastic leukemia. J Clin Invest. 1998;102(2):455–462. doi: 10.1172/JCI3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yotnda P, Firat H, Garcia-Pons F, et al. Cytotoxic T cell response against the chimeric p210 BCR-ABL protein in patients with chronic myelogenous leukemia. J Clin Invest. 1998;101(10):2290–2296. doi: 10.1172/JCI488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fisk B, Blevins TL, Wharton JT, Ioannides CG. Identification of an immunodominant peptide of HER-2/neu protooncogene recognized by ovarian tumor-specific cytotoxic T lymphocyte lines. J Exp Med. 1995;181(6):2109–2117. doi: 10.1084/jem.181.6.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kraus MH, Popescu NC, Amsbaugh SC, King CR. Overexpression of the EGF receptor-related proto-oncogene erbB-2 in human mammary tumor cell lines by different molecular mechanisms. EMBO J. 1987;6(3):605–610. doi: 10.1002/j.1460-2075.1987.tb04797.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Slamon DJ, Godolphin W, Jones LA, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244(4905):707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- 76.Gaugler B, Brouwenstijn N, Vantomme V, et al. A new gene coding for an antigen recognized by autologous cytolytic T lymphocytes on a human renal carcinoma. Immunogenetics. 1996;44(5):323–330. doi: 10.1007/BF02602776. [DOI] [PubMed] [Google Scholar]
- 77.Barker CF, Billingham RE. Immunologically privileged sites. Adv Immunol. 1977;25:1–54. [PubMed] [Google Scholar]
- 78.Griffith TS, Brunner T, Fletcher SM, et al. Fas ligand-induced apoptosis as a mechanism of immune privilege. Science. 1995;270(5239):1189–1192. doi: 10.1126/science.270.5239.1189. [DOI] [PubMed] [Google Scholar]
- 79.Abi-Hanna D, Wakefield D, Watkins S. HLA antigens in ocular tissues. I. In vivo expression in human eyes. Transplantation. 1988;45(3):610–613. doi: 10.1097/00007890-198803000-00021. [DOI] [PubMed] [Google Scholar]
- 80.Kessler JH, Beekman NJ, Bres-Vloemans SA, et al. Efficient identification of novel HLA-A(*)0201-presented cytotoxic T lymphocyte epitopes in the widely expressed tumor antigen PRAME by proteasome-mediated digestion analysis. J Exp Med. 2001;193(1):73–88. doi: 10.1084/jem.193.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmitz M, Diestelkoetter P, Weigle B, et al. Generation of survivin-specific CD8+ T effector cells by dendritic cells pulsed with protein or selected peptides. Cancer Res. 2000;60(17):4845–4849. [PubMed] [Google Scholar]
- 82.Schmidt SM, Schag K, Muller MR, et al. Survivin is a shared tumor-associated antigen expressed in a broad variety of malignancies and recognized by specific cytotoxic T cells. Blood. 2003;102(2):571–576. doi: 10.1182/blood-2002-08-2554. [DOI] [PubMed] [Google Scholar]
- 83.Ropke M, Hald J, Guldberg P, et al. Spontaneous human squamous cell carcinomas are killed by a human cytotoxic T lymphocyte clone recognizing a wild-type p53-derived peptide. Proc Natl Acad Sci USA. 1996;93(25):14704–14707. doi: 10.1073/pnas.93.25.14704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barfoed AM, Petersen TR, Kirkin AF, et al. Cytotoxic T-lymphocyte clones, established by stimulation with the HLA-A2 binding p5365-73 wild type peptide loaded on dendritic cells In vitro, specifically recognize and lyse HLA-A2 tumour cells overexpressing the p53 protein. Scand J Immunol. 2000;51(2):128–133. doi: 10.1046/j.1365-3083.2000.00668.x. [DOI] [PubMed] [Google Scholar]
- 85.Ohminami H, Yasukawa M, Fujita S. HLA class I-restricted lysis of leukemia cells by a CD8(+) cytotoxic T-lymphocyte clone specific for WT1 peptide. Blood. 2000;95(1):286–293. [PubMed] [Google Scholar]
- 86.Asemissen AM, Keilholz U, Tenzer S, et al. Identification of a highly immunogenic HLA-A*01-binding T cell epitope of WT1. Clin Cancer Res. 2006;12(24):7476–7482. doi: 10.1158/1078-0432.CCR-06-1337. [DOI] [PubMed] [Google Scholar]
- 87.De Plaen E, Lurquin C, Van Pel A, et al. Immunogenic (tum-) variants of mouse tumor P815: cloning of the gene of tum-antigen P91A and identification of the tum-mutation. Proc Natl Acad Sci USA. 1988;85(7):2274–2278. doi: 10.1073/pnas.85.7.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sibille C, Chomez P, Wildmann C, et al. Structure of the gene of tum-transplantation antigen P198: a point mutation generates a new antigenic peptide. J Exp Med. 1990;172(1):35–45. doi: 10.1084/jem.172.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Szikora JP, Van Pel A, Brichard V, et al. Structure of the gene of tum-transplantation antigen P35B: presence of a point mutation in the antigenic allele. EMBO J. 1990;9(4):1041–1050. doi: 10.1002/j.1460-2075.1990.tb08208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boon T, Van Pel A. T cell-recognized antigenic peptides derived from the cellular genome are not protein degradation products but can be generated directly by transcription and translation of short subgenic regions. A hypothesis. Immunogenetics. 1989;29(2):75–79. doi: 10.1007/BF00395854. [DOI] [PubMed] [Google Scholar]
- 91.Guilloux Y, Lucas S, Brichard VG, et al. A peptide recognized by human cytolytic T lymphocytes on HLA-A2 melanomas is encoded by an intron sequence of the N-acetylglucosaminyltransferase V gene. J Exp Med. 1996;183(3):1173–1183. doi: 10.1084/jem.183.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Van den Eynde BJ, Gaugler B, Probst-Kepper M, et al. A new antigen recognized by cytolytic T lymphocytes on a human kidney tumor results from reverse strand transcription. J Exp Med. 1999;190(12):1793–1800. doi: 10.1084/jem.190.12.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Robbins PF, El-Gamil M, Li YF, et al. The intronic region of an incompletely spliced gp100 gene transcript encodes an epitope recognized by melanoma-reactive tumor-infiltrating lymphocytes. J Immunol. 1997;159(1):303–308. [PubMed] [Google Scholar]
- 94.Lupetti R, Pisarra P, Verrecchia A, et al. Translation of a retained intron in tyrosinase-related protein (TRP) 2 mRNA generates a new cytotoxic T lymphocyte (CTL)-defined and shared human melanoma antigen not expressed in normal cells of the melanocytic lineage. J Exp Med. 1998;188(6):1005–1016. doi: 10.1084/jem.188.6.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Topalian SL, Solomon D, Avis FP, et al. Immunotherapy of patients with advanced cancer using tumor-infiltrating lymphocytes and recombinant interleukin-2: a pilot study. J Clin Oncol. 1988;6(5):839–853. doi: 10.1200/JCO.1988.6.5.839. [DOI] [PubMed] [Google Scholar]
- 96.Wang RF, Johnston SL, Zeng G, et al. A breast and melanoma-shared tumor antigen: T cell responses to antigenic peptides translated from different open reading frames. J Immunol. 1998;161(7):3598–3606. [PubMed] [Google Scholar]
- 97.Aarnoudse CA, van den Doel PB, Heemskerk B, Schrier PI. Interleukin-2-induced, melanoma-specific T cells recognize CAMEL, an unexpected translation product of LAGE-1. Intl J Cancer. 1999;82(3):442–448. doi: 10.1002/(sici)1097-0215(19990730)82:3<442::aid-ijc19>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 98.Ronsin C, Chung-Scott V, Poullion I, et al. A non-AUG-defined alternative open reading frame of the intestinal carboxyl esterase mRNA generates an epitope recognized by renal cell carcinoma-reactive tumor-infiltrating lymphocytes in situ. J Immunol. 1999;163(1):483–490. [PubMed] [Google Scholar]
- 99.Probst-Kepper M, Stroobant V, Kridel R, et al. An alternative open reading frame of the human macrophage colony-stimulating factor gene is independently translated and codes for an antigenic peptide of 14 amino acids recognized by tumor-infiltrating CD8 T lymphocytes. J Exp Med. 2001;193(10):1189–1198. doi: 10.1084/jem.193.10.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Probst-Kepper M, Hecht HJ, Herrmann H, et al. Conformational restraints and flexibility of 14-meric peptides in complex with HLA-B*3501. J Immunol. 2004;173(9):5610–5616. doi: 10.4049/jimmunol.173.9.5610. [DOI] [PubMed] [Google Scholar]
- 101.Godet Y, Moreau-Aubry A, Guilloux Y, et al. MELOE-1 is a new antigen overexpressed in melanomas and involved in adoptive T cell transfer efficiency. J Exp Med. 2008;205(11):2673–2682. doi: 10.1084/jem.20081356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Godet Y, Moreau-Aubry A, Mompelat D, et al. An additional ORF on meloe cDNA encodes a new melanoma antigen, MELOE-2, recognized by melanoma-specific T cells in the HLA-A2 context. Cancer Immunol Immunother. 2010;59(3):431–439. doi: 10.1007/s00262-009-0762-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rosenberg SA, Tong-On P, Li Y, et al. Identification of BING-4 cancer antigen translated from an alternative open reading frame of a gene in the extended MHC class II region using lymphocytes from a patient with a durable complete regression following immunotherapy. J Immunol. 2002;168(5):2402–2407. doi: 10.4049/jimmunol.168.5.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schiavetti F, Thonnard J, Colau D, et al. A human endogenous retroviral sequence encoding an antigen recognized on melanoma by cytolytic T lymphocytes. Cancer Res. 2002;62(19):5510–5516. [PubMed] [Google Scholar]
- 105.Bullock TN, Eisenlohr LC. Ribosomal scanning past the primary initiation codon as a mechanism for expression of CTL epitopes encoded in alternative reading frames. J Exp Med. 1996;184(4):1319–1329. doi: 10.1084/jem.184.4.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shastri N, Schwab S, Serwold T. Producing nature’s gene-chips: the generation of peptides for display by MHC class I molecules. Annu Rev Immunol. 2002;20:463–493. doi: 10.1146/annurev.immunol.20.100301.064819. [DOI] [PubMed] [Google Scholar]
- 107.Dolstra H, Fredrix H, Maas F, et al. A human minor histocompatibility antigen specific for B cell acute lymphoblastic leukemia. J Exp Med. 1999;189(2):301–308. doi: 10.1084/jem.189.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264(9):5031–5035. [PubMed] [Google Scholar]
- 109.Malarkannan S, Horng T, Shih PP, et al. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity. 1999;10(6):681–690. doi: 10.1016/s1074-7613(00)80067-9. [DOI] [PubMed] [Google Scholar]
- 110.Schwab SR, Li KC, Kang C, Shastri N. Constitutive display of cryptic translation products by MHC class I molecules. Science. 2003;301(5638):1367–1371. doi: 10.1126/science.1085650. [DOI] [PubMed] [Google Scholar]
- 111.Schwab SR, Shugart JA, Horng T, et al. Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol. 2004;2(11):e366. doi: 10.1371/journal.pbio.0020366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Starck SR, Ow Y, Jiang V, et al. A distinct translation initiation mechanism generates cryptic peptides for immune surveillance. PloS One. 2008;3(10):e3460. doi: 10.1371/journal.pone.0003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moreau-Aubry A, Le Guiner S, Labarriere N, et al. A processed pseudogene codes for a new antigen recognized by a CD8(+) T cell clone on melanoma. J Exp Med. 2000;191(9):1617–1624. doi: 10.1084/jem.191.9.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 115.Ghislain M, Udvardy A, Mann C. S. cerevisiae 26S protease mutants arrest cell division in G2/metaphase. Nature. 1993;366(6453):358–362. doi: 10.1038/366358a0. [DOI] [PubMed] [Google Scholar]
- 116.Palombella VJ, Rando OJ, Goldberg AL, Maniatis T. The ubiquitin-proteasome pathway is required for processing the NF-kappa B1 precursor protein and the activation of NF-kappa B. Cell. 1994;78(5):773–785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- 117.Rock KL, Goldberg AL. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 118.Groll M, Huber R. Substrate access and processing by the 20S proteasome core particle. Internatl J Biochem Cell Biol. 2003;35(5):606–616. doi: 10.1016/s1357-2725(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 119.Lowe J, Stock D, Jap B, et al. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science. 1995;268(5210):533–539. doi: 10.1126/science.7725097. [DOI] [PubMed] [Google Scholar]
- 120.Groll M, Ditzel L, Lowe J, et al. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature. 1997;386(6624):463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 121.Unno M, Mizushima T, Morimoto Y, et al. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002;10(5):609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- 122.Seemuller E, Lupas A, Stock D, et al. Proteasome from Thermoplasma acidophilum: a threonine protease. Science. 1995;268(5210):579–582. doi: 10.1126/science.7725107. [DOI] [PubMed] [Google Scholar]
- 123.Orlowski M. The multicatalytic proteinase complex, a major extralysosomal proteolytic system. Biochemistry. 1990;29(45):10289–10297. doi: 10.1021/bi00497a001. [DOI] [PubMed] [Google Scholar]
- 124.Heinemeyer W, Gruhler A, Mohrle V, et al. PRE2, highly homologous to the human major histocompatibility complex-linked RING10 gene, codes for a yeast proteasome subunit necessary for chrymotryptic activity and degradation of ubiquitinated proteins. J Biol Chem. 1993;268(7):5115–5120. [PubMed] [Google Scholar]
- 125.Enenkel C, Lehmann H, Kipper J, et al. PRE3, highly homologous to the human major histocompatibility complex-linked LMP2 (RING12) gene, codes for a yeast proteasome subunit necessary for the peptidylglutamyl-peptide hydrolyzing activity. FEBS Lett. 1994;341(2–3):193–196. doi: 10.1016/0014-5793(94)80455-9. [DOI] [PubMed] [Google Scholar]
- 126.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86(6):961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 127.Heinemeyer W, Fischer M, Krimmer T, et al. The active sites of the eukaryotic 20 S proteasome and their involvement in subunit precursor processing. J Biol Chem. 1997;272(40):25200–25209. doi: 10.1074/jbc.272.40.25200. [DOI] [PubMed] [Google Scholar]
- 128.Dick TP, Nussbaum AK, Deeg M, et al. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J Biol Chem. 1998;273(40):25637–25646. doi: 10.1074/jbc.273.40.25637. [DOI] [PubMed] [Google Scholar]
- 129.Gueckel R, Enenkel C, Wolf DH, Hilt W. Mutations in the yeast proteasome beta-type subunit Pre3 uncover position-dependent effects on proteasomal peptidase activity and in vivo function. J Biol Chem. 1998;273(31):19443–19452. doi: 10.1074/jbc.273.31.19443. [DOI] [PubMed] [Google Scholar]
- 130.Nussbaum AK, Dick TP, Keilholz W, et al. Cleavage motifs of the yeast 20S proteasome beta subunits deduced from digests of enolase 1. Proc Natl Acad Sci USA. 1998;95(21):12504–12509. doi: 10.1073/pnas.95.21.12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Toes RE, Nussbaum AK, Degermann S, et al. Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J Exp Med. 2001;194(1):1–12. doi: 10.1084/jem.194.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Michalek MT, Grant EP, Gramm C, et al. A role for the ubiquitin-dependent proteolytic pathway in MHC class I-restricted antigen presentation. Nature. 1993;363(6429):552–554. doi: 10.1038/363552a0. [DOI] [PubMed] [Google Scholar]
- 133.Rock KL, Gramm C, Rothstein L, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78(5):761–771. doi: 10.1016/s0092-8674(94)90462-6. [DOI] [PubMed] [Google Scholar]
- 134.Morel S, Levy F, Burlet-Schiltz O, et al. Processing of some antigens by the standard proteasome but not by the immunoproteasome results in poor presentation by dendritic cells. Immunity. 2000;12(1):107–117. doi: 10.1016/s1074-7613(00)80163-6. [DOI] [PubMed] [Google Scholar]
- 135.Ottaviani S, Zhang Y, Boon T, van der Bruggen P. A MAGE-1 antigenic peptide recognized by human cytolytic T lymphocytes on HLA-A2 tumor cells. Cancer Immunol Immunother. 2005;54(12):1214–1220. doi: 10.1007/s00262-005-0705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Parmentier N, Stroobant V, Colau D, et al. Production of an antigenic peptide by insulin-degrading enzyme. Nat Immunol. 2010;11(5):449–454. doi: 10.1038/ni.1862. [DOI] [PubMed] [Google Scholar]
- 137.Stohwasser R, Standera S, Peters I, et al. Molecular cloning of the mouse proteasome subunits MC14 and MECL-1: reciprocally regulated tissue expression of interferon-gamma-modulated proteasome subunits. Eur J Immunol. 1997;27(5):1182–1187. doi: 10.1002/eji.1830270520. [DOI] [PubMed] [Google Scholar]
- 138.Macagno A, Gilliet M, Sallusto F, et al. Dendritic cells up-regulate immunoproteasomes and the proteasome regulator PA28 during maturation. Eur J Immunol. 1999;29(12):4037–4042. doi: 10.1002/(SICI)1521-4141(199912)29:12<4037::AID-IMMU4037>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 139.Driscoll J, Brown MG, Finley D, Monaco JJ. MHC-linked LMP gene products specifically alter peptidase activities of the proteasome. Nature. 1993;365(6443):262–264. doi: 10.1038/365262a0. [DOI] [PubMed] [Google Scholar]
- 140.Gaczynska M, Rock KL, Goldberg AL. Gamma-interferon and expression of MHC genes regulate peptide hydrolysis by proteasomes. Nature. 1993;365(6443):264–267. doi: 10.1038/365264a0. [DOI] [PubMed] [Google Scholar]
- 141.Fehling HJ, Swat W, Laplace C, et al. MHC class I expression in mice lacking the proteasome subunit LMP-7. Science. 1994;265(5176):1234–1237. doi: 10.1126/science.8066463. [DOI] [PubMed] [Google Scholar]
- 142.Van Kaer L, Ashton-Rickardt PG, Eichelberger M, et al. Altered peptidase and viral-specific T cell response in LMP2 mutant mice. Immunity. 1994;1(7):533–541. doi: 10.1016/1074-7613(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 143.Van den Eynde BJ, Morel S. Differential processing of class-I-restricted epitopes by the standard proteasome and the immunoproteasome. Curr Opin Immunol. 2001;13(2):147–153. doi: 10.1016/s0952-7915(00)00197-7. [DOI] [PubMed] [Google Scholar]
- 144.Chapiro J, Claverol S, Piette F, et al. Destructive cleavage of antigenic peptides either by the immunoproteasome or by the standard proteasome results in differential antigen presentation. J Immunol. 2006;176(2):1053–1061. doi: 10.4049/jimmunol.176.2.1053. [DOI] [PubMed] [Google Scholar]
- 145.Schultz ES, Chapiro J, Lurquin C, et al. The production of a new MAGE-3 peptide presented to cytolytic T lymphocytes by HLA-B40 requires the immunoproteasome. J Exp Med. 2002;195(4):391–399. doi: 10.1084/jem.20011974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chapatte L, Ayyoub M, Morel S, et al. Processing of tumor-associated antigen by the proteasomes of dendritic cells controls in vivo T-cell responses. Cancer Res. 2006;66(10):5461–5468. doi: 10.1158/0008-5472.CAN-05-4310. [DOI] [PubMed] [Google Scholar]
- 147.Guillaume B, Chapiro J, Stroobant V, et al. Two abundant proteasome subtypes that uniquely process some antigens presented by HLA class I molecules. Proc Natl Acad Sci USA. 2010;107(43):18599–18604. doi: 10.1073/pnas.1009778107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Serwold T, Gonzalez F, Kim J, et al. ERAAP customizes peptides for MHC class I molecules in the endoplasmic reticulum. Nature. 2002;419(6906):480–483. doi: 10.1038/nature01074. [DOI] [PubMed] [Google Scholar]
- 149.Saric T, Chang SC, Hattori A, et al. An IFN-gamma-induced aminopeptidase in the ER, ERAP1, trims precursors to MHC class I-presented peptides. Nat Immunol. 2002;3(12):1169–1176. doi: 10.1038/ni859. [DOI] [PubMed] [Google Scholar]
- 150.York IA, Chang SC, Saric T, et al. The ER aminopeptidase ERAP1 enhances or limits antigen presentation by trimming epitopes to 8–9 residues. Nat Immunol. 2002;3(12):1177–1184. doi: 10.1038/ni860. [DOI] [PubMed] [Google Scholar]
- 151.Saveanu L, Carroll O, Lindo V, et al. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol. 2005;6(7):689–697. doi: 10.1038/ni1208. [DOI] [PubMed] [Google Scholar]
- 152.Hammer GE, Gonzalez F, Champsaur M, et al. The aminopeptidase ERAAP shapes the peptide repertoire displayed by major histocompatibility complex class I molecules. Nat Immunol. 2006;7(1):103–112. doi: 10.1038/ni1286. [DOI] [PubMed] [Google Scholar]
- 153.Hammer GE, Gonzalez F, James E, et al. In the absence of aminopeptidase ERAAP, MHC class I molecules present many unstable and highly immunogenic peptides. Nat Immunol. 2007;8(1):101–108. doi: 10.1038/ni1409. [DOI] [PubMed] [Google Scholar]
- 154.Geier E, Pfeifer G, Wilm M, et al. A giant protease with potential to substitute for some functions of the proteasome. Science. 1999;283(5404):978–981. doi: 10.1126/science.283.5404.978. [DOI] [PubMed] [Google Scholar]
- 155.Glas R, Bogyo M, McMaster JS, et al. A proteolytic system that compensates for loss of proteasome function. Nature. 1998;392(6676):618–622. doi: 10.1038/33443. [DOI] [PubMed] [Google Scholar]
- 156.Princiotta MF, Schubert U, Chen W, et al. Cells adapted to the proteasome inhibitor 4-hydroxy- 5-iodo-3-nitrophenylacetyl-Leu-Leu-leucinal-vinyl sulfone require enzymatically active proteasomes for continued survival. Proc Natl Acad Sci USA. 2001;98(2):513–518. doi: 10.1073/pnas.021132398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kessler B, Hong X, Petrovic J, et al. Pathways accessory to proteasomal proteolysis are less efficient in major histocompatibility complex class I antigen production. J Biol Chem. 2003;278(12):10013–10021. doi: 10.1074/jbc.M211221200. [DOI] [PubMed] [Google Scholar]
- 158.York IA, Bhutani N, Zendzian S, et al. Tripeptidyl peptidase II is the major peptidase needed to trim long antigenic precursors, but is not required for most MHC class I antigen presentation. J Immunol. 2006;177(3):1434–1443. doi: 10.4049/jimmunol.177.3.1434. [DOI] [PubMed] [Google Scholar]
- 159.Basler M, Groettrup M. No essential role for tripeptidyl peptidase II for the processing of LCMV-derived T cell epitopes. Eur J Immunol. 2007;37(4):896–904. doi: 10.1002/eji.200636372. [DOI] [PubMed] [Google Scholar]
- 160.Firat E, Huai J, Saveanu L, et al. Analysis of direct and cross-presentation of antigens in TPPII knockout mice. J Immunol. 2007;179(12):8137–8145. doi: 10.4049/jimmunol.179.12.8137. [DOI] [PubMed] [Google Scholar]
- 161.Marcilla M, Villasevil EM, de Castro JA. Tripeptidyl peptidase II is dispensable for the generation of both proteasome-dependent and proteasome-independent ligands of HLA-B27 and other class I molecules. Eur J Immunol. 2008;38(3):631–639. doi: 10.1002/eji.200737444. [DOI] [PubMed] [Google Scholar]
- 162.Kawahara M, York IA, Hearn A, et al. Analysis of the role of tripeptidyl peptidase II in MHC class I antigen presentation in vivo. J Immunol. 2009;183(10):6069–6077. doi: 10.4049/jimmunol.0803564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Seifert U, Maranon C, Shmueli A, et al. An essential role for tripeptidyl peptidase in the generation of an MHC class I epitope. Nat Immunol. 2003;4(4):375–379. doi: 10.1038/ni905. [DOI] [PubMed] [Google Scholar]
- 164.Guil S, Rodriguez-Castro M, Aguilar F, et al. Need for tripeptidyl-peptidase II in major histocompatibility complex class I viral antigen processing when proteasomes are detrimental. J Biol Chem. 2006;281(52):39925–39934. doi: 10.1074/jbc.M608522200. [DOI] [PubMed] [Google Scholar]
- 165.Diekmann J, Adamopoulou E, Beck O, et al. Processing of two latent membrane protein 1 MHC class I epitopes requires tripeptidyl peptidase II involvement. J Immunol. 2009;183(3):1587–1597. doi: 10.4049/jimmunol.0803441. [DOI] [PubMed] [Google Scholar]
- 166.Preta G, Marescotti D, Fortini C, et al. Inhibition of serine-peptidase activity enhances the generation of a survivin-derived HLA-A2-presented CTL epitope in colon-carcinoma cells. Scand J Immunol. 2008;68(6):579–588. doi: 10.1111/j.1365-3083.2008.02175.x. [DOI] [PubMed] [Google Scholar]
- 167.Beninga J, Rock KL, Goldberg AL. Interferon-gamma can stimulate post-proteasomal trimming of the N terminus of an antigenic peptide by inducing leucine aminopeptidase. J Biol Chem. 1998;273(30):18734–18742. doi: 10.1074/jbc.273.30.18734. [DOI] [PubMed] [Google Scholar]
- 168.Stoltze L, Schirle M, Schwarz G, et al. Two new proteases in the MHC class I processing pathway. Nat Immunol. 2000;1(5):413–418. doi: 10.1038/80852. [DOI] [PubMed] [Google Scholar]
- 169.Towne CF, York IA, Neijssen J, et al. Leucine aminopeptidase is not essential for trimming peptides in the cytosol or generating epitopes for MHC class I antigen presentation. J Immunol. 2005;175(10):6605–6614. doi: 10.4049/jimmunol.175.10.6605. [DOI] [PubMed] [Google Scholar]
- 170.Towne CF, York IA, Watkin LB, et al. Analysis of the role of bleomycin hydrolase in antigen presentation and the generation of CD8 T cell responses. J Immunol. 2007;178(11):6923–6930. doi: 10.4049/jimmunol.178.11.6923. [DOI] [PubMed] [Google Scholar]
- 171.Towne CF, York IA, Neijssen J, et al. Puromycin-sensitive aminopeptidase limits MHC class I presentation in dendritic cells but does not affect CD8 T cell responses during viral infections. J Immunol. 2008;180(3):1704–1712. doi: 10.4049/jimmunol.180.3.1704. [DOI] [PubMed] [Google Scholar]
- 172.Kim E, Kwak H, Ahn K. Cytosolic aminopeptidases influence MHC class I-mediated antigen presentation in an allele-dependent manner. J Immunol. 2009;183(11):7379–7387. doi: 10.4049/jimmunol.0901489. [DOI] [PubMed] [Google Scholar]
- 173.Levy F, Burri L, Morel S, et al. The final N-terminal trimming of a subaminoterminal proline-containing HLA class I-restricted antigenic peptide in the cytosol is mediated by two peptidases. J Immunol. 2002;169(8):4161–4171. doi: 10.4049/jimmunol.169.8.4161. [DOI] [PubMed] [Google Scholar]
- 174.Saric T, Beninga J, Graef CI, et al. Major histocompatibility complex class I-presented antigenic peptides are degraded in cytosolic extracts primarily by thimet oligopeptidase. J Biol Chem. 2001;276(39):36474–36481. doi: 10.1074/jbc.M105517200. [DOI] [PubMed] [Google Scholar]
- 175.York IA, Mo AX, Lemerise K, et al. The cytosolic endopeptidase, thimet oligopeptidase, destroys antigenic peptides and limits the extent of MHC class I antigen presentation. Immunity. 2003;18(3):429–440. doi: 10.1016/s1074-7613(03)00058-x. [DOI] [PubMed] [Google Scholar]
- 176.Neefjes J, Gottfried E, Roelse J, et al. Analysis of the fine specificity of rat, mouse and human TAP peptide transporters. Eur J Immunol. 1995;25(4):1133–1136. doi: 10.1002/eji.1830250444. [DOI] [PubMed] [Google Scholar]
- 177.Geiss-Friedlander R, Parmentier N, Moller U, et al. The cytoplasmic peptidase DPP9 is rate-limiting for degradation of proline-containing peptides. J Biol Chem. 2009;284(40):27211–27219. doi: 10.1074/jbc.M109.041871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lonchay C, van der Bruggen P, Connerotte T, et al. Correlation between tumor regression and T cell responses in melanoma patients vaccinated with a MAGE antigen. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14631–14638. doi: 10.1073/pnas.0405743101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fagan JM, Waxman L. Purification of a protease in red blood cells that degrades oxidatively damaged haemoglobin. Biochem J. 1991;277(Pt 3):779–786. doi: 10.1042/bj2770779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kurochkin IV, Goto S. Alzheimer’s beta-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett. 1994;345(1):33–37. doi: 10.1016/0014-5793(94)00387-4. [DOI] [PubMed] [Google Scholar]
- 181.Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19(5):608–624. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- 182.Morita M, Kurochkin IV, Motojima K, et al. Insulin-degrading enzyme exists inside of rat liver peroxisomes and degrades oxidized proteins. Cell Struct Funct. 2000;25(5):309–315. doi: 10.1247/csf.25.309. [DOI] [PubMed] [Google Scholar]
- 183.Kirschner RJ, Goldberg AL. A high molecular weight metalloendoprotease from the cytosol of mammalian cells. J Biol Chem. 1983;258(2):967–976. [PubMed] [Google Scholar]
- 184.Hanada K, Yewdell JW, Yang JC. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature. 2004;427(6971):252–256. doi: 10.1038/nature02240. [DOI] [PubMed] [Google Scholar]
- 185.Vigneron N, Stroobant V, Chapiro J, et al. An antigenic peptide produced by peptide splicing in the proteasome. Science. 2004;304(5670):587–590. doi: 10.1126/science.1095522. [DOI] [PubMed] [Google Scholar]
- 186.Warren EH, Vigneron NJ, Gavin MA, et al. An antigen produced by splicing of noncontiguous peptides in the reverse order. Science. 2006;313(5792):1444–1447. doi: 10.1126/science.1130660. [DOI] [PubMed] [Google Scholar]
- 187.Dalet A, Vigneron N, Stroobant V, et al. Splicing of distant peptide fragments occurs in the proteasome by transpeptidation and produces the spliced antigenic peptide derived from fibroblast growth factor-5. J Immunol. 2010;184(6):3016–3024. doi: 10.4049/jimmunol.0901277. [DOI] [PubMed] [Google Scholar]
- 188.Dalet A, Stroobant V, Vigneron N, Van den Eynde BJ. Differences in the production of spliced antigenic peptides by the standard proteasome and the immunoproteasome. Eur J Immunol. 2011;41(1):39–46. doi: 10.1002/eji.201040750. [DOI] [PubMed] [Google Scholar]
- 189.Kageyama S, Tsomides TJ, Sykulev Y, Eisen HN. Variations in the number of peptide-MHC class I complexes required to activate cytotoxic T cell responses. J Immunol. 1995;154(2):567–576. [PubMed] [Google Scholar]
- 190.Irvine DJ, Purbhoo MA, Krogsgaard M, Davis MM. Direct observation of ligand recognition by T cells. Nature. 2002;419(6909):845–849. doi: 10.1038/nature01076. [DOI] [PubMed] [Google Scholar]
- 191.Moore MW, Carbone FR, Bevan MJ. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell. 1988;54(6):777–785. doi: 10.1016/s0092-8674(88)91043-4. [DOI] [PubMed] [Google Scholar]
- 192.Mosse CA, Meadows L, Luckey CJ, et al. The class I antigen-processing pathway for the membrane protein tyrosinase involves translation in the endoplasmic reticulum and processing in the cytosol. J Exp Med. 1998;187(1):37–48. doi: 10.1084/jem.187.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Mosse CA, Hsu W, Engelhard VH. Tyrosinase degradation via two pathways during reverse translocation to the cytosol. Biochem Biophys Res Commun. 2001;285(2):313–319. doi: 10.1006/bbrc.2001.5153. [DOI] [PubMed] [Google Scholar]
- 194.Altrich-VanLith ML, Ostankovitch M, Polefrone JM, et al. Processing of a class I-restricted epitope from tyrosinase requires peptide N-glycanase and the cooperative action of endoplasmic reticulum aminopeptidase 1 and cytosolic proteases. J Immunol. 2006;177(8):5440–5450. doi: 10.4049/jimmunol.177.8.5440. [DOI] [PubMed] [Google Scholar]
- 195.Ostankovitch M, Altrich-Vanlith M, Robila V, Engelhard VH. N-glycosylation enhances presentation of a MHC class I-restricted epitope from tyrosinase. J Immunol. 2009;182(8):4830–4835. doi: 10.4049/jimmunol.0802902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Godefroy E, Moreau-Aubry A, Diez E, et al. alpha v beta3-dependent cross-presentation of matrix metalloproteinase-2 by melanoma cells gives rise to a new tumor antigen. J Exp Med. 2005;202(1):61–72. doi: 10.1084/jem.20042138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev. 2002;2(3):161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 198.Khasigov PZ, Podobed OV, Ktzoeva SA, et al. Matrix metalloproteinases of normal human tissues. Biochemistry (Mosc) 2001;66(2):130–140. doi: 10.1023/a:1002879128392. [DOI] [PubMed] [Google Scholar]
- 199.Renaud V, Godefroy E, Larrieu P, et al. Folding of matrix metalloproteinase-2 prevents endogenous generation of MHC class-I restricted epitope. PloS One. 2010;5(7):e11894. doi: 10.1371/journal.pone.0011894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Fruh K, Ahn K, Djaballah H, et al. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375(6530):415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 201.Hill A, Jugovic P, York I, et al. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375(6530):411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 202.Ahn K, Gruhler A, Galocha B, et al. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity. 1997;6(5):613–621. doi: 10.1016/s1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 203.van Hall T, Laban S, Koppers-Lalic D, et al. The varicellovirus-encoded TAP inhibitor UL49.5 regulates the presentation of CTL epitopes by Qa-1b1. J Immunol. 2007;178(2):657–662. doi: 10.4049/jimmunol.178.2.657. [DOI] [PubMed] [Google Scholar]
- 204.Wei ML, Cresswell P. HLA-A2 molecules in an antigen-processing mutant cell contain signal sequence-derived peptides. Nature. 1992;356(6368):443–446. doi: 10.1038/356443a0. [DOI] [PubMed] [Google Scholar]
- 205.Hunt DF, Henderson RA, Shabanowitz J, et al. Characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science. 1992;255(5049):1261–1263. doi: 10.1126/science.1546328. [DOI] [PubMed] [Google Scholar]
- 206.Weinzierl AO, Rudolf D, Hillen N, et al. Features of TAP-independent MHC class I ligands revealed by quantitative mass spectrometry. Eur J Immunol. 2008;38(6):1503–1510. doi: 10.1002/eji.200838136. [DOI] [PubMed] [Google Scholar]
- 207.Wolfel C, Drexler I, Van Pel A, et al. Transporter (TAP)- and proteasome-independent presentation of a melanoma-associated tyrosinase epitope. Internatl J Cancer. 2000;88(3):432–438. [PubMed] [Google Scholar]
- 208.El Hage F, Stroobant V, Vergnon I, et al. Preprocalcitonin signal peptide generates a cytotoxic T lymphocyte-defined tumor epitope processed by a proteasome-independent pathway. Proc Natl Acad Sci USA. 2008;105(29):10119–10124. doi: 10.1073/pnas.0802753105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Lee N, Goodlett DR, Ishitani A, et al. HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J Immunol. 1998;160(10):4951–4960. [PubMed] [Google Scholar]
- 210.Lemberg MK, Bland FA, Weihofen A, et al. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J Immunol. 2001;167(11):6441–6446. doi: 10.4049/jimmunol.167.11.6441. [DOI] [PubMed] [Google Scholar]
- 211.Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol Cell. 2002;10(4):735–744. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 212.Wearsch PA, Cresswell P. The quality control of MHC class I peptide loading. Curr Opin Cell Biol. 2008;20(6):624–631. doi: 10.1016/j.ceb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Lehner PJ, Surman MJ, Cresswell P. Soluble tapasin restores MHC class I expression and function in the tapasin-negative cell line 220. Immunity. 1998;8(2):221–231. doi: 10.1016/s1074-7613(00)80474-4. [DOI] [PubMed] [Google Scholar]
- 214.Sadasivan B, Lehner PJ, Ortmann B, et al. Roles for calreticulin and a novel glycoprotein, tapasin, in the interaction of MHC class I molecules with TAP. Immunity. 1996;5(2):103–114. doi: 10.1016/s1074-7613(00)80487-2. [DOI] [PubMed] [Google Scholar]
- 215.Williams AP, Peh CA, Purcell AW, et al. Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity. 2002;16(4):509–520. doi: 10.1016/s1074-7613(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 216.Ortmann B, Copeman J, Lehner PJ, et al. A critical role for tapasin in the assembly and function of multimeric MHC class I-TAP complexes. Science. 1997;277(5330):1306–1309. doi: 10.1126/science.277.5330.1306. [DOI] [PubMed] [Google Scholar]
- 217.Wearsch PA, Cresswell P. Selective loading of high-affinity peptides onto major histocompatibility complex class I molecules by the tapasin-ERp57 heterodimer. Nat Immunol. 2007;8(8):873–881. doi: 10.1038/ni1485. [DOI] [PubMed] [Google Scholar]
- 218.Zernich D, Purcell AW, Macdonald WA, et al. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J Exp Med. 2004;200(1):13–24. doi: 10.1084/jem.20031680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Purcell AW, Gorman JJ, Garcia-Peydro M, et al. Quantitative and qualitative influences of tapasin on the class I peptide repertoire. J Immunol. 2001;166(2):1016–1027. doi: 10.4049/jimmunol.166.2.1016. [DOI] [PubMed] [Google Scholar]
- 220.Vigneron N, Peaper DR, Leonhardt RM, Cresswell P. Functional significance of tapasin membrane association and disulfide linkage to ERp57 in MHC class I presentation. Eur J Immunol. 2009;39(9):2371–2376. doi: 10.1002/eji.200939536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Chen M, Bouvier M. Analysis of interactions in a tapasin/class I complex provides a mechanism for peptide selection. EMBO J. 2007;26(6):1681–1690. doi: 10.1038/sj.emboj.7601624. [DOI] [PMC free article] [PubMed] [Google Scholar]