Abstract
NFE2L3 [Nuclear factor (erythroid-derived 2)-like 3] or NRF3, a member of the Cap‘n’Collar (CNC) family, is a basic-region leucine zipper (bZIP) transcription factor that was first identified over 10 years ago. Contrary to its extensively studied homolog NFE2L2 (NRF2), the regulation and function of the NFE2L3 protein have not yet attracted as much attention. Nevertheless, several recent reports have now shed light on the possible roles of NFE2L3. Structural and biochemical studies revealed a series of domains and modifications that are critical for its cellular regulation. The control of the subcellular localization of NFE2L3 appears to be essential for understanding its role in various cellular processes. Importantly, newer studies provide fascinating insights linking NFE2L3 to differentiation, inflammation, and carcinogenesis. Here, we present an overview of the current level of knowledge of NFE2L3 transcription factor biology in humans and mice. From being the Cinderella of the CNC transcription factors for many years, NFE2L3 may now rapidly come into its own.
Keywords: NFE2L3, NRF3, Cap‘n’Collar transcription factor, Carcinogenesis, Inflammation
Introduction
The Cap‘n’Collar (CNC) proteins are a subgroup of basic region-leucine zipper (bZIP) transcription factors conserved among birds, insects, worms, fish, and mammals but absent in plants and fungi [1]. Since the discovery in 1989 of the first CNC transcription factor NFE2 (Nuclear Factor-Erythroid derived 2), also known as p45 NFE2 [2], several other members have been identified. The CNC family includes the Drosophila CNC1 protein [3], Caenorhabditis elegans Skn-1 [4], and vertebrate NFE2L1 (Nuclear factor erythroid 2 related factor-1) also known as NRF1/LCRF/TCF11 [5–7], NFE2L2 (or NRF2) [8], NFE2L3 (or NRF3) [9–11], as well as the more distantly related BACH1 [12] and BACH2 [12] proteins.
Members of the CNC family are characterized by a highly conserved 43-amino-acid region (Figs. 1, 2, 3), referred to as the CNC domain, which contributes to the unique DNA-binding specificity of these transcription factors [13, 14]. CNC proteins also contain a basic region-leucine zipper motif (bZIP) consisting of a region rich in basic residues conferring DNA-binding activity, followed by six heptad repeats of hydrophobic residues forming a leucine zipper motif, which acts as a dimerization domain (Figs. 1, 2, 3) [15]. The DNA-binding domain includes a nuclear localization signal, which has been shown to be functional in human p45 NFE2 [16], NFE2L2 [17], and BACH2 [18]. BACH1 and BACH2 proteins are characterized by the presence of a BTB (Broad complex, Tramtrack, Bric-a-brac) domain required for protein–protein interaction and facilitating homo- and/or hetero-dimer formation [12, 19–21]. CNC factors function as obligate heterodimers by complexing with small Maf (musculoaponeurotic fibrosarcoma) MAFG, MAFK, and MAFF [9, 22–28] and jun proteins [29, 30] for DNA binding. The resulting complexes bind to NFE2 (Nuclear Factor-Erythroid 2)-, MARE (Maf recognition element)-, ARE (antioxidant response element)- and StreB (stress-response element)/EpRE (electrophile response element)-type DNA binding sites [9, 10, 28, 31–33].
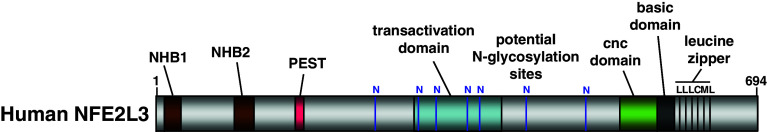
Fig. 1.
Structure of the human NFE2L3 transcription factor. NHB1 N-terminal homology box 1, NHB2 N-terminal homology box 2, PEST PEST domain, N potential N-glycosylation sites, transactivation domain, transactivation domain as defined by Gal4-luciferase reporter studies, CNC domain Cap‘n’Collar homology domain, basic domain basic DNA-binding domain, leucine zipper leucine zipper dimerization domain
Fig. 2.

Comparison of human CNC transcription factor sequences. Alignment of human p45 NFE2, NFE2L1, NFE2L2, NFE2L3, BACH1, and BACH2 protein sequences was performed using ClustalW program [117]. Identical residues are shaded in grey. The basic region (in light blue) and the CNC domain (in orange) are indicated. Asterisks indicate leucine zipper residues
Fig. 3.

Cross-species comparison of NFE2L3 sequences. Protein sequences of NFE2L3 from nine different species were aligned using ClustalW program [117]. Identical residues are shaded in grey. The basic region (in light blue), the CNC domain (in orange), the NHB1 domain (in purple), the NHB2 domain (in light green) and the PEST motif (in black) are indicated. Asterisks indicate leucine zipper residues
The role and regulation of the CNC member NFE2L2 in oxidative stress and in cancer prevention have been extensively studied and discussed in many reviews [34–59]. Furthermore, several reviews focus on the respective roles of p45 NFE2 [60–64], NFE2L1 [65], BACH1 [66–68], BACH2 [69, 70], and small MAF proteins [26, 28, 71–73]. Here, we provide the first review that focuses specifically on NFE2L3, including recent insights into its biology and its potential relevance to human diseases.
Structure of NFE2L3 gene and protein
The cloning of the human and mouse NFE2L3 genes was first reported in 1999 [9]. Fluorescent in situ hybridization (FISH) experiments mapped the human NFE2L3 gene on the chromosome 7p15-p14 [9], whereas the mouse Nfe2l3 is located on chromosome 6B3 [11]. In both species, the NFE2L3 gene maps near the HOXA gene cluster. This is similar to the genetic loci of p45 NFE2, NFE2L1, and NFE2L2, which map near the HOXC, HOXB, and HOXD genes, respectively [9, 74]. These observations support the idea that p45 NFE2, NFE2L1, NFE2L2, and NFE2L3 are derived from a single ancestral gene localized in proximity to the ancestral HOX cluster and they have diverged to give rise to four closely related transcription factors through chromosome duplication [9]. In contrast, the BACH1 and BACH2 genes seem to derive from another duplication since both genes are not associated with any HOX cluster. This observation could explain why Bach1 and Bach2 have functions distinct from the other CNC transcription factors.
The human NFE2L3 transcript encodes a 694-amino-acid protein, whereas mouse Nfe2l3 mRNA gives rise to a 660-amino-acid protein (Fig. 3) [9, 10]. Bioinformatics analysis of the NFE2L3 proteins among different species from zebrafish to human indicates a high degree of conservation through evolution of its key domains including the NHB1 (N-terminal homology box 1) domain, the NHB2 (N-terminal homology box 2) domain [75], the CNC domain, the basic region and the leucine zipper domains, supporting the importance of these domains for NFE2L3 functions (Figs. 1, 3). Intriguingly, human and mouse NFE2L3 proteins share only approximately 68% homology (Fig. 3), which is significantly less than among the orthologues of other CNC family members with 89, 97, and 80% overall identities for p45 NFE2, NFE2L1, and NFE2L2, respectively [9]. This suggests that the human and mouse NFE2L3 proteins may have acquired relatively more distinct functions when compared to the other CNC family members.
NFE2L3 expression in tissues and cells
Expression levels of NFE2L3 have been studied in both tissues and cells of human and mouse origins [9–11, 54, 76–81]. For instance, the highest levels of human NFE2L3 are found in placenta, specifically in chorionic villi from at least week 12 of gestation through term [9, 10]. Its expression has been found in primary placental cytotrophoblasts, but not in placental fibroblasts. In line with these data, NFE2L3 transcript and protein levels are high in both BeWo and JAR cell lines (Table 1), which are human choriocarcinoma cell lines derived from trophoblastic tumors of the placenta [10, 82]. Besides its expression in placenta, human NFE2L3 is expressed from intermediate to low levels in a wide variety of other tissues including heart, brain, lung, kidney, pancreas, leukocytes, colon, thymus, and spleen [9]. Barely detectable levels of human NFE2L3 were observed in human megakaryocytes and erythrocytes [80], whereas no expression was found in testis, prostate, skeletal muscle, and ovary [9]. NFE2L3 transcripts and/or proteins are expressed in a series of cell lines (Table 1) [9, 10, 75, 81–85]. Expression of Nfe2l3 in mouse tissues is broad with high levels found in thymus, brain, lung, stomach, uterus, placenta, adipose tissue, and testis [11, 76, 78].
Table 1.
List of human cell lines expressing NFE2L3 at the RNA and/or protein level
| Cell lines | NFE2L3 expression | |
|---|---|---|
| Cell/tumor type | References | |
| HL-60 | Acute promyelocytic leukemia | [9] |
| THP-1 | Acute monocytic leukemia | [9] |
| RPMI8226 | Myeloma | [9] |
| U937 | Histiocytic lymphoma | [9] |
| Raji | Burkitt lymphoma | [9] |
| BALM-2 | Burkitt lymphoma | [9] |
| NAMALWA | Burkitt lymphoma | [9] |
| HRS | Hodgkin lymphoma | [84] |
| HL | Hodgkin lymphoma | [83] |
| BeWo | Choriocarcinoma | [10] |
| JAR | Choriocarcinoma | [10, 75] |
| MDA-MB-231 | Breast cancer | [82] |
| MCF-10 | Breast cancer | [85] |
| HaCaT | Keratinocytes (non-tumorigenic) | [81] |
The expression pattern of NFE2L3 was evaluated by in situ hybridization during development of avian embryos and revealed expression in mesodermal derivatives including heart, somites, yolk sac, and kidney. Since this was different to the rather ubiquitous expression described for other CNC factors, it was suggested that NFE2L3 may play an earlier, more specific role in target gene regulation in the developing avian embryo [86].
Molecular mechanisms of NFE2L3 transactivation
During the past few years, important progress has been made in the understanding of NFE2L3 transactivation. NFE2L3 was identified as a partner of MafG in a yeast-based in vivo protein–protein interaction screen [10]. Functional studies using a Gal4-luciferase reporter revealed the presence of a potent transactivation domain in the center of human NFE2L3 protein (amino acids 298–399). It was shown that NFE2L3 can transcriptionally activate MARE-driven, β-globin gene expression in QT6 cells [9] as well as ARE-driven, NADPH dehydrogenase quinone 1 (Nqo1) gene expression in COS-1 cells [75]. Using the same reporter gene, Zhang and colleagues found that the transactivation capacity of NFE2L3 appeared to be less potent than NFE2L1 or NFE2L2 [75]. In contrast, overexpression of NFE2L3 leads to a repression of both luciferase reporter under the control of the NQO1 ARE as well as endogenous Nqo1 gene expression in human Hep-G2 cells [87] and mouse embryonic stem cells [79], indicating that NFE2L3 can either activate or repress transcription of its target genes depending on the cellular context. One may hypothesize that these differences are due to the expression levels of NFE2L3 and/or its cofactors.
Complex regulation of NFE2L3 expression
Biochemical studies revealed the existence of at least three differently migrating forms of endogenous NFE2L3, a slow migrating ‘A’ form, an intermediate ‘B’ form, and a faster migrating ‘C’ form [82]. Fractionation studies and immunofluorescence experiments showed that the various NFE2L3 versions are localized in specific subcellular compartments: ‘A’ is associated with the endoplasmic reticulum, whereas ‘B’ is mostly cytoplasmic and ‘C’ is found mainly in the nucleus (Fig. 4) [75, 82]. The ‘A’, ‘B,’ and ‘C’ forms of human NFE2L3 have short half-lives, generally less than 1 h. In accordance with the rapid turnover, a PEST motif has been identified in the NFE2L3 sequence and is conserved across different species (Fig. 3) [75, 82]. This motif has been shown to be functional and to negatively regulate its activity [75]. Studies using different proteasome inhibitors strongly suggest that the three NFE2L3 versions are degraded through the ubiquitin–proteasome pathway (Fig. 4) [82].
Fig. 4.
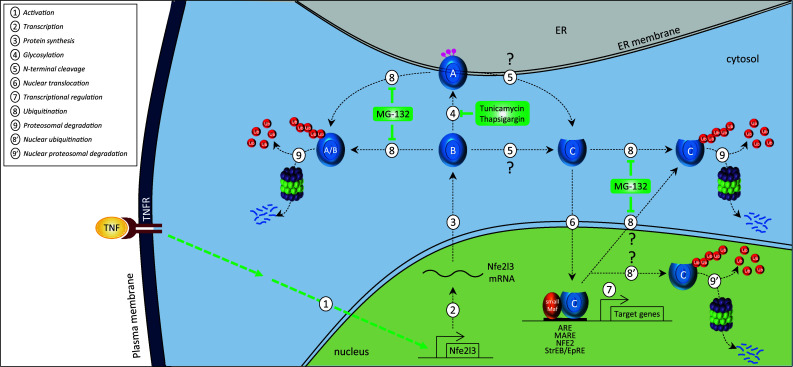
Proposed model for NFE2L3 regulation. Hypothetical model to describe the cellular regulation of NFE2L3 based on published literature. Once activated by a stimulus (e.g., TNF), transcription of NFE2L3 gene occurs in the nucleus leading to the production of NFE2L3 RNA in the nucleus and then to the translation in the cytosol into a ‘B’ form of NFE2L3 protein. The ‘B’ form can be targeted to the ER where it is N-glycosylated to become the A form of NFE2L3. The ‘A’ and/or ‘B’ forms of NFE2L3 is (are) hypothesized to be cleaved at the N-terminal end into a ‘C’ form, which is mainly found in the nucleus. The ‘C’ form is considered to be the major active form of NFE2L3 heterodimerizing with small Maf proteins. The resulting heterodimers activate transcription of their target genes through binding to specific DNA-binding sites including ARE, MARE, NFE2, StreB/EpRE sequences. The NFE2L3 ‘A’, ‘B’, and ‘C’ forms have short half-lives and are most likely degraded through the ubiquitin–proteasome pathway in the cytosol and/or in the nucleus
What distinguishes the ‘A’, ‘B’, and ‘C’ forms of NFE2L3? Using the N-linked specific deglycosylating enzymes PNGase F and EndoH, it was demonstrated that the ‘A’ form corresponds to a N-glycosylated NFE2L3 protein, whereas the ‘B’ and ‘C’ forms are unglycosylated (Fig. 4) [75, 82]. Treatment with both deglycosylation enzymes increases the ‘B’ form of NFE2L3 indicating that this version of NFE2L3 is indeed the unglycosylated form of ‘A’. Accordingly, seven potential sites for N-linked glycosylation were identified in the center portion of the NFE2L3 protein (Fig. 1). The exact nature of the ‘C’ form of NFE2L3 remains unclear. It has been speculated that it may be a truncated form of NFE2L3 lacking the N-terminal portion of the protein (Fig. 4) [82]. However, the site of cleavage generating the C form is still unidentified and one may hypothesize that this cleavage site is different from the one that releases the signal peptide targeting the protein to the ER. It has been reported that residues 40–75 of mouse NFE2L3, comprising several Site-1 protease recognition sites, control proteolytic processing that produces the ‘C’ form. Indeed, transactivation activity of NFE2L3 is inhibited by N-acetyl-l-leucyl-l-leucyl-l-norleucinal (ALLN), an inhibitor of proteasome activity, which has been shown to block the cleavage activity of the Site-1 protease [75]. However, human NFE2L3 sequence lacks a signal peptidase cleavage site [75], adding another layer of complexity to NFE2L3 regulation. With respect to mouse NFE2L3, a hypothetical model has been proposed that describes possible mechanisms that control its translocation from the ER to the nucleus, and it was also suggested that a deglycosylation event is required [75].
The NHB domains: important features of NFE2L3
Comparison between mouse NFE2L1 and NFE2L3 protein sequences revealed two conserved regions called NHB1 and NHB2 domains. The NHB1 domain, part of an ER signal peptide, has been previously shown to be required for ER targeting of the CNC factor NFE2L1 [88–90]. Of interest, the NHB1 domain of NFE2L3 is also present and highly conserved among species (Fig. 2). Recently, it has been reported that the NHB1 domain of mouse NFE2L3 is indeed necessary to target the protein to the ER and to enable the increase of NFE2L3 activity by the ER stressors tunicamycin and brefeldin A [75]. This is in contrast to the human protein, whose levels are decreased in the presence of tunicamycin [82]. The function of the NHB2 sequence of NFE2L3 still remains uncertain, but it may be involved in the control of NFE2L3 activity as well as its post-translational processing within the ER [75].
Mouse knockout models of CNC transcription factors
CNC factor deficient mice
Significant progress in the understanding of the function of the CNC transcription factor family has been made through the generation of knockout mouse models [11, 54, 91–95]. Gene targeting experiments showed that the p45 NFE2 protein is required for megakaryocyte biogenesis [96]. Homozygous Nfe2l1 null mice die in utero during mid- to late embryonic development due to a non-cell autonomous defect in definitive erythropoiesis [92, 97], whereas Nfe2l2 is dispensable for mouse development [95]. Bach1 null mice develop normally and are fertile [94]. Analysis of Bach2 null animals revealed that this factor is a key regulator of antibody class switching in B cells [93].
Analysis of Nfe2l3-deficient mice
Until recently, the physiological role of NFE2L3 remained elusive. To gain insights into the in vivo roles of NFE2L3 protein, mice deficient for this transcription factor gene were independently generated in two laboratories [11, 54]. Nfe2l3 null mice develop and grow normally under non-challenging conditions. No differences were observed between wild-type and Nfe2l3 −/− mice with respect to several blood parameters including red blood cell count, white blood cell count, erythrocyte cellular index as well as blood chemistries including glucose, blood urea, nitrogen, cholesterol, triglycerides, iron, and bilirubin [11]. Mice were infected with acute lymphocytic choriomeningitis virus and no difference was found between wild-type and Nfe2l3 null mice in the number of virus-specific CD8 and CD4 T cells as well as B-lymphocyte response [11].
To further investigate functional redundancy among CNC proteins, Nfe2l3 null mice were crossed with Nfe2l2 and/or p45 Nfe2 deficient mice. Nfe2l3 −/− :Nfe2l2 −/− and Nfe2l3 −/− :p45 Nfe2 mice develop normally and exhibit survival rates corresponding to the expected Mendelian ratio [54] and do not exhibit defects beyond those seen with the loss of p45 Nfe2 alone [11]. Triple-compound knockout p45 Nfe2 −/− :Nfe2l2 −/− :Nfe2l3 −/− mice were also generated and unexpectedly some survive to adulthood, suggesting that the CNC protein NF2L1 and/or other regulatory factors may compensate for the combined absence of p45 NFE2, NFE2L2, and NFE2L3 [11].
Linking NFE2L3 and carcinogenesis: in vivo evidence from mouse model
Recent in vivo data linked the NFE2L3 transcription factor to protection against lymphomagenesis [98]. Nfe2l3 −/− mice exposed for four consecutive weeks to benzo[a]pyrene (B[a]P), a carcinogen present in tobacco smoke [99], exhibited significantly increased mortality compared to wild-type animals. Thirty-two percent of B[a]P-treated Nfe2l3 −/− mice developed lymphoma, whereas only 6% of wild-type mice did. Pathological analyses of affected tissue sections revealed a high incidence (21%) of T-cell lymphoblastic lymphoma in B[a]P-treated Nfe2l3 −/− mice. In line with this data, high expression of Nfe2l3 transcripts was observed in the normal thymus of both human and mouse origin [9, 11]. These findings strongly suggest a protective role for NFE2L3 transcription factor in carcinogen-induced lymphomagenesis.
A series of genechip array data further hints at a possible role of NFE2L3 in various malignancies. Human NFE2L3 transcript levels are increased in different types of lymphoma including Hodgkin lymphoma [83, 84], non-Hodgkin cell lineages [83], as well as Mantle cell lymphoma specimens [100–103]. In addition to its overexpression in human lymphoma samples, NFE2L3 mRNA levels were found to be upregulated in human breast cancer cells [85] and testicular carcinoma tissue samples [104, 105]. Nevertheless, these gene profiling results strictly reflect mRNA levels and it is not known whether the NFE2L3 transcripts induced in these human samples code for functional or mutated versions of the transcription factor.
NFE2L3 and inflammation
In addition to its function in carcinogenesis, there is evidence that NFE2L3 may have a role in inflammation. Nfe2l3 null mice have been shown to be highly sensitive to exposure to the antioxidant butylated hydroxytoluene (BHT), which provokes acute lung injury after a single administration in mice [106]. BHT-treated Nfe2l3 −/− mice exhibit respiratory distress as well as increased body weight loss when compared to their wild-type counterparts [78]. At the molecular level, BHT treatment decreases both Nfe2l1 and Nfe2l3 transcripts in the lung of wild-type mice, whereas Nfe2l2 mRNA levels are upregulated under similar conditions. In addition, basal gene expression of the gene coding for Pparγ2 (peroxisome proliferator-activated receptor gamma 2), a protein possessing anti-inflammatory properties, was found to be increased in white adipose tissue and lung of Nfe2l3-deficient mice, suggesting a role for this factor in its transcriptional regulation [107]. Contrary to the highly toxic BHT, butylated hydroxyanisole (BHA), another phenolic antioxidant, is well tolerated in mice [108]. Exposure of wild-type animals to BHA induces hepatic Nqo1 gene expression; similar results were observed in BHA-treated Nfe2l3-deficient mice [54], indicating that expression of Nfe2l3 gene is dispensable, at least in liver, for the regulation of Nqo1 gene expression. This may be due to the fact that Nfe2l3 is not (or only minimally) expressed in the liver [11]. In wound healing experiments, it has been shown that Nfe2l2 −/− mice exhibit increased infiltration of macrophages, resulting in prolonged inflammation [81]. Furthermore, an upregulation of Nfe2l3 mRNA levels was reported in both unwounded and early wounded skin of Nfe2l2 null animals, suggesting that NFE2L3 may compensate in this tissue for the absence of NFE2L2. In addition, the authors proposed that, as demonstrated for NFE2L2, NFE2L3 may be under the control of KGF (Keratinocyte growth factor) in the human keratinocyte cell line HaCaT [81].
Additional support for the importance of NFE2L3 regulation following specific inflammatory cytokines comes from gene regulation studies. It was found that transcripts as well as protein levels of NFE2L3 are upregulated by the cytokine TNF in JAR choriocarcinoma cells (Fig. 4) [10], while genechip array studies showed that interferon-γ increases NFE2L3 mRNA levels in human uterine endothelial cells [109]. However, not all inflammatory cytokines induce expression of NFE2L3 transcripts since interleukin 1beta does not modulate NFE2L3 mRNA levels in human myometrial PHM1-31 cells.
Target genes of NFE2L3
Identification of NFE2L3 target genes is essential for a full understanding of its roles, its regulation, and to identify the pathways regulated by NFE2L3. Although several potential target genes of NFE2L3 have been reported in the literature, no clear evidence has been provided for an in vivo relevance. For instance, reporter assay studies identified NFE2L3 as a negative regulator of the Prdx6 promoter in human pulmonary A549 cells [79, 110]. Hence, it is likely that Prdx6 is a target gene of NFE2L3, at least in an in vitro context. Another candidate gene regulated by NFE2L3 may be Nqo1, since, overexpression of NFE2L3 leads to decreased Nqo1expression in Hep-G2 [87] and smooth muscle cells [79]. The repression requires DNA binding of NFE2L3 to the ARE of the Nqo1promoter, but not the transcriptional activation domain of NFE2L3 [87]. Besides the Nqo1 gene, it was also proposed that the smooth muscle cell marker genes SMαA, SM22α, calponin and Nox4 could be potential targets of NFE2L3 [79]. Finally, the Pparg2 gene and more unpredictably the Nfe2l2 gene may be targets of NFE2L3 as recently suggested [78]. However, further experiments are required to confirm that all genes mentioned above are genuine targets of NFE2L3 in vivo.
Novel possible avenues for NFE2L3 function
A few recent reports shed light on possible novel functions of the NFE2L3 protein. For instance, NFE2L3 was proposed to be used as a stemness marker gene since its mRNA levels had the highest fold change (>300-fold change) when comparing rhesus macaque fibroblasts to pluripotent stem cells [111, 112]. This interesting result was confirmed by other laboratories using rhesus monkey embryonic stem cells transduced with specific transcription factors [113, 114].
Additionally, Pepe and colleagues [79] described a role for NFE2L3 in mouse smooth muscle cell (SMC) differentiation from stem cells. Unfortunately, it is not clear whether the effects observed in smooth muscle cells are due to the ‘A’, ‘B’, or ‘C’ forms of NFE2L3 as the authors observed only one form, for which the molecular weight was not specified, as opposed to the multiple forms of NFE2L3 seen in earlier reports [75, 82].
Genome-wide association studies recently identified NFE2L3 locus as potentially associated with human endometriosis, a common gynecological disease [115] and with waist-to-hip ratio, a measurement of obesity and an indicator for developing serious health conditions including diabetes [116]. Although somewhat unanticipated, these findings will require further attention in order to understand other potential hidden facets of NFE2L3.
Conclusions and perspectives
Recent studies provide exciting novel data that advance our understanding of the NFE2L3 transcription factor at the cellular level as well as in vivo. Indeed, several reports demonstrate that NFE2L3 is a stringently regulated and a post-translationally modified transcription factor [9–11, 75, 78, 79, 82, 86, 87]. Specifically, glycosylation of NFE2L3 appears to be an important modification targeting one form of the transcription factor to the endoplasmic reticulum. One cannot exclude that NFE2L3 protein undergoes additional post-translational modifications such as phosphorylation, sumoylation, and/or ubiquitination. Although it is hypothesized that the nuclear NFE2L3 is cleaved at the N-terminal end, the specific cleavage site still remains unknown [75, 82].
It would certainly be of interest to identify the regulators of NFE2L3 expression. Besides the proinflammatory cytokine TNF and interferon-γ, no other cellular stimuli have yet been described as controlling NFE2L3 levels. In addition, it will be a major challenge to identify the pathways that regulate the activity of this transcription factor and to determine the role played by NFE2L3 in the ER.
Our understanding of NFE2L3 biology has been improved through the analysis of cellular models and the generation of mice deficient in this transcription factor. Absence of NFE2L3 renders the mice susceptible to carcinogen-induced lymphomagenesis [98], suggesting a possible role for NFE2L3 in the regulation of T cells. The fact that NFE2L3 transcripts have been found to be elevated in many different cancer types in gene expression profiling studies [83–85, 100–105] suggests a link of NFE2L3 to human cancer. Hence, NFE2L3 and associated pathways may represent novel potential targets for therapeutic treatment of cancer patients.
What is the role of NFE2L3 in stem cell differentiation? Although several reports have identified Nfe2l3 as a stemness marker gene, because of its early upregulation during stem cell differentiation, its precise role is still unresolved specifically because research on stem cells is as complex as it is fascinating and hopeful. Further efforts need to be devoted to this important area of research.
Finally, identification of the bona fide target genes of NFE2L3 is essential for a full understanding of its functions, its regulation, and to find the pathways regulated by NFE2L3. Although several potential target genes of NFE2L3 have been reported in the literature including Prdx6 [79, 110], Nqo1 [79, 87], SMαA [79], SM22α [79], NOX4 [79], Pparγ 2 [78], and Nfe2l2 [78], no clear evidence has been provided for the in vivo relevance of these data.
Undoubtedly, the most exciting time for research on NFE2L3 is yet to come. There are still many aspects of its regulation and function that need to be better understood, including the control of its subcellular regulation, the range of target genes as well as its precise role in tumorigenesis. NFE2L3 has been the Cinderella of the CNC factors for many years; it is now time for this transcription regulator to come out into the spotlight.
Acknowledgments
We would like to thank Zaynab Nouhi, Meenakshi Kannan, Anna Derjuga, Jadwiga Gasiorek, and Koren K. Mann for critical reading of the manuscript and fruitful discussions. This work was supported by a postdoctoral fellowship of the Fonds de la recherche en santé du Québec (FRSQ) to GC and a grant from the Canadian Institute of Health Research (MOP-97932) to VB.
References
- 1.Sykiotis GP, Bohmann D. Stress-activated Cap‘n’Collar transcription factors in aging and human disease. Sci Signal. 2010;3(112):re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mignotte V, Wall L, deBoer E, Grosveld F, Romeo PH. Two tissue-specific factors bind the erythroid promoter of the human porphobilinogen deaminase gene. Nucleic Acids Res. 1989;17(1):37–54. doi: 10.1093/nar/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mohler J, Vani K, Leung S, Epstein A. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech Dev. 1991;34(1):3–9. doi: 10.1016/0925-4773(91)90086-L. [DOI] [PubMed] [Google Scholar]
- 4.Bowerman B, Eaton BA, Priess JR. skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell. 1992;68(6):1061–1075. doi: 10.1016/0092-8674(92)90078-Q. [DOI] [PubMed] [Google Scholar]
- 5.Chan JY, Han XL, Kan YW. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc Natl Acad Sci USA. 1993;90(23):11371–11375. doi: 10.1073/pnas.90.23.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caterina JJ, Donze D, Sun CW, Ciavatta DJ, Townes TM. Cloning and functional characterization of LCR-F1: a bZIP transcription factor that activates erythroid-specific, human globin gene expression. Nucleic Acids Res. 1994;22(12):2383–2391. doi: 10.1093/nar/22.12.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luna L, Johnsen O, Skartlien AH, Pedeutour F, Turc-Carel C, Prydz H, Kolsto AB. Molecular cloning of a putative novel human bZIP transcription factor on chromosome 17q22. Genomics. 1994;22(3):553–562. doi: 10.1006/geno.1994.1428. [DOI] [PubMed] [Google Scholar]
- 8.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci USA. 1994;91(21):9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi A, Ito E, Toki T, Kogame K, Takahashi S, Igarashi K, Hayashi N, Yamamoto M. Molecular cloning and functional characterization of a new Cap‘n’Collar family transcription factor Nrf3. J Biol Chem. 1999;274(10):6443–6452. doi: 10.1074/jbc.274.10.6443. [DOI] [PubMed] [Google Scholar]
- 10.Chenais B, Derjuga A, Massrieh W, Red-Horse K, Bellingard V, Fisher SJ, Blank V. Functional and placental expression analysis of the human NRF3 transcription factor. Mol Endocrinol. 2005;19(1):125–137. doi: 10.1210/me.2003-0379. [DOI] [PubMed] [Google Scholar]
- 11.Derjuga A, Gourley TS, Holm TM, Heng HH, Shivdasani RA, Ahmed R, Andrews NC, Blank V. Complexity of CNC transcription factors as revealed by gene targeting of the Nrf3 locus. Mol Cell Biol. 2004;24(8):3286–3294. doi: 10.1128/MCB.24.8.3286-3294.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oyake T, Itoh K, Motohashi H, Hayashi N, Hoshino H, Nishizawa M, Yamamoto M, Igarashi K. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16(11):6083–6095. doi: 10.1128/mcb.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kerppola TK, Curran T. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene. 1994;9(3):675–684. [PubMed] [Google Scholar]
- 14.Toki T, Itoh J, Kitazawa J, Arai K, Hatakeyama K, Akasaka J, Igarashi K, Nomura N, Yokoyama M, Yamamoto M, Ito E. Human small Maf proteins form heterodimers with CNC family transcription factors and recognize the NF-E2 motif. Oncogene. 1997;14(16):1901–1910. doi: 10.1038/sj.onc.1201024. [DOI] [PubMed] [Google Scholar]
- 15.Landschulz WH, Johnson PF, McKnight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240(4860):1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 16.Perdomo J, Fock EL, Kaur G, Yan F, Khachigian LM, Jans DA, Chong BH. A monopartite sequence is essential for P45 NF-E2 nuclear translocation, transcriptional activity and platelet production. J Thromb Haemost. 2010;8:2542–2553. doi: 10.1111/j.1538-7836.2010.04058.x. [DOI] [PubMed] [Google Scholar]
- 17.Theodore M, Kawai Y, Yang J, Kleshchenko Y, Reddy SP, Villalta F, Arinze IJ. Multiple nuclear localization signals function in the nuclear import of the transcription factor Nrf2. J Biol Chem. 2008;283(14):8984–8994. doi: 10.1074/jbc.M709040200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshino H, Kobayashi A, Yoshida M, Kudo N, Oyake T, Motohashi H, Hayashi N, Yamamoto M, Igarashi K. Oxidative stress abolishes leptomycin B-sensitive nuclear export of transcription repressor Bach2 that counteracts activation of Maf recognition element. J Biol Chem. 2000;275(20):15370–15376. doi: 10.1074/jbc.275.20.15370. [DOI] [PubMed] [Google Scholar]
- 19.Ohira M, Seki N, Nagase T, Ishikawa K, Nomura N, Ohara O. Characterization of a human homolog (BACH1) of the mouse Bach1 gene encoding a BTB-basic leucine zipper transcription factor and its mapping to chromosome 21q22.1. Genomics. 1998;47(2):300–306. doi: 10.1006/geno.1997.5080. [DOI] [PubMed] [Google Scholar]
- 20.Blouin JL, Duriaux Sail G, Guipponi M, Rossier C, Pappasavas MP, Antonarakis SE. Isolation of the human BACH1 transcription regulator gene, which maps to chromosome 21q22.1. Hum Genet. 1998;102(3):282–288. doi: 10.1007/s004390050692. [DOI] [PubMed] [Google Scholar]
- 21.Ito N, Watanabe-Matsui M, Igarashi K, Murayama K. Crystal structure of the Bach1 BTB domain and its regulation of homodimerization. Genes Cells. 2009;14(2):167–178. doi: 10.1111/j.1365-2443.2008.01259.x. [DOI] [PubMed] [Google Scholar]
- 22.Itoh K, Igarashi K, Hayashi N, Nishizawa M, Yamamoto M. Cloning and characterization of a novel erythroid cell-derived CNC family transcription factor heterodimerizing with the small Maf family proteins. Mol Cell Biol. 1995;15(8):4184–4193. doi: 10.1128/mcb.15.8.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnsen O, Skammelsrud N, Luna L, Nishizawa M, Prydz H, Kolsto AB. Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1. Nucleic Acids Res. 1996;24(21):4289–4297. doi: 10.1093/nar/24.21.4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnsen O, Murphy P, Prydz H, Kolsto AB. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 1998;26(2):512–520. doi: 10.1093/nar/26.2.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marini MG, Chan K, Casula L, Kan YW, Cao A, Moi P. hMAF, a small human transcription factor that heterodimerizes specifically with Nrf1 and Nrf2. J Biol Chem. 1997;272(26):16490–16497. doi: 10.1074/jbc.272.26.16490. [DOI] [PubMed] [Google Scholar]
- 26.Motohashi H, Shavit JA, Igarashi K, Yamamoto M, Engel JD. The world according to Maf. Nucleic Acids Res. 1997;25(15):2953–2959. doi: 10.1093/nar/25.15.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD. The Cap‘n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001;61(8):3299–3307. [PubMed] [Google Scholar]
- 28.Blank V. Small Maf proteins in mammalian gene control: mere dimerization partners or dynamic transcriptional regulators? J Mol Biol. 2008;376:913–925. doi: 10.1016/j.jmb.2007.11.074. [DOI] [PubMed] [Google Scholar]
- 29.Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17(24):3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 30.Jeyapaul J, Jaiswal AK. Nrf2 and c-Jun regulation of antioxidant response element (ARE)-mediated expression and induction of gamma-glutamylcysteine synthetase heavy subunit gene. Biochem Pharmacol. 2000;59(11):1433–1439. doi: 10.1016/s0006-2952(00)00256-2. [DOI] [PubMed] [Google Scholar]
- 31.Motohashi H, O’Connor T, Katsuoka F, Engel J, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294(1–2):1. doi: 10.1016/S0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 32.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22(11):437–441. doi: 10.1016/S0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 33.Rushmore TH, Morton MR, Pickett CB. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J Biol Chem. 1991;266(18):11632–11639. [PubMed] [Google Scholar]
- 34.Klaunig JE, Kamendulis LM, Hocevar BA. Oxidative stress and oxidative damage in carcinogenesis. Toxicol Pathol. 2010;38(1):96–109. doi: 10.1177/0192623309356453. [DOI] [PubMed] [Google Scholar]
- 35.Kundu JK, Surh YJ. Nrf2-Keap1 signaling as a potential target for chemoprevention of inflammation-associated carcinogenesis. Pharm Res. 2010;27(6):999–1013. doi: 10.1007/s11095-010-0096-8. [DOI] [PubMed] [Google Scholar]
- 36.Martin-Montalvo A, Villalba JM, Navas P, de Cabo R. NRF2, cancer and calorie restriction. Oncogene. 2010;30:505–520. doi: 10.1038/onc.2010.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kensler TW, Wakabayashi N. Nrf2: friend or foe for chemoprevention? Carcinogenesis. 2010;31(1):90–99. doi: 10.1093/carcin/bgp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284(20):13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W, Kong AN. Molecular mechanisms of Nrf2-mediated antioxidant response. Mol Carcinog. 2009;48(2):91–104. doi: 10.1002/mc.20465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwak MK, Kensler TW. Targeting NRF2 signaling for cancer chemoprevention. Toxicol Appl Pharmacol. 2010;244(1):66–76. doi: 10.1016/j.taap.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beyer TA, Auf dem Keller U, Braun S, Schafer M, Werner S. Roles and mechanisms of action of the Nrf2 transcription factor in skin morphogenesis, wound repair and skin cancer. Cell Death Differ. 2007;14(7):1250–1254. doi: 10.1038/sj.cdd.4402133. [DOI] [PubMed] [Google Scholar]
- 42.Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev. 2006;38(4):769–789. doi: 10.1080/03602530600971974. [DOI] [PubMed] [Google Scholar]
- 43.Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD. Dual roles of Nrf2 in cancer. Pharmacol Res. 2008;58(5–6):262–270. doi: 10.1016/j.phrs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Motohashi H, Yamamoto M. Nrf2-Keap1 defines a physiologically important stress response mechanism. Trends Mol Med. 2004;10(11):549–557. doi: 10.1016/j.molmed.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 45.Hayes JD, McMahon M. NRF2 and KEAP1 mutations: permanent activation of an adaptive response in cancer. Trends Biochem Sci. 2009;34(4):176–188. doi: 10.1016/j.tibs.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Hayes JD, McMahon M. Molecular basis for the contribution of the antioxidant responsive element to cancer chemoprevention. Cancer Lett. 2001;174(2):103–113. doi: 10.1016/S0304-3835(01)00695-4. [DOI] [PubMed] [Google Scholar]
- 47.Maher J, Yamamoto M. The rise of antioxidant signaling—the evolution and hormetic actions of Nrf2. Toxicol Appl Pharmacol. 2010;244(1):4–15. doi: 10.1016/j.taap.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi M, Yamamoto M. Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv Enzyme Regul. 2006;46:113–140. doi: 10.1016/j.advenzreg.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Itoh K. Disease regulation by Nrf2 antioxidant system. Seikagaku. 2009;81(6):447–455. [PubMed] [Google Scholar]
- 50.Jaiswal AK. Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med. 2004;36(10):1199–1207. doi: 10.1016/j.freeradbiomed.2004.02.074. [DOI] [PubMed] [Google Scholar]
- 51.Kaspar JW, Niture SK, Jaiswal AK. Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic Biol Med. 2009;47(9):1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niture SK, Kaspar JW, Shen J, Jaiswal AK. Nrf2 signaling and cell survival. Toxicol Appl Pharmacol. 2010;244(1):37–42. doi: 10.1016/j.taap.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong KI, Kobayashi A, Katsuoka F, Yamamoto M. Two-site substrate recognition model for the Keap1-Nrf2 system: a hinge and latch mechanism. Biol Chem. 2006;387(10–11):1311–1320. doi: 10.1515/BC.2006.164. [DOI] [PubMed] [Google Scholar]
- 54.Kobayashi A, Ohta T, Yamamoto M. Unique function of the Nrf2-Keap1 pathway in the inducible expression of antioxidant and detoxifying enzymes. Methods Enzymol. 2004;378:273–286. doi: 10.1016/S0076-6879(04)78021-0. [DOI] [PubMed] [Google Scholar]
- 55.Florczyk U, Loboda A, Stachurska A, Jozkowicz A, Dulak J. Role of Nrf2 transcription factor in cellular response to oxidative stress. Postepy Biochem. 2010;56(2):147–155. [PubMed] [Google Scholar]
- 56.Men’shikova EB, Tkachev VO, Zenkov NK. Redox-dependent signaling system Nrf2/are in inflammation. Mol Biol (Mosk) 2010;44(3):389–404. [PubMed] [Google Scholar]
- 57.Jaganjac M. Possible involvement of granulocyte oxidative burst in Nrf2 signaling in cancer. Indian J Med Res. 2010;131:609–616. [PubMed] [Google Scholar]
- 58.Boutten A, Goven D, Boczkowski J, Bonay M. Oxidative stress targets in pulmonary emphysema: focus on the Nrf2 pathway. Expert Opin Ther Targets. 2010;14(3):329–346. doi: 10.1517/14728221003629750. [DOI] [PubMed] [Google Scholar]
- 59.Jung KA, Kwak MK. The Nrf2 system as a potential target for the development of indirect antioxidants. Molecules. 2010;15(10):7266–7291. doi: 10.3390/molecules15107266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrews NC. Erythroid transcription factor NF-E2 coordinates hemoglobin synthesis. Pediatr Res. 1994;36(4):419–423. doi: 10.1203/00006450-199410000-00001. [DOI] [PubMed] [Google Scholar]
- 61.Shivdasani RA. The role of transcription factor NF-E2 in megakaryocyte maturation and platelet production. Stem Cells. 1996;14(Suppl 1):112–115. doi: 10.1002/stem.5530140714. [DOI] [PubMed] [Google Scholar]
- 62.Andrews NC. The NF-E2 transcription factor. Int J Biochem Cell Biol. 1998;30(4):429–432. doi: 10.1016/S1357-2725(97)00135-0. [DOI] [PubMed] [Google Scholar]
- 63.Itou E. The role of NF-E2 related transcription factors in hematopoiesis. Rinsho Ketsueki. 1999;40(4):280–283. [PubMed] [Google Scholar]
- 64.Shivdasani RA. Molecular and transcriptional regulation of megakaryocyte differentiation. Stem Cells. 2001;19(5):397–407. doi: 10.1634/stemcells.19-5-397. [DOI] [PubMed] [Google Scholar]
- 65.Biswas M, Chan JY. Role of Nrf1 in antioxidant response element-mediated gene expression and beyond. Toxicol Appl Pharmacol. 2010;244(1):16–20. doi: 10.1016/j.taap.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Igarashi K, Ota K, Nakame A. Regulation of cellular senescence by Bach1. Nippon Rinsho. 2009;67(7):1423–1428. [PubMed] [Google Scholar]
- 67.Hira S, Tomita T, Matsui T, Igarashi K, Ikeda-Saito M. Bach1, a heme-dependent transcription factor, reveals presence of multiple heme binding sites with distinct coordination structure. IUBMB Life. 2007;59(8–9):542–551. doi: 10.1080/15216540701225941. [DOI] [PubMed] [Google Scholar]
- 68.Igarashi K, Sun J. The heme-Bach1 pathway in the regulation of oxidative stress response and erythroid differentiation. Antioxid Redox Signal. 2006;8(1–2):107–118. doi: 10.1089/ars.2006.8.107. [DOI] [PubMed] [Google Scholar]
- 69.Muto A. Bach2 orchestrates the transcriptional programme of B cell activation. Seikagaku. 2005;77(5):427–431. [PubMed] [Google Scholar]
- 70.Igarashi K, Ochiai K, Muto A. Architecture and dynamics of the transcription factor network that regulates B-to-plasma cell differentiation. J Biochem. 2007;141(6):783–789. doi: 10.1093/jb/mvm106. [DOI] [PubMed] [Google Scholar]
- 71.Motohashi H, Yamamoto M. Carcinogenesis and transcriptional regulation through Maf recognition elements. Cancer Sci. 2007;98(2):135–139. doi: 10.1111/j.1349-7006.2006.00358.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Motohashi H. Small Maf proteins as transcription factors regulating maturation and maintenance of the cell. Seikagaku. 2003;75(9):1193–1201. [PubMed] [Google Scholar]
- 73.Motohashi H. Small Maf proteins in bZip transcription factor net-work. Seikagaku. 2000;72(4):291–295. [PubMed] [Google Scholar]
- 74.Chan JY, Cheung MC, Moi P, Chan K, Kan YW. Chromosomal localization of the human NF-E2 family of bZIP transcription factors by fluorescence in situ hybridization. Hum Genet. 1995;95(3):265–269. doi: 10.1007/BF00225191. [DOI] [PubMed] [Google Scholar]
- 75.Zhang Y, Kobayashi A, Yamamoto M, Hayes JD. The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J Biol Chem. 2009;284(5):3195–3210. doi: 10.1074/jbc.M805337200. [DOI] [PubMed] [Google Scholar]
- 76.Funatsu N, Inoue T, Nakamura S. Gene expression analysis of the late embryonic mouse cerebral cortex using DNA microarray: identification of several region- and layer-specific genes. Cereb Cortex. 2004;14(9):1031–1044. doi: 10.1093/cercor/bhh063. [DOI] [PubMed] [Google Scholar]
- 77.Kokot A, Metze D, Mouchet N, Galibert MD, Schiller M, Luger TA, Bohm M. Alpha-melanocyte-stimulating hormone counteracts the suppressive effect of UVB on Nrf2 and Nrf-dependent gene expression in human skin. Endocrinology. 2009;150(7):3197–3206. doi: 10.1210/en.2008-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chevillard G, Nouhi Z, Anna D, Paquet M, Blank V. Nrf3-deficient mice are not protected against acute lung and adipose tissue damages induced by butylated hydroxytoluene. FEBS Lett. 2010;584(5):923–928. doi: 10.1016/j.febslet.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 79.Pepe AE, Xiao Q, Zampetaki A, Zhang Z, Kobayashi A, Hu Y, Xu Q. Crucial role of nrf3 in smooth muscle cell differentiation from stem cells. Circ Res. 2010;106(5):870–879. doi: 10.1161/CIRCRESAHA.109.211417. [DOI] [PubMed] [Google Scholar]
- 80.Terui K, Takahashi Y, Kitazawa J, Toki T, Yokoyama M, Ito E. Expression of transcription factors during megakaryocytic differentiation of CD34+ cells from human cord blood induced by thrombopoietin. Tohoku J Exp Med. 2000;192(4):259–273. doi: 10.1620/tjem.192.259. [DOI] [PubMed] [Google Scholar]
- 81.Braun S, Hanselmann C, Gassmann MG, auf dem Keller U, Born-Berclaz C, Chan K, Kan YW, Werner S. Nrf2 transcription factor, a novel target of keratinocyte growth factor action which regulates gene expression and inflammation in the healing skin wound. Mol Cell Biol. 2002;22(15):5492–5505. doi: 10.1128/MCB.22.15.5492-5505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nouhi Z, Chevillard G, Derjuga A, Blank V. Endoplasmic reticulum association and N-linked glycosylation of the human Nrf3 transcription factor. FEBS Lett. 2007;581(28):5401–5406. doi: 10.1016/j.febslet.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 83.Willenbrock K, Kuppers R, Renne C, Brune V, Eckerle S, Weidmann E, Brauninger A, Hansmann ML. Common features and differences in the transcriptome of large cell anaplastic lymphoma and classical Hodgkin’s lymphoma. Haematologica. 2006;91(5):596–604. [PubMed] [Google Scholar]
- 84.Kuppers R, Klein U, Schwering I, Distler V, Brauninger A, Cattoretti G, Tu Y, Stolovitzky GA, Califano A, Hansmann ML, Dalla-Favera R. Identification of Hodgkin and Reed-Sternberg cell-specific genes by gene expression profiling. J Clin Invest. 2003;111(4):529–537. doi: 10.1172/JCI16624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Rhee DK, Park SH, Jang YK. Molecular signatures associated with transformation and progression to breast cancer in the isogenic MCF10 model. Genomics. 2008;92(6):419–428. doi: 10.1016/j.ygeno.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 86.Etchevers HC. The Cap‘n’Collar family member NF-E2-related factor 3 (Nrf3) is expressed in mesodermal derivatives of the avian embryo. Int J Dev Biol. 2005;49(2–3):363–367. doi: 10.1387/ijdb.041942he. [DOI] [PubMed] [Google Scholar]
- 87.Sankaranarayanan K, Jaiswal AK. Nrf3 negatively regulates antioxidant-response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. J Biol Chem. 2004;279(49):50810–50817. doi: 10.1074/jbc.M404984200. [DOI] [PubMed] [Google Scholar]
- 88.Wang W, Chan JY. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J Biol Chem. 2006;281(28):19676–19687. doi: 10.1074/jbc.M602802200. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Y, Crouch DH, Yamamoto M, Hayes JD. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem J. 2006;399(3):373–385. doi: 10.1042/BJ20060725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Y, Lucocq JM, Yamamoto M, Hayes JD. The N-terminal homology box 1 (NHB1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also to enable its Asn-glycosylation. Biochem J. 2007;408:161–172. doi: 10.1042/BJ20070786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shivdasani RA, Orkin SH. Erythropoiesis and globin gene expression in mice lacking the transcription factor NF-E2. Proc Natl Acad Sci USA. 1995;92(19):8690–8694. doi: 10.1073/pnas.92.19.8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Farmer SC, Sun CW, Winnier GE, Hogan BL, Townes TM. The bZIP transcription factor LCR-F1 is essential for mesoderm formation in mouse development. Genes Dev. 1997;11(6):786–798. doi: 10.1101/gad.11.6.786. [DOI] [PubMed] [Google Scholar]
- 93.Muto A, Tashiro S, Nakajima O, Hoshino H, Takahashi S, Sakoda E, Ikebe D, Yamamoto M, Igarashi K. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429(6991):566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 94.Sun J, Hoshino H, Takaku K, Nakajima O, Muto A, Suzuki H, Tashiro S, Takahashi S, Shibahara S, Alam J, Taketo MM, Yamamoto M, Igarashi K. Hemoprotein Bach1 regulates enhancer availability of heme oxygenase-1 gene. EMBO J. 2002;21(19):5216–5224. doi: 10.1093/emboj/cdf516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci USA. 1996;93(24):13943–13948. doi: 10.1073/pnas.93.24.13943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, Jackson CW, Hunt P, Saris CJ, Orkin SH. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell. 1995;81(5):695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 97.Chan JY, Kwong M, Lu R, Chang J, Wang B, Yen TS, Kan YW. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 1998;17(6):1779–1787. doi: 10.1093/emboj/17.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chevillard G, Paquet M, Blank V. Nfe2l3 (Nrf3) deficiency predisposes mice to T-cell lymphoblastic lymphoma. Blood. 2011;117(6):2005–2008. doi: 10.1182/blood-2010-02-271460. [DOI] [PubMed] [Google Scholar]
- 99.Pfeifer GP, Denissenko MF, Olivier M, Tretyakova N, Hecht SS, Hainaut P. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21(48):7435–7451. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 100.Thieblemont C, Nasser V, Felman P, Leroy K, Gazzo S, Callet-Bauchu E, Loriod B, Granjeaud S, Gaulard P, Haioun C, Traverse-Glehen A, Baseggio L, Bertucci F, Birnbaum D, Magrangeas F, Minvielle S, Avet-Loiseau H, Salles G, Coiffier B, Berger F, Houlgatte R. Small lymphocytic lymphoma, marginal zone B-cell lymphoma, and mantle cell lymphoma exhibit distinct gene-expression profiles allowing molecular diagnosis. Blood. 2004;103(7):2727–2737. doi: 10.1182/blood-2003-06-2160. [DOI] [PubMed] [Google Scholar]
- 101.Obrador-Hevia A, Fernandez de Mattos S, Villalonga P, Rodriguez J. Molecular biology of mantle cell lymphoma: from profiling studies to new therapeutic strategies. Blood Rev. 2009;23(5):205–216. doi: 10.1016/j.blre.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 102.Rizzatti EG, Falcao RP, Panepucci RA, Proto-Siqueira R, Anselmo-Lima WT, Okamoto OK, Zago MA. Gene expression profiling of mantle cell lymphoma cells reveals aberrant expression of genes from the PI3K-AKT, WNT and TGFbeta signalling pathways. Br J Haematol. 2005;130(4):516–526. doi: 10.1111/j.1365-2141.2005.05630.x. [DOI] [PubMed] [Google Scholar]
- 103.Ortega-Paino E, Fransson J, Ek S, Borrebaeck CA. Functionally associated targets in mantle cell lymphoma as defined by DNA microarrays and RNA interference. Blood. 2008;111(3):1617–1624. doi: 10.1182/blood-2007-02-068791. [DOI] [PubMed] [Google Scholar]
- 104.Almstrup K, Ottesen AM, Sonne SB, Hoei-Hansen CE, Leffers H, Rajpert-De Meyts E, Skakkebaek NE. Genomic and gene expression signature of the pre-invasive testicular carcinoma in situ. Cell Tissue Res. 2005;322(1):159–165. doi: 10.1007/s00441-005-1084-x. [DOI] [PubMed] [Google Scholar]
- 105.Almstrup K, Leffers H, Lothe RA, Skakkebaek NE, Sonne SB, Nielsen JE, Rajpert-De Meyts E, Skotheim RI. Improved gene expression signature of testicular carcinoma in situ. Int J Androl. 2007;30(4):292–302. doi: 10.1111/j.1365-2605.2007.00758.x. [DOI] [PubMed] [Google Scholar]
- 106.Witschi H, Malkinson AM, Thompson JA. Metabolism and pulmonary toxicity of butylated hydroxytoluene (BHT) Pharmacol Ther. 1989;42(1):89–113. doi: 10.1016/0163-7258(89)90023-5. [DOI] [PubMed] [Google Scholar]
- 107.Paola RD, Cuzzocrea S. Peroxisome proliferator-activated receptors and acute lung injury. PPAR Res. 2007;2007:63745. doi: 10.1155/2007/63745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miyakawa Y, Takahashi M, Furukawa F, Toyoda K, Sato H, Hayashi Y. Pneumotoxicity of butylated hydroxytoluene applied dermally to CD-1 mice. Toxicol Lett. 1986;34(1):99–105. doi: 10.1016/0378-4274(86)90151-7. [DOI] [PubMed] [Google Scholar]
- 109.Kitaya K, Yasuo T, Yamaguchi T, Fushiki S, Honjo H. Genes regulated by interferon-gamma in human uterine microvascular endothelial cells. Int J Mol Med. 2007;20(5):689–697. [PubMed] [Google Scholar]
- 110.Chowdhury I, Mo Y, Gao L, Kazi A, Fisher AB, Feinstein SI. Oxidant stress stimulates expression of the human peroxiredoxin 6 gene by a transcriptional mechanism involving an antioxidant response element. Free Radic Biol Med. 2009;46(2):146–153. doi: 10.1016/j.freeradbiomed.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ben-Yehudah A, CAt Easley, Hermann BP, Castro C, Simerly C, Orwig KE, Mitalipov S, Schatten G. Systems biology discoveries using non-human primate pluripotent stem and germ cells: novel gene and genomic imprinting interactions as well as unique expression patterns. Stem Cell Res Ther. 2010;1(3):24. doi: 10.1186/scrt24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450(7169):497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 113.Sritanaudomchai H, Ma H, Clepper L, Gokhale S, Bogan R, Hennebold J, Wolf D, Mitalipov S. Discovery of a novel imprinted gene by transcriptional analysis of parthenogenetic embryonic stem cells. Hum Reprod. 2010;25(8):1927–1941. doi: 10.1093/humrep/deq144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3(6):587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 115.Painter JN, Anderson CA, Nyholt DR, Macgregor S, Lin J, Lee SH, Lambert A, Zhao ZZ, Roseman F, Guo Q, Gordon SD, Wallace L, Henders AK, Visscher PM, Kraft P, Martin NG, Morris AP, Treloar SA, Kennedy SH, Missmer SA, Montgomery GW, Zondervan KT. Genome-wide association study identifies a locus at 7p15.2 associated with endometriosis. Nat Genet. 2011;43(1):51–54. doi: 10.1038/ng.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Heid IM, Jackson AU, Randall JC, Winkler TW, Qi L, Steinthorsdottir V, Thorleifsson G, Zillikens MC, Speliotes EK, Magi R, Workalemahu T, White CC, Bouatia-Naji N, Harris TB, Berndt SI, Ingelsson E, Willer CJ, Weedon MN, Luan J, Vedantam S, Esko T, Kilpelainen TO, Kutalik Z, Li S, Monda KL, Dixon AL, Holmes CC, Kaplan LM, Liang L, Min JL, Moffatt MF, Molony C, Nicholson G, Schadt EE, Zondervan KT, Feitosa MF, Ferreira T, Allen HL, Weyant RJ, Wheeler E, Wood AR, Estrada K, Goddard ME, Lettre G, Mangino M, Nyholt DR, Purcell S, Smith AV, Visscher PM, Yang J, McCarroll SA, Nemesh J, Voight BF, Absher D, Amin N, Aspelund T, Coin L, Glazer NL, Hayward C, Heard-Costa NL, Hottenga JJ, Johansson A, Johnson T, Kaakinen M, Kapur K, Ketkar S, Knowles JW, Kraft P, Kraja AT, Lamina C, Leitzmann MF, McKnight B, Morris AP, Ong KK, Perry JR, Peters MJ, Polasek O, Prokopenko I, Rayner NW, Ripatti S, Rivadeneira F, Robertson NR, Sanna S, Sovio U, Surakka I, Teumer A, van Wingerden S, Vitart V, Zhao JH, Cavalcanti-Proenca C, Chines PS, Fisher E, Kulzer JR, Lecoeur C, Narisu N, Sandholt C, Scott LJ, Silander K, Stark K, Tammesoo ML, Teslovich TM, Timpson NJ, Watanabe RM, Welch R, Chasman DI, Cooper MN, Jansson JO, Kettunen J, Lawrence RW, Pellikka N, Perola M, Vandenput L, Alavere H, Almgren P, Atwood LD, Bennett AJ, Biffar R, Bonnycastle LL, Bornstein SR, Buchanan TA, Campbell H, Day IN, Dei M, Dorr M, Elliott P, Erdos MR, Eriksson JG, Freimer NB, Fu M, Gaget S, Geus EJ, Gjesing AP, Grallert H, Grassler J, Groves CJ, Guiducci C, Hartikainen AL, Hassanali N, Havulinna AS, Herzig KH, Hicks AA, Hui J, Igl W, Jousilahti P, Jula A, Kajantie E, Kinnunen L, Kolcic I, Koskinen S, Kovacs P, Kroemer HK, Krzelj V, Kuusisto J, Kvaloy K, Laitinen J, Lantieri O, Lathrop GM, Lokki ML, Luben RN, Ludwig B, McArdle WL, McCarthy A, Morken MA, Nelis M, Neville MJ, Pare G, Parker AN, Peden JF, Pichler I, Pietilainen KH, Platou CG, Pouta A, Ridderstrale M, Samani NJ, Saramies J, Sinisalo J, Smit JH, Strawbridge RJ, Stringham HM, Swift AJ, Teder-Laving M, Thomson B, Usala G, van Meurs JB, van Ommen GJ, Vatin V, Volpato CB, Wallaschofski H, Walters GB, Widen E, Wild SH, Willemsen G, Witte DR, Zgaga L, Zitting P, Beilby JP, James AL, Kahonen M, Lehtimaki T, Nieminen MS, Ohlsson C, Palmer LJ, Raitakari O, Ridker PM, Stumvoll M, Tonjes A, Viikari J, Balkau B, Ben-Shlomo Y, Bergman RN, Boeing H, Smith GD, Ebrahim S, Froguel P, Hansen T, Hengstenberg C, Hveem K, Isomaa B, Jorgensen T, Karpe F, Khaw KT, Laakso M, Lawlor DA, Marre M, Meitinger T, Metspalu A, Midthjell K, Pedersen O, Salomaa V, Schwarz PE, Tuomi T, Tuomilehto J, Valle TT, Wareham NJ, Arnold AM, Beckmann JS, Bergmann S, Boerwinkle E, Boomsma DI, Caulfield MJ, Collins FS, Eiriksdottir G, Gudnason V, Gyllensten U, Hamsten A, Hattersley AT, Hofman A, Hu FB, Illig T, Iribarren C, Jarvelin MR, Kao WH, Kaprio J, Launer LJ, Munroe PB, Oostra B, Penninx BW, Pramstaller PP, Psaty BM, Quertermous T, Rissanen A, Rudan I, Shuldiner AR, Soranzo N, Spector TD, Syvanen AC, Uda M, Uitterlinden A, Volzke H, Vollenweider P, Wilson JF, Witteman JC, Wright AF, Abecasis GR, Boehnke M, Borecki IB, Deloukas P, Frayling TM, Groop LC, Haritunians T, Hunter DJ, Kaplan RC, North KE, O’Connell JR, Peltonen L, Schlessinger D, Strachan DP, Hirschhorn JN, Assimes TL, Wichmann HE, Thorsteinsdottir U, van Duijn CM, Stefansson K, Cupples LA, Loos RJ, Barroso I, McCarthy MI, Fox CS, Mohlke KL, Lindgren CM. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42(11):949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]