Abstract
The INhibitor of Growth (ING) proteins belong to a well-conserved family which presents in diverse organisms with several structural and functional domains for each protein. The ING family members are found in association with many cellular processes. Thus, the ING family proteins are involved in regulation of gene transcription, DNA repair, tumorigenesis, apoptosis, cellular senescence and cell cycle arrest. The ING proteins have multiple domains that are potentially capable of binding to many partners. It is conceivable, therefore, that such proteins could function similarly within protein complexes. In this case, within this family, each function could be attributed to a specific domain. However, the role of ING domains is not definitively clear. In this review, we summarize recent advances in structure–function relationships in ING proteins. For each domain, we describe the known biological functions and the approaches utilized to identify the functions associated with ING proteins.
Keywords: ING, Tumor suppressor, Protein domain, PHD, NLS
Introduction
The first member of INhibitor of Growth (ING) family was found in 1996 by a strategy based on subtractive hybridization between cDNAs from a normal mammary cell line and seven breast cancer cell lines followed by a subsequent in vivo selection for cDNA fragments capable of promoting neoplastic transformation [1]. This gene is called ING1 which encodes a 33-kDa protein (p33ING1b). Fluorescent in situ hybridization and radiation hybrid mapping linked ING1 to the cytogenetic marker SHGC-5819 at 13q34 [2, 3]. The ING1 gene has three exons and can be alternatively spliced to generate p47ING1a, p33ING1b, p27ING1d, and p24ING1c; the last of these results from an internal initiation at the first ATG within exon 2 [4–8]. Since this discovery, four additional ING genes (ING2-5) encoding proteins and several splicing isoforms of ING2 [9] and ING4 [10] have been identified. By homology search of p33ING1b complementary cDNA sequence, the ING2 (known as ING1L) gene was cloned [11]. In 2003, ING3 encoding a 46.8-kDa protein (p47ING3) with a C-terminal plant homeodomain (PHD) finger motif was subsequently identified through a computational domain search [12]. The same year, the two newest members of the ING family, ING4 and ING5 were identified through a computational sequence homology search for expressed sequence tag clones with a PHD finger motif [13].
ING family and biological functions
The ING family proteins regulate a wide variety of cellular processes. The inhibition of ING1 using antisense expression constructs promotes cell transformation in cell lines and tumor formation in vivo, and blocks cells in the G1 phase of the cell cycle when expressed ectopically [1]. Also, studies have indicated that ING proteins are involved in cell cycle checkpoints and cell cycle progression [14, 15].
ING1 expression is significantly repressed in 44% of human primary breast cancers and 100% of established breast cancer cell lines [16]. Decreased ING1 expression has been found in many other forms of solid and blood tumors [17–26]. Similarly, the expression of ING2, ING3 and ING4 is reduced in human melanoma [27–29]. All ING family proteins have been shown to cooperate with p53 to induce apoptosis and cellular senescence [12, 13, 30–33], and accordingly the notion that the ING family proteins act as class II tumor suppressors has emerged. In addition, suppression of ING proteins has been shown to increase cell migration and to relieve contact inhibition [10, 34, 35].
In addition, many studies using different model systems have implicated the ING family proteins in promotion of apoptosis [30, 31, 36, 37], DNA damage repair [38–40], control of cellular aging [41], negative regulation of cell proliferation [1, 42], chromatin remodeling [43, 44], hormone responses [45, 46] and regulation of tumor growth via NF-κB [47] and hypoxia inducible factor pathways [48, 49]. Several types of tumors have been found to have either altered ING protein subcellular localization, ING mutations, or deletions [28, 50–53]. Various studies have suggested that most of the ING proteins are required for proper p53 function [14, 15], although more recent mouse model experiments indicate otherwise [54].
ING family proteins in chromatin remodeling and gene transcription
The ING proteins have been found in chromatin remodeling complexes [55], indicating that ING proteins may act in the nucleus to regulate transcription [56, 57]. The chromatin structure is very dynamic and is affected by multiple modifications of chromatin-associated proteins, including, but not limited to, histones and remodeling cofactors within particular chromatin regions. Indeed, chromosomal DNA and its associated proteins undergo dramatic alterations in structure during normal cellular processes such as DNA synthesis, transcription and repair [58, 59]. Conversely, it is known that DNA damage leads to changes in gene expression [60–62], and it is now clear that mechanisms that affect directly upon higher-order chromatin structure regulate cellular metabolic processes such as transcription, DNA replication and DNA repair.
Chromatin structure is increasingly being attributed to modification of the basic histones, the subunits of nucleosomes. Histones are positively charged, low molecular weight DNA scaffolding proteins that are subject to numerous posttranslational modifications including acetylation, methylation, phosphorylation, SUMOylation and ubiquitination [63, 64]. These modifications play diverse roles in modulating chromatin structure and have been linked to the regulation of gene transcription [65]. Histone acetylation neutralizes the charge of basic (positively charged) lysine residues within histone proteins. Consequently, there is destabilization of the binding of histones to the negatively charged DNA so that other enzymes/protein complexes are capable of unwinding the chromatin, accessing the DNA at selective sites and transcribing target genes. In other words, the dynamic modification of histones through the enzymatic actions of histone acetyltransferase (HAT) and histone deacetylase (HDAC) protein complexes modifies nucleosome structure, altering the degree of DNA relaxation and subsequently modifying the accessibility of regions of DNA to transcription factors [66].
Not surprisingly, HAT and HDAC protein complex activity must be tightly regulated in order to maintain the appropriate level of histone acetylation in a given cellular environment. In fact, deacetylation of histone residues by HDAC can tighten a DNA strand because of the electric charge change of the histone tails; positively charged histone tails, which have high affinity for negatively charged DNA, can be neutralized by acetylation, causing DNA relaxation. Several HAT/HDAC coactivators and corepressors have been identified. The ING family proteins are involved in chromatin remodeling, and bind to and affect the activity of both HAT and HDAC protein complexes. In fact, ING1 induces histone acetylation, promotes DNA repair and interacts with proliferating cell nuclear antigen (PCNA) [39, 67]. ING2 is also implicated in the initial DNA damage sensing and chromatin remodeling in the nucleotide excision repair (NER) process [38, 68], and recently a new function of ING2 in the control of DNA replication has been found [69].
The functional domains of ING family proteins
ING proteins are a well-conserved family which are present throughout eukaryotic proteomes [70]. Phylogenetic analyses have identified new ING family members in diverse organisms, including rats, frogs, fish, mosquitoes, fruit flies, worms, fungi and plants [70]. All ING genes with the exception of ING3 are found near the ends of chromosomes, and the function and expression of ING5 could be affected by telomere erosion [70]. ING1 has four protein isoforms with identical C-terminus parts containing a conserved PHD finger motif [14]. p33ING2 shares 60% identity with p33ING1b and encodes a 33-kDa protein [11]. Compared to ING1b, ING2 contains an extra and unique leucine zipper domain which is thought to mediate hydrophobic protein–protein interactions [56]. p29ING4 and p28ING5 are highly homologous with 72.8% identity [13].
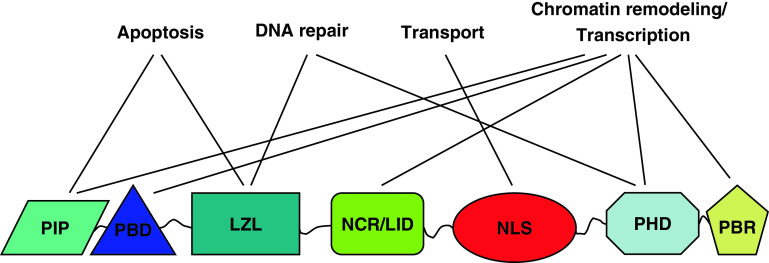
Various studies have suggested that ING family proteins exert their biological functions through their associations with specific molecular partners (Table 1). These associations are possible through various structural and functional domains present in proteins. Thus, they allow the assembly and regulate the subnuclear localization of distinct complexes consisting of different combinations of proteins and interactors. In fact, the ING family members share a highly conserved PHD at their C-termini, a conserved central region containing the nuclear localization signal (NLS) and a variable N-terminal region. Thus, within ING proteins there are a number of distinct domains including PCNA-interacting protein (PIP) box, partial bromodomain (PBD), leucine zipper-like (LZL) domain, novel conserved region/lamin interaction domain (NCR/LID), NLS, PHD, and polybasic region (PBR) (Fig. 1).
Table 1.
Protein or complex reported to bind to ING family protein domain
| Protein | Domain | Protein/complex involved | Reference |
|---|---|---|---|
| ING1 | PHD | ARF | [119] |
| PHD | Brg1, BAF47/53/60/155/170/250 | [44] | |
| PHD | DMAP-1 | [120] | |
| N/A | GADD45 | [40] | |
| PHD | H3K4me2/3 | [103, 105, 121] | |
| PHD | HDAC1 | [122] | |
| N/A | HMT activity | [123] | |
| N/A | hSir2 | [124] | |
| NLS | Karyopherin α,β | [78] | |
| LID | Lamin A | [74] | |
| PHD | mSin3, HDAC1/2, SAP30, RbAp46/48 | [44, 125] | |
| N/A | p15 (PAF) | [126] | |
| N/A | p42, p35 | [125] | |
| N/A | p53 | [127] | |
| PHD | p300 | [122] | |
| PIP | PCNA | [67] | |
| N/A | RBP1 | [125] | |
| N/A | SIRT1 | [128] | |
| N/A | TRRAP, PCAF, CBP | [122] | |
| N/A | 14-3-3 | [129] | |
| ING2 | PHD | H3K4me2/3 | [103, 105] |
| N/A | BAF47/53a/155/170 | [55] | |
| N/A | HMT activity | [123] | |
| PHD | mSin3A, HDAC1/2, RbAp46/48 | [55] | |
| PHD | p300, p300/p53 | [32, 73] | |
| PHD | PtdIns5P | [33] | |
| PHD | SNON | [130] | |
| NCR | PCNA | [69] | |
| N/A | RBP1/RBP1-like | [55] | |
| N/A | SAP30, SAP130, SDS3, BRMS1/BRMS1-like | [131] | |
| N/A | SIRT1 | [128] | |
| ING3 | N/A | AcK5-H4, AcK8-H4, AcK12-H4 | [55] |
| N/A | DMAP1, RUVBL1/2, MRG15, hEaf6, BAF53a | [55] | |
| N/A | GAS41 | [55] | |
| PHD | H3K4me2/3 | [103, 105] | |
| PHD | TIP60, p400, TRRAP, Brd8, EPC1/2 | [55] | |
| ING4 | N/A | AcK5-H4, AcK8-H4, AcK12-H4 | [55] |
| N/A | G3BP2a | [10, 35] | |
| PHD | H3K4me1/2/3 | [103, 105] | |
| PHD | HBO1 | [55] | |
| N/A | hEaf6 | [55] | |
| N/A | HPH-2 | [48] | |
| N/A | JADE1/2/3 | [55] | |
| NCR | Liprin alpha 1 | [10, 35] | |
| N/A | NF-κB p65 | [47, 132] | |
| PHD | p53 | [13, 77] | |
| PHD | p300 | [13] | |
| ING5 | N/A | AcK5-H4, AcK8-H4, AcK12-H4, AcK14-H4 | [55] |
| N/A | BRPF1/2/3 | [55] | |
| PHD | H3K4me1/2/3 | [103, 105, 133] | |
| PHD | HBO1 | [55] | |
| N/A | JADE1/2/3 | [55] | |
| N/A | MCM2/4/6 | [55] | |
| PHD | MOZ/MORF | [55] | |
| PHD | p53 | [13] | |
| PHD | p300 | [13] |
N/A not available
Fig. 1.
The ING family protein domains and their functions. The PIP box binds to PCNA and promotes ING1-mediated apoptosis. The PBD and NCR/LID binds to SAP30, HDAC and HAT, and regulates their activities. The LZL domain of ING2 is required for DNA repair and induction of apoptosis. The NLS binds to the karyopherin-α and β transporter proteins for targeting ING proteins to their functional site, the nucleus and/or the nucleolus for ING1. It has also been reported to mediate interaction with p53 [77]. The PHD motif plays a role in HAT and/or HDAC activity, which can then regulate transcription at specific loci. Both PHD and PBR bind to PtdInsPs, suggesting that PBR may also be involved in chromatin remodeling and transcription regulation
The N-terminus of ING family proteins
The N-termini of ING proteins are more variable and mediate the majority of reported protein–protein interactions and functions as a protein-binding domain that targets distinct nuclear components and chromatin-remodeling complexes [14]. It contains a LZL domain and a NCR [71]. Also, a functionally defined domain, called SAID (SAP30-interacting domain) has been reported for ING1b at the N-terminus [44]. This domain, which was defined as the region of ING1b that directly interacts with SAP30 (the sin3-associated protein 30), is also suspected to be present on ING2, since both of these proteins directly interact with SAP30 [44, 72]. This interaction is thought to bridge ING1 and ING2 to SAP30-containing HDAC1/2 complexes.
ING1 also has in its N-terminus a PIP box through which it binds PCNA in a DNA damage-inducible manner [67]. Since PCNA is an essential factor for DNA replication and repair, ING1 may act to couple these processes to chromatin remodeling. The interaction of this domain is specifically induced upon UV damage [67] and has been hypothesized to switch PCNA activity away from DNA replication towards DNA repair. Among ING members, the PIP box is unique to ING1b. Bioinformatics analysis has revealed a second domain present only in ING1b and called PBD (partial bromodomain because of its sequence homology to bromodomains). The PBD binds SAP30 of mSin3A-HDAC1 which might target HDAC, and possibly HAT activity for some ING proteins [44].
The LZL domain is found in the N-terminus of all ING proteins, except ING1. This domain consists of four to five conserved leucine or isoleucine residues spanning every seven amino acids (forming a hydrophobic patch near the N-terminus) with a similar leucine distribution for ING3 to ING5 [15, 70]. However, little is known about the function of LZL and it been has reported that the LZL domain of ING2 is required for the induction of apoptosis and NER [73]. Truncated ING2 mutants lacking the LZL domain do not display elevated apoptosis following UV exposure [73], suggesting that this domain is required for ING2-mediated apoptosis. RNAi-mediated knockdown of ING2 has also been found to abrogate the NER capacity of melanoma cells [38]. Interestingly, the NER ability of ING2 has been found to require the LZL domain [73]. In 2005, He et al. [70] proposed that this region is responsible for homo- and hetero-oligomerization between the members of the family.
The NCR is found in all ING proteins [70]. It was identified by sequence analyses and constitutes the second most highly conserved domain in the ING family proteins. The NCR domain is now known as LID. This N-terminal region of ING1 has been found to interact directly with lamin A [74]. The NCR/LID domain is found only in ING proteins, through which they bind to and colocalize with lamin A [74], suggesting that the association with nuclear lamina is a common feature of this family. The NCR/LID domain has been speculated to be another region of ING proteins to which HAT and HDAC complexes, including the SAP30 protein, bind by its KIQI or KVQL sequence [15].
The central nuclear localization signal
All ING family proteins contain an NLS with an additional NLS for ING4 and ING5. Recently, many studies have been conducted for ING1 to understand the role of NLS [37, 75], and have shown that NLS deletion results in cytoplasmic accumulation of the protein. The nuclear localization of ING proteins appear critical for their functions, as is evident by the observation of loss of nuclear ING1 staining in a number of cancers [76], and that deleting the entire NLS of ING4 results in a protein that can no longer bind to p53 in cotransfection experiments [77]. Also, the NLS of ING1 contains two copies of a putative nucleolar translocation signal which interacts with the proteins karyopherin-α and -β for nuclear import [78]. The nucleolar translocation of ING1 after exposure to UV light appears to be required for ING-associated apoptosis [37].
The C-terminus plant homeodomain
Among ING family proteins, the greatest homology occurs within the PHD motif [70]. This highly conserved motif is found throughout eukaryotic proteomes, predominantly on chromatin-associated proteins [79]. Structurally, the PHD motif is close to RING (Really Interesting New Gene) and LIM (Lin-11/Isl-1/Mec-3) domains, which contain a zinc-binding domains that ligate two zinc ions [80]. The PHD motif which has been associated with SUMOylation is found in more than 400 eukaryotic proteins and has recently emerged as a chromatin recognition motif that reads the methylation state of histones. The PHD motif comprises approximately 60 amino acids with a C4HC3 signature and belongs to the treble class of zinc-binding domains, containing two zinc ions bound in a cross-braced topology [81–83]. Zinc coordination by PHD fingers is achieved via ligation of zinc atoms to alternating pairs of residues from the consensus Cys4-His-Cys3 sequence distribution: zinc one is bound by Cys1, Cys2, His and Cys6, whereas zinc two is bound by Cys3, Cys4, Cys7 and Cys8 [82, 84–86]. Beyond the conservation of zinc-coordinating residues, approximately 150 distinct PHD-bearing proteins have been predicted to occur in humans [81]. PHD fingers display substantial diversity in their sequences, particularly between Cys6 and Cys7, suggesting that the biological activity of PHD fingers might similarly be diverse [85].
There is much evidence that PHD fingers mediate important physiological functions [81]. Mutations within the PHD fingers of numerous proteins have been implicated in tumorigenesis, as well as the pathogenesis of immunodeficiency syndromes, autoimmune syndromes, and several other genetic disorders [82, 87–89]. Many of these mutations occur at zinc-coordinating residues, indicating that zinc ligation and hence integrity of the PHD finger fold are critical for the function of PHD finger-containing proteins. A second class of disease-linked PHD finger mutations do not disrupt zinc coordination but rather are located between the sixth and seventh zinc-coordinating residues, a segment which, based on known PHD finger structures and structural modeling, is thought to be at or near the surface of the domain. Some have postulated that this surface forms a molecular interaction interface and that mutations within this region might disrupt this activity and in doing so manifest the disease phenotype [85, 90]. In fact, substitution of the basic residues between the sixth and seventh zinc-coordinating residues into alanines disrupts binding of the PHD fingers of ING1, ING2, ATP-dependent chromatin remodeling factor, and recombination activating gene 2 (RAG2) to phosphatidylinositol phosphates (PtdInsPs) [89]. Further, such mutations render ING2 and RAG2 largely inactive [33, 89]. The PHD motif closely resembles a canonical RING domain but lacks the RING E2 ubiquitin ligase activity [83]. Insight into the biological function of PHD fingers comes in part from studies of the structurally related FYVE and RING finger modules [82–84, 86, 91, 92]. The FYVE finger is a well-characterized PtdInsP-binding module, and RING fingers function as components of E3 ubiquitin ligase enzymes [93, 94]. Both of these functions have been reported for PHD fingers from different proteins [33, 95–98], though recent analyses argue that putative PHD fingers with E3-ubiquitin ligase activity are more likely to be RING finger variants rather than true PHD fingers [99, 100].
The phosphoinositide signaling pathways regulate a diverse array of cellular processes including actin polymerization, cell migration, and vesicular trafficking. These phosphoinositide-dependent processes are modulated by the tightly regulated synthesis and metabolism of mono- and polyphosphorylated phosphoinosotide species at discrete subcellular sites [101]. A nuclear phosphoinositide signaling was reported in 2003 by Gozani et al. [33] who identified a physiologically important interaction between PtdIns5P and ING2 mediated by the ING2 PHD motif. In addition, PHD fingers have also been reported to be involved in other protein–protein interactions [91, 102] and to interact with nucleosomes by a direct link to methylated histones (Table 2), specifically H3K4me2 and H3K4me3 [103, 104], supporting a functional role for the PHD motif.
Table 2.
Common function of the PHD of ING proteins in histone H3 tail modification. The ING1–5 PHD domain, known to bind H3K4me2/3, contains conserved Cys and His residues required for the coordination of two zinc ions, residues that make up the aromatic cage and residues that form specific hydrogen bond contacts to the H3K4me3 peptide. In column three, the chromatin remodeling complexes associated with ING proteins that recognize H3K4me are listed. Column four indicates the method used to analyze the interaction between the ING PHD domain and H3K4me peptide
| Protein | H3K4 modification | Associated complex | Method | Reference |
|---|---|---|---|---|
| ING1 | H3K4me2/3 | CBP/p300 HATS | Fluorescence | [105] |
| H3K4me3 | mSin3/HDAC1/2 | Fluorescence | [121] | |
| ING2 | H3K4me2/3 | p300 HAT | Fluorescence | [105] |
| H3K4me3 | mSin3/HDAC1/2 | IP | [103] | |
| ING3 | H3K4me2/3 | TIP60 HAT | Fluorescence | [105] |
| ING4 | H3K4me2/3 | HBO1 HAT | Fluorescence | [105, 134] |
| H3K4me3 | N/A | Fluorescence | [135] | |
| H3K4me3 | N/A | NMR | [114] | |
| H3K4me3 | N/A | NMR | [136] | |
| ING5 | H3K4me2/3 | MOZ/MORF HAT | Fluorescence | [105] |
| H3K4me3 | HBO1 HAT | Fluorescence | [133, 134] |
N/A not available
In fact, the PHD motif, which is the most conserved region within the ING family showing sequence homology greater than 78%, has been found in all human ING proteins to preferentially bind di- and trimethylated H3K4 and repress gene transcription [103, 105, 106]. Mutations in the PHD motif of p33ING1b and a region found to interact with SAP30 (Sin3A Associated Protein 30), a component of Sin3A corepressor complexes [107], abrogate the enhancement of p33ING1b in DNA repair in host cell reactivation assays and radioimmunoassays [50, 108]. Furthermore, the p33ING1b variant is not recruited to UV-induced DNA lesions, but enhances NER in XPC-proficient cells possibly due to its ability to bind XPA [39]. Since XPC/hHR23B acts as the first step of the NER pathway by recognizing helix-distorting DNA lesions and XPA acts to stabilize the resulting open DNA structure [109], p33ING1b may play an essential role in the early steps of the NER pathway, possibly by facilitating access of the NER machinery to chromatin.
Additionally, two p33ING1b PHD mutations (R102L and N260S) were detected in 20% of 46 tested melanomas [50], and either of these alterations proved to be as detrimental as deletion of the entire PHD motif for the enhancement of NER mediated by p33ING1b in host-cell-reactivation assays and radioimmunoassays. Furthermore, those patients bearing an ING1 codon 102 or 260 mutation had a reduced 5-year survival rate (50 vs. 82%) [50]. These findings highlight the importance of the PHD motif in ING1 function and tumor suppression, as loss of NER activity would likely facilitate tumorigenesis by increasing genomic instability. In agreement with this proposal, other reports have indicated the presence of ING1 mutations in the coding region for the PHD motif or the NLS in melanoma, head and neck squamous cell carcinoma, esophageal squamous cell carcinoma, breast cancer, pancreatic cancer, and in colon cancer [14], supporting a role for ING PHD motif in epigenetic regulation of gene expression.
The polybasic region
ING1 and ING2, which are evolutionarily and functionally close [55, 70, 110], contain a short region called the PBR in their C-terminus part, adjacent to the PHD motif. Although the biological functions of this region are not well understood, it has been reported that PtdIns5P bind to the PHD motif of ING2 [33, 111]. Later, it was found that PtdIns3P and PtdIns4P can bind to the PBR [112]. These authors showed that when exchanged between different PHD motif, the PBR is a strong determinant of the binding specificity of PtdInsPs [112]. These findings establish the PBR as a phosphoinositide-binding module and suggest that the PHD domains function downstream of phosphoinositide signaling triggered by the interaction between PBRs and phosphoinositides.
Conclusion
Numerous studies have been done to elucidate the functional mechanisms of ING family proteins. To date, two mechanisms have been clearly identified in ING family proteins that regulate the major biological processes. These are the interaction of the ING PHD domain with methylated histone tails [103, 105, 113, 114], and the binding of ING (ING1-2) proteins to bioactive signaling phospholipids [33, 112, 115] to function as nuclear PtdIns receptors [101, 116, 117]. PtdIns have an important role in mediating a variety of biological processes, including the response to stress, and because they regulate essential cellular functions, PtdIns metabolism is tightly regulated at the subcellular level [118]. Therefore, it is possible that the ING proteins transduce stress signals by binding phospholipids, targeting to chromatin, and reading the local histone code, which subsequently contributes to epigenetic regulation. However, these mechanisms cannot by themselves explain all the events in which ING proteins are involved. This is reinforced by the existence within this family of various functional domains, first among the proteins, then among the isoforms of some proteins. Thus, for a better understanding of the precise function of each domain, it would be necessary to pursue a motif–function relationship study for each member and each isoform of ING family proteins. A systematic functional analysis of different domains to understand their inactivation mechanism and the role in tumor suppression should help to define the functional differences between members of the ING family proteins.
Acknowledgments
This work was supported by the Canadian Institutes of Health Research (MOP-84559 and MOP-93810) and the Canadian Dermatology Foundation to G. Li. R.P.C.W. is a recipient of a Terry Fox Foundation Research Studentship from Canadian Cancer Society Research Institute.
References
- 1.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 2.Garkavtsev I, Demetrick D, Riabowol K. Cellular localization and chromosome mapping of a novel candidate tumor suppressor gene (ING1) Cytogenet Cell Genet. 1997;76:176–178. doi: 10.1159/000134539. [DOI] [PubMed] [Google Scholar]
- 3.Zeremski M, Horrigan SK, Grigorian IA, Westbrook CA, Gudkov AV. Localization of the candidate tumor suppressor gene ING1 to human chromosome 13q34. Somat Cell Mol Genet. 1997;23:233–236. doi: 10.1007/BF02721376. [DOI] [PubMed] [Google Scholar]
- 4.Garkavtsev II. Suppression of the novel growth inhibitor p33ING1 promotes neoplastic transformation. Nat Genet. 1999;23:373. doi: 10.1038/15566. [DOI] [PubMed] [Google Scholar]
- 5.Jager D, Stockert E, Scanlan MJ, Gure AO, Jager E, Knuth A, Old LJ, Chen YT. Cancer-testis antigens and ING1 tumor suppressor gene product are breast cancer antigens: characterization of tissue-specific ING1 transcripts and a homologue gene. Cancer Res. 1999;59:6197–6204. [PubMed] [Google Scholar]
- 6.Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res. 2000;60:3143–3146. [PubMed] [Google Scholar]
- 7.Saito A, Furukawa T, Fukushige S, Koyama S, Hoshi M, Hayashi Y, Horii A. p24/ING1-ALT1 and p47/ING1-ALT2, distinct alternative transcripts of p33/ING1. J Hum Genet. 2000;45:177–181. doi: 10.1007/s100380050206. [DOI] [PubMed] [Google Scholar]
- 8.Cheung KJ, Jr, Li G. The tumor suppressor ING1: structure and function. Exp Cell Res. 2001;268:1–6. doi: 10.1006/excr.2001.5258. [DOI] [PubMed] [Google Scholar]
- 9.Wagner MJ, Helbing CC. Multiple variants of the ING1 and ING2 tumor suppressors are differentially expressed and thyroid hormone-responsive in Xenopus laevis. Gen Comp Endocrinol. 2005;144:38–50. doi: 10.1016/j.ygcen.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Unoki M, Shen JC, Zheng ZM, Harris CC. Novel splice variants of ING4 and their possible roles in the regulation of cell growth and motility. J Biol Chem. 2006;281:34677–34686. doi: 10.1074/jbc.M606296200. [DOI] [PubMed] [Google Scholar]
- 11.Shimada Y, Saito A, Suzuki M, Takahashi E, Horie M. Cloning of a novel gene (ING1L) homologous to ING1, a candidate tumor suppressor. Cytogenet Cell Genet. 1998;83:232–235. doi: 10.1159/000015188. [DOI] [PubMed] [Google Scholar]
- 12.Nagashima M, Shiseki M, Pedeux RM, Okamura S, Kitahama-Shiseki M, Miura K, Yokota J, Harris CC. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene. 2003;22:343–350. doi: 10.1038/sj.onc.1206115. [DOI] [PubMed] [Google Scholar]
- 13.Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y, Appella E, Yokota J, Harris CC. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–2378. [PubMed] [Google Scholar]
- 14.Campos EI, Chin MY, Kuo WH, Li G. Biological functions of the ING family tumor suppressors. Cell Mol Life Sci. 2004;61:2597–2613. doi: 10.1007/s00018-004-4199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32:509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 16.Toyama T, Iwase H, Watson P, Muzik H, Saettler E, Magliocco A, DiFrancesco L, Forsyth P, Garkavtsev I, Kobayashi S, Riabowol K. Suppression of ING1 expression in sporadic breast cancer. Oncogene. 1999;18:5187–5193. doi: 10.1038/sj.onc.1202905. [DOI] [PubMed] [Google Scholar]
- 17.Ohmori M, Nagai M, Tasaka T, Koeffler HP, Toyama T, Riabowol K, Takahara J. Decreased expression of p33ING1 mRNA in lymphoid malignancies. Am J Hematol. 1999;62:118–119. doi: 10.1002/(SICI)1096-8652(199910)62:2<118::AID-AJH11>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 18.Oki E, Maehara Y, Tokunaga E, Kakeji Y, Sugimachi K. Reduced expression of p33(ING1) and the relationship with p53 expression in human gastric cancer. Cancer Lett. 1999;147:157–162. doi: 10.1016/S0304-3835(99)00288-8. [DOI] [PubMed] [Google Scholar]
- 19.Tokunaga E, Maehara Y, Oki E, Kitamura K, Kakeji Y, Ohno S, Sugimachi K. Diminished expression of ING1 mRNA and the correlation with p53 expression in breast cancers. Cancer Lett. 2000;152:15–22. doi: 10.1016/S0304-3835(99)00434-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen L, Matsubara N, Yoshino T, Nagasaka T, Hoshizima N, Shirakawa Y, Naomoto Y, Isozaki H, Riabowol K, Tanaka N. Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Res. 2001;61:4345–4349. [PubMed] [Google Scholar]
- 21.Krishnamurthy J, Kannan K, Feng J, Mohanprasad BK, Tsuchida N, Shanmugam G. Mutational analysis of the candidate tumor suppressor gene ING1 in Indian oral squamous cell carcinoma. Oral Oncol. 2001;37:222–224. doi: 10.1016/S1368-8375(00)00081-6. [DOI] [PubMed] [Google Scholar]
- 22.Bromidge T, Lynas C. Relative levels of alternative transcripts of the ING1 gene and lack of mutations of p33/ING1 in haematological malignancies. Leuk Res. 2002;26:631–635. doi: 10.1016/S0145-2126(01)00185-0. [DOI] [PubMed] [Google Scholar]
- 23.Gunduz M, Ouchida M, Fukushima K, Ito S, Jitsumori Y, Nakashima T, Nagai N, Nishizaki K, Shimizu K. Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31, in human head and neck cancers. Oncogene. 2002;21:4462–4470. doi: 10.1038/sj.onc.1205540. [DOI] [PubMed] [Google Scholar]
- 24.Ito K, Kinjo K, Nakazato T, Ikeda Y, Kizaki M. Expression and sequence analyses of p33(ING1) gene in myeloid leukemia. Am J Hematol. 2002;69:141–143. doi: 10.1002/ajh.10031. [DOI] [PubMed] [Google Scholar]
- 25.Nouman GS, Anderson JJ, Wood KM, Lunec J, Hall AG, Reid MM, Angus B. Loss of nuclear expression of the p33(ING1b) inhibitor of growth protein in childhood acute lymphoblastic leukaemia. J Clin Pathol. 2002;55:596–601. doi: 10.1136/jcp.55.8.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen B, Campos EI, Crawford R, Martinka M, Li G. Analyses of the tumour suppressor ING1 expression and gene mutation in human basal cell carcinoma. Int J Oncol. 2003;22:927–931. [PubMed] [Google Scholar]
- 27.Lu F, Dai DL, Martinka M, Ho V, Li G. Nuclear ING2 expression is reduced in human cutaneous melanomas. Br J Cancer. 2006;95:80–86. doi: 10.1038/sj.bjc.6603205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, Dai DL, Martinka M, Li G. Prognostic significance of nuclear ING3 expression in human cutaneous melanoma. Clin Cancer Res. 2007;13:4111–4116. doi: 10.1158/1078-0432.CCR-07-0408. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis. 2008;29:1373–1379. doi: 10.1093/carcin/bgn086. [DOI] [PubMed] [Google Scholar]
- 30.Cheung KJ, Jr, Li G. p33(ING1) enhances UVB-induced apoptosis in melanoma cells. Exp Cell Res. 2002;279:291–298. doi: 10.1006/excr.2002.5610. [DOI] [PubMed] [Google Scholar]
- 31.Chin MY, Ng KC, Li G. The novel tumor suppressor p33ING2 enhances UVB-induced apoptosis in human melanoma cells. Exp Cell Res. 2005;304:531–543. doi: 10.1016/j.yexcr.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 32.Pedeux R, Sengupta S, Shen JC, Demidov ON, Saito S, Onogi H, Kumamoto K, Wincovitch S, Garfield SH, McMenamin M, Nagashima M, Grossman SR, Appella E, Harris CC. ING2 regulates the onset of replicative senescence by induction of p300-dependent p53 acetylation. Mol Cell Biol. 2005;25:6639–6648. doi: 10.1128/MCB.25.15.6639-6648.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gozani O, Karuman P, Jones DR, Ivanov D, Cha J, Lugovskoy AA, Baird CL, Zhu H, Field SJ, Lessnick SL, Villasenor J, Mehrotra B, Chen J, Rao VR, Brugge JS, Ferguson CG, Payrastre B, Myszka DG, Cantley LC, Wagner G, Divecha N, Prestwich GD, Yuan J. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell. 2003;114:99–111. doi: 10.1016/S0092-8674(03)00480-X. [DOI] [PubMed] [Google Scholar]
- 34.Kim S, Chin K, Gray JW, Bishop JM. A screen for genes that suppress loss of contact inhibition: identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc Natl Acad Sci U S A. 2004;101:16251–16256. doi: 10.1073/pnas.0407158101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen JC, Unoki M, Ythier D, Duperray A, Varticovski L, Kumamoto K, Pedeux R, Harris CC. Inhibitor of growth 4 suppresses cell spreading and cell migration by interacting with a novel binding partner, liprin alpha1. Cancer Res. 2007;67:2552–2558. doi: 10.1158/0008-5472.CAN-06-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Helbing CC, Veillette C, Riabowol K, Johnston RN, Garkavtsev I. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res. 1997;57:1255–1258. [PubMed] [Google Scholar]
- 37.Scott M, Boisvert FM, Vieyra D, Johnston RN, Bazett-Jones DP, Riabowol K. UV induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucleic Acids Res. 2001;29:2052–2058. doi: 10.1093/nar/29.10.2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Chin MY, Li G. The novel tumor suppressor p33ING2 enhances nucleotide excision repair via inducement of histone H4 acetylation and chromatin relaxation. Cancer Res. 2006;66:1906–1911. doi: 10.1158/0008-5472.CAN-05-3444. [DOI] [PubMed] [Google Scholar]
- 39.Kuo WH, Wang Y, Wong RP, Campos EI, Li G. The ING1b tumor suppressor facilitates nucleotide excision repair by promoting chromatin accessibility to XPA. Exp Cell Res. 2007;313:1628–1638. doi: 10.1016/j.yexcr.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 40.Cheung KJ, Jr, Mitchell D, Lin P, Li G. The tumor suppressor candidate p33(ING1) mediates repair of UV-damaged DNA. Cancer Res. 2001;61:4974–4977. [PubMed] [Google Scholar]
- 41.Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagashima M, Shiseki M, Miura K, Hagiwara K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K, Harris CC. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc Natl Acad Sci U S A. 2001;98:9671–9676. doi: 10.1073/pnas.161151798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/MCB.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1) Mol Cell Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner MJ, Gogela-Spehar M, Skirrow RC, Johnston RN, Riabowol K, Helbing CC. Expression of novel ING variants is regulated by thyroid hormone in the Xenopus laevis tadpole. J Biol Chem. 2001;276:47013–47020. doi: 10.1074/jbc.M106965200. [DOI] [PubMed] [Google Scholar]
- 46.Toyama T, Iwase H, Yamashita H, Hara Y, Sugiura H, Zhang Z, Fukai I, Miura Y, Riabowol K, Fujii Y. p33(ING1b) stimulates the transcriptional activity of the estrogen receptor alpha via its activation function (AF) 2 domain. J Steroid Biochem Mol Biol. 2003;87:57–63. doi: 10.1016/S0960-0760(03)00388-1. [DOI] [PubMed] [Google Scholar]
- 47.Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428:328–332. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- 48.Ozer A, Wu LC, Bruick RK. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF) Proc Natl Acad Sci U S A. 2005;102:7481–7486. doi: 10.1073/pnas.0502716102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Piche B, Li G. Inhibitor of growth tumor suppressors in cancer progression. Cell Mol Life Sci. 2010;67:1987–1999. doi: 10.1007/s00018-010-0312-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campos EI, Martinka M, Mitchell DL, Dai DL, Li G. Mutations of the ING1 tumor suppressor gene detected in human melanoma abrogate nucleotide excision repair. Int J Oncol. 2004;25:73–80. [PubMed] [Google Scholar]
- 51.Vieyra D, Senger DL, Toyama T, Muzik H, Brasher PM, Johnston RN, Riabowol K, Forsyth PA. Altered subcellular localization and low frequency of mutations of ING1 in human brain tumors. Clin Cancer Res. 2003;9:5952–5961. [PubMed] [Google Scholar]
- 52.Gunduz M, Nagatsuka H, Demircan K, Gunduz E, Cengiz B, Ouchida M, Tsujigiwa H, Yamachika E, Fukushima K, Beder L, Hirohata S, Ninomiya Y, Nishizaki K, Shimizu K, Nagai N. Frequent deletion and down-regulation of ING4, a candidate tumor suppressor gene at 12p13, in head and neck squamous cell carcinomas. Gene. 2005;356:109–117. doi: 10.1016/j.gene.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 53.Cengiz B, Gunduz E, Gunduz M, Beder LB, Tamamura R, Bagci C, Yamanaka N, Shimizu K, Nagatsuka H (2010) Tumor-specific mutation and down-regulation of ING5 detected in oral squamous cell carcinoma. Int J Cancer. doi: 10.1002/ijc.25224 [DOI] [PubMed]
- 54.Coles AH, Liang H, Zhu Z, Marfella CG, Kang J, Imbalzano AN, Jones SN. Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res. 2007;67:2054–2061. doi: 10.1158/0008-5472.CAN-06-3558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doyon Y, Cayrou C, Ullah M, Landry AJ, Cote V, Selleck W, Lane WS, Tan S, Yang XJ, Cote J. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 56.Feng X, Hara Y, Riabowol K. Different HATS of the ING1 gene family. Trends Cell Biol. 2002;12:532–538. doi: 10.1016/S0962-8924(02)02391-7. [DOI] [PubMed] [Google Scholar]
- 57.Berardi P, Russell M, El-Osta A, Riabowol K. Functional links between transcription, DNA repair and apoptosis. Cell Mol Life Sci. 2004;61:2173–2180. doi: 10.1007/s00018-004-4179-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cunliffe VT. Memory by modification: the influence of chromatin structure on gene expression during vertebrate development. Gene. 2003;305:141–150. doi: 10.1016/S0378-1119(03)00386-X. [DOI] [PubMed] [Google Scholar]
- 60.Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/MCB.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amundson SA, Bittner M, Meltzer P, Trent J, Fornace AJ., Jr Induction of gene expression as a monitor of exposure to ionizing radiation. Radiat Res. 2001;156:657–661. doi: 10.1667/0033-7587(2001)156[0657:IOGEAA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 62.Sesto A, Navarro M, Burslem F, Jorcano JL. Analysis of the ultraviolet B response in primary human keratinocytes using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2002;99:2965–2970. doi: 10.1073/pnas.052678999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wood C, Snijders A, Williamson J, Reynolds C, Baldwin J, Dickman M. Post-translational modifications of the linker histone variants and their association with cell mechanisms. FEBS J. 2009;276:3685–3697. doi: 10.1111/j.1742-4658.2009.07079.x. [DOI] [PubMed] [Google Scholar]
- 64.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta. 2009;1789:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 65.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–599. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 66.Wu RS, Panusz HT, Hatch CL, Bonner WM. Histones and their modifications. CRC Crit Rev Biochem. 1986;20:201–263. doi: 10.3109/10409238609083735. [DOI] [PubMed] [Google Scholar]
- 67.Scott M, Bonnefin P, Vieyra D, Boisvert FM, Young D, Bazett-Jones DP, Riabowol K. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci. 2001;114:3455–3462. doi: 10.1242/jcs.114.19.3455. [DOI] [PubMed] [Google Scholar]
- 68.Li J, Wang Y, Wong RP, Li G. The role of ING tumor suppressors in UV stress response and melanoma progression. Curr Drug Targets. 2009;10:455–464. doi: 10.2174/138945009788185031. [DOI] [PubMed] [Google Scholar]
- 69.Larrieu D, Ythier D, Binet R, Brambilla C, Brambilla E, Sengupta S, Pedeux R. ING2 controls the progression of DNA replication forks to maintain genome stability. EMBO Rep. 2009;10:1168–1174. doi: 10.1038/embor.2009.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He GH, Helbing CC, Wagner MJ, Sensen CW, Riabowol K. Phylogenetic analysis of the ING family of PHD finger proteins. Mol Biol Evol. 2005;22:104–116. doi: 10.1093/molbev/msh256. [DOI] [PubMed] [Google Scholar]
- 71.Ythier D, Larrieu D, Brambilla C, Brambilla E, Pedeux R. The new tumor suppressor genes ING: genomic structure and status in cancer. Int J Cancer. 2008;123:1483–1490. doi: 10.1002/ijc.23790. [DOI] [PubMed] [Google Scholar]
- 72.Loewith R, Smith JS, Meijer M, Williams TJ, Bachman N, Boeke JD, Young D. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J Biol Chem. 2001;276:24068–24074. doi: 10.1074/jbc.M102176200. [DOI] [PubMed] [Google Scholar]
- 73.Wang Y, Wang J, Li G. Leucine zipper-like domain is required for tumor suppressor ING2-mediated nucleotide excision repair and apoptosis. FEBS Lett. 2006;580:3787–3793. doi: 10.1016/j.febslet.2006.05.065. [DOI] [PubMed] [Google Scholar]
- 74.Han X, Feng X, Rattner JB, Smith H, Bose P, Suzuki K, Soliman MA, Scott MS, Burke BE, Riabowol K. Tethering by lamin A stabilizes and targets the ING1 tumour suppressor. Nat Cell Biol. 2008;10:1333–1340. doi: 10.1038/ncb1792. [DOI] [PubMed] [Google Scholar]
- 75.Ha S, Park S, Yun CH, Choi Y. Characterization of nuclear localization signal in mouse ING1 homolog protein. Biochem Biophys Res Commun. 2002;293:163–166. doi: 10.1016/S0006-291X(02)00224-3. [DOI] [PubMed] [Google Scholar]
- 76.Gong W, Suzuki K, Russell M, Riabowol K. Function of the ING family of PHD proteins in cancer. Int J Biochem Cell Biol. 2005;37:1054–1065. doi: 10.1016/j.biocel.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 77.Zhang X, Wang KS, Wang ZQ, Xu LS, Wang QW, Chen F, Wei DZ, Han ZG. Nuclear localization signal of ING4 plays a key role in its binding to p53. Biochem Biophys Res Commun. 2005;331:1032–1038. doi: 10.1016/j.bbrc.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 78.Russell MW, Soliman MA, Schriemer D, Riabowol K. ING1 protein targeting to the nucleus by karyopherins is necessary for activation of p21. Biochem Biophys Res Commun. 2008;374:490–495. doi: 10.1016/j.bbrc.2008.07.076. [DOI] [PubMed] [Google Scholar]
- 79.Sutherland JJ, Higgs RE, Watson I, Vieth M. Chemical fragments as foundations for understanding target space and activity prediction. J Med Chem. 2008;51:2689–2700. doi: 10.1021/jm701399f. [DOI] [PubMed] [Google Scholar]
- 80.Matthews JM, Bhati M, Lehtomaki E, Mansfield RE, Cubeddu L, Mackay JP. It takes two to tango: the structure and function of LIM, RING, PHD and MYND domains. Curr Pharm Des. 2009;15:3681–3696. doi: 10.2174/138161209789271861. [DOI] [PubMed] [Google Scholar]
- 81.Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/S0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 82.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J Mol Biol. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 83.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 84.Capili AD, Schultz DC, Rauscher IF, Borden KL. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 2001;20:165–177. doi: 10.1093/emboj/20.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kwan AH, Gell DA, Verger A, Crossley M, Matthews JM, Mackay JP. Engineering a protein scaffold from a PHD finger. Structure. 2003;11:803–813. doi: 10.1016/S0969-2126(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 86.Bottomley MJ, Stier G, Pennacchini D, Legube G, Simon B, Akhtar A, Sattler M, Musco G. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease. J Biol Chem. 2005;280:11505–11512. doi: 10.1074/jbc.M413959200. [DOI] [PubMed] [Google Scholar]
- 87.Gibbons RJ, Bachoo S, Picketts DJ, Aftimos S, Asenbauer B, Bergoffen J, Berry SA, Dahl N, Fryer A, Keppler K, Kurosawa K, Levin ML, Masuno M, Neri G, Pierpont ME, Slaney SF, Higgs DR. Mutations in transcriptional regulator ATRX establish the functional significance of a PHD-like domain. Nat Genet. 1997;17:146–148. doi: 10.1038/ng1097-146. [DOI] [PubMed] [Google Scholar]
- 88.Saugier-Veber P, Drouot N, Wolf LM, Kuhn JM, Frebourg T, Lefebvre H. Identification of a novel mutation in the autoimmune regulator (AIRE-1) gene in a French family with autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. Eur J Endocrinol. 2001;144:347–351. doi: 10.1530/eje.0.1440347. [DOI] [PubMed] [Google Scholar]
- 89.Elkin SK, Ivanov D, Ewalt M, Ferguson CG, Hyberts SG, Sun ZY, Prestwich GD, Yuan J, Wagner G, Oettinger MA, Gozani OP. A PHD finger motif in the C terminus of RAG2 modulates recombination activity. J Biol Chem. 2005;280:28701–28710. doi: 10.1074/jbc.M504731200. [DOI] [PubMed] [Google Scholar]
- 90.Gozani O, Field SJ, Ferguson CG, Ewalt M, Mahlke C, Cantley LC, Prestwich GD, Yuan J. Modification of protein sub-nuclear localization by synthetic phosphoinositides: evidence for nuclear phosphoinositide signaling mechanisms. Adv Enzyme Regul. 2005;45:171–185. doi: 10.1016/j.advenzreg.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 91.Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, Yan AM, Eberharter A, Gibson TJ, Becker PB, Aasland R. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 92.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 93.Misra S, Miller GJ, Hurley JH. Recognizing phosphatidylinositol 3-phosphate. Cell. 2001;107:559–562. doi: 10.1016/S0092-8674(01)00594-3. [DOI] [PubMed] [Google Scholar]
- 94.Stenmark H, Aasland R, Driscoll PC. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett. 2002;513:77–84. doi: 10.1016/S0014-5793(01)03308-7. [DOI] [PubMed] [Google Scholar]
- 95.Coscoy L, Sanchez DJ, Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol. 2001;155:1265–1273. doi: 10.1083/jcb.200111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu Z, Xu S, Joazeiro C, Cobb MH, Hunter T. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol Cell. 2002;9:945–956. doi: 10.1016/S1097-2765(02)00519-1. [DOI] [PubMed] [Google Scholar]
- 97.Jones DR, Divecha N. Linking lipids to chromatin. Curr Opin Genet Dev. 2004;14:196–202. doi: 10.1016/j.gde.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 98.Uchida D, Hatakeyama S, Matsushima A, Han H, Ishido S, Hotta H, Kudoh J, Shimizu N, Doucas V, Nakayama KI, Kuroda N, Matsumoto M. AIRE functions as an E3 ubiquitin ligase. J Exp Med. 2004;199:167–172. doi: 10.1084/jem.20031291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aravind L, Iyer LM, Koonin EV. Scores of RINGS but no PHDs in ubiquitin signaling. Cell Cycle. 2003;2:123–126. doi: 10.4161/cc.2.2.335. [DOI] [PubMed] [Google Scholar]
- 100.Scheel H, Hofmann K. No evidence for PHD fingers as ubiquitin ligases. Trends Cell Biol. 2003;13:285–287. doi: 10.1016/S0962-8924(03)00102-8. [DOI] [PubMed] [Google Scholar]
- 101.Bunce MW, Bergendahl K, Anderson RA. Nuclear PI(4,5)P(2): a new place for an old signal. Biochim Biophys Acta. 2006;1761:560–569. doi: 10.1016/j.bbalip.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 102.Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, Lacoste N, Cayrou C, Davrazou F, Saha A, Cairns BR, Ayer DE, Kutateladze TG, Shi Y, Cote J, Chua KF, Gozani O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature05140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pena PV, Musselman CA, Kuo AJ, Gozani O, Kutateladze TG. NMR assignments and histone specificity of the ING2 PHD finger. Magn Reson Chem. 2009;47:352–358. doi: 10.1002/mrc.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718–722. doi: 10.1038/nature06034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Laherty CD, Billin AN, Lavinsky RM, Yochum GS, Bush AC, Sun JM, Mullen TM, Davie JR, Rose DW, Glass CK, Rosenfeld MG, Ayer DE, Eisenman RN. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol Cell. 1998;2:33–42. doi: 10.1016/S1097-2765(00)80111-2. [DOI] [PubMed] [Google Scholar]
- 108.Campos EI, Xiao H, Li G. Generation of a polyclonal antibody specifically against the p33(ING1b) tumor suppressor. J Immunoassay Immunochem. 2004;25:71–80. doi: 10.1081/ias-120027227. [DOI] [PubMed] [Google Scholar]
- 109.Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: directional binding of the XPC complex and DNA strand scanning. Mol Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 110.Feng X, Bonni S, Riabowol K. HSP70 induction by ING proteins sensitizes cells to tumor necrosis factor alpha receptor-mediated apoptosis. Mol Cell Biol. 2006;26:9244–9255. doi: 10.1128/MCB.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Huang W, Zhang H, Davrazou F, Kutateladze TG, Shi X, Gozani O, Prestwich GD. Stabilized phosphatidylinositol-5-phosphate analogues as ligands for the nuclear protein ING2: chemistry, biology, and molecular modeling. J Am Chem Soc. 2007;129:6498–6506. doi: 10.1021/ja070195b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kaadige MR, Ayer DE. The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. J Biol Chem. 2006;281:28831–28836. doi: 10.1074/jbc.M605624200. [DOI] [PubMed] [Google Scholar]
- 113.Martin DG, Baetz K, Shi X, Walter KL, MacDonald VE, Wlodarski MJ, Gozani O, Hieter P, Howe L. The Yng1p plant homeodomain finger is a methyl-histone binding module that recognizes lysine 4-methylated histone H3. Mol Cell Biol. 2006;26:7871–7879. doi: 10.1128/MCB.00573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Palacios A, Garcia P, Padro D, Lopez-Hernandez E, Martin I, Blanco FJ. Solution structure and NMR characterization of the binding to methylated histone tails of the plant homeodomain finger of the tumour suppressor ING4. FEBS Lett. 2006;580:6903–6908. doi: 10.1016/j.febslet.2006.11.055. [DOI] [PubMed] [Google Scholar]
- 115.Jones DR, Bultsma Y, Keune WJ, Halstead JR, Elouarrat D, Mohammed S, Heck AJ, D’Santos CS, Divecha N. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell. 2006;23:685–695. doi: 10.1016/j.molcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 116.Shi X, Gozani O. The fellowships of the INGs. J Cell Biochem. 2005;96:1127–1136. doi: 10.1002/jcb.20625. [DOI] [PubMed] [Google Scholar]
- 117.Bunce MW, Gonzales ML, Anderson RA. Stress-ING out: phosphoinositides mediate the cellular stress response. Sci STKE. 2006;2006:pe46. doi: 10.1126/stke.3602006pe46. [DOI] [PubMed] [Google Scholar]
- 118.Irvine RF. Nuclear lipid signalling. Nat Rev Mol Cell Biol. 2003;4:349–360. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- 119.Gonzalez L, Freije JM, Cal S, Lopez-Otin C, Serrano M, Palmero I. A functional link between the tumour suppressors ARF and p33ING1. Oncogene. 2006;25:5173–5179. doi: 10.1038/sj.onc.1209526. [DOI] [PubMed] [Google Scholar]
- 120.Xin H, Yoon HG, Singh PB, Wong J, Qin J. Components of a pathway maintaining histone modification and heterochromatin protein 1 binding at the pericentric heterochromatin in mammalian cells. J Biol Chem. 2004;279:9539–9546. doi: 10.1074/jbc.M311587200. [DOI] [PubMed] [Google Scholar]
- 121.Pena PV, Hom RA, Hung T, Lin H, Kuo AJ, Wong RP, Subach OM, Champagne KS, Zhao R, Verkhusha VV, Li G, Gozani O, Kutateladze TG. Histone H3K4me3 binding is required for the DNA repair and apoptotic activities of ING1 tumor suppressor. J Mol Biol. 2008;380:303–312. doi: 10.1016/j.jmb.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vieyra D, Loewith R, Scott M, Bonnefin P, Boisvert FM, Cheema P, Pastyryeva S, Meijer M, Johnston RN, Bazett-Jones DP, McMahon S, Cole MD, Young D, Riabowol K. Human ING1 proteins differentially regulate histone acetylation. J Biol Chem. 2002;277:29832–29839. doi: 10.1074/jbc.M200197200. [DOI] [PubMed] [Google Scholar]
- 123.Goeman F, Otto K, Kyrylenko S, Schmidt O, Baniahmad A. ING2 recruits histone methyltransferase activity with methylation site specificity distinct from histone H3 lysines 4 and 9. Biochim Biophys Acta. 2008;1783:1673–1680. doi: 10.1016/j.bbamcr.2008.04.019. [DOI] [PubMed] [Google Scholar]
- 124.Kataoka H, Bonnefin P, Vieyra D, Feng X, Hara Y, Miura Y, Joh T, Nakabayashi H, Vaziri H, Harris CC, Riabowol K. ING1 represses transcription by direct DNA binding and through effects on p53. Cancer Res. 2003;63:5785–5792. [PubMed] [Google Scholar]
- 125.Skowyra D, Zeremski M, Neznanov N, Li M, Choi Y, Uesugi M, Hauser CA, Gu W, Gudkov AV, Qin J. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J Biol Chem. 2001;276:8734–8739. doi: 10.1074/jbc.M007664200. [DOI] [PubMed] [Google Scholar]
- 126.Simpson F, Lammerts van Bueren K, Butterfield N, Bennetts JS, Bowles J, Adolphe C, Simms LA, Young J, Walsh MD, Leggett B, Fowles LF, Wicking C. The PCNA-associated factor KIAA0101/p15(PAF) binds the potential tumor suppressor product p33ING1b. Exp Cell Res. 2006;312:73–85. doi: 10.1016/j.yexcr.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 127.Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV. The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature. 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 128.Binda O, Nassif C, Branton PE. SIRT1 negatively regulates HDAC1-dependent transcriptional repression by the RBP1 family of proteins. Oncogene. 2008;27:3384–3392. doi: 10.1038/sj.onc.1211014. [DOI] [PubMed] [Google Scholar]
- 129.Gong W, Russell M, Suzuki K, Riabowol K. Subcellular targeting of p33ING1b by phosphorylation-dependent 14-3-3 binding regulates p21WAF1 expression. Mol Cell Biol. 2006;26:2947–2954. doi: 10.1128/MCB.26.8.2947-2954.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sarker KP, Kataoka H, Chan A, Netherton SJ, Pot I, Huynh MA, Feng X, Bonni A, Riabowol K, Bonni S. ING2 as a novel mediator of transforming growth factor-beta-dependent responses in epithelial cells. J Biol Chem. 2008;283:13269–13279. doi: 10.1074/jbc.M708834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smith KT, Martin-Brown SA, Florens L, Washburn MP, Workman JL. Deacetylase inhibitors dissociate the histone-targeting ING2 subunit from the Sin3 complex. Chem Biol. 2008;17(1):65–74. doi: 10.1016/j.chembiol.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Nozell S, Laver T, Moseley D, Nowoslawski L, De Vos M, Atkinson GP, Harrison K, Nabors LB, Benveniste EN. The ING4 tumor suppressor attenuates NF-kappaB activity at the promoters of target genes. Mol Cell Biol. 2008;28:6632–6645. doi: 10.1128/MCB.00697-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Champagne KS, Saksouk N, Pena PV, Johnson K, Ullah M, Yang XJ, Cote J, Kutateladze TG. The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide. Proteins. 2008;72:1371–1376. doi: 10.1002/prot.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Saksouk N, Avvakumov N, Champagne KS, Hung T, Doyon Y, Cayrou C, Paquet E, Ullah M, Landry AJ, Cote V, Yang XJ, Gozani O, Kutateladze TG, Cote J. HBO1 HAT complexes target chromatin throughout gene coding regions via multiple PHD finger interactions with histone H3 tail. Mol Cell. 2009;33:257–265. doi: 10.1016/j.molcel.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hung T, Binda O, Champagne KS, Kuo AJ, Johnson K, Chang HY, Simon MD, Kutateladze TG, Gozani O. ING4 mediates crosstalk between histone H3 K4 trimethylation and H3 acetylation to attenuate cellular transformation. Mol Cell. 2009;33:248–256. doi: 10.1016/j.molcel.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Palacios A, Munoz IG, Pantoja-Uceda D, Marcaida MJ, Torres D, Martin-Garcia JM, Luque I, Montoya G, Blanco FJ. Molecular basis of histone H3K4me3 recognition by ING4. J Biol Chem. 2008;283:15956–15964. doi: 10.1074/jbc.M710020200. [DOI] [PMC free article] [PubMed] [Google Scholar]