Abstract
Protein post-translational modifications diversify the proteome and install new regulatory levels that are crucial for the maintenance of cellular homeostasis. Over the last decade, the ubiquitin-like modifying peptide small ubiquitin-like modifier (SUMO) has been shown to regulate various nuclear processes, including transcriptional control. In plants, the sumoylation pathway has been significantly implicated in the response to environmental stimuli, including heat, cold, drought, and salt stresses, modulation of abscisic acid and other hormones, and nutrient homeostasis. This review focuses on the emerging importance of SUMO in the abiotic stress response, summarizing the molecular implications of sumoylation and emphasizing how high-throughput approaches aimed at identifying the full set of SUMO targets will greatly enhance our understanding of the SUMO–abiotic stress association.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1094-2) contains supplementary material, which is available to authorized users.
Keywords: Abiotic stress, Arabidopsis, Post-translational modification, SUMO
Introduction
Modulation of protein activity is essential for the functioning of a living organism, particularly during rapid environmental changes, when physiological responses must often occur both quickly and reversibly. This modulation can take place by the addition of small molecules to the target proteins, a process known as post-translation modification (PTM). Important modifiers of proteins include not only phosphate, methyl, acetyl, lipids, and sugars, but also small peptides [1, 2]. Ubiquitin is the foremost example of the latter, but a series of similar ubiquitin-like modifiers (UBLs) have also been described that share analogous structural conformation and conjugation machinery [1, 3, 4]. One such UBL, the small ubiquitin-like modifier (SUMO), is an essential factor in development processes in eukaryotic organisms, being implicated in several cellular mechanisms such as the maintenance of genome integrity, subcellular trafficking, transcription modulation, and regulation of the cell cycle [5, 6]. Unlike ubiquitin, SUMO is not traditionally associated to protein degradation, rather to the control of the target’s conformation, which interferes with protein activity and creates or blocks interacting interfaces depending on the target at hand [7, 8]. Since sumoylation and ubiquitination target the same type of amino acid, they were initially suggested to be antagonistic processes. This notion is currently evolving, as recruitment of ubiquitin by SUMO chains has been shown to occur in humans and yeast via SUMO-targeted ubiquitin ligases (STUbLs) [9]. SUMO may therefore act as a positive regulator of the ubiquitin proteasome system (UPS), though STUbL plant homologs have yet to be established. In support of this mechanism, heat shock has been found to induce the formation of mixed SUMO/ubiquitin chains in Arabidopsis [10].
One unique characteristic of SUMO is environmental stress challenges induce a drastic increase in SUMO-conjugates; this increase seems to be preserved among eukaryotic organisms [11–15]. In the model plant Arabidopsis, SUMO is specifically involved in a plethora of abiotic stress responses, including those to extreme temperatures, water-availability, salinity, oxidative stress, and nutrient imbalance [15–27]. In addition, it is involved in plant development and the response to pathogens [28–30]. Many of the known SUMO targets are related to RNA- and DNA-associated processes, namely transcription factors (TFs) and chromatin-remodeling components [10, 11, 31]. SUMO can be removed from conjugates by SUMO proteases, with the protein then returning to its non-modified state. Thus, the balance between the conjugated/deconjugated forms is a major determinant in the modulation of SUMO-target function [11, 15]. These highly reversible and transient modifications place SUMO as a rapid transcriptional regulator in response to stress.
This review focuses on recent advances regarding the ever-growing link between PTM by SUMO and plant responses to environmental challenges. We also demonstrate how new information on the full range of SUMO targets may bring new insights into the modulation of the plant stress response.
A primer of the sumoylation pathway
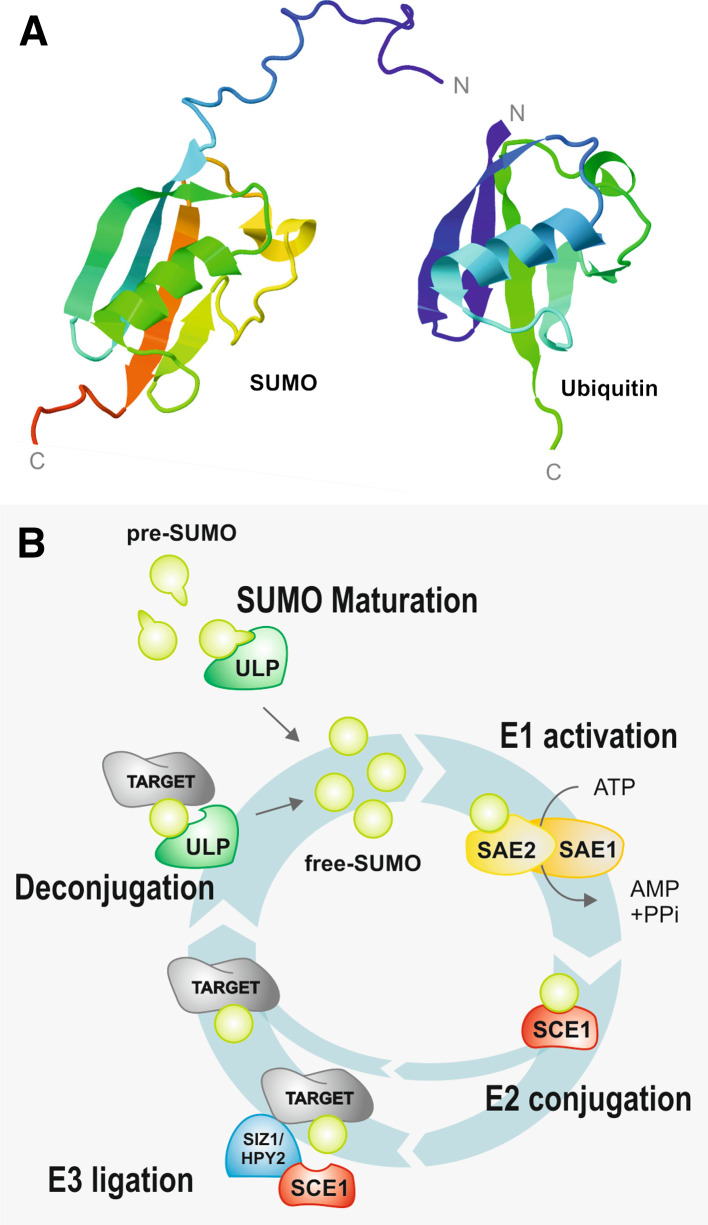
SUMO is a small protein of approximately 100–115 amino acids. Despite its relatively reduced homology to other UBLs, it shares a similar ubiquitin-like structural conformation characterized by a β-grasp fold that seems to act as a multi-functional scaffold (Fig. 1a) [3, 32]. Unlike ubiquitin, SUMO possesses a flexible amino acid extension in its N-terminal end, and its topology is differently charged [3, 33]. The Arabidopsis genome contains eight putative SUMO copies, but only four paralogs have confirmed gene expression (SUM1 ~ SUM2 > SUM3 ~ SUM5) [27]. At least three SUMOs can be found in Oryza sativa and four in Populus trichocarpa [34, 35]. Arabidopsis SUM1 and -2 (SUM1/2) are functionally equivalent [27] and in planta, the SUM1, -3 and -5 isoforms have been shown to conjugate with high-molecular-weight target proteins [36]. SUMO isoforms display different conjugation profiles, and not all isoforms are capable of forming poly-SUMO chains (SUM1/2, but not SUM3) [15, 27, 30, 37, 38]. SUMO profiles show that SUM1/2 and SUM3 have different specificities and possibly different targets. In vitro, conjugation rates are highest for SUM1 and SUM2 ≫ SUM3 > SUM5, possibly because differences in the residues are important for the interaction with the E1 activating enzyme [30, 38].
Fig. 1.
The sumoylation pathway. a Three-dimensional (3D) structure of human small ubiquitin-like modifier (SUMO) 1 (acc. no. 1A5R) and ubiquitin (acc. no. 1UBQ), obtained from the Protein Data Bank (www.rcsb.org/pdb/home/home.do/) and visualized using Jmol, an open-source Java viewer for chemical structures in 3D (www.jmol.org/). b The sumoylation cycle is a conserved five-step pathway (involving maturation, E1-activation, E2-conjugation, E3-ligation, deconjugation) and mediates the balance between the conjugated/deconjugated forms of a target protein. SUMO isoforms encode a pre-SUMO peptide that undergoes maturation by ubiquitin-like proteases (ULP). These SUMO-specific cysteine endopeptidases cleave the C-terminal end, exposing a di-glycine (GG) motif. In the presence of ATP, heterodimeric E1 SUMO-activating enzymes 1 and 2 (SAE1, SAE2) promote the C-terminal binding of SUMO to AMP (SUMO-AMP). A SUMO glycine (G) residue is also coupled to a cysteine (C) of the SAE2, through a high-energy thioester bond. The peptide is then conjugated to an E2 SUMO-conjugating enzyme (SCE1), through transesterification of a C residue in the E2. E2s are subsequently capable of transferring SUMO to a target protein. This step is mostly mediated by SUMO E3 ligases, even though E3-independent transfer is possible. An isopeptide bond is generated between the SUMO G residue and the ε-amino group of a lysine (K) side chain in the target protein’s sumoylation consensus motif ψKXE (ψ, large hydrophobic residue; K, lysine; X, any amino acid; E, glutamic acid), although alternative sumoylation sites also exist. ULPs display isopeptidase in addition to endopeptidase activity, deconjugating SUMO from the target. This final step recycles SUMO and, most significantly, mediates the balance between the target’s conjugated/deconjugated forms
SUMO ubiquitin-like proteases (ULP), also designated sentrin/SUMO-specific proteases (SENP), process pre-SUMOs by removing C-terminal amino acids, exposing a di-glycine motif. Sumoylation by which the maturated SUMO is covalently attached to a target protein occurs through a three-step cascade (E1, E2, E3) similar to the ubiquitin pathway (Fig. 1b). The E1 (SUMO activating enzyme: SAE1–SAE2 heterodimer) promotes the ATP-dependent activation of SUMO, while the E2 (SUMO conjugating enzyme: SCE) mediates conjugation of SUMO to a target protein. SUMO E3 ligases enhance the conjugation step. SUMO can be removed by the action of SUMO proteases, thereby recycling free SUMO into the pathway (Fig. 1b). Conjugation traditionally occurs in a lysine residue of the target protein, within a sumoylation consensus motif ψKXE (ψ, large hydrophobic residue; K, lysine; X, any amino acid; E, glutamic acid). Several alternative SUMO-conjugation sites have also been described, including the inverted consensus motif, hydrophobic cluster motif, phosphorylation-dependent SUMO motif (PDSM), and the negatively charged amino acid-dependent SUMO motif (NDSM) [2, 39]. Positioning of the motif within the target is extremely important. Most validated SUMO consensus sites tend to be placed in extended loops or intrinsically disordered regions of the substrate outside of its globular fold, since the motif adopts an extended conformation to interact effectively with the E2. In addition, SUMO interacting motifs (SIMs) mediate non-covalent interactions between SUMO and various different SIM-containing proteins, adding complexity to the network of SUMO-dependent protein interactions. SIMs are traditionally composed of a short stretch of hydrophobic amino acids (V/I)X(V/I)(V/I), flanked by acidic residues [39].
Orthologs for the full scope of the SUMO pathway components can be found in plant genomes. Genomic studies in Arabidopsis thaliana have validated the existence of a functional SUMO pathway in plants, revealing the important role of this pathway in developmental processes and the plant’s response to external stimuli (Table 1). Mutations that disrupt the main conjugation machinery, i.e., SUMO peptides (SUM1/2), the SAE2 subunit of the E1 heterodimer, and/or the SUMO E2 conjugation enzyme SCE1, result in developmental arrest at the early stages of embryogenesis [27]; a similar finding has been observed in other models [40]. However, over-expression of SUMOs results in growth-defective plants [30, 36]. To date, two E3 ligases have been characterized in Arabidopsis, the SIZ/PIAS-type SAP and Miz 1 (SIZ1) and the NSE2/MMS21-type High ploidy 2 (HPY2), both with pleiotropic phenotypes in loss-of-function mutants, evidencing the importance of E3s within the pathway [16, 21, 29, 41–43]. SUMO proteases are more abundant in the genome than any other SUMO pathway component, resulting in a high degree of redundancy [37, 44, 45]. Mutants also display developmental phenotypes: Early in Short Days 4 (ESD4) mutants are severely dwarfed, and their developmental defects are incremented by the over-expression of SUM1 [46]; ULP1c and ULP1d, also designated Overly Tolerant to Salt 2 and -1 (OTS2 and -1), respectively, act redundantly to regulate flowering and rosette growth (Castro et al., unpublished data) [26]. More information can be found in a series of excellent reviews that recently addressed the diversity of the plant SUMO machinery and its impact on plant development [4, 45, 47].
Table 1.
Expressed Arabidopsis small ubiquitin-like modifier pathway components
| SUMO pathway component (AGI code) | Loss- or gain-of-function allele | Developmental phenotype | Abiotic stress-related phenotype | Reference |
|---|---|---|---|---|
| SUMO peptide | ||||
| SUM1 (At4g26840) | sum1-1 | Wild type | [27] | |
| 35S∷SUM1 | Early flowering short petioles | Lower ABA root growth inhibition; decreased acquired thermotolerance | [27, 30, 55, 76] | |
| SUM2 (At5g55160) | sum2-1 | Wild-type | [27] | |
| 35S∷SUM2 | Early flowering short petioles | Lower ABA root growth inhibition | [30, 76] | |
| sum1-1 sum2-1 | Embryo lethal | [27] | ||
| sum1-1 amiR-SUM2 | Pleiotropic | [30] | ||
| SUM3 (At5g55170) | sum3-1 | Late flowering | [30] | |
| 35S∷SUM3 | Early flowering | [30] | ||
| SUM5 (At2g32765) | n.d | n.d | ||
| E1 (activation) | ||||
| SAE1a (At4g24940) | sae1a-1 | Wild-type | [27] | |
| SAE1b (At5g50580) | n.d | n.d | ||
| SAE2 (At2g21470) | sae2-1 | Embryo lethal | [27] | |
| E2 (conjugation) | ||||
| SCE1 (At3g57870) | sce1-5, sce1-6 | Embryo lethal | [27] | |
| co-SCE1a a | n.d | Higher ABA root growth inhibition | [76] | |
| E3 (ligation) | ||||
| HPY2/MMS21 (At3g15150) | hpy2-1, hpy2-2, mms21-1 | Pleiotropic | [42, 43] | |
| SIZ1 (At5g60410) | siz1-1, siz1-2, siz1-3 | Pleiotropic | Sensitivity to extreme temperatures, drought and copper excess; abnormal Pi-starvation responses; higher ABA-induced inhibition of germination and root growth; impaired in N-metabolism; tolerance to salt | [16–25] |
| Protease | ||||
| ESD4 (At4g15880) | esd4-1, esd4-2 | Pleiotropic | [46, 99] | |
| 35S∷ESD4 | Wild-type | [46] | ||
| esd4-1 35S∷SUM1,2,3 | Pleiotropic | [46] | ||
| esd4-1 35S∷preSUM1,2,3 | Pleiotropic | [46] | ||
| ULP1a/ELS1 (At3g06910) | els1-1, els1-2 | Slightly smaller | [100] | |
| esd4-2 els1-1 | Pleiotropic | [100] | ||
| ULP1b (At4g00690) | n.d | n.d | ||
| ULP1c/OTS2 (At1g10570) | ots2-1 | Wild-type | [26] | |
| ULP1d/OTS1 (At1g60220) | ots1-1 | Wild-type | [26] | |
| 35S∷OTS1 | Salt tolerance | [26] | ||
| ots1-1 ots2-1 | Early flowering | Salt sensitivity | [26] | |
| ots1-1 ots2-1 35S∷HA:SUM1 | Smaller rosette | [101] | ||
| ULP2a (At4g33620) | n.d | n.d | ||
| ULP2b (At1g09730) | n.d | n.d | ||
SUMO small ubiquitin-like modifier, n.d not determined, ABA abscisic acid, Pi inorganic phosphate
aCo-supression line
The SUMO–abiotic stress association
The accumulation of SUMO-conjugates during stress is ubiquitous in eukaryotes [11, 14, 15]. In plants it has been observed in rice, poplar and, more frequently, Arabidopsis following heat shock [15, 16, 22, 23, 27, 30], cold shock [17, 48], drought [21], salt stress [26], exposure to excessive copper [25], and incubation with hydrogen peroxide, ethanol, and canavanine [15]. Conjugation is accompanied by a decrease in the pool of free SUMOs and correlates with the duration and intensity of the stress [15, 49]. In the absence of the stimulus, SUMO-conjugate levels decrease within hours or even minutes, suggesting that sumoylation acts transiently [11, 15].
Functional approaches using Arabidopsis thaliana knockout mutants have implicated various SUMO pathway components in abiotic stress responses (Table 1). The lethality of the SUM1/2, E1, and E2 knockouts has meant that most evidence has been obtained in E3 and ULP mutants. Null SIZ1 alleles (siz1-1, siz1-2, and siz1-3) display a series of abiotic stress-related phenotypes, including sensitivity to extreme temperatures, drought stress, and excess copper, altered phosphate-starvation responses, reduced nitrogen (N) assimilation, and salt tolerance (Table 1) [16–25]. SIZ/PIAS family members are composed of different regulatory domains [50], and directed mutation studies have implicated the SIZ1 SP-RING domain (essential for SUMO conjugation and nuclear localization) in heat shock sensitivity during germination [23]. In rice, the two SIZ1 orthologs (OsSIZ1/2) are involved in heat stress-induced sumoylation, but they can only partially complement the Arabidopsis siz1 mutant [51], suggesting that OsSIZ1 and -2 have slightly different functions. The accumulation of SUMO-conjugate levels during heat, cold, and drought stress and following exposure to excess copper has been shown to be essentially SIZ1 dependent, although the slight but visible presence of stress-responsive SUMO-conjugates in siz1 suggests either alternative E3s or E3-independent conjugation [16, 17, 21, 25, 27]. HPY2, an E3 ligase that also displays an SP-RING domain, has been mainly associated with the regulation of cell cycle division, and no role in abiotic stress resistance has yet been attributed to this ligase [42, 43]. There are a number of other genes in the Arabidopsis genome possessing an SP-RING domain which are potential SUMO E3 ligases, including PIAS-like 1 (At1g08910) and PIAS-like 2 (At5g41580) proposed by Novatchkova and co-workers [52]. Interestingly, PIAS-like 2 has been found to be modified by SUM1 [10], although its involvement in stress-responses has yet to be reported.
Relative to other SUMO pathway components, there are a larger number of plant SUMO proteases and these have different SUMO isoform discrimination and enzymatic activities [37, 44]. Plant SUMO proteases display some degree of functional redundancy which has delayed their characterization. The fact that SUMO targets seem to be conjugated transiently following stress imposition implicates ULP-dependent deconjugation in the abiotic stress response. The identification of abiotic stress-related phenotypes has been limited to the redundant gene pair ULP1c/OTS2 and ULP1d/OTS1. Conti and co-workers [26] reported that this ULP1 pair is a determinant of salt tolerance, and subsequent evidence suggests they also act as negative regulators of drought tolerance (Castro et al., unpublished data).
Identification of SUMO targets
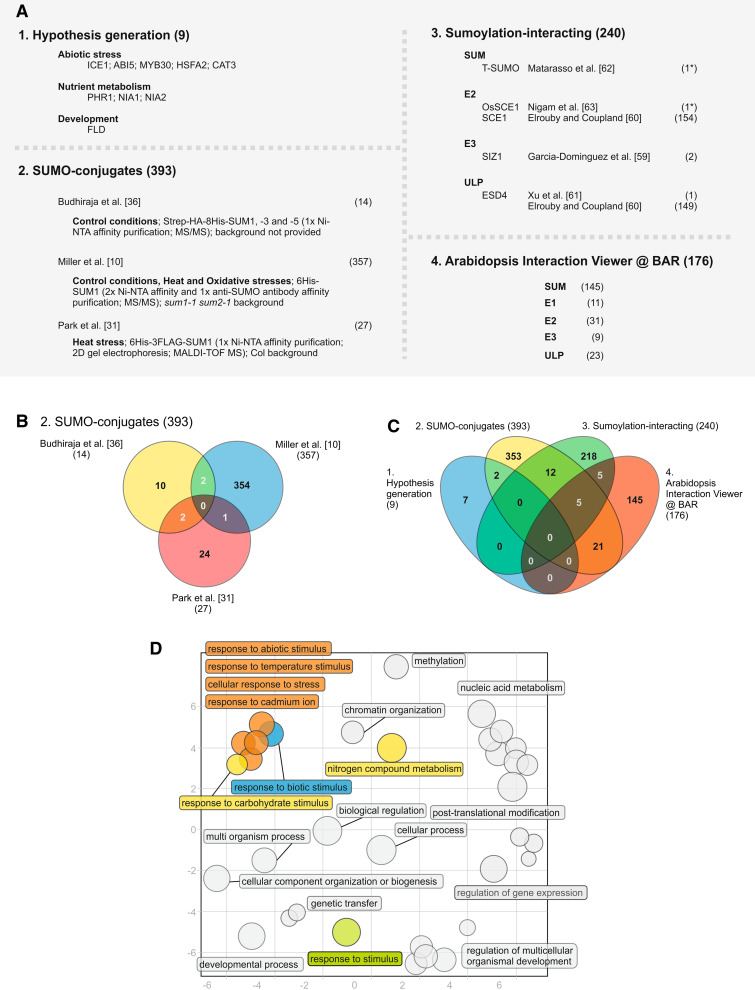
Identification of the full set of sumoylated proteins is a major objective of current SUMO research, as it provides a molecular link between SUMO function and the numerous phenotypes displayed by SUMO pathway components. In non-plant models, various strategies have been employed to screen for SUMO targets, namely, purification of epitope-tagged SUMO, use of anti-SUMO antibodies, or isolation through SIMs [2, 53]. In plants, initial approaches relied on hypothesis generation to identify candidate genes, based on phenotypic evidence and literature mining, and resulted in the identification of nine proteins that are sumoylated [Fig. 2a, subset 1; Electronic Supplementary Material (ESM) File 1] [16–18, 24, 38, 41, 54, 55]. Candidate genes were validated through a series of in bacteria, in planta, or in vitro studies. The majority of proteins play a regulatory role in gene expression, which is consistent with traditional SUMO function [56–58]. Importantly, most proteins are involved in abiotic stress responses, thereby validating the physiological and functional data in support of a major role for sumoylation in abiotic stress resistance. However, the discovery rate using candidate gene approaches is slow when the large number of hypothesized sumoylation targets within the plant proteome is taken into account. This limitation has led to a recent series of systematic functional genomics approaches being used to identify SUMO targets (Fig. 2a; ESM File 1). These approaches can be categorized into the in planta screening of Tag-SUMO-conjugates coupled with peptide sequencing (herein designated SUMO-conjugates) [10, 31, 36] or the identification of protein–protein interaction (PPI) partners of the sumoylation machinery (herein designated Sumoylation-interacting) [59–63].
Fig. 2.
Annotation and characterization of the predicted plant SUMO targets. a The four major strategies adopted for identifying plant SUMO targets have rendered a total of 768 proteins. b Venn diagram analysis of the three existing SUMO-conjugate studies. c Venn diagram analysis of the four subsets of strategies used to identify SUMO targets. d Scatterplot of enriched gene ontology (GO) terms (biological process) for the subset of SUMO-conjugates. GO functional categorization was performed using VirtualPlant 1.2 software (http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/), using the BioMaps function with a 0.01 p-value cutoff [102]. Exclusion of GO term redundancy and subsequent scatterplot analysis were performed using the REVIGO tool (http://revigo.irb.hr/), with a 0.5 C-value [65]. Bubble size indicates the frequency of the GO term, colored circles indicate GO terms related to stress or nutritional stimuli. The scatterplot represents the cluster representatives in a 2D space (x- and y-axis) derived by applying multidimensional scaling to a matrix of the GO terms’ semantic similarities [65]. # Number of genes within the subset, asterisk non-Arabidopsis genes, MALDI–TOF MS matrix-assisted laser desorption/ionization–time of flight mass spectrometry
In plants, mass identification of SUMO-conjugates (Fig. 2a, subset 2) was first performed by Budhiraja and co-workers [36] through in vivo expression of HIS-tagged SUM1, -3, and -5. Single-step enrichment by affinity column chromatography was used before mass spectrometric protein identification, revealing 14 putative SUMO targets. Five of the candidates were subsequently shown to be sumoylated in vitro. Most targets are involved in DNA-related or RNA-dependent processes, namely, the regulation of chromatin structure, splicing, translation, and assembly and dis-assembly [36]. The highest rending SUMO-conjugate assay was performed by Miller and co-workers [10], who developed a stringent method to isolate a total of 357 His-SUM1-conjugating proteins from Arabidopsis. Given the known involvement of SUMO in abiotic stress, Arabidopsis plants were subjected to heat and oxidative stresses in addition to the control treatment. Once more, the majority of targets consisted of nuclear proteins involved in chromatin remodeling/repair, transcription, RNA metabolism, and protein trafficking. Interestingly, many were condition specific, which supports a stress-specific modulation of the pool of SUMO-conjugates. Park and co-workers [31] used two-dimensional (2D) gel electrophoresis to screen for SUMO targets following heat stress imposition and identified a total of 27 proteins involved in DNA- or RNA-related metabolism, signaling pathways, and general metabolism. The seemingly deficient coverage of SUMO targets evidenced by Budhiraja et al. [36] and Park et al. [31] may be due to the use of overextended tags, which have been shown to compromise SUMO function in Arabidopsis [10]. For instance, 6His-FLAG3-SUM1 proteins failed to identify SUMO-conjugates under conditions of no stress, when SUMO conjugation is lowest [31]. Tagged SUMOs may also compete deficiently with the native peptide, a problem that was overcome by Miller and co-workers’ [10] use of a sum1-1 sum2-1 background. As a result there is no significant overlap between the three sets of SUMO-conjugates, as evidenced by Venn diagram analysis (Fig. 2b).
In a sumoylation-interacting approach (Fig. 2a, subset 3), a high-throughput strategy aimed at identifying SUMO targets was carried out by Elrouby and Coupland [60], who used a yeast two-hybrid (Y2H) system to identify 238 interactors of SUMO pathway components SCE1 and/or ESD4. An Escherichia coli-based sumoylation system was used to test a substantial number of targets, indicating that approximately half are bona fide SUMO substrates. Proteins involved in stress responses, namely temperature stress, were shown to be over-represented within Y2H interactors. A similar screening using SIZ1 as bait resulted in the identification of GTE3 and GTE5, members of global transcription factor group E that contain a bromodomain that is possibly involved in binding to acetylated histones [59]. Other Y2H interactions have been reported, including the interaction of Nuclear Pore Anchor (NUA) protein with ESD4. In other models, tomato Cys protease LeCp interacted with the SUM1/2 ortholog T-SUMO, and rice OsFKB20, a stress-inducible FK506-binding protein, interacted with OsSCE1 [61–63]. As an additional source of potential SUMO targets, we used the Arabidopsis Interactions Viewer function from BAR [64], a database of almost 105 predicted and confirmed Arabidopsis interacting proteins, to identify estimated interactors for all components of the sumoylation machinery (Fig. 2a, subset 4; ESM File 1). Our analysis rendered a total of 176 predicted interactors, mostly associated with SUMO peptides.
We cross-referenced all predicted plant SUMO targets in order to obtain an overview of all four subsets of proteins (Fig. 2c; ESM File 1). Not surprisingly, four out of the five most over-represented proteins included SUM1, SAE2, SCE1, and SUMO E3 ligase candidate PIAS-like 2, which validates the current analysis. However, there was still no significant overlap between subsets, similar to an analogous study of yeast SUMO targets [53]. This limited overlap suggests that saturation is far from being achieved; however, it may also reflect the different methodologies employed, particularly considering that PPI-based approaches (subsets 3 and 4) may detect non-covalent interactions mediated by SIMs rather than the bona fide sumoylation of substrates. Since SUMO-conjugate genes provide the highest confidence candidates, we analyzed gene ontology (GO) term enrichment for this subset (Fig. 2d). The REVIGO tool was used to exclude redundant GO terms, as redundancy tends to confound interpretation and inflate the perceived number of biologically relevant results [65]. As expected, functional categorization of biological processes revealed standard roles in SUMO function. However, over-represented GO terms also included stimuli that have been physiologically and functionally associated with the sumoylation pathway, namely, abiotic stress and nutrient-related stimuli. Using a detailed GO term categorization of the subset of 393 SUMO-conjugates, we identified 52 abiotic stress-related proteins (ESM File 2). These form a core of highly likely SUMO targets that link SUMO function to a wide range of abiotic stress responses. In non-plant models, many known targets are regulators of expression (acting as transcription factors, co-activators, or repressors) [40]. A detailed analysis of the 52 genes we identified reveals a strong involvement in transcriptional regulation and nucleic acid binding activities, concomitant with the role for SUMO in the control of transcription during environmental challenges already envisaged for known plant SUMO targets [16, 17, 55].
Molecular basis of SUMO regulation of abiotic stress tolerance
Extreme temperatures
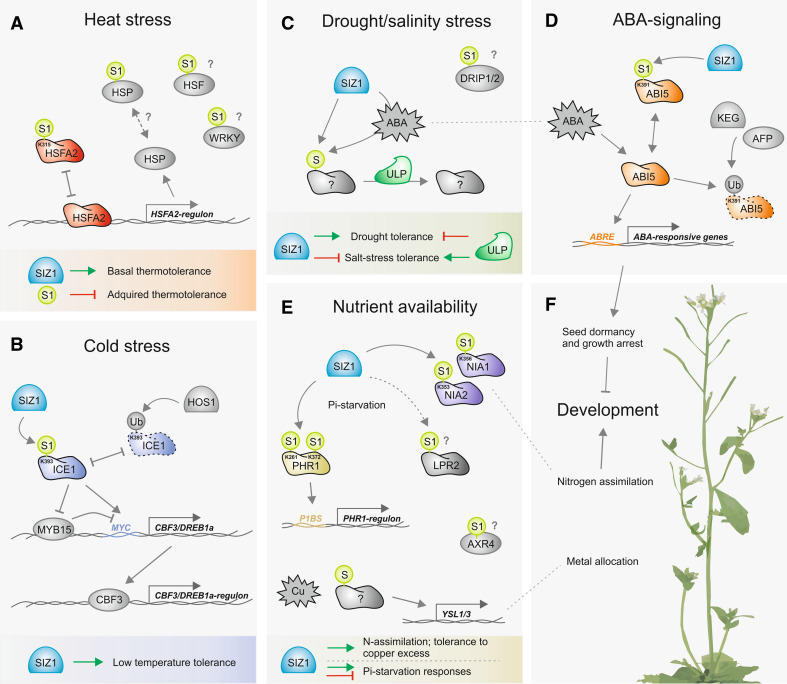
During heat stress, protein stability is compromised, which affects cellular structures and organelles, including the nucleus [66]. The best documented resistance proteins comprise transiently expressed heat shock proteins (HSPs) which act as molecular chaperones of the native protein structure [66, 67], as well as heat shock factors (HSFs) that function as key signaling effectors, modulating the transcription of heat-responsive genes [67]. Both types of proteins can be abundantly found in confirmed or predicted SUMO conjugates, including HSFA1D, HSFA2, HSFB2B, HSP70-1/HSC70-1, HSP17.4, HSC70-3/HSP70-3, HSP17.6C-CI, and HSP70. HSP70 proteins are particularly over-represented in the different subsets of sumoylated proteins (ESM File 1), which is consistent with their central role in protein folding processes, namely, during external stress [68]. Interestingly, over-expression of HSC70 results in a reduced accumulation of SUM1/2 conjugates following heat shock [15]. The impact of sumoylation on these targets is unresolved, with the exception of the Arabidopsis transcription factor HSFA2 [55] (Fig. 3a). HSFA2 is a key element in acquired thermotolerance [69], and its activity in the nucleus seems to be repressed by SUM1 at position K315 [55]. Over-expression of SUM1 in seedlings results in a reduced tolerance to repeated heat, implying that sumoylation acts negatively upon acquired thermotolerance [55]. Conversely, SIZ1 seems to be a positive regulator of basal responses (acting independently of salicylic acid), but not of acquired thermotolerance [22, 27], which suggests the involvement of a SIZ1-independent pathway in the control of acquired thermotolerance. The seemingly antagonistic effect of SUMO pathway components on the different heat stress responses reflects the complex nature of these mechanisms. It also supports the idea that modulation of SUMO-conjugate steady-state levels during heat stress represents a dynamic and precisely fine-tuned process [70]. A microarray analysis study revealed that in the siz1 mutant, eight HSPs and HSFs (e.g. HSFA7A and HSF4/HSFB1) were up-regulated under standard growth conditions, while no down-regulated HSPs and HSFs were observed [21]. Similarly, sHSP-CI is consistently down-regulated following SUM1 over-expression [55]. Experimental evidence corroborates the notion that sumoylation acts mainly as an inhibitor of transcription [56]. Apart from HSFs, other heat-related TFs are predicted to be sumoylated in association to heat stress, namely WRKY3 and WRKY4, two Group 1 members of the large WRKY TF family associated with numerous stress stimuli [10, 71].
Fig. 3.
Molecular aspects of the SUMO-abiotic stress association in Arabidopsis thaliana. a SIZ1 is a positive regulator of basal thermotolerance. Heat shock likely induces sumoylation of several heat shock factors (HSFs), heat shock proteins (HSPs), and WRKYs. Sumoylation of HSFA2 blocks its activity and consequently down-regulates acquired thermotolerance. b Cold stress regulates the transcription factor (TF) ICE1 through SIZ1-dependent sumoylation, antagonizing HOS1-dependent ubiquitination (Ub) and the degradation of ICE1. Sumoylation activates ICE1 inhibiting MYB15 expression and activating the CBF3/DREB1a-regulon. c Salt and drought stress responses seem to be antagonistically regulated by SIZ1 and ULP1c/d. SIZ1 sumoylates and exerts a positive effect on key regulators of the drought response, while ULP1c/d may counteract this effect by removing SUMO from the target. d ABI5, a key TF in the abscisic acid (ABA) signaling pathway, is sumoylated by SIZ1, which antagonizes ABI5-ubiquitination but also inactivates ABI5 TF activity. e Nutrient availability can be controlled by SUMO. SIZ1 sumoylates nitrate reductases NIA1 and NIA2, contributing positively to nitrogen (N) assimilation. In response to inorganic phosphate (Pi) starvation, SIZ1 bi-sumoylates PHR1 and possibly LPR2, activating the expression of the PHR1-regulon and blocking LPR2 function in the remodeling of root architecture under conditions of Pi starvation. In response to excess copper (Cu), SIZ1 sumoylates an unknown target that directly or indirectly regulates expression of YSL1/3, important for metal re-allocation. f Sumoylation impacts on development at various levels, including ABI5-mediated seed dormancy and growth arrest, nutrient homeostasis, and allocation of metal ions
In addition to heat shock, SIZ1 is also important for cold acclimation and tolerance to freezing and chilling. More specifically, Miura and co-workers [17] found that upon cold imposition, SIZ1 positively affects the expression of the C-repeat Binding Factor 3/Dehydration Responsive Element Binding factor 1a (CBF3/DREB1a) TF and, consequently, its regulon. The CBF3/DREB1a regulator Inducer of CBF Expression 1 (ICE1) was shown to be sumoylated by SIZ1 at position K393, which does not seems to impact on ICE1 TF activity, rather it counteracts polyubiquitination by the E3 ubiquitin ligase HOS1, decreasing ICE1-degradation and allowing CBF3/DREB1a-regulon expression (Fig. 3b). ICE1 sumoylation can also negatively regulate MYB15, a repressor of the CBF3/DREB1a-regulon that binds to MYB elements in the promoter of several cold-inducible genes (Fig. 3b) [17]. It is likely that other SUMO substrates are involved in the response to cold, since the transgenic line ICE1(K393R) displays less sensitivity to freezing than the siz1 mutant. Also, SUMO-conjugates increase drastically after cold imposition, indicating that numerous proteins are SUMO modified upon challenge. We identified various cold-related proteins within the subset of abiotic stress-related SUMO-conjugates (see ESM File 2), namely, Stabilized 1 (STA1) and the components of transcriptional coactivator complexes ADA2a, ADA2b, and GCN5.
Drought and salt stresses
Drought and salt stresses have a tremendous impact on plant growth and development, significantly affecting crop yield. Plants cope with water limitation using complex physiological and molecular strategies that can be generally grouped within the categories of escaping, avoiding, or tolerating the stress [72]. Drought induces SUMO-conjugate accumulation in Arabidopsis, a process partially dependent on the activity of the E3 ligase SIZ1 [21]. SIZ1 seems to act positively on drought tolerance since the siz1 mutant shows drought sensitivity to short- and long-term dehydration. In addition, microarray data indicates that an extensive number of drought-responsive genes are significantly de-regulated in the siz1 mutant [21]. In terms of the stress hormone abscisic acid (ABA), there is sufficient evidence to suggest that both ABA-dependent and -independent mechanisms are involved in the SUMO–drought association (Fig. 3c). In support of ABA-independent mechanisms, no significant difference in the sumoylation pattern following drought imposition was observed between wild-type and aba2 (a mutant impaired in ABA biosynthesis) plants [21]. The authors of this study suggest that SIZ1 participates in ABA-independent pathways mediated by TFs other than ERD1 and DREB2A, since their regulons are not transcriptionally affected in the siz1 mutant. On the other hand, sumoylation may control the activity of DREB2A by regulating DREB2A-Interacting Protein 1 and -2 (DRIP1/2), predicted to be a SUM1 target by Miller and co-workers [10]. These two proteins contain C3HC4 RING domains functioning as E3 ubiquitin ligases that target DREB2A for proteolysis [73], therefore acting as negative regulators of drought responses.
In contrast, rice seedlings treated with ABA were shown to accumulate SUMO-conjugates [51, 74]. Most significantly, de-regulated genes in siz1-3 during drought have been found to have a 41 % overlap with ABA-responsive genes, and under normal growth conditions, genes of the ABA biosynthetic pathway (namely ABA1 and NCED3) are also de-regulated [21, 75]. Developmentally, the siz1 mutant displays ABA hypersensitivity in cotyledon greening after germination, functionally associated to the SP-RING domain responsible for the ligase activity of SIZ1 [23]. Over-expression of SUM1/2 was observed to attenuate ABA-mediated growth inhibition while SCE1a-co-suppressed lines displayed the opposite phenotype [76]. It is likely that ABA-signaling changes the sumoylation pattern of at least a small number of targets, enough to exert a phenotypical effect on the plant. A suitable target is the homeobox leucine zipper TF ATHB6, a SUMO-conjugate candidate that negatively regulates ABA responses [77]. Strong evidence supporting the SUMO–ABA relationship, albeit distinct from the drought response, is the demonstrated sumoylation of ABA Insensitive 5 (ABI5), a bZIP TF that positively regulates ABA-dependent seed germination and desiccation via binding of the ABA-responsive element (ABRE, ACGTGG/TC) cis-element (Fig. 3d) [18]. SIZ1 knockout does not affect ABI5 expression but enhances that of its regulon. The K391 residue of ABI5 is sumoylated in vivo and in vitro in a SIZ1-dependent fashion, rendering ABI5 inactive. In addition, sumoylation may also stabilize ABI5 by counteracting ubiquitin-dependent degradation mediated by the ubiquitin E3 ligase Keep On Going (KEG) [18].
In contrast to the positive regulation of drought-stress responses, SIZ1 acts as a negative regulator of high salinity responses (Fig. 3c). In fact, siz1 was first isolated from a second mutation screening that suppressed the sos3 salt-sensitivity phenotype [16], and siz1 seedlings are tolerant to salt. In parallel, the double knockout mutant for SUMO proteases ULP1c/OTS2 and ULP1d/OTS1 displays sensitivity to salt stress, while over-expression of ULP1d/OTS1 increases salt tolerance [26]. The mutant ots1 ots2 disrupts SUMO deconjugation constitutively, increasing the accumulation of SUM1/2-conjugated proteins (but not SUM3), particularly in response to salt stress [26]. Miura and co-workers [19] recently found that siz1 accumulates less sodium (Na) and more potassium (K) in shoots compared to the wild type, suggesting the involvement of ionic adjustments. Salt stress has also been shown to negatively modulate ULP1d/OTS1 (and probably ULP1c/OTS2) abundance via the ubiquitin–proteasome system rather than through transcription [26]. Thus, it is possible that, at least partially, the increment of SUM1/2-conjugates during stress is due to the turnover of SUMO proteases, implying a new level of regulation in the sumoylation pathway.
Nutrient imbalance
Nutrient deficiency is a type of stress that severely conditions plant growth and development. To circumvent nutritional scarcity plants possess a wide range of strategies, involving morphological, biochemical and transcriptional remodeling. Sumoylation, by controlling the homeostasis of essential nutrients such as N, inorganic phosphate (Pi), and copper (Cu), is emerging as a hub in nutritional sensing and response processes in plants (Fig. 3e). Under low Pi conditions, the siz1 mutant shows exacerbated Pi-starvation responses, such as inhibition of primary root growth, extensive lateral root and root hair development, increased root-to-shoot ratio, and anthocyanin accumulation, suggesting that this E3 acts as a negative regulator [16, 19, 20]. Remodeling of the root architecture during Pi-deficiency involves an altered auxin pattern, with SIZ1 acting as a negative regulator in the transcription of a series of auxin-responsive genes [20]. This regulation may involve the sumoylation of Auxin-Resistant 4 (AXR4, present in the list of abiotic stress-related SUMO-conjugates). AXR4 is involved in auxin redistribution and re-modulates root architecture in response to Pi starvation [78]. Miura and co-workers [16] found that Phosphate Starvation Response 1 (PHR1), a key transcription factor in several Pi-starvation responses, is positively regulated by SIZ1-dependent sumoylation at positions K261 and K372 (Fig. 3e). In support of this finding, SIZ1 appears to positively regulate Pi-starvation genes such as IPS1 and RNS1, which are part of the PHR1-regulon [16]. Also, PHR1 expression is not significantly induced nor is its subcellular localization affected by Pi-starvation [79], suggesting modulation at the PTM level.
Unlike siz1, no differences in root hair length and number have been observed in the phr1 mutant [16, 79, 80], suggesting the existence of additional pathways regulated by SIZ1/SUMO in response to Pi-starvation. One plausible candidate found in the SUMO-conjugate list by Miller et al. [10] is Low Phosphate Root 2 (LPR2). LPR2 and its paralog LPR1 are multicopper oxidases that positively control the decrease in primary root length and increase in the number of lateral roots upon Pi-starvation [81]. Since the lpr2 mutant seems to be insensitive (while siz1 is hypersensitive) to Pi-starvation, sumoylation may have a negative effect on LPR2 function. This antagonistic role is supported by the intermediate phenotype displayed by the lpr1 siz1 double mutant in terms of root architecture, anthocyanin content, and regulation of the Pi-starvation-responsive genes PAP2, IPS1 and PT2 [82].
SIZ1-dependent sumoylation also controls N homeostasis in Arabidopsis, positively regulating the catalytic activity of nitrate reductases NIA1 and NIA2 [24]. These two enzymes are important for N-assimilation, explaining why siz1 displays a low N content. Moreover, the siz1 pleiotropic phenotype is reverted by exogenous ammonium but not nitrate, reinforcing the notion that deficient N reduction is one of the main determinants of the siz1 pleiotropic phenotype (Fig. 3e, f) [24].
Nutrient availability is essential for normal growth, yet an excess of nutrients may lead to detrimental effects. For example, Cu is a crucial factor in multiple biological processes, but an overabundance induces reactive oxygen species (ROS) production and results in toxicity due to its high redox activity [83]. The involvement of SIZ1 in the control of Cu level and distribution was suggested by Chen and co-workers [25], who showed that under conditions of excess Cu, the mutant siz1 accumulated this nutrient in the aerial organs and showed Cu hypersensitivity. These phenotypes could be partially explained by the observed induction of the metal transporters Yellow Stripe-Like 1 and -3 (YSL1/3). Since sumoylated proteins increase in a Cu dose-dependent fashion, SUMO is likely to block the transcription of YSL1/3 (Fig. 3e) [25]. YSL transporters have also been associated to iron and zinc remobilization [84], and Chen and co-workers [25] observed that manganese, zinc, and Pi also accumulate in the siz1 mutant while the accumulation of potassium decreases, suggesting that sumoylation is closely involved in the allocation and homeostasis of metal ions as well as other nutrients.
Additional insights into SUMO function and regulation by stress
In plants, SUMO seems to take part in the interplay between normal development and abiotic-stress coping modes. Hormones are important factors in many tolerance responses [85, 86], and should play a key role in the SUMO–abiotic stress association since mutants for SUMO pathway components have been shown to de-regulate the metabolism/homeostasis of salicylic acid (SA), ABA, auxins, ethylene, brassinosteroids, jasmonic acid, and cytokinins [10, 18, 20, 21, 28, 29, 41–43, 62, 76]. The foremost example is SA, which accumulates considerably in sum1-1 amiR-SUM2, and siz1 mutants. Inhibiting SA levels in siz1 mutants by mutating PAD4 or ectopically expressing the bacterial salicylate hydrolase transgene NahG largely reverts its pleiotropic phenotype [28]. This includes the SIZ1-dependent response to cold, but not that to basal thermotolerance, highlighting an underlying complexity [22, 48].
SUMO modulation of abiotic stress responses occurs primarily at the nuclear level. Saracco and co-workers [27] observed that sumoylated proteins concentrate in the nucleus, while part of the free SUMO is cytoplasmic, suggesting that SUMO exerts a function in the regulation and remodeling of the nuclear proteome. In agreement with this function, isolated SUMO targets are mainly nuclear proteins [10, 31, 36]. In general, SUMO is assumed to be a repressor of transcription, namely by modification of chromatin-remodeling complexes and more specifically by the promotion of histone deacetylation [87, 88]. Not surprisingly, chromatin remodeling is also a critical aspect of plant abiotic stress responses [89], and we have identified several chromatin-associated proteins, such as GCN5, ADA2a, and ADA2b, within the subset of abiotic stress-related SUMO-conjugates (see ESM File 2). A functional correlation is now emerging between sumoylation and mRNA fate in the nucleus (particularly in response to abiotic stress), since in non-plant models, sumoylation candidates are involved in all steps of mRNA processing and export from the nucleus [90]. In support of this functional correlation, Arabidopsis ESD4, the first SUMO protease described in plants, is preferentially located in the nuclear periphery, associated to the nuclear pore complex component NUA [61] and possibly to the nucleoporin NUP160 [91]. Mutants of these components accumulate SUMO-conjugates and Poly(A) + RNA in the nucleus [61, 91]. The E3 ligase siz1 mutant displays similar mRNA retention in the nucleus, while evidencing decreased SUMO levels, particularly in response to stress [91]. It would appear that the disruption of SUMO homeostasis leads to mRNA accumulation in the nucleus, a phenomenon that can also be observed following abiotic stress [91].
Perhaps the most intriguing enigma lays in the regulation of the SUMO pathway. Part of the answer may reside in the fact that the sumoylation machinery itself is a target of SUMO modification. For example, upon being exposed to heat stress, the E1 subunit SAE1 and E2 SCE1 undergo reduced sumoylation while the sumoylation of SIZ1 increases substantially [49]. Moreover, SUMO components may themselves be susceptible to temperature changes, as suggested by Castaño-Miquel and co-workers [38] who showed that sumoylation is enhanced by high temperatures. Interestingly, SIZ1 is a target of multimeric sumoylation in lysines K100, K479 (a non-consensus site) and K488, the first also being induced by oxidative stress [10]. In mammals, low physiological concentrations of H2O2 inhibit SUMO conjugation by inducing the formation of a disulfide bond between the catalytic cysteines of the E1 and E2 enzymes [92], whereas higher ROS levels inhibit SUMO proteases, leading to increased conjugation [93]. Modulation of sumoylation by the redox status of the cell is an interesting concept, given that most environmental stimuli trigger ROS signaling events in a wave-like manner [94], consistent with the transient nature of the sumoylation/desumoylation cycle. Interestingly, siz1 mutants display increased H2O2 levels [95]. Ascorbate Peroxidase 1 (APX1) and Catalase 3 (CAT3), two important H2O2 scavengers and modulators of the celular redox status [96, 97], are also likely to be sumoylated [10, 38]. Future research efforts should not overlook the interplay between SUMO and ROS homeostasis.
An increasing focus of attention is the cross-talk between diverse PTMs [2, 39]. An attractive prospect is the identification in plants of human and yeast STUbL orthologs that would link the sumoylation of a target to its ubiquitin-dependent protein degradation [9]. Acetylation can also target the same lysine residue as SUMO and ubiquitin [40], and future focus on the three competing PTMs should be important. In non-plant models, sumoylation was also shown be both positively and negatively regulated by substrate phosphorylation [40]. In Arabidopsis, cross-talk between MAP Kinase 3/6/4 signaling and sumoylation has been suggested, with one example being the common targeting of WRKY TFs [88], opening up new possibilities for SUMO–abiotic stress interplay in plants.
Final considerations and future perspectives
A strong correlation between sumoylation and abiotic stress tolerance seems to be conserved among eukaryotic organisms [98], and SUMO has clearly emerged as a heavyweight PTM contender in the regulation of plant development, hormonal metabolism, resistance to pathogen challenge and, particularly, the response to environmental stimuli. Many SUMO targets act as key hubs in abiotic stress responses, yet in vivo, SUMO substrates are modified at very low steady states, a clear contradiction to the drastic phenotypes of mutants with altered SUMO pathways. One possible explanation for this paradox is that SUMO may be a PTM as common as phosphorylation. A first glimpse at the rapidly expanding number of SUMO targets suggests as much, with sumoylation candidates implicating this PTM in key abiotic stress responses. Future gene-centered approaches will be pivotal to confirm these hypotheses at a molecular level. Studies of SUMO pathway components should also be addressed. The E3 ligase SIZ1 is clearly a major abiotic stress determinant, but solving SUMO protease function and specificity will shed new light on the dynamics of SUMO conjugation/deconjugation cycles. Most significantly, future research should address the mechanistic influence of SUMO on target molecules, including chromatin remodeling and RNA-fate mechanisms. The use of high-throughput strategies, such as that of Miller et al. [10], to accelerate the discovery of SUMO conjugates and map them to different environmental challenges is now an attractive prospect, particularly when coupled with the use of null mutants of SUMO pathway components. It is clear that understanding the full impact of SUMO on the proteome during abiotic stress will be a demanding yet exciting challenge in forthcoming years.
Electronic supplementary material
Below is the link to the electronic supplementary material.
References
- 1.Kerscher O, Felberbaum R, Hochstrasser M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol. 2006;22:159–180. doi: 10.1146/annurev.cellbio.22.010605.093503. [DOI] [PubMed] [Google Scholar]
- 2.Vertegaal AC. Uncovering ubiquitin and ubiquitin-like signaling networks. Chem Rev. 2011;111:7923–7940. doi: 10.1021/cr200187e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Downes B, Vierstra RD. Post-translational regulation in plants employing a diverse set of polypeptide tags. Biochem Soc Trans. 2005;33:393–399. doi: 10.1042/BST0330393. [DOI] [PubMed] [Google Scholar]
- 4.Miura K, Hasegawa PM. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010;20:223–232. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Lomeli H, Vazquez M. Emerging roles of the SUMO pathway in development. Cell Mol Life Sci. 2011;68:4045–4064. doi: 10.1007/s00018-011-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meulmeester E, Melchior F. Cell biology: SUMO. Nature. 2008;452:709–711. doi: 10.1038/452709a. [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson KA, Henley JM. Mechanisms, regulation and consequences of protein SUMOylation. Biochem J. 2010;428:133–145. doi: 10.1042/BJ20100158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geoffroy MC, Hay RT. An additional role for SUMO in ubiquitin-mediated proteolysis. Nat Rev Mol Cell Biol. 2009;10:564–568. doi: 10.1038/nrm2707. [DOI] [PubMed] [Google Scholar]
- 10.Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Golebiowski F, Matic I, Tatham MH, Cole C, Yin Y, Nakamura A, Cox J, Barton GJ, Mann M, Hay RT. System-wide changes to SUMO modifications in response to heat shock. Sci Signal. 2009;2:ra24. doi: 10.1126/scisignal.2000282. [DOI] [PubMed] [Google Scholar]
- 12.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H. Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol. 2008;10:547–555. doi: 10.1038/ncb1717. [DOI] [PubMed] [Google Scholar]
- 13.Manza LL, Codreanu SG, Stamer SL, Smith DL, Wells KS, Roberts RL, Liebler DC. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem Res Toxicol. 2004;17:1706–1715. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- 14.Zhou W, Ryan JJ, Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae. Induction of protein sumoylation by cellular stresses. J Biol Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurepa J, Walker JM, Smalle J, Gosink MM, Davis SJ, Durham TL, Sung DY, Vierstra RD. The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem. 2003;278:6862–6872. doi: 10.1074/jbc.M209694200. [DOI] [PubMed] [Google Scholar]
- 16.Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, Yun DJ, Hasegawa PM. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci USA. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miura K, Jin JB, Lee J, Yoo CY, Stirm V, Miura T, Ashworth EN, Bressan RA, Yun DJ, Hasegawa PM. SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell. 2007;19:1403–1414. doi: 10.1105/tpc.106.048397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc Natl Acad Sci USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miura K, Sato A, Ohta M, Furukawa J. Increased tolerance to salt stress in the phosphate-accumulating Arabidopsis mutants siz1 and pho2. Planta. 2011;234:1191–1199. doi: 10.1007/s00425-011-1476-y. [DOI] [PubMed] [Google Scholar]
- 20.Miura K, Lee J, Gong Q, Ma S, Jin JB, Yoo CY, Miura T, Sato A, Bohnert HJ, Hasegawa PM. SIZ1 regulation of phosphate starvation-induced root architecture remodeling involves the control of auxin accumulation. Plant Physiol. 2011;155:1000–1012. doi: 10.1104/pp.110.165191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catala R, Ouyang J, Abreu IA, Hu Y, Seo H, Zhang X, Chua NH. The Arabidopsis E3 SUMO ligase SIZ1 regulates plant growth and drought responses. Plant Cell. 2007;19:2952–2966. doi: 10.1105/tpc.106.049981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo CY, Miura K, Jin JB, Lee J, Park HC, Salt DE, Yun DJ, Bressan RA, Hasegawa PM. SIZ1 small ubiquitin-like modifier E3 ligase facilitates basal thermotolerance in Arabidopsis independent of salicylic acid. Plant Physiol. 2006;142:1548–1558. doi: 10.1104/pp.106.088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheong MS, Park HC, Hong MJ, Lee J, Choi W, Jin JB, Bohnert HJ, Lee SY, Bressan RA, Yun DJ. Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol. 2009;151:1930–1942. doi: 10.1104/pp.109.143719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park BS, Song JT, Seo HS. Arabidopsis nitrate reductase activity is stimulated by the E3 SUMO ligase AtSIZ1. Nat Commun. 2011;2:400. doi: 10.1038/ncomms1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen C, Chen Y, Tang I, Liang H, Lai C, Chiou J, Yeh K. Arabidopsis SUMO E3 ligase SIZ1 is involved in excess copper tolerance. Plant Physiol. 2011;156:2225–2234. doi: 10.1104/pp.111.178996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conti L, Price G, O’Donnell E, Schwessinger B, Dominy P, Sadanandom A. Small ubiquitin-like modifier proteases OVERLY TOLERANT TO SALT1 and -2 regulate salt stress responses in Arabidopsis. Plant Cell. 2008;20:2894–2908. doi: 10.1105/tpc.108.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of sumoylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee J, Nam J, Park HC, Na G, Miura K, Jin JB, Yoo CY, Baek D, Kim DH, Jeong JC, Kim D, Lee SY, Salt DE, Mengiste T, Gong Q, Ma S, Bohnert HJ, Kwak SS, Bressan RA, Hasegawa PM, Yun DJ. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- 29.Miura K, Lee J, Miura T, Hasegawa PM. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 2010;51:103–113. doi: 10.1093/pcp/pcp171. [DOI] [PubMed] [Google Scholar]
- 30.van den Burg HA, Kini RK, Schuurink RC, Takken FL. Arabidopsis small ubiquitin-like modifier paralogs have distinct functions in development and defense. Plant Cell. 2010;22:1998–2016. doi: 10.1105/tpc.109.070961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park HC, Choi W, Park HJ, Cheong MS, Koo YD, Shin G, Chung WS, Kim WY, Kim MG, Bressan RA, Bohnert HJ, Lee SY, Yun DJ. Identification and molecular properties of SUMO-binding proteins in Arabidopsis. Mol Cells. 2011;32:143–151. doi: 10.1007/s10059-011-2297-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burroughs AM, Balaji S, Iyer LM, Aravind L. Small but versatile: the extraordinary functional and structural diversity of the beta-grasp fold. Biol Direct. 2007;2:18. doi: 10.1186/1745-6150-2-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayer P, Arndt A, Metzger S, Mahajan R, Melchior F, Jaenicke R, Becker J. Structure determination of the small ubiquitin-related modifier SUMO-1. J Mol Biol. 1998;280:275–286. doi: 10.1006/jmbi.1998.1839. [DOI] [PubMed] [Google Scholar]
- 34.Miura K, Jin JB, Hasegawa PM. Sumoylation, a post-translational regulatory process in plants. Curr Opin Plant Biol. 2007;10:495–502. doi: 10.1016/j.pbi.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Reed JM, Dervinis C, Morse AM, Davis JM. The SUMO conjugation pathway in Populus: genomic analysis, tissue-specific and inducible SUMOylation and in vitro de-SUMOylation. Planta. 2010;232:51–59. doi: 10.1007/s00425-010-1151-8. [DOI] [PubMed] [Google Scholar]
- 36.Budhiraja R, Hermkes R, Muller S, Schmidt J, Colby T, Panigrahi K, Coupland G, Bachmair A. Substrates related to chromatin and to RNA-dependent processes are modified by Arabidopsis SUMO isoforms that differ in a conserved residue with influence on desumoylation. Plant Physiol. 2009;149:1529–1540. doi: 10.1104/pp.108.135053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colby T, Matthai A, Boeckelmann A, Stuible HP. SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol. 2006;142:318–332. doi: 10.1104/pp.106.085415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Castano-Miquel L, Segui J, Lois LM. Distinctive properties of Arabidopsis SUMO paralogues support the in vivo predominant role of AtSUMO1/2 isoforms. Biochem J. 2011;436:581–590. doi: 10.1042/BJ20101446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bossis G, Melchior F. SUMO: regulating the regulator. Cell Div. 2006;1:13. doi: 10.1186/1747-1028-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin JB, Jin YH, Lee J, Miura K, Yoo CY, Kim WY, Van Oosten M, Hyun Y, Somers DE, Lee I, Yun DJ, Bressan RA, Hasegawa PM. The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 2008;53:530–540. doi: 10.1111/j.1365-313X.2007.03359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang L, Yang S, Zhang S, Liu M, Lai J, Qi Y, Shi S, Wang J, Wang Y, Xie Q, Yang C. The Arabidopsis SUMO E3 ligase AtMMS21, a homologue of NSE2/MMS21, regulates cell proliferation in the root. Plant J. 2009;60:666–678. doi: 10.1111/j.1365-313X.2009.03992.x. [DOI] [PubMed] [Google Scholar]
- 43.Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell. 2009;21:2284–2297. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chosed R, Mukherjee S, Lois LM, Orth K. Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem J. 2006;398:521–529. doi: 10.1042/BJ20060426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lois LM. Diversity of the SUMOylation machinery in plants. Biochem Soc Trans. 2010;38:60–64. doi: 10.1042/BST0380060. [DOI] [PubMed] [Google Scholar]
- 46.Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G. A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell. 2003;15:2308–2319. doi: 10.1105/tpc.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ. SUMO and SUMOylation in plants. Mol Cells. 2011;32:305–316. doi: 10.1007/s10059-011-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miura K, Ohta M. SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J Plant Physiol. 2010;167:555–560. doi: 10.1016/j.jplph.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Miller MJ, Vierstra RD. Mass spectrometric identification of SUMO substrates provides insights into heat stress-induced SUMOylation in plants. Plant Signal Behav. 2011;6:130–133. doi: 10.4161/psb.6.1.14256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rytinki MM, Kaikkonen S, Pehkonen P, Jaaskelainen T, Palvimo JJ. PIAS proteins: pleiotropic interactors associated with SUMO. Cell Mol Life Sci. 2009;66:3029–3041. doi: 10.1007/s00018-009-0061-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park HC, Kim H, Koo SC, Park HJ, Cheong MS, Hong H, Baek D, Chung WS, Kim DH, Bressan RA, Lee SY, Bohnert HJ, Yun DJ. Functional characterization of the SIZ/PIAS-type SUMO E3 ligases, OsSIZ1 and OsSIZ2 in rice. Plant Cell Environ. 2010;33:1923–1934. doi: 10.1111/j.1365-3040.2010.02195.x. [DOI] [PubMed] [Google Scholar]
- 52.Novatchkova M, Budhiraja R, Coupland G, Eisenhaber F, Bachmair A. SUMO conjugation in plants. Planta. 2004;220:1–8. doi: 10.1007/s00425-004-1370-y. [DOI] [PubMed] [Google Scholar]
- 53.Makhnevych T, Sydorskyy Y, Xin X, Srikumar T, Vizeacoumar FJ, Jeram SM, Li Z, Bahr S, Andrews BJ, Boone C, Raught B. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol Cell. 2009;33:124–135. doi: 10.1016/j.molcel.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 54.Okada S, Nagabuchi M, Takamura Y, Nakagawa T, Shinmyozu K, Nakayama J, Tanaka K. Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 2009;50:1049–1061. doi: 10.1093/pcp/pcp056. [DOI] [PubMed] [Google Scholar]
- 55.Cohen-Peer R, Schuster S, Meiri D, Breiman A, Avni A. Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotholerance. Plant Mol Biol. 2010;74:33–45. doi: 10.1007/s11103-010-9652-1. [DOI] [PubMed] [Google Scholar]
- 56.Gill G. Something about SUMO inhibits transcription. Curr Opin Genet Dev. 2005;15:536–541. doi: 10.1016/j.gde.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 57.Lyst MJ, Stancheva I. A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans. 2007;35:1389–1392. doi: 10.1042/BST0351389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta. 2009;1789:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Dominguez M, March-Diaz R, Reyes JC. The PHD domain of plant PIAS proteins mediates sumoylation of bromodomain GTE proteins. J Biol Chem. 2008;283:21469–21477. doi: 10.1074/jbc.M708176200. [DOI] [PubMed] [Google Scholar]
- 60.Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci USA. 2010;107:17415–17420. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu XM, Rose A, Muthuswamy S, Jeong SY, Venkatakrishnan S, Zhao Q, Meier I. NUCLEAR PORE ANCHOR,, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development. Plant Cell. 2007;19:1537–1548. doi: 10.1105/tpc.106.049239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Matarasso N, Schuster S, Avni A. A novel plant cysteine protease has a dual function as a regulator of 1-aminocyclopropane-1-carboxylic acid synthase gene expression. Plant Cell. 2005;17:1205–1216. doi: 10.1105/tpc.105.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nigam N, Singh A, Sahi C, Chandramouli A, Grover A. SUMO-conjugating enzyme (Sce) and FK506-binding protein (FKBP) encoding rice (Oryza sativa L.) genes: genome-wide analysis, expression studies and evidence for their involvement in abiotic stress response. Mol Genet Genomics. 2008;279:371–383. doi: 10.1007/s00438-008-0318-5. [DOI] [PubMed] [Google Scholar]
- 64.Geisler-Lee J, O’Toole N, Ammar R, Provart NJ, Millar AH, Geisler M. A predicted interactome for Arabidopsis. Plant Physiol. 2007;145:317–329. doi: 10.1104/pp.107.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Richter K, Haslbeck M, Buchner J. The heat shock response: life on the verge of death. Mol Cell. 2010;40:253–266. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 67.Kotak S, Larkindale J, Lee U, von Koskull-Doring P, Vierling E, Scharf KD. Complexity of the heat stress response in plants. Curr Opin Plant Biol. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 68.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol. 2007;143:251–262. doi: 10.1104/pp.106.091322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Anckar J, Hietakangas V, Denessiouk K, Thiele DJ, Johnson MS, Sistonen L. Inhibition of DNA binding by differential sumoylation of heat shock factors. Mol Cell Biol. 2006;26:955–964. doi: 10.1128/MCB.26.3.955-964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Eulgem T, Rushton PJ, Robatzek S, Somssich IE. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 72.Verslues PE, Juenger TE. Drought, metabolites, and Arabidopsis natural variation: a promising combination for understanding adaptation to water-limited environments. Curr Opin Plant Biol. 2011;14:240–245. doi: 10.1016/j.pbi.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 73.Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, Tanokura M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20:1693–1707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chaikam V, Karlson DT. Response and transcriptional regulation of rice SUMOylation system during development and stress conditions. BMB Rep. 2010;43:103–109. doi: 10.5483/BMBRep.2010.43.2.103. [DOI] [PubMed] [Google Scholar]
- 75.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 76.Lois LM, Lima CD, Chua NH. Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell. 2003;15:1347–1359. doi: 10.1105/tpc.009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Himmelbach A, Hoffmann T, Leube M, Hohener B, Grill E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002;21:3029–3038. doi: 10.1093/emboj/cdf316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nacry P, Canivenc G, Muller B, Azmi A, Van Onckelen H, Rossignol M, Doumas P. A role for auxin redistribution in the responses of the root system architecture to phosphate starvation in Arabidopsis. Plant Physiol. 2005;138:2061–2074. doi: 10.1104/pp.105.060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rubio V, Linhares F, Solano R, Martin AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nilsson L, Muller R, Nielsen TH. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana . Plant Cell Environ. 2007;30:1499–1512. doi: 10.1111/j.1365-3040.2007.01734.x. [DOI] [PubMed] [Google Scholar]
- 81.Svistoonoff S, Creff A, Reymond M, Sigoillot-Claude C, Ricaud L, Blanchet A, Nussaume L, Desnos T. Root tip contact with low-phosphate media reprograms plant root architecture. Nat Genet. 2007;39:792–796. doi: 10.1038/ng2041. [DOI] [PubMed] [Google Scholar]
- 82.Wang X, Du G, Meng Y, Li Y, Wu P, Yi K. The function of LPR1 is controlled by an element in the promoter and is independent of SUMO E3 Ligase SIZ1 in response to low Pi stress in Arabidopsis thaliana . Plant Cell Physiol. 2010;51:380–394. doi: 10.1093/pcp/pcq004. [DOI] [PubMed] [Google Scholar]
- 83.Cuypers A, Smeets K, Ruytinx J, Opdenakker K, Keunen E, Remans T, Horemans N, Vanhoudt N, Van Sanden S, Van Belleghem F, Guisez Y, Colpaert J, Vangronsveld J. The cellular redox state as a modulator in cadmium and copper responses in Arabidopsis thaliana seedlings. J Plant Physiol. 2011;168:309–316. doi: 10.1016/j.jplph.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 84.Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, Misson J, Schikora A, Czernic P, Mari S. Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot. 2009;103:1–11. doi: 10.1093/aob/mcn207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 86.Qin F, Shinozaki K, Yamaguchi-Shinozaki K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011;52:1569–1582. doi: 10.1093/pcp/pcr106. [DOI] [PubMed] [Google Scholar]
- 87.van den Burg HA, Takken FL. Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci. 2009;14:286–294. doi: 10.1016/j.tplants.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 88.van den Burg HA, Takken FL. SUMO-, MAPK-, and resistance protein-signaling converge at transcription complexes that regulate plant innate immunity. Plant Signal Behav. 2010;5:1597–1601. doi: 10.4161/psb.5.12.13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kim JM, To TK, Nishioka T, Seki M. Chromatin regulation functions in plant abiotic stress responses. Plant Cell Environ. 2010;33:604–611. doi: 10.1111/j.1365-3040.2009.02076.x. [DOI] [PubMed] [Google Scholar]
- 90.Meier I. mRNA export and sumoylation-Lessons from plants. Biochim Biophys Acta. 2012;1819:531–537. doi: 10.1016/j.bbagrm.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 91.Muthuswamy S, Meier I. Genetic and environmental changes in SUMO homeostasis lead to nuclear mRNA retention in plants. Planta. 2011;233:201–208. doi: 10.1007/s00425-010-1278-7. [DOI] [PubMed] [Google Scholar]
- 92.Bossis G, Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 93.Xu Z, Chan HY, Lam WL, Lam KH, Lam LS, Ng TB, Au SW. SUMO proteases: redox regulation and biological consequences. Antioxid Redox Signal. 2009;11:1453–1484. doi: 10.1089/ars.2008.2182. [DOI] [PubMed] [Google Scholar]
- 94.Mittler R, Vanderauwera S, Suzuki N, Miller G, Tognetti VB, Vandepoele K, Gollery M, Shulaev V, Van Breusegem F. ROS signaling: the new wave? Trends Plant Sci. 2011;16:300–309. doi: 10.1016/j.tplants.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 95.Kim MG. Alerted defense system attenuates hypersensitive response-associated cell death in arabidopsis siz1 mutant. J Plant Biol. 2010;53:70–78. doi: 10.1007/s12374-009-9089-8. [DOI] [Google Scholar]
- 96.Mhamdi A, Queval G, Chaouch S, Vanderauwera S, Van Breusegem F, Noctor G. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J Exp Bot. 2010;61:4197–4220. doi: 10.1093/jxb/erq282. [DOI] [PubMed] [Google Scholar]
- 97.Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses. Plant Physiol. 2007;144:1777–1785. doi: 10.1104/pp.107.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tempe D, Piechaczyk M, Bossis G. SUMO under stress. Biochem Soc Trans. 2008;36:874–878. doi: 10.1042/BST0360874. [DOI] [PubMed] [Google Scholar]
- 99.Reeves PH, Murtas G, Dash S, Coupland G. Early in short days 4, a mutation in Arabidopsis that causes early flowering and reduces the mRNA abundance of the floral repressor FLC. Development. 2002;129:5349–5361. doi: 10.1242/dev.00113. [DOI] [PubMed] [Google Scholar]
- 100.Hermkes R, Fu YF, Nurrenberg K, Budhiraja R, Schmelzer E, Elrouby N, Dohmen RJ, Bachmair A, Coupland G. Distinct roles for Arabidopsis SUMO protease ESD4 and its closest homolog ELS1. Planta. 2011;233:63–73. doi: 10.1007/s00425-010-1281-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Conti L, Kioumourtzoglou D, O’Donnell E, Dominy P, Sadanandom A. OTS1 and OTS2 SUMO proteases link plant development and survival under salt stress. Plant Signal Behav. 2009;4:225–227. doi: 10.4161/psb.4.3.7867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Katari MS, Nowicki SD, Aceituno FF, Nero D, Kelfer J, Thompson LP, Cabello JM, Davidson RS, Goldberg AP, Shasha DE, Coruzzi GM, Gutierrez RA. VirtualPlant: a software platform to support systems biology research. Plant Physiol. 2010;152:500–515. doi: 10.1104/pp.109.147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.