Abstract
Firefly luciferase-catalyzed reaction proceeds via the initial formation of an enzyme-bound luciferyl adenylate intermediate. The chemical origin of the color modulation in firefly bioluminescence has not been understood until recently. The presence of the same luciferin molecule, in combination with various mutated forms of luciferase, can emit light at slightly different wavelengths, ranging from red to yellow to green. A historical perspective of development in understanding of color emission mechanism is presented. To explain the variation in the color of the bioluminescence, different factors have been discussed and five hypotheses proposed for firefly bioluminescence color. On the basis of recent results, light-color modulation mechanism of firefly luciferase propose that the light emitter is the excited singlet state of OL− [1(OL−)*], and light emission from 1(OL−)* is modulated by the polarity of the active-site environment at the phenol/phenolate terminal of the benzothiazole fragment in oxyluciferin.
Keywords: Bioluminescence, Firefly luciferase, Color emission, Imaging, Red-emitter
Introduction
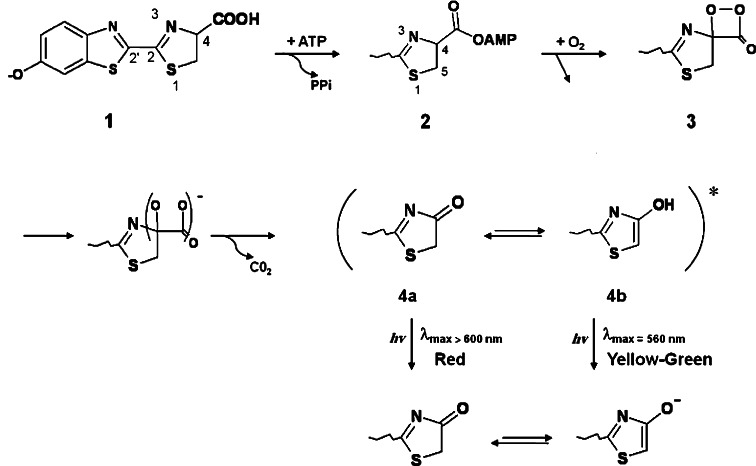
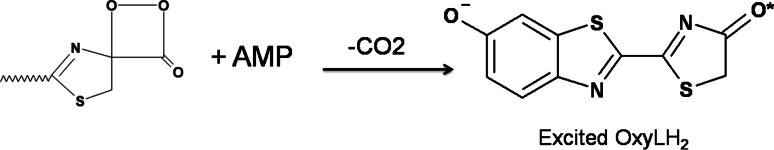
Bioluminescence, the phenomenon of light emission in living organisms, has been observed in wide ranges of luminous organisms (both prokaryotes and eukaryotes) including bacteria, fungi, algae, fish, squid, shrimp, and insects [1–5]. Luminous organisms produce light by an enzymatic reaction of a luciferase with a luciferin substrate. Luminescent reactions are quite distinct among luminous organisms, but in each case the reaction is an oxidation process with molecular oxygen and is a conversion of chemical energy into light [6–11]. In insects, the luminous species are mainly found in three families: fireflies (Lampyridae), railroad worms (Phengodidae), and click beetles (Elateridae) [12–14]. The bioluminescence systems of these insects are essentially the same with identical substrates (luciferin, ATP, and Mg2+) and with almost similar luciferases. Firefly luciferases catalyze a two-step oxidation of luciferin to produce light, oxyluciferin, CO2, and AMP as described in Fig. 1 [15, 16]. The most important characteristics of the firefly bioluminescence system are the requirement of ATP, large quantum yields of light production, and photon generation over a wide range of colors from green to red (emission maxima in the range 530–640 nm) [17–21]. According to these important properties, applications of firefly (beetle) bioluminescence in biological investigation are growing [22–25]. For further application, its fundamental chemistry should be investigated. Some important properties, including the quantum yield [26] and the complex structures of luciferase and substrates [27, 28] have recently been clarified. Moreover, regeneration of luciferin in light-emitting organs of fireflies (lantern) has been shown [29, 30]. In contrast, the light color modulation mechanism is still unclear.
Fig. 1.
Two-step oxidation of luciferin during firefly luciferase reaction to produce light, oxyluciferin, CO2, and AMP. Oxyluciferin emits light through keto or enol tautomer
Since even a few photons can be detected using available light measuring technology (such as a luminometer or charged-coupled devices), bioluminescence-based assays have been exploited as a powerful technology for numerous applications in different aspects of biotechnology and biological investigation over the last decade. They have been used as sensitive tools for monitoring gene expression, protein localization, and protein–protein interactions, detection of infections, monitoring of cell death and apoptosis, tumor growth and metastasis in whole animals [31, 32], reporter gene assays in a wide area of application [33], protein trafficking [32], drug screening [34], and detection of environmental contamination [35, 36]. The major advantages of techniques based on bioluminescence are the very low background in biological systems, versatility, noninvasiveness, reproducibility, high rate, and ease of assay performance with high sensitivity and low cost [31–33].
Luciferase-based in vivo imaging (bioluminescence imaging) enables and facilitates real-time analysis of disease development at the molecular level in living organisms, monitoring throughout the course of disease, and progression and tracking of infection. In addition, serial quantification of biological processes in intact animals is possible by bioluminescence imaging [31, 37]. Lack of intrinsic bioluminescence in mammalian tissues makes bioluminescence imaging a sensitive tool for in vivo imaging [31]. Photons are both scattered and absorbed as they exceed through mammalian tissues. Shorter wavelengths of light (blue and green) are largely absorbed by tissues, whereas longer wavelengths (red light) are less affected [37, 38]. Furthermore, hemoglobin is the main absorber of light in the body, with strong absorption peaks in the blue and green part of the visible spectrum, but absorption is reduced for wavelengths longer than 600 nm (red region) [39]. Therefore, red light can be transmitted through several centimeters of tissue and be detected externally more easily than absorbed blue or green light [31].
This property is an important consideration in the selection of appropriate luciferases for use in bioluminescence imaging [31]. Moreover, red-emitting variants of luciferase are also desirable for multiple tagging systems in whole cells as well as for dual-reporter systems [32, 33, 40]. Sample medium or environmental conditions produce high variability in the response, which is the main negative aspect encountered in an assay using luciferase as a reporter. To prevail over this problem, dual-reporter systems are designed in which, rather than a main reporter (e.g., a green emitting luciferase), a second control reporter (e.g., a red-emitting luciferase or another luciferase from other bioluminescent systems) to improve the analytical signal and distinguish the main signal from non-specific interference signals is used [33]. Consequently, red-emitting luciferase is also appropriate for multiplex analysis and dual-reporter systems in biosensor application and for minimizing light absorption by tissue in whole animals. Due to the importance of color variation of bioluminescence systems in wide ranges of applications in biotechnology and bioscience research, the mechanism of color variation has attracted much interest.
The bioluminescence color of fireflies has a broad range from green to red, depending on the luciferase species: fireflies emit in the yellow–green light [41, 42], click beetles emit in the green–orange [42, 43], and rail-road worms emit in a wide range from green to red [44, 45]. Even though chemical reaction catalyzed by all beetle luciferases and the used substrate (luciferin) are identical; still these varieties in color of emission are observed [46]. One of the main basis for differences in the luciferases bioluminescence color is the property of the emitter microenvironment localized in the enzyme active site. The observed bioluminescence color of pH-sensitive luciferases (but not in click beetle and rail-road worm luciferases) is performed with a clear shift to the red region in acidic conditions [47, 48]. The variety of bioluminescence color is also attributed to the luciferase structure [49]. The construction of chimeric proteins using click beetle luciferases [50] and firefly luciferase [51, 52], site-directed and random mutagenesis studies have revealed some important residues and regions involved in bioluminescence color [46, 47, 51, 53–55]. Moreover, in spite of the presence of an identical substrate in different mutant form and emission of different color, much attention has been paid to the structure and properties of the emitter.
The proposed candidates for the light-emitter structure are the keto form of oxyluciferin phenolate anion (OL−), its enol isomer, and the dianion having phenolate and enolate moieties [15].
To reveal the light-color modulation mechanism with reference to these previous studies, much attention has been paid to the fluorescence properties of OL− [56–60]. Although the elucidation of the spectroscopic properties of OL− may contribute to understanding the light-color modulation mechanism, its spectroscopic properties have not been investigated directly because of its low thermal stability.
The majority of critical residues in color determination and also substrate-binding residues are conserved among firefly luciferases. In substitution of three specific amino acids using site-specific mutagenesis in Lampyris turkestanicus luciferase [61], the color of emitted light was changed to red concomitant with a decreasing decay rate [62]. Different specific mutations (H245N, S284T, and H431Y) led to changes in the bioluminescence color. Mutation of the corresponding residues in other luciferases brought about a similar color shift in bioluminescence emission spectra [63]. Change in the color of emitted light indicates the critical role of some conserved residues in bioluminescence color determination among firefly luciferases.
Knowing these results, the mechanism of color shift in firefly luciferase has always been considered as an enigma in the bioluminescence story [64]. Based on all experimental observations, different mechanisms have been proposed to make clear color variations in beetle luciferases.
Enol-keto tautomerization of oxyluciferin
Original observation in color shift of firefly luciferase was found under application of broad ranges of pH for assay medium. It was based on analogy to the chemiluminescence of active esters in solutions. According to changes in the bioluminescence color of firefly luciferase at different pH, it was proposed that the excited state of the keto-form of the OxyLH2 anion can relax by emitting red light, while the excited state of the enol-form of the OxyLH2 anion emits yellow–green light [15, 65]. It was suggested that the resulting color would then depend on the keto-enol equilibrium, as Fig. 1 shows. It was proposed that red light emission from Luc at acidic pHs consequences from the keto form of the emitter oxyluciferin (Fig. 1). On the other hand, yellow–green emission of native firefly luciferases (λmax at 560 nm) was attributed to the enol tautomer of excited-state oxyluciferin formed by a presumed Luc-assisted tautomerization. Among the beetle luciferases, pH-sensitivity of bioluminescence color observed in the firefly does not occur in the families of click beetles or railroad worms. Color modulation of the tautomeric emitters, which represent the approximate extremes in wavelength, may occur through changes in the polarity of the emitter binding site. In fact, the pioneering studies by Seliger and coworkers [66] and DeLuca [65] emphasize that the polarity of the active-site environment is operative in modulating the bioluminescence color. However, as will be discussed later, recent experiments and theoretical calculation [67, 68] demonstrated that the multicolor luminescence requires only the keto-form OxyLH2. Additionally, small alteration or lack of spectral shifts have been observed for firefly luciferase mutants and variants with many residue differences [69–71], which could not be explained by the mechanism proposed by White et al. Meanwhile, the role of active site polarity in bioluminescence color has recently been considered as one of the most influent environmental factors in charge stabilization on phenolate ion of oxyluciferin. According to recent experimental observation and theoretical calculations, the light emitter of all color is the excited singlet state of the keto form of oxyluciferin phenolate anion [68]. However, the active site polarity as the main source of enol-keto tautomerization equilibrium is the common part of White hypothesis and recent findings.
Twisted intramolecular charge-transfer mechanism
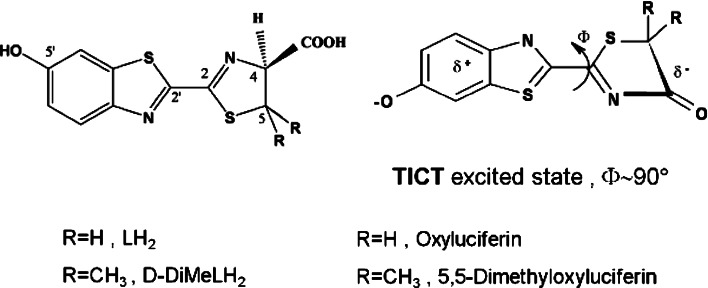
On the basis of theoretical calculations and semi-empirical investigation, another mechanism was proposed by McCapra et al. [72]. In spite of the White group, they had proposed that the presence of only one emitter form (keto tautomer) is sufficient to support emission of both green and red color emission. It was suggested that color variation is mainly associated with conformations of the keto form of excited-state oxyluciferin related by rotation about the C2–C2′ bond (Fig. 2). At the extremes, red emission was attributed to a minimum energy conformation of a twisted intramolecular charge-transfer (TICT) excited state with Ø = 90° (Fig. 2) and green to a higher-energy conformer with Ø < 90°. According to their postulation, the keto-form OxyLH2 in the first singlet excited state (S1) perform a twisted structure rather than a planar one. The planar species is regarded as a saddle point on the S1 potential energy surface. The color of the light emission should depend on the rotation around the C–C bond of the –N=C–C=N– moiety. In addition, it has been proposed that the excited singlet state of OL− [1(OL−)*] has a twisted structure with intramolecular charge transfer (ICT) character, where the benzothiazole and thiazolone rings are perpendicular as a result of rotation around the C2–C2′ bond.
Fig. 2.
TICT excited state of oxyluciferin with φ = 90° (right). The structure of dimethyl oxyluciferin (left)
This hypothesis was found mystifying, since the environment around the C–C bond is strongly conjugative and this bond should have a certain amount of double bond character (a partial double bond) on the S1 potential energy surface [67]. Therefore, due to less freedom of rotation, the turning around of this C–C bond has to override a large barrier. It was confirmed that the planar keto-form and enol-form OxyLH2 are minima on the S1 potential energy surface by means of semiempirical and configuration interaction singles (CIS) calculations, respectively [73, 74]. Therefore, it seems that a mechanism involving ‘twisted’ conformation to explain color changes in the firefly bioluminescence is non-feasible. Further, luciferase–emitter interactions are proposed to maintain specific conformers of the excited state, thus influencing color [72]. Since the minimum energy conformer is the red emitter, perturbations that disrupt luciferase–luciferin interactions through changes in protein tertiary structure, e.g., acid or heat denaturation, would be expected to give red light. However, recent findings support the presence of only a keto form in the bioluminescence reaction but without rotation about the C2–C2′ bond [68].
Polarization of the OxyLH2 microenvironment
Comparison of the bioluminescence spectra of WT and mutant forms of L. mingrelica luciferase and their dependence on the pH lead to explanation of a hypothesis. Accordingly, the shifts in the spectral maximum are either due to proton transfer between the phenolate group of OxyLH2 and the positively charged arginine residue (Arg218), or to the keto-enol-enolate equilibrium of the hydroxythiazole ring (Fig. 3) directed by interaction with His247 and Thr345 [75]. It has also been reported [76] that green bioluminescence of in situ produced DMOxyLH, which is constrained to the keto form, can emit both red and yellow–green light. This observation was later employed by Hirano et al. [68] to develop a more explicit model of the color change based on the interaction with the environment. This result is in agreement with the TICT mechanism, but as carefully shown by the Branchini results, the keto-enol tautomerization mechanism of White is not required to explain red and green firefly bioluminescence [77].
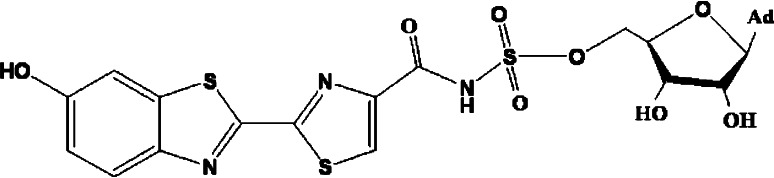
Fig. 3.
Structure of a stable analogue of luciferin adenylate intermediate structure; 5-O-[N-(dehydroluciferyl) sulfamoyl] adenosine (DLSA)
In fact, this hypothesis assumes that the color of the bioluminescence depends on the extent of polarization of the OxyLH2 in the microenvironment of the enzyme–OxyLH2 complex; that is to say, larger red shift of bioluminescence color depends on the higher polarizability [72, 75, 76]. This hypothesis is apparently supported by the time-dependent density functional theory (TD-DFT) calculations [74]. According to the computed data, the various enol-form of the oxyluciferin anions, enol-s-trans(−1), enol-s-trans(−2), and enol-s-trans(−1)’ (Figs. 7 and 8 in Ref. [74]), are responsible for the emitted light change from green to red, which is the beetle's naturally displayed light [78], whereas, the keto-s-trans, keto-s-cis(−1), and keto-s-trans(−1) forms of oxyluciferin (Fig. 10 in Ref. [74] and the corresponding structures in Chart 1 in Ref. [74]) emit light from violet to blue. This is not in agreement with the experimental conclusion that the multicolor luminescence can be obtained only from the keto-forms [68, 77]. It was finally concluded that the participation of the enol forms of oxyluciferin in bioluminescence is plausible but not required to explain the multicolor emission [74]. However, in spite of the good contribution of this hypothesis in explaining the environmental effect on bioluminescence color, it failed to explain the role of the chemical origin of multicolor bioluminescence.
Resonance-structure mechanism
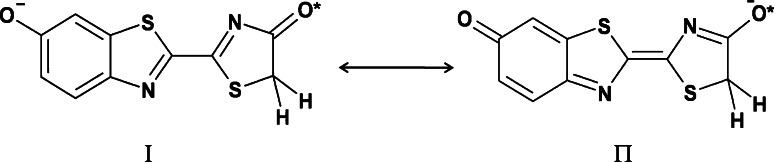
According to a recently elucidated mechanism, luciferase modulates emission color by controlling the resonance-based charge delocalization of the anionic keto form of the oxyluciferin excited state [79]. Resonance stabilization of the anion I (Scheme 1) would extend the π-electron skeleton, producing a resonance hybrid of the excited state of lower energy than if the oxyluciferin anion were constrained as species I. A resonance stabilized excited state more nearly resembling species II would be the red emitter. According to this hypothesis, the appearance of bimodal spectra of bioluminescence for some luciferase mutant or under acidic pH would be described by resonance hybrids comprised of structures resembling species I and II in the approximate proportion that are found in the respective emission spectra. This proposal is quite similar to the keto-enol mechanism of White, with the recognition that species I alone have the same degree of stabilization as does the enol form of oxyluciferin that was previously identified as the green light emitter. In fact, one excited state showing a –N=C–C=N– moiety would relax by emitting green light, another one showing a –N–C=C–N– moiety would emit red light. This solution for the problem of the light-emitter structure was reported by Branchini et al. [79]. The hypothesis was raised when they found that the bioluminescence of the adenylate of 5,5-dimethylluciferin can generate various colors of light. The product of the bioluminescence reaction of 5,5-dimethylluciferin adenylate must be 5,5-dimethyloxyluciferin (1-OH), which is unable to provide its enol and enolate species. On the basis of this finding, 1(OL−)* becomes the most plausible candidate for the light-emitter structure in the excited singlet state during firefly (beetle) bioluminescence. This conclusion is also consistent with the reaction mechanism in which 1(OL−)* is the primary product of the chemiexcitation process from the anionic dioxetanone intermediate (Dx−), as shown in Scheme 2 [80–83]. It has also been proposed that the position of a basic moiety (such as an arginine residue) in the vicinity of the excited singlet state of OL− [1(OL−)*] is a factor affecting the resonance properties of OL− [79].
Scheme 1.
Charge delocalization of the anionic oxyluciferin
Scheme 2.
Chemiexcitation process of the anionic dioxetanone intermediate of oxyluciferin
Therefore, it is reasonable to postulate that the color variation originates from modulation of the emission properties of 1(OL−)* in the active site of firefly luciferase. Changes in the 1(OL−)* emission properties have been explained by the use of the resonance structures of OL−, which have electron conjugation between the benzothiazole and thiazolone rings [79]. Furthermore, environmental factors surrounding 1(OL−)* must be considered.
Size of the luciferase protein cavity
The first crystal structure of Photinus pyralis firefly luciferase has been solved in the absence of substrate or transition state analogues [27, 84]. A 2.0 Ǻ cocrystal structure of P. pyralis firefly luciferase with an unusually potent inhibitor (PTC124) revealed the inhibitor to be the acyl-AMP mixed-anhydride adduct PTC124-AMP [85]. The crystal structures of wild-type and red mutant (S286N) luciferases from the Japanese Genji-botaru (Luciola cruciata) in complex with a high-energy intermediate analogue, 5-O-[N-(dehydroluciferyl)-sulfamoyl] adenosine (DLSA) have been obtained [86]. Comparing these structures to those of the wild-type luciferase complexed with AMP plus oxyluciferin (products) reveals a significant conformational change in the wild-type enzyme but not in the red mutant. They replaced luciferyl-AMP by the stable analogue 5-O-[N-(dehydroluciferyl) sulfamoyl] adenosine (DLSA, see Fig. 3) and captured three key ‘snapshots’ of the reaction: (1) luciferase bound to the reactants (ATP-Mg2+), (2) luciferase bound to the DLSA, and (3) luciferase bound to the products oxyluciferin-AMP (OxyLH2-AMP). The structures of luciferase bound to the reactants and the products were found to be similar: both possess an active site that the authors qualified as “open” since the substrate is less packed by the environment surrounding the protein compared to that for the luciferase-DLSA complex. When the protein is bound to DLSA, Ser286 in L. cruciata luciferase (corresponding to Ser284 of P. pyralis luciferase) forms hydrogen bonds with Tyr257 and Asn231 via a water molecule. This conformational change is also accompanied by a 6° rotation of the side chain of Phe249 toward DLSA and rotation of the side chain of Ile288 by 131° (see Fig. 3a in Ref. [86]). The result is a “closed” form of the active site, creating a structure in which the benzothiazole ring of DLSA is tightly sandwiched in a hydrophobic pocket including Ile288. Nakatsu et al. have proposed that controlling the hydrophobic microenvironment of the first excited single state (S1) of the oxyluciferin (OxyLH2) molecule [1(OL−)*] is a key to understanding the bioluminescence color variations. To confirm this, they have linked the observed color modulation to the size of the protein pocket surrounding OxyLH2. They have suggested that the “open” form allows some energy loss in the OxyLH2 excited state; leading to the emission of low-energy red light, while the “closed” form, with its much more rigid microenvironment, minimizes this energy loss, thereby emitting a yellow–green light.
This conformational change involves movement of the hydrophobic side chain of Ile288 towards the benzothiazole ring of DLSA. According to crystallographic data, it has been postulated that the degree of vibrational energy loss of 1(OL−)*, which is controlled by a transient movement of Ile288, determines the color of bioluminescence during the emission reaction. They have suggested a control mechanism of the amount of energy loss based on the size of the luciferase protein cavity. In this case, a non-relaxed form of keto OxyLH2 should emit yellow–green light, while after geometrical relaxation it should emit red light. The geometrical relaxation is controlled by the size of the cavity of luciferase, which has been site-specific modified. It remains to be demonstrated if the different cavity allows more freedom to the luciferin substrate, or constrains it in a different structure. This hypothesis leads to new challenges in the color emission hypothesis and leads to recent suggestions and experiments. It should be noted that the solvent perturbation method (solvent engineering) usually makes proteins more rigid and stable. However, more rigid protein induced by the presence of osmolytes [87, 88] or immobilization [89] of luciferase did not bring any shift in the bioluminescence color of firefly luciferase. That is to say, if local rigidity of luciferase cavity was the reason, at least minor shifts in spectra due to global stabilization would have been observed. This study and related crystal structures revealed important issues involving luciferin–luciferase intermediate structures that have not been reported earlier and will be discussed later.
Role of capping the flexible loop: a lesson from nature
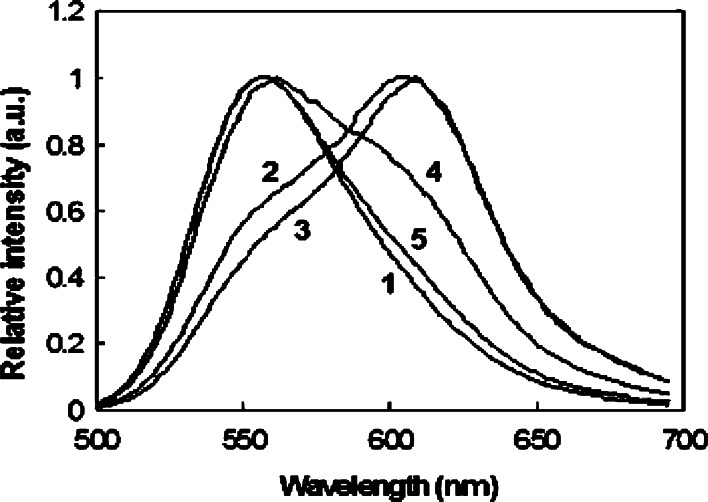
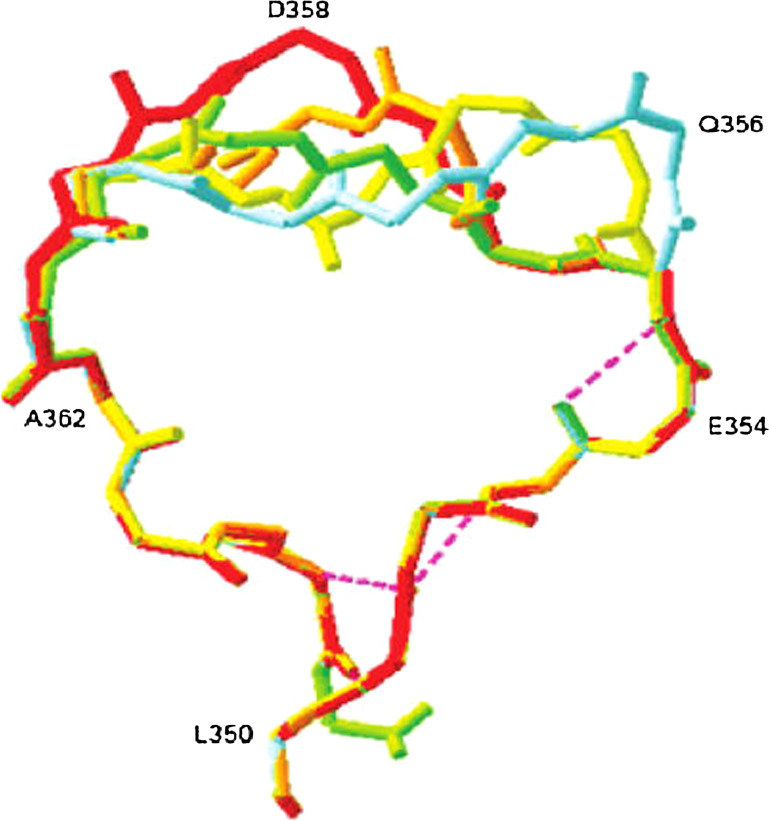
Among all reported beetle luciferases [90, 91], the Phrixotrix rail-road worm luciferase is the only luciferase that naturally emits red light through two cephalic lanterns (λmax = 628 nm) in addition to the yellow–green bioluminescence (λmax = 542 nm) emitted through 11 pairs of lateral lanterns along the body. Phrixothrix hirtus red (PhRE) luciferase shows 46–49% identity with firefly luciferases. PhRE luciferase with naturally red-emitting ability is a unique model to investigate the relationship of luciferase structure with BL color [44, 92]. The sequence multiple alignment among P. hirtus, the only native species with red bioluminescence, and the other green-emitter luciferases showed multiple differences. Among them, in green-emitter luciferases an Arg residue (Arg356 in P. pyralis luciferase) corresponding to Arg353 in P. hirtus is missed. It is located in an important flexible loop capping the active site. Insertion of this residue (Arg356) in a green-emitter luciferase (Lampyris turkestanicus) brought about with change of emitted color from green to red and increase of optimum temperature [93]. For further clarification of the effect of this position and geometry of loop-containing residues from 352 to 359 on the light color, four different residues including Arg, Lys, Glu, Gln were inserted at position 356 of P. pyralis luciferase [94]. These residues were considered similar in side-chain length, but different charges at optimum pH of luciferase activity (pH 7.8). The comparison of bioluminescence emission spectra revealed that insertion of positive residues (Arg356, Lys356) had more effect in bioluminescence color shift to red, and insertion of these residues resulted in a spectrum with a major peak in 608 nm and a minor shoulder in 557 nm (Fig. 4). Meanwhile, insertion of Glu354 in Photinus pyralis luciferase leads to a minor shift in bioluminescence spectra. The emission spectrum of Glu354 was bimodal with a major and minor peak at 557 nm and 591 nm, respectively. The insertion of Gln354 did not have an effect on emission spectra and its bioluminescence spectra were similar to native (Ppy) luciferase. According to the loop conformation (based on a model, Fig. 5), it was concluded that the insertion of positive residues (Arg356, Lys356) could induce new ionic and H-bonds and as a result displace the flexible loop and exposes the active site to water and therefore result in red light emission, and a similar result was obtained for L. turkestanicus luciferase [93]. This is closer to reality, as according to the hypothesis of long range interaction in production of phenolate anion of oxyluciferin, the key luciferase residues for the luciferin binding site are absolutely conserved among the luciferases, and therefore have major roles in this mechanism. The natural red bioluminescence of P. hirtus is interesting, as this luciferase also contains all of the essential proposed residues for green light emission and even deletion of the mentioned critical residue (Arg353) did not have an effect on the its emission color [95]. Insertion of these residues plus the previous findings may explain the importance of this position in diversion of luciferases to red and green emitters.
Fig. 4.
Bioluminescence emission spectra produced by the native form Photinus pyralis (1) and mutant (2, Arg356; 3, Lys356; 4, Glu356; 5, Gln356) luciferases by a luciferase-catalyzed reaction at pH 7.8. Reprinted from [94] with permission from American Chemical Society (ACS)
Fig. 5.
Superposition of flexible loop (residues 350–365) models from native (green) and mutant (Lys356, red; Arg356, orange; Glu356, yellow; Gln356, blue) luciferases. The insertion of new residues forms a longer flexible loop in comparison to native luciferase. Reprinted from [94] with permission from American Chemical Society (ACS)
In order to better understand the importance of the mentioned flexible loop in color emission, mutagenesis of L. turkestanicus luciferase at position 354 along with insertion of Arg356 were performed [96]. E354R and E354K exhibit a bimodal bioluminescence spectrum with a maximum in the red region and a smaller shoulder in the green region, while E354K/Arg356 and E354Q/Arg356 display a bimodal spectrum with a maximum in the green region and a smaller shoulder in the red region. A similar spectrum was also reported for a similar mutant of Hotaria parvula luciferase at pH 7.2, and pH 9 suggested that a single substitution at this position (corresponding to the E354 in L. turkestanicus) is sufficient to cause a shift to the red region of the spectrum in firefly luciferase. It is worthwhile to note that mutant of L. turkestanicus luciferase (with insertion of Arg356) performed a bimodal spectrum with a major peak in the red region, but introduction of two Arg in the same protein (E354R and Arg insertion at 356) produced a bioluminescence spectra with a single peak at the red region. It should be noted that addition of two Arg in the same loop brought about with decrease in decay time of luminescence. A similar result was obtained for a single mutant of P. pyralis firefly luciferase. That is to say, in most of the reported mutations, a relation between color shift to red and decrease of decay rate have been seen. From the above-mentioned investigation, it may be concluded that conformational displacement of a loop containing residues 352 to 359 due to positive charge saturation make emitter site more solvent accessible which is suggested in similar reports [22, 68].
Crystal structure, absorption, and emission spectra of oxyluciferin
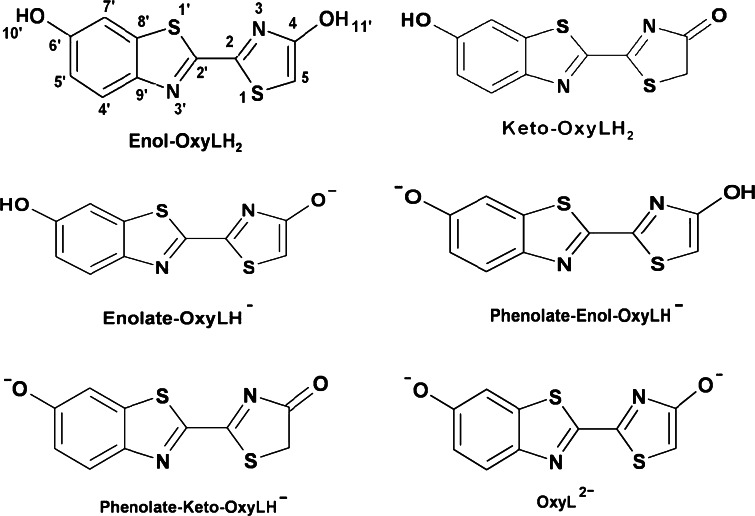
The structure of the pure luciferase substrate, D(−)–LH2, was determined about 30 years ago [97–99], but due to marked instability and associated impediments with purification and crystallization [100] of the real light-emitting molecule, OxyLH2, very little is known about its structure, and its crystal structure has not been determined until recently [101]. With consideration of only the more stable trans conformations around the C2–C2′ bond, by means of triple chemical equilibrium (deprotonation of the two hydroxyl groups and keto-enol tautomerism of the 4-hydroxythiazole ring) OxyLH2 can exist as six chemical forms, as shown in Fig. 6 [101]. The detailed systematic investigation of the solvent/pH effects on the absorption and emission spectra of OxyLH2 was not reported. In fact, the exact chemical form in which OxyLH2 exists at various conditions during the chemiluminescence and bioluminescence reactions, as well as the structures, absorption, and emission spectra of its forms outlined in Fig. 6 have long remained a subject of speculation. Moreover, the ultrafast dynamics of the first singlet excited state (S1) of the emitter and its ions as free species or as a complex with luciferase were also not investigated in detail either, although the spectroscopy of the reactant LH2 was reported [102–104].
Fig. 6.
Possible trans-C2–C2′ bond chemical forms of the firefly emitter oxyluciferin, OxyLH2
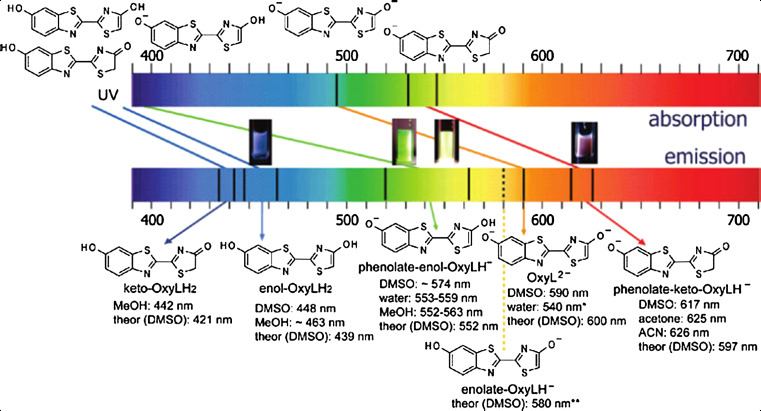
According to a detailed and comprehensive study on the absorption and emission spectra, and the crystal structure of oxyluciferin, it was shown that the triple chemical equilibrium of the unsubstituted emitter can provide a range of emission energies, spanning wavelengths from the blue to the red region of the visible spectrum, without the necessity of constraining its molecular structure to the keto form [101]. Based on experimental and theoretical spectroscopic data, assignment of the absorption and emission spectra of the firefly emitter oxyluciferin at different wavelengths are shown in Fig. 7.
Fig. 7.
Assignment of the absorption and emission spectra of the firefly emitter oxyluciferin, based on experimental and theoretical spectroscopic data (*Unstable species; **Difficult to be determined experimentally because of the small concentration in mixture with other species). The figures in the middle show the color change of oxyluciferin (from left to right) in DMSO without added base, in water with and without base, and in DMSO in presence of base. Reprinted from [101] with permission from American Chemical Society (ACS)
According to information obtained through steady-state and time-resolved spectral analysis, the structure of pure oxyluciferin and its 5-methyl analogue indicate that the enol group of the hydroxythiazole ring can be notably stabilized in the protein scaffold, and the phenolate ion of the enol form was considered as the closest species to the properties of the green–yellow emission observed in the wild-type luciferase (yellow–green-emitting system). Evaluation of the spectrum–pH relationship showed that within a relatively narrow pH region, including conditions that are close to the physiological conditions, a complex mixture of several species with distinct spectral properties exist, which interpret severe shifts in the equilibrium and the emitted wavelength in small pH changes. This important finding is in favor of original observation and suggestions raised by White et al. [15]. As discussed earlier, according to the appearance of red–green emitter forms of firefly luciferase under different pH, the presence of enol-keto tautomer form were interpreted [15, 79]. In addition to pH, the polarity of the environment, presence of ionic species in close proximity to the emitter, intramolecular charge redistribution caused by directional intermolecular interactions, and π–π stacking have been considered as the main effectors on the interplay among the three equilibrium states [101]. Therefore, it may be suggested that due to the environmental condition, preference to one of those structures under real biological conditions may occur. Having considered the possible interplay of multiple factors based on model solutions of oxyluciferin or its analogues, a clear preference to any of these factors as the primary controlling aspect to apply to the real biological system in vivo has not been found [101].
Solvent-critical effect on light-color modulation
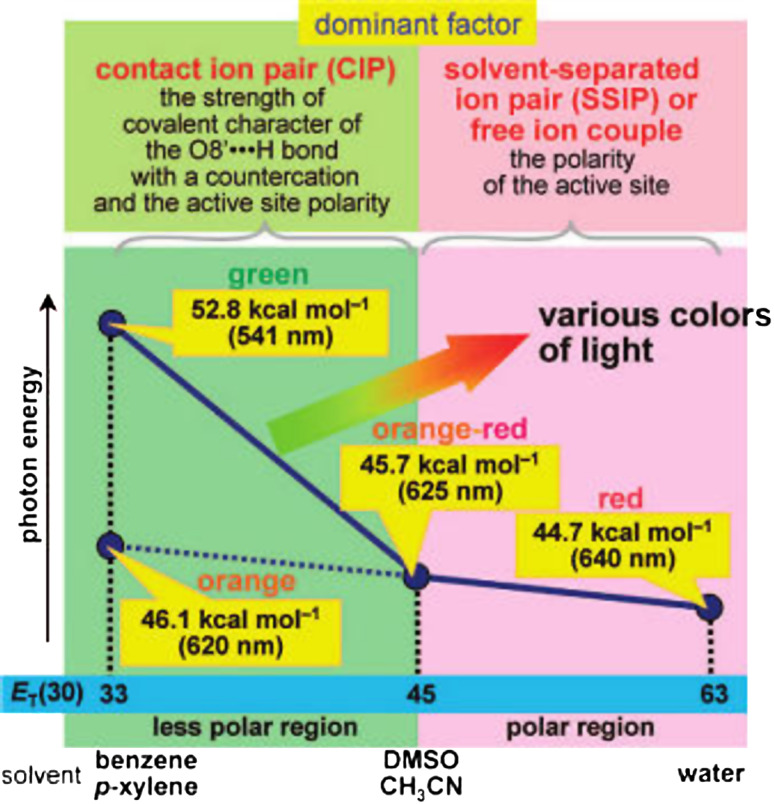
On the other hand, with consideration of the spectroscopic properties of excited state of oxyluciferin in different organic solvents, multicolor behavior of bioluminescence by considering only one species in the excited singlet state, 1(OL−)* is explained [68]. Phenolate anion 1-O− was used as a model compound of the keto form of wild-type oxyluciferin phenolate anion (OL−), which is postulated to be the most plausible candidate for the structure of the light emitter of the bioluminescence [78]. The spectroscopic properties of the neutral form 1-OH and the methyl ether derivative 1-OCH3 were also investigated for comparison. It was found that the fluorescence emission maximum (λF) of 1-O− varied in the range from 541 to 640 nm depending on the solvent polarity and the bonding interaction between 1(1-O−)* and a countercation (the conjugate acid of the organic base). In a polar solvent, 1(1-O−)* and the countercation form an SSIP (solvent separated ion-pair) or a free ion couple, and the polarity of the solvent modulates the λF values of 1-O− in the range from 625 to 640 nm. In a less polar solvent, a CIP (contact ion pair) of 1(1-O−)* and the countercation was formed. In the CIP, the solvent polarity and the strength of the covalent character of the O8′· · ·H bond between 1(1-O−)* and the countercation modulate the λF value of 1-O− in the range from 540 to 625 nm [68].
According to this study, it was concluded that: (1) the light emitter is the excited singlet state of the keto form of oxyluciferin phenolate anion, 1(OL−)*, and (2) the emission maximum of OL− is modulated by two main factors: the polarity of the active-site environment of the luciferase surrounding 1(OL−)* and the properties of the bonding interaction between the O8′ atom of 1(OL−)* and a hydrogen atom of a countercation, which will be the protonated form of a basic moiety of an amino acid residue in the active site. Meanwhile, an important recent study claims the phenolate ion of the enol form was considered as the closest species to the properties of the green–yellow emission observed in the wild-type luciferase (yellow–green-emitting system) [101]. It should be noted, pioneering studies were emphasized on the polarity of the active-site environment in modulating the bioluminescence color [16]. In fact, color modulation of the tautomeric emitters, which represent the approximate extremes in wavelength, may occur through changes in the polarity of the emitter binding site [15]. On the other hand, this postulation agreed with experimental observations that implicate displacement of a flexible loop in firefly luciferase, which cause more solvent accessibility to the emitter binding site [93–96].
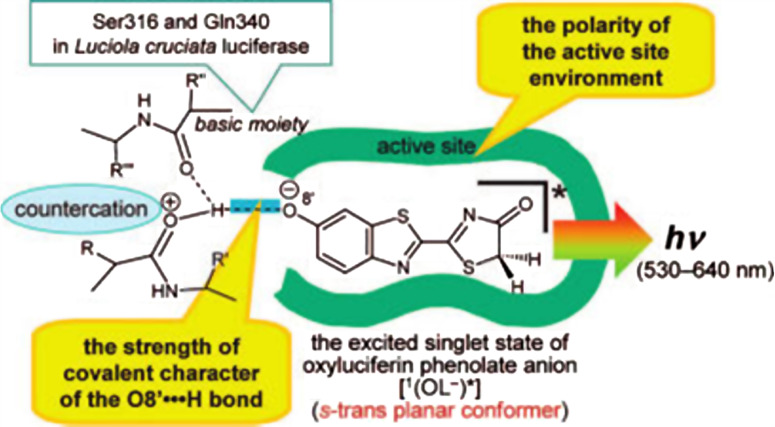
On the basis of the spectroscopic properties of 1-O−, a road map for understanding the bioluminescence color variation as a function of a solvent-polarity parameter, ET(30), was obtained (Fig. 8). According to the proposed road map, the mechanism of the in vivo bioluminescence colors was attributed to the polarities of the active-site environments of luciferases and the bonding characters of the interactions between 1(OL−)* and protonated basic moieties. It was predicted that stronger acidity of the protonated basic moiety in a luciferase leads to expansion of the color variation range. The potential basic moiety may consist of multiple amide carbonyls in the active site. For example, in L. cruciata luciferase, the active site has a non-polar character similar to p-xylene and benzene solutions, and the amide carbonyls of Ser316 and Gln340 (corresponding to Gln338 of P. pyralis) play an important role as basic moieties (Fig. 9). Additionally, the structure of 1(OL−)* for the firefly (beetle) bioluminescence is postulated to be the s-trans planar conformer.
Fig. 8.
Road map for understanding the bioluminescence color variation as a function of a solvent-polarity parameter. Reprinted from [68] with permission from American Chemical Society (ACS)
Fig. 9.
The active site model of firefly luciferase. The amide carbonyls of Ser316 and Gln340 play an important role as basic moieties similar to non-polar solvents. Reprinted from [68] with permission from American Chemical Society (ACS)
QM/MM calculations and classical molecular dynamics simulations
The result of both recent experimental studies [68, 101] was further supported by an important theoretical investigation by Navizet et al. [105]. In a recent study, quantum mechanics/molecular mechanics (QM/MM) calculations and classical molecular dynamics (MD) simulations based on the “open” and “closed” x-ray structures of firefly L. cruciata luciferase was used. It should be noted that the crystal structures of native firefly L. cruciata luciferase and its red mutant have been obtained in combination with an intermediate state (DLSA). According to QM/MM studies, the proposed mechanism of controlling the amount of energy loss based on the size of the luciferase protein cavity (in firefly L. cruciata luciferase) contrasts with the conclusion of recent experimental reports [68, 101] in which the color modulation of the light emission as a function of various base/solvent combinations of an OxyLH2 analog has been studied. It has been suggested that the color tuning might not be regulated by the rigidity of the active site but rather by the changes of polarity near the phenolate oxygen of the keto form of OxyLH2. It should be noted that according to a recent report, it was concluded that the enol form of OxyLH2 can play a role in the emission and that several factors (pH, polarity of the microenvironment, presence of ionic species in the cavity, etc.) can act collectively in the protein scaffold during the tuning of the emission color [101].
It is noteworthy that most of the earlier theoretical studies dealing with the emission spectrum of the OxyLH2 have been reported in vacuo without attention to the steric and electrostatic contributions arising from the protein [67, 68, 75, 106–109]. Some of them were done at the density functional level of theory [68, 75, 107, 109] using standard functional which are unable to treat correctly the charge-transfer states [110, 111]. Some theoretical studies have already been performed using the x-ray structures of firefly luciferase from Photinus pyralis determined without substrates and Luciola cruciata as discussed earlier [86].
The two proposed models for the active site in P. pyralis, introduced by the Branchini [80] and Ugarova groups [59], were later confirmed by their similarities to the more current L. cruciata data. Two quantum-mechanics/molecular mechanics (QM/MM) studies have recently been reported [107–112] and in both cases, single reference methods were employed for the QM subsection, and the red-to-green shift was not well reproduced. Furthermore, the microenvironmental contributions were not analyzed in detail with respect to the H-bonding network around OxyLH2.
According to QM/MM data of luciferase reaction two important findings were obtained: (a) the phenomenon of multicolor bioluminescence is mainly related to the polarization of the close surroundings of OxyLH2 (in support of other experimental findings) [68, 101] and (b) the deduced luciferase cavity size (according to crystal structure of Japanese firefly luciferase) does not force the relative structures of the oxyluciferin substrate sufficiently to account for the observed color modulation [105]. The emphasis is on the importance of the H-bond networks formed between the water molecules, keto-1, and the residues involved in the protein cavity. Keto-1 is actually the same structure reported as 1(OL−)* in excited state in another recent study [68]. All energy calculations were performed on the keto-1 form of OxyLH2, which is broadly considered to be the species responsible for light emission inside the wild firefly.
It should be noted that all mutations in a flexible loop of firefly luciferases were accompanied with displacement of the loop and probably higher solvent accessibility to benzothiazole moiety of oxyluciferin. Formation of new ionic and H-bonds were also reported. Therefore, it may be suggested that the mutations of the residues involved in the H-bond network can lead to different polarizations acting on the benzothiazole moiety and change the color of the emitted light accordingly.
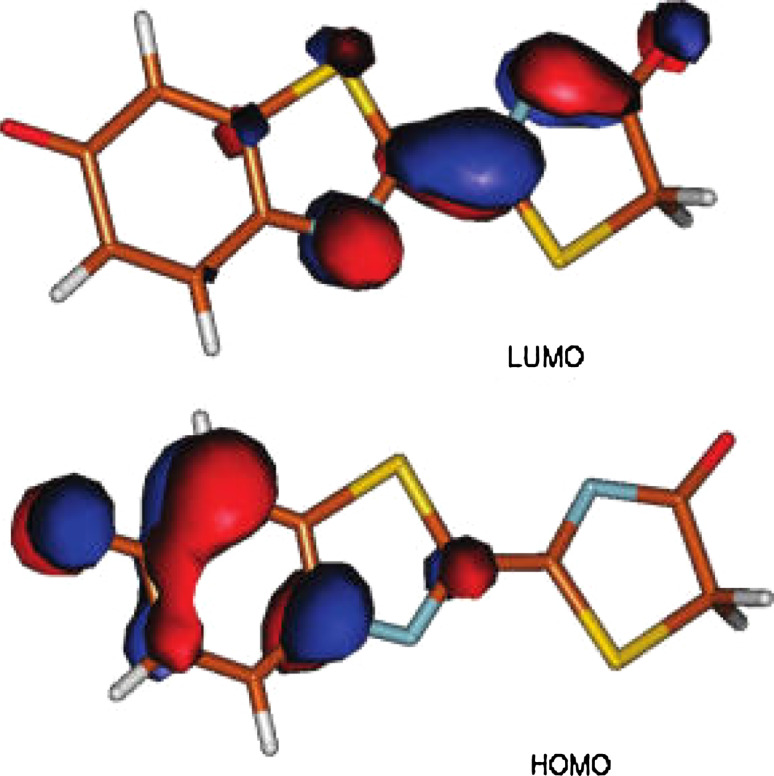
According to molecular modeling, transition from S1 to S0 leads to an internal negative charge transfer from the thiazolone ring near the AMP to the benzothiazole ring (phenolate) (see graphic HOMO and LUMO model in Fig. 10). Hence, any factor favoring the stabilization of the charge on the benzothiazole moiety will induce a stabilization of the ground state relative to the excited state, i.e., blue shifting of the transition energy.
Fig. 10.
Graphic HOMO and LUMO model of oxyluciferin. According to molecular modeling, transition from S1 to S0 leads to an internal negative charge transfer from the thiazolone ring near the AMP to the benzothiazole ring. Reprinted from [105] with permission from American Chemical Society (ACS)
On the other hand, water orientation and residue arrangements have different external electrostatic potentials on the QM keto-1 moiety, particularly the benzothiazole fragment. Therefore, if the external potential on the benzothiazole part stabilizes a charge there, due to luciferase conformational changes, then the transition energy should increase. However, the extension of the QM/MM studies to the other conformers suggested by recent experiments [101] and also mutant proteins will be suitable for more analysis.
Conclusions
In spite of the long history of work on firefly luciferase and the extensive experimental work towards production and application of red-emitter firefly luciferases, the chemical origin of multicolor bioluminescence have not been clearly understood through experimental investigation. Besides the classical observation and original interpretation, in recent years, combinations of theoretical and experimental investigations have shed light on the origin of color differences in firefly emission.
Considering all recent experimental and theoretical investigation and original observations, to explain the color tuning of the bioluminescence, the limitation of the environmental effects to any single factor would represent an oversimplification. This is an important issue when minor modification in luciferase primary structure is accompanied by global conformational changes, and especially considering that even the presence of a single water molecule in the protein scaffold could result in drastic change of the properties of the environment. Moreover, unlike other fluorescent proteins, most notably the green fluorescent protein, where the emitter is attached to the protein, in the case of the firefly luciferase the emitter has larger freedom of movement inside the active pocket, and thus the dynamic aspects of the emitter-environment interaction need to be considered for color tuning mechanism. Instead, it seems viable to accept that several factors can act collectively in the protein scaffold during the tuning of the emission color, until direct evidence is obtained about the structural changes that occur upon the color change. Therefore, it may be concluded that all point mutations of firefly luciferase with changes of color emission have effective roles in redirection of oxyluciferin or dynamic of water molecules around the emitter site. On the other hand, the interplay among luciferase structure rigidity, protein stability, and conformational changes should also be considered effective in controlling the water content and proximity around the oxyluciferin emitter site [113]. Meanwhile, in vivo bioluminescence color tuning may be attributed to some other factors which are mainly out of control dissimilar to in vitro conditions. Among those, matrix protein and protein–protein interactions, the presence of proteases may be mentioned, as their effect on luciferase structural stability and flexibility has been shown [114–116]. Decrease of protease-prone regions of firefly luciferase by site-directed mutagenesis brought about with more structural stability, higher and more stable bioluminescence signal in cell culture [117].
Overall, according to different investigations, seeking the origin of the different bioluminescence colors includes two different points: identical emitter structure and environmental factors that affect the deexcitation energy of the emitter. Environmental factors make a series of intramolecular properties of luciferase structure and solution properties. Both factors have been discussed and considered clearly [101]. The neutral form of oxyluciferin is a blue emitter under all conditions where it remains neutral in S0 as well as in S1 [101]. Therefore, due to the lack of blue emission in the real biological system, the emitter in the excited state should be ionized. The titration results showed larger acidity of the phenol hydroxyl group, and thus the excited emitter exists as a phenolate ion.
This does not rule out the possibility that the ion is created in the excited state, after a proton transfer has occurred from the neutral form. However, the comparative study of the emitter fluorescence in aqueous solutions and emitter-luciferase bioluminescence spectra confirmed that the emitting species exists as phenolate ion even in the ground state.
This is followed by another question on the emitting phenolate ion if it exists as enol or keto form. Although the data obtained on molecules similar to OxyLH2 are very useful, it seems difficult to answer this question solely from the spectra-structural data obtained from model compounds other than the real emitter. Fluorescence and bioluminescence spectra of the un-substituted emitter in various solvents, the pH dependence of the spectra, the crystal structures of the emitter, and the model theoretical calculations are all consistent with the yellow–green emitter in the luciferase existing as the phenolate ion of the enol form, phenolate-enol-OxyLH− [101]. On the other hand, according to another recent experimental study, the light emitter is the excited singlet state of the keto form of oxyluciferin phenolate anion, 1(OL−)* and the emission maximum of OL− is controlled by polarity of the active-site environment of the luciferase surrounding 1(OL−)* and the properties of the bonding interaction between 1(OL−)* and a countercation like a basic amino acid [68]. However, based on quantum mechanics/molecular mechanics (QM/MM) calculations and classical molecular dynamics (MD) simulations based on the “open” and “closed” x-ray structures of firefly L. cruciata luciferase the emission of various colors by only a keto form was confirmed [105].
Moreover, the red shift observed upon interaction with luciferase shows in the active pocket, the ground-state phenolate ion is enclosed in a less polar environment than the aqueous solution [101]. This implies that there are no water molecules proximate to the phenolate group of the emitter, which would otherwise greatly affect the environment polarity, or that the effect from the eventual water molecule(s) to the overall polarity is overweighed by interaction with hydrophobic protein residues, including alkyl residues (Ile) and phenyl rings (Phe).
The excited state of the phenolate ion decays before significant reorganization of the protein matrix has occurred, that is, the protein dynamics are slower relative to the deexcitation. A second proton transfer from the hydroxythiazole ring is not likely to occur. Thus, the emitter appears as a monoanion, a conclusion which is in agreement with the above discussion and is also supported by the comparison of the fluorescence lifetimes.
As discussed earlier, the color tuning phenomenon of the firefly luciferase is affected by intramolecular or intermolecular factors. Intramolecular factors, mainly the TICT mechanism, were ruled out by theoretical models [107, 118]. Therefore, only the intermolecular factors (luciferase–emitter interactions) are mainly considered. The crystal structure of L. cruciata luciferase in complex with an intermediate structure of emitter (DLSA) provided valuable information into the nature of the binding site of the emitter [86]. Accordingly, it was concluded that the color change can be caused by reversible closing/opening of the pocket around the emitter by conformational adjust of the proximate isoleucine residue (Ile288), which is induced by point mutation [86]. Conclusion was ruled out by recent QM/MM studies, which implicates that the deduced luciferase cavity size does not oblige the relative structures of the oxyluciferin substrate sufficiently to account for the observed color modulation [105].
Meanwhile, the crystal structure of the unbound emitter (of L. cruciata) in the same pocket shows that in the WT enzyme, the emitter does not slip far from the product AMP. There are several possible factors that are responsible of the ligand remaining close to the AMP in the active pocket, and each of these could change during the color tuning. The most apparent reason is the hydrogen bond of the thiazole oxygen atom to a water molecule that further bonds to the protein scaffold (Gly318 and Lys531; corresponding to Lys 529 of P. pyralis) through two additional hydrogen bonds [86].
Moreover, according to the mentioned crystal structure, the thiazole oxygen is close to the negative phosphate residue and fixed by a strong hydrogen bond to the phosphate thereby exist as enol form. Displacement of AMP relative to the emitter and the water molecule would weaken or destroy these hydrogen bonds, which would destabilize the enol form and the emitter could switch to its keto form [101]. The corresponding change of the hydrogen-bonding network will unavoidably result in redistribution of the negative charge within the ion and thus it will affect the emitted color. However, as it is recently stated, the excited singlet state of the keto form of oxyluciferin phenolate anion, 1(OL−)* could produce all emitted colors from green to red [68, 105].
Additional detail that had not been pointed out in previous studies, but became apparent from crystal structure, is that the phenyl ring from the residue Phe 249 (from L. cruciata luciferase) approaches the planar molecule laterally and could π–π interact with its π system. Being attached by a flexible bond, this phenyl group could reversibly approach or retreat from the ligand during the bioluminescence reaction. Changes of the π–π stacking distance as small as 0.02–0.10 Å are expected to result in significant shifts in the emission maxima by modulation of the deexcitation pathways. In turn, the orange emitter OxyL2−, the yellow–orange emitter enolate-OxyLH−, or perhaps even the yellow–green emitter phenolate-enol-OxyLH− could shift their emission to the orange or red regions. Moreover, a dynamic water molecule that comes close to the phenolate group of the emitter during the process may change the overall situation radically [101]. The critical role of dynamic water molecule were further credited by the effect of solvent polarity on the bioluminescence color, which shows that more polar region is responsible for red-color emission and that less polar region can interpret production of various colors of light from green to orange–red [68]. In fact, insertion of positive side-chain residues into a flexible loop of P. pyralis luciferase [94] and L. turkestanicus luciferase [93] brought about with displacement of the loop and probably more solvent accessibility and red-color emission.
The preceding discussion generally implies that the emitter is maintained close to the site of its creation by collective intermolecular interactions, which include hydrogen bonding (with water molecules, amino acid residues and/or AMP), Coulomb interactions (with the phosphate of the AMP), and π–π interactions (with a phenyl ring). Then, which of these factors is the most important for the color tuning? Effective factors will be ultimately determined by the relative positions of the ligand in the yellow–green– and red-emitting states, and therefore by the conformational change of the whole protein network [101]. Moreover, microenvironmental modifications (through the above interactions) may destabilize or stabilize the charge localized on the benzothiazole moiety. The same modifications can be induced by mutating some residues during experimental studies or by controlling the number of water molecules inside the cavity while performing in silico investigations [105]. At the present, the structure of the free (unattached) emitter only in the WT green-emitting protein is known. However, because the protein framework acts as a cooperative ensemble, it seems more sensible to suggest that it will affect the structure of the emitter in a multiple ways, with the specific and nonspecific interactions participating simultaneously. However, according to recent developments, the H-bond networks in the luciferase cavity involve water molecules and protein residues that mainly affects the charge redistribution in the oxyluciferin emitter during the S1 to S0 transition. This process is experimentally observed as the color modulation [105].
The effects of environmental polarity on the enolization of the keto form and the deprotonation of the enol, and the role of the neutral and ionized 6′-OH group in the fluorescence of the firefly emitter, oxyluciferin, has been reported through a detailed study of the structure and absorption and fluorescence spectra of its 6′-dehydroxylated analogue [119]. It has been shown that the deprotonated 6′-O− group is a necessary factor in accounting for the observed yellow–green and red emissions of oxyluciferin. Its negative charge is essential for effective excited-state charge transfer, which lowers the emission energy and broadens the emission spectrum. Deprotonation effect of the 6′-OH group on the emission energy from blue- to red in different tautomeric form has been shown [119]. However, the specific and non-specific interactions of protein framework as a cooperative ensemble in the light emission should be considered.
Concluding remark: is it possible to consider the mystery of firefly luciferase being resolved? In fact, according to recent developments it may be concluded that one of the main oxyluciferin emitter forms of luciferase reaction is the oxyluciferin keto form of the phenolate anion and the color of emitted light exclusively depends on the polarity of the surrounding area, which is solely attributed to water orientation. Is it the end of story?
Acknowledgments
Research council of Tarbiat Modares University (Prof. Y. Fathollahi) for financial support of bioluminescence research program is acknowledged. I thank Ms. Farangis Ataei for technical assistance.
References
- 1.Harvey EN. Bioluminescence. New York: Academic; 1952. [Google Scholar]
- 2.Haneda Y, Johnson FH, editors. Bioluminescence in progress. New Jersey: Princeton University Press; 1966. [Google Scholar]
- 3.Herring PJ, editor. Bioluminescence in action. New York: Academic; 1978. [Google Scholar]
- 4.Campbell AK. Chemiluminescence: principle and applications in biology and medicine. New York: VCH; 1988. [Google Scholar]
- 5.Shimomura O. Bioluminescence: chemical principles and methods. Singapore: World Scientific; 2006. [Google Scholar]
- 6.Shimomura O. Bioluminescence in the sea: photoprotein systems. Soc Exp Biol Symp. 1985;39:351–372. [PubMed] [Google Scholar]
- 7.Hastings JW, Morin JG. Bioluminescence. In: Prosser CL, editor. Neural and integrative animal physiology. New York: Wiley; 1991. pp. 131–170. [Google Scholar]
- 8.Meighen EA, Dunlap PV. Physiological, biochemical and genetic control of bacterial bioluminescence. Adv Microbial Physiol. 1993;34:1–67. doi: 10.1016/S0065-2911(08)60027-2. [DOI] [PubMed] [Google Scholar]
- 9.Wilson T, Hastings JW. Bioluminescence. Annu Rev Cell Dev Biol. 1998;14:197–230. doi: 10.1146/annurev.cellbio.14.1.197. [DOI] [PubMed] [Google Scholar]
- 10.Bronstein I, Fortin J, Stanley PE, Stewart GS, Kricka LJ. Chemiluminescent and bioluminescent reporter gene assays. Anal Biochem. 1994;219:169–181. doi: 10.1006/abio.1994.1254. [DOI] [PubMed] [Google Scholar]
- 11.Greer LF, 3rd, Szalay AA. Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence. 2002;17:43–73. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- 12.Lloyd JE. Bioluminescence and communication in insects. Annu Rev Entomol. 1983;28:131–160. doi: 10.1146/annurev.en.28.010183.001023. [DOI] [Google Scholar]
- 13.Wood KV. The chemical mechanism and evolutionary development of beetle bioluminescence. Photochem Photobiol. 1995;62:662–673. doi: 10.1111/j.1751-1097.1995.tb08714.x. [DOI] [Google Scholar]
- 14.Viviani VR. The origin, diversity, and structure function relationships of insect luciferases. Cell Mol Life Sci. 2002;59:1833–1850. doi: 10.1007/PL00012509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.White EH, Rapaport E, Seliger HH, Hopkins TA. The chemi- and bioluminescence of firefly luciferin: an efficient chemical production of electronically excited states. Bioorg Chem. 1971;1:92–122. doi: 10.1016/0045-2068(71)90009-5. [DOI] [Google Scholar]
- 16.DeLuca M. Firefly luciferase. Adv Enzymol Relat Areas Mol Biol. 1976;44:37–68. doi: 10.1002/9780470122891.ch2. [DOI] [PubMed] [Google Scholar]
- 17.McElroy WD. The energy source for bioluminescence in an isolated system. Proc Natl Acad Sci USA. 1947;33:342–345. doi: 10.1073/pnas.33.11.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seliger HH, McElroy WD. Quantum yield in the oxidation of firefly luciferin. Biochem Biophys Res Commun. 1959;1:21–24. doi: 10.1016/0006-291X(59)90082-8. [DOI] [Google Scholar]
- 19.Seliger HH, McElroy WD. Spectral emission and quantum yield of firefly bioluminescence. Arch Biochem Biophys. 1960;88:136–141. doi: 10.1016/0003-9861(60)90208-3. [DOI] [PubMed] [Google Scholar]
- 20.Wood KV, Lam YA, Seliger HH, McElroy WD. Complementary DNA coding click beetle luciferases can elicit bioluminescence of different colors. Science. 1989;244:700–703. doi: 10.1126/science.2655091. [DOI] [PubMed] [Google Scholar]
- 21.de Wet JR, Wood KV, Helinski DR, DeLuca M. Cloning firefly luciferase. Methods Enzymol. 1986;133:3–14. doi: 10.1016/0076-6879(86)33050-7. [DOI] [PubMed] [Google Scholar]
- 22.DeLuca M, McElroy WD. Purification and properties of firefly luciferase. Methods Enzymol. 1978;57:3–15. doi: 10.1016/0076-6879(78)57003-1. [DOI] [Google Scholar]
- 23.de Wet JR, Wood KV, Helinski DR, DeLuca M. Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli . Proc Natl Acad Sci USA. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greer LF, Szalay AA. Imaging of light emission from the expression of luciferases in living cells and organisms: a review. Luminescence. 2002;17:43–74. doi: 10.1002/bio.676. [DOI] [PubMed] [Google Scholar]
- 25.Contag CH, Bachmann MH. Advances in in vivo bioluminescence imaging of gene expression. Annu Rev Biomed Eng. 2002;4:235–260. doi: 10.1146/annurev.bioeng.4.111901.093336. [DOI] [PubMed] [Google Scholar]
- 26.Ando Y, Niwa K, Yamada N, Irie T, Enomoto T, Kubota H, Ohmiya Y, Akiyama H. Development of a quantitative bio/chemiluminescence spectrometer determining quantum yields: re-examination of the aqueous luminol chemiluminescence standard. Photochem Photobiol. 2007;83:1205–1210. doi: 10.1111/j.1751-1097.2007.00140.x. [DOI] [PubMed] [Google Scholar]
- 27.Conti E, Franks NP, Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/S0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 28.Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006;440:372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- 29.Gomi K, Kajiyama N. Oxyluciferin, a luminescence product of firefly luciferase, is enzymatically regenerated into luciferin. J Biol Chem. 2001;276:36508–36513. doi: 10.1074/jbc.M105528200. [DOI] [PubMed] [Google Scholar]
- 30.Emamzadeh R, Hosseinkhani S, Hemati R, Sadeghizadeh M. RACE-based amplification of cDNA and expression of a luciferin-regenerating enzyme (LRE): An attempt towards persistent bioluminescent signal. Enzyme Microb Technol. 2010;47:159–165. doi: 10.1016/j.enzmictec.2010.05.008. [DOI] [Google Scholar]
- 31.Doyle TC, Burns SM, Contag CH. In vivo bioluminescence imaging for integrated studies of infection. Cell Microbiol. 2004;6:303–317. doi: 10.1111/j.1462-5822.2004.00378.x. [DOI] [PubMed] [Google Scholar]
- 32.Roda A, Pasini P, Mirasoli M, Michelini E, Guardigli M. Biotechnological application of bioluminescence and chemiluminescence. Trends Biotechnol. 2004;22:295–303. doi: 10.1016/j.tibtech.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Branchini BR, Southworth TR, Khattak NF, Michelini E, Roda A. Red and green emitting firefly luciferase mutants for bioluminescent reporter application. Anal Biochem. 2005;345:140–148. doi: 10.1016/j.ab.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 34.Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 35.Billard P, DuBow MS. Bioluminescence-based assay for detection and characterization of bacteria and chemicals in clinical laboratories. Clin Biochem. 1998;31:1–14. doi: 10.1016/S0009-9120(97)00136-7. [DOI] [PubMed] [Google Scholar]
- 36.Bitton G, Koopman B. Bacterial and enzymatic bioassays for toxicity testing in the environment. Rev Environ Contam Toxicol. 1992;125:1–22. doi: 10.1007/978-1-4612-2890-5_1. [DOI] [PubMed] [Google Scholar]
- 37.Cheong WF, Prahl SA, Welch AJ. A review of the optical properties of biological tissues. IEEE J Quantum Electron. 1990;26:2166–2185. doi: 10.1109/3.64354. [DOI] [Google Scholar]
- 38.Rice BW, Cable MD, Nelson MB. In vivo imaging of light-emitting probes. J Biomed Opt. 2001;6:432–440. doi: 10.1117/1.1413210. [DOI] [PubMed] [Google Scholar]
- 39.Tromberg BJ, Shah N, Lanning R, Cerussi A, Espinoza J, Pham T, Svaasand L, Butler J. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia. 2000;2:26–40. doi: 10.1038/sj.neo.7900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shapiro E, Lu C, Baneyx F. A set of multicolored Photinus pyralis luciferase mutants for in vivo bioluminescence applications. Protein Eng Des Sel. 2005;18:581–587. doi: 10.1093/protein/gzi066. [DOI] [PubMed] [Google Scholar]
- 41.Seliger HH, Buck JB, Fastie WG, McElroy WD. The spectral distribution of firefly light. J Gen Physiol. 1964;48:95–104. doi: 10.1085/jgp.48.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biggley WH, Lioyd JE, Seliger HH. The spectral distribution of firefly light II. J Gen Physiol. 1967;50:1681–1692. doi: 10.1085/jgp.50.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colepicolo NP, Costa C, Bechara EJH. Brazilian species of luminescent Elateridae: Luciferin identification and bioluminescence spectra. Insect Biochem. 1986;16:803–810. doi: 10.1016/0020-1790(86)90117-4. [DOI] [Google Scholar]
- 44.Viviani VR, Bechara EJH. Biophysical and biochemical aspects of Phengodid (Railroad-Worm) bioluminescence. Photochem Photobiol. 1993;58:615–622. doi: 10.1111/j.1751-1097.1993.tb04941.x. [DOI] [Google Scholar]
- 45.Viviani VR, Bechara EJH. Bioluminescence and biological aspects of Brazilian railroad-worms (Coleoptera: Phengodidae) Ann Entomol Soc Am. 1997;90:389–398. [Google Scholar]
- 46.Ugarova NN, Brovko LY. Relationship between the structure of the protein globule and bioluminescence spectra of firefly luciferase. Russ Chem Bull. 2001;50:1752–1761. doi: 10.1023/A:1014365609421. [DOI] [Google Scholar]
- 47.Viviani VR, Bechara EJH. Bioluminescence of Brazilian fireflies (Coleoptera, Lampyridae): spectral distribution and pH effect on Luciferase-elicited colors. Comparison with Elaterid and Phengodid Luciferases. Photochem Photobiol. 1995;62:490–495. doi: 10.1111/j.1751-1097.1995.tb02373.x. [DOI] [Google Scholar]
- 48.Seliger HH, McElroy WD. The colors of firefly bioluminescence: enzyme configuration and species specificity. Proc Natl Acad Sci USA. 1964;52:75–81. doi: 10.1073/pnas.52.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viviani V, Uchida A, Suenaga N, Ryufuku M, Ohmiya Y. Thr226 is a key residue for bioluminescence spectra determination in beetle luciferases. Biochem Biophys Res Commun. 2001;280:1286–1291. doi: 10.1006/bbrc.2001.4254. [DOI] [PubMed] [Google Scholar]
- 50.Wood KV. Luc genes: introduction of color into bioluminescence assays. J Biolumin Chemilum. 1990;5:107–110. doi: 10.1002/bio.1170050206. [DOI] [PubMed] [Google Scholar]
- 51.Ohmiya Y, Hirano T, Ohashi M. The structural origin of the color differences in the bioluminescence of firefly luciferase. FEBS Lett. 1996;384:83–86. doi: 10.1016/0014-5793(96)00288-8. [DOI] [PubMed] [Google Scholar]
- 52.Viviani VR, Joaquim da silva Neto A, Ohmiya Y. The influence of the region between residues 220 and 344 and beyond in Phrixotrix railroad worm luciferases green and red bioluminescence. Protein Eng Des Sel. 2004;17:113–117. doi: 10.1093/protein/gzh016. [DOI] [PubMed] [Google Scholar]
- 53.Viviani VR, Uchida A, Viviani W, Ohimiya Y. The influence of Ala243 (Gly247), Arg215 and Thr226 (Asn230) on the bioluminescence spectra and pH-sensitivity of railroad worm, click beetle and firefly luciferases. Photochem Photobiol. 2002;76:538–544. doi: 10.1562/0031-8655(2002)076<0538:TIOAGA>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 54.Kajiyama N, Nakano E. Isolation and characterization of mutants of firefly luciferase which produce different colors of light. Protein Eng. 1991;4:691–693. doi: 10.1093/protein/4.6.691. [DOI] [PubMed] [Google Scholar]
- 55.Mamaev SV, Laikhter AL, Arslan T, Hecht SM. Firefly luciferase: alteration of the color of emitted light resulting from substitutions at position 286. J Am Chem Soc. 1996;118:7243–7244. doi: 10.1021/ja961053c. [DOI] [Google Scholar]
- 56.White EH, Roswell DF. Analogs and derivatives of firefly oxyluciferin, the light emitter in firefly bioluminescence. Photochem Photobiol. 1991;53:131–136. doi: 10.1111/j.1751-1097.1991.tb08478.x. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki N, Sato M, Nishikawa K, Goto T. Firefly bioluminescence II. Identification of a 2-(6-hydroxybenzothiazol-2′-YL-)-4-hydroxybenzothiazole as a product in the firefly lanterns and as a product in the chemiluminescence of firefly luciferin in DMSO. Tetrahedron Lett. 1969;10:4683–4684. doi: 10.1016/S0040-4039(01)88782-7. [DOI] [Google Scholar]
- 58.Suzuki N, Sato M, Okada K, Goto T. Studies on firefly bioluminescence-I: Synthesis and spectral properties of firefly oxyluciferin, a possible emitting species in firefly bioluminescence. Tetrahedron. 1972;28:4065–4074. doi: 10.1016/S0040-4020(01)93637-3. [DOI] [Google Scholar]
- 59.Gandelman OA, Brovko LY, Ugarova NN, Shchegolev AA. The bioluminescence system of firefly A fluorescence spectroscopy study of the interaction of the reaction product, oxyluciferin, and its analogs with luciferase. Biochemistry (Moscow) 1990;55:785–789. [Google Scholar]
- 60.Gandelman OA, Brovko LY, Ugarova NN, Chikishev AY, Shkurimov APJ. Oxyluciferin fluorescence is a model of native bioluminescence in the firefly luciferin-luciferase system. Photochem Photobiol B. 1993;19:187–191. doi: 10.1016/1011-1344(93)87083-Y. [DOI] [Google Scholar]
- 61.Alipour BS, Hosseinkhani S, Nikkhah M, Naderi-Manesh H, Chaichi MJ, Osaloo SK. Molecular cloning, sequence analysis, and expression of a cDNA encoding the luciferase from the glow-worm, Lampyris turkestanicus . Biochem Biophys Res Commun. 2004;325:215–222. doi: 10.1016/j.bbrc.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 62.Tafreshi NK, Sadeghizadeh M, Emamzadeh R, Ranjbar B, Naderi-Manesh H, Hosseinkhani S. Site-directed mutagenesis of firefly luciferase: implication of conserved residue(s) in bioluminescence emission spectra among firefly luciferases. Biochem J. 2008;412:27–33. doi: 10.1042/BJ20070733. [DOI] [PubMed] [Google Scholar]
- 63.Caysa H, Jacob R, Müther N, Branchini B, Messerle M, Söling A. A red shifted codon-optimized firefly luciferase is a sensitive reporter for bioluminescence imaging. Photochem Photobiol Sci. 2009;8:52–56. doi: 10.1039/b814566k. [DOI] [PubMed] [Google Scholar]
- 64.Baldwin TO. Firefly luciferase: the structure is known, but the mystery remains. Structure. 1996;4:223–228. doi: 10.1016/S0969-2126(96)00026-3. [DOI] [PubMed] [Google Scholar]
- 65.DeLuca M. Hydrophobic nature of the active site of firefly luciferase. Biochemistry. 1969;8:160–166. doi: 10.1021/bi00829a023. [DOI] [PubMed] [Google Scholar]
- 66.Morton RA, Hopkins TA, Seliger HH. The spectroscopic properties of firefly luciferin and related compounds. An approach to product emission. Biochemistry. 1969;8:1598–1607. doi: 10.1021/bi00832a041. [DOI] [PubMed] [Google Scholar]
- 67.Liu YJ, De Vico L, Lindh R. Ab initio investigation on the chemical origin of the firefly bioluminescence. Photochem Photobiol A. 2008;194:261–267. doi: 10.1016/j.jphotochem.2007.08.022. [DOI] [Google Scholar]
- 68.Hirano T, Hasumi Y, Ohtsuka K, Maki S, Niwa H, Yamaji M, Hashizume D. Spectroscopic studies of the light-color modulation mechanism of firefly (beetle) bioluminescence. J Am Chem Soc. 2009;131:2385–2396. doi: 10.1021/ja808836b. [DOI] [PubMed] [Google Scholar]
- 69.Imani M, Hosseinkhani S, Ahmadian S, Nazari M. Design and introduction of a disulfide bridge in firefly luciferase: increase of thermostability and decrease of pH sensitivity. Photochem Photobiol Sci. 2010;9:1167–1177. doi: 10.1039/c0pp00105h. [DOI] [PubMed] [Google Scholar]
- 70.Sala-Newby GB, Thomson CM, Campbell AK. Sequence and Biochemical similarities between the luciferases of the glow-worm Lampyris noctiluca and the firefly luciferase from Photinus pyralis . Biochem J. 1996;313:761–767. doi: 10.1042/bj3130761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mortazavi M, Hosseinkhani S, Khajeh K, Ranjbar B, Emamzadeh AR. Spectroscopic and functional characterization of Lampyris turkestanicus luciferase: a comparative study. Acta Biochem Biophys Sin. 2008;40:365–374. doi: 10.1111/j.1745-7270.2008.00411.x. [DOI] [PubMed] [Google Scholar]
- 72.McCapra F, Gilfoyle DJ, Young DW, Church NJ, Spencer P. In: Bioluminescence and chemiluminescence: fundamental and applied aspects. Campbell AK, Kricka LJ, Stanley PE, editors. Chichester: Wiley; 1994. pp. 387–391. [Google Scholar]
- 73.Brovko LY, Gandelman OA, Savich WI. In: Bioluminescence and chemiluminescence: fundamental and applied aspects. Campbell AK, Kricka LJ, Stanley PE, editors. Chichester: Wiley; 1994. pp. 525–527. [Google Scholar]
- 74.Orlova G, Goddard JD, Brovko LY. Theoretical study of the amazing firefly bioluminescence: the formation and structures of the light emitters. J Am Chem Soc. 2003;125:6962–6971. doi: 10.1021/ja021255a. [DOI] [PubMed] [Google Scholar]
- 75.Ugarova NN, Brovko LY. Relationship between the structure of the protein globule and bioluminescence spectra of firefly luciferase. Russ Chem Bull. 2001;50:1752–1761. doi: 10.1023/A:1014365609421. [DOI] [Google Scholar]
- 76.Gandelman OA, Brovko LY, Ugarova NN, Chikishev AY, Shkurimov AP. Oxyluciferin fluorescence is a model of native bioluminescence in the firefly luciferin-luciferase system. J Photochem Photobiol B. 1993;19:187–191. doi: 10.1016/1011-1344(93)87083-Y. [DOI] [Google Scholar]
- 77.Branchini BR, Murtiashaw MH, Magyar RA, Portier NC, Ruggiero MC, Stroh JG. Yellow-green and red firefly bioluminescence from 5, 5-dimethyloxyluciferin. J Am Chem Soc. 2002;124:2112–2113. doi: 10.1021/ja017400m. [DOI] [PubMed] [Google Scholar]
- 78.Hastings JW. Chemistries and colors of bioluminescent reactions: a review. Gene. 1996;173:5–11. doi: 10.1016/0378-1119(95)00676-1. [DOI] [PubMed] [Google Scholar]
- 79.Branchini BR, Southworth TL, Murtiashaw MH, Magyar RA, Gonzalez SA, Ruggiero MC, Stroh JG. An alternative mechanism of bioluminescence color determination in firefly luciferase. Biochemistry. 2004;43:7255–7262. doi: 10.1021/bi036175d. [DOI] [PubMed] [Google Scholar]
- 80.Hopkins TA, Seliger HH, White EH, Cass MH. The chemiluminescence of firefly luciferin. A model for the bioluminescent reaction and identification of the product excited state. J Am Chem Soc. 1967;89:7148–7150. doi: 10.1021/ja01002a076. [DOI] [PubMed] [Google Scholar]
- 81.McCapra F, Chang YC, Francois VP (1968) The chemiluminescence of a firefly luciferin analogue. Chem Commun 22–23
- 82.Shimomura O, Goto T, Johnson FH. Source of oxygen in the CO2 produced in the bioluminescent oxidation of firefly luciferin. Proc Natl Acad Sci USA. 1977;74:2799–2802. doi: 10.1073/pnas.74.7.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wannlund J, DeLuca M, Stempel K, Boyer PD. Use of 14C-carboxyl-luciferin in determining the mechanism of the firefly luciferase catalyzed reactions. Biochem Biophys Res Commun. 1978;81:987–992. doi: 10.1016/0006-291X(78)91448-1. [DOI] [PubMed] [Google Scholar]
- 84.Conti E, Stachelhaus T, Marahiel MA, Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Auld DS, Lovell S, Thorne N, Lea WA, Maloney DJ, Shen M, Rai G, Battaile KP, Thomas CJ, Simeonov A, Hanzlik RP, Inglese J. Molecular basis for the high-affinity binding and stabilization of firefly luciferase by PTC124. Proc Natl Acad Sci USA. 2010;107:4878–4883. doi: 10.1073/pnas.0909141107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nakatsu T, Ichiyama S, Hiratake J, Saldanha A, Kobashi N, Sakata K, Kato H. Structural basis for the spectral difference in luciferase bioluminescence. Nature. 2006;440:372–376. doi: 10.1038/nature04542. [DOI] [PubMed] [Google Scholar]
- 87.Mehrabi M, Hosseinkhani S, Ghobadi S. Stabilization of firefly luciferase against thermal stress by osmolytes. Int J Biol Macromol. 2008;43:187–191. doi: 10.1016/j.ijbiomac.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 88.Ganjalikhany MR, Ranjbar B, Hosseinkhani S, Khalifeh K, Hassani L. Roles of trehalose and magnesium sulfate on structural and functional stability of firefly luciferase. J Mol Catal B Enzym. 2010;62:127–132. doi: 10.1016/j.molcatb.2009.09.015. [DOI] [Google Scholar]
- 89.Yousefi-Nejad M, Hosseinkhani S, Khajeh K, Ranjbar B. Expression, purification and immobilization of firefly luciferase on alkyl-substituted Sepharose 4B. Enzyme Microb Technol. 2007;40:740–746. doi: 10.1016/j.enzmictec.2006.06.023. [DOI] [Google Scholar]
- 90.Emamzadeh AR, Hosseinkhani S, Sadeghizadeh M, Nikkhah M, Chaichi MJ, Mortazavi M. cDNA cloning, expression and homology modeling of a luciferase from the firefly Lampyroidea maculata . J Biochem Mol Biol. 2006;39:578–585. doi: 10.5483/bmbrep.2006.39.5.578. [DOI] [PubMed] [Google Scholar]
- 91.Inouye S. Firefly luciferase: an adenylate-forming enzyme for multicatalytic functions. Cell Mol Life Sci. 2010;67:387–404. doi: 10.1007/s00018-009-0170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Viviani VR, Bechara EJ, Ohmiya Y. Cloning, sequence analysis, and expression of active Phrixothrix railroad-worms luciferases: relationship between bioluminescence spectra and primary structures. Biochemistry. 1999;38:8271–8279. doi: 10.1021/bi9900830. [DOI] [PubMed] [Google Scholar]
- 93.Tafreshi NKh, Hosseinkhani S, Sadeghizadeh M, Sadeghi M, Ranjbar B, Naderi-Manesh H. The influence of insertion of a critical residue (Arg356) in structure and bioluminescence spectra of firefly luciferase. J Biol Chem. 2007;282:8641–8647. doi: 10.1074/jbc.M609271200. [DOI] [PubMed] [Google Scholar]
- 94.Moradi A, Hosseinkhani S, Naderi-Manesh H, Sadeghizadeh M, Alipour BS. Effect of charge distribution in a flexible loop on the bioluminescence color of firefly luciferases. Biochemistry. 2009;48:575–582. doi: 10.1021/bi802057w. [DOI] [PubMed] [Google Scholar]
- 95.Viviani VR, Arnoldi FG, Ogawa FT, Brochetto-Braga M. Few substitutions affect the bioluminescence spectra of Phrixothrix (Coleoptera: Phengodidae) luciferases: a site-directed mutagenesis survey. Luminescence. 2007;22:362–369. doi: 10.1002/bio.972. [DOI] [PubMed] [Google Scholar]
- 96.Alipour BS, Hosseinkhani S, Ardestani SK, Moradi A. The effective role of positive charge saturation in bioluminescence color and thermostability of firefly luciferase. Photochem Photobiol Sci. 2009;8:847–855. doi: 10.1039/b901938c. [DOI] [PubMed] [Google Scholar]
- 97.Dennis D, Stanford RH. The crystal and molecular structure of firefly D(−)-luciferin. Acta Crystallogr B. 1973;29:1053–1058. doi: 10.1107/S0567740873003845. [DOI] [Google Scholar]
- 98.Blank GE, Pletcher J, Sax M. On the crystal structure of firefly D(−)-luciferin. Acta Crystallogr B. 1974;30:2525. doi: 10.1107/S0567740874007527. [DOI] [Google Scholar]
- 99.Blank GE, Pletcher J, Sax M. The molecular structure of firefly D-(−)-luciferin: a single crystal x-ray analysis. Biochem Biophys Res Commun. 1971;42:583–588. doi: 10.1016/0006-291X(71)90411-6. [DOI] [PubMed] [Google Scholar]
- 100.Suzuki N, Sato M, Okada K, Goto T. Studies on firefly bioluminescence-I: synthesis and spectral properties of firefly oxyluciferin, a possible emitting species in firefly bioluminescence. Tetrahedron. 1972;28:4065–4074. doi: 10.1016/S0040-4020(01)93637-3. [DOI] [Google Scholar]
- 101.Naumov P, Ozawa Y, Ohkubo K, Fukuzumi S. Structure and spectroscopy of oxyluciferin, the light emitter of the firefly bioluminescence. J Am Chem Soc. 2009;131:11590–11605. doi: 10.1021/ja904309q. [DOI] [PubMed] [Google Scholar]
- 102.Wada N, Shibata R. Absorption spectral changes of firefly luciferin in deoxygenated dimethyl sulfoxide. J Phys Soc Jpn. 1997;66:3312–3313. doi: 10.1143/JPSJ.66.3312. [DOI] [Google Scholar]
- 103.Wada N, Mitsuta K, Kohno M, Suzuki N. ESR studies of firefly D-(−)-luciferin chemiluminescence in dimethyl sulfoxide. J Phys Soc Jpn. 1989;58:3501–3504. doi: 10.1143/JPSJ.58.3501. [DOI] [Google Scholar]
- 104.Brovko LY, Cherednikova EY, Chikishev AY, Chudinova EA. Effect of microenvironment on spectral-kinetic properties of firefly luciferin. Moscow Univ Trans Phys Astron Ser. 1998;3:26–29. [Google Scholar]
- 105.Navizet I, Liu YJ, Ferré N, Xiao HY, Fang WH, Lindh R. Color-tuning mechanism of firefly investigated by multi-configurational perturbation method. J Am Chem Soc. 2010;132:706–712. doi: 10.1021/ja908051h. [DOI] [PubMed] [Google Scholar]
- 106.Li Zw, Ren Am, Guo JF, Yang T, Goddard JD, Feng JK. Color-tuning mechanism in firefly luminescence: theoretical studies on fluorescence of oxyluciferin in aqueous solution using time dependent density functional theory. J Phys Chem A. 2008;112:9796–9800. doi: 10.1021/jp8014047. [DOI] [PubMed] [Google Scholar]
- 107.Yang T, Goddard JD. Predictions of the geometries and fluorescence emission energies of oxyluciferins. J Phys Chem A. 2007;111:4489–4497. doi: 10.1021/jp068542b. [DOI] [PubMed] [Google Scholar]
- 108.Ren A, Guo J, Feng J, Zou L, Li Z, Goddard JD. TDDFT study of the electronic structure, absorption and emission spectra of the light emitters of the amazing firefly bioluminescence and solvation effects on the spectra. Chin J Chem. 2008;26:55–64. doi: 10.1002/cjoc.200890038. [DOI] [Google Scholar]
- 109.Nakatani N, Hasegawa JY, Nakatsuji H. Artificial color tuning of firefly luminescence: theoretical mutation by tuning electrostatic interactions between protein and luciferin. Chem Phys Lett. 2009;469:191–194. doi: 10.1016/j.cplett.2008.12.062. [DOI] [Google Scholar]
- 110.Kobayashi R, Amos RD. The application of CAM-B3LYP to the charge-transfer band problem of the zincbacteriochlorin–bacteriochlorin complex. Chem Phys Lett. 2006;420:106–109. doi: 10.1016/j.cplett.2005.12.040. [DOI] [Google Scholar]
- 111.Tozer DJ, Amos RD, Handy NC, Roos BO, Serrano-Andre′s L. Does density functional theory contribute to the understanding of excited states of unsaturated organic compounds? Mol Phys. 1999;97:859–868. doi: 10.1080/00268979909482888. [DOI] [Google Scholar]
- 112.Tagami A, Ishibashi N, Kato DI, Taguchi N, Mochizuki Y, Watanabe H, Ito M, Tanaka S. Ab initio quantum-chemical study on emission spectra of bioluminescent luciferases by fragment molecular orbital method. Chem Phys Lett. 2009;472:118–123. doi: 10.1016/j.cplett.2009.02.076. [DOI] [Google Scholar]
- 113.Maghami P, Ranjbar B, Hosseinkhani S, Ghasemi A, Moradi A, Gill P. Relationship between stability and bioluminescence color of firefly luciferase. Photochem Photobiol Sci. 2010;9:376–383. doi: 10.1039/b9pp00161a. [DOI] [PubMed] [Google Scholar]
- 114.Ataei F, Hosseinkhani S, Khajeh K. Limited proteolysis of luciferase as a reporter in nanosystem biology: a comparative study. Photochem Photobiol. 2009;85:1162–1167. doi: 10.1111/j.1751-1097.2009.00583.x. [DOI] [PubMed] [Google Scholar]
- 115.Ataei F, Hosseinkhani S, Khajeh K. Luciferase protection against proteolytic degradation: a key for improving signal in nano-system biology. J Biotechnol. 2009;144:83–88. doi: 10.1016/j.jbiotec.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 116.Thompson JF, Geoghegan KF, Lloyd DB, Lanzetti AJ, Magyar RA, Anderson SM, Branchini BR. Mutation of a protease-sensitive region in firefly luciferase alters light emission properties. J Biol Chem. 1997;272:18766–18771. doi: 10.1074/jbc.272.30.18766. [DOI] [PubMed] [Google Scholar]
- 117.Riahi-Madvar A, Hosseinkhani S. Design and characterization of novel trypsin-resistant firefly luciferases by site-directed mutagenesis. Protein Eng Des Sel. 2009;22:655–663. doi: 10.1093/protein/gzp047. [DOI] [PubMed] [Google Scholar]
- 118.Nakatani N, Hasegawa J, Nakatsuji H. Red light in chemiluminescence and yellow-green light in bioluminescence:# color-tuning mechanism of firefly, Photinus pyralis, studied by the symmetry-adapted cluster–configuration interaction method. J Am Chem Soc. 2007;129:8756–8765. doi: 10.1021/ja0611691. [DOI] [PubMed] [Google Scholar]
- 119.Naumov P, Kochunnoonny M. Spectral-structural effects of the keto-enol-enolate and phenol-phenolate equilibria of oxyluciferin. J Am Chem Soc. 2010;132:11566–11579. doi: 10.1021/ja102885g. [DOI] [PubMed] [Google Scholar]