Abstract
Recent research into the role of microRNA (miR) in the immune system has identified the miR-29 family as critical regulators of key processes in adaptive immunity. The miR-29 family consists of four members with shared regulatory capacity, namely miR-29a, miR-29b-1, miR-29b-2 and miR-29c. Being expressed in both T and B cells, as well as the main accessory cell types of thymic epithelium and dendritic cells, the miR-29 family has been identified as a putative regulator of immunity due to the predicted suppression of key immunological pathways. The generation of a series of in vivo molecular tools targeting the miR-29 family has identified the critical role of these miR in setting the molecular threshold for three central events in adaptive immunity: (1) control over thymic production of T cells by modulating the threshold for infection-associated thymic involution, (2) creating a neutral threshold for T cell polarization following activation, and (3) setting the threshold for B cell oncogenic transformation. These results identify the miR-29 family as potent immune modulators which have already been exploited through the evolution of a viral mimic and could potentially be exploited further for therapeutic intervention.
Keywords: MicroRNA, Thymus, T cells, B cells, Immunology, Leukemia
Introduction
MicroRNA (miR) are small noncoding RNA with the capacity to interfere in the expression of protein-coding mRNA. The regulation of mRNA by miR has a potent effect on many cellular functions. Within the adaptive immune system, several families of miR have been identified to be of elevated importance, due to functions in regulating key immunological pathways [1]. Recent studies have added the miR-29 family to the list of key miR in the adaptive immune system. The miR-29 family consists of four closely related members, miR-29a, miR-29b-1, miR-29b-2 and miR-29c. Each member is characterized by the same “seed region” (positions two to eight of the 5′ end) and hence heavily overlap in their predicted mRNA targeting. There are two bi-cistronic clusters of miR-29, the miR-29a/b-1 cluster and the miR-29b-2/c cluster, which have arisen by gene duplication [2, 3]. The miR-29a/b-1 cluster is located on the antisense strand of chromosome 7 of the human genome and chromosome 6 of the mouse genome, while the miR-29b-2/c cluster is located on the antisense strand of the human genome and the sense strand of the mouse genome, and on chromosome 1 in both species. The mature sequences of miR-29b-1 and miR-29b-2 are identical, while miR-29a and miR-29c are more distantly removed from miR-29b and are distinguished from each other by a difference in only a single nucleotide outside the seed sequence [4, 5]. Despite similar sequences, the miR have different subcellular compartmentalization, as miR-29a is mainly cytoplasmic, while miR-29b and miR-29c are concentrated in the nucleus [3, 6]. In the case of miR-29b the nuclear localization is due to a hexanucleotide motif in the 3′ end, allowing shuttling via CRM1 [2, 3, 7]. The mature human and mouse miR-29 sequences are identical, but the regulation of mRNA by miR-29 may vary across species based on expression patterns and the presence of the anti-sense seed in the target mRNA [8, 9].
Expression and regulation of the miR-29 family
The miR-29 family shows broad expression, being ubiquitously detected at the organ level in both humans [10] and mice [11, 12]. The highest expression at the organ level in humans is in the brain and heart [13], and in mice in the brain [11, 14]. In the adaptive immune system, the miR-29 family is highly expressed in both T cells and B cells, although relative subset expression still needs to be elucidated, as well as the key accessory cell types of dendritic cells and thymic epithelial cells [5, 15]. The transcription factor binding sites in the promoters of the miR-29 genes driving this gene expression pattern are conserved between human and mouse, suggesting an explanation for the cross-species expression concordance [2, 8, 9, 16]. There are binding sites for the transcription factor Myc in the promoter region of both clusters [2]. Eyholzer et al. [17] describe the regulation of miR-29a/b-1 expression through CEBPA and Mott et al. [16] reported and confirmed the binding sites for c-Myc and further identified a Gli binding site, involved in hedgehog signaling, as well as multiple NF-κB sites. The miR-29a/b-1 promoter region also includes two binding sites for TCF/LEF, factors involved in canonical Wnt signaling [18]. In the upstream region of the miR-29b-2/c cluster, there are binding sites for the YY1 transcription factor controlled by the activation of NF-κB [8]. Different splice variants have been reported for the two miR-29 primary transcripts, but the control mechanisms, expression patterns and biological effects remain unclear [2, 16].
Differences between the promoter regulation of the miR-29a/b-1 and miR-29b-2/c clusters may be responsible for the inconsistent ratio of miR-29a, miR-29b and miR-29c expression across tissues [13]. However, miR-29 is also regulated in a post-transcriptional manner. A study by Hwang et al. in HeLa cells demonstrated that while the clusters of miR-29a/b-1 and miR-29b-2/c are cotranscribed, the mature miR show differential expression. MiR-29a was found to be present in all stages of the cell cycle, while miR-29b was detected at high levels only during mitosis and was characterized by rapid decay in all other stages; by contrast mature miR-29c was not detected [3]. Zhang et al. used a pulse-chase experiment to accurately measure the turnover rate of miR. This study showed that the uracils that miR-29b possesses in its 9–11 nucleotide positions are responsible for its observed short life-span, and that the differences between miR-29a and miR-29c result in the relatively quicker decay of miR-29c [19]. The stable expression levels of miR-29a, miR-29b and miR-29c in individual cell types is therefore likely to depend on the expression of the two clusters, alternative splicing of primary RNA and differential decay, all factors which may vary in a cell-specific manner.
Biological functions of the miR-29a family
Based on seed sequence prediction, the miR-29 family has up to 6,000 predicted targets, largely overlapping between the different members [20]. While only about 50 of the predicted targets have been experimentally validated, analysis of the preliminary list suggests a number of processes in which the miR-29 family may be important. A list of about 1,000 predicted targets with a high degree of conservation indicates that the miR-29 family is likely to have a significant impact on three gene networks: (1) cellular processes and connective tissues, (2) nervous and cardiovascular systems, and (3) cancer and hematological function (Table 1). The unsupervised association with connective tissue disorder has been experimentally demonstrated, with predicted targets validated from within both constituent proteins (elastin, fibrillin, fibrinogen and peroxidasin) [12, 21–28] and extracellular matrix-modifying enzymes (ADAM12, BMP1 and MMP2) [29, 30]. Furthermore, reduced miR-29 expression is associated with fibrosis [12, 24, 31] and forced expression protects against fibrosis [32]. The role of the miR-29 family in the nervous and cardiovascular systems is under-investigated, but it is notable that the brain and heart are the tissues with the highest expression of miR-29, and several key neurological genes, including BACE-1 [33], Arpc3 [34] and PGRN [35], have been validated as direct targets. The third network, cancer and hematological function, is the most relevant to this review, as it suggests the potential for enrichment of immunological pathway genes. Indeed, when canonical signaling pathways are tested for enrichment of predicted miR-29 target genes, of the ten pathways with the most significant enrichment for predicted targets, five have a critical role in the adaptive immune system (Table 2).
Table 1.
Biological pathways enriched for predicted miR-29 family targets
| Network | Predicted miR-29 targets within network |
|---|---|
| Cellular processes and connective tissues | ADAM19, ADAMTS2, ADAMTS5, AGTR2, AHR, AKT3, ANK3, ARNT, BAK1, BBC3, BCL11A, BTG2, CAMKK2, CCND2, CDK6, COL15A1, COL16A1, COL1A2, COL1A1,COL2A1, COL3A1, COL4A1, COL4A3, COL4A2, COL5A1, COL5A2, COL6A3, COL7A1, CX3CL1, DICER1, DNMT3A, DNMT3B, DUSP2, FKBP4, FOS, FOXO3, FOXO4, FSTL1, G6PC, GAB1, GLIS2, GSK3B, HIF3A, HIP1, HMGCR, ICOS, IGF1, IL1RAP, IRS1, ITGB1, JARID2, KDM5A, LEP, LIF, LOX, LPL, LRP6, MAP2K4, MAPK10, MCL1, MMP2, MSTN, MYBL2, MYCN, NCOA3, NFATC4, PDGFA, PDGFB, PDGFRB, PIK3R1, PMP22, POU2F2, PPARD, PPM1D, PRKG1, PTEN, PTX3, SERPINH1, SOCS7, SP1, SPARC, TBX21, TFEC, TNFRSF1A, TP53INP1, TPM1, TRAF3, TRIM63, VCL, VEGFA, WISP1, YWHAE, YY1, ZFP36 |
| Nervous and cardiovascular systems | ADCYAP1R1, AHR, ATP1B1, BDH1, CCDC88A, CNOT6, COL11A1, COL9A1, CSPG4, DDX3X, DPYSL5, E2F7, ELN, ENHO, EPHB3, ERCC6, FBN2, FRAS1, FREM1, GAS7, GNG2, GRIP1, HEXA, HSPG2, IREB2, ISLR2, KCTD20, KIF3B, KLF4, LAMC1, LARP4, MAP2K4, MAP2K6, MAT1A, MCF2L, NFIB, NPAS3, NRAS, PER1, PER3, PGAP1, PIK3R3, PURA, QKI, RAB12, RAB30, RARB, RORA, RPS6KA3, SH3RF3, SHB, SLC1A2, STMN2, STX16, SYNCRIP, TIAM1, TNFAIP1, TRAF4, TRIB2, TSPAN4, TUBB2A, VAMP3, VPS26B, ZFX |
| Cancer, hematological function | AKAP13, ANTXR2, ARPP19, ASIC1, ATP7A, BAK, BMP1, BMPR1A, CCDC80, CDC42BPA, CDC7, CLDN1, COL6A2, CYP21A2, DIABLO, DNAJB2, DOT1L, DPP4, DTX4, EIF2S2, ELF2, ELL2, EN1, ETV6, FERMT2, FGA, FRMD4A, GPCPD1, HAPLN1, HTR7, IFI30, IFNAR2, IGF1, ITGA11, KCNMA1, KDM6B, KPNA1, LIF, LOX, MARK3, MOG, NCOR2, NFATC3, OXTR, PDGFC, PITPNA, PLP1, PPIC, PPP1R13B, PRELP, PRKRA, PTHLH, REST, REV3L, SCMH1, SCN3B, SGK1, SYT7, TGFB2, TLL1, TSC22D3, ZNF346 |
IPA (Ingenuity® Systems, http://www.ingenuity.com) was used to analyze 919 mRNA with a predicted conserved binding site for miR-29 (TargetScan) to detect disproportionate representation of target genes within biological networks. The three most enriched networks and the predicted miR-29 targets within these networks are shown.
Table 2.
Canonical signaling pathways enriched for predicted miR-29 family targets
| Canonical pathway | p value | Enrichment | Predicted targets |
|---|---|---|---|
| April signaling | 0.0004 | 8/40 (20 %) | MAP2K4, FOS, TRAF3, NFAT5, NFATC3, MAPK10, NFATC4, ELK1, |
| IL-6 signaling | 0.0004 | 15/118 (13 %) | MAP2K4, MAP2K6, NRAS, TNFRSF1A, PIK3R1, MAP4K4, VEGFA, PIK3R3, FOS, COL1A1, MAPK10, AKT3, ELK1, IL1RAP, MCL1 |
| BAFF signaling | 0.0005 | 8/42 (19 %) | MAP2K4, FOS, TRAF3, NFAT5, NFATC3, MAPK10, NFATC4, ELK1 |
| Glioma signaling | 0.0005 | 13/102 (13 %) | NRAS, CAMK1D, PDGFA, PIK3R1 CDK6, PDGFC, PDGFB, PTEN, PIK3R3, IGF1, AKT3, PDGFRB, CAMK2G |
| Axonal guidance signaling | 0.0006 | 33/398 (8 %) | DPYSL2, NFATC3, PDGFA, PIK3R1 ROBO1 PDGFC, TUBB2B, VEGFA, NFAT5, IGF1, PLXNA1, EFNA5, BAIAP2, ADAM19, AKT3, SRGAP2, GSK3B, GNG12, BMP1, EFNA2, ITGB1, NRAS, EPHA1, TUBB2A, DPYSL5, NFATC4, PDGFB, PIK3R3, GLIS2, ADAM12, EPHB3, FZD5, GNG2 |
| TR/RXR activation | 0.0006 | 12/87 (14 %) | PIK3R3, COL6A3, PIK3R1, SLC16A2, AKT3, NCOA4, SYT2, G6PC, NCOR2, FGA, DIO2, NCOA3 |
| BCR signaling | 0.001 | 16/143 (11 %) | MAP2K4, MAP2K6, NRAS, POU2F2, NFATC3, PIK3R1, NFATC4, CREB5, PTEN, PIK3R3, NFAT5, GAB1, AKT3, GSK3B, ELK1, CAMK2G |
| Intrinsic prothrombin activation pathway | 0.002 | 6/29 (21 %) | COL1A2, COL1A1, COL5A3, COL2A1, FGA, COL3A1 |
| PDGF signaling | 0.002 | 10/72 (14 %) | MAP2K4.PIK3R3.FOS.NRAS.PDGFA.PIK3R1.ELK1.PDGFC.PDGFB.PDGFRB |
| Estrogen-dependent breast cancer signaling | 0.002 | 9/62 (15 %) | PIK3R3, FOS, NRAS, IGF1, SP1, PIK3R1, AKT3, CREB5, ELK1 |
IPA (Ingenuity® Systems, http://www.ingenuity.com) was used to analyze 919 mRNA with a predicted conserved binding site for miR-29 (TargetScan) to detect disproportionate representation of target genes within canonical signaling pathways. The ten most significantly enriched canonical pathways, p value of enrichment, representation of miR-29 targets within the pathway and the list of predicted targets are shown.
While the miR-29 family is highly expressed in adaptive immune cells, and predicted miR-29 target genes cluster within key signaling pathways of the adaptive immune system, the physiological role of any miR depends not just on molecular capacity to inhibit, but also on the coexpression of miR and the mRNA target, the biological relevance of the inhibition degree and any network effects which may neutralize or exacerbate individual effects. A key step in understanding the effects of the miR-29 family has thus been the development of multiple in vivo molecular tools to access gain-of-function or loss-of-function in mouse models. Gain-of-function models have been developed where miR-29 family members are overexpressed, through a transgenic model, such as the B cell-specific overexpression of the miR-29a/b-1 cluster under the VH promoter-IgH-Eμ enhancer [25], a viral transfection model, such as the retroviral transfection of bone-marrow stem cells with miR-29a [36] or sleeping beauty-mediated transfection of lung epithelium [32], or systemic delivery of miR-29a [37]. Loss-of-function models have been developed as classical knockout mice of the miR-29a/b-1 cluster [15], a Cre-Lox-inducible knockout of the miR-29a/b-1 cluster [38] or the expression of a miR-29 “sponge” sequence (either by transgene or lentivirus), capable of acting as a decoy to preserve the expression of bona fide miR-29 targets [39]. These in vivo tools have allowed the discovery of three key roles of the miR-29 family in the adaptive immune system: setting the threshold in thymic involution, helper T cell differentiation and lymphocyte oncogenesis.
Setting the threshold for thymic involution
Analysis of miR-29a/b-1-deficient mice indicates that miR-29a/b-1 are not essential for the T cell-intrinsic differentiation pathways, with lineage commitment, β-selection and positive selection all intact in knockout mice [15]. Nevertheless, miR-29a/b-1 does have a critical function in supporting T cell production in a T cell-extrinsic manner, namely a function in preventing inappropriate thymic atrophy.
While the thymus is essential for T cell differentiation and the production of a normal peripheral T cell repertoire, constitutive function is not required. Atrophy of the thymus, termed thymic involution, results in a >90 % reduction in thymus size and a corresponding reduction in T cell generation. There are multiple triggers for thymic involution, including infection, age, pregnancy and stress, and no consensus on whether the physiological function is primarily immunological (i.e., to halt T cell differentiation) or metabolic (i.e., to shut down a metabolically expensive process [40–42]. Despite this, the molecular mechanisms that underpin thymic involution are becoming increasingly well defined [43]. One of the key mechanisms by which thymic involution is driven during infection is through type I interferons (IFNα and IFNβ) produced by the pathogen-sensing pathway.
During an infection, type I IFN is produced in response to pathogen-associated molecular patterns (PAMPs), via several discrete pathways including the Toll-like receptor (TLR) family, the nucleotide-oligomerization domain (NOD)-like receptors and the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs). Following production, type I IFN signaling occurs through a common heterodimeric receptor, known as the IFNα/β receptor (IFNAR), which is expressed by nearly all cell types. Binding to IFNAR activates the Jak/Stat pathway, leading to the induction of interferon response factors (IRFs), which protect the cell against infection [44]. In addition to the system-wide anti-infection function, the type I IFN pathway has been coopted by the thymus as the trigger for thymic involution. Thymic epithelial cells detect type I IFN produced during an infection, and drive a coordinated process of thymic involution, keeping thymic size and function at a minimum until the infection resolves [15, 45].
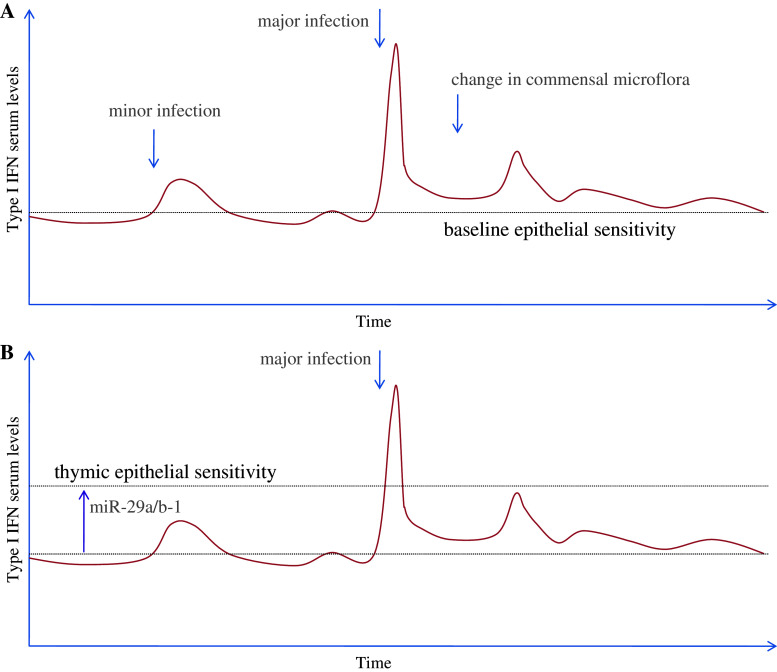
Utilization of the type I IFN pathway allows the thymic epithelium to respond to infections by minimizing thymic function, either to prevent the creation of immunological tolerance against the infectious organism or to divert metabolic responses from development to immunity [46]. However, the exploitation of an evolutionarily conserved pathway creates the potential issue of perpetual thymic involution. Epithelial cells outside the thymus are exquisitely sensitive to type I IFN allowing rapid protection against infection. However, the thymus produces a basal level of IFNα independent of infection, which is thought to be important for T cell differentiation [47]. Furthermore, commensal bacteria can trigger low levels of type I IFN. Thus, thymic epithelial cells are under opposing forces to ensure rapid involution during a high-risk infection, while “ignoring” the basal production of type I IFN. The mechanism for tuning type I IFN signaling in thymic epithelial cells is set by miR-29a/b-1 expression, which inhibits the production of IFNAR1 [15]. Through the expression of miR-29a/b-1, sensitivity to type I IFN is reduced in the thymus to the point where involution does not occur in response to baseline or commensal production, but the capacity to appropriately respond to major infections is maintained (Fig. 1). When miR-29a/b-1 is deleted in thymic epithelial cells, IFNAR expression and signaling is increased to the point where the thymus undergoes chronic involution, preventing the future production of T cells [15]. Notably, loss of the entire miR network through Dicer deletion in thymic epithelial cells phenocopies miR-29a/b-1, indicating that miR-29a/b-1 is the dominant miR in this pathway [15].
Fig. 1.
Setting the threshold of thymic involution. a Epithelial cells outside the thymus maintain a high sensitivity to type I IFN, responding to minor and major infections and changes in commensal microflora. Replicated in the thymus this would result in chronic thymic involution, preventing normal T cell production. b To establish a level of sensitivity appropriate for thymic involution, thymic epithelial cells express miR-29a/b-1, reducing sensitivity to type I IFN. This elevates the threshold for thymic involution to the point where major infectious events can trigger involution, but normal function is maintained throughout minor infectious events and changes in commensal microflora
Setting the threshold for T cell polarization
Following T cell maturation, the miR-29 family has a critical T cell-intrinsic function in setting the threshold for polarization into different effector T cell fates. Upon CD4 T cell activation, the lineage fate decision is critical to determine the function of the activated T cell, with one of the most fundamental decisions being between the fates of Th1 cellular immunity, with strong IFNγ production, or Th2 humoral immunity, with strong IL-4 production. The balance of IFNγ production is of particular importance, due to the role of IFNγ in stabilizing Th1 and inhibiting Th2 differentiation [48], and directly increasing resistance against intracellular infections during cellular immunity [49]. MiR play a critical role in this cell fate decision, as Dicer-deficient T cells show an innate polarization towards IFNγ-producing Th1 cells in vitro [50]. Surprisingly, this innate polarization is reversed in vivo, where Dicer-deficient T cells show reduced entry into the IFNγ-producing Th1 lineage [51]. Nevertheless, individual miR can have effects opposite to the network as a whole.
In a recent study, Steiner et al. used an innovative genetic strategy to determine the miR responsible for the Th1 polarization in miR-deficient T cells in vitro. This study used a miR replacement approach, starting with DGCR8 −/− miR-deficient CD4 T cells (with the same innate in vitro polarization to the Th1 lineage as Dicer-deficient cells) and screening pools of miR for the capacity to restore the miR-deficient phenotype back to the wild-type phenotype [52]. This screen identified miR-29a and miR-29b as capable of correcting the Th1 bias of DGCR8 −/− miR-deficient CD4 T cells, thus identifying the miR-29 family as critical suppressors of the Th1 cell fate. A concurrent study assessed the role of the miR-29 family in vivo, through the transgenic expression of a miR-29 “sponge” sequence. Building on the in vitro results, these mice, with a loss of miR-29-dependent inhibition, showed a large increase in the number of Th1 cells and IFNγ production [39]. The miR-29 family likely suppresses entry into the Th1 fate by regulating several key targets. Steiner et al. [52] identified T-bet and Eomes as validated direct targets of miR-29. Ma et al. [39], by contrast, did not see increased T-bet and Eomes, and instead demonstrated that IFNγ is a direct target of the miR-29 family. It is likely that miR-29 suppresses the Th1 fate by targeting all three genes—IFNγ, T-bet and Eomes—and that enhanced expression of T-bet and Eomes largely functions to reduce the threshold of Th1 induction, with expression levels normalized following polarization by feedback mechanisms.
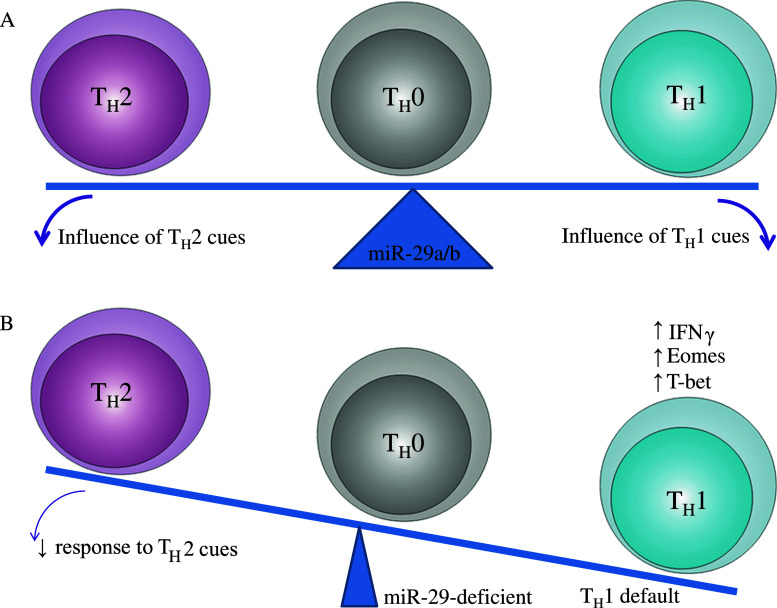
Together, these two studies suggest three functions of miR-29a/b in mature T cells. Firstly, the expression of miR-29a/b in mature CD4 T cells is critical for setting the threshold for the Th1/Th2 cell fate decision. In the presence of miR-29a/b, the cell fate decision is finely balanced, allowing microenvironmental influences to determine the effector cell type produced, while in the absence of miR-29a/b, the cell fate decision is skewed towards the Th1 lineage (Fig. 2). Secondly, downregulation of miR-29a/b following exposure to intracellular bacteria removes this counterbalancing force and initiates a positive feedback loop of enhanced IFNγ production and increased resistance to infection [39]. Thirdly, miR-29a/b likely plays a similar role in the CD8 T cell and NK cell lineages, as both of these cell types reduce miR-29a/b following exposure to intracellular bacteria and have enhanced IFNγ production in miR-29 “sponge” mice [39].
Fig. 2.
Setting the threshold for Th1 polarization. a Through the expression of miR-29a/b, T cells are finely balanced in the Th1-Th2 cell fate decision. This fine balance allows external microenvironmental factors to influence the cell fate decision, allowing the adaptive immune response to be highly responsive to the context of infection. b In the absence of miR-29a/b, IFNγ, Eomes and T-bet are derepressed, allowing stochastic effects to drive the cell fate decision towards the Th1 fate. This induction of a strong default reduces the capacity of the undifferentiated T cell to respond to appropriate microenvironmental cues, resulting in a less context-dependent immune response
Setting the threshold for lymphoid oncogenesis
The miR-29 family has important functions in B cells, as suggested by the significant enrichment of B cell signaling pathways among miR-29 targets (Table 2). Unlike the other cellular lineages of the adaptive immune system, no obvious phenotype has yet been observed for B cells in miR-29-deficient mice [15, 39]. By contrast, an important phenotype in the B cell lineage has been observed in cases where the miR-29 family is overexpressed—the onset of oncogenesis.
MiR-29a is upregulated in aggressive B cell chronic lymphocytic leukemia (B-CLL), and further upregulated in indolent B-CLL [25], compared to nontransformed B cells. This upregulation is likely to be a key event in transformation, as transgenic mice overexpressing miR-29a/b-1 in B cells show an expansion of CD5+CD19+IgM+ B cells that is similar to the findings in indolent B-CLL [25]. In a fascinating “natural experiment” of ectopic expression, the bovine leukemia virus, which causes a B-CLL-like leukemia, expresses a viral miR, BLV-miR-B4, with an identical seed region to the miR-29 family [53]. While these overexpression events both suggest an oncogenic function for miR-29 in B cells, Tcl1, an important oncogene in aggressive B-CLL, is a direct target of miR-29, which would be more consistent with a tumor suppressor function of miR-29 [54]. Indeed, miR-29 downregulation is a poor prognostic marker in aggressive B-CLL [55], although notably this level of expression is still higher than that of normal B cells [25].
The seemingly paradoxical association of elevated expression with both tumor formation and nonaggressive growth indicates the complex nature of the miR-29 family in B cell oncogenesis. One plausible explanation lies in the high levels of expression of miR-29a in hematopoietic stem cells. Elevated expression in mature B cells may thus replicate the hematopoietic stem cell phenotype; indeed, ectopic expression can promote a stem cell-like proliferative and self-renewal capacity [36]. The initiation of a stem cell-like program would act as an oncogenic event, but the controlled proliferative nature of stem cells would result in a nonaggressive tumor, such as indolent B-CLL. Alternative oncogenic events may have the effect of freeing up the tumor cell line from a dependence on a miR-29-driven stem cell-like phenotype, at which point miR-29 expression would become a limiting factor in proliferation through downregulation of oncogenes such as Bcl1-2, Mcl1, Tcl1 and SKI [54, 56, 57] and the activation of the p53 pathway through the repression of p85α and CDC42 [58]. B cells that transformed into miR-29-independent cancers would therefore demonstrate selective pressure to downregulate miR-29 expression. This complex relationship would explain why ectopic expression of miR-29 can drive oncogenesis in B cells [25] and myeloid cells [36], yet downregulation is associated with more aggressive forms of cancer in both lineages [55, 59–61].
Beyond the adaptive immune system, it is worth noting that the function of the miR-29 family can act in either an oncogenic or a tumor suppressor fashion. For example, miR-29 acts as oncomirs in B cells, as described above, and cervical epithelium [62], while acting as a tumor suppressor miR in hepatocytes, skin epithelium [63] and gastric epithelium [64]. The confluence of capacity to repress both oncogenes, as described above, and proapoptotic genes such as Bak, Bim, Bmf, Hrk and Puma [65], mean that the tumorigenic function of miR-29 needs to be experimentally determined in each cell lineage.
Concluding remarks
The miR-29 family has been implicated in regulation of the adaptive immune system by virtue of its expression in all its cellular constituents and the enrichment of adaptive immune pathways in the unbiased analysis of predicted targets. Through the recent development of a raft of different molecular tools, important functions of the miR-29 family have been uncovered, identifying miR-29 as a crucial regulator of thymic function, T cell polarization and B cell oncogenesis. Additional functions of miR-29 in adaptive immunity are bound to be discovered through further research. Perhaps the best illustration of the importance of a mere 22 base pairs is the evolution of a viral miR-29 mimic to exploit its function in setting thresholds for the adaptive immune system.
Acknowledgments
This work was funded by grants from KU Leuven, the VIB and Fonds voor Wetenschappelijk (FWO). A.L. was supported by a Juvenile Diabetes Research Foundation (JDRF) Career Development Award and a European Research Council (ERC) Start Grant. D.D.A. was supported by an Interfacultaire Raad voor Ontwikkelingssamenwerking (IRO) fellowship from KU Leuven.
References
- 1.Liston A, Linterman M, Lu LF. MicroRNA in the adaptive immune system, in sickness and in health. J Clin Immunol. 2010;30:339–346. doi: 10.1007/s10875-010-9378-5. [DOI] [PubMed] [Google Scholar]
- 2.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 4.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 5.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao JY, Ma LM, Guo YH, Zhang YC, Zhou H, Shao P, Chen YQ, Qu LH. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castanotto D, Lingeman R, Riggs AD, Rossi JJ. CRM1 mediates nuclear-cytoplasmic shuttling of mature microRNAs. Proc Natl Acad Sci U S A. 2009;106:21655–21659. doi: 10.1073/pnas.0912384106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber MJ. New human and mouse microRNA genes found by homology search. FEBS J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 10.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 12.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/S0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 15.Papadopoulou AS, Dooley J, Linterman MA, Pierson W, Ucar O, Kyewski B, Zuklys S, Hollander GA, Matthys P, Gray DH, De Strooper B, Liston A. The thymic epithelial microRNA network elevates the threshold for infection-associated thymic involution via miR-29a mediated suppression of the IFN-alpha receptor. Nat Immunol. 2012;13:181–187. doi: 10.1038/ni.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mott JL, Kurita S, Cazanave SC, Bronk SF, Werneburg NW, Fernandez-Zapico ME. Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J Cell Biochem. 2010;110:1155–1164. doi: 10.1002/jcb.22630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eyholzer M, Schmid S, Wilkens L, Mueller BU, Pabst T. The tumour-suppressive miR-29a/b1 cluster is regulated by CEBPA and blocked in human AML. Br J Cancer. 2010;103:275–284. doi: 10.1038/sj.bjc.6605751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem. 2010;285:25221–25231. doi: 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z, Zou J, Wang GK, Zhang JT, Huang S, Qin YW, Jing Q. Uracils at nucleotide position 9–11 are required for the rapid turnover of miR-29 family. Nucleic Acids Res. 2011;39:4387–4395. doi: 10.1093/nar/gkr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fort A, Borel C, Migliavacca E, Antonarakis SE, Fish RJ, Neerman-Arbez M. Regulation of fibrinogen production by microRNAs. Blood. 2010;116:2608–2615. doi: 10.1182/blood-2010-02-268011. [DOI] [PubMed] [Google Scholar]
- 22.Maegdefessel L, Azuma J, Toh R, Merk DR, Deng A, Chin JT, Raaz U, Schoelmerich AM, Raiesdana A, Leeper NJ, McConnell MV, Dalman RL, Spin JM, Tsao PS. Inhibition of microRNA-29b reduces murine abdominal aortic aneurysm development. J Clin Invest. 2012;122:497–506. doi: 10.1172/JCI61598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ott CE, Grunhagen J, Jager M, Horbelt D, Schwill S, Kallenbach K, Guo G, Manke T, Knaus P, Mundlos S, Robinson PN. MicroRNAs differentially expressed in postnatal aortic development downregulate elastin via 3′ UTR and coding-sequence binding sites. PLoS One. 2011;6:e16250. doi: 10.1371/journal.pone.0016250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roderburg C, Urban GW, Bettermann K, Vucur M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi M, Tacke F, Trautwein C, Luedde T. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 25.Santanam U, Zanesi N, Efanov A, Costinean S, Palamarchuk A, Hagan JP, Volinia S, Alder H, Rassenti L, Kipps T, Croce CM, Pekarsky Y. Chronic lymphocytic leukemia modeled in mouse by targeted miR-29 expression. Proc Natl Acad Sci U S A. 2010;107:12210–12215. doi: 10.1073/pnas.1007186107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengupta S, den Boon JA, Chen IH, Newton MA, Stanhope SA, Cheng YJ, Chen CJ, Hildesheim A, Sugden B, Ahlquist P. MicroRNA 29c is down-regulated in nasopharyngeal carcinomas, up-regulating mRNAs encoding extracellular matrix proteins. Proc Natl Acad Sci U S A. 2008;105:5874–5878. doi: 10.1073/pnas.0801130105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soci UP, Fernandes T, Hashimoto NY, Mota GF, Amadeu MA, Rosa KT, Irigoyen MC, Phillips MI, Oliveira EM. MicroRNAs 29 are involved in the improvement of ventricular compliance promoted by aerobic exercise training in rats. Physiol Genomics. 2011;43:665–673. doi: 10.1152/physiolgenomics.00145.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. Inhibition of miR-29 by TGF-beta-Smad3 signaling through dual mechanisms promotes transdifferentiation of mouse myoblasts into myofibroblasts. PLoS One. 2012;7:e33766. doi: 10.1371/journal.pone.0033766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fang JH, Zhou HC, Zeng C, Yang J, Liu Y, Huang X, Zhang JP, Guan XY, Zhuang SM. MicroRNA-29b suppresses tumor angiogenesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 30.Luna C, Li G, Qiu J, Epstein DL, Gonzalez P. Role of miR-29b on the regulation of the extracellular matrix in human trabecular meshwork cells under chronic oxidative stress. Mol Vis. 2009;15:2488–2497. [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B, Komers R, Carew R, Winbanks CE, Xu B, Herman-Edelstein M, Koh P, Thomas M, Jandeleit-Dahm K, Gregorevic P, Cooper ME, Kantharidis P. Suppression of microRNA-29 expression by TGF-beta1 promotes collagen expression and renal fibrosis. J Am Soc Nephrol. 2011;23:252–265. doi: 10.1681/ASN.2011010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao J, Meng XM, Huang XR, Chung AC, Feng YL, Hui DS, Yu CM, Sung JJ, Lan HY. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther. 2012;20:1251–1260. doi: 10.1038/mt.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A, De Strooper B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci U S A. 2008;105:6415–6420. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lippi G, Steinert JR, Marczylo EL, D’Oro S, Fiore R, Forsythe ID, Schratt G, Zoli M, Nicotera P, Young KW. Targeting of the Arpc3 actin nucleation factor by miR-29a/b regulates dendritic spine morphology. J Cell Biol. 2011;194:889–904. doi: 10.1083/jcb.201103006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiao J, Herl LD, Farese RV, Gao FB. MicroRNA-29b regulates the expression level of human progranulin, a secreted glycoprotein implicated in frontotemporal dementia. PLoS One. 2010;5:e10551. doi: 10.1371/journal.pone.0010551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Zhou L, Jiang P, Lu L, Chen X, Lan H, Guttridge DC, Sun H, Wang H. Loss of miR-29 in myoblasts contributes to dystrophic muscle pathogenesis. Mol Ther. 2012;20:1222–1233. doi: 10.1038/mt.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braconi C, Kogure T, Valeri N, Huang N, Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM, Patel T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene. 2011;30:4750–4756. doi: 10.1038/onc.2011.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma F, Xu S, Liu X, Zhang Q, Xu X, Liu M, Hua M, Li N, Yao H, Cao X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12:861–869. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 40.Aronson M. Hypothesis: involution of the thymus with aging – programmed and beneficial. Thymus. 1991;18:7–13. [PubMed] [Google Scholar]
- 41.Dowling MR, Hodgkin PD. Why does the thymus involute? A selection-based hypothesis. Trends Immunol. 2009;30:295–300. doi: 10.1016/j.it.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Shanley DP, Aw D, Manley NR, Palmer DB. An evolutionary perspective on the mechanisms of immunosenescence. Trends Immunol. 2009;30:374–381. doi: 10.1016/j.it.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Dooley J, Liston A. Molecular control over thymic involution: from cytokines and microRNA to aging and adipose tissue. Eur J Immunol. 2012;42:1073–1079. doi: 10.1002/eji.201142305. [DOI] [PubMed] [Google Scholar]
- 44.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Anz D, Thaler R, Stephan N, Waibler Z, Trauscheid MJ, Scholz C, Kalinke U, Barchet W, Endres S, Bourquin C. Activation of melanoma differentiation-associated gene 5 causes rapid involution of the thymus. J Immunol. 2009;182:6044–6050. doi: 10.4049/jimmunol.0803809. [DOI] [PubMed] [Google Scholar]
- 46.King CC, Jamieson BD, Reddy K, Bali N, Concepcion RJ, Ahmed R. Viral infection of the thymus. J Virol. 1992;66:3155–3160. doi: 10.1128/jvi.66.5.3155-3160.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colantonio AD, Epeldegui M, Jesiak M, Jachimowski L, Blom B, Uittenbogaart CH. IFN-alpha is constitutively expressed in the human thymus, but not in peripheral lymphoid organs. PLoS One. 2011;6:e24252. doi: 10.1371/journal.pone.0024252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Annu Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 49.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian L, De Hertogh G, Fedeli M, Staats KA, Schonefeldt S, Humblet-Baron S, Van Den Bosch L, Dellabona P, Dooley J, Liston A. Loss of T cell microRNA provides systemic protection against autoimmune pathology in mice. J Autoimmun. 2012;38:39–48. doi: 10.1016/j.jaut.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 52.Steiner DF, Thomas MF, Hu JK, Yang Z, Babiarz JE, Allen CD, Matloubian M, Blelloch R, Ansel KM. MicroRNA-29 regulates T-box transcription factors and interferon-gamma production in helper T cells. Immunity. 2011;35:169–181. doi: 10.1016/j.immuni.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kincaid RP, Burke JM, Sullivan CS. RNA virus microRNA that mimics a B-cell oncomiR. Proc Natl Acad Sci U S A. 2012;109:3077–3082. doi: 10.1073/pnas.1116107109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, Volinia S, Alder H, Liu CG, Rassenti L, Calin GA, Hagan JP, Kipps T, Croce CM. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 55.Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113:5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- 56.Teichler S, Illmer T, Roemhild J, Ovcharenko D, Stiewe T, Neubauer A. MicroRNA29a regulates the expression of the nuclear oncogene Ski. Blood. 2011;118:1899–1902. doi: 10.1182/blood-2010-09-306258. [DOI] [PubMed] [Google Scholar]
- 57.Xiong Y, Fang JH, Yun JP, Yang J, Zhang Y, Jia WH, Zhuang SM. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2009;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 58.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–29. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 59.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang F, Wang XS, Yang GH, Zhai PF, Xiao Z, Xia LY, Chen LR, Wang Y, Wang XZ, Bi LX, Liu N, Yu Y, Gao D, Huang BT, Wang J, Zhou DB, Gong JN, Zhao HL, Bi XH, Yu J, Zhang JW. miR-29a and miR-142-3p downregulation and diagnostic implication in human acute myeloid leukemia. Mol Biol Rep. 2011;39:2713–2722. doi: 10.1007/s11033-011-1026-5. [DOI] [PubMed] [Google Scholar]
- 61.Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, Dessureault S, Moscinski LC, Rezania D, Dalton WS, Sotomayor E, Tao J, Cheng JQ. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Wang F, Xu J, Ye F, Shen Y, Zhou J, Lu W, Wan X, Ma D, Xie X. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J Pathol. 2011;224:484–495. doi: 10.1002/path.2873. [DOI] [PubMed] [Google Scholar]
- 63.Nguyen T, Kuo C, Nicholl MB, Sim MS, Turner RR, Morton DL, Hoon DS. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2010;6:388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cui Y, Su WY, Xing J, Wang YC, Wang P, Chen XY, Shen ZY, Cao H, Lu YY, Fang JY. MiR-29a inhibits cell proliferation and induces cell cycle arrest through the downregulation of p42.3 in human gastric cancer. PLoS One. 2011;6:e25872. doi: 10.1371/journal.pone.0025872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kole AJ, Swahari V, Hammond SM, Deshmukh M. miR-29b is activated during neuronal maturation and targets BH3-only genes to restrict apoptosis. Genes Dev. 2011;25:125–130. doi: 10.1101/gad.1975411. [DOI] [PMC free article] [PubMed] [Google Scholar]