Abstract
In eukaryotes, cellular energy in the form of ATP is produced in the cytosol via glycolysis or in the mitochondria via oxidative phosphorylation and, in photosynthetic organisms, in the chloroplast via photophosphorylation. Transport of adenine nucleotides among cell compartments is essential and is performed mainly by members of the mitochondrial carrier family, among which the ADP/ATP carriers are the best known. This work reviews the carriers that transport adenine nucleotides into the organelles of eukaryotic cells together with their possible functions. We focus on novel mechanisms of adenine nucleotide transport, including mitochondrial carriers found in organelles such as peroxisomes, plastids, or endoplasmic reticulum and also mitochondrial carriers found in the mitochondrial remnants of many eukaryotic parasites of interest. The extensive repertoire of adenine nucleotide carriers highlights an amazing variety of new possible functions of adenine nucleotide transport across eukaryotic organelles.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-010-0612-3) contains supplementary material, which is available to authorized users.
Keywords: Mitochondria, Adenine nucleotides, Mitochondrial carriers, Transport, Organelles
Introduction
In most eukaryotic organisms, ATP is synthesized both in the cytosol through glycolysis and in the mitochondria through oxidative phosphorylation. While only two ATP molecules are generated by glycolysis per glucose molecule, around 30 ATP molecules are generated by oxidative phosphorylation, which is dependent on the presence of oxygen. Thus, the mitochondria are considered the power plants of the cell. Mitochondria fulfill many roles in addition to energy production: calcium-handling, heat production, amino acid and fatty acid metabolism, synthesis of iron-sulfur clusters, and apoptosis regulation.
Photosynthetic organisms, such as plants and algae, also produce ATP during the light reactions of photosynthesis in the chloroplasts. Furthermore, plastids also participate in starch metabolism and de novo synthesis of nucleotides in plants.
Transport of adenine nucleotides among cell compartments is essential. Adenine nucleotides (especially ATP) must be transported from the compartments that produce them to the compartments that use them. A decade ago, only the mitochondrial ADP/ATP carriers (AAC) and the ADP/ATP transporters in the plastid envelope were known [1]. However, in the last years many carrier proteins in the mitochondria and in other organelles have been identified at the biochemical and molecular level. Most of them belong to the mitochondrial carrier family (MCF, SLC25 transport family) of proteins [2–4].
There are 34 mitochondrial carriers in Saccharomyces cerevisiae [5], around 50 in mammals [2], and 58 in Arabidopsis thaliana [3]. They are not only located to the mitochondria, but also to the peroxisomes and chloroplasts. Mitochondrial carriers share a common structure of around 300 amino acids and six transmembrane helixes (for review [2–4]) and can be divided into three types according to the substrate that they transport: carriers of amino acids, carriers of ketoacids, and carriers of nucleotides (which are further subdivided into carriers of mononucleotides and carriers of dinucleotides) [6–8]. The uncoupling proteins (UCPs; for review [9]) are phylogenetically included among the ketoacid carriers group, although their real substrate is unknown. This work focuses on the transporters of adenine nucleotides and is aimed at bringing together the classical adenine nucleotide transporters of mitochondria with newly identified nucleotide carriers. The reader is referred to [10, 11] for reviews on the classical AACs.
We review the adenine nucleotide carriers of model organisms belonging to the five eukaryotic clades [(Fig. S1) in the Electronic Supplementary Material (ESM)] [12, 13]: Homo sapiens and S. cerevisiae as opisthokonts, A. thaliana as a plantae, Dictyostelium discoideum as an amoebozoan, Trypanosoma brucei as an excavate, and Plasmodium falciparum as a chromalveolate. We also review the adenine nucleotide carriers of some amitochondriate parasite organisms (Fig. S1 in ESM).
Structure and transport mechanism of the mitochondrial carriers
Structure
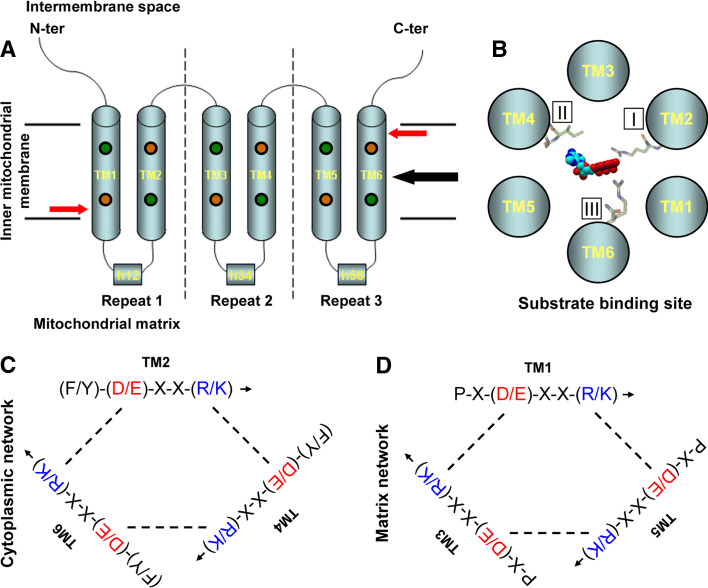
All mitochondrial carriers have a similar structure. They have a small molecular mass of around 30 kDa and contain three homologous repeats of around 100 amino acids, each of them consisting of two transmembrane helixes connected by a long loop that faces the mitochondrial matrix, whereas the repeats are linked by shorter loops that face the intermembrane space (Fig. 1a). Despite a low sequence conservation, a consensus sequence, P-h-(D/E)-X-X-(K/R), has been determined [2–4].
Fig. 1a–d.
Structure and transport mechanism of the mitochondrial carriers. a Mitochondrial carriers contain three repetitions of 100 amino acids (separated by dashed lines), and each consists of two α-helixes connected by a segment that faces the mitochondrial matrix and makes a short helix parallel to the membrane plane. The substrate binding site of the carrier is located at the midpoint of the membrane (black arrow). The location of the matrix and cytoplasmic salt-bridge networks is indicated by red arrows. Conserved prolines are indicated by orange circles and conserved glycines are indicated by green circles. TM1–TM6 indicates the transmembrane helixes, and h12, h34, and h56 indicate the short helixes. b Putative substrate binding site viewed from the intermembrane space: the substrate binds to three binding points, I, II, and III, located in helixes 2, 4, and 6, respectively. In adenine nucleotide carriers, the adenine ring binds to contact point II, a hydrophobic pocket formed by G-[IVLM], whereas the phosphate groups bind to contact point I and contact point III, which are normally [RK]. c, d Cytoplasmic network of salt bridges (located in even-numbered helixes close to the intermembrane space) and matrix network of salt bridges (located in odd-numbered helixes close to the mitochondrial matrix). The positively and negatively charged residues of the salt bridges are shown in blue and red, respectively. The matrix network blocks the access to the matrix when the carrier is in the c conformation, whereas the cytoplasmic network blocks the access to the intermembrane space when the carrier is in the m conformation. Binding of the substrate to the carrier disrupts the salt bridges and triggers its conversion to the other state
The only mitochondrial carrier whose structure is known is that of the AAC bound to its specific inhibitor carboxyatractyloside (CAT), which has been determined by X-ray crystallography [14]. This information together with previous structural information has led to structural predictions of other mitochondrial carriers. Although for a long time these transporters have been thought to function as dimers [15–17], recent studies have suggested that the functional unit may be a monomer (for review, [18]). Mitochondrial carriers can be electroneutral or electrogenic and, whereas most of them are strict exchangers, a few can function both as uniporters and as exchangers, or even as strict uniporters [4, 8].
Transport mechanism
Knowledge of the structure of the AAC [14] and bioinformatic studies [6–8] have allowed the identification of a putative common binding site and mechanism of transport.
According to the proposed model, the substrate binds the mitochondrial carrier to three binding points, numbered I, II, and III, located at equivalent positions in transmembrane helixes 2, 4, and 6, respectively (Fig. 1b). These contact points are highly conserved among ortholog proteins in different organisms. Contact point II discriminates among different substrate types. Amino acid transporters contain the motif R-[DE], nucleotide transporters contain the motif G-[IVLM], and ketoacid transporters contain the motif R-[QHNT]. Contact point I discriminates among different substrates of the same type. For instance, carboxyl or phosphate groups bind to [RK], amino groups bind to [DE], aromatic rings interact with [FY], and hydrophobic groups bind via van der Waals interactions to [ILV]. Contact point III is often [RK] and binds phosphate (in nucleotides) or carboxyl groups (in amino acids and ketoacids).
There are two groups of conserved amino acids in symmetrical positions in all mitochondrial carriers. The first one is located in the odd helixes, close to the mitochondrial matrix, and consists of the P-X-(D/E)-X-X-(R/K) motif (the mitochondrial carrier signature). The charged residues of this motif generate a network of salt bridges that blocks the access to the matrix when the carrier is in the c (cytoplasm-facing) conformation [8]. Interestingly, the proline of the mitochondrial carrier signature (Fig. 1a) is known to sharply kink the odd helixes when the carrier is in the c conformation [14], and this would reorient the P-X-(D/E)-X-X-(R/K) motifs to form the network of salt bridges. The second group of conserved amino acids is located in the even helixes, close to the intermembrane space, and consists of the (F/Y)-(D/E)-X-X-(R/K) motif. The charged residues of this motif do not have an obvious function in the c conformation, but it has been proposed that they could generate a network of salt bridges that would block the access to the intermembrane space when the carrier is in the m (matrix-facing) conformation [8]. A conserved proline (Fig. 1a), which is located close to this network, has also been identified in the even helixes and has been proposed to generate a kink in the even helixes when the carrier is in the m conformation, and this would bring together the (F/Y)-(D/E)-X-X-(R/K) motifs to form the network of salt bridges [19].
These two groups of conserved amino acids have been named the matrix and cytoplasmic network, respectively (Fig. 1c, d). Binding of the substrate to the binding site could disturb the network and would trigger the opening of the carrier and its conversion to the other state [8]. The conserved prolines, along with well conserved glycines in odd and even helixes that are aligned with the prolines (Fig. 1a), would act as hinges to open or close the carrier on the matrix or cytosolic side [19]. This model of carrier opening may be able to explain why some carriers function as uniporters. For example, in the glutamate carriers the cytoplasmic network does not contain the conserved charged residues, and thus the salt bridges do not form when the carrier is in the m conformation; this would allow the opening of the carrier even in the absence of internal substrate, and the transition back to the c conformation [8]. This model, however, does not agree with all available experimental data as, for instance, the GDP/GTP carrier shows a weak cytoplasmic network and would be expected to show some degree of uniporter activity [8], but has been shown to be a strict exchanger [20].
Mitochondrial adenine nucleotide carriers
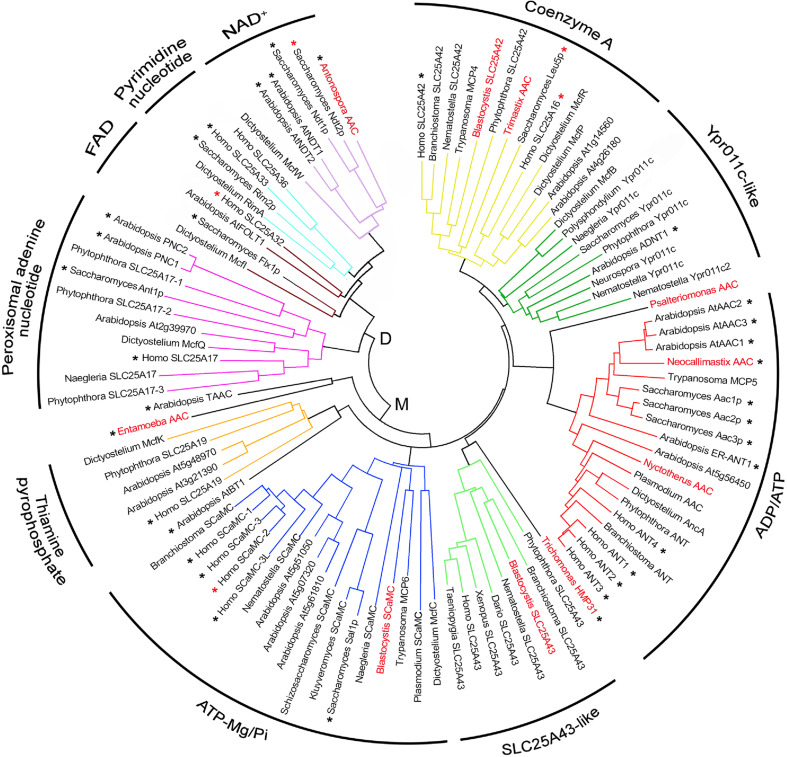
There are four groups of mitochondrial adenine nucleotide carriers present in the five eukaryotic clades: the AAC (see section “ADP/ATP carrier”), the ATP-Mg/Pi carrier [“Short calcium binding mitochondrial carrier (SCaMC) or ATP-Mg/Pi carrier”], the coenzyme A (CoA) carrier (“Coenzyme A carrier”) and the uncharacterized carrier Ypr011c (“Uncharacterized mitochondrial nucleotide transporters”; including the AMP/ATP carrier of plants, “Mitochondrial AMP/ATP transporter”). Some of them may have disappeared in some subgroups inside a major clade (especially in amitochondriate organisms, “Hydrogenosomal and mitosomal adenine nucleotide carriers”). In addition, there is also a peroxisomal adenine nucleotide carrier present in the five eukaryotic clades [“Peroxisomal adenine nucleotide transporter (pANT)”]. The five clusters of adenine nucleotide carriers are shown in Table 1 and Fig. 2.
Table 1.
Mitochondrial and peroxisomal adenine nucleotide carriers. The ADP/ATP carriers, ATP-Mg/Pi carriers, coenzyme A carriers, Ypr011c-like carriers, and peroxisomal AMP/ATP carriers of different species (S. cerevisiae, H. sapiens, A. thaliana, D. discoideum, T. brucei, and P. falciparum) are shown. The consensus sequences of the putative substrate binding sites I, II, and III are shown. Contact point II of most adenine nucleotide carriers binds the adenine ring and contains G-[IVLM]. Contact point III binds phosphate groups and is normally [RK]. Also, the highly conserved RRRMMM motif of the ADP/ATP carrier and its deviation in the other nucleotide carriers are indicated
| AdN carriers | Species | Contact points | Motif | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S. cerevisiae | H. sapiens | A. thaliana | D. discoideum | T. brucei | P. falciparum | I | II (adenine) | III | RRRMMM | |
| AAC (ADP/ATP) | Aac1p (YMR056c) Aac2p (YBL030c) Aac3p (YBR085w) |
ANT1 (SLC25A4) ANT2 (SLC25A5) ANT3 (SLC25A6) ANT4 (SLC25A31) |
AAC1 (At3g08580) AAC2 (At5g13490) AAC3 (At4g28390) |
AncA | MCP5 (Tb10.61.1830) | CAA58541.1 | R T N | GI | R | RRR MMM |
| SCaMC (ATP-Mg/Pi) | Sal1p (YNL083w) |
SCaMC-1 (SLC25A24) SCaMC-2 (SLC25A25) SCaMC-3 (SLC25A23) SCaMC-3like (SLC25A41) |
At5g61810 At5g07320 At5g51050 |
McfC | MCP6 (Tb927.4.1660) | XP_001351960.1 | KE K | GI | K | RTR LQAa |
| Coenzyme A | Leu5p (YHR002w) |
SLC25A42 SLC25A16 |
At4g26180 At1g14560 |
McfP, McfR | MCP4 (Tb10.70.2290) | Absentb | R Y (K/Q/H) | G(M/V/I/S) | R/K | RRR LQVa |
| Ypr011c-like | Ypr011c (YPR011c) | Absent | ADNT1 (At4g01100) | McfB | Absentc | Absentd | R (Y/N) (Q/K) | GV | K | RRR FQVa |
| Peroxisomal AMP/ATP | Ant1p (YPR128c) | SLC25A17 |
PNC1 (At3g05290) PNC2 (At5g27520) |
McfQ | ?e | Absentf | (A/S/Q) (Q/S) Y | L (T/V) | K/Q | KAM LQSa |
| Number of MC | 34 | 48 | 58 | 31 | 24 | 13 | ||||
| Clade | Opisthokonts | Plantae | Amoebozoans | Excavates | Chromalveolates | |||||
aOnly the yeast sequence is shown
bPresent in the chromalveolate Phytophthora infestans (XP_002897719.1)
cPresent in the excavate Naegleria gruberi (XP_002674890.1)
dPresent in Phytophthora infestans (XP_002901049.1)
ePresent in Naegleria gruberi (XP_002678224.1)
fPresent in Phytophthora infestans (XP_002900592.1, XP_002902667.1, and XP_002909633.1)
Fig. 2.
Phylogenetic tree of MCF members that transport nucleotides. Nucleotide carriers are subdivided into dinucleotide carriers (D; NAD+ carriers, pyrimidine nucleotide carriers, FAD carriers, and peroxisomal adenine nucleotide carriers) and mononucleotide carriers (M; thiamine pyrophosphate carriers, ATP-Mg/Pi carriers, SLC25A43-like carriers, ADP/ATP carriers, Ypr011c-like carriers, and coenzyme A carriers). Carriers characterized by direct transport measurements are marked with a black asterisk. Carriers whose function has been inferred by genetic studies or complementation assays are marked with a red asterisk. Adenine nucleotide carriers from amitochondriate organisms mentioned in the text are in red. Protein sequence multiple alignment was performed by Clustal W software. The phylogenetic unrooted tree was obtained by neighbor-joining based on the amino acid pairwise distance with the Poisson-correction method using the Mega4.0 software
ADP/ATP carrier
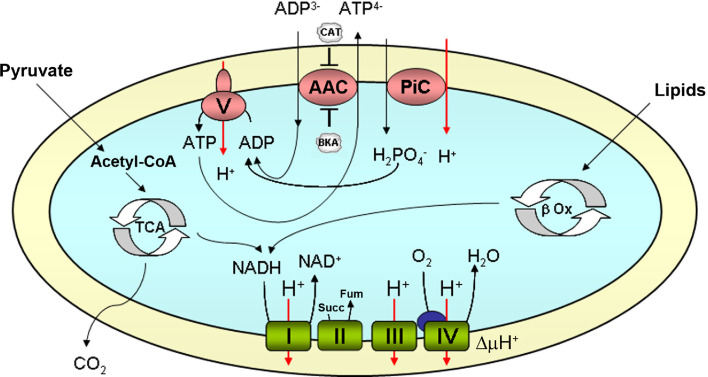
The mitochondrial ADP/ATP carrier (AAC) performs an electrogenic exchange of ADP3− with ATP4− between the cytosol and the mitochondrial matrix [10]. It is essential for oxidative phosphorylation because, in mitochondria energized by respiration, it normally exports ATP synthesized in the mitochondria for cytosolic ADP (Fig. 3). An important characteristic of this carrier is that it is unable to modify the net content of adenine nucleotides (ATP + ADP + AMP), as it always exchanges one nucleotide for another [10]. The affinity of the AACs for adenine nucleotides is very high, with a KM of around 2 μM in yeast and around 4–8 μM in humans [21].
Fig. 3.
Function of the mitochondrial ADP/ATP carrier. Schematic representation of the oxidative phosphorylation process. The NADH generated by lipid oxidation (β Ox), in the Krebs cycle (TCA) and other reactions, and succinate (Succ) generated in the TCA are oxidated by the respiratory chain complexes (complexes I, II, III, and IV in green, cytochrome C in blue), and protons are pumped to the intermembrane space. The proton electrochemical gradient (∆μH+) is used by the H+-ATP synthase (complex V) to generate ATP from ADP and phosphate (Pi). Pi is imported into the mitochondrial matrix in symport with a proton through its own transporter (PiC, SLC25A3). The ATP generated is transported to the cytosol in exchange for ADP through the ADP/ATP carrier (AAC). The AAC may be inhibited by CAT and BKA, which act on opposite sides of the membrane
Several inhibitors of this carrier are known. CAT binds to the AAC in its cytoplasmic side and fixes it in the c (cytoplasm-facing) conformation [10]. On the other hand, the membrane-permeant bongkrekic acid (BKA) binds to the carrier in its mitochondrial side and fixes it in the m (matrix-facing) conformation [10].
The putative substrate binding site of the AACs is highly conserved [6, 7]. The adenine ring of ADP binds to contact point II, a hydrophobic pocket formed by G-I (G183-I184 in human ANT1), whereas the phosphate groups bind to contact point I, formed by R (R80 in human ANT1), and contact point III, formed by R (R280 in human ANT1) [6, 7]. The motif RRRMMM (R235-R236-R237-M238-M239-M240 in human ANT1) is highly conserved in all AACs.
AAC in S. cerevisiae
The genome of S. cerevisiae contains three AAC genes: AAC1, AAC2, and AAC3 (ORFs YMR056c, YBL030c, and YBR085w, respectively). Aac1p and Aac2p show 77% identity and 88% similarity. Aac1p and Aac3p show 73% identity and 88% similarity. Aac2p and Aac3p show 89% identity and 97% similarity. For further information, see ESM.
AAC in mammals
The genome of mammals contains four AAC genes: ANT1, ANT2, ANT3, and ANT4 (carriers SLC25A4, SLC25A5, SLC25A6, and SLC25A31, respectively). Rodents are an exception, as they only have three AACs, those equivalent to ANT1, ANT2, and ANT4 (slc25a4, slc25a5, and slc25a31). Ant1 and Ant2 show 88% identity and 94% similarity. Ant1 and Ant3 show 87% identity and 94% similarity. Ant1 and Ant4 show 73% identity and 84% similarity. Ant2 and Ant3 show 92% identity and 97% similarity. Ant2 and Ant4 show 72% identity and 83% similarity. Ant 3 and Ant4 show 72% identity and 84% similarity. For further information, see ESM.
AAC in A. thaliana
The genome of A. thaliana contains three mitochondrial AAC genes: AtAAC1, AtAAC2, and AtAAC3 (genes At3g08580, At5g13490, and At4g28390, respectively). AtAac1 and AtAac2 show 86% identity and 93% similarity. AtAac1 and AtAac3 show 74% identity and 82% similarity. AtAac2 and AtAac3 show 74% identity and 81% similarity.
The amino acid sequences of all three Aac proteins contain a cleavable N-terminal transit peptide [22], which enhances the import of these proteins into the inner mitochondrial membrane compared to the import of other mitochondrial carriers (such as the oxoglutarate/malate carrier) that do not contain a cleavable extension.
AAC in D. discoideum
The genome of this slime mold contains 31 putative mitochondrial carriers, and a single AAC gene, ancA. It codes for a protein of 309 amino acids [23].
AAC in T. brucei
The genome of this kinetoplastid parasite contains 24 putative mitochondrial carriers, and a single AAC gene, MCP5. It codes for a protein of 307 amino acids [24].
This parasite has a complex life cycle that alternates between two stages. Oxidative phosphorylation takes place mainly in procyclic-form trypanosomes (found in the midgut of the tse-tse fly), and the AAC works to export the ATP produced in the mitochondria in exchange for cytosolic ADP, as in respiring yeast or mammalian cells (see Fig. S2A in ESM). On the other hand, in bloodstream-form trypanosomes (found in the blood and tissue fluids of mammals), with highly simplified mitochondria, ATP is obtained by substrate-level phosphorylation during glycolysis, and it must be imported into the mitochondria. In this stage, the mitochondrial membrane potential is not generated by respiration, but by the electrogenic exchange of cytosolic ATP4− for mitochondrial ADP3− through the AAC coupled to H+ pumping through the F0F1 ATP synthase working in reverse [25], as in anaerobic yeast (see ESM, Fig. S2B).
The related species T. evansi, which only exists in the bloodstream form, lacks mitochondrial DNA (i.e., it is a dyskinetoplastic species) [25]. In this species, the mitochondrial membrane potential is always generated by the electrogenic exchange of cytosolic ATP4− for mitochondrial ADP3− through the AAC coupled to ATP hydrolysis by F1 ATPase (the F0 fragment is absent, as some of its subunits are coded by mitochondrial DNA), as in ρ0 yeast or ρ0 mammalian cells (see ESM, Fig. S2C) [25].
AAC in P. falciparum
The genome of this apicomplexan parasite contains only 13 putative mitochondrial carriers, and a single AAC gene that codes for a protein of 301 amino acids (CAA58541.1).
In this organism, the mitochondria are highly simplified and the metabolism is highly glycolytic [26]. Their small mitochondrial DNA encodes only three proteins (three subunits of the respiratory chain, but no subunits of the F0F1-ATP synthase), and it does not encode any tRNAs, suggesting that they are imported from the cytosol [26]. The electron transport chain is present, but all the complexes have lost some subunits compared to other eukaryotes, and complex I is substituted by an alternative non-proton-pumping NADH dehydrogenase, as in yeast. The sensitivity of the parasite to inhibitors of the respiratory chain indicates that it is indispensible, probably due to its requirement for the regeneration of ubiquinone as electron acceptor for dihydroorotate dehydrogenase [27], an essential enzyme in de novo pyrimidine biosynthesis (there is no salvage pathway in this organism). The mitochondrial electron transport chain can become dispensable upon acquisition of an alternative means to synthesize pyrimidines independently of ubiquinone regeneration by the electron transport chain [27]. The mitochondrial membrane potential is not abolished by electron transport chain inhibitors, and thus it might be generated by the electrogenic exchange of cytosolic ATP4− for mitochondrial ADP3− through the AAC coupled to ATP hydrolysis by F1-ATPase (some subunits of the F0 fragment appear to be absent in this organism), as in ρ0 yeast or ρ0 mammalian cells (see ESM, Fig. S2C). The absence of a complete F0F1-ATP synthase in the apicomplexans suggests that they may have lost the capacity for H+-coupled ATP synthesis and hydrolysis [26] and argues against the use of the respiratory chain as a means of generating membrane potential and ATP through oxidative phosphorylation. In contrast, it has been demonstrated that oligomycin-sensitive oxidative phosphorylation is able to take place in mitochondria of a related species, P. berghei [28], and this suggests that apicomplexans might employ highly divergent or novel subunits to fulfill the functions of the F0 fragment [26].
Short calcium binding mitochondrial carrier (SCaMC) or ATP-Mg/Pi carrier
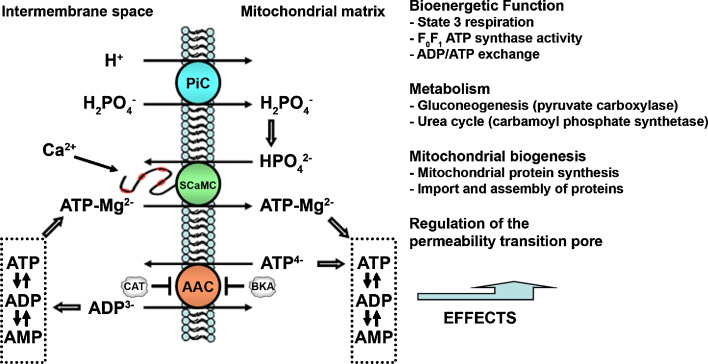
The mitochondrial ATP-Mg/Pi carrier was functionally identified by Aprille’s group in the 1980s in rat liver mitochondria (for review [29, 30]), but the corresponding genes have been identified only very recently [31–34]. It performs an electroneutral exchange of ATP-Mg2− (or HADP2−) with HPO4 2− between the cytosol and mitochondrial matrix (Fig. 4). It is able to modify the net content of mitochondrial adenine nucleotides (ATP + ADP + AMP). The direction and magnitude of net adenine nucleotide transport between the cytosol and mitochondria depend on the relative ATP-Mg and Pi concentration gradients. The ATP-Mg/Pi carrier is insensitive to CAT and BKA, the classic inhibitors of the AAC [29, 30, 32].
Fig. 4.
Function of the mitochondrial ATP-Mg/Pi carrier. The ATP-Mg/Pi carrier (SCaMC) exchanges ATP-Mg/Pi for Pi (as HPO4 2−) between the cytosol and the mitochondria. The electron transport chain generates a ΔpH, which in turn generates a Pi gradient through the Pi carrier (PiC). This gradient allows the entry of ATP-Mg into the mitochondria, even against concentration gradient, whenever the carrier is activated by a Ca2+ signal in the intermembrane space. The ATP-Mg/Pi carrier contributes to the regulation of the net content of matrix adenine nucleotides (ATP + ADP + AMP). The ADP/ATP carrier (AAC) is unable to do so, as it always exchanges one nucleotide for another. Changes in the matrix adenine nucleotide pool regulate the reactions shown in the figure (Adapted from [29, 30])
This carrier possesses a novel feature among mitochondrial carriers consisting of an N-terminal extension with EF-hand calcium-binding motifs that face the intermembrane space [31, 32]. This short N-terminal domain is homologous to calmodulin. The presence of the EF-hand motifs suggests that the transport activity of these proteins might be regulated by changes in the cytosolic calcium concentration, as has been observed experimentally [29, 30, 33). Furthermore, it could provide a new way to transduce calcium signal to the mitochondria without the need for calcium entry into the organelle (for review [35]).
The putative substrate binding site of the SCaMCs is also highly conserved [6, 7]. The adenine ring of ATP or ADP binds to contact point II, formed by G-I, as in the AAC (G416-I417 in yeast Sal1p), whereas the phosphate groups bind to contact point I, formed by K (K314 in yeast Sal1p), and contact point III, formed by K (K523 in yeast Sal1p) [6, 7]. Contact point I contains a negatively charged residue (E318 in yeast Sal1p) that would repel the phosphate groups of ATP or ADP. The Mg2+ that comes with ATP, or the H+ that comes with ADP are thought to bind to that residue, and this would shield the negative charges [6, 7]. For that reason, the cations are fundamental for transport through the SCaMCs, and ATP alone is not transported. On the other hand, in the AAC, the equivalent residue is T (T84 in human ANT1), and it does not coordinate Mg2+, explaining why this carrier does not require the cation. The RRRMMM motif of the AACs is not conserved in the SCaMCs.
SCaMC in S. cerevisiae
The genome of S. cerevisiae contains only one SCaMC gene, SAL1 (ORF YNL083w). Sal1p, with 545 amino acids, shows 24, 25, and 26% identity and 42, 43, and 43% similarity with Aac1p, Aac2p, and Aac3p, respectively. It shows 31% identity and 50% similarity with the unidentified carrier Ypr011c, and 22% identity and 41% similarity with the CoA carrier Leu5p.
Sal1p exchanges ATP-Mg2− with HPO4 2− or HADP2− with HPO4 2− between the cytosol and mitochondrial matrix [33, 36, 37], and transport is absolutely dependent on the presence of extramitochondrial Ca2+, with an S0.5 of 15 μM [33, 36]. Ca2+ modifies the V max of the carrier, but not the KM, which is around 0.2 mM for both ADP and ATP-Mg [33], two orders of magnitude higher than that of the AACs, which is around 2 μM [21].
Viability (V) function of the SCaMC
The only essential mitochondrial functions in yeast, mitochondrial protein import and assembly, and iron-sulfur cluster biogenesis, require ATP in the mitochondrial matrix (for review [38, 39]). However, a yeast mutant of the W303 strain lacking all three AAC isoforms is able to grow on glucose in aerobiosis [40], and thus the group of Kolarov proposed the existence of another carrier responsible for adenine nucleotide transport in the triple aac1,2,3 mutant [40]. According to that hypothesis, the absence of this unknown carrier would result in lethality of the triple aac1,2,3 mutant.
Attempts to disrupt the main AAC gene, AAC2, in other genetic backgrounds such as the FY1679 strain, were found to be lethal [41]. Chen discovered that gene YNL083w was mutated in these strains, and renamed it SAL1 (suppressor of a ac2 lethality). SAL1 was then found to code for a member of the SCaMC subfamily of mitochondrial carriers [41] and to be a calcium-dependent mitochondrial ATP-Mg/Pi carrier [36]. As expected by the Kolarov group’s hypothesis, the combination of disruptions in AAC2 and SAL1 is lethal in the W303 strain [36, 41], and this lethality is suppressed by expression of AAC2, AAC3, or SAL1, but not AAC1, from monocopy or multicopy expression vectors [41]. Chen suggested that Aac2p and Aac3p, but not Aac1p, are bifunctional molecules [41]; in addition to their role in respiratory growth, Aac2p and Aac3p would also support the cell’s viability on fermentable carbon sources.
These two distinct roles of AAC were designated as R (for respiration) and V (for viability) functions, respectively [41]. Accordingly, Aac1p would only have the R function, while Sal1p would only have the V function [41]. Further work by Thorsness’s group clearly demonstrated that Aac1p is also able to suppress the lethality of the double mutant, when expressed at the AAC2 locus in the form of a knock-in strain, although with less efficiency than Aac2p and Aac3p [42]. According to our present interpretation, the V function of the AACs or Sal1p is to provide ATP for the protein import system and for iron-sulfur cluster biogenesis [11].
Disruption mutants
The sal1 mutant shows no growth defects in the situations tested so far, and the SAL1 gene appeared to be dispensable in respiratory and fermentative growth, in anaerobiosis, and in the absence of mitochondrial DNA. Subsequent studies have discovered that sal1 mutants indeed show a phenotype, which is a higher rate of spontaneous petite formation (mitochondrial DNA instability). In a sal1 null mutant of the D273-10B strain, the rate is 9.5% compared to 1% in the wild type strain [42]. In the S288C strain, SAL1 is one of the genes implicated in the very high rate of spontaneous petite formation, so that a sal1 loss of function mutation raises the rate from 26 to 79% [43]. Thus, the loss of Sal1p destabilizes mtDNA.
On the other hand, as has been indicated, the SCaMC was found to be essential in yeast cells lacking the main isoform of the ADP/ATP carrier, AAC2 [36, 41], and also in yeast cells if the AAC is inhibited by BKA [37]. A sal1 mutant with inactivated EF-hands behaves exactly as a null mutant [41], proving that calcium binding is essential for the carrier function, as we have observed [33]. Chen’s group found that aac2, sal1 double mutants rapidly lose mitochondrial DNA [44], and loss of mitochondrial DNA is always lethal in aac2 mutants [45], as they lose their only way to generate a mitochondrial membrane potential (for review [11]). The loss of mitochondrial DNA was not caused by a loss of dATP levels or a decrease in mitochondrial transcription, but mitochondrial synthesis of proteins was significantly reduced [44], and this resulted in mtDNA depletion. Thus, a reduced import of ATP in mitochondria caused by lack of Aac2p and Sal1p is a likely explanation for these results.
SCaMC in other yeast
In the yeast Schizosaccharomyces pombe there is a single SCaMC gene. It codes for a protein of 426 amino acids, shorter than that of other yeasts, as it lacks the N-terminal extension with EF-hand calcium-binding motifs [31].
In the yeast Kluyveromyces lactis there is a single SCaMC gene. It codes for a protein of 517 amino acids. Compared to mammalian SCaMCs, Sal1p contains an insertion of 40 amino acids between EF-hands 3 and 4 in the N-terminal domain. However, the corresponding insertion in K. lactis contains only 12 amino acids.
SCaMC in mammals
The genome of mammals contains four SCaMC genes [31, 32, 34, 46]: SCaMC-1(APC1), SCaMC-2 (APC3), SCaMC-3 (APC2), and SCaMC-3like (carriers SLC25A24, SLC25A25, SLC25A23, and SLC25A41, respectively). SCaMC-1 and SCaMC-2 show 61–66% identity and 77–83% similarity. SCaMC-1 and SCaMC-3 show 61% identity and 79–80% similarity. SCaMC-1 and SCaMC-3L show 54% identity and 71% similarity. SCaMC-2 and SCaMC-3 show 64–70% identity and 75–82% similarity. SCaMC-2 and SCaMC-3L show 56% identity and 71% similarity. SCaMC-3 and SCaMC-3L show 64% identity and 77% similarity.
SCaMC-1 and SCaMC-3 have been reconstituted into proteoliposomes and shown to perform an electroneutral exchange of ATP-Mg or ADP for Pi [32]. In SCaMC-1, the KM for both ADP and ATP-Mg is around 0.2 mM [32], similar to that of the yeast protein Sal1p [33]. On the other hand, in SCaMC-3, the KM for ATP-Mg is 0.2 mM, but the KM for ADP is higher, around 0.5 mM [32]. The transport activity of SCaMC-2 has not been tested so far. SCaMC-3L is an exception among the SCaMCs, as it lacks the N-terminal extension with EF-hand calcium-binding motifs, and thus its activity is not regulated by Ca2+ [34]. The KM for ATP-Mg is 0.4 mM and for ADP is 0.9 mM [34]. Thus, as in its closest paralog, SCaMC-3, the affinity for both nucleotides is different in this isoform.
Isoforms generated by alternative splicing
In humans, SCaMC-1 and SCaMC-2 each give rise to several transcripts by use of alternative promoters (for review [35]), resulting in proteins that differ in their first exon, which codes for the first EF-hand. In contrast, in humans, SCaMC-3 gives rise to several C-terminal variants by alternative splicing [47]. Only the SCaMC-3a variant is complete, as the rest are truncated and lack the last transmembrane helix (for review [35]). They are probably not functional.
The SCaMC subfamily of mitochondrial carriers is thus the most complex group of mitochondrial carriers. The large number of isoforms and spliced variants might provide different calcium sensitivities to the transport activity of these carriers.
Expression
The mRNAs of human SCaMCs are ubiquitous. Most human tissues such as pancreas, kidney, skeletal muscle, lung, brain, or heart express more than one isoform [31, 32]. Only the mRNAs of SCaMC-1a and SCaMC-3L show a restricted distribution in testis, and that of SCaMC-2c in brain [31].
At the protein level in mouse, SCaMC-1 is mainly expressed in the small intestine, lung, and colon; SCaMC-2 is expressed at very low levels in some tissues such as heart or brain; SCaMC-3 is expressed exclusively in liver and brain (unpublished observations); and SCaMC-3L is expressed mainly in testis [34, 46]. SCaMC-1 is also highly expressed in human proliferating and tumor cell lines (unpublished observations).
Function of the SCaMC
The SCaMCs are probably activated in vivo by calcium-mobilizing hormones, such as vasopressin and glucagon in mammals [29, 30]. They exchange ATP-Mg for Pi and participate in the net uptake and net loss of adenine nucleotides [29, 30]. By changing the matrix adenine nucleotide content, the SCaMC play an important role in the regulation of the mitochondrial metabolic pathways that have adenine nucleotide-dependent enzymes (Fig. 4). These include state 3 respiration, gluconeogenesis, urea cycle, mitochondrial protein synthesis, and import of nuclear encoded proteins into mitochondria [29, 30].
The SCaMCs appears to be important for mitochondrial maturation after birth. In fact, the mitochondrial adenine nucleotide pool has been found to increase several-fold in newborn rat liver mitochondria within 3 h after birth, and this increase coincides with the maturation of mitochondrial respiration [29, 30]. On the other hand, the mitochondrial adenine nucleotide pool also increases in adult liver mitochondria upon fasting or glucagon treatment and decreases after hypoxia/ischemia or during hibernation [29, 30].
SCaMC in A. thaliana
The genome of A. thaliana contains three SCaMC genes: At5g61810, At5g07320, and At5g51050 [31]. The subcellular localization and organ distribution of these proteins, and whether there are splicing variants as in mammals, are currently unknown.
SCaMC in D. discoideum
The genome of D. discoideum contains four genes that have been reported to be similar to SCaMCs [23]. Protein McfC contains an N-terminal domain with four EF hands, and its substrate binding site is identical to the canonical one for SCaMCs, with the exception of contact point II, which is A344-T345, instead of GI. Protein McfB is, on the contrary, similar to the uncharacterized yeast protein Ypr011c (see “Uncharacterized mitochondrial nucleotide transporters”). Proteins McfA and McfV lack calcium-binding motifs and are also close to Ypr011c and SCaMCs, but their substrate binding site is largely modified. They probably also transport adenine nucleotides or related compounds.
SCaMC in T. brucei
The genome of this human pathogen codes for a protein that is very similar to the SCaMCs. MCP6 is a mitochondrial carrier of 385 amino acids [48], and remarkably, it lacks an N-terminal extension with EF-hand calcium-binding motifs, and thus, transport is probably Ca2+-independent.
In bloodstream-form trypanosomes (found in the blood and tissue fluids of mammals), the protein is predominantly glycosomal, whereas in the procyclic form (found in the midgut of the tse-tse fly), it is found mainly in the mitochondria [48]. Glycosomes are peroxisome-like organelles that contain most of the glycolytic enzymes in both life forms of the trypanosome [48]. Thus, MCP6 could link the nucleotide pools from the different compartments, mitochondria, cytosol, or glycosomes. Surprisingly, it was shown not to exchange ADP/ATP or ATP-Mg/Pi when expressed in Escherichia coli [48]. Nevertheless, the same authors classified it as an SCaMC in a later study [24]. The putative substrate binding site is practically identical to that of SCaMCs, but for K118 E122 V126 instead of K E K in contact point I. Also, the presence of a negatively charged residue in contact point I (E122) suggests that the substrate is transported with a cation, as in SCaMCs. It is essential for survival of T. brucei, as depletion of MCP6 probably inhibits mitochondrial DNA (kinetoplast) replication [48]. This is reminiscent of the mtDNA instability of the sal1 mutant in yeast.
SCaMC in P. falciparum
The genome of this pathogen codes for a single putative SCaMC protein of 590 amino acids (XP_001351960.1). It has a long N-terminal domain of around 150 amino acids that lacks EF-hand calcium-binding motifs, and thus, transport is probably Ca2+-independent. The putative substrate binding site is identical to that of SCaMCs, but for G325-V326 instead of GI in contact point II. Nevertheless, GV is present in many other adenine nucleotide carriers, and thus it probably binds the adenine group as efficiently.
Coenzyme A carrier
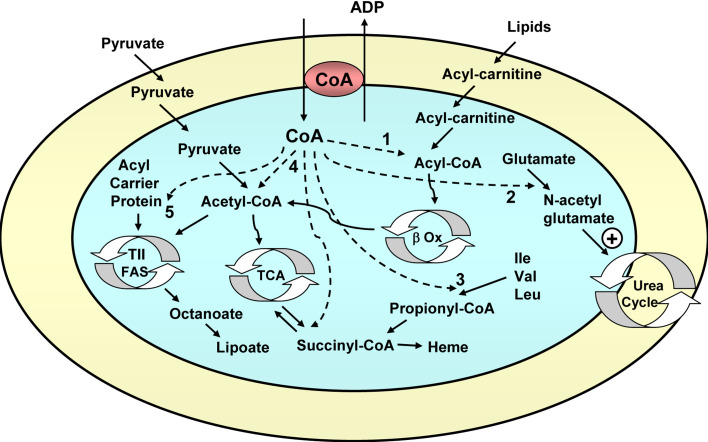
CoA is an essential cofactor that is synthesized in the cytosol of the cell and must be transported to the mitochondrial matrix, where it has a role in the Krebs cycle (pyruvate dehydrogenase and α-ketoglutarate dehydrogenase reactions), β-oxidation of fatty acids, synthesis of heme, and other mitochondrial reactions (Fig. 5).
Fig. 5.
Function of the mitochondrial coenzyme A carrier. The CoA carrier exchanges cytosolic CoA for mitochondrial ADP. CoA is used: 1 in the β-oxidation of fatty acids (β Ox); 2 in the synthesis of N-acetylglutamate, which is an activator of the urea cycle; 3 in the catabolism of branched amino acids (leucine, isoleucine, and valine), generating succinyl-CoA, which goes into the Krebs cycle; 4 in the Krebs cycle (TCA) at the level of pyruvate dehydrogenase (to generate acetyl-CoA) and α-ketoglutarate dehydrogenase (to generate succinyl-CoA, which may also be used for heme synthesis); 5 to provide the mitochondrial acyl carrier protein, a key component of the mitochondrial type II fatty acid synthase (TII FAS, involved in the generation of octanoate, a precursor of the key mitochondrial cofactor lipoate), with its prosthetic group, phosphopantetheine, with production of adenosine 3′,5′-diphosphate, which is exported to the cytosol by the CoA carrier instead of ADP
The genome of S. cerevisiae contains a single CoA carrier gene [49], LEU5 (ORF YHR002w). The leu5 yeast mutant grows poorly in respiratory (nonfermentable) carbon sources and shows a 15-fold reduction in mitochondrial (but not cytosolic) CoA levels.
The genome of mammals contains two CoA carrier genes: SLC25A42 and SLC25A16 [49, 50]. SLC25A42 has been shown to be a strict exchanger of CoA with adenine nucleotides (probably ADP) and is able to complement the leu5 yeast mutant. The KM of SLC25A42 for its substrates is around 50–100 μM [50]. Although SLC25A16 has not been reconstituted in proteoliposomes yet, it is also able to complement the leu5 yeast mutant, suggesting that its substrate specificity must be similar [49].
The CoA carrier has not been identified in A. thaliana yet. Putative orthologs are At4g26180 and At1g14560 (Fig. 2). Putative CoA carriers have also been identified in D. discoideum (proteins McfP and McfR, Fig. 2) [23] and T. brucei (protein MCP4, Fig. 2) [24], but seem to be absent in the malaria parasite P. falciparum. The absence of the carrier in this parasite is not surprising, as it lacks pyruvate dehydrogenase, the β-oxidation pathway, and other pathways that require CoA [26]. However, the carrier is present in other chromalveolates, such as Phytophthora infestans (XP_002897719.1, Fig. 2).
The putative substrate binding site of CoA carriers is conserved [6, 7]. The adenine ring of CoA could bind to contact point II, formed by G-(V/M) (G213-M214 in yeast Leu5p), whereas the sulphur of the pantothenate group could bind to contact point I, formed by (K/Q) (K112 in yeast Leu5p), and the phosphate groups to contact point III, formed by (R/K) (K336 in yeast Leu5p) [8]. The RRRMMM motif of the AACs is not conserved in CoA carriers.
Mitochondrial AMP/ATP transporter
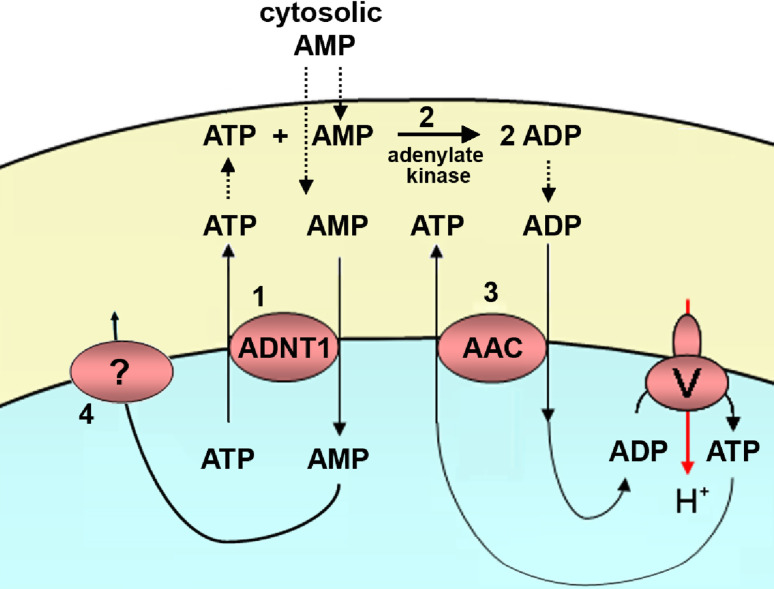
The mitochondrial AMP/ATP (ADNT1) carrier has only been identified in plants and performs an exchange of AMP with ATP between the cytosol and mitochondrial matrix, which is insensitive to BKA and CAT [51]. The genome of A. thaliana contains one mitochondrial ADNT1 gene: At4g01100. It is expressed strongly in seedlings and to a lesser extent in leaves and flowers [51]. It codes for a protein of 352 amino acids.
Its primary function is probably to catalyze the exchange between cytosolic AMP and intramitochondrial ATP. The affinity of this carrier for adenine nucleotides is around one order of magnitude higher than that of the AACs, with a KM of 26 μM for AMP and ATP, and 48 μM for ADP [51]. An ADNT1-mediated AMP/ATP exchange is likely to occur across the inner mitochondrial membrane when AMP is the predominant adenine nucleotide present in the cytosol (Fig. 6). It is known that cytosolic AMP increases in plant tissues during emergence from dormancy and during stresses such as anoxia. In those conditions, catalytic amounts of ATP exported from the mitochondria via ADNT1 and cytosolic AMP would be converted by the intermembrane space adenylate kinase into two ADPs. These ADP molecules would then reenter the mitochondrial matrix via the AAC, and there they would be converted to ATP by oxidative phosphorylation (Fig. 6). These previous steps before oxidative phosphorylation (AMP/ATP exchange and adenylate kinase reaction) could explain the lag phase observed in isolated plant mitochondria before maximum rates of respiration and ATP synthesis are achieved after AMP addition [52]. Accordingly, ADNT1 knock-out plants showed slightly shorter roots than those of wild-type plants and a restricted rate of root respiration during the first days after seed germination [51].
Fig. 6.
Function of the Arabidopsis mitochondrial AMP/ATP carrier. In plants that emerge from dormancy or in plants under anoxic stress, AMP is the predominant nucleotide in the cytosol. Cytosolic AMP is exchanged for catalytic amounts of mitochondrial ATP through the AMP/ATP carrier (1). The ATP that appears in the intermembrane space reacts with another AMP (2) and generates two molecules of ADP (adenylate kinase reaction). The newly formed ADP is exchanged with mitochondrial ATP through the ADP/ATP carrier (AAC), and this allows oxidative phosphorylation and respiration to start (3). In order for the AMP/ATP carrier reaction to proceed, AMP must somehow leave the mitochondrial matrix (4)
This carrier seems to be absent from mammals, but Ypr011c (see “Uncharacterized mitochondrial nucleotide transporters”) could be the yeast ortholog [51]. In the putative substrate binding site of the AMP/ATP carrier, the adenine ring of the nucleotide probably binds to contact point II, formed by G209-M210, as in other nucleotide carriers, whereas the phosphate groups could bind to contact point I, formed by R105 or K113, or contact point III, formed by K325. The RRRMMM motif of the AACs is only slightly modified to R269-R270-R271-M272-Q273-M274. The cytoplasmic and mitochondrial salt bridge networks are basically complete, and this suggests that this carrier is an exchanger, as has been observed experimentally [51].
Uncharacterized mitochondrial nucleotide transporters
Ypr011c-like carriers
In yeast there is a single member in the cluster of adenine nucleotide transporters that remains to be characterized, Ypr011c (ORF YPR011c). It is likely to be an exchanger of nucleotides, with a minor net import activity, which does not require cations such as Mg2+ or H+ [8]. Ypr011c-like carriers are very close to CoA carriers (with a very similar substrate binding site, see Tables 1 and 2) and SCaMCs, with around 30% identity and 50% similarity.
Table 2.
Other mitochondrial nucleotide carriers: SLC25A43 (mammals), Ypr011c (yeast), and ADNT1 (A. thaliana). The consensus sequences of the putative substrate binding sites I, II, and III are shown, and also the deviation from the RRRMMM motif of the ADP/ATP carrier. Note the difference in the substrate binding sites between Ypr011c and ADNT1, which belong to the same cluster (Ypr011c-like carriers). Numbers indicate identity/similarity with other known mitochondrial carriers: ADP/ATP carriers, ATP-Mg/Pi carriers, CoA carriers, and carriers from plants
| Identity/similarity to | Contact points | Motif | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. cerevisiae | H. sapiens | A. thaliana | I | II (adenine) | III | RRRMMM | |||||||
| Aac2p | Sal1p | Leu5p | Ypr011c | ANT1 | SCaMC-1 | SLC25A42 | SLC25A43 | Most similar | |||||
| Hs: SLC25A43 | 25/44 | 23/47 | 24/40 | 31/50a | 27/46 | 28/49 | 28/50 | – | ADNT1 31/51 | R C Q | GA | K | KRK MQA |
| Sc: YPR011c | 31/47 | 30/50 | 31/50 | – | 30/48 | 33/52a | 33/51 | 31/50 | ADNT1 41/55 | R Y Q | GV | K | RRR FQV |
| At: ADNT1 | 28/47 | 28/48 | 32/51 | 41/55a | 25/42 | 33/53 | 25/45 | 23/46 | – | R N K | GV | K | RRR MQM |
aHighest identity/similarity
It could be the yeast ortholog of the AMP/ATP carrier ADNT1 of A. thaliana [51], as it shows 41% identity and 55% similarity, although the putative substrate binding site is different (Table 2). There are also orthologs in members of all the other eukaryotic clades (Fig. 2): the invertebrate metazoan Nematostella vectensis (44/57% identity/similarity, XP_001626526.1), the chromalveolate P. infestans (42/56% identity/similarity, XP_002901049.1; but it is absent in P. falciparum), the excavate Naegleria gruberi (42/59% identity/similarity, XP_002674890.1; but it is absent in T. brucei), and the amoebozoan D. discoideum (44/58% identity/similarity, McfB). The ortholog of D. discoideum, McfB, is interesting and unique, as it contains an N-terminal extension with two EF-hands, similarly to the SCaMCs [23], and this is a novel feature in this kind of carriers. Surprisingly, this carrier is absent in chordates (including vertebrates), where SLC25A43 is present instead.
SLC25A43-like carriers
In some chordates (including all vertebrates), there is also a single member in the cluster of adenine nucleotide transporters that remains to be characterized, SLC25A43 (Fig. 2). It is absent in nonchordates, with the surprising exception of the invertebrate N. vectensis (XP_001627437.1) and the chromalveolates P. infestans (XP_002895040.1) and Blastocystis hominis (see “SCaMC/CoA/Ypr011c-like adenine nucleotide carriers”). This suggests that it has either disappeared in most of the major eukaryotic clades, or it has appeared in late metazoans and has been laterally transferred to these exceptions. The most similar protein in yeast is Ypr011c, in mammals the CoA carrier SLC25A42, and in plants the AMP/ATP carrier ADNT1.
In placental mammals, contact point I of the putative substrate binding site contains R C Q (R78 C82 Q86 in H. sapiens), unique among nucleotide carriers (Table 2), whereas in the rest of the chordates the consensus sequence is R Y Q as in the CoA carrier or Ypr011c. Contact point II is unique among nucleotide carriers in all vertebrates (Table 2) and contains G-A (G173-A174 in H. sapiens), whereas it contains GV in the basal chordate Branchiostoma floridae (the amphioxus or lancet, XP_002611712.1) as in the CoA carrier or Ypr011c. It has been predicted to be a uniporter of nucleotides that does not require cations such as Mg2+ or H+ [8].
Other mitochondrial carriers that transport nucleotides
Mimivirus mitochondrial carrier (VMC1): the DNA of the giant virus Mimiviridae mimivirus encodes a mitochondrial carrier of 237 amino acids. It is the smallest carrier identified so far [53]. It transports dATP and dTTP and functions as an exchanger, possibly with ADP. It is insensitive to BKA and CAT. It has been speculated that it would take mitochondrial dATP and dTTP from the host for the replication of its large genome [53]. The substrate binding site resembles that of ketoacid carriers and uncoupling proteins [53].
FAD carrier: Flx1p (YIL134w) in S. cerevisiae [54], SLC25A32 in mammals [55], AtFOLT11 (At5g66380) in A. thaliana (located in the chloroplast envelope) [56], McfI (and perhaps McfM) in D. discoideum [23]. Probably absent in P. falciparum. It belongs to the dinucleotide carriers group (Fig. 2) [8].
NAD+ carrier: Ndt1p (YIL006w) and Ndt2p (YEL006w) in S. cerevisiae [57], NDT1 (At2g47490) and NDT2 (At1g25380) in A. thaliana (located in the chloroplast envelope and in the mitochondria, respectively) [58], McfW in D. discoideum [23]. Absent in mammals and probably absent in P. falciparum and T. brucei. It belongs to the dinucleotide carriers group (Fig. 2) [8].
Pyrimidine nucleotide carrier: Rim2p (YBR192w) in S. cerevisiae [59], SLC25A33 [60] and SLC25A36 in mammals, RimA in D. discoideum [23], and possibly MCP23 (Tb927.5.1550) in T. brucei [24]. Absent in plants and probably absent in P. falciparum. It belongs to the dinucleotide carriers group, even though it transports mononucleotides (Fig. 2) [8].
Thiamine pyrophosphate carrier: Tpc1p (YGR096w) in S. cerevisiae [61], SLC25A19 in mammals [62], At3g21390 and At5g48970 in A. thaliana, McfK in D. discoideum [23], and possibly MCP7 (Tb10.61.0610) in T. brucei [24] and XP_001350397.1 in P. falciparum. It belongs to the mononucleotide carriers group (Fig. 2) [8].
S-adenosyl-methionine carrier: Sam5p (YNL003c) in S. cerevisiae [63], SLC25A26 in mammals [64], SAMC1 (At4g39460) and SAMC2 (At1g34065) in A. thaliana [65], and possibly MCP20 (Tb10.61.2510) in T. brucei [24] and XP_001350804.1 in P. falciparum. Absent in D. discoideum [23]. It belongs to the amino acid carriers group, even though S-adenosyl-methionine contains an adenine group [8].
GDP/GTP carrier: Ggc1p (YDL198c) in S. cerevisiae [20] and MCP13 (Tb927.2.2970) in T. brucei [24]. Absent in P. falciparum, mammals and plants. It belongs to the ketoacid carriers group, even though it transports nucleotides [8].
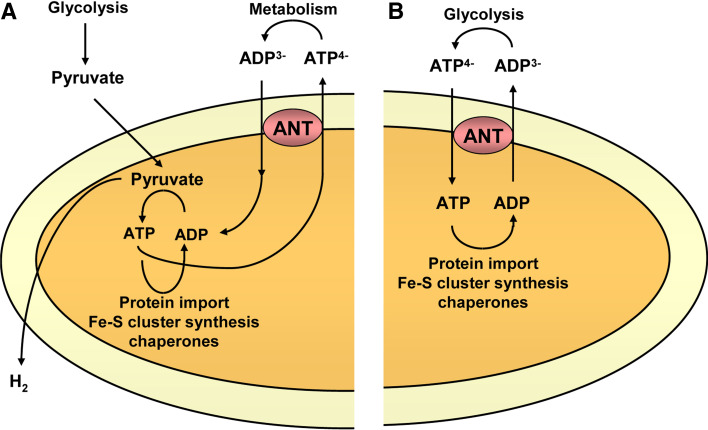
Hydrogenosomal and mitosomal adenine nucleotide carriers
A few groups of unicellular eukaryotes appear to lack mitochondria and contain instead double-membrane-enclosed organelles called hydrogenosomes or mitosomes, depending on whether or not they generate molecular hydrogen [66]. They are thought to have evolved from mitochondria by the concomitant loss of classic mitochondrial features, most notably electron transport chain proteins and the mitochondrial genome. Hydrogenosomes produce ATP by oxidation of pyruvate using a pyruvate-ferredoxin oxidoreductase and molecular hydrogen as a waste product using a hydrogenase. Mitosomes, on the other hand, are even more simplified and have no obvious role in ATP production. All of these simplified organelles always contain at least one mitochondrial chaperone and some elements of the iron-sulfur cluster assembly machinery [66]. Phylogenetic analyses support a common origin for mitochondria, hydrogenosomes, and mitosomes, and these mitochondrial remnants probably arose from mitochondria independently several times during evolution (see Fig. S1 in ESM), as a result of movement into anaerobic habitats. Recently, even anaerobic metazoans of the phylum Loricifera have been shown to lack mitochondria and contain hydrogenosome-like organelles [67].
The mitochondrial carrier repertoire in these mitochondrial remnants is often simplified to a single member, which is always an adenine nucleotide transporter. The compulsory presence of an adenine nucleotide transporter in mitochondrial remnants is reminiscent of a related phenomenon in yeast mitochondria, where ATP import either by the AACs or Sal1p is required for viability [11]. ATP is essential in these organelles for the protein import system and for the assembly of iron-sulfur clusters (Fig. 7). There are several types of nucleotide transporters in these organelles (Table 3). They appear to have obtained adenine nucleotide carriers independently from different sources (Fig. 2): canonical AACs (“Canonical ADP/ATP carriers”), Ypr011c/CoA/SCaMC-like carriers (“SCaMC/CoA/Ypr011c-like adenine nucleotide carriers”), modified NAD+ carriers (“NAD+-like adenine nucleotide carriers”), or even by lateral gene transfer from prokaryotes (“Adenine nucleotide carriers not belonging to the mitochondrial carrier family”). Even close relatives, such as the microsporidians Antonospora locustae and Encephalitozoon cuniculi, may contain different types of adenine nucleotide transporters. Thus, different organisms with simplified mitochondria have recruited different carriers to maintain adenine nucleotide transport.
Fig. 7a, b.
Function of hydrogenosomal and mitosomal adenine nucleotide transporters. a Hydrogenosomes generate H2 and ATP by substrate-level phosphorylation. The ATP is used for protein import, chaperone function, or synthesis of Fe-S clusters, and is also exported to the cytosol in exchange for ADP by an adenine nucleotide transporter (ANT). b Mitosomes do not produce ATP, and thus it must be imported from the cytosol to support mitosomal reactions by an adenine nucleotide transporter (ANT)
Table 3.
Hydrogenosomal/mitosomal adenine nucleotide carriers. The carriers are divided in four groups: similar to AACs, similar to SCaMC/CoA/Ypr011c carriers, similar to NAD+ carriers, and carriers that do not belong to the mitochondrial carrier family. The eukaryotic clade of the organism, the type of organelle (aM anaerobic mitochondria, Hy hydrogenosome, Mt mitosome), and the type of metabolism of the organelle (I, imports ATP; II, generates ATP) are indicated. Nucleotide carriers that do not belong to the mitochondrial carrier family are not compared with other proteins. Numbers indicate identity/similarity with other known mitochondrial carriers as in Table 2, and putative substrate binding sites are indicated. Contact point II of most of them contains G-[IV] and contact point III contains [RK]
| Clade | Organelle | Type | Identity/similarity to | Contact points | Motif | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AACs | SCaMCs | CoA | Ypr011c | SLC25A43 | Most similar in plants | I | II (adenine) | III | RRRMMM | |||||
| Similar to AAC | ||||||||||||||
| Nyctotherus ovalis | Chromalveolate | Hy | II | 63/78 ANT1a | 30/50 SCaMC-1 | 27/47 SLC25A42 | 31/49 | 28/47 | AAC1 51/65 | R T N | GI | R | RRR LMM | |
| Neocallimastix frontalis | Opisthokont | Hy | II | 65/79 Aac2pa | 28/50 SCaMC-1 | 27/45 SLC25A42 | 30/44 | 26/45 | AAC1 68/81 | R T N | GI | R | RRR MMM | |
| Similar to SCaMC/CoA | ||||||||||||||
| Blastocystis hominis | Chromalveolate | aM | II | Contains several adenine nucleotide carriers | ||||||||||
| Trichomonas vaginalis | Excavate | Hy | II | 28/47 Aac2p ANT1 | 35/57 SCaMC-1a | 34/55 SLC25A42 | 33/52 | 30/54 | AtBT1 31/57 | R Q K | GV | K | RKR MML | |
| Entamoeba histolytica | Amoebozoan | Mt | I | 26/45 ANT1 | 28/48 SCaMC-1 | 30/47 SLC25A42 | 29/49a | 26/45 | AtBT 30/54 | K T K | SV | K | KRK LLA | |
| Trimastix pyriformis | Excavate | aM | II | 29/46 Aac2p | 26/44Sal1p SCaMC-1 | 31/47 SLC25A42a | 26/44 | 25/45 | At5g07320 (SCaMC) 29/49 | R F R | GQ | K | KRR CLV | |
| Psalteriomonas lanterna | Excavate | Hy | II | 42/57 ANT1a | 36/52 SCaMC-1 | 34/54 SLC25A42 | 34/55 | 29/49 | At5g56450 35/55 | R N K | GI | R | RRR MQM | |
| Similar to NAD+ carriers | ||||||||||||||
| Antonospora locustae | Opisthokont | Mt | I | 24/46 Aac2p | 25/46 SCaMC-1 | 29/46 SLC25A42 | 29/48 | – | NDT1 37/53 | G T Y | GV | R | RIR QQM | |
| Identity/similarity to other carriers | ScNdt1p/2p: 39/52a 37/54 | HsSLC25A32 32/53 | ||||||||||||
| Not MC | ||||||||||||||
| Encephalitozoon cuniculi | Opisthokont | Mt | I | Does not belong to the MCF | ||||||||||
aHighest identity/similarity
Canonical ADP/ATP carriers
Those adenine nucleotide carriers from anaerobic ciliates and anaerobic chytridiomycete fungi are true AACs (Fig. 2) and are accordingly inhibited by CAT and BKA.
Nyctotherus ovalis
The genome of this gut-dwelling anaerobic ciliate codes for two mitochondrial transporters [68]. One is a putative S-adenosylmethionine carrier and the other is a transporter of 308 amino acids that clusters with true AAC sequences. The hydrogenosome of this organism has retained a rudimentary genome that encodes subunits of mitochondrial complex I, and these proteins cluster with their homologues from aerobic ciliates, such as Paramecium or Tetrahymena [69]. Complex I is probably involved in the generation of a hydrogenosomal membrane potential [69]. Two nuclear-encoded complex II subunits are present, but complexes III and IV are absent.
Neocallimastix patriciarum and Neocallimastix frontalis
The genome of these anaerobic fungi codes for a single transporter of 308 amino acids [68, 70]. Its sequence clusters with fungal AAC sequences. It is able to complement growth in respiratory carbon sources of the aac2 S. cerevisiae mutant, and this demonstrates that the hydrogenosomal AAC of Neocallimastix is recognized by the mitochondrial protein import machinery of S. cerevisiae and a conservation of function.
SCaMC/CoA/Ypr011c-like adenine nucleotide carriers
Those carriers of anaerobic flagellates, anaerobic stramenopiles, or microsporidians are closer to the SCaMCs/CoA/Ypr011c-like proteins than to the AACs. However, none of them has an amino-terminal extension with Ca2+ binding domains as do the SCaMCs. The precise carrier from which each of them has evolved is not clear, as the percentage of identity/similarity to SCaMCs, CoA carriers, or Ypr011c-like carriers is very similar (Table 3). Apparently, those that have been tested in transport essays are not inhibited by CAT or BKA.
Blastocystis hominis
The genome of this opportunistic human parasite belonging to the stramenopiles codes for several mitochondrial carriers. It was previously reported that there were only 9 transporters [71], but in blast searches on B. hominis protein sequences in NCBI, using several mitochondrial carriers as query, we have found at least 20 carriers, which is consistent with their complex metabolism [71]. There are several adenine nucleotide carriers such as a CoA carrier, an SCaMC, and an SLC25A43-like carrier, but interestingly no AACs (Fig. 2). The organelle of this organism contains a circular genome that encodes subunits of mitochondrial complex I [72] and also possesses a membrane potential [71]. Four nuclear-encoded complex II subunits are present, but complexes III and IV are absent. There is no evidence of hydrogen production yet, but there is a hydrogenase in the organelle. These organelles are considered as anaerobic mitochondria.
Trichomonas vaginalis
The genome of this anaerobic flagellate belonging to the parabasalids codes for five mitochondrial carriers, among them a transporter of 316 amino acids [68]. This protein, Hmp31, contains a cleavable N-terminal transit peptide that is not essential for import but is able to replace a hydrogenosomal matrix presequence when fused to a different sequence. It transports ADP and ATP, operating as an exchanger [73]. The hydrogenosome of Trichomonas contains the NADH dehydrogenase module of mitochondrial complex I [74] and also possesses a membrane potential. Complexes II, III, and IV are absent.
Entamoeba histolytica
The genome of this human parasitic amoebozoan codes for two mitochondrial carriers. One of them is a highly modified phosphate carrier [75]. The other is an adenine nucleotide carrier of 276 amino acids [76]. It is targeted to the mitosome and it transports ATP, ADP, and AMP, operating as an electroneutral exchanger. Interestingly, Entamoeba has lost mitochondrial-type iron-sulfur cluster assembly proteins and possesses instead an analogous bacterial-type system acquired by lateral gene transfer [77]. However, there are conflicting data on whether the system is located in the cytosol, the mitosome, or both [75, 77].
Trimastix pyriformis
The genome of this free-living anaerobic flagellate belonging to the excavates codes for three mitochondrial carriers [78]. One of them clusters with FAD and dicarboxylate transporters, another has no obvious similarity with any carrier, and the third one, with 270 amino acids, clusters with adenine nucleotide carriers [78]. There is no evidence of hydrogen production yet, but there is a hydrogenase gene. These organelles are considered to be anaerobic mitochondria.
Psalteriomonas lanterna
The genome of this microaerophilic amoeboflagellate belonging to the heteroloboseans codes for a single mitochondrial carrier of 298 amino acids, which is probably targeted to the hydrogenosome [79]. The hydrogenosome contains the NADH dehydrogenase module of mitochondrial complex I [79]. Complexes II, III, and IV are absent.
NAD+-like adenine nucleotide carriers
The genome of the microsporidian A. locustae codes for a single mitochondrial carrier of 299 amino acids whose localization is currently unknown, but it is targeted to the mitochondria when expressed in S. cerevisiae [80]. Thus, it is probably targeted to the mitosome in Antonospora [80]. It was shown to transport ATP and ADP [80], preferentially as an exchanger. Surprisingly, this carrier is phylogenetically close to the NAD+ transporter of S. cerevisiae or A. thaliana (Fig. 2), and not to adenine nucleotide carriers, and thus it must have altered its function at some point in its evolution [80].
Adenine nucleotide carriers not belonging to the mitochondrial carrier family
There are four ATP/ADP transporters in the microsporidian E. cuniculi that are not related to the mitochondrial carrier family (EcNTT1–EcNTT4) [81]. Their amino acid sequence exhibits a high degree of similarity to the ATP/ADP transporter from the obligate intracellular gram-negative bacterium Rickettsia prowazekii and to the ATP/ADP transporter of the plastid envelope (see “Plastidic adenine nucleotide carriers not belonging to the mitochondrial carrier family”). This suggests that these proteins have been obtained by lateral gene transfer, probably from prokaryotes. The carriers contain 12 putative transmembrane helixes and a molecular weight of around 60 kDa. They are insensitive to the AAC inhibitors BKA and CAT.
EcNTT3 is located to the mitosome of E. cuniculi, and it is thought to provide it with ATP [81]. On the other hand, EcNTT1, EcNTT2, and EcNTT4 are located in the surface of the parasite, but only when it is living inside host cells, and would function to steal ATP from them [81]. Phylogenetic and immunofluorescence data indicate that NTTs are also present in other microsporidians, such as Paranosema grylli and Trachipleistophora hominis [81].
Plastidic adenine nucleotide carriers
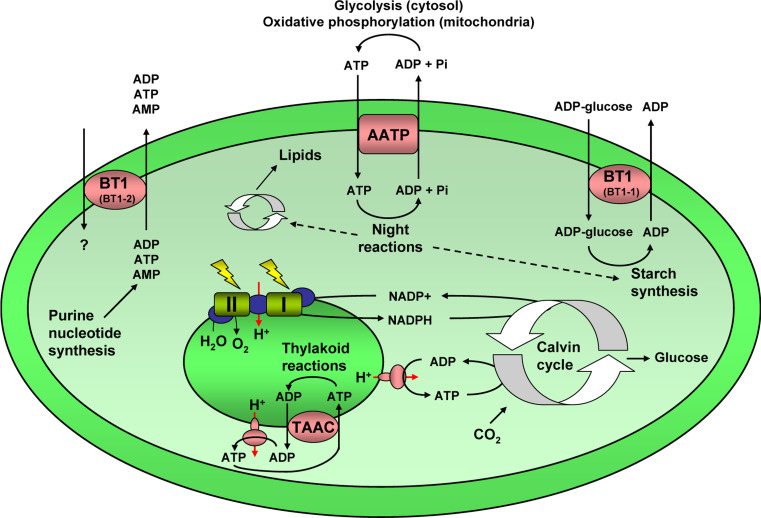
Plant chloroplasts possess an outer membrane and an envelope (inner membrane), and between these two layers is the intermembrane space. The material within the chloroplast is called the stroma, which contains the DNA, ribosomes, etc. In the stroma there are sub-organelles called thylakoids, where the light reactions of photosynthesis and ATP synthesis take place, inside of which there is a lumen. Thus, in the chloroplast, there must be transport systems that connect the cytosol with the stroma, and the stroma with the thylakoid lumen (Fig. 8; Table 4).
Fig. 8.
Plastidic adenine nucleotide transporters. In plastids there are several adenine nucleotide transporters. The plastid envelope adenine nucleotide carriers (AATP) exchange cytosolic ATP for mitochondrial ADP and Pi, and this ATP is used for night reactions, including synthesis of lipids and starch synthesis. The thylakoid adenine nucleotide carriers (TAAC) exchange stromal ATP (generated by photophosphorylation in the plastid ATP synthase) for thylakoidal ADP, and this ATP is used for thylakoid reactions, such as protein phosphorylation, folding, import, and degradation, turnover and repair of the photosystem II complex. The ubiquitous BT1 (BT1-2) transporter exports adenine nucleotides, which are exclusively synthesized de novo in plastids, to the cytosol. The related BT1 (BT1-1) transporter, which is only present in monocotyledonous plants, imports cytosolic ADP-glucose, required for starch synthesis, in exchange for plastidic ADP
Table 4.
Extramitochondrial adenine nucleotide carriers in plants. ADP/ATP exchangers in the thylakoid membrane and in the plastid envelope, adenine nucleotide uniporters in the plastid envelope, ADP/ATP carrier in the ER, and uncharacterized carriers that are similar to them are shown. Plastidic adenine nucleotide carriers that do not belong to the mitochondrial carrier family are not compared with other proteins. Numbers indicate identity/similarity with other known mitochondrial carriers as in Table 2, and putative substrate binding sites are indicated. Contact point II of most of them contains G-[IVM] and contact point III contains [RK]
| Identity/similarity to | Contact points | Motif | |||||||
|---|---|---|---|---|---|---|---|---|---|
| AACs | SCaMCs | CoA | Ypr011 | SLC25A43 | I | II (adenine) | III | RRRMMM | |
| ADP/ATP exchangers in the thylakoid membrane | |||||||||
| At: TAAC (At5g01500) | 31/46 ANT1 | 33/56 SCaMC-1 | 36/54 SLC25A42a | 32/50 | 30/49 | R Y Q | S I | K | RRQ MQL |
| At3g51870 | 31/47 ANT1 | 33/54 SCaMC-1 | 36/54 SLC25A42a | 30/49 | 29/48 | R Y Q | GI | K | RRQ MQM |
| Adenine nucleotide uniporters in the plastid envelope | |||||||||
| AtBT1 (At4g32400) | 27/46 ANT1 Aac2p | 35/56 SCaMC-1 | 35/57 SLC25A42a | 37/55 | 30/51 | R S E | GV | K | RKH MQV |
| At3g20240 | 23/44 ANT1 | 30/51 SCaMC-1a | 29/51 SLC25A42 | 27/49 | 26/47 | R T E | GM | K | RKR LMV |
| ADP/ATP exchangers in the plastid envelope | |||||||||
| AATP1, AATP2 | Do not belong to the MCF | ||||||||
| Endoplasmic reticulum ADP/ATP carrier | |||||||||
| At: ER-ANT1 (At5g17400) | 62/77 Aac2pa | 28/46 SCaMC-1 | 29/44 SLC25A42 | 28/44 | 26/47 | R T N | GI | L | RRR MML |
| At5g56450 | 44/63 ANT1a | 29/49 SCaMC-1 | 29/45 SLC25A42 | 31/47 | 27/50 | R S N | GV | R | RRR IMM |
aHighest identity/similarity
Plastidic adenine nucleotide carriers not belonging to the mitochondrial carrier family
There are two ATP/ADP transporters in the plastid envelope of A. thaliana that are not related to the mitochondrial carrier family (AATP1 and AATP2) [82]. They catalyze the electroneutral exchange of cytosolic ATP4− for stromal ADP3− and H2PO4 − [83]. Their amino acid sequence exhibits a high degree of similarity to the ATP/ADP transporter from the obligate intracellular gram-negative bacterium R. prowazekii, and they may have been obtained by lateral gene transfer. These proteins contain 578 and 569 amino acids, respectively, 12 putative transmembrane helixes, and a molecular weight of around 60 kDa. They may be members of the 12-TM family of transport systems. They are insensitive to the AAC inhibitors BKA and CAT.
The main function of the plastidic ATP/ADP transporter is the supply of the stroma with cytosolic ATP required for various important anabolic reactions (Fig. 8). For example, starch, amino acid, and fatty acid synthesis in heterotrophic plastids is strictly dependent upon the exogenous supply of ATP. Also, nocturnal ATP import is crucial for controlled chlorophyll biosynthesis and contributes to the prevention of light-induced cell damage induced by accumulation of photoreactive tetrapyrrole intermediates [84]. Accordingly, double AATP1 and AATP2 knock-out plants show a dwarf phenotype and necrotic lesions in the leaves when grown in low light/short day conditions, where they accumulate the photoreactive intermediate Proto IX. This does not take place in high light/long day conditions because in these situations the plastids are able to regenerate stromal ATP by substrate-level phosphorylation produced by increased turnover of transitory starch, and this ATP prevents Proto IX accumulation and thus oxidative damage [84].
Plastidic adenine nucleotide carriers belonging to the mitochondrial carrier family
ADP/ATP exchangers
There is an AAC in the plastid thylakoid of A. thaliana [85], and it belongs to the mitochondrial carrier family (TAAC, At5g01500 gene). It contains 415 amino acids and an N-terminal chloroplast transit peptide. It exchanges stromal ATP for thylakoid lumen ADP.
ATP is produced during the light-dependent photosynthetic reactions on the stromal side of the thylakoid membrane. Besides its utilization during CO2 fixation in the stroma, ATP drives many energy-dependent processes in thylakoids (Fig. 8), including protein phosphorylation, folding, import, and degradation [85]. It is also converted to GTP, which is involved in the turnover and repair of the photosystem II complex, which is mainly inactivated under high light stress conditions [86]. TAAC knock-out plants displayed lower shoot weight when reaching full development and were less tolerant of high light conditions in comparison with the wild type, apparently by a deficient repair of photosystem II under light stress in the mutants [86]. Mutants also show a 40% reduction in the thylakoid content, resulting in pale green leaves, and uptake of ATP into isolated thylakoids is reduced around 30–40% [85]. This residual ATP uptake activity suggests that there must be an alternative carrier for ATP in thylakoids that compensates for the absence of TAAC. Indeed, the gene At3g51870 codes for an uncharacterized carrier that is very similar to TAAC, and it could perform this function.
Adenine nucleotide uniporters
The protein Brittle 1 (AtBT1, gene At4g32400) in A. thaliana is a transporter of AMP, ADP, and ATP (Fig. 8) and is able to perform nucleotide uniport [87]. The homologous protein of Solanum tuberosum, StBT1, has also been characterized [88]. It was proposed to provide the cytosol and other compartments with adenine nucleotides exclusively synthesized de novo inside plastids [88]. The highly specific inhibitors of mitochondrial AACs, BKA and CAT, showed no inhibitory effect.
AtBT1 knock-out plants are unable to develop [87], suggesting that the absence of Brittle 1 is lethal, and that no other carrier or metabolic pathway can compensate for the loss of this transporter. On the other hand, plants with a reduced expression of the protein survive but show impaired growth [87]. Addition of adenosine increases growth in these mutants, and this is explained because plants contain a nucleotide salvage pathway in the cytosol.
It was previously thought that Brittle 1 in Zea mays (Zmbt1) encoded an ADP-glucose transporter, residing in the inner membrane of maize endosperm plastids [89]. This apparent paradox was later explained in further phylogenetic and biochemical analyses, which classify the BT1 family into two distinct groups [89]. One group is restricted to cereals, where they mediate ADP-glucose transport in exchange for ADP for starch synthesis (BT1). Indeed, whereas ADP-glucose is synthesized in the plastid of most plants, it is synthesized in the cytosol of monocotyledonous plants, and thus, it must be imported into the chloroplast (Fig. 8). The other group, in which AtBT1 is included, occurs in all plants and is involved in the uniport of adenine nucleotides (BT1-2). It is surprising that, despite showing 79% identity and 89% similarity in the transport domain and identical substrate contact points between them, the substrates of both groups are different. The presence of a negatively charged residue in contact point I (Table 4) suggests that transport may require a cation, as do SCaMCs.
The gene At3g20240 codes for an uncharacterized protein that is highly similar to Brittle 1, but whose subcellular localization is currently unknown.
Adenine nucleotide carriers in other organelles
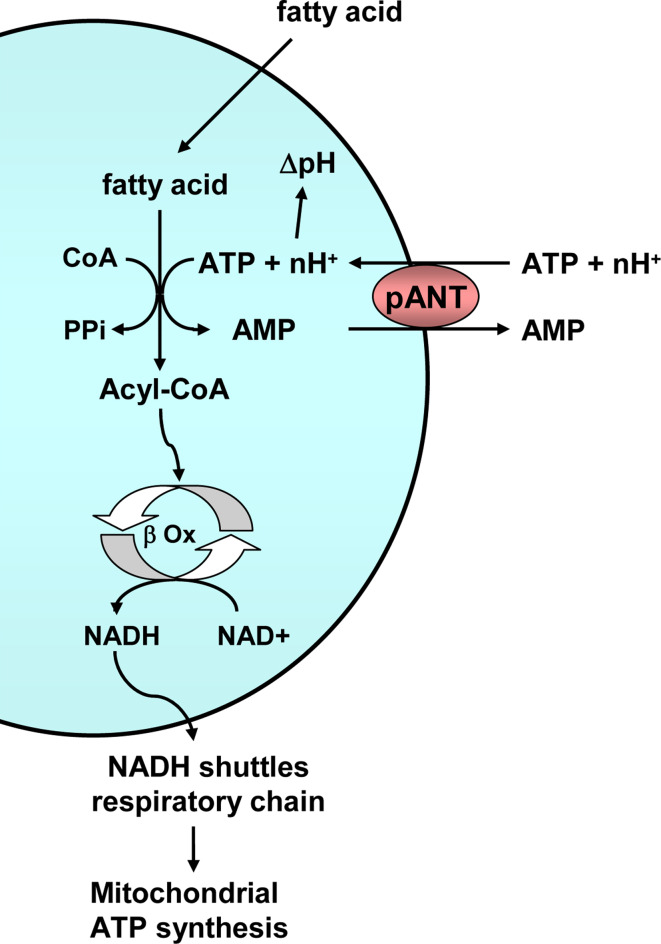
Peroxisomal adenine nucleotide transporter (pANT)
The peroxisomal adenine nucleotide carrier performs an electroneutral exchange of cytosolic ATP for peroxisomal AMP, which is H+ compensated (Fig. 9). The carrier is an important supply of ATP for the activation of fatty acids for peroxisomal β-oxidation, for the generation of an H+ gradient [90], and possibly for peroxisomal biogenesis. Peroxisomal carriers are more similar to pyrimidine nucleotide, NAD+, and FAD carriers (dinucleotide carriers, Fig. 2) than to adenine nucleotide carriers [8].
Fig. 9.
Function of the peroxisomal adenine nucleotide transporter. The peroxisomal adenine nucleotide transporter (pANT) exchanges cytosolic ATP and an as yet undetermined number of protons for peroxisomal AMP. ATP is used in the activation of fatty acids for peroxisomal β oxidation (β Ox), and the protons generate a peroxisomal ∆pH
There is a single gene in yeast, ANT1 (ORFs YPR128c), and its disruption results in impaired growth on laurate, a medium-chain fatty acid, as a single carbon source, whereas normal growth was observed with long-chain fatty acids [91]. β-Oxidation of medium-chain fatty acids in S. cerevisiae takes place in the peroxisomes, a process that is dependent on intraperoxisomal ATP.
SLC25A17 is the peroxisomal AMP/ATP carrier in mammals. In humans, the peroxisomal β-oxidation system is required for chain shortening of certain branched chain fatty acids and very long chain fatty acids, and therefore it needs ATP in the peroxisomal matrix for their activation [92]. On the other hand, medium-chain fatty acids are oxidized in the mitochondria.
In A. thaliana there are three peroxisomal mitochondrial carriers, At2g39970, At3g05290, and At5g27520, but only At3g05290 (PNC1) and At5g27520 (PNC2) are thought to be AMP/ATP transporters. Knock-out plants for both genes are impaired in seedling growth, which depends on fatty acid oxidation in the peroxisomes [93]. The function of At2g39970 is unknown, but it clusters with the rest of peroxisomal adenine nucleotide transporters.
McfQ is the putative peroxisomal adenine nucleotide transporter in D. discoideum (Fig. 2) [23]. The carrier is probably absent in P. falciparum, but present in other chromalveolates such as P. infestans (XP_002900592.1, XP_002902667.1, and XP_002909633.1, Fig. 2). In T. brucei there are several candidates [24], MCP1 (Tb09.211.3200), MCP2 (Tb11.01.5960), and MCP22 (Tb11.01.5950), but they do not really cluster with the rest of peroxisomal transporters. In addition, MCP2 and MCP22 are apparently located to the mitochondria, whereas the localization of MCP1 is still unknown [24], and this leaves MCP1 as the only probable peroxisomal candidate. On the other hand, β-oxidation in this organism probably takes place in glycosomes [94], peroxisome-like organelles in which there is already a nucleotide transporter (the SCaMC MCP6) [48], and thus a peroxisomal nucleotide transporter may not be required. This carrier is nevertheless present in other excavates such as N. gruberi (XP_002678224.1, Fig. 2).
Endoplasmic reticulum adenine nucleotide transporter (ER-ANT)
The endoplasmic reticulum of A. thaliana contains the product of an AAC gene [95], ER-ANT1 (At5g17400). It shows 61, 61, and 63% identity and 77, 77, and 79% similarity with AtAac1, AtAac2, and AtAac3, respectively. In contrast to them, ER-ANT1 does not possess a putative N-terminal transit peptide. It supplies the ER lumen with ATP for BiP chaperones, calreticulin chaperones, and Ca2+-dependent protein kinase [95]. Remarkably, it is insensitive to BKA and CAT.
ER-ANT knock-out plants exhibited a dramatic growth retardation and impaired root and seed development [95]. In addition, the levels of several endoplasmic reticulum proteins that are dependent on a sufficient ATP supply were substantially decreased.
The ER-ANT is absent in yeast or mammals, and it is unknown which protein is responsible for ATP uptake in the ER of these organisms. The gene At5g56450 in Arabidopsis codes for an uncharacterized protein that is similar to ER-ANT1, but whose subcellular localization is currently unknown.
Vesicular nucleotide transporter
The vesicular nucleotide transporter does not belong to the mitochondrial carrier family (SLC25) but to the SLC17 transporter family [96]. This family comprises nine genes in mammals [96] and includes Na+/PO4 2− cotransporters (SLC17A1, SLC17A2, ALC17A3, and SLC17A4), the transporter of H+-sialic acid or aspartate (SLC17A5) [97], and glutamate transporters (SLC17A6, SLC17A7, and SLC17A8).
The vesicular nucleotide transporter (VNUT, SLC17A9) mediates the accumulation of ATP in secretory vesicles such as synaptic vesicles and adrenal chromaffin granules, and it is important for the exocytosis of ATP. It contains 430 amino acids and 12 putative transmembrane helixes, and it is present in various vertebrates and invertebrates [96]. It was shown to transport ATP, ADP, and GTP by using membrane potential as the driving force. Transport requires chloride anions, as do some other members of this family [97], and surprisingly, it is inhibited by atractyloside, but only in the presence of Mg2+ [96].
Concluding remarks
Mitochondrial carriers are present in all five eukaryotic clades (opisthokonts, plantae, amoebozoans, excavates, and chromalveolates), suggesting that they were fundamental for the development of eukaryotes and that mitochondria are now essential components of eukaryotic cells. They are not only located to mitochondria, but to other organelles such as peroxisomes, endoplasmic reticulum, and plastids.
Although some eukaryotes appear to lack mitochondria and were thought to be primitive and to have separated from other eukaryotes before the mitochondrial endosymbiosis, recent studies suggest that this is a derived feature. In these amitochondriate species, hydrogenosomes or mitosomes have appeared by a dramatic reductive evolution of the former mitochondria. Mitochondrial carriers are, as the rest of the mitochondrial components (respiratory complexes, H+-ATP synthase, mitochondrial DNA, enzymes), also reduced and often a single member remains, which is always an adenine nucleotide transporter. Thus, ATP seems to be always required for the function of these mitochondrial homologues. The only common metabolic function identified for mitochondria, hydrogenosomes, and mitosomes is iron-sulfur cluster biogenesis [66]. Some enzymes responsible for this pathway require ATP, and all of them must be imported into the organelle using the ATP-requiring protein import system and properly folded using ATP-requiring chaperones. This explains the strict requirement of ATP, and thus the obligatory presence of an adenine nucleotide carrier in these extremely simplified organelles. Indeed, whereas mitochondrial DNA, oxidative phosphorylation, and many other mitochondrial processes are dispensable in yeast or even in some mammalian cells in culture, ATP is still required for the protein import system, chaperones and iron-sulfur cluster biogenesis, and thus either an AAC or an SCaMC is essential for viability in yeast [11, 36, 41]. There may be, however, a significant difference between mitochondria and mitochondrial remnants: the mitochondrial protein import system requires, in addition to ATP, a membrane potential, whereas, to our knowledge, not all types of mitosomes or hydrogenosomes have been shown to display a membrane potential yet. Perhaps some mitosomes and hydrogenosomes have bypassed the membrane potential requirement as they employ a highly modified and reduced protein import system [75]. To sum up, an adenine nucleotide carrier was probably the first mitochondrial carrier that appeared in evolution, and one of the key proteins that allowed a stable endosymbiosis to occur.
Studying these adenine nucleotide transport systems in organelles is interesting, as many of these organisms with simplified mitochondria, such as E. histolytica, Giardia intestinalis, T. vaginalis, and microsporidia, are human parasites. As these carriers are often highly modified, the discovery of specific inhibitors of mitosomal or hydrogenosomal adenine nucleotide carriers can be helpful when dealing with human diseases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was supported by grants from Ministerio de Educación y Ciencia (BFU2008-04084/BMC), Comunidad de Madrid (S-GEN-0269-2006 MITOLAB-CM), the European Union (LSHM-CT-2006-518153), and CIBERER (an initiative of the ISCIII) to J.S.; by grants from the ISCIII (PI080610) to A. del A. and by an institutional grant from the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa. J.T. is a recipient of a fellowship from the Comunidad de Madrid.
Abbreviations
- MCF
Mitochondrial carrier family
- CAT
Carboxyatractyloside
- BKA
Bongkrekic acid
- AAC
ADP/ATP carrier
- SCaMC
Short calcium-binding mitochondrial carrier
- CoA
Coenzyme A
- ORF
Open-reading frame
References
- 1.Winkler HH, Neuhaus HE. Non-mitochondrial ATP transport. Trends Biochem Sci. 1999;24:64–68. doi: 10.1016/S0968-0004(98)01334-6. [DOI] [PubMed] [Google Scholar]
- 2.del Arco A, Satrústegui J. New mitochondrial carriers: an overview. Cell Mol Life Sci. 2005;62:2204–2227. doi: 10.1007/s00018-005-5197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picault N, Hodges M, Palmieri L, Palmieri F. The growing family of mitochondrial carriers in Arabidopsis . Trends Plant Sci. 2004;9:138–146. doi: 10.1016/j.tplants.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Palmieri F, Pierri CL. Mitochondrial metabolite transport. Essays Biochem. 2010;47:37–52. doi: 10.1042/bse0470037. [DOI] [PubMed] [Google Scholar]
- 5.Palmieri F, Agrimi G, Blanco E, Castegna A, Di Noia MA, Iacobazzi V, Lasorsa FM, Marobbio CM, Palmieri L, Scarcia P, Todisco S, Vozza A, Walker J. Identification of mitochondrial carriers in Saccharomyces cerevisiae by transport assay of reconstituted recombinant proteins. Biochim Biophys Acta. 2006;1757:1249–1262. doi: 10.1016/j.bbabio.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 6.Kunji ER, Robinson AJ. The conserved substrate binding site of mitochondrial carriers. Biochim Biophys Acta. 2006;1757:1237–1248. doi: 10.1016/j.bbabio.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 7.Robinson AJ, Kunji ER. Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci USA. 2006;103:2617–2622. doi: 10.1073/pnas.0509994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson AJ, Overy C, Kunji ER. The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc Natl Acad Sci USA. 2008;105:17766–17771. doi: 10.1073/pnas.0809580105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rial E, Zardoya R. Oxidative stress, thermogenesis and evolution of uncoupling proteins. J Biol. 2009;8:58. doi: 10.1186/jbiol155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klingenberg M. The ADP and ATP transport in mitochondria and its carrier. Biochim Biophys Acta. 2008;1778:1978–2021. doi: 10.1016/j.bbamem.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Traba J, Satrústegui J, del Arco A. Transport of adenine nucleotides in the mitochondria of Saccharomyces cerevisiae: interactions between the ADP/ATP carriers and the ATP-Mg/Pi carrier. Mitochondrion. 2009;9:79–85. doi: 10.1016/j.mito.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 12.Fritz-Laylin LK, Prochnik SE, Ginger ML, Dacks JB, Carpenter ML, Field MC, Kuo A, Paredez A, Chapman J, Pham J, Shu S, Neupane R, Cipriano M, Mancuso J, Tu H, Salamov A, Lindquist E, Shapiro H, Lucas S, Grigoriev IV, Cande WZ, Fulton C, Rokhsar DS, Dawson SC. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 13.Adl SM, Leander BS, Simpson AG, Archibald JM, Anderson OR, Bass D, Bowser SS, Brugerolle G, Farmer MA, Karpov S, Kolisko M, Lane CE, Lodge DJ, Mann DG, Meisterfeld R, Mendoza L, Moestrup Ø, Mozley-Standridge SE, Smirnov AV, Spiegel F. Diversity, nomenclature, and taxonomy of protists. Syst Biol. 2007;56:684–689. doi: 10.1080/10635150701494127. [DOI] [PubMed] [Google Scholar]
- 14.Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trézéguet V, Lauquin GJ, Brandolin G. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature. 2003;426:39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- 15.Hatanaka T, Hashimoto M, Majima E, Shinohara Y, Terada H. Functional expression of the tandem-repeated homodimer of the mitochondrial ADP/ATP carrier in Saccharomyces cerevisiae . Biochem Biophys Res Commun. 1999;262:726–730. doi: 10.1006/bbrc.1999.1283. [DOI] [PubMed] [Google Scholar]
- 16.Huang SG, Odoy S, Klingenberg M. Chimers of two fused ADP/ATP carrier monomers indicate a single channel for ADP/ATP transport. Arch Biochem Biophys. 2001;394:67–75. doi: 10.1006/abbi.2001.2520. [DOI] [PubMed] [Google Scholar]
- 17.Phelps A, Wohlrab H. Homodimeric mitochondrial phosphate transport protein. Transient subunit/subunit contact site between the transport relevant transmembrane helices A. Biochemistry. 2004;43:6200–6207. doi: 10.1021/bi036148n. [DOI] [PubMed] [Google Scholar]
- 18.Kunji ER, Crichton PG. Mitochondrial carriers function as monomers. Biochim Biophys Acta. 2010;1797:817–831. doi: 10.1016/j.bbabio.2010.03.023. [DOI] [PubMed] [Google Scholar]
- 19.Palmieri F, Pierri CL. Structure and function of mitochondrial carriers—role of the transmembrane helix P and G residues in the gating and transport mechanism. FEBS Lett. 2010;584:1931–1939. doi: 10.1016/j.febslet.2009.10.063. [DOI] [PubMed] [Google Scholar]
- 20.Vozza A, Blanco E, Palmieri L, Palmieri F. Identification of the mitochondrial GTP/GDP transporter in Saccharomyces cerevisiae . J Biol Chem. 2004;279:20850–20857. doi: 10.1074/jbc.M313610200. [DOI] [PubMed] [Google Scholar]
- 21.De Marcos Lousa C, Trézéguet V, Dianoux AC, Brandolin G, Lauquin GJ. The human mitochondrial ADP/ATP carriers: kinetic properties and biogenesis of wild-type and mutant proteins in the yeast S. cerevisiae . Biochemistry. 2002;41:14412–14420. doi: 10.1021/bi0261490. [DOI] [PubMed] [Google Scholar]
- 22.Murcha MW, Millar AH, Whelan J. The N-terminal cleavable extension of plant carrier proteins is responsible for efficient insertion into the inner mitochondrial membrane. J Mol Biol. 2005;351:16–25. doi: 10.1016/j.jmb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 23.Satre M, Mattei S, Aubry L, Gaudet P, Pelosi L, Brandolin G, Klein G. Mitochondrial carrier family: repertoire and peculiarities of the cellular slime mould Dictyostelium discoideum . Biochimie. 2007;89:1058–1069. doi: 10.1016/j.biochi.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Colasante C, Peña Diaz P, Clayton C, Voncken F. Mitochondrial carrier family inventory of Trypanosoma brucei brucei: identification, expression and subcellular localisation. Mol Biochem Parasitol. 2009;167:104–117. doi: 10.1016/j.molbiopara.2009.05.004. [DOI] [PubMed] [Google Scholar]
- 25.Schnaufer A, Clark-Walker GD, Steinberg AG, Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vaidya AB, Mather MW. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol. 2009;63:249–267. doi: 10.1146/annurev.micro.091208.073424. [DOI] [PubMed] [Google Scholar]
- 27.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum . Nature. 2007;446:88–91. doi: 10.1038/nature05572. [DOI] [PubMed] [Google Scholar]
- 28.Uyemura SA, Luo S, Moreno SN, Docampo R. Oxidative phosphorylation, Ca(2+) transport, and fatty acid-induced uncoupling in malaria parasites mitochondria. J Biol Chem. 2000;275:9709–9815. doi: 10.1074/jbc.275.13.9709. [DOI] [PubMed] [Google Scholar]
- 29.Aprille JR. Regulation of the mitochondrial adenine nucleotide pool size in liver: mechanism and metabolic role. FASEB J. 1988;2:2547–2556. doi: 10.1096/fasebj.2.10.3290024. [DOI] [PubMed] [Google Scholar]
- 30.Aprille JR. Mechanism and regulation of the mitochondrial ATP-Mg/P(i) carrier. J Bioenerg Biomembr. 1993;25:473–481. doi: 10.1007/BF01108404. [DOI] [PubMed] [Google Scholar]
- 31.del Arco A, Satrústegui J. Identification of a novel human subfamily of mitochondrial carriers with calcium-binding domains. J Biol Chem. 2004;279:24701–24713. doi: 10.1074/jbc.M401417200. [DOI] [PubMed] [Google Scholar]
- 32.Fiermonte G, De Leonardis F, Todisco S, Palmieri L, Lasorsa FM, Palmieri F. Identification of the mitochondrial ATP-Mg/Pi transporter. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem. 2004;279:30722–30730. doi: 10.1074/jbc.M400445200. [DOI] [PubMed] [Google Scholar]
- 33.Traba J, Froschauer EM, Wiesenberger G, Satrústegui J, del Arco A. Yeast mitochondria import ATP through the calcium-dependent ATP-Mg/Pi carrier Sal1p, and are ATP consumers during aerobic growth in glucose. Mol Microbiol. 2008;69:570–585. doi: 10.1111/j.1365-2958.2008.06300.x. [DOI] [PubMed] [Google Scholar]
- 34.Traba J, Satrústegui J, del Arco A. Characterization of SCaMC-3-like/slc25a41, a novel calcium-independent mitochondrial ATP-Mg/Pi carrier. Biochem J. 2009;418:125–133. doi: 10.1042/BJ20081262. [DOI] [PubMed] [Google Scholar]
- 35.Satrústegui J, Pardo B, del Arco A. Mitochondrial transporters as novel targets for intracellular calcium signaling. Physiol Rev. 2007;87:29–67. doi: 10.1152/physrev.00005.2006. [DOI] [PubMed] [Google Scholar]
- 36.Cavero S, Traba J, del Arco A, Satrústegui J. The calcium-dependent ATP-Mg/Pi mitochondrial carrier is a target of glucose-induced calcium signalling in Saccharomyces cerevisiae . Biochem J. 2005;392:537–544. doi: 10.1042/BJ20050806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laco J, Zeman I, Pevala V, Polčic P, Kolarov J. Adenine nucleotide transport via Sal1 carrier compensates for the essential function of the mitochondrial ADP/ATP carrier. FEMS Yeast Res. 2010;10:290–306. doi: 10.1111/j.1567-1364.2010.00606.x. [DOI] [PubMed] [Google Scholar]
- 38.Rehling P, Brandner K, Pfanner N. Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- 39.Lill R, Mühlenhoff U. Maturation of iron-sulfur proteins in eukaryotes: mechanisms, connected processes, and diseases. Annu Rev Biochem. 2008;77:669–700. doi: 10.1146/annurev.biochem.76.052705.162653. [DOI] [PubMed] [Google Scholar]
- 40.Drgon T, Sabova L, Nelson N, Kolarov J. ADP/ATP translocator is essential only for anaerobic growth of yeast Saccharomyces cerevisiae . FEBS Lett. 1991;289:159–162. doi: 10.1016/0014-5793(91)81059-H. [DOI] [PubMed] [Google Scholar]
- 41.Chen XJ. Sal1p, a calcium-dependent carrier protein that suppresses an essential cellular function associated with the Aac2 isoform of ADP/ATP translocase in Saccharomyces cerevisiae . Genetics. 2004;167:607–617. doi: 10.1534/genetics.103.023655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith CP, Thorsness PE. The molecular basis for relative physiological functionality of the ADP/ATP carrier isoforms in Saccharomyces cerevisiae . Genetics. 2008;179:1285–1299. doi: 10.1534/genetics.108.087700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dimitrov LN, Brem RB, Kruglyak L, Gottschling DE. Polymorphisms in multiple genes contribute to the spontaneous mitochondrial genome instability of Saccharomyces cerevisiae S288C strains. Genetics. 2009;183:365–383. doi: 10.1534/genetics.109.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kucejova B, Li L, Wang X, Giannattasio S, Chen XJ. Pleiotropic effects of the yeast Sal1 and Aac2 carriers on mitochondrial function via an activity distinct from adenine nucleotide transport. Mol Genet Genomics. 2008;280:25–39. doi: 10.1007/s00438-008-0342-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen XJ, Clark-Walker GD. The petite mutation in yeasts: 50 years on. Int Rev Cytol. 1999;194:197–238. doi: 10.1016/S0074-7696(08)62397-9. [DOI] [PubMed] [Google Scholar]
- 46.Haitina T, Lindblom J, Renstrom T, Fredriksson R. Fourteen novel human members of mitochondrial solute carrier family 25 (SLC25) widely expressed in the central nervous system. Genomics. 2006;88:779–790. doi: 10.1016/j.ygeno.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 47.del Arco A. Novel variants of human SCaMC-3, an isoform of the ATP-Mg/P(i) mitochondrial carrier, generated by alternative splicing from 3′-flanking transposable elements. Biochem J. 2005;389:647–655. doi: 10.1042/BJ20050283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colasante C, Alibu VP, Kirchberger S, Tjaden J, Clayton C, Voncken F. Characterization and developmentally regulated localization of the mitochondrial carrier protein homologue MCP6 from Trypanosoma brucei . Eukaryot Cell. 2006;5:1194–1205. doi: 10.1128/EC.00096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prohl C, Pelzer W, Diekert K, Kmita H, Bedekovics T, Kispal G, Lill R. The yeast mitochondrial carrier Leu5p and its human homologue Graves’ disease protein are required for accumulation of coenzyme A in the matrix. Mol Cell Biol. 2001;21:1089–1097. doi: 10.1128/MCB.21.4.1089-1097.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fiermonte G, Paradies E, Todisco S, Marobbio CM, Palmieri F. A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme a and adenosine 3′, 5′-diphosphate in human mitochondria. J Biol Chem. 2009;284:18152–18159. doi: 10.1074/jbc.M109.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Palmieri L, Santoro A, Carrari F, Blanco E, Nunes-Nesi A, Arrigoni R, Genchi F, Fernie AR, Palmieri F. Identification and characterization of ADNT1, a novel mitochondrial adenine nucleotide transporter from Arabidopsis . Plant Physiol. 2008;148:1797–1808. doi: 10.1104/pp.108.130310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts J, Aubert S, Gout E, Bligny R, Douce R. Cooperation and competition between adenylate kinase, nucleoside diphosphokinase, electron transport and ATP synthase in plant mitochondria studied by 31P-nuclear magnetic resonance. Plant Physiol. 1997;113:191–199. doi: 10.1104/pp.113.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monné M, Robinson AJ, Boes C, Harbour ME, Fearnley IM, Kunji ER. The mimivirus genome encodes a mitochondrial carrier that transports dATP and dTTP. J Virol. 2007;81:3181–3186. doi: 10.1128/JVI.02386-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tzagoloff A, Jang J, Glerum DM, Wu M. FLX1 codes for a carrier protein involved in maintaining a proper balance of flavin nucleotides in yeast mitochondria. J Biol Chem. 1996;271:7392–7397. doi: 10.1074/jbc.271.13.7392. [DOI] [PubMed] [Google Scholar]
- 55.Spaan AN, Ijlst L, van Roermund CW, Wijburg FA, Wanders RJ, Waterham HR. Identification of the human mitochondrial FAD transporter and its potential role in multiple acyl-CoA dehydrogenase deficiency. Mol Genet Metab. 2005;86:441–447. doi: 10.1016/j.ymgme.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 56.Bedhomme M, Hoffmann M, McCarthy EA, Gambonnet B, Moran RG, Rébeillé F, Ravanel S. Folate metabolism in plants: an Arabidopsis homolog of the mammalian mitochondrial folate transporter mediates folate import into chloroplasts. J Biol Chem. 2005;280:34823–34831. doi: 10.1074/jbc.M506045200. [DOI] [PubMed] [Google Scholar]
- 57.Todisco S, Agrimi G, Castegna A, Palmieri F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae . J Biol Chem. 2006;281:1524–1531. doi: 10.1074/jbc.M510425200. [DOI] [PubMed] [Google Scholar]
- 58.Palmieri F, Rieder B, Ventrella A, Blanco E, Do PT, Nunes-Nesi A, Trauth AU, Fiermonte G, Tjaden J, Agrimi G, Kirchberger S, Paradies E, Fernie AR, Neuhaus HE. Molecular identification and functional characterization of Arabidopsis thaliana mitochondrial and chloroplastic NAD+ carrier proteins. J Biol Chem. 2009;284:31249–31259. doi: 10.1074/jbc.M109.041830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marobbio CM, Di Noia MA, Palmieri F. Identification of a mitochondrial transporter for pyrimidine nucleotides in Saccharomyces cerevisiae: bacterial expression, reconstitution and functional characterization. Biochem J. 2006;393:441–446. doi: 10.1042/BJ20051284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Floyd S, Favre C, Lasorsa FM, Leahy M, Trigiante G, Stroebel P, Marx A, Loughran G, O’Callaghan K, Marobbio CM, Slotboom DJ, Kunji ER, Palmieri F, O’Connor R. The insulin-like growth factor-I-mTOR signaling pathway induces the mitochondrial pyrimidine nucleotide carrier to promote cell growth. Mol Biol Cell. 2007;18:3545–5355. doi: 10.1091/mbc.E06-12-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marobbio CM, Vozza A, Harding M, Bisaccia F, Palmieri F, Walker JE. Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J. 2002;21:5653–5661. doi: 10.1093/emboj/cdf583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lindhurst MJ, Fiermonte G, Song S, Struys E, De Leonardis F, Schwartzberg PL, Chen A, Castegna A, Verhoeven N, Mathews CK, Palmieri F, Biesecker LG. Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc Natl Acad Sci USA. 2006;103:15927–15932. doi: 10.1073/pnas.0607661103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marobbio CM, Agrimi G, Lasorsa FM, Palmieri F. Identification and functional reconstitution of yeast mitochondrial carrier for S-adenosylmethionine. EMBO J. 2003;22:5975–5982. doi: 10.1093/emboj/cdg574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agrimi G, Di Noia MA, Marobbio CM, Fiermonte G, Lasorsa FM, Palmieri F. Identification of the human mitochondrial S-adenosylmethionine transporter: bacterial expression, reconstitution, functional characterization and tissue distribution. Biochem J. 2004;379:183–190. doi: 10.1042/BJ20031664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Palmieri L, Arrigoni R, Blanco E, Carrari F, Zanor MI, Studart-Guimaraes C, Fernie AR, Palmieri F. Molecular identification of an Arabidopsis S-adenosylmethionine transporter. Analysis of organ distribution, bacterial expression, reconstitution into liposomes, and functional characterization. Plant Physiol. 2006;142:855–865. doi: 10.1104/pp.106.086975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2010;365:713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Danovaro R, Dell’anno A, Pusceddu A, Gambi C, Heiner I, Kristensen RM. The first metazoa living in permanently anoxic conditions. BMC Biol. 2010;8:30. doi: 10.1186/1741-7007-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Voncken F, Boxma B, Tjaden J, Akhmanova A, Huynen M, Verbeek F, Tielens AG, Haferkamp I, Neuhaus HE, Vogels G, Veenhuis M, Hackstein JH. Multiple origins of hydrogenosomes: functional and phylogenetic evidence from the ADP/ATP carrier of the anaerobic chytrid Neocallimastix sp. Mol Microbiol. 2002;44:1441–1454. doi: 10.1046/j.1365-2958.2002.02959.x. [DOI] [PubMed] [Google Scholar]
- 69.Boxma B, de Graaf RM, van der Staay GW, van Alen TA, Ricard G, Gabaldón T, van Hoek AH, Moon-van der Staay SY, Koopman WJ, van Hellemond JJ, Tielens AG, Friedrich T, Veenhuis M, Huynen MA, Hackstein JH. An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–79. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- 70.van der Giezen M, Slotboom DJ, Horner DS, Dyal PL, Harding M, Xue GP, Embley TM, Kunji ER. Conserved properties of hydrogenosomal and mitochondrial ADP/ATP carriers: a common origin for both organelles. EMBO J. 2002;21:572–579. doi: 10.1093/emboj/21.4.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stechmann A, Hamblin K, Pérez-Brocal V, Gaston D, Richmond GS, van der Giezen M, Clark CG, Roger AJ. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–585. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perez-Brocal V, Clark CG. Analysis of two genomes from the mitochondrion-like organelle of the intestinal parasite Blastocystis: complete sequences, gene content, and genome organization. Mol Biol Evol. 2008;25:2475–2482. doi: 10.1093/molbev/msn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tjaden J, Haferkamp I, Boxma B, Tielens AG, Huynen M, Hackstein JH. A divergent ADP/ATP carrier in the hydrogenosomes of Trichomonas gallinae argues for an independent origin of these organelles. Mol Microbiol. 2004;51:1439–1446. doi: 10.1111/j.1365-2958.2004.03918.x. [DOI] [PubMed] [Google Scholar]
- 74.Hrdý I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Embley TM. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- 75.Dolezal P, Dagley MJ, Kono M, Wolynec P, Likić VA, Foo JH, Sedinová M, Tachezy J, Bachmann A, Bruchhaus I, Lithgow T. The essentials of protein import in the degenerate mitochondrion of Entamoeba histolytica . PLoS Pathog. 2010;6:e1000812. doi: 10.1371/journal.ppat.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan KW, Slotboom DJ, Cox S, Embley TM, Fabre O, van der Giezen M, Harding M, Horner DS, Kunji ER, León-Avila G, Tovar J. A novel ADP/ATP transporter in the mitosome of the microaerophilic human parasite Entamoeba histolytica . Curr Biol. 2005;15:737–742. doi: 10.1016/j.cub.2005.02.068. [DOI] [PubMed] [Google Scholar]
- 77.Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, Henze K, Tovar J. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell Microbiol. 2010;12:331–342. doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- 78.Hampl V, Silberman JD, Stechmann A, Diaz-Triviño S, Johnson PJ, Roger AJ. Genetic evidence for a mitochondriate ancestry in the ‘amitochondriate’ flagellate Trimastix pyriformis . PLoS ONE. 2008;3:e1383. doi: 10.1371/journal.pone.0001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Graaf RM, Duarte I, van Alen TA, Kuiper JW, Schotanus K, Rosenberg J, Huynen MA, Hackstein JH. The hydrogenosomes of Psalteriomonas lanterna . BMC Evol Biol. 2009;9:287. doi: 10.1186/1471-2148-9-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Williams BA, Haferkamp I, Keeling PJ. An ADP/ATP-specific mitochondrial carrier protein in the microsporidian Antonospora locustae . J Mol Biol. 2008;375:1249–1257. doi: 10.1016/j.jmb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 81.Tsaousis AD, Kunji ER, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi . Nature. 2008;453:553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- 82.Möhlmann T, Tjaden J, Schwöppe C, Winkler HH, Kampfenkel K, Neuhaus HE. Occurrence of two plastidic ATP/ADP transporters in Arabidopsis thaliana L.—molecular characterisation and comparative structural analysis of similar ATP/ADP translocators from plastids and Rickettsia prowazekii . Eur J Biochem. 1998;252:353–359. doi: 10.1046/j.1432-1327.1998.2520353.x. [DOI] [PubMed] [Google Scholar]
- 83.Trentmann O, Jung B, Neuhaus HE, Haferkamp I. Nonmitochondrial ATP/ADP transporters accept phosphate as third substrate. J Biol Chem. 2008;283:36486–36493. doi: 10.1074/jbc.M806903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reinhold T, Alawady A, Grimm B, Beran KC, Jahns P, Conrath U, Bauer J, Reiser J, Melzer M, Jeblick W, Neuhaus HE. Limitation of nocturnal import of ATP into Arabidopsis chloroplasts leads to photooxidative damage. Plant J. 2007;50:293–304. doi: 10.1111/j.1365-313X.2007.03049.x. [DOI] [PubMed] [Google Scholar]
- 85.Thuswaldner S, Lagerstedt JO, Rojas-Stütz M, Bouhidel K, Der C, Leborgne-Castel N, Mishra A, Marty F, Schoefs B, Adamska I, Persson BL, Spetea C. Identification, expression, and functional analyses of a thylakoid ATP/ADP carrier from Arabidopsis . J Biol Chem. 2007;282:8848–8859. doi: 10.1074/jbc.M609130200. [DOI] [PubMed] [Google Scholar]
- 86.Yin L, Lundin B, Bertrand M, Nurmi M, Solymosi K, Kangasjärvi S, Aro EM, Schoefs B, Spetea C. Role of thylakoid ATP/ADP carrier in photoinhibition and photoprotection of photosystem II in Arabidopsis . Plant Physiol. 2010;153:666–677. doi: 10.1104/pp.110.155804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kirchberger S, Tjaden J, Neuhaus HE. Characterization of the Arabidopsis Brittle1 transport protein and impact of reduced activity on plant metabolism. Plant J. 2008;56:51–63. doi: 10.1111/j.1365-313X.2008.03583.x. [DOI] [PubMed] [Google Scholar]
- 88.Leroch M, Kirchberger S, Haferkamp I, Wahl M, Neuhaus HE, Tjaden J. Identification and characterization of a novel plastidic adenine nucleotide uniporter from Solanum tuberosum . J Biol Chem. 2005;280:17992–18000. doi: 10.1074/jbc.M412462200. [DOI] [PubMed] [Google Scholar]
- 89.Kirchberger S, Leroch M, Huynen MA, Wahl M, Neuhaus HE, Tjaden J. Molecular and biochemical analysis of the plastidic ADP-glucose transporter (ZmBT1) from Zea mays . J Biol Chem. 2007;282:22481–22491. doi: 10.1074/jbc.M702484200. [DOI] [PubMed] [Google Scholar]
- 90.Lasorsa FM, Scarcia P, Erdmann R, Palmieri F, Rottensteiner H, Palmieri L. The yeast peroxisomal adenine nucleotide transporter: characterization of two transport modes and involvement in DeltapH formation across peroxisomal membranes. Biochem J. 2004;381:581–585. doi: 10.1042/BJ20040856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Palmieri L, Rottensteiner H, Girzalsky W, Scarcia P, Palmieri F, Erdmann R. Identification and functional reconstitution of the yeast peroxisomal adenine nucleotide transporter. EMBO J. 2001;20:5049–5059. doi: 10.1093/emboj/20.18.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Visser WF, van Roermund CW, Waterham HR, Wanders RJ. Identification of human PMP34 as a peroxisomal ATP transporter. Biochem Biophys Res Commun. 2002;299:494–497. doi: 10.1016/S0006-291X(02)02663-3. [DOI] [PubMed] [Google Scholar]
- 93.Linka N, Theodoulou FL, Haslam RP, Linka M, Napier JA, Neuhaus HE, Weber AP. Peroxisomal ATP import is essential for seedling development in Arabidopsis thaliana . Plant Cell. 2008;20:3241–3257. doi: 10.1105/tpc.108.062042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Michels PA, Bringaud F, Herman M, Hannaert V. Metabolic functions of glycosomes in trypanosomatids. Biochim Biophys Acta. 2006;1763:1463–1477. doi: 10.1016/j.bbamcr.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 95.Leroch M, Neuhaus HE, Kirchberger S, Zimmermann S, Melzer M, Gerhold J, Tjaden J. Identification of a novel adenine nucleotide transporter in the endoplasmic reticulum of Arabidopsis . Plant Cell. 2008;20:438–451. doi: 10.1105/tpc.107.057554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sawada K, Echigo N, Juge N, Miyaji T, Otsuka M, Omote H, Yamamoto A, Moriyama Y. Identification of a vesicular nucleotide transporter. Proc Natl Acad Sci USA. 2008;105:5683–5686. doi: 10.1073/pnas.0800141105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Miyaji T, Echigo N, Hiasa M, Senoh S, Omote H, Moriyama Y. Identification of a vesicular aspartate transporter. Proc Natl Acad Sci USA. 2008;105:11720–11724. doi: 10.1073/pnas.0804015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.