Abstract
Several recent publications have described examples of physical and functional interations between tetraspanins and specific membrane proteases belonging to the TM-MMP and α-(ADAMs) and γ-secretases families. Collectively, these examples constitute an emerging body of evidence supporting the notion that tetraspanin-enriched microdomains (TEMs) represent functional platforms for the regulation of key cellular processes including the release of surface protein ectodomains ("shedding"), regulated intramembrane proteolysis ("RIPing") and matrix degradation and assembly. These cellular processes in turn play a crucial role in an array of physiological and pathological phenomena. Thus, TEMs may represent new therapeutical targets that may simultaneously affect the proteolytic activity of different enzymes and their substrates. Agonistic or antagonistic antibodies and blocking soluble peptides corresponding to tetraspanin functional regions may offer new opportunities in the treatment of pathologies such as chronic inflammation, cancer, or Alzheimer's disease. In this review article, we will discuss all these aspects of functional regulation of protease activities by tetraspanins.
Keywords: Tetraspanins, MMP, ADAMS, α-secretases, Shedding, RIP
Introduction
Tetraspanins are a large family of small proteins spanning four times the cellular membranes, forming a small and a large extracellular loop (termed SEL or EC1 and LEL or EC2, respectively) and leaving C- and N-terminal intracellular tails. In addition, tetraspanins share a number of structural features, including 4–8 conserved extracellular cysteines (two of them in the characteristic CCG motif found in the mid-region of the LEL domain in all tetraspanins) that are engaged in the formation of 2–4 disulfide bonds that maintain the three-dimensional conformation of the LEL. These proteins are widely expressed in multicellular eukaryotic organisms ranging from plants to fungi, sponges, worms, insects, and mammals. A striking feature of tetraspanins is their tendency to associate with other membrane proteins, including other tetraspanins, different members of the integrin and immunoglobulin superfamilies, growth factor receptors, and GPCRs. Tetraspanins also associate with lipids, such as cholesterol and gangliosides, and with cytosolic signaling and cytoskeletal proteins giving rise to the formation of higher-order lipid–protein membrane complexes termed TEMs (tetraspanin-enriched microdomains) which work as specialized organizational units. It should be noted that although TEMs share some properties with other types of membrane microdomains, lipid-rafts, they are physically and functionally distinct entities. Through their role in the organization and dynamics of TEMs, tetraspanins are involved in a multitude of physiological and pathological processes such as parasite and viral infections, fertilization, maintenance of epithelial integrity, formation of neuromuscular junctions, induction of immune responses, platelet aggregation, and tumor progression. Excellent reviews covering the general structural and functional aspects of tetraspanins have been published in recent years and readers are referred to them for reference on the biology of these proteins [1–4].
Several recent reports have described examples of physical association and functional interplay between tetraspanins and specific membrane proteases supporting the notion that TEMs represent functional platforms for the regulation of cellular processes including the release of surface protein ectodomains ("shedding"), regulated intramembrane proteolysis ("RIPing") and matrix degradation and assembly. In the present review, we will discuss these examples of functional regulation of protease activities by tetraspanins.
Functional regulation of α-secretases (sheddases ADAM10 and ADAM17) by tetraspanins
Protein "shedding" is a term coined to refer to the proteolytic release of the ectodomains of transmembrane proteins, and represents an important mechanism that endows cells with the capacity to rapidly change their surface phenotype, generating at the same time soluble mediators with the potential to signal and influence cell behavior in an autocrine, juxtacrine, paracrine, or even endocrine manner. Accordingly, ectodomain shedding plays a key regulatory role in cellular processes such as adhesion, migration, invasion, cell fusion, proliferation, and signaling, which in turn have a strong repercussion in different biological phenomena, including development, fertilization, angiogenesis, leukocyte extravasation and inflammatory responses, tumor cell growth and dissemination, and brain pathology. Members of the ADAM (A Disintegrin And Metalloproteinase) family of metalloproteinases are key "sheddases", i.e. enzymes responsible for the shedding of a great variety of transmembrane proteins (see [5–8] for excellent reviews on ADAMs).
Two closely related members of the ADAM family, ADAM10 (also known as Kuzbanian from its ortholog gene -kuz- in Drosophila) and ADAM17 (also termed TACE: TNF-alpha converting enzyme), are key sheddases responsible for the release of the ectodomains of a multitude of protein substrates [7, 9] including growth factors, cytokines, receptors, adhesion proteins, and the amyloid precursor protein. A large proportion of these substrates are shared between ADAM10 and ADAM17, which leads to the assumption that the activities of these two metalloproteinases can be sometimes complementary or redundant [5, 7, 9]. No consensus sequences for specific ADAM10 or ADAM17 proteolytic cleavage have been identified in their substrates and it seems that the main common feature of this type of cleavage is that it occurs at a juxtamembranal extracellular location. Both ADAM10 and ADAM17 play a major role in the proteolytic release of specific EGFR ligands regulating the subsequent EGFR-dependent signaling, which is of great relevance in development and cancer (reviewed in [6]). In this regard, while ADAM10 is involved in the shedding of EGF and betacellulin, ADAM17 is largely responsible for cleavage of TGFα, HB-EGF, epiregulin, and amphiregulin. Adam17-/- mice display developmental defects similar to those observed in TGFα, HB-EGF, amphiregulin, or EGFR-deficient animals [10, 11], clearly evidencing the in vivo relevance of ADAM17 in EGFR signaling.
It is of note that ADAM10 and/or ADAM17-mediated release of specific EFGR ligands has been observed to mediate transactivation of EGFR signaling after activation of a variety of GPCRs by agonists like thrombin, angiotensin-II, or endothelin-1 through a cross-talk mechanism termed TMPS ("triple membrane-passing signaling") [5, 12–15]. The broad relevance of TMPS has been demonstrated for various families of GPCRs and found to have implications in medically relevant processes like cardiac hypertrophy, mucus production in lungs, and the migration, survival, proliferation, and invasion of different types of cancer cells [13].
The large variety of ADAM10 and ADAM17 substrates, and their potential physiological and pathological relevance strongly suggest that the proteolytic activity of these enzymes must be subject to fine mechanisms of control. It is currently accepted that in many cell types, the constitutive or basal ectodomain release of most substrates, as well as that induced by calcium ionophores, is carried out by ADAM10, while that induced after short-term stimulation with phorbol esters is mostly dependent on ADAM17 [16]. Different reports have shown that the cytoplasmic domain of ADAM17 is phosphorylated upon cell treatment with PMA [17–19] but, on the other hand, a truncated form of ADAM17 lacking the intracytoplasmic domain is still activated by phorbol esters [7, 11]. Some studies have shown that phorbol ester stimulation increases the intracellular processing and maturation of ADAM17 by furin as well as its transport to the cell surface [19, 20] while other studies indicate that increased transport to the cell surface is not associated with enhanced ADAM17 activation [16]. The mechanisms by which GPCRs induce the activation of ADAM10 and/or ADAM17 in the TMPS process remain equally obscure for the moment [13, 14]. Interaction with specific regulatory proteins involved in cell signaling and substrate accessibility control have been proposed as modulatory mechanisms of ADAM activity (reviewed in [21]).
In this regard, several studies have identified specific members of the tetraspanin family as modulators of ADAM activities. Through controlling the inclusion in––and/or the exclusion from––discrete TEMs of both ADAMs and their protein substrates, tetraspanins may act as important controllers of ectodomain shedding, regulating both the accessibility of substrates and their effective cleavage rates.
One of the first studies suggesting a functional association between a tetraspanin and an ADAM sheddase was published by Yan et al. [15] who demonstrated that stimulation of GPCRs with bombesin caused transactivation of EGFR via the ADAM10/Kuzbanian-dependent release of HB-EGF ligand, representing a typical example of TMPS as described above. Interestingly, ectopically expressed ADAM10 was shown to interact with tetraspanin CD9 and the extent of this association was increased after bombesin stimulation. Furthermore, the potency of HB-EGF in stimulating cell growth correlated with its association to CD9. The authors concluded that GPCR regulates ADAM10-dependent activation of HB-EGF by promoting the binding of ADAM10 to molecular complexes centered on CD9.
More recently, Arduise et al. [22] reported that ADAM10 is associated within the context of TEMs with several tetraspanins, including CD9, CD81, and CD82. Specific antibodies directed to these tetraspanins rapidly stimulated the ADAM10-mediated release of TNF-α and EGF, without altering ADAM10-tetraspanin interactions. These authors found that the stimulatory effects of anti-tetraspanin antibodies did neither require tyrosine phosphorylation nor PKC signaling, but in contrast, an intact MEK/Erk pathway was required. Tetraspanins could also directly regulate ADAM10 activity via their association at the membrane, since the stimulatory effect of anti-tetraspanin antibodies correlated with their ability to induce the redistribution of ADAM10 into patches enriched in tetraspanin molecules. The authors proposed that tetraspanins exerted a negative regulation on ADAM10 sheddase activity so that the local increase in ADAM10 concentration would overcome this inhibitory effect.
Thereafter, ADAM10 was found to associate with a different tetraspanin, namely TSPAN12 [23]. Authors employed a mass-spectrometry approach to search for TSPAN12-associated proteins and identified peptides corresponding to both ADAM10 and ADAM17 sheddases, but only the association of ADAM10 with TSPAN12 was further confirmed by reciprocal co-immunoprecipitation in different tumor cell lines. The large extracellular loop, as well as the intracellular C-terminal domain and the palmitoylation sites, were identified as the three regions of TSPAN12 involved in the association with ADAM10. Using complementary molecular approaches, such as overexpression, RNA silencing, and mutation, the functional relevance of TSPAN12/ADAM10 complexes in the processing of the amyloid precursor protein (APP) was evidenced. These authors postulated that association with TSPAN12 lead to the stabilization of active ADAM10 on the cell surface and/or to the acceleration of ADAM10 activation by prodomain convertases. As a result of this enhanced maturation/activation of ADAM10, the α-secretase activity was increased.
In the studies by Arduise et al. and Xu et al., the authors did not observe an association of tetraspanins with the second relevant sheddase ADAM17. A possible explanation is that, on the cells employed, ADAM17 was expressed at a much lower level compared to ADAM10, making it difficult to reveal the interaction. Direct association of the tetraspanin CD9 with ADAM17 on the surface of leukocytes and endothelial cells has been recently described by our group [24]. This interaction was established using a combination of imaging and biochemical approaches based on co-localization, in situ proximity ligation assays, co-immunoprecipitation, cross linking, and pull-down experiments. Treatment of cells with agonistic anti-CD9 antibodies and neoexpression and silencing of this tetraspanin, clearly demonstrated that CD9 regulates negatively the ADAM17-mediated shedding of TNF-α and ICAM-1. This negative modulation of ADAM17 by CD9-specific antibodies was in contrast with the results of Arduise et al. [22], who found that treatment of cells expressing both ADAM10 and ADAM17 with anti-CD9 antibodies, in the absence of phorbol esters, increased the ADAM10-mediated release of TNF-α. We compared side by side the effects of a group of CD9-specific mAbs on the constitutive (non-stimulated, ADAM10-mediated) and phorbol ester-stimulated (ADAM17-mediated) release of TNF-α from CD9-expressing Raji B cells, and confirmed that they exerted a positive effect on ADAM10 proteolytic activity while inhibiting that of ADAM17. These results provide an excellent example indicating that, although many of the substrate proteins may be shared by ADAM10 and ADAM17, the association of these metalloproteinases on the cell surface with specific tetraspanins like CD9 within the context of TEMs, may represent an important regulatory mechanism for the selective and even opposing regulation of their individual activities (Fig. 1).
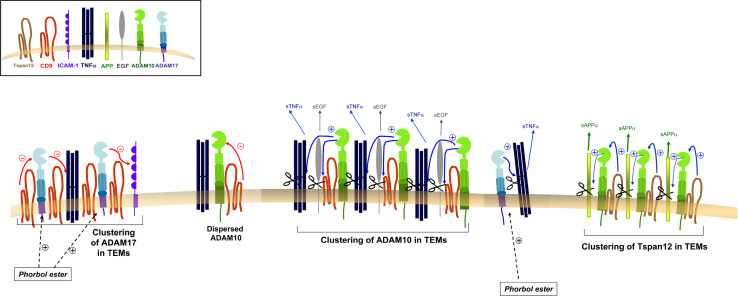
Fig. 1.
Functional regulation of sheddases ADAM10 and ADAM17 by tetraspanins. A fraction of ADAM10 is constitutively associated with tetraspanin CD9 (and also with tetraspanins CD81, CD82) in dispersed complexes on the cell surface in which tetraspanins exert an inhibitory effect on the sheddase activity of ADAM10 against transmembrane substrates TNFα and EGF. Upon antibody engagement of tetraspanins, associated ADAM10 molecules and substrates are recruited and clustered into TEMs and this increase in their local concentration overcomes the inhibitory effect of tetraspanins on ADAM10 activity. Similarly, overexpression of TSPAN12 favors the recruitment of ADAM10 into TEMs and stimulates its α-secretase activity against the APP substrate. The sheddase activity of dispersed ADAM17 is stimulated by phorbol esters resulting in the release of the ectodomains of its substrates TNFα and ICAM-1. Recruitment of ADAM17 into CD9-centered TEMs is induced by overexpression or antibody engagement of this tetraspanin, which exerts negative regulation on the ADAM17 sheddase activity, resulting in reduced ectodomain release of TNFα and ICAM-1
Although protein ectodomain shedding has been long assumed to take place only at the cell surface, several studies have shown that this might not be the case, as active ADAM10 and ADAM17 have been found in exosomes from different sources, including tumor cells [25–28]. Exosomes are 40–100 nm vesicles that derive from multivesicular bodies and are released by many cells upon fusion of these endocytic organelles with the plasma membrane. In vivo, exosomes can be detected in a variety of body fluids, including blood, urine, ascites, and amniotic fluid [29, 30]. Exosomes seem to work as an important mechanism of intercellular communication as vehicles for the transference of subsets of proteins, mRNA and microRNAs between exosome-producing (donor) and exosome-receiving (target) cells. This type of intercellular communication has revealed its importance in processes like the spreading of infectious agents, immune responses and tumor progression [31]. Tetraspanins are not only expressed on the plasma membrane of cells but are also present in various types of intracellular vesicles of the endocytic pathway and particularly in exosomes. In fact, different tetraspanins (CD9, CD63, CD81, and CD82) are enriched in the membrane of exosomes and are often used as the best markers for these extracellular vesicles. Constitutive and stimulus-induced cleavage of the ectodomains of TNF-α or of the adhesion molecules L1 and CD44 by ADAM10 and ADAM17 in exosomes released by ovarian cancer or melanoma cells has been demonstrated, suggesting that these vesicles may also act as vehicles for cellular export of soluble molecules. In fact, the uptake by T lymphocytes of melanoma-derived exosomes containing cleaved TNF-α and ADAM17 has been shown to induce oxidative stress in the target cells and could be responsible for the melanoma-induced inhibition of effector T cell function [28]. Thus, exosomal TEMs may represent important alternative platforms for controlling the efficiency of cleavage and the targeted delivery of a variety of biologically important soluble ectodomains, with relevance in cancer progression and immune system regulation.
Relation of tetraspanins with other members of the ADAM family
ADAMs are modular transmembrane proteins consisting of an N-terminal prodomain, followed by metalloprotease, disintegrin, cysteine-rich, EGF, transmembrane, and C-terminal cytosolic domains. However, of the 21 ADAM members identified in the human genome, only 13 have an intact metalloprotease domain including the HEXXH consensus motif required for catalytic activity [21]. Functional associations of tetraspanins with other members of the ADAM family, namely ADAM1 (fertilin α) and ADAM2 (fertilin β), have been described. However, in this case, the functional effects resulting from these interactions cannot be ascribed to alterations in proteolytic activity since ADAM1 and ADAM2 do not belong to the group of proper sheddases endowed with proteolytic activity [5].
Blocking anti-CD9 mAbs were shown to inhibit the sperm-egg binding and fusion [32]. CD9 was thought to affect the binding of α6β1 integrin on the oocyte to ADAM-2 on the sperm surface [33, 34] although thereafter this integrin was found not to be essential for sperm-egg binding and fusion [35]. Nevertheless, the CD9 and CD81 knock-out female mice showed that these tetraspanins are indeed required for the specific step of sperm-egg membrane fusion [36–41]. More recently, CD9 was demonstrated to act as a receptor for PSG17 (pregnancy-specific glycoprotein 17) [42], opening the possibility of CD9 acting as a receptor for some member of this family expressed on the sperm [43]. Moreover, the oocyte was shown to release CD9-enriched vesicles, which are incorporated to the membrane of sperm cell and could act as fusogenic factors [44], although these results could not be reproduced in a different laboratory [45].
Tetraspanins and γ-secretases
The proteolytic cascade termed "regulated intramembrane proteolysis" (RIP) controls the processing and subsequent intracellular signaling mediated by several transmembrane proteins. The main characteristic of this process is that the cleavage of the substrates is performed intramembranously by a group of proteases called I-CLiPs, which generate intracellular domain fragments (ICD) that in most cases are intermediates destined for lysosome or proteasome-mediated degradation, but for an increasing number of "RIPed" proteins can also translocate to the nucleus and regulate transcriptional activation of target genes (see [5–7] for reviews). Several physiological processes are regulated by RIP including cholesterol homeostasis, immune surveillance, cellular signaling, and amyloid formation in Alzheimer’s disease. An increasing number of proteins have been identified as substrates for RIP including the amyloid precursor protein (APP), whose cleavage generates amyloid β peptides (Aβ), a major component of senile neuritic extracellular plaques deposited in the brains of Alzheimer’s disease (AD) patients [46], which is the most common cause of dementia in aging populations; the adhesion molecules CD44, N-cadherin, E-cadherin and L1, as well as members of the EGFR (m80-ErbB-4) family, the p75 neurotrophin receptor (p75NTR), and Notch receptors and their ligands (Delta, Jagged).
There are four known families of I-CLiP proteases: (1) the site-2 protease (S2P); (2) the aspartyl proteases γ-secretase complex, also called presenilin complex, involved in the cleavage of truncated type I transmembrane proteins; (3) the signal peptide peptidase (SPP) acting in this case on truncated type II transmembrane proteins, and (4) the rhomboid serine proteases. Although all of them share the feature of the intramembrane proteolytic mechanism, the cleavage of substrates by the SPP metalloproteases and the γ-secretase families needs to be preceded by an "ectodomain" shedding step carried out by the α or β-secretases. ADAM10/Kuzbanian and ADAM17/TACE are the main sheddases responsible for the α-secretase activity [47–50]. The γ-secretase activity is absolutely dependent on the assembly of a multimeric complex including the catalytic subunit presenilin and nicastrin (NCT), Aph-1, Pen-2 membrane proteins [51].
In a recent paper, Wakabayashi et al. suggested that additional factors could be involved the regulation of the activity of the γ-secretase complex [52]. Using a proteomic approach, they found several different proteins interacting with presenilin1 and presenilin2. Biochemical association was found with the proteins of the tetraspanin family CD81, CD9, and UpK1b as well as with the tetraspanin-associated proteins EWI-2, EWI-F, integrins, and CD98. Knockdown of CD81, EWI-F, or CD98 had no effect on the expression of the γ-secretase components but significantly inhibited γ-secretase activity with a concomitant decrease in the release of the amyloid β-peptide (Aβ), while overexpression of the tetraspanin-associated proteins enhanced Aβ secretion. On the contrary, tetraspanin CD9 overexpression lead to a significant decrease in the Aβ generation. The functional relevance of the interaction between the γ-secretase complex and proteins from the tetraspanin-enriched microdomains was also demonstrated using cells derived from CD81- and CD9-knockout mice where the γ-secretase activity was significantly impaired. For some cellular processes, treatment with monoclonal antibodies against tetraspanin molecules has been shown to have either an agonist or antagonist activity [22, 24, 53]. In this study, γ-secretase activity was also a target for modulation using antibodies against the CD81 and CD9 tetraspanins, which resulted in a disruption, more evident for CD9, of the Aβ generation, thus implicating TEMs in the endogenous regulation of γ-secretase [52].
Another well-known substrate of the RIP process is the Notch receptor, a critical component of signaling mechanisms that regulates diverse processes during development such as cell fate specification and differentiation, neurogenesis, neuritic growth and neural stem cell maintenance, synaptic plasticity and long-term memory, which have been related to cognitive function in the adult brain [54–57]. Notch has also been postulated to be involved in Alzheimer's disease and an increase of its levels in patients suffering AD has been described [58]. Interaction of this receptor at the cell surface with its ligands on adjacent cells triggers a proteolytic sequence leading to enhanced γ-secretase activity and the generation of Notch intracellular domain (NICD) as a final product that is able to translocate to the nucleus modulating the transcription of different target genes. In this sequence, the initial cleavage has been shown to be performed by ADAM10 and/or ADAM 17 in in vitro and in vivo models [47, 48].
A recent study using different approaches such as the Caenorhabditis elegans genetic model and human cellular cultures reports that the worm tetraspanin TSP-12-and its human ortholog TSPAN33-facilitate Notch signaling by enhancing γ-secretase activity so that the reduction (by small interfering RNA) or loss (a null allele) of these tetraspanins suppress the effects of constitutively active mutant Notch proteins and the signaling mediated by a transmembrane form of activated Notch that requires γ-secretase activity. These results support the conclusion that these tetraspanins promote γ-secretase processing of Notch rather than promoting other signaling events occurring downstream of γ-secretase cleavage [59]. In this report, TSP-12 and TSP-14 were found to have redundant functions in C. elegans, while TSPAN33 and TSPAN5 synergistically regulated Notch in human cancers. In contrast, CD81 or CD9 tetraspanins did not seem to play a role in this context.
Tetraspanins and matrix metalloproteinases
Many tetraspanins were initially described as molecules induced in tumors that regulate the capacity of tumor cells to migrate and metastasize. Much of the evidence for this came from survival studies in cancer patients and the ability of tetraspanin-specific antibodies to reduce tumor metastases in animal models [1, 2, 60, 61]. Tetraspanins might also influence not only invasion but also matrix deposition [62] and degradation. Supporting this, some phenotypes of tetraspanin-deficient mice, such as the disorganized basement membrane in the renal glomeruli [63] and skin [64] of CD151-KO mice, are consistent with a defect in extracellular matrix organization.
The first evidence that related tetraspanins to matrix degradation came from the study by Sugiura and Berditchevski that demonstrated that antibody-mediated crosslinking of several tetraspanins induced MMP-2 secretion in a PI3 K-dependent manner [65]. Thereafter, crosslinking of CD81 by the hepatitis C virus E2 glycoprotein was shown to upregulate MMP2 in hepatoma cell lines [66]. CD151 overexpression induces MMP-9 expression in human melanoma cells [67] and in hepatocellular carcinoma, promoting neoangiogenesis [68]. CD9 expression induces MMP-2 in melanoma [69], while its depletion augments MMP2 secretion in small cell lung cancer cells [70] and mouse blastocysts [71]. Overexpression of CD81 or CD82 in several myeloma cells reduces MMP2 expression [72].
In addition to regulating the gene expression of some soluble metalloproteinases, tetraspanins may also anchor them to the plasma membrane or may modulate their activity through association with regulatory molecules. Thus, CD151 has been shown to bind MMP7 [73], while CD63 interacts with the soluble metalloprotease inhibitor TIMP-1 [74].
Tetraspanins can also directly associate with the membrane-anchored metalloproteinase MT1-MMP/MMP14 in different cell types [75–77]. MT1-MMP is a fundamental protease for matrix proteolysis during cancer invasion, angiogenesis, and bone function [78]. MT1-MMP can itself proteolyse several components of the extracellular matrix and, in addition, it is the most common activator of pro-MMP2. Mice deficient for MT1-MMP [79], MMP2 [80], or both [81] demonstrate a genetic synergism of both enzymes, since the double knock-out animals display the most severe phenotype and die immediately after birth with respiratory failure, abnormal blood vessels and immature muscle fibers [81].
Maturation of MT1-MMP takes place intracellularly, so that MT1-MMP activity at the plasma membrane needs to be finely regulated by a strict control of its internalization rate, membrane compartmentalization and subcellular localization. MT1-MMP can be internalized by both clathrin-coated vesicles or incorporated into caveolae [82, 83]. Thereafter, it may recycle to the plasma membrane through a Rab8 compartment [84]. Overexpression of tetraspanin CD63, increased MT1-MMP degradation at lysosomes [76]. This effect was also observed in tumor cells upon tetraspanin CD81, CD9, and TSPAN12 gene silencing [75].
At the plasma membrane, MT1-MMP regulation comprises autocatalytic events, inhibition by TIMP-2, and interactions with other plasma membrane molecules, such as CD44, that dictate its subcellular localization or substrate accessibility [78]. In endothelial cells, MT1-MMP was found to be directly associated with CD151, as demonstrated by co-immunoprecipitation and FRET experiments [77]. Insertion of MT1-MMP into TEMs was shown to be crucial for its association and functional coordination with integrins. Hence, endothelial cells depleted of CD151 or derived from CD151-deficient mouse presented aberrant collagenolysis [77] because of disruption of the ternary CD151/MT1-MMP/alpha3beta1 integrin complex (Fig. 2).
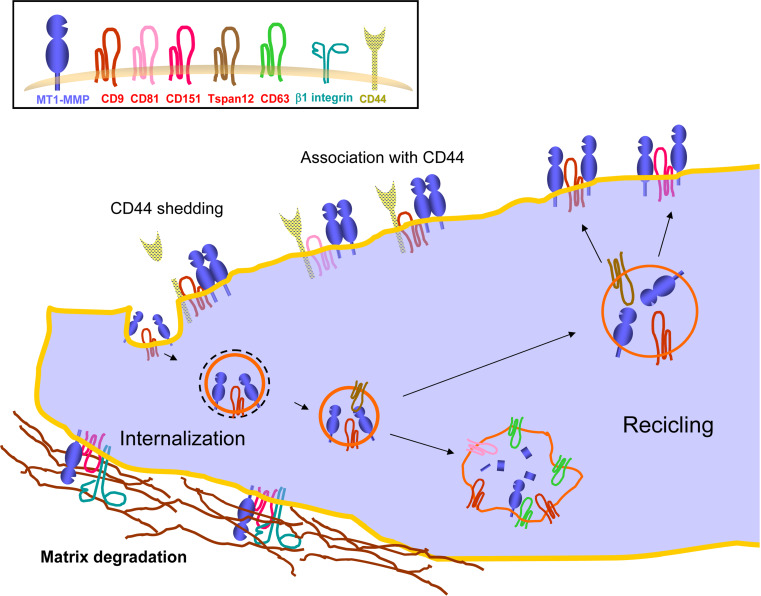
Fig. 2.
Functional regulation of MT1-MMP/MMP14 by tetraspanins. Tetraspanins directly associate with the membrane-anchored metalloproteinase MT1-MMP/MMP14 in different cell types. Association with tetraspanins regulates the rate of MT1-MMP degradation at lysosomes. At the plasma membrane tetraspanins may function as a molecular link with other transmembrane proteins. Thus, in endothelial cells, CD151 spatiotemporally coordinates MT1-MMP with integrins for extracellular matrix degradation. MT1-MMP subcellular localization is also dependant on its association with CD44, which is in turn shedded by the protease, and is also a component of TEMs
The stoichiometries of tetraspanin-MT1-MMP complexes can be in turn regulated by EWI-2 [85]. EWI-2 is a member of the Ig superfamily receptor, tightly associated with tetraspanins CD9 and CD81, whose re-expression in glioblastoma cells causes an increase in CD9-CD81 associations and disturbs that of tetraspanins with MT1-MMP impairing tumor cell invasivity.
Tetraspanins and other proteases
Some experimental evidence exists on the regulation of other proteases by tetraspanins. The proteolytic function of the urokinase receptor was shown to be regulated by CD82 expression levels [86]. Although this tetraspanin did not associate directly with the protease, it induced its relocalization to focal adhesions, where it associated with α5β1 integrin. This subcellular relocalization was concomitant with an inhibition of the proteolytic activity [86]. In neutrophils, tetraspanin CD63 is fundamental for the targeting of elastase to secretory granules [87]. The dipeptidyl peptidase IV/CD26 was found to be inserted into tetraspanin-enriched microdomains by means of a proteomic screening [88]. Proteomics also revealed the presence of CD13 ectopeptidase in TSPAN12 immunoprecipitates, although this result could not be confirmed by the authors in Western-blot experiments [23].
Regulation of proteolysis at the substrate level
As pointed out above, the number of substrates for ADAM10 and ADAM17 is rapidly growing and many of these substrate proteins can be cleaved in vitro by both ADAM10 and ADAM17. An attractive possibility is that in vivo tetraspanins may control the substrate specificity for each ADAM on the cell surface by regulating the segregation of substrates into distinct tetraspanin microdomains; antibody engagement could bring different microdomains together, promoting substrate cleavage. Interestingly, many substrates of ADAM10 or ADAM17 are in turn associated with tetraspanins. The tetraspanin CD9 has been shown to interact with the transmembrane precursors of HB-EGF, TGFα, epiregulin, and amphiregulin, markedly enhancing the juxtacrine signaling mediated by these transmembrane ligands of EGFR [89, 90]. In fact, CD9 has been reported to strongly decrease the PMA-induced proteolytic conversion of transmembrane to soluble TGF-α [89]. Moreover, the adhesive function of ICAM-1 and VCAM-1 during leukocyte transendothelial migration is functionally regulated by endothelial tetraspanins, including CD9 [91]. ICAM-1 and VCAM-1 are key endothelial adhesion molecules mediating leukocyte extravasation, which are also substrates of ADAM17 [92, 93], and elevated soluble levels of these proteins have been found in several inflammatory and tumoral pathologies [94, 95]. Hence, CD9 silencing on stimulated HUVEC results in a marked decrease in the membrane level of ICAM-1 reflecting the release from the inhibitory effect exerted by CD9 on ADAM17 activity [24]. These results also provide a mechanism by which CD9 regulates the levels of ICAM-1 on endothelial cells and reinforces the subsequent consequences in terms of leukocyte adhesion and transmigration [91, 96].
The reported inhibitory effect of CD9 on ADAM17 activity [24] would explain the important role of this tetraspanin as an enhancer of EGFR juxtacrine signaling by increasing the surface levels of these transmembrane ligands. CD9 could also potentially inhibit the reported crosstalk between GPCRs and EGFR signaling, which requires the release of EGFR ligands by ADAM10 and ADAM17, and has been shown to contribute to tumorigenesis, migration, and invasion in different tumor cell lines (reviewed in [5, 7]). It remains to be established whether the tetraspanin-mediated compartmentalization of substrates coordinates the activity of α- and γ-secretases in the sequential steps of RIPing.
The same principle may apply to MT1-MMP. MT1-MMP subcellular localization is also dependant on its association with CD44 [97], which is in turn shedded by the protease [98]. Different substrates of MT1-MMP are also inserted into TEMs (i.e., CD44, ICAM-1) [2], while the ternary complex involving MT1-MMP/CD151/α3β1 integrin would spatially direct the proteolysis towards the extracellular matrix substrates that are ligands of the integrin. Thus, this compartmentalization of substrates into specialized membrane microdomains might impose a new level of regulation on the activity of different membrane proteases.
Therapeutic interventions based on tetraspanins
The broad regulatory effects exerted by tetraspanins on the activity of different relevant proteases discussed above, open several potential opportunities for therapeutic intervention in pathological processes including Alzheimer’s disease, cancer progression, and inflammation. It seems likely that the large variety of tools that have been developed for the study of tetraspanins, including monoclonal antibodies, recombinant functional domains and interfering RNAs, could have significant therapeutic potential [60, 99]. In fact some of these reagents have already been tested for this purpose, including antibodies specific for CD81 in multiple sclerosis [100], for CD9 in gastric cancer [101] and for CD37 in B cell malignancies [102].
Regarding the mechanisms of etiopathogenesis of Alzheimer’s disease at the molecular level, the β-amyloid precursor protein is normally found throughout the body and may be cleaved first by either α- (ADAM10 or ADAM17) or by β-secretases, which is a pre-requisite for the subsequent action of the γ-secretase. When the sequential cleavage of β-amyloid precursor protein (APP) is carried out by α- followed by γ-secretase activities, it generates a peptide (P3) that precludes the generation of the neurotoxic amyloid β-peptide (Aβ), and constitutes a non-amyloidogenic pathway. In contrast, the amyloidogenic pathway implies the sequential action of the β-secretase (BACE1: β-site amyloid precursor protein-cleaving enzyme,) followed by the γ-secretase, generating the highly hydrophobic and insoluble amyloid β-peptide (Aβ), which is then secreted and aggregates extracellularly, forming the characteristic pathologic amyloid neuritic plaques deposited in the brains of AD patients. Accordingly, ADAM10 overexpression prevents amyloid plaque formation and deposition and lessens cognitive deficits in a transgenic murine model of Alzheimer’s disease [103]. Regarding ADAM17/TACE, Tachida et al. [104] found that the proinflammatory cytokine interleukin IL-1β produced a significantly enhanced α-cleavage pattern, resulting in decreased Aβ production in neuroblastoma cell lines or mouse primary cultured neurons. These effects were mediated through up-regulation and increased maturation of ADAM17/TACE. Moreover, IL-1Rα, the physiological antagonist for the IL-1 receptor, reversed the effects of IL-1β. Therefore, these studies suggest that strategies aimed at increasing expression and activity of either ADAM10 or ADAM17 may be beneficial in AD pathology. In contrast, in another study, the highly selective ADAM17/TACE inhibitor BMS-561392 caused a significant reduction in the levels of secreted APPα (the soluble product of APP cleavage by α-secretases) but without a corresponding increase in Aβ production in vitro or in vivo when infused into the brains of human APP-expressing transgenic and wild-type mice [105].
Taking into account the regulatory effect of TSPAN12 as an enhancer of the activity of ADAM10 [23], which is a major α-secretase responsible for APP processing along the non-amyloidogenic pathway [106, 107], it was proposed that TSPAN12 could induce a therapeutically desirable reduction in brain deposits of Aβ peptide. Therefore, as seen for other tetraspanins, the use of TSPAN12-specific reagents with agonistic effects such as its soluble LEL domain or specific stimulatory anti-TSPAN12 antibodies could potentially be therapeutically beneficial in Alzheimer’s disease. The potential regulatory effects exerted by TSPAN12 or by other tetraspanins on the activity of ADAM17 (the second relevant sheddase responsible for α-secretase activity), or on the activity of the γ-secretase complex that are crucial in the processing of APP remain to be explored and may reveal additional molecular targets for therapeutic intervention.
In other pathological processes however it may be desirable to inhibit the proteolytic activity of ADAM sheddases. This could be the case in processes like tumor progression and metastasis. It is worth keeping in mind that various cell surface substrates of ADAM10 and ADAM17 are key growth factors or cell adhesion molecules with demonstrated implication in cancer progression and in fact an association between ADAM10 and ADAM17 expression and activity and these processes has been established in various types of cancer [8, 9, 108, 109]. Following the same line of reasoning as indicated above for AD, targeting TSPAN12 with antagonistic approaches such as expression of dominant-negative TSPAN12 mutants, RNA silencing, inhibitory antibodies or inhibitory soluble LELs, could have a beneficial effect derived from down-modulation of ADAM10 maturation and/or activity in tumor progression and metastasis.
It is also well established the tumor and metastasis-suppressor role of CD9 in a variety of carcinoma types [1]. The presence of CD9 impairs survival, invasion, and metastasis in many tumor cell types (Ikeyama 1993, Takeda 2007; Saito 2006) and therefore this tetraspanin may represent an important target for development of new approaches in cancer therapy. We found that neoexpression of CD9 or treatment with agonistic anti-CD9 mAbs increased the membrane expression of TNF-α on different human colon carcinoma cells thus affecting their survival [53], consistent with the recent results from our group that demonstrate that tetraspanin CD9 negatively regulates ADAM17 activity [24]. To what extent some of these suppressor effects of CD9 in cancer progression can be attributed to or are related to regulation of the shedding of important adhesion molecules or growth factors through its inhibitory effects on ADAM17 activity opens a whole new field of research that remains to be explored.
The results published by Dunn et al. [59] implicating some tetraspanins in promoting constitutive Notch signaling again open interesting possibilities from a potential therapeutic perspective, since targeting these specific tetraspanin members might partially reduce constitutive Notch activity without causing the major side-effects associated with strongly and globally reducing Notch or presenilin activity using γ-secretase inhibitors. As suggested by these authors, depending on the specific interactions in putative complexes comprising γ-secretase, specific tetraspanin(s) and Notch, it could be possible to design specific small molecule or peptide inhibitors with the ability to interfere these interactions. Furthermore, if different tetraspanins promote the selective γ-secretase-mediated cleavage of distinct substrates, it could be possible to develop γ-secretase inhibitors that are disease-specific. For example, such inhibitors could distinctly target Alzheimer’s disease or specifically treat Notch-driven tumors.
Angiogenesis represents another pathologically relevant process in which the modulation of protease activity by tetraspanins could be interesting from a therapeutic perspective. Antibodies against CD9, CD81, or CD151 affect α3β1 or α6β1 integrin-dependent endothelial migration during wound healing and angiotube formation [110–114] and CD151-α3β1 complexes are key regulators of the angiotensin-II induced angiogenic response [115]. Moreover, the proposed use of anti-CD9 antibodies in gastric cancer therapy is partially based on their inhibitory effect on tumor angiogenesis [101]. A role in retinal vascular development in mice has recently been described for association between Tspan12 and the Norrin receptor [116].
The tetraspanin CD151 is important in endothelial cell biology and CD151-null mice are defective in angiogenesis in vivo under certain pathological conditions such as matrigel plug or tumor implantation [117]. Endothelial cells derived from these CD151 deficient mice show defects in migration, spreading, invasion, matrigel contraction, tube formation, and spheroid sprouting [117]. They also present selective signaling defects when grown on laminin substrates [117] and impaired collagenolysis as a result of disruption of the complex formed by the protease MT1-MMP with CD151 and α3β1 integrin [77]. Therefore, given the importance of this tetraspanin in endothelial migration and angiogenesis, strategies designed to target CD151 could be beneficial in pathological situations where inhibition of angiogenesis (like tumor progression or retinopathies) or induction of angiogenesis (like myocardial infarction) may be desirable. In fact, it has been shown that CD151 overexpression increases angiogenesis and blood reperfusion after a myocardial infarction [118].
Conclusions and further perspectives
Insertion into tetraspanin-enriched microdomains regulates the enzymatic activity of several proteases from different families. They may affect the rate of lysosomal degradation versus recycling or their incorporation into specialized intracellular vesicles. Alternatively, they may functionally connect the proteases with regulatory molecules such as integrins or CD44. Insertion into TEMs of both the proteases and their substrates may also dynamically regulate substrate accessibility. Future research would have to determine whether tetraspanins also affect other regulatory mechanisms such as dimerization or autocatalytic processing. Thus, TEM appear as a new therapeutical target that may simultaneously affect the proteolytic activity of different enzymes and their substrates. Therefore, agonistic or antagonistic antibodies, blocking soluble peptides based on tetraspanin functional regions at the large extracellular loop (LEL) offer new opportunities for the treatment of diseases such as inflammation, cancer, and Alzheimer's disease.
Acknowledgments
This work was supported by grants BFU2007-66443/BMC and BFU2010-19144/BMC from Ministerio de Ciencia e Innovación, a grant from Fundación de Investigación Médica Mutua Madrileña and by the RETICS Program RD08/0075-RIER (Red de Inflamación y Enfermedades Reumáticas) from Instituto de Salud Carlos III (to C.C.), a grant from Fundación de Investigación Médica Mutua Madrileña (to M.D.G.L.), and the grant PI080794 from Instituto de Salud Carlos III (to M.Y-M).
Abbreviations
- AD
Alzheimer's disease
- ADAM
A disintegrin and metalloprotease domain
- APP
Amyloid precursor protein
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- GPCRs
G protein-coupled receptors
- HB-EGF
Heparin-binding epidermal growth factor
- ICAM-1
Intercellular adhesion molecule-1
- mAbs
Monoclonal antibodies
- MT1-MMP
Membrane-type-1-matrix metalloprotease
- PMA
Phorbol-12-myristate-13-acetate
- RIP
Regulated intramembrane proteolysis
- TEMs
Tetraspanin-enriched microdomains
- TMPS
Triple membrane-passing signaling
- TNFα
Tumor necrosis factor-α
Contributor Information
María Yáñez-Mó, Email: myanez.hlpr@salud.madrid.org.
Maria Dolores Gutiérrez-López, Email: mdgutier@med.ucm.es.
Carlos Cabañas, Email: ccabanas@cbm.uam.es.
References
- 1.Charrin S, le Naour F, Silvie O, Milhiet PE, Boucheix C, Rubinstein E. Lateral organization of membrane proteins: tetraspanins spin their web. Biochem J. 2009;420:133–154. doi: 10.1042/BJ20082422. [DOI] [PubMed] [Google Scholar]
- 2.Zoller M. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat Rev Cancer. 2009;9:40–55. doi: 10.1038/nrc2543. [DOI] [PubMed] [Google Scholar]
- 3.Yanez-Mo M, Barreiro O, Gordon-Alonso M, Sala-Valdes M, Sanchez-Madrid F. Tetraspanin-enriched microdomains: a functional unit in cell plasma membranes. Trends Cell Biol. 2009;19:434–446. doi: 10.1016/j.tcb.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Hemler ME. Tetraspanin functions and associated microdomains. Nature Rev. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 5.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Aspects Med. 2008;29:258–289. doi: 10.1016/j.mam.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blobel CP. ADAMs: key components in EGFR signalling and development. Nature Rev. 2005;6:32–43. doi: 10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 7.Reiss K, Saftig P. The "a disintegrin and metalloprotease" (ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol. 2009;20:126–137. doi: 10.1016/j.semcdb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Murphy G. The ADAMs: signalling scissors in the tumour microenvironment. Nat Rev Cancer. 2008;8:929–941. doi: 10.1038/nrc2459. [DOI] [PubMed] [Google Scholar]
- 9.Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol. 2009;20:164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Jackson LF, Qiu TH, Sunnarborg SW, Chang A, Zhang C, Patterson C, Lee DC. Defective valvulogenesis in HB-EGF and TACE-null mice is associated with aberrant BMP signaling. EMBO J. 2003;22:2704–2716. doi: 10.1093/emboj/cdg264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sternlicht MD, Sunnarborg SW. The ADAM17-amphiregulin-EGFR axis in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2008;13:181–194. doi: 10.1007/s10911-008-9084-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer OM, Hart S, Gschwind A, Ullrich A. EGFR signal transactivation in cancer cells. Biochem Soc Trans. 2003;31:1203–1208. doi: 10.1042/bst0311203. [DOI] [PubMed] [Google Scholar]
- 13.Liebmann C. EGF receptor activation by GPCRs: an universal pathway reveals different versions. Mol Cell Endocrinol. 2010;331:222–231. doi: 10.1016/j.mce.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Ohtsu H, Dempsey PJ, Eguchi S. ADAMs as mediators of EGF receptor transactivation by G protein-coupled receptors. Am J Physiol. 2006;291:C1–C10. doi: 10.1152/ajpcell.00620.2005. [DOI] [PubMed] [Google Scholar]
- 15.Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J Cell Biol. 2002;158:221–226. doi: 10.1083/jcb.200112026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiuchi K, Le Gall S, Schulte M, Yamaguchi T, Reiss K, Murphy G, Toyama Y, Hartmann D, Saftig P, Blobel CP. Substrate selectivity of epidermal growth factor-receptor ligand sheddases and their regulation by phorbol esters and calcium influx. Mol Biol Cell. 2007;18:176–188. doi: 10.1091/mbc.E06-01-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diaz-Rodriguez E, Montero JC, Esparis-Ogando A, Yuste L, Pandiella A. Extracellular signal-regulated kinase phosphorylates tumor necrosis factor alpha-converting enzyme at threonine 735: a potential role in regulated shedding. Mol Biol Cell. 2002;13:2031–2044. doi: 10.1091/mbc.01-11-0561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan H, Turck CW, Derynck R. Characterization of growth factor-induced serine phosphorylation of tumor necrosis factor-alpha converting enzyme and of an alternatively translated polypeptide. J Biol Chem. 2003;278:18617–18627. doi: 10.1074/jbc.M300331200. [DOI] [PubMed] [Google Scholar]
- 19.Soond SM, Everson B, Riches DW, Murphy G. ERK-mediated phosphorylation of Thr735 in TNFalpha-converting enzyme and its potential role in TACE protein trafficking. J Cell Sci. 2005;118:2371–2380. doi: 10.1242/jcs.02357. [DOI] [PubMed] [Google Scholar]
- 20.Nagano O, Murakami D, Hartmann D, De Strooper B, Saftig P, Iwatsubo T, Nakajima M, Shinohara M, Saya H. Cell-matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca(2+) influx and PKC activation. J Cell Biol. 2004;165:893–902. doi: 10.1083/jcb.200310024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy G. Regulation of the proteolytic disintegrin metalloproteinases, the ‘Sheddases’. Semin Cell Dev Biol. 2009;20:138–145. doi: 10.1016/j.semcdb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Arduise C, Abache T, Li L, Billard M, Chabanon A, Ludwig A, Mauduit P, Boucheix C, Rubinstein E, Le Naour F. Tetraspanins regulate ADAM10-mediated cleavage of TNF-alpha and epidermal growth factor. J Immunol. 2008;181:7002–7013. doi: 10.4049/jimmunol.181.10.7002. [DOI] [PubMed] [Google Scholar]
- 23.Xu D, Sharma C, Hemler ME. Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. Faseb J. 2009;23:3674–3681. doi: 10.1096/fj.09-133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutiérrez-López M, Gilsanz A, Yánez-Mó M, Ovalle S, Lafuente M, Domínguez C, Monk P, González-Alvaro I, Sánchez-Madrid F, Cabañas C (2011) The sheddase activity of ADAM17/TACE is regulated by the tetraspanin CD9. Cell Mol Life Sci. doi:10.1007/s00018-011-0639-0 [DOI] [PMC free article] [PubMed]
- 25.Gutwein P, Mechtersheimer S, Riedle S, Stoeck A, Gast D, Joumaa S, Zentgraf H, Fogel M, Altevogt DP. ADAM10-mediated cleavage of L1 adhesion molecule at the cell surface and in released membrane vesicles. Faseb J. 2003;17:292–294. doi: 10.1096/fj.02-0430fje. [DOI] [PubMed] [Google Scholar]
- 26.Stoeck A, Keller S, Riedle S, Sanderson MP, Runz S, Le Naour F, Gutwein P, Ludwig A, Rubinstein E, Altevogt P. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem J. 2006;393:609–618. doi: 10.1042/BJ20051013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keller S, Konig AK, Marme F, Runz S, Wolterink S, Koensgen D, Mustea A, Sehouli J, Altevogt P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009;278:73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 28.Soderberg A, Barral AM, Soderstrom M, Sander B, Rosen A. Redox-signaling transmitted in trans to neighboring cells by melanoma-derived TNF-containing exosomes. Free Radic Biol Med. 2007;43:90–99. doi: 10.1016/j.freeradbiomed.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 29.Thery C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 30.Simons M, Raposo G. Exosomes–vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Zoller M. Gastrointestinal tumors: metastasis and tetraspanins. Z Gastroenterol. 2006;44:573–586. doi: 10.1055/s-2006-926795. [DOI] [PubMed] [Google Scholar]
- 32.Chen MS, Tung KS, Coonrod SA, Takahashi Y, Bigler D, Chang A, Yamashita Y, Kincade PW, Herr JC, White JM. Role of the integrin-associated protein CD9 in binding between sperm ADAM 2 and the egg integrin alpha6beta1: implications for murine fertilization. Proc Natl Acad Sci USA. 1999;96:11830–11835. doi: 10.1073/pnas.96.21.11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ziyyat A, Rubinstein E, Monier-Gavelle F, Barraud V, Kulski O, Prenant M, Boucheix C, Bomsel M, Wolf JP. CD9 controls the formation of clusters that contain tetraspanins and the integrin alpha 6 beta 1, which are involved in human and mouse gamete fusion. J Cell Sci. 2006;119:416–424. doi: 10.1242/jcs.02730. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi Y, Bigler D, Ito Y, White JM. Sequence-specific interaction between the disintegrin domain of mouse ADAM 3 and murine eggs: role of beta1 integrin-associated proteins CD9, CD81, and CD98. Mol Biol Cell. 2001;12:809–820. doi: 10.1091/mbc.12.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller BJ, Georges-Labouesse E, Primakoff P, Myles DG. Normal fertilization occurs with eggs lacking the integrin alpha6beta1 and is CD9-dependent. J Cell Biol. 2000;149:1289–1296. doi: 10.1083/jcb.149.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C. Severely reduced female fertility in CD9-deficient mice. Science. 2000;287:319–321. doi: 10.1126/science.287.5451.319. [DOI] [PubMed] [Google Scholar]
- 37.Kaji K, Oda S, Shikano T, Ohnuki T, Uematsu Y, Sakagami J, Tada N, Miyazaki S, Kudo A. The gamete fusion process is defective in eggs of Cd9-deficient mice. Nat Genet. 2000;24:279–282. doi: 10.1038/73502. [DOI] [PubMed] [Google Scholar]
- 38.Miyado K, Yamada G, Yamada S, Hasuwa H, Nakamura Y, Ryu F, Suzuki K, Kosai K, Inoue K, Ogura A, Okabe M, Mekada E. Requirement of CD9 on the egg plasma membrane for fertilization. Science. 2000;287:321–324. doi: 10.1126/science.287.5451.321. [DOI] [PubMed] [Google Scholar]
- 39.Rubinstein E, Ziyyat A, Prenant M, Wrobel E, Wolf JP, Levy S, Le Naour F, Boucheix C. Reduced fertility of female mice lacking CD81. Dev Biol. 2006;290:351–358. doi: 10.1016/j.ydbio.2005.11.031. [DOI] [PubMed] [Google Scholar]
- 40.Kaji K, Oda S, Miyazaki S, Kudo A. Infertility of CD9-deficient mouse eggs is reversed by mouse CD9, human CD9, or mouse CD81; polyadenylated mRNA injection developed for molecular analysis of sperm-egg fusion. Dev Biol. 2002;247:327–334. doi: 10.1006/dbio.2002.0694. [DOI] [PubMed] [Google Scholar]
- 41.Zhou GB, Liu GS, Meng QG, Liu Y, Hou YP, Wang XX, Li N, Zhu SE. Tetraspanin CD9 in bovine oocytes and its role in fertilization. J Reprod Dev. 2009;55:305–308. doi: 10.1262/jrd.20099. [DOI] [PubMed] [Google Scholar]
- 42.Waterhouse R, Ha C, Dveksler GS. Murine CD9 is the receptor for pregnancy-specific glycoprotein 17. J Exp Med. 2002;195:277–282. doi: 10.1084/jem.20011741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ellerman DA, Ha C, Primakoff P, Myles DG, Dveksler GS. Direct binding of the ligand PSG17 to CD9 requires a CD9 site essential for sperm–egg fusion. Mol Biol Cell. 2003;14:5098–5103. doi: 10.1091/mbc.E03-04-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyado K, Yoshida K, Yamagata K, Sakakibara K, Okabe M, Wang X, Miyamoto K, Akutsu H, Kondo T, Takahashi Y, Ban T, Ito C, Toshimori K, Nakamura A, Ito M, Miyado M, Mekada E, Umezawa A. The fusing ability of sperm is bestowed by CD9-containing vesicles released from eggs in mice. Proc Natl Acad Sci USA. 2008;105:12921–12926. doi: 10.1073/pnas.0710608105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta S, Primakoff P, Myles DG. Can the presence of wild-type oocytes during insemination rescue the fusion defect of CD9 null oocytes? Mol Reprod Dev. 2009;76:602. doi: 10.1002/mrd.21040. [DOI] [PubMed] [Google Scholar]
- 46.Woo HN, Baik SH, Park JS, Gwon AR, Yang S, Yun YK, Jo DG. Secretases as therapeutic targets for Alzheimer’s disease. Biochem Biophys Res Commun. 2011;404:10–15. doi: 10.1016/j.bbrc.2010.11.132. [DOI] [PubMed] [Google Scholar]
- 47.Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens JR, Cumano A, Roux P, Black RA, Israel A. A novel proteolytic cleavage involved in Notch signaling: the role of the disintegrin-metalloprotease TACE. Molecular cell. 2000;5:207–216. doi: 10.1016/s1097-2765(00)80417-7. [DOI] [PubMed] [Google Scholar]
- 48.Mumm JS, Schroeter EH, Saxena MT, Griesemer A, Tian X, Pan DJ, Ray WJ, Kopan R. A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Molecular cell. 2000;5:197–206. doi: 10.1016/s1097-2765(00)80416-5. [DOI] [PubMed] [Google Scholar]
- 49.Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- 50.Srinivasan B, Wang Z, Brun-Zinkernagel AM, Collier RJ, Black RA, Frank SJ, Barker PA, Roque RS. Photic injury promotes cleavage of p75NTR by TACE and nuclear trafficking of the p75 intracellular domain. Mol Cell Neurosci. 2007;36:449–461. doi: 10.1016/j.mcn.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 51.Spasic D, Annaert W. Building gamma-secretase: the bits and pieces. J Cell Sci. 2008;121:413–420. doi: 10.1242/jcs.015255. [DOI] [PubMed] [Google Scholar]
- 52.Wakabayashi T, Craessaerts K, Bammens L, Bentahir M, Borgions F, Herdewijn P, Staes A, Timmerman E, Vandekerckhove J, Rubinstein E, Boucheix C, Gevaert K, De Strooper B. Analysis of the gamma-secretase interactome and validation of its association with tetraspanin-enriched microdomains. Nat Cell Biol. 2009;11:1340–1346. doi: 10.1038/ncb1978. [DOI] [PubMed] [Google Scholar]
- 53.Ovalle S, Gutierrez-Lopez MD, Olmo N, Turnay J, Lizarbe MA, Majano P, Molina-Jimenez F, Lopez-Cabrera M, Yanez-Mo M, Sanchez-Madrid F, Cabanas C. The tetraspanin CD9 inhibits the proliferation and tumorigenicity of human colon carcinoma cells. Int J Cancer. 2007;121:2140–2152. doi: 10.1002/ijc.22902. [DOI] [PubMed] [Google Scholar]
- 54.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 55.Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poirazi P, Mel BW. Impact of active dendrites and structural plasticity on the memory capacity of neural tissue. Neuron. 2001;29:779–796. doi: 10.1016/s0896-6273(01)00252-5. [DOI] [PubMed] [Google Scholar]
- 57.Shors TJ, Miesegaes G, Beylin A, Zhao M, Rydel T, Gould E. Neurogenesis in the adult is involved in the formation of trace memories. Nature. 2001;410:372–376. doi: 10.1038/35066584. [DOI] [PubMed] [Google Scholar]
- 58.Berezovska O, Xia MQ, Hyman BT. Notch is expressed in adult brain, is coexpressed with presenilin-1, and is altered in Alzheimer disease. J Neuropathol Exp Neurol. 1998;57:738–745. doi: 10.1097/00005072-199808000-00003. [DOI] [PubMed] [Google Scholar]
- 59.Dunn CD, Sulis ML, Ferrando AA, Greenwald I. A conserved tetraspanin subfamily promotes Notch signaling in Caenorhabditis elegans and in human cells. Proc Natl Acad Sci USA. 2010;107:5907–5912. doi: 10.1073/pnas.1001647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hemler ME. Targeting of tetraspanin proteins–potential benefits and strategies. Nat Rev Drug Discov. 2008;7:747–758. doi: 10.1038/nrd2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lazo PA. Functional implications of tetraspanin proteins in cancer biology. Cancer Sci. 2007;98:1666–1677. doi: 10.1111/j.1349-7006.2007.00584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cook GA, Wilkinson DA, Crossno JT, Jr, Raghow R, Jennings LK. The tetraspanin CD9 influences the adhesion, spreading, and pericellular fibronectin matrix assembly of Chinese hamster ovary cells on human plasma fibronectin. Exp Cell Res. 1999;251:356–371. doi: 10.1006/excr.1999.4596. [DOI] [PubMed] [Google Scholar]
- 63.Sachs N, Kreft M, h Weerman MA, Beynon AJ, Peters TA, Weening JJ, Sonnenberg A. Kidney failure in mice lacking the tetraspanin CD151. J Cell Biol. 2006;175:33–39. doi: 10.1083/jcb.200603073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cowin AJ, Adams D, Geary SM, Wright MD, Jones JC, Ashman LK. Wound healing is defective in mice lacking tetraspanin CD151. J Invest Dermatol. 2006;126:680–689. doi: 10.1038/sj.jid.5700142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugiura T, Berditchevski F. Function of alpha3beta1-tetraspanin protein complexes in tumor cell invasion. Evidence for the role of the complexes in production of matrix metalloproteinase 2 (MMP-2) J Cell Biol. 1999;146:1375–1389. doi: 10.1083/jcb.146.6.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mazzocca A, Sciammetta SC, Carloni V, Cosmi L, Annunziato F, Harada T, Abrignani S, Pinzani M. Binding of hepatitis C virus envelope protein E2 to CD81 up-regulates matrix metalloproteinase-2 in human hepatic stellate cells. J Biol Chem. 2005;280:11329–11339. doi: 10.1074/jbc.M410161200. [DOI] [PubMed] [Google Scholar]
- 67.Hong IK, Jin YJ, Byun HJ, Jeoung DI, Kim YM, Lee H. Homophilic interactions of Tetraspanin CD151 up-regulate motility and matrix metalloproteinase-9 expression of human melanoma cells through adhesion-dependent c-Jun activation signaling pathways. J Biol Chem. 2006;281:24279–24292. doi: 10.1074/jbc.M601209200. [DOI] [PubMed] [Google Scholar]
- 68.Shi GM, Ke AW, Zhou J, Wang XY, Xu Y, Ding ZB, Devbhandari RP, Huang XY, Qiu SJ, Shi YH, Dai Z, Yang XR, Yang GH, Fan J. CD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinoma. Hepatology (Baltimore, MD) 2010;52:183–196. doi: 10.1002/hep.23661. [DOI] [PubMed] [Google Scholar]
- 69.Hong IK, Kim YM, Jeoung DI, Kim KC, Lee H. Tetraspanin CD9 induces MMP-2 expression by activating p38 MAPK, JNK and c-Jun pathways in human melanoma cells. Exp Mol Med. 2005;37:230–239. doi: 10.1038/emm.2005.31. [DOI] [PubMed] [Google Scholar]
- 70.Saito Y, Tachibana I, Takeda Y, Yamane H, He P, Suzuki M, Minami S, Kijima T, Yoshida M, Kumagai T, Osaki T, Kawase I. Absence of CD9 enhances adhesion-dependent morphologic differentiation, survival, and matrix metalloproteinase-2 production in small cell lung cancer cells. Cancer Res. 2006;66:9557–9565. doi: 10.1158/0008-5472.CAN-06-1131. [DOI] [PubMed] [Google Scholar]
- 71.Liu WM, Cao YJ, Yang YJ, Li J, Hu Z, Duan EK. Tetraspanin CD9 regulates invasion during mouse embryo implantation. J Mol Endocrinol. 2006;36:121–130. doi: 10.1677/jme.1.01910. [DOI] [PubMed] [Google Scholar]
- 72.Tohami T, Drucker L, Shapiro H, Radnay J, Lishner M. Overexpression of tetraspanins affects multiple myeloma cell survival and invasive potential. FASEB J. 2007;21:691–699. doi: 10.1096/fj.06-6610com. [DOI] [PubMed] [Google Scholar]
- 73.Shiomi T, Inoki I, Kataoka F, Ohtsuka T, Hashimoto G, Nemori R, Okada Y. Pericellular activation of proMMP-7 (promatrilysin-1) through interaction with CD151. Lab Investig J Tech Methods Pathol. 2005;85:1489–1506. doi: 10.1038/labinvest.3700351. [DOI] [PubMed] [Google Scholar]
- 74.Jung K, Liu X, Chirco R, Fridman R, Kim H. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 2006;25:3934–3942. doi: 10.1038/sj.emboj.7601281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lafleur MA, Xu D, Hemler ME. Tetraspanin proteins regulate membrane type-1 matrix metalloproteinase-dependent pericellular proteolysis. Mol Biol Cell. 2009;20:2030–2040. doi: 10.1091/mbc.E08-11-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takino T, Miyamori H, Kawaguchi N, Uekita T, Seiki M, Sato H. Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane-type 1 matrix metalloproteinase. Biochem Biophys Res Commun. 2003;304:160–166. doi: 10.1016/s0006-291x(03)00544-8. [DOI] [PubMed] [Google Scholar]
- 77.Yanez-Mo M, Barreiro O, Gonzalo P, Batista A, Megias D, Genis L, Sachs N, Sala-Valdes M, Alonso MA, Montoya MC, Sonnenberg A, Arroyo AG, Sanchez-Madrid F. MT1-MMP collagenolytic activity is regulated through association with tetraspanin CD151 in primary endothelial cells. Blood. 2008;112:3217–3226. doi: 10.1182/blood-2008-02-139394. [DOI] [PubMed] [Google Scholar]
- 78.Sato H, Takino T. Coordinate action of membrane-type matrix metalloproteinase-1 (MT1-MMP) and MMP-2 enhances pericellular proteolysis and invasion. Cancer Sci. 2010;101:843–847. doi: 10.1111/j.1349-7006.2010.01498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Holmbeck K, Bianco P, Caterina J, Yamada S, Kromer M, Kuznetsov SA, Mankani M, Robey PG, Poole AR, Pidoux I, Ward JM, Birkedal-Hansen H. MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell. 1999;99:81–92. doi: 10.1016/s0092-8674(00)80064-1. [DOI] [PubMed] [Google Scholar]
- 80.Itoh T, Tanioka M, Yoshida H, Yoshioka T, Nishimoto H, Itohara S. Reduced angiogenesis and tumor progression in gelatinase A-deficient mice. Cancer Res. 1998;58:1048–1051. [PubMed] [Google Scholar]
- 81.Oh J, Takahashi R, Adachi E, Kondo S, Kuratomi S, Noma A, Alexander DB, Motoda H, Okada A, Seiki M, Itoh T, Itohara S, Takahashi C, Noda M. Mutations in two matrix metalloproteinase genes, MMP-2 and MT1-MMP, are synthetic lethal in mice. Oncogene. 2004;23:5041–5048. doi: 10.1038/sj.onc.1207688. [DOI] [PubMed] [Google Scholar]
- 82.Galvez BG, Matias-Roman S, Yanez-Mo M, Vicente-Manzanares M, Sanchez-Madrid F, Arroyo AG. Caveolae are a novel pathway for membrane-type 1 matrix metalloproteinase traffic in human endothelial cells. Mol Biol Cell. 2004;15:678–687. doi: 10.1091/mbc.E03-07-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gingras D, Beliveau R. Emerging concepts in the regulation of membrane-type 1 matrix metalloproteinase activity. Biochim Biophys Acta. 2010;1803:142–150. doi: 10.1016/j.bbamcr.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 84.Bravo-Cordero JJ, Marrero-Diaz R, Megias D, Genis L, Garcia-Grande A, Garcia MA, Arroyo AG, Montoya MC. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007;26:1499–1510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kolesnikova TV, Kazarov AR, Lemieux ME, Lafleur MA, Kesari S, Kung AL, Hemler ME. Glioblastoma inhibition by cell surface immunoglobulin protein EWI-2, in vitro and in vivo. Neoplasia. 2009;11:77–86. doi: 10.1593/neo.81180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bass R, Werner F, Odintsova E, Sugiura T, Berditchevski F, Ellis V. Regulation of urokinase receptor proteolytic function by the tetraspanin CD82. J Biol Chem. 2005;280:14811–14818. doi: 10.1074/jbc.M414189200. [DOI] [PubMed] [Google Scholar]
- 87.Kallquist L, Hansson M, Persson AM, Janssen H, Calafat J, Tapper H, Olsson I. The tetraspanin CD63 is involved in granule targeting of neutrophil elastase. Blood. 2008;112:3444–3454. doi: 10.1182/blood-2007-10-116285. [DOI] [PubMed] [Google Scholar]
- 88.Le Naour F, Andre M, Boucheix C, Rubinstein E. Membrane microdomains and proteomics: lessons from tetraspanin microdomains and comparison with lipid rafts. Proteomics. 2006;6:6447–6454. doi: 10.1002/pmic.200600282. [DOI] [PubMed] [Google Scholar]
- 89.Shi W, Fan H, Shum L, Derynck R. The tetraspanin CD9 associates with transmembrane TGF-alpha and regulates TGF-alpha-induced EGF receptor activation and cell proliferation. J Cell Biol. 2000;148:591–602. doi: 10.1083/jcb.148.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Higashiyama S, Iwamoto R, Goishi K, Raab G, Taniguchi N, Klagsbrun M, Mekada E. The membrane protein CD9/DRAP 27 potentiates the juxtacrine growth factor activity of the membrane-anchored heparin-binding EGF-like growth factor. J Cell Biol. 1995;128:929–938. doi: 10.1083/jcb.128.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Barreiro O, Yanez-Mo M, Sala-Valdes M, Gutierrez-Lopez MD, Ovalle S, Higginbottom A, Monk PN, Cabanas C, Sanchez-Madrid F. Endothelial tetraspanin microdomains regulate leukocyte firm adhesion during extravasation. Blood. 2005;105:2852–2861. doi: 10.1182/blood-2004-09-3606. [DOI] [PubMed] [Google Scholar]
- 92.Garton KJ, Gough PJ, Raines EW. Emerging roles for ectodomain shedding in the regulation of inflammatory responses. J Leukoc Biol. 2006;79:1105–1116. doi: 10.1189/jlb.0106038. [DOI] [PubMed] [Google Scholar]
- 93.Tsakadze NL, Sithu SD, Sen U, English WR, Murphy G, D’Souza SE. Tumor necrosis factor-alpha-converting enzyme (TACE/ADAM-17) mediates the ectodomain cleavage of intercellular adhesion molecule-1 (ICAM-1) J Biol Chem. 2006;281:3157–3164. doi: 10.1074/jbc.M510797200. [DOI] [PubMed] [Google Scholar]
- 94.Gearing AJ, Newman W. Circulating adhesion molecules in disease. Immunol Today. 1993;14:506–512. doi: 10.1016/0167-5699(93)90267-O. [DOI] [PubMed] [Google Scholar]
- 95.van Kilsdonk JW, van Kempen LC, van Muijen GN, Ruiter DJ, Swart GW. Soluble adhesion molecules in human cancers: sources and fates. Eur J Cell Biol. 2010;89:415–427. doi: 10.1016/j.ejcb.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 96.Barreiro O, Zamai M, Yanez-Mo M, Tejera E, Lopez-Romero P, Monk PN, Gratton E, Caiolfa VR, Sanchez-Madrid F. Endothelial adhesion receptors are recruited to adherent leukocytes by inclusion in preformed tetraspanin nanoplatforms. J Cell Biol. 2008;183:527–542. doi: 10.1083/jcb.200805076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mori H, Tomari T, Koshikawa N, Kajita M, Itoh Y, Sato H, Tojo H, Yana I, Seiki M. CD44 directs membrane-type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin-like domain. EMBO J. 2002;21:3949–3959. doi: 10.1093/emboj/cdf411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Marrero-Diaz R, Bravo-Cordero JJ, Megias D, Garcia MA, Bartolome RA, Teixido J, Montoya MC. Polarized MT1-MMP-CD44 interaction and CD44 cleavage during cell retraction reveal an essential role for MT1-MMP in CD44-mediated invasion. Cell Motil Cytoskeleton. 2009;66:48–61. doi: 10.1002/cm.20325. [DOI] [PubMed] [Google Scholar]
- 99.Stipp CS. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev Mol Med. 2010;12:e3. doi: 10.1017/S1462399409001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dijkstra S, Kooij G, Verbeek R, van der Pol SM, Amor S, Geisert EE, Jr, Dijkstra CD, van Noort JM, Vries HE. Targeting the tetraspanin CD81 blocks monocyte transmigration and ameliorates EAE. Neurobiol Dis. 2008;31:413–421. doi: 10.1016/j.nbd.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 101.Nakamoto T, Murayama Y, Oritani K, Boucheix C, Rubinstein E, Nishida M, Katsube F, Watabe K, Kiso S, Tsutsui S, Tamura S, Shinomura Y, Hayashi N. A novel therapeutic strategy with anti-CD9 antibody in gastric cancers. J Gastroenterol. 2009;44:889–896. doi: 10.1007/s00535-009-0081-3. [DOI] [PubMed] [Google Scholar]
- 102.Robak T, Robak P, Smolewski P. TRU-016, a humanized anti-CD37 IgG fusion protein for the potential treatment of B-cell malignancies. Curr Opin Investig Drugs. 2009;10:1383–1390. [PubMed] [Google Scholar]
- 103.Postina R, Schroeder A, Dewachter I, Bohl J, Schmitt U, Kojro E, Prinzen C, Endres K, Hiemke C, Blessing M, Flamez P, Dequenne A, Godaux E, van Leuven F, Fahrenholz F. A disintegrin-metalloproteinase prevents amyloid plaque formation and hippocampal defects in an Alzheimer disease mouse model. J Clin Invest. 2004;113:1456–1464. doi: 10.1172/JCI20864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tachida Y, Nakagawa K, Saito T, Saido TC, Honda T, Saito Y, Murayama S, Endo T, Sakaguchi G, Kato A, Kitazume S, Hashimoto Y. Interleukin-1 beta up-regulates TACE to enhance alpha-cleavage of APP in neurons: resulting decrease in Abeta production. J Neurochem. 2008;104:1387–1393. doi: 10.1111/j.1471-4159.2007.05127.x. [DOI] [PubMed] [Google Scholar]
- 105.Kim ML, Zhang B, Mills IP, Milla ME, Brunden KR, Lee VM. Effects of TNFalpha-converting enzyme inhibition on amyloid beta production and APP processing in vitro and in vivo. J Neurosci. 2008;28:12052–12061. doi: 10.1523/JNEUROSCI.2913-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chow VW, Mattson MP, Wong PC, Gleichmann M. An overview of APP processing enzymes and products. Neuromolecular Med. 2009;12:1–12. doi: 10.1007/s12017-009-8104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Woo HN, Park JS, Gwon AR, Arumugam TV, Jo DG. Alzheimer’s disease and Notch signaling. Biochem Biophys Res Commun. 2009;390:1093–1097. doi: 10.1016/j.bbrc.2009.10.093. [DOI] [PubMed] [Google Scholar]
- 108.Moss ML, Stoeck A, Yan W, Dempsey PJ. ADAM10 as a target for anti-cancer therapy. Curr Pharm Biotechnol. 2008;9:2–8. doi: 10.2174/138920108783497613. [DOI] [PubMed] [Google Scholar]
- 109.Arribas J, Bech-Serra JJ, Santiago-Josefat B. ADAMs, cell migration and cancer. Cancer Metastasis Rev. 2006;25:57–68. doi: 10.1007/s10555-006-7889-6. [DOI] [PubMed] [Google Scholar]
- 110.Klein-Soyer C, Azorsa DO, Cazenave JP, Lanza F. CD9 participates in endothelial cell migration during in vitro wound repair. Arterioscler Thromb Vasc Biol. 2000;20:360–369. doi: 10.1161/01.atv.20.2.360. [DOI] [PubMed] [Google Scholar]
- 111.Sincock PM, Fitter S, Parton RG, Berndt MC, Gamble JR, Ashman LK. PETA-3/CD151, a member of the transmembrane 4 superfamily, is localised to the plasma membrane and endocytic system of endothelial cells, associates with multiple integrins and modulates cell function. J Cell Sci. 1999;112:833–844. doi: 10.1242/jcs.112.6.833. [DOI] [PubMed] [Google Scholar]
- 112.Deissler H, Kuhn EM, Lang GE, Deissler H. Tetraspanin CD9 is involved in the migration of retinal microvascular endothelial cells. Int J Mol Med. 2007;20:643–652. [PubMed] [Google Scholar]
- 113.Zhang XA, Kazarov AR, Yang X, Bontrager AL, Stipp CS, Hemler ME. Function of the tetraspanin CD151-alpha6beta1 integrin complex during cellular morphogenesis. Mol Biol Cell. 2002;13:1–11. doi: 10.1091/mbc.01-10-0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yanez-Mo M, Alfranca A, Cabanas C, Marazuela M, Tejedor R, Ursa MA, Ashman LK, de Landazuri MO, Sanchez-Madrid F. Regulation of endothelial cell motility by complexes of tetraspan molecules CD81/TAPA-1 and CD151/PETA-3 with alpha3 beta1 integrin localized at endothelial lateral junctions. J Cell Biol. 1998;141:791–804. doi: 10.1083/jcb.141.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dominguez-Jimenez C, Yanez-Mo M, Carreira A, Tejedor R, Gonzalez-Amaro R, Alvarez V, Sanchez-Madrid F. Involvement of alpha3 integrin/tetraspanin complexes in the angiogenic response induced by angiotensin II. FASEB J. 2001;15:1457–1459. doi: 10.1096/fj.00-0651fje. [DOI] [PubMed] [Google Scholar]
- 116.Junge HJ, Yang S, Burton JB, Paes K, Shu X, French DM, Costa M, Rice DS, Ye W. TSPAN12 regulates retinal vascular development by promoting Norrin-but not Wnt-induced FZD4/beta-catenin signaling. Cell. 2009;139:299–311. doi: 10.1016/j.cell.2009.07.048. [DOI] [PubMed] [Google Scholar]
- 117.Takeda Y, Kazarov AR, Butterfield CE, Hopkins BD, Benjamin LE, Kaipainen A, Hemler ME. Deletion of tetraspanin Cd151 results in decreased pathologic angiogenesis in vivo and in vitro. Blood. 2007;109:1524–1532. doi: 10.1182/blood-2006-08-041970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zuo H, Liu Z, Liu X, Yang J, Liu T, Wen S, Zhang XA, Cianflone K, Wang D. CD151 gene delivery after myocardial infarction promotes functional neovascularization and activates FAK signaling. Molecular Med (Cambridge, Mass) 2009;15:307–315. doi: 10.2119/molmed.2009.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]