Abstract
Tlx3 (HOX11L2) is regarded as one of the selector genes in excitatory versus inhibitory fate specification of neurons in distinct regions of the nervous system. Expression of Tlx3 in a post-mitotic immature neuron favors a glutamatergic over GABAergic fate. The factors that regulate Tlx3 have immense importance in the fate specification of glutamatergic neurons. Here, we have shown that Notch target gene, Hes-1, negatively regulates Tlx3 expression, resulting in decreased generation of glutamatergic neurons. Down-regulation of Hes-1 removed the inhibition on Tlx3 promoter, thus promoting glutamatergic differentiation. Promoter–protein interaction studies with truncated/mutated Hes-1 protein suggested that the co-repressor recruitment mediated through WRPW domain of Hes-1 has contributed to the repressive effect. Our results clearly demonstrate a new and unique role for canonical Notch signaling through Hes-1, in neurotransmitter/subtype fate specification of neurons in addition to its known functional role in proliferation/maintenance of neural progenitors.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-011-0765-8) contains supplementary material, which is available to authorized users.
Keywords: Notch signaling, Tlx3, Glutamatergic differentiation, Hes-1, Neural differentiation
Introduction
Excitatory versus inhibitory fate specification of neural progenitors during development/neurogenesis is a very tightly regulated process. The process of fate specification is regulated by various factors including bHLH and homeodomain transcription factors along with extra cellular environment [1–5]. Combinatorial expression of transcription factors is one of the mechanisms indicated in the generation of excitatory versus inhibitory fate specification of nascent neurons. The expression of these transcription factors is controlled by selector genes, which could induce/control a particular fate and at the same time suppress an alternative fate. During dorsal spinal cord neurogenesis, the homeodomain transcription factors Tlx3 and Tlx1 specify a glutamatergic over GABAergic fate, whereas Pax2 in combination with Lbx1 specifies a GABAergic fate [6, 7]. Tlx3 is also able to reverse the effect of Pax2 in GABAergic determination. However, Ptf1α suppresses Tlx3 through Notch signaling in an RBPJk-independent manner and induces GABAergic fate [8]. Also, over-expression of Tlx3 in chick spinal cord induces a glutamatergic fate. Similar results were demonstrated with ES cells, where over-expression of Tlx3 leads to glutamatergic fate specification by increasing glutamatergic marker genes and transporters [7]. Therefore, it is confirmed that Tlx3 can act as a selector gene in excitatory versus inhibitory neural fate determination. In addition to Tlx3 , many pro-neural genes such as Ngn1/2, Mash-1, and Math1–3 also influence the excitatory/inhibitory fate specification in distinct/different regions of brain. Ngn1 is known to specify a glutamatergic fate in dorsal telencephalon, whereas Mash-1 directs the progenitors towards a GABAergic fate [9]. At the cellular level, correlation expression analyses have shown that during the early phase of neurogenesis, Mash-1 induces GSH1/2, which in turn induces Tlx3 [10]. However, during the late phase of neurogenesis, Mash-1 induces Ptf1α, which in turn suppresses GSH1/2, thereby promoting GABAergic neuron differentiation [10]. These studies have shown that Mash-1 has a context-dependent role in the regulation of Tlx3. Another ubiquitous transcription factor NFY, is also known to induce constitutive expression of Tlx3 [11].
Extracellular signaling mechanisms such as Notch signaling also influence the subtype specification in epiphysis, dorsal spinal cord, and ventral spinal cord, where it regulates the excitatory versus inhibitory neuronal fate specification [12–15]. Peng et al. [14] have clearly demonstrated that the induction of Notch signaling in a cell leads to Scl-dependent activation of inhibitory neuronal differentiation program, whereas the adjacent cell with attenuated Notch leads to a glutamatergic fate. Evidence has also emerged regarding the functional role of Notch signaling in neurotransmitter fate specification by controlling transcription factor expression [10, 16].
From various reports it appears that the expression of Tlx3 has a major role in defining the excitatory versus inhibitory fate of neural progenitors. Even though Tlx3 has been shown to play an important role in excitatory versus inhibitory fate specification, the actual mechanism for regulation of Tlx3 is not clearly understood. Therefore, understanding the mechanisms involved in the regulation of Tlx3 could shed light into its role in excitatory versus inhibitory fate specification of neurons. Here, we have shown for the first time that Tlx3 promoter can be regulated by Hes-1, a downstream component of Notch signaling, thereby regulating the excitatory versus inhibitory fate of neural progenitors. We have used two cell types to understand the regulation of Tlx3 and fate specification. IMR32, a human neuroblastoma cell line, was used to study the regulatory mechanism of Tlx3, since Tlx3 is constitutively expressed in IMR32 cell line. Here, perturbation of Hes-1 indicated its role in regulating Tlx3 expression and was able to directly repress Tlx3 promoter by recruitment of co-repressors and interaction with Tlx3 promoter. Further, the regulatory role of Hes-1 on Tlx3 expression and excitatory versus inhibitory fate specification was demonstrated in embryonic stem cell-derived neural progenitors (ES-NPs), which is an excellent system to study fate specification. Our results demonstrate that Hes-1, which is known to be involved in proliferation of neural progenitors [17], plays an important role in deciding the excitatory versus inhibitory fate of neural progenitors.
Materials and methods
Plasmids and constructs
The Tlx3 promoter (1,310 bp) was PCR amplified (Table 1) from the genomic DNA of human blood and initially cloned into TA cloning system (pTlx3 1310-TA, Supplementary Fig. 1). Tlx3 promoter luciferase (pTlx3 1310-luc) was constructed by directionally cloning Sac1 and Xho1 digested 1,310-bp promoter fragment from pTlx3-TA into promoter-less pGL2 basic vector. Similarly, pTlx3 1310-EGFP was constructed by directionally cloning Sac1 and Xho1-digested promoter fragment from pTlx3-1310 TA in SacI/SalI digested promoter-less EGFP vector. In order to generate the pTlx3-CREM construct, a promoter-less CREM vector was generated by deleting the CMV promoter from CMV-CREM (Addgene # 8395) [18]. Further, the Kpn1-Age1-digested promoter fragment from pTlx3-EGFP was directionally cloned into CREM vector digested with Kpn1 and partially digested with Age1. To specifically analyze the regulatory regions in Tlx3 promoter regulation, deletions were made in the promoter using restriction digestion and PCR amplification. pTlx3 del-592 (−592 to +272 bp) with 864 bp having −592 to +272 bp of promoter sequence having one proximal C site was constructed by removing MluI/SacI fragment from pTlx3 1310-luc and subsequent religation after end filling with Klenow enzyme. pTlx3 del-323 (−323 to +272 bp) having no C sites was constructed by Sma1 digestion of pTlx3 1310-luc and subsequent re-ligation of the ends. pTlx3 del-202 (−202 to +272 bp) was made by blunt-end cloning of 474 bp AfeI/EcoRV-digested promoter fragment from pTlx3-TA into HindIII-digested and end-filled pGL2 basic vector. The orientation and sequence of the insert in the vector was confirmed by restriction digestion and DNA sequencing.
Table 1.
List of primers used
| Gene name | Primer sequence (5′–3′) | Ann temp. (°C) | Product size (bp) | Accession no. |
|---|---|---|---|---|
| β-actin | F-AGACTTCGAGCAGGAGATG | 56 | 322 | NM_007393.2 |
| R-CTTGATCTTCATGGTGCTAGG | ||||
| β-III-tubulin | F-CAACCAGATAGGGGCCAAGTTC | 55 | 290 | NM_023279.2 |
| R-GGCCTGAATAGGTGTCCAAAGG | ||||
| Hes-1 | F-TCAACACGACACCGGACAAAC | 56 | 295 | NM_008235.2 |
| R-TTCATGCACTCGCTGAAGCC | ||||
| vGLUT2 | F-TCGGACAGATCTACAGGGTG | 56 | 345 | NM_080853.3 |
| R-GCGTGATGATATAGCCCCAG | ||||
| Ngn1 | F- GCTTCAGAAGACTTCACCTATG | 56 | 303 | NM_010896 |
| R-TGGAGAAATAGACCGAGGG | ||||
| Mash-1 | F-GAAGATGAGCAAGGTGGAG | 56 | 158 | NM_008553 |
| R-CATAGAGTTCAAGTCGTTGGAG | ||||
| Viaat | F-CATCTCCATTGGCATCATCG | 56 | 276 | NM_009508.2 |
| R-AAGAAGGGCAACGGATAGG | ||||
| Tlx3 promoter amplification | F-AGCTGTGCTTCCCTTGAACTCTCAAAGCC | 57 | 1,310 | – |
| R-GGAAATAGGAGCTTAGGGACTGTTCCAAGGTGAC | ||||
| Tlx3 gene amplification | F-GAACTCGAGATGGAGGCGCCCGCCAGCGCGCAGAC | 57 | 1,000 | – |
| R-GGGCGGCCGCTCACACCAGGGAGGTGACAGCGG |
pCI-Hes-1 was a kind gift from Dr. R. Kageyama (Kyoto University, Kyoto, Japan) [19], pFLAG-NICD was a generous gift from Dr. R. Kopan (Washington School of Medicine, St. Louis, MO, USA) [20] and pFLAG-dn-Hes-1(B*ΔSHes-1) was from Dr. Anderstrom (Karolinska Institute, Sweden) [21]. pFLAG-dn-Hes-1 has E43A, K44A, R47A mutations and the C-terminal truncation was made using an internal SmaI site, and acts as a dominant negative Hes-1. pBDMHes-1 GFP(B*Hes-1), with basic domain mutated Hes-1, was purchased from Addgene (Addgene #15134) [22] with three basic domain mutations E43A, K44A, and R47A, which makes it unable to bind DNA. pΔWRPW Hes-1 was a gift from Dr. Minato, (Kyoto University, Kyoto, Japan) [23] having truncated C-terminal WRPW domain with all other domains intact. Wild-type Hes-1 and B*ΔS Hes-1 was PCR amplified and sub-cloned into Xho1/EcoRV sites of pCAGIG expression vector containing IRES-EGFP (Addgene#11159) [24] in order to track the transfected cells. Similarly, the Tlx3-expressing construct, pCAGIG-Tlx3 was made by PCR amplification of Tlx3 from HeLa cDNA and cloned into the EcoR1 site of pCAGIG expression vector.
Embryonic stem cell culture and neural differentiation
Mouse D3 ES cells (ATCC) were cultured and EBs were generated as described previously [17, 25]. Briefly, proliferating ES cells were grown in 0.1% gelatin-coated plates with DMEM high glucose (Invitrogen) supplemented with 15% defined FBS (Hyclone), 2 mM l-glutamine (Invitrogen), 1× Nucleosides, 0.1 mM β-mercaptoethanol and 1,000 U/ml LIF (Chemicon). The cells were passaged at confluency of about 50% and for neural induction, and embryoid bodies (EB) were generated by RA induction. For neuronal differentiation, RA-induced EBs were plated on poly-d-Lysine (150 μg/ml) and laminin (1 μg/ml)-coated plates in neuronal differentiation medium (DMEM/F12 supplemented with 1% N2 supplement (Invitrogen), 0.5% FBS, Heparin (2 μg/ml) and FGF2 (10 ng/ml) (Chemicon). For ES-NP generation, the cells were allowed to differentiate for 2 days and the partially differentiated EBs were further trypsinized and plated onto uncoated 6-well plates (~1.5 × 106 cells/well) in ES-NP proliferation medium consisting of DMEM/F12 supplemented with 1% N2 supplement, heparin (2 μg/ml) and FGF2 (20 ng/ml; Sigma-Aldrich) [17]. The neurospheres generated were used for all further transfection experiments. Transfection was done using Lipofectamine LTX (Invitrogen) according to the manufacturer’s protocol with ~20% transfection efficiency. The trypsinized cells were incubated with transfection complex for 10 min at room temperature and plated on PDL/laminin-coated cover glasses or 24-well plates for 7 days for differentiation and neuronal fate specification was studied by immunocytochemical or by RT-PCR analysis. Stable Tlx3-Luc-expressing ES-cell line was generated for assaying Tlx3 expression in ES-NPs.
IMR32 neuroblastoma cell culture
IMR32 cells were obtained from Riken BRC Cell Bank, Japan, and expanded in DMEM (Invitrogen) medium supplemented with 10% FBS (Sigma) and 1% non-essential amino acids at 37°C with 5% CO2. The cells were trypsinized at about 70–80% confluency with 0.05% Trypsin. The cells were grown in 24-well plates for luciferase assay and in six-well plates for FACS analysis. Transfections were performed using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s instructions.
Immunofluorescence analysis
Immunocytochemical analysis was done after 7 days of differentiation in neuronal differentiation medium. The cells were washed once in 1× PBS, and fixed in 4% paraformaldehyde for 15 min at 4°C followed by blocking in 5% NGS (Normal Goat Serum; Sigma-Aldrich). The cells were permeabilized with 0.1% Triton-X100 for vGlut2 (1:200, Chemicon) and GABA (1:300, Chemicon) and in 0.2% Triton-X100 for β-III tubulin (1:200, Chemicon) and 0.4% for anti-Tlx3 antibody (1:2,000, gift from Dr. Carmen Birchmeier, Germany), followed by an overnight incubation in primary antibodies at 4°C [17] Cells were examined for epifluorescence following incubation with appropriate secondary antibody conjugated to Cy3/FITC (1:400 Jackson Immunoresearch) in an upright fluorescent microscope (Olympus BX61) and images were captured using a cooled CCD camera (Andor 885). Hes-1 and dnHes-1 transfected cells expressing GFP and having neuronal morphology were selected for analysis. The percentage of GABA/vGlut2-positive cells per transfected GFP expressing cell was quantified by a "blind count" method where the positive/negative cells in a particular field were counted in an unbiased way by two different persons. For statistical analysis, more than ten fields were counted and a graph was plotted to represent the percentage of positive or negative cells. pCAG-EGFP-transfected cells were used as a control to determine the basal differentiation of GABA/vGlut2 neurons.
RT-PCR analysis
For RT-PCR analysis, total RNA was isolated using the Qiagen RNA easy kit (Qiagen). The isolated RNA was treated with DNAse to avoid any DNA contamination and ~2 μg of RNA was reverse transcribed into cDNA using superscript RT as described previously [26]. cDNA of different samples were normalized using β-actin and the specific products were amplified using specific primers (Table 1) on a RoboCycler Gradient 96 (Stratagene, La Jolla, CA, USA).
Dual-luciferase assay
IMR32 cells in 24-well plates were transfected with respective plasmid using Lipofectamine 2000 (Invitrogen) in OPTIMEM medium as per the manufacturer’s instructions. Each transfection was done in triplicate and for co-transfections, the DNA concentration in each tube was normalized by adding control pCI vector. After 8 h of transfection, the medium was replaced with fresh IMR32 medium and incubated at 37°C for a further 48 h. The cells were then lysed as per the manufacturer’s protocol (Promega) and luciferase assay was performed in a luminometer (TD 20/20 Luminometer) with dual luciferase mode. Each experiment was done in triplicate and the firefly luciferase values were normalized using Renilla luciferase values and a graph was plotted with these normalized values [17].
Measurement of histone-deacetylase activity
Trichostatin A (TSA) was used to inhibit histone deacetylase activity. For this Hes-1 transfected IMR32 cells were treated with Trichostatin A (0.05 μM), an HDAC inhibitor, for 8 h before luciferase assay to examine the effect of histone deacetylation. The following constructs were used to measure histone deacetylase activity. (a) pTlx3 1310-Luc alone (b) pTlx3 1310-Luc + pCI-Hes-1 and (c) pTlx3 1310-Luc + pCI-Hes-1 + TSA. Luciferase activity was measured and a graph was plotted for each plasmid transfection combination and TSA treatment.
FACS analysis for Tlx3 promoter activity
The Tlx3 promoter-driven d2EGFP (destabilized EGFP) reporter system was used for FACS analysis of Tlx3. The half-life of d2EGFP was less than 2 h, and thus the changes in the activation or repression of the promoter could be studied using this reporter system. For FACS analysis, IMR-32 cells were seeded on six-well plates and cells were transfected using Lipofectamine 2000 as described previously. To examine the effect of other genes, 3 μg of expression plasmids such as pCI-Hes-1, pFLAG-NICD was co-transfected along with 1 μg of pTlx3 1310-d2EGFP. The amount of transfected DNA was normalized in the Tlx3 promoter control wells by co-transfecting with the empty expression vector. After 72 h of transfection, the cells were trypsinized and analyzed for EGFP expression in a BD FACS Aria Flow Cytometer (BD Biosciences, USA). Un-transfected IMR32 cells were used as the negative control and transfected cells with constitutive GFP expression (GFP under the control of CAG promoter) were used as the positive control and accordingly quadrants were selected for analysis [17].
Statistical analysis
Statistical significance between the groups was calculated by independent Student's t test assuming equal variance. Values with p < 0.05 were considered as statistically significant.
Results
Tlx3 favors glutamatergic over GABAergic differentiation in ES cell-derived neural progenitors
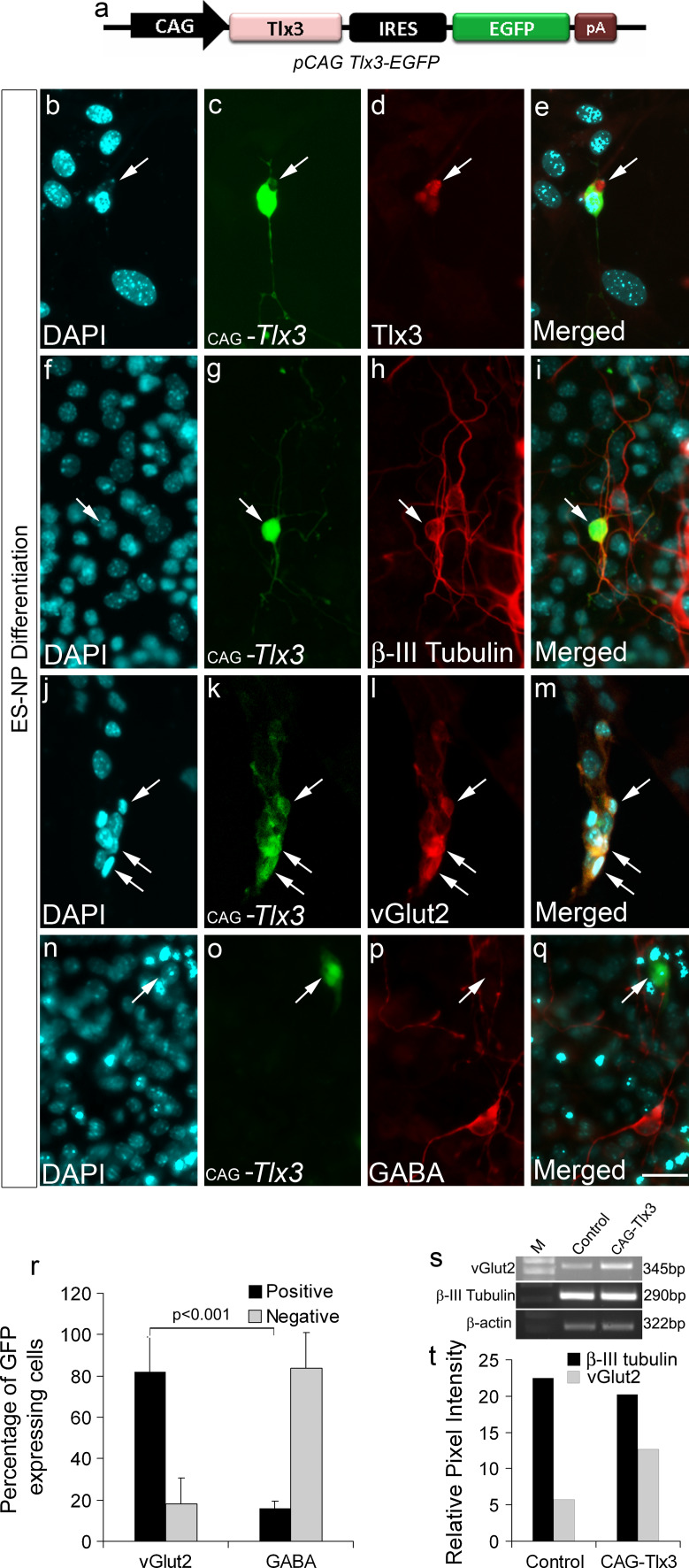
Tlx3 is a selector gene known to induce glutamatergic fate in developing neurons in the nervous system along with simultaneous down regulation of GABAergic differentiation [7, 27, 28]. The function of Tlx3 in excitatory fate specification has been characterized in detail, but the molecular determinants causing its activation or repression are not yet clearly understood. Since Tlx3 is known to play a crucial role in neuronal sub-type specification, our interest was to understand how Tlx3 is regulated during neural differentiation. Therefore, we first went ahead and confirmed the role of Tlx3 in determination of excitatory versus inhibitory fate in ES-NPs. For this, ES-NPs were generated from ES cells as previously described [17, 25] (see "Materials and methods"). For over-expressing Tlx3, we cloned the coding region of Tlx3 gene under the control of CAG promoter in an EGFP expression vector (Fig. 1a, pCAG-Tlx3-EGFP). The pCAG Tlx3-EGFP construct was transiently transfected into ES-NPs and allowed to differentiate for 7 days as described earlier (see "Materials and methods"). Expression of Tlx3 in transfected cells was confirmed with anti-Tlx3 antibody (Fig. 1b–e). Our results clearly show that Tlx3-expressing cells differentiated into neurons and expressed immature neural differentiation marker, β-III tubulin (Fig. 1f–i). Further up-regulation of Tlx3 induced glutamatergic differentiation (86.95 ± 12.60%) as evidenced with vGlut2 expression (Fig. 1j–m, r). These results were further confirmed with RT-PCR analysis (Fig. 1s, t). The majority of Tlx3-expressing cells were negative for GABA (87.61 ± 17.03%; Fig. 1n–q, r). In our experiment, we have also included pCAG-EGFP transfection as a control to check the percentage of glutamatergic and GABAergic neurons generated under normal differentiation conditions. Our results showed that pCAG-EGFP transfected ES-NPs differentiated into equal proportion of glutamatergic and GABAergic neurons, respectively (Supplementary Fig. 2). Therefore, from the above data, it was confirmed that Tlx3 was able to promote glutamatergic differentiation and decrease GABAergic differentiation from ES-NPs, similar to that observed in the nervous system.
Fig. 1.
Up-regulation of Tlx3 in ES-NPs promotes a glutamatergic fate: a Schematic of Tlx3 expression construct, where Tlx3 is expressed under the control of CAG promoter with IRES-EGFP so that the transfected cells will be expressing GFP. b–e Authenticity of Tlx3 expression construct was validated by anti-Tlx3 immunostaining which showed Tlx3 expression in transfected cells. f–i Tlx3-expressing cells were positive for neuronal marker β-III tubulin, thus confirming their neuronal nature. j–q Tlx3-expressing cells were vGlut2-positive and were negative for GABA. r Quantitative immunocytochemical analysis of vGlut2 and GABA in Tlx3 over-expressed cells showed a significantly high percentage (p < 0.001) of cells undergoing glutamatergic differentiation compared to GABAergic differentiation. s–t RT-PCR analysis showed increased vGlut2 expression in Tlx3 over-expressed ES-NPs. Data are expressed as mean ± SD of triplicates (n = 3) from three different experiments. Scale bar = 50 µM
Hes-1-binding C sites are critical for the regulation of Tlx3 promoter
In order to understand the molecular regulation of the Tlx3 promoter, we first conducted an in silico analysis of the Tlx3 promoter. The Tlx3 gene was found to have three exons and two introns [10] with the transcription start site lying 272 bp upstream of ATG (Fig. 2a). A putative TATA box was also identified upstream of Transcription Start Site (TSS) by neural network promoter prediction software (http://www.fruitfly.org/seq_tools/promoter.html). Sequence analysis with TRANSFAC Version 1.3 motif finder indicated the presence of binding sites for transcription factors such as NFY, Nkx2.5, AP-1, VMyb, NF-1 and Pbx-1. A very interesting finding was that in addition to these sites, the promoter region also possessed four E boxes, one homeodomain binding ATTA sequence [29], two N boxes, and three C sites (Fig. 2a). These N boxes and class C sites are known possible Hes-1 binding sites in Hes-1 target genes such as Mash-1 and NeuroD [30] and E boxes are possible bHLH activator-binding sites. These possible binding sites in Tlx3 promoter suggested that it can be regulated both positively by bHLH activators and negatively by Hes-1, a downstream component of Notch signaling.
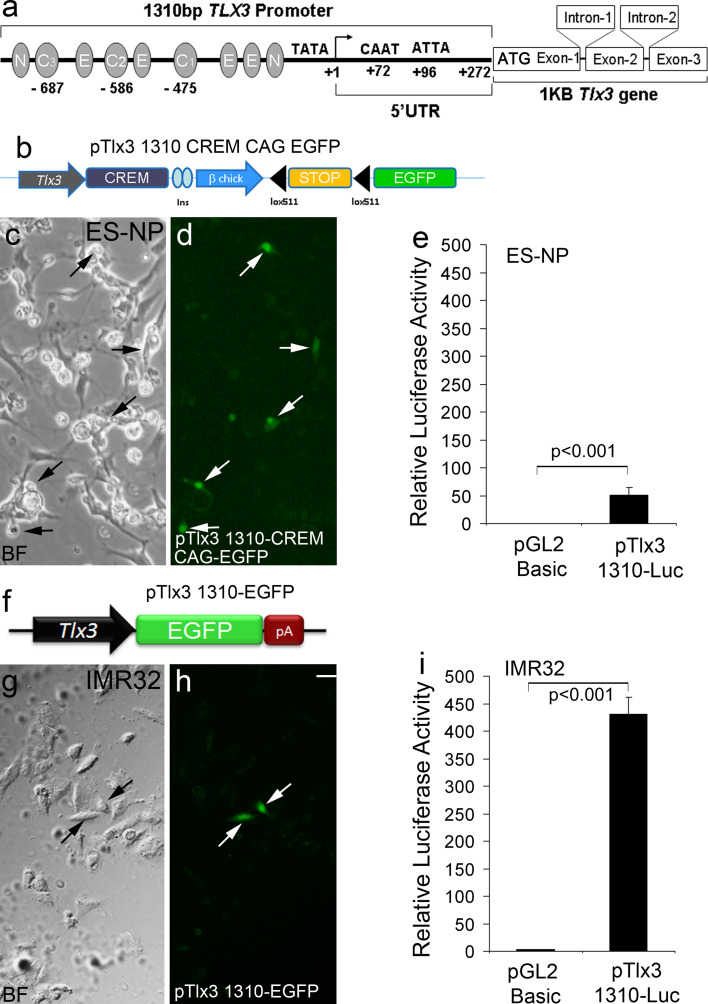
Fig. 2.
Functional analysis of the Tlx3 promoter in ES-NPs and IMR32 cell lines: a Schematic of the Tlx3 promoter with different transcription factor-binding sites. b Tlx3-driven GFP expression cassette used to transfect ES-NPs. Since the level of expression of Tlx3 is low, we enhanced the visualization of Tlx3 expression using a Cre-lox construct, pTlx3 1310-CREM-CAG-EGFP. c–d ES-NPs upon differentiation showed GFP expression indicating functional activity of the cloned Tlx3 promoter. e Luciferase activity in ES-NPs upon transient transfection of pTlx3-1310-Luc in differentiated ES-NPs showed a significant increase (p < 0.001) compared to control. f Schematic of Tlx3 promoter-GFP reporter construct used to study Tlx3 expression in the IMR32 cell line. g–h Transient transfections with pTlx3-1310-EGFP construct showed GFP-positive cells, indicating Tlx3 expression in IMR32 cell line. i This was again confirmed by assaying Tlx3 promoter activity, which showed a significant increase (p < 0.001) in luciferase expression compared to the control. Significantly high promoter activation indicates a higher level of expression of Tlx3 in IMR32 cell line compared to ES-NPs. Data are expressed as mean ± SD of triplicates (n = 3) from three different experiments. Scale bar = 20 µM
With this information, we went ahead and cloned a 1,310-bp Tlx3 promoter region upstream of ATG from human blood (Supplementary Fig. 1). The amplified promoter was cloned into TA cloning vector and further sub-cloned into pGL2 and pEGFP vectors. To check the activity of the cloned 1,310-bp Tlx3 promoter, we transfected the pTlx3 1310-GFP vector into ES-NPs, since we did not see any expression of EGFP that led us to speculate that Tlx3 might be expressed at a very low level or in a very narrow window in post-mitotic neurons that are just entering differentiation. Therefore, to amplify the weak GFP expression from the Tlx3 promoter, we cloned the 1,310-bp Tlx3 promoter into pTlx3 CREM-EGFP vector and used this to transfect ES-NPs. Here, the very low level of activation of Tlx3 promoter will induce the expression of Cre, which will loop out the STOP sequence flanked by lox sites, thereby constitutively expressing EGFP under the control of CAG promoter (Fig. 2b). Therefore, the cells that express any small level of Tlx3 during a small window will express EGFP thereafter. Our results showed the expression of EGFP in ES-NPs transfected with this construct (Fig. 2c–d), thereby confirming the functional integrity of the cloned Tlx3 promoter and its expression in ES-NPs. However, when we analyzed the luciferase activity after transfecting ES-NPs with pTlx3 1310-luc, we found a significant increase (p < 0.001) in activity compared to the basal level (pTlx3 1310-luc, 50.66 ± 13.29; pGL2, 0.20 ± 0.05; Fig. 2e). From these results, it is clear that the level of Tlx3 expression in ES-NPs is not always the same in all the cells and it would be ideal to have a constant expression of Tlx3 in a cell system to study the regulation of the Tlx3 promoter. Therefore, for the functional characterization, and to study the regulatory motifs in Tlx3 promoter, we used a human neuroblastoma cell line, IMR32 that had a constitutively high Tlx3 expression compared to ES-NPs (IMR32, 431.40 ± 30.54; ES-NP, 50.66 ± 13.29) and would be an ideal system to study the promoter regulation (Fig. 2f–i).
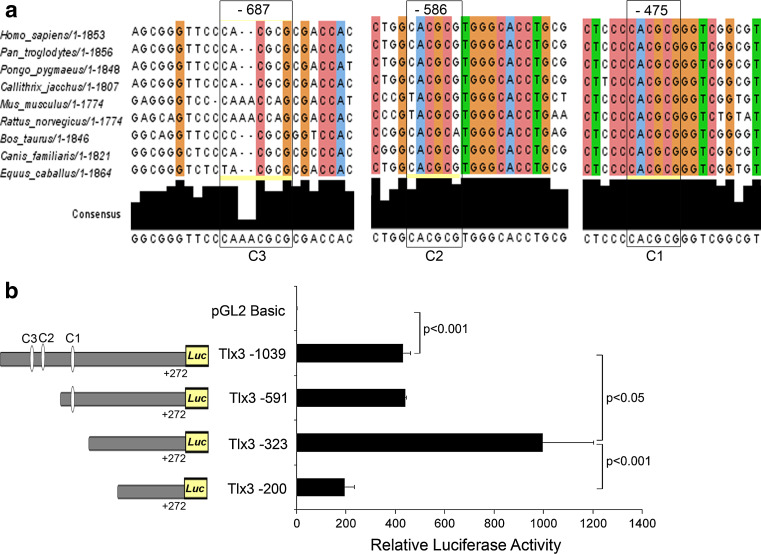
Since our initial in silico analysis indicated the presence of Hes-1-binding sites in Tlx3 promoter, we further checked the degree of conservation of the C-sites among different species. Subsequent analysis showed that Hes-1-binding C sites were conserved among various mammalian species (Fig. 3a). Of the different Hes-1-binding C sites, the proximal C1 site at −475 showed 100% conservation among different species analyzed all though the other C sites were also found to be conserved to a lesser extent among different species. The C2 site at −586 position showed 66.6% and C3 site at −687 showed 55.5% conservation (Fig. 3a). The higher degree of conservation of these C sites indicates that Hes-1 may have a conserved role in Tlx3 regulation. We further made 5′ deletion constructs with and without the C-sites in Tlx3 promoter. All deleted fragments of Tlx3 promoter were analyzed for luciferase activity in IMR32 neuroblastoma cell line, where Tlx3 is constitutively expressed. Our results showed that Del-591, devoid of the distal 447 bp including two C sites (C2 and C3), did not show any significant difference in the activity compared to the full-length 1,310-bp promoter (Tlx3 1,310, 431.40 ± 30.54 and Tlx3 del-591, 441.15 ± 4.03; Fig. 3b). Interestingly, truncation of an additional 268 bp (Tlx3 del-323), which deleted the proximal C1 site (−475 bp), showed significantly higher luciferase activity (Tlx3 1310-luc, 431.40 ± 30.54; Tlx3 del-323, 997.35 ± 205.55; p < 0.05; Fig. 3b) when compared to the full-length promoter. However, Tlx3 del-200 with a further deletion of 123 bp showed a significant reduction in promoter activity compared to the previous deletions (Tlx3 del-200, 194.74 ± 3 8.75; Tlx3 del-323, 997.35 ± 205.55; p < 0.01; Fig. 3b), since this region was highly conserved among species and a reduction of the promoter activity indicated the presence of positive regulatory elements in this region. In silico analysis of the 123-bp fragment deleted from Del-200 showed the presence of binding sites for NFY, which is a known constitutive activator of Tlx3. These results showed that Tlx3 promoter has both positive and negative regulatory regions. Since the promoter activity was increasing upon deletion of the proximal C1 site, we assume that this proximal C1 site may be the physiologically active binding site for Hes-1 involved in the repression of Tlx3 promoter. Therefore, we assume that the Tlx3 promoter is maintained in a constitutively active manner by NFY and Hes-1 is able to negatively regulate its expression by binding to the C1 site.
Fig. 3.
Hes-1-binding C sites are involved in regulation of Tlx3 promoter: a C1, C2 and C3 sites showed 100, 66.6, and 55.5% consensus among different mammalian species. Consensus was analyzed using the software "Jalview version 2". b Tlx3 1310 promoter with all the three C sites showed a significant increase (p < 0.001) in luciferase activity compared to the control. Deletion of C2 and C3 sites did not show any significant reduction in luciferase activity, but deletion of the proximal C1 site (Del-323) significantly increased (p < 0.05) the luciferase activity indicating that this C-site is critical for the repression of the Tlx3 promoter by Hes-1. Further deletion of 123-bp (Del-200) resulted in a significant reduction (p < 0.001) in activity compared to Del-323, indicating that this 123-bp region might be involved in the possible activation of Tlx3. Data are expressed as mean ± SD of triplicates (n = 3) from three different experiments
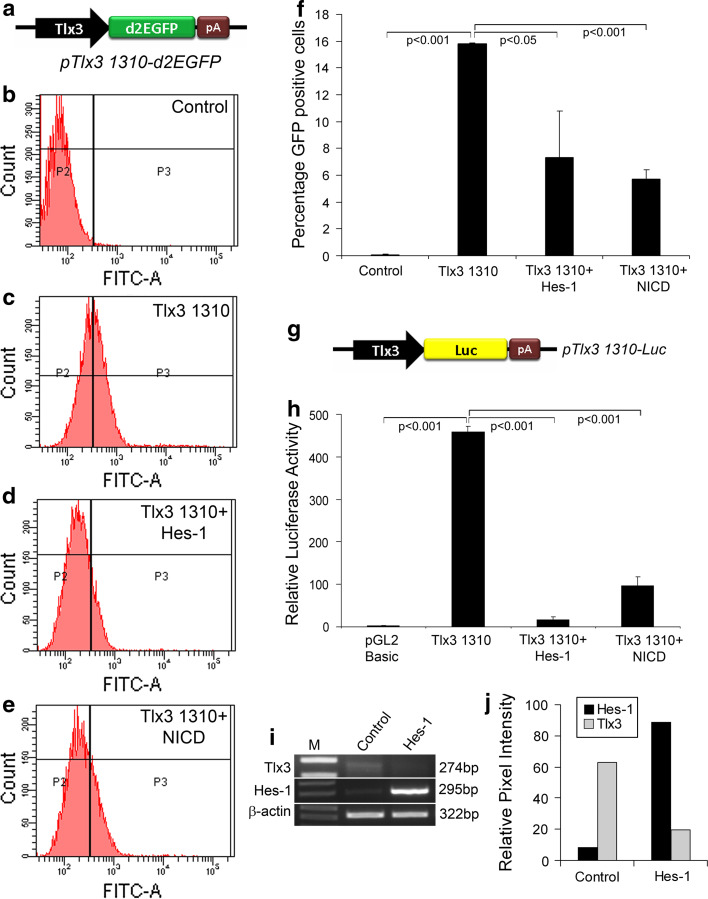
Hes-1 negatively regulates Tlx3 promoter activity
In order to further analyze the regulatory role of Hes-1 on Tlx3 promoter, we transiently over-expressed Hes-1 (pCI-Hes-1) in the IMR32 cell line along with Tlx3 promoter reporter system (pTlx3 1310-d2EGFP; Fig. 4a). The transfected cells were further subjected to FACS analysis. Here, d2EGFP was used as a reporter, since it has a half-life of less than 2 h, and will effectively show any short-term variation in expression of Tlx3. FACS analysis of cells transfected with pTlx3-d2EGFP showed a drastic increase in cells expressing GFP compared to the controls (control, 0.07 ± 0.06 and pTlx3 1310-d2EGFP, 15.8 ± 0.05, p < 0.001; Fig. 4b–c, f). The expression of GFP was significantly reduced with co-expression of Hes-1 compared to those transfected with pTlx3 1310-d2EGFP alone (pTlx3 1310-d2EGFP, 15.8 ± 0.05 and pTlx3 1310-d2EGFP + Hes-1, 7.30 ± 3.40, p < 0.001; Fig. 4d, f). Since Hes-1 is a direct target gene of Notch signaling, we next wanted to know whether activation of Notch signaling itself could down-regulate Tlx3 expression. Therefore, we activated Notch by constitutively over-expressing Notch-Intra cellular domain (NICD) along with the Tlx3 promoter construct. As expected, NICD significantly reduced the number of GFP-expressing cells compared to those transfected with the Tlx3 promoter alone (pTlx3 1310-d2EGFP, 15.8 ± 0.05 and pTlx3 1310-d2EGFP + NICD, 5.70 ± 0.69, p < 0.001; Fig. 4e, f). This was further confirmed with luciferase assay for which the IMR32 cell line was transfected with pTlx3 1310-Luc alone (Fig. 4g), and in combination with Hes-1 and NICD. To rule out any possible promoter competition, all samples were transfected with an empty vector. The results obtained from luciferase assay were exactly the same as those obtained from our FACS analysis showing a significant increase in Tlx3 promoter activity compared to the control (control, 3.32 ± 0.15; pTlx3 1310-Luc, 459.38 ± 12.20, p < 0.001; Fig. 4h). The increased Tlx3 promoter activity was significantly reduced when the cells were co-transfected either with Hes-1 or NICD (pTlx3 1310-Luc, 459.38 ± 12.20; pTlx3 1310-Luc + Hes-1, 17.88 ± 5.50; pTlx3 1310-Luc + NICD, 97.50 ± 19.95, p < 0.001; Fig. 4h). These results were further corroborated with RT-PCR analysis, which showed a significant reduction in expression of Tlx3 in Hes-1 over-expressed cells (Fig. 4i, j). Thus, our data clearly suggest that Hes-1 through canonical Notch signaling is able to negatively regulate the expression of Tlx3.
Fig. 4.
Hes-1 acts as a repressor of Tlx3 promoter: a Schematic of Tlx3 promoter-driven d2EGFP having a half-life of 2 h so that the change in Tlx3 expression is accurately reflected by the GFP expression. b–e FACS analysis of IMR32 cells transfected with pTlx3 1310-d2GFP in the presence or absence of Hes-1 or NICD. f Graph depicting the percentage of GFP-positive cells obtained with FACS analysis. Transfection with pTlx3-1310-d2EGFP alone significantly increased (p < 0.001) the percentage of GFP-expressing cells, whereas co-transfection with Hes-1 and NICD significantly reduced (p < 0.05 and p < 0.001, respectively) the percentage of GFP-expressing cells. g Schematic of Tlx3 promoter-driven luciferase reporter system. h Luciferase assay of Tlx3 promoter showed a significant increase (p < 0.001) in activity compared to control. Expression of Hes-1 alone with Tlx3 promoter showed a significant decrease (p < 0.001) in Tlx3 promoter activity. i–j RT-PCR analysis of Tlx3 in IMR32 cells showed a reduction in Tlx3 expression upon Hes-1 transfection. Data are expressed as mean ± SD of triplicates (n = 3) from three different experiments
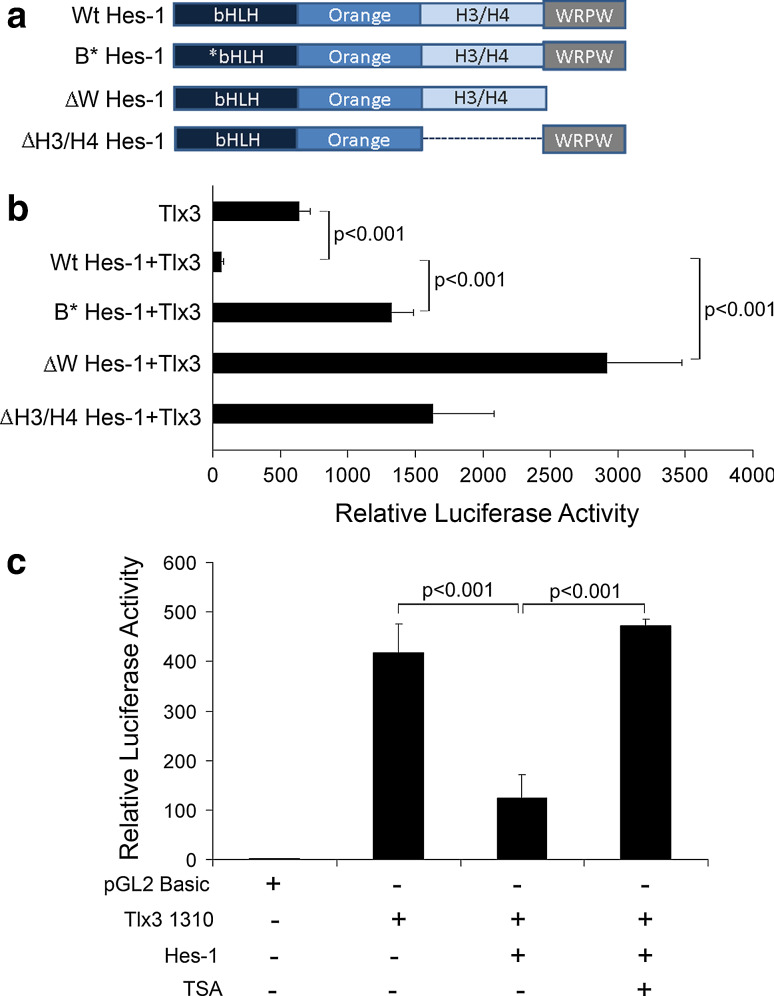
Co-repressor recruitment through WRPW-domain along with DNA binding regulates Tlx3 promoter
From our previous results, it was clear that Hes-1 negatively regulates the expression of Tlx3 gene, possibly by binding to the C1 site (−475) in the Tlx3 promoter. To understand the actual mechanism of repression by Hes-1, we generated a series of deletion constructs of Hes-1, which will determine the functional domains of Hes-1 required for interaction with Tlx3 promoter (Fig. 5a). Wild-type Hes-1 has different functional domains such as the basic HLH domain, the orange domain (H3/H4 domain), and the WRPW domain. The functional role of different domains of Hes-1 was analyzed by luciferase assay in IMR32 cells transfected with mutated/truncated Hes-1 constructs along with Tlx3-Luc vector and Tlx3 promoter activity was measured and compared with the wild-type Hes-1.
Fig. 5.
WRPW domain mediated-protein interaction along with DNA- binding and histone deacetylase activity is involved in regulation of Tlx3 promoter: a Schematic representation of different functional domains of Wt type Hes-1 and different truncated/mutated Hes-1 proteins used in this study. B*Hes-1 represents basic domain-mutated Hes-1 and thus is unable to bind target DNA. However, all of the other domains are intact so that it can interact with other proteins and is able to recruit co-repressor proteins and can dimerize with other bHLH factors. ∆W Hes-1 represents WRPW domain truncation with deletion of extreme C-terminal four amino acids. ∆W Hes-1 is able to carry out all functions of Hes-1 except co-repressor recruitment. ∆H3/H4 Hes-1 indicates H3/H4 domain truncated Hes-1, where protein–protein interactions may be affected. b Luciferase activity was measured with co-transfection of the above-mentioned Hes-1 constructs along with pTlx3 1310-Luc to study the contribution of different functional domains of Hes-1 in Tlx3 promoter regulation. The Wt type Hes-1 significantly repressed (p < 0.001) Tlx3 promoter activity, whereas B*Hes-1 significantly reduced (p < 0.001) the repression compared to Wt type, indicating a function role of DNA binding in Tlx3 promoter regulation. Again, ∆W Hes-1 with WRPW domain truncation showed a significant activation/de-repression (p < 0.001) of the promoter, indicating an active co-repressor recruitment role in Tlx3 promoter regulation. ∆H3/H4 domain truncated Hes-1 also did not show any repression indicating functional protein interactions even with co-repressors in Hes-1-mediated repression. c Treatment with deacetylase inhibitor, Trichostatin A (TSA) resulted in the abolishment of inhibition caused by Hes-1. These results indicated that histone deacetylase activity is one of the mechanisms through which Hes-1 represses the Tlx3 promoter
First, we looked at the role of basic domain on repression of Tlx3 promoter. For this, the basic DNA-binding domain of Hes-1 was mutated, leaving the rest of the domains intact. We observed a significant de-repression of Tlx3 promoter activity compared to Wt type Hes-1-transfected cells (control, 643.55 ± 78.51, Wt Hes-1, 63.11 ± 13.46, and B*Hes-1, 1327.15 ± 156.9, p < 0.001; Fig. 5b). These results suggested that DNA-binding is required for the repression of Tlx3 expression by Hes-1, similar to that reported with other Hes-1 target genes such as NeuroD and Mash1 etc. [30, 31]. These results are in agreement with our previous results (Fig. 3b) that showed increased promoter activity with deletion of Hes-1-binding C1 site on Tlx3 promoter.
Next, we co-transfected WRPW domain truncated Hes-1 (∆W Hes-1) along with pTlx3 1310-Luc. Interestingly, the absence of WRPW domain of Hes-1 resulted in a significant increase in luciferase activity, which was more than that of control (∆W Hes-1, 2,920.95 ± 552.9; Tlx3 promoter alone control, 643.55 ± 78.50, p < 0.001; Fig. 5b). WRPW (Trp–Arg–Pro–Trp) domain of Hes-1 is known to interact with or recruit TLE class of co-repressors for interaction with the promoter DNA [32]. These results indicated that recruitment of TLE co-repressors through protein–protein interaction is extremely critical for the complete repression of Tlx3 promoter. TLE classes of co-repressors are known to exert their repressive effects only in a context-dependent interaction with DNA-binding proteins [33]. Though absence of repression might be due to the inability of this truncated Hes-1 to recruit the co-repressors, the mechanism behind the increased activity of the promoter was not clear.
We also looked at the role of the H3/H4 domain, which is also reported to have a significant role in recruiting co-repressors through protein–protein interactions and dimerization. Our results indicated that H3/H4 domain truncated Hes-1 (∆H3/H4 Hes-1) did not show any repression of Tlx3 promoter compared to the control (∆H3/H4 Hes-1,1, 634.25 ± 452.65; Tlx3 promoter alone control, 643.55 ± 78.50; Fig. 5b), even though the bHLH domain and WRPW domain were intact, indicating that the H3/H4 domain also has an important functional role in Tlx3 repression. Therefore, our results suggested that Hes-1 is capable of carrying out repression of the Tlx3 gene by binding to Tlx3 promoter along with recruitment of TLE co-repressors, as indicated with repression of other Hes-1-target genes [23, 34, 35].
Since the mechanism of repression in the Tlx3 promoter also involved protein interaction with TLE class of co-repressors, we further wanted to know whether histone modification functions of TLE can regulate the expression of Tlx3. Previous reports have shown that TLE-mediated repression of target genes can happen due to histone deacetylase (HDAC) activity [36].
Therefore, we further analyzed the histone deacetylase modification during Hes-1-mediated repression of Tlx3 promoter by inhibiting HDACs with 0.05 µM TSA for 8 h followed by luciferase assay. Our results showed that repression caused by Hes-1 was abolished by TSA treatment (Tlx3 promoter control, 417.46 ± 58.55; Hes-1, 124.69 ± 46.39; Hes-1 + TSA 472.60 ± 13.18; Fig. 5c). These results suggested that HDAC, a member of co-repressor complex recruited by Hes-1, play an important role in Tlx3 repression. By recruiting HDACs, Hes-1 modifies histones to keep the chromatin in a closed confirmation so that the transcriptional machinery is not able to access the promoter. These results also point out that the increased promoter activity with ΔWRPW domain may be due to absence of WRPW, which was deficient in recruiting HDAC. Thus, ΔWRPW Hes-1 over-expression may remove the very low endogenous level of deacetylation of the promoter by endogenous Hes-1. Tlx3 is supposed to be constitutively activated by NFY-mediated basal transcriptional machinery, but at some point, histone deacetylase activity mediated by Hes-1 makes Tlx3 promoter inaccessible for the general transcriptional machinery. Hence, we assume that tissue-specific expression of Tlx3 occurs by the reversal of this protein DNA complex. These complex mechanisms may also involve other factors and pathways, which have to be investigated further.
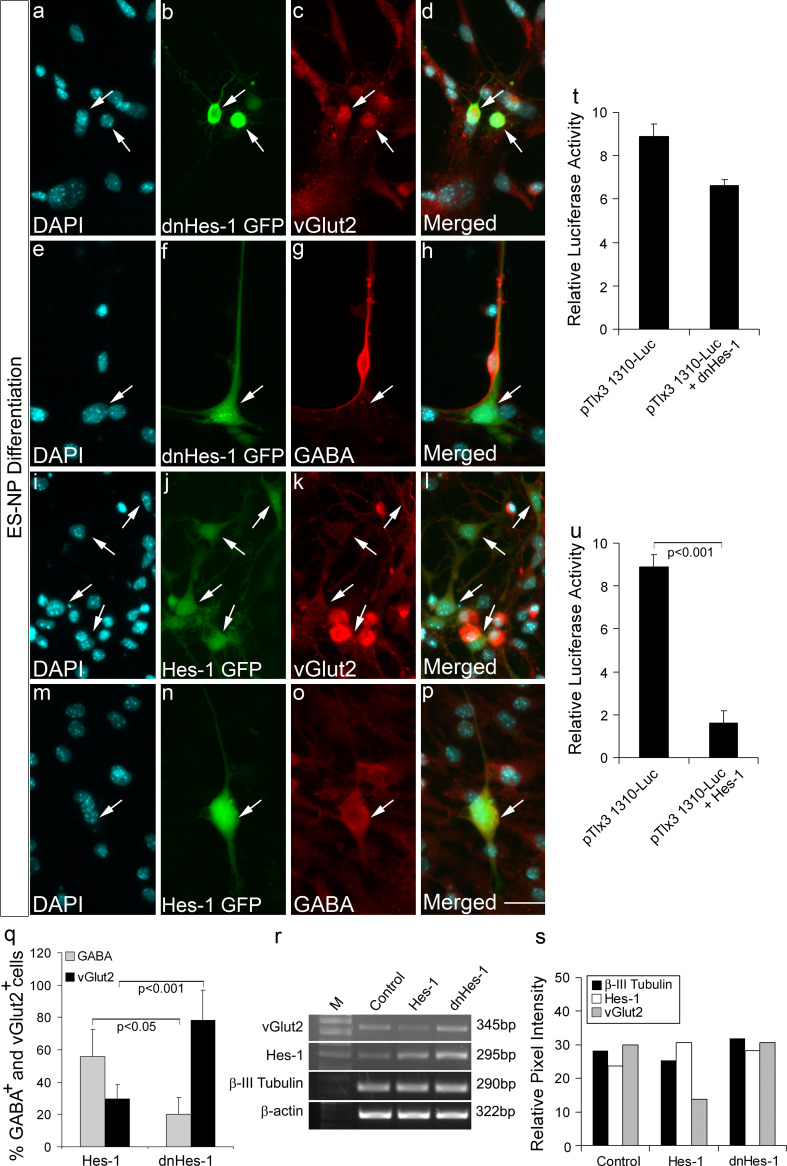
Hes-1 suppresses glutamatergic fate and favors GABAergic fate in ES-NPs during differentiation
Since we now know that Hes-1 can repress the Tlx3 promoter, we next wanted to confirm the role of Hes-1 in suppressing the excitatory fate of ES-NPs. From our previous results, it was clear that Tlx3 over-expression could cause a shift to glutamatergic fate in ES-NPs (Fig. 1j–m, r–t). Therefore, we next analyzed the functional role of Hes-1 in fate specification of ES-NPs using transient transfection with Hes-1 and dnHes-1 constructs and the neurotransmitter fate was analyzed after differentiation. For this, the coding sequence of Hes-1 and dominant negative Hes-1 was amplified and cloned into pCAGIG expression vector under the control of CAG promoter with IRES-GFP cassette to track the transfected cells. The potential of dnHes-1 construct to down-regulate Hes-1 activity was previously confirmed in ES-NPs (Supplementary Fig. 3). We further analyzed whether the transfected cells are differentiating into neurons by immunohistochemical analysis with anti β-III tubulin antibody. We observed that both dnHes-1 and Hes-1-transfected cells differentiated into almost equal proportion of β-III tubulin-positive cells (Supplementary Fig. 4). We further analyzed the transfected cells for expression of glutamatergic or GABAergic markers after the cells were differentiated into neurons. The majority of the dnHes-1-transfected GFP-expressing cells differentiated into vGlut2 immunoreactive cells (Fig. 6a–d, q; 78.08 ± 18.87%) along with a significant reduction in GABA-expressing cells (Fig. 6e–h, q; 20.23 ± 10.05%). Therefore, it appears that down-regulation of Hes-1 expression removed the repression on Tlx3 promoter (Fig. 6t), thereby inducing a significant increase in vGlut2-positive cells (p < 0.001; Fig. 6q) compared to those with increased Hes-1 expression. However, the majority of the differentiated Hes-1-expressing cells were positive for GABA (Fig. 6m–p, q; 56.00 ± 16.45%) with reduced vGlut2-positive cells (Fig. 6i–l, q; 29.50 ± 8.81%). To check the expression of Tlx3, we carried out Tlx3 promoter analysis as ascertained by luciferase activity, which suggested that the expression of Hes-1 significantly repressed Tlx3 promoter (p < 0.001; Fig. 6u) and significantly increased the percentage of GABA-positive cells (p < 0.05; Fig. 6q), compared to cells with reduced Hes-1 expression. We have used luciferase assay for determining the expression of Tlx3 in presence of Hes-1 and dnHes-1 since we assume that the expression of Tlx3 varies in the ES-NPs depending on the stage of differentiation, moreover the expression level of Tlx3 in these cells is very low. Therefore, to have a robust assessment of the Tlx3 expression in ES-NPs, we used Tlx3 promoter-driven luciferase assay to confirm the repression of Tlx3 by Hes-1. Further, the immunocytochemical results were corroborated by RT-PCR analysis, which showed a significant decrease in the expression of transcripts corresponding to vGlut2 in Hes-1 over-expressed cells with an increase in the vGlut2 expression in dnHes-1 over-expressed cells (Fig. 6r, s). The amplification of Hes-1 was increased by transfection of Hes-1 and dnHes-1 since our Hes-1 primer was designed to amplify both Hes-1 and dnHes-1 thereby confirming its up-regulation in the transfected cells (Fig. 6r, s; Supplementary Fig. 6). Further, neural differentiation of Hes-1 and dnHes-1 transfected cells was assessed by the expression of Ngn1 and Mash-1 and that of GABAergic differentiation by the expression of Viaat (Supplementary Fig. 6). Here, we have shown an increase in Mash-1 and Viaat expression in cells transfected with Hes-1 (Supplementary Fig. 6). The increase in expression of Hes-1 could probably block the expression of Tlx3 through a parallel pathway, where the increased Mash-1 could induce Ptf1α, which in turn could suppress GSH1/2, thereby reducing Tlx3 and promoting GABAergic differentiation [10]. Our results also show a discrepancy in the endogenous expression of Hes-1 (Fig. 6r) and the repression (not significant) found with dnHes-1 transfection (Fig. 6t). Ideally, there should not be any repression of Tlx3 promoter when transfected with dnHes-1 (Fig. 6t). The repression that we see may be due to the fact that Hes-1 is not constitutively expressed in ES cells (even though we have Hes-1 expression in control; Fig. 6r). Hes-1 expression undergoes a periodic oscillation in about ~2 h interval by an auto-feedback regulation at the single-cell level. Therefore, in our ES-NPs also, all the cells may not have the same level of Hes-1 expression. This could be a reason for the repression seen with transfection of dnHes-1 compared to the control, which definitely needs further investigation. Further, the results on fate specification of ES-NPs were exactly in corroboration with our previous results with IMR32 cell line. We suggest that down-regulation of endogenous Hes-1 will de-repress the Tlx3 promoter, thereby favoring a glutamatergic fate, whereas up-regulation of Hes-1 will repress Tlx3 promoter leading to the down-regulation of glutamatergic fate favoring GABAergic differentiation.
Fig. 6.
Functional role of Hes-1 in glutamatergic versus GABAergic fate specification of neural progenitors: dnHes-1 GFP and Hes-1 GFP constructs were transfected in ES-NPs and were allowed to differentiate further for 6 days. a–h dnHes-1 GFP-transfected cells showed robust glutamatergic differentiation as evidenced by expression of vGlut2 and were negative for GABA. i–p The majority of Hes-1 GFP-transfected cells were negative for vGlut2 and were positive for GABA. These results show that down-regulation of Hes-1 removes the repression on Tlx3 promoter leading to robust glutamatergic differentiation. q Graph represents quantitative analysis of percentage of glutamatergic and GABAergic neurons differentiated in the presence of Hes-1 and dnHes-1. ES-NPs transfected with dnHes-1 showed a significant increase (p < 0.001) in vGlut2-positive cells compared to those transfected with Hes-1, whereas ES-NPs transfected with Hes-1 showed a significant increase (p < 0.05) in GABA-positive cells compared to those transfected with dnHes-1. r–s RT-PCR analysis showed decreased vGlut2 expression in the presence of Hes-1 compared to the control and enhanced Hes-1 expression in the presence of dnHes-1 in differentiated ES-NPs. The expression of Hes-1 was increased by transfection of Hes-1 and dnHes-1 since our Hes-1 primer was designed to amplify both Hes-1 and dnHes-1, there by confirming its up-regulation in the transfected cells. Expression of β-III tubulin in all the conditions indicates robust neuronal differentiation. t Luciferase activity in ES-NPs transfected with dnHes-1 does not show a significant decrease in Tlx3 expression compared to the control. The ES-NPs used for this experiment had stable integration of pTlx3 1310-Luc. u Luciferase activity in ES-NPs transfected with Hes-1 showed a significant decrease (p < 0.001) in Tlx3 expression compared to control. In panel a–r the progenitors were transfected with Hes-1 and dnHes-1 alone, whereas in panels t–u the progenitors having stable integration of Tlx3 promoter-Luciferase construct were transfected with Hes-1 and dnHes-1 vectors. Data are expressed as mean ± SD of triplicates (n = 3) from three different experiments. Scale bar = 50 µM
In addition to Tlx3 and Hes-1, there may be other factors that are responsible for pushing the cells toward a GABAergic fate. The different transcription factors involved along with Tlx3 in this process have to be characterized further in order to get a full picture of glutamatergic versus GABAergic differentiation. These results obtained with ES cells can be extrapolated to embryonic neurogenesis, since ES-NPs are very similar to primary neurospheres [37]. The elucidation of actual mechanism of glutamatergic versus GABAergic differentiation will lead to excellent therapies against various neurological disorders with imbalance in glutamatergic and GABAergic neurons. These results also allow fate-specific neuronal differentiation from in vitro neurosphere cultures, which can again be used for therapeutic purposes. In conclusion, we have shown for the first time that the Notch target gene Hes-1, which is well known for its role in neuronal progenitor proliferation and maintenance, has a unique regulatory role in excitatory versus inhibitory fate specification.
Discussion
Glutamatergic/GABAergic differentiation is a fine-tuned process occurring along with neurogenesis during neuronal differentiation [4]. This process could either be modulated or selected by several factors including proneural genes such as Ngn1, Mash1, or/and by homeodomain transcription factors such as Tlx3, Ptf1α, Pax2, Lbx1, and Lhx1 [7, 10], which act as post-mitotic regulators in glutamatergic/GABAergic differentiation. The factors behind these processes are different and may vary at different regions of the brain [4, 7] but the advantage of having different mechanisms in different regions of the nervous system is less understood. It is known that Tlx3, a post-mitotic homeodomain transcription factor, promotes glutamatergic differentiation in distinct regions of the nervous system [27, 28] Elucidation of molecular regulation of Tlx3 gene may reveal upstream key players of glutamatergic versus GABAergic specification in different regions of the brain. Even though the role of Tlx3 is confirmed in glutamatergic fate specification, the actual mechanism by which Tlx3 promoter is regulated is not clearly understood. There are no reports regarding the regulation of Tlx3 promoter except one where Ptf1α was shown to suppress Tlx3 expression leading to specification of GABAergic fate [10]. Our results clearly indicate that Hes-1, an upstream regulator of neurogenesis, has a predominant role in Tlx3 expression. Tlx3 promoter-driven GFP expression analysis during ES cell differentiation showed that it is expressed in progenitors/cells that had entered differentiation. Evidence from various sources suggested that Tlx3 is expressed post-mitotically and will be expressed until terminal differentiation [7, 27]. It is also well known that Hes-1 is expressed during proliferation of neural progenitors and is down-regulated during neuronal differentiation [38]. From temporal expression patterns, and also considering the repressive effect of Hes-1 on Tlx3 promoter (Figs. 4, 6u), we can assume that Hes-1 keeps the expression of Tlx3 in a down-regulated state until the progenitor/precursors enters neuronal differentiation. If this timing is seen with reference to classical Notch signaling, then one of the neighboring daughter cells that has entered differentiation must have down-regulated Hes-1 with a glutamatergic fate due to increased expression of Tlx3. Since all the differentiating cells do not always differentiate into glutamatergic neurons and generate GABAergic neurons also, it appears that Tlx3 expression may be promoting a glutamatergic fate but may not have any influence on the differentiation of GABAergic neurons. The GABAergic neurons may be generated through the activation of other signaling pathways. It is known that Mash-1 can induce Ptf1α which in turn suppresses GSH1/2 and represses Tlx3 leading to GABAergic differentiation [10].
Another interesting observation was the negligible expression of Hes-1 in IMR32 cell line, which has a constitutive expression of Tlx3, supporting the idea of negative regulation of Tlx3 by Hes-1 [29, 39]. A very recent report showed that Tlx3 is also negatively regulated by c-Jun in response to endogenous calcium spike activity in Xenopus [28]. The question that now arises is whether a similar Tlx3 pathway is involved in Glutamatergic versus GABAergic fate specification in the CNS. Previous reports have shown the involvement of Tlx3 in Glutamatergic/GABAergic fate specification in the spinal cord and hindbrain [40]. Tlx3 mutant mice having defects in ventral medulla show improper development of somatic sensory neurons in the dorsal spinal cord and sensory neurons in the brain stem, indicating its role in developmental neurogenesis [41]. Most importantly, Tlx3 null mice die immediately after birth due to excessive inhibition caused by an excess GABAergic input resulting in central respiratory failure [7, 42]. Also, over-expression of Tlx3 in chick spinal cord and in embryonic stem cells resulted in excess glutamatergic differentiation with less or repressed GABAergic differentiation [7, 27]. We also found increased expression of Tlx3 in epileptic hippocampus (data not shown), which may lead to excessive generation of glutamatergic neurons at the expense of GABAergic neurons leading to seizures. Thus, our observation suggests that Tlx3 is one of the major regulators of excitatory neurogenesis in the CNS.
Our results also showed that Tlx3 over-expression and inhibition of Hes-1 using dnHes1 caused considerably increased glutamatergic neuronal differentiation evidenced by increased vGlut2 immunoreactivity. Further, Hes-1 over-expression generated more GABAergic neurons than glutamatergic neurons. Our results also indicated that classical Notch signaling has a powerful role in the specification of neurotransmitter choice. Precursor cells that are just entering differentiation will have down-regulated Hes-1, and may go towards a glutamatergic fate while the neighboring daughter cell may adopt a GABAergic fate. This mechanism is applicable only in a cell that undergoes symmetric division and generated both glutamatergic and GABAergic neurons. However, in another scenario, where glutamatergic and GABAergic neurons emerge from different precursors, the mechanism of neurotransmitter regulation may be different, and needs to be investigated further.
It also appears that WRPW domain-mediated co-repressor recruitment may be the prominent regulatory mechanism of Hes-1 in Tlx3 promoter (Fig. 5b) as seen with other target genes [34, 35]. Co-repressors execute most of the repressor functions through HDACs [43] and analysis with an HDAC inhibitor, TSA, showed that Hes-1 mediates its regulatory effect partly through the activity of HDACs on Tlx3 promoter (Fig. 5c). NFY is also known to bind at the CAAT box on Tlx3 promoter and through acetylation maintains an open configuration [44]. Therefore, it is logical to assume that Hes-1 mediates its regulation through the antagonistic effect of acetylation of promoter and maintain a closed confirmation and, thus is inaccessible for the general transcriptional machinery (Fig. 5c). This has been proven by the mutation of CAAT-binding sites [45]. Based on previous reports, we assume that NFY is able to constitutively activate Tlx3 promoter by binding to the +72 CAAT-binding site in the 5′UTR region. We observed a complete loss of Tlx3 expression in constructs with deleted 5′UTR regions (Supplementary Fig. 5). We also found that WRPW-truncated Hes-1 was unable to repress Tlx3 promoter, which may be due to the inability of this truncated Hes-1 to recruit co-repressors. Analysis using B*Hes-1 and deletion analysis of Hes-1-binding sites in the Tlx3 promoter indicates a DNA-binding-mediated regulation of Tlx3 expression by Hes-1 similar to that reported with other Hes-1 target genes (Fig. 5b). Therefore, it can be concluded that Hes-1 negatively regulates Tlx3 promoter by recruiting TLE co-repressors as indicated by the repression of other genes [34, 35]. Increasing evidence is emerging regarding the role of chromatin re-modulation in the context-dependent regulation of fate-specifying genes during development [46–49]. Our results also indicate towards the notion that Hes-1 regulates the expression of Tlx3, a key player of glutamatergic specification, through its chromatin-modifying effects.
However, while interpreting our data, we have not avoided the possibility of regulatory effects of Hes-1 on other target genes also, since Hes-1 is one of the upstream players of neurogenesis [38]. The fate specification observed during ES cell differentiation may be a combined effect of action on different target genes. Therefore, different transcription factors involved along with Tlx3 in this process has to be further characterized in order to get a full picture of glutamatergic versus GABAergic differentiation, which is still one of the major basic mysteries in neuroscience research.
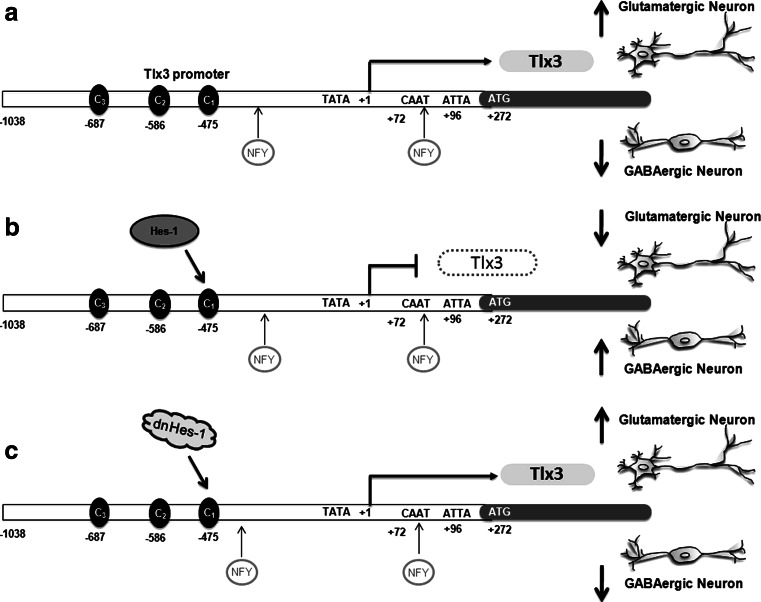
In conclusion, we have found a unique role for Hes-1, a Notch target gene, in cell fate determination of excitatory and inhibitory neurons, by regulating Tlx3 gene expression. Tlx3, a known selector gene for excitatory versus inhibitory cell fate determination, is constitutively expressed through NFY, which as such promotes neural precursors to differentiate along glutamatergic fate rather than selecting a GABAergic fate (Fig. 7a). However, in the presence of Hes-1, which represses Tlx3, the cells are inhibited from being differentiated into glutamatergic neurons and preferably differentiate along GABAergic lineage (Fig. 7b). This was further confirmed with dnHes-1 experiments, where there was no repression of Tlx3 and cell were differentiated preferentially into glutamatergic neurons (Fig. 7c). Our findings shed light on the complex regulation of cell fate determination and the involvement of different signaling molecules in bringing the right proportion of cell population in a given tissue environment and at a given context. The elucidation of the mechanism of glutamatergic versus GABAergic differentiation will lead to excellent therapies against various neurological disorders with an imbalance in glutamatergic and GABAergic neurons.
Fig. 7.
Schematic showing the regulation of Tlx3 by Hes-1 during neuronal fate specification: a Expression of Tlx3 through NFY promotes glutamatergic fate over its complementary GABAergic fate. b Expression of Hes-1 represses Tlx3 by binding to the C1 site, thereby suppressing glutamatergic fate and increasing the GABAergic neurons. c Down-regulation of Hes-1 expression by dnHes-1 removes the repression on the Tlx3 promoter, thus enhancing the differentiation of glutamatergic neurons
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (TIFF 61606 kb)
Supplementary material 2 (TIFF 39296 kb)
Supplementary material 3 (TIFF 31560 kb)
Supplementary material 4 (TIFF 40967 kb)
Supplementary material 5 (TIFF 62432 kb)
Supplementary material 6 (TIFF 31492 kb)
Acknowledgments
This work was supported by research grants from the Department of Science & Technology and the Department of Biotechnology, Government of India, and Intra mural grants from RGCB to JJ; CLI, RS, MSD, VAR and SBD were supported by research fellowships from CSIR & ICMR, Government of India. The authors thank Dr. R Kageyama, Dr. R Kopan, Dr. Anderstrom, Dr. Minato and Addgene for generously providing us the plasmid constructs.
Footnotes
C. L. Indulekha and T. S. Divya contributed equally to this work.
References
- 1.Jan YN, Jan LY. HLH proteins, fly neurogenesis, and vertebrate myogenesis. Cell. 1993;75:827–830. doi: 10.1016/0092-8674(93)90525-U. [DOI] [PubMed] [Google Scholar]
- 2.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 3.Kintner C. Neurogenesis in embryos and in adult neural stem cells. J Neurosci. 2002;22:639–643. doi: 10.1523/JNEUROSCI.22-03-00639.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Q. Transcriptional regulation of neuronal phenotype in mammals. J Physiol. 2006;575:379–387. doi: 10.1113/jphysiol.2006.113449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guillemot F. Spatial and temporal specification of neural fates by transcription factor codes. Development. 2007;134:3771–3780. doi: 10.1242/dev.006379. [DOI] [PubMed] [Google Scholar]
- 6.Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/S0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- 7.Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, Chen CL, Busslinger M, Goulding M, Onimaru H, Ma Q. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- 8.Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- 9.Schuurmans C, Armant O, Nieto M, Stenman JM, Britz O, Klenin N, Brown C, Langevin LM, Seibt J, Tang H, Cunningham JM, Dyck R, Walsh C, Campbell K, Polleux F, Guillemot F. Sequential phases of cortical specification involve Neurogenin-dependent and -independent pathways. EMBO J. 2004;23:2892–2902. doi: 10.1038/sj.emboj.7600278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- 11.Borghini S, Vargiolu M, Di Duca M, Ravazzolo R, Ceccherini I. Nuclear factor Y drives basal transcription of the human TLX3, a gene overexpressed in T-cell acute lymphocytic leukemia. Mol Cancer Res. 2006;4:635–643. doi: 10.1158/1541-7786.MCR-05-0250. [DOI] [PubMed] [Google Scholar]
- 12.Yun K, Fischman S, Johnson J, De Angelis MH, Weinmaster G, Rubenstein JL. Modulation of the notch signaling by Mash1 and Dlx1/2 regulates sequential specification and differentiation of progenitor cell types in the subcortical telencephalon. Development. 2002;129:5029–5040. doi: 10.1242/dev.129.21.5029. [DOI] [PubMed] [Google Scholar]
- 13.Del Barrio MG, Taveira-Marques R, Muroyama Y, Yuk DI, Li S, Wines-Samuelson M, Shen J, Smith HK, Xiang M, Rowitch D, Richardson WD. A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development. 2007;134:3427–3436. doi: 10.1242/dev.005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng CY, Yajima H, Burns CE, Zon LI, Sisodia SS, Pfaff SL, Sharma K. Notch and MAML signaling drives Scl-dependent interneuron diversity in the spinal cord. Neuron. 2007;53:813–827. doi: 10.1016/j.neuron.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Batista MF, Jacobstein J, Lewis KE. Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev Biol. 2008;322:263–275. doi: 10.1016/j.ydbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 16.Cau E, Blader P. Notch activity in the nervous system: to switch or not switch? Neural Dev. 2009;4:36. doi: 10.1186/1749-8104-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanalkumar R, Indulekha CL, Divya TS, Divya MS, Anto RJ, Vinod B, Vidyanand S, Jagatha B, Venugopal S, James J. ATF2 maintains a subset of neural progenitors through CBF1/Notch independent Hes-1 expression and synergistically activates the expression of Hes-1 in Notch-dependent neural progenitors. J Neurochem. 2010;113:807–818. doi: 10.1111/j.1471-4159.2010.06574.x. [DOI] [PubMed] [Google Scholar]
- 18.Kaczmarczyk SJ, Green JE. A single vector containing modified cre recombinase and LOX recombination sequences for inducible tissue-specific amplification of gene expression. Nucleic Acids Res. 2001;29:E56. doi: 10.1093/nar/29.12.e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshiura S, Ohtsuka T, Takenaka Y, Nagahara H, Yoshikawa K, Kageyama R. Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc Natl Acad Sci USA. 2007;104:11292–11297. doi: 10.1073/pnas.0701837104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ong CT, Cheng HT, Chang LW, Ohtsuka T, Kageyama R, Stormo GD, Kopan R. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J Biol Chem. 2006;281:5106–5119. doi: 10.1074/jbc.M506108200. [DOI] [PubMed] [Google Scholar]
- 21.Muller P, Kietz S, Gustafsson JA, Strom A. The anti-estrogenic effect of all-trans-retinoic acid on the breast cancer cell line MCF-7 is dependent on HES-1 expression. J Biol Chem. 2002;277:28376–28379. doi: 10.1074/jbc.C200340200. [DOI] [PubMed] [Google Scholar]
- 22.Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL. rax, Hes1, and notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron. 2000;26:383–394. doi: 10.1016/S0896-6273(00)81171-X. [DOI] [PubMed] [Google Scholar]
- 23.Murata K, Hattori M, Hirai N, Shinozuka Y, Hirata H, Kageyama R, Sakai T, Minato N. Hes1 directly controls cell proliferation through the transcriptional repression of p27Kip1. Mol Cell Biol. 2005;25:4262–4271. doi: 10.1128/MCB.25.10.4262-4271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuda T, Cepko CL. Electroporation and RNA interference in the rodent retina in vivo and in vitro. Proc Natl Acad Sci USA. 2004;101:16–22. doi: 10.1073/pnas.2235688100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanalkumar R, Vidyanand S, Lalitha Indulekha C, James J. Neuronal vs. glial fate of embryonic stem cell-derived neural progenitors (ES-NPs) is determined by FGF2/EGF during proliferation. J Mol Neurosci. 2010;42:17–27. doi: 10.1007/s12031-010-9335-z. [DOI] [PubMed] [Google Scholar]
- 26.James J, Das AV, Bhattacharya S, Chacko DM, Zhao X, Ahmad I. In vitro generation of early-born neurons from late retinal progenitors. J Neurosci. 2003;23:8193–8203. doi: 10.1523/JNEUROSCI.23-23-08193.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kondo T, Sheets PL, Zopf DA, Aloor HL, Cummins TR, Chan RJ, Hashino E. Tlx3 exerts context-dependent transcriptional regulation and promotes neuronal differentiation from embryonic stem cells. Proc Natl Acad Sci USA. 2008;105:5780–5785. doi: 10.1073/pnas.0708704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marek KW, Kurtz LM, Spitzer NC. cJun integrates calcium activity and tlx3 expression to regulate neurotransmitter specification. Nat Neurosci. 2010;13:944–950. doi: 10.1038/nn.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borghini S, Bachetti T, Fava M, Di Duca M, Cargnin F, Fornasari D, Ravazzolo R, Ceccherini I. The TLX2 homeobox gene is a transcriptional target of PHOX2B in neural-crest-derived cells. Biochem J. 2006;395:355–361. doi: 10.1042/BJ20051386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giagtzoglou N, Alifragis P, Koumbanakis KA, Delidakis C. Two modes of recruitment of E(spl) repressors onto target genes. Development. 2003;130:259–270. doi: 10.1242/dev.00206. [DOI] [PubMed] [Google Scholar]
- 31.Chen H, Thiagalingam A, Chopra H, Borges MW, Feder JN, Nelkin BD, Baylin SB, Ball DW. Conservation of the Drosophila lateral inhibition pathway in human lung cancer: a hairy-related protein (HES-1) directly represses achaete-scute homolog-1 expression. Proc Natl Acad Sci USA. 1997;94:5355–5360. doi: 10.1073/pnas.94.10.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/S0378-1119(00)00161-X. [DOI] [PubMed] [Google Scholar]
- 33.Buscarlet M, Perin A, Laing A, Brickman JM, Stifani S. Inhibition of cortical neuron differentiation by Groucho/TLE1 requires interaction with WRPW, but not Eh1, repressor peptides. J Biol Chem. 2008;283:24881–24888. doi: 10.1074/jbc.M800722200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher AL, Ohsako S, Caudy M. The WRPW motif of the hairy-related basic helix–loop–helix repressor proteins acts as a 4-amino-acid transcription repression and protein–protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross DA, Hannenhalli S, Tobias JW, Cooch N, Shiekhattar R, Kadesch T. Functional analysis of Hes-1 in preadipocytes. Mol Endocrinol. 2006;20:698–705. doi: 10.1210/me.2005-0325. [DOI] [PubMed] [Google Scholar]
- 36.Nuthall HN, Joachim K, Stifani S. Phosphorylation of serine 239 of Groucho/TLE1 by protein kinase CK2 is important for inhibition of neuronal differentiation. Mol Cell Biol. 2004;24:8395–8407. doi: 10.1128/MCB.24.19.8395-8407.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouhon IA, Joannides A, Kato H, Chandran S, Allen ND. Embryonic stem cell-derived neural progenitors display temporal restriction to neural patterning. Stem Cells. 2006;24:1908–1913. doi: 10.1634/stemcells.2006-0031. [DOI] [PubMed] [Google Scholar]
- 38.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Two mammalian helix–loop–helix factors structurally related to Drosophila hairy and enhancer of split. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 39.Jogi A, Ora I, Nilsson H, Lindeheim A, Makino Y, Poellinger L, Axelson H, Pahlman S. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc Natl Acad Sci USA. 2002;99:7021–7026. doi: 10.1073/pnas.102660199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatano M, Iitsuka Y, Yamamoto H, Dezawa M, Yusa S, Kohno Y, Tokuhisa T. Ncx, a Hox11-related gene, is expressed in a variety of tissues derived from neural crest cells. Anat Embryol (Berl) 1997;195:419–425. doi: 10.1007/s004290050061. [DOI] [PubMed] [Google Scholar]
- 41.Qian Y, Shirasawa S, Chen CL, Cheng L, Ma Q. Proper development of relay somatic sensory neurons and D2/D4 interneurons requires homeobox genes Rnx/Tlx-3 and Tlx-1. Genes Dev. 2002;16:1220–1233. doi: 10.1101/gad.982802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirasawa S, Arata A, Onimaru H, Roth KA, Brown GA, Horning S, Arata S, Okumura K, Sasazuki T, Korsmeyer SJ. Rnx deficiency results in congenital central hypoventilation. Nat Genet. 2000;24:287–290. doi: 10.1038/73516. [DOI] [PubMed] [Google Scholar]
- 43.Sekiya T, Zaret KS. Repression by Groucho/TLE/Grg proteins: genomic site recruitment generates compacted chromatin in vitro and impairs activator binding in vivo. Mol Cell. 2007;28:291–303. doi: 10.1016/j.molcel.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Currie RA. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 45.Bolognese F, Pitarque-Marti M, Lo Cicero V, Mantovani R, Maier JA. Characterization of the human EDF-1 minimal promoter: involvement of NFY and Sp1 in the regulation of basal transcription. Gene. 2006;374:87–95. doi: 10.1016/j.gene.2006.01.030. [DOI] [PubMed] [Google Scholar]
- 46.Hsieh J, Gage FH. Epigenetic control of neural stem cell fate. Curr Opin Genet Dev. 2004;14:461–469. doi: 10.1016/j.gde.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 47.Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 48.Namihira M, Kohyama J, Abematsu M, Nakashima K. Epigenetic mechanisms regulating fate specification of neural stem cells. Philos Trans R Soc Lond B Biol Sci. 2008;363:2099–2109. doi: 10.1098/rstb.2008.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee S, Lee SK. Crucial roles of histone-modifying enzymes in mediating neural cell-type specification. Curr Opin Neurobiol. 2010;20:29–36. doi: 10.1016/j.conb.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1 (TIFF 61606 kb)
Supplementary material 2 (TIFF 39296 kb)
Supplementary material 3 (TIFF 31560 kb)
Supplementary material 4 (TIFF 40967 kb)
Supplementary material 5 (TIFF 62432 kb)
Supplementary material 6 (TIFF 31492 kb)