Abstract
Nanostructures appear to be promising for a number of applications in molecular diagnostics, mainly due to the increased surface-to-volume ratio they can offer, the very low limit of detection achievable, and the possibility to fabricate point-of-care diagnostic devices. In this paper, we review examples of the use of nanostructures as diagnostic tools that bring in marked improvements over prevalent classical assays. The focus is laid on the various sensing paradigms that possess the potential or have demonstrated the capability to replace or augment current analytical strategies. We start with a brief introduction of the various types of nanostructures and their physical properties that determine the transduction principle. This is followed by a concise collection of various functionalization protocols used to immobilize biomolecules on the nanostructure surface. The sensing paradigms are discussed in two contexts: the nanostructure acting as a label for detection, or the nanostructure acting as a support upon which the molecular recognition events take place. In order to be successful in the field of molecular diagnostics, it is important that the nanoanalytical tools be evaluated in the appropriate biological environment. The final section of the review compiles such examples, where the nanostructure-based diagnostic tools have been tested on realistic samples such as serum, demonstrating their analytical power even in the presence of complex matrix effects. The ability of nanodiagnostic tools to detect ultralow concentrations of one or more analytes coupled with portability and the use of low sample volumes is expected to have a broad impact in the field of molecular diagnostics.
Keywords: Nanoparticles, Carbon nanotubes, Nanowires, Quantum dots, Electrical detection, Electrochemical detection, Surface plasmon resonance, Biobarcode assay, Immunoassay, Biochips, Nucleic acid assays, Label-free detection, Biosensing, Sandwich assays
Introduction
The domain of Life Sciences is undergoing a dramatic transformation, wherein molecular biology and biotechnology are aiming at the understanding of biological processes as a whole. This has led to the emergence of a number of fields such as genomics, proteomics, metabolomics, and transcriptomics [1, 2]. The availability of a massive data set from the human genome project [3] has motivated the thorough investigation of the molecular basis for various diseases. Towards this purpose, it is important to analyze nucleotide polymorphisms or study genotypes across the entire human genome [4]. Continuous improvements in nucleic acid assays are essential in order to achieve this goal. Furthermore, it has also been inferred that it is not only the genomic sequence that determines the biological processes in an individual [1]. The same part of the genomic sequence can code for different proteins depending on how the pre-mRNA is spliced. Added to this, post-transcriptional modifications bring in a further range of variability. Thus, the use of proteins as biomarkers for diseases or disease states is becoming increasingly important [5, 6]. While the availability of a single biomarker for the screening of a specific disease is not yet well established [7, 8], the potential for the diagnosis based on a multitude of biomarkers presents an exciting possibility [9–11]. Another related application involves the direct detection and identification of emerging pathogenic threats such as viruses and bacteria [12–14]. These require the development of ultrasensitive detection platforms with minimal fast negatives. The success of in vitro molecular diagnostics will thus rely in the future on novel biosensing platforms that can bring in significant improvements in detection limits, sensitivity, and parallelism [15].
In order to appreciate the need for such novel platforms, it is worth having a closer look at assays that are currently well established. Current nucleic acid assays have been mainly based on the polymerase chain reaction (PCR) [16]. PCR is a target amplification technique and represents the ultimate in terms of sensitivity, but has certain drawbacks including complexity, cost, and lack of portability. It is also challenging to implement multiplexed detection in order to detect multiple targets with a single assay [4]. Despite these aspects, PCR has been widely successful in the analysis of ultralow concentrations of DNA. While such a target amplification strategy is available for the analysis of nucleic acids, there is no such comparable technique for the amplification of proteins. As a consequence, the analytical tools must be highly sensitive in order to detect ultralow concentrations of proteins [17].
The use of nanostructures as diagnostic tools aims to address these concerns namely improving sensitivity, attaining the lowest possible detection limits, and bringing in multiplexing capabilities for parallel recognition [18, 19]. Due to their small sizes, nanostructures can detect very few molecules in solution offering very low limits of detection (LOD). On the other hand, supports decorated with nanostructures provide for a much larger surface, thereby improving sensitivity. Furthermore, conventional labels (such as fluorescent dyes) can be immobilized on nanostructures to significantly enhance the detection signal allowing for better sensitivity [20, 21]. Another important aspect while using nanostructures for detection is the absence of the necessity for labeling in most situations [22–25]. Lastly, the miniaturized nature of these sensors allows for integration in a small chip showing promise for point-of-care medical diagnostics––tests that can be carried out directly at the bedside of a patient [17, 21, 26, 27].
The review is organized as follows. We start with a description of various types of nanostructures and indicate the advantages and disadvantages over traditional materials that are used in their place for biosensing. We discuss the physical properties of the nanostructures that form the basis for the appropriate transduction principle. In order to deploy the nanostructure as a diagnostic tool, it is necessary to functionalize their surfaces with biomolecules. A concise description of standard biofunctionalization techniques that are specific to nanostructures is presented next. Following this, various detection paradigms are discussed. Most of the nanostructure-based diagnostic tools also rely on traditional biosensing principles such as receptor–ligand, antibody–antigen, or nucleic acid interactions. In some cases, the detection format introduces new molecular recognition paradigms that are unique for nanostructures. Since the techniques are rather generic, they are applicable in both nucleic acid assays and immunoassays. A distinction between the two is made only in the final section where we discuss examples from the literature that implement these sensing paradigms and demonstrate clearly the advantages over standard techniques. In the case of immunoassays, we discuss examples carried out on realistic biological samples, comparing their capabilities to the gold standard for protein analysis, namely ELISA (enzyme-linked immunosorbent assay) [28]. When discussing nucleic acid assays, we focus on techniques that appear to be competitive to PCR. Furthermore, the use of nanostructures for improving techniques such as polymorphism detection and sequencing is presented. This section also includes a discussion of multiplexing strategies utilizing nanostructures. The review concludes with a summary and implications and hurdles for the use of nanostructures for in vitro diagnostics.
It is worth mentioning here that there is a plethora of possibilities that promises improvements while using nanostructures for applications in molecular diagnostics. Not all of them have really demonstrated an advantage against classical assays. We will focus here mainly on examples that have the scope for in vitro molecular diagnostics with or without the possibility to implement in a point-of-care format. Furthermore, single molecule approaches that require micro- to nanomolar concentrations of the analyte are not typically viewed as high-sensitivity methods in the context of medical diagnostics [17] and hence will not be discussed here. Exceptions are sequencing techniques that bring in additional information that is not obtainable by standard assays.
Nanostructures
Nanostructures refer to materials whose sizes measure <100 nm in at least one dimension. Accordingly, they can be classified as zero- (0D), one- (1D), and two-(2D) dimensional systems. A diagnostic assay usually involves at least one receptor onto which the analyte binds and a label whose physical property (such as fluorescence) is used to detect the binding event. In heterogeneous formats, a support might be used to immobilize the receptor. Nanostructures are deployed either as a label or as a support, depending on their physical and chemical properties. To be used as labels, they need to be stable in relevant buffers or biological fluids, detectable in a required environment, and possess functional groups that can be used for conjugation with biomolecules. Towards this purpose, the surface is functionalized with ligands that ensure a stable aqueous dispersion of the nanoparticles. In this section, we provide an overview of the major nanostructures (see Fig. 1) that are used in diagnostic assays. It also includes a discussion of the physical properties of the respective nanostructures, which determine their role either as a label or as a support. The various transduction principles specific to the nanostructures are summarized in Table 1.
Fig. 1.
An overview of various nanostructures (NSs) that are used in diagnostic assays. NSs with a blue background are used as supports to immobilize receptors or labels, while those with a yellow background function as labels. NSs in a green background function both as labels and as supports. The insets in white background show the major bioconjugation protocol used to immobilize receptors on the corresponding nanomaterial. The green antibody in the insets represents a generic biomolecule
Table 1.
Sensor-transduction principles specific to various nanostructures
| Nanostructure | Optical | Electrical | Electrochemical | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Fluorescence | Colorimetry absorption | SPR, LSPR | Raman, SERS | Resistance, impedance | Field-effect | Stripping voltammetry | Amperometry, potentiometry | |||
| 0D | Metallic NPs | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ♦ | ||
| Quantum Dots | ♦ | ♦ | ||||||||
| Silica NSs* | ||||||||||
| Magnetic NPs* | ||||||||||
| 1D | Carbon nanotubes | ♦ | ♦ | ♦ | ♦ | ♦ | ||||
| Silicon nanowires | ♦ | ♦ | ||||||||
| Nanopores | ♦ | ♦ | ||||||||
| Waveguides | ♦ | |||||||||
| 2D | Metal films | ♦ | ♦ | |||||||
| Nanostructured surfaces | ♦ | ♦ | ♦ | ♦ | ||||||
| Graphene | ♦ | ♦ | ||||||||
NP Nanoparticles, NS nanospheres, (L)SPR (local) surface plasmon resonance, SERS surface-enhanced Raman spectroscopy
* Silica nanospheres and magnetic nanoparticles are mainly used as supports for immobilizing labels; they are usually not detected directly
0D Nanostructures
In 0D nanostructures (see Fig. 1––0D) the size is <100 nm in all the three dimensions. Examples of such systems include nanoparticles and nanospheres. Depending on their electrical properties, they can be classified as metallic, semiconducting, insulating, or magnetic nanoparticles.
Metallic nanoparticles (e.g., gold and silver) have been widely used in numerous bioassays [29–31]. Due to their unique physical properties they can be detected either by optical or by electrical methods and hence serve as good labels. They exhibit a bright color due to the presence of a plasmon absorption band (also called local surface plasmon resonance––LSPR) and hence can be probed using simple methods such as colorimetry or absorption spectroscopy [20, 32, 33]. In addition, they produce a characteristic Raman signal [34] and hence surface-enhanced Raman spectroscopy (SERS) can be used to probe the nanoparticles [35–37]. The color or the Raman spectrum of the nanoparticles changes as a function of their size and the presence or absence of biomolecules in their vicinity. Since they are nanoscale particles they scatter light and resonant light scattering (RLS) is used to identify their presence [38, 39]. Metal nanoparticles possess redox properties and hence are sensitive to various electrochemical methods [29, 40] such as stripping voltammetry [41]. They have been used as nanoscaffolds to increase the sensitivity of electrochemical assays [40–44]. They are stabilized in solutions using thiol-based surfactants. In addition to being used as labels, metal NPs serve as supports to carry receptors.
Semiconducting nanoparticles or quantum dots (QDs) refer to fluorescent nanocrystals made mainly from compounds of group II and group VI elements of the periodic table. They serve as labels in bioassays due to their excellent fluorescence properties [33, 45–47]. Typically, quantum dots are composed of a core semiconductor (for example CdSe) and a shell semiconductor with a larger spectral band gap (e.g., ZnS). In order to disperse them in solutions, QDs are silanized or covered with polymers. They are an ideal substitute for classical dyes or fluorophores since they exhibit very narrow, symmetric, bright and size-dependent emission and broad absorption spectra [48]. They are more resistant to photobleaching compared to many other fluorophores. However, there are some limitations such as toxicity, inability to have perfect control over their size, agglomeration, surface oxidation, and non-specific binding [26]. Quantum dots are used as very efficient donors of energy to dye acceptors in fluorescence resonance energy transfer (FRET)-based assays [49–51]. Analogous to metal NPs, QDs can also be detected through electrochemical methods such as stripping voltammetry [52, 53].
Silica nanospheres are insulating 0D structures and they do not have measurable electrical or optical properties [54]. Hence, they are mainly used as a support for carrying optical labels in order to amplify the detection signal. Usually they consist of a silica matrix doped with many organic or inorganic dye molecules [25, 55, 56]. Typical examples of inorganic dyes include lanthanide-based [25, 57] and ruthenium-based [55, 58] chelates. Inorganic dye-loaded silica particles have favorable optical properties similar to QDs, such as good photostability, sharp emission peaks, and long fluorescence lifetimes. Due to their hydrophilic surface, they can be directly dispersed in aqueous solutions.
Magnetic nanoparticles are 0D nanostructures made of a magnetic material such as iron or an alloy of nickel and iron [25, 45, 56, 59]. They are deployed as support to carry receptors. The advantage of using magnetic nanoparticles is that the receptor-analyte complex can be manipulated using a simple external magnet. In this manner, the binding takes place in solution, while the detection takes place after immobilizing the analyte-receptor complex on a substrate [59]. This overcomes limitations to sensitivity brought by other factors such as diffusion, convection etc. [60].
1D Nanostructures
1D nanostructures (see Fig. 1––1D) include materials whose size is <100 nm in two dimensions. They can be considered as nanoparticles elongated in one dimension. Typical 1D nanostructures include carbon nanotubes, nanowires, nanopores, and waveguides.
Carbon nanotubes (CNTs) are rolled up sheets of carbon that can be either metallic or semiconducting. CNTs are produced in a number of synthesis methods and can be procured commercially in powder form [61]. Their average diameter is around 1 nm (single-walled CNTs) and hence they possess a very high surface-to-volume ratio. Due to this, their electrical properties (e.g., resistance) are very sensitive to chemical and biological species in their surroundings. Based on this property, CNTs are widely used as supports functioning as label-free electrical detectors in heterogeneous assays [21, 24]. Furthermore, CNTs have favorable optical properties such as a characteristic Raman signal and are used as labels [62]. Moreover, they act as a support to carry a payload of labels [63] similar to the case of silica nanospheres.
Silicon nanowires (SiNWs) require high temperatures to be synthesized and are usually prepared on silicon wafers [64, 65]. They are mainly used as supports. Due to their semiconducting properties, they are useful for label-free electrical detection in a manner similar to that of CNTs.
Nanopores are 1D nanostructures where a hole of diameter <100 nm is extended in one dimension and find application in DNA sequencing. Typical examples are protein channels (e.g., α-hemolysin [66] or another kind of porin [67]) incorporated into lipid bilayers/synthetic membranes or holes in solid-state membranes (e.g., silicon nitride, silicon oxide, or graphene) [68–70]. Solid-state nanopores are quite robust and can be fabricated with a high level of precision using ion-beam [68] or electron-beam [71] techniques.
Waveguides are 1D nanostructures made of silica, which are used as a support to monitor biomolecular recognition events on their surface. They are prepared in such a way that the properties of the light passing through the waveguide are sensitive to biochemical events happening on the surface [72]. The use of waveguides in diagnostic assays is still in its infancy [15].
2D Nanostructures
2D nanostructures (see Fig. 1––2D) refer to structures where only one dimension has a size <100 nm. All 2D nanostructures discussed here serve as supports to immobilize receptors. They are usually sheets of a certain material, which have special properties different from that of the corresponding bulk material.
Metal films with a thickness <100 nm absorb light at specific wavelengths due to the presence of surface plasmons [73]. The absorption wavelength is a function of the dielectric constant around the surface of the metallic layer. This aspect has been utilized in the fabrication of label-free surface plasmon resonance (SPR) biosensors [73].
Nanostructured surfaces refer to electrodes where the surface is decorated with nanotubes, nanoparticles, or nanopyramids [21, 74–76]. Nanocomposites can also be prepared on such surfaces by mixing electroactive materials with nanostructures [21, 77–82]. Such surfaces have varied surface chemistry depending on the kind of nanostructure present in the composite.
Graphene is an ideal 2D surface comprising just one layer of carbon atoms [83]. Hence, diagnostic assays based on graphene are expected to deliver very high sensitivity [83].
Rationale behind the use of nanostructures in diagnostic assays
The first and foremost feature that nanostructures offer is their size (a few to 1,000 Å), which is in the range of the size of various biomolecules such as nucleic acids, small proteins, and viruses. The small size brings a tremendous increase in the surface-to-volume ratio that is essential for maximizing sensitivity. A major advantage of using nanostructures in a label-free paradigm is that the amount of receptors immobilized on the detector surface can be as low as a single molecule [65, 84, 85]. As a result, just a few analyte molecules are sufficient to cause a measurable signal providing for very low limits of detection (LOD). The lower the LOD, the earlier a biomarker characteristic of a disease state can be identified [15].
Secondly nanostructures exhibit specialized physical and chemical properties that are generally not available in the bulk. For example, the binding of analytes to receptors immobilized on carbon nanotubes brings in significant changes in the resistance of the CNT that is used as a sensor signal [86, 87]. Such a phenomenon does not occur in bulk metals. Furthermore, surface plasmons in thin metal films (2D) and nanoparticles (0D) [73] are characteristic properties at the nanoscale that are otherwise not available in bulk microscale materials. These unusual properties arise due to the fact that the electrons are confined in one to three dimensions. Another related aspect is the improvement in robustness of a certain physical property such as fluorescence of quantum dots due to their 0D nanostructure [45, 47]. While nanostructures exhibit favorable properties in a standalone manner, they also help augment transduction characteristics of bulk materials. A typical example is the improvement in electrochemical detection achievable by nanostructuring of the electrode surface [79–82, 88] due to increase in surface area.
A key aspect for the success of nanostructures is the tunable fabrication of the materials or the ability to tailor the chemical and physical properties. For example, the emission wavelength of QDs can be tuned by just varying their size [45, 47]. Through various chemical and bio-functionalization protocols (as outlined in the next section), the surface of the nanostructures is optimized for the detection of desired analytes. It is important to note that selectivity towards a certain analyte is determined mainly by biochemistry and this requires an appropriate coupling of the receptor to the nanostructure surface with minimal loss in biomolecule activity.
Although optical methods have been quite successful in achieving very low LODs, electrical methods have the exclusive advantage that they are portable, and hence more promising for applications in the point-of-care. Furthermore, such one-step methods are likely to be less prone to matrix effects due to the fewer number of analytical steps needed and hence show good promise for on-chip point-of-care applications. Other advantages of nanostructures include the ability to miniaturize the diagnostic tool and the increase in speed, reducing reagent, and sample consumption [65, 85].
In spite of these favorable aspects, there are a number of challenges, which are still to be overcome [84]. It is worth mentioning that a relatively high proportion of reported analytical bioassays describe only the basis of the method but lack the corresponding application to the analysis of real samples [26]. The suitability of these assays in the presence of sample matrices has not been established, because there are still several limiting factors, such as non-specific adsorption or binding, size variation, aggregation and lack of stability, and solubility [26]. Furthermore, the extension of these technologies to the parallelity of the 2D microarray format is usually complicated. One challenge is the optical or electrical addressing of the individual reaction sites with sub-micrometer spacing. Another major challenge is the capability to target and confine chemical functionalization to the sensor site itself [15]. For example, sensors that probe only a small area but require functionalization of a larger one (say due to the use of traditional spotting techniques or functionalization of silicon (Si) nanowires on a silicon wafer) do not really improve the LOD as binding occurs everywhere and not preferentially at the sensor site [84].
Biofunctionalization of nanostructures
The first step towards the detection of a biomolecule is the preparation of a support with an appropriate receptor or the availability of an appropriate nanobioconjugate. Towards this purpose, biomolecules need to be immobilized on the surface of a desired nanostructure [89]. The biomolecules include nucleotides, antibodies, and other ligands (such as aptamers, synthetic receptors) acting as receptors or enzymes that work as reporters. Here we present the common bioconjugate techniques that are deployed to achieve this immobilization, whereby the biomolecule is either attached covalently or non-covalently. In homogeneous formats, a covalent strategy is widely used. On the other hand, in heterogeneous formats, both covalent and non-covalent strategies have been successfully demonstrated. A collection of the various functionalization strategies is presented in Fig. 1.
One of the generic protocols used for biofunctionalization in a heterogeneous format is spotting of biomolecules [90]. This is performed using array spotters analogous to the fabrication of microarrays or other lithographic methods such as dip-pen lithography [90, 91]. The coupling is mostly non-covalent. However, by allowing for temperature and humidity control and appropriate solution processing capabilities, covalent attachment can also be achieved [90]. Other strategies are specific to the nanomaterial that needs to be coupled with the biomolecule [92]. Metallic surfaces such as gold are decorated in a facile manner with thiol-modified biomolecules [93]. The thiol groups of the biomolecule react with the metal surface resulting in covalent bonds to the surface of the metal nanoparticle or metal film. Silica surfaces (such as silicon nanowires or silica nanospheres) contain silanol groups [94, 95]. Here, heterobifunctional linkers are deployed, which contain silane groups on one end and a carboxyl or epoxide group on the other. The silane groups link to the nanoparticle surface while the carboxyl or epoxide groups are used to attach biomolecules using standard coupling chemistries [92, 95]. Similar procedures have been reported for the functionalization of magnetic nanoparticles [96]. Quantum dots are usually functionalized with ligands in order to stabilize them in aqueous solutions or organic solvents. Biomolecules are coupled using a ligand-exchange procedure [97]. On the other hand, quantum dot-biomolecule conjugates are readily available for a number of primary or secondary receptors from companies such as Invitrogen [98].
Carbon surfaces are easily oxidized by gentle oxygen plasma or heating in weak acids [99, 100]. This renders the surface with carboxyl, epoxide, or hydroxyl groups, which are then directly used to attach amine-functionalized biomolecules using carbodiimide or other coupling chemistries. The chemistry of carbon is quite rich and this has been utilized in a range of methods to functionalize carbon nanotubes [100]. The most successful of these strategies appears to be electrochemical functionalization, which provides the attachment of biomolecules onto carbon surfaces in a very versatile manner in a covalent or non-covalent fashion [78, 86, 87, 101]. Other possibilities include thermally activated chemistry or photochemistry [100]. Protocols based on heterobifunctional linkers are also utilized to couple biomolecules on to the nanotube surface [99].
Detection paradigms
The availability of a broad range of nanostructures has also triggered an avalanche of detection modalities that have been successful for the detection of biomolecules. Here we focus mainly on those strategies that show promise for an improvement in LOD or are easily amenable to on-chip analytical systems that can be used for point-of-care diagnostics. The whole range of detection strategies can be broadly classified under labeled or label-free detection. While in the former case the analyte needs to be labeled during or before the detection cycle, the label-free methods do not require the analyte to be labeled (see Fig. 2).
Fig. 2.
Mind-map of the various nanostructure-based biomolecule detection paradigms
Labeled detection
The simplest way to detect an analyte is to attach a label to it and capture it using a receptor. Figure 3a shows a schematic of this strategy in comparison to nanostructure-based protocols (Fig. 3b–d). In the former case, organic dyes are typically used as labels with fluorescence serving as the sensor signal. An example is real-time PCR [16, 27]. In nanostructure-based protocols, quantum dots, carbon nanotubes, or metal nanoparticles serve as labels (Fig. 3b, c). The coupling of the nanostructure to the analyte is carried out using the functionalization schemes outlined before. Various read-out methods are deployed as shown in Table 1 depending on the type of nanostructure. In this strategy (Fig. 3b, c), a characteristic physical property of the nanostructure is utilized. For example, quantum dots have improved fluorescent properties with respect to organic fluorophores bringing in an improvement in the sensor signal [33, 46, 48]. On the other hand, metal nanoparticles can be detected using simple techniques such as colorimetry, absorption spectrometry, or electrochemistry [20, 29, 38, 102]. Carbon nanotubes serve mainly as Raman labels [36, 37].
Fig. 3.
Schematics of labeled biomolecule detection. NP and CNT refer to 0D and 1D nanostructure respectively. a A classical approach involving a label (such as an organic dye) directly attached to the analyte. Nanostructure-based approaches: b a carbon nanotube (CNT) or c a nanoparticle acting as a label; d a nanoparticle loaded with dye molecules acting as a label
Another aspect of nanostructures is their capability of providing a large surface, which is judiciously utilized to increase the number of label molecules attached per analyte (see Fig. 3d). Typical examples include dye-loaded silica nanospheres [25, 55]. The improvement in sensor signal is apparent here when one considers that for every analyte molecule there is a multitude of labels, which produces a form of intrinsic signal amplification.
Label-free detection
Label-free methods can be classified broadly under three categories namely sandwich, direct, and mediator-based assays (see Fig. 2). Below we describe the various strategies and the advantages arising due to the use of nanostructures in these paradigms.
Sandwich assays
Sandwich assays are very efficient since they avoid direct labeling of the analyte but allow for a similar detection strategy and comparable or better sensitivity. Nanostructures have been utilized in a number of sandwich assays. The general architecture of a sandwich assay (such as in ELISA) as shown in Fig. 4a comprises of an immobilized primary receptor that binds the analyte. Subsequently, a secondary receptor binds to the free portion of the bound analyte. The secondary receptor carries a reporter or a label, which is used for the detection of the analyte using optical or electrochemical methods. In ELISA, the receptors are antibodies, while the reporter is an enzyme [28]. In nucleic acid assays, the receptors are probe sequences and the reporter is mostly a fluorophore [27]. The sandwich assays utilizing nanostructures are also based on this basic principle (see Fig. 4b–f).
Fig. 4.
Schematics of major label-free sandwich assays. a A classical approach––enzyme-linked immunosorbent assay (ELISA) showing the analyte sandwiched between an immobilized primary receptor and a labeled secondary receptor. Nanostructure-based approaches: b CNTs or NPs loaded with reporters (enzymes or dye molecules) act as labels; c the NPs attached to secondary receptors act as labels themselves. After electroless deposition of silver, they are detected by colorimetry (Ag) or by measuring the resistance (R) between two electrodes; d increasing the density of primary receptors by immobilizing them on nanoparticles or nanostructured surfaces; e Biobarcode assay: every analyte molecule is translated into numerous (n) oligonucleotides; f Nanobarcode assay: every analyte molecule is translated into a metal nanowire carrying a specific code. In both barcode assays, a unique code is deployed (Code A or Code B) for the corresponding analyte (A or B)
Nanostructures attached to secondary receptors As in the case of labeled detection, the availability of a large surface has been utilized to increase the loading of the secondary receptor with a high density of reporters (see Fig. 4b). Since every analyte molecule corresponds to a large number of reporter molecules on the secondary receptor, a great enhancement in the sensor signal is achieved. A typical example of this strategy is presented by dye-loaded silica nanospheres [103, 104]. Another example is the use of a nanotube as the support for enzyme molecules such as alkaline phosphatase, which leads to a tremendous increase in the signal upon formation of the sandwich [63]. The advantage of this strategy is that diagnostic analyzers already available in analytical labs can be utilized, with improved sensitivity or a lower LOD.
It is not always necessary to use immobilized reporters on the nanostructure. The nanoparticles (attached to the secondary receptor) themselves can serve as reporters. The detection of the nanoparticle label is carried out in many ways with or without amplification. Direct detection of the nanoparticles without amplification is carried out through resonant light scattering or electrochemical methods (see Table 1) [20]. In order to amplify the presence of nanoparticles, a simple chemical strategy has been widely successful. This involves the electroless deposition of silver onto the immobilized nanoparticles [105] (see Fig. 4c). To perform this, the chip is left in a solution containing a silver salt and an appropriate reducing agent. After a few minutes, silver deposits onto the metal nanoparticles. A simple document scanner is used to read out the amount of silver deposited, which is proportional to the analyte concentration. Alternatively, the chip is provided with electrode gaps and the formation of silver is detected electrically, which gives a detection limit of 500 fM [106].
Nanostructures attached to primary receptors. To facilitate detection in a sandwich assay, the sandwich along with the label needs to be immobilized on an appropriate substrate. Typically, the primary receptors are fixed on a glass substrate. In order to increase sensitivity further, the glass substrate itself is nanostructured (see Fig. 4d). Through this dual amplification strategy [18, 19, 63, 107, 108], a high label loading and a high density of primary receptors on the support are simultaneously achieved. The nanostructuring of the substrate is achieved before or after formation of the sandwich. In the former case, magnetic nanoparticles decorated with primary receptors are used to capture the analyte [18, 19, 107, 108]. Then the secondary receptors coupled to labeled nanostructures are allowed to bind to the free portion of the analyte. Finally, after the sandwich is formed the magnetic nanoparticles holding the sandwich are immobilized by applying a magnetic field. The unbound molecules are washed away using a rigorous washing step. Alternatively, the surface is initially nanostructured by coating it with nanoparticles or nanotubes. This allows for a huge surface and a large amount of primary receptors can be immobilized [82, 109]. The advantage of using magnetic nanoparticles is that the analytes are captured in solution and it is more efficient than a completely heterogeneous format, wherein the analyte has to locate the receptor and bind to it.
Barcodes. The use of barcodes is unique to the area of nanostructure-based diagnostics. There are two kinds of barcodes namely biobarcodes and nanobarcodes. Biobarcodes refer to biomolecules, mainly DNA, that have a specific sequence that forms the code [18, 19]. Nanobarcodes refer to nanostructures that constitute a code due to their composition [45, 108, 110, 111]. In the former case, the analyte is effectively amplified while in the latter the target is translated into a nanostructure on a one-to-one basis (see Fig. 4e, f). We will discuss these two cases separately below.
The principle behind biobarcode recognition is outlined in Fig. 4e. Here the secondary receptor is attached to a nanoparticle, which contains a large number of specific nucleotide sequences. Every different secondary receptor is attached to a unique nucleotide sequence and in this manner the binding of a certain analyte is encoded. Since every nanoparticle is loaded with a large number of nucleotides––around 228 for a 30-nm particle while using 64 mer oligonucleotide codes [112]––one analyte molecule is amplified to around 228 oligonucleotide sequences. After the binding event, the bound oligonucleotides are let free and are subsequently detected in one of the many methods outlined above. This is similar to performing a seven-step PCR (to get 2^8 or 256 copies) in one go. By comparison, a seven-step PCR requires, on average, 30 min, while the one-step biobarcode amplification requires around 90 min [18, 19]. However, the number of particles obtained can be varied by tuning the nanoparticle size. While this might not appear advantageous for the detection of DNA, it attains LOD in the attomolar range for the detection of proteins [19].
In nanobarcode immunoassays (see Fig. 4f), the nanostructure attached to the secondary antibody acts as a code. Examples are coded metallic nanowires [113] or microbeads carrying quantum dots in varying proportions [114]. Metallic nanobarcodes are prepared by electroplating metals such as Au or Ag into templates (such as alumina pores) and releasing the resulting striped nanorods. Unlike biobarcodes, the nanobarcodes do not amplify the amount of analyte. Every analyte molecule translates to one nanostructure corresponding to a specific code. The power of this assay is apparent mainly in multiplexing applications as discussed in “Multiplexing”.
Direct assays
Direct assays are the simplest methods in terms of detection complexity and processing steps. In these assays, a small amount of biological fluid is spotted on a nanostructure sensor surface that is functionalized with an appropriate receptor. The selective binding of the desired analyte on to the immobilized receptor causes a very sensitive change, which is efficiently detected through optical or electrical means (see Fig. 5). Due to their portability, electrical assays show the highest promise for entry into the field of point-of-care diagnostics. In comparison to sandwich assays, which require multiple steps requiring at least two binding events and one reporter event, a direct detection cycle requires only one step, yielding results much faster.
Fig. 5.
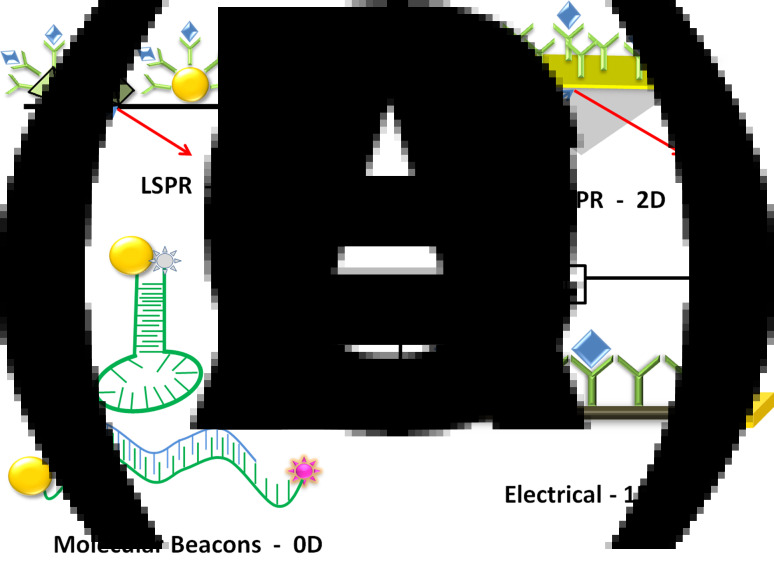
Schematics of label-free direct assays (unique for nanostructures). In general, the receptors are immobilized on the nanostructures. The binding of the analyte induces changes in some physical property of the nanostructure that is sensitively measured. a Local surface plasmon resonance (LSPR) on nanoparticles or nanopyramids and b surface plasmon resonance (SPR) on metal films detected by measuring the absorption spectrum or the angle of the reflected light. c Molecular beacons, where nanoparticles quench the fluorescence of an acceptor dye. d Changes in resistance (R), impedance (Z) or current (I) of 1D nanostructures (carbon nanotubes or silicon nanowires) measured electrically
Direct label-free detection of biomolecules on a nanostructure is possible due to characteristic properties of nanostructures that are not possible in the bulk. Towards optical detection, surface plasmons in 2D nanofilms and in 0D nanoparticles [73, 115, 116] have been widely deployed as the sensor signal (see Fig. 5a, b). The binding of the analyte to immobilized receptors leads to a change in dielectric properties and results in a shift in the optical intensity or reflection angle. This principle is widely used in surface plasmon resonance (SPR) biosensors. Limits of detection in nanomolar concentrations are typical for this technique. This principle has also been commercialized, and a number of companies are offering SPR instrumentation [116, 117]. The major advantage of using SPR is the possibility to perform kinetic studies at low concentration ranges [73, 115, 116]. Another characteristic of metallic nanoparticles is that they quench the fluorescence of organic dyes and quantum dots. Thus they are used in molecular beacons as a quencher [51] (see Fig. 5c). Nanoparticles (as acceptors) and quantum dots (as donors) have been used as a couple in FRET-based DNA detection [50].
In direct assays utilizing electrical detection, 1D nanostructures such as silicon nanowires and carbon nanotubes are widely deployed. The principle used here is a change in conductivity upon binding of the analyte to the functionalized receptor sites on the nanostructure surface (see Fig. 5d) [65, 118, 119]. The disadvantage here is that every sample requires a detailed procedure involving expensive serial techniques such as electron beam lithography. Moreover, the calibration of such sensors has been an issue since the baseline varies in every sensor trial [84, 120]. To overcome some of these disadvantages, a reference electrode is used as a gate and the sensor signal is measured in an ion-selective field-effect transistor configuration (ISFET). Carbon nanotube-based devices have been realized in such an ISFET configuration without the use of any expensive serial processing technique [84, 87, 101]. The sensors can be prepared on a large scale and the receptors are attached exclusively on the surface of the nanotubes. In this manner, LODs in the attomolar range for DNA have been achieved. In order to be applicable for realistic diagnostic devices, testing on realistic biological samples has yet to be demonstrated. Other 1D nanostructures such as ZnO nanowires also appear promising for direct assays [121].
Mediator-based assays
Mediator-based assays utilizing nanostructures have mainly been formulated in an electrochemical detection format mostly for the detection of DNA [21, 40, 41, 44, 122]. A key contribution of the nanostructures in these assays is the increase in surface area for immobilization of receptors. The assay is based on the use of an electroactive mediator that binds to duplex DNA and not to single-stranded DNA [21]. The probe DNA is immobilized on the nanostructured electrode. The binding of the analyte in the presence of the mediator results in an electrochemical current that varies as a function of the analyte concentration [40]. Due to the nanostructuring of the electrode surface, the electron transfer kinetics are improved, resulting in an improved sensor signal [52, 65, 84]. Typical nanostructures used include nanotubes and nanoparticles [21, 24, 40]. Due to the large combination of electrodes that are prepared by mixing a broad range of materials, there is a huge amount of literature available for this strategy. The reader is referred to other extensive works available [24, 29, 40, 41, 95, 109, 122, 123]. These sensors, however, show promise for point-of-care diagnostics, since they are easily realized in the format of well-known glucose strips [124].
Towards in vitro molecular diagnostics
Based on the principles outlined above, we present in this section several examples where the nanosensing paradigms have been successfully implemented, highlighting methods that have delivered sensitivities competitive to PCR and ELISA.
Immunoassays
Nanostructure-based immunoassays have been mainly designed for tumor or cancer markers, the most common of them being prostate specific antigen (PSA) [125]. Other commonly investigated proteins include immunoglobulins and other antigens implied in pathogenic diseases. Unlike nucleic acid assays, an immunoassay must be sensitive in the concentration range of interest, rather than providing the ability to detect as low a concentration as possible. On the other hand, the ability to detect concentrations lower than the currently achievable limit (in the pg/ml range when using ELISA [28]) can open up new vistas for therapy and diagnostics [17]. In the case of PSA, the relevant threshold for cancer diagnostics is 4 ng/ml [125]. A number of nanostructure-based immunoassays have shown LODs well below this limit [26].
The simplest immunoassays involving nanoparticles are similar to that of classical immunoprecipitation or immunocoagulation [126]. Here, the primary and secondary antibodies are coupled to nanoparticles and are subsequently used in a sandwich assay [127]. The formation of the sandwich leads to coagulation. Due to the special optical properties of nanoparticles, simple absorption spectroscopy is used to visualize the coagulate with LODs in the ng/ml range for IgG. Many sandwich assays based on nanoparticles for tumor markers such as PSA, α-fetoprotein (AFP) and carcinoembryonic antigen (CEA) have demonstrated LODs in the pg/ml range in serum [26, 37, 128, 129]. The ultimate detection limit has been achieved by biobarcode assays that have demonstrated ag/ml LOD for PSA [19]. This is comparable to the best established sensitivities of Immuno-PCR [130] and single-molecule ELISA [131]. This has motivated the commercialization of the biobarcode sensor platform, which has acquired FDA (Food and Drug Administration) approval and is in the clinical testing phase [17].
Direct detection systems based on conductivity changes in Si nanowires and carbon nanotubes offer LODs in the ng/ml level [22, 65, 132]. Moreover, they have mainly been experimented with test solutions and rarely with serum samples. Surface plasmon resonance-based immunosensors usually offer higher LODs [116, 117]. Mediator-based electrochemical assays incorporating nanotubes and nanoparticles have been successful in achieving much lower detection limits in the fg/ml range [122] and have even been tested on realistic serum samples [25, 125]. An alternative to detect proteins is to use aptamers in place of primary antibodies. Aptamers are short nucleic acid sequences that are designed or engineered to bind specific targets. Direct detection techniques might prove advantageous here since the use of a sandwich strategy with aptamers is not straightforward. Aptamer-immobilized CNTs have demonstrated LODs for IgE in the pg/ml range [133].
Nucleic acid assays
The standard assay for the detection of nucleic acids is based on PCR [16], which has been extremely successful in nucleic acid diagnostics due to its incredible power in amplifying a specific target sequence. The sensitivity is so high that commercial kits are available that can efficiently amplify and detect as few as 5–10 copies per μl [134]. In order for nanostructure-based assays to be competitive and viable, it is necessary to demonstrate sensitivity and versatility in relation to PCR. Among the various paradigms mentioned above, biobarcodes have demonstrated this capability. Using this simple technology, detection of DNA in zeptomolar range (corresponding to 10 molecules in 30 μl) has been successfully shown [18]. The simplicity of this assay has been utilized to probe genomic DNA at femtomolar levels in an array format [38, 135].
Although PCR and biobarcodes achieve very high sensitivity, they require a bulky optical reader to analyze the binding events. Hence, they are not directly amenable to on-chip detection for point-of-care diagnostics. For this purpose, direct label-free detection using 1D-nanostructure-based sensors appear promising [84]. In these systems, it is only necessary to drop the test solution on the sensor surface and the analyte binding is sensitively recorded by the probe DNA-functionalized nanostructures. Deploying carbon nanotubes, a detection limit in the attomolar range (corresponding to 1,800 molecules in 30 μl) has been reported in buffer solutions [101]. It was possible to achieve this sensitivity even in a heterogeneous solution mixture, where the target DNA comprised only 2% of the total DNA concentration. With silicon nanowires [118, 119] and with mediator-based electrochemical assays [122] femtomolar LODs have been reported.
The use of nanostructures brings in other advantages, which go beyond just achieving a low LOD. We will discuss two important applications here, namely single-nucleotide polymorphism (SNP) and DNA sequencing.
SNP––With the availability of the human genome sequence [136, 137], a major focus is devoted to studying the genetic variability such as SNPs and other mutations. They are expected to serve as markers in pharmacogenomics and disease diagnosis [4, 138]. The detection of SNPs through the measurement of melting curves is a key procedure in a number of genotyping assays [139]. Even in the initial experiments it was observed that the use of nanoparticles resulted in much sharper melting curves in comparison to standard fluorophores [102]. The sharper transition in the melting curves provides for a higher signal-to-noise ratio, paving the way for the detection of base-pair mismatches at very low concentrations of the analyte molecules. Additionally, a broad range of kinetic studies can be efficiently carried out utilizing nanoparticles as labels [140]. QDs either as a label [141] or as a quencher in a molecular beacon [142] have also been successfully utilized to probe SNPs. With QD labels, mutations in the human oncogene p53 could be detected at nanomolar concentrations [141].
Sequencing––Sequencing of DNA is an important step in nucleic acid diagnostics as was apparent in the deciphering of the human genome [3]. From a diagnostic point of view, sequencing a target nucleic acid can directly decipher its composition thereby avoiding the need for hybridization-based assays. A number of protocols have been demonstrated for sequencing long oligonucleotides, some of them being tedious and expensive [143]. It would be ideal to have a nanoscale reader that identifies the analyte DNA or RNA by just reading out its sequence composition. Nanopore-based methods are aimed at achieving this. This technique does not require amplification of starting material or addition of modified nucleotide phosphates or expensive reading instrumentation [66, 67, 144]. The sequencing is performed by measuring the ionic current through the nanopore as the analyte molecules are electrophoretically driven through it [66]. Depending on the number of strands or composition of DNA present in the pore, the level of current blockage is different [144]. Protein nanopores can be genetically modified in a controllable manner (e.g., by increasing recognition sites) in order to optimize the detection signal [145, 146]. Controllable single-site modifications of solid-state nanopores are, however, much more difficult. The oligonucleotides of interest can also be modified before sequencing for slower movement through the pore [67]. Well-resolved signals representing the sequence including the possibility to detect methylcytosine was demonstrated using a hemolysin mutant nanopore [147].
Multiplexing
Multiplexing refers to the capability to detect more than one analyte at once in a heterogeneous sample. Nucleic acid microarrays or protein microarrays have been widely successful in multiplexed detection offering high detection throughput [148, 149]. Multiplexing is usually achieved by the use of a range of fluorophores, each capable of detecting a specific target either in a labeled or in a label-free format [27]. Replacing the organic fluorophores with nanostructures extends the range of detection techniques and brings down detection limits. A simple strategy is to substitute the dyes with quantum dots of different colors [150–152] or nanoparticles of different sizes [20]. Using multicolor QDs, SNP mutations in the human oncogene p53 and of human hepatitis B and C viruses was reported [141]. With Au nanoparticles of two different sizes, simultaneous detection and kinetics of two DNA targets could be observed in the nanomolar range [34, 153]. Multicolor silica nanospheres were deployed in order to detect three species of bacteria (E. coli, S. Typhimurium and S. aureus) simultaneously [104]. A related multiplex strategy involves the use of electrochemical stripping voltammetry to identify different quantum dots that are used as labels in a sandwich configuration. Examples include the detection of three different species of DNA sequences in the nanomolar range [52] or the detection of three different proteins namely β-microglobulin, BSA, and IgG in the ng/ml level [53].
Biobarcodes and nanobarcodes represent an efficient strategy for multiplexed analyte detection. Here, every secondary receptor targeting a different analyte is coupled to a unique code. Oligonucleotide biobarcodes were utilized to demonstrate the simultaneous identification of four synthetic DNA targets associated with hepatitis B surface antigen, Variola virus, Ebola virus, and HIV at sub-picomolar concentrations in buffer solutions [108]. Although multiplexing is also possible with nanobarcodes, biobarcodes are more successful due to the fact that they can perform multiplexing and amplification simultaneously in just one step. Hence, metallic nanobarcodes usually require an amplification step before use. Using multiplexed PCR products of 20 genomic DNA samples, a metallic nanobarcode-based assay showed simultaneous genotyping of 15 SNPs demonstrating a capability to distinguish 30 sequences [110].
In heterogeneous formats, multiplexing is achieved by controlling the positioning of the receptors at a desired location. For example, in order to detect four analytes simultaneously, it is required to position four receptors at four different locations on the same chip. Two strategies appear promising for achieving such a site-specific immobilization: spotting and magnetic manipulation. Spotting is performed by traditional microarray spotters [27]. Multiplexed detection using two or three different spotted antibodies on silicon nanowires could detect PSA at pg/ml levels [22]. Magnetic manipulation involves the preparation of solutions of magnetic nanoparticles, each of them functionalized with a different receptor. Using an external magnetic field, they are subsequently immobilized at a desired location on a chip. Using this strategy, multiplex detection of four different tumor markers AFP, CEA, CA125, and CA15-3 could be demonstrated, albeit in the μg/ml range [59]. The assay was evaluated with serum specimens and compared favorably with results from ELISA with minimal crosstalk between the four detectors.
Conclusions
To summarize, it is clear that nanostructure-based assays are opening up numerous attractive possibilities for the detection of biomarkers and are hence promising for applications such as disease diagnosis, proteomics, genomics, and pharmacological investigations. The results presented here are just a selection from the whole range of published results and is not intended to be a complete collection. The review grouped the available results into specific detection paradigms, in order to facilitate easy association of a specific assay to a certain detection strategy. Various biofunctionalization approaches specific to the nanostructure of interest were also presented. The success of the assays will heavily depend on how they compare with conventional assays in terms of various analytical parameters [26].
Despite these promises, there are a number of hurdles still to be overcome. Some are specific to nanostructures such as efficient dispersion of nanoparticles, optimal chemical functionalization of nanostructure surfaces simultaneously retaining the activity of biomolecules, reproducible fabrication, and high toxicity of QDs etc. [154]. Others involve additional complexities that arise when making a transition to the real world. Most of the assays based on nanostructures have been demonstrated in homogeneous media, without consideration of matrix effects. However, the ultimate sensitivity of these techniques will be determined by their selectivity in complex media. Furthermore, in most cases, some kind of sample handling and human interfaces will be essential. These system aspects will bring in a number of challenges that need to be optimized before the nanoassays are ready for field applications. Microfluidic platforms can help in bridging these gaps to some extent [85]. As new diagnostic tools based on these nanostructures continue to be developed, it will be important to drive the established strategies to reach the clinical stage [17]. The transition of these tools from the laboratory to the clinic is still in its infancy. The potential for such assays to become part of routine medical testing will also depend on technological, financial, and policy factors. Once these challenges are addressed and the efficient methodologies established, it is hoped that nanoscale diagnostic tools will help us to elucidate unidentified pathways associated with diseases [4] or diagnose diseases in one click with minimal amount of physiological sample.
Acknowledgments
This project was funded by the German Federal Ministry of Education and Research (BMBF) under project ID O3X5516.
References
- 1.Veenstra TD, Yates JR. Proteomics for biological discovery. New Jersey: Wiley; 2006. [Google Scholar]
- 2.Campbell MA, Heyer LJ. Discovering genomics, proteomics and bioinformatics. New Jersey: Benjamin Cummings; 2002. [Google Scholar]
- 3.The human genome project, http://www.ornl.gov/sci/techresources/Human_Genome/home.shtml (21 March 2011)
- 4.Kirk BW, Feinsod M, Favis R, Kliman RM, Barany F. Single-nucleotide polymorphism seeking long term association with complex disease. Nucl Acid Res. 2002;30:3295–3311. doi: 10.1093/nar/gkf466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sander C. Genomic medicine and the future of health care. Science. 2000;287:1977–1978. doi: 10.1126/science.287.5460.1977. [DOI] [PubMed] [Google Scholar]
- 6.Srinivas PR, Verma M, Zhao YM, Srivastave S. Proteomics for cancer biomarker discovery. Clin Chem. 2002;48:1160–1169. [PubMed] [Google Scholar]
- 7.Master SR. Diagnostic proteomics: back to the basics? Clin Chem. 2005;51:1333–1334. doi: 10.1373/clinchem.2005.053686. [DOI] [PubMed] [Google Scholar]
- 8.Issaq HJ, Conrads TP, Prieto DA, Tirumalai R, Veenstra TD. SELDI-TOF-MS for diagnostic proteomics. Anal Chem. 2003;75:149A–155A. doi: 10.1021/ac031249c. [DOI] [PubMed] [Google Scholar]
- 9.Issaq HJ, Xiao Z, Veenstra TD. Serum and plasma proteomics. Chem Rev. 2007;107:3601–3620. doi: 10.1021/cr068287r. [DOI] [PubMed] [Google Scholar]
- 10.Sidransky D. Emerging molecular markers of cancer. Nat Rev Cancer. 2002;2:210–219. doi: 10.1038/nrc755. [DOI] [PubMed] [Google Scholar]
- 11.Wulfkuhle JD, Liotta LA, Petricoin EF. Proteomic applications for the early detection of cancer. Nat Rev Cancer. 2003;3:267–275. doi: 10.1038/nrc1043. [DOI] [PubMed] [Google Scholar]
- 12.Dawson ED, Moore CL, Smagala JA, Dankbar DM, Mehlmann M, Townsend MB, Smith CB, Cox NJ, Kuchta RD, Rowlen KL. MChip: a tool for influenza surveillance. Anal Chem. 2006;78:7610–7615. doi: 10.1021/ac061739f. [DOI] [PubMed] [Google Scholar]
- 13.Lu PS. Early diagnosis of avian influenza. Science. 2006;312:337–341. doi: 10.1126/science.1128199. [DOI] [PubMed] [Google Scholar]
- 14.Guzman MG, Kouri G. Dengue diagnosis, advances and challenges. Int J Infect Dis. 2004;8:69–80. doi: 10.1016/j.ijid.2003.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Erickson D, Mandal S, Yang AHJ, Cordovez B. Nanobiosensors: optofluidic, electrical and mechanical approaches to biomolecular detection at the nanoscale. Microfluid Nanofluid. 2008;4:33–52. doi: 10.1007/s10404-007-0198-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mullis KB. The polymerase chain reaction. Heidelberg: Birkhaeuser Press; 1994. [Google Scholar]
- 17.Giljohann DA, Mirkin CA. Drivers of biodiagnostic development. Nature. 2009;462:461–464. doi: 10.1038/nature08605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nam J-M, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J Am Chem Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 19.Nam J-M, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 20.Tansil NC, Gao Z. Nanoparticles in biomolecular detection. Nano Today. 2006;1:28–37. doi: 10.1016/S1748-0132(06)70020-2. [DOI] [Google Scholar]
- 21.Balasubramanian K, Burghard M. Biosensors based on carbon nanotubes. Anal Bioanal Chem. 2006;385:452–468. doi: 10.1007/s00216-006-0314-8. [DOI] [PubMed] [Google Scholar]
- 22.Zheng GF, Patolsky F, Cui Y, Wang WU, Lieber CM. Multiplexed electrical detection of cancer markers with nanowire sensor arrays. Nat Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 23.Drummond TG, Hill MG, Barton JK. Electrochemical DNA sensors. Nat Biotechnol. 2003;21:1192–1199. doi: 10.1038/nbt873. [DOI] [PubMed] [Google Scholar]
- 24.Gruner G. Carbon nanotube transistors for biosensing applications. Anal Bioanal Chem. 2006;384:322–335. doi: 10.1007/s00216-005-3400-4. [DOI] [PubMed] [Google Scholar]
- 25.Yan J, Estevez MC, Smith JE, Wang K, He X, Wang L, Tan W. Dye-doped nanoparticles for bioanalysis. Nano Today. 2007;2:44–50. doi: 10.1016/S1748-0132(07)70086-5. [DOI] [Google Scholar]
- 26.Gomez-Henz A, Fernandez-Romero JM, Aguilar-Caballos MP. Nanostructures as analytical tools in bioassays. Tr Anal Chem. 2008;27:394–406. doi: 10.1016/j.trac.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dill K, Liu R, Grodzinski P, editors. Microarrays: preparation, microfluidics, detection methods, and biological applications. Berlin Heidelberg New York: Springer; 2009. [Google Scholar]
- 28.Engvall E, Perlmann P. Enzyme-linked immunosorbent assay (ELISA). Quantitative assay of immunoglobulin G. Immunochemistry. 1971;8:871–874. doi: 10.1016/0019-2791(71)90454-X. [DOI] [PubMed] [Google Scholar]
- 29.Merkoci A. Nanoparticles-based strategies for the detection of DNA, proteins and cells. Biosens Bioelectron. 2010;26:1164–1177. doi: 10.1016/j.bios.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 30.Sperling RA, Gil PR, Zhang M, Zanella M, Parak WJ. Biological applications of gold nanoparticles. Chem Soc Rev. 2008;37:1896–1908. doi: 10.1039/b712170a. [DOI] [PubMed] [Google Scholar]
- 31.Giljohann DA, Seferos DS, Daniel WL, Massich D, Patel PC, Mirkin CA. Gold nanoparticles for biology and medicine. Angew Chem Int Ed. 2010;49:3280–3294. doi: 10.1002/anie.200904359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eustis S, El-Sayed MA. Why gold nanoparticles are more precious than pretty gold: noble metal surface plasmon resonance and its enhancement of the radiative and nonradiative properties of nanocrystals of different shapes. Chem Soc Rev. 2006;35:209–217. doi: 10.1039/b514191e. [DOI] [PubMed] [Google Scholar]
- 33.Seydack M. Nanoparticle labels in immunosensing using optical detection methods. Biosens Bioelectron. 2005;20:2454–2469. doi: 10.1016/j.bios.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 34.Cao YC, Jin R, Mirkin CA. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–1540. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 35.Cui Y, Ren B, Yao JL, Gu RA, Tian ZQ. Multianalyte immunoassay based on surface-enhanced Raman spectroscopy. J Raman Spectrosc. 2007;38:896–902. doi: 10.1002/jrs.1742. [DOI] [Google Scholar]
- 36.Grubisha DS, Lipert RJ, Park HY, Driskell J, Porter MD. Femtomolar detection of prostate-specific antigen: an immunoassay based on surface-enhanced Raman scattering and immunogold labels. Anal Chem. 2003;75:5936–5943. doi: 10.1021/ac034356f. [DOI] [PubMed] [Google Scholar]
- 37.Gong JL, Liang Y, Huang Y, Chen JW, Jiang JH, Shen GL, Yu RQ. Ag/SiO2 core-shell nanoparticle-based surface-enhanced Raman probes for immunoassay of cancer marker using silica-coated magnetic nanoparticles as separation tools. Biosens Bioelectron. 2007;22:1501–1507. doi: 10.1016/j.bios.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 38.Storhoff JJ, Marla SS, Bao P, Hagenow S, Mehta H, Lucas A, Garimella V, Patno T, Buckingham W, Cork W, Mueller UR. Gold nanoparticle-based detection of genomic DNA targets on microarrays using a novel optical detection system. Biosens Bioelectron. 2004;19:875–883. doi: 10.1016/j.bios.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Darbha GK, Rai US, Singh AK, Ray PC. Gold-nanorod-based sensing of sequence specific HIV-1 virus DNA by using hyper-Rayleigh scattering spectroscopy. Chem Eur J. 2008;14:3896–3903. doi: 10.1002/chem.200701850. [DOI] [PubMed] [Google Scholar]
- 40.Pumera M, Sanchez S, Ichinose I, Tang J. Electrochemical nanobiosensors. Sens Actuat B. 2007;123:1195–1205. doi: 10.1016/j.snb.2006.11.016. [DOI] [Google Scholar]
- 41.Castaneda MT, Alegret S, Merkoci A. Electrochemical sensing of DNA using gold nanoparticles. Electroanalysis. 2007;19:743–753. doi: 10.1002/elan.200603784. [DOI] [Google Scholar]
- 42.Li H, Sun Z, Zhong W, Hao N, Xu D, Chen HY. Ultrasensitive electrochemical detection for DNA arrays based on silver nanoparticle aggregates. Anal Chem. 2010;82:5477–5483. doi: 10.1021/ac101193e. [DOI] [PubMed] [Google Scholar]
- 43.Merkoci A. Electrochemical biosensing with nanoparticles. FEBS J. 2007;274:310–316. doi: 10.1111/j.1742-4658.2006.05603.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang J. Nanoparticle-based electrochemical bioassays of proteins. Electroanalysis. 2007;19:769–776. doi: 10.1002/elan.200603789. [DOI] [Google Scholar]
- 45.Ozkan M. Quantum dots and other nanoparticles: what can they offer to drug discovery? Drug Discov Today. 2004;9:1065–1071. doi: 10.1016/S1359-6446(04)03291-X. [DOI] [PubMed] [Google Scholar]
- 46.Sapsford KE, Pons T, Medintz IL, Mattoussi H. Biosensing with luminescent semiconductor quantum dots. Sensors. 2006;6:925–953. doi: 10.3390/s6080925. [DOI] [Google Scholar]
- 47.Costa-Fernandez JM, Pereiro R, Sanz Medel A. The use of luminescent quantum dots for optical sensing. Tr Anal Chem. 2006;25:207–218. doi: 10.1016/j.trac.2005.07.008. [DOI] [Google Scholar]
- 48.Ekimov AI, Onushchenko AA. Quantum size effect in three-dimensional microscopic semiconductor crystals. JETP Lett. 1981;34:345–349. [Google Scholar]
- 49.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Single quantum-dot-based DNA nanosensor. Nat Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 50.Zhao D, Jime Z, Quanxi D, Ning D, Shichao X, Bo S, Yuehua B. Adaptation of Au nanoparticles and CdTe quantum dots in DNA detection. Clin J Chem Eng. 2007;15:791–794. [Google Scholar]
- 51.Maxwell DJ, Taylor JR, Nie S. Self-assembled nanoparticle probes for recognition and detection of biomolecules. J Am Chem Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Liu G, Merkoci A. Electrochemical coding technology for simultaneous detection of multiple DNA targets. J Am Chem Soc. 2003;125:3214–3215. doi: 10.1021/ja029668z. [DOI] [PubMed] [Google Scholar]
- 53.Liu G, Wang J, Kim J, Jan MR. Electrochemical coding for multiplexed immunoassays of proteins. Anal Chem. 2004;76:7126–7130. doi: 10.1021/ac049107l. [DOI] [PubMed] [Google Scholar]
- 54.Tallury P, Payton K, Santra S. Silica-based multimodal/multifunctional nanoparticles for bioimaging and biosensing applications. Nanomedicine. 2008;3:579–592. doi: 10.2217/17435889.3.4.579. [DOI] [PubMed] [Google Scholar]
- 55.Lian W, Litherland SA, Badrane H, Tan W, Wu D, Baker HV, Gulig PA, Lim DV, Jin S. Ultra-sensitive detection of biomolecules with fluorescent dye-doped nanoparticles. Anal Bioanal Chem. 2004;334:135–144. doi: 10.1016/j.ab.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 56.Smith JE, Wang L, Tan W. Bioconjugated silica-coated nanoparticles for bioseparation and bioanalysis. Tr Anal Chem. 2006;25:848–855. doi: 10.1016/j.trac.2006.03.008. [DOI] [Google Scholar]
- 57.Hemmilae I, Laitala V. Progress in lanthanides as luminescent probes. J Fluoresc. 2005;15:529–542. doi: 10.1007/s10895-005-2826-6. [DOI] [PubMed] [Google Scholar]
- 58.Hempen C, Karst U. Labeling strategies for bioassays. Anal Bioanal Chem. 2006;384:572–583. doi: 10.1007/s00216-005-3392-0. [DOI] [PubMed] [Google Scholar]
- 59.Tang D, Yuan R, Chai Y. Magnetic control of an electrochemical microfluidic device with an arrayed immunosensor for simultaneous multiple immunoassays. Clin Chem. 2007;53:1323–1329. doi: 10.1373/clinchem.2006.085126. [DOI] [PubMed] [Google Scholar]
- 60.Squires TM, Messinger RJ, Manalis SR. Making it stick: convection, reaction and diffusion in surface-based biosensors. Nat Biotechnol. 2008;26:417–426. doi: 10.1038/nbt1388. [DOI] [PubMed] [Google Scholar]
- 61.Jorio A, Dresselhaus G, Dresselhaus MS. Carbon nanotubes: advanced topics in synthesis, structure, properties and applications. Berlin Heidelberg New York: Springer; 2008. [Google Scholar]
- 62.Chen Z, Tabakman SM, Goodwin AP, Kattah MG, Daranciang D, Wang X, Zhang G, Li X, Liu Z, Utz PJ, Jiang K, Fan S, Dai H. Protein microarrays with carbon nanotubes as multicolor Raman labels. Nat Biotechnol. 2008;26:1285–1292. doi: 10.1038/nbt.1501. [DOI] [PubMed] [Google Scholar]
- 63.Yu X, Munge B, Patel V, Jensen G, Bhirde A, Gong JD, Kim SN, Gillespie J, Gutkind JS, Papadimtrakopoulos F, Rusling JF. Carbon nanotube amplification strategies for highly sensitive immunodetection of cancer biomarkers. J Am Chem Soc. 2006;128:11199–11205. doi: 10.1021/ja062117e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Duan XF, Lieber CM. General synthesis of compound semiconductor nanowires. Adv Mater. 2000;12:298–302. doi: 10.1002/(SICI)1521-4095(200002)12:4<298::AID-ADMA298>3.0.CO;2-Y. [DOI] [Google Scholar]
- 65.Roy S, Gao Z. Nanostructure-based electrical biosensors. Nano Today. 2009;4:318–324. doi: 10.1016/j.nantod.2009.06.003. [DOI] [Google Scholar]
- 66.Kasianowicz JJ, Brandin E, Branton D, Deamer WW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad USA. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Derrington IM, Butler TZ, Collins MD, Manrao E, Pavlenok M, Niederweis M, Gundlach JH. Nanopore DNA sequencing with MspA. Proc Natl Acad USA. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J, Stein D, McMullan C, Branton D, Aziz MJ, Golovchenko JA. Ion-beam sculpting at nanometre length scales. Nature. 2001;412:166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 69.Merchant CA, Healy K, Wanunu M, Ray V, Peterman N, Bartel J, Fischbein MD, Venta K, Luo Z, Johnson AT, Drndić M. DNA translocation through graphene nanopores. Nano Lett. 2010;10:3163–3167. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 70.Nelson T, Zhang B, Prezhdo OV. Detection of nucleic acids with graphene nanopores: ab initio characterization of a novel sequencing device. Nano Lett. 2010;10:3237–3242. doi: 10.1021/nl9035934. [DOI] [PubMed] [Google Scholar]
- 71.Kim MJ, Wanunu M, Bell DC, Meller A. Rapid fabrication of uniformly sized nanopores and nanopore arrays for parallel DNA analysis. Adv Mater. 2006;18:3149–3153. doi: 10.1002/adma.200601191. [DOI] [Google Scholar]
- 72.Horvath R, Cottier K, Pedersen HC, Ramsden JJ. Multidepth screening of living cells using optical waveguides. Biosens Bioelectron. 2008;24:799–804. doi: 10.1016/j.bios.2008.06.059. [DOI] [PubMed] [Google Scholar]
- 73.Homola Jiri., editor. Surface plasmon resonance based sensors. Berlin Heidelberg New York: Springer; 2006. [Google Scholar]
- 74.Walcarius A. Ordered porous thin films in electrochemical analysis. Tr Anal Chem. 2008;27:593–603. doi: 10.1016/j.trac.2008.03.011. [DOI] [Google Scholar]
- 75.Xu M, Wang J, Zhou F. Nanomaterial modified electrodes. Curr Topics Electrochem. 2006;11:57–70. [Google Scholar]
- 76.Wang J. Electrochemical biosensors: towards point-of-care cancer diagnostics. Biosens Bioelectron. 2006;21:1887–1892. doi: 10.1016/j.bios.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 77.Shenhar R, Norsten TB, Rotello VM. Polymer-mediated nanoparticle assembly: structural control and applications. Adv Mater. 2005;17:657–669. doi: 10.1002/adma.200401291. [DOI] [Google Scholar]
- 78.Balasubramanian K, Burghard M. Electrochemically functionalized carbon nanotubes for device applications. J Mater Chem. 2008;18:3071–3083. doi: 10.1039/b718262g. [DOI] [Google Scholar]
- 79.Xu K, Huang JR, Ye ZZ, Ying YB, Li YB. Recent development of nano-materials used in DNA biosensors. Sensors. 2009;9:5534–5557. doi: 10.3390/s90705534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu C, Hu S (2009) Carbon nanotube-based electrochemical sensors: principles and applications in biomedical systems. J Sens 187615. doi:10.1155/2009/187615
- 81.Vaddiraju S, Tomazos I, Burgess DJ, Jain FC, Papadimitrakopoulos F. Emerging synergy between nanotechnology and implantable biosensors: a review. Biosens Bioelectron. 2010;25:1553–1565. doi: 10.1016/j.bios.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yogeswaran U, Chen SM. A review on the electrochemical sensors and biosensors composed of nanowires as sensing material. Sensors. 2008;8:290–313. doi: 10.3390/s8010290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yang W, Ratinac KR, Ringer SP, Thordarson P, Gooding JJ, Braet F. Carbon nanomaterials in biosensors: should you use nanotubes or graphene? Angew Chem Int Ed. 2010;49:2114–2138. doi: 10.1002/anie.200903463. [DOI] [PubMed] [Google Scholar]
- 84.Balasubramanian K. Challenges in the use of 1D nanostructures for on-chip biosensing and diagnostics. Biosens Bioelectron. 2010;26:1195–1204. doi: 10.1016/j.bios.2010.07.041. [DOI] [PubMed] [Google Scholar]
- 85.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 86.Maroto A, Balasubramanian K, Burghard M, Kern K. Functionalized metallic carbon nanotube devices for pH sensing. Chem PhysChem. 2007;8:220–223. doi: 10.1002/cphc.200600498. [DOI] [PubMed] [Google Scholar]
- 87.Vlandas A, Kurkina T, Ahmad A, Kern K, Balasubramanian K. Enzyme-free sugar sensing in microfluidic channels with an affinity-based carbon nanotube sensor. Anal Chem. 2010;82:6090–6097. doi: 10.1021/ac1007656. [DOI] [PubMed] [Google Scholar]
- 88.Siqueira JR, Caseli L, Cresphilo FN, Zucolotto V, Oliviera ON. Immobilization of biomolecules on nanostructured films for biosensing. Biosens Bioelectron. 2010;25:1254–1263. doi: 10.1016/j.bios.2009.09.043. [DOI] [PubMed] [Google Scholar]
- 89.Tallury P, Malhotra A, Byrne LM, Santra S. Nanobioimaging and sensing of infectious diseases. Adv Drug Deliv Rev. 2010;62:424–437. doi: 10.1016/j.addr.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rozkiewicz DI, Ravoo BJ, Reinhoudt DN. Chapter 14. Immobilization and patterning of biomolecules on surfaces. In: Rurack K, Martinez-Manez R, editors. The supramolecular chemistry of organic-inorganic hybrid materials. New Jersey: Wiley; 2010. [Google Scholar]
- 91.Jain KK. Nanobiotechnology in molecular diagnostics: current techniques and applications. Norwich (UK): Horizon Bioscience; 2006. [Google Scholar]
- 92.Huo Q. A perspective on bioconjugated nanoparticles and quantum dots. Coll Surf B: Biointerfaces. 2007;59:1–10. doi: 10.1016/j.colsurfb.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 93.Subbiah R, Veerapandian M, Yun KS. Nanoparticles: functionalization and multifunctional applications in biomedical sciences. Curr Med Chem. 2010;17:4559–4577. doi: 10.2174/092986710794183024. [DOI] [PubMed] [Google Scholar]
- 94.Xing ZC, Chang YM, Kang IK. Immobilization of biomolecules on the surface of inorganic nanoparticles for biomedical applications. Sci Technol Adv Mater. 2010;11:014101. doi: 10.1088/1468-6996/11/1/014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gavalas Chopra N, VG Hinds BJ, Bachas LG. Functional one-dimensional nanomaterials: applications in nanoscale biosensors. Anal Lett. 2007;40:2067–2096. doi: 10.1080/00032710701567170. [DOI] [Google Scholar]
- 96.Hao R, Xing RJ, Xu ZC, Hou YL, Gao S, Sun SH. Synthesis, functionalization and biomedical applications of multifunctional magnetic nanoparticles. Adv Mater. 2010;22:2729–2742. doi: 10.1002/adma.201000260. [DOI] [PubMed] [Google Scholar]
- 97.Ma QA, Su XG. Near-infrared quantum dots: synthesis, functionalization and analytical applications. Analyst. 2010;135:1867–1877. doi: 10.1039/c0an00233j. [DOI] [PubMed] [Google Scholar]
- 98.Invitrogen Corp. http://www.invitrogen.com/ (21 March 2011)
- 99.Wu H-C, Chang X, Liu L, Zhao F, Zhao Y. Chemistry of carbon nanotubes in biomedical applications. J Mater Chem. 2010;20:1036–1052. doi: 10.1039/b911099m. [DOI] [Google Scholar]
- 100.Balasubramanian K, Burghard M. Chemically functionalized carbon nanotubes. Small. 2005;1:180–192. doi: 10.1002/smll.200400118. [DOI] [PubMed] [Google Scholar]
- 101.Kurkina T, Vlandas A, Ahmad A, Kern K, Balasubramanian K. Label-free detection of few copies of DNA using carbon nanotube impedance biosensors. Angew Chem Intl Ed. 2011;50:3710–3714. doi: 10.1002/anie.201006806. [DOI] [PubMed] [Google Scholar]
- 102.Elghanian R, Storhoff JJ, Mucic RC, Letsinger RL, Mirkin CA. Selective colorimetric detection of polynucleotides based on the distance-dependent optical properties of gold nanoparticles. Science. 1997;277:1078–1081. doi: 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- 103.Zhao XJ, Hilliard LR, Mechery SJ, Wang YP, Bagwe RP, Jin SG, Tan WH. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticles. Proc Natl Acad Sci USA. 2004;101:15027–15032. doi: 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang L, Zhao W, O’Donoghue B, Tan W. Fluorescent nanoparticles for multiplexed bacteria monitoring. Bioconjugate Chem. 2007;18:297–301. doi: 10.1021/bc060255n. [DOI] [PubMed] [Google Scholar]
- 105.Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1756–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 106.Park S-J, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1066348. [DOI] [PubMed] [Google Scholar]
- 107.Wang J, Liu G, Jan MR. Ultrasensitive electrical biosensing of proteins and DNA: carbon-nanotube derived amplification of the recognition and transduction events. J Am Chem Soc. 2004;126:3010–3011. doi: 10.1021/ja031723w. [DOI] [PubMed] [Google Scholar]
- 108.Stoeva SI, Lee J-S, Thaxton S, Mirkin CA. Multiplexed DNA detection with biobarcoded nanoparticle probes. Angew Chem Int Ed. 2006;45:3303–3306. doi: 10.1002/anie.200600124. [DOI] [PubMed] [Google Scholar]
- 109.Wanekaya AK, Chen W, Myung NV, Mulchandani A. Nanowire-based electrochemical biosensors. Electroanalysis. 2006;18:533–550. doi: 10.1002/elan.200503449. [DOI] [Google Scholar]
- 110.Sha MY, Walton SM, Norton SM, Taylor M, Yamanaka M, Natan MJ, Xu C, Drmanac S, Huang A, Borcherding R, Penn SG. Multiplexed SNP genotyping using nanobarcode particle technology. Anal Bioanal Chem. 2006;384:658–666. doi: 10.1007/s00216-005-0225-0. [DOI] [PubMed] [Google Scholar]
- 111.Tok JBH, Chuang FYS, Kao MC, Rose KA, Pannu SS, Sha MY, Chakarova G, Penn SG, Dougherty GM. Metallic striped nanowires as multiplexed immunoassay platforms for pathogen detection. Angew Chem Int Ed. 2006;45:6900–6904. doi: 10.1002/anie.200601104. [DOI] [PubMed] [Google Scholar]
- 112.Ki E-Y, Stanton J, Vega RA, Kunstman KJ, Mirkin CA, Wolinsky SM. A real-time PCR-based method for determining the surface coverage of thiol-capped oligonucleotides bound onto gold nanoparticles. Nucl Acids Res. 2006;34:e54. doi: 10.1093/nar/gkl135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nicewarner-Pena SR, Freeman RG, Reiss BD, He L, Pena DJ, Walton ID, Cromer R, Keating CD, Natan MJ. Submicrometer metallic barcodes. Science. 2001;294:137–141. doi: 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- 114.Han M, Gao X, Su JZ, Nie S. Quantum dot-tagged microbeads for multiplexed optical coding of biomolecules. Nat Biotechnol. 2001;19:631–635. doi: 10.1038/90228. [DOI] [PubMed] [Google Scholar]
- 115.Haes AJ, Chang L, Klein WL, van Duyne RP. Detection of a biomarker for Alzheimer’s disease from synthetic and clinical samples using a nanoscale optical biosensor. J Am Chem Soc. 2005;127:2264–2271. doi: 10.1021/ja044087q. [DOI] [PubMed] [Google Scholar]
- 116.Schasfoort RBM, Tudos AJ. Handbook of surface plasmon resonance. London: Royal Society of Chemistry; 2008. [Google Scholar]
- 117.Xia X, Ye ZZ, Wu J, Yi-Bin Y. Application and research development of surface plasmon resonance-based immunosensors for protein detection. Chin J Anal Chem. 2010;38:1052–1059. doi: 10.1016/S1872-2040(09)60059-1. [DOI] [Google Scholar]
- 118.Gao Z, Agarwal A, Trigg AD, Singh N, Fang C, Tung CH, Fan Y, Buddharaju KD, Kong J. Silicon nanowire arrays for label-free detection of DNA. Anal Chem. 2007;79:3291–3297. doi: 10.1021/ac061808q. [DOI] [PubMed] [Google Scholar]
- 119.Hahm J, Lieber CM. Direct ultrasensitive electrical detection of DNA and DNA sequence variations using nanowire nanosensors. Nano Lett. 2004;4:51–54. doi: 10.1021/nl034853b. [DOI] [Google Scholar]
- 120.Star A, Tu E, Niemann J, Gabriel JCP, Joiner CS, Valcke C. Label-free detection of DNA hybridization using carbon nanotube network field-effect transistors. Proc Natl Acad Sci USA. 2006;103:921–926. doi: 10.1073/pnas.0504146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pachauri V, Vlandas A, Kern K, Balasubramanian K. Site-specific self-assembled liquid-gated ZnO nanowire transistors for sensing applications. Small. 2010;6:589–594. doi: 10.1002/smll.200900876. [DOI] [PubMed] [Google Scholar]
- 122.Jacobs CB, Peairs MJ, Venton BJ. Review: carbon nanotube-based electrochemical sensors for biomolecules. Anal Chim Acta. 2010;662:105–127. doi: 10.1016/j.aca.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 123.Sadik OA, Aluoch AO, Zhou AL. Status of biomolecular recognition using electrochemical techniques. Biosens Bioelectron. 2009;24:2749–2765. doi: 10.1016/j.bios.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 124.Wei D, Bailey MJA, Andrew P, Ryhanen T. Electrochemical biosensors at the nanoscale. Lab Chip. 2009;9:2123–2131. doi: 10.1039/b903118a. [DOI] [PubMed] [Google Scholar]
- 125.Wu J, Fu Z, Yan F, Ju H. Biomedical and clinical applications of immunoassays and immunosensors for tumor markers. Tr Anal Chem. 2007;26:679–688. doi: 10.1016/j.trac.2007.05.007. [DOI] [Google Scholar]
- 126.Holme D, Peck H. Analytical biochemistry. New Jersey: Prentice Hall; 1998. [Google Scholar]
- 127.Hirsch LR, Jackson JB, Lee A, Halas NJ, West JL. A whole blood immunoassay using gold nanoshells. Anal Chem. 2003;75:2377–2381. doi: 10.1021/ac0262210. [DOI] [PubMed] [Google Scholar]
- 128.Kerman K, Endo T, Tsukamoto M, Chikae M, Takamura Y, Tamiya E. Quantum dot–based immunosensor for the detection of prostate-specific antigen using fluorescence microscopy. Talanta. 2007;71:1494–1499. doi: 10.1016/j.talanta.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 129.Tan M, Wang G, Hai X, Ye Z, Yuan J. Development of functionalized fluorescent europium nanoparticles for biolabeling and time-resolved fluorometric applications. J Mater Chem. 2004;14:2896–2901. doi: 10.1039/b407535h. [DOI] [Google Scholar]
- 130.Sano T, Smith CL, Cantor CR. Immuno-PCR: very sensitive antigen detection by means of specific antibody-DNA conjugates. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 131.Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stern E, Klemic JF, Routenberg DA, Wyrembak PN, Turner-Evans DB, Hamilton AD, LaVan DA, Fahmy TM, Reed MA. Label-free immunodetection with CMOS-compatible semiconducting nanowires. Nature. 2007;445:519–522. doi: 10.1038/nature05498. [DOI] [PubMed] [Google Scholar]
- 133.Maehashi K, Matsumoto K, Takamura Y, Tamiya E. Aptamer-based label-free immunosensors using carbon nanotube field-effect transistors. Electroanalysis. 2009;21:1285–1290. doi: 10.1002/elan.200804552. [DOI] [Google Scholar]
- 134.QuantiTect Multiplex PCR Kits, Qiagen Inc. http://www.qiagen.com/ (21 March 2011)
- 135.Hill HD, Vega RA, Mirkin CA. Nonenzymatic detection of bacterial genomic DNA using the bio bar code assay. Anal Chem. 2007;79:9218–9233. doi: 10.1021/ac701626y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 137.Morgan MJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 138.LaFramboise T. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucl Acids Res. 2009;37:4181–4193. doi: 10.1093/nar/gkp552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gilman B, Schaffner S, Van Etten WJ, Reich D, Higgins J, Daly MJ, Blumenstiel B, Baldwin J, Stange-Thomann N, Zody MC, Linton L, Lander ES, Altshuler D. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 140.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J Am Chem Soc. 1998;120:1959–1964. doi: 10.1021/ja972332i. [DOI] [Google Scholar]
- 141.Gerion D, Chen F, Kannan B, Fu A, Parak WJ, Chen DJ, Majumdar A, Alivisatos AP. Room-temperature single-nucleotide polymorphism and multiallele DNA detection using fluorescent nanocrystals and microarrays. Anal Chem. 2003;75:4766–4772. doi: 10.1021/ac034482j. [DOI] [PubMed] [Google Scholar]
- 142.Dubertret B, Calame M, Libchaber AJ. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 143.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 144.Akeson M, Branton D, Kasianowicz JJ, Brandin E, Deamer DW. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wu HC, Astier Y, Maglia G, Mikhailova E, Bayley H. Protein nanopores with covalently attached molecular adapters. J Am Chem Soc. 2007;129:16142–16148. doi: 10.1021/ja0761840. [DOI] [PubMed] [Google Scholar]
- 147.Clarke J, Wu H-C, Jayasinghe L, Patel A, Reid S, Bayley H. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotech. 2009;4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 148.Bubendorf L. High throughput microarray technologies: from genomics to clinics. Eur Urol. 2001;40:231–238. doi: 10.1159/000049777. [DOI] [PubMed] [Google Scholar]
- 149.Campas M, Katakis I. DNA biochip arraying, detection and amplification strategies. Tr Anal Chem. 2004;23:49–62. doi: 10.1016/S0165-9936(04)00104-9. [DOI] [Google Scholar]
- 150.Smith AM, Dave S, Nie S, True L, Gao X. Multicolor quantum dots for molecular diagnostics of cancer. Expert Rev Mol Diagn. 2006;6:231–244. doi: 10.1586/14737159.6.2.231. [DOI] [PubMed] [Google Scholar]
- 151.Russ AW, Kurll UJ. Developing mixed films of immobilized oligonucleotides and quantum dots for the multiplexed detection of nucleic acid hybridization using a combination of fluorescence resonance energy transfer and direct excitation of fluorescence. Langmuir. 2010;26:6041–6047. doi: 10.1021/la903751m. [DOI] [PubMed] [Google Scholar]
- 152.Zhang CY, Hu J. Single Quantum Dot-based nanosensor for multiple DNA detection. Anal Chem. 2010;82:1921–1927. doi: 10.1021/ac9026675. [DOI] [PubMed] [Google Scholar]
- 153.Taton TA, Lu G, Mirkin CA. Two-color labeling of oligonucleotide arrays via size-selective scattering of nanoparticle probes. J Am Chem Soc. 2001;123:5164–5165. doi: 10.1021/ja0102639. [DOI] [PubMed] [Google Scholar]
- 154.Azzazy HME, Mansour MMH. In vitro diagnostic prospects of nanoparticles. Clin Chim Acta. 2009;403:1–8. doi: 10.1016/j.cca.2009.01.016. [DOI] [PubMed] [Google Scholar]