Abstract
mRNA localization is a mechanism used by various organisms to control the spatial and temporal production of proteins. This process is a highly regulated event that requires multiple cis- and trans-acting elements that mediate the accurate localization of target mRNAs. The intrinsic nature of localization elements, together with their interaction with different RNA-binding proteins, establishes control mechanisms that can oversee the transcript from its birth in the nucleus to its specific final destination. In this review, we aim to summarize the different mechanisms of mRNA localization, with a particular focus on the various control mechanisms that affect the localization of mRNAs in the cytoplasm.
Keywords: Cytoplasmic RNA transport, RNA localization, Localization elements, RNA-binding proteins
Introduction
The cytoplasmic transport and localization of mRNAs is a post-transcriptional mechanism that restricts the synthesis of proteins to specific sites within cells. For years, studies on mRNA localization were limited to a few localized transcripts in a small number of model systems. Now, this process has been observed in several branches of the eukaryotic tree, from fungi to mammals and plants [1–3]. Recent evidence even points to translation-independent mRNA localization in eubacteria [4]. Moreover, in the past few years, large scale screens based on biochemical, genomic, or cytological assays identified subpopulations of mRNAs that are localized to several subcellular domains or organelles, such as the endoplasmic reticulum [5], mitochondria [6], the bud of yeast cells [7], Drosophila blastoderm embryo [8], the mitotic microtubules [9], dendrites [10], axons [11, 12], and pseudopodia [13]. These studies revealed that mRNA localization is more widespread and involves a larger set of transcripts than previously thought. Typical examples of mRNA localization are presented in Fig. 1.
Fig. 1a–e.
Examples of localized mRNAs. a Localization of the ASH1 mRNA (red) at the bud tip of the budding yeast Saccharomyces cerevisiae (left panel). Nuclear DNA is in blue. Nomarski image is shown in the right panel. Image taken from [59]. b Localization of Vg1 localization element (VLE) RNA (red) at the vegetal pole of Xenopus oocyte. The image was provided by James Gagnon and Kim Mowry, Brown University. c Localization of bicoid mRNA (green) at the anterior pole of a Drosophila embryo. Red DAPI. d Localization of nanos mRNA at the posterior pole of a Drosophila embryo. Red DAPI. The images were provided by Éric Lécuyer, Institut de Recherche Clinique de Montréal. e CamKIIa mRNA granules (red) in dendrites of cultured hippocampal neuron. White square presents higher magnification of RNA granules in dendrites (showed in the upper right panel). The image was provided by Sharon Swanger and Gary Bassell, Emory University [189]
Why do cells use this mechanism to regulate gene expression? One reason is that mRNA localization limits the expression of a protein to a specific time and place. Indeed, several localized mRNAs encode proteins involved in asymmetric cell division or cell fate determination, which requires a delicate spatiotemporal regulation of these factors [14]. This mechanism also facilitates protein sorting to organelles, as several mRNAs coding for mitochondrial, endoplasmic reticulum (ER), and even peroxisome proteins are enriched at these organelles [15, 16]. Another function of mRNA localization is to restrict the expression of potentially toxic proteins. A good example is the mRNA of myelin basic protein MBP, which is localized to myelinating oligodendrocyte processes [17]. Disruption in the localization of MBP mRNA leads to ectopic MBP expression and aberrant myelination in vivo [18].
In this article, we review the main mechanisms of mRNA localization with a special focus on the different levels of control of the localization of mRNAs. We will pay attention to three levels of control: (1) the nuclear events leading to the recognition of localized transcripts by their localization factors, (2) the role of cis-acting elements in modulating mRNA localization, and (3) the extracellular signaling pathways that regulate mRNA localization. We will not cover the translational control of localized mRNA since excellent reviews on this subject have been published recently [19–21].
Mechanisms of mRNA localization
For several transcripts, their localization depends on the presence of a peptide sequence in the emerging peptidic chain that targets the translated mRNA to its site of localization. This mechanism has been well described for transcripts encoding secreted or membrane proteins that are localized to the ER, for instance [22]. Nevertheless, a large number of transcripts do not depend on translation for their localization, as they are frequently transported in a translationally repressed form. A common theme for these localized mRNAs is that they contain cis-acting elements in their sequences, called zipcodes or localization elements, which are recognized by specific trans-acting RNA-binding proteins [23]. These localization elements are commonly found in the 3′ untranslated region (UTR) of transcripts [23]. However, in several cases, localization elements have been identified in the 5′UTR or coding region of mRNAs [24, 25]. The RNA-binding proteins associated with a transcript determine the mechanism used for its localization.
Diffusion and entrapment of mRNA
One of the simplest ways to localize an mRNA is for the transcript to diffuse in the cytoplasm and then become trapped at a specific location via binding to anchors. During late stages of oogenesis in Drosophila, Nanos is localized at the posterior pole of embryos, where it acts as a translational repressor of hunchback mRNA, thereby allowing expression of genes involved in abdominal development [26]. Posterior localization of this protein is also essential for germ cell development [27]. However, only 4% of total nanos mRNA is localized at the posterior and the rest can be found throughout the cytoplasm [28]. Live cell imaging of endogenous nanos mRNA by GFP-tagging revealed a motor-independent mechanism for localization of this transcript at the oocyte posterior [29]. When oocytes are treated with microtubule depolymerization agents, nanos mRNA can still localize to the posterior pole, but since cytoplasmic streaming is reduced, less nanos mRNA gets to the pole [29]. This suggests that nanos mRNA is localized mainly by a diffusion-based mechanism. On the other hand, while actin filaments are not involved in nanos mRNA transport, they are necessary for its anchoring at the posterior pole [29, 30].
Another well studied example of the diffusion-entrapment mechanism is the Xenopus Xcat-2 mRNA, which encodes a Nos related zinc-finger RNA-binding protein [31]. During the early stages of Xenopus oogenesis, Xcat-2 mRNA is restricted to a specific structure in the cytoplasm called the mitochondrial cloud (MC). The mitochondrial cloud, also called Balbiani body, consists mostly of mitochondria and small vesicles, and is the source of germinal granule material [32]. The movement of the MC in the cytoplasm results in the localization of the Xcat-2 mRNA at the vegetal cortex. Transfer of this mRNA to the vegetal pole is suggested to occur through a region of the MC called the Message transport organizing center (METRO). Xcat-2 mRNA accumulates within this region and subsequently localizes to the vegetal cortex (Fig. 2a) [33, 34]. Direct evidence that supports the diffusion and entrapment model for this mRNA came from a study in which the authors used time-lapse confocal microscopy and FRAP analysis to monitor the movement of injected fluorescent Xcat-2 RNA construct. Upon injection, the Xcat-2 mRNA diffuses into the cytoplasm and subsequently aggregates in the METRO region of the MC [35]. This pathway is motor-independent since depolymerization of the microtubules has no effect on the localization of Xcat-2 in MC [35]. However, a recent study reported that Xcat-2 mRNA localization during early stages of oogenesis is reduced by inhibition of Kinesin II, suggesting that the migration of Xcat-2 mRNA to the MC depends to some extent on this molecular motor [36]. Finally, anchoring of the Xcat-2 transcript upon arrival to the MC is suggested to occur via its association with a distinct domain of the ER in the MC region and is independent of germinal granule formation [35]. However, factors mediating this association still remain to be determined.
Fig. 2a–e.
Schematic presentation of different mechanisms of mRNA localization. a Diffusion and entrapment of Xcat2 mRNA in the mitochondrial cloud during early oogenesis in Xenopus. The movement of the mitochondrial cloud toward the vegetal cortex during stages 2–4 (indicated by a thick arrow) leads to the localization of this mRNA at the vegetal pole. b Selective stabilization of Hsp83 mRNA at the posterior pole of Drosophila embryo. Smaug protein triggers the degradation of unlocalized Hsp83 transcripts elsewhere in the embryo. c Localization of the ASH1 mRNA to the bud tip of yeast cells. Upon transcription and nuclear export, this mRNA is actively transported by a myosin-based mechanism on actin filaments. d Localization of oskar and bicoid mRNA in Drosophila oocytes. During Drosophila oogenesis, oskar and bicoid mRNA are synthesized in the nurse cells, transported into the ooplasm, and localize to the posterior and anterior poles of the oocyte, respectively. Arrows indicate the movement and direction of RNP complexes along the microtubules. e In immature neurons, β-actin mRNA is actively transported to the neuronal growth cone through microtubule-based transport
Selective degradation and stabilization of mRNA
Localized stabilization of a transcript is another mechanism by which an mRNA can be subcellularly targeted. In this case, an mRNA is rapidly degraded in most parts of the cell, but it is protected from degradation at a specific location. The hsp83 mRNA, which encodes a heat shock protein in Drosophila, is a well-characterized example of this kind of localization (Fig. 2b). This transcript is localized at the posterior pole of the early Drosophila embryo by the selective stabilization of the mRNA at the posterior pole and degradation of the transcript elsewhere in the cytoplasm [37]. The level of hsp83 mRNA, which is a maternally encoded transcript, decreases more rapidly in embryos than in unfertilized eggs, which suggests that two separate mechanisms control the stability of this transcript [38]. These two independent pathways, which are called “maternal” and “zygotic” pathways, use maternally and embryonic encoded proteins, respectively, to degrade the hsp83 transcript [38]. By analyzing the 3′UTR of hsp83 mRNA, a region from nucleotides 253–349 was identified as the Hsp83 degradation element (HDE), which directs the destabilization of this mRNA in unfertilized eggs. However, this region has no effect in the zygotic degradation pathway, and transcripts without the HDE domain are subject to degradation by the embryonic degradation machinery [38]. The hsp83 ORF has also been shown to affect the stability of the transcript. A region at the 3′ end of the ORF, which comprises 615 nucleotides, has been found to be responsible for this destabilization, and was consequently called Hsp38 instability element (HIE) [39]. This region, which has the major effect in the destabilization of the transcript, functions together with the HDE for complete degradation. The HIE domain contains six stem-loop structures that are recognized by the maternally encoded RNA-binding protein Smaug [39, 40]. It was shown that in Smaug mutants, degradation and thus localization of hsp83 mRNA are impaired. Smaug recruits the CCR4/POP2/NOT deadenylase complex, triggering deadenylation and thus degradation of the hsp83 transcript [40]. Although Smaug is present throughout the pole plasm, the hsp83 mRNA is protected from Smaug action at the posterior pole. This protection is related to a 57 nt region in the 3′UTR (nucleotides 351–407) downstream of HDE, which is called HPE (Hsp83 protection element). HPE is sufficient to confer stability to an unstable transcript at the pole plasm [40]. The mechanism by which this domain functions is not clear, and may include interaction of trans-acting factors that block the availability of the transcript to Smaug.
Smaug is involved in the degradation of several maternal mRNAs in Drosophila, and its expression is regulated by the PAN GU kinase (PNG), which controls smaug mRNA translation [41]. Among the maternal transcripts whose selective degradation by the Smaug/CCR4 complex restricts their localization at the posterior pole is the nanos mRNA. As mentioned in the previous section on diffusion and entrapment of mRNA, only 4% of total nanos mRNA is localized at the posterior pole, while the bulk of nanos mRNA is distributed in the cytoplasm in a translationally repressed complex and is actively degraded. Degradation of nanos mRNA depends on the CCR4/NOT deadenylase complex and is recruited on this transcript by Smaug [42]. Deadenylation of nanos mRNA by CCR4/NOT also contributes to the translational repression of this transcript. Only a fraction of nanos mRNA is stabilized at the posterior pole of the embryo, and this occurs via the action of Oskar, which prevents the binding of Smaug to the nanos mRNA [42]. However, Smaug is not the only activator of nanos mRNA decay, and a recent study reported that the piRNA pathway is also involved in nanos mRNA degradation [43]. In Drosophila, piRNAs or Piwi-interacting RNAs, are small RNAs of 22–30 nucleotides that are abundant in the germ line and which mostly derive from transposons or other repeated elements (reviewed in [44]). These RNAs are associated with PIWI-types Argonaute proteins such as Piwi, Aubergine, and Ago3, and they participate in the silencing of transposons by directing the cleavage of transposon transcripts. Simonelig and colleagues found that mutants of factors involved in piRNAs biogenesis (such as Armitage, Spindle-E, and Squash) or in the Piwi-type Argonaute proteins Aubergine and Ago3, which are associated with piRNAs, result in the stabilization of nanos mRNA and ectopic expression of Nanos protein throughout the embryo [43]. Deletion of putative piRNA binding sites in the 3′UTR of nanos mRNA increases the stability of this transcript and results in embryo patterning defects. Interestingly, Aubergine, Ago3, Smaug, CCR4, and nanos mRNA are present in the same complex, suggesting that Smaug/CCR4 and the piRNA pathway cooperate to regulate nanos mRNA deadenylation and degradation.
Active transport
The motor-based transport of transcripts is used by most cell types to actively localize mRNAs to specific cellular subregions [45]. In the cytoplasm, the ribonucleoprotein (RNP) complex associates with molecular motors that are required for active transport along the cytoskeleton [46]. Microtubule-dependent motors, such as the minus-end dynein or the plus-end kinesin motors, are commonly used for the transport of mRNA over long distances, either in fungi, Drosophila, Xenopus, or mammalian cells (see examples below) [46, 47]. The non-canonical type V myosin is also used as a molecular motor to carry transcripts on the actin cytoskeleton, usually over short distances (see examples below) [48, 49].
Nuclear events that control cytoplasmic mRNA localization
It is well established that nuclear processing of nascent mRNA affects its cytoplasmic fate by controlling different mechanisms, including translation and degradation [50]. The possibility that nuclear events also affect the cytoplasmic localization of an mRNA emerged from the finding that several RNA-binding proteins involved in mRNA localization are either nuclear residents or shuttle between the nucleus and the cytoplasm [51]. Indeed, several lines of evidence now suggest that localized mRNAs are “marked” in the nucleus prior to their export to the cytoplasm. Different mechanisms leading to the “marking” of localized mRNAs in the nucleus have been uncovered in the past few years.
Cotranscriptional recruitment of mRNA localization factors
The observation that mRNAs can be “marked” for cytoplasmic localization in the nucleus raises the possibility that these events may occur on the nascent transcripts, during transcription. The ZBP1 protein1, which is involved in the localization of β-actin mRNA to the leading edge of chicken embryo fibroblasts, is predominately cytoplasmic, but it shuttles between the nucleus and cytoplasm and is recruited cotranscriptionally to the β-actin mRNA [52]. Another zipcode-binding protein, ZBP22, is also involved in β-actin mRNA localization [53]. ZBP2 is a predominately nuclear protein, which is also recruited cotranscriptionally to the nascent β-actin mRNA [54]. During β-actin mRNA synthesis, ZBP2 binds to the zipcode before ZBP1, and its binding favors ZBP1 recruitment. However, both proteins recognize distinct but adjacent motifs of the β-actin zipcode. It was suggested that upon ZBP1 binding, ZBP2 dissociates from the zipcode, thus the mRNA that leaves the nucleus contains only ZBP1 [54]. This differential binding of ZBP2 and ZBP1 therefore mediates the assembly of the β-actin mRNA localization complex before its export to the cytoplasm.
How important is the nuclear recognition of localized mRNAs by their localization factors and to what extent does the transcription machinery participate in this process? Evidence from the budding yeast suggests that early nuclear events are crucial to ultimately define the cytoplasmic localization of a transcript. The yeast RNA-binding protein She2 interacts directly with cis-acting elements of bud-localized transcripts and directs them to the localization machinery [55, 56]. She2, unlike other proteins of the locasome (i.e., the multiproteic complex involved in the transport and targeting of bud localized mRNAs in S. cerevisiae, see more details below), is present in both the nucleus and cytoplasm. Indeed, it was shown that She2 shuttles between the cytoplasm and nucleus, and that nuclear export of She2 depends on binding to RNA, suggesting that it recognizes its target mRNAs in the nucleus [57]. She2 shuttling depends on a non-classical nuclear localization signal (NLS) and mutations in this motif lead to impaired localization of ASH1 mRNA to the bud tip, suggesting that nuclear shuttling of She2p is important for ASH1 mRNA sorting [58, 59]. Recently it was shown that She2 interacts with the elongating RNA polymerase II via the transcription elongation factor Spt4-Spt5/DSIF [60]. This interaction leads to the co-transcriptional recruitment of She2 to bud-localized transcripts, and mutants of SPT4 and SPT5 were found to display defective ASH1 mRNA localization. This finding reveals an important role for the transcription machinery in the very first steps of localization events in the nucleus and shows that the association of She2 with the transcription machinery promotes the recruitment of this factor on nascent transcripts [61].
Splicing and mRNA localization
Splicing of pre-mRNA is a mechanism for the excision of introns from the protein-coding exonic sequences. A pre-mRNA can also give rise to different transcripts by alternative splicing of its exons, coding for different variants of a protein [62]. Splicing variants can result in the retention or elimination of a localization element in an mRNA, leading to various transcripts that can localize to different parts of a cell. An example is the stardust A (sdt) mRNA, which localizes at the apical pole of Drosophila follicle cell epithelium, leading to the apical localization of the Sdt protein, a regulator of the Crb complex [63]. Localization of sdt mRNA is regulated during development and depends on the alternative splicing of coding exon 3, which is thought to contain a localization element [63]. While exon 3 is present during early oogenesis, leading to the localization of sdt mRNA to the apical pole, exclusion of this exon by alternative splicing at stage 10 results in unlocalized sdt mRNA. Other localized mRNAs are known to be regulated via alternative splicing of their localization elements, such as the cyclin B mRNA in Drosophila [64]. Surprisingly, intronic sequences can also mediate mRNA localization. Indeed, a recent study revealed that retention of specific introns in neuronal transcripts could be involved in their localization at dendrites [65]. These cytoplasmic intron sequence-retaining transcripts (CIRTS) are present in mammalian neurons and contain introns with short interspersed repetitive elements (SINE) derived from the BC1 RNA. These elements are thought to be responsible for the localization of the non-coding BC1 RNA to dendrites in rodents [66]. However, recent in vivo studies in mice were unable to confirm the role of these elements in dendritic mRNA localization [67].
The splicing reaction itself can be an important factor for cytoplasmic localization of an mRNA. In metazoans, splicing of pre-mRNA leaves a specific complex on sequences located upstream of the exon–exon junction: the exon junction complex (EJC). This complex is exported to the cytoplasm with the mature mRNA and remains associated until the first round of translation [68]. The EJC plays an important role in mRNA export, translation, and quality control [69]. Surprisingly, a role for the EJC in the localization of oskar mRNA at the posterior pole of Drosophila oocyte has also been uncovered. Indeed, mutants of components of the EJC, such as eIF4AIII, Mago Nashi, and Tsunagi/Y14 disrupt oskar mRNA localization [70–72]. Interestingly, only the splicing of the first intron is important for proper localization, as its deletion leads to impaired localization at the posterior pole. However, oskar transgenes in which the first intron was substituted by another intron sequence still localized their mRNA normally, suggesting that it is the position of the EJC complex that is important for localization [70]. The mechanism by which nuclear splicing is coupled to cytoplasmic localization of oskar mRNA is probably via an interaction with Barentz (Btz), a cytoplasmic protein essential for oskar localization [73]. Btz interacts with eIF4AIII and is subsequently recruited to the oskar mRNA localization complex [74]. Since eIF4AIII is a general factor present in the EJC of all spliced mRNAs, it is not clear how Btz would recognize only the oskar mRNA.
Nuclear assembly of the mRNA localization complex
As mentioned above, some of the trans-acting factors involved in mRNA localization shuttle between the nucleus and the cytoplasm, suggesting that the mRNA localization process is initiated in the nucleus. In Xenopus, the localization of maternal transcripts such as the Vg1 and VegT mRNAs to the vegetal pole of oocytes depends on a multiprotein complex that contains the RNA-binding proteins hnRNP I and the Xenopus homolog of ZBP1, the Vg1RBP/Vera protein [75–77]. The vegetal localization of Vg1 is mediated by localization elements in the 3′UTR of the transcript [78]. The Vera protein recognizes and specifically binds the localization elements of Vg1 and VegT mRNAs in the oocytes and directs their localization via formation of an mRNP complex [79, 80]. By analyzing RNA–protein interactions of oocyte mRNP complexes, Kress and colleagues [81] found that hnRNP I and Vera associate with Vg1 and VegT mRNAs in both the nucleus and the cytoplasm, indicating that the very first step of cytoplasmic localization of maternal mRNAs in the oocytes starts in the nucleus. Upon nuclear export, additional factors such as xStau, the Xenopus homolog of Staufen, and Prrp are recruited to the mRNP complex and direct their vegetal localization during mid-oogenesis [82, 83]. Moreover, nuclear RNA-binding proteins, such as the yeast Loc1 protein [84] or the vertebrate ZBP2 protein [53], bind and promote the cytoplasmic localization of ASH1 and β-actin mRNA, respectively. This further supports the idea of a nuclear “marking” of mRNAs destined for a particular cytoplasmic localization.
Cis-acting localization elements and the control of mRNA localization
Once in the cytoplasm, marked mRNAs are targeted for their specific localization. As mentioned above, the presence of a cis-acting localization element is essential for the recognition and targeting of an mRNA via a specific localization pathway. For some localized transcripts, the presence of a single localization element is sufficient to allow efficient localization [85]. However, the analysis of localization elements in several transcripts revealed a more complex picture, in which redundant or different localization elements are required for optimal localization of an mRNA.
Redundant localization elements can act cooperatively to promote mRNA localization
Having several copies of the same localization element in an mRNA is a way to ensure the efficient transport and localization of a transcript and can be a consequence of the type of molecular motor used for its transport. Indeed, association with a poorly processive motor may require the recruitment of several copies of this motor to a single mRNA in order to maintain continuous transport. A good example is the ASH1 mRNA in the budding yeast Saccharomyces cerevisiae, which is localized at the bud tip during late anaphase of the cell cycle [86, 87]. Localization of ASH1 mRNA is essential for the asymmetric distribution of Ash1, which acts as a transcriptional repressor of the HO endonuclease and results in inhibition of mating-type switching in daughter cells [88, 89]. The ASH1 mRNA contains four localization elements, three in the coding sequence (E1, E2A, and E2B) and one overlapping the end of the coding sequence and the 3′UTR (E3) [25, 90]. While the presence of these four elements leads to an optimal localization, deletion analysis revealed that each element is sufficient for localization of a reporter mRNA to the bud. When each of these elements was inserted in multiple copies in the 3′UTR, the new constructs showed nearly normal localization. However, for these mRNAs, the asymmetric distribution of Ash1 was impaired, suggesting that the position of these elements is important for Ash1 sorting but not for ASH1 mRNA localization [91]. Although the primary sequences of the four ASH1 localization elements are different, they all fold into a stem-loop structure that contains a few conserved nucleotides [92, 93]. All four elements interact with the same RNA-binding protein called She2, which is involved in the localization of bud-localized mRNAs in S. cerevisiae [7, 55, 56]. She2 forms a tetramer under physiological conditions, and mutations that disrupt this tetrameric state abolish its RNA-binding capacity and impair She2-dependent localization to the bud tip [94]. She2 interacts directly with the C-terminal domain of She3, an adaptor protein that links the She2–mRNA complex to the molecular motor Myo4 (Fig. 2c) [55, 95, 96]. Recent evidence also suggests that She3, besides its role in connecting the She2–RNA complex to Myo4, is itself able to bind RNA and acts synergistically with She2 to increase the affinity and specificity of RNA binding [97].
Recent studies on Myo4 helped to explain why multiple localization elements are required for proper ASH1 mRNA localization. Myo4 is a class V myosin whose main function is the transport of mRNAs to the bud tip using actin filaments [98–100]. Myo4, unlike other type V myosins, is a nonprocessive monomer in vivo, but it becomes processive when present in the form of oligomers [101, 102]. Purification of the localization complex associated with a single localization element revealed that multiple copies of Myo4 are associated with this RNA [103]. Moreover, increasing the number of Myo4 attached to the ASH1 mRNA increased the efficiency of localization of this transcript. These results suggest that each localization element interacts with higher order protein complexes in which a She2 tetramer may recruit multiple copies of Myo4, thus ensuring a continuous and processive movement of the mRNP complex into the bud. Moreover, it is possible that a She2 tetramer binds simultaneously to the localization elements of a single transcript or, alternatively, to those of different mRNAs. This would bring multiple mRNAs together within a single complex in which several Myo4 molecules modulate their transport to the bud tip [103].
Even if the localization of a transcript occurs independently of a molecular motor, redundant elements may still be required for efficient localization. An example is the Drosophila nanos mRNA, which is localized by a diffusion-based mechanism at the posterior pole during late stages of oogenesis, allowing abdominal and germ cell development (see “mRNA diffusion and entrapment” section above) [26–29]. Nanos mRNA contains cis-acting elements in its 3′UTR that are sufficient for posterior localization [104]. These localization elements are located in a large and complex region of 547 nucleotides, which consists of partially redundant sequence elements, named +1 to +4. The combination of these four localization elements is necessary for complete localization of nanos mRNA [105]. However, a 41 nt region within element +2′, called the minimal element (ME), which is highly conserved between D. melanogaster and D. virilis, was shown to be sufficient for localization when present in three copies [106]. So far, two trans-acting factors have been found to interact with the nanos mRNA localization elements. Rumpelstiltskin (Rump), a Drosophila homolog of hnRNP M, which was previously described as a protein of 75 kD (p75), interacts directly and specifically with the 5′ region of the +2′ element in vitro and in vivo [106, 107]. Mutations that disrupt the binding of Rump abolish the localization capacity of the +2′ element, suggesting a role for this protein in the localization of nanos mRNA. However, the fact that some mutations disrupt localization but not Rump binding suggests that other factors are also needed for proper nanos mRNA localization. Indeed, recent evidence suggest that Aubergine, a protein previously shown to be required for oskar mRNA localization and translational control, is also involved in nanos mRNA localization. However Aubergine’s effect on nanos seems to be independent of its role in RNA silencing (see above) and oskar mRNA localization [108, 109].
Different localization elements act together to promote mRNA localization
The presence of several localization elements in the same transcript can be a way to recruit different trans-acting factors, each one having a specific role in the localization process or both having redundant functions. In chicken embryo fibroblasts (CEFs), the β-actin mRNA localizes at the lamellipodia, which results in fibroblast’s polarity and motility [110]. Localization of this transcript depends on active transport on the actin cytoskeleton and is directed by a 54 nt zipcode element that is situated in the 3′UTR of the mRNA, immediately downstream of the stop codon [111]. In the absence of the 54 nt element, another element of 43 nt was found to have a suboptimal activity for the localization process. The two localization elements share two submotifs, GGACU and AAUGC, which are both necessary for the localization. However, a high A/C content region in the 54 nt zipcode, which is absent in the 43 nt element, was suggested to confer a maximum localization activity to the transcript [111]. The 54 nt zipcode serves as a binding site for the Zipcode binding protein 1 (ZBP1) [112]. Both the GGACU and A/C rich motifs of this zipcode play an important role in this interaction, as revealed by in vitro selection and mutagenesis [111, 113]. In contrast, the distal 43 nt zipcode does not interact with ZBP1, suggesting that other trans-acting factors may interact with the 43 nt element and mediate mRNA localization.
In the central nervous system, the Myelin basic protein (MBP) mRNA is localized at the myelin compartment of oligodendrocytes, a process essential for myelin biogenesis [114]. Transport and localization of MBP mRNA is directed by two distinct regions located in the 3′UTR of the transcript. One is an element called the RNA transport signal (RTS), which mediates MBP mRNA transport along oligodendrocytes to the myelin sheath [115]. The RTS element is composed of 21 nucleotides and serves as the binding site for the heterogeneous nuclear ribonucleoprotein (hnRNP) A2 [116]. In addition, localization and anchoring of MBP mRNA to the myelin compartment is mediated by another region called the RNA localization region (RLR). This region of 340 nucleotides forms stable secondary structures that are conserved across rat, mouse, and human MBP mRNA. Interestingly, the RLR is only necessary for localization of mRNA containing coding sequences. In reporter RNA constructs in which the coding region is deleted, RTS can direct both transport and localization of the transcript, suggesting that different pathways are involved in the localization of coding and non-coding mRNA constructs [115].
The Vg1 mRNA is another good example of the role of different and redundant localization elements in mediating mRNA localization. The Vg1 maternal mRNA, which encodes a member of the TGF-β family, is localized to the vegetal cortex of Xenopus embryos during mid-oogenesis [117, 118]. This transcript requires a 340 nucleotide localization element in its 3′UTR to promote localization to the vegetal pole of embryos [78]. Characterization of this localization element revealed the presence of four repeated sequence elements, termed E1 to E4, present in two to five copies in the 3′UTR, and another element, termed VM1, present in three copies [76, 119]. Element E2 was found to interact with the RNA-binding protein Vera/Vg1 RBP, which is involved in Vg1 mRNA localization [76, 79]. Deletion of elements E1, E4, and particularly E2, decreases the interaction with Vera and either impairs or abolishes Vg1 mRNA localization. The element VM1 was found to recruit the RNA-binding protein PTB/hnRNP I, which is involved in remodeling the interaction between Vg1 mRNA and Vera/Vg1 RBP [119, 120]. Interestingly, duplication of the VM1 element is sufficient to promote the localization of a reporter mRNA to the vegetal pole of Xenopus laevis embryos [119]. However, clusters of five VM1 elements or E2 elements from the 3′UTR of the Xenopus borealis Vg1 mRNA are insufficient to promote mRNA localization separately, suggesting that clusters of E2 and VM1 elements are both necessary for vegetal localization [121].
Different localization elements act at various steps of the localization pathway
The presence of different localization elements within a transcript may be necessary to determine different steps of a pathway leading to the final localization of the mRNA. In this case, each localization element would specify a particular step in the localization process, and each step would be coupled to the preceding one. The combination of all these elements together would result in a specific localization pathway. For instance, during early stages of oogenesis in Xenopus, the Xcat-2 mRNA localizes at the vegetal cortex of oocytes via a diffusion and entrapment mechanism (see above) [31]. As mentioned above, Xcat-2 mRNA first accumulates within a region of the MC called the METRO and subsequently localizes at the vegetal cortex [33, 34]. Xcat-2 mRNA localization requires two distinct cis-acting elements: a 250 nt localization signal called Mitochondrial Cloud Localization Element (MCLE), which is located at the 5′ end of the 3′UTR and is responsible for the entrapment of Xcat-2 in the MC [122], and a 164 nt germinal granule localization element (GGLE), situated immediately downstream of MCLE, which further directs the localization of Xcat-2 to the germinal granules within the MC [123]. Interestingly, fusion of the Xcat-2 GGLE motif to the mitochondrial cloud localized mRNA Xlsirt resulted in the accumulation of the Xlsirt chimera in germinal granules, suggesting that the combination of both MC localization element and the GGLE is required for localization at the germinal granules [123].
In Drosophila, several transcripts contain multiple subdomains in their 3′UTR that act as a localization element at different stages of development. For example, during oogenesis, oskar is synthesized in nurse cells and subsequently transferred and localized at the posterior pole of adjacent oocytes in the egg chamber (Fig. 2d) [124]. Proper localization of oskar mRNA is essential for posterior body patterning and germ cell determination. Different cis-acting elements within the 3′UTR of this transcript are implicated in different steps of this process: (1) movement of the transcript from the nurse cells to the oocyte, (2) transient accumulation at the anterior pole, and finally (3) posterior pole localization [125]. Mutations in a region of the oskar 3′UTR that disrupts early localization steps affect subsequent steps, suggesting that localization of oskar mRNA takes place in distinct stages, and early localization events are needed for later events to proceed normally [125].
The bicoid mRNA is transcribed in the nurse cells during stages 4 and 5 of oogenesis, transported into the oocyte, and becomes localized to the anterior pole of the oocyte during late oogenesis [126]. This transport requires an intact microtubule network and dynein [127]. The bicoid transcript contains several cis-acting elements residing in its 3′UTR that are required for both transfer from nurse cells to the oocyte and its proper anterior localization. A region of 625 nt that is capable of forming secondary structures consisting of multiple stem-loops is necessary and sufficient for this localization [128, 129]. By using several transgenes containing a small deletion in the 3′UTR, an element of 50 nt named BLE1 (bicoid localization element 1) was found to be essential for the early steps of the localization process [130]. This localization element is composed of a stem-loop structure in which the specific nucleotide sequence is not important for activity, but rather it is the secondary structure that plays a crucial role. Adjacent to the stem-loop are nucleotides whose identities are important for proper localization [131]. Several other secondary structures in the 3′UTR of this transcript direct localization of the bicoid mRNA in later steps of localization. For instance, a stem-loop in Domain V of bicoid 3′UTR interacts with Vps36, a member of the ESCRT-II complex (endosomal sorting complex required for transport), which is composed of three subunits: Vps22, Vps25, and Vps36 [132]. This interaction is important for bicoid mRNA localization, as mutants of ESCRT-II display impaired localization of this transcript to the anterior pole in late stages. Interestingly, dimerization of bicoid mRNA is proposed to be important for these later localization events. This phenomenon is directed by a stem-loop situated in Domain III of the 3′UTR which is suggested to form a recognition site for trans-acting factors such as Staufen [133]. Interaction of Staufen with the extensive stem-loop structure situated in the 3′UTR of bicoid leads to formation of an mRNP complex that localizes at the anterior pole in a microtubule-dependent manner [134]. A recent study reported that Staufen and dynein are present in the same particle as bicoid mRNA at the anterior pole, supporting a role of Staufen in the localization of this transcript [135]. While Staufen and ESCRT-II are involved in bicoid mRNA localization in late phases of development (S11 and onward), the protein Exuperantia is required during earlier phases, for transport from nurse cells to anterior pole localization following transfer into the oocyte [136].
The gurken mRNA is a transcript localized at the posterior pole of Drosophila oocytes during early stages of oogenesis (stages 1–6), and to the dorsal anterior corner during later stages (stages 8–9) [137, 138]. The localization elements of this transcript are found in both 5′ and 3′ untranslated regions, as well as in some parts of the coding sequence [24, 139]. These multiple elements are needed for the two-step localization pattern of this mRNA at the different stages of oogenesis. The 5′UTR contains signals that direct mRNA localization in the early stages, while a region in the 5′ coding sequence promotes accumulation and/or stabilization of the transcript in the anterior cortical ring during mid-oogenesis [24]. Further analysis of the coding sequence showed that a short region of 64 nt that is capable of forming a stem-loop is sufficient for apical localization [140]. Interestingly, this element is also found in the I factor RNA, a non-LTR retrotransposon that shows the same two-step localization pattern as gurken mRNA [140]. The 3′UTR of gurken mRNA also contains cis-acting elements that are necessary to restrict this transcript to the dorsal anterior corner during later stages of oogenesis [24].
Control of molecular motors by localization elements
Besides their role in defining the final destination of an mRNA, localization elements can also modulate the activity or identity of molecular motors associated with a transcript. Evidence for this mechanism came from live cell imaging of the Drosophila hairy mRNA, one of the pair-rule transcripts such as fushi tarazu (ftz), runt, and wingless that localize apically of the peripheral nuclei in the Drosophila syncytial stage embryos [141]. Hairy mRNA localization depends on the following: two partially redundant stem-loop structures, SL1 and SL2a, present in its 3′UTR [142]; the proteins Egalitarian (Egl) and Bic-D [143, 144]; and the minus-end molecular motor dynein [145]. Microinjection of fluorescently labeled RNA in the Drosophila blastoderm revealed that hairy mRNA, like non-localizing transcripts, displays bidirectional transport, suggesting that motors with opposite polarity associate with this transcript [146]. However, unlike non-localized transcripts, hairy mRNA spends more time undergoing minus-end transport toward the apical pole. Interestingly, increasing the number of localization elements in hairy mRNA doubles the speed of apical trafficking compared to wild-type mRNA, suggesting that these elements increase the number of minus-end motors recruited to the transcript [146]. Recently, work by Bullock and colleagues [147] revealed that these stem-loops are bound by the RNA-binding protein Egalitarian (Egl), which interacts with the dynein cofactor Bic-D to recruit the molecular motor dynein. Since Egl also binds to dynein light-chain (Dlc), a regulator of dynein’s motor activity, this suggests a model in which Egl can recruit an mRNA to dynein via Bic-D and stimulate the activity of this motor via Dlc [147].
An additional mechanism through which a zipcode can modulate mRNA localization is by the ability to recruit different molecular motors. For instance, localization of the β-actin mRNA to the leading edge of fibroblasts depends on the actin cytoskeleton, suggesting a myosin-based transport, while the localization of the same transcript at the growth cone of neurons is microtubule dependent, pointing instead toward kinesin-based transport [148, 149]. Since both localization events depend on the ZBP1 protein, this suggests that this factor can recruit different motors according to the cellular context. Another example was observed in Xenopus oocytes, in which localization of the Vg1 mRNA to the vegetal pole depends on both kinesin-1 and kinesin-2. While kinesin-1 interacts with the Vg1 localization element (VLE), kinesin-1 also interacts with kinesin-2, and both motors carry out overlapping functions in RNA transport [150].
Extracellular signaling and control of mRNA localization
Another mechanism by which cells can control the cytoplasmic localization of specific mRNA is via signal transduction pathways, especially through extracellular signaling. The first evidence that signal transduction pathways control mRNA localization came from the study of β-actin mRNA localization in fibroblast [151, 152]. Indeed, while β-actin mRNA normally localizes at the leading edge of chicken fibroblasts, it displays a diffuse cytoplasmic distribution in serum-starved fibroblasts. Interestingly, addition of serum results in a rapid relocalization of this transcript to the leading edge of the cells. Addition of growth factors, such as PDGF or lysophosphatidic acid (LPA), has the same effect of promoting β-actin mRNA localization [151, 152], suggesting that growth signaling pathways induce β-actin mRNA localization at the leading edge. However, these growth factors are also known to promote the polymerization of the actin cytoskeleton via the small GTPases Rac and Rho [153]. Therefore, it is not clear if these signaling pathways act directly on the mRNA localization machinery or indirectly by promoting stress fiber formation or via both mechanisms.
Most evidence supporting a role of signaling pathways in promoting mRNA localization comes from studies in neurons, in which several transcripts are localized to dendrites or in the axon [21, 154]. The adult neuron is constituted of dendritic trees that contain over 10,000 dendritic spines, each corresponding to a single excitatory synapse. In these synapses, hundreds of different proteins are known to be present. Long-term potentiation (LTP) or long-term depression (LTD), two phenomena at the base of memory, are induced by high frequency stimulation or low frequency stimulation (respectively) of excitatory synapses. These stimulations result in the activation of specific receptors at the synapses, such as the metabotropic glutamate (mGluR) or N-methyl d-aspartate (NMDAR) receptors, which trigger downstream signaling cascades [155]. Maintenance of LTP or LTD depends on gene expression and local translation of transcripts localized at the excited synapses [21]. Therefore, synaptic activation in neurons is an excellent system to study the role of signaling pathways in promoting the transport and localization of mRNAs.
Seminal studies on the Arc/Arg3.1 mRNA provided the first evidence that synaptic activity can promote the localization of a specific mRNA in dendrites. Arc is among the immediate-early genes activated following seizures, learning experiences, or induction of LTP (reviewed in [156]). Using high frequency activation of perforant path synapses in vivo, Steward and colleagues [157] observed that newly transcribed Arc mRNA localizes at the activated synapses, suggesting that extracellular signaling promotes both transcription and transport of this transcript. Interestingly, when Arc mRNA is induced in the absence of a localized synaptic signal (in the case of a generalized seizure for instance), this mRNA was uniformly distributed in dendrites. Only the subsequent activation of synapses led to the transport and localization of Arc mRNA to the activated synapses [157]. This selective targeting of Arc mRNA was shown to depend on the activation of the N-methyl d-aspartate receptor (NMDAR) [158], suggesting that a specific signaling pathway can control the transport and localization of mRNA to synapses. Besides Arc, several other transcripts, including BDNF and TrkB [159], CamKIIα [160], β-actin [161], and AMPA receptor subunits GluR1 and GluR2 [162], were also found to localize in dendrites of cultured neurons following synapse activation.
Other studies revealed that not only synaptic activity but also nerve growth factors stimulate mRNA localization in neurons. Axon outgrowth responds to attractive signals, i.e., neurotrophic factors, in order to guide the axonal growth cone and establish wiring between neurons [163]. Among the intracellular responses to these signals, local translation of mRNA localized at the growth cone participates in the movement and directional steering of the axon [164]. For instance, neurotrophin-3 (NT-3) was initially found to induce localization of the β-actin mRNA to the growth cones of forebrain neurons [149, 165]. Other growth factors, such as the brain derived neurotrophic factor (BDNF) or Netrin-1, also induce β-actin mRNA localization to the growth cones of X. laevis neurons [166, 167] and in rat cortical neurons [168].
Axonal localization of specific mRNAs can also be controlled by growth factors, such as the myo-inositol monophosphate 1 (Impa1) mRNA, which is induced to localize to distal axons by nerve growth factor (NGF) [12]. Interestingly, mRNA profiling in axons revealed that neurotrophins can specifically increase or decrease the levels of individual mRNAs in the axon [11]. For instance, neurotrophins that favor axon growth (e.g., NGF, BDNF, and NT-3) regulate the axonal level of a number of mRNAs. However, factors that act as inhibitors of axon growth (e.g., myelin-associated glycoprotein and semaphorin 3A) regulate a distinct set of axonal mRNAs and induce an opposite effect on the transcripts regulated by NGF, BDNF, and NT-3 [11]. Importantly, these effects occurred on pre-existing pools of mRNA, since an inhibitor of RNA polymerase II was included during the treatments with growth factors. These results suggest that the regulation of mRNA localization is very specific at the level of individual mRNAs and for the different extracellular cues that activate similar signaling pathways.
Controlling the motility and directionality of RNA granules
A question raised by these results is how a signaling pathway can control the localization of a specific mRNA. Live-cell imaging of neuronal cells provided some insights into possible mechanisms behind the effect of signaling pathways on mRNA transport and localization. Initial live-cell imaging studies using the nucleic acid dye SYTO 14 revealed the presence of mobile RNA granules in the dendrites of cultured neurons [169]. Upon addition of neurotrophin-3, the number and motility of these RNA granules rapidly increased within 5–15 min post-stimulation [170]. By coexpression of a reporter mRNA containing the 3′UTR of the CamKIIα mRNA and MS2 stem-loops with the green fluorescent protein fused to the MS2 RNA-binding protein, Rook and colleagues [171] were able to visualize GFP-tagged CamKII 3′UTR RNA granules in the dendrites of cultured neurons. In the absence of neuronal activation, RNA granules exhibited either oscillatory, anterograde (away from the cell body), or retrograde (toward the cell body) movements. Interestingly, neuronal depolarization using KCl resulted in a shift toward anterograde movement of the RNA granules within dendrites. Similar effects were observed using a GFP-tagged ZBP1, which binds the β-actin mRNA zipcode and promotes localization of this transcript to the growth cones of neurons [165]. GFP-ZBP1 forms granules that contain β-actin mRNA and display bidirectional movements in neurites [165]. However, 15 min after KCl exposure of neurons, the level of GFP-ZBP1 particles in dendrites was significantly increased, suggesting a rapid localization of a pre-existing somatic pool of ZBP1 molecules instead of de novo synthesis of GFP-ZBP1 [161].
Altogether, these results suggest that localized transcripts are packed into granules containing both microtubule plus-end and minus-end molecular motors, i.e., kinesin and dynein. Biochemical studies support these results, as large RNA granules (over 1,000S) from mouse brain were purified by affinity chromatography using the C-terminal cargo-binding domain of the kinesin-1 Kif5 as bait [172]. These granules were found to contain the mRNA CamKIIα and Arc, and RNA-binding proteins associated with mRNA transport (Purα, staufen, FMRP), protein synthesis, and RNA metabolism. Another approach to purify RNA transport granules from developing rat brain by biochemical fractionations revealed the presence of the kinesin Kif5, dynein, ZBP1, and β-actin mRNA in these granules, but not the CamKIIα mRNA [173]. Both results suggest that RNA granules are heterogeneous and that their content can vary depending on the stage of development. Nevertheless, the presence of both kinesin and dynein in the same granule could result in a “tug-of-war” between both molecular motors, leading to the oscillatory or bidirectional movement observed for the RNA granules by live-cell imaging [171]. This is supported by the observation of similar bidirectional transport of RNA granules containing GFP-labeled Kif5 [172], ZBP1 [165], or Staufen [174].
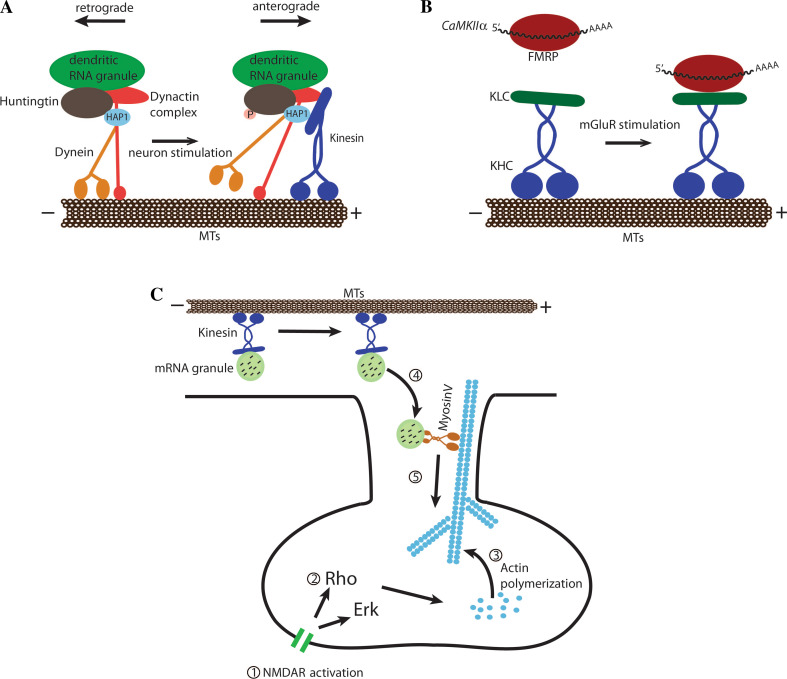
The observation that synaptic activity promotes anterograde movement of RNA granules suggests that, upon activation of a signaling pathway, subsequent phosphorylation of a specific factor in the RNA granules may favor anterograde over retrograde transport. Evidence for such a “molecular switch” mechanism has been obtained for vesicular transport in neurons [175]. In these vesicles, the huntingtin protein acts as a scaffold that recruits both dynein and kinesin. Huntingtin binds directly to the dynein motor, but indirectly with the p150Glued subunit of dynactin and kinesin-1 via the huntingtin-associated protein-1 (HAP1) [176, 177]. In its unphosphorylated form, kinesin-1 interacts weakly with the motor complex associated with huntingtin, and vesicles are prone to retrograde transport. Upon phosphorylation of huntingtin at Ser421 by the kinase Akt, the interaction between kinesin-1 and the dynactin motor complex increases, favoring anterograde transport. Since huntingtin has been recently found to be associated with dendritic RNA granules and is involved in their transport [178], it is possible that huntingtin may play a similar role in dendritic mRNA localization (Fig. 3a).
Fig. 3a–c.
Signal transduction and mRNA localization. a Model of huntingtin phosphorylation upon neuron stimulation. In the nonphosphorylated state, huntingtin associates with dynein and more weakly with kinesin, which results in either bidirectional or retrograde transport of cargoes associated with huntingtin. In its phosphorylated state, huntingtin binds more strongly to kinesin via HAP1 and dynactin, which leads to anterograde transport of its cargoes, including dendritic mRNAs. MTs Microtubules. b In dendrites, FMRP-associated mRNAs are transported by kinesin upon mGluR activation. mGluR stimulation increases the interaction between kinesin light chain (KLC) and FMRP. KHC Kinesin heavy chain. c Arc/Arg3.1 mRNA localization to synapses following NMDA receptor (NMDAR) stimulation. Upon NMDAR stimulation (1), the Erk and Rho kinases are activated (2), promoting actin polymerization in the synapse (3). mRNA granules in a translational-inactive state are transported in dendrites along the microtubules by kinesin. Actin polymerization leads to the transfer of RNA granules into the synapse, via the myosin V molecular motor (4), which transports its cargo into activated synapses (5)
Modulation of the interaction between molecular motors and RNA-binding proteins
Posttranslational modifications induced by the activated signaling pathway can also modulate the association between a molecular motor and the RNA-binding protein that recruits the localized mRNA. Evidence for such a mechanism comes from studies on the RNA-binding Fragile X mental retardation protein (FMRP), which participates both in dendritic mRNA transport and translational repression of localized mRNA [179]. FMRP is involved in the transport of the MAP1B and CamKIIα mRNAs into dendrites following stimulation of the glutamate receptor mGluR5 [180–182]. A recent study by Dictenberg and colleagues [183] revealed that FMRP is associated with Kif5 via the kinesin light chain (KLC) in neurons. Interestingly, upon stimulation of glutamate receptor, the amount of FMRP associated with Kif5 increases, while the total amount of FMRP does not change, suggesting that activation of the glutamate signaling pathway stimulates the association between FMRP and the kinesin complex (Fig. 3b). However, the molecular mechanism behind this process is still unknown.
Local reorganization of the cytoskeleton
Finally, another mechanism by which signaling pathways can control mRNA localization is by promoting a local reorganization of the cytoskeleton. Evidence for this mechanism comes from the work of Steward and colleagues [184], who reported that polymerization of actin at activated synapses is important for local Arc mRNA targeting. The authors observed that inhibition of actin polymerization by a Rho kinase inhibitor, or latrunculin B, blocks Arc mRNA targeting to the activated synapse but does not block Arc mRNA transport into dendrites. Interestingly, both Arc mRNA localization and actin polymerization at activated synapses depend on NMDA receptor activity, suggesting that the signaling pathway activated by this receptor coordinates both processes. Finally, the authors also found that, besides actin polymerization, activation of the Erk kinase via the MAP kinase pathway is required for Arc mRNA localization. These results suggest that following the long distance, microtubule-based transport of Arc mRNA into dendrites, the local polymerization of actin at an activated synapse is required to either anchor Arc mRNA at this synapse or transfer this mRNA to an actin-based motor for terminal localization at spines inside the synapse (Fig. 3c) [184]. This last step may require the actin-dependent molecular motor Myosin-Va, which has been reported to transport mRNA/protein complexes in dendritic spines [185]. Surprisingly, the Arc protein is required to stabilize F-actin at active synapses [186], suggesting a positive feedback between Arc protein localization and actin polymerization at the synapse.
Conclusions
In this review, we have followed the travels of localized mRNAs from their transcription in the nucleus, the recruitment of localization factors to their zipcodes, their encounter with protein motors that can speed them on their way, the extracellular signaling cascades that may change their fate, and the arrival of these transcripts at their final destination. We have reviewed in detail various mechanisms by which trans-acting RNA-binding proteins and cis-acting RNA sequences control localized transcripts in different organisms and cell types. Although we have underlined the importance of cis-acting localization elements in this process, few of them have been well characterized and we still know little about their interactions with RNA-binding proteins. Furthermore, most of the information on the control of mRNA localization comes from in-depth studies on a select group of transcripts. The advent of genome-wide studies, which lead to the identification of numerous examples of localized mRNAs to various cytoplasmic locations or organelles, opens the possibility that some of these transcripts may use novel mechanisms of localization and that control of their localization may occur in different ways than the currently studied localized mRNAs. This will require the identification of localization elements in these transcripts and the trans-acting binding factors that recognize these elements. Moreover, for most of these mRNAs, the biological function of their localization is still unclear. Finally, the development of new methods to study mRNA localization in real time has greatly increased our knowledge in this field [187]. However, most of these studies have been performed in cultured cells or embryo, and little is known about mRNA localization in a whole animal during its development or pathogenesis. The recent development of transgenic mice for in vivo detection of an endogenous mRNA at high resolution opens the possibility of dealing with such questions [188]. These are only some of the new challenges that await the field for the future.
Acknowledgments
We thank Emmanuelle Querido and Éric Lécuyer for critical reading of the manuscript, and Kim Mowry, Gary Bassell, and Éric Lécuyer for providing images. This work was supported by the Grant MOP43855 from the Canadian Institutes for Health Research (CIHR). P.C is a Senior Scholar from the Fond de Recherche en Santé du Québec (FRSQ).
Footnotes
The ZBP1, Vg1 RBP/Vera, IMP1, 2, 3, CRD-BP, and KOC proteins are part of the same family of RNA-binding proteins that contain two RNA recognition motifs (RRM) followed by four hnRNP K Homology (KH) domains. ZBP1 is the chicken ortholog of the mammalian IGF-II RNA binding protein 1/IMP1, a member of a family of three highly related paralogs, IMP1 to 3. Xenopus Vg1 RBP/Vera is the ortholog of mammalian IMP3.
ZBP2, KSRP, FBP1, 2, 3, and MARTA are part of the same family of RNA-binding proteins that contain four KH domains. ZBP2 is the chicken ortholog of FBP2, a member of the three mammalian paralogs FBP1-3.
References
- 1.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326(5957):1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zarnack K, Feldbrügge M. mRNA trafficking in fungi. Mol Gen Genomics. 2007;278(4):347–359. doi: 10.1007/s00438-007-0271-8. [DOI] [PubMed] [Google Scholar]
- 3.Okita TW, Choi S-B. mRNA localization in plants: targeting to the cell’s cortical region and beyond. Curr Opin Plant Biol. 2002;5(6):553–559. doi: 10.1016/S1369-5266(02)00304-7. [DOI] [PubMed] [Google Scholar]
- 4.Nevo-Dinur K, Nussbaum-Shochat A, Ben-Yehuda S, Amster-Choder O. Translation-independent localization of mRNA in E. coli . Science. 2011;331(6020):1081–1084. doi: 10.1126/science.1195691. [DOI] [PubMed] [Google Scholar]
- 5.Pyhtila B, Zheng T, Lager PJ, Keene JD, Reedy MC, Nicchitta CV. Signal sequence- and translation-independent mRNA localization to the endoplasmic reticulum. RNA. 2008;14(3):445–453. doi: 10.1261/rna.721108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marc P, Margeot A, Devaux F, Blugeon C, Corral-Debrinski M, Jacq C. Genome-wide analysis of mRNA targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Nat Acad Sci USA. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131(1):174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Blower MD, Feric E, Weis K, Heald R. Genome-wide analysis demonstrates conserved localization of messenger RNAs to mitotic microtubules. J Cell Biol. 2007;179(7):1365–1373. doi: 10.1083/jcb.200705163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberwine J, Belt B, Kacharmina JE, Miyashiro K. Analysis of subcellularly localized mRNAs using in situ hybridization, mRNA amplification, and expression profiling. Neurochem Res. 2002;27(10):1065–1077. doi: 10.1023/A:1020956805307. [DOI] [PubMed] [Google Scholar]
- 11.Willis DE, van Niekerk EA, Sasaki Y, Mesngon M, Merianda TT, Williams GG, Kendall M, Smith DS, Bassell GJ, Twiss JL. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J Cell Biol. 2007;178(6):965–980. doi: 10.1083/jcb.200703209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreassi C, Zimmermann C, Mitter R, Fusco S, Devita S, Saiardi A, Riccio A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat Neurosci. 2010;13(3):291–301. doi: 10.1038/nn.2486. [DOI] [PubMed] [Google Scholar]
- 13.Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453(7191):115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du T-G, Schmid M, Jansen R-P. Why cells move messages: the biological functions of mRNA localization. Semin Cell Dev Biol. 2007;18(2):171–177. doi: 10.1016/j.semcdb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Corral-Debrinski M, Blugeon C, Jacq C. In yeast, the 3′ untranslated region or the presequence of ATM1 is required for the exclusive localization of its mRNA to the vicinity of mitochondria. Mol Cell Biol. 2000;20(21):7881–7892. doi: 10.1128/MCB.20.21.7881-7892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zipor G, Haim-Vilmovsky L, Gelin-Licht R, Gadir N, Brocard C, Gerst JE. Localization of mRNAs coding for peroxisomal proteins in the yeast, Saccharomyces cerevisiae . Proc Nat Acad Sci USA. 2009;106(47):19848–19853. doi: 10.1073/pnas.0910754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ainger K, Avossa D, Morgan F, Hill S, Barry C, Barbarese E, Carson J. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123(2):431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyons DA, Naylor SG, Scholze A, Talbot WS. Kif1b is essential for mRNA localization in oligodendrocytes and development of myelinated axons. Nat Genet. 2009;41(7):854–858. doi: 10.1038/ng.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Besse F, Ephrussi A. Translational control of localized mRNAs: restricting protein synthesis in space and time. Nat Rev Mol Cell Biol. 2008;9(12):971–980. doi: 10.1038/nrm2548. [DOI] [PubMed] [Google Scholar]
- 20.Kindler S, Wang H, Richter D, Tiedge H. RNA transport and local control of translation. Annu Rev Cell Dev Biol. 2005;21(1):223–245. doi: 10.1146/annurev.cellbio.21.122303.120653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci. 2007;8(10):776–789. doi: 10.1038/nrn2150. [DOI] [PubMed] [Google Scholar]
- 22.Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up! Trends Biochem Sci. 2010;35(8):459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Jambhekar A, DeRisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA. 2007;13(5):625–642. doi: 10.1261/rna.262607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thio GL, Ray RP, Barcelo G, Schupbach T. Localization of gurken RNA in Drosophila oogenesis requires elements in the 5′ and 3′ regions of the transcript. Dev Biol. 2000;221(2):435–446. doi: 10.1006/dbio.2000.9690. [DOI] [PubMed] [Google Scholar]
- 25.Chartrand P, Meng X-H, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9(6):333–336. doi: 10.1016/S0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- 26.Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98(2):81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 27.Gavis ER, Chatterjee S, Ford NR, Wolff LJ. Dispensability of nanos mRNA localization for abdominal patterning but not for germ cell development. Mech Dev. 2008;125(1–2):81–90. doi: 10.1016/j.mod.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergsten SE, Gavis ER. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development. 1999;126(4):659–669. doi: 10.1242/dev.126.4.659. [DOI] [PubMed] [Google Scholar]
- 29.Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila . Curr Biol. 2003;13(14):1159–1168. doi: 10.1016/S0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- 30.Lantz VA, Clemens SE, Miller KG. The actin cytoskeleton is required for maintenance of posterior pole plasm components in the Drosophila embryo. Mech Dev. 1999;85(1–2):111–122. doi: 10.1016/S0925-4773(99)00096-9. [DOI] [PubMed] [Google Scholar]
- 31.Mosquera L, Forristall C, Zhou Y, King ML. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development. 1993;117(1):377–386. doi: 10.1242/dev.117.1.377. [DOI] [PubMed] [Google Scholar]
- 32.Heasman J, Quarmby J, Wylie CC. The mitochondrial cloud of Xenopus oocytes: the source of germinal granule material. Dev Biol. 1984;105(2):458–469. doi: 10.1016/0012-1606(84)90303-8. [DOI] [PubMed] [Google Scholar]
- 33.Forristall C, Pondel M, Chen L, King ML. Patterns of localization and cytoskeletal association of two vegetally localized RNAs, Vg1 and Xcat-2. Development. 1995;121(1):201–208. doi: 10.1242/dev.121.1.201. [DOI] [PubMed] [Google Scholar]
- 34.Kloc M, Etkin LD. Two distinct pathways for the localization of RNAs at the vegetal cortex in Xenopus oocytes. Development. 1995;121(2):287–297. doi: 10.1242/dev.121.2.287. [DOI] [PubMed] [Google Scholar]
- 35.Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell. 2004;15(10):4669–4681. doi: 10.1091/mbc.E04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinrich B, Deshler JO. RNA localization to the Balbiani body in Xenopus oocytes is regulated by the energy state of the cell and is facilitated by kinesin II. RNA. 2009;15(4):524–536. doi: 10.1261/rna.975309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bashirullah A, Cooperstock RL, Lipshitz HD. Spatial and temporal control of RNA stability. Proc Nat Acad Sci USA. 2001;98(13):7025–7028. doi: 10.1073/pnas.111145698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD. Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster . EMBO J. 1999;18(9):2610–2620. doi: 10.1093/emboj/18.9.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Semotok JL, Luo H, Cooperstock RL, Karaiskakis A, Vari HK, Smibert CA, Lipshitz HD. Drosophila maternal Hsp83 mRNA destabilization is directed by multiple SMAUG recognition elements in the open reading frame. Mol Cell Biol. 2008;28(22):6757–6772. doi: 10.1128/MCB.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol. 2005;15(4):284–294. doi: 10.1016/j.cub.2005.01.048. [DOI] [PubMed] [Google Scholar]
- 41.Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12(1):143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development. 2006;133(22):4573–4583. doi: 10.1242/dev.02649. [DOI] [PubMed] [Google Scholar]
- 43.Rouget C, Papin C, Boureux A, Meunier A-C, Franco B, Robine N, Lai EC, Pelisson A, Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467(7319):1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khurana JS, Theurkauf W. piRNAs, transposon silencing, and Drosophila germline development. J Cell Biol. 2010;191(5):905–913. doi: 10.1083/jcb.201006034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bullock SL. Translocation of mRNAs by molecular motors: think complex? Semin Cell Dev Biol. 2007;18(2):194–201. doi: 10.1016/j.semcdb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 47.Zarnack K, Feldbrugge M. Microtubule-dependent mRNA transport in fungi. Eukaryot Cell. 2010;9(7):982–990. doi: 10.1128/EC.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/S1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- 49.Krauss J, Lûpez de Quinto S, Nüsslein-Volhard C, Ephrussi A. Myosin-V regulates oskar mRNA localization in the Drosophila oocyte. Curr Biol. 2009;19(12):1058–1063. doi: 10.1016/j.cub.2009.04.062. [DOI] [PubMed] [Google Scholar]
- 50.Kuersten S, Goodwin EB. Linking nuclear mRNP assembly and cytoplasmic destiny. Biol Cell. 2005;97(6):469–478. doi: 10.1042/BC20040106. [DOI] [PubMed] [Google Scholar]
- 51.Farina KL, Singer RH. The nuclear connection in RNA transport and localization. Trends Cell Biol. 2002;12(10):466–472. doi: 10.1016/S0962-8924(02)02357-7. [DOI] [PubMed] [Google Scholar]
- 52.Oleynikov Y, Singer RH. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr Biol. 2003;13(3):199–207. doi: 10.1016/S0960-9822(03)00044-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol. 2002;156(1):41–52. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pan F, Huttelmaier S, Singer RH, Gu W. ZBP2 facilitates binding of ZBP1 to β-actin mRNA during transcription. Mol Cell Biol. 2007;27(23):8340–8351. doi: 10.1128/MCB.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bohl F, Kruse C, Frank A, Ferring D, Jansen R-P. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19(20):5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p-She3p complex to ASH1 mRNA. EMBO J. 2000;19(23):6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kruse C, Jaedicke A, Beaudouin J, Bohl F, Ferring D, Guttler T, Ellenberg J, Jansen RP. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J Cell Biol. 2002;159(6):971–982. doi: 10.1083/jcb.200207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Du TG, Jellbauer S, Müller M, Schmid M, Niessing D, Jansen RP. Nuclear transit of the RNA-binding protein She2 is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9:781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen Z, Paquin N, Forget A, Chartrand P. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell. 2009;20(8):2265–2275. doi: 10.1091/mbc.E08-11-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Z, St-Denis A, Chartrand P. Cotranscriptional recruitment of She2p by RNA pol II elongation factor Spt4-Spt5/DSIF promotes mRNA localization to the yeast bud. Genes Dev. 2010;24(17):1914–1926. doi: 10.1101/gad.1937510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Forget A, Chartrand P. Cotranscriptional assembly of mRNP complexes that determine the cytoplasmic fate of mRNA. Transcription. 2011;2:86–90. doi: 10.4161/trns.2.2.14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Horne-Badovinac S, Bilder D. Dynein regulates epithelial polarity and the apical localization of stardust A mRNA. PLoS Genet. 2008;4(1):e8. doi: 10.1371/journal.pgen.0040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dalby B, Glover DM. 3′ non-translated sequences in Drosophila cyclin B transcripts direct posterior pole accumulation late in oogenesis and peri-nuclear association in syncytial embryos. Development. 1992;115(4):989–997. doi: 10.1242/dev.115.4.989. [DOI] [PubMed] [Google Scholar]
- 65.Buckley PT, Lee MT, Sul J-Y, Miyashiro KY, Bell TJ, Fisher SA, Kim J, Eberwine J. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69(5):877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muslimov IA, Santi E, Homel P, Perini S, Higgins D, Tiedge H. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J Neurosci. 1997;17(12):4722–4733. doi: 10.1523/JNEUROSCI.17-12-04722.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khanam T, Raabe CA, Kiefmann M, Handel S, Skryabin BV, Brosius J. Can ID repetitive elements serve as cis-acting dendritic targeting elements? An in vivo study. PLoS One. 2007;2(9):e961. doi: 10.1371/journal.pone.0000961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tange Tÿ, Nott A, Moore MJ. The ever-increasing complexities of the exon junction complex. Curr Opin Cell Biol. 2004;16(3):279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 69.Giorgi C, Moore MJ. The nuclear nurture and cytoplasmic nature of localized mRNPs. Semin Cell Dev Biol. 2007;18(2):186–193. doi: 10.1016/j.semcdb.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 70.Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428(6986):959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- 71.Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, González-Reyes A, St Johnston D. The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila . Curr Biol. 1997;7(7):468–478. doi: 10.1016/S0960-9822(06)00218-1. [DOI] [PubMed] [Google Scholar]
- 72.Hachet O, Ephrussi A. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol. 2001;11(21):1666–1674. doi: 10.1016/S0960-9822(01)00508-5. [DOI] [PubMed] [Google Scholar]
- 73.van Eeden FJM, Palacios IM, Petronczki M, Weston MJD, St Johnston D. Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior pole. J Cell Biol. 2001;154(3):511–524. doi: 10.1083/jcb.200105056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palacios IM, Gatfield D, St Johnston D, Izaurralde E. An eIF4AIII-containing complex required for mRNA localization and nonsense-mediated mRNA decay. Nature. 2004;427(6976):753–757. doi: 10.1038/nature02351. [DOI] [PubMed] [Google Scholar]
- 75.Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell. 1999;4(3):431–437. doi: 10.1016/S1097-2765(00)80345-7. [DOI] [PubMed] [Google Scholar]
- 76.Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 1997;276(5315):1128–1131. doi: 10.1126/science.276.5315.1128. [DOI] [PubMed] [Google Scholar]
- 77.Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 1998;12(11):1593–1598. doi: 10.1101/gad.12.11.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340 nt RNA sequence element in Xenopus oocytes. Science. 1992;255(5047):991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- 79.Deshler JO, Highett MI, Abramson T, Schnapp BJ. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol. 1998;8(9):489–496. doi: 10.1016/S0960-9822(98)70200-3. [DOI] [PubMed] [Google Scholar]
- 80.Kwon S, Abramson T, Munro TP, John CM, Kohrmann M, Schnapp BJ. UUCAC- and Vera-dependent localization of VegT RNA in Xenopus oocytes. Curr Biol. 2002;12(7):558–564. doi: 10.1016/S0960-9822(02)00740-6. [DOI] [PubMed] [Google Scholar]
- 81.Kress TL, Yoon YJ, Mowry KL. Nuclear RNP complex assembly initiates cytoplasmic RNA localization. J Cell Biol. 2004;165(2):203–211. doi: 10.1083/jcb.200309145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allison R, Czaplinski K, Git A, Adegbenro E, Stennard F, Houliston E, Standart N. Two distinct staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA. 2004;10(11):1751–1763. doi: 10.1261/rna.7450204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yoon YJ, Mowry KL. Xenopus staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131(13):3035–3045. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]
- 84.Long RM, Gu W, Meng X, Gonsalvez G, Singer RH, Chartrand P. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J Cell Biol. 2001;153(2):307–318. doi: 10.1083/jcb.153.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Serano TL, Cohen RS. A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121(11):3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- 86.Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen R-P. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277(5324):383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 87.Takizawa PA, Sil A, Swedlow J, Herskowitz I, Vale R. Actin-dependent localization of an mRNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 88.Sil A, Herskowitz I. Identification of an asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84(5):711–722. doi: 10.1016/S0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- 89.Bobola N, Jansen R-P, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84(5):699–709. doi: 10.1016/S0092-8674(00)81048-X. [DOI] [PubMed] [Google Scholar]
- 90.Gonzalez I, Buonomo SBC, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol. 1999;9(6):337–340. doi: 10.1016/S0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- 91.Chartrand P, Meng X, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of Ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell. 2002;10:1319–1330. doi: 10.1016/S1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- 92.Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol. 2005;25(11):4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jambhekar A, McDermott K, Sorber K, Shepard KA, Vale RD, Takizawa PA, DeRisi JL. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae . Proc Nat Acad Sci USA. 2005;102(50):18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Müller M, Richter K, Heuck A, Kremmer E, Buchner J, Jansen R-P, Niessing D. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA. 2009;15(11):2002–2012. doi: 10.1261/rna.1753309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Munchow S, Sauter C, Jansen R. Association of the class V myosin Myo4p with a localised messenger RNA in budding yeast depends on She proteins. J Cell Sci. 1999;112(10):1511–1518. doi: 10.1242/jcs.112.10.1511. [DOI] [PubMed] [Google Scholar]
- 96.Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Nat Acad Sci USA. 2000;97(10):5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Müller M, Heym RG, Mayer A, Kramer K, Schmid M, Cramer P, Urlaub H, Jansen R-P, Niessing D. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol. 2011;9(4):e1000611. doi: 10.1371/journal.pbio.1000611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84(5):687–697. doi: 10.1016/S0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- 99.Haarer BK, Petzold A, Lillie SH, Brown SS. Identification of MYO4, a second class V myosin gene in yeast. J Cell Sci. 1994;107(Pt 4):1055–1064. doi: 10.1242/jcs.107.4.1055. [DOI] [PubMed] [Google Scholar]
- 100.Reck-Peterson SL, Tyska MJ, Novick PJ, Mooseker MS. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J Cell Biol. 2001;153(5):1121–1126. doi: 10.1083/jcb.153.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Heuck A, Du T-G, Jellbauer S, Richter K, Kruse C, Jaklin S, Müller M, Buchner J, Jansen R-P, Niessing D. Monomeric myosin V uses two binding regions for the assembly of stable translocation complexes. Proc Nat Acad Sci USA. 2007;104(50):19778–19783. doi: 10.1073/pnas.0706780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dunn BD, Sakamoto T, Hong M-SS, Sellers JR, Takizawa PA. Myo4p is a monomeric myosin with motility uniquely adapted to transport mRNA. J Cell Biol. 2007;178(7):1193–1206. doi: 10.1083/jcb.200707080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chung S, Takizawa PA. Multiple Myo4 motors enhance ASH1 mRNA transport in Saccharomyces cerevisiae . J Cell Biol. 2010;189(4):755–767. doi: 10.1083/jcb.200912011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gavis ER, Lehmann R. Localization of nanos RNA controls embryonic polarity. Cell. 1992;71(2):301–313. doi: 10.1016/0092-8674(92)90358-J. [DOI] [PubMed] [Google Scholar]
- 105.Gavis ER, Curtis D, Lehmann R. Identification of cis-acting sequences that control nanos RNA localization. Dev Biol. 1996;176:36–50. doi: 10.1006/dbio.1996.9996. [DOI] [PubMed] [Google Scholar]
- 106.Bergsten SE, Huang T, Chatterjee S, Gavis ER. Recognition and long-range interactions of a minimal nanos RNA localization signal element. Development. 2001;128(3):427–435. doi: 10.1242/dev.128.3.427. [DOI] [PubMed] [Google Scholar]
- 107.Jain RA, Gavis ER. The Drosophila hnRNP M homolog Rumpelstiltskin regulates nanos mRNA localization. Development. 2008;135(5):973–982. doi: 10.1242/dev.015438. [DOI] [PubMed] [Google Scholar]
- 108.Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER. Aubergine is a component of a nanos mRNA localization complex. Dev Biol. 2011;349(1):46–52. doi: 10.1016/j.ydbio.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wilson JE, Connell JE, Macdonald PM. Aubergine enhances oskar translation in the Drosophila ovary. Development. 1996;122(5):1631–1639. doi: 10.1242/dev.122.5.1631. [DOI] [PubMed] [Google Scholar]
- 110.Lawrence JB, Singer RH. Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell. 1986;45(3):407–415. doi: 10.1016/0092-8674(86)90326-0. [DOI] [PubMed] [Google Scholar]
- 111.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ross A, Oleynikov Y, Kislauskis E, Taneja K, Singer R. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol. 1997;17(4):2158–2165. doi: 10.1128/mcb.17.4.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of b-actin zipcode induces RNA looping. Genes Dev. 2010;24(2):148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Carson JH, Kwon S, Barbarese E. RNA trafficking in myelinating cells. Curr Opin Neurobiol. 1998;8(5):607–612. doi: 10.1016/S0959-4388(98)80088-3. [DOI] [PubMed] [Google Scholar]
- 115.Ainger K, Avossa D, Diana AS, Barry C, Barbarese E, Carson JH. Transport and localization elements in myelin basic protein mRNA. J Cell Biol. 1997;138(5):1077–1087. doi: 10.1083/jcb.138.5.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hoek KS, Kidd GJ, Carson JH, Smith R. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 1998;37(19):7021–7029. doi: 10.1021/bi9800247. [DOI] [PubMed] [Google Scholar]
- 117.Tannahill D, Melton DA. Localized synthesis of the Vg1 protein during early Xenopus development. Development. 1989;106(4):775–785. doi: 10.1242/dev.106.4.775. [DOI] [PubMed] [Google Scholar]
- 118.Melton DA. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 1987;328(6125):80–82. doi: 10.1038/328080a0. [DOI] [PubMed] [Google Scholar]
- 119.Gautreau D, Cote CA, Mowry KL. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development. 1997;124(24):5013–5020. doi: 10.1242/dev.124.24.5013. [DOI] [PubMed] [Google Scholar]
- 120.Lewis RA, Gagnon JA, Mowry KL. PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol Cell Biol. 2008;28(2):678–686. doi: 10.1128/MCB.00999-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lewis RA, Kress TL, Cote CA, Gautreau D, Rokop ME, Mowry KL. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech Dev. 2004;121(1):101–109. doi: 10.1016/j.mod.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 122.Zhou Y, King ML. Localization of Xcat-2 RNA, a putative germ plasm component, to the mitochondrial cloud in Xenopus stage I oocytes. Development. 1996;122(9):2947–2953. doi: 10.1242/dev.122.9.2947. [DOI] [PubMed] [Google Scholar]
- 123.Kloc M, Bilinski S, Pui-Yee Chan A, Etkin LD. The targeting of Xcat2 mRNA to the germinal granules depends on a cis-acting germinal granule localization element within the 3′UTR. Dev Biol. 2000;217:221–229. doi: 10.1006/dbio.1999.9554. [DOI] [PubMed] [Google Scholar]
- 124.Lehmann R, Nusslein-Volhard C. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila . Cell. 1986;47(1):141–152. doi: 10.1016/0092-8674(86)90375-2. [DOI] [PubMed] [Google Scholar]
- 125.Kim-Ha J, Webster PJ, Smith JL, Macdonald PM. Multiple RNA regulatory elements mediate distinct steps in localization of oskar mRNA. Development. 1993;119:169–178. doi: 10.1242/dev.119.1.169. [DOI] [PubMed] [Google Scholar]
- 126.Berleth T, Burri M, Thoma G, Bopp D, Richstein S, Frigerio G, Noll M, Nusslein-Volhard C. The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 1988;7(6):1749–1756. doi: 10.1002/j.1460-2075.1988.tb03004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schnorrer F, Bohmann K, Nusslein-Volhard C. The molecular motor dynein is involved in targeting swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat Cell Biol. 2000;2(4):185–190. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- 128.MacDonald PM. Bicoid mRNA localization signal: phylogenetic conservation of function and RNA secondary structure. Development. 1990;110(1):161–171. doi: 10.1242/dev.110.1.161. [DOI] [PubMed] [Google Scholar]
- 129.Macdonald PM, Struhl G. Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature. 1988;336(6199):595–598. doi: 10.1038/336595a0. [DOI] [PubMed] [Google Scholar]
- 130.Macdonald PM, Kerr K, Smith JL, Leask A. RNA regulatory element BLE1 directs the early steps of bicoid mRNA localization. Development. 1993;118(4):1233–1243. doi: 10.1242/dev.118.4.1233. [DOI] [PubMed] [Google Scholar]
- 131.Macdonald PM, Kerr K. Mutational analysis of an RNA recognition element that mediates localization of bicoid mRNA. Mol Cell Biol. 1998;18(7):3788–3795. doi: 10.1128/mcb.18.7.3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Irion U, St Johnston D. Bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature. 2007;445(7127):554–558. doi: 10.1038/nature05503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ferrandon D, Koch I, Westhof E, Nusslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16(7):1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ferrandon D, Elphick L, Nusslein-Volhard C, St Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79(7):1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 135.Weil TT, Xanthakis D, Parton R, Dobbie I, Rabouille C, Gavis ER, Davis I. Distinguishing direct from indirect roles for bicoid mRNA localization factors. Development. 2010;137(1):169–176. doi: 10.1242/dev.044867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Cha B-J, Koppetsch BS, Theurkauf WE. In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell. 2001;106(1):35–46. doi: 10.1016/S0092-8674(01)00419-6. [DOI] [PubMed] [Google Scholar]
- 137.Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGF alpha-like protein. Cell. 1993;75(1):165–174. [PubMed] [Google Scholar]
- 138.Neuman-Silberberg FS, Schupbach T. The Drosophila TGF-alpha-like protein Gurken: expression and cellular localization during Drosophila oogenesis. Mech Dev. 1996;59(2):105–113. doi: 10.1016/0925-4773(96)00567-9. [DOI] [PubMed] [Google Scholar]
- 139.Saunders C, Cohen RS. The role of oocyte transcription, the 5′UTR, and translation repression and derepression in Drosophila gurken mRNA and protein localization. Mol Cell. 1999;3(1):43–54. doi: 10.1016/S1097-2765(00)80173-2. [DOI] [PubMed] [Google Scholar]
- 140.Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I. Gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev Cell. 2005;9(1):51–62. doi: 10.1016/j.devcel.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 141.Davis I, Ish-Horowicz D. Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67(5):927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- 142.Bullock SL, Zicha D, Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 2003;22(10):2484–2494. doi: 10.1093/emboj/cdg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila . Genes Dev. 1997;11(4):423–435. doi: 10.1101/gad.11.4.423. [DOI] [PubMed] [Google Scholar]
- 144.Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414(6864):611–616. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- 145.Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105(2):209–219. doi: 10.1016/S0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 146.Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol. 2006;16(14):1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 147.Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23(13):1546–1558. doi: 10.1101/gad.531009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sundell CL, Singer RH. Requirement of microfilaments in sorting of actin messenger RNA. Science. 1991;253:1275–1277. doi: 10.1126/science.1891715. [DOI] [PubMed] [Google Scholar]
- 149.Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of b-actin mRNA and protein localization within growth cones. J Cell Biol. 1999;147(1):59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell. 2008;15(3):426–436. doi: 10.1016/j.devcel.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hill MA, Schedlich L, Gunning P. Serum-induced signal transduction determines the peripheral location of beta-actin mRNA within the cell. J Cell Biol. 1994;126(5):1221–1229. doi: 10.1083/jcb.126.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Latham VM, Kislauskis EH, Singer RH, Ross AF. Beta-actin mRNA localization is regulated by signal transduction mechanisms. J Cell Biol. 1994;126(5):1211–1219. doi: 10.1083/jcb.126.5.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9(9):690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 154.Sossin WS, DesGroseillers L. Intracellular trafficking of RNA in neurons. Traffic. 2006;7(12):1581–1589. doi: 10.1111/j.1600-0854.2006.00500.x. [DOI] [PubMed] [Google Scholar]
- 155.Newpher TM, Ehlers MD. Spine microdomains for postsynaptic signaling and plasticity. Trends Cell Biol. 2009;19(5):218–227. doi: 10.1016/j.tcb.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 156.Bramham C, Alme M, Bittins M, Kuipers S, Nair R, Pai B, Panja D, Schubert M, Soule J, Tiron A, Wibrand K. The arc of synaptic memory. Exp Brain Res. 2010;200(2):125–140. doi: 10.1007/s00221-009-1959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21(4):741–751. doi: 10.1016/S0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 158.Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30(1):227–240. doi: 10.1016/S0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 159.Tongiorgi E, Righi M, Cattaneo A. Activity-dependent dendritic targeting of BDNF and TrkB mRNAs in hippocampal neurons. J Neurosci. 1997;17(24):9492–9505. doi: 10.1523/JNEUROSCI.17-24-09492.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Burgin KE, Waxham MN, Rickling S, Westgate SA, Mobley WC, Kelly PT. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J Neurosci. 1990;10(6):1788–1798. doi: 10.1523/JNEUROSCI.10-06-01788.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ. Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta -actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci. 2003;23(8):3251–3261. doi: 10.1523/JNEUROSCI.23-08-03251.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Grooms SY, Noh K-M, Regis R, Bassell GJ, Bryan MK, Carroll RC, Zukin RS. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J Neurosci. 2006;26(32):8339–8351. doi: 10.1523/JNEUROSCI.0472-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.O’Donnell M, Chance RK, Bashaw GJ. Axon growth and guidance: receptor regulation and signal transduction. Annu Rev Neurosci. 2009;32(1):383–412. doi: 10.1146/annurev.neuro.051508.135614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yoon B, Zivraj K, Holt C (2009) Local translation and mRNA trafficking in axon pathfinding. In: Koenig E (ed) Cell biology of the axon, vol 48. Results and problems in cell differentiation. Springer, Berlin Heidelberg New York, pp 269–288 [DOI] [PMC free article] [PubMed]
- 165.Zhang HL, Eom T, Oleynikov Y, Shenoy S, Liebelt D, Dictenberg J, Singer R, Bassell G. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/S0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 166.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat Neurosci. 2006;9(10):1265–1273. doi: 10.1038/nn1773. [DOI] [PubMed] [Google Scholar]
- 167.Leung K-M, van Horck FPG, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9(10):1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sasaki Y, Welshhans K, Wen Z, Yao J, Xu M, Goshima Y, Zheng JQ, Bassell GJ. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local beta-actin synthesis and growth cone turning. J Neurosci. 2010;30(28):9349–9358. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci. 1996;16(24):7812–7820. doi: 10.1523/JNEUROSCI.16-24-07812.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Knowles RB, Kosik KS. Neurotrophin-3 signals redistribute RNA in neurons. Proc Nat Acad Sci USA. 1997;94(26):14804–14808. doi: 10.1073/pnas.94.26.14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Rook MS, Lu M, Kosik KS. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci. 2000;20(17):6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43(4):513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 173.Elvira G, Wasiak S, Blandford V, Tong X-K, Serrano A, Fan X, del Rayo Sanchez-Carbente M, Servant F, Bell AW, Boismenu D, Lacaille J-C, McPherson PS, DesGroseillers L, Sossin WS. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5(4):635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 174.Kohrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell. 1999;10(9):2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Colin E, Zala D, Liot G, Rangone H, Borrell-Pages M, Li X-J, Saudou F, Humbert S. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 2008;27(15):2124–2134. doi: 10.1038/emboj.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Engelender S, Sharp AH, Colomer V, Tokito MK, Lanahan A, Worley P, Holzbaur ELF, Ross CA. Huntingtin-associated protein 1 (HAP1) interacts with the p150Glued subunit of dynactin. Hum Mol Genet. 1997;6(13):2205–2212. doi: 10.1093/hmg/6.13.2205. [DOI] [PubMed] [Google Scholar]
- 177.Li S-H, Gutekunst C-A, Hersch SM, Li X-J. Interaction of huntingtin-associated protein with dynactin P150Glued. J Neurosci. 1998;18(4):1261–1269. doi: 10.1523/JNEUROSCI.18-04-01261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Savas JN, Ma B, Deinhardt K, Culver BP, Restituito S, Wu L, Belasco JG, Chao MV, Tanese N. A role for huntington disease protein in dendritic RNA granules. J Biol Chem. 2010;285(17):13142–13153. doi: 10.1074/jbc.M110.114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bagni C, Greenough WT. From mRNP trafficking to spine dysmorphogenesis: the roots of fragile X syndrome. Nat Rev Neurosci. 2005;6(5):376–387. doi: 10.1038/nrn1667. [DOI] [PubMed] [Google Scholar]
- 180.Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4(6):350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 181.Kao D-I, Aldridge GM, Weiler IJ, Greenough WT. Altered mRNA transport, docking, and protein translation in neurons lacking fragile X mental retardation protein. Proc Nat Acad Sci USA. 2010;107(35):15601–15606. doi: 10.1073/pnas.1010564107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile X mental retardation protein and Fmr1 mRNA localization differentially in dendrites and at synapses. J Neurosci. 2004;24(11):2648–2655. doi: 10.1523/JNEUROSCI.0099-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell. 2008;14(6):926–939. doi: 10.1016/j.devcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Huang F, Chotiner JK, Steward O. Actin polymerization and ERK phosphorylation are required for Arc/Arg3.1 mRNA targeting to activated synaptic sites on dendrites. J Neurosci. 2007;27(34):9054–9067. doi: 10.1523/JNEUROSCI.2410-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yoshimura A, Fujii R, Watanabe Y, Okabe S, Fukui K, Takumi T. Myosin-va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr Biol. 2006;16(23):2345–2351. doi: 10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 186.Messaoudi E, Kanhema T, Soulé J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 synthesis controls long-term potentiation consolidation through regulation of local actin polymerization in the dentate gyrus in vivo. J Neurosci. 2007;27(39):10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Weil TT, Parton RM, Davis I. Making the message clear: visualizing mRNA localization. Trends Cell Biol. 2010;20(7):380–390. doi: 10.1016/j.tcb.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lionnet T, Czaplinski K, Darzacq X, Shav-Tal Y, Wells AL, Chao JA, Park HY, de Turris V, Lopez-Jones M, Singer RH. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nat Meth. 2011;8(2):165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Swanger SA, Bassell GJ, Gross C. High-resolution fluorescence in situ hybridization to detect mRNAs in neuronal compartments in vitro and in vivo. Methods Mol Biol. 2011;714:103–123. doi: 10.1007/978-1-61779-005-8_7. [DOI] [PubMed] [Google Scholar]