Abstract
MicroRNAs are short endogenous RNA molecules that are able to regulate (mainly inhibiting) gene expression at the post-transcriptional level. The MicroRNA expression profile is cell-specific, but it is sensitive to perturbations produced by stresses and diseases. Endothelial cells subjected to metabolic stresses, such as calorie restriction, nutrients excess (glucose, cholesterol, lipids) and hypoxia may alter their functionality. This is predictive for the development of pathologies like atherosclerosis, diabetes, and hypertension. Moreover, cancer cells can activate a resting endothelium by secreting pro-angiogenic factors, in order to promote neoangiogenesis, which is essential for tumor growth. Endothelial altered phenotype is mirrored by altered mRNA, microRNA, and protein expression, with a microRNA being able to control pathways by regulating the expression of multiple mRNAs. In this review we will consider the involvement of microRNAs in modulating the response of endothelial cells to metabolic stresses and their role in promoting or halting angiogenesis.
Keywords: Endothelial cells, Angiogenesis, MicroRNAs, Metabolic stresses, Metabolic disorders, Cancer
MicroRNAs
MicroRNAs are a class of endogenous 22–25 nt non-coding single-stranded RNA molecules that regulate gene expression post-transcriptionally. MicroRNAs can affect multiple cell processes, like proliferation, apoptosis, differentiation, and also development [1].
MicroRNAs are transcribed by RNA polymerase II as longer molecules (pri-miRNAs). The ribonuclease Drosha maturates them into hairpin structure (pre-miRNAs), then exported into the cytoplasm, where another ribonuclease, Dicer, cuts the loop, forming a double-stranded RNA molecule. One of the two strands is degraded, whereas the other, the mature microRNA, is incorporated into RISC (RNA-induced silencing complex), which mediates its action [2]. MicroRNAs generally bind to the 3′ UTR of target genes and inhibit their translation [3], but in other instances they have also been shown to destabilize the targeted mRNA [4]. Each microRNA can potentially bind several (hundreds) target messenger RNAs through a very short (down to six nucleotides) complementary region, called “seed”, which perfectly pairs with the mRNA 3′ UTR sequence.
The most challenging issue about microRNA study is the recognition mechanism and so the identification of mRNA targets. Informatic algorithms (TargetScans, PicTar, Miranda, to name a few) help to find probable targets, but the binding needs always experimental validation for each candidate target. Besides, since these algorithms are based on different criteria, they give different output targets and each of them has a high probability to produce either false-positive or -negative results.
So far, several experimental approaches for the identifications of microRNA targets have been developed [5]. They are based on transcriptome analysis [4], protein analysis [6], or biochemical approaches [7, 8].
The cell dynamics, arising from the connectivity of genes and proteins, is extremely complex, and unpredictable, although deterministic. As a consequence of the imperfect base-pairing mechanism, the same miRNA, in different contexts, is able to drive the cell towards different phenotypes. For that, miRNAs are natural candidates to a central role in cellular adaptability mechanisms.
When deciphering a phenomenon like a metabolic stress or tumor progression, microRNA expression profiling often reveals, as for mRNA expression profiling, tens of microRNAs differently expressed. Indeed, microRNA expression has been found so distinguishing in pathological states that microRNA profiling has been suggested as a biomarker to classify human cancers [9]. Moreover, Gallagher et al. [10] analyzed microRNAs and mRNAs of skeletal muscle insulin-resistant patients and used a ranking system to identify pathways affected by the disease.
The regulation of gene expression by microRNAs is fine and complex at the same time. Each miRNA potentially interacts with several mRNAs so that miRNA (interference) network and gene expression network are embedded to each other [11].
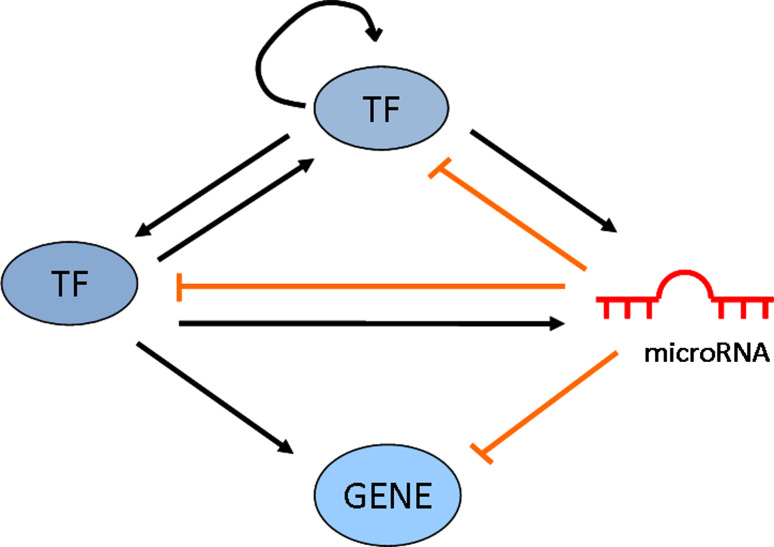
Both microRNAs and TFs are metazoan gene regulators, sharing common features like pleiotropy, combinatorial and cooperative mechanism of action. MicroRNAs and TFs are also involved in coregulational network motifs (Fig. 1).
Fig. 1.
Schematic representation of a network comprising TFs, microRNAs, and target genes with the possible regulatory motifs
MicroRNA depletion, for example miR-221 and miR-222 depletion, besides changing mRNA expression, induced changes in microRNA expression, with nine microRNAs up-regulated and 23 microRNAs down-regulated [12]. This finding envisages a model in which a microRNA can affect the expression of transcription factors (TFs) or components of the microRNA processing machinery, so that the overall microRNAs are in turn affected.
This issue was further developed by means of computational models to assess the extent of regulation of gene expression mediated by combinations of TFs and microRNAs [13], inferring that genes are more likely to be co-regulated by pairs of TFs or pairs of miRNAs than by pairs of TF-miRNA, perhaps due to a higher probability of evolutionary duplication events of shorter DNA sequences.
Chen et al. [14] analyzed the biological processes enriched for different regulation types and they found that two biological processes (pigmentation and reproduction) require a coordinate TFs-microRNA coregulation. On the other hand, it turned out that biological adhesion, developmental and cellular processes are enriched in microRNA–microRNA coregulation types, suggesting that microRNAs should carefully coordinate to regulate these processes.
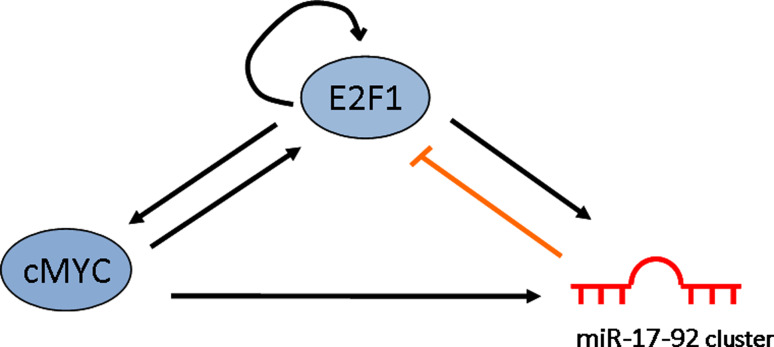
The computational model by Tsang et al. [15] revealed the existence of two classes of circuits, respectively, negative and positive transcriptional coregulation of a miRNA and its targets. These circuits can lead to a feedback and feedforward loop of regulation. One main example is represented by the MYC-miR-17-92 cluster-E2F1 circuit, in which MYC induces expression of both E2F1 and the miR-17-92 cluster, the latter in turn represses E2F1 expression (Fig. 2) [16]. This “genetic buffering” seems intended to minimize noise in the level of E2F1 protein. A similar kind of circuit can be seen for other microRNAs, specifically for those with antithetic functions displayed in different contexts. MiR-20a, belonging to miR-17-92 cluster, is part of the MYC-miR-17-92 cluster-E2F1self-regulating and has a double role in tumorigenesis and senescence, according to the endogenous level of E2F1 [17].
Fig. 2.
Schematic representation of a network motif with a feedforward loop mediated by cMYC to E2F1 and a negative feedback regulatory loop mediated by miR-17-92 cluster to E2F1
The bioinformatic research for microRNAs targeting VEGF exemplifies the presence of regulatory modules. As hypoxia induces VEGF, microRNAs down-regulated by hypoxia were evaluated for their direct regulation of VEGF, according to the presence of binding site in the VEGF mRNA sequence. VEGF was regulated by multiple microRNAs (miR-15b, miR-16, miR-20a, miR-20b) that displayed co-regulatory effects on other angiogenic factors in CNE cells (nasopharyngeal carcinoma line) and also a combinatorial mechanism of action [18]. This mechanism envisages the possibility that microRNAs could compete for a binding site (multimiRNA binding site) or they cooperate by synergistically reinforcing the inhibition of the same target, binding it in several points. This has been shown, for example, for miR-15b, miR-24, miR-25, and miR-141, whose joint reduction is necessary to increase MKK4 abundance in replicative senescence [19]. More recently, a cooperative microRNA network (comprising miR-19b, miR-20a, miR-26a, miR-92, and miR-223) has been claimed responsible to inhibit tumor suppressor genes and promote T cell acute lymphoblastic leukemia [20].
Endothelial cells
Blood vascular endothelial cells (ECs) line the inner part of the blood vessels and thus interface the blood with the rest of the tissues. This position enables ECs to mediate several functions. In concert with the immune cells, they play a role in the inflammation process: by modulating the intercellular junctions they can become more or less permeable and therefore promote inflammation and edema, while the surface molecules expressed on EC membrane permits leukocytes (neutrophils) adhesion and migration. ECs regulate the coagulation process by secreting von Willebrand factor; they also control blood flow and pressure by mediating vasodilation or vasoconstriction through the secretion of paracrine factors acting on smooth muscle cells [21, 22].
ECs are normally quiescent, with a turnover of several months [23], but they can be activated by pro-angiogenic factors released by damaged tissues during physiological processes like regeneration in wound healing or during the menstrual cycle in the endometrium. Pro-angiogenic factors, like VEGF (vascular endothelial growth factor) and bFGF (basic fibroblast growth factor), mediate the formation of new vessels from pre-existing ones, a phenomenon called angiogenesis. ECs, activated in response to these stimuli, divide and migrate to sprout new vessels. While this phenomenon is to be promoted after an ischemic stress in order to form collateral vessels that can again provide nutrients and oxygen to the tissues, it should be counteracted in a tumor context. Cancer cells, in fact, secrete growth factors to sustain blood supply for their nutriment and also migration. Thus, the resulting imbalance between pro-angiogenic and antiangiogenic stimuli (thrombospondin and endothelin, for instance) determines the phenotype of an EC: if the balance tips towards pro-angiogenic factors, an angiogenic switch occurs.
ECs are directly subjected to physico-chemical stresses (shear stress, hypoxia, glucose, cholesterol, lipids), which are able to alter gene expression, interfere with cellular metabolism, induce molecular modifications, and enhance reactive oxygen species (ROS) production. The result is that the response to pro- or antiangiogenic factors of ECs are deeply modified. From a certain perspective, metabolic stresses themselves could be seen as (mainly) antiangiogenic stimuli when applied to ECs because they imbalance the EC intracellular context and, as a consequence, they modify EC response towards the extracellular environment.
The endothelial dysfunction is defined as the incapacity of endothelium to maintain vascular homeostasis, mainly due to the interference with the synthesis of NO (nitric oxide), which is the principal vasodilating factor produced by ECs [24]. This condition is linked to the development of diseases as atherosclerosis [25], hypertension, diabetes, and cardio vascular diseases [26]. This is translated into modified expression of endothelial markers and impaired angiogenic properties that in vitro are measured as the ability to migrate, proliferate, and form tube-like structures in response to pro-angiogenic stimuli. In molecular terms, these alterations in endothelial functionality are the results of complex interactions between transcriptional and translational processes. Potential mediators of these processes are microRNAs that modulate and are modulated by TFs, thus functioning as regulatory molecules of gene expression networks.
Metabolism of endothelial cells
ECs line all the vases, from the capillaries to the venous and arterial compartments. The venous or arterial fate of ECs is regulated during vasculogenesis [21] and ECs maintain their diversity in the adult gene expression profiles (reviewed in [27]). This accounts for the different morphology and functions of the two compartments, principally related to the O2 partial pressure and hemodynamics. The arteries and arterioles are responsible for the vascular tone and are subjected to shear stress; on the other hand, the veins are larger but with thinner walls, so that the blood flows slower and is less oxygenated and more prone to leukocyte trafficking [21, 28].
In the endothelium, ECs line according to the blood flow and are resting cells [23, 29] adapted to survive in hypoxic condition and in the presence of ROS [30]. This was further supported by the finding that ECs stabilize the transcription factor HIF1α (hypoxia-induced factor 1 α, a transcription factor activated at low O2 levels that up-regulates genes involved in glycolysis and angiogenesis) at a lower O2 concentration than other cells [31].
Peters et al. [29] compared the metabolic capacities of freshly isolated HUVEC with in vitro cultivated ones (three passages) and, according to others [30, 32], they found “aerobic glycolysis” in ECs. In fact, cell cultivation, leading to increased proliferation, enhances glycolysis as well as other pathways (tricarboxylic acid cycle, fatty oxidization, pentose phosphate cycle) still active in ECs but at a less extent compared to glycolysis. Thus, it appears that mitochondria in ECs act as signaling organelles more than source of ATP. Chronic, small increases in NO levels stimulate mitochondrial biogenesis in diverse cell types [33], conversely, higher levels of NO produced by ECs inhibit cell respiration by binding to cytochrome c oxidase. The inhibition of oxidative phosphorylation by NO increases ROS production, which can function activating AMPK (5′ adenosine monophosphate protein kinase) [31]; in turn, O2 is diverted from the mitochondria towards the cytosol where it activates prolyl hydroxylases (O2 sensing enzymes that inactivate HIFs), therefore the cells do not register hypoxia [34].
MicroRNAs involved in metabolic stresses and disorders
Hypoxia
Hypoxia, which means diminished oxygen concentration, can occur during ischemia, stroke, tumor growth, and at high altitude. Oxygen-sensing enzymes, like prolyl hydroxylases, promote HIF1α proteasomal degradation at normal O2 concentration; conversely, when O2 concentration drops, HIF1α is free to enter the nucleus, heterodimerize with HIF1β, and act as a transcription factor. The pathways activated by HIF1α promote EC proliferation, migration, and angiogenesis, but may also mediate cell death, if the hypoxic stimulus has been too severe [35].
Hypoxia mediates HIF1α-dependent up-regulation of miR-210 both in primary ECs and in cancer cell lines [36–38]. MiR-210 was found to stimulate tubulogenesis and chemotaxis of ECs by inhibiting Ephrin A3 [38] and to modulate mitochondrial function by decreasing iron-sulfur cluster assembly proteins ISCU1/2 [37, 39] and COX10 [39] (Table 1), key factors of the mitochondrial electron transport chain and tricarboxylic acid cycle. This has important consequences: mitochondrial respiration activity is inhibited, whereas ROS are generated. This environment favors the metabolic switch from respiration to glycolysis (the Warburg effect), which is typical of cancer cells adapted to a hypoxic condition. Recently, miR-200b, which directly target Ets-1, an important angiogenesis-related transcription factor, has been shown to be downregulated by hypoxia. Its downregulation promotes migration and tube formation of ECs [40].
Table 1.
MicroRNAs regulating angiogenic properties in response to metabolic stresses
| Stress | MicroRNA | Targeta | MicroRNA functional impact on angiogenic properties | Cell type | References |
|---|---|---|---|---|---|
| Hypoxia | ↑miR-210 | Ephrin A3 | Promotion of migration and tube-like structure formation | Human ECs (HUVEC) | [38] |
| ISCU1/2 COX10 | Inhibition of mitochondrial respiration | Human ECs (HPAEC, HUVEC, HAEC), colon cancer cell lines | [37, 39] | ||
| Hypoxia | ↓miR-200b | Ets-1 | Inhibition of migration and tube-like structure formation | Human ECs (HMEC) | [40] |
| Population doubling | ↑miR-217 | SIRT1 | Promotion of senescence | Human ECs (HUVEC) | [103] |
| Inhibition of tube formation | |||||
| Population doubling | ↑miR-34 | SIRT1 |
Promotion of senescence Inhibition of proliferation |
Human ECs (HUVEC) | [105] |
| High glucose, hyperglycemia | ↑miR-320 | IGF-1 | Inhibition of proliferation and migration | GK rat ECs (MMVEC) | [65] |
| CD71 | Inhibition of proliferation | Human leukemia cell line (HL-60) | [66] | ||
| PIK3R2 (p85) | Insulin resistance | Mouse adipocytes (3T3-L1) | [67] | ||
| HSP20 | Apoptosis | Rat cardiomyocytes | [68] | ||
| High glucose | ↑miR-221 | c-kit | Inhibition of proliferation and migration | Human ECs (HUVEC) | [75] |
| High glucose, AGEs | ↓miR-221 |
P27KIP1 P57KIP2 |
Promotion of proliferation | Human ECs (HUVEC) | [77] |
| CCND1 | Inhibition of proliferation | Human head and neck carcinoma cell line UMSCC10B | [80] | ||
| High glucose/low growth factor | ↑miR-503 | Cdc25 | G1 phase cell cycle arrest | Mouse C2C12 myoblast | 81] |
| Cdc25, CCNE1 | Inhibition of proliferation and tube-like structure formation | Human ECs (HUVEC) | [79] | ||
| Low cellular sterol | T miR-33 |
ABCA1 ABCG1 NPC1 |
Inhibition of cholesterol efflux | Human ECs (Ea-hy), human and mouse hepatocyte and monocyte cell lines mouse hepatocyte and monocyte cell lines human hepatocyte and monocyte cell lines | [91] |
| Ox-LDL | ↑miR-125a-5p | PreproET-1 | Control vasoconstriction |
Murine cardiac microvascular ECs (H5V) murine brain microvascular ECs (b.END.3) |
[95] |
| ↓miR-125b |
aTarget validated by gene reporter assay
ROS, as superoxide and hydrogen peroxide, are by-products of cellular metabolism being able to damage lipids, proteins, and nucleic acids. Excessive or sustained ROS production are implicated in atherosclerosis, hypertension, diabetic cardiovascular complications, and ischemia–reperfusion injury. Otherwise, an emerging body of evidence has been showing that in moderate concentration, ROS are able to regulate cellular functions. In vascular ECs, the principal enzymes responsible for ROS generation are NAD(P)H oxidase, xanthine oxidase, uncoupled eNOS (endothelial nitric oxide synthase), and mitochondrial electron leakage, while conditions like hypoxia stimulate ROS production. The produced ROS (above all mitochondrial ROS) are able to regulate vascular tone, oxygen sensing, cell growth and proliferation, apoptosis, and inflammatory responses [41].
Hyperglycemia
Hyperglycemia is recognized as a major determinant causing vascular problems in diabetes mellitus, obesity, and metabolic syndrome that may be traced back to endothelial dysfunction. The latter is strictly linked to insulin resistance, because insulin is able to activate eNOS [42, 43] and also because endothelial dysfunction and insulin resistance share common factors triggering their onset: glucotoxicity, lipotoxicity, and inflammation [44]. Four main molecular mechanisms have been implicated in glucose-mediated vascular damage: (i) increased flux through the polyol pathway; (ii) protein kinase C (PKC) activation; (iii) increased hexosamine pathway activity; (iv) increased production of advanced glycation end-products (AGEs). In fact, the more glucose that enters the cells and the tricarboxylic acid cycle, the more electron donors (NADH and FADH2) are generated and therefore electrons tend to block in the complex III of the mitochondrial electron transport chain and are donated to O2 one at a time, forming superoxide. ROS activate the poly(ADP-ribose) polymerase (PARP), which in turn inhibits the glycolytic enzyme glyceraldehyde-3 phosphate dehydrogenase (GAPDH). This inhibition directly causes activation of AGE and PKC pathways, while the accumulation of upstream glycolytic metabolites, fructose-6 phosphate and glucose itself, causes the activation of the hexosamine pathway and polyol pathway, respectively [45]. Increased oxidative stress is the final outcome and hence the leading feature that underlies endothelial dysfunction onset and progression [46, 47].
ECs cultured in high glucose to mimic hyperglycemic conditions have a reduced proliferation rate and a pronounced increase in apoptosis compared to cells grown in normal glucose [48–50]. An inhibitory effect of high glucose on migration and angiogenic potential [51–54], an impairment of eNOS functionality and expression [42, 55–58] and an increased expression of the adhesion molecules ICAM-1, ELAM, VCAM [59–62] have also been reported.
ECs use mainly GLUT1 as a glucose transporter. At normal glucose concentration, this transporter is not up-regulated by insulin, but an insulin-stimulated GLUT1 up-regulation and consequent glucose uptake has been reported at high glucose concentration [63]. ECs are not able to regulate efficiently the transport of glucose to maintain its intracellular concentration constant [64] and hence, unlike other cell types, they suffer damage from hyperglycemia.
The first microRNA reported being mis-expressed in ECs exposed to hyperglycemia is miR-320. It was found to be increased, among other microRNAs, in micro vascular ECs (MMVEC) isolated from type 2 diabetes Goto Kakizaki (GK) rats compared to control Wistar rats. Its inhibition promoted proliferation and migration of GK MMVEC, presumably by targeting IGF-1 [65]. Other direct targets of miR-320 have been identified and validated: the transferrin receptor CD71, with a role in cell proliferation in a leukemia cell line [66]; PI3 K subunit p85 in insulin-resistant adipocytes [67]; the heat shock protein HSP20 in cardiomyocytes [68]. An inverse correlation (suggesting at least an indirect regulation) was observed between the expression of miR-320 and cyclin-dependent kinases 6 (CDK6) in primary murine bronchial epithelial cells [69], and Mcl-1 in cholangiocytes [70]. On the whole, in these cell lines miR-320 has an anti-proliferative role. This finding was further supported by other works, where miR-320 is listed in the down-regulated microRNAs in colon cancer [71], breast cancer [72], cholangiocarcinoma [70], cell lung carcinoma [73], and epigenetically regulated in pancreatic cancer [74].
Li et al. [75] researched in high glucose-treated HUVEC the EC specific miR-221, whose recognized target is c-kit [76], and found it over-expressed, with a role in inhibiting cell migration. Different results were obtained by Togliatto et al. [77], who found miR-221 and miR-222 over-expressed in ECs treated with high glucose or AGEs. They demonstrated also in vivo that impaired vessel formation mediated by high glucose and AGEs is controlled by miR-221 and miR-222 by targeting the cell cycle regulators P27KIP1 and P57KIP2. MiR-320 and miR-221 were also found over-expressed in type 2 GK rat livers (miR-221) [78], and in 3T3 adipocytes rendered insulin resistant through high glucose and high insulin treatment (miR-320) [67], suggesting that these microRNAs may have a broad involvement in glucose homeostasis in insulin-targeted tissues. miR-221-222 have also a role in tumor progression and in angiogenesis (see later).
The expression of miR-503 was found increased in myocardial ECs from type 2 GK rats [65], in 3T3-L1 insulin-resistant adipocytes [67] and also in muscle of type 2 diabetes and insulin-resistant human patients [10]. MiR-503 is up-regulated in HUVEC grown in high glucose/low growth factors-containing medium (to mimic diabetes and starving condition caused by ischemia); in limb muscle and plasma of diabetic patients with critical limb ischemia and in the ischemic muscle and ECs of diabetic mice [79]. MiR-503 down-regulates the expression of the cell cycle regulators cyclin D1 (CCND1) [80], cdc25a [79, 81] and cyclin E1 (CCNE1) [79]. In HUVEC, miR-503 forced expression led to impaired migration, adhesion, network formation capacities, and impaired proliferation due to an increase of G0/G1-arrested cells. MiR-503 also reduced vascular smooth muscle cells proliferation and migration, which are instrumental for arteriogenesis. Conversely, miR-503 inhibition restored normal proliferation and in vitro angiogenesis of ECs grown in high glucose/low growth factors-containing medium, without affecting EC functions under normal culture conditions [79]. In diabetic mice with induced limb ischemia, the inhibition of miR-503 by injecting an adenoviral vector containing a decoy sequence for miR-503 normalized post-ischemic blood flow and muscular neovascularization, as well as cdc25a and CCNE1 expression [79] (Table 1). These findings suggest that miR-503 antagonizes post-ischemic neovascularization in diabetes mellitus and thus it can be considered a potential therapeutic target to improve healing of diabetic ischemic tissues.
Hypertriglyceridemia and hypercholesterolemia
It is well known that chronic hypertriglyceridemia and hypercholesterolemia are risk factors in the development of atherosclerosis. It is widely accepted that hypertriglyceridemia leads to endothelial activation and dysfunction and it is associated to superoxide anion production and decrease in NO bioavailability [82, 83]. In fact, the release of free fatty acids during triglyceride hydrolysis of chylomicrons and VLDL (very low-density lipoprotein), mediated by EC membrane lipoprotein lipase, may cause endothelial cell injury and initiate thrombotic events [84, 85].
Superoxide anion in ECs is mainly produced by enzymatic oxidants: uncoupled eNOS (mainly due to decrease in the cofactor tetrahydrobiopterin, BH4) and by NADPH oxidase [26, 86, 87]. The mechanism of LDL oxidation is not well clarified, but it seems to involve the endothelium itself. ROS in the sub-endothelial space can oxidize free fatty acids and low-density lipoprotein (ox-LDL). Also phospholipase A2 and lipoxygenase are involved in LDL lipoperoxidazion. Ox-LDL are highly immunogenic, attack the arterial intima and activate ECs and monocytes, increasing proinflammatory gene activity and uptake by macrophages. This is the initial step in the formation of fatty streaks that finally turn in the atheromatous plaque [88]. ECs treated with ox-LDL in vitro show decreased expression of eNOS [89], increased proliferation, and hypertrophy [90].
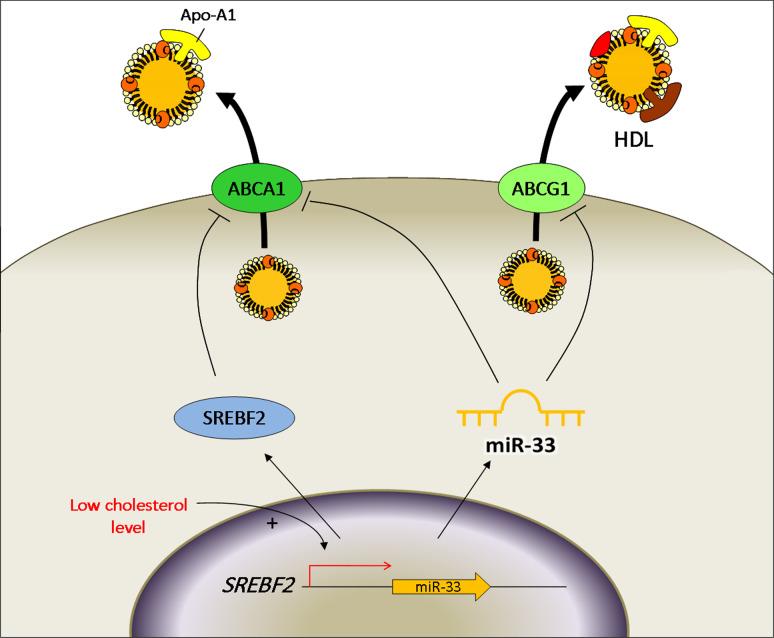
Lipid and cholesterol metabolism is regulated mainly in liver and adipose tissue. The most expressed microRNAs in liver is miR-122, whose inhibition results in lowering cholesterol plasma levels both in mammals and non-human primates and also in the treatment of chronic hepatitis C virus infection. MiR-33 is also involved in cholesterol metabolism. This microRNA, besides being expressed in hepatocytes and macrophages, is expressed in ECs (Ea-hy cell line) [91] and it represses genes involved in cholesterol mobilization: ABCA1 (in human and mouse cells), ABCG1 (in mouse cells), NPC1 (in human cells). MiR-33 acts in a synergistic manner with its host gene SREBF2 [91, 92], a transcription factor which regulates the expression of genes involved in cholesterol biosynthesis and uptake. Moreover, SREBF2 in turn down-regulates ABCA1 in vascular ECs (Fig. 3) [93]. In this view, miR-33 and SREBF2 regulate cholesterol homeostasis and reverse cholesterol transport (the mechanism that regulates circulating levels of HDL) in several tissues: under low cellular sterol concentration miR-33 and SREBF2 are up-regulated and inhibit cholesterol efflux through apolipoprotein A1 by ABCA1 and through HDL by ABCG1; vice versa, when the cellular cholesterol concentration is high, they are down-regulated to allow the cholesterol import through the membrane transporters. Mice transgenically deficient for miR-33 [94] or inhibited for miR-33 expression by tail injection of LNA miR-33 antisense [92] or by infection with lentivirus coding for antimiR-33 [91] result in the over-expression of ABCA1 and increased plasma HDL levels. Thus miR-33 inhibition appears to be a valuable strategy to rise HDL levels, but the cellular level of cholesterol might be impaired.
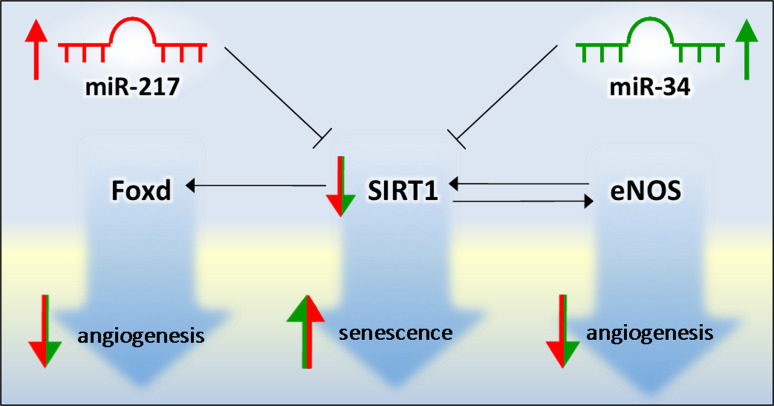
Fig. 3.
Schematic representation of a network comprising miR-217 and miR-34 and SIRT1 that promotes senescence of ECs
There are not many studies about microRNA dysregulated in the endothelium of animals fed with high-fat diets or in ECs challenged with high cholesterol or high fat. A recent work shows that miR-125a-5p is increased after 6 h of ECs stimulation with ox-LDL, whereas miR-125b is constitutively decreased. These microRNAs are both decreased in aorta of stroke-prone spontaneously hypertensive (SHR-SP) rats, where they are associated with up-regulation of preproET-1 (the precursor of endothelin 1) expression, which is their validated target [95] (Table 1). Conversely, miR-125a-5p is up-regulated in ox-LDL-treated monocytes where it mediates lipid uptake and decreases the secretion of some inflammatory cytokines (interleukin-2, interleukin-6, tumor necrosis factor-α, transforming growth factor-β) and ORP9 (oxysterol-binding protein-like 9) in monocyte-derived macrophages [96].
Calorie restriction
Calorie-restriction diets have been associated to increased life span in yeast, mice, and also in non-human primates [97]. The overall mechanism is not yet well understood, but it has been hypothesized that caloric and metabolic restriction causes lifespan extension by decreasing the target of the rapamycin (TOR) pathway, which is the most sensitive nutrient pathway in cells, which would lead to a decrease in protein synthesis and a promotion of autophagy [98]. It has also been found that calorie restriction induces increase of eNOS expression, which is responsible for mitochondrial biogenesis and in part for sirtuin 1 (SIRT1) expression [99]. In fact, another consequence of calorie restriction is the increased ratio [NAD+/NADH] that promotes the activation of NAD+-dependent protein deacetylase sirtuins [100], which are able to epigenetically regulate gene expression and are associated with increased lifespan. Moreover, SIRT1 was shown to prevent stress-induced senescence in ECs [101] and control angiogenic functions through the transcription factor FOXO1 and eNOS [102].
It has been reported that miR-217 expression increases in senescent human ECs in vitro (late passages HUVEC; high population doubling) and in human atherosclerotic plaques, promoting SIRT1 inhibition through a direct binding [103]. Also, miR-34 (already known for its tumor-suppressor actions) [104], was found to regulate senescence in ECs by targeting SIRT1 [105] (Fig. 4).
Fig. 4.
Schematic representation of cholesterol efflux. When intracellular cholesterol is low, miR-33 and its host gene SREBF2 are up-regulated and inhibit cholesterol efflux through ABCA1 (in human and mouse cells) and ABCG1 (in mouse cells)
Angiogenesis
The formation of new blood vessels from pre-existing ones is called angiogenesis. The process is active in the embryo, during the menstrual cycle, in the placenta, and during wound healing [106, 107]. Angiogenesis plays a role in adulthood pathologies, like autoimmune diseases, angioproliferative disease, degenerative and metabolic diseases and cardiovascular diseases associated to tissue ischemia. Moreover, a great number of neoplasia is able to induce the formation of new vessels needed for the growth of the tumor bulk and for its metastatic diffusion (cancer neoangiogenesis). From human to animal cancer models arises the evidence that angiogenesis can be switched on at different stages of tumor progression, according to the cancer type and its microenvironment. Human cancers arise in the absence of angiogenic activity and can lie dormant for months and years, before the rise of the angiogenic and then malignant phenotype. The angiogenic switch activation has been ascribed to the synthesis and releases of pro- angiogenic factors by the tumor itself [108–110]. The balance hypothesis for the angiogenic switch [110] states that the level of inhibitors and activators governs the passage from EC quiescence to angiogenesis. This balance is altered through an increased expression of activators or a reduced concentration of endogenous inhibitors.
ECs have a pivotal role in angiogenesis, thanks to the interaction between the receptors on their membrane and the pro-angiogenic factors. The triggered signal transduction pathways promote the proliferation, migration, and morphologic organization of ECs in tight connection with the surrounding environment. The formation of new vessels goes through several well-defined stages characterized by modifications of both the endothelium and the extracellular matrix. In the first stage, there is the destabilization of a pre-existing vessel with increase permeability and loss of intercellular junctions. In the second stage, migration and proliferation of ECs occur, helped by the release of proteolytic enzymes to degrade the extracellular matrix, thus favoring migration. Highly motile filopodia-extending endothelial “tip cells” stay at the forefront of a vessel sprout, guiding it towards the angiogenic stimulus. Behind them there are the proliferating “stalk cells” that elongate the vessel [111]. In the third stage, there is the differentiation of ECs: they stop proliferating, form primitive capillaries, and return quiescent. In this phase, the recruitment of supporting peri-endothelial cells, namely pericytes and smooth muscular cells, as well as the reorganization of intercellular interactions, occur [112].
Several pro-angiogenic factors have been identified, among which VEGF, TGFβ (transforming growth factor beta), IL-8 (interleukin 8), PDGF (platelet-derived growth factor). Other factors have instead an inhibitory role, as TSP-1, INFβ (interferon beta). Also, proteases control the availability of inhibitors or promoters: bFGF for instance can be sequestered in the extracellular matrix and be released after the matrix proteolytic degradation [113]; plasminogen, involved in the coagulation cascade, gives rise to plasmin and also angiostatin, a potent angiogenesis inhibitor [114]. Also, the signal of adhesion molecule is important: quiescent vessels express a set of integrins, whereas nascent capillaries express another set [115].
Tumor growth exceeding 1 mm3 demands glucose and O2, which are insufficiently provided by the already-present vasculature. Cancer cells secrete factors promoting angiogenesis, but the new vessels are characterized by an altered morphology and structure (functional shunting is present [116]) and are composed by tumor ECs, loosely attached pericytes and abnormal basement membrane.
The mRNA transcription profile characterizing the angiogenic switch has been analyzed by comparing human microvascular ECs treated with endostatin, a potent angiogenesis inhibitor, or VEGF and/or bFGF, potent angiogenesis activators [117]. Cluster analysis revealed more than 2,000 inversely regulated genes, among which “hub” genes, nodal genes in the angiogenesis pathway, were identified, namely PPARδ(peroxisome proliferative activated receptor δ), STAT3, MMP1, c-FLIP, ROBO/SLIT1.
A similar analysis has been made comparing the transcription profiles of four types of dormant tumors (breast carcinoma, glioblastoma, liposarcoma, and osteosarcoma) to corresponding fast-growing tumors. The latter showed a pronounced angiogenic phenotype, confirmed also at transcriptional levels, while dormant tumor expressed a high level of angiogenesis inhibitor, like TSP1 and also novel markers of tumor dormancy were identified, like Ephrin receptor A5 and H2BK [118].
MicroRNAs involved in inhibiting/promoting angiogenesis
Dicer ablation
The initial works on microRNA function were designed as loss-of-function experiments, where it was tried to decrease all the microRNAs either in organisms or in cultured cells. This was done by targeting Dicer and Drosha, the enzymes devoted to microRNA synthesis and maturation.
The first evidence about an involvement of microRNAs in mammalian angiogenesis came in 2005, when Yang et al. [119] reported about mice severely hypomorphic for the dicer allele (dicerex1/2), the terminal nuclease responsible for the maturation of microRNAs. Mice died between E12.5 and E14.5 and showed a primary vascular structure, testifying that vasculogenesis was not affected, whereas angiogenesis was totally impaired, despite high levels of VEGF and its receptors [119]. Other Dicer hypomorphic mice (generated by gene-trap) were viable, but female were infertile, due to the impaired vessel formation in the ovary and consequent corpus luteum insufficiency [120]. Dicer silencing, performed in ECs in vitro using a siRNA, impaired EC migration, proliferation and tube formation, both in vitro and in vivo (Matrigel plug assay) [121, 122]. The gene expression pattern of Dicer-depleted cells is somewhat surprising: several pro-angiogenic cytokines and growth factor receptors were up-regulated, as well as eNOS, Tie2, and AKT (but its phosphorylation was inhibited) [121, 122]. Conversely, TSP1 expression was up-regulated [122]. Drosha silencing induced minor phenotypic alterations and it was not effective in vivo [122]. It is important to say that in these works, Dicer and Drosha silencing brought to a small decrease in microRNAs (about 30%, with some microRNA more decreased than others) and this could have rearranged the balance between microRNAs and their targets. Moreover, the hypoxic environment that was formed could have promoted the expression of pro-angiogenic factors.
It is worth saying that Dicer-silenced HMEC, despite having high levels of VEGF, produced less basal pro-angiogenic ROS through NADPH oxidase when activated with VEGF, phorbol ester (phorbol myristate acetate, PMA) and TNFα, because of a reduction in the p47phox NADPH oxidase subunity [123]. This was independent from VEGF, suggesting that the Dicer-dependent miRNAs regulate the angiogenic response downstream of VEGF.
Antiangiogenic microRNAs
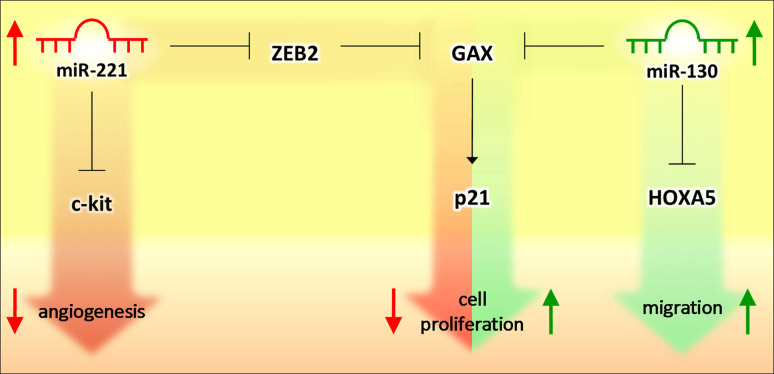
Among the first microRNAs with a reported role in angiogenesis were miR-221 and 222. They were shown to be highly expressed in HUVEC [121, 122] and, according to bioinformatic tools, they were predicted and then validated with GFP reporter assay to target the c-kit receptor [76]. This receptor binds the SCF ligand to promote in vitro tube formation, proliferation, and migration, which are then inhibited by miR-221-222. This observation was also made by Chen et al. [124] who reported another pathway regulated by miR-221 in ECs. They showed that miR-221 promotes GAX expression by directly down-regulating the repressor ZEB2. GAX expression, as miR-221, was high in quiescent vascular cells and in serum-depleted cells. GAX acted inhibiting EC proliferation by promoting p21 transcription [125] and angiogenesis through inhibition of NF-κB pathway [126]. Besides miR-221, miR-130a was demonstrated to down-regulate GAX and HOXA5 (an antiangiogenic homeobox gene) expression in ECs (Fig. 5) [127]. In this case, miR-130a is up-regulated by serum and pro-angiogenic factors and, once over-expressed in ECs, counteracted the antiangiogenic activity of GAX and HOXA5 that normally keep ECs in their resting state. MiR-222 (but not miR-221) was also down-regulated after pro-inflammatory stimuli (IL-3, bFGF) that promote inflammation-mediated neoangiogenesis and intraplaque vessel formation and it was found to directly target STAT5A, a transcription factor known to be activated by IL3 and bFGF signaling [128].
Fig. 5.
Schematic representation of opposite networks comprising miRNAs and TFs. When miR-221 is high (in quiescent ECs), it up-regulates the TF GAX by inhibiting the repressor ZEB2. Consequently, p21 is up-regulated and cell proliferation is inhibited. On the contrary, serum and pro-angiogenic factors increase miR-130 expression, leading to the down-regulation of its direct target, GAX, so that cell proliferation and angiogenesis are favored
MiR-15a/16-1 cluster is composed by miR-15a and miR-16 and it has been found lost in several lymphoid malignancies, like chronic lymphocytic leukemia (CLL) and multiple myeloma (MM), whereas a role as tumor suppressor has been ascribed to this cluster. Gatt et al. [129] reported about the knock down of the cluster for loss-of-function studies by using a lentiviral system to stably transduce MM cells with the miR-16 “sponge”, a sequence able to sequester and inhibit both miR-16 and its seed family member miR-15a. The resulting MM cells were more proliferating and, once injected in recipient mice, could increase tumor load and angiogenesis. The analysis of the gene expression profiling of cluster-depleted cells helped to find out targeted genes, among which appeared FGFR1, PI3KCa, MDM4, and VEGFa (Table 2). It is noteworthy that miR-16 was demonstrated to pair to the 3′UTR binding site and inhibit translation driven by the VEGF IRES-B, but not IRES-A [130]. Cellular IRES (internal ribosome entry sites) are normally found in genes encoding critical growth regulatory genes, like VEGF. VEGF is translated from two start codons, each of which is regulated by an independent IRES, leading to the production of different VEGF isoforms. The finding that microRNAs are able to control gene expression also through IRES is important because it adds another level of microRNA regulation on protein expression.
Table 2.
MicroRNAs involved in angiogenesis driven by pro/antiangiogenic factors or tumor context
| Pro/antiangiogenic stimulus | microRNA | Targeta | MicroRNA function | Cell type | References |
|---|---|---|---|---|---|
| SCF | ↓miR-221 | c-kit | Inhibition of angiogenesis | Human ECs (HUVEC) | [76] |
| Quiescent, serum-depleted cells | ↑miR-221 | ZEB2 | Inhibition of proliferation | Human ECs (HUVEC) | [124] |
| IL-3 bFGF | ↓miR-221 | STAT5A | Inhibition of angiogenesis | Human ECs (HUVEC) | [128] |
| Serum, VEGF, bFGF | ↑miR-130a | GAX | Regulation of angiogenesis | Human ECs (HUVEC) | [127] |
| HOXA5 | |||||
| Cancer | ↓miR-15a-16 | FGFR1 | Inhibition of proliferation and angiogenesis | Multiple myeloma cell line | [129] |
| PI3KCa | |||||
| MDM4 | |||||
| VEGFa | |||||
| Hind-limb ischemia model, acute myocardial infarction | ↑miR-92 | ITGA5 | Inhibition of angiogenesis | Human ECs (HUVEC) | [131] |
| VEGF, bFGF cancer | ↑miR-132 | P120RasGAP | Promotion of angiogenesis | Human ECs (HUVEC), human embryonic stem cell vasculogenesis model | [132] |
| Cancer | ↑miR-296 | HGS | Promotion of angiogenesis | Human ECs (HBMVEC) | [133] |
| Cancer | ↑miR-93 | Integrin-p8 | Promotion of angiogenesis | Glioma cell line (U87) + ECs (Ypen) | [134] |
| Cancer | ↑miR-378 |
SuFu Fus-1 |
Promotion of proliferation and angiogenesis | Glioma cell line (U87) | [135] |
| K-Ras, Myc over-expressed | miR-17-92 | TSP1 | Promotion of angiogenesis | Mouse colonocytes | [138] |
| ↑miR-18 | SMAD4 | [139] | |||
| ↑miR-19 | CTGF | [138] | |||
| ↑miR-20 | TGFbR2 | [139] | |||
| TNF-a VEGF, bFGF | ↑miR-126 | VCAM1 | Leukocyte adhesion | Human ECs (HUVEC) | [141] |
| PIK3R2 (p85) | Promotion of angiogenesis | Human ECs (HUVEC) | [142, 143] | ||
| SPRED1 | Promotion of angiogenesis | Human ECs (HUVEC, HAEC) | [142–144] | ||
| PAK1 | Regulation of vascular integrity | Human ECs (HUVEC) | [145] | ||
| Cancer | IRS1 | Inhibition of proliferation (G0/G1 arrest) | Human breast cancer (MCF7), HEK293 | [149] | |
| VEGFA | Inhibition of proliferation (G0/G1 arrest) | Lung cancer cell lines | [150] | ||
| Human breast cancer (MCF7) | [151] | ||||
| CRK | Inhibition of adhesion, invasion, proliferation | Lung cancer cell lines | [152] | ||
| Gastric cancer cell line | [153] | ||||
| EGFL7 | Inhibition of proliferation | Lung cancer cell line | [154] |
aTarget validated by gene reporter assay
MiR-92a, a component of miR-17-92 cluster, contrary to the other members, conferred antiangiogenic properties to HUVEC in vitro and in vivo (Matrigel plug assay) and its over-expression induced defects in intersegmental vessel formation in zebrafish [131]. MiR-92a expression was increased in a hind-limb ischemia model and after induction of acute myocardial infarction in mice. Systemic injection of antagomir-92a reduced necrosis and improved perfusion and recovery of ischemic limb and enhanced recovery after myocardial infarction. eNOS expression and integrin subunits alfa5 (ITGA5) were inversely related to miR-92 expression and ITGA5 was proposed as a direct target of miR-92a [131].
Pro-angiogenic microRNAs
MiR-132 is a microRNA able to turn on the angiogenic switch. Anand et al. [132] identified miR-132 in two models of activated ECs, namely HUVEC treated with either VEGF or bFGF, and in a human embryonic stem cell vasculogenesis model. An up-regulation of miR-132 has been found in HUVEC treated with conditioned media from breast and pancreatic tumor cell lines, in the endothelium of human breast tumors, in hemangioma and in the endothelium of murine pancreatic tumors. The miR-132 expression is inversely correlated with the one of its validated target, p120RasGAP (encoded by Rasa1 gene), a negative regulator of Ras. An anti-miR-132 treatment inhibited Matrigel plug angiogenesis, but it was not effective in mice with an inducible deletion for Rasa1. Importantly, the authors exploited an innovative technology based on integrin ανβ3-targeted nanoparticles, to selectively deliver anti-miR-132 into tumor endothelium. The systemic administration of the nanoparticles was able to increase p120RasGAP in tumor vasculature and significantly decrease tumor burden and angiogenesis of an orthotopic xenograft mouse model of human breast carcinoma [132]. Another microRNA, miR-296 was demonstrated to be up-regulated in HBMVEC (human brain microvascular ECs) cocultured with a tumor cell line (human U87 glioma cells) or treated with medium from U87 cells or just with VEGF or bFGF and also in tumor blood vessels from highly angiogenic glioma specimens from human patients [133]. MiR-296 was shown to prevent the degradation of the growth factor receptors VEGFR2 and PDGFRβ by decreasing HGS (hepatocyte growth factor-regulated tyrosine kinase substrate), which is involved in the regulation of the levels of these receptors. Also in this case, as for miR-132, miR-296 expression promoted angiogenesis, while its inhibition by cholesterol-conjugated antagomir-296 antisense oligonucleotide reduced tumor neovascularization in mice.
Conversely, miR-93 seems to play a slightly different role in the angiogenic switch. When it was over-expressed in U87 it did not promote cell proliferation; nevertheless U87-derived tumor over-expressing miR-93 were bigger and displayed more blood vessels. MiR-93 seems to have a role in promoting tumor cell survival through the inhibition of integrin-β8. The overall mechanisms led to an increase in angiogenesis and a better tumor nutrition supply. These findings were further supported by U87-EC coculture experiments [134].
Another microRNA that promotes tumor growth, cell survival and angiogenesis is miR-378 [135]. It was found over-expressed in several tumors and it promoted the formation of larger vessels. It acts by targeting the tumor suppressor SuFu and Fus-1.
In the human genome, the miR-17-92 cluster encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1), which are tightly grouped within an 800-bp region of human chromosome 13. These microRNAs have been often found to play a role in cancer and development [136] and in postnatal angiogenesis, where their induction in Dicer-silenced mice promoted TSP1 down-regulation and consequent promotion of cell proliferation and angiogenesis [137]. The cluster has been also involved in augmentation of tumor angiogenesis in a model of p53 null, K-Ras and Myc over-expressing colonocytes engrafted into syngenic mice. The cluster over-expression, which recapitulated Myc-induced phenotype (in the p53 null and Ras over-expressing context), did not promote cell proliferation in vitro, nor in vivo, until the threshold tumor size to promote the angiogenic switch was reached. The microarray analysis did not reveal the induction of pro-angiogenic factors, but antiangiogenic factors, like TSP1 and proteins with TSP1 repeats, as CTGF and clusterin were in the list of Myc-downregulated genes. TSP1 and CTGF have been validated as miR-17-92 targets (miR-18 is primarily responsible for TSP1 and miR-19 for CTGF down-regulation) [138], while clusterin is indirectly regulated by miR-17-92, which acts on clusterin-activating TGFβ signaling; specifically, miR-20a down-regulates TGFβ receptor type 2 and miR-18 down-regulates Smad 4 expression [139].
MiR-126 is among the most expressed microRNA in ECs. It is encoded by an intron of Egfl7 gene and for that miR-126 and Egfl7 are cotranscribed [140]. This microRNA was shown to regulate the TNF-α-stimulated expression of VCAM1 (vascular cell adhesion molecule 1) and therefore to mediate leukocyte adhesion. The transfection of an antisense miR-126 promoted VCAM1 expression and leukocyte binding to HUVEC, whether the over-expression of miR-126 decreased the protein expression and leukocytes adhesion [141]. MiR-126 was also found to have a role in vascular integrity and angiogenesis. Fish et al. [142] reported that HUVEC transfected (nucleoporated) with a morpholino antisense to miR-126 showed reduced migration and reduced proliferation, due to an increase in apoptosis, in response to VEGF and bFGF. The knocking down of miR-126 in fertilized zebrafish eggs produced no differences in morphology or vascular patterning, but the zebrafish transgenic line Tg (gata1: dsRed), which expresses dsRed in blood cells, revealed severe cranial hemorrhages. The proposed mechanism of action is that miR-126 controls the VEGF downstream pathways in two ways: by inhibiting PIK3R2 (p85β) expression [142, 143], which negatively regulates the activity of PI3 kinase, and by inhibiting SPRED1 (Sprouty-related EVH1 domain-containing protein 1) [142–144], which represses the activation on the MAP kinase. Moreover, p21-activated kinase1 (pak1), whose over-expression causes cranial hemorrhage, was shown to be a target of both miR-126a (named miR-126 in previous literature) and miR-126b (which differs from miR-126a by just one nucleotide in the mature sequence) [145]. MiR-126-deficient mice were created by homologous recombination (miR-126−/−) [144] and by Cre-lox system, producing miR-126Δ/Δ mice [143]. These types of transgenic mice presented, respectively, about 40 and 50% of embryonic lethality due to severe systemic edema, multifocal hemorrhages and leaky vessels, while the surviving neonates exhibited delayed postnatal retinal angiogenesis. Surprisingly, these phenotypes were previously reported for Egfl7 gene-trap and lacZ knock-in mice [146], but Egfl7Δ/Δ mice, wherein miR-126 was not eliminated, were phenotypically normal [143]. This suggests that the altered phenotypes previously attributed to Egfl7 were actually due to miR-126 deletion. MiR-126 is also involved in vascular remodeling guided by blood flow in the aortic arch ECs. MiR-126 expression was shown to be promoted by the flow-induced transcription factor klf2, therefore linking hemodynamic forces to VEGF signaling [147].
Besides regulating angiogenesis and vascular integrity, miR-126 was found under-expressed in several microRNA cancer profiles, like breast and lung cancers (for a review see [148]). These findings addressed studies on miR-126 as a cell cycle regulator and revealed that miR-126 inhibits proliferation of cancer cells by promoting an arrest of cells at G0/G1. This seems to be reached in several ways. MiR-126, once over-expressed, appears able to target IRS1 protein in MCF7 and HEK293 [149]; to inhibit VEGFA in lung cancer cell lines [150] and breast cancer lines [151]; to inhibit CRK in lung cancer cell lines [152] and in gastric cancer [153]; to inhibit EGFL7 in lung cancer cell lines [154] (Table 2). Nevertheless, other studies revealed a malignant role for miR-126 in promoting tumor angiogenesis. Donnem et al. [155] found negative prognosis for non-small cell lung carcinoma tumors wherein miR-126 was over-expressed and saw a weak but significant correlation between miR-126 and VEGF expressions. When stratified, the prognostic impact was highly associated with squamous cell carcinoma patients or those patients with nodal metastasis (compared to adenocarcinoma and large-cell carcinoma patients). Another study reported that miR-126 is highly over-expressed in lung metastases, compared with primary lung tumors [156]. This would suggest that the principal pathways regulated by miR-126 would lead to promotion of angiogenesis during development, and possibly in metastatic malignant tumors, while the high level of miR-126 in ECs would help to maintain a resting state by controlling cell cycle. More studies are needed to properly assign a role for miR-126 in different contexts.
Secretory microRNAs
There are new emerging concepts about a systemic signaling based on microRNAs. The discovery of microRNAs in biological fluids [157] is opening new frontiers on the study of microRNAs. Zampetaki et al. [158] isolated apoptotic bodies and microparticles from plasma of patients with type 2 diabetes and control subjects. Once having extracted and analyzed microRNA expression, they found lower plasma levels of miR-20b, miR-21, miR-24, miR-15a, miR-126, miR-191, miR-197, miR-223, miR-320, and miR-486 in diabetic patients. In particular, miR-126, which is highly expressed in ECs, was consistently under-represented also in apoptotic bodies shed from ECs treated with high glucose. Since apoptotic bodies and microparticles can be transferred to other cell types, the authors suggested that low miR-126 plasma levels may reduce the delivery of miR-126 to monocytes and contribute to endothelial dysfunction.
Human ECs in vitro and murine endothelium in vivo were in turn demonstrated to internalize microvesicles (exosomes) secreted by monocytes (THP-1 cell line) or isolated from plasma of atherosclerosis patients. These microvesicles contained high levels of an exogenous microRNA, miR-150, which was able to trigger EC migration by reducing endothelial c-MYB protein levels [159].
Currently, microRNAs are no longer seen just as endogenous regulators of gene expression, but also as reliable biomarkers for pathologies and secreted factors able to mediate cell-to-cell communication.
Acknowledgments
This work was supported by Associazione Italiana per la Ricerca sul Cancro, AIRC (Project number 4753) and by Istituto Superiore di Sanità, ISS (Project number 527/A/3A/4). We thank Mrs. Penelope Ivall Garrud for careful reading of the manuscript and Serena Lucotti for drawing most of the figures.
Abbreviations
- ECs
Endothelial cells
- VEGF
Vascular endothelial growth factor
- bFGF
Basic fibroblast growth factor
- ROS
Reactive oxygen species
- NO
Nitric oxide
- eNOS
Endothelial nitric oxide synthase
- TFs
Transcription factors
References
- 1.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 4.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433(7027):769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 5.Orom UA, Lund AH. Experimental identification of microRNA targets. Gene. 2010;451(1–2):1–5. doi: 10.1016/j.gene.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Vinther J, Hedegaard MM, Gardner PP, Andersen JS, Arctander P. Identification of miRNA targets with stable isotope labeling by amino acids in cell culture. Nucleic Acids Res. 2006;34(16):e107. doi: 10.1093/nar/gkl590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karginov FV, Conaco C, Xuan Z, Schmidt BH, Parker JS, Mandel G, Hannon GJ. A biochemical approach to identifying microRNA targets. Proc Natl Acad Sci USA. 2007;104(49):19291–19296. doi: 10.1073/pnas.0709971104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orom UA, Lund AH. Isolation of microRNA targets using biotinylated synthetic microRNAs. Methods San Diego Calif. 2007;43(2):162–165. doi: 10.1016/j.ymeth.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Gallagher IJ, Scheele C, Keller P, Nielsen AR, Remenyi J, Fischer CP, Roder K, Babraj J, Wahlestedt C, Hutvagner G, Pedersen BK, Timmons JA. Integration of microRNA changes in vivo identifies novel molecular features of muscle insulin resistance in type 2 diabetes. Genome Med. 2010;2(2):9. doi: 10.1186/gm130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitto L, Ripoli A, Cremisi F, Simili M, Rainaldi G. microRNA(interference) networks are embedded in the gene regulatory networks. Cell cycle Georgetown Tex. 2008;7(16):2458–2461. doi: 10.4161/cc.7.16.6455. [DOI] [PubMed] [Google Scholar]
- 12.Tuccoli A, Poliseno L, Rainaldi G. miRNAs regulate miRNAs: coordinated transcriptional and post-transcriptional regulation. Cell cycle (Georgetown. Tex) 2006;5(21):2473–2476. doi: 10.4161/cc.5.21.3422. [DOI] [PubMed] [Google Scholar]
- 13.Zhou Y, Ferguson J, Chang JT, Kluger Y. Inter- and intra-combinatorial regulation by transcription factors and microRNAs. BMC Genom. 2007;8:396. doi: 10.1186/1471-2164-8-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CY, Chen ST, Fuh CS, Juan HF, Huang HC. Coregulation of transcription factors and microRNAs in human transcriptional regulatory network. BMC Bioinform. 2011;12(Suppl 1):41. doi: 10.1186/1471-2105-12-S1-S41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26(5):753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coller HA, Forman JJ, Legesse-Miller A. “Myc’ed messages”: myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3(8):e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rizzo M, Mariani L, Pitto L, Rainaldi G, Simili M. miR-20a and miR-290, multi-faceted players with a role in tumourigenesis and senescence. J Cell Mol Med. 2010;14(11):2633–2640. doi: 10.1111/j.1582-4934.2010.01173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS ONE. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marasa BS, Srikantan S, Masuda K, Abdelmohsen K, Kuwano Y, Yang X, Martindale JL, Rinker-Schaeffer CW, Gorospe M. Increased MKK4 abundance with replicative senescence is linked to the joint reduction of multiple microRNAs. Sci Signal. 2009;2(94):ra69. doi: 10.1126/scisignal.2000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mavrakis KJ, Van Der Meulen J, Wolfe AL, Liu X, Mets E, Taghon T, Khan AA, Setti M, Rondou P, Vandenberghe P, Delabesse E, Benoit Y, Socci NB, Leslie CS, Van Vlierberghe P, Speleman F, Wendel HG (2011) A cooperative microRNA-tumor suppressor gene network in acute T cell lymphoblastic leukemia (T-ALL). Nat Genet. doi:10.1038/ng.858 [DOI] [PMC free article] [PubMed]
- 21.Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA, McEver RP, Pober JS, Wick TM, Konkle BA, Schwartz BS, Barnathan ES, McCrae KR, Hug BA, Schmidt AM, Stern DM. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91(10):3527–3561. [PubMed] [Google Scholar]
- 22.Sumpio BE, Riley JT, Dardik A. Cells in focus: endothelial cell. Int J Biochem Cell Biol. 2002;34(12):1508–1512. doi: 10.1016/S1357-2725(02)00075-4. [DOI] [PubMed] [Google Scholar]
- 23.Hobson B, Denekamp J. Endothelial proliferation in tumours and normal tissues: continuous labelling studies. Br J Cancer. 1984;49(4):405–413. doi: 10.1038/bjc.1984.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tabit CE, Chung WB, Hamburg NM, Vita JA. Endothelial dysfunction in diabetes mellitus: molecular mechanisms and clinical implications. Rev Endocr Metab Disord. 2010;11(1):61–74. doi: 10.1007/s11154-010-9134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Ming XF. Recent advances in understanding endothelial dysfunction in atherosclerosis. Clin Med Res. 2006;4(1):53–65. doi: 10.3121/cmr.4.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Esper RJ, Nordaby RA, Vilarino JO, Paragano A, Cacharron JL, Machado RA. Endothelial dysfunction: a comprehensive appraisal. Cardiovasc Diabetol. 2006;5:4. doi: 10.1186/1475-2840-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson MR, Lai X, Witzmann FA, Yoder MC. Venous and arterial endothelial proteomics: mining for markers and mechanisms of endothelial diversity. Expert Rev Proteomics. 2010;7(6):823–831. doi: 10.1586/epr.10.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.dela Paz NG, D’Amore PA. Arterial versus venous endothelial cells. Cell Tissue Res. 2009;335(1):5–16. doi: 10.1007/s00441-008-0706-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peters K, Kamp G, Berz A, Unger RE, Barth S, Salamon A, Rychly J, Kirkpatrick CJ. Changes in human endothelial cell energy metabolic capacities during in vitro cultivation. The role of “aerobic glycolysis” and proliferation. Cell Physiol Biochem. 2009;4(5):483–492. doi: 10.1159/000257490. [DOI] [PubMed] [Google Scholar]
- 30.Dobrina A, Rossi F. Metabolic properties of freshly isolated bovine endothelial cells. Biochim Biophys Acta. 1983;762(2):295–301. doi: 10.1016/0167-4889(83)90084-8. [DOI] [PubMed] [Google Scholar]
- 31.Quintero M, Colombo SL, Godfrey A, Moncada S. Mitochondria as signaling organelles in the vascular endothelium. Proc Natl Acad Sci USA. 2006;103(14):5379–5384. doi: 10.1073/pnas.0601026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krutzfeldt A, Spahr R, Mertens S, Siegmund B, Piper HM. Metabolism of exogenous substrates by coronary endothelial cells in culture. J Mol Cell Cardiol. 1990;22(12):1393–1404. doi: 10.1016/0022-2828(90)90984-A. [DOI] [PubMed] [Google Scholar]
- 33.Nisoli E, Carruba MO. Nitric oxide and mitochondrial biogenesis. J Cell Sci. 2006;119(Pt 14):2855–2862. doi: 10.1242/jcs.03062. [DOI] [PubMed] [Google Scholar]
- 34.Hagen T, Taylor CT, Lam F, Moncada S. Redistribution of intracellular oxygen in hypoxia by nitric oxide: effect on HIF1alpha. Science. 2003;302(5652):1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 35.Fraisl P, Mazzone M, Schmidt T, Carmeliet P. Regulation of angiogenesis by oxygen and metabolism. Dev Cell. 2009;16(2):167–179. doi: 10.1016/j.devcel.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 36.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27(5):1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10(4):273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine-kinase ligand Ephrin-A3. J Biol Chem. 2008;283(23):15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29(30):4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 40.Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets Ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286(3):2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang DX, Gutterman DD. Mitochondrial reactive oxygen species-mediated signaling in endothelial cells. Am J Physiol Heart Circ Physiol. 2007;292(5):H2023–H2031. doi: 10.1152/ajpheart.01283.2006. [DOI] [PubMed] [Google Scholar]
- 42.Ding Y, Vaziri ND, Coulson R, Kamanna VS, Roh DD. Effects of simulated hyperglycemia, insulin, and glucagon on endothelial nitric oxide synthase expression. Am J Physiol Endocrinol Metab. 2000;279(1):E11–E17. doi: 10.1152/ajpendo.2000.279.1.E11. [DOI] [PubMed] [Google Scholar]
- 43.Du X, Edelstein D, Obici S, Higham N, Zou MH, Brownlee M. Insulin resistance reduces arterial prostacyclin synthase and eNOS activities by increasing endothelial fatty acid oxidation. J Clin Investig. 2006;116(4):1071–1080. doi: 10.1172/JCI23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim JA, Montagnani M, Koh KK, Quon MJ. Reciprocal relationships between insulin resistance and endothelial dysfunction: molecular and pathophysiological mechanisms. Circulation. 2006;113(15):1888–1904. doi: 10.1161/CIRCULATIONAHA.105.563213. [DOI] [PubMed] [Google Scholar]
- 45.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54(6):1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 46.Nishikawa T, Edelstein D, Du XL, Yamagishi S, Matsumura T, Kaneda Y, Yorek MA, Beebe D, Oates PJ, Hammes HP, Giardino I, Brownlee M. Normalizing mitochondrial superoxide production blocks three pathways of hyperglycaemic damage. Nature. 2000;404(6779):787–790. doi: 10.1038/35008121. [DOI] [PubMed] [Google Scholar]
- 47.Quagliaro L, Piconi L, Assaloni R, Da Ros R, Szabo C, Ceriello A. Primary role of superoxide anion generation in the cascade of events leading to endothelial dysfunction and damage in high glucose treated HUVEC. Nutr Metab Cardiovasc Dis. 2007;17(4):257–267. doi: 10.1016/j.numecd.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 48.Kamal K, Du W, Mills I, Sumpio BE. Antiproliferative effect of elevated glucose in human microvascular endothelial cells. J Cell Biochem. 1998;71(4):491–501. doi: 10.1002/(SICI)1097-4644(19981215)71:4<491::AID-JCB4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 49.Varma S, Lal BK, Zheng R, Breslin JW, Saito S, Pappas PJ, Hobson RW, 2nd, Duran WN. Hyperglycemia alters PI3 k and Akt signaling and leads to endothelial cell proliferative dysfunction. Am J Physiol. 2005;289(4):H1744–H1751. doi: 10.1152/ajpheart.01088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piconi L, Quagliaro L, Assaloni R, Da Ros R, Maier A, Zuodar G, Ceriello A. Constant and intermittent high glucose enhances endothelial cell apoptosis through mitochondrial superoxide overproduction. Diabetes/Metab Res Rev. 2006;22(3):198–203. doi: 10.1002/dmrr.613. [DOI] [PubMed] [Google Scholar]
- 51.Yu P, Yu DM, Qi JC, Wang J, Zhang QM, Zhang JY, Tang YZ, Xing QL, Li MZ. High d-glucose alters PI3 K and Akt signaling and leads to endothelial cell migration, proliferation and angiogenesis dysfunction. Zhonghua yi xue za zhi. 2006;86(48):3425–3430. [PubMed] [Google Scholar]
- 52.Hamuro M, Polan J, Natarajan M, Mohan S. High glucose induced nuclear factor kappa B mediated inhibition of endothelial cell migration. Atherosclerosis. 2002;162(2):277–287. doi: 10.1016/S0021-9150(01)00719-5. [DOI] [PubMed] [Google Scholar]
- 53.Ho FM, Lin WW, Chen BC, Chao CM, Yang CR, Lin LY, Lai CC, Liu SH, Liau CS. High glucose-induced apoptosis in human vascular endothelial cells is mediated through NF-kappaB and c-Jun NH2-terminal kinase pathway and prevented by PI3 K/Akt/eNOS pathway. Cell Signal. 2006;18(3):391–399. doi: 10.1016/j.cellsig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Sheu ML, Ho FM, Yang RS, Chao KF, Lin WW, Lin-Shiau SY, Liu SH. High glucose induces human endothelial cell apoptosis through a phosphoinositide 3-kinase-regulated cyclooxygenase-2 pathway. Arterioscler Thromb Vasc Biol. 2005;25(3):539–545. doi: 10.1161/01.ATV.0000155462.24263.e4. [DOI] [PubMed] [Google Scholar]
- 55.Guo X, Chen LW, Liu WL, Guo ZG. High glucose inhibits expression of inducible and constitutive nitric oxide synthase in bovine aortic endothelial cells. Acta Pharmacol Sin. 2000;21(4):325–328. [PubMed] [Google Scholar]
- 56.Salt IP, Morrow VA, Brandie FM, Connell JM, Petrie JR. High glucose inhibits insulin-stimulated nitric oxide production without reducing endothelial nitric-oxide synthase Ser1177 phosphorylation in human aortic endothelial cells. J Biol Chem. 2003;278(21):18791–18797. doi: 10.1074/jbc.M210618200. [DOI] [PubMed] [Google Scholar]
- 57.Schnyder B, Pittet M, Durand J, Schnyder-Candrian S. Rapid effects of glucose on the insulin signaling of endothelial NO generation and epithelial Na transport. Am J Physiol Endocrinol Metab. 2002;282(1):E87–E94. doi: 10.1152/ajpendo.00050.2001. [DOI] [PubMed] [Google Scholar]
- 58.Srinivasan S, Hatley ME, Bolick DT, Palmer LA, Edelstein D, Brownlee M, Hedrick CC. Hyperglycaemia-induced superoxide production decreases eNOS expression via AP-1 activation in aortic endothelial cells. Diabetologia. 2004;47(10):1727–1734. doi: 10.1007/s00125-004-1525-1. [DOI] [PubMed] [Google Scholar]
- 59.Altannavch TS, Roubalova K, Kucera P, Andel M. Effect of high glucose concentrations on expression of ELAM-1, VCAM-1 and ICAM-1 in HUVEC with and without cytokine activation. Physiol Res. 2004;53(1):77–82. [PubMed] [Google Scholar]
- 60.Haubner F, Lehle K, Munzel D, Schmid C, Birnbaum DE, Preuner JG. Hyperglycemia increases the levels of vascular cellular adhesion molecule-1 and monocyte-chemoattractant-protein-1 in the diabetic endothelial cell. Biochem Biophys Res Commun. 2007;360(3):560–565. doi: 10.1016/j.bbrc.2007.06.044. [DOI] [PubMed] [Google Scholar]
- 61.Kado S, Wakatsuki T, Yamamoto M, Nagata N. Expression of intercellular adhesion molecule-1 induced by high glucose concentrations in human aortic endothelial cells. Life Sci. 2001;68(7):727–737. doi: 10.1016/S0024-3205(00)00968-1. [DOI] [PubMed] [Google Scholar]
- 62.Quagliaro L, Piconi L, Assaloni R, Da Ros R, Maier A, Zuodar G, Ceriello A. Intermittent high glucose enhances ICAM-1, VCAM-1 and E-selectin expression in human umbilical vein endothelial cells in culture: the distinct role of protein kinase C and mitochondrial superoxide production. Atherosclerosis. 2005;183(2):259–267. doi: 10.1016/j.atherosclerosis.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 63.Gosmanov AR, Stentz FB, Kitabchi AE. De novo emergence of insulin-stimulated glucose uptake in human aortic endothelial cells incubated with high glucose. Am J Physiol Endocrinol Metab. 2006;290(3):E516–E522. doi: 10.1152/ajpendo.00326.2005. [DOI] [PubMed] [Google Scholar]
- 64.Mann GE, Yudilevich DL, Sobrevia L. Regulation of amino acid and glucose transporters in endothelial and smooth muscle cells. Physiol Rev. 2003;83(1):183–252. doi: 10.1152/physrev.00022.2002. [DOI] [PubMed] [Google Scholar]
- 65.Wang XH, Qian RZ, Zhang W, Chen SF, Jin HM, Hu RM. MicroRNA-320 expression in myocardial microvascular endothelial cells and its relationship with insulin-like growth factor-1 in type 2 diabetic rats. Clin Exp Pharmacol Physiol. 2009;36(2):181–188. doi: 10.1111/j.1440-1681.2008.05057.x. [DOI] [PubMed] [Google Scholar]
- 66.Schaar DG, Medina DJ, Moore DF, Strair RK, Ting Y. miR-320 targets transferrin receptor 1 (CD71) and inhibits cell proliferation. Exp Hematol. 2009;37(2):245–255. doi: 10.1016/j.exphem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 67.Ling HY, Ou HS, Feng SD, Zhang XY, Tuo QH, Chen LX, Zhu BY, Gao ZP, Tang CK, Yin WD, Zhang L, Liao DF. Changes in microRNA profile and effects of miR-320 in insulin-resistant 3T3-L1 adipocytes. Clin Exp Pharmacol Physiol. 2009;36(9):e32–e39. doi: 10.1111/j.1440-1681.2009.05207.x. [DOI] [PubMed] [Google Scholar]
- 68.Ren XP, Wu J, Wang X, Sartor MA, Qian J, Jones K, Nicolaou P, Pritchard TJ, Fan GC. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation. 2009;119(17):2357–2366. doi: 10.1161/CIRCULATIONAHA.108.814145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Duan H, Jiang Y, Zhang H, Wu Y. MiR-320 and miR-494 affect cell cycles of primary murine bronchial epithelial cells exposed to benzo[a]pyrene. Toxicol In Vitro. 2009;24(3):928–935. doi: 10.1016/j.tiv.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 70.Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, Li L, Huang DD, Ding J, Shen F, Zhou WP, Wu MC, Wang HY. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009;50(2):358–369. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 71.Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjot L, Wiuf C, Sorensen FJ, Kruhoffer M, Laurberg S, Kauppinen S, Orntoft TF, Andersen CL. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68(15):6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 72.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA (New York NY) 2008;14(11):2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gao W, Shen H, Liu L, Xu J, Shu Y (2010) MiR-21 overexpression in human primary squamous cell lung carcinoma is associated with poor patient prognosis. J Cancer Res Clin Oncol. doi:10.1007/s00432-010-0918-4 [DOI] [PMC free article] [PubMed]
- 74.Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, Goggins MG, Mendell JT, Maitra A. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9(3):293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Y, Song YH, Li F, Yang T, Lu YW, Geng YJ. MicroRNA-221 regulates high glucose-induced endothelial dysfunction. Biochem Biophys Res Commun. 2009;381(1):81–83. doi: 10.1016/j.bbrc.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, Mercatanti A, Hammond S, Rainaldi G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068–3071. doi: 10.1182/blood-2006-01-012369. [DOI] [PubMed] [Google Scholar]
- 77.Togliatto G, Trombetta A, Dentelli P, Rosso A, Brizzi MF. MIR221/MIR222-driven post-transcriptional regulation of P27KIP1 and P57KIP2 is crucial for high-glucose- and AGE-mediated vascular cell damage. Diabetologia. 2011;54(7):1930–1940. doi: 10.1007/s00125-011-2125-5. [DOI] [PubMed] [Google Scholar]
- 78.Herrera BM, Lockstone HE, Taylor JM, Wills QF, Kaisaki PJ, Barrett A, Camps C, Fernandez C, Ragoussis J, Gauguier D, McCarthy MI, Lindgren CM. MicroRNA-125a is over-expressed in insulin target tissues in a spontaneous rat model of type 2 diabetes. BMC Med Genomics. 2009;2:54. doi: 10.1186/1755-8794-2-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Caporali A, Meloni M, Vollenkle C, Bonci D, Sala-Newby GB, Addis R, Spinetti G, Losa S, Masson R, Baker AH, Agami R, le Sage C, Condorelli G, Madeddu P, Martelli F, Emanueli C (2011) Deregulation of microRNA-503 contributes to diabetes mellitus-induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. doi:10.1161/CIRCULATIONAHA.110.952325 [DOI] [PubMed]
- 80.Jiang Q, Feng MG, Mo YY. Systematic validation of predicted microRNAs for cyclin D1. BMC Cancer. 2009;9:194. doi: 10.1186/1471-2407-9-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sarkar S, Dey BK, Dutta A. MiR-322/424 and -503 are induced during muscle differentiation and promote cell cycle quiescence and differentiation by down-regulation of Cdc25A. Mol Biol Cell. 2010;21(13):2138–2149. doi: 10.1091/mbc.E10-01-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vogel RA, Corretti MC, Gellman J. Cholesterol, cholesterol lowering, and endothelial function. Prog Cardiovasc Dis. 1998;41(2):117–136. doi: 10.1016/S0033-0620(98)80008-X. [DOI] [PubMed] [Google Scholar]
- 83.Hennig B, Toborek M, McClain CJ. High-energy diets, fatty acids and endothelial cell function: implications for atherosclerosis. J Am Coll Nutr. 2001;20(2 Suppl):97–105. doi: 10.1080/07315724.2001.10719021. [DOI] [PubMed] [Google Scholar]
- 84.Zilversmit DB. Atherogenic nature of triglycerides, post-prandial lipidemia, and triglyceride-rich remnant lipoproteins. Clin Chem. 1995;41(1):153–158. [PubMed] [Google Scholar]
- 85.Wang L, Gill R, Pedersen TL, Higgins LJ, Newman JW, Rutledge JC. Triglyceride-rich lipoprotein lipolysis releases neutral and oxidized FFAs that induce endothelial cell inflammation. J Lipid Res. 2009;50(2):204–213. doi: 10.1194/jlr.M700505-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kalinowski L, Malinski T. Endothelial NADH/NADPH-dependent enzymatic sources of superoxide production: relationship to endothelial dysfunction. Acta Biochim Pol. 2004;51(2):459–469. [PubMed] [Google Scholar]
- 87.Kawashima S, Yokoyama M. Dysfunction of endothelial nitric oxide synthase and atherosclerosis. Arterioscler Thromb Vasc Biol. 2004;24(6):998–1005. doi: 10.1161/01.ATV.0000125114.88079.96. [DOI] [PubMed] [Google Scholar]
- 88.Witztum JL. The role of oxidized LDL in atherosclerosis. Adv Exp Med Biol. 1991;285:353–365. doi: 10.1007/978-1-4684-5904-3_43. [DOI] [PubMed] [Google Scholar]
- 89.Liao JK, Shin WS, Lee WY, Clark SL. Oxidized low-density lipoprotein decreases the expression of endothelial nitric oxide synthase. J Biol Chem. 1995;270(1):319–324. doi: 10.1074/jbc.270.1.319. [DOI] [PubMed] [Google Scholar]
- 90.Seibold S, Schurle D, Heinloth A, Wolf G, Wagner M, Galle J. Oxidized LDL induces proliferation and hypertrophy in human umbilical vein endothelial cells via regulation of p27Kip1 expression: role of RhoA. J Am Soc Nephrol. 2004;15(12):3026–3034. doi: 10.1097/01.ASN.0000146425.58046.6A. [DOI] [PubMed] [Google Scholar]
- 91.Rayner KJ, Suarez Y, Davalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernandez-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science (New York, NY) 2010;328(5985):1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Naar AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science (New York NY) 2010;328(5985):1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zeng L, Liao H, Liu Y, Lee TS, Zhu M, Wang X, Stemerman MB, Zhu Y, Shyy JY. Sterol-responsive element-binding protein (SREBP) 2 down-regulates ATP-binding cassette transporter A1 in vascular endothelial cells: a novel role of SREBP in regulating cholesterol metabolism. J Biol Chem. 2004;279(47):48801–48807. doi: 10.1074/jbc.M407817200. [DOI] [PubMed] [Google Scholar]
- 94.Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci USA. 2010;107(40):17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Li D, Yang P, Xiong Q, Song X, Yang X, Liu L, Yuan W, Rui YC (2010) MicroRNA-125a/b-5p inhibits endothelin-1 expression in vascular endothelial cells. J Hypertens. doi:10.1097/HJH.0b013e32833a4922 [DOI] [PubMed]
- 96.Chen T, Huang Z, Wang L, Wang Y, Wu F, Meng S, Wang C. MicroRNA-125a–5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc Res. 2009;83(1):131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 97.Smith DL, Jr, Nagy TR, Allison DB. Calorie restriction: what recent results suggest for the future of ageing research. Eur J Clin Invest. 2010;40(5):440–450. doi: 10.1111/j.1365-2362.2010.02276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blagosklonny MV. Calorie restriction: decelerating mTOR-driven aging from cells to organisms (including humans) Cell Cycle. 2010;9(4):683–688. doi: 10.4161/cc.9.4.10766. [DOI] [PubMed] [Google Scholar]
- 99.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310(5746):314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 100.Ghosh S, George S, Roy U, Ramachandran D, Kolthur-Seetharam U. NAD: a master regulator of transcription. Biochim Biophys Acta. 2010;1799(10–12):681–693. doi: 10.1016/j.bbagrm.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 101.Ota H, Akishita M, Eto M, Iijima K, Kaneki M, Ouchi Y. Sirt1 modulates premature senescence-like phenotype in human endothelial cells. J Mol Cell Cardiol. 2007;43(5):571–579. doi: 10.1016/j.yjmcc.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 102.Potente M, Ghaeni L, Baldessari D, Mostoslavsky R, Rossig L, Dequiedt F, Haendeler J, Mione M, Dejana E, Alt FW, Zeiher AM, Dimmeler S. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21(20):2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Menghini R, Casagrande V, Cardellini M, Martelli E, Terrinoni A, Amati F, Vasa-Nicotera M, Ippoliti A, Novelli G, Melino G, Lauro R, Federici M. MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation. 2009;120(15):1524–1532. doi: 10.1161/CIRCULATIONAHA.109.864629. [DOI] [PubMed] [Google Scholar]
- 104.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17(2):193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 105.Ito T, Yagi S, Yamakuchi M. MicroRNA-34a regulation of endothelial senescence. Biochem Biophys Res Commun. 2010;398(4):735–740. doi: 10.1016/j.bbrc.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 106.Ferrara N, Chen H, Davis-Smyth T, Gerber HP, Nguyen TN, Peers D, Chisholm V, Hillan KJ, Schwall RH. Vascular endothelial growth factor is essential for corpus luteum angiogenesis. Nat Med. 1998;4(3):336–340. doi: 10.1038/nm0398-336. [DOI] [PubMed] [Google Scholar]
- 107.Risau W. Mechanisms of angiogenesis. Nature. 1997;386(6626):671–674. doi: 10.1038/386671a0. [DOI] [PubMed] [Google Scholar]
- 108.Ribatti D, Nico B, Crivellato E, Roccaro AM, Vacca A. The history of the angiogenic switch concept. Leukemia. 2007;21(1):44–52. doi: 10.1038/sj.leu.2404402. [DOI] [PubMed] [Google Scholar]
- 109.Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z, Hanahan D. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat Cell Biol. 2000;2(10):737–744. doi: 10.1038/35036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanahan D, Folkman J. Patterns and emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 1996;86(3):353–364. doi: 10.1016/S0092-8674(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 111.Gerhardt H, Golding M, Fruttiger M, Ruhrberg C, Lundkvist A, Abramsson A, Jeltsch M, Mitchell C, Alitalo K, Shima D, Betsholtz C. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. J Cell Biol. 2003;161(6):1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bussolino F, Mantovani A, Persico G. Molecular mechanisms of blood vessel formation. Trends Biochem Sci. 1997;22(7):251–256. doi: 10.1016/S0968-0004(97)01074-8. [DOI] [PubMed] [Google Scholar]
- 113.Whitelock JM, Murdoch AD, Iozzo RV, Underwood PA. The degradation of human endothelial cell-derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin, and heparanases. J Biol Chem. 1996;271(17):10079–10086. doi: 10.1074/jbc.271.17.10079. [DOI] [PubMed] [Google Scholar]
- 114.Gately S, Twardowski P, Stack MS, Cundiff DL, Grella D, Castellino FJ, Enghild J, Kwaan HC, Lee F, Kramer RA, Volpert O, Bouck N, Soff GA. The mechanism of cancer-mediated conversion of plasminogen to the angiogenesis inhibitor angiostatin. Proc Natl Acad Sci USA. 1997;94(20):10868–10872. doi: 10.1073/pnas.94.20.10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ruegg C, Alghisi GC. Vascular integrins: therapeutic and imaging targets of tumor angiogenesis. Recent Results Cancer Res. 2010;180:83–101. doi: 10.1007/978-3-540-78281-0_6. [DOI] [PubMed] [Google Scholar]
- 116.Pries AR, Hopfner M, le Noble F, Dewhirst MW, Secomb TW. The shunt problem: control of functional shunting in normal and tumour vasculature. Nat Rev. 2010;10(8):587–593. doi: 10.1038/nrc2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Abdollahi A, Schwager C, Kleeff J, Esposito I, Domhan S, Peschke P, Hauser K, Hahnfeldt P, Hlatky L, Debus J, Peters JM, Friess H, Folkman J, Huber PE. Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proc Natl Acad Sci USA. 2007;104(31):12890–12895. doi: 10.1073/pnas.0705505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Almog N, Ma L, Raychowdhury R, Schwager C, Erber R, Short S, Hlatky L, Vajkoczy P, Huber PE, Folkman J, Abdollahi A. Transcriptional switch of dormant tumors to fast-growing angiogenic phenotype. Cancer Res. 2009;69(3):836–844. doi: 10.1158/0008-5472.CAN-08-2590. [DOI] [PubMed] [Google Scholar]
- 119.Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280(10):9330–9335. doi: 10.1074/jbc.M413394200. [DOI] [PubMed] [Google Scholar]
- 120.Otsuka M, Zheng M, Hayashi M, Lee JD, Yoshino O, Lin S, Han J. Impaired microRNA processing causes corpus luteum insufficiency and infertility in mice. J Clin Investig. 2008;118(5):1944–1954. doi: 10.1172/JCI33680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer-dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100(8):1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- 122.Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101(1):59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- 123.Shilo S, Roy S, Khanna S, Sen CK. Evidence for the involvement of miRNA in redox regulated angiogenic response of human microvascular endothelial cells. Arterioscler Thromb Vasc Biol. 2008;28(3):471–477. doi: 10.1161/ATVBAHA.107.160655. [DOI] [PubMed] [Google Scholar]
- 124.Chen Y, Banda M, Speyer CL, Smith JS, Rabson AB, Gorski DH. Regulation of the expression and activity of the anti-angiogenic homeobox gene GAX/MEOX2 by ZEB2 and microRNA-221. Mol Cell Biol. 2010;30(15):3902–3913. doi: 10.1128/MCB.01237-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gorski DH, Leal AJ. Inhibition of endothelial cell activation by the homeobox gene Gax. J Surg Res. 2003;111(1):91–99. doi: 10.1016/S0022-4804(03)00042-8. [DOI] [PubMed] [Google Scholar]
- 126.Patel S, Leal AD, Gorski DH. The homeobox gene Gax inhibits angiogenesis through inhibition of nuclear factor-kappaB-dependent endothelial cell gene expression. Cancer Res. 2005;65(4):1414–1424. doi: 10.1158/0008-5472.CAN-04-3431. [DOI] [PubMed] [Google Scholar]
- 127.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates anti-angiogenic homeobox genes GAX and HOXA5. Blood. 2008;111(3):1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, Brizzi MF. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol. 2010;30(8):1562–1568. doi: 10.1161/ATVBAHA.110.206201. [DOI] [PubMed] [Google Scholar]
- 129.Gatt ME, Zhao JJ, Ebert MS, Zhang Y, Chu Z, Mani M, Gazit R, Carrasco DE, Dutta-Simmons J, Adamia S, Minvielle S, Tai YT, Munshi NC, Avet-Loiseau H, Anderson KC, Carrasco DR (2010) MicroRNAs 15a/16-1 function as tumor suppressor genes in multiple myeloma. Blood. doi:10.1182/blood-2009-11-253294 [DOI] [PubMed]
- 130.Karaa ZS, Iacovoni JS, Bastide A, Lacazette E, Touriol C, Prats H. The VEGF IRESes are differentially susceptible to translation inhibition by miR-16. RNA. 2009;15(2):249–254. doi: 10.1261/rna.1301109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bonauer A, Carmona G, Iwasaki M, Mione M, Koyanagi M, Fischer A, Burchfield J, Fox H, Doebele C, Ohtani K, Chavakis E, Potente M, Tjwa M, Urbich C, Zeiher AM, Dimmeler S. MicroRNA-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science (New York, NY) 2009;324(5935):1710–1713. doi: 10.1126/science.1174381. [DOI] [PubMed] [Google Scholar]
- 132.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, King PD, Weis SM, Cheresh DA. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16(8):909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wurdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, Weissleder R, Breakefield XO, Krichevsky AM. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14(5):382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fang L, Deng Z, Shatseva T, Yang J, Peng C, Du WW, Yee AJ, Ang LC, He C, Shan SW, Yang BB (2010) MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-beta8. Oncogene. doi:10.1038/onc.2010.465 [DOI] [PubMed]
- 135.Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci USA. 2007;104(51):20350–20355. doi: 10.1073/pnas.0706901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133(2):217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Suarez Y, Fernandez-Hernando C, Yu J, Gerber SA, Harrison KD, Pober JS, Iruela-Arispe ML, Merkenschlager M, Sessa WC. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci USA. 2008;105(37):14082–14087. doi: 10.1073/pnas.0804597105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38(9):1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, Cleary MA, Thomas-Tikhonenko A. The Myc-miR-17 92 axis blunts TGFβ signaling and production of multiple TGFβ-dependent anti-angiogenic factors. Cancer Res. 2010;70(20):8233–8246. doi: 10.1158/0008-5472.CAN-10-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Saito Y, Friedman JM, Chihara Y, Egger G, Chuang JC, Liang G. Epigenetic therapy upregulates the tumor suppressor microRNA-126 and its host gene EGFL7 in human cancer cells. Biochem Biophys Res Commun. 2009;379(3):726–731. doi: 10.1016/j.bbrc.2008.12.098. [DOI] [PubMed] [Google Scholar]
- 141.Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc Natl Acad Sci USA. 2008;105(5):1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fish JE, Santoro MM, Morton SU, Yu S, Yeh RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY, Srivastava D. miR-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kuhnert F, Mancuso MR, Hampton J, Stankunas K, Asano T, Chen CZ, Kuo CJ. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135(24):3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 144.Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel-Duby R, Olson EN. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zou J, Li WQ, Li Q, Li XQ, Zhang JT, Liu GQ, Chen J, Qiu XX, Tian FJ, Wang ZZ, Zhu N, Qin YW, Shen B, Liu TX, Jing Q. Two functional microRNA-126 s repress a novel target gene p21-activated kinase 1 to regulate vascular integrity in zebrafish. Circ Res. 2011;108(2):201–209. doi: 10.1161/CIRCRESAHA.110.225045. [DOI] [PubMed] [Google Scholar]
- 146.Schmidt M, Paes K, De Maziere A, Smyczek T, Yang S, Gray A, French D, Kasman I, Klumperman J, Rice DS, Ye W. EGFL7 regulates the collective migration of endothelial cells by restricting their spatial distribution. Development. 2007;134(16):2913–2923. doi: 10.1242/dev.002576. [DOI] [PubMed] [Google Scholar]
- 147.Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNA-mediated integration of haemodynamics and VEGF signalling during angiogenesis. Nature. 2010;464(7292):1196–1200. doi: 10.1038/nature08889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Meister J, Schmidt MH. miR-126 and miR-126*: new players in cancer. ScientificWorld Journal. 2010;10:2090–2100. doi: 10.1100/tsw.2010.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang J, Du YY, Lin YF, Chen YT, Yang L, Wang HJ, Ma D. The cell growth suppressor, mir-126, targets IRS-1. Biochem Biophys Res Commun. 2008;377(1):136–140. doi: 10.1016/j.bbrc.2008.09.089. [DOI] [PubMed] [Google Scholar]
- 150.Liu B, Peng XC, Zheng XL, Wang J, Qin YW. MiR-126 restoration down-regulate VEGF and inhibit the growth of lung cancer cell lines in vitro and in vivo. Lung Cancer. 2009;66(2):169–175. doi: 10.1016/j.lungcan.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 151.Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, Bai Y, Shen Y, Yuan W, Jing Q, Qin Y. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351(1–2):157–164. doi: 10.1007/s11010-011-0723-7. [DOI] [PubMed] [Google Scholar]
- 152.Crawford M, Brawner E, Batte K, Yu L, Hunter MG, Otterson GA, Nuovo G, Marsh CB, Nana-Sinkam SP. MicroRNA-126 inhibits invasion in non-small cell lung carcinoma cell lines. Biochem Biophys Res Commun. 2008;373(4):607–612. doi: 10.1016/j.bbrc.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 153.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298(1):50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 154.Sun Y, Bai Y, Zhang F, Wang Y, Guo Y, Guo L. miR-126 inhibits non-small cell lung cancer cells proliferation by targeting EGFL7. Biochem Biophys Res Commun. 2010;391(3):1483–1489. doi: 10.1016/j.bbrc.2009.12.098. [DOI] [PubMed] [Google Scholar]
- 155.Donnem T, Lonvik K, Eklo K, Berg T, Sorbye SW, Al-Shibli K, Al-Saad S, Andersen S, Stenvold H, Bremnes RM, Busund LT (2011) Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer. doi:10.1002/cncr.25907 [DOI] [PubMed]
- 156.Barshack I, Lithwick-Yanai G, Afek A, Rosenblatt K, Tabibian-Keissar H, Zepeniuk M, Cohen L, Dan H, Zion O, Strenov Y, Polak-Charcon S, Perelman M (2010) MicroRNA expression differentiates between primary lung tumors and metastases to the lung. Pathol Res Pract. pii:S0344-0338(10)00087-7 [DOI] [PubMed]
- 157.Gilad S, Meiri E, Yogev Y, Benjamin S, Lebanony D, Yerushalmi N, Benjamin H, Kushnir M, Cholakh H, Melamed N, Bentwich Z, Hod M, Goren Y, Chajut A. Serum microRNAs are promising novel biomarkers. PLoS ONE. 2008;3(9):e3148. doi: 10.1371/journal.pone.0003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, Mayr A, Weger S, Oberhollenzer F, Bonora E, Shah A, Willeit J, Mayr M. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 159.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, Sun F, Lu J, Yin Y, Cai X, Sun Q, Wang K, Ba Y, Wang Q, Wang D, Yang J, Liu P, Xu T, Yan Q, Zhang J, Zen K, Zhang CY. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39(1):133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]