Abstract
Inherited cardiomyopathies are caused by point mutations in sarcomeric gene products, including α-cardiac muscle actin (ACTC1). We examined the biochemical and cell biological properties of the α-cardiac actin mutations Y166C and M305L identified in hypertrophic cardiomyopathy (HCM). Untagged wild-type (WT) cardiac actin, and the Y166C and M305L mutants were expressed by the baculovirus/Sf9-cell system and affinity purified by immobilized gelsolin G4–6. Their correct folding was verified by a number of assays. The mutant actins also displayed a disturbed intrinsic ATPase activity and an altered polymerization behavior in the presence of tropomyosin, gelsolin, and Arp2/3 complex. Both mutants stimulated the cardiac β-myosin ATPase to only 50 % of WT cardiac F-actin. Copolymers of WT and increasing amounts of the mutant actins led to a reduced stimulation of the myosin ATPase. Transfection of established cell lines revealed incorporation of EGFP- and hemagglutinin (HA)-tagged WT and both mutant actins into cytoplasmic stress fibers. Adenoviral vectors of HA-tagged WT and Y166C actin were successfully used to infect adult and neonatal rat cardiomyocytes (NRCs). The expressed HA-tagged actins were incorporated into the minus-ends of NRC thin filaments, demonstrating the ability to form hybrid thin filaments with endogenous actin. In NRCs, the Y166C mutant led after 72 h to a shortening of the sarcomere length when compared to NRCs infected with WT actin. Thus our data demonstrate that a mutant actin can be integrated into cardiomyocyte thin filaments and by its reduced mode of myosin interaction might be the basis for the initiation of HCM.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-012-1030-5) contains supplementary material, which is available to authorized users.
Keywords: Cardiac actin, Actin expression, Hypertrophic cardiomyopathy, Myosin, Tropomyosin
Introduction
Familial or inherited cardiomyopathies are caused by point mutations in genes encoding almost solely contractile proteins and are therefore regarded as diseases of the cardiac sarcomere. They are transmitted in an autosomal dominant manner occurring with a prevalence of about 1:500. The two most frequent forms are hypertrophic and dilative cardiomyopathy (HCM and DCM). Both are dependent on particular single point mutations in one of the many proteins building the sarcomere. DCM is characterized by an enlargement of particularly the left ventricle that finally impairs pumping of blood into the circulation. The resulting cardiac insufficiency can only be effectively cured by cardiac transplantation. HCM is characterized by an asymmetrically increased muscle mass of the left ventricle or the interventricular septum. HCM occurs with a prevalence of 0.2 % [1] and often remains unrecognized until it causes sudden cardiac death among young, apparently healthy persons [2, 3]; particularly dramatic are cases where young athletes succumb to sudden death during sportive action. At later stages, when diastolic filling of the left ventricle becomes impaired, HCM-affected patients suffer from exercise limitation, chest pain, dyspnea, syncopes, vertigo, and ventricular arrhythmias.
Meanwhile, over 450 mutations are known in myofilament-related genes [4], which are responsible for 80 % of all HCM cases [5, 6], whereas only two non-sarcomeric proteins carrying mutations have been reported [7, 8]. Animal models have shown that the effects of most mutations are not due to haploinsufficiency, but rather to a dominant negative type of action [9]. It has been hypothesized that the mutant protein has a changed or novel function and thereby interferes as a “poisonous peptide” with the protein produced from the healthy allele. Although the molecular mechanisms behind the changes in cardiac morphology are only poorly understood, it has been suggested that mutations in sarcomeric genes affect a number of different signaling pathways that contribute to the generation of cardiomyocyte hypertrophy and the alteration in cardiac architecture [3, 10–12]. In general, HCM is believed to arise as a compensation to overcome an insufficiency of contractile force [13, 14], whereas DCM is believed to be the consequence of a loss of sarcomeric stability or tensile resistance [15, 16].
In order to fully understand the mechanisms leading to the different phenotypes of familial cardiomyopathies, it is essential to investigate the consequences of these point mutations in the biochemical and cell biological context. In the past, the phenotypic changes caused by cardiac actin mutations were analyzed by transient expression of the actin mutants in established cell lines like non-muscle COS-7 cells [17] or in myoblasts/myotubes [18]. Since the physiological relevance of expressing cardiac actin mutants in these cells is limited, it is necessary to analyze cardiac actin mutants in cardiomyocytes. E1/E3-deleted adenoviral vectors have been used to infect primary cardiomyocytes with various sarcomeric proteins [19–22], but to date there are no data available about the effects of adenoviral-induced expression of α-cardiac actin mutants in primary cardiomyocytes. For a detailed biochemical analysis, it is furthermore crucial to express these actin mutants in abundant quantities. A number of attempts have been made to overexpress recombinant or endogenous actin in different organisms [23–26] or in a cell-free coupled translation system [17], all with limitations in solubility, sequence homology, or yield. Recently, the expression of soluble mammalian actin variants in adequate amounts succeeded in the baculovirus/Sf9-cell system [27–30], which includes the essential eukaryotic chaperonin TRiC/CCT [31] and is capable of posttranslational modifications of eukaryotic proteins.
Actin, an ubiquitous protein found in every eukaryotic cell, plays a pivotal role in a vast number of processes such as maintenance of cell shape, cell motility, intracellular transport processes, mitosis, and shortening of muscles [32–34]. In higher eukaryotes, there are six isoforms of actin: β-cytoplasmic, γ-cytoplasmic, α-vascular, γ-enteric, α-skeletal muscle, and α-cardiac muscle, which are completely conserved across species from avians to mammals [35]. Isoactins differ among each other by only a few amino-acid exchanges, localized mostly in the N-terminal region. Each actin isoform is designed to fulfill particular tissue-specific functions, even though the molecular basis of these functional specificities is not yet understood. Although over 80 3D-structures of actin have been reported (for reviews, see [36, 37]), and their influence on the interaction potential with a number of actin binding proteins is well documented [38], only sparse insights have been obtained into the structural consequences of isoform-specific exchanges or mutations on F-actin filament architecture. Mutations in actin-coding genes lead to serious disturbances in numerous cellular processes. Typical diseases correlated with point mutations of actin isoforms include cardiac and skeletal muscle myopathies [28, 29], atrial septal defects [39], sensory hearing loss, or dystonia [40–42]. Gene targeting experiments in mice have shown that deletion of the α-cardiac actin isoform leads to cardiac failure and death shortly before birth, despite a compensatory upregulation of α-skeletal and/or vascular α-smooth muscle actins [43]. Interestingly, more than 100 mutations within the α-skeletal muscle actin gene (ACTA1) are known and are responsible for 20 % of the congenital myopathies, whereas only 12 mutations within the ACTC1 (α-cardiac actin) gene were identified and correlated with diseases, such as cardiac myopathies and atrial septal defects (ASD). Nine of them cause HCM (H90Y, R97C, E99K, P164A, Y166C, A232V, A295S, M305L, A331P; [44–48]), two of them lead to DCM (R312H and E361G; [15]), and one was found in ASD (M123V; [39]).
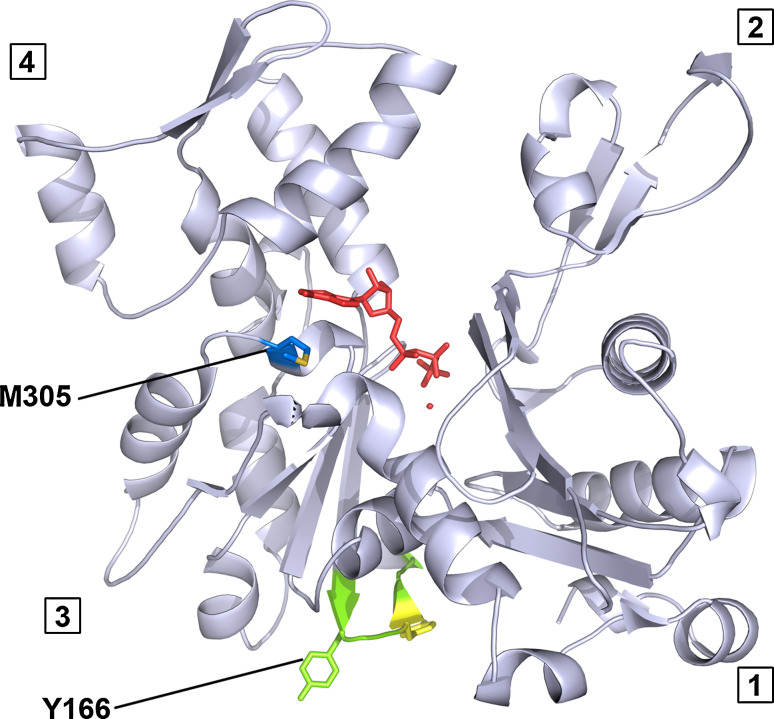
The current study investigates the biochemical and cell biology consequences of the α-cardiac actin mutations Tyr166Cys (Y166C) and Met305Leu (M305L) (see Fig. 1). The mutation at position 166 leads to removal of an aromatic ring. Y166 is part of a sequence in the so-called W-loop (residues 165–172), which has been suggested to act as a nucleotide state-sensing region of actin [49]. This region undergoes subtle conformational changes from an unstructured coil to a β-turn motif upon ATP hydrolysis resulting from the formation of backbone hydrogen bonds between Y166 and Y169 [50]. In addition, the W-loop and thus Y166 has been shown to participate in the interface of the long-pitch actin–actin contacts in F-actin [51–53]. The mutation M305L results in the elimination of a thioether group. M305 is located close to the nucleotide-binding site, in particular underneath the adenine ring binding region [51], and also part of a prominent loop–helix–loop structure in subdomain 3 (300–326 sequence). This motif is exposed on the surface of the F-actin filament and has been implicated in the binding of divalent cations [54].
Fig. 1.
Location of the cardiomyopathic actin mutations Y166C and M305L. Atomic structure of skeletal muscle α-actin as determined in complex with DNase I ([51]; pdb: 1ATN). The model was modified using PyMOL 1.4 highlighting the residues Y166 in green and M305 in blue. The four different subdomains are numbered. The methionine on position 305 is located close to the nucleotide ATP (red). The tyrosine on position 166 is located in the so-called W-loop (subdomain 3), supposed to participate in long pitch helical actin–actin interactions and to couple with Y169 (yellow) in a nucleotide-dependent manner [49]
We expressed the human α-cardiac actin mutants Y166C and M305L and the wild-type (WT) control by means of the baculovirus/Sf9-system. Affinity purification of tag-free recombinant actins was achieved by using a gelsolin deletion construct comprising segments 4 to 6 (G4–6) [55, 56], and their biochemical properties were analyzed in in vitro experiments. Combined with this approach, we examined the intracellular expression and localization of cardiac actin variants in transfected cell lines and primary rat cardiomyocytes. In particular, we focused on their incorporation into thin filaments and their effects on sarcomere organization and structure in neonatal rat cardiomyocytes.
Materials and methods
cDNA cloning
The pcDNA3.1/NT-GFP-TOPO®-WT-α-cardiac actin and its two mutants Y166C and M305L were donated by Dipl.-biol. S. Fister and Dr. H. Milting (Herz- und Diabeteszentrum NRW, Universitätsklinik der Ruhr-Universität Bochum, Bad Oeynhausen, Germany). In this construct, the green fluorescent protein [57] was fused to the N-terminus of the α-cardiac actins [58]. These plasmids served as templates for cloning the actin versions into p3xHA-C1 plasmid. The p3xHA-C1 plasmid was a kind gift from Dr. T. Engel (Leibniz-Institut für Arteriosklerosis, Münster, Germany), who deleted the cDNA of EGFP from pEGFP-C1 plasmid (Clontech, Heidelberg, Germany) and instead cloned into this plasmid the cDNA of a three times repeated hemagglutinin-tag (HA). The primers used for amplifying the actin cDNAs were as follows: 5′-GTTATGTGTGACGACGAGGAGACC-3′ and 5′-ATTGCCCTTTTAGAAGCATTTGCG-3′. PCR inserts were cloned into p3xHA-C1 using XbaI and XhoI sites.
Gelsolin expression plasmid
The deletion construct of human gelsolin G4–6 (used for recombinant actin purification) was kindly supplied by Dr. A.G. Weeds (LMB-MRC, Cambridge, UK) and subcloned from shuttle vector pKN172 into the cold-shock expression plasmid pCOLD II (TAKARA BIO, Paris, France) using the restriction sites for BamHI and HindIII (enzymes from Fermentas, St.-Leon-Rot, Germany). The pCOLD II plasmid provides a His-Tag sequence fused to the N-terminus allowing affinity chromatography.
Actin Sf9-expression plasmids
The DNA from human cardiac actin WT and the M305L and Y166C mutants were amplified by PCR from the p3xHA plasmids with a pair of oligonucleotides: 5′-GCATGCATT TAGAAGCATTTGCGGTGGAC-3′ and 5′-GCCTCGAGATGTGTGACGACGAGGAGACC-3′. The genes were cloned into pFastBac Dual vectors by usage of the restriction enzymes XhoI and NsiI (Fermentas) under the p10 promoter. To verify correct cloning and the absence of induced errors due to DNA amplification, all constructs were double-strand sequenced.
Expression and purification of actin binding proteins (ABPs)
Full-length human cytoplasmic gelsolin, and its N-terminal segment G1 and N-terminal half G1-3 were recombinantly expressed in E. coli and purified from bacterial homogenates as detailed previously [55]. The C-terminal gelsolin half G4–6 was expressed and purified similarly [55, 56]. The yield was typically 130 mg gelsolin G4–6/l of bacterial culture and its purification resulted in >98 % homogeneity (see Fig. 2a). Arp2/3 complex was purified from Acanthameba castellani [59], together with VCA peptide kindly provided by Prof. D.L. Barber and Dr. A. Schoenichen (University of California, San Fransisco, USA). Thymosin β4 was commercially obtained from Bachem (Bubendorf; Switzerland). Purification of tropomyosin (TM) was performed according to Coulton et al. [60]. The expression construct of Mus musculus smooth muscle tropomyosin α1-chain in pJC20 vector was kindly provided by Dr. Anja Schmidtmann (University of Kent, Canterbury, UK). The α1-chain clone differs by only one amino acid exchange from cardiac α-TM and contains an alanine–serine (AS) N-terminal extension to mimic acetylation, essential for its native behavior [60].
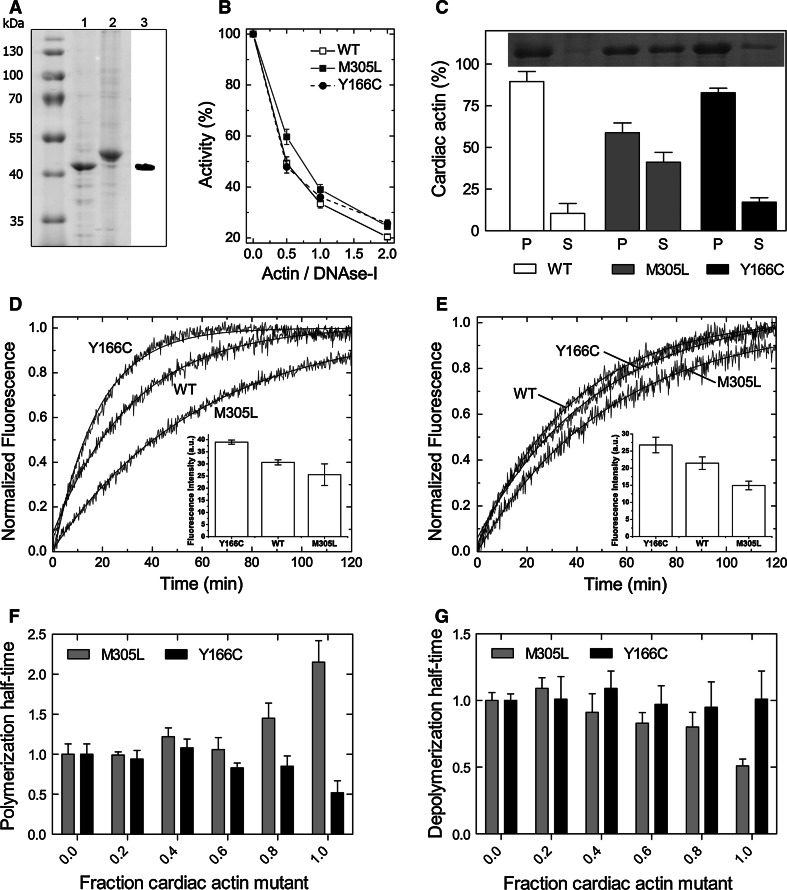
Fig. 2.
Purification, folding and polymerization of recombinant WT and mutant actin constructs. a Purified untagged human α-cardiac actin (1) after elution from immobilized gelsolin G4–6 (2) with Ca2+-chelating buffer (G4-6 control, lane 2). Expression of α-cardiac actin was confirmed by immunoblotting (3). b DNase I inhibition test. Data are presented as mean ± SD, n = 3. c Sedimentation of WT F-actin and the Y166C and M305L mutants. Fractions of cardiac actin in the pellets (P) and supernatants (S) were visualized by SDS-PAGE (n = 3, error bars SD). d, e Polymerization of the cardiac actin variants (7.5 μM) in the absence (d) and presence (e) of TM (added in a molar ratio of 1:7 to actin). Polymerization courses were normalized to show differences in polymerization rates, insets show the fluorescence amplitudes expressed in arbitrary units (AU). f Polymerization half-times of mixed ratios of WT and mutant actin (n = 4). Half-times were normalized to a WT content of 100 % within the actin filament. g Depolymerization half-times of WT F-actin including different fractions of mutant actin (n = 4). Half-times were normalized to the pure WT actin filament
Expression and purification of recombinant actin
The human α-cardiac muscle WT actin and both mutants were expressed in the baculovirus/Sf9-system. The preparation of recombinant baculoviruses was performed according to the manufacturer’s recommendation (Invitrogen, Karlsruhe, Germany) by transforming the pFastBac Dual vectors into DH10Bac E.coli cells, bacmid isolation and transfection into Sf9 insect cells using Cellfectin II (Invitrogen). Infected Sf9-cells were harvested after 3 days and stored at −80 °C. Purification of the tag-free WT cardiac actin and both mutants was performed by affinity chromatography using the gelsolin deletion mutant G4–6 as recently described by Ohki et al. [56]. The proteins were supplemented with 3 % sucrose, flash frozen in liquid N2 and stored at −80 °C. Protein purity was confirmed by SDS-PAGE and western blot analysis (Fig. 2a). The actin preparations showed no contamination by the G4–6 as tested by SDS-PAGE (10 % polyacrylamide), which clearly separated both proteins (Fig. 2a). The residual actin content within the G4–6 fraction was usually <5 %. As a precaution, the G4–6 batches were reused for the purification of only the same actin variant.
Purification of native cardiac β-myosin from Sus scrofa (Ss) domestica
To purify cardiac β-myosin from adult domestic pig (Ss) protocols by Jacques et al. [61] and Pant et al. [62] were modified as follows. One gram of heart tissue from the left ventricular muscle, frozen in liquid N2, was minced in a liquid nitrogen-cooled mortar and suspended in 7.5 ml high salt extraction buffer (150 mM K2HPO4, 10 mM Na4P2O7, 300 mM KCl, 2 mM MgCl2, 1 mM Na2ATP, 10 mM DTT, proteinase inhibitors chymostatin, E64, pepstatin, leupeptin each 3 μg/ml, pH 6.8). The homogenate was extracted for 1 h at 4 °C on a rotating mixer and centrifuged at 61,000g for 30 min. The supernatant was precipitated for 1 h with 230 ml ice-cold 5 mM DTT, followed by centrifugation at 75,000g for 30 min. The precipitated myosin pellet was washed and dissolved in 1.0 ml resuspension buffer (25 mM Hepes, pH 7.4, 300 mM KCl, 1 mM EGTA, 4 mM MgCl2, 10 mM DTT, 0.015 μg each of chymostatin, E64, pepstatin and leupeptin). Cardiac myosin was stored in 50 % glycerol at −20 °C. The typical yield was 10–15 mg.
Gel electrophoresis
Polyacrylamide gel (10 or 12 %) electrophoresis in the presence of SDS (SDS-PAGE) was performed according to [63]. Native gel electrophoresis (NGE) was performed as described previously [64].
Sedimentation assay
An amount of 50 μM cardiac actin in G-buffer (10 mM Tris–HCl, pH 8.0, 0.2 mM CaCl2, 7 mM β-mercaptoethanol, 1 mM ATP) was polymerized by the addition of 2 mM MgCl2 and 0.1 M KCl for at least 2 h at 21 °C. F-actin was sedimented by centrifugation at 100,000g for 45 min and the supernatant was carefully separated. The sedimented actin was dissolved in an equal volume and the samples were visualized by SDS-PAGE followed by densitometric analysis.
Thermal denaturation
Denaturation of monomeric actin (0.05 mg/ml) was measured by monitoring the thermal dependence change of ellipticity at 222 nm in a temperature controlled π*-180 Spectrometer equipped with a circular dichroism unit (Applied Photophysics, Leatherhead, UK). Measurements were performed in a 0.3-cm path length cells with a temperature increase of 2 °C min−1 in 5 mM Tris-buffer (pH 8.0) and 0.1 mM ATP [65, 66], and the transition midpoint of the unfolding reaction was calculated by fitting a Boltzmann function to the experimental data.
DNase I inhibition assay
The DNase I inhibition assay was performed as described by Mannherz et al. [67] using salmon sperm DNA (D1626; Sigma-Aldrich, Germany) at 50 μg/ml final concentration in 10 mM Tris–HCl, pH 8.0, 1 mM MgCl2, and 0.1 mM CaCl2. To determine the effect of the actin variants on the DNase I endonuclease activity, 3.2 μM DNase I from bovine pancreas (DN25; Sigma-Aldrich) was pre-incubated with 0.0–6.4 μM of the G-actin variant in G-buffer at 21 °C for 20 min. Thereafter, 10 μl aliquots were added to 0.8 ml of the DNA-containing test solution and the absorbance was immediately monitored at 260 nm for 15 min at 21 °C using a spectrophotometer (DU 800; Beckman Coulter). The percentage of DNase I inhibition was determined from the ratio of the initial linear rates in the presence and absence of actin.
Actin polymerization assay
Actin polymerization rates were determined by the increase in fluorescence caused by incorporation of pyrene-labeled actin into actin filaments [68]. Pyrene-labeled α-skeletal actin was pre-cleared by dialysis against G-buffer and centrifugation at 100,000g for 30 min. Pyrene-actin (5 % of total actin concentration) was mixed with globular α-cardiac actin in G-buffer. Actin concentration of homopolymers was adjusted between 1 and 10 μM, and polymerization of copolymers with different fractions of WT and mutant protein was measured with 7.5 μM F-actin. Polymerization was induced by the addition of 2 mM MgCl2 and 0.1 M KCl. The increase of pyrene-fluorescence with an excitation wavelength of 365 nm was monitored at 385 nm using a Schimadzu RF5001PC using 100 μl containing micro-cuvettes or at 407 nm using a Varian Cary Eclipse spectrofluorometer in a total volume of 50 μl on a microplate (384 well, Greiner bio-one) at 21 °C. For determination of the apparent half-times of polymerization, the kinetic traces were approximated by a single exponential function and the half-times were calculated from the rate constant.
Actin depolymerization assay
An amount of 20 μM actin with different ratios of WT and mutant actin was polymerized in the presence of 5 % pyrene-actin by the addition of 2 mM MgCl2 and 0.1 M KCl for at least 3 h at 21 °C; subsequently, incubation was extended on ice overnight. After equilibration to 21 °C and dilution of F-actin 1:100 into G-buffer, the decrease in fluorescence at the wavelength settings indicated above was monitored (Varian Cary Eclipse spectrofluorometer) at 21 °C.
Critical concentration
To determine the critical concentration of actin polymerization, varying concentrations of the cardiac actins supplemented with 5 % pyrene-α-skeletal actin were polymerized with 2 mM MgCl2 and 0.1 M KCl. The steady-state fluorescence of polymerized actin was plotted versus actin concentration, and the critical concentration was calculated from the intersections of a linear fit with the abscissa.
Myosin ATPase assay
Steady-state ATPase activities of 1 μM Ss-β-cardiac myosin stimulated by WT α-cardiac actin, the HCM mutants or copolymers of WT and the mutants were performed at 25 °C using a modified version of NADH-coupled assay [69] in ATPase standard buffer containing 40 mM Hepes, pH 7.4, 25 mM KCl, 2 mM MgCl2, 0.5 mM DTT, 0.2 mM NADH and an ATP regeneration system consisting of 0.05 mg/ml pyruvate kinase, 0.5 mM PEP, and 0.02 mg/ml lactate dehydrogenase. Actins were polymerized by the addition of 2 mM MgCl2 and 0.1 M KCl for at least 3 h at 21 °C and used in the assay or kept on ice overnight. F-actin concentration of homopolymers was adjusted between 0 and 30 μM, activation of myosin ATPase by copolymers of WT and mutant actin was assayed with 15 μM F-actin. The reaction was performed in UV transparent half-area 96 well microtiter plates (Corning, New York, USA) with a sample volume of 50 μl. The reaction was started by automated injection of 1 mM ATP in a temperature-controlled plate-reader (SPECTROstar Omega; BMG LABTECH, Ortenberg, Germany). NADH oxidation was followed by measuring the decrease in absorption at 340 nm (ε = 6,220 M−1 cm−1) and ATPase rates were determined by linear curve fitting. Values for K app and V max were calculated from fitting the data to the Michaelis–Menten equation.
Actin ATPase assay
Intrinsic steady-state ATPase-activities of 10 μM monomeric cardiac actin variants in a buffer containing 10 mM Tris–HCl (pH 7.4), 0.2 mM MgCl2 and 0.2 mM DTT or treadmilling rates of 10 μM F-actin [70] in standard ATPase buffer were measured at 25 °C. Measurements were performed as described for the myosin ATPase with the NADH-coupled assay and a sample volume of 100 μl. Actin variants were polymerized for at least 3 h at 21 °C and used in the assay or kept on ice overnight for determination of the treadmilling rates.
Cell culture
NIH 3T3 (murine fibroblastic) and HEK293 (human embryonic kidney) cells were obtained from DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) and cultured in DMEM medium containing 0.45 g/l glucose, 1 % penicillin/streptomycin, 1 % glutamine, 0.5 % sodium pyruvate, and 10 % fetal calf serum. HL-1 (cardiomyocyte-like) cells, a kind gift of Prof. Dr. W. Claycomb (New Orleans, USA), were kept in Claycomb medium enriched with 1 % penicillin/streptomycin, 1 % glutamine, 100 μM norepinephrine, and 10 % fetal calf serum [71]. Cells were cultured at 37 °C in 5 % CO2 and 90 % humidified air and split twice weekly, using 0.25 % trypsin/0.05 % EDTA solution.
Isolation of adult rat cardiomyocytes (ARCs)
Adult Wistar Kyoto rats were euthanized in accordance with the guidelines of the European community (86/609/EEC), and cardiomyocytes were isolated as originally described by Bechem et al. [72]. For details, see Online Resource 1.
Isolation of neonatal rat cardiomyocytes (NRCs)
Neonatal rats were sacrificed in accordance with the guidelines of the European community (86/609/EEC). Cardiomyocytes were isolated according to Przygodzki et al. [73]. For details, see Online Resource 1.
Cell transfection
Cells were seeded on glass coverslips in 6-well plates (3 × 105 cells/well) or in 6-cm culture dishes and transfected after 24 h with 3 or 5 μg of DNA encoding GFP-actins or HA-actins using MATra-A® transfection reagent (Iba, Munich, Germany) After 24–48 h, the cells were either fixed with 4 % paraformaldehyde (PFA) or harvested in lysis buffer using a rubber policeman.
Generation of recombinant adenoviruses
Recombinant adenoviruses (Ad) were produced using the AdEasy™ kit (Qbiogene, Heidelberg, Germany). DNA sequences encoding N-terminally HA-tagged WT- and Y166C-, and M305L-α-cardiac actins were amplified by PCR with the primers: 5′-ATCATGGATTACCCATACGATGTTC-3′ and 5′-ATCGCCCTTTTAGAAGCATTTGCG-3′. PCR products were cloned into the EcoRV site of the pAdTrack-CMV shuttle plasmid. Electrocompetent E. coli BJ5183 were simultaneously transformed with PmeI linearized shuttle plasmid and adenoviral AdEasy-1 DNA backbone. Following homologous recombination in bacteria, clones were screened by restriction digestion with the PacI enzyme that in the case of positive clones resulted in two fragments of 33 and 4.5 kb. For adenovirus production, HEK293 cells were transfected with linearized WT-, Y166C- or M305L-α-cardiac actin pAdEasy-1 constructs using Lipofectamine™ (Invitrogen). HEK293 cells were re-infected twice with recombinant adenoviruses in order to obtain higher amounts of viral particles. For more details concerning the structure of recombinant adenoviral DNA and the steps of recombinant adenoviruses generation, see [74]. All DNA constructs were verified by sequencing.
Infection of ARCs and NRCs with recombinant adenoviruses
Twenty-four hours after plating, ARCs were infected with 1 ml culture medium containing approximately 104 infectious particles. Generally 24 h after infection, 70–80 % of cells gave an EGFP fluorescence signal. The ARCs were incubated for 4 days after infection. NRCs were infected 24 or 72 h after isolation with 20 μl of adenoviruses (~104 infectious particles) added to 2 ml of medium. Assuming that each cardiomyocyte was infected by one adenoviral particle, the effectiveness of infection of NRCs with Ad + WT-α-cardiac actin and Ad + Y166C-α-cardiac actin was about 92 and 96 %, respectively. Unfortunately, infection of NRCs using the AD-M305L α-cardiac actin construct was not achieved. Then, 24 or 72 h after infection, the cells were either fixed with warm (37 °C) 4 % PFA for immunocytochemistry or harvested with the help of rubber policeman for western blotting. Cells infected with only GFP-encoding viruses were used as controls.
Immunoblotting
Cell pellets were resuspended in lysis buffer (10 mM Tris–HCl, pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 20 mM Na4P2O7, 2 mM Na3VO4, 1 % Triton X-100, 10 % glycerol, 0.1 % SDS, 0.5 % deoxycholate), vortexed for 30 s and frozen. After thawing, extracts were vortexed and centrifuged at 20,817g at 4 °C for 5 min. The protein concentration of the supernatant was determined using the Bio-Rad protein assay (Bio-Rad, Munich, Germany). Then, 30 μg of protein were separated on 10 or 12.5% SDS-PAGE gels, blotted, probed with the respective antibodies, and finally developed with the enhanced chemiluminescence detection system (ECL; GE Amersham) [64]. For re-probing, the membranes were stripped and immunostained for total actin or α-actinin. Blotting of purified recombinant actin was performed similarly.
Antibodies
Monoclonal anti-β-actin (clone AC-15), mouse anti-α-sarcomeric actinin, rabbit anti-all isoactins (clone C11) and rabbit anti-goat-biotin antibodies were obtained from Sigma-Aldrich. Donkey anti-mouse-Alexa Fluor®-488, donkey anti-mouse-Alexa Fluor®-568, donkey anti-rabbit-Alexa Fluor®-488, donkey anti-rabbit-Alexa Fluor®-568, and donkey anti-goat-Alexa Fluor®-633 antibodies as well as Alexa Fluor®-568-labeled DNase I were from Molecular Probes (Eugene, OR, USA). Goat anti-HA (clone Y-11) antibody was obtained from Santa Cruz Biotechnology (Heidelberg, Germany) and mouse anti-α-cardiac actin was purchased at Progen Biotechnik (Heidelberg, Germany) or Sigma-Aldrich (A9357). Monoclonal anti-emerin was obtained from Novocastra Laboratories (Newcastle, UK). Monoclonal anti-myomesin (clone B4) antibody [75] was a kind gift from Dr. E. Ehler (King’s College London, UK.).
Confocal microscopy
Control cells, transfected cells, and those infected with denoviruses were fixed with warm (37 °C) 4 % PFA for 20 min at RT and permeabilized with 0.1 % Triton X-100 in PBS for 6 min. For staining with β-actin antibody, we fixed the cells with ice-cold methanol for 6 min at 4 °C. After fixation, the coverslips or plastic dishes were blocked for 30 min with 3 % BSA in PBS. All antibodies were diluted in 3 % BSA in PBS. We immunostained cells either with goat anti-HA IgGs, rabbit anti-cofilin, or with monoclonal antibodies directed against anti-β-actin, anti-α-cardiac actin, anti-sarcomeric α-actinin, anti-emerin and anti-myomesin. The secondary IgGs were conjugated either with Alexa Fluor®-488, Alexa Fluor®-568, or Alexa Fluor®-633. For double immunostaining, donkey anti-mouse IgGs were used in order to avoid cross-reactivity. F-actin was visualized by staining with TRITC-conjugated phalloidin (Sigma-Aldrich) and nuclei were stained with Hoechst 33342 (Riedel-de-Haen, Seelze, Germany). The coverslips or plastic dishes were washed several times with PBS for 5 min between incubation steps. Finally, cells were mounted in DAKO cytomatic fluorescent mounting medium (DAKO, Copehagen, Denmark). Immunofluorescence microscopy was performed using a Zeiss LSM 510 confocal laser scanning microscope. For documentation, at least five cells were photographed from three independent experiments and a representative image is presented. The length of the sarcomeres was measured with the help of LSM Image Browser (Carl Zeiss Vision, Göttingen, Germany) as the distance between two adjacent Z-discs visualized by anti-α-actinin staining. For each condition shown in Fig. 9 (see "Infection and expression of exogenous actins leads to alterations in sarcomere lengths", the lengths of 7 sarcomeres of 10 different NRCs each (total 70 sarcomeres) were determined.
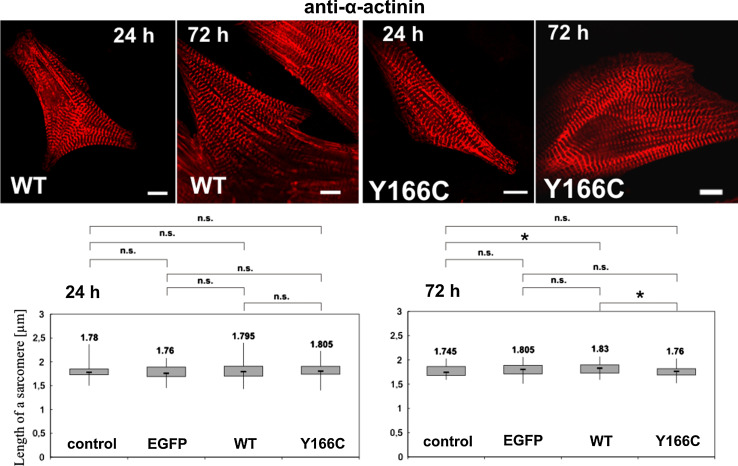
Fig. 9.
Determination of sarcomere width of NRCs 24 and 72 h after infection. Sarcomere widths were determined from confocal images immunostained with anti-α-actinin (red). Upper row images of NRCs 24 and 72 h after infection with the adenoviral vectors leading to expression of WT or Y166C mutant actin. Bars 10 μm. Lower row the determined sarcomere widths for uninfected control and NRCs infected with empty adenoviral vector leading to expression of only EGFP, with vector for WT, or the Y166C mutant actin. *Statistical significance as determined by the Student’s t test (P < 0.05); n.s. not significant. For details, see text
Electron microscopy
For negative staining, F-actin samples were diluted to 0.1 mg/ml and adsorbed to freshly glow-discharged carbon-coated grids. After negative staining with 0.5 % uranyl formate, the samples were examined in a Jeol JEM1400 electron microscope (Jeol, Tokyo, Japan) operated at 120 kV.
Computational and statistical analysis
Graphs were plotted in Excel 2007 (Microsoft®) or in Origin 8.5 (OriginLab). DNA sequences were analyzed in DNAstar Lasergene software (DNASTAR). Student’s unpaired t test was employed to analyze densitometric data as well as lengths of sarcomeres between non-infected and infected NRCs, and the means were regarded as statistically different when p < 0.05. Densitometric analysis of bands was performed using ImageJ (NIH, USA) or Ultra Quant 6.0 (Ultra-Lum, Ontario, Canada) software.
Results
Affinity purification of recombinant human α-cardiac actins using gelsolin G4–6
Figure 1 gives the positions of the mutated residues Y166C and M305L investigated in this study within the 3D-structure of actin. For a detailed analysis of the functional consequences of these cardiac actin mutations causing HCM (see Fig. 1), we expressed WT and both mutants in Sf9-insect cells as untagged proteins. The C-terminal fragment (G4–6) of human gelsolin, which was immobilized via an N-terminal His6-tag to Ni-NTA columns, was used to purify the recombinant cardiac actins by affinity chromatography. Elution of the actins was achieved by Tris–HCl buffer supplemented with the Ca2+-chelator EDTA and resulted in highly pure protein (about 95 % homogeneity) as determined by SDS-PAGE and further confirmed by immunoblotting using an antibody against cardiac actin (as shown for WT in Fig. 2a). All three actin variants were isolated to similar purity with a yield of around 5 mg/3 × 109 Sf9-cells.
Assessing the native functionality of affinity-purified cardiac actins
Thermal stability
The thermal unfolding of G-actin is an irreversible process, which results in the loss of approximately 30 % of the α-helical structure [76]. We analyzed the unfolding reaction of the cardiac actin variants by circular dichroism spectroscopy (Online Resource 2). The transition midpoints were comparable for WT (50.3 ± 1.2 °C) and the mutant actins M305L and Y166C (50.4 ± 1.5 and 52.0 ± 1.2 °C; n = 4), suggesting that the point mutations do not alter the intrinsic stability of the recombinantly expressed cardiac actin variants.
DNase I inhibition
The DNase I inhibition assay is an alternative method to probe the proper folding of monomeric actin. Only properly folded, full-length actin is capable to bind DNase I with high affinity [76, 77]. G-actin binds DNase I mainly via the D- (DNase I-binding) loop (residues 38–51, located in subdomain 2 [51]) and an additional region in subdomain 4 thereby inhibiting its endonuclease activity. Compared to WT actin, the endonuclease inhibiting activity of the mutant actins was not considerably changed (Fig. 2b), albeit the M305L mutation showed a 10 % lower inhibitory effect.
Native gel electrophoresis
Additionally, the ability of the actin variants to interact with several ABPs was tested by NGE [78]. The interaction with DNase I, thymosin β4, gelsolin G1 and G1–3 led to identical mobility shifts as the skeletal muscle actin reference (Online Resource 3). This finding provides further evidence that the recombinantly expressed cardiac actin variants adopted a fully native state after purification.
Polymerization and depolymerization behaviour
Initial sedimentation experiments of the F-actin variants demonstrated markedly perturbed polymerization ability for the M305L mutant. In contrast, almost identical amounts of WT and Y166C actin were detected in the respective pellets, indicating a similar extent of polymerization (Table 1; Fig. 2c). Polymerization kinetics of WT and the mutant actins were subsequently analyzed using pyrene-labelled actin. The M305L mutant actin showed substantial reduced polymerization behavior compared to WT (twofold increased half-time), whereas the Y166C mutation led to an enhancement (Fig. 2d). The apparent half-times of polymerization and the critical concentration (see Online Resource 4) of the three cardiac actin variants are summarized in Table 1. WT and M305L mutant exhibited a Cc of 1.0 μM under the given assay conditions, whereas the Cc for the Y166C mutant was 0.5 μM. Furthermore, the polymerization behavior was tested in the presence of tropomyosin (TM) composed of two α1-chains (α-TM homodimer). Although TM compensated the difference between WT and Y166C actin, it did not restore the polymerization ability of M305L to WT level (Table 1; Fig. 2e). Polymerization experiments of mixtures of WT and the mutant actins were performed to assess the influence of the pathological actin variants within WT filaments (Fig. 2f). Addition of the Y166C mutant to WT-cardiac actin at increasing ratios initially showed little effect on the rate of polymerization of these mixtures. Similar results were obtained for the M305L mutant; however, when comprising 60 % and more of the total actin, the M305L mutant clearly reduced, whereas the Y166C mutant increased the rate of polymerization of these mixtures (Fig. 2f).
Table 1.
Summary of the polymerization experiments and intrinsic kinetic constants of the human α-cardiac actin variants in G- and F-form
| Constant | Human α-cardiac actin variant | ||
|---|---|---|---|
| Wild-type | M305L | Y166C | |
| Polymerization half-timea | |||
| t 1/2 (min) | 23.8 ± 3.1 | 51.2 ± 6.3 | 12.4 ± 3.5 |
| t 1/2, tropomyosin (min) | 26.8 ± 3.5 | 36.9 ± 4.8 | 33.5 ± 4.4 |
| Sedimentationb | |||
| Pellet (%) | 89.6 ± 6.0 | 58.9 ± 5.8 | 82.9 ± 2.7 |
| Supernatant (%) | 10.4 ± 6.0 | 41.1 ± 5.8 | 17.1 ± 2.7 |
| Critical concentrationc | |||
| Cc (μM) | 1.0 ± 0.1 | 1.0 ± 0.1 | 0.5 ± 0.1 |
| Actin ATPase activityd | |||
| G-actin ATPase (h−1) | 0.64 ± 0.09 | 2.32 ± 0.15 | 0.03 ± 0.01 |
| F-actin ATPase (h−1) | 2.38 ± 0.36 | 0.74 ± 0.21 | 0.57 ± 0.03 |
Data are taken from Online Resource 4. Means ± SD
aPolymerization half-time of 7.5 μM cardiac actin (n = 3)
bSedimentation of human α-cardiac actins. Fractions of total actin in pellets and supernatants are given in % of total actin (n = 3)
cCritical concentration of polymerization of the cardiac actin variants
dIntrinsic ATPase activities of 10 μM actin variants in G- and Mg2+-form and treadmilling rate of 10 μM filamentous actin (n = 4)
Table 2.
Summary of Michaelis–Menten parameters of actin-activated Sus scrofa domestica cardiac β-myosin
| Constant | Human α-cardiac actin variant | ||
|---|---|---|---|
| Wild-type | M305L | Y166C | |
| ATPase of Ss β-cardiac myosin | |||
| K app (μM) | 13.7 ± 3.1 | 11.3 ± 2.7 | 10.4 ± 3.9 |
| V max (s−1) | 0.052 ± 0.006 | 0.028 ± 0.002 | 0.024 ± 0.001 |
Depolymerization experiments were performed with copolymers composed of different fractions of WT and a mutant actin. The data showed no change in the depolymerization half-time for increasing ratios of the Y166C mutant. However, the M305L mutant led to a decrease of the depolymerization half-time, i.e. an increased rate of depolymerization, when the amount of this mutant was larger than 50 % compared with WT (Fig. 2g). These data suggest that the WT actin is able to maintain filament integrity over a broad range of mutant content.
Determination of the intrinsic and treadmilling ATPase rates of cardiac actin variants
To analyze the reason for the differences in the polymerization behavior, we examined the ATP turnover rates of monomeric WT and the Y166C and M305L mutants in Mg2+-form (i.e. in the presence of 0.2 mM MgCl2). For WT cardiac actin, we determined an ATPase rate of 0.64 ± 0.09 h−1 (Table 1), which is comparable to the rate of bovine cytoplasmatic G-actin (0.6 ± 0.11 h−1) as given by Schüler et al. [79]. We observed an increased ATPase rate (about 3.5-fold increase) for the M305L mutant and a considerably reduced rate (about 20-fold) for the Y166C mutant (Table 1).
When polymerized to F-actin filaments by addition of 2 mM MgCl2 and 0.1 M KCl, we obtained an ATPase rate of 2.38 ± 0.36 h−1 for WT actin, but for the Y166C and M305L mutants the rates were reduced to 0.57 ± 0.03 h−1 and 0.74 ± 0.21 h−1, respectively. These differences in the intrinsic G- and F-form ATPases indicate considerable deviations in the cycling ability of cardiac actin variants. “Premature” ATP-hydrolysis by the M305L variant in the monomeric state might interfere with its polymerization efficiency and cause a lower stability of M305L filaments, whereas the reduced ATPase in both G- and F-form might indicate that Y166C is able to form more stable filaments. Indeed, it has been shown that the ATPase rate of F-actin is a direct measure of the rate of actin treadmilling or cycling [70, 80]. Following this reasoning, our data suggest that the cycling rates for both mutant F-actin variants are reduced (three- to fourfold) compared to WT α-cardiac actin (Table 1), albeit due to different mechanisms. Surprisingly, the ATPase rate of M305L decreased under polymerizing conditions, whereas it increased for WT and Y166C as expected (see Table 1).
Polymerization of the cardiac actins in the presence of nucleating proteins
Under steady state conditions, new actin subunits predominantly associate to the plus-ends of F-actin filaments and migrate through the filament, until they dissociate from the slow-growing or minus-ends [81]. Due to the differences in the ATPase activity of filaments of WT, Y166C, and M305L actin, we measured their polymerization time courses separately either with gelsolin-capped plus-ends or minus-ends capped by the Arp2/3 complex (Fig. 3).
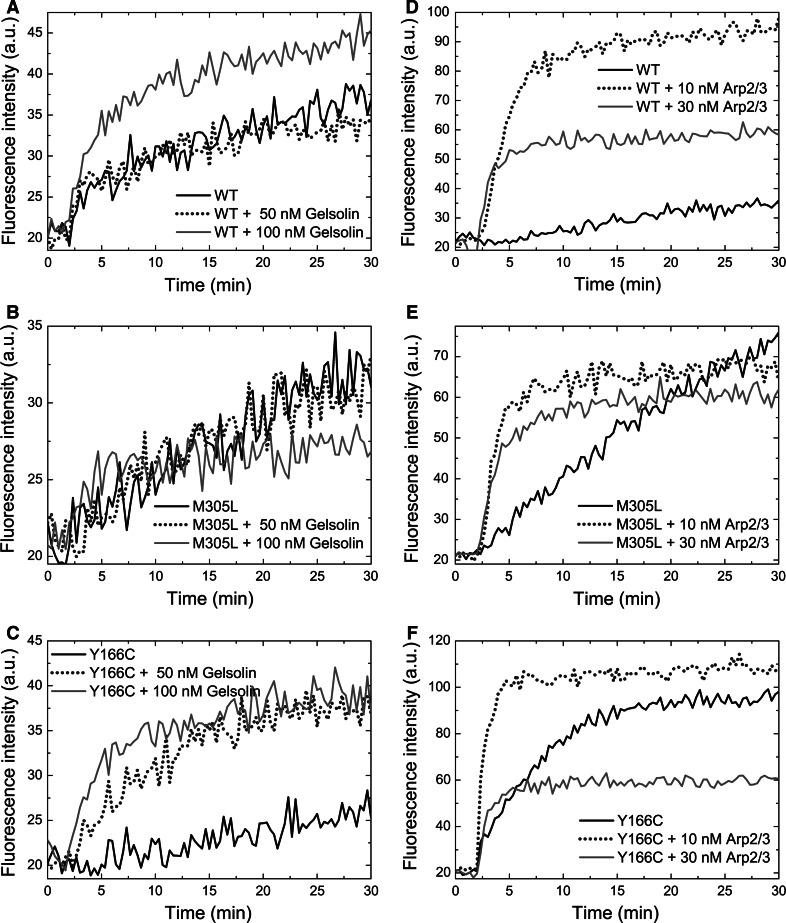
Fig. 3.
Effect of plus and minus end capping ABPs on the polymerization behaviour of cardiac actins. a–c Polymerization kinetics of 5 μM cardiac actin variants in the presence of intact cytoplasmic gelsolin. Gelsolin was added to actin in molar ratios of 1:100 (+50 nM gelsolin) and 1:50 (+100 nM gelsolin). d–f Identical experiments in the presence of Arp2/3 complex in molar ratios to actin of 1:500 (+10 nM Arp2/3) and 1:166 (+30 nM Arp2/3). Polymerization was initiated at t = 2 min. Arp2/3 complex was activated at t = 1 min by addition of 100 nM VCA peptide. a, d WT actin; b, e M305L actin; c, f Y166C actin. Fluorescence intensities are expressed in arbitrary units (AU)
Gelsolin nucleates actin polymerization by binding two actin molecules and thus forms polymerization competent nuclei [82]. Since it blocks the plus-ends, filament elongation can only occur at the minus-ends [83]. The addition of gelsolin to actin in a molar ratio of 1:50 led to an increased rate and extent of polymerization for WT and the Y166C mutant. In contrast, polymerization of M305L was largely unaffected, suggesting that monomers of this variant did not associate to the minus-end nuclei and therefore appeared to possess a significantly decreased affinity for the minus-ends of their F-actin filaments (Fig. 3a–c).
The Arp2/3 complex is responsible for the formation of branched F-actin filament networks [84, 85]. By capping the minus-end, activated Arp2/3 complex nucleates filament outgrowth by allowing subunit addition to the plus-ends. We tested the effect of Arp2/3 complex at 1:500 and 1:166 molar ratios to the actins (Fig. 3d–f). For WT cardiac actin, we observed considerable nucleation as revealed by an increased rate and extent of polymerization (Fig. 3d). Polymerization of M305L was only slightly stimulated, whereas the behavior of the Y166C mutant was intermediate (Fig. 3e, f). These data indicate a considerably reduced ability of M305L to associate to both plus- and minus-ends. In contrast, Y166C appeared to possess an increased ability to associate to the minus-end. Combined with the perturbed actin ATPase rates, these results show that M305L actin only weakly assembles at either end and thus might form less stable filaments. The Y166C actin filaments apparently exhibit a higher stability with a significantly reduced ATP turnover.
Ultrastructure of the human cardiac F-actin filaments
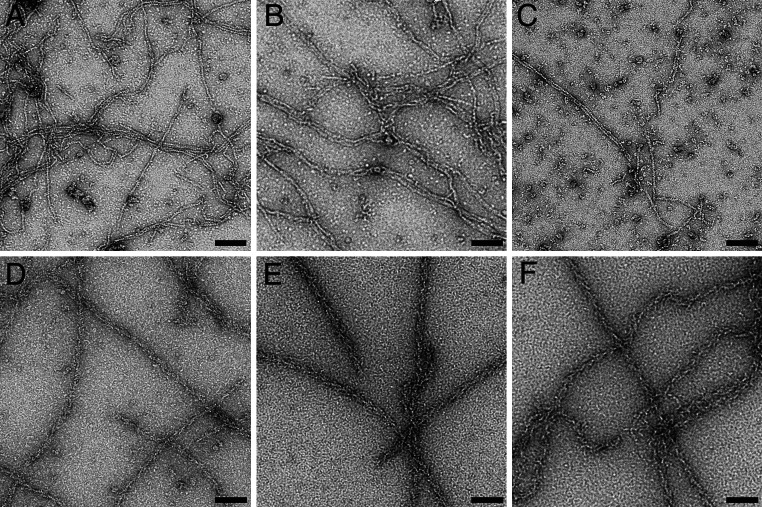
We performed transmission electron microscopy (EM) of negatively stained filaments polymerized from the cardiac actin variants alone and after decoration with TM and rabbit skeletal muscle myosin subfragment 1 (S1) (Fig. 4). WT and Y166C actin formed F-actin filaments of normal and similar appearance. However, the M305L mutant formed fewer and shorter filaments (Fig. 4c) corroborating our biochemical data. Interestingly, decoration with TM and skeletal muscle myosin-subfragment 1 (S1) stabilized the filaments, resulting in long filaments with the typical arrowhead appearance for all three cardiac actin variants (Fig. 4d–f).
Fig. 4.
Electron microscopic images of F-actin filaments of the cardiac actin variants after negative staining. a–c F-actins without tropomyosin; d–f plus tropomyosin and rabbit skeletal muscle subfragment 1. a, d WT actin; b, e Y166C mutant; c, f M305L mutant actin. Bars 100 nm
Reduced stimulation of the Mg2+-dependent ATPase of β-cardiac myosin by both mutant actin variants
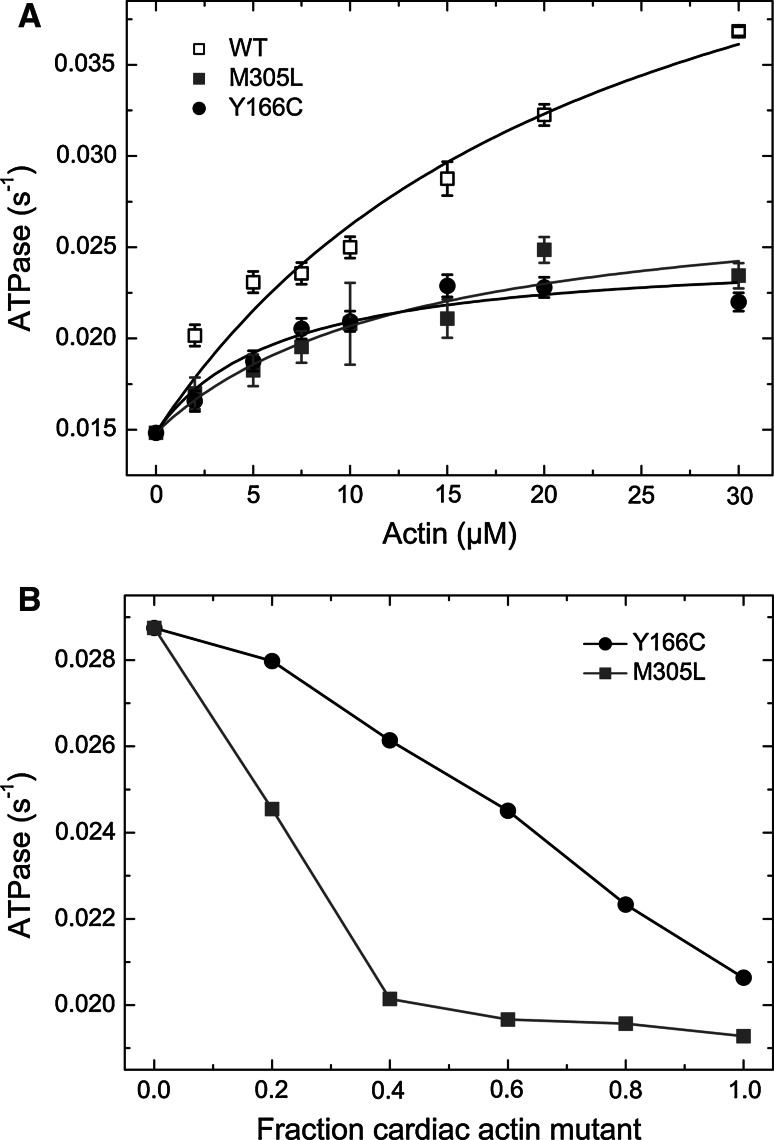
Impairment of the force productive interaction between F-actin and myosin is regarded as one of the main causes in the development of HCM. As a measure of the actomyosin interaction, we determined the stimulation of the Mg2+-dependent ATPase activity of Ss-β-cardiac myosin by increasing concentrations of the three cardiac F-actins (Fig. 5a). The observed rates were plotted against the actin concentration and fitted to a hyperbola [86]. The maximal stimulation of the ATPase rate of Ss-β-cardiac myosin (V max) by WT actin was 0.052 ± 0.006 s−1 and its apparent affinity (K app) was 13.7 ± 3.1 μM, comparable to previously determined rates of β-cardiac pig heavy meromyosin activated by rabbit α-skeletal F-actin (V max = 0.08 ± 0.01 s−1, K app = 8.6 ± 0.79 μM; [87, 88]). Both actin mutants led to a significant decrease in V max (0.028 s−1 and 0.024 s−1 for Y166C and M305L, respectively; Table 2). The apparent affinity of β-cardiac myosin to the cardiac actin mutants (K app) was in the same range as WT actin (~10 μM). Therefore, both mutant actins stimulated the myosin ATPase with about 50 % lower efficiency. Binding of TM to filamentous actin, composed of mutant actin, did not affect their reduced capability to stimulate β-cardiac myosin (data not shown).
Fig. 5.
Actin activated ATPase of β-cardiac myosin. a Stimulation of the Ss-β-cardiac myosin ATPase by pure α-cardiac actin WT and the mutants Y166C and M305L. Data from the fit are presented in Table 2. b Stimulation of the cardiac β-myosin ATPase by copolymers of WT and the mutant actins at increasing ratios
It has been proposed that cardiomyopathies are caused by a dominant negative effect of the mutated contractile protein (for reviews; see [89, 90])—also known as the poisonous peptide hypothesis [9]. Therefore, we tested the ability of mixtures of WT cardiac actin with each mutant to stimulate the ATPase of β-cardiac myosin. For this purpose, we copolymerized WT cardiac actin and a mutant at increasing ratios (0, 0.2, 0.4, 0.6, 0.8; and 1.0). At a final actin concentration of 15 μM we observed with increasing amounts of both mutant actins a steadily reduced ability to stimulate the cardiac β-myosin ATPase rate (Fig. 5b). The fact that already small amounts of mutant actin perturb the actomyosin coupling, strengthens the hypothesis of the dominant negative (“poisonous”) effect for the Y166C and M305L mutants in HCM.
Transfection of the cardiac actin variants into established cell lines
For transfection, we initially generated vectors containing WT cardiac actin and the mutants Y166C and M305L as N-terminally tagged EGFP-constructs (see “Materials and methods”). Examples of transfected NIH 3T3 fibroblasts and cardiomyocyte-derived HL-1 cells [71] are shown in Online Resource 5. In NIH 3T3 cells, WT and M305L actin are incorporated into stress fibers, whereas Y166C remained diffusely distributed throughout the cytoplasm and was surprisingly abundant in the nucleus (Online Resource 5). In HL-1 cells, all three actin variants localized in stress fibers or in occasionally detected sarcomere-like structures (large arrows in Online Resource 5). In a number of NIH 3T3 transfectants, we observed for all three EGFP-tagged variants intranuclear aggregates or rods as verified by anti-emerin staining (lower row in Online Resource 5). Most likely, these rods represent deposits of excess EGFP-actin in complex with cofilin as shown by double immunolabeling (lower row in Online Resource 5).
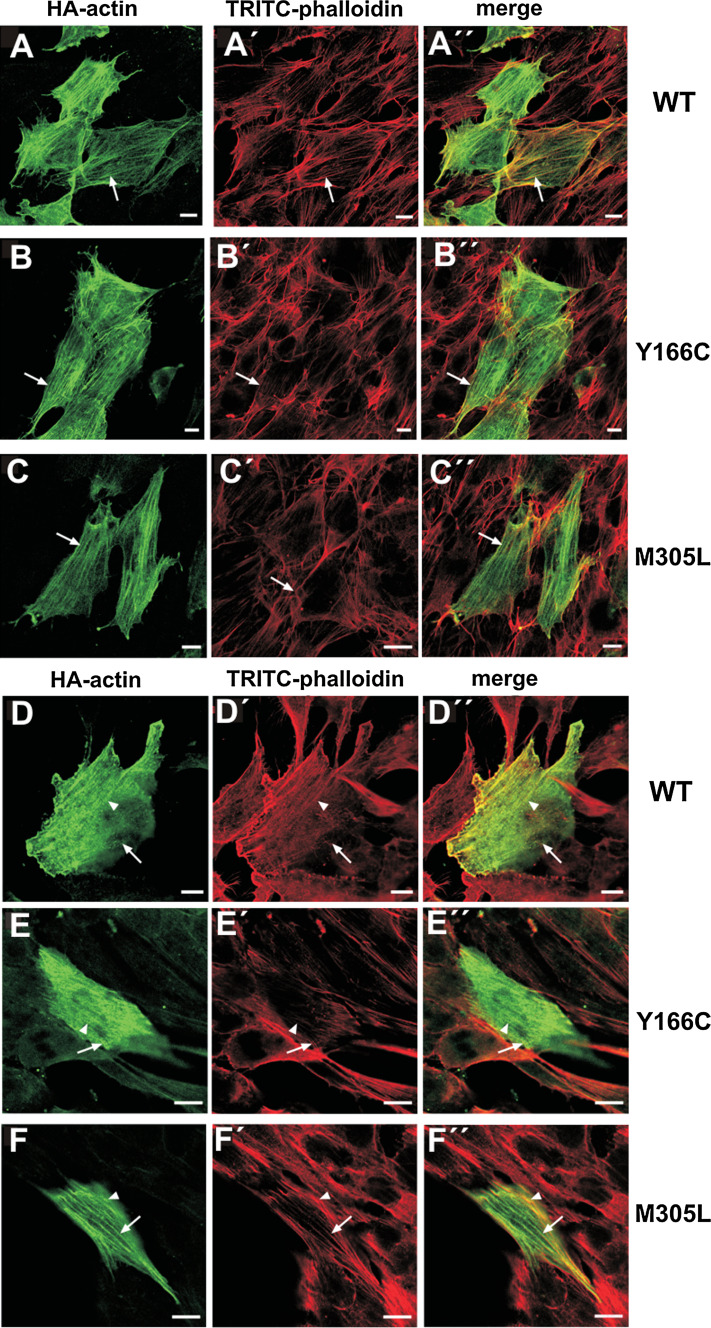
Because the large N-terminal EGFP-tag might hinder incorporation of the transfected cardiac actins into cellular structures, we generated corresponding vectors with a 27 amino acid triple HA-tag at the N-terminus. Transfection of NIH 3T3 (Fig. 6a–c) and HL-1 cells (Fig. 6d–f) with HA-tagged constructs gave an identical distribution for WT and M305L. In addition, the HA-tagged Y166C mutant incorporated into stress fibers of NIH 3T3 cells. We did not observe intranuclear rods of the HA-tagged actins. In summary, these data demonstrate that, dependent on the size of the N-terminal tag, all three cardiac actin variants were incorporated into the cytoskeleton, forming copolymers with endogenous cytoplasmic actin (NIH3T3 cells) and even incorporated into sarcomere-like structure of HL-1 cells.
Fig. 6.
Expression of HA-tagged cardiac actins in established cell lines. Confocal images of NIH 3T3 and HL-1 cells transfected with clones of the three cardiac actin variants. a–c gives NIH 3T3 cells and d–f cardiomyocyte-like HL-1 cells transfected with HA-tagged cardiac actins as indicated at the margins. The cells were immunostained with anti-HA-antibody (green) and counterstained with TRITC-phalloidin (red). For details, see “Materials and methods”. Scale bars 10 μm
Infection experiments of primary adult rat cardiomyocytes
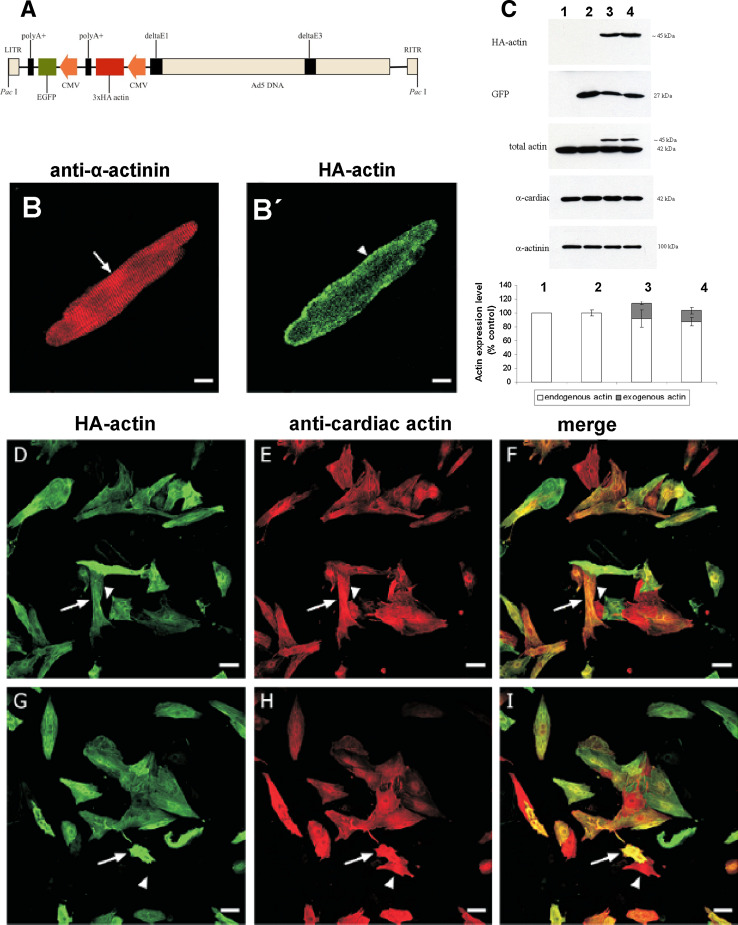
Next, we investigated the possibility to transfect primary rat cardiomyocytes with these cardiac actin variant constructs. However, it was found impossible to achieve transfection employing the techniques used for tissue culture cells. Therefore, adenoviral constructs were generated, which contained the three cardiac actin variants with an N-terminal HA-tag and a polycistronic EGFP-sequence in order to identify transfected cells (see “Materials and methods”; Fig. 7a). First, we attempted to achieve expression of WT cardiac actin into cardiomyocytes isolated from adult rats. It was possible to infect these cells as verified by EFGP expression (not shown). Immunostaining with the anti-HA antibody indicated expression of WT cardiac actin, which was localized and appeared incorporated into sarcomeres (as indicated by anti-α-actinin counterstain) throughout the whole cell, although it appeared concentrated in the submembranous zone (Fig. 7b, b′).
Fig. 7.
Infection of cardiomyocytes with adenoviral constructs containing mutant and WT actin. a Bicistronic adenoviral construct containing cardiac actin variant and EGFP. b Western blots of cell homogenates of control NRCs (lane 1); infected with empty virus particles (lane 2); containing WT- (lane 3); and Y166C adenoviral constructs (lane 4) immunostained with anti-HA to visualize expressed cardiac actin variants; with anti-GFP to visualize expressed EGFP; with an antibody (C11) recognizing total actin; with anti-α-cardiac actin (which was found to only recognize endogenous cardiac actin); and anti-α-actinin to verify equal loading. b, b′ ARC infected with WT-adenoviral constructs: b immunostained with anti-α-actinin and b′′ EGFP-fluorescence. Scale bars 10 μm. d–i NRCs infected with WT- (d–f) and Y166C-construct (g–i). Note that only cells expressing endogenous cardiac actin were infected with the cardiac variants visualized by anti-HA staining (for further details, see text). Scale bars (d–i) 50 μm
Infection experiments using neonatal rat cardiomyocytes
We repeated the infection experiments with cardiomyocytes isolated from newborn rats. Three days after isolation and maintenance in culture (see “Materials and methods”), the newborn rat cardiomyocytes (NRCs) were infected with the adenoviral vectors of WT cardiac actin and the Y166C and M305L mutants. After 24 h, successful infection was verified by EGFP fluorescence. After additional 48 h in culture, the NRCs were collected for homogenization or fixed with 4 % paraformaldehyde (PFA) for immunostaining.
Expression of the transfected cardiac actins was verified by western blotting of homogenates using anti-HA antibody (Fig. 7c). The data obtained demonstrated expression of HA-tagged WT actin and Y166C mutant but not of the M305L mutant. The reason for the failure to express the M305L mutant is presently not understood (see later). Staining of the immunoblot with a general anti-actin antibody, which stained both endogenous and exogenous actin (identified by its slightly higher molecular mass), and subsequent densitometry showed that the transfected WT and Y166C mutant actin were expressed to about 22 and 16 % of the total actin, respectively (Fig. 7c). In contrast, the anti-cardiac actin specific antibody recognized only the endogenous cardiac actin on immunoblots, but not the transfected HA-tagged actins probably due to masking of the N-terminus by the HA-tag, since this antibody is directed against the N-terminus of cardiac actin. Western blots with anti-GFP showed that EGFP was expressed in amounts similar to the HA-actins, indicating that the differences in the expression of the exogenous actins were due to differences in their translation, in particular since immunostaining with anti-α-actinin indicated identical loading of the homogenates (Fig. 7c).
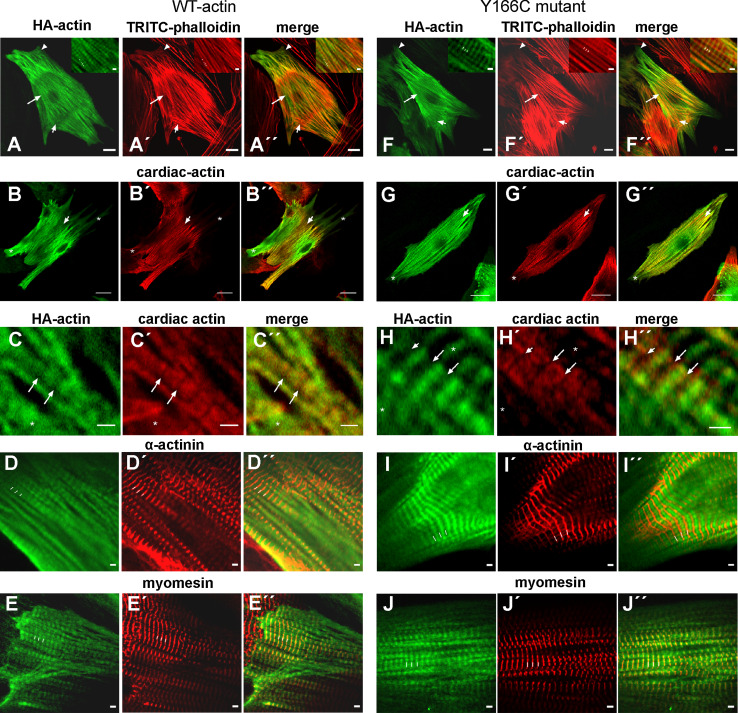
Immunostaining of infected NRCs with anti-HA-antibody (green) and anti-cardiac actin (red) allowed the differentiation of non-infected from infected cells. All cells positively stained with anti-HA antibody (WT: Fig. 7d–f; Y166C: Fig. 7g–i) were also stained with anti-cardiac actin, indicating that only NRCs had been infected to a high percentage (about 90 %; arrows in Fig. 7 d–i). Non-infected NRCs were stained only by anti-cardiac actin (arrowheads in Fig. 7d–i). Residual mesenchymal cells could only be visualized by nuclear staining with Hoechst 33342, but were anti-HA and anti-cardiac actin negative.
Exogenous cardiac actins are incorporated into sarcomeric thin filaments at their minus ends
Immunostaining of NRCs with anti-HA (green) and TRITC-phalloidin demonstrated that both exogenous WT and the Y166C mutant actin were incorporated in long F-actin filament bundles and in sarcomeric structures (Fig. 8a, f), which were often only observed in the center of the cells. Towards the cell periphery, the sarcomeric structures converged to thick bundles of stress fiber-like appearance, which further extended into lamellipodia-like protrusions of the plasma membrane (arrowheads in Fig. 8a, f). These fiber bundles were most probably sarcomere precursors, since they were also immunostained by anti-sarcomeric α-actinin. We often detected an abrupt alteration in the thin filament organization appearing as a clear borderline between sarcomeres and stress fiber bundles (Fig. 8a, f). Both structures were stained by TRITC-phalloidin and indicated that the stress fiber bundles were obviously running continuously through the central sarcomeric regions. It is well known that the templates for sarcomerogenesis are stress fiber bundles [91, 92]. It was, however, noted that in both WT- and Y166C-infected NRCs there was not always complete colocalization of the TRITC-phalloidin with the anti-HA staining as indicated by the arrows in Fig. 8a, f: long arrows give regions with sarcomeric organization and predominantly anti-HA-positive and short arrows regions of stress fiber bundles predominantly stained by TRITC-phalloidin. Surprisingly, more sarcomeric structures were detected by anti-HA- than phalloidin-staining, suggesting that the HA-tag diminished binding of TRITC-phalloidin to these actins. In spite of these staining differences, we also observed regions of colocalization of both stains, demonstrating that within these regions the exogenous actin variant co-polymerized with the endogenous actin (see the magnified insets in the images in Fig. 8a, f). Peripheral straight actin bundles became visible by both anti-HA and TRITC-phalloidin staining, although again there was not always complete colocalization (see merged images of Fig. 8a′′, f′′).
Fig. 8.
Confocal images of NRCs producing endogenous and HA-tagged exogenous actin. Confocal images of NRCs infected with WT (a–e) and Y166C (f–j) cardiac actin constructs. After infection, the or exogenous cardiac actins were visualized by anti-HA staining (green) and counterstained by TRITC-phalloidin (a, f′), anti-α-cardiac actin to visualize the endogenous cardiac actin (b′, c′, g′, h′), anti-α-actinin (d′, i′), or anti-myomesin (e′, j′. For details, see text. Bars (a, f) 10 μm; (b, g) 20 μm; bars in all insets 2 μm
Staining with anti-cardiac actin antibody again showed the presence of filamentous and sarcomeric structures and regions where both structures were built exclusively from exogenous actins (marked by small asterisks in Fig. 8b, c, g, h), whereas in other regions there were sarcomeres obviously containing both exo- and endogenous actins (short arrow in Fig. 8b, g).
Higher magnification indicated that the HA-positive bands were wider for exogenous WT- than Y166C-actin (see long arrows in Fig. 8c, h, which point to the middle of the gaps between the stained bands). Within the sarcomeres stained by both antibodies, there was no complete overlap between anti-HA and anti-cardiac actin stain (Fig. 8c, h). Indeed, the merged images (Fig. 8c′′, h′′) showed that the endogenous actin (red) was in many instances localized towards the dark gaps adjacent to the broader anti-HA-positive bands of exogenous actins. Therefore, the merged images at higher magnification (Fig. 8c′′, h′′) suggested that regions of endogenous (red) and exogenous actin (green) existed within the sarcomeres in an alternating sequence.
In order to identify the filamentous polarity of these regions and thus the side of incorporation of the expressed cardiac actins, we counterstained with anti-α-actinin (Fig. 8d, i) and -myomesin (Fig. 8e, j). Anti-α-actinin staining showed that the Z-line was localized between the anti-HA-positive bands suggesting that incorporation of the exogenous actins had not occurred adjacent to the Z-lines. In contrast, anti-myomesin staining demonstrated positive staining (red) in the middle of the HA-positive bands (green) indicating that the M-line was within the HA-positive band. Indeed, it has often been observed that the free thin filament ends come close to each other or even overlap in NRCs [92]. Thus, these data suggested that the incorporation of both expressed actins preferentially occurred at the thin filament minus-ends, in agreement with published data for skeletal and cardiac muscle cells [91, 93].
Infection and expression of exogenous actins leads to alterations in sarcomere lengths
The width of the anti-HA-positive bands after immunostaining (Fig. 8c, h) indicated that WT-cardiac actin was slightly more efficiently incorporated into the sarcomeres than the Y166C mutant. In addition, we observed differences in sarcomere length when comparing non-transfected control NRCs to NRCs transfected with empty vector and to NRCs transfected with WT cardiac actin or the Y166C mutant (see Fig. 8). Therefore, we determined the sarcomere lengths of these differently treated NRCs by measuring the distances between Z-lines as visualized by anti-α-actinin staining (Fig. 9). To this aim, NRCs were infected 24 h after their isolation and fixed and immunostained with anti-α-actinin 24 or 72 h after infection. The data obtained and shown in Fig. 9 repeatedly indicated an increased sarcomere width after transfection. After 24 h, the sarcomeres of all the infected cells appeared equally increased in width as compared to uninfected control NRCs. Seventy-two hours after infection, this increase was identical for the NRCs transfected with empty vector expressing only EGFP and the one leading to expression of WT actin, but absent for the Y166C mutant cardiac actin (Fig. 9). The sarcomeres containing Y166C actin were shortened by ~0.2 μm compared to those infected with WT actin, and reached the width of uninfected control NRCs (Fig. 9).
Discussion
Here, we demonstrate the expression of WT and the HCM-related Y166C and M305L mutants of cardiac actin in the baculovirus/Sf9 insect cell system and their affinity purification using the C-terminal half of gelsolin G4–6 [56]. The amounts obtained were sufficiently high to allow an extensive comparative biochemical study of these actins undisturbed by affinity tags. Although insect cells are capable of post-translational modifications, mass spectrometry analyses of the recombinant actins revealed that the unusual methylation of His73 does not occur, implying that the intrinsic properties might be varied as previously described for the β-actin isoform by Nyman et al. [94]. However, it has been reported [56] that recombinant actin, expressed in insect cells and purified by G4–6, possesses the same functionality as actin obtained from other sources when tested in standard assays, e.g., in polymerization experiments or in the actomyosin coupling [56].
Various tests like thermal stability, DNase I-inhibition, and the interaction with several ABPs, including thymosin β4 and gelsolin deletion constructs, did not indicate folding defects. Thus, our results strongly indicate that the expressed cardiac actins attained their regular native configuration. Two previous studies proposed that an improper fold of actin molecules caused myopathies [17, 95]. Vang and co-workers [17] studied the folding properties of eight cardiomyopathy-related actin mutations, synthesized in an in vitro cell-free coupled transcription/translation system derived from reticulocyte extracts. They found a significantly reduced protease resistance and strong impairment in attaining the native conformation for three out of six HCM mutations (E99K, P164A, and M305L), when assayed by TRiC- or DNase I-binding. Our results agree only partially with their observation, since the ability of the M305L mutant prepared by our procedure inhibited DNase I albeit with slightly reduced efficiency. Furthermore, its binding to DNase I was not impaired when assayed by NGE. Since expression in Sf9-cells is usually performed at 27 °C, it is conceivable that improper folding was less pronounced as compared to actins synthesized in a cell-free coupled transcription/translation system at 37 °C [17]. Furthermore, differences in the protein processing and folding machinery between insect cells and reticulocytes cannot be fully excluded and might explain the discrepant reports on the behavior of the M305L mutant. We are therefore confident that the purified cardiac actin variants used in our study possess the natural fold, and that the observed differences are most likely not due to the expression and purification procedures.
Both HCM actin mutants evidently affect the actin–actin interaction. The polymerization behavior was perturbed in a divergent fashion. Compared to WT, the Y166C mutant showed an increased, and the M305L mutant a reduced, rate and extent of polymerization. The divergent polymerization behavior appears to be due to differences in their intrinsic G- and F-actin ATPase activities. Within copolymers, however, the altered actin–actin interactions appeared to be compensated by WT actin unless the mutant fraction within the filament exceeded 60 % of total actin.
In order to mimic physiological conditions, we also tested the stabilizing effect of TM. The differences between Y166C and WT actin were diminished in the presence of TM, but the polymerization behavior of the M305L mutation was not restored to the WT level. Tyr166 is located at the bottom of subdomain three within the W-loop and directly involved in actin–actin intrastrand interactions [52]. As described in the “Introduction”, it has been postulated that the W-loop is involved in “nucleotide sensing” by hydrogen-bond formation between Y166 and Y169 [49, 50]. Our results suggest that the described local conformational change of the W-loop is not possible in Y166C-actin, and therefore this mutant might have a higher tendency to remain in the ATP-bound form. Indeed, the Y166C mutant possesses considerably lower G- and F-actin ATPase rates, suggesting that even after polymerization the incorporated subunits may remain more firmly bound to the minus-ends leading to reduced treadmilling.
On the other hand, the M305L mutation has a direct effect on nucleotide hydrolysis, as the adenine moiety of ATP or ADP fits into a hydrophobic pocket between subdomains 3 and 4, which includes the Met305 residue [51, 96]. This mutation alters the hydrolysis rate of the actin bound ATP substantially, as shown by the enhanced G-actin ATPase, which will cause a large proportion of monomeric M305L actin to contain ADP, and consequently lead to a reduced ability to polymerize and/or to dissociate more readily from filament ends, since the affinity of ADP-G-actin to filament ends is generally reduced (see also [80, 97]). This assumption is supported by the observed lower ATPase rate of polymerized M305L actin and its reduced rate to associate to both filament ends. In agreement with this assumption, we observed an increased tendency of M305L F-actin to depolymerize, which within the sarcomere might lead to thin filament instability and decreased force generation.
Both cardiomyopathic mutant actins showed a 50 % reduced ability to stimulate the ATPase activity of cardiac β-myosin. This observation is consistent with the assumption that an insufficient actomyosin interaction causes a reduced contractile force and might result in stimuli for the induction of an adaptive hypertrophy finally leading to HCM [13, 14]. Both mutations are localized on the surface of subdomain 3 (Fig. 1; [51]), and available structural data of EM-reconstructions of the acto–myosin interface demonstrate that their side chains are situated in close proximity to residues directly involved in myosin motor domain binding [98–100]. In addition, it has been shown that the myosin-actin interface comprises two adjacent actin subunits along the long-pitch helix including residue Y166 of the “upper” actin [98].
We also analyzed the ability of hybrid F-actin filaments to stimulate the myosin ATPase and observed a clear suppressive effect even for low fractions of mutated protein. Copolymers of WT cardiac actin with increasing ratios of the Y166C or M305L mutant led to a gradual decrease in the β-myosin ATPase stimulating capacity, consistent with the proposal that mutations of sarcomeric proteins exert a dominant negative (=poisonous) effect in familial cardiomyopathies. Hence, a mixture of normal and abnormal components appears to be sufficient to disrupt the integrity of the overall structure based on a “weak links in a chain” principle [89–91]. The mutant-specific magnitude of the suppressive effect correlates with observations showing that carriers of the Y166C mutation have a lower penetrance and much less severe disease symptoms than carriers of the M305L mutation [47].
Finally, we analyzed the behavior of the actin variants under in vivo conditions by transfection experiments. First, we employed a number of established fibroblastic cell lines and the cardiomyocyte HL-1 line [71] in transfection experiments and demonstrated incorporation of the EGFP-tagged WT and M305L variants into stress fibers or sarcomere-like structures, whereas the EGFP-Y166C mutant remained diffusely within the cytoplasm. When HA-tagged, all three actin variants were incorporated into stress fibers, indicating that the size of the N-terminal tag influences the intracellular distribution of the transfected actin. The transfected cardiac actins formed copolymers with the endogenous β-actin as verified by anti-β-actin-specific antibodies. In transfected cardiomyocyte HL-1 cells, which occasionally form sarcomeric structures [71], we again observed incorporation of the three HA-tagged cardiac actins into their stress fibers and also the sarcomeric structures.
To investigate the behavior of the mutants in a more relevant cell system, we infected primary rat cardiomyocytes isolated from adult (ARCs) or newborn (NRCs) rats with adenoviral constructs transferring these variants as HA-tagged actins. In adult cardiomyocytes, we obtained expression of WT actin, which was incorporated into sarcomeres predominantly located at the cell periphery. In contrast, in infected NRCs, the expressed actins were distributed throughout the entire cell. We succeeded to infect NRCs with HA-tagged WT and the Y166C actin, but unfortunately not with the M305L mutant. The exact reason for this failure is still unclear. We suspect that shortly after expression the M305L mutant was degraded; however, this hypothesis was not directly tested.
Both WT and the Y166C-mutant were incorporated into stress fiber-like bundles and sarcomeric thin filaments as verified by double immunostaining with anti-HA and TRITC-phalloidin. Double staining of the endogenous actin by an anti-cardiac actin antibody and of exogenous by anti-HA demonstrated the formation of hybrid filaments, although double staining showed that a number of filamentous and sarcomeric structures were solely composed of the exogenous actins. Nevertheless, a number of sarcomeres were detected, which were stained with both anti-HA and -cardiac actin antibodies in an alternating manner. Thus, our data give for the first time evidence that a mutated cardiac actin can also form hybrid filaments with the endogenous cardiac actin in intact cardiomyocytes. Examination of these sarcomeres by double immunostaining with anti-HA and with anti-α-actinin or -myomesin showed incorporation of the transfected actin at the middle of the sarcomeres, i.e. at the thin filament minus-ends, in agreement with published reports for skeletal and cardiac muscle cells [93].
Surprisingly, we observed differences in sarcomere lengths in infected NRCs. The extent of sarcomere lengthening was identical 24 h after infection with empty vector only expressing EGFP and transfection with both HA-actins. It therefore appears possible that mere expression of EGFP might have altered the ionic balance of the myofilaments and induced a kind of sarcomere stretching. Seventy-two hours after transfection, we found a significant decrease in sarcomere length back to the control width only for the Y166C-variant. At present, we are unable to give an explanation for these actin variant-dependent changes in sarcomere lengths. We hope that future electron microscopic analyses of infected NRCs will shed light on this issue.
In summary, we studied by biochemical and cell biological methods the impact of two cardiac actin mutations that are known to occur in HCM patients. Our data emphasize the complexity of thin filament organization and suggest explanations for the differing phenotypes associated with the Y166C and M305L mutations. The primary defect will most probably be due to the mutation-specific disturbances in the actin–actin and acto–myosin interactions. These disturbances are most likely the molecular basis for the subsequent initiation of the hypertrophic pathway finally leading to the pathogenesis of HCM. We are, however, well aware of the fact that, within patients, a number of other factors like the genetic background, environmental factors, and other diseases will greatly influence the penetrance and severity of inherited myopathies. Therefore, future progress should also include in vivo studies using animal models [101], and integrate pharmacological approaches such as RNAi therapy.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank Prof. Pott (Bochum) for supplying ARCs and Dipl. Biol. S. Fister and Dr. H. Milting (Herz- und Diabeteszentrum NRW, Bad Oeyenhausen) for the human cardiac actin clones, and Prof. Claycomb (New Orleans, Lousiana, USA) for supplying the HL-1 cells. Mass spectrometry analysis was kindly performed at the Hannover Medical School (MHH) Core Unit Mass Spectrometry/Proteomics by A. Pich. This work was supported by the ‘Deutsche Forschungsgemeinschaft’ Grant MA 807/17-2 (to H.G.M.), MA 1081/11-2 (to D.J.M), RA 1781/1-1 (to S.R.), the ‘Fonds der Chemischen Industrie’ Grant 684052 (to E.B.), the Hannover Biomedical Research School (to M.B.R.), the Deutsche Akademische Ausstauschdienst (DAAD) and FoRUM der Ruhr-Universität Bochum (to A.J.M.), the Schweizer National Fond (to C.A.S.), and the Max-Planck Society (to S.R. and E.B.).
Conflict of interest
None.
Footnotes
M. Müller and A. J. Mazur contributed equally to this work.
References
- 1.Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.CIR.92.4.785. [DOI] [PubMed] [Google Scholar]
- 2.Maron BJ. Hypertrophic cardiomyopathy. Lancet. 1997;350:127–133. doi: 10.1016/S0140-6736(97)01282-8. [DOI] [PubMed] [Google Scholar]
- 3.Seidman JG, Seidman C. The genetic basis for cardiomyopathy: from mutation identification to mechanistic paradigms. Cell. 2001;104:557–567. doi: 10.1016/S0092-8674(01)00242-2. [DOI] [PubMed] [Google Scholar]
- 4.Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol. 2008;19:104–110. doi: 10.1111/j.1540-8167.2007.00965.x. [DOI] [PubMed] [Google Scholar]
- 5.Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 6.Marian AJ, Roberts R. The molecular genetic basis for hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2001;33:655–670. doi: 10.1006/jmcc.2001.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arad M, Seidman JG, Seidman CE. Phenotypic diversity in hypertrophic cardiomyopathy. Hum Mol Genet. 2002;11:2499–2506. doi: 10.1093/hmg/11.20.2499. [DOI] [PubMed] [Google Scholar]
- 8.Geier C, Perrot A, Ozcelik C, Binner P, Counsell D, Hoffmann K, Pilz B, Martiniak Y, Gehmlich K, van der Ven PF, Furst DO, Vornwald A, von Hodenberg E, Nurnberg P, Scheffold T, Dietz R, Osterziel KJ. Mutations in the human muscle LIM protein gene in families with hypertrophic cardiomyopathy. Circulation. 2003;107:1390–1395. doi: 10.1161/01.CIR.0000056522.82563.5F. [DOI] [PubMed] [Google Scholar]
- 9.Yang Q, Sanbe A, Osinska H, Hewett TE, Klevitsky R, Robbins J. A mouse model of myosin binding protein C human familial hypertrophic cardiomyopathy. J Clin Invest. 1998;102:1292–1300. doi: 10.1172/JCI3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown JH, Del Re DP, Sussman MA. The Rac and Rho hall of fame: a decade of hypertrophic signaling hits. Circ Res. 2006;98:730–742. doi: 10.1161/01.RES.0000216039.75913.9e. [DOI] [PubMed] [Google Scholar]
- 11.Hannigan GE, Coles JG, Dedhar S. Integrin-linked kinase at the heart of cardiac contractility, repair, and disease. Circ Res. 2007;100:1408–1414. doi: 10.1161/01.RES.0000265233.40455.62. [DOI] [PubMed] [Google Scholar]
- 12.Barry SP, Davidson SM, Townsend PA. Molecular regulation of cardiac hypertrophy. Int J Biochem Cell Biol. 2008;40:2023–2039. doi: 10.1016/j.biocel.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Lankford EB, Epstein ND, Fananapazir L, Sweeney HL. Abnormal contractile properties of muscle fibers expressing beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J Clin Invest. 1995;95:1409–1414. doi: 10.1172/JCI117795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watkins H, Seidman CE, Seidman JG, Feng HS, Sweeney HL. Expression and functional assessment of a truncated cardiac troponin T that causes hypertrophic cardiomyopathy. Evidence for a dominant negative action. J Clin Invest. 1996;98:2456–2461. doi: 10.1172/JCI119063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 16.Mogensen J, Klausen IC, Pedersen AK, Egeblad H, Bross P, Kruse TA, Gregersen N, Hansen PS, Baandrup U, Borglum AD. Alpha-cardiac actin is a novel disease gene in familial hypertrophic cardiomyopathy. J Clin Invest. 1999;103:R39–R43. doi: 10.1172/JCI6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vang S, Corydon TJ, Borglum AD, Scott MD, Frydman J, Mogensen J, Gregersen N, Bross P. Actin mutations in hypertrophic and dilated cardiomyopathy cause inefficient protein folding and perturbed filament formation. FEBS J. 2005;272:2037–2049. doi: 10.1111/j.1742-4658.2005.04630.x. [DOI] [PubMed] [Google Scholar]
- 18.Bathe FS, Rommelaere H, Machesky LM. Phenotypes of myopathy-related actin mutants in differentiated C2C12 myotubes. BMC Cell Biol. 2007;8:2. doi: 10.1186/1471-2121-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzger JM, Michele DE, Rust EM, Borton AR, Westfall MV. Sarcomere thin filament regulatory isoforms. Evidence of a dominant effect of slow skeletal troponin I on cardiac contraction. J Biol Chem. 2003;278:13118–13123. doi: 10.1074/jbc.M212601200. [DOI] [PubMed] [Google Scholar]
- 20.Sussman MA, Baque S, Uhm CS, Daniels MP, Price RL, Simpson D, Terracio L, Kedes L. Altered expression of tropomodulin in cardiomyocytes disrupts the sarcomeric structure of myofibrils. Circ Res. 1998;82:94–105. doi: 10.1161/01.RES.82.1.94. [DOI] [PubMed] [Google Scholar]
- 21.Marian AJ, Yu QT, Mann DL, Graham FL, Roberts R. Expression of a mutation causing hypertrophic cardiomyopathy disrupts sarcomere assembly in adult feline cardiac myocytes. Circ Res. 1995;77:98–106. doi: 10.1161/01.RES.77.1.98. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Moncman CL, Winkelmann DA. Mutations in the motor domain modulate myosin activity and myofibril organization. J Cell Sci. 2003;116:4227–4238. doi: 10.1242/jcs.00709. [DOI] [PubMed] [Google Scholar]
- 23.Frankel S, Condeelis J, Leinwand L. Expression of actin in Escherichia coli. Aggregation, solubilization, and functional analysis. J Biol Chem. 1990;265:17980–17987. [PubMed] [Google Scholar]
- 24.Aspenstrom P, Karlsson R. Interference with myosin subfragment-1 binding by site-directed mutagenesis of actin. Eur J Biochem. 1991;200:35–41. doi: 10.1111/j.1432-1033.1991.tb21045.x. [DOI] [PubMed] [Google Scholar]
- 25.Johara M, Toyoshima YY, Ishijima A, Kojima H, Yanagida T, Sutoh K. Charge-reversion mutagenesis of Dictyostelium actin to map the surface recognized by myosin during ATP-driven sliding motion. Proc Natl Acad Sci USA. 1993;90:2127–2131. doi: 10.1073/pnas.90.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noguchi TQ, Kanzaki N, Ueno H, Hirose K, Uyeda TQ. A novel system for expressing toxic actin mutants in Dictyostelium and purification and characterization of a dominant lethal yeast actin mutant. J Biol Chem. 2007;282:27721–27727. doi: 10.1074/jbc.M703165200. [DOI] [PubMed] [Google Scholar]
- 27.Joel PB, Fagnant PM, Trybus KM. Expression of a nonpolymerizable actin mutant in Sf9 cells. Biochemistry. 2004;43:11554–11559. doi: 10.1021/bi048899a. [DOI] [PubMed] [Google Scholar]
- 28.Bookwalter CS, Trybus KM. Functional consequences of a mutation in an expressed human alpha-cardiac actin at a site implicated in familial hypertrophic cardiomyopathy. J Biol Chem. 2006;281:16777–16784. doi: 10.1074/jbc.M512935200. [DOI] [PubMed] [Google Scholar]
- 29.Miller BM, Trybus KM. Functional effects of nemaline myopathy mutations on human skeletal alpha-actin. J Biol Chem. 2008;283:19379–19388. doi: 10.1074/jbc.M801963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwasa M, Maeda K, Narita A, Maeda Y, Oda T. Dual roles of Gln137 of actin revealed by recombinant human cardiac muscle alpha-actin mutants. J Biol Chem. 2008;283:21045–21053. doi: 10.1074/jbc.M800570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dunn AY, Melville MW, Frydman J. Review: cellular substrates of the eukaryotic chaperonin TRiC/CCT. J Struct Biol. 2001;135:176–184. doi: 10.1006/jsbi.2001.4380. [DOI] [PubMed] [Google Scholar]
- 32.Kodama A, Lechler T, Fuchs E. Coordinating cytoskeletal tracks to polarize cellular movements. J Cell Biol. 2004;167:203–207. doi: 10.1083/jcb.200408047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sheterline P, Clayton J, Sparrow J. Actin. Protein Profile. 1995;2:1–103. [PubMed] [Google Scholar]
- 34.Huxley HE. Fifty years of muscle and the sliding filament hypothesis. Eur J Biochem. 2004;271:1403–1415. doi: 10.1111/j.1432-1033.2004.04044.x. [DOI] [PubMed] [Google Scholar]
- 35.Vandekerckhove J, Weber K. At least six different actins are expressed in a higher mammal: an analysis based on the amino acid sequence of the amino-terminal tryptic peptide. J Mol Biol. 1978;126:783–802. doi: 10.1016/0022-2836(78)90020-7. [DOI] [PubMed] [Google Scholar]
- 36.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schoenenberger CA, Mannherz HG, Jockusch BM. Actin: from structural plasticity to functional diversity. Eur J Cell Biol. 2011;90:797–804. doi: 10.1016/j.ejcb.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Pfaendtner J, Lyman E, Pollard TD, Voth GA. Structure and dynamics of the actin filament. J Mol Biol. 2010;396:252–263. doi: 10.1016/j.jmb.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matsson H, Eason J, Bookwalter CS, Klar J, Gustavsson P, Sunnegardh J, Enell H, Jonzon A, Vikkula M, Gutierrez I, Granados-Riveron J, Pope M, Bu’Lock F, Cox J, Robinson TE, Song F, Brook DJ, Marston S, Trybus KM, Dahl N. Alpha-cardiac actin mutations produce atrial septal defects. Hum Mol Genet. 2008;17:256–265. doi: 10.1093/hmg/ddm302. [DOI] [PubMed] [Google Scholar]
- 40.Zhu M, Yang T, Wei S, DeWan AT, Morell RJ, Elfenbein JL, Fisher RA, Leal SM, Smith RJ, Friderici KH. Mutations in the gamma-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26) Am J Hum Genet. 2003;73:1082–1091. doi: 10.1086/379286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Wijk E, Krieger E, Kemperman MH, De Leenheer EM, Huygen PL, Cremers CW, Cremers FP, Kremer H. A mutation in the gamma actin 1 (ACTG1) gene causes autosomal dominant hearing loss (DFNA20/26) J Med Genet. 2003;40:879–884. doi: 10.1136/jmg.40.12.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Procaccio V, Salazar G, Ono S, Styers ML, Gearing M, Davila A, Jimenez R, Juncos J, Gutekunst CA, Meroni G, Fontanella B, Sontag E, Sontag JM, Faundez V, Wainer BH. A mutation of beta -actin that alters depolymerization dynamics is associated with autosomal dominant developmental malformations, deafness, and dystonia. Am J Hum Genet. 2006;78:947–960. doi: 10.1086/504271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar A, Crawford K, Close L, Madison M, Lorenz J, Doetschman T, Pawlowski S, Duffy J, Neumann J, Robbins J, Boivin GP, O’Toole BA, Lessard JL. Rescue of cardiac alpha-actin-deficient mice by enteric smooth muscle gamma-actin. Proc Natl Acad Sci USA. 1997;94:4406–4411. doi: 10.1073/pnas.94.9.4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morita H, Rehm HL, Menesses A, McDonough B, Roberts AE, Kucherlapati R, Towbin JA, Seidman JG, Seidman CE. Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med. 2008;358:1899–1908. doi: 10.1056/NEJMoa075463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olson TM, Doan TP, Kishimoto NY, Whitby FG, Ackerman MJ, Fananapazir L. Inherited and de novo mutations in the cardiac actin gene cause hypertrophic cardiomyopathy. J Mol Cell Cardiol. 2000;32:1687–1694. doi: 10.1006/jmcc.2000.1204. [DOI] [PubMed] [Google Scholar]
- 46.Monserrat L, Hermida-Prieto M, Fernandez X, Rodriguez I, Dumont C, Cazon L, Cuesta MG, Gonzalez-Juanatey C, Peteiro J, Alvarez N, Penas-Lado M, Castro-Beiras A. Mutation in the alpha-cardiac actin gene associated with apical hypertrophic cardiomyopathy, left ventricular non-compaction, and septal defects. Eur Heart J. 2007;28:1953–1961. doi: 10.1093/eurheartj/ehm239. [DOI] [PubMed] [Google Scholar]
- 47.Mogensen J, Perrot A, Andersen PS, Havndrup O, Klausen IC, Christiansen M, Bross P, Egeblad H, Bundgaard H, Osterziel KJ, Haltern G, Lapp H, Reinecke P, Gregersen N, Borglum AD. Clinical and genetic characteristics of alpha cardiac actin gene mutations in hypertrophic cardiomyopathy. J Med Genet. 2004;41:e10. doi: 10.1136/jmg.2003.010447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation. 2003;108:445–451. doi: 10.1161/01.CIR.0000080896.52003.DF. [DOI] [PubMed] [Google Scholar]
- 49.Kudryashov DS, Grintsevich EE, Rubenstein PA, Reisler E. A nucleotide state-sensing region on actin. J Biol Chem. 2010;285:25591–25601. doi: 10.1074/jbc.M110.123869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zheng X, Diraviyam K, Sept D. Nucleotide effects on the structure and dynamics of actin. Biophys J. 2007;93:1277–1283. doi: 10.1529/biophysj.107.109215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 52.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 53.Lorenz M, Popp D, Holmes KC. Refinement of the F-actin model against X-ray fiber diffraction data by the use of a directed mutation algorithm. J Mol Biol. 1993;234:826–836. doi: 10.1006/jmbi.1993.1628. [DOI] [PubMed] [Google Scholar]
- 54.Houmeida A, Bennes R, Benyamin Y, Roustan C. Sequences of actin implicated in the polymerization process: a simplified mathematical approach to probe the role of these segments. Biophys Chem. 1995;56:201–214. doi: 10.1016/0301-4622(95)00038-Y. [DOI] [PubMed] [Google Scholar]
- 55.Way M, Gooch J, Pope B, Weeds AG. Expression of human plasma gelsolin in Escherichia coli and dissection of actin binding sites by segmental deletion mutagenesis. J Cell Biol. 1989;109:593–605. doi: 10.1083/jcb.109.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohki T, Ohno C, Oyama K, Mikhailenko SV, Ishiwata S. Purification of cytoplasmic actin by affinity chromatography using the C-terminal half of gelsolin. Biochem Biophys Res Commun. 2009;383:146–150. doi: 10.1016/j.bbrc.2009.03.144. [DOI] [PubMed] [Google Scholar]
- 57.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 58.Fister S (2004) Expression von humanem kardialem alpha-Aktin zur Untersuchung von Mutationen der hypertrophen Kardiomyopathie. Diploma thesis, Universität Bielefeld, Bielefeld
- 59.Zalevsky J, Lempert L, Kranitz H, Mullins RD. Different WASP family proteins stimulate different Arp2/3 complex-dependent actin-nucleating activities. Curr Biol. 2001;11:1903–1913. doi: 10.1016/S0960-9822(01)00603-0. [DOI] [PubMed] [Google Scholar]
- 60.Coulton A, Lehrer SS, Geeves MA. Functional homodimers and heterodimers of recombinant smooth muscle tropomyosin. Biochemistry. 2006;45:12853–12858. doi: 10.1021/bi0613224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jacques AM, Briceno N, Messer AE, Gallon CE, Jalilzadeh S, Garcia E, Kikonda-Kanda G, Goddard J, Harding SE, Watkins H, Esteban MT, Tsang VT, McKenna WJ, Marston SB. The molecular phenotype of human cardiac myosin associated with hypertrophic obstructive cardiomyopathy. Cardiovasc Res. 2008;79:481–491. doi: 10.1093/cvr/cvn094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pant K, Watt J, Greenberg M, Jones M, Szczesna-Cordary D, Moore JR. Removal of the cardiac myosin regulatory light chain increases isometric force production. FASEB J. 2009;23:3571–3580. doi: 10.1096/fj.08-126672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 64.Mannherz HG, Ballweber E, Galla M, Villard S, Granier C, Steegborn C, Schmidtmann A, Jaquet K, Pope B, Weeds AG. Mapping the ADF/cofilin binding site on monomeric actin by competitive cross-linking and peptide array: evidence for a second binding site on monomeric actin. J Mol Biol. 2007;366:745–755. doi: 10.1016/j.jmb.2006.11.100. [DOI] [PubMed] [Google Scholar]
- 65.Ajtai K, Venyaminov S. CD study of the actin DNase I complex. FEBS Lett. 1983;151:94–96. doi: 10.1016/0014-5793(83)80350-0. [DOI] [PubMed] [Google Scholar]
- 66.Greenfield NJ. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat Protoc. 2006;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mannherz HG, Leigh JB, Leberman R, Pfrang H. A specific 1:1 G-actin:DNAase i complex formed by the action of DNAase I on F-actin. FEBS Lett. 1975;60:34–38. doi: 10.1016/0014-5793(75)80412-1. [DOI] [PubMed] [Google Scholar]
- 68.Kouyama T, Mihashi K. Fluorimetry study of N-(1-pyrenyl)iodoacetamide-labelled F-actin. Local structural change of actin protomer both on polymerization and on binding of heavy meromyosin. Eur J Biochem. 1981;114:33–38. doi: 10.1111/j.1432-1033.1981.tb06167.x. [DOI] [PubMed] [Google Scholar]
- 69.Mannherz HG, Brehme H, Lamp U. Depolymerisation of F-actin to G-actin and its repolymerisation in the presence of analogs of adenosine triphosphate. Eur J Biochem. 1975;60:109–116. doi: 10.1111/j.1432-1033.1975.tb20981.x. [DOI] [PubMed] [Google Scholar]
- 70.Mazur AJ, Gremm D, Dansranjavin T, Litwin M, Jockusch BM, Wegner A, Weeds AG, Mannherz HG. Modulation of actin filament dynamics by actin-binding proteins residing in lamellipodia. Eur J Cell Biol. 2010;89:402–413. doi: 10.1016/j.ejcb.2009.10.017. [DOI] [PubMed] [Google Scholar]
- 71.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci USA. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bechem M, Pott L, Rennebaum H. Atrial muscle cells from hearts of adult guinea-pigs in culture: a new preparation for cardiac cellular electrophysiology. Eur J Cell Biol. 1983;31:366–369. [PubMed] [Google Scholar]
- 73.Przygodzki T, Sokal A, Bryszewska M. Calcium ionophore A23187 action on cardiac myocytes is accompanied by enhanced production of reactive oxygen species. Biochim Biophys Acta. 2005;1740:481–488. doi: 10.1016/j.bbadis.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 74.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grove BK, Kurer V, Lehner C, Doetschman TC, Perriard JC, Eppenberger HM. A new 185,000-dalton skeletal muscle protein detected by monoclonal antibodies. J Cell Biol. 1984;98:518–524. doi: 10.1083/jcb.98.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schüler H, Lindberg U, Schutt CE, Karlsson R. Thermal unfolding of G-actin monitored with the DNase I-inhibition assay stabilities of actin isoforms. Eur J Biochem. 2000;267:476–486. doi: 10.1046/j.1432-1327.2000.01023.x. [DOI] [PubMed] [Google Scholar]
- 77.Mannherz HG, Goody RS, Konrad M, Nowak E. The interaction of bovine pancreatic deoxyribonuclease I and skeletal muscle actin. Eur J Biochem. 1980;104:367–379. doi: 10.1111/j.1432-1033.1980.tb04437.x. [DOI] [PubMed] [Google Scholar]
- 78.Safer D. An electrophoretic procedure for detecting proteins that bind actin monomers. Anal Biochem. 1989;178:32–37. doi: 10.1016/0003-2697(89)90351-5. [DOI] [PubMed] [Google Scholar]
- 79.Schüler H, Karlsson R, Schutt CE, Lindberg U (2006) The connection between actin ATPase and polymerization. In: Advances in Molecular and Cell Biology, vol 37. Elsevier, New York, pp 49–66
- 80.Carlier MF. Actin polymerization and ATP hydrolysis. Adv Biophys. 1990;26:51–73. doi: 10.1016/0065-227X(90)90007-G. [DOI] [PubMed] [Google Scholar]
- 81.Wegner A. Head to tail polymerization of actin. J Mol Biol. 1976;108:139–150. doi: 10.1016/S0022-2836(76)80100-3. [DOI] [PubMed] [Google Scholar]
- 82.Bryan J, Kurth MC. Actin-gelsolin interactions. Evidence for two actin-binding sites. J Biol Chem. 1984;259:7480–7487. [PubMed] [Google Scholar]
- 83.Janmey PA, Chaponnier C, Lind SE, Zaner KS, Stossel TP, Yin HL. Interactions of gelsolin and gelsolin-actin complexes with actin. Effects of calcium on actin nucleation, filament severing, and end blocking. Biochemistry. 1985;24:3714–3723. doi: 10.1021/bi00335a046. [DOI] [PubMed] [Google Scholar]
- 84.Kelleher JF, Atkinson SJ, Pollard TD. Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J Cell Biol. 1995;131:385–397. doi: 10.1083/jcb.131.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/S0092-8674(03)00120-X. [DOI] [PubMed] [Google Scholar]
- 86.Michaelis L, Menten ML. Die Kinetik der Invertin Wirkung. Biochem Z. 1913;49:333–369. [Google Scholar]
- 87.Taylor RS, Weeds AG. The magnesium-ion-dependent adenosine triphosphatase of bovine cardiac Myosin and its subfragment-1. Biochem J. 1976;159:301–315. doi: 10.1042/bj1590301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pereira JS, Pavlov D, Nili M, Greaser M, Homsher E, Moss RL. Kinetic differences in cardiac myosins with identical loop 1 sequences. J Biol Chem. 2001;276:4409–4415. doi: 10.1074/jbc.M006441200. [DOI] [PubMed] [Google Scholar]
- 89.Wilkie AO. The molecular basis of genetic dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Schwartz K. Familial hypertrophic cardiomyopathy. Nonsense versus missense mutations. Circulation. 1995;91:2865–2867. doi: 10.1161/01.CIR.91.12.2865. [DOI] [PubMed] [Google Scholar]
- 91.Clark KA, McElhinny AS, Beckerle MC, Gregorio CC. Striated muscle cytoarchitecture: an intricate web of form and function. Annu Rev Cell Dev Biol. 2002;18:637–706. doi: 10.1146/annurev.cellbio.18.012502.105840. [DOI] [PubMed] [Google Scholar]
- 92.Ehler E, Gautel M. The sarcomere and sarcomerogenesis. Adv Exp Med Biol. 2008;642:1–14. doi: 10.1007/978-0-387-84847-1_1. [DOI] [PubMed] [Google Scholar]
- 93.Littlefield R, Almenar-Queralt A, Fowler VM. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol. 2001;3:544–551. doi: 10.1038/35078517. [DOI] [PubMed] [Google Scholar]
- 94.Nyman T, Schuler H, Korenbaum E, Schutt CE, Karlsson R, Lindberg U. The role of MeH73 in actin polymerization and ATP hydrolysis. J Mol Biol. 2002;317:577–589. doi: 10.1006/jmbi.2002.5436. [DOI] [PubMed] [Google Scholar]
- 95.Costa CF, Rommelaere H, Waterschoot D, Sethi KK, Nowak KJ, Laing NG, Ampe C, Machesky LM. Myopathy mutations in alpha-skeletal-muscle actin cause a range of molecular defects. J Cell Sci. 2004;117:3367–3377. doi: 10.1242/jcs.01172. [DOI] [PubMed] [Google Scholar]
- 96.Kabsch W, Holmes KC. The actin fold. FASEB J. 1995;9:167–174. doi: 10.1096/fasebj.9.2.7781919. [DOI] [PubMed] [Google Scholar]
- 97.Pollard TD. Polymerization of ADP-actin. J Cell Biol. 1984;99:769–777. doi: 10.1083/jcb.99.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Milligan RA. Protein-protein interactions in the rigor actomyosin complex. Proc Natl Acad Sci USA. 1996;93:21–26. doi: 10.1073/pnas.93.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lorenz M, Holmes KC. The actin-myosin interface. Proc Natl Acad Sci USA. 2010;107:12529–12534. doi: 10.1073/pnas.1003604107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.dos Remedios CG, Moens PD. Actin and the actomyosin interface: a review. Biochim Biophys Acta. 1995;1228:99–124. doi: 10.1016/0005-2728(94)00169-6. [DOI] [PubMed] [Google Scholar]
- 101.Song W, Dyer E, Stuckey DJ, Copeland O, Leung MC, Bayliss C, Messer A, Wilkinson R, Tremoleda JL, Schneider MD, Harding SE, Redwood CS, Clarke K, Nowak K, Monserrat L, Wells D, Marston SB. Molecular mechanism of the E99K mutation in cardiac actin (ACTC Gene) that causes apical hypertrophy in man and mouse. J Biol Chem. 2011;286:27582–27593. doi: 10.1074/jbc.M111.252320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.