Abstract
This study aims to define the function of Slug transcription factor in human normal osteoblasts (hOBs). To date, Slug is considered exclusively a marker of malignancy in bone tissue. Here, we identified, for the first time, a role for Slug in hOBs using a knockdown approach. We demonstrated that Slug is positively correlated with osteoblast markers, including Runx2, osteopontin, osteocalcin, Collagen type 1, Wnt/β-catenin signaling mediators, and mineral deposition. At the same time, Slug silencing potentiates the expression of Sox-9, a factor indispensable for chondrogenic development. These data, with the finding that Slug is in vivo recruited by the promoters of Runx2 and Sox-9 genes, suggest that, in hOBs, Slug may act both as positive and negative transcriptional regulator of Runx2 and Sox-9 genes, respectively. In summary, our results support the hypothesis that Slug functions as a novel regulator of osteoblast activity and may be considered a new factor required for osteoblast maturation.
Electronic supplementary material
The online version of this article (doi:10.1007/s00018-009-0149-5) contains supplementary material, which is available to authorized users.
Keywords: Human osteoblasts, Slug, Wnt signaling, Runx2, Sox-9
Introduction
Osteoblasts, bone producing cells, originate from multi-potent mesenchymal stem cells and are responsible for the secretion of the organic extracellular matrix of bone, both during development and later during the remodeling of mature bone [1]. Osteoblast differentiation and activity are regulated by multiple signaling pathways and specific factors that, by influencing gene expression, cell proliferation, and migration, control bone mass homeostasis. Among these factors, Wingless-type proteins (Wnts) and bone morphogenetic proteins (BMP) drive early events, while the helix-loop-helix proteins Twist and Id maintain proliferation [2–5]. Subsequently, transcriptional regulation of osteoblast differentiation, maturation, and activity is mainly due to key regulators including Runx2, Msx1 and 2, Osterix, NFAT2, c-fos, c-jun, ATF4, fra-2, jun D, Dlx3 and 5 [5–12]. In recent years, the knowledge of the molecular mechanisms that drive the function of these factors in the regulation of bone cell biology has greatly improved; nevertheless, many aspects have not yet been investigated. In particular, the intricate cross-talk among the different pathways is only partially known. In addition, the expression and function of the previously mentioned factors in osteoblasts have been extensively analyzed in experimental animal models, tumor cells, or cellular lines, whereas data about these molecules in human primary osteoblasts cultures are very scarce.
Accumulated evidence has shown that lymphocyte enhancer binding factor 1/T cell factor (LEF1/TCF) transcription factors, the nuclear effectors of the Wnt/β-catenin signaling pathway, influence osteoblast proliferation, function, and regeneration, enhancing expression of Runx2, the master transcription regulator of osteoblasts [13]. Nevertheless, most downstream bone-specific target genes of this pathway are only partially known. Among these, Slug has been recently implicated in osteosarcoma progression as a Wnt-responsive molecule strongly correlated with a loss of tumor suppressors such as E-cadherin [14, 15]. We recently demonstrated that Slug gene is also expressed in normal human osteoblasts, and Lef/Tcf cis elements present on its promoter act in regulating its transcription through a direct binding of Lef-1, TCF-1, and TCF-4 in vivo (submitted manuscript).
Human Slug, also named Snail 2, belongs to the Snail family of genes encoding zinc-finger transcription factors [14]. It is expressed at different stages of development in different tissues, and mediates epithelial–mesenchymal transition [14–16]. Moreover, Slug is involved in a broad spectrum of biological functions, such as cell differentiation, cell motility, cell-cycle regulation, and apoptosis [17]. Slug is also expressed in most normal adult human tissues, but little is known about its potential functions.
In order to identify new potential osteoblast-specific proteins, in the current study we investigated the role of Slug in relation to the expression of Wnt/β-catenin signaling mediators and bone-related genes in human mature osteoblasts (hOBs). For this purpose, we evaluated the effects of Slug gene knockdown on osteoblast maturation by using siRNA strategy. In addition, in order to correlate the expression level of Slug and its activity in regulating the transcription of genes that are crucial for osteoblast differentiation, we investigated Slug in vivo occupancy of the E boxes regulatory sites present in the Runx2 and Sox-9 gene promoters.
Our findings describe an unknown regulatory function of Slug and provide clear evidence for a pivotal role of Slug in regulating osteoblast maturation in human.
Materials and methods
Isolation and culture of osteoblasts and chondrocytes
Bone tissues were harvested from different patients undergoing total knee replacement for osteoarthritis (mean ± SD age 65 ± 10 years). The study was approved by the Istituto Ortopedico Rizzoli (Bologna, Italy) ethical committee and informed consent was obtained from each patient. Trabecular bone was obtained from the inner portion of the tibial plateau. Bone chips were removed from the tibial plateau, collected in a V-glass tube containing 1.5 ml of 1:1 mixture of DMEM/Ham’s F12 K no calcium (Gibco, Invitrogen, Paisley, Scotland, UK) and supplemented with antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin), 25 μg/ml ascorbic acid, 4 mM glutamine (Sigma–Aldrich, St. Louis, MO, USA) and 2 mM calcium chloride (referred to as enzyme medium) as previously reported [18] and according to the methods described by Robey and Termine [19].
Primary chondrocytes were obtained from patients with osteoarthritis undergoing knee arthroplasty. The chondrocytes were isolated from minced tissue by sequential enzymatic digestion as previously described [20].
Immunocytochemistry
A sample of 104 human osteoblasts (at passage 2) were seeded in 8-well chamber slide and allowed to adhere for 96 h. Human osteoblasts were fixed in 4% PFA for 20 min at room temperature and then hydrated with TBS 1% BSA for 5 min at room temperature. The slides were incubated with monoclonal antibodies anti-human -CD45 (Dako Cytomation, Glostrup, Denmark), -CD146 (Nocastra, Newcastle, UK), -CD105 (produced from the hybridoma cell line, clone SH2; ATCC, Rockville, MD), -Runx-2, -osteocalcin, (all purchased from R&D Systems, Minneapolis, MN), -alkaline phosphatase, -collagen type 1 (both obtained from DSHB, Department of Biological Sciences, Iowa city, IA), -bone sialoprotein, (Fisher Scientific,Pittsburg, PA, USA), -estrogen receptor alpha (Upstate Biotechnology, Lake Placid, NY), and -collagen type 2 (Millipore, 900 Middlesex Tpk Billerica, USA) for 1 h at room temperature. The slides were washed three times with TBS 1% BSA and then sequentially incubated with multilinker biotinilated secondary antibody and alkaline phosphatase-conjugated streptavidin (Kit BioGenex, San Ramon, CA, USA) at room temperature for 20 min. The slides were developed using fast red as substrate, counterstained with haematoxylin, mounted with glycerol jelly, and evaluated in a brightfield microscope. Negative and isotype matched controls were performed. Positive cells were manually counted by two evaluators blinded to marker evaluated. For each well, we randomly selected 20 fields at high magnification (×40). Results were expressed as the percentage of positive cells on the total number of cells counted.
Analysis of osteoblast activity
For alkaline phosphatase staining, the Alkaline phosphatase (ALP) Leukocyte kit (Sigma) was used. To perform the test, prefixed mono-layered cells were incubated at room temperature in a solution containing naphthol AS-BI phosphate and freshly prepared fast blue BB salt buffered at pH 9.5 with 2-amino-2-methyl-1,3-propanediol (AMPD). The presence of sites of ALP activity appeared as blue cytoplasmatic staining.
ALP activity was evaluated in hOBs by the hydrolysis of p-nitrophenylphosphate (PNPP). The enzyme activity, expressed as μmol/min/μg of protein, was evaluated 6 days after siRNA/Slug2 treatment. One unit was defined as the amount of enzyme which hydrolyzed 1 nmol/PNPP per minute.
The extent of mineralized matrix in the plates was determined by Alizarin Red S staining (Sigma) in the cells cultured for up to 21 days in osteogenic medium consisting in DMEM, high-glucose, supplemented with 10% FBS, 10 mM β-glycerophosphate, 0.1 mM dexamethasone, and 50 mM ascorbate. In the committed cells, the osteogenic medium was changed every 3 days as well as RNA interference treatment where indicated. The cells were then fixed in 70% ethanol for 1 h at room temperature, washed with PBS, stained with 40 mM AR-S (pH 4.2) for 10 min at room temperature, washed five times with deionized water, and incubated in PBS for 15 min to eliminate non-specific staining. The stained matrix was observed at different magnifications using a Leitz microscope. Matrix mineralization was quantified by measuring the number and surface of mineralized nodules using a digital image analyzer (“Quantity one” software, Biorad). The surface and the number of all mineralized nodules were quantified in 2 wells per condition at day 14 and 21 of culture.
Small interfering RNA (siRNA) transfection
Three sets of Stealth RNAi duplexes and corresponding Stealth control were synthesized by Invitrogen Life Technologies (Carlsbad, CA, USA). Stealth RNAi compounds are 25 mer dsRNA containing proprietary chemical modifications that enhance nuclease stability and reduce off-target effects.
The following Stealth RNAi sequences were used: siRNA/Slug1 sense: 5′-CCGUAUCUCUAUGAGAGUUACUCCA-3′, antisense: 5′-UGGAGUAACUCUCAUAGAGAUACGG-3′; siRNA/Slug2 sense: 5′-CCCUGGUUGCUUCAAGGACACAUUA-3′, antisense: 5′-UAAUGUGUCCUUGAAGCAACCAGGG-3′; siRNA/Slug3 sense: 5′-GGCUCAUCUGCAGACCCAUUCUGAU-3′, antisense: 5′-AUCAGAAUGGGUCUGCAGAUGAGCC-3′.
The most effective fragments used for targeting human Slug were siRNA/Slug2.
Twenty-four hours before siRNA transfection, hOBs were seeded in triplicate at density of 16 × 103/cm2 in DMEM with 10% FBS. Cells were transfected with 30 nM siRNA using Lipofectamine RNAiMAX (Invitrogen Life Technologies) according to the manufacturer’s instructions. Transfected cells were incubated for 6 days at 37°C before gene silencing analysis. As a negative control for the siRNA treatment, Medium GC Stealth RNAi Negative Control Duplex (Invitrogen) was used. Knockdown of Slug expression was verified by Real-Time RT-PCR.
Real-time quantitative RT-PCR
Cells from three wells were harvested and total RNA was extracted using an RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction and as previously described [21]. Real-time PCR was carried out using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA). TaqMan technology, the Assays-On-Demand kit for human Slug, Lef-1, β-catenin, Runx2, Sox-9, OPN, OC, Col1a1, RANKL, and c-myc, were used. The mRNA levels of target genes were corrected for GAPDH mRNA levels (endogenous control). All PCR reactions were performed in triplicate for each sample and were repeated three times. All experimental data were expressed as the mean ± SEM.
Western blotting
For western blot analysis, the cells were washed twice with ice-cold PBS and cell lysates were prepared as previously reported [22]. Then, 10 μg of each sample was electrophoresed on a 12% SDS-polyacrylamide gel. The proteins were then transferred onto an Immobilon-P PVDF membrane (Millipore, Billerica, USA). After blocking with PBS-0.05% Tween 20 and 5% dried milk, the membrane was probed with the following antibodies: Slug (L40C6) from Cells Signaling Technology (Danvers, CA, USA), Runx2 (sc-10758) and Sox-9 (sc-20095), from Santa Cruz Biotechnology (Santa Cruz, CA, USA), Lef-1 (L7901) from Sigma Aldrich, IP3 K (06-195), and Active-β-Catenin (05-665) from Upstate Biotechnology (Lake Placid, NY).
After washing with PBS-Tween, the membranes were incubated with peroxidase-conjugated anti-rabbit antibody (1:50000) or anti-mouse (1:2000) (Dako, Glostrup, Denmark) in 5% non-fat milk. Immunocomplexes were detected using Supersignal West Femto Substrate (Pierce, Rockford, IL, USA). Anti-IP(3)K was used to confirm equal protein loading.
Osteocalcin assay
Osteocalcin secretion was measured in cell culture supernatants collected from osteoblasts plated in 24-well dishes and cultured with D-MEM, 10% FCS in presence or in absence of 30 nM siRNA/Slug2 for 6 days. Prior to measurements, the media were collected, centrifuged at 1,300g for 5 min, and tested by using a human osteocalcin enzyme-linked immunosorbent assay (ELISA) kit, according to manufacturer’s instructions (DRG Diagnostics, Germany). Osteocalcin levels were corrected with total protein content and expressed as nanograms per micrograms of cell protein and each treatment was performed in duplicate.
Chromatin immunoprecipitation (ChIP) assay
The ChIP assay was carried out as previously described [21] using the standard protocol supplied by Upstate Biotechnology with their ChIP assay reagents.
The cells were cross-linked with 1% formaldehyde for 10 min at 37°C, washed in ice-cold PBS, and suspended in SDS lysis buffer for 10 min on ice. Samples were sonicated, diluted 10-fold in dilution buffer supplemented with protease inhibitors, and precleared with 80 μl of DNA-coated protein A-agarose; the supernatant was used directly for immunoprecipitation with 5 μg of anti- Slug, (sc-10436) (Santa Cruz Biotechnology), overnight at 4°C. Immunocomplexes were mixed with 80 μl of DNA-coated protein A-agarose followed by incubation for 1 h at 4°C. Beads were collected and sequentially washed 5 times with 1 ml each of the following buffers: low salt wash buffer (0.1% SDS,1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH 8.1, 150 mM NaCl), high salt wash buffer (0.1% SDS,1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl pH-8.1, 500 mM NaCl), LiCl wash buffer (0.25 mM LiCl, 1% IGEPAL-CA630, 1% deoxycholic acid, 1 mM EDTA, 10 mM Tris-pH 8.1), and TE buffer. The immunocomplexes were eluted two times by adding a 250-μl aliquot of a freshly prepared solution of 1% SDS, 0.1 M NaHCO3 and the cross-linking reactions were reversed by incubation at 65°C for 4 h. Further, the samples were digested with proteinase K (10 mg/ml) at 42°C for 1 h, DNA was recovered by phenol/chloroform extractions, ethanol precipitated using 1 μl of 20 mg/ml glycogen as the carrier, and suspended in sterile water. For PCR analysis, aliquots of chromatin before immunoprecipitation were saved (input). PCR was performed to analyze the presence of DNA precipitated by Slug-specific antibody, and by using specific primers (Table 1) to amplify fragments of the Runx2 and Sox-9 gene promoters.
Table 1.
Immunocytochemical analysis of human primary osteoblasts (hOBs)
| Markers | Percentage of positive cells | ||||
|---|---|---|---|---|---|
| hOB1 | hOB2 | hOB3 | hOB4 | hOB5 | |
| CD45 | Negative | Negative | Negative | Negative | Negative |
| ALP | 5 | 15 | 15 | 12 | 15 |
| CD146 | 2 | 2 | 4 | 2 | 4 |
| CD105 | 100 | 100 | 100 | 100 | 100 |
| Col1a1 | 50 | 60 | 60 | 60 | 60 |
| Runx2 | 80 | 80 | 80 | 80 | 80 |
| BSP | 100 | 100 | 100 | 100 | 100 |
| OC | 60 | 80 | 90 | 100 | 80 |
| ER-α | 100 | 100 | 100 | 100 | 100 |
Each PCR reaction was performed with 5 μl of the bound DNA fraction or 2 μl of the input. The PCR was performed as follows: preincubation at 95°C for 5 min, 30 cycles of 1 min denaturation at 95°C, 1 min annealing at 62°C, and 1 min at 72°C, with one final incubation at 72°C for 5 min. No-antibody control was included in each experiment.
Statistical analysis
Data are presented as the mean ± SEM from at least three independent experiments. Statistical analysis was performed by one-way analysis of variance and the Student’s t test. A P value <0.05 was considered statistically significant.
Results
Phenotypical characterization and osteogenic potential of human osteoblasts
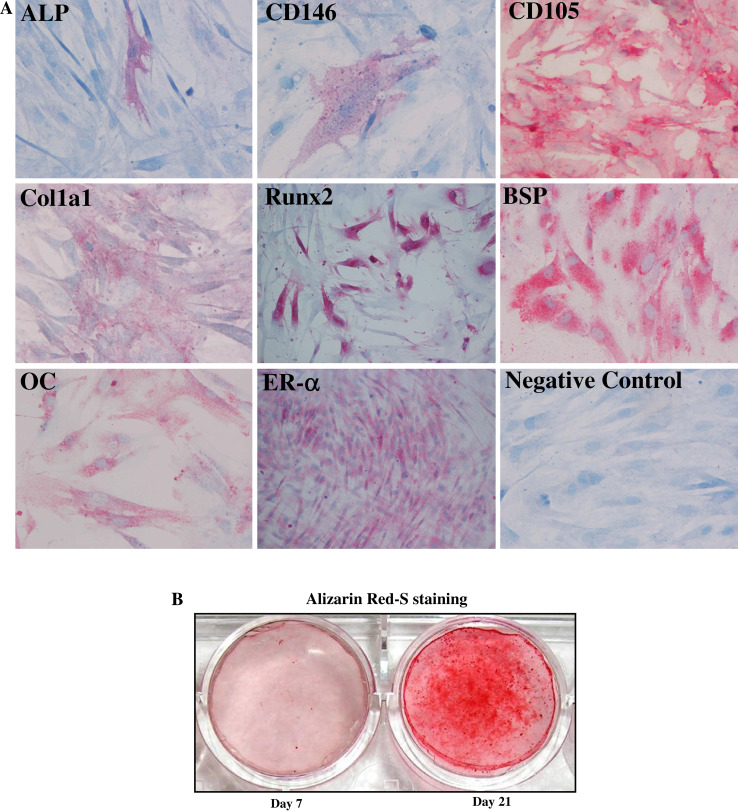
Human primary osteoblast cultures (hOBs) were generated from bone chips removed from the tibial plateau as previously described [18] and as reported in “Materials and methods”. We first analyzed a panel of nine phenotypic markers in cells at the second passage in culture. All osteoblasts were highly positive for the typical osteogenic markers, including Runx2, collagen type 1 (Col1a1), bone sialoprotein (BSP), and osteocalcin (OC), and weakly positive to ALP. In addition, the cells were positive for estrogen receptor alpha (ERα), a protein that is known to be associated to osteoblast differentiation. The samples were negative for a typical hematopoietic marker (CD45), only partially positive for a mesenchymal marker such as CD146, and positive for CD105. After this analysis, the cells that we used may be considered mature osteoblasts because they express low levels of CD146 and ALP, and high levels of CD105, as previously reported [18]. The percentage of positive cells for the markers analyzed by immunocytochemistry in five hOB samples is shown in Table 1, and the immunocytochemical staining of a representative sample is reported in Fig. 1a. These immunocytochemical data have also been confirmed by FACS analysis (data not shown). Next, the cells were characterized for their osteogenic capacity. All hOBs exhibited an evident extracellular matrix mineralization after 21 days of culture under osteogenic conditions (a representative sample is shown in Fig. 1b).
Fig. 1.
Phenotypical characterization of hOBs. Five hOB samples were subjected to immunocytochemical analysis for ALP, CD146, BSP, OC, CD105, Col1a1, Runx2, and ER-α phenotypical markers. a The staining showed the local expression of the markers analyzed in a representative sample (×20 magnification). b The formation of extracellular matrix by cells treated with β-glycerophosphate, ascorbic acid, and dexamethasone. Mineral formation was examined by Alizarin Red S staining. The deposition of calcium salts was observed in osteogenic cultures at day 21
Slug knockdown and Wnt signaling
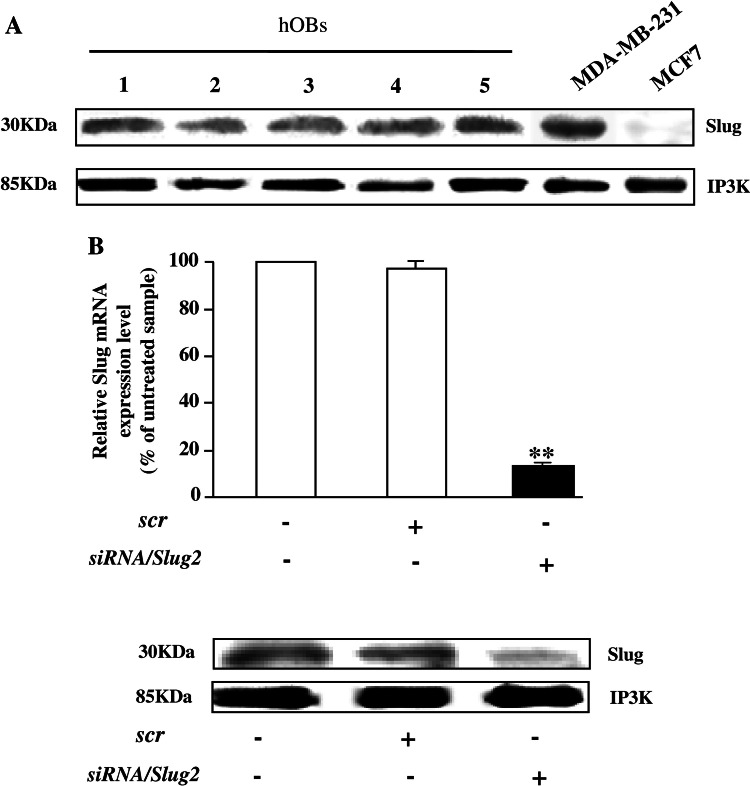
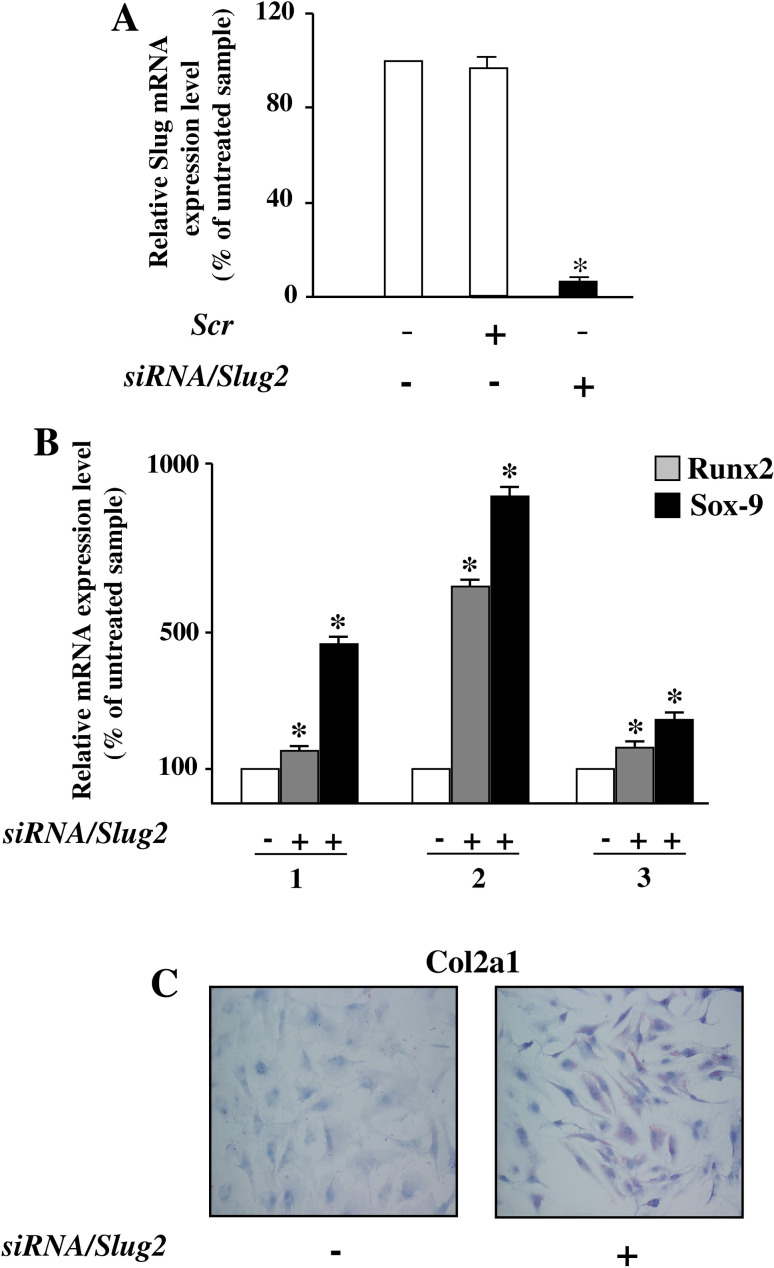
We firstly confirmed by western blot analysis on whole cell extracts (Fig. 2a) that all hOB samples express Slug protein at comparable levels, approximately the same that were found in Slug-positive MDA-MB-231 breast cancer cells [23]. Then, to test whether Slug expression may be correlated with osteoblast phenotype, we used a Slug knockdown approach using siRNA. For this purpose, three siRNAs against Slug were designed, siRNA/Slug1, siRNA/Slug2, and siRNA/Slug3, in order to down-regulate Slug expression. Transfection of hOBs and osteosarcoma SaOS2 cell line with these siRNAs resulted in the down-regulation of Slug transcript by 30% (siRNA/Slug1 and siRNA/Slug3) or 80% (siRNA/Slug2) (electronic supplementary material, Fig. 1). Therefore, we blocked Slug endogenous production treating the cells for 6 days with siRNA/Slug2 30 nM and obtaining a strong inhibition of Slug mRNA and protein, as revealed by quantitative RT-PCR and western blot, respectively (Fig. 2b).
Fig. 2.
Silencing of Slug gene expression by siRNA/Slug2 in hOBs. a Western blot analysis of endogenous Slug expression in hOBs. 10 μg of whole cell lysates from five hOBs samples were assayed on a 12% SDS-polyacrylamide gel. The proteins were visualized using Supersignal Femto Substrate (Pierce). Size markers are reported (KDa). IP3 K was used as a loading control. Positive and negative controls for Slug band (Slug-positive MDA-MB-231 breast cancer cells, and Slug-negative MCF7 breast cancer cells, respectively) are shown. b hOBs were transfected with siRNA/Slug2 or a non-relevant siRNA (scr). Slug expression was determined both at mRNA and protein level, and revealed by quantitative RT-PCR and western blot analysis, respectively. RT-PCR results, after correction to GAPDH content, are expressed as siRNA/Slug2 over control ratio. Results represent means ± SEM of five hOBs samples (**P < 0.01). At the bottom, representative western blot of siRNA/Slug2 treated cells shows a specific decrease of endogenous Slug protein level
In order to establish a role of Slug in osteoblasts, we analyzed the effects of Slug silencing on the expression of mediators of a central pathway in bone metabolism, such as Wnt signaling.
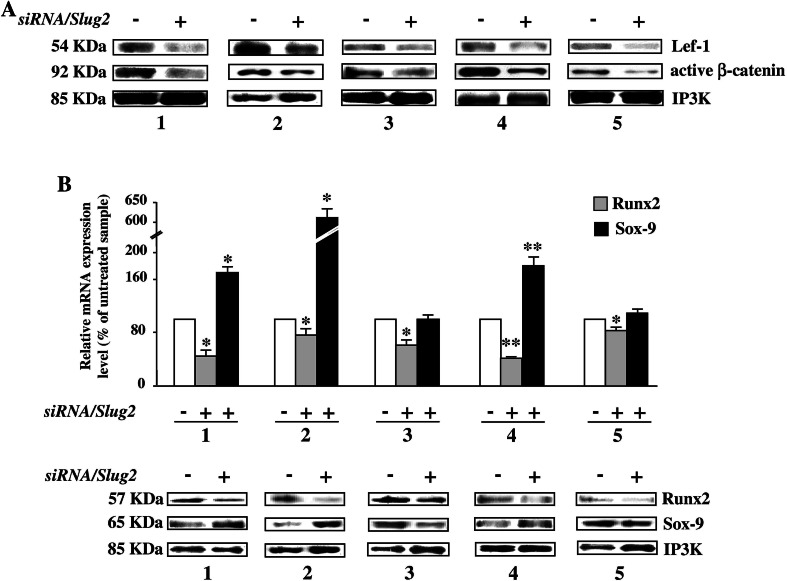
As shown in Fig. 3a, Slug knockdown significantly reduced the protein levels of two important Wnt mediators, Lef-1 and β-catenin, in all the analyzed samples. Attempts to detect a corresponding reduction of mRNA expression level of Lef-1 and β-catenin met with variable success, and we found approximately the same Lef-1 and β-catenin mRNA levels both in siRNA-transfected cells and in control cells (data not shown). This suggests that, for what concerns these two genes, the regulation mediated by Slug silencing is mainly at the protein level that is independent of mRNA. Therefore, we hypothesized that the levels of Slug may interfere with Wnt signaling modulating the levels of Lef-1 and β-catenin, and that Slug may consequently act by controlling the expression of specific genes in osteoblasts.
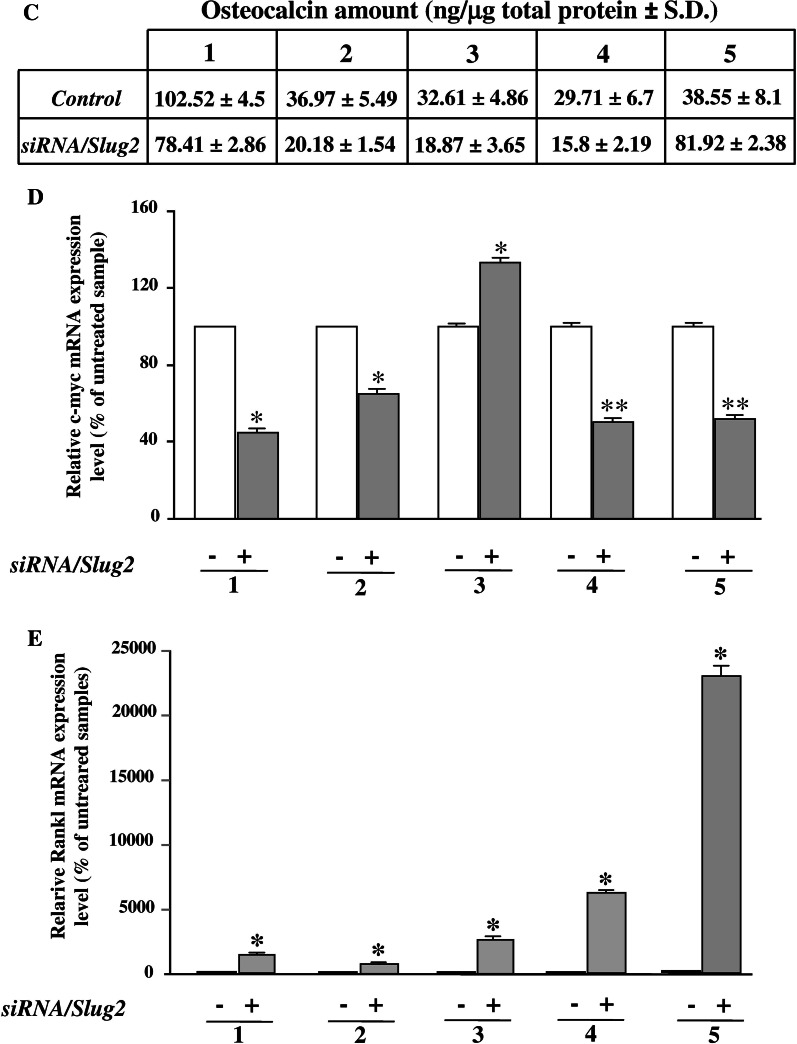
Fig. 3.
Effect of Slug interference on Wnt signaling target genes in hOBs. After siRNA/Slug2 treatment the expression of bone-related Wnt target genes was analyzed in five hOB samples. a Western blot analysis of Lef-1 and active β-catenin protein levels. Size markers are reported (KDa). IP3 K was used as a loading control. b Runx2 and Sox-9 expression analysis in siRNA/Slug2 treated cells. mRNA and protein levels were determined by quantitative RT-PCR and western blot analysis, respectively. c Osteocalcin, c-myc, and RANKL expression analysis in siRNA/Slug2 treated cells. Osteocalcin protein was measured in cell culture supernatants by ELISA. The level of c-myc and RANKL expression was determined by quantitative RT-PCR. In all quantitative RT-PCR experiments the results, after correction to GAPDH content, are expressed as siRNA/Slug2 over control ratio. Results represent means ± SEM of triplicate determinations (*P < 0.05, **P < 0.01)
In order to strengthen this hypothesis, the expression of downstream target genes of canonical Wnt signaling such as Runx2, Sox-9, osteocalcin, RANKL, and c-myc were examined in Slug silenced cells. As shown in Fig. 3b, the expression of Runx2, the master transcription regulator of osteoblasts previously identified as a Lef-1/β-catenin target gene [13], was markedly decreased in Slug silenced cells compared to untreated osteoblasts. On the contrary, Slug knockdown induced expression of Sox-9, a factor indispensable for chondrogenic development [24], in four out five samples. In this case, the effects of Slug silencing were both at mRNA and protein level. Accordingly, in four out five samples, the amount of secreted osteocalcin, which is a marker of late osteoblast differentiation positively modulated by Wnt signaling [25], was significantly reduced by Slug knockdown (Fig. 3c), as well as the expression of c-myc which is induced in response of activation of Wnt signaling [26] (Fig. 3d). On the contrary, mRNA levels of the receptor activator of NFkB ligand (RANKL), the expression of which is repressed by Wnt signaling [27], was significantly increased in the siRNA/Slug2 hOBs-treated samples (Fig. 3e). As a whole, these results suggest that Slug knockdown affects Wnt signaling and consequently its downstream target genes in human osteoblasts.
Slug interacts in vivo with the promoters of Runx2 and Sox-9
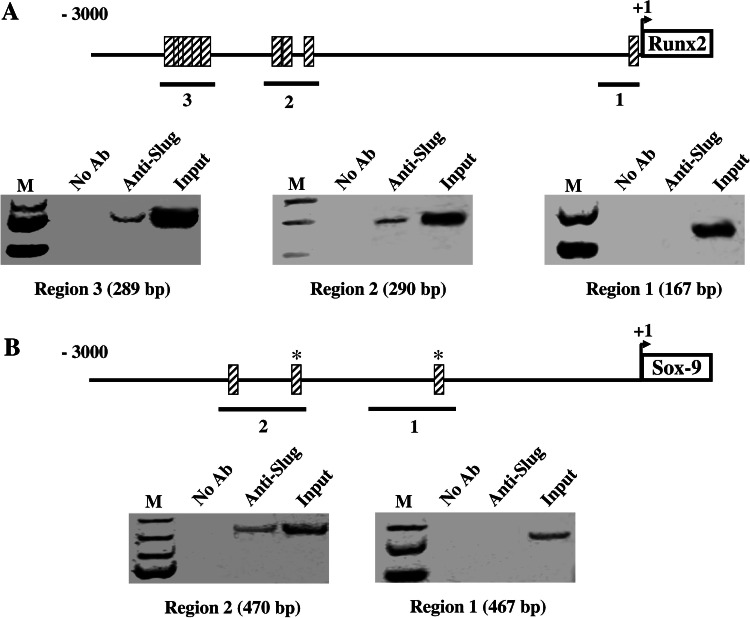
Investigations were then performed to address whether Slug may be directly involved in the control of specific gene transcription in hOBs. For this purpose, the promoters of Runx2 and Sox-9 were chosen for chromatin immunoprecipitation (ChIP) analysis. At first, we searched putative Slug binding sites, named E boxes [28], in the promoter regions of human Runx2 and Sox-9 genes by using a program for predicting transcription factor binding sites (TFSEARCH, www.cbrc.jp/research/db/TFSEARCH.html). As shown in Fig. 4, many E boxes were found in the promoter of both genes; their functionality was then investigated, analyzing the in vivo association between Slug and the promoter sequences by ChIP binding assays (Fig. 4). To this aim, human primary osteoblasts were exposed to formaldehyde to cross-link proteins and DNA, and sonicated to fragment the chromatin. Specific antibody against Slug was used to immunoprecipitate the protein–DNA complexes. The presence of the promoter-specific DNA region before immunoprecipitation was confirmed by PCR (input). After immunoprecipitation, DNA was extracted from the beads and used as a template to generate specific PCR products spanning the putative Slug binding sites from −3,000 bp to +1 bp in the promoter of both genes. Slug recruitment was assessed at the different promoter regions, as indicated in Fig. 4, by using specific sets of primers (Table 2). Slug occupancy was detected at the region 2 and 3 of Runx2 gene (Fig. 4a) and at the region 2 of Sox-9 gene (Fig. 4b).
Fig. 4.
In vivo recruitment of Slug on Runx2 and Sox-9 gene promoters. Protein–DNA complexes were formaldehyde-cross-linked in hOBs in vivo. Chromatin fragments from these cells were subjected to immunoprecipitation with antibody against Slug. After cross-link reversal, the coimmunoprecipitated DNA was amplified by PCR using the primers reported in Table 2, and specific for the indicated promoter regions. PCR fragments were resolved in 1.5% agarose gels. Aliquots of chromatin taken before immunoprecipitation were used as Input positive controls whereas chromatin eluted from immunoprecipitations lacking antibody were used as no antibody (NoAb) controls. a The immunoprecipitates were subjected to PCR analysis using primer pairs spanning the reported regions of Runx2 promoter. b The immunoprecipitates were subjected to PCR analysis using primer pairs spanning the reported regions of Sox-9 promoter. The specific molecular weights of PCR fragments are shown in parentheses. The relative positions of Slug putative binding sites (striped boxes) are indicated. *Sites showing 100% homology with consensus-binding site (CAGGTG)
Table 2.
PCR primers used for chromatin immunoprecipitation assay (ChIP)
| Gene | Primer sequences | Product size (bp) |
|---|---|---|
| Runx2 | Forward F1:5′-ATATCCTTCTGGATGCCAGG-3′ | 167 |
| Reverse R1:5′-AAGCACTATTACTGGAGAGGC-3′ | ||
| Forward F2:5′-GTTTCAGTGAATGCTAATGTAG-3′ | 290 | |
| Reverse R2:5′-AAGCGTTCATTTAACATGCAG-3′ | ||
| Forward F3:5′-CAAGAGCTTTATTTGCATTGAC-3′ | 282 | |
| Reverse R3:5′-TTGTCCTCTGTGAGGCCTAT-3′ | ||
| Sox9 | Forward F1:5′-GATAGTGTCCTCACTTCGCA-3′ | 467 |
| Reverse R1:5′-TCCACTCTGGCGGAGTCATG-3′ | ||
| Forward F2:5′-CAGCCACCACCATCCAAGTT-3′ | 470 | |
| Reverse R2:5′-GAAGGGCATTGTGTGTACAG-3′ |
These ChIP experiments were repeated four times with identical results and demonstrate that both Runx2 and Sox-9 are Slug target genes in human osteoblasts.
Slug knockdown inhibits maturation of osteoblasts and supports differentiation of chondrocytes
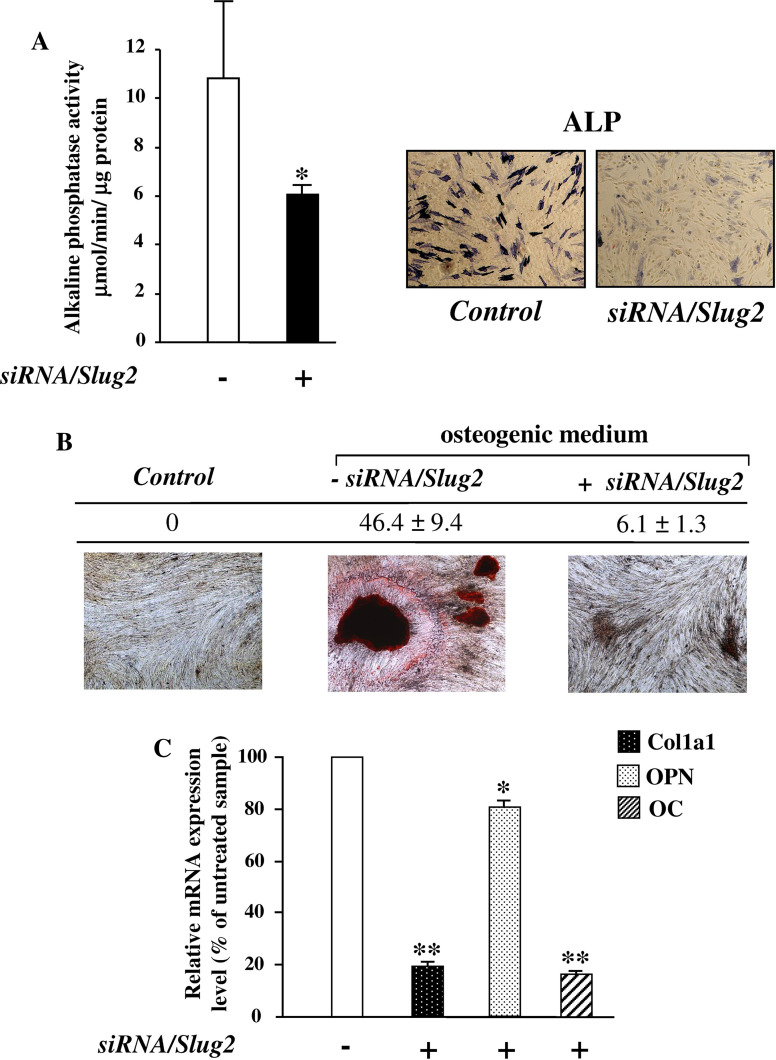
Afterwards, the effect of Slug silencing on osteoblast maturation was examined analyzing ALP activity and the formation of mineralized matrix. As shown in Fig. 5a, there was a significative decrease of ALP activity after 6 days of siRNA/Slug2 treatment. Accordingly, Slug silenced cells showed a reduced mineralization ability; in fact, 14 days after the initiation of osteoblast induction in osteogenic medium, a reduction up to 86% of mineral deposition was observed in Slug silenced cells in comparison with non-silenced cells (Fig. 5b). This decrease in osteoblast maturation was accompanied by a significative decrease of other correlated markers including Col1a1 and classical Runx2 targets such as osteopontin and osteocalcin (Fig. 5c).
Fig. 5.
Effect of Slug interference on osteoblast maturation. After siRNA/Slug2 treatment the cells were analyzed for the presence of alkaline phosphatase activity (ALP), formation of mineralized matrix and expression of osteoblast maturation markers. a Alkaline phosphatase activity was evaluated by PNPP hydrolysis and ALP Leukocyte kit. The presence of sites of ALP activity appeared as blue cytoplasmic staining as shown in the reported representative sample (×20 magnification). b Mineral formation was examined by Alizarin Red S staining in the cells cultured with β-glycerophosphate, ascorbic acid, and dexamethasone (osteogenic medium). The deposition of calcium salts was observed in osteogenic cultures at day 14, but not in control cells (not cultured in osteogenic medium), and was quantified by measuring the number and surface of mineralized nodules using a digital image analyzer (“Quantity one” software, Biorad). The ratio of the surface to the number of nodules in a representative hOB sample is reported. c The expression of Col1a1, osteopontin (OPN) and osteocalcin (OC) was determined by quantitative RT-PCR in the cells cultured in osteogenic medium. The results, after correction to GAPDH content, are expressed as siRNA/Slug2 over control ratio. Results represent means ± SEM of triplicate determinations (*P < 0.05, **P < 0.01)
Since we have here demonstrated that Slug knockdown-induced expression of Sox-9 in hOBs, investigations were then performed to evaluate the potential pro-chondrogenic effect of Slug knockdown in human undifferentiated chondrocytes at passage 6–8. In these conditions, the cells express very low level of Collagen type 2 [29]. Slug mRNA expression was blocked in these cells with the same approach and effectiveness as in hOBs (Fig. 6a). Runx2 and Sox-9 expression was then tested. As shown in Fig. 6b, a significative upregulation of both these genes, required for chondrocyte maturation [30], was found in all analyzed samples after Slug silencing. This effect was accompanied by a significant increase of Collagen type 2 evaluated by immunocytochemistry (Fig. 6c).
Fig. 6.
Silencing of Slug gene expression by siRNA/Slug2 in undifferentiated chondrocytes. a Undifferentiated chondrocytes were transfected with siRNA/Slug2 or a non-relevant siRNA (scr). Slug expression was determined at mRNA, and revealed by quantitative RT-PCR. RT-PCR results, after correction to GAPDH content, are expressed as siRNA/Slug2 over control ratio. Results represent means ± SEM of three chondrocytes samples (*P < 0.05). b Runx2 and Sox-9 expression analysis in siRNA/Slug2 treated cells. mRNA levels were determined by quantitative RT-PCR analysis. c Collagen type 2 expression analysis in siRNA/Slug2 treated cells. The presence of protein was determined by immunocytochemistry and appeared as violet cytoplasmic staining, as shown in the reported representative sample (×20 magnification)
These preliminary findings demonstrate that when endogenous Slug levels are suppressed chondrocytes are inclined to differentiate, supporting the idea that Slug may play a critical role as pro-chondrogenic factor.
Discussion
To our knowledge, this is the first study to examine the role of Slug on normal human primary osteoblasts. Much is known about the role of Slug during development and its action in malignant progression [14–17, 31]; nevertheless, its expression and function in normal adult tissues has not been elucidated. To date, for what concerns bone tissue, Slug is considered exclusively a marker of malignancy and, consequently, an attractive potential target for therapeutic modulation of bone metastasis and osteosarcoma invasiveness [31], blocking its expression and potentially repressing any detrimental downstream effects.
However, there are no data about Slug expression and regulation in human normal osteoblasts. Here, we identified a role for Slug in normal human osteoblast maturation. Gene expression analysis showed that Slug is positively correlated with osteoblast markers, such as Runx2, and Wnt/β-catenin signaling. A requirement for Slug in osteoblast maturation is supported by Slug knockdown data. In fact, the suppression of Slug mediated by siRNA weakens Wnt/β-catenin signaling, decreases ALP activity, as well as osteoblast mineralization, osteopontin, osteocalcin, and Col1a1, demonstrating that Slug may be considered a novel osteogenic factor. At the same time, Slug silencing does not reflect a global suppression of gene expression. In fact, the expression of RANKL which is suppressed by Wnt signaling in osteoblasts [27] was enhanced in response to Slug knockdown. Furthermore, we observed that Slug silencing potentiates the expression of Sox-9 both in hOBs and in undifferentiated chondrocytes. This is particularly interesting in the context of maturation and differentiation of these cells because Sox-9 is an indispensable factor for chondrogenic development [24, 30] activating cartilage-specific genes, but also acts as a transcriptional repressor for osteoblast differentiation [32] interacting with Runx2 and repressing its function. Therefore, our findings suggest that Slug is required for supporting osteogenic maturation, and its suppression also has a potential pro-chondrogenic effect. Thus, Slug could affect phenotypical changes in response to alteration of its expression levels favoring the dominance of Runx2 function over Sox-9 or vice versa. This is in agreement with several observations demonstrating that, in order to achieve differentiation towards a desired lineage, it is important to direct the stem cell differentiation with correct levels of transcription factors [2, 33]. Additional investigations will be required to understand if Slug plays a role in cell fate determination between chondrocytes and osteoblasts that are derived from common mesenchymal progenitors [34].
Our observation that the Slug gene silencing may decrease Runx2 and increase Sox-9 expression in hOBs was further validated by the in vivo occupancy of the E boxes regulatory sites present in the Runx2 and Sox-9 gene promoters. This suggests that Slug may act, at the same time, both as positive and negative transcriptional regulator of Runx2 and Sox-9 genes, respectively, in human osteoblasts. This supports a role for Slug in maintaining high levels of Runx2 and low levels of Sox-9 to promote osteoblast maturation. Nevertheless, further studies will be needed to characterize the action of Slug on the promoter sequences of these genes, and to establish whether Slug interacts with other transcription factors both in osteoblasts and their precursors.
Several observations indicate that Sox-9 and Runx2 are targets of one of the most crucial signaling for normal skeletogenesis, Wnt/β-catenin pathway [13, 24, 35–38]. Interestingly, we demonstrated that the levels of β-catenin and Lef-1 proteins, two important mediators of Wnt/β-catenin signaling, correlate with Slug expression levels and significantly decrease in Slug silenced cells. Consistent with these findings, we have recently shown that Slug gene expression is positively regulated by Lef-1 which directly binds to Lef/Tcf cis elements present on its promoter (submitted manuscript).
Previous studies have established that Wnt/β-catenin signaling events are mediated by the existence of a large signalosome in which inputs from Wnt signaling, steroid receptors, BMPs, and kinases converge to induce differentiation of osteoblast precursors [39, 40]. However, there are still many unresolved issues about osteoblast regulation through these different pathways. In this scenario, Slug might represent a new regulation factor required for human osteoblast differentiation and maturation. Recent data indicate that, in addition to Slug, another member of the zinc-finger Snail family, Snail1, is involved in bone cell differentiation, but with opposite effect [41]. In fact, in this study, the authors demonstrated that Snail1 controls bone mass by repressing the transcription of Runx2 and vitamin D receptor genes in murine osteoblasts. This suggests that Snail proteins may be involved in the complex dinamics of osteoblast differentiation and maturation processes through different mechanisms. Alternatively, it is plausible that the effects of a transcription factor on the regulation of bone specific genes are different in murine and human cells, and depend on different moments of osteoblastic maturation. This concept may explain the different results from another group demonstrating that Snail enhances expression of osteoblast markers in the MC3T3-E1 cell line [42]. In this study, in addition to Snail, the authors investigated the contribution of many helix-loop-helix (HLH) transcription factors such as Twist proteins that compete with all the Snail factors for the same E-box motifs on the target genes. In agreement with several lines of evidence, the authors indicate that Twist proteins inhibit osteoblast differentiation by interfering with Runx2 function. The evidence that we here obtained on Slug in human osteoblasts supports the conclusions of this study, revealing that the integrated activities of negative and positive E-box-related regulatory factors may control osteoblast differentiation. Therefore, even if the gene regulatory networks are highly intricate, it is possible that further characterization of the role of Snail together with HLH transcriptional regulators in human bone differentiation may provide new insights into the discovery of new molecular targets to use in bone repair and engineering [43].
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This research was supported by grants from MIUR COFIN-2007, Regione Emilia Romagna, Programma di Ricerca Regione Universita’ 2007–2009, the Fondazione Cassa di Risparmio di Padova e Rovigo. E.L. is a recipient of a fellowship from the Fondazione Cassa di Risparmio di Cento.
References
- 1.Karsenty G, Wagner EF. Reaching a genetic and molecular understanding of skeletal development. Dev Cell. 2002;2:389–406. doi: 10.1016/S1534-5807(02)00157-0. [DOI] [PubMed] [Google Scholar]
- 2.Soltanoff CS, Yang S, Chen W, Li YP. Signaling networks that control the lineage commitment and differentiation of bone cells. Crit Rev Eukaryot Gene Expr. 2009;19:1–46. doi: 10.1615/critreveukargeneexpr.v19.i1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartmann C. A Wnt canon orchestrating osteoblastogenesis. Trends Cell Biol. 2006;16:151–158. doi: 10.1016/j.tcb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Wu X, Shi W, Cao X. Multiplicity of BMP signaling in skeletal development. Ann N Y Acad Sci. 2007;1116:29–49. doi: 10.1196/annals.1402.053. [DOI] [PubMed] [Google Scholar]
- 5.Hayashi M, Nimura K, Kashiwagi K, Harada T, Takaoka K, Kato H, Tamai K, Kaneda Y. Comparative roles of Twist-1 and Id1 in transcriptional regulation by BMP signaling. J Cell Sci. 2007;120:1350–1357. doi: 10.1242/jcs.000067. [DOI] [PubMed] [Google Scholar]
- 6.Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 7.Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. doi: 10.1615/CritRevEukaryotGeneExpr.v14.i12.10. [DOI] [PubMed] [Google Scholar]
- 8.Xiao G, Jiang D, Ge C, Zhao Z, Lai Y, Boules H, Phimphilai M, Yang X, Karsenty G, Franceschi RT. Cooperative interactions between activating transcription factor 4 and Runx2/Cbfa1 stimulate osteoblast-specific osteocalcin gene expression. J Biol Chem. 2005;280:30689–30696. doi: 10.1074/jbc.M500750200. [DOI] [PubMed] [Google Scholar]
- 9.Hoffmann HM, Catron KM, van Wijnen AJ, McCabe LR, Lian JB, Stein GS, Stein JL. Transcriptional control of the tissue-specific, developmentally regulated osteocalcin gene requires a binding motif for the Msx family of homeodomain proteins. Proc Natl Acad Sci USA. 1994;91:12887–12891. doi: 10.1073/pnas.91.26.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, Stein GS, Stein JL, Lian JB. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24:9248–9261. doi: 10.1128/MCB.24.20.9248-9261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng ZL, Sharff KA, Tang N, Song WX, Luo J, Luo X, Chen J, Bennett E, Reid R, Manning D, Xue A, Montag AG, Luu HH, Haydon RC, He TC. Regulation of osteogenic differentiation during skeletal development. Front Biosci. 2008;13:2001–2021. doi: 10.2741/2819. [DOI] [PubMed] [Google Scholar]
- 12.Machwate M, Jullienne A, Moukhtar M, Lomri A, Marie PJ. c- fos protooncogene is involved in the mitogenic effect of transforming growth factor-beta in osteoblastic cells. Mol Endocrinol. 1995;9:187–198. doi: 10.1210/me.9.2.187. [DOI] [PubMed] [Google Scholar]
- 13.Gaur T, Lengner CJ, Hovhannisyan H, Bhat RA, Bodine PV, Komm BS, Javed A, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 14.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 15.Conacci-Sorrell M, Simcha I, Ben-Yedidia T, Blechman J, Savagner P, Ben-Ze’ev A. Autoregulation of E-cadherin expression by cadherin-cadherin interactions: the roles of beta-catenin signaling, Slug, and MAPK. J Cell Biol. 2003;163:847–857. doi: 10.1083/jcb.200308162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hajra KM, Chen DY, Fearon ER. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 17.Barrallo-Gimeno A, Nieto MA. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development. 2005;132:3151–3161. doi: 10.1242/dev.01907. [DOI] [PubMed] [Google Scholar]
- 18.Tonnarelli B, Manferdini C, Piacentini A, Codeluppi K, Zini N, Ghisu S, Facchini A, Lisignoli G. Surface-dependent modulation of proliferation, bone matrix molecules, and inflammatory factors in human osteoblasts. J Biomed Mater Res. 2008;89:687–696. doi: 10.1002/jbm.a.32019. [DOI] [PubMed] [Google Scholar]
- 19.Robey PG, Termine JD. Human bone cells in vitro. Calcif Tissue Int. 1985;37:453–460. doi: 10.1007/BF02557826. [DOI] [PubMed] [Google Scholar]
- 20.Grigolo B, Lisignoli G, Piacentini A, Fiorini M, Gobbi P, Mazzotti G, Duca M, Pavesio A, Facchini A. Evidence for redifferentiation of human chondrocytes grown on a hyaluronan-based biomaterial (HYAff 11): molecular, immunohistochemical and ultrastructural analysis. Biomaterials. 2002;23:1187–1195. doi: 10.1016/S0142-9612(01)00236-8. [DOI] [PubMed] [Google Scholar]
- 21.Lambertini E, Tavanti E, Torreggiani E, Penolazzi L, Gambari R, Piva R. ER alpha and AP-1 interact in vivo with a specific sequence of the F promoter of the human ER alpha gene in osteoblasts. J Cell Physiol. 2008;216:101–110. doi: 10.1002/jcp.21379. [DOI] [PubMed] [Google Scholar]
- 22.Lambertini E, Penolazzi L, Tavanti E, Schincaglia GP, Zennaro M, Gambari R, Piva R. Human estrogen receptor alpha gene is a target of Runx2 transcription factor in osteoblasts. Exp Cell Res. 2007;313:1548–1560. doi: 10.1016/j.yexcr.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 23.Tripathi MK, Misra S, Khedkar SV, Hamilton N, Irvin-Wilson C, Sharan C, Sealy L, Chaudhuri G. Regulation of BRCA2 gene expression by the SLUG repressor protein in human breast cells. J Biol Chem. 2005;280:17163–17171. doi: 10.1074/jbc.M501375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu ED, Demay MB, Gori F. Wdr5 is essential for osteoblast differentiation. J Biol Chem. 2008;283:7361–7367. doi: 10.1074/jbc.M703304200. [DOI] [PubMed] [Google Scholar]
- 26.Katoh M. WNT signaling in stem cell biology and regenerative medicine. Curr Drug Targets. 2008;9:565–570. doi: 10.2174/138945008784911750. [DOI] [PubMed] [Google Scholar]
- 27.Spencer GJ, Utting JC, Etheridge SL, Arnett TR, Genever PG. Wnt signalling in osteoblasts regulates expression of the receptor activator of NFkappaB ligand and inhibits osteoclastogenesis in vitro. J Cell Sci. 2006;119:1283–1296. doi: 10.1242/jcs.02883. [DOI] [PubMed] [Google Scholar]
- 28.Mittal MK, Myers JN, Misra S, Bailey CK, Chaudhuri G. In vivo binding to and functional repression of the VDR gene promoter by SLUG in human breast cells. Biochem Biophys Res Commun. 2008;372:30–34. doi: 10.1016/j.bbrc.2008.04.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okazaki K, Li J, Yu H, Fukui N, Sandell LJ. CCAAT/enhancer-binding proteins beta and delta mediate the repression of gene transcription of cartilage-derived retinoic acid-sensitive protein induced by interleukin-1 beta. J Biol Chem. 2002;277:31526–31533. doi: 10.1074/jbc.M202815200. [DOI] [PubMed] [Google Scholar]
- 30.Komori T (2009) Regulation of bone development and extracellular matrix protein genes by RUNX2. Cell Tissue Res. doi 10.1007/s00441-009-0832-8 [DOI] [PubMed]
- 31.Guo Y, Zi X, Koontz Z, Kim A, Xie J, Gorlick R, Holcombe RF, Hoang BH. Blocking Wnt/LRP5 signaling by a soluble receptor modulates the epithelial to mesenchymal transition and suppresses met and metalloproteinases in osteosarcoma Saos-2 cells. J Orthop Res. 2007;25:964–971. doi: 10.1002/jor.20356. [DOI] [PubMed] [Google Scholar]
- 32.Zhou G, Zheng Q, Engin F, Munivez E, Chen Y, Sebald E, Krakow D, Lee B. Dominance of Sox9 function over Runx2 during skeletogenesis. Proc Natl Acad Sci USA. 2006;103:19004–19009. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davis LA, Zur Nieden NI. Mesodermal fate decisions of a stem cell: the Wnt switch. Cell Mol Life Sci. 2008;65:2658–2674. doi: 10.1007/s00018-008-8042-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou L, Zou X, Li H, Mygind T, Zeng Y, Lü N, Bünger C. Molecular mechanism of osteochondroprogenitor fate determination during bone formation. Adv Exp Med Biol. 2006;585:431–441. doi: 10.1007/978-0-387-34133-0_28. [DOI] [PubMed] [Google Scholar]
- 35.Topol L, Chen W, Song H, Day TF, Yang Y. Sox9 inhibits Wnt signaling by promoting beta-catenin phosphorylation in the nucleus. J Biol Chem. 2009;284:3323–3333. doi: 10.1074/jbc.M808048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamidouche Z, Haÿ E, Vaudin P, Charbord P, Schüle R, Marie PJ, Fromigué O. FHL2 mediates dexamethasone-induced mesenchymal cell differentiation into osteoblasts by activating Wnt/beta-catenin signaling-dependent Runx2 expression. FASEB J. 2008;22:3813–3822. doi: 10.1096/fj.08-106302. [DOI] [PubMed] [Google Scholar]
- 37.Dong YF, Soung do Y, Schwarz EM, O’Keefe RJ, Drissi H. Wnt induction of chondrocyte hypertrophy through the Runx2 transcription factor. J Cell Physiol. 2006;208:77–86. doi: 10.1002/jcp.20656. [DOI] [PubMed] [Google Scholar]
- 38.Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell. 2005;8:739–750. doi: 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 39.Kousteni S, Almeida M, Han L, Bellido T, Jilka RL, Manolagas SC. Induction of osteoblast differentiation by selective activation of kinase-mediated actions of the estrogen receptor. Mol Cell Biol. 2007;27:1516–1530. doi: 10.1128/MCB.01550-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodine PV, Komm BS. Wnt signaling and osteoblastogenesis. Rev Endocr Metab Disord. 2006;7:33–39. doi: 10.1007/s11154-006-9002-4. [DOI] [PubMed] [Google Scholar]
- 41.de Frutos CA, Dacquin R, Vega S, Jurdic P, Machuca-Gayet I, Nieto MA. Snail1 controls bone mass by regulating Runx2 and VDR expression during osteoblast differentiation. EMBO J. 2009;28:686–696. doi: 10.1038/emboj.2009.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Hassan MQ, Li ZY, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Intricate gene regulatory networks of helix-loop-helix (HLH) proteins support regulation of bone-tissue related genes during osteoblast differentiation. J Cell Biochem. 2008;105:487–496. doi: 10.1002/jcb.21844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leucht P, Minear S, Ten Berge D, Nusse R, Helms JA. Translating insights from development into regenerative medicine: the function of Wnts in bone biology. Semin Cell Dev Biol. 2008;19:434–443. doi: 10.1016/j.semcdb.2008.09.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.