Abstract
Homing endonucleases (HE) are double-stranded DNAses that target large recognition sites (12–40 bp). HE-encoding sequences are usually embedded in either introns or inteins. Their recognition sites are extremely rare, with none or only a few of these sites present in a mammalian-sized genome. However, these enzymes, unlike standard restriction endonucleases, tolerate some sequence degeneracy within their recognition sequence. Several members of this enzyme family have been used as templates to engineer tools to cleave DNA sequences that differ from their original wild-type targets. These custom HEs can be used to stimulate double-strand break homologous recombination in cells, to induce the repair of defective genes with very low toxicity levels. The use of tailored HEs opens up new possibilities for gene therapy in patients with monogenic diseases that can be treated ex vivo. This review provides an overview of recent advances in this field.
Keywords: Homing endonucleases, Protein engineering, Monogenic diseases, Double-strand break, DNA repair, Gene therapy, Protein structure, Protein–DNA interaction
Homing endonucleases basic mechanisms
Homing is a gene conversion process, whereby a mobile sequence is copied and inserted into a cognate allele that lacks this sequence. This relies on site-specific double-strand cleavage in the target allele, catalyzed by a nuclease encoded within the mobile sequence. Homing was discovered in the early 1970s upon the identification of the intron-encoded “homing endonuclease (HE)” responsible for the significant polarity of recombination observed for rRNA gene markers in genetic crosses between yeast mitochondria [1]. HEs are usually encoded within protein-encoding genes, by self-splicing introns within the RNA (group I, group II and Archaeal introns) or by inteins that self-splice at the protein level (Fig. 1). HEs may also be present as freestanding elements between individual genes. Homing is widespread and HEs or their homologs have been found in all phyla. HEs recognize very long DNA sequences (12–40 bp); hence they are also known as meganucleases. They tolerate certain substitutions within the target site, resulting in the effective recognition and cleavage of families of sequences. However, the long target sites occur only rarely in a whole genome. For instance, I-SceI recognizes only one such site in the ~13,000-kbp yeast genome. The relative rarity of HE target sites means that HEs pose a smaller threat to the integrity of the host DNA than restriction endonucleases (RE). Additionally, the corresponding mobile DNA elements do not require reversible methylation by methylases to protect the host DNA from multiple rounds of cleavage. Instead, intron- and intein-encoded HEs irreversibly disrupt the target site by inserting the DNA of its mobile element. The insertion sites of freestanding HEs encoded by T-even phages are, however, spatially separated (up to hundreds of base pairs) from the cleavage sites [2]. Thus, potential cleavage sites for the freestanding HEs exist in both the host (self) and target (non-self) DNA. In this context, the exact mechanism underlying protection of the self-DNA from cleavage remains unknown. Nucleotide polymorphism may provide a means of distinguishing between the self and non-self DNA. The cleavage-proficient self DNA variants would be counter-selected and eliminated, and the resistant variants would be preferentially inherited in the progeny phage [3].
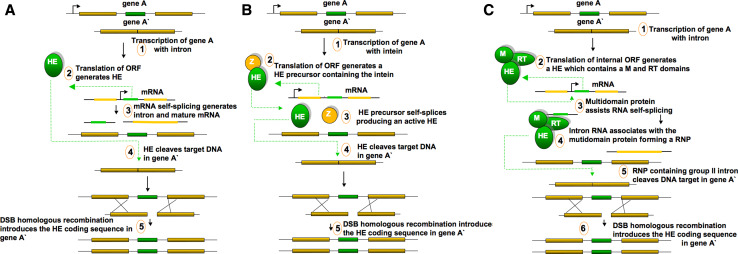
Fig. 1.
Homing mechanisms for group I introns (a), inteins (b) and group II introns (c). The homing endonuclease coding sequence of gene A (green) is duplicated in its cognate allele, gene A′. Mobile ORFs and encoded products are green; host gene exons and products are yellow; the other nucleic acid sequence is black, DNA is coloured with a 3D effect and RNA is flat. The ORF position is indicated with a black arrow. HE Homing endonuclease, M maturase, RT reverse-transcriptase, RNP ribonucleoprotein. The different stages of each mechanism are numbered in the scheme. The diagram showing the mechanism of action for group II introns is highly simplified. More detailed figures of their homing mechanisms can be found in specific reviews [158]
HEs can be considered as selfish nucleases that promote their own proliferation by cleaving the foreign DNA, thereby inducing DNA repair by recombination, which in turn increases the chances of duplication of their genes. HEs often modify the target sites to minimize the destruction of their own DNA. This is achieved through either irreversible disruption of the target DNA during the process of their own proliferation or reversible modification catalyzed by another enzyme [4, 5].
The nomenclature of HEs resembles that of REs, with additional prefixes indicating different classes: I, for intron-encoded enzymes (e.g. I-CreI); PI, for intein-encoded enzymes (usually with an additional protein splicing activity, e.g. PI-SceI); and F, for freestanding enzymes [6]. The homing mechanisms used by the three types of HEs are summarized in Fig. 1. HEs encoded by group I and Archaeal introns or inteins exhibit similar mechanisms of homing (Fig. 1a, b). Their mechanism of action relies solely on the endonuclease, which cleaves the target site in the intron-less allele to generate recombinogenic ends. This leads to strictly DNA-dependent homologous recombination, resulting in the duplication of the intron (or the intein coding sequence) at the cleaved site. The only difference observed is the mechanism of action adopted by these HEs: in the case of group I introns, mRNA self-splices, and in the case of group II, the translated polypeptide self-processes to generate the enzyme (Fig. 1a, b). A similar mechanism is exhibited by freestanding HEs, which cause the duplication of the corresponding gene at a site located at some distance from the cleavage site [3]. The homing of group II introns may or may not involve DNA homologous recombination, but is always initiated by a complex formed by the protein encoded by the intron ORF and the excised intron (Fig. 1c). This ribonucleoprotein (RNP) displays endonuclease, reverse transcriptase and maturase activity (binding their host intron and facilitating RNA-catalyzed splicing). The RNP initiates homing by reverse splicing directly into a DNA target site and is then reverse transcribed by the intron-encoded protein. After DNA insertion, the introns remove themselves by protein-assisted, autocatalytic RNA splicing, thereby minimizing host damage [7].
Homing endonucleases are highly specific and can induce homologous recombination in different types of cells, including mammalian cells. These enzymes are therefore useful templates for the development of custom tools for gene targeting [8]. Such tools may be used for a number of potential biomedical and biotechnological applications, ranging from genome editing to gene therapy. However, the enzyme selected as a template must first be engineered to recognize the new DNA target sequence with high specificity. Prior understanding of the structure–function relationship of these enzymes with their DNA target is therefore essential for the design and development of custom enzymes that target a DNA sequence of interest, at the same time avoiding deleterious side effects when produced in cells.
Homing endonuclease families
The different mechanisms of recognition and cleavage exhibited by group I and group II intron HEs and the existence of distinct families of HEs suggest that different homing endonucleases evolved independently. HEs have been grouped into five families (LAGLIDADG, HNH, GIY-YIG, His-Cys box and PD-(D/E)XK; see Table 1), classified by their conserved active site core motifs [9].
Table 1.
Summary of crystallographic structures of the different homing endonuclease families
| Family representative member | Oligomeric state | DNA bound? no. of bp | DNA curvature | DNA binding mode | Active site ions (no.) | No. of bp overhang after cleavage | Fold/catalytic residues | PDB code |
|---|---|---|---|---|---|---|---|---|
| LAGLIDADG (I-CreI & I-DmoI) | Dimer (I-CreI) |
Yes 24-mer |
45° | Endothermic [160] | 3 Ca2+/3 Mg2+/3 Mn2+ | 4 bp | LAGLIDADG fold/acidic residues coordinate metal ion, metal dependent hydrolysis following an in-line displacement mechanism | 1AF5 (apo) [161], 1BP7 (DNA bound) [21], 1g9z (Mg2+) & 1g9y (Ca2+) [31] |
| Monomer (I-DmoI) |
Yes 25-mer |
40° | – | 1 Ca2+/2 Mn2+ | 4 bp | 1B24 (Apo [162]), 2VS7 (Ca2+), 2VS8 (Mn2+) [22] | ||
| HNH (I-HmuI) | Monomer |
Yes 36-mer |
40° 4–5 bp downstream to the cleavage site | Endothermic [160] | 1 Mn2+ | Nick | HNH fold/D74, N96, N99, H75/H75 is general base, ion is charge stabilizer | 1U3E (Mn2+) [55] |
| His-Cys Box (I-PpoI) | Dimer |
Yes 21-mer |
55° across the central 4 bp | Exothermic [160] | Mg2+, Mn2+, Na+/1 ion per active site | 4 (5 [65, 66]) | HNH fold/D119, H98, C105, R61/H98 is general base, ion is charge stabilizer | 1A73 (Mg2+), 1A74 (no ion) [68], 1CZ0 (Na+), 1CYQ (H98A), [70] 1EVX (apo), 1EVW(L116A) [71], 2O6 M (H98Q) [72] |
| GIY-YIG (I-TevI) | Catalytic domain Monomer | No | −40° toward the major groove after bottom strand cleavage [87] | NA | Mn2+/one ion per active site | 2 | Novel fold/E75, R27, Y17 | 1LN0, 1MKO (E75A mutant) [80] |
| DNA-binding domain Monomer |
Yes 21-mer |
Mainly undistorted | NA | – | – | Zn finger- NUMOD3-IENR1 domains define cleavage site/ | 1I3 J [91], 1T2T [163] | |
| PD-(D/E)XK (I-SspI) | Tetramer (only two active subunits) |
Yes 29-mer |
25° around the central bases | Exothermic [160] | Ca2+, Mg2+/one ion (Ca2+) per active site in mutant structure, wild type might have two ions per active site | 3 | PD-(D/E)-XK nuclease fold/acidic residues coordinate metal ion, metal dependent hydrolysis following an in-line displacement mechanism | 2OST (Ca2+) [97] |
A new HE family was recently predicted from an analysis of microbial environmental metagenomic sequence data [10]. The authors found a novel genomic arrangement of certain genes, whereby the coding region of a conserved enzyme was divided into two by a split-intein, with an endonuclease gene in between the two coding regions. These freestanding endonuclease genes were found to belong to different families, the GIY-YIG family or the novel very-short patch repair (Vsr)-like family of endonucleases. Sequence-to-sequence or sequence-to-multiple sequence alignment comparisons showed similarities between these endonuclease-encoding regions and Vsr DNA G:T-mismatch endonucleases [11]. Broader sequence similarity searches showed that this family of HEs is also present in group I introns and as freestanding inserts in the absence of surrounding intervening sequences [10].
As well as their conserved sequence motifs, the HE families also display differences in their catalytic mechanism, biological and genomic distribution, and in their similarities with non-homing endonucleases. The structure–function relationships of the different families, with some representative examples, are described below and summarized in Table 1. The families described are all encoded within group I introns or inteins, and some of them are also associated with maturase activity. No structural data are currently available for group II intron HEs, which function as part of a RNP complex with its own excised intron RNA.
The LAGLIDADG family
These endonucleases contain the LAGLIDADG sequence motif. They are the best characterized of the HE families. Several hundred intron-encoded nucleases have been identified and their endonuclease activity demonstrated experimentally in many cases. They have a widespread distribution, being present in the genomes of plant and algal chloroplasts, fungal and protozoan mitochondria, and Archaea [12]. On the basis of primary sequence homology and additional information from structural studies of some of its members, the LAGLIDADG family can be divided into two subfamilies. Homing endonucleases belonging to the first subfamily function as homodimers and bind consensus DNA targets displaying palindromic or near-palindromic symmetry. Each monomer contains a single copy of the LAGLIDADG motif that forms the core of the dimer interface, with the conserved acidic residues forming part of the active site. Examples from this family are I-CreI, I-CeuI and I-MsoI (Fig. 2) [13–16]. The second subfamily consists of proteins with two LAGLIDADG motifs, one in each of the two similar domains of the monomeric enzyme. These monomeric pseudosymmetric enzymes recognize and cleave non-palindromic DNA sites. Examples from this family are I-AniI, I-DmoI or I-SceI (Fig. 2) [17–19].
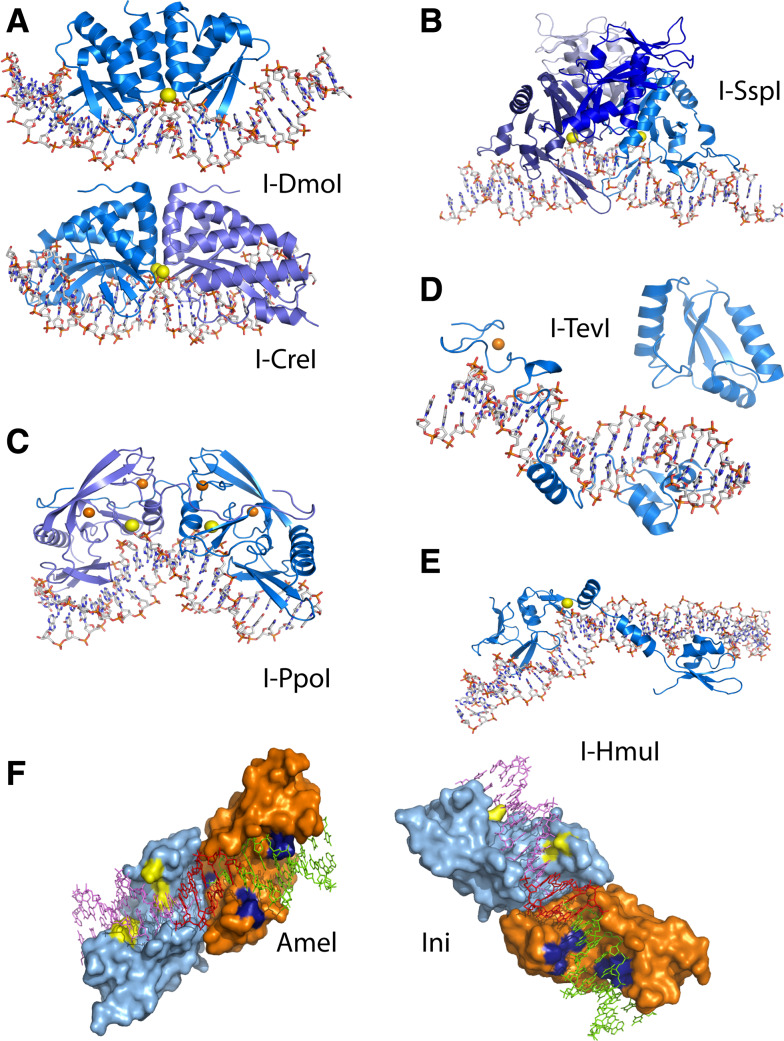
Fig. 2.
Crystallographic structures of representative members of the five HE families. a LAGLIDADG family: monomeric I-DmoI and homodimeric I-CreI. b PD-D/E-XK family: the tetrameric I-SspI. c His-Cys Box family: the homodimeric I-PpoI. d GIY-YIG family: I-TevI catalytic and DNA binding domains. e HNH family: the monomeric I-HmuI. f Engineered heterodimeric variants Amel and Ini based on the I-CreI template. a–e The enzymes are shown in cartoon representation with the bound target site in stick representation. Catalytic ions are shown as yellow spheres and structural Zn ions are shown as orange spheres
LAGLIDADG HEs recognize long DNA sites (22 bp and longer) and cleave along the minor groove to generate cohesive 4 bp 3′-OH overhangs. X-ray structures obtained for several LAGLIDADG proteins from the two subfamilies (I-CreI, I-MsoI and I-CeuI, and I-AniI, I-SceI, I-DmoI and PI-SceI) bound to their DNA targets have provided insight into their mechanisms of DNA recognition, binding and catalysis [13, 18–24].
The first two artificially engineered meganuclease chimeras, H-DreI and DmoCre [25, 26], were generated using templates from the LAGLIDADG family, by fusing the N-terminal domain of I-DmoI to I-CreI. Different studies have shown that single-chain variants of I-CreI (whose native form is homodimeric) retain their catalytic activity and can be further modified to recognize non-palindromic DNA sequences [26, 27].
The LAGLIDADG enzymes are quite flexible in terms of target DNA recognition and can tolerate individual polymorphisms without considerable loss of binding or cleavage efficiency [13, 19, 23]. The extended substrate-binding cleft allows direct protein-DNA contact, facilitating docking before and during catalysis. This family of enzymes forces an acute twist of the DNA to bring the scissile phosphodiester bonds of both strands together at the narrow catalytic site. The catalytic mechanism resembles the canonical two-metal-ion mechanism, previously described for phosphoryl transferases and RE [28–30]. In-depth studies of the mechanisms of catalysis for both subfamilies have been based on structural data obtained for I-CreI and I-DmoI bound with their DNA targets [20, 22, 31]. Catalysis requires the two conserved acidic residues at the carboxyl-termini of the LAGLIDADG helices in the active site and coordination of the divalent metal ions. The structures of uncleaved substrate complexes (determined in the presence of non-activating Ca2+ ions) for I-CreI and I-SceI, show the presence of three Ca2+ ions at the active site. Similarly, the corresponding cleaved substrate structures had three Mg2+/Mn2+ ions in the active sites [31]. However, the structures of I-CeuI and I-DmoI show the unambiguous presence of a single Ca2+ ion [22, 24], although in non-equivalent positions within the active site. The cleaved substrate structure of I-DmoI shows the presence of only two Mn2+ cations, with a water molecule modelled at the central position. Anomalous signal was not observed at this central position in the Mn2+ anomalous difference Fourier electron density map, that corresponds to the third metal in other enzymes [22]. This is also seen in the structures of I-AniI and PI-SceI [18, 19, 23], which do not show any ion or water molecules in the shared central position. It is possible that this third metal site is transiently occupied during catalysis. The presence of the single Ca2+ ion in the I-DmoI substrate bound structure is indicative of asymmetry during catalysis, suggesting a preference in the cleavage of one of the DNA strands [17, 22], as previously suggested for I-SceI [22, 32].
A subset of HEs act as maturases, these enzymes help in the splicing of their host intron by promoting a conformation that favors RNA-catalyzed splicing. The majority of known group I maturases are members of the LAGLIDADG protein family, acting as both homing endonucleases and RNA-splicing maturases. I-AniI is a good example of an enzyme that acts both as a highly specific endonuclease and as an RNA maturase [18, 33]. RNA maturase activity is required for normal expression of the mitochondrial apocytochrome b oxidase gene [33, 34]. It thus confers a selective advantage to the host and the LAGLIDADG protein-fold is thereby maintained.
HEs from the LAGLIDADG family have been engineered to specifically target new DNA sequences and are emerging as a powerful tool for gene targeting (see “Gene therapy applications”). Some have been designed using a combinatorial approach for binding and cleaving of specific DNA sequences [35–38] (Fig. 2).
The HNH family
The distinguishing signature motif of this family of HEs is their HNH nuclease motif, in which the last histidine can sometimes be substituted by asparagine. HNH motifs appear to have diverged from a common ancestor, adopting a variety of biological roles that require the action of a nuclease. Indeed, this motif is found in a wide range of enzymes including HEs coded by group II introns, intein-associated HEs, non-specific bacterial and fungal nucleases, such as the colicin bacterial enzyme family [39, 40], DNA-acting enzymes such as transposases, restriction endonucleases (such as KpnI, HpHI, Hind4II, Eco31I), DNA packaging factors, and bacterial factors involved in developmentally controlled DNA rearrangements [12, 41–45]. The HNH family has been divided into at least eight subfamilies based on the sequence of the nuclease core containing the HNH motif and the presence or absence of distinctive conserved flanking residues, such as cysteine pairs that may bind to additional metal ions [41]. Unlike members of the other families, some of the HNH HEs, such as I-HmuI, generate single-strand breaks in the target DNA [46].
Well characterized members of the HNH family encoded within group I introns include I-HmuI and I-HmuII from the B. subtilis bacteriophage SPO1 and SP83 introns [47–50], I-BasI from Bastille bacteriophage [51], phage T4 endonuclease I-TevIII [52], the I-CmoeI endonuclease from the psbA gene of the C. moewusii chloroplast [53] and the related ORF from the same gene in C. reinhardtii [54]. So far, the only solved structure in this family is that of I-HmuI bound to its double-stranded DNA target (Fig. 2). The structural data obtained showed the cleavage of a single strand of its target, forming a nick in the DNA [49, 55]. The only bound metal ion binds the free 5′-phosphate, the 3′-OH, and a pair of enzyme side-chains. The elongated structure of I-HmuI shows a series of continuous different structural domains and motifs that perfectly intercalate and distort the DNA chain. The number of specific (protein-base) contacts within its binding site is small (14 out of 25 bp, 2 flanking the site of the strand break) [55]. For this reason, although I-HmuI, and probably its analogous HNH endonucleases, can accommodate a much longer target than known members of the LAGLIDADG and His-Cys box families, its overall specificity is low.
His-Cys box family
The host genes for all known His-Cys box family members have been identified as nuclear ribosomal DNA loci in a variety of eukaryotic organisms. This family has a conserved region of around 100 amino acids that contains conserved His and Cys residues [56, 57]. Amino-acid sequence beyond this motif is not significantly conserved across the family. Several family members, such as I-PpoI, I-DirI and I-DirII, have been studied. I-PpoI, the most fully characterized member, is a 163 amino acid protein encoded within the mobile intron PpLSU3, found in the extrachromosomal rDNA of the myxomycete P. polycephalum [58–60]. I-DirI and I-DirII are around 260 amino acids long. They were identified within group I introns at position 956 in the SSU rDNA of two closely related isolates of the plasmodial slime mould D. iridis [61–63]. Recent reports (reviewed in [64]) shed some light on the expression of these rDNA-encoded proteins, highlighting the important role of catalytic RNAs.
Members of the His-Cys box family of HEs recognize and cleave intronless homing sites of around 15–19 bp. The homing sites are palindromic or pseudopalindromic. Similarly to other HE enzymes, their DNA target sites may be modified at many positions—multiple substitutions—but still be recognized and cleaved. However, insertions and deletions are not tolerated and prevent cleavage [60, 65, 66]. The regions flanking the homing site come into contact with the enzyme—the footprint of I-PpoI extends to 23 bp, whereas its homing site is only 15 bp—and can affect catalysis, independently of the reaction conditions. Most members of this family generate 4 bp 3′-OH overhangs [67]. However, the Naeglia sp. nucleases I-NanI, I-NitI and I-NgrI were found to generate 5 bp 3′-OH staggered ends, differing from all other known HEs [65–67]. The oligomeric state of the His-Cys box family members has only been directly observed for I-PpoI, which forms a dimer [68] (Fig. 2). The sequence outside the His-Cys motif is not sufficiently conserved across family members to allow their oligomeric state to be predicted. However, the palindromic nature of their homing sites is indicative of a homodimeric scaffold, although not necessarily resembling the I-PpoI dimer interface.
The structure of I-PpoI in its apo form and bound to its homing site has been studied extensively by X-ray crystallography [68–72]. Many divalent cations can be used by I-PpoI, although there is a preference for those that can adopt a octahedral geometry [59]. The I-PpoI dimer displays a mixed α/β topology, resulting in an elongated molecule. In contrast to LAGLIDADG HEs, the I-PpoI dimer interface is small, highly solvated and more loosely packed. The C-terminal end residues are domain-swapped and contribute to the dimer interface. Cys or His residues in the His-Cys box motif form two novel Zn binding folds, which have a structural and stabilizing role. I-PpoI significantly distorts the homing-site DNA, rendering the scissile phosphodiester bonds accessible to the two separate active sites. Structure-guided protein engineering has been used to alter the DNA recognition specificity of I-PpoI [73]. These studies found little correlation between the binding, selectivity and cleavage properties of individual variant proteins.
GIY-YIG family
Most GIY-YIG family enzymes are complex multidomain proteins that perform various functions by combining the characteristic GIY-YIG catalytic domain with other non-catalytic domains responsible for DNA binding specifity [74]. Therefore, due to their non-catalytic function, the regions outside this domain are not conserved across family members, and vary from enzyme to enzyme. Multiple sequence alignments have been used to further examine the catalytic domain [75]. This domain ranges between 70 and 100 amino acids in length and contains five conserved sequence motifs. Not all the motifs are necessarily present in all the proteins in the family. One of these is the GIY-YIG motif which gives the family its name. The catalytic residues are not present in this characteristic sequence but are found in the other four conserved motifs within the catalytic domain. HEs belonging to this family have been characterized by the presence of the conserved sequence, “GIY-10/11 residues-YIG”. However, recent observations have shown that at least one family member, I-BathII, has an unconventional GIY-(X)8-YIG motif [76]. The GIY-YIG domain is found in proteins encoded by group I introns in the mitochondria and chloroplast genomes of fungi, algae and liverworts and in proteins encoded within the genomes of several bacteriophages (T4 and T4-related phages), in intergenic regions of bacteria, phage and viruses, and in the UvrC subunits of bacterial and archaeal (A)BC excinucleases [75].
The most extensively studied member of this family, I-TevI, consists of an N-terminal GIY-YIG catalytic domain and a C-terminal DNA-binding domain, which are joined by a 75 amino acid linker containing a Zinc-finger [77–79] (Fig. 2). The catalytic domain encompasses the five motifs conserved by this family and has a novel α/β fold with a central three-stranded anti-parallel beta sheet flanked by three helices [80]. The C-terminal domain itself is composed of a minor groove-binding α-helix (‘NUMOD3’ motif) and a helix–turn–helix (HTH) motif also referred to as ‘IENR1’ domain [81]. It resembles the C-terminal DNA-binding domain of the HNH HE I-HmuI. As mentioned above, the DNA binding domain is an unusually extended structure that wraps around the DNA, lining the minor groove of the DNA-duplex. The interdomain segment of I-TevI has a complex function and has been studied in depth [75, 82, 83]. It has essential roles in determining distance along the sequence when selecting the cleavage site; indeed, the catalytic domain on its own makes non-specific cuts. Mutations in the Zn-finger and in an unstructured segment show that the linker is essential for normal I-TevI function.
These HEs generate double-strand breaks (DSB) in a divalent ion-dependent manner, producing 2 bp 3′-OH overhangs. Mg2+ is most commonly used for functional experiments but these enzymes can use other cations [84]. It remains unclear how a monomeric enzyme with one active site makes two cuts in the DNA strand. These enzymes may act as monomers, although there are no structural data available for the full-length protein bound to DNA. They cleave the two strands of the DNA by independent nicking reactions; thus, there is either a conformational change in the DNA to accommodate both strands in one active site or the two catalytic domains may oligomerize to form a transient complex with the DNA [85]. This issue has been addressed in biochemical studies of double-strand break formation by I-TevI and I-BmoI [85–87]. Additionally, a role for monomer–dimer transition has been demonstrated in the cleavage of double-stranded DNA by the Eco29kl endonuclease, another GIY-YIG superfamily member [88].
The GIY-YIG HEs recognize long DNA targets (around 40 bp). The C-terminal DNA binding domain recognizes a region of approximately 20 bp around the intron insertion site and the N-terminal catalytic domain cleaves approximately 25 bp upstream [89–92]. In general, GIY-YIG HEs cleave their intronless homing sites with relaxed specificity. I-TevI, for example, tolerates several mutations, as well as insertions of up to 5 bp and deletions of up to 16 bp between the intron insertion site and the cleavage site [93, 94].
The PD-(D/E)XK family
This family of HE belongs to the PD-(D/E)XK superfamily of nucleases. The first member of this family to be identified was I-Ssp6803I, or I-SspI for simplicity. It was the first example of a chromosomally encoded group I intron endonuclease in bacteria [95]. I-SspI is encoded by a self-splicing intron in the tRNAfMet gene of Synechocystis PCC6803 [95, 96].
I-SspI is a very small protein, only 150 amino acids long, making it the smallest HE characterized to date. Sequence comparison of I-SspI with the Hjc resolvase family revealed several conserved residues indicative of a canonical PD-(D/E)-XK active site within this HE [97, 98]. The PD-(D/E)-XK nuclease fold locates the catalytic residues at the concave surface of a curved β-sheet, facing an α-helix. This fold is typically found in most known RE and in a wide variety of DNA repair enzymes, all of which act on short DNA targets with strict fidelity. I-SspI was the first HE to be identified with this fold, which had previously only been found in REs. Similarly, the LAGLIDADG fold has only been found in HEs. I-SspI is unique in that it supports the PD-(D/E)-XK fold, but recognizes a long site: 23 bp, of which the central 17 bp show particularly high specificity [95, 97, 99]. This target site is pseudopalindromic and encompasses the entire anticodon stem and loop of the tRNAfMet gene.
I-SspI forms a tetramer that binds one DNA molecule (Fig. 2), consistent with solution studies [97]. This tetrameric arrangement allows the coding gene I-SspI to be small. This minimizes its interference with the folding and splicing of its host intron and creates an elongated DNA-binding surface that is able to recognize a long target site. At the same time, two of the subunits become separated active centers that cleave each strand of the double helix, while the two other subunits in the tetramer stabilize the whole assembly.
This enzyme cuts its target DNA on both strands and, in contrast to other HEs, cleaves one DNA strand precisely at the position of intron insertion. The DNA is cleaved across the minor groove producing 3′-OH single-strand overhangs. These overhangs are the result of a metal-dependent in-line displacement mechanism, characteristic of the PD-(D/E)XK fold. However, I-SspI is unique in that the extensions are three bases long rather than the two or four bp overhangs generated by most other HEs. The enzyme generates transient nicked intermediates during DNA cleavage under certain conditions [99]. DNA base-pair mutations at several positions show a strong correlation between cleavage and binding specificities; some mutations however affect cleavage but not binding [99].
Mechanisms of double-stranded break repair
Homologous recombination (HR) results in the exchange of genetic information between two endogenous DNA molecules or between an endogenous and exogenous DNA molecule. It may be useful in gene therapy for the exchange between an endogenous defective chromosomal sequence and an exogenous repair DNA construct. HR requires a few hundred base pairs of homology between both the targeting and the targeted DNA. The presence of free DNA ends in the exogenous construct is sufficient to stimulate this process [100]. These free DNA ends are resected to generate 3′ single-stranded overhangs and to form a nucleofilament with proteins from the RecA/Rad51 family [101, 102]. This nucleofilament then invades the homologous duplex, resulting in strand transfer [101, 103] (Fig. 3).
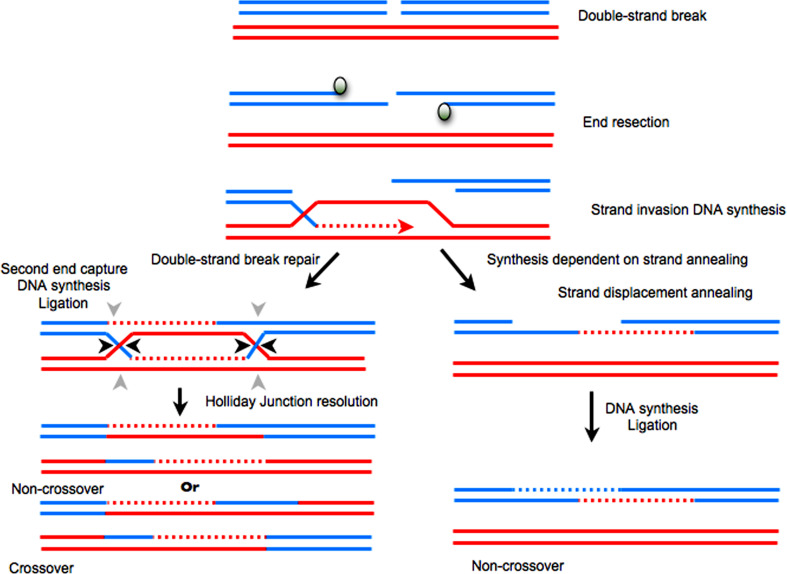
Fig. 3.
Double-strand breaks (DSBs) can be repaired by several homologous recombination (HR)-mediated pathways, including double-strand break repair (DSBR) and synthesis-dependent strand annealing (SDSA). Upper In both pathways, repair is initiated by resection of a DSB to provide 3′ single-stranded DNA (ssDNA) overhangs. Strand invasion at these 3′ ssDNA overhangs into a homologous sequence is followed by DNA synthesis at the invading end. Lower left After strand invasion and synthesis, the second DSB end can be captured to form an intermediate with two Holliday junctions (HJs). After gap-repair DNA synthesis and ligation, the resolved structure at the HJs may be a non-crossover (black arrowheads at both HJs) or crossover product (green and black arrowheads). Lower right Alternatively, the reaction may proceed to SDSA by strand displacement and the annealing of the extended single-strand end to the ssDNA at the other break end, followed by gap-filling DNA synthesis and ligation. The repair product from SDSA is always non-crossover
Non-homologous end joining (NHEJ) and HR are two competing DSB repair pathways. NHEJ is used for DSB repair in the absence of a homologous repair sequence [102–104]. The most frequent outcome of NHEJ is the perfect joining of the broken ends; however, imperfect rejoining may result in the addition or deletion of base pairs, inactivating the targeted open reading frame.
Strategies for gene repair using meganucleases
Depending on the strategy chosen to repair or edit a given gene, different options are available (Table 2). In many gene repair applications, mainly for therapeutic purposes, endonuclease-induced breaks must be repaired in a conservative manner to maintain the information contained in the DNA sequence. DSB can be repaired by either HR or by NHEJ. HR occurs without loss of sequence information. However, NHEJ usually results in the loss of sequence at the repair junction and can promote chromosome translocations at DSBs, leading to genomic instability [105]. Recent studies have shown that the use of a nicking enzyme rather than a DSB nuclease might stimulate HR while reducing genomic instability associated with the alternative NHEJ repair of DSBs [106, 107]. Alternatively, these single-strand breaks can be solved by proteins involved in downstream steps of the base excission repair pathway [108, 109]. However, when single-strand breaks are not rapidly or properly repaired, they can collapse replication forks, block transcription or promote excessive activation of the single-strand break sensor protein poly(ADP-ribose) polymerase 1 (PARP1) with dangerous consequences for the cell [110]. Taking this into account, together with the structure–function relationships described above, members of the HNH and GIY-YIG families seem to be appropriate candidates for use as templates to engineer a nicking enzyme that promotes DSB repair using the HR pathway. Additionally, they bind non-palindromic DNA targets, widening the range of potential sequences that can be targeted. However, they lack specific protein-DNA contacts, consequently lowering their natural specificity and increasing the risk of deleterious off-site cleavage. Similarly, although the PD-D/E-XK family displays a high specificity for their targets, the tetrameric oligomerization state, observed in I-SspI, presents an extra challenge in redesigning the DNA recognition properties of members of this family. Members of the His-Cys and LAGLIDADG families may therefore be considered as more suitable templates. Both of these types of HE are highly specific. Attempts at the structure-based protein engineering of I-PpoI have yielded ambiguous results [73]. However, newly designed variants of I-CreI, a representative homodimeric member of the LAGLIDADG family, have been able to target DNA sequences that are completely different from its wild-type target sequences [38]. The LAGLIDADG family also has the advantage of providing two scaffolds, monomeric and homodimeric, widening the range of sequences, palindromic and non-palindromic, that can be targeted (Table 2).
Table 2.
Schematic summary of the strategies employed to generate HE variants with novel target specificities
| Method | Cluster mutagenesis | Directed evolution | Chimeras | Single-chain variants | Computational redesign | Semi-rational design | Semi-rational genomic target design |
|---|---|---|---|---|---|---|---|
| Homing Endonuclease |
I-CreI I-CreI I-CreI I-Ssp6803I |
I-SceI PI-SceI I-ScaI PI-SceI I-SceI |
I-DmoI_I-CreI I-DmoI_I-CreI PI-SceI I-DmoI |
I-CreI I-MsoI I-CreI |
I-MsoI I-CreI |
I-CreI | I-CreI |
| Structural information required |
Yes Yes Yes Yes |
No Yes Yes |
Yes Yes Yes Yes |
Yes Yes Yes |
Yes Yes |
Yes Yes |
Yes Yes Yes |
| Variant selection |
Two-plasmid selection in E. coli High-throughput fluorescence competition-binding assay |
Two-plasmid (two-hybrid) selection in E. coli | In vitro cleavage |
In vivo cleavage Two-plasmid selection in E. coli In vitro and in vivo cleavage and reparation |
In vitro cleavage | High-throughput screening in yeast | Combinatorial generation of variants and high-throughput screening of variant/target in yeast |
| Variant/DNA crystal structures |
No Yes Yes Yes |
No No No |
Yes No No No |
No Yes Yes (Unpublished) |
Yes No |
No No |
No No Yes |
| References |
[164] [165] [166] [99] |
[167] [168] [125] |
[25] [26] [169] [170] |
[26] [171] [27] |
[128] [172] |
[126] [36] |
[173] [35] [38] |
Gene therapy applications
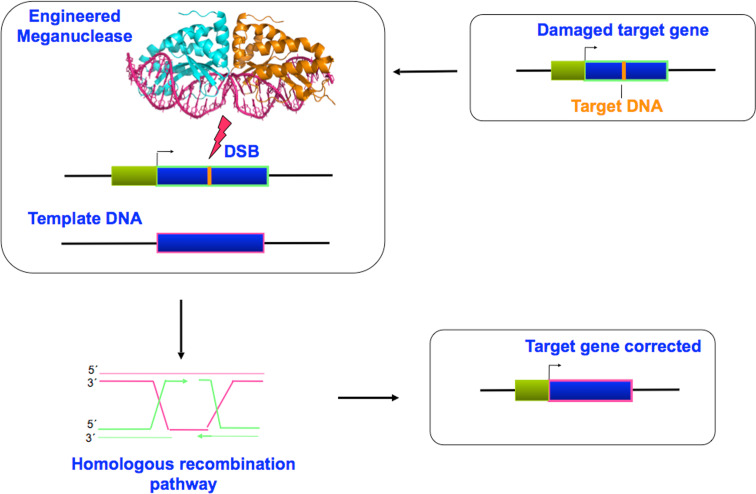
Cell and gene therapy are emerging as fundamental tools, fighting genetic diseases by attacking at the origin of their cause, through the repair of or compensation for the defective gene. However, most strategies tested so far have relied on retroviral or lentiviral vectors to deliver the corrected gene. Due to the semi-random viral integration throughout the genome, these strategies have several potential limitations: transgene silencing, disruption of endogenous genes and the transcriptional activation of neighboring genes. An alternative approach involves use of the correct DNA copy to specifically target the mutated gene for correction of the defect. Gene repair does not simply restore a physiological pathway or function, but directly erases the deleterious mutation (Fig. 4). This brings significant advantages, elimintating the risk of oncogene activation, and alleviating the recurring problem of transgene silencing.
Fig. 4.
Diagram showing the ex vivo approach. The location and characterization of chromosomal damage is followed by the introduction of the engineered HE in the isolated cells population together with the correcting template DNA matrix. The mutated gene, upon DBS-induced HR, is repaired and, after selection, cells with a normal gene are recovered
Engineering meganucleases for therapeutic purposes
The use of HEs in therapeutic applications is dependent on the capacity of a DSB to actively induce HR [100, 111]. The potential repair of a particular mutated gene requires the generation of an engineered meganuclease that forms a DSB as close as possible to the mutated gene and triggers HR (Figs. 5, 6). Unfortunately, difficulties in modifying the specificity of DNA recognition has limited the engineering of DNA-binding proteins [112, 113]. However, recent progress has allowed the production of nucleases that initiate DSB-induced recombination at a chosen locus. A canonical zinc finger (ZF) DNA binding motif has been engineered and fused to the catalytic domain of FokI to produce zinc finger nucleases (ZFNs) composed of two subunits, which assemble to form dimers at the cleavage site [114, 115]. Engineered ZFNs, together with a chromosomal repair matrix, can induce gene correction in the Drosophila yellow gene, in the germ line as well as in somatic tissues [116]. Several studies in HEK-293 cells have demonstrated that ZFNs can induce 0.4, 0.5 and 1% efficiency of maximal gene correction [117–119]. A similar experimental design was also successfully applied to tobacco plants [120]. However, high levels of cytotoxicity, potentially hampering their therapeutic use, were observed in Drosophila and mammalian cells, presumably due to off-target cleavage. In another study ZFNs were redesigned to correct mutations in the human ILR2G-encoding gene. This work observed gene correction with up to 18 and 5% efficiency in K562 cells and in human CD4 + T cells, respectively, with reduced toxicity [121]. In silico studies, using protein modelling and energy calculations, have allowed further development of ZFNs to reduce non-specific cleavage [122].
Fig. 5.
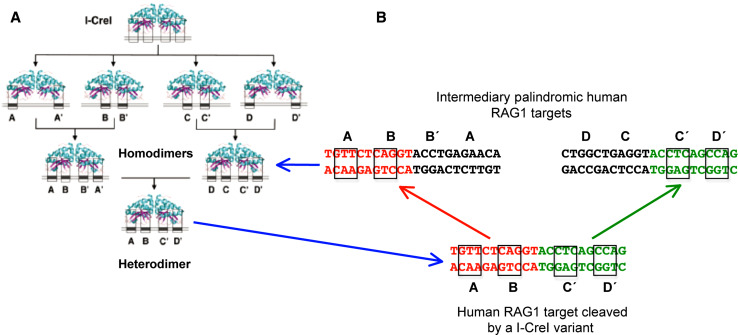
Strategy for the making of redesigned HEs. a General strategy. I-CreI variant libraries with locally altered specificity are generated. These mutants were assembled into homodimeric and heterodimeric proteins using a combinatorial approach, generating meganucleases with fully redesigned specificity. b The RAG1 series of targets. Two intermediary palindromic targets were derived from the non-palindromic human gene target. These were used to select for the homodimeric I-CreI variants that served as scaffold for the specific heterodimer. The two 3 bp segments used in the library screening are boxed
Fig. 6.
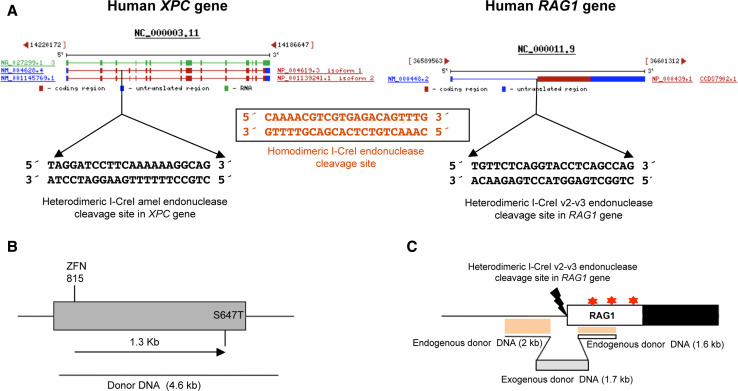
Targeted genes by modified endonucleases. a Human XPC (left panel) and RAG1 (right panel) genes showing the sequence recognized and cleaved by I-CreI amel [38] and I-CreI v2-v3 [27] variants which differ in 15 bp and 16 bp respectively from the original I-CreI 22 bp target (black box) [31]. The gene information was retrieved from http://www.ncbi.nlm.nih.gov/. b Tobacco acetolactate synthase gene showing the ZFN DSB site and the maximum endogenous repaired distance obtained by HR at a frequency of 2% [145]. c Human RAG1 gene showing the I-CreI v2-v3 DSB site and the donor DNA construct used to verify the 6% frequency of exogenous DNA integration, as a marker, in the genomic locus [27]. Red stars show clusters of mutations in the active core of the RAG1 protein that cause T-B-severe combined immune deficiency or Omenn syndrome [159]
HEs have emerged as very promising tools to promote DNA DSB for gene targeting due to their low cleavage frequency in eukaryotic genomes. Although several hundreds of HEs have been identified, the repertoire of cleavable sequences available to target any gene of interest is still very limited. Known structures can thus be used as a basis to modify the specificity of known HEs and create new custom variants (Table 2). I-CreI is a well-characterised homodimeric member of the LAGLIDADG family of HEs (LHE). Mutations in the DNA-interacting regions of this enzyme allow the cleavage of novel targets or lead to loss of cleavage activity at the I-CreI natural site [123, 124]. The DNA-binding residues of I-SceI have also been modified to obtain further variants with altered binding specificity [125].
The crystal structure of I-CreI with its natural DNA reveals two groups of residues that make specific contacts with two 3-bp segments of one half of the pseudo-palindromic target sequence [31]. Residues in each group were randomly mutated to generate different libraries of I-CreI variants (Fig. 5). These libraries were tested for cleavage against all 64 possible targets originated from changing each of the three bases in the corresponding segment to the four nucleotides. A high-throughput screening (HTS) method was used to test a large number of mutant-target combinations [36, 126]. As described below, it is based on inducing HR by DSB in yeast. The I-CreI library variants were cloned into a replicative yeast expression vector and transformed in a selective S. cerevisiae strain. The 64 combinatorial target DNAs were cloned in the LacZ-based yeast reporter vector, disrupting the LacZ gene, and trasformed in a complementary yeast strain. After mating of the two yeast strains, positive events were detected by selection of blue colonies due to the generation of the complete LacZ gene after I-CreI variant DSB cleavage and recombination. Millions of combinations were screened, leading to the identification of several hundreds of mutants with independent altered local specificity. These mutants were then used in a combinatorial screen, which was expanded to include homodimeric and heterodimeric variants of I-CreI (Fig. 5a). In vitro studies confirmed that many of the active variants maintained the essential properties of the wild-type I-CreI molecules, including structure, stability, cleavage efficiency and narrow specificity. Computational analysis, based on energy calculations, helped to improve the active variants by highlighting key residues [36, 127, 128].
The general redesign approach, based on the methods described above, can be summarised in two main steps: firstly, semi-rational mutagenesis followed by high-throughput screening generates I-CreI variants with changes in specific regions to alter their ability to recognize and cleave target DNA; secondly, a combinatorial strategy is used to join the altered domains to generate new HEs with changes in the DNA-binding domain, allowing recognition of different DNA targets. This approach also offers the considerable advantage over computational design of being able to select HEs to target a sequence that differs greatly from the natural target sequence (Figs. 5b, 6).
This general approach was used for the successful design of new I-CreI variants that cleave the human XPC (Xeroderma pigmentosum group C) gene [35]. Xeroderma pigmentosum (XP) is a monogenic disease associated with an extreme sensitivity to sunlight (Table 3). XP patients are at an increased risk from skin cancer and, in some cases, develop neurological defects [129, 130]. There is no efficient treatment available to XP patients, apart from repeated surgery to remove affected skin areas. The majority of patients die due to metastasis before reaching adulthood. Attempts have been made to replace affected areas of skin with skin from another site, which has not previously been exposed to the sun, from the same patient [131, 132]. However, the skin grafts are also sun-sensitive and the benefit to the patients is limited to a few years. XP diseases (Table 3) are good candidates for cell therapy with HEs, as they are monogenic and amenable to ex vivo treatment (Fig. 4). Cells from the skin lineage can be easily manipulated ex vivo, and then used to reconstruct functional skin [133, 134]. I-CreI has been succesfully engineered to recognize two sequences from the XPC gene [35]. Two derivatives of the modified I-CreI, recognizing a specific sequence in the XPC1 gene, have been found to induce high levels of specific gene targeting in mammalian cells with no obvious signs of genotoxicity [38] (Table 2).
Table 3.
Summary of monogenic diseases that may be amenable to treatment based on homing endonuclease-induced double-strand break homologous recombination
| Disease | Gene | OMIMa | Chromosome | Frequency | Inheritance | References |
|---|---|---|---|---|---|---|
| Acute intermittent porphyria | Porphobilinogen deaminase | 609806 | 11q23 | >104 | Autosomal dominant | [174] |
| Congenital erythropoietic porphyria | Uroporphyrinogen cosynthase | 263700 | 10q25 | >105 | Autosomal recessive | [175, 176] |
| Porphyria cutanea tarda | Uroporphyrinogen decarboxylase | 176100 | 1p34 | <104 | Autosomal dominant | [177] |
| Hereditary coproporphyria | Coproporphyrinogen oxidase | 612732 | 3q12 | >105 | Autosomal dominant | [178] |
| Variegate porphyria | Protoporphyrinogen oxidase | 600923 | 1q22 | >105 | Autosomal dominant | [179] |
| Erythropoietic protoporphyria | Ferrochelatase | 612386 | 18q21 | >105 | Autosomal dominant | [180] |
| α Thalassemia | α Hemoglobin (HBA1) | 141800 | 16p13 | <104 | Autosomal recessive | [181] |
| α Thalassemia | α Hemoglobin (HBA2) | 141850 | 16p13 | <104 | Autosomal recessive | [182] |
| β Thalassemia | β Hemoglobin (HBB) | 141900 | 11p15 | <104 | Autosomal recessive | [183] |
| Sickle cell anemia | β Hemoglobin (HBB) | 603903 | 11p15 | <104 | Autosomal recessive | [184] |
| Familial mediterranean fever | Familial mediterranean fever (MEFV) | 608107 | 16p13 | <104 In Mediterranean people | Autosomal dominant | [185] |
| Severe combined immunodeficiency (SCID) | Adenosine deaminase (ADA) | 608958 | 20q13 | <105 | Autosomal recessive | [186, 187] |
| Severe combined immunodeficiency (SCID) | Interleukin 2 receptor gamma (IL2RG) | 308380 | Xq13 | >105 | X-linked | [188] |
| Severe combined immunodeficiency (SCID). Omenn syndrome | Recombination-activating gene (RAG1) | 179615 | 11p13 | >104 | Autosomal recessive | [189] |
| Severe combined immunodeficiency (SCID). Omenn syndrome | Recombination-activating gene (RAG2) | 179616 | 11p13 | >104 | Autosomal recessive | [190] |
| Severe combined immunodeficiency (SCID) | Janus kinase 3 (JAK3) | 600173 | 19p13 | <104 | Autosomal recessive | [191] |
| Chronic myeloid leukemia | Breakpoint cluster region | 151410 | 22q11 | <105 | Somatic | [192] |
| Chronic granulomatous disease | CYBB (gp91-phox), cytochrome b(-245) beta subunit | 300481 | Xp21 | <105 | X-linked | [193] |
| Chronic granulomatous disease | CYBA (p22-phox), cytochrome b(-245) alpha subunit | 608508 | 16q24 | <105 | Autosomal recessive | [194] |
| Chronic granulomatous disease | NCF1 (p47-phox), neutrophil cytosolic factor 1 | 608512 | 7q11 | <105 | Autosomal recessive | [195] |
| Chronic granulomatous disease | NCF2 (p67-phox), neutrophil cytosolic factor 2 | 233710 | 1q25 | <105 | Autosomal recessive | [196] |
| Hemophilia A | Coagulation factor VIII | 306700 | Xq28 | <104 | X-linked | [197] |
| Hemophilia B | Coagulation factor IX | 300746 | Xq27 | <105 | X-linked | [198] |
| Fanconi anemia | FANCA-N, fanconi anemia complementation group | 227650 | 16q24, Xp22, 9q22, 13q12, 3p25, 6p22, 11p15 | >105 | Autosomal recessive, X-linked (FANCB) | [199] |
| Paroxysmal nocturnal hemoglobinuria | Phosphatidylinositol glycan, class A (PIGA) | 311770 | Xp22 | Unknown | Somatic (do not inherited) | [200] |
| Immunodeficiency with hyper-IgM | CD40 antigen ligand | 300386 | Xq26 | <105 | X-linked | [201] |
| Autoimmune polyendocrinopathy ectodermal dystrophy | Autoimmune regulator (AIRE) | 607358 | 21q22 | >105 | Autosomal recessive | [202, 203] |
| Malignant melanoma | Cyclin-dependent kinase inhibitor 2A (CDKN2A) | 600160 | 9p21 | >105 | Autosomal recessive | [204, 205] |
| Xeroderma pigmentosum | Xeroderma pigmentosum, complementation group C (XPC) | 278720 | 3p25 | <105 | Autosomal recessive | [206, 207] |
| Xeroderma pigmentosum | Xeroderma pigmentosum, complementation group E (XPE) | 278740 | 11p12 | <105 | Autosomal recessive | [208] |
| Familial hypercholesterolemia | Low density lipoprotein receptor (LDLR) | 606945 | 19p13 | >104 | Autosomal dominant | [209] |
| Urea cycle disorder | Ornithine carbamoyltransferase (OTC) | 300461 | Xp21 | >105 | X-linked | [210] |
| Lesch-Nyhan syndrome | Hypoxanthine guanine phosphoribosyltransferase 1 (HPRT1) | 308000 | Xq26 | <105 | X-linked | [211] |
| Human tumors | Tumor protein p53 (TP53) | 191170 | 17p13 | >104 | Somatic | [152, 153] |
aThe Online Mendelian Inheritance in Man (OMIM) (http://www.ncbi.nlm.nih.gov/sites/entrez?db=OMIM&itool=toolbar) number is included for more information
A similar study was carried out with the human RAG1 gene [37], mutation of which is responsible for a severe combined immunodeficiency disorder (Table 3; Fig. 5). The sequence of the RAG1 gene was scanned searching for any 22-bp sequence that may be cleaved by a I-CreI variant obtained through the combinatorial approach. Eighteen potential sequences were identified, the most promising of which was chosen to screen combinatorial libraries of variant I-CreI. A set of active heterodimeric I-CreI variants that cleave this sequence and display high levels of activity in yeast and CHO cells was identified. This was the first LHE to be entirely redesigned to cleave a naturally occurring sequence [37] (Figs. 5 and 6). Furthermore, heterodimeric and single chain I-CreI variants that cleave the RAG1 gene are as active as I-SceI in mammalian cells, with no detectable cell toxicity [27]. These findings show that modified HEs from the LAGLIDADG family may be used as effective and safe tools in gene therapy. The methods described can thus be used as a proof of principle for future therapeutic applications.
The efficacy of gene correction is an important and complex consideration in gene therapy. It is affected by several factors, such as transfection rate for the HEs and repair sequences, location at the nucleus, expression levels, HR machinery, chromatin state and cell cycle. Several studies have found the frequency of successful induced gene targeting (GT) events to range from 10−5 to 10−1 when using a chromosomal reporter system and either I-SceI or ZFN target sites in embryonic stem or HEK-293 cells, respectively [118, 135]. Given the number of parameters involved, and the fact that they vary from one study to another (including cell type, locus, reporter system, correction or insertion, repair matrix, protocols and manipulation), it is difficult to explain these differences. Redesigned HEs have also been shown to induce GT with a similar efficiency as that obtained with I-SceI [38]. All these studies were carried out with artificial DNA constructs, and most of them with immortalised cell lines. Recombination efficiency may vary for endogenous genes in primary cells. Studies showing that HR machinery levels are increased in the late S and G2 phases of the cell cycle suggest that GT may be more efficient in rapidly dividing cells [136, 137]. Cell types differ in their ability to undergo HR, in part because most cells are no longer dividing in differentiated tissues. Another aspect to take into account is chromatin accessibility, which may affect the efficiency of DSB formation in particular DNA regions. One report shows a surprisingly high efficiency of DSB-induced recombination for the endogenous human ILR2G gene in primary CD4 + T cells [121].
It is pointless enhancing the DSB activity of HEs if the lesion is repaired by NHEJ, which is often frequently undertaken in mammalian cells. Several laboratories have tried to enhance HR or inactivate NHEJ, to improve gene targeting in mammalian cells [138–140]. However, no clear solutions have emerged. One study has reported a 60-fold enhancement of GT in plants, by over-expression of Rad54 [141]. Another study has shown that Rad 18 enhances HR-dependent DSB repair in chicken and human cells [142]. A recent insightful study has shown that the use of an enzyme that generates nicks rather than DSBs may promote HR, at the same time minimising the genomic instability associated with DSB-induced NHEJ [107]. A deeper understanding of recombination mechanisms will be needed for further advances in this field, with a view to achieving the ultimate goal: a gene targeting event in every transfected cell.
It is important to consider that the frequency of correction decreases as a function of increasing distance from the cleavage site, with a four-fold drop in frequency after the first 100 bp, and no detectable recombination events occurring beyond 500 bp from the DSB [143]. However, in one study, 13% of conversion events were observed up to about 4 kbp away from the initial DSB site [144]. Similarly, a recent study using ZFNs to cleave endogenous plant genes reported over 2% of transformed cells displaying mutations up to 1.3 kbp from the ZFN cleavage site [145] (Fig. 6b). Again, the efficacy depends on various factors such as the locus, the sequence and the design of the repaired sequence. Overall, these findings show that, for optimal gene correction efficiency, the DSB must be generated as close as possible to the target sequence. The application of modified HEs in human gene therapy would thus require the sequencing of the damaged gene to localise the mutated nucleotides. This strategy would allow the selection of the closest target sequence that best matches the specificity of modified HEs from an existing library. These HEs could then serve as a template for further modification to improve binding to and cleavage of the target site of interest (Fig. 6c).
Current limitations are associated with low efficacy, possible cytotoxic effects and vectorisation. The ex vivo treatment of patient cells bearing the damaged gene with engineered HEs would allow corrected clones carrying the repaired gene to be selected for therapeutic use. Only a low gene correction rate has been obtained from in vivo viral-based treatments due to low infection rates and low expression levels. Ex vivo treatment using recombinant proteins, as previously shown for stem cell generation [146], would eliminate the need for plasmids and viruses to correct target genes (Fig. 4).
Monogenic diseases and ex vivo treatment
Monogenic diseases result from a change in a single gene, occurring in all cells of the body. Though relatively rare, they affect millions of people worldwide. It is currently estimated that over 10,000 of human diseases are monogenic. The nature of the disease depends on the function of the gene that has been modified. Single-gene or monogenic diseases can be divided into several categories: autosomal dominant disorders, in which only one allele of the gene is mutated in affected patients; autosomal recessive disorders, in which both alleles of the gene are mutated; X-linked, which are caused by mutations in genes on the X chromosome; Y-linked, caused by mutations in genes on the Y chromosome; and mitochondrial diseases, caused by modification of genes in mitochondrial DNA. Regarding these genetic conditions, gene therapy in some of these diseases, using modified HEs, will need the correct gene to be supplied as a template for the proper repair of the mutated gene, mediated by HR. Ex vivo treatment with modified HEs is limited to monogenic hematopoietic diseases, monogenic skin diseases, some monogenic metabolic disorders and potentially some kinds of cancer (Table 3; Fig. 4).
Only a few cell types are able to support ex vivo manipulation and re-implantation. The most promising of these, due to their plasticity, are hematopoietic stem cells, which can differentiate to form all the blood cell types. They are found in the adult bone marrow [147] and blood [148]. Hematopoietic stem-cell transplantation is used primarily for hematological and lymphoid cancers, but also for many other disorders [149]. Mesenchymal stem cells (MSC) can be expanded ex vivo, are easily extracted, and differentiate into multiple cell lineages. These cells thus provide a new clinical tool for cell and gene therapy. Several groups have isolated MSCs from bone marrow, umbilical cord blood and adipose tissue. MSCs seem to have different properties, depending on their origin [150].
Mutations in the TP53 gene
As well as their use in the treatment of monogenic disorders, the DSB activity of HEs could be used for the correction of somatic mutations. Mutation of the TP53 gene is the most frequent genetic change in human cancer, with one in two tumours bearing a p53 mutation. Most mutations are clustered in the DNA binding domain [151]. The importance of the role of p53 in tumour suppression is reflected by the fact that this protein is defective in virtually all human cancers. In about 50% of cases, it is inactivated by mutations; in other cases, p53 activity is suppressed, leading to disruption of its associated pathways. Studies on transgenic mice, in which p53 expression is reversibly switched on and off, show that restoration of p53 function can lead to tumour regression in vivo; thus restoration of p53 activity may be a promising therapeutic strategy [152, 153]. The activity of p53 could potentially be restored by the repair of the mutations that disrupt its normal function, through specifically induced DSB. However, the state of the art currently only allows for the treatment of certain types of cancer that are amenable to ex vivo treatment.
Concluding remarks
This review discusses the growing interest in the use of highly specific engineered HEs as a possible therapeutic tool to correct defects in different types of diseases. The redesign of these enzymes requires in-depth understanding of their physicochemical properties to elucidate mechanisms underlying their protein-DNA recognition and their mode of action. Depending on the molecular mechanisms mediating DNA repair, different outcomes of the action of these meganucleases may be foreseen. Whether these tools can provide the basis for a general approach in gene repair in monogenic human diseases remains unclear. Aspects such as repair efficiency and the selection of clones carrying the repaired gene must be addressed to determine whether this technology can be used in patients. However, it is beyond doubt that the use of this technology has considerable advantages over alternative methods. If successful, the use of these tools would circumvent the use of viral vectors for gene therapy, diminishing the risk of deleterious effects caused by DNA-based approaches [154–157]. The use of this technology in a multigenic disease such as cancer remains a matter of debate. However, key genes, such as TP53, have been identified to have major roles in tumour development. The further study of potential therapeutic approaches involving the repair of such genes is thus needed and should be of particular interest.
Acknowledgments
This work was supported European Union MEGATOOLS (LSHG-CT-2006-037226), the Ministerio de Ciencia e Inovación (MICINN) Grant BFU2007-30703-E and BFU2008-01344 to G.M., and the ETORTEK-2008 programme, at the Structural Biology Unit of CIC bioGUNE. M.J.M. holds a Juan de la Cierva contract from the Spanish MICINN.
Contributor Information
Jesús Prieto, Phone: +34-912246900, FAX: +34-912246976, Email: jprieto@cnio.es.
Guillermo Montoya, Phone: +34-912246900, FAX: +34-912246976, Email: gmontoya@cnio.es.
References
- 1.Dujon B. Homing nucleases and the yeast mitochondrial omega locus: a historical perspective. Berlin: Springer; 2005. [Google Scholar]
- 2.Liu Q, Belle A, Shub DA, Belfort M, Edgell DR. SegG endonuclease promotes marker exclusion and mediates co-conversion from a distant cleavage site. J Mol Biol. 2003;334:13–23. doi: 10.1016/j.jmb.2003.09.027. [DOI] [PubMed] [Google Scholar]
- 3.Edgell DR. Free-standing homing endonucleases of T-even phage: freeloaders or functionaries? Berlin: Springer; 2005. [Google Scholar]
- 4.Edgell DR. Selfish DNA: new abode for homing endonucleases. Curr Biol. 2002;12:R276–R278. doi: 10.1016/s0960-9822(02)00799-6. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev S, Dryden DT, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 8.Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoddard BL. Homing endonuclease structure and function. Q Rev Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 10.Dassa B, London N, Stoddard BL, Schueler-Furman O, Pietrokovski S. Fractured genes: a novel genomic arrangement involving new split inteins and a new homing endonuclease family. Nucleic Acids Res. 2009;37:2560–2573. doi: 10.1093/nar/gkp095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsutakawa SE, Morikawa K. The structural basis of damaged DNA recognition and endonucleolytic cleavage for very short patch repair endonuclease. Nucleic Acids Res. 2001;29:3775–3783. doi: 10.1093/nar/29.18.3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalgaard JZ, Klar AJ, Moser MJ, Holley WR, Chatterjee A, Mian IS. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 1997;25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevalier B, Turmel M, Lemieux C, Monnat RJ, Jr, Stoddard BL. Flexible DNA target site recognition by divergent homing endonuclease isoschizomers I-CreI and I-MsoI. J Mol Biol. 2003;329:253–269. doi: 10.1016/s0022-2836(03)00447-9. [DOI] [PubMed] [Google Scholar]
- 14.Thompson AJ, Yuan X, Kudlicki W, Herrin DL. Cleavage and recognition pattern of a double-strand-specific endonuclease (I-creI) encoded by the chloroplast 23S rRNA intron of Chlamydomonas reinhardtii. Gene. 1992;119:247–251. doi: 10.1016/0378-1119(92)90278-w. [DOI] [PubMed] [Google Scholar]
- 15.Turmel M, Otis C, Cote V, Lemieux C. Evolutionarily conserved and functionally important residues in the I-CeuI homing endonuclease. Nucleic Acids Res. 1997;25:2610–2619. doi: 10.1093/nar/25.13.2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Kim HH, Yuan X, Herrin DL. Purification, biochemical characterization and protein-DNA interactions of the I-CreI endonuclease produced in Escherichia coli. Nucleic Acids Res. 1997;25:3767–3776. doi: 10.1093/nar/25.19.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aagaard C, Awayez MJ, Garrett RA. Profile of the DNA recognition site of the archaeal homing endonuclease I-DmoI. Nucleic Acids Res. 1997;25:1523–1530. doi: 10.1093/nar/25.8.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolduc JM, Spiegel PC, Chatterjee P, Brady KL, Downing ME, Caprara MG, Waring RB, Stoddard BL. Structural and biochemical analyses of DNA and RNA binding by a bifunctional homing endonuclease and group I intron splicing factor. Genes Dev. 2003;17:2875–2888. doi: 10.1101/gad.1109003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moure CM, Gimble FS, Quiocho FA. The crystal structure of the gene targeting homing endonuclease I-SceI reveals the origins of its target site specificity. J Mol Biol. 2003;334:685–695. doi: 10.1016/j.jmb.2003.09.068. [DOI] [PubMed] [Google Scholar]
- 20.Chevalier B, Sussman D, Otis C, Noel AJ, Turmel M, Lemieux C, Stephens K, Monnat RJ, Jr, Stoddard BL. Metal-dependent DNA cleavage mechanism of the I-CreI LAGLIDADG homing endonuclease. Biochemistry. 2004;43:14015–14026. doi: 10.1021/bi048970c. [DOI] [PubMed] [Google Scholar]
- 21.Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA recognition and cleavage by the LAGLIDADG homing endonuclease I-CreI. Mol Cell. 1998;2:469–476. doi: 10.1016/s1097-2765(00)80146-x. [DOI] [PubMed] [Google Scholar]
- 22.Marcaida MJ, Prieto J, Redondo P, Nadra AD, Alibes A, Serrano L, Grizot S, Duchateau P, Paques F, Blanco FJ, et al. Crystal structure of I-DmoI in complex with its target DNA provides new insights into meganuclease engineering. Proc Natl Acad Sci USA. 2008;105:16888–16893. doi: 10.1073/pnas.0804795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moure CM, Gimble FS, Quiocho FA. Crystal structure of the intein homing endonuclease PI-SceI bound to its recognition sequence. Nat Struct Biol. 2002;9:764–770. doi: 10.1038/nsb840. [DOI] [PubMed] [Google Scholar]
- 24.Spiegel PC, Chevalier B, Sussman D, Turmel M, Lemieux C, Stoddard BL. The structure of I-CeuI homing endonuclease: evolving asymmetric DNA recognition from a symmetric protein scaffold. Structure. 2006;14:869–880. doi: 10.1016/j.str.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Chevalier BS, Kortemme T, Chadsey MS, Baker D, Monnat RJ, Stoddard BL. Design, activity, and structure of a highly specific artificial endonuclease. Mol Cell. 2002;10:895–905. doi: 10.1016/s1097-2765(02)00690-1. [DOI] [PubMed] [Google Scholar]
- 26.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Paques F, Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grizot S, Smith J, Prieto J, Daboussi F, Redondo P, Merino N, Villate M, Thomas S, Lemaire L, Montoya G, et al. (2009) Efficient targeting of a SCID gene by an engineered single chain homing endonuclease. Nucleic Acids Res (in press) [DOI] [PMC free article] [PubMed]
- 28.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci USA. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kostrewa D, Winkler FK. Mg2 + binding to the active site of EcoRV endonuclease: a crystallographic study of complexes with substrate and product DNA at 2 A resolution. Biochemistry. 1995;34:683–696. doi: 10.1021/bi00002a036. [DOI] [PubMed] [Google Scholar]
- 30.Viadiu H, Aggarwal AK. The role of metals in catalysis by the restriction endonuclease BamHI. Nat Struct Biol. 1998;5:910–916. doi: 10.1038/2352. [DOI] [PubMed] [Google Scholar]
- 31.Chevalier BS, Monnat RJ, Jr, Stoddard BL. The homing endonuclease I-CreI uses three metals, one of which is shared between the two active sites. Nat Struct Biol. 2001;8:312–316. doi: 10.1038/86181. [DOI] [PubMed] [Google Scholar]
- 32.Moure CM, Gimble FS, Quiocho FA. Crystal structures of I-SceI complexed to nicked DNA substrates: snapshots of intermediates along the DNA cleavage reaction pathway. Nucleic Acids Res. 2008;36:3287–3296. doi: 10.1093/nar/gkn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ho Y, Kim SJ, Waring RB. A protein encoded by a group I intron in Aspergillus nidulans directly assists RNA splicing and is a DNA endonuclease. Proc Natl Acad Sci USA. 1997;94:8994–8999. doi: 10.1073/pnas.94.17.8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ho Y, Waring RB. The maturase encoded by a group I intron from Aspergillus nidulans stabilizes RNA tertiary structure and promotes rapid splicing. J Mol Biol. 1999;292:987–1001. doi: 10.1006/jmbi.1999.3070. [DOI] [PubMed] [Google Scholar]
- 35.Arnould S, Perez C, Cabaniols JP, Smith J, Gouble A, Grizot S, Epinat JC, Duclert A, Duchateau P, Paques F. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J Mol Biol. 2007;371:49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 36.Arnould S, Chames P, Perez C, Lacroix E, Duclert A, Epinat JC, Stricher F, Petit AS, Patin A, Guillier S, et al. Engineering of large numbers of highly specific homing endonucleases that induce recombination on novel DNA targets. J Mol Biol. 2006;355:443–458. doi: 10.1016/j.jmb.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 37.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, Bravo J, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Redondo P, Prieto J, Munoz IG, Alibes A, Stricher F, Serrano L, Cabaniols JP, Daboussi F, Arnould S, Perez C, et al. Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases. Nature. 2008;456:107–111. doi: 10.1038/nature07343. [DOI] [PubMed] [Google Scholar]
- 39.Friedhoff P, Franke I, Meiss G, Wende W, Krause KL, Pingoud A. A similar active site for non-specific and specific endonucleases. Nat Struct Biol. 1999;6:112–113. doi: 10.1038/5796. [DOI] [PubMed] [Google Scholar]
- 40.Kuhlmann UC, Moore GR, James R, Kleanthous C, Hemmings AM. Structural parsimony in endonuclease active sites: should the number of homing endonuclease families be redefined? FEBS Lett. 1999;463:1–2. doi: 10.1016/s0014-5793(99)01499-4. [DOI] [PubMed] [Google Scholar]
- 41.Mehta P, Katta K, Krishnaswamy S. HNH family subclassification leads to identification of commonality in the His-Me endonuclease superfamily. Protein Sci. 2004;13:295–300. doi: 10.1110/ps.03115604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jakubauskas A, Giedriene J, Bujnicki JM, Janulaitis A. Identification of a single HNH active site in type IIS restriction endonuclease Eco31I. J Mol Biol. 2007;370:157–169. doi: 10.1016/j.jmb.2007.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Azarinskas A, Maneliene Z, Jakubauskas A. Hin4II, a new prototype restriction endonuclease from Haemophilus influenzae RFL4: discovery, cloning and expression in Escherichia coli. J Biotechnol. 2006;123:288–296. doi: 10.1016/j.jbiotec.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 44.Saravanan M, Vasu K, Kanakaraj R, Rao DN, Nagaraja V. R.KpnI, an HNH superfamily REase, exhibits differential discrimination at non-canonical sequences in the presence of Ca2 + and Mg2+ Nucleic Acids Res. 2007;35:2777–2786. doi: 10.1093/nar/gkm114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cymerman IA, Obarska A, Skowronek KJ, Lubys A, Bujnicki JM. Identification of a new subfamily of HNH nucleases and experimental characterization of a representative member, HphI restriction endonuclease. Proteins. 2006;65:867–876. doi: 10.1002/prot.21156. [DOI] [PubMed] [Google Scholar]
- 46.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodrich-Blair H, Scarlato V, Gott JM, Xu MQ, Shub DA. A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell. 1990;63:417–424. doi: 10.1016/0092-8674(90)90174-d. [DOI] [PubMed] [Google Scholar]
- 48.Goodrich-Blair H, Shub DA. The DNA polymerase genes of several HMU-bacteriophages have similar group I introns with highly divergent open reading frames. Nucleic Acids Res. 1994;22:3715–3721. doi: 10.1093/nar/22.18.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goodrich-Blair H, Shub DA. Beyond homing: competition between intron endonucleases confers a selective advantage on flanking genetic markers. Cell. 1996;84:211–221. doi: 10.1016/s0092-8674(00)80976-9. [DOI] [PubMed] [Google Scholar]
- 50.Landthaler M, Lau NC, Shub DA. Group I intron homing in Bacillus phages SPO1 and SP82: a gene conversion event initiated by a nicking homing endonuclease. J Bacteriol. 2004;186:4307–4314. doi: 10.1128/JB.186.13.4307-4314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landthaler M, Shub DA. The nicking homing endonuclease I-BasI is encoded by a group I intron in the DNA polymerase gene of the Bacillus thuringiensis phage Bastille. Nucleic Acids Res. 2003;31:3071–3077. doi: 10.1093/nar/gkg433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eddy SR, Gold L. The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev. 1991;5:1032–1041. doi: 10.1101/gad.5.6.1032. [DOI] [PubMed] [Google Scholar]
- 53.Drouin M, Lucas P, Otis C, Lemieux C, Turmel M. Biochemical characterization of I-CmoeI reveals that this H-N-H homing endonuclease shares functional similarities with H-N-H colicins. Nucleic Acids Res. 2000;28:4566–4572. doi: 10.1093/nar/28.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holloway SP, Deshpande NN, Herrin DL. The catalytic group-I introns of the psbA gene of chlamydomonas reinhardtii : core structures, ORFs and evolutionary implications. Curr Genet. 1999;36:69–78. doi: 10.1007/s002940050474. [DOI] [PubMed] [Google Scholar]
- 55.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J Mol Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 56.Johansen S, Embley TM, Willassen NP. A family of nuclear homing endonucleases. Nucleic Acids Res. 1993;21:4405. doi: 10.1093/nar/21.18.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muscarella DE, Vogt VM. A mobile group I intron from Physarum polycephalum can insert itself and induce point mutations in the nuclear ribosomal DNA of saccharomyces cerevisiae. Mol Cell Biol. 1993;13:1023–1033. doi: 10.1128/mcb.13.2.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muscarella DE, Ellison EL, Ruoff BM, Vogt VM. Characterization of I-Ppo, an intron-encoded endonuclease that mediates homing of a group I intron in the ribosomal DNA of Physarum polycephalum. Mol Cell Biol. 1990;10:3386–3396. doi: 10.1128/mcb.10.7.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wittmayer PK, Raines RT. Substrate binding and turnover by the highly specific I-PpoI endonuclease. Biochemistry. 1996;35:1076–1083. doi: 10.1021/bi952363v. [DOI] [PubMed] [Google Scholar]
- 60.Wittmayer PK, McKenzie JL, Raines RT. Degenerate DNA recognition by I-PpoI endonuclease. Gene. 1998;206:11–21. doi: 10.1016/s0378-1119(97)00563-5. [DOI] [PubMed] [Google Scholar]
- 61.Johansen S, Vogt VM. An intron in the nuclear ribosomal DNA of Didymium iridis codes for a group I ribozyme and a novel ribozyme that cooperate in self-splicing. Cell. 1994;76:725–734. doi: 10.1016/0092-8674(94)90511-8. [DOI] [PubMed] [Google Scholar]
- 62.Johansen S, Elde M, Vader A, Haugen P, Haugli K, Haugli F. In vivo mobility of a group I twintron in nuclear ribosomal DNA of the myxomycete Didymium iridis. Mol Microbiol. 1997;24:737–745. doi: 10.1046/j.1365-2958.1997.3921743.x. [DOI] [PubMed] [Google Scholar]
- 63.Haugen P, Wikmark OG, Vader A, Coucheron DH, Sjottem E, Johansen SD. The recent transfer of a homing endonuclease gene. Nucleic Acids Res. 2005;33:2734–2741. doi: 10.1093/nar/gki564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johansen SD, Haugen P, Nielsen H. Expression of protein-coding genes embedded in ribosomal DNA. Biol Chem. 2007;388:679–686. doi: 10.1515/BC.2007.089. [DOI] [PubMed] [Google Scholar]
- 65.Elde M, Haugen P, Willassen NP, Johansen S. I-NjaI, a nuclear intron-encoded homing endonuclease from Naegleria, generates a pentanucleotide 3’ cleavage-overhang within a 19 base-pair partially symmetric DNA recognition site. Eur J Biochem. 1999;259:281–288. doi: 10.1046/j.1432-1327.1999.00035.x. [DOI] [PubMed] [Google Scholar]
- 66.Elde M, Willassen NP, Johansen S. Functional characterization of isoschizomeric His-Cys box homing endonucleases from Naegleria. Eur J Biochem. 2000;267:7257–7266. doi: 10.1046/j.1432-1327.2000.01862.x. [DOI] [PubMed] [Google Scholar]
- 67.Ellison EL, Vogt VM. Interaction of the intron-encoded mobility endonuclease I-PpoI with its target site. Mol Cell Biol. 1993;13:7531–7539. doi: 10.1128/mcb.13.12.7531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Flick KE, Jurica MS, Monnat RJ, Jr, Stoddard BL. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 69.Flick KE, McHugh D, Heath JD, Stephens KM, Monnat RJ, Jr, Stoddard BL. Crystallization and preliminary X-ray studies of I-PpoI: a nuclear, intron-encoded homing endonuclease from Physarum polycephalum. Protein Sci. 1997;6:2677–2680. doi: 10.1002/pro.5560061226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Galburt EA, Chevalier B, Tang W, Jurica MS, Flick KE, Monnat RJ, Jr, Stoddard BL. A novel endonuclease mechanism directly visualized for I-PpoI. Nat Struct Biol. 1999;6:1096–1099. doi: 10.1038/70027. [DOI] [PubMed] [Google Scholar]
- 71.Galburt EA, Chadsey MS, Jurica MS, Chevalier BS, Erho D, Tang W, Monnat RJ, Jr, Stoddard BL. Conformational changes and cleavage by the homing endonuclease I-PpoI: a critical role for a leucine residue in the active site. J Mol Biol. 2000;300:877–887. doi: 10.1006/jmbi.2000.3874. [DOI] [PubMed] [Google Scholar]
- 72.Eastberg JH, Eklund J, Monnat R, Jr, Stoddard BL. Mutability of an HNH nuclease imidazole general base and exchange of a deprotonation mechanism. Biochemistry. 2007;46:7215–7225. doi: 10.1021/bi700418d. [DOI] [PubMed] [Google Scholar]
- 73.Eklund JL, Ulge UY, Eastberg J, Monnat RJ., Jr Altered target site specificity variants of the I-PpoI His-Cys box homing endonuclease. Nucleic Acids Res. 2007;35:5839–5850. doi: 10.1093/nar/gkm624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dunin-Horkawicz S, Feder M, Bujnicki JM. Phylogenomic analysis of the GIY-YIG nuclease superfamily. BMC Genomics. 2006;7:98. doi: 10.1186/1471-2164-7-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kowalski JC, Belfort M, Stapleton MA, Holpert M, Dansereau JT, Pietrokovski S, Baxter SM, Derbyshire V. Configuration of the catalytic GIY-YIG domain of intron endonuclease I-TevI: coincidence of computational and molecular findings. Nucleic Acids Res. 1999;27:2115–2125. doi: 10.1093/nar/27.10.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nord D, Sjoberg BM. Unconventional GIY-YIG homing endonuclease encoded in group I introns in closely related strains of the Bacillus cereus group. Nucleic Acids Res. 2008;36:300–310. doi: 10.1093/nar/gkm1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Derbyshire V, Kowalski JC, Dansereau JT, Hauer CR, Belfort M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J Mol Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- 78.Bujnicki JM, Rotkiewicz P, Kolinski A, Rychlewski L. Three-dimensional modeling of the I-TevI homing endonuclease catalytic domain, a GIY-YIG superfamily member, using NMR restraints and Monte Carlo dynamics. Protein Eng. 2001;14:717–721. doi: 10.1093/protein/14.10.717. [DOI] [PubMed] [Google Scholar]
- 79.Liu Q, Derbyshire V, Belfort M, Edgell DR. Distance determination by GIY-YIG intron endonucleases: discrimination between repression and cleavage functions. Nucleic Acids Res. 2006;34:1755–1764. doi: 10.1093/nar/gkl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Roey P, Meehan L, Kowalski JC, Belfort M, Derbyshire V. Catalytic domain structure and hypothesis for function of GIY-YIG intron endonuclease I-TevI. Nat Struct Biol. 2002;9:806–811. doi: 10.1038/nsb853. [DOI] [PubMed] [Google Scholar]
- 81.Sitbon E, Pietrokovski S. New types of conserved sequence domains in DNA-binding regions of homing endonucleases. Trends Biochem Sci. 2003;28:473–477. doi: 10.1016/S0968-0004(03)00170-1. [DOI] [PubMed] [Google Scholar]
- 82.Dean AB, Stanger MJ, Dansereau JT, Van Roey P, Derbyshire V, Belfort M. Zinc finger as distance determinant in the flexible linker of intron endonuclease I-TevI. Proc Natl Acad Sci USA. 2002;99:8554–8561. doi: 10.1073/pnas.082253699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu Q, Dansereau JT, Puttamadappa SS, Shekhtman A, Derbyshire V, Belfort M. Role of the interdomain linker in distance determination for remote cleavage by homing endonuclease I-TevI. J Mol Biol. 2008;379:1094–1106. doi: 10.1016/j.jmb.2008.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brok-Volchanskaya VS, Kadyrov FA, Sivogrivov DE, Kolosov PM, Sokolov AS, Shlyapnikov MG, Kryukov VM, Granovsky IE. Phage T4 SegB protein is a homing endonuclease required for the preferred inheritance of T4 tRNA gene region occurring in co-infection with a related phage. Nucleic Acids Res. 2008;36:2094–2105. doi: 10.1093/nar/gkn053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Carter JM, Friedrich NC, Kleinstiver B, Edgell DR. Strand-specific contacts and divalent metal ion regulate double-strand break formation by the GIY-YIG homing endonuclease I-BmoI. J Mol Biol. 2007;374:306–321. doi: 10.1016/j.jmb.2007.09.027. [DOI] [PubMed] [Google Scholar]
- 86.Bryk M, Quirk SM, Mueller JE, Loizos N, Lawrence C, Belfort M. The td intron endonuclease I-TevI makes extensive sequence-tolerant contacts across the minor groove of its DNA target. EMBO J. 1993;12:4040–4041. doi: 10.1002/j.1460-2075.1993.tb06082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mueller JE, Smith D, Bryk M, Belfort M. Intron-encoded endonuclease I-TevI binds as a monomer to effect sequential cleavage via conformational changes in the td homing site. EMBO J. 1995;14:5724–5735. doi: 10.1002/j.1460-2075.1995.tb00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ibryashkina EM, Sasnauskas G, Solonin AS, Zakharova MV, Siksnys V. Oligomeric structure diversity within the GIY-YIG nuclease family. J Mol Biol. 2009;387:10–16. doi: 10.1016/j.jmb.2009.01.048. [DOI] [PubMed] [Google Scholar]
- 89.Chu FK, Maley G, Pedersen-Lane J, Wang AM, Maley F. Characterization of the restriction site of a prokaryotic intron-encoded endonuclease. Proc Natl Acad Sci USA. 1990;87:3574–3578. doi: 10.1073/pnas.87.9.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bell-Pedersen D, Quirk SM, Bryk M, Belfort M. I-TevI, the endonuclease encoded by the mobile td intron, recognizes binding and cleavage domains on its DNA target. Proc Natl Acad Sci USA. 1991;88:7719–7723. doi: 10.1073/pnas.88.17.7719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. EMBO J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Edgell DR, Shub DA. Related homing endonucleases I-BmoI and I-TevI use different strategies to cleave homologous recognition sites. Proc Natl Acad Sci USA. 2001;98:7898–7903. doi: 10.1073/pnas.141222498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bryk M, Belisle M, Mueller JE, Belfort M. Selection of a remote cleavage site by I-tevI, the td intron-encoded endonuclease. J Mol Biol. 1995;247:197–210. doi: 10.1006/jmbi.1994.0133. [DOI] [PubMed] [Google Scholar]
- 94.Edgell DR, Stanger MJ, Belfort M. Coincidence of cleavage sites of intron endonuclease I-TevI and critical sequences of the host thymidylate synthase gene. J Mol Biol. 2004;343:1231–1241. doi: 10.1016/j.jmb.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 95.Bonocora RP, Shub DA. A novel group I intron-encoded endonuclease specific for the anticodon region of tRNA(fMet) genes. Mol Microbiol. 2001;39:1299–1306. doi: 10.1111/j.1365-2958.2001.02318.x. [DOI] [PubMed] [Google Scholar]
- 96.Biniszkiewicz D, Cesnaviciene E, Shub DA. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao L, Bonocora RP, Shub DA, Stoddard BL. The restriction fold turns to the dark side: a bacterial homing endonuclease with a PD-(D/E)-XK motif. EMBO J. 2007;26:2432–2442. doi: 10.1038/sj.emboj.7601672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Orlowski J, Boniecki M, Bujnicki JM. I-Ssp6803I: the first homing endonuclease from the PD-(D/E)XK superfamily exhibits an unusual mode of DNA recognition. Bioinformatics. 2007;23:527–530. doi: 10.1093/bioinformatics/btm007. [DOI] [PubMed] [Google Scholar]
- 99.Zhao L, Pellenz S, Stoddard BL. Activity and specificity of the bacterial PD-(D/E)XK homing endonuclease I-Ssp6803I. J Mol Biol. 2009;385:1498–1510. doi: 10.1016/j.jmb.2008.10.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 101.Sung P, Klein H. Mechanism of homologous recombination: mediators and helicases take on regulatory functions. Nat Rev Mol Cell Biol. 2006;7:739–750. doi: 10.1038/nrm2008. [DOI] [PubMed] [Google Scholar]
- 102.Brugmans L, Kanaar R, Essers J. Analysis of DNA double-strand break repair pathways in mice. Mutat Res. 2007;614:95–108. doi: 10.1016/j.mrfmmm.2006.01.022. [DOI] [PubMed] [Google Scholar]
- 103.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- 105.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci. 2009;34:264–272. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 106.Lee GS, Neiditch MB, Salus SS, Roth DB. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell. 2004;117:171–184. doi: 10.1016/s0092-8674(04)00301-0. [DOI] [PubMed] [Google Scholar]
- 107.McConnell Smith A, Takeuchi R, Pellenz S, Davis L, Maizels N, Monnat RJ, Jr, Stoddard BL. Generation of a nicking enzyme that stimulates site-specific gene conversion from the I-AniI LAGLIDADG homing endonuclease. Proc Natl Acad Sci USA. 2009;106:5099–5104. doi: 10.1073/pnas.0810588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fortini P, Dogliotti E. Base damage and single-strand break repair: mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 2007;6:398–409. doi: 10.1016/j.dnarep.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 109.Dianov GL, Parsons JL. Co-ordination of DNA single strand break repair. DNA Repair (Amst) 2007;6:454–460. doi: 10.1016/j.dnarep.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 110.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 111.Johnson RD, Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem Soc Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- 112.Rimseliene R, Maneliene Z, Lubys A, Janulaitis A. Engineering of restriction endonucleases: using methylation activity of the bifunctional endonuclease Eco57I to select the mutant with a novel sequence specificity. J Mol Biol. 2003;327:383–391. doi: 10.1016/s0022-2836(03)00142-6. [DOI] [PubMed] [Google Scholar]
- 113.Voziyanov Y, Konieczka JH, Stewart AF, Jayaram M. Stepwise manipulation of DNA specificity in Flp recombinase: progressively adapting Flp to individual and combinatorial mutations in its target site. J Mol Biol. 2003;326:65–76. doi: 10.1016/s0022-2836(02)01364-5. [DOI] [PubMed] [Google Scholar]
- 114.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 116.Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- 117.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 118.Alwin S, Gere MB, Guhl E, Effertz K, Barbas CF, 3rd, Segal DJ, Weitzman MD, Cathomen T. Custom zinc-finger nucleases for use in human cells. Mol Ther. 2005;12:610–617. doi: 10.1016/j.ymthe.2005.06.094. [DOI] [PubMed] [Google Scholar]
- 119.Porteus MH. Mammalian gene targeting with designed zinc finger nucleases. Mol Ther. 2006;13:438–446. doi: 10.1016/j.ymthe.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 120.Wright DA, Townsend JA, Winfrey RJ, Jr, Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF. High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J. 2005;44:693–705. doi: 10.1111/j.1365-313X.2005.02551.x. [DOI] [PubMed] [Google Scholar]
- 121.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 122.Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nat Biotechnol. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- 123.Seligman LM, Stephens KM, Savage JH, Monnat RJ., Jr Genetic analysis of the Chlamydomonas reinhardtii I-CreI mobile intron homing system in Escherichia coli . Genetics. 1997;147:1653–1664. doi: 10.1093/genetics/147.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: perspectives for gene therapy. Curr Gene Ther. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 125.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J Am Chem Soc. 2006;128:2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 126.Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonucleases using double-strand break induced homologous recombination. Nucleic Acids Res. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chica RA, Doucet N, Pelletier JN. Semi-rational approaches to engineering enzyme activity: combining the benefits of directed evolution and rational design. Curr Opin Biotechnol. 2005;16:378–384. doi: 10.1016/j.copbio.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 128.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat RJ, Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stary A, Sarasin A. The genetics of the hereditary xeroderma pigmentosum syndrome. Biochimie. 2002;84:49–60. doi: 10.1016/s0300-9084(01)01358-x. [DOI] [PubMed] [Google Scholar]
- 130.Cleaver JE. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat Rev Cancer. 2005;5:564–573. doi: 10.1038/nrc1652. [DOI] [PubMed] [Google Scholar]
- 131.Aslan G, Karacal N, Gorgu M. New tumor formation on split-thickness skin grafted areas in xeroderma pigmentosum. Ann Plast Surg. 1999;43:657–660. [PubMed] [Google Scholar]
- 132.Sonmez Ergun S. Resurfacing the dorsum of the hand in a patient with Xeroderma pigmentosum. Dermatol Surg. 2003;29:782–784. doi: 10.1046/j.1524-4725.2003.29198.x. [DOI] [PubMed] [Google Scholar]
- 133.Asselineau D, Bernhard B, Bailly C, Darmon M. Epidermal morphogenesis and induction of the 67 kD keratin polypeptide by culture of human keratinocytes at the liquid-air interface. Exp Cell Res. 1985;159:536–539. doi: 10.1016/s0014-4827(85)80027-6. [DOI] [PubMed] [Google Scholar]
- 134.Arnaudeau-Begard C, Brellier F, Chevallier-Lagente O, Hoeijmakers J, Bernerd F, Sarasin A, Magnaldo T. Genetic correction of DNA repair-deficient/cancer-prone xeroderma pigmentosum group C keratinocytes. Hum Gene Ther. 2003;14:983–996. doi: 10.1089/104303403766682241. [DOI] [PubMed] [Google Scholar]
- 135.Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Res. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 138.Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yanez RJ, Porter AC. Differential effects of Rad52p overexpression on gene targeting and extrachromosomal homologous recombination in a human cell line. Nucleic Acids Res. 2002;30:740–748. doi: 10.1093/nar/30.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Di Primio C, Galli A, Cervelli T, Zoppe M, Rainaldi G. Potentiation of gene targeting in human cells by expression of Saccharomyces cerevisiae Rad52. Nucleic Acids Res. 2005;33:4639–4648. doi: 10.1093/nar/gki778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shaked H, Melamed-Bessudo C, Levy AA. High-frequency gene targeting in Arabidopsis plants expressing the yeast RAD54 gene. Proc Natl Acad Sci USA. 2005;102:12265–12269. doi: 10.1073/pnas.0502601102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Saberi A, Hochegger H, Szuts D, Lan L, Yasui A, Sale JE, Taniguchi Y, Murakawa Y, Zeng W, Yokomori K, et al. RAD18 and poly(ADP-ribose) polymerase independently suppress the access of nonhomologous end joining to double-strand breaks and facilitate homologous recombination-mediated repair. Mol Cell Biol. 2007;27:2562–2571. doi: 10.1128/MCB.01243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Elliott B, Richardson C, Winderbaum J, Nickoloff JA, Jasin M. Gene conversion tracts from double-strand break repair in mammalian cells. Mol Cell Biol. 1998;18:93–101. doi: 10.1128/mcb.18.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Donoho G, Jasin M, Berg P. Analysis of gene targeting and intrachromosomal homologous recombination stimulated by genomic double-strand breaks in mouse embryonic stem cells. Mol Cell Biol. 1998;18:4070–4078. doi: 10.1128/mcb.18.7.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009;459:442–445. doi: 10.1038/nature07845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, et al. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Muller-Sieburg CE, Cho RH, Thoman M, Adkins B, Sieburg HB. Deterministic regulation of hematopoietic stem cell self-renewal and differentiation. Blood. 2002;100:1302–1309. [PubMed] [Google Scholar]
- 148.Cutler C, Antin JH. Peripheral blood stem cells for allogeneic transplantation: a review. Stem Cells. 2001;19:108–117. doi: 10.1634/stemcells.19-2-108. [DOI] [PubMed] [Google Scholar]
- 149.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354:1813–1826. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 150.Kern S, Eichler H, Stoeve J, Kluter H, Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- 151.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 152.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323–1334. doi: 10.1016/j.cell.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 153.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 154.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 155.Fischer A, Abina SH, Thrasher A, von Kalle C, Cavazzana-Calvo M. LMO2 and gene therapy for severe combined immunodeficiency. N Engl J Med. 2004;350:2526–2527. doi: 10.1056/NEJM200406103502422. [DOI] [PubMed] [Google Scholar]
- 156.Abbott A. Questions linger about unexplained gene-therapy trial death. Nat Med. 2006;12:597. doi: 10.1038/nm0606-597a. [DOI] [PubMed] [Google Scholar]
- 157.Thrasher AJ, Gaspar HB, Baum C, Modlich U, Schambach A, Candotti F, Otsu M, Sorrentino B, Scobie L, Cameron E, et al. Gene therapy: X-SCID transgene leukaemogenicity. Nature. 2006;443:E5–E6. doi: 10.1038/nature05219. [DOI] [PubMed] [Google Scholar]
- 158.Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 159.Corneo B, Moshous D, Gungor T, Wulffraat N, Philippet P, Le Deist FL, Fischer A, de Villartay JP. Identical mutations in RAG1 or RAG2 genes leading to defective V(D)J recombinase activity can cause either T-B-severe combined immune deficiency or Omenn syndrome. Blood. 2001;97:2772–2776. doi: 10.1182/blood.v97.9.2772. [DOI] [PubMed] [Google Scholar]
- 160.Eastberg JH, McConnell Smith A, Zhao L, Ashworth J, Shen BW, Stoddard BL. Thermodynamics of DNA target site recognition by homing endonucleases. Nucleic Acids Res. 2007;35:7209–7221. doi: 10.1093/nar/gkm867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Heath PJ, Stephens KM, Monnat RJ, Jr, Stoddard BL. The structure of I-Crel, a group I intron-encoded homing endonuclease. Nat Struct Biol. 1997;4:468–476. doi: 10.1038/nsb0697-468. [DOI] [PubMed] [Google Scholar]
- 162.Silva GH, Dalgaard JZ, Belfort M, Van Roey P. Crystal structure of the thermostable archaeal intron-encoded endonuclease I-DmoI. J Mol Biol. 1999;286:1123–1136. doi: 10.1006/jmbi.1998.2519. [DOI] [PubMed] [Google Scholar]
- 163.Edgell DR, Derbyshire V, Van Roey P, LaBonne S, Stanger MJ, Li Z, Boyd TM, Shub DA, Belfort M. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nat Struct Mol Biol. 2004;11:936–944. doi: 10.1038/nsmb823. [DOI] [PubMed] [Google Scholar]
- 164.Seligman LM, Chisholm KM, Chevalier BS, Chadsey MS, Edwards ST, Savage JH, Veillet AL. Mutations altering the cleavage specificity of a homing endonuclease. Nucleic Acids Res. 2002;30:3870–3879. doi: 10.1093/nar/gkf495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sussman D, Chadsey M, Fauce S, Engel A, Bruett A, Monnat R, Jr, Stoddard BL, Seligman LM. Isolation and characterization of new homing endonuclease specificities at individual target site positions. J Mol Biol. 2004;342:31–41. doi: 10.1016/j.jmb.2004.07.031. [DOI] [PubMed] [Google Scholar]
- 166.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Gruen M, Chang K, Serbanescu I, Liu DR. An in vivo selection system for homing endonuclease activity. Nucleic Acids Res. 2002;30:e29. doi: 10.1093/nar/30.7.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gimble FS, Moure CM, Posey KL. Assessing the plasticity of DNA target site recognition of the PI-SceI homing endonuclease using a bacterial two-hybrid selection system. J Mol Biol. 2003;334:993–1008. doi: 10.1016/j.jmb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 169.Steuer S, Pingoud V, Pingoud A, Wende W. Chimeras of the homing endonuclease PI-SceI and the homologous Candida tropicalis intein: a study to explore the possibility of exchanging DNA-binding modules to obtain highly specific endonucleases with altered specificity. Chembiochem. 2004;5:206–213. doi: 10.1002/cbic.200300718. [DOI] [PubMed] [Google Scholar]
- 170.Silva GH, Belfort M, Wende W, Pingoud A. From monomeric to homodimeric endonucleases and back: engineering novel specificity of LAGLIDADG enzymes. J Mol Biol. 2006;361:744–754. doi: 10.1016/j.jmb.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 171.Li H, Pellenz S, Ulge U, Stoddard BL, Monnat RJ., Jr Generation of single-chain LAGLIDADG homing endonucleases from native homodimeric precursor proteins. Nucleic Acids Res. 2009;37:1650–1662. doi: 10.1093/nar/gkp004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fajardo-Sanchez E, Stricher F, Paques F, Isalan M, Serrano L. Computer design of obligate heterodimer meganucleases allows efficient cutting of custom DNA sequences. Nucleic Acids Res. 2008;36:2163–2173. doi: 10.1093/nar/gkn059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Smith JG, Sylvestre, Arnould S, Duclert A, Epinat J-C, Prieto J, Redondo P, Blanco F, Bravo J, Montoya G, Pâques F, Duchateau P (2006) A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res [DOI] [PMC free article] [PubMed]
- 174.Ulbrichova D, Hrdinka M, Saudek V, Martasek P. Acute intermittent porphyria–impact of mutations found in the hydroxymethylbilane synthase gene on biochemical and enzymatic protein properties. Febs J. 2009;276:2106–2115. doi: 10.1111/j.1742-4658.2009.06946.x. [DOI] [PubMed] [Google Scholar]
- 175.Wiederholt T, Poblete-Gutierrez P, Gardlo K, Goerz G, Bolsen K, Merk HF, Frank J. Identification of mutations in the uroporphyrinogen III cosynthase gene in German patients with congenital erythropoietic porphyria. Physiol Res. 2006;55(Suppl 2):S85–S92. doi: 10.33549/physiolres.930000.55.S2.85. [DOI] [PubMed] [Google Scholar]
- 176.Romeo G, Levin EY. Uroporphyrinogen 3 cosynthetase in human congenital erythropoietic porphyria. Proc Natl Acad Sci USA. 1969;63:856–863. doi: 10.1073/pnas.63.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Mendez M, Sorkin L, Rossetti MV, Astrin KH, del CBAM, Parera VE, Aizencang G, Desnick RJ (1998) Familial porphyria cutanea tarda: characterization of seven novel uroporphyrinogen decarboxylase mutations and frequency of common hemochromatosis alleles. Am J Hum Genet 63:1363–1375 [DOI] [PMC free article] [PubMed]
- 178.Schmitt C, Gouya L, Malonova E, Lamoril J, Camadro JM, Flamme M, Rose C, Lyoumi S, Da Silva V, Boileau C, et al. Mutations in human CPO gene predict clinical expression of either hepatic hereditary coproporphyria or erythropoietic harderoporphyria. Hum Mol Genet. 2005;14:3089–3098. doi: 10.1093/hmg/ddi342. [DOI] [PubMed] [Google Scholar]
- 179.Roberts AG, Puy H, Dailey TA, Morgan RR, Whatley SD, Dailey HA, Martasek P, Nordmann Y, Deybach JC, Elder GH. Molecular characterization of homozygous variegate porphyria. Hum Mol Genet. 1998;7:1921–1925. doi: 10.1093/hmg/7.12.1921. [DOI] [PubMed] [Google Scholar]
- 180.Rufenacht UB, Gouya L, Schneider-Yin X, Puy H, Schafer BW, Aquaron R, Nordmann Y, Minder EI, Deybach JC. Systematic analysis of molecular defects in the ferrochelatase gene from patients with erythropoietic protoporphyria. Am J Hum Genet. 1998;62:1341–1352. doi: 10.1086/301870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Higgs DR, Vickers MA, Wilkie AO, Pretorius IM, Jarman AP, Weatherall DJ. A review of the molecular genetics of the human alpha-globin gene cluster. Blood. 1989;73:1081–1104. [PubMed] [Google Scholar]
- 182.Weiss I, Cash FE, Coleman MB, Pressley A, Adams JG, Sanguansermsri T, Liebhaber SA, Steinberg MH. Molecular basis for alpha-thalassemia associated with the structural mutant hemoglobin Suan-Dok (alpha 2 109leu—arg) Blood. 1990;76:2630–2636. [PubMed] [Google Scholar]
- 183.Tassiopoulos S, Deftereos S, Konstantopoulos K, Farmakis D, Tsironi M, Kyriakidis M, Aessopos A. Does heterozygous beta-thalassemia confer a protection against coronary artery disease? Ann NY Acad Sci. 2005;1054:467–470. doi: 10.1196/annals.1345.068. [DOI] [PubMed] [Google Scholar]
- 184.Blouin MJ, Beauchemin H, Wright A, De Paepe M, Sorette M, Bleau AM, Nakamoto B, Ou CN, Stamatoyannopoulos G, Trudel M. Genetic correction of sickle cell disease: insights using transgenic mouse models. Nat Med. 2000;6:177–182. doi: 10.1038/72279. [DOI] [PubMed] [Google Scholar]
- 185.Chae JJ, Komarow HD, Cheng J, Wood G, Raben N, Liu PP, Kastner DL. Targeted disruption of pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol Cell. 2003;11:591–604. doi: 10.1016/s1097-2765(03)00056-x. [DOI] [PubMed] [Google Scholar]
- 186.Tzall S, Ellenbogen A, Eng F, Hirschhorn R. Identification and characterization of nine RFLPs at the adenosine deaminase (ADA) locus. Am J Hum Genet. 1989;44:864–875. [PMC free article] [PubMed] [Google Scholar]
- 187.Hirschhorn R, Vawter GF, Kirkpatrick JA, Jr, Rosen FS. Adenosine deaminase deficiency: frequency and comparative pathology in autosomally recessive severe combined immunodeficiency. Clin Immunol Immunopathol. 1979;14:107–120. doi: 10.1016/0090-1229(79)90131-4. [DOI] [PubMed] [Google Scholar]
- 188.Speckmann C, Pannicke U, Wiech E, Schwarz K, Fisch P, Friedrich W, Niehues T, Gilmour K, Buiting K, Schlesier M, et al. Clinical and immunologic consequences of a somatic reversion in a patient with X-linked severe combined immunodeficiency. Blood. 2008;112:4090–4097. doi: 10.1182/blood-2008-04-153361. [DOI] [PubMed] [Google Scholar]
- 189.Santagata S, Gomez CA, Sobacchi C, Bozzi F, Abinun M, Pasic S, Cortes P, Vezzoni P, Villa A. N-terminal RAG1 frameshift mutations in Omenn’s syndrome: internal methionine usage leads to partial V(D)J recombination activity and reveals a fundamental role in vivo for the N-terminal domains. Proc Natl Acad Sci USA. 2000;97:14572–14577. doi: 10.1073/pnas.97.26.14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tabori U, Mark Z, Amariglio N, Etzioni A, Golan H, Biloray B, Toren A, Rechavi G, Dalal I. Detection of RAG mutations and prenatal diagnosis in families presenting with either T-B- severe combined immunodeficiency or Omenn’s syndrome. Clin Genet. 2004;65:322–326. doi: 10.1111/j.1399-0004.2004.00227.x. [DOI] [PubMed] [Google Scholar]
- 191.Notarangelo LD, Mella P, Jones A, de Saint Basile G, Savoldi G, Cranston T, Vihinen M, Schumacher RF. Mutations in severe combined immune deficiency (SCID) due to JAK3 deficiency. Hum Mutat. 2001;18:255–263. doi: 10.1002/humu.1188. [DOI] [PubMed] [Google Scholar]
- 192.Saglio G, Storlazzi CT, Giugliano E, Surace C, Anelli L, Rege-Cambrin G, Zagaria A, Jimenez Velasco A, Heiniger A, Scaravaglio P, et al. A 76-kb duplicon maps close to the BCR gene on chromosome 22 and the ABL gene on chromosome 9: possible involvement in the genesis of the Philadelphia chromosome translocation. Proc Natl Acad Sci USA. 2002;99:9882–9887. doi: 10.1073/pnas.152171299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Rae J, Newburger PE, Dinauer MC, Noack D, Hopkins PJ, Kuruto R, Curnutte JT. X-Linked chronic granulomatous disease: mutations in the CYBB gene encoding the gp91-phox component of respiratory-burst oxidase. Am J Hum Genet. 1998;62:1320–1331. doi: 10.1086/301874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li A, Prasad A, Mincemoyer R, Satorius C, Epstein N, Finkel T, Quyyumi AA. Relationship of the C242T p22phox gene polymorphism to angiographic coronary artery disease and endothelial function. Am J Med Genet. 1999;86:57–61. doi: 10.1002/(sici)1096-8628(19990903)86:1<57::aid-ajmg11>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 195.Roos D, de Boer M, Koker MY, Dekker J, Singh-Gupta V, Ahlin A, Palmblad J, Sanal O, Kurenko-Deptuch M, Jolles S, et al. Chronic granulomatous disease caused by mutations other than the common GT deletion in NCF1, the gene encoding the p47phox component of the phagocyte NADPH oxidase. Hum Mutat. 2006;27:1218–1229. doi: 10.1002/humu.20413. [DOI] [PubMed] [Google Scholar]
- 196.Nunoi H, Iwata M, Tatsuzawa S, Onoe Y, Shimizu S, Kanegasaki S, Matsuda I. AG dinucleotide insertion in a patient with chronic granulomatous disease lacking cytosolic 67-kD protein. Blood. 1995;86:329–333. [PubMed] [Google Scholar]
- 197.Santacroce R, Acquila M, Belvini D, Castaldo G, Garagiola I, Giacomelli SH, Lombardi AM, Minuti B, Riccardi F, Salviato R, et al. Identification of 217 unreported mutations in the F8 gene in a group of 1, 410 unselected Italian patients with hemophilia A. J Hum Genet. 2008;53:275–284. doi: 10.1007/s10038-007-0238-y. [DOI] [PubMed] [Google Scholar]
- 198.Ljung R, Petrini P, Tengborn L, Sjorin E. Haemophilia B mutations in Sweden: a population-based study of mutational heterogeneity. Br J Haematol. 2001;113:81–86. doi: 10.1046/j.1365-2141.2001.02759.x. [DOI] [PubMed] [Google Scholar]
- 199.Rogatko A, Auerbach AD. Segregation analysis with uncertain ascertainment: application to Fanconi anemia. Am J Hum Genet. 1988;42:889–897. [PMC free article] [PubMed] [Google Scholar]
- 200.Hillmen P, Lewis SM, Bessler M, Luzzatto L, Dacie JV. Natural history of paroxysmal nocturnal hemoglobinuria. N Engl J Med. 1995;333:1253–1258. doi: 10.1056/NEJM199511093331904. [DOI] [PubMed] [Google Scholar]
- 201.Allen RC, Armitage RJ, Conley ME, Rosenblatt H, Jenkins NA, Copeland NG, Bedell MA, Edelhoff S, Disteche CM, Simoneaux DK, et al. CD40 ligand gene defects responsible for X-linked hyper-IgM syndrome. Science. 1993;259:990–993. doi: 10.1126/science.7679801. [DOI] [PubMed] [Google Scholar]
- 202.Stolarski B, Pronicka E, Korniszewski L, Pollak A, Kostrzewa G, Rowinska E, Wlodarski P, Skorka A, Gremida M, Krajewski P, et al. Molecular background of polyendocrinopathy-candidiasis-ectodermal dystrophy syndrome in a Polish population: novel AIRE mutations and an estimate of disease prevalence. Clin Genet. 2006;70:348–354. doi: 10.1111/j.1399-0004.2006.00690.x. [DOI] [PubMed] [Google Scholar]
- 203.Ahonen P. Autoimmune polyendocrinopathy–candidosis–ectodermal dystrophy (APECED): autosomal recessive inheritance. Clin Genet. 1985;27:535–542. doi: 10.1111/j.1399-0004.1985.tb02037.x. [DOI] [PubMed] [Google Scholar]
- 204.Robertson KD, Jones PA. Tissue-specific alternative splicing in the human INK4a/ARF cell cycle regulatory locus. Oncogene. 1999;18:3810–3820. doi: 10.1038/sj.onc.1202737. [DOI] [PubMed] [Google Scholar]
- 205.Kannengiesser C, Dalle S, Leccia MT, Avril MF, Bonadona V, Chompret A, Lasset C, Leroux D, Thomas L, Lesueur F, et al. New founder germline mutations of CDKN2A in melanoma-prone families and multiple primary melanoma development in a patient receiving levodopa treatment. Genes Chromosomes Cancer. 2007;46:751–760. doi: 10.1002/gcc.20461. [DOI] [PubMed] [Google Scholar]
- 206.Lynch HT, Fusaro RM, Johnson JA. Xeroderma pigmentosum. Complementation group C and malignant melanoma. Arch Dermatol. 1984;120:175–179. doi: 10.1001/archderm.120.2.175. [DOI] [PubMed] [Google Scholar]
- 207.Cleaver JE, Thompson LH, Richardson AS, States JC. A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum Mutat. 1999;14:9–22. doi: 10.1002/(SICI)1098-1004(1999)14:1<9::AID-HUMU2>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 208.Nichols AF, Ong P, Linn S. Mutations specific to the xeroderma pigmentosum group E Ddb- phenotype. J Biol Chem. 1996;271:24317–24320. doi: 10.1074/jbc.271.40.24317. [DOI] [PubMed] [Google Scholar]
- 209.Defesche JC, Schuurman EJ, Klaaijsen LN, Khoo KL, Wiegman A, Stalenhoef AF. Silent exonic mutations in the low-density lipoprotein receptor gene that cause familial hypercholesterolemia by affecting mRNA splicing. Clin Genet. 2008;73:573–578. doi: 10.1111/j.1399-0004.2008.00999.x. [DOI] [PubMed] [Google Scholar]
- 210.Yamaguchi S, Brailey LL, Morizono H, Bale AE, Tuchman M. Mutations and polymorphisms in the human ornithine transcarbamylase (OTC) gene. Hum Mutat. 2006;27:626–632. doi: 10.1002/humu.20339. [DOI] [PubMed] [Google Scholar]
- 211.Nyhan WL, Wong DF. New approaches to understanding Lesch-Nyhan disease. N Engl J Med. 1996;334:1602–1604. doi: 10.1056/NEJM199606133342411. [DOI] [PubMed] [Google Scholar]