Abstract
In the three domains of life, the Sec, YidC/Oxa1, and Tat translocases play important roles in protein translocation across membranes and membrane protein insertion. While extensive studies have been performed on the endoplasmic reticular and Escherichia coli systems, far fewer studies have been done on archaea, other Gram-negative bacteria, and Gram-positive bacteria. Interestingly, work carried out to date has shown that there are differences in the protein transport systems in terms of the number of translocase components and, in some cases, the translocation mechanisms and energy sources that drive translocation. In this review, we will describe the different systems employed to translocate and insert proteins across or into the cytoplasmic membrane of archaea and bacteria.
Keywords: Archaea, Gram-positive, SecYEG, Tat, YidC
Introduction
All living cells are compartmentalized, irrespective of whether they belong to the eukaryotic, prokaryotic, or archaeal domains of life. The most complex cells are found in eukaryotes, and these contain a number of membrane-bound organelles, such as the endoplasmic reticulum (ER), nucleus, mitochondrion, peroxisome, golgi, nucleus, and lysosome. In total, there are over ten aqueous compartments and ten membrane subcellular locations in eukaryotic cells, each containing a unique set of proteins. Cells of bacteria and archaea are relatively simpler, with fewer aqueous compartments and intracellular membranes. Gram-negative bacteria contain at least four subcellular locations (cytoplasm, inner membrane, periplasm, and outer membrane), while Gram-positive bacteria and archaea each contain at least three subcellular locations (cytoplasm, membrane, and cell wall).
Proteins are sorted to their correct intracellular destinations or the extracellular space from their site of synthesis, which is typically in the cytoplasm. This process can be very complex, as in the case of a chloroplast protein localized to the thylakoid lumen, which involves protein import into the chloroplast across the outer and inner membranes, followed by translocation across the internal thylakoid membrane.
Approximately 25% of all proteins in a cell must cross at least one membrane to be properly localized, and roughly 20–25% of all proteins are membrane proteins that must insert into the lipid bilayer. Archaea, bacteria, and eukaryotes possess specialized translocases and insertases that catalyze the translocation of proteins into the membrane. The Sec machinery operates to insert proteins into the ER of eukaryotes and into the cytoplasmic membrane (plasma membrane) of bacteria and archaea, but it cannot translocate folded substrates across the membrane. In contrast, the Tat (twin-arginine translocation) system functions to translocate folded proteins across the cytoplasmic membrane in bacteria and archaea as well as across the chloroplast thylakoid membrane in eukaryotes. The YidC/Oxa1 insertase inserts proteins into the membrane in mitochondria, chloroplasts, bacteria and, possibly, certain archaea. Whereas the Sec system can function both in the insertion of proteins into the membrane and the translocation of proteins across membranes, the YidC system is dedicated primarily to inserting proteins into the membrane, while the Tat system functions to translocate proteins both into and across membranes.
The focus of this review is to discuss our current state of knowledge regarding how proteins are transported in bacteria and archaea. Here, we review the mechanisms by which proteins are selected for protein translocation and targeted to the translocase in the membrane. We also discuss the insertases used to insert membrane proteins and the different translocation complexes used to transport folded and unfolded substrates while maintaining the membrane permeability barrier. The transport of proteins in the ER and Escherichia coli systems are only briefly discussed (see [1–5] for recent reviews of these systems).
Membrane proteins
Integral membrane proteins come in two basic structures: helix bundle and β-barrel configurations. The α-helical integral membrane proteins can span the membrane with different numbers of transmembrane helices and in different orientations relative to the membrane. Transmembrane segments can vary in length, but they are typically 20–30 residues long, and the hydrophilic loops connecting the transmembrane regions are usually short [6]. The orientation of integral membrane proteins tends to follow the “positive inside rule” with lysine and arginine residues enriched in cytoplasmic rather than extracytoplasmic regions. This “positive inside rule” is obeyed by bacterial inner membrane proteins, eukaryotic plasma membrane proteins, thylakoid membrane proteins, and mitochondrial inner membrane proteins [7–9]. Positively charged residues flanking the transmembrane helices are determinants of membrane protein topology, i.e., the topology can be manipulated by changing the location of the positively charged residues bordering the transmembrane segments (for review, see Dalbey [10]). The other basic structure of integral membrane proteins is encountered in β-barrel proteins. These proteins contain β-sheets arranged in such a way that two or more peptide backbones are placed side by side and stabilized by hydrogen bonding. β-Barrel proteins are found mostly in the bacterial outer membrane and the mitochondrial outer membrane. The reader is referred to Ruiz et al. [11] for a review of the assembly of β-barrel proteins into the outer membrane of bacteria.
Topology prediction programs
The experimental determination of the three-dimensional (3D) structure of membrane proteins is very difficult, primarily because procedures for producing large quantities of properly folded membrane proteins and for subsequent crystallization are often not successful. Consequently, the development of computational methods for predicting the topology of membrane proteins has been of great importance in attempts to obtain a better understanding of the function(s) of membrane proteins.
The first prediction programs were based on hydrophobicity plots, as the membrane spanning α-helices generally consists of stretches of hydrophobic amino acids [12–15]. This strategy was improved by the inclusion of the “positive inside rule” in the predictions, which enables the prediction of the in–out topology [16]. Based on these residue-based principles, several prediction programs have been developed (e.g., Toppred [12, 16]).
The second generation of prediction programs considers the protein sequence as a whole, instead of as single residues. Several successful prediction programs (e.g., TMHMM, HMMTOP) are based on hidden Markov Models (HMM) [17–19], but there are also many other successful model-based approaches (e.g., MEMSAT, ToppredII, TMAP, PHDtm) [19–21]. All of these programs calculate the most probable topology of a protein based on the odds of each amino acid of the protein being positioned at a certain location (intra- or extracytoplasmic or in the membrane). In general, these model-based prediction programs score better than the residue-based prediction programs [22]. Two new prediction programs have recently been introduced that are not based on the statistical chances that residues are localized at a certain site but, rather, on the energetics of membrane insertion and the “positive inside rule” (SCAMPI, TopPredΔG) [23].
Further improvements of the model-based predictions have been achieved through the use of artificial neural networks (MEMSAT 3, PHDtm) and by the inclusion of HMM models for signal peptide predictions to discriminate signal peptides from transmembrane helices (Phobius, MEMSAT 3) [21, 24–27]. Additionally, increasingly more programs have been developed to include evolutionary information; for example, predictions based on the recognition of evolutionary conserved profiles or on multiple-sequence alignments of homologs (e.g., Polyphobius, PRODIV-TMHMM, PHDtm) [24, 28].
All of the above-mentioned topology prediction programs are based on the assumption that a membrane protein consists of intra- and extracellular loops with alternating α-helices and that these α-helices have to completely span the membrane in an orientation that is largely perpendicular to the membrane surface. However, in reality, protein structure is often more complex than is assumed by the prediction program and includes short membrane spans and long tilted membrane spans, helices that only partially insert into the membrane, and kinked helices. The recently developed program OCTOPUS is currently the only available online membrane protein topology prediction program that can predict some of these more complex features of membrane-inserted helices [29, 30].
Signal recognition particle targeting pathway
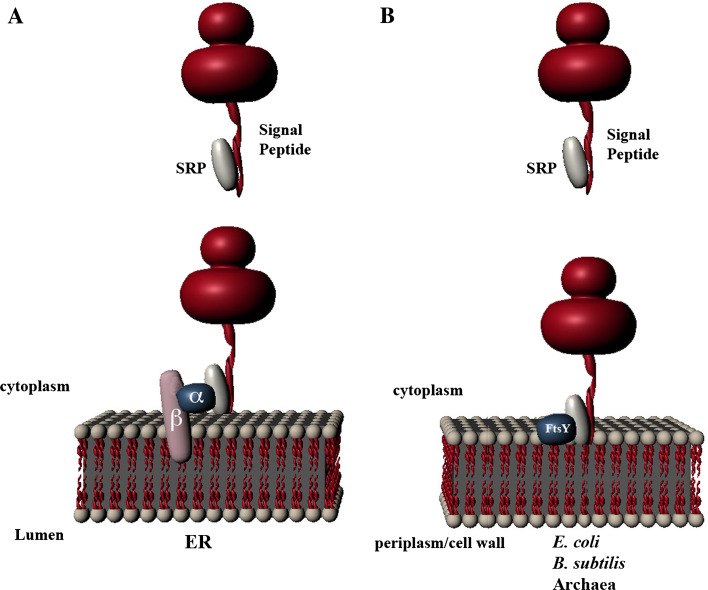
The first step in the biogenesis of membrane proteins is the targeting of proteins to the membrane. In this step, the signal recognition particle (SRP) system plays a primary role (Fig. 1). This SRP targeting system was initially discovered through studies on the ER system. The SRP is composed of six protein subunits and a 7S RNA (Table 1) [31]. It binds to the ribosome-bound nascent chain when the signal peptide emerges from the ribosome. The binding of SRP arrests protein synthesis [32], thereby enabling the nascent chain/SRP complex to be targeted to the membrane before the nascent chain grows too large and achieves a folded conformation, thus rendering it incapable of being translocated across the membrane. Targeting of the nascent chain to the membrane is achieved by recognition of the SRP-nascent chain-ribosome complex by the membrane-associated SRP receptor. The ER SRP receptor (SR) is a dimer composed of a membrane-embedded subunit (SRβ) and a peripherally membrane-associated subunit (SRα) [33]. Both the SR and SRP have GTPase activity [31]. The interaction of SRP with its receptor occurs only when both have bound guanosine-5'-triphosphate (GTP). Binding of the ribosome to the Sec61 channel for protein translocation across the membrane causes the release of SRP from the ribosome-nascent chain complex and stimulates the hydrolysis of GTP which, in turn, promotes the dissociation of SRP from the SR [34, 35]. It has recently been shown that the SRP RNA can stimulate the interaction of SRP with the SR when the SRP is bound to a signal sequence [36]. In this fashion, RNA stimulates the GTPase activities of the SRP and SR complex.
Fig. 1.
Signal recognition particle (SRP) targeting pathway. a SRP binds to the ribosome-nascent chain complex when the signal peptide emerges from the ribosome and targets the complex to the membrane by binding to its receptor on the membrane. The endoplasmic reticulum (ER) SRP receptor is a dimer with a peripheral membrane protein subunit (SRα) and an integral membrane protein subunit (SRβ). b In bacteria and archaea, the SRP receptor FtsY contains only one protein subunit, which is homologous to subunit SRα in the ER
Table 1.
The translocation and targeting components in the ER, Archaeal and Bacterial plasma membrane
| Translocation and targeting components | Endoplasmic reticulum | Archaea | Escherichia coli | Bacillus subtilis |
|---|---|---|---|---|
| SRP RNA | 7S | 7S | 4.5S | scRNA |
| SRP protein subunits | SRP54, SRP19, SRP9/14, SRP 68/72 | Ffh, SRP19 | Ffh | Ffh, HBsu |
| SR | α, β | FtsY | FtsY | FtsY |
| Sec translocase | Sec61αβγ | SecYEβ | SecYEG | SecYEG |
| Associated subunits | Sec62/63, Sec71/72, TRAM | SecDF | SecDFYajC | SecDFYajC |
| Motor ATPase | Bip | / | SecA | SecA |
| YidC system | / | YidC | YidC | SpoIIIJ, YqjG |
| Tat translocase | / | TatA, TatC | TatA, TatB, TatE, TatC | TatAdCd, TatAyCy |
SRP, Signal recognition particle; SR, Endoplasmic reticulum (ER) SRP receptor; scRNA, small cytoplasmic RNA
In E. coli, the SRP targeting system is critical for the membrane insertion of many—but not all—inner membrane proteins [37]. The system is much simpler in E. coli than in eukaryotes. The E. coli SRP is composed of Ffh (54 homologs) and a 4.5S RNA; the E. coli SRP receptor is FtsY, which is homologous to the SRP receptor (SRα) (Fig. 1; Table 1) (for review, see [38]). In E. coli, Ffh, 4.5S RNA, and FtsY are all essential for cellular growth [2, 39]. Many secretory proteins, on the other hand, are targeted to the membrane by the SecB pathway. These proteins bind to SecB in the cytoplasm and are then targeted to SecA, localized at the inner membrane surface.
SRP membrane targeting in Gram-positive bacteria
Not surprisingly, the SRP system which targets proteins for secretion in Bacillus subtilis and other Gram-positive bacteria is homologous to the system in E. coli (Fig. 1b). However, E. coli proteins can be targeted to the Sec translocase either via the SRP pathway or by SecB targeting. This latter pathway is absent from all Gram-positive bacteria. Proteomic studies have indicated that many secretory proteins in B. subtilis are likely to be targeted to the Sec translocase by the SRP pathway [40]. Interestingly, the extracellular accumulation of individual proteins was found to be affected to different extents by depletion of Ffh or FtsY, and the observed SRP dependence of certain secretory proteins did not seem to correlate with signal peptide length or hydrophobicity. This is a remarkable finding because the SRP of B. subtilis is known to have a clear preference for the most hydrophobic signals [41]. These findings suggest that other, yet unidentified, determinants in secretory proteins are probably also important in terms of SRP dependence. The high-level production of secretory proteins also resulted in elevated cellular Ffh and FtsY levels, which is due to post-transcriptional regulation [40].
The SRP in B. subtilis consists of Ffh, the small cytoplasmic (sc)RNA, and the non-specific DNA-binding protein HBsu, and it can bind to the SRP receptor FtsY (Table 1) [42–45]. Ffh and FtsY are paralogs sharing a homologous GTPase domain. Its scRNA of 271 nucleotides contains the three conserved domains (I, II, and IV) of the eukaryotic 7S RNA [46]. The ffh, hbs, ftsY, and scr genes are essential in B. subtilis [47, 48] as well as in other Gram-positives, such as Staphylococcus aureus and Streptococcus pneumoniae [49, 50]. However, it should be noted that Ffh and FtsY can be depleted to sub-detection levels without any major effects on growth and cell viability when B. subtilis cells are grown in broth [40]. A third paralog of Ffh and FtsY, named FlhF, is present in B. subtilis and a number of other Gram-positive and Gram-negative bacteria (but not in E. coli) [51]. FlhF has been implicated in type III secretion system (T3SS)-like flagellar protein targeting systems. Indeed, the mutation of flhF in Bacillus cereus results in strong motility and secretion phenotypes since FlhF is involved in assembly of the flagellum, cell motility, and regulation of protein secretion [52]. The structure of B. subtilis FlhF has been determined [53], and unlike the situation in B. cereus, B. subtilis FlhF does not seem to be involved in protein secretion and is even dispensable for cell motility [54].
Strikingly, Streptococcus mutans can survive without ffh, ftsY, and scr, although the mutant strains are acid- and salt-sensitive, which is probably due to inefficient integration of the F1F0 ATPase into the membrane [55–57]. The levels of several important chaperones and proteases are increased in these mutants, whereas protein synthesis seems to be down-regulated, which suggests that the S. mutans cells can somehow adapt to the absence of the SRP pathway [58].
The SRP pathway in archaea
In archaea, the SRP targeting pathway represents an intermediate between the SRP system of bacteria and Eukarya. Just like the SRP pathway, which is essential for the insertion of membrane proteins in E. coli and B. subtilis, archaeal SRP has been proven to be essential by showing that the ffh gene is indispensable for cell viability [59].
The archaeal SRP is composed of SRP54/Ffh, SRP19 homologs, and a 7S-like SRP RNA, which works as a framework for the assembly of SRP protein components (Table 1) [60]. Despite the overall lack of sequence conservation, archaeal SRP RNA has a very similar secondary structure to its counterpart in Eukarya, except for the presence of an additional helix (helix 1) formed by pairing the 5′ end and 3′ end of the RNA and the absence of helix 6 that is found in the eukaryotic molecule [61]. The structure of the SRP complex in Methanococcus jannaschii is consistent with the view that the association of SRP RNA with the SPR54 NG domain plays a prime role in regulating the ordered sequence of events in protein targeting [62].
SRP54/Ffh performs the main function in the SRP pathway since it is responsible for the binding of the nascent chain and SPR receptor [63, 64]. Unlike the eukaryotic SRP54, some archaeal SRP54/Ffh proteins are missing the conserved threonine at the GTP binding site, which suggests a mechanism of GTP hydrolysis that differs from the eukaryotic SRP54 [60]. The targeting of ribosome-nascent chain complexes to the membrane is mediated by the interaction between the NG domain of SRP54 and SR. It has been shown that these two GTPases interact tightly through a “twinning” of their GTP substrates [65, 66].
SRP19 is found in all archaea, which suggests that SRP19 may play a role in the archaeal SRP system. In Eukarya, SRP19 is involved in SRP assembly and facilitates the binding of SRP54 to the SRP RNA by interacting with SRP RNA helix 8 [67–70]. Nevertheless, it has been shown that, unlike in Eukarya, archaeal SRP19 is not strictly required for the binding of archaeal SRP RNA to SRP54/Ffh [67, 71, 72]. The SRP19-independent binding of archaeal SRP RNA and SRP54/Ffh seems to reflect the stability of the SRP complex, which may be required for growth at extreme temperatures or pH values or in highly saline environments [68]. The structure of the SRP complex from Pyrococcus furiosus [73] revealed that the SRP54 signal sequence-binding domain (M domain) is linked to the GTPase domain (NG domain) by a flexible α-helix, which likely helps its scanning for newly synthesized signal sequences.
Archaea do not have the eukaryotic SRP components SRP9/14, which are responsible for the translational pause in Eukarya, or the components SRP68/72, which are involved in the docking of nascent chain-SRP complexes to the ER membrane in Eukarya [74–76]. It is thus possible that the tRNA-like structure of the archaeal SRP RNA Alu domain could directly contact the ribosome without SRP9/14 [77].
All archaea contain the SRP receptor FtsY (Table 1). This SRP receptor has a C-terminal nucleotide-binding domain that is highly conserved in the SRα of Eukarya and the bacterial FtsY [78]. In contrast, the N-terminal region of the archaeal FtsY has no sequence similarity to its eukaryotic and bacterial counterparts. However, most archaeal FtsY proteins contain clusters of positively charged residues. For the E. coli FtsY, it has been suggested that these charged residues facilitate the binding to negatively charged head groups of the phospholipids at the surface of the membrane [79–81]. Between these charged motifs in the archaeal FtsY are hydrophobic residues that are implicated in interactions with the hydrophobic core of the lipid bilayer. Interestingly, the structure of FtsY in its free and GDP·magnesium-bound form differs from the structure of bacterial FtsY as it contains a long N-terminal helix [82, 83].
Functionally, the FtsY in archaea is more similar to its bacterial counterparts than to the eukaryotic SRP. As in bacteria [2], the archaeal FtsY can be detected both in a soluble cytoplasmic state and a membrane-bound state [84, 85]. Additionally, the archaeal FtsY also works alone as an SRP receptor without any inner membrane partner [86].
The Sec pathway
The main machinery for protein export and membrane protein insertion in E. coli is the Sec translocase-mediated pathway [1, 87]. The Sec translocase is comprised of the SecYEG translocation channel and the accessory components SecA, SecDFYajC, and YidC (Fig. 2; Table 1). SecA is the motor ATPase and is essential for driving the export of proteins across the inner membrane in E. coli [88] and for translocating hydrophilic domains of certain inner membrane proteins [89]. Exported proteins are translocated through the center of the Sec channel as a loop, with the N- and C-terminal ends located at the cytoplasmic surface of the membrane. SecA is believed to promote protein export by driving the substrate into the SecYEG channel through a power-stroke or Brownian ratchet mode-of-action [1]. The protein chain is moved across the channel in steps of 20–25 residues as SecA undergoes a series of conformational changes through cycles of ATP binding and hydrolysis at the membrane surface [90–92]. SecDF improves protein export and membrane protein insertion efficiency in vivo although these components are not absolutely essential [93, 94]. YidC is required for membrane protein insertion of certain Sec-dependent substrates and for membrane insertion of Sec-independent substrates (for review, see Kiefer and Kuhn [95]). YidC is particularly important for the biogenesis of inner membrane respiratory chain complexes [96, 97], but it typically does not play a critical role in the export of the protein across the inner membrane [98].
Fig. 2.
The translocation machineries of the Sec pathway. a In Escherichia coli, the translocation apparatus comprises SecYEG, SecDF, SecA, and YidC. b In the ER, the Sec translocase comprises the Sec61αβγ translocation channel, Sec62/Sec63 complex, containing Sec62, Sec63, Sec71, and Sec72 components, and chaperone Bip. c In Bacillus subtilis, the Sec translocase components are SecYEG, SecDF and SecA. d In archaea, the Sec translocase components are SecYEβ and SecDF
The translocation components in the ER are the Sec 61αβγ translocation channel, Sec62/Sec63/Sec71/Sec72 components, translocation-associated membrane protein (TRAM), in some cases, Bip (Fig. 2b; Table 1) [1, 87]. Sec61α and Sec61γ are homologous to the bacterial SecY and SecE, respectively [99]. Sec61αβγ and, in some cases, TRAM facilitate co-translational protein translocation. It is believed that an SRP-targeted protein is pushed through the channel in an N- and C-terminal direction as the protein is being synthesized one amino acid at a time. For post-translational protein export, the Secαβγ channel mediates the insertion of the exported protein across the membrane, but this process also requires the Sec62/Sec71/Sec72 components for protein targeting to the translocation site [100]. The ATPase Bip in the ER lumen drives the polypeptide through the Secαβγ channel [101]. Thus, unlike the SecA ATPase in E. coli, which functions from the cytoplasmic side of the membrane, Bip acts from the extracytoplasmic side of the membrane.
As in E. coli, the most important machinery for the secretion of proteins and insertion of membrane proteins in Gram-positive bacteria is the Sec machinery. Similar to E. coli and the ER system, proteins are targeted to the Sec-translocase by a signal peptide, consisting of a positively charged N-terminus (N-region), a hydrophobic core (H-region), and a cleavage site (C-region). In general, signal peptides in Gram-positive bacteria are longer and more hydrophobic than E. coli signal peptides [102]. The increased hydrophobicity of B. subtilis signal peptides has been shown to be critical for efficient targeting to the Sec translocase in B. subtilis, whereas in E. coli this high hydrophobicity is not required [41].
Again identical to the situation in E. coli, SecA, SecY, and SecE are essential for the survival of B. subtilis (Fig. 2c; Table 1). SecG is dispensable, although its absence results in cold sensitivity and secretion defects [47, 103]. In accordance with the fact that SecG is not essential for growth, the sequence of SecG is much less conserved among Gram-positive and Gram-negative bacteria, and SecG is even totally absent from several Gram-positive species. SecE is substantially smaller in Gram-positive bacteria than in Gram-negative ones, with the SecE of the former consisting of only one membrane span, which corresponds to the C-terminal membrane span of the E. coli SecE. In E. coli, the two N-terminal membrane spans of SecE are important in preventing the degradation of SecE by FtsH [104], but they are dispensable for protein translocation. The major components of the Sec-translocases in E. coli and B. subtilis are quite similar, and hybrid translocases (combining SecY, SecE, SecG, and SecA from both bacteria) can form stable complexes; however, these hybrid complexes are very inefficient in terms of substrate recognition and translocation in E. coli [105].
Differences found in the Gram-positive Sec pathway
Some Gram-positive bacteria and mycobacteria (mostly pathogenic species) contain two secA genes. Interestingly, usually only the most conserved secA gene is essential for transport of the vast majority of exported proteins and cell survival, while the other copy is involved in the secretion of a specific subset of proteins (for reviews, see Rigel and Braunstein [106] and Sibbald et al. [107]). One exception is Corynebacterium glutamicum, for which both copies of secA are essential for survival [108]. A second secY gene has also been found in several Gram-positive bacteria with two secA genes [106, 109, 110]. In a number of these species, such as Streptococcus gordonii, the secY2 gene is located in an operon containing other genes that are required for the translocation of the SecA2/SecY2-dependent proteins [109–111]. In S. gordonii, two of these genes encode SecE and SecG paralogs, respectively [112]. This finding suggests that the other genes may also encode proteins that are part of a novel accessory Sec translocase that functions in parallel or in concert with the canonical Sec translocase.
Proteins secreted by SecA2 or SecA2/SecY2 are often involved in virulence [109, 113]. Some of these proteins are totally dependent on secretion by the accessory Sec translocase [109, 113, 114], whereas other proteins can be secreted either by the accessory or the canonical Sec pathway [113]. Although it seems that glycosylation in the cytoplasm can prevent proteins from being translocated by the canonical Sec translocase, studies on the GspB glycoprotein from S. gordonii have shown that such proteins can be translocated by the accessory Sec translocases [112, 115]. Just how proteins are specifically targeted to the accessory secretion pathway is currently unknown, but Sec pathway specificity is at least in part determined by subtle differences in the SecA and SecA2 subunits, as unique contacts between SecA2 and the other components of the accessory Sec system seem to preclude cross-functioning with the canonical Sec system [116]. Some of the proteins that depend on this pathway have a conventional signal peptide, whereas other proteins either have a signal peptide with an atypically long N-region, or they seem to lack a signal sequence altogether [106, 112, 114, 115, 117, 118].
Homologs of the SecDFYajC complex are also present in B. subtilis and other Gram-positive bacteria. Interestingly, in B. subtilis, SecD and SecF are present as a ‘Siamese twin’, i.e., as a fusion of these two homologous proteins, called SecDF [119]. This Siamese twin unit seems to be ubiquitous among the Firmicutes, but it can also be found in several other Gram-positive and Gram-negative species. Knockout of secDF in B. subtilis results in cold sensitivity and reduces the capacity to secrete overproduced proteins [119]. Therefore, it is believed that SecDF increases the efficiency of protein translocation by an as yet unknown mechanism.
While most studies on the Sec pathway in Gram-positive bacteria have dealt with exported proteins, very few studies have addressed the requirement of the Sec pathway for membrane protein insertion [120]. Notably, however, good progress has been made in the analysis of the membrane proteome of Gram-positive bacteria, such as B. subtilis, and in this context Bunai and co-workers were first to report the SecA requirement of 11 integral membrane proteins with one or two transmembrane segments on the basis of proteomic studies [121]. As new and improved tools for the analysis of membrane proteins by proteomics are becoming available, it can be anticipated that they will soon be applied to the dissection of the roles of Sec pathway components in membrane protein biogenesis in organisms such as B. subtilis and S. aureus [122–125].
Localization and structure of SecYEG complexes
The subcellular localization of SecA and SecY in B. subtilis was investigated using green fluorescent protein (GFP) fusions and also by immunofluorescence for SecY. The results of these studies indicated that the membrane-bound Sec translocase (as in E. coli) may be present in a spiral-like structure along the cell [126]. Interestingly, in the coccus-shaped Gram-positive bacterium Streptococcus pyogenes, the Sec translocase was found to be located at only one specific site, the Exportal, which has been defined as a microdomain specialized in secretion [127, 128]. The mechanisms by which these distributions are accomplished and the purpose of these arrangements are still poorly understood, although it has been proposed that the concentrated secretion at specific sites might enable bacteria to coordinate protein translocation and subsequent folding [128]. Consistent with this idea, the HtrA protease, which is involved in the maturation of a secreted cysteine protease (SpeB), was localized exclusively to the ExPortal of S. pyogenes [128]. However, it remains to be shown whether extracytoplasmic folding catalysts that assist in the folding of Sec-dependently translocated proteins are also localized at the Exportal.
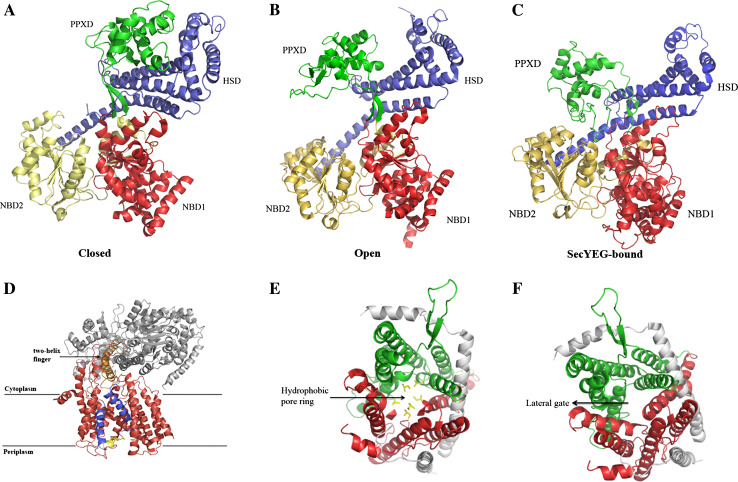
To understand how SecA functions in protein export, the structure of this protein was determined by X-ray crystallography. More than five structures of SecA have been identified from a number of Gram-positive (B. subtilus and Mycobacterium tuberculosis) and Gram-negative bacteria (Thermus thermophilus, E. coli, and Thermotoga maritima) [129–134]. Most of the SecA structures are in the dimeric state, but there are also several structures of monomeric SecA. Briefly, SecA is composed of five functional regions (for review, see Papanikou et al. [135]): two nucleotide binding domains (NBD1 and NBD2), a pre-protein crosslinking domain (PPXD), a α-helical scaffold domain (HSD), and a C-terminal translocation domain (HWD and IRA1). The SecA structure can take two different conformations based on the distance between PPXD and HWD: one in the closed state (Fig. 3a) and one in the open state (Fig. 3b). While Osborne et al. [131] have proposed that the groove between the PPXD and HSD is the substrate binding region, Cooper et al. [136] have proposed—on the basis of electron paramagnetic resonance (EPR) studies—that this binding site is located on the opposite side of PPXD. However, it should be noted that these structures were determined in the absence of the Sec translocation channel. In 2008, the structure of the SecA/SecYEG complex from T. maritima, a Gram-negative bacterium, was solved at a resolution of 4.5 Å [134]. The structure of the SecA/SecYEG complex showed one SecA bound to one copy of SecYEG. This structure revealed a SecA clamp region formed by PPXD, NBD2, and part of the HSD that may be involved in capturing the substrate (Fig. 3c). When SecA binds to the SecYEG complex, SecYEG undergoes a conformational change that primes the channel for translocation (Fig. 3d). The SecA binding also causes the lateral gate of the SecYEG channel to open up towards the center of the lipid bilayer and the plug to move away from the center of the channel toward the periplasmic side of the membrane (Fig. 3d). Interestingly, there is a two-helix finger in SecA near the entrance of the SecYEG channel (Fig. 3d) which the authors suggest moves up and down in each ATP hydrolysis cycle to drive the exported protein through the SecYEG complex. Subsequent studies with SecA, SecYEG, and a substrate trapped in the channel using disulfide crosslinking were able to identify which region of the two-helix finger of SecA can contact the peptide substrate during protein translocation [137].
Fig. 3.
Structure of the SecA and the SecY (Sec61) channel. a–c Different conformations of SecA with the view from the cytosol: a the closed conformation, b the open conformation, c the SecY-binding conformation. The pre-protein crosslinking domain (PPXD) is shown in green, the α-helical scaffold domain (HSD) in blue, the nucleotide binding domains (NBD2 and NBD1) in yellow and red, respectively. d The two-helix finger of SecA at the cytoplasmic entrance of the SecYEG complex. The finger is highlighted in orange and indicated by an arrow. SecYEG is colored red, except for the TM2 and TM7 segments, which are in blue and the “plug” in yellow. e The hydrophobic pore ring within the center of the Sec61αβγ channel. The view is from cytosol. The N- and C-terminal halves are in red and green, respectively. The residues comprising the hydrophobic pore ring are highlighted in yellow and indicated by an arrow. f The lateral gate region within the Sec61αβγ complex is indicated by an arrow at the interface of TM2b and TM7. The view is from cytosol, as in d. The figures were constructed using the coordinates from the protein data base. PDB accession numbers: a 1M6N, b 1TF2, c, d 3DIN, e, f 1RH5
The structure of the Sec channel (SecYE complex) from the Gram-negative bacterium T. thermophilus was recently solved at a resolution of 3.2 Å with bound Fab fragments of the monoclonal antibody [138]. Tsukazaki et al. [138] found that the transmembrane helices TM6, TM8, and TM9 of the SecYE complex from T. thermophilus were shifted at about 10 Å relative to the Methanococcus jannaschii SecYEβ complex that lacks the Fab fragment and SecA. They proposed that the structure of the SecYE channel is in the pre-open state that may mimic the conformation when SecA is bound; the Fab fragment contacts cytoplasmic loops C4 and C5 of SecY, which is the same region to which SecA can be crosslinked. The region of SecA that contacts SecYE in T. thermophilus has been named motif IV [138].
Despite recent progress, a number of issues still remain to be addressed in the structural area of the Sec complex. For example, the structure of the SecA/SecYEG complex suggests that a two-helix finger is important as it is localized at the entrance of the SecYEG translocation channel. The hypothesis is that the SecA two-helix finger moves back and forth during the ATPase cycle and that these movements are involved in translocation of the substrate. This hypothesis needs to be tested. Another important question requiring an answer is whether the SecA/SecYEG structure corresponds to the state following capture of a substrate in the respective organism from which this translocase has been derived. The determination of the structure of SecA/SecYEG with a bound substrate would be helpful in proving this. Also, more experimental data are needed to establish the substrate binding site of SecA. The determination of the structure of a SecA/substrate complex would be helpful to show whether the groove between PPXD, NBD2, and part of HSD is the binding site for the substrate.
The archaeal Sec translocase
As in bacteria and eukaryotes, archaea use the Sec translocase to export secretory proteins across the membrane and to insert proteins into the membrane. The archaeal Sec translocase consists of SecY, SecE, and Sec61β (Fig. 2d; Table 1) SecY genes are found in all archaeal genomes and, like its eukaryal and bacterial counterparts, the SecY protein of archaea spans the membrane ten times [139–141]. Complementation experiments using a conditionally lethal E. coli SecY mutant transformed with an archaeal secY gene suggest that the archaeal SecY can functionally replace the bacterial SecY [139]. The SecY in archaea is, however, more similar to the eukaryotic Sec61α than the bacterial SecY [140]. Likewise, the archaeal SecE is more like eukaryotic Sec61γ in that it has a single transmembrane domain at the C terminus [140]. In contrast to the clear homology between Sec61αγ in eukaryotes and SecYE in bacteria, Sec61β and SecG do not resemble each other at all [140]. In archaea, the Sec61β homolog was first found by PSI-BLAST searches [140, 142].
The structure of the archaeal Sec translocase was solved in 2004 in M. jannaschii at a resolution of 3.2 Å [143]. This first structure of a protein-conducting channel was a landmark achievement. The structure showed that the Sec translocase forms a clamshell-like structure within the membrane. SecY forms the core of the channel, while SecE and Sec61β help to stabilize it in the membrane. In the center of the channel is a constriction point comprised of a pore ring, 3–5 Å in size, through which the hydrophilic region of the exported protein is believed to pass (Fig. 3e). The translocation channel also has an opening on one side via the TM2b/TM7 interface region, which the authors called the lateral gate (Fig. 3f). In an earlier study, this region had been shown to bind the signal peptide [144]. Based on these results, in 2004, van den Berg et al. proposed that the signal peptide initially binds to the lateral gate prior to exiting the channel and integrating into the lipid bilayer [143].
Based on the structure of the Sec translocation channel, it was hypothesized that the transmembrane segments of membrane proteins would exit the channel through the lateral gate. While the lateral gate is closed in the M. jannaschii Sec channel, it is believed to open up by a “breathing action” to allow the transmembrane segments to integrate into the lipid bilayer. The structure of the T. maritima SecYEG/SecA complex showed that the lateral gate is more open toward the center [134]. Currently, there is no evidence that this TM2b/TM7 interface region undergoes structural changes or that the opening is triggered by the incoming transmembrane segment.
While the same SecYEβ/Sec61αβγ complex is used in archaea and ER, the eukaryotic associated subunits Sec62/63, Sec71/72, and TRAM have not yet been identified in archaea [140, 145]. At the same time, bacterial associated subunits, such as SecD and SecF, are found in many archaea with high conservation, while homologs of SecA have not yet been discovered. The apparent absence of SecA implies that archaea may use an accessory protein, such as an ATPase, that is different from SecA as an energy-transducing source for protein transport or that these organisms may even rely entirely on the ion gradient across the membrane to drive protein translocation. Another possibility is that the energy source is derived from GTP hydrolysis, with the protein pushed through the Sec translocation channel as the protein is being synthesized, as is proposed in the ER system for the SRP-targeted proteins.
Although the archaeal SecDF shows high similarity to bacterial SecDF in terms of membrane topology and positioning of the conserved sequence elements, there are differences in the large extracytoplasmic domains of these proteins, especially in the SecD protein [146]. These differences may be due to the function of the loop in bacteria, which seems to modulate SecA during the protein translocation step [147–149]. Accordingly, there is no need for conservation of this loop region in archaea due to the absence of SecA [86, 150].
YidC-pathway
Operating in parallel with the Sec pathway is the evolutionarily conserved YidC pathway, which is found not only in bacteria and some archaea (Table 1), but also in the mitochondria and chloroplasts of eukaryotes [151, 152]. YidC is an essential protein in E. coli that is required for the membrane insertion of proteins previously thought to insert spontaneously [98]. YidC functions as a membrane insertase to mediate membrane protein insertion [153] (Fig. 4a). It can also function cooperatively with the SecYEG translocase to mediate the membrane integration of proteins [154, 155]. In the Sec pathway, SecDFYajC physically links YidC to the Sec translocase [156]. Interestingly, residues 215–265 within the periplasmic domain of the E. coli YidC are required for YidC to interact with SecF [157].
Fig. 4.
The YidC pathway. a YidC can function on its own to mediate membrane protein insertion. b Membrane topology of YidC in E. coli with six transmembrane segments and a large periplasmic domain between transmembrane helices 1 and 2. c Membrane topology of YidC in B. subtilis. d Predicted membrane topology of YidC in Euryarchaeota
A number of Sec-dependent and Sec-independent substrates have been shown to require YidC for insertion, including subunit 2 of cytochrome bo3 oxidase (CyoA) and subunits a and c (of the F1Fo ATP synthase) [95, 158]. This explains to a large extent why YidC depletion causes a large perturbation in the assembly and activity of the F1Fo ATP synthase and cytochrome bo oxidase [96, 159, 160].
Structural studies on the E. coli YidC
In terms of structural studies on YidC, all we know so far is that the E. coli YidC, similar to the YidC homologs from most Gram-negative bacteria, spans the membrane six times with a large periplasmic domain located between the first two transmembrane segments (Fig. 4b; [161]). The structure of the periplasmic domain has been determined by X-ray crystallography and found to form a β-super sandwich fold [162, 163]. Crosslinking methods have revealed that TM3 of YidC is in the proximity of the substrate during membrane biogenesis [164, 165], while intrinsic fluorescence spectroscopy studies showed that substrate binding to YidC causes a conformational change in YidC [166]. A debatable issue is whether YidC functions as a monomer or oligomer during membrane protein insertion. Purified YidC appears as a monomer and dimer on a blue native polyacrylamide gel, and purified Oxa1, the mitochondrial homolog, as a tetramer [167, 168].
Based on cryo-electron microscopy and crosslinking studies, Kohler et al. [169] recently proposed that YidC forms dimeric insertion pores on translating ribosomes. In their model, the insertion pore is formed at the interface of two YidC molecules with transmembrane segment (TS) 2 and 3 of one YidC subunit interacting with TS2 and TS3 of the other subunit. Supporting evidence for this model is provided by the fact that a dimeric YidC or a dimeric Oxa1 can be formed via disulfide crosslinking by adding an oxidizing agent to the YidC sample; YidC contains a single cysteine at position 423 in TM3, and Oxa1 contains a single cysteine in TM1 corresponding to TM2 of YidC. Likewise, a critical role of TM2 and TM3 in YidC function is supported by genetic studies [170, 171]. Clearly, it is important to provide further biochemical and structural data to support this model. It will be necessary to obtain a high-resolution structure of YidC in order to determine the structural basis of the YidC insertase and chaperone functions. Knowledge of the structure will reveal whether YidC has a lateral gate, as seen with the Sec translocation channel.
Gram-positive YidC
All Gram-positive bacteria have at least one gene encoding a YidC homolog within their genome, but most also contain a second gene for a YidC homolog. Generally, the YidC proteins of Gram-positive bacteria are shorter than their homologs in Gram-negative bacteria [145, 172]. Typically, many of the former span the membrane five times (Fig. 4c) and are synthesized with a cleavable signal peptide that is processed by the lipoprotein signal peptidase. The C-terminal domain contains five transmembrane helices that are conserved in all bacterial, mitochondrial, and chloroplasts members [172]. The transmembrane segments of the B. subtilis YidC homologs are numbered 2–6 since they correspond to their respective transmembrane segments in the Gram-negative bacterial YidC. In B. subtilis, two paralogs of YidC are present: SpoIIIJ and YqjG. The presence of either one of these proteins is sufficient for cell survival, indicating that they can complement each other, at least partially [173, 174]. Consistent with their role in membrane protein integration, depletion of SpoIIIJ in cells lacking YqjG leads to a decreased stability of specific membrane proteins, such as CtaC and FtsH, although the stability of the other membrane proteins tested to date remains unaffected [173]. In addition, SpoIIIJ-depleted cells lacking YqjG have been found to display a post-translocational defect in protein secretion. This phenotype is most likely indirectly caused by a defect in membrane protein biogenesis.
A sporulation defect is observed in the absence of SpoIIIJ. This defect cannot be complemented by native YqjG, but it can be complemented by some mutated forms of YqjG [175]. This sporulation defect is possibly caused by impaired or incorrect integration of the SpoIIIAE protein into the membrane, while the correct integration of this protein is required for completion of the sporulation process [175, 176]. These findings demonstrate that SpoIIIJ and YqjG have only partially overlapping substrate specificities [173–175] and cannot fully complement for each other.
Analogous to B. subtilis, two paralogs of YidC have been identified in S. mutans: YidC1 and YidC2 [56, 177]. Attempts to simultaneously delete yidC1 and yidC2 have all failed, indicating that the presence of at least one of these proteins is required for cell survival. Interestingly, the deletion of yidC2 (but not of yidC1) in S. mutans leads to a similar phenotype as the deletion of members of the SRP pathway. In one study, combining a yidC2 deletion with deletions in genes for the SRP pathway resulted in barely viable strains [56], which suggests that YidC2 can compensate for the absence of the SRP pathway, thus enabling the survival of S. mutans without a functional SRP. This result also implied that YidC2 in S. mutans may have a similar function as its homolog Oxa1p in yeast mitochondria. In these organelles, the missing SRP pathway is functionally replaced by Oxa1p to which the mitochondrial ribosomes bind directly for cotranslational insertion of proteins into the inner membrane. Support for this view was recently obtained by studies showing that YidC2 from S. mutans and Oxa1p from yeast mitochondria can reciprocally complement each other in vivo [178].
YidC in Euryarchaeota
Genes for YidC homologs have been found in the genome sequences of Euryarchaeota, but not in the genome sequences of other archaea [145, 172]. The euryarchaeotal YidC homologs are predicted to lack two of the transmembrane segments (corresponding to transmembrane 4 and 5 of the E. coli YidC) that are present in the bacterial, mitochondrial, and chloroplast homologs (Fig. 4d) [145]. Transmembrane segments 2, 3, and 4 are homologous to transmembrane segments 2, 3, and 6 of the Gram-negative bacterial YidC, respectively. No functional studies have been reported yet on the archaeal YidC homologs. Therefore, their relevance for membrane protein integration remains to be established.
Tat pathway
A third widely conserved translocation system is the twin-arginine translocation (Tat) system. The Tat translocase can transport fully folded proteins across membranes, whereas the Sec translocase can only transport proteins that are in an unfolded state [1, 179–182]. The Tat translocase is found in many bacteria, archaea, and plant chloroplasts. Usually, only a small subset of proteins are transported via the Tat system to the inner membrane, the periplasm, or the extracellular environment [183]. However, in some archaea, almost all proteins appear to be secreted via the Tat system [59, 184]. The Tat pathway also seems to be responsible for the transport of many proteins in Streptomyces and Mycobacterium species [185–189]. Importantly, the Tat pathway that is found in several bacterial pathogens is required for their virulence in plants and humans [190–195].
Signal peptides targeting proteins to the Sec or Tat translocases are quite similar in overall structure. However, the N-domain of Tat signal peptides contains a typical twin-arginine motif, ZRRXϕϕ, where Z designates a hydrophilic residue, X can be any residue, and ϕ is a hydrophobic residue [179]. The H-domain is usually less hydrophobic than that in Sec signal peptides [196]. A prerequisite for a protein to be directed to the Tat system is the presence of the Tat signal peptide, as the twin-arginine residues facilitate recognition by the Tat translocase (Fig. 5a) [196]. Nevertheless, in many cases, the Tat-signal peptide by itself cannot determine pathway selectivity. Certain Tat signal peptides can target fused Sec substrates to the Sec pathway [197]. However, the introduction of positive charges at the N-terminus of the mature protein to be exported prevents translocation by the Sec pathway, but not the Tat pathway [197]. Consistently, the C-region of Tat signal peptides often contains positively charged residues that help avoid translocation via the Sec translocase [198–201]. Based on the known properties of Tat signal peptides, the Tatfind algorithm was developed for systematic whole-genome searches to identify Tat-dependent transported proteins [59].
Fig. 5.
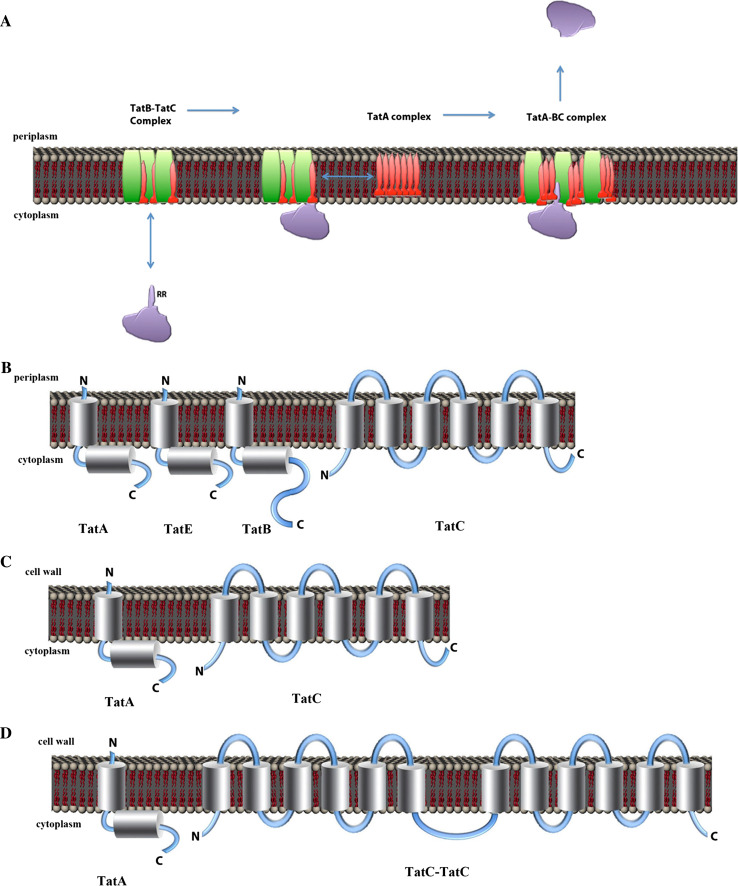
Protein export by the twin-arginine translocation (Tat) pathway. a Proposed mechanism of export by the Tat pathway. RR Twin-arginine motif in the signal peptide of a protein translocated via Tat. b Membrane topology for TatA, TatE, TatB, and TatC in E. coli. c Membrane topology for TatA and TatC in B. subtilis. d The membrane topology of TatA and TatC in the Archaeon Halobacterium sp. NRC-1. N N-terminus, C C-region
The composition of the Tat translocase is variable. In Gram-negative bacteria, three distinct proteins are required for Tat-mediated translocation: TatA, TatB, and TatC. Escherichia coli additionally contains a TatA paralog named TatE, which is dispensable for protein transport via the Tat pathway (Fig. 5b; Table 1). Notably, TatB is absent in most Gram-positive bacteria and archaea (Fig. 5c and d) [202]. Thus, the minimal Tat translocase is composed of one TatA and one TatC subunit. However, additional copies of TatA and/or TatC are often found (e.g., B. subtilis contains three TatA homologs and two TatC homologs, but no TatB homolog).
Structural features of the Tat complexes and the translocation pore
TatA and TatB are predicted to consist of a transmembrane helix, a hinge-region, a cytoplasmic amphipathic helix, and a cytoplasmic domain of variable length (Fig. 5b). Although both proteins are related, the overall sequence conservation is poor (only one amino acid is invariably conserved). The cytoplasmic domain is larger in TatB proteins than in TatA proteins [202]. TatC contains six transmembrane spans, with both the – and C-terminus facing the cytoplasm [203, 204]. The amino acid sequence conservation in TatC protein is better than in the TatA/B-like proteins but, even so, there are only 15 well-conserved amino acids in TatC.
Some structural information is currently available on the translocation pore of the Tat-translocase in E. coli, which probably consists of a large ring-shaped multimer of TatA proteins of variable size [205–207]. TatB and TatC are involved in signal peptide recognition and the targeting of proteins to the TatA pore (Fig. 5a) [207–210]. TatC seems to be mainly involved in the initial signal peptide recognition, whereas TatB seems to be required for the transfer of the protein from TatC to TatA [208]. The exact mechanism of signal recognition, docking to the TatA-pore, and subsequent translocation remains to be elucidated.
Notably, the exact mechanism by which the Tat translocase facilitates translocation is a matter of debate [180, 181]. Although likely, it is not known with absolute certainty whether TatA is the pore [206] or whether the pore comprises all the components (TatABC) [181]. The variously shaped TatA multimers that have been observed were isolated after detergent treatment and may not actually occur in the membrane. To help understand the mechanism by which proteins are translocated by the Tat translocase, researchers need to obtain an atomic structure of the Tat machinery as this structure would clarify how the Tat machinery transports differently sized folded substrates while preventing the leakage of solutes and ions across the membrane.
Multiple Tat translocases in the Gram-positive B. subtilis
Interestingly, although TatA and TatB have different functions within Gram-negative Tat translocases, mutations in TatA in E. coli have been found to enable Tat-mediated transport in the absence of TatB. This finding indicates that TatA is intrinsically capable of performing the function of TatB [211]. Additionally, the TatAd protein from B. subtilis can complement for the absence of either TatAE or TatB in E. coli [212]. These findings, plus the fact that TatB is missing from most Gram-positive bacteria, indicate that the Gram-positive version of TatA performs the functions of both TatA and TatB in organisms lacking a tatB gene [213].
Two independently operating Tat translocases exist in B. subtilis, namely, TatAdCd and TatAyCy (Fig. 5c; Table 1). Despite this abundance of Tat translocases, however, only two proteins have been identified that are exclusively transported by these translocases: the Dyp-type peroxidase YwbN is secreted via TatAyCy, and the phosphodiesterase PhoD is secreted by TatAdCd [214, 215]. A third Tat substrate, the esterase LipA, can be secreted both via the Sec and Tat pathways, with Tat-dependent secretion being most evident under conditions of LipA hyper-production [216]. Tat-dependent LipA secretion seems to be facilitated both by TatAdCd and TatAyCy. These findings indicate that the two Tat translocases of B. subtilis have overlapping but non-identical substrate specificities. The function of a third TatA subunit in B. subtilis, denoted TatAc, is not clear, since no proteins have been found that are secreted in a TatAc-dependent manner [214, 215].
The archaeal Tat pathway
Little is currently known about the archaeal Tat pathway, and most information has been derived from genomic sequencing. The Tat components (TatA and TatC) in archaea are similar to those found in E. coli and chloroplasts. Similar to most Gram-positive bacteria, TatB is not detectable in archaea, although it is clearly required for Tat translocation in E. coli [217]. The TatC in archaea is homologous to the TatC in bacteria and chloroplasts, but only a limited number of residues are well-conserved. Remarkably, most sequenced haloarchaea have a Tat component that is a natural fusion of two TatC subunits (Fig. 5d) [184, 218, 219]. The function of the TatC–TatC fusion protein may be related to the Tat translocation under high salt conditions, since this component seems to be specific to haloarchaea.
As observed in many Gram-positive bacteria, there can be multiple copies of the Tat components in archaea. The haloarchaeon, Haloferax volcanii, has been shown to have two TatA homologs and two TatC homologs, with three of these being essential for cell viability [184]. At the same time, the number of these components is not related to the number of the Tat substrates. For example, Sulfolobus solfataricus, which has only five predicted Tat substrates, has three copies of TatA and two copies of TatC. In comparison, Halobacterium sp. NRC-1, which encodes 68 putative Tat substrates, has only one copy of TatA and two copies of TatC. One possible explanation is that this observed bias towards Tat-dependent protein transport in haloarchaea, such as Halobacterium, may relate to the fact that many secretory proteins have to fold rapidly to prevent aggregation under highly saline conditions. Therefore, these proteins would fold in the cytoplasm and be transported by the Tat pathway [59, 218]. Interestingly, the bias towards Tat-dependent protein transport in the haloarchaeon H. volcanii seems so strong that it even exports soluble C-terminally anchored membrane proteins and lipoproteins via Tat [220]. However, many of the observed putative substrates for the Tat translocase in archaea, in general, are cofactor-binding proteins, and these need to be folded before translocation to allow for cofactor assembly. For example, all of the predicted Tat substrates in T. acidophilum and T. volcanicum are Rieske iron-sulphur proteins.
A recent study in the haloarchaeon Haloarcula hispanica showed that the secretion of a Tat-dependent substrate, α-amylase, does not depend on the proton motive force as it does in the chloroplasts and bacterial system [221, 222], but rather on the sodium-motive force [223]. A similar electrical gradient may drive transport in the halophile Halobacterium sp. NRC-1, which is predicted to use the Tat pathway for the translocation of more than 90% of its secreted proteins [59, 184]. One possible explanation may be the high extracellular salt concentration imposed by its particular lifestyle. Interestingly, Tat-dependent translocation in E. coli has been shown recently to be able to rely on the electrical transmembrane gradient, independent of the pH gradient [224].
Summary
The emerging studies reviewed in this paper reveal remarkable differences in the ways a number of bacteria and archaea insert and translocate proteins across the cell membrane as compared to the well-established E. coli and ER systems. These differences illustrate the importance of examining protein export and membrane biogenesis in a variety of different organisms with the aim of searching for conserved principles. A common theme is that there are multiple copies of the translocation components in Gram-positive bacteria and archaea. The presence of multiple copies of the translocation components, such as SecY, SecA, YidC, TatA, or TatC, may have to do with different substrate specificities making protein export or membrane protein insertion more efficient for certain substrates. Furthermore, the expression of differently multiplied translocation components can be regulated in different ways, providing the cells with the possibility to fine-tune their translocation machinery to particular needs, such as those as dictated by environmental conditions. In many cases, certain translocation components can contribute to the virulence of pathogenic bacteria. Also, in contrast to the situation found in the widely studied bacteria, such as E. coli and B. subtilis, the Tat translocation system can play a mainstream role for the export of proteins in certain other Gram-positive bacteria and archaea.
One of the most important recent developments in the field is the elucidation of structures of the Sec complex from the Archaeon M. jannaschii and the Gram-negative bacteria Thermotoga maritima and Thermus thermophilus and of a SecA/SecYEG complex from T. maritima. These structures as well as recent biochemical studies reveal how the translocase can function to move polypeptides through the center of the channel via the pore ring and how it allows the exit of transmembrane segments into the lipid bilayer. Elucidation of the SecA/SecYEG structure revealed a two-helix finger that may be involved in translocating the substrate through the channel.
Taken together, the observations reviewed in this paper indicate that there are still plenty of opportunities for new discoveries in the field of protein insertion and translocation across cell membranes, in particular in organisms that live in extreme and challenging ecological niches. Studies on protein translocation in these organisms are likely to deepen our insights in the structure and function of protein translocation machineries that are conserved in all three domains of life.
Acknowledgments
This work was supported by the National Institutes of Health grant GM63862 (to R.E. D.), EU grants LSHM-CT-2006-019064 and PITN-GA-2008-215524, the transnational SysMO initiative through project BACELL SysMO, the European Science Foundation under the EUROCORES Programme EuroSCOPE, and grant 04-EScope 01-011 from the Research Council for Earth and Life Sciences of the Netherlands Organization for Scientific Research (to J.M.v.D.).
Footnotes
J. Yuan and J. C. Zweers contributed equally to this work.
Contributor Information
Jan Maarten van Dijl, Phone: +31-50-3615187, FAX: +31-50-3619105, Email: J.M.van.Dijl@med.umcg.nl.
Ross E. Dalbey, Phone: +614-292-2384, FAX: +614-292-1532, Email: dalbey@chemistry.ohio-state.edu
References
- 1.Driessen AJ, Nouwen N. Protein translocation across the bacterial cytoplasmic membrane. Annu Rev Biochem. 2008;77:643–667. doi: 10.1146/annurev.biochem.77.061606.160747. [DOI] [PubMed] [Google Scholar]
- 2.Luirink J, ten Hagen-Jongman CM, van der Weijden CC, Oudega B, High S, Dobberstein B, Kusters R. An alternative protein targeting pathway in Escherichia coli: studies on the role of FtsY. EMBO J. 1994;13:2289–2296. doi: 10.1002/j.1460-2075.1994.tb06511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mainprize IL, Vulcu F, Andrews DW. The signal recognition particle and its receptor in the ER protein targeting in the enzymes vol 25, 3rd edn. In: Dalbey RE, Koehler CM, Tamanoi F, editors. Molecular machines involved in protein transport across cellular membranes. San Diego: Academic Press; 2007. pp. 177–206. [Google Scholar]
- 4.Alken M, Hegde R. The translocation apparatus of the endoplasmic reticulum in the enzymes, vol 25, 3rd edn. In: Dalbey RE, Koehler CM, Tamanoi F, editors. Molecular machines involved in protein transport across cellular membranes. San Diego: Academic Press; 2007. pp. 207–243. [Google Scholar]
- 5.Nakatsukasa K, Brodsky JL. The role of Bip/Kar2p in the translocation of proteins across the ER membrane in the enzymes, vol 25, 3rd edn. In: Dalbey RE, Koehler CM, Tamanoi F, editors. Molecular machines involved in protein transport across cellular membranes. San Diego: Academic Press; 2007. pp. 245–273. [Google Scholar]
- 6.von Heijne G. Recent advances in the understanding of membrane protein assembly and structure. Q Rev Biophys. 1999;32:285–307. doi: 10.1017/s0033583500003541. [DOI] [PubMed] [Google Scholar]
- 7.Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem. 1988;174:671–678. doi: 10.1111/j.1432-1033.1988.tb14150.x. [DOI] [PubMed] [Google Scholar]
- 9.Gavel Y, von Heijne G. The distribution of charged amino acids in mitochondrial inner-membrane proteins suggests different modes of membrane integration for nuclearly and mitochondrially encoded proteins. Eur J Biochem. 1992;205:1207–1215. doi: 10.1111/j.1432-1033.1992.tb16892.x. [DOI] [PubMed] [Google Scholar]
- 10.Dalbey RE. Positively charged residues are important determinants of membrane protein topology. Trends Biochem Sci. 1990;15:253–257. doi: 10.1016/0968-0004(90)90047-f. [DOI] [PubMed] [Google Scholar]
- 11.Ruiz N, Kahne D, Silhavy TJ. Advances in understanding bacterial outer-membrane biogenesis. Nat Rev Microbiol. 2006;4:57–66. doi: 10.1038/nrmicro1322. [DOI] [PubMed] [Google Scholar]
- 12.von Heijne G. Membrane proteins: the amino acid composition of membrane-penetrating segments. Eur J Biochem. 1981;120:275–278. doi: 10.1111/j.1432-1033.1981.tb05700.x. [DOI] [PubMed] [Google Scholar]
- 13.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 14.Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg D, Weiss RM, Terwilliger TC. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982;299:371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- 16.von Heijne G. Membrane protein structure prediction Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 17.Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 18.Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–182. [PubMed] [Google Scholar]
- 19.Persson B, Argos P. Prediction of membrane protein topology utilizing multiple sequence alignments. J Protein Chem. 1997;16:453–457. doi: 10.1023/a:1026353225758. [DOI] [PubMed] [Google Scholar]
- 20.Jones DT, Taylor WR, Thornton JM. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 1994;33:3038–3049. doi: 10.1021/bi00176a037. [DOI] [PubMed] [Google Scholar]
- 21.Rost B, Casadio R, Fariselli P, Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- 23.Bernsel A, Viklund H, Falk J, Lindahl E, von Heijne G, Elofsson A. Prediction of membrane-protein topology from first principles. Proc Natl Acad Sci USA. 2008;105:7177–7181. doi: 10.1073/pnas.0711151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Viklund H, Elofsson A. Best alpha-helical transmembrane protein topology predictions are achieved using hidden Markov models and evolutionary information. Protein Sci. 2004;13:1908–1917. doi: 10.1110/ps.04625404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones DT. Do transmembrane protein superfolds exist? FEBS Lett. 1998;423:281–285. doi: 10.1016/s0014-5793(98)00095-7. [DOI] [PubMed] [Google Scholar]
- 26.Jones DT. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics. 2007;23:538–544. doi: 10.1093/bioinformatics/btl677. [DOI] [PubMed] [Google Scholar]
- 27.Kall L, Krogh A, Sonnhammer EL. A combined transmembrane topology and signal peptide prediction method. J Mol Biol. 2004;338:1027–1036. doi: 10.1016/j.jmb.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 28.Kall L, Krogh A, Sonnhammer EL (2005) An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21[Suppl 1]:i251–i257 [DOI] [PubMed]
- 29.Viklund H, Elofsson A. OCTOPUS: improving topology prediction by two-track ANN-based preference scores and an extended topological grammar. Bioinformatics. 2008;24:1662–1668. doi: 10.1093/bioinformatics/btn221. [DOI] [PubMed] [Google Scholar]
- 30.Bernsel A, Viklund H, Hennerdal A, Elofsson A (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res 37[Suppl 2]:W465–W468. doi: 10.1093/nar/gkp363 [DOI] [PMC free article] [PubMed]
- 31.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 32.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III Signal recognition protein (SRP) causes signal sequence-dependent and site- specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajima S, Lauffer L, Rath VL, Walter P. The signal recognition particle receptor is a complex that contains two distinct polypeptide chains. J Cell Biol. 1986;103:1167–1178. doi: 10.1083/jcb.103.4.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valent QA, Scotti PA, High S, de Gier JW, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prinz A, Behrens C, Rapoport TA, Hartmann E, Kalies KU. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal RNA. EMBO J. 2000;19:1900–1906. doi: 10.1093/emboj/19.8.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradshaw N, Neher SB, Booth DS, Walter P. Signal sequences activate the catalytic switch of SRP RNA. Science. 2009;323:127–130. doi: 10.1126/science.1165971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ulbrandt ND, Newitt JA, Bernstein HD. The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 38.Luirink J, Sinning I. SRP-mediated protein targeting: structure and function revisited. Biochim Biophys Acta. 2004;1694:17–35. doi: 10.1016/j.bbamcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 39.Phillips GJ, Silhavy TJ. The E. coli ffh gene is necessary for viability and efficient protein export. Nature. 1992;359:744–746. doi: 10.1038/359744a0. [DOI] [PubMed] [Google Scholar]
- 40.Zanen G, Antelmann H, Meima R, Jongbloed JD, Kolkman M, Hecker M, van Dijl JM, Quax WJ. Proteomic dissection of potential signal recognition particle dependence in protein secretion by Bacillus subtilis. Proteomics. 2006;6:3636–3648. doi: 10.1002/pmic.200500560. [DOI] [PubMed] [Google Scholar]
- 41.Zanen G, Houben EN, Meima R, Tjalsma H, Jongbloed JD, Westers H, Oudega B, Luirink J, van Dijl JM, Quax WJ. Signal peptide hydrophobicity is critical for early stages in protein export by Bacillus subtilis. FEBS J. 2005;272:4617–4630. doi: 10.1111/j.1742-4658.2005.04777.x. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura K, Nishiguchi M, Honda K, Yamane K. The Bacillus subtilis SRP54 homologue, Ffh, has an intrinsic GTPase activity and forms a ribonucleoprotein complex with small cytoplasmic RNA in vivo. Biochem Biophys Res Commun. 1994;199:1394–1399. doi: 10.1006/bbrc.1994.1385. [DOI] [PubMed] [Google Scholar]
- 43.Oguro A, Kakeshita H, Honda K, Takamatsu H, Nakamura K, Yamane K. srb: a Bacillus subtilis gene encoding a homologue of the alpha-subunit of the mammalian signal recognition particle receptor. DNA Res. 1995;2:95–100. doi: 10.1093/dnares/2.2.95. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura K, Yahagi S, Yamazaki T, Yamane K. Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J Biol Chem. 1999;274:13569–13576. doi: 10.1074/jbc.274.19.13569. [DOI] [PubMed] [Google Scholar]
- 45.Struck JC, Vogel DW, Ulbrich N, Erdmann VA. The Bacillus subtilis scRNA is related to the 4.5S RNA from Escherichia coli. Nucleic Acids Res. 1988;16:2719. doi: 10.1093/nar/16.6.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishiguchi M, Honda K, Amikura R, Nakamura K, Yamane K. Structural requirements of Bacillus subtilis small cytoplasmic RNA for cell growth, sporulation, and extracellular enzyme production. J Bacteriol. 1994;176:157–165. doi: 10.1128/jb.176.1.157-165.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, Bron S, Bunai K, Chapuis J, Christiansen LC, Danchin A, Debarbouille M, Dervyn E, Deuerling E, Devine K, Devine SK, Dreesen O, Errington J, Fillinger S, Foster SJ, Fujita Y, Galizzi A, Gardan R, Eschevins C, Fukushima T, Haga K, Harwood CR, Hecker M, Hosoya D, Hullo MF, Kakeshita H, Karamata D, Kasahara Y, Kawamura F, Koga K, Koski P, Kuwana R, Imamura D, Ishimaru M, Ishikawa S, Ishio I, Le Coq D, Masson A, Mauel C, Meima R, Mellado RP, Moir A, Moriya S, Nagakawa E, Nanamiya H, Nakai S, Nygaard P, Ogura M, Ohanan T, O’Reilly M, O’Rourke M, Pragai Z, Pooley HM, Rapoport G, Rawlins JP, Rivas LA, Rivolta C, Sadaie A, Sadaie Y, Sarvas M, Sato T, Saxild HH, Scanlan E, Schumann W, Seegers JF, Sekiguchi J, Sekowska A, Seror SJ, Simon M, Stragier P, Studer R, Takamatsu H, Tanaka T, Takeuchi M, Thomaides HB, Vagner V, van Dijl JM, Watabe K, Wipat A, Yamamoto H, Yamamoto M, Yamamoto Y, Yamane K, Yata K, Yoshida K, Yoshikawa H, Zuber U, Ogasawara N. Essential Bacillus subtilis genes. Proc Natl Acad Sci USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakamura K, Imai Y, Nakamura A, Yamane K. Small cytoplasmic RNA of Bacillus subtilis: functional relationship with human signal recognition particle 7S RNA and Escherichia coli 4.5S RNA. J Bacteriol. 1992;174:2185–2192. doi: 10.1128/jb.174.7.2185-2192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, King CKGP, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu ZY, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol. 2002;43:1387–1400. doi: 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 50.Thanassi JA, Hartman-Neumann SL, Dougherty TJ, Dougherty BA, Pucci MJ. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 2002;30:3152–3162. doi: 10.1093/nar/gkf418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carpenter PB, Hanlon DW, Ordal GW. flhF, a Bacillus subtilis flagellar gene that encodes a putative GTP-binding protein. Mol Microbiol. 1992;6:2705–2713. doi: 10.1111/j.1365-2958.1992.tb01447.x. [DOI] [PubMed] [Google Scholar]
- 52.Salvetti S, Ghelardi E, Celandroni F, Ceragioli M, Giannessi F, Senesi S. FlhF, a signal recognition particle-like GTPase, is involved in the regulation of flagellar arrangement, motility behaviour and protein secretion in Bacillus cereus. Microbiology. 2007;153:2541–2552. doi: 10.1099/mic.0.2006/005553-0. [DOI] [PubMed] [Google Scholar]
- 53.Bange G, Petzold G, Wild K, Parlitz RO, Sinning I. The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound. GTP Proc Natl Acad Sci USA. 2007;104:13621–13625. doi: 10.1073/pnas.0702570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zanen G, Antelmann H, Westers H, Hecker M, van Dijl JM, Quax WJ. FlhF, the third signal recognition particle-GTPase of Bacillus subtilis, is dispensable for protein secretion. J Bacteriol. 2004;186:5956–5960. doi: 10.1128/JB.186.17.5956-5960.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crowley PJ, Svensater G, Snoep JL, Bleiweis AS, Brady LJ. An ffh mutant of Streptococcus mutans is viable and able to physiologically adapt to low pH in continuous culture. FEMS Microbiol Lett. 2004;234:315–324. doi: 10.1016/j.femsle.2004.03.043. [DOI] [PubMed] [Google Scholar]
- 56.Hasona A, Crowley PJ, Levesque CM, Mair RW, Cvitkovitch DG, Bleiweis AS, Brady LJ. Streptococcal viability and diminished stress tolerance in mutants lacking the signal recognition particle pathway or YidC2. Proc Natl Acad Sci USA. 2005;102:17466–17471. doi: 10.1073/pnas.0508778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kremer BH, van der Kraan M, Crowley PJ, Hamilton IR, Brady LJ, Bleiweis AS. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J Bacteriol. 2001;183:2543–2552. doi: 10.1128/JB.183.8.2543-2552.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasona A, Zuobi-Hasona K, Crowley PJ, Abranches J, Ruelf MA, Bleiweis AS, Brady LJ. Membrane composition changes and physiological adaptation by Streptococcus mutans signal recognition particle pathway mutants. J Bacteriol. 2007;189:1219–1230. doi: 10.1128/JB.01146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose RW, Bruser T, Kissinger JC, Pohlschroder M. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol. 2002;45:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 60.Zwieb C, Eichler J. Getting on target: the archaeal signal recognition particle. Archaea. 2002;1:27–34. doi: 10.1155/2002/729649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaine BP. Structure of the archaebacterial 7S RNA molecule. Mol Gen Genet. 1990;221:315–321. doi: 10.1007/BF00259394. [DOI] [PubMed] [Google Scholar]
- 62.Hainzl T, Huang S, Sauer-Eriksson AE. Interaction of signal-recognition particle 54 GTPase domain and signal-recognition particle RNA in the free signal-recognition particle. Proc Natl Acad Sci USA. 2007;104:14911–14916. doi: 10.1073/pnas.0702467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Model for signal sequence recognition from amino-acid sequence of 54 K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 64.Romisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54 K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 65.Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 66.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhuiyan SH, Gowda K, Hotokezaka H, Zwieb C. Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res. 2000;28:1365–1373. doi: 10.1093/nar/28.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Diener JL, Wilson C. Role of SRP19 in assembly of the Archaeoglobus fulgidus signal recognition particle. Biochemistry. 2000;39:12862–12874. doi: 10.1021/bi001180s. [DOI] [PubMed] [Google Scholar]
- 69.Tozik I, Huang Q, Zwieb C, Eichler J. Reconstitution of the signal recognition particle of the halophilic archaeon Haloferax volcanii. Nucleic Acids Res. 2002;30:4166–4175. doi: 10.1093/nar/gkf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hainzl T, Huang S, Sauer-Eriksson AE. Structural insights into SRP RNA: an induced fit mechanism for SRP assembly. RNA. 2005;11:1043–1050. doi: 10.1261/rna.2080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moll R, Schmidtke S, Schafer G. Domain structure, GTP-hydrolyzing activity and 7S RNA binding of Acidianus ambivalens ffh-homologous protein suggest an SRP-like complex in archaea. Eur J Biochem. 1999;259:441–448. doi: 10.1046/j.1432-1327.1999.00065.x. [DOI] [PubMed] [Google Scholar]
- 72.Yurist S, Dahan I, Eichler J. SRP19 is a dispensable component of the signal recognition particle in Archaea. J Bacteriol. 2007;189:276–279. doi: 10.1128/JB.01410-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Egea PF, Napetschnig J, Walter P, Stroud RM (2008) Structures of SRP54 and SRP19, the two proteins that organize the ribonucleic core of the signal recognition particle from Pyrococcus furiosus. PLoS ONE 3:e3528 [DOI] [PMC free article] [PubMed]
- 74.Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- 75.Lutcke H. Signal recognition particle (SRP) a ubiquitous initiator of protein translocation. Eur J Biochem. 1995;228:531–550. doi: 10.1111/j.1432-1033.1995.tb20293.x. [DOI] [PubMed] [Google Scholar]
- 76.Stroud RM, Walter P. Signal sequence recognition and protein targeting. Curr Opin Struct Biol. 1999;9:754–759. doi: 10.1016/s0959-440x(99)00040-8. [DOI] [PubMed] [Google Scholar]
- 77.Chang DY, Newitt JA, Hsu K, Bernstein HD, Maraia RJ. A highly conserved nucleotide in the Alu domain of SRP RNA mediates translation arrest through high affinity binding to SRP9/14. Nucleic Acids Res. 1997;25:1117–1122. doi: 10.1093/nar/25.6.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zwieb C, Samuelsson T. SRPDB (signal recognition particle database) Nucleic Acids Res. 2000;28:171–172. doi: 10.1093/nar/28.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de Leeuw E, Poland D, Mol O, Sinning I, ten Hagen-Jongman CM, Oudega B, Luirink J. Membrane association of FtsY, the E. coli SRP receptor. FEBS Lett. 1997;416:225–229. doi: 10.1016/s0014-5793(97)01238-6. [DOI] [PubMed] [Google Scholar]
- 80.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zelazny A, Seluanov A, Cooper A, Bibi E. The NG domain of the prokaryotic signal recognition particle receptor, FtsY, is fully functional when fused to an unrelated integral membrane polypeptide. Proc Natl Acad Sci USA. 1997;94:6025–6029. doi: 10.1073/pnas.94.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Egea PF, Tsuruta H, de Leon GP, Napetschnig J, Walter P, Stroud RM (2008) Structures of the signal recognition particle receptor from the archaeon Pyrococcus furiosus: implications for the targeting step at the membrane. PLoS ONE 3:e3619 [DOI] [PMC free article] [PubMed]
- 83.Gawronski-Salerno J, Coon JST, Focia PJ, Freymann DM. X-ray structure of the T. aquaticus FtsY:GDP complex suggests functional roles for the C-terminal helix of the SRP GTPases. Proteins. 2007;66:984–995. doi: 10.1002/prot.21200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moll R, Schmidtke S, Schaefer G. A putative signal recognition particle receptor alpha subunit (SR alpha) homologue is expressed in the hyperthermophilic crenarchaeon Sulfolobus acidocaldarius. FEMS Microbiol Lett. 1996;137:51–56. doi: 10.1111/j.1574-6968.1996.tb08081.x. [DOI] [PubMed] [Google Scholar]
- 85.Moll R, Schmidtke S, Petersen A, Schafer G. The signal recognition particle receptor alpha subunit of the hyperthermophilic archaeon Acidianus ambivalens exhibits an intrinsic GTP-hydrolyzing activity. Biochim Biophys Acta. 1997;1335:218–230. doi: 10.1016/s0304-4165(96)00141-9. [DOI] [PubMed] [Google Scholar]
- 86.Eichler J. Archaeal protein translocation crossing membranes in the third domain of life. Eur J Biochem. 2000;267:3402–3412. doi: 10.1046/j.1432-1327.2000.01396.x. [DOI] [PubMed] [Google Scholar]
- 87.Rapoport TA. Protein translocation across the eukaryotic endoplasmic reticulum and bacterial plasma membranes. Nature. 2007;450:663–669. doi: 10.1038/nature06384. [DOI] [PubMed] [Google Scholar]
- 88.Vrontou E, Economou A. Structure and function of SecA, the preprotein translocase nanomotor. Biochim Biophys Acta. 2004;1694:67–80. doi: 10.1016/j.bbamcr.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 89.Deitermann S, Sprie GS, Koch HG. A dual function for SecA in the assembly of single spanning membrane proteins in Escherichia coli. J Biol Chem. 2005;280:39077–39085. doi: 10.1074/jbc.M509647200. [DOI] [PubMed] [Google Scholar]
- 90.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 91.Schiebel E, Driessen AJ, Hartl FU, Wickner W. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 92.van der Wolk JP, de Wit JG, Driessen AJ. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pogliano JA, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chen M, Xie K, Yuan J, Yi L, Facey SJ, Pradel N, Wu LF, Kuhn A, Dalbey RE. Involvement of SecDF and YidC in the membrane insertion of M13 procoat mutants. Biochemistry. 2005;44:10741–10749. doi: 10.1021/bi047418k. [DOI] [PubMed] [Google Scholar]
- 95.Kiefer D, Kuhn A. YidC as an essential and multifunctional component in membrane protein assembly. Int Rev Cytol. 2007;259:113–138. doi: 10.1016/S0074-7696(06)59003-5. [DOI] [PubMed] [Google Scholar]
- 96.Van Der Laan M, Urbanus ML, Ten Hagen-Jongman CM, Nouwen N, Oudega B, Harms N, Driessen AJ, Luirink J. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc Natl Acad Sci USA. 2003;100:5801–5806. doi: 10.1073/pnas.0636761100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Price CE, Driessen AJ. YidC is involved in the biogenesis of anaerobic respiratory complexes in the inner membrane of Escherichia coli. J Biol Chem. 2008;283:26921–26927. doi: 10.1074/jbc.M804490200. [DOI] [PubMed] [Google Scholar]
- 98.Samuelson JC, Chen M, Jiang F, Moller I, Wiedmann M, Kuhn A, Phillips GJ, Dalbey RE. YidC mediates membrane protein insertion in bacteria. Nature. 2000;406:637–641. doi: 10.1038/35020586. [DOI] [PubMed] [Google Scholar]
- 99.Jungnickel B, Rapoport TA, Hartmann E. Protein translocation: common themes from bacteria to man. FEBS Lett. 1994;346:73–77. doi: 10.1016/0014-5793(94)00367-x. [DOI] [PubMed] [Google Scholar]
- 100.Lyman SK, Schekman R. Binding of secretory precursor polypeptides to a translocon subcomplex is regulated by BiP. Cell. 1997;88:85–96. doi: 10.1016/s0092-8674(00)81861-9. [DOI] [PubMed] [Google Scholar]
- 101.Sanders SL, Whitfield KM, Vogel JP, Rose MD, Schekman RW. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell. 1992;69:353–365. doi: 10.1016/0092-8674(92)90415-9. [DOI] [PubMed] [Google Scholar]
- 102.von Heijne G, Abrahmsen L. Species-specific variation in signal peptide design Implications for protein secretion in foreign hosts. FEBS Lett. 1989;244:439–446. doi: 10.1016/0014-5793(89)80579-4. [DOI] [PubMed] [Google Scholar]
- 103.van Wely KH, Swaving J, Broekhuizen CP, Rose M, Quax WJ, Driessen AJ. Functional identification of the product of the Bacillus subtilis yvaL gene as a SecG homologue. J Bacteriol. 1999;181:1786–1792. doi: 10.1128/jb.181.6.1786-1792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schatz PJ, Bieker KL, Ottemann KM, Silhavy TJ, Beckwith J. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 1991;10:1749–1757. doi: 10.1002/j.1460-2075.1991.tb07699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Swaving J, van Wely KH, Driessen AJ. Preprotein translocation by a hybrid translocase composed of Escherichia coli and Bacillus subtilis subunits. J Bacteriol. 1999;181:7021–7027. doi: 10.1128/jb.181.22.7021-7027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rigel NW, Braunstein M. A new twist on an old pathway—accessory Sec [corrected] systems. Mol Microbiol. 2008;69:291–302. doi: 10.1111/j.1365-2958.2008.06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sibbald MJ, Ziebandt AK, Engelmann S, Hecker M, de Jong A, Harmsen HJ, Raangs GC, Stokroos I, Arends JP, Dubois JY, van Dijl JM. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol Mol Biol Rev. 2006;70:755–788. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Caspers M, Freudl R. Corynebacterium glutamicum possesses two secA homologous genes that are essential for viability. Arch Microbiol. 2008;189:605–610. doi: 10.1007/s00203-008-0351-0. [DOI] [PubMed] [Google Scholar]
- 109.Bensing BA, Sullam PM. An accessory sec locus of Streptococcus gordonii is required for export of the surface protein GspB and for normal levels of binding to human platelets. Mol Microbiol. 2002;44:1081–1094. doi: 10.1046/j.1365-2958.2002.02949.x. [DOI] [PubMed] [Google Scholar]
- 110.Wu H, Bu S, Newell P, Chen Q, Fives-Taylor P. Two gene determinants are differentially involved in the biogenesis of Fap1 precursors in Streptococcus parasanguis. J Bacteriol. 2007;189:1390–1398. doi: 10.1128/JB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takamatsu D, Bensing BA, Sullam PM. Genes in the accessory sec locus of Streptococcus gordonii have three functionally distinct effects on the expression of the platelet-binding protein GspB. Mol Microbiol. 2004;52:189–203. doi: 10.1111/j.1365-2958.2004.03978.x. [DOI] [PubMed] [Google Scholar]
- 112.Bensing BA, Takamatsu D, Sullam PM. Determinants of the streptococcal surface glycoprotein GspB that facilitate export by the accessory Sec system. Mol Microbiol. 2005;58:1468–1481. doi: 10.1111/j.1365-2958.2005.04919.x. [DOI] [PubMed] [Google Scholar]
- 113.Chen Q, Wu H, Fives-Taylor PM. Investigating the role of secA2 in secretion and glycosylation of a fimbrial adhesin in Streptococcus parasanguis FW213. Mol Microbiol. 2004;53:843–856. doi: 10.1111/j.1365-2958.2004.04116.x. [DOI] [PubMed] [Google Scholar]
- 114.Gibbons HS, Wolschendorf F, Abshire M, Niederweis M, Braunstein M. Identification of two Mycobacterium smegmatis lipoproteins exported by a SecA2-dependent pathway. J Bacteriol. 2007;189:5090–5100. doi: 10.1128/JB.00163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bensing BA, Siboo IR, Sullam PM. Glycine residues in the hydrophobic core of the GspB signal sequence route export toward the accessory Sec pathway. J Bacteriol. 2007;189:3846–3854. doi: 10.1128/JB.00027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bensing BA, Sullam PM. Characterization of Streptococcus gordonii SecA2 as a paralogue of SecA. J Bacteriol. 2009;191:3482–3491. doi: 10.1128/JB.00365-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Braunstein M, Espinosa BJ, Chan J, Belisle JT, Jacobs WR., Jr SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:453–464. doi: 10.1046/j.1365-2958.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- 118.Chen Q, Sun B, Wu H, Peng Z, Fives-Taylor PM. Differential roles of individual domains in selection of secretion route of a Streptococcus parasanguinis serine-rich adhesin, Fap1. J Bacteriol. 2007;189:7610–7617. doi: 10.1128/JB.00748-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bolhuis A, Broekhuizen CP, Sorokin A, van Roosmalen ML, Venema G, Bron S, Quax WJ, van Dijl JM. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J Biol Chem. 1998;273:21217–21224. doi: 10.1074/jbc.273.33.21217. [DOI] [PubMed] [Google Scholar]
- 120.Zweers JC, Barak I, Becher D, Driessen AJ, Hecker M, Kontinen VP, Saller MJ, Vavrova L, van Dijl JM. Towards the development of Bacillus subtilis as a cell factory for membrane proteins and protein complexes. Microb Cell Fact. 2008;7:10. doi: 10.1186/1475-2859-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bunai K, Nozaki M, Kakeshita H, Nemoto T, Yamane K. Quantitation of de novo localized (15)N-labeled lipoproteins and membrane proteins having one and two transmembrane segments in a Bacillus subtilis secA temperature-sensitive mutant using 2D-PAGE and MALDI-TOF MS. J Proteome Res. 2005;4:826–836. doi: 10.1021/pr049755l. [DOI] [PubMed] [Google Scholar]
- 122.Bunai K, Yamane K. Effectiveness and limitation of two-dimensional gel electrophoresis in bacterial membrane protein proteomics and perspectives. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:227–236. doi: 10.1016/j.jchromb.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 123.Dreisbach A, Otto A, Becher D, Hammer E, Teumer A, Gouw JW, Hecker M, Volker U. Monitoring of changes in the membrane proteome during stationary phase adaptation of Bacillus subtilis using in vivo labeling techniques. Proteomics. 2008;8:2062–2076. doi: 10.1002/pmic.200701081. [DOI] [PubMed] [Google Scholar]
- 124.Wolff S, Hahne H, Hecker M, Becher D. Complementary analysis of the vegetative membrane proteome of the human pathogen Staphylococcus aureus. Mol Cell Proteomics. 2008;7:1460–1468. doi: 10.1074/mcp.M700554-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hahne H, Wolff S, Hecker M, Becher D. From complementarity to comprehensiveness–targeting the membrane proteome of growing Bacillus subtilis by divergent approaches. Proteomics. 2008;8:4123–4136. doi: 10.1002/pmic.200800258. [DOI] [PubMed] [Google Scholar]
- 126.Campo N, Tjalsma H, Buist G, Stepniak D, Meijer M, Veenhuis M, Westermann M, Muller JP, Bron S, Kok J, Kuipers OP, Jongbloed JD. Subcellular sites for bacterial protein export. Mol Microbiol. 2004;53:1583–1599. doi: 10.1111/j.1365-2958.2004.04278.x. [DOI] [PubMed] [Google Scholar]
- 127.Rosch J, Caparon M. A microdomain for protein secretion in Gram-positive bacteria. Science. 2004;304:1513–1515. doi: 10.1126/science.1097404. [DOI] [PubMed] [Google Scholar]
- 128.Rosch JW, Caparon MG. The ExPortal: an organelle dedicated to the biogenesis of secreted proteins in Streptococcus pyogenes. Mol Microbiol. 2005;58:959–968. doi: 10.1111/j.1365-2958.2005.04887.x. [DOI] [PubMed] [Google Scholar]
- 129.Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 130.Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR, Jr, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA. 2003;100:2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA . Proc Natl Acad Sci USA. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Vassylyev DG, Mori H, Vassylyeva MN, Tsukazaki T, Kimura Y, Tahirov TH, Ito K. Crystal structure of the translocation ATPase SecA from Thermus thermophilus reveals a parallel, head-to-head dimer. J Mol Biol. 2006;364:248–258. doi: 10.1016/j.jmb.2006.09.061. [DOI] [PubMed] [Google Scholar]
- 133.Papanikolau Y, Papadovasilaki M, Ravelli RB, McCarthy AA, Cusack S, Economou A, Petratos K. Structure of dimeric SecA, the Escherichia coli preprotein translocase motor. J Mol Biol. 2007;366:1545–1557. doi: 10.1016/j.jmb.2006.12.049. [DOI] [PubMed] [Google Scholar]
- 134.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Papanikou E, Karamanou S, Economou A. Bacterial protein secretion through the translocase nanomachine. Nat Rev Microbiol. 2007;5:839–851. doi: 10.1038/nrmicro1771. [DOI] [PubMed] [Google Scholar]
- 136.Cooper DB, Smith VF, Crane JM, Roth HC, Lilly AA, Randall LL. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J Mol Biol. 2008;382:74–87. doi: 10.1016/j.jmb.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Erlandson KJ, Miller SB, Nam Y, Osborne AR, Zimmer J, Rapoport TA. A role for the two-helix finger of the SecA ATPase in protein translocation. Nature. 2008;455:984–987. doi: 10.1038/nature07439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Tsukazaki T, Mori H, Fukai S, Ishitani R, Mori T, Dohmae N, Perederina A, Sugita Y, Vassylyev DG, Ito K, Nureki O. Conformational transition of Sec machinery inferred from bacterial SecYE structures. Nature. 2008;455:988–991. doi: 10.1038/nature07421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Auer J, Spicker G, Bock A. Presence of a gene in the archaebacterium Methanococcus vannielii homologous to secY of eubacteria. Biochimie. 1991;73:683–688. doi: 10.1016/0300-9084(91)90048-6. [DOI] [PubMed] [Google Scholar]
- 140.Cao TB, Saier MH., Jr The general protein secretory pathway: phylogenetic analyses leading to evolutionary conclusions. Biochim Biophys Acta. 2003;1609:115–125. doi: 10.1016/s0005-2736(02)00662-4. [DOI] [PubMed] [Google Scholar]
- 141.Irihimovitch V, Eichler J. Post-translational secretion of fusion proteins in the halophilic archaea Haloferax volcanii. J Biol Chem. 2003;278:12881–12887. doi: 10.1074/jbc.M210762200. [DOI] [PubMed] [Google Scholar]
- 142.Kinch LN, Saier MH, Jr, Grishin NV. Sec61beta—a component of the archaeal protein secretory system. Trends Biochem Sci. 2002;27:170–171. doi: 10.1016/s0968-0004(01)02055-2. [DOI] [PubMed] [Google Scholar]
- 143.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 144.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast. ER Membrane Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 145.Pohlschroder M, Hartmann E, Hand NJ, Dilks K, Haddad A. Diversity and evolution of protein translocation. Annu Rev Microbiol. 2005;59:91–111. doi: 10.1146/annurev.micro.59.030804.121353. [DOI] [PubMed] [Google Scholar]
- 146.Eichler J. Evolution of the prokaryotic protein translocation complex: a comparison of archaeal and bacterial versions of SecDF. Mol Phylogenet Evol. 2003;27:504–509. doi: 10.1016/s1055-7903(03)00015-0. [DOI] [PubMed] [Google Scholar]
- 147.Duong F, Wickner W. The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J. 1997;16:4871–4879. doi: 10.1093/emboj/16.16.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Economou A, Pogliano JA, Beckwith J, Oliver DB, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 149.Nouwen N, Piwowarek M, Berrelkamp G, Driessen AJ. The large first periplasmic loop of SecD and SecF plays an important role in SecDF functioning. J Bacteriol. 2005;187:5857–5860. doi: 10.1128/JB.187.16.5857-5860.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Pohlschroder M, Prinz WA, Hartmann E, Beckwith J. Protein translocation in the three domains of life: variations on a theme. Cell. 1997;91:563–566. doi: 10.1016/s0092-8674(00)80443-2. [DOI] [PubMed] [Google Scholar]
- 151.Yi L, Dalbey RE. Oxa1/Alb3/YidC system for insertion of membrane proteins in mitochondria, chloroplasts and bacteria (review) Mol Membr Biol. 2005;22:101–111. doi: 10.1080/09687860500041718. [DOI] [PubMed] [Google Scholar]
- 152.Henry R, Goforth RL, Schunemann (2007) Chloroplast SRP/FtsY and Alb3 in protein integration into the thylakoid membrane. in the enzymes, vol 25, 3rd edn. In: Dalbey RE, Koehler CM Tamanoi F (eds) Molecular machines involved in protein transport across cellular membranes. Academic Press, San Diego, pp 493–521
- 153.Serek J, Bauer-Manz G, Struhalla G, Van Den Berg L, Kiefer D, Dalbey R, Kuhn A. Escherichia coli YidC is a membrane insertase for Sec-independent proteins. EMBO J. 2004;23:294–301. doi: 10.1038/sj.emboj.7600063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Samuelson JC, Jiang F, Yi L, Chen M, Gier JW, Kuhn A, Dalbey RE. Function of YidC for the insertion of M13 procoat protein in E. coli: translocation of mutants that show differences in their membrane potential dependence and Sec- requirement. J Biol Chem. 2001;16:16. doi: 10.1074/jbc.M105793200. [DOI] [PubMed] [Google Scholar]
- 156.Nouwen N, Driessen AJ. SecDFyajC forms a heterotetrameric complex with YidC. Mol Microbiol. 2002;44:1397–1405. doi: 10.1046/j.1365-2958.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 157.Xie K, Kiefer D, Nagler G, Dalbey RE, Kuhn A. Different regions of the nonconserved large periplasmic domain of Escherichia coli YidC are involved in the SecF interaction and membrane insertase activity. Biochemistry. 2006;45:13401–13408. doi: 10.1021/bi060826z. [DOI] [PubMed] [Google Scholar]
- 158.Xie K, Dalbey RE. Inserting proteins into the bacterial cytoplasmic membrane using the Sec and YidC translocases. Nat Rev Microbiol. 2008;6:234–244. doi: 10.1038/nrmicro3595. [DOI] [PubMed] [Google Scholar]
- 159.Kol S, Nouwen N, Driessen AJ (2008) Mechanisms of YidC-mediated insertion and assembly of multimeric membrane protein complexes. J Biol Chem 3505–3525 [DOI] [PubMed]
- 160.van der Laan M, Nouwen NP, Driessen AJ. YidC–an evolutionary conserved device for the assembly of energy-transducing membrane protein complexes. Curr Opin Microbiol. 2005;8:182–187. doi: 10.1016/j.mib.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 161.Saaf A, Monne M, de Gier JW, von Heijne G. Membrane topology of the 60-kDa Oxa1p homologue from Escherichia coli. J Biol Chem. 1998;273:30415–30418. doi: 10.1074/jbc.273.46.30415. [DOI] [PubMed] [Google Scholar]
- 162.Oliver DC, Paetzel M. Crystal structure of the major periplasmic domain of the bacterial membrane protein assembly facilitator YidC. J Biol Chem. 2008;283:5208–5216. doi: 10.1074/jbc.M708936200. [DOI] [PubMed] [Google Scholar]
- 163.Ravaud S, Stjepanovic G, Wild K, Sinning I. The crystal structure of the periplasmic domain of the Escherichia coli membrane protein insertase YidC contains a substrate binding cleft. J Biol Chem. 2008;283:9350–9358. doi: 10.1074/jbc.M710493200. [DOI] [PubMed] [Google Scholar]
- 164.Yu Z, Koningstein G, Pop A, Luirink J. The conserved third transmembrane segment of YidC contacts nascent Escherichia coli inner membrane proteins. J Biol Chem. 2008;283:34635–34642. doi: 10.1074/jbc.M804344200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Klenner C, Yuan J, Dalbey RE, Kuhn A. The Pf3 coat protein contacts TM1 and TM3 of YidC during membrane biogenesis. FEBS Lett. 2008;582:3967–3972. doi: 10.1016/j.febslet.2008.10.044. [DOI] [PubMed] [Google Scholar]
- 166.Winterfeld S, Imhof N, Roos T, Bar G, Kuhn A, Gerken U. Substrate-induced conformational change of the Escherichia coli membrane insertase YidC. Biochemistry. 2009;48:6684–6691. doi: 10.1021/bi9003809. [DOI] [PubMed] [Google Scholar]
- 167.van der Laan M, Houben EN, Nouwen N, Luirink J, Driessen AJ. Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Rep. 2001;2:519–523. doi: 10.1093/embo-reports/kve106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nargang FE, Preuss M, Neupert W, Herrmann JM. The Oxa1 protein forms a homooligomeric complex and is an essential part of the mitochondrial export translocase in Neurospora crassa. J Biol Chem. 2002;277:12846–12853. doi: 10.1074/jbc.M112099200. [DOI] [PubMed] [Google Scholar]
- 169.Kohler R, Boehringer D, Greber B, Bingel-Erlenmeyer R, Collinson I, Schaffitzel C, Ban N. YidC and Oxa1 form dimeric insertion pores on the translating ribosome. Mol Cell. 2009;34:344–353. doi: 10.1016/j.molcel.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 170.Jiang F, Chen M, Yi L, de Gier JW, Kuhn A, Dalbey RE. Defining the regions of Escherichia coli YidC that contribute to activity. J Biol Chem. 2003;278:48965–48972. doi: 10.1074/jbc.M307362200. [DOI] [PubMed] [Google Scholar]
- 171.Yuan J, Phillips GJ, Dalbey RE. Isolation of cold-sensitive yidC mutants provides insights into the substrate profile of the YidC insertase and the importance of transmembrane 3 in YidC function. J Bacteriol. 2007;189:8961–8972. doi: 10.1128/JB.01365-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yen MR, Harley KT, Tseng YH, Saier MH., Jr Phylogenetic and structural analyses of the oxa1 family of protein translocases. FEMS Microbiol Lett. 2001;204:223–231. doi: 10.1111/j.1574-6968.2001.tb10889.x. [DOI] [PubMed] [Google Scholar]
- 173.Tjalsma H, Bron S, van Dijl JM. Complementary impact of paralogous Oxa1-like proteins of Bacillus subtilis on post-translocational stages in protein secretion. J Biol Chem. 2003;278:15622–15632. doi: 10.1074/jbc.M301205200. [DOI] [PubMed] [Google Scholar]
- 174.Murakami T, Haga K, Takeuchi M, Sato T. Analysis of the Bacillus subtilis spoIIIJ gene and its paralogue gene, yqjG. J Bacteriol. 2002;184:1998–2004. doi: 10.1128/JB.184.7.1998-2004.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008;69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Serrano M, Vieira F, Moran CP, Jr, Henriques AO. Processing of a membrane protein required for cell-to-cell signaling during endospore formation in Bacillus subtilis. J Bacteriol. 2008;190:7786–7796. doi: 10.1128/JB.00715-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dong Y, Palmer SR, Hasona A, Nagamori S, Kaback HR, Dalbey RE, Brady LJ. Functional overlap but lack of complete cross-complementation of Streptococcus mutans and Escherichia coli YidC orthologs. J Bacteriol. 2008;190:2458–2469. doi: 10.1128/JB.01366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Funes S, Hasona A, Bauerschmitt H, Grubbauer C, Kauff F, Collins R, Crowley PJ, Palmer SR, Brady LJ, Herrmann JM. Independent gene duplications of the YidC/Oxa/Alb3 family enabled a specialized cotranslational function. Proc Natl Acad Sci USA. 2009;106:6656–6661. doi: 10.1073/pnas.0809951106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Natale P, Bruser T, Driessen AJ. Sec- and Tat-mediated protein secretion across the bacterial cytoplasmic membrane–distinct translocases and mechanisms. Biochim Biophys Acta. 2008;1778:1735–1756. doi: 10.1016/j.bbamem.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 180.Palmer T, Sargent F, Berks BC. Export of complex cofactor-containing proteins by the bacterial Tat pathway. Trends Microbiol. 2005;13:175–180. doi: 10.1016/j.tim.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 181.Robinson C, Bolhuis A. Tat-dependent protein targeting in prokaryotes and chloroplasts. Biochim Biophys Acta. 2004;1694:135–147. doi: 10.1016/j.bbamcr.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 182.Sargent F. The twin-arginine transport system: moving folded proteins across membranes. Biochem Soc Trans. 2007;35:835–847. doi: 10.1042/BST0350835. [DOI] [PubMed] [Google Scholar]
- 183.Hatzixanthis K, Palmer T, Sargent F. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol. 2003;49:1377–1390. doi: 10.1046/j.1365-2958.2003.03642.x. [DOI] [PubMed] [Google Scholar]
- 184.Dilks K, Gimenez MI, Pohlschroder M. Genetic and biochemical analysis of the twin-arginine translocation pathway in halophilic archaea. J Bacteriol. 2005;187:8104–8113. doi: 10.1128/JB.187.23.8104-8113.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Widdick DA, Dilks K, Chandra G, Bottrill A, Naldrett M, Pohlschroder M, Palmer T. The twin-arginine translocation pathway is a major route of protein export in Streptomyces coelicolor. Proc Natl Acad Sci USA. 2006;103:17927–17932. doi: 10.1073/pnas.0607025103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Widdick DA, Eijlander RT, van Dijl JM, Kuipers OP, Palmer T. A facile reporter system for the experimental identification of twin-arginine translocation (Tat) signal peptides from all kingdoms of life. J Mol Biol. 2008;375:595–603. doi: 10.1016/j.jmb.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 187.Saint-Joanis B, Demangel C, Jackson M, Brodin P, Marsollier L, Boshoff H, Cole ST. Inactivation of Rv2525c, a substrate of the twin arginine translocation (Tat) system of Mycobacterium tuberculosis, increases beta-lactam susceptibility and virulence. J Bacteriol. 2006;188:6669–6679. doi: 10.1128/JB.00631-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Posey JE, Shinnick TM, Quinn FD. Characterization of the twin-arginine translocase secretion system of Mycobacterium smegmatis. J Bacteriol. 2006;188:1332–1340. doi: 10.1128/JB.188.4.1332-1340.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.McDonough JA, Hacker KE, Flores AR, Pavelka MS, Jr, Braunstein M. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J Bacteriol. 2005;187:7667–7679. doi: 10.1128/JB.187.22.7667-7679.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Bronstein PA, Marrichi M, Cartinhour S, Schneider DJ, DeLisa MP. Identification of a twin-arginine translocation system in Pseudomonas syringae pv. tomato DC3000 and its contribution to pathogenicity and fitness. J Bacteriol. 2005;187:8450–8461. doi: 10.1128/JB.187.24.8450-8461.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Caldelari I, Mann S, Crooks C, Palmer T. The Tat pathway of the plant pathogen Pseudomonas syringae is required for optimal virulence. Mol Plant Microbe Interact. 2006;19:200–212. doi: 10.1094/MPMI-19-0200. [DOI] [PubMed] [Google Scholar]
- 192.Lavander M, Ericsson SK, Broms JE, Forsberg A. The twin arginine translocation system is essential for virulence of Yersinia pseudotuberculosis. Infect Immun. 2006;74:1768–1776. doi: 10.1128/IAI.74.3.1768-1776.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.De Buck E, Maes L, Meyen E, Van Mellaert L, Geukens N, Anne J, Lammertyn E. Legionella pneumophila Philadelphia-1 tatB and tatC affect intracellular replication and biofilm formation. Biochem Biophys Res Commun. 2005;331:1413–1420. doi: 10.1016/j.bbrc.2005.04.060. [DOI] [PubMed] [Google Scholar]
- 194.Rossier O, Cianciotto NP. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect Immun. 2005;73:2020–2032. doi: 10.1128/IAI.73.4.2020-2032.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.De Buck E, Lammertyn E, Anne J. The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol. 2008;16:442–453. doi: 10.1016/j.tim.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 196.Cristobal S, de Gier JW, Nielsen H, von Heijne G. Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Tullman-Ercek D, DeLisa MP, Kawarasaki Y, Iranpour P, Ribnicky B, Palmer T, Georgiou G. Export pathway selectivity of Escherichia coli twin arginine translocation signal peptides. J Biol Chem. 2007;282:8309–8316. doi: 10.1074/jbc.M610507200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Blaudeck N, Kreutzenbeck P, Freudl R, Sprenger GA. Genetic analysis of pathway specificity during posttranslational protein translocation across the Escherichia coli plasma membrane. J Bacteriol. 2003;185:2811–2819. doi: 10.1128/JB.185.9.2811-2819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bogsch E, Brink S, Robinson C. Pathway specificity for a delta pH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J. 1997;16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Cristobal S, Scotti P, Luirink J, von Heijne G, de Gier JW. The signal recognition particle-targeting pathway does not necessarily deliver proteins to the sec-translocase in Escherichia coli. J Biol Chem. 1999;274:20068–20070. doi: 10.1074/jbc.274.29.20068. [DOI] [PubMed] [Google Scholar]
- 201.Ize B, Gerard F, Wu LF. In vivo assessment of the Tat signal peptide specificity in Escherichia coli. Arch Microbiol. 2002;178:548–553. doi: 10.1007/s00203-002-0488-1. [DOI] [PubMed] [Google Scholar]
- 202.Wu LF, Ize B, Chanal A, Quentin Y, Fichant G. Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J Mol Microbiol Biotechnol. 2000;2:179–189. [PubMed] [Google Scholar]
- 203.Behrendt J, Standar K, Lindenstrauss U, Bruser T. Topological studies on the twin-arginine translocase component TatC. FEMS Microbiol Lett. 2004;234:303–308. doi: 10.1016/j.femsle.2004.03.048. [DOI] [PubMed] [Google Scholar]
- 204.Ki JJ, Kawarasaki Y, Gam J, Harvey BR, Iverson BL, Georgiou G. A periplasmic fluorescent reporter protein and its application in high-throughput membrane protein topology analysis. J Mol Biol. 2004;341:901–909. doi: 10.1016/j.jmb.2004.05.078. [DOI] [PubMed] [Google Scholar]
- 205.Sargent F, Gohlke U, De Leeuw E, Stanley NR, Palmer T, Saibil HR, Berks BC. Purified components of the Escherichia coli Tat protein transport system form a double-layered ring structure. Eur J Biochem. 2001;268:3361–3367. doi: 10.1046/j.1432-1327.2001.02263.x. [DOI] [PubMed] [Google Scholar]
- 206.Gohlke U, Pullan L, McDevitt CA, Porcelli I, de Leeuw E, Palmer T, Saibil HR, Berks BC. The TatA component of the twin-arginine protein transport system forms channel complexes of variable diameter. Proc Natl Acad Sci USA. 2005;102:10482–10486. doi: 10.1073/pnas.0503558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Oates J, Barrett CM, Barnett JP, Byrne KG, Bolhuis A, Robinson C. The Escherichia coli twin-arginine translocation apparatus incorporates a distinct form of TatABC complex, spectrum of modular TatA complexes and minor TatAB complex. J Mol Biol. 2005;346:295–305. doi: 10.1016/j.jmb.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 208.Alami M, Luke I, Deitermann S, Eisner G, Koch HG, Brunner J, Muller M. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell. 2003;12:937–946. doi: 10.1016/s1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 209.Bolhuis A, Mathers JE, Thomas JD, Barrett CM, Robinson C. TatB and TatC form a functional and structural unit of the twin-arginine translocase from Escherichia coli. J Biol Chem. 2001;276:20213–20219. doi: 10.1074/jbc.M100682200. [DOI] [PubMed] [Google Scholar]
- 210.Holzapfel E, Eisner G, Alami M, Barrett CM, Buchanan G, Luke I, Betton JM, Robinson C, Palmer T, Moser M, Muller M. The entire N-terminal half of TatC is involved in twin-arginine precursor binding. Biochemistry. 2007;46:2892–2898. doi: 10.1021/bi062205b. [DOI] [PubMed] [Google Scholar]
- 211.Blaudeck N, Kreutzenbeck P, Muller M, Sprenger GA, Freudl R. Isolation and characterization of bifunctional Escherichia coli TatA mutant proteins that allow efficient tat-dependent protein translocation in the absence of TatB. J Biol Chem. 2005;280:3426–3432. doi: 10.1074/jbc.M411210200. [DOI] [PubMed] [Google Scholar]
- 212.Barnett JP, Eijlander RT, Kuipers OP, Robinson C. A minimal Tat system from a gram-positive organism: a bifunctional TatA subunit participates in discrete TatAC and TatA complexes. J Biol Chem. 2008;283:2534–2542. doi: 10.1074/jbc.M708134200. [DOI] [PubMed] [Google Scholar]
- 213.Jongbloed JD, van der Ploeg R, van Dijl JM. Bifunctional TatA subunits in minimal Tat protein translocases. Trends Microbiol. 2006;14:2–4. doi: 10.1016/j.tim.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 214.Jongbloed JD, Grieger U, Antelmann H, Hecker M, Nijland R, Bron S, van Dijl JM. Two minimal Tat translocases in Bacillus. Mol Microbiol. 2004;54:1319–1325. doi: 10.1111/j.1365-2958.2004.04341.x. [DOI] [PubMed] [Google Scholar]
- 215.Jongbloed JD, Antelmann H, Hecker M, Nijland R, Bron S, Airaksinen U, Pries F, Quax WJ, van Dijl JM, Braun PG. Selective contribution of the twin-arginine translocation pathway to protein secretion in Bacillus subtilis. J Biol Chem. 2002;277:44068–44078. doi: 10.1074/jbc.M203191200. [DOI] [PubMed] [Google Scholar]
- 216.Kouwen TR, van der Ploeg R, Antelmann H, Hecker M, Homuth G, Mader U, van Dijl JM. Overflow of a hyper-produced secretory protein from the Bacillus Sec pathway into the Tat pathway for protein secretion as revealed by proteogenomics. Proteomics. 2009;9:1018–1032. doi: 10.1002/pmic.200800580. [DOI] [PubMed] [Google Scholar]
- 217.Sargent F, Stanley NR, Berks BC, Palmer T. Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J Biol Chem. 1999;274:36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 218.Bolhuis A. Protein transport in the halophilic archaeon Halobacterium sp. NRC-1: a major role for the twin-arginine translocation pathway? Microbiology. 2002;148:3335–3346. doi: 10.1099/00221287-148-11-3335. [DOI] [PubMed] [Google Scholar]
- 219.Thomas JR, Bolhuis A. The tatC gene cluster is essential for viability in halophilic archaea. FEMS Microbiol Lett. 2006;256:44–49. doi: 10.1111/j.1574-6968.2006.00107.x. [DOI] [PubMed] [Google Scholar]
- 220.Gimenez MI, Dilks K, Pohlschroder M. Haloferax volcanii twin-arginine translocation substates include secreted soluble, C-terminally anchored and lipoproteins. Mol Microbiol. 2007;66:1597–1606. doi: 10.1111/j.1365-2958.2007.06034.x. [DOI] [PubMed] [Google Scholar]
- 221.Mould RM, Robinson C. A proton gradient is required for the transport of two lumenal oxygen-evolving proteins across the thylakoid membrane. J Biol Chem. 1991;266:12189–12193. [PubMed] [Google Scholar]
- 222.Braun NA, Davis AW, Theg SM. The chloroplast Tat pathway utilizes the transmembrane electric potential as an energy source. Biophys J. 2007;93:1993–1998. doi: 10.1529/biophysj.106.098731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Kwan DC, Thomas JR, Bolhuis A. Bioenergetic requirements of a Tat-dependent substrate in the halophilic archaeon Haloarcula hispanica. FEBS J. 2008;275:6159–6167. doi: 10.1111/j.1742-4658.2008.06740.x. [DOI] [PubMed] [Google Scholar]
- 224.Bageshwar UK, Musser SM. Two electrical potential-dependent steps are required for transport by the Escherichia coli Tat machinery. J Cell Biol. 2007;179:87–99. doi: 10.1083/jcb.200702082. [DOI] [PMC free article] [PubMed] [Google Scholar]