Abstract
The Alzheimer’s amyloid precursor protein (APP) belongs to a conserved gene family that also includes the mammalian APLP1 and APLP2, the Drosophila APPL, and the C. elegans APL-1. The biological function of APP is still not fully clear. However, it is known that the APP family proteins have redundant and partly overlapping functions, which demonstrates the importance of studying all APP family members to gain a more complete picture. When APP was first cloned, it was speculated that it could function as a receptor. This theory has been further substantiated by studies showing that APP and its homologues bind both extracellular ligands and intracellular adaptor proteins. The APP family proteins undergo regulated intramembrane proteolysis (RIP), generating secreted and cytoplasmic fragments that have been ascribed different functions. In this review, we will discuss the APP family with focus on biological functions, binding partners, and regulated processing.
Keywords: Alzheimer’s disease, Amyloid-β, APP family, APP intracellular domain, Insulin-like growth factor, Secretases, Regulated intramembrane proteolysis
Introduction
More than 20 years have passed since the molecular cloning of the human amyloid precursor protein (APP). Soon after its discovery, APP [1] was found to be evolutionary highly conserved. In addition, two mammalian genes encoding the homologous proteins APP-like protein-1 and -2 (APLP1 and APLP2) were identified [2, 3]. Research on the APP family and in particular on APP itself has led to evolvement of the concept of presenilin-dependent regulated intramembrane proteolysis (RIP). RIP is important not only in the proteolytic processing of the APP family but also for the processing and function of other proteins. This includes proteins that can be classified as signal transduction-associated receptors and latent transcription factors, such as Notch. The APP family proteins are intriguing with many different suggested functions. It is still not fully clear whether the proteins function as bona fide signaling receptors and/or adhesion molecules or whether the physiological function is primarily mediated by a shedded soluble fragment. However, it is clear that the APP family has redundant and at least partly overlapping functions. Therefore, a comparison of the members will give a more complete picture. Homologues in other species also include the APP-like proteins APPL [4] and APL-1 [5] in fruit fly (Drosophila melanogaster) and worm (Caenorhabditis elegans), respectively. In particular, experiments conducted in fruit flies have been of value in shedding light on possible functions of the APP family.
APP is the most well studied member of the APP family due to its role in Alzheimer’s disease (AD). This neurodegenerative disorder is characterized by the presence of neurofibrillary tangles and senile plaques in the brain. The major constituent of amyloid plaques is the hydrophobic peptide amyloid β (Aβ) [6, 7], that can occur in different lengths (39–43 amino acids). Aβ40 (40 amino acids long) is the most abundant form. However, a slightly longer form, Aβ42, which is produced in much lower quantities, is more prone to aggregate and to form amyloid fibrils [8, 9]. Sequencing of Aβ subsequently led to the molecular cloning of the longer precursor protein APP [1]. APP was originally proposed to be a receptor, and there is increasing evidence supporting this view, as will be discussed in more detail below.
The accumulation of Aβ, which is produced through sequential cleavage of APP (reviewed in [10]), is considered to cause AD pathology and to induce the formation of neurofibrillary tangles, cell loss, vascular damage, and dementia [11]. The hypothesis is supported by the fact that the mutations causing familial early onset forms of AD occur either in APP or in presenilin-1 or -2 (PS1 or PS2), which are involved in the processing of APP. Most of these mutations increase the production and/or accumulation of, in particular, Aβ42. It is also known that patients with Down’s syndrome, caused by trisomy of chromosome 21, which contains the APP gene, develop AD-like pathology [12].
The APP family
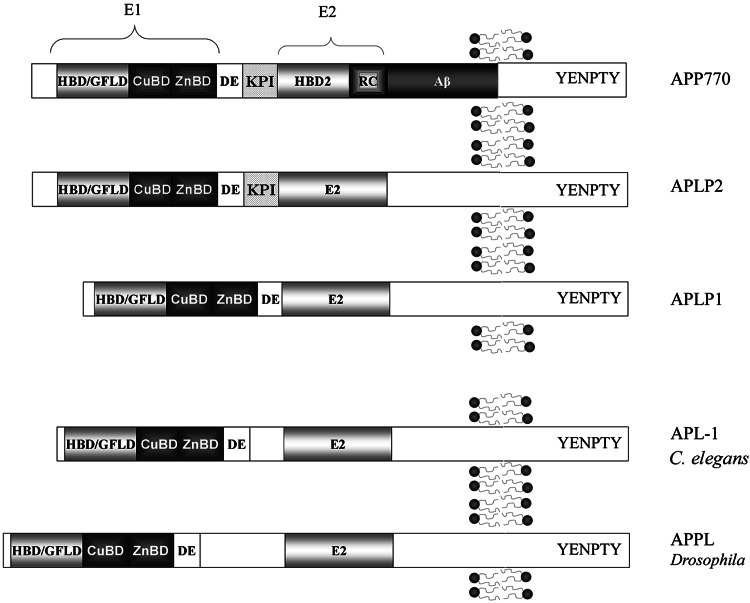
All APP family members are type 1 integral membrane proteins, with a single membrane-spanning domain, a large ectoplasmic N-terminal region and a shorter cytoplasmic C-terminal region (cf. [1, 13]). The APP sequence can be divided into multiple distinct domains (Fig. 1). The ectoplasmic region of APP, which constitutes the major part of the protein, can be divided into the E1 and E2 domains (reviewed in [14]). The E1 domain can be further divided into a number of subdomains, including a heparin-binding/growth-factor-like domain (HFBD/GFLD), a copper-binding domain (CuBD) and a zinc-binding domain (ZnBD). The E1 domain is followed by an acidic region rich in aspartic acid and glutamic acid (DE) and a Kunitz protease inhibitor domain (KPI; not present in APP695). The E2 region consists of another HFBD/GFLD and a random coil (RC) region. The cytoplasmic region of APP contains a protein interaction motif, namely the YENPTY sequence (including the NPXY internalization signal), which is conserved in all APP homologues. The sequences of all APP homologues can be divided into similar domain structures as APP (Fig. 1).
Fig. 1.
Schematic illustration of the domain organization of APP and its homologues. All members of the APP family contain heparin-binding/growth-factor-like domains (HBD/GFLD), copper- and zinc-binding domains (CuBD and ZnBD), an acidic domain (DE), and a protein interaction motif (YENPTY) in the C-terminal
There are three major isoforms of mammalian APP: APP695, APP751, and APP770. Alternative splicing of APLP2 also produces multiple protein isoforms, while only one form of APLP1 has been detected. The main difference between the APP isoforms is the presence or absence of the KPI domain and a chondroitin sulfate glycosaminoglycan (CS GAG) attachment site. This is also the case for the APLP2 isoforms. APLP1 consists of 650 amino acids and lacks both the KPI domain and the CS GAG attachment site [2, 15]. APP and APLP2 are ubiquitously expressed. However, the different isoforms can be preferentially expressed in different cell types, such as the 695-amino-acid-long isoform of APP, which is mainly found in cells of neuronal origin. APLP1 expression has been reported to be restricted to the nervous system [16, 17]. The C. elegans homologue APL-1 is expressed in multiple tissues [18], whereas the Drosophila APPL protein is exclusively expressed in the nervous system [19].
Proteolytic processing of APP and formation of Aβ
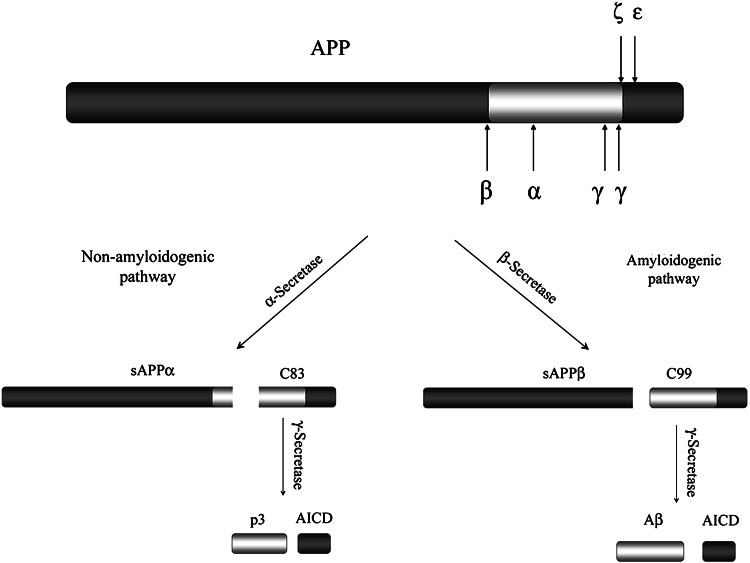
The proteolytic processing of APP can be divided into two different pathways (Fig. 2), the amyloidogenic pathway, which leads to generation of Aβ, and the nonamyloidogenic pathway. Both pathways are mediated by at least three cleavage events. The amyloidogenic processing of APP is initiated through cleavage by β-secretase, which leads to secretion of the large N-terminal ectodomain, sAPPβ. The remaining C-terminal stub of 99 amino acids (C99) can then be further processed by γ-secretase, generating Aβ and a free APP intracellular domain (AICD). AICD can translocate into the nucleus where it may function as a transcription factor [20]. γ-Secretase cleavage between Val637 (in the APP695 isoform) and Ile638 generates Aβ40, and cleavage after Ala639 results in production of Aβ42 (reviewed in [10]). The remaining C-terminal fragments after γ-secretase cleavage were expected to be 57 or 59 amino acids long (C57 and C59). However, instead a 50-amino-acid-long C-terminal fragment (C50) has been identified as AICD [21]. Consequently, the final processing step has been suggested to be a result of three cleavage events. The C99 fragment is first cleaved at the ε-site at the Leu646-Val647 bond [22, 23], followed by cleavage at the ζ-site between Val643 and Ile644, and finally the peptide is cut at the γ-site [24].
Fig. 2.
Schematic illustration of APP processing. The major cleavage of APP in the ectoplasmic domain occurs at the α- and β-site, resulting in the secretion of sAPPα or sAPPβ. The remaining C-terminal membrane-bound fragments, C83 or C99, are subsequently cleaved within the transmembrane region by γ-secretase, releasing AICD and p3 or Aβ
In the nonamyloidogenic pathway, generation of Aβ is precluded since APP is initially cleaved by α-secretase within the Aβ sequence near the ectoplasmic side of the plasma membrane [25, 26]. The α-secretase cleavage of APP releases the N-terminal ectodomain sAPPα from the cell surface leaving an 83-amino-acid-long C-terminal membrane-bound fragment (C83). This fragment can be further processed by γ-secretase in a similar way as in the amyloidogenic pathway, giving rise to the small peptide p3 and AICD.
Although APP processing has been studied extensively, there is not yet a complete picture of the processing events and the enzymes involved. α-Secretase has been characterized as a zinc metalloproteinase that cleaves APP at the Lys613-Leu614 bond [27]. Several α-secretase candidates exist, and they are all members of the ADAM (a disintegrin and metalloproteinase) family. The two most likely candidates are ADAM10 and ADAM17, also known as TACE (tumor necrosis factor-α converting enzyme), but also ADAM9 has been proposed to play a role. ADAMs are type 1 integral membrane proteins with a multidomain structure, including signal peptide, pro-domain, catalytic metalloprotease domain, disintegrin/cystein-rich domain, transmembrane domain, and a short cytoplasmic domain [28].
β-Secretase is an aspartyl protease that cleaves APP at the Met597-Asp598 bond, which constitutes the first step towards Aβ production. Two β-secretases have been identified: BACE (β-site APP-cleaving enzyme; also known as BACE1) and BACE2 [29–32]. BACE is more widely expressed in the brain, and BACE knock-out animals do not produce any detectable levels of Aβ [33, 34]. BACE is a single domain integral protein with its active site located on the ectoplasmic side of the membrane [29, 31]. The optimal pH of BACE activity is approximately 4.5, indicating that the β-site cleavage of APP takes place in more acidic cellular compartments, such as the endosomes [31]. BACE cleavage has also been suggested to occur in lipid rafts [35].
γ-Secretase is an aspartyl protease with low sequence specificity that cleaves the substrate within its transmembrane domain. The enzyme is a protein complex that consists of anterior pharyx defective 1 (APH-1), nicastrin, PS1 or PS2, and presenilin enhancer-2 (PEN-2) [36–39]. PS1 is a nine transmembrane domain (TM) protein and is considered to harbor the active site of the enzyme [40]. Two highly conserved aspartate residues (Asp257 and Asp385 in human PS1) within TM6 and TM7 constitute the core of the catalytic site [41]. Nicastrin has one TM and a large ectodomain, proposed to function as a gatekeeper to the PS1 active site [42]. Nicastrin also functions as a substrate receptor and is important for the assembly process of the γ-secretase complex. The functions of APH1 and PEN2 are not yet fully understood. However, APH1 has been implicated in stabilization of PS1 and plays an important part during assembly of the complex, whereas PEN2 stabilizes the final complex and is involved in endoproteolysis of PS1 (reviewed in [43, 44]).
Proteolytic processing of APP homologues
Although the sequences in APP, where the various secretases cleave, are very different in the two mammalian homologous, both APLP1 and APLP2 are believed to be proteolytically processed in a similar way as APP. The first indication that APLPs were cleaved by γ-secretase was the detection of elevated levels of an APLP1 membrane-bound C-terminal fragment in brain tissues from animals deficient in PS1 [45]. Cleavage of APLPs by γ-secretase generates not only fragments corresponding to membrane-bound APP C-terminal fragments, but also to AICDs, denoted ALID1 and 2 (APP-like intracellular domains 1 and 2) [46, 47]. Substantial evidence shows that APLP2 is processed by α- and β-secretase, while the processing events are less certain for APLP1 [48, 49]. The exact cleavage sites have still not been determined. The C. elegans APP homologue, APL-1, has been shown to be cleaved within its ectoplasmic domain, releasing a soluble form of the protein [5, 19]. Further, APL-1 has been suggested to be cleaved by the C. elegans PS1 homologue, SEL-12 [18]. The Drosophila APP homologue, APPL, is also known to release a soluble form [19]. In addition, homologues to ADAM10 [50], BACE [51], and presenilin [52, 53] have been identified in Drosophila (Kuzbanian, dBACE, and DPS, respectively). In a recent study, the processing of APPL was further studied [51]. APPL was found to be cleaved by Kuzbanian and Dps. Even though the Aβ sequence in APP is not conserved in APPL, both dBACE and human BACE were able to cleave APPL. Interestingly, co-expression of APPL and dBACE resulted in generation of Drosophila Aβ (dAβ) aggregates, which caused spongiform lesions, neuronal cell death, and behavioral deficits.
Posttranslational modifications of the APP family
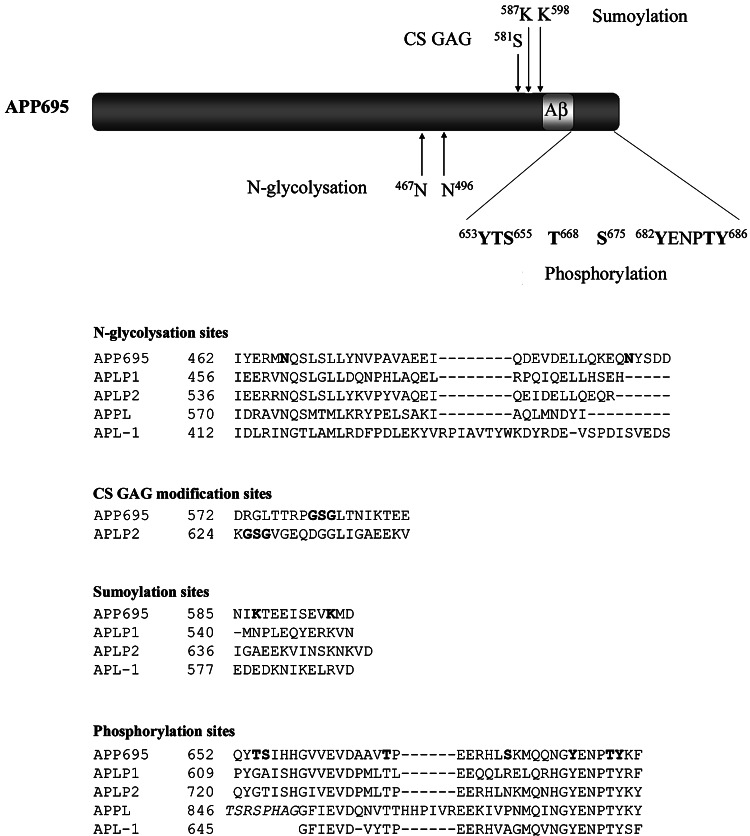
The APP family proteins are posttranslationally modified in several different ways (Fig. 3). All three mammalian members, as well as APPL and APL-1, undergo N-glycosylation [5, 13, 19, 48, 54]. Both APP and APLP2 have been shown to be O-linked glycosylated [55, 56], sialylated [56], and CS GAG modified [57, 58]. N-glycosylation of APLP1 was shown to be regulated by factors affecting neuronal differentiation [47]. In addition, in a study by Eggert et al. N-glycosylation was shown to affect the processing of APLP1 [48]. Treatment with tunicamycin, which blocks N-glycosylation generated additional APLP1 fragments, whereas the processing of APLP2 and APP was unaffected. However, APP processing was observed to be affected by sialylation, since overexpression of sialyltransferase enhanced secretion of Aβ [59]. APP can also undergo sumoylation at lysines 587 and 595 [60].
Fig. 3.
Posttranslational modification sites in APP and its homologues. Schematic illustration of posttranslational modification sites in APP. Note, APP also undergoes O-glycosylation, but the site has still not been determined. Alignment of the APP modification sequences with its homologues. The APP N 467 N-glycosylation site is conserved in all homologues, whereas the APP K 695 sumoylation site is only conserved within the mammalian homologues. Three known in vivo phosphorylation sites (T 668, Y 682, T 686, and Y 687) are conserved in all homologues and one additional site (Y 653) in APLP1 and APLP2. The proteins were aligned using the BLASTp Blossum62 matrix
APP family members have also been shown to be phosphorylated. APP has several sites in its intracellular domain that can be phosphorylated in vitro [61–63]. Among these, Thr654, Ser655, and Thr668 (Fig. 3) have been demonstrated to be phosphorylated in adult rat brain [63] [64], and Tyr653, Ser655, Thr668, Ser675, Tyr682, Thr686 and Tyr687 (Fig. 3) were found to be phosphorylated in postmortem brain tissue of patients with AD [65]. In addition, Thr668 phosphorylated APP has been found to be enriched in endocytic compartments and to colocalize with BACE in AD brains [65]. The Thr668 phosphorylation of APP has been suggested to be mediated by different enzymes. Cdk1/cdc2 kinase was shown to phosphorylate APP at Thr668 in dividing cells [63], whereas c-Jun N-terminal kinase 3 (JNK3) seems to be responsible for Thr668 phosphorylation during neuronal differentiation [66]. In postmitotic neurons, both cyclin-dependent kinase 5 (cdk5) and glycogen synthase kinase-3β (GSK-3β) have been implicated to phosphorylate APP at Thr668 [67, 68]. Thr668-phosphorylated APP has been demonstrated to form complexes with JNK-interacting protein-1 (JIP-1) which are then transported to neurites [69]. Furthermore, Thr668 phosphorylation has been proposed to affect the processing of APP. In a study by Lee et al., it was shown that Aβ production was reduced in rat embryonic cortical neurons when treated with a cdk5 inhibitor, which reduced Thr668 phosphorylation, or when transfected with Thr668Ala mutant APP [65]. However, a similar study with Thr668Ala mutant APP mice showed identical levels and distribution of all APP metabolic products as in wild-type mice [70]. In yet another study, Thr668 phosphorylation of APP was reported to induce increased β-cleavage, but decreased γ-cleavage, resulting in decreased Aβ production [71]. Other phosphorylation sites than Thr668 have also been implicated in the regulation of APP processing. In a recent study, phosphorylation of APP at Tyr687 was suggested to regulate α-cleavage of APP since overexpression of the mutant Tyr687Ala decreased the level of the C83 fragment [72].
Synthetic peptides corresponding to the cytoplasmic domain of APLP1 and APLP2 have been shown to be phosphorylated by protein kinase C (PKC) [73], as previously shown for APP [61]. APLP2, but not APLP1, was further demonstrated to be phosphorylated by cdc2 kinase [73]. Alignment of the APP family members shows that the APP Thr668 phosphorylation site is conserved in all APP homologues; APLP1 (Thr624), APLP2 (Thr636), APPL (Thr855), and APL-1 (Thr660) (Fig. 3). APLP2 as well as APP was observed in a study by Taru and Suzuki to be phosphorylated at this site by JNK1 and JNK2 in response to cellular stress [74]. Three other in vivo phosphorylated sites in APP, Tyr682, Thr686, and Tyr687 (which are part of the YENPTY motif), are also conserved in all APP homologues. Tyr653, another in vivo phosphorylated site in APP, is conserved in both mammalian homologues, but not in APPL or APL-1.
Phosphorylation of APP has also been shown to regulate intracellular signaling via adaptor proteins, which will be discussed later. Thr668 phosphorylation was shown to induce large conformational changes of the intracellular domain [75]. It was later shown that the peptidyl-, prolyl cis/trans isomerase Pin1 binds to Thr668-phosphorylated APP both in vitro and in vivo and thereby accelerates the production of the trans conformation of Pro669 [76]. This proline residue is conserved in APLP2 (Pro737) and APL-1 (Pro662), but not in APLP1 or APPL. It was further demonstrated that the Pin1-catalyzed isomerization of APP regulated its processing, since overexpression or knockout of Pin1 reduced or increased the secretion of Aβ, respectively. It has further been shown that Pin1 knockout in mice resulted in age-dependent neuropathy [77].
Function of the APP family
The exact biological function of APP and its homologues is still unknown. However, several in vitro and in vivo studies have yielded strong evidence for roles of APP, both in the developing and adult nervous system, in cell adhesion, neuronal survival, neurite outgrowth, synaptogenesis, vesicular transport, neuronal migration, modulation of synaptic plasticity, and insulin and glucose homeostasis. Whether these functions are mediated through receptor- or ligand-like properties of APP is in most cases still not known. However, there is evidence suggesting that APP may function as both a ligand and a receptor. Studies with knockout mice have shown that single disruptions of APP, APLP1, or APLP2 only caused minor abnormalities [78–80]. In contrast, APLP2−/−/APP−/− mice and APLP2−/−/APLP1−/− both showed a lethal phenotype, whereas APLP1−/−/APP−/− mice were viable [80]. Triple knockout mice showed a 100% lethal phenotype [81]. These results suggest that APLP2 has a key physiological role among the mammalian family members and that there seems to be functional redundancy among the homologues. Little is known about the function of APLP1 and APLP2, as well as the nonmammalian homologues. However, the functional overlap with APP has been demonstrated in several studies, providing evidence for roles of the homologues in neurite outgrowth, cell adhesion, and neuronal migration.
Cell adhesion, neurite outgrowth, and synaptogenesis
Many studies have indicated that APP participates in cell adhesion. Schubert et al. observed that APP can modulate the adhesion of PC12 cells to the substrata [82]. APP has also been shown to bind to several extracellular matrix molecules, such as laminin and heparin sulphate proteoglycans [83]. In addition, X-ray crystallography analysis has shown that the ectodomain of APP can form antiparallel dimers [84]. It was later shown that APP was able to form both homodimers and heterodimers with APLP1 and APLP2, which in cell culture studies were able to induce cell–cell adhesion [85]. The study further showed that endogenous APLP2 was required for cell–cell adhesion in mouse embryonic fibroblasts. However, in a recent study it was observed that although all mammalian homologues could form cis interactions, trans interactions could only be formed by APLP1 [86]. Thus, this latter study suggests a specific role of APLP1 in cell adhesion.
The binding of APP to heparin sulphate proteoglycans was reported by Small et al. to stimulate neurite outgrowth in cultured chick sympathetic and mouse hippocampal neurons [87]. APP and the secreted fragment sAPPα have in several studies been shown to regulate neurite elongation and branching. Milward et al. showed that an 18-h treatment of both soluble and membrane-associated APP increased neurite length and branching in PC12 cells without affecting the number of neurites per cell [88]. It was also suggested that APP could mediate NGF-induced neurite outgrowth, since antibodies against APP specifically diminished the NGF effect on neurite length and branching. In a study by Perez et al., hippocampal neurons from APP knock-out mice displayed shorter axonal outgrowth after 1 day in culture, compared to their wild-type counterparts [89]. These results are in agreement with the Milward study. However, Perez et al. also showed that after 3 days in culture, the APP knock-out neurons displayed longer axons, suggesting that APP initially stimulates neurite outgrowth but over time inhibits it. The effect of secreted APP was also determined by growing hippocampal neurons with astrocytes from wild-type and APP knock-out mice. Surprisingly, it was found that co-culture with APP knock-out astrocytes increased neurite outgrowth of both wild-type and APP knock-out neurons. The mechanism of APP-regulated neurite outgrowth was further elucidated by a study by Young-Pearse and collaborators in which antibodies directed against integrin β1 were shown to inhibit the neurite elongation stimulated by sAPPα [90]. Interestingly, it was also shown that the secreted fragment sAPPα was unable to induce neurite outgrowth in the absence of cellular APP expression and that sAPPα interfered with the interaction between APP and integrin β1. The findings suggest that sAPPα enhances neurite outgrowth by inhibiting the function of full-length APP. The secreted forms of APLP1 and APLP2 were both also shown to induce neurite outgrowth through similar interaction with integrin β1 as sAPPα, further illustrating the functional overlap of the mammalian APP family proteins. The Drosophila homologue APPL has also been shown to be involved in neurite outgrowth. In contrast to APP, deletion of APPL resulted in shorter neurites after 6 days in culture, while overexpressing APPL increased the neurite length [91].
Roch and collaborators demonstrated that a 17-mer sAPP peptide (amino acids 319–335) was able to induce cellular and behavioral changes when infused into adult rat brains [92]. A 2-week period of peptide infusion resulted in an 18% increase of synaptic density in the frontoparietal cortex, as well as enhanced memory retention. Moya et al. showed that APP is developmentally regulated and that an increased expression was observed at the time of synaptogenesis [93]. Further evidence for a role of APP during neuronal development was demonstrated in a study by Young-Pearse and collaborators, in which in utero electroporation of shRNAs into the developing cortex was used to knock-down APP in rodents. The study showed that APP was required for correct migration of neuronal precursor cells into the cortical plate [94]. In addition, overexpression of APLP1 or APLP2 was shown to rescue the neuronal migration defect caused by the APP knock-out. The mechanism involved in APP modulation of neurogenesis was further described in a recent paper by Ma et al. [95]. In this study it was demonstrated that APP was able to interact with the GPI-linked recognition molecule TAG1, which increased AICD production. Neuronal precursor cells from APP−/−/TAG1−/− mice had enhanced neurogenesis, indicating that a TAG1-APP signaling pathway negatively modulates neurogenesis.
APP has also been suggested to have a role in axonal outgrowth after traumatic brain injury. In a study by Leyssen et al., it was shown that overexpression of APP or the Drosophila homologue APPL in adult Drosophila brain neurons promoted post-developmental axonal arborization [96]. APPL was further shown to be up-regulated as a response to brain injury in adult flies.
Neuronal survival and apoptosis
The secreted APP fragment sAPPα has also been shown to enhance long-term neuronal survival when added to the medium of rat cortical cell cultures [97] and to prevent neuronal death in human cortical cell cultures deprived of glucose or exposed to excitotoxins [98]. Further, sAPPα has even been shown to protect against Aβ toxicity [99]. Survival of rat hippocampal neurons was reduced to 40% after a 4-day exposure to Aβ, whereas 75% of the neurons survived the same exposure when co-treated with sAPPα.
While sAPPα has been shown to have neuroprotective properties, AICD has been suggested to be involved in apoptosis. Apoptosis was induced by overexpression of AICD in human neuroglioma cells and was shown to be dependent on AICD nuclear translocation and interaction with the histone acetyltransferase Tip60 (Tat-interacting protein 60 kDa) [100]. In a recent study, AICD-induced apoptosis was proposed to be neuron-specific, since overexpression of AICD induced apoptosis in P19 cells differentiated into a neuronal phenotype, while it had no effect on undifferentiated P19 cells and P19 cells differentiated into muscle cells [101]. The mechanism behind AICD-induced apoptosis is still not known, but has been suggested to involve AICD-dependent upregulation of glycogen synthase kinase-3β [102]. In another study, the apoptosis triggering mechanism of AICD was shown to involve enhancement of p53-mediated apoptosis [103]. Recently, a study by Tang et al. demonstrated that the APLP1 gene is a direct transcriptional target of p53 [104]. It was further shown that knock-down of APLP1 reduced stress-induced apoptosis in neuroblastoma cells, whereas overexpression of APLP1 enhanced it. The authors speculated that induction of APLP1 expression by p53, which is activated in acute neuronal injury as well as chronic neurodegenerative disorders, may enhance neuronal apoptosis.
Modulation of synaptic plasticity and axonal transport
Several studies in both cell culture and in vivo systems have demonstrated a role of APP, and especially sAPPα, in neuronal excitability and synaptic plasticity. Studies in cultured hippocampal neurons showed that sAPPα had a suppressive effect on whole cell currents induced by NMDA (N-methyl-d-aspartate) [105]. This effect was rapid and reversible and was also shown to involve activation of receptors linked to cGMP production and K+ channels [105, 106]. The Drosophila APPL protein has also been shown to affect the neural excitability, possibly through activation of K+ channels by secreted APPL [91]. The involvement of APP in memory formation was demonstrated by injecting antibodies directed against APP into the brains of adult rats, which impaired behavioral performance on different learning and memory tasks [107, 108]. sAPPα has also been shown to increase in an NMDA receptor-dependent manner in adult rats after induction of long-term potentiation (LTP) [109]. Aβ may also affect learning and memory. This was demonstrated in a study by Nabeshima and Nitta in which Aβ was continuously injected into the lateral ventricles of adult rats for 2 weeks [110]. The Aβ treatment impaired the performance of the rats in the water maze task as well as in the passive avoidance task. In addition, Aβ has been shown to significantly impair LTP [111]. A recent study by Puzzo et al. proposed that Aβ has detrimental effects only at high concentrations (nM range), whereas low concentrations (pM range) positively modulate synaptic plasticity and memory [112].
Full-length APP was shown early on to undergo fast axonal transport to synaptic sites of human and rat brain, implying a role of APP in synaptic activity [113, 114]. It was later shown by co-immunoprecipitation, fractionation by sucrose gradient, and through direct in vitro binding that APP interacted with kinesin-1 by binding to the kinesin light chain subunit [115]. The suggestion that APP functions as a vesicular receptor for kinesin-1 was further supported by in vivo studies in Drosophila, demonstrating disruption of axonal transport caused by APPL mutations [116]. Despite these extensive and convincing studies by the Goldstein group, several independent groups were unable to reproduce these results [117], making the hypothesis that APP serves as a kinesin-1 receptor still somewhat controversial.
Additional functions
In addition to the effects on neuronal function and survival, APP and its homologues have been implicated in other physiological functions not restricted to the nervous system. Of particular interest is a recent study by Needham et al. that revealed a possible new function of the APP family, namely as modulators of insulin and glucose homeostasis [118]. The study correlated the postnatal lethality in the APP−/−/APLP2−/− knock-out mice to hyperinsulinemia and low glucose levels. In addition, another study suggested that APP may be involved in obesity-related insulin resistance, since the expression levels of APP in adipocytes increased in obesity and correlated with insulin resistance [119].
Interactions with intracellular adaptor proteins
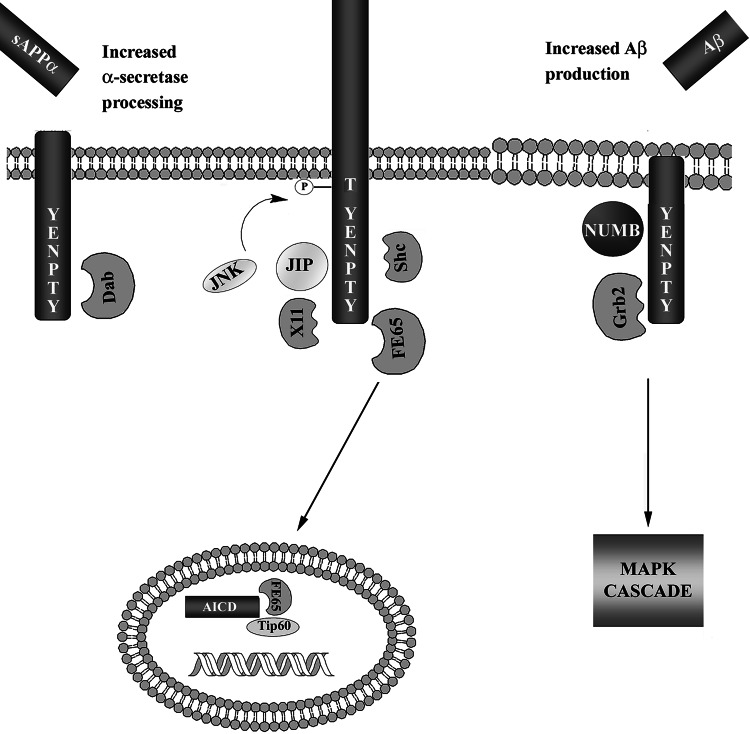
The intracellular domain of APP contains the YENPTY motif, which has been shown to interact with several adaptor proteins, including FE65, X11, JIP, Shc, Grb2, Dab1, and Numb (Fig. 4). The interaction of adaptor proteins with the APP homologues is not as well studied. However, since the intracellular domain of APP is highly conserved in all APP homologues, and they all contain the YENPTY motif (Fig. 3), it is likely that all APP homologues signal through similar adaptor proteins. In fact, some of the APP-interacting proteins have also been demonstrated to interact with APP homologues. The interaction between APP and most adaptor proteins is dependent on phosphorylation of the intracellular domain of APP, either at Thr668 or Tyr682. This suggests that the phosphorylation status of APP regulates downstream signal transduction and could result in altered APP trafficking and processing.
Fig. 4.
Schematic illustration of proposed interactions between APP and adaptor proteins. APP interacts with several adaptor proteins through the YENPTY motif in its intracellular domain. The interactions between APP and the adaptor proteins affect the metabolism of APP. Dab1 interaction increases α-secretase processing, while Grb2 and Numb increase β-secretase processing. JIP, X11, and Shc stabilize full-length APP. Note that β-secretase cleavage of APP is considered to take place in endosomes or lipid rafts, as illustrated by the different appearance of the membrane. JIP and X11 both regulate the phosphorylation at Thr668 through JNK, thereby regulating the interaction with other adaptor proteins. Grb2 interaction with APP results in activation of the MAPK cascade, similar to Grb2 interaction with tyrosine kinase receptors. The γ-secretase generated fragment AICD has been demonstrated to translocate into the nucleus where it forms a complex with FE65 and Tip60. The complex has been suggested to regulate gene transcription (see the text for references)
FE65
The first protein identified as an APP adaptor protein was FE65 [120]. The interaction between the two proteins was detected using the phosphotyrosine interaction/phosphotyrosine binding (PID/PTB) domain of FE65 as bait to screen a human brain cDNA library in yeast using the two-hybrid system. FE65 belongs to a protein family that also includes FE65-like 1 (FE65L1) and FE65-like 2 (FE65L2), all of which bind to APP [121, 122]. FE65 has also been shown to bind to APLP1 and APLP2 [75, 123]. FEH-1 is a FE65 homologue in C. elegans that has been found to bind to the APP homologue APL-1 [124]. A role of FE65 family proteins for the function of APP family proteins is indicated by the fact that mice deficient in APP, APLP1, and APLP2 share developmental abnormalities with mice deficient in FE65 and FE65L1 [81, 125].
FE65 was shown to bind to the YENPTY motif of APP [120, 126]. The interaction was further shown to affect APP trafficking and metabolism, since overexpression of FE65 increased the amount of APP at the plasma membrane and enhanced both Aβ and sAPPα secretion [127]. In contrast, the Suzuki group demonstrated that overexpression of FE65 suppressed the secretion of Aβ [75]. In a study by Hu et al., it was shown that FE65 can be cleaved from a 97-kDa protein into a 65-kDa fragment. Co-expression of the 65-kDa FE65 fragment and APP695 in HEK293 cells strongly reduced the secretion of sAPPα compared to cells transfected with only APP695, whereas the 97-kDa fragment had little effect [128]. This suggests that the cleavage of FE65 into the 65-kDa fragment regulates sAPPα production. In a study by Cao and Südhof, it was found that FE65 formed a complex with AICD and the histone acetyl transferase Tip60, and together induced gene transactivation [20]. The mechanism of FE65 and AICD nuclear translocation seems to be regulated by phosphorylation of APP at Thr668, but there are contradictory reports. In a study by Chang et al. it was suggested that APP Thr668 phosphorylation was essential for binding to FE65 and for nuclear localization of AICD [129]. In contrast, several studies by the Suzuki group have demonstrated that FE65 binds to APP and is released upon APP Thr668 phosphorylation, whereby it translocates into the nucleus [75, 130]. Further, they also showed that AICD was predominantly found in a nonphosphorylated state in the nucleus in vivo, and that it seemed to translocate into the nucleus independently of FE65 [131]. Their results were confirmed by a study by Radzimanowski et al. in which phosphorylated AICD was found to have a sevenfold reduced binding to FE65, compared to nonphosphorylated AICD [132]. Circular dichroism (CD) spectra analysis of the cytoplasmic domain of APP revealed that the percentage of the α-helix of the Thr668 phosphorylated peptide was significantly reduced compared to nonphosphorylated peptide. It was concluded that Thr668 phosphorylation induces large conformational changes, affecting the conformation of the YENPTY motif and thereby suppressing the interaction with FE65 [75].
X11/Mint
Members of the X11, also called munc-18-interacting protein (Mint), family are also known to interact with the cytosolic domain of APP [126, 133]. X11/Mints, as FE65, bind to the YENPTY motif via their PID/PTB domain [126]. The interaction seems to stabilize APP and inhibit Aβ production in cultured cells [134, 135]. These results were confirmed in an in vivo study by Lee et al. that showed that transgenic mice overexpressing APP with a familial AD-linked mutation (APPswe, i.e., APP with the Swedish mutation; Lys670Asn and Met671Leu) displayed decreased Aβ production and plaque formation when X11/X11α/Mint1 (referred to as X11) or X11L/X11β/Mint2 (referred to as X11L), the two neuronal X11s, were overexpressed [136, 137]. The mechanism involved in X11 regulation of APP metabolism was further elucidated in a recent study by Saito et al. in which an accumulation of APP and C99 was detected in lipid rafts of X11−/−/X11L−/− knock-out mice [138]. This suggests that X11s interact with APP and inhibit its translocation into lipid rafts, where it would be processed by BACE, and thereby lowering Aβ production.
APP and APLP2 are phosphorylated at Thr668 and Thr736, respectively, by JNK in response to cellular stress [74]. X11L was found to enhance the stress-induced phosphorylation of both APP and APLP2. Since the Thr668 phosphorylation of APP has been found to influence the interaction with several adaptor proteins, these results suggests that X11L can regulate the interaction of APP with other proteins. X11L has been shown to bind to the type 1 transmembrane protein Alcadein (Alzheimer-related cadherin-like protein) through its PTB domain [139]. X11L was further demonstrated to join APP and Alcadein together, forming a tripartite complex. The presence of Alcadein enhanced the X11L-mediated stabilization of APP. X11 family members also contain at least one PDZ domain through which X11 has been shown to interact with several proteins involved in synaptic functions (reviewed in [140]). A Drosophila homologue to X11, dX11, has been shown to interact with APPL [141].
JIP
Another family of PTB-containing proteins, which was discovered to interact with APP using the yeast two-hybrid system, is the JNK-interacting protein (JIP) family [142, 143]. The JIP family includes JIP-1a, JIP-1b [also known as Islet-Brain1 (IB1)] and JIP-2. The interaction between JIP proteins and APP is dependent on the YENPTY motif. However, the JIP interaction with APLP1 and APLP2 was shown to be much weaker, indicating that other structural elements might affect the interaction [142]. It has further been demonstrated that JIP-1b regulates the metabolism of APP [144]. Pulse-chase studies of metabolically radio-labeled N2a cells co-expressing APP and JIP-1b showed that JIP-1b stabilized immature APP and decreased sAPPα and Aβ secretion. JIP-1a (which is a splice variant lacking a complete PTB domain) and JIP-2 bound only weakly to APP and did not influence the processing of APP. JIP-1b is known to scaffold APP into complex with JNK [142, 144], and the association induces phosphorylation of APP at Thr668 [144, 145]. However, the regulation of APP metabolism was suggested to be independent of JNK activation [144]. Although the APP interaction to the kinesin light chain (KLC) is considered somewhat controversial [115, 117], Inomata et al. demonstrated that a small amount of APP was co-immunoprecipitated with KLC, and this interaction was strongly increased by co-expression of JIP-1b [145]. The Drosophila APPL protein has been shown to interact with APLIP1 (APP-like protein interacting protein 1), which is highly homologous to JIP-1b [144]. Recent reports further support that JIP-1b/APLIP1 is involved in axonal transport of APP/APPL-containing vesicles [146, 147].
Shc and Grb2
Shc (Src homology collagen-like) A, a member of the cytoplasmic adaptor protein family also including Shc C, was initially reported by Borg et al. not to interact with APP [126]. However, in a study by Tarr et al., it was demonstrated that both Shc A and Shc C could in fact interact with APP, but only when APP was phosphorylated at Tyr682 [148]. Phosphorylation at Thr668 was proposed to modulate the interaction, since the APP Thr668Glu/Ala mutations reduced APP and Shc A co-immunoprecipitation. It was further established that the interaction modulates APP processing, since RNAi against Shc C decreased the levels of secreted Aβ [149]. In a study by Russo et al., tyrosine-phosphorylated β-secretase-derived C99 in human brain extracts was found in complexes with Shc A and growth factor receptor-bound protein 2 (Grb2) [150]. In contrast, α-secretase-derived C83 was not tyrosine phosphorylated and did not interact with Shc A or Grb2. Interestingly, both Grb2 and ShcA have been shown to be enriched in AD brains [150]. It was later demonstrated that Grb2, through its Src homology 2 (SH2) domain, binds to the YENPTY motif, only when Tyr682 is phosphorylated [151]. Grb2, like Shc A and Shc C, has also been demonstrated to affect APP metabolism, since overexpression of Grb2 increased the generation of Aβ in HEK293 cells overexpressing APP [151]. Grb2 has also been shown to interact with PS1 [152]. It was further shown that the interaction among Grb2, APP, and PS1 could activate the mitogen-activated protein kinase (MAPK) pathway [150, 152].
Dab1
All members of the mammalian APP family and especially APLP1 interact with the Disabled-1 (Dab1) protein, which is important for nervous-system development [153, 154]. Overexpression of APP, APLP1, and APLP2 was also shown to increase serine phosphorylation of Dab1. Dab1, as other adaptor proteins, can also affect the metabolism of APP. Hoe and collaborators found that expression of Dab1 increased α-secretase cleavage of APP and decreased Aβ levels [155]. This could be a result of altered APP trafficking since the APP levels increased at the surface, where α-secretase cleavage preferentially occurs. This was later suggested to be due to a synergistic effect of Dab1 and the Src family tyrosine kinase Fyn [156]. Fyn was suggested to phosphorylate the YENPTY motif of APP at Tyr682, which may increase the interaction between Dab1 and APP.
Numb
APP has been shown to interact with Numb, a cytosolic protein that regulates the endocytosis and degradation of Notch [157]. Numb was shown to interact with the YENPTY motif of APP through its PTB domain. The function of the Numb-APP interaction was further analyzed by Kyriazis et al. who discovered that Numb could regulate APP metabolism and transport [158]. Different isoforms of Numb, differing in the length of the PTB domain, were shown to have opposite effects on APP. Stable expression of short-PTB (SPTB) Numb increased APP accumulation in early endosomes and enhanced Aβ secretion, whereas long-PTB (LPTB) Numb reduced the amount of APP and Aβ secretion. The effect of Numb on APP metabolism was suggested to be a result of altered APP trafficking.
PAT1
Another adaptor protein that regulates APP transport is PAT1 (protein interacting with APP tail 1), which was identified and characterized by Zheng et al. as a microtubule-binding protein that interacts with the basolateral sorting signal (BaSS) in the cytoplasmic domain of APP [159]. The PAT1 homologue, PAT1a, was later shown to interact with all mammalian APP family members in vivo, with the highest affinity to APLP2 [160]. The interaction between APP/APLPs and PAT1/PAT1a was further shown to influence the intracellular transport and promote processing of APP/APLPs [159, 160].
Regulated intramembrane proteolysis
As described above, the APP family proteins undergo sequential processing. They are initially cleaved in the extracellular domain, either by α- or β-secretase, followed by PS1-dependent cleavage in the plane of the membrane, liberating a cytosolic fragment (AICD or AICD-like fragments). Another PS1 substrate is the Notch receptor. Notch is a plasma membrane receptor that signals through RIP. Notch, like the APP family, is processed by a two-step cleavage mechanism, resulting in PS1-dependent release of a cytosolic fragment (Notch intracellular domain; NICD). NICD translocates to the nucleus where it controls transcription of genes involved in cell-fate decisions (reviewed in [161]). Due to the similarities in the processing of Notch, it has been suggested that the processing of APP family proteins is also a part of a signaling pathway, making them cell-signaling receptors.
Effects on gene transcription
The first suggestion that AICD, like NICD, was involved in gene transcription came from a study by Cao and Südhof. They used an APP-Gal4 fusion protein, which induced luciferase expression in cells co-transfected with a Gal4-dependent reporter plasmid [20]. The transactivation was very low. However, when co-transfected with FE65 the activity was greatly increased. It was further shown that the mechanism involved activation of the histone acetyl transferase Tip60, possibly through direct interaction with FE65 and AICD. APLP1 and APLP2 also undergo presenilin-dependent RIP, generating the AICD-like fragments ALID1 and ALID2, respectively [123]. ALID1 and 2, like AICD, are stabilized by FE65 and are suggested to regulate gene transcription.
Several possible AICD target genes have been suggested, for example, KAI1, GSK-3β, APP, FE65, Tip60, Neprilysin, p53, and EGFR (reviewed in [162]). In a study by Baek and collaborators, it was shown that when FE65, APP, and Tip60 were co-transfected, they formed a trimeric complex that was recruited to the KAI1 promoter and induced expression [163]. Treatment with a γ-secretase inhibitor blocked the induced expression. Furthermore, both protein and mRNA levels of KAI1 were shown to increase in APP-overexpressing mice. Kim et al. showed that overexpression of APP770, C99, or APPswe all caused increased AICD levels in the nucleus as well as increased expression of GSK-3β [102]. The expression of neprilysin, a putative Aβ-degrading enzyme was reduced in PS1−/−/PS2−/− double-knockout mice, and the neprilysin gene promoters were shown to be transactivated by the intracellular domains from all three mammalian APP family proteins [164]. Even though several putative AICD target genes have been identified, these data are still somewhat controversial due to conflicting results. For example, a study by Hébert et al. showed that the protein levels of APP, KAI1, GSK-3β, and neprilysin were unaffected in PS double-knockout fibroblasts, and that AICD, even in the presence of FE65, only induced a very weak activation of the KAI1 promoter [165].
Proteolysis-dependent signaling
APP is often compared to Notch since they both undergo PS-dependent RIP that could result in effects on gene transcription. Notch signaling is initiated through interaction with the transmembrane proteins Delta, Jagged, or Serrate (reviewed in [161]). It is tempting to speculate that signaling by APP and its homologues also can be regulated through a similar mechanism. In fact, a recent study by Ma et al. identified the glycophosphatidylinositol (GPI)-linked recognition molecule TAG-1 as an extracellular binding partner [95]. The interaction between TAG-1 and APP increased the PS-dependent release of AICD, suggesting that TAG-1 is a functional APP ligand. The mammalian APP family proteins have also been shown to form cis- and trans-, homo- and heterocomplexes through their E1 domains [85]. The APP dimerization was further investigated in a study by Kaden et al. [86]. They showed that APP dimerization affected its processing, since a 21-amino-acid-long peptide corresponding to the E1 sequence between the GFLD and CuBD domains selectively increased β-secretase cleavage. In addition, the extracellular matrix protein F-spondin has previously been proposed to function as an APP ligand [166, 167]. F-spondin was shown to induce processing and signaling of APP, strengthening the theory that APP can act as a receptor.
In addition to ligand interactions, several other factors are known to induce APP processing. Early studies demonstrated that PKC phosphorylation regulated the APP metabolism by increasing α-site cleavage of APP while Aβ generation was decreased [168–170]. In these studies PKC was activated by phorbol esters, but activation of G-protein-coupled receptors activating phospholipase C [171, 172] and activation of growth factor receptors [173, 174] could also increase α-site cleavage of APP in a PKC-dependent manner. Studies with synthetic peptides corresponding to the cytoplasmic domain of APP suggested that PKC could phosphorylate APP [61]. However, activation of PKC induced APP processing even in the absence of the phosphorylation sites [175]. Furthermore, in a later in vivo study, PKC activation failed to increase APP phosphorylation [176], indicating that the effect of PKC on APP metabolism is not mediated through direct phosphorylation of APP, but perhaps through regulation of APP-cleaving enzymes. This theory was also supported by studies in our laboratory [177] showing that TACE was upregulated in a PKC-dependent manner in response to retinoic acid, which induces neuronal differentiation of human neuroblastoma cells concomitant with increased APP α-site cleavage.
Insulin-like growth factor (IGF-1) is a member of the insulin family of hormones and is gaining increasing attention for its role in brain function (e.g., cognition) and disease (reviewed in [178]). IGF-1 has been suggested to play a part in Aβ clearance, and serum levels have been shown to decrease with age. IGF-1 is also known to induce α-secretase processing of APP [179]. We have further investigated the intracellular signaling pathways involved in IGF-1-increased α-secretase processing of APP. However, these studies showed that PKC-inhibition only partly inhibited IGF-1-induced sAPPα production, indicating that several signaling pathways could be involved in regulating α-secretase cleavage of APP (Jacobsen et al., unpublished data). Phosphatidylinositol 3-kinase (PI3K) has previously been suggested also to be involved in regulating APP processing [180]. In fact, PI3K inhibition completely blocked IGF-1-induced sAPPα secretion [179].
We have also observed that APLP1 and APLP2 processing is induced by IGF-1. However, the signaling pathways involved in the regulation of the proteolysis of the three homologous proteins seem to differ (Fig. 5). In contrast to APP, IGF-1-induced APLP1 processing was shown to be dependent on MAPK in addition to PI3K and PKC [179] (Jacobsen et al., unpublished data). Furthermore, IGF-1-induced APLP2 processing was independent of PI3K and MAPK but was completely blocked by PKC inhibitors (Jacobsen et al., unpublished data). It still remains to be determined how these kinases regulate the processing of the APP family proteins. The effect could be mediated by phosphorylation of the APP family of proteins or the processing enzymes, making the protein a better substrate, altering the enzyme activity, or affecting the localization of the substrate or enzyme. From these studies, we conclude that there are clear differences in the regulated processing of the three mammalian homologues, which opens the possibility to selectively modulate APP processing.
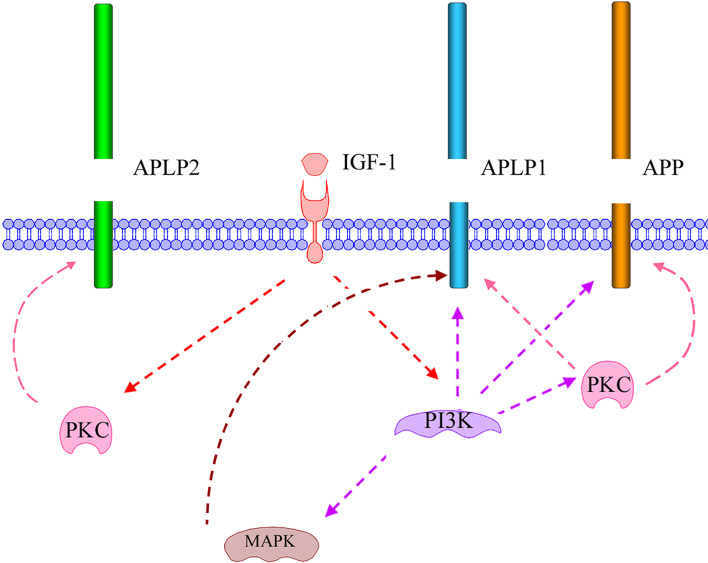
Fig. 5.
Schematic illustration of IGF-1-induced signaling resulting in increased processing of the mammalian APP family proteins. IGF-1-induced processing of APP and APLP1 is almost completely dependent on activation of PI3K, but is also affected by PKC activation. This indicates that PKC acts downstream of PI3K. In contrast, IGF-1-induced processing of APLP2 is independent of PI3K activation but dependent on PKC activation, indicating that PKC also is activated independently of PI3K during IGF-1 stimulation. Note that different PKC isoforms could be involved. Only the IGF-1-induced processing of APLP1 was shown to be dependent on MAPK activation. A model is proposed where IGF-1 binds and activates membrane-bound IGF-1 receptors, resulting in activation of PI3K and PKC, independently of each other
Although APP processing has been extensively studied, it is not yet clear how different stimuli result in altered processing. Here we propose a model of how the processing of APP or APP family members may be regulated (Fig. 6). The signal transduction in this model is initiated either by direct interaction with a ligand or by extracellular stimuli. The extracellular stimuli can, via membrane-bound receptors, activate downstream protein kinases. In turn, this can lead to phosphorylation of the cytoplasmic domain of the APP family proteins or of proteins involved in their proteolytic processing. We speculate that the ligand interaction induces a conformation change of the intracellular domain of the APP family protein. Conformational change and phosphorylation status may affect the processing of the APP family protein due to altered affinity for interacting proteins.
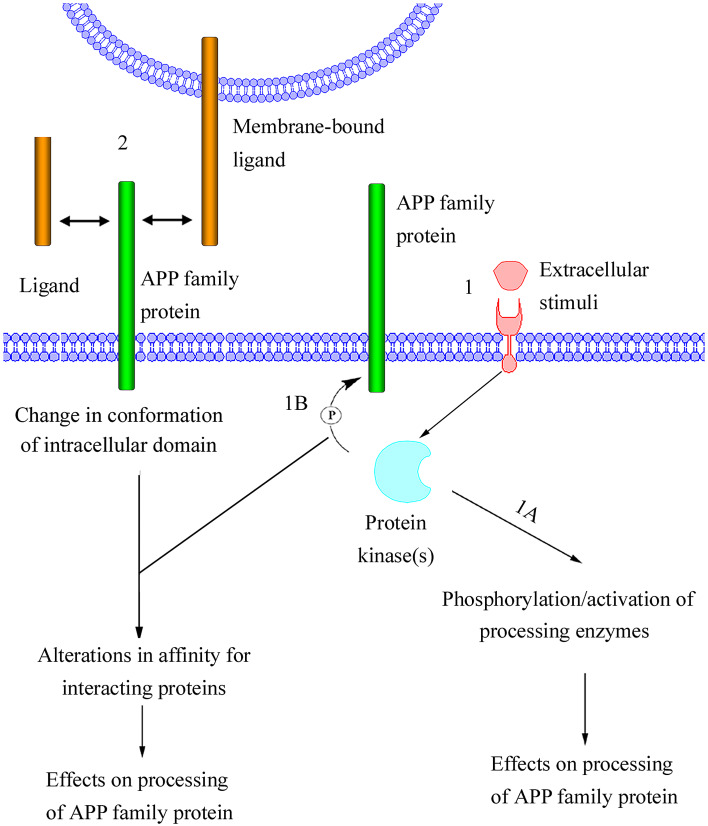
Fig. 6.
Proposed model of regulated processing of APP family proteins. In this model, processing is induced either (1) by an extracellular stimuli (e.g., growth factors) or (2) through direct interaction with a soluble ligand (e.g., sAPPα), an extracellular matrix protein (e.g., F-spondin; not shown) or a membrane-bound ligand (e.g., TAG-1, APP, APLP1, or APLP2). The extracellular stimuli activate membrane-bound receptors, resulting in activation of cytoplasmic protein kinases. One possible effect of the activated protein kinases is phosphorylation of proteins involved in cleavage of APP family proteins (1A). This could result in altered activity or intracellular localization of the enzymes, thereby affecting the processing of APP family proteins. Alternatively, the activated protein kinases could phosphorylate APP family proteins (1B). Phosphorylation of the cytosolic domain of APP family proteins would most likely affect the affinity for interacting proteins. These interacting proteins could be adaptor proteins, which regulate APP family protein localization, resulting in altered proximity of APP family proteins to processing enzymes. The phosphorylation of the intracellular domain could also turn APP family proteins into better substrates for enzymatic cleavage. It is likely that the interaction of APP family proteins with soluble or membrane-bound ligands would affect the conformation of the intracellular domain of APP family proteins. In this model, the conformational change, like the extracellular stimuli-induced phosphorylation of APP family proteins is thought to alter the affinity for interacting proteins, thereby affecting the processing of APP family proteins
Conclusion
Ever since the molecular cloning of APP, the proposal that it may function as a receptor has been debated. Increasing evidence indicates that all members of the APP family can transduce a signal from the outside of the cell. This proposed signaling seems to be dependent on proteolysis. In addition to ligand-induced processing, there clearly is a cross-talk with signaling pathways induced by other cell-surface receptors. It is likely that this indirect regulation of the proteolytic processing of APP family proteins will also affect signaling mediated by secreted and/or cytoplasmic fragments. It can be speculated that signaling induced by ligand binding to APP family proteins and signaling induced by other types of receptors converge by means of effects on APP/APP-like protein processing and interactions with adaptor proteins. It is still not fully clear whether the physiological functions of the APP family are mediated mainly through ectodomain shedding or if transactivation involving AICD or AICD-like fragments play a more important role. One important issue that also remains to be clarified is how the alternative proteolytic pathways (that exist at least for APP) are linked to effects on cell signaling.
References
- 1.Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Müller-Hill B. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature. 1987;325:733–736. doi: 10.1038/325733a0. [DOI] [PubMed] [Google Scholar]
- 2.Wasco W, Bupp K, Magendantz M, Gusella JF, Tanzi RE, Solomon F. Identification of a mouse brain cDNA that encodes a protein related to the Alzheimer disease-associated amyloid beta protein precursor. Proc Natl Acad Sci USA. 1992;89:10758–10762. doi: 10.1073/pnas.89.22.10758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wasco W, Gurubhagavatula S, Paradis MD, Romano DM, Sisodia SS, Hyman BT, Neve RL, Tanzi RE. Isolation and characterization of APLP2 encoding a homologue of the Alzheimer’s associated amyloid beta protein precursor. Nat Genet. 1993;5:95–100. doi: 10.1038/ng0993-95. [DOI] [PubMed] [Google Scholar]
- 4.Rosen DR, Martin-Morris L, Luo LQ, White K. A Drosophila gene encoding a protein resembling the human β-amyloid protein precursor. Proc Natl Acad Sci USA. 1989;86:2478–2482. doi: 10.1073/pnas.86.7.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daigle I, Li C. apl-1, a Caenorhabditis elegans gene encoding a protein related to the human β-amyloid protein precursor. Proc Natl Acad Sci USA. 1993;90:12045–12049. doi: 10.1073/pnas.90.24.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glenner GG, Wong CW, Quaranta V, Eanes ED. The amyloid deposits in Alzheimer’s disease: their nature and pathogenesis. Appl Pathol. 1984;2:357–369. [PubMed] [Google Scholar]
- 7.Masters CL, Simms G, Weinman NA, Multhaup G, McDonald BL, Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc Natl Acad Sci USA. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burdick D, Soreghan B, Kwon M, Kosmoski J, Knauer M, Henschen A, Yates J, Cotman C, Glabe C. Assembly and aggregation properties of synthetic Alzheimer’s A4/β amyloid peptide analogs. J Biol Chem. 1992;267:546–554. [PubMed] [Google Scholar]
- 9.Jarrett JT, Berger EP, Lansbury PT., Jr The carboxy terminus of the β amyloid protein is critical for the seeding of amyloid formation: implications for the pathogenesis of Alzheimer’s disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 10.Esler WP, Wolfe MS. A portrait of Alzheimer secretases—new features and familiar faces. Science. 2001;293:1449–1454. doi: 10.1126/science.1064638. [DOI] [PubMed] [Google Scholar]
- 11.Hardy JA, Higgins GA. Alzheimer’s disease: the amyloid cascade hypothesis. Science. 1992;256:184–185. doi: 10.1126/science.1566067. [DOI] [PubMed] [Google Scholar]
- 12.Mann DM, Yates PO, Marcyniuk B. Alzheimer’s presenile dementia, senile dementia of Alzheimer type and Down’s syndrome in middle age form an age related continuum of pathological changes. Neuropathol Appl Neurobiol. 1984;10:185–207. doi: 10.1111/j.1365-2990.1984.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 13.Dyrks T, Weidemann A, Multhaup G, Salbaum JM, Lemaire HG, Kang J, Müller-Hill B, Masters CL, Beyreuther K. Identification, transmembrane orientation and biogenesis of the amyloid A4 precursor of Alzheimer’s disease. EMBO J. 1988;7:949–957. doi: 10.1002/j.1460-2075.1988.tb02900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gralle M, Ferreira ST. Structure and functions of the human amyloid precursor protein: The whole is more than the sum of its parts. Prog Neurobiol. 2007;82:11–32. doi: 10.1016/j.pneurobio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Paliga K, Peraus G, Kreger S, Dürrwang U, Hesse L, Multhaup G, Masters CL, Beyreuther K, Weidemann A. Human amyloid precursor-like protein 1—cDNA cloning, ectopic expression in COS-7 cells and identification of soluble forms in the cerebrospinal fluid. Eur J Biochem. 1997;250:354–363. doi: 10.1111/j.1432-1033.1997.0354a.x. [DOI] [PubMed] [Google Scholar]
- 16.Slunt HH, Thinakaran G, Von Koch C, Lo AC, Tanzi RE, Sisodia SS. Expression of a ubiquitous, cross-reactive homologue of the mouse β-amyloid precursor protein (APP) J Biol Chem. 1994;269:2637–2644. [PubMed] [Google Scholar]
- 17.Lorent K, Overbergh L, Moechars D, De Strooper B, Van Leuven F, Van den Berghe H. Expression in mouse embryos and in adult mouse brain of three members of the amyloid precursor protein family, of the α2-macroglobulin receptor/low density lipoprotein receptor-related protein and of its ligands apolipoprotein E, lipoprotein lipase, α2-macroglobulin and the 40,000 molecular weight receptor-associated protein. Neuroscience. 1995;65:1009–1025. doi: 10.1016/0306-4522(94)00555-J. [DOI] [PubMed] [Google Scholar]
- 18.Hornsten A, Lieberthal J, Fadia S, Malins R, Ha L, Xu X, Daigle I, Markowitz M, O’Connor G, Plasterk R, Li C. APL-1, a Caenorhabditis elegans protein related to the human β-amyloid precursor protein, is essential for viability. Proc Natl Acad Sci USA. 2007;104:1971–1976. doi: 10.1073/pnas.0603997104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo L, Martin-Morris L, White K. Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J Neurosci. 1990;10:3849–3861. doi: 10.1523/JNEUROSCI.10-12-03849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao X, Südhof TC. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 21.Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron MM, Teplow DB, Haass C. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep. 2001;2:835–841. doi: 10.1093/embo-reports/kve180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu C, Kim SH, Ikeuchi T, Xu H, Gasparini L, Wang R, Sisodia SS. Characterization of a presenilin-mediated amyloid precursor protein carboxyl-terminal fragment γ. Evidence for distinct mechanisms involved in γ-secretase processing of the APP and Notch1 transmembrane domains. J Biol Chem. 2001;276:43756–43760. doi: 10.1074/jbc.C100410200. [DOI] [PubMed] [Google Scholar]
- 23.Weidemann A, Eggert S, Reinhard FB, Vogel M, Paliga K, Baier G, Masters CL, Beyreuther K, Evin G. A novel ε-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry. 2002;41:2825–2835. doi: 10.1021/bi015794o. [DOI] [PubMed] [Google Scholar]
- 24.Zhao G, Mao G, Tan J, Dong Y, Cui MZ, Kim SH, Xu X. Identification of a new presenilin-dependent ζ-cleavage site within the transmembrane domain of amyloid precursor protein. J Biol Chem. 2004;279:50647–50650. doi: 10.1074/jbc.C400473200. [DOI] [PubMed] [Google Scholar]
- 25.Esch FS, Keim PS, Beattie EC, Blacher RW, Culwell AR, Oltersdorf T, McClure D, Ward PJ. Cleavage of amyloid-β peptide during constitutive processing of its precursor. Science. 1990;248:1122–1124. doi: 10.1126/science.2111583. [DOI] [PubMed] [Google Scholar]
- 26.Sisodia SS, Koo EH, Beyreuther K, Unterbeck A, Price DL. Evidence that β-amyloid protein in Alzheimer’s disease is not derived by normal processing. Science. 1990;248:492–495. doi: 10.1126/science.1691865. [DOI] [PubMed] [Google Scholar]
- 27.Roberts SB, Ripellino JA, Ingalls KM, Robakis NK, Felsenstein KM. Non-amyloidogenic cleavage of the β-amyloid precursor protein by an integral membrane metalloendopeptidase. J Biol Chem. 1994;269:3111–3116. [PubMed] [Google Scholar]
- 28.Howard L, Lu X, Mitchell S, Griffiths S, Glynn P. Molecular cloning of MADM: a catalytically active mammalian disintegrin-metalloprotease expressed in various cell types. Biochem J. 1996;317:45–50. doi: 10.1042/bj3170045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hussain I, Powell D, Howlett DR, Tew DG, Meek TD, Chapman C, Gloger IS, Murphy KE, Southan CD, Ryan DM, Smith TS, Simmons DL, Walsh FS, Dingwall C, Christie G. Identification of a novel aspartic protease (Asp 2) as β-secretase. Mol Cell Neurosci. 1999;14:419–427. doi: 10.1006/mcne.1999.0811. [DOI] [PubMed] [Google Scholar]
- 30.Sinha S, Anderson JP, Barbour R, Basi GS, Caccavello R, Davis D, Doan M, Dovey HF, Frigon N, Hong J, Jacobson-Croak K, Jewett N, Keim P, Knops J, Lieberburg I, Power M, Tan H, Tatsuno G, Tung J, Schenk D, Seubert P, Suomensaari SM, Wang S, Walker D, Zhao J, McConlogue L, John V. Purification and cloning of amyloid precursor protein β-secretase from human brain. Nature. 1999;402:537–540. doi: 10.1038/990114. [DOI] [PubMed] [Google Scholar]
- 31.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. β-secretase cleavage of Alzheimer’s amyloid precursor protein by the transmembrane aspartic protease BACE. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 32.Yan R, Bienkowski MJ, Shuck ME, Miao H, Tory MC, Pauley AM, Brashier JR, Stratman NC, Mathews WR, Buhl AE, Carter DB, Tomasselli AG, Parodi LA, Heinrikson RL, Gurney ME. Membrane-anchored aspartyl protease with Alzheimer’s disease β-secretase activity. Nature. 1999;402:533–537. doi: 10.1038/990107. [DOI] [PubMed] [Google Scholar]
- 33.Luo Y, Bolon B, Kahn S, Bennett BD, Babu-Khan S, Denis P, Fan W, Kha H, Zhang J, Gong Y, Martin L, Louis JC, Yan Q, Richards WG, Citron M, Vassar R. Mice deficient in BACE1, the Alzheimer’s β-secretase, have normal phenotype and abolished β-amyloid generation. Nat Neurosci. 2001;4:231–232. doi: 10.1038/85059. [DOI] [PubMed] [Google Scholar]
- 34.Roberds SL, Anderson J, Basi G, Bienkowski MJ, Branstetter DG, Chen KS, Freedman SB, Frigon NL, Games D, Hu K, Johnson-Wood K, Kappenman KE, Kawabe TT, Kola I, Kuehn R, Lee M, Liu W, Motter R, Nichols NF, Power M, Robertson DW, Schenk D, Schoor M, Shopp GM, Shuck ME, Sinha S, Svensson KA, Tatsuno G, Tintrup H, Wijsman J, Wright S, McConlogue L. BACE knockout mice are healthy despite lacking the primary beta-secretase activity in brain: implications for Alzheimer’s disease therapeutics. Hum Mol Genet. 2001;10:1317–1324. doi: 10.1093/hmg/10.12.1317. [DOI] [PubMed] [Google Scholar]
- 35.Ehehalt R, Keller P, Haass C, Thiele C, Simons K. Amyloidogenic processing of the Alzheimer β-amyloid precursor protein depends on lipid rafts. J Cell Biol. 2003;160:113–123. doi: 10.1083/jcb.200207113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goutte C, Hepler W, Mickey KM, Priess JR. aph-2 encodes a novel extracellular protein required for GLP-1-mediated signaling. Development. 2000;127:2481–2492. doi: 10.1242/dev.127.11.2481. [DOI] [PubMed] [Google Scholar]
- 37.Yu G, Nishimura M, Arawaka S, Levitan D, Zhang L, Tandon A, Song YQ, Rogaeva E, Chen F, Kawarai T, Supala A, Levesque L, Yu H, Yang DS, Holmes E, Milman P, Liang Y, Zhang DM, Xu DH, Sato C, Rogaev E, Smith M, Janus C, Zhang Y, Aebersold R, Farrer LS, Sorbi S, Bruni A, Fraser P, St George-Hyslop P. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and βAPP processing. Nature. 2000;407:48–54. doi: 10.1038/35024009. [DOI] [PubMed] [Google Scholar]
- 38.Francis R, McGrath G, Zhang J, Ruddy DA, Sym M, Apfeld J, Nicoll M, Maxwell M, Hai B, Ellis MC, Parks AL, Xu W, Li J, Gurney M, Myers RL, Himes CS, Hiebsch R, Ruble C, Nye JS, Curtis D. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev Cell. 2002;3:85–97. doi: 10.1016/S1534-5807(02)00189-2. [DOI] [PubMed] [Google Scholar]
- 39.Edbauer D, Winkler E, Regula JT, Pesold B, Steiner H, Haass C. Reconstitution of γ-secretase activity. Nat Cell Biol. 2003;5:486–488. doi: 10.1038/ncb960. [DOI] [PubMed] [Google Scholar]
- 40.Yu G, Chen F, Levesque G, Nishimura M, Zhang DM, Levesque L, Rogaeva E, Xu D, Liang Y, Duthie M, St George-Hyslop PH, Fraser PE. The presenilin 1 protein is a component of a high molecular weight intracellular complex that contains beta-catenin. J Biol Chem. 1998;273:16470–16475. doi: 10.1074/jbc.273.26.16470. [DOI] [PubMed] [Google Scholar]
- 41.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature. 1999;398:513–517. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 42.Shah S, Lee SF, Tabuchi K, Hao YH, Yu C, LaPlant Q, Ball H, Dann CE, Sudhof T, Yu G. Nicastrin functions as a gamma-secretase substrate receptor. Cell. 2005;122:435–447. doi: 10.1016/j.cell.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 43.Verdile G, Gandy SE, Martins RN. The role of presenilin and its interacting proteins in the biogenesis of Alzheimer’s beta amyloid. Neurochem Res. 2006;32:609–623. doi: 10.1007/s11064-006-9131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wolfe MS. The γ-secretase complex: membrane-embedded proteolytic ensemble. Biochemistry. 2006;45:7931–7939. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 45.Naruse S, Thinakaran G, Luo JJ, Kusiak JW, Tomita T, Iwatsubo T, Qian X, Ginty DD, Price DL, Borchelt DR, Wong PC, Sisodia SS. Effects of PS1 deficiency on membrane protein trafficking in neurons. Neuron. 1998;21:1213–1221. doi: 10.1016/S0896-6273(00)80637-6. [DOI] [PubMed] [Google Scholar]
- 46.Gu Y, Misonou H, Sato T, Dohmae N, Takio K, Ihara Y. Distinct intramembrane cleavage of the β-amyloid precursor protein family resembling γ-secretase-like cleavage of Notch. J Biol Chem. 2001;276:35235–35238. doi: 10.1074/jbc.C100357200. [DOI] [PubMed] [Google Scholar]
- 47.Holback S, Adlerz L, Iverfeldt K. Increased processing of APLP2 and APP with concomitant formation of APP intracellular domains in BDNF and retinoic acid-differentiated human neuroblastoma cells. J Neurochem. 2005;95:1059–1068. doi: 10.1111/j.1471-4159.2005.03440.x. [DOI] [PubMed] [Google Scholar]
- 48.Eggert S, Paliga K, Soba P, Evin G, Masters CL, Weidemann A, Beyreuther K. The proteolytic processing of the amyloid precursor protein gene family members APLP-1 and APLP-2 involves α-, β-, γ-, and ε-like cleavages: modulation of APLP-1 processing by N-glycosylation. J Biol Chem. 2004;279:18146–18156. doi: 10.1074/jbc.M311601200. [DOI] [PubMed] [Google Scholar]
- 49.Endres K, Postina R, Schroeder A, Mueller U, Fahrenholz F. Shedding of the amyloid precursor protein-like protein APLP2 by disintegrin-metalloproteinases. FEBS J. 2005;272:5808–5820. doi: 10.1111/j.1742-4658.2005.04976.x. [DOI] [PubMed] [Google Scholar]
- 50.Rooke J, Pan D, Xu T, Rubin GM. KUZ, a conserved metalloprotease-disintegrin protein with two roles in Drosophila neurogenesis. Science. 1996;273:1227–1231. doi: 10.1126/science.273.5279.1227. [DOI] [PubMed] [Google Scholar]
- 51.Carmine-Simmen K, Proctor T, Tschäpe J, Poeck B, Triphan T, Strauss R, Kretzschmar D. Neurotoxic effects induced by the Drosophila amyloid-β peptide suggest a conserved toxic function. Neurobiol Dis. 2008;33:274–281. doi: 10.1016/j.nbd.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hong CS, Koo EH. Isolation and characterization of Drosophila presenilin homolog. Neuroreport. 1997;8:665–668. doi: 10.1097/00001756-199702100-00017. [DOI] [PubMed] [Google Scholar]
- 53.Boulianne GL, Livine-Bar I, Humphreys JM, Liang Y, Lin C, Rogaev E, George-Hyslop PH. Cloning and characterization of the Drosophila presenilin homologue. Neuroreport. 1997;8:1025–1029. doi: 10.1097/00001756-199703030-00041. [DOI] [PubMed] [Google Scholar]
- 54.Sprecher CA, Grant FJ, Grimm G, O’Hara PJ, Norris F, Norris K, Foster DC. Molecular cloning of the cDNA for a human amyloid precursor protein homolog: evidence for a multigene family. Biochemistry. 1993;32:4481–4486. doi: 10.1021/bi00068a002. [DOI] [PubMed] [Google Scholar]
- 55.Weidemann A, König Gr, Bunke D, Fischer P, Salbaum JM, Masters CL, Beyreuther K. Identification, biogenesis, and localization of precursors of Alzheimer’s disease A4 amyloid protein. Cell. 1989;57:115–126. doi: 10.1016/0092-8674(89)90177-3. [DOI] [PubMed] [Google Scholar]
- 56.Lyckman AW, Confaloni AM, Thinakaran G, Sisodia SS, Moya KL. Post-translational processing and turnover kinetics of presynaptically targeted amyloid precursor superfamily proteins in the central nervous system. J Biol Chem. 1998;273:11100–11106. doi: 10.1074/jbc.273.18.11100. [DOI] [PubMed] [Google Scholar]
- 57.Pangalos MN, Shioi J, Robakis NK. Expression of the chondroitin sulfate proteoglycans of amyloid precursor (appican) and amyloid precursor-like protein 2. J Neurochem. 1995;65:762–769. doi: 10.1046/j.1471-4159.1995.65020762.x. [DOI] [PubMed] [Google Scholar]
- 58.Thinakaran G, Sisodia SS. Amyloid precursor-like protein 2 (APLP2) is modified by the addition of chondroitin sulfate glycosaminoglycan at a single site. J Biol Chem. 1994;269:22099–22104. [PubMed] [Google Scholar]
- 59.Nakagawa K, Kitazume S, Oka R, Maruyama K, Saido TC, Sato Y, Endo T, Hashimoto Y. Sialylation enhances the secretion of neurotoxic amyloid-β peptides. J Neurochem. 2006;96:924–933. doi: 10.1111/j.1471-4159.2005.03595.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhang YQ, Sarge KD. Sumoylation of amyloid precursor protein negatively regulates Aβ aggregation levels. Biochem Biophys Res Commun. 2008;374:673–678. doi: 10.1016/j.bbrc.2008.07.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gandy S, Czernik AJ, Greengard P. Phosphorylation of Alzheimer disease amyloid precursor peptide by protein kinase C and Ca2+/calmodulin-dependent protein kinase II. Proc Natl Acad Sci USA. 1988;85:6218–6221. doi: 10.1073/pnas.85.16.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Knops J, Gandy S, Greengard P, Lieberburg I, Sinha S. Serine phosphorylation of the secreted extracellular domain of APP. Biochem Biophys Res Commun. 1993;197:380–385. doi: 10.1006/bbrc.1993.2490. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki T, Oishi M, Marshak DR, Czernik AJ, Nairn AC, Greengard P. Cell cycle-dependent regulation of the phosphorylation and metabolism of the Alzheimer amyloid precursor protein. EMBO J. 1994;13:1114–1122. doi: 10.1002/j.1460-2075.1994.tb06360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oishi M, Nairn AC, Czernik AJ, Lim GS, Isohara T, Gandy SE, Greengard P, Suzuki T. The cytoplasmic domain of Alzheimer’s amyloid precursor protein is phosphorylated at Thr654, Ser655, and Thr668 in adult rat brain and cultured cells. Mol Med. 1997;3:111–123. [PMC free article] [PubMed] [Google Scholar]
- 65.Lee MS, Kao SC, Lemere CA, Xia W, Tseng HC, Zhou Y, Neve R, Ahlijanian MK, Tsai LH. APP processing is regulated by cytoplasmic phosphorylation. J Cell Biol. 2003;163:83–95. doi: 10.1083/jcb.200301115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kimberly WT, Zheng JB, Town T, Flavell RA, Selkoe DJ. Physiological regulation of the beta-amyloid precursor protein signalling domain byc-Jun N-terminal kinase JNK3 during neuronal differentiation. J Neurosci. 2005;25:5533–5543. doi: 10.1523/JNEUROSCI.4883-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Iijima K, Ando K, Takeda S, Satoh Y, Seki T, Itohara S, Greengard P, Kirino Y, Nairn AC, Suzuki T. Neuron-specific phosphorylation of Alzheimer’s β-amyloid precursor protein by cyclin-dependent kinase 5. J Neurochem. 2000;75:1085–1091. doi: 10.1046/j.1471-4159.2000.0751085.x. [DOI] [PubMed] [Google Scholar]
- 68.Aplin AE, Gibb GM, Jacobsen JS, Gallo JM, Anderton BH. In vitro phosphorylation of the cytoplasmic domain of the amyloid precursor protein by glycogen synthase kinase-3β. J Neurochem. 1996;67:699–707. doi: 10.1046/j.1471-4159.1996.67020699.x. [DOI] [PubMed] [Google Scholar]
- 69.Muresan Z, Muresan V. Coordinated transport of phosphorylated amyloid-β precursor protein and c-Jun NH2 terminal kinase-interacting protein-1. J Cell Biol. 2005;171:615–625. doi: 10.1083/jcb.200502043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sano Y, Nakaya T, Pedrini S, Takeda S, Iijima-Ando K, Iijima K, Mathews PM, Shigeyoshi I, Gandy S, Suzuki T. Physiological mouse brain Aβ levels are not correlated to the phosphorylation state of threonine-668 of Alzheimer’s APP. PLoS ONE. 2006;1:e51. doi: 10.1371/journal.pone.0000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Feyt C, Pierrot N, Tasiaux B, Hees JV, Kienlen-Campard P, Courtoy PJ, Octave JN. Phosphorylation of APP695 at Thr668 decreases γ-cleavage and extracellular Aβ. Biochem Biophys Res Commun. 2007;357:1004–1010. doi: 10.1016/j.bbrc.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 72.Takahashi K, Niidome T, Akaike A, Kihara T, Sugimoto H. Phosphorylation of amyloid protein (APP) at Tyr687 regulates APP processing by α- and γ-secretase. Biochem Biophys Res Commun. 2008;377:544–549. doi: 10.1016/j.bbrc.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki T, Ando K, Isohara T, Oishi M, Lim GS, Satoh Y, Wasco W, Tanzi RE, Nairn AC, Greengard P, Gandy SE, Kirino Y. Phosphorylation of Alzheimer β-amyloid precursor-like proteins. Biochemistry. 1997;36:4643–4649. doi: 10.1021/bi962618k. [DOI] [PubMed] [Google Scholar]
- 74.Taru H, Suzuki T. Facilitation of stress-induced phosphorylation of beta-amyloid precursor protein family members by X11-like/Mint2 protein. J Biol Chem. 2004;279:21628–21636. doi: 10.1074/jbc.M312007200. [DOI] [PubMed] [Google Scholar]
- 75.Ando K, Iijima K, Elliott JI, Kirino Y, Suzuki T. Phosphorylation-dependent regulation of the interaction of amyloid precursor protein with Fe65 affects the production of β-amyloid. J Biol Chem. 2001;276:40353–40361. doi: 10.1074/jbc.M106460200. [DOI] [PubMed] [Google Scholar]
- 76.Pastorino L, Sun A, Lu PJ, Zhou XZ, Balastik M, Finn G, Wulf G, Lim J, Li SH, Li X, Xia W, Nicholson LK, Lu KP. The prolyl isomerase Pin1 regulates amyloid precursor protein processing and amyloid-β production. Nature. 2006;440:528–533. doi: 10.1038/nature04543. [DOI] [PubMed] [Google Scholar]
- 77.Liou YC, Sun A, Ryo A, Zhou XZ, Yu ZX, Huang HK, Uchida T, Bronson R, Bing G, Li X, Hunter T, Lu KP. Role of the isomerase Pin1 in protecting against age-dependent neurododegeneration. Nature. 2003;424:556–561. doi: 10.1038/nature01832. [DOI] [PubMed] [Google Scholar]
- 78.Zheng H, Jiang M, Trumbauer ME, Sirinathsinghji DJ, Hopkins R, Smith DW, Heavens RP, Dawson GR, Boyce S, Conner MW, Stevens KA, Slunt HH, Sisoda SS, Chen HY, Van der Ploeg LH. β-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell. 1995;81:525–531. doi: 10.1016/0092-8674(95)90073-X. [DOI] [PubMed] [Google Scholar]
- 79.von Koch CS, Zheng H, Chen H, Trumbauer M, Thinakaran G, van der Ploeg LH, Price DL, Sisodia SS. Generation of APLP2 KO mice and early postnatal lethality in APLP2/APP double KO mice. Neurobiol Aging. 1997;18:661–669. doi: 10.1016/S0197-4580(97)00151-6. [DOI] [PubMed] [Google Scholar]
- 80.Heber S, Herms J, Gajic V, Hainfellner J, Aguzzi A, Rülicke T, von Kretzschmar H, von Koch C, Sisodia S, Tremml P, Lipp HP, Wolfer DP, Müller U. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J Neurosci. 2000;20:7951–7963. doi: 10.1523/JNEUROSCI.20-21-07951.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Herms J, Anliker B, Heber S, Ring S, Fuhrmann M, Kretzschmar H, Sisodia S, Müller U. Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. EMBO J. 2004;23:4106–4115. doi: 10.1038/sj.emboj.7600390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schubert D, Jin LW, Saitoh T, Cole G. The regulation of amyloid beta protein precursor secretion and its modulatory role in cell adhesion. Neuron. 1989;3:689–694. doi: 10.1016/0896-6273(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 83.Narindrasorasak S, Lowery DE, Altman RA, Gonzalez-DeWhitt P, Greenberg B, Kisilevsky R. Characterization of high affinity binding between laminin and Alzheimer’s disease amyloid precursor proteins. Lab Invest. 1992;67:643–652. [PubMed] [Google Scholar]
- 84.Wang Y, Ha Y. X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol Cell. 2004;15:343–353. doi: 10.1016/j.molcel.2004.06.037. [DOI] [PubMed] [Google Scholar]
- 85.Soba P, Eggert S, Wagner K, Zentgraf H, Siehl K, Kreger S, Löwer A, Langer A, Merdes G, Paro R, Masters CL, Müller U, Kins S, Beyreuther K. Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J. 2005;24:3624–3634. doi: 10.1038/sj.emboj.7600824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kaden D, Munter L, Joshi M, Treiber C, Weise C, Bethge T, Voigt P, Schaefer M, Beyermann M, Reif B, Multhaup G. Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect β-secretase cleavage of APP. J Biol Chem. 2008;283:7271–7279. doi: 10.1074/jbc.M708046200. [DOI] [PubMed] [Google Scholar]
- 87.Small D, Nurcombe V, Reed G, Clarris H, Moir R, Beyreuther K, Masters CL. A heparin-binding domain in the amyloid protein precursor of Alzheimer’s disease is involved in the regulation of neurite outgrowth. J Neurosci. 1994;14:2117–2127. doi: 10.1523/JNEUROSCI.14-04-02117.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Milward EA, Papadopoulos R, Fuller SJ, Moir RD, Small D, Beyreuther K, Masters CL. The amyloid protein precursor of Alzheimer’s disease is a mediator of the effects of nerve growth factor on neurite outgrowth. Neuron. 1992;9:129–137. doi: 10.1016/0896-6273(92)90228-6. [DOI] [PubMed] [Google Scholar]
- 89.Perez RG, Zheng H, Van der Ploeg LH, Koo EH. The β-amyloid precursor protein of Alzheimer’s disease enhances neuron viability and modulates neuronal polarity. J Neurosci. 1997;17:9407–9414. doi: 10.1523/JNEUROSCI.17-24-09407.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Young-Pearse TL, Chen AC, Chang R, Marquez C, Selkoe DJ. Secreted APP regulates the function of full-length APP in neurite outgrowth through interactions with integrin beta 1. Neural Develop. 2008;3:1–13. doi: 10.1186/1749-8104-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Y, Liu T, Peng Y, Yuan C, Guo A. Specific functions of Drosophilia amyloid precursor-like protein in the development of nervous sydtem and nonneuronal tissues. J Neurobiol. 2004;61:343–358. doi: 10.1002/neu.20048. [DOI] [PubMed] [Google Scholar]
- 92.Roch JM, Masliah E, Roch-Levecq AC, Sundsmo MP, Otero DA, Veinbergs I, Saitoh T. Increase of synaptic density and memory retention by a peptide representing the trophic domain of the amyloid β/A4 protein precursor. Proc Natl Acad Sci USA. 1994;91:7450–7454. doi: 10.1073/pnas.91.16.7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Moya KL, Benowitz LI, Schneider GE, Allinquant B. The amyloid precursor protein is developmentally regulated and correlated with synaptogenesis. Dev Biol. 1994;161:597–603. doi: 10.1006/dbio.1994.1055. [DOI] [PubMed] [Google Scholar]
- 94.Young-Pearse TL, Bai J, Chang R, Zheng JB, LoTurco JJ, Selkoe DJ. A critical function for beta-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J Neurosci. 2007;27:14459–14469. doi: 10.1523/JNEUROSCI.4701-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ma QH, Futagawa T, Yang WL, Jiang XD, Zeng L, Takeda Y, Xu RX, Bagnard D, Schachner M, Furley AJ, Karagogeos D, Watanabe K, Dawe GS, Xiao ZC. A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis. Nat Cell Biol. 2008;10:283–294. doi: 10.1038/ncb1690. [DOI] [PubMed] [Google Scholar]
- 96.Leyssen M, Ayaz D, Hebert SS, Reeve S, De Strooper B, Hassan BA. Amyloid precursor protein promotes post-developmental neurite arborization in the Drosophila brain. EMBO J. 2005;24:2944–2955. doi: 10.1038/sj.emboj.7600757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Araki W, Kitaguchi N, Tokushima Y, Ishii K, Aratake H, Shimohama S, Nakamura S, Kimura J. Trophic effect of β-amyloid precursor protein on cerebral cortical neurons in culture. Biochem Biophys Res Commun. 1991;181:265–271. doi: 10.1016/S0006-291X(05)81412-3. [DOI] [PubMed] [Google Scholar]
- 98.Mattson MP, Cheng B, Culwell AR, Esch FS, Lieberburg I, Rydel RE. Evidence for excitoprotective and intraneuronal calcium-regulating roles for secreted forms of the β-amyloid precursor protein. Neuron. 1993;10:243–254. doi: 10.1016/0896-6273(93)90315-I. [DOI] [PubMed] [Google Scholar]
- 99.Goodman Y, Mattson MP. Secreted forms of beta-amyloid precursor protein protect hippocampal neurons against amyloid beta-peptide-induced oxidative injury. Exp Neurol. 1994;128:1–12. doi: 10.1006/exnr.1994.1107. [DOI] [PubMed] [Google Scholar]
- 100.Kinoshita A, Whelan CM, Berezovska O, Hyman BT. The γ secretase carboxyl-terminal domain of the amyloid precursor protein induces apoptrosis via Top60 in H4 cells. J Biol Chem. 2002;277:28530–28536. doi: 10.1074/jbc.M203372200. [DOI] [PubMed] [Google Scholar]
- 101.Nakayama K, Ohkawara T, Hiratochi M, Koh CS, Nagase H. The intracellular domain of amyloid precursor protein induces neuron-specific apoptosis. Neurosci Lett. 2008;44:127–131. doi: 10.1016/j.neulet.2008.08.034. [DOI] [PubMed] [Google Scholar]
- 102.Kim HS, Kim EM, Lee JP, Park CH, Kim S, Seo JH, Chang KA, Yu E, Jeong SJ, Chong YH, Suh YH. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3β expression. FASEB J. 2003;17:1951–1953. doi: 10.1096/fj.03-0106fje. [DOI] [PubMed] [Google Scholar]
- 103.Ozaki T, Li Y, Kikuchi H, Tomita T, Iwatsubo T, Nakagawara A. The intracellular domain of the amyloid precursor protein (AICD) enhances the p53-mediated apoptosis. Biochem Biophys Res Commun. 2006;351:57–63. doi: 10.1016/j.bbrc.2006.09.162. [DOI] [PubMed] [Google Scholar]
- 104.Tang X, Milayavsky M, Goldfinger N, Rotter V. Amyloid-beta precursor-like protein APLP1 is a novel p53 transcriptional target gene that augments neuroblastoma cell death upon genotoxic stress. Oncogene. 2007;26:7302–7312. doi: 10.1038/sj.onc.1210542. [DOI] [PubMed] [Google Scholar]
- 105.Furukawa K, Mattson MP. Secreted amyloid precursor protein alpha selectively suppresses NMDA currents in hippocampal neurons: involvement of cyclic GMP. Neuroscience. 1997;83:429–438. doi: 10.1016/S0306-4522(97)00398-9. [DOI] [PubMed] [Google Scholar]
- 106.Furukawa K, Barger SW, Blalock EM, Mattson MP. Activation of K+ channels and suppression of neuronal activity by secreted β-amyloid-precursor protein. Nature. 1996;379:74–78. doi: 10.1038/379074a0. [DOI] [PubMed] [Google Scholar]
- 107.Doyle E, Bruce MT, Breen KC, Smith DC, Anderton B, Regan CM. Intraventricular infusions of antibodies to amyloid-beta-protein precursor impair the acquisition of a passive avoidance response in the rat. Neurosci Lett. 1990;115:97–102. doi: 10.1016/0304-3940(90)90524-D. [DOI] [PubMed] [Google Scholar]
- 108.Huber G, Martin JR, Loffler J, Moreau JL. Involvement of amyloid precursor protein in memory formation in the rat: an indirect antibody approach. Brain Res. 1993;603:348–352. doi: 10.1016/0006-8993(93)91261-P. [DOI] [PubMed] [Google Scholar]
- 109.Fazeli MS, Breen K, Errington ML, Bliss TV. Increase in extracellular NCAM and amyloid precursor protein following induction of long-term potentiation in the dentate gyrus of anaesthetized rats. Neurosci Lett. 1994;169:77–80. doi: 10.1016/0304-3940(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 110.Nabeshima T, Nitta A. Memory impairment and neuronal dysfunction induced by beta-amyloid protein in rats. J Exp Med. 1994;174:241–249. doi: 10.1620/tjem.174.241. [DOI] [PubMed] [Google Scholar]
- 111.Chen QS, Kagan BL, Hirakura Y, Xie CW. Impairment of hippocampal long-term potentiation by Alzheimer amyloid beta-peptides. J Neurosci Res. 2000;60:65–72. doi: 10.1002/(SICI)1097-4547(20000401)60:1<65::AID-JNR7>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 112.Puzzo D, Privitera L, Leznik E, Fa M, Staniszewski A, Palmeri A, Arancio O. Picomolar amyloid-beta positively modulates synaptic plasticity and memory in hippocampus. J Neurosci. 2008;28:14537–14545. doi: 10.1523/JNEUROSCI.2692-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Koo EH, Sisodia SS, Archer DR, Martin LJ, Weidemann A, Beyreuther K, Fischer P, Masters CL, Price DL. Precursor of amyloid protein in Alzheimer disease undergoes fast anterograde axonal transport. Proc Natl Acad Sci USA. 1990;87:1561–1565. doi: 10.1073/pnas.87.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schubert W, Prior R, Weidemann A, Dircksen H, Multhaup G, Masters CL, Beyreuther K. Localization of Alzheimer ßA4 amyloid precursor protein at central and peripheral synaptic sites. Brain Res. 1990;563:184–194. doi: 10.1016/0006-8993(91)91532-6. [DOI] [PubMed] [Google Scholar]
- 115.Kamal A, Stokin GB, Yang Z, Xia CH, Goldstein LS. Axonal transport of amyloid precursor protein is mediated by direct binding to the kinesin light chain subunit of kinesin-I. Neuron. 2000;28:449–459. doi: 10.1016/S0896-6273(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 116.Gunawardena S, Goldstein LS. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophilia . Neuron. 2001;32:389–401. doi: 10.1016/S0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- 117.Lazarov O, Morfini GD, Lee EB, Farah MH, Szodorai A, DeBoer SR, Koliatsos VE, Kins S, Lee VM, Wong PC, Price DL, Brady ST, Sisoda SS. Axonal transport, amyloid precursor protein, kinesin-1, and the processing apparatus: revisited. J Neurosci. 2005;25:2386–2395. doi: 10.1523/JNEUROSCI.3089-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Needham BE, Wlodek ME, Ciccotosto GD, Fam BC, Masters CL, Proietto J, Andrikopoulos S, Cappai R. Identification of the Alzheimer’s disease amyloid precursor protein (APP) and its homologue APLP2 as essential modulators of glucose and insulin homeostasis and growth. J Pathol. 2008;215:155–163. doi: 10.1002/path.2343. [DOI] [PubMed] [Google Scholar]
- 119.Lee Y, Tharp WG, Maple RL, Nair S, Permana PA, Pratley RE. Amyloid precursor protein expression is upregulated in adipocytes in obesity. Obesity. 2008;16:1493–1500. doi: 10.1038/oby.2008.267. [DOI] [PubMed] [Google Scholar]
- 120.Fiore F, Zambrano N, Minopoli G, Donini V, Duilio A, Russo T. The regions of the Fe65 protein homologous to the phosphotyrosine interaction/phosphotyrosine binding domain of Shc bind the intracellular domain of the Alzheimer’s amyloid precursor protein. J Biol Chem. 1995;270:30853–30856. doi: 10.1074/jbc.270.52.30853. [DOI] [PubMed] [Google Scholar]
- 121.Guénette SY, Chen J, Jondro PD, Tanzi RE. Association of a novel human FE65-like protein with the cytoplasmic domain of the β-amyloid precursor protein. Proc Natl Acad Sci USA. 1996;93:10832–10837. doi: 10.1073/pnas.93.20.10832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Duilio A, Faraonio R, Minopoli G, Zambrano N, Russo T. Fe65L2: a new member of Fe65 protein family interacting with the intracellular domain of Alzheimer’s β-amyloid precursor protein. Biochem J. 1998;330:513–519. doi: 10.1042/bj3300513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Scheinfeld MH, Ghersi E, Laky K, Fowlkes BJ, D’Adamio L. Processing of β-amyloid precursor-like protein-1 and -2 by γ-secretase regulates transcription. J Biol Chem. 2002;277:44195–44201. doi: 10.1074/jbc.M208110200. [DOI] [PubMed] [Google Scholar]
- 124.Zambrano N, Bimonte M, Arbucci S, Gianni D, Russo T, Bazzicalupo P. feh-1 and apl-1, the Caenorhabditis elegans orthologues of mammalian Fe65 and β-amyloid precursor protein genes, are involved in the same pathway that controls nematode pharyngeal pumping. J Cell Sci. 2002;115:1411–1422. doi: 10.1242/jcs.115.7.1411. [DOI] [PubMed] [Google Scholar]
- 125.Guénette S, Chang Y, Hiesberger T, Richardson JA, Eckman CB, Eckman EA, Hammer RE, Herz J. Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development. EMBO J. 2006;25:420–431. doi: 10.1038/sj.emboj.7600926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Borg JP, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domain of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sabo SL, Lanier LM, Ikin AF, Khorkova O, Sahasrabudhe S, Greengard P, Buxbaum JD. Regulation of β-amyloid secretion by FE65, an amyloid protein precursor-binding protein. J Biol Chem. 1999;274:7952–7957. doi: 10.1074/jbc.274.12.7952. [DOI] [PubMed] [Google Scholar]
- 128.Hu Q, Wang L, Yang Z, Cool BH, Zitnik G, Martin GM. Endoproteolytic cleavage of FE65 converts the adaptor protein to a potent suppressor of the sAPPα pathway in primates. J Biol Chem. 2005;280:12548–12558. doi: 10.1074/jbc.M411855200. [DOI] [PubMed] [Google Scholar]
- 129.Chang KA, Kim HS, Ha TY, Ha JW, Shin KY, Jeong YH, Lee JP, Park CH, Kim S, Baik TK, Suh YH. Phosphorylation of amyloid precursor protein (APP) at Thr668 regulates the nuclear translocation of the APP intracellular domain and induces neurodegeneration. Mol Cell Biol. 2006;26:4327–4338. doi: 10.1128/MCB.02393-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nakaya T, Kawai T, Suzuki T. Regulation of FE65 nuclear translocation and function by amyloid β-protein precursor in osmotically stressed cells. J Biol Chem. 2008;283:19119–19131. doi: 10.1074/jbc.M801827200. [DOI] [PubMed] [Google Scholar]
- 131.Nakaya T, Suzuki T. Role of APP phosphorylation in FE65-dependent gene transactivation mediated by AICD. Genes Cells. 2006;11:633–645. doi: 10.1111/j.1365-2443.2006.00968.x. [DOI] [PubMed] [Google Scholar]
- 132.Radzimanowski J, Simon B, Sattler M, Beyreuther K, Sinning I, Wild K. Structure of the intracellular domain of the amyloid precursor protein in complex with Fe65-PTB2. EMBO Rep. 2008;9:1134–1140. doi: 10.1038/embor.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tanahashi H, Tabira T. X11L2, a new member of the X11 protein family, interacts with Alzheimer’s β-amyloid precursor protein. Biochem Biophys Res Commun. 1999;255:663–667. doi: 10.1006/bbrc.1999.0265. [DOI] [PubMed] [Google Scholar]
- 134.Borg JP, Yang Y, De Taddéo-Borg M, Margolis B, Turner RS. The X11α protein slows cellular amyloid precursor protein processing and reduces Aβ40 and Aβ42 secretion. J Biol Chem. 1998;273:14761–14766. doi: 10.1074/jbc.273.24.14761. [DOI] [PubMed] [Google Scholar]
- 135.Sastre M, Turner RS, Levy E. X11 interaction with β-amyloid precursor protein modulates its cellular stabilization and reduces amyloid β-protein secretion. J Biol Chem. 1998;273:22351–22357. doi: 10.1074/jbc.273.35.22351. [DOI] [PubMed] [Google Scholar]
- 136.Lee JH, Lau KF, Perkinton MS, Standen CL, Shemilt SJA, Mercken L, Cooper JD, McLoughlin DM, Miller CCJ. The neuronal adaptor protein X11α reduces Aβ levels in the brains of Alzheimer’s APPswe Tg2576 transgenic mice. J Biol Chem. 2003;278:47025–47029. doi: 10.1074/jbc.M300503200. [DOI] [PubMed] [Google Scholar]
- 137.Lee JH, Lau KF, Perkinton MS, Standen CL, Rogelj B, Falinska A, McLoughlin DM, Miller CCJ. The neuronal adaptor protein X11β reduces amyloid β-protein levels and amyloid plaque formation in the brains of transgenic mice. J Biol Chem. 2004;279:49099–49104. doi: 10.1074/jbc.M405602200. [DOI] [PubMed] [Google Scholar]
- 138.Saito Y, Sano Y, Vassar R, Gandy S, Nakaya T, Yamamoto T, Suzuki T. X11 proteins regulate the translocation of APP into detergent resistant membrane and suppress the amyloidgenic cleavage of APP by BACE in brain. J Biol Chem. 2008;283:35763–35771. doi: 10.1074/jbc.M801353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Araki Y, Tomita S, Yamaguchi H, Miyagi N, Sumioka A, Kirino Y, Suzuki T. Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid beta-protein precursor metabolism. J Biol Chem. 2003;278:49448–49458. doi: 10.1074/jbc.M306024200. [DOI] [PubMed] [Google Scholar]
- 140.King GD, Turner S. Adaptor protein interactions: modulators of amyloid precursor protein metabolism and Alzheimer’s disease risk? Exp Neurol. 2004;185:208–219. doi: 10.1016/j.expneurol.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 141.Hase M, Yagi Y, Taru H, Tomita S, Sumioka A, Hiori K, Miyamoto K, Sasamura T, Nakamura M, Matsuno K, Suzuki T. Expression and characterization of the Drosophilia X11-like/Mint protein during neural development. J Neurochem. 2002;81:1223–1232. doi: 10.1046/j.1471-4159.2002.00911.x. [DOI] [PubMed] [Google Scholar]
- 142.Matsuda S, Yasukawa T, Homma Y, Ito Y, Niikura T, Hiraki T, Hirai S, Ohno S, Kita Y, Kawasumi M, Kouyama K, Yamamoto T, Kyriakis JM, Nishimoto I. c-Jun N-terminal kinase (JNK)-interacting protein-1b/islet-brain-1 scaffolds Alzheimer’s amyloid precursor protein with JNK. J Neurosci. 2001;21:6597–6607. doi: 10.1523/JNEUROSCI.21-17-06597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Scheinfeld MH, Roncarati R, Vito P, Lopez PA, Abdallah M, D’Adamio L. Jun NH2-terminal kinase (JNK) interacting protein 1 (JIP1) binds the cytoplasmic domain of the Alzheimer’s β-amyloid precursor protein (APP) J Biol Chem. 2002;277:3767–3775. doi: 10.1074/jbc.M108357200. [DOI] [PubMed] [Google Scholar]
- 144.Taru H, Kirino Y, Suzuki T. Differential roles of JIP scaffold proteins in the modulation of amyloid precursor protein metabolism. J Biol Chem. 2002;277:27567–27574. doi: 10.1074/jbc.M203713200. [DOI] [PubMed] [Google Scholar]
- 145.Inomata H, Nakamura Y, Hayakawa A, Takata H, Suzuki T, Miyazawa K, Kitamura N. A scaffold protein JIP-1b enhances amyloid precursor protein phosphorylation by JNK and its association with kinesin light chain 1. J Biol Chem. 2003;278:22946–22955. doi: 10.1074/jbc.M212160200. [DOI] [PubMed] [Google Scholar]
- 146.Araki Y, Kawano T, Taru H, Saito Y, Wada S, Miyamoto K, Kobayashi H, Ishikawa HO, Ohsugi Y, Yamamoto T, Matsuno K, Kinjo M, Suzuki T. The novel cargo Alcadein induces vesicle association of kinesin-1 motor components and activates axonal transport. EMBO J. 2007;26:1475–1486. doi: 10.1038/sj.emboj.7601609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Horiuchi D, Collins CA, Bhat P, Barkus RV, Diantonio A, Saxton WM. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr Biol. 2007;17:1313–1317. doi: 10.1016/j.cub.2007.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tarr PE, Roncarati R, Pelicci G, Pelicci PG, D’Adamio L. Tyrosine phosphorylation of the β-amyloid precursor protein cytoplasmic tail promotes interaction with Shc. J Biol Chem. 2002;277:16798–16804. doi: 10.1074/jbc.M110286200. [DOI] [PubMed] [Google Scholar]
- 149.Xie Z, Dong Y, Maeda U, Xia W, Tanzi RE. RNA interference silencing of the adaptor molecules ShcC and Fe65 differentially affect amyloid precursor protein processing and Aβ generation. J Biol Chem. 2007;282:4318–4325. doi: 10.1074/jbc.M609293200. [DOI] [PubMed] [Google Scholar]
- 150.Russo C, Dolcini V, Salis S, Venezia V, Zambrano N, Russo T, Schettini G. Signal transduction through tyrosine-phosphorylated C-terminal fragments of amyloid precursor protein via an enhanced interaction with Shc/Grb2 adaptor proteins in reactive astrocytes of Alzheimer’s diease brain. J Biol Chem. 2002;277:35282–35288. doi: 10.1074/jbc.M110785200. [DOI] [PubMed] [Google Scholar]
- 151.Zhou D, Noviello C, D’Ambrosio C, Scaloni A, D’Adamio L. Growth factor receptor-bound protein 2 interaction with the tyrosine-phosphorylated tail of amyloid β precursor protein is mediated by its Src homology 2 domain. J Biol Chem. 2004;279:25374–25380. doi: 10.1074/jbc.M400488200. [DOI] [PubMed] [Google Scholar]
- 152.Nizzari M, Venezia V, Repetto E, Caorsi V, Magrassi R, Gagliani MC, Carlo P, Florio T, Schettini G, Tacchetti C, Russo T, Diaspro A, Russo C. Amyloid precursor protein and presenelin 1 interact with the adaptor GRB2 and modulate Erk1, 2 signaling. J Biol Chem. 2007;282:13833–13844. doi: 10.1074/jbc.M610146200. [DOI] [PubMed] [Google Scholar]
- 153.Howell BW, Lanier LM, Frank R, Gertler FB, Cooper JA. The disabled 1 phosphotyrosine-binding domain binds to the internalization signals of transmembrane glycoproteins and to phospholipids. Mol Cell Biol. 1999;19:5179–5188. doi: 10.1128/mcb.19.7.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Homayouni R, Rice DS, Sheldon M, Curran T. Disabled-1 binds to the cytoplasmic domain of amyloid precursor-like protein 1. J Neurosci. 1999;19:7507–7515. doi: 10.1523/JNEUROSCI.19-17-07507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hoe HS, Train TS, Matsuoka Y, Howell BW, Rebeck W. DAB1 and Reelin effects on amyloid precursor protein and ApoE receptor 2 trafficking and processing. J Biol Chem. 2006;281:35176–35185. doi: 10.1074/jbc.M602162200. [DOI] [PubMed] [Google Scholar]
- 156.Hoe HS, Minami SS, Makarova A, Lee J, Hyman BT, Matsuoka Y, Rebeck GW. Fyn modulation of Dab1 effects on amyloid precursor protein and ApoE receptor 2 processing. J Biol Chem. 2008;283:6288–6299. doi: 10.1074/jbc.M704140200. [DOI] [PubMed] [Google Scholar]
- 157.Roncarati R, Sestan N, Scheinfeld MH, Berechid BE, Lopez PA, Meucci O, McGlade JC, Rakic P, D’Adamio L. The γ-secretase-generated intracellular domain of β-amyloid precursor protein binds Numb and inhibits Notch signaling. Proc Natl Acad Sci USA. 2002;99:7102–7107. doi: 10.1073/pnas.102192599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kyriazis GA, Wei Z, Vandermey M, Jo DG, Xin O, Mattson MP, Chan SL. Numb endocytic adapter proteins regulate the transport and processing of the amyloid precursor protein in an isoform-dependent manner. J Biol Chem. 2008;283:25492–25502. doi: 10.1074/jbc.M802072200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Zheng P, Eastman J, Pol SV, Pimplikar SW. PAT1, a microtubule-interacting protein, recognizes the basolateral sorting signal of amyloid precursor protein. Proc Natl Acad Sci USA. 1998;95:14745–14750. doi: 10.1073/pnas.95.25.14745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kuan YH, Gruebl T, Soba P, Eggert S, Nesic I, Back S, Kirsch J, Beyreuther K, Kins S. PAT1a modulates intracellular transport and processing of amyloid precursor protein (APP), APLP1 and APLP2. J Biol Chem. 2006;281:40114–40123. doi: 10.1074/jbc.M605407200. [DOI] [PubMed] [Google Scholar]
- 161.Gordon WR, Arnett KL, Blacklow SC. The molecular logic of Notch signaling–a structural and biochemical perspective. J Cell Sci. 2008;121:3109–3119. doi: 10.1242/jcs.035683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Müller T, Meyer HE, Egensperger R, Marcus K. The amyloid precursor protein intracellular domain (AICD) as a modulator of gene transcription, apoptosis, and cytoskeletal dynamics—relevance for Alzheimer’s disease. Prog Neurobiol. 2008;85:393–406. doi: 10.1016/j.pneurobio.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 163.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/S0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 164.Pardossi-Piquard R, Dunys J, Kawarai T, Sunyach C, Ring S, D’Adamio L, Shen J, Müller U, Hyslop P, Checler F. Presenelin-dependent transcriptional control of the Aβ-degrading enzyme neprilysin by intracellular domains of βAPP and APLP. Neuron. 2005;46:541–554. doi: 10.1016/j.neuron.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 165.Hébert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Müller U, De Strooper B. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep. 2006;7:737–745. doi: 10.1038/sj.embor.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ho A, Sudhof T. Binding of F-spondin to amyloid-beta precursor protein: a candidate amyloid-beta precursor protein ligand that modulates amyloid-beta precursor protein cleavage. Proc Natl Acad Sci USA. 2004;101:2548–2553. doi: 10.1073/pnas.0308655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hoe HS, Wessner D, Beffert U, Becker AG, Matsuoka Y, Rebeck GW. F-spondin interaction with the apolipoprotein E receptor ApoEr2 affects processing of amyloid precursor protein. Mol Cell Biol. 2005;25:9259–9268. doi: 10.1128/MCB.25.21.9259-9268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Buxbaum JD, Gandy SE, Cicchetti P, Ehrlich ME, Czernik AJ, Fracasso RP, Ramabhadran TV, Unterbeck AJ, Greengard P. Processing of Alzheimer βA4 amyloid precursor protein: modulation by agents that regulate protein phosphorylation. Proc Natl Acad Sci USA. 1990;87:6003–6006. doi: 10.1073/pnas.87.15.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Caporaso GL, Gandy SE, Buxbaum JD, Ramabhadran TV, Greengard P. Protein phosphorylation rgeulates secretion of Alzheimer β/A4 amyloid precursor protein. Proc Natl Acad Sci USA. 1992;89:3055–3059. doi: 10.1073/pnas.89.7.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Buxbaum JD, Koo EH, Greengard P. Protein phosphorylation inhibits production of Alzheimer amyloid β/A4 peptide. Proc Natl Acad Sci USA. 1993;90:9195–9198. doi: 10.1073/pnas.90.19.9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nitsch RM, Slack BE, Wurtman RJ, Growdon JH. Release of Alzheimer amyloid precursor derivatives stimulated by activation of muscarinic acetylcholine receptors. Science. 1992;258:304–307. doi: 10.1126/science.1411529. [DOI] [PubMed] [Google Scholar]
- 172.Lee RK, Wurtman RJ, Cox AJ, Nitsch RM. Amyloid precursor protein processing is stimulated by metabotropic glutamate receptors. Proc Natl Acad Sci USA. 1995;92:8083–8087. doi: 10.1073/pnas.92.17.8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Slack BE, Breu J, Muchnicki L, Wurtman RJ. Rapid stimulation of amyloid precursor protein release by epidermal growth factor: role of protein kinase C. Biochem J. 1997;327:245–249. doi: 10.1042/bj3270245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Slack BE, Ma LK, Seah CC. Constitutive shedding of the amyloid precursor protein ectodomain is up-regulated by tumour necrosis factor-α converting enzyme. Biochem J. 2001;357:787–794. doi: 10.1042/0264-6021:3570787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.da Cruz e Silva OA, Iverfeldt K, Oltersdorf T, Sinha S, Lieberburg I, Ramabhadran TV, Suzuki T, Sisodia S, Gandy S, Greengard P. Regulated cleavage of Alzheimer beta-amyloid precursor protein in the absence of the cytoplasmic tail. Neuroscience. 1993;57:873–877. doi: 10.1016/0306-4522(93)90031-A. [DOI] [PubMed] [Google Scholar]
- 176.Hung AY, Selkoe DJ. Selective ectodomain phosphorylation and regulated cleavage of β-amyloid precursor protein. EMBO J. 1994;13:534–542. doi: 10.1002/j.1460-2075.1994.tb06291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Holback S, Adlerz L, Gatsinzi T, Jacobsen KT, Iverfeldt K. PI3-K- and PKC-dependent up-regulation of APP processing enzymes by retinoic acid. Biochem Biophys Res Commun. 2007;365:298–303. doi: 10.1016/j.bbrc.2007.10.167. [DOI] [PubMed] [Google Scholar]
- 178.Torres-Aleman I. Targeting insulin-like growth factor-1 to treat Alzheimer’s disease. Expert Opin Ther Targets. 2007;12:1535–1542. doi: 10.1517/14728222.11.12.1535. [DOI] [PubMed] [Google Scholar]
- 179.Adlerz L, Holback S, Multhaup G, Iverfeldt K. IGF-1-induced processing of the amyloid precursor protein family is mediated by different signaling pathways. J Biol Chem. 2007;282:10203–10209. doi: 10.1074/jbc.M611183200. [DOI] [PubMed] [Google Scholar]
- 180.Petanceska S, Gandy S. The phosphatidylinositol 3-kinase inhibitor Wortmannin alters the metabolism of the Alzheimer’s amyloid precursor protein. J Neurochem. 1999;73:2316–2320. doi: 10.1046/j.1471-4159.1999.0732316.x. [DOI] [PubMed] [Google Scholar]