Abstract
In epithelial cells, sorting of membrane proteins to the basolateral surface depends on the presence of a basolateral sorting signal (BaSS) in their cytoplasmic domain. Amyloid precursor protein (APP), a basolateral protein implicated in the pathogenesis of Alzheimer’s disease, contains a tyrosine-based BaSS, and mutation of the tyrosine residue results in nonpolarized transport of APP. Here we report identification of a protein, termed PAT1 (protein interacting with APP tail 1), that interacts with the APP-BaSS but binds poorly when the critical tyrosine is mutated and does not bind the tyrosine-based endocytic signal of APP. PAT1 shows homology to kinesin light chain, which is a component of the plus-end directed microtubule-based motor involved in transporting membrane proteins to the basolateral surface. PAT1, a cytoplasmic protein, associates with membranes, cofractionates with APP-containing vesicles, and binds microtubules in a nucleotide-sensitive manner. Cotransfection of PAT1 with a reporter protein shows that PAT1 is functionally linked with intracellular transport of APP. We propose that PAT1 is involved in the translocation of APP along microtubules toward the cell surface.
The surface of epithelial cells is divided into apical and basolateral domains. They maintain their cell-surface polarity by intracellular sorting and polarized delivery of proteins and lipids to appropriate domains. In Madin-Darby canine kidney cells (MDCK), the newly synthesized apical and basolateral membrane proteins are sorted in the trans-Golgi network (TGN) into specific transport vesicles, which are then directly delivered to the appropriate domains. The basolateral sorting of cell surface proteins depends on the presence of peptide-based basolateral sorting signals (BaSS) in the cytoplasmic domains of proteins (1–3). Comparison of the known BaSS sequences indicates that these signals are nonidentical but structurally similar. Although many BaSS sequences have been characterized in detail, little is known about the cellular proteins that recognize BaSS. The identity of the putative basolateral sorting receptor that specifically binds the BaSS and sorts proteins to the basolateral surface is unknown.
We have been interested in understanding the mechanism (4) and regulation (5, 6) of polarized protein sorting in epithelial cells. The current study was undertaken to identify proteins that bind the BaSS of amyloid precursor protein (APP), a cell surface protein implicated in the pathogenesis of Alzheimer’s disease. APP is expressed ubiquitously by neuronal and non-neuronal cells and is sorted to axons in neurons (7) and the basolateral surface in epithelial cells (8–10). The basolateral sorting information of APP is contained in the 15-amino acid juxtamembrane segment of the cytoplasmic tail. This region contains a single tyrosine residue that, when mutated to alanine, results in nonpolarized distribution of APP in MDCK cells (9).
We report here the identification of a novel protein termed PAT1 (protein interacting with APP tail 1) that recognizes the APP-BaSS. PAT1 binds microtubules in a nucleotide-sensitive manner and appears to be involved in APP trafficking. Because microtubules are oriented with their plus ends toward the basolateral surface, PAT1 may play a role in the basolateral sorting of APP.
MATERIALS AND METHODS
Molecular Biology.
Standard methods were used for plasmid construction. Details of the yeast two-hybrid protocol, plasmid vectors, and cells are described in ref. 11. For two-hybrid screening, L40A5 yeast cells were transformed with a bait vector (p1979A) containing the wild-type APP-BaSS (KKKQYTSIHHG) fused to the DNA-binding domain of LexA and mated with EGY48 yeast cells transformed with a HeLa cDNA library in pJG4–5 plasmid. The positive clones were grown in the presence of 5-fluoroorotic acid to eliminate the bait plasmid, and the cells were retransformed with a second bait plasmid containing the mutant APP-BaSS (KKKQATSIHHG). The full-length PAT1 was obtained by 5′ rapid amplification of cDNA ends, and the insert in PAT1 was sequenced from both strands. The PAT1 ORF was amplified by PCR and cloned in the pFLAG-CMV-2 (Kodak) expression vector. For antisense PAT1 expression, a 2.07-kb SmaI–EcoRI fragment containing the entire coding region was cloned in the reverse orientation in the same vector. pCB6-APP was obtained by cloning the HindIII fragment containing the entire APP coding region from pCEP4-APP (a kind gift from Steve Younkin, Mayo Clinic, Jacksonville) into pCB6 vector. Horseradish peroxidase (HRP)/βA4/TM/CD in pSG5, a fusion construct in which the extracellular domain of APP is replaced by HRP, was a gift from Bart De Strooper (KUL, Leuven, Belgium) and is described in ref. 12.
Cell Culture and Biochemical Procedures.
COS-1 cells in a 6-well plate were transfected with PAT1 tagged with the FLAG-epitope at the N terminus in pFLAG-CMV-2 (Kodak; 1 μg per well) by using DOTAP transfection reagent (Boehringer Mannheim), and the cells were used 48–72 h after transfection. Cells were homogenized in 80 mM Pipes, pH 6.8/5 mM EGTA/1 mM MgCl2/250 mM sucrose by passage through a 27-gauge needle. The homogenate was centrifuged at 150,000 × g for 1 h in a TLA 120.2 rotor (Beckman) to separate cytosol from the membranes. For gel-filtration experiments, 0.5 ml of cytosol was injected onto a Superose-12 HR 10/30 FPLC column (Pharmacia) equilibrated with PBS. The fractions (0.2 ml) were Western blotted with anti-FLAG M5 antibody (Kodak). The ScintiStrip binding assay was performed as described (13). Briefly, ScintiStrip (Wallac, Oy, Finland) wells were coated with 0.5 μg of BSA, glutathione S-transferase (GST), or GST fused with the APP cytoplasmic tail containing the juxtamembrane 11-, 29-, or 47-amino acid (whole tail) fragment. PAT1 or luciferase proteins were in vitro translated, 0.05 μCi (1 Ci = 37 GBq) of labeled proteins was added to the ScintiStrip wells, unbound materials were removed, and the bound radioactivity was counted.
Sucrose Density Gradient Floatation.
A postnuclear supernatant from transfected COS-1 cells in 10 mM Hepes, pH 7.0/1 mM EDTA/0.5 mM MgCl2 was made 1.6 M sucrose by adding enough 2.3 M sucrose. In a TLS-55 tube (Beckman), 700 μl of postnuclear supernatant in 1.6 M sucrose was overlaid with 500 μl of 1.4 M sucrose, 500 μl of 1.2 M sucrose, and 300 μl of 1.0 M sucrose and centrifuged for 3 h at 44,000 × g in a TLX-120 ultracentrifuge (Beckman). Fractions (100 μl) were collected from the top, subjected to acetone precipitation, and analyzed by SDS/PAGE and Western blotting.
Chemical Cross-Linking.
Transfected cells were rinsed with cold PBS 48 h posttransfection and treated with 10 mM dimethyl pimelimidate (Pierce) in 100 mM Hepes buffer (pH 8.0) for 1 h at 4°C. The reaction was terminated with 100 mM Tris (pH 7.4) in PBS, and the cells were lysed directly in SDS/PAGE sample buffer.
Antibodies and Fluorescence Microscopy.
Anti-kinesin antibody (MC44) was a gift from M. McNiven (Mayo Clinic, Rochester, MN) and has been described previously (14). Anti-APP antibody (90/3) was a gift from J. Culvenor and C. Masters (University of Melbourne, Melbourne) and has been described (15). Monoclonal anti-tubulin antibody (T-6793) was purchased from Sigma, and anti-kinesin light chain (KLC) antibody (L1) was from Chemicon. Monoclonal anti-PAT1 antibodies were raised against the GST–PAT1 fusion protein. Hybridoma culture supernatants were screened by ELISA and Western blot analysis against the purified recombinant PAT1. Monoclonal antibody 26 (mAb26) recognized the GST–PAT1 fusion protein and the purified recombinant protein but not GST alone. Cells were fixed and permeabilized simultaneously with 3% paraformaldehyde/0.3% Triton X-100, quenched with 50 mM NH4Cl, and incubated overnight with the mAb26 hybridoma culture supernatant. Coverslips were processed for direct-TSA (Tyramide Signal Amplification; NEN Life Sciences Products) system according to the manufacturer’s instructions and visualized with Cy3-tyramide. For double labeling with tubulin, coverslips were incubated with anti-tubulin for 60 min and visualized with secondary antibody conjugated with fluorescein isothiocyanate. Some cells (see Fig. 4 d–f) were fixed in acetone for 30 s at −20°C and processed for conventional immunofluorescence without TSA.
Figure 4.
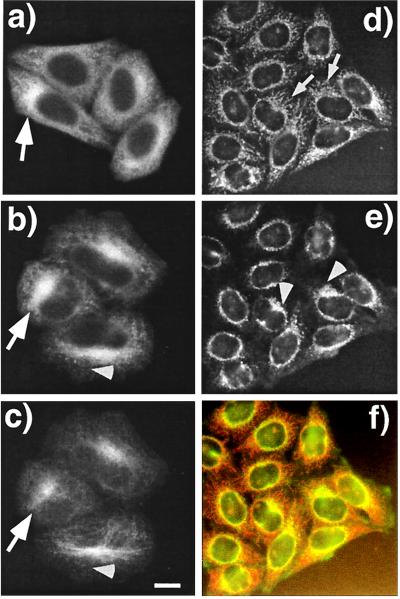
PAT1 is a cytoplasmic protein enriched in the perinuclear region. (a–c) Paraformaldehyde-fixed HeLa cells were labeled with mAb26 (a and b) and visualized by using the TSA system with HRP-conjugated secondary antibody and Cy3-tyramide. Cells in b are were also double-labeled with anti-tubulin antibody and visualized with fluorescein isothiocyanate-labeled secondary antibody (c). Note that PAT1 gives a punctate cytoplasmic staining, and in some cells the staining is clearly enriched in the perinuclear region (arrows in a and b) which colocalilizes with the microtubule organization center (MTOC) as detected by heavily concentrated microtubules (arrow in c). (Bar = 10 μm.) (d–f) Acetone-fixed HeLa cells were double-labeled with mAb26 (d) and anti-APP antibody (e) and visualized without TSA technique with rhodamin and fluorescein isothiocyanate-labeled secondary antibodies, respectively. The filamentous labeling of PAT1 is more obvious in these cells (arrows), which overlap with APP (arrowhead). The overlap of APP and PAT1 distribution is clearly seen as the orange signal in the merged image (f). The data showing the antibody specificity are presented on the PNAS web site (www.pnas.org) as Fig. 4 g and h.
Microtubule Binding Assay.
The assay was performed as described (16) ATP-depleted cytosol from transfected COS-1 cells was incubated for 15 min. at room temperature with taxol-stabilized microtubules in the presence of 1 mM AMP-PNP or 10 mM Mg-ATP. The reaction mixture was centrifuged through a 25% sucrose cushion at 166,000 × g for 30 min in a TLS-55 rotor (Beckman). Microtubule-associated proteins in the pellet were analyzed by SDS/PAGE and Western blotting.
Cotransfection of COS-1 Cells with PAT1 and Reporter Proteins.
COS-1 cells were cotransfected with 0.045 μg of HRP/βA4/TM/CD (in pSG5) and 1 μg of PAT1 plasmid (in sense or antisense orientation in pFLAG-CMV-2) or the parental plasmid (pFLAG-CMV-2) per well in a 6-well plate by using DOTAP as described above. Under these conditions, more than 80% of the HRP-positive cells were cotransfected with PAT1 as determined by double immunolabeling, and the transfection efficiency of HRP/βA4/TM/CD as judged by immunofluorescence was similar in cells cotransfected with either control or FLAG-PAT1 plasmid (not shown). After 36–48 h, transfected cells were rinsed and the HRP activity released in the medium was assayed as described (12).
RESULTS
Identification and Cloning of PAT1.
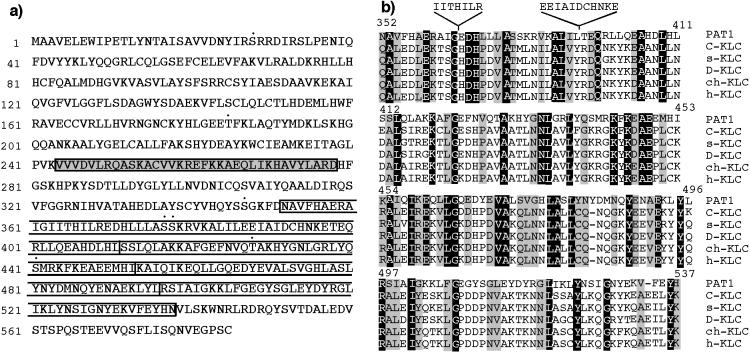
We used the APP-BaSS (amino acid sequence in one letter code KKKQYTSIHHG) fused to the DNA-binding domain of LexA as a bait and isolated nine positive clones (PAT1–9; Protein interacting with APP Tail 1–9) from a HeLa cDNA library (11). The bait plasmid from the positive clones was removed and the yeast cells were retransformed with a mutant APP-BaSS bait (where the critical tyrosine is replaced by an alanine residue; KKKQATSIHHG) to select the clones that fail to interact with the mutant bait. PAT1 was analyzed in further detail because the mutation of the critical tyrosine residue in the APP-BaSS sequence either abolished or significantly reduced interaction with the PAT1 clone (see below). The prey plasmid from the PAT1 clone contained a cDNA insert of 2,005 bp and the sequence was found to be 99.7% identical to an uncharacterized putative ORF (KIAA 0228; GenBank accession no. D86981; ref. 17). The full-length 2,385-bp cDNA clone (GenBank accession no. AF017782) was obtained by performing the 5′ rapid amplification of cDNA ends protocol. The predicted protein sequence consists of 585 amino acids (Fig. 1a) with a calculated molecular mass of 66.9 kDa and a predicted isoelectric point of 6.9. Antibodies raised against PAT1 recognized predominantly a single polypeptide of ≈70 kDa [see Fig. 4g, which is published as supplemental data on the PNAS web site (www.pnas.org)], confirming that our cDNA encoded a full-length protein of 70 kDa. Structural analysis of the sequence shows that the protein is hydrophilic with no obvious signal sequence or membrane-spanning domains. The N-terminal and C-terminal regions are predicted to form globular structures and a stretch of 35 amino acids is predicted to form 5 heptad repeats that are likely to form a coiled coil (Fig. 1e; an appendix with additional information on the sequence analysis is published as supplemental data on the PNAS web site, www.pnas.org). Four imperfect tandem repeats are found toward the C-terminal end of the protein (open boxes in Fig. 1). There are several putative protein kinase C phosphorylation sites (indicated by •). Search of public databases revealed significant homology to KLC extending over the entire (see Fig. 1c, which is published as supplemental data on the PNAS web site, www.pnas.org). The region of maximum homology between PAT1 and KLC contains the characteristic four 42-amino acid imperfect tandem repeats conserved in all species of KLCs (Fig. 1b). Several residues in each repeat are conserved between the KLC and PAT1 sequences; however, the first repeat contains two insertions of 7 and 11 amino acids.
Figure 1.
(a) One-letter amino acid sequence of PAT1. The region of PAT1 predicted to form a coiled-coil is shown in the shaded box, and the four imperfect KLC-like tandem repeats are in the open boxes. Putative protein kinase C phosphorylation sites are denoted by •. (b) Alignment of the KLC-like repeats. The four 42-amino acid, imperfect KLC-like tandem repeats in PAT1 aligned with KLC repeats from various organisms. The numbers indicate the position of amino acids in PAT1 sequence. C-KLC , Caenorhabditis elegans (accession no. P46822); s-KLC, squid (accession no. P46825); D-KLC, Drosophila (accession no. P46824); ch-KLC, chicken (accession no. 1208772); h-KLC, human (accession no. Q07866). An appendix with information on sequence analysis, alignments, etc., is published as supplemental data on the PNAS web site (www.pnas.org).
PAT1 Binds Specifically to the Tyrosine-Containing APP-BaSS.
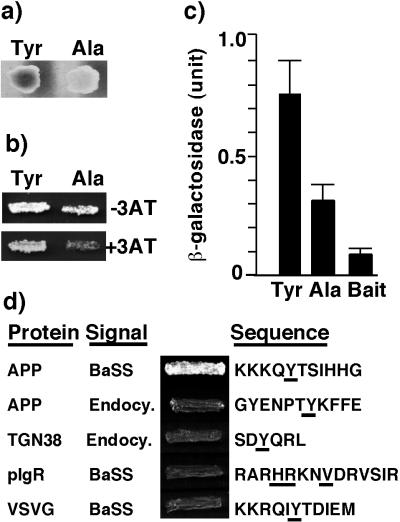
The specificity of the interaction of PAT1 with APP-BaSS was examined in the yeast two-hybrid assay by mutating the critical tyrosine residue that renders the BaSS nonfunctional (9). Interactions were monitored by the expression of β-galactosidase activity and by growth on histidine-deficient plates. Interaction of PAT1 with mutant APP-BaSS was abolished in the color assay (Fig. 2a) and severely reduced in the growth assay (Fig. 2b). A quantitative β-galactosidase assay showed significant reduction in the interaction of PAT1 with the mutant APP-BaSS (Fig. 2c). The interaction of PAT1 with APP-BaSS as evidenced in the yeast two-hybrid system was corroborated by testing whether in vitro translated [35S]methionine-labeled PAT1 was able to interact with APP-BaSS fused to GST. Table 1 shows that PAT1 directly bound to the 11-amino acid-long APP-BaSS and to the complete cytoplasmic domain of APP. Reduced binding to the longer constructs may be caused by steric hindrance.
Figure 2.
Interaction of PAT1 prey with APP-BaSS and various baits. (a) β-Galactosidase color assay: Yeast diploid cells (L40A5xEGY48) containing prey vector (pJG4–5) expressing PAT1 and bait vectors (p1979A) containing either the wild-type APP-BaSS (Tyr) or the mutant APP-BaSS (Ala) were grown on a galactose + histidine plate in the presence of -bromo-4-chloro-3-indolyl β-d-galactoside. These and other data are obtained from a pooled population of at least 5 colonies. (b) Growth assay: Yeast diploid cells obtained as above were grown on galactose plates lacking histidine with or without 20 μM 3-amino-1, 2,4-triazole (3-AT). (c) Quantitative β-galactosidase assay: Yeast diploid cells obtained as above were grown in galactose-containing liquid culture for 12–18 h, and the β-galactosidase activity was determined and normalized for cell density. Data represent average results from five individual colonies. Error bars indicate the standard deviation. Yeast cells (L40A5) transformed with the wild-type APP-BaSS bait vector alone were used as a negative control. (d) YSV90 yeast cells were cotransformed with PAT1 prey vector (in pJG4–5) and indicated bait vectors (in p1979A), and the transformants were grown on galactose-lacking leucine for 2–3 days. The names of proteins and the signaling activity of the sequence (BaSS or endocytic) are given on the left. Signal sequences fused to the DNA-binding domain of LexA are given on the right, and the residues crucial for signaling activity are underlined.
Table 1.
In vitro binding of PAT1 to APP cytoplasmic tail fusion proteins
| Immobilized ligands | Bound radioactivity, cpm
|
|
|---|---|---|
| PAT1 | Luciferase | |
| BSA | 2,108 ± 174 | 566 ± 44 |
| GST | 3,046 ± 135 | 872 ± 10 |
| GST-C11 | 15,894 ± 64 | 1,202 ± 181 |
| GST-C29 | 7,434 ± 234 | 917 ± 22 |
| GST-C47 | 5,434 ± 109 | 1,108 ± 17 |
Data shown are the mean and the range of duplicate values from one experiment, which was representative of three experiments.
PAT1 Interacts Selectively with APP-BaSS.
Because PAT1 recognizes the tyrosine-based BaSS, we examined whether it also interacts with tyrosine-based endocytic signals. The cytoplasmic domain of APP harbors an NPxY sequence motif that is responsible for its endocytosis. Both in the growth assay (Fig. 2d) and the β-galactosidase assay (not shown), PAT1 was found not to interact with the NPxY endocytic signal from APP or with the SDYQRL endocytic motif from the cytoplasmic tail of protein TGN38. PAT1 also failed to interact with the non-tyrosine-based BaSS from the polymeric Ig receptor or with the tyrosine-based BaSS of vesicular stomatitis virus G protein. Together, these results show that the interaction of PAT1 with APP-BaSS is specific and selective.
PAT1, a Cytosolic Protein, Is also Found Associated with Membranes.
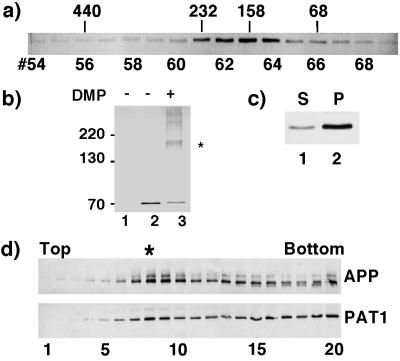
The presence of heptad repeats suggests that PAT1 interacts with itself or with other proteins. To test this, COS-1 cells transiently transfected with a pFLAG-CMV-2 vector expressing PAT1 were homogenized 48 h posttransfection, cytosol (150,000 × g supernatant) was separated on a Superose-12 column, and fractions were analyzed by Western blotting (Fig. 3a) by using anti-FLAG antibody (M5, Kodak). Cytosolic PAT1 eluted as a single species with an approximate size of 160–180 kDa. A monomer of PAT1 is expected to elute at 65–70 kDa, suggesting that PAT1 associates with a protein(s) of approximately 110 kDa. This was confirmed by using an independent chemical cross-linking approach. COS-1 cells transfected with the PAT1-pFLAG-CMV-2 expression vector were treated with the membrane-permeable cross-linker dimethyl pimelimidate . The cells were lysed in SDS/PAGE sample buffer, and the total lysate was analyzed by SDS/PAGE and Western blotting. The presence of a predominant cross-linked band of approximately 160–180 kDa (Fig. 3b, lane 3) suggests that PAT1 associates with a 110-kDa protein. Together, these observations suggest that in vivo PAT1 exists as a complex with a 110-kDa protein.
Figure 3.
PAT1 exists as an oligomeric complex, associates with the membrane and cofractionates with APP. (a) Gel filtration: Cytosol prepared from COS-1 cells transiently transfected with PAT1 in pFLAG-CMV-2 vector was separated on a Superose 12 gel filtration column. The fractions were Western blotted and probed with anti-FLAG M5 antibody and detected by alkaline phosphatase reaction. The elution positions of protein standards (in kDa) are indicated. (b) Chemical cross-linking: COS-1 cells mock-transfected (lane 1) or transiently transfected with FLAG-PAT1 in (lanes 2 and 3) were treated without (lane 2) or with (lane 3) 10 mM dimethyl pimelimidate (DMP, Pierce) at 4°C for 60 min. After the reaction was quenched, the cells were directly lysed in SDS/PAGE sample buffer and analyzed by SDS/PAGE and Western blotting as above. ∗ indicates the cross-linked band of 160–180 kDa detected by M5 antibody. The additional bands may be higher oligomers. (c) Subcellular fractionation: FLAG-PAT1 transfected COS-1 cells were homogenized and centrifuged at 150,000 × g (TLA 120.2, Beckman), and the supernatant (lane 1) and the pellet (lane 2) fractions were analyzed by SDS/PAGE and Western blotting. Proportion of PAT1 in pellet fraction varied between 50% and 75%. (d) Cofractionation of PAT1 and APP. A postnuclear supernatant from COS-1 cells transfected with FLAG-PAT1 and APP was fractionated by sucrose gradient centrifugation, and the fractions were analyzed by SDS/PAGE and Western blotting and probed with anti-APP (22C11; upper panel) or anti-FLAG antibody (lower panel). Note that a portion of PAT1 cofractionates with APP (fraction 8, indicated by ∗). The quantitation of these data is presented on the PNAS web site (www.pnas.org) as Fig. 3e.
When a homogenate from COS-1 cells expressing FLAG-PAT1 was fractionated by differential sedimentation, PAT1 was found to be present both in 150,000 × g supernatant (cytosol) and pellet (membrane) fractions (Fig. 3c), suggesting that like many proteins involved in vesicular trafficking, PAT1 is present in both cytosolic and membrane-associated pools. Association of PAT1 with membranes was confirmed by suspending the homogenate in 2.3 M sucrose and subjecting it to floatation gradient centrifugation. The majority of PAT1 was recovered in 1.0–1.6 M sucrose fractions (not shown), as are many membrane-associated proteins.
Membrane-Associated PAT1 Cofractionates with APP.
We next determined whether the membrane associated PAT1 distributes with APP. A postnuclear supernatant from COS-1 cells cotransfected with PAT1-pFLAG-CMV-2 and pCB6-APP was fractionated by sucrose density gradient floatation as described in Materials and Methods, and the fractions were analyzed by SDS/PAGE and Western blotting and probed with anti-APP antibody (22C11) or with anti-FLAG antibody (Fig. 3d). A large proportion of APP migrated as a major peak at fraction 8 of the gradient (1.2/1.4 M sucrose interface) with the remainder appearing in the heavier fractions. A figure showing quantitation from Fig. 3d is published as supplemental data (Fig. 3e) on the PNAS web site (www.pnas.org). A portion of PAT1 also peaked in fraction 8 and the rest was detected in the bottom fractions (“load”) together with soluble proteins and other dense membrane-bound compartments (fractions 14–20). PAT1 is cytosolic and its appearance in the bottom fractions is not unexpected. These data show that membrane-associated PAT1 cofractionates with APP containing vesicles.
PAT1 Is Concentrated in the Golgi Region and Partially Codistributes with APP.
We raised monoclonal antibodies against the PAT1–GST fusion protein. Western blot analysis showed that mAb26 reacted with purified recombinant PAT1 and recognized predominantly a single polypeptide of ≈70 kDa in a HeLa cell lysate (see Fig. 4g, which is published as supplemental data on the PNAS web site, www.pnas.org). This antibody was therefore used to localize PAT1 by immunofluorescence microscopy. In paraformaldehyde-fixed HeLa cells (Fig. 4 a–c), this antibody stained punctate structures distributed throughout the cytoplasm (Fig. 4a) although staining was often seen to be concentrated in the perinuclear region (indicated by an arrow). A monoclonal antibody against GST protein did not stain these structures (see Fig. 4h, which is published as supplemental data on the PNAS web site, www.pnas.org). PAT1-containing structures were abundant in the perinuclear region (Fig. 4b; arrow), and mAb26-staining frequently aligned with microtubules as detected with an anti-tubulin antibody (Fig. 4e, arrowheads). The filamentous staining of PAT1 was more obvious in cells fixed in acetone and viewed by conventional immunofluorescence without TSA (Fig. 4d). Unlike paraformaldehyde, acetone fixation preserves the cytoskeletal structures better than the membranes and cytosol. When double-labeled with anti-APP antibody (90/3) under these conditions (Fig. 4e), a partial overlap of APP and PAT1 was seen, which is more obvious in the merged image (Fig. 4f). In some cells, PAT1-positive filamentous structures (arrows) were seen to emanate from the APP-positive Golgi complex (arrowheads; ref. 15). Together with the fractionation data, these observations show that PAT1 is predominantly present in the Golgi region and overlaps with APP in its distribution, consistent with a possible role in APP transport.
PAT1 Binds Microtubules and Is Involved in APP Secretion.
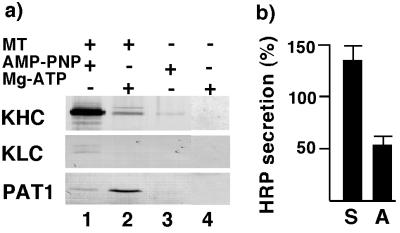
When PAT1-expressing COS-1 cells were extracted with nonionic detergent, approximately 50% of PAT1 was insoluble (not shown), indicating that PAT1 may associate with the cytoskeleton. This observation and the homology with KLC suggested that PAT1 interacts with microtubules. To investigate this possibility, we used the known property of kinesin: that it stably binds microtubules in the presence of AMP-PNP (a nonhydrolyzable analog of ATP) but not in the presence of ATP (18). Cytosol prepared from transfected COS-1 cells was incubated with taxol-stabilized microtubules in the presence of AMP-PNP or Mg-ATP and the microtubule-bound material was analyzed by SDS/PAGE and Western blotting and probed with anti-kinesin heavy chain (KHC), anti-KLC, or anti-FLAG antibodies. Fig. 5a shows that, as expected, KHC and KLC bound microtubules in the presence of AMP-PNP (lane 1) but not when Mg-ATP was present (lane 2). PAT1 showed limited but significant binding (lane 1 vs. lane 3) to microtubules in the presence of AMP-PNP (lane 1). Interestingly, this binding was stimulated 5- to 10-fold in the presence of Mg-ATP (lane 2). These data show that PAT1 interacts with microtubules and that this association may be regulated by posttranslational modifications.
Figure 5.
PAT1 interacts with microtubules and affects APP trafficking. (a) Microtubule binding assay: Cytosol from transfected COS-1 cells was incubated with (lanes 1 and 2) or without (lane 3 and 4) taxol-stabilized microtubules in the presence of 1 mM AMP-PNP (lanes 1 and 3) or 10 mM Mg-ATP (lanes 2 and 4). Microtubules were pelleted through a 25% sucrose cushion, and the pellet was analyzed by SDS/PAGE and Western blotting as described in Fig. 3a and probed with anti-KHC antibody, anti-KLC antibody, and with M5 antibody. (b) COS-1 cells were cotransfected with HRP/βA4/TM/CD plasmid and control-plasmid or sense-plasmid (S) to overexpress and antisense-plasmid (A) to down-regulate cellular PAT1 levels. After 36–48 h, HRP activity released in the medium was assayed as described (12). HRP activity is expressed as percent of control and the results are from duplicate measurements of four independent experiments. Error bars show standard deviation.
We next determined whether PAT1 was functionally involved in APP trafficking. We used a transient cotransfection assay similar to that used by Echard et al. (19) to examine the effects of alteration in PAT1 levels on the transport of a reporter fusion protein, HRP-APP (12). This reporter protein contains HRP fused to the C-terminal fragment of APP containing the βA4-amyloid region, the transmembrane, and the cytoplasmic tail domains (HRP/βA4/TM/CD). In COS-1 cells, the chimeric reporter protein shows the transport characteristics of wild-type APP and is processed by α-secretase, which results in the release of HRP activity in culture medium with kinetics identical to that observed for the secretion of wild-type APP (12). Fig. 5b shows that COS-1 cells expressing HRP/βA4/TM/CD showed enhanced HRP secretion when cotransfected with PAT1 sense-plasmid and significantly reduced HRP secretion when cotransfected with PAT1 antisense-plasmid compared with cells cotransfected with control plasmid (pFLAG-CMV-2 without PAT1). Cells cotransfected without the reporter fusion protein released no detectable HRP activity in the medium. The increased secretion of HRP in PAT1 overexpressing cells most likely reflects increased intracellular transport of the reporter protein to the cell surface. Because the α-secretase cleavage occurs at or near the plasma membrane in COS-1 cells (12, 20), increased transport of the reporter protein to the cell surface is expected to result in enhanced secretion of HRP in the medium. These data provide evidence that PAT1 is functionally associated with some aspect of APP trafficking/processing.
DISCUSSION
PAT1, a Microtubule-Binding Protein That Recognizes APP-BaSS.
We have identified and characterized PAT1, a protein that specifically recognizes the APP-BaSS and binds microtubules. The interaction of PAT1 with APP-BaSS is nearly abolished when the critical tyrosine residue is mutated to alanine, a single mutation that causes the nonpolarized delivery of APP in MDCK cells. Importantly, PAT1 does not bind the tyrosine-containing endocytic signal of APP. These observations show that PAT1 interacts specifically with APP-BaSS. PAT1 also interacts with the whole cytoplasmic domain of APP although with somewhat reduced affinity probably due to steric hindrance. Weaker binding with longer peptide has also been observed in other cases (21).
PAT1 is a cytosolic protein and like many proteins involved in membrane trafficking, fractionates into a soluble and a membrane-associated pool. In addition, PAT1 is also capable of binding microtubules, indicating that in vivo PAT1 probably recycles among these three pools. Immunofluorescent localization studies revealed that PAT1 was distributed throughout the cytoplasm in a punctate or filamentous pattern. Often, the PAT1-positive structures were most abundant in the perinuclear region of the cell, where microtubules are highly concentrated, where Golgi-complex and the TGN reside, and where the majority of APP is localized (15). These data were confirmed biochemically by showing that PAT1 cofractionates with APP-containing membrane organelles. It should be noted that kinesin in cultured cells, like PAT1, is also concentrated in the perinuclear region and is associated with the Golgi complex (14) and with microtubules. Thus, our observations are consistent with a role for PAT1 in some aspect of APP transport involving microtubules and Golgi complex.
Sequence comparison shows that PAT1 shares significant homology with KLC. PAT1 contains four 42-amino acid-long tandem-repeats that are also found in KLC from all the species studied to date. However, the function of these repeats or that of the KLC is not clear, and antibody inhibition studies indicate that they seem to be involved in some aspect of vesicular transport (22). Earlier reports have suggested that the KLC may be involved in binding the cargo vesicles and/or in regulating the ATPase activity of KHC. Biochemical fractionation and cross-linking data show that PAT1 is present in a complex with a protein of the size of KHC (≈110 kDa). However, the 110-kDa protein was not recognized by antibodies to KHC, and its identity remains unknown.
Possible Functions of PAT1.
The interaction of PAT1 with the APP-tail, its subcellular location, association with membranes, and binding to microtubules indicate that PAT1 is well suited to be involved in APP trafficking along the microtubules. Indeed, our data suggest that PAT1 plays a functional role in APP trafficking/processing. Many transport events are mediated by microtubules and the associated motor proteins (23, 24). Based on the ability of PAT1 to bind both the APP-tail and microtubules, we favor a model that PAT1 binds to APP cytoplasmic domain in a newly formed vesicle and helps them “dock” at microtubules. It is also possible that PAT1 binding to APP cytoplasmic domain triggers their inclusion into specific transport vesicles budding from the TGN. In nonepithelial cells, microtubules are arranged with minus ends near the Golgi complex and plus ends radiating toward the cell surface. Thus, PAT1 may mediate the transport of Golgi-derived APP-containing vesicles to the cell surface. Kinesin has been shown to be required for APP transport toward the plus ends of microtubules in neurons (7). In vitro studies have shown that “accessory proteins” are required for the movement of kinesin along microtubules (25), and it is possible that PAT1 may be one such accessory protein.
In addition, PAT1 may function in the so-called “cognate-polarity” pathways in nonepithelial cells. Recent studies have suggested the existence of cognate-apical and cognate-basolateral pathways in nonepithelial cells (26, 27). In the absence of distinct domains in their cell surface, these two pathways in nonepithelial cells lead to a single destination, the plasma membrane. This viewpoint suggests that in HeLa cells APP is transported to the cell surface along the “cognate-basolateral” pathway. By Western blot analysis, PAT1 was found to be present in the MDCK and HEK-293 cell lines and in human and mouse brain and kidney (data not shown). By Northern blot analysis, Nagase et al. (17) have shown that PAT1 message (referred as KIAA 0228) is expressed in several tissues and nonepithelial cell lines. Thus, PAT1 or PAT1-like proteins together with microtubules could provide a molecular basis for the cognate basolateral pathway observed in nonpolarized cells.
The importance of microtubules in basolateral protein transport in epithelial cells has been demonstrated (23, 28). Does PAT1 play a role in sorting and delivery of APP to the basolateral surface of epithelial cells? While awaiting an experimental proof, it is interesting to note that in epithelial cells, microtubules are arranged with minus ends near the Golgi-complex (near the apical cell surface) and plus ends directed toward the basolateral cell surface (29, 30). Thus, in epithelial cells PAT1-mediated transport of APP vesicles along the microtubules would lead the vesicles to the basolateral surface. Selective recognition of the APP-BaSS by PAT1 and the lack of interaction with mutant BaSS support the possible role of PAT1 in basolateral sorting of APP.
In conclusion, PAT1 is a protein that specifically recognizes APP-BaSS and seems important for APP trafficking. Its subcellular localization and ability to interact with microtubules are consistent with its role in transport of APP to the cell surface. The characteristic arrangement of microtubules in epithelial cells indicates that PAT1 may be involved in basolateral delivery of APP.
Supplementary Material
Acknowledgments
We thank M. McNiven, S. Hamm-Alvarez, J. Culvenor, and B. De Strooper for the gift of reagents; G. Matera for help with taking immunofluorescence images; and A. Tartakoff and M. Lamm for comments on the manuscript. This work was supported by grants from the National Institutes of Health, the Human Frontier Science Program, and the Alzheimer’s foundation to S.W.P. and by a National Institutes of Health grant to S.V.P.
ABBREVIATIONS
- MDCK
Madin-Darby canine kidney cells
- TGN
trans-Golgi network
- BaSS
basolateral sorting signal
- APP
amyloid precursor protein
- PAT1
protein interacting with APP tail 1
- GST
glutathione S-transferase
- HRP
horseradish peroxidase
- KLC
kinesin light chain
- KHC
kinesin heavy chain
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF017782).
References
- 1.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Mostov K, Apodaca G, Aroeti B, Okamoto C. J Cell Biol. 1992;116:577–583. doi: 10.1083/jcb.116.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matter K, Mellman I. Curr Opin Cell Biol. 1994;6:545–554. doi: 10.1016/0955-0674(94)90075-2. [DOI] [PubMed] [Google Scholar]
- 4.Pimplikar S W, Ikonen E, Simons K. J Cell Biol. 1994;125:1025–1035. doi: 10.1083/jcb.125.5.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pimplikar S W, Simons K. Nature (London) 1993;362:456–458. doi: 10.1038/362456a0. [DOI] [PubMed] [Google Scholar]
- 6.Pimplikar S W, Simons K. J Biol Chem. 1994;269:19054–19059. [PubMed] [Google Scholar]
- 7.Ferreira A, Caceres A, Kosik K S. J Neurosci. 1993;13:3112–3123. doi: 10.1523/JNEUROSCI.13-07-03112.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haass C, Koo E H, Teplow D B, Selkoe D J. Proc Natl Acad Sci USA. 1994;91:1564–1568. doi: 10.1073/pnas.91.4.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haass C, Koo E H, Capell A, Teplow D B, Selkoe D J. J Cell Biol. 1995;128:537–547. doi: 10.1083/jcb.128.4.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Strooper B, Craessaerts K, Dewachter I, Moechars D, Greenberg B, Van Leuven F, Van Den Berghe H. J Biol Chem. 1995;270:4058–4065. doi: 10.1074/jbc.270.8.4058. [DOI] [PubMed] [Google Scholar]
- 11.Vande Pol S B, Brown M C, Turner C E. Oncogene. 1998;16:43–52. doi: 10.1038/sj.onc.1201504. [DOI] [PubMed] [Google Scholar]
- 12.De Strooper B, Craessaerts K, Van Leuven F, Van Den Berghe H. J Biol Chem. 1995;270:30310–30314. doi: 10.1074/jbc.270.51.30310. [DOI] [PubMed] [Google Scholar]
- 13.Zhong P, Chen Y A, Tam D, Chung D, Scheller R H, Miljanich G P. Biochemistry. 1997;36:4317–4326. doi: 10.1021/bi9625408. [DOI] [PubMed] [Google Scholar]
- 14.Marks D L, Larkin J M, McNiven M A. J Cell Sci. 1994;107:2417–2426. doi: 10.1242/jcs.107.9.2417. [DOI] [PubMed] [Google Scholar]
- 15.Culvenor J G, Friedhuber A, Fuller S J, Beyreuther K, Masters C L. Exp Cell Res. 1995;220:474–481. doi: 10.1006/excr.1995.1339. [DOI] [PubMed] [Google Scholar]
- 16.McIlvain J J, Burkhardt J K, Hamm A S, Argon Y, Sheetz M P. J Biol Chem. 1994;269:19176–19182. [PubMed] [Google Scholar]
- 17.Nagase T, Seki N, Ishikawa K, Ohira M, Kawarabayasi Y, Ohara O, Tanaka A, Kotani H, Miyajima N, Nomura N. DNA Res. 1996;3:321–329. doi: 10.1093/dnares/3.5.321. [DOI] [PubMed] [Google Scholar]
- 18.Vale R D, Reese T S, Sheetz M P. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Echard A, Jollivet F, Martinez O, Lacapere J J, Rousselet A, Janoueix-Lerosey I, Goud B. Science. 1998;279:580–585. doi: 10.1126/science.279.5350.580. [DOI] [PubMed] [Google Scholar]
- 20.Sisodia S S. Proc Natl Acad Sci USA. 1992;89:6075–6079. doi: 10.1073/pnas.89.13.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartsch D, Ghirardi M, Skehel P A, Karl K A, Herder S P, Chen M, Bailey C H, Kandel E R. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 22.Stenoinen D L, Brady S T. Mol Biol Cell. 1997;8:675–689. doi: 10.1091/mbc.8.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafont F, Burkhardt J K, Simons K. Nature (London) 1994;372:801–803. doi: 10.1038/372801a0. [DOI] [PubMed] [Google Scholar]
- 24.Bloom G S, Goldstein L S B. J Cell Biol. 1998;140:1277–1280. doi: 10.1083/jcb.140.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroer T A, Schnapp B J, Reese T J, Sheetz M P. J Cell Biol. 1988;107:1785–1792. doi: 10.1083/jcb.107.5.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muesch A, Xu H, Shield D, Rodriguez-Boulan E. J Cell Biol. 1996;133:543–558. doi: 10.1083/jcb.133.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshimori T, Keller P, Roth M G, Simons K. J Cell Biol. 1996;133:247–256. doi: 10.1083/jcb.133.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pous C, Chabin K, Drechou A, Barbot L, Phung-Koskas T, Settegrana C, Bourguet-Kondracki M L, Maurice M, Cassio D, Guyot M, et al. J Cell Biol. 1998;142:153–165. doi: 10.1083/jcb.142.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bacallao R, Antony C, Dotti C, Karsenti E, Stelzer E H, Simons K. J Cell Biol. 1989;109:2817–2832. doi: 10.1083/jcb.109.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ihrke G, Neufeld E B, Meads T, Shanks M R, Cassio D, Laurent M, Schroer T A, Pagano R E, Hubbard A L. J Cell Biol. 1993;123:1761–1775. doi: 10.1083/jcb.123.6.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.