Abstract
Nonsense-mediated decay is well known by the lucid definition of being a RNA surveillance mechanism that ensures the speedy degradation of mRNAs containing premature translation termination codons. However, as we review here, NMD is far from being a simple quality control mechanism; it also regulates the stability of many wild-type transcripts. We summarise the abundance of research that has characterised each of the NMD factors and present a unified model for the recognition of NMD substrates. The contentious issue of how and where NMD occurs is also discussed, particularly with regard to P-bodies and SMG6-driven endonucleolytic degradation. In recent years, the discovery of additional functions played by several of the NMD factors has further complicated the picture. Therefore, we also review the reported roles of UPF1, SMG1 and SMG6 in other cellular processes.
Keywords: NMD, Nonsense mRNA surveillance, Post-transcriptional gene regulation, PTC, mRNA turnover
Introduction
The cascade of events during gene expression, from transcription of the DNA encoded genetic information to the eventual protein synthesis, arguably represents some of the most influential biochemical pathways for a living organism. Naturally, diverse regulation mechanisms have evolved to ensure the accuracy of gene expression at multiple levels, amongst them is a process commonly referred to as nonsense-mediated mRNA decay (NMD) or mRNA surveillance. Thirty years ago, it was observed in Saccharomyces cerevisiae that nonsense codons truncating the open reading frame (ORF) of the ura3 mRNA dramatically reduced the RNA’s half-life [1] and that in β0-thalassemic patients homozygous for a nonsense mutation in the β-globin gene, the β-globin mRNA was subjected to rapid degradation [2]. Fast degradation of mRNAs harbouring truncated ORFs due to premature translation termination codons (PTCs) was subsequently documented in many other organisms, and it is believed to occur in most if not all eukaryotes (reviewed in [3]). The names coined to describe this PTC-associated mRNA turnover pathway, NMD and mRNA surveillance, emphasise its quality control function in preventing the production of potentially deleterious C-terminally truncated proteins translated from PTC-containing mRNAs. However, it has become clear during recent years that many physiological mRNAs are also NMD substrates, indicating a role for NMD beyond mRNA quality control as a translation-dependent post-transcriptional regulator of gene expression (reviewed in [4]).
Herein, we describe different types of NMD substrates and review recent literature that has led to a unified molecular model for the identification of NMD eliciting mRNAs. Additionally, we summarise our knowledge about the trans-acting NMD factors, their biochemical features and the intriguing double-life of several of these factors.
NMD targets both aberrant and physiological transcripts
One important group of NMD substrates comprise transcripts harbouring a PTC that truncates their ORFs. If not detected and degraded, the PTC-containing (PTC+) transcripts can result in the accumulation of potentially harmful C-terminally truncated proteins. PTC+ transcripts arise either by mutations (or rearrangements in the case of immunoglobulin genes) at the DNA level, or at the RNA level due to errors in transcription, pre-mRNA processing and in particular by aberrant alternative splicing (Table 1).
Table 1.
Transcripts that can be targeted for NMD in mammalian cells
| PTC+ | PTCs arising at DNA level | Nonsense mutations that directly generate PTCs |
| Nucleotide insertions and deletions that shift the reading frame | ||
| Mutations leading to splice signal alterations | ||
| DNA rearrangements of immunoglobulin and T-cell receptor genes | ||
| PTCs arising at RNA level | Transcription errors | |
| Faulty or alternative splicing | ||
| Pre-mRNAs that escaped nuclear retention | ||
| Programmed frameshifts | ||
| PTC− | Physiological mRNAs | mRNAs with uORF |
| mRNAs with introns in the 3′ UTR | ||
| mRNAs with long 3′ UTRs | ||
| Selenoprotein mRNAs | ||
| mRNAs of transposons, retroviruses and pseudogenes |
On the DNA level, direct nonsense mutations and more frequently frame-shifting deletions and insertions generate PTCs. Moreover, mutations at splice sites or splicing regulatory sequences can result in aberrantly spliced PTC+ transcripts. The programmed DNA rearrangements occurring in the immunoglobulin and T-cell receptor genes during lymphocyte maturation generate PTCs at a high frequency, due to random deletions and the addition of non-templated nucleotides at the recombination sites (reviewed in [5]). Hence, NMD is very important for the differentiation and maintenance of hematopoietic cells [6].
Errors during transcription can also produce PTC+ transcripts, albeit less frequently. In contrast, unproductive alternative splicing is believed to constitute a major source of PTC+ mRNAs in mammals. As much as 95% of multi-exon human genes are alternatively spliced [7], and the average number of alternatively spliced mRNA isoforms per gene is approximately 3.5 in humans [8]. Using bioinformatics approaches, it was proposed that about one-third of the alternatively spliced human mRNAs contain a PTC, implying a widespread coupling of alternative splicing and NMD [9]. For example, several studies have revealed that some alternative splicing (AS) events exploit NMD for post-transcriptional regulation (termed regulated unproductive splicing and translation: RUST or AS-NMD [9]). This type of regulation is common for many splicing regulators, including SR proteins and hnRNPs, which indicates an important feedback regulation of splicing (reviewed in [10, 11]). Interestingly, such PTC-introducing exons often coincide with ultra-conserved genomic elements, suggesting a key role for RUST in vertebrate biology. However, a microarray profiling study determining the relative levels of PTC+ compared to the PTC-free splice variants in a variety of mammalian tissue types proposed that most PTC-generating alternative splicing events located within the ORF of mammalian genes produce PTC+ mRNAs that are not under strong positive selection pressure and hence are unlikely to have important functional roles [12]. Altogether, this advocates that cells produce a large number of faulty PTC+ mRNAs that are recognised and eliminated by NMD. Nonetheless, several studies have inferred an even broader role of the NMD pathway in muting the “transcriptional noise” of supposedly non-functional RNAs such as transcribed pseudogenes, ancient transposons or mRNA-like non-protein coding RNAs from intergenic regions [13–15].
Another group of NMD substrates includes physiological RNA transcripts that encode functional full-length proteins. A crucial role of NMD, not only as a vacuum cleaner for aberrant transcripts but also as a regulator of physiological mRNA abundance, became apparent due to several microarray studies in different organisms (reviewed in [16]). Transcriptome-profiling with NMD-deficient Saccharomyces cerevisiae, Drosophila melanogaster and Homo sapiens cells revealed that NMD directly and indirectly controls the abundance of 3–10% of all mRNAs in the respective cells [14, 17–21]. Several features of physiological mRNAs can render them NMD-sensitive, whereas some mRNAs have evolved stabilising elements protecting them from NMD. Notably, introns in the 3′ untranslated regions (3′ UTR), ORFs located upstream of the main coding region (uORFs), programmed frameshifts and long 3′ UTRs all can elicit NMD. Messenger RNAs containing UGA triplet(s) that direct selenocysteine incorporation represent an interesting case of NMD substrates; when selenium is abundant, UGA codes for selenocysteine, but it is interpreted as a PTC when the selenium concentration is low [22] (see "In selenium-deprived cells, NMD reduces the selenoprotein encoding mRNA" for a discussion of selenoproteins and NMD). The large and diverse repertoire of transcripts controlled by NMD reflects the significant influence of NMD on the metabolism of the cell and consequently in many human diseases.
NMD is implicated in the modulation of many genetic diseases
There are numerous examples of human diseases associated with mutations that result in PTCs [23–25]. If translated, the PTC+ mRNAs would give rise to truncated proteins that have either completely lost their function, are still functional, have acquired dominant-negative function or have gained new functions. As a consequence of these different possibilities, NMD has a double-edged effect on the manifestation of a disease: NMD is detrimental if it prevents the production of proteins with some residual function, but it is beneficial if it prevents the synthesis of toxic truncated proteins. Hence, NMD represents a crucial modulator of the clinical outcome of many genetic diseases.
The majority of PTC+ disease-associated alleles exert their negative effects due to insufficient production of a functional protein. An example where NMD aggravates the clinical outcome is provided by several disease phenotypes caused by mutations in the dystrophin gene. While most of the truncating mutations in the dystrophin gene are associated with a similar phenotype, the rare truncating mutations that occur near the 3′ end of the dystrophin gene can result in extremely variable phenotypes. It has been suggested that all truncated proteins encoded by genes with mutations near the 3′ end would in theory be capable of rescuing the DMD phenotype, but when NMD prevents their synthesis, the clinical manifestations of the disease are aggravated [26]. Conversely, NMD has a well-documented beneficial role in the degradation of PTC+ β-globin mRNA, thereby preventing the synthesis of C-terminally truncated β-globin that would otherwise cause toxic precipitation together with surplus α-globin chains. In a heterozygote context, the second wild-type allele supports almost normal levels of β-globin synthesis, contributing to the correct haemoglobin assembly, which is reflected in the recessive inheritance of this β-thalassemia type. However, rare NMD-insensitive PTCs are responsible for the dominant form of β-thalassemia [27, 28].
Given the general inspection and clean-up role of NMD, it is not surprising that diseases associated with premature translation termination are remarkably diverse. Furthermore, as alluded to above, the PTC position determines if NMD ensues and this contributes to the severity of the clinical manifestations. To list a few examples of this diversity: (1) the disease severity in the connective tissue disorder Marfan syndrome correlates with the abundance of the PTC+ fibrillin 1 (FBN1) mRNA [29], (2) PTCs at different positions in CFTR (cystic fibrosis transmembrane-conductance regulator) can cause mild to severe cystic fibrosis [30], and (3) truncated forms of IFNGR1 (interferon gamma receptor 1) can result in recessively or dominantly inherited susceptibility to mycobacterial infections [31, 32].
NMD also appears to play a prominent role in carcinogenesis. PTCs in several tumour suppressor genes (BRCA1, p53, WT1) have been reported to result in reduced abundance of their mRNAs due to NMD [33–36]. Indeed, a strategy referred to as “gene identification by NMD inhibition” (GINI) has been successfully used to identify tumour suppressor genes [37, 38], pointing to a crucial role of NMD in eliminating faulty tumour suppressor transcripts and thus protecting cells from malignant growth.
In addition to diseases that result from nonsense mutations, defects in NMD factors can also cause disease. A recent study identified mutations in the NMD core factor UPF3B to be responsible for X-linked mental retardation [39]. Furthermore, there is a growing body of evidence that many physiological transcripts are subject to NMD regulation. It is likely that the impairment of NMD-dependent regulation of the abundance of these mRNAs is a yet to be identified cause of various human diseases.
For many disorders caused by PTC-generating mutations there are no effective treatments yet available. However, in instances where NMD eliminates transcripts that, despite the PTC, would still encode a functional protein, promising results have been obtained with PTC read-through approaches. Remarkably, as little as 1–5% of normal protein levels can greatly reduce or eliminate the principal manifestations of PTC-associated diseases such as cystic fibrosis and Hurler syndrome [40–42]. Aminoglycoside antibiotics (e.g. gentamicin) have been shown in vitro to suppress nonsense mutations by promoting read-through of termination codons and have improved CFTR function in clinical trials with cystic fibrosis patients [43, 44]. However, very high aminoglycoside concentrations are required and the associated toxic side effects have limited their clinical use. Instead, a new suppressor of PTCs was recently reported that selectively induces ribosomal read-through only at PTCs and not at natural termination codons [45]. This small molecular compound, called Ataluren (formerly PTC124), rescued striated muscle function in mdx mice (a model for muscular dystrophy) and has been tested in phase II clinical trials on cystic fibrosis patients. The oral administration of the drug reduced the epithelial electrophysiological abnormalities caused by CFTR dysfunction. Ataluren was generally well tolerated with infrequent and mild to moderate adverse effects [46]. These results are encouraging and the application of Ataluren might well be extended to other diseases such as DMD.
The parts list: human NMD factors
The first trans-acting factors required for NMD were identified through genetic screens in S. cerevisiae (called Upfs, for up-frameshift; [47–50]) and in C. elegans (called SMGs, for suppressor of morphological defects on genitalia; [51–53]). The human orthologues were later identified based on sequence similarities [54–61]. The NMD pathway in human cells comprises the factors UPF1, UPF2, UPF3A, UPF3B, SMG1, SMG5, SMG6 and SMG7, with UPF1, UPF2 and UPF3 (A and B) being the homologues of C. elegans SMG2, SMG3 and SMG4, respectively (reviewed in [3]; Table 2). More recently, four additional NMD factors have been described: NAG, DHX34, SMG8 and SMG9. RNAi-mediated depletion of NAG and DHX34, the human homologues of C. elegans SMGL-1 and SMGL-2, increased the abundance of a NMD reporter mRNA [62]. The two factors SMG8 and SMG9 were shown to regulate SMG1 kinase activity in human cells and their knockdowns moderately stabilised a PTC+ β-globin reporter transcript [63]. An interspecies comparison of NMD factors revealed a number of interesting differences with probable mechanistic consequences (see below). While SMG7 is present in mammals and C. elegans, D. melanogaster appears to lack a SMG7 homologue [64], and in contrast to metazoans, S. cerevisiae only requires UPF1, UPF2 and UPF3 for NMD. SMG5 and SMG6 homologues are not present in S. cerevisiae and deletion of the SMG7 homologue Ebs1p has been shown to only slightly increase the abundance of endogenous NMD targets [65].
Table 2.
Human NMD factors
| Protein | Alternative name/name in other species | Characteristics | Direct interaction partners | Cellular localisation | References |
|---|---|---|---|---|---|
| UPF1 | RENT1 (human, mouse) | RNA helicase, nucleic acid-dependent ATPase and RNA-binding protein | UPF2 | Shuttling protein; at steady-state predominantly in the cytoplasm | [48–50, 54, 56, 58–61, 66–71, 73, 74, 80, 83, 92, 96, 102, 103, 188, 225–227] |
| SMG2 (C. elegans) | SMG1 | ||||
| Phosphorylated at serine residues in C-terminal SQ motifs | SMG5a | ||||
| SMG6 | |||||
| SMG7 | |||||
| eRF1 | |||||
| eRF3 | |||||
| PP2Ab | |||||
| BTZe | |||||
| UPF2 | RENT2 (human, mouse) | Phosphoprotein, phosphorylated at serine residues in N-terminal part | UPF1 | Cytoplasmic; mainly perinuclear | [20, 49, 50, 56, 57, 60, 70, 71, 74, 79, 82, 83, 85, 89, 92, 226, 228, 229] |
| Nmd2p (S. cerevisiae) | UPF3A UPF3B | ||||
| SMG3 (C. elegans) | Promotes phosphorylation of UPF1 | eRF3c | |||
| SMG1 | |||||
| UPF3A and UPF3B | UPF3X (= UPF3B) | Associates with the EJC | UPF2 | Shuttling protein; at steady-state primarily in the nucleus (UPF3B) | [39, 49, 56, 60, 79, 83, 85–89, 92, 93, 226, 228] |
| SMG4 (C. elegans) | Promotes phosphorylation of UPF1 | eRF3c | |||
| Y14d | |||||
| MAGOHd | |||||
| BTZe | |||||
| eIF4A3e | |||||
| SMG1 | ATX | Ser/Thr-kinase of PIKK family, phosphorylates UPF1 | UPF1 | Cytoplasm and nucleus | [55, 61, 63, 74, 75, 94, 106, 222, 224] |
| UPF2 | |||||
| SMG8f | |||||
| SMG9f | |||||
| SMG5 | EST1B | Required for UPF1 dephosphorylation, directs PP2A to phosphorylated UPF1 | UPF1a | Cytoplasm; co-localises with SMG7 to P-bodies | [58, 64, 80, 96, 97, 99, 202, 203] |
| SMG7 | |||||
| PP2A | |||||
| SMG6 | EST1A | Ribonuclease with endonucleolytic activity. Required for UPF1 dephosphorylation | UPF1 | Cytoplasm | [64, 81, 96, 99–101, 202, 203] |
| PP2Af | |||||
| SMG7 | EST1C | Required for UPF1 dephosphorylation | UPF1 | Cytoplasm; co-localises with SMG5 to P-bodies | [53, 58, 64, 65, 96, 97, 202, 203] |
| Ebs1p (S. cerevisiae) | SMG5 | ||||
| PP2Af | |||||
| SMG8 | – | Forms a complex with SMG1 | SMG1f | Not analysed | [63] |
| Inhibits SMG1 kinase activity | |||||
| SMG9 | – | Forms a complex with SMG1 | SMG1f | Not analysed | [63] |
| NAG | SMGL1 (C. elegans) | Essential for NMD in humans and C. elegans | Not analysed | Not analysed | [62] |
| DHX34 | SMGL2 (C. elegans) | RNA helicase activity. Essential for NMD in humans and C. elegans | Not analysed | Not analysed | [62] |
aProof for direct interaction missing, shown by immunoprecipitation and mutational analysis (deletion of UPF1’s N-terminal 63 amino acids [58])
bDirect interaction only shown in C. elegans [80]
cDirect interaction only shown in S. cerevisiae [226]
dProof for direct interaction missing; interaction to UPF3B shown by immunoprecipitation and mutational analysis (MAGOH D66A–E68A and Y14 E82A–E83A [92])
eProof for direct interaction missing; detected in complex with UPF3B or UPF1, respectively
fProof for direct interaction missing; detected in complex with indicated factor
The UPF proteins constitute the core NMD machinery
Of all the UPF genes, UPF1 is functionally the most important factor for NMD and hence is the most conserved [3, 66]. UPF1 is a group 1 RNA helicase and nucleic acid-dependent ATPase. The ATPase activity resides in two of the seven helicase motifs in the middle section of the protein and is linked to the 5′-to-3′ helicase activity [67, 68]. ATP hydrolysis provides the energy to facilitate modulations in the structure of RNA or RNA–protein complexes [69]. UPF1 interacts with UPF2 [56, 60, 70] through its N-terminal cysteine- and histidine-rich (CH) domain, which displays a unique combination of three zinc-binding motifs arranged in two tandem modules [71]. UPF1 also interacts with the eukaryotic translation release factors eRF1 and eRF3 [72, 73], and Kashima et al. reported based on immunoprecipitation experiments that UPF1 forms a complex with SMG1, eRF1 and eRF3 (called SURF complex [74]). UPF1 is a phosphoprotein and sequential phosphorylation/dephosphorylation cycles are essential for NMD in mammals and C. elegans because they contribute to remodelling of the mRNA surveillance complex [58]. The C-terminal serine-glutamine (SQ) motifs of UPF1 are targets for phosphorylation by SMG1 [55, 61, 75]. In addition to the SMG1 kinase, UPF1 phosphorylation was shown to require UPF2 and UPF3 [66, 74, 75], but more recent studies have provided evidence for both UPF2-independent and UPF3-independent NMD pathways [73, 76–79]. The phosphorylation of UPF1 probably induces the dissociation of eRF3 from UPF1, since it was found that the over-expression of a SMG1 mutant deficient for its kinase activity strongly increased UPF1 co-immunoprecipitation with eRF3 [74]. Phosphorylated UPF1 interacts with SMG5, SMG6 and SMG7, which in turn promotes dephosphorylation of UPF1 by the protein phosphatase PP2A [58, 80, 81].
Similar to UPF1, UPF2 is also a phosphoprotein, both in mammals [81] and in S. cerevisiae [82]. Phosphorylation of the yeast Upf2p occurs at serine residues in its N-terminal domain. Together with other specific amino acids in the N-terminal region, these serine residues are responsible for eliciting NMD and for the interaction of Upf2p with Hrp1p, an RNA-binding protein implicated in yeast NMD [82]. The N-terminal region also contains several nuclear localisation signals (NLS), yet the protein resides predominantly in the cytoplasm [56, 57, 60]. Besides binding to UPF1, UPF2 also interacts through a separate surface with UPF3, thereby acting as a bridge between these two proteins [56, 60, 83, 84]. The two regions of human UPF2 that contact UPF1 have been mapped to amino acids 94–133 and 1,085–1,124/1,167–1,194, with the C-terminal region of UPF2 contributing more to the interaction [60]. The precise interaction of the C-terminal region of UPF2 with the CH domain of UPF1 has recently been determined in a structural study [70]. Using highly conserved, mainly negatively-charged, residues in the last of its three MIF4G (middle portion of eIF4G) domains, UPF2 interacts with a mainly positively-charged β-sheet surface of the RNP domain (ribonucleoprotein-type RNA-binding domain) of UPF3B [85]. Notably, UPF2 alone and the UPF2-UPF3B complex, but not UPF3B alone, bind to RNA in vitro [85].
UPF3 is the least conserved of the UPF proteins [3]. In contrast to yeast, nematodes and flies, the human genome encodes two UPF3 genes: UPF3A on chromosome 13 and UPF3B (also called UPF3X) on the X chromosome [56, 60]. Both genes generate two alternatively spliced mRNAs, resulting in four UPF3 isoforms. In UPF3B, skipping of exon 8 leads to a protein lacking amino acids 270–282 of the otherwise 483 amino acids containing polypeptide, and exon 4 skipping of UPF3A results in a 420 amino acids containing UPF3AS protein, lacking amino acids 117–149 of the 452 amino acids long UPF3AL isoform [60]. Two proteins corresponding in mass to UPF3AS and UPF3AL have been detected using an UPF3A-specific anti-serum [86], but the short form is probably not ubiquitously produced [87]. UPF3A and UPF3B share an overall similarity of 60%, with the N-terminal amino acids 38–236 (comprising the RNP domain and a nuclear export signal) being the most conserved (86% similarity). The C-terminal halves (amino acids 202–453) are considerably more divergent and harbour one or several NLSs [56, 60]. Despite the extensive similarity, tethering assays have shown that UPF3B is more effective than UPF3A at triggering NMD and stimulating translation [88]. High activity in NMD correlates with a short C-terminal sequence motif that is well conserved in UPF3B of different species, but not in UPF3A proteins [88]. Recently, it was shown that UPF3A and UPF3B compete for binding to UPF2 and consequently, when UPF3B levels are low, more UPF3A can bind to UPF2, which stabilises UPF3A because UPF3A alone is likely to be inherently unstable. On the other hand, when UPF3B levels are high, less UPF3A can bind to UPF2 and therefore the UPF3A levels decrease. Most likely it is crucial that UPF3A is quickly degraded when UPF3B levels are high because UPF3A is a less efficient NMD activator [87]. The UPF3 proteins are components of the exon-junction complex (EJC) [89]. The C-terminal region of UPF3 interacts with a composite binding site of the EJC core [90, 91] comprising parts of Y14, MAGOH and eIF4A3 [88, 92, 93].
The SMG proteins determine the phosphorylation status of UPF1
As previously mentioned, SMG1 is a protein kinase that can phosphorylate UPF1 [55, 61]. SMG1 belongs to the phosphatidylinositol 3-kinase-related protein kinase (PIKK) super family and functions specifically as a serine-threonine kinase (reviewed in [94]). Regulation of UPF1’s phosphorylation state during NMD is probably limited to metazoans because S. cerevisiae appear to lack orthologues of SMG1, SMG5 and SMG6, and NMD is only moderately affected by a deletion of the SMG7 homologue Ebs1p [65]. Biochemical studies have revealed SMG1 in different multi-protein complexes: in addition to the SURF complex, where SMG1 is associated with UPF1, eRF1 and eRF3, SMG1 also co-immunoprecipitates with EJC components (eIF4A3, Y14, MAGOH) and NMD factors (UPF1, UPF2, UPF3A, UPF3B and SMG7) in HeLa cell extracts [74]. There is evidence that UPF1 phosphorylation requires the association between the SURF complex and the EJC, and that this association is mediated by direct interactions of SMG1 with UPF2 and/or Y14 [74].
C. elegans mutated for SMG5, SMG6 or SMG7 are deficient in NMD and accumulate hyper-phosphorylated SMG2 (the UPF1 orthologue), suggesting that these factors are required for dephosphorylation of SMG2 [66]. Moreover, SMG5 interacts specifically with phosphorylated SMG2, with SMG7, and with the structural and catalytic subunits of protein phosphatase 2A (PP2A), inferring that SMG5 may give PP2A specificity for UPF1 [80]. Similarly, human SMG6 was also shown to co-purify with the catalytic subunit of PP2A, SMG1, UPF1, UPF2 and UPF3B, and also to specifically target dephosphorylation of UPF1, but not of UPF2 [81]. (Please note that SMG6 is termed Smg5/7a in this study.)
SMG5, SMG6 and SMG7 share a similar domain organisation: all contain two tetratricopeptide (TPR) repeats, either at the N-terminus (SMG5 and SMG7) or in the middle section (SMG6). TPR-containing domains consist of 34 amino acids long TPR repeats that usually function as mediators of protein–protein interactions [95]. For SMG7, the TPRs were shown to adopt a similar fold to 14-3-3, which is a signal-transduction protein that binds phosphoserine-containing polypeptides. Sequence similarities suggest conservation of this 14-3-3-like domain structure in SMG5 and SMG6 [96], and in SMG7 this domain is responsible for binding to UPF1 [96] as well as contributing to an interaction with the corresponding 14-3-3-like domain of SMG5 [97].
SMG5 and SMG6 contain a PIN-like domain (for PilT N terminus) in their C-termini. Generally, PIN-like domains function as phosphodiesterases and often exhibit nuclease activity [98]. Despite the fact that the PIN-like domains of SMG5 and SMG6 adopt a similar overall fold that is related to ribonucleases of the RNase H family, SMG6 harbours the canonical triad of acidic residues crucial for RNase H activity, whereas SMG5 lacks two of these three key catalytic residues [99]. These structural differences are reflected at the molecular level, as only the PIN domain of SMG6 has nuclease activity on single-stranded RNA in vitro [99], and it was recently demonstrated in human and Drosophila cells that SMG6 is the endonuclease that can initiate cleavage of nonsense mRNA near the PTC [100, 101] (see below).
Intracellular localization of NMD factors
Reliable determination of the intracellular distribution of any protein by immunofluorescence microscopy depends on the availability of highly specific antibodies. Moreover, plasmid-based expression of tagged proteins or GFP fusion proteins can produce non-physiological localisation patterns. With this caveat in mind, immunolocalisation studies of NMD factors in human cells revealed that UPF1 localises predominantly to the cytoplasm [56, 60]. However, biochemical evidence implies that UPF1 shuttles between the cytoplasm and the nucleus [102], which is consistent with its role in both nuclear and cytoplasmic processes ([103, 104] and see below). UPF2 exhibits perinuclear cytoplasmic localisation and UPF3 is a nucleo-cytoplasmic shuttling protein with steady-state nuclear localisation [56, 60]. The SMG5, SMG6 and SMG7 proteins are all localised to the cytoplasm. Over-expressed SMG5 and SMG7, together with UPF1, co-localise with P-bodies, and the C-terminus of SMG7 has been shown to be required for this localisation [96, 97, 105]. In contrast, it was shown that over-expressed SMG6 accumulated in separate cytoplasmic foci [97]. Biochemical fractionation and immunofluorescence microscopy revealed that SMG1 localises to both the cytoplasm and the nucleus [106].
Intriguingly, in Arabidopsis thaliana, most of UPF3 and a substantial fraction of UPF2 were recently reported to localise to the nucleolus, and nucleolar fractions were highly enriched in aberrant mRNAs that are NMD substrates [107].
Distinguishing between proper and aberrant translation termination
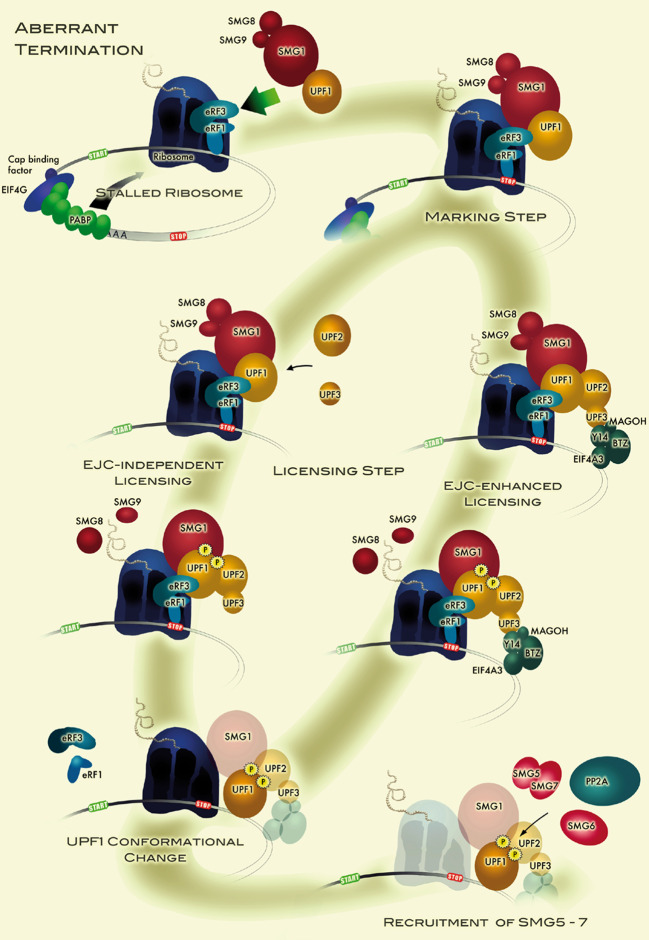
NMD strictly depends on reading frame recognition and therefore on translation. The decision of whether NMD is to be initiated or not is made when a ribosome stalls at any of the three termination codons (UAG, UGA and UAA). The local mRNP environment of the termination codon influences the process of translation termination (reviewed in [108–110]). During normal translation termination, eRF1 recognises the termination codon in the A-site of the stalling ribosome and forms a complex with the GTPase eRF3 to catalyse peptide release [111–113]. Through its C-terminal region eRF3 interacts with the C-terminus of eRF1, while the N-terminal region of eRF3 interacts with the C-terminal domain of poly(A)-binding protein (PABP) [114–116], which is believed to stimulate proper and efficient translation termination [117] (Fig. 1). It has been shown in yeast that when the interaction between the eRF3 orthologue Sup35 and Pab1p is impaired, the terminating ribosome cannot efficiently dissociate from the mRNA [118]. Consistent with the corresponding role of mammalian cytoplasmic poly(A) binding protein 1 (PABPC1) in stimulating translation termination, it was recently demonstrated that mammalian cells lacking PABPC1 exhibited increased read-through of termination codons [73]. In the case of transcripts that are subjected to NMD, the interaction between eRF3 and PABPC1 is less efficient and is antagonised by UPF1 recruitment [119] (Fig. 2). In S. cerevisiae, D. melanogaster and human cells, NMD can be suppressed by tethering PABP near the PTC [73, 118–122], which further supports the NMD antagonising function of PABP.
Fig. 1.

Schematic model of efficient translation termination in the proper mRNP environment. The model postulates that normal translation termination involves an interaction between PABP and eRF3, which by a currently unknown mechanism promotes fast polypeptide release, disassembly of the ribosomal subunits and re-initiation of the ribosome at the start codon. The proper termination-stimulating mRNP environment is characterised by a protein complex involving PABP, eIF4G, the cap binding factor (eIF4E or CBP80/CBP20) and additional factors that bring the 5′ and the 3′ ends of the mRNA in close proximity and constrain the mRNP in a circular structure
Fig. 2.
Model for aberrant translation termination, which leads to the assembly of a mRNA surveillance complex that marks the mRNA for subsequent degradation. When the stop codon is located in an mRNP environment where it fails to receive the PABP-mediated termination-stimulating signal, the ribosome stalls for a prolonged period of time at the stop codon, which allows binding of UPF1 to eRF3. The assembly of this SURF complex marks the mRNA for NMD. The model further postulates that this marking step is still reversible and that the mRNA is only irreversibly committed to NMD after UPF1 phosphorylation (licensing step). UPF2 and/or UPF3 are necessary for SMG1-mediated UPF1 phosphorylation. The presence of an EJC in the 3′ UTR serves as a strong enhancer of NMD (EJC-enhanced licensing), because UPF2 and/or UPF3 interaction with the SURF is greatly facilitated by virtue of their close proximity, whereas UPF2 and/or UPF3 recruitment take longer in the absence of an EJC (EJC-independent licensing), resulting in overall less efficient NMD. Following phosphorylation and possibly induced by ATP hydrolysis, UPF1 undergoes a conformational change that increases its affinity for RNA and is then ready for interaction with SMG5–7, which initiate mRNA degradation (see Fig. 3)
The key NMD factor UPF1 has been shown to interact with eRF3 and eRF1 [72], but it is currently unknown how UPF1 is recruited to the terminating ribosome, and if it is present in all termination events. There is evidence that the decision of whether NMD is to be triggered or not relies upon competition between UPF1 and PABP for binding to eRF3 on the terminating ribosome [119]. The outcome of this competition is largely influenced by the structure of the mRNP [121]. Extensive research using a variety of model systems has provided a multitude of data regarding the NMD-triggering characteristics of mRNPs. One clearly defined NMD-eliciting feature is the length of the 3′ UTR, which proposes that the physical distance between the termination codon and the poly(A) tail of the transcript determines the fate of the mRNA. Experiments carried out in yeast, worms, plants, human and fly cell lines revealed that termination codons are recognised by the NMD machinery when they are situated too far upstream from the poly(A) tail [52, 62, 118–121, 123–126]. The “faux 3′ UTR model” was first coined to describe NMD in yeast, proposing that premature termination is intrinsically abnormal since the long 3′ UTR is not properly configured to bring Pab1p into the proximity of termination codon to ensure efficient translation termination [118]. Based on the aforementioned studies conducted in a variety of organisms, we advocate extension of the “faux 3′ UTR model” to a unified model that explains the basic mechanism for recognition of NMD substrates in all organisms [4] (Fig. 2). In the situation of the NMD-inhibiting signal from PABP being absent, then UPF2 and/or UPF3 can bind to UPF1. In mammals, UPF1 is present in the SURF complex and UPF2 and/or UPF3B are thought to promote SMG1-mediated phosphorylation of UPF1 [74]. Subsequently, SMG5, SMG6 and/or SMG7 proteins can bind to phosphorylated UPF1, eventually leading to the degradation of the transcript. The results from Kashima et al. argue that phosphorylation of UPF1 requires both UPF2 and UPF3B [74]. Yet, recent data from other laboratories unveiled the existence of both UPF2-independent and UPF3-independent branches of the NMD pathway [73, 76–79].
Numerous investigations have shown that not only the physical distance of the 3′ UTR but also the specific RNA sequences that it contains can determine the fate of the mRNA. For example, it was proposed that the surveillance complex scans and identifies a downstream sequence element (DSE) that stimulates NMD in yeast, but this element has remained poorly defined [127, 128]. Despite the fact that similar sequence elements have not been found in mammals, it is possible that a longer 3′ UTR provides a better platform for binding of trans-acting factors that improve NMD efficiency. The average 3′ UTR length in humans is 700–800 nucleotides, yet experiments with reporter mRNAs harbouring 3′ UTRs that are only 200–300 nucleotides in length were already able to display some destabilisation due to NMD in human cell culture [121]. As proposed by Singh et al., mRNAs containing long 3′ UTRs have perhaps evolved mechanisms to evade NMD [119]. The stability can possibly be achieved by highly structured 3′ UTRs, where RNA base pairing and internal loops can bring the poly(A) tail closer to the termination codon [121]. Furthermore, specific RNA sequences or secondary structures might recruit NMD antagonising factors to the vicinity of the stop codon.
Mammalian tissue culture studies have established the splicing-dependent EJC [129, 130] as another important trans-acting component for NMD initiation. The mammalian transcriptome is complex with a large number of multi-exon genes and extensive alternative splicing. One-third of all alternatively spliced transcripts are expected to be NMD substrates [9]. The coupling of a splicing-dependent signal to NMD facilitates efficient recognition of this group of NMD substrates and increases the accuracy of gene expression [131–133]. Indeed, if a termination codon occurs prematurely in the mRNA, it is probable that an exon–exon junction will be present downstream of the PTC, and this probability increases the further upstream the PTC is located. During pre-mRNA splicing, EJCs are deposited 20–24 nucleotides upstream of exon–exon junctions [129]. The EJC is a dynamic structure with a heterogeneous protein composition, wherein the proteins Y14, MAGOH, eIF4A3 and Barentsz (Btz) remain associated with mRNA after export into the cytoplasm and constitute the stable core of the EJC [90, 91]. The NMD factors UPF2 and UPF3 have been found associated with EJC, which hinted for a role of the EJC in NMD [89]. It is thought that EJCs located within an ORF are removed by elongating ribosomes in a process that involves the ribosome-associated protein PYM [134]. In contrast, EJCs located downstream of the termination codon remain associated with the mRNP. The presence of an EJC downstream of the SURF complex greatly facilitates UPF2 and/or UPF3 interacting with the SURF by virtue of their close proximity. According to this mechanistic model, the presence of an EJC downstream of the termination codon serves as a potent enhancer of NMD (Fig. 2; reviewed in [110]).
Another interesting observation with regards to NMD efficiency as a function of PTC position is that in several mRNAs PTCs close to the translation initiation codon fail to elicit efficient NMD [36, 121, 135–138]. It is known that PABPC1 can bind to eukaryotic initiation factor 4G (eIF4G) which acts to bridge the 3′ and 5′ ends of the mRNA into a circular structure that enhances translation [139, 140]. In this closed-loop structure, the NMD suppressor PABPC1 would also be in close distance to the 5′ region of an mRNA where it can antagonise NMD initiation at early PTCs, regardless of the presence of downstream EJCs [122, 141]. In addition, the ribosome could initiate at an in-frame AUG downstream of the early PTC and so remove the remaining EJCs from the mRNA, which would eliminate a potent promoter of NMD.
In mammalian cells, NMD has been proposed to occur during the so-called pioneer round of translation, before eIF4E replaces the cap-binding complex (CBC) [142]. CBP80, which is a constituent of the CBC, has been shown to interact with UPF1 and the knockdown of CBP80 reversed NMD, but not the repression of steady-state translation through the translation inhibitor 4E-BP1 ([143, 144], reviewed in [10]). In contrast, NMD occurs mainly on eIF4E-bound mRNAs in S. cerevisiae: CBC-bound transcripts are largely insensitive to NMD and the yeast homologue of CBP80 is dispensable for NMD [145–147].
In summary, we believe that most of the currently available experimental data support a NMD model in which the competitive interaction between UPF1 and PABP with the ribosome-bound release factors is the key determinant for NMD initiation. Alternative models that remain to be experimentally tested were recently proposed by Brogna and Wen [148]. It is not yet possible to unambiguously predict which mRNPs are able to elicit NMD and to explain each of the discussed NMD-triggering features of mRNPs. A variety of studies have shown that certain cis-acting elements, such as long 3′ UTRs or the presence of EJCs downstream of the termination codon, are sufficient on their own to trigger NMD, probably because, in both cases, UPF1 binding to eRF3 and subsequently to UPF2 and/or UPF3 is facilitated. Nonetheless, neither a long 3′ UTR, a downstream EJC nor any other reported NMD triggering feature on mRNA is absolutely required for NMD. Therefore, the only certain requirement for NMD so far is the translation-dependent recruitment of UPF1 to the mRNA.
Degradation of NMD targets
Evidence for endonucleolytic and exonucleolytic decay pathways
Messenger RNA turnover involves an important and tightly regulated set of ribonuclease-mediated degradation pathways used by eukaryotic cells to regulate normal gene expression, to get rid of aberrant mRNAs (e.g. those containing PTCs) and to eliminate parasitic mRNA species (viral mRNAs, transposons). Due to their critical functions, many of the ribonucleases responsible for RNA degradation are highly conserved amongst eukaryotes [149–152]. Degradation of normal mRNAs is generally initiated by the removal of the poly(A) tail from the 3′ end of mRNAs. In yeast, the deadenylation reaction is catalysed by the Ccr4p/Caf1p complex, while in mammals the poly(A) tail is shortened by the consecutive action of two different complexes: the PAN2/PAN3 complex first shortens full-length poly(A) tails of approximately 200 bases to approximately 110 bases. These intermediate poly(A) tails are subsequently targeted by the CCR4/CAF1 complex that removes the remaining adenines [153] and followed by exonucleolytic degradation of the transcript in 3′-to-5′ direction by the exosome [154]. Concomitant with CCR4/CAF1-mediated deadenylation, the decapping enzyme DCP1/DCP2 removes the 7-methyl guanosine (m7G) cap structure, leaving an unprotected 5′ end that is accessible for rapid degradation by the abundant 5′-to-3′ exonuclease XRN1.
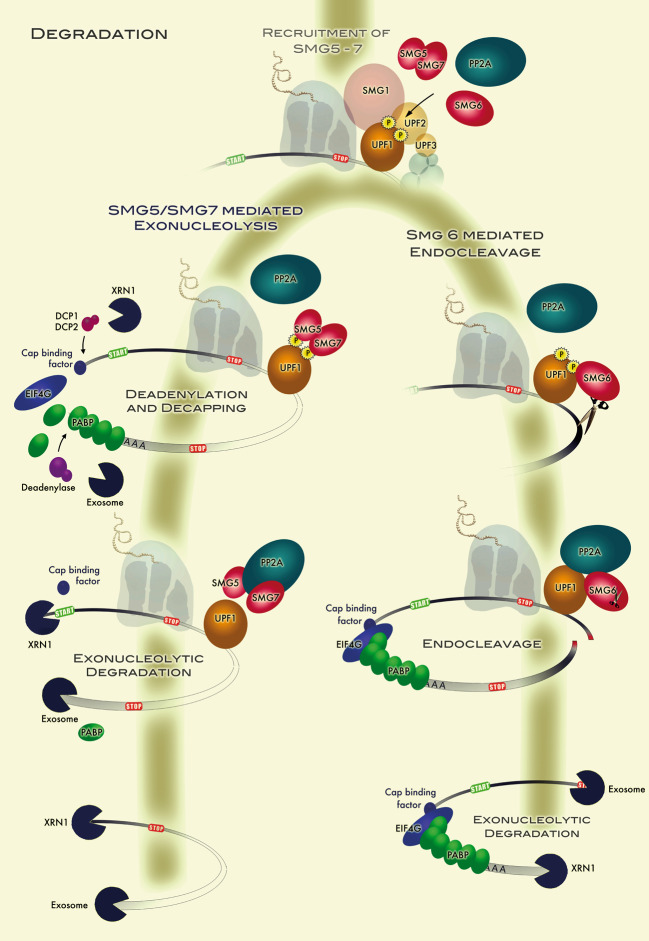
An important question to address is what pathway of degradation NMD follows. For instance, are the general mRNA turnover pathways utilised after improper translation termination, or is NMD initiated by special nucleases? In S. cerevisiae, degradation of PTC+ mRNAs has been shown to rely on the general mRNA turnover pathway and a modification thereof that is typified by deadenylation-independent removal of the m7G-cap structure, followed by subsequent Xrn1p-mediated 5′-to-3′ exonucleolytic decay [155, 156]. However, NMD substrates were also found to be channelled into a degradation pathway that involves deadenylation followed by 3′-to-5′ exonucleolysis by the exosome and Ski complexes [155, 157]. In contrast, NMD in D. melanogaster is elicited by endonucleolytic cleavage of the PTC+ mRNA in the vicinity of the PTC. The resulting 5′ and 3′ decay intermediates are then rapidly degraded in the 3′-to-5′ direction by the exosome and in the 5′-to-3′ direction by XRN1, respectively, and can only be detected in cells depleted for the aforementioned exonucleases [158]. In human cells, the situation is less clear and probably more complex. Degradation of NMD reporter transcripts via the conventional mRNA turnover pathway, starting with deadenylation, followed by decapping and XRN1-mediated exonucleolytic decay, has been reported [159–161] (Fig. 3; SMG5/SMG7 mediated exonucleolysis). Consistent with a role of decapping in human NMD, UPF1 and DCP1a have recently been documented to interact via a protein called PNRC2 [162]. On the other hand, endonucleolytic cleavage near the PTC has recently been demonstrated in human cells depleted for XRN1 [100]. Moreover, the PIN domain of SMG6 was shown to possess the endonuclease activity responsible for initiating NMD in both Drosophila and human cells [100, 101]. Therefore, it seems that mammalian NMD targets can be degraded by both a SMG6-dependent endonucleolytic pathway and a deadenylation- and decapping-dependent exonucleolytic pathway (Fig. 3; SMG6 mediated endocleavage), whereas Drosophila NMD is confined to the former and yeast NMD to the latter decay pathway. Consistent with this conclusion, mammals possess both SMG6 and SMG7, whereas Drosophila lacks a SMG7 homologue and yeast has no SMG6 equivalent. Further work is required to determine the relative contributions of the two decay pathways involved in mammalian NMD and to understand what determines which decay route is taken by the different types of mRNAs directed to the NMD pathway.
Fig. 3.
Model for degradation of NMD substrates. The model posits that UPF1-bound mRNAs can be degraded by two different pathways, depending on whether the SMG5/SMG7 heterodimer or the endonuclease SMG6 binds to phosphorylated UPF1. Interaction of SMG5/SMG7 with phospho-UPF1 promotes deadenylation followed by decapping and exonucleolytic RNA decay from both ends (left branch). Interaction of SMG6 with phospho-UPF1 leads to a SMG6-mediated endonucleolytic cleavage near the aberrant termination site, followed by the exonucleolytic degradation of the two RNA fragments from the initial cleavage site
Does human NMD take place in P-bodies?
Prominent cytoplasmic foci called processing bodies (P-bodies, also called DCP1- or GW-bodies) have received ample attention during the last few years because they seem to represent important sites for translational repression and mRNA decay [163–165]. These highly dynamic granules are enriched for components of the decapping and 5′-to-3′ degradation machinery, including DCP1/DCP2, the decapping activators RCK/p54, Hedls/Ge-1, EDC3 and Pat1, the Lsm1-7 complex and the 5′-to-3′ exonuclease XRN1. Mammalian P-bodies also contain the deadenylases CCR4/CAF1 and PAN2/PAN3 [166], as well as components of the miRNA pathway [164]. Additionally, it has been found that, under certain conditions, the NMD factors UPF1, UPF2, UPF3, SMG5 and SMG7 are localised in P-bodies [97, 105, 162, 167]. The co-localisation of the mRNA degradation machinery and of various NMD factors in P-bodies has stimulated research on the functional significance of P-bodies in NMD. In yeast cells, Upf1p, Upf2p and Upf3p accumulate in P-bodies in Δdcp1, Δdcp2 or Δxrn1 strains [167]. Furthermore, a NMD reporter transcript was shown to localise to P-bodies in an Upf1p-dependent manner and more NMD reporter transcript accumulated in P-bodies of Δupf2 or Δupf3 strains [167]. These observations argue that NMD in yeast occurs within complexes that can accumulate into P-bodies. In contrast, NMD was not affected in Drosophila and human cells by RNAi-mediated depletion of GW182 and Ge-1, a treatment that prevented the formation of any microscopically visible P-bodies [168, 169]. These results indicate that NMD does not require the presence of P-bodies in metazoans. Moreover, SMG6 does not co-localise with P-bodies [97], which is consistent with the view that most NMD in human cells might occur outside of P-bodies where it is initiated by a decapping-independent SMG6-mediated endonucleolytic cleavage of the substrate RNA [100]. Conversely, this observation does not rule out the possibility that, under physiological conditions, NMD may occur to some extent in P-bodies, in keeping with the reported localisation of several NMD factors and a NMD reporter transcript to P-bodies in over-expression conditions [97, 105, 162].
An extensive array of phenotypes are observed upon inactivation of NMD effectors
While the mechanistic details of NMD are being gradually unravelled, the physiological role of NMD still remains largely unknown. The phenotypes observed upon inactivation of NMD effectors vary considerably amongst different organisms. NMD mutants in S. cerevisiae only show a partial impairment in respiratory growth which is enhanced at low temperatures [48], whereas C. elegans mutants have defects in the male bursa and the hermaphrodite vulva [51, 52]. In contrast, UPF1 and UPF2 are essential for the viability of D. melanogaster larvae [170], and depletion of NMD factors impairs proliferation of Drosophila embryo-derived cells (S2 cells) [19]. Likewise in zebrafish, UPF1, UPF2, SMG5 and SMG6 were shown to be essential for embryonic development and survival [171]. In mice, knockout of UPF1 is embryonic lethal and isolated blastocysts undergo apoptosis after only a few days in culture [172]. Likewise, no UPF2 knockout mouse embryos could be detected at day 9.5 post coitus, and conditional knockouts of UPF2 in hematopoietic precursors led to the complete extinction of hematopoietic stem and progenitor cells [6]. Similarly, in Arabidopsis thaliana, UPF1 is necessary for seedling growth [173].
Collectively, the severity of the phenotype resulting from inactivation of the various NMD factors correlates with the overall complexity of the organism and its extent of alternative splicing. This allows for the speculation that the accumulation of aberrantly spliced mRNAs and the resulting production of detrimental C-terminally truncated proteins may be the cause of the observed phenotypes. On the other hand, it may be that NMD regulates the expression of an essential protein in mice and flies but not in yeast and worms. Moreover, several of the proteins characterised as NMD factors have also been reported to function in cellular processes seemingly unrelated to NMD, and it is therefore equally plausible that the observed phenotypes are the consequence of disrupting these NMD-independent mechanisms.
In selenium-deprived cells, NMD reduces the selenoprotein encoding mRNA
Selenium (Se) is an essential micronutrient that is linked to many aspects of human health. A unique class of proteins called selenoproteins, most of which are involved in protecting the cell from oxidative stress, incorporate selenocysteine (Sec or U) at in-frame UGA codons in archaea, prokaryotes and eukaryotes. Unlike the other amino acids, Sec is made on its transfer RNA (tRNA) and selenocysteyl-tRNA[ser]sec is delivered to the A-site of the ribosome by a specific elongation factor called EFsec [174]. The fact that the UGA codon specifies Sec incorporation rather than translation termination is chiefly due to the presence of specific secondary structures in selenoprotein-encoding mRNAs termed selenocysteine insertion (SECIS) elements, and the trans-acting factors that associate with these SECIS elements. In eukaryotes, the SECIS element is located in the 3′ UTR (reviewed in [175]). A protein called SECIS binding protein 2 (SBP2) binds to SECIS elements and to the large ribosomal subunit and recruits the eEFsec-selenocysteyl-tRNA[ser]sec complex to the ribosome, ensuring efficient Sec incorporation in vivo and in vitro [175, 176]. Moreover, SBP2 can actually suppress the termination promoting function of eRF1 [177].
The efficiency of Sec incorporation varies depending on the nature of the SECIS element, the position of the UGA in the ORF [178] and the nucleotides surrounding it [177, 179], as well as the intracellular selenium concentration [22]. It was observed that Se deficiency decreases the abundance of mRNAs encoding the selenoprotein, glutathione peroxidase 1, and that this is possibly mediated by the NMD pathway [22]. The mRNA coding for phospholipid hydroperoxide glutathione peroxidase (PHGPx) was also reported to be an NMD target in NIH3T3 fibroblasts or H35 hepatocytes under Se-deprived conditions [180], but no corresponding change in PHGPx mRNA abundance in the liver or testis of Se-deficient rats was observed [181]. It was suggested that a mechanism absent in mouse NIH3T3 fibroblasts and rat H35 hepatocytes masks the NMD of PHGPx mRNA in the rat liver and testis [180]. Given the importance of selenoproteins to the viability of the animal and their specialised functions, the fact that these proteins are highly conserved and ancient, the complex cis-acting elements and trans-acting factors that ensure that the UGA codon is not read as a stop codon and their intricate hierarchy of expression which matches the Se intake, it may be that, under physiological conditions, NMD in combination with additional, presently unknown processes acts to regulate the stability of selenoprotein encoding mRNAs.
The multiple personalities of the NMD factors UPF1, SMG1 and SMG6
Staufen-1-mediated mRNA decay (SMD) and NMD are competitive pathways
The double-stranded RNA binding protein Staufen was originally identified as a maternal factor required for the correct formation of the anterioposterior axis in the D. melanogaster embryo [182, 183]. Staufen is principally known to be involved in the transport of mRNAs to ultimately achieve their localised translation [184, 185], in mice [186] and in humans [186, 187]. Mammalian Staufen protein has four double-strand RNA binding domains (dsRBDs) and contains a putative microtubule-binding domain of microtubule-associated protein 1B in its C-terminus that is not present in the Staufen protein of D. melanogaster or C. elegans [187].
In addition to Staufen’s previously characterised functions, mammalian Staufen was found to play a role in mRNA decay [188]. Yeast two-hybrid analysis revealed that Staufen-1 (STAU1) interacts with UPF1, and this interaction was confirmed by GST pull-downs and far western blotting (Table 3). Tethering of STAU1 to a reporter mRNA can induce UPF1-dependent and translation-dependent mRNA degradation called SMD (for STAU1-mediated mRNA decay) [188]. Natural targets of SMD are expected to bind STAU1 downstream of the translation termination codon and, upon depletion of either STAU1 or UPF1, the mRNA should be stabilised. In HeLa cells depleted for STAU1, approximately 1% of the transcriptome was more than twofold up-regulated whilst approximately 1% was more than twofold down-regulated, inferring that STAU1 potentially acts to regulate a multitude of functionally unrelated physiological transcripts and can influence many metabolic pathways [189].
Table 3.
Summary of additional NMD independent functions for UPF1, SMG1 and SMG6 in different cellular pathways
| Factor | Reported roles | Notes of interest | References |
|---|---|---|---|
| UPF1 | Staufen-mediated decay pathway: STAU1 binds the 3’ UTR and recruits UPF1 to elicit decay of the mRNA | NMD and SMD are competitive pathways, UPF1 binds either UPF2 or STAU1 but not to both simultaneously | [188, 189, 193] |
| DNA replication, repair and stability pathways: UPF1 contributes to the completion of DNA replication and proper cell cycle progression | UPF1 depletion results in an early S-phase arrest. UPF1 associates with chromatin in a cell cycle regulated manner and when ATR is depleted, chromatin loading of UPF1 is impaired. γ-irradiation increases UPF1 chromatin association. UPF1 co-immunoprecipitates with p66 subunit and p125 subunits of DNA polymerase δ | [103, 104, 207, 208] | |
| Replication dependent histone mRNA metabolism: UPF1 is important for histone mRNA degradation | UPF1 depletion stabilises histone mRNA after HU treatment and at the end of S-phase. Immunoprecipitation showed that UPF1 interacts with SLBP/HBP | [214] | |
| HIV-1 metabolism: possibly UPF1 stabilises HIV-1 RNA in infected cells | UPF1 is a component of the HIV-1 RNP and its function requires its ATPase activity. Its recruitment may be mediated by STAU1. over-expression of UPF1 results in up-regulation of HIV-1 expression at the RNA and protein level | [194] | |
| Telomere metabolism: possibly UPF1 facilitates TERRA release from telomeres | Detected in telomeric chromatin fractions. Depletion of UPF1 increases TERRA at telomeres and leads to telomere damage | [103, 104, 202, 203] | |
| SMG1 | Protection of genomic stability: SMG1 initiates cellular stress responses when genome integrity, mRNA translation or nutrient availability is compromised | SMG1 is activated by DNA damage and stimulated by UV or γ-radiation. Depletion of SMG1 results in DNA damage and increased sensitivity to radiation. Phosphorylates p53 in vitro and upon γ-radiation in vivo. Along with ATM, SMG1 can phosphorylate UPF1 in response to dsDNA breaks caused by γ-radiation. SMG1 is important for oxidative stress protection in C. elegans and protects against TNF-α induced apoptosis in human cells. SMG1 along with ATM can modulate p21 levels to inhibit cell division, induce DNA repair or block apoptosis | [106, 222, 224] |
| Telomere metabolism: possibly SMG1 facilitates TERRA release from telomeres | Detected in telomeric chromatin fractions. Depletion of SMG1 increases TERRA signals and leads to telomere damage | [103, 104, 202, 203] | |
| SMG6 | Telomere metabolism: possibly SMG6 facilitates TERRA release from telomeres | Detected in telomeric chromatin fractions. Physically interacts with telomerase. Depletion of SMG6 increases TERRA signals and leads to telomere damage. Over-expression results in end-to-end chromosome fusions and altered telomere lengths | [103, 104, 202–204] |
In addition to STAU1, there is a second Staufen protein called STAU2 that is encoded by a different gene and shares 51% amino acid identity with STAU1 [190]. Considering the high sequence similarity between STAU1 and STAU2 and also the evidence that STAU2 co-immunoprecipitates with mRNA from human cells [190, 191], a genome-wide approach was undertaken to examine if STAU1 and STAU2 can bind to and regulate a similar collection of mRNAs. Distinct but overlapping subsets of cellular mRNAs were found to associate with STAU1 and STAU2 containing RNP complexes [190]. To further elucidate the involvement of the Staufen proteins in mammalian RNA metabolism, the structure of the STAU1 and STAU2 binding sites and their position relative to the translation start and stop codons were examined. The position of the STAU1 binding site is critical as it determines whether the mRNA is targeted for enhanced translation [192] or if it is to be degraded by SMD [188]. It appears that SMD is a mechanism that depends on translation and is elicited when STAU1 binds to its hairpin binding site approximately 25–30 nucleotides downstream from the translation termination codon. Further analysis has indicated that the STAU1 binding site requires more elaborate secondary structure than a single hairpin element [189]. STAU1 is thought to recruit UPF1 to the 3′ UTR via direct interaction [188] and, therefore, SMD may be triggered by UPF1 interacting with the eRFs at the translation termination codon analogous to NMD. With this in mind, it will be insightful to determine if SMD also involves the UPF1 binding endonuclease SMG6, when it elicits degradation of its target transcripts. Interestingly, the STAU1 and UPF2-binding sites within UPF1 have recently been shown to overlap, rendering STAU1 and UPF2 binding to UPF1 mutually exclusive [193]. During the differentiation of C2C12 myoblasts to myotubes, NMD and SMD pathways appear to be in competition: SMD activity increases while the activity of the UPF2-dependent NMD pathway decreases [193]. In support of the physiological importance of this competition, SMD targets PAX3 mRNA whose decay promotes myogenesis, while the mRNA encoding for myogenin, a protein required for myogenesis, is an NMD target [193].
STAU1 and UPF1 are both involved in HIV-1 metabolism
UPF1 and STAU1 were also reported to function together in HIV-1 RNA metabolism [194–196] (Table 3). STAU1 was shown to associate with HIV-1 genomic RNA and two to five STAU1 proteins are incorporated per HIV-1 virion [197]. STAU1 over-expression can increase the abundance of HIV-1 genomic RNA and of STAU1 protein packaged into the virion [196]. RNAi-mediated depletion of STAU1 resulted in a substantial decrease of viral infectivity [195]. Moreover, STAU1 was shown in an RNA-dependent manner to directly interact with the nucleocapsid domain of the pr55Gag, a key mediator of HIV-1 genomic RNA encapsidation [195]. Therefore, the HIV-1 RNP contains the major structural protein pr55Gag, viral genomic RNA and the host protein STAU1, and it has been proposed that STAU1 together with pr55Gag plays an important role in viral assembly, genomic RNA encapsidation and the generation of infectious viral particles [195].
Recently, UPF1 was also identified as a component of the HIV-1 RNP [194]. The presence of UPF1 with the HIV-1 RNP is most likely mediated by STAU1 and it does not require any interactions between UPF2 and UPF3. Knockdown of UPF1 led to a large reduction in steady-state HIV-1 RNA and pr55Gag protein levels, and over-expression of UPF1 resulted in up-regulation of HIV-1 expression at the level of both RNA and protein. The effects of UPF1 on HIV-1 RNA stability were dependent on its ATPase activity and required ongoing translation [194]. It was suggested that targeting of UPF1 function might represent a suitable approach to arrest HIV-1 late in the replication cycle [194]. Further work will shed light on how UPF1 is recruited to the HIV-1 RNP, its exact role and also if UPF1 function is utilised by other retroviruses.
NMD factors function at telomeres
Telomeres are the heterochromatic structures located at the termini of linear chromosomes. They compensate for incomplete semi-conservative DNA replication and also protect the chromosomal ends against recombination with each other and with double-strand breaks (DSBs) inside the chromosomes. While repetitive sequences cover an average of 350 base pairs in yeast telomeres, human telomeres exceed several kilobases composed of TTAGGG repeats [198, 199].
The first connection between NMD factors and telomere function was reported when it was discovered that mutations of Upf1p, Upf2p and Upf3p in yeast led to telomere shortening and de-repression of silenced telomeric loci [200, 201]. Consistent with the idea that NMD affects the expression of genes important for telomere function, it was found that NMD mutant strains had increased levels of mRNAs encoding the telomerase catalytic subunit (Est2p), regulators of telomerase (Est1p, Est3p, Stn1p, Ten1p) and proteins implicated in regulation of telomeric chromatin structure (Sas2p, Orc5p) [200]. However, the picture became more complicated when a bioinformatics search for human homologues of the S. cerevisiae ever shorter telomeres 1 (Est1) gene identified the same three proteins that were independently identified as human orthologues of the C. elegans NMD factors SMG5 (= EST1B), SMG6 (= EST1A), and SMG7 (= EST1C) [202, 203]. Moreover, EST1A/SMG6 and EST1B/SMG5 were found to associate with telomerase activity in human cell extracts and over-expression of EST1A/SMG6 led to end-to-end chromosome fusions and altered telomere lengths [202, 203] (Table 3). EST1A/SMG6 interacts with telomerase by binding to the telomerase RNA with high affinity but low specificity, and also by making protein–protein contacts with telomerase reverse transcriptase (TERT) [204].
For a long time, telomeres were believed to be transcriptionally inactive. However, recently, a set of pol II transcripts called telomeric repeat-containing RNA (TERRA), also known as TelRNA, have been discovered in human [104, 205] and yeast cells [206] (reviewed in [207]). Fascinatingly, enrichment of NMD factors was detected in telomeric chromatin fractions, and the depletion of UPF1, SMG1 and EST1A/SMG6 (and to a lesser extent UPF2 and EST1C/SMG7) increased TERRA signals and triggered telomere damage, including complete telomere loss [103, 104]. Despite the presence of one UAG stop codon in each telomeric repeat sequence, it seems that the role of the NMD factors is most likely not to stimulate TERRA degradation but rather to reduce its association with telomeres [104]. If the increased level of TERRA at telomeres is the cause of the telomere de-protection observed in NMD-deficient cells, this would suggest that TERRA is unfavourable for telomere stability. On the other hand, if NMD-depletion triggers telomere damage independently of an effect on TERRA, the increased association of TERRA with telomeres could be stimulated by the cell in an attempt to strengthen telomere protection. Similarly, the increased levels of TERRA observed with thermal shock may help to protect telomeres against stress-mediated damage [205]. TERRA might also be involved in the regulation of telomerase activity. EST1A/SMG6 physically interacts with telomerase [204] and, in vitro, TERRA probably inhibits telomerase by RNA duplex formation in the template region of the telomerase RNA component (TERC) [205]. The effect of TERRA on telomerase remains to be tested in vivo.
The roles of UPF1 in DNA replication and repair
Evidence from several different studies has converged in recent years to show that UPF1 plays a significant role in DNA and RNA maintenance pathways that are necessary for the cell to accomplish DNA replication (Table 3). UPF1 depletion was reported to result in an early S-phase arrest, in which the cells could fire the start of DNA replication but were unable to complete DNA replication, and induced an ATR-dependent DNA-damage response [103]. The cells accumulated nuclear foci comprising the S-phase marker proliferation cell nuclear antigen (PCNA), indicating that UPF1 is essential for accomplishing DNA replication during S-phase of the cell cycle. Notably, knocking down UPF2 caused no adverse effect to S-phase progression, suggesting that UPF1’s function in DNA replication is unrelated to NMD [103]. It was further observed that hyper-phosphorylated UPF1 was associated with chromatin and that the amount of chromatin-associated UPF1 vastly increased during S-phase and also upon γ-irradiation [103]. When ATR was depleted, chromatin loading of UPF1 was impaired, whereas NMD was not affected under such conditions [103]. This implies that, either during DNA replication and/or a DNA repair response, ATR phosphorylates UPF1 leading to its association with chromatin, or alternatively that UPF1 is present on the chromatin during replication and becomes phosphorylated by ATR in response to DNA damage. Evidence for the involvement of UPF1 in DNA replication and repair is compounded further by the finding that UPF1 co-immunoprecipitates with the p66 subunit and p125 catalytic subunit of the replicative DNA polymerase δ. In contrast, UPF2 was not able to co-immunoprecipitate with p125 DNA polymerase δ [103, 208]. The function of UPF1 in DNA replication and cell cycle progression becomes even more intricate by evidence showing that UPF1 is involved in the degradation of replication dependent histone transcripts upon DNA replication inhibition and at the end of S-phase [209].
UPF1 acts in the degradation of replication-dependent histone mRNAs
The replication-dependent histone genes provide the large amount of histone proteins (108 molecules of each core histone protein) required for genome duplication [210]. Histone proteins are produced during S-phase of the cell cycle and their expression is co-ordinately regulated and finely balanced with DNA replication. Three major processes, transcription, mRNA 3′ end processing and mRNA stability control, contribute to the complex regulation of histone gene expression during S-phase [211]. This combination of transcriptional and post-transcriptional mechanisms acts to regulate the 35-fold increase of histone mRNA levels as cells progress from G1- into S-phase and ensures that the mRNA abundance returns to baseline levels as the cells exit S-phase. The half-life of histone mRNAs decreases from 45–60 min during S-phase to approximately 10 min at the end of S-phase due to a regulatory mechanism that acts to rapidly eliminate histone mRNAs from the cytoplasm when DNA synthesis is completed or inhibited. Since histone gene transcription is only increased three to fivefold during S-phase, the post-transcriptional regulation accounts for the remaining five to sixfold increase in histone mRNA levels and for their rapid elimination when DNA replication is completed or inhibited (reviewed in [211–213].
Histone production and DNA synthesis are intimately coupled by a poorly understood mechanism involving checkpoint kinases, such as ATR and DNA-activated protein kinase (DNA-PK) [214, 215]. Additionally, UPF1 has also been reported to play a role in histone mRNA destabilisation [214–216] (Table 3). RNAi-mediated knockdown of UPF1 (but not of UPF2) or over-expression of dominant-negative UPF1 mutants (K498A and R843C) resulted in an increased histone H2A mRNA level after inhibition of DNA synthesis by hydroxyurea (HU) treatment or at the end of S-phase [214]. Furthermore, a weak interaction between UPF1 and the histone stem-loop binding protein (SLBP; also called hairpin binding protein, HBP) was detected in immunoprecipitation experiments. Recently, it has been revealed that the degradation of histone mRNAs requires many of the factors involved in degradation of polyadenylated mRNA, such as LSM1, DCP2, XRN1 and the exosome components PM/Scl-100 (RRP6) and EXOSC4 (RRP41) [217]. Furthermore, it has been reported that histone mRNA degradation begins with the addition of 8–12 uridines by uridylyl transferases (TUTases) to the 3′ end of the histone mRNA [217]. The concentration of oligouridylated histone mRNAs increased strongly 15 min after HU treatment, and knockdown of TUTases 1 and 3 reduced the rate of histone mRNA degradation [217]. UPF1 has been proposed to be involved in the recruitment of the TUTase 1 and 3 to the histone mRNA [217]. Alternatively, it has also been suggested that the RNA helicase activity of UPF1 may remodel the mRNP complex to allow the TUTases to bind the 3′ end of the mRNA [218]. So far, there is no direct evidence for either of these roles by UPF1, and its function in histone mRNA metabolism remains unclear.
As mentioned, the coupling of DNA replication and histone mRNA stability is thought to be mediated by PIKKs acting upon downstream effectors. In addition to SMG1 (also called ATX), the mammalian PIKKs include ATM, ATR, mTOR/FRAP and DNA-PK. Checkpoint regulation in mammals is intricate and ultimately unevenly divided between ATM and ATR [219]. ATM, ATR and DNA-PK are activated by various forms of DNA damage. ATR is activated by aberrant DNA structures induced by UV light or replicational stress caused by DNA replication inhibitors, both leading to stalling of the replication fork. ATM is mainly activated by DSBs predominately after exposure to ionising radiation. DNA-PK is required for DSB repair by non-homologous end-joining (NHEJ) and telomere maintenance. In a recent study, Kaygun and Marzluff reported that ATR but not ATM is required for histone mRNA degradation after inhibition of DNA replication [209]. Müller et al. reported that ATR is not functionally limiting for the rate or extent of histone mRNA decay induced by replication stress and cannot fully account for the coupling between DNA replication and histone mRNA stability. Exposure of cells to an inhibitor of DNA-PK unveiled DNA-PK also to be involved in linking histone mRNA abundance with DNA replication [215]. DNA-PK is activated during replication stress and the DNA-PK pathway is enhanced when ATR signalling fails, highlighting the complex nature of such signalling pathways and showing that, in vivo, the relative contribution from each signalling pathway would be based on the nature of the DNA lesion generating the replication stress. Since SMG1 interacts with and phosphorylates UPF1, and because of its resemblance to these other PIKKs with documented roles in regulation of cell cycle and DNA replication, it is tempting to speculate that SMG1 may also contribute to the complex signalling involved in the coupling of DNA replication and histone mRNA stability.
A role for SMG1 in safeguarding genome integrity
The human members of the PIKK family all comprise FAT, FATC and PI3K domains. SMG1 differs from the other PIKKs because the PI3K domain is separated from the FATC domain by a large insert of over 1,000 amino acids, whereas only approximately 100 amino acids separate these two domains in the other PIKKs [61, 75, 220]. In C. elegans SMG1, this 1,000 amino acids section does not exist. Possibly, this portion appeared during metazoan evolution and allows SMG1 to interact with a broader range of upstream regulatory proteins and downstream targets [221]. In human cells, depletion of SMG1 results in spontaneous DNA damage and a hugely increased sensitivity to ionising radiation [106] (Table 3). Exposure of cells to UV light or ionising radiation stimulates SMG1 kinase activity. Moreover, like ATM, SMG1 is able to phosphorylate p53 at serine 15 and expression of SMG1 is required for optimal p53 activation after genotoxic stress. Therefore, similar to ATM and ATR, SMG1 is central to the regulation of the genotoxic stress response machinery in mammalian cells. Recent reports have also implicated SMG1 to be important in the response to oxidative stress [222]. SMG1 has been observed to initiate p53 phosphorylation during the early stages of oxidative stress, and ATM acts to maintain the p53 phosphorylation over time. Currently, it is not clear why the activation of SMG1 precedes that of ATM in hyperoxia conditions. Experiments using cells depleted of ATM also indicated that SMG1 and ATM act sequentially and independently of each other to regulate the G1-checkpoint during prolonged oxidative stress [222]. Potentially, SMG1 and ATM may recognise and respond to different lesions that are produced due to prolonged hyperoxia. Phosphorylated p53 activates transcription of p21 and, interestingly, it was shown that the destabilisation of p21 during hyperoxia could be restored by treatment with wortmannin, implying the involvement of PIKKs [222]. Human SMG1 and ATM also act, independently of p53, to target p21 for degradation by the proteasome. Hence, this dual function of SMG1 and ATM which can regulate both the synthesis and degradation of p21, thereby finely controlling the p21 levels necessary to inhibit cell proliferation, induce DNA repair and block apoptosis [222].
Similarly SMG1 was also found to be important in responding to oxidative stress in C. elegans. In a study investigating lifespan regulation in C. elegans, SMG1 was identified in a screen for genes that prolong lifespan in a daf-18-dependent manner [223]. Further analysis revealed that the SMG1 function in lifespan control requires cep-1, the C. elegans orthologue of p53. Moreover, the role of SMG1 in lifespan control is due to its function in oxidative stress response. Interestingly, it was also reported that in S. pombe, Upf1p and Upf2p are required for survival of oxidative stress [21]. To investigate the possibility that SMG1 may have a function in stress signalling induced by cytokines, a wide range of agents that can induce cell death were examined in SMG1-, ATM-, ATR-, UPF1- or UPF2-depleted cells [224]. Only depletion of SMG1, but not of the other factors, substantially increased the rate and the extent of apoptotic cell death mediated by tumour necrosis factor-alpha (TNF-α) [224]. SMG1, like other PIKKs, functions to initiate cellular stress responses when genome integrity, mRNA translation or nutrient availability is compromised, and the role of SMG1 in NMD represents simply just one of its many roles.
Conclusion and future directions
Apart from increasing our understanding of the fundamental molecular mechanisms that control the extent and accuracy of the expression of our genetic information, a principal goal of studies on NMD is to eventually utilise the knowledge for curing diseases and improving health. In spite of significant advances during recent years, we are still far from understanding the complete details regarding how mRNAs are recognised as NMD targets, their subsequent degradation and the physiological importance of NMD factors regulating up to 10% of the transcriptome.
The unified NMD model makes many testable predictions and will hopefully provide a useful framework for future mechanistic investigations. The model emphasises the requirement of a properly configured 3′ UTR for normal translation termination. The presence of PABP nearby a terminating ribosome is one crucial characteristic for a properly configured 3′ UTR, but additional features are expected to exist. Deviations from this 3′ UTR configuration or specific factors that prohibit the termination-promoting interaction of PABP with the translation termination complex signals an anomaly in the mRNA molecule and triggers NMD. The mechanistic details of UPF1 recruitment onto the SURF complex and its subsequent assembly with the ribosome remain to be worked out. In addition, several open questions regarding the degradation pathway(s) of NMD substrates need to be addressed. For example, does mammalian NMD really comprise two independent ways to initiate RNA decay: an endonucleolytic SMG6-dependent route and a decapping-dependent exonucleolytic route?
Finally, it has come to light in recent years that several factors which were initially identified and characterised for their roles in the NMD pathway have highly important functions in the cell that are independent of NMD. It is becoming more evident that UPF1 plays many roles with regards to the regulation of S-phase progression, considering it has been reported to be involved in replication-dependent histone mRNA metabolism, the replication machinery, and in conjunction with SMG6 in telomere maintenance. Additionally, there is an increased association of UPF1 onto chromatin during S-phase and depletion of UPF1 leads to an early S-phase arrest. Likewise, the recently reported functions of SMG1 also indicate this protein to be ultimately required for genome stability. It is more difficult to relate the function of UPF1 in SMD and in HIV-1 RNA metabolism to its S-phase and telomere functions. It is attractive to speculate in an evolutionary context about when UPF1 acquired these functions in addition to its role in NMD. Replication-dependent histones in yeast are polyadenylated and do not contain SLBP/HBP; perhaps this is why depletion of UPF1 is detrimental to human cells but not to yeast cells. An important goal for the future is to begin to decipher what constitutes a direct effect and an indirect effect when the NMD factors are depleted and genome-wide microarray profiling is conducted. The future will also be about determining the parameters for each of the many different roles that UPF1 carries out, for instance its cellular location, its role in cell cycle control, where and when it is phosphorylated, whether there is a specific phosphorylation pattern directing UPF1 to its function, its stability and where and when its many interaction partners bind. Making sense of all the molecular functions of UPF1 will not only be critical to understanding the mechanism of NMD, but also to elucidation of all of the other processes where UPF1 has been found to play a role.
Acknowledgments
The research of the authors is supported by grants from the European Research Council, the Swiss National Science Foundation, the Novartis Foundation for Biomedical Research, the Helmut Horten Foundation and also by the Kanton Bern. O.M. is a fellow of the Max Cloëtta Foundation and R.Z.O. is supported by a fellowship from CONACYT México.
References
- 1.Losson R, Lacroute F. Interference of nonsense mutations with eukaryotic messenger RNA stability. Proc Natl Acad Sci USA. 1979;76:5134–5137. doi: 10.1073/pnas.76.10.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maquat LE, Kinniburgh AJ, Rachmilewitz EA, Ross J. Unstable beta-globin mRNA in mRNA-deficient beta o thalassemia. Cell. 1981;27:543–553. doi: 10.1016/0092-8674(81)90396-2. [DOI] [PubMed] [Google Scholar]
- 3.Culbertson MR, Leeds PF. Looking at mRNA decay pathways through the window of molecular evolution. Curr Opin Genet Dev. 2003;13:207–214. doi: 10.1016/S0959-437X(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 4.Stalder L, Muhlemann O. The meaning of nonsense. Trends Cell Biol. 2008;18:315–321. doi: 10.1016/j.tcb.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Wilkinson MF. Nonsense surveillance in lymphocytes? Immunity. 1998;8:135–141. doi: 10.1016/S1074-7613(00)80466-5. [DOI] [PubMed] [Google Scholar]
- 6.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, Jacobsen SE, Nerlov C, Porse BT. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 8.Kim H, Klein R, Majewski J, Ott J. Estimating rates of alternative splicing in mammals and invertebrates. Nat Genet. 2004;36:915–916. doi: 10.1038/ng0904-915. [DOI] [PubMed] [Google Scholar]
- 9.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet. 2008;9:699–712. doi: 10.1038/nrg2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: what is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Pan Q, Saltzman AL, Kim YK, Misquitta C, Shai O, Maquat LE, Frey BJ, Blencowe BJ. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurihara Y, Matsui A, Hanada K, Kawashima M, Ishida J, Morosawa T, Tanaka M, Kaminuma E, Mochizuki Y, Matsushima A, Toyoda T, Shinozaki K, Seki M. Genome-wide suppression of aberrant mRNA-like noncoding RNAs by NMD in Arabidopsis . Proc Natl Acad Sci USA. 2009;106:2453–2458. doi: 10.1073/pnas.0808902106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 15.Mitrovich QM, Anderson P. mRNA surveillance of expressed pseudogenes in C. elegans . Curr Biol. 2005;15:963–967. doi: 10.1016/j.cub.2005.04.055. [DOI] [PubMed] [Google Scholar]
- 16.Rehwinkel J, Raes J, Izaurralde E. Nonsense-mediated mRNA decay: target genes and functional diversification of effectors. Trends Biochem Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/S1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 19.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez-Gabriel MA, Watt S, Bahler J, Russell P. Upf1, an RNA helicase required for nonsense-mediated mRNA decay, modulates the transcriptional response to oxidative stress in fission yeast. Mol Cell Biol. 2006;26:6347–6356. doi: 10.1128/MCB.00286-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriarty PM, Reddy CC, Maquat LE. Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA. Mol Cell Biol. 1998;18:2932–2939. doi: 10.1128/mcb.18.5.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 24.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 25.Kuzmiak HA, Maquat LE. Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol Med. 2006;12:306–316. doi: 10.1016/j.molmed.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Kerr TP, Sewry CA, Robb SA, Roberts RG. Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum Genet. 2001;109:402–407. doi: 10.1007/s004390100598. [DOI] [PubMed] [Google Scholar]
- 27.Hall GW, Thein S. Nonsense codon mutations in the terminal exon of the beta-globin gene are not associated with a reduction in beta-mRNA accumulation: a mechanism for the phenotype of dominant beta-thalassemia. Blood. 1994;83:2031–2037. [PubMed] [Google Scholar]
- 28.Thein SL, Hesketh C, Taylor P, Temperley IJ, Hutchinson RM, Old JM, Wood WG, Clegg JB, Weatherall DJ. Molecular basis for dominantly inherited inclusion body beta-thalassemia. Proc Natl Acad Sci USA. 1990;87:3924–3928. doi: 10.1073/pnas.87.10.3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietz HC, McIntosh I, Sakai LY, Corson GM, Chalberg SC, Pyeritz RE, Francomano CA. Four novel FBN1 mutations: significance for mutant transcript level and EGF-like domain calcium binding in the pathogenesis of Marfan syndrome. Genomics. 1993;17:468–475. doi: 10.1006/geno.1993.1349. [DOI] [PubMed] [Google Scholar]
- 30.Salvatore F, Scudiero O, Castaldo G. Genotype-phenotype correlation in cystic fibrosis: the role of modifier genes. Am J Med Genet. 2002;111:88–95. doi: 10.1002/ajmg.10461. [DOI] [PubMed] [Google Scholar]
- 31.Jouanguy E, Altare F, Lamhamedi S, Revy P, Emile JF, Newport M, Levin M, Blanche S, Seboun E, Fischer A, Casanova JL. Interferon-gamma-receptor deficiency in an infant with fatal bacille Calmette-Guerin infection. N Engl J Med. 1996;335:1956–1961. doi: 10.1056/NEJM199612263352604. [DOI] [PubMed] [Google Scholar]
- 32.Jouanguy E, Lamhamedi-Cherradi S, Lammas D, Dorman SE, Fondaneche MC, Dupuis S, Doffinger R, Altare F, Girdlestone J, Emile JF, Ducoulombier H, Edgar D, Clarke J, Oxelius VA, Brai M, Novelli V, Heyne K, Fischer A, Holland SM, Kumararatne DS, Schreiber RD, Casanova JL. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 33.Cardinali M, Kratochvil FJ, Ensley JF, Robbins KC, Yeudall WA. Functional characterization in vivo of mutant p53 molecules derived from squamous cell carcinomas of the head and neck. Mol Carcinog. 1997;18:78–88. doi: 10.1002/(SICI)1098-2744(199702)18:2<78::AID-MC3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 34.Englert C, Vidal M, Maheswaran S, Ge Y, Ezzell RM, Isselbacher KJ, Haber DA. Truncated WT1 mutants alter the subnuclear localization of the wild-type protein. Proc Natl Acad Sci USA. 1995;92:11960–11964. doi: 10.1073/pnas.92.26.11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reddy JC, Morris JC, Wang J, English MA, Haber DA, Shi Y, Licht JD. WT1-mediated transcriptional activation is inhibited by dominant negative mutant proteins. J Biol Chem. 1995;270:10878–10884. doi: 10.1074/jbc.270.41.24502. [DOI] [PubMed] [Google Scholar]
- 36.Perrin-Vidoz L, Sinilnikova OM, Stoppa-Lyonnet D, Lenoir GM, Mazoyer S. The nonsense-mediated mRNA decay pathway triggers degradation of most BRCA1 mRNAs bearing premature termination codons. Hum Mol Genet. 2002;11:2805–2814. doi: 10.1093/hmg/11.23.2805. [DOI] [PubMed] [Google Scholar]
- 37.Bloethner S, Mould A, Stark M, Hayward NK. Identification of ARHGEF17, DENND2D, FGFR3, and RB1 mutations in melanoma by inhibition of nonsense-mediated mRNA decay. Genes Chromosomes Cancer. 2008;47:1076–1085. doi: 10.1002/gcc.20598. [DOI] [PubMed] [Google Scholar]
- 38.Ivanov I, Lo KC, Hawthorn L, Cowell JK, Ionov Y. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene. 2007;26:2873–2884. doi: 10.1038/sj.onc.1210098. [DOI] [PubMed] [Google Scholar]
- 39.Tarpey PS, Lucy Raymond F, Nguyen LS, Rodriguez J, Hackett A, Vandeleur L, Smith R, Shoubridge C, Edkins S, Stevens C, O’Meara S, Tofts C, Barthorpe S, Buck G, Cole J, Halliday K, Hills K, Jones D, Mironenko T, Perry J, Varian J, West S, Widaa S, Teague J, Dicks E, Butler A, Menzies A, Richardson D, Jenkinson A, Shepherd R, Raine K, Moon J, Luo Y, Parnau J, Bhat SS, Gardner A, Corbett M, Brooks D, Thomas P, Parkinson-Lawrence E, Porteous ME, Warner JP, Sanderson T, Pearson P, Simensen RJ, Skinner C, Hoganson G, Superneau D, Wooster R, Bobrow M, Turner G, Stevenson RE, Schwartz CE, Andrew Futreal P, Srivastava AK, Stratton MR, Gecz J. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerem E. Pharmacologic therapy for stop mutations: how much CFTR activity is enough? Curr Opin Pulm Med. 2004;10:547–552. doi: 10.1097/01.mcp.0000141247.22078.46. [DOI] [PubMed] [Google Scholar]
- 41.Ramalho AS, Beck S, Meyer M, Penque D, Cutting GR, Amaral MD. Five percent of normal cystic fibrosis transmembrane conductance regulator mRNA ameliorates the severity of pulmonary disease in cystic fibrosis. Am J Respir Cell Mol Biol. 2002;27:619–627. doi: 10.1165/rcmb.2001-0004OC. [DOI] [PubMed] [Google Scholar]
- 42.Stephenson J. Antibiotics show promise as therapy for genetic disorders. JAMA. 2001;285:2067–2068. doi: 10.1001/jama.285.16.2067. [DOI] [PubMed] [Google Scholar]
- 43.Clancy JP, Bebok Z, Ruiz F, King C, Jones J, Walker L, Greer H, Hong J, Wing L, Macaluso M, Lyrene R, Sorscher EJ, Bedwell DM. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am J Respir Crit Care Med. 2001;163:1683–1692. doi: 10.1164/ajrccm.163.7.2004001. [DOI] [PubMed] [Google Scholar]
- 44.Wilschanski M, Famini C, Blau H, Rivlin J, Augarten A, Avital A, Kerem B, Kerem E. A pilot study of the effect of gentamicin on nasal potential difference measurements in cystic fibrosis patients carrying stop mutations. Am J Respir Crit Care Med. 2000;161:860–865. doi: 10.1164/ajrccm.161.3.9904116. [DOI] [PubMed] [Google Scholar]
- 45.Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, Wilde RG, Karp G, Takasugi J, Chen G, Jones S, Ren H, Moon YC, Corson D, Turpoff AA, Campbell JA, Conn MM, Khan A, Almstead NG, Hedrick J, Mollin A, Risher N, Weetall M, Yeh S, Branstrom AA, Colacino JM, Babiak J, Ju WD, Hirawat S, Northcutt VJ, Miller LL, Spatrick P, He F, Kawana M, Feng H, Jacobson A, Peltz SW, Sweeney HL. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. [DOI] [PubMed] [Google Scholar]
- 46.Kerem E, Hirawat S, Armoni S, Yaakov Y, Shoseyov D, Cohen M, Nissim-Rafinia M, Blau H, Rivlin J, Aviram M, Elfring GL, Northcutt VJ, Miller LL, Kerem B, Wilschanski M. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet. 2008;372:719–727. doi: 10.1016/S0140-6736(08)61168-X. [DOI] [PubMed] [Google Scholar]
- 47.Culbertson MR, Underbrink KM, Fink GR. Frameshift suppression in Saccharomyces cerevisiae. II. Genetic properties of group II suppressors. Genetics. 1980;95:833–853. doi: 10.1093/genetics/95.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 49.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae . Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He F, Jacobson A. Identification of a novel component of the nonsense-mediated mRNA decay pathway by use of an interacting protein screen. Genes Dev. 1995;9:437–454. doi: 10.1101/gad.9.4.437. [DOI] [PubMed] [Google Scholar]
- 51.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. A new kind of informational suppression in the nematode Caenorhabditis elegans . Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pulak R, Anderson P. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 53.Cali BM, Kuchma SL, Latham J, Anderson P. smg-7 is required for mRNA surveillance in Caenorhabditis elegans . Genetics. 1999;151:605–616. doi: 10.1093/genetics/151.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Applequist SE, Selg M, Raman C, Jack HM. Cloning and characterization of HUPF1, a human homolog of the Saccharomyces cerevisiae nonsense mRNA-reducing UPF1 protein. Nucleic Acids Res. 1997;25:814–821. doi: 10.1093/nar/25.4.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denning G, Jamieson L, Maquat LE, Thompson EA, Fields AP. Cloning of a novel phosphatidylinositol kinase-related kinase: characterization of the human SMG-1 RNA surveillance protein. J Biol Chem. 2001;276:22709–22714. doi: 10.1074/jbc.C100144200. [DOI] [PubMed] [Google Scholar]
- 56.Lykke-Andersen J, Shu MD, Steitz JA. Human Upf proteins target an mRNA for nonsense-mediated decay when bound downstream of a termination codon. Cell. 2000;103:1121–1131. doi: 10.1016/S0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 57.Mendell JT, Medghalchi SM, Lake RG, Noensie EN, Dietz HC. Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes. Mol Cell Biol. 2000;20:8944–8957. doi: 10.1128/MCB.20.23.8944-8957.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohnishi T, Yamashita A, Kashima I, Schell T, Anders KR, Grimson A, Hachiya T, Hentze MW, Anderson P, Ohno S. Phosphorylation of hUPF1 induces formation of mRNA surveillance complexes containing hSMG-5 and hSMG-7. Mol Cell. 2003;12:1187–1200. doi: 10.1016/S1097-2765(03)00443-X. [DOI] [PubMed] [Google Scholar]
- 59.Perlick HA, Medghalchi SM, Spencer FA, Kendzior RJ, Jr, Dietz HC. Mammalian orthologues of a yeast regulator of nonsense transcript stability. Proc Natl Acad Sci USA. 1996;93:10928–10932. doi: 10.1073/pnas.93.20.10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Serin G, Gersappe A, Black JD, Aronoff R, Maquat LE. Identification and characterization of human orthologues to Saccharomyces cerevisiae Upf2 protein and Upf3 protein (Caenorhabditis elegans SMG-4) Mol Cell Biol. 2001;21:209–223. doi: 10.1128/MCB.21.1.209-223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamashita A, Ohnishi T, Kashima I, Taya Y, Ohno S. Human SMG-1, a novel phosphatidylinositol 3-kinase-related protein kinase, associates with components of the mRNA surveillance complex and is involved in the regulation of nonsense-mediated mRNA decay. Genes Dev. 2001;15:2215–2228. doi: 10.1101/gad.913001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Longman D, Plasterk RH, Johnstone IL, Caceres JF. Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 2007;21:1075–1085. doi: 10.1101/gad.417707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamashita A, Izumi N, Kashima I, Ohnishi T, Saari B, Katsuhata Y, Muramatsu R, Morita T, Iwamatsu A, Hachiya T, Kurata R, Hirano H, Anderson P, Ohno S. SMG-8 and SMG-9, two novel subunits of the SMG-1 complex, regulate remodeling of the mRNA surveillance complex during nonsense-mediated mRNA decay. Genes Dev. 2009;23:1091–1105. doi: 10.1101/gad.1767209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gatfield D, Unterholzner L, Ciccarelli FD, Bork P, Izaurralde E. Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 2003;22:3960–3970. doi: 10.1093/emboj/cdg371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Luke B, Azzalin CM, Hug N, Deplazes A, Peter M, Lingner J. Saccharomyces cerevisiae Ebs1p is a putative ortholog of human Smg7 and promotes nonsense-mediated mRNA decay. Nucleic Acids Res. 2007;35:7688–7697. doi: 10.1093/nar/gkm912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Page MF, Carr B, Anders KR, Grimson A, Anderson P. SMG-2 is a phosphorylated protein required for mRNA surveillance in Caenorhabditis elegans and related to Upf1p of yeast. Mol Cell Biol. 1999;19:5943–5951. doi: 10.1128/mcb.19.9.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW. Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA. 2000;6:1226–1235. doi: 10.1017/S1355838200000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Czaplinski K, Weng Y, Hagan KW, Peltz SW. Purification and characterization of the Upf1 protein: a factor involved in translation and mRNA degradation. RNA. 1995;1:610–623. [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng Z, Muhlrad D, Lim MK, Parker R, Song H. Structural and functional insights into the human Upf1 helicase core. EMBO J. 2007;26:253–264. doi: 10.1038/sj.emboj.7601464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Clerici M, Mourao A, Gutsche I, Gehring NH, Hentze MW, Kulozik A, Kadlec J, Sattler M, Cusack S. Unusual bipartite mode of interaction between the nonsense-mediated decay factors, UPF1 and UPF2. EMBO J. 2009;28:2293–2306. doi: 10.1038/emboj.2009.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kadlec J, Guilligay D, Ravelli RB, Cusack S. Crystal structure of the UPF2-interacting domain of nonsense-mediated mRNA decay factor UPF1. RNA. 2006;12:1817–1824. doi: 10.1261/rna.177606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Czaplinski K, Ruiz-Echevarria MJ, Paushkin SV, Han X, Weng Y, Perlick HA, Dietz HC, Ter-Avanesyan MD, Peltz SW. The surveillance complex interacts with the translation release factors to enhance termination and degrade aberrant mRNAs. Genes Dev. 1998;12:1665–1677. doi: 10.1101/gad.12.11.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, Ohno M, Dreyfuss G, Ohno S. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Grimson A, O’Connor S, Newman CL, Anderson P. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans . Mol Cell Biol. 2004;24:7483–7490. doi: 10.1128/MCB.24.17.7483-7490.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, 2nd, Wilkinson MF. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, Kulozik AE. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 78.Gehring NH, Lamprinaki S, Hentze MW, Kulozik AE. The hierarchy of exon-junction complex assembly by the spliceosome explains key features of mammalian nonsense-mediated mRNA decay. PLoS Biol. 2009;7:e1000120. doi: 10.1371/journal.pbio.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saltzman AL, Kim YK, Pan Q, Fagnani MM, Maquat LE, Blencowe BJ. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol Cell Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Anders KR, Grimson A, Anderson P. SMG-5, required for C.elegans nonsense-mediated mRNA decay, associates with SMG-2 and protein phosphatase 2A. EMBO J. 2003;22:641–650. doi: 10.1093/emboj/cdg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiu SY, Serin G, Ohara O, Maquat LE. Characterization of human Smg5/7a: A protein with similarities to Caenorhabditis elegans SMG5 and SMG7 that functions in the dephosphorylation of Upf1. RNA. 2003;9:77–87. doi: 10.1261/rna.2137903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang W, Cajigas IJ, Peltz SW, Wilkinson MF, Gonzalez CI. Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:3390–3400. doi: 10.1128/MCB.26.9.3390-3400.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Weng Y, Czaplinski K, Peltz SW. Identification and characterization of mutations in the UPF1 gene that affect nonsense suppression and the formation of the Upf protein complex but not mRNA turnover. Mol Cell Biol. 1996;16:5491–5506. doi: 10.1128/mcb.16.10.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kadlec J, Izaurralde E, Cusack S. The structural basis for the interaction between nonsense-mediated mRNA decay factors UPF2 and UPF3. Nat Struct Mol Biol. 2004;11:330–337. doi: 10.1038/nsmb741. [DOI] [PubMed] [Google Scholar]
- 86.Schell T, Kocher T, Wilm M, Seraphin B, Kulozik AE, Hentze MW. Complexes between the nonsense-mediated mRNA decay pathway factor human upf1 (up-frameshift protein 1) and essential nonsense-mediated mRNA decay factors in HeLa cells. Biochem J. 2003;373:775–783. doi: 10.1042/BJ20021920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, Wilkinson MF. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 88.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. Rna. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–4997. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Andersen CB, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CL, Pedersen JS, Seraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 91.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 92.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 93.Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon–exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 94.Yamashita A, Kashima I, Ohno S. The role of SMG-1 in nonsense-mediated mRNA decay. Biochim Biophys Acta. 2005;1754:305–315. doi: 10.1016/j.bbapap.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 95.D’Andrea LD, Regan L. TPR proteins: the versatile helix. Trends Biochem Sci. 2003;28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 96.Fukuhara N, Ebert J, Unterholzner L, Lindner D, Izaurralde E, Conti E. SMG7 is a 14–3-3-like adaptor in the nonsense-mediated mRNA decay pathway. Mol Cell. 2005;17:537–547. doi: 10.1016/j.molcel.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 97.Unterholzner L, Izaurralde E. SMG7 acts as a molecular link between mRNA surveillance and mRNA decay. Mol Cell. 2004;16:587–596. doi: 10.1016/j.molcel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 98.Clissold PM, Ponting CP. PIN domains in nonsense-mediated mRNA decay and RNAi. Curr Biol. 2000;10:R888–R890. doi: 10.1016/S0960-9822(00)00858-7. [DOI] [PubMed] [Google Scholar]
- 99.Glavan F, Behm-Ansmant I, Izaurralde E, Conti E. Structures of the PIN domains of SMG6 and SMG5 reveal a nuclease within the mRNA surveillance complex. EMBO J. 2006;25:5117–5125. doi: 10.1038/sj.emboj.7601377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 101.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mendell JT, ap Rhys CM, Dietz HC. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- 103.Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–439. doi: 10.1016/j.cub.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 104.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 105.Durand S, Cougot N, Mahuteau-Betzer F, Nguyen CH, Grierson DS, Bertrand E, Tazi J, Lejeune F. Inhibition of nonsense-mediated mRNA decay (NMD) by a new chemical molecule reveals the dynamic of NMD factors in P-bodies. J Cell Biol. 2007;178:1145–1160. doi: 10.1083/jcb.200611086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Brumbaugh KM, Otterness DM, Geisen C, Oliveira V, Brognard J, Li X, Lejeune F, Tibbetts RS, Maquat LE, Abraham RT. The mRNA surveillance protein hSMG-1 functions in genotoxic stress response pathways in mammalian cells. Mol Cell. 2004;14:585–598. doi: 10.1016/j.molcel.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 107.Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsiss nucleolus. Plant Cell. 2009;21:2045–2057. doi: 10.1105/tpc.109.067736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hilleren P, Parker R. mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA. 1999;5:711–719. doi: 10.1017/S1355838299990519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Amrani N, Sachs MS, Jacobson A. Early nonsense: mRNA decay solves a translational problem. Nat Rev Mol Cell Biol. 2006;7:415–425. doi: 10.1038/nrm1942. [DOI] [PubMed] [Google Scholar]
- 110.Muhlemann O, Eberle AB, Stalder L, Zamudio Orozco R. Recognition and elimination of nonsense mRNA. Biochim Biophys Acta. 2008;1779:538–549. doi: 10.1016/j.bbagrm.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 111.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae . EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M, Justesen J, Kisselev L. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 114.Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, Zhouravleva G. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol Cell. 2002;94:205–216. doi: 10.1016/S0248-4900(02)01194-2. [DOI] [PubMed] [Google Scholar]
- 115.Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3’-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem. 1999;274:16677–16680. doi: 10.1074/jbc.274.24.16677. [DOI] [PubMed] [Google Scholar]
- 116.Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci USA. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 119.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. EMBO J. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Eberle AB, Stalder L, Mathys H, Orozco RZ, Muhlemann O. Posttranscriptional gene regulation by spatial rearrangement of the 3′ untranslated region. PLoS Biol. 2008;6:e92. doi: 10.1371/journal.pbio.0060092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Silva AL, Ribeiro P, Inacio A, Liebhaber SA, Romao L. Proximity of the poly(A)-binding protein to a premature termination codon inhibits mammalian nonsense-mediated mRNA decay. RNA. 2008;14:563–576. doi: 10.1261/rna.815108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kebaara BW, Atkin AL. Long 3′-UTRs target wild-type mRNAs for nonsense-mediated mRNA decay in Saccharomyces cerevisiae . Nucleic Acids Res. 2009;37:2771–2778. doi: 10.1093/nar/gkp146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kerenyi Z, Merai Z, Hiripi L, Benkovics A, Gyula P, Lacomme C, Barta E, Nagy F, Silhavy D. Inter-kingdom conservation of mechanism of nonsense-mediated mRNA decay. EMBO J. 2008;27:1585–1595. doi: 10.1038/emboj.2008.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kertesz S, Kerenyi Z, Merai Z, Bartos I, Palfy T, Barta E, Silhavy D. Both introns and long 3’-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–6157. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muhlrad D, Parker R. Aberrant mRNAs with extended 3 ‘UTRs are substrates for rapid degradation by mRNA surveillance. RNA. 1999;5:1299–1307. doi: 10.1017/S1355838299990829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Peltz SW, Brown AH, Jacobson A. mRNA destabilization triggered by premature translational termination depends on at least three cis-acting sequence elements and one trans-acting factor. Genes Dev. 1993;7:1737–1754. doi: 10.1101/gad.7.9.1737. [DOI] [PubMed] [Google Scholar]
- 128.Ruiz-Echevarria MJ, Gonzalez CI, Peltz SW. Identifying the right stop: determining how the surveillance complex recognizes and degrades an aberrant mRNA. EMBO J. 1998;17:575–589. doi: 10.1093/emboj/17.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Le Hir H, Izaurralde E, Maquat LE, Moore MJ. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev. 2000;14:1098–1108. [PMC free article] [PubMed] [Google Scholar]
- 131.Thermann R, Neu-Yilik G, Deters A, Frede U, Wehr K, Hagemeier C, Hentze MW, Kulozik AE. Binary specification of nonsense codons by splicing and cytoplasmic translation. EMBO J. 1998;17:3484–3494. doi: 10.1093/emboj/17.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang J, Sun X, Qian Y, La Duca JP, Maquat LE. At least one intron is required for the nonsense-mediated decay of triosephosphate isomerase mRNA: a possible link between nuclear splicing and cytoplasmic translation. Mol Cell Biol. 1998;18:5272–5283. doi: 10.1128/mcb.18.9.5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of beta-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/S1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gehring NH, Lamprinaki S, Kulozik AE, Hentze MW. Disassembly of exon junction complexes by PYM. Cell. 2009;137:536–548. doi: 10.1016/j.cell.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 135.Buzina A, Shulman MJ. Infrequent translation of a nonsense codon is sufficient to decrease mRNA level. Mol Biol Cell. 1999;10:515–524. doi: 10.1091/mbc.10.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Inacio A, Silva AL, Pinto J, Ji X, Morgado A, Almeida F, Faustino P, Lavinha J, Liebhaber SA, Romao L. Nonsense mutations in close proximity to the initiation codon fail to trigger full nonsense-mediated mRNA decay. J Biol Chem. 2004;279:32170–32180. doi: 10.1074/jbc.M405024200. [DOI] [PubMed] [Google Scholar]
- 137.Romao L, Inacio A, Santos S, Avila M, Faustino P, Pacheco P, Lavinha J. Nonsense mutations in the human beta-globin gene lead to unexpected levels of cytoplasmic mRNA accumulation. Blood. 2000;96:2895–2901. [PubMed] [Google Scholar]
- 138.Zhang J, Maquat LE. Evidence that translation reinitiation abrogates nonsense-mediated mRNA decay in mammalian cells. EMBO J. 1997;16:826–833. doi: 10.1093/emboj/16.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 140.Tarun SZ, Jr, Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc Natl Acad Sci USA. 1997;94:9046–9051. doi: 10.1073/pnas.94.17.9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Silva AL, Romao L. The mammalian nonsense-mediated mRNA decay pathway: to decay or not to decay! Which players make the decision? FEBS Lett. 2009;583:499–505. doi: 10.1016/j.febslet.2008.12.058. [DOI] [PubMed] [Google Scholar]
- 142.Ishigaki Y, Li X, Serin G, Maquat LE. Evidence for a pioneer round of mRNA translation. mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/S0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 143.Chiu SY, Lejeune F, Ranganathan AC, Maquat LE. The pioneer translation initiation complex is functionally distinct from but structurally overlaps with the steady-state translation initiation complex. Genes Dev. 2004;18:745–754. doi: 10.1101/gad.1170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hosoda N, Kim YK, Lejeune F, Maquat LE. CBP80 promotes interaction of Upf1 with Upf2 during nonsense-mediated mRNA decay in mammalian cells. Nat Struct Mol Biol. 2005;12:893–901. doi: 10.1038/nsmb995. [DOI] [PubMed] [Google Scholar]
- 145.Gao Q, Das B, Sherman F, Maquat LE. Cap-binding protein 1-mediated and eukaryotic translation initiation factor 4E-mediated pioneer rounds of translation in yeast. Proc Natl Acad Sci USA. 2005;102:4258–4263. doi: 10.1073/pnas.0500684102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Maderazo AB, Belk JP, He F, Jacobson A. Nonsense-containing mRNAs that accumulate in the absence of a functional nonsense-mediated mRNA decay pathway are destabilized rapidly upon its restitution. Mol Cell Biol. 2003;23:842–851. doi: 10.1128/MCB.23.3.842-851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kuperwasser N, Brogna S, Dower K, Rosbash M. Nonsense-mediated decay does not occur within the yeast nucleus. RNA. 2004;10:1907–1915. doi: 10.1261/rna.7132504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16:107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- 149.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 150.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 151.Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 152.Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 153.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 154.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 155.Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/S0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 156.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 157.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′->5′ degradation. Molecular Cell. 2003;11:1405–1413. doi: 10.1016/S1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 158.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila . Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 159.Couttet P, Grange T. Premature termination codons enhance mRNA decapping in human cells. Nucleic Acids Res. 2004;32:488–494. doi: 10.1093/nar/gkh218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Lejeune F, Li X, Maquat LE. Nonsense-mediated mRNA decay in mammalian cells involves decapping, deadenylating, and exonucleolytic activities. Mol Cell. 2003;12:675–687. doi: 10.1016/S1097-2765(03)00349-6. [DOI] [PubMed] [Google Scholar]
- 161.Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23:4805–4813. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cho H, Kim KM, Kim YK. Human proline-rich nuclear receptor coregulatory protein 2 mediates an interaction between mRNA surveillance machinery and decapping complex. Mol Cell. 2009;33:75–86. doi: 10.1016/j.molcel.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 163.Franks TM, Lykke-Andersen J. The control of mRNA decapping and P-body formation. Mol Cell. 2008;32:605–615. doi: 10.1016/j.molcel.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Eulalio A, Behm-Ansmant I, Izaurralde E. P bodies: at the crossroads of post-transcriptional pathways. Nat Rev Mol Cell Biol. 2007;8:9–22. doi: 10.1038/nrm2080. [DOI] [PubMed] [Google Scholar]
- 165.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 166.Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sheth U, Parker R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Stalder L, Muhlemann O. Processing bodies are not required for mammalian nonsense-mediated mRNA decay. RNA. 2009;15:1265–1273. doi: 10.1261/rna.1672509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Metzstein MM, Krasnow MA. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet. 2006;2:e180. doi: 10.1371/journal.pgen.0020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wittkopp N, Huntzinger E, Weiler C, Sauliere J, Schmidt S, Sonawane M, Izaurralde E. Nonsense-mediated mRNA decay effectors are essential for zebrafish embryonic development and survival. Mol Cell Biol. 2009;29:3517–3528. doi: 10.1128/MCB.00177-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 173.Yoine M, Nishii T, Nakamura K. Arabidopsis UPF1 RNA helicase for nonsense-mediated mRNA decay is involved in seed size control and is essential for growth. Plant Cell Physiol. 2006;47:572–580. doi: 10.1093/pcp/pcj035. [DOI] [PubMed] [Google Scholar]
- 174.Baron C, Heider J, Bock A. Interaction of translation factor SELB with the formate dehydrogenase H selenopolypeptide mRNA. Proc Natl Acad Sci USA. 1993;90:4181–4185. doi: 10.1073/pnas.90.9.4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Caban K, Copeland PR. Size matters: a view of selenocysteine incorporation from the ribosome. Cell Mol Life Sci. 2006;63:73–81. doi: 10.1007/s00018-005-5402-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Donovan J, Caban K, Ranaweera R, Gonzalez-Flores JN, Copeland PR. A novel protein domain induces high affinity selenocysteine insertion sequence binding and elongation factor recruitment. J Biol Chem. 2008;283:35129–35139. doi: 10.1074/jbc.M806008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gupta M, Copeland PR. Functional analysis of the interplay between translation termination, selenocysteine codon context, and selenocysteine insertion sequence-binding protein 2. J Biol Chem. 2007;282:36797–36807. doi: 10.1074/jbc.M707061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wen W, Weiss SL, Sunde RA. UGA codon position affects the efficiency of selenocysteine incorporation into glutathione peroxidase-1. J Biol Chem. 1998;273:28533–28541. doi: 10.1074/jbc.273.43.28533. [DOI] [PubMed] [Google Scholar]
- 179.Howard MT, Moyle MW, Aggarwal G, Carlson BA, Anderson CB. A recoding element that stimulates decoding of UGA codons by Sec tRNA[Ser]Sec. RNA. 2007;13:912–920. doi: 10.1261/rna.473907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Sun X, Li X, Moriarty PM, Henics T, LaDuca JP, Maquat LE. Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues. Mol Biol Cell. 2001;12:1009–1017. doi: 10.1091/mbc.12.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bermano G, Nicol F, Dyer JA, Sunde RA, Beckett GJ, Arthur JR, Hesketh JE. Tissue-specific regulation of selenoenzyme gene expression during selenium deficiency in rats. Biochem J. 1995;311:425–430. doi: 10.1042/bj3110425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.St Johnston D, Beuchle D, Nusslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-O. [DOI] [PubMed] [Google Scholar]
- 183.St Johnston D. The beginning of the end. EMBO J. 2001;20:6169–6179. doi: 10.1093/emboj/20.22.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ferrandon D, Elphick L, Nusslein-Volhard C, St Johnston D. Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell. 1994;79:1221–1232. doi: 10.1016/0092-8674(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 185.Kim-Ha J, Kerr K, Macdonald PM. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- 186.Marion RM, Fortes P, Beloso A, Dotti C, Ortin J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2212–2219. doi: 10.1128/mcb.19.3.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wickham L, Duchaine T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol. 1999;19:2220–2230. doi: 10.1128/mcb.19.3.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 189.Kim YK, Furic L, Parisien M, Major F, DesGroseillers L, Maquat LE. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Furic L, Maher-Laporte M, DesGroseillers L. A genome-wide approach identifies distinct but overlapping subsets of cellular mRNAs associated with Staufen1- and Staufen2-containing ribonucleoprotein complexes. RNA. 2008;14:324–335. doi: 10.1261/rna.720308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Monshausen M, Gehring NH, Kosik KS. The mammalian RNA-binding protein Staufen2 links nuclear and cytoplasmic RNA processing pathways in neurons. Neuromolecular Med. 2004;6:127–144. doi: 10.1385/NMM:6:2-3:127. [DOI] [PubMed] [Google Scholar]
- 192.Dugre-Brisson S, Elvira G, Boulay K, Chatel-Chaix L, Mouland AJ, DesGroseillers L. Interaction of Staufen1 with the 5′ end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ajamian L, Abrahamyan L, Milev M, Ivanov PV, Kulozik AE, Gehring NH, Mouland AJ. Unexpected roles for UPF1 in HIV-1 RNA metabolism and translation. RNA. 2008;14:914–927. doi: 10.1261/rna.829208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Chatel-Chaix L, Clement JF, Martel C, Beriault V, Gatignol A, DesGroseillers L, Mouland AJ. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol Cell Biol. 2004;24:2637–2648. doi: 10.1128/MCB.24.7.2637-2648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mouland AJ, Mercier J, Luo M, Bernier L, DesGroseillers L, Cohen EA. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: evidence for a role in genomic RNA encapsidation. J Virol. 2000;74:5441–5451. doi: 10.1128/JVI.74.12.5441-5451.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Falcon AM, Fortes P, Marion RM, Beloso A, Ortin J. Interaction of influenza virus NS1 protein and the human homologue of Staufen in vivo and in vitro. Nucleic Acids Res. 1999;27:2241–2247. doi: 10.1093/nar/27.11.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/S0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 199.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 200.Dahlseid JN, Lew-Smith J, Lelivelt MJ, Enomoto S, Ford A, Desruisseaux M, McClellan M, Lue N, Culbertson MR, Berman J. mRNAs encoding telomerase components and regulators are controlled by UPF genes in Saccharomyces cerevisiae . Eukaryotic Cell. 2003;2:134–142. doi: 10.1128/EC.2.1.134-142.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lew JE, Enomoto S, Berman J. Telomere length regulation and telomeric chromatin require the nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1998;18:6121–6130. doi: 10.1128/mcb.18.10.6121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Reichenbach P, Hoss M, Azzalin CM, Nabholz M, Bucher P, Lingner J. A human homolog of yeast est1 associates with telomerase and uncaps chromosome ends when overexpressed. Curr Biol. 2003;13:568–574. doi: 10.1016/S0960-9822(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 203.Snow BE, Erdmann N, Cruickshank J, Goldman H, Gill RM, Robinson MO, Harrington L. Functional conservation of the telomerase protein Est1p in humans. Curr Biol. 2003;13:698–704. doi: 10.1016/S0960-9822(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 204.Redon S, Reichenbach P, Lingner J. Protein RNA and protein interactions mediate association of human EST1A/SMG6 with telomerase. Nucleic Acids Res. 2007;35:7011–7022. doi: 10.1093/nar/gkm724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 206.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae . Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 207.Chawla R, Azzalin CM. The telomeric transcriptome and SMG proteins at the crossroads. Cytogenet Genome Res. 2008;122:194–201. doi: 10.1159/000167804. [DOI] [PubMed] [Google Scholar]
- 208.Carastro LM, Tan CK, Selg M, Jack HM, So AG, Downey KM. Identification of delta helicase as the bovine homolog of HUPF1: demonstration of an interaction with the third subunit of DNA polymerase delta. Nucleic Acids Res. 2002;30:2232–2243. doi: 10.1093/nar/30.10.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Kaygun H, Marzluff WF. Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol. 2005;25:6879–6888. doi: 10.1128/MCB.25.16.6879-6888.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Marzluff WF. Metazoan replication-dependent histone mRNAs: a distinct set of RNA polymerase II transcripts. Curr Opin Cell Biol. 2005;17:274–280. doi: 10.1016/j.ceb.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 211.Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- 212.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Nicholson P, Muller B. Post-transcriptional control of animal histone gene expression–not so different after all. Mol Biosyst. 2008;4:721–725. doi: 10.1039/b802133c. [DOI] [PubMed] [Google Scholar]
- 214.Kaygun H, Marzluff WF. Regulated degradation of replication-dependent histone mRNAs requires both ATR and Upf1. Nat Struct Mol Biol. 2005;12:794–800. doi: 10.1038/nsmb972. [DOI] [PubMed] [Google Scholar]
- 215.Muller B, Blackburn J, Feijoo C, Zhao X, Smythe C. DNA-activated protein kinase functions in a newly observed S phase checkpoint that links histone mRNA abundance with DNA replication. J Cell Biol. 2007;179:1385–1398. doi: 10.1083/jcb.200708106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Su C, Gao G, Schneider S, Helt C, Weiss C, O’Reilly MA, Bohmann D, Zhao J. DNA damage induces downregulation of histone gene expression through the G(1) checkpoint pathway. EMBO J. 2004;23:1133–1143. doi: 10.1038/sj.emboj.7600120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wilusz CJ, Wilusz J. New ways to meet your (3’) end oligouridylation as a step on the path to destruction. Genes Dev. 2008;22:1–7. doi: 10.1101/gad.1634508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 220.Morita T, Yamashita A, Kashima I, Ogata K, Ishiura S, Ohno S. Distant N- and C-terminal domains are required for intrinsic kinase activity of SMG-1, a critical component of nonsense-mediated mRNA decay. J Biol Chem. 2007;282:7799–7808. doi: 10.1074/jbc.M610159200. [DOI] [PubMed] [Google Scholar]
- 221.Abraham RT. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 222.Gehen SC, Staversky RJ, Bambara RA, Keng PC, O’Reilly MA. hSMG-1 and ATM sequentially and independently regulate the G1 checkpoint during oxidative stress. Oncogene. 2008;27:4065–4074. doi: 10.1038/onc.2008.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Masse I, Molin L, Mouchiroud L, Vanhems P, Palladino F, Billaud M, Solari F. A novel role for the SMG-1 kinase in lifespan and oxidative stress resistance in Caenorhabditis elegans . PLoS One. 2008;3:e3354. doi: 10.1371/journal.pone.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Oliveira V, Romanow WJ, Geisen C, Otterness DM, Mercurio F, Wang HG, Dalton WS, Abraham RT. A protective role for the human SMG-1 kinase against tumor necrosis factor-alpha-induced apoptosis. J Biol Chem. 2008;283:13174–13184. doi: 10.1074/jbc.M708008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Pal M, Ishigaki Y, Nagy E, Maquat LE. Evidence that phosphorylation of human Upfl protein varies with intracellular location and is mediated by a wortmannin-sensitive and rapamycin-sensitive PI 3-kinase-related kinase signaling pathway. RNA. 2001;7:5–15. doi: 10.1017/S1355838201000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wang W, Czaplinski K, Rao Y, Peltz SW. The role of Upf proteins in modulating the translation read-through of nonsense-containing transcripts. EMBO J. 2001;20:880–890. doi: 10.1093/emboj/20.4.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Azzalin CM, Lingner J. The double life of UPF1 in RNA and DNA stability pathways. Cell Cycle. 2006;5:1496–1498. doi: 10.4161/cc.5.14.3093. [DOI] [PubMed] [Google Scholar]
- 228.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Molecular Cell. 2003;11:939–949. doi: 10.1016/S1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 229.He F, Brown AH, Jacobson A. Interaction between Nmd2p and Upf1p is required for activity but not for dominant-negative inhibition of the nonsense-mediated mRNA decay pathway in yeast. RNA. 1996;2:153–170. [PMC free article] [PubMed] [Google Scholar]