Abstract
The stereochemistry of the inositol backbone provides a platform on which to generate a vast array of distinct molecular motifs that are used to convey information both in signal transduction and many other critical areas of cell biology. Diphosphoinositol phosphates, or inositol pyrophosphates, are the most recently characterized members of the inositide family. They represent a new frontier with both novel targets within the cell and novel modes of action. This includes the proposed pyrophosphorylation of a unique subset of proteins. We review recent insights into the structures of these molecules and the properties of the enzymes which regulate their concentration. These enzymes also act independently of their catalytic activity via protein–protein interactions. This unique combination of enzymes and products has an important role in diverse cellular processes including vesicle trafficking, endo- and exocytosis, apoptosis, telomere length regulation, chromatin hyperrecombination, the response to osmotic stress, and elements of nucleolar function.
Keywords: Inositol pyrophosphate, Diphosphoinositol phosphate, Inositol phosphate, Inositol hexakisphosphate kinase, Diphosphoinositol pentakisphosphate kinase, Diphosphoinositol polyphosphate phosphohydrolase
Introduction
The cyclitol myo-inositol has served as an extraordinary building block on which the substitution of simple mono-ester phosphate groups has enabled the creation of a vast repertoire of unique molecules with diverse cellular functions including signal transduction, vesicle trafficking, gene regulation, cell division, apoptosis, and metal ion chelation [1–9]. The possibility of this enormous family is grounded in the stereochemistry of myo-inositol itself. The orientation of the hydroxyls around the specifically myo version of inositol allows for the phosphorylation of the simple cyclohexane ring in a series of combinations which can theoretically yield up to 63 stereochemically unique forms. Of course, the phosphate head group has also found its way into an impressive subset of phospholipids too [7–9]. The growing recognition that cells had exploited many of these possible combinations, and the fact that few had recognised functions, poorly prepared those in the field for a jump beyond the fully occupied inositol ring, represented by inositol hexakisphosphate (InsP 6). However, many of us had noted peaks more polar than InsP 6 on our HPLC runs and had speculated that there maybe inositol pyrophosphate derivatives present in cells [10–13]. The existence of such forms should come as no surprise, as sugars with di- and triester phosphate groups are part of the common molecular currency of the cell, e.g., ADP, ATP. The final characterization of the observed ‘extra’ peaks came from three separate laboratories, two working together. A collaboration between Stephens’ and Mayr’s groups studying Dictyostelium inositol phosphates [14, 15] and Shears’ group studying mammalian cells [16] revealed that inositol pyrophosphates did indeed exist. Moreover, the inclusion of fluoride, which acted as a phosphatase inhibitor, led to a dramatic accumulation of the low abundance inositol pyrophosphates [16]. This observation revealed that the pyrophosphates underwent rapid metabolic cycles with the parent inositol phosphates. For example, a substantial proportion of the InsP 6 pool fluxes through the pyrophosphate pool in an hour or so [16]. This discovery in turn served as a corrective against the view that InsP 6 itself was metabolically inert cellular “wallpaper”, an indignity which it had suffered for several years. An initial and critical misapprehension that has often dogged the inositide field is to earmark a newly discovered inositol phosphate as an intracellular signal like inositol 1,4,5-trisphosphate, Ins(1,4,5)P 3) [1], rather than some other kind of regulator. However, an early idea expressed by several groups was that inositol pyrophosphates could be a new high-energy compound similar to ATP or creatine phosphate, e.g., [14, 17]. Whether this is the case or not is still disputed—due to the relative cellular concentrations of ATP compared to inositol pyrophosphates in cells. Nonetheless, an intriguing variant of this idea, the pyrophosphorylation of proteins, is gaining ground and will be considered later. However, this is only one of the areas that inositol pyrophosphates have impacted. In this review, we will start with the latest information on the inositol pyrophosphate structures themselves and then move on to the enzymes that regulate them. Finally, we will examine more recent studies in which functional information regarding these pyrophosphates has emerged.
Inositol pyrophosphates: structure
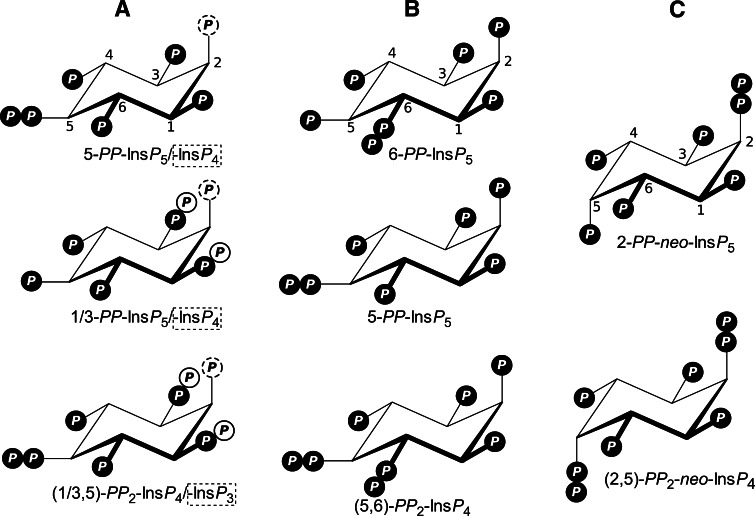
In order to fully appreciate the structures of the inositol pyrophosphates a more detailed understanding of the potential substitution of the inositol ring is required. Figure 1 acts as an important guide for the following discussion on structures. The most abundant inositol (1,2,3,4,5,6-cyclohexanol) is myo-inositol, which has 5 equatorial hydroxyl groups and one axial (the 2-hydroxyl). If one looks at the myo-inositol ring with the 2-hydroxyl pointing at the viewer, the numbering of the hydroxyl groups is counterclockwise [18]. To help memorization, this structure is often compared to a turtle [19]: The axial 2-hydroxyl corresponds to the head sticking up, 1- and 3-hydroxyls correspond to the front right and left flipper, respectively, 6- and 4-hydroxyls to the back right and left flipper, respectively, and the tail to the 5-position. This configuration has a plane of symmetry through the 2- and the 5-position and makes substitutions on the 1- or 3-position and on the 4/6-position enantiomers. These enantiomers cannot be distinguished with chemical methods such as NMR or HPLC, but only enzymatically, if appropriate and well-characterised enzymes are available. The chemical structures of several inositol pyrophosphates are shown in Fig. 1.
Fig. 1.
Structures of inositol pyrophosphates in different organisms. Phosphates are symbolised as a circled P. a Mammalians and yeast and some dictyostelids. The underlying structure is myo-inositol with the one axial hydroxyl, at the 2-position (stereochemical numbering according to NC-IUB). The phosphate at the 2-position is dashed to indicate that both InsP6 and Ins(1,3,4,5,6)P 5 are common substrates for pyrophosphorylation. Pyrophosphorylations are on the 5- and 1/3-positions. β-Phosphates on the 1/3-position are on white background to indicate that it has not been resolved which enantiomer is present. 5-PP-InsP 5 is the more abundant PP-InsP 5. b Inositol pyrophosphates in dictyostelids. Based on myo-inositol. Pyrophosphorylation are on the 5- and 6-position. 6-PP-InsP5 is the most abundant PP-InsP 5. c Entamoeba histolytica and Phreatamoeba balamuthi. These are based on neo-inositol with two opposite axial hydroxyls, at the 2- and 5-position (numbering analogous to myo-inositol for comparison), on which the pyrophosphorylations occur. Because of the symmetry of neo-inositol only one PP-InsP 5 exists, 2-PP-InsP 5
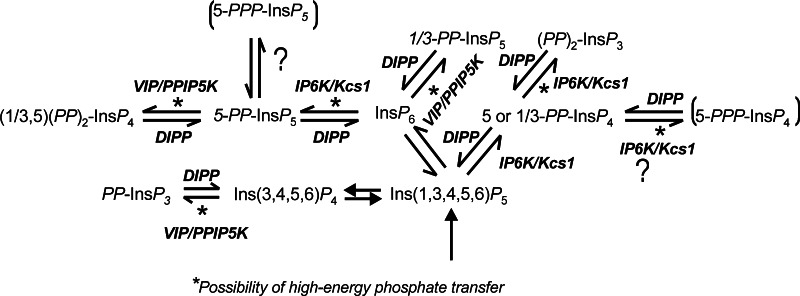
In mammalians and yeast, inositol pyrophosphates result from pyrophosphorylation of the 5-hydroxyl [20] and the 1/3-hydroxyl [21, 22] of inositol hexakisphosphate (InsP 6) by the inositol hexakisphosphate kinases (IP6Ks) and Vip1/PPIP5Ks, respectively (Fig. 2). Originally, the pyrophosphorylation sites on inositol by Vip1 were provisionally assigned to the 4/6-positions [21], but it was recently correctly determined to be at the 1/3-position [22]. In addition IP6Ks can phosphorylate 1,3,4,5,6-pentakisphosphate (Ins(1,3,4,5,6)P 5/InsP5) in both the 5- and the (1/3) positions [23]. Inositol pyrophosphates originating from InsP 4 and other InsP 5 isomers have been described in yeast defective for Ins(1,3,4,5,6)P 5 and InsP 6 synthesis [24, 25]. This both demonstrates the range of substrates that can be pyrophosphorylated when the more abundant physiological substrates InsP 5 and InsP 6 are missing and that the pyrophosphate derivatives of these other substrates are unlikely to have a physiological role unless concentrations of InsP 5 and InsP 6 are significantly depleted (see Fig. 2).
Fig. 2.
Metabolic interrelationships of inositol pyrophosphates. Inositol pyrophosphates are involved in complex metabolic interrelationships both with their precursors and each other. Inositol pyrophosphates in parentheses denote structures that are known products of the kinase in vitro but await convincing demonstration in intact, unmodified cells
All naturally occurring inositol pyrophosphates are diphospho- and bisdiphosphoinositol polyphosphates, no trisdiphosphoinositol polyphosphate has been seen so far. However, tri- and possibly tetraphosphoinositol polyphosphates have been observed in yeast after overexpression of IP6Ks and can also be produced in vitro after longer incubation with mammalian IP6Ks [23]. The PP-InsP 5 isomer in mammalian and yeast is mostly 5-PP-InsP 5 [20], with minor 1/3-PP-InsP 5 [21, 22], and the structure of the bisdiphosphoinositol tetrakisphosphate is (1/3,5)-(PP)2-InsP 4 [22] (Fig. 1a). This differs from Dictyostelium discoideum, which has 6-PP-InsP 5 as its major PP-InsP 5, but also minor amounts of 5-PP-InsP 5 [20] and (5,6)-(PP)2-InsP 4 with two vicinal pyrophosphate groups [26] (Fig. 1b). The assignment of one pyrophosphate group to the 6-position and not to the 4-position was done enzymatically, by two different approaches, one using a kinase and one using a phosphatase. In the first case, purified 6-PP-InsP 5 5-kinase from D. discoideum phosphorylates only 6-PP-InsP 5 (either chemically synthesized or purified from D. discoideum) but not 4-PP-InsP 5 on the 5-position [26]. Secondly, multiple inositol polyphosphate phosphatase (MIPP) (see also section "Diphosphoinositol-polyphosphate phosphatases") is inhibited by different PP-InsP 5 isomers with distinct potencies and the potency of PP-InsP 5 purified from D. discoideum is more consistant with 6-PP-InsP 5 than with 4-PP-InsP 5 [20]. Other Dictyostelids such as Polysphondylium species have more 5-PP-InsP 5 than 6-PP-InsP 5 and apart from (5,6)-(PP)2-InsP 4 also (1/3,5)-(PP)2-InsP 4 [27]. In Entamoeba histolytica and Phreatamoeba balamuthi, inositol pyrophosphates based on neo-inositol, not myo-inositol, have been described and identified as 2-PP-neo-InsP 5 and (2,5)-PP 2-neo-InsP 4 [28] (Fig. 1c).
The fact that the Dictyostelium (PP)2-InsP 4 has the 4/6 pyrophosphate moiety rather than the 1/3 one may have important functional consequences. This is because the steric hinderance generated by the close proximity of the 4/6 to the 5-position will mean that the pyrophosphate will have a greater free-energy of hydroysis than that found in yeasts and mammalian cells, eluding, perhaps, to a different functional role.
Enzymes responsible for formation and degradation of inositol pyrophosphates
Inositol hexakisphosphate kinases
The first inositol hexakisphosphate kinase (IP6K) to be isolated was originally termed PiUS (phosphate uptake stimulator) highlighting a connection between pyrophosphate metabolism with phosphate homeostasis. However, its IP6K activity was not originally recognized [29]. Snyder’s group showed that PiUS was an InsP 6 kinase [30] as did Irvine’s group working with the original discoverers of PiUS [31]. Snyder’s group renamed it IP6K2 [30] as they had also discovered an additional IP6K which they named IP6K1. In mammalian cells, three isoforms now exist whereas in yeast only one is found [30–32]. We will discuss the most recent information on these kinases, further details can be found in older reviews [33, 34]. At the outset, it is important to say that “InsP 6 kinase” is a misnomer because the major cellular InsP 5, Ins(1,3,4,5,6)P 5, is also a physiological substrate [23, 30]. Furthermore, in addition to the IP6Ks, the inositol phosphate multikinase (IPMK/ipk2) [30] can convert Ins(1,3,4,5,6)P 5 to its pyrophosphate derivative, at least in vitro [35, 36]. The slow conversation rate indicates a lack of physiological importance and its presence cannot rescue defective phenotypes (e.g., telomere length regulation) in which the conventional means of generating PP-InsP 4 (via kcs1) is removed [37, 38]. Although all isoforms studied can use both Ins(1,3,4,5,6)P 5 and InsP 6 as substrates (Fig. 2), the relative affinities for these two polyphosphates differs between the IP6Ks. These are reported in Table 1 and discussed in the context of the individual isoforms. The inositol pyrophosphate pattern derived from the existing mammalian IP6Ks is identical, e.g., [23], explained by the high amino acid similarity among them (~60%) which becomes much higher (~80%) at the catalytic site (similarities determined by EMBOSS Pairwise Alignment Algorithms, from EMBL-EBI website).
Table 1.
Kinetic properties of IP6K’s
| Enzyme | Substrate | K m | V max (μmol/mg per min) | References |
|---|---|---|---|---|
| IP6K1 | Ins(1,3,4,5,6)P 5 | 6.7 | 0.26 | [30, 48] |
| InsP 6 | 1.2 | 0.31 | [30, 48] | |
| IP6K2 | Ins(1,3,4,5,6)P 5 | 8.4 | 0.07 | [30, 48] |
| InsP 6 | 0.43 | 0.07 | [30, 48] | |
| IP6K3 | Ins(1,3,4,5,6)P 5 | 5.5 | 0.8 | [32] |
| InsP 6 | 0.9 | 0.6 | [32] | |
| Kcs1 | Ins(1,3,4,5,6)P 5 | 1.2 | 2.0 | [30, 48] |
| InsP 6 | 3.3 | 2.0 | [30, 48] |
IP6K1
IP6K1, originally cloned by Snyder’s group [30], has high amino acid similarity in a number of different species (99% between mouse and rat; 95% between rat–human and mouse–human; using EMBOSS algorithm above). The human gene is located on chromosome 3 (3p21.3) and it codes a 50-kDa protein. Northern blot analysis of mouse tissue reveals a 5-Kb transcript which has the highest level of expression in brain and testis, with weak expression in heart, kidney, liver, lung, and spleen [30]. Interestingly, there is a smaller 2-Kb transcript highly expressed in testis alone. Subsequent studies have revealed robust levels of expression in pancreatic β-cells [39], a neuroendocrine cell sharing several common phenotypic characteristics with neuronal cells. The testis result, together with a mass screen for novel alternative splicings [40], in which the IP6K1 data is found at the authors’ website, suggests that alternative splicing is possible in the mouse IP6K1. Also, at least one alternative splice variant of human IP6K1 has been described [41] in which a region of the N-terminal part of the protein is spliced out, giving a protein with a lower predicted MW of around 31 kDa. An important question is whether there is evidence for proteins associated with such transcripts? Our experience in pancreatic β-cells is that using western blotting lower molecular weight bands with IP6K immunoreactivity (the antibody does not select between IP6K1 and IP6K2) exist in cell homogenates of β-cells. At least one of these is, however, likely to be a proteolysis product, as withdrawal of protease inhibitors increases its amount (R. Fiume, C.J Barker and P–O. Berggren, unpublished observations). Indeed, β-cells as well as neuronal cells possess active proprotein convertases [42, 43], proteases involved in the processing of peptide hormones. In IP6K1, for example, there is consensus site for such a protease which is in a region of low order, making this putative cleavage site more likely, and so IP6Ks with lower molecular weights must be viewed with care. One function of the alternatively spliced variants could be to spacially segregate the kinase in different cellular compartments.
Screening the expressed protein sequence using, for example, ELM databases suggests many potential post-translation modifications, but there is little current information about which are relevant, with two exceptions. A screen of phosphorylated proteins in mitosis reported phosphorylation of a peptide derived from human IP6K1 on serine 151 [44]. This sits in a group IV WW motif. The phosphorylation of this type of WW motif regulates subsequent protein–protein interactions; for example, phosphorylation of this motif in the mitotic protein Pin1 regulates its cellular localization and substrate preferences [45]. The fact this phosphorylation of IP6K1 occurred in a mitotic-dependent screen is interesting, as PP-InsP 5’s concentration changes during cell cycle progression [46]. Both reports are consistent with inositol pyrophosphates having cell cycle-dependent roles. There are also many potential CK2 phosphorylation sites, and we have noted that the protein can be phosphorylated in vitro by CK2, although this apparently does not change enzyme activity (C. Illies, P-O. Berggren, C.J. Barker, unpublished data). Close association of IP6Ks with CK2 could be important for other reasons, as CK2 prephosphorylation is a prerequisite of the protein pyrophosphorylation carried out by inositol pyrophosphates [47].
In the case of IP6K1 (and IP6K3), the enzyme phosphorylates both InsP 6 and InsP 5 with only a fivefold preference for InsP 6 [30, 48]. IP6K2, however, has a significantly higher affinity for InsP 6 over InsP 5 [30] (see Table 1). However, in yeast and several mammalian cell types [49, 50], InsP 6 is the dominant inositol phosphate of the two, making the generation of InsP 5 pyrophosphates less favorable.
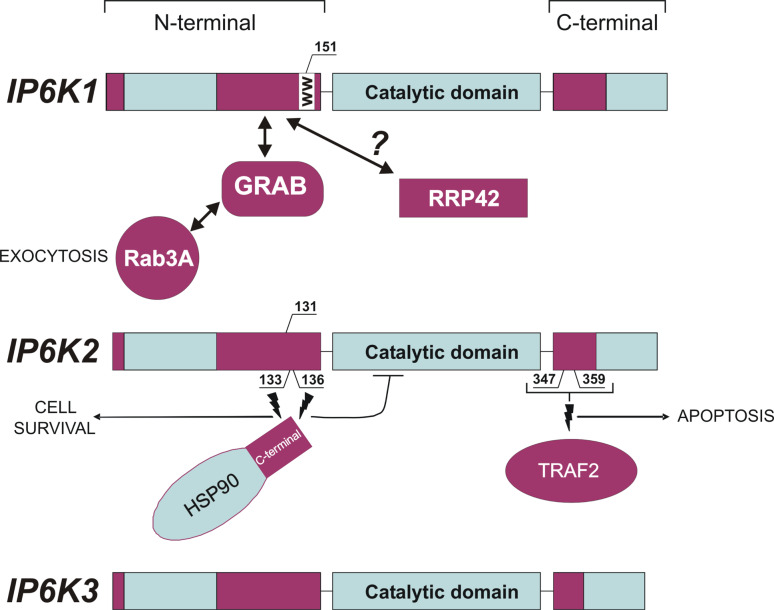
Besides a difference in substrate preference, one other major difference between IP6K1 and IP6K2 is the ability to associate with different binding partners. In the case of IP6K1, it associates with a guanine nucleotide exchange factor (GEF) protein called GRAB [51]. GRAB’s GEF activity is directed towards Rab3A, but it can also associate with other Rab3 proteins; however, the functional consequences of these other interactions was not explored [51]. One possible role for the interaction of GRAB and Rab3A is the promotion of exocytosis in neuronal cells (see later). Despite many sequence similarities between IP6K1 and IP6K2, IP6K2 does not associate with GRAB [51]. This difference probably correlates with an as yet undefined part of the N-terminal end of IP6K1 (see Fig. 3). Therefore, it is worth repeating that this N-terminus is deleted in the splice variant of IP6K1, described above, suggesting at least one functional difference that alternative splicing could confer on IP6K1, the loss of GRAB interaction. Although GRAB mRNA was found outside the CNS [51], antibodies against the protein localized it fairly exclusively to neuronal tissues. We wondered whether the coiled-coil domain of the GRAB was similar to others in the databases, thus identifying GRAB structural homologues and therefore other possible interaction partners for IP6K1, and of course Rab3A. In collaboration with a structural biologist, we used threading protocols to search for similar coiled-coil domains in other proteins. We only found one significant match and that was to the APC protein critical in mitogenesis and predisposition to bowel cancer [52] (B.M. Hallberg, personal communication). Whether this protein can actually interact with IP6K1 remains to be determined experimentally.
Fig. 3.
IP6Ks showing basic organization of proteins. The figure illustrates the three human IP6Ks (IP6K1, IP6K2, and IP6K3) highlighting their similarities (Blue) and differences (Red) and location of important protein–protein interactions. IP6K1 It is not clear where GRAB binds. However, it is most likely towards the N-terminal as this is a unique region. Rab3A and IP6K1 are both binding partners of GRAB and their hydrophobicity plots bear a passing similarity in this region, too. Proteomic studies have described serine phosphorylation in a WW motif which could indicate interactions with ubiquitin ligase and/or a mitotic protein, and there is a hint of an interaction with the protein RRP42 (see main text). IP6K2 The two distinct regions of IP6K2 serve as unique binding domains for HSP90 and TRAF2. In the case of HSP90 association mutation of either serine 133 or 136 is sufficient to disrupt the association. In contrast, mutations of both serines 347 and 349 are required to prevent TRAF2 association. IP6K3 Apart from its catalytic site there is nothing known about functional motifs in this protein or of any potential interacting partners. Interesting, proteomic studies have revealed phosphorylation of succession of a serine-242, threonine-243 and serine-244. This is in no recognized protein phosphorylation motif, so its function is unclear. However, it is in the region of the catalytic site so may act to regulate enzyme activity
The GRAB/IP6K1 interaction highlights an important aspect of IP6K function, namely that the protein also mediates important cellular effects that are not dependent on catalytic activity. The same bifunctionality is also found in IP6K2. Are there other proteins that could potentially interact with IP6K1? Examination of the human interactome reveals a possible interaction with EXOSC7/RRP42 (exosome component 7 or ribosomal RNA-processing protein 42). The exosome is an RNA-processing complex thought to be involved in RNA quality control [53], and whilst present in both cytosol and nucleus, it has a key role in the nucleolus [53]. The evidence for IP6K1 association is based on a high-throughput yeast two-hybrid screen [54]. The possibility of this interaction is worth seriously considering on two counts. Firstly, several of the proteins that have the specific pyrophosphorylation modification are nucleolar proteins, e.g., Nucleolin, Srp40p, Nopp140 and TCOF1 [55]. Secondly, our own unpublished data from pancreatic β-cells detects an undefined IP6K in the nucleolar region of the nucleus (Fiume et al., unpublished data).
IP6K1 was the first gene involved in inositol pyrophosphate metabolism to be knocked out in a mouse. The global knockout of IP6K1 has been described both in an NIH-funded mouse knockout database [56] and in a published report [57], although only the latter reported the phenotype in detail. However, a common phenotype between the two knockouts, which were accomplished using different strategies, is a reduction in plasma insulin levels. This supports but does not directly prove the idea that the gene is important in insulin secretion (see functional discussions later).
IP6K2
As we have already noted, IP6K2 (or IHPK2) is also known as inorganic phosphate uptake stimulator (PiUS), which was identified in 1997 from a rabbit duodenal cDNA library [29]. After cRNA-transcription and oocyte injection, its cDNA stimulated Na+-dependent and -independent inorganic phosphate uptake. It was identified as an IP6K in 1999 [30, 31]. Human IP6K2 has been mapped to chromosome 3 (3p21.31). Northern blot analysis has shown a tissue distribution which is highest in brain and lung and lower in liver, kidney, and testis [30]. It has also been found in intestine, heart [29], and islets of Langerhans or pancreatic β-cells [39]. Its gene encodes for a protein of around 49 kD which has been assumed as the canonical sequence or isoform 1. In fact, alternative splicing has been found through database analysis (UniProtKB/Swiss-Prot Q9UHH9), which results in two more transcript variants encoding for shorter isoforms (97 and 70 amino acids) that lack most of the C-terminal part of the whole protein. So far, there is no experimental evidence for these transcript variants.
IP6K2, as is also true for the other IP6Ks, is both an Ins(1,3,4,5,6)P 5 and a InsP 6 kinase; it has also been implicated to have in vitro activities towards the Ins(1,3,4,5,6)P 5 pyrophosphates, diphosphoinositol tetrakisphosphate (PP-InsP 4) and bis-diphosphoinositol trisphosphate [(PP)2-InsP 3] [23, 48]. In contrast to IP6K1 and IP6K3, IP6K2 has been shown to have a 20-fold lower affinity for Ins(1,3,4,5,6)P 5 compared with InsP 6 and a 7-fold lower activity for PP-InsP 4 compared to IP6K1. These catalytic differences [48] could bias IP6K2 towards generating InsP 6-based pyrophosphates which may be important in its unique physiological role in apoptosis (see later section). However, so far, there is no direct experimental evidence to support this notion. No enzymatic activity has been shown with Ins(1,2)P 2, Ins(1,4,5)P 3, or Ins(1,3,4,5)P 4; however, there is a low activity towards Ins(3,4,5,6)P 4 [30, 48].
Recently, Podobnik’s group has structurally characterized an expanded number of pyrophosphates that can be derived in vitro by IP6K2 using Ins(1,3,4,5,6)P 5 and InsP 6 as substrates [23]. They suggest that these products may also occur in intact yeast, but this only convincingly occurs after intervention, either by overexpression or gene deletion [23]. This is the most informative study so far about the catalytic site of the IP6Ks and IP6K2 in particular, and so at this point we will examine the information in greater depth for IP6K2 than for the other kinases. The study suggests that IP6K2 may also synthesize an “InsP 8” from InsP 6, which has been established to be an inositol pentakisphosphate with a triphosphate group in the 5 position (5-PPP-InsP 5). This “InsP 8” produced from InsP 6 is a minor species compared to PP-InsP 5. Further enzyme incubation shows a later eluting compound with higher polarity, suggested to be an “InsP 9”, but which has not been characterized due to its low abundance. Because “InsP 8” and “InsP 9” occur after a significant decrease of InsP 6, it seems that substrate specificity is a rate-limiting step for these reactions. Polarity, steric factors, and the speculated lower solubility of the higher inositol pyrophosphate may play an important role for the substrate preference at the catalytic active site [23]. The employment of Ins(1,3,4,5,6)P 5 as a substrate indicates the importance of the 2-axial phosphate group in directing the pyrophosphorylation position. This is because amongst the products of Ins(1,3,4,5,6)P 5 pyrophosphorylation mediated by IP6K2 are structures that contain pyrophosphate groups at both the 5- and the 1/3 positions, rather than just the 5-position observed when using InsP 6 as a substrate.
It may be asked, what is the physiological relevance of the studies above describing the unusual products of IP6K activity? Whilst the jury is out on whether triphosphoinositol phosphates and other exotic pyrophosphate derivatives are generated under normal conditions, similar materials are produced when the IP6K’s are overexpressed in yeast, e.g., [23]. This has also been our experience with some pancreatic β-cell lines in which overexpression of IP6Ks lead to a large accumulation of pyrophosphates with the polarity of “InsP 8”, “InsP 9” and beyond (C. Illies, P-O. Berggren, C.J. Barker, unpublished observations). Since these products are likely to be tri-, tetra- and even penta-ester phosphates, and not the conventional pyrophosphates found in cells, two possible difficulties in interpretation may arise. Firstly, the exotic products could mask real changes in bona fide inositol pyrophosphates when measuring by HPLC. Secondly, in a cell system, these products may interfere with normal cell function. One further practical note in regard to overexpression studies is that the catalytic activity of the kinases is not only influenced by the buffer environment [23] but also by the kind of tag attached [58]. The GST-IP6K1 fusion protein has different catalytic properties compared to the HIS-tagged protein [58]. Because of these complexities, overexpression studies should always be supported by silencing or other gene-deletion approaches.
Besides its substrate specificity, the physiological role of IP6K2 may also be explained by its unique primary and tertiary protein structure which can lead to either different localization or a different pattern of protein–protein interactions. Among IP6Ks, the difference in the amino acid sequence is mostly confined to the N-terminal and to a section of 40 residues in the C-terminal (see Fig. 3). Through GFP-tagged protein, localization of IP6K2 by confocal microscope has been described to be mostly in the nuclear compartment [32], the other IP6Ks are also distributed in the cytoplasm. A bipartite nuclear localization signal (NLS) has been found in IP6K2’s sequence (298-YLRRELLGPVLKKLTEL-314) which is not completely shared with other IP6Ks. In NIH-OVCAR-3 cells, IP6K2 resides mainly in the cytoplasm and perinuclear region [59]. Interferon-β (IFN-β) treatment induces translocation of IP6K2 to the nucleus. Replacement of bipartite NLS with alanine inhibits the capability of IP6K2 to translocate into the nucleus after IFN-β treatment [59]. Translocation from the nucleus to mitochondria has also been described following cytotoxic stimulation [60]. The C-terminal region of IP6K2 has been shown to contain two functional SXXE motifs (346-DSDAEDLEDLSEESADES-363), putative binding sites for tumor necrosis factor receptor-associated factor 2 (TRAF2), which drives a physiological role for IP6K2 that is independent of its catalytic activity. Combinations of single point mutation at serines 347 and 359 to alanine reduce TRAF2 binding activity [61]. Once again, these consensus sequences are unique to IP6K2. A more recent study employing tandem mass spectrometric analysis revealed that IP6K2 may bind heat-shock protein 90 (HPS90) [62]. Deletion mapping of both HSP90 and IP6K2 has identified the binding site for IP6K2 at the N-terminal between 132 and 143 amino acids (132-VRQHRKEEKMKS-143), while the HSP90 region involved occurs at the C-terminal. Single point mutation of arginine to alanine either 133 or 136 abolishes IP6K2–HSP90 binding. Binding to HSP90 inhibits IP6K2 catalytic activity leading to marked reduction in PP-InsP 5 levels. This mechanism has been suggested to occur through the ability of HSP90 to mask the catalytic active site of IP6K2 [62]. Overall, all these specific interactions along with the kinase ability to synthesize inositol pyrophosphates has committed IP6K2 as a key player in the process of apoptosis, which we will consider in depth later. Therefore, the role of IP6K2, like IP6K1, does not completely depend on its enzyme activity but also on specific protein–protein interactions within the cellular environment (Fig. 3).
Note. During the revision of this manuscript a report describing a mouse knockout model of IP6K2 was published online [63]. The phenotype was significantly different from that of the IP6K1 knockout mouse [57]. In particular, there was no effect on growth or fertility and notably no decrease in plasma insulin concentrations: features of the IP6K1 knockout mouse. This report highlights the significance of context for the production of PP-InsP 5 and possibly the importance of the unique protein:protein interations and/or localization displayed by the different IP6K isoforms. The data also underscore that the effect on insulin secretion is specific to IP6K1 [39, 57].
IP6K3
IP6K3 was isolated from brain [32] and is largely restricted to neuronal tissues, with some expression in the kidney and considerably less in other tissues. Certainly, it is not even found in neuroendocrine cells like pancreatic β-cells [39]. In the human genome, in contrast to IP6K1 and IP6K2, which are found on chromosome 3, IP6K3 is found on chromosome 6 (6p21.31). There is some evidence for alternatively spliced variants in both human and mouse. The original northern blots reveal strong bands of 6 and 2 Kb, with 2 other fainter bands, consistent with this [32]. The strongest expression is found in the 6-Kb band in the cerebellum, interestingly the 2-Kb band is dominant in the hippocampus, suggesting that the alternative splicing links to different brain regions. Its molecular weight (46 kDa) is lower than both IP6K1 and IP6K2, and a lower theoretical pI makes it significantly more acidic than the other two proteins. The pI of a protein can have important implications in the protein–protein, protein–substrate, and protein–membrane interactions [64]. It has no clear distinguishing function, though its dominant distribution in the CNS does suggest it may have a neuronal-specific one. Although IP6K3 and IP6K1 have similar kinetic properties and substrate preferences which are distinct from IP6K2, there are differences too, for example, the Vmax of the IP6K3 is double that of the IP6K1 (Table 1). GFP-tagged IP6K3 seems to preferentially localize in the cytosol, in contrast to GFP-tagged versions of both IP6K1 and 2 which show different levels of nuclear association [32].
Kcs1 and dInsP6K/I6KA
In yeast and Dictyostelium, there is only one isoform of IP6K, Kcs1 [30] and dInsP6K/I6KA [65], respectively. This immediately highlights the fact that in mammalian cells there is a likely specialization of IP6Ks which may be, in part, due to the different protein–protein interactions they can engage in, as described above. The yeast enzyme also has an additional functional module distinct from its catalytic activity [24, 30, 66]. It possesses two leucine heptad repeat domains not obvious in its mammalian counterparts. This suggests it, too, has a specialized function in yeast not replicated in mammalian cells, and indeed this protein domain is apparently important for some aspects of cell wall and vacuolar regulation [24]. Evidence that Kcs1, like IP6K1 and IP6K2, is engaged in protein–protein interactions comes from yeast two-hybrid studies in which Bmh2 [67] was found as a putative partner. This yeast protein is a homologue of the mammalian 14-3-3 family with diverse roles in cell signaling [68]. Interaction of Kcs1 with 14-3-3 proteins may not be unique to yeast, however, as both IP6K1 and 2 have consensus motifs for the binding of 14-3-3 proteins. The Dictyostelium kinase is an important player linking pyrophosphates to the regulation of a Dictyostelium specific PtdIns(3,4,5)P 3 binding domain which we will discuss later [65].
One oddity, which results from the silencing of Kcs1 [48] and both IP6K1 and IP6K2 (C. Illies, C.J. Barker, P-O. Berggren, unpublished data), is that the concentration of Ins(1,3,4,5,6)P 5 is decreased. The explanation for this observation is unclear, as it is counter-intuitive. It implies more complex metabolic relationships between pyrophosphorylated and the non-pyrophosphorylated inositol phosphates than is currently understood. It also may mean that the phenotype of the IP6K knockouts at both the cellular and whole animal level could be due to the reduced levels of Ins(1,3,4,5,6)P 5 as well as those of the pyrophosphates. One simple explanation is that an InsP 4 pyrophosphate (PP-InsP 3) could be hidden under the Ins(1,3,4,5,6)P 5 peak. In mammalian cells, this could be a pyrophosphate derivative of Ins(3,4,5,6)P 4 [48]. The issue may be more important in yeast as reduction in the Ins(1,3,4,5,6)P 5 is much greater than mammalian cells.
InsP6/PP-InsP5 kinases: VIP/PPIP5K
There had been independent indirect [69] and direct [70] evidence for an enzyme distinct from the IP6K family that would phosphorylate PP-InsP 5 to “an InsP 8” (PP)2-InsP 4. An enzyme activity had even been purified from mammalian cells that was able to carry out this phosphorylation step [71]. However, the final molecular identity of this enzyme emerged from studies in yeast [21] and mammalian cells [72, 73]. In yeast, an additional, genetically distinct IP6K activity (and not IPMK) was observed in cells in which kcs1, the yeast homologue to mammalian IP6Ks, had been deleted [21]. The enzyme was identical to Vip1 and Asp1 from S. cervicea and S. pombe, respectively. The mammalian ortholog was also characterized, cloned and sequenced and called either VIP1 [72] or, PPIP5K [73]. Two forms exist, PPIP5K1/VIP1, a 160-kDa protein, and the shorter PPIP5K2/VIP2 (138 kDa) [21, 72, 73]. The biggest differences between the two isoforms lie in the C-terminal of the protein. These kinases are widely expressed with a high concentration in skeletal muscle, heart, and brain [72]. The PPIP5K/VIP1/2 proteins had two domains which were of immediate interest to those in the inositide field. These were an ATP-grasp domain similar to that found in some other inositide kinases [74], and a histidine acid phosphatase-like domain found in many histidine acid phosphatases and phytases, including MIPP, the main enzyme responsible for the dephosphorylation of InsP 5 and InsP 6 [75]. Interestingly, so far there is no evidence that this PPIP5K/VIP phosphatase domain is catalytically active [21, 72, 73] (at least against inositol polyphosphates), perhaps reflecting a different amino acid composition at a critical domain in the active site [21, 72, 73]. The mammalian enzyme was discovered to be inhibited by InsP 6, a fact that emerged during its purification [73]. This might suggest, given the substrate preferences of other histidine acid phosphatases (e.g., MIPP [75]), that the second domain of the protein may serve as a regulatory domain with InsP 6 acting as a negative allosteric regulator. Indeed, a truncated form of this protein without the acid phosphatase domain is apparently more active [21] or at least makes a bigger impact on the (PP)2-InsP 4 levels when overexpressed in cells [21]. However, this may also reflect differences in expression levels. One clear function that was confirmed by the Shears group [73] is a role in osmotic stress and this will be discussed later. The phenotypes for the deletion of VIP1 and asp1 are different. The growth of asp1 deletion mutant is temperature sensitive, but the equivalent mutant in VIP1 only exhibits temperature sensitivity when combined with a deletion of Las17 (the yeast WASP protein), perhaps reflecting differences in the N-terminal sequence [21].
Both InsP 6 and PP-InsP 5 are substrates for this enzyme producing a new form of PP-InsP 5 and (PP)2-InsP 4 [21, 72, 73], but neither Ins(1,3,4,5,6)P 5 nor its pyrophosphate derivatives (PP-InsP 4 or (PP)2-InsP 3) [73] are good substrates. As discussed above, the original provisional structural assignment for these molecules was revealed to be incorrect when a more discriminatory 2D-NMR technique was applied [22]. The enzyme phosphorylates the mono-ester phosphate group on the 1/3- not the 4/6-position of the inositol ring. An important question is whether, like the IP6Ks, the PPIP5K/VIPs have a role independent of their catalytic activity. So far, there are no conclusively identified partners for these enzymes; however, inspection of a yeast interactome reveals many potential binding partners. These assignments must be regarded with a great deal of caution until further confirmatory work can be carried out. However, like IP6K1, human proteomic approaches found phosphorylation of serines 475 [76] and 1152 [77], which are a putative CK2 phosphorylation site and a WW domain, respectively. Interestingly, similar phosphorylations are also found on IP6K1 (see above).
Diphosphoinositol-polyphosphate phosphatases
Even though the diphosphoinositol-polyphosphate phosphohydrolases (DIPPs) were first cloned [78] before the IP6Ks [30, 31], and their identification has connected highly phosphorylated inositol phosphates to the similarly highly phosphorylated diadenosine polyphosphates [79], there is today only little information on their physiological role.
DIPPs belong to the Nudix hydrolase family, characterised by the nudix motif, or, as it was formerly called, the MutT motif [80]. This protein motif was first described on the Escherichia coli protein MutT, which protects from DNA mutations by degrading dGTP to dGMP and PP i, thus being a pyrophosphohydrolase. Later, it was discovered that many proteins containing the MutT motif hydrolyse a wide variety of substrates consisting of a nucleoside diphosphate linked to some other moiety X, and the MutT family was renamed to Nudix [80]. With the realization that inositol pyrophosphates, which do not contain a nucleoside, are also substrates for Nudix hydrolases, the designation Nudt for Nudix-type was chosen, including the adoption of NUDT as the gene symbol by HUGO Gene Nomenclature Committee [81].
In human and mouse, there are 4 DIPP genes, encoding 5 proteins. DIPP1 is the product of the NUDT3 gene. DIPP2α and DIPP2β, which only differ by one amino acid, the glutamine Q86, which is inserted in DIPP2β, are both products of the NUDT4 by alternative usage of an intron boundary [82]. NUDT10 and NUDT11 are arranged in tandem on the X chromosome and give rise to DIPP3α (also called hAps2) and DIPP3β (hAps1), respectively [83, 84]. In humans, DIPP3α differs from DIPP3β by just one amino acid [83], whereas in mouse they are identical [85]. While DIPP1 [78, 86] and DIPP2 [87, 88] are expressed in a broad range of tissues, the expression of DIPP3 is more restricted [84], with DIPP3α expressed in testis, liver, and kidney, and DIPP3β in brain [85]. In yeasts, there is only one DIPP, Ddp1p/Yor163w in Saccharomyces cerevisiae and Aps1 in Schizosaccharomyces pombe [89–91].
DIPPs are proteins with a molecular weight of about 20 kDa. They have a common catalytic site [81] for hydrolysing inositol pyrophosphates such as PP-InsP 4, PP-InsP 5, and (PP)2-InsP 4 [87] (see Fig. 2), diadenosine polyphosphates, especially the higher Ap5A and Ap6A [89, 90] and (5-phosphoribosyl 1-pyrophosphate pyrophosphatase) PRPP [92], although PRPP is probably not a substrate for DIPPs in vivo [84]. DIPPs are inhibited by fluoride with a K i around 10 μM [78, 83]. Despite their range of different substrates, they show catalytic specificity in that they hydrolyze the 5-β-phosphate from PP-InsP 5, whereas on (1/3,5)-(PP)2-InsP 4 the 1/3-β-phosphate is hydrolysed [70].
DIPP1 is the most catalytically active of the three DIPPs [85, 87]. It has a much lower K m for inositol pyrophosphates than for ApnAs and PRPP [78, 85, 92], making inositol pyrophosphates their preferred substrates. In fact, its specificity constant for these compounds is remarkably high, close to the diffusion limit [93]. DIPP2α and DIPP2β differ by a single amino acid (Q86), but this glutamine has a strong influence on catalytic function [87], DIPP2α is catalytically more active than DIPP2β towards PP-InsP5 and (PP)2-InsP 4, but does not seem to metabolize Ap5A and Ap6A at all [87]. DIPP3α and β are the least active among DIPPs towards inositol pyrophosphates [83–85]. They depend on millimolar manganese concentrations [83] for efficient hydrolysis of diadenosine polyphosphates, which makes these compounds unlikely substrates because total cellular Mn2+ is around 30 μM with free Mn2+ < 1 μM [94].
Another enzyme that can degrade inositol pyrophosphates, but is unrelated to the nudix-type family, is the multiple inositol polyphosphate phosphatase (MIPP). It also degrades, among other inositol phosphates, PP-InsP 5 and PP 2-InsP 4 [70, 75]. It differs from the DIPPs in that it specifically removes the 5-β-phosphate from both PP-InsP 5 and (PP)2-InsP 4. MIPP is located in the endoplasmic reticulum [95] and does not seem to play a role in inositol pyrophosphate degradation in vivo [70].
As mentioned above, there are only a few functional studies on DIPPs and their role in signaling. Because their putative functions are diverse, we will tackle them here in this section rather than the section dealing with functional aspects of diphosphoinositol phosphates. DIPPs are highly active enzymes, especially DIPP1 and DIPP2α, which have high specificity constants, and their reactions with inositol pyrophosphates are close to the diffusion limit. With this in mind, one can wonder why we have such high inositol pyrophosphate levels at all [34]. The high activity of DIPPs is underscored by the rapid increase of inositol pyrophosphates after fluoride inhibition of DIPPs [96]. Of course, the rapid cycling between PP-InsP 5 and InsP 6 is the result of active phosphatases and kinases, and, so far, only evidence for the regulation of kinases [62] but not DIPPs has been found.
DIPP2 mRNA is increased in rat brain after prolonged lithium therapy [88]. Lithium is used to treat bipolar mood disorder and interferes with inositol metabolism [97]. This finding suggests that DIPPs might be a relevant target of lithium therapy.
One viral protein (g5RP/D250) of the African swine fever virus (ASFV) was found to encode a nudix-type hydrolase with activity towards PP-InsP 5 and (PP)2-InsP 4 [98]. Infection with ASFV leads to a reduction of inositol pyrophosphates, but also to reduced GTP. In contrast to other DIPPs, g5Rp removes the 5-β-phosphate from both PP-InsP 5 and PP-InsP 4.
Another attempt to determine DIPP substrates in vivo and its function was done in S. pombe [99]. Disruption of aps1, the only DIPP in S. pombe, raised intracellular PP-InsP 5, but did not affect Ap5A. This indicates that the inositol pyrophosphates and not diadenosine polyphosphates are the natural substrates for aps1. Overexpression of aps1 on the other hand did not change inositol pyrophosphate levels and even counter-intuitively increased Ap5A. This paradoxical increase of Ap5A could be secondary to the strongly swollen phenotype that overexpression of aps1 caused.
Ectopic expression of murine DIPP1 results in inhibition of signaling through the ERK1/2 pathway [86], and leads to reduced ERK1/2 phosphorylation and inhibition of EGF receptor signalling. This effect seems to be independent of DIPP1’s catalytic activity, as a kinase-inactivated mutant of DIPP has the same effect. The exact site of DIPP1 action was not identified, but the authors suggest it to be at or downstream of MEK1 activation. This last observation leads to a growing conviction that most of the enzymes that modulate inositol pyrophosphates exert their effect in both a catalytical-dependent and -independent manner.
Functions of inositol pyrophosphates
(see Figure 4.)
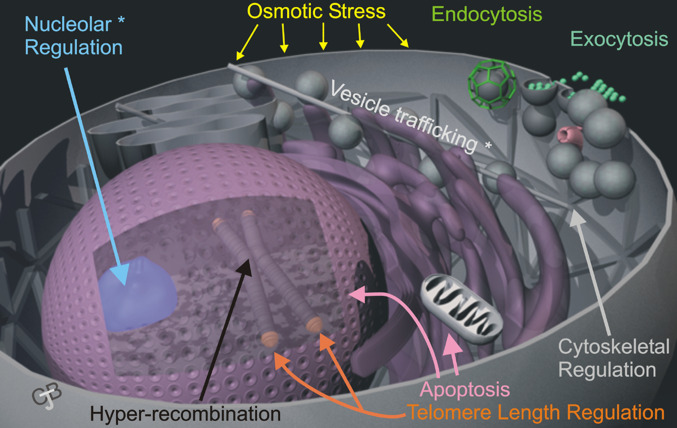
Fig. 4.
Cellular functions of inositol pyrophosphates. The stylized figure of the cell illustrates the areas in which inositol pyrophosphates and/or the enzymes that metabolise them play a role. The asterix denotes areas of cell function that may be dependent on the ability of the inositol pyrophosphates to “pyrophosphorylate” proteins. The nucleous is highlighted, not because any specific functions have been elucidated, but because several pyrophosphorylated proteins are thought to be resident there. In addition to the areas highlighted, inositol pyrophosphates regulate phosphate homeostasis and cyclin-complexes in yeast
Protein pyrophosphorylation
The idea that diphosphoinositol polyphosphates, in analogy to other high-energy-phosphate containing molecules such as ATP, could donate their β-phosphate to other molecules in a phosphorylation reaction came up soon after the first description of these molecules. Indeed, the IP6K reaction is fully reversible and PP-InsP 5 can donate its β-phosphate back to ADP [17]. More recently, it was shown that PP-InsP 5 can also donate its β-phosphate to proteins [55], at least in vitro.
The phosphorylation of proteins by PP-InsP 5 has many notable features. It is in fact a pyrophosphorylation because the β-phosphate from PP-InsP 5 is transferred to a phospho-serine that was previously phosphorylated by CK2 [47]. This transfer does not require an additional enzyme, but depends in the intact structure of the target protein, as denatured proteins are not subject to this modification [55]. The required secondary structure cannot be large, as a small peptide of 25 amino acids, derived from Nopp140 (see below), undergoes phosphorylation. This process is not specific to the position of the β-phosphate, as not only 5-PP-InsP 5 but also 1/3-PP-InsP 5 and (1/3,5)(PP)2-InsP 4 can donate their β-phosphates [47].
The proteins known to be pyrophosphorylated by PP-InsP 5 are the yeast proteins Nsr1p (the yeast homologue of nucleolin), Srp40p, and Ygr130cp, and the mammalian proteins Nopp140 (homologue of Srp40p), TCOF1, and adaptor complex AP-3β1/2 [55]. They share a characteristic sequence rich in serines and acidic amino acids forming phosphorylation sites for CK2 [100] close to a stretch of basic amino acids. Nucleolin, Srp40p, Nopp140, and TCOF1 are nucleolar proteins and known substrates for CK2 [101–104]. Nucleolin and Nopp140 can be highly phosphorylated, and the phosphorylation process of Nopp140 by CK2 occurs in a cooperative manner with phospho-serines produced by CK2, creating new CK2 phosphorylation sites [103]. Interestingly, Nopp140 has been associated with another higher inositol polyphosphate, InsP 6, in an independent interaction with CK2. Nopp140 binds to and inhibits CK2, and this binding can be released by InsP 6 [105, 106]. Since InsP 6 is also in a dynamic exchange with PP-InsP 5, this suggests a tight regulatory integration between InsP 6, PP-InsP 5, and CK2. Furthermore, the pyrophosphorylated proteins Nopp140/Srp40p, nucleolin/Nsr1p, and TCOF1 all play a role in ribosome biogenesis [102, 107–109], and a recent study in yeast demonstrates a function of Kcs1p in ribosome biogenesis as well [110].
The process of PP-InsP 5-mediated protein pyrophosphorylation has so far only been demonstrated to occur in vitro. However, cells contain 20–50 times more InsP 6 than PP-InsP 5. Inclusion of 50-fold excess of InsP 6 over PP-InsP 5 strongly inhibits protein pyrophosphorylation in vitro, but does not completely abolish it [55]. The evidence that protein pyrophosphorylation may occur in intact cells comes from two experiments. Firstly, in vitro phosphorylation of overexpressed NSR1 was enhanced in yeast lacking IP6K compared to wild-type yeast [55]. An interpretation of this observation is that the lack of IP6K1 reduces the pyrophosphorylation of the NSR1. Therefore, when the NSR1 is extracted and pyrophosphorylation studied in vitro, it is more amenable to receive the pyrophosphorylation compared to the wild-type which is already pyrophosphorylated. Secondly, enhanced PP-InsP 5 and (PP)2-InsP 4 concentrations occur in yeast strains lacking NSR1. [55]. However, an indirect effect of NSR1 deletion might be possible. Also, yeast strains lacking SRP40, another pyrophosphorylated protein, did not show altered PP-InsP 5 or PP-InsP 4 levels.
A demonstration that pyrophosphorylation of target proteins has a functional consequence is still missing. So far, there is only one negative study, which investigated the connection between osmotically induced rise in (PP)2-InsP 4 and translocation of nucleolin [111]. Nucleolin is the mammalian homologue of the yeast protein NSR1, which is known to be pyrophosphorylated by PP-InsP 5 in vitro [55]. No correlation between (PP)2-InsP 4 levels and osmotically induced translocation of nucleolin could be found in intact cells [111].
Vesicle trafficking
Vesicle trafficking was the first cellular function that inositol pyrophosphates were associated with. The evidence came from in vitro binding to vesicle coat proteins, following up studies that showed binding of InsP 6 to the clathrin assembly protein AP-2 [112, 113]. PP-InsP 5, together with InsP 6, was shown to bind to coatomer in mammals [114] and yeast [115], and to block its ion channel activity [114]. The clathrin assembly protein AP180 (also called AP-3, but not to be confused with the clathrin assembly complex AP-3) was also shown to bind InsP 6 [116] and PP-InsP 5 [117, 118]. Binding of InsP 6 and PP-InsP 5 to AP180 inhibited clathrin assembly. The work on PP-InsP 5 binding to coatomer, AP-2 and AP180 was not followed up after it became clear that phosphatidylinositol 3,4,5-trisphosphate (PtdIns(3,4,5)P 3/PIP3) is a natural ligand for AP-2 [119], AP180 [120], and coatomer [121]. But further exploration of these issues was hampered due to the limited availability of inositol pyrophosphates for binding studies.
The cloning of IP6Ks and the ensuing availability of molecular-biological tools led to new evidence on inositol pyrophosphate involvement in vesicle trafficking. The first identified role was in exocytosis via an interaction with GRAB (see section "IP6K1") [51]. This binding is specific to IP6K isoform 1 and does not occur with IP6K2. However, the production of inositol pyrophosphates is not necessary for this effect, as a kinase-inactivated IP6K1 mutant (IP6K1-K/A) still binds GRAB. Overexpression of IP6K1-wt or IP6K1-K/A and antisense RNA against GRAB enhance exocytosis for PC12 cells and bovine adrenal chromaffin cells, whereas overexpression of Rab3A or GRAB has a negative effect on stimulated secretion [51]. These findings are in accordance with a limiting function on exocytosis by Rab3A and an inhibitory function on Rab3A by the IP6K1 protein.
In kcs1Δ yeast, which lacks their only IP6K, Kcs1, a fragmented vacuole was noticed [48]. Further investigation showed that interfering with inositol pyrophosphate production by either deleting Kcs1p (yeast IP6K) or Arg82p (yeast IPMK) impaired trafficking from endosomes to the vacuole [24, 118]. There was a suggestion of an endocytosis defect [118] which may also represent a disruption of the TGN. However, the effect seen on vesicle trafficking was not specific to PP-InsP 5 and (PP)2-InsP 4 (the inositol pyrophosphates based on InsP 6), since PP-InsP 4 and (PP)2-InsP 3 [the pyrophosphates based on Ins(1,3,4,5,6)P5] are equally as active [24, 118].
Evidence for direct role of inositol pyrophosphates on exocytosis comes from studies on pancreatic β cells [39]. These cells have rather high 5-PP-InsP 5 concentrations, in the range of 5 μM, corresponding to 5–10% of InsP 6. Secretion from the readily releasable pool (RRP) could be increased by overexpression of all three isoforms of IP6K, but kinase-inactivated mutants were without effect. This suggested that production of inositol pyrophosphates was responsible for the enhanced secretion in this system, and indeed, direct application with a patch pipette of 5-PP-InsP 5 to the cells increased secretion with a half maximal concentration of about 1 μM, within the estimated physiological concentration range of 5-PP-InsP 5 of 0.1–2 μM [34]. Other PP-InsP 5 isomers were also able to enhance secretion, but dose–response curves were not established. Neither (PP)2-InsP 4 or PP-InsP 4 were tested.
These findings are reminiscent of the above mentioned result [24, 118], where different inositol pyrophosphates were able to substitute for each other. In contrast to overexpression of IP6Ks, siRNA of IP6Ks showed isoform specificity. Only siRNA for IP6K1, but not IP6K2, resulted in a reduction of secretion from the RRP [39]. This effect was not caused by the absence of IP6K1 protein, because exogenously added 5-PP-InsP 5 completely reversed the effect of the silencing. It seems, thus, that IP6K1 and IP6K2 produce separately functioning pools of inositol pyrophosphates. We have subsequently confirmed this result in a beta cell line where silencing of IP6K2 reduced PP-InsP 5 concentrations but did not significantly affect secretion (G.C. Gaboardi, C. Illies, C.J. Barker, P-O. Berggren, unpublished observations). A similar result showing an isoform selective effect with silencing, but not overexpression, of IP6Ks was obtained when studying apoptosis [60], only that here it was the siRNA of IP6K2 that was effective (see also section "Inositol pyrophosphates and apoptosis").
A study in IP6K1 -/- mice showed reduced blood insulin levels in these animals, supporting the importance of inositol pyrophosphate for insulin secretion [57]. Furthermore, a recent study using a specific IP6K inhibitor confirmed that IP6K activity is required for glucose-stimulated insulin secretion in MIN6 insulin secreting cells [122].
In contrast to the clearly defined role of IP6K1 for Rab3A activity leading to exocytosis [51], which is independent of catalytic activity, the mechanism of inositol pyrophosphate action on exocytosis [39, 122] is still unclear. The ability of different inositol pyrophosphate isomers to substitute for each other might hint towards protein pyrophosphorylation [47, 55], but evidence for this is still lacking.
Osmotic stress
One cellular response that offers a distinct role for (PP)2-InsP 4, compared to both PP-InsP 5 isoforms and the Ins(1,3,4,5,6)P 5 pyrophosphates, is that (1/3,5)-(PP)2-InsP 4 dramatically increases in response to hyperosmotic stress [73, 111, 123, 124]. The cloning and sequencing of the enzyme that forms it (VIP/PPIP5K) has confirmed that this previously observed phenomenon [73] results from the direct activation of PPIP5K/VIP. Two important questions arise when considering these studies. Firstly, the specific role the increase of (PP)2-InsP 4 plays in the stress-response, and secondly, the physiological relevance of such a response in a mammal. Is the generation of (PP)2-InsP 4 in the osmotic response simply a phenotypic change evoked by the significant cellular stress, or does it have a more profound signaling role? The fact that substantial energy expenditure is required to form it and the rise in its concentration is not simply an inhibition of its degradation, but an activation of its formation, suggests its generation is not artifactual, but a deliberate strategy of the cell. One could postulate that this may lead either to prevention of the stress itself or to deal with the damaging after effects of such an environmental insult. Both osmotic and heat stress seem to activate the kinase activity. There is an interesting coincidence here as it has long been documented that highly phosphorylated diadenosine polyphosphates are increased under similar stress conditions [79, 125], and both these families of molecules are degraded by the DIPPs (see section "Diphosphoinositol-polyphosphate phosphatases") [81–92]. It is conceivable that increases in (PP)2-InsP 4 could lead to parallel increases in diadenosine polyphosphates by direct competition, or vice versa. How might the production of (PP)2-InsP 4 be involved in the cell stress reaction? Work from the S. pombe version of the kinase (Asp1) indicates its catalytic activity, and hence the production of inositol pyrophosphates is required for phenotypic changes which may involve the actin-related protein (Arp) complexes [21] important in the control of the actin cytoskeleton. Furthermore, Arp2/3 are known to be involved in the cytoskeleton’s response to osmotic stress [126], indicating that these players could integrate (PP)2-InsP 4 production into a protective response for the cell at the level of the cytoskeleton. Clearly, direct evidence linking all these players together in mammalian cells is an important future goal of pyrophosphate research.
Cell studies whilst being important in defining biochemical mechanisms need to be considered alongside the effect on the whole organism. So the second important question is whether the osmotic and temperature stresses are relevant in a mammal and of course in human beings? At first sight, the osmotic stresses required to elevate inositol pyrophosphates may seem to be the most difficult to marry to the experiences of the intact organism. However, it has recently been appreciated that hyperosmotic stress of the magnitude that could influence (PP)2-InsP 4 levels is encountered by mammalian cells in vivo at various stages of normal physiology, including during lymphocyte development and bone formation, as well as several other scenarios [111]. Of course, hyperosmolarity is also associated with pathology, e.g. diabetes in its extreme form leads to the hyperglycemic hyperosmolar state [127]. (PP)2-InsP 4 concentrations are elevated 3–4 fold in response to the comparatively more modest lowered or raised temperatures. In humans, the effect of heat and cold, especially at the level of the dermis or during high fever [128], may be an important arena in which changes in inositol pyrophosphates play a physiological role.
Inositol pyrophosphates and apoptosis
Inositol pyrophosphates influence the process of apoptosis through the IP6K2. Apoptosis is a form of programmed cell death which involves a series of biochemical events leading disposal of cellular debris without resulting in damage to the organism. Apoptosis itself is implicated in a variety of diseases such as cancer and ischemic damage. Two groups have been instrumental in elucidating the role of IP6K2 in apoptosis. Because the types of stimuli, signaling pathways, cell types, and mode of operation of IP6K2 are different in the studies carried out between the two laboratories, we will examine them separately.
In initial studies using an antisense knockout approach, Lindner and co-workers first characterized IP6K2 as regulator of interferon-induced death [129]. Their studies mainly focused on NIH-OVCAR-3 human ovarian carcinoma cells (which lack a functional p53 gene) and the response of these cells to interferon and cytokines. Following IFN-β treatment, both IP6K2 protein level and overall IP6K activity increased without any change in mRNA content. These early findings demonstrated that overexpression of IP6K2 enhances IFN-β-induced apoptosis, while the expression of antisense IP6K2 mRNA led the carcinoma cells to be highly resistant to IFN-β. A catalytic inactive form of IP6K2 also impaired the IFN-β apoptotic effect showing that the kinase dead may act as dominant negative [129]. Further studies suggested that IP6K2 could be also be involved in IFN-α2-mediated cell death, although this pathway is a much less effective apoptotic signal than IFN-β. However, in cells stably transfected with IP6K2, the apoptotic effect of IFN-α2 was enhanced to the same extent of IFN-β. Τhese results have been correlated with the induction of Apo2 ligand/tumor necrosis factor-related apoptosis-inducing ligand (Apo2L/TRAIL). A similar IP6K2 pro-apoptotic effect has been described for γ-radiation-induced cell death. Both IFN-β and γ-radiation treatment lead to increase caspase 8, either mRNA or activity, which becomes even higher after overexpression of IP6K2 [59, 130].
A change in IP6K’s cellular localization has also been observed following IFN-β treatment, showing that endogenous IP6K2 may translocate to the nucleus. However, this movement does not occur with IFN-α2 stimulation, unless the kinase is overexpressed. An NLS mutant of IP6K2, which is unable to enter the nucleus, acts as a dominant negative creating a greater resistance to apoptosis and suggesting that nuclear IP6K2 localization is also required for it to exert its pro-apoptotic effect in interferon-induced death [59].
Snyder and co-workers [60] have extended the role of IP6K2 as mediator of cell death to different types of cell lines in response to different apoptotic agents. They also showed that overexpression of both IP6K1 and IP6K3 was as effective at producing cell death as IP6K2. This underscores that PP-InsP 5 (or other upstream products of the IP6K2) are important mediators of death. On the other hand, only silencing endogenous IP6K2 and not IP6K1 or 3 curtails apoptosis, establishing a physiological role of IP6K2 in cell viability [60].
In contrast to the nuclear translocation described for IP6K2 by the Lindner group, Snyder’s group suggests a re-distribution of IP6K2 after apoptotic stimulus from the nucleus to mitochondria which are positive for Bax [60]. Bax is a known apoptotic protein that translocates to the mitochondria during apoptosis. The different translocation seen by the Lindner and Snyder groups could be explained either by different cell death agents being employed or the different time courses of apoptotic onset considered in the experiments.
Mechanistically, inositol pyrophosphates might act either as high-energy phosphate donors which may occur in protein pyrophosphorylation [47, 55] or through binding to specific target involved in cell survival like PKB, in which PP-InsP 5 might compete with PtdIns (3,4,5)P 3 for its PH domain which is required for the activation of PKB [65] (see section "Regulation of PH domains" for detailed discussions of the controversy surrounding this).
IP6Ks appear to have a duality in function in which involves both catalytic activity and protein–protein interactions (see the role of IP6K1 in exocytosis above). In the case of IP6K2, both cytokine-mediated apoptosis [61] and the more general cytotoxic stresses [60, 62] are mediated by IP6K2 associating with other proteins rather than just by the production of inositol pyrophosphates. Co-immunoprecipitation studies have shown that IP6K2 may associate with TRAF2 which is an adapter protein for the tumor necrosis factor receptor (TNF-R) and interleukin-1 receptor superfamily [131]. Their association increases after IFN-β treatment, and this association is largely blocked by a combined mutation of serine 347 and 359 on IP6K2. Cells stably transfected with IP6K2 mutated in the binding site for TRAF2 are no longer able to enhance apoptosis induced by TNF-α. Binding to TRAF2 has also been shown to attenuate the phosphorylation of the TRAF2 downstream transforming growth factor β-activated kinase 1 (TAK1) and the protein kinase B (PKB) which leads to NF-κB inhibition [61]. Because PP-InsP 5 has been described to compete with PtdIns(3,4,5)P 3 for the PH domain of PKB/Akt [65], IP6K2 might be able to increase local PP-InsP 5 concentration through binding to TRAF2. This high local PP-InsP 5 then may displace PKB from PtdIns(3,4,5)P 3 preventing the recruitment of PKB to the plasma membrane and its activation by PDKs (see section "Regulation of PH domains").
More recently, an association between IP6K2 and HSP90 in the mediation of apoptosis has been described [62]. HSP90 is a chaperone protein that has the ability to control stability, conformation, activation, and distribution of numerous proteins involved in cell growth, differentiation, and survival [132]. Some anticancer drugs are already known to target HSP90 leading to inhibition of its chaperone activity [133, 134]. The interaction between the two proteins has been elucidated through mutating the arginine 133 and 136 of IP6K2 and the C-terminal of HSP90. It has been shown that HSP90 binding decreases IP6K2 catalytic activity. It is proposed that HSP90 masks a region of IP6K2 important for InsP 6 binding, since the IP6K2 binding region for HSP90 seems also to be important for the kinase activity [62]. Pro-apoptotic drugs like the DNA-binding cisplatin that target the C-terminal region of HSP90 inhibit the binding to IP6K2 and increase the IP6K activity. According to these studies, the depletion of HSP90 and overexpression of IP6K2-mutant binding site for HSP90 decrease cell survival, the latter mutant more than the wild-type isoform [62]. Finally, there is evidence that PKB may be stabilized by HSP90 [135]. Therefore, HSP90, along with its ability to inhibit IP6K2 catalytic activity, may lead to a stronger concerted PKB pathway activation, important for cell survival [135] and for integrating both cell death and growth promotion.
There is one further documented protein–protein interaction of IP6K2 which is relevant for a role in apoptosis. In a high-throughput yeast two-hybrid system [54], IP6K2 has been shown to interact with phospholipid scramblase 1 (PLSCR1), which is well known to be involved in destroying plasma membrane phospholipid asymmetry at critical cellular events like cell activation, injury, and apoptosis. Particularly in the latter context, after interferon treatment, PLSCR1 increases and translocates to the nucleus mimicking the observed movement of IP6K2 [59, 136].
In contrast to the pro-apoptotic role of IP6K2, in primary chicken embryo fibroblasts, a correlation between transforming activity of β-catenin mutants and the upregulation of IP6K2 mRNA has been shown [137], although this does not directly prove that IP6K2 drives the oncogenic activity. However, in general, IP6K2 is an important mediator of apoptosis, both by its production of diphosphoinositol polyphosphates and protein–protein interactions with other proteins.
What insights does the newly characterized IP6K2 knockout mouse [63] bring to the role of IP6K2 apoptosis? In practical terms, the knockout mice become more susceptible to chemically-induced aerodigestive tract carcinomas and more resistant to death from ionizing radiation. Both these observations are consistent with the data garnered from cell studies and support a physiological role for IP6K2 in apoptosis.
Telomere length and hyperrecombination in yeast
The regulation of telomere length has attracted a lot of attention, principally due to the link between length and longevity [138]. Two groups initially reported the involvement of inositol pyrophosphates in telomere length regulation in yeast [37, 38]; an equivalent study in mammalian cells is still lacking. Such a study would be attractive as there are mammalian homologues to the yeast players. The overexpression of the yeast IP6K, kcs1, or its deletion, promoted telomere shortening or lengthening, respectively. One of the groups [38] also showed that the catalytic activity and hence the pyrophosphate product was critical. This is important because, as we indicated in the section on kcs1, this enzyme also has functional roles independent of its kinase activity. Kcs1 in common with IP6K1 and IP6K3 is equally happy phosphorylating either InsP 6 or InsP 5, generating two distinct families of inositol pyrophosphates, with or without a phosphate group in the 2-position of the ring, respectively. The elimination of Ipk1, the kinase which converts InsP 5 to InsP 6, and thus preventing InsP 6 pyrophosphate formation, led to increased levels of the InsP 5 pyrophosphate PP-InsP 4 and the concomitant decrease in telomere length [37, 38]. This indicates a distinct role for the InsP 5 pyrophosphates. Unfortunately, no physiological mechanism regulating the concentration of these pyrophosphates has been reported and, as suggested by others, [139] the modulation by gene deletion or overexpression can hardly be considered physiological. It is conceivable that, due to the rapid turnover of the pyrophosphates, changes in cellular energy metabolism might impinge on at least the turnover, if not the mass, of the InsP 5 pyrophosphates. A family of PI3-kinase-related enzymes are suggested to be involved in this process [37], although this may be more contentious [139].
The yeast IP6K is known as Kcs1, and the gene KCS1 (kinase C supressor-1) was identified by the observation that mutations in yeast PKCs led to hyper-recombination of chromosomes [66]. This dramatic increase in homologous recombination was reversed by a mutation in Kcs1 [140]. The stimulation of recombination in yeast is attributed to a number of events: DNA-damage, enhanced transcription, or mutations in cyclins which lead to prolongation of S-phase. The catalytic activity of Kcs1 was deemed essential for the effect [140], but since both PP-InsP 5 and (PP)2-InsP 4 are generated it is difficult to say which pyrophosphate may be responsible, or indeed whether allosteric regulation or pyrophosphoryation is the mode of action. With the advent of VIP1 and its homologues, the PP-InsP 5/VIP kinases [21, 72, 73], it is now possible to examine the relative importance of these pyrophosphates, but these experiments have yet to be carried out.
Phosphate metabolism and regulation of yeast cyclin complexes
The first discovered IP6K, PiUS (IP6K2) linked cellular phosphate metabolism to inositol pyrophosphates, [29–31]. An additional study has established that the intestinal mRNA level of IP6K2/PiUS is increased in parallel to an increase in Na+-dependent inorganic phosphate co-transport activity when rats are fed with low inorganic phosphate diet [141]. Furthermore, kcs1, the yeast IP6K, has also been linked to phosphate metabolism [34]. Nonetheless, a recent study in yeast (cerevisiae) directly ties diphosphoinositol metabolism to phosphate metabolism, and this relates to the IP6K-like activity of Vip1(PPIP5K) [142]. In this study, a cyclin/cyclin-dependent kinase (cdk) and cyclin-dependent kinase inhibitor (cdki) known as Pho 80, Pho 85, and Pho81, respectively, are regulated by the inositol pyrophosphate now understood to be 1/3-PP-InsP 5 [22] (but described as the 4/6-isoform in the original paper) [142]. Under high phosphate conditions, the Pho80–Pho85 complex regulates the transcription factor, Pho4, which is exported from nucleus to cytoplasm. Phosphate limitation leads to the association of the cdk inhibitor, Pho81, which inhibits the complex. 1/3-PP-InsP 5 generated by Vip1 in yeast was found to be a factor which inhibited the kinase activity in a Pho81-dependent manner. Interestingly, only the 1/3-PP-InsP 5 produced by Vip1, and not the 5-PP-InsP 5 produced by Kcs1, was able to exert an inhibitory effect. This suggests a stereospecific role and thus points to a mechanism of allosteric regulation rather than pyrophosphorylation. Further studies confirmed this conclusion in greater detail [143]. One area of contention is the link between the level of phosphate and PP-InsP 5. In the original work, phosphate starvation was linked with increased PP-InsP 5, thus fitting the model. However, another group in an admittedly different yeast strain with slightly different conditions have found that phosphate starvation leads to a decreased rather than increased level of PP-InsP 5 [144].
An important question is whether such interactions have any role in mammalian cell systems. Although, changes in inositol pyrophosphates are associated with cell cycle progression [46], the particular yeast cyclin system, which regulates phosphate homeostasis whilst integrated into cell growth regulation [145], is not part of the classic cyclin/cdk components of the cell cycle machinery. The homologous system in mammalian cells would be Cdk5 and its regulators [146]. This is interesting as Cdk5 has several important roles in mammalian cells [146], not least in the regulation of insulin secretion [147], a process regulated by inositol pyrophosphates in pancreatic β-cells [39, 122]. Therefore, it would be interesting to see whether the novel (1/3)-PP-InsP 5 has a role in the regulation of the mammalian cdks.
Regulation of PH domains
A study on Dictyostelium has opened a potentially exciting new vista on inositol pyrophosphate function: that is, the allosteric regulation of signal transduction pathways by binding of inositol pyrophosphates to plextrin homology (PH) domains [65]. PH domains confer on proteins the ability to associate with other proteins or more commonly phosphorylated lipids like the phosphoinositides [148]. A classic example is the recruitment of PH-domain proteins like Akt/PKB to membranes where they are subsequently activated and carry out important downstream signal transduction events [149]. In mammalian cells, inositol pentakisphosphates, Ins(1,3,4,5,6)P 5 and Ins(1,2,3,5,6)P 5, may complex to PH domains from PKB/Akt and Plextrin, respectively [150, 151]. In Dictyostelium, an organism in which the inositol pyrophosphate concentrations are substantially higher than in mammalian cells, PP-InsP 5 binds to the PH domain of a protein (Crac) involved in cAMP-driven chemotaxis [65]. The pyrophosphate binding to the PH domain disrupts the binding of the PtdIns(3,4,5)P 3, the normal ligand for the domain, and thus prevents the recruitment of the protein and the subsequent chemotactic response. Deletion of the Dictyostelium IP6K leads to both increased aggregation and cAMP sensitivity. In addition, stimulation of Dictyostelium by cAMP leads to substantial elevation of both PP-InsP 5 and (PP)2-InsP 4, indicating a dynamic regulation of Crac by inositol pyrophosphates is possible [65].
An important question is whether such a mechanism exists in mammalian cells? In the published study, Snyder’s group showed that the PH domain from PKB/Akt, as well as other proteins, can also bind PP-InsP 5 [65]. Furthermore, as we have hinted at in the section "Inositol pyrophosphates and apoptosis", such a mechanism may be implied in studies of the interaction of TRAF2 and IP6K2. However, two contrary factors need to be borne in mind. Firstly, the PP-InsP 5 binding to PKB/Akt is disputed by another group [152], and secondly, the relative concentrations of the inositol lipids known to bind the PKB/Akt PH domain versus the cellular PP-InsP 5 concentration require a careful examination. The main biological caveat is that in mammalian cells the concentration of PP-InsP 5 is considerably lower than in Dictyostelium (low vs high micromolar); thus at first sight it is rather unlikely that this low concentration of PP-InsP 5 can compete with the recognized inositol lipid ligands of PKB/Akt [e.g., PtdIns(3,4)P 2/PtdIns(3,4,5)P 3]. Nonetheless, recent work from the Snyder laboratory (Chakraborty and Snyder, personal communication) now establishes a possible link between PKB/Akt and inositol pyrophosphates. They have discovered that the IP6K1 knockout mouse described previously [57] has a generalized upregulation of the downstream effectors of PKB/Akt, suggesting that reduced levels of pyrophosphates lead to a reciprocal increase in PKB/Akt activity. The mouse data are at least consistent with the idea that PP-InsP 5 could bind to PKB/Akt. Furthermore, their unpublished observations suggest that this interference in PKB/Akt signaling might be happening at the level of activation of PKB/Akt by PDKs. Mechanistically, they suggest that 5-PP-InsP 5 very potently inhibits PDK1 activity towards Akt T308 without affecting PDK1 activity in general; that is, 5-PP-InsP 5 does not directly inhibit PDK1 actions on artificial substrates. This is consistent with an earlier report suggesting only a very weak binding of 5-PP-InsP 5 to PDK1 [153]. 5-PP-InsP 5 also efficiently inhibits PtdIns(3,4,5)P 3-dependent activation of Akt T308 phosphorylation in vitro when preincubated with Akt. 5-PP-InsP 5’s inhibitory effect is lost if ΔPH-Akt is used as PDK1 substrate, further indicating that 5-PP-InsP 5’s inhibitory action on PKB/Akt is via its PH-domain. The inhibition of PDK1-dependent phosphorylation occurs with 10–100 nM 5-PP-InsP 5 but not with the 1/3-PP-InsP 5 isomer made by VIP1. This suggests specificity at a concentration that is compatible with a physiological role in mammalian cells. The combination of the two PP-InsP 5 effects, i.e. (1) inhibition of PtdIns(3,4,5)P 3’s stimulatory effect on Akt, and (2) direct inhibition of PDK1 phosphorylation of T308 in the absence of PtdIns(3,4,5)P 3, might lead to a stronger concerted inactivation of the PKB/Akt signaling. Another important finding in the study reveals that PP-InsP 5’s inhibitory effect on PtdIns(3,4,5)P 3-dependent stimulation of Akt phosphorylation is largely abolished when PKB/Akt is pre-exposed to the phospholipid. This observation might clarify the physiological scenario by indicating that PKB/Akt-bound to PP-InsP 5 is no longer available to PtdIns(3,4,5)P 3 at the plasma membrane. On the other hand, PKB/Akt already translocated to plasma membrane is resistant to PP-InsP 5-mediated inactivation. In summary, the authors focused on the inhibition of Akt phosphorylation by PP-InsP 5 both in vitro and under in vivo conditions using IP6K1 knockout mice, data which strongly suggest an impact of PP-InsP 5 on Akt/PKB signaling.
What about the conflicting information that the PKB/Akt PH domain does not bind PP-InsP 5 [152]? One concern is that this latter study was also unable to detect significant binding of Ins(1,3,4,5)P 4 at concentrations up to 100 μM. Ins(1,3,4,5)P 4 has been previously found to bind the PH domain of PKB/Akt [65, 154, 155]. We believe a closer examination of how these binding assays are carried out is helpful. Many of the binding assays rely on displacement of a previously bound PtdIns(3,4,5)P 3,. including the study which reveals no binding of PP-InsP 5 to the PH domain of PKB/Akt [152]. These data are thus actually consistent with the data from the Snyder laboratory as they noted (see previous section) that bound PtdIns(3,4,5)P 3 prevented PP-InsP 5 inhibition. However, as discussed above, this is only one of the possible physiological scenarios, as PP-InsP 5 is likely to be already bound to the PKB/Akt PH domain before it ever sees PtdIns(3,4,5)P 3 at the plasma membrane. So PP-InsP 5 may not be able to displace PtdIns(3,4,5)P 3 once it has bound but rather interferes with the complex choreography between PKB/Akt and PDK before lipid binding. One reason why PtdIns(3,4,5)P 3 binding may prevent further interaction is that the polar head group of PtdIns(3,4,5)P 3 has been shown either to induce a large conformational change in the PH domain of PKB/Akt or to modulate its kinase activity [155, 156].
These data indicate the possibility that the insights gained from the studies in Dictyostelium may have a direct bearing on the role of inositol pyrophosphates in mammalian signal transduction, thus forming an important dynamic cross-talk between the lipid products of PI3K and the water soluble inositol pyrophosphates.
The above exploration of the function of inositol pyrophosphates in cellular regulation highlights two important dimensions. Firstly, the fact that both the pyrophosphates themselves, and the kinases that make them, have important roles, and secondly, that both pyrophosphorylation and non-covalent binding are mechanisms by which the pyrophosphates themselves exert their influence. The generation of detailed mechanistic information relating to pyrophosphate-mediated allosteric regulation and the functional outcome of the pyrophosphorylation process are immediate and obvious next steps in the development of this exciting field.
Acknowledgments
We thank S.H. Snyder and A. Chakraborty for providing unpublished work and B.M. Hallberg, for structural insights. The 3-D figure was constructed in trueSpace 7.6, Caligari Inc. This work was supported by grants from Karolinska Institutet, Novo Nordisk Foundation, the Swedish Research Council, the Swedish Diabetes Association, EFSD, The Family Erling-Persson Foundation, Berth von Kantzow’s Foundation and EuroDia (LSHM-CT-2006-518153).
References
- 1.Berridge MJ. Inositol trisphosphate and calcium oscillations. Biochem Soc Symp. 2007;7:1–7. doi: 10.1042/BSS0740001. [DOI] [PubMed] [Google Scholar]
- 2.Michell RH. Inositol derivatives: evolution and functions. Nat Rev Mol Cell Biol. 2008;9:151–161. doi: 10.1038/nrm2334. [DOI] [PubMed] [Google Scholar]
- 3.Irvine RF. Inositide evolution—towards turtle domination? J Physiol. 2005;566:295–300. doi: 10.1113/jphysiol.2005.087387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeds AM, Frederick JP, Tsui MM, York JD. Roles for inositol polyphosphate kinases in the regulation of nuclear processes and developmental biology. Adv Enzym Regul. 2007;47:10–25. doi: 10.1016/j.advenzreg.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shears SB. How versatile are inositol phosphate kinases? Biochem J. 2004;377:265–280. doi: 10.1042/BJ20031428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcázar-Román AR, Wente SR. Inositol polyphosphates: a new frontier for regulating gene expression. Chromosoma. 2008;117:1–13. doi: 10.1007/s00412-007-0126-4. [DOI] [PubMed] [Google Scholar]
- 7.Duronio V. The life of a cell: apoptosis regulation by the PI3K/PKB pathway. Biochem J. 2008;415:333–344. doi: 10.1042/BJ20081056. [DOI] [PubMed] [Google Scholar]
- 8.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vicinanza M, D’Angelo G, Di Campli A, De Matteis MA. Phosphoinositides as regulators of membrane trafficking in health and disease. Cell Mol Life Sci. 2008;65:2833–2841. doi: 10.1007/s00018-008-8353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Europe-Finner GN, Ludérus ME, Small NV, Van Driel R, Reymond CD, Firtel RA, Newell PC. Mutant ras gene induces elevated levels of inositol tris- and hexakisphosphates in Dictyostelium . J Cell Sci. 1988;89:13–20. doi: 10.1242/jcs.89.1.13. [DOI] [PubMed] [Google Scholar]
- 11.Europe-Finner GN, Gammon B, Newell PC. Accumulation of [3H]-inositol into inositol polyphosphates during development of Dictyostelium . Biochem Biophys Res Commun. 1991;181:191–196. doi: 10.1016/s0006-291x(05)81400-7. [DOI] [PubMed] [Google Scholar]
- 12.Oliver KG, Putney JW, Jr, Obie JF, Shears SB. The interconversion of inositol 1, 3, 4, 5, 6-pentakisphosphate and inositol tetrakisphosphates in AR4–2J cells. J Biol Chem. 1992;267:21528–21534. [PubMed] [Google Scholar]
- 13.Wong NS, Barker CJ, Morris AJ, Craxton A, Kirk CJ, Michell RH. The inositol phosphates in WRK1 rat mammary tumour cells. Biochem J. 1992;286:459–468. doi: 10.1042/bj2860459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayr GW, Radenberg T, Thiel U, Vogel G, Stephens LR. Phosphoinositol diphosphates: non-enzymic formation in vitro and occurrence in vivo in the cellular slime mold Dictyostelium . Carbohydr Res. 1992;234:247–262. [Google Scholar]
- 15.Stephens L, Radenberg T, Thiel U, Vogel G, Khoo KH, Dell A, Jackson TR, Hawkins PT, Mayr GW. The detection, purification, structural characterization, and metabolism of diphosphoinositol pentakisphosphate(s) and bisdiphosphoinositol tetrakisphosphate(s) J Biol Chem. 1993;268:4009–4015. [PubMed] [Google Scholar]
- 16.Menniti FS, Miller RN, Putney JW, Shears SB. Turnover of inositol polyphosphate pyrophosphates in pancreatoma cells. J Biol Chem. 1993;268:3850–3856. [PubMed] [Google Scholar]
- 17.Voglmaier SM, Bembenek ME, Kaplin AI, Dormán G, Olszewski JD, Prestwich GD, Snyder SH. Purified inositol hexakisphosphate kinase is an atp synthase: diphosphoinositol pentakisphosphate as a high-energy phosphate donor. Proc Natl Acad Sci USA. 1996;93:4305–4310. doi: 10.1073/pnas.93.9.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nomenclature Committee of the International Union of Biochemistry Numbering of atoms in myo-inositol recommendations. 1988: nomenclature committee of the international union of biochemistry. Biochem J. 1988;258:1–2. [PMC free article] [PubMed] [Google Scholar]
- 19.Agranoff BW. Cyclitol confusion. Trends Biochem Sci. 1978;3:N283–N285. [Google Scholar]
- 20.Albert C, Safrany ST, Bembenek ME, Reddy KM, Reddy K, Falck J, Bröcker M, Shears SB, Mayr GW. Biological variability in the structures of diphosphoinositol polyphosphates in Dictyostelium discoideum and mammalian cells. Biochem J. 1997;327:553–560. doi: 10.1042/bj3270553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulugu S, Bai W, Fridy PC, Bastidas RJ, Otto JC, Dollins DE, Haystead TA, Ribeiro AA, York JD. A conserved family of enzymes that phosphorylate inositol hexakisphosphate. Science. 2007;316:106–109. doi: 10.1126/science.1139099. [DOI] [PubMed] [Google Scholar]
- 22.Lin H, Fridy PC, Ribeiro AA, Choi JH, Barma DK, Vogel G, Falck JR, Shears SB, York JD, Mayr GW. Structural analysis and detection of biological inositol pyrophosphates reveals that the vip/ppip5k family are 1/3-kinases. J Biol Chem. 2008;284:1863–1872. doi: 10.1074/jbc.M805686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Draskovic P, Saiardi A, Bhandari R, Burton A, Ilc G, Kovacevic M, Snyder SH, Podobnik M. Inositol hexakisphosphate kinase products contain diphosphate and triphosphate groups. Chem Biol. 2008;15:274–286. doi: 10.1016/j.chembiol.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Dubois E, Scherens B, Vierendeels F, Ho MMW, Messenguy F, Shears SB. In Saccharomyces cerevisiae, the inositol polyphosphate kinase activity of kcs1p is required for resistance to salt stress, cell wall integrity, and vacuolar morphogenesis. J Biol Chem. 2002;277:23755–23763. doi: 10.1074/jbc.M202206200. [DOI] [PubMed] [Google Scholar]
- 25.Seeds AM, Bastidas RJ, York JD. Molecular definition of a novel inositol polyphosphate metabolic pathway initiated by inositol 1, 4, 5-trisphosphate 3-kinase activity in Saccharomyces cerevisiae . J Biol Chem. 2005;280:27654–27661. doi: 10.1074/jbc.M505089200. [DOI] [PubMed] [Google Scholar]
- 26.Laussmann T, Reddy KM, Reddy KK, Falck JR, Vogel G. Diphospho-myo-inositol phosphates from Dictyostelium identified as d-6-diphospho-myo-inositol pentakis-phosphate and d-5, 6-bisdiphospho-myo-inositol tetrakisphosphate. Biochem J. 1997;322:31–33. doi: 10.1042/bj3220031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laussmann T, Hansen A, Reddy KM, Reddy KK, Falck JR, Vogel G. Diphospho-myo-inositol phosphates in Dictyostelium and Polysphondylium: identification of a new bisdiphospho-myo-inositol tetrakisphosphate. FEBS Lett. 1998;426:145–150. doi: 10.1016/s0014-5793(98)00329-9. [DOI] [PubMed] [Google Scholar]
- 28.Martin JB, Laussmann T, Bakker-Grunwald T, Vogel G, Klein G. Neo-inositol polyphosphates in the amoeba Entamoeba histolytica . J Biol Chem. 2000;275:10134–10140. doi: 10.1074/jbc.275.14.10134. [DOI] [PubMed] [Google Scholar]
- 29.Norbis F, Boll M, Stange G, Markovich D, Verrey F, Biber J, Murer H. Identification of a cDNA/protein leading to an increased Pi-uptake in Xenopus laevis oocytes. J Membr Biol. 1997;156:19–24. doi: 10.1007/s002329900183. [DOI] [PubMed] [Google Scholar]
- 30.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 31.Schell MJ, Letcher AJ, Brearley CA, Biber J, Murer H, Irvine RF. PiUS (Pi uptake stimulator) is an inositol hexakisphosphate kinase. FEBS Lett. 1999;461:169–172. doi: 10.1016/s0014-5793(99)01462-3. [DOI] [PubMed] [Google Scholar]
- 32.Saiardi A, Nagata E, Luo HR, Snowman AM, Snyder SH. Identification and characterization of a novel inositol hexakisphosphate kinase. J Biol Chem. 2001;276:39179–39185. doi: 10.1074/jbc.M106842200. [DOI] [PubMed] [Google Scholar]
- 33.Bennett M, Onnebo SM, Azevedo C, Saiardi A. Inositol pyrophosphates: metabolism and signaling. Cell Mol Life Sci. 2006;63:552–564. doi: 10.1007/s00018-005-5446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shears SB. Understanding the biological significance of diphosphoinositol polyphosphates (‘inositol pyrophosphates’) Biochem Soc Symp. 2007;7:211–221. doi: 10.1042/BSS0740211. [DOI] [PubMed] [Google Scholar]
- 35.Saiardi A, Nagata E, Luo HR, Sawa A, Luo X, Snowman AM, Snyder SH. Mammalian inositol polyphosphate multikinase synthesizes inositol 1, 4, 5-trisphosphate and an inositol pyrophosphate. Proc Natl Acad Sci USA. 2001;98:2306–2311. doi: 10.1073/pnas.041614598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T, Caffrey JJ, Shears SB. The transcriptional regulator, Arg82, is a hybrid kinase with both monophosphoinositol- and diphosphoinositol-polyphosphate synthase activity. FEBS Lett. 2001;494:208–212. doi: 10.1016/s0014-5793(01)02351-1. [DOI] [PubMed] [Google Scholar]
- 37.Saiardi A, Resnick AC, Snowman AM, Wendland B, Snyder SH. Inositol pyrophosphates regulate cell death and telomere length through phosphoinositide 3-kinase-related protein kinases. Proc Natl Acad Sci USA. 2005;102:1911–1914. doi: 10.1073/pnas.0409322102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- 39.Illies C, Gromada J, Fiume R, Leibiger B, Yu J, Juhl K, Yang S-N, Barma DK, Falck JR, Saiardi A, Barker CJ, Berggren P-O. Requirement of inositol pyrophosphates for full exocytotic capacity in pancreatic beta cells. Science. 2007;318:1299–1302. doi: 10.1126/science.1146824. [DOI] [PubMed] [Google Scholar]
- 40.Watahiki A, Waki K, Hayatsu N, Shiraki T, Kondo S, Nakamura M, Sasaki D, Arakawa T, Kawai J, Harbers M, Hayashizaki Y, Carninci P. Libraries enriched for alternatively spliced exons reveal splicing patterns in melanocytes and melanomas. Nat Methods. 2004;1:233–239. doi: 10.1038/nmeth719. [DOI] [PubMed] [Google Scholar]
- 41.Maruyama Y, Wakamatsu A, Kawamura Y, Kimura K, Yamamoto J, Nishikawa T, Kisu Y, Sugano S, Goshima N, Isogai T, Nomura N. Human gene and protein database (HGPD): a novel database presenting a large quantity of experiment-based results in human proteomics. Nucl Acids Res. 2009;37:D762–D766. doi: 10.1093/nar/gkn872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scamuffa N, Calvo F, Chrétien M, Seidah NG, Khatib AM. Proprotein convertases: lessons from knockouts. FASEB J. 2006;20:1954–1963. doi: 10.1096/fj.05-5491rev. [DOI] [PubMed] [Google Scholar]
- 43.Tomita T. Immunocytochemical localization of prohormone convertase 1/3 and 2 in pancreatic islet cells and islet cell tumors. Pancreas. 2001;23:172–176. doi: 10.1097/00006676-200108000-00007. [DOI] [PubMed] [Google Scholar]
- 44.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J Biol Chem. 2002;277:2381–2384. doi: 10.1074/jbc.C100228200. [DOI] [PubMed] [Google Scholar]
- 46.Barker CJ, Wright J, Hughes PJ, Kirk CJ, Michell RH. Complex changes in cellular inositol phosphate complement accompany transit through the cell cycle. Biochem J. 2004;380:465–473. doi: 10.1042/BJ20031872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhandari R, Saiardi A, Ahmadibeni Y, Snowman AM, Resnick AC, Kristiansen TZ, Molina H, Pandey A, Werner JK, Juluri KR, Xu Y, Prestwich GD, Parang K, Snyder SH. Protein pyrophosphorylation by inositol pyrophosphates is a posttranslational event. Proc Natl Acad Sci USA. 2007;104:15305–15310. doi: 10.1073/pnas.0707338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saiardi A, Caffrey JJ, Snyder SH, Shears SB. The inositol hexakisphosphate kinase family: catalytic flexibility and function in yeast vacuole biogenesis. J Biol Chem. 2000;275:24686–24692. doi: 10.1074/jbc.M002750200. [DOI] [PubMed] [Google Scholar]
- 49.Larsson O, Barker CJ, Sjöholm A, Carlqvist H, Michell RH, Bertorello A, Nilsson T, Honkanen RE, Mayr GW, Zwiller J, Berggren PO. Inhibition of phosphatases and increased Ca2+ channel activity by inositol hexakisphosphate. Science. 1997;278:471–474. doi: 10.1126/science.278.5337.471. [DOI] [PubMed] [Google Scholar]
- 50.Michell RH, King CE, Piper CJ, Stephens LR, Bunce CM, Guy GR, Brown G. Inositol lipids and phosphates in erythrocytes and HL60 cells. Soc Gen Physiol Ser. 1988;43:345–355. [PubMed] [Google Scholar]
- 51.Luo HR, Saiardi A, Nagata E, Ye K, Yu H, Jung TS, Luo X, Jain S, Sawa A, Snyder SH. Grab: a physiologic guanine nucleotide exchange factor for rab3a, which interacts with inositol hexakisphosphate kinase. Neuron. 2001;31:439–451. doi: 10.1016/s0896-6273(01)00384-1. [DOI] [PubMed] [Google Scholar]
- 52.Tickenbrock L, Cramer J, Vetter IR, Muller O. The coiled coil region (amino acids 129–250) of the tumor suppressor protein adenomatous polyposis coli (APC): its structure and its interaction with chromosome maintenance region 1 (Crm-1) J Biol Chem. 2002;277:32332–32338. doi: 10.1074/jbc.M203990200. [DOI] [PubMed] [Google Scholar]
- 53.Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 55.Saiardi A, Bhandari R, Resnick AC, Snowman AM, Snyder SH. Phosphorylation of proteins by inositol pyrophosphates. Science. 2004;306:2101–2105. doi: 10.1126/science.1103344. [DOI] [PubMed] [Google Scholar]
- 56.Lexicon knockout mouse NIH-0750, mouse genome database (MGD), mouse genome informatics Web Site, http://www.informatics.jax.org/external/ko/lexicon/1223.html (18 July 2006)
- 57.Bhandari R, Juluri KR, Resnick AC, Snyder SH. Gene deletion of inositol hexakisphosphate kinase 1 reveals inositol pyrophosphate regulation of insulin secretion, growth, and spermiogenesis. Proc Natl Acad Sci USA. 2008;105:2349–2353. doi: 10.1073/pnas.0712227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Azevedo C, Burton A, Bennett M, Onnebo SMN, Saiardi A (2010) Synthesis of InsP7 by the inositol hexakisphosphate kinase 1 (IP6K1) In: Barker CJ (ed) Methods in molecular biology. Preliminary entry 2203, 2010, ISBN 978-1-60327-174-5, Hardcover, Due: July 2010 (in press) [DOI] [PubMed]
- 59.Morrison BH, Tang Z, Jacobs BS, Bauer JA, Lindner DJ. Apo2L/TRAIL induction and nuclear translocation of inositol hexakisphosphate kinase 2 during IFN-beta-induced apoptosis in ovarian carcinoma. Biochem J. 2005;385:595–603. doi: 10.1042/BJ20040971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagata E, Luo HR, Saiardi A, Bae BI, Suzuki N, Snyder SH. Inositol hexakisphosphate kinase-2, a physiologic mediator of cell death. J Biol Chem. 2005;280:1634–1640. doi: 10.1074/jbc.M409416200. [DOI] [PubMed] [Google Scholar]
- 61.Morrison BH, Bauer JA, Lupica JA, Tang Z, Schmidt H, DiDonato JA, Lindner DJ. Effect of inositol hexakisphosphate kinase 2 on transforming growth factor beta-activated kinase 1 and NF-kappaB activation. J Biol Chem. 2007;282:15349–15356. doi: 10.1074/jbc.M700156200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakraborty A, Koldobskiy MA, Sixt KM, Juluri KR, Mustafa AK, Snowman AM, van Rossum DB, Patterson RL, Snyder SH. Hsp90 regulates cell survival via inositol hexakisphosphate kinase-2. Proc Natl Acad Sci USA. 2008;105:1134–1139. doi: 10.1073/pnas.0711168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morrison BH, Haney R, Lamarre E, Drazba J, Prestwich GD, Lindner DJ. Gene deletion of inositol hexakisphosphate kinase 2 predisposes to aerodigestive tract carcinoma. Oncogene. 2009;28:2383–2392. doi: 10.1038/onc.2009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xuhua, X (2007) Bioinformatics and the cell : modern computational approaches in genomics, proteomics and transcriptomics. Springer, New York
- 65.Luo HR, Huang YE, Chen JC, Saiardi A, Iijima M, Ye K, Huang Y, Nagata E, Devreotes P, Snyder SH. Inositol pyrophosphates mediate chemotaxis in Dictyostelium via pleckstrin homology domain-PtdIns(3, 4, 5)P3 interactions. Cell. 2003;114:559–572. doi: 10.1016/s0092-8674(03)00640-8. [DOI] [PubMed] [Google Scholar]
- 66.Huang KN, Symington LS. Suppressors of a Saccharomyces cerevisiae pkc1 mutation identify alleles of the phosphatase gene PTC1 and of a novel gene encoding a putative basic leucine zipper protein. Genetics. 1995;141:1275–1285. doi: 10.1093/genetics/141.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Uetz P, Glot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emill A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae . Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 68.Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16:162–172. doi: 10.1016/j.semcancer.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 69.York SJ, Armbruster BN, Greenwell P, Petes TD, York JD. Inositol diphosphate signaling regulates telomere length. J Biol Chem. 2005;280:4264–4269. doi: 10.1074/jbc.M412070200. [DOI] [PubMed] [Google Scholar]
- 70.Shears SB, Ali N, Craxton A, Bembenek ME. Synthesis and metabolism of bis-diphosphoinositol tetrakisphosphate in vitro and in vivo. J Biol Chem. 1995;270:10489–10497. doi: 10.1074/jbc.270.18.10489. [DOI] [PubMed] [Google Scholar]
- 71.Huang CF, Voglmaier SM, Bembenek ME, Saiardi A, Snyder SH. Identification and purification of diphosphoinositol pentakisphosphate kinase, which synthesizes the inositol pyrophosphate bis(diphospho)inositol tetrakisphosphate. Biochemistry. 1998;37:14998–15004. doi: 10.1021/bi981920l. [DOI] [PubMed] [Google Scholar]
- 72.Fridy PC, Otto JC, Dollins DE, York JD. Cloning and characterization of two human VIP1-like inositol hexakisphosphate and diphosphoinositol pentakisphosphate kinases. J Biol Chem. 2007;282:30754–30762. doi: 10.1074/jbc.M704656200. [DOI] [PubMed] [Google Scholar]
- 73.Choi JH, Williams J, Cho J, Falck JR, Shears SB. Purification, sequencing, and molecular identification of a mammalian PP-InsP5 kinase that is activated when cells are exposed to hyperosmotic stress. J Biol Chem. 2007;282:30763–30775. doi: 10.1074/jbc.M704655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grishin NV. Phosphatidylinositol phosphate kinase: a link between protein kinase and glutathione synthase folds. J Mol Biol. 1999;291:239–247. doi: 10.1006/jmbi.1999.2973. [DOI] [PubMed] [Google Scholar]
- 75.Craxton A, Caffrey JJ, Burkhart W, Safrany ST, Shears SB. Molecular cloning and expression of a rat hepatic multiple inositol polyphosphate phosphatase. Biochem J. 1997;328:75–81. doi: 10.1042/bj3280075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 77.Cantin GT, Yi W, Lu B, Park SK, Xu T, Lee JD, Yates JR., III Combining protein-based IMAC, peptide-based IMAC, and MudPIT for efficient phosphoproteomic analysis. J Proteome Res. 2008;7:1346–1351. doi: 10.1021/pr0705441. [DOI] [PubMed] [Google Scholar]
- 78.Safrany ST, Caffrey JJ, Yang X, Bembenek ME, Moyer MB, Burkhart WA, Shears SB. A novel context for the ‘mutt’ module, a guardian of cell integrity, in a diphosphoinositol polyphosphate phosphohydrolase. EMBO J. 1998;17:6599–6607. doi: 10.1093/emboj/17.22.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McLennan AG. Dinucleoside polyphosphates-friend or foe? Pharmacol Ther. 2000;87:73–89. doi: 10.1016/s0163-7258(00)00041-3. [DOI] [PubMed] [Google Scholar]
- 80.Bessman MJ, Frick DN, O’Handley SF. The mutt proteins or “nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 81.Yang X, Safrany ST, Shears SB. Site-directed mutagenesis of diphosphoinositol polyphosphate phosphohydrolase, a dual specificity nudt enzyme that attacks diadenosine polyphosphates and diphosphoinositol polyphosphates. J Biol Chem. 1999;274:35434–35440. doi: 10.1074/jbc.274.50.35434. [DOI] [PubMed] [Google Scholar]
- 82.Caffrey JJ, Shears SB. Genetic rationale for microheterogeneity of human diphosphoinositol polyphosphate phosphohydrolase type 2. Gene. 2001;269:53–60. doi: 10.1016/s0378-1119(01)00446-2. [DOI] [PubMed] [Google Scholar]
- 83.Leslie NR, McLennan AG, Safrany ST. Cloning and characterisation of haps1 and haps2, human diadenosine polyphosphate-metabolising nudix hydrolases. BMC Biochem. 2002;3:20. doi: 10.1186/1471-2091-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hidaka K, Caffrey JJ, Hua L, Zhang T, Falck JR, Nickel GC, Carrel L, Barnes LD, Shears SB. An adjacent pair of human nudt genes on chromosome x are preferentially expressed in testis and encode two new isoforms of diphosphoinositol polyphosphate phosphohydrolase. J Biol Chem. 2002;277:32730–32738. doi: 10.1074/jbc.M205476200. [DOI] [PubMed] [Google Scholar]
- 85.Hua LV, Hidaka K, Pesesse X, Barnes LD, Shears SB. Paralogous murine nudt10 and nudt11 genes have differential expression patterns but encode identical proteins that are physiologically competent diphosphoinositol polyphosphate phosphohydrolases. Biochem J. 2003;373:81–89. doi: 10.1042/BJ20030142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chu C, Alapat D, Wen X, Timo K, Burstein D, Lisanti M, Shears S, Kohtz DS. Ectopic expression of murine diphosphoinositol polyphosphate phosphohydrolase 1 attenuates signaling through the erk1/2 pathway. Cell Signal. 2004;16:1045–1059. doi: 10.1016/j.cellsig.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 87.Caffrey JJ, Safrany ST, Yang X, Shears SB. Discovery of molecular and catalytic diversity among human diphosphoinositol-polyphosphate phosphohydrolases an expanding nudt family. J Biol Chem. 2000;275:12730–12736. doi: 10.1074/jbc.275.17.12730. [DOI] [PubMed] [Google Scholar]
- 88.Hua LV, Green M, Warsh JJ, Li PP. Molecular cloning of a novel isoform of diphosphoinositol polyphosphate phosphohydrolase: a potential target of lithium therapy. Neuropsychopharmacology. 2001;24:640–651. doi: 10.1016/S0893-133X(00)00233-5. [DOI] [PubMed] [Google Scholar]
- 89.Cartwright JL, McLennan AG. The Saccharomyces cerevisiae yor163w gene encodes a diadenosine 5′, 5″‘-p1, p6-hexaphosphate (ap6a) hydrolase member of the mutt motif (nudix hydrolase) family. J Biol Chem. 1999;274:8604–8610. doi: 10.1074/jbc.274.13.8604. [DOI] [PubMed] [Google Scholar]
- 90.Ingram SW, Stratemann SA, Barnes LD. Schizosaccharomyces pombe aps1, a diadenosine 5′, 5′ “-p1, p6-hexaphosphate hydrolase that is a member of the nudix (mutt) family of hydrolases: cloning of the gene and characterization of the purified enzyme. Biochemistry. 1999;38:3649–3655. doi: 10.1021/bi982951j. [DOI] [PubMed] [Google Scholar]
- 91.Safrany ST, Ingram SW, Cartwright JL, Falck JR, McLennan AG, Barnes LD, Shears SB. The diadenosine hexaphosphate hydrolases from schizosaccharomyces pombe and Saccharomyces cerevisiae are homologues of the human diphosphoinositol polyphosphate phosphohydrolase: overlapping substrate specificities in a mutt-type protein. J Biol Chem. 1999;274:21735–21740. doi: 10.1074/jbc.274.31.21735. [DOI] [PubMed] [Google Scholar]
- 92.Fisher DI, Safrany ST, Strike P, McLennan AG, Cartwright JL. Nudix hydrolases that degrade dinucleoside and diphosphoinositol polyphosphates also have 5-phosphoribosyl 1-pyrophosphate (prpp) pyrophosphatase activity that generates the glycolytic activator ribose 1, 5-bisphosphate. J Biol Chem. 2002;277:47313–47317. doi: 10.1074/jbc.M209795200. [DOI] [PubMed] [Google Scholar]
- 93.Fersht A. Structure and mechanism in protein science. New York: Freeman; 1998. [Google Scholar]
- 94.Ash DE, Schramm VL. Determination of free and bound manganese(ii) in hepatocytes from fed and fasted rats. J Biol Chem. 1982;257:9261–9264. [PubMed] [Google Scholar]
- 95.Ali N, Craxton A, Shears SB. Hepatic ins(1, 3, 4, 5)p4 3-phosphatase is compartmentalized inside endoplasmic reticulum. J Biol Chem. 1993;268:6161–6167. [PubMed] [Google Scholar]
- 96.Glennon MC, Shears SB. Turnover of inositol pentakisphosphates, inositol hexakisphosphate and diphosphoinositol polyphosphates in primary cultured hepatocytes. Biochem J. 1993;293(Pt 2):583–590. doi: 10.1042/bj2930583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harwood AJ. Lithium and bipolar mood disorder: the inositol-depletion hypothesis revisited. Mol Psychiatry. 2005;10:117–126. doi: 10.1038/sj.mp.4001618. [DOI] [PubMed] [Google Scholar]
- 98.Cartwright JL, Safrany ST, Dixon LK, Darzynkiewicz E, Stepinski J, Burke R, McLennan AG. The g5r (d250) gene of african swine fever virus encodes a nudix hydrolase that preferentially degrades diphosphoinositol polyphosphates. J Virol. 2002;76:1415–1421. doi: 10.1128/JVI.76.3.1415-1421.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ingram SW, Safrany ST, Barnes LD. Disruption and overexpression of the schizosaccharomyces pombe aps1 gene, and effects on growth rate, morphology and intracellular diadenosine 5′, 5″‘-p1, p5-pentaphosphate and diphosphoinositol polyphosphate concentrations. Biochem J. 2003;369:519–528. doi: 10.1042/BJ20020733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pinna LA. Casein kinase 2: an ‘eminence grise’ in cellular regulation? Biochim Biophys Acta. 1990;1054:267–284. doi: 10.1016/0167-4889(90)90098-x. [DOI] [PubMed] [Google Scholar]
- 101.Caizergues-Ferrer M, Belenguer P, Lapeyre B, Amalric F, Wallace MO, Olson MO. Phosphorylation of nucleolin by a nucleolar type nii protein kinase. Biochemistry. 1987;26:7876–7883. doi: 10.1021/bi00398a051. [DOI] [PubMed] [Google Scholar]
- 102.Meier UT. Comparison of the rat nucleolar protein nopp140 with its yeast homolog srp40: differential phosphorylation in vertebrates and yeast. J Biol Chem. 1996;271:19376–19384. [PubMed] [Google Scholar]
- 103.Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell. 1992;70:127–138. doi: 10.1016/0092-8674(92)90539-o. [DOI] [PubMed] [Google Scholar]
- 104.Jones NC, Farlie PG, Minichiello J, Newgreen DF. Detection of an appropriate kinase activity in branchial arches i and ii that coincides with peak expression of the treacher collins syndrome gene product, treacle. Hum Mol Genet. 1999;8:2239–2245. doi: 10.1093/hmg/8.12.2239. [DOI] [PubMed] [Google Scholar]
- 105.Kim Y-K, Lee KJ, Jeon H, Yu YG. Protein kinase ck2 is inhibited by human nucleolar phosphoprotein p140 in an inositol hexakisphosphate-dependent manner. J Biol Chem. 2006;281:36752–36757. doi: 10.1074/jbc.M604785200. [DOI] [PubMed] [Google Scholar]
- 106.Lee W-K, Lee S-Y, Kim W-I, Rho Y-H, Bae Y-S, Lee C, Kim IY, Yu YG. Characterization of the insp6-dependent interaction between ck2 and nopp140. Biochem Biophys Res Commun. 2008;376:439–444. doi: 10.1016/j.bbrc.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 107.Valdez BC, Henning D, So RB, Dixon J, Dixon MJ. The treacher collins syndrome (tcof1) gene product is involved in ribosomal dna gene transcription by interacting with upstream binding factor. Proc Natl Acad Sci USA. 2004;101:10709–10714. doi: 10.1073/pnas.0402492101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ginisty H, Serin G, Ghisolfi-Nieto L, Roger B, Libante V, Amalric F, Bouvet P. Interaction of nucleolin with an evolutionarily conserved pre-ribosomal rna sequence is required for the assembly of the primary processing complex. J Biol Chem. 2000;275:18845–18850. doi: 10.1074/jbc.M002350200. [DOI] [PubMed] [Google Scholar]
- 109.Lee WC, Zabetakis D, Mélèse T. Nsr1 is required for pre-rrna processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992;12:3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Horigome C, Ikeda R, Okada T, Takenami K, Mizuta K. Genetic interaction between ribosome biogenesis and inositol polyphosphate metabolism in Saccharomyces cerevisiae . Biosci Biotechnol Biochem. 2009;73:443–446. doi: 10.1271/bbb.80599. [DOI] [PubMed] [Google Scholar]
- 111.Yang L, Reece JM, Cho J, Bortner CD, Shears SB. The nucleolus exhibits an osmotically regulated gatekeeping activity that controls the spatial dynamics and functions of nucleolin. J Biol Chem. 2008;283:11823–11831. doi: 10.1074/jbc.M800308200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Beck KA, Keen JH. Interaction of phosphoinositide cycle intermediates with the plasma membrane-associated clathrin assembly protein ap-2. J Biol Chem. 1991;266:4442–4447. [PubMed] [Google Scholar]
- 113.Voglmaier SM, Keen JH, Murphy JE, Ferris CD, Prestwich GD, Snyder SH, Theibert AB. Inositol hexakisphosphate receptor identified as the clathrin assembly protein ap-2. Biochem Biophys Res Commun. 1992;187:158–163. doi: 10.1016/s0006-291x(05)81473-1. [DOI] [PubMed] [Google Scholar]
- 114.Fleischer B, Xie J, Mayrleitner M, Shears SB, Palmer DJ, Fleischer S. Golgi coatomer binds, and forms k(+)-selective channels gated by, inositol polyphosphates. J Biol Chem. 1994;269:17826–17832. [PubMed] [Google Scholar]
- 115.Ali N, Duden R, Bembenek ME, Shears SB. The interaction of coatomer with inositol polyphosphates is conserved in Saccharomyces cerevisiae . Biochem J. 1995;310(Pt 1):279–284. doi: 10.1042/bj3100279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Norris FA, Ungewickell E, Majerus PW. Inositol hexakisphosphate binds to clathrin assembly protein 3 (ap-3/ap180) and inhibits clathrin cage assembly in vitro. J Biol Chem. 1995;270:214–217. doi: 10.1074/jbc.270.1.214. [DOI] [PubMed] [Google Scholar]
- 117.Ye W, Ali N, Bembenek ME, Shears SB, Lafer EM. Inhibition of clathrin assembly by high affinity binding of specific inositol polyphosphates to the synapse-specific clathrin assembly protein ap-3. J Biol Chem. 1995;270:1564–1568. [PubMed] [Google Scholar]
- 118.Saiardi A, Sciambi C, McCaffery JM, Wendland B, Snyder SH. Inositol pyrophosphates regulate endocytic trafficking. Proc Natl Acad Sci USA. 2002;99:14206–14211. doi: 10.1073/pnas.212527899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gaidarov I, Chen Q, Falck JR, Reddy KK, Keen JH. A functional phosphatidylinositol 3,4,5-trisphosphate/phosphoinositide binding domain in the clathrin adaptor ap-2 alpha subunit: implications for the endocytic pathway. J Biol Chem. 1996;271:20922–20929. doi: 10.1074/jbc.271.34.20922. [DOI] [PubMed] [Google Scholar]
- 120.Hao W, Tan Z, Prasad K, Reddy KK, Chen J, Prestwich GD, Falck JR, Shears SB, Lafer EM. Regulation of ap-3 function by inositides: identification of phosphatidylinositol 3,4,5-trisphosphate as a potent ligand. J Biol Chem. 1997;272:6393–6398. doi: 10.1074/jbc.272.10.6393. [DOI] [PubMed] [Google Scholar]
- 121.Chaudhary A, Gu QM, Thum O, Profit AA, Qi Y, Jeyakumar L, Fleischer S, Prestwich GD. Specific interaction of golgi coatomer protein alpha-cop with phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:8344–8350. doi: 10.1074/jbc.273.14.8344. [DOI] [PubMed] [Google Scholar]
- 122.Padmanabhan U, Dollins DE, Fridy PC, York JD, Downes CP. Characterization of a selective inhibitor of inositol hexakisphosphate kinases: use in defining biological roles and metabolic relationships of inositol pyrophosphates. J Biol Chem. 2009;284:10571–10582. doi: 10.1074/jbc.M900752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pesesse X, Choi K, Zhang T, Shears SB. Signaling by higher inositol polyphosphates: synthesis of bisdiphosphoinositol tetrakisphosphate (“InsP8”) is selectively activated by hyperosmotic stress. J Biol Chem. 2004;279:43378–43381. doi: 10.1074/jbc.C400286200. [DOI] [PubMed] [Google Scholar]
- 124.Choi K, Mollapour E, Shears SB. Signal transduction during environmental stress: InsP8 operates within highly restricted contexts. Cell Signal. 2005;17:1533–1541. doi: 10.1016/j.cellsig.2005.03.021. [DOI] [PubMed] [Google Scholar]
- 125.Baxi MD, Vishwanatha JK. Diadenosine polyphosphates: their biological and pharmacological significance. J Pharmacol Toxicol Methods. 1995;33:121–128. doi: 10.1016/1056-8719(94)00127-p. [DOI] [PubMed] [Google Scholar]
- 126.Di Ciano C, Nie Z, Szászi K, Lewis A, Uruno T, Zhan X, Rotstein OD, Mak A, Kapus A. Osmotic stress-induced remodeling of the cortical cytoskeleton. Am J Physiol Cell Physiol. 2002;283:C850–C865. doi: 10.1152/ajpcell.00018.2002. [DOI] [PubMed] [Google Scholar]
- 127.Chiasson JL, Aris-Jilwan N, Bélanger R, Bertrand S, Beauregard H, Ekoé JM, Fournier H, Havrankova J. Diagnosis and treatment of diabetic ketoacidosis and the hyperglycemic hyperosmolar state. Can Med Assoc J. 2003;168:859–866. [PMC free article] [PubMed] [Google Scholar]
- 128.Sonna LA, Fujita J, Gaffin SL, Lilly CM. Invited review: effects of heat and cold stress on mammalian gene expression. J Appl Physiol. 2002;92:1725–1742. doi: 10.1152/japplphysiol.01143.2001. [DOI] [PubMed] [Google Scholar]
- 129.Morrison BH, Bauer JA, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 mediates growth suppressive and apoptotic effects of interferon-beta in ovarian carcinoma cells. J Biol Chem. 2001;276:24965–24970. doi: 10.1074/jbc.M101161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Morrison BH, Bauer JA, Hu J, Grane RW, Ozdemir AM, Chawla-Sarkar M, Gong B, Almasan A, Kalvakolanu DV, Lindner DJ. Inositol hexakisphosphate kinase 2 sensitizes ovarian carcinoma cells to multiple cancer therapeutics. Oncogene. 2002;21:1882–1889. doi: 10.1038/sj/onc/1205265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Arch RH, Gedrich RW, Thompson CB. Tumor necrosis factor receptor-associated factors (TRAFs)—a family of adapter proteins that regulates life and death. Genes Dev. 1998;12:2821–2830. doi: 10.1101/gad.12.18.2821. [DOI] [PubMed] [Google Scholar]
- 132.Barginear MF, Van Poznak C, Rosen N, Modi S, Hudis CA, Budman DR. The heat shock protein 90 chaperone complex: an evolving therapeutic target. Curr Cancer Drug Targets. 2008;8:522–532. doi: 10.2174/156800908785699379. [DOI] [PubMed] [Google Scholar]
- 133.Sharp S, Workman P. Inhibitors of the HSP90 molecular chaperone: current status. Adv Cancer Res. 2006;95:323–348. doi: 10.1016/S0065-230X(06)95009-X. [DOI] [PubMed] [Google Scholar]
- 134.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 135.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem. 2002;277:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 136.Sahu SK, Gummadi SN, Manoj N, Aradhyam GK. Phospholipid scramblases: an overview. Arch Biochem Biophys. 2007;462:103–114. doi: 10.1016/j.abb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 137.Aoki M, Sobek V, Maslyar DJ, Hecht A, Vogt PK. Oncogenic transformation by beta-catenin: deletion analysis and characterization of selected target genes. Oncogene. 2002;21:6983–6991. doi: 10.1038/sj.onc.1205796. [DOI] [PubMed] [Google Scholar]
- 138.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 139.Shears SB. Telomere maintenance by intracellular signals: new kid on the block? Proc Natl Acad Sci USA. 2005;102:1811–1812. doi: 10.1073/pnas.0409801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Luo HR, Saiardi A, Yu H, Nagata E, Ye K, Snyder SH. Inositol pyrophosphates are required for DNA hyperrecombination in protein kinase c1 mutant yeast. Biochemistry. 2002;41:2509–2515. doi: 10.1021/bi0118153. [DOI] [PubMed] [Google Scholar]
- 141.Katai K, Miyamoto K, Kishida S, Segawa H, Nii T, Tanaka H, Tani Y, Arai H, Tatsumi S, Morita K, Taketani Y, Takeda E. Regulation of intestinal Na+-dependent phosphate co-transporters by a low-phosphate diet and 1,25-dihydroxyvitamin D3. Biochem J. 1999;343:705–712. [PMC free article] [PubMed] [Google Scholar]
- 142.Lee YS, Mulugu S, York JD, O’Shea EK. Regulation of a cyclin-CDK-CDK inhibitor complex by inositol pyrophosphates. Science. 2007;316:109–112. doi: 10.1126/science.1139080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lee YS, Huang K, Quiocho FA, O’Shea EK. Molecular basis of cyclin-CDK-CKI regulation by reversible binding of an inositol pyrophosphate. Nat Chem Biol. 2008;4:25–32. doi: 10.1038/nchembio.2007.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Onnebo SM, Saiardi A. Inositol pyrophosphates get the vip1 treatment. Cell. 2007;129:647–649. doi: 10.1016/j.cell.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 145.Pyerin W, Barz T, Ackermann K. Protein kinase CK2 in gene control at cell cycle entry. Mol Cell Biochem. 2005;274:189–200. doi: 10.1007/s11010-005-2951-1. [DOI] [PubMed] [Google Scholar]
- 146.Dhariwala FA, Rajadhyaksha MS. An unusual member of the Cdk family: Cdk5. Cell Mol Neurobiol. 2008;28:351–369. doi: 10.1007/s10571-007-9242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lilja L, Yang SN, Webb DL, Juntti-Berggren L, Berggren PO, Bark C. Cyclin-dependent kinase 5 promotes insulin exocytosis. J Biol Chem. 2001;276:34199–341205. doi: 10.1074/jbc.M103776200. [DOI] [PubMed] [Google Scholar]
- 148.Maffucci T, Falasca M. Specificity in pleckstrin homology (PH) domain membrane targeting: a role for a phosphoinositide-protein co-operative mechanism. FEBS Lett. 2001;506:173–179. doi: 10.1016/s0014-5793(01)02909-x. [DOI] [PubMed] [Google Scholar]
- 149.Chan TO, Rittenhouse SE, Tsichlis PN. AKT/PKB and other D3 phosphoinositide-regulated kinases: kinase activation by phosphoinositide-dependent phosphorylation. Annu Rev Biochem. 1999;68:965–1014. doi: 10.1146/annurev.biochem.68.1.965. [DOI] [PubMed] [Google Scholar]
- 150.Piccolo E, Vignati S, Maffucci T, Innominato PF, Riley AM, Potter BV, Pandolfi PP, Broggini M, Iacobelli S, Innocenti P, Falasca M. Inositol pentakisphosphate promotes apoptosis through the PI 3-K/Akt pathway. Oncogene. 2004;23:1754–1765. doi: 10.1038/sj.onc.1207296. [DOI] [PubMed] [Google Scholar]
- 151.Jackson SG, Zhang Y, Haslam RJ, Junop MS. Structural analysis of the carboxy terminal PH domain of pleckstrin bound to D-myo-inositol 1, 2, 3, 5, 6-pentakisphosphate. BMC Struct Biol. 2007;7:80. doi: 10.1186/1472-6807-7-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Downes CP, Gray A, Fairservice A, Safrany ST, Batty IH, Fleming I. The regulation of membrane to cytosol partitioning of signalling proteins by phosphoinositides and their soluble headgroups. Biochem Soc Trans. 2005;33:1303–1307. doi: 10.1042/BST0331303. [DOI] [PubMed] [Google Scholar]
- 153.Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Downes CP, Safrany ST, Alessi DR, van Aalten DM. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J. 2004;23:3918–3928. doi: 10.1038/sj.emboj.7600379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Takeuchi H, Kanematsu T, Misumi Y, Sakane F, Konishi H, Kikkawa U, Watanabe Y, Katan M, Hirata M. Distinct specificity in the binding of inositol phosphates by pleckstrin homology domains of pleckstrin, RAC-protein kinase, diacylglycerol kinase and a new 130 kDa protein. Biochim Biophys Acta. 1997;1359:275–285. doi: 10.1016/s0167-4889(97)00109-2. [DOI] [PubMed] [Google Scholar]
- 155.Frech M, Andjelkovic M, Ingley E, Reddy KK, Falck JR, Hemmings BA. High affinity binding of inositol phosphates and phosphoinositides to the pleckstrin homology domain of RAC/protein kinase B and their influence on kinase activity. J Biol Chem. 1997;272:8474–8481. doi: 10.1074/jbc.272.13.8474. [DOI] [PubMed] [Google Scholar]
- 156.Milburn CC, Deak M, Kelly SM, Price NC, Alessi DR, Van Aalten DM. Binding of phosphatidylinositol 3,4,5-trisphosphate to the pleckstrin homology domain of protein kinase B induces a conformational change. Biochem J. 2003;375:531–538. doi: 10.1042/BJ20031229. [DOI] [PMC free article] [PubMed] [Google Scholar]