Abstract
The Rh (Rhesus) genes encode a family of conserved proteins that share a structural fold of 12 transmembrane helices with members of the major facilitator superfamily. Interest in this family has arisen from the discovery of Rh factor’s involvement in hemolytic disease in the fetus and newborn, and of its homologs widely expressed in epithelial tissues. The Rh factor and Rh-associated glycoprotein (RhAG), with epithelial cousins RhBG and RhCG, form four subgroups conferring upon vertebrates a genealogical commonality. The past decade has heralded significant advances in understanding the phylogenetics, allelic diversity, crystal structure, and biological function of Rh proteins. This review describes recent progress on this family and the molecular insights gleaned from its gene evolution, membrane biology, and disease association. The focus is on its long evolutionary history and surprising structural conservation from prokaryotes to humans, pointing to the importance of its functional role, related to but distinct from ammonium transport proteins.
Keywords: Rh protein family, Plasma membrane, Epithelia, Erythrocytes, Channels and transporters, Systemic pH, Gene evolution, Molecular genetics, Disease association
Introduction
Most of us become aware of the “Rh (Rhesus) factor” at a blood bank or sense its importance in a hospital where we see “Rh” along with “ABO” as labels on all bags of blood to be transfused into patients. At other times we may hear the words “Rh-positive” and “Rh-negative” or learn touching stories from friends or relatives about pregnant women who needed clinical care owing to a mismatch with their baby’s Rh blood type. All these now routine medical practices rest on the genotype-phenotype principles of human genetics, as illuminated by Landsteiner, Levine, and Weiner in their seminal discoveries of the ABO and Rh blood group substances [1–4]. Indeed, the Rh and ABO antigens are still the clinically most significant [5] and genetically most polymorphic of all human blood group systems to date [6]. However, ABO are carbohydrate antigens [7] depending on the enzymatic activity and specificity of allelic glycosyltransferases [8], whereas Rh antigens are protein motifs [9, 10], whose surface expression entails an interaction of two genetic loci [11, 12]. The protein nature endows Rh antigens, particularly the more recently evolved D antigen, with the inherent ability to mount potent alloimmune reactions to counteract such conflicting situations as fetal-maternal incompatibility.
As a model system that has been well studied for seven decades, Rh proteins have generated many exciting moments of discovery in the disciplines of hematology, biochemistry, and human genetics. The foundation of Rh phenotypic variation and population genetics was laid out in the first 35 years [13], culminating in the development of prophylaxis therapy for hemolytic disease of the fetus and newborn (HDFN) in the 1960s [14, 15]. A series of studies in the 1980s established Rh antigens and Rh-associated glycoprotein (RhAG) as integral membrane proteins [7, 8, 16]. Such endeavors led to the partially determined protein sequences [17–19] that paved the way for cloning of Rh30 (RHCE and RHD) and RhAG (RHAG) in the early 1990s [20–24]. Ensuing studies resulted in definition of the common D and CcEe antigens in molecular terms [25] and compilation of a thorough compendium of human Rh allelic diversification [6, 9, 26–30]. The nature of RhAG as a genetic regulator for Rh antigen expression was also verified via the identification at the RHAG locus of mutations that cause Rh-deficiency syndrome [31–37].
Despite their folding into 12 transmembrane helices (TMH) common to members of the major facilitator superfamily (MFS) [38], the red cell Rh proteins have been notoriously refractory to functional definition [39, 40], let alone specification of their elusive substrate. The cloning of epithelial RhBG and RhCG cousins and their remote relatives has extended the erythroid paradigm and opened new avenues of research in nonerythroid tissues and model organisms [41–43]. The functional importance of such nonerythroid homologs in their natural settings has been established in the unicellular green alga C. reinhardtii [44–46], the worm C. elegans [47, 48], and the mouse as a mammal [49]. Now Rh30, RhAG, RhBG, and RhCG together define the four subgroups of the family and grant vertebrates a genealogical commonality [50–52]. The past decade has heralded a new era for our understanding of the Rh family in terms of its genetic diversity, protein evolution, three-dimensional (3D) structure, and biological function. Despite the debate on their substrate specificities as CO2 and/or ammonia, Rh proteins have emerged as a key class of plasma-membrane proteins playing important roles in maintaining systemic pH at the organismal level. This review provides an update of recent findings on the Rh family and focuses on multifaceted insights newly gained from its molecular evolution, structural conservation, membrane biology, and disease association.
Genetic structure and molecular evolution of the Rh protein family
Origin and taxonomic distribution
The Rh family is now believed to have arisen from a common ancestor of prokaryotic origins, based on our increasing awareness of the presence of its homologs in bacteria [52] (Table 1). Despite their absence in sequenced archaeal genomes [46], Rh homologs are present as single-copy loci in certain bacterial taxa dwelling in soils, waters or subsurface and showing unique metabolic features, slower cell cycles, or more elaborate intracellular partitions [53–61]. Of the ammonia-oxidizing and CO2-fixing bacteria, three are aerobic lithoautotrophs [53, 54] and one is an anaerobic anammox [55]. Four are rod-shaped free-living Clostridia species that are obligate anaerobes capable of sporulating and acetate-oxidation or sulfate-reduction along with CO2-fixation [58, 59]. Geobacter sp. M21 belongs to clade 1 of the four Geobacteraceae clades sampled from Fe(III)-reducing subsurface regions [60], whereas the Ellin345 isolate of A. bacteria is a highly capsulated aerobic heterotroph widely found in soils [61]. The open reading frames (ORF) of these bacterial genes encode Rh proteins with a notable sequence identity to one another and to human proteins, particularly the RhAG, RhBG, and RhCG glycoproteins (Table 1). The common features dictate an orthologous relationship between bacterial and human Rh proteins implicating a conserved function across enormously distant phylogenies.
Table 1.
Comparison between bacterial and human Rh proteins
| Taxonomic name | Bacterial Rh homolog | Human Rh protein family | |||||
|---|---|---|---|---|---|---|---|
| ORF (aa) | % Id/Sm (aa align) | % Id/Sm (aa align) | |||||
| RhAG | RhBG | RhCG | RhCE | RhD | |||
| 409 | 458 | 479 | 417 | 417 | |||
| Proteobacteria β | |||||||
| Nitrosomonas europaea | 425 | 100/100 (425) | 37/58 (354) | 36/51 (407) | 34/53 (387) | 29/48 (318) | 31/49 (277) |
| Nitrosomonas sp. AL212 | 457 | 70/83 (401) | 38/58 (357) | 39/55 (407) | 33/50 (430) | 29/48 (314) | 33/51 (283) |
| Nitrosomonas multiformis | 407 | 73/84 (407) | 36/57 (355) | 41/59 (340) | 36/55 (313) | 28/47 (313) | 29/50 (317) |
| Proteobacteria δ | |||||||
| Geobacter sp. M21 | 403 | 51/66 (362) | 36/55 (342) | 37/55 (301) | 33/51 (336) | 29/48 (284) | 29/48 (286) |
| Planctomycetes | |||||||
| Kuenenia stuttgartiensis | 585 | 50/67 (360) | 33/55 (368) | 39/59 (313) | 34/57 (296) | 25/46 (321) | 25/46 (338) |
| Firmicutes-Clostridia | |||||||
| Clostridium carboxidivorans | 404 | 50/70 (345) | 35/55 (376) | 36/56 (346) | 34/55 (313) | 26/49 (311) | 28/52 (277) |
| Clostridium cellulovorans | 401 | 50/70 (345) | 36/58 (345) | 36/60 (319) | 30/50 (384) | 27/47 (318) | 28/52 (309) |
| Clostridium papyrosolvens | 391 | 63/78 (356) | 37/57 (352) | 38/58 (306) | 32/53 (379) | 27/48 (340) | 28/49 (335) |
| Desulfotomaculum acetoxidans | 400 | 51/68 (346) | 38/57 (341) | 35/56 (347) | 34/55 (292) | 27/50 (304) | 30/53 (273) |
| Acidobacteria | |||||||
| Acidobacteria bacterium | 390 | 61/75 (388) | 36/56 (345) | 36/54 (342) | 34/54 (312) | 27/46 (332) | 29/49 (290) |
| Archaea | |||||||
| Archaeoglobus fulgidus Amt1 | 391 | 26/44 (222) | 27/44 (240) | 28/43 (216) | 28/43 (153) | 41/48 (97) | 28/44 (225) |
% Id/Sm Percent identity/similarity based on pairwise alignment of protein sequences. N. europaea Rh (top) and A. fulgidus Amt1 (bottom) are used for comparison since their 3D structures are known. % Id/Sm between each bacterial Rh and human Rh proteins (RhAG to RhD) is shown (left to right). The number in parenthesis is the total amino acid sites aligned. Note that three β-proteobacteria have Rh only, whereas the remaining bacteria retain both Rh and Amt genes
ORF Open reading frame in total amino acids (aa). The length of bacterial and human Rh proteins falls in the range from 390–585 to 409–479 residues, respectively
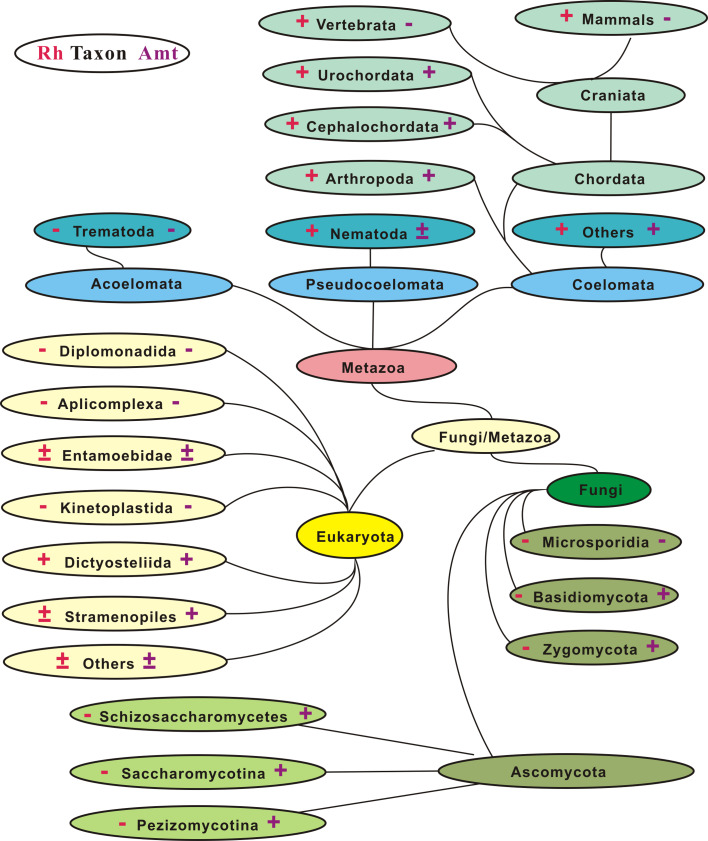
In contrast to their prokaryotic rarity, members of the Rh family show a much broader distribution in eukaryotes (Fig. 1). It appears that Rh genes were first dispersed among unicellular eukaryotes and then became ubiquitous in the animal kingdom from primitive metazoans to invertebrates [52]. Ultimately, the Rh family genesis arrived at its prominence and steady state in vertebrates, with definitive subgroups emerging from fish to mammals, as typified by the four loci in humans (Table 1). Of unicellular eukaryotes, most Rh-harboring species are free-living microbes in soils and/or waters, such as social amoebae slime molds (myxomycetes), water molds (oomycetes), or marine diatoms (stramenophiles). Rh genes are also present in the green alga C. reinhardtii [44], but so far not in other nonvascular plants, i.e., the Bryophyta (mosses), Marchantiophyta (liverworts), Anthocerotophyta (hornworts), or vascular (seed) plants. Strikingly Rh genes are absent or lost all at once in the morphologically similar but phylogenetically dissimilar fungi and in certain parasites such as the obligate parasitic protists, Apicomplexa and Kinetoplastida (Fig. 1).
Fig. 1.
Distribution of Rh genes and their coexistence with Amt genes in eukaryotes. Plus and minus indicate presence and absence, respectively, of Rh (left) or Amt genes (right). The results were obtained from tblastn/blastp search using Rh or Amt protein queries against genome databases. Taxonomic divisions are modified from the eukaryotic tree (www.ncbi.nlm.nih.gov/sutils/genom_tree.cgi). Certain taxa lack both Rh and Amt
Rh gene gains and losses in eukaryotic evolution
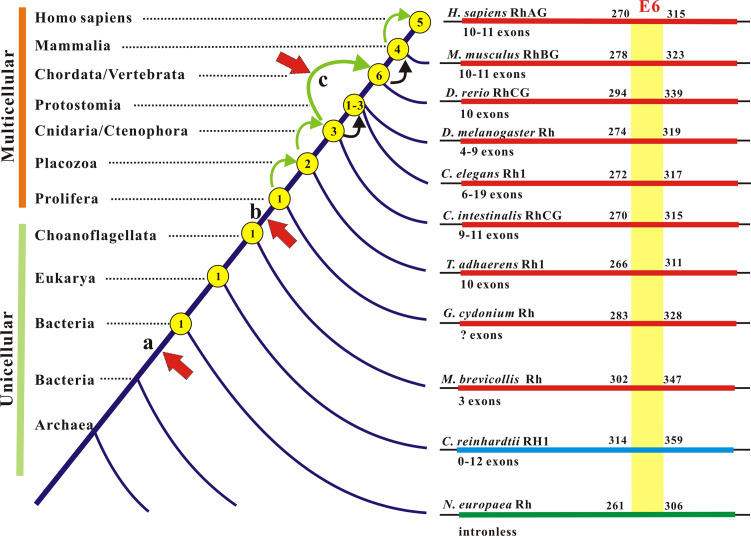
Gene duplication is a critical selection force that gives rise to raw materials for neofunctionalization and subfunctionalization of protein families [62]. As to this novelty, several rounds of gene duplication involving the Rh family have occurred and may account for its presence in extant organisms [50]. A careful inspection of copy number, exon–intron junction, and physical location of RH loci discloses hallmarks that recapitulate their expansion and contraction in the eukaryotic life domain (Fig. 2). Together, the evolutionary events have shaped the conservation and diversification of the Rh family in transitions from single-copy to multi-copies, from intronless to multi-introns, from unicellularity to multicellularity, and from epithelial cells (nonerythroid) to non-epithelial (erythroid) cells. Thus the Rh family is an excellent model system, on a large taxonomic scale, to look into the pattern of gene gain and gene loss throughout molecular evolution (Fig. 2).
Fig. 2.
Gene gain and gene loss in the genesis and evolution of the Rh protein family. Left Gene gain and loss are denoted by copy numbers (circled) on the tree trunk (not to scale). Bending arrows indicate gene duplications (green) or gene contractions (black). The increase from three to six genes may arise by a genome-wide duplication. Vertical arrows to the truck point to evolutionary events with which Rh gene duplications coincide: a origin of an ancient Rh gene and its branch off Amt genes in Bacteria and Archaea below the arrow; b origin of epithelia; c origin of erythrocyte. Right Exon remodeling in Rh family genes from representative taxa is shown. E6 (exon 6), the most conserved exon encoding 46 amino acids in metazoans, is used as a reference
Unicellular eukaryotic microbes may carry one (common) to three genes (rare). Such Rh duplicates seem to have arisen by adaptation, as their intraspecific identity generally exceeds their interspecific identity [50, 52]. Metazoan Rh members are also duplicates of a common ancestor but display more complex gene-gain and gene-loss patterns (Fig. 2). Supporting evidence includes a single-copy gene in the bona fide outgroup M. brevicollis (a unicellular choanoflagellate) [63]; one to three in invertebrates (water flea D. duplex is special, harboring six); six in teleost fish; and four in frogs, birds, and mammals [50]. While gene duplication apparently occurred twice in invertebrates, the doubling of genes from three in lancelet [64] or tunicate [65] to six in teleost fish [50] likely resulted from whole-genome duplication through tetraploidization [66]. This genome-wide duplication had reset the stage to place Rh genes on different chromosomes. The subsequent loss of two genes finalized four loci, i.e., RhAG, RhBG, RhCG, and Rh30, which together define the four distinct subgroups and confer upon vertebrate animals a genealogical commonality [52]. In recent primate to human evolution, the division of RH into RHCE and RHD via tandem gene duplication reformed the Rh blood group system [11, 12]. Gene duplications in metazoans went through chaotic changes in exon–intron organization of Rh genes in the wide variety of phylogenies. Although Rh genes are intronless in bacteria and certain eukaryotes, they keep multiple introns in all metazoans (Fig. 2). Depending on species, these introns invade the protein-coding sequence at different positions and thus break exons with changing numbers (Fig. 2); yet, Rh proteins still maintain their conserved features, thereby sharing astounding structural homologies.
Negative versus positive selection on subgroups
Congruent with species orders, Rh homologs cluster in subgroups [50] suggesting that natural selection has acted differently on these subgroups. The Rh family as a whole is highly conserved, but its subgroups vary in divergence rate and sites thereby relating functional specification to species adaptation [52]. One key advance from unicellularity to multicellularity is the origin of epithelia; the duplication of Rh gene from one in sponge [67] to two in the Placozoa [68], the basal metazoans with epithelia [69, 70], coincides with this morphological innovation (Fig. 2). Invertebrate Rh proteins comprise a large group having endured a long period of negative selection and a similar degree of sequence identity to the individual vertebrate subgroups, RhBG > RhCG > RhAG > Rh30 [52]. Hence the Rh ancestors born with epithelia must have branched to engender those homologs now expressed in invertebrate hemocytes and vertebrate red cells. In vertebrates, negative selection continues on epithelial homologs, but positive selection occurs to erythroid Rh proteins. The Rh30 subgroup has diverged rapidly and steadily after its origin [71–74], whereas the RhAG subgroup has experienced two phases of selection, negatively in lower vertebrates but positively in mammals parallel to Rh30 selection [51]. The fast co-evolution of Rh30/RhAG matches with the timeline of morphological transition of red cells from elliptical to biconcave through enucleation.
Distant relatedness of Rh to Amt
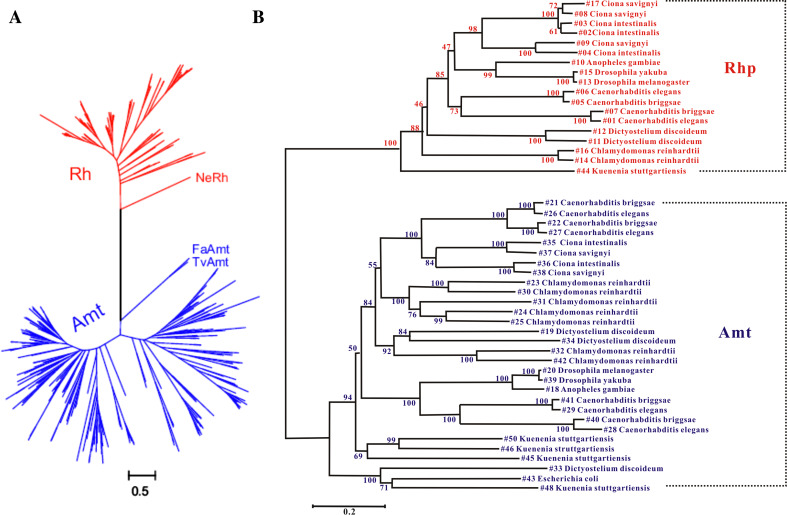
Ammonia transporters (Amt) are thought to be members of the MFS club [38]; they and Rh proteins form the only known related families [46, 75]. Our knowledge of their differences and similarities in organism distribution, molecular evolution, and protein structure has dramatically increased since the report linking human erythroid Rh proteins to Amt proteins [75]. The two families not only show an opposite pattern of distribution but coexist in a great variety of organisms (Fig. 1). Apparently Rh ancestors had already branched off from Amt ancestors in prokaryotes and the two had gone through their respective evolutionary pathways after separation [50, 51] (Fig. 3a). In addition, the Rh proteins and Amt proteins in those organisms from bacteria to invertebrates that have both types of genes also cluster independently and are separated by an unpredictable distance because of the long period of divergent evolution (Fig. 3b). These findings pinpoint a distant and paralogous relatedness arguing for a functional distinction between the Rh family and the Amt family [46, 52].
Fig. 3.
Rh and Amt are distantly related families but have gone through divergent and independent evolution. a The maximum likelihood (ML) joint tree of 111 Rh proteins (red) and 260 Amt (blue) proteins. Bacterial NeRh and archaeal FaAmt and TvAmt are at the base of the respective families. b The ML joint protein tree of 18 Rh and 30 Amt in species that harbor both types of genes. The values at nodes are the bootstrap proportion from ML. This expanded analysis covers species from bacteria to invertebrates
Primary structure, transembrane fold, and 3D structure of Rh proteins
Biochemical features of primary structures
With the primary structures of Rh proteins known in human red cells [20–24], epithelial cells, and other species [41, 42], a consensus has emerged as to the index of conservation and diversification for the Rh family [50, 51]. Analysis of selected members from bacteria to humans reinforces such common features (Table 2). The total length of Rh proteins differs considerably depending on species, e.g., it is 390 aa for the A. bacteria Rh protein and 958 aa for the Rh2 homolog of N. gruberi (a soil/water amoeboflagellate), but the vast majority have a size of 400–500 aa (Table 2). The primary sequences that define TM helices are most conserved and thus hydrophobic residues account for two-thirds or more of total residues. For the TM helices, the ratio of hydrophobic/polar residues is constant (3:1) and so is the Gly/Pro ratio (4–5:1) and strict His occurrence (Table 2). The patterns of charge and size are distinct among intracellular loops (ICL), extracellular loops (ECL), and C-tails. C-tail is most diverged in sequence and size between subgroups [43]. Paradoxical to this variation is the invariant retention of charged residues in N and C-termini, although not at the same sites. Negative charges show a greater presence in the C-tail of RhAG, RhBG, or RhCG and positive ones in the N-terminus of Rh30, manifesting a bi-modularity pI profile as acidic and basic, respectively (Table 2). As little variability occurs in TMH and ICL, the C-tail contributes a net negative charge to Rh glycoproteins, a feature also pertaining to most Amt proteins [51]. Hence, the variable C-tail and the tightly packed TM helices consolidate Rh function and regulation in a single adaptable modular design in the membrane.
Table 2.
Primary composition and biochemical features of Rh proteins
| Name | ORF (aa) | pI | TM% | Hydro/polar | TM | ECL (aa) | ICL (aa) | Total (aa) (Nt/Ct) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Overall | Tm | G/P | H | |||||||
| Human | ||||||||||
| RhAG | 409 | 6.66 | 64.8 | 182/96 | 159/52 | 31/7 | 5 | 68 | 42 | 25 (4/21) |
| RhBG | 458 | 6.70 | 57.9 | 203/99 | 151/53 | 31/7 | 8 | 96 | 43 | 55 (13/42) |
| RhCG | 479 | 6.20 | 55.3 | 209/108 | 150/63 | 27/7 | 5 | 100 | 42 | 72 (10/62) |
| RhCE | 417 | 9.41 | 63.5 | 202/98 | 164/57 | 22/5 | 2 | 69 | 44 | 39 (11/28) |
| RhD | 417 | 8.51 | 63.5 | 201/98 | 161/59 | 23/5 | 2 | 69 | 44 | 39 (11/28) |
| Eukaryotes | ||||||||||
| DmRh | 449 | 5.74 | 59.9 | 203/105 | 164/56 | 25/6 | 3 | 103 | 43 | 38 (9/29) |
| CeRh1 | 463 | 6.21 | 58.1 | 198/114 | 155/58 | 23/7 | 7 | 96 | 41 | 61 (10/51) |
| TaRh1 | 445 | 5.36 | 59.6 | 204/108 | 155/59 | 22/6 | 4 | 88 | 41 | 52 (7/45) |
| TaRh2 | 459 | 5.73 | 57.7 | 213/116 | 158/65 | 20/4 | 6 | 102 | 41 | 54 (8/46) |
| GcRh | 523 | 4.79 | 51.4 | 225/129 | 153/58 | 28/7 | 3 | 129 | 40 | 87 (6/81) |
| MbRh | 480 | 5.51 | 55.2 | 210/119 | 155/56 | 29/6 | 6 | 115 | 48 | 48 (5/43) |
| Bacteria | ||||||||||
| NeRh | 425 | 6.33 | 62.4 | 214/89 | 163/52 | 28/7 | 3 | 61 | 39 | 60 (4/56) |
| NiRh | 457 | 5.85 | 58.0 | 209/103 | 160/49 | 30/8 | 6 | 60 | 39 | 93 (4/89) |
| NmRh | 407 | 5.60 | 65.1 | 200/89 | 167/47 | 27/7 | 4 | 61 | 39 | 41 (4/37) |
| GeRh | 403 | 4.77 | 65.8 | 200/83 | 158/54 | 24/6 | 3 | 57 | 38 | 43 (4/39) |
| KsRh | 585 | 6.85 | 45.3 | 248/134 | 157/56 | 24/6 | 4 | 260 | 38 | 22 (3/19) |
| CcRh | 404 | 8.03 | 65.6 | 192/96 | 160/53 | 23/9 | 3 | 75 | 38 | 26 (4/22) |
| CIRh | 401 | 5.22 | 66.1 | 193/90 | 157/56 | 26/8 | 3 | 60 | 39 | 23 (4/19) |
| CpRh | 391 | 6.34 | 67.8 | 197/79 | 160/54 | 28/6 | 4 | 75 | 38 | 27 (4/23) |
| DaRh | 400 | 7.25 | 66.2 | 187/96 | 158/57 | 25/8 | 3 | 75 | 38 | 22 (4/18) |
| AaRh | 390 | 6.85 | 67.9 | 186/87 | 157/56 | 27/7 | 3 | 60 | 42 | 23 (4/19) |
| Archaea | ||||||||||
| AfAmt1a | 391 | 5.90 | 66.0 | 208/71 | 171/43 | 25/4 | 3 | 63 | 43 | 28 (3/25) |
TM % Percent of TMH residues to total residues of a full-length protein, Hydro/Polar hydrophobic vs. polar residues in full-length protein (overall) or in TMH (Tm), G/P and H Gly/Pro and His residues assigned to transmembrane segments, ECL and ICL total size of extracellular and intracellular loops in residue numbers, Nt/Ct the total and respective size of N- and C-terminal regions in residue numbers. The data were from structure-based sequence alignments and are available upon request
aAfAmt1 lacks TMH0 or signal peptide and thus has 11 TM helices. This Amt is chosen for comparison because it gives rise to a better alignment with Rh proteins than EcAmtB
Fold of transmembrane helices
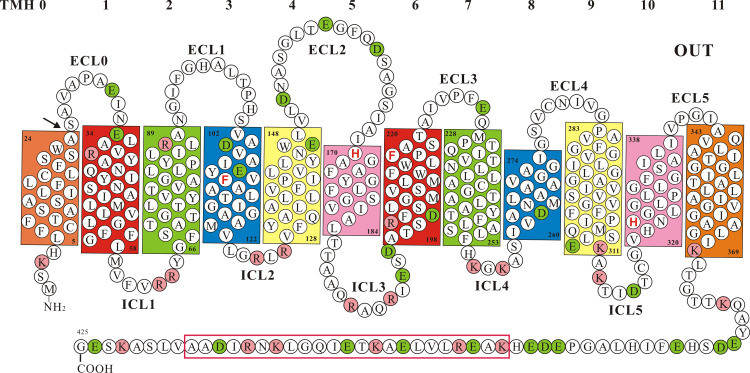
Rh proteins are predicted to have 12 transmembrane helices [40, 43], a condition which has now been observed in the 3D structure of NeRh, the N. europaea Rh, and human RhCG [76–78]. Figure 4 shows a 2D topology of NeRh with its short N-terminus and long C-tail facing the cytoplasm. This model and Table 2 illuminate some identifying features, both general and particular to Rh proteins. The two 6-TMH halves (TMH0-5 and 6–11) grossly appear as “self-images” given their patched identities and shared motifs. For Rh glycoprotein homologs TMH1, 3, 5, 6, 8, and 10 are conserved with key amino acids, e.g., twin-His and twin-Phe. TMH0 is peripheral [78] as suggested [79, 80], but TMH1:6, 2:7, 3:8, 4:9, and 5:10 each form a pair of TM domains packed close in the membrane [76–78]. The surface-charge across the lipid bilayer is asymmetric because ECL and ICL are coated by net negative and net positive charges, respectively (Fig. 4). ICL are short and invariant in size (Table 2); of these, ICL3 between TMH5 and 6 is the longest allowing 6-TM halves limited mobility, and others are just long enough for helix turns. ECL changes are largely confined to elongation or truncation of ECL0 and ECL5. The strict total length of ICL as a feature common to Rh and Amt (Table 2) makes them unique within the MFS club. It is tempting to speculate that oscillation of TMH5 plays a crucial role in triggering the Rh channel function as in the AmtB protein [81].
Fig. 4.
Membrane topology of N. europaea Rh protein as a model for the Rh family. TMH0 is absent in the crystals and is based on hydropathy plot, whereas TMH 1-11 are from the 3D structure. Closely packed TMH are colored the same except TMH0. The residues positioned at the bilayer leaflets are numbered. Twin-His H170/H324 and twin-Phe F110/F218 are bold (red). Positive K/R charges are colored pink and negative D/E charges green. The α-helical bundle-forming sequence in the C-tail is boxed
Of note is TMH0, an integral part of Rh proteins [82, 83] but a cleavable signal peptide (SP) either present [84–86] or absent in Amt proteins [87]. Thus unlike the Rh proteins, membrane-bound Amt proteins lack the TMH0 but have the N-terminal end from part of ECL0 float in the periplasmic space. In the heterologous E. coli cell NeRh-TMH0 was likely recognized as SP and removed [76, 77]; but whether it is so processed in the native host N. europaea is unclear [88, 89]. Perhaps different from Amt Rh-TMH0 is a hybrid motif of SP and TMH given its possible origin from an SP-like segment. As a key issue related to intracellular routing and membrane assembly, the dual role of Rh-TMH0 in both prokaryotes and eukaryotes needs to be verified. The absence of TMH0 in NeRh crystals [76, 77] but presence in human Rh proteins [82, 83] also raises the question of what effects TMH0 has on protein oligomerization and intramembrane folding [79, 80].
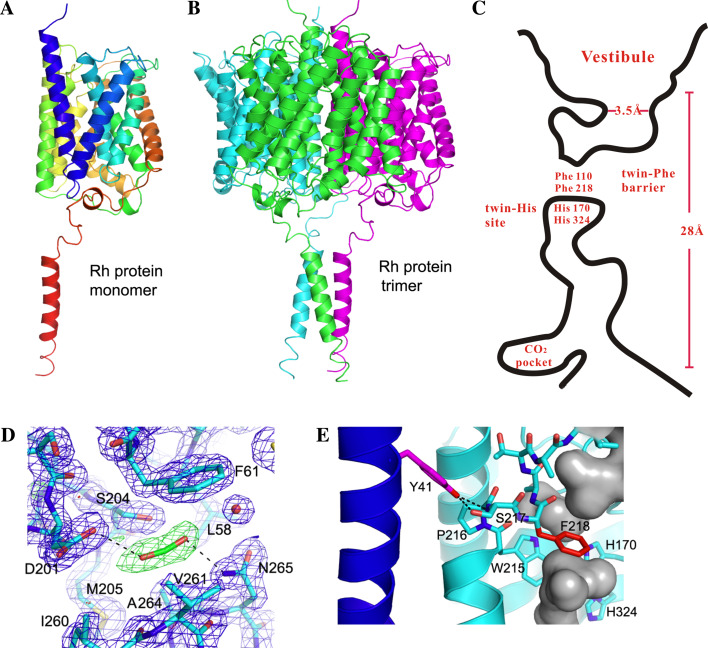
3D structure of NeRh protein and its implications for function
The high-resolution 3D structures of E. coli AmtB (EcAmtB) and A. fulgidus Amt1 (AfAmt1) have provided fresh insights into the mechanism of ammonia conduction [85–87]. EcAmtB and AfAmt1 both fold as homotrimers with each monomer forming a vestibule for NH4 +/NH3 binding and a long narrow hydrophobic channel for ammonia transport. Mechanistically, both Amt proteins appear to operate as a gas channel that mediates the passage of neutral species of ammonia, but the conduit of proton transfer/movement as a prerequisite for gas conduction is only partially understood [90–92]. Given the known relatedness, the 3D structures of Amt proteins had been used as a template for homology modeling of human Rh proteins to gain insight into their function [79, 80, 93, 94]. The studies suggest that Rh glycoproteins and Rh30 proteins both adopt a 3D fold similar to the two Amt proteins, but Rh30 lacks the conserved His and/or Phe residues important for substrate conduction.
The diffraction of NeRh crystals has laid a solid foundation to relate the 3D structure to the function of Rh family proteins [76, 77]. As compared to the Amt structures [85–87], the overall fold and membrane topology of NeRh are similar. Due to TMH0 removal each subunit is composed of 11 TMH with pseudo two-fold symmetry enclosing a central pore presumably mediating gas transport (Fig. 5a). The NeRh structure is also an α3-homotrimer generated by a crystallographic three-fold passing through the center of the three monomers (Fig. 5b). A cartoon shows the extracellular vestibule, central channel, and other notable features in one subunit (Fig. 5c). Besides the general folding pattern, a prominent conservation is the twin-His site, namely His170/His324 in NeRh [76, 77], His170/His320 in EcAmtB [85, 86], and His157/His305 in AfAmt1 protein [87]. Although controversial, this twin-His site is thought to mediate NH4 + ion deprotonation [85] and is crucial for function [95]. In both Rh and Amt structures, the twin-His site lies just below the twin-Phe barrier (Fig. 5c); the two His adopt an unusual coplanar orientation and situate in a fairly hydrophobic environment. The coplanar orientation may enable the two His residues to stabilize a proton between them, and the hydrophobic milieu may shield them from approaching water molecules and thereby foster anhydrous proton migration. Together with twin-Phe, twin-His may also add in selectivity by blocking the passage of small cations such as Na+ and K+, while allowing the passage of neutral gases such as CO2 or NH3 [76, 77]. Notably, the twin-His site is conserved in Rh glycoproteins, but one or both residues are mutated in Rh30 homologs [50, 79, 80], supporting that the Rh30 group has evolved to gain a different function [11, 12, 27, 30].
Fig. 5.
3D structure of the N. europaea Rh protein. Each drawing is oriented such that the periplasm is above and cytoplasm is below. a Ribbon diagram of a monomer. b Ribbon diagram of a homotrimer. c Diagram of the putative central channel in a monomer. The constriction of extracellular vestibule (3.5 Å) and membrane-crossing (28 Å) are denoted. Twin-Phe barrier, twin-His site, and CO2-binding site are illustrated. d Atomic details of CO2-binding residues in the pocket. e Interactions between Tyr41 and TMH1. Potential hydrogen bonds between Tyr41 and Ser217 are shown as black dotted lines
Structural comparison also reveals critical differences between Rh and Amt crystals. (1) Most notably, NeRh has a C-tail α-helix directed along the three-fold axis but away from the trimer proper [76, 77] (Fig. 5a). The three helices come close to form a left-handed three-helix bundle, a fairly mobile part with much higher average residue B factor (76 Å2) than the rest of the protein (20 Å2). (2) NeRh has a CO2-binding pocket, a unique structure formed by fairly conserved residues when native crystals were pressurized under CO2 and then flash cooled in liquid nitrogen [76]. This pocket is located within a deep cavity near the channel exit to the cytoplasm (Fig. 5d); its role is presently unclear but could be to promote CO2 movement in and out of the pore as a secondary site. (3) NeRh has a higher presence of prolines in TMH like its homologs (Table 2). Such internal prolines can lead to distortions or kinks in TM helices [96] and thereby impact helix packing or protein function (e.g., dictating the hinge sites for a conformational change). (4) NeRh lacks the extracellular vestibule π-cation binding site formed by three aromatic residues thought to be key to NH4 +/NH3 recruitment in EcAmtB [84]. (5) The twin-Phe are conserved in NeRh (Phe110/Phe218) (Fig. 5c), EcAmtB (Phe107/Phe215), or AfAmt1 (Phe96/Phe204) but show altered orientation. In Amt, the two Phe adopt positions that block substrate passage through the channel [85–87] as important functional sites [97]. In NeRh Phe218 appears to be the major barrier, since Phe110 adopts a different orientation [76, 77]. (6) Although the role of twin-His may be similar, one difference in potential functional impact is that in NeRh the Nε2 atoms of His170 and His324 form hydrogen bonds to neighboring water molecules. In contrast, His320 of EcAmtB or His305 of AfAmt1 has only one hydrogen-bonding partner. The structural differences between Rh and Amt have implications for function and suggest important targets for future studies.
A putative gating mechanism for opening of Rh channel proteins
The α-helical bundle found in NeRh C-tail suggests that it may mediate a functionally relevant protein–protein interaction [76, 77]. The 3D structures of AmtB-GlnK protein complexes yield clues to this view in that GlnK inhibits ammonia uptake via binding to the ICL face [98, 99]. If the C-tail of NeRh binds to a protein, it could similarly block substrate passage by linking such an interaction to the putative CO2 binding site in two ways [76]: (1) substrate transfer is facilitated by the binding partner; (2) substrate-induced protein recruitment is executed as a sensing mechanism, as has been proposed for A. brasilense AmtB [100]. Currently, the solved 3D structures of Amt and Rh proteins are all in a channel-closed configuration [76, 77, 85–87], where twin-Phe separates the channel into two parts and thereby shuts off transport (Fig. 5c). To open the channel passage through NeRh, a C-tail-mediated partner binding must disengage this twin-Phe block, for example, by altering its orientation.
There are three key structural elements that support this potential form of regulation. (1) The C-tail α-helix has a glutamate that is salt bridged to Arg63 and Arg64 on ICL1 between TMH1 and TMH2 (Fig. 4). (2) Tyr41 on TMH1 is hydrogen bonded to the main-chain NH and side-chain-OH of Ser217 on TMH6 of an adjacent subunit (Fig. 5e). (3) Ser217 lies in between the internal Pro216 that induces a helical kink in the TMH6 helix (Fig. 4) and the Phe218 that serves as a major steric barrier for transport (Fig. 5c). Thus changes in the interactions between Tyr41 and Ser217 could affect the magnitude of TMH6 tilting, which in turn impacts the movement of Phe218 to enable channel opening or closing [76, 77].
The mechanism envisaged in which pore opening of the Rh protein is dictated by C-tail-mediated interactions begs for binding partners. Potential candidates include carbonic anhydrase (CAH) or RuBisCO if Rh proteins conduct CO2 or ammonia monoxygenase (AMO) and glutamine synthetase if Rh proteins were Amt equivalents. Of special note is the α-CAH enzyme, for its distribution is totally correlated with that of Rh proteins in all organisms whose genome sequences have been determined [50]. Given this phylogenetic coexistence, it is of great interest to test out whether α-CAH could be docked onto NeRh trimer and, if so, whether the interaction occurs in biochemical and functional forms in vivo [101]. It is also of interest to determine how C-tail-mediated regulations cope with the great C-tail sequence diversity in functional adaptations of Rh subgroups.
Substrate specificity
Rh proteins were inferred to participate in ammonia transport by sequence relatedness to Amt proteins [75], a view evidenced by human RhAG expression in a yeast mep mutant showing growth on ammonia as the only nitrogen source [102]. Follow-up studies in oocyte, yeast, or other cell types indicated that Rh proteins mediated the passage of both charged and uncharged species [103–114]. NeRh protein also seemed to have a role in NH3 transport [88, 89], as [14C]-methylamine uptake was competitively inhibited by NH3 and NeRh expression improved growth of the yeast mep mutant on limited ammonium. However, one challenge to the view of Rh proteins being ammonia channels is that AMO, the chief site of ammonia oxidation in N. europaea, is an integral membrane protein. Although acetylene did not affect ammonia passage [88], it inhibits the active site but not the substrate channel of AMO that might facilitate NH3 transport. These observations would make ammonia transport into the cell less important.
There is also evidence supporting an alternate view for the function of Rh proteins as a CO2 channel. Studies of Rh1 in C. reinhardtii showed its strong up-regulation upon high [CO2] induction [44]. Further studies of the phenotypic properties of C. reinhardtii strains depleted of Rh1 expression by RNAi provide evidence for its role in CO2 transport [45]. Under a high [CO2] condition, the rh1-deficient mutants grow more slowly than wild-type cells. Moreover, arguing against its role in ammonia transport, these rh1 mutants accumulate [14C]-methylamine normally when grown with either arginine (high accumulation) or ammonia (low accumulation) as the nitrogen source [45]. Recently it was found that the rate of CO2 transport is much lower in Rhnull cells lacking the Rh proteins than in normal red cells [115, 116]. Together these data corroborate the view that Rh proteins have a distinct functional role in mediating CO2 passage as compared to Amt proteins [46].
Expression of Rh genes and proteins: location and induction
Tissue and cell-specific expression
Rh antigens are abundant non-glycosylated proteins of the red cell membrane [9, 10] with ~1 × 105 copies per cell [40] and their glycosylated partner RhAG may be of similar quantity [16, 19]. In humans and mice, Rh30 and RhAG promoters contain cis-acting regulatory elements to direct erythroid-specific expression [32, 117–120]. RhAG is also expressed in human esophageal epithelia [121] and in mouse brain [122] independently of Rh30. RhCG and RhBG are highly expressed in kidney, testis, liver, brain, or skin and variably in many other tissues [41, 42] with RhCG being a most abundant renal transcript based on quantitative RNA profiling [123]. The 5′-regions of RhCG and RhBG are highly C/G-rich, thus acting as general promoters to govern their broad epithelial expression [41, 42]. In the case of C. reinhardtii [44] and C. elegans [47], the level of Rh expression is also high. In C. elegans, the Rh1 gene is driven by a strong promoter and is highly expressed in multiple sites mimicking the pattern of mammals. The high-level expression of Rh proteins and their slow evolution may be correlated with the demand for precise protein folding [124].
Location and induction of expression
As in humans, Rh proteins from C. elegans and C. reinhardtii are also routed to the plasma membrane [47, 125]. The only exception is RhgA from D. discoideum, which resides in contractile vacuoles [126], organelles that play a key role in osmoregulation [127]. A stretch of negatively charged residues clustered in the C-tail of RhgA appears crucial for this targeting [128]. In human red cells, Rh30 and RhAG are vertically linked to the cytoskeleton through their C-tails. Whereas Rh30 is present in both ankyrin R and protein 4.1R-based complexes [129], in an ankyrin-based assembly RhAG associates Rh30 and band three as a macrocomplex, which is thought to be an integral gas exchange metabolon for CO2 transport [129–131]. In contrast, RhCG and RhBG are found in nearly all types of epithelia and are targeted to distinct membrane domains: RhCG is mainly apical and RhBG mainly basolateral. For RhBG, its C-tail tyrosine-sorting signal appears crucial for basolateral location and its C-tail may attach to the ankyrin G-based network in MDCK cells [132]. However, RhCG and RhBG may not always occur in the same cell type or the same tubular structure. In mice, RhBG is found in periveinous hepatocytes [133] but coexpressed with RhCG in gastrointestinal tracts [134]. In rats, Rhbg is ubiquitously expressed in the alimentary tract, i.e., esophagus, stomach, duodenum, jejunum, ileum, and colon, and Rhcg is coexpressed with Rhbg in these tissues except the stomach and colon [135]. In human RhCG is widely found in esophageal epithelia [121] and various renal tubules [136, 137], but RhBG is barely detected in kidneys [138]. The differences may reflect organ-specific physiology or species-specific adaptation.
The plasma-membrane homing means Rh proteins face external challenges directly in unicellular eukaryotes and indirectly in multicellular animals. Thus Rh genes are prone to induction by developmental cues and environmental perturbations. In C. reinhardtii, the expression of Rh1 gene or protein is specifically induced by high CO2 [44]. In C. elegans, Rh genes show increased, differential expression in a stage-specific manner as required for embryonic and adult development [47]. In mammals, Rh genes in epithelial tissues and erythroid cells show an elevated expression in a stage-specific pattern [43, 118]. In whole animals such as rodents, epithelial Rh expression also responds to changes in internal milieu and food sources. In rats the expression of Rhbg and Rhcg responds to metabolic acidosis [139]. In mice, depleting potassium from food alters Rh expression in the renal collecting duct, resulting in Rhcg up-regulation and Rhbg down-regulation, respectively [140].
The Rh gene family and human disease associations
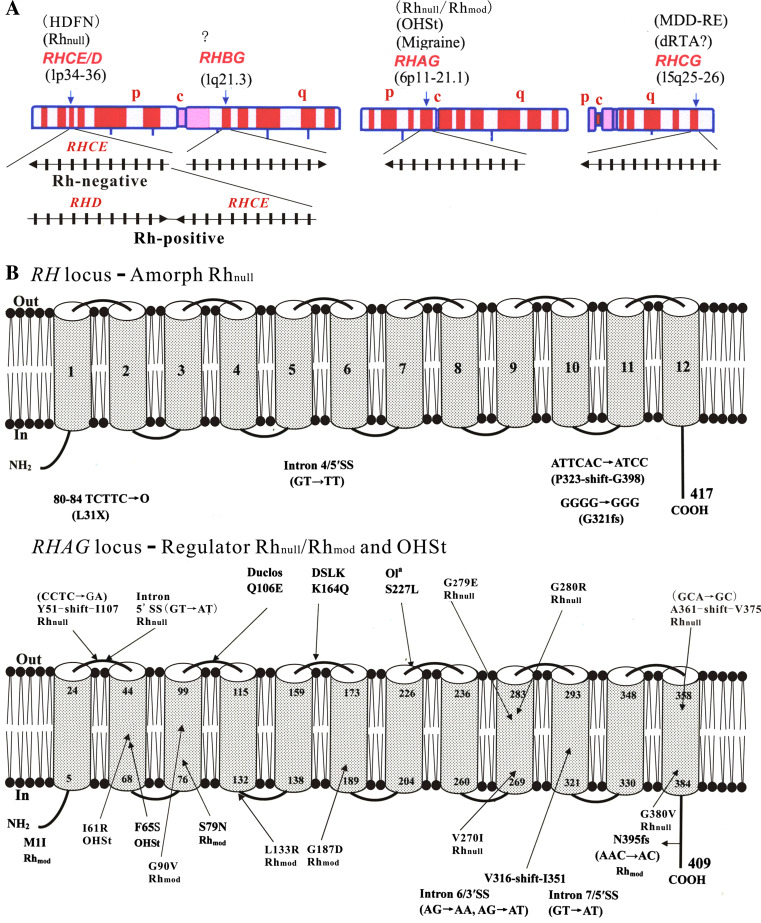
The genes of the human Rh family are located on three chromosomes (Fig. 6a); they share synteny with mouse homologs in both exon–intron structure and the linkage map that characterize the subgroups [11, 43]. The association of erythroid Rh proteins with red cell disorders has been well established [11, 12, 27]. Rh antigens, particularly the D antigen, play a major part in HDFN due to the incompatibility between Rh-negative and Rh-positive blood [3, 4] (Fig. 6a, left). Hence accurate RH genotyping forms a core of HDFN management and transfusion therapy in clinical settings [141–143]. The complete absence of red-cell Rh antigens caused by complex mutations in the RH locus defines the amorph type of Rhnull syndrome with mild phenotypes [144, 145] (Fig. 6b, upper). In contrast, genetic mutations of RHAG (Fig. 6b, lower) cause recessive Rh deficiency syndrome (regulator type Rhnull or Rhmod) [31–37, 146], or dominant over-hydrated hereditary stomatocytosis (OHSt) showing increased permeability to nomovalent cations [147]. The two genetic disorders show mild chronic hemolytic anemia and share some phenotypic features with other forms of hereditary stomatocytosis [148]. Such changes indicate that ablation or disruption of the RhAG function also affects red cell integrity.
Fig. 6.
Chromosomal location and disease association of human Rh family genes. a RH, RHBG, RHAG, and RHCG reside in chromosomes 1, 6, and 15. The exon–intron structure and orientation of each gene as well as Rh-negative and Rh-positive haplotypes are shown. c Centromere, p short arm, q long arm, HDFN hemolytic disease of the fetus and newborn, OHSt over-hydrated hereditary stomatocytosis, MDD-RE recurrent early-onset major depressive disorders, dRTA distal renal tubular acidosis. Question mark denotes unknown. b Diagram of mutations of RH and RHAG genes. Amorph type Rhnull (upper); regulator Rhnull, Rhmod, and OHSt (lower). The 12-TMH of Rh and RhAG is based on NeRh (Fig. 4). Duclos, DSLK, and Ola located on the ECLs are point changes of RhAG likely to be neutral antigenic polymorphisms
On the other hand, negative selection has placed epithelial RhBG and RhCG under highly conserved evolution [50], accounting for the current lack of their genetic mutations in humans. However, their physiologic importance has been shown by the knockdown phenotypes of orthologs from primitive species such as C. reinhardtii and C. elegans [45, 47, 48]. RhBG and RhCG have been implied as potential tumor suppressors given their sharp down-regulation in human esophageal squamous epithelial cancers [121] and mouse brain tumors [149]. Lately the physiological importance of mammalian Rh proteins has been shown in Rhcg knockout mice, which have pH perturbations in urine and epididymal fluids [49], indicating a deficit in pH balance that is primary to renal NH4 + excretion. In contrast to the negative data observed in Rhbg knockout mice [150], this work implies that the human RhCG gene, when mutated, may result in distal renal tubular acidosis and male infertility [49]. Notably human RHCG has also been identified as a candidate gene for early-onset major depressive disorder [151], and human RHAG is linked to a subtype of migraine [152] (Fig. 6a). It is hoped that these studies will stimulate future efforts to decipher the role of Rh glycoprotein genes in human diseases.
Conclusion and outlook
The last decade has seen much progress in research on the Rh family while a debate goes on regarding its substrate specificity. Current evidence suggests a role for Rh proteins as dual channels for CO2 and ammonia, whether in neutral forms or as charged species (CO2/HCO3 − vs. NH3/NH4 +). This review highlights their long history of evolution and their surprising structural conservation from prokaryotes to humans and elicits a challenging question as to their biological function: why would nature give rise to two proteins, Rh and Amt, to coexist and fulfill the same function across a wide range of organisms? The evidence accumulated is compelling that the functional role of Rh proteins is related to but distinct from Amt proteins. A mechanism is required to reconcile the substrate duality given the differences and similarities of CO2 versus NH3 in physicochemical properties. Of note is that pH is an intimate factor linked to the two molecules, but its role in transport may be overlooked in studies of Rh proteins. Thorough studies of the pH effect on the transport process of Rh and Amt may hold a key to our understanding of their function.
Acknowledgments
The authors’ work was supported by NIH grants HL54459, HD62704, and HL66274, and funds from the New York Blood Center. We are grateful to Drs. Olga Blumenfeld for helpful comments, Jason Peng for the expanded analysis of Rh and Amt in Fig. 3b, and Michael Chan and Xin Li for Fig. 5. We thank Drs. Sydney Kustu, Michael Chan, and Jonathan Kaunitz for preprints and personal communications. We also want to express our gratitude to Dr. Mohandas Narla for support and interest in the Rh topics.
References
- 1.Landsteiner K. Zur Kenntnis der antifermentativen, lytischen und agglutinierenden Wirkungen des Blutserums und der Lymphe. Zentbl Bakt Orig. 1900;27:357–362. [Google Scholar]
- 2.Landsteiner K. Ueber Agglutinationserscheinungen normalen menschlichen Blutes. Wien Klin Wochenschr. 1901;14:1132–1134. [PubMed] [Google Scholar]
- 3.Levine P, Stetson RE. An unusual case of intragroup agglutination. J Am Med Assoc. 1939;113:126–127. [Google Scholar]
- 4.Landsteiner K, Weiner AS. An agglutinable factor in human blood recognized by immune sera for Rhesus blood. Proc Soc Exp Biol Med. 1940;43:223. [Google Scholar]
- 5.Klein HG, Anstee DJ. Mollison’s blood transfusion in clinical medicine. Oxford: Blackwell; 2005. [Google Scholar]
- 6.Blumenfeld OO, Patnaik SK. Allelic genes of blood group antigens: a source of human mutations and cSNPs documented in the Blood Group Antigen Gene Mutation Database. Hum Mutat. 2004;23:8–16. doi: 10.1002/humu.10296. [DOI] [PubMed] [Google Scholar]
- 7.Watkins WM. Blood-group substances. Science. 1966;152:172–181. doi: 10.1126/science.152.3719.172. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto F, Clausen H, White T, Marken J, Hakomori S. Molecular genetic basis of the histo-blood group ABO system. Nature. 1990;345:229–233. doi: 10.1038/345229a0. [DOI] [PubMed] [Google Scholar]
- 9.Moore S, Woodrow CF, McClelland DB. Isolation of membrane components associated with human red cell antigens Rh(D), (c), (E) and Fy. Nature. 1982;295:529–531. doi: 10.1038/295529a0. [DOI] [PubMed] [Google Scholar]
- 10.Gahmberg CG. Molecular identification of the human Rho (D) antigen. FEBS Lett. 1982;140:93–97. doi: 10.1016/0014-5793(82)80528-0. [DOI] [PubMed] [Google Scholar]
- 11.Huang C-H, Liu PZ, Cheng JG. Molecular biology and genetics of the Rh blood group system. Semin Hematol. 2000;37:150–165. doi: 10.1016/s0037-1963(00)90040-4. [DOI] [PubMed] [Google Scholar]
- 12.Le Van Kim C, Colin Y, Cartron JP. Rh proteins: key structural and functional components of the red cell membrane. Blood Rev. 2006;20:93–110. doi: 10.1016/j.blre.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Race RR, Sanger R (1975) Blood groups in man. Blackwell, Oxford
- 14.Clark CA, Donohoe WTA, Durkin C, et al. Prevention of Rh-haemolytic disease: results of the clinical trial: a combined study from centres in England and Baltimore. Br Med J. 1966;2:907–914. doi: 10.1136/bmj.2.5519.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollack W, Gorman JG, Freda VJ, Ascari WQ, Allen AE, Baker WJ. Results of clinical trials of RhoGAM in women. Transfusion. 1968;8:151–153. doi: 10.1111/j.1537-2995.1968.tb04895.x. [DOI] [PubMed] [Google Scholar]
- 16.Moore S, Green C. The identification of specific Rhesus-polypeptide-blood-group-ABH-active-glycoprotein complexes in the human red-cell membrane. Biochem J. 1987;244:735–741. doi: 10.1042/bj2440735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saboori AM, Smith BL, Agre P. Polymorphism in the Mr 32,000 Rh protein purified from Rh(D)-positive and -negative erythrocytes. Proc Natl Acad Sci USA. 1988;85:4042–4045. doi: 10.1073/pnas.85.11.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloy C, Blanchard D, Dahr W, Beyreuther K, Salmon C, Cartron JP. Determination of the N-terminal sequence of human red cell Rh(D) polypeptide and demonstration that the Rh(D), (c), and (E) antigens are carried by distinct polypeptide chains. Blood. 1988;72:661–666. [PubMed] [Google Scholar]
- 19.Avent ND, Ridgwell K, Mawby WJ, Tanner MJ, Anstee DJ, Kumpel B. Protein-sequence studies on Rh-related polypeptides suggest the presence of at least two groups of proteins which associate in the human red-cell membrane. Biochem J. 1988;256:1043–1046. doi: 10.1042/bj2561043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avent ND, Ridgwell K, Tanner MJ, Anstee DJ. cDNA cloning of a 30 kDa erythrocyte membrane protein associated with Rh (Rhesus)-blood-group-antigen expression. Biochem J. 1990;271:821–825. doi: 10.1042/bj2710821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cherif-Zahar B, Bloy C, Le Van Kim C, Blanchard D, Bailly P, Hermand P, Salmon C, Cartron JP, Colin Y. Molecular cloning and protein structure of a human blood group Rh polypeptide. Proc Natl Acad Sci USA. 1990;87:6243–6247. doi: 10.1073/pnas.87.16.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Van Kim C, Mouro I, Cherif-Zahar B, Raynal V, Cherrier C, Cartron JP, Colin Y. Molecular cloning and primary structure of the human blood group RhD polypeptide. Proc Natl Acad Sci USA. 1992;89:10925–10929. doi: 10.1073/pnas.89.22.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arce MA, Thompson ES, Wagner S, Coyne KE, Ferdman BA, Lublin DM. Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood. 1993;82:651–655. [PubMed] [Google Scholar]
- 24.Ridgwell K, Spurr NK, Laguda B, MacGeoch C, Avent ND, Tanner MJ. Isolation of cDNA clones for a 50 kDa glycoprotein of the human erythrocyte membrane associated with Rh (rhesus) blood-group antigen expression. Biochem J. 1992;287(Pt 1):223–228. doi: 10.1042/bj2870223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouro I, Colin Y, Cherif-Zahar B, Cartron JP, Le Van Kim C. Molecular genetic basis of the human Rhesus blood group system. Nat Genet. 1993;5:62–65. doi: 10.1038/ng0993-62. [DOI] [PubMed] [Google Scholar]
- 26.Huang C-H. Molecular insights into the Rh protein family and associated antigens. Curr Opin Hematol. 1997;4:94–103. doi: 10.1097/00062752-199704020-00004. [DOI] [PubMed] [Google Scholar]
- 27.Avent ND, Reid ME. The Rh blood group system: a review. Blood. 2000;95:375–387. [PubMed] [Google Scholar]
- 28.Wagner FF, Flegel WA. Review: the molecular basis of the Rh blood group phenotypes. Immunohematology. 2004;20:23–36. [PMC free article] [PubMed] [Google Scholar]
- 29.Avent ND, Madgett TE, Lee ZE, Head DJ, Maddocks DG, Skinner LH. Molecular biology of Rh proteins and relevance to molecular medicine. Expert Rev Mol Med. 2006;8:1–20. doi: 10.1017/S1462399406010969. [DOI] [PubMed] [Google Scholar]
- 30.Westhoff CM. The structure and function of the Rh antigen complex. Semin Hematol. 2007;44:42–50. doi: 10.1053/j.seminhematol.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherif-Zahar B, Raynal V, Gane P, Mattei MG, Bailly P, Gibbs B, Colin Y, Cartron JP. Candidate gene acting as a suppressor of the RH locus in most cases of Rh-deficiency. Nat Genet. 1996;12:168–173. doi: 10.1038/ng0296-168. [DOI] [PubMed] [Google Scholar]
- 32.Huang C-H. The human Rh50 glycoprotein gene. Structural organization and associated splicing defect resulting in Rh(null) disease. J Biol Chem. 1998;273:2207–2213. doi: 10.1074/jbc.273.4.2207. [DOI] [PubMed] [Google Scholar]
- 33.Huang C-H, Liu Z, Cheng G, Chen Y. Rh50 glycoprotein gene and Rh-null disease: a silent splice donor is trans to a Gly279 --> Glu missense mutation in the conserved transmembrane segment. Blood. 1998;92:1776–1784. [PubMed] [Google Scholar]
- 34.Hyland CA, Cherif-Zahar B, Cowley N, Raynal V, Parkes J, Saul A, Cartron JP. A novel single missense mutation identified along the RH50 gene in a composite heterozygous Rhnull blood donor of the regulator type. Blood. 1998;91:1458–1463. [PubMed] [Google Scholar]
- 35.Cherif-Zahar B, Matassi G, Raynal V, Gane P, Delaunay J, Arrizabalaga B, Cartron JP. Rh-deficiency of the regulator type caused by splicing mutations in the human RH50 gene. Blood. 1998;92:2535–2540. [PubMed] [Google Scholar]
- 36.Huang C-H, Cheng GJ, Reid ME, Chen Y. Rhmod syndrome: a family study of the translation-initiator mutation in the Rh50 glycoprotein gene. Am J Hum Genet. 1999;64:108–117. doi: 10.1086/302215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C-H, Cheng G, Liu Z, Chen Y, Reid ME, Halverson G, Okubo Y. Molecular basis for Rh(null) syndrome: identification of three new missense mutations in the Rh50 glycoprotein gene. Am J Hematol. 1999;62:25–32. doi: 10.1002/(sici)1096-8652(199909)62:1<25::aid-ajh5>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 38.Busch W, Saier MH., Jr The transporter classification (TC) system. Crit Rev Biochem Mol Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 39.Agre P, Cartron JP. Molecular biology of the Rh antigens. Blood. 1991;78:551–563. [PubMed] [Google Scholar]
- 40.Anstee DJ, Tanner MJ. Biochemical aspects of the blood group Rh (rhesus) antigens. Baillieres Clin Haematol. 1993;6:401–422. doi: 10.1016/s0950-3536(05)80152-0. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z, Chen Y, Mo R, Hui C, Cheng JF, Mohandas N, Huang C-H. Characterization of human RhCG and mouse Rhcg as novel nonerythroid Rh glycoprotein homologues predominantly expressed in kidney and testis. J Biol Chem. 2000;275:25641–25651. doi: 10.1074/jbc.M003353200. [DOI] [PubMed] [Google Scholar]
- 42.Liu Z, Peng J, Mo R, Hui C, Huang C-H. Rh type B glycoprotein is a new member of the Rh superfamily and a putative ammonia transporter in mammals. J Biol Chem. 2001;276:1424–1433. doi: 10.1074/jbc.M007528200. [DOI] [PubMed] [Google Scholar]
- 43.Huang C-H, Liu PZ. New insights into the Rh superfamily of genes and proteins in erythroid cells and nonerythroid tissues. Blood Cells Mol Dis. 2001;27:90–101. doi: 10.1006/bcmd.2000.0355. [DOI] [PubMed] [Google Scholar]
- 44.Soupene E, King N, Feild E, Liu P, Niyogi KK, Huang C-H, Kustu S. Rhesus expression in a green alga is regulated by CO2 . Proc Natl Acad Sci USA. 2002;99:7769–7773. doi: 10.1073/pnas.112225599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soupene E, Inwood W, Kustu S. Lack of the Rhesus protein Rh1 impairs growth of the green alga Chlamydomonas reinhardtii at high CO2 . Proc Natl Acad Sci USA. 2004;101:7787–7792. doi: 10.1073/pnas.0401809101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kustu S, Inwood W. Biological gas channels for NH3 and CO2: evidence that Rh (Rhesus) proteins are CO2 channels. Transfus Clin Biol. 2006;13:103–110. doi: 10.1016/j.tracli.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 47.Ji Q, Hashmi S, Liu Z, Zhang J, Chen Y, Huang C-H. CeRh1 (rhr-1) is a dominant Rhesus gene essential for embryonic development and hypodermal function in Caenorhabditis elegans . Proc Natl Acad Sci USA. 2006;103:5881–5886. doi: 10.1073/pnas.0600901103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharabi K, Hurwitz A, Simon AJ, Beitel GJ, Morimoto RI, Rechavi G, Sznajder JI, Gruenbaum Y. Elevated CO2 levels affect development, motility, and fertility and extend life span in Caenorhabditis elegans . Proc Natl Acad Sci USA. 2009;106:4024–4029. doi: 10.1073/pnas.0900309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biver S, Belge H, Bourgeois S, Van Vooren P, Nowik M, Scohy S, Houillier P, Szpirer J, Szpirer C, Wagner CA, Devuyst O, Marini AM. A role for Rhesus factor Rhcg in renal ammonium excretion and male fertility. Nature. 2008;456:339–343. doi: 10.1038/nature07518. [DOI] [PubMed] [Google Scholar]
- 50.Huang C-H, Peng J. Evolutionary conservation and diversification of Rh family genes and proteins. Proc Natl Acad Sci USA. 2005;102:15512–15517. doi: 10.1073/pnas.0507886102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peng J, Huang C-H. Rh proteins versus Amt proteins: an organismal and phylogenetic perspective on CO2 and NH3 gas channels. Transfus Clin Biol. 2006;13:85–94. doi: 10.1016/j.tracli.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 52.Huang C-H. Molecular origin and variability of the Rh gene family: an overview of evolution, genetics and function (The 13th Congress of the European Haematology Association, Copenhagen, Denmark) Haematologica. 2008;2:149–157. [Google Scholar]
- 53.Ramos JL. Lessons from the genome of a lithoautotroph: making biomass from almost nothing. J Bacteriol. 2003;185:2690–2691. doi: 10.1128/JB.185.9.2690-2691.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norton JM, Klotz MG, Stein LY, Arp DJ, Bottomley PJ, Chain PS, Hauser LJ, Land ML, Larimer FW, Shin MW, Starkenburg SR. Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl Environ Microbiol. 2008;74:3559–3572. doi: 10.1128/AEM.02722-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Medigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJ, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MS, Wagner M, Le Paslier D. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- 56.Lindsay MR, Webb RI, Strous M, Jetten MS, Butler MK, Forde RJ, Fuerst JA. Cell compartmentalisation in planctomycetes: novel types of structural organisation for the bacterial cell. Arch Microbiol. 2001;175:413–429. doi: 10.1007/s002030100280. [DOI] [PubMed] [Google Scholar]
- 57.Fuerst JA. Intracellular compartmentation in planctomycetes. Annu Rev Microbiol. 2005;59:299–328. doi: 10.1146/annurev.micro.59.030804.121258. [DOI] [PubMed] [Google Scholar]
- 58.Drake HL, Gossner AS, Daniel SL. Old acetogens, new light. Ann N Y Acad Sci. 2008;1125:100–128. doi: 10.1196/annals.1419.016. [DOI] [PubMed] [Google Scholar]
- 59.Widdel F, Pfennig N. Sporulation and further nutritional characteristics of Desulfotomaculum acetoxidans . Arch Microbiol. 1981;129:401–402. doi: 10.1007/BF00406471. [DOI] [PubMed] [Google Scholar]
- 60.Holmes DE, O’Neil RA, Vrionis HA, N’Guessan L, A, Ortiz-Bernad I, Larrahondo MJ, Adams LA, Ward JA, Nicoll JS, Nevin KP, Chavan MA, Johnson JP, Long PE, Lovley DR (2007) Subsurface clade of Geobacteraceae that predominates in a diversity of Fe(III)-reducing subsurface environments. ISME J 1:663–677 [DOI] [PubMed]
- 61.Hugenholtz P, Goebel BM, Pace NR. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ohno S. Evolution by gene duplication. New York: Springer; 1970. [Google Scholar]
- 63.King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, Marr M, Pincus D, Putnam N, Rokas A, Wright KJ, Zuzow R, Dirks W, Good M, Goodstein D, Lemons D, Li W, Lyons JB, Morris A, Nichols S, Richter DJ, Salamov A, Sequencing JG, Bork P, Lim WA, Manning G, Miller WT, McGinnis W, Shapiro H, Tjian R, Grigoriev IV, Rokhsar D. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, Benito-Gutierrez EL, Dubchak I, Garcia-Fernandez J, Gibson-Brown JJ, Grigoriev IV, Horton AC, de Jong PJ, Jurka J, Kapitonov VV, Kohara Y, Kuroki Y, Lindquist E, Lucas S, Osoegawa K, Pennacchio LA, Salamov AA, Satou Y, Sauka-Spengler T, Schmutz J, Shin IT, Toyoda A, Bronner-Fraser M, Fujiyama A, Holland LZ, Holland PW, Satoh N, Rokhsar DS. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 65.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 66.Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- 67.Seack J, Pancer Z, Muller IM, Muller WE. Molecular cloning and primary structure of a Rhesus (Rh)-like protein from the marine sponge Geodia cydonium . Immunogenet. 1997;46:493–498. doi: 10.1007/s002510050310. [DOI] [PubMed] [Google Scholar]
- 68.Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, Signorovitch AY, Moreno MA, Kamm K, Grimwood J, Schmutz J, Shapiro H, Grigoriev IV, Buss LW, Schierwater B, Dellaporta SL, Rokhsar DS. The Trichoplax genome and the nature of placozoans. Nature. 2008;454:955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- 69.Leys S, Nichols SA, Adams EDM. Epithelia and integration in sponges. Integr Comp Biol. 2009;49:167–177. doi: 10.1093/icb/icp038. [DOI] [PubMed] [Google Scholar]
- 70.Schierwater B, de Jong D, Desalle R. Placozoa and the evolution of Metazoa and intrasomatic cell differentiation. Int J Biochem Cell Biol. 2009;41:370–379. doi: 10.1016/j.biocel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 71.Kitano T, Sumiyama K, Shiroishi T, Saitou N. Conserved evolution of the Rh50 gene compared to its homologous Rh blood group gene. Biochem Biophys Res Commun. 1998;249:78–85. doi: 10.1006/bbrc.1998.9074. [DOI] [PubMed] [Google Scholar]
- 72.Matassi G, Cherif-Zahar B, Pesole G, Raynal V, Cartron JP. The members of the RH gene family (RH50 and RH30) followed different evolutionary pathways. J Mol Evol. 1999;48:151–159. doi: 10.1007/pl00006453. [DOI] [PubMed] [Google Scholar]
- 73.Huang C-H, Liu Z, Apoil PA, Blancher A. Sequence, organization, and evolution of Rh50 glycoprotein genes in nonhuman primates. J Mol Evol. 2000;51:76–87. doi: 10.1007/s002390010068. [DOI] [PubMed] [Google Scholar]
- 74.Kitano T, Saitou N. Evolutionary history of the Rh blood group-related genes in vertebrates. Immunogenet. 2000;51:856–862. doi: 10.1007/s002510000202. [DOI] [PubMed] [Google Scholar]
- 75.Marini AM, Urrestarazu A, Beauwens R, Andre B. The Rh (rhesus) blood group polypeptides are related to NH4 + transporters. Trends Biochem Sci. 1997;22:460–461. doi: 10.1016/s0968-0004(97)01132-8. [DOI] [PubMed] [Google Scholar]
- 76.Li X, Jayachandran S, Nguyen HH, Chan MK. Structure of the Nitrosomonas europaea Rh protein. Proc Natl Acad Sci USA. 2007;104:19279–19284. doi: 10.1073/pnas.0709710104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lupo D, Li XD, Durand A, Tomizaki T, Cherif-Zahar B, Matassi G, Merrick M, Winkler FK. The 1.3-A resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. Proc Natl Acad Sci USA. 2007;104:19303–19308. doi: 10.1073/pnas.0706563104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gruswitz F, Chaudhary S, Ho JD, Pezeshki B, Ho C-M, Stroud R (2009) Crystal structure of the human Rhesus glycoprotein RhCG, PDB|3HD6|A
- 79.Burton NM, Anstee DJ. Structure, function and significance of Rh proteins in red cells. Curr Opin Hematol. 2008;15:625–630. doi: 10.1097/MOH.0b013e328311f422. [DOI] [PubMed] [Google Scholar]
- 80.Conroy MJ, Bullough PA, Merrick M, Avent ND. Modelling the human rhesus proteins: implications for structure and function. Br J Haematol. 2005;131:543–551. doi: 10.1111/j.1365-2141.2005.05786.x. [DOI] [PubMed] [Google Scholar]
- 81.Inwood WB, Hall JA, Kim KS, Fong R, Kustu S (2009) Genetic evidence for an essential oscillation of transmembrane spanning segment 5 in the Escherichia coli ammonium channel AmtB. Genetics. doi:10.1534/genetics.109.109579 [DOI] [PMC free article] [PubMed]
- 82.Avent ND, Butcher SK, Liu W, Mawby WJ, Mallinson G, Parsons SF, Anstee DJ, Tanner MJ. Localization of the C termini of the Rh (rhesus) polypeptides to the cytoplasmic face of the human erythrocyte membrane. J Biol Chem. 1992;267:15134–15139. [PubMed] [Google Scholar]
- 83.Eyers SA, Ridgwell K, Mawby WJ, Tanner MJ. Topology and organization of human Rh (rhesus) blood group-related polypeptides. J Biol Chem. 1994;269:6417–6423. [PubMed] [Google Scholar]
- 84.von Wirén N, Merrick M. Regulation and function of ammonium carriers in bacteria, fungi and plants. Topics Curr Genet. 2004;9:95–120. [Google Scholar]
- 85.Khademi S, O’Connell J, 3rd, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 86.Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli . Proc Natl Acad Sci USA. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus . Proc Natl Acad Sci USA. 2005;102:14994–14999. doi: 10.1073/pnas.0506254102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Weidinger K, Neuhauser B, Gilch S, Ludewig U, Meyer O, Schmidt I. Functional and physiological evidence for a rhesus-type ammonia transporter in Nitrosomonas europaea . FEMS Microbiol Lett. 2007;273:260–267. doi: 10.1111/j.1574-6968.2007.00805.x. [DOI] [PubMed] [Google Scholar]
- 89.Cherif-Zahar B, Durand A, Schmidt I, Hamdaoui N, Matic I, Merrick M, Matassi G. Evolution and functional characterization of the RH50 gene from the ammonia-oxidizing bacterium Nitrosomonas europaea . J Bacteriol. 2007;189:9090–9100. doi: 10.1128/JB.01089-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Khademi S, Stroud RM. The Amt/MEP/Rh family: structure of AmtB and the mechanism of ammonia gas conduction. Physiology (Bethesda) 2006;21:419–429. doi: 10.1152/physiol.00051.2005. [DOI] [PubMed] [Google Scholar]
- 91.Javelle A, Lupo D, Li XD, Merrick M, Chami M, Ripoche P, Winkler FK. Structural and mechanistic aspects of Amt/Rh proteins. J Struct Biol. 2007;158:472–481. doi: 10.1016/j.jsb.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 92.Fong RN, Kim KS, Yoshihara C, Inwood WB, Kustu S. The W148L substitution in the Escherichia coli ammonium channel AmtB increases flux and indicates that the substrate is an ion. Proc Natl Acad Sci USA. 2007;104:18706–18711. doi: 10.1073/pnas.0709267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Callebaut I, Dulin F, Bertrand O, Ripoche P, Mouro I, Colin Y, Mornon JP, Cartron JP. Hydrophobic cluster analysis and modeling of the human Rh protein three-dimensional structures. Transfus Clin Biol. 2006;13:70–84. doi: 10.1016/j.tracli.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 94.Merrick M, Javelle A, Durand A, Severi E, Thornton J, Avent ND, Conroy MJ, Bullough PA. The Escherichia coli AmtB protein as a model system for understanding ammonium transport by Amt and Rh proteins. Transfus Clin Biol. 2006;13:97–102. doi: 10.1016/j.tracli.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 95.Javelle A, Lupo D, Zheng L, Li XD, Winkler FK, Merrick M. An unusual twin-his arrangement in the pore of ammonia channels is essential for substrate conductance. J Biol Chem. 2006;281:39492–39498. doi: 10.1074/jbc.M608325200. [DOI] [PubMed] [Google Scholar]
- 96.Chamberlain AK, Faham S, Yohannan S, Bowie JU. Construction of helix-bundle membrane proteins. Adv Protein Chem. 2003;63:19–46. doi: 10.1016/s0065-3233(03)63002-0. [DOI] [PubMed] [Google Scholar]
- 97.Javelle A, Lupo D, Ripoche P, Fulford T, Merrick M, Winkler FK. Substrate binding, deprotonation, and selectivity at the periplasmic entrance of the Escherichia coli ammonia channel AmtB. Proc Natl Acad Sci USA. 2008;105:5040–5045. doi: 10.1073/pnas.0711742105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Conroy MJ, Jamieson SJ, Blakey D, Kaufmann T, Engel A, Fotiadis D, Merrick M, Bullough PA. Electron and atomic force microscopy of the trimeric ammonium transporter AmtB. EMBO Rep. 2004;5:1153–1158. doi: 10.1038/sj.embor.7400296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gruswitz F, O’Connell J, Stroud RM. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. Proc Natl Acad Sci USA. 2007;104:42–47. doi: 10.1073/pnas.0609796104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Huergo LF, Chubatsu LS, Souza EM, Pedrosa FO, Steffens MB, Merrick M. Interactions between PII proteins and the nitrogenase regulatory enzymes DraT and DraG in Azospirillum brasilense. FEBS Lett. 2006;580:5232–5236. doi: 10.1016/j.febslet.2006.08.054. [DOI] [PubMed] [Google Scholar]
- 101.Chan MK (2009) Personal communication
- 102.Marini AM, Matassi G, Raynal V, Andre B, Cartron JP, Cherif-Zahar B. The human Rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat Genet. 2000;26:341–344. doi: 10.1038/81656. [DOI] [PubMed] [Google Scholar]
- 103.Westhoff CM, Ferreri-Jacobia M, Mak DO, Foskett JK. Identification of the erythrocyte Rh blood group glycoprotein as a mammalian ammonium transporter. J Biol Chem. 2002;277:12499–12502. doi: 10.1074/jbc.C200060200. [DOI] [PubMed] [Google Scholar]
- 104.Hemker MB, Cheroutre G, van Zwieten R, Maaskant-van Wijk PA, Roos D, Loos JA, van der Schoot CE, von dem Borne AE. The Rh complex exports ammonium from human red blood cells. Br J Haematol. 2003;122:333–340. doi: 10.1046/j.1365-2141.2003.04425.x. [DOI] [PubMed] [Google Scholar]
- 105.Ripoche P, Bertrand O, Gane P, Birkenmeier C, Colin Y, Cartron JP. Human Rhesus-associated glycoprotein mediates facilitated transport of NH3 into red blood cells. Proc Natl Acad Sci USA. 2004;101:17222–17227. doi: 10.1073/pnas.0403704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Benjelloun F, Bakouh N, Fritsch J, Hulin P, Lipecka J, Edelman A, Planelles G, Thomas SR, Cherif-Zahar B. Expression of the human erythroid Rh glycoprotein (RhAG) enhances both NH3 and NH4 + transport in HeLa cells. Pflugers Arch. 2005;450:155–167. doi: 10.1007/s00424-005-1381-y. [DOI] [PubMed] [Google Scholar]
- 107.Ludewig U. Electroneutral ammonium transport by basolateral rhesus B glycoprotein. J Physiol (London) 2004;559:751–759. doi: 10.1113/jphysiol.2004.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bakouh N, Benjelloun F, Hulin P, Brouillard F, Edelman A, Cherif-Zahar B, Planelles G. NH3 is involved in the NH4 + transport induced by the functional expression of the human Rh C glycoprotein. J Biol Chem. 2004;279:15975–15983. doi: 10.1074/jbc.M308528200. [DOI] [PubMed] [Google Scholar]
- 109.Nakhoul NL, Dejong H, Abdulnour-Nakhoul SM, Boulpaep EL, Hering-Smith K, Hamm LL. Characteristics of renal Rhbg as an NH4 + transporter. Am J Physiol Renal Physiol. 2005;288:F170–F181. doi: 10.1152/ajprenal.00419.2003. [DOI] [PubMed] [Google Scholar]
- 110.Zidi-Yahiaoui N, Mouro-Chanteloup I, D’Ambrosio AM, Lopez C, Gane P, Le van Kim C, Cartron JP, Colin Y, Ripoche P. Human Rhesus B and Rhesus C glycoproteins: properties of facilitated ammonium transport in recombinant kidney cells. Biochem J. 2005;391:33–40. doi: 10.1042/BJ20050657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mak DO, Dang B, Weiner ID, Foskett JK, Westhoff CM. Characterization of ammonia transport by the kidney Rh glycoproteins RhBG and RhCG. Am J Physiol Renal Physiol. 2006;290:F297–F305. doi: 10.1152/ajprenal.00147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mayer M, Schaaf G, Mouro I, Lopez C, Colin Y, Neumann P, Cartron JP, Ludewig U. Different transport mechanisms in plant and human AMT/Rh-type ammonium transporters. J Gen Physiol. 2006;127:133–144. doi: 10.1085/jgp.200509369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ludewig U. Ion transport versus gas conduction: function of AMT/Rh-type proteins. Transfus Clin Biol. 2006;13:111–116. doi: 10.1016/j.tracli.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 114.Zidi-Yahiaoui N, Callebaut I, Genetet S, Le Van Kim C, Cartron JP, Colin Y, Ripoche P, Mouro-Chanteloup I. Functional analysis of human RhCG: comparison with E. coli ammonium transporter reveals similarities in the pore and differences in the vestibule. Am J Physiol Cell Physiol. 2009;297:C537–C547. doi: 10.1152/ajpcell.00137.2009. [DOI] [PubMed] [Google Scholar]
- 115.Endeward V, Cartron JP, Ripoche P, Gros G. Red cell membrane CO2 permeability in normal human blood and in blood deficient in various blood groups, and effect of DIDS. Transfus Clin Biol. 2006;13:123–127. doi: 10.1016/j.tracli.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 116.Endeward V, Cartron JP, Ripoche P, Gros G. RhAG protein of the Rhesus complex is a CO2 channel in the human red cell membrane. FASEB J. 2008;22:64–73. doi: 10.1096/fj.07-9097com. [DOI] [PubMed] [Google Scholar]
- 117.Cherif-Zahar B, Le Van Kim C, Rouillac C, Raynal V, Cartron JP, Colin Y. Organization of the gene (RHCE) encoding the human blood group RhCcEe antigens and characterization of the promoter region. Genomics. 1994;19:68–74. doi: 10.1006/geno.1994.1014. [DOI] [PubMed] [Google Scholar]
- 118.Liu Z, Huang C-H. The mouse Rhl1 and Rhag genes: sequence, organization, expression, and chromosomal mapping. Biochem Genet. 1999;37:119–138. doi: 10.1023/a:1018726303397. [DOI] [PubMed] [Google Scholar]
- 119.Iwamoto S, Suganuma H, Kamesaki T, Omi T, Okuda H, Kajii E. Cloning and characterization of erythroid-specific DNase I-hypersensitive site in human rhesus-associated glycoprotein gene. J Biol Chem. 2000;275:27324–27331. doi: 10.1074/jbc.M003297200. [DOI] [PubMed] [Google Scholar]
- 120.Mouro-Chanteloup I, D’Ambrosio AM, Gane P, Le Van Kim C, Raynal V, Dhermy D, Cartron JP, Colin Y. Cell-surface expression of RhD blood group polypeptide is posttranscriptionally regulated by the RhAG glycoprotein. Blood. 2002;100:1038–1047. [PubMed] [Google Scholar]
- 121.Chen BS, Xu ZX, Xu X, Cai Y, Han YL, Wang J, Xia SH, Hu H, Wei F, Wu M, Wang MR. RhCG is downregulated in oesophageal squamous cell carcinomas, but expressed in multiple squamous epithelia. Eur J Cancer. 2002;38:1927–1936. doi: 10.1016/s0959-8049(02)00190-9. [DOI] [PubMed] [Google Scholar]
- 122.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 123.Son CG, Bilke S, Davis S, Greer BT, Wei JS, Whiteford CC, Chen QR, Cenacchi N, Khan J. Database of mRNA gene expression profiles of multiple human organs. Genome Res. 2005;15:443–450. doi: 10.1101/gr.3124505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yoshihara C, Inoue K, Schichnes D, Ruzin S, Inwood W, Kustu S. An Rh1-GFP fusion protein is in the cytoplasmic membrane of a white mutant strain of Chlamydomonas reinhardtii . Mol Plant. 2008;1:1007–1020. doi: 10.1093/mp/ssn074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Benghezal M, Gotthardt D, Cornillon S, Cosson P. Localization of the Rh50-like protein to the contractile vacuole in Dictyostelium . Immunogenet. 2001;52:284–288. doi: 10.1007/s002510000279. [DOI] [PubMed] [Google Scholar]
- 127.Kessin RH. Dictyostelium: evolution, cell biology, and the development of multicellularity. Cambridge, New York: Cambridge University Press; 2001. [Google Scholar]
- 128.Mercanti V, Blanc C, Lefkir Y, Cosson P, Letourneur F. Acidic clusters target transmembrane proteins to the contractile vacuole in Dictyostelium cells. J Cell Sci. 2006;119:837–845. doi: 10.1242/jcs.02808. [DOI] [PubMed] [Google Scholar]
- 129.Salomao M, Zhang X, Yang Y, Lee S, Hartwig JH, Chasis JA, Mohandas N, An X. Protein 4.1R-dependent multiprotein complex: new insights into the structural organization of the red blood cell membrane. Proc Natl Acad Sci USA. 2008;105:8026–8031. doi: 10.1073/pnas.0803225105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nicolas V, Le Van Kim C, Gane P, Birkenmeier C, Cartron JP, Colin Y, Mouro-Chanteloup I. Rh-RhAG/ankyrin-R, a new interaction site between the membrane bilayer and the red cell skeleton, is impaired by Rh(null)-associated mutation. J Biol Chem. 2003;278:25526–25533. doi: 10.1074/jbc.M302816200. [DOI] [PubMed] [Google Scholar]
- 131.Bruce LJ, Beckmann R, Ribeiro ML, Peters LL, Chasis JA, Delaunay J, Mohandas N, Anstee DJ, Tanner MJ. A band 3-based macrocomplex of integral and peripheral proteins in the RBC membrane. Blood. 2003;101:4180–4188. doi: 10.1182/blood-2002-09-2824. [DOI] [PubMed] [Google Scholar]
- 132.Lopez C, Metral S, Eladari D, Drevensek S, Gane P, Chambrey R, Bennett V, Cartron JP, Le Van Kim C, Colin Y. The ammonium transporter RhBG: requirement of a tyrosine-based signal and ankyrin-G for basolateral targeting and membrane anchorage in polarized kidney epithelial cells. J Biol Chem. 2005;280:8221–8228. doi: 10.1074/jbc.M413351200. [DOI] [PubMed] [Google Scholar]
- 133.Weiner ID, Miller RT, Verlander JW. Localization of the ammonium transporters, Rh B glycoprotein and Rh C glycoprotein, in the mouse liver. Gastroenterology. 2003;124:1432–1440. doi: 10.1016/s0016-5085(03)00277-4. [DOI] [PubMed] [Google Scholar]
- 134.Handlogten ME, Hong SP, Zhang L, Vander AW, Steinbaum ML, Campbell-Thompson M, Weiner ID. Expression of the ammonia transporter proteins Rh B glycoprotein and Rh C glycoprotein in the intestinal tract. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1036–G1047. doi: 10.1152/ajpgi.00418.2004. [DOI] [PubMed] [Google Scholar]
- 135.Kaunitz J (2009) Personal communication
- 136.Han KH, Croker BP, Clapp WL, Werner D, Sahni M, Kim J, Kim HY, Handlogten ME, Weiner ID. Expression of the ammonia transporter, Rh C glycoprotein, in normal and neoplastic human kidney. J Am Soc Nephrol. 2006;17:2670–2679. doi: 10.1681/ASN.2006020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Quentin F, Eladari D, Cheval L, Lopez C, Goossens D, Colin Y, Cartron JP, Paillard M, Chambrey R. RhBG and RhCG, the putative ammonia transporters, are expressed in the same cells in the distal nephron. J Am Soc Nephrol. 2003;14:545–554. doi: 10.1097/01.asn.0000050413.43662.55. [DOI] [PubMed] [Google Scholar]
- 138.Brown AC, Hallouane D, Mawby WJ, Karet FE, Saleem MA, Howie AJ, Toye AM. RhCG is the major putative ammonia transporter expressed in the human kidney, and RhBG is not expressed at detectable levels. Am J Physiol Renal Physiol. 2009;296:F1279–F1290. doi: 10.1152/ajprenal.00013.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Seshadri RM, Klein JD, Kozlowski S, Sands JM, Kim YH, Han KH, Handlogten ME, Verlander JW, Weiner ID. Renal expression of the ammonia transporters, Rhbg and Rhcg, in response to chronic metabolic acidosis. Am J Physiol Renal Physiol. 2006;290:F397–F408. doi: 10.1152/ajprenal.00162.2005. [DOI] [PubMed] [Google Scholar]
- 140.Cheval L, Duong Van Huyen JP, Bruneval P, Verbavatz JM, Elalouf JM, Doucet A. Plasticity of mouse renal collecting duct in response to potassium depletion. Physiol Genomics. 2004;19:61–73. doi: 10.1152/physiolgenomics.00055.2004. [DOI] [PubMed] [Google Scholar]
- 141.Avent ND. Large-scale blood group genotyping: clinical implications. Br J Haematol. 2009;144:3–13. doi: 10.1111/j.1365-2141.2008.07285.x. [DOI] [PubMed] [Google Scholar]
- 142.Anstee DJ. Red cell genotyping and the future of pretransfusion testing. Blood. 2009;114:248–256. doi: 10.1182/blood-2008-11-146860. [DOI] [PubMed] [Google Scholar]
- 143.Illanes S, Soothill P. Current aspects of the clinical management of haemolytic disease of the newborn and fetus (The 13th Congress of the European Haematology Association, Copenhagen, Denmark) Haematologica. 2008;2:175–178. [Google Scholar]
- 144.Huang C-H, Chen Y, Reid ME, Seidl C. Rhnull disease: the amorph type results from a novel double mutation in RhCe gene on D-negative background. Blood. 1998;92:664–671. [PubMed] [Google Scholar]
- 145.Cherif-Zahar B, Matassi G, Raynal V, Gane P, Mempel W, Perez C, Cartron JP. Molecular defects of the RHCE gene in Rh-deficient individuals of the amorph type. Blood. 1998;92:639–646. [PubMed] [Google Scholar]
- 146.Tilley L, Green C, Poole J, Gaskell A, Ridgwell K, Burton NM, Uchikawa M, Tsuneyama H, Ogasawara K, Akkok CA, Daniels G (2009) A new blood group system, RHAG: three antigens resulting from amino acid substitutions in the Rh-associated glycoprotein. Vox Sang. doi:10.1111/j.1423-0410.2009.01243.x [DOI] [PubMed]
- 147.Bruce LJ, Guizouarn H, Burton NM, Gabillat N, Poole J, Flatt JF, Brady RL, Borgese F, Delaunay J, Stewart GW. The monovalent cation leak in overhydrated stomatocytic red blood cells results from amino acid substitutions in the Rh-associated glycoprotein. Blood. 2009;113:1350–1357. doi: 10.1182/blood-2008-07-171140. [DOI] [PubMed] [Google Scholar]
- 148.Bruce LJ. Hereditary stomatocytosis and cation-leaky red cells–recent developments. Blood Cells Mol Dis. 2009;42:216–222. doi: 10.1016/j.bcmd.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 149.Johansson FK, Brodd J, Eklof C, Ferletta M, Hesselager G, Tiger CF, Uhrbom L, Westermark B. Identification of candidate cancer-causing genes in mouse brain tumors by retroviral tagging. Proc Natl Acad Sci USA. 2004;101:11334–11337. doi: 10.1073/pnas.0402716101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chambrey R, Goossens D, Bourgeois S, Picard N, Bloch-Faure M, Leviel F, Geoffroy V, Cambillau M, Colin Y, Paillard M, Houillier P, Cartron JP, Eladari D. Genetic ablation of Rhbg in the mouse does not impair renal ammonium excretion. Am J Physiol Renal Physiol. 2005;289:F1281–F1290. doi: 10.1152/ajprenal.00172.2005. [DOI] [PubMed] [Google Scholar]
- 151.Verma R, Holmans P, Knowles JA, Grover D, Evgrafov OV, Crowe RR, Scheftner WA, Weissman MM, DePaulo JR, Jr, Potash JB, Levinson DF. Linkage disequilibrium mapping of a chromosome 15q25–26 major depression linkage region and sequencing of NTRK3. Biol Psychiatry. 2008;63:1185–1189. doi: 10.1016/j.biopsych.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Norberg A, Forsgren L, Holmberg D, Holmberg M. Exclusion of the juvenile myoclonic epilepsy gene EFHC1 as the cause of migraine on chromosome 6, but association to two rare polymorphisms in MEP1A and RHAG. Neurosci Lett. 2006;396:137–142. doi: 10.1016/j.neulet.2005.11.039. [DOI] [PubMed] [Google Scholar]