Abstract
The last several decades have seen an explosion of knowledge in the field of structural biology. With critical advances in spectroscopic techniques in examining structures of biomacromolecules, in maturation of molecular biology techniques, as well as vast improvements in computation prowess, protein structures are now being elucidated at an unprecedented rate. In spite of all the recent advances, the protein folding puzzle remains as one of the fundamental biochemical challenges. A facet to this empiric problem is the structural determinants of protein folding. What are the driving forces that pivot a polypeptide chain to a specific conformation amongst the vast conformation space? In this review, we shall discuss some of the structural determinants to protein folding that have been identified in the recent decades.
Keywords: Protein folding, Folding models, Folding propensity, Structural determinants, Posttranslational modifications, Secondary structures
Introduction
Protein folding is one of the most fundamental and yet most intriguing questions of structural biology. As aptly described by Honig et al., the protein folding puzzle is a “classical challenge in which the problem is easy to state, existence of an answer is apparent, and yet the solution remains elusive” [1]. Simply stated, how does a linear polypeptide chain of scrambled amino acid residues fold into a native distinctive conformation with pinpoint accuracy?
The impetus for in vitro protein folding research stems from two of the most well-known theories of protein folding: Anfinsen et al. [2, 3] on the folding of ribonuclease A, and the Levinthal Paradox by Cyprus Levinthal [4, 5]. In 1969, Anfinsen et al. demonstrated that ribonuclease A can spontaneously refold into its native conformation in the in vitro setting, without assistance from biological systems or other biomacromolecules (Fig. 1). These findings led to the next question: is this native conformation achieved by a random search of the vast conformational space that is available to the linear polypeptide? It was subsequently pointed out by Cyprus Levinthal that such a random search for the native conformational state of a protein would require an infinitesimally long time. For example, a 100-residue protein taking a mere 10−11 s per configuration, will take 1052 years to search through the entire set of possible conformations [5, 6]! This is in stark contrast to the millisecond time scale that a protein typically takes to fold to its native conformation. Therefore, it is apparent that there exist specific folding intermediates or pathways through which the proteins can adopt the final destined conformation. The implications from these theories triggered extensive research on the identification of folding intermediates [7], real-time folding studies [8], computation modeling and simulations [9, 10], and energy landscape analysis [11]. From these pioneering works, several models on mechanism of protein folding were conceptualized [12].
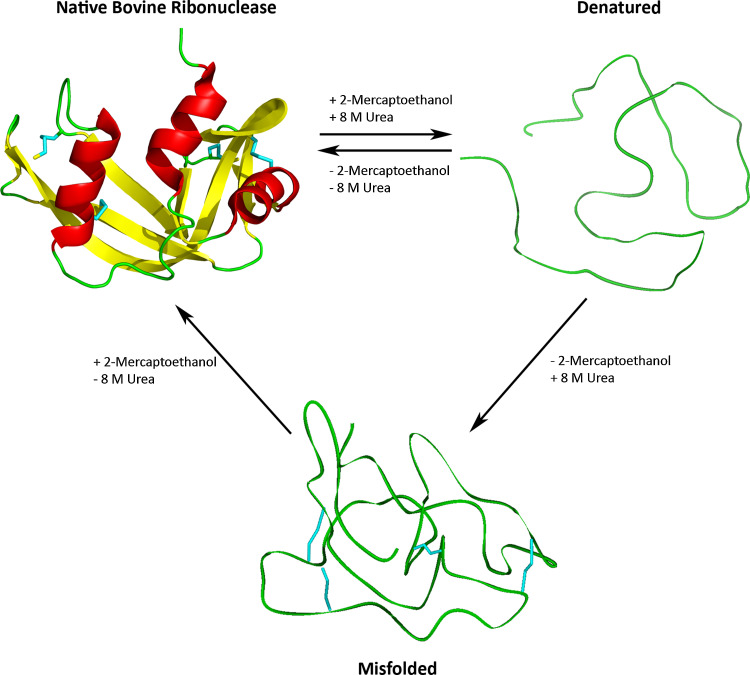
Fig. 1.
Spontaneous refolding of bovine ribonuclease. Christian Anfinson et al. demonstrated that native bovine ribonuclease (PDB ID 7RSA) denatured with 8 M urea and 2-mercaptoethanol spontaneously refolds to the fully active refolded conformation upon removal of the denaturing reagents. Oxidation of the fully denatured polypeptide in the presence of 8 M urea in the absence of sulfhydryl group containing 2-mercaptoethanol results in a “scrambled” mixture of misfolded disulfide isomers. Subsequent removal of urea and addition of 2-mercaptoethanol facilitated disulfide rearrangement of the misfolded protein to the native conformation and restoration of full enzymatic activity [2, 3]
One of the first models of protein folding was the nucleation growth model, in which a predominant structural nucleus is formed, and the secondary and tertiary structures of the protein propagate from this nucleus [13]. However, this mechanism was subsequently less favored due to the inability to account for folding intermediates present in folding pathways [14]. The next theoretical mechanism for protein folding was suggested by Oleg Ptitsyn, who proposed that protein folding occurs through a sequential on-pathway process with the formation of secondary structures in an initial “molten globular” state, with each stage of the dynamic protein folding process stabilized by the former stage [14, 15]. This model was subsequently formulated as the diffusion–collision model and the related framework model, which theorized that structural elements rapidly assemble independently before “colliding” to coalesce and stabilize by interactions between secondary structures to form the final tertiary form [16, 17]. A third model of protein folding was the hydrophobic collapse model, which proposed that the collapse of the primary structure around a hydrophobic core reduces the scope of search for the native conformation [18]. Subsequently, it was suggested that secondary and tertiary structures in the framework model were formed in conjunction with the hydrophobic collapse of the polypeptide chain, leading to unification of the two folding models to the nucleation–condensation model [19, 20]. However, the various models are not without problems. For example, the hydrophobic interactions in the hydrophobic collapse model may indicate difficulties in structural reorganization of the polypeptide chain along the folding pathway. Further, it is unlikely that the unfolded state of a polypeptide exists in the form of a completely unstructured chain [21]. By examining the denatured states of a small model protein, Shortle et al. [22] further proposed that long range ordering of the polypeptide segments could very well occur before emergence of defined structural elements of the protein.
One of the critical advances in the understanding of folding pathways was spearheaded through the development of Φ-value analysis by Fersht et al. [23]. For this, the kinetics of mutants, in which deliberate perturbations were made to specific interactions in the folding pathway, were measured so as to examine the folding pathways concerned. In effect, these Φ-values are structural indices for specific residues in the folding intermediate [24]. Numerous recent innovative experimental applications or strategies have subsequently been devised to aid the understanding of the protein folding problem. These strategies include, but are not limited to, NMR spectroscopy [25, 26], single molecule studies with Förster resonance energy transfer (FRET) or atomic force microscopy [27, 28], UV Raman spectroscopy [29], computational approaches and molecular dynamics simulations [30, 31], as well as theoretical analysis [32]. Through these methods, novel information that was previously inaccessible could now be obtained and even utilized in the reengineering of ultrafast folding proteins [33]. Complemented with classical strategies such as the Φ-values, these methods have been instrumental in the recent advancement of knowledge of protein folding pathways.
Although much of the specifics to folding pathways suggested by these folding models are still ambiguous, it is now known that folding occurs via a series of folding intermediates, through either a two-state (nucleation–condensation model) or three-state (diffusion–collision model) mechanism [24, 34, 35]. However, there are indications that the two-state and three-state folding models are, in fact, extremes of a common folding mechanism [21, 35], and the fundamental difference between the two routes lies in the relative stability of the folding intermediates [36]. Indeed, these two extremes of the folding mechanism have been demonstrated on single protein models, indicative of a unifying mechanism of protein folding that incorporates aspects of both two-state and three-state models [37]. Variations between proteins in following a single pathway or multiple different pathways could also be attributed to differences in the number of nucleation “foldon” motifs, as well as conservation of transition state structures within the final protein structures [38, 39]. Numerous reviews pertaining to the various experimental and theoretical aspects of the protein folding problem are available, and will not be further detailed in this review.
Although much advancement of knowledge has been made over the four decades since research into the protein folding problem intensified, this biochemical puzzle is far from being solved. It is now apparent that the various proposed models of protein folding are not likely to operate in a mutually exclusive fashion, and that the folding mechanisms are likely to vary between proteins [40]. Whilst these proposed folding models may be conceptually different, a commonality seen across the models is the formation of secondary structure elements.
Despite the fact that several groups have been successful in designing small protein molecules to specific conformations, prediction of the structure of native structures from the amino acid sequence remains a challenge [14, 41]. It is now clear that the various associated interactions arising from the functional side chains, albeit a few weaker than others, contribute to the protein stability and protein folding process [12, 42, 43]. These forces include electrostatic interactions [44], hydrogen bonding interactions [45], van der Waals interactions [46], and hydrophobic interactions [42]. Through the use of an all-atom ab initio simulation, Yang et al. demonstrated that it is likely a combination of the multiple interacting forces that drives the protein folding process [43]. Indeed, these interacting forces are the fundamental interactions on which most, if not all, of the structural determinants of protein folding rely. These contributing forces of interaction implicated in protein folding were reviewed nearly two decades ago [42, 47].
We now have a much better grasp of the mechanisms of protein folding, and the possibility of overcoming this challenge seems increasingly promising [12]. To an extent, we also understand the various in vivo [48] and in vitro [49, 50] parameters that influence the folding profile and thermodynamic stability of these sensitive biomacromolecules. The classical experiment by Christian Anfinsen reiterates the point that the native conformation of a protein is likely to be dictated by the primary structure of the molecule [2, 51]. Therefore, it is interesting to examine the structural determinants of the polypeptide chain that influence the folding pathway and final confirmation of the mature protein. In this review, we shall discuss some of the currently known or suggested structural determinants of protein folding.
Letters of the protein language alphabet
With merely 20 amino acids in the standard polypeptide alphabet, nature has conjured an impressive array of proteins. Over 50,000 known protein structures across the various genera have been deposited in the protein data bank (http://www.rcsb.org), and an exponentially higher number of distinct protein sequences with unique three-dimensional structures remain to be elucidated [52, 53]. Such diverse permutations arise from the assorted properties of the amino acid residues with structurally and biochemically distinct side chains.
It is the diverse physicochemical properties of these structural building blocks that endow distinctive characteristics to a polypeptide chain. Further, the functional side chains of the amino acids are at least as important as the backbone atoms in laying the platform for the multiple interacting forces critical for structure, function, and stability of proteins. Indeed, the side chain “appendages” to the polypeptide backbone have been so intriguing that a sidechain-only model of protein folding has been previously proposed [54]. Through the analysis and extrapolation of information derived from databases and literature on protein folding and conformation, it was also previously suggested that certain amino acids exert specific influence on the local conformation [55]. We shall discuss how the physicochemical properties of the various amino acids preferentially result in specific structural conformations.
Propensity of forming secondary structures
The right-handed α-helix is the most commonly observed secondary structure in proteins with 3.6 residues per turn and translation of 1.5 Å per residue [56]. The geometry of a classical α-helix involves the carbonyl group hydrogen bonding to the amide proton of the i + 4 residue of the polypeptide, with side chains projecting away from the helix backbone.
The propensity of a polypeptide to form an α-helix has been shown to be dependent on various factors [57]. The most prominent factor is the amino acid constituents in the helix-forming segment, and the consequent interactions that are sterically permitted. As early as the 1960s, several other groups presented predictive methodologies for forecasting potential secondary structures based on the characteristics of the various residues [10, 58, 59]. Although specific potentials of all 20 amino acids in forming secondary structures were not consistently predicted, the most favorable and least favorable helix formers were generally in agreement. For example, aliphatic residues with branched Cβ side chains (Val, Ile, Thr) were deemed unfavorable due to the large volume of the functional group that results in steric hindrances [60]. On the other hand, alanine residues have long been recognized to possess a high helix-forming propensity. Polyalanine, polyglutamine, polylysine, and polyaspartate peptides were also shown to be able to form stable helices in solution [61]. The alignment of periodic repeats of multiple backbone hydrogen bonds running parallel to the helix axis results in a net cumulative dipole along the termini of the helix [60]. In contrast, glycines have the highest helix destabilizing effect due to the lack of a side chain functional group that results in inherent flexibility of the peptide bond [62]. Due to the lack of an amide proton, the imino acid residue proline has also been noted to be a helix breaker [63]. In view of its unique physicochemical properties, proline will be further discussed in the next section.
In addition to the hydrogen bond interactions of backbone atoms, formation of helix-stabilizing hydrogen bond interactions between side chains has also been noted to contribute to the helix formation and propagation. One of the early examples identified was the helix-stabilizing interactions between Glu2–Arg10 and Phe8–His12 in the N-terminal helical segment of RNase S that reinforced the multitude of hydrogen-bonds of the helix [64]. Incidentally, the exact interactions were also observed in the isolated corresponding C-peptide fragment of RNase S, indicating the polypeptide’s capability to autonomously nucleate into a helix [8]. Conversely, an unfavorable side chain interaction such as His12–Asp14 in the S-peptide of RNAse could present as a helix stop signal by preventing the propagation of the helix beyond residue 13 [65]. Further work by Shoemaker et al. on C-peptide variants indicated that the charge–dipole interaction of constituent charged residues with the macrodipole of the helix contributes to the enthalpy change for helix formation: negatively charged residues such as glutamate and aspartate occur preferentially near the positive N-terminal dipole of the helix, while positively charged residues such as lysine, arginine, and histidine commonly reside at the negatively charged C-terminal dipole of the helix [64, 66]. These findings were supported by similar observations in other helical peptides [67].
In the bid to decipher the protein folding problem, one of the fundamental questions is to comprehend the factors that influence short amino acid sequences to fold into various conformations in solution. Early predictive algorithms involved basic parameters such as nucleation and elongation factors [68], and subsequently incorporated contributions of side chain interactions to improve the prediction accuracy of helix formation [69]. However, these algorithms were still unable to predict helix-coil transition in solution. The comparatively most accurate predictive algorithm for helix formation of various residues in non-repetitive sequences is the AGADIR algorithm developed by Muñoz et al. [70–72] The AGADIR algorithm incorporated several of the contributions discussed above, including specific amino acid-dependent entropic cost, hydrogen bonds interactions between main chain and side chains, as well as capping and dipole effects. Through correlation with NMR analysis, AGADIR was shown to be capable of accurately predicting helical behavior of peptides under various experimental conditions [70].
The other secondary structure element commonly observed in protein structures is the β-sheet. The distinctive feature of the β-sheet is the ability to form either parallel, or anti-parallel, sheets capable of bringing residues that are distant in sequence into close proximity of one another. Unlike the periodic backbone hydrogen bonding in α-helices, the backbone hydrogen bonding pattern in β-sheets is dependent on the orientation of the β-strands [73].
The propensity of a polypeptide chain to form a β-sheet is also influenced by the specific amino acid constituents. Through the statistical analysis of known protein structures and experimental measurements of individual amino acids in the “host–guest” method initially utilized for analyzing helices [62], residues with Cβ-branched side chains or aromatic side chains were noted to be most favorable in the formation of β-sheets [10, 73]. Although these findings could indicate these amino acids’ importance and tendency of forming β-sheets, the frequent observation of β-sheets in hydrophobic cores of proteins suggests their concurrent importance in hydrophobic interactions [73]. As with α-helices, flexible or disorder-promoting residues (glycine and proline) are similarly disfavored in the formation of β-sheet structures. However, statistical analyses have shown that these flexible residues and certain residues with small polar side chains, such as asparagine, aspartic acid, and serine, are favorable for forming the β-turns by satisfying supporting hydrogen bonding requirements of the main chain amide [73–75]. Residues that resulted in stabilizing interactions were also more frequently located in β-sheet structures. For example, Phe-Phe, Phe-Tyr, Glu-Arg, and Glu-Lys interactions were identified by experimental and statistical methods to occur most frequently in β-sheets, while Thr-Val and Thr-Trp interactions were the least frequently observed interacting pair [73].
The importance of specific amino acid composition in determining secondary structure elements can be further illustrated by intrinsically unstructured proteins. Unlike globular proteins that typically form hydrophobic cores, intrinsically disordered proteins possess a comparatively lower proportion of “order-promoting” bulky hydrophobic residues and aromatic residues, but are significantly richer in “disorder-promoting” residues including proline and glycine [76]; the reverse of the requirements for forming α-helical and β-sheeted elements discussed earlier.
In addition to influencing the specific hydrogen-bonding network within the respective secondary structure element, distal side chain interactions of constituting residues could also have a significant influence on the final structure of the protein. The classical illustration would be the amphipathic characteristic of α-helices, which exhibit predominant nonpolar side chains on one surface and polar side chains on the opposite surface of the helical cylinder [77]. Such a polarized arrangement of residues with opposing physicochemical properties formed the basis of tertiary structural frameworks such as the α-helical coiled coil, in which the specific heptad repeat motif of hydrophobic residues at helix interfaces allowed meshing of side chains in a knob-into-hole packing of helical bundles [78]. While most methodical analysis of protein folding were based on independent folding domains, the vast majority of eukaryotic proteins are likely to possess a multiple independent folding domain that could also exhibit cooperativity in the folding process [79].
Imino acid proline
Of the 20 standard amino acids that are found in nature, proline is without doubt one of the most unusual: the fusion of the backbone with the side chain atom forms a unique pyrolidine ring, and at the same time eliminates the amide proton. Both of these features confer conformation limitation and distinctive structural features to the polypeptide chain in which the imino residue resides [80]. The first consequence of the pyrolidine ring of proline residues is the introduction of a kink to the polypeptide backbone [60, 81]. While placement of a proline at the N-terminal of an α-helix highly stabilizes the helix, insertion of prolines at any other positions are generally considered to be highly disruptive to the formation and propagation of α-helices or β-sheets, as well as having possible structural impacts on the conformation of the protein. For this reason, statistical data have shown that proline residues are most frequently located within the first helical turn as a helix stabilizer, or immediately at the C-terminal of the helix as a capping residue [10, 80, 81].
The second structural consequence of proline’s pyrolidine ring arises from the lack of amide proton which impairs the imino acid’s participation in formation of classical secondary structural elements due to the inability to act as hydrogen bond donor to carbonyl groups of other residues. However, the CδH2 protons of proline can form weaker hydrogen bonds with amide carbonyls of i − 2 or i − 4 residues in a helix, justifying for certain degrees of tolerance to proline residues in a number of globular and transmembrane proteins [82]. In other cases, conserved proline residues have also been identified in the middle of helices, possibly upholding specific structural or functional roles by inducing kinks in the length of the helices concerned [83].
In order to illustrate the structural implications of proline on protein folding, we will discuss a few examples of animal toxins, in which the influence of various structural features either in isolation or in combination can be clearly visualized on these highly conserved and compact structural frameworks [84]. In this case, the disulfide-pairing arrangement and consequent conformation of maurotoxin, a 34-mer potassium channel blocker isolated from scorpion venom, was shown to be drastically influenced by the presence of proline residues [85]. Despite possessing a conserved cysteine framework of the four-disulfide-bridged scorpion toxin, maurotoxin adopts a novel cysteine-pairing pattern (C1–C5, C2–C6, C3–C4, and C7–C8) that is distinct from the classical cysteine-pairing framework (C1–C5, C2–C6, C3–C7, and C4–C8) seen in Pi1 and HsTx1 toxins, the other two members of the same family. Sequence analysis of maurotoxin revealed two prolines flanking the non-conventional C3–C4 disulfide bridge. Substitution of these proline residues with alanines resulted in a conformational switch to the conventional cysteine pairing seen in Pi1 and HsTx1 toxins.
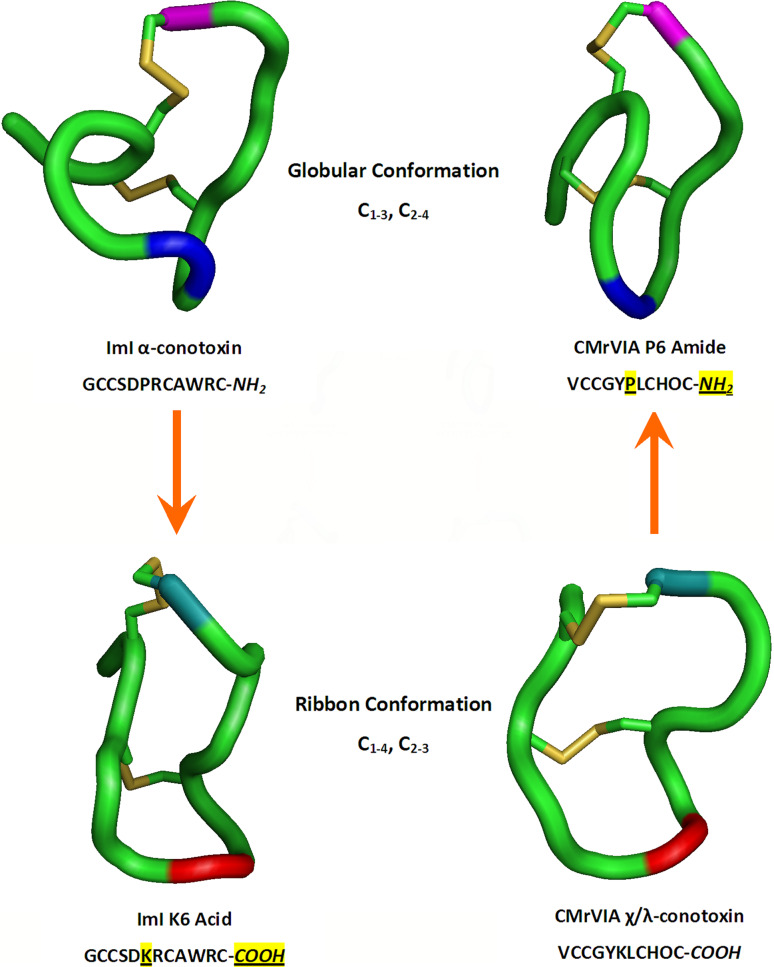
As a further illustration on the role of proline as a structural determinant of protein folding, the conserved proline residue in the first intercysteine loop of ImI α-conotoxin was previously identified by our group to have profound implications on the folding of this 12-mer peptide toxin. α-conotoxins and χ/λ-conotoxins share similar cysteine arrangement framework of 4 cysteine residues, but natively fold with distinct disulfide pairing patterns into the globular (C1–C3, C2–C4) and ribbon conformation (C1–C4, C2–C3), respectively [86, 87]. By substituting Pro6 with a conserved lysine residue from the corresponding position in χ/λ-conotoxins, it was possible to switch the native globular conformation of ImI α-conotoxin to the non-native ribbon conformation. These findings were corroborated by switching CMrVIA χ/λ-conotoxin’s native ribbon conformation to the non-native globular conformation with a reciprocal substitution of Lys6 with a proline residue (Fig. 2) [88]. It was thus proposed that proline’s tendency to bring two segments of the peptide backbone into juxtaposition acted as a conformational switch resulting in the preferential disulfide connectivity seen in ImI α-conotoxin. However, it is important to note at this juncture that changes of conformation might not necessarily imply changes in the folding pathway.
Fig. 2.
Role of C-terminal amidation and proline residue on the folding of ImI α-conotoxin and CMrVIA χ/λ-conotoxin. Proline and lysine residues manipulated within the first intercysteine loop of the conotoxins are represented in blue and red, respectively. C-terminal amide and carboxylic acid are presented in magenta and teal, respectively
A similar importance of proline was also observed in the folding of larger proteins such as cytochrome P450, in which alanine substitutions of Pro30 and Pro33 in the conserved proline-rich region in CYP2C11 resulted in mutants that were misfolded and biologically inactive [89]. The purpose of the proline-rich region was hypothesized to function as a hydrophobic knob for partial insertion of the protein into the membrane and to create a stable nucleus for initiation of protein folding.
Another unique structural characteristic of proline residues is the comparatively higher intrinsic propensity to adopt either the trans or cis peptide bond conformation with the preceding residue in the polypeptide chain [80]. It has been suggested that the ratio of the cis:trans prolyl peptide bond may be influenced by the residue following the proline residue and immediate environment of the proline residue [80]. While the precise reason for adopting the cis prolyl peptide bond are varied and not necessarily definitive, the increased propensity of proline residues to adopt the cis peptide bond is nonetheless significant, considering the fact that the majority of peptide bonds exist in the trans conformation [90].
Structurally, cis-trans isomerization of the prolyl peptide bond has been shown to be important for a variety of proteins such as Stefin B, a 11 kDA protein which forms amyloid fibrils by domain swapping between two dimers. The domain swapping involves extensive intermolecular contact as well as concurrent trans-cis isomerization. By substituting prolines for serines, it was shown that the conserved trans Pro74 in the monomeric and dimeric Stefin B is involved in the isomerization into the cis conformation to facilitate tetramerization that is crucial for amyloid fibril formation [91]. Functionally, the rate of cis-trans isomerization of prolyl peptide bond aids in the regulation of a variety of biological processes. For example, the rate of isomerization Pro213 in the hinge subdomain of N1–N2 domain of the gene-3-protein of phage fd has been shown to act as a kinetic block to the domain reassembly and conformational switch of the gene-3-protein, thereby regulating the rate of phage infection [92, 93]. The work by Kamen et al. [94] also indicated that prolyl cis-trans isomerization is implicated in the folding kinetics of proteins such as pectate lyase C.
Prolyl cis-trans isomerization is also a critical conversion in numerous cellular processes and is catalyzed by the four families of peptidyl prolyl cis-trans isomerases (PPIases). The rate of catalysis of prolyl cis-trans isomerization by these ubiquitous enzymes in turn act as molecular timers of cellular processes [95]. In fact, two of these PPIases are targets of immunosuppressant drugs [96, 97].
In this section, we have discussed the general propensities of the various amino acids in forming secondary structural elements. However, departures from the predicted propensities are not uncommon. For example, identical sequence segments have been identified in both β-sheeted and α-helical structures [98]. Peptide fragments termed “chameleon sequences” have also been reported in which the peptide segments are essentially ambivalent and the conformations of the segments concerned are largely dependent on the local structure of the proteins [99]. In the earlier sections, we mentioned that the conformational restraining proline residue is a well-recognized helix breaker. Interestingly, polypeptide segments containing consecutive proline residues have also been reported as adopting a distinct nonconventional secondary structure termed the poly-L-proline helix (PPII) that presents as an all-trans helix with a left-handed turn. Unlike canonical α-helices, PPII helices are not held together by hydrogen bonds between i and i + 4 residues, but merely because of the steric constraints and physical rigidity of prolines in the helices. This further explains the left-handedness of PPII helices compared to right-handed canonical α-helices. The PPII helices have been reported to be important in diverse structural and biological functions including the formation of collagen [100], signal transduction, transcription, and immunological reactions [101].
Sequence-specific side chain aromatic interactions, such as π–π, cation–π, amide–π, and aryl–sulfur interactions, have also been implicated to be important contributors to various secondary structures such as β-turns [102, 103]. Engineered non-native aromatic side chain interactions have been shown to divert the conformation populations of ubiquitin into a complex folding landscape [104]. In other cases, specific steric arrangement of charged residues can also facilitate specific interaction and consequent conformational change. For example, the binding pockets of calcium binding proteins are coordinated by a bipyramidal or octahedral arrangement of negative charged residues, to which calcium ions facilitate conformational changes by overcoming repulsion of similar charges [105]. The importance of specific interaction can be further exemplified by the variety of exogenous stimuli such as the presence of metal ions, pesticides, variation in pH, temperature, protein concentration, and organic solvents that can result in partial protein unfolding and consequently amyloidogenesis [106, 107].
In summary, these observations indicate that, although propensities of the various amino acids can influence the structural characteristics and folding profile of the polypeptides, the context in which the residues are located, and the consequent differences in microenvironment and the addition of exogenous stimuli, can affect the protein folding.
Posttranslational modifications and folding
Posttranslational modification of proteins is nature’s strategy for widening the diversity of the repertoire of polypeptides assembled from the standard list of 20 natural amino acids. Currently, over 200 kinds of posttranslational modifications have been documented, and they may be broadly classified as (1) covalent addition/ modifications of functional group to side chains, and (2) hydrolytic cleavage of peptide bonds [53]. Most of these posttranslational modifications result in functional consequences. In addition, several forms of posttranslational modifications have structural implications for the folding and conformation of the proteins. In the first part of this section, we shall discuss the various representative types of posttranslational modifications that bear direct implications to protein folding. In the remaining parts, we will touch on several forms of posttranslational modifications that are important for modifying local conformation of the tertiary structure and biological functions of proteins, but are not generally known to impact global protein folding. This is especially since some of these modifications occur only after the protein folding process has been completed in the endoplasmic reticulum.
It should be emphasized that this section is not intended to be encyclopedic on the plethora of possible posttranslational modifications, but rather, to illustrate with selected examples on the significance and the possible impact of these modifications on protein folding (Table 1).
Table 1.
Examples on the significance and the possible impact of these modifications on protein folding
| Posttranslational modification | Consequence | Examples | Structural implications | References |
|---|---|---|---|---|
| Protein folding | ||||
| Oxidation-reduction | Formation of disulfide bridges | Insulin superfamily, animal venoms, BPTI | Structural stability | [114, 117, 126] |
| Putative role in improving protein folding rate | ||||
| Cooperativity in folding by multiple disulfide bridges | ||||
| Oxidation of methionine to methionine sulfoxide/ sulfone | Transthyretin, λ-repressor protein, calmodulin, thrombomodulin | Disruption of native non-covalent interactions | [130–134] | |
| Glycosylation | Covalent attachment of carbohydrate moiety to conserved glycosylation sites | Lysosomal hydrolases, coagulation factors, antibodies, erythropoietin, contalukin-G | Augment solubility and stability profile | [135, 136, 148, 149, 163, 238] |
| Promote folded conformation by reducing conformational space | ||||
| Formation of new interactions between glycan and polypeptide | ||||
| Hydroxylation | Stereospecific hydroxylation of proline residue | Collagen, conotoxins | Stabilization of trans isomer of hydroxyprolyl peptide bond | [160, 164] |
| Improve rate of oxidative folding in α-conotoxin model | ||||
| Carboxyamidation | Substitution of C-terminal carboxyl group with amide group | Venom proteins, as well as peptide hormones such as vasopressin, oxytocin, and calcitonin | Modification to electrostatic interactions | [163, 169, 239] |
| γ-Carboxylation | Carboxylation of glutamic acids | Osteocalcin, conantokins, as well as various other calcium binding proteins including coagulation factors and clotting inhibitors | Facilitate calcium-induced folding | [163, 177, 178] |
| Protein conformation | ||||
| Phosphorylation | Covalent attachment of phosphoryl group to serine, threonine, and tyrosine in eukaryotes, histidine and aspartate in prokaryotes | Glycogen phosphorylase, isocitrate dehydrogenase, as well as a large number of proteins involved in signal transduction and numerous critical cellular processes | Change in protein’s tertiary conformation due to the dianionic phosphoryl group participating in hydrogen bonds | [187] |
| Methylation/acetylation | Covalent attachment of methyl or acetyl group to ε-amino group of lysines or arginines | Histone proteins | Augment specific cation–π interactions | [190, 191] |
| Farnesylation | Attachment of farnesyl group to CAAX C-terminal motif | (SNARE) Ykt6 protein | Increase the α-helical content and stability of protein | [194] |
| Nitration | Attachment of nitrate group to tyrosine | Mn superoxide dismutase, α-synuclein, tyrosine hydroxylase, tau protein | Modification to hydrophobic and electrostatic interactions | [193] |
| Sulfation | Transfer of sulfuryl group to tyrosine | Glycoprotein Ibαα | Contribution of negative charges | [240] |
| Citrullation | Conversion of arginine into citrulline residue | Fillagrin, trichohyalin, histone proteins | Charge reversal results in the disruption of electrostatic interactions and hydrogen bonds | [241] |
| Polyglutamylation | Elongation of polyglutamate chain at specific glutamic residues of the protein | α-tubulin, β-tubulin, nucleosome assembly proteins NAP1 and NAP2 | Possible roles in the conformation and interaction of target proteins | [242] |
| Polyglycylation | Covalent attachment of multiple glycyl units to γ-carboxyl group of glutamate | α-tubulin, β-tubulin | Possible roles in the conformation and interaction of target proteins | [243] |
Oxidation-reduction
One of the most important and commonly observed posttranslational modifications of protein is the oxidation of the free thiol group of cysteine residues and the consequent formation of disulfide bridges. Formation of disulfide bridges is influenced by the proximity and accessibility of the thiol groups on the polypeptide chain, and catalyzed by protein disulfide isomerase and Dsb family of proteins in eukaryotic and prokaryotic systems, respectively [108]. These posttranslational covalent linkages play critical roles in the structural stability of proteins [109]. Consequently, disulfide engineering had been used as an approach to stabilize proteins [110]. Disulfide linkages are also directly implicated in biological functions, such as catalytic disulfide bridges in oxidoreductases [111] and allosteric disulfide bridges that regulate protein function by triggering conformational changes [112]. In addition, the kinetics of the protein folding process have also been suggested to be influenced by the relative location of disulfide bridges to the folding nucleus [110].
Disulfide-rich proteins are frequently chosen as models for studies of the in vitro protein folding process due to the possibility of trapping disulfide folding intermediates, thereby allowing the reconstitution of folding pathways. Further, disulfide connectivity can possibly improve the overall folding rate of proteins by the formation of productive disulfide bridges that bring about the necessary interactions for the formation of a stable conformation. In other cases, the hydrophobic disulfide bridge may also be involved in the formation of hydrophobic clusters that are necessary as the folding nucleus to initiate the folding process [110, 113]. The presence of multiple disulfide bridges has also been demonstrated to have a potential positive influence on the folding. For example, in several small disulfide-rich proteins, such as BPTI and proinsulin, the interactions between the three sets of disulfide bridges resulted in cooperativity of folding by lowering the conformational entropy necessary for interaction [114–116].
The insulin superfamily consists of small disulfide-containing globular proteins that share a common structural motif, and they have been extensively studied for the contribution of disulfide bridges to the structure and folding of proteins [117]. The insulin molecule, for example, consists of two chains that are linked by one intrachain disulfide bridge, A6–A11, and two interchain bonds, A7–B7 and A20–B19. It was shown that deletion of the intrachain disulfide bridge negated the α-helical N-terminal domain of the A–chain and the activity of insulin, while deletion of interchain disulfide A7–B7 resulted in more severe structural disturbances. However, deletion of either pair of disulfide bridges did not compromise the in vitro refolding of the proinsulin molecule, suggesting that these connectivities were non-critical in the folding of insulin molecule [118, 119]. In contrast, the interchain A20–B19 disulfide bond has been noted to be critical in stabilizing the folding intermediate [120]. Deletion of this critical disulfide bridge significantly disrupted the ordered secondary structure and increased the proinsulin’s susceptibility to proteolysis [119]. At the other extreme of the spectrum, multiple disulfide bridges have occasionally been noted to be redundant, with folding and structural stability maintained with a reduced number of native disulfide bridges [121, 122].
Due to the conformational restraint exerted by disulfide bridges, permutations of cysteine connectivity in cysteine-rich polypeptides almost invariably result in differing conformations. As such, disulfide bond connectivities are typically considered to be non-promiscuous; a trait that is critical for the accurate disulfide connectivity and conformation of numerous disulfide-rich proteins and peptides. However, there are examples in which one cysteine framework can result in more than a single possible disulfide connectivity. For example, the insulin-like growth factor I (IGF I), which is a member of the insulin superfamily discussed earlier, shares homologous structural motifs with other members of the same superfamily such as porcine insulin precursor and relaxin [120]. However, unlike other members of the superfamily, IGF I folds into two distinct conformations [123]. It was subsequently determined that sequence 1–4 and residue 9 of the B chain of IGF I were critical in determining the disulfide connectivity of the protein [124]. A further example of multiple disulfide isoforms arising from single cysteine template is that of the epidermal growth factor-like (EGF-like) domains of thrombomodulin, in which the fifth EGF-like domain exhibits a disulfide bonding pattern distinct from other EGF or EGF-like domains [125].
Non-conventional disulfide linkages have also been observed in other classes of cysteine rich peptides such as conotoxins derived from the venom of the marine predatory cone snails. Conotoxins are classified based on their highly conserved cysteine framework and their distinct biological activity. Unlike the majority of conotoxins that exhibit strict fidelity to their respective disulfide connectivity distinct biological activities, conotoxins with the fourteen-cysteine, six-cysteine and four-cysteine conotoxin scaffolds can each adopt at least two distinct conformations with differently connected disulfide bridges [126]. In our earlier work, we have shown that the conserved Pro6 and C-terminal amidation are instrumental in determining the disulfide connectivity of ImI α-conotoxin (Fig. 2) [88, 127]. In the case of GI α-conotoxin, Zhang and Snyder [128] demonstrated that the propensity of the peptide toxin to fold into the native globular conformation is largely dependent on the loop sizes of the two intercysteine loops. Similarly, the disulfide connectivity of charybdotoxin from the venom of the scorpion, Leiurus quinquestriatus, was influenced by the position of the cysteine residues and intercysteine loop spaces [122]. In an engineered barnase structure, the inclusion of disulfide bridges reduced the rate of protein unfolding [129]. Taken together, these findings show that disulfide bonds not only play an important role in the structure and function but also the folding process of the proteins.
The other residue of the 20 standard amino acids that is prone to oxidation is methionine, which also contains a polarizable sulfur atom. The thioether side chain of methionine may be oxidized to methionine sulfoxide or methionine sulfone, and has been implicated in several diseases. This includes Alzheimer’s disease and Parkinson’s disease, as well as various disease states relating to oxidative damage. Conversely, oxidation of the amyloidogenic transthyretin prevents the formation of fibril formation by disrupting the aggregation interface [130]. Oxidized methionine has been demonstrated to affect the local environment within the protein, and has also been utilized to stabilize the denatured state of monomeric λ-repressor protein, and to disrupt the conformational stability of calmodulin and thrombomodulin [131–133]. These observations indicate that oxidation of methionine potentially destabilizes non-covalent interactions that are normally critical in maintaining the stability of certain proteins. As a corollary to this, it is highly possible that methionine oxidation can potentially influence protein folding. An illustration of the influence of methionine oxidation on protein folding was demonstrated in the work by Dado et al., in which the oxidation of specific methionine residues in a designed 18-mer peptide resulted in the transition of the α-helical conformation towards the β-strand [134].
N-linked glycosylation
N-linked glycosylation is another common form of posttranslational modification, with nearly 50% of all human proteins being glycosylated [135]. Understandably, glycosylation must play an integral role in the trafficking, folding and function of eukaryotic proteins. The glycosylation process essentially occurs in three stages, in which the first phase occurs while the nascent polypeptide is still in the translocon complex and is associated to the endoplasmic reticulum-associated degradation (ERAD) system. Aside from participation in the folding of the polypeptide, the glycan indirectly influences the protein folding process by serving as an intracellular sorting signal on the folding status of the protein [136]. The second phase largely concerns the trimming of the glycan for direct targeting of the proteins, as in the case of lysosomal hydrolases [137]. The final stage of modification is effected in the target site of the protein [136]. Of particular interest in this context is the influence of glycosylation on the folding of the protein.
The carbohydrate moiety contributes to the folding of the protein in several ways. Firstly, glycosylation has been known to improve the overall stability of proteins [138], and has been utilized to improve the circulation half-life and activity of erythropoietin analogues [139] as well as selective delivery of enzymes into lysosomes of target tissues [140–144]. The large hydrophilic carbohydrate group exerts a biophysical influence on the polypeptide by increasing the solubility of the protein and augmenting the stability of the protein by covalently attaching to the protein’s surface. In silico examination of protein models also illustrates that the protein stabilizing effect stems from the destabilization of the unfolded state by the glycan moiety [145]. Secondly, although the glycan moiety does not directly result in secondary structure formation, the bulk of the relatively large glycan moiety promotes the folded conformation by decreasing the available conformational space, as well as the mobility of the unfolded conformation [136, 146, 147]. For example, sequential deglycosylation of human β2 glycoprotein I resulted in an increased change in the secondary structure of the native protein as determined by circular dichroism, reiterating the steric hindrance imposed by the carbohydrate moiety [148]. Thirdly, the addition of the large glycan moiety resulted in the formation of new interactions between the carbohydrates and amino acids and/or stabilized the protein fold by counterbalancing an unfavorable clustering of charges [149]. In addition to the global effect of improving solubility of the polypeptide, interactions between the N-acetyl group of glycans and the polypeptide have also been shown to affect the local conformation of the peptide backbone by promoting β-turns [150].
Examination of a glycosylated loop heptadecapeptide derived from the α subunit of the neuromuscular nicotinic acetylcholine receptor of Torpedo californica revealed that glycosylation of the conserved glycosylation site shifted the cis-trans equilibrium of a constituent proline residue towards the trans isomer, and the dithiol/disulfide equilibrium towards the oxidized species [151]. These observations demonstrate that steric and electrostatic hindrances conferred by the glycan can significantly affect the folding and structure of the protein. In addition to influencing the folding of proteins into secondary and tertiary structures, glycosylation has also been demonstrated to affect the quaternary assembly of protein monomers into higher order assemblies. For example, a bulky N-linked heptasaccharide binds to Asn17 of Erythrina corallodendron lectin, and forces the dimer to adopt a quaternary structure that is structurally non-identical to other non-glycosylated homologous lectins [152].
However, not all glycoproteins are equally dependent on the glycosylation for their folding and secretion [145]. While most eukaryotic proteins require glycosylation at some point in the folding process [153], the folding rate and pathway of several examples have been reported to be unaffected by the glycosylation state [154–156]. Further, not all glycosylation sites within a polypeptide chain possess the same bearing on the folding process. For example, of the five glycosylation sites on glucocerebrosidase, only four are N-glycosylated. Of these, only Asn19 was shown to be truly critical for the enzyme’s function, while deglycosylation to the other sites had comparable catalytic efficiency [140]. Similar trends were also observed in influenza hemagglutinin as well as in several lysosomal hydrolases such as α-galactosidase and α-glucosidase, in which only one of the several attached glycans is truly essential for activity [157–159]. Therefore, although the specific mechanics for predicting critical glycosylation sites that are deemed critical for the folding pathway of proteins are still ambiguous, the contribution of these covalently linked glycans to the structure and function of proteins are beyond doubt.
Hydroxylation
Although hydroxylation of amino acids is generally not as common as the other forms of posttranslational modifications, it is the most frequently observed form of posttranslational modification seen in the human, in the form of hydroxyproline in collagen [160]. Collagen is ubiquitously found in the structural tissue of vertebrates with an exceedingly high proportion of hydroxylated proline that forms the characteristic Xaa-Yaa-Gly triad repeats in its tropocollagen triple helices coiled coil. The stereoselective hydroxylation results in a (2S,4R)-4-hydroxyproline (Hyp) in the Yaa position of the triad repeat of the collagen’s primary structure [161]. While it was previously suggested that the hydroxyproline stabilizes the collagen triple helix via bridging water molecules [162], it was demonstrated through O-methylation of hydroxyproline that such conformational stability conferred by the hydroxylation was contributed by the “stereoelectronic” properties of the stereospecific hydroxyl group that promotes the proper conformation for triple helix formation [160, 161].
Apart from playing critical roles in the structural framework of collagen in vertebrates, hydroxylation of proline residues has been shown to have important influence on protein folding. For example, hydroxyproline is commonly observed across several families of conotoxins derived from the marine predatory cone snails [163]. While the functional roles of the hydroxylation remains uncertain, work by Lopez-Vera et al. demonstrated that hydroxylation of proline residues can have an important influence on protein folding rates [164]. In contrast, hydroxylation of proline not only improved the rate of oxidative folding of the α-conotoxin model, but also improved the in vitro folding rate of the ω-MVIIC that is notoriously difficult to fold [164, 165]. It has been suggested that the electron-withdrawing effect of the hydroxyl group stabilizes the trans isomer of the hydroxyproline about the peptide bond, thereby contributing to the improved folding propensity [161]. It was further proposed that proline hydroxylation and disulfide formation might occur concurrently and cooperatively within the endoplasmic reticulum [126, 166]. In another example, hydroxylation contribute to the bioactivity of μ-conotoxin GIIIA, but was not observed to be beneficial to the folding of the peptide toxin [164].
Carboxyamidation
Carboxyamidation has been noted to be an important form of posttranslational maturation of polypeptides, especially in the cases of peptide hormones [167–169]. While there are several examples in which C-terminal amidation of the peptides do not result in significant functional disparity [169, 170], non-amidated forms of several peptides have been demonstrated to suffer significant reduction in biological potency [171–175].
Despite the fact that carboxyamidation results in a subtle substitution of a negatively charged carboxylic terminal with an amide group at the distal end of the polypeptide chain, such modification has been shown to be sufficient to result in conformational changes of the peptide’s conformation. For example, in the case of eumenine mastoparan-AF (EMP-AF) isolated from the wasp venom, although both the native amidated and the synthetic deamidated forms possess amphipathic helical structures, the amidated peptide exhibited a well-defined helical structure with enhanced rigidity. In contrast, the C-terminal region of the deamidated analog exhibited higher degrees of conformational freedom due to charge repulsion and hydrogen bond disruption at the C-terminal segment of the synthetic peptide variant [169]. Similar consequence of conformational disruption of C-terminal region was also noted in the deamidated form of the crustacean hyperglycemic hormone Pej-SGP-I, in which the native amidated form was reported to be significantly more potent [171].
C-terminal amidation has also been observed in several families of the conotoxins derived from the marine predatory cone snails [163]. Unlike the previous examples in which the deamidated analogues exhibit conformational disruption at the C-terminal region, we have earlier shown that C-terminal amidation potentially determines the disulfide connectivity to specific families of these cysteine-rich peptide toxins, acting as a conformational switch. For example, the native ImI α-conotoxin folds predominantly into the globular conformation with a specific C1–3,C2–4 disulfide connectivity. In contrast, the synthetic carboxylate analog folded predominantly into the non-native ribbon conformation with C1–4,C2–3 disulfide connectivity and the consequent reduction of biological potency [88, 127]. In contrast, C-terminal amidation did not seem to influence the folding of other families of conotoxins [176]. These findings indicate that, although not consistently applicable, the modification to the electrostatic interactions brought about by C-terminal amidation can substantially influence the final conformations of proteins.
γ-Carboxyglutamic acid
γ-Carboxylation of glutamic acid is an important posttranslational modification that is particularly pertinent for hemostasis. Specifically, posttranslational carboxylation of specific glutamate residues is indispensible in the sequences of the several coagulation factors, osseous tissues, or procoagulant factors derived from venom sources [177, 178]. These γ-carboxyglutamate residues (Gla) function by acting as binding sites for calcium ions, which are a ubiquitous component of critical cellular processes [179].
Posttranslationally γ-carboxylated glutamic acids are usually observed in specific calcium-binding proteins. Despite the wide tissue distribution of the vitamin K-dependent carboxylase that is responsible for this modification, Gla residues are generally less commonly implicated to the structure or function of other proteins [180]. However, this unusual modification has been shown to be present in several peptide toxins derived from Conus venom, and, in the case of conantokins, multiple Gla residues are usually present within the short peptide sequences [181, 182]. It has been demonstrated that the binding specificity of Gla residues to calcium ions facilitates the folding of several conantokins into their mature conformations. For example, addition of calcium ions resulted in a significant increase yield and rate of folding of the “spasmodic peptide” derived from C. textile [183]. Similar calcium-induced folding was also noted in several conantokins, and is particularly significant considering the fact that the majority of conantokins lack disulfide bridges, and the presence of Gla residues is the predominant driving force for the peptides to adopt their mature helical conformation and/or self-assembly into dimeric structures [184, 185]. In the case of conantokin-P which possesses a single disulfide bridge, the addition of calcium ions to the folding buffer accelerated the oxidative folding of the peptide, reinforcing the determining influence of Gla residues on the folding of these small peptides.
Posttranslational modifications affecting conformation of mature proteins
In addition to the various forms of posttranslational modifications discussed above, a variety of other forms of modifications, though not directly implicated in protein folding, have been shown to influence the conformation and the consequent functions of mature proteins. One of the most pervasive forms of posttranslational modification is the phosphorylation of proteins executed by the ~500 specific kinases, providing an intracellular switch to diverse cellular processes [52, 53]. By covalently attaching a tetrahedral phosphoryl group to amino acids’ neutral hydroxyl side chain, phosphorylation brings about conformational changes to the proteins’ tertiary structures or assembly interfaces, thereby effecting cellular functions and processes [186]. Such conformational changes are possibly brought about by the dianionic phosphoryl group in physiological pH that can participate in hydrogen bonds, such as those with main-chain nitrogens of an α-helix or with the guanidinium side chain of arginine residues [187].
Methylation of lysine in histone proteins has been known to be involved in the chromatin condensation by interaction of the methylated lysine with three aromatic side chains. Specifically, the methylated histone tail of histone H3 inserts as a β-strand into the β-sandwich chromodomain of HP1 protein [188]. Based on this finding, a β-hairpin peptide corresponding to the histone tail was synthesized and found to be stabilized by the specific cation–π interaction between Trp2–Lys9 [189]. Further work by Hughes et al. [190, 191] indicated that N-methylation of the lysine or arginine residues augment the magnitude of this interaction, resulting in an exceptionally stable β-hairpins. Methylation of the ε-amino group of lysine altered the interactions with this polar side chain by conferring hydrophobic interactions, as well as generating a more polarized methylammonium group for cation–π interaction [188]. Another posttranslational modification known to influence the lysine residue of histone proteins is the acetylation of the ε-amino side chain [192]. Although acetylation of lysine alters the geometry of interaction and neutralizes the positive charge of lysine’s protonated side chain that is essential for cation–π interaction described earlier, amide–π interaction was found to be at least as strong as the cation–π interaction between lysine and the aromatic ring [103]. Taken together, methylation and acetylation of lysine on histone proteins illustrate the specificity of histone-DNA interaction, as well as provide insights on protein folding.
Other forms of posttranslational modifications include nitration of tyrosine residues, lipidation by farnesylation, citrullation of arginine residues, and polyglutamylation and polyglycylation of glutamate residues (Table 1). Nitration of tyrosine residues modifies the hydrophobic and electrostatic interactions of the target protein, thereby disrupting the conformation and activity of the protein [193]. It is for this reason that nitration of tyrosine residues has been implicated in a variety of neurodegenerative diseases. Lipidation by farnesylation of the soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE) Ykt6 protein was shown to increase the α-helical content of the 23 kDa protein as well as to stabilize the final conformation [194].
A minimalist approach
Inferences from Christian Anfinsen’s theory of protein folding suggested that the amino acid sequence of a polypeptide encodes for sufficient information required for the native folding of the protein. However, recent research indicates that, as much as specific-sequence information is necessary, a significant degree of sequence degeneration and redundancy exists in the amino acid syllabus and only a fraction of the encoded information is likely to be necessary for fully defining the native protein structure [195, 196]. Studies on the site-directed mutagenesis of various proteins have concurred with the theory of redundancy of protein sequence. For example, by selecting residues that were not directly involved in structural or functional roles, the exhaustive analysis of the lac repressor protein indicated that mutations made at nearly half the 142 sites on the protein were phenotypically silent [195, 197–199]. Tolerance to single or multiple substitutions was also seen in the Arc repressor protein [200], bovine pancreatic trypsin inhibitor [201, 202], and RNA-binding protein ROP [203], among others. In addition to sequence redundancy, several forms of posttranslational modifications discussed earlier also exhibited certain degrees of redundancy.
In addition to the redundancy of sequence information, it was also observed that there was a generally lower degree of conservation of surface-exposed residue side chain as compared to the buried nonpolar residues, with the former group generally being more tolerant of sequence substitution [195]. Such a difference in evolutionary conservation reiterates the importance of the nonpolar or neutral residues that make up the hydrophobic core structure of proteins [204]. Although some exceptions exist, the segregation of hydrophobic and hydrophilic residues into the buried and surface exposed positions of the final structure generally fulfils the requirement of protein solubility and facilitates the folding process of the protein [205]. Clearly, the amphipathic pattern of hydrophobic/hydrophilic residue distributions must represent some of the protein folding instructions encoded in the polypeptide [195]. To this, two general models of protein folding have been derived. The first concept of “jigsaw puzzle model” was proposed by Chothia et al. for α-helices in which the close packing and complementarity of specific side chains define the basic architecture of the backbone of the protein [206]. This model was initially supported by work examining interactions concerning hydrophobic interfaces which indicated that changes in charge and volume of residues located within the core of proteins are generally destabilizing [207, 208]. However, a number of combinations of amino acids have been substituted into core sequences and yet resulted in viable proteins [209–213]. The second model, termed “oil-droplet model”, posits that periodic binary distribution of hydrophobic and hydrophilic residues permits the formation of structural elements by hydrophobic interactions, without the need for specific inter-residue contacts [214, 215]. Nonetheless, drastic changes in shapes or volumes of the hydrophobic residues concerned can still result in changes in conformation of the tertiary structure [216]. Taken together, the hydrophobicity described by the “oil-droplet model” is recognized to be important. However, interdigitation of core residue side chains, though critical in selected models, may not necessarily be as strictly dictated as by the “jigsaw puzzle model”. Although each of the two models of protein folding can only provide a limited perspective to the folding process, they are, nonetheless, sufficient for the design and generation of empiric secondary structure elements.
One of the early designs of de novo protein using the binary distribution of hydrophilic and hydrophobic residues was accomplished by Kamtekar et al., in which a four-helix bundle protein was empirically designed [214]. The degeneracy of sequence was demonstrated by using phenylalanine, leucine, isoleucine, methionine, and valine for the nonpolar residues, while polar residues were fulfilled by using glutamine, glutamate, aspartate, asparagine, lysine, or histidine. Through the use of merely 11 out of the 20 available natural amino acid residues, the designed protein correctly folded into the intended tertiary conformation, and exhibited a native-like hydrophobic/hydrophilic residue distribution. Further work by Schafmeister et al. showed that it is possible to design a larger four-helix bundle using just seven amino acids, with the designed protein yielding crystal structure of 2.9 Å resolution [217]. The concept of a minimized alphabet for protein folding was subsequently extended to the design of a small β-sheeted protein, the SH3 domain, in which five amino acids (isoleucine, lysine, glutamate, alanine, and glycine) were sufficient for coding the 53-residue domain [218]. In a more recent article by Taylor et al., it was demonstrated that the enzyme AroQ chorismate mutase could be simplified to binary modules of four polar and four nonpolar residues [219]. It was also subsequently demonstrated that a fully functional variant of the enzyme could be constructed with a mere nine-letter amino acid alphabet [220]. Thus, the possibility of designing basic secondary structure elements using a binary approach reaffirms the importance of hydrophobic interaction in the protein folding pathway. It also suggests the possibility of designing larger and more complex protein architecture. In a bold reverse engineering attempt by Silverman et al. [221], the prototypical (β/α)8 TIM barrel domain of the triosephosphate enzyme was shown to exhibit sequence degeneracy and could be encoded with a reduced amino acid alphabet. It was determined that 142 out of the 182 examined sites on the enzyme’s ~250-residue sequence could be mutated to one of the seven-letter amino acid alphabet and result in viable enzymes (Fig. 3).
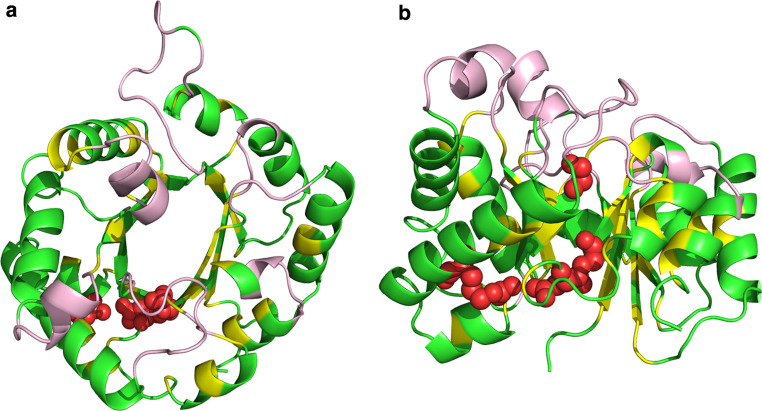
Fig. 3.
Mutagenesis analysis of yeast triosephosphate isomerase (TIM)(PDB ID 7TIM). A total of 182 out of the 250 residues of the TIM protein were systematically mutated with one of the 7-letter amino acid alphabet (FVLAKEQ) reduced from the original 20-letter alphabet. Conservative substitutions were made based on either structural features or phylogenetic conservation between various TIM analogues. The top and side views of the TIM molecule are shown in a and b, respectively. Residues highlighted in green represent the 97 singly mutated positions that resulted in minimal disruption of functional activity of the protein as well as the 45 residues of the original protein that contain letters of the 7-letter alphabet. Segments in yellow represent the 36 positions that resulted in intermediate loss of protein activity, while residues represented by red spheres are the four immutable residues R189, D227, G209, and G228. Segments in pale pink are residues not examined in the mutagenesis analysis
Having discussed the propensities of the various amino acids in influencing protein folding, the influence of posttranslational modifications, and the redundancy of sequence information, it should not be neglected that a plethora of short sequence motifs are possible with the various permutation and combinations of these residues. Specific sequence motifs have been defined for various local conformational motifs. These include, but are not limited to, “tyrosine corner” in the Greek key β-barrel [222], consensus sequences resulting in Type I β-turns [74], stacking of aromatic residues with a proline residue to form Type VI β-turn [223], packing motifs for transmembrane helices [224], and proline-rich motifs in cytochrome P450s [89], as well as numerous others [55, 81]. These specialized folding motifs exert specific structural and biological properties that fall beyond the scope of the current review.
Challenges of protein folding
Thus far, we have discussed some of the structural features of proteins that have been reported to influence the folding of the protein. While several of the features described were inherent properties of the basic amino acid sequence and concur with the theory proposed by Christian Anfinsen at a time when understanding of protein folding was still at its infancy, several structural determinants were made posttranslationally to the polypeptide chain. However, it is still not possible to conclusively claim universal applicability for these structural “determinants”. Indeed, it is the uniqueness of each polypeptide’s amino acid sequence and the subsequent maturation steps that make the protein folding problem particularly challenging to decipher. A classical illustration of the distinctiveness of proteins can be seen in amyloidogenesis: while specific proteins have been shown to be prone to fibrillation, proteins that are unrelated to any known disorder can also be converted into aggregates that exhibit similar structural features to amyloid fibrils [106, 107, 225]. It is important to emphasize that determinants discussed in this review are merely a fraction of the knowledge demanded of the intricately complex picture of protein folding. In essence, the determinants of correct folding (or in the case of amyloidogenesis, protein misfolding) are neither completely understood nor can they be simplistically limited to structural features on the sequence. Several other aspects of protein folding that are at least as important include the thermodynamics and kinetics of protein folding, folding intermediates and pathways, and computation prediction [7–11].
A principal goal of understanding the structural determinants of protein folding is essential to appreciate the “language of proteins”. From a clinical perspective, such knowledge could facilitate the understanding of conformational diseases [226], in which the misfolding of endogenous proteins resulted in pathological consequences [227–229]. From a biochemical perspective, one of the ultimate aims of protein design would be the design of tailored protein structures, with precisely tailored biological functions. Although this has yet to be achieved, the understanding of the structural determinants coupled with increased computational capabilities has facilitated the design of various de novo small proteins with specific secondary structures intended in mind [230–233]. In the work by Kuhlman et al. [232], the accuracy of fold of the designed protein could even be predicted to an impressive atomic-level of accuracy! By reducing the degeneracy of encoding residues, several natural proteins including the prototypical (β/α)8 barrel structure have been successfully redesigned with a much smaller subset of the amino acid alphabet [217–221]. Another successful approach of protein design utilized the specific coordination of negatively charged residues to form calcium-inducible folding of the target protein [105]. The current computational prowess also led to the reverse design of a zinc-free “zinc finger” domain [234]. An alternative approach to successful de novo design utilized extensive combinatorial libraries instead of the classical rational design of sequence space, through which the optimized sequence for a desired trait or conformation was identified by binary patterning [215]. Thus, it has also been recently suggested that the current state of technology can possibly facilitate the de novo design of proteins with native-like properties of up to or surpassing 100 residues by using first principles [235].
The protein folding problem is a complex topic that has consumed several decades of research. With the recent advances in structural biology and theoretical and computational approaches, as well as the various biophysical techniques, including a variety of fast spectroscopic assays that allows monitoring of the folding process in real-time, it is now often possible to accurately predict the three-dimensional structures of small proteins through template-based models [236, 237]. While it is still a challenge to accurately predict the structures of larger, multi-domain proteins from their empiric amino acid sequences [14, 41, 236], the recent advances in the various aspects of the protein folding suggest that the solution to this grand biochemical challenge may one day be in sight.
Acknowledgement
Tse Siang Kang is supported by the National University of Singapore under the Overseas Postdoctoral Fellowship.
References
- 1.Honig B, Cohen FE. Adding backbone to protein folding: why proteins are polypeptides. Fold Des. 1996;1:R17–R20. doi: 10.1016/S1359-0278(96)00005-3. [DOI] [PubMed] [Google Scholar]
- 2.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 3.Anfinsen CB, Haber E, Sela M, White FH., Jr The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci USA. 1961;47:1309–1314. doi: 10.1073/pnas.47.9.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levinthal C. Are there protein folding pathways? J Chim Phys. 1968;65:44–45. [Google Scholar]
- 5.Levinthal C. How to fold graciously. In: DeBrunner JTP, Munck E, editors. Mossbauer spectroscopy in biological systems. USA: University of Illinois ; 1969. pp. 22–24. [Google Scholar]
- 6.Karplus M. The Levinthal paradox: yesterday and today. Fold Des. 1997;2:S69–S75. doi: 10.1016/s1359-0278(97)00067-9. [DOI] [PubMed] [Google Scholar]
- 7.Creighton TE. Disulfide bonds as probes of protein folding pathways. Methods Enzymol. 1986;131:83–106. doi: 10.1016/0076-6879(86)31036-x. [DOI] [PubMed] [Google Scholar]
- 8.Baldwin RL. The search for folding intermediates and the mechanism of protein folding. Annu Rev Biophys. 2008;37:1–21. doi: 10.1146/annurev.biophys.37.032807.125948. [DOI] [PubMed] [Google Scholar]
- 9.Fine R, Dimmler G, Levinthal C. FASTRUN: a special purpose, hardwired computer for molecular simulation. Proteins. 1991;11:242–253. doi: 10.1002/prot.340110403. [DOI] [PubMed] [Google Scholar]
- 10.Chou PY, Fasman GD. Prediction of protein conformation. Biochemistry. 1974;13:222–245. doi: 10.1021/bi00699a002. [DOI] [PubMed] [Google Scholar]
- 11.Leopold PE, Montal M, Onuchic JN. Protein folding funnels: a kinetic approach to the sequence-structure relationship. Proc Natl Acad Sci USA. 1992;89:8721–8725. doi: 10.1073/pnas.89.18.8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dill KA, Ozkan SB, Shell MS, Weikl TR. The protein folding problem. Annu Rev Biophys. 2008;37:289–316. doi: 10.1146/annurev.biophys.37.092707.153558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wetlaufer DB. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc Natl Acad Sci USA. 1973;70:697–701. doi: 10.1073/pnas.70.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fersht AR. From the first protein structures to our current knowledge of protein folding: delights and scepticisms. Nat Rev Mol Cell Biol. 2008;9:650–654. doi: 10.1038/nrm2446. [DOI] [PubMed] [Google Scholar]
- 15.Ptitsyn OB. How the molten globule became. Trends Biochem Sci. 1995;20:376–379. doi: 10.1016/s0968-0004(00)89081-7. [DOI] [PubMed] [Google Scholar]
- 16.Kim PS, Baldwin RL. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- 17.Karplus M, Weaver DL. Protein folding dynamics: the diffusion-collision model and experimental data. Protein Sci. 1994;3:650–668. doi: 10.1002/pro.5560030413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ptitsyn O. How molten is the molten globule? Nat Struct Biol. 1996;3:488–490. doi: 10.1038/nsb0696-488. [DOI] [PubMed] [Google Scholar]
- 19.Fersht AR. Optimization of rates of protein folding: the nucleation–condensation mechanism and its implications. Proc Natl Acad Sci USA. 1995;92:10869–10873. doi: 10.1073/pnas.92.24.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fersht AR. Nucleation mechanisms in protein folding. Curr Opin Struct Biol. 1997;7:3–9. doi: 10.1016/s0959-440x(97)80002-4. [DOI] [PubMed] [Google Scholar]
- 21.Gianni S, Guydosh NR, Khan F, Caldas TD, Mayor U, White GW, DeMarco ML, Daggett V, Fersht AR. Unifying features in protein-folding mechanisms. Proc Natl Acad Sci USA. 2003;100:13286–13291. doi: 10.1073/pnas.1835776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shortle D, Ackerman MS. Persistence of native-like topology in a denatured protein in 8 M urea. Science. 2001;293:487–489. doi: 10.1126/science.1060438. [DOI] [PubMed] [Google Scholar]
- 23.Fersht AR, Matouschek A, Serrano L. The folding of an enzyme. I. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 24.Ivarsson Y, Travaglini-Allocatelli C, Brunori M, Gianni S. Mechanisms of protein folding. Eur Biophys J. 2008;37:721–728. doi: 10.1007/s00249-007-0256-x. [DOI] [PubMed] [Google Scholar]
- 25.Dyson HJ, Wright PE. Elucidation of the protein folding landscape by NMR. Methods Enzymol. 2005;394:299–321. doi: 10.1016/S0076-6879(05)94011-1. [DOI] [PubMed] [Google Scholar]
- 26.Dyson HJ, Wright PE. Unfolded proteins and protein folding studied by NMR. Chem Rev. 2004;104:3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 27.Borgia A, Williams PM, Clarke J. Single-molecule studies of protein folding. Annu Rev Biochem. 2008;77:101–125. doi: 10.1146/annurev.biochem.77.060706.093102. [DOI] [PubMed] [Google Scholar]
- 28.Forman JR, Clarke J. Mechanical unfolding of proteins: insights into biology, structure and folding. Curr Opin Struct Biol. 2007;17:58–66. doi: 10.1016/j.sbi.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 29.Balakrishnan G, Weeks CL, Ibrahim M, Soldatova AV, Spiro TG. Protein dynamics from time resolved UV Raman spectroscopy. Curr Opin Struct Biol. 2008;18:623–629. doi: 10.1016/j.sbi.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schaeffer RD, Fersht A, Daggett V. Combining experiment and simulation in protein folding: closing the gap for small model systems. Curr Opin Struct Biol. 2008;18:4–9. doi: 10.1016/j.sbi.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dokholyan NV. Studies of folding and misfolding using simplified models. Curr Opin Struct Biol. 2006;16:79–85. doi: 10.1016/j.sbi.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 32.Caflisch A. Network and graph analyses of folding free energy surfaces. Curr Opin Struct Biol. 2006;16:71–78. doi: 10.1016/j.sbi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Kubelka J, Hofrichter J, Eaton WA. The protein folding ‘speed limit’. Curr Opin Struct Biol. 2004;14:76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Brockwell DJ, Radford SE. Intermediates: ubiquitous species on folding energy landscapes? Curr Opin Struct Biol. 2007;17:30–37. doi: 10.1016/j.sbi.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gianni S, Ivarsson Y, Jemth P, Brunori M, Travaglini-Allocatelli C. Identification and characterization of protein folding intermediates. Biophys Chem. 2007;128:105–113. doi: 10.1016/j.bpc.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez IE, Kiefhaber T. Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J Mol Biol. 2003;325:367–376. doi: 10.1016/s0022-2836(02)01230-5. [DOI] [PubMed] [Google Scholar]
- 37.Gianni S, Geierhaas CD, Calosci N, Jemth P, Vuister GW, Travaglini-Allocatelli C, Vendruscolo M, Brunori M. A PDZ domain recapitulates a unifying mechanism for protein folding. Proc Natl Acad Sci USA. 2007;104:128–133. doi: 10.1073/pnas.0602770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zarrine-Afsar A, Larson SM, Davidson AR. The family feud: do proteins with similar structures fold via the same pathway? Curr Opin Struct Biol. 2005;15:42–49. doi: 10.1016/j.sbi.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 39.Lindberg MO, Oliveberg M. Malleability of protein folding pathways: a simple reason for complex behaviour. Curr Opin Struct Biol. 2007;17:21–29. doi: 10.1016/j.sbi.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Daggett V, Fersht AR. Is there a unifying mechanism for protein folding? Trends Biochem Sci. 2003;28:18–25. doi: 10.1016/s0968-0004(02)00012-9. [DOI] [PubMed] [Google Scholar]
- 41.Englander SW, Mayne L, Krishna MM. Protein folding and misfolding: mechanism and principles. Q Rev Biophys. 2007;40:287–326. doi: 10.1017/S0033583508004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dill KA. Dominant forces in protein folding. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 43.Yang JS, Chen WW, Skolnick J, Shakhnovich EI. All-atom ab initio folding of a diverse set of proteins. Structure. 2007;15:53–63. doi: 10.1016/j.str.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 44.Cho JH, Raleigh DP. Electrostatic interactions in the denatured state and in the transition state for protein folding: effects of denatured state interactions on the analysis of transition state structure. J Mol Biol. 2006;359:1437–1446. doi: 10.1016/j.jmb.2006.04.038. [DOI] [PubMed] [Google Scholar]
- 45.Mirsky AE, Pauling L. On the structure of native, denatured, and coagulated proteins. Proc Natl Acad Sci USA. 1936;22:439–447. doi: 10.1073/pnas.22.7.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Stites WE. Packing is a key selection factor in the evolution of protein hydrophobic cores. Biochemistry. 2001;40:15280–15289. doi: 10.1021/bi011776v. [DOI] [PubMed] [Google Scholar]
- 47.Richards FM, Lim WA. An analysis of packing in the protein folding problem. Q Rev Biophys. 1993;26:423–498. doi: 10.1017/s0033583500002845. [DOI] [PubMed] [Google Scholar]
- 48.Marin M. Folding at the rhythm of the rare codon beat. Biotechnol J. 2008;3:1047–1057. doi: 10.1002/biot.200800089. [DOI] [PubMed] [Google Scholar]
- 49.Sauer RT, Milla ME, Waldburger CD, Brown BM, Schildbach JF. Sequence determinants of folding and stability for the P22 Arc repressor dimer. Faseb J. 1996;10:42–48. doi: 10.1096/fasebj.10.1.8566546. [DOI] [PubMed] [Google Scholar]
- 50.Barrow CJ, Yasuda A, Kenny PT, Zagorski MG. Solution conformations and aggregational properties of synthetic amyloid beta-peptides of Alzheimer’s disease. Analysis of circular dichroism spectra. J Mol Biol. 1992;225:1075–1093. doi: 10.1016/0022-2836(92)90106-t. [DOI] [PubMed] [Google Scholar]
- 51.Haber E, Anfinsen CB. Regeneration of enzyme activity by air oxidation of reduced subtilisin-modified ribonuclease. J Biol Chem. 1961;236:422–424. [PubMed] [Google Scholar]
- 52.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 53.Walsh CT. Posttranslational modification of proteins. Greenwood Village, CO: Roberts; 2006. [Google Scholar]
- 54.Sali A, Shakhnovich E, Karplus M. How does a protein fold? Nature. 1994;369:248–251. doi: 10.1038/369248a0. [DOI] [PubMed] [Google Scholar]
- 55.Wilson DR, Finlay BB. The ‘Asx-Pro turn’ as a local structural motif stabilized by alternative patterns of hydrogen bonds and a consensus-derived model of the sequence Asn-Pro-Asn. Protein Eng. 1997;10:519–529. doi: 10.1093/protein/10.5.519. [DOI] [PubMed] [Google Scholar]
- 56.Pauling L, Corey RB, Branson HR. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc Natl Acad Sci USA. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feyereisen C, Morcellet M, Loucheux C. Preferential and absolute adsorption to poly[N5-(3-hydroxypropyl)-L-glutamine] in water/2-chloroethanol solvent mixtures. Macromolecules. 1977;10:485–488. doi: 10.1021/ma60056a049. [DOI] [PubMed] [Google Scholar]
- 58.Schiffer M, Edmundson AB. Use of helical wheels to represent the structures of proteins and to identify segments with helical potential. Biophys J. 1967;7:121–135. doi: 10.1016/S0006-3495(67)86579-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kotelchuck D, Scheraga HA. The influence of short-range interactions on protein onformation. II. A model for predicting the alpha-helical regions of proteins. Proc Natl Acad Sci USA. 1969;62:14–21. doi: 10.1073/pnas.62.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Creighton TE. Proteins: structures and molecular properties. New York: Freeman; 1993. [Google Scholar]
- 61.Marqusee S, Robbins VH, Baldwin RL. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci USA. 1989;86:5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakrabartty A, Schellman JA, Baldwin RL. Large differences in the helix propensities of alanine and glycine. Nature. 1991;351:586–588. doi: 10.1038/351586a0. [DOI] [PubMed] [Google Scholar]
- 63.Altmann KH, Wojcik J, Vasquez M, Scheraga HA. Helix-coil stability constants for the naturally occurring amino acids in water. XXIII. Proline parameters from random poly (hydroxybutylglutamine-co-L-proline) Biopolymers. 1990;30:107–120. doi: 10.1002/bip.360300112. [DOI] [PubMed] [Google Scholar]
- 64.Shoemaker KR, Fairman R, Kim PS, York EJ, Stewart JM, Baldwin RL. The C-peptide helix from ribonuclease A considered as an autonomous folding unit. Cold Spring Harb Symp Quant Biol. 1987;52:391–398. doi: 10.1101/sqb.1987.052.01.045. [DOI] [PubMed] [Google Scholar]
- 65.Kim PS, Baldwin RL. A helix stop signal in the isolated S-peptide of ribonuclease A. Nature. 1984;307:329–334. doi: 10.1038/307329a0. [DOI] [PubMed] [Google Scholar]
- 66.Marqusee S, Baldwin RL. Helix stabilization by Glu−···Lys + salt bridges in short peptides of de novo design. Proc Natl Acad Sci USA. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi S, Kim EH, Hibino T, Ooi T. Comparison of alpha-helix stability in peptides having a negatively or positively charged residue block attached either to the N- or C-terminus of an alpha-helix: the electrostatic contribution and anisotropic stability of the alpha-helix. Biopolymers. 1989;28:995–1009. doi: 10.1002/bip.360280507. [DOI] [PubMed] [Google Scholar]
- 68.Zimm BH, Brag JK. Theory of phase transition between helix and random coil in polypeptide chains. J Chem Phys. 1959;31:526. [Google Scholar]
- 69.Finkelstein AV, Badretdinov AY, Ptitsyn OB. Physical reasons for secondary structure stability: alpha-helices in short peptides. Proteins. 1991;10:287–299. doi: 10.1002/prot.340100403. [DOI] [PubMed] [Google Scholar]
- 70.Munoz V, Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. Nat Struct Biol. 1994;1:399–409. doi: 10.1038/nsb0694-399. [DOI] [PubMed] [Google Scholar]
- 71.Munoz V, Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. II. Helix macrodipole effects and rational modification of the helical content of natural peptides. J Mol Biol. 1995;245:275–296. doi: 10.1006/jmbi.1994.0023. [DOI] [PubMed] [Google Scholar]
- 72.Munoz V, Serrano L. Elucidating the folding problem of helical peptides using empirical parameters. III. Temperature and pH dependence. J Mol Biol. 1995;245:297–308. doi: 10.1006/jmbi.1994.0024. [DOI] [PubMed] [Google Scholar]
- 73.Smith C, Regan L. Construction and design of β-sheets. Acc Chem Res. 1997;30:153–161. [Google Scholar]
- 74.Lee J, Dubey VK, Longo LM, Blaber M. A logical OR redundancy within the Asx-Pro-Asx-Gly type I beta-turn motif. J Mol Biol. 2008;377:1251–1264. doi: 10.1016/j.jmb.2008.01.055. [DOI] [PubMed] [Google Scholar]
- 75.Chou PY, Fasman GD. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 76.Williams RM, Obradovi Z, Mathura V, Braun W, Garner EC, Young J, Takayama S, Brown CJ, Dunker AK (2001) The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput 89–100 [DOI] [PubMed]
- 77.DeGrado W, Kezdy F, Kaiser E. Design, synthesis and characterization of a cytotoxic peptide with melittin-like activity. J Am Chem Soc. 1981;103:679–681. [Google Scholar]
- 78.Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 79.Han JH, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat Rev Mol Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 80.MacArthur MW, Thornton JM. Influence of proline residues on protein conformation. J Mol Biol. 1991;218:397–412. doi: 10.1016/0022-2836(91)90721-h. [DOI] [PubMed] [Google Scholar]
- 81.Senes A, Engel DE, DeGrado WF. Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr Opin Struct Biol. 2004;14:465–479. doi: 10.1016/j.sbi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 82.Chakrabarti P, Chakrabarti S. C–H…O hydrogen bond involving proline residues in alpha-helices. J Mol Biol. 1998;284:867–873. doi: 10.1006/jmbi.1998.2199. [DOI] [PubMed] [Google Scholar]
- 83.Barlow DJ, Thornton JM. Helix geometry in proteins. J Mol Biol. 1988;201:601–619. doi: 10.1016/0022-2836(88)90641-9. [DOI] [PubMed] [Google Scholar]
- 84.Kini RM. Molecular moulds with multiple missions: functional sites in three-finger toxins. Clin Exp Pharmacol Physiol. 2002;29:815–822. doi: 10.1046/j.1440-1681.2002.03725.x. [DOI] [PubMed] [Google Scholar]
- 85.Carlier E, Fajloun Z, Mansuelle P, Fathallah M, Mosbah A, Oughideni R, Sandoz G, Di Luccio E, Geib S, Regaya I, Brocard J, Rochat H, Darbon H, Devaux C, Sabatier JM, de Waard M. Disulfide bridge reorganization induced by proline mutations in maurotoxin. FEBS Lett. 2001;489:202–207. doi: 10.1016/s0014-5793(00)02433-9. [DOI] [PubMed] [Google Scholar]
- 86.Arias HR, Blanton MP. Alpha-conotoxins. Int J Biochem Cell Biol. 2000;32:1017–1028. doi: 10.1016/s1357-2725(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 87.Balaji RA, Ohtake A, Sato K, Gopalakrishnakone P, Kini RM, Seow KT, Bay BH. λ-Conotoxins, a new family of conotoxins with unique disulfide pattern and protein folding. Isolation and characterization from the venom of Conus marmoreus . J Biol Chem. 2000;275:39516–39522. doi: 10.1074/jbc.M006354200. [DOI] [PubMed] [Google Scholar]
- 88.Kang TS, Radic Z, Talley TT, Jois SD, Taylor P, Kini RM. Protein folding determinants: structural features determining alternative disulfide pairing in alpha- and chi/lambda-conotoxins. Biochemistry. 2007;46:3338–3355. doi: 10.1021/bi061969o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kemper B. Structural basis for the role in protein folding of conserved proline-rich regions in cytochromes P450. Toxicol Appl Pharmacol. 2004;199:305–315. doi: 10.1016/j.taap.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 90.Lorenzen S, Peters B, Goede A, Preissner R, Frommel C. Conservation of cis prolyl bonds in proteins during evolution. Proteins. 2005;58:589–595. doi: 10.1002/prot.20342. [DOI] [PubMed] [Google Scholar]
- 91.Jenko Kokalj S, Guncar G, Stern I, Morgan G, Rabzelj S, Kenig M, Staniforth RA, Waltho JP, Zerovnik E, Turk D. Essential role of proline isomerization in stefin B tetramer formation. J Mol Biol. 2007;366:1569–1579. doi: 10.1016/j.jmb.2006.12.025. [DOI] [PubMed] [Google Scholar]
- 92.Martin A, Schmid FX. A proline switch controls folding and domain interactions in the gene-3-protein of the filamentous phage fd. J Mol Biol. 2003;331:1131–1140. doi: 10.1016/s0022-2836(03)00864-7. [DOI] [PubMed] [Google Scholar]
- 93.Eckert B, Martin A, Balbach J, Schmid FX. Prolyl isomerization as a molecular timer in phage infection. Nat Struct Mol Biol. 2005;12:619–623. doi: 10.1038/nsmb946. [DOI] [PubMed] [Google Scholar]
- 94.Kamen DE, Woody RW. Identification of proline residues responsible for the slow folding kinetics in pectate lyase C by mutagenesis. Biochemistry. 2002;41:4724–4732. doi: 10.1021/bi0115131. [DOI] [PubMed] [Google Scholar]
- 95.Lu KP, Finn G, Lee TH, Nicholson LK. Prolyl cis-trans isomerization as a molecular timer. Nat Chem Biol. 2007;3:619–629. doi: 10.1038/nchembio.2007.35. [DOI] [PubMed] [Google Scholar]
- 96.Takahashi N, Hayano T, Suzuki M. Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature. 1989;337:473–475. doi: 10.1038/337473a0. [DOI] [PubMed] [Google Scholar]
- 97.Harding MW, Galat A, Uehling DE, Schreiber SL. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature. 1989;341:758–760. doi: 10.1038/341758a0. [DOI] [PubMed] [Google Scholar]
- 98.Kabsch W, Sander C. On the use of sequence homologies to predict protein structure: identical pentapeptides can have completely different conformations. Proc Natl Acad Sci USA. 1984;81:1075–1078. doi: 10.1073/pnas.81.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ambroggio XI, Kuhlman B. Design of protein conformational switches. Curr Opin Struct Biol. 2006;16:525–530. doi: 10.1016/j.sbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 100.Siligardi G, Drake AF. The importance of extended conformations and, in particular, the PII conformation for the molecular recognition of peptides. Biopolymers. 1995;37:281–292. doi: 10.1002/bip.360370406. [DOI] [PubMed] [Google Scholar]
- 101.Rath A, Davidson AR, Deber CM. The structure of “unstructured” regions in peptides and proteins: role of the polyproline II helix in protein folding and recognition. Biopolymers. 2005;80:179–185. doi: 10.1002/bip.20227. [DOI] [PubMed] [Google Scholar]
- 102.Waters ML. Aromatic interactions in peptides: impact on structure and function. Biopolymers. 2004;76:435–445. doi: 10.1002/bip.20144. [DOI] [PubMed] [Google Scholar]
- 103.Hughes RM, Waters ML. Effects of lysine acetylation in a beta-hairpin peptide: comparison of an amide-pi and a cation-pi interaction. J Am Chem Soc. 2006;128:13586–13591. doi: 10.1021/ja0648460. [DOI] [PubMed] [Google Scholar]
- 104.Rea AM, Simpson ER, Meldrum JK, Williams HE, Searle MS. Aromatic residues engineered into the beta-turn nucleation site of ubiquitin lead to a complex folding landscape, non-native side-chain interactions, and kinetic traps. Biochemistry. 2008;47:12910–12922. doi: 10.1021/bi801330r. [DOI] [PubMed] [Google Scholar]
- 105.Li S, Yang W, Maniccia AW, Barrow D Jr, Tjong H, Zhou HX, Yang JJ. Rational design of a conformation-switchable Ca(2+)- and Tb(3+)-binding protein without the use of multiple coupled metal-binding sites. FEBS J. 2008;275:5048–5061. doi: 10.1111/j.1742-4658.2008.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uversky VN. Amyloidogenesis of natively unfolded proteins. Curr Alzheimer Res. 2008;5:260–287. doi: 10.2174/156720508784533312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 108.Sevier CS, Kaiser CA. Formation and transfer of disulphide bonds in living cells. Nat Rev Mol Cell Biol. 2002;3:836–847. doi: 10.1038/nrm954. [DOI] [PubMed] [Google Scholar]
- 109.Mamathambika BS, Bardwell JC. Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol. 2008;24:211–235. doi: 10.1146/annurev.cellbio.24.110707.175333. [DOI] [PubMed] [Google Scholar]
- 110.Abkevich VI, Shakhnovich EI. What can disulfide bonds tell us about protein energetics, function and folding: simulations and bioninformatics analysis. J Mol Biol. 2000;300:975–985. doi: 10.1006/jmbi.2000.3893. [DOI] [PubMed] [Google Scholar]
- 111.Nakamura H. Thioredoxin and its related molecules: update 2005. Antioxid Redox Signal. 2005;7:823–828. doi: 10.1089/ars.2005.7.823. [DOI] [PubMed] [Google Scholar]
- 112.Schmidt B, Ho L, Hogg PJ. Allosteric disulfide bonds. Biochemistry. 2006;45:7429–7433. doi: 10.1021/bi0603064. [DOI] [PubMed] [Google Scholar]
- 113.Yokota A, Izutani K, Takai M, Kubo Y, Noda Y, Koumoto Y, Tachibana H, Segawa S. The transition state in the folding-unfolding reaction of four species of three-disulfide variant of hen lysozyme: the role of each disulfide bridge. J Mol Biol. 2000;295:1275–1288. doi: 10.1006/jmbi.1999.3442. [DOI] [PubMed] [Google Scholar]
- 114.Creighton TE. Protein folding. An unfolding story. Curr Biol. 1995;5:353–356. doi: 10.1016/s0960-9822(95)00070-4. [DOI] [PubMed] [Google Scholar]
- 115.Narhi LO, Hua QX, Arakawa T, Fox GM, Tsai L, Rosenfeld R, Holst P, Miller JA, Weiss MA. Role of native disulfide bonds in the structure and activity of insulin-like growth factor 1: genetic models of protein-folding intermediates. Biochemistry. 1993;32:5214–5221. doi: 10.1021/bi00070a033. [DOI] [PubMed] [Google Scholar]
- 116.Thornton JM. Disulphide bridges in globular proteins. J Mol Biol. 1981;151:261–287. doi: 10.1016/0022-2836(81)90515-5. [DOI] [PubMed] [Google Scholar]
- 117.Guo ZY, Qiao ZS, Feng YM. The in vitro oxidative folding of the insulin superfamily. Antioxid Redox Signal. 2008;10:127–139. doi: 10.1089/ars.2007.1860. [DOI] [PubMed] [Google Scholar]
- 118.Weiss MA, Hua QX, Jia W, Chu YC, Wang RY, Katsoyannis PG. Hierarchical protein “un-design”: insulin’s intrachain disulfide bridge tethers a recognition alpha-helix. Biochemistry. 2000;39:15429–15440. doi: 10.1021/bi001905s. [DOI] [PubMed] [Google Scholar]
- 119.Chang SG, Choi KD, Jang SH, Shin HC. Role of disulfide bonds in the structure and activity of human insulin. Mol Cells. 2003;16:323–330. [PubMed] [Google Scholar]
- 120.Hua QX, Mayer JP, Jia W, Zhang J, Weiss MA. The folding nucleus of the insulin superfamily: a flexible peptide model foreshadows the native state. J Biol Chem. 2006;281:28131–28142. doi: 10.1074/jbc.M602616200. [DOI] [PubMed] [Google Scholar]
- 121.Zhu Q, Liang S, Martin L, Gasparini S, Menez A, Vita C. Role of disulfide bonds in folding and activity of leiurotoxin I: just two disulfides suffice. Biochemistry. 2002;41:11488–11494. doi: 10.1021/bi026136m. [DOI] [PubMed] [Google Scholar]
- 122.Drakopoulou E, Vizzavona J, Neyton J, Aniort V, Bouet F, Virelizier H, Menez A, Vita C. Consequence of the removal of evolutionary conserved disulfide bridges on the structure and function of charybdotoxin and evidence that particular cysteine spacings govern specific disulfide bond formation. Biochemistry. 1998;37:1292–1301. doi: 10.1021/bi9721086. [DOI] [PubMed] [Google Scholar]
- 123.Hober S, Forsberg G, Palm G, Hartmanis M, Nilsson B. Disulfide exchange folding of insulin-like growth factor I. Biochemistry. 1992;31:1749–1756. doi: 10.1021/bi00121a024. [DOI] [PubMed] [Google Scholar]
- 124.Huang QL, Zhao J, Tang YH, Shao SQ, Xu GJ, Feng YM. The sequence determinant causing different folding behaviors of insulin and insulin-like growth factor-1. Biochemistry. 2007;46:218–224. doi: 10.1021/bi0616798. [DOI] [PubMed] [Google Scholar]
- 125.White CE, Hunter MJ, Meininger DP, Garrod S, Komives EA. The fifth epidermal growth factor-like domain of thrombomodulin does not have an epidermal growth factor-like disulfide bonding pattern. Proc Natl Acad Sci USA. 1996;93:10177–10182. doi: 10.1073/pnas.93.19.10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bulaj G, Olivera BM. Folding of conotoxins: formation of the native disulfide bridges during chemical synthesis and biosynthesis of Conus peptides. Antioxid Redox Signal. 2008;10:141–155. doi: 10.1089/ars.2007.1856. [DOI] [PubMed] [Google Scholar]
- 127.Kang TS, Vivekanandan S, Jois SD, Kini RM. Effect of C-terminal amidation on folding and disulfide-pairing of alpha-conotoxin ImI. Angew Chem Int Ed Engl. 2005;44:6333–6337. doi: 10.1002/anie.200502300. [DOI] [PubMed] [Google Scholar]
- 128.Zhang RM, Snyder GH. Factors governing selective formation of specific disulfides in synthetic variants of alpha-conotoxin. Biochemistry. 1991;30:11343–11348. doi: 10.1021/bi00111a021. [DOI] [PubMed] [Google Scholar]
- 129.Clarke J, Fersht AR. Engineered disulfide bonds as probes of the folding pathway of barnase: increasing the stability of proteins against the rate of denaturation. Biochemistry. 1993;32:4322–4329. doi: 10.1021/bi00067a022. [DOI] [PubMed] [Google Scholar]
- 130.Maleknia SD, Reixach N, Buxbaum JN. Oxidation inhibits amyloid fibril formation of transthyretin. FEBS J. 2006;273:5400–5406. doi: 10.1111/j.1742-4658.2006.05532.x. [DOI] [PubMed] [Google Scholar]
- 131.Chugha P, Sage HJ, Oas TG. Methionine oxidation of monomeric lambda repressor: the denatured state ensemble under nondenaturing conditions. Protein Sci. 2006;15:533–542. doi: 10.1110/ps.051856406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao J, Yin DH, Yao Y, Sun H, Qin Z, Schoneich C, Williams TD, Squier TC. Loss of conformational stability in calmodulin upon methionine oxidation. Biophys J. 1998;74:1115–1134. doi: 10.1016/S0006-3495(98)77830-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wood MJ, Becvar LA, Prieto JH, Melacini G, Komives EA. NMR structures reveal how oxidation inactivates thrombomodulin. Biochemistry. 2003;42:11932–11942. doi: 10.1021/bi034646q. [DOI] [PubMed] [Google Scholar]
- 134.Dado GP, Gellman SH. Redox control of secondary structure in a designed peptide. J Am Chem Soc. 1993;115:12609–12610. [Google Scholar]
- 135.Wong CH. Protein glycosylation: new challenges and opportunities. J Org Chem. 2005;70:4219–4225. doi: 10.1021/jo050278f. [DOI] [PubMed] [Google Scholar]
- 136.Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 137.Kornfeld S. Trafficking of lysosomal enzymes in normal and disease states. J Clin Invest. 1986;77:1–6. doi: 10.1172/JCI112262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sola RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sinclair AM, Elliott S. Glycoengineering: the effect of glycosylation on the properties of therapeutic proteins. J Pharm Sci. 2005;94:1626–1635. doi: 10.1002/jps.20319. [DOI] [PubMed] [Google Scholar]
- 140.Berg-Fussman A, Grace ME, Ioannou Y, Grabowski GA. Human acid beta-glucosidase. N-glycosylation site occupancy and the effect of glycosylation on enzymatic activity. J Biol Chem. 1993;268:14861–14866. [PubMed] [Google Scholar]
- 141.Kobayashi T, Honke K, Jin T, Gasa S, Miyazaki T, Makita A. Components and proteolytic processing sites of arylsulfatase B from human placenta. Biochim Biophys Acta. 1992;1159:243–247. doi: 10.1016/0167-4838(92)90051-e. [DOI] [PubMed] [Google Scholar]
- 142.Millat G, Froissart R, Maire I, Bozon D. IDS transfer from overexpressing cells to IDS-deficient cells. Exp Cell Res. 1997;230:362–367. doi: 10.1006/excr.1996.3435. [DOI] [PubMed] [Google Scholar]
- 143.Scriver CR. The metabolic & molecular bases of inherited disease. New York: McGraw-Hill; 2001. [Google Scholar]
- 144.Zhao KW, Faull KF, Kakkis ED, Neufeld EF. Carbohydrate structures of recombinant human alpha-L-iduronidase secreted by Chinese hamster ovary cells. J Biol Chem. 1997;272:22758–22765. doi: 10.1074/jbc.272.36.22758. [DOI] [PubMed] [Google Scholar]
- 145.Shental-Bechor D, Levy Y. Effect of glycosylation on protein folding: a close look at thermodynamic stabilization. Proc Natl Acad Sci USA. 2008;105:8256–8261. doi: 10.1073/pnas.0801340105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wyss DF, Wagner G. The structural role of sugars in glycoproteins. Curr Opin Biotechnol. 1996;7:409–416. doi: 10.1016/s0958-1669(96)80116-9. [DOI] [PubMed] [Google Scholar]
- 147.Wormald MR, Dwek RA. Glycoproteins: glycan presentation and protein-fold stability. Structure. 1999;7:R155–R160. doi: 10.1016/s0969-2126(99)80095-1. [DOI] [PubMed] [Google Scholar]
- 148.Walsh MT, Watzlawick H, Putnam FW, Schmid K, Brossmer R. Effect of the carbohydrate moiety on the secondary structure of beta 2-glycoprotein. I. Implications for the biosynthesis and folding of glycoproteins. Biochemistry. 1990;29:6250–6257. doi: 10.1021/bi00478a020. [DOI] [PubMed] [Google Scholar]
- 149.Wyss DF, Choi JS, Li J, Knoppers MH, Willis KJ, Arulanandam AR, Smolyar A, Reinherz EL, Wagner G. Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science. 1995;269:1273–1278. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]
- 150.O’Connor SE, Imperiali B. Modulation of protein structure and function by asparagine-linked glycosylation. Chem Biol. 1996;3:803–812. doi: 10.1016/s1074-5521(96)90064-2. [DOI] [PubMed] [Google Scholar]
- 151.Rickert KW, Imperiali B. Analysis of the conserved glycosylation site in the nicotinic acetylcholine receptor: potential roles in complex assembly. Chem Biol. 1995;2:751–759. doi: 10.1016/1074-5521(95)90103-5. [DOI] [PubMed] [Google Scholar]
- 152.Shaanan B, Lis H, Sharon N. Structure of a legume lectin with an ordered N-linked carbohydrate in complex with lactose. Science. 1991;254:862–866. doi: 10.1126/science.1948067. [DOI] [PubMed] [Google Scholar]
- 153.Helenius A. How N-linked oligosaccharides affect glycoprotein folding in the endoplasmic reticulum. Mol Biol Cell. 1994;5:253–265. doi: 10.1091/mbc.5.3.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hickman S, Kornfeld S. Effect of tunicamycin on IgM, IgA, and IgG secretion by mouse plasmacytoma cells. J Immunol. 1978;121:990–996. [PubMed] [Google Scholar]
- 155.Landolfi NF, Rich RR, Cook RG. Differential glycosylation requirements for the cell surface expression of class I molecules. J Immunol. 1985;134:423–430. [PubMed] [Google Scholar]
- 156.Shackelford DA, Strominger JL. Analysis of the oligosaccharides on the HLA-DR and DC1 B cell antigens. J Immunol. 1983;130:274–282. [PubMed] [Google Scholar]
- 157.Hebert DN, Zhang JX, Chen W, Foellmer B, Helenius A. The number and location of glycans on influenza hemagglutinin determine folding and association with calnexin and calreticulin. J Cell Biol. 1997;139:613–623. doi: 10.1083/jcb.139.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hermans MM, Wisselaar HA, Kroos MA, Oostra BA, Reuser AJ. Human lysosomal alpha-glucosidase: functional characterization of the glycosylation sites. Biochem J. 1993;289:681–686. doi: 10.1042/bj2890681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ioannou YA, Zeidner KM, Grace ME, Desnick RJ. Human alpha-galactosidase A: glycosylation site 3 is essential for enzyme solubility. Biochem J. 1998;332:789–797. doi: 10.1042/bj3320789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kotch FW, Guzei IA, Raines RT. Stabilization of the collagen triple helix by O-methylation of hydroxyproline residues. J Am Chem Soc. 2008;130:2952–2953. doi: 10.1021/ja800225k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Raines RT. 2005 Emil Thomas Kaiser Award. Protein Sci. 2006;15:1219–1225. doi: 10.1110/ps.062139406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brodsky B, Ramshaw JA. The collagen triple-helix structure. Matrix Biol. 1997;15:545–554. doi: 10.1016/s0945-053x(97)90030-5. [DOI] [PubMed] [Google Scholar]
- 163.Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lopez-Vera E, Walewska A, Skalicky JJ, Olivera BM, Bulaj G. Role of hydroxyprolines in the in vitro oxidative folding and biological activity of conotoxins. Biochemistry. 2008;47:1741–1751. doi: 10.1021/bi701934m. [DOI] [PubMed] [Google Scholar]
- 165.Kubo S, Chino N, Kimura T, Sakakibara S. Oxidative folding of omega-conotoxin MVIIC: effects of temperature and salt. Biopolymers. 1996;38:733–744. doi: 10.1002/(sici)1097-0282(199606)38:6<733::aid-bip5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 166.Koivunen P, Salo KE, Myllyharju J, Ruddock LW. Three binding sites in protein-disulfide isomerase cooperate in collagen prolyl 4-hydroxylase tetramer assembly. J Biol Chem. 2005;280:5227–5235. doi: 10.1074/jbc.M412480200. [DOI] [PubMed] [Google Scholar]
- 167.Martinez A, Treston AM. Where does amidation take place? Mol Cell Endocrinol. 1996;123:113–117. doi: 10.1016/s0303-7207(96)03903-2. [DOI] [PubMed] [Google Scholar]
- 168.Merkler DJ. C-terminal amidated peptides: production by the in vitro enzymatic amidation of glycine-extended peptides and the importance of the amide to bioactivity. Enzyme Microb Technol. 1994;16:450–456. doi: 10.1016/0141-0229(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 169.Sforca ML, Oyama S, Jr, Canduri F, Lorenzi CC, Pertinhez TA, Konno K, Souza BM, Palma MS, Ruggiero Neto J, Azevedo WF, Jr, Spisni A. How C-terminal carboxyamidation alters the biological activity of peptides from the venom of the eumenine solitary wasp. Biochemistry. 2004;43:5608–5617. doi: 10.1021/bi0360915. [DOI] [PubMed] [Google Scholar]
- 170.Yasin B, Lehrer RI, Harwig SS, Wagar EA. Protegrins: structural requirements for inactivating elementary bodies of Chlamydia trachomatis . Infect Immun. 1996;64:4863–4866. doi: 10.1128/iai.64.11.4863-4866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Katayama H, Ohira T, Aida K, Nagasawa H. Significance of a carboxyl-terminal amide moiety in the folding and biological activity of crustacean hyperglycemic hormone. Peptides. 2002;23:1537–1546. doi: 10.1016/s0196-9781(02)00094-3. [DOI] [PubMed] [Google Scholar]
- 172.Mor A, Nicolas P. The NH2-terminal alpha-helical domain 1–18 of dermaseptin is responsible for antimicrobial activity. J Biol Chem. 1994;269:1934–1939. [PubMed] [Google Scholar]
- 173.Sandvik AK, Dockray GJ. Biological activity of carboxy-terminal gastrin analogs. Eur J Pharmacol. 1999;364:199–203. doi: 10.1016/s0014-2999(98)00846-2. [DOI] [PubMed] [Google Scholar]
- 174.Ali MF, Soto A, Knoop FC, Conlon JM. Antimicrobial peptides isolated from skin secretions of the diploid frog, Xenopus tropicalis (Pipidae) Biochim Biophys Acta. 2001;1550:81–89. doi: 10.1016/s0167-4838(01)00272-2. [DOI] [PubMed] [Google Scholar]
- 175.Meredith J, Ring M, Macins A, Marschall J, Cheng NN, Theilmann D, Brock HW, Phillips JE. Locust ion transport peptide (ITP): primary structure, cDNA and expression in a baculovirus system. J Exp Biol. 1996;199:1053–1061. doi: 10.1242/jeb.199.5.1053. [DOI] [PubMed] [Google Scholar]
- 176.Fuller E, Green BR, Catlin P, Buczek O, Nielsen JS, Olivera BM, Bulaj G. Oxidative folding of conotoxins sharing an identical disulfide bridging framework. FEBS J. 2005;272:1727–1738. doi: 10.1111/j.1742-4658.2005.04602.x. [DOI] [PubMed] [Google Scholar]
- 177.Shearer MJ. Vitamin K metabolism and nutriture. Blood Rev. 1992;6:92–104. doi: 10.1016/0268-960x(92)90011-e. [DOI] [PubMed] [Google Scholar]
- 178.Hauschka PV, Lian JB, Gallop PM. Direct identification of the calcium-binding amino acid, gamma-carboxyglutamate, in mineralized tissue. Proc Natl Acad Sci USA. 1975;72:3925–3929. doi: 10.1073/pnas.72.10.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Howard JB, Nelsestuen GL. Isolation and characterization of vitamin K-dependent region of bovine blood clotting factor X. Proc Natl Acad Sci USA. 1975;72:1281–1285. doi: 10.1073/pnas.72.4.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Suttie JW. Vitamin K-dependent carboxylase. Annu Rev Biochem. 1985;54:459–477. doi: 10.1146/annurev.bi.54.070185.002331. [DOI] [PubMed] [Google Scholar]
- 181.McIntosh JM, Olivera BM, Cruz LJ, Gray WR. Gamma-carboxyglutamate in a neuroactive toxin. J Biol Chem. 1984;259:14343–14346. [PubMed] [Google Scholar]
- 182.Rigby AC, Lucas-Meunier E, Kalume DE, Czerwiec E, Hambe B, Dahlqvist I, Fossier P, Baux G, Roepstorff P, Baleja JD, Furie BC, Furie B, Stenflo J. A conotoxin from Conus textile with unusual posttranslational modifications reduces presynaptic Ca2+ influx. Proc Natl Acad Sci USA. 1999;96:5758–5763. doi: 10.1073/pnas.96.10.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bulaj G, Buczek O, Goodsell I, Jimenez EC, Kranski J, Nielsen JS, Garrett JE, Olivera BM. Efficient oxidative folding of conotoxins and the radiation of venomous cone snails. Proc Natl Acad Sci USA. 2003;100(Suppl 2):14562–14568. doi: 10.1073/pnas.2335845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gowd KH, Twede V, Watkins M, Krishnan KS, Teichert RW, Bulaj G, Olivera BM. Conantokin-P, an unusual conantokin with a long disulfide loop. Toxicon. 2008;52:203–213. doi: 10.1016/j.toxicon.2008.04.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chen Z, Blandl T, Prorok M, Warder SE, Li L, Zhu Y, Pedersen LG, Ni F, Castellino FJ. Conformational changes in conantokin-G induced upon binding of calcium and magnesium as revealed by NMR structural analysis. J Biol Chem. 1998;273:16248–16258. doi: 10.1074/jbc.273.26.16248. [DOI] [PubMed] [Google Scholar]
- 186.Latzer J, Shen T, Wolynes PG. Conformational switching upon phosphorylation: a predictive framework based on energy landscape principles. Biochemistry. 2008;47:2110–2122. doi: 10.1021/bi701350v. [DOI] [PubMed] [Google Scholar]
- 187.Johnson LN, Lewis RJ. Structural basis for control by phosphorylation. Chem Rev. 2001;101:2209–2242. doi: 10.1021/cr000225s. [DOI] [PubMed] [Google Scholar]
- 188.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 189.Tatko CD, Waters ML. Comparison of C-H…pi and hydrophobic interactions in a beta-hairpin peptide: impact on stability and specificity. J Am Chem Soc. 2004;126:2028–2034. doi: 10.1021/ja038258n. [DOI] [PubMed] [Google Scholar]
- 190.Hughes RM, Waters ML. Influence of N-methylation on a cation-pi interaction produces a remarkably stable beta-hairpin peptide. J Am Chem Soc. 2005;127:6518–6519. doi: 10.1021/ja0507259. [DOI] [PubMed] [Google Scholar]
- 191.Hughes RM, Waters ML. Arginine methylation in a beta-hairpin peptide: implications for Arg-pi interactions, DeltaCp(o), and the cold denatured state. J Am Chem Soc. 2006;128:12735–12742. doi: 10.1021/ja061656g. [DOI] [PubMed] [Google Scholar]
- 192.Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J. 1997;16:2054–2060. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Reynolds MR, Berry RW, Binder LI. Nitration in neurodegeneration: deciphering the “Hows” “nYs”. Biochemistry. 2007;46:7325–7336. doi: 10.1021/bi700430y. [DOI] [PubMed] [Google Scholar]
- 194.Pylypenko O, Schonichen A, Ludwig D, Ungermann C, Goody RS, Rak A, Geyer M. Farnesylation of the SNARE protein Ykt6 increases its stability and helical folding. J Mol Biol. 2008;377:1334–1345. doi: 10.1016/j.jmb.2008.01.099. [DOI] [PubMed] [Google Scholar]
- 195.Bowie JU, Reidhaar-Olson JF, Lim WA, Sauer RT. Deciphering the message in protein sequences: tolerance to amino acid substitutions. Science. 1990;247:1306–1310. doi: 10.1126/science.2315699. [DOI] [PubMed] [Google Scholar]
- 196.Plaxco KW, Riddle DS, Grantcharova V, Baker D. Simplified proteins: minimalist solutions to the ‘protein folding problem’. Curr Opin Struct Biol. 1998;8:80–85. doi: 10.1016/s0959-440x(98)80013-4. [DOI] [PubMed] [Google Scholar]
- 197.Miller JH. Genetic studies of the lac repressor. XI. On aspects of lac repressor structure suggested by genetic experiments. J Mol Biol. 1979;131:249–258. doi: 10.1016/0022-2836(79)90075-5. [DOI] [PubMed] [Google Scholar]
- 198.Miller JH, Coulondre C, Hofer M, Schmeissner U, Sommer H, Schmitz A, Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979;131:191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- 199.Miller JH, Schmeissner U. Genetic studies of the lac repressor. X. Analysis of missense mutations in the lacI gene. J Mol Biol. 1979;131:223–248. doi: 10.1016/0022-2836(79)90074-3. [DOI] [PubMed] [Google Scholar]
- 200.Brown BM, Sauer RT. Tolerance of Arc repressor to multiple-alanine substitutions. Proc Natl Acad Sci USA. 1999;96:1983–1988. doi: 10.1073/pnas.96.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Islam MM, Sohya S, Noguchi K, Yohda M, Kuroda Y. Crystal structure of an extensively simplified variant of bovine pancreatic trypsin inhibitor in which over one-third of the residues are alanines. Proc Natl Acad Sci USA. 2008;105:15334–15339. doi: 10.1073/pnas.0802699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kuroda Y, Kim PS. Folding of bovine pancreatic trypsin inhibitor (BPTI) variants in which almost half the residues are alanine. J Mol Biol. 2000;298:493–501. doi: 10.1006/jmbi.2000.3622. [DOI] [PubMed] [Google Scholar]
- 203.Munson M, O’Brien R, Sturtevant JM, Regan L. Redesigning the hydrophobic core of a four-helix-bundle protein. Protein Sci. 1994;3:2015–2022. doi: 10.1002/pro.5560031114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Baldwin RL. Temperature dependence of the hydrophobic interaction in protein folding. Proc Natl Acad Sci USA. 1986;83:8069–8072. doi: 10.1073/pnas.83.21.8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bashford D, Chothia C, Lesk AM. Determinants of a protein fold. Unique features of the globin amino acid sequences. J Mol Biol. 1987;196:199–216. doi: 10.1016/0022-2836(87)90521-3. [DOI] [PubMed] [Google Scholar]
- 206.Chothia C, Levitt M, Richardson D. Helix to helix packing in proteins. J Mol Biol. 1981;145:215–250. doi: 10.1016/0022-2836(81)90341-7. [DOI] [PubMed] [Google Scholar]
- 207.Kellis JT, Jr, Nyberg K, Sali D, Fersht AR. Contribution of hydrophobic interactions to protein stability. Nature. 1988;333:784–786. doi: 10.1038/333784a0. [DOI] [PubMed] [Google Scholar]
- 208.Eriksson AE, Baase WA, Zhang XJ, Heinz DW, Blaber M, Baldwin EP, Matthews BW. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science. 1992;255:178–183. doi: 10.1126/science.1553543. [DOI] [PubMed] [Google Scholar]
- 209.Gassner NC, Baase WA, Matthews BW. A test of the “jigsaw puzzle” model for protein folding by multiple methionine substitutions within the core of T4 lysozyme. Proc Natl Acad Sci USA. 1996;93:12155–12158. doi: 10.1073/pnas.93.22.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Lim WA, Sauer RT. The role of internal packing interactions in determining the structure and stability of a protein. J Mol Biol. 1991;219:359–376. doi: 10.1016/0022-2836(91)90570-v. [DOI] [PubMed] [Google Scholar]
- 211.Baldwin EP, Hajiseyedjavadi O, Baase WA, Matthews BW. The role of backbone flexibility in the accommodation of variants that repack the core of T4 lysozyme. Science. 1993;262:1715–1718. doi: 10.1126/science.8259514. [DOI] [PubMed] [Google Scholar]
- 212.Pielak GJ, Auld DS, Beasley JR, Betz SF, Cohen DS, Doyle DF, Finger SA, Fredericks ZL, Hilgen-Willis S, Saunders AJ. Protein thermal denaturation, side-chain models, and evolution: amino acid substitutions at a conserved helix–helix interface. Biochemistry. 1995;34:3268–3276. doi: 10.1021/bi00010a017. [DOI] [PubMed] [Google Scholar]
- 213.Axe DD, Foster NW, Fersht AR. Active barnase variants with completely random hydrophobic cores. Proc Natl Acad Sci USA. 1996;93:5590–5594. doi: 10.1073/pnas.93.11.5590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kamtekar S, Schiffer JM, Xiong H, Babik JM, Hecht MH. Protein design by binary patterning of polar and nonpolar amino acids. Science. 1993;262:1680–1685. doi: 10.1126/science.8259512. [DOI] [PubMed] [Google Scholar]
- 215.Moffet DA, Hecht MH. De novo proteins from combinatorial libraries. Chem Rev. 2001;101:3191–3203. doi: 10.1021/cr000051e. [DOI] [PubMed] [Google Scholar]
- 216.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 217.Schafmeister CE, LaPorte SL, Miercke LJ, Stroud RM. A designed four helix bundle protein with native-like structure. Nat Struct Biol. 1997;4:1039–1046. doi: 10.1038/nsb1297-1039. [DOI] [PubMed] [Google Scholar]
- 218.Riddle DS, Santiago JV, Bray-Hall ST, Doshi N, Grantcharova VP, Yi Q, Baker D. Functional rapidly folding proteins from simplified amino acid sequences. Nat Struct Biol. 1997;4:805–809. doi: 10.1038/nsb1097-805. [DOI] [PubMed] [Google Scholar]
- 219.Taylor SV, Walter KU, Kast P, Hilvert D. Searching sequence space for protein catalysts. Proc Natl Acad Sci USA. 2001;98:10596–10601. doi: 10.1073/pnas.191159298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Walter KU, Vamvaca K, Hilvert D. An active enzyme constructed from a 9-amino acid alphabet. J Biol Chem. 2005;280:37742–37746. doi: 10.1074/jbc.M507210200. [DOI] [PubMed] [Google Scholar]
- 221.Silverman JA, Balakrishnan R, Harbury PB. Reverse engineering the (beta/alpha)8 barrel fold. Proc Natl Acad Sci USA. 2001;98:3092–3097. doi: 10.1073/pnas.041613598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hemmingsen JM, Gernert KM, Richardson JS, Richardson DC. The tyrosine corner: a feature of most Greek key beta-barrel proteins. Protein Sci. 1994;3:1927–1937. doi: 10.1002/pro.5560031104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Yao J, Dyson HJ, Wright PE. Three-dimensional structure of a type VI turn in a linear peptide in water solution. Evidence for stacking of aromatic rings as a major stabilizing factor. J Mol Biol. 1994;243:754–766. doi: 10.1016/0022-2836(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 224.Moore DT, Berger BW, DeGrado WF. Protein–protein interactions in the membrane: sequence, structural, and biological motifs. Structure. 2008;16:991–1001. doi: 10.1016/j.str.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gsponer J, Vendruscolo M. Theoretical approaches to protein aggregation. Protein Pept Lett. 2006;13:287–293. doi: 10.2174/092986606775338407. [DOI] [PubMed] [Google Scholar]
- 226.Carrell RW, Lomas DA. Conformational disease. Lancet. 1997;350:134–138. doi: 10.1016/S0140-6736(97)02073-4. [DOI] [PubMed] [Google Scholar]
- 227.Conn PM, Ulloa-Aguirre A, Ito J, Janovick JA. G protein-coupled receptor trafficking in health and disease: lessons learned to prepare for therapeutic mutant rescue in vivo. Pharmacol Rev. 2007;59:225–250. doi: 10.1124/pr.59.3.2. [DOI] [PubMed] [Google Scholar]
- 228.Brady RO. Enzyme replacement for lysosomal diseases. Annu Rev Med. 2006;57:283–296. doi: 10.1146/annurev.med.57.110104.115650. [DOI] [PubMed] [Google Scholar]
- 229.Santucci R, Sinibaldi F, Fiorucci L. Protein folding, unfolding and misfolding: role played by intermediate States. Mini Rev Med Chem. 2008;8:57–62. doi: 10.2174/138955708783331522. [DOI] [PubMed] [Google Scholar]
- 230.Dahiyat BI, Mayo SL. De novo protein design: fully automated sequence selection. Science. 1997;278:82–87. doi: 10.1126/science.278.5335.82. [DOI] [PubMed] [Google Scholar]
- 231.Kortemme T, Ramirez-Alvarado M, Serrano L. Design of a 20-amino acid, three-stranded beta-sheet protein. Science. 1998;281:253–256. doi: 10.1126/science.281.5374.253. [DOI] [PubMed] [Google Scholar]
- 232.Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- 233.Hecht MH, Richardson JS, Richardson DC, Ogden RC. De novo design, expression, and characterization of Felix: a four-helix bundle protein of native-like sequence. Science. 1990;249:884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- 234.Magliery TJ, Regan L. Combinatorial approaches to protein stability and structure. Eur J Biochem. 2004;271:1595–1608. doi: 10.1111/j.1432-1033.2004.04075.x. [DOI] [PubMed] [Google Scholar]
- 235.Baltzer L, Nilsson H, Nilsson J. De novo design of proteins—what are the rules? Chem Rev. 2001;101:3153–3163. doi: 10.1021/cr0000473. [DOI] [PubMed] [Google Scholar]
- 236.Moult J, Fidelis K, Kryshtafovych A, Rost B, Hubbard T, Tramontano A. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69(Suppl 8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Honig B. Protein folding: from the levinthal paradox to structure prediction. J Mol Biol. 1999;293:283–293. doi: 10.1006/jmbi.1999.3006. [DOI] [PubMed] [Google Scholar]
- 238.Walsh G, Jefferis R. Post-translational modifications in the context of therapeutic proteins. Nat Biotechnol. 2006;24:1241–1252. doi: 10.1038/nbt1252. [DOI] [PubMed] [Google Scholar]
- 239.Bradbury AF, Smyth DG. Peptide amidation. Trends Biochem Sci. 1991;16:112–115. doi: 10.1016/0968-0004(91)90044-v. [DOI] [PubMed] [Google Scholar]
- 240.Dong J, Ye P, Schade AJ, Gao S, Romo GM, Turner NT, McIntire LV, Lopez JA. Tyrosine sulfation of glycoprotein I(b)alpha. Role of electrostatic interactions in von Willebrand factor binding. J Biol Chem. 2001;276:16690–16694. doi: 10.1074/jbc.M101035200. [DOI] [PubMed] [Google Scholar]
- 241.Tarcsa E, Marekov LN, Mei G, Melino G, Lee SC, Steinert PM. Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem. 1996;271:30709–30716. doi: 10.1074/jbc.271.48.30709. [DOI] [PubMed] [Google Scholar]
- 242.Janke C, Rogowski K, van Dijk J. Polyglutamylation: a fine-regulator of protein function? ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:636–641. doi: 10.1038/embor.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.MacRae TH. Tubulin post-translational modifications—enzymes and their mechanisms of action. Eur J Biochem. 1997;244:265–278. doi: 10.1111/j.1432-1033.1997.00265.x. [DOI] [PubMed] [Google Scholar]