Abstract
Prion diseases are fatal neurodegenerative and infectious disorders of humans and animals, characterized by structural transition of the host-encoded cellular prion protein (PrPc) into the aberrantly folded pathologic isoform PrPSc. RNA, DNA or peptide aptamers are classes of molecules which can be selected from complex combinatorial libraries for high affinity and specific binding to prion proteins and which might therefore be useful in diagnosis and therapy of prion diseases. Nucleic acid aptamers, which can be chemically synthesized, stabilized and immobilized, appear more suitable for diagnostic purposes, allowing use of PrPSc as selection target. Peptide aptamers facilitate appropriate intracellular expression, targeting and re-routing without losing their binding properties to PrP, a requirement for potential therapeutic gene transfer experiments in vivo. Elucidation of structural properties of peptide aptamers might be used as basis for rational drug design, providing another attractive application of peptide aptamers in the search for effective anti-prion strategies.
Keywords: Prion, Prion protein, Aptamer, Peptide aptamer, Combinatorial library, Anti-prion compound
Prions and prion diseases
Prion diseases, also called transmissible spongiform encephalopathies (TSEs), are strictly fatal neurodegenerative and infectious disorders that can affect both humans and animals. Animal prion diseases comprise scrapie in sheep and goats, bovine spongiform encephalopathy (BSE) in cattle, chronic wasting disease (CWD) in elk and deer, feline spongiform encephalopathy (FSE) in domestic and wild cats, transmissible mink encephalopathy (TME) in mink, and exotic ungulate encephalopathy in zoo animals. Human prion diseases include Creutzfeldt–Jakob disease (CJD), Gerstmann–Sträussler–Scheinker (GSS) syndrome, fatal familial insomnia (FFI), kuru, and the new variants of CJD (vCJD and secondary vCJD). Prion diseases feature a rapidly progressing clinical course that leads inevitably to death, usually within months. Typically, this is preceded by a long incubation period free of symptoms, lasting for years to many decades, for example, in humans. Severe neuronal loss is characteristic for prion diseases, accompanied by strong astrogliosis and mild microglia activation. This results in a progressive severe spongiform vacuolation and degeneration of the central nervous system (CNS) manifesting itself particularly in ataxia, behavioral changes and, in the human case, in dementia, a highly progressive loss of intellectual abilities [1–5].
The protein-only hypothesis elaborated mainly by S.B. Prusiner states that these diseases are caused by prions, proteinaceous infectious particles devoid of any encoding nucleic acid [2, 6]. Prions consist mainly, if not entirely, of the abnormally folded isoform (PrPSc) of the normal, host-encoded prion protein PrPc [2–4, 6–8]. Prions have self-propagating capacities and are able to catalyze a profound conformational change of PrPc into an aggregated form resulting finally in accumulation of misfolded and aggregated PrPSc in the brain. Prion pathology therefore shares striking similarities with other protein misfolding and neurodegenerative diseases like Alzheimer’s, Huntington’s and Parkinson’s disease [9, 10]. However, prions are unique as they are not only able to replicate their conformation, using PrPc as substrate and PrPSc as template, but are also naturally and experimentally transmissible within and to a significant extent also between species [2–4]. For human prion diseases, the existence of three distinct manifestations is another unique feature [1, 2, 5, 8] (Table 1). Human prion diseases can occur endogenously, either genetically by defined germ-line mutations, or more frequently, sporadically by a still unknown mechanism. Furthermore, they can be acquired by exogenous infection, e.g., by prion-contaminated foodstuff or medically used materials [1]. Kuru reached epidemic proportions amongst the Fore people in Papua New Guinea, where it was transferred by ritualistic cannibalism, in which brains of the deceased were eaten as a sign of dead person-worship [11]. The BSE epidemics in the 1980s and the emergence of BSE-caused vCJD in the 1990s significantly raised people’s awareness of prion diseases, by realizing that acquired forms can have epidemic and zoonotic potential and can become dangerous for the human population [12–15]. In contrast to ‘classical’ human prion diseases like CJD and GSS, in vCJD the infectious agent is abundant in the lymphoreticular system and many other organs beside the CNS [16]. Interestingly, this is also true for CWD, another only recently recognized highly horizontally transmissible prion disease [17, 18]. Not unexpectedly, vCJD transmission via blood transfusion was reported and appears to be rather effective, resulting in secondary vCJD [19, 20]. This fact certainly increases the risk of horizontal spread of vCJD within humans. With regard to the exogenous infection process and the long incubation time, it also offers the perspective that post-exposure drug application in peripheral sites of the body might prevent neuroinvasion of prions and onset of disease. Of note, to date there are neither reliable pre-clinical diagnostic means nor effective and feasible therapeutic or prophylactic strategies available against prion diseases.
Table 1.
Manifestations of human prion diseases
| Manifestation | Disease | Mechanism |
|---|---|---|
| Acquired | Kuru (epidemic in the 1950s); iatrogenic Creutzfeldt–Jakob disease (iCJD), variant CJD (vCJD) (total so far >200 cases) | Infection by environmental exposure to prions; exogenous |
| Genetic | Familial or genetic CJD (~10%); Gerstmann–Sträussler–Scheinker syndrome (GSS); fatal familial insomnia (FFI) | Mutation in the PRNP gene (more than 30 different types are known); endogenous |
| Sporadic | Sporadic CJD (~1 case per million per year worldwide, ~90%) | Apparently spontaneous formation of PrPSc; endogenous |
Cell biology of PrPc and PrPSc, the prion conversion process, and putative sites for intervention
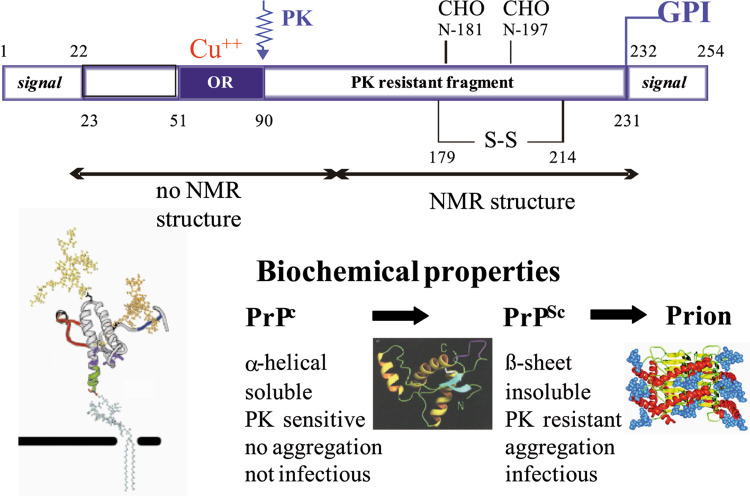
The normal host-encoded PrPc is a glycoprotein which transits the secretory pathway [21, 22]. Upon synthesis, it is co-translationally translocated into the lumen of the endoplasmic reticulum (ER), where the N-terminal signal sequence is removed (Fig. 1). Further post-translational modifications comprise the addition of carbohydrate chains at two N-glycosylation sites, the establishment of a disulfide bond, and the attachment of a glycosyl–phosphatidyl–inositol (GPI) anchor promoted by the C-terminal signal peptide which is thereby cleaved off. Properly folded PrPc exits the Golgi apparatus and is transported to the outer leaflet of the plasma membrane where it is incorporated via its GPI anchor moiety. PrPc resides mainly in lipid rafts, cholesterol and glycosphingolipid-rich domains of the plasma membrane. As these are important sites for signal transduction, it is suggestive that PrPc might have a physiological role in signaling processes. In addition, there is experimental evidence that PrPc has antioxidant properties and binds copper [23]. Overall, the function of PrPc still remains enigmatic and allows room for presumptions, although a neuroprotective role presently appears most likely [4, 23–27]. The 37-kDa/67-kDa laminin receptor (LRP/LR) is one of the described potential receptors for PrPc, prion uptake and conversion co-factor for prion formation [28, 29]. The spread of prions from cell to cell could be imparted by exosomes [30, 31].
Fig. 1.
Structural and biochemical properties of prion proteins. Schematic representation of PrPc, including primary structure and post-translational modifications. The signal peptide (SP) is removed during translocation into the ER lumen, residues 231–254 are removed in order to add a GPI anchor. The octapeptide repeat region (OR) encompasses residues 51–90. A disulfide bond and two N-glycosylation sites are present in the carboxy-terminal globular domain. The structured part of PrPc in NMR and the PK resistant part of PrPSc are indicated. At the lower left, a model of PrPc attached to the outer leaflet of the membrane via its GPI anchor is shown; in the middle, the NMR structure of PrPc [73]; and at the right, a structure model for N-terminally truncated PrPSc [75]
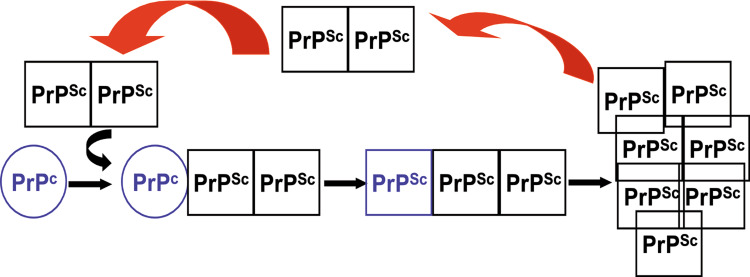
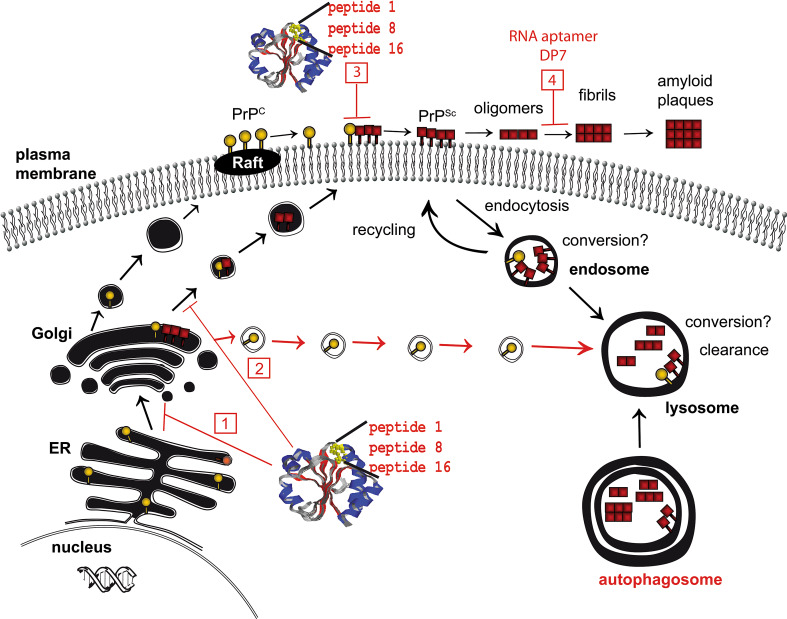
PrPc has a high content of α-helices whereas PrPSc is rich in β-sheet regions, rendering it insoluble and partially resistant to proteolytic digestion (Fig. 1) [2, 6]. The conformational conversion into PrPSc is thought to appear at the cell surface, in lipid rafts or caveolae-like domains, or along the early endocytotic pathway [32, 33], and is postulated to involve a direct physical contact between PrPc and PrPSc. The exact molecular mechanism of conversion is still the subject of intensive research. Several lines of evidence suggest a more general model in which amyloids are formed by a crystallization-like process, known as ‘seeded-nucleation’ ([34, 35]; Fig. 2). The process provides several possible targets for intervention, for most of which examples have been experimentally addressed (Fig. 3). Such targets are PrPc synthesis [36] or proper cell surface localization of PrPc [37]. Another experimental strategy might be over-stabilization of PrPc or PrPSc [38, 39], thereby impeding the nucleation process. Sterical hindrance of PrPc/PrPSc interaction is a widely used experimental approach [40–42], including interference with putative [43, 44] and yet to be characterized co-factors in the conversion process. Reducing PrPSc template by increasing the cellular degradation for prions represents a more recent target [45–48]. Absorbing prions at the periphery of the body [49, 50] before they can start replication and neuroinvasion is another strategy for preventing prion diseases.
Fig. 2.
Conversion process according to the ‘seeding-nucleation’ model. For conversion of PrPc into PrPSc, initially the interaction of both isoforms is necessary. Subsequently, PrPc adopts the PrPSc conformation, and PrPSc forms high molecular weight aggregates. Theses aggregates break up and the oligomeric PrPSc units form new seeds for recruiting PrPc molecules for conversion
Fig. 3.
Targets for therapeutic strategies based on cell biology of prion proteins. For inhibition of the formation of PrPSc either PrPc or PrPSc can be targeted. Intracellular trapping or re-routing of PrPc can be achieved by PrP binding molecules like peptide aptamers (peptides 1, 8, 16) fused to appropriate targeting signals (1, 2 numbers referring to the numbers drawn in red boxes). Peptide aptamers expressed recombinantly in E. coli and purified can inhibit prion conversion if added to the culture media of infected cells (3). Regarding PrPSc aggregate formation as a target, molecules like RNA aptamers that bind to PrPc (DP7) can be used to enable degradation of PrPSc (4)
More problems than solutions in prion disease diagnosis and therapy
As prions use host-encoded PrPc as substrate for conversion, newly generated prions always have the same primary structure and obviously post-translational modifications as host-encoded PrPc [2–4]. Due to self-tolerance, they therefore do not provoke a detectable immune reaction in infected individuals. Antibodies can be generated experimentally in appropriate animals but they do not discriminate between PrPc and PrPSc isoforms [2, 6]. Although there are reports of antibodies specific for PrPSc in the literature [51, 52], these were never used in commercial diagnostic formats. Therefore, for diagnostic purposes PrPc is usually removed, e.g., by digestion with proteinase K, or rendered inaccessible by biochemical manipulations, and the remaining pathological PrPSc is then detected by these antibodies in immunoblot, ELISA or immunocytochemistry. Another caveat is that PrPSc is in the CNS and beyond the blood–brain barrier (BBB), although with some exceptions (e.g., vCJD, scrapie and CWD), and therefore biopsy or autopsy material is needed for examination. As a result of all this, preclinical diagnosis is presently barely possible in sporadic forms of prion disease. In vCJD, one hallmark is the accumulation of PrPSc in lymphoreticular organs, e.g., tonsils and appendix, at the pre-clinical stage [53], which opens the possibility of detecting PrPSc in biopsy material. The recently described cyclic amplification of protein misfolding (PMCA) technology which uses PrPSc as template and PrPc from brain tissue as substrate has increased the limit of detection by many orders of magnitude [54, 55], although its diagnostic use is not yet guaranteed. However, a sensitive test including a PMCA amplification step combined with signal amplification by using T7 polymerase (Am-A-FACTT) has been developed [56]. This assay enabled detection of PrPSc aggregates in the blood of CWD-infected deer and elk as an example of a naturally occurring prion disease. Several other promising approaches to enable detection of PrPSc in a pre-clinical state have been described [57], and pre-clinical diagnosis might be supported by the analysis of surrogate markers indicative for prion diseases [58].
In terms of therapy and prophylaxis, no effective and feasible means against prion diseases are yet available, although some reliable proof-of-principle was obtained in a variety of experimental in vitro and animal models (for review, see [59]). One general caveat is that human prion diseases are very rare. However, numbers of sporadic CJD patients are constant with an incidence of ~1:1,000,000 per year world-wide, corresponding to >5,000 victims every year. Numbers of zoonotic (vCJD: >200 cases in total) and iatrogenic (e.g., by contaminated hGH or dura mater transplants) CJD are much lower, although they have gained much more attention in the public awareness. As preclinical diagnosis is missing, patients are diagnosed when the disease is in an already highly progressed stage with severe neurodegeneration, implying that therapeutic approaches may be too late as reversion of pathology is unlikely. However, there is very good experimental evidence from recent conditional transgenic animal models that some kind of reversion of pathologic alterations and even clinical symptoms might be achievable [60, 61]. Furthermore, prion diseases are strictly disorders of the CNS and any therapeutic compound has to effectively cross the BBB. Unfortunately, almost all compounds which showed anti-prion activity in in vitro and/or cell culture assays suffer from rather ineffective crossing of BBB. On the other hand, the long incubation period from years to decades in acquired forms may offer the potential of post-exposure intervention at peripheral sites of the body, and the compounds not able to penetrate the BBB might be effective here. Finally, in recent years, the common idea has evolved that it will not be a single compound but more the intelligent combination of compounds, resulting in additive or even synergistic anti-prion mechanisms and less pronounced side-effects.
Aptamer technology and nucleic acid aptamers
Originally, the term ‘aptamer’ designated synthetic single-stranded RNA or DNA molecules selected from combinatorial libraries for binding to a certain target molecule due to their folding into unique 3D structures. Selection requires a process called SELEX (systematic evolution of ligands by exponential enrichment) [62, 63]. Here, a chemically synthesized DNA library is amplified by PCR, or, in the case of RNA aptamers, the RNA library is generated by in vitro transcription. This pool of nucleic acid molecules is incubated with the target and then the non-binding aptamers are removed. Binding aptamers are eluted from the target, amplified and again incubated with the target. By repeating this cycle several times under increasing stringency, aptamers with high affinity and specificity for their target are enriched [62, 63]. A vast variety of molecules can serve as targets, including peptides [64] and proteins [65, 66]. RNA or DNA aptamers can be chemically synthesized in large amounts, and the problem of instability, in particular of RNA aptamers, can be overcome by introducing chemical modifications, e.g., 2′-fluoro- or 2′-amino-substituted pyrimidines [67], which mediate increased resistance to nucleases. In contrast to antibodies, their small molecule size encourages aptamers to bind to functional pockets of proteins, and they can be selected to even discriminate between conformational isoforms of proteins. These properties might be important for diagnosis or therapy of protein misfolding diseases such as prion diseases. With respect to diagnosis, aptamers specifically recognizing PrPSc and ideally binding to native PrPSc without the necessity of preceding PK treatment of samples would be desirable. In the case of therapy, both PrPc and PrPSc could serve as targets, as illustrated in Figs. 3 and 4, providing that binding to PrPc does not interfere with its still not definitely known physiological function. In therapeutic approaches, the PrP region recognized by aptamers is critical for successful inhibition of prion conversion. Using anti-PrP antibodies, Fab fragments or PrP derived peptides, several PrP domains were defined that appear to be involved in conversion processes, as covering of these specific sites negatively influenced PrPSc propagation either in in vitro assays or in prion-infected cell culture models [41, 42, 68–70]. However, it has to be taken into account that sterical hindrance of conversion by comparatively bulky molecules like IgG cannot be completely excluded.
Fig. 4.
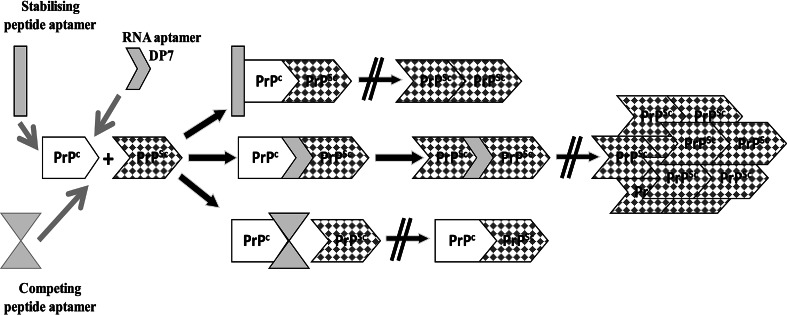
Scheme depicting proposed sites of interference of aptamers. By binding of peptide aptamers to PrPc, the interaction with PrPSc can be inhibited either by covering the binding sites or by withdrawal of PrPc from the subcellular site of conversion (competing peptide aptamer). Alternatively, PrPc–PrPSc interaction can take place despite binding of peptide aptamers, but PrPc is overstabilized and not eligible for structural transitions (stabilising peptide aptamer). In contrast, RNA aptamer DP7 binds to PrPc. It does not inhibit conversion to PrPSc; however, it might interfere with the formation of high molecular weight PrPSc aggregates
The remarkable ability of aptamers to distinguish between protein isoforms has already been described in the first study by Weiss et al. [71] reporting on RNA aptamers that specifically interacted with the cellular isoform PrPc of mice, hamster and cattle, but did not bind to disease-associated PrP27-30 in brain homogenates of prion infected mice. Mapping of the binding sites of these aptamers to PrP revealed that they recognize residues 23–52 [71], which explains the failure to interact with PrP27-30, since the N-terminal target sequence of the aptamer is removed upon PK digestion. For PrP binding aptamers in this study, structure prediction revealed a G quartet scaffold, and the formation of this structure was essential for the interaction [71]. Another RNA aptamer (DP7) revealing G quartet structure was selected against the human PrP peptide encompassing residues 90–129 [72], a highly conserved sequence covering the hydrophobic domain of PrP which is flexible in PrPc and folds into β-sheets upon conversion into PrPSc [73–75]. DP7 binds to PrPs of different species and, when applied to prion-infected cell cultures, the accumulation of PK resistant PrPSc was reduced, although initial steps of conversion appeared to occur since N-terminally truncated and insoluble PrP was present. Thus, it was suggested that RNA aptamers intercalated into PrP aggregates and might therefore prevent the formation of high-molecular weight aggregates [72]. Of note, this was the only study employing prion-infected cultured cells for investigating the conversion inhibitory activity of nucleic acid aptamers. However, novel insights into the conversion process demonstrated that the most infectious unit with the best seeding properties of RML prions appears to consist of 12–15 PrPSc molecules [76]. Therefore, the question arises whether inhibition of high molecular weight formation is indeed deleterious for prion propagation or rather promotes prion conversion over time due to an increased formation of such oligomeric PrPSc molecules.
The binding site of DP7 partially overlaps with that of RNA aptamer SAF-93 stabilized by 2′-fluoro-modification and selected against hamster-derived scrapie-associated fibrils [77]. SAF-93 in contrast to DP7 preferentially binds to β-PrP recognizing a region that at least partially is located between residues 106 and 126. However, a second binding site was mapped to the N-terminal part of PrP which appeared to be non-conformation specific since the dissociation constant (K d) was 8 μM, which is high compared to that of the conformation specific C-terminal site (16 nM). SAF-93 inhibited conversion to PrPSc with an IC50 of 40 nM, which was about 20-fold lower than that of DP7 [72]. For SAF-93, this value was achieved in in vitro conversion assays, whereas effects of DP7 were tested directly in prion-infected cultured cells. Thus, IC50 values are hardly comparable since in vitro conversion assays probably allow much more defined conditions and non-specific interactions, for example with other cellular proteins or if factors in the culture medium are excluded. As SAF-93 appears to be specific for β-sheet-rich and disease-associated PrP and recognizes PrPs of various species including bovine PrP, it could be a valuable tool for diagnostic purposes. In addition, the specificity for PrPSc did not depend on PK digestion and therefore abnormally folded forms that might be infectious should be recognized. Without affecting binding specificity and affinity, SAF-93 could be reduced from the original 116 nucleotides to a length suitable for chemical synthesis. It is possible to derivatize the aptamer with biotin without negatively influencing its binding properties, which provides the possibility for application in multiple detection formats [78]. Similarly, RNA aptamer 60-3 selected against recombinant murine PrP did not show altered binding affinities upon 2′-fluoro modification and biotinylation at its 5′ end [79]. Its PrP binding site was again located at the amino-terminal part of PrP between residues 23 and 108. However, the authors argued that the main binding site is located between residues 23 and 89 since deletion of this part reduced the affinity for recombinant mouse PrP about 10-fold. Notably, this region has been discussed to be a non-specific binding site for nucleic acids [77] and was also mapped for the RNA aptamer isolated by Weiss et al. [71]. Aptamer 60-3 recognized, in addition to recombinant mouse PrP, brain-derived PrPc and, albeit with lower affinity, recombinant bovine PrP. The decreased affinity might be due to alterations in the binding site of 60-3, since bovine PrP harbors an additional octapeptide sequence [79, 80]. As described above, so far no definite physiological function could be ascribed to PrPc, but there is evidence that prion proteins can bind nucleic acids [81, 82]. In order to identify RNA sequences that preferentially bind to susceptible allelic PrP, variant RNA aptamers were selected against ovine PrP carrying either the susceptible VRQ or the resistant ARR genotype [83]. The most frequently isolated aptamer RM312 shared striking similarity with DP7 but did not discriminate between the allelic variants of ovine PrP. However, two binding sites were mapped for RM312, spanning residues 25–34 and 101–110, respectively. The second binding site indeed correlates with parts of the sequence employed for the selection of DP7 [72]. Another motif was almost perfectly conserved with RNA aptamer SAF131 selected against hamster SAFs [77]. The amino-terminal binding site has also been described several times before [71, 77, 79]. With respect to sequence homologies with cellular RNAs, a search in Genbank database with the minimal binding motif revealed similarities with several known mRNAs which were not characterized further [83].
For diagnostic purposes, DNA aptamers might be advantageous compared to RNA aptamers in that they are more resistant against nucleases. This property might be important in cases where aptamers are applied for analyses which require incubation with enzyme-rich material like body fluids or in cases of prion diagnostics with brain or tissue homogenates. Indeed, DNA aptamers recognizing human recombinant PrP were selected by SELEX. Further evaluation revealed that aptamers interacting with recombinant PrP were also able to bind to brain- or cell culture-derived PrPc of various species including cattle, sheep and deer [84]. The binding sites located between residues 23 and 89 were similar to those of many RNA aptamers, and binding was specific for PrPc. The authors suggest that PrPc specific aptamers could be employed for pre-absorption of diagnostic material, which subsequently might allow analysis without PK digestion, assuming that residual PrP is the disease-associated isoform which did not bind to the DNA aptamers. Along this line, aptamers described by Takemura et al. [85] were analysed for their ability to bind to recombinant human PrP upon immobilization on gold-coated magnetic nanoparticles. The finding that interaction with PrP was still possible despite immobilization and modification of the aptamer might provide the background for sensitive diagnostic application of PrPSc specific nucleic acid aptamers.
To avoid interactions with N-terminal PrP, as was frequently found for RNA aptamers, DNA aptamers where selected against N-terminally truncated PrP [86]. This screen resulted in aptamers recognizing native PrPc at the surface of cultured cells. These aptamers appear to be more promising tools for diagnostic purposes, since a trivalent pool of aptamers was reactive against guanidinium-denatured PrPSc. This might enable the development of a highly sensitive PrPSc detection system with specificity for conformational differences similar to the conformation-dependent immunoassay [87]. Unfortunately, DNA aptamers described so far only recognized PrPc and denatured PrPSc, and despite discussing their potential therapeutic applications, the question whether these aptamers inhibit prion conversion at least in in vitro conversion assays or cell culture models of prion infection has not yet been addressed. As a note of caution, it has to be mentioned that nucleic acid aptamers in general might be difficult for therapeutic application in prion diseases. On the one hand, if chemically synthesized aptamers are employed, the BBB will pose a major problem, such as for most of the so far identified therapeutic compounds. On the other hand, lentiviral delivery to the brain as successfully performed with siRNA constructs targeting PrPc mRNA [88, 89] might also be problematic. Although RNA aptamers can be transcribed intracellularly from a plasmid construct [90], they are still located within the cytosol. In the case of PrP binding aptamers, this excludes binding to their target protein due to compartmentalization, since PrPc is expressed within vesicles of the secretory pathway or at the cell surface. This problem could be circumvented by using peptide aptamers, which are introduced in the following section.
Peptide aptamers can be targeted to the secretory pathway and inhibit cellular prion conversion
Peptide aptamers are the most recently described class of aptamers, and the term refers to a class of molecules consisting of a variable peptide sequence, derived from a combinatorial library and embedded within a constant scaffold protein. The variable peptide sequence is linked N- and C-terminally to the scaffold [91, 92]. Thereby, its conformational freedom is reduced compared to free peptides or peptides fused terminally to a carrier protein, leading to a more stable fold and increased affinity for their target molecules and an increased stability [93]. Peptide aptamers were used for inhibition or activation of target proteins, and the identification of protein–protein interactions [94], and they can be used as a basis for rational drug design [95]. Peptide aptamers are usually selected by yeast-two hybrid screening, providing the advantage of intracellular screening in contrast to in vitro methods like phage display. There are several scaffold proteins available, e.g., green fluorescent protein [96], the Z domain of staphylococcal protein A [97], the lipocalin fold [98], and the most frequently used E. coli Thioredoxin A (trxA). In the latter, the peptide moiety is inserted into the active site loop of the protein, thereby destroying its catalytic activity. A trxA-based peptide library has been used to select peptide aptamers targeting cyclin-dependent kinase 2 which enabled successful inhibition of the target protein [91]. Once peptide aptamers recognizing their target protein are selected, affinity and specificity can be improved by random mutagenesis of the peptide moiety [99]. One clear advantage over nucleic acid aptamers is that it is possible to create targeting and modifying peptide aptamers by fusion, e.g., to a nuclear import signal and to the active part of an ubiquitin ligase, respectively [99]. This enables modulation of the target not only by direct interaction but also by guiding proteins to certain cellular locations and functions. Although, so far, in vivo applications are highly limited, functional peptide aptamers have been generated, among others, against the hepatitis B-virus core protein [100], the human papilloma virus proteins E6 and E7 [101, 102], or the epidermal growth factor receptor [103].
In respect of prion proteins, both binding to PrP and modification of the subcellular transport of PrPc provide interesting approaches that could interfere with PrPSc formation. Our group recently demonstrated that peptide aptamers interacting with PrP that were recombinantly expressed and purified from E. coli inhibit PrPSc accumulation in prion-infected cultured cells when added to the culture medium [104]. Furthermore, in this study, for the first time trxA-based peptide aptamers were targeted to the secretory pathway which did not alter their binding properties to PrPc. Intracellular expression of these peptide aptamers fused carboxy-terminally to an ER retention motif or to a transmembrane and cytosolic domain, that induces a bypass of the plasma membrane in prion-infected cells, also resulted in inhibition of prion conversion and an impairment of primary prion infection. These observations are highly interesting since they demonstrate that peptide aptamers can be expressed intracellularly and can be targeted to the secretory pathway, in contrast to nucleic acid aptamers. It opens the possibility, on the one hand, for evaluation of peptide aptamers for therapeutic or prophylactic treatment of prion diseases when delivered directly to the CNS, e.g., by lentiviral transfer of appropriate vector constructs as already performed with recombinant viruses encoding single chain variable fragments (scFvs) against PrP [105]. On the other hand, elucidating the structure of the PrP-binding peptide moiety might help to select or synthesize small molecule drugs harboring similar binding properties as the peptide aptamer. A more rational approach in the search for peptide inhibitors of prion propagation would be to design antisense peptides. Such molecules are designed by transcription of the antisense strand of a given target protein, and it has been demonstrated by several instances that such peptides interact with the protein encoded by the sense strand [106–109]. Interestingly, the presence of naturally occurring antisense peptides encoded by the antisense strand of the PrP gene [110] or by normal cellular RNAs interacting with PrP mRNA [111] have been described, and peptides thereof might provide lead structures for designing nucleic acid or peptide aptamers. However, this strategy would limit the peptide sequence variability since only antisense-strand-derived peptides would be used. In contrast, a clear advantage of screening a combinatorial library is the huge variety of structures from which the best binding peptide can be selected.
In conclusion, RNA, DNA or peptide aptamers are interesting classes of molecules to improve diagnosis, prophylaxis or therapy of prion diseases, if reasonably applied. Due to the in vitro selection process for nucleic acid aptamers, they might be more suitable for diagnostic approaches. Only with this technology can PrPSc be used for the library screen to identify PrPSc specific molecules, which is not possible in a yeast-2-hybrid screen, the method of choice for selecting peptide aptamers. Furthermore, the possibilities to chemically synthesise large amounts of aptamers and to introduce functional groups for surface immobilisation are beyond question properties that make them ideal candidates for diagnostic purposes. In terms of prophylaxis or therapy of prion diseases, peptide aptamers might be more favorable due to the possibilities of intracellular expression and targeting without losing their binding properties, both features which might allow gene transfer experiments in vivo. Elucidating the structure of peptide aptamers in order to use them as a basis for rational drug design is another attractive and promising possible application of peptide aptamers in eventually combating prion diseases.
References
- 1.Dearmond SJ, Prusiner SB. Etiology and pathogenesis of prion diseases. Am J Pathol. 1995;146:785–811. [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weissmann C. The state of the prion. Nat Rev Microbiol. 2004;2:861–871. doi: 10.1038/nrmicro1025. [DOI] [PubMed] [Google Scholar]
- 4.Aguzzi A, Polymenidou M. Mammalian prion biology: one century of evolving concepts. Cell. 2004;116:313–327. doi: 10.1016/S0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- 5.Collinge J. Molecular neurology of prion disease. J Neurol Neurosurg Psychiatry. 2005;76:906–919. doi: 10.1136/jnnp.2004.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 7.Cohen FE, Pan KM, Huang Z, Baldwin M, Fletterick RJ, Prusiner SB. Structural clues to prion replication. Science. 1994;264:530–531. doi: 10.1126/science.7909169. [DOI] [PubMed] [Google Scholar]
- 8.Collinge J. Prion diseases of humans and animals: their causes and molecular basis. Annu Rev Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 9.Aguzzi A, Haass C. Games played by rogue proteins in prion disorders and Alzheimer’s disease. Science. 2003;302:814–818. doi: 10.1126/science.1087348. [DOI] [PubMed] [Google Scholar]
- 10.Dobson CM. Protein aggregation and its consequences for human disease. Protein Pept Lett. 2006;13:219–227. doi: 10.2174/092986606775338362. [DOI] [PubMed] [Google Scholar]
- 11.Gajdusek DC. Unconventional viruses and the origin and disappearance of kuru. Science. 1977;197:943–960. doi: 10.1126/science.142303. [DOI] [PubMed] [Google Scholar]
- 12.Collinge J, Sidle KC, Meads J, Ironside J, Hill AF. Molecular analysis of prion strain variation and the aetiology of ‘new variant’ CJD. Nature. 1996;383:685–690. doi: 10.1038/383685a0. [DOI] [PubMed] [Google Scholar]
- 13.Anderson RM, Donnelly CA, Ferguson NM, Woolhouse MEJ, Watt CJ, Udy HJ, Mawhinney S, Dunstan SP, Southwood TRE, Wilesmith JW, Ryan JBM, Hoinville LJ, Hillerton JE, Austin AR, Wells GAH (1997) Transmission dynamics and epidemiology of BSE in British cattle (vol 382, pp 779, 1996). Nature 386:302–302 [DOI] [PubMed]
- 14.Bruce ME, Will RG, Ironside JW, McConnell I, Drummond D, Suttie A, McCardle L, Chree A, Hope J, Birkett C, Cousens S, Fraser H, Bostock CJ. Transmissions to mice indicate that ‘new variant’ CJD is caused by the BSE agent. Nature. 1997;389:498–501. doi: 10.1038/39057. [DOI] [PubMed] [Google Scholar]
- 15.Hill AF, Desbruslais M, Joiner S, Sidle KC, Gowland I, Collinge J, Doey LJ, Lantos P. The same prion strain causes vCJD and BSE. Nature. 1997;389:448–450. doi: 10.1038/38925. [DOI] [PubMed] [Google Scholar]
- 16.Wadsworth JD, Joiner S, Hill AF, Campbell TA, Desbruslais M, Luthert PJ, Collinge J. Tissue distribution of protease resistant prion protein in variant Creutzfeldt–Jakob disease using a highly sensitive immunoblotting assay. Lancet. 2001;358:171–180. doi: 10.1016/S0140-6736(01)05403-4. [DOI] [PubMed] [Google Scholar]
- 17.Sigurdson CJ, Miller MW. Other animal prion diseases. Br Med Bull. 2003;66:199–212. doi: 10.1093/bmb/66.1.199. [DOI] [PubMed] [Google Scholar]
- 18.Angers RC, Browning SR, Seward TS, Sigurdson CJ, Miller MW, Hoover EA, Telling GC. Prions in skeletal muscles of deer with chronic wasting disease. Science. 2006;311:1117. doi: 10.1126/science.1122864. [DOI] [PubMed] [Google Scholar]
- 19.Peden AH, Head MW, Ritchie DL, Bell JE, Ironside JW. Preclinical vCJD after blood transfusion in a PRNP codon 129 heterozygous patient. Lancet. 2004;364:527–529. doi: 10.1016/S0140-6736(04)16811-6. [DOI] [PubMed] [Google Scholar]
- 20.Llewelyn CA, Hewitt PE, Knight RS, Amar K, Cousens S, Mackenzie J, Will RG. Possible transmission of variant Creutzfeldt–Jakob disease by blood transfusion. Lancet. 2004;363:417–421. doi: 10.1016/S0140-6736(04)15486-X. [DOI] [PubMed] [Google Scholar]
- 21.Caughey B, Raymond GJ. The scrapie-associated form of PrP is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J Biol Chem. 1991;266:18217–18223. [PubMed] [Google Scholar]
- 22.Borchelt DR, Taraboulos A, Prusiner SB. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J Biol Chem. 1992;267:16188–16199. [PubMed] [Google Scholar]
- 23.Westergard L, Christensen HM, Harris DA. The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim Biophys Acta. 2007;1772:629–644. doi: 10.1016/j.bbadis.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caughey B, Baron GS. Prions and their partners in crime. Nature. 2006;443:803–810. doi: 10.1038/nature05294. [DOI] [PubMed] [Google Scholar]
- 25.McLennan NF, Brennan PM, McNeill A, Davies I, Fotheringham A, Rennison KA, Ritchie D, Brannan F, Head MW, Ironside JW, Williams A, Bell JE. Prion protein accumulation and neuroprotection in hypoxic brain damage. Am J Pathol. 2004;165:227–235. doi: 10.1016/S0002-9440(10)63291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuwahara C, Takeuchi AM, Nishimura T, Haraguchi K, Kubosaki A, Matsumoto Y, Saeki K, Matsumoto Y, Yokoyama T, Itohara S, Onodera T. Prions prevent neuronal cell-line death. Nature. 1999;400:225–226. doi: 10.1038/22241. [DOI] [PubMed] [Google Scholar]
- 27.Chiarini LB, Freitas AR, Zanata SM, Brentani RR, Martins VR, Linden R. Cellular prion protein transduces neuroprotective signals. EMBO J. 2002;21:3317–3326. doi: 10.1093/emboj/cdf324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hundt C, Peyrin JM, Haik S, Gauczynski S, Leucht C, Rieger R, Riley ML, Deslys JP, Dormont D, Lasmezas CI, Weiss S. Identification of interaction domains of the prion protein with its 37-kDa/67-kDa laminin receptor. EMBO J. 2001;20:5876–5886. doi: 10.1093/emboj/20.21.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leucht C, Simoneau S, Rey C, Vana K, Rieger R, Lasmezas CI, Weiss S. The 37 kDa/67 kDa laminin receptor is required for PrP(Sc) propagation in scrapie-infected neuronal cells. EMBO Rep. 2003;4:290–295. doi: 10.1038/sj.embor.embor768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fevrier B, Vilette D, Archer F, Loew D, Faigle W, Vidal M, Laude H, Raposo G. Cells release prions in association with exosomes. Proc Natl Acad Sci USA. 2004;101:9683–9688. doi: 10.1073/pnas.0308413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vella LJ, Sharples RA, Lawson VA, Masters CL, Cappai R, Hill AF. Packaging of prions into exosomes is associated with a novel pathway of PrP processing. J Pathol. 2007;211:582–590. doi: 10.1002/path.2145. [DOI] [PubMed] [Google Scholar]
- 32.Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB. Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol. 1995;129:121–132. doi: 10.1083/jcb.129.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vey M, Pilkuhn S, Wille H, Nixon R, Dearmond SJ, Smart EJ, Anderson RG, Taraboulos A, Prusiner SB. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc Natl Acad Sci USA. 1996;93:14945–14949. doi: 10.1073/pnas.93.25.14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Come JH, Fraser PE, Lansbury PT., Jr A kinetic model for amyloid formation in the prion diseases: importance of seeding. Proc Natl Acad Sci USA. 1993;90:5959–5963. doi: 10.1073/pnas.90.13.5959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lansbury PT. Mechanism of scrapie replication. Science. 1994;265:1510. doi: 10.1126/science.8079159. [DOI] [PubMed] [Google Scholar]
- 36.Daude N, Marella M, Chabry J. Specific inhibition of pathological prion protein accumulation by small interfering RNAs. J Cell Sci. 2003;116:2775–2779. doi: 10.1242/jcs.00494. [DOI] [PubMed] [Google Scholar]
- 37.Gilch S, Winklhofer KF, Groschup MH, Nunziante M, Lucassen R, Spielhaupter C, Muranyi W, Riesner D, Tatzelt J, Schatzl HM. Intracellular re-routing of prion protein prevents propagation of PrP(Sc) and delays onset of prion disease. EMBO J. 2001;20:3957–3966. doi: 10.1093/emboj/20.15.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tatzelt J, Prusiner SB, Welch WJ. Chemical chaperones interfere with the formation of scrapie prion protein. EMBO J. 1996;15:6363–6373. [PMC free article] [PubMed] [Google Scholar]
- 39.Caspi S, Halimi M, Yanai A, Sasson SB, Taraboulos A, Gabizon R. The anti-prion activity of Congo red. Putative mechanism. J Biol Chem. 1998;273:3484–3489. doi: 10.1074/jbc.273.6.3484. [DOI] [PubMed] [Google Scholar]
- 40.Chabry J, Priola SA, Wehrly K, Nishio J, Hope J, Chesebro B. Species-independent inhibition of abnormal prion protein (PrP) formation by a peptide containing a conserved PrP sequence. J Virol. 1999;73:6245–6250. doi: 10.1128/jvi.73.8.6245-6250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horiuchi M, Baron GS, Xiong LW, Caughey B. Inhibition of interactions and interconversions of prion protein isoforms by peptide fragments from the C-terminal folded domain. J Biol Chem. 2001;276:15489–15497. doi: 10.1074/jbc.M100288200. [DOI] [PubMed] [Google Scholar]
- 42.Peretz D, Williamson RA, Kaneko K, Vergara J, Leclerc E, Schmitt-Ulms G, Mehlhorn IR, Legname G, Wormald MR, Rudd PM, Dwek RA, Burton DR, Prusiner SB. Antibodies inhibit prion propagation and clear cell cultures of prion infectivity. Nature. 2001;412:739–743. doi: 10.1038/35089090. [DOI] [PubMed] [Google Scholar]
- 43.Caughey B, Raymond GJ. Sulfated polyanion inhibition of scrapie-associated PrP accumulation in cultured cells. J Virol. 1993;67:643–650. doi: 10.1128/jvi.67.2.643-650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zuber C, Knackmuss S, Rey C, Reusch U, Rottgen P, Frohlich T, Arnold GJ, Pace C, Mitteregger G, Kretzschmar HA, Little M, Weiss S. Single chain Fv antibodies directed against the 37 kDa/67 kDa laminin receptor as therapeutic tools in prion diseases. Mol Immunol. 2008;45:144–151. doi: 10.1016/j.molimm.2007.04.030. [DOI] [PubMed] [Google Scholar]
- 45.Winklhofer KF, Tatzelt J. Cationic lipopolyamines induce degradation of PrPSc in scrapie-infected mouse neuroblastoma cells. Biol Chem. 2000;381:463–469. doi: 10.1515/BC.2000.061. [DOI] [PubMed] [Google Scholar]
- 46.Ertmer A, Gilch S, Yun SW, Flechsig E, Klebl B, Stein-Gerlach M, Klein MA, Schatzl HM. The tyrosine kinase inhibitor STI571 induces cellular clearance of PrPSc in prion-infected cells. J Biol Chem. 2004;279:41918–41927. doi: 10.1074/jbc.M405652200. [DOI] [PubMed] [Google Scholar]
- 47.Aguib Y, Heiseke A, Gilch S, Riemer C, Baier M, Schatzl HM, Ertmer A. Autophagy induction by trehalose counteracts cellular prion infection. Autophagy. 2009;5:361–369. doi: 10.4161/auto.5.3.7662. [DOI] [PubMed] [Google Scholar]
- 48.Heiseke A, Aguib Y, Riemer C, Baier M, Schatzl HM. Lithium induces clearance of protease resistant prion protein in prion-infected cells by induction of autophagy. J Neurochem. 2009;109:25–34. doi: 10.1111/j.1471-4159.2009.05906.x. [DOI] [PubMed] [Google Scholar]
- 49.Priola SA, Raines A, Caughey WS. Porphyrin and phthalocyanine antiscrapie compounds. Science. 2000;287:1503–1506. doi: 10.1126/science.287.5457.1503. [DOI] [PubMed] [Google Scholar]
- 50.Forloni G, Iussich S, Awan T, Colombo L, Angeretti N, Girola L, Bertani I, Poli G, Caramelli M, Grazia BM, Farina L, Limido L, Rossi G, Giaccone G, Ironside JW, Bugiani O, Salmona M, Tagliavini F. Tetracyclines affect prion infectivity. Proc Natl Acad Sci USA. 2002;99:10849–10854. doi: 10.1073/pnas.162195499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korth C, Stierli B, Streit P, Moser M, Schaller O, Fischer R, Schulz-Schaeffer W, Kretzschmar H, Raeber A, Braun U, Ehrensperger F, Hornemann S, Glockshuber R, Riek R, Billeter M, Wuthrich K, Oesch B. Prion (PrPSc)-specific epitope defined by a monoclonal antibody. Nature. 1997;390:74–77. doi: 10.1038/36337. [DOI] [PubMed] [Google Scholar]
- 52.Paramithiotis E, Pinard M, Lawton T, LaBoissiere S, Leathers VL, Zou WQ, Estey LA, Lamontagne J, Lehto MT, Kondejewski LH, Francoeur GP, Papadopoulos M, Haghighat A, Spatz SJ, Head M, Will R, Ironside J, O’Rourke K, Tonelli Q, Ledebur HC, Chakrabartty A, Cashman NR. A prion protein epitope selective for the pathologically misfolded conformation. Nat Med. 2003;9:893–899. doi: 10.1038/nm883. [DOI] [PubMed] [Google Scholar]
- 53.Hilton DA, Ghani AC, Conyers L, Edwards P, McCardle L, Ritchie D, Penney M, Hegazy D, Ironside JW. Prevalence of lymphoreticular prion protein accumulation in UK tissue samples. J Pathol. 2004;203:733–739. doi: 10.1002/path.1580. [DOI] [PubMed] [Google Scholar]
- 54.Saborio GP, Permanne B, Soto C. Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature. 2001;411:810–813. doi: 10.1038/35081095. [DOI] [PubMed] [Google Scholar]
- 55.Castilla J, Saa P, Hetz C, Soto C. In vitro generation of infectious scrapie prions. Cell. 2005;121:195–206. doi: 10.1016/j.cell.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 56.Chang B, Cheng X, Yin S, Pan T, Zhang H, Wong P, Kang SC, Xiao F, Yan H, Li C, Wolfe LL, Miller MW, Wisniewski T, Greene MI, Sy MS. Test for detection of disease-associated prion aggregate in the blood of infected but asymptomatic animals. Clin Vaccine Immunol. 2007;14:36–43. doi: 10.1128/CVI.00341-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brown P. Blood infectivity, processing and screening tests in transmissible spongiform encephalopathy. Vox Sang. 2005;89:63–70. doi: 10.1111/j.1423-0410.2005.00683.x. [DOI] [PubMed] [Google Scholar]
- 58.Dabaghian RH, Mortimer PP, Clewley JP. Prospects for the development of pre-mortem laboratory diagnostic tests for Creutzfeldt-Jakob disease. Rev Med Virol. 2004;14:345–361. doi: 10.1002/rmv.450. [DOI] [PubMed] [Google Scholar]
- 59.Gilch S, Krammer C, Schatzl HM. Targeting prion proteins in neurodegenerative disease. Expert Opin Biol Ther. 2008;8:923–940. doi: 10.1517/14712598.8.7.923. [DOI] [PubMed] [Google Scholar]
- 60.Mallucci G, Dickinson A, Linehan J, Klohn PC, Brandner S, Collinge J. Depleting neuronal PrP in prion infection prevents disease and reverses spongiosis. Science. 2003;302:871–874. doi: 10.1126/science.1090187. [DOI] [PubMed] [Google Scholar]
- 61.Mallucci GR, White MD, Farmer M, Dickinson A, Khatun H, Powell AD, Brandner S, Jefferys JG, Collinge J. Targeting cellular prion protein reverses early cognitive deficits and neurophysiological dysfunction in prion-infected mice. Neuron. 2007;53:325–335. doi: 10.1016/j.neuron.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 62.Ellington AD. RNA selection—aptamers achieve the desired recognition. Curr Biol. 1994;4:427–429. doi: 10.1016/S0960-9822(00)00093-2. [DOI] [PubMed] [Google Scholar]
- 63.Gold L. Oligonucleotides as research, diagnostic, and therapeutic agents. J Biol Chem. 1995;270:13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- 64.Proske D, Hofliger M, Soll RM, Beck-Sickinger AG, Famulok M. A Y2 receptor mimetic aptamer directed against neuropeptide Y. J Biol Chem. 2002;277:11416–11422. doi: 10.1074/jbc.M109752200. [DOI] [PubMed] [Google Scholar]
- 65.Lupold SE, Hicke BJ, Lin Y, Coffey DS. Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Res. 2002;62:4029–4033. [PubMed] [Google Scholar]
- 66.Li Y, Lee HJ, Corn RM. Detection of protein biomarkers using RNA aptamer microarrays and enzymatically amplified surface plasmon resonance imaging. Anal Chem. 2007;79:1082–1088. doi: 10.1021/ac061849m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gold L, Polisky B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 68.Enari M, Flechsig E, Weissmann C. Scrapie prion protein accumulation by scrapie-infected neuroblastoma cells abrogated by exposure to a prion protein antibody. Proc Natl Acad Sci USA. 2001;98:9295–9299. doi: 10.1073/pnas.151242598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Moroncini G, Kanu N, Solforosi L, Abalos G, Telling GC, Head M, Ironside J, Brockes JP, Burton DR, Williamson RA. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc Natl Acad Sci USA. 2004;101:10404–10409. doi: 10.1073/pnas.0403522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Solforosi L, Bellon A, Schaller M, Cruite JT, Abalos GC, Williamson RA. Toward molecular dissection of PrPC–PrPSc interactions. J Biol Chem. 2007;282:7465–7471. doi: 10.1074/jbc.M610051200. [DOI] [PubMed] [Google Scholar]
- 71.Weiss S, Proske D, Neumann M, Groschup MH, Kretzschmar HA, Famulok M, Winnacker EL. RNA aptamers specifically interact with the prion protein PrP. J Virol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Proske D, Gilch S, Wopfner F, Schatzl HM, Winnacker EL, Famulok M. Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem. 2002;3:717–725. doi: 10.1002/1439-7633(20020802)3:8<717::AID-CBIC717>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 73.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wuthrich K. NMR structure of the mouse prion protein domain PrP(121–321) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 74.Wille H, Michelitsch MD, Guenebaut V, Supattapone S, Serban A, Cohen FE, Agard DA, Prusiner SB. Structural studies of the scrapie prion protein by electron crystallography. Proc Natl Acad Sci USA. 2002;99:3563–3568. doi: 10.1073/pnas.052703499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Govaerts C, Wille H, Prusiner SB, Cohen FE. Evidence for assembly of prions with left-handed beta-helices into trimers. Proc Natl Acad Sci USA. 2004;101:8342–8347. doi: 10.1073/pnas.0402254101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silveira JR, Raymond GJ, Hughson AG, Race RE, Sim VL, Hayes SF, Caughey B. The most infectious prion protein particles. Nature. 2005;437:257–261. doi: 10.1038/nature03989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rhie A, Kirby L, Sayer N, Wellesley R, Disterer P, Sylvester I, Gill A, Hope J, James W, Tahiri-Alaoui A. Characterization of 2’-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J Biol Chem. 2003;278:39697–39705. doi: 10.1074/jbc.M305297200. [DOI] [PubMed] [Google Scholar]
- 78.Sayer NM, Cubin M, Rhie A, Bullock M, Tahiri-Alaoui A, James W. Structural determinants of conformationally selective, prion-binding aptamers. J Biol Chem. 2004;279:13102–13109. doi: 10.1074/jbc.M310928200. [DOI] [PubMed] [Google Scholar]
- 79.Sekiya S, Noda K, Nishikawa F, Yokoyama T, Kumar PK, Nishikawa S. Characterization and application of a novel RNA aptamer against the mouse prion protein. J Biochem. 2006;139:383–390. doi: 10.1093/jb/mvj046. [DOI] [PubMed] [Google Scholar]
- 80.Wopfner F, Weidenhofer G, Schneider R, von Brunn A, Gilch S, Schwarz TF, Werner T, Schatzl HM. Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol. 1999;289:1163–1178. doi: 10.1006/jmbi.1999.2831. [DOI] [PubMed] [Google Scholar]
- 81.Gabus C, Derrington E, Leblanc P, Chnaiderman J, Dormont D, Swietnicki W, Morillas M, Surewicz WK, Marc D, Nandi P, Darlix JL. The prion protein has RNA binding and chaperoning properties characteristic of nucleocapsid protein NCP7 of HIV-1. J Biol Chem. 2001;276:19301–19309. doi: 10.1074/jbc.M009754200. [DOI] [PubMed] [Google Scholar]
- 82.Gomes MP, Millen TA, Ferreira PS, Cunha E, Silva NL, Vieira TC, Almeida MS, Silva JL, Cordeiro Y. Prion protein complexed to N2a cellular RNAs through its N-terminal domain forms aggregates and is toxic to murine neuroblastoma cells. J Biol Chem. 2008;238:19616–19625. doi: 10.1074/jbc.M802102200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mercey R, Lantier I, Maurel MC, Grosclaude J, Lantier F, Marc D. Fast, reversible interaction of prion protein with RNA aptamers containing specific sequence patterns. Arch Virol. 2006;151:2197–2214. doi: 10.1007/s00705-006-0790-3. [DOI] [PubMed] [Google Scholar]
- 84.Takemura K, Wang P, Vorberg I, Surewicz W, Priola SA, Kanthasamy A, Pottathil R, Chen SG, Sreevatsan S. DNA aptamers that bind to PrP(C) and not PrP(Sc) show sequence and structure specificity. Exp Biol Med (Maywood) 2006;231:204–214. doi: 10.1177/153537020623100211. [DOI] [PubMed] [Google Scholar]
- 85.Kouassi GK, Wang P, Sreevatan S, Irudayaraj J. Aptamer-mediated magnetic and gold-coated magnetic nanoparticles as detection assay for prion protein assessment. Biotechnol Prog. 2007;23:1239–1244. doi: 10.1021/bp0602101. [DOI] [PubMed] [Google Scholar]
- 86.Bibby DF, Gill AC, Kirby L, Farquhar CF, Bruce ME, Garson JA. Application of a novel in vitro selection technique to isolate and characterise high affinity DNA aptamers binding mammalian prion proteins. J Virol Methods. 2008;151:107–115. doi: 10.1016/j.jviromet.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 87.Safar J, Wille H, Itri V, Groth D, Serban H, Torchia M, Cohen FE, Prusiner SB. Eight prion strains have PrP(Sc) molecules with different conformations. Nat Med. 1998;4:1157–1165. doi: 10.1038/2654. [DOI] [PubMed] [Google Scholar]
- 88.Pfeifer A, Eigenbrod S, Al Khadra S, Hofmann A, Mitteregger G, Moser M, Bertsch U, Kretzschmar H. Lentivector-mediated RNAi efficiently suppresses prion protein and prolongs survival of scrapie-infected mice. J Clin Invest. 2006;116:3204–3210. doi: 10.1172/JCI29236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White MD, Farmer M, Mirabile I, Brandner S, Collinge J, Mallucci GR. Single treatment with RNAi against prion protein rescues early neuronal dysfunction and prolongs survival in mice with prion disease. Proc Natl Acad Sci USA. 2008;105:10238–10243. doi: 10.1073/pnas.0802759105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Blind M, Kolanus W, Famulok M. Cytoplasmic RNA modulators of an inside-out signal-transduction cascade. Proc Natl Acad Sci USA. 1999;96:3606–3610. doi: 10.1073/pnas.96.7.3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Colas P, Cohen B, Jessen T, Grishina I, Mccoy J, Brent R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature. 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 92.Hoppe-Seyler F, Crnkovic-Mertens I, Tomai E, Butz K. Peptide aptamers: Specific inhibitors of protein function. Curr Mol Med. 2004;4:529–538. doi: 10.2174/1566524043360519. [DOI] [PubMed] [Google Scholar]
- 93.Ladner RC. Constrained peptides as binding entities. Trends Biotechnol. 1995;13:426–430. doi: 10.1016/S0167-7799(00)88997-0. [DOI] [PubMed] [Google Scholar]
- 94.Geyer CR, Colman-Lerner A, Brent R. “Mutagenesis” by peptide aptamers identifies genetic network members and pathway connections. Proc Natl Acad Sci USA. 1999;96:8567–8572. doi: 10.1073/pnas.96.15.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Baines IC, Colas P. Peptide aptamers as guides for small-molecule drug discovery. Drug Discov Today. 2006;11:334–341. doi: 10.1016/j.drudis.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 96.Peelle B, Gururaja TL, Payan DG, Anderson DC. Characterization and use of green fluorescent proteins from Renilla mulleri and Ptilosarcus guernyi for the human cell display of functional peptides. J Protein Chem. 2001;20:507–519. doi: 10.1023/A:1012514715338. [DOI] [PubMed] [Google Scholar]
- 97.Nord K, Gunneriusson E, Ringdahl J, Stahl S, Uhlen M, Nygren PA. Binding proteins selected from combinatorial libraries of an alpha-helical bacterial receptor domain. Nat Biotechnol. 1997;15:772–777. doi: 10.1038/nbt0897-772. [DOI] [PubMed] [Google Scholar]
- 98.Beste G, Schmidt FS, Stibora T, Skerra A. Small antibody-like proteins with prescribed ligand specificities derived from the lipocalin fold. Proc Natl Acad Sci USA. 1999;96:1898–1903. doi: 10.1073/pnas.96.5.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Colas P, Cohen B, Ferrigno PK, Silver PA, Brent R. Targeted modification and transportation of cellular proteins. Proc Natl Acad Sci USA. 2000;97:13720–13725. doi: 10.1073/pnas.97.25.13720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Butz K, Denk C, Fitscher B, Crnkovic-Mertens I, Ullmann A, Schroder CH, Hoppe-Seyler F. Peptide aptamers targeting the hepatitis B virus core protein: a new class of molecules with antiviral activity. Oncogene. 2001;20:6579–6586. doi: 10.1038/sj.onc.1204805. [DOI] [PubMed] [Google Scholar]
- 101.Butz K, Denk C, Ullmann A, Scheffner M, Hoppe-Seyler F. Induction of apoptosis in human papillomavirus-positive cancer cells by peptide aptamers targeting the viral E6 oncoprotein. Proc Natl Acad Sci USA. 2000;97:6693–6697. doi: 10.1073/pnas.110538897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nauenburg S, Zwerschke W, Jansen-Durr P. Induction of apoptosis in cervical carcinoma cells by peptide aptamers that bind to the HPV-16 E7 oncoprotein. FASEB J. 2001;15:592–594. doi: 10.1096/fj.00-0604fje. [DOI] [PubMed] [Google Scholar]
- 103.Buerger C, Nagel-Wolfrum K, Kunz C, Wittig I, Butz K, Hoppe-Seyler F, Groner B. Sequence-specific peptide aptamers, interacting with the intracellular domain of the epidermal growth factor receptor, interfere with Stat3 activation and inhibit the growth of tumor cells. J Biol Chem. 2003;278:37610–37621. doi: 10.1074/jbc.M301629200. [DOI] [PubMed] [Google Scholar]
- 104.Gilch S, Kehler C, Schatzl HM. Peptide aptamers expressed in the secretory pathway interfere with cellular PrPSc formation. J Mol Biol. 2007;371:362–373. doi: 10.1016/j.jmb.2007.05.052. [DOI] [PubMed] [Google Scholar]
- 105.Wuertzer CA, Sullivan MA, Qiu X, Federoff HJ. CNS delivery of vectored prion-specific single-chain antibodies delays disease onset. Mol Ther. 2008;16:481–486. doi: 10.1038/sj.mt.6300387. [DOI] [PubMed] [Google Scholar]
- 106.Siernion IZ, Cebrat M, Kluczyk A. The problem of amino acid complementarity and antisense peptides. Curr Protein Pept Sci. 2004;5:507–527. doi: 10.2174/1389203043379413. [DOI] [PubMed] [Google Scholar]
- 107.Root-Bernstein RS, Holsworth DD. Antisense peptides: a critical mini-review. J Theor Biol. 1998;190:107–119. doi: 10.1006/jtbi.1997.0544. [DOI] [PubMed] [Google Scholar]
- 108.Huang YY, Zhao R, Luo J, Xiong SX, Shangguan DH, Zhang HW, Liu GQ, Chen Y. Design, synthesis and screening of antisense peptide based combinatorial peptide libraries towards an aromatic region of SARS-CoV. J Mol Recognit. 2008;21:122–131. doi: 10.1002/jmr.880. [DOI] [PubMed] [Google Scholar]
- 109.Imai M, Baranyi L, Okada N, Okada H. Inhibition of HIV-1 infection by synthetic peptides derived CCR5 fragments. Biochem Biophys Res Commun. 2007;353:851–856. doi: 10.1016/j.bbrc.2006.12.084. [DOI] [PubMed] [Google Scholar]
- 110.Rother KI, Clay OK, Bourquin JP, Silke J, Schaffner W. Long non-stop reading frames on the antisense strand of heat shock protein 70 genes and prion protein (PrP) genes are conserved between species. Biol Chem. 1997;378:1521–1530. doi: 10.1515/bchm.1997.378.12.1521. [DOI] [PubMed] [Google Scholar]
- 111.Moser M, Oesch B, Bueler H. An anti-prion protein? Nature. 1993;362:213–214. doi: 10.1038/362213b0. [DOI] [PubMed] [Google Scholar]