Abstract
We identified a T-DNA-generated mutation in the chaperonin-60α gene of Arabidopsis that produces a defect in embryo development. The mutation, termed schlepperless (slp), causes retardation of embryo development before the heart stage, even though embryo morphology remains normal. Beyond the heart stage, the slp mutation results in defective embryos with highly reduced cotyledons. slp embryos exhibit a normal apical-basal pattern and radial tissue organization, but they are morphologically retarded. Even though slp embryos are competent to transcribe two late-maturation gene markers, this competence is acquired more slowly as compared with wild-type embryos. slp embryos also exhibit a defect in plastid development–they remain white during maturation in planta and in culture. Hence, the overall developmental phenotype of the slp mutant reflects a lesion in the chloroplast that affects embryo development. The slp phenotype highlights the importance of the chaperonin-60α protein for chloroplast development and subsequently for the proper development of the plant embryo and seedling.
Plant embryogenesis is a complex developmental process that can be divided into four conceptual phases (West and Harada, 1993; Goldberg et al., 1994). During the first phase, the body plan of the mature embryo is established and specific groups of cells give rise to the shoot meristem, cotyledons (embryonic leaves), axis (hypocotyl), radicle (embryonic root), and the root meristem. The embryo then undergoes maturation during the second phase of development, which is characterized by the deposition of storage materials (Goldberg et al., 1989). The third phase involves desiccation leading to seed dormancy. When proper conditions allow, germination follows seed dormancy leading to the development of a seedling. The cotyledons serve as a source of food reserves during germination. Each of the four phases requires coordinated expression of specific and overlapping genetic programs involving cell division, cell differentiation, and other housekeeping, cellular functions (Goldberg et al., 1989).
The molecular and cellular mechanisms that dictate the early events of embryogenesis are not yet known. One approach to address this question is the isolation and characterization of mutants that are impaired in embryo development. Several embryo-defective mutants have been isolated in Arabidopsis (Mayer et al., 1991; Meinke, 1991; Yadegari et al., 1994; Meinke, 1995), petunia (Souer et al., 1996), and corn (Clark and Sheridan, 1991). Some of the mutated genes affecting Arabidopsis embryogenesis have been isolated, including GNOM (Shevell et al., 1994), SHOOT MERISTEMLESS (STM; Long et al., 1996), MONOPTEROS (MP; Hardtke and Berleth, 1998), KNOLLE (Lukowitz et al., 1996), and EMBRYO-DEFECTIVE DEVELOPMENT1 (EDD1; Uwer et al., 1998). Despite the cloning of these genes, their specific roles in the embryogenic process remain to be elucidated. Nevertheless, the functional identities of the encoded proteins indicate that some may play roles in cellular processes that are important for controlling plant embryogenesis as well as other aspects of plant development. For instance, the gene product of MP is involved not only in the establishment of embryo axis formation (Berleth and Jürgens, 1993), but also in vascular development beyond the embryonic stage (Hardtke and Berleth, 1998). Likewise, aside from being involved in specifying the apical and basal regions of the embryos (Mayer et al., 1993; Shevell et al., 1994), GNOM may also be involved later in plant development because its gene product is essential for establishing cell polarity required for normal cell division and expansion, (Shevell et al., 1994; Grebe et al., 2000; Shevell et al., 2000). Genes that affect the function and/or development of organelles may also be playing similarly significant roles in embryogenesis. For example, a mutation in EDD1 leads to embryo arrest between globular and heart stages (Uwer et al., 1998). Because EDD1 encodes a plastidic form of glycyl-tRNA synthetase (Uwer et al., 1998), it is essential for carrying out the metabolic processes that occur within the chloroplast, which can be important for maintaining normal development of the adult plant.
In this paper, we describe the characterization of an embryo-defective mutant allele that was isolated from a population of T-DNA-mutagenized Arabidopsis lines (Yadegari et al., 1994). The embryonic cotyledons of this mutant, termed schlepperless (slp), are highly reduced and the entire embryo remains white during maturation even though the wild-type (WT) embryos turn green. The coding sequence interrupted by the T-DNA insertion corresponds to the nuclear-encoded plastid chaperonin-60α subunit gene. We named this mutant schlepperless, because it is derived from the Yiddish term “schlepper” (meaning “to carry”). Our analysis indicates that the absence of a functional chaperonin-60α protein has adverse consequences on the development of the chloroplast, and subsequently, the development of the embryo. We conclude that the chaperonin-60α protein is essential for the proper development of Arabidopsis plants and, most likely, other eukaryotic organisms.
RESULTS
schlepperless Embryos Develop Abnormally
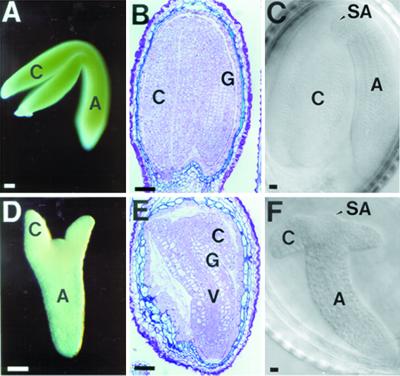
We characterized WT embryo development to establish a base line to which slp embryo development could be compared. The greening of a WT embryo started at the heart stage of embryogenesis (data not shown), which was indicative of chloroplast development from proplastid progenitors (Schultz and Jensen, 1968; Mansfield and Briarty, 1991). As the embryo matured, the hypocotyl and cotyledons of WT embryo turned green (Fig. 1A). At this stage, the cotyledons were fully expanded and the whole embryo occupied most of the space taken previously by the endosperm within the maturing seed (Fig. 1, B and C). The corresponding slp embryo (taken from the same heterozygous silique as the WT embryo) was white and did not turn green even at a mature stage (Fig. 1D).
Figure 1.
Morphology of schlepperless embryos. Mature embryos from WT (A, B, and C) and schlepperless (D, E, and F). Whole mount photographs of embryos dissected out of the same mature silique (A and D). Sections of embryos from mature silique embedded in LR White plastic resin (B and E). Nomarski photographs of embryos taken from the same mature silique (C and F). C, Cotyledon; Ep, epidermis; G, ground tissue; H, hypocotyl; SA, shoot apical meristem; V, vascular tissue. Bars = 50 μm.
In contrast, the slp embryo was not as large as the WT embryo (Fig. 1, A–C) and its cotyledons were very reduced in size (Fig. 1, D–F). However, the tissue organization in advanced stage mutant embryos was apparently normal (Fig. 1E). The vascular tissue, ground tissue, and epidermal cells were present in their proper positions relative to each other as in the WT embryos (compare Fig. 1B with 1E). The radicle and the shoot apical meristem (which appears as a dome shape) were also present and appeared normal in the slp embryo (Figs. 1, C and F).
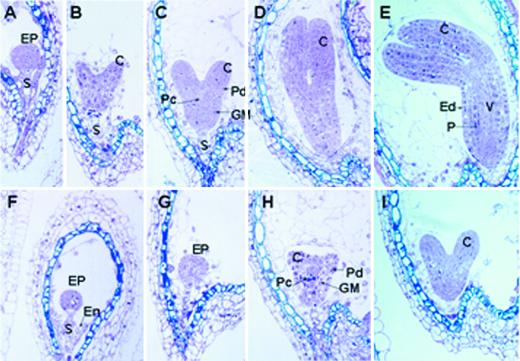
To determine at what stage of embryogenesis the schlepperless phenotype was first observed, a developmental series of slp embryo sections were obtained and were compared with those of the segregating WT embryos from a heterozygous SLP/slp plant (see “Materials and Methods”). Figure 2, A through E, show sections of WT embryos up to the early curled stage (Jürgens and Mayer, 1994) from a heterozygous plant. The morphology of the slp embryo appeared to be normal up to the heart stage of embryogenesis (Fig. 2, F–I). However, development of the mutant embryos was considerably retarded compared with WT embryos (compare Fig. 2, A–E, with Fig. 2, F–I). The WT embryos were already at the early curled stage (Fig. 2E), whereas the slp embryos were still morphologically at the heart stage (Fig. 2I). The tissue organization of the slp embryo appeared to be normal at this early stage of embryogenesis. The developing dermal, ground, and vascular tissues that were present in the WT were also apparent in the slp mutant (compare Fig. 2, A–C, with Fig. 2, F–H). Together, these data indicate that the slp mutant embryos develop more slowly than WT embryos during the early stages of seed development and have an abnormal morphology by maturity.
Figure 2.
Developmental analysis of schlepperless embryos. A developmental series of sections for Arabidopsis WT (A–E) and schlepperless (F–I) embryos embedded in LR White plastic resin. The mutant embryos were from the same silique from which the corresponding WT embryos were taken. A, Axis; C, cotyledon; EP, embryo proper; Ep, epidermis; En, endosperm; GM, ground meristem; P, storage parenchyma; Pc, procambium; Pd, protoderm; S, suspensor; V, vascular tissue.
schlepperless Embryos Germinate in Culture
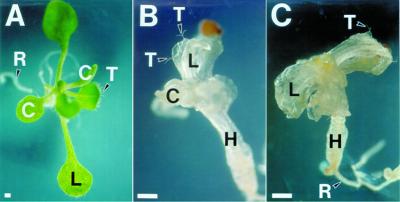
To determine if the slp mutant phenotype can be rescued by tissue culture, mature mutant embryos were dissected from seeds and germinated on media containing Suc, vitamins, and salts (see “Materials and Methods”). The slp embryos were able to germinate in the culture media, although their development was slower and abnormal compared with that of WT embryos. After 12 d in culture, WT seedlings usually possessed four green rosette leaves and a well-developed root system (Fig. 3A). The cotyledons of the seedlings expanded fully and became green (Fig. 3A). In contrast, the seedlings that developed from slp embryos were white (Fig. 3B). The undeveloped embryonic cotyledons emerged from the slp seeds but did not develop any further. No further greening was observed in the slp seedlings, even after remaining in culture for 72 d (Fig. 3C). The aerial portions of the slp seedlings were stunted and only exhibited callus-like structures after extended periods of culture (data not shown), even though the mutant seedlings were able to form normal-looking roots and root hairs (Fig. 3C). Some of the slp seedlings developed leaf-like structures that were translucent and contained trichomes, originating from the apical dome (Figs. 3, B and C).
Figure 3.
Germination of WT and schlepperless seeds in vitro. A, WT seedling after 12 d in culture; B and C, schlepperless seedling after 42 (B) and after 70 (C) d in culture. The mutant seedling shown in C was the same seedling shown in B. C, Cotyledon; H, hypocotyl; L, leaf; R, root; T, trichome. Bars = 50 μm.
Germination of desiccated (dried) mutant seeds was also tested on the same germination medium (GM). The seedlings that developed resembled those obtained from non-mature and un-desiccated embryos (data not shown), suggesting that slp mutant embryos are able to undergo a normal dessication and maturation program. Together, these results indicate that even though slp embryos are capable of germinating, they are unable to form normal seedlings and, ultimately, mature plants. In addition, our results indicate that the slp phenotype could not be rescued by components present in the tissue culture medium.
schlepperless Embryos Are Competent to Transcribe Late-Maturation Genes
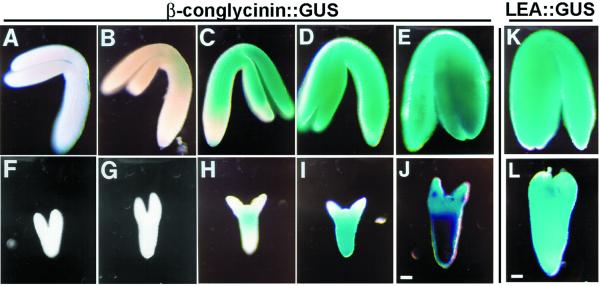
To determine if slp embryos were capable of transcribing genes that are normally expressed during the maturation phase in WT embryos, we crossed SLP/slp heterozygous plants to an Arabidopsis line transgenic for a 7S::GUS construct (Hirai et al., 1994). This construct contained 0.9 kb of the 5′ region of the β-conglycinin α-subunit gene fused to the β-glucuronidase (GUS) reporter gene (Hirai et al., 1994). Histochemical localization of GUS enzyme activity was monitored in WT and slp embryos during seed development (see “Materials and Methods”). In WT embryos, initial expression of the 7S::GUS transgene occurred at the early bent cotyledon stage (Fig. 4B). 7S::GUS expression at this stage was limited to the upper hypocotyl region and the cotyledons. In mature WT embryos, the transgene was expressed in the cotyledons and in the hypocotyl (Fig. 4E).
Figure 4.
schlepperless embryo is competent to transcribe late-maturation genes. Expression pattern of β-conglycinin::GUS transgene in WT (A–E) and schlepperless (F–J) embryos at different developmental stages. Expression pattern of LEA::GUS transgene in mature WT (K) and schlepperless (L) embryos. Corresponding WT and mutant embryos were taken from the same silique. Blue staining indicates activity of GUS. Bars = 50 μm.
In contrast, the initial expression of the 7S::GUS transgene was delayed in slp mutant embryos. Developing mutant embryos taken from the same siliques as WT embryos (Fig. 4, A–E) did not show any GUS staining at first (compare Fig. 4B with Fig. 4G). GUS activity was initially detected in the hypocotyl region of mutant embryos (Fig. 4, H and I). Only later, in more mature slp embryos, was the expression pattern extended into the short cotyledons (Fig. 4J).
Mutant slp embryo development was also analyzed using a late embryogenesis abundant (LEA) gene fusion construct (LEA::GUS; Goupil et al., 1992). In WT plants, LEA::GUS is expressed late in embryogenesis and has an expression pattern similar to the 7S::GUS gene (Goupil et al., 1992; Hirai et al., 1994; West et al., 1994; Fig. 4K). Mutant slp embryos containing the LEA::GUS construct showed a similar pattern of GUS activity as was obtained with the 7S::GUS construct (data not shown; Fig. 4L). In early slp embryos, LEA::GUS expression was limited to the hypocotyl region (data not shown). During embryo maturation, LEA::GUS gene expression extended into the highly reduced cotyledon region of the mutant slp embryos (Fig. 4L). Taken together, these data indicate that the mutant embryos are competent to transcribe genes that are normally expressed late in embryogenesis. However, the acquisition of this competence is dependent upon the developmental state of the embryos. The delay of maturation gene expression in slp embryos most likely reflects the inherent delay of slp morphological development.
schlepperless Is Tagged with T-DNA
A single SLP/slp mutant line was obtained in a screen of T-DNA-mutagenized lines of Arabidopsis (see “Materials and Methods”). Two lines of evidence led us to conclude that this mutant allele was most likely interrupted at one locus, and that this T-DNA interruption is the cause of the slp phenotype. First, the results of DNA blot analysis of 50 WT and heterozygous F1 segregants, where both the left and right borders of the T-DNA were used as probes, indicated that the T-DNA cosegregated only with the heterozygous plants and not with the WT segregants (data not shown). Second, an analysis of nearly 150 kanamycin-resistant (Kan-R) F1 progeny (indicative of the presence of neomycin phosphotransferase II gene contained within the T-DNA) showed 100% cosegregation of Kan-R and the slp phenotype (data not shown).
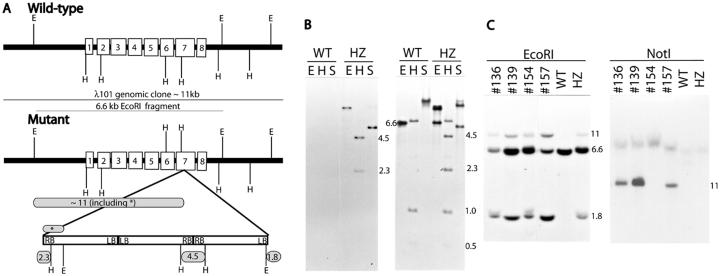
Using plasmid rescue cloning of T-DNA-flanking genomic sequences, and by constructing a genomic library of SLP/slp heterozygous plants (see “Materials and Methods”), a map of the slp mutant locus was obtained indicating that the SLP gene was interrupted in one locus with three T-DNAs arranged in a concatemer (Fig. 5A). Genomic clones corresponding to all four T-DNA junctions (left border/left border junction, the right border/right border junction, the left border/plant sequence junction, and the right border/plant sequence junction) were isolated and they were confirmed using genomic DNA-blot analysis (Fig. 5, A and B; data not shown). For example, when a blot containing digested genomic DNAs from heterozygous SLP/slp (HZ) and WT plants was hybridized with a right border T-DNA sequence, two HindIII fragments were detected (2.3 and 4.5 kb in Fig. 5B). These fragments represent the right border/plant sequence junction (2.3 kb) and the right border/right border junction (4.5 kb).
Figure 5.
The Chaperonin-60α gene is interrupted by T-DNA in slp mutant. A, Diagrammatic representation of the WT chaperonin-60α locus and the concatemer of T-DNAs inserted in slp line (not drawn to scale). Exons are shown as unshaded boxes superimposed on the heavy line. The 11-kb WT genomic clone and the 6.6-kb EcoRI sub-clone are indicated by thin lines. The insertion point of the T-DNA concatemer is indicated by two lines below the seventh exon. Left border (LB) and right border (RB) regions are indicated in the adjacent rectangles that represent the T-DNA concatemer (see Errampalli et al., 1991, for the T-DNA map). Numbers in shaded rounded rectangles indicate the expected size of some polymorphic fragments as shown in the genomic blots (B and C). Not all of the restriction sites within the T-DNA are shown. E, EcoRI; H, HindIII, S, SalI. B, DNA gel-blot analysis of WT and heterozygous (HZ) individuals using right border (first) or a BamHI fragment representing a portion of the RB-plant-flanking region (second). Relevant fragments are indicated by fragment size corresponding to the map in (A). C, Genomic DNA-blot analysis of randomly selected F2 plants used in the genetic analysis presented in Table I. The probe used was the partial pC31 cDNA clone. Genomic DNAs digested with EcoRI were electrophoresed in 1% (w/v) agarose gel, whereas those digested with NotI were electrophoresed in 0.5% (w/v) agarose gel. Samples from genotypically WT (WT) and heterozygous (HZ) individuals were included as controls.
We used the plant genomic sequences flanking the T-DNA insert to isolate the corresponding WT genomic clones from an Arabidopsis λ-library. One of the WT genomic clones isolated was λ101 with an insert size of 11 kb (Fig. 5A). Sequence analysis of the genomic clone suggested the presence of an open reading frame. A partial cDNA clone, pC31, was subsequently isolated using a 6.6-kb EcoRI fragment of λ101 as a probe (data not shown).
To determine whether the open reading frame identified represented the gene mutated in the SLP/slp line, we rescued the mutation by complementation. An 11-kb NotI fragment from the λ101 genomic clone (Fig. 5A) was sub-cloned into pGSH166N vector (see “Materials and Methods”). This fragment contained the whole chaperonin-60α gene (approximately 5.7 kb) and no other genes were present on this fragment (see Lin et al., 1999). The vector contains a hygromycin-resistant (Hyg-R) marker and, therefore, could be differentiated from the T-DNA that contained Kan-R marker used in the initial mutagenesis (Errampalli et al., 1991; Feldmann, 1991). A complementation analysis was performed (see “Materials and Methods” for details) after crossing the heterozygous mutant line (slp/SLP; Kan-R) with the lines transgenic for the genomic clone (i.e. tCpn; Hyg-R). If the complementation was successful, we expected that we would observe a new class of heterozygous individuals producing mutant seeds at a 6.25% frequency. We found this new class of F2 segregants as shown in Table I (Class III). This new class would not have been found in a non-complemented heterozygous line (slp/SLP; Kan-R) that produced mutant seeds at 25% frequency. Crosses using other independent transformants (i.e. tCpn; Hyg-R) produced similar results (data not shown).
Table I.
Summary of F2 analysis in one of the complementation crosses
| F2 Class (Description) | Expected F2 If There Is No Complementation
|
Expected F2 If There Is Complementation
|
Observed F2
|
|||
|---|---|---|---|---|---|---|
| Genotypesa | % | Genotypesa | % | % | No. (Total = 270) | |
| Class I (produce all WT seeds) | SLP/SLP (1)SLP/SLP; tCpn (2)SLP/SLP; tCpn/tCpn (1) | 33.3 | SLP/SLP (1)SLP/SLP; tCpn (2)SLP/SLP; tCpn/tCpn (1)SLP/slp; tCpn/tCpn (2)slp/slp; tCpn/tCpn (1) | 46.6 | 50 | 135 |
| Class II (produce mutant seeds at 25% frequency) | SLP/slp (2)SLP/slp; tCpn (4)SLP/slp; tCpn/tCpn (2) | 66.7 | SLP/slp (2)slp/slp; tCpn (2) | 26.7 | 28 | 76 |
| Class III (produce mutant seeds at 6.25% frequency) | – | – | SLP/slp; tCpn (4) | 26.7 | 22 | 59 |
Chi-square analysis: calculated χ2 = 3.21 < tabular χ2 = 5.99 (at 0.05 level and degrees of freedom = 2).
Nos. in parentheses indicate the no. of individuals expected to have the particular genotype out of 12 (if there is no complementation) or 15 (if there is complementation) surviving F2 individuals.
To establish cosegregation of the complementing transgene with the phenotype, genomic DNAs from randomly selected F2 plants were analyzed using DNA-blot analysis (see “Materials and Methods”). The phenotypic WT F2 segregants identified as SLP/slp heterozygotes, based on the presence of the polymorphic EcoRI fragments (11 and 1.8 kb, Fig. 5, A and C), were determined to contain the WT version of SLP on the 11-kb NotI-containing transgene inherited from the parent transformant (tCpn, Hyg-R; Fig. 5C). F2 individuals nos. 136 and 139 were examples of such WT segregants (Fig. 5C). We also identified some F2 segregants that were genotypically slp/slp homozygotes, based on testcross analysis (data not shown). F2 individual number 157 (Fig. 5C) was an example of an slp/slp homozygous segregant that should have been dead in a non-complementing background. It produced mutant seeds at about 25% frequency because it contained one copy of the transgene (tCpn, data not shown). These data indicated that an 11-kb NotI fragment containing the transgene with a WT copy of SLP was responsible for the complementation of the slp phenotype.
The slp mutation was also mapped (see “Materials and Methods”) and determined to be located between positions 40.6 and 56.1 of chromosome 2 (data not shown). This is consistent with the published sequences for chromosome 2 in which chaperonin-60α gene is located within the same physical region (Lin et al., 1999). Taken together, the genetic and molecular evidence indicated that the interruption of the SLP locus by T-DNA insertion is responsible for the embryonic abnormalities observed in the slp/slp mutant individuals.
SCHLEPPERLESS Locus Encodes a Chaperonin-60α Protein
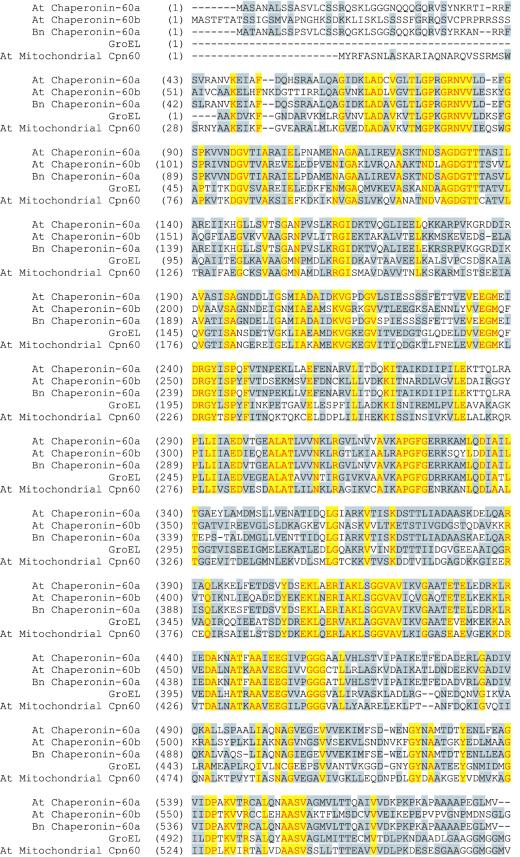
Sequencing of both the λ101 genomic clone and the partial cDNA pC31 clone showed that the interrupted open reading frame corresponded to a plastid chaperonin-60α subunit gene (Hemmingsen et al., 1988; Martel et al., 1990; Cloney et al., 1994). The exons and introns were deduced from a comparison of the genomic clone sequence to the partial cDNA sequences (data not shown; Martel et al., 1990) and to the previously published sequence of chaperonin-60α from Brassica napus (Martel et al., 1990; Cole et al., 1994). The deduced sequence of the chaperonin-60α protein of Arabidopsis has 586 amino acid residues, and has a predicted molecular mass of approximately 62 kD. The first 46 amino acids appears to constitute a transit peptide that, if cleaved at an Asn (N) residue (amino acid no. 47, Fig. 6), would yield the mature form of the protein (Martel et al., 1990).
Figure 6.
Alignment of the Arabidopsis chaperonin-60α protein with other related proteins. The Arabidopsis chaperonin-60α protein was aligned with chaperonin-60α protein from B. napus (95% similarity), Arabidopsis chaperonin-60β protein (70% similarity), the Arabidopsis mitochondrial chaperonin-60 (90% similarity), and GroEL protein from Escherichia coli (70% similarity) using the AlignX program of VectorNTI software. Amino acids that are identical in all five proteins are presented in yellow blocks with red letters. Conservative amino acids are presented in gray blocks with black letters. Sources of these proteins are cited in the text. The genomic sequence of the Arabidopsis Chaperonin-60α has the GenBank accession no. U49357.
The Arabidopsis chaperonin-60α protein shows a very high degree of conservation when compared with plastid chaperonin-60 α-subunit from other species. For example, it has 95% similarity to B. napus (Cole et al., 1994) and castor bean (Hemmingsen et al., 1988). However, it is quite divergent from other chaperonin proteins produced by Arabidopsis (e.g. the β-subunit of chaperonin-60 and chaperonin-60 mitochondrial proteins). It shows only 70% similarity to the β-subunit form (Zabaleta et al., 1992) and about 90% similarity to the chaperonin-60 protein localized in the mitochondria (Prasad and Stewart, 1992; see also accession no. AP001297 of chromosome 3 of the Arabidopsis genome). It has 70% and 72% similarity to the groEL proteins from E. coli (Hemmingsen et al., 1988) and Brucella (Roop et al., 1992), respectively. GroEL is the prokaryotic equivalent of chaperonin-60α protein. Figure 6 shows the similarity alignment of the Arabidopsis chaperonin-60α protein with other chaperonin proteins. Portions of the rescued plasmids and mutant phage clones were also sequenced to determine the exact nucleotide where T-DNA insertion interrupted the gene. This analysis showed that the concatemer of three T-DNAs was inserted in the seventh exon, thus interrupting the protein-coding sequence at amino acid 449 (Figs. 5A and 6). Therefore, an active chaperonin-60α protein product would not be present in the slp/slp embryos.
The Chaperonin-60α mRNA Is Present in Several Organs
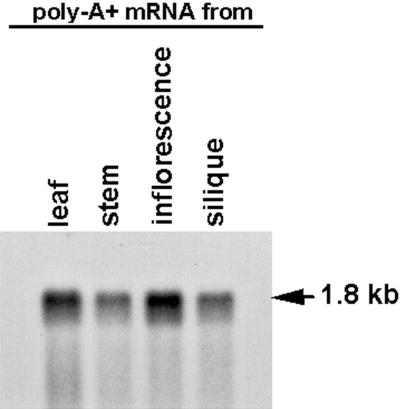
To begin to analyze the expression pattern of the chaperonin-60α gene in Arabidopsis, an RNA blot containing poly(A+) mRNA from leaf, silique, stem, and inflorescence tissues was hybridized with the 6.6-kb EcoRI fragment from the λ101 genomic clone (Fig. 5A; see “Materials and Methods”). As shown in Figure 7, the chaperonin-60α mRNA accumulates in all stages of development examined. The 1.8-kb mRNA that was detected in the four types of tissues is consistent with the expected size of the mRNA that would be encoded by the SLP gene.
Figure 7.
RNA-blot analysis of chaperonin-60α. Each lane contains 0.5 μg of poly(A+) mRNA from each organ. The RNA blot was hybridized with the 6.6-kb EcoRI fragment from λ101 genomic clone (see Fig. 5A).
Plastids Are Undeveloped in slp/slp Embryos
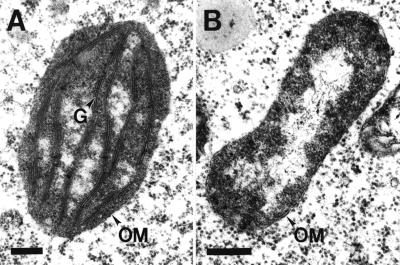
Because it is known that chaperonin-60α protein is involved in the folding and assembly of proteins (e.g. Rubisco; Goloubinoff et al., 1989a, 1989b) that are imported into the chloroplast, we determined if chloroplast development was affected in the slp/slp embryos. WT (green) and mutant (white) seeds from siliques of a heterozygous SLP/slp plant were fixed, embedded, sectioned, and analyzed for the presence and morphology of embryonic plastids using transmission electron microscopy (see “Materials and Methods”). As shown in Figure 8, the plastids in WT embryos developed into chloroplasts with well-stacked, membrane-appressed grana (Fig. 8A). In contrast, no well-developed plastids were detected in mutant slp/slp embryos–the mutant plastids contained unstacked or seemingly collapsed membrane structures (Fig. 8B). These results indicated that the differentiation of plastids into chloroplasts during Arabidopsis embryo development requires a functional chaperonin-60α protein.
Figure 8.
Transmission electron microscopy analysis of plastids. Chloroplast (A) from a WT embryo and undeveloped plastid (B) from slp embryo. G, Grana; OM, outer membrane. Bars = 0.15 μm.
DISCUSSION
The role of the chloroplast in plant embryogenesis has not been explored extensively. For the most part, the role of chloroplast has been studied only in relation to postembryonic stages of plant development. The results from several nuclear gene mutations that have detrimental effects on chloroplast biogenesis and metabolism (Tsugeki et al., 1996; Uwer et al., 1998) indicate that this organelle can be playing a significant role in plant embryogenesis. Our analysis of the SCHLEPPERLESS gene supports this conclusion.
The Arabidopsis slp mutant is defective in embryo development. The slp/slp mutant cotyledons are very short and the whole embryo is white, even at the mature stage (Fig. 1D). Mutant embryos up to the heart stage appear to be morphologically normal, although embryo development is temporally retarded in comparison with WT development (Fig. 2). The morphological defect of slp/slp embryos is manifested after the heart stage of embryogenesis (Fig. 2). In addition, the acquisition of competence to transcribe late-maturation genes is also delayed in mutant embryo and appears to depend on the developmental state of the embryo (Fig. 4). This result is in contrast with the transcriptional competence of raspberry, leafy cotyledon1, and other embryo-defective (emb) mutants that are capable of transcribing late-maturation genes in a temporally appropriate manner as WT embryos (West et al., 1994; Yadegari et al., 1994; Devic et al., 1996).
The morphology of slp/slp embryos is similar to that of edd1 mutants (Uwer et al., 1998). Both edd1 and slp seeds are able to germinate and form white plantlets when cultured in vitro (Fig. 3; Uwer et al., 1998). In contrast to some embryo-defective mutants whose chloroplasts are not affected by the mutation and are able to turn green when cultured (Franzmann et al., 1989; Patton et al., 1998), slp/slp embryos could not be rescued by the tissue culture medium (Fig. 3).
The development of chloroplast is affected in the slp/slp mutant, resulting in aberrant plastids in mature mutant embryos (Fig. 8). The fact that the embryos remain white (Fig. 1D) indicates that slp/slp mutant is most likely photosynthetic incompetent. However, in contrast with mutants that lack the photosynthetic capacity but remain morphologically normal (e.g. albino mutants), slp/slp embryos have abnormal morphology. The albino mutants have defects in chloroplast ultrastructure and are not photosynthetically competent, but are able to form morphologically normal mature adult plants (Hudson et al., 1993; Long et al., 1993; Sundberg et al., 1997). Therefore, the defect in slp mutant is more complex than just not being able to effectively carry out photosynthesis. The defect can be explained by the nature of the gene mutated in schlepperless, which appears to affect not only photosynthesis but also other processes that occur in the chloroplast and are required for proper chloroplast development.
SCHLEPPERLESS Locus Encodes Chaperonin-60α Subunit Protein
We cloned the WT SLP gene corresponding to the gene interrupted by T-DNA in the slp mutant (Fig. 5) and demonstrated that this genomic clone is capable of complementing the slp mutation (Table I and Fig. 5D). The gene encodes the plastid chaperonin-60α-subunit protein that shows significant homology with other chaperonin proteins, both from prokaryotes and eukaryotes (Hemmingsen et al., 1988; Zabaleta et al., 1992; Cole et al., 1994). The phenotypes (including white color and undeveloped plastid) that we observed for slp mutant are all consistent with the chaperonin-60α gene being mutated in this embryo-defective line.
The chaperonin-60α-subunit is considered to be the equivalent of the prokaryotic groEL protein (Hemmingsen et al., 1988). Mutation in the GroEL gene is lethal and the protein is considered essential for bacterial growth (Fayet et al., 1989). The heptameric ring structure of groEL proteins, in conjunction with heptameric ring of groES (another form of chaperonin), are involved in the folding and assembly of proteins to attain their proper and functional conformations (for review, see Ellis and van der Vies, 1991). Some details of the molecular mechanisms for this chaperonin-mediated protein folding and assembly are already known and elucidated using in vitro analysis (Weissman et al., 1994, 1996; for example, see Weissman et al., 1994, 1996; Mayhew et al., 1996; Ma and Karplus, 1998; Wang et al., 1998). In fact, Rubisco (one of the chloroplast-localized proteins) is one of the substrates used for this in vitro analysis (Goloubinoff et al., 1989a, 1989b; Gutteridge and Gatenby, 1995).
Chaperonin-60α Protein Is Required for Chloroplast Development
During chloroplast biogenesis, plastid volume and composition change as a consequence of acquisition of photosynthetic competence and activation of other biosynthetic processes (Mullet, 1988). The acquisition of this competence and the activation of the biosynthetic processes require the activation of both nuclear and chloroplast genes (Mullet, 1988). The nuclear-encoded chloroplast-localized proteins must be expressed at the proper temporal and developmental state of the chloroplast. In addition, some of these nuclear-encoded components have to be assembled inside the chloroplasts (e.g. photosystem II components that are normally assembled in the grana; Hermann et al., 1985), and the assembly requires that these components be in their proper conformation. We propose that chaperonin-60α protein is important for folding a variety of proteins essential for chloroplast development and functions.
In the slp embryos, we showed that the plastids are not fully developed and that there is no evidence for stacked or organized thylakoid membrane (Fig. 8). This indicates that functional chaperonin-60α protein is necessary for chloroplast development. Therefore, it is possible that, in the absence of functional chaperonin-60α-subunit, proteins that are imported into the chloroplasts are not folded properly and thus not functional. As such, the plastids are deprived of the necessary components for their biogenesis and/or function. These components may include proteins that are important not only for photosynthetic activity (e.g. light-harvesting chloroplast a/b protein), but other proteins (e.g. Gln synthetase) that are important for other functions of the chloroplasts (i.e. amino acid and fatty acid biosyntheses; Lubben et al., 1989). The products that are synthesized within the chloroplast (e.g. sugars, fatty acids, terpenoids, and amino acids) are not only utilized by the chloroplast organelle itself for its own development and function (for example, see Jarvis et al., 2000), but also by the cell for other metabolic processes. Fatty acids biosynthesis is considered essential for growth and its absence is lethal (Ohlrogge and Browse, 1995).
Embryo Development Is Dependent on Functional Chloroplasts
Chloroplasts in embryos are derived from undifferentiated proplastids that are normally inherited maternally by the plant zygote (Kirk and Tilney-Bassett, 1978). Greening starts in the early heart-shaped embryos and the initiation of greening is always associated with the development of granal stacks within the plastids. The plastids in globular embryos contain single lamellae, but in heart-shaped embryos, they begin to form rudimentary stacks of membrane that develop further into full grana by the torpedo stage (Schultz and Jensen, 1968; Mansfield and Briarty, 1991). There are indications that nongreen proembryos could be actively synthesizing proteins that are eventually localized in the plastids. For example, mRNAs corresponding to chloroplast psbA gene (encodes the D1 protein of photosystem II) and nuclear rbcS gene (encodes the small subunit of Rubisco) begin to accumulate in the proembryo (Degenhardt et al., 1991). Because the folding of proteins encoded by these mRNAs is chaperonin mediated, functional chaperonin-60α protein is necessary at the very early stages of embryogenesis, even before the embryo becomes green.
As shown in Figure 2, we demonstrated that slp embryo is morphologically normal up to the heart stage but its development is slower than that of WT embryos. Whatever accounts for this slow development may be related to the state of competence of the plastids. It is possible that competent proplastids contribute to the normal rate of embryonic process. It is interesting to note that cell division is extremely rapid as embryo develops from globular to heart stage and as the cotyledon initials are formed quickly (Mansfield and Briarty, 1991). It is possible that an early developed plastid starts to synthesize products that are nutritionally important and become readily available to the embryo as it develops. Otherwise, the amount of these biosynthetic products may be limited and insufficient for the slp embryo to undergo a normal rate of development. This limited amount of products may come from the proplastids probably rendered competent to some extent, albeit for a limited period during early embryogenesis, by the presence of a limited supply of chaperonin-60α protein contained within the maternally inherited proplastids. It is also possible that the folding of the chloroplast-localized proteins in the proplastids at the early stage of embryogenesis may be mediated by other chaperonins (e.g. hsp70 or chaperonin-60β; Madueño et al., 1993; Tsugeki and Nishimura, 1993, Zabaleta et al., 1994). This, perhaps, renders the proplastids functional to a limited degree at this stage of embryo development.
The data presented in this paper do not preclude the possibility that the folding of some non-chloroplastic proteins (e.g. cytoplasmic proteins) can be mediated by chaperonin-60α, similar to the groEL-mediated folding of citrate synthase, polynucleotide phosphorylase, and ketoglutarate dehydrogenase (Horwich et al., 1993) or to the chaperonin-mediated folding of actin (Gao et al., 1992; Siegers et al., 1999; Thulasiraman et al., 1999) and tubulin (Yaffe et al., 1992). If chaperonin-60α is involved in the folding of some cytoplasmic proteins in plant cells, then this particular defect may be reflected in the overall phenotype of schlepperless and is indistinguishable from the consequent defects in the chloroplast. The schlepperless mutant should allow studies to be carried out that distinguish between the effect of chaperonin-60α on chloroplast and cytoplasmic proteins.
In the developing embryo, the demand for biosynthetic products is very high, especially during the late maturation phase when macromolecules (proteins, lipids, and/or carbohydrates) are stored in the cotyledons (Goldberg et al., 1989; Shotwell and Larkins, 1989), leading to the expansion of the whole organ. Thus, it may not be surprising that in slp embryos, where plastids are undeveloped and nonfunctional as a result of mutation in the chaperonin-60α gene (Fig. 8) cotyledons do not expand fully. In a similar manner, the loss of function in other nuclear genes whose products are chloroplast localized and/or may be needed for early plastid biogenesis and competence lead also to defects in embryo development. Mutations in these genes either prevent embryo to undergo morphogenesis as in the case of raspberry mutants (Yadegari et al., 1994; N.R. Apuya, R. Yadegari, R.L. Fischer, J.J. Harada, and R.B. Goldberg, unpublished data) or prevent embryo to develop cotyledons as in the case of edd1 mutant (defective in plastidic form of glycyl-tRNA synthetase; Uwer et al., 1998). It is important to realize that mutations in these genes are important for understanding the molecular, biochemical, and cellular basis of plant embryogenesis. In this respect, the role of chloroplast in embryonic development should be explored in more detail. Characterizing other nuclear-encoded gene products that are not necessarily involved directly in photosynthesis but are important for chloroplast function during embryogenesis can elucidate this role.
In conclusion, the results of our analyses indicate that chaperonin-60α protein is essential for the growth and development in plants and the absence of this protein leads to severe defects in embryo and seedling development.
MATERIALS AND METHODS
Mutant Isolation and Genetic Analysis
The line characterized in this study was A2137, one of the 5,822 T-DNA-mutagenized lines of Arabidopsis ecotype Wassilewskija that were screened at the DuPont Experimental Station (Wilmington, DE, in November 1990) and at the University of Arizona (Tucson, AZ, in November 1991; Feldmann and Marks, 1987; Errampalli et al., 1991; Feldmann, 1991; Castle et al., 1993; Yadegari et al., 1994). The recessive embryo-defective mutation was maintained in heterozygous plants (slp/SLP) that produced WT and mutant seeds at a 3:1 ratio. The cosegregation of T-DNA and the embryo defective phenotype was analyzed by a Kan-R assay and by genomic DNA-blot analysis using the T-DNA right and left border sequences as probes. To map the chromosomal location of the mutation, heterozygous plants were crossed to the mapping lines obtained from the Arabidopsis Biological Resource Center (Columbus, OH). Initial cross was made to the mapping line CS3078 (a mapping line containing one marker in every chromosome) to narrow down the chromosomal location of the mutation. A subsequent cross was made to mapping line W6 that has cp2, cer8, and as markers in chromosome 2. Linkage analysis to the phenotypic markers was performed by characterizing 500 segregants from the F2 population and estimates of recombination were done using the RECF2 program (Koornneef and Stam, 1992).
Seed Germination in Tissue Culture
Seeds were sterilized in a commercial bleach solution for about 10 min and rinsed four times with sterile water. Sterilized seeds were subsequently plated in germination medium (GM) containing 1× Murashige and Skoog salt, 1% (w/v) Suc, 100 mg L−1 inositol, 1 mg L−1 thiamine, 0.5 mg L−1 nicotinic acid, 0.5 g L−1 MES (pH 7), and 0.5% (w/v) phytagar (Gibco, Gaithersburg, MD). Germinating mutant seeds were transferred to fresh GM plates every 10 d until termination of the experiment. For Kan-R and/or Hyg-R assay, seeds were germinated in GM containing 50 μg mL−1 kanamycin sulfate and/or 20 μg mL−1 hygromycin.
GUS Detection
Embryos resulting from the cross between the heterozygous plants and the homozygous transgenic lines containing either β-conglycinin-α′-promoter::GUS or LEA::GUS fusion constructs were assayed for GUS activity using the procedures of Jefferson et al. (1987) with some modifications. The GUS staining solution consisted of 50 mm sodium phosphate buffer (pH 7), 10 mm EDTA, 0.1% (v/v) Triton X-100, 0.5 mm potassium ferricyanide, 0.5 mm potassium ferrocyanide, and 1 mm 5-bromo-4-chloro-3-indoyl glucuronide. Dissected embryos were incubated in this solution at 37°C for either 2 h (LEA::GUS) or 5 h (β-conglycinin-α′::GUS). Embryos were destained for 5 h in 70% (v/v) ethanol and processed for dark-field microscopy.
Microscopy
Bright-Field Microscopy
Siliques from heterozygous plants were collected, cut into 2-mm pieces, fixed, and embedded in LR White plastic resin (Polysciences, Inc., Warrington, PA). Sections (1 μm thick) were made using a microtome (LKB Ultratome V; LKB, Bromma, Sweden) and stained for about 10 min with 0.5% (w/v) toluidine blue in 0.1% (w/v) borate solution. Bright-field photographs were taken with Gold 100 film (ISO 100/21°, Kodak, Rochester, NY) using a compound microscope (Olympus BH-2; Olympus Corporation, Lake Success, NY).
Nomarski Microscopy
Mutant and WT seeds were fixed in ethanol:acetic acid (9:1) solution overnight, and successively washed in 90% and 70% (v/v) ethanol for at least 30 min each. Seeds were cleared with chloral hydrate:glycerol:water solution (8:1:2, w:v:v) for at least 2 h prior to microscopy (Berleth and Jürgens, 1993). Embryos were visualized using Nomarski optics on a Zeiss Axiophot (Carl Zeiss, Inc., Oberkochlen, Germany). Photographs were taken using Kodak TMAX 100 (E.I. 100/21°) film.
Transmission Electron Microscopy
The procedures cited by Yadegari et al. (1994) were followed except LR White plastic resin was used as the embedding medium.
Whole Mount Photography
Dark-field photographs of germinating seedlings in culture were taken using a dissecting microscope (Olympus SZH, Olympus Corp.). Dark-field photographs of embryos assayed for GUS were taken using a compound microscope (Olympus BH2) using Kodak Gold 100 film (ISO 100/21°).
Genomic DNA Isolation, Restriction Analysis, DNA Blotting, and Labeling
Genomic DNA was isolated according to the procedures established by Dellaporta et al. (1983). Digestion of genomic DNA with restriction enzymes was done overnight following the conditions recommended by the manufacturers. Digested DNA was size fractionated by electrophoresis in agarose gels and then transferred to Nytran nylon membrane (Schleicher and Schuell, Keene, NH) following the recommended protocol by the manufacturer. Prehybridization and hybridization of DNA blots were done following the procedures recommended by Ausubel et al. (1992). Labeled DNA probes were synthesized using the random priming technique (Feinberg and Vogelstein, 1983).
Isolation of Mutant and WT Genomic Clones
Plasmid rescue was done following the protocol of Behringer and Medford (1992). The procedures recommended by Sambrook et al. (1989) for colony lifts and hybridization were followed. A genomic library of the heterozygous line was also constructed using the λGEM-12 vector and following the protocol recommended by Promega (Madison, WI). Screening of the library and the plasmid transformants was done using the right and left border sequences of T-DNA as probes. The procedures established by Ausubel et al. (1992) for plating and transferring bacteriophage library and for pretreatment of filters for hybridization were followed. The isolation of WT genomic clones was done following the same protocol but using the plant flanking sequences obtained from the mutant clones as probes. The Escherichia coli strain KW251 was used for all genomic phage experiments. Sub-cloning of certain fragments from rescued plasmids and phage clones was done using pGEM-3Z(f-) as vector and following the standard cloning procedures recommended by Sambrook et al. (1989).
Isolation of cDNA Clones
A cDNA library (in λZAP vector) constructed from poly(A+) mRNA from WT Arabidopsis siliques was used to isolate cDNA clones. The protocol for the isolation of phage genomic clones was essentially followed except for using XL1-Blue strain as the host bacteria. The plasmid forms of the cDNA clones were isolated from the corresponding phage cDNA clones following the in vivo excision protocol recommended by Stratagene (La Jolla, CA). The 6.6-kb EcoRI fragment from WT genomic clone λ101 (see Fig. 5A) was used as probe for the cDNA library screening.
DNA Sequencing
The sequencing of cDNA clones, and sub-clones of rescued plasmids and phage genomic clones, was done following the dideoxy-sequencing procedures recommended by United States Biochemicals (Cleveland). The sequencing of the 6.6-kb EcoRI fragment of genomic clone λ101 was done at Plant Genetics Systems using the PCR with Taq polymerase and an automated sequencer (Applied Biosystems, Inc., Foster City, CA). Analysis of sequences was done using the Genetics Computer Group software and the National Center for Biotechnology Information BLAST e-mail server.
RNA Techniques
Polysomal RNAs were isolated according to the procedures described by Cox and Goldberg (1988). Poly(A+) mRNAs were isolated using the Poly-AT Tract mRNA Isolation System (Promega) and following the recommended protocol by the manufacturer. The isolated mRNAs were electrophoresed through a formaldehyde-agarose gel, transferred to a Nytran membrane (Schleicher and Schuell), and hybridized with 32P-labeled DNA according to the procedures recommended by Ausubel et al. (1992).
Plant Transformation and Complementation Analysis
An 11-kb NotI fragment from λ101 genomic clone (see Fig. 5A) was sub-cloned into the NotI site of pGSH166N vector (courtesy of Plant Genetic Systems, Gent, Belgium). The resulting recombinant was subsequently transferred to Agrobacterium tumefaciens to be used for root transformation. The original procedures established by Valvekens et al. (1988) for Agrobacterium-mediated transformation of Arabidopsis root explants were followed. WT Arabidopsis (ecotypes C24 and Nossen) were used as recipients.
Complementation analysis was done by crossing heterozygous plants (slp/SLP; Kan-R) to transgenic lines containing the WT chaperonin-60α gene (designated as tCpn; Hyg-R). The Hyg-R and Kan-R F1 progenies from this cross were selected and allowed to grow in the greenhouse. The F2 seeds collected from the F1 plants were germinated without selection. The resulting F2 plants were phenotyped by dissecting two to three siliques from each plant and the number of mutant and WT seeds were counted for genetic analysis as presented in Table I. Depending on the percentage of mutant seeds contained within the two or three siliques, the F2 plants were grouped into three classes as listed in Table I. Segregants in class I were individuals that produced only WT embryos, whereas those in class II and class III produced mutant embryos at different frequencies. If the complementation were successful, the segregants that are either heterozygous (SLP/slp) or homozygous (slp/slp) would be included in class I as long as they are homozygous to the transgene (tCpn). The frequency of mutant embryos produced by individuals in either class II or class III depends on their genotypes as well (see Table I). For instance, if homozygous (slp/slp) segregants contain only one copy of the transgene, then we expect them to be classified in class II (i.e. producing mutant seeds at 25% frequency). However, if the heterozygous segregants (SLP/slp) contain only one copy of the transgene, then they would be classified in class III (i.e. producing mutant seeds at about 6.25% frequency), which is a class that is produced only if the complementation was successful (see Table I). Genomic DNAs from randomly selected F2 plants were isolated for DNA-blot analysis shown in Figure 5C.
The genotypes of some randomly selected F2 segregants representative of each class were determined by germinating their progeny seeds under kanamycin and hygromycin selection. Kan-R allowed us to follow the segregation of the slp mutation, whereas Hyg-R allowed us to follow the segregation of the transgene (tCpn). At the same time, testcross analysis was also done by crossing the selected individuals to WT (SLP/SLP) to determine the homozygosity or heterozygosity of the slp mutation. The seed products of the testcross were germinated under selection for the two antibiotics and the resulting plants were analyzed for an embryo-defective phenotype.
ACKNOWLEDGMENTS
We would like to acknowledge Jef Seurink (Plant Genetic Systems) for sequencing the genomic clone, Birgitta Sjostrand (University of California, Los Angeles) for help with electron microscopy, and Margaret Kowalczyk (University of California, Los Angeles) for the preparation of figures. We also acknowledge Ken Feldmann (Ceres, Inc.) for allowing us to screen the T-DNA mutants while he was at the University of Arizona, and Satoshi Naito (Hokkaido University, Japan) and Renee Sung (University of California, Berkeley) for providing the Arabidopsis lines transgenic for the GUS constructs. We extend our gratitude to all the individuals within our collaboration for incisive discussion and help in carrying out this research.
LITERATURE CITED
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocol in Molecular Biology. New York: Greene Publishing and Wiley-Interscience; 1992. [Google Scholar]
- Behringer FJ, Medford JI. A plasmid rescue technique for the recovery of plant DNA disrupted by T-DNA insertion. Plant Mol Biol Rep. 1992;10:190–198. [Google Scholar]
- Berleth T, Jürgens G. The role of monopteros in organizing the basal body region of the Arabidopsis embryo. Development. 1993;118:575–587. [Google Scholar]
- Castle LA, Errampalli D, Aterton TL, Franzmann LH, Yoon ES, Meinke DW. Genetic and molecular characterization of embryonic mutants identified following seed transformation in Arabidopsis. Mol Gen Genet. 1993;241:504–514. doi: 10.1007/BF00279892. [DOI] [PubMed] [Google Scholar]
- Clark JK, Sheridan WF. Isolation and characterization of 51 embryo-specific mutations in maize. Plant Cell. 1991;3:935–951. doi: 10.1105/tpc.3.9.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloney LP, Bekkaoui DR, Feist GI, Lane WS, Hemmingsen SM. Brassica napus plastid and mitochondrial chaperonin-60 protein contain multiple distinct polypeptides. Plant Physiol. 1994;105:233–241. doi: 10.1104/pp.105.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole KP, Blakeley SD, Dennis DT. Isolation of a full-length cDNA encoding Brassica napus plastid chaperonin-60α subunit. Plant Physiol. 1994;105:453. doi: 10.1104/pp.105.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Goldberg RB. Analysis of plant gene expression. In: Shaw CH, editor. Plant Molecular Biology: A Practical Approach. Oxford: IRL Press; 1988. pp. 1–34. [Google Scholar]
- Degenhardt J, Fiebig C, Link G. Chloroplast and nuclear transcripts for plastid proteins in Arabidopsis thaliana: tissue distribution in mature plants and during seedling development and embryogenesis. Bot Acta. 1991;104:455–463. [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;4:19–21. [Google Scholar]
- Devic M, Albert S, Delseney M. Induction and expression of seed-specific promoters in Arabidopsis embryo-defective mutants. Plant J. 1996;9:205–215. doi: 10.1046/j.1365-313x.1996.09020205.x. [DOI] [PubMed] [Google Scholar]
- Ellis RJ, van der Vies SM. Molecular chaperones. Ann Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- Errampalli D, Patton D, Castle L, Mickelson L, Hansen K, Schnall J, Feldmann K, Meinke D. Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell. 1991;3:149–157. doi: 10.1105/tpc.3.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayet O, Ziegelhoffer T, Georgopoulos C. The groES and groEL heat shock gene products of E. coli are essential for bacterial growth at all temperatures. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Feldmann K. T-DNA insertion mutagenesis in Arabidopsis: mutational spectrum. Plant J. 1991;1:71–82. [Google Scholar]
- Feldmann K, Marks MD. Agrobacterium-mediated transformation of germinating seeds of Arabidopsis: a non-tissue culture approach. Mol Gen Genet. 1987;208:1–9. [Google Scholar]
- Franzmann L, Patton DA, Meinke DW. In vitro morphogenesis of arrested embryos from lethal mutants of Arabidopsis thaliana. Theor Appl Genet. 1989;77:609–616. doi: 10.1007/BF00261231. [DOI] [PubMed] [Google Scholar]
- Gao Y, Thomas JO, Chow RL, Lee GH, Cowan NJ. A cytoplasmic chaperonin that catalyzes β-actin folding. Cell. 1992;69:1043–1050. doi: 10.1016/0092-8674(92)90622-j. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Barker SJ, Perez-Grau L. Regulation of gene expression during plant embryogenesis. Cell. 1989;56:149–160. doi: 10.1016/0092-8674(89)90888-x. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, de Paiva G, Yadegari R. Plant embryogenesis: zygote to seed. Science. 1994;266:605–614. doi: 10.1126/science.266.5185.605. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. The role of chaperonins in protein folding: the reconstitution of active dimers from urea-denatured ribulose biphosphate carboxylase depends on the chaperonin protein and MgATP. Nature. 1989a;342:884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- Goloubinoff P, Gatenby AA, Lorimer GH. GroE heat shock proteins promote assembly of foreign prokaryotic ribulose biphosphate carboxylase oligomers in E. coli. Nature. 1989b;337:44–47. doi: 10.1038/337044a0. [DOI] [PubMed] [Google Scholar]
- Goupil P, Hatzopoulos P, Franz G, Hempel FD, You R, Sung ZR. Transcriptional regulation of seed-specific carrot gene, DC8. Plant Mol Biol. 1992;18:1049–1063. doi: 10.1007/BF00047708. [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld J-U, Salchert K, Koncz C, Jürgens G. A conserved domain of the Arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutteridge S, Gatenby AA. Rubisco synthesis, assembly, mechanism, and regulation. Plant Cell. 1995;7:809–819. doi: 10.1105/tpc.7.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Berleth T. The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 1998;17:1405–1411. doi: 10.1093/emboj/17.5.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmingsen SM, Woolford C, van der Vies SM, Tilly K, Dennis DT, Georgopoulos CP, Hendrix RW, Ellis RJ. Homologous plant and bacterial protein chaperone oligomeric assembly. Nature. 1988;333:330–334. doi: 10.1038/333330a0. [DOI] [PubMed] [Google Scholar]
- Hermann RG, Westhoff P, Alt J, Tittgen J, Nelson N. Thylakoid membrane proteins and their genes. In: van Vloten-Doting L, Groot GSP, Hall TC, editors. Molecular Form and Function of the Plant Genome. Amsterdam: Plenum Press; 1985. pp. 233–256. [Google Scholar]
- Hirai MY, Fujiwara T, Goto K, Komeda Y, Chino M, Naito S. Differential regulation of soybean seed storage protein gene promoter-GUS fusions by exogenous applied methionine in transgenic Arabidopsis thaliana. Plant Cell Physiol. 1994;35:927–934. [Google Scholar]
- Horwich AL, Low KB, Fenton WA, Hirshfield IN, Furtak K. Folding in vivo of bacterial cytoplasmic proteins: role of groEL. Cell. 1993;74:909–917. doi: 10.1016/0092-8674(93)90470-b. [DOI] [PubMed] [Google Scholar]
- Hudson A, Carpenter R, Doyle S, Coen ES. Olive: a key gene required for chlorophyll biosynthesis in Antirrhinum majus. EMBO J. 1993;12:3711–3719. doi: 10.1002/j.1460-2075.1993.tb06048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J. Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase1 mutant. Plant Physiol. 2000;97:8175–8179. doi: 10.1073/pnas.100132197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens G, Mayer U. Arabidopsis. In: Bard JBL, editor. Embryos, Color Atlas of Development. London: Wolfe Publishing; 1994. pp. 7–21. [Google Scholar]
- Kirk JTO, Tilney-Bassett RAE. The plastids: their chemistry, structure, growth, and inheritance. Ed 2. Amsterdam, New York: Elsevier/North-Holland Biomedical Press; 1978. [Google Scholar]
- Koornneef M, Stam P. Genetic analysis. In: Koncz C, Chua NH, Schell J, editors. Methods in Arabidopsis Research. River Edge, MN: World Scientific Publishing Co.; 1992. pp. 83–99. [Google Scholar]
- Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, Barnstead M. Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature. 1999;402:761–768. doi: 10.1038/45471. [DOI] [PubMed] [Google Scholar]
- Long D, Martin M, Sundberg E, Swinburne J, Puangsomlee P, Coupland G. The maize transposable element Ac/Ds as a mutagen in Arabidopsis: identification of an albino mutant induced by Ds insertion. Proc Natl Acad Sci USA. 1993;90:10370–10374. doi: 10.1073/pnas.90.21.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the knotted class of homeodomain proteins encoded by the STM of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Lubben TH, Donaldson GK, Viitanen PV, Gatenby AA. Several proteins imported into chloroplasts from stable complexes with groEL-related chloroplast molecular chaperone. Plant Cell. 1989;1:1223–1230. doi: 10.1105/tpc.1.12.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;84:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Ma J, Karplus M. The allosteric mechanism of the chaperonin GroEL: a dynamic analysis. Proc Natl Acad Sci USA. 1998;95:8502–8507. doi: 10.1073/pnas.95.15.8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madueño F, Napler JA, Gray JC. Newly imported Rieske iron-sulfur protein associates with both Cpn60 and Hsp70 in the chloroplast stroma. Plant Cell. 1993;5:1865–1876. doi: 10.1105/tpc.5.12.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield SG, Briarty LG. Early embryogenesis in Arabidopsis thaliana: II. The developing embryo. Can J Bot. 1991;69:461–476. [Google Scholar]
- Martel R, Cloney LP, Petcher LE, Hemmingsen SM. Unique composition of plastid chaperonin-60: α and β polypeptide-encoding genes are highly divergent. Gene. 1990;94:181–187. doi: 10.1016/0378-1119(90)90385-5. [DOI] [PubMed] [Google Scholar]
- Mayer U, Büttner G, Jürgens G. Apical-basal pattern formation in the Arabidopsis embryo: studies on the role of the gnom gene. Development Suppl. 1993;117:149–162. [Google Scholar]
- Mayer U, Torres Ruiz RA, Berleth T, Misera S, Jürgens G. Mutations affecting body organization in the Arabidopsis embryo. Nature. 1991;353:402–407. [Google Scholar]
- Mayhew M, da Silva ACR, Martin J, Erdument-Bromage H, Tempst P, Hartl FU. Protein folding in the central cavity of the GroEL-GroES chaperonin complex. Nature. 1996;379:420–426. doi: 10.1038/379420a0. [DOI] [PubMed] [Google Scholar]
- Meinke DW. Embryonic mutants of Arabidopsis thaliana. Dev Genet. 1991;12:382–392. [Google Scholar]
- Meinke DW. Molecular genetics of plant embryogenesis. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:369–394. [Google Scholar]
- Mullet J. Chloroplast development and gene expression. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:475–502. [Google Scholar]
- Ohlrogge J, Browse J. Lipid biosynthesis. Plant Cell. 1995;7:957–970. doi: 10.1105/tpc.7.7.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton DA, Schetter AL, Franzmann LH, Nelson K, Ward ER, Meinke DW. An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 1998;116:935–946. doi: 10.1104/pp.116.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad TK, Stewart CR. cDNA clones encoding Arabidopsis thaliana and Zea mays mitochondrial chaperonin HSP60 and gene expression during seed germination and heat shock. Plant Mol Biol. 1992;18:873–885. doi: 10.1007/BF00019202. [DOI] [PubMed] [Google Scholar]
- Roop RM, Price ML, Dunn BE, Boyle SM, Srirananganathan N, Schurig GG. Molecular cloning and nucleotide sequence analysis of the gene encoding the immunoreactive Brucella abortis HSP60 protein. Microb Pathog. 1992;12:47–62. doi: 10.1016/0882-4010(92)90065-v. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schultz P, Jensen WA. Capsella embryogenesis: the early embryo. J Ultrastuct Res. 1968;22:376–392. doi: 10.1016/s0022-5320(68)90028-2. [DOI] [PubMed] [Google Scholar]
- Shevell DE, Kunkel T, Chua NH. Cell wall alterations in the Arabidopsis emb30 mutant. Plant Cell. 2000;12:2047–2059. doi: 10.1105/tpc.12.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevell DE, Leu W, Gillmor CS, Xia G, Feldmann KA, Chua NH. Emb30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to sec7. Cell. 1994;77:1051–1062. doi: 10.1016/0092-8674(94)90444-8. [DOI] [PubMed] [Google Scholar]
- Shotwell MA, Larkins BA. The molecular biology and biochemistry of seed storage proteins. In: Marcus A, editor. The Biochemistry of Plants. Vol. 15. San Diego: Academic Press; 1989. pp. 297–345. [Google Scholar]
- Siegers K, Waldmann T, Leroux MR, Green K, Shevchenko A, Schiabel E, Hartl FU. Compartmentation of protein folding in vivo: sequestration of non-native polypeptide by the chaperonin-GimC system. EMBO J. 1999;18:75–84. doi: 10.1093/emboj/18.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souer E, van Houwelingen A, Kloos D, Mol J, Koes R. The No Apical Meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell. 1996;85:159–170. doi: 10.1016/s0092-8674(00)81093-4. [DOI] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G. Albino3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell. 1997;9:717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thulasiraman V, Yang CF, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki R, Kochieva EZ, Fedoroff NV. A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 1996;10:479–489. doi: 10.1046/j.1365-313x.1996.10030479.x. [DOI] [PubMed] [Google Scholar]
- Tsugeki R, Nishimura M. Interaction of homologues of hsp70 and cpn60 with ferroxin-NADP+ reductas upon its import into chloroplasts. FEBS Lett. 1993;320:198–202. doi: 10.1016/0014-5793(93)80585-i. [DOI] [PubMed] [Google Scholar]
- Uwer U, Willmitzer L, Altmann T. Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell. 1998;10:1277–1294. doi: 10.1105/tpc.10.8.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valvekens D, Van Montagu M, Lijsebettens M. Agrobacterium tumafaciens-mediated transformation of Arabidopsis root explants using kanamycin selection. Proc Natl Acad Sci USA. 1988;85:5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Michelitsch MD, Weissman JD. GroEL-GroES mediated protein folding requires an intact central cavity. Proc Natl Acad Sci USA. 1998;95:12163–12168. doi: 10.1073/pnas.95.21.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman JS, Kashi Y, Fenton W, Horwich AL. GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell. 1994;78:693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- Weissman JS, Rye HS, Fenton W, Beecham J, Horwich AL. Characterization of the active intermediate of a GroEL-GroES-mediated protein folding reaction. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- West MAL, Harada JJ. Embryogenesis in higher plants: an overview. Plant Cell. 1993;5:1361–1369. doi: 10.1105/tpc.5.10.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Matsudaira-Yee K, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadegari R, de Paiva GR, Laux T, Koltunow AM, Apuya N, Zimmerman JL, Fischer RL, Harada JJ, Goldberg RB. Cell differentiation and morphogenesis are uncoupled in Arabidopsis raspberry embryos. Plant Cell. 1994;6:1713–1729. doi: 10.1105/tpc.6.12.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Farr GW, Miklos D, Horwich AL, Sternlicht ML, Sternlicht H. TCP1 complex is a molecular chaperone in tubulin biogenesis. Nature. 1992;358:245–248. doi: 10.1038/358245a0. [DOI] [PubMed] [Google Scholar]
- Zabaleta E, Oropeza A, Assad N, Mandel A, Salerno G, Herrera-Estrella L. Antisense expression of chaperonin 60β in transgenic tobacco plants leads to abnormal phenotypes and altered distribution of plant assimilates. Plant J. 1994;6:425–432. [Google Scholar]
- Zabaleta E, Oropeza A, Jimenez B, Salerno G, Crespo M, Herrera-Estrella L. Isolation and characterization of genes encoding chaperonin 60β from Arabidopsis thaliana. Gene. 1992;111:175–181. doi: 10.1016/0378-1119(92)90685-i. [DOI] [PubMed] [Google Scholar]