Abstract
We recently reported the cloning and characterization of an Arabidopsis (ecotype Columbia) diacylglycerol acyltransferase cDNA (Zou et al., 1999) and found that in Arabidopsis mutant line AS11, an ethyl methanesulfonate-induced mutation at a locus on chromosome II designated as Tag1 consists of a 147-bp insertion in the DNA, which results in a repeat of the 81-bp exon 2 in the Tag1 cDNA. This insertion mutation is correlated with an altered seed fatty acid composition, reduced diacylglycerol acyltransferase (DGAT; EC 2.3.1.20) activity, reduced seed triacylglycerol content, and delayed seed development in the AS11 mutant. The effect of the insertion mutation on microsomal acyl-coenzyme A-dependent DGAT is examined with respect to DGAT activity and its substrate specificity in the AS11 mutant relative to wild type. We demonstrate that transformation of mutant AS11 with a single copy of the wild-type Tag1 DGAT cDNA can complement the fatty acid and reduced oil phenotype of mutant AS11. More importantly, we show for the first time that seed-specific over-expression of the DGAT cDNA in wild-type Arabidopsis enhances oil deposition and average seed weight, which are correlated with DGAT transcript levels. The DGAT activity in developing seed of transgenic lines was enhanced by 10% to 70%. Thus, the current study confirms the important role of DGAT in regulating the quantity of seed triacylglycerols and the sink size in developing seeds.
Seed triacylglycerol (TAG) biosynthesis is located in the endoplasmic reticulum with glycerol-3-phosphate and fatty acyl-coenzyme A (CoAs) as the primary substrates. There are three acyltransferases and a phosphohydrolase involved in the plant storage lipid bioassembly, namely glycerol-3-phosphate acyltransferase (GPAT, EC 2.3.1.15), lyso-phosphatidic acid acyltransferase (LPAT, EC 2.3.1.51), phosphatidate phosphohydrolase (PAPase, EC 3.1.3.4), and diacylglycerol acyltransferase (DGAT, EC 2.3.1.20). The three acyltransferases catalyze the stepwise acylation of the glycerol backbone with the final step being the acylation of sn-1,2-diacylglycerols (DAGs) by DGAT to form TAGs, a biochemical process generally known as the Kennedy pathway (Kennedy, 1961; Barron and Stumpf, 1962; Stymne and Stobart, 1987). The acyl-CoA dependent acylation of sn-1,2-DAG as catalyzed by DGAT is the only enzyme in the traditional Kennedy pathway that is exclusively committed to TAG biosynthesis.
In developing and germinating seeds of oilseed plants, TAG accumulation and DGAT activity have been shown to associate with the endoplasmic reticulum (high-speed microsomal fraction) (Cao and Huang, 1986; Stobart et al., 1986; Stymne and Stobart, 1987; Frentzen, 1993; Settlage et al., 1995; Lacey and Hills, 1996). The biochemical properties of microsomal DGAT have been examined in a number of plant systems (Frentzen, 1993), including developing seeds (Cao and Huang, 1987; Bernerth and Frentzen, 1990; Vogel and Browse, 1996) and embryo cultures (Taylor et al., 1991, 1992; Weselake et al., 1991; Little et al., 1994) of Brassica napus. In general, studies with developing seeds indicate that DGAT activity increases rapidly during the active phase of oil accumulation and then decreases markedly as seed lipid content reaches a plateau (Tzen et al., 1993; Weselake et al., 1993).
A number of studies with both mammalian (Mayorek et al., 1989; Tijburg et al., 1989) and plant (Ichihara et al., 1988; Perry and Harwood, 1993a, 1993b; Settlage et al., 1995; Perry et al., 1999) systems have suggested that DGAT may catalyze a rate-limiting reaction in TAG bioassembly. For example, developing seeds of B. napus L., cv Shiralee, have been shown to produce significant levels of DAG during the active phase of oil accumulation (Perry and Harwood, 1993a, 1993b). More recently, using light/dark treatments in this cultivar, it has been shown that during conditions of high lipid accumulation, the amounts of Kennedy pathway intermediates phosphatidate, and diacylglycerol increase significantly. During this time, the DGAT activity is the lowest of the four Kennedy pathway enzymes. The alteration in carbon flux resulted in changes to the acyl quantity of the DAG pool but not other intermediates (Perry et al., 1999). The data collectively suggest that the DGAT reaction may regulate the flow of carbon into TAG at times of high lipid accumulation. However, this hypothesis has not been rigorously tested or reduced to practice by transgenically altering the expression of a DGAT gene in a seed-specific manner.
We previously characterized an ethyl methanesulfonate (EMS)-induced mutant of Arabidopsis, designated AS11, which displayed an altered fatty acid composition (Katavic et al., 1995). AS11 seeds have reduced levels of the very long chain fatty acid eicosenoic acid (20:1) and reduced oleic acid (18:1) and accumulate α-linolenic acid (18:3) as the major fatty acid in TAGs. The AS11 mutant has a consistently lower ratio of TAG/DAG in developing seeds, and it accumulates an elevated amount of seed DAG, which is the substrate of the DGAT. It was shown that AS11 had reduced DGAT activities throughout seed development and thus a reduced TAG content phenotype, providing some evidence that DGAT may be controlling flux into TAG biosynthesis. Genetic analysis indicated that the fatty acid phenotype in AS11 is caused by a semidominant mutation in a nuclear gene, designated Tag1. The mutation in line AS11 was characterized as a 147-bp insertion in the chromosome II DNA, which results in a repeat of the 81-bp exon 2 in the Tag1 transcript. This insertion mutation is correlated with an altered seed fatty acid composition, a reduced seed TAG content, reduced DGAT (EC 2.3.1.20) activity, and delayed seed development, characteristic of the AS11 mutant seed line. Tag1 was mapped to chromosome II (Katavic et al., 1995).
We recently reported the identification, functional assignment, and cloning of this DGAT gene, Tag1, from Arabidopsis (Zou et al., 1999; GenBank accession no. AJ238008) through a databank sequence search and assisted by map-based information. Our data demonstrated that the encoded product of the Tag1 gene is 41% identical over a stretch of more than 400 amino acids to the mouse (Cases et al., 1998; GenBank accession no. AF078752) and human (Oelkers et al., 1998; GenBank accession no. AF059202) DGATs and includes a signature putative diacylglycerol binding motif. The TAG1 gene was shown to encode an acyl-CoA-dependent DGAT by functional expression of the recombinant protein produced in yeast cells (Zou et al., 1999).
These results were in agreement with concurrent publications describing the cloning of the Arabidopsis DGAT sequence and functional expression of the recombinant protein in insect cell cultures (Hobbs et al., 1999; GenBank accession no. AJ131831) and the cloning of the Arabidopsis DGAT sequence, a similar study of the insertion mutation allele from AS11 (tag 1-1), and the description of a new deletion mutant ABX45 allele (tag 1-2) as reported by Routaboul et al. (1999). The Arabidopsis DGAT sequence is also highly homologous (approximately 90% amino acid identity) to subsequently published putative DGAT sequences from B. napus reported by Nykiforuk et al., 1999 (GenBank accession nos. AF155224 and AF164434) and cited by Weselake and Taylor (1999), and more recently, a direct GenBank submission by A.P. Brown, T.P. Schierer, and A.R. Slabas (GenBank accession no. AAF64065).
Here we report that the Arabidopsis DGAT cDNA can complement the AS11 mutant lipid phenotype, restoring the oil content and acyl composition to that of wild type (WT). Furthermore, we demonstrate that over-expression of the acyl-CoA-dependent DGAT in a seed-specific manner in wild-type plants results in augmentation of seed oil deposition and average seed weight.
RESULTS
Further Study of the AS11 Mutant Line and the Effects of the Mutation on DGAT Activity, Its Acyl-CoA Substrate Specificity, and Oil Accumulation
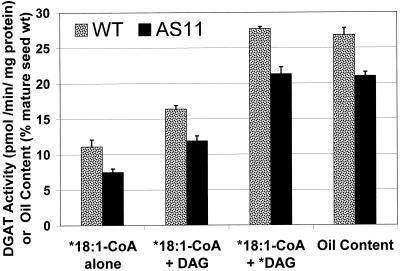
Using microsomal fractions prepared from WT and AS11 mid-green developing seed, we were able to compare the acyl-CoA-dependent DGAT (EC 2.3.1.20) activity to the resultant oil content in the mature seed of each line. As shown in Figure 1, there was a strong correlation between the reduced DGAT activity exhibited in developing seed microsomes of the AS11 mutant and the reduced oil content in the mature seed of this mutant line in comparison with WT. Furthermore, the AS11 DGAT activity relative to that of WT in developing seeds remained proportionally constant and directly correlated with mature seed oil content, regardless of whether the acyl-CoA-dependent microsomal DGAT activity was measured with only the acyl-CoA donor radiolabeled (14C 18:1-CoA; AS11 DGAT activity = 73% of WT) or if both the DAG acceptor (14C sn-1,2 diolein) and acyl-CoA donor (3H-18:1-CoA) were labeled (AS11 DGAT activity = 76% of WT).
Figure 1.
Comparisons of 18:1-CoA-dependent DGAT activity (values expressed as pmol min−1 mg−1 protein) in microsomes prepared from developing seeds at the mid-green stage of embryo development in wild-type and DGAT mutant AS11 lines of Arabidopsis with the seed oil content in WT and AS11 mature seed (values for oil content expressed as percentage of dry weight). In single label experiments, 18 μm 14C-oleoyl-CoA (10 nCi nmol −1) was the acyl donor in the absence of exogenous diolein (*18:1-CoA alone) or in the presence of exogenous 200 μm unlabeled sn-1,2 diolein (*18:1-CoA + DAG). In the double label experiment, 3H-labeled oleoyl-CoA was used at a radiospecific activity of 50 nCi nmol −1 and a final concentration of 40 μm with 14C-labeled sn-1,2 diolein provided at a specific activity of 2 nCi nmol−1 and a final concentration of 200 μm (*18:1-CoA + *DAG).
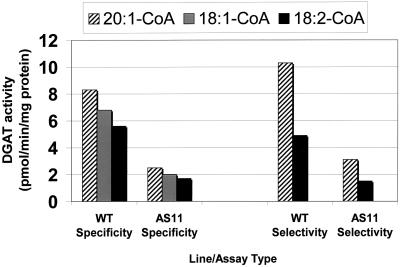
The altered fatty acid phenotype in seeds raised questions about whether the acyl-CoA specificity of the mutant enzyme is affected. A study of the DGAT acyl-CoA specificity and selectivity (as defined by Cao and Huang, 1987) in WT and AS11 developing seed is shown in Figure 2. It is clear that, although microsomal DGAT activity is reduced in AS11 compared with WT (compare to Fig. 1; for previous crude homogenate studies, see Katavic et al., 1995), the trend in relative acyl-CoA preference is identical in both WT and AS11 seed lines.
Figure 2.
Comparison of the acyl-CoA preference of the DGAT in WT and AS11 developing seed (mid-green stage). Seeds of WT and AS11 Arabidopsis were harvested, and homogenates were prepared and DGAT activity measured as described by Katavic et al. (1995). In the comparative specificity studies, the DGAT activity was assayed in the presence of 18 μm [1-14C]-labeled 18:1-CoA, 18:2-CoA, or 20:1-CoA, supplied individually, and 200 μm unlabeled sn-1,2-diolein. In the comparative selectivity study equal concentrations (18 μm) of [1-14C]-labeled 18:2-CoA and 20:1-CoA were supplied simultaneously to the seed homogenates in the presence of 200 μm unlabeled sn-1,2-diolein. Separation and measurement of the relative proportions of the radiolabeled TAG products (18:1/18:1/14C-18:1; 18:1/18:1/14C-18:2 or 18:1/18:1/14C-20:1) was conducted by reverse-phase radio-HPLC as described by Weselake et al. (1991).
Complementation of the Arabidopsis AS11 Mutant Line by Transformation with the DGAT cDNA
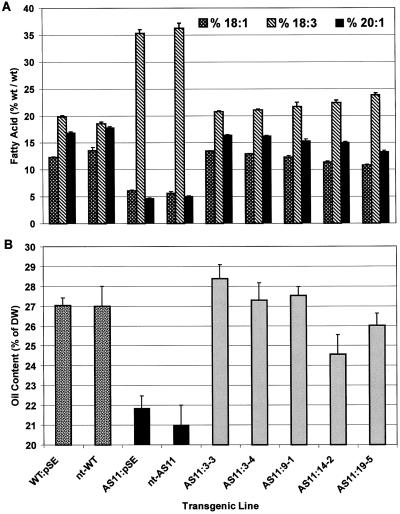
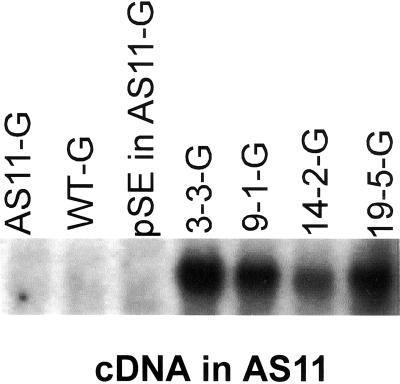
A napin:DGAT plasmid was introduced into Agrobacterium tumefaciens and used to transform Arabidopsis mutant AS11. A number of T3 single insert transgenic lines were isolated that complemented the fatty acid mutant phenotype found in AS11 (reduced 20:1 and elevated polyunsaturated fatty acids), restoring the wild-type seed fatty acid profile (Fig. 3A). In addition, many of these same AS11 transgenic lines exhibited oil contents restored to near WT values or beyond (Fig. 3B). The level of DGAT expression in the AS11 transgenic lines containing single copies of the napin-driven cDNA generally correlated well with the restoration of both the oil content and acyl composition, with line 3-3 being the best, followed by lines 9-1, 19-5, and 14-2, respectively (Fig. 4). There was no apparent advantage of multiple copy inserts in restoring AS11 seed oil phenotypes to those of WT (data not shown).
Figure 3.
Complementation of the AS11 DGAT mutation with the WT cDNA leads to a restoration of WT seed oil composition and content. A, Transformation of Arabidopsis mutant line AS11 with single copies of the DGAT cDNA under the control of a napin promoter, leads to a restoration of the WT fatty acid composition in the seed oil of the transformant lines 3-3, 3-4, 9-1, 14-2, and 19-5. Fatty acid composition (% [w/w]) was determined on the seed oil extracted from Arabidopsis WT pSE and AS11 pSE (empty plasmid) control transformants, non-transformed (nt) WT and n-t AS11 controls, and T3 seeds of napin:DGAT transgenic lines. B, Seed oil content of napin:DGAT T3 transgenic AS11 mutant seed lines containing a single insertion of the DGAT cDNA. Oil content is expressed as percentage of seed dry weight for pSE in WT (empty plasmid) and n-t WT controls (stippled bars), for pSE in AS11 (empty plasmid) and n-t AS11 controls (black bars), and napin:DGAT AS11 transgenic lines 3-3, 3-4, 9-1,14-2, and 19-5 containing a single copy of the DGAT cDNA (gray bars). se bars are indicated (n = 3–5 replicate analyses performed on seed lots from each line with 100–200 seeds analyzed/replicate).
Figure 4.
Northern analysis of TAG1 gene expression in non-transformed Arabidopsis WT and AS11 lines, a pSE (empty plasmid) AS11 control transformant, as well as napin:DGAT AS11 T3 transgenic lines 3-3, 9-1,14-2, and 19-5, each containing a single copy of the DGAT cDNA. Total RNA was extracted from siliques containing mid-green (G) developing seeds. The TAG1 DNA probe was 32P-labeled by random priming.
Over-Expression of the DGAT cDNA in Wild-Type Arabidopsis
The napin:DGAT plasmid was introduced into A. tumefaciens, used to transform wild-type Arabidopsis, and the progeny was analyzed as described in “Materials and Methods.” Based on kanamycin selection, 24 primary napin:DGAT transgenic lines were produced, the T1 plantlets grown to maturity, and T2 seeds harvested. At the same time 12 independent plasmid only control transgenic (pSE vector without DGAT insert) lines, as well as non-transformed (n-t) WT and AS11 lines were propagated and analyzed.
After analysis, seven independent T2 transgenic lines containing the napin:DGAT construct (lines 2, 9, 10, 16, 21, 23, and 24) were selected for detailed study based on increased oil deposition on a per seed basis, an increased average 1,000-seed weight, a strong linear correlation between the two traits (correlation coefficient r = 0.97), and enhanced expression of the DGAT transcript, compared with a set of pSE in WT and n-t WT control lines. On a mature seed dry weight basis, oil content increased by anywhere from 9% to 12% dry weight, representing net overall increases of 34% to 46%. Analyses of these T2 transgenic lines by 1H-NMR, a non-destructive method, confirmed a relative increase in oil on a per seed basis of 30% to 40% (data not shown).
From the T2 progeny, segregation analyses were performed on the T3 generation, and homozygous lines were identified and subjected to further analysis. The data for the napin:DGAT transgenic lines were compared with those acquired from 12 independent T3 pSE (empty plasmid) in WT control plants and an equal number of n-t WT and n-t AS11 control lines, all grown in the same growth chamber at the same time, in a random block design. As shown in Figure 5, on a mature seed weight basis, the homozygous napin: DGAT lines exhibited oil content increases ranging from 3 to 8 percentage points, representing net overall increases of 11% to 28%, compared with the range exhibited by pSE WT controls. When comparing homozygous lines containing a single insert (lines 10, 21, 23, and 24) versus multiple inserts (lines 2, 9, 16), with respect to oil content (compare to Fig. 5), it is noteworthy that the single insert lines performed quite well. For reasons that are not readily apparent, the pSE in WT transgenics displayed approximately 2% higher oil content compared with the n-t WT lines (Fig. 5). It could be that applying the kanamycin selection pressure to the pSE WT controls added some sort of additional stress that resulted in a slightly higher basal oil content. Nonetheless, we believe that for true comparison purposes with respect to oil content, the pSE in WT control lines are the better controls, because they reduce the risk of over estimating the oil content increase observed in the napin:DGAT transgenics.
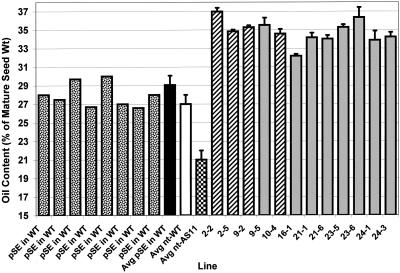
Figure 5.
Transformation of WT Arabidopsis with the DGAT cDNA under the control of a napin promoter leads to a higher seed oil content. Homozygous T3 napin:DGAT lines were sampled in triplicate, each sample consisting of 100 to 200 seeds/sample, accurately counted and weighed. For the plasmid only pSE in WT transgenic controls and nt-WT and nt-AS11controls, 10 individual transgenic plants were sampled and individual control seed lots similarly analyzed. Each error bar indicates se. Seed oil content (oil as a percentage of mature seed weight) is shown for Arabidopsis T3 seeds of pSE (empty plasmid) control WT transgenics (stippled bars show representative oil content ranges; solid black bar is the average of oil contents in 10 independent pSE in WT plasmid only controls), nt-WT controls (white bar), nt-AS11 controls (checkered bar), and homozygous napin:DGAT transgenic lines with multiple inserts 2-2, 2-5, 9-2, 9-5, and 16-1(hatched bars), and lines with single inserts 10-4, 21-1, 21-6, 23-5, 23-6, 24-1, and 24-3 (gray bars).
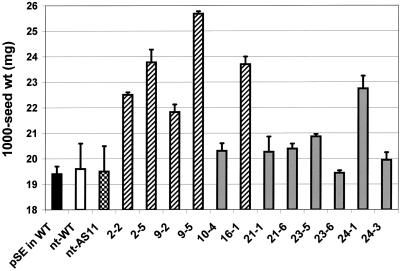
In general, the average 1,000-seed weight in the napin:DGAT homozygous transgenic lines was usually increased or, as in the case of line 23-6, not significantly affected (Fig. 6).
Figure 6.
Introduction of the napin:DGAT cDNA results in increases in the average 1,000-seed weight (expressed as milligrams of weight/1,000 seeds) of Arabidopsis T3 seeds. Homozygous T3 napin:DGAT lines were sampled in triplicate, each sample consisting of 100 to 200 seeds/sample accurately counted and weighed. For the plasmid only pSE controls, 10 individual transgenic plants were sampled and individual control seed lots similarly analyzed. Each error bar indicates se. Data are presented from pSE in WT (empty plasmid) control transgenics (solid black bar), nt-WT control lines (white bar), nt-AS11 control lines (checkered bar), homozygous napin:DGAT transgenic lines with multiple inserts 2-2, 2-5, 9-2, 9-5, and 16-1(hatched bars), and single insert lines 10-4, 21-1, 21-6, 23-5, 23-6, 24-1, and 24-3 (gray bars).
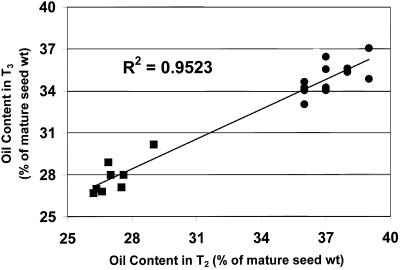
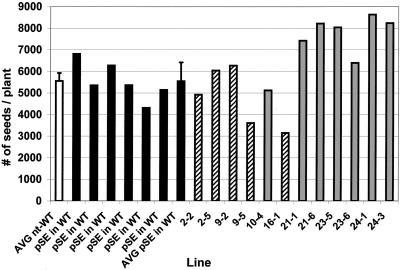
The heritability of the high oil trait from the pooled segregating T2 generation to the average of the corresponding homozygous T3 progeny is demonstrated in Figure 7 with a linear regression correlation coefficient of 0.94. In addition, the number of seeds per plant (yield) was not negatively affected in most of the homozygous T3 transgenic lines (Fig. 8). In fact, many of the lines showed significant increases in seed yield. The two exceptions to this trend were lines 9-5 and 16-1. However, when the increase in the average 1,000-seed weight for these lines was taken into account (compare with Fig. 6), the yield per plant compared with that of n-t WT plants as well as the plasmid only (pSE in WT) average was 90% and 72% for lines 9-5 and 16-1, respectively.
Figure 7.
Heritability of the high oil trait from the pooled segregating T2 generation to the average values for the corresponding selected homozygous T3 progeny, shows a strong linear correlation. ▪, Intercepts for the pSE in WT control generations; ●, intercepts for the napin:DGAT generations.
Figure 8.
Number of seeds per plant in Arabidopsis n-t WT controls (n = 6; white bar), six individual pSE in WT T3 transgenic controls and their average (black bars), versus homozygous napin:DGAT T3 transgenic lines with multiple inserts (2-2, 2-5, 9-2, 9-5, and 16-1, hatched bars), and single insert lines 10-4, 21-1, 21-6, 23-5, 23-6, 24-1, and 24-3 (gray bars).
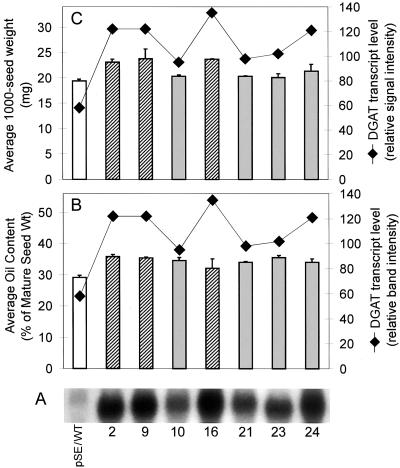
Figure 9A shows the relative DGAT expression levels in mid-developing T3 seeds (from pooled siliques following propagation of T2 lines). In general, in developing seeds of the napin:DGAT transgenics, the DGAT transcript was strongly over-expressed compared with the pSE in WT (plasmid only) control lines. The DGAT transcript intensity was generally stronger in the napin:DGAT transgenic lines containing multiple inserts (lines 2, 9, and 16) as compared with single inserts (lines 10, 21, 23, and 24) (Fig. 9A). The relative DGAT transcript level correlated quite well with the averaged values from the corresponding homozygous T3 mature seed lines with respect to the average seed weight (milligrams per 1,000 seeds, Fig. 9C), but the correlation was less distinct with oil content (Fig. 9B).
Figure 9.
A, Northern analysis of TAG1 gene expression in pooled developing T3 seed samples from Arabidopsis pSE (empty plasmid) WT control transformants, as well as developing progeny from parent transgenic lines 2, 9, 10, 16, 21, 23, and 24, transformed with the napin:DGAT construct. Total RNA was extracted from siliques containing mid-green developing seeds. The TAG1 DNA probe was 32P-labeled by random priming. B, Correlation of relative DGAT transcript level (♦) with oil content (percentage of mature seed weight); pSE control (black bar) or napin:DGAT multiple insert (hatched bars) and single insert (gray bars) transgenics. Values for transgenics represent the average from the corresponding homozygous T3 progeny as reported in Figure 5. C, Correlation of relative DGAT transcript level (♦) with average seed weight (expressed as milligram/1,000 seeds); pSE control (black bar) or napin:DGAT multiple insert (hatched bars) and single insert (gray bars) transgenics. Values for transgenics represent the average from the corresponding homozygous T3 progeny as reported in Figure 6.
DGAT activity was assessed in microsomal preparations from early-to-mid-green developing T3 seeds. In a previous study (Katavic et al., 1995) we had determined that the DGAT activity in WT seeds was highest at these developmental stages. Seed material was pooled from stage 3 to stage 6 siliques, as defined by Zou et al. (1996). Table I shows the DGAT activities measured in representative napin:DGAT homozygous multiple copy and single copy lines as well as the corresponding activity in pSE controls and compares these with the oil content measured in each line. The DGAT activity in the napin:DGAT transgenic lines was generally approximately 10% to 70% higher than that observed in the pSE controls.
Table I.
Acyl-CoA-dependent DGAT activity in microsomal fractions prepared from pooled mid-developing T3 seed produced from T2 transgenic lines of pSE control and napin:DGAT transformed Arabidopsis are compared to the seed oil content of mature T3 transgenic seed lines
| Transgenic Line | Seed Oil Content (±se) | DGAT Activitya (±se) | Relative DGAT Activity |
|---|---|---|---|
| % mature seed wt | pmol/min/mg protein | % pSE control set at 100% | |
| pSE controls | 29.1 (0.8) | 13.0 (0.6) | 100 |
| 2 (Multiple insert) | 35.9 (0.6) | 15.2 (0.6) | 117 |
| 9 (Multiple insert) | 35.4 (0.3) | 18.0 (0.5) | 138 |
| 21 (Single insert) | 34.1 (0.3) | 22.1 (3.1) | 170 |
| 23 (Single insert) | 35.5 (0.7) | 14.1 (0.6) | 109 |
| 24 (Single insert) | 34.1 (1.0) | 17.0 (1.7) | 131 |
Assays were conducted using sn-1,2 diolein and 1-14C oleoyl-CoA as the acyl acceptor and donor, respectively. Values represent the average of three independent determinations.
Total 14C-TAGs measured using TLC, scraping, and scintillation counting; in all cases, 14C-triolein was a major product.
We recognize both the advantages and limitations imposed by the use of the ARASYSTEM and concede that seed “yield” is probably more appropriately addressed in a field context in a crop plant. Nonetheless the current results with the model oilseed plant Arabidopsis suggest that in transgenic lines over-expressing the DGAT cDNA, both oil content and seed yield can be increased substantially.
Whereas wild-type lines containing over-expressed DGAT cDNA affected oil deposition, there were only small changes in the fatty acid composition, and the overall proportions of VLCFAs were not increased significantly (data not shown). Rather, there was a small but significant decrease in the proportion of total saturates and an increase in the mono-unsaturates and the 18:1/[18:2 + 18:3] index.
DISCUSSION
We recently identified a DGAT gene from Arabidopsis and cloned its corresponding cDNA. We were able to demonstrate that the lesion at a locus designated Tag1 in mutant line AS11 (Katavic et al., 1995) is an insertion mutation that results in an extra exon 2 sequence (81-bp) repeat in the AS11 DGAT transcript (Zou et al., 1999).
The fact that (a) Tag1 appears to be a single copy gene in Arabidopsis (Routaboul et al., 1999; Zou et al., 1999), (b) that there is no significant difference in DGAT transcript levels in mid- developing WT and AS11 seed (Zou et al., 1999; compare with Fig. 4), and (c) that the AS11 line exhibits reduced but significant acyl-CoA-dependent DGAT activity (compare with Fig. 1) collectively suggest that the repeat insertion in Tag1 does not totally abolish the activity of the DGAT gene product. Rather, the altered fatty acid phenotype of AS11 appears to be an indirect effect of lower DGAT activity rather than, for example, an increased preference of the DGAT for polyunsaturated C18 fatty acyl-CoAs or a reduced preference for very long chain acyl-CoAs.
As discussed previously (Zou et al., 1999), the DNA aberration observed in this mutant was unexpected since EMS generally causes point mutations, as in the case of the recently characterized mutant allele tag 1-2 from line ABX45 wherein one base (G) at position 180 in exon 1 is deleted (Routaboul et al., 1999). Although at that time we could not rule out the possibility that this AS11 mutant was the result of a spontaneous mutation event, the study published by Routaboul et al. (1999) concomittantly and independently confirmed our findings: AS11 does contain a 147-bp repeat insertion consisting of exon 2 flanked by parts of introns 1 and 2 (Zou et al., 1999). EMS-induced deletions and insertions had been reported in other systems (Mogami et al., 1986; Okagaki et al., 1991). However, Poirier et al. (1999) recently reported the characterization of two new independent EMS mutant isolates (SK353 in Columbia ecotype and SK54-3 in RLD ecotype) shown to have similar fatty acid and oil content phenotypes to that of AS11. Through complementation studies these two new isolates were shown to be new alleles of Tag1. This recent finding strongly supports the probability that the insertion event observed in mutant AS11 was EMS-induced.
It has been reported that, in addition to acyl-CoA-dependent DGAT as the terminal step in the Kennedy pathway (Kennedy, 1961; Barron and Stumpf, 1962), there are additional mechanisms for synthesizing TAGs in mammals, yeasts, and plants. A DAG transacylase reaction by which a fatty acyl group may be transferred directly from one diacylglycerol to a second diacylglycerol acceptor, has been characterized from rat intestinal microsomes (Lehner and Kuksis, 1993). In mammals, a mouse deletion mutant (Dgat−/−) devoid of DGAT still retained significant TAG synthesis activity, indicating that DGAT-independent TAG synthesis can occur (Smith et al., 2000). In the yeast Saccharomyces cerevisiae, a gene designated LRO1, a homolog of the mammalian lecithin:cholesterol acyltransferase (LCAT, EC 2.3.1.43), encodes an enzyme capable of esterifying diacylglycerol using phosphatidylcholine as the acyl donor. This novel enzymatic reaction is responsible for the majority of TAG synthesis in the yeast cell during exponential growth (Oelkers et al., 2000).
Pioneering plant research in developing oilseeds has revealed that, in addition to the traditional DGAT reaction, TAG synthesis can occur in the absence of acyl-CoA. An acyl-CoA-independent selective channeling or transfer of acyl moieties from one DAG to another to give TAG, was reported (Dahlqvist et al., 1997; Stobart et al., 1997). Dahlqvist et al. (2000) recently reported the presence of an enzyme in developing oilseeds of castor and Crepis involved in TAG synthesis via direct acyl transfer from the sn-2 position of PC to the sn-3 position of DAG yielding TAG. This enzyme has been named phospholipid:DGAT (PDAT) and was proposed to be involved in the accumulation of the high levels of ricinoleic acid and vernolic acid found in castor (Ricinus communis) and hawk's beard (Crepis palaestina) TAGs, respectively. It was demonstrated that the enzyme is present in yeast microsomes, and the PDAT-encoding gene (YNR008w) from yeast, an LCAT homolog, was identified. Whereas the authors also identified a sequence from Arabidopsis, which was more closely related to the yeast PDAT than to any of the cloned LCATs, there was no data reported on cloning or expression of the putative Arabidopsis PDAT.
In the current study, we performed an experiment wherein the microsomal fractions prepared from WT and AS11 developing seed were assayed for PDAT activity in reaction mixtures containing labeled sn-1 palmitoyl-, sn-2 [1-14C]oleoyl-PC, and sn-1,2 diolein. A small amount of radiolabeled TAG was produced. Our results with Arabidopsis are similar to those observed for sunflower microsomes by Dahlqvist et al. (2000); that is, when sn-2 14C-oleoyl-radiolabeled PC is supplied, the apparent PDAT activity is quite low. The low amount of radiolabeled TAG produced in our “PDAT” reactions precluded an accurate stereospecific analysis to assess whether there had been transfer of the sn-2 labeled oleoyl moiety from PC to the sn-3 position of DAG to give sn-3 oleoyl-labeled TAG. Furthermore, when we performed the experiment using sn-1 palmitoyl-, sn-2 [1-14C] linoleoyl-PC, we could not detect any significant PDAT activity in either WT or AS11 microsomes. Given our previous work, which showed that in AS11 TAGs, the proportion of 18:2 in the sn-3 position was 31% compared with only 8% in WT (Katavic et al., 1995), one would expect that if an alteration in a PDAT-type of reaction was responsible for the AS11 acyl composition phenotype, it would have been apparent with this experiment. Thus, whereas our results are inconclusive as to whether an acyl-CoA-independent PDAT activity exists in Arabidopsis based on our measurement of radiolabeled TAG, the putative acyl-CoA-independent PDAT activity in WT and AS11 microsomes was similar (approximately 0.7 pmol min−1 mg−1 protein) and significantly lower, by an order of magnitude, than the WT DGAT activity.
There have also been very different acyl-CoA-dependent DGATs identified from Mortierella and S. cereviseae (Lardizabal et al., 2000). These have no significant homology (6%–14% identity overall) to any of the plant DGATs isolated thus far and do not, for example, contain a signature DAG binding motif or significant stretches of conserved amino acid residues. However, an Arabidopsis homolog to the yeast gene cloned by Calgene (putative protein accession no. CAB 63016) recently has been reported, which has 21% overall identity and 29% identity over a stretch of 243 amino acids, to the yeast DGAT. To date, there have been no data reported on the relative level of expression of this “DGAT homolog” in Arabidopsis nor a confirmation of its gene product as a functional DGAT.
Nonetheless, ectopic expression of the fungal, yeast, or other plant DGATs, or manipulation of PDAT expression in certain oilseed plants may provide additional glimpses into the malleability of TAG bioassembly processes in oilseeds.
While we acknowledge that additional mechanisms for TAG synthesis (e.g. PDAT) may exist in Arabidopsis, the apparent WT and AS11 PDAT activities are essentially identical, but also very low, suggesting that there is no differential contribution of such alternate mechanisms to the phenotype observed in AS11. Rather, the results from our biochemical comparisons of the AS11 and WT microsomal acyl-CoA-dependent DGAT activity and specificity strongly suggest that the reduction in AS11 oil content is largely attributable to the reduced DGAT activity of the mutated AS11 TAG1 gene product. Previously (Zou et al., 1999), we suggested a mechanism whereby such a reduced DGAT activity in AS11 may result in the altered acyl composition observed in AS11 seed TAGs (i.e. a reduction in monounsaturated [e.g. 18:1 and 20:1] and an enrichment in polyunsaturated [e.g. 18:3] fatty acyl moieties).
The accumulated biochemical and molecular biological data known for the Arabidopsis DGAT and related acyl-CoA-dependent acyltransferases provide a glimpse of the structure-function relationships involved. In sequences of GPATs and LPATs there is a motif designated box I that contains the conserved sequence XHXXX(X)D (Frentzen and Wolter, 1998). Via site-directed mutagenesis studies in LPATs, the box I motif has been suggested to be an important part of the active site, in particular, the conserved His and Asp residues. It has been suggested that the His residue abstracts a proton from the hydroxy group of the acyl acceptor (LPA) and thereby facilitates its nucleophilic attack on the thioester bond of the acyl donor (acyl-CoA). The closely spaced Asp residue has been suggested to stabilize the positive charge on the His imidazole ring (Frentzen and Wolter, 1998). In the DGAT sequences examined, the consensus sequence N(S/A/G)R(L/V)(I/F/A)(I/L)EN(L/V) has been identified (Fig. 10). We propose that the invariant Arg (R) and Glu (E) residues in DGATs could perform similar functions to those of the basic His (H) and acidic Asp (D) residues, respectively, found in GPATs and LPATs. It is notable that the amino acid sequence predicted for human acyl-CoA:cholesterol acyltransferase genes (Yang et al., 1997; GenBank accession nos. L21934 and AF059203) also have RLXXXE motifs that align in the vicinity of the DGAT motif (data not shown).
Figure 10.
Alignment of DGAT sequences (and GenBank accession nos.) from Arabidopsis (AJ238008 and AJ131831), B. napus (AF155224), Nicotiana tabacum (AF129003), mouse (AF078752), human (AF059202), and Caenorhabditis elegans (Z75526) in the region covering the insertion repeat found in mutant AS11. The conserved R and E residues, indicated by an asterisk, are proposed to constitute key residues at the active site. A proposed “box I” type of motif, which is conserved in all DGAT sequences reported thus far (and analogous to the motif found in other acyl-CoA acyltransferases) is underlined.
It is significant that there is a concensus amino acid sequence for an acyl-CoA binding motif (116RTRESPLSSDAIFKQSHAG134) immediately upstream (and ending with the first four residues) of the AS11 exon 2 repeat insertion mutation site (Weselake et al., 2000). If our proposal is correct, the exon 2 repeat in mutant AS11 yields a protein altered with respect to its active site region. This could result in perturbed access of substrates to the active site, an altered interaction of the box I motif with the diacylglycerol binding site (Zou et al., 1999), an altered topology proximal to the acyl-oA binding site, or protein instability, and result in reduced DGAT activity. Site-directed mutagenesis of the invariant R and/or E in the DGAT sequence will help to establish if this hypothesis is correct.
To study the effect of DGAT expression in plants, we transformed wild-type and AS11 Arabidopsis plants with the DGAT cDNA. We have demonstrated that the insertion mutation in the mutant AS11 DGAT (resulting in an extra exon 2 sequence in its transcript) can be complemented by transgenically expressing a single copy of the WT cDNA in a seed-specific manner in the AS11 background. The fatty acid composition and oil contents in the AS11 DGAT transformants are restored essentially to wild-type levels. These experiments are essential to report because the findings confirm the nature of the lesion in AS11 and directly tie the AS11 lipid phenotype to this mutation.
It is interesting that even in the AS11 lines complemented with multiple copies of the napin-driven DGAT cDNA (average oil contents of 26.1% ± 0.3% dry weight; data not shown), the effect on oil content was not as strong as those observed in the WT single copy napin-driven DGAT cDNA over-expression transgenics. It may be that in the WT experiment the endogenous DGAT activity is augmented, whereas in AS11 there is significant competition for substrates between the introduced wild-type DGAT and the mutated DGAT of lower activity already resident in the AS11 mutant. This phenomenon clearly needs further investigation.
From a biotechnological viewpoint, it is important that we have demonstrated that the DGAT cDNA is useful in manipulating DGAT expression and TAG accumulation in plants. By transforming wild-type plants with a construct containing the DGAT gene in a sense orientation under the control of a seed-specific promoter (napin), DGAT activity, the accumulation of seed oil, and average seed weight were enhanced. In contrast, the use of a B. napus DGAT cDNA in an anti-sense orientation under the control of the cruciferin promoter recently has apparently resulted in a reduction in seed oil content (Shorrosh, 2000). Thus, there appears to be some correlation between DGAT expression levels and oil content. It is surprising that despite what we had predicted based on the fatty acid composition of mutant AS11 seed oil (Katavic et al., 1995), the over-expression of the DGAT and increased oil content and seed weight were not accompanied by an increase in proportions of the VLCFAs. The acyl composition of seed oil from the napin:DGAT T3 transgenics remained insignificantly different from that found in the pSE (plasmid only) controls.
To our knowledge, this is the first report of enhanced seed oil deposition and seed weight achieved by over-expression of a DGAT in plants. The degree to which oil content increases could account for the observed increases in average seed weight ranged from 40% to 100%. We previously demonstrated that transformation of Arabidopsis and rapeseed (B. napus) with a yeast sn-2 acyltransferase (LPAT) resulted in seed oils with increased proportions of 22:1 and other very long-chain fatty acids and significant increases in seed oil content and average seed weight (Zou et al., 1997; Marillia et al., 2000). These results have been confirmed in two transgenic field trials (Katavic et al., 2001). Taken together, both results suggest a certain plasticity in the developing seed with respect to increasing the quantity of TAG deposited and, in some way perhaps by signaling, the overall sink size and resultant seed weight. Clearly in many cases a proportion of the added seed weight must be due to increases in e.g. oleosin or seed storage protein biomass.
Not unexpectedly, there was not necessarily a direct linear correlation between the relative transcript level (compare with Fig. 9A), the degree of DGAT activity enhancement, and oil content (Fig. 9B; Table I). The correlation between the latter two parameters was not high (a scatter plot of the data from Table I, the r2 value = 0.11; data not shown). The relationship between these measures of altered metabolism at the transcript, enzyme activity, and oil content levels is clearly complex and will require more detailed study: There may be tight control of transcript stability that would not necessarily be evident in our sampling window. As well, it is difficult to directly determine the embryo stage for sampling. Pooling siliques 7 through 18 may dilute the peak value for DGAT activity (on a per milligram protein basis) and result in an apparently lower DGAT rate. The napin promoter may also contribute by broadening the period of DGAT expression and oil deposition. In addition, there is perhaps some degree of post-transcriptional regulation affecting DGAT activity. Nonetheless, it can be said that all lines with increased DGAT transcript did exhibit increased DGAT activity and an increase in oil content.
It is tempting, metabolically, to speculate that increases in DGAT activity may lower the size of the acyl-CoA pools, thereby signaling a need for enhanced fatty acid synthesis (e.g. enhancement of ACCase activity).
In summary, the current results in Arabidopsis and recent studies of DGAT expression in tobacco (Bouvier-Navé et al., 2000) have important implications for the modification of other crops through biotechnology by altering DGAT expression. Some of the manipulations that may be possible using DGAT genes or parts thereof include seeds with increased or decreased oil content, seeds containing oils with an enhanced diacylglycerol content, seed oils with an altered acyl composition, plants producing larger or heavier seeds, plants exhibiting an enhanced or altered capacity to accumulate TAGs, or other storage compounds in other organs (e.g. tubers, roots, leaves). Clearly the next wave of research will entail altering the expression of DGAT in commercial oilseed crops. Such studies will allow one to determine the critical role of DGAT in regulating carbon flux into TAGs and the effect of such manipulations on overall oil deposition, average seed weight, and seed yield in a field context.
MATERIALS AND METHODS
Substrates and Reagents
[1-14C]Oleic acid (56 mCi mmol−1) and [1-14C]linoleic acid (55.6 mCi mmol−1) were purchased from NEN ResearchProducts(Mississauga,Ontario,Canada). [1-14C]Eicosenoic acid (55 mCi mmol−1), 1-palmitoyl-2-oleoyl phosphatidylcholine (58 mCi mmol−1), 1-palmitoyl-2-linoleoyl [1-14C-linoleoyl] phosphatidylcholine (58 mCi mmol−1), and diolein [1-14C-oleoyl] (55 mCi mmol−1) were purchased from American Radiolabeled Chemicals (St. Louis). [9,10-3H]Oleic acid (8 Ci mmol−1) was purchased from Amersham Pharmacia Biotech (Quebec). Labeled fatty acids were converted to the corresponding acyl-CoA thioesters using the method described by Taylor et al. (1990). Specific activities were adjusted as required by diluting with authentic unlabeled standards. Unlabeled acyl-CoAs, ATP, CoASH, polar lipid standards, and most other biochemicals were purchased from Sigma (St. Louis). Neutral lipid standards were obtained from NuChek Prep (Elysian, MN), whereas FAME standards were supplied by Supelco Canada (Oakville, Ontario, Canada). The sn-1,2-diolein from 14C-labeled diolein [1-14C oleic] (55 mCi mmol−1) and unlabeled diolein (already enriched in the sn-1,2 isomer) were both further purified by thin layer chromatography (TLC) on 10% (v/v) borate-impregnated TLC plates and the isolated sn-1,2 isomer emulsified in HEPES buffer the presence of 0.2% (v/v) Tween 20 as described by Taylor et al. (1991). Mixed TAG and diacylglycerol standards for gas chromatography, which were not commercially available, were synthesized from the corresponding di- or mono-acylglycerols by condensation with the appropriate acyl chloride and purified as described by Taylor et al. (1991). HPLC-grade solvents (Omni-Solv, BDH Chemicals, Toronto) were used throughout these studies.
Plant Material
The Arabidopsis mutant line AS11 was generated and characterized relative to WT Arabidopsis ecotype Columbia as described by Katavic et al. (1995). Arabidopsis ecotype Columbia WT, mutant AS11 (AS11 seeds were deposited at the American Type Culture Collection, ATCC Deposit No. PTA 1013) and all napin:DGAT transgenic lines were grown in the same growth chamber, at the same time, at 22°C with a diurnal photoperiod of 16-h light (120 μE m−2 s−1) and 8-h dark. For consistency in comparison, especially with respect to the reproducibility of oil content and seed weight measurements, transgenic lines were always grown along with n-t WT and AS11, and plasmid only WT (pSE WT) and AS11 (pSE AS11) controls, in the same chamber and at the same time. The Arasystem (Lehle Seeds, Tuscon, AZ), a plastic cone, and cylinder support system was used throughout these studies to isolate individual transgenic lines and facilitate seed harvesting from all lines.
Lipid Analyses in AS11, WT, and DGAT Transgenic Lines
Total lipid extracts (TLEs) and lipid class analyses in WT and the AS11 mutant and determination of oil content and composition in all seed lines were performed as described previously (Taylor et al., 1991, 1992; Katavic et al., 1995; Zou et al., 1997). In all cases, the data represent the averages of four to eight determinations.
In some cases, relative seed oil content was also measured by magic angle sample spinning 1H-NMR, according to the method of Rutar (1989). Analyses were conducted with 50-seed samples of intact wild type and AS11 seeds using a Bruker AM wide-bore spectrometer (Bruker Analytische Masstechnik, Silberstreifen, Germany) operating at 360 MHz. To reduce anisotropic line broadening, the seed sample was rotated at 1 kHz in a zirconium rotor oriented 54.7° to the magnetic field. The integration response for resonances attributable to liquid-like oil were summed, and the value for transgenic seed was recorded relative to the response for the WT control seed sample, the latter set at a value of 1.00.
Preparation of Microsomal Fractions and DGAT and PDAT Assays
Two-hundred siliques (pooled silique stages 3–6 inclusive, as described by Zou et al., 1996) were harvested from n-t WT, n-t AS11, pSE in WT control, and napin:DGAT transgenic lines. Developing seeds (early-to-mid-green stage) were harvested and immediately powdered with liquid nitrogen in a mortar and pestle. Grinding medium (100 mm HEPES-KOH, pH 7.4 containing 0.32 m Suc, 1 mm EDTA, and 1 mm dithiothreitol) was immediately added, and grinding continued on ice for 3 min. The slurried cell free homogenate was filtered through two layers of Miracloth (Calbiochem, La Jolla, CA), and centrifuged at 10,000g for 20 min using a centrifuge (model RC5C, Sorvall/Mandel Scientific Co., Guelph, ON, Canada) equipped with an SS-34 rotor. The supernatant was then recentrifuged at 105,000g for 1 h on a Sorvall Ultra Pro 80 ultracentrifuge using an SW 40Ti rotor. The supernatant was discarded, the 105,000g microsomal pellet fraction was resuspended in a minimum volume of grinding medium, and the preparation was disbursed by probe sonication on ice for 2 min in 30-s cycles using a Labsonic 2,000-U probe sonicator (B. Braun Biotech, Allentown, PA) on the low setting. The concentration of protein was then determined by the method of Bradford (1976), and relative microsomal protein concentrations were normalized by adjusting the volume of each preparation with grinding medium.
Acyl-CoA-dependent DGAT activity (EC 2.3.1.20) assays were conducted at pH 7.4 with shaking at 100 revolutions min−1 in a water bath at 30°C for 30 to 60 min. Assay mixtures (500 μL final volume) contained microsomal protein (100–200 μg), 90 mm HEPES-NaOH, 0.5 mm ATP, 0.5 mm CoASH, 1 mm MgCl2, 200 μm sn-1,2 diolein in 0.02% (v/v) Tween 20, and 18 μm or 40 μm acyl-CoA. In most cases, the acyl-CoA donor was 14C-labeled and supplied at a radiospecific activity of 10 nCi nmol −1 and a final concentration of 18 μm. In double label DGAT assays, 3H-labeled oleoyl-CoA was used at a radiospecific activity of 50 nCi nmol −1 and a final concentration of 40 μm with 14C-labeled sn-1,2 diolein provided at a specific activity of 2 nCi nmol −1 and a final concentration of 200 μm.
Acyl-CoA-independent phospholipid:DGAT (PDAT) activity was tested essentially as described by Dahlqvist et al. (2000) at 30°C for 60 min in the presence of 18 μm 1-palmitoyl-2-[1-14C-oleoyl] phosphatidylcholine (58 mCi mmol−1) or 1-palmitoyl-2-[1-14C-linoleoyl] phosphatidylcholine (58 mCi mmol−1) as the acyl donor, 200 μm sn-1,2 diolein in 0.02% (v/v) Tween 20 as the acyl acceptor, and 200 μg of microsomal protein in 100 mm HEPES-NaOH, pH 7.4, containing 0.5 mm ATP, 0.5 mm CoASH, 1 mm MgCl2 in a final reaction volume of 500 μL. All assays were conducted in triplicate, and each experiment replicated twice.
All acyltransferase reactions were terminated by the addition of CH2Cl2:iso-propanol (1:2), phases were separated, and labeled TAGs were isolated and purified by TLC on siliga gel G plates developed in hexane:diethyl ether:acetic acid (70:30:1 v/v/v), as described by Taylor et al. (1991). The radiolabeled TAG spots were visualized on a Bioscan AR-2,000 radio-TLC scanner using Win-Scan 2D software (Bioscan, Washington, D.C.) and the bands scraped and quantified on a liquid scintillation counter.
General Molecular Techniques, DNA and RNA Manipulation, and Analyses
Isolation of plasmid DNA, restriction digestions, modification and ligation of DNA, PCR, agarose, PAGE, transformation and culture of Escherichia coli strains, DNA gel-blot analyses (Southern, 1975), and RNA gel-blot analyses were carried out according to standard protocols (Sambrook et al., 1989).
The Arabidopsis TAG1 DGAT amino acid sequence was compared with sequences available in databanks using the BLAST program (Altschul et al., 1990).
Construction of DGAT cDNA Transformation Vector for Seed-Specific Expression
The cloned full-length DGAT cDNA was used as a template for PCR amplification with the primers DGATXbaI (CTAGTCTAGAATGGCGATTTTGGA) and DGATXhoI (GCGCTCGAGTTTCATGACATCGA) to provide new restriction sites on each end of the sequence. The PCR profile was as follows: 94°C for 1 min; 30 cycles of 94°C for 30 s, 55°C for 30 s, 72°C for 1 min; and 72°C for 5 min. The PCR product was then ligated into the PCR-2.1 vector (Invitrogen, Carlsbad, CA). A 1.6-kb fragment was excised by a XbaI/KpnI digestion and ligated into the corresponding sites of the pSE. The plant transformation vector pSE was prepared from pRD400 (Datla et al., 1992) by introducing a HindIII/XbaI fragment containing the B. napus napin promoter (Josefsson et al., 1987) and a KpnI/EcoRI fragment containing the Agrobacterium nos terminator (Bevan, 1983). The 1.6-kb DGAT cDNA fragment was ligated into XbaI/KpnI-digested pSE in the sense orientation. The resulting plasmid was designated napin:DGAT. Hence in the napin:DGAT construct, the Arabidopsis DGAT cDNA is under the control of the napin promoter. The construct integrity was confirmed by sequencing.
Transformation of Agrobacterium with Plant DGAT Vector Constructs
Electrocompetent Agrobacterium cells, GV3101 (pMP90) strain, were prepared as follows. An Agrobacterium culture was grown 24 to 48 h in 2× YT Medium (double-strength yeast extract + tryptone; Becton Dickinson, Sparks, MD), and when the A600 reached 0.5 to 0.7, the cells were chilled on ice and pelleted by centrifugation (5,000g, 10 min in a glutamate semialdehyde rotor at 4°C). The pellet was washed in 1, 0.5, and 0.02 volumes of cold 10% (v/v) sterile glycerol and resuspended in 0.01 volume of cold 10% (v/v) glycerol. The electrocompetent cells were then frozen in liquid N2 and stored at −70°C. The Agrobacterium cells were transformed by electroporation with 20 to 50 ng of transforming DNA (napin:DGAT) according to the manufacturer's instructions, plated on a selective medium (Luria-Bertani broth with 50 μg mL−1 kanamycin), and incubated for 48 h at 28°C. Single transformed cells were grown for 16 h (28°C, 225 rpm) in 5 mL Luria-Bertani broth with 50 μg mL−1 kanamycin and 25 μg mL−1 gentamycin. DNA extraction and purification were performed with a Qiaprep Spin Miniprep kit (Qiagen, Valencia, CA). The fidelity of the construct was rechecked by DNA sequencing before plant transformation.
Transformation of Arabidopsis
Seeds of Arabidopsis ecotype Columbia WT and mutant AS11 (Katavic et al., 1995) were grown at 22°C under fluorescent illumination (120 μE m−2 s−1) in a 16-h-light/8-h-dark regime. Four to six plants typically were raised in a 10 cm2 pot in moistened Terra-lite Redi-earth (W.R. Grace and Company, Ajax, Ontario, Canada). To prevent the soil mix in the pot from falling into the inoculation media, soil was mounded as a platform with seeds sown on top, and the whole pot covered by a nylon window screen and secured by a rubber band. To grow Agrobacterium, a 5-mL suspension in Luria-Bertani medium containing 50 μg mL−1 kanamycin and 25 μg mL−1 gentamycin was cultured overnight at 28°C. The day before infiltration, this “seed culture” was divided into four flasks containing 250 mL of Luria-Bertani medium supplemented with 50 μg mL−1 kanamycin and 25 μg mL−1 gentamycin. These culture were grown overnight at 28°C. Plants were vacuum infiltrated in an Agrobacterium suspension when the first flowers started opening.
The transformation was performed as described by Clough and Bent (1998), using Silwet L-77 at a concentration of 0.005% in the dipping solution. The next day, the plants were uncovered, set upright, and allowed to grow for approximately 4 weeks in a growth chamber under continuous light conditions as described by Katavic et al. (1995). When the siliques were mature and dry, seeds were harvested and selected for positive transformants.
Selection of Putative Transformants (Transgenic Plants) and Analysis of Transgenic Plants and Seed Weights
For each construct, seeds were harvested in bulk. Seeds were surface-sterilized by submerging them in a solution containing 20% (v/v) bleach and 0.01% (v/v) Triton X-100 for 20 min, followed by three rinses with sterile water. Sterilized seeds were then plated by resuspending them in sterile 0.1% (w/v) phytagar at room temperature (approximately 1 mL phytagar for every 500–1,000 seeds), and then applying a volume containing 2,000 to 4,000 seeds onto 150 × 15-mm kanamycin selection plate. Plates were incubated for 2 d in the cold without light and then grown for 7 to 10 d in a controlled environment (22°C under fluorescent illumination [120 μE m−2 s−1] in a 16-h-light/8-h-dark regime). The selection media contained one-half Murashige Skoog Gamborg medium, 0.8% (w/v) phytagar, 3% (w/v) Suc, 50 μg mL−1 kanamycin, and 50 μg mL−1 timentin. Petri dishes and lids were sealed with a Micropore surgical tape (3M Canada, Inc., London, ON, Canada). After 7 to 10 d, drug-resistant plants that had green leaves and well established roots within the medium were identified as T1 transformants, and at the 3- to 5-leaf stage selected transformants were transplanted into flats filled with heavily moistened soil mix. Transformants were grown to maturity, and mature seeds (T2 generation as defined in Katavic et al., 1994) were harvested from individual plants and further analyzed or propagated. Segregation analyses were performed on T2 plantlets screened on kanamycin to determine whether there were single (expected ratio of resistant:susceptible plantlets = 3:1) or multiple copies of the napin:DGAT transgene. Homozygous single and multiple insert T2 lines exhibiting enhanced oil deposition and DGAT expression compared with one-dozen plasmid-only control transgenics were propagated to give T3 seed lines, for which further data on oil content, average seed weight, and yield per plant were collected. Average seed weights were determined from pooled T2 or individual T3 segregant seed lots and based upon six to eight individual samplings of 150 to 250 seeds/sample, with the seeds of each replicate being accurately counted on an Electronic Dual Light Transilluminator (Ultra Lum, Paramount, CA), using Scion Image software (Scion Corporation, Frederick, MD). Weights of these samples were then individually recorded.
DNA and RNA Isolation from Transformants: Southern and Northern Analyses
Genomic DNA was isolated from individual T1 plants following the protocol of Dellaporta et al. (1983). A PCR amplification using the paired primers described previously for the DGAT cDNA or for the DGAT gene, was performed to confirm the presence of the cDNA in the T1 transformants. Southern analyses (Southern, 1975) were performed to further confirm and select those transformants containing single or multiple copies of the inserted fragment. DNA samples were digested with restriction enzymes BglII, resolved by electrophoresis on a 1% (w/v) agarose gel, and Southern blotting performed using a nylon filter (Hybond N+, Amersham) according to Sambrook et al. (1989). The DGAT cDNA fragment, labeled with α-[32P]dCTP (NEN/Mandel Scientific Co., Guelph, ON, Canada) using the Random Primer DNA labeling kit (Life Technologies/Gibco-BRL, Cleveland) was used as a probe. Hybridization was performed at 60°C according to Church and Gilbert (1984). The filter was then exposed to X-OMAT-AR film (Kodak, Rochester, NY).
Northern analyses of DGAT transcripts in both control and transgenic lines were performed as follows: Using the method of Lindstrom and Vodkin (1991), total RNA was extracted from developing T3 seeds at the mid-green stage of development (as defined previously by Katavic et al., 1995; Zou et al., 1996). RNA samples were denatured with formaldehyde and separated on 1.2% (v/v) formaldehyde-agarose gels. Ten to 40 μg of total RNA was loaded, and the amount of RNA per lane was calibrated by the ethidium bromide-staining intensity of the rRNA bands. The DGAT DNA probe was 32P-labeled by random-priming according to protocols of the manufacturer (Life Technologies/Gibco-BRL), and the blots were hybridized with 32P-labeled Arabidopsis DGAT cDNA under stringent conditions (60°C). Relative intensities of hybridization signals in autoradiograms from northern analyses were measured with an Ultra Lum Electronic Dual Light Transilluminator using Scion Image software.
ACKNOWLEDGMENTS
The authors thank B. Panchuk, D. Schwab and Dr. L. Pelcher of the PBI DNA Technologies Unit for sequencing and primer synthesis, Dr. J. Giraudat for the gift of the Arabidopsis silique-specific cDNA library, B. Chatson for the 1H-NMR analyses, L. Steinhauer for additional technical assistance, and K. Yao and D. Hunter for constructing the pSE vector. Critical reviews of this manuscript were kindly provided by Drs. A.J. Cutler and F. Georges.
Footnotes
This work was supported by the Saskatchewan Department of Agriculture and Food (grant nos. 96000046 and 19990362). This is National Research Council of Canada publication no. 43794.
LITERATURE CITED
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barron EJ, Stumpf PK. The biosynthesis of triglycerides by avocado mesocarp enzymes. Biochem Biophys Acta. 1962;60:329–337. doi: 10.1016/0006-3002(62)90408-0. [DOI] [PubMed] [Google Scholar]
- Bernerth R, Frentzen M. Utilization of erucoyl-CoA by acyltransferases from developing seeds of Brassica napus(L.) involved in triacylglycerol biosynthesis. Plant Sci. 1990;67:21–28. [Google Scholar]
- Bevan M. Binary Agrobacterium vectors for plant transformation. Nucleic Acid Res. 1983;12:8711–8721. doi: 10.1093/nar/12.22.8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier-Navé P, Benveniste P, Oelkers P, Sturley SL, Schaller H. Expression in yeast and tobacco of plant cDNAs encoding acyl CoA:diacylglycerol acyltransferase. Eur J Biochem. 2000;267:85–96. doi: 10.1046/j.1432-1327.2000.00961.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cao Y-Z, Huang AHC. Diacylglycerol acyltransferase in maturing oil seeds of maize and other species. Plant Physiol. 1986;82:813–820. doi: 10.1104/pp.82.3.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y-Z, Huang AHC. Acyl coenzyme A preference of diacylglycerol acyltransferase from maturing seeds of Cuphea, maize, rapeseed and canola. Plant Physiol. 1987;84:762–765. doi: 10.1104/pp.84.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Smith JS, Zheng Y-W, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ. Identification of a gene encoding an acyl CoA: diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc Natl Acad Sci USA. 1998;95:13018–13023. doi: 10.1073/pnas.95.22.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Dahlqvist A, Banas A, Stymne S. Selective channelling of unusual fatty acids into triacylglycerols. In: Sánchez J, Cerdá-Olmedo E, Martinez-Force E, editors. Advances in Plant Lipid Research. Seville, Spain: Universidad de Sevilla; 1997. pp. 211–214. [Google Scholar]
- Dahlqvist A, Ståhl U, Lenman M, Banas A, Lee M, Sandager L, Ronne H, Stymne S. Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc Natl Acad Sci USA. 2000;97:6487–6492. doi: 10.1073/pnas.120067297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datla RSS, Hammerlindl JK, Panchuk B, Pelcher LE, Keller WA. Modified binary plant transformation vectors with the wild-type gene encoding NPTII. Gene. 1992;211:383–384. doi: 10.1016/0378-1119(92)90232-e. [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Frentzen M. Acyltransferases and triacylglycerols. In: Moore Jr TS, editor. Lipid Metabolism in Plants. Boca Raton, FL: CRC Press; 1993. pp. 195–230. [Google Scholar]
- Frentzen M, Wolter FP. Plant lipid biosynthesis: fundamentals and agricultural applications. In: Hardwood JL, editor. Society of Experimental Biology Seminar Series. Vol. 67. New York: Cambridge University Press; 1998. pp. 247–272. [Google Scholar]
- Hobbs HD, Chaofu L, Hills M. Cloning of a cDNA encoding acyltransferase from Arabidopsis thalianaand its functional expression. FEBS Lett. 1999;452:145–149. doi: 10.1016/s0014-5793(99)00646-8. [DOI] [PubMed] [Google Scholar]
- Ichihara K, Takahashi T, Fujii S. Diacylglycerol acyltransferase in maturing safflower seeds: its influences on the fatty acid composition of the triacylglycerol and on the rate of triacylglycerol synthesis. Biochim Biophys Acta. 1988;958:125–129. doi: 10.1016/0005-2760(88)90253-6. [DOI] [PubMed] [Google Scholar]
- Josefsson L-G, Lenman M, Ericson ML, Rask L. Structure of a gene encoding the 1.7S storage protein, napin, from Brassica napus. J Biol Chem. 1987;262:12196–12201. [PubMed] [Google Scholar]
- Katavic V, Friesen W, Barton DL, Gossen KK, Giblin EM, Luciw T, An J, Zou J-T, MacKenzie SL, Keller WA et al. (2001) Improving erucic acid content in rapeseed through biotechnology: what can the Arabidopsis FAE1 and the yeast SLC1-1 genes contribute? Crop Sci 41: (in press)
- Katavic V, Haughn GW, Reed D, Martin M, Kunst L. In planta transformation of Arabidopsis thaliana. Mol Gen Genet. 1994;245:363–370. doi: 10.1007/BF00290117. [DOI] [PubMed] [Google Scholar]
- Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J-T, MacKenzie SL, Covello PS, Kunst L. Alteration of fatty acid composition by an EMS-induced mutation in Arabidopsis thalianaaffecting diacylglycerol acyltransferase activity. Plant Physiol. 1995;108:399–409. doi: 10.1104/pp.108.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EP. Biosynthesis of complex lipids. Fed Pro Fed Am Soc Exp Biol. 1961;20:934–940. [PubMed] [Google Scholar]
- Lacey DJ, Hills MJ. Heterogeneity of the endoplasmic reticulum with respect to lipid synthesis in developing seeds of Brassica napusL. Planta. 1996;199:545–551. [Google Scholar]
- Lardizabal KD, Hawkins D, Thompson GA, inventors. January 13, 2000. Diacylglycerol acyltransferase proteins. Patent Application Nos. PCT/US99/15243 and WO 00/01713
- Lehner R, Kuksis A. Triacylglycerol synthesis by an sn-1,2 (2, 3)-diacylglycerol transacylase from rat intestinal microsomes. J Biol Chem. 1993;268:8781–8786. [PubMed] [Google Scholar]
- Lindstrom JT, Vodkin LO. A soybean cell wall protein is affected by seed color genotype. Plant Cell. 1991;3:561–571. doi: 10.1105/tpc.3.6.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little D, Weselake RJ, Pomeroy MK, Furukawa-Stoffer T, Bagu J. Solubilization and characterization of diacylglycerol acyltransferase from microspore-derived cultures of oilseed rape. Biochem J. 1994;304:951–958. doi: 10.1042/bj3040951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marillia E-F, Zou JT, Katavic V, Qi Q, Jako C, Barton DL, Friesen W, Giblin EM, Gossen KK, Kumar A. Metabolic engineering of Brassicaseeds oils: improvement of oil quality and quantity and alteration of carbon flux. In: Arencibia AD, editor. Plant Genetic Engineering: Towards the Third Millenium. New York: Elsevier Science Publishing; 2000. pp. 182–188. [Google Scholar]
- Mayorek N, Grinstein I, Bar-Tana J. Triacylglycerol synthesis in cultured rat hepatocytes: the rate-limiting role of diacylglycerol acyltransferase. Eur J Biochem. 1989;182:395–400. doi: 10.1111/j.1432-1033.1989.tb14844.x. [DOI] [PubMed] [Google Scholar]
- Mogami K, O'Donnell PT, Bernstein SI, Wright TRF, Emerson CP., Jr Mutations of the Drosophilamyosin heavy-chain gene: effects on transcription, myosin accumulation, and muscle function. Proc Natl Acad Sci USA. 1986;83:1393–1397. doi: 10.1073/pnas.83.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykiforuk C, Laroche A, Weselake RJ. Isolation and sequence analysis of a novel cDNA encoding a putative diacylglycerol acyltransferase from a microspore-derived cell suspension culture of Brassica napusL. cv Jet Neuf. Plant Physiol. 1999;120:99–123. [Google Scholar]
- Oelkers P, Behar A, Cromley D, Billheimer JT, Sturley ST. Characterization of two human genes encoding acyl coenzyme A: cholesterol acyltransferase-related enzymes. J Biol Chem. 1998;273:26765–26771. doi: 10.1074/jbc.273.41.26765. [DOI] [PubMed] [Google Scholar]
- Oelkers P, Tinkelenberg A, Erdeniz N, Cromley D, Billheimer JT, Sturley SL. A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J Biol Chem. 2000;275:15609–15612. doi: 10.1074/jbc.C000144200. [DOI] [PubMed] [Google Scholar]
- Okagaki RJ, Neuffer MG, Wessler SR. A deletion common to two independently derived Waxy mutations in maize. Genetics. 1991;128:425–431. doi: 10.1093/genetics/128.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry HY, Bligny R, Gout E, Harwood JL. Changes in Kennedy pathway intermediates associated with increased triacylglycerol synthesis in oil-seeds rape. Phytochemistry. 1999;52:799–804. [Google Scholar]
- Perry HY, Harwood JL. Changes in the lipid content of developing seeds of Brassica napus. Phytochemistry. 1993a;32:1411–1415. [Google Scholar]
- Perry HY, Harwood JL. Use of [2–3H] glycerol precursor in radiolabelling studies of acyl lipids in developing seeds of Brassica napus. Phytochemistry. 1993b;34:69–73. [Google Scholar]
- Poirier Y, Ventre G, Caldelari D. Increased flow of fatty acids toward β oxidation in developing seeds of Arabidopsisdeficient in diacylglycerol acyltransferase activity or synthesizing medium-chain-length fatty acids. Plant Physiol. 1999;121:1359–1366. doi: 10.1104/pp.121.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routaboul J-M, Benning C, Bechtold N, Caboche M, Lepiniec L. The TAG1 locus of Arabidopsisencodes for a diacylglycerol acyltransferase. Plant Physiol Biochem. 1999;37:831–840. doi: 10.1016/s0981-9428(99)00115-1. [DOI] [PubMed] [Google Scholar]
- Rutar V. Magic angle sample spinning NMR spectroscopy of liquids as a non-destructive method for studies of plant seeds. J Agric Food Chem. 1989;37:67–70. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. Ed 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Settlage SH, Wilson RF, Kwanyuen P. Localization of diacylglycerol acyltransferase to oil body associated endoplasmic reticulum. Plant Physiol Biochem. 1995;33:399–407. [Google Scholar]
- Shorrosh BS, inventor. November 9, 2000. Plant acyltransferases. International Patent Application No. WO 00/66749
- Smith SJ, Cases S, Jensen DR, Chen HC, Sande E, Tow B, Sanan DA, Raber J, Eckel RH, Farese RV., Jr Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking DGAT. Nat Genet. 2000;25:87–90. doi: 10.1038/75651. [DOI] [PubMed] [Google Scholar]
- Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stobart AK, Stymne S, Höglund S. Safflower microsomes catalyze oil accumulation in vitro: a model system. Planta. 1986;169:33–37. doi: 10.1007/BF01369772. [DOI] [PubMed] [Google Scholar]
- Stobart K, Mancha M, Lenman M, Dahlqvist A, Stymne S. Triacylglycerols are synthesized and utilized by transacylation reactions in microsomal preparations of developing safflower (Cartharmus tinctoriusL.) seeds. Planta. 1997;203:58–66. [Google Scholar]
- Stymne S, Stobart AK. Triacylglycerol biosynthesis. In: Stumpf PK, editor. The Biochemistry of Plants. Vol. 9. New York: Academic Press; 1987. pp. 175–214. [Google Scholar]
- Taylor DC, Barton DL, Rioux KP, MacKenzie SL, Reed DW, Underhill EW, Pomeroy MK, Weber N. Biosynthesis of acyl lipids containing very-long chain fatty acids in microspore-derived embryos of Brassica napusL. cv. Reston. Plant Physiol. 1992;99:1609–1618. doi: 10.1104/pp.99.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DC, Weber N, Barton DL, Underhill EW, Hogge LR, Weselake RJ, Pomeroy MK. Triacylglycerol bioassembly in microspore-derived embryos of Brassica napusL. cv Reston. Plant Physiol. 1991;97:65–79. doi: 10.1104/pp.97.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DC, Weber N, Hogge LR, Underhill EW. A simple enzymatic method for the preparation of radiolabeled erucoyl-CoA and other long-chain fatty acyl-CoAs and their characterization by mass spectrometry. Anal Biochem. 1990;184:311–316. doi: 10.1016/0003-2697(90)90686-4. [DOI] [PubMed] [Google Scholar]
- Tijburg LB, Geelen MJ, van Golde LM. Regulation of the biosynthesis of triacylglycerol, phosphatidylcholine and phosphatidylethanloamine in the liver. Biochim Biophy Acta. 1989;1004:1–19. doi: 10.1016/0005-2760(89)90206-3. [DOI] [PubMed] [Google Scholar]
- Tzen TC, Cao Y, Laurent P, Ratnayake C, Huang HC. Lipids, proteins and structures of seed oil bodies from diverse species. Plant Physiol. 1993;101:267–276. doi: 10.1104/pp.101.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel G, Browse J. Cholinephosphotransferase and diacylglycerol acyltransferase: substrate specificities at a key branch point in seed lipid metabolism. Plant Physiol. 1996;110:923–931. doi: 10.1104/pp.110.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ, Laroche A, Taylor DC, Zou J-T. Development of Canola Germplasm with Increased Oil Formation Capacity: Final Report to the Alberta Agricultural Research Institute/Farming for the Future Program. Calgary, Canada: Colin Bate Books; 2000. [Google Scholar]
- Weselake RJ, Pomeroy MK, Furukawa TL, Golden JL, Little DB, Laroche A. Developmental profile of diacylglycerol acyltransferase in maturing seeds of oilseed rape and safflower and micro-spore-derived cultures of oilseed rape. Plant Physiol. 1993;102:565–571. doi: 10.1104/pp.102.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weselake RJ, Taylor DC. The study of storage lipid biosynthesis using microspore-derived cultures of oilseed rape. Prog Lipid Res. 1999;38:401–460. doi: 10.1016/s0163-7827(99)00011-9. [DOI] [PubMed] [Google Scholar]
- Weselake RJ, Taylor DC, Pomeroy MK, Lawson SL, Underhill EW. properties of diacylglycerol acyltransferase from microspore-derived embryos of Brassica napusL. Phytochemistry. 1991;30:3533–3538. [Google Scholar]
- Yang H, Cromley D, Wang H, Billheimer JT, Sturley SL. Functional expression of a cDNA to human acyl-coenzyme A: cholesterol acyltransferase in yeast. J Biol Chem. 1997;272:3980–3985. doi: 10.1074/jbc.272.7.3980. [DOI] [PubMed] [Google Scholar]
- Zou J-T, Brokx SJ, Taylor DC. Cloning of a cDNA encoding the 21.2 kDa oleosin isoform from Arabidopsis thalianaand down-regulation of its expression in a mutant defective in diacylglycerol acyltransferase activity. Plant Mol Biol. 1996;31:429–433. doi: 10.1007/BF00021805. [DOI] [PubMed] [Google Scholar]
- Zou J-T, Katavic V, Giblin EM, Barton DL, MacKenzie SL, Keller WA, Hu X, Taylor DC. Modification of seed oil content and acyl composition in the Brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell. 1997;9:909–923. doi: 10.1105/tpc.9.6.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J-T, Wei Y, Jako C, Kumar A, Selvaraj G, Taylor DC. The Arabidopsis thaliana TAG1mutant has a mutation in a diacylglycerol acyltransferase gene. Plant J. 1999;19:645–653. doi: 10.1046/j.1365-313x.1999.00555.x. [DOI] [PubMed] [Google Scholar]