Abstract
We previously reported that Shewanella oneidensis MR-1 is highly sensitive to UVC (254 nm), UVB (290 to 320 nm), and UVA (320 to 400 nm). Here we delineated the cellular response of MR-1 to UV radiation damage by analyzing the transcriptional profile during a 1-h recovering period after UVC, UVB, and UVA exposure at a dose that yields about a 20% survival rate. Although the SOS response was observed with all three treatments, the induction was more robust in response to short-wavelength UV radiation (UVB and UVC). Similarly, more prophage-related genes were induced by short-wavelength UV radiation. MR-1 showed an active detoxification mechanism in response to UVA, which included the induction of antioxidant enzymes and iron-sequestering proteins to scavenge reactive oxygen species. In addition, a great number of genes encoding multidrug and heavy metal efflux pumps were induced following UVA irradiation. Our data suggested that activation of prophages appears the major lethal factor in MR-1 following UVC or UVB irradiation, whereas oxidative damage contributes greatly to the high UVA sensitivity in MR-1.
The deleterious effect of UV radiation (UVR) is highly dependent on the wavelength of radiation. DNA is the major chromophore following exposure to short-wavelength UVR. Both UVC (<290 nm) and UVB (290 to 320 nm) can induce the formation of cyclobutyl pyrimidine dimers (CPDs) and pyrimidine (6-4) pyrimidinone photoproducts, which are mutagenic and lethal to bacteria if unrepaired (30). Damage induced by long-wavelength UVR is more complex, since a variety of non-DNA targets with a maximum λ in the range of 290 to 400 nm are present in the cell (6, 7, 18). In addition, both UVB and UVA can produce reactive oxygen species (ROS), causing oxidative damage to a variety of molecules in the cell (6, 7).
Bacteria have evolved various mechanisms to cope with UVR-induced damage. In Escherichia coli, both photoreactivation and nucleotide excision repair (NER) are highly efficient in removing CPDs (19, 34), whereas recA-mediated recombination repair can bypass CPDs during DNA replication, thus improving DNA damage tolerance (10, 23). The LexA-RecA mediated SOS response is a global response to DNA damage involving the induction of more than 30 unlinked genes, many of which are involved in DNA replication and repair and in the control of cell division (4, 20). E. coli also possesses a variety of glycosylases to repair oxidative DNA damage through the base excision repair pathway (10). In addition, several regulatory genes are involved in protecting cells from oxidative stress. For example, OxyR, a LysR family protein, can activate the transcription of genes involved in peroxide metabolism and protection (katG, ahpC, ahpF, and dps) and in redox balance (gor, grxA, and trxC) and genes encoding regulators, such as fur and oxyS (36). The E. coli SoxRS regulon provides defense against oxidative damage caused by superoxide anions. More than 10 genes, including nfo (endonuclease IV) and sodA (Mn-superoxide dismutase), belong to the SoxRS regulon (12, 38). Sigma factor 38 (rpoS) is another important regulator in E. coli in response to oxidative stress (17). Some genes that are under the control of OxyR are also regulated by RpoS (8). Similar oxidative stress regulators have been identified in many other bacteria as well as pathogenic bacteria (3, 5, 26, 29, 33, 39).
Although extensive studies have focused on distinguishing different genes and regulons in response to far UV (UVC) and near UV (UVB and UVA), global genetic information remains limited due to the complexity of UVR-induced damage and limitations in the methods used in past studies. To our knowledge, there are no reports that compare the commonalities and the differences of biological effects induced by UVC (254 nm), a UVR wavelength that was commonly used in studying UVR-induced DNA damage but is less biologically relevant, and by UVB and UVA wavelengths, which are more biologically relevant, since both are present in natural solar UV radiation.
Shewanella oneidensis MR-1, an environmental gamma proteobacterium, can reduce a variety of compounds, including toxic metal ions and radionuclides (22, 25). Previous data indicated that MR-1 is highly sensitive to all wavelengths of UVR, solar UV, and ionizing radiation (32). However, this sensitivity could not be explained by its genome content. Similar to E. coli, which is more radiation resistant, MR-1 contains the major DNA damage repair and damage tolerance systems, including SOS response, recombination repair, mutagenic repair, nucleotide excision repair, and mismatch repair, and a DNA photolyase (14). MR-1 also contains a suite of genes, including rpoS and also a homolog of oxyR (SO1328), potentially involved in protection from oxidative stress. For scavenging ROS, MR-1 has genes encoding catalase (katB), catalase/peroxidase (katG1 and katG2), organic hydroperoxide resistance protein (ohr), alkyl hydroperoxide reductase (ahpC and ahpF), and a Dps protein (dps). For the repair of oxidative DNA damage, the MR-1 genome contains putative genes of tag, ung, mutM, mutY, mutT, nth, and xthA that are important in removing the damaged bases (14).
S. oneidensis MR-1, with its physiological response to UVR previously characterized (32) and a known phylogenetic relationship with other very-well-characterized organisms, e.g., E. coli organisms, represents an excellent model for a comprehensive analysis of the genomic response to different UVR wavelengths. Furthermore, the information obtained here will enhance our understanding of the important factors that contribute to the high UVR sensitivity of MR-1. We found that the genomic responses to UVC, UVB, and UVA during the 1-h recovery period are distinctly different. We observed similar genomic responses to UVC between MR-1 and E. coli; however, there are also distinct differences which may contribute to the increased UVC sensitivity of MR-1. In addition, the induction of genes encoding multidrug and heavy metal efflux pumps and of toxin-producing genes following UVA irradiation highlights previously unknown phenotypes in response to this stress.
Microarray experiments and data analysis.
An S. oneidensis MR-1 whole-genome cDNA array containing about 95% of the total S. oneidensis MR-1 open reading frames (ORFs) (11) was used to examine the transcriptional profiles of MR-1 following UVC, UVB, and UVA irradiation as described previously (32). Briefly, MR-1 was grown in Davis medium with 15 mM lactate as a carbon source until the optical density at 600 nm (OD600) reached 0.2 to 0.3. The culture (80 ml) was split into two parts: one was used for UVR irradiation, and the other one was used for the controls. The UVC, UVB, and UVA sources used were XX-15, XX-15 M, and XX-15L lamps (UVP Products, San Gabriel, Calif.), respectively. The energy output of each lamp was monitored with a UV-X radiometer (UVP Products) fitted with the appropriate sensor. The exposure doses were 3.3 J m−2 for UVC, 568 J m−2 for UVB, and 25 kJ m−2 for UVA, which yielded about a 20% survival rate (32). After irradiation, cells were transferred to a 100-ml flask and incubated at 30°C on a shaker. An aliquot of cells (12 ml) was collected after 5, 20, and 60 min of incubation for RNA extraction as described previously (32). Both UVC- and UVB-irradiated samples were collected in a dark room to avoid photoreactivation. Controls were treated in the same way except for UVR irradiation. Total RNA was isolated by using an RNeasy mini kit (QIAGEN, Valencia, Calif.). Prehybridization and RNA labeling were performed as described by Schroeder et al. (35) with a 2:3 ratio of 5-(3-aminoallyl)-dUTP and dTTP. Hybridization and washing were carried out as described by Hegde et al. (13). At each time point of each treatment, six hybridizations from three biological replicates and two technical replicates (dye swap) were performed. GENESPRING 6.0 software (Silicon Genetics, Redwood City, Calif.) was used to analyze all microarray hybridization data. Only those spots with more than 55% of pixels greater than background plus 2 standard deviations in either the Cy5 or Cy3 channel were used for data analysis (24). Data were normalized both per chip and per gene (LOWESS method) (43). Those genes that showed a statistically significant change in gene expression (P < 0.05) and a >2-fold change in magnitude were regarded as significant. The number of clusters for K-means analysis was determined by preanalyzing data using hierarchical cluster analysis.
Global gene expression trends in response to UVC, UVB, and UVA.
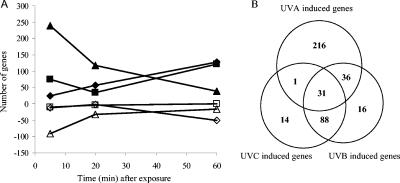
We observed distinct gene expression trends in MR-1 during the 1-h recovering period following UVC and UVA exposure. The response to UVA was fast: the differentiation peak appeared at 5 min, with a total of 239 induced genes and 92 repressed genes (Fig. 1A). In contrast, the response to UVC was much slower: the differentiation peak occurred at 60 min, with 128 induced genes and 51 repressed genes (Fig. 1A). The UVB-induced gene expression trend appeared to follow a composite pattern: two induction peaks were observed at 5 min (“UVA pattern”) and 60 min (“UVC pattern”), respectively (Fig. 1A). This unique pattern of response suggested that the shorter UVB wavelengths can cause UVC-type damage, whereas the longer wavelengths of UVB can produce UVA-type damage in MR-1. For all three treatments, more genes were induced than repressed (Fig. 2). Since our focus is primarily on the induced genes, which will probably encode proteins that are most directly involved in DNA repair and detoxification to overcome the cellular damage, we did not analyze the down-regulated genes in detail.
FIG. 1.
(A) The global gene expression trends in response to UVC, UVB, and UVA during the 1-h recovery period after exposure. Responses of up-regulated (⧫) and down-regulated (⋄) genes in response to UVC, up-regulated (▪) and down-regulated (□) genes in response to UVB, and up-regulated (▴) and down-regulated (▵) genes in response to UVA are shown. (B) Venn diagram of up-regulated genes in response to UVC, UVB, and UVA irradiation.
FIG. 2.
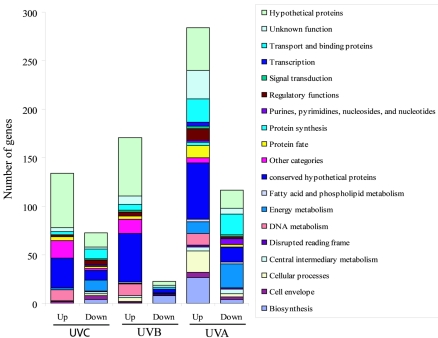
Distribution of the differentially expressed genes in various functional categories following UVC, UVB, and UVA exposure. The total numbers of induced (Up) and repressed (Down) genes were 134 and 73 for UVC, 171 and 23 for UVB, and 284 and 117 for UVA, respectively.
Totals of 134, 171, and 284 genes were induced at least at one time point examined during the 1-h recovering period following UVC, UVB, and UVA irradiation (Fig. 2). Almost 70% of the UVB-induced genes (119 genes) were up-regulated with the UVC treatment, whereas only about 40% of UVB-induced genes (67 genes) were up-regulated with the UVA treatment (Fig. 1B), which indicates that the UVB-induced stress response in MR-1 is more similar to that of UVC. A total of 31 genes were induced by all three UVR wavelengths, which have been clustered into three groups (Table 1). Cluster I (16 genes) contained SOS-responding genes, a site-specific recombinase gene, and seven hypothetical and conserved hypothetical genes. SO4604 is located in the same operon of lexA (SO4603). The coinduction of this gene with lexA indicates that it may belong to the SOS regulon in MR-1. New annotation of this ORF indicates that the gene product is possibly a SulA-like protein (M. Romine, personal communication). In E. coli, sulA expression is regulated via the SOS regulon. Accumulation of SulA causes rapid cessation of cell division and the appearance of long, nonseptate filaments. The gene product of SO4604 may have similar biological functions as the SulA of E. coli, since we observed the filamentous structure of MR-1 after UVC exposure (Fig. 3B). A putative LexA binding site was identified for SO2604 (TACTGTATATAAAAACAGTG) but not for other hypothetical or conserved hypothetical genes in this cluster. Cluster II (11 genes) contained genes involved in prophage replication or transposition. All eight hypothetical and conserved hypothetical genes in this cluster are located in the lambdaSo genome (14), which implied their potential function in replication and transcription of lambdaSo genes. Cluster III (four genes) contained an ISSo12 transposase and three conserved hypothetical genes (Table 1).
TABLE 1.
Relative expression of common up-regulated genes in response to UVC, UVB, and UVA radiationa
| Gene | Cluster groupb | Product(s) | Functionc | UVR ratios (± SD) (by min)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UVC
|
UVB
|
UVA
|
||||||||||
| 5 | 20 | 60 | 5 | 20 | 60 | 5 | 20 | 60 | ||||
| SOA0013 | I | UmuD protein | DNA metabolism | 5.1 ± 1.1 | 15.2 ± 1.7 | 16.1 ± 2.9 | 4.0 ± 1.0 | 11.0 ± 3.0 | 19.8 ± 3.5 | 5.0 ± 2.3 | 4.3 ± 1.3 | 4.3 ± 2.0 |
| SO0089 | I | Hypothetical protein | Unknown function | 3.1 ± 0.7 | 6.7 ± 1.7 | 6.9 ± 0.9 | 2.3 ± 0.4 | 5.1 ± 2.1 | 7.0 ± 2.5 | 2.3 ± 1.0 | 2.3 ± 1.1 | 1.6 ± 0.4 |
| SOA0012 | I | UmuC protein | DNA metabolism | 3.6 ± 1.3 | 8.4 ± 1.7 | 8.1 ± 0.7 | 2.6 ± 0.5 | 6.3 ± 1.9 | 8.0 ± 2.0 | 2.6 ± 0.5 | 3.3 ± 0.9 | 2.1 ± 0.5 |
| SO3061 | I | DNA topoisomerase III | DNA metabolism | 4.1 ± 0.8 | 8.0 ± 0.9 | 9.9 ± 3.3 | 2.9 ± 0.3 | 6.9 ± 0.5 | 9.8 ± 1.2 | 3.5 ± 0.2 | 3.4 ± 0.5 | 2.1 ± 0.4 |
| SO2603 | I | Conserved hypothetical proteins | Unknown function | 3.2 ± 0.2 | 5.2 ± 0.3 | 5.7 ± 0.6 | 2.6 ± 0.3 | 4.8 ± 0.3 | 5.7 ± 0.5 | 2.7 ± 0.2 | 2.4 ± 0.4 | 1.6 ± 0.3 |
| SO2604 | I | Conserved hypothetical proteins | Unknown function | 3.7 ± 0.5 | 6.1 ± 0.8 | 6.7 ± 0.9 | 3.0 ± 0.8 | 5.5 ± 1.6 | 7.3 ± 0.9 | 3.5 ± 0.6 | 3.1 ± 0.4 | 2.1 ± 0.4 |
| SO1114 | I | DNA-damage-inducible protein P | Unknown function | 3.6 ± 0.3 | 6.8 ± 0.7 | 6.2 ± 0.8 | 3.6 ± 0.6 | 5.8 ± 1.6 | 12.3 ± 1.8 | 4.2 ± 0.9 | 4.7 ± 1.3 | 2.9 ± 0.5 |
| SO3462 | I | DNA repair protein RecN | DNA metabolism | 7.9 ± 1.6 | 16.0 ± 3.7 | 15.0 ± 1.4 | 7.4 ± 1.2 | 11.5 ± 1.8 | 20.3 ± 1.0 | 6.8 ± 0.7 | 6.3 ± 1.0 | 3.7 ± 0.9 |
| SO3429 | I | Regulatory protein RecX | DNA metabolism | 4.8 ± 1.3 | 7.8 ± 0.3 | 7.4 ± 1.4 | 4.1 ± 0.7 | 6.3 ± 1.2 | 9.6 ± 1.2 | 3.5 ± 0.3 | 3.3 ± 0.2 | 2.1 ± 0.2 |
| SO4603 | I | LexA repressor | Regulatory function | 6.7 ± 0.6 | 12.2 ± 1.2 | 11.8 ± 2.1 | 8.0 ± 1.1 | 9.9 ± 2.0 | 17.6 ± 1.8 | 5.6 ± 1.8 | 6.1 ± 2.0 | 3.4 ± 0.8 |
| SO4604 | I | Conserved hypothetical protein | Unknown function | 6.9 ± 0.8 | 11.8 ± 1.9 | 12.5 ± 1.3 | 7.6 ± 1.9 | 9.5 ± 2.7 | 20.1 ± 1.7 | 7.0 ± 1.0 | 7.1 ± 2.1 | 3.8 ± 0.8 |
| SO3430 | I | RecA protein | DNA metabolism | 5.0 ± 1.2 | 8.0 ± 1.2 | 9.4 ± 2.1 | 4.0 ± 0.6 | 6.8 ± 0.9 | 10.2 ± 2.6 | 3.5 ± 0.5 | 3.6 ± 0.6 | 2.3 ± 0.2 |
| SO4605 | I | Hypothetical protein | Unknown function | 5.7 ± 1.6 | 8.3 ± 2.9 | 6.4 ± 2.0 | 5.1 ± 2.3 | 9.5 ± 4.2 | 8.7 ± 3.3 | 3.0 ± 1.7 | 3.2 ± 0.4 | 2.3 ± 0.4 |
| SO1757 | I | Conserved hypothetical protein | Unknown function | 13.6 ± 2.8 | 22.6 ± 2.9 | 16.7 ± 3.0 | 20.7 ± 8.9 | 17.2 ± 7.8 | 32.7 ± 3.9 | 23.2 ± 7.7 | 33.0 ± 18.8 | 14.5 ± 5.7 |
| SO3327 | I | Hypothetical protein | Unknown function | 2.0 ± 1.0 | 5.0 ± 1.1 | 4.5 ± 0.8 | 2.5 ± 0.6 | 2.0 ± 1.3 | 5.4 ± 3.1 | 3.8 ± 1.4 | 1.5 ± 0.5 | 1.8 ± 0.7 |
| SO2037 | I | Site-specific recombinase, phage integrase family | DNA metabolism | 1.7 ± 0.7 | 1.9 ± 0.6 | 2.4 ± 0.4 | 1.2 ± 0.4 | 1.6 ± 0.4 | 2.2 ± 0.6 | 1.8 ± 0.6 | 2.3 ± 1.0 | 1.5 ± 0.5 |
| SO2998 | II | Hypothetical protein | Unknown function | 0.9 ± 0.1 | 2.2 ± 0.5 | 15.2 ± 4.2 | 0.9 ± 0.2 | 1.3 ± 0.3 | 14.7 ± 4.6 | 0.8 ± 0.2 | 1.4 ± 0.1 | 2.2 ± 0.4 |
| SO3000 | II | Conserved hypothetical protein | Unknown function | 1.0 ± 0.4 | 3.1 ± 1.2 | 25.3 ± 5.9 | 0.9 ± 0.2 | 1.8 ± 0.5 | 37.5 ± 5.4 | 1.0 ± 0.1 | 1.5 ± 0.3 | 3.1 ± 0.8 |
| SO2993 | II | Prophage lambdaSo, type II DNA modification methyltransferase | DNA metabolism | 0.9 ± 0.3 | 3.0 ± 1.1 | 26.2 ± 4.9 | 1.0 ± 0.1 | 1.7 ± 0.1 | 35.6 ± 5.4 | 1.6 ± 0.2 | 1.5 ± 0.3 | 2.9 ± 0.9 |
| SO2997 | II | Hypothetical protein | Unknown function | 0.9 ± 0.2 | 2.6 ± 0.6 | 17.3 ± 3.7 | 0.9 ± 0.2 | 1.5 ± 0.5 | 19.5 ± 5.7 | 1.0 | 1.2 ± 0.2 | 2.5 ± 0.5 |
| SO2988 | II | Conserved hypothetical protein | Unknown function | 1.1 ± 0.2 | 2.5 ± 0.5 | 21.0 ± 3.7 | 0.9 ± 0.1 | 1.3 ± 0.2 | 21.6 ± 3.9 | 1.2 ± 0.4 | 1.4 ± 0.4 | 2.1 ± 0.6 |
| SO3001 | II | Hypothetical protein | Unknown function | 1.0 ± 0.1 | 2.3 ± 0.6 | 23.1 ± 10.7 | 0.8 ± 0.1 | 1.3 ± 0.2 | 19.1 ± 8.7 | 0.8 ± 0.3 | 1.3 ± 0.1 | 2.2 ± 0.7 |
| SO3002 | II | Conserved hypothetical protein | Unknown function | 1.1 ± 0.3 | 2.2 ± 0.7 | 21.5 ± 3.8 | 0.9 ± 0.1 | 1.4 ± 0.3 | 24.8 ± 8.4 | 0.9 ± 0.2 | 1.4 ± 0.2 | 2.1 ± 0.3 |
| SO2984 | II | Conserved hypothetical protein | Unknown function | 1.0 ± 0.2 | 1.8 ± 0.3 | 14.3 ± 1.9 | 1.0 ± 0.2 | 1.3 ± 0.2 | 14.8 ± 1.0 | 1.0 ± 0.2 | 1.0 | 2.2 ± 0.5 |
| SO2985 | II | Prophage lambdaSo, replication protein O | Other categories | 1.1 ± 0.4 | 2.0 ± 0.6 | 15.8 ± 2.8 | 1.0 ± 0.1 | 1.3 ± 0.2 | 15.9 ± 2.8 | 1.2 ± 0.2 | 1.4 ± 0.5 | 2.2 ± 0.4 |
| SO2983 | II | Hypothetical protein | Unknown function | 1.1 ± 0.1 | 1.8 ± 0.4 | 16.0 ± 3.7 | 0.8 ± 0.2 | 1.3 ± 0.2 | 17.3 ± 1.0 | 1.3 ± 0.1 | 1.1 ± 0.2 | 2.1 ± 0.3 |
| SO0644 | II | Prophage MuSol, DNA transpo- sition protein, putative | Other categories | 1.0 ± 0.1 | 2.2 ± 0.6 | 13.3 ± 1.2 | 1.0 ± 0.4 | 1.4 ± 0.4 | 19.0 ± 2.0 | 2.3 ± 0.7 | 1.2 ± 0.5 | 1.3 ± 0.1 |
| SO1759 | III | Conserved hypothetical proteins | Unknown function | 10.0 ± 4.6 | 10.7 ± 1.1 | 6.7 ± 1.0 | 13.3 ± 3.5 | 11.3 ± 4.2 | 11.3 ± 2.5 | 11.1 ± 3.7 | 8.3 ± 3.2 | 4.4 ± 2.4 |
| SO3849 | III | Conserved hypothetical proteins | Unknown function | 9.7 ± 3.9 | 9.8 ± 1.2 | 5.2 ± 0.5 | 11.1 ± 3.8 | 11.9 ± 8.5 | 9.3 ± 1.4 | 7.2 ± 1.9 | 5.5 ± 3.1 | 1.7 ± 0.3 |
| SO1761d | III | Conserved hypothetical proteins | Unknown function | 2.8 ± 1.4 | 3.4 ± 2.4 | 2.2 ± 0.7 | 5.3 ± 3.4 | 4.0 ± 2.2 | 2.8 ± 1.1 | 4.3 ± 2.3 | 3.9 ± 2.1 | 3.9 ± 0.8 |
| SO3854 | III | ISSo12, transposase | Other categories | 8.0 ± 3.5 | 8.0 ± 3.6 | 3.8 ± 1.0 | 3.0 ± 1.0 | 5.7 ± 2.0 | 4.3 ± 2.0 | 2.5 ± 1.2 | 1.5 ± 0.6 | 1.1 ± 0.2 |
The data presented here are the ratios of UVR-irradiated samples to the controls.
Cluster analysis was performed using either standard correlation or distance to calculate the similarities.
The functional groups are based on the published genome annotation (14).
The only gene that showed different clusters (cluster I or cluster III), by use of two methods.
FIG. 3.
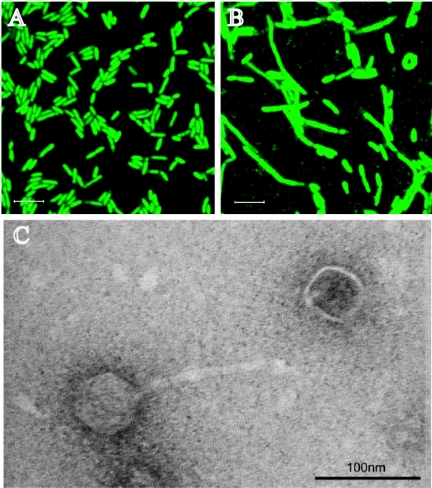
SYBR green I staining of MR-1 (A), MR-1 irradiated by UVC (B), and the TEM images for phage isolated from UVC-irradiated MR-1 (C). The scan zoom was 4.0 for the images in panels A and B.
Approximately 40% of annotated ORFs in MR-1 belong to either conserved hypothetical protein (871 ORFs) or hypothetical protein (1,161 ORFs) (14). A total of 181 of those ORFs were induced under our experimental conditions, among which 18 were induced in response to all three wavelength groups of UVR, 61 were induced in response to both UVB and UVC, and 19 were induced in response to both UVB and UVA (see Fig. S2 in the supplemental material). There are 8, 12, and 64 ORFs induced specifically by UVC, UVB, and UVA irradiation, respectively (see Fig. S2 and Table S7 in the supplemental material). In conjunction with other approaches, we should be able to gain a better understanding of their potential biological functions in MR-1.
Gene expression profile following UVC irradiation.
Based on The Institute for Genomic Research annotation (14), the UVC-induced genes were grouped into 11 functional categories, of which both “hypothetical proteins” (41.8%) and “conserved hypothetical protein” (23.1%) were dominant (UVC; Fig. 2). Other large groups included the “other categories” (13.4%), which mainly are prophage-related genes and transposases; “DNA metabolism” (8.2%); “protein fate” (3%); and “unknown function” (3%) groups. Since the K-means analysis is a particularly useful method to identify unique classes of genes that are differentially expressed in a time-dependent manner, we performed a K-means analysis of UVR-induced genes and examined each cluster in detail (see Fig. S1 in the supplemental material). Three major clusters were revealed for UVC-induced genes (see Fig. S1 in the supplemental material). The first cluster (39 genes) contained immediate-responding genes: the induction started at 5 min and continued to 20 or 60 min. The representative genes were those involved in DNA replication, recombination, and repair (see Fig. S1 and Table S4 in the supplemental material). The second cluster (42 genes) contained intermediate-responding genes. The induction began at 20 min and continued to 60 min. The representative genes were those involved in replication and transcription of prophage genes (see Fig. S1 and Table S4 in the supplemental material). The third cluster (52 genes) contained late-responding genes: no induction occurred until 60 min. The representative genes were those encoding the prophage structural proteins, such as major and minor tail proteins, tail assembly protein, and major head subunit (see Fig. S1 and Table S4 in the supplemental material).
Induction of DNA damage repair genes after UVC exposure.
Strong inductions of recA and lexA were observed following UVC exposure, which indicated the induction of the SOS response in MR-1 (Table 1). In addition, we observed similar strong inductions of recN, recX, topB, dinP, and the umuDC operon. Induction of the umuDC operon correlated very well with our prior observation of increased mutability in MR-1 following UVC exposure (32). Although no induction was observed for ruvAB, a weak induction of recG (2- to 2.5-fold) was observed. This result suggests that recombination repair is functional for MR-1 (see Table S4 in the supplemental material). We previously demonstrated that, unlike for E. coli, the expression of NER component genes, e.g., uvrA, uvrB, and uvrD, is not damage inducible (32). Here we attempted to quantify the expression levels of the three genes in both UVC-irradiated and nonirradiated samples by quantitative reverse transcription-PCR (Q RT-PCR). recA was used as a positive control (induced), and radC was used as a negative control (noninduced). A detailed description of Q RT-PCR conditions is in the supplemental text. A consistent result was observed by Q RT-PCR, except the induction increase (n-fold) for recA measured by microarray hybridization was lower than that measured by Q RT-PCR assay (Table 1; see Table S2 in the supplemental material). The correlation obtained using ldhA as an internal control (R2, 0.9478) was better than that with the 16S rrn gene (R2, 0.7394). The mean basal expression levels ± standard deviations were 472 ± 231, 205 ± 123, and 160 ± 85 copies in 500 pg of total cDNA for uvrA, uvrB, and uvrD, respectively, which were lower than that of recA (2,312 ± 1,272).
Possession of efficient DNA repair capacity following UV radiation stress is essential for the survival of all organisms. We observed a strong SOS induction, similar to that of E. coli; however, some damage-inducible genes, such as ruvAB, recF, xthA, and mutM, did not show any induction under any of our experimental conditions, as we observed for NER component genes. Furthermore, no homologs of dinI, dinD, nfo, nfi, vsr, and alkA were found in the MR-1 genome (14). It appeared that an alteration in gene regulation and an alteration in gene content have occurred in S. oneidensis MR-1, an organism not known so far to live currently in environments exposed to light.
Induction of prophage-related genes by UVR.
It is well known that short-wavelength UVR can induce the lytic cycle of lysogenic bacteriophage. We observed the induction for a great number of prophage-related genes in MR-1 after UVC exposure, with the largest percentage (74.7%) of genes induced from the lambdaSo genome (Table 2). In addition, induction of early genes which are involved in lambdaSo replication and transcription was observed from 5 to 20 min, whereas induction of later genes which encode phage structural proteins was observed only at 60 min. A similar expression pattern was observed for prophage MuSo1 but not for MuSo2. A total of 15 genes (SO0643 through SO0652 and SO0674 through SO0678) were induced from the MuSo1 genome. The activation of genes responsible for transposition and a positive regulator of later transcription (SO0643 through SO0652) indicated a potential activation of phage MuSo1. Indeed, gene products of SO0674 through SO0678 are structural proteins of MuSo1. A total of 16 genes (SO2653 through SO2668) were induced from the MuSo2 genome, which include genes responsible for transposition and a positive regulator of later transcription. However, no genes encoding structural proteins of phage MuSo2 were induced (see Table S4 in the supplemental material).
TABLE 2.
Numbers of induced prophage-related genes following UVC, UVB, and UVA exposure
| Prophage | Length (bp)a | Total ORFs | Total induced ORFs
|
No. of indicated proteins
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hypothetical proteins
|
Conserved hypothetical proteins
|
Others
|
|||||||||||||||
| Total | Induced by:
|
Total | Induced by:
|
Total | Induced by:
|
||||||||||||
| UVC | UVB | UVA | UVC | UVB | UVA | UVC | UVB | UVA | UVC | UVB | UVA | ||||||
| lambdaSo | 51,857 | 75 | 56 | 51 | 11 | 38 | 29 | 26 | 5 | 16 | 14 | 14 | 4 | 21 | 13 | 11 | 2 |
| MuSo1 | 34,551 | 42 | 15 | 15 | 1 | 21 | 6 | 6 | 0 | 8 | 4 | 4 | 0 | 14 | 5 | 5 | 1 |
| MuSo2 | 35,666 | 53 | 16 | 15 | 2 | 20 | 9 | 9 | 0 | 17 | 3 | 3 | 1 | 16 | 4 | 3 | 1 |
According to Heidelberg et al. (14).
The effect of UVB on phage gene induction was comparable to that of UVC for all three MR-1 prophages (Table 2). In contrast, UVA exposure induced the expression of few genes, including only 11 of 75 genes of lambdaSo and 1 of 42 and 2 of 53 genes in MuSo1 and MuSo2, respectively (Table 2).
Gene expression data strongly suggested that UVC may induce the lytic cycle of lambdaSo in MR-1 (Table 2). Using SYBR green I staining, we observed phage particles in the cultures exposed to UVC (Fig. 3B) but not in control cultures (Fig. 3A). In addition, cells exposed to UVC were enlarged (Fig. 3B) greatly compared to the control samples (Fig. 3A). This observation is consistent with the previous observation that the inhibition of cell division is a consequence of the UVC-induced stress response for many bacteria (15) (Table 1). We observed, in a suspension from UVC-irradiated MR-1 cells, phage with a head and tail structure by transmission electron microscopy (TEM) (Fig. 3C). Similar phage particles were seen in the UVB-irradiated samples (data not shown). A detailed description of the detection of UVC-induced prophage particles by SYBR green I staining and examination of UVC-induced prophage by TEM is in the supplemental text.
Gene expression profile following UVA irradiation.
Unlike the UVC gene expression profile, the UVA-induced genes were distributed in 16 functional categories more evenly (Fig. 2). The top six large groups were the conserved hypothetical proteins (19.7%), hypothetical proteins (15.5%), “biosynthesis” (11.6%), unknown functions (10.2%), “transport and binding proteins” (8.8%), and “cellular process” (7.8%) (Fig. 2) groups. Compared to the UVC transcriptional profile, genes in the DNA metabolism and other categories groups reduced from 8.2 and 13.4% to 4.2 and 1.8%, respectively, whereas genes in the “regulatory function,” “signal transduction,” “transcription,” and “metabolism” categories showed slight increases in percentage (Fig. 2).
UVA-induced DNA damage.
We observed the induction of several key genes of the SOS regulon in MR-1 following UVA irradiation, although the increase in induction (n-fold) was less substantial than either UVC or UVB (Table 1). In addition, we observed a strong induction (20-fold) of phrB, which encodes a DNA photolyase mediating photoreactivation, and a weak induction of mutL (2.1-fold), which encodes a component of DNA mismatch repair. This result indicated that DNA is one of the targets for UVA.
Scavenging of UVA-induced reactive oxygen species.
The removal of reactive molecules that result from photooxidation is a challenge faced by organisms in coping with UVA-induced stress. The induction of antioxidant enzymes and proteins is a common strategy of bacteria to scavenge ROS. In S. oneidensis MR-1, we observed at 5 min the induction of katG1 (SO0725, 3.8-fold), alkyl hydroperoxide reductase subunit C (SO0958, 4.3-fold), a cytochrome c551 peroxidase (SO2178, 2.8-fold), an organic hydroperoxide resistance protein (SO0976, 8.7-fold), and a putative glutathione peroxidase (SO1563, 4.7-fold) (see Fig. S1 and Table S5 in the supplemental material). In addition, we observed the strong induction of SO1773 (8.0-fold), which encodes a catalase-related protein, and SO3349 (11.9-fold), which encodes a second putative glutathione peroxidase. Induction of these two ORFs occurred at 5 min and lasted until 20 min (see Fig. S1 and Table S5 in the supplemental material). Although MR-1 possesses a katB (SO1070) and another katG (SO4405), no induction of either gene was observed. Since the overall hybridization signals of katB were lower than most of the spots on the array, Q RT-PCR was performed. Similar results were observed (see Table S3 in the supplemental material).
The intracellular iron pool plays an important role in near-UVR-induced damage. First, iron-containing proteins may act as chromophores, becoming excited and thereby being damaged directly (6, 18). Ferrous iron can catalyze the formation of hydroxyl radicals through the Fenton reaction, influencing the generation of ROS following UVA irradiation (16, 31). Hence, regulation of iron uptake and metabolism and iron sequestration are important protection mechanisms against UVA-induced oxidative damage. Indeed, we observed the induction of several iron-sequestering protein-encoding genes, such as SO1158 (ferritin-like Dps protein, 3.6-fold), bcp (bacterioferritin comigratory protein, 7.0-fold), and hemH (ferrochelatase, 10.4-fold), which encodes the enzyme that inserts iron into protoporphyrin IX to make heme. Correspondingly, genes involved in iron uptake were strongly repressed at 5 min after irradiation (for SO3669 through SO3675, 0.25-, 0.37-, 0.27-, 0.38-, 0.46-, and 0.30-fold, respectively) (see Fig. S1 and Table S5 in the supplemental material). Also, the expression of SO4077, which encodes a putative TonB-dependent receptor, was repressed more than threefold during the 1-h recovery period. The expression levels of SO3669 (hugA), SO3670 (tonB1), and SO3671 (exbB1) increased slightly at 20 min (2.3-, 2.2-, and 2.3-fold, respectively), which may indicate the requirement of iron for the synthesis of new proteins in MR-1 following UVA irradiation (see Table S5 in the supplemental material).
Induction of toxin and toxin secretion-related genes by UVA irradiation.
The MR-1 genome contains a putative pore-forming RTX (repeats in toxin) toxin operon (SO4146 through SO4149) and a gene cluster (SO4317 through SO4319) that is related to RTX production and secretion. MR-1 also contains a gene encoding a putative hemolysin (SO1354). Hemolysin can bind to and lyse mammalian cell membranes and, at low concentrations, perturb cell signal transduction, causing the release of inflammatory mediators (37, 41, 42). We observed the induction of SO4149, which encodes an RTX (2.0-fold); SO4148 (4.9-fold), which encodes a HlyD family secretion protein involved in secretion of toxin; and SO1354 (2.6-fold) (see Table S5 in the supplemental material).
Secretion of RTX toxins requires three gene products in E. coli: HlyB, HlyD, and TolC. Both HlyB and HlyD are inner membrane proteins, functioning as an ATPase (HlyB) and an adaptor (HlyD), whereas TolC is an outer membrane exit-duct protein (2, 9, 40). This tripartite machinery transports toxins directly across the entire cell envelope. Interestingly, MR-1 is highly redundant in hlyD. There are a total of 17 ORFs encoding HlyD family proteins, of which six (SO1881, SO1925, SO3483, SO4015, SO4327, and SO4693) are located close to genes coding for RND (the resistance-nodulation-cell division) antiporter AcrB/AcrD/AcrF family protein. High induction was observed for MR-1 following UVA irradiation for SO1925 (5.4-fold) and SO4327 (10.0-fold) (see Table S5 in the supplemental material).
Induction of multidrug and heavy metal efflux pumps by UVA irradiation.
Similar to HlyD, MR-1 is also highly redundant (nine copies) in genes encoding AcrB/AcrD/AcrF family protein (14) and has a gene (SO4328) encoding a truncated AcrB/AcrD/AcrF family protein (629 amino acids) due to an authentic frameshift. In E. coli, AcrAB-TolC is a major, constitutively expressed multidrug efflux pump that provides resistance to structurally unrelated noxious molecules (1, 28). AcrB functions as antiporter which uses proton flux as the source of energy, whereas AcrA functions as an adaptor and TolC works in the same way as it does in the type I secretion pathway (HlyBD-TolC) (1, 44). Strong inductions of SO1923 (7.8-fold), SO1924 (10.2-fold), and SO4328 (10.2-fold) were observed after exposure to UVA (see Fig. S1 and Table S5 in the supplemental material). In addition, SO0525, which encodes an EmrB/QacA family protein, showed a 4.5-fold induction. EmrB of E. coli is an integral membrane translocase which mediates drug extrusion (21). MR-1 also carries a chromosome-borne heavy metal efflux pump (SO4597 and SO4598) and a plasmid-borne heavy metal efflux pump (SOA0153 and SOA0154). Both SO4598 and SOA0153 encode a CzcA family protein, which is a cation/proton antiporter of the RND family protein, whereas both SO4597 and SOA0154 encode a putative heavy metal efflux membrane fusion protein (M. Romine, personal communication). CzcA, along with CzcB, a membrane fusion protein, and CzcC, an outer membrane protein, confers resistance to cobalt, zinc, and cadmium ions (27). Strong induction (6.0- to 7.0-fold) was observed for all four ORFs after UVA exposure (see Fig. S1 and Table S5 in the supplemental material). These data suggest that heavy metal and multidrug efflux pumps may function as a method of detoxification in UVA-irradiated MR-1 cells.
Due to the highly homologous motifs between or among the ORFs encoding the same or similar products, we may have observed cross-hybridization in our microarray-based gene expression experiments. To validate our observations, we designed gene-specific primers for four genes (SO1923, SO1924, SO4328, and SOA0154) that encode the heavy metal and multidrug efflux pumps described above (see Table S1 in the supplemental material). We also included ohr (highly induced), recA (moderately induced), and radC (no induction) in the Q RT-PCR analysis for validation and comparison. Consistent results were observed by the Q RT-PCR assay (see Table S3 in the supplemental material). Again, the correlation obtained using ldhA as the internal control (R2, 0.8953) was better than that with the 16S rrn genes (R2, 0.8000). We also confirmed less induction of recA than that with UVC and no induction of radC in UVA-irradiated samples (see Table S3 in the supplemental material).
Induction of other stress-related genes by UVA irradiation.
Other stress-related genes that were induced by UVA exposure included those that were involved in cell motility (SO1989, 5.5-fold; SO3247, 2.8-fold; SO3248, 6.1-fold; SO3282, 2.4-fold; SO3241, 2.1-fold), in cell signaling (SO4170, 13.3-fold), and in producing antibiotic resistance (SO4299, 2.8-fold; SO0837, 2.1-fold). We also observed a slight induction of some heat shock and chaperone proteins, such as HslU (SO4160, 2.1-fold), HtpG (SO2016, 2.1-fold), and DnaK (SO1126, 2.2-fold) (see Table S5 in the supplemental material).
Gene expression profile following UVB irradiation.
The UVB-induced genes were distributed in 14 functional categories, which was more than that of UVC (11) but less than that of UVA (16). Similar to the UVC transcriptional profile, both hypothetical proteins (35.1%) and conserved hypothetical proteins (29.2%) were dominant. In addition, the number of genes in the DNA metabolism and other categories decreased slightly, whereas the numbers of genes in the cellular processes, transporter and binding proteins, and regulatory function groups increased slightly compared to the UVC profile (Fig. 2). Those changes indicated a shift in response to damage induced by short-wavelength UVR to long-wavelength UVR, from direct DNA damage and activation of prophages to global photooxidative damage.
Genes induced by UVB could be roughly divided into UVC-pattern genes and UVA-pattern genes. UVC-pattern genes were mainly distributed in clusters I, III, and IV, whereas UVA-pattern genes were mainly distributed in cluster II (see Fig. S1 and Table S6 in the supplemental material). A strong SOS induction was observed following UVB irradiation, which indicated that, similar to the case with UVC, photons at UVB wavelengths can cause direct DNA damage in MR-1 (Table 1). Similar to UVA, we observed the induction of genes encoding antioxidant enzyme (SO3349, 9.3-fold), iron sequestration (SO3348, 10.7-fold), multidrug efflux pumps (SO4328, 4.9-fold), and the production of toxin and resistance traits (SO4170, 6.9-fold; SO4327, 4.3-fold) although the number of induced genes in each category was less than that for UVA (Table S6 in the supplemental material). This result confirmed our previous observation that the UVB-induced stress response was more similar to that of UVC (32).
Summary.
This study systematically investigated and compared the genomic responses to the three important wavelength groups of UV radiation: UVC, UVB, and UVA. The genomic response to UVA was the greatest, with about 8% of the genome expressed differentially, and immediate, with most of the genes showing induction at 5 min. These responding characteristics reflected the UVA-induced damaging effects: oxidative damage was global and required repair immediately. In contrast, only about 4% of genome showed differential expression following UVC exposure, with most of the genes showing induction at 60 min. These responding characteristics indicate that the UVC-induced cellular damage is less global with DNA as a major target. However, due to the harboring of three prophages by MR-1, a great number of prophages were induced by UVC in a time-dependent manner (activation of the early gene at 20 min and activation of the late gene at 60 min). The UVB-induced genomic responding pattern was the reflection of its wavelength composition (290 to 320 nm): the short wavelength of UVB has UVC effects, such as direct damage of DNA and activation of prophage, whereas the long wavelength of UVB has UVA effects, such as production of oxidative damage. Although DNA damage is a common effect of all three wavelengths of UVR, the damage induced by either UVC or UVB is stronger than that induced by UVA. However, DNA damage may or may not be the major lethal factor in MR-1 following UVC or UVB irradiation, considering the amount of DNA damage induced by 3.3 J m−2 of UVC or 568 J m−2 of UVB and an active DNA-repairing mechanism we observed. Hence, the activation of prophage appears the major lethal factor in MR-1 following UVC or UVB irradiation. Although MR-1 possesses a very active detoxification mechanism which includes the traditional strategies for defending against oxidative stress, e.g., production of antioxidant enzymes and proteins, sequestration of transition metals, and activation of degradative pathways, etc., as well as novel strategies, such as the activation of multidrug and heavy metal efflux pumps, it still failed in overcoming the UVA-induced oxidative stress. MR-1 is one of the most UVA-sensitive bacteria known so far. As a respiratory generalist, it is rich in cytochromes and iron-containing proteins (14). Cytochromes, along with other respiratory-chain components, such as flavins and quinones, are potential targets for long-wavelength UVR. UVR-rich targets may be the intrinsic causes of this bacterium's high susceptibility to UVA-induced oxidative stress in S. oneidensis MR-1.
Supplementary Material
Acknowledgments
We thank Jeff Landgraf and Annette Thelen at the Genomic Technology Support Facility of Michigan State University for their helpful discussions on microarray data analysis and Q RT-PCR and Shirley Owens and Alicia Pastor at the Michigan State University Center for Advanced Microscopy for their assistance with phage detection. We thank the members of the Shewanella Federation for helpful discussions on the biology and genomics of these organisms.
This research was supported by The United States Department of Energy under the Genomics:GTL (grant DE-FG-02-02ER63342) and Microbial Genome Programs of the Office of Biological and Environmental Research in the Office of Science. Oak Ridge National Laboratory is managed by University of Tennessee-Battelle LLC for the Department of Energy under contract DE-AC05-00OR22725.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andersen, C. 2003. Channel-tunnels: outer membrane components of type I secretion systems and multidrug efflux pumps of Gram-negative bacteria. Rev. Physiol. Biochem. Pharmacol. 147:122-165. [DOI] [PubMed] [Google Scholar]
- 2.Buchanan, S. K. 2001. Type I secretion and multidrug efflux: transport through the TolC channel-tunnel. Trends Biochem. Sci. 26:3-6. [DOI] [PubMed] [Google Scholar]
- 3.Christman, M. F., R. W. Morgan, F. S. Jacobson, and B. N. Ames. 1985. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41:753-762. [DOI] [PubMed] [Google Scholar]
- 4.Courcelle, J., A. Khodursky, B. Peter, P. B. Brown, and P. C. Hanawalt. 2001. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics 158:41-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eiamphungporn, W., K. Nakjarung, B. Prapagdee, P. Vattanaviboon, and S. Mongkolsuk. 2003. Oxidant-inducible resistance to hydrogen peroxide killing in Agrobacterium tumefaciens requires the global peroxide sensor-regulator OxyR and KatA. FEMS Microbiol. Lett. 225:167-172. [DOI] [PubMed] [Google Scholar]
- 6.Eisenstark, A. 1987. Mutagenic and lethal effects of near-ultraviolet radiation (290-400 nm) on bacteria and phage. Environ. Mol. Mutagen. 10:317-337. [DOI] [PubMed] [Google Scholar]
- 7.Eisenstark, A. 1989. Bacterial genes involved in response to near-ultraviolet radiation. Adv. Genet. 26:99-147. [DOI] [PubMed] [Google Scholar]
- 8.Eisenstark, A. 1998. Bacterial gene products in response to near-ultraviolet radiation. Mutat. Res. 422:85-95. [DOI] [PubMed] [Google Scholar]
- 9.Felmlee, T., S. Pellett, E.-Y. Lee, and R. A. Welch. 1985. Escherichia coli hemolysin is released extracellularly without cleavage of a signal peptide. J. Bacteriol. 163:88-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis, p. 14-19, 135-181, 435-440. American Society for Microbiology, Washington, D.C.
- 11.Gao, H., Y. Wang, X. Liu, T. Yan, L. Wu, E. Alm, A. Arkin, D. K. Thompson, and J. Zhou. 2004. Global transcriptome analysis of the heat shock response of Shewanella oneidensis. J. Bacteriol. 186:7796-7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenberg, J. T., P. Monach, J. H. Chou, P. D. Josephy, and B. Demple. 1990. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents. Proc. Natl. Acad. Sci. USA 87:6181-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hegde, P., R. Qi, K. Abernathy, C. Gay, S. Dharap, R. Gaspard, J. E. Hughes, E. Snesrud, N. Lee, and J. Quackenbush. 2000. A concise guide to cDNA microarray analysis. BioTechniques 29:548-562. [DOI] [PubMed] [Google Scholar]
- 14.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, R. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 15.Huisman, O., R. D'Ari, and S. Gottesman. 1984. Cell division control in Escherichia coli: specific induction of the SOS sfiA protein is sufficient to block septation. Proc. Natl. Acad. Sci. USA 81:4490-4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imlay, J. A., S. M. Chin, and S. Linn. 1988. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science 240:640-642. [DOI] [PubMed] [Google Scholar]
- 17.Ivanova, A. B., G. V. Glinsky, and A. Eisenstark. 1997. Role of RpoS regulon in resistance to oxidative stress and near-UV radiation in ΔOxyR suppressor mutants of Escherichia coli. Free Radic. Biol. Med. 23:627-636. [DOI] [PubMed] [Google Scholar]
- 18.Jagger, J. 1983. Physiological effects of near-ultraviolet radiation on bacteria. Photochem. Photobiol. Rev. 7:1-75. [Google Scholar]
- 19.Kim, S. T., and A. Sancar. 1993. Photochemistry, photophysics, and mechanism of pyrimidine dimer repair by DNA photolyase. Photochem. Photobiol. 57:895-904. [DOI] [PubMed] [Google Scholar]
- 20.Little, J. W., and D. W. Mount. 1982. The SOS regulatory system of Escherichia coli. Cell 29:11-22. [DOI] [PubMed] [Google Scholar]
- 21.Lomovskaya, O., and K. Lewis. 1992. emr, an Escherichia coli locus for multidrug resistance. Proc. Natl. Acad. Sci. USA 89:8938-8942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Middleton, S. S., R. B. Latmani, M. R. Mackey, M. H. Ellisman, B. M. Tebo, and C. S. Criddle. 2003. Cometabolism of Cr (VI) by Shewanella oneidensis MR-1 produces cell-associated reduced chromium and inhibits growth. Biotechnol. Bioeng. 83:627-637. [DOI] [PubMed] [Google Scholar]
- 23.Miller, R. V., and T. A. Kokjohn. 1990. General microbiology of recA: environmental and evolutionary significance. Annu. Rev. Microbiol. 44:365-394. [DOI] [PubMed] [Google Scholar]
- 24.Murray, A. E., D. Lies, G. Li, K. Nealson, J. Zhou, and J. M. Tiedje. 2001. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc. Natl. Acad. Sci. USA 98:9853-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Myers, C. R., and K. H. Nealson. 1988. Bacterial manganese reduction and growth with manganese oxide as the sole electron acceptor. Science 240:1319-1321. [DOI] [PubMed] [Google Scholar]
- 26.Nachin, L., M. E. Hassouni, L. Loiseau, D. Expert, and F. Barras. 2001. SoxR-dependent response to oxidative stress and virulence of Erwinia chrysanthemi: the key role of SufC, an orphan ABC ATPase. Mol. Microbiol. 39:960-972. [DOI] [PubMed] [Google Scholar]
- 27.Nies, D. H., A. Nies, L. Chu, and S. Silver. 1989. Expression and nucleotide sequence of a plasmid-determined divalent cation efflux system from Alcaligenes eutrophus. Proc. Natl. Acad. Sci. USA 86:7351-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido, H. 1998. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 27:S32-S41. [DOI] [PubMed] [Google Scholar]
- 29.Ochsner, U. A., M. L. Vasil, E. Alsabbagh, K. Parvatiyar, and D. J. Hassett. 2000. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J. Bacteriol. 182:4533-4544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pfeifer, G. P. 1997. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem. Photobiol. 65:270-283. [DOI] [PubMed] [Google Scholar]
- 31.Pomposiello, J. P., and B. Demple. 2002. Global adjustment of microbial physiology during free radical stress. Adv. Microb. Physiol. 46:320-327. [DOI] [PubMed] [Google Scholar]
- 32.Qiu, X., G. W. Sundin, B. Chai, and J. M. Tiedje. 2004. Survival of Shewanella oneidensis MR-1 after UV radiation exposure. Appl. Environ. Microbiol. 70:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rocha, E. R., G. Owens, and C. J. Smith. 2000. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 182:5059-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sancar, A. 1996. DNA excision repair. Annu. Rev. Biochem. 65:43-81. [DOI] [PubMed] [Google Scholar]
- 35.Schroeder, R. G., L. M. Peterson, and R. D. Fleischmann. 2002. Improved quantitation and reproducibility in Mycobacterium tuberculosis DNA microarrays. J. Mol. Microbiol. Biotechnol. 4:123-126. [PubMed] [Google Scholar]
- 36.Storz, G., and M. Zheng. 2000. Oxidative stress, p. 47-59. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. American Society for Microbiology, Washington, D.C.
- 37.Trent, M. S., L. M. Worsham, and M. L. Ernst-Fonberg. 1998. The biochemistry of hemolysin toxin activation: characterization of HlyC, an internal protein acyltransferase. Biochemistry 37:4644-4652. [DOI] [PubMed] [Google Scholar]
- 38.Tsaneva, I. R., and B. Weiss. 1990. soxR, a locus governing a superoxide response regulation in Escherichia coli K-12. J. Bacteriol. 172:4197-4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueshima, J., M. Shoji, D. B. Ratnayake, K. Abe, S. Yoshida, K. Yamamoto, and K. Nakayama. 2003. Purification, gene cloning, gene expression and mutants of Dps from the obligate anaerobe Porphyromonas gingivalis. Infect. Immun. 71:1170-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wandersman, C., and P. Delepelaire. 1990. TolC, an Escherichia coli outer membrane protein required for hemolysin secretion. Proc. Natl. Acad. Sci. USA 87:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welch, R. A. 1991. Pore-forming cytolysins of gram negative bacteria. Mol. Microbiol. 5:521-528. [DOI] [PubMed] [Google Scholar]
- 42.Welch, R. A., and S. Pellett. 1988. Transcriptional organization of the Escherichia coli hemolysin genes. J. Bacteriol. 170:1622-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang, Y. H., S. Dudoit, P. Luu, D. M. Lin, V. Peng, J. Ngai, and T. P. Speed. 2002. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zgurskaya, H. I., and H. Nikaido. 1999. Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA 96:7190-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.