Abstract
Cysteine and methionine availability influences many processes in the cell. In bacteria, transcription of the specific genes involved in the synthesis of these two amino acids is usually regulated by different mechanisms or regulators. Pathways for the synthesis of cysteine and methionine and their interconversion were experimentally determined for Lactococcus lactis, a lactic acid bacterium commonly found in food. A new gene, yhcE, was shown to be involved in methionine recycling to cysteine. Surprisingly, 18 genes, representing almost all genes of these pathways, are under the control of a LysR-type activator, FhuR, also named CmbR. DNA microarray experiments showed that FhuR targets are restricted to this set of 18 genes clustered in seven transcriptional units, while cysteine starvation modifies the transcription level of several other genes potentially involved in oxidoreduction processes. Purified FhuR binds a 13-bp box centered 46 to 53 bp upstream of the transcriptional starts from the seven regulated promoters, while a second box with the same consensus is present upstream of the first binding box, separated by 8 to 10 bp. O-Acetyl serine increases FhuR binding affinity to its binding boxes. The overall view of sulfur amino acid metabolism and its regulation in L. lactis indicates that CysE could be a master enzyme controlling the activity of FhuR by providing its effector, while other controls at the enzymatic level appear to be necessary to compensate the absence of differential regulation of the genes involved in the interconversion of methionine and cysteine and other biosynthesis genes.
Sulfur is a constituent of many indispensable components of the cell, such as cysteine, methionine, thiamine, biotin, lipoic acid, coenzyme A, etc. Among these compounds, cysteine has a central role, since its de novo synthesis represents the main pathway of sulfur acquisition in microorganisms and plants. Cysteine is thus the principal metabolite from which most sulfur-containing compounds are built. It is also an essential amino acid of the catalytic domain of universal proteins having iron-sulfur clusters, such as cytochromes and aconitase. Cysteine also plays a central role in protein folding, assembly, and stability via the formation of disulfide bounds. Moreover, cysteine-derived proteins, such as thioredoxin and glutathione, play a central role in the protection against oxidative stress. The second sulfur amino acid, methionine, is a key compound controlling the initiation of translation and is crucial to a variety of methyltransferase reactions.
As a result of the essential roles in metabolism, cysteine and methionine supply might be limiting for bacterial growth in different situations of importance for humans, such as pathogenic events or fermentation processes. For example, methionine availability is limiting for growth of group B streptococci in plasma (65). Sulfur amino acid biosynthesis genes have been characterized as virulence factors in Brucella melitensis (48), Haemophilus parasuis (36), and Salmonella enterica (18). In Mycobacterium tuberculosis, the cysDNC operon involved in the sulfate activation pathway forms a stress-induced operon (60), while many thiol and cysteine metabolism genes are part of the sigH regulon necessary for optimal survival of the bacterium in macrophages (52). Cysteine metabolism is also involved in the regulation of toxin in Bordetella pertussis (4), and autoinducer 2, a signaling molecule derivative of the sulfur metabolism conserved in both gram-positive and gram-negative bacteria, is involved in interspecies communication and regulation of virulence factors (54, 68). Lastly, methionine is one of the key amino acids limiting the growth of Lactococcus lactis in dairy fermentations.
In spite of their central importance, biosynthesis and regulation of cysteine and methionine have not been extensively studied in gram-positive organisms other than Bacillus subtilis (29). In this model bacterium, several genes of the methionine and cysteine biosynthesis pathway seem to be regulated by premature termination of transcription, using the S-box or T-box mechanism. The S-box regulon is composed of several transcriptional units mostly involved in cysteine and methionine metabolism and was also found in Staphylococcus aureus and Clostridium acetobutylicum (29). Promoter regions of these operons contain a conserved leader sequence motif, a transcriptional terminator, and a mutually exclusive structure designated antiterminator. The formation of these alternative structures is modulated by the leader RNA in function of S-adenosylmethionine availability (55). In addition to S-box regulation, the cysES operon, encoding serine acetyltransferase and cysteinyl-tRNA synthetase, is regulated by the T-box mechanism, which involves the uncharged cysteinyl-tRNA as effector that triggers the antitermination mechanism (28, 35). Finally, a LysR-type transcriptional regulator (LTTR), CysL, has been identified to be involved in the control of expression of the cysJI operon, which encodes sulfite reductase (34). It is noteworthy that this scheme of regulation characterized for B. subtilis is totally different from the Escherichia coli regulatory network, which involves three LTTRs and a repressor (27, 45). Relatively little is known about regulation of sulfur metabolism in the family of the Streptococcaceae. MtaR (formerly CpsY), an LTTR, might control the expression of a methionine permease in group B streptococci, but neither the signal nor the molecular targets of MtaR were characterized (65). By a comparative genomics approach, a potential DNA-binding MET-box for MtaR has been proposed (61). However, in L. lactis only one potential MET-box is present upstream of the metE gene, suggesting that other control pathways exist in this bacterium. In L. lactis subsp. cremoris, CmbR, the closest MtaR homologous regulator (31% identity), has been characterized as the activator of the metC-cysK operon (19). Furthermore, it has been suggested that CmbR (also annotated FhuR in the genome sequence from L. lactis IL1403 [5]) also regulates metA (19) and the fhu operon encoding a ferrichrome ABC transporter (19, 22). Consequently, it appears that CmbR may control genes involved in different metabolisms related to sulfur amino acid biosynthesis and iron metabolism. Interestingly, two observations indicate that FhuR is produced at a relatively high level in the cell, a feature that was shown to be important for a regulator to have substantial impact on gene regulation over a wide range of targets (26): (i) the fhuR codon index is very biased and comparable to those of many highly expressed genes (P. Renault, unpublished data), and (ii) FhuR is among the only seven regulators detected by two-dimensional gel electrophoresis (33).
These data prompted us to study extensively the targets of the FhuR/CmbR regulator in order to have a new insight in the sulfur metabolism and its implications in L. lactis. We searched by transcriptome analysis the complete set of genes regulated by FhuR or induced during cysteine starvation, and we found that FhuR activates seven transcriptional units encoding the complete set of L. lactis genes responsible for cysteine and methionine supply. Purified FhuR was shown to bind specifically to the promoter region of all these transcriptional units. Addition of O-acetyl serine (OAS), the first intermediate of cysteine biosynthesis, to purified FhuR protein modified significantly its binding to target promoters, suggesting that FhuR activity is triggered by OAS. This property has led us to propose a unique regulatory scheme for sulfur metabolism in L. lactis, where CysE, the single enzyme involved in OAS synthesis from serine and acetyl-CoA, is central for the activation of the full set of genes responsible for their supply. Lastly, the DNA-binding sequence motif recognized by FhuR was characterized and shares the typical features of DNA boxes recognized by LTTRs.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The bacterial strains used in this study are listed in Table 1. E. coli strain LLB1 was used for plasmid propagation. E. coli was grown in Luria-Bertani medium at 37°C (53). L. lactis IL1403 strains were grown at 30°C in M17 glucose medium (M17) or in chemically defined medium (CDM) (67, 72) containing trehalose (0.5 g · liter−1) as carbon source and 18 amino acids (all except aspartic acid and glutamic acid). When specified, CDM was prepared without cysteine (CDM-Cys). The effects of OAS and NAS on gene expression were evaluated by addition of 1 mM to 5 mM OAS or 4 mM NAS to exponentially growing cells (optical density at 600 nm [OD600] of 0.2) in M17 medium. When required, 5-bromo-4-chloro-3-indolyl-β-d-galactoside (40 μg · ml−1), erythromycin (5 μg · ml−1 for L. lactis, 100 μg · ml−1 for E. coli), chloramphenicol (10 μg · ml−1 for L. lactis), isopropyl-1-thio-β-d-galactopyranoside (IPTG; 40 μg · ml−1), tetracycline (5 μg · ml−1 for L. lactis), or ampicillin (100 μg · ml−1 for E. coli) was added to the culture medium.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant markers, phenotypes, and characteristics | Reference or source |
|---|---|---|
| E. coli strains | ||
| LLB1 | supE Δthi (lac-proAB) hsdD5 (F′+traD36 proAB lacIqZΔM15) pcnB | 47 |
| ER2566 | F− λ−fhuA2 [lon] ompT lacZ::T7 gene1 gal sulA11 Δ(mcrC-mrr) 114::IS10 R(mcr-73::miniTn10-TetS)2 R(zgb-210::Tn10)(TetS) endA1 [dcm] | NEB |
| JIM8551 | Apr, ER2566 containing pJIM5740 for FhuR protein expression | This work |
| L. lactis strains | ||
| IL1403 | L. lactis subsp. lactis, His− Iso− Leu− Val− | 14 |
| JIM8475 | Emr, IL1403 containing pJIM1062 integrated at the PpurH locus, His− Iso− Leu− Val− | This work |
| JIM8494 | IL1403 ΔfhuR after homologous recombination of pJIM5702, His− Iso− Leu− Val− | This work |
| JIM8499 | Emr, IL1403 containing pJIM5706 integrated at the PmetB2 locus, His− Iso− Leu− Val− | This work |
| JIM8500 | Emr, IL1403 containing pJIM5707 integrated at the PfhuR locus, His− Iso− Leu− Val− | This work |
| JIM8539 | Emr, IL1403 containing pJIM5724 integrated at the PyriD locus, His− Iso− Leu− Val− | This work |
| JIM8543 | Emr, IL1403 containing pJIM5728 integrated at the PyjgC locus, His− Iso− Leu− Val− | This work |
| JIM8545 | Emr, IL1403 containing pJIM5737 integrated at the PyhcE locus, His− Iso− Leu− Val− | This work |
| JIM8546 | Emr, IL1403 metB2 after homologous recombination of pJIM5732, His− Iso− Leu− Val− | This work |
| JIM8547 | Emr, IL1403 cysK after homologous recombination of pJIM5733, His− Iso− Leu− Val− | This work |
| JIM8657 | Emr, IL1403 containing pJIM5751 integrated at the PcysM locus, His− Iso− Leu− Val− | This work |
| JIM8658 | Emr, IL1403 containing pJIM5752 integrated at the PplpA locus, His− Iso− Leu− Val− | This work |
| JIM8659 | Emr, IL1403 containing pJIM5754 integrated at the PcysD locus, His− Iso− Leu− Val− | This work |
| JIM8662 | Emr, IL1403 yhcE after homologous recombination of pJIM5750, His− Iso− Leu− Val− | This work |
| JIM8681 | Emr, IL1403 containing pJIM5760 integrated at the PcysE locus, His− Iso− Leu− Val− | This work |
| Plasmids | ||
| pGEM-T easy | Apr, M13ori pBR322ori, linear T-overhangs vector | Promega |
| pTYB1 | Apr, expression vector used in the IMPACT-CN protein purification system | NEB |
| pGhost 8 | Tetr, thermosensitive replicative plasmid in L. lactis | 51 |
| pGhost 9 | Emr, thermosensitive replicative plasmid in L. lactis | 51 |
| pJIM2242 | Emr, ori+ ΔrepA, integrative vector in L. lactis | 32 |
| pJIM2374 | Emr, ori+ ΔrepA, integrative promoter probe vector containing luxAB genes | 15 |
| Plasmid for protein expression | ||
| pJIM5740 | 942-bp fragment carrying the fhuR gene cloned into NdeI-SapI sites of pTYB1 | This work |
| Plasmid for gene deletion | ||
| pJIM5702 | SalI fusion of pGhost 9 and pGEM-T easy containing 713-bp fragment upstream and 746-bp fragment downstream fhuR gene ligated in BamHI | This work |
| Plasmids for promoter-lux fusion | ||
| pJIM1062 | PpurH-lux, SalI fusion of pJIM2374 and pGEM-T easy containing PpurH on a 442-bp fragment | This work |
| pJIM5706 | PmetB2-lux, SalI fusion of pJIM2374 and pGEM-T easy containing PmetB2 on a 680-bp fragment | This work |
| pJIM5707 | PfhuR-lux, SalI fusion of pJIM2374 and pGEM-T easy containing PfhuR on a 471-bp fragment | This work |
| pJIM5724 | PyriD-lux, SalI fusion of pJIM2374 and pGEM-T easy containing PyriD on a 715-bp fragment | This work |
| pJIM5728 | PyjgC-lux, NcoI fusion of pJIM2374 and pGEM-T easy containing PyjgC on a 692-bp fragment | This work |
| pJIM5737 | PyhcE-lux, SalI fusion of pJIM2374 and pGEM-T easy containing PyhcE on a 675-bp fragment | This work |
| pJIM5751 | PcysM-lux, SalI fusion of pJIM2374 and pGEM-T easy containing PcysM on a 691-bp fragment | This work |
| pJIM5752 | PplpA-lux, NcoI fusion of pJIM2374 and pGEM-T easy containing PplpA on a 829-bp fragment | This work |
| pJIM5754 | PcysD-lux, SalI fusion of pJIM2374 and pGEM-T easy containing PcysD on a 723-bp fragment | This work |
| pJIM5760 | PcysE-lux, SphI fusion of pJIM2374 and pGEM-T easy containing PcysE on a 697-bp fragment | This work |
| Plasmids for gene inactivation | ||
| pJIM5732 | SalI fusion of pJIM2242 and pGEM-T easy containing 668-bp metB2 internal fragment | This work |
| pJIM5733 | SalI fusion of pJIM2242 and pGEM-T easy containing 602-bp cysK internal fragment | This work |
| pJIM5750 | SalI fusion of pJIM2242 and pGEM-T easy containing 694-bp yhcE internal fragment | This work |
DNA manipulation procedures.
Plasmids and total DNA were prepared as described previously (50, 53, 66). Procedures for DNA manipulations and E. coli transformation were performed as described by Maniatis et al. (53). All enzymes for DNA technology were used according to the manufacturers' specifications. Electrotransformation of L. lactis was performed as described by Holo and Nes (38). The oligonucleotides used in this work (Table 2) were synthesized by Sigma Genosys. DNA sequencing was performed on both strands using a fluorescent sequencing procedure (Perkin-Elmer Biosystem).
TABLE 2.
Primers used for PCR amplificationa
| Primer group and gene name | Primer pair sequences (5′-3′) |
|---|---|
| Primers for fhuR gene deletion | |
| fhuR up | CCTGATTTGATTATTACAGTTGATAAAG, TGGCGACGAGGATCCGTAATTGTTTAAT |
| fhuR down | GAAGTCTACTTTGGATCCTATGAATTTTA, GGACTGATTGCTACTAGTATTCTTGTTCC |
| Primers for fhuR gene amplification | |
| fhuR | TAAGGACTTTATCATATGAATATTAAACAATTACG, GAGTATTTATTTTAGCTCTTCCGCAAAATTCATAG |
| Primers for promoter amplification | |
| cysD | CTTCGGGTCATCTGGATTGA, AATGTTGCTCGTAAACATCC |
| cysE | GGTAACTCCATGATAAAGCT, CAATCAGGACAATGAAGAAT |
| cysM | CCGCAGCGTTAATAATTTCTT, CATTCCACCACTAGCGGGTG |
| fhuR | GCTTAGGAGAATTTGATGCAG, CGCCTTCAATAGTTAAAGTCGC |
| metB2 | GGTGGTTCAGGAGGAGCATTAGC, GAACCGCATGAATTCCAGCAAG |
| plpA | GCAGCGATAGCAACAATTAC, CCGTTAATAAACGGACATTG |
| purH | ACCAAGTGGTCGTAAAGTTGA, ACGGCTTTCGTTCCACCCGTTG |
| yhcE | CAGCATTTGAACCTGAACGAGT, CTAAGTGCCACCACGAACGATT |
| yjgC | GAAGTTCAGTCACGAGCTGACA, CCGCACGAGCAACCTCAATATC |
| yriD | GCGATTAAGGCAATTGCAAGTG, AATGACTGGACTTGTCAACTCA |
| Primers for gene inactivation | |
| cysK | GATTCGGCAAGCCGAAGCAG, GCGGTCTCTAAAGCCTCATC |
| metB2 | GACTTGCTGGAATTCATGCGG, TCATCCCTCCAAAGGCAGAC |
| yhcE | CGTTCGTGGTGGCACTTAGA, CCTAACACAAGCAAGACATC |
| Primers for Q-PCRb | |
| cadA | AGATAAGGTGGTCGATTTACCG, CGTATCAAGTACCACTGCCTCA |
| cysD | CAGCGACTCAAGCGATGTACTA, TACTCCCAAGTGTTCCCAAGTG |
| guaC | TATGGGAGCAACAATGGTAATG, TTTGATATTCTGAAGCCGAACC |
| mesJ | AATGCACGTGATTTCCGTTATC, CCCTGAAGACTTCTCAAAGGAC |
| metA | CCTTGATAAACCGCATAATTCA, GAAGGACTGAAACCCAATCTTC |
| metB1 | GTTGCTAGAGCAAATTGTCCTG, CAACGACCTATCAACATCCAGA |
| metB2 | GCCTTGGAAGAATTAATTGCAG, TTCCACCATAAACATCATCAGC |
| tuf | CGCGAACGTGGTATCACA, GTCCATTTGGGCAGCACC |
| ydcD | TAGTGAACCGGCAAATAAGGTC, GAGCCAGATTTGGTTCTGGTAG |
| yjiB | ACATTGAAGCCGGAAATACTTG, AGTGGCCTCGATAATTTGACTG |
| ykhH | GGAGCAATCAGAAACCAAGAAC, CAATACCCACAAGAACAAGCAG |
| ymcF | ATGCTCAAGGTTCTGTTTCGAG, TTCGCTAAGTCAACGGTTGTAG |
| yrfD | GGTTGCTGCTATTGGTAACTCG, AATCATACAGTCCACCCTCACC |
| yrjB | TTTAGGCCTTGAAGATCTTGGT, TTCAAGCCCTTTGACTTTATCC |
| ytjE | CAATTCCTGGAACAACTCCTTC, CGCAGACTATGGAGTTTATGGA |
| Primers for start transcription mapping | |
| cysD | CAATGGCATCATAATCAATC, CCCGGATTGCCCAAAGTTTC |
| cysM | GCATCAATAATCTCAGCTCC, CATGAATTTCAGGATTTGCG |
| fhuR | CTAATTCAAGTTTATTGAGC, GAGCGATTTTGTTTATTGAG |
| metA | ACCATAGCGAGCATAAAGTG, CAACAAATATGCAGACTTGA |
| metB2 | CAGTGTTAAGGCATCATGAGC, GCTGAAATTTCCTTAATGTCC |
| plpA | GACGAATTGGTGAAATTGTTG, CCAATTGCTTTCAAATCACC |
| yhcE | GCCAACGGTCTGAACGACCATC, GGAATTAACGAAGGTGAAGG |
| yjgC | GTTCCAATCTTGAATTGAC, GCTGGATTACCAATTGTCG |
| Primers for gel retardation probe | |
| cysD | GATAGAGCGGAACGGCGCGT, AATGTTGCTCGTAAACATCC |
| cysM | GTTGAAGTTTAACAATTGGAG, CCGCAGCGTTAATAATTTCTT |
| fhuR | GCTTAGGAGAATTTGATGCAG, CGCCTTCAATAGTTAAAGTCGC |
| metA | CGCGGCATGAGATTGACCACC, GATTAGCCTGGATTCCCCTAG |
| metB2 | CACGTCGTGATGAAGCGGCT, CGGAACAGATACTGCTCCTG |
| plpA | CCTACGCTGGCATTACCCAG, CGAGAACCATTCCGTCAAGA |
| purH (int.) | CGCATTCCTTGTTTAAGGTCG, GTCACATGAAATCAATGACAG |
| yhcE | CCTTTGTATCAACTGGATTG, GTTGCCCTTCTGAATAAGCC |
| yjgC | CAAGTCGGACTTGACCAAGC, GAACCTGATGAGCATGCAAC |
| Primers for substitution analysis of metB2 promoter FhuR box | |
| metB2 (A) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (B) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTCTAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (C) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTACAAGATTTTTTTATAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (D) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTCCCCATAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (E) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGACCCTTTTATAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| Primers for fhuR gene deletion | |
| metB2 (F) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATCCACTTTTTTTATAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (G) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGAATTAACTTTTTTTATGTATACAAAAACG |
| metB2 (H) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGAATTAACTTTTTTTATGTATCAAAAAACG |
| metB2 (I) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGAATTAACTTTTTTTATACCCAAAAAAACG |
| metB2 (J) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGAATTAACTTTTTTTCTGTATAAAAAAACG |
| metB2 (K) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGAATTAACTTTCCCCATGTATAAAAAAACG |
| metB2 (L) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGAATTCCACTTTTTTATGTATAAAAAAACG |
| metB2 (M) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGAACTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (N) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCAGACTTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (O) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGCCACATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (P) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAGAAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (Q) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATAACAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (R) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTATCGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (S) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTTACAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (T) | CACGTCGTGATGAAGCGGCT,TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTTCATAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (U) | CACGTCGTGATGAAGCGGCT,TAAAGATATCACGAAAGTGTGCCCAGTGCTATAAGATTTTTCTATAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| metB2 (V) | CACGTCGTGATGAAGCGGCT, TAAAGATATCACGAAAGTGTGCCCAGTGCTCTAAGATTTTTTTATAGCAGAATTAACTTTTTTTATGTATAAAAAAACG |
| Primers for substitution analysis of cysD promoter FhuR box | |
| cysD (A) | CATGGACTTGCAAAGTGTCG, CAACCTTGAGTTGTATTTTTTTATTTCTACTTTTAAAAGTTCTATCAGCTCCATTAAAAAATCATATCGATAG |
| cysD (B) | CATGGACTTGCAAAGTGTCG, CAACCTTGAGTTGTATTTTTTTATTTCTACTTTTAAAAGTTCTATCAGCTCCAGTAAAAAATCATATCGATAG |
| cysD (C) | CATGGACTTGCAAAGTGTCG, CAACCTTGAGTTGTATTTTTTTATTTCTACTTTTAAAAGTTCTATCAGCTCCATTAAAAAATCATGTCGATAG |
| cysD (D) | CATGGACTTGCAAAGTGTCG, CAACCTTGAGTTGTATTTTTTTATTTCTACTTTTAAAAGTTCTATCAGCTCCATGGGGAAATCATATCGATAG |
| cysD (E) | CATGGACTTGCAAAGTGTCG, CAACCTTGAGTTGTATTTTTTTATTTCTACTTTTAAAAGTTCTATCAGCTCCATTAAAGGGTCATATCGATAG |
| cysD (F) | CATGGACTTGCAAAGTGTCG, CAACCTTGAGTTGTATTTTTTTATTTCTACTTTTAAAAGTTCTATCAGCTCCATTAAAAAAGTGGATCGATAG |
The BamHI restriction sites designed for fhuR gene deletion in L. lactis IL1403 strain are in bold. The NdeI-SapI restriction sites designed for fhuR gene amplification used to purify the protein are in bold. int., internal fragment of the purH gene. The substitution bases designed into the FhuR box of the metB2 and cysD genes are underlined. (A), probe control without substitution; (B-V), probes with substitutions in the FhuR box.
Q-PCR, quantitative PCR.
Gene inactivation in L. lactis.
The chromosomal fhuR gene was deleted in L. lactis strain IL1403 as follows. DNA fragments of 713 bp and 746 bp carrying, respectively, the upstream and the downstream regions of the fhuR gene were generated by PCR. These fragments contain a BamHI restriction site at their 3′ and 5′ ends, respectively, introduced in fhuR up and fhuR down primers (Table 2). The PCR-amplified products were digested with BamHI, ligated, and cloned into the pGEM-T easy cloning vector (Promega). The absence of PCR-induced mutations in the insert corresponding to the fused upstream and downstream regions of the fhuR gene was verified by sequencing. This plasmid was fused to the lactococcal thermosensitive vector pGhost 9 in SalI to yield pJIM5702. In L. lactis IL1403, the fhuR gene was deleted using pJIM5702 by double crossing over as described by Biswas et al. (3) to give the JIM8494 strain. PCR amplification and DNA sequencing were performed to verify the effective fhuR gene deletion in the JIM8494 strain.
The metB2, cysK, and yhcE genes were inactivated by the insertion of pJIM5732, pJIM5733, and pJIM5750, respectively, containing internal fragments of these genes. An internal fragment of the metB2, cysK, and yhcE genes was amplified by PCR (Table 2), cloned into the pGEM-T easy vector, and then fused with the L. lactis integration vector pJIM2242 as described in Table 1. The resulting plasmids were integrated into the chromosome of L. lactis IL1403 by single crossing over as described earlier (32), yielding strains JIM8546, JIM8547, and JIM8662 inactivated for the metB2, cysK, and yhcE genes, respectively.
Construction of lux transcriptional fusions and determination of activities in L. lactis.
The reporter luxAB genes were placed under the control of several potential promoter regions and integrated at their respective loci as described earlier (31). Briefly, chromosomal DNA from L. lactis IL1403 was used as template to amplify 440-bp to 830-bp DNA fragments containing the potential promoters of the cysD, cysE, cysM, fhuR, metB2, plpA, yhcE, and yjgC genes. The PCR fragments were cloned into pGEM-T Easy vector, sequenced, and fused with the lactococcal integrative vector pJIM2374 (Table 1). The resulting plasmids were integrated by homologous recombination in the L. lactis IL1403 chromosome by single crossing over using the pGhost 8 as helper (24). Strains carrying single and a nontandem copy of the integrated plasmid were screened by PCR with specific primers. The resulting strains (Table 1) contained the lux genes downstream of the cloned promoter region, followed by a copy of the intact gene. Luciferase assays were carried out as described by Guédon et al. (31). Luciferase activity was measured throughout the culture growth, and values reported below in Table 3 correspond to the mean of the luciferase activity determined at an OD600 of 0.4 from at least three independent measures.
TABLE 3.
Genes significantly induced by the FhuR regulatora
| Gene | Function(s) | Induction WT vs ΔfhuR
|
Induction +/− cysteine
|
Transcriptional organization | ||||
|---|---|---|---|---|---|---|---|---|
| Array | QRT-PCR | Lux | Array | QRT-PCR | Lux | |||
| cysD | O-Acetylhomoserine/O-acetylserine sulfhydrylase | 2.4 | 4.1 | /b | 13.1 | / | 23 | cysD |
| cysM | Cysteine synthase | 2.5 | / | / | 10.8 | / | 76 | cysM |
| metB2 | Cystathionine beta/gamma-lyase | 44 | 63.4 | 83 | 12.1 | / | 34 | metB2-cysK |
| cysK | Cysteine synthase | 29 | / | / | 11.4 | / | / | metB2-cysK |
| metA | Homoserine O-succinyltransferase | 2.4 | 2.9 | / | 5.2 | / | / | metA-metB1-ytjE |
| ytjE | Aspartate aminotransferase | 2.4 | 3.8 | / | 4.7 | / | / | metA-metB1-ytjE |
| yhcE | Homocysteine methyltransferase | 12 | / | 45 | 5.9 | / | 9 | yhcE |
| plpA | Methionine ABC transporter (substrate-binding protein) | 26.8 | / | / | 5.8 | / | 8 | plpA-plpB-plpC-plpD-ydcB-ydcC-ydcD |
| plpB | Methionine ABC transporter (substrate-binding protein) | 29.9 | / | / | 4.5 | / | / | plpA-plpB-plpC-plpD-ydcB-ydcC-ydcD |
| plpC | Methionine ABC transporter (substrate-binding protein) | 22.5 | / | / | 4.6 | / | / | plpA-plpB-plpC-plpD-ydcB-ydcC-ydcD |
| plpD | Methionine ABC transporter (substrate-binding protein) | 6.2 | / | / | 3.7 | / | / | plpA-plpB-plpC-plpD-ydcB-ydcC-ydcD |
| ydcB | Methionine ABC transporter (ATP-binding protein) | 1.8 | / | / | 3.0 | / | / | plpA-plpB-plpC-plpD-ydcB-ydcC-ydcD |
| ydcD | Cobalt transport protein CbiQ | 2.2 | 2.6 | / | 2.6 | / | / | plpA-plpB-plpC-plpD-ydcB-ydcC-ydcD |
| yjgC | Cystine or cysteine ABC transporter (substrate-binding protein) | 3.0 | / | 8 | 5.1 | / | 22 | yjgC-yjgD-yjgE |
| yjgD | Cystine or cysteine ABC transporter (permease protein) | —c | / | / | 2.8 | / | / | yjgC-yjgD-yjgE |
| yjgE | Cystine or cysteine ABC transporter (ATP-binding protein) | — | / | / | 2.9 | / | / | yjgC-yjgD-yjgE |
| purC | Phosphoribosylaminoimidazole-succinocarboxamide synthetase | 3.5 | / | / | — | / | / | purC |
| purD | Phosphoribosylamine-glycine ligase | 3.0 | 1.0 | / | — | / | / | purD-purE-purK |
| purH | Bifunctional purine biosynthesis protein PurH | 3.0 | 0.9 | / | — | / | 1.1 | purH |
| yriD | Hypothetical protein | 2.1 | / | / | — | / | 1.1 | yriD |
| guaC | GMP reductase | 2.1 | / | / | — | / | / | guaC |
| malQ | 4-alpha-glucanotransferase (amylomaltase) | 4.2 | / | / | — | / | / | ygjD-malQ-glgC-glgD-glgA-glgP-amyX |
| tra905 | Transposase of IS905 | 0.2 | / | / | — | / | / | ymcF |
| ymcF | Unknown protein | 0.1 | 0.11 | / | — | / | / | tra905 |
| ykhH | Unknown protein | 0.3 | 0.86 | / | — | / | / | ykhG-ykhH |
| ysdC | Hypothetical protein | — | / | / | 0.02 | / | / | ysdC |
| yrjB | Oxidoreductase | — | / | / | 0.3 | 0.46 | / | yrjB-yrjC-yrjD |
| yrjC | Iron-binding oxidase subunit | — | / | / | 0.3 | / | / | yrjB-yrjC-yrjD |
| yrjD | Hypothetical protein | — | / | / | 0.3 | / | / | yrjB-yrjC-yrjD |
| nifJ | Pyruvate-flavodoxin oxidoreductase | — | / | / | 0.4 | / | / | nifJ |
| yajH | Unknown protein | — | / | / | 0.4 | / | / | yajH |
| cadA | Cadmium efflux ATPase | — | / | / | 0.5 | 0.53 | / | cadA |
| yqeI | Cation transport protein | — | / | / | 2.9 | / | / | rarA-yqeI |
| yjiB | Amino acid aminohydrolase | — | / | / | 3.1 | 4.46 | / | yjiB |
| yrfD | Amino acid antiporter | — | / | / | 3.4 | 1.51 | / | octA-yrfD |
| mesJ | Cell cycle protein MesJ | — | / | / | 3.9 | 1.28 | / | yacG-mesJ-hpT |
| yliF | Unknown protein | — | / | / | 3.5 | / | / | yliC-yliD-yliE-yliF |
| pi230 | Prophage pi2 protein 30, terminase | — | / | / | 4.3 | / | / | unit pi2 prophage |
| pi307 | Prophage pi3 protein 07 | — | / | / | 2.5 | / | / | unit pi3 prophage |
| ps105 | Prophage ps1 protein 05, DNA primase | — | / | / | 2.8 | / | / | unit ps1 prophage |
| ps106 | Prophage ps1 protein 06 | — | / | / | 2.6 | / | / | unit ps1 prophage |
| ps107 | Prophage ps1 protein 07 | — | / | / | 3.1 | / | / | unit ps1 prophage |
| ps114 | Prophage ps1 protein 014 | — | / | / | 3.6 | / | / | unit ps1 prophage |
Results show genes that were induced in CDM trehalose (induction of wild type [WT] versus ΔfhuR) or in absence of cysteine in CDM trehalose medium (induction +/− cysteine) during exponential growth of L. lactis IL1403 as revealed by transcriptome (array column), real-time quantitative RT-PCR (QRT-PCR column), and luciferase activity analysis (Lux column).
/, not determed.
—, not significantly regulated.
RNA isolation.
Total RNA was isolated from IL1403 wild type and JIM8494 fhuR mutant strains cultivated in CDM and from IL1403 wild-type strain cultivated in CDM with or without cysteine. When the OD600 reached 0.4, cells from 25-ml portions of the cultures were pelleted, rapidly frozen in liquid nitrogen, and stored at −80°C. Frozen pellets were resuspended in solution containing water (400 μl) and phenol-chloroform (500 μl; 5/1; Sigma) and transferred in tubes containing 500 mg of glass beads (Sigma), sodium dodecyl sulfate (SDS; 30 μl; 10%), and Na-acetate (30 μl; pH 4.8; 3 M). Cells were broken in a Savant FastPrep FP120 apparatus (40 seconds at 5.0), and the mixtures were centrifuged to remove beads and cellular solid fractions (13,000 rpm, 10 min, 4°C). Total RNA from the upper liquid phase was extracted using a classical phenol-chloroform extraction method (23). To obtain very pure RNA containing low amounts of small stable RNA molecules (tRNAs and 5S rRNA), additional purification steps were carried out, as instructed by the manufacturer, using the High Pure RNA isolation kit (Roche). Moreover, eventual residual chromosomal DNA was removed by treating total RNA preparations with RNase-free DNase I (Roche) according to the manufacturer's protocol. After a precipitation step, the RNA concentration was determined by absorption at 260 nm, and quality of RNA preparations was checked (i) by determining the absorbance ratio at 260 and 280 nm, (ii) by visualizing the integrity of the 23S and 16S rRNA bands on an agarose gel, and (iii) by verifying the absence of DNA contamination by PCR.
Transcriptional start mapping.
Starts of transcription of the cysD, cysM, fhuR, metA, metB2, plpA, yhcE, and yjgC genes were determined using the 5′/3′ RACE kit (Roche) as recommended by the supplier with RNA preparations previously described as template. Primers used are listed in Table 2.
DNA microarray analysis.
To determine the lactococcal fhuR/cysteine regulon, genome-wide expression profiles were established, using a commercial DNA microarray (HO50C; Eurogentec) containing most of the L. lactis IL1403 genes (2,072 open reading frames spotted in duplicate). Synthesis of CyDye fluorescently labeled cDNAs was carried out with the CyScribe cDNA postlabeling kit (Amersham) essentially according to the manufacturer's protocol. cDNA synthesis was carried out with 20-μg total RNA samples at 42°C during 12 h in a 20-μl reaction mixture containing RNA, random nonamer primers, reverse transcriptase buffer, dithiothreitol (DTT), nucleotide mix, aminoallyl-dUTP, and Superscript III reverse transcriptase (Invitrogen). RNA templates were then degraded by NaOH. Prior to purification with a CyScribe GFX column, aminoallyl-modified cDNA solutions were neutralized with HEPES. Labeling of aminoallyl-modified cDNAs with Cy3 or Cy5 dye and purification of the CyDye fluorescently labeled cDNAs were carried out following the manufacturer's protocol. The Cy3- and Cy5-labeled cDNAs were then pooled and dried to obtain, finally, 10 μl of probe. The efficiency of labeling was monitored by measuring the probe absorbance at 260 nm, 550 nm, and 650 nm.
DNA microarrays were prehybridized for 1 h at 42°C with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% SDS, and 0.1 mg · ml−1 bovine serum albumin, washed, and dried. Prior to hybridization, 9 μl of pooled Cy3- and Cy5-labeled cDNAs and 5 μl of heat-denatured salmon sperm DNA (10 mg · ml−1) were heated at 98°C for 2 min and rapidly cooled on ice. Microarrays were hybridized with solutions containing 40 μl of EGT hybridization buffer (Eurogentec), Cy3- and Cy5-labeled cDNAs, and heat-denatured salmon sperm DNA. After depositing the hybridization mixtures onto the microarrays, a coverslip was applied and the arrays were placed in hybridization chambers (CMT; Corning Inc., Corning, NY) for 16 h at 37°C. After hybridization, the microarrays were washed in 0.2× SSC and 0.1% SDS (10 min with agitation), rinsed in 0.2× SSC (5 min with agitation), subsequently rinsed in water, and finally dried. DNA microarrays were scanned through two channels for the respective fluorescent dyes using a confocal laser scanner (Virtek) to monitor the fluorescence intensities. For each spot, signal and local background intensities were determined with Imagene software (version 5.1; BioDiscovery, Inc.). A local background-subtracted signal value was calculated for each spot. Raw data were normalized with the preP software using a local normalization (lowess method) to reduce fluorescence bias introduced by the labeling reactions as well as by the differences in fluorescence intensity between the two dyes. A dye ratio corresponding to the mean of the normalized Cy3 and Cy5 signals and a z-test on the mean of normalized signals was carried out to compare expression profiles of IL1403 and its derivative fhuR mutant or those of IL1403 cells grown with or without cysteine, as described by Garcia de la Nava et al. (21). Genes considered significantly differentially expressed corresponded to those for which (i) the P value was lower than 0.01 and (ii) the expression level difference was higher than 2 (induced genes) or lower than 0.5 (repressed genes). Full data sets are available at http://genome.jouy.inra.fr/efp/base/www/.
Real-time quantitative RT-PCR amplification.
The quantitative reverse transcription-PCR (RT-PCR) approach was carried out using first-strand cDNA as template. cDNA was synthesized from 20-μg RNA samples as described in the previous section but without using aminoallyl-dUTP. Specific primers for each gene (Table 2) were designed using eprimers software (EMBOSS). PCR was carried out in a 25-μl volume containing 10 μl cDNA (diluted at 1/5,000), specific primers (0.2 μM each), and 12.5 μl ABsolute QPCR SYBR Green mix (ABgene). Reactions were run on an ABI 7700 instrument (Applied Biosystems) using the following cycling parameters: DNA polymerase activation at 95°C for 15 min, 40 cycles of denaturation at 94°C for 15 s, and extension at 60°C for 1 min. Measures were taken for each strain from cDNA synthesized from RNA extracted from three independent cultures and performed in triplicate for each gene (Perkin). The results were normalized by using the L. lactis tuf gene, coding for the elongation factor TU, as control.
Protein purification.
FhuR protein was purified using the IMPACT system (New England BioLabs [NEB]). The fhuR gene from L. lactis IL1403 was amplified by PCR using fhuR primers (Table 2) and fused in NdeI/SapI sites to the intein-CBD sequence in the pTYB1 vector (NEB) to give pJIM5740. Integrity of the cloned fragment was verified by DNA sequence. FhuR protein was expressed from pJIM5740 in E. coli strain ER2566 (NEB) grown at 37°C in 1 liter of LB containing ampicillin at 100 μg · ml−1. Protein expression was induced overnight at 16°C by adding 1 mM IPTG. Harvested cells were resuspended at 4°C in 50 ml of lysis buffer L (20 mM Tris [pH 9.0], 500 mM NaCl, 1 mM EDTA). The cells were then disrupted by sonication (30 min) with a Vibra cell disrupter (Bioblock Scientific). The lysate was centrifuged at 18,000 × g at 4°C for 30 min, and the supernatant was loaded onto a chitin column equilibrated in buffer L. FhuR protein was eluted in cleavage buffer (20 mM Tris [pH 9.0], 500 mM NaCl, 1 mM EDTA, 50 mM DTT). Finally, the purified protein was stored at −20°C in 30% glycerol. The purity of FhuR was confirmed by 10% SDS-polyacrylamide gel electrophoresis, and the protein concentration (20 μg · ml−1) was determined by the Bradford method (Bio-Rad) as recommended by the supplier.
Gel retardation assay.
DNA probes of about 400 bp corresponding to the promoter regions of cysD, cysM, fhuR, metA, metB2, plpA, yhcE, and yjgC were generated by PCR using specific primers (Table 2) and labeled at the 5′ end with [γ-32P]ATP by the T4 polynucleotide kinase (NEB). Unincorporated nucleotides were removed with the NucleoSpin PCR purification kit (Macherey Nagel). FhuR-DNA complexes were formed in 10 μl by incubating the 32P-labeled probes (10 × 10−15 mole of DNA) with different amounts of FhuR purified protein (0.6 μM to 3.6 μM) in binding buffer (20 mM Tris [pH 7.8], 100 mM KCl, 0.5 mM EDTA, 2 mM MgSO4, 1 mM DTT, 10% glycerol) in the presence of 0.1 μg · μl−1 of poly(dI-dC). When required, OAS (Sigma) was added to the binding reaction mixtures at 60 mM (final). Reaction mixtures were incubated at 25°C for 15 min, and electrophoresis was carried out at 4°C in TAM buffer (6 mM Tris [pH 7.8], 10 mM NaOAc, 4 mM MgOAc, 1 mM EDTA) as described by Xu and Marians (78). metB2 and cysD promoter fragments with specific substitutions were obtained by PCR using mutated primers listed in Table 2.
RESULTS
Determination of FhuR/cysteine-regulated genes by DNA microarray analysis.
In order to identify genes regulated specifically by FhuR or by cysteine depletion, the expression of 2,072 genes was followed by the use of DNA microarrays. Two sets of experiments were carried out. The first compared the transcription profiles in L. lactis IL1403 wild type and its derivative with fhuR deleted, grown in CDM in the presence of cysteine. The second compared the profiles of the wild-type strain grown in CDM with and without cysteine. In Table 3 are reported the genes whose expression levels statistically and significantly differed by at least a factor of 2, a list established by filtering the results as described in Materials and Methods.
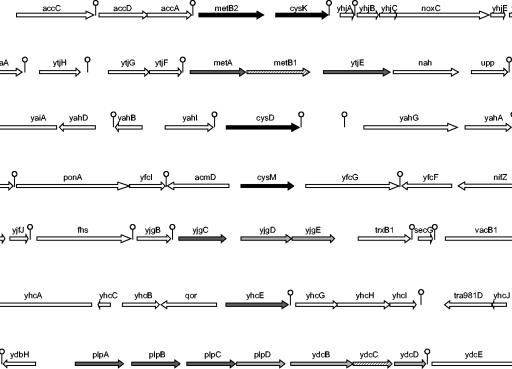
Comparison of transcription profiles from the fhuR mutant with the wild-type strain revealed that 23 genes were significantly differentially expressed, 3 of them being repressed and the others being activated by FhuR. Similar analysis of the effect of cysteine depletion indicated that 34 genes were affected by environmental cysteine availability and, in particular, 27 genes were differentially induced in response to the absence of this amino acid in the growth medium. A set of 14 genes is common to the two experiments. These 14 genes are clustered in seven different locations in the chromosome and may thus reflect the differential expression of seven transcriptional units (Fig. 1). All genes from a cluster were regulated in a similar way. A search for the potential functions of these genes strongly suggests their involvement in methionine and cysteine metabolism. In addition to the already-studied metB2-cysK operon in L. lactis, cysD, cysM, metA, metB1, and ytjE have orthologous counterparts well characterized in several bacteria. YhcE belongs to a protein family of approximately 350 amino acids that is homologous to the carboxy-terminal domain of the approximately 750-amino-acid-long homocysteine methyl transferases (MetE family). However, no enzyme of this family has been biochemically studied yet. Lastly, the two remaining transcriptional units, plpABCD-ydcBD and yjgCDE, encode transport systems likely involved in methionine and cysteine or cystine uptake, respectively, as suggested by their homology to YmtK and MetQ of B. subtilis (8, 39).
FIG. 1.
Organization of the L. lactis genes activated by FhuR and upon cysteine starvation. Activated genes are presented as black arrows (more than 10-fold modulation) or dark and light gray arrows (5- to 10-fold and 3- to 5-fold, respectively). Hatched arrows are genes not present on the DNA microarrays. Lollypops are predicted terminators. Two of the genes (yjgDE) were activated by FhuR but not by cysteine starvation.
In addition to the genes differentially expressed in the two experiments, several genes appeared to be specifically regulated either by FhuR or by cysteine absence: (i) in the absence of FhuR, genes involved in purine metabolism (purC, purD, purH, and guaC), glycogen metabolism (malQ), and an unknown protein (yriD) were down-regulated, suggesting their activation by FhuR. Conversely, genes coding for two unknown proteins (ymcF and ykhH) and a transposase (tra905) were repressed by FhuR. (ii) In the absence of cysteine, 11 genes appeared to be induced, including genes encoding an amino acid antiporter (yrfD), an amino acid aminohydrolase (yjiB), a cation transporter (yqeI), a protein potentially implicated in the cell cycle control (mesJ), and several unknown and phage proteins. Moreover, seven genes might be repressed in the absence of cysteine, including genes involved in cation metabolism and oxidoreduction (yrjBCD, nifJ, and cadA) and two genes of unknown function.
Analysis of FhuR/cysteine-regulated genes by real-time quantitative RT-PCR and transcriptional fusions.
The results obtained by the transcriptome analysis were checked by two approaches, real-time quantitative RT-PCR and reporter gene fusions.
First, RNAs extracted from independent bacterial cultures of the wild-type (IL1403) and fhuR (JIM8494) strains grown in CDM or the wild-type strain grown in CDM with and without cysteine were used as template in the quantitative RT-PCR (Table 3). This analysis confirmed that the levels of transcription of cysD, metA, metB1, metB2, ydcD, and ytjE genes were higher in the wild type than in the fhuR mutant strains, while that of the ymcF messenger was lower. Furthermore, the ratios calculated by quantitative RT-PCR were generally close (within a factor of 2) to those obtained with the DNA microarray technique. However, purD, purH, and ykhH genes were not found to be differentially expressed in these experiments and were thus not analyzed further.
Second, luciferase activity produced from transcriptional fusions of the luxAB genes under the control of cysD, cysE, cysM, metB2, plpA, purH, yhcE, yjgC, and yriD promoters was measured from IL1403 cells cultivated in CDM without versus with cysteine (Table 3). As expected from DNA microarray experiments, the cysD, cysM, metB2, plpA, yhcE, and yjgC genes were expressed at a higher level in the absence of cysteine, while cysE, purH, and yriD were constitutively expressed (Table 3 and data not shown). These results confirmed that the transcription of most genes implicated in methionine and cysteine supply, including uptake system and biosynthesis pathways, is controlled by a cysteine-dependent signal and by the FhuR regulator.
Functional analysis of several FhuR-regulated genes.
In order to better define the role of FhuR, we have further characterized the role of several genes of its regulon. For this purpose, we inactivated yhcE, metB2 (orthologue of metC of L. lactis MG1363), and cysK by insertion of a plasmid carrying an internal fragment of these genes. Washed precultures of these mutants were then streaked on CDM-agar plates complemented with different sulfur sources (methionine, cysteine, and/or sulfide). First, none of the strains was able to grow in the absence of methionine, as is the case for most dairy lactococcal strains (12). Second, metB2 and cysK mutants were unable to grow on methionine as sole sulfur source, indicating that these genes are essential for the conversion of methionine to cysteine. The cellular function of MetB2 and CysK would be similar to that of YrhA and YrhB of B. subtilis, their best homologues in B. subtilis, where they are involved in the recycling of methionine to cysteine (1; I. Martin-Verstraete, personal communication). Interestingly, these two mutants could grow on cysteine-depleted CDM if sulfide was added, indicating that sulfide could be used effectively for de novo cysteine synthesis. Lastly, the yhcE mutant was able to grow on medium depleted of cysteine, but its growth was significantly affected if 0.5 mM of sulfide was added to methionine. Addition of higher concentrations of sulfide (2 mM) totally impaired its growth, suggesting that YhcE is necessary for cysteine synthesis under this particular condition.
FhuR-specific binding on promoter region.
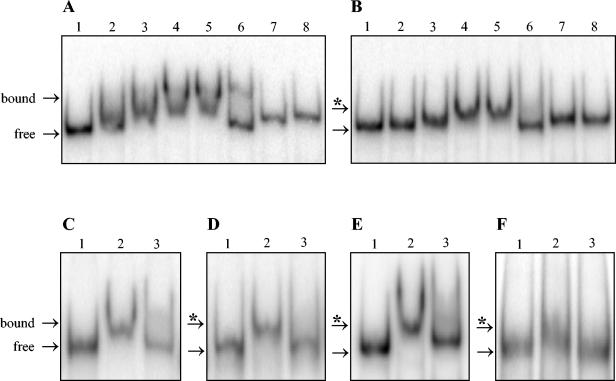
To test whether FhuR directly regulates transcription of the cysteine and methionine biosynthetic and transport genes, we carried out gel retardation experiments with purified FhuR protein (see Materials and Methods). We first tested the promoter region of metB2-cysK that appears to be the most efficiently regulated gene (Table 3). A 390-bp radiolabeled PCR fragment corresponding to the metB2 promoter region was incubated with an increasing concentration of the FhuR protein. As shown in Fig. 2A (lanes 1 to 5), the metB2 probe gave a distinct retarded band in the presence of the FhuR protein. The addition of an excess of metB2 unlabeled probe dramatically decreased FhuR binding to the same labeled DNA fragment (Fig. 2A, lane 6). Finally, no band shift could be detected if an internal purH gene fragment was used as a probe, even with an excess of FhuR protein (Fig. 2A, lanes 7 and 8), confirming the specificity of FhuR binding to the metB2 promoter region.
FIG. 2.
Gel mobility shift assay for FhuR binding to different regulated gene promoter regions. The promoter regions of FhuR target genes were PCR amplified and labeled. Radiolabeled DNA probes (10 × 10−15 mole) for metB2 (A), plpA (B), yjgC (C), cysM (D), cysD (E), and metA (F) were incubated with purified FhuR protein at various concentrations and analyzed on nondenaturing polyacrylamide gel electrophoresis (see Materials and Methods for details). (A and B) Lane 1, no protein; lanes 2 to 5, 0.6, 1.2, 2.4, and 3.6 μM final concentrations of the FhuR protein, respectively; lane 6, 3.6 μM final concentration of the FhuR protein with an excess of unlabeled DNA probe (10 × 10−13 mole). As a negative control, lanes 7 and 8 contain no protein and 3.6 μM (final) of the FhuR protein, respectively, with a purH gene 32P-labeled internal fragment as DNA probe. (C to F) Lanes 1 and 2, no protein and 3.6 μM (final) of the FhuR protein, respectively; lane 3, 3.6 μM (final) of the FhuR protein with an excess of unlabeled DNA probe (10 × 10−13 mole). In all panels, the bands for the probe (free) and the FhuR-probe complex (bound) are indicated by an arrow and an arrow with an asterisk, respectively.
Next, we similarly tested whether FhuR could also form specific DNA complexes with the promoter regions of cysD, cysM, yhcE, yjgC-yjgD-yjgE, metA-metB1-ytjE, and plpA-plpB-plpC-plplD-ydcB-ydcC-ydcD clusters (Fig. 2B to F). The specificity of these interactions was also verified by using an excess of the corresponding unlabeled probes. FhuR was found to clearly and specifically shift DNA probes generated from the promoter regions of cysD, cysM, metA, plpA, and yjgC, but not of yhcE. Nevertheless, a slight and specific mobility shift of the yhcE probe could be observed with 3.7 μM FhuR protein (data not shown). Lastly, no shifts were obtained when the potential promoter region of the fhuR gene was used as probe. These results demonstrate that FhuR can interact specifically with the promoter regions of each FhuR/cysteine-regulated cluster of genes and may thus directly activate their transcription.
Effect of OAS on FhuR-dependent regulation.
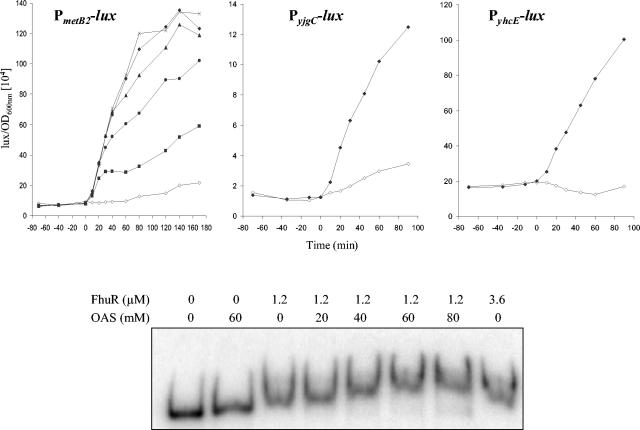
OAS supplementation in culture medium containing cysteine was shown to increase by approximately 3.5-fold the cysthathionine lyase activity in L. lactis MG1363, suggesting that OAS could be an effector of the regulation by CmbR (19). Moreover, those authors suggested that NAS (N-acetyl serine, made spontaneously from OAS) could be the inducer, in a similar way as E. coli CysB (43, 45). To investigate if OAS or NAS are inducers of the FhuR-dependent regulation in L. lactis IL1403, the effect of their addition on transcription of metB2-cysK during growth of L. lactis in the presence of cysteine (noninduction condition for FhuR regulation) was studied. Luciferase activities from the JIM8499 strain carrying the PmetB2::lux fusion were measured without or with OAS added at different concentrations (1 mM to 5 mM) to exponentially growing cells (OD600 = 0.2) in M17 medium (Fig. 3). Expression of the fusion increased within 5 min upon OAS addition to reach a maximal level after 1 h. Addition of 1 mM, 2 mM, 3 mM, and 4 mM of OAS increased by 3-, 6-, 8-, and 10-fold, respectively, the transcription of the metB2-cysK operon, but higher OAS concentrations (5 mM to 20 mM) did not further increase metB2-cysK transcription. No increase of luciferase activities was obtained when 4 mM OAS was added to cultures of the fhuR mutant strain, indicating that OAS induction depends on the presence of FhuR. Lastly, a similar experiment carried out in the presence of 4 mM NAS did not lead to any induction of metB2-cysK transcription, suggesting that NAS is not the FhuR effector.
FIG. 3.
In vivo and in vitro effects of OAS on FhuR-dependent regulation in L. lactis. Upper panel: effect of OAS on expression of FhuR-regulated genes of L. lactis. Luciferase activities (104 lux/OD600) measured from JIM8499, JIM8543, and JIM8545 strains carrying the PmetB2-lux, PyjgC-lux, and PyhcE-lux fusions, respectively, grown in M17 medium with or without OAS. The x axis shows time of incubation relative to the addition of OAS (time zero). Symbols: ⋄, without OAS; ▪, with OAS at a final concentration of 1 mM; •, at 2 mM; ▴, at 3 mM; ♦, at 4 mM; ×, at 5 mM. Lower panel: OAS effect on FhuR-metB2 complex migration. The labeled DNA fragment containing the metB2 promoter region (10 × 10−15 mole) was incubated with different concentrations of purified FhuR protein (0 μM, 1.2 μM, and 3.6 μM) with an increasing concentration of OAS (0 mM, 20 mM, 40 mM, 60 mM, and 80 mM) and analyzed on nondenaturing polyacrylamide gel electrophoresis.
To confirm that OAS induction was also effective on other genes of the FhuR regulon, luciferase activities were measured from fusions with the cysD, cysM, plpABCD, yhcE, and yjgC FhuR-dependent promoters and with the fhuR and cysE FhuR-independent promoters. Whereas the expression from the cysD, cysM, plpABCD, yhcE, and yjgC luciferase fusion was induced 3.5-, 28-, 10-, 3.5-, and 6-fold, respectively, in the presence of 4 mM OAS, the expression of cysE and fhuR remained unchanged (Fig. 3), showing that addition of OAS induces specifically the transcription of the FhuR regulon.
To directly demonstrate the role of OAS on FhuR activity, we compared the mobility of the metB2-cysK promoter fragment with 1.2 μM of FhuR in the presence of an increasing concentration of OAS or NAS as control. At this concentration, FhuR produced an intermediary shift between free and fully bound fragment in the absence of effector (Fig. 2A). When 20 mM of OAS was added, a slight increase of the mobility of the FhuR-metB2 promoter fragment complex was observed, and a further increase was obtained with the addition of OAS up to 60 mM (Fig. 3). A similar effect was observed on the mobility of the FhuR-plpA promoter fragment complex (data not shown). In contrast to OAS, NAS did not change the FhuR-DNA complex mobility even at a high concentration (tested up to 40 mM) (data not shown). These experiments show that OAS modifies the affinity of FhuR for its targets in vitro and leads to an increase of the transcription from the FhuR-dependent genes in vivo, suggesting that OAS is the effector for the FhuR regulation.
Identification of the FhuR box.
To characterize the targets of FhuR, we first searched for motifs conserved in the seven promoter regions and carried out validation experiments by band shift assay on two promoter regions carrying modified sequences.
The identification of the potential motifs was done by following two independent methods: the first was unsupervised and used probabilistic routine by a dedicated software, and the second was based on educated guesses founded on prior knowledge in relation to LysR-type regulator motifs. The statistical approach to search for motif sequences that are common to this set of promoters was performed with the MEME algorithm (2). DNA sequences of 250 bp present upstream of the start codon of metB2, metA, yhcE, cysD, cysM, plpA, and yjgC were used as the training set. The MEME algorithm displayed a 20-bp motif with CNATAAAWWTTTYTWAKNGC as the consensus (the T-N11-A is underlined), with an elevated score and an acceptable log e-value.
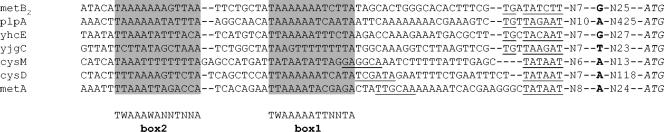
The second approach to propose a FhuR-binding motif was based on the fact that this regulator belongs to the LysR-type prokaryotic transcriptional regulators. This type of regulator binds to sequences of the general structure T-N11-A, displaying a partial dyad symmetry and located in a region comprised between −20 and −80 from the +1 transcriptional start site (25). We first characterized the promoters present upstream of metB2, plpA, yhcE, yjgC, cysM, cysD, and metA genes by transcriptional start mapping (Fig. 4). Analysis of the upstream region allowed us to propose consensus sequences for these promoters (Fig. 4). It is noteworthy that metB2, plpA, yhcE, and yjgC gene promoters have a −10 extended promoter consensus sequence (TG upstream of the −10 box) but a degenerated −10 box and a very poorly consensual −35 box, while the cysD, cysM, and metA gene promoters have a canonical −10 box and a recognizable −35 box (at least three out of six conserved positions). Interestingly, two potential T-N11-A boxes could be located, centered 46 to 53 bp (box 1) and 69 to 73 bp (box 2) upstream of the transcriptional starts (Fig. 4). The consensus sequences of box 1 and box 2, TWAAAAATTNNTA and TWAAAWANNTNNA, respectively, are relatively similar, and their locations are in good agreement with the usual location of LTTR DNA-binding sequences. Furthermore, box 1 is included in the central part of the motif proposed for metB2, yjgC, yhcE, and cysD by the unsupervised method, while box 2 is located within the unsupervised method motif found upstream of cysM. The good convergence of these two analyses prompted us to experimentally test the potential FhuR-binding boxes.
FIG. 4.
Sequence alignments of promoters controlled by FhuR. The ATG start codons of each gene are indicated in italics. The experimental sites of transcription initiation are given in bold letters. The extend −10, −10, and −35 corresponding regions for each promoter are underlined. The potential FhuR box 1 and box 2 are shaded.
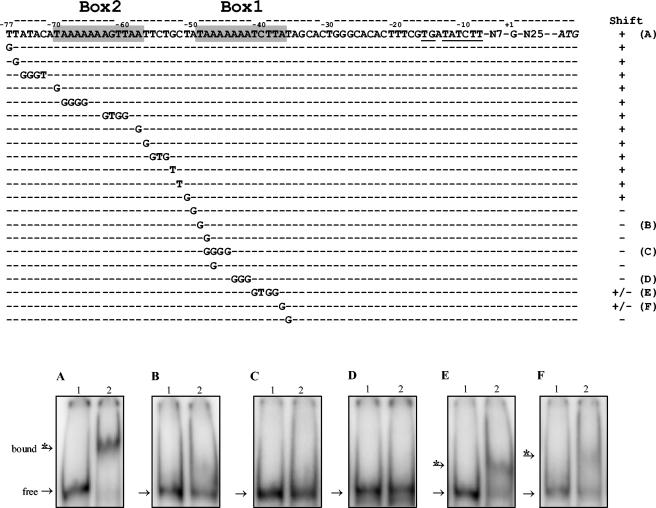
The proposed FhuR boxes of the metB2 and cysD gene promoter regions were mutated by specific substitutions and tested by gel shift with purified FhuR (Fig. 5 and 6). First, single base substitutions at the extremities of the metB2 FhuR box (T→G at −49 and A→G at −37) corresponding to the conserved bases for LTTR affected the FhuR binding. This binding was completely abolished when the DNA fragment carried the base substitutions affecting the left side of box 1 (Fig. 5B) and was significantly reduced by substitutions affecting the right side of that box (Fig. 5F). Mutations within box 1 also modified the pattern of the band shift assay. The single A→G substitution at position −48 or −47 decreased the amount of shifted DNA (data not shown). More drastic modifications, such as four base substitutions (A→G at positions −48 to −45) or three base substitutions (A→G at positions −44 to −42) completely abolished FhuR-DNA complex formation (Fig. 5, C, and D, respectively). Finally, substitutions of four residues at position −41 to −38 also decreased, but only partially, the amount of shifted probe (Fig. 5E). In conclusion, overall the changes tested in the proposed recognition sequence for FhuR modified significantly the mobility of the probe, suggesting that the box 1 sequence is necessary for FhuR binding to the metB2 promoter region. Single and multiple substitutions outside of box 1 from the metB2 promoter were also tested. Among the 14 substitutions, including multiple changes in the box 2 consensus, only 2 single substitutions produced a modification of the pattern of the band shift assay. These two substitutions (A→G at −50 or T→G at −36), flanking box 1, abolished completely the formation of a FhuR-DNA complex and produced patterns similar to those obtained with the changes in the adjacent T at −49 and A at −37 bases bordering box 1 (Fig. 5).
FIG. 5.
Substitution analysis of the metB2 promoter region. Upper panel: sequence of metB2 wild-type and mutated promoter region. Position +1 corresponds to the experimental transcriptional start. The deduced extended −10 box is underlined, and putative FhuR-DNA-binding box 1 and box 2 are shaded. Shifts + and − indicate the observation of a band shift or not, respectively, of the corresponding DNA fragment in the presence of the purified FhuR protein. Letters A to F on the right refer to the corresponding lower panel. Lower panels: gel mobility shift assay for FhuR binding to metB2 wild-type and mutated promoter regions. The corresponding fragments were PCR amplified and labeled. DNA probes (10 × 10−15 mole) were incubated without (lane 1) or with 3.6 μM FhuR (lane 2) in the presence of 60 mM OAS and analyzed by nondenaturing polyacrylamide gel electrophoresis. (A) metB2 wild-type promoter region. (B to F) Mutated sequences indicated on the right column of the upper panel. In each binding assay, the bands for the probe (free) and the FhuR-probe complex (bound) are indicated by an arrow and an arrow with an asterisk, respectively.
FIG. 6.
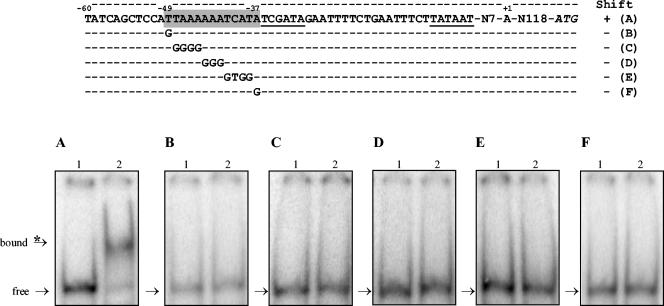
Substitution analysis of the cysD promoter region. Upper panel: sequence of cysD wild-type and mutated promoter region. Position +1 corresponds to the experimental transcriptional start. The deduced −10 and −35 boxes are underlined, and the putative FhuR DNA-binding box 1 is shaded. Shifts + and − indicate the observation of a band shift or not, respectively, of the corresponding DNA fragment in the presence of the purified FhuR protein. Letters A to F on the right refer to the corresponding lower panel. Lower panel: gel mobility shift assay corresponding to the cysD promoter region as indicated in the upper panel. The experimental procedures are the same as those presented for Fig. 5. (A) cysD wild-type promoter region. (B to F) Mutated sequences indicated on the right column of the upper panel. In each binding assay, the bands for the probe (free) and the FhuR-probe complex (bound) are indicated by an arrow and an arrow with an asterisk, respectively.
Similar modifications were carried out on the predicted cysD gene FhuR box. Single base substitution at the extremities of the cysD FhuR box (T→G at −49 and A→G at −37), corresponding to the conserved bases for LTTR, abolished completely the FhuR-DNA complex formation (Fig. 6B and F). Multiple modifications within the motif also abolished the binding of FhuR to the cysD promoter region (Fig. 6C, D, and E). These biochemical experiments led us to conclude that the predicted T-N11-A motifs are necessary for the formation of the FhuR-DNA complex. Therefore, they are the best candidates to be the DNA-binding motifs specifically recognized by FhuR in these promoter regions.
DISCUSSION
In this work, our goal was to determine the set of genes controlled by FhuR, an LTTR for which previous results suggested that it could be a pleitropic regulator, involved at least in sulfur amino acid biosynthesis, iron metabolism, and heme-dependent respiration (19, 22). We found that FhuR regulates many L. lactis genes involved in sulfur metabolism, but none in iron metabolism. The set of genes that are differentially expressed in an fhuR mutant is composed of seven transcriptional units, including two permeases for sulfur amino acids or related compounds and all genes likely involved in methionine and cysteine synthesis except cysE and metEF. This result was confirmed by searching the set of differentially expressed genes in the absence of cysteine, which revealed essentially the same set of genes with few additional transcriptional units. These data allowed us to provide a global scheme for sulfur metabolism in L. lactis and its regulation. Moreover, we have provided molecular data allowing us to better define the FhuR regulation mechanism on its target sequence genes.
Sulfur metabolism in L. lactis.
The two sets of transcriptome experiments we carried out should allow us to define three classes of genes: (i) those differentially expressed by the FhuR activator in response to cysteine depletion, and those differentially expressed only (ii) in the absence of cysteine or (iii) in the fhuR mutant. Interestingly, the first class is the most important and contains most sulfur metabolic genes from L. lactis (see below) and the second class consists mostly of genes of unknown or putative functions, whereas only ymcF appears to belong to the third class.
Genes differentially expressed specifically upon cysteine starvation (class II, FhuR-independent and cysteine-dependent regulation) might be controlled by signals related to cellular functions perturbed by cysteine starvation or by its synthesis. Among these genes, yrjBCD, nifJ, and cadA, which share significant similarity to iron-dependent oxidoreductase, pyruvate-flavodoxin oxidoreductase, and ion efflux ATPase, respectively, have a reduced expression in the absence of cysteine. On the other hand, yqeI, yjiB, yrfD, and pi and ps genes, which share homology to cation transport protein, amino acid aminohydrolase, antiporter, and prophage genes have an increased transcription rate. Interestingly, many of these genes are related to ion metabolism and oxidoreduction mechanisms, confirming the importance of cysteine availability for these functions. Phage induction might be a side effect of some general perturbation in the cell, although none of the general stress-induced genes involved in functions such as heat shock, proteases, and the SOS-like response were found to be differentially expressed.
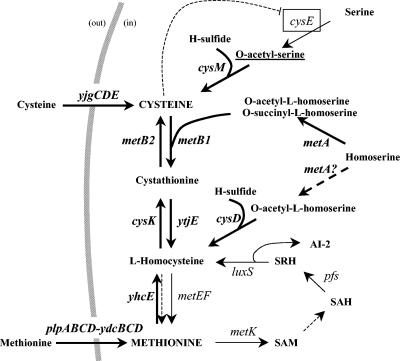
The set of genes common to the two transcriptome experiments are directly involved in uptake and de novo synthesis of methionine and cysteine and their interconversion. First, two transporters encoded by the yjgCDE and plpABCD-ydcBCD transcription units are induced by FhuR in the absence of cysteine. These transporters are likely involved in cysteine (or cystine) and methionine uptake, respectively, in regard to the homology of their substrate-binding components, YjgC and PlpABCD, to YtmJK and MetQ in B. subtilis (8, 39). The presence of four potential amino acid-binding subunits (PlpABCD) is intriguing and appears to be unique to L. lactis. This feature might reflect the possibility for L. lactis to import different methionine derivatives or sulfur-containing compounds in its original ecological niche, which is likely vegetal (16). Second, the set of FhuR-regulated genes, completed by a focused search of sulfur metabolism genes in the genome of L. lactis IL1403, allowed us to propose a new scheme for sulfur metabolism (Fig. 7). Several physiological experiments were carried out to consolidate this scheme.
FIG. 7.
Cysteine and methionine biosynthesis pathway and its regulation by FhuR in L. lactis. Genes regulated by FhuR or by cysteine starvation are indicated in bold and with thick arrows. Dashed arrows indicate that the reactions could not be fully ascertained from the literature or our experiments. The boxed cysE and the dashed line indicate that CysE is likely subject to feedback control by cysteine. The underlining of O-acetyl-serine emphasizes the fact that this compound might trigger the activity of FhuR. Abbreviations: SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; SRH, S-ribosylhomocysteine; AI-2, autoinducer 2.
The set of L. lactis genes involved in methionine and cysteine metabolism is different from those well characterized in B. subtilis and E. coli. An important difference is the lack of genes corresponding to the assimilation of sulfate, suggesting that L. lactis uses other sulfur-donor compounds in its environment. Two key enzymes might allow the assimilation of sulfide, CysM and CysD, which are potentially O-acetyl-l-serine (thiol)-lyase and O-acetyl-l-homoserine (thiol)-lyase, respectively, leading thus to the synthesis of cysteine and homocysteine. CysK from the metB2-cysK operon was previously proposed to be the L. lactis cysteine synthase. However, a cysK mutant is still able to grow in the absence of cysteine when sulfide is provided as sole sulfur source, indicating that CysM alone can perform this reaction. An alternative role for CysK will be discussed below. OAS, the CysM substrate, would be produced by CysE, whereas the enzyme producing O-acetyl-l-homoserine, the substrate of CysD, could be MetA by analogy to B. subtilis (7), notwithstanding that this protein belongs to the homoserine transsuccinylase family of enzymes, producing O-succinyl-l-homoserine (6). Two operons might then be involved in the interconversion of methionine and cysteine, metA-B1-ytjE and metB2-cysK. MetB1 and YtjE are encoded in a single cluster in other streptococci such as Streptococcus anginosus, where their products have been characterized to be a cystathionine γ-synthase and a cystathionine β-lyase, respectively (79-81). The metB2-cysK operon (metC-cysK in L. lactis strain MG1363) has been extensively studied in L. lactis, where MetC was characterized as a cystathionine β-lyase and CysK as a potential cysteine synthase, respectively (20). However, we have shown here that inactivation of metB2 or cysK in L. lactis IL1403 leads to cysteine auxotrophy in the presence of methionine as sole sulfur donor, indicating that these two genes are involved in the interconversion of methionine to cysteine. Therefore, it is likely that CysK and MetB2 are cystathionine β-synthase and γ-lyase, respectively, as proposed for the products of the yrhAB operon in B. subtilis (the best lactococcal homologues, which share 63% and 28% identity with MetB2 and CysK). Moreover, the fact that the metB2-cysK operon is the most strongly induced transcription unit in CDM containing methionine but not cysteine supports a role for these genes in the conversion of methionine to cysteine. Finally, yhcE, encoding a product homologous to homocysteine methyltransferase, could be a B12-independent methionine synthase in L. lactis, in addition to MetE. However, its inactivation leads to cysteine auxotrophy in the presence of methionine and high sulfide content in the medium. This result indicates that methionine might enter the interconversion pathway to cysteine in two different ways. The first, which is sulfide sensitive, would involve MetK, Pfs, and LuxS, a pathway found in various bacteria (77). The second, which is sulfide resistant, would involve YhcE, which would function either reversibly or in the opposite direction than the classical methionine synthase MetE (27). Lastly, yhcE transcription is activated in the absence of cysteine, contrarily to metE, a datum in agreement with a role for YhcE in methionine conversion to cysteine in L. lactis.
Regulation of sulfur metabolism in L. lactis.
FhuR appears to play a central role in the regulation of sulfur metabolism in L. lactis, as it activates the transcription of seven clusters encoding 18 sulfur metabolism genes in L. lactis (Fig. 1). Only cysE (first step of cysteine biosynthesis), metEF (last step of methionine biosynthesis), metK, pfs, and luxS (S-adenosylmethionine and autoinducer 2 synthesis), and yfaB (sulfate permease) are not controlled by FhuR. Since this regulator was previously proposed to be involved in the control of the fhu operon, we performed real-time quantitative RT-PCR experiments to compare its transcription in IL1403 and its fhuR derivative. These experiments did not allow us to detect any significant change in the expression of the fhu operon, confirming it is not regulated by FhuR in L. lactis IL1403 (data not shown). We also designed several experiments to check the possible transcriptional regulation of fhuR, either by itself or by cysteine depletion. The lack of binding of FhuR to its own promoter suggests the absence of retrocontrol. Moreover, a gene fusion with luciferase genes did not allow us to find conditions leading to a significant change in the reporter activity. In particular, we did not confirm that FhuR is modulated by catabolic repression, as suggested earlier for L. lactis subsp. cremoris MG1363 (22). These results indicate that FhuR is expressed constitutively. Although many LTTRs negatively control their own transcription, FhuR might be among exceptions to this general rule. Possibly, the lack of retrocontrol of fhuR gene transcription is correlated to high abundance of its product in the cell, clearly identifiable by two-dimensional gel electrophoresis (33).
FhuR was shown to activate several gene transcription levels and by gel mobility shift assays to bind to sites located at bp −46 to −53 from the transcriptional start, a position very close to that of the −35 promoter boxes. The consensus of the FhuR DNA-binding sequence has been deduced from the comparative analysis of the seven promoters and confirmed by biochemical experiments. A second consensual box appears to be present 25 bp upstream of the previously defined DNA-binding box. Although this second box is not necessary for in vitro binding of FhuR to the metB2 promoter, it may be involved in transcriptional activation in vivo. Indeed, several LysR-type transcriptional regulators require both a DNA-binding box and an activating box separated by one helix turn (64). With the hypothesis that FhuR DNA-binding boxes follow the general rule for LysR-type regulators, a T-N11-A palindromic sequence, the expected FhuR box consensus sequence would be TAAAAWWWTTTTA. However, most of the boxes are not perfectly conserved, the best-fitting one being the metB2 box (one mismatch) and then plpA and yjgC (two mismatches), yhcE (three mismatches) and, finally, metA, cysD, and cysM boxes (four mismatches). In addition to the box, the flanking bases might also participate in the recognition by FhuR, since these bases are well conserved and their replacement by other bases decreases FhuR binding affinity to the metB2 promoter. The FhuR box consensus is different from the one of the recently proposed MET box (TATGTTtnaAACTATA [lowercase letters indicate lesser conserved nucleotides) found by computing methionine biosynthesis genes in Streptococcaceae (61). Since FhuR is activated in the absence of cysteine (see below), the newly characterized box in L. lactis might be considered a CYS box, although it regulates an extended set of genes in L. lactis.
In vivo activation of FhuR occurs in the absence of cysteine. Here, we show that a high concentration of OAS modifies the pattern of the band shift assays carried out with purified FhuR and several promoters, suggesting that OAS is the effector for FhuR. The amount of OAS used in the in vitro experiment is likely not the in vivo working concentration for this compound, since 50 mM might be at least 10-fold higher than what is physiologically expected. This result could be an artifact due, for example, to the pH used in band shift experiments, which is almost 2 pH units higher than the intracellular pH of L. lactis. Such a change between in vivo and in vitro pH will modify the balance between the different ionic forms of OAS and, thus, might decrease the amount of its active form on FhuR. Importantly, the fact that, in the presence of cysteine, addition of physiological concentrations of OAS activates the transcription of at least six FhuR-activated promoters in L. lactis IL1403 (this work) and of metC-cysK in the MG1363 strain (19) confirms that OAS is the effector of FhuR. However, it could not be excluded that the effector is a derivative of OAS that remains to be determined. To our knowledge, OAS could be transformed spontaneously into NAS or into cysteine by CysM. We have shown that none of them is a cofactor of FhuR since (i) NAS does not induce FhuR-dependent gene expression and does not modify the affinity of FhuR for its targets and (ii) cysteine leads to repression of the FhuR regulon but does not inhibit its induction by OAS in vivo. Therefore, OAS is likely the cofactor of the FhuR-dependent regulation in L. lactis.
A general view of sulfur amino acid gene regulation could be drawn from the results of this work. Interestingly, almost all genes involved in the supply of these amino acids are controlled by FhuR and OAS. We point out that eventual additional systems of regulation similar to those already found in gram-positive bacteria, such as T-box and S-box attenuation systems responding to uncharged cysteyl- or methionyl-tRNA and S-adenosylmethionine, were not successful (13, 30, 55). In E. coli, three activators, CysB, Cbl, and MetR, and one repressor, MetJ, control sulfur metabolism with, respectively, NAS, adenosine phosphosulfate, homocysteine, and S- adenosylmethionine as effectors (9-11, 45, 62, 74, 75). Although it cannot be excluded that such other transcriptional regulators may exist, FhuR is probably the key control for sulfur metabolism in L. lactis. In E. coli as well as in B. subtilis, the diversity of regulators and signals allows us to control independently genes for de novo synthesis of cysteine, methionine, and the two interconversion pathways, leading thus to a fine control of synthesis in function of the need of the cell. Curiously, FhuR activates the complete set of genes, including metB2-cysK and metA-metB1-ytjE that function in opposite directions, in cysteine and methionine interconversion. If none of these enzymes was subjected to a feedback control, this would lead to a futile cycle.
Since FhuR is produced constitutively, the activation of its regulon would directly rely on the production of its effector, OAS, which is a product of CysE, the serine acetyltransferase, performing the first step of cysteine synthesis. CysE has been shown to be feedback inhibited by cysteine in different bacteria (17, 41, 49, 76), in yeast (71), in protists (57, 58), and in plants (56, 63, 73). To our knowledge, serine acetyltransferases (SAT) naturally insensitive to feedback inhibition have only been found in particular locales in plants (44) and likely in endosymbiont bacteria (46). The molecular basis for cysteine feedback inhibition has been established in SAT from different origins (17, 37, 40-42, 59, 69). Interestingly, amino acids involved in cysteine feedback of E. coli CysE are conserved in the primary sequence of L. lactis CysE, suggesting that the constitutively expressed L. lactis CysE could also be feedback inhibited by cysteine (data not shown). Interestingly, expression of SAT desensitized to feedback inhibition by cysteine led to an overproduction of cysteine in E. coli (17, 70), Corynebacterium glutamicum (76), or plants (56, 73), suggesting that cysteine control on this enzyme is important in most organisms. In L. lactis, CysE would thus be the key enzyme controlling both (i) the transcription of the FhuR regulon and (ii) de novo cysteine synthesis.
Conclusion.
Regulation of sulfur amino acid biosynthesis in bacteria appears to be rather diverse and to involve different mechanisms of regulation. In the present work, we have shown that in L. lactis a single regulator, FhuR, activates principal genes for methionine and cysteine metabolism in the function of cysteine availability. This new regulatory scheme appears to be fairly minimal, compared to those already well determined in E. coli and B. subtilis. However, this apparent simplicity suggests additional levels of regulation, probably at the enzyme level, to coordinate methionine and cysteine supply and autoinducer 2 signaling pathways. Understanding the reasons for the use of different regulatory mechanisms for the same amino acid metabolism in different bacteria is an exciting future challenge.
Acknowledgments
B. Sperandio had a grant from Ministère de la recherche et de l'Education Nationale.
We thank I. Guillouard and C. Delorme for critically reading of the manuscript, J. P. Furet for his assistance and helpful advice on quantitative RT-PCR experiments, and N. Pons for providing the beta version of the motif research software using the MEME algorithm. Finally, we thank I. Martin-Verstraete for her helpful advice on sulfur metabolism and, in particular, on the role of metB2-cysK genes.
REFERENCES
- 1.Auger, S., A. Danchin, and I. Martin-Verstraete. 2002. Global expression profile of Bacillus subtilis grown in the presence of sulfate or methionine. J. Bacteriol. 184:5179-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 3.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogdan, J. A., J. Nazario-Larrieu, J. Sarwar, P. Alexander, and M. S. Blake. 2001. Bordetella pertussis autoregulates pertussis toxin production through the metabolism of cysteine. Infect. Immun. 69:6823-6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Born, T. L., and J. S. Blanchard. 1999. Enzyme-catalyzed acylation of homoserine: mechanistic characterization of the Escherichia coli metA-encoded homoserine transsuccinylase. Biochemistry 38:14416-14423. [DOI] [PubMed] [Google Scholar]
- 7.Brush, A., and H. Paulus. 1971. The enzymic formation of O-acetylhomoserine in Bacillus subtilis and its regulation by methionine and S-adenosylmethionine. Biochem. Biophys. Res. Commun. 45:735-741. [DOI] [PubMed] [Google Scholar]
- 8.Burguiere, P., S. Auger, M. F. Hullo, A. Danchin, and I. Martin-Verstraete. 2004. Three different systems participate in l-cystine uptake in Bacillus subtilis. J. Bacteriol. 186:4875-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bykowski, T., J. R. van der Ploeg, R. Iwanicka-Nowicka, and M. M. Hryniewicz. 2002. The switch from inorganic to organic sulphur assimilation in Escherichia coli: adenosine 5′-phosphosulphate (APS) as a signalling molecule for sulphate excess. Mol. Microbiol. 43:1347-1358. [DOI] [PubMed] [Google Scholar]
- 10.Cai, X. Y., M. E. Maxon, B. Redfield, R. Glass, N. Brot, and H. Weissbach. 1989. Methionine synthesis in Escherichia coli: effect of the MetR protein on metE and metH expression. Proc. Natl. Acad. Sci. USA 86:4407-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai, X. Y., B. Redfield, M. Maxon, H. Weissbach, and N. Brot. 1989. The effect of homocysteine on MetR regulation of metE, metR and metH expression in vitro. Biochem. Biophys. Res. Commun. 163:79-83. [DOI] [PubMed] [Google Scholar]
- 12.Chopin, A. 1993. Organization and regulation of genes for amino acid biosynthesis in lactic acid bacteria. FEMS Microbiol. Rev. 12:21-37. [DOI] [PubMed] [Google Scholar]
- 13.Chopin, A., V. Biaudet, and S. D. Ehrlich. 1998. Analysis of the Bacillus subtilis genome sequence reveals nine new T-box leaders. Mol. Microbiol. 29:662-664. [DOI] [PubMed] [Google Scholar]
- 14.Chopin, A., M. C. Chopin, A. Moillo-Batt, and P. Langella. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260-263. [DOI] [PubMed] [Google Scholar]
- 15.Delorme, C., S. D. Ehrlich, and P. Renault. 1999. Regulation of expression of the Lactococcus lactis histidine operon. J. Bacteriol. 181:2026-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delorme, C., J. J. Godon, S. D. Ehrlich, and P. Renault. 1993. Gene inactivation in Lactococcus lactis: histidine biosynthesis. J. Bacteriol. 175:4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denk, D., and A. Bock. 1987. l-Cysteine biosynthesis in Escherichia coli: nucleotide sequence and expression of the serine acetyltransferase (cysE) gene from the wild-type and a cysteine-excreting mutant. J. Gen. Microbiol. 133:515-525. [DOI] [PubMed] [Google Scholar]
- 18.Ejim, L. J., V. M. D'Costa, N. H. Elowe, J. C. Loredo-Osti, D. Malo, and G. D. Wright. 2004. Cystathionine beta-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 72:3310-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez, M., M. Kleerebezem, O. P. Kuipers, R. J. Siezen, and R. van Kranenburg. 2002. Regulation of the metC-cysK operon, involved in sulfur metabolism in Lactococcus lactis. J. Bacteriol. 184:82-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez, M., W. van Doesburg, G. A. Rutten, J. D. Marugg, A. C. Alting, R. van Kranenburg, and O. P. Kuipers. 2000. Molecular and functional analyses of the metC gene of Lactococcus lactis, encoding cystathionine beta-lyase. Appl. Environ. Microbiol. 66:42-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia de la Nava, J., S. van Hijum, and O. Trelles. 2003. PreP: gene expression data pre-processing. Bioinformatics 19:2328-2329. [DOI] [PubMed] [Google Scholar]
- 22.Gaudu, P., G. Lamberet, S. Poncet, and A. Gruss. 2003. CcpA regulation of aerobic and respiration growth in Lactococcus lactis. Mol. Microbiol. 50:183-192. [DOI] [PubMed] [Google Scholar]
- 23.Glatron, M. F., and G. Rapoport. 1972. Biosynthesis of the parasporal inclusion of Bacillus thuringiensis: half-life of its corresponding messenger RNA. Biochimie 54:1291-1301. [DOI] [PubMed] [Google Scholar]
- 24.Godon, J. J., C. J. Pillidge, K. Jury, C. A. Shearman, and M. J. Gasson. 1995. Molecular analysis of the Lactococcus lactis sex factor. Dev. Biol. Stand. 85:423-430. [PubMed] [Google Scholar]
- 25.Goethals, K., P. Mergaert, M. Gao, D. Geelen, M. Van Montagu, and M. Holsters. 1992. Identification of a new inducible nodulation gene in Azorhizobium caulinodans. Mol. Plant-Microbe Interact. 5:405-411. [DOI] [PubMed] [Google Scholar]
- 26.Gorke, B., L. Fraysse, and A. Galinier. 2004. Drastic differences in Crh and HPr synthesis levels reflect their different impacts on catabolite repression in Bacillus subtilis. J. Bacteriol. 186:2992-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greene, R. C. 1996. Biosynthesis of methionine, p. 542-560. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 28.Grundy, F. J., and T. M. Henkin. 1993. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell 74:475-482. [DOI] [PubMed] [Google Scholar]
- 29.Grundy, F. J., and T. M. Henkin. 1998. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 30:737-749. [DOI] [PubMed] [Google Scholar]
- 30.Grundy, F. J., and T. M. Henkin. 2003. The T box and S box transcription termination control systems. Front. Biosci. 8:d20-d31. [DOI] [PubMed] [Google Scholar]
- 31.Guedon, E., P. Renault, S. D. Ehrlich, and C. Delorme. 2001. Transcriptional pattern of genes coding for the proteolytic system of Lactococcus lactis and evidence for coordinated regulation of key enzymes by peptide supply. J. Bacteriol. 183:3614-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guedon, E., P. Serror, S. D. Ehrlich, P. Renault, and C. Delorme. 2001. Pleiotropic transcriptional repressor CodY senses the intracellular pool of branched-chain amino acids in Lactococcus lactis. Mol. Microbiol. 40:1227-1239. [DOI] [PubMed] [Google Scholar]
- 33.Guillot, A., C. Gitton, P. Anglade, and M. Y. Mistou. 2003. Proteomic analysis of Lactococcus lactis, a lactic acid bacterium. Proteomics 3:337-354. [DOI] [PubMed] [Google Scholar]
- 34.Guillouard, I., S. Auger, M. F. Hullo, F. Chetouani, A. Danchin, and I. Martin-Verstraete. 2002. Identification of Bacillus subtilis CysL, a regulator of the cysJI operon, which encodes sulfite reductase. J. Bacteriol. 184:4681-4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henkin, T. M. 1994. tRNA-directed transcription antitermination. Mol. Microbiol. 13:381-387. [DOI] [PubMed] [Google Scholar]
- 36.Hill, C. E., D. S. Metcalf, and J. I. MacInnes. 2003. A search for virulence genes of Haemophilus parasuis using differential display RT-PCR. Vet. Microbiol. 96:189-202. [DOI] [PubMed] [Google Scholar]
- 37.Hindson, V. J. 2003. Serine acetyltransferase of Escherichia coli: substrate specificity and feedback control by cysteine. Biochem. J. 375:745-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holo, H., and I. F. Nes. 1989. High-frequency transformation by electroporation of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hullo, M. F., S. Auger, E. Dassa, A. Danchin, and I. Martin-Verstraete. 2004. The metNPQ operon of Bacillus subtilis encodes an ABC permease transporting methionine sulfoxide, d- and l-methionine. Res. Microbiol. 155:80-86. [DOI] [PubMed] [Google Scholar]
- 40.Inoue, K., M. Noji, and K. Saito. 1999. Determination of the sites required for the allosteric inhibition of serine acetyltransferase by l-cysteine in plants. Eur. J. Biochem. 266:220-227. [DOI] [PubMed] [Google Scholar]
- 41.Johnson, C. M., B. Huang, S. L. Roderick, and P. F. Cook. 2004. Kinetic mechanism of the serine acetyltransferase from Haemophilus influenzae. Arch. Biochem. Biophys. 429:115-122. [DOI] [PubMed] [Google Scholar]
- 42.Johnson, C. M., S. L. Roderick, and P. F. Cook. 2005. The serine acetyltransferase reaction: acetyl transfer from an acylpantothenyl donor to an alcohol. Arch. Biochem. Biophys. 433:85-95. [DOI] [PubMed] [Google Scholar]
- 43.Jovanovic, M., M. Lilic, D. J. Savic, and G. Jovanovic. 2003. The LysR-type transcriptional regulator CysB controls the repression of hslJ transcription in Escherichia coli. Microbiology 149:3449-3459. [DOI] [PubMed] [Google Scholar]
- 44.Kawashima, C. G., O. Berkowitz, R. Hell, M. Noji, and K. Saito. 2005. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiol. 137:220-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kredich, N. M. 1996. Biosynthesis of cysteine, p. 514-527. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 46.Lai, C. Y., and P. Baumann. 1992. Sequence analysis of a DNA fragment from Buchnera aphidicola (an endosymbiont of aphids) containing genes homologous to dnaG, rpoD, cysE, and secB. Gene 119:113-118. [DOI] [PubMed] [Google Scholar]
- 47.Le Bourgeois, P. 1993. Ph.D. thesis. Univesrité Paul Sabatier, Toulouse, France.
- 48.Lestrate, P., R. M. Delrue, I. Danese, C. Didembourg, B. Taminiau, P. Mertens, X. De Bolle, A. Tibor, C. M. Tang, and J. J. Letesson. 2000. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol. Microbiol. 38:543-551. [DOI] [PubMed] [Google Scholar]
- 49.Leu, L. S., and P. F. Cook. 1994. Kinetic mechanism of serine transacetylase from Salmonella typhimurium. Biochemistry 33:2667-2671. [DOI] [PubMed] [Google Scholar]
- 50.Loureiro Dos Santos, A. L., and A. Chopin. 1987. Shotgun cloning in Streptococcus lactis. FEMS Microbiol. Lett. 42:209-212. [Google Scholar]
- 51.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor σH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 53.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 54.Marouni, M. J., and S. Sela. 2003. The luxS gene of Streptococcus pyogenes regulates expression of genes that affect internalization by epithelial cells. Infect. Immun. 71:5633-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDaniel, B. A., F. J. Grundy, I. Artsimovitch, and T. M. Henkin. 2003. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. USA 100:3083-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noji, M., and K. Saito. 2002. Molecular and biochemical analysis of serine acetyltransferase and cysteine synthase towards sulfur metabolic engineering in plants. Amino Acids 22:231-243. [DOI] [PubMed] [Google Scholar]
- 57.Nozaki, T., T. Asai, L. B. Sanchez, S. Kobayashi, M. Nakazawa, and T. Takeuchi. 1999. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar. Regulation and possible function of the cysteine biosynthetic pathway in Entamoeba. J. Biol. Chem. 274:32445-32452. [DOI] [PubMed] [Google Scholar]
- 58.Nozaki, T., Y. Shigeta, Y. Saito-Nakano, M. Imada, and W. D. Kruger. 2001. Characterization of transsulfuration and cysteine biosynthetic pathways in the protozoan hemoflagellate, Trypanosoma cruzi. Isolation and molecular characterization of cystathionine beta-synthase and serine acetyltransferase from Trypanosoma. J. Biol. Chem. 276:6516-6523. [DOI] [PubMed] [Google Scholar]
- 59.Olsen, L. R., B. Huang, M. W. Vetting, and S. L. Roderick. 2004. Structure of serine acetyltransferase in complexes with CoA and its cysteine feedback inhibitor. Biochemistry 43:6013-6019. [DOI] [PubMed] [Google Scholar]
- 60.Pinto, R., Q. X. Tang, W. J. Britton, T. S. Leyh, and J. A. Triccas. 2004. The Mycobacterium tuberculosis cysD and cysNC genes form a stress-induced operon that encodes a tri-functional sulfate-activating complex. Microbiology 150:1681-1686. [DOI] [PubMed] [Google Scholar]
- 61.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2004. Comparative genomics of the methionine metabolism in gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 32:3340-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saint-Girons, I., N. Duchange, G. N. Cohen, and M. M. Zakin. 1984. Structure and autoregulation of the metJ regulatory gene in Escherichia coli. J. Biol. Chem. 259:14282-14285. [PubMed] [Google Scholar]
- 63.Saito, K., H. Yokoyama, M. Noji, and I. Murakoshi. 1995. Molecular cloning and characterization of a plant serine acetyltransferase playing a regulatory role in cysteine biosynthesis from watermelon. J. Biol. Chem. 270:16321-16326. [DOI] [PubMed] [Google Scholar]
- 64.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 65.Shelver, D., L. Rajagopal, T. O. Harris, and C. E. Rubens. 2003. MtaR, a regulator of methionine transport, is critical for survival of group B streptococcus in vivo. J. Bacteriol. 185:6592-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simon, D., and A. Chopin. 1988. Construction of a vector plasmid family and its use for molecular cloning in Streptococcus lactis. Biochimie 70:559-566. [DOI] [PubMed] [Google Scholar]
- 67.Sissler, M., C. Delorme, J. Bond, S. D. Ehrlich, P. Renault, and C. Francklyn. 1999. An aminoacyl-tRNA synthetase paralog with a catalytic role in histidine biosynthesis. Proc. Natl. Acad. Sci. USA 96:8985-8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sperandio, V., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Takagi, H., N. Awano, S. Kobayashi, M. Noji, K. Saito, and S. Nakamori. 1999. Overproduction of l-cysteine and l-cystine by expression of genes for feedback inhibition-insensitive serine acetyltransferase from Arabidopsis thaliana in Escherichia coli. FEMS Microbiol. Lett. 179:453-459. [DOI] [PubMed] [Google Scholar]
- 70.Takagi, H., C. Kobayashi, S. Kobayashi, and S. Nakamori. 1999. PCR random mutagenesis into Escherichia coli serine acetyltransferase: isolation of the mutant enzymes that cause overproduction of l-cysteine and l-cystine due to the desensitization to feedback inhibition. FEBS Lett. 452:323-327. [DOI] [PubMed] [Google Scholar]
- 71.Takagi, H., K. Yoshioka, N. Awano, S. Nakamori, and B. Ono. 2003. Role of Saccharomyces cerevisiae serine O-acetyltransferase in cysteine biosynthesis. FEMS Microbiol. Lett. 218:291-297. [DOI] [PubMed] [Google Scholar]
- 72.Terzaghi, B., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Urano, Y., T. Manabe, M. Noji, and K. Saito. 2000. Molecular cloning and functional characterization of cDNAs encoding cysteine synthase and serine acetyltransferase that may be responsible for high cellular cysteine content in Allium tuberosum. Gene 257:269-277. [DOI] [PubMed] [Google Scholar]
- 74.Urbanowski, M. L., and G. V. Stauffer. 1989. Genetic and biochemical analysis of the MetR activator-binding site in the metE metR control region of Salmonella typhimurium. J. Bacteriol. 171:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Urbanowski, M. L., and G. V. Stauffer. 1989. Role of homocysteine in metR-mediated activation of the metE and metH genes in Salmonella typhimurium and Escherichia coli. J. Bacteriol. 171:3277-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wada, M., N. Awano, K. Haisa, H. Takagi, and S. Nakamori. 2002. Purification, characterization and identification of cysteine desulfhydrase of Corynebacterium glutamicum, and its relationship to cysteine production. FEMS Microbiol. Lett. 217:103-107. [DOI] [PubMed] [Google Scholar]
- 77.Winzer, K., K. R. Hardie, N. Burgess, N. Doherty, D. Kirke, M. T. Holden, R. Linforth, K. A. Cornell, A. J. Taylor, P. J. Hill, and P. Williams. 2002. LuxS: its role in central metabolism and the in vitro synthesis of 4-hydroxy-5-methyl-3(2H)-furanone. Microbiology 148:909-922. [DOI] [PubMed] [Google Scholar]
- 78.Xu, L., and K. J. Marians. 2000. Purification and characterization of DnaC810, a primosomal protein capable of bypassing PriA function. J. Biol. Chem. 275:8196-8205. [DOI] [PubMed] [Google Scholar]
- 79.Yoshida, Y., Y. Nakano, A. Amano, M. Yoshimura, H. Fukamachi, T. Oho, and T. Koga. 2002. lcd from Streptococcus anginosus encodes a C-S lyase with alpha, beta-elimination activity that degrades l-cysteine. Microbiology 148:3961-3970. [DOI] [PubMed] [Google Scholar]
- 80.Yoshida, Y., M. Negishi, A. Amano, T. Oho, and Y. Nakano. 2003. Differences in the βC-S lyase activities of viridans group streptococci. Biochem. Biophys. Res. Commun. 300:55-60. [DOI] [PubMed] [Google Scholar]
- 81.Yoshida, Y., M. Negishi, and Y. Nakano. 2003. Homocysteine biosynthesis pathways of Streptococcus anginosus. FEMS Microbiol. Lett. 221:277-284. [DOI] [PubMed] [Google Scholar]