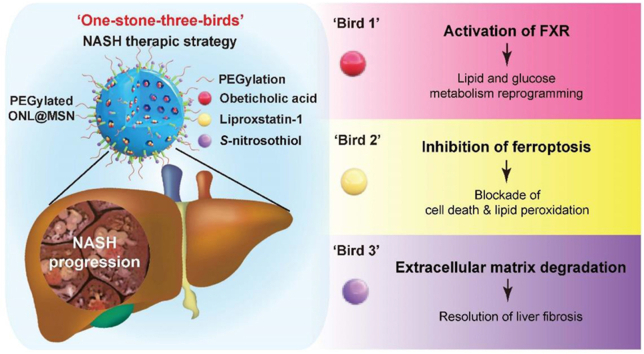
Abstract
Obeticholic acid (OCA), a farnesoid X receptor (FXR) agonist with favorable effects on fatty and glucose metabolism, has been considered the leading candidate drug for nonalcoholic steatohepatitis (NASH) treatment. However, its limited effectiveness in resolving liver fibrosis and lipotoxicity-induced cell death remains a major drawback. Ferroptosis, a newly recognized form of cell death characterized by uncontrolled lipid peroxidation, is involved in the progression of NASH. Nitric oxide (NO) is a versatile biological molecule that can degrade extracellular matrix. In this study, we developed a PEGylated thiolated hollow mesoporous silica nanoparticles (MSN) loaded with OCA, as well as a ferroptosis inhibitor liproxsatin-1 and a NO donor S-nitrosothiol (ONL@MSN). Biochemical analyses, histology, multiplexed flow cytometry, bulk-tissue RNA sequencing, and fecal 16S ribosomal RNA sequencing were utilized to evaluate the effects of the combined nanoparticle (ONL@MSN) in a mouse NASH model. Compared with the OCA-loaded nanoparticles (O@MSN), ONL@MSN not only protected against hepatic steatosis but also greatly ameliorated fibrosis and ferroptosis. ONL@MSN also displayed enhanced therapeutic actions on the maintenance of intrahepatic macrophages/monocytes homeostasis, inhibition of immune response/lipid peroxidation, and correction of microbiota dysbiosis. These findings present a promising synergistic nanotherapeutic strategy for the treatment of NASH by simultaneously targeting FXR, ferroptosis, and fibrosis.
Key words: Nonalcoholic steatohepatitis, Nanotherapy, Farnesoid X receptor, Ferroptosis, Fibrosis, Obeticholic acid, S-Nitrosothiol, Liproxstatin-1
Graphical abstract
The PEGylated and thiolated hollow mesoporous silica nanoparticles (MSN), loaded with OCA, S-nitrosothiol, and liproxstatin-1, exhibit potent therapeutic action against NASH by simultaneously targeting FXR, ferroptosis, and fibrosis.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) affects approximately one-quarter of the global adult population and poses a significant burden on both health and economy1. Hepatic abnormalities in NAFLD encompass a wide array of the clinicopathological spectrum of liver histology ranging from nonalcoholic fatty liver (NAFL) to nonalcoholic steatohepatitis (NASH) and steatofibrosis, ultimately resulting in cirrhosis, liver failure, and hepatocellular carcinoma. While the simple hepatic steatosis pathologies in NAFL stage are reversible, the inflammation and fibrosis in NASH stage can only occasionally be reversed, and progression to steatofibrosis, liver cirrhosis, and hepatocellular carcinoma is more frequent2. To date, the treatment of NAFLD is mainly based on lifestyle management such as dietary adjustments, physical exercise, and psychological intervention, all with the aim of reducing and controlling body weight as the primary objective. Notably, lifestyle management is most effective during NAFL but less effective in liver fibrosis and later stages2. There are no approved therapies by the European Medicines Agency or the American Food and Drug Administration to address this issue. Some pharmacological agents such as Vitamin E, glucagon-like peptide-1 (GLP-1) receptor agonists, and the proliferator-activated receptor gamma (PPARγ) ligand pioglitazone, are recommended for selected NASH patients3,4. These drugs primarily offer the specific advantage of reducing body weight and providing cardiovascular benefits. Despite these advancements, the prevention and treatment of NASH remain unsatisfactory, and there is a demand for better therapeutic options.
Liver fibrosis, a progressive pathological process that leads to the accumulation of excess extracellular matrix (ECM) in the liver, is considered a major histological feature of NASH. In NASH, there is an imbalance between fibrogenesis and fibrolysis, resulting in the excessive deposition of ECM. This is accompanied by a decrease in the secretion and activity of matrix metalloproteinases (MMPs) and an increase in the tissue inhibitors of MMPs (TIMPs), particularly TIMP-1, which is the primary physiological inhibitor of most MMPs5. The resolution of liver fibrosis is a crucial endpoint for assessing the effectiveness of NASH treatment.
Obeticholic acid (OCA), an analog of the natural bile acid chenodeoxycholic acid, acts as an agonist of the farnesoid X receptor (FXR), which has a significant impact on lipid and glucose homeostasis6. Obeticholic acid (OCA), an agonist of FXR which was developed by Intercept Pharmaceuticals, showed its potent alleviation on liver fibrosis and inflammation in hundreds of investigations7. An important impulse to identify OCA able to treat chronic liver diseases, and NASH in particular, was provided in 2014 by the results of the FLINT trial, showing the capacity of OCA to improve histological features of NASH, including hepatic steatosis, hepatocyte ballooning, inflammation, and fibrosis8. Unfortunately, it was rejected by the FDA in 2020 after a world-scale phase 3 double-blind, randomized, placebo-controlled, and multicenter trial because the trial did not totally meet its primary histological endpoint in advanced NASH patients, although OCA displayed moderate therapeutic action against fibrosis and inflammation during NAFLD9. It should be noted that fibrosis is an extremely difficult problem to overcome in almost all organs.
Ferroptosis is a newly recognized form of iron-dependent regulated cell death characterized by uncontrolled lipid peroxidation10, 11, 12, 13. It is distinct from other well-known types of cell death such as necrosis and apoptosis in terms of morphology, biochemistry, and genetics. In NASH, where lipotoxicity induced by excessive accumulation of intrahepatic lipids is a primary driver, the ferroptosis-associated with lipid peroxidation is considered a critical cause of hepatocyte death and inflammation. As a result, extensive research has been conducted to investigate the detrimental role of ferroptosis in NASH by mediating cell death and inflammation14, 15, 16, 17. Based on the results from these studies, including our recent works18, 19, 20, inhibition of ferroptosis has been recently confirmed as a potential approach to slow down the progression of NASH. Additionally, aside from its role in inducing cell death, ferroptosis has been reported to be a significant contributor to liver fibrosis induced by iron overload or chemical toxins21,22. Therefore, blocking ferroptosis appears to be a promising therapeutic strategy for the treatment of NASH. In addition, nitric oxide (NO), a pluripotent biological molecule involved in NAFLD progression23. also contributes to extracellular matrix degradation. Recently, an engineered PEGylated mesoporous silica nanoparticles (@MSN) loaded with NO donor was shown to activate pro-MMPs and deplete intratumor collagen fibrils24. Moreover, due to the potent anti-oxidant activity of NO, NO donor was delivered into the liver via nanoparticles showed protection against oxidative stress-associated pathology in NASH25. Thus, ferroptosis blocker and NO donor may have the ability to inhibit cell death and resolve liver fibrosis in NASH condition.
In this study, we developed a novel coordinative nanomedicine based on PEGylated thiolated hollow MSNs loaded with FXR agonist OCA, ferroptosis inhibitor liproxsatin-1, and NO donor S-nitrosothiol. To evaluate the therapeutic potential of this nanomedicine, we utilized a well-established animal model of NASH by feeding mice with a methionine/choline-deficient (MCD) diet. Various techniques, including histology, biochemical analysis, multicolor flow cytometry, bulk-tissue RNA sequencing (RNA-seq), and 16S ribosomal RNA sequencing (16S-rRNA-seq) of bacterial communities in fecal samples, were employed. The results demonstrated that this multifunctional nanotherapy exhibited a potent therapeutic effect against NASH by simultaneously targeting FXR, ferroptosis, and fibrosis. Consequently, it presents a promising synergistic strategy for the treatment of NASH.
2. Materials and methods
2.1. Materials
Obeticholic acid, liproxstatin-1, tetraethyl orthosilicate (TEOS, 98%), hexadecyltrimethylammonium bromide (CTAB, 99%), 2,3-diaminonaphthalene (2,3-DAN), diethylenetriaminepentaacetic acid (DTPA), and 3-(mercaptopropyl) trimethoxysilane (MPTMS) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Methoxy-PEG-maleimide (MW 5 kDa) was purchased from Sunny-Biotech (Shanghai, China). Dulbecco's modified Eagle medium (DMEM), fetal bovine serum (FBS), penicillin, streptomycin, trypsin, and DiD probe were obtained from Thermo‒Fisher Scientific (Northumberland, UK). Cell counting kit-8 (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan). One Step PrimeScript™ III RT-Qpcr Mix, RNAiso Plus reagent, and RNase-free DNase I were purchased from Takara (Tokyo, Japan). The primer oligonucleotides were purchased from Sangon Biological Engineering Technology & Co., Ltd. (Shanghai, China). TruStain FcXTM anti-CD16/32 antibody, rat monoclonal anti-CD45-BV605 (#103139), rat monoclonal anti-CD11b-FITC (#101205), rat monoclonal anti-TIM4-PerCP-Cy5 (#130019), mouse monoclonal anti-CX3CR1-BV421 (#49023), rat monoclonal anti-Ly6C-PE (#128007), mouse monoclonal anti-F4/80-APC-R700 (#565787), rat monoclonal anti-CD163-PE/Cyanine7 (#155319), rat monoclonal anti-CCR2-BV650 (#150613), Mouse monoclonal anti-CD64-BV510, APC anti-mouse MERTK (#151508) and propidium iodide solution were purchased from BioLegend (San Diego, USA). Primary antibodies against aSMA, TFR1, GPX4, and b-actin were purchased from Abcam (Cambridge, UK). Primary antibodies against TIMP1, TIMP-2, CPT1a, CXCL1, IL-6, and ACSL4 were purchased from ProteinTech (Wuhan, China). The goat polyclonal IRDye 800CW anti-mouse IgG antibody and IRDye 800CW Goat anti-rabbit IgG antibody were purchased from LI-COR Biosciences (Lincoln, NE). The serum ALT, AST, urea nitrogen, and creatinine assay kits were purchased from Cayman Chemical (Ann Arbor, MI, USA). The methionine and choline-deficient diet (MCD) was purchased from Research Diet (#A02082002B). Collagenase IV was purchased from Worthington Biochemical Corporation (New Jersey, US).
2.2. Mice
Wild-type control mice (C57BL/6J), aged 8 weeks, were procured from Sino-British SIPPR/BK Lab Animal Ltd. (Shanghai, China). All experimental procedures involving mice were reviewed and approved by the Institutional Animal Care and Use Committee of Naval Medical University and followed the Principles of Laboratory Animal Care published by the National Institutes of Health (NIH publication 86-23 revised 1985) and ARRIVE guidelines. The mice were housed in a temperature-controlled environment (23 ± 2 °C) with free access to water and chow, under a 12/12-h light/dark cycle, and every effort was made to minimize their use and discomfort. The animal health status, including body weight measurement, behavior observation, and responses to external stimuli, was monitored daily.
2.3. Preparation of nanoparticles
Mesoporous silica nanoparticles (MSN) were synthesized according to the method described24. Specifically, 0.5 g of CTAB and 1.75 mL of NaOH (2 mol/L) were dispersed in 250 mL of deionized water. The mixture was heated to 70 °C under vigorous stirring and maintained for 30 min. To the mixture, 2.2 mL of TEOS was rapidly added via injection for 1 min while stirring. Then, 3 mL of ethyl acetate was mixed into the mixture and vigorously stirred for 30 s. The reaction continued for 2 h to age the MSN. Mesoporous silica nanoparticles were collected by centrifugation (12,500 rpm, 25 min; Eppendorf, 5424R, Hamburg, Germany) and washed with ethanol three times. To modify mercapto on the surface of MSN, 3 mL of MPTMS was added to the mixture of 250 mg of MSN in 50 mL of ethanol under stirring. The solution was refluxed at 80 °C for 24 h under argon gas protection, and then the mercapto-modified MSN was collected by centrifugation (12,500 rpm, 25 min; Eppendorf) and rinsed with methanol. To remove CTAB (as the surfactant template), MSN-SH with CTAB was sonicated in an acidic ethanol solution under a 60 °C water bath, and MSN-SH was collected by centrifugation (12,500 rpm, 25 min; Eppendorf). The mercapto groups on the MSN-SH surface were quantified by Ellman's reagent. To modify PEG on MSN-SH, MSN-SH (100 mg) and methoxy-PEG aleimide (40 mg) (molar ratio of mercapto to maleimide groups 10:1) were dispersed into 100 mL HEPES buffer (0.01 mol/L, pH 7.2). The reaction was conducted by stirring at room temperature for 6 h under argon protection. The reaction product was centrifuged (12,500 rpm, 25 min; Eppendorf) to collect PEG-MSN-SH. To modify nitrosothiol on PEG-MSN-SH, 100 mg PEG-MSN-SH was dispersed in a 40 mL mixture of methanol and water (v/v, 1/1) in an ice bath. 5 mL of hydrochloric acid (6 mol/L) was then added dropwise with stirring for 15 min. Then, 2 mL of sodium nitrite solution (8 mol/L, containing 1 mmol/L DTPA) was added to the mixture. After reacting for 3 h, the solution was centrifuged (12,500 rpm, 25 min; Eppendorf) to collect N@MSN (S-nitrosothiol-loaded PEG-MSN). The loading capacity and encapsulation efficiency of the OCA and liproxsatin-1 were illustrated in Supporting Information Table S1.
2.4. Drug loading and DiD labelling
To load the drug, 80 mg of either obeticholic acid or liproxstatin-1 was dissolved in 800 μL of DMF. Then, 240 mg of S-nitrosothiol-loaded PEG-MSN was dispersed in the solution and shaken at 4 °C for 12 h. The drug-loaded MSN were collected by centrifugation (12,500 rpm, 25 min; Eppendorf). Then, the drug-loaded MSNs were dispersed in 40 mL of HEPES buffer (1 mmol/L, pH 8.5). When used, the solution was diluted with sterile water for injection twice.
2.5. Characterization of the nanoparticles in vitro
The ultrastructure analysis of both mesoporous silica nanoparticles (MSN) and drug-loaded MSN was conducted employing field-emission transmission electron microscopy (TEM). To facilitate TEM imaging, MSN and drug-loaded MSN were first dispersed in deionized water at a concentration of 5 mg/mL. Subsequently, 5 mL of this prepared solution was carefully transferred onto a perforated carbon-coated copper TEM grid. Following air drying, a state-of-the-art high-resolution TEM instrument (FEI TALOS L120C) was employed to capture detailed images of the samples, operating at an accelerating voltage of 200 kV. For a comprehensive understanding of the nanoparticles’ characteristics, the size and zeta potential were meticulously determined utilizing a ZetaSizer Nano instrument manufactured by Malvern Instruments Ltd., UK. To prepare the samples for analysis, 200 mL of the particle suspension (at a concentration of 25 mg/mL) was meticulously diluted in 1.8 mL of deionized water (H2O). The resulting solutions were then meticulously transferred into 1 mL polystyrene cuvettes, ensuring precise conditions for subsequent analysis.
2.6. Assay of drugs loading in nanoparticles
The drug loading for the nanoparticles was measured by an indirect method that measures the absorbance of excessive drugs in the supernatant by reverse-phase HPLC (LC-20A, Shimadzu Inc., Japan). 80 mg of either obeticholic acid or liproxstatin-1 was dissolved in 800 μL of DMF. Then, 240 mg of MSN (PEG-MSN, or N@MSN) was dispersed in the solution and shaken at 4 °C for 12 h. The supernatant was collected by centrifugation (12,500 rpm, 25 min; Eppendorf) and fixed in volume up to 100 mL with methanol.
2.7. Drug release examination in vitro
To determine the in vitro drug release from the nanoparticles at neutral or acidic pH (7.4 or 5.5), 100 μL of solutions of drug and DiD loaded nanoparticles were placed in 2 mL ofuge tubes with 1 mL of 0.1% SDS PBS (pH 7.4 or 5.5) shaken in the dark at 250 rpm at 37 °C. At different time points (0, 1, 2, 4, 8, 12, 24, and 48 h), the solutions were centrifuged (12,500 rpm, 25 min; Eppendorf), and the supernatants were collected. The precipitated nanoparticles were added to an equal volume (1 mL) of fresh suspended buffer (PBS containing 0.1% SDS). The absorbance of drugs in the supernatant was measured by the reverse-phase HPLC as described above. As shown in Eq. (1), the release rate was calculated using the formula, where Wi is the amount of accumulative released drugs, and Wtotal is the total amount of drugs.
| (1) |
2.8. Mouse NASH model
The mouse NASH model was induced by feeding the 8-week-old mice with a methionine/choline-deficient (MCD) diet for 4 weeks as described previously18,19.
2.9. Cell culture
AML12 cell line was acquired from the Cell Bank of the Shanghai Chinese Academy of Sciences (Shanghai, China). The cells were cultured using DMEM medium supplemented with 10% fetal bovine serum under a temperature of 37 °C and 5% CO2 concentration. For evaluating cell uptake of nanoparticles, the DiD-labeled nanoparticles and Hoechst 33342 were added into the culture of AML12 cells. Thirty minutes later, the cells were visualized using a fluorescence microscope (Olympus, IX71).
2.10. Cell viability assay
AML12 cells were seeded in 96-well plates and stimulated with different agents for 48 h, followed by incubation with 10 μL of CCK8 at a temperature of 37 °C for 40 min. The absorbance was measured at 450 nm using a microplate reader.
2.11. Distribution of nanoparticles using in vivo imaging system
NASH mice were depilated in advance with hair removal cream. The drug labeled with the fluorescent probe DiD was injected into the mice via the tail vein. The five groups of drugs administered included physiological saline, MSN, O@MSN, ON@MSN, and ONL@MSN. After inducing 5% isoflurane anesthesia, the mice were placed in a Tanon ABL-X5 (Shanghai Tanon Science & Technology Co., Ltd.) for imaging, capturing the background light and red fluorescence. For organ imaging, the mice were euthanized, and their organs (liver, spleen, lungs, kidneys, and inguinal fat) were isolated and the fluorescence was captured.
2.12. Histological analysis
Paraffin-embedded liver tissues were fixed in formalin and 5-μm thick sections were cut for H&E, Masson's trichrome, and Sirius red staining. For tyramide signal amplification (TSA)-based multiplex-color immunofluorescence staining in the liver, the DiD dye-labeled nanoparticles were injected into the tail vein of mice. Twenty-four hours later, the liver tissue of mice was isolated and cut into pieces. TSA multiplex-color immunofluorescence staining was performed using a commercial kit (abs50028-20T, Absin, Shanghai, China) according to the manufacturer's instructions.
2.13. NAFLD activity score
To differentiate the severity of NAFLD, histological specimens in mice were scored according to the NAFLD activity score, with systematic evaluation of hepatocellular ballooning, lobular inflammation, and fibrosis with the recommendations from NAFLD Clinical Research Network26. The amount of steatosis (percentage of hepatocytes containing fat droplets) was scored as 0 (<5%), 1 (5%–33%), 2 (>33%–66%), and 3 (>66%) with Image J software (NIH). Hepatocyte ballooning was classified as 0 (none), 1 (few), or 2 (many cells/prominent ballooning). Foci of lobular inflammation were scored as 0 (no foci), 1 (<2 foci per 200 × field), 2 (2–4 foci per 200 × field), and 3 (>4 foci per 200 × field). We isolated the same liver lobe to reduce the discrepancy among mice and always observed 8–10 different visual fields for each mouse.
2.14. Real-time quantitative PCR
Real-time quantitative PCR was conducted using a CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA). Total RNA was extracted from tissues using RNAiso Plus reagent and RNase-free DNase I was used at 37 °C for 30 min to reduce the risk of genomic DNA contamination. The RNA sample was subjected to the One Step PrimeScript™ III RT-Qpcr Mix with specific primers. The primer oligonucleotides (listed in Supporting Information Table S2) were purchased from Sangon Biological Engineering Technology & Co., Ltd. (Shanghai, China) and purified using high-performance liquid chromatography. Each PCR reaction was performed in triplicate, and the comparative analysis was performed using the 2‒ΔΔCt method27.
2.15. Western blot experiment
Tissues were homogenized in the RIPA buffer in the presence of a protease inhibitor cocktail at 4 °C. The samples were then centrifuged at 15,000 × g in 4 °C for 20 min. Samples were boiled and then loaded in SDS–PAGE. After electrophoresis, the samples were transferred to a nitrocellulose membrane in a transfer buffer. Thereafter, the nitrocellulose membrane was blocked in 5% non-fat milk (w/v) dissolved in phosphate-buffered saline with Tween 20 and then incubated with specific primary antibodies for 4–6 h and followed by secondary antibodies for 2 h at room temperature. Images were captured and quantitative analysis was performed using the Odyssey imaging system (Li-Cor, NE, USA).
2.16. Isolation of liver cells
The liver was perfused for 2 min with HBSS (Hank's balanced salt solution) at a rate of 10 mL/min via portal vein until the liver turned pale. Afterward, the liver was perfused with HBSS containing 1 mg/mL Collagenase IV at 3 mL/min for 10–15 min. Livers were then removed, minced, and incubated for 20 min with 0.5 mg/mL Collagenase IV and 0.2 mg/mL DNase on a shaker (100 rpm) at 37 °C for 30 min. All subsequent procedures were performed at 4 °C. Samples were filtered over a 100 mm mesh filter and red blood cells were lysed. The cells were again filtered over a 40 mm mesh filter and centrifugated for 1 min at 50 × g to isolate hepatocytes. The remaining non-parenchymal cells were centrifuged at 500 × g for 10 min at 4 °C before proceeding to antibody.
2.17. Flow cytometry
To block Fc receptors, non-parenchymal cells (1 × 106) were incubated on ice with 0.025 μg of TruStain FcXTM anti-CD16/32 antibody (BioLegend) for 10 min. Then, appropriate antibodies were added and incubated for 45 min in the dark at 4 °C. Dead cells were excluded by staining with propidium iodide. Multiplexing flow cytometry was used to cluster leukocytes (CD45+) with antibodies against CD11b, F4/80, Ly6C, CD64, CX3CR1, CCR2, CD163, and TIM4. Samples were acquired using CytoFLEX S (Beckman) and analyzed with FlowJo software (version 10; Tree Star). The flow cytometry antibodies used are listed in the Supporting Information.
2.18. Bulk-tissue RNA sequencing
Total RNA was extracted from the tissue using RNAiso Reagent according to the manufacturer's instructions and genomic DNA was removed using DNase I. Then RNA quality was determined by 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). Libraries were size selected for cDNA target fragments of 300 bp on 2% Low Range Ultra Agarose followed by PCR amplified using Phusion DNA polymerase (NEB) for 15 PCR cycles. After quantified by TBS380, the paired-end RNA-seq sequencing library was sequenced with the Illumina HiSeq 4000 sequencer. For bioinformatics analyses, KEGG pathway analysis was carried out by KOBAS (http://kobas.cbi.pku.edu.cn/home.do). The raw data was deposited in the NCBI BioSample database (BioSample ID: SAMN35682223).
2.19. Fecal 16S rRNA sequencing
For 16S rRNA sequencing and transcriptome sequencing, microbial DNA and mRNA were extracted from specific tissues or cells of interest at a selected time point. Illumina platform-based sequencing was carried out using the NovaSeq Reagent Kit method for library construction, and bioinformatics analysis was performed using advanced computational tools to analyze the sequencing data. For eukaryotic mRNA sequencing, Illumina sequencing technology was employed to sequence all transcripts of interest in a specific tissue or cell at a certain time point. The sequencing experiment was performed using the Illumina NovaSeq Reagent Kit method for library construction, and analysis of the sequencing data was carried out using a bioinformatics platform. The raw data was deposited in the NCBI BioProject database (BioProject ID: PRJNA981653).
2.20. Statistical analysis
All results were presented as mean ± standard error of mean (SEM). The distribution of data within all individual groups was verified by the Shapiro–Wilk test of normality. The statistical analysis was performed by analysis of variance (ANOVA) followed by the Tukey-hoc test. The number of animals/samples in each group is indicated in the figure legend. Unless otherwise stated, the statistical significance level was set at 0.05. All statistical analyses were performed using GraphPad Prism 8.
3. Results and discussion
3.1. Synthesis and in vitro stability of nanoparticles
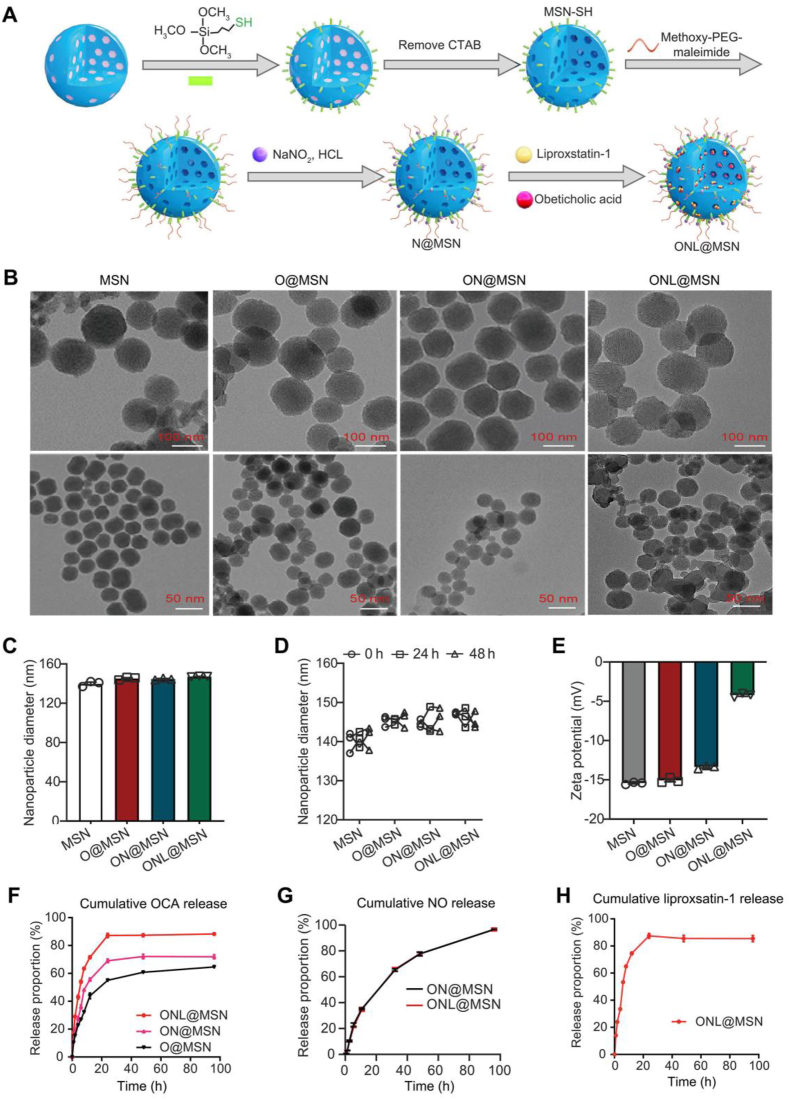
The drug-loaded PEGylated thiolated MSNs were synthesized following previously described methods with some modifications24. Thiol groups were introduced on the nanoparticle surface through the modification of mercaptopropyl-trimethoxysilane (MPTMS), as shown in Fig. 1A. The surfactant template, cetrimonium bromide (CTAB), was removed to generate empty nanoparticles with a surface modified with SH (MSN-SH). Methoxy-PEG aleimide was then applied to confer PEGylation of the nanoparticle surface (PEG-MSN-SH). The remaining thiols were transformed into NO donor S-nitrosothiols by reacting with NaNO2. Lastly, OCA and liproxstatin-1 were loaded into the nanoparticles to form ONL@MSN. Field-emission transmission electron microscopy (TEM) was used to visualize these drug-loaded nanoparticles (Fig. 1B). The diameters and zeta potentials of the nanoparticles were determined. The diameters of MSN, O@MSN, ON@MSN, and ONL@MSN were approximately 140 nm (Fig. 1C). The colloidal stability of the nanoparticles (@MSN, O@MSN, ON@MSN, and ONL@MSN) in 10% FBS-supplemented PBS at 37 °C was well maintained for at least 48 h (Fig. 1D). The zeta potentials were −15.4, −14.7, and −13.3 mV in MSN, O@MSN, and ON@MSN, respectively, while the zeta potential in ONL@MSN was −4.59 mV (Fig. 1E). The cumulative release of OCA in ONL@MSN at 6 h is about 54%, but 27% and 36% in O@MSN and OCA in ON@MSN, respectively (Fig. 1F). There is no difference in NO release between the ON@MSN and ONL@MSN (Fig. 1G). The cumulative release of liproxsatin-1 in ONL@MSN is about 53% at 6 h (Fig. 1H).
Figure 1.
Preparation and characterization of nanoparticles. (A) A schematic illustration depicts the process for creating PEGylated thiolated hollow mesoporous silica nanoparticles (MSN) loaded with OCA, NO donor S-nitrosothiol, and ferroptosis inhibitor liproxsatin-1 (referred as ONL@MSN). (B) Field-emission transmission electron microscopy showing the ultrastructure of synthesized nanoparticles. Scale bar = 100 nm. (C) The diameter of the synthesized nanoparticles in normal condition. (D) The diameter of the synthesized nanoparticles in 10% FBS supplemented PBS at 37 °C for 48-h incubation. (E) The zeta potential of the synthesized nanoparticles. (F) Cumulative OCA release from O@MSN, ON@MSN, and ONL@MSN in PBS (pH 6.8). (G) Cumulative NO release from ON@MSN and ONL@MSN in PBS (pH 6.8). (H) Cumulative liproxstain-1 release from ONL@MSN in PBS (pH 6.8).
3.2. Evaluation of distributed DiD-labeled nanoparticles in hepatocytes in vitro and in vivo
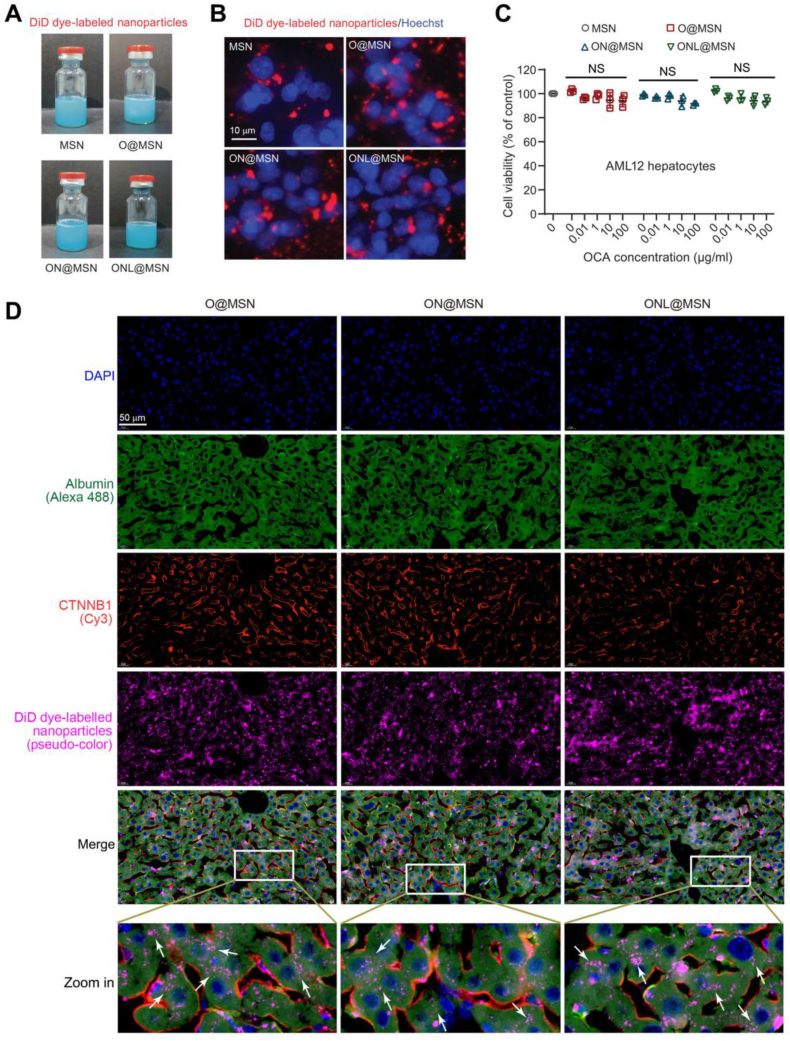
No apparent differences were observed visually among the synthesized ONL@MSN, as well as O@MSN and ON@MSN when examined with the naked eye (Fig. 2A). In AML12 cells, a normal mouse hepatocyte cell line, DiD-labeled nanoparticles were absorbed by the cells and predominantly located in the cytosol (Fig. 2B). This observation aligns with the findings of a prior study28. To assess the cytotoxicity of these nanoparticles, a CCK-8 assay was conducted to measure the inhibitory effects of varying concentrations of nanoparticles on AML12 cells. The cell viability assay indicated that these nanoparticles exhibited low cytotoxicity across the OCA concentration range of 0.01–100 μg/mL (Fig. 2C). To demonstrate the entry of nanoparticles into target cells, a TSA-based multiplex immunofluorescence staining was employed to elucidate the distribution of DiD-labeled nanoparticles in mouse livers. In mouse livers, it was evident that a majority of DiD-labeled nanoparticles were primarily distributed in hepatocytes (white arrows in zoom-in images, Fig. 2D). These findings suggest the anticipated absorption of nanoparticles by hepatocytes.
Figure 2.
Evaluation of the distribution of prepared nanoparticles within hepatocytes in vitro and in vivo. (A) Photographs of the DiD-dye labeled synthesized nanoparticles in aqueous solution. (B) Cultured AML12 hepatocytes uptake the nanoparticles labeled with the DiD probe (red). The nucleus was labeled with Hoechst33342 (blue). The images were captured using fluorescence microscope. Scale bar = 10 μm. (C) Cell viability of AML12 hepatocytes incubating with MSN, O@MSN, ON@MSN, and ONL@MSN at the indicated concentration of OCA (0.01–100 μg/mL) for 48 h. (D) A tyramide signal amplification (TSA)-based multiplex-color immunofluorescence staining showing the distribution of DiD dye-labeled nanoparticles in mice liver. The DiD-dye labeled nanoparticles were injected into the tail vein of mice, and the liver tissue of mice was isolated and cut into pieces 24 h later. The TSA multiplex-color immunofluorescence staining was performed and the images were captured using the confocal laser scanning microscope. Albumin (green) indicates hepatocytes. CTNNB1 (orange) indicates the cell membrane. DiD (pink) indicates the nanoparticles absorbed by liver cells. Scale bar = 50 μm.
3.3. Liver-targeted biodistribution of nanoparticles
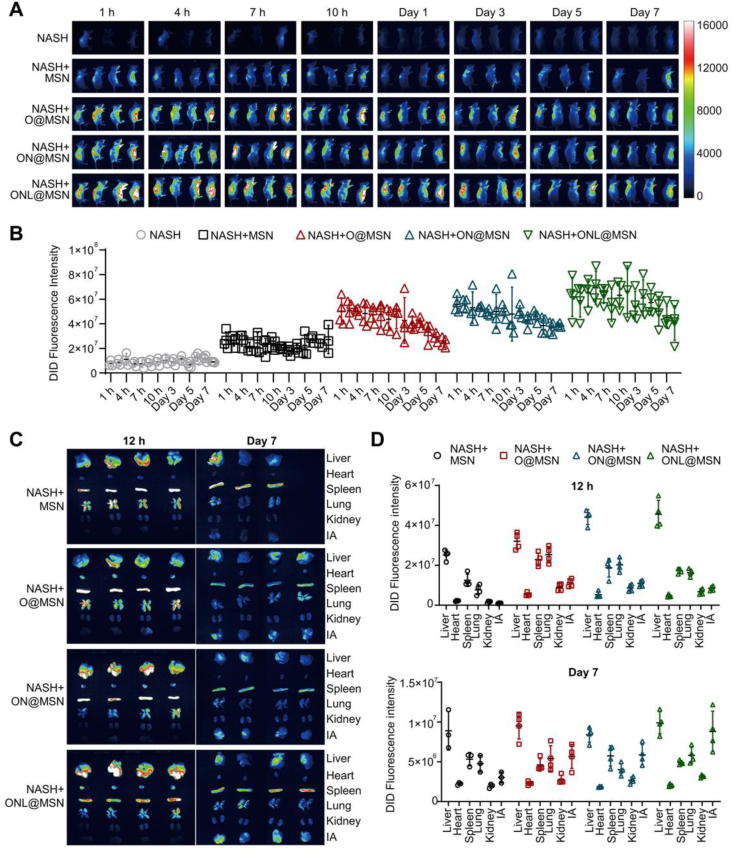
To investigate the in vivo distribution of nanoparticles, NASH mice were injected with DiD-labeled nanoparticles via the tail vein, including a saline control and four types of nanoparticles (MSN, O@MSN, ON@MSN, and ONL@MSN). Minimal fluorescence was observed in NASH mice injected with saline, whereas NASH mice injected with MSN showed a significantly higher fluorescence signal (Fig. 3A). The fluorescence intensity was further enhanced in NASH mice injected with O@MSN, ON@MSN, and ONL@MSN, with the strongest intensity observed in mice injected with ONL@MSN (Fig. 3A). The nanoparticles primarily accumulated in the visceral organs and maintained a relatively high level for one week (Fig. 3B).
Figure 3.
Analysis of in vivo distribution of DiD-labeled nanoparticles in NASH mice. (A) Images were captured at different time intervals (1, 4, 7, 10 h, Day 1, Day 3, Day 5, and Day 7) after tail intravenous injection of the nanoparticles into NASH mice using an in vivo imaging system (n = 4 per group). (B) The fluorescence intensity in the abdomen region of NASH mice receiving injections of normal saline, MSN, O@MSN, ON@MSN, and ONL@MSN nanoparticles was measured at 1 h, 4, 7, 10 h, Days 1, 3, 5, and 7 post-injection. (C) The fluorescence images of organs were isolated at 12 h and Day 7 post DiD-labeled nanoparticle injection. (D) The fluorescence intensity of organs isolated at 12 h and on Day 7 post DiD-labeled nanoparticle injection.
While it is widely recognized that the liver is the main accumulation site for nanoparticles, we aimed to determine the primary target organs of our synthesized nanoparticles. Major organs including the liver, heart, spleen, lung, kidney, and inguinal fat (IA) were collected for fluorescence imaging at 12 h and 7 days after injection. DiD-labeled nanoparticles were found to mainly accumulate in the liver (Fig. 3D). The nanoparticle also deposited in the spleen and lung to some extent (Fig. 3D). Interestingly, the IA appeared to be a significant deposit site for nanoparticles at 7 days post-injection, in addition to the liver, spleen, and lung (Fig. 3D). The fluorescence intensity was highest in the liver at both time points (Fig. 3E). These results indicate that the nanoparticles synthesized in this study, loaded with drugs, primarily accumulate in the liver.
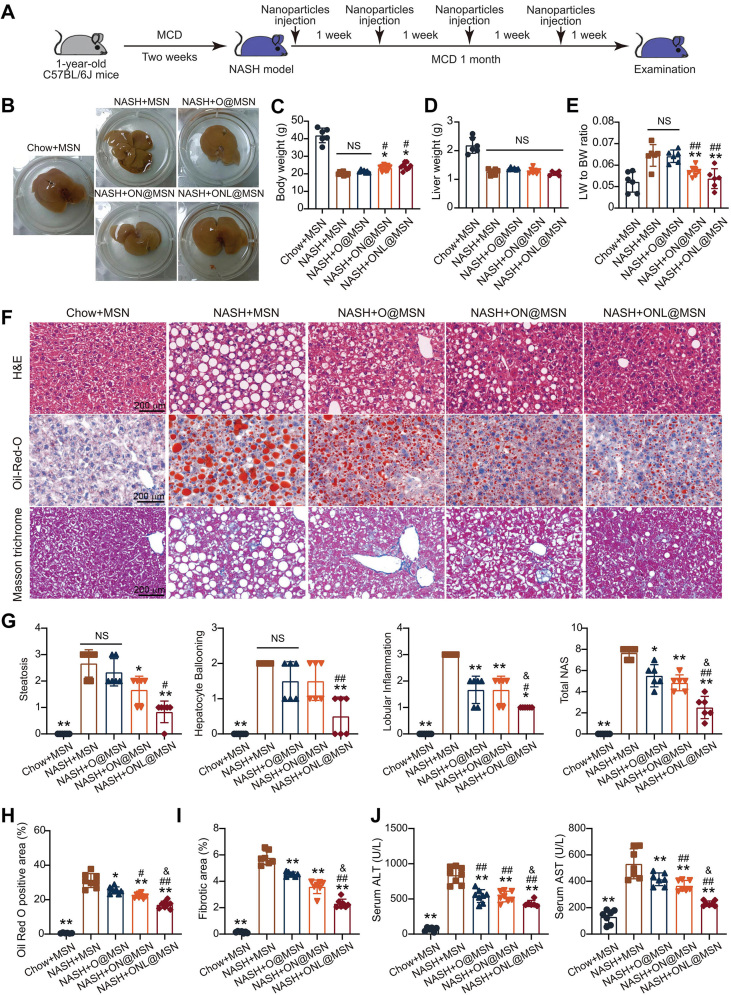
3.4. Synthesized nanoparticles alleviate NASH pathologies in mice
To assess the therapeutic effect of the synthesized nanoparticles, we utilized a mouse model of NASH induced by feeding an MCD diet (Fig. 4A). OCA is always used at the dose of ∼10 mg/kg in rodent animal models with NASH28,29. At the experimental endpoint, liver tissue isolated from NASH mice injected with MSN (NASH + MSN) exhibited evident cirrhosis, while this was not observed in mice injected with the other three types of nanoparticles (NASH + O@MSN, NASH + ON@MSN, and NASH + ONL@MSN) at 3 mg/kg. These findings suggest that the synthesized nanoparticles provide protection against NASH pathologies (Fig. 4B). The body weight of NASH mice significantly decreased, but ON@MSN and ONL@MSN slightly increased the body weight (Fig. 4C). The nanoparticles did not alter liver weight (Fig. 4D). However, the ratio of liver weight to body weight, which was significantly increased in NASH mice, was suppressed by ON@MSN and ONL@MSN, but not O@MSN (Fig. 4E). Histological examinations including hematoxylin and eosin (HE) staining, Oil Red O staining, and Masson trichrome staining were performed to evaluate pathological changes in the liver (Fig. 4F). Based on HE staining, the NAFLD activity score (NAS) was calculated, taking into account steatosis, hepatocyte ballooning, and lobular inflammation (Fig. 4G). All three types of nanoparticles, O@MSN, ON@MSN, and ONL@MSN, successfully inhibited pathological NASH changes, with the most significant improvement observed in ONL@MSN-injected mice (Fig. 4G). Similarly, Oil Red O staining and Masson trichrome staining revealed that ONL@MSN exhibited the highest reduction in lipid deposits and fibrotic areas in liver tissue, respectively (Figure 3, Figure 4H and I). Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) activities, which were significantly elevated in NASH mice, were suppressed by all three types of nanoparticles, with ONL@MSN demonstrating the strongest effect (Fig. 4J). These results collectively demonstrate that the synthesized nanoparticles, especially ONL@MSN, effectively alleviate hepatic steatosis in NASH mice.
Figure 4.
ONL@MSN alleviates inflammation, steatosis, and fibrosis in a NASH mouse model. (A) Experimental design for the administration of nanoparticles in a mouse NASH model. (B) Representative images of liver tissue from different groups, including the mice treated with Chow + MSN (chow-fed mice receiving MSN injection), NASH + MSN group (NASH mice receiving MSN injection), NASH + O@MSN group (NASH mice receiving O@MSN injection), NASH + ON@MSN group (NASH mice receiving ON@MSN injection), and NASH + ONL@MSN group (NASH mice receiving ONL@MSN injection). (C–E) The effect of drug-loaded nanoparticle treatment on body weight (C), liver weight (D), and the liver weight-to-body weight ratio (E). (F) Representative images of liver sections stained with H&E, Oil Red O, and Masson's trichrome. Scale bar = 200 μm. (G) Statistical indexes of steatosis, hepatocyte ballooning, and lobular inflammation, and the calculated NAFLD activity score (NAS). (H) Quantification of lipid accumulation in the liver by Oil Red O staining. (I) Quantification of liver fibrosis by Masson's staining. (J) Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels in mice. Data are presented as mean ± SEM and analyzed using one-way ANOVA followed by Tukey's analysis. ∗P < 0.05, ∗∗P < 0.01 vs. NASH + MSN. #P < 0.05, ##P < 0.01 vs. NASH + O@MSN. &P < 0.05 vs. NASH + ON@MSN. NS, no significance. n = 6 per group.
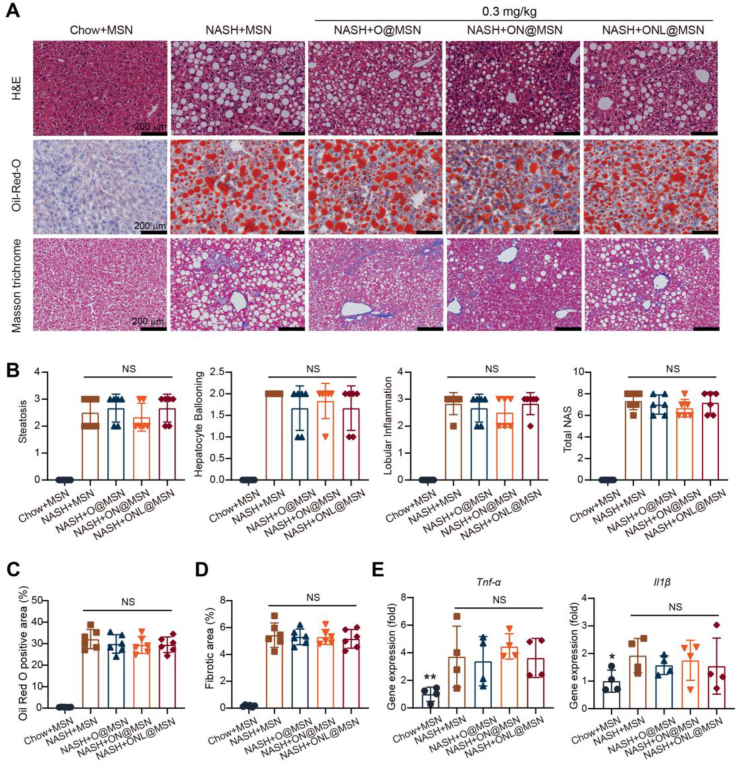
3.5. Synthesized nanoparticles showed limited efficacy in alleviating NASH pathology at a low dose
The impact of these nanoparticles was also examined at a relatively low dose (0.3 mg/kg). However, histological analyses (H&E, Masson's trichrome, αSMA immunohistochemistry) (Fig. 5A) revealed that none of the three nanoparticles exhibited efficacy in mitigating NAFLD activity score (Fig. 5B), lipid accumulation (Fig. 5C), or liver fibrosis (Fig. 5D). Consistently, the drug-loaded nanoparticles did not lead to the downregulation of mRNA expressions of pro-inflammatory factors, including Tnf-α and Il1β (Fig. 5E).
Figure 5.
Combined nanoparticles fail to provide protection against NASH at a relatively low dose. (A) Representative liver sections stained with H & E, Oil Red O, and Masson's trichrome in mice from various groups (Chow + MSN, NASH + MSN, NASH + O@MSN, NASH + ON@MSN, and NASH + ONL@MSN). A dose of 0.3 mg/kg was administered in these experiments. Scale bar = 200 μm. (B) Statistical parameters for steatosis, hepatocyte ballooning, lobular inflammation, and the calculated NAFLD activity score (NAS). n = 6 per group. (C) Quantification of lipid accumulation in the liver through Oil Red O staining. n = 6 per group. (D) Quantification of liver fibrosis via Masson's staining. n = 6 per group. (E) Quantitative analysis of the gene expression of pro-inflammatory factors (Tnf-α and Il1β) in the liver of mice treated with nanoparticles. n = 4 per group. Data are presented as mean ± SEM and analyzed using one-way ANOVA followed by Tukey's analysis. NS, no significance.
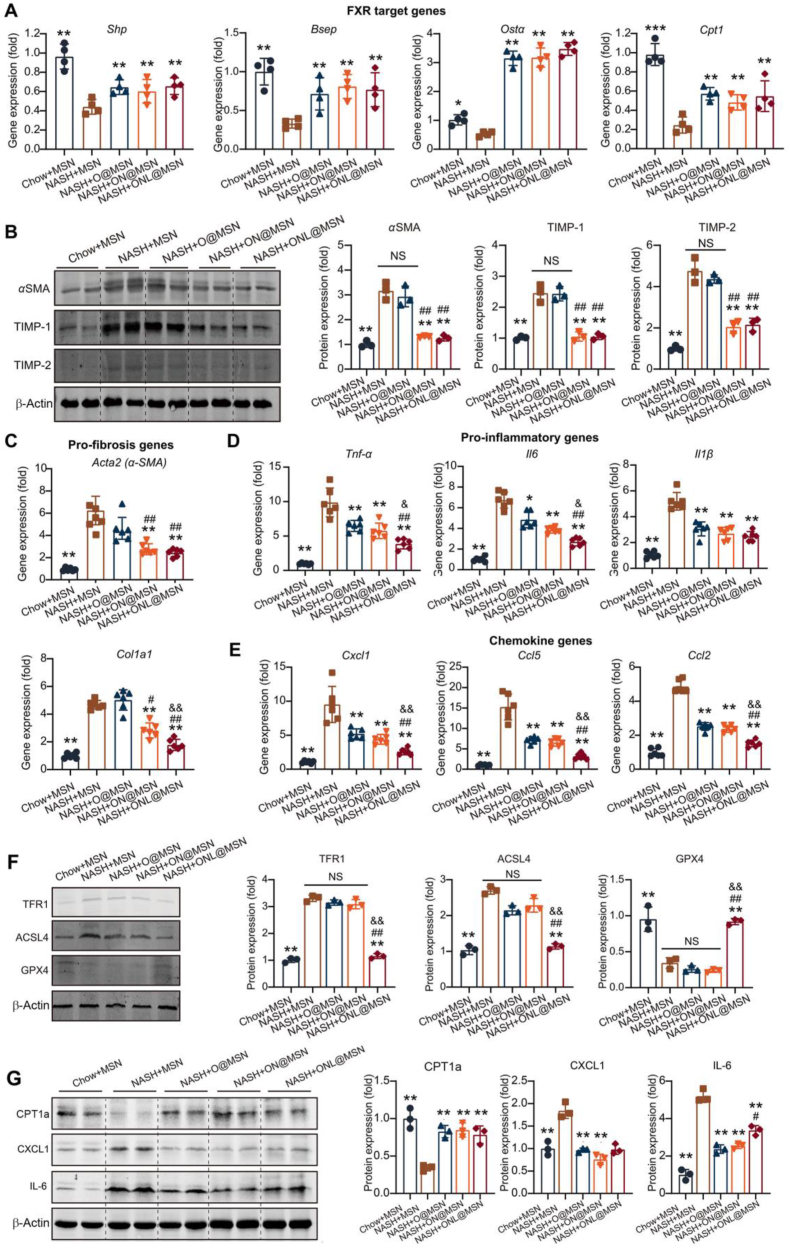
3.6. ONL@MSN successfully exerts combined effects on FXR, ferroptosis, and fibrosis in NASH mice
Following the confirmation of the nanoparticles’ therapeutic action against NASH pathologies in NASH mice, we conducted further investigations to determine whether the nanoparticles targeted FXR, ferroptosis, and fibrosis as intended. The expression of FXR-targeted genes, including small heterodimer partner (Shp), bile salt export pump (Bsep), organic solute transporter-α (Ostα), and carnitine palmitoyltransferase 1 (Cpt1), was downregulated in the liver tissues of NASH mice. However, O@MSN, ON@MSN, and ONL@MSN significantly upregulated these FXR-targeted genes, indicating the activation of released OCA from these nanoparticles on hepatic FXR. There was no difference in the FXR activation among the three groups of nanoparticles (Fig. 6A).
Figure 6.
ONL@MSN exerts combined effects on FXR, ferroptosis, and fibrosis in NASH mice. (A) Quantitative analysis of gene expression of FXR-targeted genes (Shp, Bsep, Osta, and Cpt1) by Real-Time PCR in the liver of mice with treatment of nanoparticles. The three types of synthesized nanoparticles (O@MSN, ON@MSN, and ONL@MSN) regulated FXR-targeted genes to the same extent. n = 4 per group. (B) Representative immunoblotting images and quantitative analysis of fibrosis-related proteins (aSMA, TIMP-1, and TIMP-2) in the liver of mice with the treatment of nanoparticles. Only ON@MSN and ONL@MSN suppressed the protein expression of fibrosis-related proteins. n = 3 per group. (C) Quantitative analysis of gene expression of fibrosis-related genes (Acta2 and Col1a1) by Real-Time PCR in the liver of mice with the treatment of nanoparticles. Only ON@MSN and ONL@MSN inhibited the gene expression of Acta2 and Col1a1. n = 6 per group. (D, E) Quantitative analysis of gene expression of proinflammatory factors (Tnf-α, Il6 and Il1b, D) and chemokines (Cxcl1, Ccl5 and Ccl2, E) in the liver of mice with the treatment of nanoparticles. The three types of synthesized nanoparticles (O@MSN, ON@MSN, and ONL@MSN) inhibited their expression to different extents. n = 6 per group. (F) Representative immunoblotting images and quantitative analysis of ferroptosis-related proteins (TFR1, ACSL4, and GPX4) in the liver of mice with the treatment of nanoparticles. Only ONL@MSN suppressed the protein expression of pro-ferroptotic factors TFR1 and ACSL4, as well as rescued the protein expression of anti-ferroptotic factor GPX4. n = 3 per group. (G) Representative immunoblotting images and quantitative analysis of protein levels of CPT1a (representative of FXR target genes), CXCL1 (representative of chemokines), and IL-6 (representative of pro-inflammatory factors) in the liver of mice with the treatment of nanoparticles. n = 3 per group. Data are presented as mean ± SEM and analyzed using one-way ANOVA followed by Tukey's analysis. ∗P < 0.05, ∗∗P < 0.01 vs. NASH + MSN. #P < 0.05, ##P < 0.01 vs. NASH + O@MSN. &P < 0.05 vs. NASH + ON@MSN. NS, no significance.
Next, we assessed the fibrosis in the mice. The protein expression of α-smooth muscle actin (αSMA), a fibrosis marker, was highly induced in NASH mice. However, ON@MSN and ONL@MSN, but not O@MSN, inhibited the upregulation of αSMA (Fig. 6B). Similar changes were observed in TIMP-1 and TIMP-2. Accordingly, the mRNA expressions of Acta2 (encoding αSMA protein) and Col1α1 (encoding type I collagen protein) were triggered in NASH mice but suppressed by ON@MSN and ONL@MSN, but not O@MSN (Fig. 6C). The increased mRNA expressions of pro-inflammatory factors, including tumor necrosis factor-α (Tnf-α), interleukin-6 (Il6), and Il1β, in NASH mice liver were inhibited by O@MSN, ON@MSN, and ONL@MSN, with ONL@MSN showing the most significant inhibitory action (Fig. 6D). Similar patterns were observed in chemokines, including C-X-C motif chemokine ligand 1 (Cxcl1), C–C motif chemokine ligand 5 (Ccl5), and Ccl2 (Fig. 6E).
Finally, we examined the ferroptosis status in the mice's liver tissues. In the livers of NASH mice, the pro-ferroptosis proteins, including transferrin receptor (TFR1)30 and acyl-CoA synthetase long-chain family member 6 (ACSL4)31, were induced, while the anti-ferroptosis protein GPX412 was depleted (Fig. 6F). These changes were only reversed by treatment with ONL@MSN, but not O@MSN and ON@MSN (Fig. 6F), indicating the specific action of the ferroptosis inhibitor liproxsatin-1. Finally, we assessed the protein levels of SHP (representative of FXR target genes), CXCL1 (representative of chemokines), and IL-6 (representative of pro-inflammatory factors), confirming consistent alterations in these proteins (Fig. 6G). Overall, these findings demonstrate that the multifunctional ONL@MSN successfully activates FXR, ameliorates hepatic fibrosis, and inhibits ferroptosis in NASH mice, surpassing the efficiency of O@MSN and ON@MSN as intended.
3.7. ONL@MSN modulates the balance between hepatic inflammation-resolving and inflammation-inducing macrophages/monocytes in NASH mice
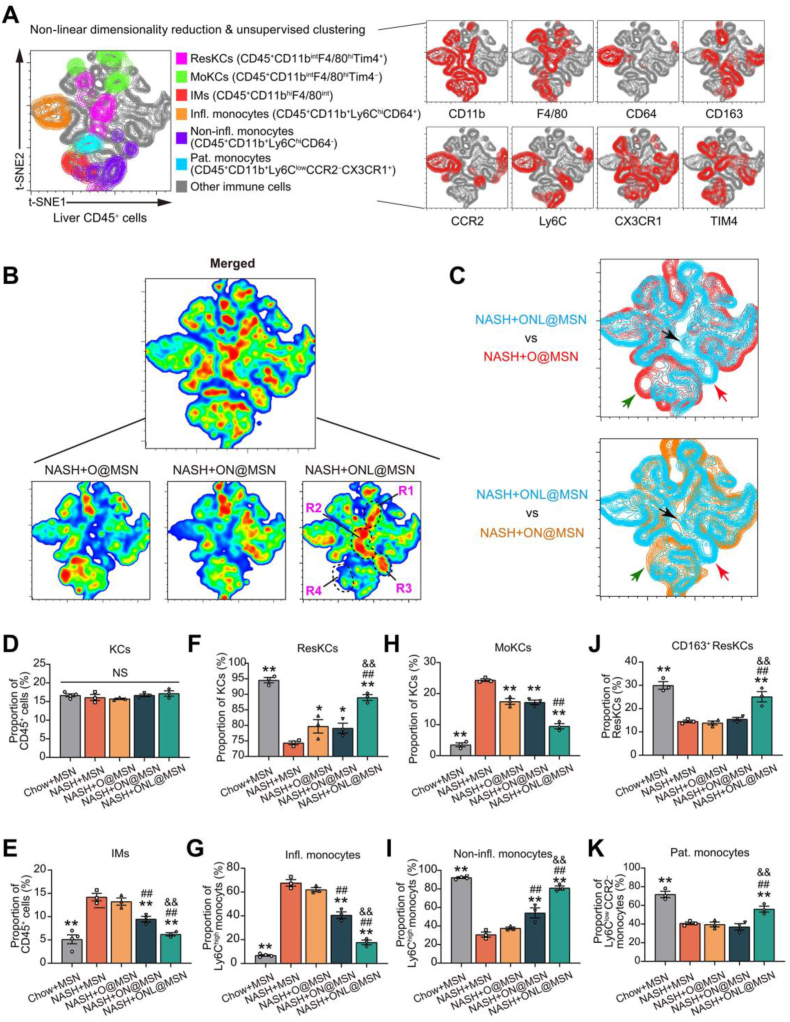
The hepatic macrophages/monocytes, primarily composed of Kupffer cells (KCs) and infiltrating monocytes (IMs), play crucial and diverse roles in the progression of NASH progression. These macrophages/monocytes have distinct functions in NASH that go beyond the commonly held belief that they are solely pro-inflammatory and detrimental32. KCs can be categorized into two groups: resident KCs (ResKCs) derived from the embryo (yolk sac) and monocyte-derived KCs (MoKCs) that infiltrate from circulation33, 34, 35. ResKCs resolve inflammation and protect against NASH, but their self-renewal is impaired during NASH, leading to their replacement by infiltrated MoKCs36,37. The IMs can be further identified as two distinct subsets: Ly6Chi monocytes and restorative Ly6Clo monocytes. The Ly6Chi monocytes can be categorized as inflammatory monocytes (CD45+CD11b+Ly6ChiCD64+), and non-inflammatory monocytes (CD45+CD11b+Ly6ChiCD64-). Additionally, there is a long-lived subset of blood patrolling monocytes that keep under surveillance and maintain the integrity of the vasculature and healthy tissues. When the tissue is injured, the patrolling monocytes enter the damaged tissue for surveillance and repair32.
Next, we applied multiplexed flow cytometry to investigate the advantage of ONL@MSN compared to two other nanoparticles in regulating hepatic macrophages/monocytes populations. We isolated liver leukocytes (CD45+ cells) from NASH mice treated with O@MSN, ON@MSN, and ONL@MSN, and used multiplexed flow cytometry to determine the clusters based on their differential expression of CD45, CD11b, F4/80, CD64, CD163, CCR2, Ly6C, CX3CR1 and TIM4 with an unsupervised machine learning algorithm with non-linear high-dimensional analysis approach. With t-distributed stochastic neighbor embedding (t-SNE)-guided gating, several macrophage/monocyte clusters, including ResKCs (F4/80hiCD11bintTIM4hi), MoKCs (F4/80hiCD11bintTIM4lo), IMs (CD11bhiF4/80int), inflammatory monocytes (CD45+CD11b+Ly6ChiCD64+), non-inflammatory monocytes (CD45+CD11b+Ly6ChiCD64−) and patrolling monocytes (CD45+CD11b+Ly6ClowCCR2−CX3CR1+) were identified (Fig. 7A). Comparing the distribution and frequencies of immune cells among the different treatments, we observed significant differences in the heatmap analysis (Fig. 7B). Four regions were identified in the heat map (Fig. 7B). Specifically, mice treated with ONL@MSN showed decreased IMs (green arrow) and increased non-inflammatory monocytes (red arrow) and ResKCs (black arrow) compared to O@MSN and ON@MSN (Fig. 7C).
Figure 7.
Multiplexed flow cytometry of hepatic macrophages/monocytes in NASH mice with the treatment of nanoparticles. (A) High-dimensional analysis of multiplexed flow cytometry using an unbiased nonlinear dimensionality reduction algorithm (t-distributed stochastic neighbor embedding, t-SNE) to identify clustering of subpopulations in hepatic CD45+ leukocyte cells. The live CD45+ leukocyte cells were gated with macrophages/monocytes-related markers (CD11b, F4/80, CD64, CD163, CCR2, Ly6C, CX3CR1 and TIM4). The multiplexed flow cytometry results were concatenated, transformed, and plotted in 2D t-SNE plots using R software. The contour plots show the relative expression of the indicated markers in CD45+ leukocyte cells. (B) A heatmap was generated to display the hepatic CD45+ leukocyte cells in NASH mice receiving treatment with O@MSN, ON@MSN, and ONL@MSN, as well as the emerged pool. (C) Comparisons of t-SNE dimensionality reduction and embedding of CD45+ leukocytes from NASH mice receiving treatment of ONL@MSN (blue) and those from NASH mice receiving treatment of O@MSN (blue) or ON@MSN (orange) respectively. The green, red, and black arrows indicate the distinct areas in leukocytes between ONL@MSN and the other two nanoparticles. (D–E) Proportions of F4/80hiCD11bint Kupffer cells (KCs, D) and infiltrating monocytes (IMs, E) in total CD45+ cells in liver tissue of NASH mice treated with different nanoparticles were determined based on manual gating analysis in multiplexed flow cytometry. n = 3 per group. (F, G) Proportions of TIM4hi resident KCs (ResKCs, F) and TIM4low monocytes-derived KCs (MoKCs, G) in total KCs in liver tissue of NASH mice treated with different nanoparticles were determined based on manual gating analysis in multiplexed flow cytometry. n = 3 per group. (H) Proportions of CD163+ ResKCs in total ResKCs in liver tissue of NASH mice treated with different nanoparticles were determined based on manual gating analysis in multiplexed flow cytometry. n = 3 per group. (I, J) Proportions of inflammatory monocytes (CD45+CD11b+Ly6ChiCD64+) and non-inflammatory monocytes (CD45+CD11b+Ly6ChiCD64-) in monocytes in liver tissue of NASH mice treated with different nanoparticles were determined based on manual gating analysis in multiplexed flow cytometry. n = 3 per group. (K) Proportions of patrolling monocytes (CD45+CD11b+Ly6ClowCCR2−CX3CR1+) were determined based on manual gating analysis in multiplexed flow cytometry. n = 3 per group. Data are presented as mean ± SEM and analyzed using one-way ANOVA followed by Tukey's analysis. ∗P < 0.05, ∗∗P < 0.01 vs. NASH + MSN. #P < 0.05, ##P < 0.01 vs. NASH + O@MSN. &P < 0.05 vs. NASH + ON@MSN. NS, no significance.
We also measured the proportions of macrophages/monocytes using manual gating. There were no significant differences in the proportions of total KCs among the five groups of mice (Fig. 7D and Supporting Information Fig. S1). By contrast, ONL@MSN greatly reduced the proportion of IMs (Fig. 7E and Fig. S1). When the KCs were divided into ResKCs and MoKCs, we found ResKCs were largely depleted, while MoKCs were induced in NASH mice liver (Fig. 7F and G, Supporting Information Fig. S2). These changes were abolished by the nanoparticles, especially by ONL@MSN (Fig. 7F and G). When the ResKCs were further divided into CD163+ ResKCs and CD163- ResKCs, it was found that only ONL@MSN increased the proportion of CD163+ ResKCs (Fig. 7H and Supporting Information Fig. S3). The IMs were divided into Ly6Chi and Ly6Clow monocytes and then the inflammatory monocytes (CD45+CD11b+Ly6ChiCD64+) and non-inflammatory monocytes (CD45+CD11b+Ly6ChiCD64−) were identified. O@MSN did not alter the proportions of inflammatory and non-inflammatory monocytes; however, ON@MSN and ONL@MSN significantly changed their proportions (Fig. 7I‒J and Supporting Information Fig. S4). Notably, the action of ONL@MSN was more potent than ON@MSN (Fig. 7I and J). Finally, the patrolling monocytes which are essential for liver integrity, were depleted in NASH mice and only reversed by ONL@MSN (Fig. 7K and Supporting Information Fig. S5). Together, the results in multiplexed flow cytometry suggest that ONL@MSN preserves inflammation-resolving macrophages/monocytes and inhibits inflammatory macrophages/monocytes in NASH mice liver.
3.8. Transcriptome analysis reveals the combined therapeutic action of ONL@MSN
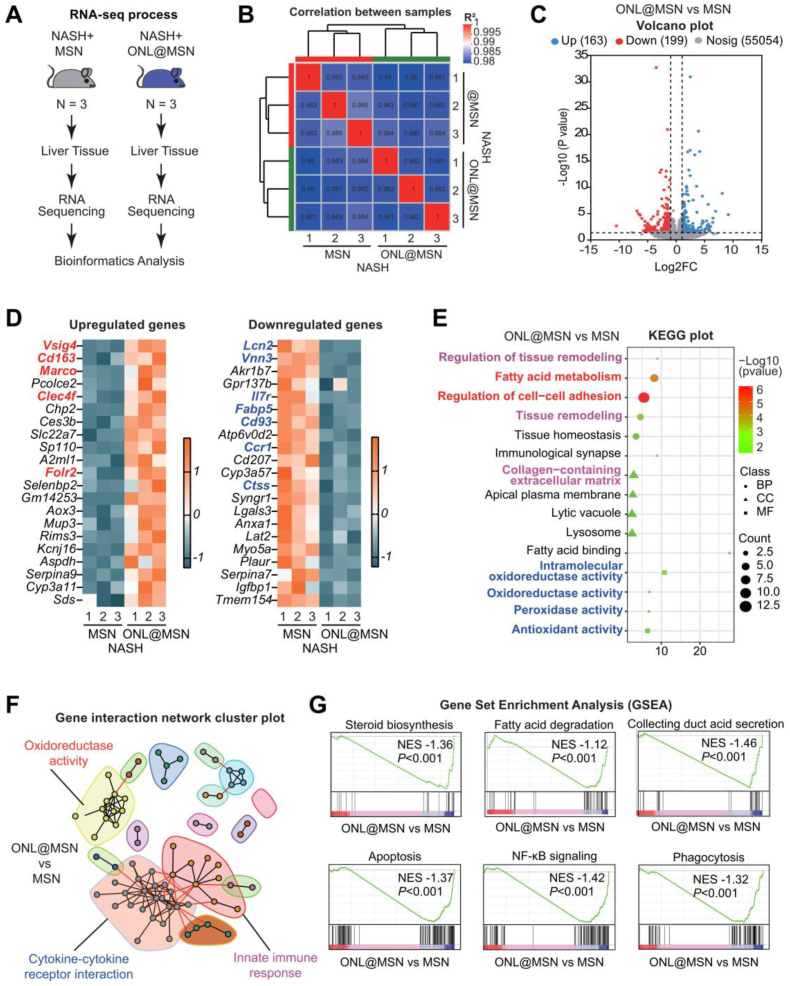
We employed RNA sequencing to analyze the changes in the liver tissue transcriptome of NASH mice receiving ONL@MSN treatment compared to NASH mice receiving MSN (Fig. 8A). The transcriptomes of the two groups exhibited a strong correlation (Fig. 8B). Volcano plots illustrated the differentially expressed genes (DEGs), revealing 163 upregulated DEGs and 199 downregulated DEGs (Fig. 8C). The top upregulated and downregulated DEGs are depicted in Fig. 8D. Notably, the most significantly upregulated DEGs included Vsig4, Cd163, Marco, Clec4f, and Folr2, which are gene signatures specific for inflammation-resolving ResKCs in NASH38. On the other hand, the most significantly downregulated DEGs included Lcn2, Vnn3, Il7r, Fabp5, Ccr1, Ctss, etc., which are gene signatures for moKCs that promote NASH39. Further analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) revealed that the effects of ONL@MSN on the transcriptome were enriched in processes such as fatty acid metabolism, regulation of cell-cell adhesion, collagen-containing extracellular matrix, oxidoreductase activity, peroxidase activity, and antioxidant activity (Fig. 8E). Gene interaction network analysis demonstrated that oxidoreductase activity, cytokine-cytokine receptor interaction, and innate immune response constituted the major cluster groups involved in the comparison between ONL@MSN and MSN (Fig. 8F). Additionally, Gene Set Enrichment Analysis (GSEA) indicated that ONL@MSN significantly inhibited biological processes including steroid biosynthesis, fatty acid degradation, collecting duct acid secretion, apoptosis, NF-κB signaling, and phagocytosis (Fig. 8G). Overall, these transcriptome analysis results provide clear evidence of the combined effects of ONL@MSN on FXR-regulated fatty acid metabolism, liver fibrosis-related extracellular matrix, peroxidation stress associated with ferroptosis, and immune modulation.
Figure 8.
Bulk-tissue RNA sequencing reveals the effects of ONL@MSN on the liver tissue transcriptome in NASH mice. (A) The experimental design involved performing bulk-tissue RNA sequencing on liver tissues from mice treated with ONL@MSN or MSN (control). (B) The correlation between samples from the two groups was assessed. (C) A volcano plot was generated to display the upregulated DEGs (n = 163) and downregulated DEGs (n = 199) in the liver tissue of mice treated with ONL@MSN compared to mice treated with MSN. (D) A heatmap plot was generated to show the top 20 upregulated and downregulated DEGs, respectively. (E) A KEGG plot was created to illustrate the main processes altered by ONL@MSN compared to MSN in terms of molecular function (MF), biological process (BP), and cell component (CC). (F) A gene interaction network cluster plot was generated to display the major important modules interacting with other genes. (G) A Gene Set Enrichment Analysis (GSEA) plot was generated to demonstrate several enriched signaling pathways resulting from the treatment of ONL@MSN compared to MSN.
We also conducted RNA-seq to assess the effects of O@MSN or ON@MSN on the liver tissue transcriptome of NASH mice. In the liver tissue of NASH mice, O@MSN resulted in the upregulation of 238 DEGs and downregulation of 275 DEGs (Supporting Information Fig. S6A), with the majority of these DEGs enriched in the fatty acid metabolic process (Fig. S6B and S6C). Gene interaction network analysis revealed that major cluster groups involved in the comparison between O@MSN and MSN included lipid metabolic process, superoxide anion generation, and metal ion binding (Fig. S6D). GSEA analysis demonstrated the inhibition of glycerolipid metabolism and lipid storage, as well as the induction of respiratory chain complex I (Fig. S6E‒S6G). In the liver tissue of NASH mice, ON@MSN led to the upregulation of 158 DEGs and downregulation of 59 DEGs (Supporting Information Fig. S7A), with a significant enrichment of DEGs in the Fatty acid derivative biosynthetic process and collagen metabolic process (Fig. S7B and S7C). Gene interaction network analysis revealed that major cluster groups involved in the comparison between ON@MSN and MSN included cytokine-cytokine receptor interaction and tissue remodeling (Fig. S7D). GSEA analysis indicated the induction of the AMPK signaling pathway and PPARγ signaling pathway, while Nitrogen metabolism was inhibited (Fig. S7E‒S7G). These findings suggest that compared to ONL@MSN, the nanoparticles O@MSN and ON@MSN exhibit less modulation of immune-related response and lipid peroxidation.
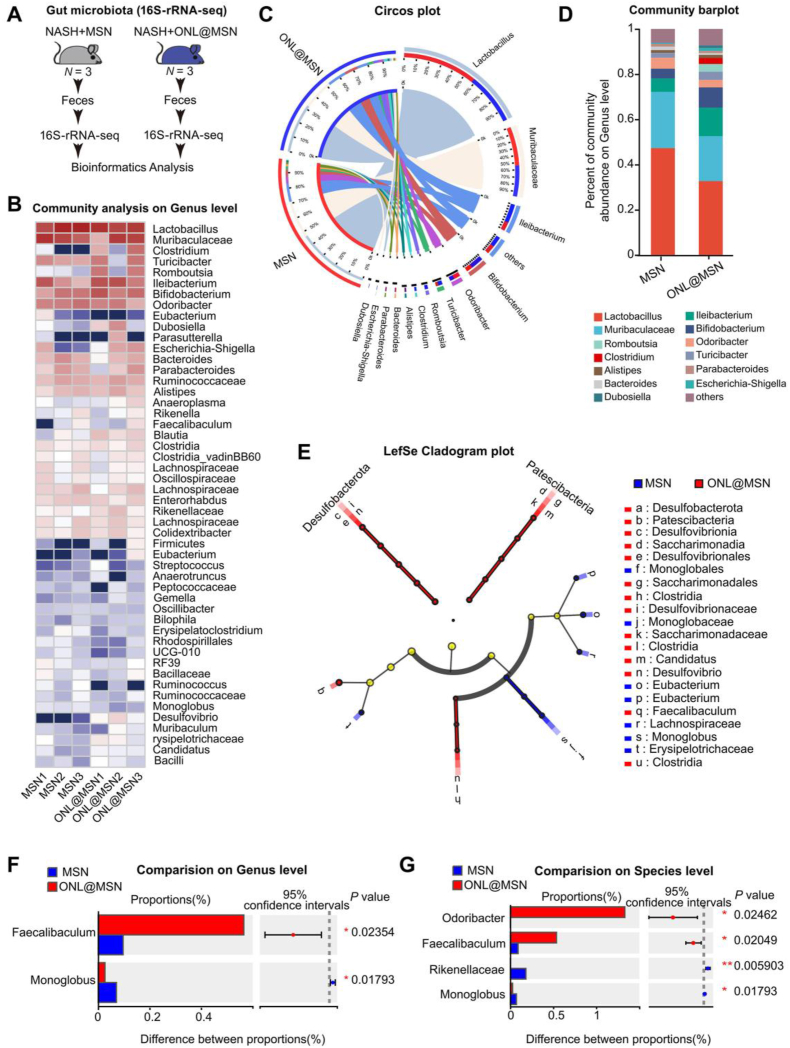
3.9. ONL@MSN improves gut microbiota dysbiosis in NASH mice
Microbes in the intestine maintain liver homeostasis and serve as a source of pathogens and molecules that contribute to NASH40. Using 16S-ribosomal RNA (rRNA) sequencing, we compared the fecal microbiota communities between the mice treated with MSN and ONL@MSN (Fig. 9A). As shown in Fig. 9B, ONL@MSN greatly changed the microbe community. Alpha diversity analysis showed that ONL@MSN did not alter the diversity of microbiota (Supporting Information Fig. S8A). Beta diversity analysis showed that the microbiota in mice treated with ONL@MSN showed good similarity (Fig. S8B). The correlation between samples within two groups (MSN vs. ONL@MSN) was confirmed (Fig. S8C). Circos plot and Community barplot showed that ONL@MSN reduced proportions of Lactobacillus Muribaculacea and Odoribacter, but increased proportions of Ileibacterium, Romboutsia, and Bifidobacterium (Fig. 9C and D). Among these microbes, Ileibacterium and Bifidobacterium were previously reported to have beneficial effects on NASH or metabolic diseases41,42. We used the LEfSe multi-level species difference discrimination method to further analyze the microbial phylotypes of the fecal microbiota and found that at the phylum level, Desulfobacterota and Patescibacteria were two major microbes induced by ONL@MSN treatment (Fig. 9E). The detailed results of LEfSe analysis are illustrated in Fig. S8D. Faecalibaculum was induced whereas Monoglobus was reduced by ONL@MSN treatment (Fig. 9F). The Odoribacter and Faecalibaculum43 were induced whereas the Monoglobus44 and Rikenellaceae were reduced by ONL@MSN treatment (Fig. 9G). These results indicate that ONL@MSN improves gut dysbiosis in NASH mice.
Figure 9.
Fecal 16S-rRNA sequencing shows the effects of ONL@MSN on the gut microbiota community in NASH mice. (A) The experimental design involved conducting fecal 16S-rRNA sequencing on the gut microbiota in the feces of mice treated with ONL@MSN or MSN (control). (B) A heatmap was generated to display the gut microbiota community in the feces of the two groups of mice. (C) A Circos plot was created to illustrate the abundance of dominant microbiota in the feces of the two groups of mice. (D) A community barplot was generated to show the composition of the dominant microbiota in the feces of the two groups of mice. (E) A cladogram was plotted using LEfSe analysis to visualize the phylogenetic distribution of bacterial lineages between the gut microbiota in the feces of mice treated with ONL@MSN and MSN. (F, G) A comparison was made at the genus (F) and species (G) levels to assess the differences in gut microbiota between the feces of mice treated with ONL@MSN and MSN. The Wilcoxon Rank Sum Test was used to compare the proportions of gut microbiota, and the corresponding P values were indicated in the figure.
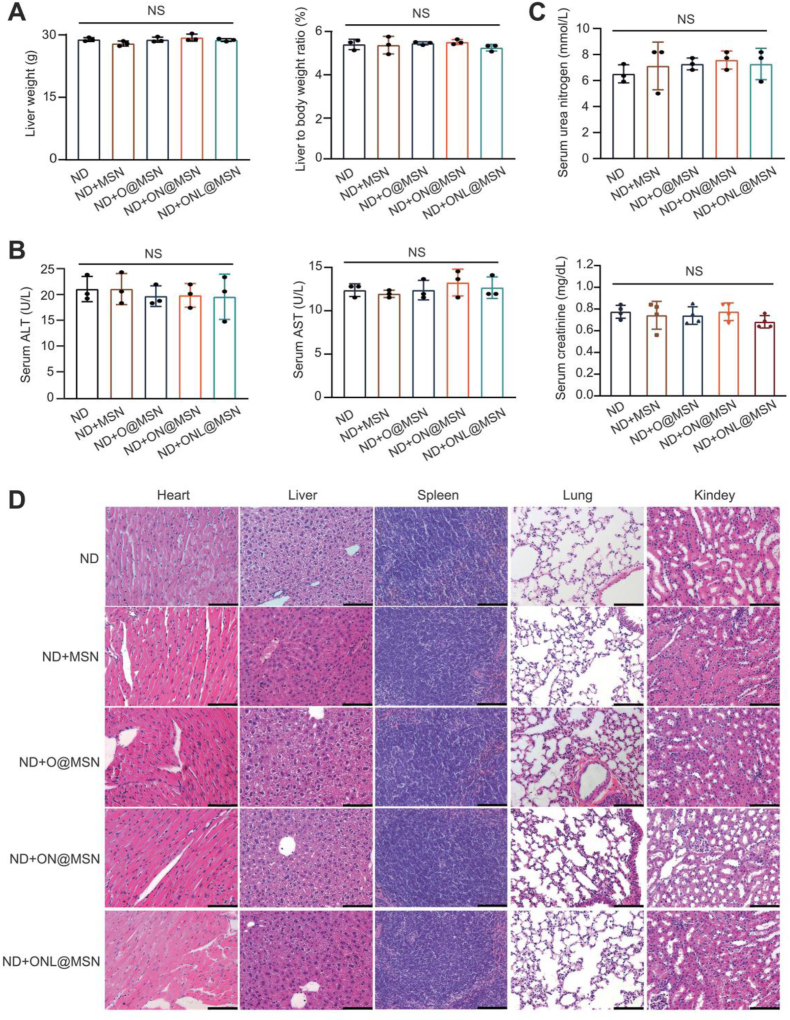
3.10. Long-term treatment of nanoparticles did not induce systemic toxicity in mice
The systemic toxicity of the synthesized nanoparticles was assessed considering the relatively long-term treatment period of NAFLD. Following a three-month intravenous administration, no significant changes in body weight or liver weight to body weight ratio were observed between the mice receiving a normal diet (ND) and those receiving ND plus treatment with O@MSN, ON@MSN, or ONL@MSN (Fig. 10A). The treatment of O@MSN, ON@MSN, and ONL@MSN did not alter the serum ALT and AST activities, indicating that liver function was unaffected (Fig. 10B). Similarly, the treatment did not cause any changes in the serum urea nitrogen and creatinine levels, which are markers of kidney function (Fig. 10C). Histopathology analysis of major organs in mice was conducted, and no evident histological abnormalities were observed in the major organs of O@MSN- and ON@MSN-treated mice, as well as ONL@MSN-treated mice (Fig. 10D). In conclusion, these results demonstrate the considerable safety and biocompatibility of these nanoparticles, making them suitable for use as a safe and effective liver-targeting treatment strategy for alleviating NASH.
Figure 10.
Long-term nanoparticle injection does not induce systemic toxicity in normal mice. (A) Assessment of liver weight, body weight, and liver-to-body weight ratio in mice fed with a normal diet (ND) three months after intravenous administration of synthesized nanoparticles. N = 3 per group. (B) Measurement of serum ALT and AST activities in mice fed with a normal diet (ND) three months after intravenous administration of synthesized nanoparticles. N = 3 per group. (C) Evaluation of serum urea nitrogen and creatinine levels in mice fed with a normal diet (ND) three months after intravenous administration of synthesized nanoparticles. N = 4 per group. (D) Presentation of H&E staining images of major organs (heart, liver, spleen, lung, and kidney) three months after intravenous administration of synthesized nanoparticles. Scale bar, 200 mm.
4. Conclusions
In this study, we demonstrate an enhanced therapy for NASH by utilizing a multifunctional nanoparticle that targets FXR, fibrosis, and ferroptosis. Although OCA, a bile acid-derived FXR agonist, is the most clinically advanced candidate drug for NASH treatment, it has shown unsatisfactory anti-fibrotic effects and a low response rate in recent phase III clinical trials9. Therefore, combining OCA with other active compounds may offer greater benefits to address this crucial issue in NASH treatment. In our study, we incorporated an NO donor and a ferroptosis inhibitor to degrade liver fibrosis and suppress hepatic ferroptotic cell death, respectively. We found that this multifunctional nanoparticle (ONL@MSN) exhibited potent therapeutic action against NASH. In addition to the canonical effects of FXR activation on lipid and glucose metabolism, ONL@MSN also effectively inhibited fibrosis and hepatic ferroptotic cell death. The significantly enhanced therapeutic efficacy of ONL@MSN in the experimental NASH model may be attributed to its preservation of inflammation-resolving KCs and inhibition of infiltrating macrophages/monocytes in NASH mice. Furthermore, our RNA-seq analysis revealed that ONL@MSN had a greater inhibitory effect on immune-related responses and lipid peroxidation compared to O@MSN and ON@MSN. Moreover, ONL@MSN significantly improved gut dysbiosis, which is an emerging potential pharmacological target for NASH intervention. Overall, this study presents a promising synergistic nanomedicine approach that simultaneously targets FXR, ferroptosis, and fibrosis, providing a therapeutic strategy.
Author contributions
Pei Wang, Ying Lu and Jiajun Xu designed the research. Jiangtao Fu, Pingping Zhang, Zhiguo Sun and Guodong Lu carried out the experiments and performed data analysis. Qi Cao, Yiting Chen, Wenbin Wu and Jiabao Zhang participated part of the experiments. Zhiguo Sun, Chunlin Zhuang and Chunquan Sheng provided experimental drugs and quality control. Pei Wang wrote the manuscript. Pei Wang and Jiangtao Fu revised the manuscript. All of the authors have read and approved the final manuscript.
Conflicts of interest
The authors have no conflicts of interest to declare.
Acknowledgments
This work was supported by the grants from National Natural Science Foundation of China (82073915 and 82104530), Shanghai Natural Science Foundation (23ZR1477500, China), Shanghai Science and Technology Commission (21XD1424900, China) and Shanghai Shuguang Program (19SG32, China).
Footnotes
Peer review under the responsibility of Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences.
Supporting data to this article can be found online at https://doi.org/10.1016/j.apsb.2024.02.017.
Contributor Information
Jiajun Xu, Email: xujiajun920@163.com.
Ying Lu, Email: acuace@163.com.
Pei Wang, Email: pwang@smmu.edu.cn.
Appendix A. Supporting information
The following is the Supporting data to this article.
References
- 1.Riazi K., Azhari H., Charette J.H., Underwood F.E., King J.A., Afshar E.E., et al. The prevalence and incidence of NAFLD worldwide: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2022;7:851–861. doi: 10.1016/S2468-1253(22)00165-0. [DOI] [PubMed] [Google Scholar]
- 2.Llovet J.M., Willoughby C.E., Singal A.G., Greten T.F., Heikenwalder M., El-Serag H.B., et al. Nonalcoholic steatohepatitis-related hepatocellular carcinoma: pathogenesis and treatment. Nat Rev Gastroenterol Hepatol. 2023;20:487–503. doi: 10.1038/s41575-023-00754-7. [DOI] [PubMed] [Google Scholar]
- 3.Chalasani N., Younossi Z., Lavine J.E., Diehl A.M., Brunt E.M., Cusi K., et al. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American association for the study of liver diseases, American college of gastroenterology, and the American gastroenterological association. Hepatology. 2012;55:2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 4.European Association for the Study of the L, European Association for the Study of D, European Association for the Study of O EASL-EASD-EASO Clinical Practice Guidelines for the management of non-alcoholic fatty liver disease. J Hepatol. 2016;64:1388–1402. doi: 10.1016/j.jhep.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Schuppan D., Surabattula R., Wang X.Y. Determinants of fibrosis progression and regression in NASH. J Hepatol. 2018;68:238–250. doi: 10.1016/j.jhep.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Teodoro J.S., Rolo A.P., Palmeira C.M. Hepatic FXR: key regulator of whole-body energy metabolism. Trends Endocrinol Metabol. 2011;22:458–466. doi: 10.1016/j.tem.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 7.Sun L., Cai J., Gonzalez F.J. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat Rev Gastroenterol Hepatol. 2021;18:335–347. doi: 10.1038/s41575-020-00404-2. [DOI] [PubMed] [Google Scholar]
- 8.Neuschwander-Tetri B.A., Loomba R., Sanyal A.J., Lavine J.E., Van Natta M.L., Abdelmalek M.F., et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Younossi Z.M., Ratziu V., Loomba R., Rinella M., Anstee Q.M., Goodman Z., et al. Obeticholic acid for the treatment of non-alcoholic steatohepatitis: interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2019;394:2184–2196. doi: 10.1016/S0140-6736(19)33041-7. [DOI] [PubMed] [Google Scholar]
- 10.Dixon S.J., Lemberg K.M., Lamprecht M.R., Skouta R., Zaitsev E.M., Gleason C.E., et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang W.S., SriRamaratnam R., Welsch M.E., Shimada K., Skouta R., Viswanathan V.S., et al. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156:317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedmann Angeli J.P., Schneider M., Proneth B., Tyurina Y.Y., Tyurin V.A., Hammond V.J., et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–1191. doi: 10.1038/ncb3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J.B., Tong J., Sun D.Y., Fu J.T., Li D.J., Wang P. Targeting ferroptosis in cardio-metabolic-diseases: mechanisms and therapeutic prospects. Med Res Rev. 2023;43:683–712. doi: 10.1002/med.21934. [DOI] [PubMed] [Google Scholar]
- 14.Qi J., Kim J.W., Zhou Z., Lim C.W., Kim B. Ferroptosis affects the progression of nonalcoholic steatohepatitis via the modulation of lipid peroxidation-mediated cell death in mice. Am J Pathol. 2020;190:68–81. doi: 10.1016/j.ajpath.2019.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Li X., Wang T.X., Huang X., Li Y., Sun T., Zang S., et al. Targeting ferroptosis alleviates methionine-choline deficient (MCD)-diet induced NASH by suppressing liver lipotoxicity. Liver Int. 2020;40:1378–1394. doi: 10.1111/liv.14428. [DOI] [PubMed] [Google Scholar]
- 16.Tsurusaki S., Tsuchiya Y., Koumura T., Nakasone M., Sakamoto T., Matsuoka M., et al. Hepatic ferroptosis plays an important role as the trigger for initiating inflammation in nonalcoholic steatohepatitis. Cell Death Dis. 2019;10:449. doi: 10.1038/s41419-019-1678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang W., Chai L., Chen X., Li Z., Feng L., Hu W., et al. Imaging changes in the polarity of lipid droplets during NAFLD-Induced ferroptosis via a red-emitting fluorescent probe with a large Stokes shift. Biosens Bioelectron. 2023;231 doi: 10.1016/j.bios.2023.115289. [DOI] [PubMed] [Google Scholar]
- 18.Tong J., Li D., Meng H., Sun D., Lan X., Ni M., et al. Targeting a novel inducible GPX4 alternative isoform to alleviate ferroptosis and treat metabolic-associated fatty liver disease. Acta Pharm Sin B. 2022;12:3650–3666. doi: 10.1016/j.apsb.2022.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tong J., Lan X.T., Zhang Z., Liu Y., Sun D.Y., Wang X.J., et al. Ferroptosis inhibitor liproxstatin-1 alleviates metabolic dysfunction-associated fatty liver disease in mice: potential involvement of PANoptosis. Acta Pharmacol Sin. 2023;44:1014–1028. doi: 10.1038/s41401-022-01010-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji Q.X., Zeng F.Y., Zhou J., Wu W.B., Wang X.J., Zhang Z., et al. Ferroptotic stress facilitates smooth muscle cell dedifferentiation in arterial remodelling by disrupting mitochondrial homeostasis. Cell Death Differ. 2023;30:457–474. doi: 10.1038/s41418-022-01099-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi J., Wu S., Tan S., Qin Y., Wang X., Jiang J., et al. Berberine alleviates liver fibrosis through inducing ferrous redox to activate ROS-mediated hepatic stellate cells ferroptosis. Cell Death Dis. 2021;7:374. doi: 10.1038/s41420-021-00768-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu A., Feng B., Yu J., Yan L., Che L., Zhuo Y., et al. Fibroblast growth factor 21 attenuates iron overload-induced liver injury and fibrosis by inhibiting ferroptosis. Redox Biol. 2021;46 doi: 10.1016/j.redox.2021.102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwakiri Y., Kim M.Y. Nitric oxide in liver diseases. Trends Pharmacol Sci. 2015;36:524–536. doi: 10.1016/j.tips.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dong X., Liu H.J., Feng H.Y., Yang S.C., Liu X.L., Lai X., et al. Enhanced drug delivery by nanoscale integration of a nitric oxide donor to induce tumor collagen depletion. Nano Lett. 2019;19:997–1008. doi: 10.1021/acs.nanolett.8b04236. [DOI] [PubMed] [Google Scholar]
- 25.Maeda H., Ishima Y., Saruwatari J., Mizuta Y., Minayoshi Y., Ichimizu S., et al. Nitric oxide facilitates the targeting Kupffer cells of a nano-antioxidant for the treatment of NASH. J Control Release. 2022;341:457–474. doi: 10.1016/j.jconrel.2021.11.039. [DOI] [PubMed] [Google Scholar]
- 26.Brunt E.M., Kleiner D.E., Wilson L.A., Belt P., Neuschwander-Tetri B.A., Network N.C.R. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 2011;53:810–820. doi: 10.1002/hep.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi C., Fu H., Li Y.H., Zhang G.Y., Zeng F.Y., Ji Q.X., et al. Exerkine fibronectin type-III domain-containing protein 5/irisin-enriched extracellular vesicles delay vascular ageing by increasing SIRT6 stability. Eur Heart J. 2022;43:4579–4595. doi: 10.1093/eurheartj/ehac431. [DOI] [PubMed] [Google Scholar]
- 28.Morrison M.C., Verschuren L., Salic K., Verheij J., Menke A., Wielinga P.Y., et al. Obeticholic acid modulates serum metabolites and gene signatures characteristic of human NASH and attenuates inflammation and fibrosis progression in Ldlr–/–.Leiden mice. Hepatol Commun. 2018;2:1513–1532. doi: 10.1002/hep4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou J., Cui S., He Q., Guo Y., Pan X., Zhang P., et al. SUMOylation inhibitors synergize with FXR agonists in combating liver fibrosis. Nat Commun. 2020;11:240. doi: 10.1038/s41467-019-14138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng H., Schorpp K., Jin J., Yozwiak C.E., Hoffstrom B.G., Decker A.M., et al. Transferrin receptor Is a specific ferroptosis marker. Cell Rep. 2020;30:3411–3423.e7. doi: 10.1016/j.celrep.2020.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doll S., Proneth B., Tyurina Y.Y., Panzilius E., Kobayashi S., Ingold I., et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol. 2017;13:91–98. doi: 10.1038/nchembio.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guilliams M., Scott C.L. Liver macrophages in health and disease. Immunity. 2022;55:1515–1529. doi: 10.1016/j.immuni.2022.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Tran S., Baba I., Poupel L., Dussaud S., Moreau M., Gelineau A., et al. Impaired Kupffer cell self-Renewal alters the liver response to lipid overload during non-alcoholic steatohepatitis. Immunity. 2020;53:627–640.e5. doi: 10.1016/j.immuni.2020.06.003. [DOI] [PubMed] [Google Scholar]
- 34.Remmerie A., Martens L., Thone T., Castoldi A., Seurinck R., Pavie B., et al. Osteopontin expression Identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity. 2020;53:641–657.e14. doi: 10.1016/j.immuni.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daemen S., Gainullina A., Kalugotla G., He L., Chan M.M., Beals J.W., et al. Dynamic shifts in the composition of resident and recruited macrophages influence tissue remodeling in NASH. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2020.108626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bleriot C., Dupuis T., Jouvion G., Eberl G., Disson O., Lecuit M. Liver-resident macrophage necroptosis orchestrates type 1 microbicidal inflammation and type-2-mediated tissue repair during bacterial infection. Immunity. 2015;42:145–158. doi: 10.1016/j.immuni.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 37.Theurl I., Hilgendorf I., Nairz M., Tymoszuk P., Haschka D., Asshoff M., et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat Med. 2016;22:945–951. doi: 10.1038/nm.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guilliams M., Bonnardel J., Haest B., Vanderborght B., Wagner C., Remmerie A., et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–396.e38. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye D., Yang K., Zang S., Lin Z., Chau H.T., Wang Y., et al. Lipocalin-2 mediates non-alcoholic steatohepatitis by promoting neutrophil-macrophage crosstalk via the induction of CXCR2. J Hepatol. 2016;65:988–997. doi: 10.1016/j.jhep.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 40.Lang S., Schnabl B. Microbiota and fatty liver disease-the known, the unknown, and the future. Cell Host Microbe. 2020;28:233–244. doi: 10.1016/j.chom.2020.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y., Ablimit N., Zhang Y., Li J., Wang X., Liu J., et al. Novel beta-mannanase/GLP-1 fusion peptide high effectively ameliorates obesity in a mouse model by modifying balance of gut microbiota. Int J Biol Macromol. 2021;191:753–763. doi: 10.1016/j.ijbiomac.2021.09.150. [DOI] [PubMed] [Google Scholar]
- 42.Malaguarnera M., Vacante M., Antic T., Giordano M., Chisari G., Acquaviva R., et al. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig Dis Sci. 2012;57:545–553. doi: 10.1007/s10620-011-1887-4. [DOI] [PubMed] [Google Scholar]
- 43.Zagato E., Pozzi C., Bertocchi A., Schioppa T., Saccheri F., Guglietta S., et al. Endogenous murine microbiota member Faecalibaculum rodentium and its human homologue protect from intestinal tumour growth. Nat Microbiol. 2020;5:511–524. doi: 10.1038/s41564-019-0649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang T., Yang S., Zhao J., Wang P., Li S., Jin Y., et al. Comprehensive analysis of gut microbiota and fecal bile acid profiles in children with biliary atresia. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.914247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.