Abstract
In the realm of genetically transformed crops, the process of plant regeneration holds utmost significance. However, the low regeneration efficiency of several wheat varieties currently restricts the use of genetic transformation for gene functional analysis and improved crop production. This research explores overexpression of TaLAX PANICLE1 (TaLAX1), which markedly enhances regeneration efficiency, thereby boosting genetic transformation and genome editing in wheat. Particularly noteworthy is the substantial increase in regeneration efficiency of common wheat varieties previously regarded as recalcitrant to genetic transformation. Our study shows that increased expression of TaGROWTH-REGULATING FACTOR (TaGRF) genes, alongside that of their co-factor, TaGRF-INTERACTING FACTOR 1 (TaGIF1), enhances cytokinin accumulation and auxin response, which may play pivotal roles in the improved regeneration and transformation of TaLAX1-overexpressing wheat plants. Overexpression of TaLAX1 homologs also significantly increases the regeneration efficiency of maize and soybean, suggesting that both monocot and dicot crops can benefit from this enhancement. Our findings shed light on a gene that enhances wheat genetic transformation and elucidate molecular mechanisms that potentially underlie wheat regeneration.
Key words: plant regeneration, genetic transformation, TaLAX1, TaGRF4–TaGIF1, cytokinin, auxin, wheat
TaLAX1 can promote shoot regeneration, thereby boosting genetic transformation and genome editing in wheat by activating TaGRFs and TaGIF1, cytokinin biosynthesis genes, and auxin-response genes. Homologs of TaLAX1 can also enhance the regeneration capacity of maize and soybean.
Introduction
Recalcitrance to genetic transformation significantly limits the potential for transgenesis and genome editing in various plant species, including wheat, maize, barley, indica rice, and soybean (Shrawat and Lorz, 2006; Hiei et al., 2014). Reliable plant regeneration from transformed tissues is of paramount importance for efficient genetic transformation. However, the inefficiency of regeneration in many crops restricts transformation techniques such as Agrobacterium-mediated transformation and particle bombardment to a limited range of crop genotypes (Altpeter et al., 2016). Thus, regeneration represents a significant bottleneck to crop transformation. One recent advance in this field is the development of the PureWheat technique for Agrobacterium-mediated wheat transformation, which utilizes a modified tissue culture medium and an innovative transformant selection strategy (Ishida et al., 2015). Although PureWheat has demonstrated improvements in transformation efficiency for the model wheat genotype Fielder, it still remains ineffective for numerous commercial wheat cultivars (Wang et al., 2017). Consequently, there is a continued need for development of broadly applicable methods to enhance regeneration and transformation in wheat and other crop species.
Traditionally, Agrobacterium-mediated transformation and particle bombardment have been performed on immature embryos and other cultured tissues because of their efficient regeneration capabilities. Regeneration induction can be achieved by manipulating the expression of factors involved in somatic embryogenesis such as LEAFY COTYLEDON1 (LEC1), WUSCHEL (WUS), BABY BOOM (BBM), AGAMOUS-LIKE15 (AGL15), and SOMATIC EMBRYOGENESIS RECEPTOR KINASE (SERK) (Feher, 2015; Ikeuchi et al., 2016; Tang et al., 2020). These factors can reprogram somatic cells into totipotent cells, thereby enhancing regeneration efficiency in various plant genotypes (Lotan et al., 1998; Zuo et al., 2002a, 2002b; Boutilier et al., 2002; Bouchabke-Coussa et al., 2013; Florez et al., 2015; Lowe et al., 2016, 2018). Consequently, the regulated expression of such factors has been used to enhance crop transformation efficiency.
Overexpression of the maize WUS2 and BBM genes has been shown to significantly enhance Agrobacterium-mediated transformation of previously non-transformable maize inbred lines. Similar improvements have also been observed in other recalcitrant crops such as rice, sorghum, and sugarcane (Lowe et al., 2016). However, overexpression of BBM and WUS2 genes can lead to undesirable morphological abnormalities in transgenic maize plants, including thick and short roots, stunted and twisted phenotypes, and sterility. As a result, there is a demand for novel approaches or identification of new developmental regulators that can alleviate the pleiotropic effects associated with overexpression of these genes in regenerated plants.
Recently, a breakthrough approach that uses conditional expression drivers was reported to increase transformation efficiency without causing detrimental effects. Specifically, implementation of the maize phospholipid transferase protein promoter to drive expression of the BBM gene, in conjunction with an auxin-inducible WUS2 gene, resulted in efficient transformation without any phenotypic abnormalities or sterility (Lowe et al., 2018; Gao et al., 2020). Furthermore, by linking the phospholipid transferase protein promoter with three viral enhancer elements, the expression of WUS2 was amplified in somatic cells. This innovative system facilitates somatic embryogenesis without integration of WUS2 upon infection with a mixture of Agrobacterium strains. Termed “altruistic transformation,” this method generates transgenic tissues capable of plant regeneration without the need for WUS2 integration (Hoerster et al., 2020).
Expression of a fusion protein that combines wheat GROWTH-REGULATING FACTOR 4 (GRF4) and its co-factor GRF-INTERACTING FACTOR 1 (GIF1) has been used to enhance regeneration efficiency in non-transformable wheat genotypes (Debernardi et al., 2020; Qiu et al., 2022). In addition, overexpression of wheat TaWOX5 has been found to significantly increase transformation efficiency not only in wheat but also in five other cereal species (Wang et al., 2022a).
Axillary meristems (AMs) play a crucial role in establishing plant architecture (McSteen and Leyser, 2005; Schmitz and Theres, 2005), and formation of AMs appears to be regulated by a conserved mechanism across different plant species (Woods et al., 2011). The rice OsLAX PANICLE1 (OsLAX1) gene and the maize ZmBARREN STALK1 (ZmBA1) gene encode non-canonical basic helix–loop–helix transcription factors that are orthologous to each other (Komatsu et al., 2003; Gallavotti et al., 2004). OsLAX1 is essential for AM formation in rice, and mutants of ZmBA1 in maize have unbranched and shortened tassels (Gallavotti et al., 2004). Similarly, mutations in the Arabidopsis ortholog AtREGULATOR OF AXILLARY MERISTEM FORMATION (AtROX) lead to impaired axillary bud formation during vegetative shoot development (Yang et al., 2012). However, it remains unclear whether a wheat ortholog of these genes is linked to AM formation. Although the protein sequence of wheat TaLAX1 is homologous to those of OsLAX1 and ZmBA1, its function differs from that of these orthologs. Mutants in Talax1-aabbdd exhibit a compact spike phenotype associated with the regulation of wheat domestication traits (He et al., 2021).
In this study, we present findings on the role of the TaLAX1 transcription factor in enhancing regeneration efficiency, genetic transformation, and genome editing in wheat. We demonstrate that overexpression of TaLAX1-A leads to upregulation of wheat TaGRFs, their co-factor TaGIF1, and cytokinin-biosynthesis and auxin-response genes. In addition, we provide evidence that TaLAX1 homologs can stimulate regeneration in maize and soybean, suggesting the potential applicability of this approach to both monocot and dicot crops.
Results
TaLAX1 overexpression enhances regeneration efficiency in the wheat genotype Fielder
In previous studies, OsLAX1 and ZmBA1, two non-canonical basic helix–loop–helix transcription factors, have been identified as crucial regulators of AM formation during vegetative and inflorescence development in rice and maize, respectively. These findings suggest that homologs of these genes in different plant species may share similar functions (Komatsu et al., 2003; Gallavotti et al., 2004). To identify wheat homologs of OsLAX1, denoted TaLAX1s, and investigate their role in wheat AM formation, we performed sequence analysis and examined the effects of overexpression. Through a BLAST search, we identified three homologous TaLAX1 proteins (TaLAX1-A, TaLAX1-B, and TaLAX1-D; Supplemental Figure 1A and 1B) in the common wheat line Chinese Spring (Triticum aestivum, AABBDD, 2n = 42). These proteins exhibited significant amino acid identity with OsLAX1 (TaLAX1-A, 51.74%; TaLAX1-B, 53.91%; and TaLAX1-D, 52.81%).
To investigate the expression pattern of TaLAX1, we performed RNA in situ hybridization of wheat spike tissue sections (Supplemental Figure 1C–1F). TaLAX1 transcripts initially accumulated in the spikelet meristem initiation sites (Supplemental Figure 1C) and later became restricted to a boundary region within the spikelet meristems (Supplemental Figure 1D and 1E). No TaLAX1 mRNA was detected in floret tissues (Supplemental Figure 1F). These results suggest that TaLAX1 may play a role in regulating meristematic cell proliferation, which is crucial for the development of spikelet meristems in the wheat spike.
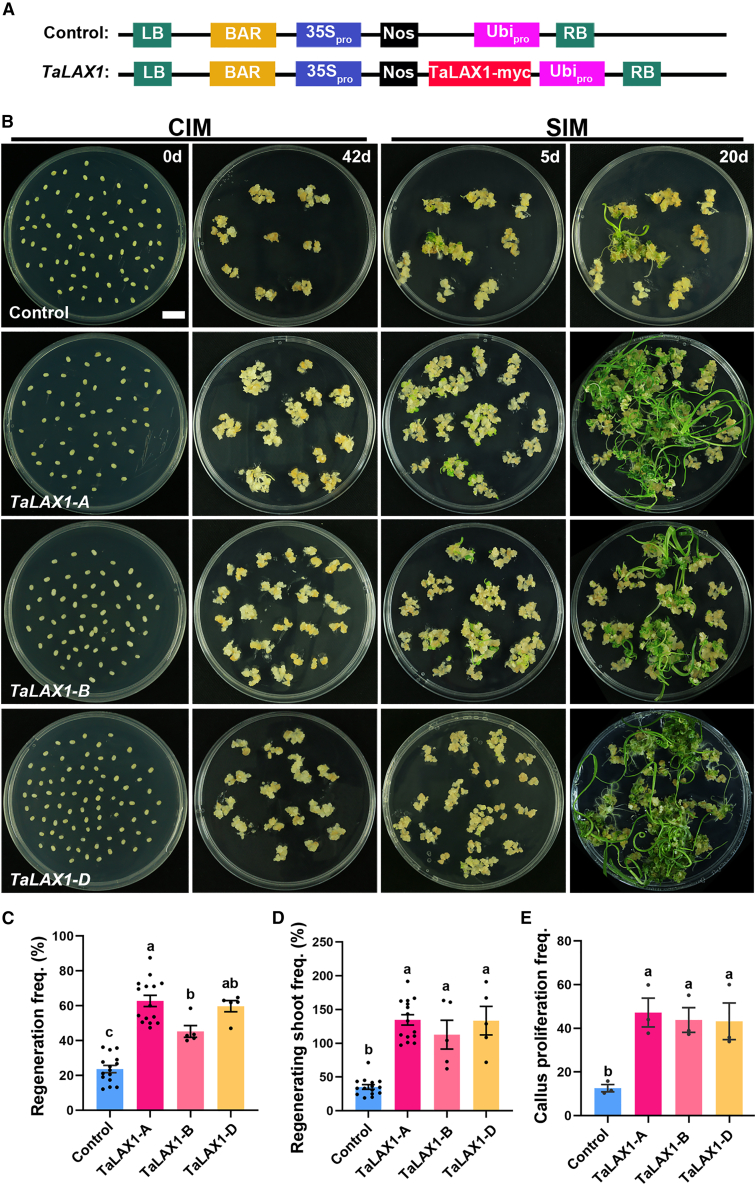
To elucidate the function of TaLAX1 in wheat, we created myc-tagged genomic constructs of TaLAX1-A, TaLAX1-B, and TaLAX1-D and inserted them into the PC186 expression vector (Hao et al., 2018) under the control of the maize ZmUbi promoter and Nos terminator. These constructs (PC186-TaLAX1-myc; Figure 1A) were introduced into the wheat genotype Fielder using the PureWheat Agrobacterium-mediated transformation method (Ishida et al., 2015). Interestingly, we observed a significant enhancement of shoot regeneration throughout the transformation process upon overexpression of TaLAX1 (Figure 1B), consistent with its proposed role in initiation of meristematic cells. We calculated regeneration frequency as the number of calli that showed at least one regenerating shoot relative to the total number of inoculated embryos (Figure 1C). Regenerating shoot frequency was calculated as the number of regenerating shoots per total number of inoculated embryos (Figure 1D). The average regeneration frequency was significantly higher in TaLAX1 transgenic calli (TaLAX1-A, 62.76% ± 3.24%; TaLAX1-B, 45.26% ± 3.37%; TaLAX1-D, 59.77% ± 3.22%) than in calli with the PC186 empty vector control (23.65% ± 2.08%; Figure 1C and Supplemental Data 1). Similarly, the regenerating shoot frequency was significantly higher in TaLAX1 transgenic calli than in the control (TaLAX1-A, 134.63% ± 7.75%; TaLAX1-B, 112.73% ± 21.33%; TaLAX1-D, 133.51% ± 21.10% vs. empty vector control, 35.20% ± 3.39%) (Figure 1D and Supplemental Data 1). Moreover, TaLAX1 overexpression also accelerated callus proliferation, as indicated by the greater increase in weight of transgenic calli after induction (Figure 1E and Supplemental Data 2). Among the TaLAX1 homologs, TaLAX1-A exhibited the highest regeneration frequency (Figure 1C) and transcript levels at each stage of regeneration (Supplemental Figure 2), leading us to focus on TaLAX1-A for further analysis.
Figure 1.
TaLAX1 promotes shoot regeneration in the common wheat variety Fielder.
(A) Schematic representation of the PC186-TaLAX1-myc vector with the ZmUbi promoter and Nos terminator; the PC186 empty vector was used as a control.
(B) Shoot regeneration phenotypes of immature embryos infected with the empty vector (control) or PC186-TaLAX1-A/B/D-myc vector. CIM, callus induction medium (including WLS-AS, WLS-Res, WLS-P5, and WLS-P10 media in the PureWheat transformation); SIM, shoot induction medium (LSZ-P5 in the PureWheat transformation); 0d and 42d, immature embryos were placed on CIM for 0 or 42 days; 5d and 20d, immature embryos were placed on CIM for 42 days and then transferred to SIM for 5 days or 20 days. Scale bar, 1 cm.
(C) Regeneration frequencies of immature embryos infected with control or PC186-TaLAX1-A/B/D-myc vector. Regeneration frequency = no. of calli showing at least one regenerating shoot/no. of inoculated embryos × 100%.
(D) Regenerating shoot frequencies of immature embryos infected with control or PC186-TaLAX1-A/B/D-myc vector. Regenerating shoot frequency = no. of regenerating shoots/no. of inoculated embryos × 100%.
(E) Callus proliferation frequencies of immature embryos infected with control or PC186-TaLAX1-A/B/D-myc vector. Callus proliferation frequency = increased weight of callus after induction on CIM for 42 days/weight of immature embryos before induction. Values in (C–E) are means ± SEM from at least three independent experiments. Black points are the results from individual experiments. One-way ANOVA and Tukey’s multiple comparison tests were performed. Different lowercase letters indicate statistically significant differences (P < 0.05).
TaLAX1-A promotes shoot regeneration in different wheat genotypes
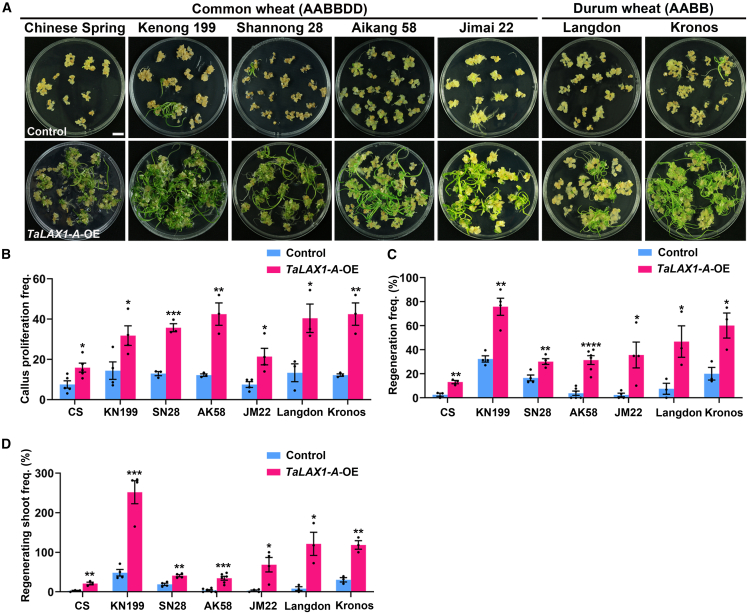
To explore the potential impact of TaLAX1 on shoot regeneration, we introduced the PC186-TaLAX1-A-myc (TaLAX1-A-OE) vector into five common wheat varieties (Chinese Spring, Kenong 199, Shannong 28, Aikang 58, and Jimai 22) and two durum wheat varieties (Langdon and Kronos), including varieties known to be recalcitrant to transformation (Figure 2A). We observed a significant enhancement of regeneration frequency, regenerating shoot frequency, and callus proliferation frequency in TaLAX1-A-OE transgenic calli across all varieties compared with the empty vector control (Figure 2B–2D and Supplemental Data 3 and 4). Notably, Aikang 58 and Jimai 22, which are widely cultivated in China, with a planting area exceeding 2 million hectares each, displayed poor callus quality and were recalcitrant to regeneration in the control groups. However, overexpression of TaLAX1-A dramatically improved callus proliferation in both Aikang 58 and Jimai 22 (Figure 2B and Supplemental Data 3). The regeneration frequency significantly increased from 3.83% ± 1.82% to 31.34% ± 3.67% in Aikang 58 and from 2.22% ± 1.48% to 35.62% ± 10.76% in Jimai 22 in response to TaLAX1-A overexpression (Figure 2C and Supplemental Data 4). In addition, there was a notable increase in the number of regenerating shoots per immature embryo in these varieties (Figure 2D and Supplemental Data 4).
Figure 2.
Overexpression of TaLAX1-A promotes shoot regeneration in different wheat genotypes.
(A) Shoot regeneration phenotypes of immature wheat embryos of different genotypes infected with empty vector (control) or TaLAX1-A-OE vector. Scale bar, 1 cm.
(B) Callus proliferation frequencies of wheat immature embryos of different genotypes infected with control or TaLAX1-A-OE vector.
(C) Regeneration frequencies of wheat immature embryos of different genotypes infected with control or TaLAX1-A-OE vector.
(D) Regenerating shoot frequencies of wheat immature embryos of different genotypes infected with control or TaLAX1-A-OE vector. CS, Chinese Spring; KN199, Kenong 199; SN28, Shannong 28; AK58, Aikang 58; JM22, Jimai 22. Values in (B–D) are means ± SEM from at least three independent experiments. Black points are the results from individual experiments. ∗∗∗∗P < 0.0001; ∗∗∗P < 0.001; ∗∗P < 0.01; ∗P < 0.05 (Student’s t-test, two-tailed).
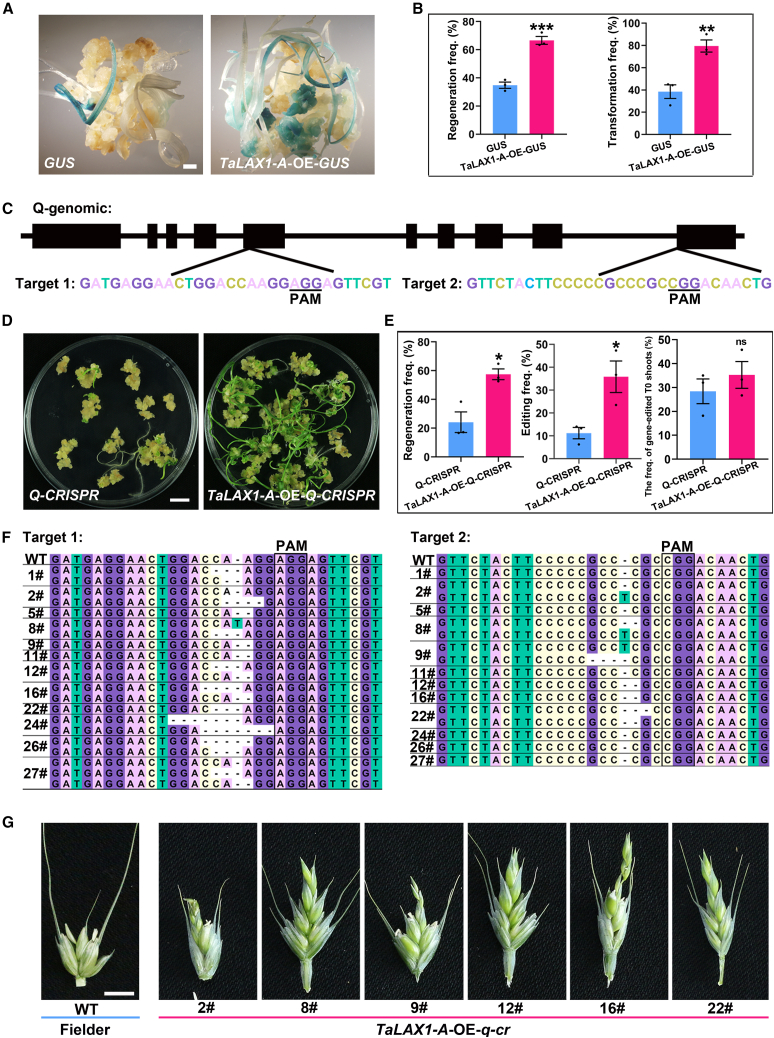
Figure 4.
TaLAX1-A overexpression improves transformation and gene editing frequencies in the common wheat variety Fielder.
(A) Shoot regeneration and transformation phenotypes of immature embryos infected with the Ubipro:GUS (GUS) or TaLAX1-A-OE-GUS vector. Scale bar, 2 mm.
(B) Regeneration and transformation frequencies of immature embryos infected with the GUS or TaLAX1-A-OE-GUS vector.
(C) Regions of the Q gene targeted with guide RNAs. PAM, protospacer-adjacent motif.
(D) Shoot regeneration phenotypes of immature embryos infected with the Q-CRISPR or TaLAX1-A-OE-Q-CRISPR vector. Scale bar, 1 cm.
(E) Regeneration frequencies, editing frequencies, and frequencies of gene-edited T0 shoots of immature embryos infected with the Q-CRISPR or TaLAX1-A-OE-Q-CRISPR vector. Editing frequency = no. of T0 plants with Q editing/no. of inoculated embryos × 100%. Frequency of gene-edited T0 shoots = no. of Q-gene-edited T0 plants/total no. of T0 plants × 100%.
(F) Twelve transgenic T0 plants with Q-edited genomic sequences. The 2#, 8#, 9#, 12#, 16#, and 22# lines carry two target mutations.
(G) Edited T0 plants showing increased numbers of florets per spikelet. Scale bar, 0.5 cm. Values in (B) and (E) are means ± SEM from three independent experiments. Black points are the results from individual experiments. ∗∗∗P < 0.001; ∗∗P < 0.01; ∗P < 0.05; ns, not significant (Student’s t-test, two-tailed).
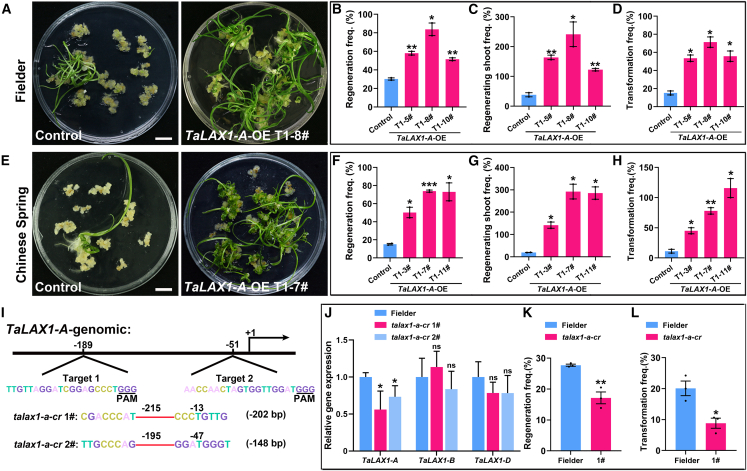
For the T1 progeny of transgenic lines overexpressing TaLAX1-A in Fielder and Chinese Spring, genotyping analyses confirmed the presence of the transgene, and the plants were found to be fertile, with increased effective tiller numbers in both varieties (Supplemental Figure 3 and Supplemental Data 5). Consistent with previous findings, overexpression of TaLAX1-A resulted in significant increases in spike length and plant height, which resembled the phenotypes observed in transgenic lines overexpressing TaLAX1-D (He et al., 2021). We measured the regeneration frequencies and regenerating shoot frequencies of embryos from T1 transgenic lines and found significant increases in both frequencies compared with the control non-transgenic lines (Figure 3A–3C and 3E–3G and Supplemental Data 6). To investigate the specific effect of reduced TaLAX1-A expression on wheat regeneration while keeping TaLAX1-B/D expression levels unchanged, we selected the TaLAX1-A promoter region, which has relatively low similarity to the TaLAX1-B/TaLAX1-D promoter sequences (TaLAX1-Apro vs. TaLAX1-Bpro, 48.38%; TaLAX1-Apro vs. TaLAX1-Dpro, 70.98%), rather than the highly similar gene regions (TaLAX1-A vs. TaLAX1-B, 88.78%; TaLAX1-A vs. TaLAX1-D, 93.63%), as the target for CRISPR–Cas9. We obtained two independent lines (talax1-a-cr 1# and 2#) with deletions in the TaLAX1-A promoter that resulted in reduced TaLAX1-A expression compared with wild-type Fielder; the expression levels of TaLAX1-B and TaLAX1-D showed no significant changes (Figure 3I and 3J). Significantly lower regeneration frequency was observed in the T1 progeny of the talax1-a-cr lines compared with wild-type Fielder (Figure 3K and Supplemental Data 7). These findings demonstrate the heritability of TaLAX1-A function in improving shoot regeneration.
Figure 3.
The function of TaLAX1-A in regeneration is heritable.
(A) Shoot regeneration phenotypes in Fielder T1 progeny of non-transgenic lines (control) or the TaLAX1-A-OE T1-8# transgenic line infected with the Ubipro:GUS vector. Scale bar, 1 cm.
(B) Regeneration frequencies of control and TaLAX1-A-OE T1-5#, TaLAX1-A-OE T1-8#, and TaLAX1-A-OE T1-10# lines in Fielder.
(C) Regenerating shoot frequencies of control and TaLAX1-A-OE T1-5#, TaLAX1-A-OE T1-8#, and TaLAX1-A-OE T1-10# lines in Fielder.
(D) Transformation frequencies of control and TaLAX1-A-OE T1-5#, TaLAX1-A-OE T1-8#, and TaLAX1-A-OE T1-10# lines in Fielder. Transformation frequency = no. of transgenic shoots with GUS signals/no. of inoculated embryos × 100%.
(E) Shoot regeneration phenotypes in Chinese Spring T1 progeny of non-transgenic lines (control) or the TaLAX1-A-OE T1-7# transgenic line infected with the Ubipro:GUS vector. Scale bar, 1 cm.
(F) Regeneration frequencies of control and TaLAX1-A-OE T1-3#, TaLAX1-A-OE T1-7#, and TaLAX1-A-OE T1-11# lines in Chinese Spring.
(G) Regenerating shoot frequencies of control and TaLAX1-A-OE T1-3#, TaLAX1-A-OE T1-7#, and TaLAX1-A-OE T1-11# lines in Chinese Spring.
(H) Transformation frequencies of control and TaLAX1-A-OE T1-3#, TaLAX1-A-OE T1-7#, and TaLAX1-A-OE T1-11# lines in Chinese Spring.
(I) Generation of TaLAX1-A knockdown lines by CRISPR–Cas9. Two target sites in the promoter of TaLAX1-A are shown. PAM, protospacer-adjacent motif.
(J) Relative expression of TaLAX1-A/B/D in the wild type and two talax1-a-cr transgenic lines of Fielder.
(K) Regeneration frequencies of the wild type and talax1-a-cr 1# transgenic line in Fielder.
(L) Transformation frequencies of the wild type and talax1-a-cr 1# transgenic line in Fielder. Values in (B)–(D), (F)–(H), (K), and (L) are means ± SEM; values in (J) are means ± SD. All experiments were performed at least two times. Black points are the results from individual experiments. ∗∗∗P < 0.001; ∗∗P < 0.01; ∗P < 0.05; ns, not significant (Student’s t-test, two-tailed).
To further test the broad-spectrum regenerative effects of TaLAX1-A, we introduced TaLAX1-A-OE and empty control vectors into wild-type Fielder using traditional particle bombardment-mediated transformation. The plants transformed with TaLAX1-A showed higher regeneration frequencies (37.83% ± 1.46%) than the controls (17.58% ± 2.42%; Supplemental Figure 4A–4C and Supplemental Data 8). Similarly, the regenerating shoot frequency was significantly higher in plants transformed with TaLAX1-A than in controls (Supplemental Figure 4D and Supplemental Data 8). Collectively, these results demonstrate the ability of TaLAX1-A overexpression to improve wheat regeneration in different varieties and with different protocols.
TaLAX1-A improves transformation and gene editing efficiency in wheat
Efficient regeneration is crucial for successful plant genetic transformation. To assess the effect of TaLAX1-A on the transformation frequency of wheat, we generated a compound expression vector (TaLAX1-A-OE-GUS) by combining Ubipro:TaLAX1 with Ubipro:GUS cassettes. Transformation experiments were performed in the common wheat variety Fielder, and the regeneration and transformation frequencies were estimated. The wheat TaLAX1-A-OE-GUS transgene significantly increased regeneration frequency compared with the control (Ubipro:GUS alone; Figure 4A and 4B and Supplemental Data 9). Consistent with this finding, the transformation frequency with the TaLAX1-A-OE-GUS expression cassette (79.52% ± 5.43%) was significantly higher than that of the control (38.46% ± 6.17%; Figure 4B and Supplemental Data 9). Improved regeneration and transformation frequencies were also observed when a mixture of Agrobacterium strains was used, one carrying the TaLAX1-A-OE expression cassette and the other carrying the Ubipro:GUS marker cassette (Supplemental Figure 5). Transgenic plants with GUS expression signals revealed higher efficiency in the TaLAX1-A-OE and GUS co-transformation experiments than in co-transformation experiments with the empty vector and GUS vector (Supplemental Figure 5 and Supplemental Data 10), suggesting that TaLAX1-A overexpression enhanced transformation frequency.
Next, the transformation frequencies of the TaLAX1-A-OE and talax1-a-cr lines were evaluated using the Ubipro:GUS vector. The TaLAX1-A-OE transgenic lines showed significantly higher transformation frequencies than the non-transgenic lines (Figure 3D and 3H and Supplemental Data 6), whereas the talax1-a-cr transgenic lines exhibited much lower transformation frequencies than wild-type Fielder (Figure 3L and Supplemental Data 7). These results indicate that TaLAX1-A overexpression not only enhances shoot regeneration but also improves transformation efficiency in wheat.
The effect of TaLAX1-A-improved transformation on CRISPR–Cas9-mediated genome editing frequency was also investigated and compared with conventional transformation. The Q gene, which encodes an AP2-like transcription factor, is essential in wheat domestication and affects the number of florets in spikelets (Zhang et al., 2011). A binary vector containing a cassette with TaLAX1-A-OE, Cas9, and a guide RNA targeting the Q gene (Figure 4C) was generated for use in Agrobacterium transformation (TaLAX1-A-OE-Q-CRISPR). The Q-CRISPR construct served as the control. As expected, regeneration frequency was significantly higher in the TaLAX1-A-OE group than in the control (Figure 4D and 4E and Supplemental Data 11). The genome editing frequency, calculated as the number of T0 plants with an edited Q gene relative to the total number of inoculated embryos, was significantly higher in the TaLAX1-A-OE context (35.88% ± 6.90%) than in the control (11.19% ± 2.48%; Figure 4E and Supplemental Data 11). However, the proportion of Q-gene-edited T0 shoots in the total number of T0 plants did not differ significantly from that of the control (Figure 4E and Supplemental Data 11). This suggests that the enhanced editing frequencies resulting from TaLAX1-A can be attributed to improved rates of regeneration. In addition, successful Cas9-induced Q-gene editing was detected in 12 independent TaLAX1-A-OE-q-cr T0 lines, and 6 lines exhibited clear mutant q-null phenotypes (Figure 4F and 4G and Supplemental Data 11 and 12). Overall, TaLAX1-A overexpression significantly enhanced gene editing frequency. Because both the Q-CRISPR and the TaLAX1-A-OE sequences can be segregated out after editing the desired genomic region, the TaLAX1 strategy is ideal for extending genome editing technology to wheat varieties with low regeneration efficiency.
TaLAX1-A promotes expression of TaGRF and TaGIF genes during wheat regeneration
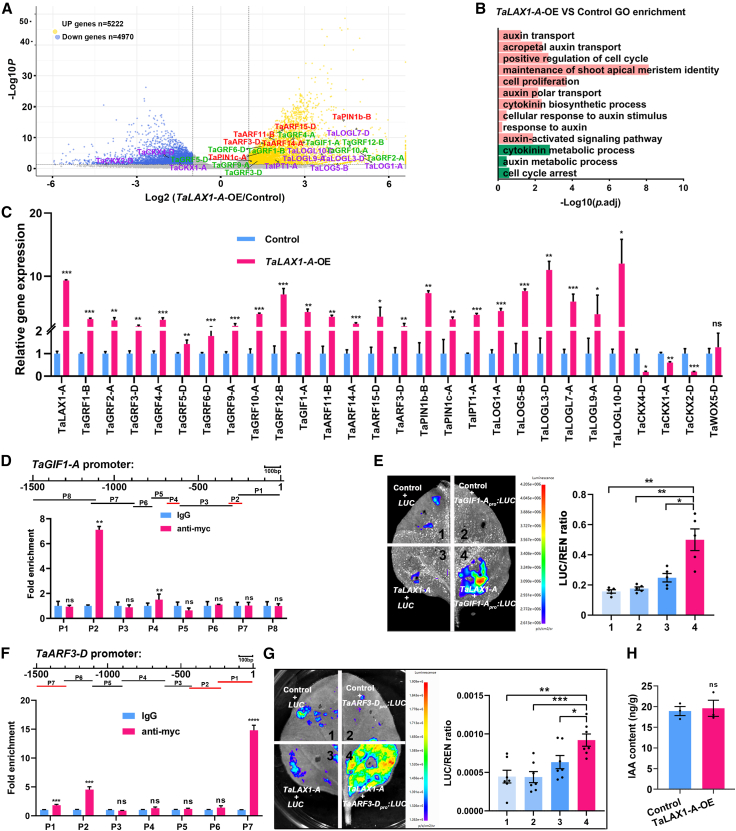
To investigate the potential regulatory roles of TaLAX1-A during wheat regeneration, we performed RNA-sequencing (RNA-seq) analysis of Chinese Spring calli 5 days after transfer to shoot-induction medium. This specific stage was chosen because of the presence of morphologically regenerated shoot primordia (Supplemental Figure 6) and the highest TaLAX1-A expression during the shoot regeneration process (Supplemental Figure 2). Analysis of the RNA-seq data revealed 10 192 differentially expressed genes (DEGs) in TaLAX1-A-OE calli compared with the empty vector controls (Figure 5A). Among these DEGs, 5222 were upregulated, and 4970 were downregulated. The upregulated genes in TaLAX1-A-OE calli, which may be activated by TaLAX1-A, were enriched in functional annotations associated with cell proliferation, maintenance of shoot apical meristem identity, cytokinin biosynthetic process, and auxin responses and transport (Figure 5B). The downregulated genes in TaLAX1-A-OE calli were enriched in cytokinin metabolic process (Figure 5B).
Figure 5.
RNA-seq analysis of TaLAX1-A-OE transgenic calli and activation of TaGIF1-A and TaARF3-D by TaLAX1-A.
(A) Volcano plot of up- and downregulated genes in TaLAX1-A-OE transgenic calli vs. empty vector (control) transgenic calli of Chinese Spring. Purple, cytokinin-related genes; red, auxin-related genes; green, TaGRFs and TaGIF1-A.
(B) Gene Ontology (GO) enrichment analysis of up- and downregulated genes from TaLAX1-A-OE transgenic calli vs. empty vector (control) transgenic calli. Red, GO terms of genes upregulated in TaLAX1-A-OE vs. control; green, GO terms of genes downregulated in TaLAX1-A-OE vs. control.
(C) Relative expression levels of selected genes regulated by TaLAX1-A in TaLAX1-A-OE transgenic calli and empty vector transgenic calli.
(D) ChIP–qPCR of the TaGIF1-A promoter using an anti-myc antibody in the TaLAX1-A-OE transgenic lines. TaLAX1-A-OE samples with IgG antibody were used as negative controls. TaLAX1-A binds to the P2 and P4 regions of the TaGIF1-A promoter.
(E) Transient expression of TaLAX1-A protein and TaGIF1-Apro:LUC reporter in tobacco leaves (left) and statistics of luciferase activity (right).
(F) ChIP–qPCR of the TaARF3-D promoter using an anti-myc antibody in the TaLAX1-A-OE transgenic lines; TaLAX1-A-OE samples with IgG antibody were used as a negative control. TaLAX1-A binds to the P1, P2, and P7 regions of the TaARF3-D promoter.
(G) Transient expression of TaLAX1-A protein and TaARF3-Dpro:LUC reporter in tobacco leaves (left) and statistics of luciferase activity (right).
(H) Endogenous IAA content in TaLAX1-A-OE transgenic calli and empty vector (control) transgenic calli. Values in (C), (D), and (F) are means ± SD; values in (E), (G), and (H) are means ± SEM. All experiments were performed at least three times. Black points are the results from individual experiments. ∗∗∗∗P < 0.0001; ∗∗∗P < 0.001; ∗∗P < 0.01; ∗P < 0.05; ns, not significant (Student’s t-test, two-tailed).
In previous studies, overexpression of TaGRF4 in combination with TaGIF1 or TaWOX5 has been shown to significantly increase regeneration efficiency in wheat and expand the number of transformable wheat genotypes (Debernardi et al., 2020; Wang et al., 2022a). Interestingly, the TaGRF genes and their co-factor, TaGIF1, were upregulated in TaLAX1-A-OE-transformed calli compared with the controls (Figure 5A). We further examined the transcript levels of these genes in TaLAX1-A-OE calli, which showed a substantial increase in TaGRF and TaGIF1 expression (Figure 5C), supporting a role for TaLAX1-A in promoting expression of TaGRFs and TaGIF1 during wheat regeneration. However, the transcript levels of TaWOX5 did not differ significantly between TaLAX1-A-OE and control calli (Figure 5C).
To determine whether TaLAX1-A acts as a direct regulator of TaGRF4 and TaGIF1 transcription, we performed chromatin immunoprecipitation followed by quantitative polymerase chain reaction (ChIP–qPCR) experiments. We observed enrichment of specific fragments of the TaGIF1-A promoter (P2, −319 bp to −239 bp, and P4, −698 bp to −618 bp) (Figure 5D). By contrast, no enrichment of TaGRF4-A promoter fragment sequences was detected (Supplemental Figure 7). Luciferase activation assays further confirmed that TaLAX1-A binds directly to the TaGIF1-A promoter region and activates its expression (Figure 5E). Collectively, these findings strongly suggest that TaLAX1-A functions as a direct regulator of TaGIF1 transcription during wheat regeneration, at least in part by binding to the TaGIF1 promoter and enhancing TaGIF1 expression, which may subsequently affect TaGRF activity.
TaLAX1-A overexpression enhances auxin response and cytokinin biosynthesis in wheat regeneration
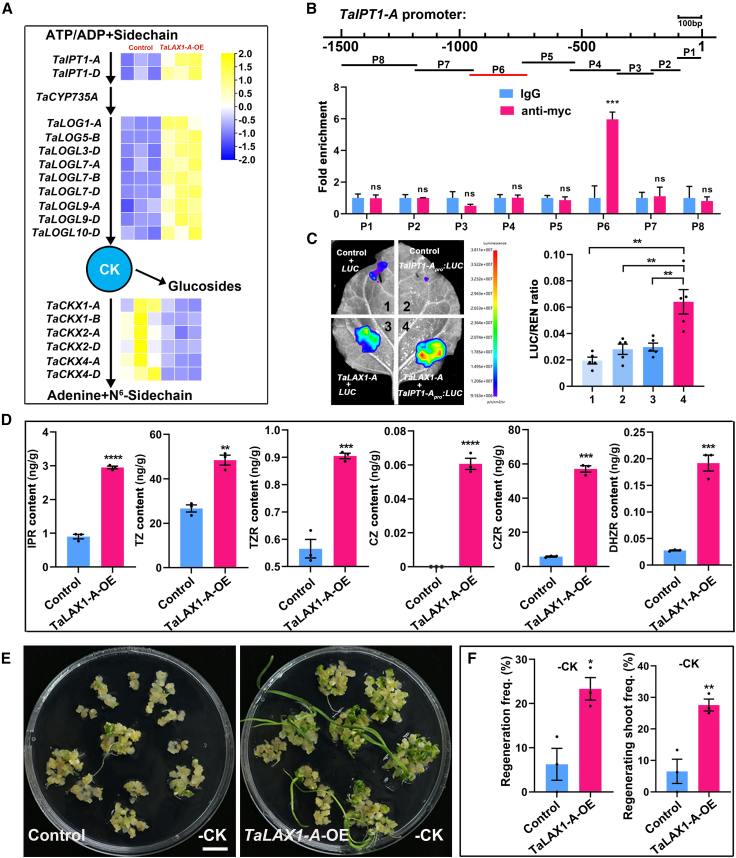
Our transcriptomic analysis revealed significant upregulation of the TaAUXIN RESPONSE FACTOR (TaARF) genes TaARF3, TaARF11, TaARF14, and TaARF15 and the auxin-transport-related TaPINFORMED1 (TaPIN) genes TaPIN1b and TaPIN1c in TaLAX1-A-OE calli compared with controls (Figure 5A and 5C). We performed ChIP–qPCR and luciferase activation assays and found that TaLAX1-A binds to the promoter of TaARF3-D, which plays a crucial role in the plant regeneration process (Cheng et al., 2013), to activate its transcription (Figure 5F and 5G). IAA content did not differ significantly between the TaLAX1-A-OE transgenic calli and the controls (Figure 5H), suggesting that auxin biosynthesis may not be affected by TaLAX1-A overexpression. Transcription of cytokinin biosynthesis genes such as TaISOPENTENYL TRANSFERASE 1 (TaIPT1), the TaLONELY GUY (TaLOG) genes TaLOG1 and TaLOG5, and the TaLONELY GUY-LIKE PROTEIN (TaLOGL) genes TaLOGL3, TaLOGL7, TaLOGL9, and TaLOGL10 was enhanced in TaLAX1-A-OE calli. Conversely, expression of the cytokinin metabolic process–related TaCYTOKININ OXIDASE (TaCKX) genes TaCKX1, TaCKX2, and TaCKX4 was downregulated in TaLAX1-A-OE-transformed calli (Figures 5A, 5C, and 6A). We performed ChIP–qPCR and luciferase activation assays and found that TaLAX1-A binds to the TaIPT1-A promoter to activate its transcription (Figure 6B and 6C). This transcriptional activation was consistent with the significantly enhanced levels of endogenous cytokinin in the TaLAX1-A-OE transgenic calli (Figure 6D). These findings suggest that TaLAX1-A may play a role in regulating cytokinin accumulation by modulating the expression of genes related to cytokinin biosynthesis and metabolism, as well as influencing auxin responses and transport.
Figure 6.
TaLAX1-A activates cytokinin biosynthesis during the process of shoot regeneration.
(A) Expression heatmap of genes related to cytokinin biosynthetic and metabolic processes in control (empty vector) transgenic calli and TaLAX1-A-OE transgenic calli.
(B) ChIP–qPCR of the TaIPT1-A promoter using an anti-myc antibody in the TaLAX1-A-OE transgenic lines; TaLAX1-A-OE samples with IgG antibody were used as a negative control. TaLAX1-A binds to the P6 region of the TaIPT1-A promoter.
(C) Transient expression of TaLAX1-A protein and TaIPT1-Apro:LUC reporter in tobacco leaves (left) and statistics of luciferase activity (right).
(D) Endogenous cytokinin content (IPR, TZ, TZR, CZ, CZR, and DHZR) in TaLAX1-A-OE transgenic calli and empty vector (control) transgenic calli.
(E) Shoot regeneration phenotypes of Fielder embryos transformed with the empty vector (control) or TaLAX1-A-OE after incubation on CIM for 42 days and then on SIM (without exogenous cytokinin) for 20 days. Scale bar, 1 cm.
(F) Regeneration frequencies and regenerating shoot frequencies of Fielder immature embryos infected with control or TaLAX1-A-OE after incubation on CIM for 42 days and then on SIM (without exogenous cytokinin) for 20 days. Values in (B) are means ± SD; values in (C), (D), and (F) are means ± SEM. All experiments in (B)–(D) and (F) were performed at least three times. Black points are the results from individual experiments. ∗∗∗∗P < 0.0001; ∗∗∗P < 0.001; ∗∗P < 0.01; ∗P < 0.05; ns, not significant (Student’s t-test, two-tailed).
In PureWheat-mediated transformation, the presence of both auxin and cytokinin is necessary to induce callus formation and promote shoot regeneration (Ishida et al., 2015). Interestingly, in Fielder embryos inoculated with Agrobacterium carrying TaLAX1-A-OE, rapid shoot regeneration was observed even in medium without exogenously added cytokinin (Figure 6E and 6F and Supplemental Data 13). Moreover, the regeneration frequency upon TaLAX1-A overexpression in the absence of cytokinin was comparable to that of the empty vector control in medium containing cytokinin (Figures 1C and 6F and Supplemental Data 1 and 13). We next assessed the regeneration frequency and regenerating shoot frequency of immature Fielder embryos transformed with TaLAX1-A-OE or the empty vector in the absence of exogenous cytokinin and auxin. Regeneration frequency and regenerating shoot frequency were significantly higher in embryos transformed with TaLAX1-A-OE than in controls (Supplemental Figure 8 and Supplemental Data 14). Using the Ubipro:GUS vector, we compared the regeneration and transformation frequencies of stable TaLAX1-A-OE T1 transgenic lines and non-transgenic lines in the absence of cytokinin or in the absence of both cytokinin and auxin. Under these conditions, the regeneration and transformation frequencies of TaLAX1-A-OE T1 transgenic plants were significantly higher than those of the non-transgenic lines (Supplemental Figures 9 and 10 and Supplemental Data 15 and 16). These results suggest that TaLAX1-A overexpression can promote shoot regeneration and partially compensate for a lack of exogenous cytokinin and auxin supplementation.
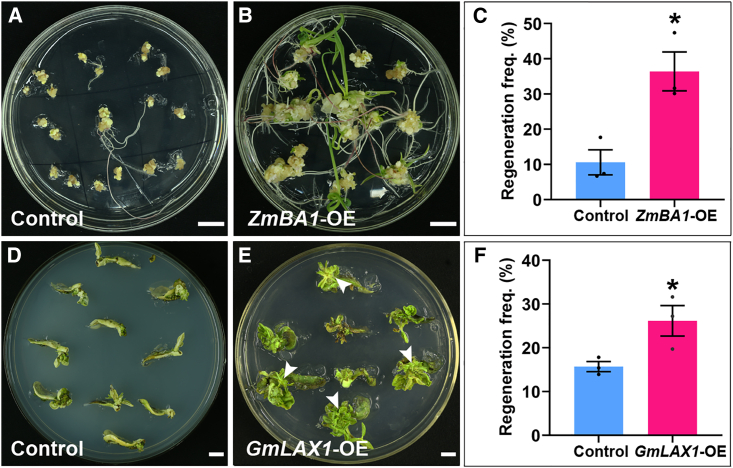
TaLAX1 homologs in maize and soybean promote shoot regeneration
To investigate the potential of TaLAX1 homologs to promote transformation efficiency in other crop species, we examined their effects in the monocot maize and the dicot soybean. Transformation of both maize and soybean is currently limited to a narrow range of genotypes (Jia et al., 2015; Lowe et al., 2016; Wang et al., 2022a, 2022b). Through phylogenetic analysis, we identified ZmBA1 in maize and GmLAX1 in soybean as homologs of wheat TaLAX1 (Supplemental Figure 1A). Overexpression of ZmBA1 significantly increased the regeneration frequency of maize B104 lines from 10.59% ± 3.55% to 36.40% ± 5.52% (Figure 7A–7C and Supplemental Data 17). Similarly, overexpression of GmLAX1 in the soybean variety Dongnong-50 significantly increased regeneration frequency from 15.68% ± 1.08% to 26.18% ± 3.48% (Figure 7D–7F and Supplemental Data 18). We performed further cultivation of the regenerated shoots derived from immature maize embryos transformed with ZmBA1-OE and soybean cotyledonary nodes transformed with GmLAX1-OE, and these regenerated shoots successfully developed into complete plants (Supplemental Figure 11A and 11C). Several of these plants were identified as overexpression lines through real-time qPCR analyses (Supplemental Figure 11B and 11D). These findings demonstrate that TaLAX1 homologs positively enhance regeneration in the monocot and dicot species tested in this study. Therefore, we propose that the benefits of LAX1 technology may be extended to improve transformation efficiency in other crop varieties.
Figure 7.
Transformation of TaLAX1 homologs into maize and soybean.
(A) Shoot regeneration phenotypes of maize (B104) transformed with the empty vector (control, p3300-CUB). Scale bar, 1 cm.
(B) Shoot regeneration phenotypes of maize (B104) transformed with ZmBA1-OE. Scale bar, 1 cm.
(C) Regeneration frequencies of B104 immature embryos infected with control or ZmBA1-OE. Regeneration frequency = no. of calli showing at least one regenerating shoot/no. of inoculated embryos × 100%.
(D) Shoot regeneration phenotypes of soybean (Dongnong-50) transformed with the empty vector (control, 35S-PBI106). Scale bar, 1 cm.
(E) Shoot regeneration phenotypes of soybean (Dongnong-50) transformed with GmLAX1-OE. Regenerated shoots are indicated with white arrows. Scale bar, 1 cm.
(F) Regeneration frequencies of Dongnong-50 cotyledonary nodes infected with control or GmLAX1-OE. Regeneration frequency = no. of explants showing at least one regenerating shoot/no. of inoculated explants × 100%. Values in (C) and (F) are means ± SEM from three independent experiments. Black points are the results from individual experiments. ∗P < 0.05 (Student’s t-test, two-tailed).
Discussion
Wheat TaLAX1 exhibits homology to OsLAX1 in rice and ZmBA1 in maize, based on their amino acid sequence similarities (Komatsu et al., 2003; Gallavotti et al., 2004). In rice, strong alleles of lax1 mutants display severe defects in initiation of lateral spikelets and panicle branches (Komatsu et al., 2003). Similarly, maize mutants with the ba1 allele fail to initiate lateral meristems during both vegetative and reproductive development, resulting in unbranched, short, and sterile tassels without spikelets (Ritter et al., 2002; Gallavotti et al., 2004; Skirpan et al., 2008; Yao et al., 2019). However, wheat Talax1-aabbdd mutants exhibit a compact spike phenotype (He et al., 2021), which is distinct from the reduced lateral branching observed in rice lax1 and maize ba1 mutants. These different phenotypes indicate that rice and maize homologs have undergone functional diversification. In this study, we revealed a novel role for TaLAX1 in the regulation of wheat regeneration and transformation. These proteins and their homologs show potential as enhancers to improve genetic transformation and genome editing efficiency in various crop species.
The low regeneration capability and genotype-specific growth requirements of explant embryos have posed challenges to achieving high transformation efficiency in wheat. Despite efforts to develop efficient transformation protocols, many wheat varieties, including Aikang 58 and Jimai 22, remain recalcitrant to existing methods (Wang et al., 2017, 2022a). Susceptibility to Agrobacterium infection and efficiency of regeneration often exhibit notable genotype specificity, limiting the applicability of transformation protocols to only a few wheat genotypes (Wang et al., 2017, 2022a). Recent studies using multiomics data have uncovered a transcriptional regulatory network involved in wheat regeneration, highlighting the sequential expression of genes responsible for cell fate reprogramming, which is triggered by auxin and coordinated with changes in chromatin accessibility and histone modifications (Liu et al., 2023). Several wheat factors involved in developmental reprogramming have been identified as critical regulators that can transform somatic cells into embryogenic cells, and these factors have been designated “boosters” for regeneration efficiency in transformed wheat embryos. Overexpression of these boosters, such as the fusion protein TaGRF4–TaGIF1 or TaWOX5, has been shown to enhance regeneration efficiency with reduced dependence on specific genotypes (Debernardi et al., 2020; Wang et al., 2022a; Qiu et al., 2022).
Interestingly, we also observed that overexpression of TaLAX1 significantly improved transformation efficiency in wheat. Transgenic plants overexpressing TaLAX1 were fertile and did not exhibit obvious developmental or reproductive defects. These findings distinguish TaLAX1 from other regeneration-related factors that may introduce additional plant phenotypes, which can pose challenges in certain experimental applications. Furthermore, TaLAX1-A overexpression enhances regeneration efficiency in previously recalcitrant wheat varieties such as Aikang 58 and Jimai 22. These varieties, known for their low regeneration efficiency, have been difficult to transform and genome edit using existing methods. Therefore, the introduction of TaLAX1-A overcomes such genotype limitations and expands the available resources for transformation.
The process of shoot regeneration in a wide variety of crops relies on the addition of appropriate plant hormones, particularly auxin and cytokinin, to the regeneration medium. Targeted manipulation of hormone signaling pathways has been shown to activate the accumulation of auxin and cytokinin, enhancing shoot regeneration even in recalcitrant crop varieties (Paek and Hahn, 2000; Cheng et al., 2013; Ishida et al., 2015; Li et al., 2017b; Meng et al., 2017). In our study, we collected calli for transcriptomic analysis 5 days after transfer to shoot induction medium because this stage is likely critical for shoot apical meristem formation: morphologically regenerated shoot primordia are present, and the highest TaLAX1 expression is observed at this stage. Through RNA-seq analysis, we identified significant upregulation of auxin response genes (TaARFs), auxin transport genes (TaPINs), and cytokinin biosynthesis genes (TaIPT1, TaLOGs, and TaLOGLs) in TaLAX1-A-overexpressing transformed tissues. Consistent with these findings, endogenous cytokinin levels were significantly higher in TaLAX1-A-overexpressing calli than in controls. Furthermore, Fielder embryos infected with the TaLAX1-A-OE vector were able to rapidly regenerate green shoots when cultured in medium without cytokinin, confirming that TaLAX1-A promotes endogenous cytokinin accumulation. Moreover, we found that TaLAX1-A directly binds to the promoter regions of both TaIPT1-A and TaARF3-D to activate their transcription. These findings suggest that TaLAX1-A may play a role in regulating shoot regeneration by modulating cytokinin biosynthesis and metabolism, as well as auxin responses and transport.
Even in the absence of exogenous cytokinin and auxin, the calli derived from Fielder embryos infected with the empty vector retained some regenerative capacity. This finding could be attributed to the intrinsic regenerative ability of the Fielder genotype, which has shown stronger regenerative capacity than other genotypes used in the PureWheat technique (Wang et al., 2017; Debernardi et al., 2020). In addition, the cutting processes involved in the regeneration protocol may induce wounding signals that trigger callus and shoot formation, leading to accumulation of hormones such as auxin, cytokinin, and jasmonate, which promote plant regeneration (Ikeuchi et al., 2017; Zhang et al., 2019). Transcriptional regulators such as ETHYLENE RESPONSE FACTOR 115 (ERF115), WOUND-INDUCED DEDIFFERENTIATION1 (WIND1), ENHANCER OF SHOOT REGENERATION 1 (ESR1), PLETHORA3 (PLT3), PLT5, and PLT7 are also upregulated by wounding and play a role in promoting plant regeneration (Ikeuchi et al., 2017; Iwase et al., 2017; Chen et al., 2022).
We found that TaGRFs and TaGIF1 were upregulated by TaLAX1-A overexpression and that TaLAX1-A directly regulated the expression of TaGIF1. These results indicate that TaLAX1-A may regulate shoot regeneration through the same pathway as TaGRF4–TaGIF1, suggesting a regulatory network involving auxin, cytokinin, and related transcription factors like TaGRFs and TaGIF1, which are responsible for cell division, cell fate transition, and ultimate shoot regeneration.
Compared with the gene combinations of BBM–WUS2 and GRF4–GIF1, overexpression of a single gene, LAX1, significantly increased crop regeneration frequency. Moreover, transgenic plants overexpressing LAX1 were fertile and did not exhibit obvious developmental or reproductive defects. These findings distinguish LAX1 from other regeneration-related factors that may introduce additional plant phenotypes, which could pose challenges in certain experimental applications. Thus, combining LAX1 technology with other approaches such as “altruistic transformation” (Hoerster et al., 2020) or the use of conditional expression drivers (Lowe et al., 2018; Gao et al., 2020) is recommended to avoid such effects. Another strategy involves linking regeneration-promoting gene elements with the CRISPR–Cas9 system (Gao et al., 2016; Lu et al., 2017; Chen et al., 2019).
In conclusion, the regeneration step is the main bottleneck for efficient transformation and genome editing in wheat. Previous studies have attempted to enhance transformation efficiency in wheat by making use of genes that promote regeneration during tissue culture (Debernardi et al., 2020; Wang et al., 2022a; Qiu et al., 2022). However, effective methods for regenerating transformed somatic cells into whole plants are still limited. In our study, we discovered that overexpression of TaLAX1 significantly increases regeneration efficiency in certain wheat varieties. Our findings demonstrate that this increased efficiency is associated with the upregulation of TaGRF and TaGIF genes, as well as genes related to auxin response and transport and cytokinin biosynthesis. Overexpression of TaLAX1 homologs also improved the formation of transgenic plants in soybean and maize, suggesting that TaLAX1 may be a promising strategy for overcoming the transformation barrier in economically important crops.
Methods
Plant materials and growth conditions
The hexaploid wheat varieties used in the present work included Fielder, Chinese Spring, Kenong 199, Shannong 28, Jimai 22, and Aikang 58. The tetraploid wheat varieties used were Langdon and Kronos. Fielder seeds were generously provided by Japan Tobacco. All wheat varieties used in the regeneration and transformation experiments were grown in growth chambers under long-day conditions (16-h light/8-h dark) at 22°C. Facultative or winter varieties were maintained at 4°C for 30 days for vernalization before being transplanted to soil. Plants used for growth and developmental phenotype analyses were planted at the experimental station of Shandong Agricultural University (E 117°, N 36°).
Plasmid construction
We generated the PC186-TaLAX1-myc construct using the Gateway system (Invitrogen, Carlsbad, CA, USA). The sequence of myc and several restriction enzyme cutting sites (GCGGCCGCatggaacagaaactgatctctgaagaagatctgGAATTCCATATGGGATCCCTGCAGGGTACCATCAGTAAGCTTCCATGGGAGCTCATCGATGTCGACTCTAGACTCGAGGGCGCGCC) was synthesized and inserted into the pENTR vector to generate the myc-mcs-ENTR vector. The TaLAX1-A/B/D genomic sequences were amplified using 2×Phanta Max Master Mix (Vazyme, Nanjing, China; P515-01) and inserted into the myc-mcs-ENTR vector between EcoRI and HindIII. myc-TaLAX1-ENTR was then recombined into the PC186 vector through Gateway LR recombination. For the TaLAX1-A-OE-GUS construct, we amplified the Nos terminator from the PC186 plasmid and inserted it into the myc-TaLAX1-A-ENTR vector between SacI and ClaI. The Ubipro:GUS module was amplified from the Ubipro:GUS plasmid and inserted into the myc-TaLAX1-A-Nos Terminator-ENTR vector between XhoI and XbaI, and myc-TaLAX1-A-Nos Terminator-Ubipro:GUS-ENTR was then recombined into the PC186 vector through Gateway LR recombination.
To generate the TaLAX1-A-CRISPR and Q-CRISPR constructs, two single-guide RNA sequences were designed to target the TaLAX1-A promoter and the coding sequence of the Q gene. In accordance with the methods described by Xing et al. (2014), the single-guide RNAs were inserted into the PBUE413 vector. To generate the TaLAX1-A-OE-Q-CRISPR construct, Ubipro:TaLAX1-A-Nos Terminator synthetic fragments were digested with PmeI and then inserted into the Q-CRISPR vector.
To generate the ZmBA1-OE construct, we amplified the ZmBA1 genomic sequence and inserted it into the p3300-CUB vector (Cambia) between SmaI and SacI. To produce the GmLAX1-OE construct, we amplified the 35S promoter sequence from the PROK II vector and inserted it into the PBI106 vector between EcoRI and BamHI. GmLAX1 genomic sequences were then amplified and inserted into the 35S-PBI106 vector between XhoI and SacI. The primers used for plasmid construction are listed in Supplemental Data 19.
Plant transformation
To perform wheat transformation, we obtained immature embryos (approximately 14 days post anthesis) from the indicated wheat lines. The transformation procedure followed the protocols outlined in a previous study (Ishida et al., 2015). The Agrobacterium tumefaciens strain EHA105 was used to introduce the PC186-TaLAX1-myc, PC186 empty vector, TaLAX1-A-OE-GUS, Ubipro:GUS, TaLAX1-A-CRISPR, TaLAX1-A-OE-Q-CRISPR, and Q-CRISPR vectors into the immature embryos. After Agrobacterium infection, the immature embryos were initially cultured on WLS-AS medium (1/100 volume of 10× LS major salts, 1/1000 volume of 100× FeEDTA, 1/1000 volume of 100× LS minor salts, 1/1000 volume of 100× MS vitamins, 8 g/l agarose, 10 mg/l glucose, 0.5 g/l MES, 0.85 mg/l AgNO3, 100 μM AS, 1.25 mg/l CuSO4·5H2O) for 2 days. The embryo axis was then excised from the embryos and transferred to WLS-Res medium (1/10 volume of 10× LS major salts, 1/100 volume of 100× FeEDTA, 1/10 volume of 100× LS minor salts, 0.5 mg/l 2,4-D, 1/100 volume of 100× MS vitamins, 2.2 mg/l picloram, 0.75 g/l MgCl2·6H2O, 0.5 g/l glutamine, 0.1 g/l casein hydrolysate, 40 g/l maltose, 1.95 g/l MES, 5 g/l agarose, 250 mg/l carbenicillin, 100 mg/l ascorbic acid, 0.85 mg/l AgNO3, 100 mg/l cefotaxime) for 5 days. The explants were next moved to WLS-P5 medium (1/10 volume of 10× LS major salts, 1/100 volume of 100× FeEDTA, 0.5 mg/l 2,4-D, 1/10 volume of 100× LS minor salts, 1/100 volume of 100× MS vitamins, 2.2 mg/l picloram, 0.1 g/l casein hydrolysate, 0.5 g/l glutamine, 0.75 g/l MgCl2·6H2O, 5 g/l agarose, 40 g/l maltose, 1.95 g/l MES, 100 mg/l ascorbic acid, 250 mg/l carbenicillin, 5 mg/l phosphinothricin, 0.85 mg/l AgNO3) and cultured for 14 days. Each explant was then divided into two parts and transferred to WLS-P10 medium (WLS-P5 supplemented with 5 mg/l phosphinothricin) for 21 days to induce callus formation. The proliferated calli were transferred to LSZ-P5 medium (1/10 volume of 10× LS major salts, 1/100 volume of 100× FeEDTA, 1/100 volume of 100× LS minor salts, 20 g/l sucrose, 1/100 volume of 100× modified LS vitamins, 5 mg/l zeatin, 0.5 g/l MES, 2.5 mg/l CuSO4·5H2O, 250 mg/l carbenicillin, 8 g/l agar, 100 mg/l cefotaxime, 5 mg/l phosphinothricin) to induce shoot regeneration. The regenerated shoots were transferred to LSF-P5 medium (1/10 volume of 10× LS major salts, 1/100 volume of 100× FeEDTA, 1/100 volume of 100× LS minor salts, 0.5 g/l MES, 1/100 volume of 100× modified LS vitamins, 15 g/l sucrose, 3 g/l Gelrite, 0.2 mg/l IBA, 250 mg/l carbenicillin, 5 mg/l phosphinothricin) until root elongation. In this study, the abbreviations used for the respective media were CIM (callus induction medium) for WLS-AS, WLS-Res, WLS-P5, and WLS-P10 and SIM (shoot induction medium) for LSZ-P5.
To perform maize transformation, ZmBA1-OE and p3300-CUB empty vectors were transformed into A. tumefaciens EHA105. Immature maize zygotic embryos (12 days after pollination) from the inbred line B104 were used for Agrobacterium-mediated transformation according to a previously described protocol (Frame et al., 2011) with slight modifications. In brief, the infected immature embryos were initially cultured on co-cultivation medium (0.7 g/l L-proline, 4.3 g/l MS salts, 100 mg/l casein hydrolysate, 0.5 ml/l dicamba, 100 mg/l myo-inositol, 2.3 g/l Gelrite, 30 g/l sucrose, 1 ml/l MS vitamin stock, 88 mM silver nitrate, 100 mM AS, 300 mg/l L-cysteine). After 16 h, the explants were transferred to selection medium (0.7 g/l L-proline, 0.5 g/l MES, 4.3 g/l MS salts, 100 mg/l casein hydrolysate, 0.5 ml/l dicamba, 30 g/l sucrose, 100 mg/l myo-inositol, 2.3 g/l Gelrite, 1 ml/l MS vitamin stock, 88 mM silver nitrate, 250 mg/l carbenicillin, 1 mg/l bialaphos) for 14 days. The explants were then transferred to regeneration medium (1 ml/l modified MS vitamin stock, 4.3 g/l MS salts, 60 g/l sucrose, 100 mg/l myo-inositol, 3 g/l Gelrite, 6 mg/l glufosinate ammonia, 100 mg/l cefotaxime) for 4–8 weeks, with regular transfers to fresh regeneration medium every 2 weeks. The regenerated shoots were transferred to rooting medium (1 ml/l modified MS vitamin stock, 4.3 g/l MS salts, 30 g/l sucrose, 100 mg/l myo-inositol, 3 g/l Gelrite, 6 mg/l glufosinate ammonia, 100 mg/l cefotaxime) until root elongation.
For soybean transformation, GmLAX1-OE and PBI106 empty vectors were introduced into Dongnong-50 using Agrobacterium (EHA105)-mediated genetic transformation of soybean cotyledonary nodes as described in a previous study (Li et al., 2017b). Cotyledonary node explants were infected with Agrobacterium and placed on co-cultivation medium (3.9 g/l B5 salts, 30 g/l sucrose, 3.9 g/l MES, 8 g/l agar powder, 400 mg/l cysteine, 154.2 mg/l DTT, 0.25 mg/l GA3, 1.67 mg/l 6-BA, 40 mg/l AS) for 3 days. The explants, with hypocotyls removed, were then cultured in shoot induction medium (3.9 g/l B5 salts, 30 g/l sucrose, 0.59 g/l MES, 8 g/l agar powder, 1.67 mg/l 6-BA, 250 mg/l Timentin, 100 mg/l cefotaxime, 45 mg/l kanamycin). After 14 days, the explants were transferred to fresh shoot induction medium without kanamycin. After 28 days of cultivation in shoot induction medium, explants with regenerated shoots were transferred to shoot elongation medium (4.3 mg/l MS, 30 g/l sucrose, 0.59 g/l MES, 8 g/l agar powder, 50 mg/l asparagine, 50 mg/l L-pyroglutamic acid, 0.1 mg/l IAA, 0.5 mg/l GA3, 1 mg/l zeatin-R, 250 mg/l Timentin, 100 mg/l cefotaxime) and cultured for 4 weeks. Finally, the regenerated shoots were transferred to rooting medium (4.3 mg/l MS, 30 g/l sucrose, 0.59 g/l MES, 8 g/l agar powder, 50 mg/l asparagine, 50 mg/l L-pyroglutamic acid) and cultured for 2 weeks to produce complete plants.
Regenerative phenotype calculations
In wheat, callus proliferation frequency was determined by measuring the weights of immature embryos cultured on CIM for 42 days. The regeneration frequencies, regenerating shoot frequencies, transformation frequencies, gene editing frequencies, and frequencies of gene-edited T0 shoots were calculated after the immature embryos had been cultured on CIM for 42 days and then on SIM for 20 days. For the calculation of regenerated shoots, only those >2 cm in length were considered, as they exhibited a higher potential for development into fully grown seedlings and plants compared with smaller shoots.
In maize, regeneration frequencies of elongated regenerating shoots were calculated before transfer to root induction medium. Only shoots >2 cm were considered and included in the calculation.
In soybean, regeneration frequencies were determined after the explants had been cultured on shoot induction medium for 28 days. Only shoots >2 cm were considered for the calculation, with particular emphasis on identifying regenerated shoots that possessed a shoot meristem.
Total RNA isolation and real-time qPCR analysis
We extracted total RNA from the indicated tissues using the CWBio Ultrapure RNA Kit. The FastKing RT Kit (Tiangen Biotech, Beijing, China) was used to synthesize first-strand cDNA. SYBR Green Real-Time PCR Master Mix (Tiangen Biotech) was used to perform qPCR analysis. We used the SYBR Green Master Mix to dilute each cDNA. Relative transcript levels were normalized to that of TaACTIN, ZmACTIN, or GmACTIN. All measurements were carried out with three biological replicates. We used the comparative CT method to determine expression values. The primers used for qPCR analysis are listed in Supplemental Data 19.
Transcriptome analysis
For transcriptome sequencing, we collected calli induced from de novo-transformed immature embryos transformed with the empty vector or TaLAX1-A-OE and cultured on CIM for 42 days and SIM for 5 days. Transcriptomics analysis was performed using previously described protocols (Li et al., 2021). Three independent biological replicates were performed for each specimen; sequencing was performed by OE Biotech (Shanghai, China). Clean reads were mapped to the wheat genome (version IWGSC RefSeq 1.1). Genes with a greater than two-fold expression change and a false discovery rate of less than 0.05 were considered to be DEGs.
RNA in situ hybridization
TaLAX1-A cDNA was amplified and ligated into the pEasy-Blunt3 vector, and the insertion direction of the fragments was verified by sequencing. RNA in situ hybridization was performed with digoxigenin-labeled sense and antisense probes on 8-μm sections of the indicated tissues as described previously (Su et al., 2020).
GUS staining
Specimens were placed in GUS staining solution (10 mM EDTA, 1 mM 5-bromo-4-chloro-3-indolyl-b-D-glucuronic acid, 3 mM each K3Fe(CN)6/K4Fe(CN)6, 100 mM Na3PO4 buffer, and 0.1% Nonidet P-40) and incubated at 37°C overnight. After staining, we used 95% ethanol to remove chlorophyll.
Chromatin immunoprecipitation analysis
TaLAX1-A-OE lines (15 days old) were harvested for ChIP analysis according to previously described protocols (Li et al., 2017a) with minor modifications. The DNA fragments combined with TaLAX1-A protein were co-immunoprecipitated using anti-myc antibody (1:500, 06-340; Sigma-Aldrich). TaLAX1-A-OE samples with IgG antibody (1:500, 12-371; Merck) were used as negative controls. For each ChIP assay, we performed three biological repeats. All primers used for the ChIP assays are listed in Supplemental Data 19.
Luciferase assay
The 1500-bp TaGIF1-A, TaIPT1-A, and TaARF3-D promoters were individually fused with the luciferase reporter gene via the XhoI and BamHI sites of the pGreenII 0800-LUC vector (Hellens et al., 2005). The coding region of the TaLAX1-A gene was fused to the pGreenII 62-SK vector downstream of the 35S promoter between the BamHI and XhoI sites. The resulting plasmids were transformed into A. tumefaciens strain EHA105. Nicotiana benthamiana leaves were infiltrated with different mixed combinations of Agrobacterium and collected for transient transcription assays as described previously (Su et al., 2020).
Quantification of cytokinin
We collected calli induced from immature embryos that had been transformed with the empty vector or TaLAX1-A-OE and cultured on CIM for 42 days. Endogenous cytokinin levels were measured by Wuhan Greensword Creation Technology using LC–MS/MS analysis as reported previously (Liu et al., 2010).
Free IAA measurements
Calli induced from immature embryos that had been transformed with the empty vector or TaLAX1-A-OE and cultured on CIM for 11 days were collected for free IAA measurements. Free IAA measurements were performed as reported previously (Li et al., 2021).
Sequence alignment and phylogenetic analysis
We used MEGA 11 to perform sequence alignment and construct a phylogenetic tree of OsLAX1 and its homologous proteins in the indicated species using the neighbor-joining method and 1000 bootstrap replicates.
Paraffin sections
Chinese Spring materials were collected at various stages of wheat regeneration during tissue culture. The specimens were preserved by immersion in a 4% glutaraldehyde solution at 4°C overnight. The fixed specimens were then dehydrated in a series of ethanol concentrations ranging from 10% to 100%. After an 8-h xylene treatment, the samples were immersed in liquid paraffin for 3–5 days and then fixed in paraffin blocks. Sections (8 μm) were stained with toluidine blue O (T3260; Sigma-Aldrich) for 1 min.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 8. All data were analyzed using Student’s t-test (two-tailed) or one-way ANOVA with Tukey’s multiple comparisons test, and P < 0.05 was considered significant.
Data and code availability
The RNA-seq data discussed in this study have been deposited at the National Genomics Data Center Genome Sequence Archive (GSA; https://ngdc.cncb.ac.cn/gsa/) under accession no. CRA008766. All source data for the statistics shown in the figures are provided in Supplemental Data 1–19. Gene sequence data mentioned in this study can be found at EnsemblPlants (https://plants.ensembl.org/index.html) under the following accession numbers: TaLAX1-A (TraesCS3A02G350600), TaLAX1-B (TraesCS3B02G383000), TaLAX1-D (TraesCS3D02G344600), ZmBA1 (Zm00001d042989), GmLAX1 (GLYMA_18G204700), Q (TraesCS5A01G473800), TaGIF1-A (TraesCS4A02G250600), TaGRF4-A (TraesCS6A02G269600), TaIPT1-A (TraesCS1A02G376300), and TaARF3-D (TraesCS3D02G292100).
Funding
This research was funded by the National Key Research and Development Program of China (2022YFF1002902), the National Natural Science Foundation of China (31730008, 32070199), and the Natural Science Foundation of Shandong Province (ZR2022JQ12).
Author contributions
Y.H.S. and X.S.Z. designed the research. Y.Y., H.Y., J.P., W.J.Y., F.L.Z., S.R.W., Y.Z., and X.Y.Z. performed the research. Y.Y. and Y.P.W. analyzed the data. Y.H.S. and X.S.Z. wrote the paper.
Acknowledgments
We would like to thank Prof. Jiajie Wu and Prof. Fei Ni (College of Agronomy, Shandong Agricultural University) for providing the PBUE413, Ubipro:GUS, and PC186 vectors, as well as the Kronos and Langdon wheat seeds. We thank Prof. Dajian Zhang (College of Agronomy, Shandong Agricultural University) for providing Dongnong-50 soybean seeds. We also thank Prof. Jian Xu (Radboud Institute for Biological and Environmental Sciences, Radboud University) for providing the PBI106 vector. No conflict of interest is declared.
Published: October 28, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Xian Sheng Zhang, Email: zhangxs@sdau.edu.cn.
Ying Hua Su, Email: suyh@sdau.edu.cn.
Supplemental information
References
- Altpeter F., Springer N.M., Bartley L.E., Blechl A.E., Brutnell T.P., Citovsky V., Conrad L.J., Gelvin S.B., Jackson D.P., Kausch A.P., et al. Advancing Crop Transformation in the Era of Genome Editing. Plant Cell. 2016;28:1510–1520. doi: 10.1105/tpc.16.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchabke-Coussa O., Obellianne M., Linderme D., Montes E., Maia-Grondard A., Vilaine F., Pannetier C. Wuschel overexpression promotes somatic embryogenesis and induces organogenesis in cotton (Gossypium hirsutum L.) tissues cultured in vitro. Plant Cell Rep. 2013;32:675–686. doi: 10.1007/s00299-013-1402-9. [DOI] [PubMed] [Google Scholar]
- Boutilier K., Offringa R., Sharma V.K., Kieft H., Ouellet T., Zhang L., Hattori J., Liu C.M., van Lammeren A.A.M., Miki B.L.A., et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z.J., Wang L., Sun W., Zhang Y., Zhou C., Su Y.H., Li W., Sun T.T., Zhao X.Y., Li X.G., et al. Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 2013;161:240–251. doi: 10.1104/pp.112.203166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Wang Y., Zhang R., Zhang H., Gao C. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- Chen Z., Debernardi J.M., Dubcovsky J., Gallavotti A. Recent advances in crop transformation technologies. Nat. Plants. 2022;8:1343–1351. doi: 10.1038/s41477-022-01295-8. [DOI] [PubMed] [Google Scholar]
- Debernardi J.M., Tricoli D.M., Ercoli M.F., Hayta S., Ronald P., Palatnik J.F., Dubcovsky J. A GRF-GIF chimeric protein improves the regeneration efficiency of transgenic plants. Nat. Biotechnol. 2020;38:1274–1279. doi: 10.1038/s41587-020-0703-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehér A. Somatic embryogenesis - Stress-induced remodeling of plant cell fate. Biochim. Biophys. Acta. 2015;1849:385–402. doi: 10.1016/j.bbagrm.2014.07.005. [DOI] [PubMed] [Google Scholar]
- Florez S.L., Erwin R.L., Maximova S.N., Guiltinan M.J., Curtis W.R. Enhanced somatic embryogenesis in Theobroma cacao using the homologous BABY BOOM transcription factor. BMC Plant Biol. 2015;15:121. doi: 10.1186/s12870-015-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame B., Main M., Schick R., Wang K. Genetic transformation using maize immature zygotic embryos. Methods Mol. Biol. 2011;710:327–341. doi: 10.1007/978-1-61737-988-8_22. [DOI] [PubMed] [Google Scholar]
- Gallavotti A., Zhao Q., Kyozuka J., Meeley R.B., Ritter M.K., Doebley J.F., Pè M.E., Schmidt R.J. The role of barren stalk1 in the architecture of maize. Nature. 2004;432:630–635. doi: 10.1038/nature03148. [DOI] [PubMed] [Google Scholar]
- Gao H., Gadlage M.J., Lafitte H.R., Lenderts B., Yang M., Schroder M., Farrell J., Snopek K., Peterson D., Feigenbutz L., et al. Superior field performance of waxy corn engineered using CRISPR-Cas9. Nat. Biotechnol. 2020;38:579–581. doi: 10.1038/s41587-020-0444-0. [DOI] [PubMed] [Google Scholar]
- Gao X., Chen J., Dai X., Zhang D., Zhao Y. An Effective Strategy for Reliably Isolating Heritable and Cas9-Free Arabidopsis Mutants Generated by CRISPR/Cas9-Mediated Genome Editing. Plant Physiol. 2016;171:1794–1800. doi: 10.1104/pp.16.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Q., Wang W., Han X., Wu J., Lyu B., Chen F., Caplan A., Li C., Wu J., Wang W., et al. Isochorismate-based salicylic acid biosynthesis confers basal resistance to Fusarium graminearum in barley. Mol. Plant Pathol. 2018;19:1995–2010. doi: 10.1111/mpp.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G., Zhang Y., Liu P., Jing Y., Zhang L., Zhu Y., Kong X., Zhao H., Zhou Y., Sun J. The transcription factor TaLAX1 interacts with Q to antagonistically regulate grain threshability and spike morphogenesis in bread wheat. New Phytol. 2021;230:988–1002. doi: 10.1111/nph.17235. [DOI] [PubMed] [Google Scholar]
- Hellens R.P., Allan A.C., Friel E.N., Bolitho K., Grafton K., Templeton M.D., Karunairetnam S., Gleave A.P., Laing W.A. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005;1:13. doi: 10.1186/1746-4811-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y., Ishida Y., Komari T. Progress of cereal transformation technology mediated by Agrobacterium tumefaciens. Front. Plant Sci. 2014;5:628. doi: 10.3389/fpls.2014.00628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoerster G., Wang N., Ryan L., Wu E., Anand A., McBride K., Lowe K., Jones T., Gordon-Kamm B. Use of non-integrating Zm-Wus2 vectors to enhance maize transformation. Vitro Cell Dev. Biol. Plant. 2020;56:265–279. [Google Scholar]
- Ikeuchi M., Iwase A., Rymen B., Lambolez A., Kojima M., Takebayashi Y., Heyman J., Watanabe S., Seo M., De Veylder L., et al. Wounding Triggers Callus Formation via Dynamic Hormonal and Transcriptional. Plant Physiol. 2017;175:1158–1174. doi: 10.1104/pp.17.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi M., Ogawa Y., Iwase A., Sugimoto K. Plant regeneration: cellular origins and molecular mechanisms. Development. 2016;143:1442–1451. doi: 10.1242/dev.134668. [DOI] [PubMed] [Google Scholar]
- Ishida Y., Tsunashima M., Hiei Y., Komari T. Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 2015;1223:189–198. doi: 10.1007/978-1-4939-1695-5_15. [DOI] [PubMed] [Google Scholar]
- Iwase A., Harashima H., Ikeuchi M., Rymen B., Ohnuma M., Komaki S., Morohashi K., Kurata T., Nakata M., Ohme-Takagi M., et al. WIND1 Promotes Shoot Regeneration through Transcriptional Activation of ENHANCER OF SHOOT REGENERATION1 in Arabidopsis. Plant Cell. 2017;29:54–69. doi: 10.1105/tpc.16.00623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Yao X., Zhao M., Zhao Q., Du Y., Yu C., Xie F. Comparison of Soybean Transformation Efficiency and Plant Factors Affecting Transformation during the Agrobacterium Infection Process. Int. J. Mol. Sci. 2015;16:18522–18543. doi: 10.3390/ijms160818522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu K., Maekawa M., Ujiie S., Satake Y., Furutani I., Okamoto H., Shimamoto K., Kyozuka J. LAX and SPA: major regulators of shoot branching in rice. Proc. Natl. Acad. Sci. USA. 2003;100:11765–11770. doi: 10.1073/pnas.1932414100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Ye K., Shi Y., Cheng J., Zhang X., Yang S. BZR1 Positively Regulates Freezing Tolerance via CBF-Dependent and CBF-Independent Pathways in Arabidopsis. Mol. Plant. 2017;10:545–559. doi: 10.1016/j.molp.2017.01.004. [DOI] [PubMed] [Google Scholar]
- Li S., Cong Y., Liu Y., Wang T., Shuai Q., Chen N., Gai J., Li Y. Optimization of Agrobacterium-Mediated Transformation in Soybean. Front. Plant Sci. 2017;8:246. doi: 10.3389/fpls.2017.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.J., Yu Y., Liu X., Zhang X.S., Su Y.H. The Arabidopsis MATERNAL EFFECT EMBRYO ARREST45 protein modulates maternal auxin biosynthesis and controls seed size by inducing AINTEGUMENTA. Plant Cell. 2021;33:1907–1926. doi: 10.1093/plcell/koab084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Bie X.M., Lin X., Li M., Wang H., Zhang X., Yang Y., Zhang C., Zhang X.S., Xiao J. Uncovering the transcriptional regulatory network involved in boosting wheat regeneration and transformation. Nat. Plants. 2023;9:908–925. doi: 10.1038/s41477-023-01406-z. [DOI] [PubMed] [Google Scholar]
- Liu Z., Wei F., Feng Y.-Q. Determination of cytokinins in plant samples by polymer monolith microextraction coupled with hydrophilic interaction chromatography-tandem mass spectrometry. Anal. Methods. 2010;2:1676–1685. [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Lowe K., La Rota M., Hoerster G., Hastings C., Wang N., Chamberlin M., Wu E., Jones T., Gordon-Kamm W. Rapid genotype "independent" Zea mays L. (maize) transformation via direct somatic embryogenesis. Vitro Cell Dev. Biol. Plant. 2018;54:240–252. doi: 10.1007/s11627-018-9905-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe K., Wu E., Wang N., Hoerster G., Hastings C., Cho M.J., Scelonge C., Lenderts B., Chamberlin M., Cushatt J., et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell. 2016;28:1998–2015. doi: 10.1105/tpc.16.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H.P., Liu S.M., Xu S.L., Chen W.Y., Zhou X., Tan Y.Y., Huang J.Z., Shu Q.Y. CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol. J. 2017;15:1371–1373. doi: 10.1111/pbi.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McSteen P., Leyser O. Shoot branching. Annu. Rev. Plant Biol. 2005;56:353–374. doi: 10.1146/annurev.arplant.56.032604.144122. [DOI] [PubMed] [Google Scholar]
- Meng W.J., Cheng Z.J., Sang Y.L., Zhang M.M., Rong X.F., Wang Z.W., Tang Y.Y., Zhang X.S. Type-B ARABIDOPSIS RESPONSE REGULATORs Specify the Shoot Stem Cell Niche by Dual Regulation of WUSCHEL. Plant Cell. 2017;29:1357–1372. doi: 10.1105/tpc.16.00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paek K.Y., Hahn E.J. Cytokinins, auxins and activated charcoal affect organogenesis and anatomical characteristics of shoot-tip cultures of Lisianthus [Eustomagrandiflorum (Raf.) Shinn]. In Vitro Cell. Dev. Biol.-Plant. 2000;36:128–132. [Google Scholar]
- Qiu F., Xing S., Xue C., Liu J., Chen K., Chai T., Gao C. Transient expression of a TaGRF4-TaGIF1 complex stimulates wheat regeneration and improves genome editing. Sci. China Life Sci. 2022;65:731–738. doi: 10.1007/s11427-021-1949-9. [DOI] [PubMed] [Google Scholar]
- Ritter M.K., Padilla C.M., Schmidt R.J. The maize mutant barren stalk1 is defective in axillary meristem development. Am. J. Bot. 2002;89:203–210. doi: 10.3732/ajb.89.2.203. [DOI] [PubMed] [Google Scholar]
- Schmitz G., Theres K. Shoot and inflorescence branching. Curr. Opin. Plant Biol. 2005;8:506–511. doi: 10.1016/j.pbi.2005.07.010. [DOI] [PubMed] [Google Scholar]
- Shrawat A.K., Lörz H. Agrobacterium-mediated transformation of cereals: a promising approach crossing barriers. Plant Biotechnol. J. 2006;4:575–603. doi: 10.1111/j.1467-7652.2006.00209.x. [DOI] [PubMed] [Google Scholar]
- Skirpan A., Wu X., McSteen P. Genetic and physical interaction suggest that BARREN STALK 1 is a target of BARREN INFLORESCENCE2 in maize inflorescence development. Plant J. 2008;55:787–797. doi: 10.1111/j.1365-313X.2008.03546.x. [DOI] [PubMed] [Google Scholar]
- Su Y.H., Zhou C., Li Y.J., Yu Y., Tang L.P., Zhang W.J., Yao W.J., Huang R., Laux T., Zhang X.S. Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA. 2020;117:22561–22571. doi: 10.1073/pnas.2015248117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L.P., Zhang X.S., Su Y.H. Regulation of cell reprogramming by auxin during somatic embryogenesis. aBIOTECH. 2020;1:185–193. doi: 10.1007/s42994-020-00029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Liu H., Du L., Ye X. Generation of marker-free transgenic hexaploid wheat via an Agrobacterium-mediated co-transformation strategy in commercial Chinese wheat varieties. Plant Biotechnol. J. 2017;15:614–623. doi: 10.1111/pbi.12660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Shi L., Liang X., Zhao P., Wang W., Liu J., Chang Y., Hiei Y., Yanagihara C., Du L., et al. The gene TaWOX5 overcomes genotype dependency in wheat genetic transformation. Nat. Plants. 2022;8:110–117. doi: 10.1038/s41477-021-01085-8. [DOI] [PubMed] [Google Scholar]
- Wang Y., He S., Long Y., Zhang X., Zhang X., Hu H., Li Z., Hou F., Ge F., Gao S., et al. Genetic variations in ZmSAUR15 contribute to the formation of immature embryo-derived embryonic calluses in maize. Plant J. 2022;109:980–991. doi: 10.1111/tpj.15609. [DOI] [PubMed] [Google Scholar]
- Woods D.P., Hope C.L., Malcomber S.T. Phylogenomic analyses of the BARREN STALK1/LAX PANICLE1 (BA1/LAX1) genes and evidence for their roles during axillary meristem development. Mol. Biol. Evol. 2011;28:2147–2159. doi: 10.1093/molbev/msr036. [DOI] [PubMed] [Google Scholar]
- Xing H.L., Dong L., Wang Z.P., Zhang H.Y., Han C.Y., Liu B., Wang X.C., Chen Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F., Wang Q., Schmitz G., Müller D., Theres K. The bHLH protein ROX acts in concert with RAX1 and LAS to modulate axillary meristem formation in Arabidopsis. Plant J. 2012;71:61–70. doi: 10.1111/j.1365-313X.2012.04970.x. [DOI] [PubMed] [Google Scholar]
- Yao H., Skirpan A., Wardell B., Matthes M.S., Best N.B., McCubbin T., Durbak A., Smith T., Malcomber S., McSteen P. The barren stalk2 Gene Is Required for Axillary Meristem Development in Maize. Mol. Plant. 2019;12:374–389. doi: 10.1016/j.molp.2018.12.024. [DOI] [PubMed] [Google Scholar]
- Zhang G., Zhao F., Chen L., Pan Y., Sun L., Bao N., Zhang T., Cui C.X., Qiu Z., Zhang Y., et al. Jasmonate-mediated wound signalling promotes plant regeneration. Nat. Plants. 2019;5:491–497. doi: 10.1038/s41477-019-0408-x. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Belcram H., Gornicki P., Charles M., Just J., Huneau C., Magdelenat G., Couloux A., Samain S., Gill B.S., et al. Duplication and partitioning in evolution and function of homoeologous Q loci governing domestication characters in polyploid wheat. Proc. Natl. Acad. Sci. USA. 2011;108:18737–18742. doi: 10.1073/pnas.1110552108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Ikeda Y., Chua N.H. Marker-free transformation: increasing transformation frequency by the use of regeneration-promoting genes. Curr. Opin. Biotechnol. 2002;13:173–180. doi: 10.1016/s0958-1669(02)00301-4. [DOI] [PubMed] [Google Scholar]
- Zuo J., Niu Q.W., Frugis G., Chua N.H. The WUSCHEL gene promotes vegetative-to-embryonic transition in Arabidopsis. Plant J. 2002;30:349–359. doi: 10.1046/j.1365-313x.2002.01289.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data discussed in this study have been deposited at the National Genomics Data Center Genome Sequence Archive (GSA; https://ngdc.cncb.ac.cn/gsa/) under accession no. CRA008766. All source data for the statistics shown in the figures are provided in Supplemental Data 1–19. Gene sequence data mentioned in this study can be found at EnsemblPlants (https://plants.ensembl.org/index.html) under the following accession numbers: TaLAX1-A (TraesCS3A02G350600), TaLAX1-B (TraesCS3B02G383000), TaLAX1-D (TraesCS3D02G344600), ZmBA1 (Zm00001d042989), GmLAX1 (GLYMA_18G204700), Q (TraesCS5A01G473800), TaGIF1-A (TraesCS4A02G250600), TaGRF4-A (TraesCS6A02G269600), TaIPT1-A (TraesCS1A02G376300), and TaARF3-D (TraesCS3D02G292100).