Abstract
The 3C-like proteinase (3CLpro) of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) is one of the most promising targets for anti-SARS-CoV drugs due to its crucial role in the viral life cycle. In this study, a database containing structural information of more than 8,000 existing drugs was virtually screened by a docking approach to identify potential binding molecules of SARS-CoV 3CLpro. As a target for screening, both a homology model and the crystallographic structure of the binding pocket of the enzyme were used. Cinanserin (SQ 10,643), a well-characterized serotonin antagonist that has undergone preliminary clinical testing in humans in the 1960s, showed a high score in the screening and was chosen for further experimental evaluation. Binding of both cinanserin and its hydrochloride to bacterially expressed 3CLpro of SARS-CoV and the related human coronavirus 229E (HCoV-229E) was demonstrated by surface plasmon resonance technology. The catalytic activity of both enzymes was inhibited with 50% inhibitory concentration (IC50) values of 5 μM, as tested with a fluorogenic substrate. The antiviral activity of cinanserin was further evaluated in tissue culture assays, namely, a replicon system based on HCoV-229E and quantitative test assays with infectious SARS-CoV and HCoV-229E. All assays revealed a strong inhibition of coronavirus replication at nontoxic drug concentrations. The level of virus RNA and infectious particles was reduced by up to 4 log units, with IC50 values ranging from 19 to 34 μM. These findings demonstrate that the old drug cinanserin is an inhibitor of SARS-CoV replication, acting most likely via inhibition of the 3CL proteinase.
Severe acute respiratory syndrome (SARS) emerged as a communicable human disease in November 2002 and rapidly spread throughout the world. The epidemic lasted until July 2003 and affected 29 countries (28, 37, 49). The cumulative number of probable SARS cases eventually reached 8,096, with 774 deaths (3). A new coronavirus, called SARS-associated coronavirus (SARS-CoV), was identified as the causative agent (8, 25, 36). An effective treatment for SARS is still not available, although recent experiments with animal models point to a beneficial effect of neutralizing antibody preparations (46, 48).
SARS-CoV belongs to the Coronaviridae. This virus family comprises further human pathogens causing less severe respiratory or gastrointestinal disease: human coronavirus 229E (HCoV-229E), human coronavirus OC43, and the recently discovered human coronaviruses NL63 and HKU1 (11, 50, 52).
The SARS-CoV genome was sequenced shortly after identification of the virus and found to contain 11 to 14 major open reading frames (30, 39, 43, 47). They encode, among others, the replicase polyproteins, the spike protein, the small envelope protein, the membrane protein, and the nucleocapsid protein. A 3C-like proteinase (3CLpro) is part of the replicase polyproteins. As deduced from the function of the 3CLpro of other coronaviruses, this enzyme plays a central role in SARS-CoV replication, as it is responsible for the proteolytic release of replicative proteins from their replicase precursor polyproteins (57). Upon host cell infection, the SARS-CoV 3CLpro is predicted to cleave the large replicase polyproteins at 11 cleavage sites, releasing the nonstructural proteins 5 to 16 (nsp5 to nsp16) (14, 30, 39, 43, 47). These proteins include the 3CLpro itself (nsp5), an RNA-binding protein (nsp9) (9, 45), an RNA-dependent RNA polymerase (nsp12), an NTPase/RNA 5′-triphosphatase/helicase (nsp13) (22), a nidoviral uridylate-specific endoribonuclease (nsp15) (21), and two proteins containing a predicted exonuclease activity (nsp14) and an S-adenosylmethionine-dependent ribose 2′-O-methyltransferase activity (nsp16). The replicase gene-encoded nonstructural proteins are predicted to form an active replication/transcription complex that is responsible for replicating the viral genome as well as generating a nested set of subgenomic transcripts encoding the structural proteins and several SARS-CoV-specific proteins that might be associated with pathogenicity. Accordingly, inhibitors that block the cleavage function of 3CLpro can be expected to inhibit virus replication, making this enzyme one of the most attractive targets for anti-SARS-CoV drugs (2, 23, 53, 54, 56).
While experimental screening generally provides clear-cut data on the inhibitory potential of a compound, it is a slow and laborious procedure. Drug libraries must be available, as must biosafety level 3 facilities, if infectious SARS-CoV is used for testing. To expedite the screening process in view of the ongoing SARS epidemic, we have chosen a virtual screening strategy (19, 42) to narrow down the number of potential candidates before experimental testing. Furthermore, the screening process was confined to drugs that have been or currently are in clinical use. The identification of an inhibitor could thus have immediate consequences for treatment of patients. At the beginning of the study, the three-dimensional (3D) structure of SARS-CoV 3CLpro as determined by X-ray crystallography was not yet available. Therefore, we searched the drug database of Molecular Design Limited against a 3D model of the SARS-CoV 3CLpro (54) built on the basis of the 3CLpro X-ray structure of a related coronavirus (2). Several candidate inhibitors of SARS-CoV 3CLpro were identified. The data obtained with the model were confirmed when the real 3D structure of SARS-CoV 3CLpro became available (55). One of the most promising compounds, the well-characterized drug cinanserin [2′-(3-dimethylaminopropylthio) cinnamanilide] (40), was chosen for detailed experimental evaluation. We show here that cinanserin binds to SARS-CoV 3CLpro, inhibits its enzymatic activity, interferes with coronavirus replication in a replicon RNA-based assay system, and most importantly, strongly reduces SARS-CoV replication in cell culture. Furthermore, cinanserin is active against the related HCoV-229E.
MATERIALS AND METHODS
Virtual screening procedure.
The 3D model of SARS-CoV 3CLpro (54) was used as a target for screening the Comprehensive Medicinal Chemistry database of Molecular Design Limited (MDL-CMC) using a docking approach (42). MDL-CMC contains the structural and pharmacological information of more than 8,000 compounds used or evaluated as therapeutic agents in humans. The program DOCK 4.0 (10) was used for primary screening. Residues within a radius of 6 Å around the catalytic center (His41 and Cys145) were used for constructing the grids for the docking screen. The resulting substructure included all residues of the binding pocket. During the docking calculations, Kollman-all-atom charges (6) were assigned to the protein, and Geisterger-Hückel charges (15, 31, 38) were assigned to the small molecules of the MDL-CMC database due to lack of proper Kollman charges. Conformational flexibility of the compounds from the database was considered in the docking search. In DOCK simulation, the ligand-receptor binding energy was approximated by the sum of the van der Waals and electrostatic interaction energies. After an initial evaluation of orientation and scoring, a grid-based rigid body minimization was carried out for the ligand to locate the nearest local energy minimum within the receptor binding site. The position and conformation of each docked molecule were optimized using the single-anchor search and torsion minimization method of DOCK 4.0. The 100 molecules with the highest score as obtained by DOCK search were rescored by CScore (5) and the scoring function of AutoDock 3.0 (34). The virtual screening was performed on a 64-processor SGI Origin 3800 supercomputer.
Synthesis of cinanserin.
For initial experiments, 2′-(3-dimethylaminopropylthio) cinnamanilide (cinanserin, SQ 10,643) was kindly provided by G. Jin (Shanghai Institute of Materia Medica, Chinese Academy of Sciences). Subsequently, cinanserin and its hydrochloride were synthesized in large scale (see Appendix SI in the supplemental material) for the performance of further bioassays. For selected experiments, cinanserin hydrochloride was purchased from MP Biomedicals (catalog number 159758).
Expression of recombinant SARS-CoV 3CLpro, HCoV-229E 3CLpro, and HRV-14 3Cpro.
Expression and purification of SARS-CoV 3CLpro was performed as described previously (44). The DNA fragment coding for HCoV-229E 3CLpro was derived from cDNA clone pFM1. The coding region was amplified by PCR (primers ATATGGATCCGCTGGTTTGCGCAAAA and ATATGTCGACTCATTGCAGGTTAACACC, BamHI and SalI sites are underlined). The PCR product was digested with BamHI and SalI and cloned into glutathione S-transferase expression vector pGEX 4T-1 (Amersham), resulting in plasmid pGEX 4T-1-HCoV-229E 3CLpro. The DNA fragment coding for human rhinovirus 14 (HRV-14) 3Cpro was synthesized with an Applied Biosystems DNA synthesizer (Shanghai Sangon Biological Engineering and Technology and Service Co. Ltd.) and cloned via BamHI and SalI sites into pGEX 4T-1 to construct plasmid pGEX 4T-1-HRV-14 3Cpro. The correct sequence of the inserts was confirmed by sequencing. Escherichia coli BL21(DE3) cells (Promega) were transformed with the expression plasmids and grown at 37°C in LB medium containing 100 μg/ml ampicillin until the optical density at 600 nm reached 0.2 to 0.8. Protein was expressed for 5 h at 22°C after induction with 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation, resuspended in 25 ml of precooled 1× phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 [pH 7.3]), and disrupted by sonication. The lysate was loaded onto a glutathione-Sepharose 4B column (Amersham-Pharmacia) equilibrated with 1× PBS at 4°C. The column was washed with 1× PBS. The glutathione S-transferase fusion proteins were digested on the column with 50 U thrombin (Amersham-Pharmacia) for 16 h. To reduce protein degradation, all procedures were performed at 4°C in the presence of 0.5 mM dithiothreitol.
Binding assays.
The binding affinity of cinanserin to SARS-CoV 3CLpro, HCoV-229E 3CLpro, and HRV-14 3Cpro in vitro was determined using the surface plasmon resonance (SPR) biosensor technology. The measurement was performed using the dual flow cell Biacore 3000 instrument (Biacore AB, Uppsala, Sweden). Immobilization of the proteins to the hydrophilic carboxymethylated dextran matrix of the sensor chip CM5 (Biacore) was carried out by the standard primary amine coupling reaction. SARS-CoV 3CLpro and HCoV-229E 3CLpro to be covalently bound to the matrix were diluted in 10 mM sodium acetate buffer (pH 4.3) to final concentrations of 25 μg/ml and 10 μg/ml. HRV-14 3Cpro was diluted in 10 mM sodium acetate buffer (pH 5.02) to a final concentration of 12 μg/ml. Equilibration of the baseline was completed by a continuous flow of HBS-EP running buffer (10 mM HEPES, 150 mM NaCl, 3.4 mM EDTA, and 0.005% [vol/vol] surfactant P20, pH 7.4) through the chip for 1 to 2 h. Biacore data were collected at 25°C with HBS-EP as the running buffer at a constant flow of 20 μl/min. Sensorgrams were processed by using automatic correction for nonspecific bulk refractive index effects. The equilibrium dissociation constants (KD) evaluating the protein-ligand binding affinity were determined by the steady-state affinity fitting analysis of the Biacore data.
Enzymatic activity assays.
The proteolytic activities of SARS-CoV 3CLpro and HCoV-229E 3CLpro were measured by a fluorescence resonance energy transfer (FRET)-based assay using a substrate labeled with 5-[(2′-aminoethyl)-amino] naphthelenesulfonic acid (EDANS) and 4-[[4-(dimethylamino) phenyl] azo] benzoic acid (Dabcyl) as the energy transfer pairs. The fluorogenic substrate EDANS-Val-Asn-Ser-Thr-Leu-Gln-Ser-Gly-Leu-Arg-Lys-(Dabcyl)-Met was prepared as described previously (26, 29). The enzyme activity was demonstrated by monitoring the increase of the emission fluorescence at a wavelength of 490 nm upon excitation at 340 nm (slit width, 10 nm) on a HITACHI F-2500 fluorescence spectrophotometer connected with a thermostat. Inhibition kinetics was determined at a constant substrate concentration with different concentrations of cinanserin. The enzymes were preincubated for 2 h at 4°C with 0 to 0.2 mM cinanserin. The final reaction mix contained 1 μM purified enzyme, 0 to 0.1 mM cinanserin, 100 mM NaCl, 20 mM phosphate buffer (pH 7.4), and 10 μM substrate. Reactions were run at 25°C with continuous monitoring of fluorescence for 60 min. The proteolytic activity of HRV-14 3Cpro and the effect of cinanserin was measured as described previously (7, 51). The remaining enzymatic activity was calculated, and the 50% inhibitory concentration (IC50) of cinanserin was determined according to the following logistic derivative equation:
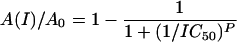 |
where A0 is the enzyme activity without inhibitor, A(I) is the enzyme activity with various levels of inhibitor, I is the inhibitor concentration, and P is the μ factor.
HCoV-229E replicase inhibition assay.
The inhibition of coronavirus replicase function was performed using BHK-Rep-1 cells as described previously (17). Briefly, BHK-Rep-1 cells contain an autonomously replicating, HCoV-229E-based replicon RNA. Green fluorescent protein (GFP) expression in BHK-Rep-1 cells is replicon mediated and serves as a marker for coronavirus replication. Different concentrations of compounds were added on monolayers of BHK-Rep-1 cells in 96-well plates, and an untreated well served as control. Three days later, the cells were analyzed by fluorescence microscopy (Leica DM R fluorescence microscope and Leica IM 1000 software) and flow cytometry (FACSCalibur and CellQuest software; BD Pharmingen). Four duplicate tests were performed. Inhibition of reporter gene expression was calculated as the reduction of GFP-expressing cells by setting the number of GFP-expressing untreated cells at 100%. In parallel, the cytotoxicity of compounds was assayed on parental BHK-21 cells using the CellTitre 96 Aqueous One kit (Promega).
SARS-CoV and HCoV-229E infection assays.
The SARS-CoV inhibition assay was performed as described previously (16). In brief, Vero cells in 24-well plates were infected in the biosafety level 4 laboratory with SARS-CoV (Frankfurt isolate) at a multiplicity of infection (MOI) of 0.01. The inoculum was removed after 1 h and replaced with fresh medium complemented with different concentrations of compound. The virus RNA concentration in the supernatant was measured by real-time PCR after 2 days. RNA was prepared from 140 μl supernatant using diatomaceous silica (4). Quantitative real-time reverse transcription-PCR (RT-PCR) was performed with the purified RNA according to a published protocol (8). In vitro transcripts of the target region were used in the PCR to generate standard curves for quantification of the virus RNA.
Infectious SARS-CoV particles in the supernatant were quantified by immunofocus assay. Vero cells in 24-well plates were inoculated with serial 10-fold dilutions of supernatant. The inoculum was removed after 1 h and replaced with a 1% methylcellulose medium overlay. After 5 days of incubation, cells were fixed with 4% formaldehyde, permeabilized with 0.5% Triton X-100, blocked with 10% fetal calf serum, and washed. Infected cell foci were detected with a reconvalescent-phase serum sample from a patient with SARS. After washing and incubation with peroxidase-labeled anti-human immunoglobulin G antibody (Dianova), foci were stained with 3,3′,5,5′-tetramethylbenzidine and counted.
Cell growth inhibition or cytotoxicity was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazoliumbromide (MTT) method. MTT solution (50 μl, 5 mg/ml PBS) was added to the cells from which an aliquot of supernatant was taken for measuring the virus RNA or infectious particle concentration. The plate was incubated for 90 min at 37°C. The supernatant was completely removed, and the cells were fixed with 4% formaldehyde for 30 min. Tetrazolium crystals were dissolved in 1 ml ethanol for 10 min, and the extinction was measured at 560 nm.
HCoV-229E infection experiments were done with MRC-5 cells at 33°C. Cells in 24-well plates were infected with an MOI of 0.1. Subsequently, cinanserin hydrochloride was added. After 2 days, the 50% tissue culture infectious dose units in the supernatant were determined. The virus RNA concentration in the supernatant was quantified by dual-probe real-time RT-PCR (C. Drosten, unpublished) based on the Access one-step RT-PCR kit (Promega, Karlsruhe, Germany) with coronavirus-specific primers (TACTATGACTGGCAGAATGTTTCA [200 nM], TACTATGACTACTAGACAGTTTCA [200 nM], GGCATAGCACTATCACACTTTGG [400 nM]) and probes (ATGCCACCGTTGTTATCGGCACTACCAA-fluorescein [40 nM] and LCred640-TTTATGGCGGGTGGGATAATATGTT-phosphate [40 nM]). Thermal cycling in a LightCycler (Roche) consisted of 42°C for 30 min, 94°C for 2 min and 45 cycles of 94°C for 5 s, 50°C for 10 s, and 72°C for 20 s. The input RNA was quantified using in vitro-transcribed HCoV-229E RNA as a standard. The cytotoxicity of compounds was assayed on MRC-5 cells with the CellTitre 96 Aqueous One kit.
The concentrations required to inhibit virus replication by 50% (IC50) or 90% (IC90) were calculated by fitting a dose-response curve to the data following logarithmic transformation of the drug concentration using Statgraphics plus 5.0 software (Statistical Graphics, Inc.).
RESULTS
Identification of cinanserin by virtual screening.
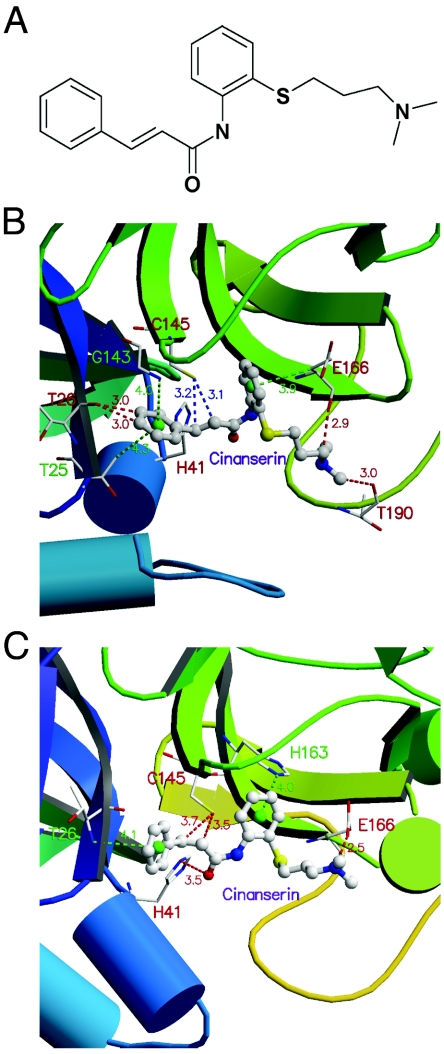
The substrate-binding pocket formed by residues within a radius of 6 Å around the catalytic center (His41 and Cys145) of the model structure of SARS-CoV 3CLpro (54) was used as the target site for virtual screening. The MDL-CMC database was searched for potential binding molecules using the program DOCK for primary screening, and the top 100 molecules were rescored using CScore (5), a consensus scoring method that integrates five popular scoring functions. The 10 compounds that showed the highest scores and were positively evaluated by at least four of the five scoring functions of CScore are shown with their clinical applications in Table 1 (see Fig. S1 in the supplemental material for their structures). The binding affinities (Ki values) of these compounds to SARS-CoV 3CLpro were predicted by AutoDock (34) (Table 1). When the crystallographic data of SARS-CoV 3CLpro became available (55), the docking simulation was repeated with these data for the top 10 compounds. The root-mean-square deviation (RMSD) between the model and crystal structure for the residues composing the substrate-binding pocket was only 0.18 Å (see Fig. S2 in the supplemental material). Accordingly, the calculations with the real structure confirmed the data obtained with the model (Table 1). In this study, we focus on 1 of the 10 compounds, namely cinanserin, as it was immediately available for experimentation and showed promising results in preliminary tests. The interaction models of cinanserin with the model structure (54) and the X-ray crystal structure of SARS-CoV 3CLpro (55) are shown in Fig. 1. Docking simulations with the crystal structures of the 3CL proteinases of HCoV-229E and porcine transmissible gastroenteritis coronavirus (2) predicted that cinanserin also fits into the active sites of these enzymes (see Fig. S3 and Table S1 in the supplemental material).
TABLE 1.
Potential binding molecules of SARS-CoV 3CLpro identified by virtual screening of the MDL-CMC database (top 10 compounds)
| Compound no. | MDL no. (MCMC 0000) | Name | SARS-CoV 3CLpro model
|
SARS-CoV 3CLpro crystal structure
|
Clinical application | ||
|---|---|---|---|---|---|---|---|
| DOCKa | AutoDockb | DOCKa | AutoDockb | ||||
| 1 | 0101 | Benzpiperylon | −30.99 | 0.08 | −29.78 | 0.02 | Connective tissue disorders |
| 2 | 1593 | Cinanserin | −37.06 | 0.36 | −32.20 | 0.39 | 5-HT receptor antagonist |
| 3 | 3048 | Fentiazac | −35.09 | 0.50 | −36.44 | 0.39 | Antiinflammatory |
| 4 | 3248 | Feclobuzone | −35.52 | 0.39 | −24.50 | 0.35 | Antiinflammatory |
| 5 | 3266 | Ridiflone | −32.52 | 0.10 | −25.51 | 0.0076 | Anxiolytic |
| 6 | 3406 | Halofenate | −34.60 | 2.82 | −29.84 | 0.38 | Hypolipidemic |
| 7 | 3612 | Trifezolac | −31.98 | 0.16 | −39.52 | 0.12 | Analgesic |
| 8 | 4719 | Butoconazole | −33.72 | 0.43 | −29.97 | 0.36 | Antifungal |
| 9 | 4945 | Pirazolac | −37.45 | 0.12 | −34.79 | 0.67 | Antirheumatic |
| 10 | 5501 | Flavoneacetic acid | −36.06 | 6.46 | −34.90 | 2.29 | Antineoplastic |
Interaction energy in kcal/mol.
Binding (dissociation) constant in μM.
FIG. 1.
Chemical structure of cinanserin and model of interaction of cinanserin with SARS-CoV 3CLpro. (A) Chemical structure of cinanserin. Predicted interaction of cinanserin with the homology model (B) and with the X-ray crystal structure (C). The green lines represent -NH · · · π or -CH · · · π hydrogen bonds, the red lines denote -CH · · · O hydrogen bonds, and the blue lines show the SH group of C145 of SARS-CoV 3CLpro pointing towards the C=C bond of cinanserin.
Binding affinity of cinanserin to SARS-CoV 3CLpro, HCoV-229E 3CLpro, and HRV-14 3Cpro.
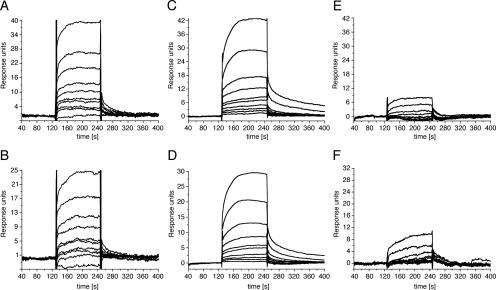
The binding of cinanserin to the bacterially expressed proteinases in vitro was determined using SPR biosensor technology. For kinetic analysis on the Biacore 3000 instrument, various concentrations of cinanserin were injected for 120 min at a flow rate of 20 μl/min to allow for interaction with the enzymes immobilized on the surface. The interaction kinetics with cinanserin and its hydrochloride are shown in Fig. 2. A significant and dose-dependent increase in the SPR response was seen if the sensor chip was coated with SARS-CoV 3CLpro and HCoV-229E 3CLpro (Fig. 2A to D). Cinanserin and its hydrochloride presented characteristic square-wave binding curves, indicating a rapidly formed but unstable complex. The concentration series were fitted to a steady-state affinity model and a 1:1 Langmuir binding model for KD determination by Biacore 3000 evaluation software, resulting in KD values of 49.4 μM/78.0 μM (cinanserin/cinanserin hydrochloride) for SARS-CoV 3CLpro and 18.2 μM/36.6 μM for HCoV-229E 3CLpro. In contrast, neither cinanserin nor cinanserin hydrochloride showed binding affinity to HRV-14 3Cpro (Fig. 2E and F).
FIG. 2.
Binding of cinanserin and cinanserin hydrochloride to SARS-CoV 3CLpro, HCoV-229E 3CLpro, and HRV-14 3Cpro as determined by SPR. Representative sensorgrams obtained with cinanserin and its hydrochloride at concentrations of 100, 70, 49, 34.3, 24.01, 16.8, 11.8, 8.24, 5.76, and 4.04 μM (curves from top to bottom) are shown. The compounds were injected for 120 s, and dissociation was monitored for more than 150 s. (A) Interaction of cinanserin with SARS-CoV 3CLpro. (B) Interaction of cinanserin hydrochloride with SARS-CoV 3CLpro. (C) Interaction of cinanserin with HCoV-229E 3CLpro. (D) Interaction of cinanserin hydrochloride with HCoV-229E 3CLpro. (E) Interaction of cinanserin with HRV-14 3Cpro. (F) Interaction of cinanserin hydrochloride with HRV-14 3Cpro.
Inhibitory activity of cinanserin on SARS-CoV 3CLpro, HCoV-229E 3CLpro, and HRV-14 3Cpro.
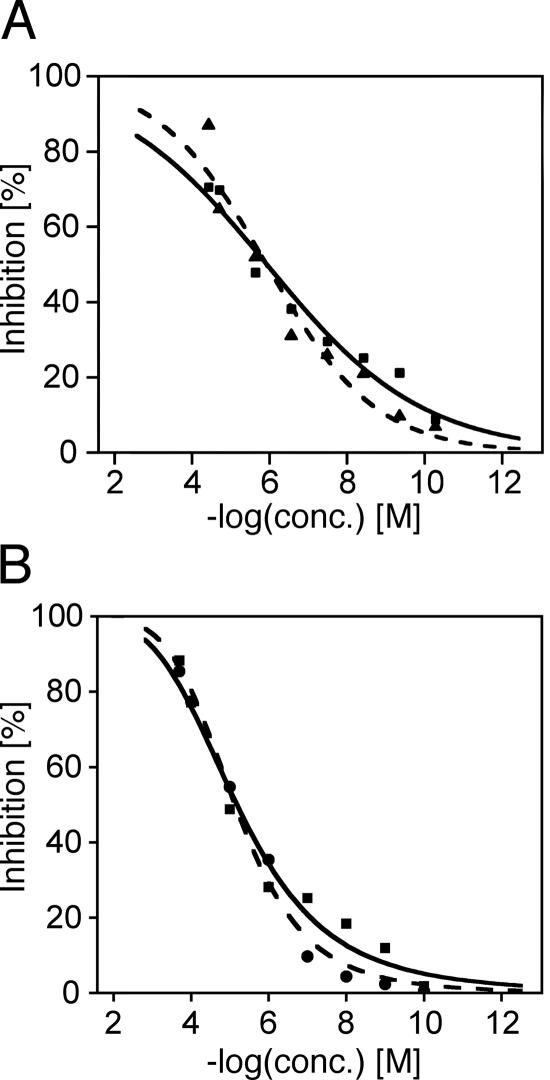
The inhibitory activity of cinanserin on the proteolytic activity of the three enzymes was measured by FRET using a peptide substrate labeled with a pair of fluorogenic dyes. The inhibition of SARS-CoV 3CLpro and HCoV-229E 3CLpro slowly increased in the concentration range from 10−10 to 10−6 M cinanserin and cinanserin hydrochloride (Fig. 3). The curves finally reached a maximum of 70 to 90% inhibition at 50 to 100 μM for both compounds. For IC50 determination, the inhibition data were fitted to a dose-response curve using a logistic derivative equation. The IC50 values of cinanserin and cinanserin hydrochloride for inhibiting the catalytic activity of SARS-CoV 3CLpro were calculated as 4.92 μM and 5.05 μM, respectively, The corresponding IC50 values for HCoV-229E 3CLpro were 4.68 μM and 5.68 μM. None of the compounds had inhibitory activity against HRV-14 3Cpro at concentrations up to 200 μM (data not shown).
FIG. 3.
Inhibitory activity of cinanserin and its hydrochloride on the proteolytic activity of SARS-CoV 3CLpro (A) and HCoV-229E 3CLpro (B). Inhibition of cleavage was measured by FRET using a peptide substrate labeled with a pair of fluorogenic dyes. Cinanserin, solid line (data points are shown as squares); cinanserin hydrochloride, dashed line (data points are shown as triangles or circles); conc., concentration.
Inhibition of coronavirus replicase.
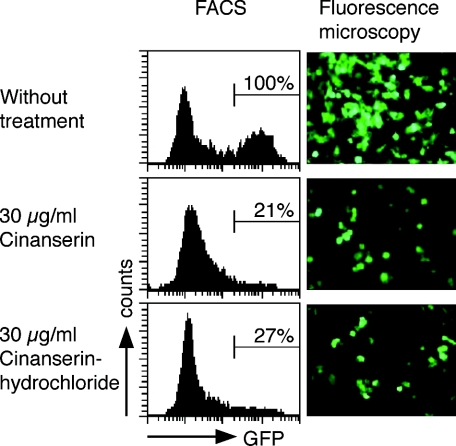
After having shown that cinanserin displays inhibitory activity against 3CLpro, we reasoned that this should result in inhibition of coronavirus replicase function. To test this, we made use of a recently established assay system based on a stable cell line, BHK-Rep-1, that contains an autonomously replicating HCoV-229E replicon RNA (17). The BHK-Rep-1 cell line displays replicon-mediated expression of GFP as a marker for coronavirus replication. Replicon-containing BHK-Rep-1 cells were treated with different concentrations of cinanserin as well as cinanserin hydrochloride. After 3 days, the cells were analyzed for GFP expression by fluorescence microscopy and fluorescence-activated cell sorter (FACS) analysis. In parallel, the cytotoxicity was assessed using parental BHK-21 cells. As shown in Fig. 4, the inhibitory effect of cinanserin and cinanserin hydrochloride was readily visible by fluorescence microscopy, and GFP-expressing cells were reduced to 21% and 27% at concentrations of 30 μg/ml (78 μM) cinanserin or cinanserin hydrochloride, respectively, as determined by FACS analysis. No cytotoxicity was observed at these concentrations.
FIG. 4.
Inhibition of coronavirus replicative function by cinanserin and cinanserin hydrochloride. BHK-Rep-1 cells containing HCoV-229E-based replicon RNA that mediates GFP expression were used to assess the inhibitory effect of the compounds by FACS analysis and fluorescence microscopy. One representative experiment out of four is shown. Bars indicate GFP-expressing BHK-Rep-1 cells in FACS analyses. The GFP expression of untreated cells was set at 100%.
Inhibition of SARS-CoV and HCoV-229E replication in cell culture.
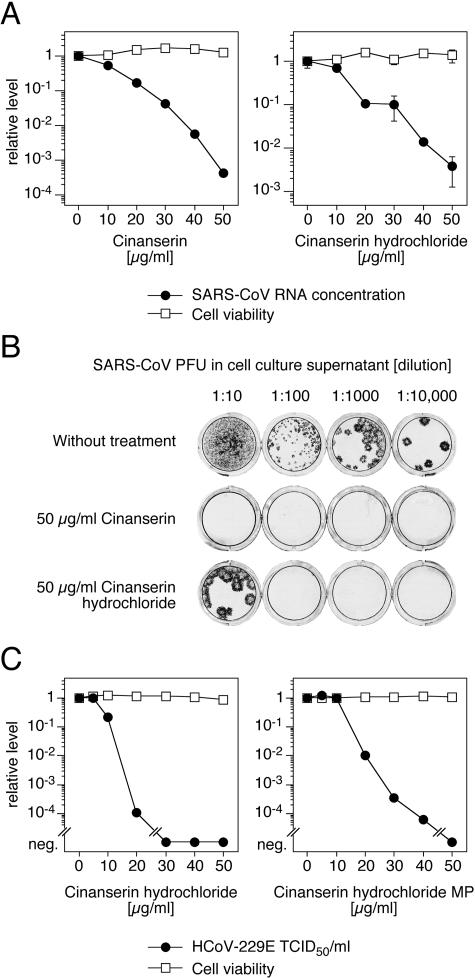
Since cinanserin inhibited the 3CLpro of SARS-CoV and HCoV-229E as well as HCoV-229E replicon activity, we wondered whether the compound also inhibits virus replication in cell culture. Vero cells were infected with the SARS-CoV isolate Frankfurt. As described previously, growth of the virus in our Vero cells is not associated with a cytopathic effect (16). Cells were treated with different concentrations of cinanserin as well as cinanserin hydrochloride, and the virus RNA concentration in the supernatant was measured by real-time PCR 2 days postinfection. In addition, the titer of infectious particles in the supernatant was measured by immunofocus assay in selected experiments. Treatment of the cells with 50 μg/ml (134 μM) cinanserin or cinanserin hydrochloride reduced the SARS-CoV RNA concentration by more than 3 log units and 2 log units, respectively (Fig. 5A). The reduction of the titer of infectious particles exactly corresponded to the reduction in virus RNA concentration (Fig. 5B). There was no evidence for toxicity of the compounds in the concentration range tested as measured with the MTT test (Fig. 5A). The IC50 and IC90 values were 11 μg/ml (31 μM) and 23 μg/ml (66 μM) for cinanserin and 13 μg/ml (34 μM) and 25 μg/ml (67 μM) for cinanserin hydrochloride.
FIG. 5.
Inhibition of SARS-CoV and HCoV-229E replication by cinanserin and cinanserin hydrochloride. (A) Reduction of SARS-CoV RNA concentration in cell culture supernatant. Vero cells were infected with SARS-CoV at an MOI of 0.01, and virus RNA concentration was measured by real-time PCR after 2 days. The influence of the compounds on cell viability was measured by the MTT test. The virus RNA concentration of untreated cells (4 × 107 RNA copies/ml) and the corresponding cell viability value were defined as 1. Means and ranges of duplicate tests are shown. (B) Reduction of SARS-CoV infectious particles in supernatant. Supernatant of infected cells treated with 50 μg/ml compound and of cells that were left untreated were harvested 2 days postinfection, and the numbers of PFU were determined by immunofocus assay. Cell culture wells inoculated with dilutions of the supernatant are shown. (C) Reduction of HCoV-229E titer in cell culture supernatant by cinanserin hydrochloride synthesized in this study (left) or purchased from MP Biomedicals (right). MRC-5 cells were infected with HCoV-229E at an MOI of 0.1, and titers of infectious particles were determined after 2 days. The influence of the compounds on cell viability was measured with CellTitre 96 Aqueous One kit. The virus titer of untreated cells (3 × 105 50% tissue culture infectious dose units/ml) and the corresponding cell viability value were defined as 1. Means of triplicate tests are shown. neg., negative.
The effect of cinanserin on HCoV-229E replication was assayed in MRC-5 cells. Cinanserin hydrochloride reduced the virus titer in the supernatant by more than 4 log units, with an IC50 value of 9.3 μg/ml (25 μM) (Fig. 5C left). The measurement of virus RNA concentration by real-time PCR confirmed the inhibitory activity of the compound (IC50 value of 7.2 μg/ml [19 μM]). The experiment was repeated with commercially available cinanserin hydrochloride (MP Biomedicals). The same effect was observed (Fig. 5C, right). There was no measurable toxicity of both compounds on MRC-5 cells in the test range.
DISCUSSION
We describe here that cinanserin, a well-characterized serotonin receptor antagonist, is an inhibitor of SARS-CoV and HCoV-229E. After the initial identification of cinanserin as a potential binding molecule of the SARS-CoV 3CLpro catalytic pocket by virtual screening, we systematically assessed its efficacy in biochemical and tissue culture-based assays. We demonstrate that cinanserin (i) binds and inhibits recombinant 3CLpro of SARS-CoV and HCoV-229E, (ii) inhibits coronavirus replicase function using a replicon assay based on HCoV-229E, and (iii) inhibits SARS-CoV and HCoV-229E replication in tissue culture.
The SARS-CoV 3CLpro was chosen as the target enzyme for several reasons. First, crystal structures for a number of coronavirus 3CLpro have been determined (2, 55), a prerequisite for the approach taken in this study. Second, it is by far the best-studied enzyme of coronaviruses. 3CLpro of several coronaviruses, including SARS-CoV 3CLpro, can be expressed in bacterial systems, and functional assays to assess its proteolytic activity in the presence of candidate inhibitors are available (26, 44). Finally, its functional importance in the coronavirus life cycle makes it an ideal target for antiviral intervention (57). It is encoded by the coronavirus replicase gene and is responsible for the proteolytic processing of replicase precursor polyproteins resulting in the formation of an active replication complex. Therefore, it is reasonable to consider the coronavirus 3CLpro indispensable for virus replication.
Cinanserin was discovered as a candidate inhibitor of SARS-CoV 3CLpro by virtual screening using a 3D model of SARS-CoV 3CLpro (54) before its X-ray crystal structure (55) was determined. The accuracy of our 3D model was confirmed by comparison with the crystal structure of SARS-CoV 3CLpro: the RMSD of all heavy atoms between these two structures was 3.11 Å (see Fig. S2 in the supplemental material). The major contribution to this value results from domain III, which is less conserved among the coronaviruses and far away from the active site. The RMSD of the binding pocket was only 0.18 Å. Accordingly, the top 10 candidates also fit into the X-ray structure (Table 1). In particular, cinanserin is predicted to interact with the model and the crystal structure in a similar way (Fig. 1). Considering the experimental data for cinanserin, our study suggests that virtual screening is a useful approach to extract potential inhibitors from available drug databases. However, since experimental confirmation was provided only for cinanserin, it is not clear how reliably virtual screening predicts the inhibitory potential of a compound.
The experimental tests demonstrated that cinanserin binds to recombinant SARS-CoV 3CLpro and inhibits its proteolytic activity, as predicted. Comparable effects were observed with HCoV-229E 3CLpro. In contrast, cinanserin neither bound to nor inhibited the activity of HRV-14 3Cpro. This suggests that the inhibitory activity of cinanserin is specific for the coronavirus 3CLpro. A particular requirement for antivirals, such as protease inhibitors, is the ability to penetrate the cell membrane to access their targets. Furthermore, the inhibitory activity should be specific for the viral target and not affect cellular host functions, at least at the inhibitory concentration. We have addressed these issues in tissue culture-based assays. First, a cell line (BHK-Rep-1) containing an autonomously replicating RNA (replicon RNA) derived from HCoV-229E was used (17). This cell line has been particularly established for the assessment of inhibitors targeting coronavirus replicative functions. GFP expression in BHK-Rep-1 cells is dependent on an active coronavirus replication complex. More than 70% of cinanserin and cinanserin hydrochloride-treated cells lost GFP expression, further supporting the idea that inhibition of 3CLpro results in loss of coronavirus replicase function. Second, it was demonstrated that cinanserin greatly reduces SARS-CoV and HCoV-229E replication in cell culture. The virus level in the supernatant was reduced by 3 to 4 log units, while there was no evidence of cytotoxicity at the inhibitory concentrations. Cinanserin was more active than its hydrochloride. It is less hydrophilic than the latter and may therefore penetrate the cell membrane more efficiently. The magnitude of inhibition observed with cinanserin by far exceeds the reductions of SARS-CoV RNA that were found with the same assay with nucleoside analogues (16). Similarly, cinanserin inhibited HCoV-229E replicon activity much more strongly than alpha interferon (17). Taken together, cinanserin is able to access its target within the cell and specifically inhibits coronavirus replicase function. The data obtained with recombinant 3CLpro suggest that the inhibitory effect seen in the cellular systems is at least partially attributable to the inhibition of the coronavirus proteinase. However, since binding of the drug to recombinant 3CLpro was not very strong and the enzyme was not completely inhibited at the maximum drug concentration, it is conceivable that additional drug effects contribute to the strong virus reduction in cell culture. Further experiments such as selection and characterization of drug-resistant virus are under way to prove these hypotheses.
In contrast to purely experimental compounds, cinanserin (SQ 10,643) is a drug that has already undergone clinical evaluation. It had been synthesized and characterized as a serotonin inhibitor by The Squibb Institute for Medical Research, New Brunswick, New Jersey, in the 1960s (40). Antiserotonin effects in peripheral organs, effects on the central nervous system (12, 40), and immunosuppressive (1, 27, 35) and antiphlogistic (41) activity were demonstrated in laboratory animals. In dogs, the main side effect was hepatotoxicity at repeated oral doses of >40 mg/kg of body weight per day (35), and the lethal dose was 100 mg/kg upon intravenous infusion of the drug at a rate of 0.5 mg/kg per minute (40). Small-scale clinical trials have been performed with patients with psychiatric disorders such as schizophrenia and mania (13, 18, 20, 24) and in patients with carcinoid syndrome (33). Beneficial effects were observed in patients with mania and carcinoid syndrome. Patients were treated for several weeks with oral doses of 600 to 800 mg/day (∼10 mg/kg), while during short-term treatment, maximum daily doses of 1,200 mg (∼20 mg/kg) were reached (13, 18, 20). Although no remarkable side effects were observed with humans, further clinical testing was suspended, since after prolonged treatment of rats at a high dosage level (120 mg/kg daily for 59 to 81 weeks), malignant hepatoma was found (see notice in reference 20). Nevertheless, in view of the anti-SARS-CoV activity of the drug demonstrated here, it might be worthwhile to reevaluate the existing pharmacological data and to test the drug with suitable animal models of coronavirus infection, particularly SARS (32, 46). On the other hand, cinanserin may serve as a lead substance for the design of more active inhibitors of 3CL proteinase with reduced toxicity and antiserotonin activity.
Supporting information.
For additional information, see Table S1, Fig. S1 to S3, and Appendix SI in the supplemental material.
Supplementary Material
Acknowledgments
We thank Beate Becker-Ziaja for excellent technical assistance.
This work was supported by the State Key Program of Basic Research of China (grants 2002CB512802, 2002CB512807, 2003CB514125, 2003CB514124, 2003CB514125, and 2002CB512807), the National Natural Science Foundation of China (grants 20372069, 29725203, and 20072042), the 863 Hi-Tech Program (grants 2001AA235051, 2001AA235071, and 2002AA233011), Sino-European Project on SARS Diagnostics and Antivirals (proposal/contract no. 003831), Shanghai Basic Research Project from the Shanghai Science and Technology Commission (grants 02DJ14070, 03DZ19212, and 03DZ19228), and the special programs of oppugning SARS from the Ministry of Science and Technology, Chinese Academy of Sciences, National Natural Science Foundation of China, and Shanghai Science and Technology Commission. This work was further supported by grant E/B41G/1G309/1A403 from the Bundesamt für Wehrtechnik und Beschaffung (to S.G.). The Bernhard Nocht Institute is supported by the Bundesministerium für Gesundheit and the Freie und Hansestadt Hamburg. The work done at the Kantonal Hospital in St. Gallen, Switzerland, was supported by the Gebert Rüf Foundation and the Suisse National Science Foundation.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Amundsen, B., and W. H. Lakey. 1970. The effect of 2′-(3-dimethylaminopropylthio) cinnamanilide on dog renal allograft survival. Surg. Forum 21:254-256. [PubMed] [Google Scholar]
- 2.Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous. 2004. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003. World Health Organization, Geneva, Switzerland. [Online.] http://www.who.int/csr/sars/country/table2004_04_21/en/.
- 4.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark, R. D., A. Strizhev, J. M. Leonard, J. F. Blake, and J. B. Matthew. 2002. Consensus scoring for ligand/protein interactions. J. Mol. Graph. Model. 20:281-295. [DOI] [PubMed] [Google Scholar]
- 6.Cornell, W. D., P. Cieplak, C. I. Bayly, I. R. Gould, K. M. Merz, D. M. Ferguson, D. C. Spellmeyer, T. Fox, J. W. Caldwell, and P. A. Kollman. 1995. A second generation force field for the simulation of proteins, nucleic acids and organic molecules. J. Am. Chem. Soc. 117:5179-5197. [Google Scholar]
- 7.Davis, G. J., Q. M. Wang, G. A. Cox, R. B. Johnson, M. Wakulchik, C. A. Dotson, and E. C. Villarreal. 1997. Expression and purification of recombinant rhinovirus 14 3CD proteinase and its comparison to the 3C proteinase. Arch. Biochem. Biophys. 346:125-130. [DOI] [PubMed] [Google Scholar]
- 8.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 9.Egloff, M. P., F. Ferron, V. Campanacci, S. Longhi, C. Rancurel, H. Dutartre, E. J. Snijder, A. E. Gorbalenya, C. Cambillau, and B. Canard. 2004. The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world. Proc. Natl. Acad. Sci. USA 101:3792-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ewing, T. J. A., and I. D. Kuntz. 1997. Critical evaluation of search algorithms for automated molecular docking and database screening. J. Comput. Chem. 18:1175-1189. [Google Scholar]
- 11.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. de Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furgiuele, A. R., J. P. High, and Z. P. Horovitz. 1965. Some central effects of SQ 10,643 [2′3-(-dimethlaminopropylthio) cinnamanilide hydrochloride], a potent serotonin antagonist. Arch. Int. Pharmacodyn. Ther. 155:225-235. [PubMed] [Google Scholar]
- 13.Gallant, D. M., and M. P. Bishop. 1968. Cinanserin (SQ. 10,643): a preliminary evaluation in chronic schizophrenic patients. Curr. Ther. Res. Clin. Exp. 10:461-463. [PubMed] [Google Scholar]
- 14.Gao, F., H. Y. Ou, L. L. Chen, W. X. Zheng, and C. T. Zhang. 2003. Prediction of proteinase cleavage sites in polyproteins of coronaviruses and its applications in analyzing SARS-CoV genomes. FEBS Lett. 553:451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gasteiger, J., and M. Marsili. 1980. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron 36:3219-3228. [Google Scholar]
- 16.Günther, S., M. Asper, C. Röser, L. K. Luna, C. Drosten, B. Becker-Ziaja, P. Borowski, H.-M. Chen, and R. S. Hosmane. 2004. Application of real-time PCR for testing antiviral compounds against Lassa virus, SARS coronavirus and Ebola virus in vitro. Antiviral Res. 63:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hertzig, T., E. Scandella, B. Schelle, J. Ziebuhr, S. G. Siddell, B. Ludewig, and V. Thiel. 2004. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J. Gen. Virol. 85:1717-1725. [DOI] [PubMed] [Google Scholar]
- 18.Holden, J. M., A. Keskiner, and P. Gannon. 1971. A clinical trial of an antiserotonin compound, cinanserin, in chronic schizophrenia. J. Clin. Pharmacol. New Drugs 11:220-226. [PubMed] [Google Scholar]
- 19.Hou, T., and X. Xu. 2004. Recent development and application of virtual screening in drug discovery: an overview. Curr. Pharm. Des. 10:1011-1033. [DOI] [PubMed] [Google Scholar]
- 20.Itil, T. M., N. Polvan, and J. M. Holden. 1971. Clinical and electroencephalographic effects of cinanserin in schizophrenic and manic patients. Dis. Nerv. Syst. 32:193-200. [PubMed] [Google Scholar]
- 21.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 101:12694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 78:5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenwitheesuk, E., and R. Samudrala. 2003. Identifying inhibitors of the SARS coronavirus proteinase. Bioorg. Med. Chem. Lett. 13:3989-3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane, F. J., Jr. 1970. Treatment of mania with cinanserin, an antiserotonin agent. Am. J. Psychiatry 126:1020-1023. [DOI] [PubMed] [Google Scholar]
- 25.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 26.Kuo, C. J., Y. H. Chi, J. T. Hsu, and P. H. Liang. 2004. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 318:862-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lakey, W. H., and J. M. Leonard. 1970. Prolongation of skin homografts with 2′-(3-dimethylaminopropylthio) cinnamanilide. Surg. Forum 21:287-288. [PubMed] [Google Scholar]
- 28.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 29.Maggiora, L. L., C. W. Smith, and Z. Y. Zhang. 1992. A general method for the preparation of internally quenched fluorogenic protease substrates using solid-phase peptide synthesis. J. Med. Chem. 35:3727-3730. [DOI] [PubMed] [Google Scholar]
- 30.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 31.Marsili, M., and J. Gasteiger. 1980. Pi-charge distributions from molecular topology and pi-orbital electronegativity. Croat. Chem. Acta 53:601-614. [Google Scholar]
- 32.Martina, B. E., B. L. Haagmans, T. Kuiken, R. A. Fouchier, G. F. Rimmelzwaan, G. Van Amerongen, J. S. Peiris, W. Lim, and A. D. Osterhaus. 2003. Virology: SARS virus infection of cats and ferrets. Nature 425:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mengel, C. E., and C. A. Lotito. 1968. A new antiserotonin in the carcinoid syndrome. Arch. Intern. Med. 121:507-510. [PubMed] [Google Scholar]
- 34.Morris, G. M., D. S. Goodsell, R. S. Halliday, R. Huey, W. E. Hart, R. K. Belew, and A. J. Olson. 1998. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19:1639-1662. [Google Scholar]
- 35.Murphy, G. P., H. D. Brede, H. W. Weber, J. H. Groenewald, and E. A. Mirand. 1970. The effect of cinanserin (antiserotonin agent) on canine renal allografts. J. Surg. Oncol. 2:145-154. [DOI] [PubMed] [Google Scholar]
- 36.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poutanen, S. M., D. E. Low, B. Henry, S. Finkelstein, D. Rose, K. Green, R. Tellier, R. Draker, D. Adachi, M. Ayers, A. K. Chan, D. M. Skowronski, I. Salit, A. E. Simor, A. S. Slutsky, P. W. Doyle, M. Krajden, M. Petric, R. C. Brunham, and A. J. McGeer. 2003. Identification of severe acute respiratory syndrome in Canada. N. Engl. J. Med. 348:1995-2005. [DOI] [PubMed] [Google Scholar]
- 38.Purcell, W. P., and J. A. Singer. 1967. Brief review and table of semiempirical parameters used in the Hückel molecular orbital method. J. Chem. Eng. Data 12:235-246. [Google Scholar]
- 39.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 40.Rubin, B., J. J. Piala, J. C. Burke, and B. N. Craver. 1964. A new, potent and specific serotonin inhibitor (Sq 10,643) 2′-(3-dimethylaminopropylthio) cinnamanilide hydrochloride: antiserotonin activity on uterus and on gastrointestinal, vascular, and respiratory systems of animals. Arch. Int. Pharmacodyn. Ther. 152:132-143. [PubMed] [Google Scholar]
- 41.Rubin, B., and M. H. Waugh. 1965. Antiphogistic effects of antiserotonin (Sq 10,643) and aminopyrine in rats versus endotoxin and other agents. Proc. Soc. Exp. Biol. Med. 119:438-443. [DOI] [PubMed] [Google Scholar]
- 42.Shoichet, B. K., S. L. McGovern, B. Wei, and J. J. Irwin. 2002. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 6:439-446. [DOI] [PubMed] [Google Scholar]
- 43.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun, H., H. Luo, C. Yu, T. Sun, J. Chen, S. Peng, J. Qin, J. Shen, Y. Yang, Y. Xie, K. Chen, Y. Wang, X. Shen, and H. Jiang. 2003. Molecular cloning, expression, purification, and mass spectrometric characterization of 3C-like protease of SARS coronavirus. Protein Expr. Purif. 32:302-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure (Cambridge) 12:341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.ter Meulen, J., A. B. Bakker, E. N. van den Brink, G. J. Weverling, B. E. Martina, B. L. Haagmans, T. Kuiken, J. de Kruif, W. Preiser, W. Spaan, H. R. Gelderblom, J. Goudsmit, and A. D. Osterhaus. 2004. Human monoclonal antibody as prophylaxis for SARS coronavirus infection in ferrets. Lancet 363:2139-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 48.Traggiai, E., S. Becker, K. Subbarao, L. Kolesnikova, Y. Uematsu, M. R. Gismondo, B. R. Murphy, R. Rappuoli, and A. Lanzavecchia. 2004. An efficient method to make human monoclonal antibodies from memory B cells: potent neutralization of SARS coronavirus. Nat. Med. 10:871-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsang, K. W., P. L. Ho, G. C. Ooi, W. K. Yee, T. Wang, M. Chan-Yeung, W. K. Lam, W. H. Seto, L. Y. Yam, T. M. Cheung, P. C. Wong, B. Lam, M. S. Ip, J. Chan, K. Y. Yuen, and K. N. Lai. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1977-1985. [DOI] [PubMed] [Google Scholar]
- 50.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, Q. M., R. B. Johnson, J. D. Cohen, G. T. Voy, J. M. Richardson, and L. N. Jungheim. 1997. Development of a continuous fluorescence assay for rhinovirus-14 3C protease using synthetic peptides. Antivir. Chem. Chemother. 8:303-310. [Google Scholar]
- 52.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, C. Y., J. T. Jan, S. H. Ma, C. J. Kuo, H. F. Juan, Y. S. Cheng, H. H. Hsu, H. C. Huang, D. Wu, A. Brik, F. S. Liang, R. S. Liu, J. M. Fang, S. T. Chen, P. H. Liang, and C. H. Wong. 2004. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. USA 101:10012-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiong, B., C. S. Gui, X. Y. Xu, C. Luo, J. Chen, H. B. Luo, L. L. Chen, G. W. Li, T. Sun, C. Y. Yu, L. D. Yue, W. H. Duan, J. K. Shen, L. Qin, T. L. Shi, Y. X. Li, K. X. Chen, X. M. Luo, X. Shen, J. H. Shen, and H. L. Jiang. 2003. A 3D model of SARS-CoV 3CL proteinase and its inhibitors design by virtual screening. Acta Pharmacol. Sin. 24:497-504. [PubMed] [Google Scholar]
- 55.Yang, H., M. Yang, Y. Ding, Y. Liu, Z. Lou, Z. Zhou, L. Sun, L. Mo, S. Ye, H. Pang, G. F. Gao, K. Anand, M. Bartlam, R. Hilgenfeld, and Z. Rao. 2003. The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor. Proc. Natl. Acad. Sci. USA 100:13190-13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, X. W., and Y. L. Yap. 2004. Exploring the binding mechanism of the main proteinase in SARS-associated coronavirus and its implication to anti-SARS drug design. Bioorg. Med. Chem. 12:2219-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.