Abstract
We have investigated regulatory sequences in noncoding human DNA that are associated with repression of an integrated human immunodeficiency virus type 1 (HIV-1) promoter. HIV-1 integration results in the formation of precise and homogeneous junctions between viral and host DNA, but integration takes place at many locations. Thus, the variation in HIV-1 gene expression at different integration sites reports the activity of regulatory sequences at nearby chromosomal positions. Negative regulation of HIV transcription is of particular interest because of its association with maintaining HIV in a latent state in cells from infected patients. To identify chromosomal regulators of HIV transcription, we infected Jurkat T cells with an HIV-based vector transducing green fluorescent protein (GFP) and separated cells into populations containing well-expressed (GFP-positive) or poorly expressed (GFP-negative) proviruses. We then determined the chromosomal locations of the two classes by sequencing 971 junctions between viral and cellular DNA. Possible effects of endogenous cellular transcription were characterized by transcriptional profiling. Low-level GFP expression correlated with integration in (i) gene deserts, (ii) centromeric heterochromatin, and (iii) very highly expressed cellular genes. These data provide a genome-wide picture of chromosomal features that repress transcription and suggest models for transcriptional latency in cells from HIV-infected patients.
The position of genes within chromosomes is known to modulate their rate of transcription (48), but relatively few studies have systematically compared regulation at multiple chromosomal sites. Of these, most have focused on identifying positively acting promoters and enhancers by “enhancer trapping” or related approaches (16, 31). Here we have used human immunodeficiency virus (HIV) integration to identify negatively acting chromosomal features, an issue of interest both in understanding global control of transcription and in assessing HIV transcriptional latency in patients.
Retroviral model systems provide a tractable means of studying the influence of chromosomal context on transcription. Each integrated provirus is joined to flanking cellular DNA at exactly the same points at the ends of the viral DNA, but integration takes place at many different sites in the host cell chromosomes. Thus, the viral genome provides a homogeneous transcription template that can be analyzed at different chromosomal locations, allowing the influence of flanking chromosomal features to be assessed.
Early during HIV gene expression, transcription is initiated by polymerase II from the viral long terminal repeat (LTR) under the control of cellular factors, including NF-κB, SP1, NFAT, and others (12, 15). Most of the resulting transcripts terminate within 100 nucleotides of the transcription initiation site (30). A low level of full-length transcripts is nevertheless synthesized, and a portion of these are spliced to yield the mRNA encoding Tat. In the late phase of viral transcription, Tat accumulates in the host cell and binds to the TAR site on the viral RNA, recruiting the cyclin T-CDK9 complex and facilitating transcriptional elongation (18, 47).
HIV transcription is known to be sensitive to the chromosomal environment at the site of integration (27, 28). In one example of such regulation, Jordan et al. found that proviruses integrated into centromeric heterochromatin had undetectable levels of basal transcription. However, activation of transcription by treatment with tumor necrosis factor alpha (TNF-α) or 12-O-tetradecanoylphorbol 13-acetate (TPA), both of which induce the NF-κB pathway, allowed activation of such proviruses (27, 28). Additional factors proposed to affect HIV transcription are reviewed in references 15 and 18.
Chromosomal features repressing HIV gene expression are of particular interest due to their possible influence on clinical latency in HIV infection. For many HIV-infected patients, treatment with highly active antiretroviral therapy can reduce viral loads to undetectable levels but, unfortunately, cells persist long term that harbor integrated proviruses capable of reseeding virus production after cessation of therapy. One well-characterized reservoir is in resting CD4-positive T cells (9, 14, 49). A low percentage of these cells harbor transcriptionally inactive HIV proviruses which may be induced to produce HIV upon T-cell activation. The finding that centromeric heterochromatin represses HIV gene expression, along with other known mechanisms for down-modulating HIV gene expression (1, 15, 18, 42, 45), provides candidate explanations connecting transcriptional repression to clinical latency.
To study how expression from the HIV type 1 (HIV-1) promoter is affected by the integration site of the provirus, we isolated cells containing stably expressed and inducible proviruses, determined integration sites by sequencing 971 host-virus DNA junctions, and then asked what identifiable features were enriched in each population. Several notable biases were found, suggesting potential mechanisms by which the chromosomal environment may modulate HIV transcription.
MATERIALS AND METHODS
Vector preparation and infections.
To produce the Tat and green fluorescent protein (GFP)-transducing HIV-based vector, 293T cells were cotransfected with pEV731 (LTR-Tat-IRES-GFP) (28), the packaging construct pCMVdeltaR8.91, and the vesicular stomatitis virus G protein-producing pMD.G construct (36). Viral supernatant was harvested 48 h later and filtered through a 0.45-μm filter unit. Vector titer was determined by infection of 6 × 105 Jurkat cells with various amounts of vector supernatant and 4 μg/ml Polybrene (hexadimethrine bromide; Sigma). Cells were harvested 96 h after infection and analyzed by fluorescence-activated cell sorting for GFP expression.
Jurkat cells were cultured at a density of 3 × 105 to 1 × 106 cells/ml in RPMI 1640 medium with 10% fetal bovine serum, 100 U/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine at 37°C. Cells were infected at a multiplicity of infection of 0.1 with 4 μg/ml Polybrene for cloning integration sites and at 1.0 for analysis by transcriptional profiling. To date, comparisons between integration site data sets made with HIV-based vectors (40, 50) have not shown any differences with integration sites made with authentic HIV (5, 50).
Acquisition of stably bright and inducible cell populations.
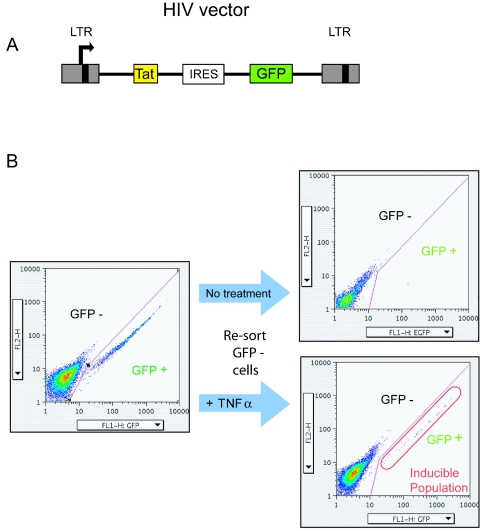
Jurkat cells were fluorescence-activated cell sorter (FACS) analyzed into GFP-positive and GFP-negative populations 2 to 4 days postinfection as described elsewhere (27, 28). At this stage, about 7% of cells were GFP positive. The GFP-positive cells were sorted for GFP expression a second time 2 weeks postinfection, and DNA was extracted (QIAgen DNeasy tissue kit), yielding stably expressed proviruses. At this stage, about 90% of cells were GFP positive (geometric mean of GFP fluorescence measured in FL1 from a representative experiment was 215). GFP-negative Jurkat cells were sorted twice more for lack of GFP expression and then cultured with TNF-α for 17 h prior to sorting. After induction, approximately 0.25% of cells became GFP positive (geometric mean, 63.3, when analyzed 4 days after sorting). Note that the absolute level of the fluorescent signal measured in FL1 varied depending on the instrument used and the gate drawn compared to the uninfected control. The cells that were inducibly GFP positive were collected and DNA was extracted, yielding the inducible sample. The inducible cells became dark upon withdrawal of TNF-α (over 90% became dim 2 weeks after removal of TNF), indicating dependence of expression on the inducing agent. The fraction of inducible cells seen in this study was similar to that reported in reference 27.
Integration site cloning and mapping to the genome.
DNA from stably expressed and inducible populations was digested with three restriction endonucleases with six-base recognition sites (NheI, SpeI, and XbaI, essentially as described in reference 40) or with MseI (which has a four-base recognition site, as described in reference 50). Digested DNA was then ligated to the appropriate adapter and amplified by nested PCR as described previously (40). Oligonucleotides used are listed in Table S1 of the supplemental material. Integration site sequences were determined to be authentic if they began at the junction with the HIV LTR, had a sequence identity of >98%, and yielded a unique best hit when mapped to the human genome using BLAT (University of California, Santa Cruz).
A small data set (20 sites) was also generated using TPA as an inducing agent and analyzed. This set was biased in favor of integration in genes, and 2/20 were in alphoid repeats, paralleling sites analyzed after induction with TNF-α (data not shown).
Expression analysis.
A total of 3 × 106 Jurkat cells (in triplicate per treatment group) were plated and either left untreated in culture, infected with the vesicular stomatitis virus G protein-pseudotyped LTR-Tat-IRES-GFP HIV-based vector (with 4 μg/ml Polybrene) at a multiplicity of infection of 1 for 24 h, or treated with 10 ng/ml TNF-α for 17 h. Cells were harvested, and total RNA was extracted using the QIAgen RNeasy kit. Labeling and hybridization of RNA to Affymetrix HG-U133A arrays was performed using the Affymetrix protocol. Analysis used Affymetrix Microarray Analysis suite 5.1 software. Changes in transcriptional activity were quantified using EASE and significance analysis of microarrays (SAM) to determine the false discovery rate. For the comparison of untreated Jurkat cells to HIV-infected cells, 575 genes were found to change at least twofold in activity (accepting a 1% false discovery rate). For the comparison of untreated cells to TNF-α-treated cells, 10 genes were found to be upregulated and 32 were downregulated under the same criteria.
Statistical analysis.
A detailed statistical analysis is presented in the supplemental material. An analysis of the randomly selected genes yielded a surprising result which suggested that the bias for favored integration in active genes (see Fig. 4, below) is stronger than the figure may suggest. Randomly selected sites that were mapped to genes were distributed into classes by expression level as in Fig. 4, below, and analyzed. The random sites did not yield a uniform distribution in each expression class, but instead revealed a bias in favor of the least-well-expressed genes (values were as follows: class 1, 15.1 to 16.1%; class 2, 14.6 to 15.7%; class 3, 15.1 to 15.3%; class 4, 12.8 to 13.4%; class 5, 11.4 to 11.6%; class 6, 11.7 to 12.1%; class 7, 10.8 to 11.2%; class 8, 6.2 to 6.7%; P < 0.0001 by chi-square; the range is for all three data sets in Fig. 4A to C, below). This is probably explained by the finding that highly expressed genes tend to have shorter introns (7) and so are smaller targets for integration. This emphasizes that the tendency to integrate in active genes is likely stronger than previously appreciated, because active genes are typically smaller than poorly expressed genes.
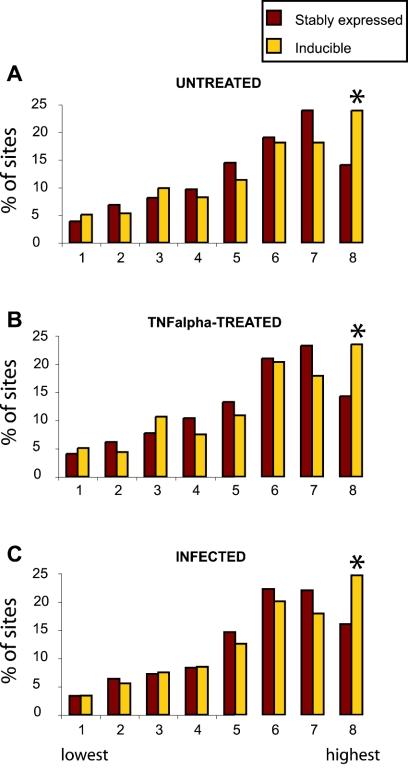
FIG. 4.
Inducible proviruses are found more commonly in very highly active genes. Expression levels were assayed in Jurkat cells (three independent Affymetrix HU133A microarrays for each condition) and scored using the Affymetrix Microarry suite 5.1 software package. To classify the expression levels of genes hosting integration events, class boundaries were first generated by dividing all the genes on the array into eight classes according to their relative level of expression. Genes that hosted integration events were then distributed into the classes defined by these boundaries, summed, and expressed as a percentage of the total number of integration sites in genes on the array. The leftmost class in each panel contains the 1/8 most weakly expressed genes, and the rightmost class contains the 1/8 most highly expressed. The highest signal value represented in each expression bin (for untreated Jurkat cells) was as follows: bin 1, 9.2; bin 2, 20.6; bin 3, 38.6; bin 4, 66; bin 5, 117; bin 6, 227; bin 7, 488; bin 8, 12050. Integration sites were analyzed using data from untreated Jurkat cells (A), TNF-treated Jurkat cells (B), or HIV-Tat-GFP-infected Jurkat cells (C) (P < 0.003; chi-square test). Inducible proviruses in the eighth class (most highly expressed) accounted for about 17% of the total.
For the Mann-Whitney test to compare expression signals for the stably expressed and inducible proviruses, the data were filtered to remove noise by analyzing only genes that were called “present” on at least two out of three arrays.
Nucleotide sequence accession numbers
The sequences for the integration sites newly determined in this study have been deposited at NCBI and assigned accession numbers CZ442176 to CZ443146. Microarray data have been deposited at the NCBI GEO repository under accession numbers GSE2504.
RESULTS
Isolation of integration sites from cells containing stably expressed and inducible proviruses.
To acquire cells containing stably expressed or weakly expressed proviruses, Jurkat cells (a CD4+ T-cell line) were infected with an HIV-based vector that encoded the HIV transcriptional activator Tat and GFP (LTR-Tat-IRES-GFP) (28) (Fig. 1A). Cells were infected at a low multiplicity of infection (0.1) to minimize the fraction harboring more than one provirus. Cells were then separated several times by FACS into GFP-expressing and nonexpressing populations (Fig. 1B). The GFP-negative population was treated with TNF-α, an agent that is known to activate LTR transcription (39) and thereby to activate transcription from silent proviruses. Cells were then sorted to obtain the induced GFP-positive population. Previous studies using this model have shown that most of these inducible proviruses are silent due to integration in chromosomal sites unfavorable for gene expression (27, 28). In addition, focusing on the inducible fraction minimizes possible complications resulting from the inactivation of viral genomes by mutation. Integrated proviruses that were not expressed and were uninducible were not studied.
FIG. 1.
Acquisition of cells containing stably expressed and inducible proviruses. (A) Tat-transducing HIV-based vector used in this study. Tat, HIV-encoded transcriptional activator; IRES, internal ribosome entry site. Transcription initiates within the left LTR. (B) Acquisition of cells containing stably expressed and inducible proviruses by FACS. Cells were infected at a multiplicity of about 0.1 and sorted for GFP-positive and -negative cells (left side). GFP-positive cells were collected and then sorted a second time to isolate a stably bright fraction. The GFP-negative (dark) population was sorted twice, and the dark cells were collected each time. The stably dark cells were then treated with TNF-α, and the resulting bright cells were collected (right side).
Chromosomal integration sites from cells in the stably expressed and inducible populations were then cloned using ligation-mediated PCR and sequenced (40, 50). The chromosomal distributions of these sites were compared to two data sets generated by infection of lymphoid cells (SupT1 cells or primary peripheral blood mononuclear cells) with HIV-based vectors (34, 40). The cells in these studies were not fractionated by the level of proviral gene expression, and so these data sets provide an overview of integration site selection by HIV. A set of 10,000 random sites in the human genome generated in silico was also included for comparison (Table 1).
TABLE 1.
Integration site data sets used in this study
| Data set | Vector | Cell type | No. of integration sites | Source or reference |
|---|---|---|---|---|
| Stably expressed | HIV: LTR-Tat-IRES-GFP | Jurkat | 587 | This report |
| Inducible | HIV: LTR-Tat-IRES-GFP | Jurkat | 384 | This report |
| HIV/SupT1 | HIV p156 (CMV-GFP) | SupT1 | 493 | 40 |
| HIV/PBMC | HIV p156 (CMV-GFP) | PBMCa | 550 | 34 |
| Random | 10,000 | This report |
PBMC, peripheral blood mononuclear cells.
Frequency of integration in genes.
Since the complement of human genes has not been fully clarified, we used four different gene catalogs to analyze the frequency of integration in transcription units (Table 2). For all sets of HIV integration sites and all types of gene calls, integration was strongly biased in favor of transcription units (34, 40, 50). For example, using the well-characterized RefSeq genes for comparison, the human genome contains 31.1% genes, while HIV integration site data sets showed frequencies of integration in genes from 66.1% (SupT1 cells) to 73.4% (Jurkat cells, inducible integration sites). The stably expressed and inducible populations of proviruses both showed similar high frequencies of integration in genes (see p. 3-9 of the statistical information provided in the supplement material).
TABLE 2.
Integration in transcription unitsa
| Chromosomal feature | Frequency (%) of transcription units at integration sites in:
|
||||
|---|---|---|---|---|---|
| Human genome (random sites) | Stably expressed sites, HIV/Jurkat | Inducible sites, HIV/Jurkat | HIV/SupT1 | HIV/PBMC | |
| Acembly | 49.2 | 87.6 | 89.1 | 83.2 | 87.8 |
| GenScan | 64.3 | 78.4 | 78.6 | 76.1 | 79.5 |
| RefSeq | 31.1 | 71.2 | 73.4 | 66.1 | 69.1 |
| UniGene | 50.8 | 79.2 | 80.7 | 72.6 | 75.1 |
All comparisons to random show P < 0.0001.
Primary sequences at integration sites.
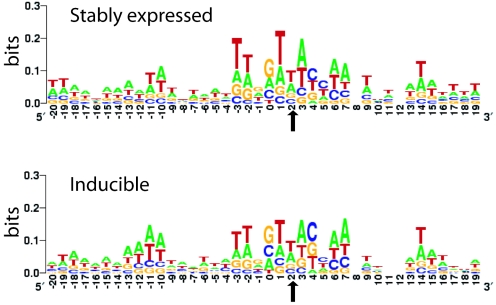
The primary sequences that served as integration targets were analyzed separately for the stably expressed and inducible proviruses (Fig. 2). The sequences from both data sets showed inverted repeat symmetry centered on the sequence 5′GT(A/T)AC3′ as previously reported (2, 5, 43). The more detailed analysis reported here also shows the presence of a longer consensus, with notable conservation about one turn of the helix in either direction out from the conserved sequences. No binding sites for known transcription factors were significantly enriched in either data set (data not shown). Thus, we could not detect any clear differences between the two data sets in the local sequences at integration sites.
FIG. 2.
Primary sequences surrounding the stably expressed and inducible proviruses. The weak consensus sequence seen at the stably expressed (top) and inducible (bottom) proviruses was rendered so that the degree of conservation is proportional to the height of each letter, using LOGO (http://weblogo.Berkeley.edu/logo.cgi). The y axis reflects the information content at each base, so that perfect conservation would have a score of 2 bits. The points of joining between the HIV and human DNA lie between −1 and 0 (for the sequenced HIV DNA end) and between 4 and 5 on the other strand for the other end of the HIV DNA. Thus, the points of joining, and the integration consensus sequence, are symmetric around position 2 (arrow).
Integration in repeated sequences: inducible proviruses are more frequently found in alphoid repeats.
Despite these similarities between the stably expressed and inducible integration sites, three features were found to differ. Each suggests a chromosomal feature disfavoring HIV transcription. The first involved the frequency of integration in repeated sequences (Table 3).
TABLE 3.
Integration in repeated sequencesa
| Chromosomal feature | Frequency (%) of repeated sequences at integration sites in:
|
||||
|---|---|---|---|---|---|
| Human genome (random sites) | Stably expressed sites, HIV/Jurkat | Inducible sites, HIV/Jurkat | HIV/SupT1 | HIV/PBMC | |
| SINES | |||||
| Alu | 9.4 | 9.1 (0.8325) | 9.5 (0.9002) | 17.6 (<0.0001) | 10.1 (0.5246) |
| MIR | 2.5 | 3.0 (0.4186) | 1.7 (0.3087) | 1.5 (0.107) | 3.2 (0.2713) |
| DNA elements | 2.7 | 2.1 (0.3491) | 3.9 (0.1207) | 2.4 (0.6898) | 3.9 (0.0844) |
| LTR elements (HERV) | 7.7 | 5.1 (0.0124) | 3.5 (0.0007) | 4.5 (0.0035) | 2.5 (<0.0001) |
| LINE | 18.0 | 21.2 (0.0368) | 15.2 (0.1207) | 19.2 (0.4347) | 15.5 (0.132) |
| Alpha satellite | 0.3 | 0.1 (0.5807) | 4.3 (<0.0001) | 0.5 (0.2987) | 0.0 (0.2142) |
The percentages are relative to all sites in the data set; values in parentheses are P values (chi-square) compared to random sites.
The frequency of integration in alphoid repeats was 4.3% in the inducible Jurkat sites but only 0% to 0.5% in the other HIV data sets. Alphoid repeats are mostly found in centromeres, and packaging of DNA in centromeric heterochromatin is known to repress transcription of many genes (41, 46). These data support the idea that HIV DNA embedded in centromeric heterochromatin is poorly expressed, so that enriching for poorly expressed proviruses enriched for those in alphoid repeats (27, 28).
A small number of integration sites (20 total) were isolated from cells after induction with TPA instead of TNF-α. Of these, two were in alphoid repeats, paralleling results with TNF-α induction (data not shown).
All HIV integration site data sets showed that human endogenous retroviruses (HERVs) are significantly disfavored targets (P < 0.013), as reported previously for the SupT1 data set (40). HERVs are enriched outside transcription units, while HIV integration is favored within transcription units, accounting for the observed bias.
Inducible proviruses are more frequently found in gene deserts.
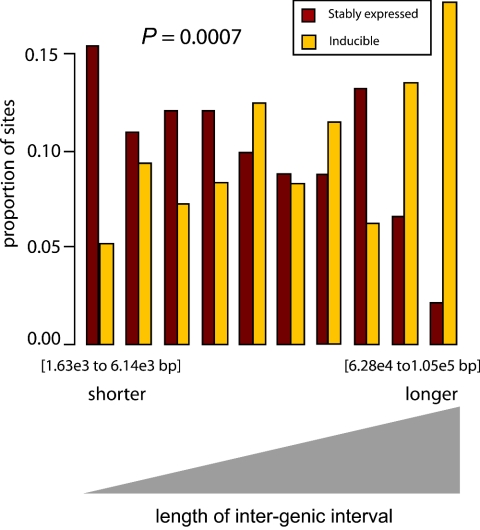
A second difference was found in an analysis of the positions of stably expressed and inducible proviruses in intergenic regions. The stably expressed proviruses were more frequently found in short intergenic regions, indicative of favored integration in gene-rich chromosomal domains, as seen previously (34, 40). In contrast, the inducible proviruses were much more frequently found in long intergenic regions or “gene deserts” (Fig. 3) (P < 0.0007, regardless of gene call used for the analysis) (see p. 67-79 of the statistical information provided in the supplement material).
FIG. 3.
Frequency of stably expressed or inducible proviruses in intergenic regions of different lengths. Shorter intergenic regions are shown to the left, and longer ones are to the right. Genscan genes were used for this analysis, though the conclusions were similar for other gene sets as well (see p. 67-79 of the statistical information provided in the supplement material). The P value is obtained from the logistic regression of event type (stable or inducible) on a cubic B-spline basis (i.e., a third-order polynomial) for intergenic distance. The units on the x axis indicate lengths of intergenic regions, in base pairs. Lengths of intergenic regions for each category were defined by the following boundaries (from left to right, in bp): 1,627, 6,135, 10,506, 14,900, 21,907, 28,989, 36,333, 43,531, 62,837, 104,802, and 3,182,720. The inducible proviruses in the rightmost five bins accounted for 14% of all inducible proviruses.
This finding was reinforced by an analysis of the density of integration events compared to the density of CpG islands, which are more common in gene-dense regions. The stably expressed proviruses were found more commonly in regions of high CpG island density, whereas the inducible sites were enriched in regions of low density (P = 0.002) (see p. 10-14 of the statistical information provided in the supplement material). This indicates that the inducible proviruses are enriched in long intergenic regions that are depleted of both genes and CpG islands.
Inducible proviruses are more frequently found in very highly expressed cellular genes.
A third chromosomal feature correlating with inducible HIV gene expression was identified by transcriptional profiling analysis of the Jurkat target cells. The expression signals of cellular genes hosting integration events were tabulated for the stably expressed and inducible proviruses. The median for both groups of genes was found to be higher than the median of all the probe sets on the HU133A microarrays used (stably expressed = 152, inducible = 177, all genes on the array = 66; units are “signal,” as defined by Affymetrix MAS 5.1). Genes in both the stably expressed and inducible populations were also more active than genes from the random control population in Table 1 (random = 57; P < 0.0001 for comparison to either the stably expressed or inducible populations; Mann-Whitney test). This broadly parallels previous studies of HIV, which revealed that active genes were favored as integration targets (34, 40, 50).
Thus, it was unexpected that the stably expressed and inducible data sets differ from each other. The median expression value for genes hosting inducible proviruses was found to be significantly higher than the median of genes hosting stably expressed proviruses (P = 0.0004; Mann-Whitney test).
To analyze this issue in more detail, expression signals of genes hosting integration events were divided into classes by their signal values and the distribution was examined (Fig. 4A). As with previous studies, genes hosting integration events were found more commonly in the more highly expressed genes. The inducible proviruses were more frequently found in the highest expression class: 24% of inducible integration sites (in genes represented on the array) compared to 14% for the stably expressed set (P = 0.003; chi-square test). In previous studies, genes in the highest expression class (eighth bin) were consistently found to be less favorable for integration (34, 40); here, this is seen as well for the stably bright population but not the inducible population. Thus, we infer that integration in the very highly expressed genes was associated with the inducible phenotype and, specifically, that the transcription level in bin 8 is disfavorable for HIV transcription. Inducible proviruses in highly expressed genes were found in both orientations relative to the direction of host gene transcription (data not shown). An analysis of the placement of integration sites within genes showed no obvious bias; for example, the inducible sites in the most highly transcribed genes (eighth bin) were not clustered near the start site of transcription (data not shown).
The relationship between integration targeting and host cell transcription was probed further by repeating the transcriptional profiling measurements under two additional conditions. Jurkat cells were infected with the HIV-Tat-GFP vector prior to RNA isolation, or cells were treated with 10 ng/ml TNF and RNA was isolated subsequently. These manipulations caused clearly detectable changes in transcription. Notably, infection with the Tat-transducing vector caused down-modulation of a large family of genes involved in signal transduction and immune responses (Fig. 5), potentially a biologically significant activity of Tat involved in evasion of the host immune response (11, 24, 29). Treatment with TNF resulted in induction of a number of previously characterized TNF-inducible genes. Though these changes were readily detectable, overall transcription in the cell types studied was still quite similar (correlation coefficients for pair-wise comparisons of any two microarrays showed R > 0.98). Analysis of genes hosting integration events using these transcriptional profiling data sets also indicated that very highly transcribed cellular genes were more common targets in the inducible data set (Fig. 4B and C).
FIG. 5.
Tat down-modulates host cell genes important in signal transduction and immune responses. Signal intensities from Affymetrix HU133A microarrays were analyzed by SAM (http://www-stat.Stanford.EDU/∼tibs/SAM/) to identify significantly affected genes and then clustered according to gene ontology using EASE (http://david.niaid.nih.gov/david/ease.htm). The three left columns show results from uninfected cells, and the three right columns show results from cells infected with the Tat-transducing HIV-based vector. A large set of Tat-repressed genes (115 probe sets corresponding to 108 different genes) was identified as overrepresented compared to all genes queried by the microarray in the “signal transducer activity” category (P = 1.16 × 10−5; Fisher exact test with Bonferroni correction for multiple comparisons). Expression values were normalized by dividing by the mean. In cases where multiple probe sets queried the activities of a single gene, the values were found to be closely similar and a single representative probe set was used for the figure. Gray tiles indicate negative values. All genes called by EASE in the “signal transducer activity” category are shown, except for six olfactory receptors and one taste receptor.
Jurkat cells as model HIV target cells: assessment using transcriptional profiling.
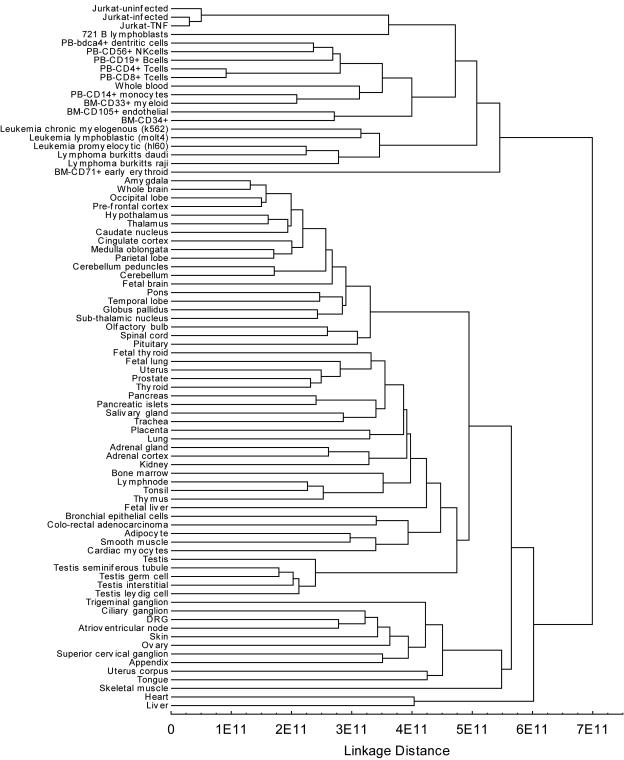
The transcriptional profiling data on Jurkat cells could be used to investigate how closely the Jurkat cell line models the primary cells normally targeted by HIV infection in vivo. Transcriptional profiles of uninfected Jurkat cells were compared to 79 transcriptional profiles of human cells and tissues (data from Hogenesch and coworkers [44]). A cluster analysis is shown in Fig. 6. Transcriptional profiles of Jurkat cells clustered with profiles of a collection of leukocytes, including CD4+ T cells. Jurkat cell transcription did differ somewhat from CD4+ T cells, however, which could be due to the transformed state of Jurkat cells or to differences in the execution of the microarray experiments. Inspection of the Jurkat transcriptional profiles indicates that many of the genes expected to be active in CD4+ T cells are indeed robustly expressed (Fig. 5 and data not shown), consistent with previous studies in which Jurkat cells were shown to be active in assays of T-cell function (e.g., references 17 and 32). In summary, transcription in the Jurkat cell clusters with authentic CD4+ T cells, helping to validate the use of Jurkat cells as a model of infection in vivo.
FIG. 6.
Clustering of transcriptional profiles from Jurkat cells with human leukocytes. Data for human tissues are from reference 44. All analyses used Affymetrix HU133A microarrays. Transcription signal values were averaged between replicates and ranked prior to clustering. Squared Euclidean distance and unweighted pair-group average linkage (also know as UPGMA) cluster analysis of the transcriptional profiles was carried out using Statistica 7.0.
DISCUSSION
Here we compared the chromosomal placement of HIV proviruses that were stably expressed after integration to proviruses that were poorly expressed but inducible upon treatment of cells with TNF-α. Three chromosomal features correlated with inducible expression: centromeric heterochromatin, gene deserts, and highly active host transcription units. Each of these is discussed below. However, only about 40% of the inducible proviruses were associated with one of these three features, and so further chromosomal environments unfavorable for expression may yet be found. In addition, studies from others using this model suggest that low-level GFP expression may also result from stochastic fluctuations in Tat levels. For cells expressing low levels of Tat protein, fluctuations in Tat concentration may extinguish LTR-driven transcription, and this may become “locked in” because Tat protein is required to activate its own expression (D. Schaffer and coworkers, personal communication).
Silencing HIV proviruses by transcriptional interference.
A significantly greater proportion of the inducible proviruses were found in the most highly expressed fraction of host genes (Fig. 4), suggesting that very-high-level host gene transcription interferes with transcription of an integrated provirus. Many studies have established that transcriptional interference can repress gene expression (4, 10, 19, 20, 22, 33), and a model HIV promoter has previously been shown to be sensitive to transcriptional interference in HeLa cells (20). For a provirus in the same orientation as the host cell gene, read-through transcription may repress by blocking access of factors to the downstream promoter or by actively dislodging bound proteins (4, 19, 20, 22, 33). In the HeLa cell model, read-through transcription was found to repress HIV transcription by dislodging bound Sp1 (20). A provirus in an orientation opposite that of the host gene may be silenced by the above mechanisms, or by transcriptional “trainwrecking” whereby two RNA polymerase complexes collide during convergent elongation. Convergent transcription could also result in transcription of both DNA strands and formation of double-stranded RNA, which might silence proviral transcription via RNA interference (reviewed in references 23 and 37), RNA-directed DNA methylation (35), induction of the interferon response (13), or generation of antisense RNA (38).
Inducible proviruses are integrated more commonly in gene deserts.
A strong trend was seen involving integration sites outside genes, in which long intergenic regions or gene deserts more frequently hosted inducible proviruses. Short intergenic regions more commonly hosted stably expressed proviruses. A similar trend was also seen comparing the frequency of integration in CpG islands, which are known to be associated with genes. A variety of mechanisms could account for this bias, none mutually exclusive. Gene deserts may be heterochromatic, and so packaged in proteins unfavorable for efficient transcription (25, 26, 46). Gene deserts may be enriched in binding sites for transcriptional silencer proteins, though no candidate binding sites emerged from our analysis of primary sequences at integration sites. Intranuclear positioning of gene deserts could also be a factor (3, 6, 8). A recent study suggested that activation of genes in yeast can be accompanied by translocation of the genes to a nuclear pore complex (6). Thus, proviruses integrated into gene-sparse regions may be localized within nuclear domains that are unfavorable for transcription.
Integration in centromeric heterochromatin disfavors HIV gene expression.
Repression of HIV expression after integration in alphoid repeats was previously observed by Eric Verdin and colleagues using the Jurkat model (27, 28). Heterochromatin adopts a condensed structure that blocks access of the transcriptional machinery (41, 46). Thus, a simple model to explain our results is that wrapping of the proviral DNA in heterochromatin blocks access of the transcriptional machinery and thereby represses transcription.
Models for the mechanism of transcriptional latency in patients.
HIV-infected patients on successful long-term antiretroviral therapy nevertheless harbor cells containing latent proviruses, and after cessation of treatment HIV from these cells can reinitiate active replication (9, 14, 21, 49). Our findings reveal mechanisms by which the surrounding chromosomal environment may silence some integrated proviruses while leaving them inducible by TNF-α treatment. The data presented here suggest that proviruses integrated in centromeric heterochromatin, gene deserts, and highly transcribed genes may contribute to the latent population.
Direct studies of integration sites from latently infected cells in patients have been challenging. One report investigated the distribution of HIV integration sites in resting CD4+ lymphocytes of patients on effective highly active antiretroviral therapy (21). However, this work was complicated by the fact that defective proviruses greatly outnumber latent proviruses in patient cells (9, 14, 49). Han et al. cloned 74 integration sites and found that 93% of the proviruses were integrated within active transcription units (21). If these sites are representative of latent integration sites in patients, then the transcriptional interference model may be the most attractive based on our data.
Supplementary Material
Acknowledgments
We thank Mark Ptashne, Robert Doms, and members of the Bushman laboratory for helpful discussions.
This work was supported by NIH grants AI52845 and AI34786, the James B. Pendleton Charitable Trust, Robin and Frederic Withington (F.D.B.), and the Fritz B. Burns Foundation (to J.R.E.).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Blankson, J. N., D. Persaud, and R. F. Siliciano. 2002. The challenge of viral reservoirs in HIV-1 infection. Annu. Rev. Med. 53:557-593. [DOI] [PubMed] [Google Scholar]
- 2.Bor, Y.-C., M. Miller, F. Bushman, and L. Orgel. 1996. Target sequence preferences of HIV-1 integration complexes in vitro. Virology 222:238-242. [DOI] [PubMed] [Google Scholar]
- 3.Boyle, S., S. Gilchrist, J. M. Bridger, N. L. Mahy, J. A. Ellis, and W. A. Bickmore. 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum. Mol. Genet. 10:211-219. [DOI] [PubMed] [Google Scholar]
- 4.Callen, B. P., K. E. Shearwin, and J. B. Egan. 2004. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol. Cell 14:647-656. [DOI] [PubMed] [Google Scholar]
- 5.Carteau, S., C. Hoffmann, and F. D. Bushman. 1998. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol. 72:4005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casolari, J. M., C. R. Brown, S. Komili, J. West, H. Hieronymus, and P. A. Silver. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117:427-439. [DOI] [PubMed] [Google Scholar]
- 7.Castillio-Davis, C. I., S. L. Mekhedov, D. L. Hartl, E. Koonin, and F. A. Kondrashov. 2002. Selection for short introns in highly expressed genes. Nat. Genet. 31:415-418. [DOI] [PubMed] [Google Scholar]
- 8.Chubb, J. R., and W. A. Bickmore. 2003. Considering nuclear compartmentalization in light of nuclear dynamics. Cell 112:403-406. [DOI] [PubMed] [Google Scholar]
- 9.Chun, T.-W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. M. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193-13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cullen, B. R., P. T. Lomedico, and G. Ju. 1984. Transcriptional interference in avian retroviruses—implications for the promoter insertion model of leukaemogenesis. Nature 307:241-245. [DOI] [PubMed] [Google Scholar]
- 11.de la Fuente, C., F. Santiago, L. Deng, C. Eadie, I. Zilberman, K. Kehn, A. Maddukuri, S. Baylor, K. Wu, C. G. Lee, A. Pumfery, and F. Kashanchi. 2002. Gene expression profile of HIV-1 Tat expressing cells: a close interplay between proliferative and differentiation signals. BMC Biochem. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emerman, M., and M. H. Malim. 1998. HIV-1 regulatory/accessory genes: keys to unraveling viral and host cell biology. Science 280:1880-1884. [DOI] [PubMed] [Google Scholar]
- 13.Fields, B. N., and D. M. Kinpe. 1996. Virology. Raven Press, New York, N.Y.
- 14.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 15.Freed, E. O. 2004. HIV-1 and the host cell: an intimate association. Trends Microbiol. 12:170-177. [DOI] [PubMed] [Google Scholar]
- 16.Friddle, C. J., et al. 2003. High-throughput mouse knockouts provide a functional analysis of the genome. Cold Spring Harbor Symp. Quant. Biol. 68:311-315. [DOI] [PubMed] [Google Scholar]
- 17.Frumento, G., A. Corradi, G. B. Ferrara, and A. Rubartelli. 1997. Activation-related differences in HLA class-I bound peptides: presentation of an IL-1 receptor antagonist-derived peptide by activated, but not resting, CD4+ T lymphocytes. J. Immunol. 159:5993-5999. [PubMed] [Google Scholar]
- 18.Garber, M., and K. A. Jones. 1999. HIV-1 Tat: coping with negative elongation factors. Curr. Opin. Immunol. 11:460-465. [DOI] [PubMed] [Google Scholar]
- 19.Greger, I. H., A. Aranda, and N. J. Proudfoot. 2000. Balancing transcriptional interference and initiation on the GAL7 promoter of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 97:8415-8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greger, I. H., F. Demarchi, M. Giacca, and N. J. Proudfoot. 1998. Transcriptional interference perturbs the binding of Sp1 to the HIV-1 promoter. Nucleic Acids Res. 26:1294-1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han, Y., K. Lassen, D. Monie, A. R. Sedaghat, S. Shimoji, S. Liu, T. C. Pierson, J. B. Margolick, R. F. Siliciano, and J. D. Siliciano. 2004. Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78:6122-6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hausler, B., and R. L. Somerville. 1979. Interaction in vivo between strong closely spaced constitutive promoters. J. Mol. Biol. 127:353-356. [DOI] [PubMed] [Google Scholar]
- 23.Hu, W. Y., F. D. Bushman, and A. C. Siva. 2004. RNA interference against retroviruses. Virus Res. 102:59-64. [DOI] [PubMed] [Google Scholar]
- 24.Izmailova, E., F. M. Bertley, Q. Huang, N. Makori, C. J. Miller, R. A. Young, and A. Aldovini. 2003. HIV-1 Tat reprograms immature dendritic cells to express chemoattractants for activated T cells and macrophages. Nat. Med. 9:191-197. [DOI] [PubMed] [Google Scholar]
- 25.Jenuwein, T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11:266-273. [DOI] [PubMed] [Google Scholar]
- 26.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 27.Jordan, A., D. Bisgrove, and E. Verdin. 2003. HIV reproducibly establishes a latent infection after acute infection of T cells in vitro. EMBO J. 22:1868-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 30.Kao, S. Y., A. F. Calman, P. A. Luciw, and B. M. Peterlin. 1987. Anti-termination of transcription within the long terminal repeat of HIV-1 by tat gene product. Nature 330:489-493. [DOI] [PubMed] [Google Scholar]
- 31.Lukacsovich, T., and D. Yamomoto. 2001. Trap a gene and find out its function: toward functional genomics in Drosophila. J. Neurogenet. 15:147-168. [DOI] [PubMed] [Google Scholar]
- 32.Manger, B., A. Weiss, K. J. Hardy, and J. D. Stobo. 1986. A transferrin receptor antibody represents one signal for the induction of IL 2 production by a human T cell line. J. Immunol. 136:532-538. [PubMed] [Google Scholar]
- 33.Martens, J. A., L. Laprade, and F. Winston. 2004. Intergenic transcription is required to repress the Saccharomyes cerevisiae SER3 gene. Nature 429:571-574. [DOI] [PubMed] [Google Scholar]
- 34.Mitchell, R., B. Beitzel, A. Schroder, P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. D. Bushman. 2004. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biol. 2:E234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morris, K. V., S. W. Chan, S. E. Jacobsen, and D. J. Looney. 2004. Small interfering RNA-induced transcriptional gene silencing in human cells. Science 305:1289-1292. [DOI] [PubMed] [Google Scholar]
- 36.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272:263-267. [DOI] [PubMed] [Google Scholar]
- 37.Plasterk, R. H. 2002. RNA silencing: the genome's immune system. Science 296:1263-1265. [DOI] [PubMed] [Google Scholar]
- 38.Scherer, L. J., and J. J. Rossi. 2003. Approaches for the sequence-specific knockdown of mRNA. Nat. Biotechnol. 21:1457-1465. [DOI] [PubMed] [Google Scholar]
- 39.Schmid, R. M., N. D. Perkins, C. S. Duckett, P. C. Andrews, and G. J. Nabel. 1991. Cloning of an NF-kappa-B subunit which stimulates HIV transcription in synergy with p65. Nature 352:733-736. [DOI] [PubMed] [Google Scholar]
- 40.Schroder, A., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. D. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 41.She, X., J. E. Horvath, Z. Jiang, G. Liu, T. S. Furey, L. Christ, R. Clark, T. Graves, C. L. Gulden, C. Alkan, J. A. Bailey, C. Sahinalp, M. Rocchi, D. Haussler, R. K. Wilson, W. Miller, S. Schwartz, and E. E. Eichler. 2004. The structure and evolution of centromeric transition regions within the human genome. Nature 430:857-864. [DOI] [PubMed] [Google Scholar]
- 42.Sheridan, P. L., T. P. Mayall, E. Verdin, and K. A. Jones. 1997. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 24:3327-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stevens, S. W., and J. D. Griffith. 1996. Sequence analysis of the human DNA flanking sites of human immunodeficiency virus type 1 integration. J. Virol. 70:6459-6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su, A. I., T. Wiltshire, S. Batalov, H. Lapp, K. A. Ching, D. Block, J. Zhang, R. Soden, M. Hayakawa, G. Kreiman, M. P. Cooke, J. R. Walker, and J. B. Hogenesch. 2004. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA 101:6062-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verdin, E. 1991. DNase I-hypersensitive sites are associated with both long terminal repeats and with the intragenic enhancer of integrated human immunodeficiency virus type 1. J. Virol. 65:6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wallrath, L. L. 1998. Unfolding the mysteries of heterochromatin. Curr. Opin. Genet. Dev. 8:147-153. [DOI] [PubMed] [Google Scholar]
- 47.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 48.Wolffe, A. P. 1998. Chromatin, 3rd ed. Academic Press, San Diego, Calif.
- 49.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged supression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 50.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration. Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.