Abstract
Infection with oncogenic human papillomaviruses (HPVs), typified by HPV type 16 (HPV16), is a necessary cause of cervical cancer. Prophylactic vaccination with HPV16 L1 virus-like particles (VLPs) provides immunity. HPV16 VLPs activate dendritic cells and a potent neutralizing immunoglobulin G (IgG) response, yet many cervical cancer patients fail to generate detectable VLP-specific IgG. Therefore, we examined the role of the innate recognition of HPV16 L1 in VLP-induced immune responses and its evasion during carcinogenesis. Nonconservative mutations within HPV16 L1 have been described in isolates from cervical cancer and its precursor, high-grade cervical intraepithelial neoplasia (CIN). We determined the effect of mutations in L1 upon in vitro self-assembly into VLPs and their influence upon the induction of innate and adaptive immune responses in mice. Several nonconservative mutations in HPV16 L1 isolated from high-grade CIN or cervical carcinoma prevent self-assembly of L1 VLPs. Intact VLPs, but not assembly-defective L1, activate dendritic cells to produce proinflammatory factors, such as alpha interferon, that play a critical role in inducing adaptive immunity. Indeed, effective induction of L1-specific IgG1 and IgG2a was dependent upon intact VLP structure. Dendritic cell activation and production of virus-specific neutralizing IgG by VLPs requires MyD88-dependent signaling, although the L1 structure that initiates MyD88-mediated signaling is distinct from the neutralizing epitopes. We conclude that innate recognition of the intact L1 VLP structure via MyD88 is critical in the induction of high-titer neutralizing IgG. Tumor progression is associated with genetic instability and L1 mutants. Selection for assembly-deficient L1 mutations suggests the evasion of MyD88-dependent immune control during cervical carcinogenesis.
Human papillomavirus type 16 (HPV16) is the primary etiologic agent of cervical cancer. However, high-risk HPV infections result in progression to cervical carcinoma in only a small percentage of infected women. Clearly, additional events are required for progression, such as viral integration and other mutations that reflect the genomic instability engendered by E6 and E7 expression (44). The immune system can limit disease by clearing pre-existing lesions, as evidenced by the transience of most HPV16 infections, associations between particular HLA types and risk for cervical cancer, and elevated incidence of HPV-related cancers in immunocompromised individuals. Conversely, the numerous decoy strategies employed by papillomavirus suggest that escape from immune surveillance is critical to viral persistence and cancer progression (12). Indeed, despite the potent immunogenicity of the papillomavirus capsid, only half of cervical cancer patients generate capsid-specific immunoglobulin G (IgG) (28).
Dendritic cells (DCs) are the most efficient antigen-presenting cells and are central to the induction of antigen-specific immunity against tumors and pathogens. DCs employ pattern recognition receptors such as the Toll-like receptor (TLR) family to sense infection. Indeed, they play a critical role in regulating the induction and nature of adaptive immune responses (39). In response to a pathogen-associated molecular pattern, TLRs signal primarily via the adaptor molecule MyD88 to activate innate responses. These innate responses include the production of a number of cytokines and chemokines, which play a key role in the induction of adaptive immunity and in regulating T-helper type 1 (Th1)/Th2 bias and isotype class switch recombination (1). The major papillomavirus capsid protein L1 self-assembles to form virus-like particles (VLPs) (21). We recently demonstrated that activation of DCs and induction of HPV16 L1-specific Th1 responses are dependent upon MyD88, indicating that HPV16 L1 VLPs represent a pathogen-associated molecular pattern that is recognized by the TLR (41). Interestingly, HPV16 VLPs activate human myeloid DCs but not Langerhans cells (11, 24).
Vaccination with HPV16 L1 VLP protects patients from persistent HPV16 infection and the development of CIN, the precursor lesion of cervical cancer (22). Protection by VLP vaccination is mediated by neutralizing IgG (2, 38). Comparison of the L1 amino acid sequences from >100 HPV genotypes reveals regions of strong homology comprising predominantly internal capsid structures, punctuated by highly divergent surface-exposed residues (5) that include the immunodominant neutralizing epitopes. The variation of capsid surface-exposed residues reflects evasion of neutralizing antibody responses.
Here, we demonstrate the influence upon capsid assembly and immune recognition of multiple mutations in the otherwise highly conserved structural residues present within independent HPV16 L1 isolates derived from a subset of cervical carcinoma and high-grade cervical intraepithelial neoplasia (CIN). We identify several mutant HPV19 L1 isolates carrying genes encoding proteins that neither assemble nor activate VLP-dependent innate and adaptive immune responses orchestrated by DC. This may represent evasion of innate immune recognition during cervical carcinogenesis.
MATERIALS AND METHODS
Generation of recombinant baculoviruses.
Recombinant baculoviruses for HPV16 L1 114/K, D202H, and Zaire 1194 have been described previously (6, 19, 21). The L1 isolates from Yamada et al. (40) and Pushko et al. (31) were subcloned into pFB-1 (Invitrogen). Recombinant baculoviruses were generated using the BAC-to-BAC system (Invitrogen).
Immunoprecipitation and Western blot analysis.
Dishes (100 mm) of Sf9 insect cells were infected at a high multiplicity of infection with recombinant baculovirus expressing HPV16 L1 derived from either the 114/K or prototype HPV16 isolates and incubated for 3 days at 27°C in Grace's medium-10% fetal calf serum (FCS)-penicillin (100 U/ml) and streptomycin (100 μg/ml), as described in reference 21. The cells were scraped from the dish into the medium and harvested by centrifugation (5 min, 600 × g, room temperature). After the medium was aspirated, the cell pellet was lysed in 2 ml of phosphate-buffered saline (PBS) supplemented with 0.5 M NaCl and 1% (vol/vol) NP-40. The detergent lysate was centrifuged at 14,000 × g for 10 min at 4°C. The supernatant was divided into aliquots and 2 μl of the monoclonal antibody (MAb) ascites added to each aliquot. Camvir-1 was purchased from Pharmingen, and H16.V5, H16.U4, and H16.V5 neat ascites were the kind gift of Neil Christensen, Hershey, PA. The samples were tumbled end-over-end for an hour at 4°C. Next, 100 μl of 50% protein A-Sepharose slurry was added to each sample and the tubes were tumbled end-over-end for another hour at 4°C. The Sepharose beads were collected by 10-s centrifugation and washed three times with 1 ml of ice-cold lysis buffer. The immunoprecipitates were boiled for 5 min in 100 μl of gel sample buffer, and proteins were separated by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and transferred to a membrane for Western blot analysis. Western blot analysis was performed using 1:2,000 rabbit anti-HPV16 L1 (114/K) VLP and peroxidase-conjugated goat anti-rabbit immunoglobulin and developed using LumiGlo (KPL).
Hemagglutination assay with purified HPV16 L1.
HPV16 L1 preparations were analyzed for the presence of HPV16 L1 by SDS-10% PAGE and Western blot analysis using 1:2,000 monoclonal antibody Camvir-1 and peroxidase-conjugated goat anti-mouse immunoglobulin and developed using LumiGlo. The ability of a twofold dilution series of each L1 preparation to agglutinate mouse red blood cells was tested as described previously (34, 35).
Mice.
129/Sv/C57BL/6/Myd88−/− and 129/Sv/C57BL/6/Myd88+/+ mice, 129-IFN-α/β−/− and 129-IFN-α/β+/+ mice (B&K Universal), 129S6-CD4TM1KNW−/− and 129S6-CD4TM1KNW+/+ mice, and C57BL/6J mice (Jackson Laboratory) were maintained in a pathogen-free animal facility at least 1 week before use. Mice were vaccinated three times at weekly intervals intravenously (i.v.) with 10 μg of purified HPV16 L1 in PBS without adjuvant. Experiments were performed in accordance with institutional guidelines.
Preparation of HPV16 L1 in Escherichia coli.
Mutant L1 were inserted into pGEX-6P-2. E. coli BL21 cells transformed with pGEX plasmid (pGEX16L1-L1) and were grown at room temparature in Luria-Bertani medium containing 100 μg/ml ampicillin. At an optical density at 600 nm of 0.3, recombinant protein expression was induced by adding 0.25 mM isopropyl-β-D-thiogalactopyranoside (IPTG) to the 0.5l 2YT culture at 30°C (4). Soluble glutathione S-transferase (GST)-L1 fusion protein was purified on a 1-ml glutathione-Sepharose Fast Flow (Amersham) column according to the manufacturer's instructions. The column was incubated overnight at 4°C with PreScission protease to release L1 from the column. Protein content was evaluated using a microBCA kit (Pierce) and SDS-PAGE. Samples were absorbed onto carbon-coated grids and stained with 1% uranyl acetate. The grids were examined with a Philips CM120 transmission electron microscope operating at 80 KV.
Preparation and flow cytometry of BMDCs.
Bone marrow-derived dendritic cells (BMDCs) were prepared from bone marrow cells collected by removing the femur bones of mice, cutting off each end, and flushing out the bone marrow with RPMI1640 medium using a syringe. The pooled cells were harvested by centrifugation at 1,600 rpm for 10 min and resuspended in 2 ml ACK buffer (0.156 M NH4Cl, 10 mM KHCO3, 0.1 mM EDTA [pH 7.3]; Biosource) for 5 min at room temperature to lyse the red blood cells. Remaining cells were washed in medium and cultured in RPMI medium containing 500 U/ml recombinant murine granulocyte-macrophage colony-stimulating factor (rmGM-CSF) for 6 days prior to analysis. For FACScan analysis of BMDCs, cells were collected in ice-cold PBS and surface marker phenotypes were analyzed with the following antibodies: fluorescein isothiocyanate-conjugated anti-CD11C (N418), anti-mouse CD86 (GL1), anti-mouse CD80 (16-10A1), anti-mouse CD40 (3/23), and phycoerythrin-labeled anti-CD4 (L3T4), anti-mouse CD8α(53-6.6), anti-mouse CD45 R/B220 (RA36B2), anti-mouse CD11b (M1/70) as well as purified anti-mouse I-Ab (25-9-17), and H-2Db (KH95). All of these antibodies were purchased from PharMingen. Single or double staining was performed using different MAbs. Cells were then washed twice before being resuspended in PBS containing 1% paraformadehyde and 1% FCS and kept at 4°C prior to flow cytometric analysis (FAScan; Becton Dickinson). For each analysis, isotype-matched control MAb was used a negative control.
Reverse transcription (RT)-PCR analysis.
Total cellular RNA was prepared using TRIzol reagent (Invitrogen), followed by RNA cleanup with the RNeasy Mini kit (QIAGEN). The primers used were as follows: for alpha interferon 4 (IFN-α4), sense, 5′ ATGGCTAGGCTCTGTGCT, and antisense, 5′ CTCCTTCTCCTCACTCAG; for interleukin-12b (IL-12b), sense, 5′ ATGTGTCCTCAGAAGCTAACC, and antisense, 5′ GGATCGGACCCTGCAGGGAACACATGC; for IL-10, sense, 5′ ATGCCTGGCTCAGCACTGCTATGC, and antisense, 5′ TTAGCTTTTCATTTTGATCATCATG; for GAPDH, sense, 5′ ATGGTGAAGGTCGGTGTGAACGGATTTGGC, and antisense, 5′ CATCGAAGGTGGAAGAGTGGGAGTTGCTGT; for MyD88, sense, 5′ ATGTCTGCGGGAGACCCCCGCG, and antisense, 5′ TCAGGGCAGGGACAAAGCCTTGGC.
Generation of MyD88 knockdown cell lines.
Oligonucleotide pairs 5′ GATCCCCGGAGCTGAAGTCGCGCATCTTCAAGAGAGATGCGCGACTTCAGCTCCTTTTTGGAAA and 5′ AGCTTTTCCAAAAAGGAGCTGAAGTCGCGCATCTCTCTTGAAGATGCGCGACTTCAGCTCCGGG were annealed and inserted between the BglII and HindIII sites of pSuper (kindly provided by T. R. Brummelkamp, The Netherlands), modified to carry hygromycin B resistance, and then transfected into macrophage cell line RAW264.7. The RAW264.7 MyD88 knockdown cell lines were maintained in Dulbecco's modified Eagle medium-10% FCS-hygromycin B.
Transfection and reporter assays.
pNF-κB-SEAP, pAP1-SEAP, and positive-control pSEAP2 as well as negative-control pTAL-SEAP reporter constructs (Clontech) were individually transfected into mouse RAW-264.7 macrophage cells with Lipofectamine 2000 (Invitrogen) in 24-well plates. After 24 h, the cells were stimulated using 25 μg/ml VLP (1.05 mg/ml stock). The supernatants were collected at the different time points and analyzed by the chemiluminescence-secreted alkaline phosphatase assay according to the manufacturer's protocol (Clontech).
ELISA.
Commercial sandwich enzyme-linked immunosorbent assay (ELISA) kits were used for the quantitation of mouse IFN-α (PBL Biomedical Laboratories), mouse IL-12 p70 (R&D), and mouse IL-10 (Pierce). The optical density of each of the sample was measured at 450 nm using a SpectraMax 190 ELISA plate reader. Cytokine levels were quantified from two to three titrations using standard curves and expressed in pg/ml. For antibody isotype analysis, sera were pooled from six mice per group and ELISAs were performed using a mouse hybridoma isotyping kit (Biomeda) as recommended by the manufacturer. The plates were coated with 100 ng/ml HPV16 L1.
In vitro neutralization assay.
Antibody neutralization of human papillomavirus type 16 pseudovirus was performed according to the method of Pastrana et al. (30).
RESULTS
Mutations in HPV16 L1 isolated from cervical carcinoma and precursor lesions compromise capsid assembly.
Variations in conserved internal residues were described in a prior survey of HPV16 L1 sequences (40) isolated from specimens of cervical carcinoma and its precursor, high-grade CIN, namely, D202H from invasive cervical carcinoma WV2916 (37); I398T; K430E from invasive cervical carcinoma 426-10 and N81D, D223G, N327S; and F446S from three independent isolates (T3, T17, and T49) with the same sequence from patients in Trinidad with high-grade CIN (8, 31). Since these changes were absent from normal variants, we hypothesized that these mutations reflect genetic instability in (pre)malignant cells and selection for escape from innate and/or adaptive immune recognition of cervical lesions by negating particle assembly. To address this hypothesis, we initially compared the ability of these L1 mutants to assemble into VLP with other nonconservative mutations in conserved internal residues that were generated either randomly during PCR amplification or which represent authentic sequences of individual genomes generated during in vivo viral replication (Table 1).
TABLE 1.
Assembly, hemagglutinating, surface binding and immunoreactive properties of HPV16 L1 isolates and variants.legendlegend
| Source of clone | Changes with respect to HPV16 L1 114/Kg | 3mersh | VLPi | HAl | H16.V5m | H16.E70 | H16.U4 | FACSo |
|---|---|---|---|---|---|---|---|---|
| Variant 114/Ba | V194I, T266A | + | + | + | ND | ND | ND | ND |
| Variant Zaire1194c | H76Y, T176N, N181T, T266A, S282P, T353P, L474F | + | + | + | + | +h | + | + |
| Variant OR4541f | T176N, N181T, T266A, T353P, T389S, L474F | + | + | + | + | + | + | + |
| Variant OR9237f | T266A, K443Q | + | + | + | + | + | + | + |
| Variant NMT455f | I159L | + | + | + | + | + | + | + |
| Cancer WV2916d | D202H | − | −d | − | − | − | − | − |
| Cancer 426-10f | T266A, I398T, K430E | + | + | + | + | + | + | + |
| CINIII T3 isolateb | N81D, D223G, N327S, I381M, A427V, F446S | − | − | − | − | − | − | − |
| T3 derivative | N81D, T266A | + | + | + | + | + | + | + |
| T3 derivative | D223G, T266A | − | − | − | − | − | + | − |
| T3 derivative | T266A, N327S, I381M | − | − | − | − | − | − | − |
| T3 derivative | T266A, A427V | + | + | + | + | + | + | + |
| T3 derivative | T266A, F446S | − | 5mersj | − | + | + | + | − |
| PCR errore | Q196R | + | + | + | + | + | + | + |
| PCR errore | T176N, T266A, N290D | + | + | + | + | + | + | + |
| PCR errore | S280A, E400G | + | + | + | + | + | + | + |
| PCR errore | T176N, N181T, T266A, T353P, T389S, L474F, N285S | + | + | + | + | + | + | + |
| PCR errore | T176N, N177S, T266A, L474F | + | + | + | + | + | + | + |
| PCR errore | T266A, K443Q, T488R | + | NDk | + | + | + | + | + |
| PCR errore | T266A, V220A, F256S, K443Q | + | + | + | + | + | + | + |
| PCR errore | H76Y, T176N, T266A, L474F, S495L | + | + | + | + | + | + | + |
| PCR errore | K162E, T176N, T266A, N290D | + | + | + | + | + | + | + |
| PCR errore | D303G | + | + | + | + | + | + | + |
| PCR errore | Q461R | + | + | + | + | + | + | + |
Variant and baculovirus described in reference 21 from condylomata accuminata. This clone supports the production of infectious virus (33).
Isolate same as T3, T17, and T49 from women with CINIII, as described in reference 31 except that the clone provided to us had an additional I381 M change.
Variant and baculovirus described in references 6 and 17 isolated from the CINIII lesion of an HIV-1 positive woman. This clone supports the production of infectious virus (32).
Isolate and baculovirus described in references 19 and 37. This HPV16 L1 clone assembles approximately 1,000-fold less efficiently into VLP than the 114/K variant when expressed in insect cells (19, 21) and does not support the production of infectious virus (33).
In an HPV16 variant survey (40) in women with predominantly normal cervical cytology, L1 genes were PCR amplified using the proofreading Thermus thermophilus DNA polymerase and cloned. Sequences were identified with mutations that did not occur in all of the three clones and are probably the result of amplification errors, although coinfection with more than a single variant or development of mutants in situ are possibilities.
Variant described in (40)
Changes with respect to the 114/K variant in conserved residues are bold. Changes in poorly conserved positions that are present in numerous variants are lightface
Presence or absence of L1 trimers after separation of gradient-purified L1 preparations by SDS-PAGE and analysis by Western blotting using monoclonal antibody CamVir-1.
Presence or absence of 55- to 60-nm capsid-like structures upon analysis of gradient-purified L1 preparations by transmission electron microscopy.
Numerous capsomers (pentamers of L1) were observed.
ND, not done.
Gradient-purified L1 preparations were tested for their ability to agglutinate mouse erythrocytes as described in reference 34.
Ability of monoclonal antibody H16.V5 to immunoprecipitate HPV16 L1 mutants and variants from lysates of baculovirus-infected insect cells.
nThe S282P mutation significantly reduced H16.E70 binding as measured by ELISA or neutralization (32), but this was not reflected in the highly sensitive immunoprecipitation and Western blot analysis.
Ability of gradient-purified HPV16 L1 to bind to the surface of SiHa cells at 4°C as detected by flow cytometric staining with the H16.V5 monoclonal antibody.
pIsolated from a cervical cancer patient in Spain and kindly provided by J. Icenogle.
Since HPV16 L1 isolates of viable variants efficiently assemble into VLP when expressed in Sf9 insect cells using recombinant baculovirus, we overexpressed these L1 isolates by the same strategy and screened for the presence of VLPs on the basis of five criteria: (i) reactivity with conformationally dependent neutralizing monoclonal antibodies (H16.V5, H16.E70, and H16.U4) against distinct L1 epitopes (32), (ii) the presence of L1 trimers by Western blot analysis (36), (iii) the presence of ∼50-nm particles by transmission electron microscopy (19), (iv) the ability to agglutinate mouse erythrocytes (34), and (v) binding to the surfaces of CaSki cells as detected by indirect immunofluorescence staining with H16.V5 and flow cytometry. Preparations of all L1 isolates with random changes in conserved residues tested, but not the isolates from invasive cervical carcinoma WV2916 (D202H) and patient T3, were recognized by the three conformationally dependent antibodies, exhibited L1 trimers by Western blot analysis, and contained VLPs that agglutinated mouse erythrocytes, bound to CaSki cells, and were visible by transmission electron microscopy (Table 1). This suggests a selection for mutations that compromise capsid assembly in cervical carcinoma and even its precursor, CINIII.
T3 L1 differs from the reference 114/K sequence by N81D, D223G, N327S, A427V, and F446S (8, 31) as well as I381 M in the clone provided to us. Therefore, we examined which of these changes in L1 could prevent VLP assembly. L1 N81D and A427V single mutants assembled efficiently. L1 F446S was recognized by the conformationally dependent antibodies, but it predominantly formed capsomers and failed to hemagglutinate. Interestingly, the D223G single mutant and the N327S I381 M double mutant failed to assemble into VLP, except that the D223G L1 was immunoprecipitated by one of three conformationally dependent neutralizing antibodies, H16.U4 (Table 1).
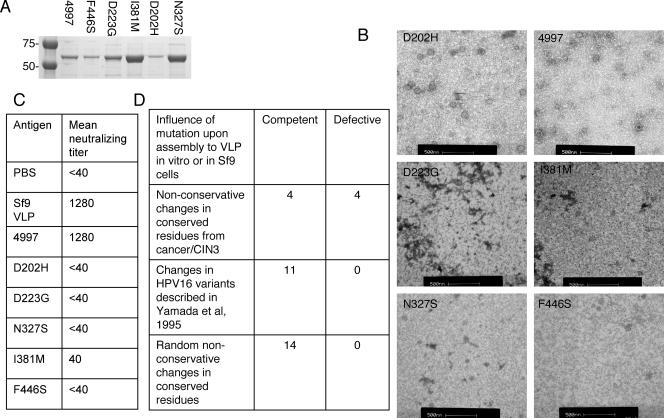
Recombinant HPV16 L1 purified from E. coli self-assembles to form VLP in vitro (4). Since the analysis of several L1 point mutants was hampered by low yield from Sf9 cells, we performed transmission electron microscopy on preparations of L1 purified from E. coli (Fig. 1). While we observed VLP in preparations of HPV16 L1 variant 4997 (which is T266A with respect to the reference 114/K HPV16 L1) and single mutants D202H, I381 M and F446S, none were detected for D223G or N327S (Fig. 1), indicating that the D202H and F446S mutants assembled more efficiently when prepared in E. coli (Fig. 1), compared to Sf9 insect cells (Table 1). Figure 1D summarizes the source of the HPV16 L1 mutants tested and their influence on VLP assembly as described above.
FIG. 1.
Assembly of VLP from HPV16 L1 mutants. (A) HPV16 L1 mutants were expressed as GST fusions in E. coli and affinity purified. The GST tag was removed from HPV16L1-GST fusion proteins by PreScission protease digestion and passaged over glutathione-Sepharose and the L1 protein analyzed by SDS-PAGE and Coomassie staining. (B) Transmission electron micrographs of in vitro-assembled L1 preparations for variant 4997 and the D202H, D223G, N327S, I381 M, and F446S mutants. The samples were absorbed onto carbon-coated grids and stained with 1% uranyl acetate and examined with a Philips CM120 transmission electron microscope operating at 80 kV. (C) Neutralizing titers of mouse antisera to in vitro-assembled preparations of each HPV16 L1 mutant. The mice (n = 6) were immunized i.v. with 10 μg of HPV16 L1 in PBS on day 0, day 7, and day 14. The sera were collected at 10 days after final vaccination. Fourfold pooled serum dilutions were analyzed in two independent experiments for their ability to neutralize HPV16 (114/K) pseudovirions, and geometric mean titers are presented. Dilutions of neutralizing ascites of monoclonal antibody H16.V5 and mouse antisera to HPV16 (114/K) L1 VLP prepared in Sf9 insect cells were included as positive controls and sera of mice vaccinated with PBS alone as a negative control for the neutralization assay. (D) Influence of L1 mutations upon VLP assembly. HPV16 L1 genes containing nonconservative changes in conserved residues from cancer/high-grade CIN isolates, changes in HPV16 variants described in Yamada et al. (40), or random nonconservative changes in conserved residues (Table 1) were tested for their ability to assemble when overexpressed in Sf9 insect cells using recombinant baculovirus and screened for the presence of VLP on the basis of five criteria: (i) reactivity with conformationally dependent neutralizing monoclonal antibodies (H16.V5, H16.E70, and H16.U4) against distinct L1 epitopes (32), (ii) the presence of L1 trimers by Western blot analysis (36), (iii) the presence of 55-nm to 60-nm particles by transmission electron microscopy (19), (iv) the ability to agglutinate mouse erythrocytes (34), and (v) binding to the surfaces of CaSki cells as detected by indirect immunofluorecence staining with H16.V5 and flow cytometry (Table 1). Mutations found to influence assembly in insect cells were retested in vitro (B). Mutations that behaved as for the reference isolate in all assays were scored as competent and the remainder as defective.
L1 compromised for particle assembly fails to induce neutralizing antibody.
To address the influence of each mutation upon the induction of virus-specific neutralizing antibody, we vaccinated C57BL/6 mice i.v. with preparations of each HPV16 L1 mutant prepared in E. coli cells in PBS without adjuvant and tested for neutralizing activity against HPV16 pseudovirions (Fig. 1C). Sera of mice vaccinated with variant 4997 L1 and the I381 M mutant neutralized HPV16 pseudovirions with geometric mean titers of 1,280 and 40 respectively, whereas the L1 D202H, D223G, N327S, and F446S single mutants behaved as for the sera of nonimmunized mice, suggesting that these mutations significantly alter L1 conformation and that correct particle assembly is required to induce neutralizing antibody.
Neutralization is associated with particular antibody isotypes.
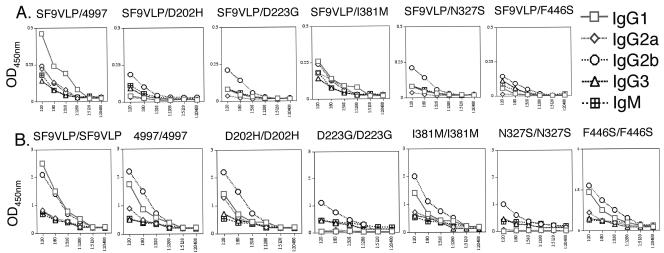
ELISAs using HPV16 L1 VLP purified from Sf9 insect cells detect predominantly conformation-dependent antibody (20). ELISA reactivity of the L1 mutant antisera with Sf9-derived HPV16 L1 VLP was detected using isotype-specific secondary antibodies. The L1 4997 variant and I381 M mutant that induce neutralizing antibody exhibit strong VLP-specific IgG1 and Ig2a responses, whereas the other assembly-compromised mutants showed minimal reactivity (Fig. 2A). Conversely, VLP-specific Ig2b and IgG3 responses were similar for all six L1 preparations (Fig. 2A). Since only sera with high levels of IgG1 and IgG2a from L1 4997 and I381 M vaccination (Fig. 1C) neutralized viruses, these may represent the primary isotypes of neutralizing antibodies. Furthermore, this suggests distinct mechanisms that trigger class switch recombination to IgG1 and IgG2a compared with VLP-specific Ig2b and IgG3.
FIG. 2.
Isotype analysis of specific reactivity in antisera to HPV16 L1 mutants. (A) Reactivity of sera from mice immunized by various HPV16L1 in vitro-assembled preparations in an HPV16 VLP ELISA. Microtiter plates were coated with 100 ng of HPV16 L1 VLP prepared in Sf9 insect cells. Sera of mice (n = 6) immunized with in vitro-assembled preparations of each HPV16 L1 mutant were diluted fourfold and reacted with the plates. Binding of specific antibody analyzed by ELISA using a mouse antibody isotyping kit is shown. (B) Same as above, but the microtiter plates were coated with in vitro-assembled preparations of each HPV16 L1 mutant after affinity purification from E. coli. Representative results from two experiments are shown.
Assembly state of L1 influences class switch recombination.
ELISA using the original L1 immunogen prepared in E. coli cells will detect all L1-specific antibody regardless of the conformational state. Therefore, to confirm the efficacy of vaccination with the capsid assembly mutants and their influence upon class switch recombination, we repeated the ELISA using the original immunogens (Fig. 2B), as well as other L1 mutants prepared in E. coli (not shown). This assay revealed significant differences in the reactivity to conformationally correct VLP and the L1 mutants. As before, L1 4997 and I381 M, which induce neutralizing antibody, also exhibited strong IgG1 and IgG2a L1-specific antibodies, whereas assembly-defective L1 D223G and N327S did not. Again, significant L1-specific IgG2b and IgG3 responses were detected regardless of mutation. Interestingly, the L1 D202H and F446S mutants that demonstrated significant assembly in vitro (Fig. 1) did produce strong L1-specific IgG1 and IgG2a responses (Fig. 2B), albeit nonneutralizing (Fig. 1C). Since IgG1 and IgG2a in the L1 D202H and F446S antisera fail to recognize conformationally correct VLP or virions, this demonstrates that the surface conformation of such mutant particles is altered.
VLP assembly is required for MyD88-dependent activation of BMDCs.
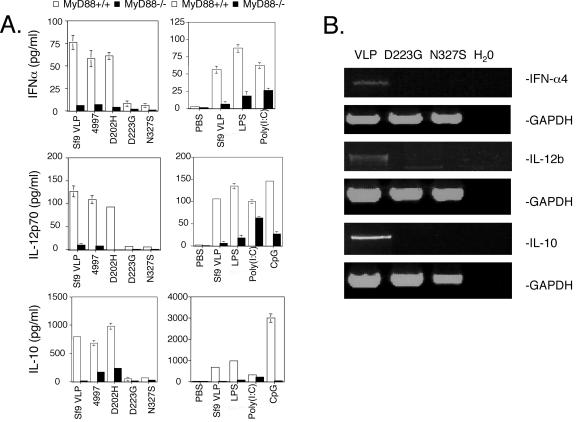
Since DCs regulate humoral responses and L1 assembly state influences BMDC activation (23), we investigated the response of BMDCs to different mutant L1 preparations. Testing of the bacterially expressed HPV16 L1 in BMDCs requires the removal of contaminating endotoxin, which itself can activate BMDCs. Thus, a polymyxin B column was employed to deplete the level of endotoxin in each HPV16 L1 preparation to <0.058 EU/ml VLP (Limulus assay E-Toxate; Sigma), a level that does not activate BMDCs (29). BMDCs stimulated with HPV16 L1 VLP prepared in Sf9 insect cells or bacteria produced similar levels of IFN-α, IL-12p70, and IL-10 (Fig. 3). Interestingly, HPV16 L1 D202H particles, but not assembly-deficient D223G and N327S L1, induced BMDC to produce IFN-α. RT-PCR analysis revealed that IFN-α4 is the predominant form upregulated by HPV16 L1 VLP but that L1 D223G and N327S preparations failed to upregulate IFN-α4 mRNA in BMDCs (Fig. 3B). Similarly, HPV16 L1 VLP of natural variants, as well as the HPV16 L1 D202H mutant, induced BMDCs to produce IL-12 and IL-10 (Fig. 3A). Again, preparations of the D223G and N327S L1 mutants failed to activate BMDCs to produce IL-12 or IL-10 and this was confirmed at the transcript level (Fig. 3). Finally, unlike HPV16 L1 VLP, the D223G and N327S L1 mutants did not upregulate the expression of costimulatory molecules CD40, CD80, and CD86 (not shown) on the surfaces of BMDCs. Thus, the data suggest that activation of BMDCs to produce IFN-α and cytokines requires VLP assembly but not the presentation of neutralizing epitopes. Furthermore, the activation of IFN-α, IL-12p70, and IL-10 production (Fig. 3) in response to assembled HPV16 L1 was abolished in MyD88−/− BMDCs, consistent with VLP-dependent signaling via the adaptor protein MyD88.
FIG. 3.
(A) Mutant L1 capsomers did not activate DCs to produce the cytokines. Production of IFN-α, IL-10, and IL-12p70 by DCs at 24 h after exposure to the following is shown: HPV16 L1 VLP from the 114/K variant prepared in Sf9 insect cells, HPV16 L1 from the 4997 variant or the D202H, and D223G or N327S mutants after in vitro assembly and treatment with polymyxin B (10 μg/ml), LPS (026:B6 at 1 μg/ml), CpG (5 μM), or poly(I:C) at 25 μg/ml. Cytokine concentrations were determined by quantitative capture ELISA. Results from three experiments are shown. (B) Relative expression of IFN-α, IL-12, and IL-10 mRNAs in BMDCs assessed by RT-PCR 12 h after stimulation with 10 μg/ml in vitro-assembled 4997, D223G, or N327S HPV16 L1 mutant after polymyxin B treatment.
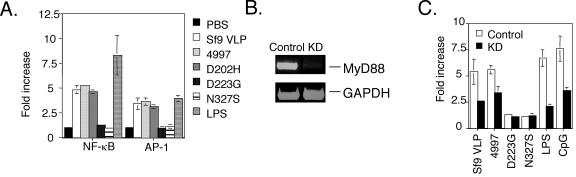
Activation of NF-κB via MyD88 requires intact VLP.
HPV16 L1 VLP prepared in insect cells activated NF-κB and AP-1-dependent transcription in the mouse macrophage cell line RAW264.7 (Fig. 4A). Both bacterially derived polymyxin B-treated HPV16 L1 and D202H L1 VLP, but neither D223G nor N327S L1 preparations, activated NF-κB and AP-1-dependent transcription in RAW264.7 cells (Fig. 4A), demonstrating a requirement for VLP assembly. The TLR4- and TLR9-dependent ligands lipopolysaccharide (LPS) and CpG, respectively, activate NF-κB-dependent transcription via MyD88 (1). Expression of an MyD88 small interfering RNA (siRNA) specifically reduces MyD88 transcript levels (Fig. 4B) and inhibits these responses, demonstrating functional MyD88 knockdown. Furthermore, activation of NF-κB-dependent transcription in RAW264.7 cells by HPV16 L1 and D202H L1 VLP is inhibited upon knockdown of MyD88 transcripts by siRNA.
FIG. 4.
VLP, but not assembly-defective HPV16 L1, mutants activate NF-κB- and AP-1-dependent transcription via MyD88. (A) The influence of in vitro-assembled HPV16 L1 mutant preparations upon NF-κB- and AP-1-dependent transcription in the macrophage cell line RAW264.7. Fold increase in chemiluminescence intensity after stimulation compared to unstimulated cells is plotted. (B) Assessment of the mRNA level of MyD88 in RAW264.7 cells by RT-PCR after stable transfection with siRNA constructs targeting MyD88 mRNA. (C) MyD88 knockdown (KD) blunts the NF-κB-dependent transcriptional response to TLR ligands LPS and CpG as well as HPV16 VLP. Results from three experiments are shown.
Induction of VLP-specific IgG1 and IgG2a is dependent upon MyD88 and IFN-α signaling.
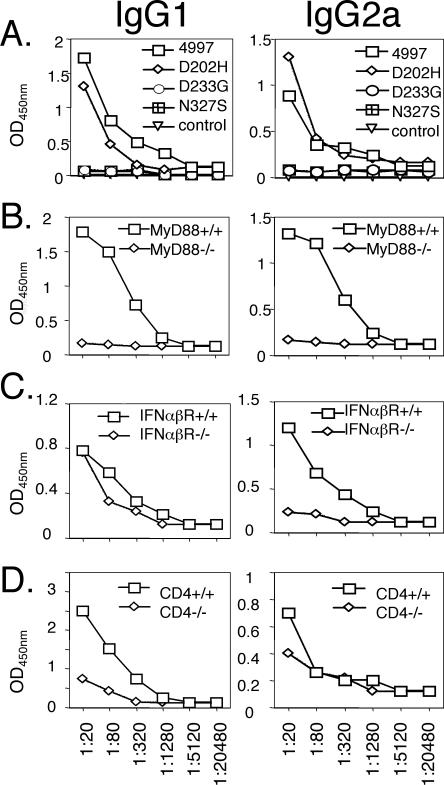
The correlation between VLP assembly, activation of DC via MyD88 (Fig. 3), and induction of strong L1-specific IgG1 and IgG2a responses (Fig. 5A) suggests a critical role for DCs in the switching to IgG1 and IgG2a VLP-specific antibody. Indeed, vaccination of MyD88−/− mice with HPV16 L1 VLP fails to induce specific IgG1 or IgG2a (Fig. 5B). IFN-α enhances antibody responses and is produced by BMDC in response to HPV16 L1 VLP (Fig. 3A). IFN-α/βR−/− mice exhibited a marked reduction in their VLP-specific IgG2a, but not IgG1, response to vaccination (Fig. 5C), suggesting that the production of IFN-α/β contributes to the development of VLP-specific IgG2a responses. Since VLPs direct the production of IFN-α by BMDCs, it is possible that T help is dispensable for the production of Ig2a. To assess the role of T-helper responses in the production of specific IgG1 and IgG2a, we vaccinated CD4−/− and CD4+/+ mice with HPV16 L1 VLP. VLP-specific IgG1 responses, for which IFN-α/β signaling was not critical, were strongly dependent upon CD4+ T help. Conversely, IgG2a responses to VLP vaccination were dependent upon IFN-α/β signaling but showed little difference between CD4−/− and CD4+/+ mice (Fig. 5D).
FIG. 5.
Mechanisms driving the induction of HPV16 L1 VLP-specific IgG1 and IgG2a. (A) ELISA of HPV16 L1 VLP-specific IgG1 and IgG2a titers in the pooled sera of C57BL/6 mice (n = 6) immunized with in vitro-assembled preparations of each HPV16 L1 mutant. (B) Production of HPV16 L1 VLP-specific IgG1 and IgG2a in MyD88-deficient and control mice. (C) Production of HPV16 L1 VLP-specific IgG1 and IgG2a in IFN-α/β receptor-deficient and control mice. (D) Production of HPV16 L1 VLP-specific IgG1 and IgG2a in CD4-deficient and control mice. The mice were immunized with HPV16 L1 VLP on day 0, day 7, and day 14. The sera were collected at 10 days after final vaccination and analyzed using a mouse isotyping kit. ELISA plates were coated with 100 ng/well of the immunogen, and representative results from three experiments are shown.
DISCUSSION
Immune evasion by cervical tumor cells through selective loss of VLP assembly function of L1.
Activation of DCs by HPV capsids to produce proinflammatory factors has several consequences for immunity against cervical cancer. First, intact L1 structure is critical to produce an effective adaptive immune response (2). Second, it can assist the induction of other HPV-specific responses, such as E6- and E7-specific T-cell immune responses (13). Third, it can create environmental “danger” signals that are critical for immune responses against cancer. Our data indicate a new mechanism during cervical carcinogenesis to avoid DC activation by selective mutation of L1 such that it can no longer assemble into particles capable of activating MyD88-dependent innate responses.
HPV VLPs induce robust immune responses outside of the epidermis, including high-titer neutralizing antibody and cellular immune responses upon parenteral vaccination (15). Indeed, intramuscular vaccination of genital warts patients with HPV6 L1 VLP may enhance regression (43). Similarly, vaccination of women with HPV16 L1 VLP was 91.2% effective (confidence interval, 80% to 97%) in protecting against transient HPV16 infection but 100% effective (90% to 100%) in protecting from persistent HPV16 infection, suggesting possible L1-mediated immune clearance of breakthrough infection (22). Furthermore, a single HPV16 L1 VLP vaccination without adjuvant protected mice in a CD8-dependent manner from challenge with a tumor cell line that was derived by transformation with the HPV16 genome (9). Thus, expression of L1 VLPs in tumor cells should evoke potent L1-specific immunity upon metastasis beyond the epithelium and among interstitial dendritic cells. The weak or absent virus-specific humoral and cellular immunity mounted by cervical cancer patients is suggestive of immune evasion (28, 42). Nonnenmacher et al. detected HPV16 VLP seropositivity in 73 to 81% of HPV16 DNA+ CINIII cervical cancers but only 51 to 59% of HPV16 DNA+ cervical cancers (28). The reasons for the numerous nonresponders and reduction in responders among cancer patients despite persistent infection are unclear. Possible factors include absent or low-level L1 expression or even mutation of L1 in cervical carcinoma cells or a protective effect of L1 immune responses. However, the initial infection must occur with a wild-type virion (and L1) and which can potentially prime a wild-type anti-VLP response. Therefore, the level of VLP-specific antibody may reflect the initial viral load. Additional serological studies are required to distinguish the relative contributions of these factors and determine how mutation of L1 influences the capsid antibody response in cervical cancer patients.
Viruses evade immune surveillance via numerous strategies to complete their life cycle (12). For example, the divergence of surface exposed L1 residues is consistent with immune evasion by alteration of neutralizing epitopes (5). Similarly, high-level L1 expression is restricted to the immunologically privileged upper layers of the epithelium where HPV16 L1 capsids fail to activate Langerhans cells (11). Cervical carcinogenesis may involve selection against formation of L1 VLP by disruption of the gene or mutation of L1 such that it can no longer signal MyD88-dependent activation of dendritic cells. Indeed, 4/8 cancer or CINIII-derived mutations in conserved residues reduced the efficiency of VLP assembly (Fig. 1D), but 0/11 changes compared to the prototype sequence in natural variants (P = 0.0181, Fisher's exact P value) or in 0/14 nonconservative mutations generated PCR errors (P = 0.0096, Fisher's exact P value). For example, HPV16 L1 isolated from the invasive cervical carcinoma WV2916 assembles with low efficiency in Sf9 insect cells and fails to activate murine BMDCs (23). More dramatic disruptions of the L1 gene, including deletions, insertions, and frameshifts that would prevent L1 expression, have been described in HPV16 isolates from cervical carcinomas (7, 16). We also defined two mutations present within independent HPV16 L1 isolates derived from three CINIII patients (31) that prevent VLP assembly as well as BMDC activation. This suggests that pressure to avoid MyD88-dependent immune recognition occurs even in the precursor lesion of cervical cancer.
Immunohistochemical studies detect L1 expression in CIN lesions but not cervical carcinoma. This is generally considered to reflect a lack of differentiation-dependent signals in the cervical carcinoma. However, it may also reflect an immunologic selection against L1 expression outside of the immunologically privileged epidermis (11). While loss of L1 expression can reflect disruption of the L1 gene during viral genome integration, the L1 gene remains intact in many carcinomas (14, 27). For carcinomas containing the complete L1 gene, the failure to detect L1 by immunohistochemistry may reflect an absence of transcription. The murine C3 tumor cell line, which was transformed by the HPV16 genome, expresses detectable levels of L1 mRNA (9). Although L1 expression in C3 cells was undetectable by Western blot analysis, C3 cells express sufficient L1 for immune recognition by CD8+ cytotoxic T cells and clearance after a single vaccination with HPV16 L1 VLP (9). Therefore, while epithelial differentiation is critical for high-level L1 expression, these studies indicate that cervical carcinomas with an intact L1 gene express sufficient L1 for recognition by L1-specific cytotoxic T cells.
In silico analysis of structural changes wrought by mutation of HPV16 L1.
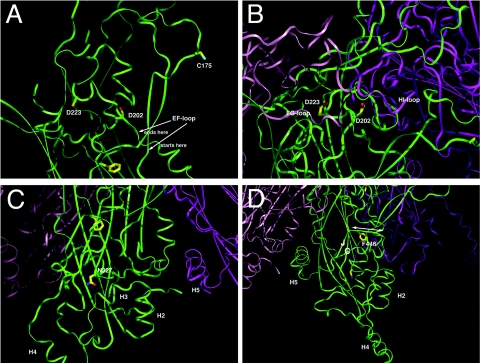
L1 structure profoundly influences immunogenicity (2, 19). Indeed, vaccination of mice with the assembly-defective D223G, N327S, and F446S L1 mutants failed to induce neutralizing antibodies. Interestingly, the HPV16 L1 D202H mutant assembles in E. coli to particles that activate BMDCs and a potent L1-specific humoral response but also fails to induce neutralizing antibodies. This suggests that the neutralizing epitopes are distinct from the capsid structures required for the MyD88-dependent activation of BMDCs but that the surface conformation of these particles is altered by this mutation. In silico analysis of the 3.5 Å resolution X-ray crystallographic structure of HPV16 L1 pentamers provides insight into key interactions that mediate particle assembly (5) when combined with the biological properties of the individual mutations in L1 (Fig. 6).
FIG. 6.
In silico analysis of the contribution of residues D202, D223, N327 and F446 to the HPV16 L1 capsomer structure. (A) The D202H mutation may affect the stability of this inner EF loop, which in turn affects the position of the outer EF loop and the C175 position that is implicated in particle assembly. D202H may also influence pentamer conformation by affecting the interface angle between two neighboring molecules in a pentamer, which could also reduce the efficiency of particle assembly in vivo (21). Interestingly, this mutant assembled efficiently in vitro in the absence of chaperones. Thus, chaperones may recognize this mutant in vivo as improperly folded and prevent capsid assembly. (B) Like D202, D223 is also located in the inner EF loop (Fig. 6A). In a pentamer, the surface FG loop from the left neighboring monomer is directly positioned on the top of the D223 and other residues of the inner EF loop. A change of D223G may influence the interactions of the FG loop of the neighboring monomer and affect the pentamer conformation (e.g., the surface loop positioning and the pentamer width), thus reducing the efficiency of particle assembly. However, the C-terminal tail which links capsomers and contains the H16.U4 neutralizing epitope (3) folds such that reactivity with this monoclonal antibody is retained. (C) N327 is located at a turn at the end of the GH loop and at the beginning of the H strand. Its interactions with the surrounding residues help anchoring the GH loop that interacts with H3 (helix 3) and H5 of the neighboring molecule. Both H3 and H5 are important for particle assembly. N327S mutation may lead to a less stabile GH loop, which may affect H3 and H5 interactions and thus assembly. (D) F446 is important for anchoring the stretch of residues of 444 to 447 (indicated by a double arrow) to the core of L1. Residues 444 to 447 are between strands J and H4, both of which are important elements for particle assembly. F446S results in much weaker interactions between the core and residues 444 to 447, which may account for the reduced efficiency of particle assembly.
Regulation of class switch recombination by innate recognition of VLP.
Innate recognition of HPV16 L1 VLP signals antigen-presenting cells (APCs) to express costimulatory molecules and secrete cytokines (10, 11, 23, 24). These APC-derived signals can drive the polarization of naive CD4+ helper T cells toward the Th2 or the Th1 phenotype (18). Th1 cells produce IFN and tumor necrosis factor (TNF) to regulate B cells to produce antigen-specific IgG2a. Th2 cells express interleukin 4 (IL-4), IL-5, IL-9, and IL-13 and can promote IgG1 and IgE class switching. Therefore, we examined the contribution of CD4+ T-helper cells to the regulation of L1-specific antibody isotype switching and production. Induction of VLP-specific IgG1 was largely dependent upon CD4+ T help. However, reduced but significant levels of L1-specific IgG2a are induced upon vaccination of CD4−/− mice with HPV16 L1 VLP, suggesting that both CD4+ T-cell-dependent and -independent mechanisms regulate IgG2a responses to HPV16 L1 VLP and that the latter make a significant contribution. BMDC produce both IL-10 and IFN-α in response to HPV16 L1 VLP, and IFN-α/βR−/− mice show decreased class switching upon HPV16 L1 VLP vaccination compared to control mice. Interestingly, mice vaccinated with mutants of L1 that fail to induce MyD88-dependent IL-10 and IFN-α production by BMDCs generate minimal specific IgG1 and IgG2a responses. These findings are consistent with a role for DC-directed class switch recombination in the CD4+ T-cell-independent response to HPV16 L1 VLP (25, 26).
Acknowledgments
We thank Mike Delannoy of the Microscopy Facility, Johns Hopkins School of Medicine, for expert confocal and electron microscopy; Chiara Fiorillo, William H. Yutzy IV, and Catherine Barden for technical support; and Douglas Lowy for comments on the manuscript.
R.B.S.R. is a paid consultant of Knobbe, Martens, Olson and Bear, LLC.
This research was supported by grants to RBSR from the National Institutes of Health (PO1 AI 48203 and P50 CA098252).
REFERENCES
- 1.Akira, S. 2003. Toll-like receptor signaling. J. Biol. Chem. 278:38105-38108. [DOI] [PubMed] [Google Scholar]
- 2.Breitburd, F., R. Kirnbauer, N. L. Hubbert, B. Nonnenmacher, C. Trin-Dinh-Desmarquet, G. Orth, J. T. Schiller, and D. R. Lowy. 1995. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J. Virol. 69:3959-3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter, J. J., G. C. Wipf, S. F. Benki, N. D. Christensen, and D. A. Galloway. 2003. Identification of a human papillomavirus type 16-specific epitope on the C-terminal arm of the major capsid protein L1. J. Virol. 77:11625-11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, X. S., G. Casini, S. C. Harrison, and R. L. Garcea. 2001. Papillomavirus capsid protein expression in Escherichia coli: purification and assembly of HPV11 and HPV16 L1. J. Mol. Biol. 307:173-182. [DOI] [PubMed] [Google Scholar]
- 5.Chen, X. S., R. L. Garcea, I. Goldberg, G. Casini, and S. C. Harrison. 2000. Structure of small virus-like particles assembled from the L1 protein of human papillomavirus 16. Mol. Cell 5:557-567. [DOI] [PubMed] [Google Scholar]
- 6.Cheng, G., J. P. Icenogle, R. Kirnbauer, N. L. Hubbert, M. E. St. Louis, C. Han, E. I. Svare, S. K. Kjaer, D. R. Lowy, and J. T. Schiller. 1995. Divergent human papillomavirus type 16 variants are serologically cross- reactive. J. Infect. Dis. 172:1584-1587. [DOI] [PubMed] [Google Scholar]
- 7.Choo, K. B., H. H. Lee, C. C. Pan, S. M. Wu, L. N. Liew, W. F. Cheung, and S. H. Han. 1988. Sequence duplication and internal deletion in the integrated human papillomavirus type 16 genome cloned from a cervical carcinoma. J. Virol. 62:1659-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cuzick, J., G. Terry, L. Ho, T. Hollingworth, and M. Anderson. 1992. Human papillomavirus type 16 in cervical smears as predictor of high-grade cervical intraepithelial neoplasia. Lancet 339:959-960. [DOI] [PubMed] [Google Scholar]
- 9.De Bruijn, M. L., H. L. Greenstone, H. Vermeulen, C. J. Melief, D. R. Lowy, J. T. Schiller, and W. M. Kast. 1998. L1-specific protection from tumor challenge elicited by HPV16 virus-like particles. Virology 250:371-376. [DOI] [PubMed] [Google Scholar]
- 10.Fausch, S. C., D. M. Da Silva, and W. M. Kast. 2003. Differential uptake and cross-presentation of human papillomavirus virus-like particles by dendritic cells and Langerhans cells. Cancer Res. 63:3478-3482. [PubMed] [Google Scholar]
- 11.Fausch, S. C., D. M. Da Silva, M. P. Rudolf, and W. M. Kast. 2002. Human papillomavirus virus-like particles do not activate Langerhans cells: a possible immune escape mechanism used by human papillomaviruses. J. Immunol. 169:3242-3249. [DOI] [PubMed] [Google Scholar]
- 12.Frazer, I. H., R. Thomas, J. Zhou, G. R. Leggatt, L. Dunn, N. McMillan, R. W. Tindle, L. Filgueira, P. Manders, P. Barnard, and M. Sharkey. 1999. Potential strategies utilised by papillomavirus to evade host immunity. Immunol. Rev. 168:131-142. [DOI] [PubMed] [Google Scholar]
- 13.Greenstone, H. L., J. D. Nieland, K. E. de Visser, M. L. De Bruijn, R. Kirnbauer, R. B. Roden, D. R. Lowy, W. M. Kast, and J. T. Schiller. 1998. Chimeric papillomavirus virus-like particles elicit antitumor immunity against the E7 oncoprotein in an HPV16 tumor model. Proc. Natl. Acad. Sci. USA 95:1800-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall, W. S., R. Goto-Mandeville, H. A. Shih, P. R. Shank, and L. Braun. 1997. Molecular analysis of episomal human papillomavirus type 16 DNA in a cervical carcinoma cell line. Virus Res. 51:183-195. [DOI] [PubMed] [Google Scholar]
- 15.Harro, C. D., Y. Y. Pang, R. B. Roden, A. Hildesheim, Z. Wang, M. J. Reynolds, T. C. Mast, R. Robinson, B. R. Murphy, R. A. Karron, J. Dillner, J. T. Schiller, and D. R. Lowy. 2001. Safety and immunogenicity trial in adult volunteers of a human papillomavirus 16 L1 virus-like particle vaccine. J. Natl. Cancer Inst. 93:284-292. [DOI] [PubMed] [Google Scholar]
- 16.Ho, L., S. Y. Chan, R. D. Burk, B. C. Das, K. Fujinaga, J. P. Icenogle, T. Kahn, N. Kiviat, W. Lancaster, P. Mavromara-Nazos, et al. 1993. The genetic drift of human papillomavirus type 16 is a means of reconstructing prehistoric viral spread and the movement of ancient human populations. J. Virol. 67:6413-6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Icenogle, J. P., K. A. Clancy, and S. Y. Lin. 1995. Sequence variation in the capsid protein genes of human papillomavirus type 16 and type 31. Virology 214:664-669. [DOI] [PubMed] [Google Scholar]
- 18.Kapsenberg, M. L. 2003. Dendritic-cell control of pathogen-driven T-cell polarization. Nat. Rev. Immunol. 3:984-993. [DOI] [PubMed] [Google Scholar]
- 19.Kirnbauer, R., F. Booy, N. Cheng, D. R. Lowy, and J. T. Schiller. 1992. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc. Natl. Acad. Sci. USA 89:12180-12184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirnbauer, R., N. L. Hubbert, C. M. Wheeler, T. M. Becker, D. R. Lowy, and J. T. Schiller. 1994. A virus-like particle enzyme-linked immunosorbent assay detects serum antibodies in a majority of women infected with human papillomavirus type 16. J. Natl. Cancer Inst. 86:494-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirnbauer, R., J. Taub, H. Greenstone, R. Roden, M. Durst, L. Gissmann, D. R. Lowy, and J. T. Schiller. 1993. Efficient self-assembly of human papillomavirus type 16 L1 and L1-L2 into virus-like particles. J. Virol. 67:6929-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koutsky, L. A., K. A. Ault, C. M. Wheeler, D. R. Brown, E. Barr, F. B. Alvarez, L. M. Chiacchierini, K. U. Jansen, et al. 2002. A controlled trial of a human papillomavirus type 16 vaccine. N. Engl. J. Med. 347:1645-1651. [DOI] [PubMed] [Google Scholar]
- 23.Lenz, P., P. M. Day, Y. Y. Pang, S. A. Frye, P. N. Jensen, D. R. Lowy, and J. T. Schiller. 2001. Papillomavirus-like particles induce acute activation of dendritic cells. J. Immunol. 166:5346-5355. [DOI] [PubMed] [Google Scholar]
- 24.Lenz, P., C. D. Thompson, P. M. Day, S. M. Bacot, D. R. Lowy, and J. T. Schiller. 2003. Interaction of papillomavirus virus-like particles with human myeloid antigen-presenting cells. Clin. Immunol. 106:231-237. [DOI] [PubMed] [Google Scholar]
- 25.Litinskiy, M. B., B. Nardelli, D. M. Hilbert, B. He, A. Schaffer, P. Casali, and A. Cerutti. 2002. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat. Immunol. 3:822-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacLennan, I., and C. Vinuesa. 2002. Dendritic cells, BAFF, and APRIL: innate players in adaptive antibody responses. Immunity 17:235-238. [DOI] [PubMed] [Google Scholar]
- 27.Meissner, J. D. 1999. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J. Gen. Virol. 80:1725-1733. [DOI] [PubMed] [Google Scholar]
- 28.Nonnenmacher, B., N. L. Hubbert, R. Kirnbauer, K. V. Shah, N. Munoz, F. X. Bosch, S. de Sanjose, R. Viscidi, D. R. Lowy, and J. T. Schiller. 1995. Serologic response to human papillomavirus type 16 (HPV-16) virus-like particles in HPV-16 DNA-positive invasive cervical cancer and cervical intraepithelial neoplasia grade III patients and controls from Colombia and Spain. J. Infect. Dis. 172:19-24. [DOI] [PubMed] [Google Scholar]
- 29.Öhlschläger, P., W. Osen, K. Dell, S. Faath, R. L. Garcea, I. Jochmus, M. Müller, M. Pawlita, K. Schäfer, P. Sehr, C. Staib, G. Sutter, and L. Gissmann. 2003. Human papillomavirus type 16 L1 capsomeres induce L1-specific cytotoxic T lymphocytes and tumor regression in C57BL/6 mice. J. Virol. 77:4635-4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pastrana, D. V., C. B. Buck, Y. Y. Pang, C. D. Thompson, P. E. Castle, P. C. FitzGerald, S. Kruger Kjaer, D. R. Lowy, and J. T. Schiller. 2004. Reactivity of human sera in a sensitive, high-throughput pseudovirus-based papillomavirus neutralization assay for HPV16 and HPV18. Virology 321:205-216. [DOI] [PubMed] [Google Scholar]
- 31.Pushko, P., T. Sasagawa, J. Cuzick, and L. Crawford. 1994. Sequence variation in the capsid protein genes of human papillomavirus type 16. J. Gen. Virol. 75:911-916. [DOI] [PubMed] [Google Scholar]
- 32.Roden, R. B., A. Armstrong, P. Haderer, N. D. Christensen, N. L. Hubbert, D. R. Lowy, J. T. Schiller, and R. Kirnbauer. 1997. Characterization of a human papillomavirus type 16 variant-dependent neutralizing epitope. J. Virol. 71:6247-6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roden, R. B., H. L. Greenstone, R. Kirnbauer, F. P. Booy, J. Jessie, D. R. Lowy, and J. T. Schiller. 1996. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J. Virol. 70:5875-5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roden, R. B., N. L. Hubbert, R. Kirnbauer, F. Breitburd, D. R. Lowy, and J. T. Schiller. 1995. Papillomavirus L1 capsids agglutinate mouse erythrocytes through a proteinaceous receptor. J. Virol. 69:5147-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roden, R. B., N. L. Hubbert, R. Kirnbauer, N. D. Christensen, D. R. Lowy, and J. T. Schiller. 1996. Assessment of the serological relatedness of genital human papillomaviruses by hemagglutination inhibition. J. Virol. 70:3298-3301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sapp, M., C. Fligge, I. Petzak, J. R. Harris, and R. E. Streeck. 1998. Papillomavirus assembly requires trimerization of the major capsid protein by disulfides between two highly conserved cysteines. J. Virol. 72:6186-6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seedorf, K., G. Krammer, M. Durst, S. Suhai, and W. G. Rowekamp. 1985. Human papillomavirus type 16 DNA sequence. Virology 145:181-185. [DOI] [PubMed] [Google Scholar]
- 38.Suzich, J. A., S. J. Ghim, F. J. Palmer-Hill, W. I. White, J. K. Tamura, J. A. Bell, J. A. Newsome, A. B. Jenson, and R. Schlegel. 1995. Systemic immunization with papillomavirus L1 protein completely prevents the development of viral mucosal papillomas. Proc. Natl. Acad. Sci. USA 92:11553-11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeda, K., T. Kaisho, and S. Akira. 2003. Toll-like receptors. Annu. Rev. Immunol. 21:335-376. [DOI] [PubMed] [Google Scholar]
- 40.Yamada, T., C. M. Wheeler, A. L. Halpern, A. C. Stewart, A. Hildesheim, and S. A. Jenison. 1995. Human papillomavirus type 16 variant lineages in United States populations characterized by nucleotide sequence analysis of the E6, L2, and L1 coding segments. J. Virol. 69:7743-7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang, R., F. M. Murillo, H. Cui, R. Blosser, S. Uematsu, K. Takeda, S. Akira, R. P. Viscidi, and R. B. S. Roden. 2004. Papillomavirus-like particles stimulate murine bone marrow-derived dendritic cells to produce alpha interferon and Th1 immune responses via MyD88. J. Virol. 78:11152-11160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youde, S. J., P. R. Dunbar, E. M. Evans, A. N. Fiander, L. K. Borysiewicz, V. Cerundolo, and S. Man. 2000. Use of fluorogenic histocompatibility leukocyte antigen-A*0201/HPV 16 E7 peptide complexes to isolate rare human cytotoxic T-lymphocyte-recognizing endogenous human papillomavirus antigens. Cancer Res. 60:365-371. [PubMed] [Google Scholar]
- 43.Zhang, L. F., J. Zhou, S. Chen, L. L. Cai, Q. Y. Bao, F. Y. Zheng, J. Q. Lu, J. Padmanabha, K. Hengst, K. Malcolm, and I. H. Frazer. 2000. HPV6b virus like particles are potent immunogens without adjuvant in man. Vaccine 18:1051-1058. [DOI] [PubMed] [Google Scholar]
- 44.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]